

Research Methods
- Getting Started
- What is Research Design?
- Research Approach
- Research Methodology
- Data Collection
- Data Analysis & Interpretation
- Population & Sampling
- Theories, Theoretical Perspective & Theoretical Framework
- Useful Resources
Further Resources
Data Analysis & Interpretation
- Quantitative Data
Qualitative Data
- Mixed Methods
You will need to tidy, analyse and interpret the data you collected to give meaning to it, and to answer your research question. Your choice of methodology points the way to the most suitable method of analysing your data.

If the data is numeric you can use a software package such as SPSS, Excel Spreadsheet or “R” to do statistical analysis. You can identify things like mean, median and average or identify a causal or correlational relationship between variables.
The University of Connecticut has useful information on statistical analysis.
If your research set out to test a hypothesis your research will either support or refute it, and you will need to explain why this is the case. You should also highlight and discuss any issues or actions that may have impacted on your results, either positively or negatively. To fully contribute to the body of knowledge in your area be sure to discuss and interpret your results within the context of your research and the existing literature on the topic.
Data analysis for a qualitative study can be complex because of the variety of types of data that can be collected. Qualitative researchers aren’t attempting to measure observable characteristics, they are often attempting to capture an individual’s interpretation of a phenomena or situation in a particular context or setting. This data could be captured in text from an interview or focus group, a movie, images, or documents. Analysis of this type of data is usually done by analysing each artefact according to a predefined and outlined criteria for analysis and then by using a coding system. The code can be developed by the researcher before analysis or the researcher may develop a code from the research data. This can be done by hand or by using thematic analysis software such as NVivo.
Interpretation of qualitative data can be presented as a narrative. The themes identified from the research can be organised and integrated with themes in the existing literature to give further weight and meaning to the research. The interpretation should also state if the aims and objectives of the research were met. Any shortcomings with research or areas for further research should also be discussed (Creswell,2009)*.
For further information on analysing and presenting qualitative date, read this article in Nature .
Mixed Methods Data
Data analysis for mixed methods involves aspects of both quantitative and qualitative methods. However, the sequencing of data collection and analysis is important in terms of the mixed method approach that you are taking. For example, you could be using a convergent, sequential or transformative model which directly impacts how you use different data to inform, support or direct the course of your study.
The intention in using mixed methods is to produce a synthesis of both quantitative and qualitative information to give a detailed picture of a phenomena in a particular context or setting. To fully understand how best to produce this synthesis it might be worth looking at why researchers choose this method. Bergin**(2018) states that researchers choose mixed methods because it allows them to triangulate, illuminate or discover a more diverse set of findings. Therefore, when it comes to interpretation you will need to return to the purpose of your research and discuss and interpret your data in that context. As with quantitative and qualitative methods, interpretation of data should be discussed within the context of the existing literature.
Bergin’s book is available in the Library to borrow. Bolton LTT collection 519.5 BER
Creswell’s book is available in the Library to borrow. Bolton LTT collection 300.72 CRE
For more information on data analysis look at Sage Research Methods database on the library website.
*Creswell, John W.(2009) Research design: qualitative, and mixed methods approaches. Sage, Los Angeles, pp 183
**Bergin, T (2018), Data analysis: quantitative, qualitative and mixed methods. Sage, Los Angeles, pp182
- << Previous: Data Collection
- Next: Population & Sampling >>
- Last Updated: Sep 7, 2023 3:09 PM
- URL: https://tudublin.libguides.com/research_methods
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
Methodology
Research Methods | Definitions, Types, Examples
Research methods are specific procedures for collecting and analyzing data. Developing your research methods is an integral part of your research design . When planning your methods, there are two key decisions you will make.
First, decide how you will collect data . Your methods depend on what type of data you need to answer your research question :
- Qualitative vs. quantitative : Will your data take the form of words or numbers?
- Primary vs. secondary : Will you collect original data yourself, or will you use data that has already been collected by someone else?
- Descriptive vs. experimental : Will you take measurements of something as it is, or will you perform an experiment?
Second, decide how you will analyze the data .
- For quantitative data, you can use statistical analysis methods to test relationships between variables.
- For qualitative data, you can use methods such as thematic analysis to interpret patterns and meanings in the data.
Table of contents
Methods for collecting data, examples of data collection methods, methods for analyzing data, examples of data analysis methods, other interesting articles, frequently asked questions about research methods.
Data is the information that you collect for the purposes of answering your research question . The type of data you need depends on the aims of your research.
Qualitative vs. quantitative data
Your choice of qualitative or quantitative data collection depends on the type of knowledge you want to develop.
For questions about ideas, experiences and meanings, or to study something that can’t be described numerically, collect qualitative data .
If you want to develop a more mechanistic understanding of a topic, or your research involves hypothesis testing , collect quantitative data .
| Qualitative | to broader populations. . | |
|---|---|---|
| Quantitative | . |
You can also take a mixed methods approach , where you use both qualitative and quantitative research methods.
Primary vs. secondary research
Primary research is any original data that you collect yourself for the purposes of answering your research question (e.g. through surveys , observations and experiments ). Secondary research is data that has already been collected by other researchers (e.g. in a government census or previous scientific studies).
If you are exploring a novel research question, you’ll probably need to collect primary data . But if you want to synthesize existing knowledge, analyze historical trends, or identify patterns on a large scale, secondary data might be a better choice.
| Primary | . | methods. |
|---|---|---|
| Secondary |
Descriptive vs. experimental data
In descriptive research , you collect data about your study subject without intervening. The validity of your research will depend on your sampling method .
In experimental research , you systematically intervene in a process and measure the outcome. The validity of your research will depend on your experimental design .
To conduct an experiment, you need to be able to vary your independent variable , precisely measure your dependent variable, and control for confounding variables . If it’s practically and ethically possible, this method is the best choice for answering questions about cause and effect.
| Descriptive | . . | |
|---|---|---|
| Experimental |
Receive feedback on language, structure, and formatting
Professional editors proofread and edit your paper by focusing on:
- Academic style
- Vague sentences
- Style consistency
See an example
| Research method | Primary or secondary? | Qualitative or quantitative? | When to use |
|---|---|---|---|
| Primary | Quantitative | To test cause-and-effect relationships. | |
| Primary | Quantitative | To understand general characteristics of a population. | |
| Interview/focus group | Primary | Qualitative | To gain more in-depth understanding of a topic. |
| Observation | Primary | Either | To understand how something occurs in its natural setting. |
| Secondary | Either | To situate your research in an existing body of work, or to evaluate trends within a research topic. | |
| Either | Either | To gain an in-depth understanding of a specific group or context, or when you don’t have the resources for a large study. |
Your data analysis methods will depend on the type of data you collect and how you prepare it for analysis.
Data can often be analyzed both quantitatively and qualitatively. For example, survey responses could be analyzed qualitatively by studying the meanings of responses or quantitatively by studying the frequencies of responses.
Qualitative analysis methods
Qualitative analysis is used to understand words, ideas, and experiences. You can use it to interpret data that was collected:
- From open-ended surveys and interviews , literature reviews , case studies , ethnographies , and other sources that use text rather than numbers.
- Using non-probability sampling methods .
Qualitative analysis tends to be quite flexible and relies on the researcher’s judgement, so you have to reflect carefully on your choices and assumptions and be careful to avoid research bias .
Quantitative analysis methods
Quantitative analysis uses numbers and statistics to understand frequencies, averages and correlations (in descriptive studies) or cause-and-effect relationships (in experiments).
You can use quantitative analysis to interpret data that was collected either:
- During an experiment .
- Using probability sampling methods .
Because the data is collected and analyzed in a statistically valid way, the results of quantitative analysis can be easily standardized and shared among researchers.
| Research method | Qualitative or quantitative? | When to use |
|---|---|---|
| Quantitative | To analyze data collected in a statistically valid manner (e.g. from experiments, surveys, and observations). | |
| Meta-analysis | Quantitative | To statistically analyze the results of a large collection of studies. Can only be applied to studies that collected data in a statistically valid manner. |
| Qualitative | To analyze data collected from interviews, , or textual sources. To understand general themes in the data and how they are communicated. | |
| Either | To analyze large volumes of textual or visual data collected from surveys, literature reviews, or other sources. Can be quantitative (i.e. frequencies of words) or qualitative (i.e. meanings of words). |
If you want to know more about statistics , methodology , or research bias , make sure to check out some of our other articles with explanations and examples.
- Chi square test of independence
- Statistical power
- Descriptive statistics
- Degrees of freedom
- Pearson correlation
- Null hypothesis
- Double-blind study
- Case-control study
- Research ethics
- Data collection
- Hypothesis testing
- Structured interviews
Research bias
- Hawthorne effect
- Unconscious bias
- Recall bias
- Halo effect
- Self-serving bias
- Information bias
Quantitative research deals with numbers and statistics, while qualitative research deals with words and meanings.
Quantitative methods allow you to systematically measure variables and test hypotheses . Qualitative methods allow you to explore concepts and experiences in more detail.
In mixed methods research , you use both qualitative and quantitative data collection and analysis methods to answer your research question .
A sample is a subset of individuals from a larger population . Sampling means selecting the group that you will actually collect data from in your research. For example, if you are researching the opinions of students in your university, you could survey a sample of 100 students.
In statistics, sampling allows you to test a hypothesis about the characteristics of a population.
The research methods you use depend on the type of data you need to answer your research question .
- If you want to measure something or test a hypothesis , use quantitative methods . If you want to explore ideas, thoughts and meanings, use qualitative methods .
- If you want to analyze a large amount of readily-available data, use secondary data. If you want data specific to your purposes with control over how it is generated, collect primary data.
- If you want to establish cause-and-effect relationships between variables , use experimental methods. If you want to understand the characteristics of a research subject, use descriptive methods.
Methodology refers to the overarching strategy and rationale of your research project . It involves studying the methods used in your field and the theories or principles behind them, in order to develop an approach that matches your objectives.
Methods are the specific tools and procedures you use to collect and analyze data (for example, experiments, surveys , and statistical tests ).
In shorter scientific papers, where the aim is to report the findings of a specific study, you might simply describe what you did in a methods section .
In a longer or more complex research project, such as a thesis or dissertation , you will probably include a methodology section , where you explain your approach to answering the research questions and cite relevant sources to support your choice of methods.
Is this article helpful?
Other students also liked, writing strong research questions | criteria & examples.
- What Is a Research Design | Types, Guide & Examples
- Data Collection | Definition, Methods & Examples
More interesting articles
- Between-Subjects Design | Examples, Pros, & Cons
- Cluster Sampling | A Simple Step-by-Step Guide with Examples
- Confounding Variables | Definition, Examples & Controls
- Construct Validity | Definition, Types, & Examples
- Content Analysis | Guide, Methods & Examples
- Control Groups and Treatment Groups | Uses & Examples
- Control Variables | What Are They & Why Do They Matter?
- Correlation vs. Causation | Difference, Designs & Examples
- Correlational Research | When & How to Use
- Critical Discourse Analysis | Definition, Guide & Examples
- Cross-Sectional Study | Definition, Uses & Examples
- Descriptive Research | Definition, Types, Methods & Examples
- Ethical Considerations in Research | Types & Examples
- Explanatory and Response Variables | Definitions & Examples
- Explanatory Research | Definition, Guide, & Examples
- Exploratory Research | Definition, Guide, & Examples
- External Validity | Definition, Types, Threats & Examples
- Extraneous Variables | Examples, Types & Controls
- Guide to Experimental Design | Overview, Steps, & Examples
- How Do You Incorporate an Interview into a Dissertation? | Tips
- How to Do Thematic Analysis | Step-by-Step Guide & Examples
- How to Write a Literature Review | Guide, Examples, & Templates
- How to Write a Strong Hypothesis | Steps & Examples
- Inclusion and Exclusion Criteria | Examples & Definition
- Independent vs. Dependent Variables | Definition & Examples
- Inductive Reasoning | Types, Examples, Explanation
- Inductive vs. Deductive Research Approach | Steps & Examples
- Internal Validity in Research | Definition, Threats, & Examples
- Internal vs. External Validity | Understanding Differences & Threats
- Longitudinal Study | Definition, Approaches & Examples
- Mediator vs. Moderator Variables | Differences & Examples
- Mixed Methods Research | Definition, Guide & Examples
- Multistage Sampling | Introductory Guide & Examples
- Naturalistic Observation | Definition, Guide & Examples
- Operationalization | A Guide with Examples, Pros & Cons
- Population vs. Sample | Definitions, Differences & Examples
- Primary Research | Definition, Types, & Examples
- Qualitative vs. Quantitative Research | Differences, Examples & Methods
- Quasi-Experimental Design | Definition, Types & Examples
- Questionnaire Design | Methods, Question Types & Examples
- Random Assignment in Experiments | Introduction & Examples
- Random vs. Systematic Error | Definition & Examples
- Reliability vs. Validity in Research | Difference, Types and Examples
- Reproducibility vs Replicability | Difference & Examples
- Reproducibility vs. Replicability | Difference & Examples
- Sampling Methods | Types, Techniques & Examples
- Semi-Structured Interview | Definition, Guide & Examples
- Simple Random Sampling | Definition, Steps & Examples
- Single, Double, & Triple Blind Study | Definition & Examples
- Stratified Sampling | Definition, Guide & Examples
- Structured Interview | Definition, Guide & Examples
- Survey Research | Definition, Examples & Methods
- Systematic Review | Definition, Example, & Guide
- Systematic Sampling | A Step-by-Step Guide with Examples
- Textual Analysis | Guide, 3 Approaches & Examples
- The 4 Types of Reliability in Research | Definitions & Examples
- The 4 Types of Validity in Research | Definitions & Examples
- Transcribing an Interview | 5 Steps & Transcription Software
- Triangulation in Research | Guide, Types, Examples
- Types of Interviews in Research | Guide & Examples
- Types of Research Designs Compared | Guide & Examples
- Types of Variables in Research & Statistics | Examples
- Unstructured Interview | Definition, Guide & Examples
- What Is a Case Study? | Definition, Examples & Methods
- What Is a Case-Control Study? | Definition & Examples
- What Is a Cohort Study? | Definition & Examples
- What Is a Conceptual Framework? | Tips & Examples
- What Is a Controlled Experiment? | Definitions & Examples
- What Is a Double-Barreled Question?
- What Is a Focus Group? | Step-by-Step Guide & Examples
- What Is a Likert Scale? | Guide & Examples
- What Is a Prospective Cohort Study? | Definition & Examples
- What Is a Retrospective Cohort Study? | Definition & Examples
- What Is Action Research? | Definition & Examples
- What Is an Observational Study? | Guide & Examples
- What Is Concurrent Validity? | Definition & Examples
- What Is Content Validity? | Definition & Examples
- What Is Convenience Sampling? | Definition & Examples
- What Is Convergent Validity? | Definition & Examples
- What Is Criterion Validity? | Definition & Examples
- What Is Data Cleansing? | Definition, Guide & Examples
- What Is Deductive Reasoning? | Explanation & Examples
- What Is Discriminant Validity? | Definition & Example
- What Is Ecological Validity? | Definition & Examples
- What Is Ethnography? | Definition, Guide & Examples
- What Is Face Validity? | Guide, Definition & Examples
- What Is Non-Probability Sampling? | Types & Examples
- What Is Participant Observation? | Definition & Examples
- What Is Peer Review? | Types & Examples
- What Is Predictive Validity? | Examples & Definition
- What Is Probability Sampling? | Types & Examples
- What Is Purposive Sampling? | Definition & Examples
- What Is Qualitative Observation? | Definition & Examples
- What Is Qualitative Research? | Methods & Examples
- What Is Quantitative Observation? | Definition & Examples
- What Is Quantitative Research? | Definition, Uses & Methods
What is your plagiarism score?
- Open access
- Published: 07 September 2020
A tutorial on methodological studies: the what, when, how and why
- Lawrence Mbuagbaw ORCID: orcid.org/0000-0001-5855-5461 1 , 2 , 3 ,
- Daeria O. Lawson 1 ,
- Livia Puljak 4 ,
- David B. Allison 5 &
- Lehana Thabane 1 , 2 , 6 , 7 , 8
BMC Medical Research Methodology volume 20 , Article number: 226 ( 2020 ) Cite this article
42k Accesses
61 Citations
60 Altmetric
Metrics details
Methodological studies – studies that evaluate the design, analysis or reporting of other research-related reports – play an important role in health research. They help to highlight issues in the conduct of research with the aim of improving health research methodology, and ultimately reducing research waste.
We provide an overview of some of the key aspects of methodological studies such as what they are, and when, how and why they are done. We adopt a “frequently asked questions” format to facilitate reading this paper and provide multiple examples to help guide researchers interested in conducting methodological studies. Some of the topics addressed include: is it necessary to publish a study protocol? How to select relevant research reports and databases for a methodological study? What approaches to data extraction and statistical analysis should be considered when conducting a methodological study? What are potential threats to validity and is there a way to appraise the quality of methodological studies?
Appropriate reflection and application of basic principles of epidemiology and biostatistics are required in the design and analysis of methodological studies. This paper provides an introduction for further discussion about the conduct of methodological studies.
Peer Review reports
The field of meta-research (or research-on-research) has proliferated in recent years in response to issues with research quality and conduct [ 1 , 2 , 3 ]. As the name suggests, this field targets issues with research design, conduct, analysis and reporting. Various types of research reports are often examined as the unit of analysis in these studies (e.g. abstracts, full manuscripts, trial registry entries). Like many other novel fields of research, meta-research has seen a proliferation of use before the development of reporting guidance. For example, this was the case with randomized trials for which risk of bias tools and reporting guidelines were only developed much later – after many trials had been published and noted to have limitations [ 4 , 5 ]; and for systematic reviews as well [ 6 , 7 , 8 ]. However, in the absence of formal guidance, studies that report on research differ substantially in how they are named, conducted and reported [ 9 , 10 ]. This creates challenges in identifying, summarizing and comparing them. In this tutorial paper, we will use the term methodological study to refer to any study that reports on the design, conduct, analysis or reporting of primary or secondary research-related reports (such as trial registry entries and conference abstracts).
In the past 10 years, there has been an increase in the use of terms related to methodological studies (based on records retrieved with a keyword search [in the title and abstract] for “methodological review” and “meta-epidemiological study” in PubMed up to December 2019), suggesting that these studies may be appearing more frequently in the literature. See Fig. 1 .
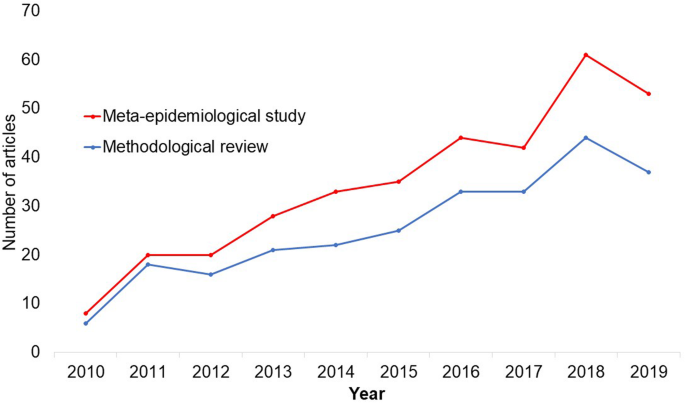
Trends in the number studies that mention “methodological review” or “meta-
epidemiological study” in PubMed.
The methods used in many methodological studies have been borrowed from systematic and scoping reviews. This practice has influenced the direction of the field, with many methodological studies including searches of electronic databases, screening of records, duplicate data extraction and assessments of risk of bias in the included studies. However, the research questions posed in methodological studies do not always require the approaches listed above, and guidance is needed on when and how to apply these methods to a methodological study. Even though methodological studies can be conducted on qualitative or mixed methods research, this paper focuses on and draws examples exclusively from quantitative research.
The objectives of this paper are to provide some insights on how to conduct methodological studies so that there is greater consistency between the research questions posed, and the design, analysis and reporting of findings. We provide multiple examples to illustrate concepts and a proposed framework for categorizing methodological studies in quantitative research.
What is a methodological study?
Any study that describes or analyzes methods (design, conduct, analysis or reporting) in published (or unpublished) literature is a methodological study. Consequently, the scope of methodological studies is quite extensive and includes, but is not limited to, topics as diverse as: research question formulation [ 11 ]; adherence to reporting guidelines [ 12 , 13 , 14 ] and consistency in reporting [ 15 ]; approaches to study analysis [ 16 ]; investigating the credibility of analyses [ 17 ]; and studies that synthesize these methodological studies [ 18 ]. While the nomenclature of methodological studies is not uniform, the intents and purposes of these studies remain fairly consistent – to describe or analyze methods in primary or secondary studies. As such, methodological studies may also be classified as a subtype of observational studies.
Parallel to this are experimental studies that compare different methods. Even though they play an important role in informing optimal research methods, experimental methodological studies are beyond the scope of this paper. Examples of such studies include the randomized trials by Buscemi et al., comparing single data extraction to double data extraction [ 19 ], and Carrasco-Labra et al., comparing approaches to presenting findings in Grading of Recommendations, Assessment, Development and Evaluations (GRADE) summary of findings tables [ 20 ]. In these studies, the unit of analysis is the person or groups of individuals applying the methods. We also direct readers to the Studies Within a Trial (SWAT) and Studies Within a Review (SWAR) programme operated through the Hub for Trials Methodology Research, for further reading as a potential useful resource for these types of experimental studies [ 21 ]. Lastly, this paper is not meant to inform the conduct of research using computational simulation and mathematical modeling for which some guidance already exists [ 22 ], or studies on the development of methods using consensus-based approaches.
When should we conduct a methodological study?
Methodological studies occupy a unique niche in health research that allows them to inform methodological advances. Methodological studies should also be conducted as pre-cursors to reporting guideline development, as they provide an opportunity to understand current practices, and help to identify the need for guidance and gaps in methodological or reporting quality. For example, the development of the popular Preferred Reporting Items of Systematic reviews and Meta-Analyses (PRISMA) guidelines were preceded by methodological studies identifying poor reporting practices [ 23 , 24 ]. In these instances, after the reporting guidelines are published, methodological studies can also be used to monitor uptake of the guidelines.
These studies can also be conducted to inform the state of the art for design, analysis and reporting practices across different types of health research fields, with the aim of improving research practices, and preventing or reducing research waste. For example, Samaan et al. conducted a scoping review of adherence to different reporting guidelines in health care literature [ 18 ]. Methodological studies can also be used to determine the factors associated with reporting practices. For example, Abbade et al. investigated journal characteristics associated with the use of the Participants, Intervention, Comparison, Outcome, Timeframe (PICOT) format in framing research questions in trials of venous ulcer disease [ 11 ].
How often are methodological studies conducted?
There is no clear answer to this question. Based on a search of PubMed, the use of related terms (“methodological review” and “meta-epidemiological study”) – and therefore, the number of methodological studies – is on the rise. However, many other terms are used to describe methodological studies. There are also many studies that explore design, conduct, analysis or reporting of research reports, but that do not use any specific terms to describe or label their study design in terms of “methodology”. This diversity in nomenclature makes a census of methodological studies elusive. Appropriate terminology and key words for methodological studies are needed to facilitate improved accessibility for end-users.
Why do we conduct methodological studies?
Methodological studies provide information on the design, conduct, analysis or reporting of primary and secondary research and can be used to appraise quality, quantity, completeness, accuracy and consistency of health research. These issues can be explored in specific fields, journals, databases, geographical regions and time periods. For example, Areia et al. explored the quality of reporting of endoscopic diagnostic studies in gastroenterology [ 25 ]; Knol et al. investigated the reporting of p -values in baseline tables in randomized trial published in high impact journals [ 26 ]; Chen et al. describe adherence to the Consolidated Standards of Reporting Trials (CONSORT) statement in Chinese Journals [ 27 ]; and Hopewell et al. describe the effect of editors’ implementation of CONSORT guidelines on reporting of abstracts over time [ 28 ]. Methodological studies provide useful information to researchers, clinicians, editors, publishers and users of health literature. As a result, these studies have been at the cornerstone of important methodological developments in the past two decades and have informed the development of many health research guidelines including the highly cited CONSORT statement [ 5 ].
Where can we find methodological studies?
Methodological studies can be found in most common biomedical bibliographic databases (e.g. Embase, MEDLINE, PubMed, Web of Science). However, the biggest caveat is that methodological studies are hard to identify in the literature due to the wide variety of names used and the lack of comprehensive databases dedicated to them. A handful can be found in the Cochrane Library as “Cochrane Methodology Reviews”, but these studies only cover methodological issues related to systematic reviews. Previous attempts to catalogue all empirical studies of methods used in reviews were abandoned 10 years ago [ 29 ]. In other databases, a variety of search terms may be applied with different levels of sensitivity and specificity.
Some frequently asked questions about methodological studies
In this section, we have outlined responses to questions that might help inform the conduct of methodological studies.
Q: How should I select research reports for my methodological study?
A: Selection of research reports for a methodological study depends on the research question and eligibility criteria. Once a clear research question is set and the nature of literature one desires to review is known, one can then begin the selection process. Selection may begin with a broad search, especially if the eligibility criteria are not apparent. For example, a methodological study of Cochrane Reviews of HIV would not require a complex search as all eligible studies can easily be retrieved from the Cochrane Library after checking a few boxes [ 30 ]. On the other hand, a methodological study of subgroup analyses in trials of gastrointestinal oncology would require a search to find such trials, and further screening to identify trials that conducted a subgroup analysis [ 31 ].
The strategies used for identifying participants in observational studies can apply here. One may use a systematic search to identify all eligible studies. If the number of eligible studies is unmanageable, a random sample of articles can be expected to provide comparable results if it is sufficiently large [ 32 ]. For example, Wilson et al. used a random sample of trials from the Cochrane Stroke Group’s Trial Register to investigate completeness of reporting [ 33 ]. It is possible that a simple random sample would lead to underrepresentation of units (i.e. research reports) that are smaller in number. This is relevant if the investigators wish to compare multiple groups but have too few units in one group. In this case a stratified sample would help to create equal groups. For example, in a methodological study comparing Cochrane and non-Cochrane reviews, Kahale et al. drew random samples from both groups [ 34 ]. Alternatively, systematic or purposeful sampling strategies can be used and we encourage researchers to justify their selected approaches based on the study objective.
Q: How many databases should I search?
A: The number of databases one should search would depend on the approach to sampling, which can include targeting the entire “population” of interest or a sample of that population. If you are interested in including the entire target population for your research question, or drawing a random or systematic sample from it, then a comprehensive and exhaustive search for relevant articles is required. In this case, we recommend using systematic approaches for searching electronic databases (i.e. at least 2 databases with a replicable and time stamped search strategy). The results of your search will constitute a sampling frame from which eligible studies can be drawn.
Alternatively, if your approach to sampling is purposeful, then we recommend targeting the database(s) or data sources (e.g. journals, registries) that include the information you need. For example, if you are conducting a methodological study of high impact journals in plastic surgery and they are all indexed in PubMed, you likely do not need to search any other databases. You may also have a comprehensive list of all journals of interest and can approach your search using the journal names in your database search (or by accessing the journal archives directly from the journal’s website). Even though one could also search journals’ web pages directly, using a database such as PubMed has multiple advantages, such as the use of filters, so the search can be narrowed down to a certain period, or study types of interest. Furthermore, individual journals’ web sites may have different search functionalities, which do not necessarily yield a consistent output.
Q: Should I publish a protocol for my methodological study?
A: A protocol is a description of intended research methods. Currently, only protocols for clinical trials require registration [ 35 ]. Protocols for systematic reviews are encouraged but no formal recommendation exists. The scientific community welcomes the publication of protocols because they help protect against selective outcome reporting, the use of post hoc methodologies to embellish results, and to help avoid duplication of efforts [ 36 ]. While the latter two risks exist in methodological research, the negative consequences may be substantially less than for clinical outcomes. In a sample of 31 methodological studies, 7 (22.6%) referenced a published protocol [ 9 ]. In the Cochrane Library, there are 15 protocols for methodological reviews (21 July 2020). This suggests that publishing protocols for methodological studies is not uncommon.
Authors can consider publishing their study protocol in a scholarly journal as a manuscript. Advantages of such publication include obtaining peer-review feedback about the planned study, and easy retrieval by searching databases such as PubMed. The disadvantages in trying to publish protocols includes delays associated with manuscript handling and peer review, as well as costs, as few journals publish study protocols, and those journals mostly charge article-processing fees [ 37 ]. Authors who would like to make their protocol publicly available without publishing it in scholarly journals, could deposit their study protocols in publicly available repositories, such as the Open Science Framework ( https://osf.io/ ).
Q: How to appraise the quality of a methodological study?
A: To date, there is no published tool for appraising the risk of bias in a methodological study, but in principle, a methodological study could be considered as a type of observational study. Therefore, during conduct or appraisal, care should be taken to avoid the biases common in observational studies [ 38 ]. These biases include selection bias, comparability of groups, and ascertainment of exposure or outcome. In other words, to generate a representative sample, a comprehensive reproducible search may be necessary to build a sampling frame. Additionally, random sampling may be necessary to ensure that all the included research reports have the same probability of being selected, and the screening and selection processes should be transparent and reproducible. To ensure that the groups compared are similar in all characteristics, matching, random sampling or stratified sampling can be used. Statistical adjustments for between-group differences can also be applied at the analysis stage. Finally, duplicate data extraction can reduce errors in assessment of exposures or outcomes.
Q: Should I justify a sample size?
A: In all instances where one is not using the target population (i.e. the group to which inferences from the research report are directed) [ 39 ], a sample size justification is good practice. The sample size justification may take the form of a description of what is expected to be achieved with the number of articles selected, or a formal sample size estimation that outlines the number of articles required to answer the research question with a certain precision and power. Sample size justifications in methodological studies are reasonable in the following instances:
Comparing two groups
Determining a proportion, mean or another quantifier
Determining factors associated with an outcome using regression-based analyses
For example, El Dib et al. computed a sample size requirement for a methodological study of diagnostic strategies in randomized trials, based on a confidence interval approach [ 40 ].
Q: What should I call my study?
A: Other terms which have been used to describe/label methodological studies include “ methodological review ”, “methodological survey” , “meta-epidemiological study” , “systematic review” , “systematic survey”, “meta-research”, “research-on-research” and many others. We recommend that the study nomenclature be clear, unambiguous, informative and allow for appropriate indexing. Methodological study nomenclature that should be avoided includes “ systematic review” – as this will likely be confused with a systematic review of a clinical question. “ Systematic survey” may also lead to confusion about whether the survey was systematic (i.e. using a preplanned methodology) or a survey using “ systematic” sampling (i.e. a sampling approach using specific intervals to determine who is selected) [ 32 ]. Any of the above meanings of the words “ systematic” may be true for methodological studies and could be potentially misleading. “ Meta-epidemiological study” is ideal for indexing, but not very informative as it describes an entire field. The term “ review ” may point towards an appraisal or “review” of the design, conduct, analysis or reporting (or methodological components) of the targeted research reports, yet it has also been used to describe narrative reviews [ 41 , 42 ]. The term “ survey ” is also in line with the approaches used in many methodological studies [ 9 ], and would be indicative of the sampling procedures of this study design. However, in the absence of guidelines on nomenclature, the term “ methodological study ” is broad enough to capture most of the scenarios of such studies.
Q: Should I account for clustering in my methodological study?
A: Data from methodological studies are often clustered. For example, articles coming from a specific source may have different reporting standards (e.g. the Cochrane Library). Articles within the same journal may be similar due to editorial practices and policies, reporting requirements and endorsement of guidelines. There is emerging evidence that these are real concerns that should be accounted for in analyses [ 43 ]. Some cluster variables are described in the section: “ What variables are relevant to methodological studies?”
A variety of modelling approaches can be used to account for correlated data, including the use of marginal, fixed or mixed effects regression models with appropriate computation of standard errors [ 44 ]. For example, Kosa et al. used generalized estimation equations to account for correlation of articles within journals [ 15 ]. Not accounting for clustering could lead to incorrect p -values, unduly narrow confidence intervals, and biased estimates [ 45 ].
Q: Should I extract data in duplicate?
A: Yes. Duplicate data extraction takes more time but results in less errors [ 19 ]. Data extraction errors in turn affect the effect estimate [ 46 ], and therefore should be mitigated. Duplicate data extraction should be considered in the absence of other approaches to minimize extraction errors. However, much like systematic reviews, this area will likely see rapid new advances with machine learning and natural language processing technologies to support researchers with screening and data extraction [ 47 , 48 ]. However, experience plays an important role in the quality of extracted data and inexperienced extractors should be paired with experienced extractors [ 46 , 49 ].
Q: Should I assess the risk of bias of research reports included in my methodological study?
A : Risk of bias is most useful in determining the certainty that can be placed in the effect measure from a study. In methodological studies, risk of bias may not serve the purpose of determining the trustworthiness of results, as effect measures are often not the primary goal of methodological studies. Determining risk of bias in methodological studies is likely a practice borrowed from systematic review methodology, but whose intrinsic value is not obvious in methodological studies. When it is part of the research question, investigators often focus on one aspect of risk of bias. For example, Speich investigated how blinding was reported in surgical trials [ 50 ], and Abraha et al., investigated the application of intention-to-treat analyses in systematic reviews and trials [ 51 ].
Q: What variables are relevant to methodological studies?
A: There is empirical evidence that certain variables may inform the findings in a methodological study. We outline some of these and provide a brief overview below:
Country: Countries and regions differ in their research cultures, and the resources available to conduct research. Therefore, it is reasonable to believe that there may be differences in methodological features across countries. Methodological studies have reported loco-regional differences in reporting quality [ 52 , 53 ]. This may also be related to challenges non-English speakers face in publishing papers in English.
Authors’ expertise: The inclusion of authors with expertise in research methodology, biostatistics, and scientific writing is likely to influence the end-product. Oltean et al. found that among randomized trials in orthopaedic surgery, the use of analyses that accounted for clustering was more likely when specialists (e.g. statistician, epidemiologist or clinical trials methodologist) were included on the study team [ 54 ]. Fleming et al. found that including methodologists in the review team was associated with appropriate use of reporting guidelines [ 55 ].
Source of funding and conflicts of interest: Some studies have found that funded studies report better [ 56 , 57 ], while others do not [ 53 , 58 ]. The presence of funding would indicate the availability of resources deployed to ensure optimal design, conduct, analysis and reporting. However, the source of funding may introduce conflicts of interest and warrant assessment. For example, Kaiser et al. investigated the effect of industry funding on obesity or nutrition randomized trials and found that reporting quality was similar [ 59 ]. Thomas et al. looked at reporting quality of long-term weight loss trials and found that industry funded studies were better [ 60 ]. Kan et al. examined the association between industry funding and “positive trials” (trials reporting a significant intervention effect) and found that industry funding was highly predictive of a positive trial [ 61 ]. This finding is similar to that of a recent Cochrane Methodology Review by Hansen et al. [ 62 ]
Journal characteristics: Certain journals’ characteristics may influence the study design, analysis or reporting. Characteristics such as journal endorsement of guidelines [ 63 , 64 ], and Journal Impact Factor (JIF) have been shown to be associated with reporting [ 63 , 65 , 66 , 67 ].
Study size (sample size/number of sites): Some studies have shown that reporting is better in larger studies [ 53 , 56 , 58 ].
Year of publication: It is reasonable to assume that design, conduct, analysis and reporting of research will change over time. Many studies have demonstrated improvements in reporting over time or after the publication of reporting guidelines [ 68 , 69 ].
Type of intervention: In a methodological study of reporting quality of weight loss intervention studies, Thabane et al. found that trials of pharmacologic interventions were reported better than trials of non-pharmacologic interventions [ 70 ].
Interactions between variables: Complex interactions between the previously listed variables are possible. High income countries with more resources may be more likely to conduct larger studies and incorporate a variety of experts. Authors in certain countries may prefer certain journals, and journal endorsement of guidelines and editorial policies may change over time.
Q: Should I focus only on high impact journals?
A: Investigators may choose to investigate only high impact journals because they are more likely to influence practice and policy, or because they assume that methodological standards would be higher. However, the JIF may severely limit the scope of articles included and may skew the sample towards articles with positive findings. The generalizability and applicability of findings from a handful of journals must be examined carefully, especially since the JIF varies over time. Even among journals that are all “high impact”, variations exist in methodological standards.
Q: Can I conduct a methodological study of qualitative research?
A: Yes. Even though a lot of methodological research has been conducted in the quantitative research field, methodological studies of qualitative studies are feasible. Certain databases that catalogue qualitative research including the Cumulative Index to Nursing & Allied Health Literature (CINAHL) have defined subject headings that are specific to methodological research (e.g. “research methodology”). Alternatively, one could also conduct a qualitative methodological review; that is, use qualitative approaches to synthesize methodological issues in qualitative studies.
Q: What reporting guidelines should I use for my methodological study?
A: There is no guideline that covers the entire scope of methodological studies. One adaptation of the PRISMA guidelines has been published, which works well for studies that aim to use the entire target population of research reports [ 71 ]. However, it is not widely used (40 citations in 2 years as of 09 December 2019), and methodological studies that are designed as cross-sectional or before-after studies require a more fit-for purpose guideline. A more encompassing reporting guideline for a broad range of methodological studies is currently under development [ 72 ]. However, in the absence of formal guidance, the requirements for scientific reporting should be respected, and authors of methodological studies should focus on transparency and reproducibility.
Q: What are the potential threats to validity and how can I avoid them?
A: Methodological studies may be compromised by a lack of internal or external validity. The main threats to internal validity in methodological studies are selection and confounding bias. Investigators must ensure that the methods used to select articles does not make them differ systematically from the set of articles to which they would like to make inferences. For example, attempting to make extrapolations to all journals after analyzing high-impact journals would be misleading.
Many factors (confounders) may distort the association between the exposure and outcome if the included research reports differ with respect to these factors [ 73 ]. For example, when examining the association between source of funding and completeness of reporting, it may be necessary to account for journals that endorse the guidelines. Confounding bias can be addressed by restriction, matching and statistical adjustment [ 73 ]. Restriction appears to be the method of choice for many investigators who choose to include only high impact journals or articles in a specific field. For example, Knol et al. examined the reporting of p -values in baseline tables of high impact journals [ 26 ]. Matching is also sometimes used. In the methodological study of non-randomized interventional studies of elective ventral hernia repair, Parker et al. matched prospective studies with retrospective studies and compared reporting standards [ 74 ]. Some other methodological studies use statistical adjustments. For example, Zhang et al. used regression techniques to determine the factors associated with missing participant data in trials [ 16 ].
With regard to external validity, researchers interested in conducting methodological studies must consider how generalizable or applicable their findings are. This should tie in closely with the research question and should be explicit. For example. Findings from methodological studies on trials published in high impact cardiology journals cannot be assumed to be applicable to trials in other fields. However, investigators must ensure that their sample truly represents the target sample either by a) conducting a comprehensive and exhaustive search, or b) using an appropriate and justified, randomly selected sample of research reports.
Even applicability to high impact journals may vary based on the investigators’ definition, and over time. For example, for high impact journals in the field of general medicine, Bouwmeester et al. included the Annals of Internal Medicine (AIM), BMJ, the Journal of the American Medical Association (JAMA), Lancet, the New England Journal of Medicine (NEJM), and PLoS Medicine ( n = 6) [ 75 ]. In contrast, the high impact journals selected in the methodological study by Schiller et al. were BMJ, JAMA, Lancet, and NEJM ( n = 4) [ 76 ]. Another methodological study by Kosa et al. included AIM, BMJ, JAMA, Lancet and NEJM ( n = 5). In the methodological study by Thabut et al., journals with a JIF greater than 5 were considered to be high impact. Riado Minguez et al. used first quartile journals in the Journal Citation Reports (JCR) for a specific year to determine “high impact” [ 77 ]. Ultimately, the definition of high impact will be based on the number of journals the investigators are willing to include, the year of impact and the JIF cut-off [ 78 ]. We acknowledge that the term “generalizability” may apply differently for methodological studies, especially when in many instances it is possible to include the entire target population in the sample studied.
Finally, methodological studies are not exempt from information bias which may stem from discrepancies in the included research reports [ 79 ], errors in data extraction, or inappropriate interpretation of the information extracted. Likewise, publication bias may also be a concern in methodological studies, but such concepts have not yet been explored.
A proposed framework
In order to inform discussions about methodological studies, the development of guidance for what should be reported, we have outlined some key features of methodological studies that can be used to classify them. For each of the categories outlined below, we provide an example. In our experience, the choice of approach to completing a methodological study can be informed by asking the following four questions:
What is the aim?
Methodological studies that investigate bias
A methodological study may be focused on exploring sources of bias in primary or secondary studies (meta-bias), or how bias is analyzed. We have taken care to distinguish bias (i.e. systematic deviations from the truth irrespective of the source) from reporting quality or completeness (i.e. not adhering to a specific reporting guideline or norm). An example of where this distinction would be important is in the case of a randomized trial with no blinding. This study (depending on the nature of the intervention) would be at risk of performance bias. However, if the authors report that their study was not blinded, they would have reported adequately. In fact, some methodological studies attempt to capture both “quality of conduct” and “quality of reporting”, such as Richie et al., who reported on the risk of bias in randomized trials of pharmacy practice interventions [ 80 ]. Babic et al. investigated how risk of bias was used to inform sensitivity analyses in Cochrane reviews [ 81 ]. Further, biases related to choice of outcomes can also be explored. For example, Tan et al investigated differences in treatment effect size based on the outcome reported [ 82 ].
Methodological studies that investigate quality (or completeness) of reporting
Methodological studies may report quality of reporting against a reporting checklist (i.e. adherence to guidelines) or against expected norms. For example, Croituro et al. report on the quality of reporting in systematic reviews published in dermatology journals based on their adherence to the PRISMA statement [ 83 ], and Khan et al. described the quality of reporting of harms in randomized controlled trials published in high impact cardiovascular journals based on the CONSORT extension for harms [ 84 ]. Other methodological studies investigate reporting of certain features of interest that may not be part of formally published checklists or guidelines. For example, Mbuagbaw et al. described how often the implications for research are elaborated using the Evidence, Participants, Intervention, Comparison, Outcome, Timeframe (EPICOT) format [ 30 ].
Methodological studies that investigate the consistency of reporting
Sometimes investigators may be interested in how consistent reports of the same research are, as it is expected that there should be consistency between: conference abstracts and published manuscripts; manuscript abstracts and manuscript main text; and trial registration and published manuscript. For example, Rosmarakis et al. investigated consistency between conference abstracts and full text manuscripts [ 85 ].
Methodological studies that investigate factors associated with reporting
In addition to identifying issues with reporting in primary and secondary studies, authors of methodological studies may be interested in determining the factors that are associated with certain reporting practices. Many methodological studies incorporate this, albeit as a secondary outcome. For example, Farrokhyar et al. investigated the factors associated with reporting quality in randomized trials of coronary artery bypass grafting surgery [ 53 ].
Methodological studies that investigate methods
Methodological studies may also be used to describe methods or compare methods, and the factors associated with methods. Muller et al. described the methods used for systematic reviews and meta-analyses of observational studies [ 86 ].
Methodological studies that summarize other methodological studies
Some methodological studies synthesize results from other methodological studies. For example, Li et al. conducted a scoping review of methodological reviews that investigated consistency between full text and abstracts in primary biomedical research [ 87 ].
Methodological studies that investigate nomenclature and terminology
Some methodological studies may investigate the use of names and terms in health research. For example, Martinic et al. investigated the definitions of systematic reviews used in overviews of systematic reviews (OSRs), meta-epidemiological studies and epidemiology textbooks [ 88 ].
Other types of methodological studies
In addition to the previously mentioned experimental methodological studies, there may exist other types of methodological studies not captured here.
What is the design?
Methodological studies that are descriptive
Most methodological studies are purely descriptive and report their findings as counts (percent) and means (standard deviation) or medians (interquartile range). For example, Mbuagbaw et al. described the reporting of research recommendations in Cochrane HIV systematic reviews [ 30 ]. Gohari et al. described the quality of reporting of randomized trials in diabetes in Iran [ 12 ].
Methodological studies that are analytical
Some methodological studies are analytical wherein “analytical studies identify and quantify associations, test hypotheses, identify causes and determine whether an association exists between variables, such as between an exposure and a disease.” [ 89 ] In the case of methodological studies all these investigations are possible. For example, Kosa et al. investigated the association between agreement in primary outcome from trial registry to published manuscript and study covariates. They found that larger and more recent studies were more likely to have agreement [ 15 ]. Tricco et al. compared the conclusion statements from Cochrane and non-Cochrane systematic reviews with a meta-analysis of the primary outcome and found that non-Cochrane reviews were more likely to report positive findings. These results are a test of the null hypothesis that the proportions of Cochrane and non-Cochrane reviews that report positive results are equal [ 90 ].
What is the sampling strategy?
Methodological studies that include the target population
Methodological reviews with narrow research questions may be able to include the entire target population. For example, in the methodological study of Cochrane HIV systematic reviews, Mbuagbaw et al. included all of the available studies ( n = 103) [ 30 ].
Methodological studies that include a sample of the target population
Many methodological studies use random samples of the target population [ 33 , 91 , 92 ]. Alternatively, purposeful sampling may be used, limiting the sample to a subset of research-related reports published within a certain time period, or in journals with a certain ranking or on a topic. Systematic sampling can also be used when random sampling may be challenging to implement.
What is the unit of analysis?
Methodological studies with a research report as the unit of analysis
Many methodological studies use a research report (e.g. full manuscript of study, abstract portion of the study) as the unit of analysis, and inferences can be made at the study-level. However, both published and unpublished research-related reports can be studied. These may include articles, conference abstracts, registry entries etc.
Methodological studies with a design, analysis or reporting item as the unit of analysis
Some methodological studies report on items which may occur more than once per article. For example, Paquette et al. report on subgroup analyses in Cochrane reviews of atrial fibrillation in which 17 systematic reviews planned 56 subgroup analyses [ 93 ].
This framework is outlined in Fig. 2 .

