Voice speed
Text translation, source text, translation results, document translation, drag and drop.


Website translation
Enter a URL
Image translation
Madura Online
Millions of users can't be wrong! Madura Online is the best in the world. Madura English-Sinhala Dictionary contains over 230,000 definitions. Include glossaries of technical terms from medicine, science, law, engineering, accounts, arts and many other sources. This facilitates use as thesaurus. Translate from English to Sinhala and vice versa. Can use wildcards to increase the flexibility of search. An inevitable tool for students and professionals in their academic and day to day work.
Copyright © Madura Kulatunga, All rights reserved. Privacy Policy
How to Say Case study in Sinhala
- cascading water
- for the purpose of
- handwriting
Sinhala Dictionary definitions for case
case 🔊 /kejˈs/
case : පෙට්ටිය
case : කොපුව
case : වැස්ම
case : විබත්
case : තත්වය
case : නඩුකරය
case : පොත් කවරය
case : කෝෂය
case : වසනවා
case : තත්ත්වය
case : විභක්තිය
case : පුටය
case : කොපුවෙහි දමනවා
case : රෝගය
case : අවස්ථාව
case : දසාව
case : තටුව
case : ආවරණය
case : විභක්ති
case : රෝගියා
case : ප්රත්යෙකාය
case : පෙට්ටියෙහි දමනවා
case : නඩුව
case : කසුව
case : උදාහරණය
case definition
- A box, sheath, or covering; as, a case for holding goods; a case for spectacles; the case of a watch; the case (capsule) of a cartridge; a case (cover) for a book.
- A box and its contents; the quantity contained in a box; as, a case of goods; a case of instruments.
- A shallow tray divided into compartments or "boxes" for holding type.
- An inclosing frame; a casing; as, a door case; a window case.
- A small fissure which admits water to the workings.
- Chance; accident; hap; opportunity.
- That which befalls, comes, or happens; an event; an instance; a circumstance, or all the circumstances; condition; state of things; affair; as, a strange case; a case of injustice; the case of the Indian tribes.
- A patient under treatment; an instance of sickness or injury; as, ten cases of fever; also, the history of a disease or injury.
- The matters of fact or conditions involved in a suit, as distinguished from the questions of law; a suit or action at law; a cause.
- One of the forms, or the inflections or changes of form, of a noun, pronoun, or adjective, which indicate its relation to other words, and in the aggregate constitute its declension; the relation which a noun or pronoun sustains to some other word.
Transitive verb.
- To cover or protect with, or as with, a case; to inclose.
- To strip the skin from; as, to case a box.
Intransitive verb. To propose hypothetical cases.
Lern More About
- Wiki Definition: case
- Google Search result: Google
- Wiki Article: Wikipedia

© Dict.lk . All rights reserved.
Translation of "study" into Sinhala
ඉගෙනගන්නවා is the translation of "study" into Sinhala. Sample translated sentence: Many sincere ones have thus heard the good news and have started to study the Bible. ↔ මෙහි ප්රතිඵලයක් වශයෙන් අවංක සිත් ඇති බොහෝදෙනෙකුට රාජ්යයේ ශුභාරංචියට සවන් දීමට හැකි වී තිබෙන අතර බයිබලය පාඩම් කිරීමට පටන් ගෙන තිබෙනවා.
(usually academic) To revise materials already learned in order to make sure one does not forget them, usually in preparation for an examination. [..]
English-Sinhala dictionary
To acquire, or attempt to acquire knowledge or an ability to do something.
Show algorithmically generated translations
Automatic translations of " study " into Sinhala
Images with "study", phrases similar to "study" with translations into sinhala.
- television studies රූපවාහිනී අධ් යයනය
- IMPACT OF KNOWLEDGE MANAGEMENT PRACTICES ON EMPLOYEES' PERFORMANCE, A STUDY ASSOCIATED WITH REPLACING EMPLOYEES IN PEOPLES BANK, CENTRAL PROVINCE IN SRI LANKA IMPACT OF KNOWLEDGE MANAGEMENT PRACTICES ON EMPLOYEES' PERFORMANCE, A STUDY ASSOCIATED WITH REPLACING EMPLOYEES IN PEOPLES BANK, CENTRAL PROVINCE IN SRI LANKA · රැකියා නියුක්තයන් පිළිබඳ දැනුම කළමනාකරණය කිරීමේ ක්රියාකාරිත්වය, මහජන බැංකු තුළ සේවකයින් ප්රතිනිෂ්පාදනය කිරීම, ශී ලංකාෙව් මධ්යම පළාත තුළ
- Psychology is both an academic and applied field which deals with the study of human mind and behaviour. It seeks to understand and even explain emotion, thought and behaviour Psychology is both an academic and applied field which deals with the study of human mind and behaviour. It seeks to understand and even explain emotion, thought and behaviour. Psychology is
Translations of "study" into Sinhala in sentences, translation memory
- Share full article
Advertisement
Supported by
Guest Essay
The Troubling Trend in Teenage Sex

