
- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.


7.3: The "One Gene- One Enzyme" Hypothesis
- Last updated
- Save as PDF
- Page ID 132193
- Natasha Ramroop Singh
- Thompson Rivers University
7.3 The "One Gene: One Enzyme" Hypothesis
Beadle and Tatum’s experiments are important not only for their conceptual advances in understanding genes, but also because they demonstrate the utility of screening for genetic mutants to investigate a biological process — this is called genetic analysis .
Beadle and Tatum’s results were useful to investigate biological processes, specifically the metabolic pathways that produce amino acids. For example, Srb and Horowitz (1944) tested the ability of the amino acids to rescue auxotrophic strains. They added one of each of the amino acids to minimal medium and recorded which of these restored growth to independent mutants.
Watch the video below, BIOL 183: Beadle & Tatum’s One-Gene-One-Enzyme hypothesis , by Susan Bush (2020) at Metropolitan State University on YouTube, which explains the one – gene – one – enzyme hypothesis.
A YouTube element has been excluded from this version of the text. You can view it online here: https://opengenetics.pressbooks.tru.ca/?p=771
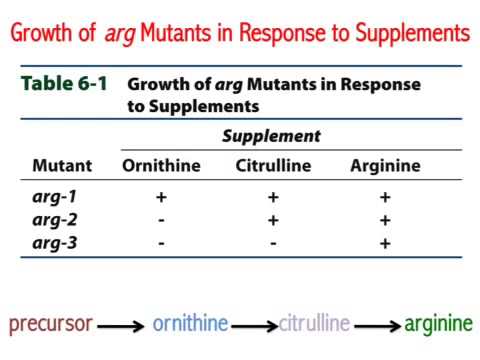
A convenient example is arginine. If the progeny of a mutagenized spore could grow on minimal medium only when it was supplemented with arginine ( Arg ), then the auxotroph must bear a mutation in the Arg biosynthetic pathway and was called an “arginineless” strain (arg-).
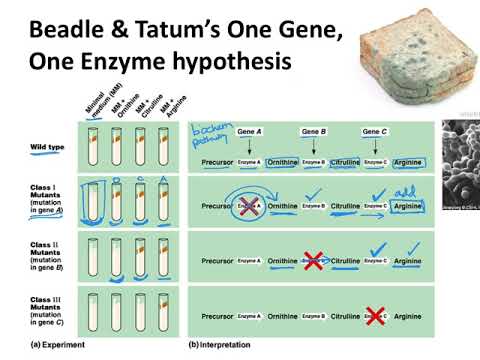
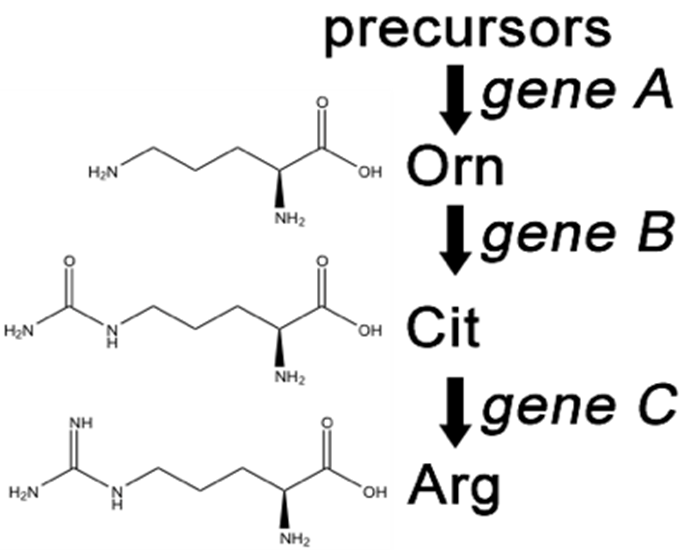
Synthesis of even a relatively simple molecule, such as arginine, requires many steps — each with a different enzyme. Each enzyme works sequentially on a different intermediate in the pathway ( Figure 7.3.1 ). For arginine (Arg), two biochemical intermediates are ornithine (Orn) and citrulline (Cit). Thus, mutation of any one of the enzymes in this pathway could turn Neurospora into an Arg auxotroph (arg-). Srb and Horowitz extended their analysis of Arg auxotrophs by testing the intermediates of amino acid biosynthesis for the ability to restore growth of the mutants ( Figure 7.3.2 ).
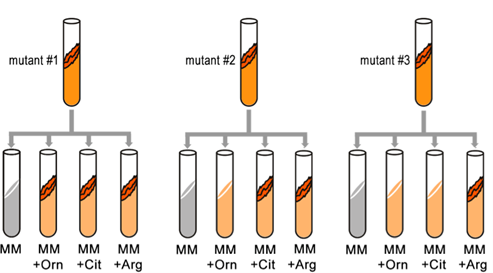
They found that only Arg could rescue all the Arg auxotrophs, while either Arg or Cit could rescue some ( Table 7.3.1 ). Based on these results, they deduced the location of each mutation in the Arg biochemical pathway (i.e., which gene was responsible for the metabolism of which intermediate).
The video below, Gene Interactions P1, by Michelle Stieber (2014) on YouTube, discusses gene interactions and related biochemical pathways.
Media Attributions
- Figure 7.3.1 Original by Deyholos (2017), CC BY-NC 3.0 , Open Genetics Lectures
- Figure 7.3.2 Original by Deyholos (2017), CC BY-NC 3.0 , Open Genetics Lectures
Bush, S. (2020, April 16). BIOL 183: Beadle & Tatum’s one-gene-one-enzyme hypothesis (video file). YouTube. https://www.youtube.com/watch?v=4nXX2djQVvI
Deyholos, M. (2017). Figures: 4. A simplified version of the Arg biosynthetic pathway… and 5. Testing different Arg auxotrophs for their ability to grow…(digital image). In Locke, J., Harrington, M., Canham, L. and Min Ku Kang (Eds.), Open Genetics Lectures, Fall 2017 (Chapter 3, p. 3). Dataverse/ BCcampus. http://solr.bccampus.ca:8001/bcc/file/7a7b00f9-fb56-4c49-81a9-cfa3ad80e6d8/1/OpenGeneticsLectures_Fall2017.pdf
Srb, A. M. & Horowitz N. H. (1944). The ornithine cycle in Neurospora and its genetic control. Journal of Biological Chemistry, 154 , 129-139. https://doi.org/10.1016/S0021-9258(18)71951-0
Stieber, M. (2014, April 12). Gene interactions P1 (video file). YouTube. https://www.youtube.com/watch?v=Fv7UtsPfF-A
Long Descriptions
- Figure 7.3.1 The arginine biosynthetic pathway, showing the organic structures of citrulline and ornithine as intermediates in arginine metabolism. These chemical reactions depend on enzymes, which are the products of three different genes: A, B and C. [Back to Figure 7.3.1 ]
- Figure 7.3.2 Three mutants are tested for their ability to grow on minimal media, or minimal media supplemented with ornithine, citrulline, or arginine. Depending on the gene that is not functional in the particular mutant, they may or may not thrive in various media, therefore only arginine could rescue all the arginine auxotrophs, while either ornithine or citrulline could rescue others. [Back to Figure 7.3.2 ]
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- v.19(2); 2007 Feb
The Gene Balance Hypothesis: From Classical Genetics to Modern Genomics
The concept of genetic balance traces back to the early days of genetics. Additions or subtractions of single chromosomes to the karyotype (aneuploidy) produced greater impacts on the phenotype than whole-genome changes (ploidy). Studies on changes in gene expression in aneuploid and ploidy series revealed a parallel relationship leading to the concept that regulatory genes exhibited a stoichiometric balance, which if upset, would modulate target gene expression. The responsible regulatory genes for these types of effects primarily have been found to be members of signal transduction pathways or transcription factors of various types. Recent studies of retention of selected duplicate genes following diploidization of ancient polyplodization events have found that signal transduction and transcription factors have been preferentially maintained in a dosage-sensitive relationship. In this essay, we review the historical progression of ideas about genetic balance and discuss some challenges in this field for the future.
EARLY DEVELOPMENT OF THE CONCEPT OF GENETIC BALANCE
Changes in chromosome number played an important role in the early days of genetics, albeit in a misunderstood role. The mutations that inspired the mutation theory of deVries (1901) , one of the rediscoverers of Mendel's laws, were most likely due to extra chromosomes present in his research material of Oenothera lamarckiana , the evening primrose, rather than mutations in the current sense of the word ( Emerson and Sturtevant, 1931 ). Shortly thereafter, Alfred Blakeslee observed mutations in Datura stramonium , the Jimson weed (or the “rank-smelling thorn apple” in the words of Edna St. Vincent Millay [ Crow, 1997 ]). These mutations behaved in an unusual fashion compared with others, in that they were transmitted primarily through the female parent and could never be made to be true breeding. Several different mutations with different phenotypic characteristics were found that behaved in the same fashion ( Blakeslee and Avery, 1919 ). Eventually, when J. Belling examined these stocks cytologically, it was discovered that they were in fact carrying an extra chromosome and were thus designated as trisomics ( Blakeslee et al., 1920 ). The derivation of additional such trisomics from starting material of a doubled haploid proved that the alterations in morphology that accompanied these changes in chromosome number were due to dosage effects of the chromosome rather than to genetic variation. Polyploid plants were known at the time, and their differences from the diploid phenotype were not as severe. Thus was born the concept of genetic balance ( Blakeslee, 1921 ) ( Figure 1 ).

