| Word Tools | | Finders & Helpers | | Apps | | More | | Synonyms | | | | | | |
| | Copyright WordHippo © 2024 | Help | Advanced Search Computer Science > Computation and LanguageTitle: a case study on english-malayalam machine translation. Abstract: In this paper we present our work on a case study on Statistical Machine Translation (SMT) and Rule based machine translation (RBMT) for translation from English to Malayalam and Malayalam to English. One of the motivations of our study is to make a three way performance comparison, such as, a) SMT and RBMT b) English to Malayalam SMT and Malayalam to English SMT c) English to Malayalam RBMT and Malayalam to English RBMT. We describe the development of English to Malayalam and Malayalam to English baseline phrase based SMT system and the evaluation of its performance compared against the RBMT system. Based on our study the observations are: a) SMT systems outperform RBMT systems, b) In the case of SMT, English - Malayalam systems perform better than that of Malayalam - English systems, c) In the case RBMT, Malayalam to English systems are performing better than English to Malayalam systems. Based on our evaluations and detailed error analysis, we describe the requirements of incorporating morphological processing into the SMT to improve the accuracy of translation. | Comments: | This paper contains 10 pages with 4 figures and 10 tables | | Subjects: | Computation and Language (cs.CL) | | Cite as: | [cs.CL] | | | (or [cs.CL] for this version) | | | Focus to learn more arXiv-issued DOI via DataCite | Submission historyAccess paper:. References & Citations- Google Scholar
- Semantic Scholar
DBLP - CS BibliographyBibtex formatted citation.  Bibliographic and Citation ToolsCode, data and media associated with this article, recommenders and search tools. arXivLabs: experimental projects with community collaboratorsarXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website. Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them. Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs . Subscribe to the PwC NewsletterJoin the community, edit social preview.  Add a new code entry for this paperRemove a code repository from this paper, mark the official implementation from paper authors, add a new evaluation result row. | TASK | DATASET | MODEL | METRIC NAME | METRIC VALUE | GLOBAL RANK | REMOVE | - MACHINE TRANSLATION
- TRANSLATION
Remove a task Add a methodRemove a method, edit datasets, a case study on english-malayalam machine translation. 27 Feb 2017 · Sreelekha. S , Pushpak Bhattacharyya · Edit social preview In this paper we present our work on a case study on Statistical Machine Translation (SMT) and Rule based machine translation (RBMT) for translation from English to Malayalam and Malayalam to English. One of the motivations of our study is to make a three way performance comparison, such as, a) SMT and RBMT b) English to Malayalam SMT and Malayalam to English SMT c) English to Malayalam RBMT and Malayalam to English RBMT. We describe the development of English to Malayalam and Malayalam to English baseline phrase based SMT system and the evaluation of its performance compared against the RBMT system. Based on our study the observations are: a) SMT systems outperform RBMT systems, b) In the case of SMT, English - Malayalam systems perform better than that of Malayalam - English systems, c) In the case RBMT, Malayalam to English systems are performing better than English to Malayalam systems. Based on our evaluations and detailed error analysis, we describe the requirements of incorporating morphological processing into the SMT to improve the accuracy of translation. Code Edit Add Remove Mark officialTasks edit add remove, datasets edit, results from the paper edit add remove, methods edit add remove. Cloud EnablementCloud Consulting Cloud-Native Apps Cloud Migration Strategies Cloud Migration Services Cloud Monitoring Cloud Security Posture Cloud Cost Optimization Private Cloud ChatGPT Integration Data Science Artificial Intelligence Data Engineering Generative AI Development Big Data Processing Machine Learning Data Management Computer Vision Data Storage Video Analytics Data Visualization Robotic Process Automation DigitalizationExtended Reality Web Development Internet of Things E-Learning Portals E-Commerce Sites Intelligent Document Processing Product Information Management Enterprise Asset Management Digital Experience Platform Customer Data Platform Microservice Architecture UX/UI Design QA Automation Cybersecurity Performance Monitoring Frontend Monitoring API Management Compliance Consulting Digital MarketingMarketing Automation Visualization Programmatic Advertising Paid Advertising Email Marketing Content Marketing Social Media Microsoft Solutions - Manufacturing
- Engineering
- Financial Services
- Life Insurance
- Hospitality
- Real Estate
SlashQ Digital queue management ContextIQ Recommendation engine Patient Transporter Management RTLS for intra hospital transfers TalQ Call accounting system Rehash Low-code insurance platform ProductivityNotification App Notification builder Kever Project management tool QuickPicks Salesforce widget Cross-IndustryIIoT Platform IoT solutions for industries App DevelopmentOpen Source Works Shared with the community Testing & MonitoringWebWatch24x7 Server monitoring tool Case Studies White Papers - Get in Touch
- Cloud Enablement Data & AI Digitalization End-to-End Digital Marketing SaaS
- Retail Healthcare Hospitality Insurance Productivity Cross-Industry App Development Testing & Monitoring
- About QBurst
- Leadership Team
- Corporate Responsibility
- Our Locations
- Privacy Policy
- Architecture
- Development
- Tools & Lab
- Delivery Model
- Open Positions
- Entry-Level Opportunities
- Submit Resume
- Company Profile
- Mid-Career Opportunities
 START A CONVERSATIONShare your requirements and we'll get back to you with how we can help. Please accept the terms to proceed. - All Categories Artificial Intelligence Big Data Business Intelligence Cloud Computer Vision Design DevOps eCommerce General IoT Marketing Automation Mobile Web
 Generating Malayalam Word Embeddings: A Case StudyResearch shows that children primarily learn languages by observing patterns in the words they hear. Computer scientists are taking a similar approach to train computers to process human language. Computers cannot process words directly. Vector representations of words, known as embeddings, are used to teach machines to make sense of them. Vectors encapsulate the properties of words in them, such as semantics or relations between words. Embeddings can also contain syntactic information and can have similar vectors for various forms of words, such as, he, him, his, etc. To obtain these vectors, models are trained with large volumes of text and the embeddings are learned during the process. This is an example of the GloVe embedding of the word “rain” (with an embedding vector size of 300).  Overview of Static EmbeddingsWord embeddings can be broadly classified into two: static and dynamic. Static embeddings have only one embedding per word regardless of the context in which they appear. These embeddings will thus be a mix of the various meanings of the word. Dynamic contextual embeddings consider the whole sentence before generating the word embedding. This makes it possible to provide different embeddings for words such as “bank” when they occur in varying contexts such as “river bank” or “bank loan”. Even with its limitations, static word embeddings have found widespread use in many NLP applications such as classification, POS tagging, sentiment analysis, etc. This section provides a high-level picture of some of the most popular static embeddings. Word2vec is one of the first word embeddings to find widespread popularity. It gives impressive results on word analogy tasks, such as finding words similar to a query word in a document. It works by training a shallow neural net to either learn to predict a missing word given the surrounding words (CBOW method) or the surrounding words given a word as input (skip-gram). CBOW is suitable for small datasets while skip-gram is more suitable for larger datasets. Global Vectors (GloVe)GloVe embeddings are generated on the basis of co-occurrence of words over the entire training dataset. They encapsulate the probability of two words occurring together. They are optimized directly on the dataset so that the dot product of two word embeddings gives the log of the count of the co-occurrence of the two words. For example, if the two words “mountain” and “river” occur in the context of each other, say 20 times within a window of 10 words in the dataset, then the generated embeddings will satisfy: Vector(mountain) . Vector(river) = log(20) fastText is an improvement over Word2vec and GloVe in that it can support out-of-vocabulary words. During its training, fastText splits each word into an n-gram of characters. Consider the word “petrol” with an n-gram length of 5. The fastText representation of this word will be <petr, petro, etrol, trol>. The fastText model then generates embeddings for each of these n-grams. This model can make sense of parts of words and allow embeddings for suffixes and prefixes. Once the words have been represented using character n-grams, a skip-gram model is trained to learn the embeddings. fastText works well with rare words. So even if a word wasn’t seen during training, it can be broken down into n-grams to get its embeddings. In the above example, the learned n-gram “petro” can be used for generating embeddings for words such as “petrochemical” or “petroleum” even when the model has not seen them before. This should be especially useful for complex agglutinative languages such as Malayalam. Challenges Specific to Malayalam Word EmbeddingsWidely spoken languages such as English and Chinese have well-developed word embeddings. Malayalam is an exceptionally difficult language for generating word embeddings as most of the words can occur in different forms in a sentence. It is a highly inflectional and agglutinative language. As an example, take the word, നീലപ്പീലിക്കണ്ണും . It is made by agglutinating നീല + പീലി + കണ്ണ് + ഉം . A large number of variations are also possible for each word. For example, take the word പാലക്കാട്. A large number of words can be derived from it, such as പാലക്കാടിൻ്റെ. പാലക്കാടിലെ and so on by modifying the ending. This makes it difficult for models like Word2vec since they cannot handle out-of-vocabulary words. Dravidian languages, including Malayalam, are morphologically very rich—there is a large number of possible inflections for each word. All this has the consequence of increasing the word vocabulary size and reduces the number of observed instances of a given token in a dataset. This adversely affects the quality of the generated word embeddings. The first thing we had to do was to find a large enough collection of Malayalam text. Currently, many regional languages such as Malayalam have a relatively smaller online presence. We initially scraped the web using tools such as Beautiful Soup and Lucene. However, Malayalam language content was hard to come by and older archives were not publicly accessible in many of the news websites. Even Wikipedia had only limited resources in Malayalam. So, we began exploring alternative avenues and came upon Common Crawl archives, which has crawl data for a large number of languages including Malayalam. Most of this data is obtained from news sites and blog posts periodically crawled over the last decade. Please check our previous blog post to learn more about how we extracted the Malayalam data from Common Crawl archives. Evaluation of Word EmbeddingsOnce the word embeddings are trained, we need to have a way of knowing how good they are. So having a proper evaluation metric is important. Evaluation metrics can be broadly categorized into two: intrinsic and extrinsic. - Intrinsic : This type of evaluation takes into account the intrinsic properties of words and their relation to each other to directly get a measure of the quality of the embeddings themselves. It is more intuitive and speeds up the whole process of building quality embeddings.
Some types of intrinsic evaluation tasks are : - Word Similarity / Relatedness : In this task, a dataset containing word pairs and manual similarity judgments is created. Using the learned word embeddings, a similarity metric between each word pair is created, such as cosine similarity between the embeddings. The correlation between the manually judged similarity scores and the ones provided by the embeddings defines the embedding quality.
- Word Analogy : Given a pair of words (A, B) with a particular relationship and a word C of the same type as A, the word analogy task involves predicting the word D that holds the same relationship with C as B holds with A. This task utilizes the linear relationship between word embeddings to search for the missing word. Word analogy datasets can capture semantic as well as syntactic relationships depending on the tuples included in the query inventory. For example: Given two related words, man and woman, the task will be to predict to which words a new word such as “King” would map to under the same relation. A good model will be able to predict the word “Queen”.This can be expressed algebraically as King - Man + Woman = Queen. In other words, the vector difference between King and Queen should capture the concept of gender.
- Extrinsic : This type of evaluation measures the utility of the embeddings in another downstream NLP application like sentiment analysis, machine translation, named entity recognition, parts of speech tagging, news/article classification, etc.