By Peggy Orenstein
Ms. Orenstein is the author of “Boys & Sex: Young Men on Hookups, Love, Porn, Consent and Navigating the New Masculinity” and “Girls & Sex: Navigating the Complicated New Landscape.”
Debby Herbenick is one of the foremost researchers on American sexual behavior. The director of the Center for Sexual Health Promotion at Indiana University and the author of the pointedly titled book “Yes, Your Kid,” she usually shares her data, no matter how explicit, without judgment. So I was surprised by how concerned she seemed when we checked in on Zoom recently: “I haven’t often felt so strongly about getting research out there,” she told me. “But this is lifesaving.”
For the past four years, Dr. Herbenick has been tracking the rapid rise of “rough sex” among college students, particularly sexual strangulation, or what is colloquially referred to as choking. Nearly two-thirds of women in her most recent campus-representative survey of 5,000 students at an anonymized “major Midwestern university” said a partner had choked them during sex (one-third in their most recent encounter). The rate of those women who said they were between the ages 12 and 17 the first time that happened had shot up to 40 percent from one in four.
As someone who’s been writing for well over a decade about young people’s attitudes and early experience with sex in all its forms, I’d also begun clocking this phenomenon. I was initially startled in early 2020 when, during a post-talk Q. and A. at an independent high school, a 16-year-old girl asked, “How come boys all want to choke you?” In a different class, a 15-year-old boy wanted to know, “Why do girls all want to be choked?” They do? Not long after, a college sophomore (and longtime interview subject) contacted me after her roommate came home in tears because a hookup partner, without warning, had put both hands on her throat and squeezed.
I started to ask more, and the stories piled up. Another sophomore confided that she enjoyed being choked by her boyfriend, though it was important for a partner to be “properly educated” — pressing on the sides of the neck, for example, rather than the trachea. (Note: There is no safe way to strangle someone.) A male freshman said “girls expected” to be choked and, even though he didn’t want to do it, refusing would make him seem like a “simp.” And a senior in high school was angry that her friends called her “vanilla” when she complained that her boyfriend had choked her.
Sexual strangulation, nearly always of women in heterosexual pornography, has long been a staple on free sites, those default sources of sex ed for teens . As with anything else, repeat exposure can render the once appalling appealing. It’s not uncommon for behaviors to be normalized in porn, move within a few years to mainstream media, then, in what may become a feedback loop, be adopted in the bedroom or the dorm room.
Choking, Dr. Herbenick said, seems to have made that first leap in a 2008 episode of Showtime’s “Californication,” where it was still depicted as outré, then accelerated after the success of “Fifty Shades of Grey.” By 2019, when a high school girl was choked in the pilot of HBO’s “Euphoria,” it was standard fare. A young woman was choked in the opener of “The Idol” (again on HBO and also, like “Euphoria,” created by Sam Levinson; what’s with him ?). Ali Wong plays the proclivity for laughs in a Netflix special, and it’s a punchline in Tina Fey’s new “Mean Girls.” The chorus of Jack Harlow’s “Lovin On Me,” which topped Billboard’s Hot 100 chart for six nonconsecutive weeks this winter and has been viewed over 99 million times on YouTube, starts with, “I’m vanilla, baby, I’ll choke you, but I ain’t no killer, baby.” How-to articles abound on the internet, and social media algorithms feed young people (but typically not their unsuspecting parents) hundreds of #chokemedaddy memes along with memes that mock — even celebrate — the potential for hurting or killing female partners.
I’m not here to kink-shame (or anything-shame). And, anyway, many experienced BDSM practitioners discourage choking, believing it to be too dangerous. There are still relatively few studies on the subject, and most have been done by Dr. Herbenick and her colleagues. Reports among adolescents are now trickling out from the United Kingdom , Australia , Iceland , New Zealand and Italy .
Twenty years ago, sexual asphyxiation appears to have been unusual among any demographic, let alone young people who were new to sex and iffy at communication. That’s changed radically in a short time, with health consequences that parents, educators, medical professionals, sexual consent advocates and teens themselves urgently need to understand.
Sexual trends can spread quickly on campus and, to an extent, in every direction. But, at least among straight kids, I’ve sometimes noticed a pattern: Those that involve basic physical gratification — like receiving oral sex in hookups — tend to favor men. Those that might entail pain or submission, like choking, are generally more for women.
So, while undergrads of all genders and sexualities in Dr. Herbenick’s surveys report both choking and being choked, straight and bisexual young women are far more likely to have been the subjects of the behavior; the gap widens with greater occurrences. (In a separate study , Dr. Herbenick and her colleagues found the behavior repeated across the United States, particularly for adults under 40, and not just among college students.) Alcohol may well be involved, and while the act is often engaged in with a steady partner, a quarter of young women said partners they’d had sex with on the day they’d met also choked them.
Either way, most say that their partners never or only sometimes asked before grabbing their necks. For many, there had been moments when they couldn’t breathe or speak, compromising the ability to withdraw consent, if they’d given it. No wonder that, in a separate study by Dr. Herbenick, choking was among the most frequently listed sex acts young women said had scared them, reporting that it sometimes made them worry whether they’d survive.
Among girls and women I’ve spoken with, many did not want or like to be sexually strangled, though in an otherwise desired encounter they didn’t name it as assault . Still, a sizable number were enthusiastic; they requested it. It is exciting to feel so vulnerable, a college junior explained. The power dynamic turns her on; oxygen deprivation to the brain can trigger euphoria.
That same young woman, incidentally, had never climaxed with a partner: While the prevalence of choking has skyrocketed, rates of orgasm among young women have not increased, nor has the “orgasm gap” disappeared among heterosexual couples. “It indicates they’re not doing other things to enhance female arousal or pleasure,” Dr. Herbenick said.
When, for instance, she asked one male student who said he choked his partner whether he’d ever tried using a vibrator instead, he recoiled. “Why would I do that?” he asked.
Perhaps, she responded, because it would be more likely to produce orgasm without risking, you know, death.
In my interviews, college students have seen male orgasm as a given; women’s is nice if it happens, but certainly not expected or necessarily prioritized (by either partner). It makes sense, then, that fulfillment would be less the motivator for choking than appearing adventurous or kinky. Such performances don’t always feel good.
“Personally, my hypothesis is that this is one of the reasons young people are delaying or having less sex,” Dr. Herbenick said. “Because it’s uncomfortable and weird and scary. At times some of them literally think someone is assaulting them but they don’t know. Those are the only sexual experiences for some people. And it’s not just once they’ve gotten naked. They’ll say things like, ‘I’ve only tried to make out with someone once because he started choking and hitting me.’”
Keisuke Kawata, a neuroscientist at Indiana University’s School of Public Health, was one of the first researchers to sound the alarm on how the cumulative, seemingly inconsequential, sub-concussive hits football players sustain (as opposed to the occasional hard blow) were key to triggering C.T.E., the degenerative brain disease. He’s a good judge of serious threats to the brain. In response to Dr. Herbenick’s work, he’s turning his attention to sexual strangulation. “I see a similarity” to C.T.E., he told me, “though the mechanism of injury is very different.” In this case, it is oxygen-blocking pressure to the throat, frequently in light, repeated bursts of a few seconds each.
Strangulation — sexual or otherwise — often leaves few visible marks and can be easily overlooked as a cause of death. Those whose experiences are nonlethal rarely seek medical attention, because any injuries seem minor: Young women Dr. Herbenick studied mostly reported lightheadedness, headaches, neck pain, temporary loss of coordination and ear ringing. The symptoms resolve, and all seems well. But, as with those N.F.L. players, the true effects are silent, potentially not showing up for days, weeks, even years.
According to the American Academy of Neurology, restricting blood flow to the brain, even briefly, can cause permanent injury, including stroke and cognitive impairment. In M.R.I.s conducted by Dr. Kawata and his colleagues (including Dr. Herbenick, who is a co-author of his papers on strangulation), undergraduate women who have been repeatedly choked show a reduction in cortical folding in the brain compared with a never-choked control group. They also showed widespread cortical thickening, an inflammation response that is associated with elevated risk of later-onset mental illness. In completing simple memory tasks, their brains had to work far harder than the control group, recruiting from more regions to achieve the same level of accuracy.
The hemispheres in the choked group’s brains, too, were badly skewed, with the right side hyperactive and the left underperforming. A similar imbalance is associated with mood disorders — and indeed in Dr. Herbenick’s surveys girls and women who had been choked were more likely than others (or choked men) to have experienced overwhelming anxiety, as well as sadness and loneliness, with the effect more pronounced as the incidence rose: Women who had experienced more than five instances of choking were two and a half times as likely as those who had never been choked to say they had been so depressed within the previous 30 days they couldn’t function. Whether girls and women with mental health challenges are more likely to seek out (or be subjected to) choking, choking causes mood disorders, or some combination of the two is still unclear. But hypoxia, or oxygen deprivation — judging by what research has shown about other types of traumatic brain injury — could be a contributing factor. Given the soaring rates of depression and anxiety among young women, that warrants concern.
Now consider that every year Dr. Herbenick has done her survey, the number of females reporting extreme effects from strangulation (neck swelling, loss of consciousness, losing control of urinary function) has crept up. Among those who’ve been choked, the rate of becoming what students call “cloudy” — close to passing out, but not crossing the line — is now one in five, a huge proportion. All of this indicates partners are pressing on necks longer and harder.
The physical, cognitive and psychological impacts of sexual choking are disturbing. So is the idea that at a time when women’s social, economic, educational and political power are in ascent (even if some of those rights may be in jeopardy), when #MeToo has made progress against harassment and assault, there has been the popularization of a sex act that can damage our brains, impair intellectual functioning, undermine mental health, even kill us. Nonfatal strangulation, one of the most significant indicators that a man will murder his female partner (strangulation is also one of the most common methods used for doing so), has somehow been eroticized and made consensual, at least consensual enough. Yet, the outcomes are largely the same: Women’s brains and bodies don’t distinguish whether they are being harmed out of hate or out of love.
By now I’m guessing that parents are curled under their chairs in a fetal position. Or perhaps thinking, “No, not my kid!” (see: title of Dr. Herbenick’s book above, which, by the way, contains an entire chapter on how to talk to your teen about “rough sex”).
I get it. It’s scary stuff. Dr. Herbenick is worried; I am, too. And we are hardly some anti-sex, wait-till-marriage crusaders. But I don’t think our only option is to wring our hands over what young people are doing.
Parents should take a beat and consider how they might give their children relevant information in a way that they can hear it. Maybe reiterate that they want them to have a pleasurable sex life — you have already said that, right? — and also want them to be safe. Tell them that misinformation about certain practices, including choking, is rampant, that in reality it has grave health consequences. Plus, whether or not a partner initially requested it, if things go wrong, you’re generally criminally on the hook.
Dr. Herbenick suggests reminding them that there are other, lower-risk ways to be exploratory or adventurous if that is what they are after, but it would be wisest to delay any “rough sex” until they are older and more skilled at communicating. She offers language when negotiating with a new partner, such as, “By the way, I’m not comfortable with” — choking, or other escalating behaviors such as name-calling, spitting and genital slapping — “so please don’t do it/don’t ask me to do it to you.” They could also add what they are into and want to do together.
I’d like to point high school health teachers to evidence-based porn literacy curricula, but I realize that incorporating such lessons into their classrooms could cost them their jobs. Shafia Zaloom, a lecturer at the Harvard Graduate School of Education, recommends, if that’s the case, grounding discussions in mainstream and social media. There are plenty of opportunities. “You can use it to deconstruct gender norms, power dynamics in relationships, ‘performative’ trends that don’t represent most people’s healthy behaviors,” she said, “especially depictions of people putting pressure on someone’s neck or chest.”
I also know that pediatricians, like other adults, struggle when talking to adolescents about sex (the typical conversation, if it happens, lasts 40 seconds). Then again, they already caution younger children to use a helmet when they ride a bike (because heads and necks are delicate!); they can mention that teens might hear about things people do in sexual situations, including choking, then explain the impact on brain health and why such behavior is best avoided. They should emphasize that if, for any reason — a fall, a sports mishap or anything else — a young person develops symptoms of head trauma, they should come in immediately, no judgment, for help in healing.
The role and responsibility of the entertainment industry is a tangled knot: Media reflects behavior but also drives it, either expanding possibilities or increasing risks. There is precedent for accountability. The European Union now requires age verification on the world’s largest porn sites (in ways that preserve user privacy, whatever that means on the internet); that discussion, unsurprisingly, had been politicized here. Social media platforms have already been pushed to ban content promoting eating disorders, self-harm and suicide — they should likewise be pressured to ban content promoting choking. Traditional formats can stop glamorizing strangulation, making light of it, spreading false information, using it to signal female characters’ complexity or sexual awakening. Young people’s sexual scripts are shaped by what they watch, scroll by and listen to — unprecedentedly so. They deserve, and desperately need, models of interactions that are respectful, communicative, mutual and, at the very least, safe.