A proposed framework for methodological studies
Conclusions
Methodological studies have examined different aspects of reporting such as quality, completeness, consistency and adherence to reporting guidelines. As such, many of the methodological study examples cited in this tutorial are related to reporting. However, as an evolving field, the scope of research questions that can be addressed by methodological studies is expected to increase.
In this paper we have outlined the scope and purpose of methodological studies, along with examples of instances in which various approaches have been used. In the absence of formal guidance on the design, conduct, analysis and reporting of methodological studies, we have provided some advice to help make methodological studies consistent. This advice is grounded in good contemporary scientific practice. Generally, the research question should tie in with the sampling approach and planned analysis. We have also highlighted the variables that may inform findings from methodological studies. Lastly, we have provided suggestions for ways in which authors can categorize their methodological studies to inform their design and analysis.
Availability of data and materials
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Abbreviations
Consolidated Standards of Reporting Trials
Evidence, Participants, Intervention, Comparison, Outcome, Timeframe
Grading of Recommendations, Assessment, Development and Evaluations
Participants, Intervention, Comparison, Outcome, Timeframe
Preferred Reporting Items of Systematic reviews and Meta-Analyses
Studies Within a Review
Studies Within a Trial
Chalmers I, Glasziou P. Avoidable waste in the production and reporting of research evidence. Lancet. 2009;374(9683):86–9.
PubMed Google Scholar
Chan AW, Song F, Vickers A, Jefferson T, Dickersin K, Gotzsche PC, Krumholz HM, Ghersi D, van der Worp HB. Increasing value and reducing waste: addressing inaccessible research. Lancet. 2014;383(9913):257–66.
PubMed PubMed Central Google Scholar
Ioannidis JP, Greenland S, Hlatky MA, Khoury MJ, Macleod MR, Moher D, Schulz KF, Tibshirani R. Increasing value and reducing waste in research design, conduct, and analysis. Lancet. 2014;383(9912):166–75.
Higgins JP, Altman DG, Gotzsche PC, Juni P, Moher D, Oxman AD, Savovic J, Schulz KF, Weeks L, Sterne JA. The Cochrane Collaboration's tool for assessing risk of bias in randomised trials. BMJ. 2011;343:d5928.
Moher D, Schulz KF, Altman DG. The CONSORT statement: revised recommendations for improving the quality of reports of parallel-group randomised trials. Lancet. 2001;357.
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100.
Shea BJ, Hamel C, Wells GA, Bouter LM, Kristjansson E, Grimshaw J, Henry DA, Boers M. AMSTAR is a reliable and valid measurement tool to assess the methodological quality of systematic reviews. J Clin Epidemiol. 2009;62(10):1013–20.
Shea BJ, Reeves BC, Wells G, Thuku M, Hamel C, Moran J, Moher D, Tugwell P, Welch V, Kristjansson E, et al. AMSTAR 2: a critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. Bmj. 2017;358:j4008.
Lawson DO, Leenus A, Mbuagbaw L. Mapping the nomenclature, methodology, and reporting of studies that review methods: a pilot methodological review. Pilot Feasibility Studies. 2020;6(1):13.
Puljak L, Makaric ZL, Buljan I, Pieper D. What is a meta-epidemiological study? Analysis of published literature indicated heterogeneous study designs and definitions. J Comp Eff Res. 2020.
Abbade LPF, Wang M, Sriganesh K, Jin Y, Mbuagbaw L, Thabane L. The framing of research questions using the PICOT format in randomized controlled trials of venous ulcer disease is suboptimal: a systematic survey. Wound Repair Regen. 2017;25(5):892–900.
Gohari F, Baradaran HR, Tabatabaee M, Anijidani S, Mohammadpour Touserkani F, Atlasi R, Razmgir M. Quality of reporting randomized controlled trials (RCTs) in diabetes in Iran; a systematic review. J Diabetes Metab Disord. 2015;15(1):36.
Wang M, Jin Y, Hu ZJ, Thabane A, Dennis B, Gajic-Veljanoski O, Paul J, Thabane L. The reporting quality of abstracts of stepped wedge randomized trials is suboptimal: a systematic survey of the literature. Contemp Clin Trials Commun. 2017;8:1–10.
Shanthanna H, Kaushal A, Mbuagbaw L, Couban R, Busse J, Thabane L: A cross-sectional study of the reporting quality of pilot or feasibility trials in high-impact anesthesia journals Can J Anaesthesia 2018, 65(11):1180–1195.
Kosa SD, Mbuagbaw L, Borg Debono V, Bhandari M, Dennis BB, Ene G, Leenus A, Shi D, Thabane M, Valvasori S, et al. Agreement in reporting between trial publications and current clinical trial registry in high impact journals: a methodological review. Contemporary Clinical Trials. 2018;65:144–50.
Zhang Y, Florez ID, Colunga Lozano LE, Aloweni FAB, Kennedy SA, Li A, Craigie S, Zhang S, Agarwal A, Lopes LC, et al. A systematic survey on reporting and methods for handling missing participant data for continuous outcomes in randomized controlled trials. J Clin Epidemiol. 2017;88:57–66.
CAS PubMed Google Scholar
Hernández AV, Boersma E, Murray GD, Habbema JD, Steyerberg EW. Subgroup analyses in therapeutic cardiovascular clinical trials: are most of them misleading? Am Heart J. 2006;151(2):257–64.
Samaan Z, Mbuagbaw L, Kosa D, Borg Debono V, Dillenburg R, Zhang S, Fruci V, Dennis B, Bawor M, Thabane L. A systematic scoping review of adherence to reporting guidelines in health care literature. J Multidiscip Healthc. 2013;6:169–88.
Buscemi N, Hartling L, Vandermeer B, Tjosvold L, Klassen TP. Single data extraction generated more errors than double data extraction in systematic reviews. J Clin Epidemiol. 2006;59(7):697–703.
Carrasco-Labra A, Brignardello-Petersen R, Santesso N, Neumann I, Mustafa RA, Mbuagbaw L, Etxeandia Ikobaltzeta I, De Stio C, McCullagh LJ, Alonso-Coello P. Improving GRADE evidence tables part 1: a randomized trial shows improved understanding of content in summary-of-findings tables with a new format. J Clin Epidemiol. 2016;74:7–18.
The Northern Ireland Hub for Trials Methodology Research: SWAT/SWAR Information [ https://www.qub.ac.uk/sites/TheNorthernIrelandNetworkforTrialsMethodologyResearch/SWATSWARInformation/ ]. Accessed 31 Aug 2020.
Chick S, Sánchez P, Ferrin D, Morrice D. How to conduct a successful simulation study. In: Proceedings of the 2003 winter simulation conference: 2003; 2003. p. 66–70.
Google Scholar
Mulrow CD. The medical review article: state of the science. Ann Intern Med. 1987;106(3):485–8.
Sacks HS, Reitman D, Pagano D, Kupelnick B. Meta-analysis: an update. Mount Sinai J Med New York. 1996;63(3–4):216–24.
CAS Google Scholar
Areia M, Soares M, Dinis-Ribeiro M. Quality reporting of endoscopic diagnostic studies in gastrointestinal journals: where do we stand on the use of the STARD and CONSORT statements? Endoscopy. 2010;42(2):138–47.
Knol M, Groenwold R, Grobbee D. P-values in baseline tables of randomised controlled trials are inappropriate but still common in high impact journals. Eur J Prev Cardiol. 2012;19(2):231–2.
Chen M, Cui J, Zhang AL, Sze DM, Xue CC, May BH. Adherence to CONSORT items in randomized controlled trials of integrative medicine for colorectal Cancer published in Chinese journals. J Altern Complement Med. 2018;24(2):115–24.
Hopewell S, Ravaud P, Baron G, Boutron I. Effect of editors' implementation of CONSORT guidelines on the reporting of abstracts in high impact medical journals: interrupted time series analysis. BMJ. 2012;344:e4178.
The Cochrane Methodology Register Issue 2 2009 [ https://cmr.cochrane.org/help.htm ]. Accessed 31 Aug 2020.
Mbuagbaw L, Kredo T, Welch V, Mursleen S, Ross S, Zani B, Motaze NV, Quinlan L. Critical EPICOT items were absent in Cochrane human immunodeficiency virus systematic reviews: a bibliometric analysis. J Clin Epidemiol. 2016;74:66–72.
Barton S, Peckitt C, Sclafani F, Cunningham D, Chau I. The influence of industry sponsorship on the reporting of subgroup analyses within phase III randomised controlled trials in gastrointestinal oncology. Eur J Cancer. 2015;51(18):2732–9.
Setia MS. Methodology series module 5: sampling strategies. Indian J Dermatol. 2016;61(5):505–9.
Wilson B, Burnett P, Moher D, Altman DG, Al-Shahi Salman R. Completeness of reporting of randomised controlled trials including people with transient ischaemic attack or stroke: a systematic review. Eur Stroke J. 2018;3(4):337–46.
Kahale LA, Diab B, Brignardello-Petersen R, Agarwal A, Mustafa RA, Kwong J, Neumann I, Li L, Lopes LC, Briel M, et al. Systematic reviews do not adequately report or address missing outcome data in their analyses: a methodological survey. J Clin Epidemiol. 2018;99:14–23.
De Angelis CD, Drazen JM, Frizelle FA, Haug C, Hoey J, Horton R, Kotzin S, Laine C, Marusic A, Overbeke AJPM, et al. Is this clinical trial fully registered?: a statement from the International Committee of Medical Journal Editors*. Ann Intern Med. 2005;143(2):146–8.
Ohtake PJ, Childs JD. Why publish study protocols? Phys Ther. 2014;94(9):1208–9.
Rombey T, Allers K, Mathes T, Hoffmann F, Pieper D. A descriptive analysis of the characteristics and the peer review process of systematic review protocols published in an open peer review journal from 2012 to 2017. BMC Med Res Methodol. 2019;19(1):57.
Grimes DA, Schulz KF. Bias and causal associations in observational research. Lancet. 2002;359(9302):248–52.
Porta M (ed.): A dictionary of epidemiology, 5th edn. Oxford: Oxford University Press, Inc.; 2008.
El Dib R, Tikkinen KAO, Akl EA, Gomaa HA, Mustafa RA, Agarwal A, Carpenter CR, Zhang Y, Jorge EC, Almeida R, et al. Systematic survey of randomized trials evaluating the impact of alternative diagnostic strategies on patient-important outcomes. J Clin Epidemiol. 2017;84:61–9.
Helzer JE, Robins LN, Taibleson M, Woodruff RA Jr, Reich T, Wish ED. Reliability of psychiatric diagnosis. I. a methodological review. Arch Gen Psychiatry. 1977;34(2):129–33.
Chung ST, Chacko SK, Sunehag AL, Haymond MW. Measurements of gluconeogenesis and Glycogenolysis: a methodological review. Diabetes. 2015;64(12):3996–4010.
CAS PubMed PubMed Central Google Scholar
Sterne JA, Juni P, Schulz KF, Altman DG, Bartlett C, Egger M. Statistical methods for assessing the influence of study characteristics on treatment effects in 'meta-epidemiological' research. Stat Med. 2002;21(11):1513–24.
Moen EL, Fricano-Kugler CJ, Luikart BW, O’Malley AJ. Analyzing clustered data: why and how to account for multiple observations nested within a study participant? PLoS One. 2016;11(1):e0146721.
Zyzanski SJ, Flocke SA, Dickinson LM. On the nature and analysis of clustered data. Ann Fam Med. 2004;2(3):199–200.
Mathes T, Klassen P, Pieper D. Frequency of data extraction errors and methods to increase data extraction quality: a methodological review. BMC Med Res Methodol. 2017;17(1):152.
Bui DDA, Del Fiol G, Hurdle JF, Jonnalagadda S. Extractive text summarization system to aid data extraction from full text in systematic review development. J Biomed Inform. 2016;64:265–72.
Bui DD, Del Fiol G, Jonnalagadda S. PDF text classification to leverage information extraction from publication reports. J Biomed Inform. 2016;61:141–8.
Maticic K, Krnic Martinic M, Puljak L. Assessment of reporting quality of abstracts of systematic reviews with meta-analysis using PRISMA-A and discordance in assessments between raters without prior experience. BMC Med Res Methodol. 2019;19(1):32.
Speich B. Blinding in surgical randomized clinical trials in 2015. Ann Surg. 2017;266(1):21–2.
Abraha I, Cozzolino F, Orso M, Marchesi M, Germani A, Lombardo G, Eusebi P, De Florio R, Luchetta ML, Iorio A, et al. A systematic review found that deviations from intention-to-treat are common in randomized trials and systematic reviews. J Clin Epidemiol. 2017;84:37–46.
Zhong Y, Zhou W, Jiang H, Fan T, Diao X, Yang H, Min J, Wang G, Fu J, Mao B. Quality of reporting of two-group parallel randomized controlled clinical trials of multi-herb formulae: A survey of reports indexed in the Science Citation Index Expanded. Eur J Integrative Med. 2011;3(4):e309–16.
Farrokhyar F, Chu R, Whitlock R, Thabane L. A systematic review of the quality of publications reporting coronary artery bypass grafting trials. Can J Surg. 2007;50(4):266–77.
Oltean H, Gagnier JJ. Use of clustering analysis in randomized controlled trials in orthopaedic surgery. BMC Med Res Methodol. 2015;15:17.
Fleming PS, Koletsi D, Pandis N. Blinded by PRISMA: are systematic reviewers focusing on PRISMA and ignoring other guidelines? PLoS One. 2014;9(5):e96407.
Balasubramanian SP, Wiener M, Alshameeri Z, Tiruvoipati R, Elbourne D, Reed MW. Standards of reporting of randomized controlled trials in general surgery: can we do better? Ann Surg. 2006;244(5):663–7.
de Vries TW, van Roon EN. Low quality of reporting adverse drug reactions in paediatric randomised controlled trials. Arch Dis Child. 2010;95(12):1023–6.
Borg Debono V, Zhang S, Ye C, Paul J, Arya A, Hurlburt L, Murthy Y, Thabane L. The quality of reporting of RCTs used within a postoperative pain management meta-analysis, using the CONSORT statement. BMC Anesthesiol. 2012;12:13.
Kaiser KA, Cofield SS, Fontaine KR, Glasser SP, Thabane L, Chu R, Ambrale S, Dwary AD, Kumar A, Nayyar G, et al. Is funding source related to study reporting quality in obesity or nutrition randomized control trials in top-tier medical journals? Int J Obes. 2012;36(7):977–81.
Thomas O, Thabane L, Douketis J, Chu R, Westfall AO, Allison DB. Industry funding and the reporting quality of large long-term weight loss trials. Int J Obes. 2008;32(10):1531–6.
Khan NR, Saad H, Oravec CS, Rossi N, Nguyen V, Venable GT, Lillard JC, Patel P, Taylor DR, Vaughn BN, et al. A review of industry funding in randomized controlled trials published in the neurosurgical literature-the elephant in the room. Neurosurgery. 2018;83(5):890–7.
Hansen C, Lundh A, Rasmussen K, Hrobjartsson A. Financial conflicts of interest in systematic reviews: associations with results, conclusions, and methodological quality. Cochrane Database Syst Rev. 2019;8:Mr000047.
Kiehna EN, Starke RM, Pouratian N, Dumont AS. Standards for reporting randomized controlled trials in neurosurgery. J Neurosurg. 2011;114(2):280–5.
Liu LQ, Morris PJ, Pengel LH. Compliance to the CONSORT statement of randomized controlled trials in solid organ transplantation: a 3-year overview. Transpl Int. 2013;26(3):300–6.
Bala MM, Akl EA, Sun X, Bassler D, Mertz D, Mejza F, Vandvik PO, Malaga G, Johnston BC, Dahm P, et al. Randomized trials published in higher vs. lower impact journals differ in design, conduct, and analysis. J Clin Epidemiol. 2013;66(3):286–95.
Lee SY, Teoh PJ, Camm CF, Agha RA. Compliance of randomized controlled trials in trauma surgery with the CONSORT statement. J Trauma Acute Care Surg. 2013;75(4):562–72.
Ziogas DC, Zintzaras E. Analysis of the quality of reporting of randomized controlled trials in acute and chronic myeloid leukemia, and myelodysplastic syndromes as governed by the CONSORT statement. Ann Epidemiol. 2009;19(7):494–500.
Alvarez F, Meyer N, Gourraud PA, Paul C. CONSORT adoption and quality of reporting of randomized controlled trials: a systematic analysis in two dermatology journals. Br J Dermatol. 2009;161(5):1159–65.
Mbuagbaw L, Thabane M, Vanniyasingam T, Borg Debono V, Kosa S, Zhang S, Ye C, Parpia S, Dennis BB, Thabane L. Improvement in the quality of abstracts in major clinical journals since CONSORT extension for abstracts: a systematic review. Contemporary Clin trials. 2014;38(2):245–50.
Thabane L, Chu R, Cuddy K, Douketis J. What is the quality of reporting in weight loss intervention studies? A systematic review of randomized controlled trials. Int J Obes. 2007;31(10):1554–9.
Murad MH, Wang Z. Guidelines for reporting meta-epidemiological methodology research. Evidence Based Med. 2017;22(4):139.
METRIC - MEthodological sTudy ReportIng Checklist: guidelines for reporting methodological studies in health research [ http://www.equator-network.org/library/reporting-guidelines-under-development/reporting-guidelines-under-development-for-other-study-designs/#METRIC ]. Accessed 31 Aug 2020.
Jager KJ, Zoccali C, MacLeod A, Dekker FW. Confounding: what it is and how to deal with it. Kidney Int. 2008;73(3):256–60.
Parker SG, Halligan S, Erotocritou M, Wood CPJ, Boulton RW, Plumb AAO, Windsor ACJ, Mallett S. A systematic methodological review of non-randomised interventional studies of elective ventral hernia repair: clear definitions and a standardised minimum dataset are needed. Hernia. 2019.
Bouwmeester W, Zuithoff NPA, Mallett S, Geerlings MI, Vergouwe Y, Steyerberg EW, Altman DG, Moons KGM. Reporting and methods in clinical prediction research: a systematic review. PLoS Med. 2012;9(5):1–12.
Schiller P, Burchardi N, Niestroj M, Kieser M. Quality of reporting of clinical non-inferiority and equivalence randomised trials--update and extension. Trials. 2012;13:214.
Riado Minguez D, Kowalski M, Vallve Odena M, Longin Pontzen D, Jelicic Kadic A, Jeric M, Dosenovic S, Jakus D, Vrdoljak M, Poklepovic Pericic T, et al. Methodological and reporting quality of systematic reviews published in the highest ranking journals in the field of pain. Anesth Analg. 2017;125(4):1348–54.
Thabut G, Estellat C, Boutron I, Samama CM, Ravaud P. Methodological issues in trials assessing primary prophylaxis of venous thrombo-embolism. Eur Heart J. 2005;27(2):227–36.
Puljak L, Riva N, Parmelli E, González-Lorenzo M, Moja L, Pieper D. Data extraction methods: an analysis of internal reporting discrepancies in single manuscripts and practical advice. J Clin Epidemiol. 2020;117:158–64.
Ritchie A, Seubert L, Clifford R, Perry D, Bond C. Do randomised controlled trials relevant to pharmacy meet best practice standards for quality conduct and reporting? A systematic review. Int J Pharm Pract. 2019.
Babic A, Vuka I, Saric F, Proloscic I, Slapnicar E, Cavar J, Pericic TP, Pieper D, Puljak L. Overall bias methods and their use in sensitivity analysis of Cochrane reviews were not consistent. J Clin Epidemiol. 2019.
Tan A, Porcher R, Crequit P, Ravaud P, Dechartres A. Differences in treatment effect size between overall survival and progression-free survival in immunotherapy trials: a Meta-epidemiologic study of trials with results posted at ClinicalTrials.gov. J Clin Oncol. 2017;35(15):1686–94.
Croitoru D, Huang Y, Kurdina A, Chan AW, Drucker AM. Quality of reporting in systematic reviews published in dermatology journals. Br J Dermatol. 2020;182(6):1469–76.
Khan MS, Ochani RK, Shaikh A, Vaduganathan M, Khan SU, Fatima K, Yamani N, Mandrola J, Doukky R, Krasuski RA: Assessing the Quality of Reporting of Harms in Randomized Controlled Trials Published in High Impact Cardiovascular Journals. Eur Heart J Qual Care Clin Outcomes 2019.
Rosmarakis ES, Soteriades ES, Vergidis PI, Kasiakou SK, Falagas ME. From conference abstract to full paper: differences between data presented in conferences and journals. FASEB J. 2005;19(7):673–80.
Mueller M, D’Addario M, Egger M, Cevallos M, Dekkers O, Mugglin C, Scott P. Methods to systematically review and meta-analyse observational studies: a systematic scoping review of recommendations. BMC Med Res Methodol. 2018;18(1):44.
Li G, Abbade LPF, Nwosu I, Jin Y, Leenus A, Maaz M, Wang M, Bhatt M, Zielinski L, Sanger N, et al. A scoping review of comparisons between abstracts and full reports in primary biomedical research. BMC Med Res Methodol. 2017;17(1):181.
Krnic Martinic M, Pieper D, Glatt A, Puljak L. Definition of a systematic review used in overviews of systematic reviews, meta-epidemiological studies and textbooks. BMC Med Res Methodol. 2019;19(1):203.
Analytical study [ https://medical-dictionary.thefreedictionary.com/analytical+study ]. Accessed 31 Aug 2020.
Tricco AC, Tetzlaff J, Pham B, Brehaut J, Moher D. Non-Cochrane vs. Cochrane reviews were twice as likely to have positive conclusion statements: cross-sectional study. J Clin Epidemiol. 2009;62(4):380–6 e381.
Schalken N, Rietbergen C. The reporting quality of systematic reviews and Meta-analyses in industrial and organizational psychology: a systematic review. Front Psychol. 2017;8:1395.
Ranker LR, Petersen JM, Fox MP. Awareness of and potential for dependent error in the observational epidemiologic literature: A review. Ann Epidemiol. 2019;36:15–9 e12.
Paquette M, Alotaibi AM, Nieuwlaat R, Santesso N, Mbuagbaw L. A meta-epidemiological study of subgroup analyses in cochrane systematic reviews of atrial fibrillation. Syst Rev. 2019;8(1):241.
Download references
Acknowledgements
This work did not receive any dedicated funding.
Author information
Authors and affiliations.
Department of Health Research Methods, Evidence and Impact, McMaster University, Hamilton, ON, Canada
Lawrence Mbuagbaw, Daeria O. Lawson & Lehana Thabane
Biostatistics Unit/FSORC, 50 Charlton Avenue East, St Joseph’s Healthcare—Hamilton, 3rd Floor Martha Wing, Room H321, Hamilton, Ontario, L8N 4A6, Canada
Lawrence Mbuagbaw & Lehana Thabane
Centre for the Development of Best Practices in Health, Yaoundé, Cameroon
Lawrence Mbuagbaw
Center for Evidence-Based Medicine and Health Care, Catholic University of Croatia, Ilica 242, 10000, Zagreb, Croatia
Livia Puljak
Department of Epidemiology and Biostatistics, School of Public Health – Bloomington, Indiana University, Bloomington, IN, 47405, USA
David B. Allison
Departments of Paediatrics and Anaesthesia, McMaster University, Hamilton, ON, Canada
Lehana Thabane
Centre for Evaluation of Medicine, St. Joseph’s Healthcare-Hamilton, Hamilton, ON, Canada
Population Health Research Institute, Hamilton Health Sciences, Hamilton, ON, Canada
You can also search for this author in PubMed Google Scholar
Contributions
LM conceived the idea and drafted the outline and paper. DOL and LT commented on the idea and draft outline. LM, LP and DOL performed literature searches and data extraction. All authors (LM, DOL, LT, LP, DBA) reviewed several draft versions of the manuscript and approved the final manuscript.
Corresponding author
Correspondence to Lawrence Mbuagbaw .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
DOL, DBA, LM, LP and LT are involved in the development of a reporting guideline for methodological studies.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Mbuagbaw, L., Lawson, D.O., Puljak, L. et al. A tutorial on methodological studies: the what, when, how and why. BMC Med Res Methodol 20 , 226 (2020). https://doi.org/10.1186/s12874-020-01107-7
Download citation
Received : 27 May 2020
Accepted : 27 August 2020
Published : 07 September 2020
DOI : https://doi.org/10.1186/s12874-020-01107-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Methodological study
- Meta-epidemiology
- Research methods
- Research-on-research
BMC Medical Research Methodology
ISSN: 1471-2288
- General enquiries: [email protected]
- Skip to main content
- Skip to primary sidebar
- Skip to footer
- QuestionPro