We decided to evaluate using the intrinsic metrics as they tend to give a better picture of the overall quality of the models, not just for a specific application. Building evaluation datasets is a labor-intensive and time-consuming operation. To mitigate this problem, we decided to programmatically build an evaluation benchmark. Our dataset consists of words from five categories (Languages, Animals, Places, Vegetables, Names). These categories were chosen as they appear in relatively distinct contexts in the training corpus we had. This helps the model to learn to more easily distinguish between each category. Each of these categories were then seeded with around 20 words that we picked randomly. Then we formed triplets on the condition that wherever A and B are picked from the same category, C is chosen from a different one. In the examples shown below, words in columns A and B are picked from the languages category and the column C is made up of the words from the remaining categories such as names, places, etc. Status indicates that the row satisfies our condition. 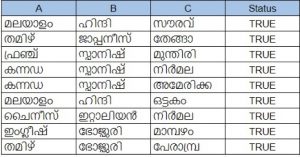 Using just 20 examples from each category, we were able to come up with around 1 lakh test cases. After a little experimentation with a smaller dataset, we decided to use it for evaluating models trained using the Common Crawl data. Experiments and ResultsOnce we established the evaluation benchmark, we trained the different models with the Malayalam text obtained from the Common Crawl archives for the month of January. The results we obtained are summarized below: 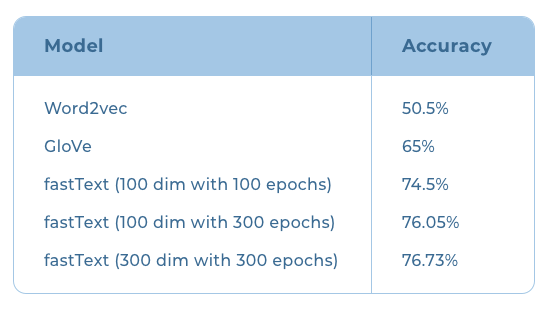 We began training with Word2vec and got an accuracy of 50.5%. Word2vec couldn’t handle many out-of-vocabulary words and hence offered low recall. GloVe embeddings couldn’t handle out-of-vocabulary words either but the accuracy improved slightly to 65% on our evaluation benchmark. The next step was to try out models that could handle out-of-vocabulary words. This led us to fastText. Facebook had released a fastText model for Malayalam trained using the Malayalam text available from the Common Crawl archives. However, the data they used were scraped way back in 2018. When we ran our evaluation on this model, we got an accuracy of 78%. Since the Common Crawl archives now had more data compared to the archives available in 2018, we decided to give training fastText a go. The same dataset obtained from the January 2020 Common Crawl archives was used for the training. After the training, the models were evaluated on our custom evaluation metric. Training the fastText model for 100 epochs with an embedding size of 100 on this dataset, we got an accuracy of 74.5% . When we continued the training to 300 epochs, the accuracy improved to 76.05% . We then increased the embedding dimension to 300 and retrained again using the same dataset for 300 epochs. This further increased our accuracy to 76.73% . Training after 300 epochs however,did not improve the results. At this point, we concluded that we need to improve the subword tokenization and text filtering to match or exceed the results of the Facebook trained fastText model—something we will have to ascertain through further exploration. Machine Translation System for Translation of Malayalam Morphological Causative Constructions into English Periphrastic Causative- First Online: 13 June 2020
Cite this chapter - T. K. Bijimol 4 ,
- John T. Abraham 5 &
- D. Jyothi Ratnam 6
Part of the book series: EAI/Springer Innovations in Communication and Computing ((EAISICC)) 517 Accesses 1 Citations Malayalam is the regional as well as the official language of Kerala, the southern state of India, and Union Territory of Lakshadweep. The Government of Kerala follows a trilingual (Malayalam, English and Hindi) educational system from primary level to graduation level. But in the context of Machine translation, Malayalam is far behind English and Hindi. Here starts the relevance of study of Malayalam-to-English Machine Translation (MT) Systems. In a sentence, the pattern of verb phrase (VP) demonstrates language-specific features, and the causative sentence construction is one among them. The part of the semantics of the verb can be represented by the causation. The sense of causation is expressed through the verb or verb phrase. Any existing online MT system has not handled causative pattern of sentence. This research studies and analyses the problems in Malayalam-to-English causative sentence translations. Based on the study, this research designs and develops a system to improve the accuracy of existing Malayalam–English by including causative sentence translation mechanism. The output was compared with the output of Google Translate and identified that the Malayalam-to-English causative sentence MT system performs translations of simple causative sentences with outstanding accuracy. This is a preview of subscription content, log in via an institution to check access. Access this chapterSubscribe and save. - Get 10 units per month
- Download Article/Chapter or Ebook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
- Durable hardcover edition
Tax calculation will be finalised at checkout Purchases are for personal use only Institutional subscriptions Similar content being viewed by others Translation Equivalence for English Periphrastic Causative Constructions into Hindi in the Context of English to Hindi Machine Translation SystemThe hindi to dogri machine translation system: grammatical perspective.  Myanmar-English Machine Translation ModelAnju, E. S., & Manoj Kumar, K. V. (2014). Malayalam to English machine translation: An EBMT system. IOSR Journal of Engineering (IOSRJEN), 4 (1). www.iosrjen.org . Auersperger, M. (2012). English causative constructions with the verbs have, get, and make, and their Czech translation counterparts . Google Scholar Banchs, R. E., D’Haro, L. F., & Li, H. (2015). Adequacy – Fluency metrics: Evaluating MT in the continuous space model framework. IEEE/ACM Transactions on Audio, Speech and Language Processing, 23 (3), 472–482. Article Google Scholar Begum, R., & Sharma, D. M. (2010). A preliminary work on Hindi causatives . In Proceedings of the eighth workshop on Asian language resources, pp. 120–128. Bharati, A., Chaitanya, V., Sangal, R., & Ramakrishnamacharyulu, K. V. (1995). Natural language processing: A Paninian perspective (pp. 65–106). New Delhi: Prentice-Hall of India. Gopinathan Nair, B. (2008). Collected papers on Malayalam language and linguistics . Thiruvanathapuram: International School of Dravidian Linguistics. Kemmer, S., & Verhagen, A. (1994). The grammar of causatives and the conceptual structure of events. Cognitive Linguistics, 5 (2), 115–156. https://doi.org/10.1515/cogl.1994.5.2.115 . Koehn, P., & Monz, C. (2006). Manual and automatic evaluation of machine translation between European languages. In Proceedings of the workshop on statistical machine translation . New York: Association for Computational Linguistics. Chapter Google Scholar Levina, B., & Hovav, M. R. (2016). A preliminary analysis of causative verbs in English . Department of Linguistics, Northwestern University, Sheridan Road, Evanston, IL, USA, Department of English, Bar Ilan University, Ramat Gan, Israel. Macketanz, V., Avramidis, E., Burchardt, A., Helcl, J., & Srivastava, A. (2017). Machine translation: Phrase-based, rule-based and neural approaches with linguistic evaluation. Cybernetics and Information Technologies, 17 (2), 1–18. Mitkov, R. (2003). The Oxford handbook of computational linguistics . Oxford: Oxford University Press. MATH Google Scholar Nadathur, P. (2017). Causative verbs: Introduction to lexical semantics , pp. 1–15. Nair, L. R., David Peter, S., & Ravindran, R. P. (2012). Design and development of a Malayalam to English translator: A transfer based approach. International Journal of Computational Linguistics (IJCL), 3 (1), 1–11. Narayanapillai, K. S. (1995). Adhunika Malayalam vyakaraNam . Thiruvananthapuram: Kerala Bhasha Institute. Rajaraja Varma, A. R. (2006). Keralapanineeyam (8th ed.). Kottayam: DC Books. Ram, K. V. (2001). Ramkumarinte Sampourna Malayala Vyakaranam . Thiruvananthapuram: Siso Books. Ratnam, J. D., Kumar, M. A., Premjith, B., Soman, K. P., & Rajendran, S. (2018). Sense disambiguation of English simple prepositions in the context of English–Hindi machine translation system. In S. Margret Anouncia & U. Wiil (Eds.), Knowledge computing and its applications . Singapore: Springer. Ratnam, J. D., Soman, K. P., Biji Mol, T. K., & Priya, M. G. (2019). Translation equivalence for English periphrastic causative constructions into Hindi in the context of English to Hindi machine translation system. In R. Kumar & U. Wiil (Eds.), Recent advances in computational intelligence. Studies in computational intelligence (Vol. 823). Cham: Springer. Ravi Sankar, S. N. (2012). A grammar of Malayalam. Journal of Language in India 12, 2012 Rev, R. C., & Nair, S. K. (1993). Draavida bhaashaa vyaakaranam (part one) (3rd ed.). Thiruvananthapuram: The State Institute of Languages. Sebastian, M. P., Sheena Kurian, K., & Santhosh Kumar, G. (2010). A classification of Sandhi rules for suffix separation in Malayalam . In 38th All India conference of dravidian linguists. Shahana, I. L., & Sharafudheen, K. A. (2014). Relative study on Malayalam-English translation using transfer based approach. International Journal of Computing and Technology, 1 (2), 24–29. Somers, H. (2005). Round-trip translation: What is it good for? In Proceedings of the Australasian language technology workshop 2005, pp. 127–133. Steven, B., Edward, L., & Ewan, K. (2009). Natural language processing with python . Sebastopol: O’Reilly Media Inc.. Download references Author informationAuthors and affiliations. Santhigiri College of Computer Science, Idukki, Kerala, India T. K. Bijimol Department of Computer Science, Bharath Matha College, Kochi, Kerala, India John T. Abraham Center for Computational Engineering & Networking (CEN), Amrita School of Engineering,Amrita Vishwa Vidyapeetham, Coimbatore, Tamil Nadu, India D. Jyothi Ratnam You can also search for this author in PubMed Google Scholar Editor informationEditors and affiliations. Department of Computer Science and Engineering, I K Gujral Punjab Technical University, Kapurthala, Punjab, India Raman Kumar Inst Politécnico de Viana do Castelo, Viana do Castelo, Portugal  Rights and permissionsReprints and permissions Copyright information© 2021 Springer Nature Switzerland AG About this chapterBijimol, T.K., Abraham, J.T., Jyothi Ratnam, D. (2021). Machine Translation System for Translation of Malayalam Morphological Causative Constructions into English Periphrastic Causative. In: Kumar, R., Paiva, S. (eds) Applications in Ubiquitous Computing. EAI/Springer Innovations in Communication and Computing. Springer, Cham. https://doi.org/10.1007/978-3-030-35280-6_11 Download citationDOI : https://doi.org/10.1007/978-3-030-35280-6_11 Published : 13 June 2020 Publisher Name : Springer, Cham Print ISBN : 978-3-030-35279-0 Online ISBN : 978-3-030-35280-6 eBook Packages : Engineering Engineering (R0) Share this chapterAnyone you share the following link with will be able to read this content: Sorry, a shareable link is not currently available for this article. Provided by the Springer Nature SharedIt content-sharing initiative Policies and ethics - Find a journal
- Track your research
Sreelekha SHomepage about me research contact me. - My Research
- My Publications
Publications- “NeuroSymbolic Integration with Uncertainty” [ Sreelekha S ] Springer Journal - Annals of Mathematics and Artificial Intelligence, 2018, Vol. 84, Issue 3–4, pp 201–220, DOI:10.1007/s10472-018-9605-y.
- “Role of Morphology Injection in SMT: A Case Study from Indian Language Perspective” [ Sreelekha S , Pushpak Bhattacharyya] ACM Transactions on Asian and Low-Resource Language Information Processing (ACM TALLIP) 2017, Vol. 17, Issue No. 1, Article 1, DOI:10.1145/3129208.
- “Statistical vs. Rule-Based Machine Translation: A Comparative Study on Indian Languages” [ Sreelekha S , Pushpak Bhattacharyya, Malathi.D] Springer Journal of Advances in Intelligence and Soft Computing, 2017, Vol. 632, DOI 10.1007/978-981-10-5520-1_60, ISBN:978-981-10-5519-5
- "NeuroSymbolic Integration with Association Rule Mining" [ Sreelekha S , Malathi.D] International Journal of Pure and Applied Mathematics, 2017, Vol. 114, Issue No:12, Pages:769-776
- “A Survey Report on Evolution of Machine Translation” [ Sreelekha S , Pushpak Bhattacharyya, Shishir K. Jha, Malathi.D] International Journal of Control Theory and Applications, 2016, Vol.9,Issue No:33, Pages:233-240
Conferences- “Morphology Injection for English-Malayalam Statistical Machine Translation” [ Sreelekha S , Pushpak Bhattacharyya]International Conference on Lexical Resources and Evaluation-LREC, Pages: 1-7, 7-12 May 2018, Miyazaki (Japan)
- "Decoding Tracer Utility" [ Sreelekha S , Pushpak Bhattacharyya] International Joint Conference on Natural Language Processing, IJCNLP-Demo, Pages: 1-4, November 27 - December 1, 2017 Taipei (Taiwan)
- “Lexical Resources to Enrich English-Malayalam MT” [ Sreelekha S , Pushpak Bhattacharyya]International Conference on Lexical Resources and Evaluation-LREC,Pages: 1-8, 23-28 May, 2016, Portorož (Slovenia)
- "NeuroSymbolic Integration with Association Rule Mining"[ Sreelekha S , Malathi.D] International Conference on Artificial Intelligence and Evolutionary Computations in Engineering Systems-ICAIECES, Pages: 1-8, 19-21 May 2016, Chennai (India)
- “Solving Data Sparsity by Morphology Injection in Factored SMT” [ Sreelekha S , Piyush Dungarwal, Pushpak Bhattacharyya, Malathi.D] ACL Anthology-Proceedings of the 12th International Conference on Natural Language Processing-ICON, Pages: 1-5, 11-13 December, 2015, Trivandrum (India).
- "Case study on English-Malayalam Machine Translation”[ Sreelekha S , Pushpak Bhattacharyya, Malathi.D] iDravidian Proceedings-ICON, Pages: 1-10, 11-13 December, 2015, Trivandrum(India).
- “Lexical Resources for Hindi-Marathi MT” [ Sreelekha S , Pushpak Bhattacharyya, D.Malathi] WILDRE Proceedings, LREC, pages-102-110, 26-31 May, 2014, Reykjavik (Iceland)
- “Comparison of SMT and RBMT, the Requirement of Hybridization for Marathi–Hindi MT” [ Sreelekha S , Raj Dabre, Pushpak Bhattacharyya] ICON- International Conference on Natural Language Processing, Pages: 1-10, 18-20 December, 2013, Noida(India).