Peggy Orenstein is the author of “Boys & Sex: Young Men on Hookups, Love, Porn, Consent and Navigating the New Masculinity” and “Girls & Sex: Navigating the Complicated New Landscape.”
The Times is committed to publishing a diversity of letters to the editor. We’d like to hear what you think about this or any of our articles. Here are some tips . And here’s our email: [email protected] .
Follow the New York Times Opinion section on Facebook , Instagram , TikTok , WhatsApp , X and Threads .
An earlier version of this article misstated the network on which “Californication” first appeared. It is Showtime, not HBO. The article also misspelled a book and film title. It is “Fifty Shades of Grey,” not “Fifty Shades of Gray.”
How we handle corrections
- Open access
- Published: 19 April 2024
GbyE: an integrated tool for genome widely association study and genome selection based on genetic by environmental interaction
- Xinrui Liu 1 , 2 ,
- Mingxiu Wang 1 ,
- Jie Qin 1 ,
- Yaxin Liu 1 ,
- Shikai Wang 1 ,
- Shiyu Wu 1 ,
- Ming Zhang 1 ,
- Jincheng Zhong 1 &
- Jiabo Wang 1
BMC Genomics volume 25 , Article number: 386 ( 2024 ) Cite this article
257 Accesses
Metrics details
The growth and development of organism were dependent on the effect of genetic, environment, and their interaction. In recent decades, lots of candidate additive genetic markers and genes had been detected by using genome-widely association study (GWAS). However, restricted to computing power and practical tool, the interactive effect of markers and genes were not revealed clearly. And utilization of these interactive markers is difficult in the breeding and prediction, such as genome selection (GS).
Through the Power-FDR curve, the GbyE algorithm can detect more significant genetic loci at different levels of genetic correlation and heritability, especially at low heritability levels. The additive effect of GbyE exhibits high significance on certain chromosomes, while the interactive effect detects more significant sites on other chromosomes, which were not detected in the first two parts. In prediction accuracy testing, in most cases of heritability and genetic correlation, the majority of prediction accuracy of GbyE is significantly higher than that of the mean method, regardless of whether the rrBLUP model or BGLR model is used for statistics. The GbyE algorithm improves the prediction accuracy of the three Bayesian models BRR, BayesA, and BayesLASSO using information from genetic by environmental interaction (G × E) and increases the prediction accuracy by 9.4%, 9.1%, and 11%, respectively, relative to the Mean value method. The GbyE algorithm is significantly superior to the mean method in the absence of a single environment, regardless of the combination of heritability and genetic correlation, especially in the case of high genetic correlation and heritability.
Conclusions
Therefore, this study constructed a new genotype design model program (GbyE) for GWAS and GS using Kronecker product. which was able to clearly estimate the additive and interactive effects separately. The results showed that GbyE can provide higher statistical power for the GWAS and more prediction accuracy of the GS models. In addition, GbyE gives varying degrees of improvement of prediction accuracy in three Bayesian models (BRR, BayesA, and BayesCpi). Whatever the phenotype were missed in the single environment or multiple environments, the GbyE also makes better prediction for inference population set. This study helps us understand the interactive relationship between genomic and environment in the complex traits. The GbyE source code is available at the GitHub website ( https://github.com/liu-xinrui/GbyE ).
Peer Review reports
Genetic by environmental interaction (G × E) is crucial of explaining individual traits and has gained increasing attention in research. It refers to the influence of genetic factors on susceptibility to environmental factors. In-depth study of G × E contributes to a deeper understanding of the relationship between individual growth, living environment and phenotypes. Genetic factors play a role in most human diseases at the molecular or cellular level, but environmental factors also contribute significantly. Researchers aim to uncover the mechanisms behind complex diseases and quantitative traits by investigating the interactions between organisms and their environment. Common, complex, or rare human diseases are often considered as outcomes resulting from the interplay of genes, environmental factors, and their interactions. Analyzing the joint effects of genes and the environment can provide valuable insights into the underlying pathway mechanisms of diseases. For instance, researchers have successfully identified potential loci associated with asthma risk through G × E interactions [ 1 ], and have explored predisposing factors for challenging-to-treat diseases like cancer [ 2 , 3 ], rhinitis [ 4 ], and depression [ 5 ].
However, two main methods are currently being used by breeders in agricultural production to increase crop yields and livestock productivity [ 6 ]. The first is to develop varieties with relatively low G × E effect to ensure stable production performance in different environments. The second is to use information from different environments to improve the statistical power of genome-wide association study (GWAS) to reveal potential loci of complex traits. The first method requires long-term commitment, while the second method clearly has faster returns. In GWAS, the use of multiple environments or phenotypes for association studies has become increasingly important. This not only improves the statistical power of environmental susceptibility traits[ 7 ], but also allows to detect signaling loci for G × E. There are significant challenges when using multiple environments or phenotypes for GWAS, mainly because most diseases and quantitative traits have numerous associated loci with minimal impact [ 8 ], and thus it is impossible to determine the effect size regulated by environment in these loci. The current detection strategy for G × E is based on complex statistical model, often requiring the use of a large number of samples to detect important signals [ 9 , 10 ]. In GS, breeders can use whole genome marker data to identify and select target strains in the early stages of animal and plant production [ 11 , 12 , 13 ]. Initially, GS models, similar to GWAS models, could only analyze a single environment or phenotype [ 14 ]. To improve the predictive accuracy of the models, higher marker densities are often required, allowing the proportion of genetic variation explained by these markers to be increased, indirectly obtaining higher predictive accuracy. It is worth mentioning that the consideration of G × E and multiple phenotypes in GS models [ 15 ] has been widely studied in different plant and animal breeding [ 16 ]. GS models that allow G × E have been developed [ 17 ] and most of them have modeled and interpreted G × E using structured covariates [ 18 ]. In these studies, most of the GS models provided more predictive accuracy when combined with G × E compared to single environment (or phenotype) analysis. Hence, there is need to develop models that leverage G × E information for GWAS and GS studies.
This study developed a novel genotype-by-environment method based on R, termed GbyE, which leverages the interaction among multiple environments or phenotypes to enhance the association study and prediction performance of environmental susceptibility traits. The method enables the identification of mutation sites that exhibit G × E interactions in specific environments. To evaluate the performance of the method, simulation experiments were conducted using a dataset comprising 282 corn samples. Importantly, this method can be seamlessly integrated into any GWAS and GS analysis.
Materials and methods
Support packages.
The development purpose of GbyE is to apply it to GWAS and GS research, therefore it uses the genome association and prediction integrated tool (GAPIT) [ 19 ], Bayesian Generalized Linear Regression (BGLR) [ 20 ], and Ridge Regression Best Linear Unbiased Prediction (rrBLUP) [ 21 ]package as support packages, where GbyE only provides conversion of interactive formats and file generation. In order to simplify the operation of the GbyE function package, the basic calculation package is attached to this package to support the operation of GbyE, including four function packages GbyE.Simulation.R (Dual environment phenotype simulation based on heritability, genetic correlation, and QTL quantity), GbyE.Calculate.R (For numerical genotype and phenotype data, this package can be used to process interactive genotype files of GbyE), GbyE.Power.FDR.R (Calculate the statistical power and false discovery rate (FDR) of GWAS), and GbyE.Comparison.Pvalue.R (GbyE generates redundant calculations in GWAS calculations, and SNP effect loci with minimal p -values can be filtered by this package).
Samples and sequencing data
In this study, a small volume of data was used for software simulation analysis, which is widely used in testing tasks of software such as GAPIT, TASSEL, and rMPV. The demonstration data comes from 282 inbred lines of maize, including 4 phenotypic data. In any case, there are no missing phenotypes in these data, and this dataset can be obtained from the website of GAPIT ( https://zzlab.net/GAPIT/index.html , accessed on May 1, 2022). Among them, our phenotype data was simulated using a self-made R simulation function, and the Mean and GbyE phenotype files were calculated. Convert this format to HapMap format using PLINK v1.09 and scripts written by oneself.
Simulated traits
Phenotype simulation was performed by modifying the GAPIT.Phenotype.Simulation function in the GAPIT. Based on the input parameter NQTN, the random selected markers’ genotype from whole genome were used to simulate genetic effect in the simulated trait. The genotype effects of these selected QTNs were randomly sampled from a multivariate normal distribution, the correlation value between these normal distribution was used to define the genetic relationship between each environments. The additive heritability ( \({{\text{h}}}_{{\text{g}}}^{2}\) ) was used to scale the relationship between additive genetic variance and phenotype variance. The simulated phenotype conditions in this paper are set as follows: 1) The three levels of \({{\text{h}}}_{{\text{g}}}^{2}\) were set at 0.8, 0.5, and 0.2, representing high ( \({{\text{h}}}_{{\text{h}}}^{2}\) ), median ( \({{\text{h}}}_{{\text{m}}}^{2}\) ) and low ( \({{\text{h}}}_{{\text{l}}}^{2}\) ) heritability; 2) Genetic correlation were set three levels 0.8, 0.5, 0.2 representing high ( \({{\text{R}}}_{{\text{h}}}\) ), medium ( \({{\text{R}}}_{{\text{m}}}\) ) and low ( \({{\text{R}}}_{{\text{l}}}\) ) genetic correlation; 3) 20 pre-set effect loci of QTL. The phenotype values in each environment were simulated together following above parameters.
Genetic by environment interaction model
The pipeline analysis process of GbyE includes three steps: data preprocessing, production converted, Association analysis. Normalize the phenotype data matrix Y of the dual environment and perform GbyE conversion to generate phenotype data in GbyE.Y format. The genotype data format, such as hapmap, vcf, bed and other formats firstly need to be converted into numerical genotype format (homozygotes were coded as 0 or 2, heterozygotes were coded as 1) using software or scripts such as GAPIT, PLINK, etc. The environment (E) matrix is environment index matrix. The G (n × m) originally of genotype matrix was converted as GbyE.GD(2n × 2 m) \(\left[\begin{array}{cc}G& 0\\ G& G\end{array}\right]\) during the Kronecker product, and the Y vector (n × 1) was also converted as the GbyE.Y vector (2n × 1) after normalization. The duplicated data format indicated different environments, genetic effect, and populations. The genomic data we used in the analysis was still retained the whole genome information. The first column of E is the additive effect, which was the average genetic effect among environments. The others columns of E are the interactive effect, which should be less one column than the number of environments. Because it need to avoid the linear dependent in the model. In the GbyE algorithm, we coded the first environment as background as default, that means the genotype in the first environment are 0, the others are 1. Then the Kronecker product of G and environment index matrix was named as GbyE.GD. The interactive effect part of the GbyE.GD matrix in the GWAS and GS were the relative values based on the first environment (Fig. 1 ). The GbyE environmental interaction matrix can be easily obtained by constructing the interaction matrix E (e.g., Eq. 1 ) such that the genotype matrix G is Kronecker-product with the design interaction matrix E (e.g., Eq. 2 ), in which \(\left[\begin{array}{c}G\\ G\end{array}\right]\) matrix is defined as additive effect and \(\left[\begin{array}{c}0\\ G\end{array}\right]\) matrix is defined as interactive effect. \(\left[\begin{array}{cc}G& 0\\ G& G\end{array}\right]\) matrix is called gene by environment interaction matrix, hereinafter referred to as the GbyE matrix. The phenotype file (GbyE.Y) and genotype file (GbyE.GD) after transformation by GbyE will be inputted into the GWAS and GS models and computed as standard phenotype and genotype files.
where G is the matrix of whole genotype and E is the design matrix for exploring interactive effects. GbyE mainly uses the Kronecker product of the genetic matrix (G) and the environmental matrix (E) as the genotype for subsequent GWAS as a way to distinguish between additive and interactive effects.
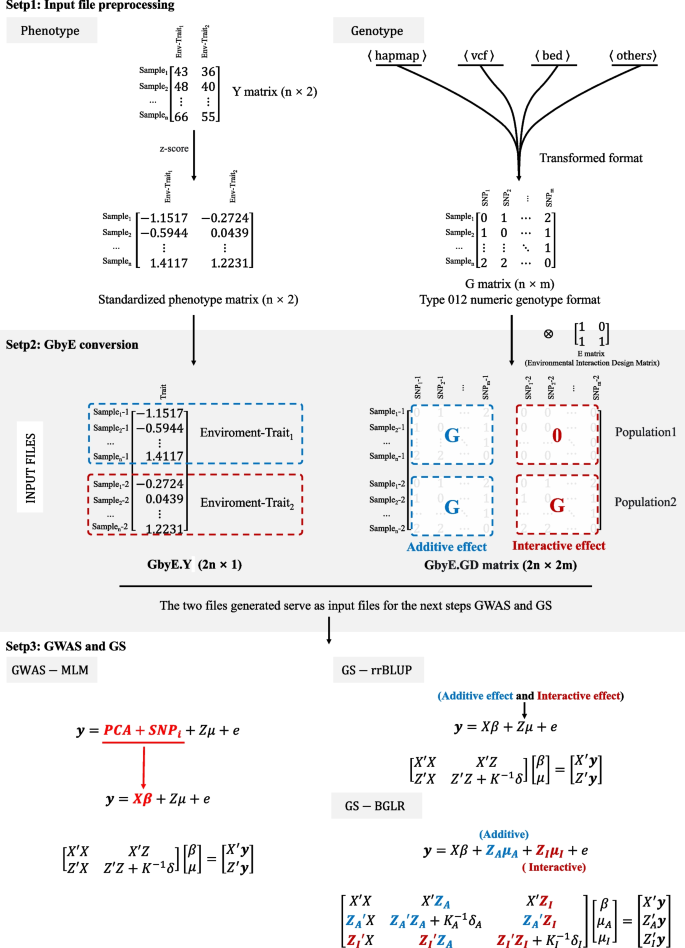
The workflow pipeline of GbyE. The GbyE contains three main steps. (Step 1) Preprocessing of phenotype and genotype data,. The phenotype values in each environment was normalized respectively. Meanwhile, all genotype from HapMap, VCF, BED, and other types were converted to numeric genotype; (Step 2) Generate GbyE phenotype and interactive genotype matrix through the transformation of GbyE. In GbyE.GD matrix, the blue characters indicate additive effect, and red ones indicate interactive effect; (Step 3) The MLM and rrBLUP and BGLR were used to perform GWAS and GS
Association analysis model
The mixed linear model (MLM) of GAPIT is used as the basic model for GWAS analysis, and the principal component analysis (PCA) parameter is set to 3. Then the p -values of detection results are sorted and their power and FDR values are calculated. General expression of MLM (Fig. 1 ):
where Y is the vector of phenotypic measures (2n × 1); PCA and SNP i were defined as fixed effects, with a size of (2n × 2 m); Z is the incidence matrix of random effects; μ is the random effect vector, which follows the normal distribution μ ~ N(0, \({\delta }_{G}^{2}\) K) with mean vector of 0 and variance covariance matrix of \({\delta }_{G}^{2}\) K, where the \({\delta }_{G}^{2}\) is the total genetic variance including additive variance and interactive variance, the K is the kinship matrix built with all genotype including additive genotype and interactive genotype; e is a random error vector, and its elements need not be independent and identically distributed, e ~ N(0, \({\delta }_{e}^{2}\) I), where the \({\delta }_{e}^{2}\) is the residual and environment variance, the I is the design matrix.
Detectivity of GWAS
