- Privacy Policy

Home » Research Techniques – Methods, Types and Examples

Research Techniques – Methods, Types and Examples
Table of Contents

Research Techniques
Definition:
Research techniques refer to the various methods, processes, and tools used to collect, analyze, and interpret data for the purpose of answering research questions or testing hypotheses.
Methods of Research Techniques
The methods of research techniques refer to the overall approaches or frameworks that guide a research study, including the theoretical perspective, research design, sampling strategy, data collection and analysis techniques, and ethical considerations. Some common methods of research techniques are:
- Quantitative research: This is a research method that focuses on collecting and analyzing numerical data to establish patterns, relationships, and cause-and-effect relationships. Examples of quantitative research techniques are surveys, experiments, and statistical analysis.
- Qualitative research: This is a research method that focuses on collecting and analyzing non-numerical data, such as text, images, and videos, to gain insights into the subjective experiences and perspectives of the participants. Examples of qualitative research techniques are interviews, focus groups, and content analysis.
- Mixed-methods research: This is a research method that combines quantitative and qualitative research techniques to provide a more comprehensive understanding of a research question. Examples of mixed-methods research techniques are surveys with open-ended questions and case studies with statistical analysis.
- Action research: This is a research method that focuses on solving real-world problems by collaborating with stakeholders and using a cyclical process of planning, action, and reflection. Examples of action research techniques are participatory action research and community-based participatory research.
- Experimental research : This is a research method that involves manipulating one or more variables to observe the effect on an outcome, to establish cause-and-effect relationships. Examples of experimental research techniques are randomized controlled trials and quasi-experimental designs.
- Observational research: This is a research method that involves observing and recording behavior or phenomena in natural settings to gain insights into the subject of study. Examples of observational research techniques are naturalistic observation and structured observation.
Types of Research Techniques
There are several types of research techniques used in various fields. Some of the most common ones are:
- Surveys : This is a quantitative research technique that involves collecting data through questionnaires or interviews to gather information from a large group of people.
- Experiments : This is a scientific research technique that involves manipulating one or more variables to observe the effect on an outcome, to establish cause-and-effect relationships.
- Case studies: This is a qualitative research technique that involves in-depth analysis of a single case, such as an individual, group, or event, to understand the complexities of the case.
- Observational studies : This is a research technique that involves observing and recording behavior or phenomena in natural settings to gain insights into the subject of study.
- Content analysis: This is a research technique used to analyze text or other media content to identify patterns, themes, or meanings.
- Focus groups: This is a research technique that involves gathering a small group of people to discuss a topic or issue and provide feedback on a product or service.
- Meta-analysis: This is a statistical research technique that involves combining data from multiple studies to assess the overall effect of a treatment or intervention.
- Action research: This is a research technique used to solve real-world problems by collaborating with stakeholders and using a cyclical process of planning, action, and reflection.
- Interviews : Interviews are another technique used in research, and they can be conducted in person or over the phone. They are often used to gather in-depth information about an individual’s experiences or opinions. For example, a researcher might conduct interviews with cancer patients to learn more about their experiences with treatment.
Example of Research Techniques
Here’s an example of how research techniques might be used by a student conducting a research project:
Let’s say a high school student is interested in investigating the impact of social media on mental health. They could use a variety of research techniques to gather data and analyze their findings, including:
- Literature review : The student could conduct a literature review to gather existing research studies, articles, and books that discuss the relationship between social media and mental health. This will provide a foundation of knowledge on the topic and help the student identify gaps in the research that they could address.
- Surveys : The student could design and distribute a survey to gather information from a sample of individuals about their social media usage and how it affects their mental health. The survey could include questions about the frequency of social media use, the types of content consumed, and how it makes them feel.
- Interviews : The student could conduct interviews with individuals who have experienced mental health issues and ask them about their social media use, and how it has impacted their mental health. This could provide a more in-depth understanding of how social media affects people on an individual level.
- Data analysis : The student could use statistical software to analyze the data collected from the surveys and interviews. This would allow them to identify patterns and relationships between social media usage and mental health outcomes.
- Report writing : Based on the findings from their research, the student could write a report that summarizes their research methods, findings, and conclusions. They could present their report to their peers or their teacher to share their insights on the topic.
Overall, by using a combination of research techniques, the student can investigate their research question thoroughly and systematically, and make meaningful contributions to the field of social media and mental health research.
Purpose of Research Techniques
The Purposes of Research Techniques are as follows:
- To investigate and gain knowledge about a particular phenomenon or topic
- To generate new ideas and theories
- To test existing theories and hypotheses
- To identify and evaluate potential solutions to problems
- To gather data and evidence to inform decision-making
- To identify trends and patterns in data
- To explore cause-and-effect relationships between variables
- To develop and refine measurement tools and methodologies
- To establish the reliability and validity of research findings
- To communicate research findings to others in a clear and concise manner.
Applications of Research Techniques
Here are some applications of research techniques:
- Scientific research: to explore, investigate and understand natural phenomena, and to generate new knowledge and theories.
- Market research: to collect and analyze data about consumer behavior, preferences, and trends, and to help businesses make informed decisions about product development, pricing, and marketing strategies.
- Medical research : to study diseases and their treatments, and to develop new medicines, therapies, and medical technologies.
- Social research : to explore and understand human behavior, attitudes, and values, and to inform public policy decisions related to education, health care, social welfare, and other areas.
- Educational research : to study teaching and learning processes, and to develop effective teaching methods and instructional materials.
- Environmental research: to investigate the impact of human activities on the environment, and to develop solutions to environmental problems.
- Engineering Research: to design, develop, and improve products, processes, and systems, and to optimize their performance and efficiency.
- Criminal justice research : to study crime patterns, causes, and prevention strategies, and to evaluate the effectiveness of criminal justice policies and programs.
- Psychological research : to investigate human cognition, emotion, and behavior, and to develop interventions to address mental health issues.
- Historical research: to study past events, societies, and cultures, and to develop an understanding of how they shape our present.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Data Collection – Methods Types and Examples

Delimitations in Research – Types, Examples and...

Research Process – Steps, Examples and Tips

Research Design – Types, Methods and Examples

Institutional Review Board – Application Sample...

Evaluating Research – Process, Examples and...
News alert: UC Berkeley has announced its next university librarian
Secondary menu
- Log in to your Library account
- Hours and Maps
- Connect from Off Campus
- UC Berkeley Home
Search form
Research methods--quantitative, qualitative, and more: overview.
- Quantitative Research
- Qualitative Research
- Data Science Methods (Machine Learning, AI, Big Data)
- Text Mining and Computational Text Analysis
- Evidence Synthesis/Systematic Reviews
- Get Data, Get Help!
About Research Methods
This guide provides an overview of research methods, how to choose and use them, and supports and resources at UC Berkeley.
As Patten and Newhart note in the book Understanding Research Methods , "Research methods are the building blocks of the scientific enterprise. They are the "how" for building systematic knowledge. The accumulation of knowledge through research is by its nature a collective endeavor. Each well-designed study provides evidence that may support, amend, refute, or deepen the understanding of existing knowledge...Decisions are important throughout the practice of research and are designed to help researchers collect evidence that includes the full spectrum of the phenomenon under study, to maintain logical rules, and to mitigate or account for possible sources of bias. In many ways, learning research methods is learning how to see and make these decisions."
The choice of methods varies by discipline, by the kind of phenomenon being studied and the data being used to study it, by the technology available, and more. This guide is an introduction, but if you don't see what you need here, always contact your subject librarian, and/or take a look to see if there's a library research guide that will answer your question.
Suggestions for changes and additions to this guide are welcome!
START HERE: SAGE Research Methods
Without question, the most comprehensive resource available from the library is SAGE Research Methods. HERE IS THE ONLINE GUIDE to this one-stop shopping collection, and some helpful links are below:
- SAGE Research Methods
- Little Green Books (Quantitative Methods)
- Little Blue Books (Qualitative Methods)
- Dictionaries and Encyclopedias
- Case studies of real research projects
- Sample datasets for hands-on practice
- Streaming video--see methods come to life
- Methodspace- -a community for researchers
- SAGE Research Methods Course Mapping
Library Data Services at UC Berkeley
Library Data Services Program and Digital Scholarship Services
The LDSP offers a variety of services and tools ! From this link, check out pages for each of the following topics: discovering data, managing data, collecting data, GIS data, text data mining, publishing data, digital scholarship, open science, and the Research Data Management Program.
Be sure also to check out the visual guide to where to seek assistance on campus with any research question you may have!
Library GIS Services
Other Data Services at Berkeley
D-Lab Supports Berkeley faculty, staff, and graduate students with research in data intensive social science, including a wide range of training and workshop offerings Dryad Dryad is a simple self-service tool for researchers to use in publishing their datasets. It provides tools for the effective publication of and access to research data. Geospatial Innovation Facility (GIF) Provides leadership and training across a broad array of integrated mapping technologies on campu Research Data Management A UC Berkeley guide and consulting service for research data management issues
General Research Methods Resources
Here are some general resources for assistance:
- Assistance from ICPSR (must create an account to access): Getting Help with Data , and Resources for Students
- Wiley Stats Ref for background information on statistics topics
- Survey Documentation and Analysis (SDA) . Program for easy web-based analysis of survey data.
Consultants
- D-Lab/Data Science Discovery Consultants Request help with your research project from peer consultants.
- Research data (RDM) consulting Meet with RDM consultants before designing the data security, storage, and sharing aspects of your qualitative project.
- Statistics Department Consulting Services A service in which advanced graduate students, under faculty supervision, are available to consult during specified hours in the Fall and Spring semesters.
Related Resourcex
- IRB / CPHS Qualitative research projects with human subjects often require that you go through an ethics review.
- OURS (Office of Undergraduate Research and Scholarships) OURS supports undergraduates who want to embark on research projects and assistantships. In particular, check out their "Getting Started in Research" workshops
- Sponsored Projects Sponsored projects works with researchers applying for major external grants.
- Next: Quantitative Research >>
- Last Updated: Apr 25, 2024 11:09 AM
- URL: https://guides.lib.berkeley.edu/researchmethods
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, automatically generate references for free.
- Knowledge Base
- Methodology
Research Methods | Definition, Types, Examples
Research methods are specific procedures for collecting and analysing data. Developing your research methods is an integral part of your research design . When planning your methods, there are two key decisions you will make.
First, decide how you will collect data . Your methods depend on what type of data you need to answer your research question :
- Qualitative vs quantitative : Will your data take the form of words or numbers?
- Primary vs secondary : Will you collect original data yourself, or will you use data that have already been collected by someone else?
- Descriptive vs experimental : Will you take measurements of something as it is, or will you perform an experiment?
Second, decide how you will analyse the data .
- For quantitative data, you can use statistical analysis methods to test relationships between variables.
- For qualitative data, you can use methods such as thematic analysis to interpret patterns and meanings in the data.
Table of contents
Methods for collecting data, examples of data collection methods, methods for analysing data, examples of data analysis methods, frequently asked questions about methodology.
Data are the information that you collect for the purposes of answering your research question . The type of data you need depends on the aims of your research.
Qualitative vs quantitative data
Your choice of qualitative or quantitative data collection depends on the type of knowledge you want to develop.
For questions about ideas, experiences and meanings, or to study something that can’t be described numerically, collect qualitative data .
If you want to develop a more mechanistic understanding of a topic, or your research involves hypothesis testing , collect quantitative data .
You can also take a mixed methods approach, where you use both qualitative and quantitative research methods.
Primary vs secondary data
Primary data are any original information that you collect for the purposes of answering your research question (e.g. through surveys , observations and experiments ). Secondary data are information that has already been collected by other researchers (e.g. in a government census or previous scientific studies).
If you are exploring a novel research question, you’ll probably need to collect primary data. But if you want to synthesise existing knowledge, analyse historical trends, or identify patterns on a large scale, secondary data might be a better choice.
Descriptive vs experimental data
In descriptive research , you collect data about your study subject without intervening. The validity of your research will depend on your sampling method .
In experimental research , you systematically intervene in a process and measure the outcome. The validity of your research will depend on your experimental design .
To conduct an experiment, you need to be able to vary your independent variable , precisely measure your dependent variable, and control for confounding variables . If it’s practically and ethically possible, this method is the best choice for answering questions about cause and effect.
Prevent plagiarism, run a free check.
Your data analysis methods will depend on the type of data you collect and how you prepare them for analysis.
Data can often be analysed both quantitatively and qualitatively. For example, survey responses could be analysed qualitatively by studying the meanings of responses or quantitatively by studying the frequencies of responses.
Qualitative analysis methods
Qualitative analysis is used to understand words, ideas, and experiences. You can use it to interpret data that were collected:
- From open-ended survey and interview questions, literature reviews, case studies, and other sources that use text rather than numbers.
- Using non-probability sampling methods .
Qualitative analysis tends to be quite flexible and relies on the researcher’s judgement, so you have to reflect carefully on your choices and assumptions.
Quantitative analysis methods
Quantitative analysis uses numbers and statistics to understand frequencies, averages and correlations (in descriptive studies) or cause-and-effect relationships (in experiments).
You can use quantitative analysis to interpret data that were collected either:
- During an experiment.
- Using probability sampling methods .
Because the data are collected and analysed in a statistically valid way, the results of quantitative analysis can be easily standardised and shared among researchers.
Quantitative research deals with numbers and statistics, while qualitative research deals with words and meanings.
Quantitative methods allow you to test a hypothesis by systematically collecting and analysing data, while qualitative methods allow you to explore ideas and experiences in depth.
In mixed methods research , you use both qualitative and quantitative data collection and analysis methods to answer your research question .
A sample is a subset of individuals from a larger population. Sampling means selecting the group that you will actually collect data from in your research.
For example, if you are researching the opinions of students in your university, you could survey a sample of 100 students.
Statistical sampling allows you to test a hypothesis about the characteristics of a population. There are various sampling methods you can use to ensure that your sample is representative of the population as a whole.
The research methods you use depend on the type of data you need to answer your research question .
- If you want to measure something or test a hypothesis , use quantitative methods . If you want to explore ideas, thoughts, and meanings, use qualitative methods .
- If you want to analyse a large amount of readily available data, use secondary data. If you want data specific to your purposes with control over how they are generated, collect primary data.
- If you want to establish cause-and-effect relationships between variables , use experimental methods. If you want to understand the characteristics of a research subject, use descriptive methods.
Methodology refers to the overarching strategy and rationale of your research project . It involves studying the methods used in your field and the theories or principles behind them, in order to develop an approach that matches your objectives.
Methods are the specific tools and procedures you use to collect and analyse data (e.g. experiments, surveys , and statistical tests ).
In shorter scientific papers, where the aim is to report the findings of a specific study, you might simply describe what you did in a methods section .
In a longer or more complex research project, such as a thesis or dissertation , you will probably include a methodology section , where you explain your approach to answering the research questions and cite relevant sources to support your choice of methods.
Is this article helpful?
More interesting articles.
- A Quick Guide to Experimental Design | 5 Steps & Examples
- Between-Subjects Design | Examples, Pros & Cons
- Case Study | Definition, Examples & Methods
- Cluster Sampling | A Simple Step-by-Step Guide with Examples
- Confounding Variables | Definition, Examples & Controls
- Construct Validity | Definition, Types, & Examples
- Content Analysis | A Step-by-Step Guide with Examples
- Control Groups and Treatment Groups | Uses & Examples
- Controlled Experiments | Methods & Examples of Control
- Correlation vs Causation | Differences, Designs & Examples
- Correlational Research | Guide, Design & Examples
- Critical Discourse Analysis | Definition, Guide & Examples
- Cross-Sectional Study | Definitions, Uses & Examples
- Data Cleaning | A Guide with Examples & Steps
- Data Collection Methods | Step-by-Step Guide & Examples
- Descriptive Research Design | Definition, Methods & Examples
- Doing Survey Research | A Step-by-Step Guide & Examples
- Ethical Considerations in Research | Types & Examples
- Explanatory Research | Definition, Guide, & Examples
- Explanatory vs Response Variables | Definitions & Examples
- Exploratory Research | Definition, Guide, & Examples
- External Validity | Types, Threats & Examples
- Extraneous Variables | Examples, Types, Controls
- Face Validity | Guide with Definition & Examples
- How to Do Thematic Analysis | Guide & Examples
- How to Write a Strong Hypothesis | Guide & Examples
- Inclusion and Exclusion Criteria | Examples & Definition
- Independent vs Dependent Variables | Definition & Examples
- Inductive Reasoning | Types, Examples, Explanation
- Inductive vs Deductive Research Approach (with Examples)
- Internal Validity | Definition, Threats & Examples
- Internal vs External Validity | Understanding Differences & Examples
- Longitudinal Study | Definition, Approaches & Examples
- Mediator vs Moderator Variables | Differences & Examples
- Mixed Methods Research | Definition, Guide, & Examples
- Multistage Sampling | An Introductory Guide with Examples
- Naturalistic Observation | Definition, Guide & Examples
- Operationalisation | A Guide with Examples, Pros & Cons
- Population vs Sample | Definitions, Differences & Examples
- Primary Research | Definition, Types, & Examples
- Qualitative vs Quantitative Research | Examples & Methods
- Quasi-Experimental Design | Definition, Types & Examples
- Questionnaire Design | Methods, Question Types & Examples
- Random Assignment in Experiments | Introduction & Examples
- Reliability vs Validity in Research | Differences, Types & Examples
- Reproducibility vs Replicability | Difference & Examples
- Research Design | Step-by-Step Guide with Examples
- Sampling Methods | Types, Techniques, & Examples
- Semi-Structured Interview | Definition, Guide & Examples
- Simple Random Sampling | Definition, Steps & Examples
- Stratified Sampling | A Step-by-Step Guide with Examples
- Structured Interview | Definition, Guide & Examples
- Systematic Review | Definition, Examples & Guide
- Systematic Sampling | A Step-by-Step Guide with Examples
- Textual Analysis | Guide, 3 Approaches & Examples
- The 4 Types of Reliability in Research | Definitions & Examples
- The 4 Types of Validity | Types, Definitions & Examples
- Transcribing an Interview | 5 Steps & Transcription Software
- Triangulation in Research | Guide, Types, Examples
- Types of Interviews in Research | Guide & Examples
- Types of Research Designs Compared | Examples
- Types of Variables in Research | Definitions & Examples
- Unstructured Interview | Definition, Guide & Examples
- What Are Control Variables | Definition & Examples
- What Is a Case-Control Study? | Definition & Examples
- What Is a Cohort Study? | Definition & Examples
- What Is a Conceptual Framework? | Tips & Examples
- What Is a Double-Barrelled Question?
- What Is a Double-Blind Study? | Introduction & Examples
- What Is a Focus Group? | Step-by-Step Guide & Examples
- What Is a Likert Scale? | Guide & Examples
- What is a Literature Review? | Guide, Template, & Examples
- What Is a Prospective Cohort Study? | Definition & Examples
- What Is a Retrospective Cohort Study? | Definition & Examples
- What Is Action Research? | Definition & Examples
- What Is an Observational Study? | Guide & Examples
- What Is Concurrent Validity? | Definition & Examples
- What Is Content Validity? | Definition & Examples
- What Is Convenience Sampling? | Definition & Examples
- What Is Convergent Validity? | Definition & Examples
- What Is Criterion Validity? | Definition & Examples
- What Is Deductive Reasoning? | Explanation & Examples
- What Is Discriminant Validity? | Definition & Example
- What Is Ecological Validity? | Definition & Examples
- What Is Ethnography? | Meaning, Guide & Examples
- What Is Non-Probability Sampling? | Types & Examples
- What Is Participant Observation? | Definition & Examples
- What Is Peer Review? | Types & Examples
- What Is Predictive Validity? | Examples & Definition
- What Is Probability Sampling? | Types & Examples
- What Is Purposive Sampling? | Definition & Examples
- What Is Qualitative Observation? | Definition & Examples
- What Is Qualitative Research? | Methods & Examples
- What Is Quantitative Observation? | Definition & Examples
- What Is Quantitative Research? | Definition & Methods
- What Is Quota Sampling? | Definition & Examples
- What is Secondary Research? | Definition, Types, & Examples
- What Is Snowball Sampling? | Definition & Examples
- Within-Subjects Design | Explanation, Approaches, Examples