- Solutions Industries Gaming Automotive Sports and events Education Government Travel & Hospitality Financial Services Healthcare Cannabis Technology Use Case AskWhy Communities Audience Contactless surveys Mobile LivePolls Member Experience GDPR Positive People Science 360 Feedback Surveys
- Resources Blog eBooks Survey Templates Case Studies Training Help center
Home Market Research
Data Analysis in Research: Types & Methods
Content Index
Why analyze data in research?
Types of data in research, finding patterns in the qualitative data, methods used for data analysis in qualitative research, preparing data for analysis, methods used for data analysis in quantitative research, considerations in research data analysis, what is data analysis in research.
Definition of research in data analysis: According to LeCompte and Schensul, research data analysis is a process used by researchers to reduce data to a story and interpret it to derive insights. The data analysis process helps reduce a large chunk of data into smaller fragments, which makes sense.
Three essential things occur during the data analysis process — the first is data organization . Summarization and categorization together contribute to becoming the second known method used for data reduction. It helps find patterns and themes in the data for easy identification and linking. The third and last way is data analysis – researchers do it in both top-down and bottom-up fashion.
LEARN ABOUT: Research Process Steps
On the other hand, Marshall and Rossman describe data analysis as a messy, ambiguous, and time-consuming but creative and fascinating process through which a mass of collected data is brought to order, structure and meaning.
We can say that “the data analysis and data interpretation is a process representing the application of deductive and inductive logic to the research and data analysis.”
Researchers rely heavily on data as they have a story to tell or research problems to solve. It starts with a question, and data is nothing but an answer to that question. But, what if there is no question to ask? Well! It is possible to explore data even without a problem – we call it ‘Data Mining’, which often reveals some interesting patterns within the data that are worth exploring.
Irrelevant to the type of data researchers explore, their mission and audiences’ vision guide them to find the patterns to shape the story they want to tell. One of the essential things expected from researchers while analyzing data is to stay open and remain unbiased toward unexpected patterns, expressions, and results. Remember, sometimes, data analysis tells the most unforeseen yet exciting stories that were not expected when initiating data analysis. Therefore, rely on the data you have at hand and enjoy the journey of exploratory research.
Create a Free Account
Every kind of data has a rare quality of describing things after assigning a specific value to it. For analysis, you need to organize these values, processed and presented in a given context, to make it useful. Data can be in different forms; here are the primary data types.
- Qualitative data: When the data presented has words and descriptions, then we call it qualitative data . Although you can observe this data, it is subjective and harder to analyze data in research, especially for comparison. Example: Quality data represents everything describing taste, experience, texture, or an opinion that is considered quality data. This type of data is usually collected through focus groups, personal qualitative interviews , qualitative observation or using open-ended questions in surveys.
- Quantitative data: Any data expressed in numbers of numerical figures are called quantitative data . This type of data can be distinguished into categories, grouped, measured, calculated, or ranked. Example: questions such as age, rank, cost, length, weight, scores, etc. everything comes under this type of data. You can present such data in graphical format, charts, or apply statistical analysis methods to this data. The (Outcomes Measurement Systems) OMS questionnaires in surveys are a significant source of collecting numeric data.
- Categorical data: It is data presented in groups. However, an item included in the categorical data cannot belong to more than one group. Example: A person responding to a survey by telling his living style, marital status, smoking habit, or drinking habit comes under the categorical data. A chi-square test is a standard method used to analyze this data.
Learn More : Examples of Qualitative Data in Education
Data analysis in qualitative research
Data analysis and qualitative data research work a little differently from the numerical data as the quality data is made up of words, descriptions, images, objects, and sometimes symbols. Getting insight from such complicated information is a complicated process. Hence it is typically used for exploratory research and data analysis .
Although there are several ways to find patterns in the textual information, a word-based method is the most relied and widely used global technique for research and data analysis. Notably, the data analysis process in qualitative research is manual. Here the researchers usually read the available data and find repetitive or commonly used words.
For example, while studying data collected from African countries to understand the most pressing issues people face, researchers might find “food” and “hunger” are the most commonly used words and will highlight them for further analysis.
LEARN ABOUT: Level of Analysis
The keyword context is another widely used word-based technique. In this method, the researcher tries to understand the concept by analyzing the context in which the participants use a particular keyword.
For example , researchers conducting research and data analysis for studying the concept of ‘diabetes’ amongst respondents might analyze the context of when and how the respondent has used or referred to the word ‘diabetes.’
The scrutiny-based technique is also one of the highly recommended text analysis methods used to identify a quality data pattern. Compare and contrast is the widely used method under this technique to differentiate how a specific text is similar or different from each other.
For example: To find out the “importance of resident doctor in a company,” the collected data is divided into people who think it is necessary to hire a resident doctor and those who think it is unnecessary. Compare and contrast is the best method that can be used to analyze the polls having single-answer questions types .
Metaphors can be used to reduce the data pile and find patterns in it so that it becomes easier to connect data with theory.
Variable Partitioning is another technique used to split variables so that researchers can find more coherent descriptions and explanations from the enormous data.
LEARN ABOUT: Qualitative Research Questions and Questionnaires
There are several techniques to analyze the data in qualitative research, but here are some commonly used methods,
- Content Analysis: It is widely accepted and the most frequently employed technique for data analysis in research methodology. It can be used to analyze the documented information from text, images, and sometimes from the physical items. It depends on the research questions to predict when and where to use this method.
- Narrative Analysis: This method is used to analyze content gathered from various sources such as personal interviews, field observation, and surveys . The majority of times, stories, or opinions shared by people are focused on finding answers to the research questions.
- Discourse Analysis: Similar to narrative analysis, discourse analysis is used to analyze the interactions with people. Nevertheless, this particular method considers the social context under which or within which the communication between the researcher and respondent takes place. In addition to that, discourse analysis also focuses on the lifestyle and day-to-day environment while deriving any conclusion.
- Grounded Theory: When you want to explain why a particular phenomenon happened, then using grounded theory for analyzing quality data is the best resort. Grounded theory is applied to study data about the host of similar cases occurring in different settings. When researchers are using this method, they might alter explanations or produce new ones until they arrive at some conclusion.
LEARN ABOUT: 12 Best Tools for Researchers
Data analysis in quantitative research
The first stage in research and data analysis is to make it for the analysis so that the nominal data can be converted into something meaningful. Data preparation consists of the below phases.
Phase I: Data Validation
Data validation is done to understand if the collected data sample is per the pre-set standards, or it is a biased data sample again divided into four different stages
- Fraud: To ensure an actual human being records each response to the survey or the questionnaire
- Screening: To make sure each participant or respondent is selected or chosen in compliance with the research criteria
- Procedure: To ensure ethical standards were maintained while collecting the data sample
- Completeness: To ensure that the respondent has answered all the questions in an online survey. Else, the interviewer had asked all the questions devised in the questionnaire.
Phase II: Data Editing
More often, an extensive research data sample comes loaded with errors. Respondents sometimes fill in some fields incorrectly or sometimes skip them accidentally. Data editing is a process wherein the researchers have to confirm that the provided data is free of such errors. They need to conduct necessary checks and outlier checks to edit the raw edit and make it ready for analysis.
Phase III: Data Coding
Out of all three, this is the most critical phase of data preparation associated with grouping and assigning values to the survey responses . If a survey is completed with a 1000 sample size, the researcher will create an age bracket to distinguish the respondents based on their age. Thus, it becomes easier to analyze small data buckets rather than deal with the massive data pile.
LEARN ABOUT: Steps in Qualitative Research
After the data is prepared for analysis, researchers are open to using different research and data analysis methods to derive meaningful insights. For sure, statistical analysis plans are the most favored to analyze numerical data. In statistical analysis, distinguishing between categorical data and numerical data is essential, as categorical data involves distinct categories or labels, while numerical data consists of measurable quantities. The method is again classified into two groups. First, ‘Descriptive Statistics’ used to describe data. Second, ‘Inferential statistics’ that helps in comparing the data .
Descriptive statistics
This method is used to describe the basic features of versatile types of data in research. It presents the data in such a meaningful way that pattern in the data starts making sense. Nevertheless, the descriptive analysis does not go beyond making conclusions. The conclusions are again based on the hypothesis researchers have formulated so far. Here are a few major types of descriptive analysis methods.
Measures of Frequency
- Count, Percent, Frequency
- It is used to denote home often a particular event occurs.
- Researchers use it when they want to showcase how often a response is given.
Measures of Central Tendency
- Mean, Median, Mode
- The method is widely used to demonstrate distribution by various points.
- Researchers use this method when they want to showcase the most commonly or averagely indicated response.
Measures of Dispersion or Variation
- Range, Variance, Standard deviation
- Here the field equals high/low points.
- Variance standard deviation = difference between the observed score and mean
- It is used to identify the spread of scores by stating intervals.
- Researchers use this method to showcase data spread out. It helps them identify the depth until which the data is spread out that it directly affects the mean.
Measures of Position
- Percentile ranks, Quartile ranks
- It relies on standardized scores helping researchers to identify the relationship between different scores.
- It is often used when researchers want to compare scores with the average count.
For quantitative research use of descriptive analysis often give absolute numbers, but the in-depth analysis is never sufficient to demonstrate the rationale behind those numbers. Nevertheless, it is necessary to think of the best method for research and data analysis suiting your survey questionnaire and what story researchers want to tell. For example, the mean is the best way to demonstrate the students’ average scores in schools. It is better to rely on the descriptive statistics when the researchers intend to keep the research or outcome limited to the provided sample without generalizing it. For example, when you want to compare average voting done in two different cities, differential statistics are enough.
Descriptive analysis is also called a ‘univariate analysis’ since it is commonly used to analyze a single variable.
Inferential statistics
Inferential statistics are used to make predictions about a larger population after research and data analysis of the representing population’s collected sample. For example, you can ask some odd 100 audiences at a movie theater if they like the movie they are watching. Researchers then use inferential statistics on the collected sample to reason that about 80-90% of people like the movie.
Here are two significant areas of inferential statistics.
- Estimating parameters: It takes statistics from the sample research data and demonstrates something about the population parameter.
- Hypothesis test: I t’s about sampling research data to answer the survey research questions. For example, researchers might be interested to understand if the new shade of lipstick recently launched is good or not, or if the multivitamin capsules help children to perform better at games.
These are sophisticated analysis methods used to showcase the relationship between different variables instead of describing a single variable. It is often used when researchers want something beyond absolute numbers to understand the relationship between variables.
Here are some of the commonly used methods for data analysis in research.
- Correlation: When researchers are not conducting experimental research or quasi-experimental research wherein the researchers are interested to understand the relationship between two or more variables, they opt for correlational research methods.
- Cross-tabulation: Also called contingency tables, cross-tabulation is used to analyze the relationship between multiple variables. Suppose provided data has age and gender categories presented in rows and columns. A two-dimensional cross-tabulation helps for seamless data analysis and research by showing the number of males and females in each age category.
- Regression analysis: For understanding the strong relationship between two variables, researchers do not look beyond the primary and commonly used regression analysis method, which is also a type of predictive analysis used. In this method, you have an essential factor called the dependent variable. You also have multiple independent variables in regression analysis. You undertake efforts to find out the impact of independent variables on the dependent variable. The values of both independent and dependent variables are assumed as being ascertained in an error-free random manner.
- Frequency tables: The statistical procedure is used for testing the degree to which two or more vary or differ in an experiment. A considerable degree of variation means research findings were significant. In many contexts, ANOVA testing and variance analysis are similar.
- Analysis of variance: The statistical procedure is used for testing the degree to which two or more vary or differ in an experiment. A considerable degree of variation means research findings were significant. In many contexts, ANOVA testing and variance analysis are similar.
- Researchers must have the necessary research skills to analyze and manipulation the data , Getting trained to demonstrate a high standard of research practice. Ideally, researchers must possess more than a basic understanding of the rationale of selecting one statistical method over the other to obtain better data insights.
- Usually, research and data analytics projects differ by scientific discipline; therefore, getting statistical advice at the beginning of analysis helps design a survey questionnaire, select data collection methods , and choose samples.
LEARN ABOUT: Best Data Collection Tools
- The primary aim of data research and analysis is to derive ultimate insights that are unbiased. Any mistake in or keeping a biased mind to collect data, selecting an analysis method, or choosing audience sample il to draw a biased inference.
- Irrelevant to the sophistication used in research data and analysis is enough to rectify the poorly defined objective outcome measurements. It does not matter if the design is at fault or intentions are not clear, but lack of clarity might mislead readers, so avoid the practice.
- The motive behind data analysis in research is to present accurate and reliable data. As far as possible, avoid statistical errors, and find a way to deal with everyday challenges like outliers, missing data, data altering, data mining , or developing graphical representation.
LEARN MORE: Descriptive Research vs Correlational Research The sheer amount of data generated daily is frightening. Especially when data analysis has taken center stage. in 2018. In last year, the total data supply amounted to 2.8 trillion gigabytes. Hence, it is clear that the enterprises willing to survive in the hypercompetitive world must possess an excellent capability to analyze complex research data, derive actionable insights, and adapt to the new market needs.
LEARN ABOUT: Average Order Value
QuestionPro is an online survey platform that empowers organizations in data analysis and research and provides them a medium to collect data by creating appealing surveys.
MORE LIKE THIS

Participant Engagement: Strategies + Improving Interaction
Sep 12, 2024

Employee Recognition Programs: A Complete Guide
Sep 11, 2024
A guide to conducting agile qualitative research for rapid insights with Digsite

Cultural Insights: What it is, Importance + How to Collect?
Sep 10, 2024
Other categories
- Academic Research
- Artificial Intelligence
- Assessments
- Brand Awareness
- Case Studies
- Communities
- Consumer Insights
- Customer effort score
- Customer Engagement
- Customer Experience
- Customer Loyalty
- Customer Research
- Customer Satisfaction
- Employee Benefits
- Employee Engagement
- Employee Retention
- Friday Five
- General Data Protection Regulation
- Insights Hub
- Life@QuestionPro
- Market Research
- Mobile diaries
- Mobile Surveys
- New Features
- Online Communities
- Question Types
- Questionnaire
- QuestionPro Products
- Release Notes
- Research Tools and Apps
- Revenue at Risk
- Survey Templates
- Training Tips
- Tuesday CX Thoughts (TCXT)
- Uncategorized
- What’s Coming Up
- Workforce Intelligence
Methodology in interpreting studies: A methodological review of evidence-based research
- January 2011
- In book: Advances in interpreting research (pp.85-119)
- Publisher: John Benjamins
- Editors: B. Nicodemus, L. Swabey

- Hong Kong Baptist University
Abstract and Figures

Discover the world's research
- 25+ million members
- 160+ million publication pages
- 2.3+ billion citations

- Sabine Braun

- Ludmila Stern

- Robert Skinner
- Robert Adam

- Franz Pöchhacker

- Miriam Shlesinger

- Yu-Hsien Chiu
- RES EVALUAT

- Stephen M. Corey

- Robert Philip Weber
- Henri C. Barik
- Recruit researchers
- Join for free
- Login Email Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google Welcome back! Please log in. Email · Hint Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google No account? Sign up
Service update: Some parts of the Library’s website will be down for maintenance on August 11.
Secondary menu
- Log in to your Library account
- Hours and Maps
- Connect from Off Campus
- UC Berkeley Home
Search form
Research methods--quantitative, qualitative, and more: overview.
- Quantitative Research
- Qualitative Research
- Data Science Methods (Machine Learning, AI, Big Data)
- Text Mining and Computational Text Analysis
- Evidence Synthesis/Systematic Reviews
- Get Data, Get Help!
About Research Methods
This guide provides an overview of research methods, how to choose and use them, and supports and resources at UC Berkeley.
As Patten and Newhart note in the book Understanding Research Methods , "Research methods are the building blocks of the scientific enterprise. They are the "how" for building systematic knowledge. The accumulation of knowledge through research is by its nature a collective endeavor. Each well-designed study provides evidence that may support, amend, refute, or deepen the understanding of existing knowledge...Decisions are important throughout the practice of research and are designed to help researchers collect evidence that includes the full spectrum of the phenomenon under study, to maintain logical rules, and to mitigate or account for possible sources of bias. In many ways, learning research methods is learning how to see and make these decisions."
The choice of methods varies by discipline, by the kind of phenomenon being studied and the data being used to study it, by the technology available, and more. This guide is an introduction, but if you don't see what you need here, always contact your subject librarian, and/or take a look to see if there's a library research guide that will answer your question.
Suggestions for changes and additions to this guide are welcome!
START HERE: SAGE Research Methods
Without question, the most comprehensive resource available from the library is SAGE Research Methods. HERE IS THE ONLINE GUIDE to this one-stop shopping collection, and some helpful links are below:
- SAGE Research Methods
- Little Green Books (Quantitative Methods)
- Little Blue Books (Qualitative Methods)
- Dictionaries and Encyclopedias
- Case studies of real research projects
- Sample datasets for hands-on practice
- Streaming video--see methods come to life
- Methodspace- -a community for researchers
- SAGE Research Methods Course Mapping
Library Data Services at UC Berkeley
Library Data Services Program and Digital Scholarship Services
The LDSP offers a variety of services and tools ! From this link, check out pages for each of the following topics: discovering data, managing data, collecting data, GIS data, text data mining, publishing data, digital scholarship, open science, and the Research Data Management Program.
Be sure also to check out the visual guide to where to seek assistance on campus with any research question you may have!
Library GIS Services
Other Data Services at Berkeley
D-Lab Supports Berkeley faculty, staff, and graduate students with research in data intensive social science, including a wide range of training and workshop offerings Dryad Dryad is a simple self-service tool for researchers to use in publishing their datasets. It provides tools for the effective publication of and access to research data. Geospatial Innovation Facility (GIF) Provides leadership and training across a broad array of integrated mapping technologies on campu Research Data Management A UC Berkeley guide and consulting service for research data management issues
General Research Methods Resources
Here are some general resources for assistance:
- Assistance from ICPSR (must create an account to access): Getting Help with Data , and Resources for Students
- Wiley Stats Ref for background information on statistics topics
- Survey Documentation and Analysis (SDA) . Program for easy web-based analysis of survey data.
Consultants
- D-Lab/Data Science Discovery Consultants Request help with your research project from peer consultants.
- Research data (RDM) consulting Meet with RDM consultants before designing the data security, storage, and sharing aspects of your qualitative project.
- Statistics Department Consulting Services A service in which advanced graduate students, under faculty supervision, are available to consult during specified hours in the Fall and Spring semesters.
Related Resourcex
- IRB / CPHS Qualitative research projects with human subjects often require that you go through an ethics review.
- OURS (Office of Undergraduate Research and Scholarships) OURS supports undergraduates who want to embark on research projects and assistantships. In particular, check out their "Getting Started in Research" workshops
- Sponsored Projects Sponsored projects works with researchers applying for major external grants.
- Next: Quantitative Research >>
- Last Updated: Sep 6, 2024 8:59 PM
- URL: https://guides.lib.berkeley.edu/researchmethods

- General Communication & Media Studies
- Communication Studies
- Communication Research Methods

Qualitative Research Methods: Collecting Evidence, Crafting Analysis, Communicating Impact, 3rd Edition
ISBN: 978-1-119-98867-0
August 2024
Wiley-Blackwell
Digital Evaluation Copy

Sarah J. Tracy
Step-by-step advice for constructing a qualitative project from beginning to end, covering both foundational theory and real-world application
Qualitative Research Methods: Collecting Evidence, Crafting Analysis, Communicating Impact guides you through sequential stages of a qualitative research project, from project design and data collection to analysis, interpretation, and presentation. Drawing on her background in qualitative research methods and human communication, Sarah J. Tracy shares personal and backstage stories while showing you how to code data, craft meaningful claims, develop theoretical explanations, and communicate research that impacts key stakeholders.
Employing a practical, problem-based contextual approach, the third edition of Qualitative Research Methods incorporates developments in textual, media, visual, arts-based, and digital analysis. New coverage includes social media data-scraping techniques, AI and ChatGPT, fieldwork and interviewing, digital ethnography, working with neurodivergent populations, adopting digital and traditional archival approaches, and much more. This edition includes a wealth of new examples, case studies, discussion questions, full-color visuals, and hands-on “Project Building Blocks” activities you can use at any stage of your qualitative research project.
Supported by a companion website containing extensive teaching and learning tools, Qualitative Research Methods: Collecting Evidence, Crafting Analysis, Communicating Impact is an indispensable resource for undergraduates, graduate students, and faculty across multiple disciplines, as well as researchers, ethnographers, and user experience professionals looking to hone their methodological practice.
SARAH J. TRACY is Professor and School Director of The Hugh Downs School of Human Communication at Arizona State University. She developed the “Big Tent” model for high-quality qualitative research and has published more than 100 scholarly monographs, in publications such as Communication Monographs, Management Communication Quarterly, and Communication Theory .

Want to create or adapt books like this? Learn more about how Pressbooks supports open publishing practices.
12 Interpretive research
Chapter 11 introduced interpretive research—or more specifically, interpretive case research. This chapter will explore other kinds of interpretive research. Recall that positivist or deductive methods—such as laboratory experiments and survey research—are those that are specifically intended for theory (or hypotheses) testing. Interpretive or inductive methods—such as action research and ethnography—one the other hand, are intended for theory building. Unlike a positivist method, where the researcher tests existing theoretical postulates using empirical data, in interpretive methods, the researcher tries to derive a theory about the phenomenon of interest from the existing observed data.
The term ‘interpretive research’ is often used loosely and synonymously with ‘qualitative research’, although the two concepts are quite different. Interpretive research is a research paradigm (see Chapter 3) that is based on the assumption that social reality is not singular or objective. Rather, it is shaped by human experiences and social contexts (ontology), and is therefore best studied within its sociohistoric context by reconciling the subjective interpretations of its various participants (epistemology). Because interpretive researchers view social reality as being embedded within—and therefore impossible to abstract from—their social settings, they ‘interpret’ the reality though a ‘sense-making’ process rather than a hypothesis testing process. This is in contrast to the positivist or functionalist paradigm that assumes that the reality is relatively independent of the context, can be abstracted from their contexts, and studied in a decomposable functional manner using objective techniques such as standardised measures. Whether a researcher should pursue interpretive or positivist research depends on paradigmatic considerations about the nature of the phenomenon under consideration and the best way to study it.
However, qualitative versus quantitative research refers to empirical or data-oriented considerations about the type of data to collect and how to analyse it. Qualitative research relies mostly on non-numeric data, such as interviews and observations, in contrast to quantitative research which employs numeric data such as scores and metrics. Hence, qualitative research is not amenable to statistical procedures such as regression analysis, but is coded using techniques like content analysis. Sometimes, coded qualitative data is tabulated quantitatively as frequencies of codes, but this data is not statistically analysed. Many puritan interpretive researchers reject this coding approach as a futile effort to seek consensus or objectivity in a social phenomenon which is essentially subjective.
Although interpretive research tends to rely heavily on qualitative data, quantitative data may add more precision and clearer understanding of the phenomenon of interest than qualitative data. For example, Eisenhardt (1989), [1] in her interpretive study of decision-making in high-velocity firms (discussed in the previous chapter on case research), collected numeric data on how long it took each firm to make certain strategic decisions—which ranged from approximately six weeks to 18 months—how many decision alternatives were considered for each decision, and surveyed her respondents to capture their perceptions of organisational conflict. Such numeric data helped her clearly distinguish the high-speed decision-making firms from the low-speed decision-makers without relying on respondents’ subjective perceptions, which then allowed her to examine the number of decision alternatives considered by and the extent of conflict in high-speed versus low-speed firms. Interpretive research should attempt to collect both qualitative and quantitative data pertaining to the phenomenon of interest, and so should positivist research as well. Joint use of qualitative and quantitative data—often called ‘mixed-mode design’—may lead to unique insights, and is therefore highly prized in the scientific community.
Interpretive research came into existence in the early nineteenth century—long before positivist techniques were developed—and has its roots in anthropology, sociology, psychology, linguistics, and semiotics. Many positivist researchers view interpretive research as erroneous and biased, given the subjective nature of the qualitative data collection and interpretation process employed in such research. However, since the 1970s, many positivist techniques’ failure to generate interesting insights or new knowledge has resulted in a resurgence of interest in interpretive research—albeit with exacting methods and stringent criteria to ensure the reliability and validity of interpretive inferences.
Distinctions from positivist research
In addition to the fundamental paradigmatic differences in ontological and epistemological assumptions discussed above, interpretive and positivist research differ in several other ways. First, interpretive research employs a theoretical sampling strategy, where study sites, respondents, or cases are selected based on theoretical considerations such as whether they fit the phenomenon being studied (e.g., sustainable practices can only be studied in organisations that have implemented sustainable practices), whether they possess certain characteristics that make them uniquely suited for the study (e.g., a study of the drivers of firm innovations should include some firms that are high innovators and some that are low innovators, in order to draw contrast between these firms), and so forth. In contrast, positivist research employs random sampling —or a variation of this technique—in which cases are chosen randomly from a population for the purpose of generalisability. Hence, convenience samples and small samples are considered acceptable in interpretive research—as long as they fit the nature and purpose of the study—but not in positivist research.
Second, the role of the researcher receives critical attention in interpretive research. In some methods such as ethnography, action research, and participant observation, the researcher is considered part of the social phenomenon, and their specific role and involvement in the research process must be made clear during data analysis. In other methods, such as case research, the researcher must take a ’neutral’ or unbiased stance during the data collection and analysis processes, and ensure that their personal biases or preconceptions do not taint the nature of subjective inferences derived from interpretive research. In positivist research, however, the researcher is considered to be external to and independent of the research context, and is not presumed to bias the data collection and analytic procedures.
Third, interpretive analysis is holistic and contextual, rather than being reductionist and isolationist. Interpretive interpretations tend to focus on language, signs, and meanings from the perspective of the participants involved in the social phenomenon, in contrast to statistical techniques that are employed heavily in positivist research. Rigor in interpretive research is viewed in terms of systematic and transparent approaches to data collection and analysis, rather than statistical benchmarks for construct validity or significance testing.
Lastly, data collection and analysis can proceed simultaneously and iteratively in interpretive research. For instance, the researcher may conduct an interview and code it before proceeding to the next interview. Simultaneous analysis helps the researcher correct potential flaws in the interview protocol or adjust it to capture the phenomenon of interest better. The researcher may even change their original research question if they realise that their original research questions are unlikely to generate new or useful insights. This is a valuable—but often understated—benefit of interpretive research, and is not available in positivist research, where the research project cannot be modified or changed once the data collection has started without redoing the entire project from the start.
Benefits and challenges of interpretive research
Interpretive research has several unique advantages. First, it is well-suited for exploring hidden reasons behind complex, interrelated, or multifaceted social processes—such as inter-firm relationships or inter-office politics—where quantitative evidence may be biased, inaccurate, or otherwise difficult to obtain. Second, it is often helpful for theory construction in areas with no or insufficient a priori theory. Third, it is also appropriate for studying context-specific, unique, or idiosyncratic events or processes. Fourth, interpretive research can also help uncover interesting and relevant research questions and issues for follow-up research.
At the same time, interpretive research also has its own set of challenges. First, this type of research tends to be more time and resource intensive than positivist research in data collection and analytic efforts. Too little data can lead to false or premature assumptions, while too much data may not be effectively processed by the researcher. Second, interpretive research requires well-trained researchers who are capable of seeing and interpreting complex social phenomenon from the perspectives of the embedded participants, and reconciling the diverse perspectives of these participants, without injecting their personal biases or preconceptions into their inferences. Third, all participants or data sources may not be equally credible, unbiased, or knowledgeable about the phenomenon of interest, or may have undisclosed political agendas which may lead to misleading or false impressions. Inadequate trust between the researcher and participants may hinder full and honest self-representation by participants, and such trust building takes time. It is the job of the interpretive researcher to ‘see through the smoke’ (i.e., hidden or biased agendas) and understand the true nature of the problem. Fourth, given the heavily contextualised nature of inferences drawn from interpretive research, such inferences do not lend themselves well to replicability or generalisability. Finally, interpretive research may sometimes fail to answer the research questions of interest or predict future behaviours.
Characteristics of interpretive research
All interpretive research must adhere to a common set of principles, as described below.
Naturalistic inquiry: Social phenomena must be studied within their natural setting.
Because interpretive research assumes that social phenomena are situated within—and cannot be isolated from—their social context, interpretations of such phenomena must be grounded within their sociohistorical context. This implies that contextual variables should be observed and considered in seeking explanations of a phenomenon of interest, even though context sensitivity may limit the generalisability of inferences.
Researcher as instrument: Researchers are often embedded within the social context that they are studying, and are considered part of the data collection instrument in that they must use their observational skills, their trust with the participants, and their ability to extract the correct information. Further, their personal insights, knowledge, and experiences of the social context are critical to accurately interpreting the phenomenon of interest. At the same time, researchers must be fully aware of their personal biases and preconceptions, and not let such biases interfere with their ability to present a fair and accurate portrayal of the phenomenon.
Interpretive analysis: Observations must be interpreted through the eyes of the participants embedded in the social context. Interpretation must occur at two levels. The first level involves viewing or experiencing the phenomenon from the subjective perspectives of the social participants. The second level is to understand the meaning of the participants’ experiences in order to provide a ‘thick description’ or a rich narrative story of the phenomenon of interest that can communicate why participants acted the way they did.
Use of expressive language: Documenting the verbal and non-verbal language of participants and the analysis of such language are integral components of interpretive analysis. The study must ensure that the story is viewed through the eyes of a person, and not a machine, and must depict the emotions and experiences of that person, so that readers can understand and relate to that person. Use of imageries, metaphors, sarcasm, and other figures of speech are very common in interpretive analysis.
Temporal nature: Interpretive research is often not concerned with searching for specific answers, but with understanding or ‘making sense of’ a dynamic social process as it unfolds over time. Hence, such research requires the researcher to immerse themself in the study site for an extended period of time in order to capture the entire evolution of the phenomenon of interest.
Hermeneutic circle: Interpretive interpretation is an iterative process of moving back and forth from pieces of observations (text), to the entirety of the social phenomenon (context), to reconcile their apparent discord, and to construct a theory that is consistent with the diverse subjective viewpoints and experiences of the embedded participants. Such iterations between the understanding/meaning of a phenomenon and observations must continue until ‘theoretical saturation’ is reached, whereby any additional iteration does not yield any more insight into the phenomenon of interest.
Interpretive data collection
Data is collected in interpretive research using a variety of techniques. The most frequently used technique is interviews (face-to-face, telephone, or focus groups). Interview types and strategies are discussed in detail in Chapter 9. A second technique is observation . Observational techniques include direct observation , where the researcher is a neutral and passive external observer, and is not involved in the phenomenon of interest (as in case research), and participant observation , where the researcher is an active participant in the phenomenon, and their input or mere presence influence the phenomenon being studied (as in action research). A third technique is documentation , where external and internal documents—such as memos, emails, annual reports, financial statements, newspaper articles, or websites—may be used to cast further insight into the phenomenon of interest or to corroborate other forms of evidence.
Interpretive research designs
Case research . As discussed in the previous chapter, case research is an intensive longitudinal study of a phenomenon at one or more research sites for the purpose of deriving detailed, contextualised inferences, and understanding the dynamic process underlying a phenomenon of interest. Case research is a unique research design in that it can be used in an interpretive manner to build theories, or in a positivist manner to test theories. The previous chapter on case research discusses both techniques in depth and provides illustrative exemplars. Furthermore, the case researcher is a neutral observer (direct observation) in the social setting, rather than an active participant (participant observation). As with any other interpretive approach, drawing meaningful inferences from case research depends heavily on the observational skills and integrative abilities of the researcher.
Action research . Action research is a qualitative but positivist research design aimed at theory testing rather than theory building. This is an interactive design that assumes that complex social phenomena are best understood by introducing changes, interventions, or ‘actions’ into those phenomena, and observing the outcomes of such actions on the phenomena of interest. In this method, the researcher is usually a consultant or an organisational member embedded into a social context —such as an organisation—who initiates an action in response to a social problem, and examines how their action influences the phenomenon, while also learning and generating insights about the relationship between the action and the phenomenon. Examples of actions may include organisational change programs—such as the introduction of new organisational processes, procedures, people, or technology or the replacement of old ones—initiated with the goal of improving an organisation’s performance or profitability. The researcher’s choice of actions must be based on theory, which should explain why and how such actions may bring forth the desired social change. The theory is validated by the extent to which the chosen action is successful in remedying the targeted problem. Simultaneous problem-solving and insight generation are the central feature that distinguishes action research from other research methods (which may not involve problem solving), and from consulting (which may not involve insight generation). Hence, action research is an excellent method for bridging research and practice.
There are several variations of the action research method. The most popular of these methods is participatory action research , designed by Susman and Evered (1978). [2] This method follows an action research cycle consisting of five phases: diagnosing, action-planning, action-taking, evaluating, and learning (see Figure 12.1). Diagnosing involves identifying and defining a problem in its social context. Action-planning involves identifying and evaluating alternative solutions to the problem, and deciding on a future course of action based on theoretical rationale. Action-taking is the implementation of the planned course of action. The evaluation stage examines the extent to which the initiated action is successful in resolving the original problem—i.e., whether theorised effects are indeed realised in practice. In the learning phase, the experiences and feedback from action evaluation are used to generate insights about the problem and suggest future modifications or improvements to the action. Based on action evaluation and learning, the action may be modified or adjusted to address the problem better, and the action research cycle is repeated with the modified action sequence. It is suggested that the entire action research cycle be traversed at least twice so that learning from the first cycle can be implemented in the second cycle. The primary mode of data collection is participant observation, although other techniques such as interviews and documentary evidence may be used to corroborate the researcher’s observations.
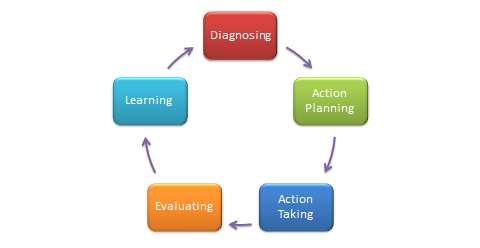
Ethnography . The ethnographic research method—derived largely from the field of anthropology—emphasises studying a phenomenon within the context of its culture. The researcher must be deeply immersed in the social culture over an extended period of time—usually eight months to two years—and should engage, observe, and record the daily life of the studied culture and its social participants within their natural setting. The primary mode of data collection is participant observation, and data analysis involves a ‘sense-making’ approach. In addition, the researcher must take extensive field notes, and narrate her experience in descriptive detail so that readers may experience the same culture as the researcher. In this method, the researcher has two roles: rely on her unique knowledge and engagement to generate insights (theory), and convince the scientific community of the transsituational nature of the studied phenomenon.
The classic example of ethnographic research is Jane Goodall’s study of primate behaviours. While living with chimpanzees in their natural habitat at Gombe National Park in Tanzania, she observed their behaviours, interacted with them, and shared their lives. During that process, she learnt and chronicled how chimpanzees seek food and shelter, how they socialise with each other, their communication patterns, their mating behaviours, and so forth. A more contemporary example of ethnographic research is Myra Bluebond-Langer’s (1996) [3] study of decision-making in families with children suffering from life-threatening illnesses, and the physical, psychological, environmental, ethical, legal, and cultural issues that influence such decision-making. The researcher followed the experiences of approximately 80 children with incurable illnesses and their families for a period of over two years. Data collection involved participant observation and formal/informal conversations with children, their parents and relatives, and healthcare providers to document their lived experience.
Phenomenology. Phenomenology is a research method that emphasises the study of conscious experiences as a way of understanding the reality around us. It is based on the ideas of early twentieth century German philosopher, Edmund Husserl, who believed that human experience is the source of all knowledge. Phenomenology is concerned with the systematic reflection and analysis of phenomena associated with conscious experiences such as human judgment, perceptions, and actions. Its goal is (appreciating and describing social reality from the diverse subjective perspectives of the participants involved, and understanding the symbolic meanings (‘deep structure’) underlying these subjective experiences. Phenomenological inquiry requires that researchers eliminate any prior assumptions and personal biases, empathise with the participant’s situation, and tune into existential dimensions of that situation so that they can fully understand the deep structures that drive the conscious thinking, feeling, and behaviour of the studied participants.
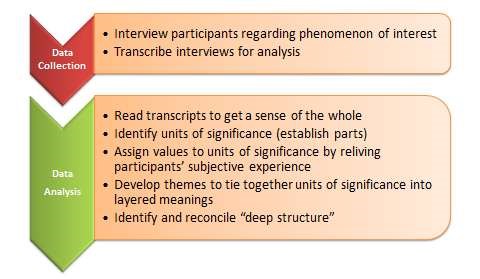
Some researchers view phenomenology as a philosophy rather than as a research method. In response to this criticism, Giorgi and Giorgi (2003) [4] developed an existential phenomenological research method to guide studies in this area. This method, illustrated in Figure 12.2, can be grouped into data collection and data analysis phases. In the data collection phase, participants embedded in a social phenomenon are interviewed to capture their subjective experiences and perspectives regarding the phenomenon under investigation. Examples of questions that may be asked include ‘Can you describe a typical day?’ or ‘Can you describe that particular incident in more detail?’. These interviews are recorded and transcribed for further analysis. During data analysis , the researcher reads the transcripts to: get a sense of the whole, and establish ‘units of significance’ that can faithfully represent participants’ subjective experiences. Examples of such units of significance are concepts such as ‘felt-space’ and ‘felt-time’, which are then used to document participants’ psychological experiences. For instance, did participants feel safe, free, trapped, or joyous when experiencing a phenomenon (‘felt-space’)? Did they feel that their experience was pressured, slow, or discontinuous (‘felt-time’)? Phenomenological analysis should take into account the participants’ temporal landscape (i.e., their sense of past, present, and future), and the researcher must transpose his/herself in an imaginary sense into the participant’s situation (i.e., temporarily live the participant’s life). The participants’ lived experience is described in the form of a narrative or using emergent themes. The analysis then delves into these themes to identify multiple layers of meaning while retaining the fragility and ambiguity of subjects’ lived experiences.
Rigor in interpretive research
While positivist research employs a ‘reductionist’ approach by simplifying social reality into parsimonious theories and laws, interpretive research attempts to interpret social reality through the subjective viewpoints of the embedded participants within the context where the reality is situated. These interpretations are heavily contextualised, and are naturally less generalisable to other contexts. However, because interpretive analysis is subjective and sensitive to the experiences and insight of the embedded researcher, it is often considered less rigorous by many positivist (functionalist) researchers. Because interpretive research is based on a different set of ontological and epistemological assumptions about social phenomena than positivist research, the positivist notions of rigor—such as reliability, internal validity, and generalisability—do not apply in a similar manner. However, Lincoln and Guba (1985) [5] provide an alternative set of criteria that can be used to judge the rigor of interpretive research.
Dependability. Interpretive research can be viewed as dependable or authentic if two researchers assessing the same phenomenon, using the same set of evidence, independently arrive at the same conclusions, or the same researcher, observing the same or a similar phenomenon at different times arrives at similar conclusions. This concept is similar to that of reliability in positivist research, with agreement between two independent researchers being similar to the notion of inter-rater reliability, and agreement between two observations of the same phenomenon by the same researcher akin to test-retest reliability. To ensure dependability, interpretive researchers must provide adequate details about their phenomenon of interest and the social context in which it is embedded, so as to allow readers to independently authenticate their interpretive inferences.
Credibility. Interpretive research can be considered credible if readers find its inferences to be believable. This concept is akin to that of internal validity in functionalistic research. The credibility of interpretive research can be improved by providing evidence of the researcher’s extended engagement in the field, by demonstrating data triangulation across subjects or data collection techniques, and by maintaining meticulous data management and analytic procedures—such as verbatim transcription of interviews, accurate records of contacts and interviews—and clear notes on theoretical and methodological decisions, that can allow an independent audit of data collection and analysis if needed.
Confirmability. Confirmability refers to the extent to which the findings reported in interpretive research can be independently confirmed by others—typically, participants. This is similar to the notion of objectivity in functionalistic research. Since interpretive research rejects the notion of an objective reality, confirmability is demonstrated in terms of ‘intersubjectivity’—i.e., if the study’s participants agree with the inferences derived by the researcher. For instance, if a study’s participants generally agree with the inferences drawn by a researcher about a phenomenon of interest—based on a review of the research paper or report—then the findings can be viewed as confirmable.
Transferability. Transferability in interpretive research refers to the extent to which the findings can be generalised to other settings. This idea is similar to that of external validity in functionalistic research. The researcher must provide rich, detailed descriptions of the research context (‘thick description’) and thoroughly describe the structures, assumptions, and processes revealed from the data so that readers can independently assess whether and to what extent the reported findings are transferable to other settings.
- Eisenhardt, K. M. (1989). Making fast strategic decisions in high-velocity environments. Academy of Management Journal , 32(3), 543–576. ↵
- Susman, G. I. and Evered, R. D. (1978) An assessment of the scientific merits of action research. Administrative Science Quarterly , 23, 582–603. ↵
- Bluebond-Langer, M. (1996). In the shadow of illness: Parents and siblings of the chronically ill child . Princeton, NJ: Princeton University Press. ↵
- Giorgi, A., & Giorgi, B. (2003). Phenomenology. In J. A. Smith (ed.), Qualitative psychology: A practical guide to research methods (pp. 25–50). London: Sage Publications ↵
- Lincoln, Y. S., & Guba, E. G. (1985). Naturalistic inquiry . Beverly Hills: Sage Publications. ↵
Social Science Research: Principles, Methods and Practices (Revised edition) Copyright © 2019 by Anol Bhattacherjee is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License , except where otherwise noted.
Share This Book

Quantitative Methodology: Measurement and Statistics, M.S.
Fall, Spring
Full-time Part-time
- September 27, 2024 (Spring 2025)
- December 3, 2024 (Fall 2025)
June 30, 2025
In-State - $12,540 Out-of-State - $26,490 More Info
This Quantitative Methodology: Measurement and Statistics, Master of Science (M.S.) program provides you with advanced training in quantitative research methods and statistical analysis. You will learn to design and conduct research studies, analyze data using sophisticated statistical techniques, and interpret and present research findings effectively. We emphasize both theoretical knowledge and practical skills, preparing you for careers in any industry. Whether pursuing further graduate studies or entering the workforce directly, you will be well-prepared to contribute to the advancement of knowledge in your chosen field.
Key Features
- Balanced Training : Gain comprehensive skills in quantitative methods suitable for various professional settings.
- Proximity to Washington, D.C. : Access diverse academic and professional opportunities in the nation's capital.
- Rigorous Core Curriculum : Master key concepts in applied measurement, statistical modeling, and evaluation methods.
- Flexibility : Choose from a range of elective courses to deepen your expertise in specific areas of interest.
- Demonstrate proficiency in applied measurement, statistical analysis, and research design.
- Apply quantitative methods to address complex research questions in diverse contexts.
- Evaluate and critique research literature and methodologies in the field of quantitative methodology.
- Communicate quantitative findings effectively to diverse audiences through written reports and presentations.
This program offers a wide range of career pathways, including:
- Research Associate
- Data Analyst
- Policy Analyst
- Evaluation Specialist
Click on admissions button below to swap url
Admission Requirements Guide to Applying
You are required to submit all required documents before submitting the application.
Program Specific Requirements
- Letters of Recommendation (3)
- Graduate Record Examination (GRE)
- Writing Sample (1)
Marieh Arnett, student, Quantitative Methodology: Measurement and Statistics
Courses in this program are carefully selected and highly customizable to give you the best possible experience. Your specific program of study will be structured to take into account your background and aspirations. Both thesis and non-thesis options are available.
QMMS Graduate Student Handbook
There is a common core of courses comprised of:
- EDMS 623 Applied Measurement: Issues and Practices (3)
- EDMS 646 General Linear Models I (3)
- EDMS 647 Causal Inference and Evaluation Methods (3)
- EDMS 651 General Linear Models II (3)
- EDMS 655 Introduction to Multilevel Modeling (3)
- EDMS 657 Exploratory Latent and Composite Variable Methods (3)
- EDMS 724 Modern Measurement Theory (3)
Additional elective coursework completes the program. A written comprehensive examination based on the first four courses of the core is required. The Graduate School allows transfer of up to six credits of appropriate prior graduate work.