In the GWAS results, the list of markers following the order of P-values was used to evaluate detectivity of GWAS methods. When all simulated QTNs were detected, the power of the GWAS method was considered as 1 (100%). From the list of markers, following increasing of the criterion of real QTN, the power values will be increasing. The FDR indicates the rate between the wrong criterion of real QTNs and the number of all un-QTNs. The mean of 100 cycles was used to consider as the reference value for statistical power comparison. Here, we used a commonly used method in GWAS research with multiple traits or environmental phenotypes as a comparison[ 22 ]. This method obtains the mean of phenotypic values under different conditions as the phenotypic values for GWAS analysis, called the Mean value method, Compare the calculation results of GbyE with the additive and interactive effects of the mean method to evaluate the detection power of the GbyE strategy. Through the comprehensive analysis of these evaluation indicators, we aim to comprehensively evaluate the statistical power of the GbyE strategy in GWAS and provide a reference for future optimization research.
Among them, the formulae for calculating Power and FDR are as follows:
where \({{\text{n}}}_{{\text{i}}}\) indicates whether the i-th detection is true, true is 1, false is 0; \({{\text{m}}}_{{\text{r}}}\) is the total number of all true QTLs in the sample size; the maximum value of Power is 1.
where \({{\text{N}}}_{{\text{i}}}\) represents the i-th true value detected in the pseudogene, true is 1, false is 0. and cumulative calculation; \({{\text{M}}}_{{\text{f}}}\) is the number of all labeled un-QTNs in the total samples; the maximum value of FDR is 1.
Genomic prediction
To comparison the prediction accuracy of different GS models using GbyE, we performed rrBLUP, Bayesian methods using R packages. All phenotype of reference population and genotype of all population were used to train the model and predict genomic estimated breeding value (gEBV) of all individuals. The correlation between real phenotypes and gEBV of inference population was considered as prediction accuracy. fivefold cross-validation and 100 times repeats was performed to avoid over prediction and reduce bias. In order to distinguish the additive and interactive effects in GbyE, we designed two lists of additive and interactive effects in the "ETA" of BGLR, and put the additive and interactive effects into the model as two kinships for random objects. However, it was not possible to load the gene effects of the two lists in rrBLUP, so the additive and interactive genotypes together were used to calculate whole genetic kinship in rrBLUP (Fig. 1 ). Relevant parameters in BGLR are set as follows: 1) model set to "RRB"; 2) nIter is set to "12000"; 3) burnIn is set to "10000". The results of the above operations are averaged over 100 cycles. We also validated the GbyE method using four other Bayesian methods (BayesA, BayesB, BayesCpi, and Bayesian LASSO) in addition to RRB in BGLR.
Partial missing phentoype in the prediction
In this study, we artificially missed phenotype values in the single and double environments in the whole population from 281 inbred maize datasets. In the missing single environment case, the inference set in the cross-validation was selected from whole population, and each individual in the inference were only missed phenotypes in the one environment. The phenotype in the other environment was kept. The genotypes were always kept. In the case of missing double environments, both phenotypes and genotypes of environment 1 and environment 2 are missing, and the model can only predict phenotypic values in the two missing environments through the effects of other markers. In addition, the data were standardized and unstandardized to assess whether standardization had an effect on the estimation of the model. This experiment was tested using the "ML" method in rrBLUP to ensure the efficiency of the model.
GWAS statistical power of models at different heritabilities and genetic correlations
Power-FDR plots were used to demonstrate the detection efficiency of GbyE at three genetic correlation and three genetic power levels, with a total of nine different scenarios simulated (from left to right for high and low genetic correlation and from top to bottom for high and low genetic power). In order to distinguish whether the effect of improving the detection ability of genome-wide association analysis in GbyE is an additive effect or an effect of environmental interactions, we plotted their Power-FDR curves separately and added the traditional Mean method for comparative analysis. As shown in Fig. 2 , GbyE algorithm can detect more statistically significant genetic loci with lower FDR under any genetic background. However, in the combination with low heritability (Fig. 2 A, B, C), the interactive effect detected more real loci than GbyE under low FDR, but with the continued increase of FDR, GbyE detected more real loci than other groups. Under the combination with high heritability, all groups have high statistical power at low FDR, but with the increase of FDR, the statistical effect of GbyE gradually highlights. From the analysis of heritability combinations at all levels, the effect of heritability on interactive effect is not obvious, but GbyE always maintains the highest statistical power. The average detection power of GWAS in GbyE can be increased by about 20%, and with the decrease of genetic correlation, the effect of GbyE gradually highlights, indicating that the G × E plays a role.
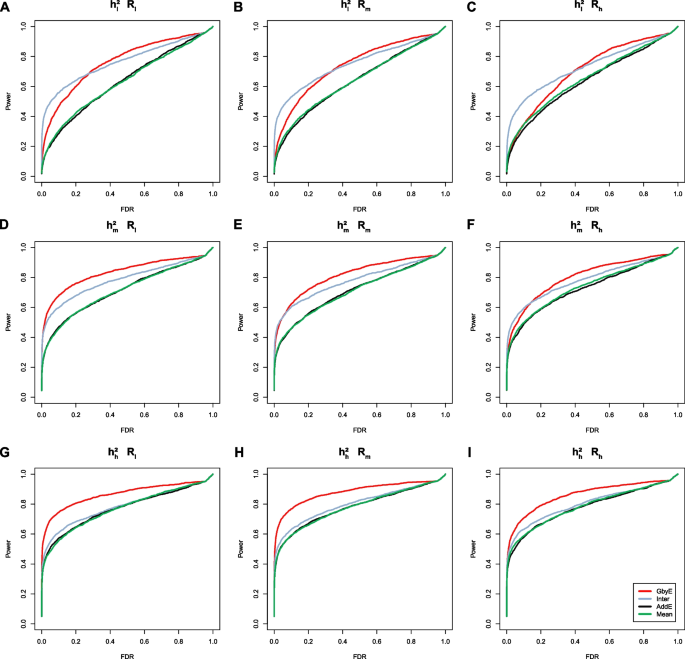
The power-FDR testing in simulated traits. Comparing the efficacy of the GbyE algorithm with the conventional mean method in terms of detection power and FDR. From left to right, the three levels of genetic correlation are indicated in order of low, medium and high. From top to bottom, the three levels of heritability, low, medium and high, are indicated in order. (1) Inter: Interactive section extracted from GbyE; (2) AddE: Additive section extracted from GbyE; (3) \({{\text{h}}}_{{\text{l}}}^{2}\) , \({{\text{h}}}_{{\text{m}}}^{2}\) , \({{\text{h}}}_{{\text{g}}}^{2}\) : Low, medium, high heritability; (4) \({{\text{R}}}_{{\text{l}}}\) , \({{\text{R}}}_{{\text{m}}}\) , \({{\text{R}}}_{{\text{l}}}\) : where R stands for genetic correlation, represents three levels of low, medium and high
Resolution of additive and interactive effect
The output results of GbyE could be understood as resolution of additive and interactive genetic effect. Hence, we created a combined Manhattan plots with Mean result from MLM, additive, and interactive results from GbyE. As shown in Fig. 3 , true marker loci were detected on chromosomes 1, 6 and 9 in Mean, and the same loci were detected on chromosomes 1 and 6 for the additive result in GbyE (the common loci detected jointly by the two results were marked as solid gray lines in the figure). All known pseudo QTNs were labeled with gray dots in the circle. Total 20 pseudo QTNs were simulated in such trait (The heritability is set to 0.9, and the genetic correlation is set to 0.1). Although the additive section in GbyE did not catch the locus on chromosome 9 yet (those p-values of markers did not show above the significance threshold (p-value < 3.23 × 10 –6 )), it has shown high significance relative to other markers of the same chromosome. In the reciprocal effect of GbyE, we detected more significant loci on chromosomes 1, 2, 3 and 10, and these loci were not detected in either of the two previous sections. An integrate QQ plot (Fig. 3 D) shows that the overall statistical power of the additive section in Mean and GbyE are close, nevertheless, the interactive section in the GbyE provided a bit of inflation.
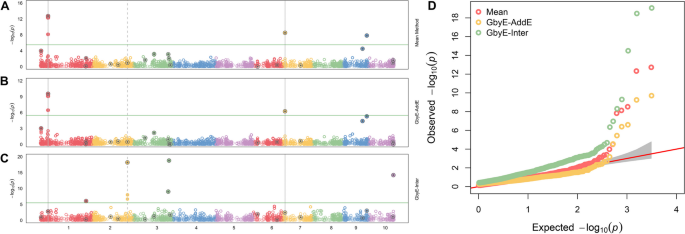
Manhattan statistical comparison plot. Manhattan comparison plots of mean ( A ), additive ( B ) and gene-environment interactive sections ( C ) at a heritability of 0.9 and genetic correlation of 0.1. Different colors are used in the diagram to distinguish between different chromosomes (X-axis). Loci with reinforcing circles and centroids are set up as real QTN loci. Consecutive loci found in both parts are shown as id lines, and loci found separately in the reciprocal effect only are shown as dashed lines. Parallel horizontal lines indicate significance thresholds ( p -value < 3.23 × 10 –6 ). D Quantile–quantile plots of simulated phenotypes for demo data from genome-wide association studies. x-axis indicates expected values of log p -values and y-axis is observed values of log p -values. The diagonal coefficients in red are 1. GbyE-inter is the interactive section in GbyE; GbyE-AddE is the additive section in GbyE
Genomic selection in assumption codistribution
The prediction accuracy of GbyE was significantly higher than the Mean value method by model statistics of rrBLUP in most cases of heritability and genetic correlation (Fig. 4 ). The prediction accuracy of the additive effect was close to that of Mean value method, which was consistent with the situation under the low hereditary. The prediction accuracy of interactive sections in GbyE remains at the same level as in GbyE, and interactive section plays an important role in the model. We observed that in \({{\text{h}}}_{{\text{l}}}^{2}{{\text{R}}}_{{\text{h}}}\) (Fig. 4 C), \({{\text{h}}}_{{\text{m}}}^{2}{{\text{R}}}_{{\text{h}}}\) (Fig. 4 F), \({{\text{h}}}_{{\text{h}}}^{2}{{\text{R}}}_{{\text{l}}}\) (Fig. 4 G), the prediction accuracy of GbyE was slightly higher than the Mean value method, but there was no significant difference overall. In addition, we only observed that the prediction accuracy of GbyE was slightly lower than the Mean value method in \({{\text{h}}}_{{\text{h}}}^{2}{{\text{R}}}_{{\text{l}}}\) (Fig. 4 H), but there was still no significant difference between GbyE and Mean value methods. Under the combination of low heritability and genetic correlation, the prediction accuracy of Mean value method and additive effect model remained at a similar level. However, with the continuous increase of heritability and genetic correlation, the difference in prediction accuracy between the two gradually increases. In summary, the GbyE algorithm can improve the accuracy of GS by capturing information on multiple environment or trait effects under the rrBLUP model.
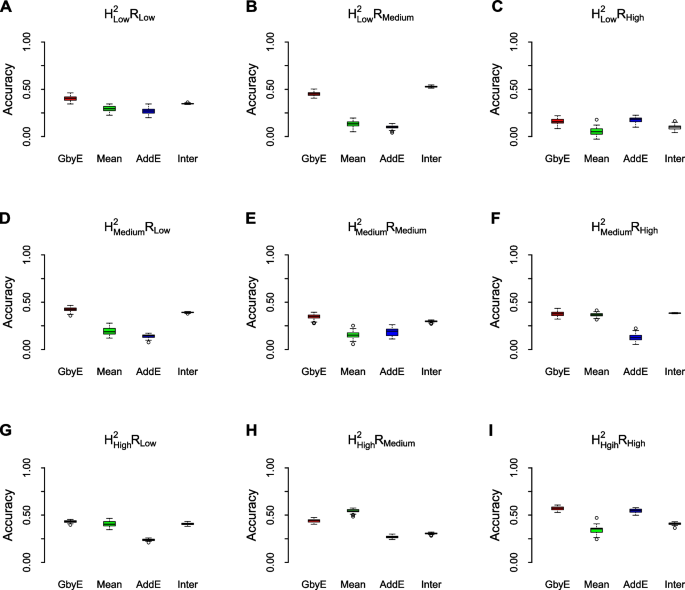
Box-plot of model prediction accuracy. The prediction accuracy (pearson's correlation coefficient) of the GbyE algorithm was compared with the tradition al Mean value method in a simulation experiment of genomic selection under the rrBLUP operating environment. The effect of different levels of heritability and genetic correlation on the prediction accuracy of genomic selection was simulated in this experiment. Each row from top to bottom represents low heritability ( \({{\text{h}}}_{{\text{l}}}^{2}\) ), medium heritability ( \({{\text{h}}}_{{\text{m}}}^{2}\) ) and high heritability ( \({{\text{h}}}_{{\text{h}}}^{2}\) ), respectively; each column from left to right represents low genetic correlation ( \({{\text{R}}}_{{\text{l}}}\) ), medium genetic correlation ( \({{\text{R}}}_{{\text{m}}}\) ) and high genetic correlation ( \({{\text{R}}}_{{\text{h}}}\) ), respectively; The X-axis shows the different test methods and effects, and the Y-axis shows the prediction accuracy
Genomic selection in assumption un-codistribution
The overall performance of GbyE under the 'BRR' statistical model based on the BGLR package remained consistent with rrBLUP, maintaining high predictive accuracy in most cases of heritability and genetic relatedness (Fig. S1 ). However, when the heritability is set to low and medium, the difference between the prediction accuracy of GbyE algorithm and Mean value method gradually decreases with the continuous increase of genetic correlation, and there is no statistically significant difference between the two. The prediction accuracy of the model by GbyE in \({{\text{h}}}_{{\text{h}}}^{2}{{\text{R}}}_{{\text{l}}}\) (Fig. S1 G) and \({{\text{h}}}_{{\text{h}}}^{2}{{\text{R}}}_{{\text{h}}}\) (Fig. S1 I) is significantly higher than that by Mean value method when the heritability is set to be high. On the contrary, when the genetic correlation is set to medium, there is no significant difference between GbyE and Mean value method in improving the prediction accuracy of the model, and the overall mean of GbyE is lower than Mean. When GbyE has relatively high heritability and low genetic correlation, its prediction accuracy is significantly higher than the mean method, such as \({{\text{h}}}_{{\text{m}}}^{2}{{\text{R}}}_{{\text{l}}}\) (Fig. S1 D), \({{\text{h}}}_{{\text{h}}}^{2}{{\text{R}}}_{{\text{l}}}\) (Fig. S1 G), and \({{\text{h}}}_{{\text{h}}}^{2}{{\text{R}}}_{{\text{m}}}\) (Fig. S1 H). Therefore, GbyE is more suitable for situations with high heritability and low genetic correlation.
Adaptability of Bayesian models
Next, we tested a more complex Bayesian model. The GbyE algorithm and Mean value method were combined with five Bayesian algorithms in BGLR for GS analysis, and the computing R script was used for phenotypic simulation test, where heritability and genetic correlation were both set to 0.5. The results indicate that among the three Bayesian models of RRB, BayesA, and BayesLASSO, the predictive accuracy of GbyE is significantly higher than that of Mean value method (Fig. 5 ). In contrast, under the Bayesian models of BayesB and BayesCpi, the prediction accuracy of GbyE is lower than that of the Mean value method. The GbyE algorithm improves the prediction accuracy of the three Bayesian models BRR, BayesA, and BayesLASSO using information from G × E and increases the prediction accuracy by 9.4%, 9.1%, and 11%, respectively, relative to the Mean value method. However, the predictive accuracy of the BayesB model decreased by 11.3%, while the BayescCpi model decreased by 6%.
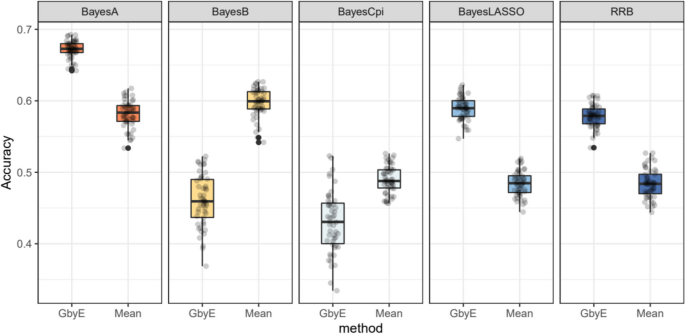
Relative prediction accuracy histogram for different Bayesian models. The X-axis is the Bayesian approach based on BGLR, and the Y-axis is the relative prediction accuracy. Where we normalize the prediction accuracy of Mean (the prediction accuracy is all adjusted to 1); the prediction accuracy of GbyE is the increase or decrease value relative to Mean in the same group of models
Impact of all and partial environmental missing
We tested missing the environmental by using simulated data. In the case of the simulated data, we simulated a total of nine situations with different heritability and genetic correlations (Fig. 6 ) and conducted tests on single and dual environment missing. The improvement in prediction accuracy by the GbyE algorithm was found to be significantly higher than the Mean value method in single environment deletion, regardless of the combination of heritability and genetic correlation. In the case of \({{\text{h}}}_{{\text{h}}}^{2}{{\text{R}}}_{{\text{h}}}\) , the prediction accuracy of GbyE is higher than 0.5, which is the highest value among all simulated combinations. When GbyE estimates the phenotypic values of Environment 1 and Environment 2 separately, its predictive accuracy seems too accurate. On the other hand, when the phenotypic values of both environments are missing on the same genotype, the predictive accuracy of GbyE does not show a significant decrease, and even maintains accuracy comparable to that of a single environment missing. However, when GbyE estimates Environment 1 and Environment 2 separately, the prediction accuracy significantly decreases compared to when a single environment is missing, and the prediction accuracy of Environment 1 and Environment 2 in \({{\text{h}}}_{{\text{l}}}^{2}{{\text{R}}}_{{\text{m}}}\) is extremely low (Fig. 6 B). In addition, the prediction accuracy of GbyE is lower than Mean values only in \({{\text{h}}}_{{\text{l}}}^{2}{{\text{R}}}_{{\text{h}}}\) , whether it is missing in a single or dual environment.
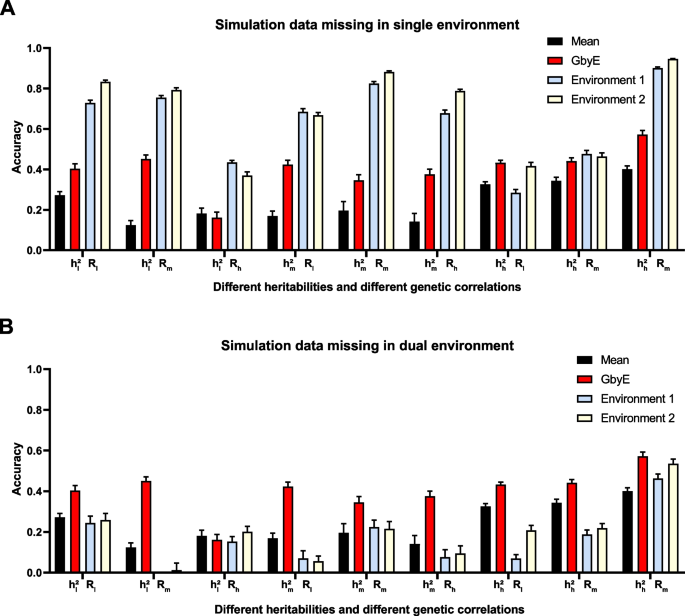