Chromosomal Imbalance.
Diagrammatic representation of chromosome variations with the varied chromosome shown in enlarged form. Full or partial monosomics with only one copy of a chromosome or chromosomal segment are usually more severely affected than haploid plants. Trisomic plants with an extra chromosome often show a greater morphological change from the diploid than observed in polyploid plants, such as tetraploids. Secondary trisomics carrying an isochromosome composed of two copies of a chromosome arm show enhanced phenotypic changes for some characteristics typical of the corresponding primary trisomic. When an isochromosome is recovered in an otherwise haploid plant, the imbalance is most severe and results in highly defective plants ( Satina et al., 1937 ). These relationships led to the concept of genetic balance.
As this work continued, trisomics were found for all of the possible 12 chromosomes of Datura , but additional forms continued to arise, each with different characteristics. Some of these new forms accentuated a portion of the phenotypic characteristics of the original set of primary trisomics but were missing other changes from the normal diploid. The basis of these forms was revealed to be an extra chromosome that possessed two identical chromosome arms derived from misdivision of the centromere, manifested by a break in the middle of a centromere with fusion of two sister chromatids, to produce mirror image chromosomes (isochromosomes) generated from a single chromosome arm ( Belling and Blakeslee, 1924 ). In these cases, the extra chromosome, called a secondary trisome, would result in a total of four copies of the respective arm, thus providing an explanation for the accentuated partial phenotype compared with the respective trisomic from which it was derived ( Figure 1 ). These results bolstered the concept that a balance of genes was important for the normal phenotype. These and other aneuploid types were summarized in an article entitled “New Jimson Weeds from Old Chromosomes” ( Blakeslee, 1934 ), which cryptically conveys the message that the dosage of chromosomes is important in this case as opposed to genetic variation. Indeed, the more extreme the altered relative dosage of chromosomes, the greater the phenotypic change. This concept is aptly illustrated by the recovery of selected isochromosomes in otherwise haploid plants that had an extreme imbalance and severe phenotype ( Satina et al., 1937 ) ( Figure 1 ).
At about the same time as the early studies of Datura were conducted by Blakeslee, Richard Goldschmidt was studying the basis of sex determination in the gypsy moth ( Lymantria dispar ) ( Goldschmidt, 1920 ). Crosses between different geographic isolates produced progeny that had differing levels of intersexuality, which is a mixture of male and female morphologies. To explain the various results, Goldschmidt hypothesized different male and female factors of varying strengths whose balance was critical for proper sex determination. Within a particular geographic isolate, the sex determination processes are normal, but crosses between different strains produced the intersexual individuals because the various factors had varied in dosage potential between the diverged populations and were no longer matched. In retrospect, this result foreshadowed the concept of Muller-Dobzhansky coevolving gene complexes that interact within a species but cause sterility or inviability in hybrids.
In further parallel studies, Calvin Bridges identified a triploid female Drosophila and among her progeny found intersexual flies that possessed a mixture of male and female parts ( Bridges, 1925 ). The karyotype of these intersexual flies was composed of two X chromosomes with an otherwise triploid complement of chromosomes. However, flies with only one X chromosome that were otherwise triploid were male-like in phenotype. Of course, among diploid flies, the males have one X chromosome and the females have two, with all other chromosomes being present in duplicate in the two sexes. By comparing the various chromosomal constitutions of males, intersexes, and diploid and triploid females, Bridges concluded that a balance between the X and the autosomes (non-sex chromosomes) was the chromosomal basis of sex determination in Drosophila . This type of mechanism for sex determination has been postulated for many plant species as well, with the most well-documented case being Rumex acetosa ( Ono, 1935 ). When the X-to-A ratio is 1, a female is produced, but when it is 0.5, a male occurs. In polyploid Rumex , X-to-A intermediate ratios produce intersexes and hermaphrodites ( Dellaporta and Calderon-Urrea, 1993 ).
Similar results of chromosome additions to produce trisomics and tetrasomics as well as the reciprocal production of monosomic individuals have recapitulated the balance conclusions in scores if not hundreds of species in both the plant and animal kingdoms (e.g., Lee et al., 1996 ; see discussion in Birchler et al., 2001 ). Although great variation occurs, in general, both monosomics and trisomics cause detrimental effects on the phenotype, with the monosomics usually being more severe. Aneuploid studies in polyploids have further supported the balance idea because there is less effect of extra or missing chromosomes when the remainder of the genome is increased in copy number. The extensive studies of aneuploids in hexaploid bread wheat by Ernie Sears exemplify these conclusions ( Sears, 1953 , 1954 ).
Over the subsequent decades, the interpretation of genetic balance split into an interesting dichotomy of enzymatic/metabolic versus gene regulatory balance that lingers today. On the one hand, the involvement with sex determination mechanisms was viewed as a regulatory balance, while on the other hand, aneuploid syndromes were viewed as perturbations of metabolism resulting from altered relationships of enzymes involved with intermediary metabolism. Indeed, the basis of sex determination in Drosophila involves a dosage relationship between transcription factors on the X chromosome relative to those on the autosomes ( Erickson and Cline, 1993 ). While aneuploid syndromes no doubt have a complicated basis, the emerging evidence suggests that it is likely that most of these balance phenomena reflect gene regulatory mechanisms in some form, although other gene products involved with macromolecular complexes are likely to exhibit a balance as well, whether or not they are directly involved with gene expression (see below).
GENE EXPRESSION IN ANEUPLOIDS
Several decades ago, one of us (J.A.B.) examined the levels of enzyme activities and proteins in aneuploid series of maize. A popular exercise at the time was the localization of the cytological position of genes encoding various enzymes by screening the genome using a set of trisomics or segmental trisomics created by overlapping translocations (e.g., Carlson, 1972 ; O'Brien and Gethman, 1973 ). Work on Datura played an inspiring role in this trend ( Carlson, 1972 ). The principle involved the concept that varying the dosage of a gene would produce a directly proportional amount of gene product ( Grell, 1962 ). Thus, reversing the procedure in a screen of trisomics covering the genome should locate the position of the structural loci for various enzymes because the increased dosage of the encoding gene would produce a greater amount of gene product. This approach was successful in some cases; the unsuccessful attempts remain unknown. Thus, to test ideas about the regulation of the Alcohol dehydrogenase-1 gene in maize ( Schwartz, 1971 ), a dosage series of the chromosome arm on which this gene is located was examined for the levels of ADH activity ( Birchler, 1979 ). Interestingly, the total amount of ADH present in a one to four dosage series was nearly equivalent to the diploid level. In other words, no gene dosage effect for Adh was found in the whole arm dosage series. However, a whole genome ploidy series involving monoploids, diploids, and tetraploids indicated that there was a directly proportional amount of ADH per cell through the series. When other enzymes encoded on other chromosomes were examined in the aneuploid series, their levels were modulated, either up or down, but the most common effect was an inverse correlation between the chromosomal dosage and the amount of activity present ( Birchler, 1979 ). These studies were extended to other proteins and other aneuploid and ploidy series with similar results ( Birchler and Newton, 1981 ). A hypothesis was formulated that the stoichiometry of regulatory genes was influential in modulating the levels of expression of the target genes studied ( Birchler and Newton, 1981 ).
The failure to find a dosage effect for ADH was referred to as dosage compensation. The basis of this response was determined to be the result of a structural gene dosage effect of Adh itself being modulated in an inverse manner by a different part of the same chromosome arm involved in the aneuploid series ( Birchler, 1981 ). In monosomics, the single Adh gene was upregulated approximately twofold. In trisomics, the three copies of Adh were each downregulated by two-thirds. Of course, dosage compensation of the X chromosome in males of Drosophila had been documented since the realization of its existence by Muller (1932) . That is, males with only one X chromosome produce about the same amount of gene product as do the two X chromosomes present in females. However, Devlin et al. (1982) found that compensation would also occur for large trisomics of autosomal chromosome arms consisting of ∼20% of the whole genome, in contrast with small trisomics surrounding a particular structural gene that vary only a very small fraction of the genome, as noted above. Later, they reported the extensive presence of the inverse dosage effect of these large trisomics on genes encoded on other chromosomes ( Devlin et al., 1988 ). The basis of autosomal dosage compensation was also found to involve the combination of a structural gene dosage effect and an inverse effect being produced simultaneously by a large trisomic segment surrounding the Adh gene in flies. In other words, a structural gene dosage effect is cancelled via an inverse effect produced by another part of the varied region ( Birchler et al., 1990 ).