Pre-print Publications- “Role of Morphology Injection in Statistical Machine Translation” [ Sreelekha S , Pushpak Bhattacharyya] CoRR abs/1709.05487 (2017)
- “Phrase Pair Mappings for Hindi-English Statistical Machine Translation” [ Sreelekha S , Pushpak Bhattacharyya] CoRR abs/1710.02100 (2017)
- “Morphology Generation for Statistical Machine Translation” [ Sreelekha S , Pushpak Bhattacharyya] CoRR abs/1710.02093 (2017)
- “Indowordnets help in Indian Language Machine Translation” [ Sreelekha S , Pushpak Bhattacharyya] CoRR abs/1710.02086 (2017)
- “Bilingual Words and Phrase Mappings for Marathi and Hindi SMT” [ Sreelekha S , Pushpak Bhattacharyya] CoRR abs/1710.02398 (2017)
- “Statistical vs. Rule-Based Machine Translation: A Case Study on Indian Language Perspective” [ Sreelekha S ] CoRR abs/1708.04559 (2017)
- “A Case study on English-Malayalam Machine Translation” [ Sreelekha S , Pushpak Bhattacharyya] CoRR abs/1702.08217 (2017)
- “Lexical Resources for Hindi-Marathi MT” [ Sreelekha S , Pushpak Bhattacharyya] CoRR abs/1703.01485 (2017)
- “Comparison of SMT and RBMT, the Requirement of Hybridization for Marathi–Hindi MT” [ Sreelekha S , Pushpak Bhattacharyya] CoRR abs/1703.03666 (2017)
- "Method for Quantifying Uncertainty in a System" [ Sreelekha S ] PCT, WIPO - World Intellectual Property Organization, 2017
- "Integrated Decision Support System and Method for Deriving Inferences from Datasets" [ Sreelekha S ] PCT, WIPO - World Intellectual Property Organization, 2017
- "Method for Quantifying Uncertainty in a System" [ Sreelekha S ] Indian Patent, 2016
- "Integrated Decision Support System and Method for Deriving Inferences from Datasets" [ Sreelekha S ] Indian Patent, 2016
Technical Systems- Decoding Tracer
- English-Malayalam StatisticalMachineTranslation System
- Malayalam-English StatisticalMachineTranslation System
- Marathi-Hindi StatisticalMachineTranslation System
- Hindi-Marathi StatisticalMachineTranslation System
- English-Hindi StatisticalMachineTranslation System
- Hindi-English StatisticalMachineTranslation System
- English-Marathi StatisticalMachineTranslation System
- Marathi-English StatisticalMachineTranslation System
- French- English StatisticalMachineTranslation System
"Satyameva Jayate" (Sanskrit: सत्यमेव जयते satyam-eva jayate; lit. "Truth alone triumphs.") A mantra from the ancient Indian scripture Mundaka Upanishad- The national motto of India In Transliteration : satyameva jayate nānṛtaṁ satyena panthā vitato devayānaḥ yenākramantyṛṣayo hyāptakāmā yatra tat satyasya paramaṁ nidhānam In English : Truth alone triumphs; not falsehood. Through truth the divine path is spread out, by which the sages whose desires have been completely fulfilled, reach where that supreme treasure of Truth resides. © Satyameva Jayate . All rights reserved. | Design by Sreelekha . How to Say Case study in Malayalam- Open access
- Published: 02 July 2024
Manufacturing mesenchymal stromal cells in a microcarrier-microbioreactor platform can enhance cell yield and quality attributes: case study for acute respiratory distress syndrome- Brandon Krupczak 1 , 4 ,
- Camille Farruggio 2 , 4 &
- Krystyn J. Van Vliet ORCID: orcid.org/0000-0001-5735-0560 2 , 3 , 4 , 5
Journal of Translational Medicine volume 22 , Article number: 614 ( 2024 ) Cite this article Metrics details Mesenchymal stem and stromal cells (MSCs) hold potential to treat a broad range of clinical indications, but clinical translation has been limited to date due in part to challenges with batch-to-batch reproducibility of potential critical quality attributes (pCQAs) that can predict potency/efficacy. Here, we designed and implemented a microcarrier-microbioreactor approach to cell therapy manufacturing, specific to anchorage-dependent cells such as MSCs. We sought to assess whether increased control of the biochemical and biophysical environment had the potential to create product with consistent presentation and elevated expression of pCQAs relative to established manufacturing approaches in tissue culture polystyrene (TCPS) flasks. First, we evaluated total cell yield harvested from dissolvable, gelatin microcarriers within a microbioreactor cassette (Mobius Breez) or a flask control with matched initial cell seeding density and culture duration. Next, we identified 24 genes implicated in a therapeutic role for a specific motivating indication, acute respiratory distress syndrome (ARDS); expression of these genes served as our pCQAs for initial in vitro evaluation of product potency. We evaluated mRNA expression for three distinct donors to assess inter-donor repeatability, as well as for one donor in three distinct batches to assess within-donor, inter-batch variability. Finally, we assessed gene expression at the protein level for a subset of the panel to confirm successful translation. Our results indicated that MSCs expanded with this microcarrier-microbioreactor approach exhibited reasonable donor-to-donor repeatability and reliable batch-to-batch reproducibility of pCQAs. Interestingly, the baseline conditions of this microcarrier-microbioreactor approach also significantly improved expression of several key pCQAs at the gene and protein expression levels and reduced total media consumption relative to TCPS culture. This proof-of-concept study illustrates key benefits of this approach to therapeutic cell process development for MSCs and other anchorage-dependent cells that are candidates for cell therapies. Cell therapy—the use of a living, manufactured cell product as a therapeutic—is well-positioned to address unmet clinical needs across a wide variety of indications that have proven refractory to conventional treatments. Within this field, mesenchymal stem and stromal cells (MSCs) have generated intense enthusiasm for decades given their innate therapeutic properties, transdifferentiation ability, capacity for allogeneic administration, and adult donor source [ 1 , 2 , 3 , 4 ]. Over one thousand clinical trials have explored or are exploring the use of MSCs to treat indications ranging from osteoarthritis to Alzheimer’s Disease, and ten MSC products have been approved by regulatory bodies around the world for clinical use [ 5 ]. Historically, much of the clinical interest in MSCs centered on their “stemness,” pursuing a hypothesis that MSCs could be administered to provide a raw material source of multipotent progenitor cells that could directly repair injury via transdifferentiation in situ to replace damaged tissue. More recently, researchers have explored indirect repair mechanisms, leveraging MSCs’ potent secretion of paracrine signals containing pro-regenerative, anti-inflammatory components such as indoleamine 2,3-dioxygenase-1 (IDO1) and interleukin-10 (IL10) [ 2 , 6 , 7 ]. Despite those putative advantages, MSC clinical translation has been limited, with no MSC products clinically approved in the United States to date. The direct repair hypothesis has come under considerable question as mounting evidence highlights the short circulation time and limited engraftment of MSCs in vivo [ 8 ]. Therapeutic efficacy of manufactured product is often inconsistent: the U.S. Food & Drug Administration (FDA) highlighted validation of manufacturing processes to ensure lot-to-lot consistency as a serious concern for Mesoblast’s remestemcel-L—an MSC product designed to treat pediatric acute steroid-refractory graft-versus-host disease—and ultimately issued the company a complete response letter for this indication [ 9 , 10 ]. It is well recognized that there exist various types of heterogeneity at the level of donors (e.g., differences in donor health and age), of tissue source (e.g., bone marrow versus umbilical cord, etc.), and of cell passage number that contribute to manufacturing difficulty even when manufacturing processes are well documented and intended to be invariant [ 11 ]. We and others have demonstrated that the conventional cell manufacturing methods of expansion in tissue culture polystyrene (TCPS) flasks can promote an emergent, phenotypic heterogeneity [ 12 , 13 , 14 , 15 ]. Flask-based culture is a form of cell culture offering modest and typically open-loop feedback on the cellular environment during expansion with a two-dimensional growth format that limits scalability of cell manufacture, though currently it remains the most commonly used approach in research settings [ 16 , 17 ]. Rennerfeldt et al. showed that time-lapsed image analysis of single cells revealed that even MSC colonies derived from a single clonal progenitor began to exhibit significant heterogeneity in cell diameter, morphological features, and proliferation capacity within four days of conventional flask culture [ 18 ]. Given these intrinsic drawbacks, there is a critical need to explore bioreactor formats that can facilitate early process development and potentially scale to at least preclinical studies. Such approaches would ideally include MSC manufacturing strategies that can simultaneously mitigate phenotypic heterogeneity to ensure batch-to-batch consistency and prime the cells to exhibit definitive therapeutic potency for a specific clinical indication. One such priming strategy to enhance MSC potency is modification of substratum stiffness. MSCs are well-known to be mechanosensitive, and therefore changes to the mechanical properties, surface topography, and ligand presentation of the substrate upon which the cells adhere differentially impact their differentiation capability and gene expression [ 19 , 20 , 21 ]. We and others have explored modifying substratum stiffness to enhance MSC potency for a given indication [ 22 , 23 ]. For example, Liu et al. demonstrated that this approach can be used successfully to alter MSC gene expression and paracrine signaling to an extent sufficient to alter outcomes in vivo [ 24 ]. Liu et al. used a mouse model of total hematopoietic failure (achieved through irradiation and uniformly lethal without treatment) and found that administration of MSCs grown on conventional TCPS with stiffness of ∼ 10 9 Pa was sufficient to rescue ∼ 20% of the mouse population (median survival time 39 days); by contrast, administration of MSCs grown on a polydimethylsiloxane (PDMS) surface with stiffness of ∼ 10 3 Pa rescued ∼ 80% of the mouse population (median survival time undefined, > 50 days) [ 24 ]. Other researchers have explored differentiated stiffness cues to enhance MSC potency for bone regeneration, inflammation reduction, treatment of cardiovascular disease, and more [ 19 , 20 , 22 ]. However, many of these strategies have relied on modifications to a 2D culture format, which carries limitations in scalability and environmental control, prompting us to explore additional stiffness-controllable platforms compatible with bioreactor culture. Microcarriers are a widely utilized technology to enable manufacture of mechanosensitive and anchorage-dependent cells such as MSCs in a bioreactor format. While a variety of commercially available microcarriers have been used for MSC culture, these microcarriers are often composed of materials with similar elastic properties to 2D TCPS flasks, including polystyrene or collagen-coated polystyrene cores. These platforms require an additional filtration step to separate the MSC product from the microcarrier base, which can dramatically impact final cell yield and can impart considerable stress to the cells as a result of prolonged exposure to enzymatic proteases and mechanical agitation [ 25 ]. Ng et al. therefore developed a dissolvable gelatin microcarrier (GMC) fabricated in a microfluidic process and stabilized with a chemical crosslinker. Those gelatin microcarriers exhibited superior diameter monodispersity, commensurate attachment efficiency, and improved cell harvest efficiency relative to commercially available controls [ 25 ]. In addition to the choice of microcarrier platform, selection of an appropriate bioreactor is important for MSC manufacture. Bioreactors with advanced process analytic technologies (PATs)—including systems to regulate temperature and pH, dynamically mix the reactor contents, and control dissolved gas content—are commercially available but often operate at volume scales ranging from tens to thousands of liters. This scale of operation can be prohibitively cost- and resource-intensive for exploratory research and process protocol optimization, while protocols optimized in a smaller-format vessel lacking these PATs (such as a spinner flask) can fail to translate reliably to larger formats. Thus, we desired a bioreactor that retained the advanced PATs of larger-format vessels in a benchtop-scale system suitable for rapid prototyping and later process parameter modulation to enhance MSC manufacture for a given indication. We selected a commercial microbioreactor (Mobius Breez, MilliporeSigma) for its suitability according to these criteria. In fact, Bower et al. compared an earlier version of this microbioreactor technology to a benchtop-scale 2-liter bioreactor using Escherichia coli cultures and demonstrated comparable cell density, metabolite profiles, and normalized plasmid DNA production in both conditions, indicating that the microbioreactor could reliably transfer process parameters and product metrics to a larger-scale system (2 L) [ 26 ]. MilliporeSigma likewise compared the Breez microbioreactor to a 3-liter bioreactor using Chinese Hamster Ovary (CHO) cells and found comparable cell density and protein product production (normalized on a per-cell basis) in both reactors, indicating that the microbioreactor is a suitable scale-down model that reliably predicts performance in larger-scale vessels for at least some metrics and cell types [ 27 ]. While neither of those protocols reflected the specific use case of therapeutic MSC manufacture (nor anchorage-dependent cell culture in general), those reports and others [ 28 , 29 ] support an expectation that process parameters identified in this microbioreactor will more reliably translate to larger-format, PAT-enabled systems than would parameters identified in conventional platforms such as TCPS or spinner flasks. However, this microbioreactor system was designed for suspension cell culture, and required substantial modifications and method development to enable culture of anchorage-dependent cells such as MSCs; we note that validation of that specific expectation of process parameter transference requires future work. Finally, we sought a “use case” or targeted clinical indication for which MSCs are plausible candidate therapies, to provide a defined objective for the processed cells’ attributes and thus the process parameter modulation in the bioreactor format. We reasoned that this indication would adhere to the following criteria: (1) the indication must predominately feature in the lungs in order to leverage MSCs’ well-characterized tendency to accumulate in this organ absent other stimuli; (2) the indication must have an inflammatory component or etiology in order to maximize MSC advantage in immunomodulatory effect; and (3) the indication must be refractory to conventional therapeutics, as cell therapies are novel products with high cost of goods and substantial regulatory burden for manufacture and approval. Acute Respiratory Distress Syndrome (ARDS) represented a confluence of these criteria as a case study for cell production processes that might be capable of priming the MSC secretome toward predictable or improved potency. ARDS