About methods, techniques, and research tools
The next step in the work of a researcher: the choice of a research method.
Let’s consider a research project as a journey: its purpose is to find answers to the (research) questions compelling to the researcher. The path to this goal is marked by the research method . Your role as a social researcher is to consciously choose the most appropriate path and follow its principles and guidelines; this oath is about choosing a research question . You also have to consider the reliability of your measurements. Remember that in the scientific community only the reliable data is considered worth analyzing, drawing conclusions and arguing about. Thus, the objectives of the research together with the research questions set the direction of your research, while the research method indicates how you will collect data. In addition, research methods imply the research tools that should be used to gather data and draw conclusions.
Note : The aim of this module is to provide practical tips and necessary knowledge for choosing the appropriate research method. During this module, you will see examples of research methods used in social sciences, but at this stage we will not discuss them in detail. You will learn more about the application of selected methods in the next section of the Toolbox: Conducting Research.
A research method is defined as a way of scientifically studying a phenomenon. It consists of specific activities within the research procedure, supplemented by a set of tools used to collect and analyze data.
The intentional , planned , and conscious choice of a research method is a guarantee of the success of the entire research project. The choice of the research method should therefore precede the research itself and lead to the consistent application of the principles of the method during its course (small lapses are possible in qualitative research, which you will learn about soon). The research method is characterized by its replicability . By this we mean that the results of the study should be verifiable and help another researcher replicate the study in another context. The diagram below presents the characteristics of research method that you should keep in mind when implementing your research project.
The concept of the research method is related to another concept that is important to us: the research technique . For each method, we can indicate many techniques that in some way detail the sequence of research activities within the selected method, while also indicating the optimal research tools. In other words, while the research method quite generally dictates the way of scientific study, the technique tells what the process should look like and with what tools it should be carried out. Usually, we are talking about a data collection technique, but its choice also determines the method of data analysis and verification of research hypotheses.
Various classifications of research methods and techniques were proposed in academic literature. Sometimes one author qualifies a procedure as a method, and another defines it as a technique. This problem is highlighted by Bäcker et al. (2016: 66-67):
in practice, the distinction between research method and research technique is vague. It is said that a method is more general than a research technique, but the line between general and specific is not set at one precise point. This distinction is somewhat intuitive (...) Ultimately, it is less important whether the set of methodological rules used in the study is called a method or a technique. It is more important that it is applied correctly and complies with the requirements.
When carrying out a research project, you will almost certainly come across terms like "methodology" or "research technique" and ‘method’. These terms look similar, but shouldn’t be used interchangeably. Their proper use proves your readiness for the role of a researcher. Sometimes, in the work of students, the word “methodology” tends to be misused. In fact, it simply means the study of scientific research methods and research procedures. You have already learned what we mean by the term "research method". Now try to intuitively link the terms from the list with their correct definition.
Top 21 must-have digital tools for researchers
Last updated
12 May 2023
Reviewed by
Jean Kaluza
Research drives many decisions across various industries, including:
Uncovering customer motivations and behaviors to design better products
Assessing whether a market exists for your product or service
Running clinical studies to develop a medical breakthrough
Conducting effective and shareable research can be a painstaking process. Manual processes are sluggish and archaic, and they can also be inaccurate. That’s where advanced online tools can help.
The right tools can enable businesses to lean into research for better forecasting, planning, and more reliable decisions.
- Why do researchers need research tools?
Research is challenging and time-consuming. Analyzing data , running focus groups , reading research papers , and looking for useful insights take plenty of heavy lifting.
These days, researchers can’t just rely on manual processes. Instead, they’re using advanced tools that:
Speed up the research process
Enable new ways of reaching customers
Improve organization and accuracy
Allow better monitoring throughout the process
Enhance collaboration across key stakeholders
- The most important digital tools for researchers
Some tools can help at every stage, making researching simpler and faster.
They ensure accurate and efficient information collection, management, referencing, and analysis.
Some of the most important digital tools for researchers include:
Research management tools
Research management can be a complex and challenging process. Some tools address the various challenges that arise when referencing and managing papers.
.css-10ptwjf{-webkit-align-items:center;-webkit-box-align:center;-ms-flex-align:center;align-items:center;background:transparent;border:0;color:inherit;cursor:pointer;-webkit-flex-shrink:0;-ms-flex-negative:0;flex-shrink:0;-webkit-text-decoration:underline;text-decoration:underline;}.css-10ptwjf:disabled{opacity:0.6;pointer-events:none;} Zotero
Coined as a personal research assistant, Zotero is a tool that brings efficiency to the research process. Zotero helps researchers collect, organize, annotate, and share research easily.
Zotero integrates with internet browsers, so researchers can easily save an article, publication, or research study on the platform for later.
The tool also has an advanced organizing system to allow users to label, tag, and categorize information for faster insights and a seamless analysis process.
Messy paper stacks––digital or physical––are a thing of the past with Paperpile. This reference management tool integrates with Google Docs, saving users time with citations and paper management.
Referencing, researching, and gaining insights is much cleaner and more productive, as all papers are in the same place. Plus, it’s easier to find a paper when you need it.
Acting as a single source of truth (SSOT), Dovetail houses research from the entire organization in a simple-to-use place. Researchers can use the all-in-one platform to collate and store data from interviews , forms, surveys , focus groups, and more.
Dovetail helps users quickly categorize and analyze data to uncover truly actionable insights . This helps organizations bring customer insights into every decision for better forecasting, planning, and decision-making.
Dovetail integrates with other helpful tools like Slack, Atlassian, Notion, and Zapier for a truly efficient workflow.
Putting together papers and referencing sources can be a huge time consumer. EndNote claims that researchers waste 200,000 hours per year formatting citations.
To address the issue, the tool formats citations automatically––simultaneously creating a bibliography while the user writes.
EndNote is also a cloud-based system that allows remote working, multiple-user interaction and collaboration, and seamless working on different devices.
Information survey tools
Surveys are a common way to gain data from customers. These tools can make the process simpler and more cost-effective.
With ready-made survey templates––to collect NPS data, customer effort scores , five-star surveys, and more––getting going with Delighted is straightforward.
Delighted helps teams collect and analyze survey feedback without needing any technical knowledge. The templates are customizable, so you can align the content with your brand. That way, the survey feels like it’s coming from your company, not a third party.
SurveyMonkey
With millions of customers worldwide, SurveyMonkey is another leader in online surveys. SurveyMonkey offers hundreds of templates that researchers can use to set up and deploy surveys quickly.
Whether your survey is about team performance, hotel feedback, post-event feedback, or an employee exit, SurveyMonkey has a ready-to-use template.
Typeform offers free templates you can quickly embed, which comes with a point of difference: It designs forms and surveys with people in mind, focusing on customer enjoyment.
Typeform employs the ‘one question at a time’ method to keep engagement rates and completions high. It focuses on surveys that feel more like conversations than a list of questions.
Web data analysis tools
Collecting data can take time––especially technical information. Some tools make that process simpler.
For those conducting clinical research, data collection can be incredibly time-consuming. Teamscope provides an online platform to collect and manage data simply and easily.
Researchers and medical professionals often collect clinical data through paper forms or digital means. Those are too easy to lose, tricky to manage, and challenging to collaborate on.
With Teamscope, you can easily collect, store, and electronically analyze data like patient-reported outcomes and surveys.
Heap is a digital insights platform providing context on the entire customer journey . This helps businesses improve customer feedback , conversion rates, and loyalty.
Through Heap, you can seamlessly view and analyze the customer journey across all platforms and touchpoints, whether through the app or website.
Another analytics tool, Smartlook, combines quantitative and qualitative analytics into one platform. This helps organizations understand user behavior and make crucial improvements.
Smartlook is useful for analyzing web pages, purchasing flows, and optimizing conversion rates.
Project management tools
Managing multiple research projects across many teams can be complex and challenging. Project management tools can ease the burden on researchers.
Visual productivity tool Trello helps research teams manage their projects more efficiently. Trello makes product tracking easier with:
A range of workflow options
Unique project board layouts
Advanced descriptions
Integrations
Trello also works as an SSOT to stay on top of projects and collaborate effectively as a team.
To connect research, workflows, and teams, Airtable provides a clean interactive interface.
With Airtable, it’s simple to place research projects in a list view, workstream, or road map to synthesize information and quickly collaborate. The Sync feature makes it easy to link all your research data to one place for faster action.
For product teams, Asana gathers development, copywriting, design, research teams, and product managers in one space.
As a task management platform, Asana offers all the expected features and more, including time-tracking and Jira integration. The platform offers reporting alongside data collection methods , so it’s a favorite for product teams in the tech space.
Grammar checker tools
Grammar tools ensure your research projects are professional and proofed.
No one’s perfect, especially when it comes to spelling, punctuation, and grammar. That’s where Grammarly can help.
Grammarly’s AI-powered platform reviews your content and corrects any mistakes. Through helpful integrations with other platforms––such as Gmail, Google Docs, Twitter, and LinkedIn––it’s simple to spellcheck as you go.
Another helpful grammar tool is Trinka AI. Trinka is specifically for technical and academic styles of writing. It doesn’t just correct mistakes in spelling, punctuation, and grammar; it also offers explanations and additional information when errors show.
Researchers can also use Trinka to enhance their writing and:
Align it with technical and academic styles
Improve areas like syntax and word choice
Discover relevant suggestions based on the content topic
Plagiarism checker tools
Avoiding plagiarism is crucial for the integrity of research. Using checker tools can ensure your work is original.
Plagiarism checker Quetext uses DeepSearch™ technology to quickly sort through online content to search for signs of plagiarism.
With color coding, annotations, and an overall score, it’s easy to identify conflict areas and fix them accordingly.
Duplichecker
Another helpful plagiarism tool is Duplichecker, which scans pieces of content for issues. The service is free for content up to 1000 words, with paid options available after that.
If plagiarism occurs, a percentage identifies how much is duplicate content. However, the interface is relatively basic, offering little additional information.
Journal finder tools
Finding the right journals for your project can be challenging––especially with the plethora of inaccurate or predatory content online. Journal finder tools can solve this issue.
Enago Journal Finder
The Enago Open Access Journal Finder sorts through online journals to verify their legitimacy. Through Engao, you can discover pre-vetted, high-quality journals through a validated journal index.
Enago’s search tool also helps users find relevant journals for their subject matter, speeding up the research process.
JournalFinder
JournalFinder is another journal tool that’s popular with academics and researchers. It makes the process of discovering relevant journals fast by leaning into a machine-learning algorithm.
This is useful for discovering key information and finding the right journals to publish and share your work in.
Social networking for researchers
Collaboration between researchers can improve the accuracy and sharing of information. Promoting research findings can also be essential for public health, safety, and more.
While typical social networks exist, some are specifically designed for academics.
ResearchGate
Networking platform ResearchGate encourages researchers to connect, collaborate, and share within the scientific community. With 20 million researchers on the platform, it's a popular choice.
ResearchGate is founded on an intention to advance research. The platform provides topic pages for easy connection within a field of expertise and access to millions of publications to help users stay up to date.
Academia is another commonly used platform that connects 220 million academics and researchers within their specialties.
The platform aims to accelerate research with discovery tools and grow a researcher’s audience to promote their ideas.
On Academia, users can access 47 million PDFs for free. They cover topics from mechanical engineering to applied economics and child psychology.
- Expedited research with the power of tools
For researchers, finding data and information can be time-consuming and complex to manage. That’s where the power of tools comes in.
Manual processes are slow, outdated, and have a larger potential for inaccuracies.
Leaning into tools can help researchers speed up their processes, conduct efficient research, boost their accuracy, and share their work effectively.
With tools available for project and data management, web data collection, and journal finding, researchers have plenty of assistance at their disposal.
When it comes to connecting with customers, advanced tools boost customer connection while continually bringing their needs and wants into products and services.
What are primary research tools?
Primary research is data and information that you collect firsthand through surveys, customer interviews, or focus groups.
Secondary research is data and information from other sources, such as journals, research bodies, or online content.
Primary researcher tools use methods like surveys and customer interviews. You can use these tools to collect, store, or manage information effectively and uncover more accurate insights.
What is the difference between tools and methods in research?
Research methods relate to how researchers gather information and data.
For example, surveys, focus groups, customer interviews, and A/B testing are research methods that gather information.
On the other hand, tools assist areas of research. Researchers may use tools to more efficiently gather data, store data securely, or uncover insights.
Tools can improve research methods, ensuring efficiency and accuracy while reducing complexity.
Should you be using a customer insights hub?
Do you want to discover previous research faster?
Do you share your research findings with others?
Do you analyze research data?
Start for free today, add your research, and get to key insights faster
Editor’s picks
Last updated: 13 April 2023
Last updated: 14 February 2024
Last updated: 27 January 2024
Last updated: 18 April 2023
Last updated: 8 February 2023
Last updated: 23 January 2024
Last updated: 30 January 2024
Last updated: 7 February 2023
Last updated: 18 May 2023
Last updated: 31 January 2024
Last updated: 13 May 2024
Latest articles
Related topics, .css-je19u9{-webkit-align-items:flex-end;-webkit-box-align:flex-end;-ms-flex-align:flex-end;align-items:flex-end;display:-webkit-box;display:-webkit-flex;display:-ms-flexbox;display:flex;-webkit-flex-direction:row;-ms-flex-direction:row;flex-direction:row;-webkit-box-flex-wrap:wrap;-webkit-flex-wrap:wrap;-ms-flex-wrap:wrap;flex-wrap:wrap;-webkit-box-pack:center;-ms-flex-pack:center;-webkit-justify-content:center;justify-content:center;row-gap:0;text-align:center;max-width:671px;}@media (max-width: 1079px){.css-je19u9{max-width:400px;}.css-je19u9>span{white-space:pre;}}@media (max-width: 799px){.css-je19u9{max-width:400px;}.css-je19u9>span{white-space:pre;}} decide what to .css-1kiodld{max-height:56px;display:-webkit-box;display:-webkit-flex;display:-ms-flexbox;display:flex;-webkit-align-items:center;-webkit-box-align:center;-ms-flex-align:center;align-items:center;}@media (max-width: 1079px){.css-1kiodld{display:none;}} build next, decide what to build next.

Users report unexpectedly high data usage, especially during streaming sessions.

Users find it hard to navigate from the home page to relevant playlists in the app.

It would be great to have a sleep timer feature, especially for bedtime listening.

I need better filters to find the songs or artists I’m looking for.
Log in or sign up
Get started for free
- U.S. Department of Health & Human Services

- Virtual Tour
- Staff Directory
- En Español
You are here
Impact of nih research.
Revolutionizing Science
Research Tools
NIH leads the charge on developing new research tools that have broad applications, pushing the boundaries on multiple research fronts.
Small Molecule Screening
Research-tools--small-molecule-screening.jpg.

Thanks to NIH, publicly funded researchers now have access to resources and tools with the capacity to screen large numbers of small molecules, helping them to more efficiently study genes and discover treatments for human diseases. Researchers used these resources to develop FDA-approved treatments for ulcerative colitis and relapsing forms of multiple sclerosis.
Image credit: National Center for Advancing Translational Sciences, NIH
- This advancement in small molecule research makes it easier for scientists to use and understand molecular compounds in basic research and drug development.
- The NIH Common Fund Molecular Libraries and Imaging Program also launched PubChem, an open chemistry database that contains information on chemical structures, properties, and biological activities of over 100 million compounds, including small molecules.
- NIH also developed tools and resources to help scientists conduct preclinical research, with a focus on small molecule screening.
Single Cell Analysis
Research-tools--single-cell-analysis.jpg.

NIH fostered a technological revolution in single cell analysis research, leading to the development of cutting-edge tools, methods, platforms, and cell atlases to identify and characterize features of single cells within a variety of human tissues. These technologies are available to the entire research community to foster additional breakthroughs in research.
Image credit: NIH
- The human body contains approximately 37 trillion cells, carefully organized in tissues to carry out the daily processes that keep the body alive and healthy. Analysis of single cells poses many technological challenges.
- Between 2012 and 2017, the NIH Common Fund Single Cell Analysis Program found a three-fold increase in the number of single cell analysis projects funded by NIH and an approximate doubling of relevant publications.
- Understanding cells at the individual level may lead to new understandings of development, health, aging, and disease.
research-tools--cryo-em.jpg

NIH funded the development and dissemination of cryo-electron microscopy (cryo-EM), a tool that enables high-resolution images of proteins and other biological structures. Cryo-EM has helped researchers identify potential new therapeutic targets for vaccines and drugs.
Image credit: Huilin Li, Brookhaven National Laboratory, and Bruce Stillman, Cold Spring Harbor Laboratory
- An NIH-funded researcher was awarded the 2017 Nobel Prize in Chemistry for their work characterizing proteins using cryo-EM.
- Since 2018, the NIH-supported National Centers for Cryo-EM enabled researchers to determine the structure of more than 300 proteins, including the SARS-CoV-2 spike protein, and trained more than 1,000 investigators in this cutting-edge technique.
Cell Culture Technology
Research-tools--cell-culture-technology.jpg.

NIH scientists created Matrigel, a specialized gel that promotes cell growth on a 3-D surface that mimics the environment within the body. Today, Matrigel is widely used in labs around the world to study cells that were previously impossible to grow and to investigate complex cell activities in a more relevant environment.
Image credit: David Sone
- Prior to this invention, scientists grew cells in a flat layer in plastic culture dishes, which was not sufficient to grow specialized cells, like stem cells.
- Using Matrigel, researchers discovered new insights into nerve growth, the formation of blood vessels, and stem and cancer cell biology. It is also being used to screen cancer drugs and to support development of artificial tissues that can mimic organ function.
- More than 13,000 scientific papers have cited the use of Matrigel in their studies.
Cancer Genome Atlas
Research-tools--cancer-genome-atlas.jpg.

The Cancer Genome Atlas (TCGA) is a landmark NIH cancer genomics program that transformed our understanding of cancer by analyzing tumors from 11,000 patients with 33 different cancer types. Findings from TCGA identified new ways to prevent, diagnose, and treat cancers, such as gliomas and stomach cancer.
Image credit: Darryl Leja, National Human Genome Research Institute, NIH
- TCGA showed that different cancers can share molecular traits regardless of the organ or tissue they are found in. This enabled the emergence of precision medicine in oncology—cancer treatment based on molecular traits rather than the tissue in the body where the cancer started.
- TCGA generated over 2.5 petabytes (1 petabyte = 500 billion pages of standard printed text!) of data on genes, proteins, and their modifications in cancer by bringing together 20 collaborating institutions across the U.S. and Canada.
Recombinant DNA
Research-tools--recombinant-dna.jpg.

Because of NIH-funded research on recombinant DNA technology, researchers developed techniques that can enable the production of large quantities of important peptides—the building blocks of proteins—which can be used to produce certain medicines.
Image credit: National Human Genome Research Institute, NIH
- Scientists use specialized molecules to snip out a specific gene from a long strand of DNA, creating recombinant DNA by inserting it into bacterial or yeast cells. These cells reproduce quickly and, following the gene’s instructions, make large amounts of the desired peptide.
- These techniques enabled the production of synthetic insulin to treat diabetes.
- Medicines produced using these techniques have been used for more than 30 years.
- In 1980, an NIH-funded researcher received a Nobel Prize for research on recombinant DNA.
Imaging Technology
Research-tools--imaging-technology.jpg.

Significant innovation in clinical imaging technology is a result of NIH-funded research. Imaging technologies now have higher resolution and greater sensitivity, with new categories of imaging, like digital 3D reconstructions, now being commonly used.
Image credit: Clinical Center, NIH
- A new type of positron emission tomography (PET) that looks for prostate cancer specific proteins has been found to be 27% more accurate than standard methods for detecting prostate cancers.
- NIH-supported improvements in PET technologies resulted in a more sensitive technology that can capture scans in under a minute and reduce the dose of dye given to patients.
- NIH-funded research led to the development of nuclear magnetic resonance imaging, which won a Nobel Prize, and is the same technique used in MRIs in clinical settings.
- Molecular Libraries and Imaging: https://commonfund.nih.gov/molecularlibraries/index
- Preclinical Research Toolbox: https://ncats.nih.gov/expertise/preclinical
- PubChem: https://pubchemdocs.ncbi.nlm.nih.gov/statistics
- Molecular Libraries and Imaging Program Highlights: https://commonfund.nih.gov/Molecularlibraries/programhighlights
- Article: Ozanimod accepted for priority review by FDA for the treatment of ulcerative colitis: https://www.scripps.edu/news-and-events/press-room/2021/20210203-rosen-roberts-ozanimod-fda-ulcerative-colitis.html
- Article: U.S. Food and Drug Administration Approves Bristol Myers Squibb’s Zeposia® (ozanimod), an Oral Treatment for Adults with Moderately to Severely Active Ulcerative Colitis: https://news.bms.com/news/corporate-financial/2021/U.S.-Food-and-Drug-Administration-Approves-Bristol-Myers-Squibbs-Zeposia-ozanimod-an-Oral-Treatment-for-Adults-with-Moderately-to-Severely-Active-Ulcerative-Colitis1/default.aspx
- NIH Single Cell Analysis Program: https://commonfund.nih.gov/singlecell
- Roy AL, et al. Sci Adv . 2018;4(8):eaat8573. PMID: 30083611 .
- The Human BioMolecular Atlas Program: https://commonfund.nih.gov/HuBMAP
- HuBMAP Data Portal: https://portal.hubmapconsortium.org/
- Cellular Senescence Network: https://commonfund.nih.gov/senescence
- LungMAP: https://www.lungmap.net/
- GenitoUrinary Development Molecular Anatomy Project: https://www.gudmap.org/
- Transformative High-Resolution Cryoelectron Microscopy Program: https://commonfund.nih.gov/CryoEM
- Cryo-Electron Microscopy Program Centers: https://www.cryoemcenters.org
- Zhang K, et al. bioRxiv [Preprint]. 2020:2020.08.11.245696. Update in: QRB Discov . 2020;1:e11. PMID: 32817943 .
- NIH Nobel Laureates: https://www.nih.gov/about-nih/what-we-do/nih-almanac/nobel-laureates
- Cressey D, et al. Nature . 2017;550(7675):167. PMID: 29022937 .
- Simian M, et al. J Cell Biol . 2017;216(1):31-40. PMID: 28031422 .
- Article: An Interview with Hynda Kleinman: https://irp.nih.gov/catalyst/v21i4/alumni-news
- Kleinman HK, et al. Semin Cancer Biol . 2005;15(5):378-86. PMID: 15975825 .
- Article: Hair today, gone tomorrow: NIDCR'S Hynda Kleinman takes off for new horizons: https://nihsearch.cit.nih.gov/catalyst/2005/05.11.01/page4.html
- The Cancer Genome Atlas Program: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga
- NIGMS-Supported Nobelists: https://www.nigms.nih.gov/pages/GMNobelists.aspx
- Article: Celebrating the discovery and development of insulin: www.niddk.nih.gov/news/archive/2021/celebrating-discovery-development-insulin
- National Institute of General Medical Sciences. The New Genetics . 2010. https://nigms.nih.gov/education/Booklets/the-new-genetics/Documents/Booklet-The-New-Genetics.pdf
- Article: Commemorating the 50th Anniversary of the National Cancer Act (NCA50): Clinical Imaging — Then and Now: https://dctd.cancer.gov/NewsEvents/20210712_NCA50.htm?cid=soc_ig_en_enterprise_nca50
- EXPLORER Total Body PET Scanner: https://health.ucdavis.edu/radiology/myexam/PET/Equipment/explorer.html
- Badawi RD, et al. J Nucl Med . 2019;60(3):299-303. PMID: 30733314 .
- Article: PSMA PET-CT Accurately Detects Prostate Cancer Spread, Trial Shows: https://www.cancer.gov/news-events/cancer-currents-blog/2020/prostate-cancer-psma-pet-ct-metastasis
- Magnetic Resonance Imaging (MRI): https://www.nibib.nih.gov/science-education/science-topics/magnetic-resonance-imaging-mri
This page last reviewed on March 1, 2023
Connect with Us
- More Social Media from NIH

Data Analysis Techniques in Research – Methods, Tools & Examples

Varun Saharawat is a seasoned professional in the fields of SEO and content writing. With a profound knowledge of the intricate aspects of these disciplines, Varun has established himself as a valuable asset in the world of digital marketing and online content creation.