In maize, a survey of aneuploids from many regions of the genome for six genes revealed that the RNA levels of any one gene could be modulated similarly by different dosage series ( Guo and Birchler, 1994 ). In other words, changes in dosage of several different segments of the genome could have the same effect on the monitored gene. The magnitude of these modulations depended on whether the tissue examined was diploid or triploid. Adding or subtracting a chromosome arm to the genotype at different ploidy levels produced an effect whose severity was coincident with the degree of dosage imbalance. Changes in gene expression in a whole genome ploidy series were not as prevalent ( Guo et al., 1996 ). Transcriptome studies of mammalian trisomics or highly aneuploid cancer cells indicate extensive transacting modulations of gene expression (e.g., Phillips et al., 2001 ; FitzPatrick et al., 2002 ; Saran et al., 2003 ; Tsafrir et al., 2006 ). Thus, the types and mode of effects parallel the classical phenotypic studies with regard to balance.
BALANCE AT THE GENE LEVEL
A simple interpretation of the cause of these transacting dosage effects is that they are caused by a gene or genes on the varied chromosome that exhibit a dosage effect themselves and that act in a regulatory fashion to modulate many targets. To screen for single gene mutations that would mimic the dosage effect, mutageneses were conducted in Drosophila to find mutations that could up- or downregulate the expression of the white eye color gene using a leaky phenotypic reporter called white-apricot . The eyes of these flies have a low level of pigment, so modulations of expression could be scored easily. An amazingly large number of mutations were recovered from these screens, totaling 47 at last count ( Birchler et al., 2001 ). The first reported example was Inverse regulator-a ( Rabinow et al., 1991 ). Mutations of this gene as a heterozygote upregulate the white gene twofold, mimicking how a monosomic condition would produce a similar aneuploid inverse effect. Trisomics of the region downregulate white . The presence of the mutation in an otherwise triploid fly upregulates white to ∼150% of the control triploids. Thus, Inr-a serves as an example of a single gene that produces a balance effect, in this case with an inverse dosage response.
The large number of modifiers of a single target gene must be understood in the context that developmental regulators often operate in a hierarchy. That is, one early developmental regulator might affect a downstream regulator and so on. If each regulator is dosage sensitive, the effect could potentially be passed along through the hierarchy. One must also appreciate that any one regulator will affect many targets, so there would be significant overlap of modifiers for different traits.
A summary of the collection of the various modifiers of white revealed that their molecular basis fell into two major classes: members of signal transduction pathways and transcription/chromatin factors ( Birchler et al., 2001 ). Thus, it was established that the dosage effects were in fact the result of regulatory processes. However, the diversity of regulatory molecules involved was mysterious at the time, and to some extent still is, but clues for why this is the case are emerging from studies of haploinsufficiency in yeast and humans (see below).
RELATIONSHIP TO QUANTITATIVE TRAITS?
Returning to the phenotypic effects of trisomics, it is noted that any one characteristic of an organism can be affected by different trisomics. This realization suggests that multiple dosage-sensitive genes might be capable of modulating a particular phenotypic characteristic. We cannot summarize the field of quantitative genetics, but we discuss some parallels as previously noted ( Guo and Birchler, 1994 ; Birchler et al., 2001 , 2005 ). For any one quantitative trait, there can often be multiple loci that affect its expression ( Tanksley, 1993 ). Crosses between varieties that differ for such traits usually exhibit an intermediate (additive) phenotype to some degree ( Tanksley, 1993 ). Thus, the multiple loci act as if they are dosage sensitive. An example involves the quantitative genetic differences between domesticated and wild sunflower ( Burke et al., 2002 ). Many (78) loci were identified that were mostly of small effect and additive in mode of action. It seems reasonable, therefore, that the effects of aneuploids and of some quantitative trait loci (QTL) have a basis in common. QTL would be expected to be a heterogeneous group, but it is reasonable that variation in regulatory genes of sundry types would be expected to be a major contributor.
Within the past half decade, it became obvious that many clinical human conditions result not necessarily from a gain-of-function mutation or a homozygous recessive, but from haploinsufficiency of particular gene products. In other words, null mutations as a heterozygote would condition syndromes in a type of dosage effect. As the molecular basis of these conditions was revealed, they consisted primarily of transcription factors. To explain these results, one of us (R.A.V.) formulated mechanistic models of transcription factor assembly into molecular complexes, noting the importance of stoichiometry of the subunits for the action of the whole ( Veitia, 2002 ). This concept has been extended to genetic and biochemical networks ( Veitia, 2003 , 2004 , 2005 ). The balance between either the subunits of a complex or between proteins with opposing actions, such as transcriptional activators and repressors, needs to be maintained to some extent to avoid negative fitness consequences.
Experimental evidence of this type of relationship has not been systematically sought in plants. However, data have been obtained from the study of haploinsufficiency in yeast ( Papp et al., 2003 ) and humans ( Kondrashov and Koonin, 2004 ). Heterozygous gene knockouts were examined in diploid yeast for growth retardation. Those that produced a haploinsufficiency were overrepresented among classes of genes whose protein products are typically involved with molecular complexes. While these classes of genes extend beyond those involved with regulatory processes to some degree, regulatory gene products typically fall under this umbrella. The human study found that members of signal transduction and transcriptional functions were overrepresented among factors causing haploinsufficiency.
The concept of dosage balance predicts a relationship between the number of interactions (connectivity) of a component and to the possibility of dosage effects when under- or overexpressed. Lemos et al. (2004) have shown that the number of interactions a protein has within a network (i.e., connectivity) constrains genetic variation of gene expression in yeast and fruitfly populations. Specifically, they found a negative correlation between the variation of gene expression and the number of protein–protein interactions. As expected, the extent of variation in expression among genes encoding interactors was smaller than that of random pairs of genes, suggesting the existence of a balance relationship. Finally, the expression levels of interactors correlated positively across strains. High coexpression for proteins in the same complex has been reported independently several times ( Jansen et al., 2002 ; Papp et al., 2003 ). These results suggest the existence of a dosage balance, which could be a force shaping gene expression even at a small evolutionary scale, that is, within populations.
Protein–protein connectivity in yeast complexes (and likely in other organisms) follows approximately a power law distribution ( Hahn et al., 2004 ), which means that most components are poorly connected. Indeed, ∼30% of genes involved in yeast complexes encode separable components (i.e., only one link with the rest of the complex, such as A or C in complex A-B-C). By comparison, only ∼10% of components have 20 or more links (analysis of data from Fraser et al., 2003 ). Mutations in the poorly connected components are more likely to lead to less pleiotropic phenotypes, with higher chances of going undetected. Moreover, these separable components are expected to be less dosage-sensitive (unless they are represented several times per macromolecule as A in A-B-A) ( Veitia, 2002 , 2003 ). This concept might explain why overexpression of subunits of yeast complexes is usually well tolerated.
This is apparent from the study of Sopko et al. (2006) , who analyzed overexpression phenotypes in a vast array of yeast strains, each containing an inducible copy of a different gene. They suggest that overexpression phenotypes in yeast reflect specific regulatory imbalances. Accordingly, they found that overexpression of periodically expressed genes (i.e., during the cell cycle) is more likely to cause cell cycle arrest or abnormal morphology than constitutively expressed cell cycle genes. This is probably so because in many cases these factors participate in network modules involving opposing forces (i.e., a kinase and a phosphatase acting on a common though differentially modified substrate). Dosage effects in these networks are predicted by dosage balance. For instance, using a complex model of the cell cycle, Chen et al. (2004) found that >70% of the parameters can be changed at least 10-fold in either direction (i.e., under- or overexpression) without preventing cycling. However, the rest of the parameters do not exhibit this flexibility, and some are very sensitive to dosage changes. For example, for the synthesis of Cdc14, the boundaries are twofold up, otherwise there will be G1 arrest, and 0.5-fold down (i.e., heterozygous deletion), and the cell faces a telophase arrest. The contrary occurs for the degradation of Cdc14. The need for balance between synthesis (copy number or expression) of Cdc14 and its degradation (i.e., copy number or expression of a protease) must be reached to avoid cell cycle arrest. Note that altering these parameters within the relevant boundaries does not produce cell cycle arrest but does induce either faster or slower cycling (a quantitative character).
BALANCE IN SEX CHROMOSOME DOSAGE COMPENSATION?