is a prevalent condition characterized by inflammation-mediated diffuse alveolar damage, fluid accumulation, and tissue fibrosis. The mortality rate is high (27–46%), there are few standard-of-care treatment options, and no cure [ 30 ]. ARDS etiologies include sepsis, head injury leading to loss of airway protection, gastric aspiration, and pulmonary infection, including most prominently by SARS-CoV-2. Prior to the COVID-19 pandemic, there were ∼ 200,000 cases of ARDS in the United States each year [ 30 ]. In order to create a suitable benchmark to define and measure any relevant effects of process parameter modulation on the MSCs, we sought to identify specific attributes of the MSC phenotype that could reasonably be expected to result in improvements to product potency when used in the treatment of ARDS. Such attributes, once fully vetted and conclusively correlated to product safety, efficacy, and quality in preclinical and clinical testing, are typically termed ‘critical quality attributes’ (CQAs). Formally, the FDA defines a CQA as “a physical, chemical, biological, or microbiological property or characteristic that should be within an appropriate limit, range, or distribution to ensure the desired product quality [ 31 ].” Given that no such CQAs for MSC products designed to treat ARDS yet exist to our knowledge, we instead developed a list of potential critical quality attributes (pCQAs) which might be reasonably expected to correlate to changes in product potency for MSC treatment of ARDS based on literature evaluation, but which will require further preclinical and clinical validation to demonstrate this outcome conclusively. We underscore that this is a case study in process development, and not a claim that the assessments here confirm any specific critical quality attribute or therapeutic use. Based on the hypothesized MSC therapeutic mechanism of paracrine signaling, we reasoned that an appropriate set of pCQAs would be genes whose expression was either correlated or anti-correlated with therapeutic efficacy for ARDS in the existing literature. We developed procedures to enable MSC culture in the microbioreactor system, utilizing our microfluidically fabricated gelatin microcarriers (GMCs) to facilitate cell harvest efficiency. We demonstrated that we could successfully seed the microbioreactor with cells and micro carriers, expand cell number, and harvest cells reliably for multiple batches from one donor as well as multiple distinct donors. We then assessed a literature-supported panel of 24 genes which served as our pCQAs and evaluated gene expression from MSCs grown on gelatin microcarriers in the microbioreactor compared to MSCs grown on conventional TCPS flasks. We also validated expression of a subset of these genes at the protein level. While this study did not explore further process development for optimization of the microbioreactor conditions or microcarrier properties, these first results demonstrate the capacity of this approach to attain viable cell yields and an enhanced MSC secretome in a process-controlled microbioreactor format. Gelatin microcarrier fabricationGelatin microcarriers were fabricated as previously reported [ 25 ]. Briefly, gelatin Type A from porcine skin (Sigma-Aldrich, G2500-100G) was measured out and mixed with sterile deionized water to a 10% weight/volume solution and heated to 50 °C for 1 h with agitation. Gelatin solution was loaded onto a syringe pump and injected into a microfluidic chip at a flow rate of 1 µL/min. Pico-Surf surfactant (Sphere Fluidics, C022) was diluted in Novec 7500 synthetic oil (Fisher Scientific NC1242157) to 0.125% and injected into the microfluidic chip at a flow rate of 15µL/min to disrupt the laminar gelatin and form droplets, which were collected in a centrifuge tube held at 4 °C to prevent droplet deformation. Microcarriers were cleaned using 1 H,1 H,2 H,2 H-Perfluoro-1-octanol (Sigma-Aldrich, 370533-25G), diluted in sterile deionized water, and crosslinked with genipin chemical crosslinking agent (Sigma-Aldrich, G4796-125MG). Crosslinked microcarriers were sterilized in ethanol and washed in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Scientific, 11885092) supplemented with 10% fetal bovine serum (FBS, Gibco, 16000-044) and 1% penicillin-streptomycin (pen-strep, Gibco, 10378-016) to prepare for cell culture. Microbioreactor operationFor the microbioreactor (Mobius Breez, MilliporeSigma), cassettes were obtained from MilliporeSigma (and initially from Erbi Biosystems). Bottle racks were filled with sterile deionized water, 150 mM NaCl phosphate buffered saline (PBS, Corning, 21-040-CV), and FBS-supplemented DMEM via syringe injection in a biosafety cabinet (BSC). Pneumatic-optical-digital controller (POD) self-tests were performed before each experiment to ensure the microbioreactor was in good working order and each cassette was setup, fluidic lines were primed, and sensors were calibrated according to the manufacturer’s instructions under normal protocol operation. Reactor settings were initially left at default values, with minimum carbon dioxide (CO 2 ) content setpoint of 5%, temperature setpoint of 37 °C, and pH setpoint of 7.40. Cell sourcesHuman bone marrow-derived MSCs were sourced from multiple commercial vendors. Each source represented a single, healthy donor of less than 50 years of age, as purified from bone marrow aspirate and supplied by the vendor. Donor 1 was acquired from RoosterBio Inc., Donor 2 from Lonza Group, and Donor 3 from ReachBio Research Labs (now Discovery Life Sciences). Cells were stored frozen in liquid nitrogen in a solution of 10% dimethyl sulfoxide (DMSO, Sigma-Aldrich, 472301), 90% FBS-supplemented DMEM until thawed for use. Cell injection and attachmentImportantly, the microbioreactor system utilizes single-use disposable cassettes to ensure sterility for each production batch; these cassettes incorporate an integrated set of microfluidic channels which are too small to allow microcarriers of the diameter used in this study to pass through. We therefore utilized cassettes modified to include a side port for initial injection of cells and microcarriers. Modified microbioreactor cassettes were incubated with anti-adherence rinse solution (Stemcell Technologies, 07010) for one hour prior to cell injection to prevent cell attachment to the polycarbonate walls of the microbioreactor. Subsequently, cassette fluidic channels were rinsed three times with PBS by using the eject-wash function. Cassettes were drained of residual media from the setup operation using the ‘reinoculate’ function and the microbioreactor system was set to the ‘manual ops’ mode to pause system activity. Cassettes were removed to a biosafety cabinet for cell injection. Cells and microcarriers suspended in 2.2mL of growth media were injected via syringe through the side port (Fig. 1 B), and any residual air in the reactor was withdrawn into the syringe (Fig. 1 C). An alternating inject-withdraw-inject strategy was most successful in delivering the full liquid contents of the syringe without overfilling the reactor and bursting the delicate gas exchange membranes; additionally, this approach allowed the excess air trapped in the syringe to be used to chase the liquid contents down the injection line and into the microbioreactor, thereby preventing accumulation in the injection line. Subsequently, the side port line was tied off with a knot to prevent the microbioreactor contents from accumulating in the line (Fig. 1 D) and the cassette was returned to the Breez POD. Notably, initial attempts to prevent fluid leakage out of the microbioreactor into the injection line using tubing clamps alone were unsuccessful, motivating the use of the tied knot. The microbioreactor was set to static mixing mode by switching to fed-batch mode, disabling intermittent mixing, and employing a custom microbioreactor control script to allow cells to attach to microcarriers without disruption while maintaining pH and CO 2 control. After 24 hours, additional R1 + R2 air resistors were added in series to the default R0 resistor to slow reactor mixing speed and reduce shear stress on the cells during culture, and static mixing mode was disabled. Perfusion was set to 0.5 vessel volumes per day (VVD, 1.0 mL media per day). 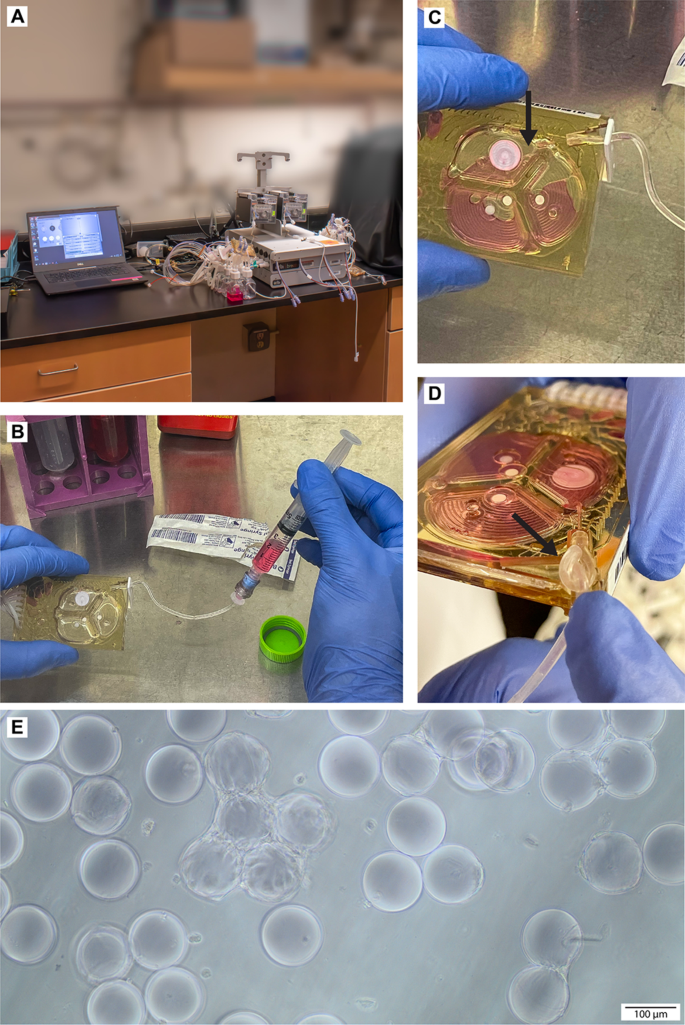 Microcarrier-microbioreactor setup and protocol. ( A ) The microbioreactor system consists of a basestation hub and CO 2 control box, occupying a modest benchtop footprint. The basestation rack can accommodate up to four PODs, which serve as the primary controller for the single-use disposable cassettes which act as the culture vessel. Input pressurized air, CO 2 , and vacuum lines connect to the basestation. The basestation connects via USB data cable to a computer which runs the microbioreactor controller software. ( B ) Anchorage-dependent cells require microcarriers to successfully grow in suspension culture, but microcarrier diameter is too large to pass through the integrated microfluidic channels at the front of the cassette. Modified single-use cassettes include a side port connected to a needleless valve to allow access directly into the reactor contents via syringe injection. ( C ) When injecting the cell-microcarrier suspension through the side port, any air that was previously in the reactor chambers will still be present. This air can be removed by tilting the cassette and withdrawing the syringe plunger until the air travels up the line and into the syringe. Subsequently, the remaining fluid can be injected. ( D ) In order to prevent cells and microcarriers from escaping into the side port during the culture period, a knot is introduced in the injection line. This knot can be secured with laboratory tape for the duration of the culture period, and then removed if needed for harvest. ( E ) Phase contrast image of MSCs growing on gelatin microcarriers while in the microbioreactor cassette Cell harvestPronase enzyme (Sigma-Aldrich, 10165921001) was diluted in PBS to a stock concentration of 10 mg/mL. For the gelatin microcarrier (GMC)-microbioreactor condition, 200 µL of Pronase stock was injected into the microbioreactor cassette through the side port and mixed into the 2 mL reactor volume to dilute the enzyme to its 1 mg/mL working concentration and allowed to incubate with mixing at 37 °C for 5 min until the microcarriers had visually digested. Subsequently, the ‘manual ops’ function was used to perform three eject-wash cycles into the waste bottle for cell harvest. In the GMC versus commercial microcarrier (CMC) experiment, 5 mL of PBS was added to the waste bottle prior to harvest in order to provide a protective cushion as the cell suspension entered the bottle, and the waste bottle was held on ice during harvest to rapidly deactivate residual Pronase enzyme. A similar protocol was employed for the CMC-microbioreactor condition, except that after 5min incubation in Pronase the reactor contents were removed via syringe through the side port due to the inability of the non-dissolvable microcarriers to pass through the microfluidic channels during an eject-wash cycle. The reactor was washed out three times with PBS and reactor contents were passed through a 70 µm filter to separate cells from microcarriers. For the TCPS condition, flask contents were aspirated and 1 mL of 1 mg/mL Pronase was added and incubated at 37°C for 5 min, followed by rinsing with 10 mL of PBS for harvest. Cell suspensions were pelleted at 300 relative centrifugal force (RCF) for 5 min, resuspended in fresh media, and diluted twofold by volume in Trypan Blue stain for counting and viability assessment on a Nexcelom automated cell counting instrument. RNA extraction and reverse transcription quantitative PCRTotal RNA content was harvested from cells and purified using an RNeasy Mini kit (Qiagen, 74106) according to the manufacturer’s instructions. Briefly, cells were pelleted and washed in PBS, resuspended in lysis buffer, and disrupted by passage through a blunt gauge needle attached to an RNase-free syringe before being deposited into an RNA purification spin column and washed with buffer. Purified total RNA was released from the column by administration of 50uL PCR-grade water, and RNA concentration and purity were measured using a Nanodrop instrument. A high-capacity cDNA reverse transcription kit (Applied Biosystems, 4368813) was used to convert the total RNA to complementary DNA. Sequence-specific exon-spanning primers were designed using the Primer-BLAST tool from the NIH and were ordered from IDT. The ΔΔCt method was used to quantify gene expression using the qBase software tool. The legacy housekeeping gene GAPDH was used alongside two MSC-specific housekeeping genes, PPIA and B2M, to normalize gene expression. Gene expression was measured in a LightCycler480 qPCR machine using SYBR-based quantitation. All RNA and DNA prep steps took place in a UV-sterilized biosafety cabinet to minimize nuclease degradation and carryover contamination from one experiment to the next. Individual genes were always measured in the same 96- or 384-well plate (e.g., EPO from Donors 1–3 and TCPS control all loaded into plate 1) to mitigate plate-to-plate variability, and all plates for each experiment were loaded and measured on the same day. Protein quantificationColorimetric sandwich ELISA kits (R&D Systems, DY286-05, DY6030B-05; Thermo Scientific, 88-7176-86, BMS2034MST; Fisher Scientific, CHC1323) were used to quantify secretome protein expression. Cell conditioned media was collected at Day 7 from each condition and diluted twofold by volume in a solution of 2% Triton-X (Sigma-Aldrich, X100) in PBS and vortexed to release vesicle-bound proteins. Remaining steps were performed according to the manufacturer’s instructions. Unconditioned media (i.e., full media unexposed to cells in culture) was used to control for background signal, and absorbance at 450 nm was measured using a Tecan plate reader with automated 570 nm optical correction. Microcarrier fabricationWe fabricated multiple batches of GMCs in a variety of diameters and crosslinking densities to evaluate microcarrier diameter polydispersity, production throughput, batch-to-batch consistency, and elastic moduli (Supplementary Fig. 1 , Additional File 1), as detailed previously by Ng et al. [ 25 ]. Each GMC batch took approximately 24 h for fabrication and utilized ∼ 1.0 mL of liquified gelatin; in the case of the 100 μm-diameter GMCs, this produced ∼ 1.2 million microcarriers for a total surface area of ∼ 360 cm 2 . We selected the 100 μm condition for use in all experiments and observed good batch-to-batch consistency in microcarrier diameter with low (< 5%) coefficient of variation (CoV) in diameter for each batch (Supplementary Fig. 1 A, Additional File 1). GMC stiffnesses ranged from ∼ 8–13 kPa (Supplementary Fig. 1D, Additional File 1) [ 32 ]. We compared our GMCs to a mean diameter-matched, commercially available microcarrier (CMC), the Sartorius Plastic P-215 series; we observed greater CoV in diameter for the CMC control (15.2%) relative to the GMCs (average 2.24% across five batches, Supplementary Fig. 1 B-C, Additional File 1). Donor-to-donor variability and batch-to-batch repeatability in microbioreactor cultureWe investigated our ability to grow MSCs in the microbioreactor on GMCs and harvest viable cells after a seven-day culture period (Fig. 1 ; see Discussion ). While the term ‘microbioreactor’ does not fully define the bioreactor working volume, this specific microbioreactor included a working volume of 2 mL of culture media under perfusion; see Methods . We sought to determine if we could successfully expand cells and obtain consistent cell yields from multiple MSC donors, as well as multiple biological replicates from one donor. We selected three donors, designated Donors 1–3, and obtained cryopreserved MSCs at a medium passage number (passage 6) for each donor. We seeded 125,000 cells from each donor into one microbioreactor cassette and one T25 flask, for a total of six growth conditions. We additionally injected GMCs with a total surface area of 50 cm 2 into the microbioreactor cassettes to provide a growth surface, reasoning that the increased nominal surface area relative to flask control was necessary to overcome the diminished contact opportunity on a microcarrier surface in suspension compared to the surface of the flask. We harvested cells after seven days as described in Methods and compared cell expansion in each microbioreactor cassette to its donor-matched TCPS flask control (Fig. 2 A). We observed statistically significant differences in cell yield between donors and also between growth platforms in this initial attempt. Cell viability was also diminished in the microbioreactor condition compared to flask control for each donor, in this early stage of the study represented in Fig. 2 . Despite these drawbacks, we nevertheless demonstrated successful MSC expansion and improved cell viability relative to initial attempts in the microbioreactor system (data not shown) for multiple cell donors. 