Data analysis techniques in research are essential because they allow researchers to derive meaningful insights from data sets to support their hypotheses or research objectives.
Data Analysis Techniques in Research : While various groups, institutions, and professionals may have diverse approaches to data analysis, a universal definition captures its essence. Data analysis involves refining, transforming, and interpreting raw data to derive actionable insights that guide informed decision-making for businesses.

A straightforward illustration of data analysis emerges when we make everyday decisions, basing our choices on past experiences or predictions of potential outcomes.
If you want to learn more about this topic and acquire valuable skills that will set you apart in today’s data-driven world, we highly recommend enrolling in the Data Analytics Course by Physics Wallah . And as a special offer for our readers, use the coupon code “READER” to get a discount on this course.
Table of Contents
What is Data Analysis?
Data analysis is the systematic process of inspecting, cleaning, transforming, and interpreting data with the objective of discovering valuable insights and drawing meaningful conclusions. This process involves several steps:
- Inspecting : Initial examination of data to understand its structure, quality, and completeness.
- Cleaning : Removing errors, inconsistencies, or irrelevant information to ensure accurate analysis.
- Transforming : Converting data into a format suitable for analysis, such as normalization or aggregation.
- Interpreting : Analyzing the transformed data to identify patterns, trends, and relationships.
Types of Data Analysis Techniques in Research
Data analysis techniques in research are categorized into qualitative and quantitative methods, each with its specific approaches and tools. These techniques are instrumental in extracting meaningful insights, patterns, and relationships from data to support informed decision-making, validate hypotheses, and derive actionable recommendations. Below is an in-depth exploration of the various types of data analysis techniques commonly employed in research:
1) Qualitative Analysis:
Definition: Qualitative analysis focuses on understanding non-numerical data, such as opinions, concepts, or experiences, to derive insights into human behavior, attitudes, and perceptions.
- Content Analysis: Examines textual data, such as interview transcripts, articles, or open-ended survey responses, to identify themes, patterns, or trends.
- Narrative Analysis: Analyzes personal stories or narratives to understand individuals’ experiences, emotions, or perspectives.
- Ethnographic Studies: Involves observing and analyzing cultural practices, behaviors, and norms within specific communities or settings.
2) Quantitative Analysis:
Quantitative analysis emphasizes numerical data and employs statistical methods to explore relationships, patterns, and trends. It encompasses several approaches:
Descriptive Analysis:
- Frequency Distribution: Represents the number of occurrences of distinct values within a dataset.
- Central Tendency: Measures such as mean, median, and mode provide insights into the central values of a dataset.
- Dispersion: Techniques like variance and standard deviation indicate the spread or variability of data.
Diagnostic Analysis:
- Regression Analysis: Assesses the relationship between dependent and independent variables, enabling prediction or understanding causality.
- ANOVA (Analysis of Variance): Examines differences between groups to identify significant variations or effects.
Predictive Analysis:
- Time Series Forecasting: Uses historical data points to predict future trends or outcomes.
- Machine Learning Algorithms: Techniques like decision trees, random forests, and neural networks predict outcomes based on patterns in data.
Prescriptive Analysis:
- Optimization Models: Utilizes linear programming, integer programming, or other optimization techniques to identify the best solutions or strategies.
- Simulation: Mimics real-world scenarios to evaluate various strategies or decisions and determine optimal outcomes.
Specific Techniques:
- Monte Carlo Simulation: Models probabilistic outcomes to assess risk and uncertainty.
- Factor Analysis: Reduces the dimensionality of data by identifying underlying factors or components.
- Cohort Analysis: Studies specific groups or cohorts over time to understand trends, behaviors, or patterns within these groups.
- Cluster Analysis: Classifies objects or individuals into homogeneous groups or clusters based on similarities or attributes.
- Sentiment Analysis: Uses natural language processing and machine learning techniques to determine sentiment, emotions, or opinions from textual data.
Also Read: AI and Predictive Analytics: Examples, Tools, Uses, Ai Vs Predictive Analytics
Data Analysis Techniques in Research Examples
To provide a clearer understanding of how data analysis techniques are applied in research, let’s consider a hypothetical research study focused on evaluating the impact of online learning platforms on students’ academic performance.
Research Objective:
Determine if students using online learning platforms achieve higher academic performance compared to those relying solely on traditional classroom instruction.
Data Collection:
- Quantitative Data: Academic scores (grades) of students using online platforms and those using traditional classroom methods.
- Qualitative Data: Feedback from students regarding their learning experiences, challenges faced, and preferences.
Data Analysis Techniques Applied:
1) Descriptive Analysis:
- Calculate the mean, median, and mode of academic scores for both groups.
- Create frequency distributions to represent the distribution of grades in each group.
2) Diagnostic Analysis:
- Conduct an Analysis of Variance (ANOVA) to determine if there’s a statistically significant difference in academic scores between the two groups.
- Perform Regression Analysis to assess the relationship between the time spent on online platforms and academic performance.
3) Predictive Analysis:
- Utilize Time Series Forecasting to predict future academic performance trends based on historical data.
- Implement Machine Learning algorithms to develop a predictive model that identifies factors contributing to academic success on online platforms.
4) Prescriptive Analysis:
- Apply Optimization Models to identify the optimal combination of online learning resources (e.g., video lectures, interactive quizzes) that maximize academic performance.
- Use Simulation Techniques to evaluate different scenarios, such as varying student engagement levels with online resources, to determine the most effective strategies for improving learning outcomes.
5) Specific Techniques:
- Conduct Factor Analysis on qualitative feedback to identify common themes or factors influencing students’ perceptions and experiences with online learning.
- Perform Cluster Analysis to segment students based on their engagement levels, preferences, or academic outcomes, enabling targeted interventions or personalized learning strategies.
- Apply Sentiment Analysis on textual feedback to categorize students’ sentiments as positive, negative, or neutral regarding online learning experiences.
By applying a combination of qualitative and quantitative data analysis techniques, this research example aims to provide comprehensive insights into the effectiveness of online learning platforms.
Also Read: Learning Path to Become a Data Analyst in 2024
Data Analysis Techniques in Quantitative Research
Quantitative research involves collecting numerical data to examine relationships, test hypotheses, and make predictions. Various data analysis techniques are employed to interpret and draw conclusions from quantitative data. Here are some key data analysis techniques commonly used in quantitative research:
1) Descriptive Statistics:
- Description: Descriptive statistics are used to summarize and describe the main aspects of a dataset, such as central tendency (mean, median, mode), variability (range, variance, standard deviation), and distribution (skewness, kurtosis).
- Applications: Summarizing data, identifying patterns, and providing initial insights into the dataset.
2) Inferential Statistics:
- Description: Inferential statistics involve making predictions or inferences about a population based on a sample of data. This technique includes hypothesis testing, confidence intervals, t-tests, chi-square tests, analysis of variance (ANOVA), regression analysis, and correlation analysis.
- Applications: Testing hypotheses, making predictions, and generalizing findings from a sample to a larger population.
3) Regression Analysis:
- Description: Regression analysis is a statistical technique used to model and examine the relationship between a dependent variable and one or more independent variables. Linear regression, multiple regression, logistic regression, and nonlinear regression are common types of regression analysis .
- Applications: Predicting outcomes, identifying relationships between variables, and understanding the impact of independent variables on the dependent variable.
4) Correlation Analysis:
- Description: Correlation analysis is used to measure and assess the strength and direction of the relationship between two or more variables. The Pearson correlation coefficient, Spearman rank correlation coefficient, and Kendall’s tau are commonly used measures of correlation.
- Applications: Identifying associations between variables and assessing the degree and nature of the relationship.
5) Factor Analysis:
- Description: Factor analysis is a multivariate statistical technique used to identify and analyze underlying relationships or factors among a set of observed variables. It helps in reducing the dimensionality of data and identifying latent variables or constructs.
- Applications: Identifying underlying factors or constructs, simplifying data structures, and understanding the underlying relationships among variables.
6) Time Series Analysis:
- Description: Time series analysis involves analyzing data collected or recorded over a specific period at regular intervals to identify patterns, trends, and seasonality. Techniques such as moving averages, exponential smoothing, autoregressive integrated moving average (ARIMA), and Fourier analysis are used.
- Applications: Forecasting future trends, analyzing seasonal patterns, and understanding time-dependent relationships in data.
7) ANOVA (Analysis of Variance):
- Description: Analysis of variance (ANOVA) is a statistical technique used to analyze and compare the means of two or more groups or treatments to determine if they are statistically different from each other. One-way ANOVA, two-way ANOVA, and MANOVA (Multivariate Analysis of Variance) are common types of ANOVA.
- Applications: Comparing group means, testing hypotheses, and determining the effects of categorical independent variables on a continuous dependent variable.
8) Chi-Square Tests:
- Description: Chi-square tests are non-parametric statistical tests used to assess the association between categorical variables in a contingency table. The Chi-square test of independence, goodness-of-fit test, and test of homogeneity are common chi-square tests.
- Applications: Testing relationships between categorical variables, assessing goodness-of-fit, and evaluating independence.
These quantitative data analysis techniques provide researchers with valuable tools and methods to analyze, interpret, and derive meaningful insights from numerical data. The selection of a specific technique often depends on the research objectives, the nature of the data, and the underlying assumptions of the statistical methods being used.
Also Read: Analysis vs. Analytics: How Are They Different?
Data Analysis Methods
Data analysis methods refer to the techniques and procedures used to analyze, interpret, and draw conclusions from data. These methods are essential for transforming raw data into meaningful insights, facilitating decision-making processes, and driving strategies across various fields. Here are some common data analysis methods:
- Description: Descriptive statistics summarize and organize data to provide a clear and concise overview of the dataset. Measures such as mean, median, mode, range, variance, and standard deviation are commonly used.
- Description: Inferential statistics involve making predictions or inferences about a population based on a sample of data. Techniques such as hypothesis testing, confidence intervals, and regression analysis are used.
3) Exploratory Data Analysis (EDA):
- Description: EDA techniques involve visually exploring and analyzing data to discover patterns, relationships, anomalies, and insights. Methods such as scatter plots, histograms, box plots, and correlation matrices are utilized.
- Applications: Identifying trends, patterns, outliers, and relationships within the dataset.
4) Predictive Analytics:
- Description: Predictive analytics use statistical algorithms and machine learning techniques to analyze historical data and make predictions about future events or outcomes. Techniques such as regression analysis, time series forecasting, and machine learning algorithms (e.g., decision trees, random forests, neural networks) are employed.
- Applications: Forecasting future trends, predicting outcomes, and identifying potential risks or opportunities.
5) Prescriptive Analytics:
- Description: Prescriptive analytics involve analyzing data to recommend actions or strategies that optimize specific objectives or outcomes. Optimization techniques, simulation models, and decision-making algorithms are utilized.
- Applications: Recommending optimal strategies, decision-making support, and resource allocation.
6) Qualitative Data Analysis:
- Description: Qualitative data analysis involves analyzing non-numerical data, such as text, images, videos, or audio, to identify themes, patterns, and insights. Methods such as content analysis, thematic analysis, and narrative analysis are used.
- Applications: Understanding human behavior, attitudes, perceptions, and experiences.
7) Big Data Analytics:
- Description: Big data analytics methods are designed to analyze large volumes of structured and unstructured data to extract valuable insights. Technologies such as Hadoop, Spark, and NoSQL databases are used to process and analyze big data.
- Applications: Analyzing large datasets, identifying trends, patterns, and insights from big data sources.
8) Text Analytics:
- Description: Text analytics methods involve analyzing textual data, such as customer reviews, social media posts, emails, and documents, to extract meaningful information and insights. Techniques such as sentiment analysis, text mining, and natural language processing (NLP) are used.
- Applications: Analyzing customer feedback, monitoring brand reputation, and extracting insights from textual data sources.
These data analysis methods are instrumental in transforming data into actionable insights, informing decision-making processes, and driving organizational success across various sectors, including business, healthcare, finance, marketing, and research. The selection of a specific method often depends on the nature of the data, the research objectives, and the analytical requirements of the project or organization.
Also Read: Quantitative Data Analysis: Types, Analysis & Examples
Data Analysis Tools
Data analysis tools are essential instruments that facilitate the process of examining, cleaning, transforming, and modeling data to uncover useful information, make informed decisions, and drive strategies. Here are some prominent data analysis tools widely used across various industries:
1) Microsoft Excel:
- Description: A spreadsheet software that offers basic to advanced data analysis features, including pivot tables, data visualization tools, and statistical functions.
- Applications: Data cleaning, basic statistical analysis, visualization, and reporting.
2) R Programming Language:
- Description: An open-source programming language specifically designed for statistical computing and data visualization.
- Applications: Advanced statistical analysis, data manipulation, visualization, and machine learning.
3) Python (with Libraries like Pandas, NumPy, Matplotlib, and Seaborn):
- Description: A versatile programming language with libraries that support data manipulation, analysis, and visualization.
- Applications: Data cleaning, statistical analysis, machine learning, and data visualization.
4) SPSS (Statistical Package for the Social Sciences):
- Description: A comprehensive statistical software suite used for data analysis, data mining, and predictive analytics.
- Applications: Descriptive statistics, hypothesis testing, regression analysis, and advanced analytics.
5) SAS (Statistical Analysis System):
- Description: A software suite used for advanced analytics, multivariate analysis, and predictive modeling.
- Applications: Data management, statistical analysis, predictive modeling, and business intelligence.
6) Tableau:
- Description: A data visualization tool that allows users to create interactive and shareable dashboards and reports.
- Applications: Data visualization , business intelligence , and interactive dashboard creation.
7) Power BI:
- Description: A business analytics tool developed by Microsoft that provides interactive visualizations and business intelligence capabilities.
- Applications: Data visualization, business intelligence, reporting, and dashboard creation.
8) SQL (Structured Query Language) Databases (e.g., MySQL, PostgreSQL, Microsoft SQL Server):
- Description: Database management systems that support data storage, retrieval, and manipulation using SQL queries.
- Applications: Data retrieval, data cleaning, data transformation, and database management.
9) Apache Spark:
- Description: A fast and general-purpose distributed computing system designed for big data processing and analytics.
- Applications: Big data processing, machine learning, data streaming, and real-time analytics.
10) IBM SPSS Modeler:
- Description: A data mining software application used for building predictive models and conducting advanced analytics.
- Applications: Predictive modeling, data mining, statistical analysis, and decision optimization.
These tools serve various purposes and cater to different data analysis needs, from basic statistical analysis and data visualization to advanced analytics, machine learning, and big data processing. The choice of a specific tool often depends on the nature of the data, the complexity of the analysis, and the specific requirements of the project or organization.
Also Read: How to Analyze Survey Data: Methods & Examples
Importance of Data Analysis in Research
The importance of data analysis in research cannot be overstated; it serves as the backbone of any scientific investigation or study. Here are several key reasons why data analysis is crucial in the research process:
- Data analysis helps ensure that the results obtained are valid and reliable. By systematically examining the data, researchers can identify any inconsistencies or anomalies that may affect the credibility of the findings.
- Effective data analysis provides researchers with the necessary information to make informed decisions. By interpreting the collected data, researchers can draw conclusions, make predictions, or formulate recommendations based on evidence rather than intuition or guesswork.
- Data analysis allows researchers to identify patterns, trends, and relationships within the data. This can lead to a deeper understanding of the research topic, enabling researchers to uncover insights that may not be immediately apparent.
- In empirical research, data analysis plays a critical role in testing hypotheses. Researchers collect data to either support or refute their hypotheses, and data analysis provides the tools and techniques to evaluate these hypotheses rigorously.
- Transparent and well-executed data analysis enhances the credibility of research findings. By clearly documenting the data analysis methods and procedures, researchers allow others to replicate the study, thereby contributing to the reproducibility of research findings.
- In fields such as business or healthcare, data analysis helps organizations allocate resources more efficiently. By analyzing data on consumer behavior, market trends, or patient outcomes, organizations can make strategic decisions about resource allocation, budgeting, and planning.
- In public policy and social sciences, data analysis is instrumental in developing and evaluating policies and interventions. By analyzing data on social, economic, or environmental factors, policymakers can assess the effectiveness of existing policies and inform the development of new ones.
- Data analysis allows for continuous improvement in research methods and practices. By analyzing past research projects, identifying areas for improvement, and implementing changes based on data-driven insights, researchers can refine their approaches and enhance the quality of future research endeavors.
However, it is important to remember that mastering these techniques requires practice and continuous learning. That’s why we highly recommend the Data Analytics Course by Physics Wallah . Not only does it cover all the fundamentals of data analysis, but it also provides hands-on experience with various tools such as Excel, Python, and Tableau. Plus, if you use the “ READER ” coupon code at checkout, you can get a special discount on the course.
For Latest Tech Related Information, Join Our Official Free Telegram Group : PW Skills Telegram Group
Data Analysis Techniques in Research FAQs
What are the 5 techniques for data analysis.
The five techniques for data analysis include: Descriptive Analysis Diagnostic Analysis Predictive Analysis Prescriptive Analysis Qualitative Analysis
What are techniques of data analysis in research?
Techniques of data analysis in research encompass both qualitative and quantitative methods. These techniques involve processes like summarizing raw data, investigating causes of events, forecasting future outcomes, offering recommendations based on predictions, and examining non-numerical data to understand concepts or experiences.
What are the 3 methods of data analysis?
The three primary methods of data analysis are: Qualitative Analysis Quantitative Analysis Mixed-Methods Analysis
What are the four types of data analysis techniques?
The four types of data analysis techniques are: Descriptive Analysis Diagnostic Analysis Predictive Analysis Prescriptive Analysis
10 Most Popular Big Data Analytics Software

Best Big Data Analytics Software: In the rapidly evolving realm of commerce, the capacity to harness the potential of data…
Best Courses For Data Analytics: Top 10 Courses For Your Career in Trend

Best courses for data analytics: If you are looking for the best courses for data analytics, then go through this…
Which Course is Best for a Data Analyst?

Looking to build your career as a Data Analyst but Don’t know how to start and where to start from?…

An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Indian J Anaesth
- v.60(9); 2016 Sep
Basic statistical tools in research and data analysis
Zulfiqar ali.
Department of Anaesthesiology, Division of Neuroanaesthesiology, Sheri Kashmir Institute of Medical Sciences, Soura, Srinagar, Jammu and Kashmir, India
S Bala Bhaskar
1 Department of Anaesthesiology and Critical Care, Vijayanagar Institute of Medical Sciences, Bellary, Karnataka, India
Statistical methods involved in carrying out a study include planning, designing, collecting data, analysing, drawing meaningful interpretation and reporting of the research findings. The statistical analysis gives meaning to the meaningless numbers, thereby breathing life into a lifeless data. The results and inferences are precise only if proper statistical tests are used. This article will try to acquaint the reader with the basic research tools that are utilised while conducting various studies. The article covers a brief outline of the variables, an understanding of quantitative and qualitative variables and the measures of central tendency. An idea of the sample size estimation, power analysis and the statistical errors is given. Finally, there is a summary of parametric and non-parametric tests used for data analysis.
INTRODUCTION
Statistics is a branch of science that deals with the collection, organisation, analysis of data and drawing of inferences from the samples to the whole population.[ 1 ] This requires a proper design of the study, an appropriate selection of the study sample and choice of a suitable statistical test. An adequate knowledge of statistics is necessary for proper designing of an epidemiological study or a clinical trial. Improper statistical methods may result in erroneous conclusions which may lead to unethical practice.[ 2 ]
Variable is a characteristic that varies from one individual member of population to another individual.[ 3 ] Variables such as height and weight are measured by some type of scale, convey quantitative information and are called as quantitative variables. Sex and eye colour give qualitative information and are called as qualitative variables[ 3 ] [ Figure 1 ].