Sep 17 Graduate Fair Expo Sep 17, 2024 4:00 – 6:00 pm
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 12 September 2024
An open-source framework for end-to-end analysis of electronic health record data
- Lukas Heumos 1 , 2 , 3 ,
- Philipp Ehmele 1 ,
- Tim Treis 1 , 3 ,
- Julius Upmeier zu Belzen ORCID: orcid.org/0000-0002-0966-4458 4 ,
- Eljas Roellin 1 , 5 ,
- Lilly May 1 , 5 ,
- Altana Namsaraeva 1 , 6 ,
- Nastassya Horlava 1 , 3 ,
- Vladimir A. Shitov ORCID: orcid.org/0000-0002-1960-8812 1 , 3 ,
- Xinyue Zhang ORCID: orcid.org/0000-0003-4806-4049 1 ,
- Luke Zappia ORCID: orcid.org/0000-0001-7744-8565 1 , 5 ,
- Rainer Knoll 7 ,
- Niklas J. Lang 2 ,
- Leon Hetzel 1 , 5 ,
- Isaac Virshup 1 ,
- Lisa Sikkema ORCID: orcid.org/0000-0001-9686-6295 1 , 3 ,
- Fabiola Curion 1 , 5 ,
- Roland Eils 4 , 8 ,
- Herbert B. Schiller 2 , 9 ,
- Anne Hilgendorff 2 , 10 &
- Fabian J. Theis ORCID: orcid.org/0000-0002-2419-1943 1 , 3 , 5
Nature Medicine ( 2024 ) Cite this article
93 Altmetric
Metrics details
- Epidemiology
- Translational research
With progressive digitalization of healthcare systems worldwide, large-scale collection of electronic health records (EHRs) has become commonplace. However, an extensible framework for comprehensive exploratory analysis that accounts for data heterogeneity is missing. Here we introduce ehrapy, a modular open-source Python framework designed for exploratory analysis of heterogeneous epidemiology and EHR data. ehrapy incorporates a series of analytical steps, from data extraction and quality control to the generation of low-dimensional representations. Complemented by rich statistical modules, ehrapy facilitates associating patients with disease states, differential comparison between patient clusters, survival analysis, trajectory inference, causal inference and more. Leveraging ontologies, ehrapy further enables data sharing and training EHR deep learning models, paving the way for foundational models in biomedical research. We demonstrate ehrapy’s features in six distinct examples. We applied ehrapy to stratify patients affected by unspecified pneumonia into finer-grained phenotypes. Furthermore, we reveal biomarkers for significant differences in survival among these groups. Additionally, we quantify medication-class effects of pneumonia medications on length of stay. We further leveraged ehrapy to analyze cardiovascular risks across different data modalities. We reconstructed disease state trajectories in patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) based on imaging data. Finally, we conducted a case study to demonstrate how ehrapy can detect and mitigate biases in EHR data. ehrapy, thus, provides a framework that we envision will standardize analysis pipelines on EHR data and serve as a cornerstone for the community.
Similar content being viewed by others

Data-driven identification of heart failure disease states and progression pathways using electronic health records
EHR foundation models improve robustness in the presence of temporal distribution shift
Harnessing EHR data for health research
Electronic health records (EHRs) are becoming increasingly common due to standardized data collection 1 and digitalization in healthcare institutions. EHRs collected at medical care sites serve as efficient storage and sharing units of health information 2 , enabling the informed treatment of individuals using the patient’s complete history 3 . Routinely collected EHR data are approaching genomic-scale size and complexity 4 , posing challenges in extracting information without quantitative analysis methods. The application of such approaches to EHR databases 1 , 5 , 6 , 7 , 8 , 9 has enabled the prediction and classification of diseases 10 , 11 , study of population health 12 , determination of optimal treatment policies 13 , 14 , simulation of clinical trials 15 and stratification of patients 16 .
However, current EHR datasets suffer from serious limitations, such as data collection issues, inconsistencies and lack of data diversity. EHR data collection and sharing problems often arise due to non-standardized formats, with disparate systems using exchange protocols, such as Health Level Seven International (HL7) and Fast Healthcare Interoperability Resources (FHIR) 17 . In addition, EHR data are stored in various on-disk formats, including, but not limited to, relational databases and CSV, XML and JSON formats. These variations pose challenges with respect to data retrieval, scalability, interoperability and data sharing.
Beyond format variability, inherent biases of the collected data can compromise the validity of findings. Selection bias stemming from non-representative sample composition can lead to skewed inferences about disease prevalence or treatment efficacy 18 , 19 . Filtering bias arises through inconsistent criteria for data inclusion, obscuring true variable relationships 20 . Surveillance bias exaggerates associations between exposure and outcomes due to differential monitoring frequencies 21 . EHR data are further prone to missing data 22 , 23 , which can be broadly classified into three categories: missing completely at random (MCAR), where missingness is unrelated to the data; missing at random (MAR), where missingness depends on observed data; and missing not at random (MNAR), where missingness depends on unobserved data 22 , 23 . Information and coding biases, related to inaccuracies in data recording or coding inconsistencies, respectively, can lead to misclassification and unreliable research conclusions 24 , 25 . Data may even contradict itself, such as when measurements were reported for deceased patients 26 , 27 . Technical variation and differing data collection standards lead to distribution differences and inconsistencies in representation and semantics across EHR datasets 28 , 29 . Attrition and confounding biases, resulting from differential patient dropout rates or unaccounted external variable effects, can significantly skew study outcomes 30 , 31 , 32 . The diversity of EHR data that comprise demographics, laboratory results, vital signs, diagnoses, medications, x-rays, written notes and even omics measurements amplifies all the aforementioned issues.
Addressing these challenges requires rigorous study design, careful data pre-processing and continuous bias evaluation through exploratory data analysis. Several EHR data pre-processing and analysis workflows were previously developed 4 , 33 , 34 , 35 , 36 , 37 , but none of them enables the analysis of heterogeneous data, provides in-depth documentation, is available as a software package or allows for exploratory visual analysis. Current EHR analysis pipelines, therefore, differ considerably in their approaches and are often commercial, vendor-specific solutions 38 . This is in contrast to strategies using community standards for the analysis of omics data, such as Bioconductor 39 or scverse 40 . As a result, EHR data frequently remain underexplored and are commonly investigated only for a particular research question 41 . Even in such cases, EHR data are then frequently input into machine learning models with serious data quality issues that greatly impact prediction performance and generalizability 42 .
To address this lack of analysis tooling, we developed the EHR Analysis in Python framework, ehrapy, which enables exploratory analysis of diverse EHR datasets. The ehrapy package is purpose-built to organize, analyze, visualize and statistically compare complex EHR data. ehrapy can be applied to datasets of different data types, sizes, diseases and origins. To demonstrate this versatility, we applied ehrapy to datasets obtained from EHR and population-based studies. Using the Pediatric Intensive Care (PIC) EHR database 43 , we stratified patients diagnosed with ‘unspecified pneumonia’ into distinct clinically relevant groups, extracted clinical indicators of pneumonia through statistical analysis and quantified medication-class effects on length of stay (LOS) with causal inference. Using the UK Biobank 44 (UKB), a population-scale cohort comprising over 500,000 participants from the United Kingdom, we employed ehrapy to explore cardiovascular risk factors using clinical predictors, metabolomics, genomics and retinal imaging-derived features. Additionally, we performed image analysis to project disease progression through fate mapping in patients affected by coronavirus disease 2019 (COVID-19) using chest x-rays. Finally, we demonstrate how exploratory analysis with ehrapy unveils and mitigates biases in over 100,000 visits by patients with diabetes across 130 US hospitals. We provide online links to additional use cases that demonstrate ehrapy’s usage with further datasets, including MIMIC-II (ref. 45 ), and for various medical conditions, such as patients subject to indwelling arterial catheter usage. ehrapy is compatible with any EHR dataset that can be transformed into vectors and is accessible as a user-friendly open-source software package hosted at https://github.com/theislab/ehrapy and installable from PyPI. It comes with comprehensive documentation, tutorials and further examples, all available at https://ehrapy.readthedocs.io .
ehrapy: a framework for exploratory EHR data analysis
The foundation of ehrapy is a robust and scalable data storage backend that is combined with a series of pre-processing and analysis modules. In ehrapy, EHR data are organized as a data matrix where observations are individual patient visits (or patients, in the absence of follow-up visits), and variables represent all measured quantities ( Methods ). These data matrices are stored together with metadata of observations and variables. By leveraging the AnnData (annotated data) data structure that implements this design, ehrapy builds upon established standards and is compatible with analysis and visualization functions provided by the omics scverse 40 ecosystem. Readers are also available in R, Julia and Javascript 46 . We additionally provide a dataset module with more than 20 public loadable EHR datasets in AnnData format to kickstart analysis and development with ehrapy.
For standardized analysis of EHR data, it is crucial that these data are encoded and stored in consistent, reusable formats. Thus, ehrapy requires that input data are organized in structured vectors. Readers for common formats, such as CSV, OMOP 47 or SQL databases, are available in ehrapy. Data loaded into AnnData objects can be mapped against several hierarchical ontologies 48 , 49 , 50 , 51 ( Methods ). Clinical keywords of free text notes can be automatically extracted ( Methods ).
Powered by scanpy, which scales to millions of observations 52 ( Methods and Supplementary Table 1 ) and the machine learning library scikit-learn 53 , ehrapy provides more than 100 composable analysis functions organized in modules from which custom analysis pipelines can be built. Each function directly interacts with the AnnData object and adds all intermediate results for simple access and reuse of information to it. To facilitate setting up these pipelines, ehrapy guides analysts through a general analysis pipeline (Fig. 1 ). At any step of an analysis pipeline, community software packages can be integrated without any vendor lock-in. Because ehrapy is built on open standards, it can be purposefully extended to solve new challenges, such as the development of foundational models ( Methods ).
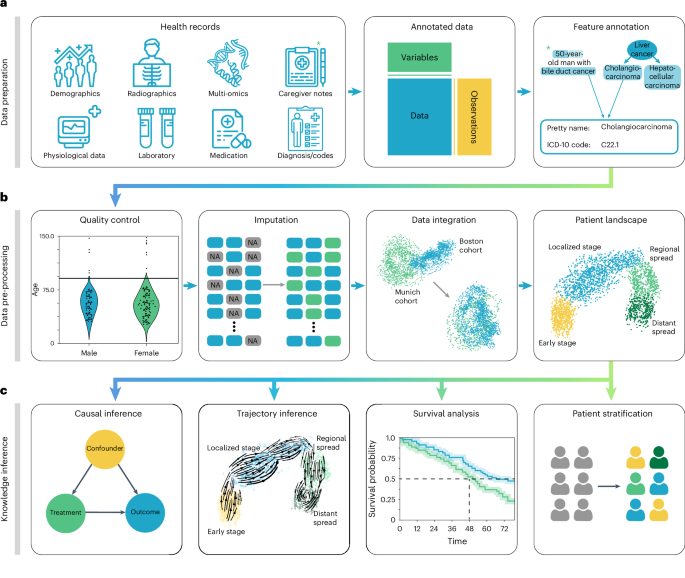
a , Heterogeneous health data are first loaded into memory as an AnnData object with patient visits as observational rows and variables as columns. Next, the data can be mapped against ontologies, and key terms are extracted from free text notes. b , The EHR data are subject to quality control where low-quality or spurious measurements are removed or imputed. Subsequently, numerical data are normalized, and categorical data are encoded. Data from different sources with data distribution shifts are integrated, embedded, clustered and annotated in a patient landscape. c , Further downstream analyses depend on the question of interest and can include the inference of causal effects and trajectories, survival analysis or patient stratification.
In the ehrapy analysis pipeline, EHR data are initially inspected for quality issues by analyzing feature distributions that may skew results and by detecting visits and features with high missing rates that ehrapy can then impute ( Methods ). ehrapy tracks all filtering steps while keeping track of population dynamics to highlight potential selection and filtering biases ( Methods ). Subsequently, ehrapy’s normalization and encoding functions ( Methods ) are applied to achieve a uniform numerical representation that facilitates data integration and corrects for dataset shift effects ( Methods ). Calculated lower-dimensional representations can subsequently be visualized, clustered and annotated to obtain a patient landscape ( Methods ). Such annotated groups of patients can be used for statistical comparisons to find differences in features among them to ultimately learn markers of patient states.
As analysis goals can differ between users and datasets, the ehrapy analysis pipeline is customizable during the final knowledge inference step. ehrapy provides statistical methods for group comparison and extensive support for survival analysis ( Methods ), enabling the discovery of biomarkers. Furthermore, ehrapy offers functions for causal inference to go from statistically determined associations to causal relations ( Methods ). Moreover, patient visits in aggregated EHR data can be regarded as snapshots where individual measurements taken at specific timepoints might not adequately reflect the underlying progression of disease and result from unrelated variation due to, for example, day-to-day differences 54 , 55 , 56 . Therefore, disease progression models should rely on analysis of the underlying clinical data, as disease progression in an individual patient may not be monotonous in time. ehrapy allows for the use of advanced trajectory inference methods to overcome sparse measurements 57 , 58 , 59 . We show that this approach can order snapshots to calculate a pseudotime that can adequately reflect the progression of the underlying clinical process. Given a sufficient number of snapshots, ehrapy increases the potential to understand disease progression, which is likely not robustly captured within a single EHR but, rather, across several.
ehrapy enables patient stratification in pneumonia cases
To demonstrate ehrapy’s capability to analyze heterogeneous datasets from a broad patient set across multiple care units, we applied our exploratory strategy to the PIC 43 database. The PIC database is a single-center database hosting information on children admitted to critical care units at the Children’s Hospital of Zhejiang University School of Medicine in China. It contains 13,499 distinct hospital admissions of 12,881 individual pediatric patients admitted between 2010 and 2018 for whom demographics, diagnoses, doctors’ notes, vital signs, laboratory and microbiology tests, medications, fluid balances and more were collected (Extended Data Figs. 1 and 2a and Methods ). After missing data imputation and subsequent pre-processing (Extended Data Figs. 2b,c and 3 and Methods ), we generated a uniform manifold approximation and projection (UMAP) embedding to visualize variation across all patients using ehrapy (Fig. 2a ). This visualization of the low-dimensional patient manifold shows the heterogeneity of the collected data in the PIC database, with malformations, perinatal and respiratory being the most abundant International Classification of Diseases (ICD) chapters (Fig. 2b ). The most common respiratory disease categories (Fig. 2c ) were labeled pneumonia and influenza ( n = 984). We focused on pneumonia to apply ehrapy to a challenging, broad-spectrum disease that affects all age groups. Pneumonia is a prevalent respiratory infection that poses a substantial burden on public health 60 and is characterized by inflammation of the alveoli and distal airways 60 . Individuals with pre-existing chronic conditions are particularly vulnerable, as are children under the age of 5 (ref. 61 ). Pneumonia can be caused by a range of microorganisms, encompassing bacteria, respiratory viruses and fungi.
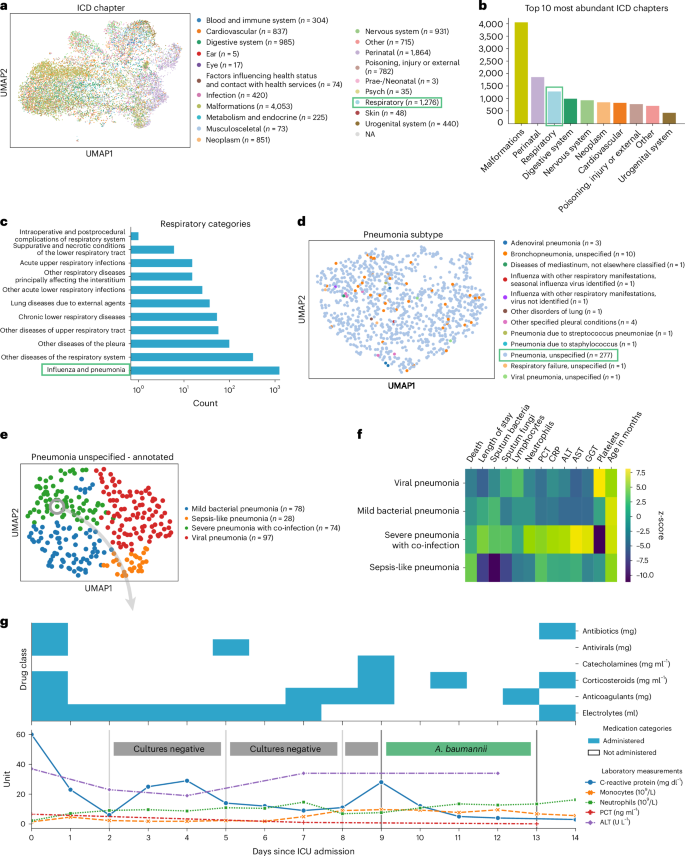
a , UMAP of all patient visits in the ICU with primary discharge diagnosis grouped by ICD chapter. b , The prevalence of respiratory diseases prompted us to investigate them further. c , Respiratory categories show the abundance of influenza and pneumonia diagnoses that we investigated more closely. d , We observed the ‘unspecified pneumonia’ subgroup, which led us to investigate and annotate it in more detail. e , The previously ‘unspecified pneumonia’-labeled patients were annotated using several clinical features (Extended Data Fig. 5 ), of which the most important ones are shown in the heatmap ( f ). g , Example disease progression of an individual child with pneumonia illustrating pharmacotherapy over time until positive A. baumannii swab.
We selected the age group ‘youths’ (13 months to 18 years of age) for further analysis, addressing a total of 265 patients who dominated the pneumonia cases and were diagnosed with ‘unspecified pneumonia’ (Fig. 2d and Extended Data Fig. 4 ). Neonates (0–28 d old) and infants (29 d to 12 months old) were excluded from the analysis as the disease context is significantly different in these age groups due to distinct anatomical and physical conditions. Patients were 61% male, had a total of 277 admissions, had a mean age at admission of 54 months (median, 38 months) and had an average LOS of 15 d (median, 7 d). Of these, 152 patients were admitted to the pediatric intensive care unit (PICU), 118 to the general ICU (GICU), four to the surgical ICU (SICU) and three to the cardiac ICU (CICU). Laboratory measurements typically had 12–14% missing data, except for serum procalcitonin (PCT), a marker for bacterial infections, with 24.5% missing, and C-reactive protein (CRP), a marker of inflammation, with 16.8% missing. Measurements assigned as ‘vital signs’ contained between 44% and 54% missing values. Stratifying patients with unspecified pneumonia further enables a more nuanced understanding of the disease, potentially facilitating tailored approaches to treatment.
To deepen clinical phenotyping for the disease group ‘unspecified pneumonia’, we calculated a k -nearest neighbor graph to cluster patients into groups and visualize these in UMAP space ( Methods ). Leiden clustering 62 identified four patient groupings with distinct clinical features that we annotated (Fig. 2e ). To identify the laboratory values, medications and pathogens that were most characteristic for these four groups (Fig. 2f ), we applied t -tests for numerical data and g -tests for categorical data between the identified groups using ehrapy (Extended Data Fig. 5 and Methods ). Based on this analysis, we identified patient groups with ‘sepsis-like, ‘severe pneumonia with co-infection’, ‘viral pneumonia’ and ‘mild pneumonia’ phenotypes. The ‘sepsis-like’ group of patients ( n = 28) was characterized by rapid disease progression as exemplified by an increased number of deaths (adjusted P ≤ 5.04 × 10 −3 , 43% ( n = 28), 95% confidence interval (CI): 23%, 62%); indication of multiple organ failure, such as elevated creatinine (adjusted P ≤ 0.01, 52.74 ± 23.71 μmol L −1 ) or reduced albumin levels (adjusted P ≤ 2.89 × 10 −4 , 33.40 ± 6.78 g L −1 ); and increased expression levels and peaks of inflammation markers, including PCT (adjusted P ≤ 3.01 × 10 −2 , 1.42 ± 2.03 ng ml −1 ), whole blood cell count, neutrophils, lymphocytes, monocytes and lower platelet counts (adjusted P ≤ 6.3 × 10 −2 , 159.30 ± 142.00 × 10 9 per liter) and changes in electrolyte levels—that is, lower potassium levels (adjusted P ≤ 0.09 × 10 −2 , 3.14 ± 0.54 mmol L −1 ). Patients whom we associated with the term ‘severe pneumonia with co-infection’ ( n = 74) were characterized by prolonged ICU stays (adjusted P ≤ 3.59 × 10 −4 , 15.01 ± 29.24 d); organ affection, such as higher levels of creatinine (adjusted P ≤ 1.10 × 10 −4 , 52.74 ± 23.71 μmol L −1 ) and lower platelet count (adjusted P ≤ 5.40 × 10 −23 , 159.30 ± 142.00 × 10 9 per liter); increased inflammation markers, such as peaks of PCT (adjusted P ≤ 5.06 × 10 −5 , 1.42 ± 2.03 ng ml −1 ), CRP (adjusted P ≤ 1.40 × 10 −6 , 50.60 ± 37.58 mg L −1 ) and neutrophils (adjusted P ≤ 8.51 × 10 −6 , 13.01 ± 6.98 × 10 9 per liter); detection of bacteria in combination with additional pathogen fungals in sputum samples (adjusted P ≤ 1.67 × 10 −2 , 26% ( n = 74), 95% CI: 16%, 36%); and increased application of medication, including antifungals (adjusted P ≤ 1.30 × 10 −4 , 15% ( n = 74), 95% CI: 7%, 23%) and catecholamines (adjusted P ≤ 2.0 × 10 −2 , 45% ( n = 74), 95% CI: 33%, 56%). Patients in the ‘mild pneumonia’ group were characterized by positive sputum cultures in the presence of relatively lower inflammation markers, such as PCT (adjusted P ≤ 1.63 × 10 −3 , 1.42 ± 2.03 ng ml −1 ) and CRP (adjusted P ≤ 0.03 × 10 −1 , 50.60 ± 37.58 mg L −1 ), while receiving antibiotics more frequently (adjusted P ≤ 1.00 × 10 −5 , 80% ( n = 78), 95% CI: 70%, 89%) and additional medications (electrolytes, blood thinners and circulation-supporting medications) (adjusted P ≤ 1.00 × 10 −5 , 82% ( n = 78), 95% CI: 73%, 91%). Finally, patients in the ‘viral pneumonia’ group were characterized by shorter LOSs (adjusted P ≤ 8.00 × 10 −6 , 15.01 ± 29.24 d), a lack of non-viral pathogen detection in combination with higher lymphocyte counts (adjusted P ≤ 0.01, 4.11 ± 2.49 × 10 9 per liter), lower levels of PCT (adjusted P ≤ 0.03 × 10 −2 , 1.42 ± 2.03 ng ml −1 ) and reduced application of catecholamines (adjusted P ≤ 5.96 × 10 −7 , 15% (n = 97), 95% CI: 8%, 23%), antibiotics (adjusted P ≤ 8.53 × 10 −6 , 41% ( n = 97), 95% CI: 31%, 51%) and antifungals (adjusted P ≤ 5.96 × 10 −7 , 0% ( n = 97), 95% CI: 0%, 0%).
To demonstrate the ability of ehrapy to examine EHR data from different levels of resolution, we additionally reconstructed a case from the ‘severe pneumonia with co-infection’ group (Fig. 2g ). In this case, the analysis revealed that CRP levels remained elevated despite broad-spectrum antibiotic treatment until a positive Acinetobacter baumannii result led to a change in medication and a subsequent decrease in CRP and monocyte levels.
ehrapy facilitates extraction of pneumonia indicators
ehrapy’s survival analysis module allowed us to identify clinical indicators of disease stages that could be used as biomarkers through Kaplan–Meier analysis. We found strong variance in overall aspartate aminotransferase (AST), alanine aminotransferase (ALT), gamma-glutamyl transferase (GGT) and bilirubin levels (Fig. 3a ), including changes over time (Extended Data Fig. 6a,b ), in all four ‘unspecified pneumonia’ groups. Routinely used to assess liver function, studies provide evidence that AST, ALT and GGT levels are elevated during respiratory infections 63 , including severe pneumonia 64 , and can guide diagnosis and management of pneumonia in children 63 . We confirmed reduced survival in more severely affected children (‘sepsis-like pneumonia’ and ‘severe pneumonia with co-infection’) using Kaplan–Meier curves and a multivariate log-rank test (Fig. 3b ; P ≤ 1.09 × 10 −18 ) through ehrapy. To verify the association of this trajectory with altered AST, ALT and GGT expression levels, we further grouped all patients based on liver enzyme reference ranges ( Methods and Supplementary Table 2 ). By Kaplan–Meier survival analysis, cases with peaks of GGT ( P ≤ 1.4 × 10 −2 , 58.01 ± 2.03 U L −1 ), ALT ( P ≤ 2.9 × 10 −2 , 43.59 ± 38.02 U L −1 ) and AST ( P ≤ 4.8 × 10 −4 , 78.69 ± 60.03 U L −1 ) in ‘outside the norm’ were found to correlate with lower survival in all groups (Fig. 3c and Extended Data Fig. 6 ), in line with previous studies 63 , 65 . Bilirubin was not found to significantly affect survival ( P ≤ 2.1 × 10 −1 , 12.57 ± 21.22 mg dl −1 ).
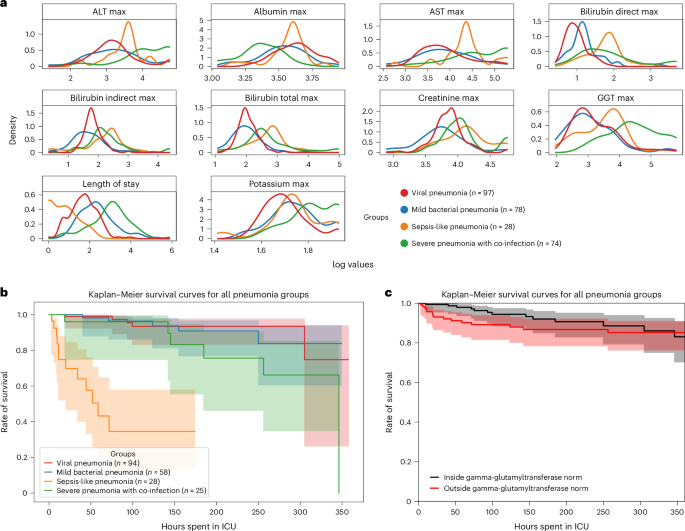
a , Line plots of major hepatic system laboratory measurements per group show variance in the measurements per pneumonia group. b , Kaplan–Meier survival curves demonstrate lower survival for ‘sepsis-like’ and ‘severe pneumonia with co-infection’ groups. c , Kaplan–Meier survival curves for children with GGT measurements outside the norm range display lower survival.