As noted above, the same dosage effects on target gene expression that result from genomic imbalance are responsible for dosage compensation of various genes in maize ( Birchler, 1979 ; Birchler and Newton, 1981 ; Guo and Birchler, 1994 ). The magnitude of expression modulation to account for dosage compensation of the various sex chromosome aneuploids in Drosophila is the inverse ratio of the X to autosome imbalance ( Birchler et al., 2006 ). Recent studies of global gene expression between males and females of Drosophila , nematodes, and mammals indicates that in each species the single X chromosome in males is upregulated approximately twofold ( Gupta et al., 2006 ), which would produce a total gene expression equivalent to the two X chromosomes in females (or hermaphrodites in nematodes). In an exhaustive comparison of average gene expression of X chromosomes and autosomes in various mammalian species and tissues ( Nguyen and Disteche, 2006 ), a potential balance relationship was revealed. In female mammals, one of the two X chromosomes is inactivated in any one developmental lineage. Nevertheless, the active X in females is upregulated approximately twofold so that on average the total expression from the single X is more or less equivalent to the average expression of the equivalent length of a pair of autosomes. The single X in males is likewise upregulated. However, in haploid tissues, the average expression of the single X chromosome per unit length is basically the same as a single autosome. Thus, it appears that the X upregulation only occurs when an X-to-autosomal imbalance is present. Sex chromosome dosage compensation has been subjected to natural selection and thus is likely to involve some modifications in mechanism to those cases observed in laboratory-constructed aneuploids ( Birchler et al., 2006 ), but some parallels do seem to exist.
BALANCE IN EVOLUTIONARY PROCESSES?
With the availability of whole-genome sequences, it has become obvious that repeated cycles of polyploidization followed by diploidization have occurred in the lineages leading to the evolutionary crown of eukaryotic organisms present today ( Wolfe, 2001 ; Simillion et al., 2002 ; Bowers et al., 2003 ). Following the production of an allotetraploid from two related species, there is gene loss that leads back to the diploid level. An analysis of the functional classes of the genes retained in duplicate in Arabidopsis and rice indicates an overrepresentation of members of signal transduction components and transcription factors ( Blanc and Wolfe, 2004 ; Maere et al., 2005 ; Chapman et al., 2006 ; Freeling and Thomas, 2006 ; Thomas et al., 2006 ). These classes of genes are similar to those that exhibit transacting dosage effects, as noted above. These findings led to the hypothesis that the duplicates are retained because they are in balance with each other and selection against deletion of one member of a pair would prevent their rapid loss ( Birchler et al., 2005 ; Freeling and Thomas, 2006 ). In other words, deletion of one member of a balanced duplicate would mimic an aneuploid effect, which would diminish reproductive success. Genes not in an interacting balance relationship would be deleted at random over evolutionary time back to the diploid level.
Indeed, gene classes preferentially found in segmental duplications are the complement of those retained from whole-genome duplications ( Davis and Petrov, 2005 ; Maere et al., 2005 ), suggesting that segmental duplications that upset a regulatory balance would be selected against because they also would mimic an aneuploid effect. Of course, genes retained in segmental duplications could be selected to condition greater increments of a particular gene product ( Sharp et al., 2005 ; Redon et al., 2006 ). Not surprisingly, members of large gene families seldom encode components of macromolecular complexes in yeast and humans. Moreover, according to recent studies, duplicability of different genes decreases as the size of the complexes increases ( Papp et al., 2003 ; Yang et al., 2003 ).
The studies on duplicate retention in Arabidopsis and rice by necessity must deal with genomes that are substantially returned to the diploid state. Thus, the possibility exists that directed elimination of members of the singleton classes occurred shortly after polyploidization to produce the observed genomic arrangement. However, a recent analysis of the genome of Paramecium tetraurelia illuminates the processes of whole-genome duplication and subsequent gene loss ( Aury et al., 2006 ). The genomic content of this meager ciliate, with a predicted number of 39,642 genes, dwarfs that of humans. The reason for this number is that the whole genome has endured three duplications. By comparing the recent, intermediate, and old events, insight into the processes of gene loss could be gained. Because many pseudogenes could be recognized in various stages of deterioration, it seems likely that the gene losses are the result of attrition over evolutionary time rather than a concerted elimination event immediately following polyploidization. Moreover, the retained duplicates tend to be classes of genes that are involved in macromolecular complexes. In addition, the retained genes show evidence of purifying selection, suggesting that mutation of one member of a duplicated pair is selected against because it upsets the correct stoichiometric relationships of subunits of such complexes. The authors thus proposed that the stoichiometry of duplicates is important for their retention.
Freeling and Thomas (2006) have suggested that the retention of duplicate regulatory genes following repeated polyploidization events holds such regulators in the evolutionary lineage for sufficient time to allow eventual divergence that fosters increasing complexity over evolutionary time. The polyploidization event proliferates the copy number of all loci. Deletions over time deteriorate the copy number of most target metabolic genes to the diploid level. However, the stoichiometric constraints on regulatory duplicates will preserve them in the lineage. Eventual divergence of function of different members of a duplicate pair of regulators will create developmental and biochemical complexity. While there is evidence that such divergence occurs, concepts of how a new balance is achieved have been little explored. However, given such changes, Freeling and Thomas argue that the repeated cycles of tetraploidization and diploidization are a contributor to driving increasing complexity during the evolution of eukaryotes.
WHY A BALANCE IN REGULATORY PROCESSES?
A regulatory system in which the components are sensitive to stoichiometric relationships provides a means for selection pressures to operate on new mutations in diploids and tetraploids while such mutations are still heterozygous. Also, a system of multiple genes affecting any one characteristic and subject to a dosage interaction will provide a means to modulate the phenotype in subtle ways via mutations in multiple genes. Because the regulatory system is dosage dependent, any new regulatory mutations have the potential to produce subtle effects as a heterozygote. If they are detrimental, they will be selected against rapidly. However, new semidominant mutations would be available in the heterozygous state for rapid adaptive changes as well. The detection of purifying selection on retained duplicates in Paramecium ( Aury et al., 2006 ) suggests that this type of situation is operating. The balance relationship provides for evolutionary changes to be of small magnitude and to result from many possible modifiers of a single phenotype, as noted for the sunflower example above. Thus, a balance relationship of regulatory complexes would optimize purifying and adaptive selection for organisms with diploid and higher levels of ploidy.
FUTURE PROBLEMS TO ADDRESS
We have summarized above the historical and recent evidence suggesting a balance relationship of regulatory genes as a consequence of their membership in macromolecular complexes or networks for which the contributing members produce a stoichiometric effect on the function of the whole. There are several exciting research directions that such a hypothesis might inspire. One possibility is to examine the kinetic and interaction properties of macromolecular complexes in an attempt to understand the basis of how varying the dosage of individual components affects the function of the complex. This area will involve examining the association parameters of multiprotein regulatory complexes, the order in which they associate, and the topological connections between the different subunits. Such studies would test the proposition that highly connected proteins in stable complexes tend to display a higher dosage sensitivity and that this sensitivity can be modulated by the specific parameters of the association process.
Also, one would want eventually to understand the impact of overall gene expression on the phenotype. Such endeavors will no doubt be quite challenging. For example, using gene chip technology, it might be possible to assess correlations between patterns of modulation of thousands of genes and their corresponding phenotypic consequences. The known parallels between multigenic aneuploid effects and quantitative traits are noted above, but the nature of this relationship is quite obscure at present. Exhaustive studies of global patterns of gene expression will need to be conducted, both on aneuploid conditions and on quantitative trait variation, to test such potential connections. Indeed, modulations of morphologies by aneuploid syndromes and QTL might involve changes in cell division patterns as much as overall changes in gene expression, complicating the detection of the responsible genes. The major challenge for molecular quantitative genetics in the future is to tease out the relationship of total genomic expression patterns to phenotypic effects.
Such relationships will be important to gain a better understanding of the role of regulatory balance in evolution. One can imagine how this balance can provide many gradual changes in phenotype in succession, but what is needed for insight into this problem is to determine to what extent multiple regulatory mutations are additive, interactive, or epistatic to each other. The inclusion of regulatory genes among the retained duplicates on the return road from polyploidy to diploidy in diverse taxa has provided an exciting development for the study of evolutionary processes. In mammals, ultraconserved noncoding DNA elements are depleted among segmentally duplicated regions of the genome ( Derti et al., 2006 ). It will be of interest to examine whether they are retained following polyploidization events and whether they are related to the balance characteristics of regulatory mechanisms.
If the stoichiometry of regulatory factors contributes to increasing complexity and speciation, the individual components must be able to escape from a preexisting balance with other factors and establish a new balance. If such divergence is a contributing factor to evolutionary change, then the processes that allow shifts from one balance relationship to another must be explored. Many classical studies of species hybrid incompatibilities have suggested the presence of coevolving gene complexes ( Dobzhansky, 1937 ; Muller, 1942 ; see discussion in Birchler et al., 2005 ). Whether interacting regulatory molecular complexes have a relationship to these mechanisms would be an interesting avenue of exploration.