Classification of variables
Quantitative variables
Quantitative or numerical data are subdivided into discrete and continuous measurements. Discrete numerical data are recorded as a whole number such as 0, 1, 2, 3,… (integer), whereas continuous data can assume any value. Observations that can be counted constitute the discrete data and observations that can be measured constitute the continuous data. Examples of discrete data are number of episodes of respiratory arrests or the number of re-intubations in an intensive care unit. Similarly, examples of continuous data are the serial serum glucose levels, partial pressure of oxygen in arterial blood and the oesophageal temperature.
A hierarchical scale of increasing precision can be used for observing and recording the data which is based on categorical, ordinal, interval and ratio scales [ Figure 1 ].
Categorical or nominal variables are unordered. The data are merely classified into categories and cannot be arranged in any particular order. If only two categories exist (as in gender male and female), it is called as a dichotomous (or binary) data. The various causes of re-intubation in an intensive care unit due to upper airway obstruction, impaired clearance of secretions, hypoxemia, hypercapnia, pulmonary oedema and neurological impairment are examples of categorical variables.
Ordinal variables have a clear ordering between the variables. However, the ordered data may not have equal intervals. Examples are the American Society of Anesthesiologists status or Richmond agitation-sedation scale.
Interval variables are similar to an ordinal variable, except that the intervals between the values of the interval variable are equally spaced. A good example of an interval scale is the Fahrenheit degree scale used to measure temperature. With the Fahrenheit scale, the difference between 70° and 75° is equal to the difference between 80° and 85°: The units of measurement are equal throughout the full range of the scale.
Ratio scales are similar to interval scales, in that equal differences between scale values have equal quantitative meaning. However, ratio scales also have a true zero point, which gives them an additional property. For example, the system of centimetres is an example of a ratio scale. There is a true zero point and the value of 0 cm means a complete absence of length. The thyromental distance of 6 cm in an adult may be twice that of a child in whom it may be 3 cm.
STATISTICS: DESCRIPTIVE AND INFERENTIAL STATISTICS
Descriptive statistics[ 4 ] try to describe the relationship between variables in a sample or population. Descriptive statistics provide a summary of data in the form of mean, median and mode. Inferential statistics[ 4 ] use a random sample of data taken from a population to describe and make inferences about the whole population. It is valuable when it is not possible to examine each member of an entire population. The examples if descriptive and inferential statistics are illustrated in Table 1 .
Example of descriptive and inferential statistics

Descriptive statistics
The extent to which the observations cluster around a central location is described by the central tendency and the spread towards the extremes is described by the degree of dispersion.
Measures of central tendency
The measures of central tendency are mean, median and mode.[ 6 ] Mean (or the arithmetic average) is the sum of all the scores divided by the number of scores. Mean may be influenced profoundly by the extreme variables. For example, the average stay of organophosphorus poisoning patients in ICU may be influenced by a single patient who stays in ICU for around 5 months because of septicaemia. The extreme values are called outliers. The formula for the mean is

where x = each observation and n = number of observations. Median[ 6 ] is defined as the middle of a distribution in a ranked data (with half of the variables in the sample above and half below the median value) while mode is the most frequently occurring variable in a distribution. Range defines the spread, or variability, of a sample.[ 7 ] It is described by the minimum and maximum values of the variables. If we rank the data and after ranking, group the observations into percentiles, we can get better information of the pattern of spread of the variables. In percentiles, we rank the observations into 100 equal parts. We can then describe 25%, 50%, 75% or any other percentile amount. The median is the 50 th percentile. The interquartile range will be the observations in the middle 50% of the observations about the median (25 th -75 th percentile). Variance[ 7 ] is a measure of how spread out is the distribution. It gives an indication of how close an individual observation clusters about the mean value. The variance of a population is defined by the following formula:

where σ 2 is the population variance, X is the population mean, X i is the i th element from the population and N is the number of elements in the population. The variance of a sample is defined by slightly different formula:

where s 2 is the sample variance, x is the sample mean, x i is the i th element from the sample and n is the number of elements in the sample. The formula for the variance of a population has the value ‘ n ’ as the denominator. The expression ‘ n −1’ is known as the degrees of freedom and is one less than the number of parameters. Each observation is free to vary, except the last one which must be a defined value. The variance is measured in squared units. To make the interpretation of the data simple and to retain the basic unit of observation, the square root of variance is used. The square root of the variance is the standard deviation (SD).[ 8 ] The SD of a population is defined by the following formula:

where σ is the population SD, X is the population mean, X i is the i th element from the population and N is the number of elements in the population. The SD of a sample is defined by slightly different formula:

where s is the sample SD, x is the sample mean, x i is the i th element from the sample and n is the number of elements in the sample. An example for calculation of variation and SD is illustrated in Table 2 .
Example of mean, variance, standard deviation

Normal distribution or Gaussian distribution
Most of the biological variables usually cluster around a central value, with symmetrical positive and negative deviations about this point.[ 1 ] The standard normal distribution curve is a symmetrical bell-shaped. In a normal distribution curve, about 68% of the scores are within 1 SD of the mean. Around 95% of the scores are within 2 SDs of the mean and 99% within 3 SDs of the mean [ Figure 2 ].

Normal distribution curve
Skewed distribution
It is a distribution with an asymmetry of the variables about its mean. In a negatively skewed distribution [ Figure 3 ], the mass of the distribution is concentrated on the right of Figure 1 . In a positively skewed distribution [ Figure 3 ], the mass of the distribution is concentrated on the left of the figure leading to a longer right tail.

Curves showing negatively skewed and positively skewed distribution
Inferential statistics
In inferential statistics, data are analysed from a sample to make inferences in the larger collection of the population. The purpose is to answer or test the hypotheses. A hypothesis (plural hypotheses) is a proposed explanation for a phenomenon. Hypothesis tests are thus procedures for making rational decisions about the reality of observed effects.
Probability is the measure of the likelihood that an event will occur. Probability is quantified as a number between 0 and 1 (where 0 indicates impossibility and 1 indicates certainty).
In inferential statistics, the term ‘null hypothesis’ ( H 0 ‘ H-naught ,’ ‘ H-null ’) denotes that there is no relationship (difference) between the population variables in question.[ 9 ]
Alternative hypothesis ( H 1 and H a ) denotes that a statement between the variables is expected to be true.[ 9 ]
The P value (or the calculated probability) is the probability of the event occurring by chance if the null hypothesis is true. The P value is a numerical between 0 and 1 and is interpreted by researchers in deciding whether to reject or retain the null hypothesis [ Table 3 ].
P values with interpretation

If P value is less than the arbitrarily chosen value (known as α or the significance level), the null hypothesis (H0) is rejected [ Table 4 ]. However, if null hypotheses (H0) is incorrectly rejected, this is known as a Type I error.[ 11 ] Further details regarding alpha error, beta error and sample size calculation and factors influencing them are dealt with in another section of this issue by Das S et al .[ 12 ]
Illustration for null hypothesis

PARAMETRIC AND NON-PARAMETRIC TESTS
Numerical data (quantitative variables) that are normally distributed are analysed with parametric tests.[ 13 ]
Two most basic prerequisites for parametric statistical analysis are:
- The assumption of normality which specifies that the means of the sample group are normally distributed
- The assumption of equal variance which specifies that the variances of the samples and of their corresponding population are equal.
However, if the distribution of the sample is skewed towards one side or the distribution is unknown due to the small sample size, non-parametric[ 14 ] statistical techniques are used. Non-parametric tests are used to analyse ordinal and categorical data.
Parametric tests
The parametric tests assume that the data are on a quantitative (numerical) scale, with a normal distribution of the underlying population. The samples have the same variance (homogeneity of variances). The samples are randomly drawn from the population, and the observations within a group are independent of each other. The commonly used parametric tests are the Student's t -test, analysis of variance (ANOVA) and repeated measures ANOVA.
Student's t -test
Student's t -test is used to test the null hypothesis that there is no difference between the means of the two groups. It is used in three circumstances:

where X = sample mean, u = population mean and SE = standard error of mean

where X 1 − X 2 is the difference between the means of the two groups and SE denotes the standard error of the difference.
- To test if the population means estimated by two dependent samples differ significantly (the paired t -test). A usual setting for paired t -test is when measurements are made on the same subjects before and after a treatment.
The formula for paired t -test is:

where d is the mean difference and SE denotes the standard error of this difference.
The group variances can be compared using the F -test. The F -test is the ratio of variances (var l/var 2). If F differs significantly from 1.0, then it is concluded that the group variances differ significantly.
Analysis of variance
The Student's t -test cannot be used for comparison of three or more groups. The purpose of ANOVA is to test if there is any significant difference between the means of two or more groups.
In ANOVA, we study two variances – (a) between-group variability and (b) within-group variability. The within-group variability (error variance) is the variation that cannot be accounted for in the study design. It is based on random differences present in our samples.
However, the between-group (or effect variance) is the result of our treatment. These two estimates of variances are compared using the F-test.
A simplified formula for the F statistic is:

where MS b is the mean squares between the groups and MS w is the mean squares within groups.
Repeated measures analysis of variance
As with ANOVA, repeated measures ANOVA analyses the equality of means of three or more groups. However, a repeated measure ANOVA is used when all variables of a sample are measured under different conditions or at different points in time.
As the variables are measured from a sample at different points of time, the measurement of the dependent variable is repeated. Using a standard ANOVA in this case is not appropriate because it fails to model the correlation between the repeated measures: The data violate the ANOVA assumption of independence. Hence, in the measurement of repeated dependent variables, repeated measures ANOVA should be used.
Non-parametric tests
When the assumptions of normality are not met, and the sample means are not normally, distributed parametric tests can lead to erroneous results. Non-parametric tests (distribution-free test) are used in such situation as they do not require the normality assumption.[ 15 ] Non-parametric tests may fail to detect a significant difference when compared with a parametric test. That is, they usually have less power.
As is done for the parametric tests, the test statistic is compared with known values for the sampling distribution of that statistic and the null hypothesis is accepted or rejected. The types of non-parametric analysis techniques and the corresponding parametric analysis techniques are delineated in Table 5 .
Analogue of parametric and non-parametric tests

Median test for one sample: The sign test and Wilcoxon's signed rank test
The sign test and Wilcoxon's signed rank test are used for median tests of one sample. These tests examine whether one instance of sample data is greater or smaller than the median reference value.
This test examines the hypothesis about the median θ0 of a population. It tests the null hypothesis H0 = θ0. When the observed value (Xi) is greater than the reference value (θ0), it is marked as+. If the observed value is smaller than the reference value, it is marked as − sign. If the observed value is equal to the reference value (θ0), it is eliminated from the sample.
If the null hypothesis is true, there will be an equal number of + signs and − signs.
The sign test ignores the actual values of the data and only uses + or − signs. Therefore, it is useful when it is difficult to measure the values.
Wilcoxon's signed rank test
There is a major limitation of sign test as we lose the quantitative information of the given data and merely use the + or – signs. Wilcoxon's signed rank test not only examines the observed values in comparison with θ0 but also takes into consideration the relative sizes, adding more statistical power to the test. As in the sign test, if there is an observed value that is equal to the reference value θ0, this observed value is eliminated from the sample.
Wilcoxon's rank sum test ranks all data points in order, calculates the rank sum of each sample and compares the difference in the rank sums.
Mann-Whitney test
It is used to test the null hypothesis that two samples have the same median or, alternatively, whether observations in one sample tend to be larger than observations in the other.
Mann–Whitney test compares all data (xi) belonging to the X group and all data (yi) belonging to the Y group and calculates the probability of xi being greater than yi: P (xi > yi). The null hypothesis states that P (xi > yi) = P (xi < yi) =1/2 while the alternative hypothesis states that P (xi > yi) ≠1/2.
Kolmogorov-Smirnov test
The two-sample Kolmogorov-Smirnov (KS) test was designed as a generic method to test whether two random samples are drawn from the same distribution. The null hypothesis of the KS test is that both distributions are identical. The statistic of the KS test is a distance between the two empirical distributions, computed as the maximum absolute difference between their cumulative curves.
Kruskal-Wallis test
The Kruskal–Wallis test is a non-parametric test to analyse the variance.[ 14 ] It analyses if there is any difference in the median values of three or more independent samples. The data values are ranked in an increasing order, and the rank sums calculated followed by calculation of the test statistic.
Jonckheere test
In contrast to Kruskal–Wallis test, in Jonckheere test, there is an a priori ordering that gives it a more statistical power than the Kruskal–Wallis test.[ 14 ]
Friedman test
The Friedman test is a non-parametric test for testing the difference between several related samples. The Friedman test is an alternative for repeated measures ANOVAs which is used when the same parameter has been measured under different conditions on the same subjects.[ 13 ]
Tests to analyse the categorical data
Chi-square test, Fischer's exact test and McNemar's test are used to analyse the categorical or nominal variables. The Chi-square test compares the frequencies and tests whether the observed data differ significantly from that of the expected data if there were no differences between groups (i.e., the null hypothesis). It is calculated by the sum of the squared difference between observed ( O ) and the expected ( E ) data (or the deviation, d ) divided by the expected data by the following formula:

A Yates correction factor is used when the sample size is small. Fischer's exact test is used to determine if there are non-random associations between two categorical variables. It does not assume random sampling, and instead of referring a calculated statistic to a sampling distribution, it calculates an exact probability. McNemar's test is used for paired nominal data. It is applied to 2 × 2 table with paired-dependent samples. It is used to determine whether the row and column frequencies are equal (that is, whether there is ‘marginal homogeneity’). The null hypothesis is that the paired proportions are equal. The Mantel-Haenszel Chi-square test is a multivariate test as it analyses multiple grouping variables. It stratifies according to the nominated confounding variables and identifies any that affects the primary outcome variable. If the outcome variable is dichotomous, then logistic regression is used.
SOFTWARES AVAILABLE FOR STATISTICS, SAMPLE SIZE CALCULATION AND POWER ANALYSIS
Numerous statistical software systems are available currently. The commonly used software systems are Statistical Package for the Social Sciences (SPSS – manufactured by IBM corporation), Statistical Analysis System ((SAS – developed by SAS Institute North Carolina, United States of America), R (designed by Ross Ihaka and Robert Gentleman from R core team), Minitab (developed by Minitab Inc), Stata (developed by StataCorp) and the MS Excel (developed by Microsoft).
There are a number of web resources which are related to statistical power analyses. A few are:
- StatPages.net – provides links to a number of online power calculators
- G-Power – provides a downloadable power analysis program that runs under DOS
- Power analysis for ANOVA designs an interactive site that calculates power or sample size needed to attain a given power for one effect in a factorial ANOVA design
- SPSS makes a program called SamplePower. It gives an output of a complete report on the computer screen which can be cut and paste into another document.
It is important that a researcher knows the concepts of the basic statistical methods used for conduct of a research study. This will help to conduct an appropriately well-designed study leading to valid and reliable results. Inappropriate use of statistical techniques may lead to faulty conclusions, inducing errors and undermining the significance of the article. Bad statistics may lead to bad research, and bad research may lead to unethical practice. Hence, an adequate knowledge of statistics and the appropriate use of statistical tests are important. An appropriate knowledge about the basic statistical methods will go a long way in improving the research designs and producing quality medical research which can be utilised for formulating the evidence-based guidelines.
Financial support and sponsorship
Conflicts of interest.
There are no conflicts of interest.

- Research Tools
- Learning Center

Discover Essential Research Tools for Effective Monitoring and Evaluation (M&E) | Boost Your M&E Process with Reliable Data Collection, Analysis, and Reporting Tools | Find the Right Instruments, Software, and Techniques to Enhance Your M&E Methods | Enhance Decision-making and Program Improvement in M&E with Powerful Research Tools.
Table of Contents
- What are research tools?x
- Research tools in M&E
Essential research tools commonly used across disciplines
How do i choose a research tool, what are methods vs tools in research, future trends and innovations in research tools for m&e, what are research tools.
Research tools refer to a wide range of resources, methods, instruments, software, or techniques that researchers use to collect , analyze , interpret, and communicate data and information during the research process.
These tools are designed to facilitate and enhance various aspects of research, such as data collection , organization, analysis, visualization, collaboration, and documentation. Research tools can be both physical (e.g., laboratory equipment, survey instruments) and digital (e.g., software, online databases).
They are essential for conducting research effectively, efficiently, and rigorously across different disciplines and research domains. Examples of research tools include laboratory equipment, survey questionnaires, statistical software, data visualization tools, literature databases, collaboration platforms, and more.
The choice of research tools depends on the specific research objectives, methods, and requirements of the study.
Optimize your resume to get more interviews: Try Our FREE Resume Scanner!
Optimize your resume for ATS with formatting, keywords, and quantified experience.
- Compare job description keywords with your resume.
- Tailor your resume to match as many keywords as possible.
- Grab the recruiter’s attention with a standout resume.