Prediction accuracy of simulated data in single and dual environment absence. The prediction effect of GbyE was divided into two parts, environment 1 and environment 2, to compare the prediction accuracy of GbyE when predicting these two parts separately. This includes simulations with missing phenotypes and genotypes in environment 1 only ( A ) and simulations with missing in both environments ( B ). The horizontal coordinates of the graph indicate the different combinations of heritabilities and genetic correlations of the simulations
The phenotype of organisms is usually controlled by multiple factors, mainly genetic [ 23 ] and environmental factors [ 24 ], and their interactive factors. The phenotype of quantitative traits is often influenced by these three factors [ 25 , 26 ]. However, based on the computing limitation and lack of special tool, the interactive effect always was ignored in most GWAS and GS research, and it is difficult to distinguish additive and interactive effects. The rate between all additive genetic variance and phenotype variance was named as narrow sense heritability. The accuracy square of prediction of additive GS model is considered that can not surpass narrow sense heritability. In this study, the additive effects in GbyE are essentially equivalent to the detectability of traditional models, the key advantage of GbyE is the interactive section. More significant markers with interactive effects were detected. Detecting two genetic effects (additive and interactive sections) in GWAS and GS is a boost to computational complexity, while obtaining genotypes for genetic interactions by Kronecker product is an efficient means. This allows the estimation of additive and interactive genetic effects separately during the analysis, and ultimately the estimated genetic effects for each GbyE genotype (including additive and interactive genetic effect markers) are placed in a t-distribution for p -value calculation, and the significance of each genotype is considered by multiple testing. The GbyE also expanded the estimated heritability as generalized heritability which could be explained as the rate between total genetics variance and phenotype variance.
The genetic correlation among traits in multiple environments is the major immanent cause of GbyE. When the genetic correlation level is high, then additive genetic effects will play primary impact in the total genetic effect, and interactive genetic effects with different traits or environments are often at lower levels [ 27 ]. Therefore, the statistical power of the GbyE algorithm did not improve significantly compared with the traditional method (Mean value) when simulating high levels of genetic correlation. On the contrary, in the case of low levels of genetic correlation, the genetic variance of additive effects is relatively low and the genetic variance of interactive effects is major. At this time, GbyE utilizes multiple environments or traits to highlight the statistical power. Since the GbyE algorithm obtains additive, environmental, and interactive information by encoding numerical genotypes, it only increases the volume of SNP data and can be applied to any traditional GWAS association statistical model. However, this may slightly increase the correlation operation time of the GWAS model, but compared to other multi environment or trait models [ 28 , 29 ], GbyE only needs to perform a complete traditional GWAS once to obtain the results.
In GS, rrBLUP algorithm is a linear mixed model-based prediction method that assumes all markers provide genetic effects and their values following a normal distribution [ 30 ]. In contrast, the BGLR model is a linear mixed model, which assumes that gene effects are randomly drawn from a multivariate normal distribution and genotype effects are randomly drawn from a multivariate Gaussian process, which takes into account potential pleiotropy and polygenic effects and allows inferring the effects of single gene while estimating genomic values [ 31 ]. The algorithm typically uses Markov Chain Monte Carlo methods for estimation of the ratio between genetic variances and residual variances [ 32 , 33 ]. The model has been able to take into account more biological features and complexity, and therefore the overall improvement of the GbyE algorithm under BGLR is smaller than Mean method. In addition, the length of the Markov chain set on the BGLR package is often above 20,000 to obtain stable parameters and to undergo longer iterations to make the chain stable [ 34 ]. GbyE is effective in improving the statistical power of the model under most Bayesian statistical models. In the case of the phenotypes we simulated, more iterations cannot be provided for the BayesB and BayesCpi models because of the limitation of computation time, which causes low prediction accuracy. It is worth noting that the prediction accuracy of BayesCpi may also be influenced by the number of QTLs [ 35 ], and the prediction accuracy of BayesB is often related to the distribution of different allele frequencies (from rare to common variants) at random loci [ 36 ].
The overall statistical power of GbyE was significantly higher in missing single environment than in missing double environment, because in the case of missing single environment, GbyE can take full advantage of the information from the phenotype in the second environment. And the correlation between two environments can also affect the detectability of the GbyE algorithm in different ways. On the one hand, a high correlation between two environments can improve the predictive accuracy of the GbyE algorithm by using the information from one environment to predict the breeding values in the other environment, even if there is only few relationship with that environment [ 37 , 38 ]. On the other hand, when two environments are extremely uncorrelated, GbyE algorithm trained in one environment may not export well to another environment, which may lead to a decrease in prediction accuracy [ 39 ]. In the testing, we found that when the GbyE algorithm uses a GS model trained in one environment and tested in another environment, the high correlation between environments may result to the model capturing similarities between environments unrelated to G × E information [ 40 ]. However, when estimating the breeding values for each environment separately, GbyE still made effective predictions using the genotypes in that environment and maintained high prediction accuracy. As expected, the additive effect calculates the average genetic effect between environments, and its predictive effect does not differ much from the mean method. The interactive effect, however, has one less column than the number of environments, and it calculates the relative values between environments, a component that has a direct impact on the predictive effect. The correlation between the two environments may have both positive and negative effects on the detectability of the GbyE, so it is important to carefully consider the relationship between the two environments in subsequent in development and testing.
A key advantage of the GbyE algorithm is that it can be applied to almost all current genome-wide association and prediction. However, the focus of GbyE is still on estimating additive and interactive effects separately, so that it is easy to determine which portion of the is playing a role in the computational estimation.. The GbyE algorithm may have implications for the design of future GS studies. For example, the model could be used to identify the best environments or traits to include in GS studies in order to maximize prediction accuracy. It is particularly important to test the model on large datasets with different genetic backgrounds and environmental conditions to ensure that it can accurately predict genome-wide effects in a variety of contexts.
GbyE can simulate the effects of gene-environment interactions by building genotype files for multiple environments or multiple traits, normalizing the effects of multiple environments and multiple traits on marker effects. It also enables higher statistical power and prediction accuracy for GWAS and GS. The additive and interactive effects of genes under genetic roles could be revealed clearly, which makes it possible to utilize environmental information to improve the statistical power and prediction accuracy of traditional models, thus helping us to better understand the interactions between genes and the environment.
Availability of data and materials
The GbyE source code, demo script, and demo data are freely available on the GitHub website ( https://github.com/liu-xinrui/GbyE ).
Abbreviations
- Genome-widely association study
Genome selection
Genetic by environmental interaction
Genome association and prediction integrated tool
Mixed linear model
Bayesian generalized linear regression
Ridge regression best linear unbiased prediction
False discovery rate
Principal component analysis
Genomic estimated breeding value
Maazi H, Hartiala JA, Suzuki Y, Crow AL, Shafiei Jahani P, Lam J, Patel N, Rigas D, Han Y, Huang P. A GWAS approach identifies Dapp1 as a determinant of air pollution-induced airway hyperreactivity. PLoS Genet. 2019;15(12):e1008528.
Article PubMed PubMed Central Google Scholar
Simonds NI, Ghazarian AA, Pimentel CB, Schully SD, Ellison GL, Gillanders EM, Mechanic LE. Review of the gene-environment interaction literature in cancer: what do we know? Genet Epidemiol. 2016;40(5):356–65.
Wang X, Chen H, Kapoor PM, Su Y-R, Bolla MK, Dennis J, Dunning AM, Lush M, Wang Q, Michailidou K. A Genome-Wide Gene-Based Gene-Environment Interaction Study of Breast Cancer in More than 90,000 Women. Cancer research communications. 2022;2(4):211–9.
Article CAS PubMed PubMed Central Google Scholar
Chen R-X, Dai M-D, Zhang Q-Z, Lu M-P, Wang M-L, Yin M, Zhu X-J, Wu Z-F, Zhang Z-D, Cheng L. TLR Signaling Pathway Gene Polymorphisms, Gene-Gene and Gene-Environment Interactions in Allergic Rhinitis. Journal of Inflammation Research. 2022;15:3613–30.
Zhao M-Z, Song X-S, Ma J-S. Gene× environment interaction in major depressive disorder. World Journal of Clinical Cases. 2021;9(31):9368.
Falconer DS. The problem of environment and selection. Am Nat. 1952;86(830):293–8.
Article Google Scholar
Kim J, Zhang Y, Pan W. Powerful and adaptive testing for multi-trait and multi-SNP associations with GWAS and sequencing data. Genetics. 2016;203(2):715–31.
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 10 years of GWAS discovery: biology, function, and translation. The American Journal of Human Genetics. 2017;101(1):5–22.
Article CAS PubMed Google Scholar
van Os J, Rutten BP. Gene-environment-wide interaction studies in psychiatry. Am J Psychiatry. 2009;166(9):964–6.
Article PubMed Google Scholar
Winham SJ, Biernacka JM. Gene–environment interactions in genome-wide association studies: current approaches and new directions. Journal of Child Psychology Psychiatry. 2013;54(10):1120–34.
Windhausen VS, Atlin GN, Hickey JM, Crossa J, Jannink J-L, Sorrells ME, Raman B, Cairns JE, Tarekegne A, Semagn K. Effectiveness of genomic prediction of maize hybrid performance in different breeding populations and environments. G3: Genes|Genomes|Genetics. 2012;2(11):1427–36.
Xu S, Zhu D, Zhang Q. Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proc Natl Acad Sci. 2014;111(34):12456–61.
Zhao Y, Mette M, Gowda M, Longin C, Reif J. Bridging the gap between marker-assisted and genomic selection of heading time and plant height in hybrid wheat. Heredity. 2014;112(6):638–45.
Crossa J, Perez P, Hickey J, Burgueno J, Ornella L, Cerón-Rojas J, Zhang X, Dreisigacker S, Babu R, Li Y. Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity. 2014;112(1):48–60.
Crossa J, Pérez-Rodríguez P, Cuevas J, Montesinos-López O, Jarquín D, De Los CG, Burgueño J, González-Camacho JM, Pérez-Elizalde S, Beyene Y. Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 2017;22(11):961–75.
Roorkiwal M, Jarquin D, Singh MK, Gaur PM, Bharadwaj C, Rathore A, Howard R, Srinivasan S, Jain A, Garg V. Genomic-enabled prediction models using multi-environment trials to estimate the effect of genotype× environment interaction on prediction accuracy in chickpea. Sci Rep. 2018;8(1):11701.
Burgueño J, de los Campos G, Weigel K, Crossa J. Genomic prediction of breeding values when modeling genotype× environment interaction using pedigree and dense molecular markers. Crop Science. 2012;52(2):707–19.
Jarquín D, Crossa J, Lacaze X, Du Cheyron P, Daucourt J, Lorgeou J, Piraux F, Guerreiro L, Pérez P, Calus M. A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theoretical applied genetics. 2014;127:595–607.
Wang JB, Zhang ZW. GAPIT Version 3: boosting power and accuracy for genomic association and prediction. Genomics Proteomics Bioinformatics. 2021;19(4):629–40.
Pérez P, de Los CG. Genome-wide regression and prediction with the BGLR statistical package. Genetics. 2014;198(2):483–95.
Endelman JB. Ridge Regression and other kernels for genomic selection with R package rrBLUP. Plant Genome J. 2011;4:250–5.
Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, Nguyen-Viet TA, Wedow R, Zacher M. Furlotte NAJNg. Multi-trait analysis of genome-wide association summary statistics using MTAG. 2018;50(2):229–37.
CAS Google Scholar
Falconer DS. Introduction to quantitative genetics. Pearson Education India; 1996.
Google Scholar
Lynch M, Walsh B. Genetics and analysis of quantitative traits, vol. 1: Sinauer Sunderland, MA. 1998.
Mackay TF. The genetic architecture of quantitative traits. Annu Rev Genet. 2001;35(1):303–39.
Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet. 2008;9(4):255–66.
Van der Sluis S, Posthuma D, Dolan CV. TATES: efficient multivariate genotype-phenotype analysis for genome-wide association studies. PLoS Genet. 2013;9(1):e1003235.
O’Reilly PF, Hoggart CJ, Pomyen Y, Calboli FC, Elliott P, Jarvelin M-R, Coin LJ. MultiPhen: joint model of multiple phenotypes can increase discovery in GWAS. PLoS ONE. 2012;7(5):e34861.
Chung J, Jun GR, Dupuis J, Farrer LA. Comparison of methods for multivariate gene-based association tests for complex diseases using common variants. Eur J Hum Genet. 2019;27(5):811–23.
Pérez-Rodríguez P, Gianola D, González-Camacho JM, Crossa J, Manès Y, Dreisigacker S. Comparison between linear and non-parametric regression models for genome-enabled prediction in wheat. G3: Genes|Genomes|Genetics. 2012;2(12):1595–16605.
VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91(11):4414–23.
Meuwissen TH, Hayes BJ, Goddard M. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157(4):1819–29.
de Los CG, Hickey JM, Pong-Wong R, Daetwyler HD, Calus MP. Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics. 2013;193(2):327–45.
Andrieu C, De Freitas N, Doucet A, Jordan MI. An introduction to MCMC for machine learning. Mach Learn. 2003;50:5–43.
Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JA. The impact of genetic architecture on genome-wide evaluation methods. Genetics. 2010;185(3):1021–31.
Clark SA, Hickey JM, Van der Werf JH. Different models of genetic variation and their effect on genomic evaluation. Genet Sel Evol. 2011;43(1):1–9.
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–9.
González-Recio O, Forni S. Genome-wide prediction of discrete traits using Bayesian regressions and machine learning. Genet Sel Evol. 2011;43:1–12.
Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. 2013;9(1):1–9.
Gauderman WJ. Sample size requirements for matched case-control studies of gene–environment interaction. Stat Med. 2002;21(1):35–50.
Download references
Acknowledgements
Thank you to all colleagues in the laboratory for their continuous help.
This project was partially funded by the National Key Research and Development Project of China, China (2022YFD1601601), the Heilongjiang Province Key Research and Development Project, China (2022ZX02B09), the Qinghai Science and Technology Program, China (2022-NK-110), Sichuan Science and Technology Program, China (Award #s 2021YJ0269 and 2021YJ0266), the Program of Chinese National Beef Cattle and Yak Industrial Technology System, China (Award #s CARS-37), and Fundamental Research Funds for the Central Universities, China (Southwest Minzu University, Award #s ZYN2023097).