ehrapy quantifies medication class effect on LOS
Pneumonia requires case-specific medications due to its diverse causes. To demonstrate the potential of ehrapy’s causal inference module, we quantified the effect of medication on ICU LOS to evaluate case-specific administration of medication. In contrast to causal discovery that attempts to find a causal graph reflecting the causal relationships, causal inference is a statistical process used to investigate possible effects when altering a provided system, as represented by a causal graph and observational data (Fig. 4a ) 66 . This approach allows identifying and quantifying the impact of specific interventions or treatments on outcome measures, thereby providing insight for evidence-based decision-making in healthcare. Causal inference relies on datasets incorporating interventions to accurately quantify effects.
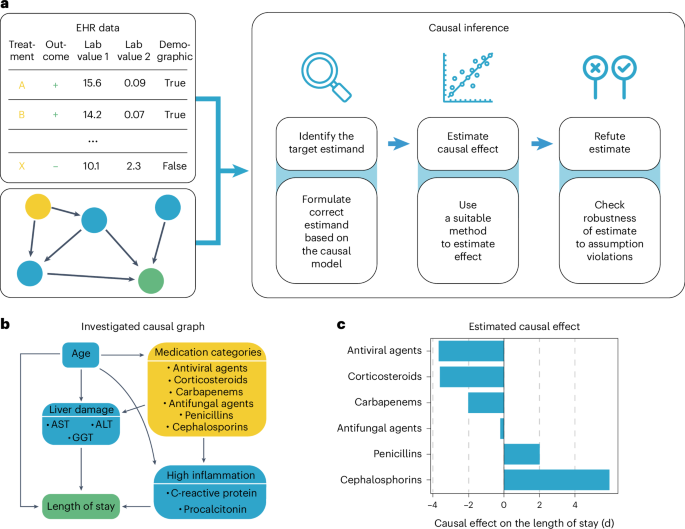
a , ehrapy’s causal module is based on the strategy of the tool ‘dowhy’. Here, EHR data containing treatment, outcome and measurements and a causal graph serve as input for causal effect quantification. The process includes the identification of the target estimand based on the causal graph, the estimation of causal effects using various models and, finally, refutation where sensitivity analyses and refutation tests are performed to assess the robustness of the results and assumptions. b , Curated causal graph using age, liver damage and inflammation markers as disease progression proxies together with medications as interventions to assess the causal effect on length of ICU stay. c , Determined causal effect strength on LOS in days of administered medication categories.
We manually constructed a minimal causal graph with ehrapy (Fig. 4b ) on records of treatment with corticosteroids, carbapenems, penicillins, cephalosporins and antifungal and antiviral medications as interventions (Extended Data Fig. 7 and Methods ). We assumed that the medications affect disease progression proxies, such as inflammation markers and markers of organ function. The selection of ‘interventions’ is consistent with current treatment standards for bacterial pneumonia and respiratory distress 67 , 68 . Based on the approach of the tool ‘dowhy’ 69 (Fig. 4a ), ehrapy’s causal module identified the application of corticosteroids, antivirals and carbapenems to be associated with shorter LOSs, in line with current evidence 61 , 70 , 71 , 72 . In contrast, penicillins and cephalosporins were associated with longer LOSs, whereas antifungal medication did not strongly influence LOS (Fig. 4c ).
ehrapy enables deriving population-scale risk factors
To illustrate the advantages of using a unified data management and quality control framework, such as ehrapy, we modeled myocardial infarction risk using Cox proportional hazards models on UKB 44 data. Large population cohort studies, such as the UKB, enable the investigation of common diseases across a wide range of modalities, including genomics, metabolomics, proteomics, imaging data and common clinical variables (Fig. 5a,b ). From these, we used a publicly available polygenic risk score for coronary heart disease 73 comprising 6.6 million variants, 80 nuclear magnetic resonance (NMR) spectroscopy-based metabolomics 74 features, 81 features derived from retinal optical coherence tomography 75 , 76 and the Framingham Risk Score 77 feature set, which includes known clinical predictors, such as age, sex, body mass index, blood pressure, smoking behavior and cholesterol levels. We excluded features with more than 10% missingness and imputed the remaining missing values ( Methods ). Furthermore, individuals with events up to 1 year after the sampling time were excluded from the analyses, ultimately selecting 29,216 individuals for whom all mentioned data types were available (Extended Data Figs. 8 and 9 and Methods ). Myocardial infarction, as defined by our mapping to the phecode nomenclature 51 , was defined as the endpoint (Fig. 5c ). We modeled the risk for myocardial infarction 1 year after either the metabolomic sample was obtained or imaging was performed.
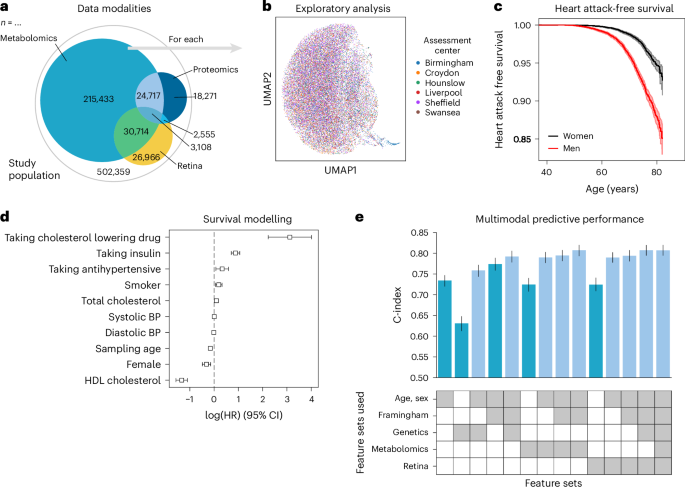
a , The UKB includes 502,359 participants from 22 assessment centers. Most participants have genetic data (97%) and physical measurement data (93%), but fewer have data for complex measures, such as metabolomics, retinal imaging or proteomics. b , We found a distinct cluster of individuals (bottom right) from the Birmingham assessment center in the retinal imaging data, which is an artifact of the image acquisition process and was, thus, excluded. c , Myocardial infarctions are recorded for 15% of the male and 7% of the female study population. Kaplan–Meier estimators with 95% CIs are shown. d , For every modality combination, a linear Cox proportional hazards model was fit to determine the prognostic potential of these for myocardial infarction. Cardiovascular risk factors show expected positive log hazard ratios (log (HRs)) for increased blood pressure or total cholesterol and negative ones for sampling age and systolic blood pressure (BP). log (HRs) with 95% CIs are shown. e , Combining all features yields a C-index of 0.81. c – e , Error bars indicate 95% CIs ( n = 29,216).
Predictive performance for each modality was assessed by fitting Cox proportional hazards (Fig. 5c ) models on each of the feature sets using ehrapy (Fig. 5d ). The age of the first occurrence served as the time to event; alternatively, date of death or date of the last record in the EHR served as censoring times. Models were evaluated using the concordance index (C-index) ( Methods ). The combination of multiple modalities successfully improved the predictive performance for coronary heart disease by increasing the C-index from 0.63 (genetic) to 0.76 (genetics, age and sex) and to 0.77 (clinical predictors) with 0.81 (imaging and clinical predictors) for combinations of feature sets (Fig. 5e ). Our finding is in line with previous observations of complementary effects between different modalities, where a broader ‘major adverse cardiac event’ phenotype was modeled in the UKB achieving a C-index of 0.72 (ref. 78 ). Adding genetic data improves predictive potential, as it is independent of sampling age and has limited prediction of other modalities 79 . The addition of metabolomic data did not improve predictive power (Fig. 5e ).
Imaging-based disease severity projection via fate mapping
To demonstrate ehrapy’s ability to handle diverse image data and recover disease stages, we embedded pulmonary imaging data obtained from patients with COVID-19 into a lower-dimensional space and computationally inferred disease progression trajectories using pseudotemporal ordering. This describes a continuous trajectory or ordering of individual points based on feature similarity 80 . Continuous trajectories enable mapping the fate of new patients onto precise states to potentially predict their future condition.
In COVID-19, a highly contagious respiratory illness caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), symptoms range from mild flu-like symptoms to severe respiratory distress. Chest x-rays typically show opacities (bilateral patchy, ground glass) associated with disease severity 81 .
We used COVID-19 chest x-ray images from the BrixIA 82 dataset consisting of 192 images (Fig. 6a ) with expert annotations of disease severity. We used the BrixIA database scores, which are based on six regions annotated by radiologists, to classify disease severity ( Methods ). We embedded raw image features using a pre-trained DenseNet model ( Methods ) and further processed this embedding into a nearest-neighbors-based UMAP space using ehrapy (Fig. 6b and Methods ). Fate mapping based on imaging information ( Methods ) determined a severity ordering from mild to critical cases (Fig. 6b–d ). Images labeled as ‘normal’ are projected to stay within the healthy group, illustrating the robustness of our approach. Images of diseased patients were ordered by disease severity, highlighting clear trajectories from ‘normal’ to ‘critical’ states despite the heterogeneity of the x-ray images stemming from, for example, different zoom levels (Fig. 6a ).
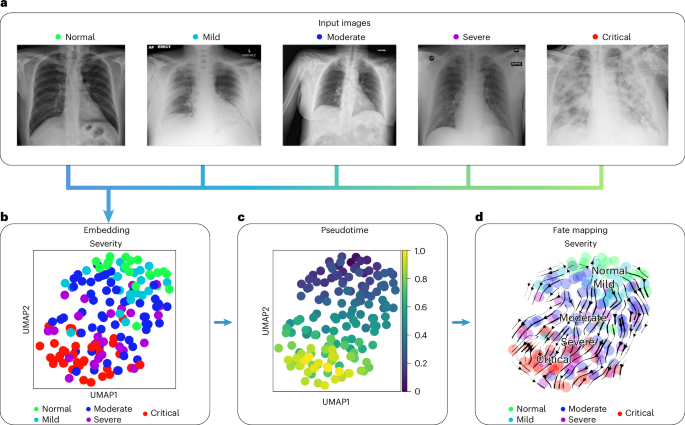
a , Randomly selected chest x-ray images from the BrixIA dataset demonstrate its variance. b , UMAP visualization of the BrixIA dataset embedding shows a separation of disease severity classes. c , Calculated pseudotime for all images increases with distance to the ‘normal’ images. d , Stream projection of fate mapping in UMAP space showcases disease severity trajectory of the COVID-19 chest x-ray images.
Detecting and mitigating biases in EHR data with ehrapy
To showcase how exploratory analysis using ehrapy can reveal and mitigate biases, we analyzed the Fairlearn 83 version of the Diabetes 130-US Hospitals 84 dataset. The dataset covers 10 years (1999–2008) of clinical records from 130 US hospitals, detailing 47 features of diabetes diagnoses, laboratory tests, medications and additional data from up to 14 d of inpatient care of 101,766 diagnosed patient visits ( Methods ). It was originally collected to explore the link between the measurement of hemoglobin A1c (HbA1c) and early readmission.
The cohort primarily consists of White and African American individuals, with only a minority of cases from Asian or Hispanic backgrounds (Extended Data Fig. 10a ). ehrapy’s cohort tracker unveiled selection and surveillance biases when filtering for Medicare recipients for further analysis, resulting in a shift of age distribution toward an age of over 60 years in addition to an increasing ratio of White participants. Using ehrapy’s visualization modules, our analysis showed that HbA1c was measured in only 18.4% of inpatients, with a higher frequency in emergency admissions compared to referral cases (Extended Data Fig. 10b ). Normalization biases can skew data relationships when standardization techniques ignore subgroup variability or assume incorrect distributions. The choice of normalization strategy must be carefully considered to avoid obscuring important factors. When normalizing the number of applied medications individually, differences in distributions between age groups remained. However, when normalizing both distributions jointly with age group as an additional group variable, differences between age groups were masked (Extended Data Fig. 10c ). To investigate missing data and imputation biases, we introduced missingness for the number of applied medications according to an MCAR mechanism, which we verified using ehrapy’s Little’s test ( P ≤ 0.01 × 10 −2 ), and an MAR mechanism ( Methods ). Whereas imputing the mean in the MCAR case did not affect the overall location of the distribution, it led to an underestimation of the variance, with the standard deviation dropping from 8.1 in the original data to 6.8 in the imputed data (Extended Data Fig. 10d ). Mean imputation in the MAR case skewed both location and variance of the mean from 16.02 to 14.66, with a standard deviation of only 5.72 (Extended Data Fig. 10d ). Using ehrapy’s multiple imputation based MissForest 85 imputation on the MAR data resulted in a mean of 16.04 and a standard deviation of 6.45. To predict patient readmission in fewer than 30 d, we merged the three smallest race groups, ‘Asian’, ‘Hispanic’ and ‘Other’. Furthermore, we dropped the gender group ‘Unknown/Invalid’ owing to the small sample size making meaningful assessment impossible, and we performed balanced random undersampling, resulting in 5,677 cases from each condition. We observed an overall balanced accuracy of 0.59 using a logistic regression model. However, the false-negative rate was highest for the races ‘Other’ and ‘Unknown’, whereas their selection rate was lowest, and this model was, therefore, biased (Extended Data Fig. 10e ). Using ehrapy’s compatibility with existing machine learning packages, we used Fairlearn’s ThresholdOptimizer ( Methods ), which improved the selection rates for ‘Other’ from 0.32 to 0.38 and for ‘Unknown’ from 0.23 to 0.42 and the false-negative rates for ‘Other’ from 0.48 to 0.42 and for ‘Unknown’ from 0.61 to 0.45 (Extended Data Fig. 10e ).
Clustering offers a hypothesis-free alternative to supervised classification when clear hypotheses or labels are missing. It has enabled the identification of heart failure subtypes 86 and progression pathways 87 and COVID-19 severity states 88 . This concept, which is central to ehrapy, further allowed us to identify fine-grained groups of ‘unspecified pneumonia’ cases in the PIC dataset while discovering biomarkers and quantifying effects of medications on LOS. Such retroactive characterization showcases ehrapy’s ability to put complex evidence into context. This approach supports feedback loops to improve diagnostic and therapeutic strategies, leading to more efficiently allocated resources in healthcare.
ehrapy’s flexible data structures enabled us to integrate the heterogeneous UKB data for predictive performance in myocardial infarction. The different data types and distributions posed a challenge for predictive models that were overcome with ehrapy’s pre-processing modules. Our analysis underscores the potential of combining phenotypic and health data at population scale through ehrapy to enhance risk prediction.
By adapting pseudotime approaches that are commonly used in other omics domains, we successfully recovered disease trajectories from raw imaging data with ehrapy. The determined pseudotime, however, only orders data but does not necessarily provide a future projection per patient. Understanding the driver features for fate mapping in image-based datasets is challenging. The incorporation of image segmentation approaches could mitigate this issue and provide a deeper insight into the spatial and temporal dynamics of disease-related processes.
Limitations of our analyses include the lack of control for informative missingness where the absence of information represents information in itself 89 . Translation from Chinese to English in the PIC database can cause information loss and inaccuracies because the Chinese ICD-10 codes are seven characters long compared to the five-character English codes. Incompleteness of databases, such as the lack of radiology images in the PIC database, low sample sizes, underrepresentation of non-White ancestries and participant self-selection, cannot be accounted for and limit generalizability. This restricts deeper phenotyping of, for example, all ‘unspecified pneumonia’ cases with respect to their survival, which could be overcome by the use of multiple databases. Our causal inference use case is limited by unrecorded variables, such as Sequential Organ Failure Assessment (SOFA) scores, and pneumonia-related pathogens that are missing in the causal graph due to dataset constraints, such as high sparsity and substantial missing data, which risk overfitting and can lead to overinterpretation. We counterbalanced this by employing several refutation methods that statistically reject the causal hypothesis, such as a placebo treatment, a random common cause or an unobserved common cause. The longer hospital stays associated with penicillins and cephalosporins may be dataset specific and stem from higher antibiotic resistance, their use as first-line treatments, more severe initial cases, comorbidities and hospital-specific protocols.
Most analysis steps can introduce algorithmic biases where results are misleading or unfavorably affect specific groups. This is particularly relevant in the context of missing data 22 where determining the type of missing data is necessary to handle it correctly. ehrapy includes an implementation of Little’s test 90 , which tests whether data are distributed MCAR to discern missing data types. For MCAR data single-imputation approaches, such as mean, median or mode, imputation can suffice, but these methods are known to reduce variability 91 , 92 . Multiple imputation strategies, such as Multiple Imputation by Chained Equations (MICE) 93 and MissForest 85 , as implemented in ehrapy, are effective for both MCAR and MAR data 22 , 94 , 95 . MNAR data require pattern-mixture or shared-parameter models that explicitly incorporate the mechanism by which data are missing 96 . Because MNAR involves unobserved data, the assumptions about the missingness mechanism cannot be directly verified, making sensitivity analysis crucial 21 . ehrapy’s wide range of normalization functions and grouping functionality enables to account for intrinsic variability within subgroups, and its compatibility with Fairlearn 83 can potentially mitigate predictor biases. Generally, we recommend to assess all pre-processing in an iterative manner with respect to downstream applications, such as patient stratification. Moreover, sensitivity analysis can help verify the robustness of all inferred knowledge 97 .
These diverse use cases illustrate ehrapy’s potential to sufficiently address the need for a computationally efficient, extendable, reproducible and easy-to-use framework. ehrapy is compatible with major standards, such as Observational Medical Outcomes Partnership (OMOP), Common Data Model (CDM) 47 , HL7, FHIR or openEHR, with flexible support for common tabular data formats. Once loaded into an AnnData object, subsequent sharing of analysis results is made easy because AnnData objects can be stored and read platform independently. ehrapy’s rich documentation of the application programming interface (API) and extensive hands-on tutorials make EHR analysis accessible to both novices and experienced analysts.
As ehrapy remains under active development, users can expect ehrapy to continuously evolve. We are improving support for the joint analysis of EHR, genetics and molecular data where ehrapy serves as a bridge between the EHR and the omics communities. We further anticipate the generation of EHR-specific reference datasets, so-called atlases 98 , to enable query-to-reference mapping where new datasets get contextualized by transferring annotations from the reference to the new dataset. To promote the sharing and collective analysis of EHR data, we envision adapted versions of interactive single-cell data explorers, such as CELLxGENE 99 or the UCSC Cell Browser 100 , for EHR data. Such web interfaces would also include disparity dashboards 20 to unveil trends of preferential outcomes for distinct patient groups. Additional modules specifically for high-frequency time-series data, natural language processing and other data types are currently under development. With the widespread availability of code-generating large language models, frameworks such as ehrapy are becoming accessible to medical professionals without coding expertise who can leverage its analytical power directly. Therefore, ehrapy, together with a lively ecosystem of packages, has the potential to enhance the scientific discovery pipeline to shape the era of EHR analysis.
All datasets that were used during the development of ehrapy and the use cases were used according to their terms of use as indicated by each provider.
Design and implementation of ehrapy
A unified pipeline as provided by our ehrapy framework streamlines the analysis of EHR data by providing an efficient, standardized approach, which reduces the complexity and variability in data pre-processing and analysis. This consistency ensures reproducibility of results and facilitates collaboration and sharing within the research community. Additionally, the modular structure allows for easy extension and customization, enabling researchers to adapt the pipeline to their specific needs while building on a solid foundational framework.
ehrapy was designed from the ground up as an open-source effort with community support. The package, as well as all associated tutorials and dataset preparation scripts, are open source. Development takes place publicly on GitHub where the developers discuss feature requests and issues directly with users. This tight interaction between both groups ensures that we implement the most pressing needs to cater the most important use cases and can guide users when difficulties arise. The open-source nature, extensive documentation and modular structure of ehrapy are designed for other developers to build upon and extend ehrapy’s functionality where necessary. This allows us to focus ehrapy on the most important features to keep the number of dependencies to a minimum.
ehrapy was implemented in the Python programming language and builds upon numerous existing numerical and scientific open-source libraries, specifically matplotlib 101 , seaborn 102 , NumPy 103 , numba 104 , Scipy 105 , scikit-learn 53 and Pandas 106 . Although taking considerable advantage of all packages implemented, ehrapy also shares the limitations of these libraries, such as a lack of GPU support or small performance losses due to the translation layer cost for operations between the Python interpreter and the lower-level C language for matrix operations. However, by building on very widely used open-source software, we ensure seamless integration and compatibility with a broad range of tools and platforms to promote community contributions. Additionally, by doing so, we enhance security by allowing a larger pool of developers to identify and address vulnerabilities 107 . All functions are grouped into task-specific modules whose implementation is complemented with additional dependencies.
Data preparation
Dataloaders.
ehrapy is compatible with any type of vectorized data, where vectorized refers to the data being stored in structured tables in either on-disk or database form. The input and output module of ehrapy provides readers for common formats, such as OMOP, CSV tables or SQL databases through Pandas. When reading in such datasets, the data are stored in the appropriate slots in a new AnnData 46 object. ehrapy’s data module provides access to more than 20 public EHR datasets that feature diseases, including, but not limited to, Parkinson’s disease, breast cancer, chronic kidney disease and more. All dataloaders return AnnData objects to allow for immediate analysis.
AnnData for EHR data
Our framework required a versatile data structure capable of handling various matrix formats, including Numpy 103 for general use cases and interoperability, Scipy 105 sparse matrices for efficient storage, Dask 108 matrices for larger-than-memory analysis and Awkward array 109 for irregular time-series data. We needed a single data structure that not only stores data but also includes comprehensive annotations for thorough contextual analysis. It was essential for this structure to be widely used and supported, which ensures robustness and continual updates. Interoperability with other analytical packages was a key criterion to facilitate seamless integration within existing tools and workflows. Finally, the data structure had to support both in-memory operations and on-disk storage using formats such as HDF5 (ref. 110 ) and Zarr 111 , ensuring efficient handling and accessibility of large datasets and the ability to easily share them with collaborators.
All of these requirements are fulfilled by the AnnData format, which is a popular data structure in single-cell genomics. At its core, an AnnData object encapsulates diverse components, providing a holistic representation of data and metadata that are always aligned in dimensions and easily accessible. A data matrix (commonly referred to as ‘ X ’) stands as the foundational element, embodying the measured data. This matrix can be dense (as Numpy array), sparse (as Scipy sparse matrix) or ragged (as Awkward array) where dimensions do not align within the data matrix. The AnnData object can feature several such data matrices stored in ‘layers’. Examples of such layers can be unnormalized or unencoded data. These data matrices are complemented by an observations (commonly referred to as ‘obs’) segment where annotations on the level of patients or visits are stored. Patients’ age or sex, for instance, are often used as such annotations. The variables (commonly referred to as ‘var’) section complements the observations, offering supplementary details about the features in the dataset, such as missing data rates. The observation-specific matrices (commonly referred to as ‘obsm’) section extends the capabilities of the AnnData structure by allowing the incorporation of observation-specific matrices. These matrices can represent various types of information at the individual cell level, such as principal component analysis (PCA) results, t-distributed stochastic neighbor embedding (t-SNE) coordinates or other dimensionality reduction outputs. Analogously, AnnData features a variables-specific variables (commonly referred to as ‘varm’) component. The observation-specific pairwise relationships (commonly referred to as ‘obsp’) segment complements the ‘obsm’ section by accommodating observation-specific pairwise relationships. This can include connectivity matrices, indicating relationships between patients. The inclusion of an unstructured annotations (commonly referred to as ‘uns’) component further enhances flexibility. This segment accommodates unstructured annotations or arbitrary data that might not conform to the structured observations or variables categories. Any AnnData object can be stored on disk in h5ad or Zarr format to facilitate data exchange.
ehrapy natively interfaces with the scientific Python ecosystem via Pandas 112 and Numpy 103 . The development of deep learning models for EHR data 113 is further accelerated through compatibility with pathml 114 , a unified framework for whole-slide image analysis in pathology, and scvi-tools 115 , which provides data loaders for loading tensors from AnnData objects into PyTorch 116 or Jax arrays 117 to facilitate the development of generalizing foundational models for medical artificial intelligence 118 .
Feature annotation
After AnnData creation, any metadata can be mapped against ontologies using Bionty ( https://github.com/laminlabs/bionty-base ). Bionty provides access to the Human Phenotype, Phecodes, Phenotype and Trait, Drug, Mondo and Human Disease ontologies.
Key medical terms stored in an AnnData object in free text can be extracted using the Medical Concept Annotation Toolkit (MedCAT) 119 .
Data processing
Cohort tracking.
ehrapy provides a CohortTracker tool that traces all filtering steps applied to an associated AnnData object. To calculate cohort summary statistics, the implementation makes use of tableone 120 and can subsequently be plotted as bar charts together with flow diagrams 121 that visualize the order and reasoning of filtering operations.
Basic pre-processing and quality control
ehrapy encompasses a suite of functionalities for fundamental data processing that are adopted from scanpy 52 but adapted to EHR data:
Regress out: To address unwanted sources of variation, a regression procedure is integrated, enhancing the dataset’s robustness.
Subsample: Selects a specified fraction of observations.
Balanced sample: Balances groups in the dataset by random oversampling or undersampling.
Highly variable features: The identification and annotation of highly variable features following the ‘highly variable genes’ function of scanpy is seamlessly incorporated, providing users with insights into pivotal elements influencing the dataset.
To identify and minimize quality issues, ehrapy provides several quality control functions:
Basic quality control: Determines the relative and absolute number of missing values per feature and per patient.
Winsorization: For data refinement, ehrapy implements a winsorization process, creating a version of the input array less susceptible to extreme values.
Feature clipping: Imposes limits on features to enhance dataset reliability.
Detect biases: Computes pairwise correlations between features, standardized mean differences for numeric features between groups of sensitive features, categorical feature value count differences between groups of sensitive features and feature importances when predicting a target variable.
Little’s MCAR test: Applies Little’s MCAR test whose null hypothesis is that data are MCAR. Rejecting the null hypothesis may not always mean that data are not MCAR, nor is accepting the null hypothesis a guarantee that data are MCAR. For more details, see Schouten et al. 122 .
Summarize features: Calculates statistical indicators per feature, including minimum, maximum and average values. This can be especially useful to reduce complex data with multiple measurements per feature per patient into sets of columns with single values.
Imputation is crucial in data analysis to address missing values, ensuring the completeness of datasets that can be required for specific algorithms. The ‘ehrapy’ pre-processing module offers a range of imputation techniques:
Explicit Impute: Replaces missing values, in either all columns or a user-specified subset, with a designated replacement value.
Simple Impute: Imputes missing values in numerical data using mean, median or the most frequent value, contributing to a more complete dataset.
KNN Impute: Uses k -nearest neighbor imputation to fill in missing values in the input AnnData object, preserving local data patterns.
MissForest Impute: Implements the MissForest strategy for imputing missing data, providing a robust approach for handling complex datasets.
MICE Impute: Applies the MICE algorithm for imputing data. This implementation is based on the miceforest ( https://github.com/AnotherSamWilson/miceforest ) package.
Data encoding can be required if categoricals are a part of the dataset to obtain numerical values only. Most algorithms in ehrapy are compatible only with numerical values. ehrapy offers two encoding algorithms based on scikit-learn 53 :
One-Hot Encoding: Transforms categorical variables into binary vectors, creating a binary feature for each category and capturing the presence or absence of each category in a concise representation.
Label Encoding: Assigns a unique numerical label to each category, facilitating the representation of categorical data as ordinal values and supporting algorithms that require numerical input.
To ensure that the distributions of the heterogeneous data are aligned, ehrapy offers several normalization procedures:
Log Normalization: Applies the natural logarithm function to the data, useful for handling skewed distributions and reducing the impact of outliers.
Max-Abs Normalization: Scales each feature by its maximum absolute value, ensuring that the maximum absolute value for each feature is 1.
Min-Max Normalization: Transforms the data to a specific range (commonly (0, 1)) by scaling each feature based on its minimum and maximum values.
Power Transformation Normalization: Applies a power transformation to make the data more Gaussian like, often useful for stabilizing variance and improving the performance of models sensitive to distributional assumptions.
Quantile Normalization: Aligns the distributions of multiple variables, ensuring that their quantiles match, which can be beneficial for comparing datasets or removing batch effects.
Robust Scaling Normalization: Scales data using the interquartile range, making it robust to outliers and suitable for datasets with extreme values.
Scaling Normalization: Standardizes data by subtracting the mean and dividing by the standard deviation, creating a distribution with a mean of 0 and a standard deviation of 1.
Offset to Positive Values: Shifts all values by a constant offset to make all values non-negative, with the lowest negative value becoming 0.
Dataset shifts can be corrected using the scanpy implementation of the ComBat 123 algorithm, which employs a parametric and non-parametric empirical Bayes framework for adjusting data for batch effects that is robust to outliers.
Finally, a neighbors graph can be efficiently computed using scanpy’s implementation.
To obtain meaningful lower-dimensional embeddings that can subsequently be visualized and reused for downstream algorithms, ehrapy provides the following algorithms based on scanpy’s implementation:
t-SNE: Uses a probabilistic approach to embed high-dimensional data into a lower-dimensional space, emphasizing the preservation of local similarities and revealing clusters in the data.
UMAP: Embeds data points by modeling their local neighborhood relationships, offering an efficient and scalable technique that captures both global and local structures in high-dimensional data.
Force-Directed Graph Drawing: Uses a physical simulation to position nodes in a graph, with edges representing pairwise relationships, creating a visually meaningful representation that emphasizes connectedness and clustering in the data.
Diffusion Maps: Applies spectral methods to capture the intrinsic geometry of high-dimensional data by modeling diffusion processes, providing a way to uncover underlying structures and patterns.
Density Calculation in Embedding: Quantifies the density of observations within an embedding, considering conditions or groups, offering insights into the concentration of data points in different regions and aiding in the identification of densely populated areas.
ehrapy further provides algorithms for clustering and trajectory inference based on scanpy:
Leiden Clustering: Uses the Leiden algorithm to cluster observations into groups, revealing distinct communities within the dataset with an emphasis on intra-cluster cohesion.
Hierarchical Clustering Dendrogram: Constructs a dendrogram through hierarchical clustering based on specified group by categories, illustrating the hierarchical relationships among observations and facilitating the exploration of structured patterns.
Feature ranking
ehrapy provides two ways of ranking feature contributions to clusters and target variables:
Statistical tests: To compare any obtained clusters to obtain marker features that are significantly different between the groups, ehrapy extends scanpy’s ‘rank genes groups’. The original implementation, which features a t -test for numerical data, is complemented by a g -test for categorical data.
Feature importance: Calculates feature rankings for a target variable using linear regression, support vector machine or random forest models from scikit-learn. ehrapy evaluates the relative importance of each predictor by fitting the model and extracting model-specific metrics, such as coefficients or feature importances.
Dataset integration
Based on scanpy’s ‘ingest’ function, ehrapy facilitates the integration of labels and embeddings from a well-annotated reference dataset into a new dataset, enabling the mapping of cluster annotations and spatial relationships for consistent comparative analysis. This process ensures harmonized clinical interpretations across datasets, especially useful when dealing with multiple experimental diseases or batches.
Knowledge inference
Survival analysis.
ehrapy’s implementation of survival analysis algorithms is based on lifelines 124 :
Ordinary Least Squares (OLS) Model: Creates a linear regression model using OLS from a specified formula and an AnnData object, allowing for the analysis of relationships between variables and observations.
Generalized Linear Model (GLM): Constructs a GLM from a given formula, distribution and AnnData, providing a versatile framework for modeling relationships with nonlinear data structures.
Kaplan–Meier: Fits the Kaplan–Meier curve to generate survival curves, offering a visual representation of the probability of survival over time in a dataset.
Cox Hazard Model: Constructs a Cox proportional hazards model using a specified formula and an AnnData object, enabling the analysis of survival data by modeling the hazard rates and their relationship to predictor variables.
Log-Rank Test: Calculates the P value for the log-rank test, comparing the survival functions of two groups, providing statistical significance for differences in survival distributions.
GLM Comparison: Given two fit GLMs, where the larger encompasses the parameter space of the smaller, this function returns the P value, indicating the significance of the larger model and adding explanatory power beyond the smaller model.
Trajectory inference
Trajectory inference is a computational approach that reconstructs and models the developmental paths and transitions within heterogeneous clinical data, providing insights into the temporal progression underlying complex systems. ehrapy offers several inbuilt algorithms for trajectory inference based on scanpy:
Diffusion Pseudotime: Infers the progression of observations by measuring geodesic distance along the graph, providing a pseudotime metric that represents the developmental trajectory within the dataset.
Partition-based Graph Abstraction (PAGA): Maps out the coarse-grained connectivity structures of complex manifolds using a partition-based approach, offering a comprehensive visualization of relationships in high-dimensional data and aiding in the identification of macroscopic connectivity patterns.
Because ehrapy is compatible with scverse, further trajectory inference-based algorithms, such as CellRank, can be seamlessly applied.
Causal inference
ehrapy’s causal inference module is based on ‘dowhy’ 69 . It is based on four key steps that are all implemented in ehrapy:
Graphical Model Specification: Define a causal graphical model representing relationships between variables and potential causal effects.
Causal Effect Identification: Automatically identify whether a causal effect can be inferred from the given data, addressing confounding and selection bias.
Causal Effect Estimation: Employ automated tools to estimate causal effects, using methods such as matching, instrumental variables or regression.
Sensitivity Analysis and Testing: Perform sensitivity analysis to assess the robustness of causal inferences and conduct statistical testing to determine the significance of the estimated causal effects.
Patient stratification
ehrapy’s complete pipeline from pre-processing to the generation of lower-dimensional embeddings, clustering, statistical comparison between determined groups and more facilitates the stratification of patients.
Visualization
ehrapy features an extensive visualization pipeline that is customizable and yet offers reasonable defaults. Almost every analysis function is matched with at least one visualization function that often shares the name but is available through the plotting module. For example, after importing ehrapy as ‘ep’, ‘ep.tl.umap(adata)’ runs the UMAP algorithm on an AnnData object, and ‘ep.pl.umap(adata)’ would then plot a scatter plot of the UMAP embedding.
ehrapy further offers a suite of more generally usable and modifiable plots:
Scatter Plot: Visualizes data points along observation or variable axes, offering insights into the distribution and relationships between individual data points.
Heatmap: Represents feature values in a grid, providing a comprehensive overview of the data’s structure and patterns.
Dot Plot: Displays count values of specified variables as dots, offering a clear depiction of the distribution of counts for each variable.
Filled Line Plot: Illustrates trends in data with filled lines, emphasizing variations in values over a specified axis.
Violin Plot: Presents the distribution of data through mirrored density plots, offering a concise view of the data’s spread.
Stacked Violin Plot: Combines multiple violin plots, stacked to allow for visual comparison of distributions across categories.
Group Mean Heatmap: Creates a heatmap displaying the mean count per group for each specified variable, providing insights into group-wise trends.
Hierarchically Clustered Heatmap: Uses hierarchical clustering to arrange data in a heatmap, revealing relationships and patterns among variables and observations.
Rankings Plot: Visualizes rankings within the data, offering a clear representation of the order and magnitude of values.
Dendrogram Plot: Plots a dendrogram of categories defined in a group by operation, illustrating hierarchical relationships within the dataset.
Benchmarking ehrapy
We generated a subset of the UKB data selecting 261 features and 488,170 patient visits. We removed all features with missingness rates greater than 70%. To demonstrate speed and memory consumption for various scenarios, we subsampled the data to 20%, 30% and 50%. We ran a minimal ehrapy analysis pipeline on each of those subsets and the full data, including the calculation of quality control metrics, filtering of variables by a missingness threshold, nearest neighbor imputation, normalization, dimensionality reduction and clustering (Supplementary Table 1 ). We conducted our benchmark on a single CPU with eight threads and 60 GB of maximum memory.
ehrapy further provides out-of-core implementations using Dask 108 for many algorithms in ehrapy, such as our normalization functions or our PCA implementation. Out-of-core computation refers to techniques that process data that do not fit entirely in memory, using disk storage to manage data overflow. This approach is crucial for handling large datasets without being constrained by system memory limits. Because the principal components get reused for other computationally expensive algorithms, such as the neighbors graph calculation, it effectively enables the analysis of very large datasets. We are currently working on supporting out-of-core computation for all computationally expensive algorithms in ehrapy.
We demonstrate the memory benefits in a hosted tutorial where the in-memory pipeline for 50,000 patients with 1,000 features required about 2 GB of memory, and the corresponding out-of-core implementation required less than 200 MB of memory.
The code for benchmarking is available at https://github.com/theislab/ehrapy-reproducibility . The implementation of ehrapy is accessible at https://github.com/theislab/ehrapy together with extensive API documentation and tutorials at https://ehrapy.readthedocs.io .
PIC database analysis
Study design.
We collected clinical data from the PIC 43 version 1.1.0 database. PIC is a single-center, bilingual (English and Chinese) database hosting information of children admitted to critical care units at the Children’s Hospital of Zhejiang University School of Medicine in China. The requirement for individual patient consent was waived because the study did not impact clinical care, and all protected health information was de-identified. The database contains 13,499 distinct hospital admissions of 12,881 distinct pediatric patients. These patients were admitted to five ICU units with 119 total critical care beds—GICU, PICU, SICU, CICU and NICU—between 2010 and 2018. The mean age of the patients was 2.5 years, of whom 42.5% were female. The in-hospital mortality was 7.1%; the mean hospital stay was 17.6 d; the mean ICU stay was 9.3 d; and 468 (3.6%) patients were admitted multiple times. Demographics, diagnoses, doctors’ notes, laboratory and microbiology tests, prescriptions, fluid balances, vital signs and radiographics reports were collected from all patients. For more details, see the original publication of Zeng et al. 43 .
Study participants
Individuals older than 18 years were excluded from the study. We grouped the data into three distinct groups: ‘neonates’ (0–28 d of age; 2,968 patients), ‘infants’ (1–12 months of age; 4,876 patients) and ‘youths’ (13 months to 18 years of age; 6,097 patients). We primarily analyzed the ‘youths’ group with the discharge diagnosis ‘unspecified pneumonia’ (277 patients).
Data collection
The collected clinical data included demographics, laboratory and vital sign measurements, diagnoses, microbiology and medication information and mortality outcomes. The five-character English ICD-10 codes were used, whose values are based on the seven-character Chinese ICD-10 codes.
Dataset extraction and analysis
We downloaded the PIC database of version 1.1.0 from Physionet 1 to obtain 17 CSV tables. Using Pandas, we selected all information with more than 50% coverage rate, including demographics and laboratory and vital sign measurements (Fig. 2 ). To reduce the amount of noise, we calculated and added only the minimum, maximum and average of all measurements that had multiple values per patient. Examination reports were removed because they describe only diagnostics and not detailed findings. All further diagnoses and microbiology and medication information were included into the observations slot to ensure that the data were not used for the calculation of embeddings but were still available for the analysis. This ensured that any calculated embedding would not be divided into treated and untreated groups but, rather, solely based on phenotypic features. We imputed all missing data through k -nearest neighbors imputation ( k = 20) using the knn_impute function of ehrapy. Next, we log normalized the data with ehrapy using the log_norm function. Afterwards, we winsorized the data using ehrapy’s winsorize function to obtain 277 ICU visits ( n = 265 patients) with 572 features. Of those 572 features, 254 were stored in the matrix X and the remaining 318 in the ‘obs’ slot in the AnnData object. For clustering and visualization purposes, we calculated 50 principal components using ehrapy’s pca function. The obtained principal component representation was then used to calculate a nearest neighbors graph using the neighbors function of ehrapy. The nearest neighbors graph then served as the basis for a UMAP embedding calculation using ehrapy’s umap function.
We applied the community detection algorithm Leiden with resolution 0.6 on the nearest neighbor graph using ehrapy’s leiden function. The four obtained clusters served as input for two-sided t -tests for all numerical values and two-sided g -tests for all categorical values for all four clusters against the union of all three other clusters, respectively. This was conducted using ehrapy’s rank_feature_groups function, which also corrects P values for multiple testing with the Benjamini–Hochberg method 125 . We presented the four groups and the statistically significantly different features between the groups to two pediatricians who annotated the groups with labels.
Our determined groups can be confidently labeled owing to their distinct clinical profiles. Nevertheless, we could only take into account clinical features that were measured. Insightful features, such as lung function tests, are missing. Moreover, the feature representation of the time-series data is simplified, which can hide some nuances between the groups. Generally, deciding on a clustering resolution is difficult. However, more fine-grained clusters obtained via higher clustering resolutions may become too specific and not generalize well enough.
Kaplan–Meier survival analysis
We selected patients with up to 360 h of total stay for Kaplan–Meier survival analysis to ensure a sufficiently high number of participants. We proceeded with the AnnData object prepared as described in the ‘Patient stratification’ subsection to conduct Kaplan–Meier analysis among all four determined pneumonia groups using ehrapy’s kmf function. Significance was tested through ehrapy’s test_kmf_logrank function, which tests whether two Kaplan–Meier series are statistically significant, employing a chi-squared test statistic under the null hypothesis. Let h i (t) be the hazard ratio of group i at time t and c a constant that represents a proportional change in the hazard ratio between the two groups, then:
This implicitly uses the log-rank weights. An additional Kaplan–Meier analysis was conducted for all children jointly concerning the liver markers AST, ALT and GGT. To determine whether measurements were inside or outside the norm range, we used reference ranges (Supplementary Table 2 ). P values less than 0.05 were labeled significant.
Our Kaplan–Meier curve analysis depends on the groups being well defined and shares the same limitations as the patient stratification. Additionally, the analysis is sensitive to the reference table where we selected limits that generalize well for the age ranges, but, due to children of different ages being examined, they may not necessarily be perfectly accurate for all children.
Causal effect of mechanism of action on LOS
Although the dataset was not initially intended for investigating causal effects of interventions, we adapted it for this purpose by focusing on the LOS in the ICU, measured in months, as the outcome variable. This choice aligns with the clinical aim of stabilizing patients sufficiently for ICU discharge. We constructed a causal graph to explore how different drug administrations could potentially reduce the LOS. Based on consultations with clinicians, we included several biomarkers of liver damage (AST, ALT and GGT) and inflammation (CRP and PCT) in our model. Patient age was also considered a relevant variable.
Because several different medications act by the same mechanisms, we grouped specific medications by their drug classes This grouping was achieved by cross-referencing the drugs listed in the dataset with DrugBank release 5.1 (ref. 126 ), using Levenshtein distances for partial string matching. After manual verification, we extracted the corresponding DrugBank categories, counted the number of features per category and compiled a list of commonly prescribed medications, as advised by clinicians. This approach facilitated the modeling of the causal graph depicted in Fig. 4 , where an intervention is defined as the administration of at least one drug from a specified category.
Causal inference was then conducted with ehrapy’s ‘dowhy’ 69 -based causal inference module using the expert-curated causal graph. Medication groups were designated as causal interventions, and the LOS was the outcome of interest. Linear regression served as the estimation method for analyzing these causal effects. We excluded four patients from the analysis owing to their notably long hospital stays exceeding 90 d, which were deemed outliers. To validate the robustness of our causal estimates, we incorporated several refutation methods:
Placebo Treatment Refuter: This method involved replacing the treatment assignment with a placebo to test the effect of the treatment variable being null.
Random Common Cause: A randomly generated variable was added to the data to assess the sensitivity of the causal estimate to the inclusion of potential unmeasured confounders.
Data Subset Refuter: The stability of the causal estimate was tested across various random subsets of the data to ensure that the observed effects were not dependent on a specific subset.
Add Unobserved Common Cause: This approach tested the effect of an omitted variable by adding a theoretically relevant unobserved confounder to the model, evaluating how much an unmeasured variable could influence the causal relationship.
Dummy Outcome: Replaces the true outcome variable with a random variable. If the causal effect nullifies, it supports the validity of the original causal relationship, indicating that the outcome is not driven by random factors.
Bootstrap Validation: Employs bootstrapping to generate multiple samples from the dataset, testing the consistency of the causal effect across these samples.
The selection of these refuters addresses a broad spectrum of potential biases and model sensitivities, including unobserved confounders and data dependencies. This comprehensive approach ensures robust verification of the causal analysis. Each refuter provides an orthogonal perspective, targeting specific vulnerabilities in causal analysis, which strengthens the overall credibility of the findings.
UKB analysis
Study population.
We used information from the UKB cohort, which includes 502,164 study participants from the general UK population without enrichment for specific diseases. The study involved the enrollment of individuals between 2006 and 2010 across 22 different assessment centers throughout the United Kingdom. The tracking of participants is still ongoing. Within the UKB dataset, metabolomics, proteomics and retinal optical coherence tomography data are available for a subset of individuals without any enrichment for specific diseases. Additionally, EHRs, questionnaire responses and other physical measures are available for almost everyone in the study. Furthermore, a variety of genotype information is available for nearly the entire cohort, including whole-genome sequencing, whole-exome sequencing, genotyping array data as well as imputed genotypes from the genotyping array 44 . Because only the latter two are available for download, and are sufficient for polygenic risk score calculation as performed here, we used the imputed genotypes in the present study. Participants visited the assessment center up to four times for additional and repeat measurements and completed additional online follow-up questionnaires.
In the present study, we restricted the analyses to data obtained from the initial assessment, including the blood draw, for obtaining the metabolomics data and the retinal imaging as well as physical measures. This restricts the study population to 33,521 individuals for whom all of these modalities are available. We have a clear study start point for each individual with the date of their initial assessment center visit. The study population has a mean age of 57 years, is 54% female and is censored at age 69 years on average; 4.7% experienced an incident myocardial infarction; and 8.1% have prevalent type 2 diabetes. The study population comes from six of the 22 assessment centers due to the retinal imaging being performed only at those.
For the myocardial infarction endpoint definition, we relied on the first occurrence data available in the UKB, which compiles the first date that each diagnosis was recorded for a participant in a hospital in ICD-10 nomenclature. Subsequently, we mapped these data to phecodes and focused on phecode 404.1 for myocardial infarction.
The Framingham Risk Score was developed on data from 8,491 participants in the Framingham Heart Study to assess general cardiovascular risk 77 . It includes easily obtainable predictors and is, therefore, easily applicable in clinical practice, although newer and more specific risk scores exist and might be used more frequently. It includes age, sex, smoking behavior, blood pressure, total and low-density lipoprotein cholesterol as well as information on insulin, antihypertensive and cholesterol-lowering medications, all of which are routinely collected in the UKB and used in this study as the Framingham feature set.
The metabolomics data used in this study were obtained using proton NMR spectroscopy, a low-cost method with relatively low batch effects. It covers established clinical predictors, such as albumin and cholesterol, as well as a range of lipids, amino acids and carbohydrate-related metabolites.
The retinal optical coherence tomography–derived features were returned by researchers to the UKB 75 , 76 . They used the available scans and determined the macular volume, macular thickness, retinal pigment epithelium thickness, disc diameter, cup-to-disk ratio across different regions as well as the thickness between the inner nuclear layer and external limiting membrane, inner and outer photoreceptor segments and the retinal pigment epithelium across different regions. Furthermore, they determined a wide range of quality metrics for each scan, including the image quality score, minimum motion correlation and inner limiting membrane (ILM) indicator.
Data analysis
After exporting the data from the UKB, all timepoints were transformed into participant age entries. Only participants without prevalent myocardial infarction (relative to the first assessment center visit at which all data were collected) were included.
The data were pre-processed for retinal imaging and metabolomics subsets separately, to enable a clear analysis of missing data and allow for the k -nearest neighbors–based imputation ( k = 20) of missing values when less than 10% were missing for a given participant. Otherwise, participants were dropped from the analyses. The imputed genotypes and Framingham analyses were available for almost every participant and, therefore, not imputed. Individuals without them were, instead, dropped from the analyses. Because genetic risk modeling poses entirely different methodological and computational challenges, we applied a published polygenic risk score for coronary heart disease using 6.6 million variants 73 . This was computed using the plink2 score option on the imputed genotypes available in the UKB.
UMAP embeddings were computed using default parameters on the full feature sets with ehrapy’s umap function. For all analyses, the same time-to-event and event-indicator columns were used. The event indicator is a Boolean variable indicating whether a myocardial infarction was observed for a study participant. The time to event is defined as the timespan between the start of the study, in this case the date of the first assessment center visit. Otherwise, it is the timespan from the start of the study to the start of censoring; in this case, this is set to the last date for which EHRs were available, unless a participant died, in which case the date of death is the start of censoring. Kaplan–Meier curves and Cox proportional hazards models were fit using ehrapy’s survival analysis module and the lifelines 124 package’s Cox-PHFitter function with default parameters. For Cox proportional hazards models with multiple feature sets, individually imputed and quality-controlled feature sets were concatenated, and the model was fit on the resulting matrix. Models were evaluated using the C-index 127 as a metric. It can be seen as an extension of the common area under the receiver operator characteristic score to time-to-event datasets, in which events are not observed for every sample and which ranges from 0.0 (entirely false) over 0.5 (random) to 1.0 (entirely correct). CIs for the C-index were computed based on bootstrapping by sampling 1,000 times with replacement from all computed partial hazards and computing the C-index over each of these samples. The percentiles at 2.5% and 97.5% then give the upper and lower confidence bound for the 95% CIs.
In all UKB analyses, the unit of study for a statistical test or predictive model is always an individual study participant.
The generalizability of the analysis is limited as the UK Biobank cohort may not represent the general population, with potential selection biases and underrepresentation of the different demographic groups. Additionally, by restricting analysis to initial assessment data and censoring based on the last available EHR or date of death, our analysis does not account for longitudinal changes and can introduce follow-up bias, especially if participants lost to follow-up have different risk profiles.
In-depth quality control of retina-derived features
A UMAP plot of the retina-derived features indicating the assessment centers shows a cluster of samples that lie somewhat outside the general population and mostly attended the Birmingham assessment center (Fig. 5b ). To further investigate this, we performed Leiden clustering of resolution 0.3 (Extended Data Fig. 9a ) and isolated this group in cluster 5. When comparing cluster 5 to the rest of the population in the retina-derived feature space, we noticed that many individuals in cluster 5 showed overall retinal pigment epithelium (RPE) thickness measures substantially elevated over the rest of the population in both eyes (Extended Data Fig. 9b ), which is mostly a feature of this cluster (Extended Data Fig. 9c ). To investigate potential confounding, we computed ratios between cluster 5 and the rest of the population over the ‘obs’ DataFrame containing the Framingham features, diabetes-related phecodes and genetic principal components. Out of the top and bottom five highest ratios observed, six are in genetic principal components, which are commonly used to represent genetic ancestry in a continuous space (Extended Data Fig. 9d ). Additionally, diagnoses for type 1 and type 2 diabetes and antihypertensive use are enriched in cluster 5. Further investigating the ancestry, we computed log ratios for self-reported ancestries and absolute counts, which showed no robust enrichment and depletion effects.
A closer look at three quality control measures of the imaging pipeline revealed that cluster 5 was an outlier in terms of either image quality (Extended Data Fig. 9e ) or minimum motion correlation (Extended Data Fig. 9f ) and the ILM indicator (Extended Data Fig. 9g ), all of which can be indicative of artifacts in image acquisition and downstream processing 128 . Subsequently, we excluded 301 individuals from cluster 5 from all analyses.
COVID-19 chest-x-ray fate determination
Dataset overview.
We used the public BrixIA COVID-19 dataset, which contains 192 chest x-ray images annotated with BrixIA scores 82 . Hereby, six regions were annotated by a senior radiologist with more than 20 years of experience and a junior radiologist with a disease severity score ranging from 0 to 3. A global score was determined as the sum of all of these regions and, therefore, ranges from 0 to 18 (S-Global). S-Global scores of 0 were classified as normal. Images that only had severity values up to 1 in all six regions were classified as mild. Images with severity values greater than or equal to 2, but a S-Global score of less than 7, were classified as moderate. All images that contained at least one 3 in any of the six regions with a S-Global score between 7 and 10 were classified as severe, and all remaining images with S-Global scores greater than 10 with at least one 3 were labeled critical. The dataset and instructions to download the images can be found at https://github.com/ieee8023/covid-chestxray-dataset .
We first resized all images to 224 × 224. Afterwards, the images underwent a random affine transformation that involved rotation, translation and scaling. The rotation angle was randomly selected from a range of −45° to 45°. The images were also subject to horizontal and vertical translation, with the maximum translation being 15% of the image size in either direction. Additionally, the images were scaled by a factor ranging from 0.85 to 1.15. The purpose of applying these transformations was to enhance the dataset and introduce variations, ultimately improving the robustness and generalization of the model.
To generate embeddings, we used a pre-trained DenseNet model with weights densenet121-res224-all of TorchXRayVision 129 . A DenseNet is a convolutional neural network that makes use of dense connections between layers (Dense Blocks) where all layers (with matching feature map sizes) directly connect with each other. To maintain a feed-forward nature, every layer in the DenseNet architecture receives supplementary inputs from all preceding layers and transmits its own feature maps to all subsequent layers. The model was trained on the nih-pc- chex-mimic_ch-google-openi-rsna dataset 130 .
Next, we calculated 50 principal components on the feature representation of the DenseNet model of all images using ehrapy’s pca function. The principal component representation served as input for a nearest neighbors graph calculation using ehrapy’s neighbors function. This graph served as the basis for the calculation of a UMAP embedding with three components that was finally visualized using ehrapy.
We randomly picked a root in the group of images that was labeled ‘Normal’. First, we calculated so-called pseudotime by fitting a trajectory through the calculated UMAP space using diffusion maps as implemented in ehrapy’s dpt function 57 . Each image’s pseudotime value represents its estimated position along this trajectory, serving as a proxy for its severity stage relative to others in the dataset. To determine fates, we employed CellRank 58 , 59 with the PseudotimeKernel . This kernel computes transition probabilities for patient visits based on the connectivity of the k -nearest neighbors graph and the pseudotime values of patient visits, which resembles their progression through a process. Directionality is infused in the nearest neighbors graph in this process where the kernel either removes or downweights edges in the graph that contradict the directional flow of increasing pseudotime, thereby refining the graph to better reflect the developmental trajectory. We computed the transition matrix with a soft threshold scheme (Parameter of the PseudotimeKernel ), which downweights edges that point against the direction of increasing pseudotime. Finally, we calculated a projection on top of the UMAP embedding with CellRank using the plot_projection function of the PseudotimeKernel that we subsequently plotted.
This analysis is limited by the small dataset of 192 chest x-ray images, which may affect the model’s generalizability and robustness. Annotation subjectivity from radiologists can further introduce variability in severity scores. Additionally, the random selection of a root from ‘Normal’ images can introduce bias in pseudotime calculations and subsequent analyses.
Diabetes 130-US hospitals analysis
We used data from the Diabetes 130-US hospitals dataset that were collected between 1999 and 2008. It contains clinical care information at 130 hospitals and integrated delivery networks. The extracted database information pertains to hospital admissions specifically for patients diagnosed with diabetes. These encounters required a hospital stay ranging from 1 d to 14 d, during which both laboratory tests and medications were administered. The selection criteria focused exclusively on inpatient encounters with these defined characteristics. More specifically, we used a version that was curated by the Fairlearn team where the target variable ‘readmitted’ was binarized and a few features renamed or binned ( https://fairlearn.org/main/user_guide/datasets/diabetes_hospital_data.html ). The dataset contains 101,877 patient visits and 25 features. The dataset predominantly consists of White patients (74.8%), followed by African Americans (18.9%), with other racial groups, such as Hispanic, Asian and Unknown categories, comprising smaller percentages. Females make up a slight majority in the data at 53.8%, with males accounting for 46.2% and a negligible number of entries listed as unknown or invalid. A substantial majority of the patients are over 60 years of age (67.4%), whereas those aged 30–60 years represent 30.2%, and those 30 years or younger constitute just 2.5%.
All of the following descriptions start by loading the Fairlearn version of the Diabetes 130-US hospitals dataset using ehrapy’s dataloader as an AnnData object.
Selection and filtering bias
An overview of sensitive variables was generated using tableone. Subsequently, ehrapy’s CohortTracker was used to track the age, gender and race variables. The cohort was filtered for all Medicare recipients and subsequently plotted.
Surveillance bias
We plotted the HbA1c measurement ratios using ehrapy’s catplot .
Missing data and imputation bias
MCAR-type missing data for the number of medications variable (‘num_medications‘) were introduced by randomly setting 30% of the variables to be missing using Numpy’s choice function. We tested that the data are MCAR by applying ehrapy’s implementation of Little’s MCAR test, which returned a non-significant P value of 0.71. MAR data for the number of medications variable (‘num_medications‘) were introduced by scaling the ‘time_in_hospital’ variable to have a mean of 0 and a standard deviation of 1, adjusting these values by multiplying by 1.2 and subtracting 0.6 to influence overall missingness rate, and then using these values to generate MAR data in the ‘num_medications’ variable via a logistic transformation and binomial sampling. We verified that the newly introduced missing values are not MCAR with respect to the ‘time_in_hospital’ variable by applying ehrapy’s implementation of Little’s test, which was significant (0.01 × 10 −2 ). The missing data were imputed using ehrapy’s mean imputation and MissForest implementation.
Algorithmic bias
Variables ‘race’, ‘gender’, ‘age’, ‘readmitted’, ‘readmit_binary’ and ‘discharge_disposition_id’ were moved to the ‘obs’ slot of the AnnData object to ensure that they were not used for model training. We built a binary label ‘readmit_30_days’ indicating whether a patient had been readmitted in fewer than 30 d. Next, we combined the ‘Asian’ and ‘Hispanic’ categories into a single ‘Other’ category within the ‘race’ column of our AnnData object and then filtered out and discarded any samples labeled as ‘Unknown/Invalid’ under the ‘gender‘ column and subsequently moved the ‘gender’ data to the variable matrix X of the AnnData object. All categorical variables got encoded. The data were split into train and test groups with a test size of 50%. The data were scaled, and a logistic regression model was trained using scikit-learn, which was also used to determine the balanced accuracy score. Fairlearn’s MetricFrame function was used to inspect the target model performance against the sensitive variable ‘race’. We subsequently fit Fairlearn’s ThresholdOptimizer using the logistic regression estimator with balanced_accuracy_score as the target object. The algorithmic demonstration of Fairlearn’s abilities on this dataset is shown here: https://github.com/fairlearn/talks/tree/main/2021_scipy_tutorial .
Normalization bias
We one-hot encoded all categorical variables with ehrapy using the encode function. We applied ehrapy’s implementation of scaling normalization with and without the ‘Age group’ variable as group key to scale the data jointly and separately using ehrapy’s scale_norm function.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Physionet provides access to the PIC database 43 at https://physionet.org/content/picdb/1.1.0 for credentialed users. The BrixIA images 82 are available at https://github.com/BrixIA/Brixia-score-COVID-19 . The data used in this study were obtained from the UK Biobank 44 ( https://www.ukbiobank.ac.uk/ ). Access to the UK Biobank resource was granted under application number 49966. The data are available to researchers upon application to the UK Biobank in accordance with their data access policies and procedures. The Diabetes 130-US Hospitals dataset is available at https://archive.ics.uci.edu/dataset/296/diabetes+130-us+hospitals+for+years+1999-2008 .
Code availability
The ehrapy source code is available at https://github.com/theislab/ehrapy under an Apache 2.0 license. Further documentation, tutorials and examples are available at https://ehrapy.readthedocs.io . We are actively developing the software and invite contributions from the community.
Jupyter notebooks to reproduce our analysis and figures, including Conda environments that specify all versions, are available at https://github.com/theislab/ehrapy-reproducibility .
Goldberger, A. L. et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101 , E215–E220 (2000).
Article CAS PubMed Google Scholar
Atasoy, H., Greenwood, B. N. & McCullough, J. S. The digitization of patient care: a review of the effects of electronic health records on health care quality and utilization. Annu. Rev. Public Health 40 , 487–500 (2019).
Article PubMed Google Scholar
Jamoom, E. W., Patel, V., Furukawa, M. F. & King, J. EHR adopters vs. non-adopters: impacts of, barriers to, and federal initiatives for EHR adoption. Health (Amst.) 2 , 33–39 (2014).
Google Scholar
Rajkomar, A. et al. Scalable and accurate deep learning with electronic health records. NPJ Digit. Med. 1 , 18 (2018).
Article PubMed PubMed Central Google Scholar
Wolf, A. et al. Data resource profile: Clinical Practice Research Datalink (CPRD) Aurum. Int. J. Epidemiol. 48 , 1740–1740g (2019).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12 , e1001779 (2015).
Pollard, T. J. et al. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci. Data 5 , 180178 (2018).
Johnson, A. E. W. et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3 , 160035 (2016).
Article CAS PubMed PubMed Central Google Scholar
Hyland, S. L. et al. Early prediction of circulatory failure in the intensive care unit using machine learning. Nat. Med. 26 , 364–373 (2020).
Rasmy, L. et al. Recurrent neural network models (CovRNN) for predicting outcomes of patients with COVID-19 on admission to hospital: model development and validation using electronic health record data. Lancet Digit. Health 4 , e415–e425 (2022).
Marcus, J. L. et al. Use of electronic health record data and machine learning to identify candidates for HIV pre-exposure prophylaxis: a modelling study. Lancet HIV 6 , e688–e695 (2019).
Kruse, C. S., Stein, A., Thomas, H. & Kaur, H. The use of electronic health records to support population health: a systematic review of the literature. J. Med. Syst. 42 , 214 (2018).
Sheikh, A., Jha, A., Cresswell, K., Greaves, F. & Bates, D. W. Adoption of electronic health records in UK hospitals: lessons from the USA. Lancet 384 , 8–9 (2014).
Sheikh, A. et al. Health information technology and digital innovation for national learning health and care systems. Lancet Digit. Health 3 , e383–e396 (2021).
Cord, K. A. M., Mc Cord, K. A. & Hemkens, L. G. Using electronic health records for clinical trials: where do we stand and where can we go? Can. Med. Assoc. J. 191 , E128–E133 (2019).
Article Google Scholar
Landi, I. et al. Deep representation learning of electronic health records to unlock patient stratification at scale. NPJ Digit. Med. 3 , 96 (2020).
Ayaz, M., Pasha, M. F., Alzahrani, M. Y., Budiarto, R. & Stiawan, D. The Fast Health Interoperability Resources (FHIR) standard: systematic literature review of implementations, applications, challenges and opportunities. JMIR Med. Inform. 9 , e21929 (2021).
Peskoe, S. B. et al. Adjusting for selection bias due to missing data in electronic health records-based research. Stat. Methods Med. Res. 30 , 2221–2238 (2021).
Haneuse, S. & Daniels, M. A general framework for considering selection bias in EHR-based studies: what data are observed and why? EGEMS (Wash. DC) 4 , 1203 (2016).
PubMed Google Scholar
Gallifant, J. et al. Disparity dashboards: an evaluation of the literature and framework for health equity improvement. Lancet Digit. Health 5 , e831–e839 (2023).
Sauer, C. M. et al. Leveraging electronic health records for data science: common pitfalls and how to avoid them. Lancet Digit. Health 4 , e893–e898 (2022).
Li, J. et al. Imputation of missing values for electronic health record laboratory data. NPJ Digit. Med. 4 , 147 (2021).
Rubin, D. B. Inference and missing data. Biometrika 63 , 581 (1976).
Scheid, L. M., Brown, L. S., Clark, C. & Rosenfeld, C. R. Data electronically extracted from the electronic health record require validation. J. Perinatol. 39 , 468–474 (2019).
Phelan, M., Bhavsar, N. A. & Goldstein, B. A. Illustrating informed presence bias in electronic health records data: how patient interactions with a health system can impact inference. EGEMS (Wash. DC). 5 , 22 (2017).
PubMed PubMed Central Google Scholar
Secondary Analysis of Electronic Health Records (ed MIT Critical Data) (Springer, 2016).
Jetley, G. & Zhang, H. Electronic health records in IS research: quality issues, essential thresholds and remedial actions. Decis. Support Syst. 126 , 113137 (2019).
McCormack, J. P. & Holmes, D. T. Your results may vary: the imprecision of medical measurements. BMJ 368 , m149 (2020).
Hobbs, F. D. et al. Is the international normalised ratio (INR) reliable? A trial of comparative measurements in hospital laboratory and primary care settings. J. Clin. Pathol. 52 , 494–497 (1999).
Huguet, N. et al. Using electronic health records in longitudinal studies: estimating patient attrition. Med. Care 58 Suppl 6 Suppl 1 , S46–S52 (2020).
Zeng, J., Gensheimer, M. F., Rubin, D. L., Athey, S. & Shachter, R. D. Uncovering interpretable potential confounders in electronic medical records. Nat. Commun. 13 , 1014 (2022).
Getzen, E., Ungar, L., Mowery, D., Jiang, X. & Long, Q. Mining for equitable health: assessing the impact of missing data in electronic health records. J. Biomed. Inform. 139 , 104269 (2023).
Tang, S. et al. Democratizing EHR analyses with FIDDLE: a flexible data-driven preprocessing pipeline for structured clinical data. J. Am. Med. Inform. Assoc. 27 , 1921–1934 (2020).
Dagliati, A. et al. A process mining pipeline to characterize COVID-19 patients’ trajectories and identify relevant temporal phenotypes from EHR data. Front. Public Health 10 , 815674 (2022).
Sun, Y. & Zhou, Y.-H. A machine learning pipeline for mortality prediction in the ICU. Int. J. Digit. Health 2 , 3 (2022).
Article CAS Google Scholar
Mandyam, A., Yoo, E. C., Soules, J., Laudanski, K. & Engelhardt, B. E. COP-E-CAT: cleaning and organization pipeline for EHR computational and analytic tasks. In Proc. of the 12th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics. https://doi.org/10.1145/3459930.3469536 (Association for Computing Machinery, 2021).
Gao, C. A. et al. A machine learning approach identifies unresolving secondary pneumonia as a contributor to mortality in patients with severe pneumonia, including COVID-19. J. Clin. Invest. 133 , e170682 (2023).
Makam, A. N. et al. The good, the bad and the early adopters: providers’ attitudes about a common, commercial EHR. J. Eval. Clin. Pract. 20 , 36–42 (2014).
Amezquita, R. A. et al. Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17 , 137–145 (2020).
Virshup, I. et al. The scverse project provides a computational ecosystem for single-cell omics data analysis. Nat. Biotechnol. 41 , 604–606 (2023).
Zou, Q. et al. Predicting diabetes mellitus with machine learning techniques. Front. Genet. 9 , 515 (2018).
Cios, K. J. & William Moore, G. Uniqueness of medical data mining. Artif. Intell. Med. 26 , 1–24 (2002).
Zeng, X. et al. PIC, a paediatric-specific intensive care database. Sci. Data 7 , 14 (2020).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562 , 203–209 (2018).
Lee, J. et al. Open-access MIMIC-II database for intensive care research. Annu. Int. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2011 , 8315–8318 (2011).
Virshup, I., Rybakov, S., Theis, F. J., Angerer, P. & Alexander Wolf, F. anndata: annotated data. Preprint at bioRxiv https://doi.org/10.1101/2021.12.16.473007 (2021).
Voss, E. A. et al. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J. Am. Med. Inform. Assoc. 22 , 553–564 (2015).
Vasilevsky, N. A. et al. Mondo: unifying diseases for the world, by the world. Preprint at medRxiv https://doi.org/10.1101/2022.04.13.22273750 (2022).
Harrison, J. E., Weber, S., Jakob, R. & Chute, C. G. ICD-11: an international classification of diseases for the twenty-first century. BMC Med. Inform. Decis. Mak. 21 , 206 (2021).
Köhler, S. et al. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 47 , D1018–D1027 (2019).
Wu, P. et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med. Inform. 7 , e14325 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19 , 15 (2018).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res . 12 , 2825–2830 (2011).
de Haan-Rietdijk, S., de Haan-Rietdijk, S., Kuppens, P. & Hamaker, E. L. What’s in a day? A guide to decomposing the variance in intensive longitudinal data. Front. Psychol. 7 , 891 (2016).
Pedersen, E. S. L., Danquah, I. H., Petersen, C. B. & Tolstrup, J. S. Intra-individual variability in day-to-day and month-to-month measurements of physical activity and sedentary behaviour at work and in leisure-time among Danish adults. BMC Public Health 16 , 1222 (2016).
Roffey, D. M., Byrne, N. M. & Hills, A. P. Day-to-day variance in measurement of resting metabolic rate using ventilated-hood and mouthpiece & nose-clip indirect calorimetry systems. JPEN J. Parenter. Enter. Nutr. 30 , 426–432 (2006).
Haghverdi, L., Büttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13 , 845–848 (2016).
Lange, M. et al. CellRank for directed single-cell fate mapping. Nat. Methods 19 , 159–170 (2022).
Weiler, P., Lange, M., Klein, M., Pe'er, D. & Theis, F. CellRank 2: unified fate mapping in multiview single-cell data. Nat. Methods 21 , 1196–1205 (2024).
Zhang, S. et al. Cost of management of severe pneumonia in young children: systematic analysis. J. Glob. Health 6 , 010408 (2016).
Torres, A. et al. Pneumonia. Nat. Rev. Dis. Prim. 7 , 25 (2021).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9 , 5233 (2019).
Kamin, W. et al. Liver involvement in acute respiratory infections in children and adolescents—results of a non-interventional study. Front. Pediatr. 10 , 840008 (2022).
Shi, T. et al. Risk factors for mortality from severe community-acquired pneumonia in hospitalized children transferred to the pediatric intensive care unit. Pediatr. Neonatol. 61 , 577–583 (2020).
Dudnyk, V. & Pasik, V. Liver dysfunction in children with community-acquired pneumonia: the role of infectious and inflammatory markers. J. Educ. Health Sport 11 , 169–181 (2021).
Charpignon, M.-L. et al. Causal inference in medical records and complementary systems pharmacology for metformin drug repurposing towards dementia. Nat. Commun. 13 , 7652 (2022).
Grief, S. N. & Loza, J. K. Guidelines for the evaluation and treatment of pneumonia. Prim. Care 45 , 485–503 (2018).
Paul, M. Corticosteroids for pneumonia. Cochrane Database Syst. Rev. 12 , CD007720 (2017).
Sharma, A. & Kiciman, E. DoWhy: an end-to-end library for causal inference. Preprint at arXiv https://doi.org/10.48550/ARXIV.2011.04216 (2020).
Khilnani, G. C. et al. Guidelines for antibiotic prescription in intensive care unit. Indian J. Crit. Care Med. 23 , S1–S63 (2019).
Harris, L. K. & Crannage, A. J. Corticosteroids in community-acquired pneumonia: a review of current literature. J. Pharm. Technol. 37 , 152–160 (2021).
Dou, L. et al. Decreased hospital length of stay with early administration of oseltamivir in patients hospitalized with influenza. Mayo Clin. Proc. Innov. Qual. Outcomes 4 , 176–182 (2020).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50 , 1219–1224 (2018).
Julkunen, H. et al. Atlas of plasma NMR biomarkers for health and disease in 118,461 individuals from the UK Biobank. Nat. Commun. 14 , 604 (2023).
Ko, F. et al. Associations with retinal pigment epithelium thickness measures in a large cohort: results from the UK Biobank. Ophthalmology 124 , 105–117 (2017).
Patel, P. J. et al. Spectral-domain optical coherence tomography imaging in 67 321 adults: associations with macular thickness in the UK Biobank study. Ophthalmology 123 , 829–840 (2016).
D’Agostino Sr, R. B. et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation 117 , 743–753 (2008).
Buergel, T. et al. Metabolomic profiles predict individual multidisease outcomes. Nat. Med. 28 , 2309–2320 (2022).
Xu, Y. et al. An atlas of genetic scores to predict multi-omic traits. Nature 616 , 123–131 (2023).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37 , 547–554 (2019).
Rousan, L. A., Elobeid, E., Karrar, M. & Khader, Y. Chest x-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 20 , 245 (2020).
Signoroni, A. et al. BS-Net: learning COVID-19 pneumonia severity on a large chest X-ray dataset. Med. Image Anal. 71 , 102046 (2021).
Bird, S. et al. Fairlearn: a toolkit for assessing and improving fairness in AI. https://www.microsoft.com/en-us/research/publication/fairlearn-a-toolkit-for-assessing-and-improving-fairness-in-ai/ (2020).
Strack, B. et al. Impact of HbA1c measurement on hospital readmission rates: analysis of 70,000 clinical database patient records. BioMed. Res. Int. 2014 , 781670 (2014).
Stekhoven, D. J. & Bühlmann, P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics 28 , 112–118 (2012).
Banerjee, A. et al. Identifying subtypes of heart failure from three electronic health record sources with machine learning: an external, prognostic, and genetic validation study. Lancet Digit. Health 5 , e370–e379 (2023).
Nagamine, T. et al. Data-driven identification of heart failure disease states and progression pathways using electronic health records. Sci. Rep. 12 , 17871 (2022).
Da Silva Filho, J. et al. Disease trajectories in hospitalized COVID-19 patients are predicted by clinical and peripheral blood signatures representing distinct lung pathologies. Preprint at bioRxiv https://doi.org/10.1101/2023.09.08.23295024 (2023).
Haneuse, S., Arterburn, D. & Daniels, M. J. Assessing missing data assumptions in EHR-based studies: a complex and underappreciated task. JAMA Netw. Open 4 , e210184 (2021).
Little, R. J. A. A test of missing completely at random for multivariate data with missing values. J. Am. Stat. Assoc. 83 , 1198–1202 (1988).
Jakobsen, J. C., Gluud, C., Wetterslev, J. & Winkel, P. When and how should multiple imputation be used for handling missing data in randomised clinical trials—a practical guide with flowcharts. BMC Med. Res. Methodol. 17 , 162 (2017).
Dziura, J. D., Post, L. A., Zhao, Q., Fu, Z. & Peduzzi, P. Strategies for dealing with missing data in clinical trials: from design to analysis. Yale J. Biol. Med. 86 , 343–358 (2013).
White, I. R., Royston, P. & Wood, A. M. Multiple imputation using chained equations: issues and guidance for practice. Stat. Med. 30 , 377–399 (2011).
Jäger, S., Allhorn, A. & Bießmann, F. A benchmark for data imputation methods. Front. Big Data 4 , 693674 (2021).
Waljee, A. K. et al. Comparison of imputation methods for missing laboratory data in medicine. BMJ Open 3 , e002847 (2013).
Ibrahim, J. G. & Molenberghs, G. Missing data methods in longitudinal studies: a review. Test (Madr.) 18 , 1–43 (2009).
Li, C., Alsheikh, A. M., Robinson, K. A. & Lehmann, H. P. Use of recommended real-world methods for electronic health record data analysis has not improved over 10 years. Preprint at bioRxiv https://doi.org/10.1101/2023.06.21.23291706 (2023).
Regev, A. et al. The Human Cell Atlas. eLife 6 , e27041 (2017).
Megill, C. et al. cellxgene: a performant, scalable exploration platform for high dimensional sparse matrices. Preprint at bioRxiv https://doi.org/10.1101/2021.04.05.438318 (2021).
Speir, M. L. et al. UCSC Cell Browser: visualize your single-cell data. Bioinformatics 37 , 4578–4580 (2021).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9 , 90–95 (2007).
Waskom, M. seaborn: statistical data visualization. J. Open Source Softw. 6 , 3021 (2021).
Harris, C. R. et al. Array programming with NumPy. Nature 585 , 357–362 (2020).
Lam, S. K., Pitrou, A. & Seibert, S. Numba: a LLVM-based Python JIT compiler. In Proc. of the Second Workshop on the LLVM Compiler Infrastructure in HPC. https://doi.org/10.1145/2833157.2833162 (Association for Computing Machinery, 2015).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17 , 261–272 (2020).
McKinney, W. Data structures for statistical computing in Python. In Proc. of the 9th Python in Science Conference (eds van der Walt, S. & Millman, J.). https://doi.org/10.25080/majora-92bf1922-00a (SciPy, 2010).
Boulanger, A. Open-source versus proprietary software: is one more reliable and secure than the other? IBM Syst. J. 44 , 239–248 (2005).
Rocklin, M. Dask: parallel computation with blocked algorithms and task scheduling. In Proc. of the 14th Python in Science Conference. https://doi.org/10.25080/majora-7b98e3ed-013 (SciPy, 2015).
Pivarski, J. et al. Awkward Array. https://doi.org/10.5281/ZENODO.4341376
Collette, A. Python and HDF5: Unlocking Scientific Data (‘O’Reilly Media, Inc., 2013).
Miles, A. et al. zarr-developers/zarr-python: v2.13.6. https://doi.org/10.5281/zenodo.7541518 (2023).
The pandas development team. pandas-dev/pandas: Pandas. https://doi.org/10.5281/ZENODO.3509134 (2024).
Weberpals, J. et al. Deep learning-based propensity scores for confounding control in comparative effectiveness research: a large-scale, real-world data study. Epidemiology 32 , 378–388 (2021).
Rosenthal, J. et al. Building tools for machine learning and artificial intelligence in cancer research: best practices and a case study with the PathML toolkit for computational pathology. Mol. Cancer Res. 20 , 202–206 (2022).
Gayoso, A. et al. A Python library for probabilistic analysis of single-cell omics data. Nat. Biotechnol. 40 , 163–166 (2022).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems 32 (eds Wallach, H. et al.). 8024–8035 (Curran Associates, 2019).
Frostig, R., Johnson, M. & Leary, C. Compiling machine learning programs via high-level tracing. https://cs.stanford.edu/~rfrostig/pubs/jax-mlsys2018.pdf (2018).
Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616 , 259–265 (2023).
Kraljevic, Z. et al. Multi-domain clinical natural language processing with MedCAT: the Medical Concept Annotation Toolkit. Artif. Intell. Med. 117 , 102083 (2021).
Pollard, T. J., Johnson, A. E. W., Raffa, J. D. & Mark, R. G. An open source Python package for producing summary statistics for research papers. JAMIA Open 1 , 26–31 (2018).
Ellen, J. G. et al. Participant flow diagrams for health equity in AI. J. Biomed. Inform. 152 , 104631 (2024).
Schouten, R. M. & Vink, G. The dance of the mechanisms: how observed information influences the validity of missingness assumptions. Sociol. Methods Res. 50 , 1243–1258 (2021).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8 , 118–127 (2007).
Davidson-Pilon, C. lifelines: survival analysis in Python. J. Open Source Softw. 4 , 1317 (2019).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57 , 289–300 (1995).
Wishart, D. S. et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34 , D668–D672 (2006).
Harrell, F. E. Jr, Califf, R. M., Pryor, D. B., Lee, K. L. & Rosati, R. A. Evaluating the yield of medical tests. JAMA 247 , 2543–2546 (1982).
Currant, H. et al. Genetic variation affects morphological retinal phenotypes extracted from UK Biobank optical coherence tomography images. PLoS Genet. 17 , e1009497 (2021).
Cohen, J. P. et al. TorchXRayVision: a library of chest X-ray datasets and models. In Proc. of the 5th International Conference on Medical Imaging with Deep Learning (eds Konukoglu, E. et al.). 172 , 231–249 (PMLR, 2022).
Cohen, J.P., Hashir, M., Brooks, R. & Bertrand, H. On the limits of cross-domain generalization in automated X-ray prediction. In Proceedings of Machine Learning Research , Vol. 121 (eds Arbel, T. et al.) 136–155 (PMLR, 2020).
Download references
Acknowledgements
We thank M. Ansari who designed the ehrapy logo. The authors thank F. A. Wolf, M. Lücken, J. Steinfeldt, B. Wild, G. Rätsch and D. Shung for feedback on the project. We further thank L. Halle, Y. Ji, M. Lücken and R. K. Rubens for constructive comments on the paper. We thank F. Hashemi for her help in implementing the survival analysis module. This research was conducted using data from the UK Biobank, a major biomedical database ( https://www.ukbiobank.ac.uk ), under application number 49966. This work was supported by the German Center for Lung Research (DZL), the Helmholtz Association and the CRC/TRR 359 Perinatal Development of Immune Cell Topology (PILOT). N.H. and F.J.T. acknowledge support from the German Federal Ministry of Education and Research (BMBF) (LODE, 031L0210A), co-funded by the European Union (ERC, DeepCell, 101054957). A.N. is supported by the Konrad Zuse School of Excellence in Learning and Intelligent Systems (ELIZA) through the DAAD program Konrad Zuse Schools of Excellence in Artificial Intelligence, sponsored by the Federal Ministry of Education and Research. This work was also supported by the Chan Zuckerberg Initiative (CZIF2022-007488; Human Cell Atlas Data Ecosystem).
Open access funding provided by Helmholtz Zentrum München - Deutsches Forschungszentrum für Gesundheit und Umwelt (GmbH).
Author information
Authors and affiliations.
Institute of Computational Biology, Helmholtz Munich, Munich, Germany
Lukas Heumos, Philipp Ehmele, Tim Treis, Eljas Roellin, Lilly May, Altana Namsaraeva, Nastassya Horlava, Vladimir A. Shitov, Xinyue Zhang, Luke Zappia, Leon Hetzel, Isaac Virshup, Lisa Sikkema, Fabiola Curion & Fabian J. Theis
Institute of Lung Health and Immunity and Comprehensive Pneumology Center with the CPC-M bioArchive; Helmholtz Zentrum Munich; member of the German Center for Lung Research (DZL), Munich, Germany
Lukas Heumos, Niklas J. Lang, Herbert B. Schiller & Anne Hilgendorff
TUM School of Life Sciences Weihenstephan, Technical University of Munich, Munich, Germany
Lukas Heumos, Tim Treis, Nastassya Horlava, Vladimir A. Shitov, Lisa Sikkema & Fabian J. Theis
Health Data Science Unit, Heidelberg University and BioQuant, Heidelberg, Germany
Julius Upmeier zu Belzen & Roland Eils
Department of Mathematics, School of Computation, Information and Technology, Technical University of Munich, Munich, Germany
Eljas Roellin, Lilly May, Luke Zappia, Leon Hetzel, Fabiola Curion & Fabian J. Theis
Konrad Zuse School of Excellence in Learning and Intelligent Systems (ELIZA), Darmstadt, Germany
Altana Namsaraeva
Systems Medicine, Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), Bonn, Germany
Rainer Knoll
Center for Digital Health, Berlin Institute of Health (BIH) at Charité – Universitätsmedizin Berlin, Berlin, Germany
Roland Eils
Research Unit, Precision Regenerative Medicine (PRM), Helmholtz Munich, Munich, Germany
Herbert B. Schiller
Center for Comprehensive Developmental Care (CDeCLMU) at the Social Pediatric Center, Dr. von Hauner Children’s Hospital, LMU Hospital, Ludwig Maximilian University, Munich, Germany
Anne Hilgendorff
You can also search for this author in PubMed Google Scholar
Contributions
L. Heumos and F.J.T. conceived the study. L. Heumos, P.E., X.Z., E.R., L.M., A.N., L.Z., V.S., T.T., L. Hetzel, N.H., R.K. and I.V. implemented ehrapy. L. Heumos, P.E., N.L., L.S., T.T. and A.H. analyzed the PIC database. J.U.z.B. and L. Heumos analyzed the UK Biobank database. X.Z. and L. Heumos analyzed the COVID-19 chest x-ray dataset. L. Heumos, P.E. and J.U.z.B. wrote the paper. F.J.T., A.H., H.B.S. and R.E. supervised the work. All authors read, corrected and approved the final paper.
Corresponding author
Correspondence to Fabian J. Theis .
Ethics declarations
Competing interests.
L. Heumos is an employee of LaminLabs. F.J.T. consults for Immunai Inc., Singularity Bio B.V., CytoReason Ltd. and Omniscope Ltd. and has ownership interest in Dermagnostix GmbH and Cellarity. The remaining authors declare no competing interests.
Peer review
Peer review information.
Nature Medicine thanks Leo Anthony Celi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary handling editor: Lorenzo Righetto, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended data fig. 1 overview of the paediatric intensive care database (pic)..
The database consists of several tables corresponding to several data modalities and measurement types. All tables colored in green were selected for analysis and all tables in blue were discarded based on coverage rate. Despite the high coverage rate, we discarded the ‘OR_EXAM_REPORTS’ table because of the lack of detail in the exam reports.
Extended Data Fig. 2 Preprocessing of the Paediatric Intensive Care (PIC) dataset with ehrapy.
( a ) Heterogeneous data of the PIC database was stored in ‘data’ (matrix that is used for computations) and ‘observations’ (metadata per patient visit). During quality control, further annotations are added to the ‘variables’ (metadata per feature) slot. ( b ) Preprocessing steps of the PIC dataset. ( c ) Example of the function calls in the data analysis pipeline that resembles the preprocessing steps in (B) using ehrapy.
Extended Data Fig. 3 Missing data distribution for the ‘youths’ group of the PIC dataset.
The x-axis represents the percentage of missing values in each feature. The y-axis reflects the number of features in each bin with text labels representing the names of the individual features.
Extended Data Fig. 4 Patient selection during analysis of the PIC dataset.
Filtering for the pneumonia cohort of the youths filters out care units except for the general intensive care unit and the pediatric intensive care unit.
Extended Data Fig. 5 Feature rankings of stratified patient groups.
Scores reflect the z-score underlying the p-value per measurement for each group. Higher scores (above 0) reflect overrepresentation of the measurement compared to all other groups and vice versa. ( a ) By clinical chemistry. ( b ) By liver markers. ( c ) By medication type. ( d ) By infection markers.
Extended Data Fig. 6 Liver marker value progression for the ‘youths’ group and Kaplan-Meier curves.
( a ) Viral and severe pneumonia with co-infection groups display enriched gamma-glutamyl transferase levels in blood serum. ( b ) Aspartate transferase (AST) and Alanine transaminase (ALT) levels are enriched for severe pneumonia with co-infection during early ICU stay. ( c ) and ( d ) Kaplan-Meier curves for ALT and AST demonstrate lower survivability for children with measurements outside the norm.
Extended Data Fig. 7 Overview of medication categories used for causal inference.
( a ) Feature engineering process to group administered medications into medication categories using drugbank. ( b ) Number of medications per medication category. ( c ) Number of patients that received (dark blue) and did not receive specific medication categories (light blue).
Extended Data Fig. 8 UK-Biobank data overview and quality control across modalities.
( a ) UMAP plot of the metabolomics data demonstrating a clear gradient with respect to age at sampling, and ( b ) type 2 diabetes prevalence. ( c ) Analogously, the features derived from retinal imaging show a less pronounced age gradient, and ( d ) type 2 diabetes prevalence gradient. ( e ) Stratifying myocardial infarction risk by the type 2 diabetes comorbidity confirms vastly increased risk with a prior type 2 (T2D) diabetes diagnosis. Kaplan-Meier estimators with 95 % confidence intervals are shown. ( f ) Similarly, the polygenic risk score for coronary heart disease used in this work substantially enriches myocardial infarction risk in its top 5% percentile. Kaplan-Meier estimators with 95 % confidence intervals are shown. ( g ) UMAP visualization of the metabolomics features colored by the assessment center shows no discernable biases. (A-G) n = 29,216.
Extended Data Fig. 9 UK-Biobank retina derived feature quality control.
( a ) Leiden Clustering of retina derived feature space. ( b ) Comparison of ‘overall retinal pigment epithelium (RPE) thickness’ values between cluster 5 (n = 301) and the rest of the population (n = 28,915). ( c ) RPE thickness in the right eye outliers on the UMAP largely corresponds to cluster 5. ( d ) Log ratio of top and bottom 5 fields in obs dataframe between cluster 5 and the rest of the population. ( e ) Image Quality of the optical coherence tomography scan as reported in the UKB. ( f ) Minimum motion correlation quality control indicator. ( g ) Inner limiting membrane (ILM) quality control indicator. (D-G) Data are shown for the right eye only, comparable results for the left eye are omitted. (A-G) n = 29,216.
Extended Data Fig. 10 Bias detection and mitigation study on the Diabetes 130-US hospitals dataset (n = 101,766 hospital visits, one patient can have multiple visits).
( a ) Filtering to the visits of Medicare recipients results in an increase of Caucasians. ( b ) Proportion of visits where Hb1Ac measurements are recorded, stratified by admission type. Adjusted P values were calculated with Chi squared tests and Bonferroni correction (Adjusted P values: Emergency vs Referral 3.3E-131, Emergency vs Other 1.4E-101, Referral vs Other 1.6E-4.) ( c ) Normalizing feature distributions jointly vs. separately can mask distribution differences. ( d ) Imputing the number of medications for visits. Onto the complete data (blue), MCAR (30% missing data) and MAR (38% missing data) were introduced (orange), with the MAR mechanism depending on the time in hospital. Mean imputation (green) can reduce the variance of the distribution under MCAR and MAR mechanisms, and bias the center of the distribution under an MAR mechanism. Multiple imputation, such as MissForest imputation can impute meaningfully even in MAR cases, when having access to variables involved in the MAR mechanism. Each boxplot represents the IQR of the data, with the horizontal line inside the box indicating the median value. The left and right bounds of the box represent the first and third quartiles, respectively. The ‘whiskers’ extend to the minimum and maximum values within 1.5 times the IQR from the lower and upper quartiles, respectively. ( e ) Predicting the early readmission within 30 days after release on a per-stay level. Balanced accuracy can mask differences in selection and false negative rate between sensitive groups.
Supplementary information
Supplementary tables 1 and 2, reporting summary, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Heumos, L., Ehmele, P., Treis, T. et al. An open-source framework for end-to-end analysis of electronic health record data. Nat Med (2024). https://doi.org/10.1038/s41591-024-03214-0
Download citation
Received : 11 December 2023
Accepted : 25 July 2024
Published : 12 September 2024
DOI : https://doi.org/10.1038/s41591-024-03214-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.
IEEE Account
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- BMC Plant Biol
- PMC11380355