Scan your resume now
Research Tools in M&E
Monitoring and Evaluation (M&E) is a crucial component of research and program evaluation . Here are some essential research tools commonly used in the field of M&E:
- Logic Models and Results Frameworks: Logic models or results frameworks are visual tools that help clarify the theory of change and establish the logical connections between project activities, outputs, outcomes, and impacts. They provide a framework for designing M&E systems and identifying key indicators.
- Key Performance Indicators (KPIs): KPIs are measurable indicators that track progress and performance toward project or program goals. They help monitor the effectiveness and efficiency of interventions. Examples of KPIs can include the number of beneficiaries reached, percentage of target achieved, or cost per output.
- Surveys and Questionnaires: Surveys and questionnaires are useful tools for collecting quantitative data in M&E. They allow you to gather information from a large number of respondents and measure variables and indicators systematically. Online survey tools like SurveyMonkey or Google Forms can simplify data collection and analysis.
- Interviews and Focus Groups: Qualitative data collection methods, such as interviews and focus groups, can provide in-depth insights into participants’ experiences, perceptions, and attitudes. These methods are particularly valuable for understanding the contextual factors and mechanisms underlying program outcomes.
- Observations and Field Notes: Direct observations and field notes are often used to collect qualitative data in real-time. They help capture detailed information about program implementation, participant behaviors, and contextual factors that might not be evident through other methods.
- Data Analysis Software: Statistical software packages like SPSS, Stata, or R are commonly used for quantitative data analysis in M&E. These tools enable researchers to clean, analyze, and interpret large datasets efficiently. Qualitative data analysis software such as NVivo or Atlas.ti can assist with organizing and analyzing qualitative data.
- Data Visualization Tools: Tools like Excel, Tableau, or Power BI allow you to create visual representations of M&E data. Visualizations help communicate complex information and findings in a clear and compelling manner to stakeholders and decision-makers.
- Geographic Information Systems (GIS): GIS tools like ArcGIS or QGIS enable researchers to analyze and visualize spatial data. They can help identify geographical patterns, hotspot analysis, and map program impact or reach.
- Evaluation Management Systems: Evaluation management systems like DevResults or DHIS2 provide a centralized platform for managing M&E data, including data entry, analysis, reporting, and visualization. These systems streamline data management processes and facilitate collaboration among evaluation team members.
- Theory-Based Evaluation Approaches: Theory-based evaluation approaches , such as the Theory of Change or Contribution Analysis, help guide the evaluation process by explicitly linking program activities to intended outcomes and impacts. These approaches provide a framework for designing evaluations and analyzing the causal mechanisms at work.
It’s important to note that the selection of research tools in M&E should align with the specific objectives, scope, and resources of the evaluation. Tailor the choice of tools to the needs of the evaluation design and ensure that they provide reliable and valid data to inform decision-making.
There are numerous research tools available to support various types of research, and the choice of tools depends on the specific field of study and research goals. However, here are some essential research tools commonly used across disciplines:
- Library Databases: Online databases such as PubMed (biomedical literature), IEEE Xplore (engineering and computer science), JSTOR (humanities and social sciences), and Scopus (multidisciplinary) provide access to a vast collection of academic journals, articles, conference papers, and other scholarly resources.
- Google Scholar : This search engine specifically focuses on scholarly literature. It allows you to find academic papers, theses, books, and conference proceedings. It’s a useful tool for accessing both open access and subscription-based scholarly content.
- ResearchGate : ResearchGate is a social networking platform for researchers. It enables collaboration, networking, and access to research publications, preprints, and datasets. Researchers can also ask and answer questions related to their field of expertise.
- Reference Management Software: Tools like Zotero, Mendeley, and EndNote help researchers organize and manage bibliographic references. They allow you to collect, store, annotate, and cite references, making the citation process more efficient and streamlined.
- Data Analysis Tools: Depending on your research field, you may need specific data analysis tools. For statistical analysis, software such as SPSS, R, or Stata is commonly used. For qualitative research, NVivo and Atlas.ti assist with analyzing textual data.
- Collaboration and Communication Tools: Tools like Slack, Microsoft Teams, or Google Workspace facilitate collaboration and communication among research teams. They provide features like file sharing, real-time editing, video conferencing, and project management.
- Data Visualization Tools: Tools like Tableau, Plotly, or Excel can help create visual representations of data. These tools make it easier to present and interpret complex data sets, enabling researchers to communicate their findings effectively.
- Online Survey Tools: Platforms like SurveyMonkey, Google Forms, or Qualtrics allow researchers to design and distribute online surveys. These tools simplify the data collection process and provide features for analyzing and visualizing survey responses.
- Reference Search and Document Delivery: Tools like interlibrary loan systems, WorldCat, or services like Unpaywall can help you access research articles and resources that may not be available in your institution’s library.
- Academic Social Networks: Platforms like Academia.edu or LinkedIn can help researchers showcase their work, connect with peers, and discover potential collaborators or mentors.
Remember that the choice of research tools may vary depending on your specific research field and requirements. It’s essential to explore and evaluate the available options to find the tools that best align with your research goals and needs.
Choosing the right research tool in Monitoring and Evaluation (M&E) requires careful consideration of various factors.
Here’s a step-by-step process to help you choose a research tool for your M&E study:
- Define Your Research Objectives: Clearly articulate the purpose and goals of your M&E study. Determine what specific information you need to collect, analyze, and communicate through the evaluation process.
- Identify Data Needs: Identify the types of data you will be working with (quantitative, qualitative, spatial) and the specific indicators or variables you need to measure. Consider the level of detail, precision, and reliability required for your data.
- Assess Available Resources: Evaluate the resources available to you, including budget, time constraints, technical expertise, and access to technology or specialized equipment. Consider the level of support you may need in terms of training, technical assistance, or collaboration.
- Research Tool Options: Conduct research to explore the range of research tools available in M&E. Consult academic literature, practitioner resources, online forums, and professional networks to identify commonly used tools in your specific field or context.
- Evaluate Tool Suitability: Evaluate each research tool option against your specific needs and constraints. Consider factors such as ease of use, data quality, scalability, compatibility with existing systems, and cost-effectiveness. Assess whether the tool aligns with the type of data you are working with and the analysis and reporting requirements of your M&E study.
- Seek Recommendations and Feedback: Consult with experts, colleagues, or M&E professionals who have experience with the tools you are considering. Seek recommendations and feedback on their effectiveness, limitations, and user-friendliness. Their insights can provide valuable perspectives in selecting the most appropriate tool.
- Trial and Testing: If feasible, conduct small-scale trials or pilot tests with a subset of your data or research participants. This allows you to assess the usability and functionality of the tool, identify any potential issues, and gain practical experience in its implementation.
- Consider Integration and Compatibility: Consider the compatibility of the research tool with other tools or systems you may be using in your M&E process. Evaluate how well the tool integrates with existing data management, analysis, or reporting systems to ensure smooth workflows and data interoperability.
- Training and Support: Assess the availability of training resources, user guides, tutorials, and technical support for the research tool. Consider the level of training required for you and your team to effectively utilize the tool and ensure proper implementation.
- Make an Informed Decision: Based on the evaluation and assessment of the above factors, make an informed decision on the research tool that best meets your M&E objectives, data requirements, available resources, and user needs.
Remember, the choice of a research tool should be driven by the specific context, research objectives, and resources available to you. It’s important to consider trade-offs and select a tool that maximizes the quality and efficiency of your M&E study.
In the context of Monitoring and Evaluation (M&E), methods and tools have similar meanings as in general research, but they are applied specifically to the M&E process :
- M&E Methods: M&E methods refer to the systematic approaches and frameworks used to assess, measure, and evaluate the effectiveness, efficiency, and impact of programs, projects, or interventions. These methods provide a structured and rigorous approach to collecting and analyzing data to inform decision-making. M&E methods may include baseline studies, surveys, interviews, focus groups, case studies, statistical analysis, impact evaluation designs, and more. They guide the overall evaluation design and determine the data collection and analysis techniques used in M&E.
- M&E Tools: M&E tools are the specific resources, instruments, software, or techniques used within the M&E methods to support the data collection, management, analysis, visualization, and reporting processes. These tools provide practical means to implement M&E methods effectively. Examples of M&E tools include data collection templates, survey questionnaires, data analysis software (e.g., SPSS, Stata, R), visualization tools (e.g., Excel, Tableau), logic models, results frameworks, evaluation management systems (e.g., DevResults, DHIS2), and more. M&E tools assist in streamlining and enhancing the efficiency and accuracy of the M&E process.
In M&E, methods establish the overall approach to evaluating and assessing programs or interventions, while tools are the specific resources or techniques used within those methods to facilitate data collection, analysis, and reporting. M&E methods guide the evaluation design and data analysis, while M&E tools provide the means to execute those methods effectively. Both methods and tools are crucial in conducting rigorous and effective M&E, ensuring that data is collected, analyzed, and interpreted in a systematic and reliable manner to inform decision-making and program improvement.
As the field of Monitoring and Evaluation (M&E) continues to evolve, researchers and practitioners are exploring new trends and innovations in research tools to enhance the effectiveness and efficiency of evaluations. Here are some emerging trends and future directions in research tools for M&E:
- Integrated Data Platforms: With the increasing volume and complexity of data generated in M&E, there is a growing need for integrated data platforms that streamline data collection, management, analysis, and reporting processes. These platforms bring together various tools and functionalities into a unified system, allowing for seamless data flow and collaboration among stakeholders.
- Artificial Intelligence (AI) and Machine Learning: AI and machine learning technologies hold great potential for automating data analysis, identifying patterns and trends, and generating insights from large datasets in M&E. By leveraging AI algorithms, researchers can gain deeper insights into program performance, identify predictive indicators, and make data-driven decisions more efficiently.
- Mobile Data Collection Tools: Mobile data collection tools are becoming increasingly popular for conducting surveys, collecting field data, and monitoring program activities in real-time. These tools enable researchers to capture data using smartphones or tablets, allowing for faster data collection, improved data quality, and enhanced accessibility in remote or resource-constrained settings.
- Blockchain Technology: Blockchain technology offers opportunities for enhancing the transparency, security, and integrity of M&E data. By leveraging blockchain-based platforms, researchers can ensure the immutability and traceability of data, reduce the risk of data manipulation or fraud, and enhance trust and accountability in the evaluation process.
- Open Data and Data Sharing Platforms: There is a growing movement towards open data and data sharing in M&E, driven by the desire for transparency, collaboration, and knowledge exchange. Open data platforms facilitate the sharing of evaluation data, findings, and resources among stakeholders, enabling greater reproducibility, accountability, and innovation in the field.
- Citizen Science and Participatory Approaches: Citizen science and participatory approaches involve engaging community members and stakeholders in the research process, from data collection to interpretation and decision-making. By involving local communities in M&E efforts, researchers can gather diverse perspectives, foster ownership, and ensure the relevance and sustainability of evaluation initiatives.
- Ethical Considerations and Data Privacy: With the increasing use of digital technologies and data-driven approaches in M&E, there is a growing awareness of the need to address ethical considerations and data privacy concerns. Researchers must prioritize ethical principles such as informed consent, data confidentiality, and protection of vulnerable populations to ensure responsible and ethical conduct of evaluations.
By embracing these emerging trends and innovations in research tools, M&E practitioners can enhance the quality, rigor, and impact of evaluations, ultimately contributing to more effective and evidence-based decision-making in development and humanitarian efforts.
Research tools play a crucial role in the field of Monitoring and Evaluation (M&E) by supporting data collection, analysis, visualization, and reporting processes . The choice of research tools should be guided by the specific objectives, context, and data requirements of the evaluation.
Essential research tools in M&E include data collection instruments (surveys, interviews, observation checklists), data analysis software (SPSS, Stata, R), data visualization tools (Excel, Tableau), logic models, KPI frameworks, GIS software, evaluation management systems, and collaboration platforms.
By selecting and utilizing appropriate research tools, M&E practitioners can enhance the efficiency , accuracy, and effectiveness of their evaluations, leading to evidence-based decision-making and program improvement.
It is important to evaluate and choose tools that align with the evaluation design, data type, available resources, and technical expertise to ensure rigorous and meaningful evaluation outcomes in M&E.
Nguen B. Riak
I found this article on Qualitative research- on research tools is so essential. I will benefit from as student taking up his academic studies.
Leave a Comment Cancel Reply
Your email address will not be published.
How strong is my Resume?
Only 2% of resumes land interviews.
Land a better, higher-paying career


Jobs for You
Research technical advisor.
- South Bend, IN, USA (Remote)
- University of Notre Dame
YMELP II Short-Term Technical Assistance (STTA)
Water, sanitation and hygiene advisor (wash) – usaid/drc.
- Democratic Republic of the Congo
Health Supply Chain Specialist – USAID/DRC
Chief of party – bosnia and herzegovina.
- Bosnia and Herzegovina
Project Manager I
- United States
Business Development Associate
Director of finance and administration, request for information – collecting information on potential partners for local works evaluation.
- Washington, USA
Principal Field Monitors
Technical expert (health, wash, nutrition, education, child protection, hiv/aids, supplies), survey expert, data analyst, team leader, usaid-bha performance evaluation consultant.
- International Rescue Committee
Services you might be interested in
Useful guides ....
How to Create a Strong Resume
Monitoring And Evaluation Specialist Resume
Resume Length for the International Development Sector
Types of Evaluation
Monitoring, Evaluation, Accountability, and Learning (MEAL)
LAND A JOB REFERRAL IN 2 WEEKS (NO ONLINE APPS!)
Sign Up & To Get My Free Referral Toolkit Now:
Research tools & techniques
Research tools and systems.
- Library Search from the Library home page
- Google Scholar
- Fryer Manuscripts
- Off-campus access
Techniques for finding information
- Library Search help
- Search techniques
- Subject guides
- Referencing style guides
- EndNote referencing software
- Digital Essentials
- Health Information and Research Essential
- Digital Researcher Lab
- Library Search
- Health Information and Research Essentials
Research Tools
This collection contains molecular, cellular, and antibody tools and methods, recombinant technologies, biosensing and analytics technologies, technologies to improve cell therapies, and more.
Recombination
Rapid, stable, high titre production of recombinant retrovirus.
- Todd Kinsella
- Garry Nolan
Somatic Recombination with MADM (Mosaic Analysis with Double Markers)
Efficient homologous recombination of large transgenes using aav donor vectors.
- Matthew Porteus
- Sriram Vaidyanathan
Recombinant Protein and Peptides to Boost Epithelial Barrier Function and Prevent Infections
- Thomas Cherpes
- Rodolfo Vicetti Miguel
- Nirk Quispe Calla
Reversal of phiC31 integrase recombination in mammalian cells
- Christopher Chavez
- Alfonso Farruggio
- Michele Calos
- Carlos Mikell
Dynamic Recombinant Hydrogels with Degradation-Independent Relaxation Kinetics
- Renato Navarro
- Michelle Huang
- Julien Roth
- Kelsea Hubka
- Sarah Heilshorn
Recombinant viruses that replicate in response to specific biochemical signals
- Michael Lin
- Hokyung Chung
Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing
- Jason Myers
- James Ferrell
Super Recombinator (SuRe), a CRISPR/Cas9-based platform for the rapid incorporation of multiple transgenes at the same genetic locus
- Mark Schnitzer
- Cheng Huang
PopTag: A modular platform for engineering function of natural and synthetic biomolecular condensates
- Lucille Shapiro
- Keren Lasker
- Steven Boeynaems
- Aaron Gitler
MAGESTIC - A High Efficiency, Massively Parallel Production of Genetically Engineered Clones for Functional Genomics and Synthetic Biology
- Lars Steinmetz
- Justin Smith
- Robert St.Onge
- James Haber
Molecular, Cellular, and Antibody Tools
Anti-canine rab9 (mouse) antibody.
- Suzanne Pfeffer
The role of IGF-1 on oligodendrocyte differentiation in the adult CNS
- Jenny Hsieh
Anti HA Monoclonal Antibody
- Brian Zabel
- Eugene Butcher
Wnt agonist and antagonists for organoid cultures
- Kenan Christopher Garcia
- David Baker
- Keunwan Park
Gelation of Uniform Interfacial Diffusant in Embedded 3D Printing
- Sungchul Shin
- Alexis Seymour
Biochemical activation of dysfunctional skeletal stem cells for skeletal regeneration
- Charles Chan
- Michael Longaker
- Thomas Ambrosi
- Owen Marecic
- Irving Weissman
- Adrian McArdle
Functionally biased ligands for the Complement 5a Receptor
- Amato Giaccia
Detection and Profiling
Single molecule sequencing and methylation profiling of cell-free dna, automated point-of-care nucleic acid amplification test.
- Ana de Olazarra
- Chengyang Yao
- Neeraja Ravi
- Katie Antilla
SPED microscopy - high-speed, cellular-resolution volumetric imaging
- Karl Deisseroth
Novel PET imaging agents for the detection of CA6 epitope in-vivo
- Sanjiv Gambhir
- Ohad Ilovich
- Richard Kimura
- Arutselvan Natarajan
- Jochen Kruip
- Susanta Sarkar
- Mathias Gebauer
- Christian Lange
- Ingo Focken
Methods for Examining Network Effects of Immune Modulation
- Michelle Atallah
- Parag Mallick
Targeted Purification and Profiling of Human Extrachromosomal DNA
- Howard Chang
Low-cost, Comprehensive Methylation Profiling Using Low DNA Inputs for Cancer Diagnostics
- Lauren Ahmann
- Yvette Ysabel Yao
Combination Nucleic Acid Cytometry with Single Cell Genomics for the Study of Rare Cell Populations
- Ansuman Satpathy
- Caleb Lareau
- Robert Stickels
Advanced Biosensing and Analysis Techniques
Method for imaging cell death in vivo.
- Francis Blankenberg
- H. William Strauss
- Allan Green
- Neil Steinmetz
Method for producing completely monodisperse, highly repetitive polypeptides for use with Free-Solution Conjugate Electrophoresis and other bioanalytical applications
- Jennifer Lin
- Annelise Barron
- Jennifer Coyne Albrecht
- Xiaoxiao Wang
A Novel Red-shifted ChannelRhodopsin - ChRmine
- Yoon Seok Kim
- Hideaki Kato
- Charu Ramakrishnan
- Susumu Yoshizawa
Quantification of Antigen Molecules Without Calibrators Using Dynamic Flow Cytometry
- Darya Orlova
- Andrei Chernyshev
- Alexander Moskalensky
- David Parks
- Leonore Herzenberg
Recruitment of donor DNA from in vivo assembled plasmids for genome editing screens
Kinase-modulated bioluminescent indicators for reporting drug activity in vivo, system to improve ease and accuracy of flow cytometry.
- Wayne Moore
Stem Cell Research
Pluripotent-specific, inducible safety switch to eliminate residual stem cells following differentiation.
- Michael Cromer
- Renata Martin
- Jonas Fowler
Purifying Human Pluripotent Stem Cell-Derived Liver Cells by Metabolic Selection
- Lay Teng Ang
Targeted integration at alpha-globin locus in human hematopoietic stem and progenitor cells
- Daniel Dever
- Joab Camarena
Efficient generation of hematopoietic stem cell (HSC)-like cells from human pluripotent stem cells: a platform to create blood and immune cells
Rapid generation of human forebrain, midbrain, and hindbrain cells from human pluripotent stem cells.
- Carolyn Dundes
- Rayyan Jokhai
- Hadia Ahsan
- Rachel Salomon-Shulman
Differential Proliferation of Human HSPCs Using Truncated Erythropoietin Receptors
Enrichment of clinically relevant cell types using receptors.
- Carsten Charlesworth
Efficient Microglia Replacement without Genetic Modification via Bone-Marrow Derived Cells
- Marius Wernig
- Yohei Shibuya
Improving Cellular Therapies
Screening for gene modifications to improve t cell function, modulating bhlhe40 in the differentiation of type 1 regulatory t cells and controlling t cell exhaustion.
- Maria-Grazia Roncarolo
- Molly Kathryn Uyeda
- Robert Freeborn
Improved adoptive cell therapy
- Crystal Mackall
- Katherine Freitas
- Elena Sotillo
Genetically modified immune cells increase potency of adoptive immunotherapy
- Dorota Klysz
Common neoantigens in EGFR and RAS mutations generate generalized cytotoxic T lymphocytes for adoptive T-cell therapies
- Hiromitsu Nakauchi
- Toshinobu Nishimura
Method to direct T-cell fate towards T stem cell memory phenotype
- Zinaida Good
- Sean Bendall
Reversal of Tumor-Induced CAR-T cell and CD8+ T-cell Exhaustion with Annexin V
- Peter Katsikis
SMASh_CARs: A "drug off" chemogenetic system for regulating CAR T-cell therapy
- Robbie Majzner
- Louai Labanieh
Optimized CAR T cell hinge regions enhance CAR functionality
- Skyler Rietberg
Enhancing Hematopoietic Stem Cell Transplantation with Genetically Modified Donor T Cells
- Alice Bertaina
- Volker Wiebking
NOT-gated CAR T Cells to Ameliorate On-target, Off-tumor Toxicity of CAR T Therapy
- Rebecca Richards
- Ravindra Majeti
Reducing neurotoxicity in CD19-directed immunotherapy
- Kevin Parker
Use of Polyvinyl Alcohol for Chimeric Antigen Receptor T-Cell Expansion
- Adam Wilkinson
- Satoshi Yamazaki
A method for developing intra-epithelial innate lymphoid cells (tissue-resident NK cells) for immunotherapy
- Uriel Moreno
- John Sunwoo
- Saumyaa Saumyaa
- June Ho Shin
- Nina Horowitz
Methods to Prevent T-cell Exhaustion and Improve CAR-T Cell Immunotherapy with Small Molecules
- Rachel Lynn
- Sanjay Malhotra
Transiently Regulated CAR-T Cells Engineered to Prevent T-cell Exhaustion and Improve Immunotherapy
- Tom Wandless
Small Molecule Modulators of Chimeric Antigen Receptors (CAR) T Cells
- Vineet Kumar
- Mallesh Pandrala
- 7 Data Collection Methods & Tools For Research

- Data Collection
The underlying need for Data collection is to capture quality evidence that seeks to answer all the questions that have been posed. Through data collection businesses or management can deduce quality information that is a prerequisite for making informed decisions.
To improve the quality of information, it is expedient that data is collected so that you can draw inferences and make informed decisions on what is considered factual.
At the end of this article, you would understand why picking the best data collection method is necessary for achieving your set objective.
Sign up on Formplus Builder to create your preferred online surveys or questionnaire for data collection. You don’t need to be tech-savvy! Start creating quality questionnaires with Formplus.
What is Data Collection?
Data collection is a methodical process of gathering and analyzing specific information to proffer solutions to relevant questions and evaluate the results. It focuses on finding out all there is to a particular subject matter. Data is collected to be further subjected to hypothesis testing which seeks to explain a phenomenon.
Hypothesis testing eliminates assumptions while making a proposition from the basis of reason.

For collectors of data, there is a range of outcomes for which the data is collected. But the key purpose for which data is collected is to put a researcher in a vantage position to make predictions about future probabilities and trends.
The core forms in which data can be collected are primary and secondary data. While the former is collected by a researcher through first-hand sources, the latter is collected by an individual other than the user.
Types of Data Collection
Before broaching the subject of the various types of data collection. It is pertinent to note that data collection in itself falls under two broad categories; Primary data collection and secondary data collection.
Primary Data Collection
Primary data collection by definition is the gathering of raw data collected at the source. It is a process of collecting the original data collected by a researcher for a specific research purpose. It could be further analyzed into two segments; qualitative research and quantitative data collection methods.
- Qualitative Research Method
The qualitative research methods of data collection do not involve the collection of data that involves numbers or a need to be deduced through a mathematical calculation, rather it is based on the non-quantifiable elements like the feeling or emotion of the researcher. An example of such a method is an open-ended questionnaire.

- Quantitative Method
Quantitative methods are presented in numbers and require a mathematical calculation to deduce. An example would be the use of a questionnaire with close-ended questions to arrive at figures to be calculated Mathematically. Also, methods of correlation and regression, mean, mode and median.