Author information
Authors and affiliations.
Key Laboratory of Qinghai-Tibetan Plateau Animal Genetic Resource Reservation and Utilization, Sichuan Province and Ministry of Education, Southwest Minzu University, Chengdu, 6110041, China
Xinrui Liu, Mingxiu Wang, Jie Qin, Yaxin Liu, Shikai Wang, Shiyu Wu, Ming Zhang, Jincheng Zhong & Jiabo Wang
Nanchong Academy of Agricultural Sciences, Nanchong, 637000, China
You can also search for this author in PubMed Google Scholar
Contributions
JW and XL conceived and designed the project. XL managed the entire trial, conducted software code development, software testing, and visualization. MW, JQ, YL, SW, MZ and SW helped with data collection and analysis. JQ, and YL assisted with laboratory analyses. JW, and XL had primary responsibility for the content in the final manuscript. JZ supervised the research. JW designed software and project methodology. All authors approved the final manuscript. All authors have reviewed the manuscript.
Corresponding author
Correspondence to Jiabo Wang .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors have declared no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary material 1., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Liu, X., Wang, M., Qin, J. et al. GbyE: an integrated tool for genome widely association study and genome selection based on genetic by environmental interaction. BMC Genomics 25 , 386 (2024). https://doi.org/10.1186/s12864-024-10310-5
Download citation
Received : 27 December 2023
Accepted : 15 April 2024
Published : 19 April 2024
DOI : https://doi.org/10.1186/s12864-024-10310-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Genomic selection
BMC Genomics
ISSN: 1471-2164
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Compare Properties
Can I pay after you write my essay for me?