Acknowledgments
Funding on this topic in the Birchler laboratory is provided by a grant from the National Science Foundation Plant Genome Program (DBI 0501712) and by the National Institutes of Health (R01 GM068042). R.A.V. is funded by the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, and the University of Paris 7.
www.plantcell.org/cgi/doi/10.1105/tpc.106.049338
- Aury, J.-M., et al. (2006). Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia . Nature 444 171–178. [ PubMed ] [ Google Scholar ]
- Belling, J., and Blakeslee, A.F. (1924). The configurations and sizes of the chromosomes in trivalents of 25-chromosome Daturas. Proc. Natl. Acad. Sci. USA 10 116–120. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Birchler, J.A. (1979). A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics 92 1211–1229. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Birchler, J.A. (1981). The genetic basis of dosage compensation of Alcohol dehydrogenase-1 in maize. Genetics 97 625–637. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Birchler, J.A., Bhadra, U., Pal-Bhadra, M., and Auger, D.L. (2001). Dosage dependent gene regulation in higher eukaryotes: Implications for dosage compensation, aneuploid syndromes and quantitative traits. Dev. Biol. 234 275–288. [ PubMed ] [ Google Scholar ]
- Birchler, J.A., Fernandez, H., and Kavi, H.H. (2006). Commonalities in compensation. Bioessays 28 565–568. [ PubMed ] [ Google Scholar ]
- Birchler, J.A., Hiebert, J.C., and Paigen, K. (1990). Analysis of autosomal dosage compensation involving the Alcohol dehydrogenase locus in Drosophila melanogaster . Genetics 124 677–686. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Birchler, J.A., and Newton, K.J. (1981). Modulation of protein levels in chromosomal dosage series of maize: The biochemical basis of aneuploid syndromes. Genetics 99 247–266. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Birchler, J.A., Riddle, N.C., Auger, D.L., and Veitia, R.A. (2005). Dosage balance in gene regulation: Biological implications. Trends Genet. 21 219–226. [ PubMed ] [ Google Scholar ]
- Blakeslee, A.F. (1921). Types of mutations and their possible significance in evolution. Am. Nat. 5 254–267. [ Google Scholar ]
- Blakeslee, A.F. (1934). New Jimson weeds from old chromosomes. J. Hered. 25 80–108. [ Google Scholar ]
- Blakeslee, A.F., and Avery, B.T. (1919). Mutations in the Jimson weed. J. Hered. 10 111–120. [ Google Scholar ]
- Blakeslee, A.F., Belling, J., and Farnham, M.E. (1920). Chromosomal duplication and Mendelian phenomena in Datura mutants. Science 52 388–390. [ PubMed ] [ Google Scholar ]
- Blanc, G., and Wolfe, K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Bowers, J.E., Chapman, B.A., Rong, J., and Paterson, A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422 433–438. [ PubMed ] [ Google Scholar ]
- Bridges, C.B. (1925). Sex in relation to chromosomes and genes. Am. Nat. 59 127–137. [ Google Scholar ]
- Burke, J.M., Tang, S., Knapp, S.J., and Rieseberg, L.H. (2002). Genetic analysis of sunflower domestication. Genetics 161 1257–1267. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Carlson, P.S. (1972). Locating genetic loci with aneuploids. Mol. Gen. Genet. 114 273–280. [ PubMed ] [ Google Scholar ]
- Chapman, B.A., Bowers, J.E., Feltus, F.A., and Paterson, A.H. (2006). Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc. Natl. Acad. Sci. USA 103 2730–2735. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Chen, K.C., Calzone, L., Csikasz-Nagy, A., Cross, F.R., Novak, B., and Tyson, J.J. (2004). Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell 15 3841–3862. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Crow, J.F. (1997). Birth defects, Jimson weeds and bell curves. Genetics 147 1–6. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Dellaporta, S.L., and Calderon-Urrea, A. (1993). Sex determination in flowering plants. Plant Cell 5 1241–1251. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Davis, J.C., and Petrov, D.A. (2005). Do disparate mechanisms of duplication add similar genes to the genome? Trends Genet. 21 548–551. [ PubMed ] [ Google Scholar ]
- Derti, A., Roth, F.P., Church, G.M., and Wu, C.T. (2006). Mammalian ultraconserved elements are strongly depleted among segmental duplications and copy number variants. Nat. Genet. 38 1216–1220. [ PubMed ] [ Google Scholar ]
- Devlin, R.H., Holm, D.G., and Grigliatti, T.A. (1982). Autosomal dosage compensation in Drosophila melanogaster strains trisomic for the left arm of chromosome 2. Proc. Natl. Acad. Sci. USA 79 1200–1204. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Devlin, R.H., Holm, D.G., and Grigliatti, T.A. (1988). The influence of whole-arm trisomy on gene expression in Drosophila. Genetics 118 87–101. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- deVries, H. (1901). Die Mutationstheorie. Bd. 1. Die Entstehung der Arten durch Mutation, Vol. 1. (Leipzig, Germany: Veit & Co.).
- Dobzhansky, T. (1937). Genetics and the Origin of Species. (New York: Columbia University Press).
- Emerson, S.H., and Sturtevant, A.H. (1931). Genetic and cytological studies of Oenothera. III. The translocation interpretation. Z. Indukt. Abstammungs-Vererbungl. 59 395–419. [ Google Scholar ]
- Erickson, J.W., and Cline, T.W. (1993). A bZIP protein, sisterless-a , collaborates with bHLH transcription factors in Drosophila development to determine sex. Genes Dev. 7 1688–1702. [ PubMed ] [ Google Scholar ]
- FitzPatrick, D.R., Ramsay, J., McGill, N.I., Shade, M., Carothers, A.D., and Hastie, N.D. (2002). Transcriptome analysis of human autosomal trisomy. Hum. Mol. Genet. 11 3249–3256. [ PubMed ] [ Google Scholar ]
- Fraser, H.B., Wall, D.P., and Hirsh, A.E. (2003). A simple dependence between protein evolution rate and the number of protein-protein interactions. BMC Evol. Biol. 3 11. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Freeling, M., and Thomas, B.C. (2006). Gene-balanced duplications, like tetraploid, provide predictable drive to increase morphological complexity. Genome Res. 16 805–814. [ PubMed ] [ Google Scholar ]
- Goldschmidt, R.B. (1920). Untersuchungen uber inter-sexualitat. Zeits. i. Abst. u. Vererb. 23 1–199. [ Google Scholar ]
- Grell, E.H. (1962). The dose effect of ma-l+ and ry+ on xanthing dehydrogenease activity in Drosophila melanogaster . Z. Vererbungsl. 93 371–377. [ Google Scholar ]
- Guo, M., and Birchler, J.A. (1994). Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 266 1999–2002. [ PubMed ] [ Google Scholar ]
- Guo, M., Davis, D., and Birchler, J.A. (1996). Dosage effects on gene expression in a maize ploidy series. Genetics 142 1349–1355. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Gupta, V., Parisi, M., Sturgill, D., Nutfall, R., Doctolero, M., Dudko, O.K., Malley, J.D., Eastman, P.S., and Oliver, B. (2006). Global analysis of X-chromosome dosage compensation. J. Biol. 5 3. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hahn, M.W., Conant, G.C., and Wagner, A. (2004). Molecular evolution in large genetic networks: Does connectivity equal constraint? J. Mol. Evol. 58 203–211. [ PubMed ] [ Google Scholar ]
- Jansen, R., Greenbaum, D., and Gerstein, M. (2002). Relating whole-genome expression data with protein-protein interactions. Genome Res. 12 37–46. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Kondrashov, F.A., and Koonin, E.V. (2004). A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 20 287–290. [ PubMed ] [ Google Scholar ]
- Lemos, B., Meiklejohn, C.D., and Hartl, D.L. (2004). Regulatory evolution across the protein interaction network. Nat. Genet. 36 1059–1060. [ PubMed ] [ Google Scholar ]
- Lee, E.A., Darrah, L.L., and Coe, E.H. (1996). Dosage effects on morphological and quantitative traits in maize aneuploids. Genome 39 898–908. [ PubMed ] [ Google Scholar ]
- Maere, S., DeBodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., and Van de Peer, Y. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102 5454–5459. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Muller, H.J. (1932). Further studies on the nature and causes of gene mutations. Proc. 6 th Int. Congr. Genetics 1 213–255. [ Google Scholar ]
- Muller, H.J. (1942). Isolating mechanisms, evolution and temperature. Biol Symp 6 71–125. [ Google Scholar ]
- Nguyen, D.K., and Disteche, C.M. (2006). Dosage compensation of the active X chromosome in mammals. Nat. Genet. 38 47–53. [ PubMed ] [ Google Scholar ]
- O'Brien, S.J., and Gethman, R.C. (1973). Segmental aneuploidy as a probe for structural genes in Drosophila: Mitochondrial membrane enzymes. Genetics 75 155–167. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ono, T. (1935). Chromosomen und sexualitat von Rumex acetosa . Sci. Rep. Tohoku Univ. (Sendai, Japan). Biology 10 41–210. [ Google Scholar ]
- Papp, B., Pal, C., and Hurst, L.D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424 194–197. [ PubMed ] [ Google Scholar ]
- Phillips, J.L., et al. (2001). The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis. Cancer Res. 61 8143–8149. [ PubMed ] [ Google Scholar ]
- Rabinow, L., Nguyen-Huynh, A.T., and Birchler, J.A. (1991). A trans-acting regulatory gene that inversely affects the expression of the white , brown and scarlet loci in Drosophila melanogaster . Genetics 129 463–480. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Redon, R., et al. (2006). Global variation in copy nuber in the human genome. Nature 444 444–454. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Saran, N.G., Pletcher, M.T., Natale, J.E., Cheng, Y., and Reeves, R.H. (2003). Global disruption of the cerebellar transcriptome in a Down syndrome mouse model. Hum. Mol. Genet. 12 2013–2019. [ PubMed ] [ Google Scholar ]
- Satina, S., Blakeslee, A.F., and Avery, A.G. (1937). Balanced and unbalanced haploids in Datura. J. Hered. 28 192–202. [ Google Scholar ]
- Schwartz, D. (1971). Genetic control of alcohol dehydrogenase–A competition model for regulation of gene action. Genetics 67 411–425. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Sears, E.R. (1953). Nullisomic analysis in wheat. Am. Nat. 87 245–252. [ Google Scholar ]
- Sears, E.R. (1954). The aneuploids of common wheat. Missouri Agric. Exp. Sta. Res. Bull. 572.