Read Also: 15 Reasons to Choose Quantitative over Qualitative Research
Secondary Data Collection
Secondary data collection, on the other hand, is referred to as the gathering of second-hand data collected by an individual who is not the original user. It is the process of collecting data that is already existing, be it already published books, journals, and/or online portals. In terms of ease, it is much less expensive and easier to collect.
Your choice between Primary data collection and secondary data collection depends on the nature, scope, and area of your research as well as its aims and objectives.
Importance of Data Collection
There are a bunch of underlying reasons for collecting data, especially for a researcher. Walking you through them, here are a few reasons;
- Integrity of the Research
A key reason for collecting data, be it through quantitative or qualitative methods is to ensure that the integrity of the research question is indeed maintained.
- Reduce the likelihood of errors
The correct use of appropriate data collection of methods reduces the likelihood of errors consistent with the results.
- Decision Making
To minimize the risk of errors in decision-making, it is important that accurate data is collected so that the researcher doesn’t make uninformed decisions.
- Save Cost and Time
Data collection saves the researcher time and funds that would otherwise be misspent without a deeper understanding of the topic or subject matter.
- To support a need for a new idea, change, and/or innovation
To prove the need for a change in the norm or the introduction of new information that will be widely accepted, it is important to collect data as evidence to support these claims.
What is a Data Collection Tool?
Data collection tools refer to the devices/instruments used to collect data, such as a paper questionnaire or computer-assisted interviewing system. Case Studies, Checklists, Interviews, Observation sometimes, and Surveys or Questionnaires are all tools used to collect data.
It is important to decide on the tools for data collection because research is carried out in different ways and for different purposes. The objective behind data collection is to capture quality evidence that allows analysis to lead to the formulation of convincing and credible answers to the posed questions.
The objective behind data collection is to capture quality evidence that allows analysis to lead to the formulation of convincing and credible answers to the questions that have been posed – Click to Tweet
The Formplus online data collection tool is perfect for gathering primary data, i.e. raw data collected from the source. You can easily get data with at least three data collection methods with our online and offline data-gathering tool. I.e Online Questionnaires , Focus Groups, and Reporting.
In our previous articles, we’ve explained why quantitative research methods are more effective than qualitative methods . However, with the Formplus data collection tool, you can gather all types of primary data for academic, opinion or product research.
Top Data Collection Methods and Tools for Academic, Opinion, or Product Research
The following are the top 7 data collection methods for Academic, Opinion-based, or product research. Also discussed in detail are the nature, pros, and cons of each one. At the end of this segment, you will be best informed about which method best suits your research.
An interview is a face-to-face conversation between two individuals with the sole purpose of collecting relevant information to satisfy a research purpose. Interviews are of different types namely; Structured, Semi-structured , and unstructured with each having a slight variation from the other.
Use this interview consent form template to let an interviewee give you consent to use data gotten from your interviews for investigative research purposes.
- Structured Interviews – Simply put, it is a verbally administered questionnaire. In terms of depth, it is surface level and is usually completed within a short period. For speed and efficiency, it is highly recommendable, but it lacks depth.
- Semi-structured Interviews – In this method, there subsist several key questions which cover the scope of the areas to be explored. It allows a little more leeway for the researcher to explore the subject matter.
- Unstructured Interviews – It is an in-depth interview that allows the researcher to collect a wide range of information with a purpose. An advantage of this method is the freedom it gives a researcher to combine structure with flexibility even though it is more time-consuming.
- In-depth information
- Freedom of flexibility
- Accurate data.
- Time-consuming
- Expensive to collect.
What are The Best Data Collection Tools for Interviews?
For collecting data through interviews, here are a few tools you can use to easily collect data.
- Audio Recorder
An audio recorder is used for recording sound on disc, tape, or film. Audio information can meet the needs of a wide range of people, as well as provide alternatives to print data collection tools.
- Digital Camera
An advantage of a digital camera is that it can be used for transmitting those images to a monitor screen when the need arises.
A camcorder is used for collecting data through interviews. It provides a combination of both an audio recorder and a video camera. The data provided is qualitative in nature and allows the respondents to answer questions asked exhaustively. If you need to collect sensitive information during an interview, a camcorder might not work for you as you would need to maintain your subject’s privacy.
Want to conduct an interview for qualitative data research or a special report? Use this online interview consent form template to allow the interviewee to give their consent before you use the interview data for research or report. With premium features like e-signature, upload fields, form security, etc., Formplus Builder is the perfect tool to create your preferred online consent forms without coding experience.
- QUESTIONNAIRES
This is the process of collecting data through an instrument consisting of a series of questions and prompts to receive a response from the individuals it is administered to. Questionnaires are designed to collect data from a group.
For clarity, it is important to note that a questionnaire isn’t a survey, rather it forms a part of it. A survey is a process of data gathering involving a variety of data collection methods, including a questionnaire.
On a questionnaire, there are three kinds of questions used. They are; fixed-alternative, scale, and open-ended. With each of the questions tailored to the nature and scope of the research.
- Can be administered in large numbers and is cost-effective.
- It can be used to compare and contrast previous research to measure change.
- Easy to visualize and analyze.
- Questionnaires offer actionable data.
- Respondent identity is protected.
- Questionnaires can cover all areas of a topic.
- Relatively inexpensive.
- Answers may be dishonest or the respondents lose interest midway.
- Questionnaires can’t produce qualitative data.
- Questions might be left unanswered.
- Respondents may have a hidden agenda.
- Not all questions can be analyzed easily.
What are the Best Data Collection Tools for Questionnaires?
- Formplus Online Questionnaire
Formplus lets you create powerful forms to help you collect the information you need. Formplus helps you create the online forms that you like. The Formplus online questionnaire form template to get actionable trends and measurable responses. Conduct research, optimize knowledge of your brand or just get to know an audience with this form template. The form template is fast, free and fully customizable.
- Paper Questionnaire
A paper questionnaire is a data collection tool consisting of a series of questions and/or prompts for the purpose of gathering information from respondents. Mostly designed for statistical analysis of the responses, they can also be used as a form of data collection.
By definition, data reporting is the process of gathering and submitting data to be further subjected to analysis. The key aspect of data reporting is reporting accurate data because inaccurate data reporting leads to uninformed decision-making.
- Informed decision-making.
- Easily accessible.
- Self-reported answers may be exaggerated.
- The results may be affected by bias.
- Respondents may be too shy to give out all the details.
- Inaccurate reports will lead to uninformed decisions.
What are the Best Data Collection Tools for Reporting?
Reporting tools enable you to extract and present data in charts, tables, and other visualizations so users can find useful information. You could source data for reporting from Non-Governmental Organizations (NGO) reports, newspapers, website articles, and hospital records.
- NGO Reports
Contained in NGO report is an in-depth and comprehensive report on the activities carried out by the NGO, covering areas such as business and human rights. The information contained in these reports is research-specific and forms an acceptable academic base for collecting data. NGOs often focus on development projects which are organized to promote particular causes.
Newspaper data are relatively easy to collect and are sometimes the only continuously available source of event data. Even though there is a problem of bias in newspaper data, it is still a valid tool in collecting data for Reporting.
- Website Articles
Gathering and using data contained in website articles is also another tool for data collection. Collecting data from web articles is a quicker and less expensive data collection Two major disadvantages of using this data reporting method are biases inherent in the data collection process and possible security/confidentiality concerns.
- Hospital Care records
Health care involves a diverse set of public and private data collection systems, including health surveys, administrative enrollment and billing records, and medical records, used by various entities, including hospitals, CHCs, physicians, and health plans. The data provided is clear, unbiased and accurate, but must be obtained under legal means as medical data is kept with the strictest regulations.
- EXISTING DATA
This is the introduction of new investigative questions in addition to/other than the ones originally used when the data was initially gathered. It involves adding measurement to a study or research. An example would be sourcing data from an archive.
- Accuracy is very high.
- Easily accessible information.
- Problems with evaluation.
- Difficulty in understanding.
What are the Best Data Collection Tools for Existing Data?
The concept of Existing data means that data is collected from existing sources to investigate research questions other than those for which the data were originally gathered. Tools to collect existing data include:
- Research Journals – Unlike newspapers and magazines, research journals are intended for an academic or technical audience, not general readers. A journal is a scholarly publication containing articles written by researchers, professors, and other experts.
- Surveys – A survey is a data collection tool for gathering information from a sample population, with the intention of generalizing the results to a larger population. Surveys have a variety of purposes and can be carried out in many ways depending on the objectives to be achieved.
- OBSERVATION
This is a data collection method by which information on a phenomenon is gathered through observation. The nature of the observation could be accomplished either as a complete observer, an observer as a participant, a participant as an observer, or as a complete participant. This method is a key base for formulating a hypothesis.
- Easy to administer.
- There subsists a greater accuracy with results.
- It is a universally accepted practice.
- It diffuses the situation of the unwillingness of respondents to administer a report.
- It is appropriate for certain situations.
- Some phenomena aren’t open to observation.
- It cannot be relied upon.
- Bias may arise.
- It is expensive to administer.
- Its validity cannot be predicted accurately.
What are the Best Data Collection Tools for Observation?
Observation involves the active acquisition of information from a primary source. Observation can also involve the perception and recording of data via the use of scientific instruments. The best tools for Observation are:
- Checklists – state-specific criteria, that allow users to gather information and make judgments about what they should know in relation to the outcomes. They offer systematic ways of collecting data about specific behaviors, knowledge, and skills.
- Direct observation – This is an observational study method of collecting evaluative information. The evaluator watches the subject in his or her usual environment without altering that environment.
FOCUS GROUPS
The opposite of quantitative research which involves numerical-based data, this data collection method focuses more on qualitative research. It falls under the primary category of data based on the feelings and opinions of the respondents. This research involves asking open-ended questions to a group of individuals usually ranging from 6-10 people, to provide feedback.
- Information obtained is usually very detailed.
- Cost-effective when compared to one-on-one interviews.
- It reflects speed and efficiency in the supply of results.
- Lacking depth in covering the nitty-gritty of a subject matter.
- Bias might still be evident.
- Requires interviewer training
- The researcher has very little control over the outcome.
- A few vocal voices can drown out the rest.
- Difficulty in assembling an all-inclusive group.
What are the Best Data Collection Tools for Focus Groups?
A focus group is a data collection method that is tightly facilitated and structured around a set of questions. The purpose of the meeting is to extract from the participants’ detailed responses to these questions. The best tools for tackling Focus groups are:
- Two-Way – One group watches another group answer the questions posed by the moderator. After listening to what the other group has to offer, the group that listens is able to facilitate more discussion and could potentially draw different conclusions .
- Dueling-Moderator – There are two moderators who play the devil’s advocate. The main positive of the dueling-moderator focus group is to facilitate new ideas by introducing new ways of thinking and varying viewpoints.
- COMBINATION RESEARCH
This method of data collection encompasses the use of innovative methods to enhance participation in both individuals and groups. Also under the primary category, it is a combination of Interviews and Focus Groups while collecting qualitative data . This method is key when addressing sensitive subjects.
- Encourage participants to give responses.
- It stimulates a deeper connection between participants.
- The relative anonymity of respondents increases participation.
- It improves the richness of the data collected.
- It costs the most out of all the top 7.
- It’s the most time-consuming.
What are the Best Data Collection Tools for Combination Research?
The Combination Research method involves two or more data collection methods, for instance, interviews as well as questionnaires or a combination of semi-structured telephone interviews and focus groups. The best tools for combination research are:
- Online Survey – The two tools combined here are online interviews and the use of questionnaires. This is a questionnaire that the target audience can complete over the Internet. It is timely, effective, and efficient. Especially since the data to be collected is quantitative in nature.
- Dual-Moderator – The two tools combined here are focus groups and structured questionnaires. The structured questionnaires give a direction as to where the research is headed while two moderators take charge of the proceedings. Whilst one ensures the focus group session progresses smoothly, the other makes sure that the topics in question are all covered. Dual-moderator focus groups typically result in a more productive session and essentially lead to an optimum collection of data.
Why Formplus is the Best Data Collection Tool
- Vast Options for Form Customization
With Formplus, you can create your unique survey form. With options to change themes, font color, font, font type, layout, width, and more, you can create an attractive survey form. The builder also gives you as many features as possible to choose from and you do not need to be a graphic designer to create a form.
- Extensive Analytics
Form Analytics, a feature in formplus helps you view the number of respondents, unique visits, total visits, abandonment rate, and average time spent before submission. This tool eliminates the need for a manual calculation of the received data and/or responses as well as the conversion rate for your poll.
- Embed Survey Form on Your Website
Copy the link to your form and embed it as an iframe which will automatically load as your website loads, or as a popup that opens once the respondent clicks on the link. Embed the link on your Twitter page to give instant access to your followers.
.png?resize=845%2C418&ssl=1)
- Geolocation Support
The geolocation feature on Formplus lets you ascertain where individual responses are coming. It utilises Google Maps to pinpoint the longitude and latitude of the respondent, to the nearest accuracy, along with the responses.
- Multi-Select feature
This feature helps to conserve horizontal space as it allows you to put multiple options in one field. This translates to including more information on the survey form.
Read Also: 10 Reasons to Use Formplus for Online Data Collection
How to Use Formplus to collect online data in 7 simple steps.
- Register or sign up on Formplus builder : Start creating your preferred questionnaire or survey by signing up with either your Google, Facebook, or Email account.

Formplus gives you a free plan with basic features you can use to collect online data. Pricing plans with vast features starts at $20 monthly, with reasonable discounts for Education and Non-Profit Organizations.
2. Input your survey title and use the form builder choice options to start creating your surveys.
Use the choice option fields like single select, multiple select, checkbox, radio, and image choices to create your preferred multi-choice surveys online.

3. Do you want customers to rate any of your products or services delivery?
Use the rating to allow survey respondents rate your products or services. This is an ideal quantitative research method of collecting data.

4. Beautify your online questionnaire with Formplus Customisation features.

- Change the theme color
- Add your brand’s logo and image to the forms
- Change the form width and layout
- Edit the submission button if you want
- Change text font color and sizes
- Do you have already made custom CSS to beautify your questionnaire? If yes, just copy and paste it to the CSS option.
5. Edit your survey questionnaire settings for your specific needs
Choose where you choose to store your files and responses. Select a submission deadline, choose a timezone, limit respondents’ responses, enable Captcha to prevent spam, and collect location data of customers.

Set an introductory message to respondents before they begin the survey, toggle the “start button” post final submission message or redirect respondents to another page when they submit their questionnaires.
Change the Email Notifications inventory and initiate an autoresponder message to all your survey questionnaire respondents. You can also transfer your forms to other users who can become form administrators.
6. Share links to your survey questionnaire page with customers.
There’s an option to copy and share the link as “Popup” or “Embed code” The data collection tool automatically creates a QR Code for Survey Questionnaire which you can download and share as appropriate.

Congratulations if you’ve made it to this stage. You can start sharing the link to your survey questionnaire with your customers.
7. View your Responses to the Survey Questionnaire
Toggle with the presentation of your summary from the options. Whether as a single, table or cards.

8. Allow Formplus Analytics to interpret your Survey Questionnaire Data

With online form builder analytics, a business can determine;
- The number of times the survey questionnaire was filled
- The number of customers reached
- Abandonment Rate: The rate at which customers exit the form without submitting it.
- Conversion Rate: The percentage of customers who completed the online form
- Average time spent per visit
- Location of customers/respondents.
- The type of device used by the customer to complete the survey questionnaire.
7 Tips to Create The Best Surveys For Data Collections
- Define the goal of your survey – Once the goal of your survey is outlined, it will aid in deciding which questions are the top priority. A clear attainable goal would, for example, mirror a clear reason as to why something is happening. e.g. “The goal of this survey is to understand why Employees are leaving an establishment.”
- Use close-ended clearly defined questions – Avoid open-ended questions and ensure you’re not suggesting your preferred answer to the respondent. If possible offer a range of answers with choice options and ratings.
- Survey outlook should be attractive and Inviting – An attractive-looking survey encourages a higher number of recipients to respond to the survey. Check out Formplus Builder for colorful options to integrate into your survey design. You could use images and videos to keep participants glued to their screens.
- Assure Respondents about the safety of their data – You want your respondents to be assured whilst disclosing details of their personal information to you. It’s your duty to inform the respondents that the data they provide is confidential and only collected for the purpose of research.
- Ensure your survey can be completed in record time – Ideally, in a typical survey, users should be able to respond in 100 seconds. It is pertinent to note that they, the respondents, are doing you a favor. Don’t stress them. Be brief and get straight to the point.
- Do a trial survey – Preview your survey before sending out your surveys to the intended respondents. Make a trial version which you’ll send to a few individuals. Based on their responses, you can draw inferences and decide whether or not your survey is ready for the big time.
- Attach a reward upon completion for users – Give your respondents something to look forward to at the end of the survey. Think of it as a penny for their troubles. It could well be the encouragement they need to not abandon the survey midway.
Try out Formplus today . You can start making your own surveys with the Formplus online survey builder. By applying these tips, you will definitely get the most out of your online surveys.
Top Survey Templates For Data Collection
- Customer Satisfaction Survey Template
On the template, you can collect data to measure customer satisfaction over key areas like the commodity purchase and the level of service they received. It also gives insight as to which products the customer enjoyed, how often they buy such a product, and whether or not the customer is likely to recommend the product to a friend or acquaintance.
- Demographic Survey Template
With this template, you would be able to measure, with accuracy, the ratio of male to female, age range, and the number of unemployed persons in a particular country as well as obtain their personal details such as names and addresses.
Respondents are also able to state their religious and political views about the country under review.
- Feedback Form Template
Contained in the template for the online feedback form is the details of a product and/or service used. Identifying this product or service and documenting how long the customer has used them.
The overall satisfaction is measured as well as the delivery of the services. The likelihood that the customer also recommends said product is also measured.
- Online Questionnaire Template
The online questionnaire template houses the respondent’s data as well as educational qualifications to collect information to be used for academic research.
Respondents can also provide their gender, race, and field of study as well as present living conditions as prerequisite data for the research study.
- Student Data Sheet Form Template
The template is a data sheet containing all the relevant information of a student. The student’s name, home address, guardian’s name, record of attendance as well as performance in school is well represented on this template. This is a perfect data collection method to deploy for a school or an education organization.
Also included is a record for interaction with others as well as a space for a short comment on the overall performance and attitude of the student.
- Interview Consent Form Template
This online interview consent form template allows the interviewee to sign off their consent to use the interview data for research or report to journalists. With premium features like short text fields, upload, e-signature, etc., Formplus Builder is the perfect tool to create your preferred online consent forms without coding experience.
What is the Best Data Collection Method for Qualitative Data?
Answer: Combination Research
The best data collection method for a researcher for gathering qualitative data which generally is data relying on the feelings, opinions, and beliefs of the respondents would be Combination Research.
The reason why combination research is the best fit is that it encompasses the attributes of Interviews and Focus Groups. It is also useful when gathering data that is sensitive in nature. It can be described as an all-purpose quantitative data collection method.
Above all, combination research improves the richness of data collected when compared with other data collection methods for qualitative data.

What is the Best Data Collection Method for Quantitative Research Data?
Ans: Questionnaire
The best data collection method a researcher can employ in gathering quantitative data which takes into consideration data that can be represented in numbers and figures that can be deduced mathematically is the Questionnaire.
These can be administered to a large number of respondents while saving costs. For quantitative data that may be bulky or voluminous in nature, the use of a Questionnaire makes such data easy to visualize and analyze.
Another key advantage of the Questionnaire is that it can be used to compare and contrast previous research work done to measure changes.
Technology-Enabled Data Collection Methods
There are so many diverse methods available now in the world because technology has revolutionized the way data is being collected. It has provided efficient and innovative methods that anyone, especially researchers and organizations. Below are some technology-enabled data collection methods:
- Online Surveys: Online surveys have gained popularity due to their ease of use and wide reach. You can distribute them through email, social media, or embed them on websites. Online surveys allow you to quickly complete data collection, automated data capture, and real-time analysis. Online surveys also offer features like skip logic, validation checks, and multimedia integration.
- Mobile Surveys: With the widespread use of smartphones, mobile surveys’ popularity is also on the rise. Mobile surveys leverage the capabilities of mobile devices, and this allows respondents to participate at their convenience. This includes multimedia elements, location-based information, and real-time feedback. Mobile surveys are the best for capturing in-the-moment experiences or opinions.
- Social Media Listening: Social media platforms are a good source of unstructured data that you can analyze to gain insights into customer sentiment and trends. Social media listening involves monitoring and analyzing social media conversations, mentions, and hashtags to understand public opinion, identify emerging topics, and assess brand reputation.
- Wearable Devices and Sensors: You can embed wearable devices, such as fitness trackers or smartwatches, and sensors in everyday objects to capture continuous data on various physiological and environmental variables. This data can provide you with insights into health behaviors, activity patterns, sleep quality, and environmental conditions, among others.
- Big Data Analytics: Big data analytics leverages large volumes of structured and unstructured data from various sources, such as transaction records, social media, and internet browsing. Advanced analytics techniques, like machine learning and natural language processing, can extract meaningful insights and patterns from this data, enabling organizations to make data-driven decisions.
Read Also: How Technology is Revolutionizing Data Collection
Faulty Data Collection Practices – Common Mistakes & Sources of Error
While technology-enabled data collection methods offer numerous advantages, there are some pitfalls and sources of error that you should be aware of. Here are some common mistakes and sources of error in data collection:
- Population Specification Error: Population specification error occurs when the target population is not clearly defined or misidentified. This error leads to a mismatch between the research objectives and the actual population being studied, resulting in biased or inaccurate findings.
- Sample Frame Error: Sample frame error occurs when the sampling frame, the list or source from which the sample is drawn, does not adequately represent the target population. This error can introduce selection bias and affect the generalizability of the findings.
- Selection Error: Selection error occurs when the process of selecting participants or units for the study introduces bias. It can happen due to nonrandom sampling methods, inadequate sampling techniques, or self-selection bias. Selection error compromises the representativeness of the sample and affects the validity of the results.
- Nonresponse Error: Nonresponse error occurs when selected participants choose not to participate or fail to respond to the data collection effort. Nonresponse bias can result in an unrepresentative sample if those who choose not to respond differ systematically from those who do respond. Efforts should be made to mitigate nonresponse and encourage participation to minimize this error.
- Measurement Error: Measurement error arises from inaccuracies or inconsistencies in the measurement process. It can happen due to poorly designed survey instruments, ambiguous questions, respondent bias, or errors in data entry or coding. Measurement errors can lead to distorted or unreliable data, affecting the validity and reliability of the findings.
In order to mitigate these errors and ensure high-quality data collection, you should carefully plan your data collection procedures, and validate measurement tools. You should also use appropriate sampling techniques, employ randomization where possible, and minimize nonresponse through effective communication and incentives. Ensure you conduct regular checks and implement validation processes, and data cleaning procedures to identify and rectify errors during data analysis.
Best Practices for Data Collection
- Clearly Define Objectives: Clearly define the research objectives and questions to guide the data collection process. This helps ensure that the collected data aligns with the research goals and provides relevant insights.
- Plan Ahead: Develop a detailed data collection plan that includes the timeline, resources needed, and specific procedures to follow. This helps maintain consistency and efficiency throughout the data collection process.
- Choose the Right Method: Select data collection methods that are appropriate for the research objectives and target population. Consider factors such as feasibility, cost-effectiveness, and the ability to capture the required data accurately.
- Pilot Test : Before full-scale data collection, conduct a pilot test to identify any issues with the data collection instruments or procedures. This allows for refinement and improvement before data collection with the actual sample.
- Train Data Collectors: If data collection involves human interaction, ensure that data collectors are properly trained on the data collection protocols, instruments, and ethical considerations. Consistent training helps minimize errors and maintain data quality.
- Maintain Consistency: Follow standardized procedures throughout the data collection process to ensure consistency across data collectors and time. This includes using consistent measurement scales, instructions, and data recording methods.
- Minimize Bias: Be aware of potential sources of bias in data collection and take steps to minimize their impact. Use randomization techniques, employ diverse data collectors, and implement strategies to mitigate response biases.
- Ensure Data Quality: Implement quality control measures to ensure the accuracy, completeness, and reliability of the collected data. Conduct regular checks for data entry errors, inconsistencies, and missing values.
- Maintain Data Confidentiality: Protect the privacy and confidentiality of participants’ data by implementing appropriate security measures. Ensure compliance with data protection regulations and obtain informed consent from participants.
- Document the Process: Keep detailed documentation of the data collection process, including any deviations from the original plan, challenges encountered, and decisions made. This documentation facilitates transparency, replicability, and future analysis.
FAQs about Data Collection
- What are secondary sources of data collection? Secondary sources of data collection are defined as the data that has been previously gathered and is available for your use as a researcher. These sources can include published research papers, government reports, statistical databases, and other existing datasets.
- What are the primary sources of data collection? Primary sources of data collection involve collecting data directly from the original source also known as the firsthand sources. You can do this through surveys, interviews, observations, experiments, or other direct interactions with individuals or subjects of study.
- How many types of data are there? There are two main types of data: qualitative and quantitative. Qualitative data is non-numeric and it includes information in the form of words, images, or descriptions. Quantitative data, on the other hand, is numeric and you can measure and analyze it statistically.
Sign up on Formplus Builder to create your preferred online surveys or questionnaire for data collection. You don’t need to be tech-savvy!