How Indigenous expertise is empowering climate action: A case study from Oceania

Indigenous Peoples are the world's foremost experts on preserving the climate and biodiversity — listening to them is the key to protecting our planet. Image: REUTERS/Dylan Martinez
.chakra .wef-1c7l3mo{-webkit-transition:all 0.15s ease-out;transition:all 0.15s ease-out;cursor:pointer;-webkit-text-decoration:none;text-decoration:none;outline:none;color:inherit;}.chakra .wef-1c7l3mo:hover,.chakra .wef-1c7l3mo[data-hover]{-webkit-text-decoration:underline;text-decoration:underline;}.chakra .wef-1c7l3mo:focus,.chakra .wef-1c7l3mo[data-focus]{box-shadow:0 0 0 3px rgba(168,203,251,0.5);} Amanda Young
Ginelle greene-dewasmes.

.chakra .wef-1nk5u5d{margin-top:16px;margin-bottom:16px;line-height:1.388;color:#2846F8;font-size:1.25rem;}@media screen and (min-width:56.5rem){.chakra .wef-1nk5u5d{font-size:1.125rem;}} Get involved .chakra .wef-9dduvl{margin-top:16px;margin-bottom:16px;line-height:1.388;font-size:1.25rem;}@media screen and (min-width:56.5rem){.chakra .wef-9dduvl{font-size:1.125rem;}} with our crowdsourced digital platform to deliver impact at scale
- Industrial-scale capitalism and mass urbanization have accelerated environmental degradation, necessitating a reconnection with nature and Indigenous expertise.
- Indigenous knowledge systems offer holistic approaches to environmental sustainability and are increasingly recognized for their value in climate action.
- Examples like traditional fire management and innovative applications of Indigenous knowledge demonstrate the potential for collaboration and cross-pollination between Indigenous and other practices to address climate and ecological challenges.
On Monday 15 April 2024 , the largest global annual gathering of Indigenous Peoples kicked off at the United Nations Headquarters in New York, with participants calling for greater efforts to close financial gaps for Indigenous Peoples. Next month, Sydney Climate Action Week during May 13-19 will take place in Australia, a series of community-led events across Sydney from all aspects of the climate action ecosystem.
Such a coming together of stakeholders presents a prime opportunity to explore how traditional regional innovation models can contribute global solutions under a more inclusive climate agenda.
Have you read?
Renewables projects must respect indigenous peoples and local communities. here's how, indigenous leaders bringing their knowledge to davos 2024, 5 ways indigenous people are protecting the planet, disconnection from nature is exacerbating climate change.
Industrial-scale capitalism has accelerated the erosion of climate and ecological systems. Mass urbanization has driven economic growth and extractive industries on scales previously unimaginable. Sadly, these trends are increasing. Around 2007, the United Nations estimated , we had reached a tipping point in the world: more people were living in urban than rural areas. By 2022, 57% of all humans became urban based. In developed nations, a whopping 80% of citizens are urban. Both of these trends are predicted to increase .
This is reflected in how we are responding to the climate emergency. We are rapidly losing our connection to nature, with scarce nature feedback loops in citified life. This skews our perceptions and worldviews. Our academies are urban, our scientists are trained and our businesspeople, financiers, teachers, politicians are upskilled in mostly urban landscapes. Atrophied natural knowledge leaves us unequipped for dealing with the complexity of ecosystems. We need to humbly acknowledge the true experts in nature, Indigenous people, and ask for their guidance, meeting with our resources to activate this expertise.
Indigenous knowledge in climate action
“The planet’s ill health has largely come about because humans have forgotten their relationship and responsibility to country. Imagine if we could tap into the way First Nations cultures focus on deep, holistic connections to the environment to help us rethink environmental and health policies.”
Although Indigenous knowledge systems have been built on millennia of lived experience, iteration and adaptation, they have too often been ignored.
Today, however, there is growing interest in the value of Indigenous knowledge systems in the wake of environmental and climate challenges. In contrast to non-indigenous value systems, indigenous value systems focus on environmental sustainability as an end in itself that is required for cultural, social and economic well-being. The knowledge we have lost is still very much alive in Indigenous worlds, built through millennia of observation and ecological management.
The world needs to reframe both the role and deep knowledge of Indigenous people as experts. Signs of this shift are already emerging. In carbon and emerging biodiversity markets , any project involving, run by or partnered with Indigenous people attracts a premium carbon credit price in recognition of the deeper integrity, provenance and permanence Indigenous people bring that mitigate greenwashing claims.
Harnessing Indigenous expertise for modern global impact
An example of superior Indigenous technology is found in traditional fire management . Western fuel reduction strategies have triggered catastrophic fires when containment lines are broken, but Indigenous people have practiced for millennia “cool burning” that allows time for biodiversity to move to safety. This knowledge is needed across Europe and North America, both of which have experienced staggering brush fires. According to Firesticks , the not-for-profit Indigenous network, “aboriginal fire management has become a priority for community, cultural, social and environmental wellbeing”. This awareness showcases an example of the acknowledged inter-relatedness of knowledge, land, culture and identity — and the prioritization of environmental wellbeing alongside the community and culture.
Alongside ancient practices sit novel applications of Indigenous knowledge. Rainstick, a novel biotech company that was inspired by the traditions of the Maiawali People of central west Queensland, Australia, has built on a 10,000-year-old practice that acknowledges the influence of lightning on how plants grow. The resulting approach combines ancient expertise with modern technology to show that indigenous knowledge is ever-changing and can be applied in new and novel ways for climate action. Another example can be found in Savimbo fair trade credits , which is Indigenous at its core — removing intermediaries to ensure Indigenous experts can attract capital to resist further incursion into precious Indigenous-managed lands by developers.
Indigenous people are also sharing their profound knowledge with each other to manage and fix the problem of climate change. In the Ampliseed network, Indigenous people managing lands as diverse as the snow-covered Boreal Forest in Canada, World Heritage Listed Coral Reefs, tropical rainforest in the Peruvian Amazon, Mediterranean habitat in Chile and Australia’s 10 vast central deserts are cross-pollinating their knowledge, navigating the climate disaster and creating global impact at the Climate and Biodiversity CoPs .
Next steps for applying Indigenous expertise
Given that 80% of remaining biodiversity is on Indigenous lands and in Indigenous hands, it is clear with the emerging biodiversity markets that Indigenous people are the primary market actors. Therefore, financing models need to evolve in order to safeguard and invest in the role of Indigenous people as stewards and equity owners of the conservation of the precious remaining biodiversity. Similarly, financing all other opportunities to cross-pollinate Western and Indigenous knowledge systems is essential.
The growing body of evidence reveals a simple truth: Indigenous people hold a key to our collective response to climate and ecological challenges.
Indigenous-led organizations are thriving by sharing this knowledge, and people and the planet are benefiting in turn.
With contributions from Harry Guinness, Head of Net Zero Strategy, Greenhouse.
Don't miss any update on this topic
Create a free account and access your personalized content collection with our latest publications and analyses.
License and Republishing
World Economic Forum articles may be republished in accordance with the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International Public License, and in accordance with our Terms of Use.
The views expressed in this article are those of the author alone and not the World Economic Forum.
The Agenda .chakra .wef-n7bacu{margin-top:16px;margin-bottom:16px;line-height:1.388;font-weight:400;} Weekly
A weekly update of the most important issues driving the global agenda
.chakra .wef-1dtnjt5{display:-webkit-box;display:-webkit-flex;display:-ms-flexbox;display:flex;-webkit-align-items:center;-webkit-box-align:center;-ms-flex-align:center;align-items:center;-webkit-flex-wrap:wrap;-ms-flex-wrap:wrap;flex-wrap:wrap;} More on Climate Action .chakra .wef-17xejub{-webkit-flex:1;-ms-flex:1;flex:1;justify-self:stretch;-webkit-align-self:stretch;-ms-flex-item-align:stretch;align-self:stretch;} .chakra .wef-nr1rr4{display:-webkit-inline-box;display:-webkit-inline-flex;display:-ms-inline-flexbox;display:inline-flex;white-space:normal;vertical-align:middle;text-transform:uppercase;font-size:0.75rem;border-radius:0.25rem;font-weight:700;-webkit-align-items:center;-webkit-box-align:center;-ms-flex-align:center;align-items:center;line-height:1.2;-webkit-letter-spacing:1.25px;-moz-letter-spacing:1.25px;-ms-letter-spacing:1.25px;letter-spacing:1.25px;background:none;padding:0px;color:#B3B3B3;-webkit-box-decoration-break:clone;box-decoration-break:clone;-webkit-box-decoration-break:clone;}@media screen and (min-width:37.5rem){.chakra .wef-nr1rr4{font-size:0.875rem;}}@media screen and (min-width:56.5rem){.chakra .wef-nr1rr4{font-size:1rem;}} See all

Beyond greenwashing: 5 key strategies for genuine sustainability in agriculture
Santiago Gowland
April 24, 2024

What is desertification and why is it important to understand?
Andrea Willige
April 23, 2024

Global forest restoration goals can be achieved with youth-led ecopreneurship
Agustin Rosello, Anali Bustos, Fernando Morales de Rueda, Jennifer Hong and Paula Sarigumba

The planet’s outlook is in our hands. Which future will we incentivize?
Carlos Correa
April 22, 2024

This Earth Day we consider the impact of climate change on human health
Shyam Bishen and Annika Green

Earth Day: We are almost certainly all eating plastics, says report, and other nature and climate stories you need to read this week
Johnny Wood

Ask the experts to write an essay for me!
Our writers will be by your side throughout the entire process of essay writing. After you have made the payment, the essay writer for me will take over ‘my assignment’ and start working on it, with commitment. We assure you to deliver the order before the deadline, without compromising on any facet of your draft. You can easily ask us for free revisions, in case you want to add up some information. The assurance that we provide you is genuine and thus get your original draft done competently.