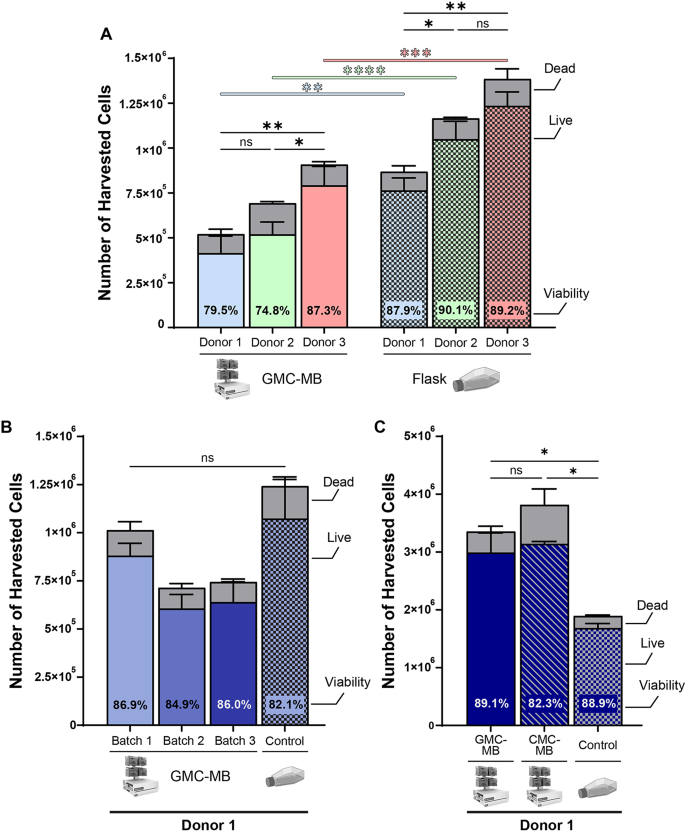 Cell harvest yield for multiple donors and growth conditions. ( A ) MSCs from three different donors were grown in microbioreactor cassettes and T25 flasks. After seven days, the cells were harvested, counted in a hemocytometer, and assessed for viability using Trypan Blue staining. MSCs were harvested from T25 flasks by incubation with Pronase digestion enzyme for five minutes at 37 C. MSCs were harvested from microbioreactor cassettes by incubation with Pronase for five minutes at 37 C to dissolve the GMCs, followed by three eject-wash cycles to collect the reactor contents. Cell counts in the microbioreactor were initially lower than their corresponding donor-matched T25 control, but cell viability was comparable and good across conditions for each donor. ( B ) MSCs from Donor 1 were grown in three separate microbioreactor cassettes and one T25 flask control to evaluate batch-to-batch variability in cell harvest yield. ( C ) MSCs were additionally grown on CMCs in the microbioreactor and a T25 flask control to compare harvest yield between GMCs and CMCs. All cell work, including counting, was performed by the same operator; improvements in cell yield between experiments likely reflect training effects and improved speed of operation for this de novo custom protocol development. Error bars represent standard deviation, SD. Significance indicators reflect results of two-way ANOVA on live cell numbers using mixed effect model and Tukey post-hoc correction for multiple comparisons. ns, not significant. *, p < 0.05. **, p < 0.005. ***, p < 0.0005. ****, p < 0.0001. GMC-MB: Gelatin microcarrier-microbioreactor We next set out to determine if we could achieve consistent cell expansion and viability across multiple biological replicates for a single donor. Using additional cryopreserved stock from Donor 1, also at passage 6—though obtained from distinct cryovials—we seeded three microbioreactor cassettes and one T25 flask with 125,000 cells each. After seven days, we harvested the cells as previously described (see Methods ) and counted the total cell yield as well as cell viability (Fig. 2 B). We found no significant difference between the cell yield of any one condition to that of any other, though the TCPS control condition maintained the highest measured cell yield. Cell yields from Donor 1 were higher in this experiment across all conditions relative to the yields obtained in Fig. 2 A, likely reflecting a training effect of improved operator speed during microbioreactor culture steps. Additionally, cell viability was similar for each group. We then sought to determine if the cell expansion, yield, and viability we observed in the microbioreactor condition were due specifically to the use of our GMCs or if similar results could be obtained with a commercially available microcarrier platform. We selected further stock of Donor 1 Passage 6 MSCs—though once again obtained from distinct cryovials relative to experiments shown in Fig. 2 A and B—and seeded 125,000 cells each into a microbioreactor cassette with 50 cm 2 GMCs, 50 cm 2 CMCs, and a T25 flask, respectively (Fig. 2 C). We harvested cells on Day 7 as described in Methods and quantified total cell yield and cell viability. We observed no statistically significant differences in cell yields or viabilities between the GMC and CMC conditions, indicating that the observed cell expansion was not dependent upon the choice of these two microcarrier identities. Further, at this stage we demonstrated significantly higher cell yield in both microbioreactor conditions relative to TCPS control. Cell viability was slightly diminished in the CMC condition relative to the other two conditions, which was expected given the more laborious and agitation-intensive harvest method required for cells expanded on those non-dissolvable microcarriers. In summary, these experiments portrayed in Fig. 2 demonstrated that we can expand MSCs in the microbioreactor condition from an initial starting culture; that we can do so for multiple donors, each with fair to good harvested cell viability; further, that we can obtain reliable growth across multiple batches for a single donor; and finally, that we can obtain higher cell counts from the microbioreactor condition compared to flask control after extensive process engineering and operator experience gain. Gene expression of potential critical quality attributesNext, we sought to determine if a combination of mechanical cues provided by the gelatin microcarrier spheres, coupled with tight regulation of the appropriate biochemical environment during the cell expansion process, could result in MSCs with an improved phenotypic profile for therapeutic use. We conducted a literature search to identify genes whose expression was either correlated or anti-correlated with therapeutic efficacy for ARDS. We identified 24 genes to serve as pCQAs and three to serve as “housekeeping” reference genes (Table 1 ). These genes and gene products, as well as citations to support their inclusion in this panel, are included in Table 1 and were selected according to one or more of the following features: (1) in vitro regulation of immune activity; (2) occurrence in an animal model of disease for the indication listed, wherein the researchers identified this gene(s) as a driving factor in therapeutic efficacy for said indication; or (3) genes identified in human ARDS patients that were correlated with either improved or worsened outcomes in those patients. We harvested total RNA content from the six growth conditions from Fig. 2 A and measured gene expression, using the TCPS flask as a normalizing control. This normalization was carried out for each respective donor, and flask controls for each donor were normalized to one and are depicted as a single striped bar (Fig. 3 ). Nine of the pCQAs were favorably modulated in all three microbioreactor conditions—in each case, upregulated gene expression is correlated to improved therapeutic effect (Fig. 3 A). Seven pCQAs were unfavorably regulated in the microbioreactor condition, either because the microbioreactor condition exhibited upregulated expression of pro-inflammatory markers (IL17, TNFα, Tissue Factor, and IL6) or downregulation of pro-therapeutic markers (DCN, FGF7, and PTX3, Fig. 3 B). The remaining eight pCQAs exhibited inconsistent modulation among donors or insufficiently high fold-change in expression to warrant further analysis and were not measured in further experiments (Fig. 3 C). 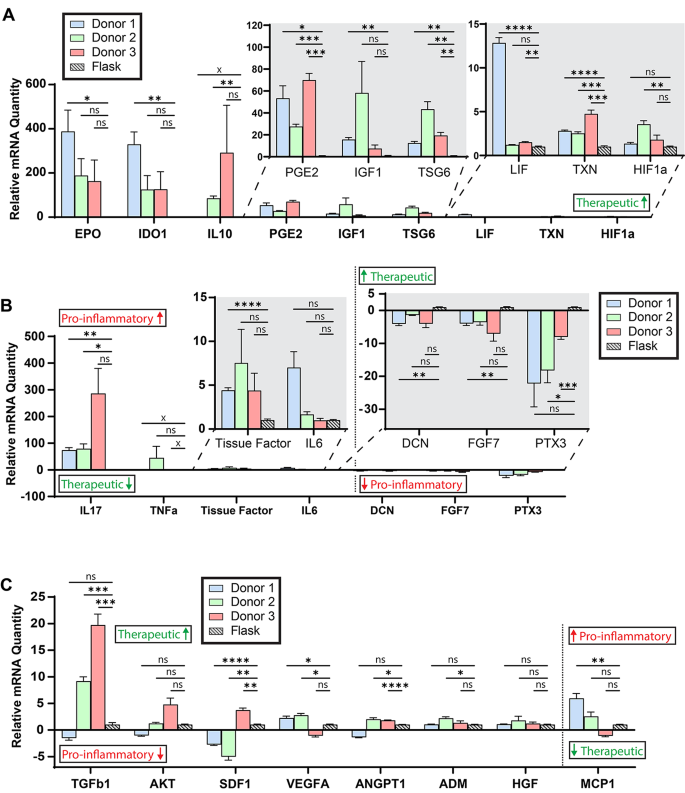 Gene Expression in gelatin microcarrier-microbioreactor MSCs Relative to Control for Three Donors. The term ‘therapeutic’ in green font (and arrow directionality) signals an anticipated correlation with improved therapeutic efficacy in ARDS, as hypothesized based on review of existing literature. The term ‘Pro-inflammatory’ in red font (and arrow directionality) signals an anticipated correlation with an anti-therapeutic effect in ARDS, as hypothesized by review of existing literature. ( A ) mRNA expression in genes promising for therapeutic efficacy in ARDS for three donors. For all genes in Panel A, higher gene expression is posited to correlate with improved therapeutic efficacy in ARDS, based on review of existing literature. Figure callouts show the same data with different axes for selected genes to enable legibility. ( B ) mRNA expression in genes which are posited to have an anti-therapeutic effect in ARDS based on existing literature. For IL17, TNFa, Tissue Factor, and IL6 in Panel B, higher expression is expected to correlate with worsened therapeutic efficacy; for DCN, FGF7, and PTX3, lower expression is expected to correlate with worsened therapeutic efficacy. ( C ) mRNA expression in genes which did not exhibit a consistent trend between donors. For the first seven genes shown from left to right in Panel C, higher expression is expected to correlate with improved therapeutic efficacy; for the eight and rightmost gene (MCP1), lower expression is expected to correlate with improved therapeutic efficacy. Each gene in the panel was measured in six technical replicates (six wells) and Ct values for individual wells are reported for each batch. An ‘x’ demarcated on the significance bar indicates no measurable expression for a given gene and donor. Gene expression was evaluated by two-step reverse-transcription qPCR using sequence-specific exon-spanning primers for each gene. Data indicate fold-change expression in GMC-microbioreactor condition relative to their donor-matched T25 control for all genes, calculated using the ΔΔCt method. Three housekeeping genes – PPIA, B2M, and GAPDH – were used to establish a baseline for comparison. Blank (water) wells and no-template controls were included on each plate to assess for contamination or primer dimer, and were always negative for fluorescence measurement. Error bars represent SEM. Significance indicators reflect results of Brown-Forsythe and Welch ANOVA tests with Dunnett T3 post-hoc correction for multiple comparisons with unequal SDs. ns, not significant. *, p < 0.05. **, p < 0.005. ***, p < 0.0005. ****, p < 0.0001. GMC: Gelatin microcarrier Having established repeatable expression of several pCQAs across multiple donors, we next sought to determine if we could obtain consistent pCQA expression across multiple biological replicates from a single donor. Similarly to the previous experiment, we harvested total RNA content from the four growth conditions depicted in Fig. 2 B and measured gene expression for the panel of pCQAs. The directionality of expression (higher or lower with respect to control conditions)—though not the magnitude in several notable cases including EPO, IDO1, and IL10—was largely consistent with that of the initial trial summarized in Fig. 3 . We observed statistically significant upregulation of six therapeutic pCQAs in at least one batch (Fig. 4 A) and significant unfavorable regulation for five pCQAs in at least one batch (Fig. 4 B). Expression of IL10 and IL6 exhibited inconsistent directionality of fold-change in this experiment. Expression of the remaining pCQAs was remarkably consistent in both directionality and magnitude of fold-change with the sole exception of IGF1, where we observed significant differences in expression fold-change relative to control for only two out of three replicates. By contrast, modulation of pro-inflammatory markers was more variable, with three genes exhibiting significant batch-to-batch differences in magnitude of fold-change expression relative to control (IL17, TNFα, and Tissue Factor). 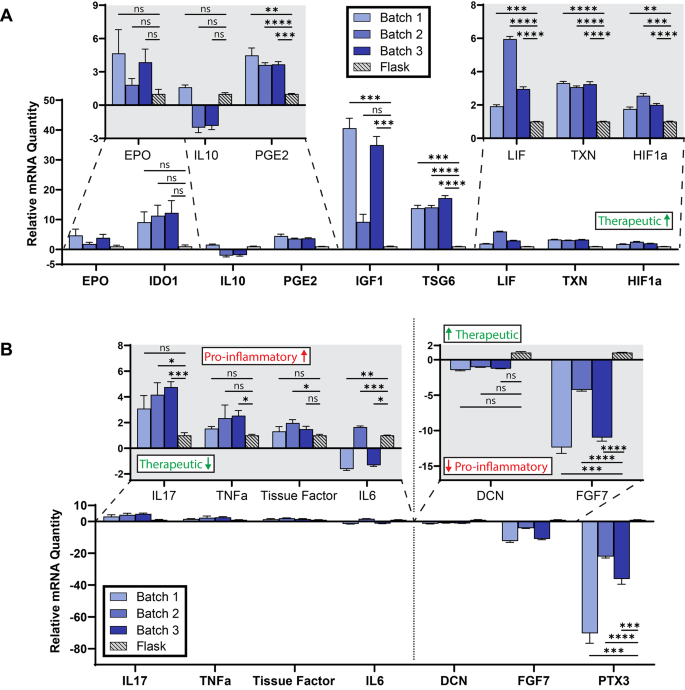 Gene Expression in gelatin microcarrier-microbioreactor MSCs Relative to Control, Three Biological Replicates. The term ‘therapeutic’ in green font (and arrow directionality) signals an anticipated correlation with improved therapeutic efficacy in ARDS, as hypothesized based on review of existing literature. The term ‘Pro-inflammatory’ in red font (and arrow directionality) signals an anticipated correlation with an anti-therapeutic effect in ARDS, as hypothesized by review of existing literature. ( A ) mRNA expression in genes promising for therapeutic efficacy in ARDS for the same donor in three different batches. For all genes in Panel A, higher gene expression is posited to correlate with improved therapeutic efficacy in ARDS, based on review of existing literature. Figure callouts show the same data with different axes for selected genes to enable legibility. A batch denotes MSCs from the same aliquot grown in a separate microbioreactor cassette. ( B ) mRNA expression in genes which are posited to have an anti-therapeutic effect in ARDS, based on existing literature. For IL17, TNFa, Tissue Factor, and IL6 in Panel B, higher expression is expected to correlate with worsened therapeutic efficacy; for DCN, FGF7, and PTX3, lower expression is expected to correlate with worsened therapeutic efficacy. Each gene in the panel was measured in six technical replicates (six wells) and Ct values for individual wells are reported for each batch. An ‘x’ demarcated on the significance bar indicates no measurable expression for a given gene and batch. Gene expression was evaluated by two-step reverse-transcription qPCR using sequence-specific exon-spanning primers for each gene. Data indicate fold-change expression in GMC-microbioreactor condition relative to their donor-matched T25 control for all genes, calculated using the ΔΔCt method. Three housekeeping genes – PPIA, B2M, and GAPDH – were used to establish a baseline for comparison. Blank (water) wells and no-template controls were included on each plate to assess for contamination or primer dimer, and were always negative for fluorescence measurement. Error bars represent SEM. Significance indicators reflect results of Brown-Forsythe and Welch ANOVA tests with Dunnett T3 post-hoc correction for multiple comparisons with unequal SDs. ns, not significant. *, p < 0.05. **, p < 0.005. ***, p < 0.0005. ****, p < 0.0001. GMC: Gelatin microcarrier Finally, we sought to confirm successful protein translation of pCQA genes for a subset of the panel. We selected three of the most-critical therapeutic pCQAs (that is, pCQAs for which there was abundant literature making strong claims for a mechanistic role in therapeutic effect for ARDS; Table 1 ) and two pro-inflammatory indicators. We harvested conditioned media on Day 7 from the four growth conditions depicted in Fig. 2 B, pooled the media from the three microbioreactor conditions, and measured secretome protein expression using unconditioned media as a background control (Fig. 5 ). We observed significantly elevated protein expression in the microbioreactor condition for two out of three therapeutic pCQA (IDO1 and IL10) and one out of two pro-inflammatory, anti-therapeutic indicator (IL17). 