Genetic diversity analysis and DNA fingerprint construction of Zanthoxylum species based on SSR and iPBS markers
Xiaoxi zhang.
1 College of Horticulture and Gardening, Yangtze University, Jingzhou, Hubei 434025 China
2 Sichuan Academy of Forestry, Chengdu, Sichuan 610081 China
Chengrong Luo
Weiwei zhang, yongling liao, associated data.
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Zanthoxylum is a versatile economic tree species utilized for its spice, seasoning, oil, medicinal, and industrial raw material applications, and it has a lengthy history of cultivation and domestication in China. This has led to the development of numerous cultivars. However, the phenomenon of mixed cultivars and confusing names has significantly obstructed the effective utilization of Zanthoxylum resources and industrial development. Consequently, conducting genetic diversity studies and cultivar identification on Zanthoxylum are crucial. This research analyzed the genetic traits of 80 Zanthoxylum cultivars using simple sequence repeat (SSR) and inter-Primer Binding Site (iPBS) molecular markers, leading to the creation of a DNA fingerprint. This study identified 206 and 127 alleles with 32 SSR markers and 10 iPBS markers, respectively, yielding an average of 6.4 and 12.7 alleles ( Na ) per marker. The average polymorphism information content ( PIC ) for the SSR and iPBS markers was 0.710 and 0.281, respectively. The genetic similarity coefficients for the 80 Zanthoxylum accessions ranged from 0.0947 to 0.9868 and from 0.2206 to 1.0000, with mean values of 0.3864 and 0.5215, respectively, indicating substantial genetic diversity. Cluster analysis, corroborated by principal coordinate analysis (PCoA), categorized these accessions into three primary groups. Analysis of the genetic differentiation among the three Zanthoxylum ( Z. bungeanum , Z. armatum , and Z. piperitum ) populations using SSR markers revealed a mean genetic differentiation coefficient ( Fst ) of 0.335 and a gene flow ( Nm ) of 0.629, suggesting significant genetic divergence among the populations. Molecular variance analysis (AMOVA) indicated that 65% of the genetic variation occurred within individuals, while 35% occurred among populations. Bayesian model-based analysis of population genetic structure divided all materials into two groups. The combined PI and PIsibs value of the 32 SSR markers were 4.265 × 10 − 27 and 1.282 × 10 − 11 , respectively, showing strong fingerprinting power. DNA fingerprints of the 80 cultivars were established using eight pairs of SSR primers, each assigned a unique numerical code. In summary, while both markers were effective at assessing the genetic diversity and relationships of Zanthoxylum species, SSR markers demonstrated superior polymorphism and cultivar discrimination compared to iPBS markers. These findings offer a scientific foundation for the conservation and sustainable use of Zanthoxylum species.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05373-1.
Introduction
Zanthoxylum L., a member of the Rutaceae family, is a small evergreen or deciduous tree, shrub, or woody vine. There are approximately 250 species worldwide, primarily found in the tropical and subtropical regions of East Asia and North America [ 1 ]. Specifically, China is home to 45 species, 13 varieties, and 2 formas distributed in both the northern and southern regions. The predominant cultivated species in China are Zanthoxylum bungeanum Maxim. and Zanthoxylum armatum DC., commonly referred to as “Huajiao” or “Chinese pepper,” which are used as edible spices [ 2 – 4 ]. Moreover, Zanthoxylum species have a wide range of applications, including in food, medicine, ornamental purposes, and soil and water conservation, demonstrating significant economic and ecological benefits.
China serves as the leading producer of Zanthoxylum , boasting the highest yield and cultivation area globally. Furthermore, China has been at the forefront of utilizing and domesticating Zanthoxylum species, with records indicating its use dating back to the 11th to 10th centuries BC [ 5 ]. Over the course of extensive cultivation and domestication, a diverse range of Zanthoxylum cultivars and types have emerged. As the cultivation area expands and the exchange of resources between different Zanthoxylum production regions becomes more frequent, the genetic background of Zanthoxylum has become increasingly complex. Additionally, varying classification criteria in different regions have contributed to issues such as cultivar confusion and name ambiguity. Consequently, instances of synonymy, homonymy, and substandard materials often arise in the cultivation and commercial circulation of Zanthoxylum . Morphological identification methods based solely on phenotypic traits prove inadequate for distinguishing these similar materials. This not only compromises the rights and interests of consumers, growers, and breeders but also hinders the development and utilization of Zanthoxylum germplasm resources and the process of cultivar selection [ 3 , 6 ]. Therefore, conducting extensive research on genetic diversity analysis, genetic map construction, and cultivar identification techniques for Zanthoxylum is highly important. This research will play a crucial role in safeguarding the development of Zanthoxylum germplasm resources and ensuring the healthy growth of the industry.
Molecular markers are extensively utilized in genetic diversity analysis, germplasm resources identification, and genetic map construction of species. Among the various molecular marker technologies available, SSR has gained wide popularity due to their high polymorphism level, reliable repeatability, codominance, and multiple allele variations. It has been chosen as the preferred method for constructing plant DNA fingerprints by the International Union for the Protection of New Plant Varieties (UPOV) [ 7 , 8 ]. In recent years, several molecular markers have been applied in the study of Zanthoxylum . Li et al. [ 9 ] conducted the first genome-wide survey of Zanthoxylum and used 36 Genomic-SSR (G-SSR) markers, which demonstrated polymorphism, to classify 15 Zanthoxylum cultivars into two categories. Using three candidate DNA barcode regions (ITS2, ETS, and trnH psbA), Zhao et al. [ 10 ] identified 69 materials representing 13 Chinese pepper species. Feng et al. on the other hand, employed SRAP [ 3 ], chloroplast DNA (cpDNA) [ 4 ], EST-SSR [ 11 ], ISSR [ 12 ], and SNP [ 13 ] markers to analyze the genetic diversity, phylogenetic relationships, and genetic structure of Zanthoxylum species. Although numerous SSR markers have been identified in Zanthoxylum species, their potential for use in identifying Zanthoxylum germplasm resources has not been validated.
iPBS (inter-Primer Binding Site), proposed in 2010 by Kalendar et al. [ 14 ], is a novel molecular marker technology for polymorphism amplification based on reverse transcription transposon sequences. Compared to other molecular marker techniques, iPBS does not require sequence information or primer design in advance. The detection of produced markers can be achieved through agarose gel electrophoresis, a simple, fast, and cost-effective method. The primers used in iPBS are versatile and can be utilized in a wide range of plants and animals. Moreover, iPBS exhibits high polymorphism and reproducibility [ 14 , 15 ]. As a result of these advantages, iPBS has been increasingly employed in plants for evaluating genetic diversity, as observed in grape [ 16 ], safflower ( Carthamus tinctorius ) [ 17 ], and bamboo [ 18 ] studies. However, so far, there are no reports on the application of iPBS as a molecular marker in Zanthoxylum . Notably, a study by Hu et al. [ 19 ] revealed that approximately 71.2% of the Z. armatum genome and 70.6% of the Z. bungeanum genome consisted of LTR-type reverse transcriptional transposons. Consequently, the reverse transcriptional transposon-based marker approach seems appealing as a tool for fingerprinting Zanthoxylum species.
In this study, we assessed the genetic diversity of 80 Zanthoxylum accessions using both SSR and iPBS molecular markers. Through this analysis, we constructed DNA fingerprints to provide a reference for the assessment of resources and cultivar identification of Zanthoxylum . Furthermore, this research endeavors to establish a scientific foundation for the utilization of Zanthoxylum resources and the protection of intellectual property rights.
Materials and methods
Plant materials and dna extraction.
Eighty plant samples including three Zanthoxylum species ( Z. bungeanum , Z. armatum , Z. piperitum ) were collected from the Zanthoxylum Germplasm Resource Bank in Hanyuan County, Sichuan Province (Table 1 ). The sampling process involved selecting well-growing Zanthoxylum species plants, randomly selecting 3 individual samples from each cultivar, and collecting fresh and pest-free Zanthoxylum leaves. These leaves were stored in a -80 °C freezer for future use.
List of 80 Zanthoxylum accessions used in the present study
| Code | Cultivar or common name | Abbreviation | Species | Provinance |
|---|---|---|---|---|
| 1 | Hanchengdangcunwuci | HCDCWC | Shanxi | |
| 2 | Hanchengxiaohongpao | HCXHP | Shanxi | |
| 3 | Hanchengyexuanyihao | HCYXYH | Shanxi | |
| 4 | Hanchengwuci | HCWC | Shanxi | |
| 5 | Hanchenghuajiao | HCHJ | Shanxi | |
| 6 | Hanchengwuciyihao | HCWCYH | Shanxi | |
| 7 | Fengxiandahongpao | FXDHP | Shanxi | |
| 8 | Gelaoxibeinongyehuajiao | GLXBNYHJ | Shanxi | |
| 9 | Fuguhuajiao | FGHJ | Shanxi | |
| 10 | Xingqinyihao | XQYH | Shanxi | |
| 11 | Xingqinerhao | XQEH | Shanxi | |
| 12 | Hanchengputaohuajiao | HCPTHJ | Shanxi | |
| 13 | Germany Huajiao | GHJ | Germany | |
| 14 | Guojiadahongpao | GJDHP | Gansu | |
| 15 | Qinanhuajiao | QAHJ | Gansu | |
| 16 | Xinongwuci | XNWC | Gansu | |
| 17 | Wududahongpao | WDDHP | Gansu | |
| 18 | Linxiamianjiao | LXMJ | Gansu | |
| 19 | Qinanyihao | QAYH | Gansu | |
| 20 | Longnanbayuejiao | LNBYJ | Gansu | |
| 21 | Nanqiangyihao | NQYH | Gansu | |
| 22 | Longnanqiyuejiao | LNQYJ | Gansu | |
| 23 | Bayuejiao | BYJ | Gansu | |
| 24 | Shizitou | SZT | Gansu | |
| 25 | Longnandahongpao | LNDHP | Gansu | |
| 26 | Baishajiao | BSJ | Hebei | |
| 27 | Doujiao | DJ | Gansu | |
| 28 | Xiheyoujiao | XHYJ | Gansu | |
| 29 | Hanyuanhuajiao | HYHJ | Sichuan | |
| 30 | Hanyuanwuci ♂ | HYWCXZ ♂ | Sichuan | |
| 31 | Hanyuanwuci ♀ | HYWCCZ ♀ | Sichuan | |
| 32 | Hanyuanzaoshu | HYZS | Sichuan | |
| 33 | Hanyuanwanshuyihao | HYWSYH | Sichuan | |
| 34 | Shujiaoerhao | SJEH | Sichuan | |
| 35 | Shujiaosanhao | SJSH | Sichuan | |
| 36 | Dahongpaowang | DHPW | Sichuan | |
| 37 | Mianyangwuciqinghuajiao | MYWCHJ | Sichuan | |
| 38 | Jinquanwuci | JQWC | Sichuan | |
| 39 | Yuexihuajiao | YXHJ | Sichuan | |
| 40 | Maoxianliuyuejiao | MXLYJ | Sichuan | |
| 41 | Maoxianqiyuejiao | MXQYJ | Sichuan | |
| 42 | Nanludahongpao | NLDHP | Sichuan | |
| 43 | Dahongpao | DHP | Sichuan | |
| 44 | Zanghongjiao | ZHJ | Sichuan | |
| 45 | Xizanghuajiao | XZHJ | Xizang | |
| 46 | Laiwuxiaohongpao | LWXHP | Shandong | |
| 47 | Laiwudahongpao | LWDHP | Shandong | |
| 48 | Jiningzouchenghuajiao | JNZCHJ | Shandong | |
| 49 | Hebeiwuci | HBWC | Hebei | |
| 50 | Hebeixinglonghuajiao | HBXLHJ | Hebei | |
| 51 | Hebeizhengluhuajiao | HBZLHJ | Hebei | |
| 52 | Linzhouhonghuajiao | LZHHJ | Henan | |
| 53 | Pingshundahongpao | PSDHP | Shanxi | |
| 54 | Ruichenghuajiao | RCHJ | Shanxi | |
| 55 | Zhenxiongxuejiao | ZXXJ | Yunnan | |
| 56 | Zhenxionghuajiao | ZXHJ | Yunnan | |
| 57 | Zhaotongdahongpao | ZTDHP | Yunnan | |
| 58 | Jinjiangyihao | JJYH | Sichuan | |
| 59 | Neijiangqinghuajiao | NJQHJ | Sichuan | |
| 60 | Meishanqinghuajiao | MSQHJ | Sichuan | |
| 61 | Hanyuanputaoqingjiao | HYPTQJ | Sichuan | |
| 62 | Pengxiqinghuajiao | PXQHJ | Sichuan | |
| 63 | Hongyatengjiao | HYTJ | Sichuan | |
| 64 | Jinyangqinghuajiao | JYQHJ | Sichuan | |
| 65 | Guanganqinghuajiao | GAQHJ | Sichuan | |
| 66 | Qingjinyihao | QJYH | Sichuan | |
| 67 | Yaojiao | YJ | Sichuan | |
| 68 | Cijiao | CJ | Sichuan | |
| 69 | Zhaotongzhuyejiao | ZTZYJ | Yunnan | |
| 70 | Wucitengjiao | WCTJ | Chongqing | |
| 71 | Jiuyeqinghuajiao | JYQHJ | Chongqing | |
| 72 | Huapinghuajiao | HPHJ | Yunnan | |
| 73 | Yongqingyihao | YQYH | Yunnan | |
| 74 | Luqingyihao | LQYH | Yunnan | |
| 75 | Putaoshanjiao | PTSJ | Japan | |
| 76 | Zhaocangshanjiao | ZCSJ | Japan | |
| 77 | Liujinshanjiao | LJSJ | Japan | |
| 78 | Japan Wuciyihao | JWCYH | Japan | |
| 79 | Huashanjiao | SHJ | Japan | |
| 80 | Zhaocangshanjiao ♂ | ZCSJ ♂ | Japan |
Following the method outlined by Porebski et al. [ 20 ], DNA was extracted using a modified CTAB method. The concentration and purity of the extracted DNA were subsequently assessed using a NanoDrop One Ultra-Micro UV Spectrophotometer (Thermo Fisher Scientific Inc., USA). The integrity of the DNA was verified through 1% agarose gel electrophoresis. The DNA was uniformly diluted to a concentration of 100 ng/µl and stored in a -40 °C refrigerator as a backup.
SSR primer screening and PCR amplification
Six hundred pairs of primers were selected from the G-SSR primers developed in the previous stage of our group, containing dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, hexanucleotide and complex types of SSR sites, and all of them were tested for specificity and synthesized by Sangon Biotech (Shanghai) Co., Ltd. These primers were used to amplify DNA from seven Zanthoxylum accessions (FGHJ, HYWCXZ ♂, SJSH, NLDHP, YJ, WCTJ, JYQHJ) (Table 1 ) that exhibited significant morphological differences. Primers with clear target bands, simple band types, and high polymorphism were selected.
PCR reaction system (25 µL): 3G Taq Master Mix for PAGE (Red Dye) (Nanjing Vazyme Biotech Co., Ltd.)12.5 µL; forward and reverse primers: 1.0 µL (10 pmol/L); DNA 100 ng; fill with ddH 2 O to 25.0 µL. PCR amplification was performed using Touchdown PCR method, with a reaction procedure of pre-denaturation at 95℃ for 6 min; denaturation at 95 °C for 15 s, annealing at 64 °C for 15 s (thereafter, cycling at 64 °C ∼ 54 °C for every 2 °C decrease until 54 °C), and extension at 72 °C for 30 s; denaturation at 95 °C for 15 s, annealing at 54 °C for 15 s, extension at 72 °C for 30 s, and cycling 25 times; extend at 72 °C for another 5 min and stored at 4 °C.
PCR products were detected by 10% nondenaturing polyacrylamide gel electrophoresis at 185 V for 130 min. After silver staining and color development, they were photographed with a camera.
iPBS primer screening and PCR amplification
Eighty-three iPBS primers published by Kalendar et al. [ 14 ] were synthesized, and these primers were amplified by PCR using DNA from Zanthoxylum accessions (FGHJ, HYWCXZ ♂, SJSH, NLDHP, YJ, WCTJ, JYQHJ) (Table 1 ) that exhibited significant morphological differences, and those with clear amplified bands, high polymorphism, and high stability were selected. 83 iPBS primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd.
PCR reaction system (25 µL): 2 × Rapid Taq Master Mix (Nanjing Vazyme Biotech Co., Ltd.) 12.5 µL, iPBS primer 1.0 µL (10 pmol/L), ddH 2 O 10.5 µL, DNA 1.0 µL. Reaction procedure: Pre-denaturation at 95 °C for 6 min; denaturation at 95 °C for 15 s, annealing at 39.0 ∼ 65.0 °C for 30 s, extension at 72 °C for 1 min, 32 cycles; complete extension at 72 °C for 5 min, stored at 4 °C.
PCR products were detected by 1.2% agarose gel electrophoresis at 100 V for 28 min, and photographed by a gel imaging system at the end of electrophoresis.
Data statistics and analysis
The bands in the SSR and iPBS electrophoresis profiles were counted using Excel 2019 and assigned corresponding “1” or “0” values based on the presence or absence of bands, respectively. These data were used to create a two-dimensional matrix of “0, 1”.
For SSR markers, the data formats were converted using DataFormater software [ 21 ]. Genetic parameters such as number of observed alleles ( Na ), number of effective alleles ( Ne ), Shannon’s information index ( I ), expected heterozygosity ( He ), observed heterozygosity ( Ho ), fixed coefficient of population genetic differentiation ( Fst ), gene flow ( Nm ), probability of identity ( PI ), and probability of identity among siblings ( PIsibs ) were computed by GenAlex 6.503 software [ 22 ]; and the test materials were principal coordinate analysis (PCoA) and analysis of molecular variance (AMOVA) were performed. The polymorphism information content ( PIC ) of SSR primers was calculated using PIC-Calc 0.6 software. Genetic similarity coefficients ( GS ) among the test materials were calculated using NTSYS-pc 2.1 software [ 23 ], and the unweighted pair-group method with arithmetic means (UPGMA) in the SAHN module was used for cluster analysis and construction of dendrograms. Population structure analysis was performed by Structure 2.3.2 software [ 24 ] with the following parameters: Length of Burin Period = 50,000, Number of MCMC Reps after Burnin = 100,000, K = 1 ∼ 10, and 5 replications for each K value; the results were uploaded to the Structure Harvester website ( https://taylor0.biology.ucla.edu/structureHarvester/ ) to determine the optimal K value; the results corresponding to the optimal K value were subsequently analyzed by repeated sampling through the CLUMPP program; and finally visualized using the distrut program.
For iPBS markers, observed alleles ( Na ), number of effective alleles ( Ne ), Shannon’ s information index ( I ), and Nei’ s gene diversity ( H ) were calculated for amplified loci and populations by PopGene 1.32 software [ 25 ]; PCoA and cluster analysis based on UPGMA method were performed using NTSYS-pc 2.1 software. Since the iPBS markers are dominant markers, the PIC was calculated with reference to the method of Hinze et al. [ 26 ]: PIC i = 1 - ( p 2 + q 2 ), where p is the frequency of “1” appearing in the i-th band of the primer and q is the frequency of “0” appearing in the i-th band of the primer; when p = q = 0.5, the PIC value of the dominant marker is the largest (0.5), and the polymorphism of the primer is the highest.
Construction of DNA fingerprint
The SSR primers for constructing fingerprints were screened according to the following conditions: (1) The amplified bands are clear, and the results are stable and reproducible; (2) Primers with high PIC and low PI values; (3) The principle of identifying the most materials with the fewest number of primers was followed; (4) Ensure the uniqueness of the fingerprint of each accession.
The band information amplified by each primer was recorded in Excel 2019 using “0”, “1”, and “9” to signify “no band,” “with band,” and “no amplification,“, respectively, to form a digital fingerprint map. Subsequently, the information (name, Latin name, cultivar types, provenance) of each Zanthoxylum accession was integrated with its fingerprint code and imported into the “Caoliao QR Code” online software ( https://cli.im/ ) to generate QR codes for the fingerprints of 80 Zanthoxylum cultivars.
SSR primer screening and genetic diversity of the markers
A total of 32 pairs of polymorphic SSR primers (Supplementary Table S1 ) were screened from 600 pairs of primers using seven Zanthoxylum accessions with significant morphological differences. These primers were subsequently used to amplify all peppercorn samples.
A total of 206 ( Na ) of the 32 pairs of SSR primers were detected in 80 Zanthoxylum accessions. The average number of alleles detected per pair of primers ranged from 3.000 (D27, T6) to 11.000 (P.17), with an average value of 6.438 (Table 2 ). This finding suggested that the tested Zanthoxylum accessions exhibit relatively abundant allelic variation. The number of effective alleles ( Ne ) varied from 1.648 (P4.2) to 6.181 (D86), with a mean value of 3.254. Observed heterozygosity ( Ho ) and expected heterozygosity ( He ) values indicate the magnitude of genetic variance for different SSR primers, with higher Ho values indicating higher heterozygosity. Among the 32 markers, the Ho values ranged from 0.225 (D112) to 0.950 (D39), and the He values ranged from 0.393 (P4.2) to 0.838 (D86). The mean values for Ho and He were 0.638 and 0.661, respectively. Shannon’s information index ( I ) varied from 0.677 (P4.2) to 1.937 (D86), with a mean value of 1.336. These results indicate that the tested Zanthoxylum materials exhibit a high degree of genetic variation and rich genetic diversity.
The genetic diversity statistics of 32 SSR markers in 80 Zanthoxylum accessions
| Marker ID | Na | Ne | I | Ho | He | Nm | PIC | PI | PIsibs |
|---|---|---|---|---|---|---|---|---|---|
| D11 | 7.000 | 4.441 | 1.597 | 0.613 | 0.775 | 0.452 | 0.827 | 0.086 | 0.384 |
| D23 | 10.000 | 4.385 | 1.749 | 0.821 | 0.772 | 0.346 | 0.706 | 0.079 | 0.384 |
| D27 | 3.000 | 1.875 | 0.686 | 0.675 | 0.467 | 0.457 | 0.657 | 0.388 | 0.614 |
| D39 | 6.000 | 3.742 | 1.496 | 0.950 | 0.733 | 1.299 | 0.735 | 0.108 | 0.411 |
| D49 | 7.000 | 2.584 | 1.240 | 0.600 | 0.613 | 1.527 | 0.738 | 0.193 | 0.492 |
| D50 | 6.000 | 3.053 | 1.329 | 0.797 | 0.672 | 0.801 | 0.710 | 0.150 | 0.451 |
| D79 | 6.000 | 2.550 | 1.205 | 0.663 | 0.608 | 0.438 | 0.649 | 0.192 | 0.494 |
| D81 | 6.000 | 4.452 | 1.611 | 0.900 | 0.775 | 3.964 | 0.793 | 0.084 | 0.383 |
| D86 | 8.000 | 6.181 | 1.937 | 0.688 | 0.838 | 0.355 | 0.785 | 0.046 | 0.342 |
| D93 | 5.000 | 3.579 | 1.385 | 0.465 | 0.721 | 0.206 | 0.813 | 0.127 | 0.421 |
| D106 | 4.000 | 2.321 | 1.062 | 0.588 | 0.569 | 0.247 | 0.665 | 0.234 | 0.524 |
| D111 | 4.000 | 2.463 | 1.027 | 0.600 | 0.594 | 0.665 | 0.621 | 0.249 | 0.515 |
| D112 | 4.000 | 1.718 | 0.750 | 0.225 | 0.418 | 0.363 | 0.484 | 0.389 | 0.638 |
| F31 | 6.000 | 3.923 | 1.520 | 0.888 | 0.745 | 0.290 | 0.777 | 0.104 | 0.404 |
| F84 | 8.000 | 2.949 | 1.336 | 0.675 | 0.661 | 0.246 | 0.819 | 0.165 | 0.461 |
| F86 | 5.000 | 2.629 | 1.139 | 0.800 | 0.620 | 0.441 | 0.722 | 0.214 | 0.494 |
| T16 | 3.000 | 2.205 | 0.886 | 0.300 | 0.547 | 0.063 | 0.649 | 0.294 | 0.550 |
| T78 | 6.000 | 3.188 | 1.375 | 0.688 | 0.686 | 0.300 | 0.709 | 0.142 | 0.442 |
| T83 | 4.000 | 2.101 | 0.951 | 0.575 | 0.524 | 0.212 | 0.795 | 0.284 | 0.559 |
| T86 | 10.000 | 3.740 | 1.660 | 0.658 | 0.733 | 0.213 | 0.710 | 0.100 | 0.409 |
| N63 | 7.000 | 3.571 | 1.401 | 0.603 | 0.720 | 0.289 | 0.800 | 0.129 | 0.422 |
| N76 | 5.000 | 2.732 | 1.194 | 0.861 | 0.634 | 0.631 | 0.656 | 0.184 | 0.479 |
| P3.16 | 6.000 | 3.236 | 1.362 | 0.550 | 0.691 | 0.301 | 0.632 | 0.147 | 0.441 |
| P4.2 | 5.000 | 1.648 | 0.677 | 0.413 | 0.393 | 1.442 | 0.400 | 0.428 | 0.660 |
| P4.11 | 9.000 | 4.967 | 1.782 | 0.608 | 0.799 | 0.368 | 0.803 | 0.069 | 0.368 |
| P4.17 | 11.000 | 4.110 | 1.774 | 0.632 | 0.757 | 0.398 | 0.746 | 0.084 | 0.393 |
| P4.19 | 10.000 | 4.778 | 1.795 | 0.538 | 0.791 | 0.289 | 0.739 | 0.072 | 0.373 |
| P5.10 | 6.000 | 3.421 | 1.399 | 0.658 | 0.708 | 0.239 | 0.572 | 0.135 | 0.430 |
| P6.20 | 7.000 | 3.365 | 1.505 | 0.411 | 0.703 | 0.154 | 0.806 | 0.119 | 0.428 |
| P6.27 | 7.000 | 2.553 | 1.294 | 0.810 | 0.608 | 1.796 | 0.703 | 0.186 | 0.492 |
| P6.30 | 6.000 | 3.353 | 1.350 | 0.808 | 0.702 | 1.131 | 0.807 | 0.143 | 0.435 |
| C6 | 9.000 | 2.325 | 1.279 | 0.367 | 0.570 | 0.206 | 0.676 | 0.209 | 0.517 |
| Total | 206.000 | 104.140 | 42.751 | 20.423 | 21.145 | 20.127 | 22.706 | 0.086 | 0.384 |
| Mean | 6.438 | 3.254 | 1.336 | 0.638 | 0.661 | 0.629 | 0.710 | 0.079 | 0.384 |
Na : Number of observed alleles; Ne : Number of effective alleles; I : Shannon’s Information Index; Ho : Observed heterozygosity; He : Expected heterozygosity; Nm : Gene flow; PIC : Polymorphic information content; PI : probability of identity; PIsibs : probability of identity among siblings
The PIC values of the 32 pairs of primers ranged from 0.400 (P4.2) to 0.827 (D11), with an average of 0.710. There were 30 pairs of primers with PIC values > 0.5, indicating that the screened primers had high polymorphism. These primers can effectively reveal the genetic diversity of the tested Zanthoxylum accessions and are suitable for DNA fingerprinting.
Genetic relationship and cluster analysis of Zanthoxylum based on SSR markers
Genetic similarity coefficients ( GS ) are commonly used to evaluate the extent of genetic similarity among individuals. In this study, the genetic similarity coefficient matrix of 80 Zanthoxylum accessions was obtained using NTSYS-pc 2.1 software (Supplementary Figure S1 ). The GS values ranged from 0.0947 to 0.9868, with an average of 0.3864, indicating noticeable variation in the genetic backgrounds of the test materials. Notably, the GS value between ‘JJYH’ and ‘ZHJ’ was the smallest (0.0947), indicating that these two plants had the highest genetic variation and the furthest genetic relationship. Conversely, the GS value between ‘LZHHJ’ and ‘BSJ’ was the largest (0.9868), indicating that these two plants had very close genetic relationships. Additionally, the frequency distribution of the 3160 GS s obtained from the two-by-two comparison of the test samples revealed that the majority of the GS s fell within the range of 0.1 to 0.5, accounting for 77.5% of the total (Supplementary Figure S2 ). Among them, the largest number of Zanthoxylum accessions had GS values ranging from 0.1 to 0.2, accounting for 26.17% of the total. Overall, these results indicate that the 80 Zanthoxylum accessions possess a diverse range of genetic characteristics and a broad genetic background.
The cluster analysis results demonstrated that using 32 SSR markers, it was possible to completely distinguish the 80 Zanthoxylum accessions (Fig. 1 ). With a GS threshold of 0.2217, the test accessions could be classified into three classes (I, II, and III). Class I consisted of 57 Z. bungeanum accessions, class II consisted of 17 Z. armatum accessions, and class III consisted of 6 Z. piperitum accessions. It is worth noting that “MYWCQHJ” (37) and “YJ” (67) in class II aggregate into a subclass at a GS value of 0.348. After calculation, it was found that the average GS values of “MYWCQHJ” and “YJ” with the other 15 Z. armatum accessions were 0.356 and 0.365, respectively, indicating that they have a distant genetic relationship with other Z. armatum accessions. Similarly, in class I, “HYWC ♂” (30) and " HYWC ♀” (31) clustered into a subclass at a GS of 0.312, showing a distant relationship with other Z. bungeanum accessions. Furthermore, we noticed that the GS values of “BSJ” (26) from Hebei and “LZHHJ” (52) from Henan amounted to 0.987, suggesting minimal genetic differences and a possible case of synonymy. Additionally, certain Zanthoxylum accessions from different source areas are clustered together, such as “LNDHP” (25) from Gansu and “MXLYJ” (40) from Sichuan, as well as “DJ” (27) from Gansu and “RCHJ” (54) from Shanxi. This clustering may be attributed to the frequent trade and introductions of Zanthoxylum between various regions. Moreover, the high correlation coefficient (0.977) calculated using the matrix comparison plot module of NTSYS-pc 2.1 software indicates the accuracy of the clustering results.