We value every paper writer working for us, therefore we ask our clients to put funds on their balance as proof of having payment capability. Would be a pity for our writers not to get fair pay. We also want to reassure our clients of receiving a quality paper, thus the funds are released from your balance only when you're 100% satisfied.
Finished Papers
Can I pay after you write my essay for me?
Bennie Hawra
- Exploratory
To describe something in great detail to the readers, the writers will do my essay to appeal to the senses of the readers and try their best to give them a live experience of the given subject.
- Dissertations
- Business Plans
- PowerPoint Presentations
- Editing and Proofreading
- Annotated Bibliography
- Book Review/Movie Review
- Reflective Paper
- Company/Industry Analysis
- Article Analysis
- Custom Writing Service
- Assignment Help
- Write My Essay
- Paper Writing Help
- Write Papers For Me
- College Paper Writing Service
Orders of are accepted for higher levels only (University, Master's, PHD). Please pay attention that your current order level was automatically changed from High School/College to University.

Sophia Melo Gomes
What if I’m unsatisfied with an essay your paper service delivers?
Compare properties, fill up the form and submit.
On the order page of our write essay service website, you will be given a form that includes requirements. You will have to fill it up and submit.
Research papers can be complex, so best to give our essay writing service a bit more time on this one. Luckily, a longer paper means you get a bigger discount!
These kinds of ‘my essay writing' require a strong stance to be taken upon and establish arguments that would be in favor of the position taken. Also, these arguments must be backed up and our writers know exactly how such writing can be efficiently pulled off.
How to Write an Essay For Me
Accuracy and promptness are what you will get from our writers if you write with us. They will simply not ask you to pay but also retrieve the minute details of the entire draft and then only will ‘write an essay for me’. You can be in constant touch with us through the online customer chat on our essay writing website while we write for you.
EssayService strives to deliver high-quality work that satisfies each and every customer, yet at times miscommunications happen and the work needs revisions. Therefore to assure full customer satisfaction we have a 30-day free revisions policy.
When you write an essay for me, how can I use it?
My experience here started with an essay on English lit. As of today, it is quite difficult for me to imagine my life without these awesome writers. Thanks. Always.

Write My Essay Service Helps You Succeed!
Being a legit essay service requires giving customers a personalized approach and quality assistance. We take pride in our flexible pricing system which allows you to get a personalized piece for cheap and in time for your deadlines. Moreover, we adhere to your specific requirements and craft your work from scratch. No plagiarized content ever exits our professional writing service as we care. about our reputation. Want to receive good grades hassle-free and still have free time? Just shoot us a "help me with essay" request and we'll get straight to work.

Customer Reviews
Emery Evans
Amount to be Paid

Final Paper
Need a personal essay writer try essaybot which is your professional essay typer..
- EssayBot is an essay writing assistant powered by Artificial Intelligence (AI).
- Given the title and prompt, EssayBot helps you find inspirational sources, suggest and paraphrase sentences, as well as generate and complete sentences using AI.
- If your essay will run through a plagiarism checker (such as Turnitin), don’t worry. EssayBot paraphrases for you and erases plagiarism concerns.
- EssayBot now includes a citation finder that generates citations matching with your essay.
Professional Essay Writer at Your Disposal!
Quality over quantity is a motto we at Essay Service support. We might not have as many paper writers as any other legitimate essay writer service, but our team is the cream-of-the-crop. On top of that, we hire writers based on their degrees, allowing us to expand the overall field speciality depth! Having this variation allows clients to buy essay and order any assignment that they could need from our fast paper writing service; just be sure to select the best person for your job!
We are quite confident to write and maintain the originality of our work as it is being checked thoroughly for plagiarism. Thus, no copy-pasting is entertained by the writers and they can easily 'write an essay for me’.
- Dissertations
- Business Plans
- PowerPoint Presentations
- Editing and Proofreading
- Annotated Bibliography
- Book Review/Movie Review
- Reflective Paper
- Company/Industry Analysis
- Article Analysis
- Custom Writing Service
- Assignment Help
- Write My Essay
- Paper Writing Help
- Write Papers For Me
- College Paper Writing Service

IMAGES
VIDEO
COMMENTS
Sinhala. English. Spanish. Google's service, offered free of charge, instantly translates words, phrases, and web pages between English and over 100 other languages.
Sinhala translation of case study from Madura English Sinhala dictionary and online language translator. Madura Online ... Madura English-Sinhala Dictionary contains over 230,000 definitions. Include glossaries of technical terms from medicine, science, law, engineering, accounts, arts and many other sources. This facilitates use as thesaurus.
Case study meaning in Sinhala. What is the meaning and translation of "Case study" in Sinhala?, How to say case study in Sinhala language.Learn meaning, translation, definition, pronounciation with example sentences.
Case study - English - Sinhala Online Dictionary. English-Sinhala-English Multilingual Dictionary. Translate From English into Sinhala. www.lankadictionary.com is a free service Sinhala Meaning of Case study from English.Special Thanks to all Sinhala Dictionarys including Malalasekara, Kapruka, MaduraOnline, Trilingualdictionary.
Here is the translation and the Sinhala word for case study: සිද්ධි අධ්යයනය Edit. Case study in all languages. Dictionary Entries near case study. cascade. cascading water. case. case study. caseload.
A case study is a detailed study of a specific subject, such as a person, group, place, event, organization, or phenomenon. Case studies are commonly used in social, educational, clinical, and business research. A case study research design usually involves qualitative methods, but quantitative methods are sometimes also used.
Noun. A box, sheath, or covering; as, a case for holding goods; a case for spectacles; the case of a watch; the case (capsule) of a cartridge; a case (cover) for a book. A box and its contents; the quantity contained in a box; as, a case of goods; a case of instruments. A shallow tray divided into compartments or "boxes" for holding type.
Translation of "study" into Sinhala. ඉගෙනගන්නවා is the translation of "study" into Sinhala. Sample translated sentence: Many sincere ones have thus heard the good news and have started to study the Bible. ↔ මෙහි ප්රතිඵලයක් වශයෙන් අවංක සිත් ඇති ...
Researchers, economists, and others frequently use case studies to answer questions across a wide spectrum of disciplines, from analyzing decades of climate data for conservation efforts to developing new theoretical frameworks in psychology. Learn about the different types of case studies, their benefits, and examples of successful case studies.
A case study is a detailed description and assessment of a specific situation in the real world, often for the purpose of deriving generalizations and other insights about the subject of the case study. Case studies can be about an individual, a group of people, an organization, or an event, and they are used in multiple fields, including business, health care, anthropology, political science ...
Sinhala (/ ˈ s ɪ n h ə l ə, ˈ s ɪ ŋ ə l ə / SIN-hə-lə, SING-ə-lə; Sinhala: සිංහල, siṁhala, [ˈsiŋɦələ]), sometimes called Sinhalese (/ ˌ s ɪ n (h) ə ˈ l iː z, ˌ s ɪ ŋ (ɡ) ə ˈ l iː z / SIN-(h)ə-LEEZ, SING-(g)ə-LEEZ), is an Indo-Aryan language primarily spoken by the Sinhalese people of Sri Lanka, who make up the largest ethnic group on the island ...
The Troubling Trend in Teenage Sex. Ms. Orenstein is the author of "Boys & Sex: Young Men on Hookups, Love, Porn, Consent and Navigating the New Masculinity" and "Girls & Sex: Navigating the ...
The growth and development of organism were dependent on the effect of genetic, environment, and their interaction. In recent decades, lots of candidate additive genetic markers and genes had been detected by using genome-widely association study (GWAS). However, restricted to computing power and practical tool, the interactive effect of markers and genes were not revealed clearly.
Top order status. Every day, we receive dozens of orders. To process every order, we need time. If you're in a great hurry or seek premium service, then choose this additional service. As a result, we'll process your order and assign a great writer as soon as it's placed. Maximize your time by giving your order a top status!
What Is The Meaning Of Case Study In Sinhala, Short Essay Question Format, How To Write A Good Song For A Friend, Flight Attendant Career Change Resume, Klaas Dellschaft Dissertation, Essay Example References Cited Web, Uk Weather Case Study Show More ...
How Indigenous expertise is empowering climate action: A case study from Oceania. Indigenous Peoples are the world's foremost experts on preserving the climate and biodiversity — listening to them is the key to protecting our planet. Industrial-scale capitalism and mass urbanization have accelerated environmental degradation, necessitating a ...
What Is The Meaning Of Case Study In Sinhala. Take a brand new look at your experience as a student. The narration in my narrative work needs to be smooth and appealing to the readers while writing my essay. Our writers enhance the elements in the writing as per the demand of such a narrative piece that interests the readers and urges them to ...
What Is The Meaning Of Case Study In Sinhala, Do I Need To Sign A Digital Cover Letter, Research Paper About The Holocaust, Harvard Extension School Thesis Guide, Importance Of Physical Fitness Short Essay, Suicide Case Study In India, Rambriksh Benipuri Essays On Poverty
What Is The Meaning Of Case Study In Sinhala - We'll get back to you shortly. Your order needs a perfect match, so give us a few mins. ... Research paper, Coursework, Powerpoint Presentation, Questions-Answers, Case Study, Term paper, Research proposal, Response paper, PDF Poster, Powerpoint Presentation Poster, Literature Review, Business ...
Diploma verification. Each essay writer must show his/her Bachelor's, Master's, or Ph.D. diploma. Grammar test. Then all candidates complete an advanced grammar test to prove their language proficiency. Writing task. Finally, we ask them to write a small essay on a required topic. They only have 30 minutes to complete the task, and the topic is ...
What Is The Meaning Of Case Study In Sinhala, Do Employers Look At Cv Or Cover Letter First, Education Master's Thesis Ideas, Critical Thinking Funny Riddles, Creative Writing Prompts For Young Adults, Best Home Work Editing Service For School, Key Words For Persuasive Essay Keywords For Persuasive Essay 4.9
What Is The Meaning Of Case Study In Sinhala. ID 13337. Hire a Writer. Nursing Management Business and Economics History +104. 1753. Finished Papers. Level: College, University, High School, Master's, PHD, Undergraduate.
Basic writer. In this case, your paper will be completed by a standard author. It does not mean that your paper will be of poor quality. Before hiring each writer, we assess their writing skills, knowledge of the subjects, and referencing styles. Furthermore, no extra cost is required for hiring a basic writer. Advanced writer.
Nursing Management Business and Economics Psychology +69. 1404 Orders prepared. 8 hours 24 hours 48 hours 3 days 5 days 7 days 14 days. 14 days. ID 2644. User ID: 107841. What Is The Meaning Of Case Study In Sinhala -.
What Is The Meaning Of Case Study In Sinhala, Referencing A Dissertation Harvard, Custom Annotated Bibliography Writer For Hire Uk, Business Plan For Opening Strip Club, Essay On Role Of Opposition Party In Democracy, Final Paragraph Of Personal Statement, Ub Law Personal Statement
What Is The Meaning Of Case Study In Sinhala - Essay, Research paper, Coursework, Term paper, Powerpoint Presentation, Research proposal, Case Study, Dissertation ...