- Sharp, A.J., et al. (2005). Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77 78–88. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Simillion, C., Vandepoele, K., Montagu, M.C., Zabeau, M., and Van de Peer, Y. (2002). The hidden duplication past of Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA 99 13627–13632. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Sopko, R., Huang, D., Preston, N., Chua, G., Papp, B., Kafadar, K., Snyder, M., Oliver, S.G., Cyert, M., Hughes, T.R., Boone, C., and Andrews, B. (2006). Mapping pathways and phenotypes by systemic gene over-expression. Mol. Cell 21 319–330. [ PubMed ] [ Google Scholar ]
- Tanksley, S.D. (1993). Mapping polygenes. Annu. Rev. Genet. 27 205–233. [ PubMed ] [ Google Scholar ]
- Thomas, B.C., Pedersen, B., and Freeling, M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16 934–946. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Tsafrir, D., et al. (2006). Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 66 2129–2137. [ PubMed ] [ Google Scholar ]
- Veitia, R.A. (2002). Exploring the etiology of haploinsufficiency. Bioessays 24 175–184. [ PubMed ] [ Google Scholar ]
- Veitia, R.A. (2003). Nonlinear effects in macromolecular assembly and dosage sensitivity. J. Theor. Biol. 220 19–25. [ PubMed ] [ Google Scholar ]
- Veitia, R.A. (2004). Gene dosage balance in cellular pathways: Implications for dominance and gene duplicability. Genetics 168 569–574. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Veitia, R.A. (2005). Gene dosage balance: Deletions, duplications and dominance. Trends Genet. 21 33–35. [ PubMed ] [ Google Scholar ]
- Yang, J., Lusk, R., and Li, W.H. (2003). Organismal complexity, protein complexity, and gene duplicability. Proc. Natl. Acad. Sci. USA 100 15661–15665. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Wolfe, K.H. (2001). Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2 333–341. [ PubMed ] [ Google Scholar ]
The Good Genes Hypothesis
- Living reference work entry
- First Online: 13 January 2023
- Cite this living reference work entry

- Urszula M. Marcinkowska 2
351 Accesses
22 Altmetric
Honest signalling
The good genes hypothesis proposes that the characteristics preferred by females are a signal of males’ ability to pass on genes which will increase the survival and reproductive success of the offspring sired with a male possessing them.
Introduction
The good genes hypothesis (GGH) was formulated by evolutionary biologist W.D. Hamilton and behavioral ecologist M. Zuk ( 1982 ). It proposes that the characteristics preferred by females are a signal of males’ ability to pass on genes (coding that certain characteristic) which will increase the survival and reproductive success of the offspring sired with a male possessing them. In such case, sexual selection would lead to the increase of the proportion of genes (coding the attractive trait) that are beneficial under prevailing conditions. In other words, good genes increase fitness of the offspring. In an evolutionary and ecological understanding individual’s fitness depends on how many copies of own...
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
Institutional subscriptions
Achorn, A. M., & Rosenthal, G. G. (2019). It’s not about him: Mismeasuring ‘good genes’ in sexual selection. Trends in Ecology & Evolution, 35 (3), 206–219. https://doi.org/10.1016/j.tree.2019.11.007
Boothroyd, L. G., Scott, I., Gray, A. W., Coombes, C. I., & Pound, N. (2013). Male facial masculinity as a Cue to health outcomes. Evolutionary Psychology, 11 (5), 1044–1058. https://doi.org/10.1177/147470491301100508
Article Google Scholar
Foo, Y. Z., Simmons, L. W., & Rhodes, G. (2017). Predictors of facial attractiveness and health in humans. Scientific Reports, 7 . https://doi.org/10.1038/srep39731
Hakkarainen, T. J., Krams, I., Coetzee, V., Skrinda, I., Kecko, S., Krama, T., … & Rantala, M. J. (2021). MHC class II heterozygosity associated with attractiveness of men and women. Evolutionary Psychology , 19 (1). https://doi.org/10.1177/1474704921991994 .
Hamilton, W. D., & Zuk, M. (1982). Heritable true fitness and bright birds: A role for parasites? Science, 218 (4570), 384–387. https://doi.org/10.1126/science.7123238
Marcinkowska, U. M., Ziomkiewicz, A., Kleisner, K., Galbarczyk, A., Klimek, M., Sancilio, A., … & Bribiescas, R. G. (2020). Oxidative stress as a hidden cost of attractiveness in postmenopausal women. Scientific Reports, 10 (1). https://doi.org/10.1038/s41598-020-76627-9 .
Prum, R. O. (2010). The Lande-Kirkpatrick mechanism is the null model of evolution by intersexual selection: Implications for meaning, honesty, and design in intersexual signals. Evolution, 64 (11), 3085–3100.
Rantala, M. J., Coetzee, V., Moore, F. R., Skrinda, I., Kecko, S., Krama, T., … & Krams, I. (2013). Adiposity, compared with masculinity, serves as a more valid Cue to immunocompetence in human mate choice. Proceedings of the Royal Society B-Biological Sciences, 280 (1751). https://doi.org/10.1098/rspb.2012.2495 .
Scott, I. M. L., Clark, A. P., Boothroyd, L. G., & Penton-Voak, I. S. (2013). Do Men’s faces really signal heritable immunocompetence? Behavioral Ecology, 24 (3), 579–589. https://doi.org/10.1093/beheco/ars092
Stephen, I. D., Hiew, V., Coetzee, V., Tiddeman, B. P., & Perrett, D. I. (2017). Facial shape analysis identifies valid Cues to aspects of physiological health in Caucasian, Asian, and African populations. Frontiers in Psychology, 8 . https://doi.org/10.3389/fpsyg.2017.01883
Weatherhead, P., & Robertson, R. (1979). Offspring quality and the polygyny threshold: “The sexy son hypothesis”. The American Naturalist, 113 (2), 201–208. https://doi.org/10.1086/283379
Zahavi, A. (1975). Mate selection—A selection for a handicap. Journal of Theoretical Biology, 53 (1), 205–214. https://doi.org/10.1016/0022-5193(75)90111-3
Zaidi, A. A., White, J. D., Mattern, B. C., Liebowitz, C. R., Puts, D. A., Claes, P., & Shriver, M. D. (2019). Facial masculinity does not appear to be a condition-dependent male ornament and does not reflect MHC heterozygosity in humans. Proceedings of the National Academy of Sciences of the United States of America, 116 (5), 1633–1638. https://doi.org/10.1073/pnas.1808659116
Download references
Author information
Authors and affiliations.
Institute of Public Health, Jagiellonian University Medical College, Cracow, Poland
Urszula M. Marcinkowska
You can also search for this author in PubMed Google Scholar

Editor information
Editors and affiliations.
Department of Psychology, Oakland University, ROCHESTER, MN, USA
Todd K. Shackelford
Section Editor information
Department of Psychology, Oakland University, Rochester, MI, USA
Oakland University, Rochester, MI, USA
Gavin Vance
Department of Psychology, Oakland University, Rochester Hills, MI, USA
Madeleine K. Meehan
Rights and permissions
Reprints and permissions
Copyright information
© 2023 Springer Nature Switzerland AG
About this entry
Cite this entry.