Connect to Formplus, Get Started Now - It's Free!
- academic research
- Data collection method
- data collection techniques
- data collection tool
- data collection tools
- field data collection
- online data collection tool
- product research
- qualitative research data
- quantitative research data
- scientific research
- busayo.longe

You may also like:
How Technology is Revolutionizing Data Collection
As global industrialization continues to transform, it is becoming evident that there is a ubiquity of large datasets driven by the need...

Data Collection Sheet: Types + [Template Examples]
Simple guide on data collection sheet. Types, tools, and template examples.
User Research: Definition, Methods, Tools and Guide
In this article, you’ll learn to provide value to your target market with user research. As a bonus, we’ve added user research tools and...
Data Collection Plan: Definition + Steps to Do It
Introduction A data collection plan is a way to get specific information on your audience. You can use it to better understand what they...
Formplus - For Seamless Data Collection
Collect data the right way with a versatile data collection tool. try formplus and transform your work productivity today..
- Open access
- Published: 27 May 2024
Current status of community resources and priorities for weed genomics research
- Jacob Montgomery 1 ,
- Sarah Morran 1 ,
- Dana R. MacGregor ORCID: orcid.org/0000-0003-0543-0408 2 ,
- J. Scott McElroy ORCID: orcid.org/0000-0003-0331-3697 3 ,
- Paul Neve ORCID: orcid.org/0000-0002-3136-5286 4 ,
- Célia Neto ORCID: orcid.org/0000-0003-3256-5228 4 ,
- Martin M. Vila-Aiub ORCID: orcid.org/0000-0003-2118-290X 5 ,
- Maria Victoria Sandoval 5 ,
- Analia I. Menéndez ORCID: orcid.org/0000-0002-9681-0280 6 ,
- Julia M. Kreiner ORCID: orcid.org/0000-0002-8593-1394 7 ,
- Longjiang Fan ORCID: orcid.org/0000-0003-4846-0500 8 ,
- Ana L. Caicedo ORCID: orcid.org/0000-0002-0378-6374 9 ,
- Peter J. Maughan 10 ,
- Bianca Assis Barbosa Martins 11 ,
- Jagoda Mika 11 ,
- Alberto Collavo 11 ,
- Aldo Merotto Jr. ORCID: orcid.org/0000-0002-1581-0669 12 ,
- Nithya K. Subramanian ORCID: orcid.org/0000-0002-1659-7396 13 ,
- Muthukumar V. Bagavathiannan ORCID: orcid.org/0000-0002-1107-7148 13 ,
- Luan Cutti ORCID: orcid.org/0000-0002-2867-7158 14 ,
- Md. Mazharul Islam 15 ,
- Bikram S. Gill ORCID: orcid.org/0000-0003-4510-9459 16 ,
- Robert Cicchillo 17 ,
- Roger Gast 17 ,
- Neeta Soni ORCID: orcid.org/0000-0002-4647-8355 17 ,
- Terry R. Wright ORCID: orcid.org/0000-0002-3969-2812 18 ,
- Gina Zastrow-Hayes 18 ,
- Gregory May 18 ,
- Jenna M. Malone ORCID: orcid.org/0000-0002-9637-2073 19 ,
- Deepmala Sehgal ORCID: orcid.org/0000-0002-4141-1784 20 ,
- Shiv Shankhar Kaundun ORCID: orcid.org/0000-0002-7249-2046 20 ,
- Richard P. Dale 20 ,
- Barend Juan Vorster ORCID: orcid.org/0000-0003-3518-3508 21 ,
- Bodo Peters 11 ,
- Jens Lerchl ORCID: orcid.org/0000-0002-9633-2653 22 ,
- Patrick J. Tranel ORCID: orcid.org/0000-0003-0666-4564 23 ,
- Roland Beffa ORCID: orcid.org/0000-0003-3109-388X 24 ,
- Alexandre Fournier-Level ORCID: orcid.org/0000-0002-6047-7164 25 ,
- Mithila Jugulam ORCID: orcid.org/0000-0003-2065-9067 15 ,
- Kevin Fengler 18 ,
- Victor Llaca ORCID: orcid.org/0000-0003-4822-2924 18 ,
- Eric L. Patterson ORCID: orcid.org/0000-0001-7111-6287 14 &
- Todd A. Gaines ORCID: orcid.org/0000-0003-1485-7665 1
Genome Biology volume 25 , Article number: 139 ( 2024 ) Cite this article
194 Accesses
1 Altmetric
Metrics details
Weeds are attractive models for basic and applied research due to their impacts on agricultural systems and capacity to swiftly adapt in response to anthropogenic selection pressures. Currently, a lack of genomic information precludes research to elucidate the genetic basis of rapid adaptation for important traits like herbicide resistance and stress tolerance and the effect of evolutionary mechanisms on wild populations. The International Weed Genomics Consortium is a collaborative group of scientists focused on developing genomic resources to impact research into sustainable, effective weed control methods and to provide insights about stress tolerance and adaptation to assist crop breeding.
Each year globally, agricultural producers and landscape managers spend billions of US dollars [ 1 , 2 ] and countless hours attempting to control weedy plants and reduce their adverse effects. These management methods range from low-tech (e.g., pulling plants from the soil by hand) to extremely high-tech (e.g., computer vision-controlled spraying of herbicides). Regardless of technology level, effective control methods serve as strong selection pressures on weedy plants and often result in rapid evolution of weed populations resistant to such methods [ 3 , 4 , 5 , 6 , 7 ]. Thus, humans and weeds have been locked in an arms race, where humans develop new or improved control methods and weeds adapt and evolve to circumvent such methods.
Applying genomics to weed science offers a unique opportunity to study rapid adaptation, epigenetic responses, and examples of evolutionary rescue of diverse weedy species in the face of widespread and powerful selective pressures. Furthermore, lessons learned from these studies may also help to develop more sustainable control methods and to improve crop breeding efforts in the face of our ever-changing climate. While other research fields have used genetics and genomics to uncover the basis of many biological traits [ 8 , 9 , 10 , 11 ] and to understand how ecological factors affect evolution [ 12 , 13 ], the field of weed science has lagged behind in the development of genomic tools essential for such studies [ 14 ]. As research in human and crop genetics pushes into the era of pangenomics (i.e., multiple chromosome scale genome assemblies for a single species [ 15 , 16 ]), publicly available genomic information is still lacking or severely limited for the majority of weed species. Recent reviews of current weed genomes identified 26 [ 17 ] and 32 weed species with sequenced genomes [ 18 ]—many assembled to a sub-chromosome level.
Here, we summarize the current state of weed genomics, highlighting cases where genomics approaches have successfully provided insights on topics such as population genetic dynamics, genome evolution, and the genetic basis of herbicide resistance, rapid adaptation, and crop dedomestication. These highlighted investigations all relied upon genomic resources that are relatively rare for weedy species. Throughout, we identify additional resources that would advance the field of weed science and enable further progress in weed genomics. We then introduce the International Weed Genomics Consortium (IWGC), an open collaboration among researchers, and describe current efforts to generate these additional resources.
Evolution of weediness: potential research utilizing weed genomics tools
Weeds can evolve from non-weed progenitors through wild colonization, crop de-domestication, or crop-wild hybridization [ 19 ]. Because the time span in which weeds have evolved is necessarily limited by the origins of agriculture, these non-weed relatives often still exist and can be leveraged through population genomic and comparative genomic approaches to identify the adaptive changes that have driven the evolution of weediness. The ability to rapidly adapt, persist, and spread in agroecosystems are defining features of weedy plants, leading many to advocate agricultural weeds as ideal candidates for studying rapid plant adaptation [ 20 , 21 , 22 , 23 ]. The insights gained from applying plant ecological approaches to the study of rapid weed adaptation will move us towards the ultimate goals of mitigating such adaptation and increasing the efficacy of crop breeding and biotechnology [ 14 ].
Biology and ecological genomics of weeds
The impressive community effort to create and maintain resources for Arabidopsis thaliana ecological genomics provides a motivating example for the emerging study of weed genomics [ 24 , 25 , 26 , 27 ]. Arabidopsis thaliana was the first flowering plant species to have its genome fully sequenced [ 28 ] and rapidly became a model organism for plant molecular biology. As weedy genomes become available, collection, maintenance, and resequencing of globally distributed accessions of these species will help to replicate the success found in ecological studies of A. thaliana [ 29 , 30 , 31 , 32 , 33 , 34 , 35 ]. Evaluation of these accessions for traits of interest to produce large phenomics data sets (as in [ 36 , 37 , 38 , 39 , 40 ]) enables genome-wide association studies and population genomics analyses aimed at dissecting the genetic basis of variation in such traits [ 41 ]. Increasingly, these resources (e.g. the 1001 genomes project [ 29 ]) have enabled A. thaliana to be utilized as a model species to explore the eco-evolutionary basis of plant adaptation in a more realistic ecological context. Weedy species should supplement lessons in eco-evolutionary genomics learned from these experiments in A. thaliana .
Untargeted genomic approaches for understanding the evolutionary trajectories of populations and the genetic basis of traits as described above rely on the collection of genotypic information from across the genome of many individuals. While whole-genome resequencing accomplishes this requirement and requires no custom methodology, this approach provides more information than is necessary and is prohibitively expensive in species with large genomes. Development and optimization of genotype-by-sequencing methods for capturing reduced representations of newly sequence genomes like those described by [ 42 , 43 , 44 ] will reduce the cost and computational requirements of genetic mapping and population genetic experiments. Most major weed species do not currently have protocols for stable transformation, a key development in the popularity of A. thaliana as a model organism and a requirement for many functional genomic approaches. Functional validation of genes/variants believed to be responsible for traits of interest in weeds has thus far relied on transiently manipulating endogenous gene expression [ 45 , 46 ] or ectopic expression of a transgene in a model system [ 47 , 48 , 49 ]. While these methods have been successful, few weed species have well-studied viral vectors to adapt for use in virus induced gene silencing. Spray induced gene silencing is another potential option for functional investigation of candidate genes in weeds, but more research is needed to establish reliable delivery and gene knockdown [ 50 ]. Furthermore, traits with complex genetic architecture divergent between the researched and model species may not be amenable to functional genomic approaches using transgenesis techniques in model systems. Developing protocols for reduced representation sequencing, stable transformation, and gene editing/silencing in weeds will allow for more thorough characterization of candidate genetic variants underlying traits of interest.
Beyond rapid adaptation, some weedy species offer an opportunity to better understand co-evolution, like that between plants and pollinators and how their interaction leads to the spread of weedy alleles (Additional File 1 : Table S1). A suite of plant–insect traits has co-evolved to maximize the attraction of the insect pollinator community and the efficiency of pollen deposition between flowers ensuring fruit and seed production in many weeds [ 51 , 52 ]. Genetic mapping experiments have identified genes and genetic variants responsible for many floral traits affecting pollinator interaction including petal color [ 53 , 54 , 55 , 56 ], flower symmetry and size [ 57 , 58 , 59 ], and production of volatile organic compounds [ 60 , 61 , 62 ] and nectar [ 63 , 64 , 65 ]. While these studies reveal candidate genes for selection under co-evolution, herbicide resistance alleles may also have pleiotropic effects on the ecology of weeds [ 66 ], altering plant-pollinator interactions [ 67 ]. Discovery of genes and genetic variants involved in weed-pollinator interaction and their molecular and environmental control may create opportunities for better management of weeds with insect-mediated pollination. For example, if management can disrupt pollinator attraction/interaction with these weeds, the efficiency of reproduction may be reduced.
A more complete understanding of weed ecological genomics will undoubtedly elucidate many unresolved questions regarding the genetic basis of various aspects of weediness. For instance, when comparing populations of a species from agricultural and non-agricultural environments, is there evidence for contemporary evolution of weedy traits selected by agricultural management or were “natural” populations pre-adapted to agroecosystems? Where there is differentiation between weedy and natural populations, which traits are under selection and what is the genetic basis of variation in those traits? When comparing between weedy populations, is there evidence for parallel versus non-parallel evolution of weediness at the phenotypic and genotypic levels? Such studies may uncover fundamental truths about weediness. For example, is there a common phenotypic and/or genotypic basis for aspects of weediness among diverse weed species? The availability of characterized accessions and reference genomes for species of interest are required for such studies but only a few weedy species have these resources developed.
Population genomics
Weed species are certainly fierce competitors, able to outcompete crops and endemic species in their native environment, but they are also remarkable colonizers of perturbed habitats. Weeds achieve this through high fecundity, often producing tens of thousands of seeds per individual plant [ 68 , 69 , 70 ]. These large numbers in terms of demographic population size often combine with outcrossing reproduction to generate high levels of diversity with local effective population sizes in the hundreds of thousands [ 71 , 72 ]. This has two important consequences: weed populations retain standing genetic variation and generate many new mutations, supporting weed success in the face of harsh control. The generation of genomic tools to monitor weed populations at the molecular level is a game-changer to understanding weed dynamics and precisely testing the effect of artificial selection (i.e., management) and other evolutionary mechanisms on the genetic make-up of populations.
Population genomic data, without any environmental or phenotypic information, can be used to scan the genomes of weed and non-weed relatives to identify selective sweeps, pointing at loci supporting weed adaptation on micro- or macro-evolutionary scales. Two recent within-species examples include weedy rice, where population differentiation between weedy and domesticated populations was used to identify the genetic basis of weedy de-domestication [ 73 ], and common waterhemp, where consistent allelic differences among natural and agricultural collections resolved a complex set of agriculturally adaptive alleles [ 74 , 75 ]. A recent comparative population genomic study of weedy barnyardgrass and crop millet species has demonstrated how inter-specific investigations can resolve the signatures of crop and weed evolution [ 76 ] (also see [ 77 ] for a non-weed climate adaptation example). Multiple sequence alignments across numerous species provide complementary insight into adaptive convergence over deeper timescales, even with just one genomic sample per species (e.g., [ 78 , 79 ]). Thus, newly sequenced weed genomes combined with genomes available for closely related crops (outlined by [ 14 , 80 ]) and an effort to identify other non-weed wild relatives will be invaluable in characterizing the genetic architecture of weed adaptation and evolution across diverse species.
Weeds experience high levels of genetic selection, both artificial in response to agricultural practices and particularly herbicides, and natural in response to the environmental conditions they encounter [ 81 , 82 ]. Using genomic analysis to identify loci that are the targets of selection, whether natural or artificial, would point at vulnerabilities that could be leveraged against weeds to develop new and more sustainable management strategies [ 83 ]. This is a key motivation to develop genotype-by-environment association (GEA) and selective sweep scan approaches, which allow researchers to resolve the molecular basis of multi-dimensional adaptation [ 84 , 85 ]. GEA approaches, in particular, have been widely used on landscape-wide resequencing collections to determine the genetic basis of climate adaptation (e.g., [ 27 , 86 , 87 ]), but have yet to be fully exploited to diagnose the genetic basis of the various aspects of weediness [ 88 ]. Armed with data on environmental dimensions of agricultural settings, such as focal crop, soil quality, herbicide use, and climate, GEA approaches can help disentangle how discrete farming practices have influenced the evolution of weediness and resolve broader patterns of local adaptation across a weed’s range. Although non-weedy relatives are not technically required for GEA analyses, inclusion of environmental and genomic data from weed progenitors can further distinguish genetic variants underpinning weed origins from those involved in local adaptation.
New weeds emerge frequently [ 89 ], either through hybridization between species as documented for sea beet ( Beta vulgaris ssp. maritima) hybridizing with crop beet to produce progeny that are well adapted to agricultural conditions [ 90 , 91 , 92 ], or through the invasion of alien species that find a new range to colonize. Biosecurity measures are often in place to stop the introduction of new weeds; however, the vast scale of global agricultural commodity trade precludes the possibility of total control. Population genomic analysis is now able to measure gene flow between populations [ 74 , 93 , 94 , 95 ] and identify populations of origin for invasive species including weeds [ 96 , 97 , 98 ]. For example, the invasion route of the pest fruitfly Drosophila suzukii from Eastern Asia to North America and Europe through Hawaii was deciphered using Approximate Bayesian Computation on high-throughput sequencing data from a global sample of multiple populations [ 99 ]. Genomics can also be leveraged to predict invasion rather than explain it. The resequencing of a global sample of common ragweed ( Ambrosia artemisiifolia L.) elucidated a complex invasion route whereby Europe was invaded by multiple introductions of American ragweed that hybridized in Europe prior to a subsequent introduction to Australia [ 100 , 101 ]. In this context, the use of genomically informed species distribution models helps assess the risk associated with different source populations, which in the case of common ragweed, suggests that a source population from Florida would allow ragweed to invade most of northern Australia [ 102 ]. Globally coordinated research efforts to understand potential distribution models could support the transformation of biosecurity from perspective analysis towards predictive risk assessment.
Herbicide resistance and weed management
Herbicide resistance is among the numerous weedy traits that can evolve in plant populations exposed to agricultural selection pressures. Over-reliance on herbicides to control weeds, along with low diversity and lack of redundancy in weed management strategies, has resulted in globally widespread herbicide resistance [ 103 ]. To date, 272 herbicide-resistant weed species have been reported worldwide, and at least one resistance case exists for 21 of the 31 existing herbicide sites of action [ 104 ]—significantly limiting chemical weed control options available to agriculturalists. This limitation of control options is exacerbated by the recent lack of discovery of herbicides with new sites of action [ 105 ].
Herbicide resistance may result from several different physiological mechanisms. Such mechanisms have been classified into two main groups, target-site resistance (TSR) [ 4 , 106 ] and non-target-site resistance (NTSR) [ 4 , 107 ]. The first group encompasses changes that reduce binding affinity between a herbicide and its target [ 108 ]. These changes may provide resistance to multiple herbicides that have a common biochemical target [ 109 ] and can be effectively managed through mixture and/or rotation of herbicides targeting different sites of action [ 110 ]. The second group (NTSR), includes alterations in herbicide absorption, translocation, sequestration, and/or metabolism that may lead to unpredictable pleotropic cross-resistance profiles where structurally and functionally diverse herbicides are rendered ineffective by one or more genetic variant(s) [ 47 ]. This mechanism of resistance threatens not only the efficacy of existing herbicidal chemistries, but also ones yet to be discovered. While TSR is well understood because of the ease of identification and molecular characterization of target site variants, NTSR mechanisms are significantly more challenging to research because they are often polygenic, and the resistance causing element(s) are not well understood [ 111 ].
Improving the current understanding of metabolic NTSR mechanisms is not an easy task, since genes of diverse biochemical functions are involved, many of which exist as extensive gene families [ 109 , 112 ]. Expression changes of NTSR genes have been implicated in several resistance cases where the protein products of the genes are functionally equivalent across sensitive and resistant plants, but their relative abundance leads to resistance. Thus, regulatory elements of NTSR genes have been scrutinized to understand their role in NTSR mechanisms [ 113 ]. Similarly, epigenetic modifications have been hypothesized to play a role in NTSR, with much remaining to be explored [ 114 , 115 , 116 ]. Untargeted approaches such as genome-wide association, selective sweep scans, linkage mapping, RNA-sequencing, and metabolomic profiling have proven helpful to complement more specific biochemical- and chemo-characterization studies towards the elucidation of NTSR mechanisms as well as their regulation and evolution [ 47 , 117 , 118 , 119 , 120 , 121 , 122 , 123 , 124 ]. Even in cases where resistance has been attributed to TSR, genetic mapping approaches can detect other NTSR loci contributing to resistance (as shown by [ 123 ]) and provide further evidence for the role of TSR mutations across populations. Knowledge of the genetic basis of NTSR will aid the rational design of herbicides by screening new compounds for interaction with newly discovered NTSR proteins during early research phases and by identifying conserved chemical structures that interact with these proteins that should be avoided in small molecule design.
Genomic resources can also be used to predict the protein structure for novel herbicide target site and metabolism genes. This will allow for prediction of efficacy and selectivity for new candidate herbicides in silico to increase herbicide discovery throughput as well as aid in the design and development of next-generation technologies for sustainable weed management. Proteolysis targeting chimeras (PROTACs) have the potential to bind desired targets with great selectivity and degrade proteins by utilizing natural protein ubiquitination and degradation pathways within plants [ 125 ]. Spray-induced gene silencing in weeds using oligonucleotides has potential as a new, innovative, and sustainable method for weed management, but improved methods for design and delivery of oligonucleotides are needed to make this technique a viable management option [ 50 ]. Additionally, success in the field of pharmaceutical drug discovery in the development of molecules modulating protein–protein interactions offers another potential avenue towards the development of herbicides with novel targets [ 126 , 127 ]. High-quality reference genomes allow for the design of new weed management technologies like the ones listed here that are specific to—and effective across—weed species but have a null effect on non-target organisms.
Comparative genomics and genome biology