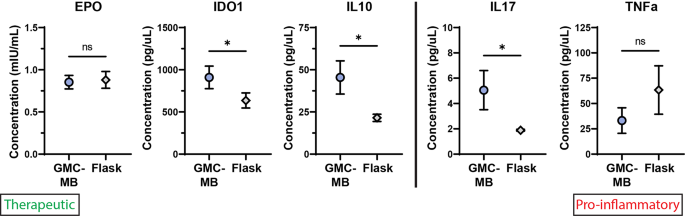 Protein Expression in gelatin microcarrier-microbioreactor and T25 Flask. The term ‘therapeutic’ in green font signals an anticipated correlation with improved therapeutic efficacy in ARDS, as hypothesized based on review of existing literature. The term ‘Pro-inflammatory’ in red font signals an anticipated correlation with an anti-therapeutic effect in ARDS, as hypothesized by review of existing literature. MSC gene expression was evaluated at the protein level for a subset of genes from Fig. 3 to confirm protein translation and evaluate the absolute concentration of effector molecule present. Protein-level data corroborated some – though not all – of the trends reported from mRNA-level data. Protein concentration was evaluated from MSC secretome by harvesting the conditioned media from culture days 5–7 and utilizing colorimetric ELISA for each analyte. Analytes were assessed at two dilutions, each in triplicate wells, for each condition. Protein concentration was normalized to cell count. Conditioned media was vortexed with 0.02% Tween20 to rupture liposomes prior to starting the ELISA protocol. Remaining steps followed the manufacturers’ protocols. Significance bars indicate results of unpaired two-tailed t-tests. *, p < 0.05. GMC-MB: Gelatin microcarrier-microbioreactor Production of cells as medicine includes challenges at the process development scale and at the manufacturing scale. These challenges include difficulties in cell yield, product heterogeneity, and product potency. Here, we describe a method of adherent cell therapy manufacturing that leverages a specific microcarrier-microbioreactor platform for enhanced control of the cellular chemical environment coupled with provision of specific physical cues to expand MSCs and modulate their secretome. We selected this particular microbioreactor system (Mobius Breez) for these experiments due to its capacity to exert tight control over the biochemical environment during cell production, with temperature regulation of ± 0.1 °C, pH control of ± 0.1 pH units, and intermittent mixing to mitigate diffusional gradients of nutrients, metabolic byproducts, and dissolved gases in reactor contents (Fig. 1 A, Supplementary Figs. 2 – 5 , Additional File 1). For example, the microbioreactor condition demonstrated > 30-fold reduction in pH drift over the seven-day culture period compared to TCPS data from literature (Supplementary Fig. 2 , Supplementary Table 1 , Additional File 1) [ 16 ]. Despite the advantageous features and appropriate scale of the microbioreactor, to date we are aware of no previous reports demonstrating production of anchorage-dependent cells such as MSCs in this system. We developed substantial modifications to default processes and protocols and worked closely with Millipore Sigma (Erbi Biosystems prior to acquisition) to implement hardware and software modifications to enable MSC manufacture; these new protocols are described in Methods. While we developed these changes for the specific use of MSCs, in principle many of the modifications we implemented could be used to enable production of other anchorage-dependent cell types, such as human induced pluripotent stem cells, in this system. With these protocol developments, we showed that we can achieve MSC expansion for multiple donors, and for multiple batches of a single donor’s cells, after extensive process parameter modification. In our multi-donor comparison, we observed substantial (and statistically significant in most cases) donor-to-donor differences in total cell yield both in the microbioreactor and in TCPS flask conditions, indicating this batch of Donor 3 cells was the most proliferative of the three selected; however, we also saw statistically significant decreases in total cell yield and substantial drops in cell viability in the microbioreactor condition relative to control for each donor, suggesting there was room for continued improvement in microbioreactor operation (Fig. 2 A). In the subsequent same-donor, multi-batch comparison, we observed higher cell yields in the microbioreactor condition across the board (Fig. 2 B); all microbioreactor settings and process parameters were held constant, leading us to hypothesize that the observed improvements reflect a combination of inherent vial-to-vial differences in this medium-passage (passage 6) of MSC stock, as well as an operator continuous improvement effect. As more experience with the modified procedures was acquired, operator speed during manual steps improved, resulting in reductions in the amount of time the microbioreactor cassettes were used in the BSC (and thus outside the biochemical control of the POD environment). In the final experiment comparing our GMC platform to a commercially available, non-dissolvable microcarrier of comparable mean diameter (Fig. 2 C), we achieved a ∼ 1.5 fold cell yield increase in the microbioreactor condition relative to the T25 control for the first time, as well as the highest absolute cell yields thus far. Once again, all microbioreactor settings and process parameters were consistent with that of the prior experiments, with the sole exception that in these experiments the cells were harvested on ice to inactivate Pronase rather than relying on dilution to mitigate compromise of cell viability. The ability to produce higher cell yields from an identical initial seeding density and culture duration in the microbioreactor was encouraging. These results by no means represent an upper bound for magnitude or reproducibility of cell yield or of product potency; we did not design this study to optimize these features but rather to demonstrate sufficient reproducibility among cell donors and batches to motivate process optimization. Future work will explore additional process parameter modifications and longer-duration culture periods in an attempt to obtain higher cell yields and improved gene product expression for this use case, which is a key aspect of process development prior to preclinical or clinical trials of potential cell therapies. In addition, while well beyond the scope of the present study, we note that the benchtop scale and closed format of this microbioreactor lends itself well to consideration of cell therapy manufacture in a point-of-care setting, potentially reducing manufacturing complexity and timelines. While challenging, process parameter modification efforts in the microbioreactor format were aided by the lower volume of media consumed in culture, which would relate to a lower cost of goods for each individual trial relative to that which would be required in a larger stirred-tank bioreactor format. In fact, operation of the microbioreactor consumed 62% less cell growth media than the T25 flask control, assuming total volume exchange every other day in the flask condition (Supplementary Fig. 6 , Supplementary Table 2 , Additional File 1). The ability to run multiple independent cassettes in parallel allowed us to conduct well-controlled experiments to assess inter- and intra-donor variation. In addition to improved biochemical control and cell yield, we further showed that this microcarrier-microbioreactor manufacturing platform modulated several key gene products that are anticipated to be correlated with therapeutic efficacy for a key example indication, ARDS. We first note that these gene products are potential CQAs and that without rigorous in vivo validation of true therapeutic activity, we cannot and do not claim that the observed changes in gene expression would correlate to improved therapeutic potency. However, one can pose a reasonable (though overly simplified) hypothesis that higher gene expression by harvested cells, for gene products that prior literature suggests a therapeutic role for ARDS, would be beneficial for therapeutic potency in this indication. At minimum, such observation would motivate further validation studies. Similarly, in an isolated context it is reasonable to expect that higher expression of pro-inflammatory gene products by MSCs would have an anti-therapeutic effect, though again this represents an overly simplistic view of the prospective role of these pCQAs in vivo. The interested reader is referred to Table 1 for a full description of the pCQAs. Building from prior work which demonstrated the importance of physical or mechanical cues for MSC gene expression and therapeutic efficacy, we showed that MSCs grown on our relatively mechanically compliant gelatin microcarriers in the microbioreactor format exhibited significantly altered gene expression of pCQAs for this indication. (Young’s elastic modulus of polystyrene is ∼ 10 9 Pa [ 33 ]; in contrast, these gelatin microcarriers exhibit stiffness of ∼ 10 4 Pa as shown in Supplementary Fig. 1 D, Additional File 1, as quantified by atomic force microscopy-enabled indentation.) We demonstrated both inter- (Fig. 3 ) and intra-donor (Fig. 4 ) repeatability for these findings. While a majority of pCQAs were favorably regulated—that is, upregulation of pro-therapeutic, immunomodulatory genes in the microcarrier-microbioreactor format relative to flask-based control—we also observed elevation of several pro-inflammatory cytokines using this novel production method. Future process modifications will determine whether further upregulation of expression of therapeutic pCQAs and downregulation of expression of inflammatory cytokines are attainable in this microcarrier-microbioreactor integration. We took rigorous precautions to ensure our gene expression data were consistent with best practices. We utilized the National Institutes of Health (NIH)’s Primer Blast tool to aid in primer design, ensuring that melting temperatures for the forward and reverse primers of each of the 27 genes surveyed were within 2 °C of one another and that each primer pair had at least one primer sequence that spanned an exon-exon gap to avoid amplifying contaminate genomic DNA. We designed multiple sets of primer pairs for each gene and validated each of the primer pairs for specificity and avoidance of primer-dimer interactions by ensuring only a single band was visible in the PCR product on an agarose gel (Supplementary Fig. 7 , Additional File 1). We also employed three housekeeping normalization genes—GAPDH as a common legacy normalization gene and two additional housekeeping genes specifically validated for MSCs—to facilitate relative quantification using the ΔΔCt method. Using three housekeeping genes instead of one provides superior accuracy for normalization [ 34 ]. Additionally, the use of stable housekeeping genes as a normalization method for qPCR has been shown to outperform normalization by total cell count or total RNA content, especially for low cell yields and low abundance genes [ 35 ]. Because expression level of each gene was assessed by its relative abundance in the microcarrier-microbioreactor condition relative to that same gene’s abundance in the control condition, on the same plate, in six technical replicates, any relative differences in primer efficiency between gene targets thus had no impact on the quantified expression. We additionally conducted a fragment analysis to assess RNA quality in both the microcarrier-microbioreactor and control conditions and found that both conditions yielded RNA with good quality (Supplementary Fig. 8 , Additional File 1). In some cases, the magnitude of fold-change in gene expression we report between the microcarrier-microbioreactor condition versus control may seem large, but we note that differences in these magnitudes are reasonable in cases for which there was little or no appreciable expression in the control condition and a limited (though appreciable) degree of expression in the microcarrier-microbioreactor cell population. In addition to evaluating mRNA content, we sought to determine if gene expression differences were also detectable at the level of functional protein (Fig. 5 ). We confirmed statistically significant changes in protein expression between conditions that mirrored the directionality of change in the mRNA data for IDO1, IL10, and IL17. For EPO and TNFα, we did not detect significantly different protein levels between conditions. Differences in magnitude between the mRNA- and protein-level data could be due to time point of collection, relative stabilities of the different proteins, and whether the protein of interest is secreted or remains internalized in the cell. Because we measured protein only in the conditioned media and not from lysed cell product (which was used to harvest total RNA instead), we would not expect to observe identically scaled protein-level changes, but the fact that we confirm significant changes in protein expression between the microcarrier-microbioreactor condition and the TCPS control and that some of these changes mirror the directionality seen in the mRNA-level data is encouraging and further validates the reliability of the qPCR measurements. ConclusionsCell therapy manufacturing in conventional tissue culture polystyrene formats carries distinct disadvantages, including open-loop or unmonitored biochemical control, provision of inappropriate physical cues, and limited scalability. Here, we report on efforts to modify a commercial microbioreactor to enable production of mesenchymal stem/stromal cells for potential therapeutic use. This microbioreactor enabled tight regulation of biochemical variables including temperature, pH, and CO 2 content in a dynamically mixed, perfusion-fed format. We utilized a custom gelatin microcarrier platform fabricated in-house to facilitate efficient cell harvest and provide mechanical cues in line with physiological values. Further, we explored whether we could use this novel manufacturing approach to prime MSCs’ expression for a panel of gene products (potential critical quality attributes) that literature suggests could be advantageous for treatment of a specific use case, acute respiratory distress syndrome. We demonstrated that we can successfully seed the microbioreactor with cells and microcarriers, enable efficient cell attachment while maintaining biochemical regulation, expand the MSC population, and reproducibly harvest cells with > 80% viability. We demonstrated this ability for multiple separate donors, as well as multiple biological replicates for one donor. After extensive process improvements and practice for fixed media composition, we achieved superior total cell yield from the microbioreactor compared to a T25 flask with identical initial seeding density. We repeated this outcome on a commercially available and non-dissolvable microcarrier, albeit with slightly diminished viability of harvested cells. Reasoning that a strong use case for demonstration of process development in a plausible indication for MSC treatment would be one that involves an inflammatory lung condition and has proven refractory to conventional therapies, we sought to gauge our manufactured cells’ therapeutic potential for treating ARDS. We analyzed the literature to build a panel of 24 genes for which expression is implicated in therapeutic efficacy for ARDS treatment (pCQAs) and assessed gene expression in our microcarrier-microbioreactor-manufactured MSCs compared to T25 flask control. We found markedly increased expression of many pCQAs in the microcarrier-microbioreactor platform, which we take to be an encouraging result, though we also found some increased expression of pro-inflammatory pCQAs and note that substantial opportunity remains for further process optimization. These trends held across multiple donors and multiple independent batches from the same donor. Finally, we quantified protein-level expression for a subset of the panel to validate successful pCQA translation and export into the cell secretome. These findings motivate future work on this approach to cell therapy manufacturing and process development in such a microbioreactor, both for enhanced cell yield and gene expression as well as validation of pCQAs. We plan to assess the impact of this cell therapy manufacturing approach on MSC intra-batch heterogeneity, which can fuel dose-to-dose variation of therapeutic potency in clinical trials and thus represents one of the substantial impediments to successful clinical translation. Figure 6 illustrates schematically the advantages of an integrated microcarrier-microbioreactor approach to cell therapy manufacture, specifically for MSC-based therapeutic process development or production. Not only does this approach enable reduced resource consumption (e.g., less media consumption, reduced operator time) and flexible manufacture (e.g., cell yield scale-up and production near point-of-care), but also the process control and biochemical/environmental control together enable rapid process development through evaluation of independent variables that may affect yield or quality attributes. For MSCs targeting specific indications via a paracrine signaling mechanism, we also showed that this approach can do more than just proliferate cells; this approach primed the MSC secretome toward improved expression of potential critical quality attributes. 