Marcinkowska, U.M. (2023). The Good Genes Hypothesis. In: Shackelford, T.K. (eds) Encyclopedia of Sexual Psychology and Behavior. Springer, Cham. https://doi.org/10.1007/978-3-031-08956-5_1081-1
Download citation
DOI : https://doi.org/10.1007/978-3-031-08956-5_1081-1
Received : 29 August 2022
Accepted : 29 August 2022
Published : 13 January 2023
Publisher Name : Springer, Cham
Print ISBN : 978-3-031-08956-5
Online ISBN : 978-3-031-08956-5
eBook Packages : Springer Reference Behavioral Science and Psychology Reference Module Humanities and Social Sciences Reference Module Business, Economics and Social Sciences
- Publish with us
Policies and ethics
- Find a journal
- Track your research
Party animal or wallflower? UC San Diego lab thinks autism-gene drug could help some thrive
Scientist says genes they're working with could also explain why wolves evolved into dogs, and why there are no chimp cities., by eric s. page • published 16 mins ago • updated 11 mins ago.
Genetic researchers studying variations in personalities may have answered an age-old question: Why are some people able to successfully navigate a cocktail party while others struggle to make the smallest of small talk?
While not able to apply their research to all party animals, the theory that researchers at UC San Diego's Sanford Stem Cell Institute are working on begins with a rare genetic condition called Williams syndrome, which is caused by the "deletion of about 25 genes in the 7q11.23 chromosomal region,' " according to a news release the university sent out, "and which is sometimes called 'the opposite of autism.' "
People with Williams are extremely social, chatty to a fault and sometimes have an oversize vocabulary that can hide what is normally perceived to be a below-average intelligence.
April is Autism Awareness Month

Missing 13-year-old boy found safe in New Mexico Walmart after 200-mile journey from Arizona

Autism Awareness Month: San Diego mother shares keys to her success raising child with autism

3 helpful resources for Autism Awareness Month and Autism Acceptance Month
Get San Diego local news, weather forecasts, sports and lifestyle stories to your inbox. Sign up for NBC San Diego newsletters.
"They have, like, a low IQ," Alysson Muotri told NBC 7. "They have developmental delays, but the vocabulary is quite enriched and above the average for people of their age. And this is independent of cultures. People in Japan with Williams syndrome behave in the same way. So the genetic here is really strong."
Another major downside: Those who are missing the 25 genes, and, specifically, GTF2I, "know no strangers, making them particularly vulnerable to abuse and bullying," as the science folks put it in their release.
On the flip side of Williams, people who have a genetic duplication in the 7q11.23 chromosomal region — known as, not surprisingly, 7q11.23 duplication syndrome — may experience autism, social phobia or, in some cases, "selective mutism."
While Williams has been studied earlier and elsewhere, researchers at UC San Diego focused on that one gene in particular — GTF2I — which they theorized was controlling the variations in personality.
Muotri is the director of the UC San Diego's Sanford Stem Cell Education and Integrated Space Stem Cell Orbital Research Center and lead author of the paper. The gregarious Brazilian ex-pat professor, who has been in the San Diego area off and on since 2002, lives in PB and loves to surf and hike when he's not also acting as the school's director of the Archealization Center and its Gene Therapy Initiative, as well as the associate director for the Center for Academic Research & Training in Anthropogeny. The guy has a resume, for sure, and he's had an effect at UC San Diego. He said about 80% of the 35 or so people working in his lab are from Brazil.
“I like to describe this gene as the gene of prejudice,” Muotri said in the news release. “Without it, everyone in the world is your friend.”
Although researchers in the past have connected many genes to autism, GTF2I “is the only gene we are aware of that regulates socialization more directly,” according to Muotri.
After the hypothesis evolved to the point it was ready to be tested, the UC San Diego lab grew "mini brains," as Muotri called them, from stem cells that would then imitate the development of a fetal brain, but one without the GTF2I gene. What researchers found was that in the absence of the gene, more cells died, there were defects in synapses and less electrical activity. All as bad as it sounds. On the other hand, genetic experiments with fruit flies yielded a surprising result.
"Fruit flies with the same alteration, genetic alteration, also become more social," Muotri told NBC 7 in a recent interview. "Usually, fruit flies prefer to eat very independently from each other, but fruit flies that [were] genetically engineered to carry this genetic alteration, they start having meals together."
Scientists are fond of fruit flies' relatively brief lifespans and simple physiological systems. With their genomes edited to include GTF2I, they eat "without the usually obligatory 'social bubble,' " and mice with the gene, are, well, friendlier, according to the UC San Diego researchers.
And then there's this bomb from the news release: "alterations to a gene that controls the function of GTF2I — potentially turning it off — may be at least partially responsible for the loving, friendly disposition of domesticated dogs compared to wild wolves." In other words, there might be no dogs if all canines had GTF2I.
"So this region in the chromosome is so important for sociability and very conserved in different species — we actually see that that region was affected during the domestication of dogs, from wolves to dogs," Muotri told NBC 7. "I mean, we see that they become more docile and they become more social. And the dogs, they even make very good eye contact compared to wolves. So this is definitely something that you see in other species."
One real-world application the research may be used for is treating people with autism.
"By studying this GTF2I gene, it allows us to dissect the molecular pathway that is inside the cells to create or to make your brain cells become highly social," Muotri told NBC 7. "By designing drugs that can modulate that pathway, we can use and apply to autism, inducing your brain to become more social."
Muotri said that design is being worked on now, in fact.
"Sometimes we team up with the pharmaceutical industry if there is a commercial interest," Muotri told NBC 7. "In the case of Williams syndrome, it's less obvious because this is a rare syndrome, but in this case, we are applying what we have learned from Williams syndrome to a more common condition: That's autism."
Muotri thinks that the GTF2I gene, or the lack thereof, could be a contributor to what distinguishes humans from their closest genetic cousin, chimpanzees, who are social animals, of course, but to much less of a degree.
"The chimpanzees, they have a social brain that is able to deal with about 15 individuals per colony, so when the colony that they live [in starts] to grow bigger, there is usually a crisis, or they start fighting, and some [of the apes] have to leave the colony," Muotri said. "We humans, we triple that social capability. So we can deal with 150 people at the same time, and we can even use hierarchical social interactions, meaning that I talk to you in a different way than I talk to the father of my wife, because I know the different hierarchy that goes to that. So the chimpanzees, they also have that, but in a less sophisticated way."
Muotri stressed that GTF2I is only one element of the equation.
"Remember, GTF2I is just one of the genes that are responsible for your social behavior," Muotri said. "There are other genes that act together with GT2I. This one is just interesting because it affects so directly how social you are that there is even what we call a polymorphism among humans, meaning that the difference in the levels of GTF2I in my brain and in your brain might be different. And depending on how much you have or I have, I'd be more or less social because of that."
This lack of GTF2I in human's brains may be profound, the professor theorized.
“It’s when we cooperate that we can put a man on the moon," Muotri said in the news release. "It’s when we cooperate that we can decode the human genome. Because we work together.”
While it seems like Planet of the Apes could be on the menu in the hands of a mad scientist, it's worth remembering that there are no dog cities. Planet of the Dogs?
Still, it's easy to follow this thread: If introducing the GTF2I gene into somebody with autism could ratchet up their level of sociability, could it have the same effect on a so-called "normal" person? Would it turn them into a social butterfly?
"You could, yeah, eventually you could," Muotri posited. "And by the way, there are drugs out there that people take to enhance their sociability. And I would say that one of the most used drugs in humanity is for that. And it's either alcohol or coffee."
But would this third option alter an average person so that they became more confident, more outgoing, have a better vocabulary?
"Well, it is a possibility that this might happen with normal people," Muotri told NBC 7. "Of course, it's not the intention of the research, but as a side-effect, it might well happen. This is what we are seeing now with some of the research in psychedelics. There are some of these psychedelic drugs that increase your serotonin level, and this is one of the neurotransmitters that makes you more social."
Roughly 1% of the population has some form of autism, according to Muotri, so a drug could have a huge pool of potential patients. It's not difficult to imagine a much larger market for people who might want to seem smarter and more confident in social settings. Still, it's not something Muotri and his lab is pursuing.
"I have not proposed that [to the pharmaceutical companies]," Muotri said. "The first thing that we need to know is if the drug has side-effects, because nobody wants to increase your sociability if we start, let's say, vomiting, right away."
Want even more science? Click here to read the full study.
This article tagged under:
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Research Briefing
- Published: 24 April 2024
Three patterns link brain organization to genes in health and disease
Nature Neuroscience ( 2024 ) Cite this article
1 Altmetric
Metrics details
- Development of the nervous system
- Genetics of the nervous system
Gene expression in the human cortex is shown to exhibit a generalizable three-component architecture that reflects neuronal, metabolic, and immune programmes of healthy brain development. The three components have distinct associations with autism spectrum disorder and schizophrenia, revealing connections between previously unrelated results from studies of case–control neuroimaging, differential gene expression, and genetic risk.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
195,33 € per year
only 16,28 € per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
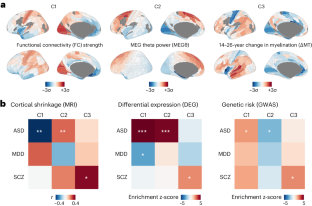
Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489 , 391–399 (2012). The original paper presenting the AHBA, in which principal components of cortical gene expression were suggested to reflect brain organization.