UPGMA clustering tree of 80 Zanthoxylum accessions based on SSR markers
Genetic diversity and differentiation of the Zanthoxylum population based on SSR markers
In this study, a total of 80 Zanthoxylum accessions were categorized into three populations based on the species: Z. bungeanum (Pop1), Z. armatum (Pop2), and Z. piperitum (Pop3). The genetic diversity analysis revealed that, among all three populations, Pop1 exhibited the highest Na , Ne , Ha , He , and I value (Table 3 ), suggesting that Pop1 possessed the highest genetic diversity. Pop2 had the second highest level, while Pop3 had the lowest. The coefficient of genetic differentiation ( Fst ) between the populations was calculated, yielding Fst values of 0.242 for Pop1 and Pop2, 0.335 for Pop1 and Pop3, and 0.429 for Pop2 and Pop3. The mean Fst was 0.335 ( Fst > 0.25), indicating significant genetic differentiation between the three populations. AMOVA further demonstrated that genetic variation in Zanthoxylum species existed mainly within individuals (65%), with relatively little variation between populations (35%) (Table 4 ). Additionally, the average Nm was 0.629 (Table 2 ), suggesting limited gene exchange among individuals within each population, potentially attributed to the phenomenon of apomixis in Zanthoxylum species.
The genetic diversity statistics among 3 populations of Zanthoxylum species
| Pop | Na | Ne | I | Ho | He |
|---|---|---|---|---|---|
| Pop1 | 4.833 | 2.447 | 1.012 | 0.654 | 0.544 |
| Pop2 | 3.694 | 1.982 | 0.769 | 0.556 | 0.415 |
| Pop3 | 1.611 | 1.342 | 0.283 | 0.227 | 0.173 |
| Total | 10.139 | 5.771 | 2.064 | 1.437 | 1.132 |
| Mean | 3.380 | 1.924 | 0.688 | 0.479 | 0.377 |
Na : Number of observed alleles; Ne : Number of effective alleles; I : Shannon’s Information Index; Ho : Observation of heterozygosity; He : Expectation of heterozygosity
The AMOVA of 3 populations of Zanthoxylum species
| Source of variance | df | SS | MS | Variance component | Variation percentage % | value |
|---|---|---|---|---|---|---|
| Among Pops | 2 | 417.851 | 208.925 | 5.702 | 35% | < 0.001 |
| Within Indiv | 80 | 857.500 | 10.719 | 10.719 | 65% | < 0.001 |
| Total | 82 | 1275.351 | - | 16.421 | 100% | - |
df: Degrees of freedom; SS: Sum of squares; MS: mean square
Furthermore, Nei’s genetic distance and genetic concordance study revealed that the genetic distance among the populations ranged from 0.854 to 1.190, with a mean value of 0.972. The genetic concordance ranged from 0.304 to 0.426, with a mean value of 0.383 (Table 5 ), indicating low genetic similarity and a high degree of genetic differentiation among the three populations. Pop2 and Pop3 exhibited the greatest genetic distance, representing the most distant relationship, whereas Pop1 and Pop2 displayed the smallest genetic distance, indicating a more recent relationship.
Unbiased estimation of Nei’s genetic distance and genetic identity in 3 populations of Zanthoxylum species
| Pop | Pop1 | Pop2 | Pop3 |
|---|---|---|---|
| Pop1 | - | 0.854 | 0.872 |
| Pop2 | 0.426 | - | 1.190 |
| Pop3 | 0.418 | 0.304 | - |
Note: The upper right data represents Nei genetic distance, while the lower left data represents Nei genetic identity
Principal coordinate analysis indicated that the first two principal coordinates accounted for 46.12% of the genetic variation among the 80 Zanthoxylum accessions. Principal coordinate 1 explained 31.71% of the variation, while principal coordinate 2 accounted for 14.41% (Fig. 2 ). The analysis classified the 80 Zanthoxylum accessions into three groups: the first group included 57 accessions of Z. bungeanum , the second group comprised 17 accessions of Z. armatum , and the third group consisted of 6 accessions of Z. piperitum . These findings were consistent with the results obtained from cluster analysis.