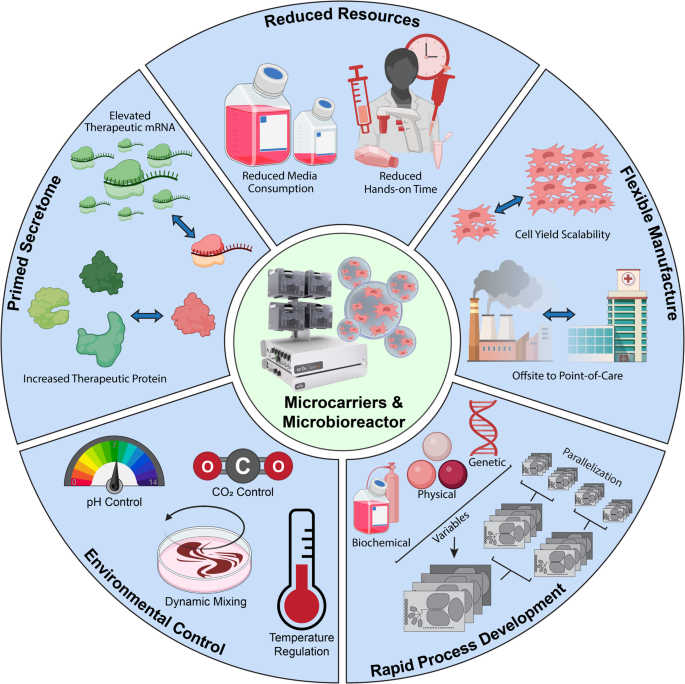 Summary Schematic of Microbioreactor-Enabled Advantages for MSC Culture This proof-of-concept study illustrates an approach to process development for a specific use case of therapeutic cell production: adherent cells for which therapeutic cell attributes may be modified by processing conditions. This approach includes closed-loop biochemical control or process analytics at the benchtop scale, with the potential to transfer these principles and practices to manufacturing scale and dosages. This microcarrier-microbioreactor integrated approach allowed us to achieve higher cell yields, comparable cell viability, reduced biochemical variation, and significantly altered gene expression with reduced growth media consumption and operator time compared to conventional flask conditions. While we find these results are promising in the context of MSC therapy for ARDS, more broadly this work also motivates investigation into additional anchorage-dependent cell types and additional target indications which would benefit from process control and quality-by-design production of cells in a research or preclinical context. Data availabilityAll data generated or analyzed during this study are included in this published article [and its supplementary information files]. AbbreviationsTwo-dimensional Three-dimensional Acute respiratory distress syndrome Biosafety cabinet Complementary DNA Chinese Hamster Ovary Commercially available microcarrier Carbon dioxide Coefficient of variation Coronavirus Disease 2019 Critical quality attribute Dulbecco’s Modified Eagle Medium Dimethyl sulfoxide Enzyme-linked immunosorbent assay Fetal Bovine Serum Food & Drug Administration Gelatin microcarrier Messenger RNA Mesenchymal stem/stromal cell National Institutes of Health Process analytic technology Phosphate buffered saline Potential critical quality attribute Polymerase chain reaction Polydimethylsiloxane Potential of hydrogen Pneumatic optical digital controller Quantitative PCR Relative centrifugal force Severe acute respiratory syndrome coronavirus 2 Tissue culture polystyrene Ultraviolet light Vessel volume per day Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 1968;6(2):230–47. Article CAS PubMed Google Scholar Roesch EA, Bonfield TL, Lazarus HM, Reese J, Hilliard K, Hilliard J, et al. A phase I study assessing the safety and tolerability of allogeneic mesenchymal stem cell infusion in adults with cystic fibrosis. J Cyst Fibros. 2023;22(3):407–13. Fu X, Liu G, Halim A, Ju Y, Luo Q, Song AG. Mesenchymal Stem Cell Migration Tissue Repair Cells. 2019;8(8). Ryan JM, Barry FP, Murphy JM, Mahon BP. Mesenchymal stem cells avoid allogeneic rejection. J Inflamm (Lond). 2005;2:8. Article PubMed Google Scholar Levy O, Kuai R, Siren EMJ, Bhere D, Milton Y, Nissar N et al. Shattering barriers toward clinically meaningful MSC therapies. Sci Adv. 2020;6. Wang J, Ren H, Yuan X, Ma H, Shi X, Ding Y. Interleukin-10 secreted by mesenchymal stem cells attenuates acute liver failure through inhibiting pyroptosis. Hepatol Res. 2018;48(3):E194–202. Ling W, Zhang J, Yuan Z, Ren G, Zhang L, Chen X, et al. Mesenchymal stem cells use IDO to regulate immunity in tumor microenvironment. Cancer Res. 2014;74(5):1576–87. Article CAS PubMed PubMed Central Google Scholar von Bahr L, Batsis I, Moll G, Hagg M, Szakos A, Sundberg B, et al. Analysis of tissues following mesenchymal stromal cell therapy in humans indicates limited long-term engraftment and no ectopic tissue formation. Stem Cells. 2012;30(7):1575–8. Article Google Scholar FDA briefing document. on remestemcel L AM session [press release]. August 13; 2020. Mesoblast Press. Release - FDA Heterogeneity Warning [press release]. September 20; 2023. Laroye C, Gauthier M, Antonot H, Decot V, Reppel L, Bensoussan D. Mesenchymal Stem/Stromal Cell Production Compliant with Good Manufacturing Practice: Comparison between Bone Marrow, the Gold Standard Adult Source, and Wharton’s Jelly, an Extraembryonic Source. J Clin Med. 2019;8(12). Rennerfeldt DA, Van Vliet KJ. Concise Review: when colonies are not clones: evidence and implications of Intracolony Heterogeneity in Mesenchymal Stem cells. Stem Cells. 2016;34(5):1135–41. Mets T, Verdonk G. Vitro Aging of Human Bone Marrow Derived Stromal cells. Mech Ageing Dev. 1981;16:81–9. Colter DCS, Prockop I. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. PNAS. 2001;98(14):7841–5. Tremain NK, Ibberson J, Kopen D, DiGirolamo GC, Phinney C. MicroSAGE Analysis of 2,353 expressed genes in a single cell-derived colony of undifferentiated human mesenchymal stem cells reveals mRNAs of multiple lineages. Stem Cells. 2001;19:408–18. Klein SG, Alsolami SM, Steckbauer A, Arossa S, Parry AJ, Ramos Mandujano G, et al. A prevalent neglect of environmental control in mammalian cell culture calls for best practices. Nat Biomed Eng. 2021;5(8):787–92. Kumar A, Starly B. Large scale industrialized cell expansion: producing the critical raw material for biofabrication processes. Biofabrication. 2015;7(4):044103. Rennerfeldt DA, Raminhos JS, Leff SM, Manning P, Van Vliet KJ. Emergent heterogeneity in putative mesenchymal stem cell colonies: single-cell time lapsed analysis. PLoS ONE. 2019;14(4):e0213452. Raman N, Imran SAM, Ahmad Amin Noordin KB, Zaman W, Nordin F. Mechanotransduction in mesenchymal stem cells (MSCs) differentiation: a review. Int J Mol Sci. 2022;23(9). Sun Y, Wan B, Wang R, Zhang B, Luo P, Wang D, et al. Mechanical stimulation on mesenchymal stem cells and surrounding microenvironments in bone regeneration: regulations and applications. Front Cell Dev Biol. 2022;10:808303. Article PubMed PubMed Central Google Scholar Werner M, Blanquer SB, Haimi SP, Korus G, Dunlop JW, Duda GN, et al. Surface curvature differentially regulates Stem Cell Migration and differentiation via altered attachment morphology and nuclear deformation. Adv Sci (Weinh). 2017;4(2):1600347. Doron G, Temenoff JS. Culture substrates for Improved manufacture of mesenchymal stromal cell therapies. Adv Healthc Mater. 2021;10(15):e2100016. Liu FD, Pishesha N, Poon Z, Kaushik T, Van Vliet KJ. Material Viscoelastic Properties modulate the mesenchymal stem cell secretome for applications in hematopoietic recovery. ACS Biomater Sci Eng. 2017;3(12):3292–306. Liu FD, Tam K, Pishesha N, Poon Z, Van Vliet KJ. Improving hematopoietic recovery through modeling and modulation of the mesenchymal stromal cell secretome. Stem Cell Res Ther. 2018;9(1):268. Ng EX, Wang M, Neo SH, Tee CA, Chen CH, Van Vliet KJ. Dissolvable gelatin-based Microcarriers generated through Droplet Microfluidics for expansion and culture of mesenchymal stromal cells. Biotechnol J. 2021;16(3):e2000048. Bower DM, Lee KS, Ram RJ, Prather KL. Fed-batch microbioreactor platform for scale down and analysis of a plasmid DNA production process. Biotechnol Bioeng. 2012;109(8):1976–86. MilliporeSigma. Mobius Breez Microbioreactor: A 2mL Automated Perfusion Cell Culture Platform. 2023. Lee HL, Boccazzi P, Ram RJ, Sinskey AJ. Microbioreactor arrays with integrated mixers and fluid injectors for high-throughput experimentation with pH and dissolved oxygen control. Lab Chip. 2006;6(9):1229–35. Lee KS, Boccazzi P, Sinskey AJ, Ram RJ. Microfluidic chemostat and turbidostat with flow rate, oxygen, and temperature control for dynamic continuous culture. Lab Chip. 2011;11(10):1730–9. Thompson BT, Chambers RC, Liu KD. Acute respiratory distress syndrome. N Engl J Med. 2017;377(6):562–72. Administration USFaD. Q8(R2) Pharmaceutical Development. In: Services USDoHaH, editor. 2009. Farruggio C. Analysis and modulation of Manufacturing conditions for Improved Cell Therapeutic Efficacy. Massachusetts Institute of Technology; 2023. O’Donnell B, White JR. Young’s modulus variation within polystyrene injection moldings. J Appl Polym Sci. 2003;47(2):189–98. Vandesompele JDP, Pattyn K, Poppe F, Van Roy B, De Paepe N, Speleman A. F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7). Erickson HS, Albert PS, Gillespie JW, Wallis BS, Rodriguez-Canales J, Linehan WM, et al. Assessment of normalization strategies for quantitative RT-PCR using microdissected tissue samples. Lab Invest. 2007;87(9):951–62. Bruno S, Chiabotto G, Camussi G. Concise review: different mesenchymal stromal/stem cell populations reside in the adult kidney. Stem Cells Transl Med. 2014;3(12):1451–5. Levy O, Kuai R, Siren EMJ, Bhere D, Milton Y, Nissar N, et al. Shattering barriers toward clinically meaningful MSC therapies. Sci Adv. 2020;6(30):eaba6884. Plotkin MD, Goligorsky MS. Mesenchymal cells from adult kidney support angiogenesis and differentiate into multiple interstitial cell types including erythropoietin-producing fibroblasts. Am J Physiol Ren Physiol. 2006;291(4):F902–12. Article CAS Google Scholar Sun C, Zhang S, Wang J, Jiang W, Xin Q, Chen X, et al. EPO enhances the protective effects of MSCs in experimental hyperoxia-induced neonatal mice by promoting angiogenesis. Aging. 2019;11(8):2477–87. Ullah M, Liu DD, Thakor AS. Mesenchymal stromal cell homing: mechanisms and strategies for improvement. iScience. 2019;15:421–38. Chu DT, Phuong TNT, Tien NLB, Tran DK, Thanh VV, Quang TL et al. An update on the progress of isolation, Culture, Storage, and clinical application of human bone marrow mesenchymal Stem/Stromal cells. Int J Mol Sci. 2020;21(3). Gonzalez-Pujana A, de Lazaro I, Vining KH, Santos-Vizcaino E, Igartua M, Hernandez RM, et al. 3D encapsulation and inflammatory licensing of mesenchymal stromal cells alter the expression of common reference genes used in real-time RT-qPCR. Biomater Sci. 2020;8(23):6741–53. Lim JY, Kim BS, Ryu DB, Kim TW, Park G, Min CK. The therapeutic efficacy of mesenchymal stromal cells on experimental colitis was improved by the IFN-gamma and poly(I:C) priming through promoting the expression of indoleamine 2,3-dioxygenase. Stem Cell Res Ther. 2021;12(1):37. Noronha NC, Mizukami A, Caliari-Oliveira C, Cominal JG, Rocha JLM, Covas DT, et al. Priming approaches to improve the efficacy of mesenchymal stromal cell-based therapies. Stem Cell Res Ther. 2019;10(1):131. Ren G, Su J, Zhang L, Zhao X, Ling W, L’Huillie A, et al. Species variation in the mechanisms of mesenchymal stem cell-mediated immunosuppression. Stem Cells. 2009;27(8):1954–62. Yin JQ, Zhu J, Ankrum JA. Manufacturing of primed mesenchymal stromal cells for therapy. Nat Biomed Eng. 2019;3(2):90–104. Godoy JAP, Paiva RMA, Souza AM, Kondo AT, Kutner JM, Okamoto OK. Clinical translation of mesenchymal stromal cell therapy for Graft Versus host disease. Front Cell Dev Biol. 2019;7:255. Li Q, Lian Y, Deng Y, Chen J, Wu T, Lai X, et al. mRNA-engineered mesenchymal stromal cells expressing CXCR2 enhances cell migration and improves recovery in IBD. Mol Ther Nucleic Acids. 2021;26:222–36. Min F, Gao F, Li Q, Liu Z. Therapeutic effect of human umbilical cord mesenchymal stem cells modified by angiotensin-converting enzyme 2 gene on bleomycin-induced lung fibrosis injury. Mol Med Rep. 2015;11(4):2387–96. Pawitan JA, Bui TA, Mubarok W, Antarianto RD, Nurhayati RW, Dilogo IH, et al. Enhancement of the therapeutic capacity of mesenchymal stem cells by genetic modification: a systematic review. Front Cell Dev Biol. 2020;8:587776. Sala E, Genua M, Petti L, Anselmo A, Arena V, Cibella J, et al. Mesenchymal stem cells reduce colitis in mice via release of TSG6, independently of their localization to the intestine. Gastroenterology. 2015;149(1):163–76. e20. Wang CL, Zhang D, Ni X, Sun Z, Zhu X. Interleukin-10-Overexpressing mesenchymal stromal cells induce a series of Regulatory effects in the Inflammatory System and promote the survival of Endotoxin-Induced Acute Lung Injury in mice Model. DNA Cell Biol. 2018;37(1):53–61. Zhu Z, Han C, Xian S, Zhuang F, Ding F, Zhang W, et al. Placental mesenchymal stromal cells (PMSCs) and PMSC-Derived extracellular vesicles (PMSC-EVs) attenuated renal fibrosis in rats with unilateral Ureteral obstruction (UUO) by regulating CD4(+) T cell polarization. Stem Cells Int. 2020;2020:2685820. Yang Z, Concannon J, Ng KS, Seyb K, Mortensen LJ, Ranganath S, et al. Tetrandrine identified in a small molecule screen to activate mesenchymal stem cells for enhanced immunomodulation. Sci Rep. 2016;6:30263. Keshtkar S, Azarpira N, Ghahremani MH. Mesenchymal stem cell-derived extracellular vesicles: novel frontiers in regenerative medicine. Stem Cell Res Ther. 2018;9(1):63. Wu H, Peng Z, Xu Y, Sheng Z, Liu Y, Liao Y, et al. Engineered adipose-derived stem cells with IGF-1-modified mRNA ameliorates osteoarthritis development. Stem Cell Res Ther. 2022;13(1):19. Kim M, Kwon JH, Bae YK, Kim GH, Um S, Ha J, et al. Soluble PTX3 of human umbilical cord blood-derived mesenchymal stem cells attenuates Hyperoxic Lung Injury by activating macrophage polarization in neonatal rat model. Stem Cells Int. 2020;2020:1802976. Qin H, Zhao A. Mesenchymal stem cell therapy for acute respiratory distress syndrome: from basic to clinics. Protein Cell. 2020;11(10):707–22. Xiao K, Hou F, Huang X, Li B, Qian ZR, Xie L. Mesenchymal stem cells: current clinical progress in ARDS and COVID-19. Stem Cell Res Ther. 2020;11(1):305. Nasef A, Mazurier C, Bouchet S, Francois S, Chapel A, Thierry D, et al. Leukemia inhibitory factor: role in human mesenchymal stem cells mediated immunosuppression. Cell Immunol. 2008;253(1–2):16–22. Suresh SC, Selvaraju V, Thirunavukkarasu M, Goldman JW, Husain A, Alexander Palesty J, et al. Thioredoxin-1 (Trx1) engineered mesenchymal stem cell therapy increased pro-angiogenic factors, reduced fibrosis and improved heart function in the infarcted rat myocardium. Int J Cardiol. 