Article CAS PubMed PubMed Central Google Scholar
Burt, J. B. et al. Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography. Nat. Neurosci. 21 , 1251–1259 (2018). This paper characterizes the first component of cortical gene expression, C1, as reflecting a neuronal hierarchy defined by tract-tracing and indexed by structural neuroimaging.
Sydnor, V. J. et al. Neurodevelopment of the association cortices: patterns, mechanisms, and implications for psychopathology. Neuron 109 , 2820–2846 (2021). This review proposes that neurodevelopment involves a ‘sensorimotor–association axis’ defined by ten brain maps, of which one is the cortical gene expression component C1.
Merikangas, A. K. et al. What genes are differentially expressed in individuals with schizophrenia? A systematic review. Mol. Psychiatry 27 , 1373–1383 (2022). This review demonstrates the lack of consistency in genes linked to schizophrenia across differential expression studies, which are also inconsistent with GWAS.
Article PubMed PubMed Central Google Scholar
Johnson, M. B. & Hyman, S. E. A critical perspective on the synaptic pruning hypothesis of schizophrenia pathogenesis. Biol. Psychiatry 92 , 440–442 (2022). This commentary calls for an understanding of synaptic pruning in schizophrenia compared with healthy adolescent neurodevelopment.
Article PubMed Google Scholar
Download references
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is a summary of: Dear, R. et al. Cortical gene expression architecture links healthy neurodevelopment to the imaging, transcriptomics and genetics of autism and schizophrenia. Nat. Neurosci . https://doi.org/10.1038/s41593-024-01624-4 (2024).
Rights and permissions
Reprints and permissions
About this article
Cite this article.
Three patterns link brain organization to genes in health and disease. Nat Neurosci (2024). https://doi.org/10.1038/s41593-024-01625-3
Download citation
Published : 24 April 2024
DOI : https://doi.org/10.1038/s41593-024-01625-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
VIDEO
COMMENTS
The 1960s definition of the gene is the one most geneticists employ today, but it is clearly out-of-date for deoxyribonucleic acid (DNA)-based organisms. ... Earlier work in the 1940s had established the connection between genes and proteins, in the "one gene-one enzyme" hypothesis of Beadle and Tatum ...
enzyme. gene. one gene-one enzyme hypothesis, idea advanced in the early 1940s that each gene controls the synthesis or activity of a single enzyme. The concept, which united the fields of genetics and biochemistry, was proposed by American geneticist George Wells Beadle and American biochemist Edward L. Tatum, who conducted their studies in ...
A unit of heredity that is passed from parent to offspring. Allele. One of different forms of a gene. Genotype. The genetic makeup of an organism (ex: TT) Phenotype. The physical characteristics of an organism (ex: tall) Dominant allele. Allele that is phenotypically expressed over another allele.
Thrifty gene hypothesis. The thrifty gene hypothesis, or Gianfranco's hypothesis[citation needed] is an attempt by geneticist James V. Neel to explain why certain populations and subpopulations in the modern day are prone to diabetes mellitus type 2. He proposed the hypothesis in 1962 to resolve a fundamental problem: diabetes is clearly a very ...
gene, unit of hereditary information that occupies a fixed position (locus) on a chromosome. Genes achieve their effects by directing the synthesis of proteins. In eukaryotes (such as animals, plants, and fungi ), genes are contained within the cell nucleus. The mitochondria (in animals) and the chloroplasts (in plants) also contain small ...
The one gene, one enzyme hypothesis is the idea that each gene encodes a single enzyme. Today, we know that this idea is generally (but not exactly) correct. Sir Archibald Garrod, a British medical doctor, was the first to suggest that genes were connected to enzymes. Beadle and Tatum confirmed Garrod's hypothesis using genetic and biochemical ...
A transcription unit is a linear sequence of DNA that extends from a transcription start site to a transcription stop site (Figure 4). Figure 4. The promoter, a DNA sequence that lies upstream of ...
7.3 The "One Gene: One Enzyme" Hypothesis. Beadle and Tatum's experiments are important not only for their conceptual advances in understanding genes, but also because they demonstrate the utility of screening for genetic mutants to investigate a biological process — this is called genetic analysis.. Beadle and Tatum's results were useful to investigate biological processes, specifically ...
In the thrifty-gene hypothesis, genetic variants that lead to the accumulation of adipose stores for preservation of nutrition until times when it would be required are under positive selection, and thus becoming more common in the population. Thus, in current times of caloric excess, previously beneficial alleles have become deleterious.
The concept of genetic balance traces back to the early days of genetics. Additions or subtractions of single chromosomes to the karyotype (aneuploidy) produced greater impacts on the phenotype than whole-genome changes (ploidy). Studies on changes in gene expression in aneuploid and ploidy series revealed a parallel relationship leading to the ...
The DNA's base pairs encode genes, which provide functions. A human DNA can have up to 500 million base pairs with thousands of genes. In biology, the word gene ( Greek: γένος, génos; [1] generation, [2] or birth, [1] or gender) has two meanings. The Mendelian gene is a basic unit of heredity.
Biology definition: A hypothesis is a supposition or tentative explanation for (a group of) phenomena, (a set of) facts, or a scientific inquiry that may be tested, verified or answered by further investigation or methodological experiment.It is like a scientific guess.It's an idea or prediction that scientists make before they do experiments. They use it to guess what might happen and then ...
Large-scale -omics studies are often described as being 'hypothesis-free'. To take one example from genomics: advances in genome-editing techniques mean that it is now possible to generate 'loss-of-function' mutants in the laboratory. Such mutations are inactivating in the sense that they lead to the loss in the function of a gene ...
"Loss of heterozygosity" is a phrase often used to describe the process that leads to the inactivation of the second copy of a tumor suppressor gene.During this process, a heterozygous cell ...
The anticodon joining hypothesis on the origin of gene-I was proposed based on the AntiC-SL tRNA hypothesis on the origin of tRNA, ... Similarly, it does not always mean that EntNew genes were created as deduced from the pan-GC-NSF(a) hypothesis, based on that the creation process could not be reasonably explained by other ideas. ...
Abstract. The epigenetic phenomenon of genomic imprinting has motivated the development of numerous theories for its evolutionary origins and genomic distribution. In this review, we examine the ...
good genes hypothesis, in biology, an explanation which suggests that the traits females choose when selecting a mate are honest indicators of the male's ability to pass on genes that will increase the survival or reproductive success of her offspring. Although no completely unambiguous examples are known, evidence supporting the good genes hypothesis is accumulating, primarily through the ...
The good genes hypothesis (GGH) was formulated by evolutionary biologist W.D. Hamilton and behavioral ecologist M. Zuk ().It proposes that the characteristics preferred by females are a signal of males' ability to pass on genes (coding that certain characteristic) which will increase the survival and reproductive success of the offspring sired with a male possessing them.
Ronald Fisher. Particulate inheritance is a pattern of inheritance discovered by Mendelian genetics theorists, such as William Bateson, Ronald Fisher or Gregor Mendel himself, showing that phenotypic traits can be passed from generation to generation through "discrete particles" known as genes, which can keep their ability to be expressed while ...
The swamping hypothesis rests on two key ideas. The first is that gene flow is asymmetric from the center of the range toward the edge, due to decreasing population sizes predicted by the abundant center hypothesis (Figure 1) [9, 12, 13].Although the abundant center hypothesis has been questioned as a general pattern [14, 15], there could still be substantial gene flow from the center of a ...
Gene-for-gene relationships have been proposed by some authors for virus-host interactions (e.g. Fraser, 1987a). A well-studied example of the gene-for-gene hypothesis applied to a plant virus and its host is the resistance of tomato to ToMV (Table 10.3). There are three resistance genes: Tm-1, Tm-2 and Tm-2 2. The virus has evolved variants ...
Two independent cases in the literature of missense mutations in SOX2 (G130A and A191T) mean the example in Fig. 6f is already a known gene for the phenotype in the OMIM 28 database. The phenotype ...
Genomic imprinting is an epigenetic phenomenon that causes genes to be expressed or not, depending on whether they are inherited from the female or male parent. Genes can also be partially imprinted. Partial imprinting occurs when alleles from both parents are differently expressed rather than complete expression and complete suppression of one parent's allele. Forms of genomic imprinting have ...
Muotri thinks that the GTF2I gene, or the lack thereof, could be a contributor to what distinguishes humans from their closest genetic cousin, chimpanzees, who are social animals, of course, but ...
Gene expression in the human cortex is shown to exhibit a generalizable three-component architecture that reflects neuronal, metabolic, and immune programmes of healthy brain development. The ...