Principal coordinate analysis of 3 populations of Zanthoxylum species based on SSR markers
Population structure analysis of Zanthoxylum based on SSR markers
In order to understand the genetic background and gene penetration of 80 Zanthoxylum accessions, the population structure of the test materials was analyzed by Structure software based on Bayesian modeling and the Q-values (Supplementary Table S2 ) (Pritchard et al., 2000) (probability that the i-th material has its genomic variation originating from the k-th subgroup) was counted. The results showed that Delta K has an optimal value when K = 2 (Fig. 3 ), therefore, the 80 Zanthoxylum accessions can be classified into 2 groups: Pop1 (blue) and Pop2 (orange) (Fig. 4 ); where Pop1 includes 63 accessions, mainly Z. bungeanum and Z. piperitum , and Pop2 includes 17 accessions, mainly Z. armatum .

Delta K values for different numbers of populations assumed (K) in the STRUCURE analysis

Population genetic structure of 80 Zanthoxylum accessions. Each rectangular column in the figure represents one accession, and the color and color scale of the columns represent the subpopulation to which it belongs and the proportion of the subpopulation it occupies (Blue represents Pop1, and Orange represents Pop2). The number on the X-axis is the accession number
Of the 80 Zanthoxylum accessions, 69 had Q-values ≥ 0.8, with a mean value of 0.99, indicating that these materials were from a single source, with a simple genetic background and a lack of genetic exchange between subgroups; 11 accessions had Q-values < 0.8 with a mean value of 0.66, suggesting that these materials possessed a mixed origin with a relatively complex genetic composition.
Fingerprinting power of SSR markers and DNA fingerprint construction
PI is an important parameter for assessing the fingerprinting power of molecular markers, with lower values indicating higher fingerprinting efficiency of the markers [ 27 ]. According to the results in Table 2 , the PI values of the 32 SSR markers ranged from 0.046 (D86) to 0.428 (P4.2), with an average value of 0.173. Assuming that all loci segregate independently, the probability of finding two random individuals with identical genotypes at the 32 marker loci is estimated to be 4.265 × 10 − 27 , i.e., it is almost impossible to find two different individuals with identical genotypes, suggesting that the markers developed in this study have strong fingerprinting power. PIsibs is considered to be the upper limit of PI [ 28 ], and the range of PIsibs values for the 32 SSR markers was 0.342 (D86) to 0.660 (P4.2), and the PIsibs value for all marker combinations was 1.282 × 10 − 11 .
Based on these results, combined with the results of primer amplification, eight SSR markers (D11, D23, D49, D81, D86, N63, P4.11, P4.17) with low PI values (the average value was 0.096) were screened to compose a core set of markers used to construct the fingerprinting of Zanthoxylum . Through the combination of these eight markers, 80 fingerprinting profiles with unique correspondences were obtained. The digital codes of 80 Zanthoxylum cultivars and their corresponding cultivar types, seed source locations and other information were merged to generate a QR code for fingerprinting (Fig. 5 ).

Fingerprint information of 80 Zanthoxylum cultivars based on SSR markers
iPBS primer screening and analysis of primer polymorphisms
Ten iPBS primers with high polymorphism and clear banding patterns were selected from a pool of 83 primers for analysis of genetic diversity in the 80 Zanthoxylum accessions (Supplementary Table S3 ).
A total of 127 bands were amplified from the ten selected primers, 120 of which were found to be polymorphic (Table 6 ). The number of bands per primer ranged from 4 to 21, with an average of 12.7 bands. The polymorphism ratio per primer ranged from 75 to 100%, with an average of 93.1%. The PIC values of the primers ranged from 0.201 to 0.324, with an average of 0.281. Notably, primer 2242 exhibited the highest level of polymorphism, with a PIC value of 0.324, while primer 2083 had the lowest level, with a PIC value of 0.201.
The amplification results and genetic diversity index of 80 Zanthoxylum accessions by 10 iPBS primers
| Primer | T | PPL (%) | PIC | Na | Ne | H | I | |
|---|---|---|---|---|---|---|---|---|
| 2083 | 7 | 7 | 100.0 | 0.201 | 2.0000 | 1.2456 | 0.1762 | 0.3030 |
| 2085 | 12 | 11 | 91.7 | 0.229 | 1.9167 | 1.2841 | 0.1928 | 0.3212 |
| 2222 | 16 | 13 | 81.3 | 0.294 | 1.8125 | 1.3923 | 0.2389 | 0.3707 |
| 2242 | 21 | 21 | 100.0 | 0.324 | 2.0000 | 1.4490 | 0.2704 | 0.4187 |
| 2243 | 16 | 16 | 100.0 | 0.312 | 2.0000 | 1.3966 | 0.2432 | 0.3804 |
| 2245 | 18 | 17 | 94.4 | 0.300 | 1.9444 | 1.4494 | 0.2737 | 0.4223 |
| 2271 | 14 | 14 | 100.0 | 0.282 | 2.0000 | 1.4129 | 0.2472 | 0.3861 |
| 2375 | 4 | 3 | 75.0 | 0.319 | 1.7500 | 1.4051 | 0.2390 | 0.3643 |
| 2380 | 9 | 8 | 88.9 | 0.287 | 1.8889 | 1.3538 | 0.2261 | 0.3579 |
| 2398 | 10 | 10 | 100.0 | 0.262 | 2.0000 | 1.3707 | 0.2381 | 0.3782 |
| Total | 127 | 120 | - | 2.811 | 19.3125 | 13.7595 | 2.3456 | 3.7028 |
| Mean | 12.7 | 12 | 93.1 | 0.281 | 1.9313 | 1.3760 | 0.2346 | 0.3703 |
T : Total number of amplified bands; N : Number of polymorphic bands; PPL : Polymorphism ratio; PIC : Polymorphic information content; Na : Number of observed alleles; Ne : Number of effective alleles; H : Nei’s genetic diversity; I : Shannon’s Information Index
Genetic diversity analysis of Zanthoxylum based on iPBS markers
The genetic diversity indices of the 80 Zanthoxylum accessions were calculated with PopGene 1.32 software (Table 6 ), and the results showed that the mean values of Na, Ne, H and I were 1.9313, 1.3760, 0.2346 and 0.3703, respectively, indicating that the genetic variation among the 80 Zanthoxylum accessions was relatively high.
Genetic similarity coefficient matrices of 80 Zanthoxylum accessions were obtained via NTSYS-pc 2.1 software (Supplementary Figure S3 ). GS varied from 0.2206 to 1.0000, with an average of 0.5215; among them, the GS values of ‘MSQHJ’ and ‘HYWC ♂’, and ‘WCTJ’ and ‘HYWC ♂’ were all 0.2206, which indicated that they were the most distantly related. There were five groups of Zanthoxylum accessions with GS values of 1; these results, in combination with the SSR marker results, indicated that these materials were very close to each other and had highly similar genetic backgrounds; on the other hand, these results also indicated that the 10 iPBS markers in this study had limited discriminatory ability. Statistics on the frequency distribution of GS values of the test materials were found (Supplementary Figure S4 ), and the GS values were mainly distributed between 0.3 and 0.7, accounting for 74.56%, with the largest number of Zanthoxylum samples with GS values between 0.3 and 0.4 accounting for 27.09%.
Cluster analysis of Zanthoxylum based on iPBS markers
Based on the matrix of genetic similarity coefficients, a dendrogram depicting iPBS marker clustering of 80 Zanthoxylum accessions was constructed using the UPGMA method (Fig. 6 ). The analysis revealed that these 80 Zanthoxylum accessions could be categorized into three distinct groups, Group I, Group II, and Group III, representing Z. bungeanum , Z. armatum , and Z. piperitum , respectively, with a GS threshold of 0.3683. Notably, ‘MYWCQHJ’ did not cluster within any group associated with Z. armatum . This phenomenon may be attributed to two factors. First, this could be due to the limited number of iPBS markers utilized in this study. Second, this difference might be attributed to the unique characteristics of the ‘MYWCQHJ’ cultivar itself, as evidenced by its separate clustering within Group I. The correlation coefficient, computed using the Matrix comparison plot module in the NTSYS-pc 2.1 software, was found to be 0.966, underscoring the high accuracy of the clustering results.

UPGMA clustering tree of 80 Zanthoxylum accessions based on iPBS markers
Furthermore, the principal coordinate analysis results concurred with the cluster analysis results. The 80 Zanthoxylum accessions were divided into three distinct categories (Fig. 7 ): the first category consisted of one accession of Z. armatum (‘MYWCQHJ’), 57 accessions of Z. bungeanum , the second category comprised 16 accessions of Z. armatum , and the third category included 6 accessions of Z. piperitum . This alignment between the two analyses strengthens the validity of the obtained classifications.

Principal coordinate analysis of 80 Zanthoxylum accessions based on iPBS markers
Genetic and cluster analysis of Zanthoxylum based on SSR + iPBS markers
The genetic similarity coefficient matrix (Supplementary Figure S5 ) and clustering tree diagram (Fig. 8 ) were constructed through the integration of SSR and iPBS molecular marker data. The finding revealed that among the 80 Zanthoxylum accessions, the GS ranged from 0.1747 to 0.9921, with an average value of 0.4422. This indicates a significant disparity in the genetic backgrounds of the accessions. It should be noted that ‘HYWC ♂’ and ‘MSQHJ’ exhibited the lowest GS values (0.1747), while ‘BSJ’ and ‘LZHHJ’ demonstrated the highest GS values (0.9921). Among the Z. bungeanum species, ‘HYWC ♂’ and ‘XZHJ’ had the smallest GS values (0.3072), while ‘BSJ’ and ‘LZHHJ’ had the highest GS values (0.9921). In the case of Z. armatum , ‘MYWCQHJ’ and ‘LQYH’ had the smallest GS values (0.3611), while ‘MSQHJ’ and ‘WCTJ’ had the largest GS values (0.9833). Finally, within the Z. piperitum category, ‘JWCYH’ and ‘HSJ’ had the lowest GS values (0.6837), while ‘ZCSJ’ and ‘ZCSJ ♂’ had the highest GS values (0.9878).

UPGMA clustering tree of 80 Zanthoxylum accessions based on SSR + iPBS markers
Upon reaching a GS of 0.2657, the 80 Zanthoxylum accessions were divided into three classes: Class I represented Z. bungeanum , Class II represented Z. armatum , and Class III represented Z. piperitum . At a GS of 0.4856, Class I could be further divided into five subclasses. The first subclass comprised 43 Z. bungeanum cultivars, including all the accessions from Shaanxi (12/12), nearly all the accessions from Gansu (13/14), and almost half of the accessions from Sichuan (8/17). These three provinces are geographically close to each other and are major areas for Zanthoxylum production. The mixing of Zanthoxylum cultivars from these regions could be attributed to frequent introductions and resource exchange. Additionally, the first subclass included three cultivars from southwestern Yunnan and a few Zanthoxylum cultivars from northern regions, such as Hebei, Henan, Shandong, and Shanxi. The second subclass comprised eight Zanthoxylum cultivars, five from Sichuan, two from Hebei, and one from Gansu. The third subclass included ‘LWDHP’ and ‘LWXHP’ from Shandong and ‘PSDHP’ from Shanxi. The fourth subclass consisted of two special cultivars, ‘HYWC ♂’ and ‘HYWC ♀’, while the remaining ‘ZHJ’ accessions formed a separate fifth subclass. In Class II, ‘MYWCQHJ’ and ‘YJ’ were found to be distantly related to the other Z. armatum accessions and clustered into separate subclasses with GS values of 0.3909 and 0.4917, respectively. Overall, the clustering results revealed that the Z. bungeanum and Z. armatum cultivars from various source locations exhibited some mixing and were not exclusively clustered based on geographic differences. Clustering analysis utilizing only SSR or iPBS markers also confirmed this phenomenon. Conversely, combining the results of both markers provided a more accurate classification and effectively represented the genetic relationships among the tested Zanthoxylum accessions.
Genetic diversity of Zanthoxylum
Genetic diversity serves as the foundation for the long-term survival and evolutionary advancement of species. The extent of genetic diversity within a species determines its evolutionary potential and ability to withstand adverse environmental factors [ 29 ]. In the case of plants, research on genetic diversity is crucial for comprehending the level of genetic variation and genetic structure within species. This serves as a significant indicator for evaluating the genetic potential of germplasm resources. Additionally, these findings could lead to resource utilization, germplasm innovation, and varietal improvement while also providing recommendations for resource conservation and management [ 30 , 31 ].
Molecular markers represent an effective method for studying species genetic diversity. There are various types of molecular markers with different characteristics. By combining different molecular markers, researchers can examine different segments of the genome, thereby enhancing the coverage and uniformity of polymorphic loci. This approach compensates for any limitations and drawbacks associated with using a single type of molecular marker, enabling researchers to gain a comprehensive understanding of the species’ genetic information and enhancing the credibility of their findings [ 32 ].
The aim of this study was to assess the genetic diversity and relatedness among 80 Zanthoxylum accessions using SSR and iPBS molecular markers. SSR molecular markers are known for their superior variability and broad distribution within the genome. They are widely utilized across numerous genetic-related fields due to their codominance, high polymorphism, reproducibility, and consistent results [ 7 ]. In this study, we identified a total of 206 allelic variations among the 80 Zanthoxylum accessions using 32 selected SSR markers. Each marker displayed an average of 6.438 alleles ( Na ), an effective number of alleles ( Ne ) of 3.254, a Shannon’s information index ( I ) of 1.336, and PIC values ranging from 0.400 to 0.827, with an average of 0.710. Notably, 30 markers exhibited high polymorphism levels ( PIC > 0.5). Among the genetic diversity indices, Na and the PIC are particularly important for assessing molecular marker polymorphisms [ 33 ]. In this study, the values for these two indices were greater than those reported by Li et al. [ 9 ] ( Na = 3.5; PIC = 0.48) and Feng et al. [ 13 ] ( Na = 4.636) in Zanthoxylum . Taken together, these findings indicate that the SSR markers employed in this study exhibited overall high polymorphism, revealing the genetic diversity of the tested Zanthoxylum accessions.
Compared to SSR molecular labeling technology, iPBS molecular labeling technology offers a simpler, faster, and more cost-effective approach. Throughout this study, 10 iPBS primers were employed to amplify a total of 127 bands across the 80 Zanthoxylum accessions. The average polymorphism rate of the primers was 93.1%. The PIC values ranged from 0.201 to 0.324, with an average of 0.281, indicating a moderate level of polymorphism, consistent with research findings in Phoenix dactylifera [ 34 ] ( PIC = 0.287) and Psidium guajava [ 35 ] ( PIC = 0.287). By combining the results of both sets of molecular markers, it was observed that the genetic diversity index obtained through iPBS markers was significantly lower than that obtained through SSR markers. This finding suggested that SSR markers possess greater polymorphism and are more suitable for analyzing the genetic diversity of Zanthoxylum germplasm resources. Such disparity is likely influenced by the number of markers used in this study; utilizing 32 SSR markers increases the likelihood of detecting greater genetic variation than does the use of only 10 iPBS markers. Moreover, SSR markers are codominant markers that distinguish between pure and heterozygous genotypes, thus conferring a greater advantage in revealing species genetic diversity than dominant markers. In summary, the utilization of both molecular markers revealed a considerable level of genetic diversity within the 80 Zanthoxylum accessions.
Genetic relationship of Zanthoxylum .
The genetic similarity coefficient is a useful tool for evaluating genetic similarity. A higher genetic similarity coefficient indicates a closer genetic relationship and greater similarity between two individuals or groups, while a lower coefficient suggests greater genetic differentiation and greater genetic diversity [ 36 ]. Among the 80 Zanthoxylum accessions, the ranges of GS values obtained through the SSR, iPBS, and SSR + iPBS methods were 0.0947 ∼ 0.9868, 0.2206 ∼ 1.0000, and 0.1747 ∼ 0.9921, respectively, with statistically significant differences. The average GS values were 0.3864, 0.5215, and 0.4422, respectively, indicating relatively rich genetic diversity and a high level of genetic variation among the tested Zanthoxylum accessions. SSR markers exhibited a wider range of GS variation and smaller average GS values than did the other markers, suggesting that SSR markers are more effective at detecting genetic variation. The genetic relationships revealed by the two marker types were consistent. For instance, in the iPBS results, GS values of 1 were obtained between ‘FXDHP’ and ‘GJDHP’, ‘BSJ’ and ‘LZHHJ’, and ‘MSQHJ’ and ‘WCTJ’. These same groups also had relatively large GS values (0.9744, 0.9868, and 0.9730) according to the SSR results, indicating very close genetic relationships. This may be attributed to inconsistent naming of the same cultivar in different regions, known as the phenomenon of synonymy. In summary, both SSR and iPBS markers can be employed to assess the phylogenetic relationships of the Zanthoxylum species. However, SSR markers showed greater diversity and a more comprehensive reflection of the phylogenetic relationships, suggesting it has greater polymorphism. Additionally, SSR + iPBS markers compensated for the limitations of iPBS markers and provided a more accurate representation of the genetic relationships among the tested Zanthoxylum accessions. The cluster analysis findings also supported these conclusions. Based on the SSR, iPBS, and SSR + iPBS markers, the 80 Zanthoxylum accessions were divided into three categories ( Z. bungeanum , Z. armatum , and Z. piperitum ), and closely related Zanthoxylum species were grouped together. However, when iPBS markers were used, ‘MYWCQHJ’, which belongs to Z. armatum , was clustered with Z. bungeanum cultivars, indicating that SSR markers provided more accurate results. Furthermore, it is possible that the unique characteristics of ‘MYWCQHJ’ contributed to this clustering result, as evidenced by the presence of multiple unique loci or band patterns (Supplementary Figure S6 ). The calculated mean GS value of ‘MYWCQHJ’ compared to those of the other 16 accessions of Z. armatum was only 0.391 (based on SSR + iPBS markers), indicating a distant relationship. These findings highlight the unique genetic variation of ‘MYWCQHJ’, which may prove valuable in future efforts related to germplasm innovation and the development of new cultivars. Additionally, on the clustering tree diagrams of both markers, it was observed that some Zanthoxylum accessions from the same region were not clustered together (Fig. 8 ). These findings suggest that long-term cultivation, domestication of Zanthoxylum species, and trading and introduction between different regions may have contributed to this phenomenon. Notably, the single Zanthoxylum accession from Germany was not grouped separately but instead clustered together with Chinese Zanthoxylum , indicating a shared origin, consistent with previous research conducted by Feng [ 37 ].
Genetic differentiation and genetic structure of Zanthoxylum
Gene differentiation ( Fst ) and gene flow ( Nm ) are crucial parameters for assessing genetic variation among populations, and they exhibit an inverse correlation wherein higher differentiation coefficients indicate lower levels of gene flow [ 38 ]. For Fst , the following categories are generally utilized: Fst ranges between 0 and 0.05, which suggests negligible genetic differentiation between populations; 0.05 and 0.15, which signifies a moderate degree of genetic differentiation; 0.15 and 0.25, which indicates a substantial degree of genetic differentiation; and Fst > 0.25, which signifies a high degree of genetic differentiation [ 39 ]. For Nm , it is generally accepted that Nm > 1 indicates that there is frequent gene exchange between populations, which prevents genetic differentiation of populations due to genetic drift and contributes to the maintenance of genetic stability of populations, while Nm < 1 indicates that gene flow is not sufficient to counteract the effects of genetic drift, thus contributing to the increase of genetic differentiation between populations [ 40 ]. In this study, we used SSR markers to analyze the genetic differentiation characteristics of three Zanthoxylum populations (Pop1, Pop2, and Pop3). The Fst values were 0.242, 0.335, and 0.429 between Pop1 and Pop2, Pop1 and Pop3, and Pop2 and Pop3, respectively, suggesting a high level of genetic differentiation among the three populations. Moreover, the mean Nm was 0.629 (< 1), indicating limited gene exchange among the populations. This can be attributed to the fusionless reproductive characteristics of Zanthoxylum species and the high levels of genetic differentiation among populations, which hinder gene flow [ 37 ]. Additionally, the AMOVA results indicated a high level of genetic differentiation among the tested Zanthoxylum accessions, with genetic variation predominantly arising within individuals (65%), while 35% of the genetic variation originated from between populations. Both cluster analysis and PCoA accurately categorized the 80 Zanthoxylum accessions into three groups corresponding to the three different Zanthoxylum species populations (Pop1, Pop2, and Pop3). The genetic analysis revealed substantial genetic distance (0.972) and low genetic concordance (0.383) among these three populations, further highlighting their high level of genetic differentiation. Geographical isolation is an important factor leading to population differentiation, due to environmental heterogeneity, genetic variation, and limited gene flow, resulting in the independent evolution of populations in different geographical regions [ 13 , 41 ]. The distinct growth environments of these three groups contributed significantly to their differentiation, with Z. armatum found in frost-free regions of southwestern China characterized by warm and humid climates; Z. bungeanum exhibiting resilience and adaptability to wide areas with harsh climates (subtropical and temperate zones); mainly distributed in northern regions of the Qinling Mountains-Huaihe River in China [ 19 ]; and Z. piperitum concentrated in certain parts of Japan. Over an extended period, the combination of natural and artificial selection has limited genetic exchange between these Zanthoxylum populations, leading to significant differentiation. Generally, higher genetic diversity indicates greater complexity of plant diversity and greater potential for environmental adaptation [ 42 ]. Among the three populations, the Z. bungeanum population (Pop1) exhibited the highest genetic diversity, while the Z. piperitum population (Pop3) displayed the lowest. This discrepancy may be attributed to the number of samples and actual cultivars, as well as the stronger environmental adaptability and wider geographic distribution of Z. bungeanum . Consequently, Z. bungeanum germplasm resources can serve as crucial genetic breeding material for future cultivar selection and breeding endeavors.
Unlike the results of UPGMA cluster analysis and PCoA, Bayesian model-based population structure analysis classified the 80 Zanthoxylum accessions into two subgroups (Fig. 4 ), of which six Z. piperitum materials were not classified into a separate category. The reasons for this discrepancy have to do with the fact that the different methods take different computational approaches or provide different amounts of information [ 37 ]; on the other hand, it may be related to the small number of Z. piperitum material used in this study. Most of the 80 Zanthoxylum accessions (86%) had a single genetic component (Q-value ≥ 0.8), and only a few materials (14%) showed a mixture of both gene pools (Q-value < 0.8), suggesting a lack of genetic exchange between Zanthoxylum subgroups, which is consistent with the results of the analysis of population genetic differentiation.
Construction of DNA fingerprint map and fingerprinting power
DNA fingerprinting is a molecular-level method used to identify different biological individuals by utilizing molecular markers. It is not influenced by environmental factors or by the developmental stage of organisms. In the case of plants, DNA fingerprinting is valuable for accurately and rapidly identifying cultivars, offering convenience for germplasm resource management, evaluation, protection of cultivar rights, and crop breeding [ 43 ]. Among several molecular markers, SSR markers are widely regarded as the preferred method for constructing plant DNA fingerprints. They have been recognized as one of the most powerful marker systems for identifying plant cultivar and have been successfully applied across multiple species [ 8 , 43 ]. For instance, He et al. [ 44 ] established the genetic fingerprints of 33 standard flue-cured tobacco varieties using 48 SSR markers and developed identification technology for new tobacco varieties based on SSR markers. Chen et al. [ 43 ] created a DNA fingerprinting database of 128 excellent oil camellia cultivar using highly variable SSR markers.
PI and PIsibs are widely used as indicators of the fingerprinting power of molecular markers in studies of fingerprinting construction [ 28 , 45 ]. In this study, the combined PI value of 32 SSR markers was 4.265 × 10 − 27 , and the low PI value showed high fingerprinting power. However, Waits et al. [ 28 ] argued that the assumption of independent segregation among sites does not hold because the substructure of plant populations is shaped by environmental and anthropogenic selection, leading to a possible overestimation of the theoretical PI , and thus PIsibs are usually used as a conservative upper limit for the PI ; specifically, PI values of 1 × 10 − 4 ∼ 1 × 10 − 2 are considered sufficient for application to the identification of individuals in natural populations. The PI and PIsibs values in this study were much lower than the putative values, indicating that the 32 SSR markers have a very high potential for fingerprinting. Therefore, we combined eight pairs of primers to construct DNA fingerprints for 80 Zanthoxylum cultivars, each of which was assigned a unique numerical code. However, it should be noted that the number of Zanthoxylum cultivars that can be identified by this fingerprint method is limited. As the number of Zanthoxylum accessions used for identification increases and new cultivars are introduced and promoted, the number of new variant sites will increase as well. In such cases, timely and periodic updates to the fingerprint will be required to ensure its ongoing role in future research and application.
In comparison to SSR markers, iPBS markers have been less frequently employed to construct DNA fingerprints. Zeng et al. [ 46 ] successfully constructed fingerprints of 85 Cymbidium goeringii germplasm resources using two iPBS primers. Demirel et al. [ 47 ] used 17 iPBS markers to fingerprint and genetically analyze 151 potato genotypes. These studies demonstrated the feasibility of constructing plant fingerprints using iPBS markers. For our study, we selected 10 iPBS primers with high polymorphism and clear amplification bands from a pool of 83 primers. However, we found that these 10 iPBS markers were not sufficient to completely differentiate the 80 Zanthoxylum cultivars.
Notably, specific bands were observed in the amplification results for SSR markers, indicating that allelic loci, such as ‘HYWC ♂’, ‘HYWC ♀’, ‘MYWCQHJ’, and ‘YJ’, can serve as important molecular traits for cultivar identification (Supplementary Figure S6 ). Considering factors such as the ease of banding, number of available markers, polymorphic information content of the primers, and amplification stability, we believe that SSR markers are more suitable for constructing DNA fingerprints of Zanthoxylum species. However, it is important to acknowledge that iPBS markers have valuable potential when genomic information is lacking for a species. Moreover, for materials that are difficult to identify using a single molecular marker, a combination of multiple markers can improve identification efficiency.
Currently, with the decreasing cost of high-throughput sequencing technology, the construction of DNA fingerprints using SSR and/or SNP markers has become the most popular choice [ 48 ]. Future research can focus on the development of these two marker types, as well as the collection of more comprehensive Zanthoxylum germplasm resources, to construct a more perfect fingerprint map. This endeavor holds significant importance for the conservation and development of Zanthoxylum germplasm resources.
Conclusions
This study aimed to assess the genetic diversity, genetic relationships, population genetic differentiation, and genetic structure of 80 Zanthoxylum accessions using 32 G-SSR markers and 10 iPBS markers. Additionally, a DNA fingerprint of Zanthoxylum cultivars was constructed. The findings of this research demonstrated that the 80 Zanthoxylum accessions exhibit a significant level of genetic diversity. Both the SSR and iPBS markers were effective at revealing the genetic relationship of Zanthoxylum species, with SSR markers providing a more comprehensive reflection of the genetic variation within the tested accessions. Moreover, limited genetic exchange was observed among the three populations of Zanthoxylum , resulting in noticeable genetic differentiation. In terms of discriminatory ability, SSR markers demonstrated greater strength than iPBS markers. Furthermore, the construction of DNA fingerprints for the 80 Zanthoxylum cultivars was achieved using eight pairs of SSR primers. These findings have significant implications for the conservation and utilization of Zanthoxylum resources, offering a valuable scientific foundation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
XZ, WZ and FX designed the experiments; XZ and WC performed the experiments; ZY and CL participated in the collection of resources; XZ, JY and YL analyzed the data; XZ wrote the manuscript. All the authors have read and agreed to the published version of the manuscript.
This work was supported by the National Key R&D Program of China (2019YFD1001200) and the Research on the Selection and Breeding of New High-Quality and Labor-Saving Cultivars of Chinese Pepper and Supporting Technology(2021YFYZ0032).
Data availability
Declarations.
The authors declare no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Weiwei Zhang, Email: moc.361@nhcgnahzww .
Feng Xu, Email: nc.ude.ueztgnay@gnefux .

IMAGES
VIDEO
COMMENTS
Data interpretation and data analysis are two different but closely related processes in data-driven decision-making. Data analysis refers to the process of examining and examining data using statistical and computational methods to derive insights and conclusions from it. It involves cleaning, transforming, and modeling the data to uncover ...
^Research may be defined as a method of studying problems whose solutions are to be derived partly or wholly from facts. _ W.S. Monroes ^Research is considered to be the more formal, systematic intensive process of carrying on the scientific method of analysis. It involves a
analysis methods, the next task is to draw Inferences from these data. In other words, Interpretation of data needs to be done, so as to derive certain conclusions, which is the whole purpose of the research study. Definition "Interpretation refers to the process of making sense of numerical data that has been collected, analysed and presented".
formulating research questions to data collection, analysis, and interpretation. It provides a step- by -step approach, ensuring that each stage of the research is
Abstract. This essay addresses a wide range of concepts related to interpretation in qualitative research, examines the meaning and importance of interpretation in qualitative inquiry, and explores the ways methodology, data, and the self/researcher as instrument interact and impact interpretive processes.
Qualitative data analysis is. concerned with transforming raw data by searching, evaluating, recogni sing, cod ing, mapping, exploring and describing patterns, trends, themes an d categories in ...
Interpretation of qualitative data can be presented as a narrative. The themes identified from the research can be organised and integrated with themes in the existing literature to give further weight and meaning to the research. The interpretation should also state if the aims and objectives of the research were met.
Textual analysis is a broad term for various research methods used to describe, interpret and understand texts. All kinds of information can be gleaned from a text - from its literal meaning to the subtext, symbolism, assumptions, and values it reveals. The methods used to conduct textual analysis depend on the field and the aims of the ...
The research methodology is an important section of any research paper or thesis, as it describes the methods and procedures that will be used to conduct the research. It should include details about the research design, data collection methods, data analysis techniques, and any ethical considerations.
Research methods are specific procedures for collecting and analyzing data. Developing your research methods is an integral part of your research design. When planning your methods, there are two key decisions you will make. First, decide how you will collect data. Your methods depend on what type of data you need to answer your research question:
data collection, analysis, and interpretation. The selection of a research approach includes the research problem or issue being addressed, the researchers' persona. experiences, and the audiences for the study. Thus, in this book, philosophical assumptions, research approaches, research designs, and research methods are four key terms ...
Methodological studies - studies that evaluate the design, analysis or reporting of other research-related reports - play an important role in health research. They help to highlight issues in the conduct of research with the aim of improving health research methodology, and ultimately reducing research waste. We provide an overview of some of the key aspects of methodological studies such ...
Definition of research in data analysis: According to LeCompte and Schensul, research data analysis is a process used by researchers to reduce data to a story and interpret it to derive insights. The data analysis process helps reduce a large chunk of data into smaller fragments, which makes sense. Three essential things occur during the data ...
Abstract. Statistical methods involved in carrying out a study include planning, designing, collecting data, analysing, drawing meaningful interpretation and reporting of the research findings. The statistical analysis gives meaning to the meaningless numbers, thereby breathing life into a lifeless data. The results and inferences are precise ...
This article is a practical guide to conducting data analysis in general literature reviews. The general literature review is a synthesis and analysis of published research on a relevant clinical issue, and is a common format for academic theses at the bachelor's and master's levels in nursing, physiotherapy, occupational therapy, public health and other related fields.
Few, though, focus on methodologi-cal issues. Methodologies for interpreting research have become increasingly diversified in recent years and warrant close examination. This method-ological ...
About Research Methods. This guide provides an overview of research methods, how to choose and use them, and supports and resources at UC Berkeley. As Patten and Newhart note in the book Understanding Research Methods, "Research methods are the building blocks of the scientific enterprise. They are the "how" for building systematic knowledge.
Data Analysis. Definition: Data analysis refers to the process of inspecting, cleaning, transforming, and modeling data with the goal of discovering useful information, drawing conclusions, and supporting decision-making. It involves applying various statistical and computational techniques to interpret and derive insights from large datasets.
Step-by-step advice for constructing a qualitative project from beginning to end, covering both foundational theory and real-world application Qualitative Research Methods: Collecting Evidence, Crafting Analysis, Communicating Impact guides you through sequential stages of a qualitative research project, from project design and data collection to analysis, interpretation, and presentation.
The term 'interpretive research' is often used loosely and synonymously with 'qualitative research', although the two concepts are quite different. Interpretive research is a research paradigm (see Chapter 3) that is based on the assumption that social reality is not singular or objective. Rather, it is shaped by human experiences and ...
Visual methodologies are used to understand and interpret images (Barbour, 2014) and include photography, film, video, painting, drawing, collage, sculpture, artwork, graffiti, advertising, and cartoons.Visual methodologies are a new and novel approach to qualitative research derived from traditional ethnography methods used in anthropology and sociology.
Abstract. This chapter addresses a wide range of concepts related to interpretation in qualitative research, examines the meaning and importance of interpretation in qualitative inquiry, and explores the ways methodology, data, and the self/researcher as instrument interact and impact interpretive processes.
This document discusses interpretation in research methodology. Interpretation involves drawing inferences from collected facts after analytical or experimental study. It has two main aspects: establishing continuity of research and explanatory concepts. Interpretation allows researchers to understand abstract principles, link findings across ...
The document discusses techniques of interpretation in research. Interpretation refers to drawing inferences from collected data and facts after analysis to understand broader meanings and establish explanatory concepts. It allows the researcher to understand abstract principles underlying findings, link results to other research, and guide future studies. Correct interpretation is an art that ...
The material covered in the Quantitative Methodology: Measurement and Statistics, Master of Science (M.S.) program is crucial in today's data-driven world, where the ability to analyze and interpret data is in demand across many industries. Ideal for individuals with a passion for research and a strong aptitude for mathematics, this program attracts students who are analytical, detail-oriented ...
Qualitative research is a type of research that aims to gather and analyse non-numerical (descriptive) data in order to gain an understanding of individuals' social reality, including understanding their attitudes, beliefs, and motivation. This type of research typically involves in-depth interviews, focus groups, or observations in order to collect data that is rich in detail and context.
As analysis goals can differ between users and datasets, the ehrapy analysis pipeline is customizable during the final knowledge inference step. ehrapy provides statistical methods for group ...
Data Analysis has become an important part of our life, as it helps us to draw useful information, decision-making conclusions on any particular raw data. One component of digital data analysis is sentiment analysis. Sentiment analysis evaluates the emotional tone of textual data and classifies it as neutral, negative, or positive. In business, this analysis is frequently utilized for market ...
Knowledge graphs (KGs), structured representations of entities and their relationships, are increasingly crucial for various Artificial Intelligence (AI) tasks. However, the complex structure and vast scale of KGs pose challenges for efficient reasoning and information retrieval. Knowledge graph embedding (KGE) is a powerful technique to bridge this gap by mapping entities and relations into ...
Therefore, conducting extensive research on genetic diversity analysis, genetic map construction, and cultivar identification techniques for Zanthoxylum is highly important. This research will play a crucial role in safeguarding the development of Zanthoxylum germplasm resources and ensuring the healthy growth of the industry.