2015;201:517–28. Cerrada I, Ruiz-Sauri A, Carrero R, Trigueros C, Dorronsoro A, Sanchez-Puelles JM, et al. Hypoxia-inducible factor 1 alpha contributes to cardiac healing in mesenchymal stem cells-mediated cardiac repair. Stem Cells Dev. 2013;22(3):501–11. Rosova I, Dao M, Capoccia B, Link D, Nolta JA. Hypoxic preconditioning results in increased motility and improved therapeutic potential of human mesenchymal stem cells. Stem Cells. 2008;26(8):2173–82. Fengyun W, LiXin Z, Xinhua Q, Bin F. Mesenchymal stromal cells attenuate infection-Induced Acute Respiratory Distress Syndrome in Animal experiments: a Meta-analysis. Cell Transpl. 2020;29:963689720969186. Laroye C, Gibot S, Huselstein C, Bensoussan D. Mesenchymal stromal cells for sepsis and septic shock: lessons for treatment of COVID-19. Stem Cells Transl Med. 2020;9(12):1488–94. Merimi M, El-Majzoub R, Lagneaux L, Moussa Agha D, Bouhtit F, Meuleman N, et al. The therapeutic potential of mesenchymal stromal cells for Regenerative Medicine: current knowledge and future understandings. Front Cell Dev Biol. 2021;9:661532. Huang P, Zhang C, Delawary M, Korchak JA, Suda K, Zubair AC. Development and evaluation of IL-6 overexpressing mesenchymal stem cells (MSCs). J Tissue Eng Regen Med. 2022;16(3):244–53. Taghavi-Farahabadi M, Mahmoudi M, Soudi S, Hashemi SM. Hypothesis for the management and treatment of the COVID-19-induced acute respiratory distress syndrome and lung injury using mesenchymal stem cell-derived exosomes. Med Hypotheses. 2020;144:109865. Kwon JH, Kim M, Bae YK, Kim GH, Choi SJ, Oh W et al. Decorin secreted by human umbilical cord blood-derived mesenchymal stem cells induces macrophage polarization via CD44 to Repair Hyperoxic Lung Injury. Int J Mol Sci. 2019;20(19). Lee JW, Fang X, Gupta N, Serikov V, Matthay MA. Allogeneic human mesenchymal stem cells for treatment of E. Coli endotoxin-induced acute lung injury in the ex vivo perfused human lung. Proc Natl Acad Sci U S A. 2009;106(38):16357–62. Cherian DS, Bhuvan T, Meagher L, Heng TSP. Biological considerations in scaling up Therapeutic Cell Manufacturing. Front Pharmacol. 2020;11:654. Wang W, Xu X, Li Z, Lendlein A, Ma N. Genetic engineering of mesenchymal stem cells by non-viral gene delivery. Clin Hemorheol Microcirc. 2014;58(1):19–48. Gnecchi M, He H, Noiseux N, Liang OD, Zhang L, Morello F, et al. Evidence supporting paracrine hypothesis for akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. FASEB J. 2006;20(6):661–9. Park JS, Suryaprakash S, Lao YH, Leong KW. Engineering mesenchymal stem cells for regenerative medicine and drug delivery. Methods. 2015;84:3–16. Reiter J, Drummond S, Sammour I, Huang J, Florea V, Dornas P, et al. Stromal derived factor-1 mediates the lung regenerative effects of mesenchymal stem cells in a rodent model of bronchopulmonary dysplasia. Respir Res. 2017;18(1):137. Yang Y, Chen QH, Liu AR, Xu XP, Han JB, Qiu HB. Synergism of MSC-secreted HGF and VEGF in stabilising endothelial barrier function upon lipopolysaccharide stimulation via the Rac1 pathway. Stem Cell Res Ther. 2015;6:250. Mei SH, McCarter SD, Deng Y, Parker CH, Liles WC, Stewart DJ. Prevention of LPS-induced acute lung injury in mice by mesenchymal stem cells overexpressing angiopoietin 1. PLoS Med. 2007;4(9):e269. Wong SW, Lenzini S, Cooper MH, Mooney DJ, Shin J-W. Soft extracellular matrix enhances inflammatory activation of mesenchymal stromal cells to induce monocyte production and trafficking. Sci Adv. 2020;6(15):eaaw0158. Mesenchymal Stem Cells. In: Scheper T, editor. Basics and clinical application I. Springer; 2013. Geven C, Kox M, Pickkers P. Adrenomedullin and Adrenomedullin-targeted therapy as treatment strategies relevant for Sepsis. Front Immunol. 2018;9:292. Maacha S, Sidahmed H, Jacob S, Gentilcore G, Calzone R, Grivel JC, et al. Paracrine mechanisms of mesenchymal stromal cells in Angiogenesis. Stem Cells Int. 2020;2020:4356359. Li X, Yang Q, Bai J, Xuan Y, Wang Y. Identification of appropriate reference genes for human mesenchymal stem cell analysis by quantitative real-time PCR. Biotechnol Lett. 2015;37(1):67–73. Download references AcknowledgementsWe thank MIT’s Koch Institute’s Robert A. Swanson (1969) Biotechnology Center for technical support, specifically the BioMicro Center for consultation on qPCR and RNA fragment analysis. We gratefully acknowledge Eric Lee for his contributions in microcarrier crosslinking analysis. This work was supported in part by the Lemelson-Vest Fund for Student Invention grant award (CFF and BK), the National Research Foundation, Prime Minister’s Office, Singapore, under its Campus for Research Excellence and Technological Enterprise (CREATE) programme through the Singapore-MIT Alliance for Research and Technology (SMART) Critical Analytics for Manufacturing Personalised-Medicine (CAMP) Interdisciplinary Research Group, and the Michael (1949) and Sonja Koerner Professorship at MIT (KJVV). Author informationAuthors and affiliations. Harvard-MIT Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, Massachusetts, 02139, USA Brandon Krupczak Department of Materials Science and Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, 02139, USA Camille Farruggio & Krystyn J. Van Vliet Department of Biological Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, 02139, USA Krystyn J. Van Vliet Singapore-MIT Alliance for Research and Technology, Critical Analytics for Manufacturing Personalised-medicine, 1 Create Way, Singapore, 138602, Singapore Brandon Krupczak, Camille Farruggio & Krystyn J. Van Vliet Departments of Materials Science & Engineering and Biomedical Engineering, Cornell University, 144 Feeney Way, Ithaca, NY, 14853, USA You can also search for this author in PubMed Google Scholar ContributionsB.K.: conception and design, collection of data, data analysis and interpretation, data visualization, manuscript writing; C.F.: provision of study material, collection of data, data analysis and interpretation; K.J.V.V.: conception and design, data analysis and interpretation, manuscript writing, financial support, final approval of manuscript. Corresponding authorCorrespondence to Krystyn J. Van Vliet . Ethics declarationsEthics approval and consent to participate. Not applicable. Consent for publicationAll authors consent for publication of this manuscript. Competing interestsThe authors declare no competing interests. Additional informationPublisher’s note. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Electronic supplementary materialBelow is the link to the electronic supplementary material. Supplementary Material 1Rights and permissions. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data. Reprints and permissions About this articleCite this article. Krupczak, B., Farruggio, C. & Van Vliet, K.J. Manufacturing mesenchymal stromal cells in a microcarrier-microbioreactor platform can enhance cell yield and quality attributes: case study for acute respiratory distress syndrome. J Transl Med 22 , 614 (2024). https://doi.org/10.1186/s12967-024-05373-7 Download citation Received : 27 March 2024 Accepted : 04 June 2024 Published : 02 July 2024 DOI : https://doi.org/10.1186/s12967-024-05373-7 Share this articleAnyone you share the following link with will be able to read this content: Sorry, a shareable link is not currently available for this article. Provided by the Springer Nature SharedIt content-sharing initiative Journal of Translational MedicineISSN: 1479-5876 - Submission enquiries: Access here and click Contact Us
- General enquiries: [email protected]
 A case study on English-Malayalam Machine Translation- Sreelekha Bombay , India Pushpak Bhattacharyya
- Published in arXiv.org 27 February 2017
- Computer Science, Linguistics
Figures and Tables from this paper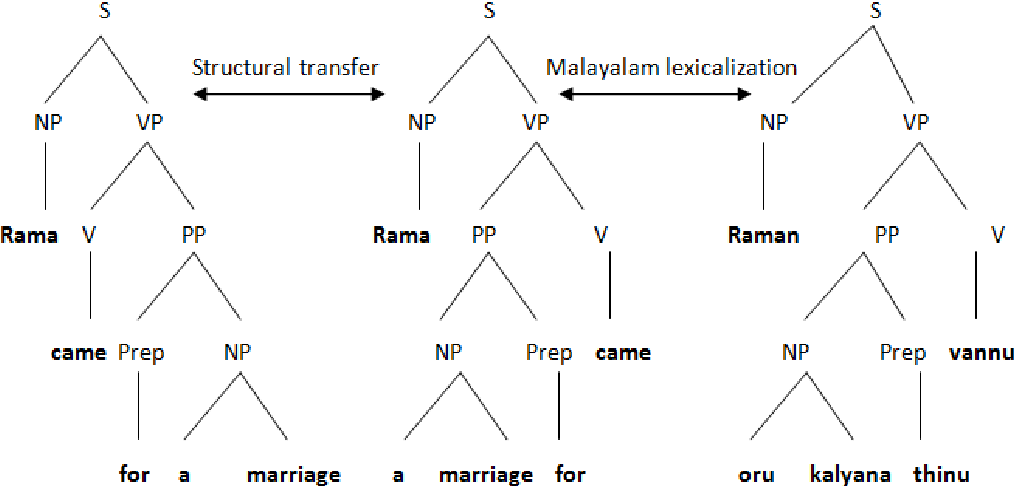 7 CitationsStatistical vs rule based machine translation; a case study on indian language perspective, lexical resources to enrich english malayalam machine translation, morphology injection for english-malayalam statistical machine translation, phrase pair mappings for hindi-english statistical machine translation, bilingual words and phrase mappings for marathi and hindi smt, role of morphology injection in smt, optimal word segmentation for neural machine translation into dravidian languages, 18 references, design and development of a malayalam to english translator-a transfer based approach, comparison of smt and rbmt; the requirement of hybridization for marathi-hindi mt, machine translation approaches and survey for indian languages, coupling statistical machine translation with rule-based transfer and generation, development of malayalam text generator for translation from english, shata-anuvadak: tackling multiway translation of indian languages, interlingua-based english–hindi machine translation and language divergence, statistical multi-source translation, decoding complexity in word-replacement translation models, lexical resources for hindi marathi mt, related papers. Showing 1 through 3 of 0 Related Papers  Published in arXiv.org 2017 Sreelekha Bombay India Pushpak Bhattacharyya  |





















IMAGES
VIDEO
COMMENTS
Sreelekha S, Pushpak Bhattacharyya. In this paper we present our work on a case study on Statistical Machine Translation (SMT) and Rule based machine translation (RBMT) for translation from English to Malayalam and Malayalam to English. One of the motivations of our study is to make a three way performance comparison, such as, a) SMT and RBMT b ...
"case study" Malayalam meaning and translation of the word. മലയാള വ്യാഖ്യാനം, അര്ഥം. അതിവേഗ ഇംഗ്ലീഷ് → മലയാളം ← മലയാളം നിഘണ്ടു
Based on our study the observations are: a) SMT systems outperform RBMT systems, b) In the case of SMT, English - Malayalam systems perform better than that of Malayalam - English systems, c) In ...
Figure 1: Structural Transfer from English - Malayalam - "A case study on English-Malayalam Machine Translation" ... Malayalam - "A case study on English-Malayalam Machine Translation" Skip to search form Skip to main content Skip to account menu. Semantic Scholar's Logo. Search 214,914,420 papers from all fields of science.
This point to the high demand of translation of. documents between English and Malayalam. The development o f proper Machine Translation. (MT) systems will help to automate the translation process ...
02/27/17 - In this paper we present our work on a case study on Statistical Machine Translation (SMT) and Rule based machine translation (RBM... AI Chat AI Image Generator AI Video Text to Speech Login. ... Login A case study on English-Malayalam Machine Translation. 02/27/2017 . ∙. by ...
A case study on English-Malayalam Machine Translation Sreelekha. S IIT Bombay India Pushpak Bhattacharyya IIT Bombay India [email protected] [email protected] Abstract machine assisted translation system or a In this paper we present our work on a case study on Statistical Machine Translation (SMT) and Rule based machine translation
വാക്കുകൾ, വാചകങ്ങൾ, വെബ് പേജുകൾ എന്നിവ മലയാളത്തിൽ നിന്ന് 100 ...
case study. Malayalam Translation. കേസ് പഠനം. kēs paṭhanaṁ. More Malayalam words for case study. കേസ് സ്റ്റഡി. kēs sṟṟaḍi case study. Find more words!
Description. A case study is an in-depth, detailed examination of a particular case within a real-world context. For example, case studies in medicine may focus on an individual patient or ailment; case studies in business might cover a particular firm's strategy or a broader market; similarly, case studies in politics can range from a narrow ...
In this paper we present our work on a case study on Statistical Machine Translation (SMT) and Rule based machine translation (RBMT) for translation from English to Malayalam and Malayalam to English. One of the motivations of our study is to make a three way performance comparison, such as, a) SMT and RBMT b) English to Malayalam SMT and Malayalam to English SMT c) English to Malayalam RBMT ...
Based on our study the observations are: a) SMT systems outperform RBMT systems, b) In the case of SMT, English - Malayalam systems perform better than that of Malayalam - English systems, c) In the case RBMT, Malayalam to English systems are performing better than English to Malayalam systems.
Generating Malayalam Word Embeddings: A Case Study. Research shows that children primarily learn languages by observing patterns in the words they hear. Computer scientists are taking a similar approach to train computers to process human language. Computers cannot process words directly. Vector representations of words, known as embeddings ...
Based on the study, this research designs and develops a system to improve the accuracy of existing Malayalam-English by including causative sentence translation mechanism. The output was compared with the output of Google Translate and identified that the Malayalam-to-English causative sentence MT system performs translations of simple ...
The requirements of incorporating morphological processing into the SMT to improve the accuracy of translation and the performance of SMT and RBMT systems are described. In this paper we present our work on a case study on Statistical Machine Translation (SMT) and Rule based machine translation (RBMT) for translation from English to Malayalam and Malayalam to English. One of the motivations of ...
Based on our study the observations are: a) SMT systems outperform RBMT systems, b) In the case of SMT, English - Malayalam systems perform better than that of Malayalam - English systems, c) In ...
78 Translation oday Use Of Language In Science Education - A Case Study Of Malayalam * V. Saratchandran Nair Abstract Language use in education has been an issue in a multilingual set up dominated by a colonial language, such as English which has become an over pervading effect because of its global stature vis-à-vis regional language/vernacular.
"Case study on English-Malayalam Machine Translation"[Sreelekha S, Pushpak Bhattacharyya, Malathi.D] iDravidian Proceedings-ICON, Pages: 1-10, 11-13 December, 2015, Trivandrum(India). "Lexical Resources for Hindi-Marathi MT" [ Sreelekha S , Pushpak Bhattacharyya, D.Malathi] WILDRE Proceedings, LREC, pages-102-110, 26-31 May, 2014 ...
Case study in Malayalam: What's Malayalam for case study? If you want to know how to say case study in Malayalam, you will find the translation here. You can also listen to audio pronunciation to learn how to pronounce case study in Malayalam and how to read it. We hope this will help you to understand Malayalam better.
114 Translation Today Globalisation, Lexical Borrowing and Language Change : A Case Study of Malayalam pookku varattu / is used, the Malayali, prefers to use / gadaagadaM/, though in Malayalam,/ pookƏ /"to go" and / varƏ/ could be used.Or, for instance the Malayali prefers to use /aakaaSavaaNi nilayaM/ for "Radio station" instead of /vaanoli nilayaM/ as in Tamil.
അതിവേഗ ഇംഗ്ലീഷ് → മലയാളം ← മലയാളം നിഘണ്ടു. ഒരു വ്യക്തിയെ ...
Table 10 : English- Malayalam SMT, RBMT- Error Analysis - "A case study on English-Malayalam Machine Translation"
Mesenchymal stem and stromal cells (MSCs) hold potential to treat a broad range of clinical indications, but clinical translation has been limited to date due in part to challenges with batch-to-batch reproducibility of potential critical quality attributes (pCQAs) that can predict potency/efficacy. Here, we designed and implemented a microcarrier-microbioreactor approach to cell therapy ...
Table 9 : Malayalam-English SMT, RBMT- Error Analysis - "A case study on English-Malayalam Machine Translation"