The genomes of weed species are as diverse as weed species themselves. Weeds are found across highly diverged plant families and often have no phylogenetically close model or crop species relatives for comparison. On all measurable metrics, weed genomes run the gamut. Some have smaller genomes like Cyperus spp. (~ 0.26 Gb) while others are larger, such as Avena fatua (~ 11.1 Gb) (Table 1 ). Some have high heterozygosity in terms of single-nucleotide polymorphisms, such as the Amaranthus spp., while others are primarily self-pollinated and quite homozygous, such as Poa annua [ 128 , 129 ]. Some are diploid such as Conyza canadensis and Echinochloa haploclada while others are polyploid such as C. sumetrensis , E. crus-galli , and E. colona [ 76 ]. The availability of genomic resources in these diverse, unexplored branches of the tree of life allows us to identify consistencies and anomalies in the field of genome biology.
The weed genomes published so far have focused mainly on weeds of agronomic crops, and studies have revolved around their ability to resist key herbicides. For example, genomic resources were vital in the elucidation of herbicide resistance cases involving target site gene copy number variants (CNVs). Gene CNVs of 5-enolpyruvylshikimate-3-phosphate synthase ( EPSPS ) have been found to confer resistance to the herbicide glyphosate in diverse weed species. To date, nine species have independently evolved EPSPS CNVs, and species achieve increased EPSPS copy number via different mechanisms [ 153 ]. For instance, the EPSPS CNV in Bassia scoparia is caused by tandem duplication, which is accredited to transposable element insertions flanking EPSPS and subsequent unequal crossing over events [ 154 , 155 ]. In Eleusine indica , a EPSPS CNV was caused by translocation of the EPSPS locus into the subtelomere followed by telomeric sequence exchange [ 156 ]. One of the most fascinating genome biology discoveries in weed science has been that of extra-chromosomal circular DNAs (eccDNAs) that harbor the EPSPS gene in the weed species Amaranthus palmeri [ 157 , 158 ]. In this case, the eccDNAs autonomously replicate separately from the nuclear genome and do not reintegrate into chromosomes, which has implications for inheritance, fitness, and genome structure [ 159 ]. These discoveries would not have been possible without reference assemblies of weed genomes, next-generation sequencing, and collaboration with experts in plant genomics and bioinformatics.
Another question that is often explored with weedy genomes is the nature and composition of gene families that are associated with NTSR. Gene families under consideration often include cytochrome P450s (CYPs), glutathione- S -transferases (GSTs), ABC transporters, etc. Some questions commonly considered with new weed genomes include how many genes are in each of these gene families, where are they located, and which weed accessions and species have an over-abundance of them that might explain their ability to evolve resistance so rapidly [ 76 , 146 , 160 , 161 ]? Weed genome resources are necessary to answer questions about gene family expansion or contraction during the evolution of weediness, including the role of polyploidy in NTSR gene family expansion as explored by [ 162 ].
Translational research and communication with weed management stakeholders
Whereas genomics of model plants is typically aimed at addressing fundamental questions in plant biology, and genomics of crop species has the obvious goal of crop improvement, goals of genomics of weedy plants also include the development of more effective and sustainable strategies for their management. Weed genomic resources assist with these objectives by providing novel molecular ecological and evolutionary insights from the context of intensive anthropogenic management (which is lacking in model plants), and offer knowledge and resources for trait discovery for crop improvement, especially given that many wild crop relatives are also important agronomic weeds (e.g., [ 163 ]). For instance, crop-wild relatives are valuable for improving crop breeding for marginal environments [ 164 ]. Thus, weed genomics presents unique opportunities and challenges relative to plant genomics more broadly. It should also be noted that although weed science at its core is an applied discipline, it draws broadly from many scientific disciplines such as, plant physiology, chemistry, ecology, and evolutionary biology, to name a few. The successful integration of weed-management strategies, therefore, requires extensive collaboration among individuals collectively possessing the necessary expertise [ 165 ].
With the growing complexity of herbicide resistance management, practitioners are beginning to recognize the importance of understanding resistance mechanisms to inform appropriate management tactics [ 14 ]. Although weed science practitioners do not need to understand the technical details of weed genomics, their appreciation of the power of weed genomics—together with their unique insights from field observations—will yield novel opportunities for applications of weed genomics to weed management. In particular, combining field management history with information on weed resistance mechanisms is expected to provide novel insights into evolutionary trajectories (e.g. [ 6 , 166 ]), which can be utilized for disrupting evolutionary adaptation. It can be difficult to obtain field history information from practitioners, but developing an understanding among them of the importance of such information can be invaluable.
Development of weed genomics resources by the IWGC
Weed genomics is a fast-growing field of research with many recent breakthroughs and many unexplored areas of study. The International Weed Genomics Consortium (IWGC) started in 2021 to address the roadblocks listed above and to promote the study of weedy plants. The IWGC is an open collaboration among academic, government, and industry researchers focused on producing genomic tools for weedy species from around the world. Through this collaboration, our initial aim is to provide chromosome-level reference genome assemblies for at least 50 important weedy species from across the globe that are chosen based on member input, economic impact, and global prevalence (Fig. 1 ). Each genome will include annotation of gene models and repetitive elements and will be freely available through public databases with no intellectual property restrictions. Additionally, future funding of the IWGC will focus on improving gene annotations and supplementing these reference genomes with tools that increase their utility.
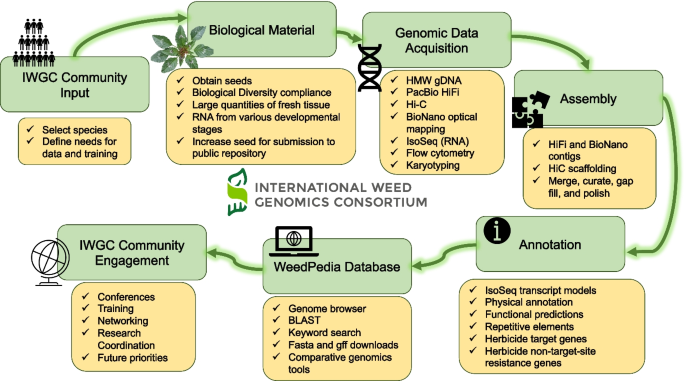
The International Weed Genomics Consortium (IWGC) collected input from the weed genomics community to develop plans for weed genome sequencing, annotation, user-friendly genome analysis tools, and community engagement
Reference genomes and data analysis tools
The first objective of the IWGC is to provide high-quality genomic resources for agriculturally important weeds. The IWGC therefore created two main resources for information about, access to, or analysis of weed genomic data (Fig. 1 ). The IWGC website (available at [ 167 ]) communicates the status and results of genome sequencing projects, information on training and funding opportunities, upcoming events, and news in weed genomics. It also contains details of all sequenced species including genome size, ploidy, chromosome number, herbicide resistance status, and reference genome assembly statistics. The IWGC either compiles existing data on genome size, ploidy, and chromosome number, or obtains the data using flow cytometry and cytogenetics (Fig. 1 ; Additional File 2 : Fig S1-S4). Through this website, users can request an account to access our second main resource, an online genome database called WeedPedia (accessible at [ 168 ]), with an account that is created within 3–5 working days of an account request submission. WeedPedia hosts IWGC-generated and other relevant publicly accessible genomic data as well as a suite of bioinformatic tools. Unlike what is available for other fields, weed science did not have a centralized hub for genomics information, data, and analysis prior to the IWGC. Our intention in creating WeedPedia is to encourage collaboration and equity of access to information across the research community. Importantly, all genome assemblies and annotations from the IWGC (Table 1 ), along with the raw data used to produce them, will be made available through NCBI GenBank. Upon completion of a 1-year sponsoring member data confidentiality period for each species (dates listed in Table 1 ), scientific teams within the IWGC produce the first genome-wide investigation to submit for publication including whole genome level analyses on genes, gene families, and repetitive sequences as well as comparative analysis with other species. Genome assemblies and data will be publicly available through NCBI as part of these initial publications for each species.
WeedPedia is a cloud-based omics database management platform built from the software “CropPedia” and licensed from KeyGene (Wageningen, The Netherlands). The interface allows users to access, visualize, and download genome assemblies along with structural and functional annotation. The platform includes a genome browser, comparative map viewer, pangenome tools, RNA-sequencing data visualization tools, genetic mapping and marker analysis tools, and alignment capabilities that allow searches by keyword or sequence. Additionally, genes encoding known target sites of herbicides have been specially annotated, allowing users to quickly identify and compare these genes of interest. The platform is flexible, making it compatible with future integration of other data types such as epigenetic or proteomic information. As an online platform with a graphical user interface, WeedPedia provides user-friendly, intuitive tools that encourage users to integrate genomics into their research while also allowing more advanced users to download genomic data to be used in custom analysis pipelines. We aspire for WeedPedia to mimic the success of other public genomic databases such as NCBI, CoGe, Phytozome, InsectBase, and Mycocosm to name a few. WeedPedia currently hosts reference genomes for 40 species (some of which are currently in their 1-year confidentiality period) with additional genomes in the pipeline to reach a currently planned total of 55 species (Table 1 ). These genomes include both de novo reference genomes generated or in progress by the IWGC (31 species; Table 1 ), and publicly available genome assemblies of 24 weedy or related species that were generated by independent research groups (Table 2 ). As of May 2024, WeedPedia has over 370 registered users from more than 27 countries spread across 6 continents.
The IWGC reference genomes are generated in partnership with the Corteva Agriscience Genome Center of Excellence (Johnston, Iowa) using a combination of single-molecule long-read sequencing, optical genome maps, and chromosome conformation mapping. This strategy has already yielded highly contiguous, phased, chromosome-level assemblies for 26 weed species, with additional assemblies currently in progress (Table 1 ). The IWGC assemblies have been completed as single or haplotype-resolved double-haplotype pseudomolecules in inbreeding and outbreeding species, respectively, with multiple genomes being near gapless. For example, the de novo assemblies of the allohexaploids Conyza sumatrensis and Chenopodium album have all chromosomes captured in single scaffolds and most chromosomes being gapless from telomere to telomere. Complementary full-length isoform (IsoSeq) sequencing of RNA collected from diverse tissue types and developmental stages assists in the development of gene models during annotation.
As with accessibility of data, a core objective of the IWGC is to facilitate open access to sequenced germplasm when possible for featured species. Historically, the weed science community has rarely shared or adopted standard germplasm (e.g., specific weed accessions). The IWGC has selected a specific accession of each species for reference genome assembly (typically susceptible to herbicides). In collaboration with a parallel effort by the Herbicide Resistant Plants committee of the Weed Science Society of America, seeds of the sequenced weed accessions will be deposited in the United States Department of Agriculture Germplasm Resources Information Network [ 186 ] for broad access by the scientific community and their accession numbers will be listed on the IWGC website. In some cases, it is not possible to generate enough seed to deposit into a public repository (e.g., plants that typically reproduce vegetatively, that are self-incompatible, or that produce very few seeds from a single individual). In these cases, the location of collection for sequenced accessions will at least inform the community where the sequenced individual came from and where they may expect to collect individuals with similar genotypes. The IWGC ensures that sequenced accessions are collected and documented to comply with the Nagoya Protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their utilization under the Convention on Biological Diversity and related Access and Benefit Sharing Legislation [ 187 ]. As additional accessions of weed species are sequenced (e.g., pangenomes are obtained), the IWGC will facilitate germplasm sharing protocols to support collaboration. Further, to simplify the investigation of herbicide resistance, the IWGC will link WeedPedia with the International Herbicide-Resistant Weed Database [ 104 ], an already widely known and utilized database for weed scientists.
Training and collaboration in weed genomics
Beyond producing genomic tools and resources, a priority of the IWGC is to enable the utilization of these resources across a wide range of stakeholders. A holistic approach to training is required for weed science generally [ 188 ], and we would argue even more so for weed genomics. To accomplish our training goals, the IWGC is developing and delivering programs aimed at the full range of IWGC stakeholders and covering a breadth of relevant topics. We have taken care to ensure our approaches are diverse as to provide training to researchers with all levels of existing experience and differing reasons for engaging with these tools. Throughout, the focus is on ensuring that our training and outreach result in impacts that benefit a wide range of stakeholders.
Although recently developed tools are incredibly enabling and have great potential to replace antiquated methodology [ 189 ] and to solve pressing weed science problems [ 14 ], specialized computational skills are required to fully explore and unlock meaning from these highly complex datasets. Collaboration with, or training of, computational biologists equipped with these skills and resources developed by the IWGC will enable weed scientists to expand research programs and better understand the genetic underpinnings of weed evolution and herbicide resistance. To fill existing skill gaps, the IWGC is developing summer bootcamps and online modules directed specifically at weed scientists that will provide training on computational skills (Fig. 1 ). Because successful utilization of the IWGC resources requires more than general computational skills, we have created three targeted workshops that teach practical skills related to genomics databases, molecular biology, and population genomics (available at [ 190 ]). The IWGC has also hosted two official conference meetings, one in September of 2021 and one in January of 2023, with more conferences planned. These conferences have included invited speakers to present successful implementations of weed genomics, educational workshops to build computational skills, and networking opportunities for research to connect and collaborate.
Engagement opportunities during undergraduate degrees have been shown to improve academic outcomes [ 191 , 192 ]. As one activity to help achieve this goal, the IWGC has sponsored opportunities for US undergraduates to undertake a 10-week research experience, which includes an introduction to bioinformatics, a plant genomics research project that results in a presentation, and access to career building opportunities in diverse workplace environments. To increase equitable access to conferences and professional communities, we supported early career researchers to attend the first two IWGC conferences in the USA as well as workshops and bootcamps in Europe, South America, and Australia. These hybrid or in-person travel grants are intentionally designed to remove barriers and increase participation of individuals from backgrounds and experiences currently underrepresented within weed/plant science or genomics [ 193 ]. Recipients of these travel awards gave presentations and gained the measurable benefits that come from either virtual or in-person participation in conferences [ 194 ]. Moving forward, weed scientists must amass skills associated with genomic analyses and collaborate with other area experts to fully leverage resources developed by the IWGC.
The tools generated through the IWGC will enable many new research projects with diverse objectives like those listed above. In summary, contiguous genome assemblies and complete annotation information will allow weed scientists to join plant breeders in the use of genetic mapping for many traits including stress tolerance, plant architecture, and herbicide resistance (especially important for cases of NTSR). These assemblies will also allow for investigations of population structure, gene flow, and responses to evolutionary mechanisms like genetic bottlenecking and artificial selection. Understanding gene sequences across diverse weed species will be vital in modeling new herbicide target site proteins and designing novel effective herbicides with minimal off-target effects. The IWGC website will improve accessibility to weed genomics data by providing a single hub for reference genomes as well as phenotypic and genotypic information for accessions shared with the IWGC. Deposition of sequenced germplasm into public repositories will ensure that researchers are able to access and utilize these accessions in their own research to make the field more standardized and equitable. WeedPedia allows users of all backgrounds to quickly access information of interest such as herbicide target site gene sequence or subcellular localization of protein products for different genes. Users can also utilize server-based tools such as BLAST and genome browsing similar to other public genomic databases. Finally, the IWGC is committed to training and connecting weed genomicists through hosting trainings, workshops, and conferences.
Conclusions
Weeds are unique and fascinating plants, having significant impacts on agriculture and ecosystems; and yet, aspects of their biology, ecology, and genetics remain poorly understood. Weeds represent a unique area within plant biology, given their repeated rapid adaptation to sudden and severe shifts in the selective landscape of anthropogenic management practices. The production of a public genomics database with reference genomes and annotations for over 50 weed species represents a substantial step forward towards research goals that improve our understanding of the biology and evolution of weeds. Future work is needed to improve annotations, particularly for complex gene families involved in herbicide detoxification, structural variants, and mobile genetic elements. As reference genome assemblies become available; standard, affordable methods for gathering genotype information will allow for the identification of genetic variants underlying traits of interest. Further, methods for functional validation and hypothesis testing are needed in weeds to validate the effect of genetic variants detected through such experiments, including systems for transformation, gene editing, and transient gene silencing and expression. Future research should focus on utilizing weed genomes to investigate questions about evolutionary biology, ecology, genetics of weedy traits, and weed population dynamics. The IWGC plans to continue the public–private partnership model to host the WeedPedia database over time, integrate new datasets such as genome resequencing and transcriptomes, conduct trainings, and serve as a research coordination network to ensure that advances in weed science from around the world are shared across the research community (Fig. 1 ). Bridging basic plant genomics with translational applications in weeds is needed to deliver on the potential of weed genomics to improve weed management and crop breeding.
Availability of data and materials
All genome assemblies and related sequencing data produced by the IWGC will be available through NCBI as part of publications reporting the first genome-wide analysis for each species.
Gianessi LP, Nathan PR. The value of herbicides in U.S. crop production. Weed Technol. 2007;21(2):559–66.
Article Google Scholar
Pimentel D, Lach L, Zuniga R, Morrison D. Environmental and economic costs of nonindigenous species in the United States. Bioscience. 2000;50(1):53–65.
Barrett SH. Crop mimicry in weeds. Econ Bot. 1983;37(3):255–82.
Powles SB, Yu Q. Evolution in action: plants resistant to herbicides. Annu Rev Plant Biol. 2010;61:317–47.
Article CAS PubMed Google Scholar
Thurber CS, Reagon M, Gross BL, Olsen KM, Jia Y, Caicedo AL. Molecular evolution of shattering loci in U.S. weedy rice. Mol Ecol. 2010;19(16):3271–84.
Article PubMed PubMed Central Google Scholar
Comont D, Lowe C, Hull R, Crook L, Hicks HL, Onkokesung N, et al. Evolution of generalist resistance to herbicide mixtures reveals a trade-off in resistance management. Nat Commun. 2020;11(1):3086.
Article CAS PubMed PubMed Central Google Scholar
Ashworth MB, Walsh MJ, Flower KC, Vila-Aiub MM, Powles SB. Directional selection for flowering time leads to adaptive evolution in Raphanus raphanistrum (wild radish). Evol Appl. 2016;9(4):619–29.
Chan EK, Rowe HC, Kliebenstein DJ. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics. 2010;185(3):991–1007.
Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94.
Harkess A, Zhou J, Xu C, Bowers JE, Van der Hulst R, Ayyampalayam S, et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun. 2017;8(1):1279.
Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L, et al. The gene Sr33 , an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science. 2013;341(6147):786–8.
Ågren J, Oakley CG, McKay JK, Lovell JT, Schemske DW. Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2013;110(52):21077–82.
Article PubMed Central Google Scholar
Schartl M, Walter RB, Shen Y, Garcia T, Catchen J, Amores A, et al. The genome of the platyfish, Xiphophorus maculatus , provides insights into evolutionary adaptation and several complex traits. Nat Genet. 2013;45(5):567–72.
Ravet K, Patterson EL, Krähmer H, Hamouzová K, Fan L, Jasieniuk M, et al. The power and potential of genomics in weed biology and management. Pest Manag Sci. 2018;74(10):2216–25.
Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou S, Liu J, et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science. 2021;373(6555):655–62.
Liao W-W, Asri M, Ebler J, Doerr D, Haukness M, Hickey G, et al. A draft human pangenome reference. Nature. 2023;617(7960):312–24.
Huang Y, Wu D, Huang Z, Li X, Merotto A, Bai L, et al. Weed genomics: yielding insights into the genetics of weedy traits for crop improvement. aBIOTECH. 2023;4:20–30.
Chen K, Yang H, Wu D, Peng Y, Lian L, Bai L, et al. Weed biology and management in the multi-omics era: progress and perspectives. Plant Commun. 2024;5(4):100816.
De Wet JMJ, Harlan JR. Weeds and domesticates: evolution in the man-made habitat. Econ Bot. 1975;29(2):99–108.
Mahaut L, Cheptou PO, Fried G, Munoz F, Storkey J, Vasseur F, et al. Weeds: against the rules? Trends Plant Sci. 2020;25(11):1107–16.
Neve P, Vila-Aiub M, Roux F. Evolutionary-thinking in agricultural weed management. New Phytol. 2009;184(4):783–93.
Article PubMed Google Scholar
Sharma G, Barney JN, Westwood JH, Haak DC. Into the weeds: new insights in plant stress. Trends Plant Sci. 2021;26(10):1050–60.
Vigueira CC, Olsen KM, Caicedo AL. The red queen in the corn: agricultural weeds as models of rapid adaptive evolution. Heredity (Edinb). 2013;110(4):303–11.
Donohue K, Dorn L, Griffith C, Kim E, Aguilera A, Polisetty CR, et al. Niche construction through germination cueing: life-history responses to timing of germination in Arabidopsis thaliana . Evolution. 2005;59(4):771–85.
PubMed Google Scholar
Exposito-Alonso M. Seasonal timing adaptation across the geographic range of Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2020;117(18):9665–7.
Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. A map of local adaptation in Arabidopsis thaliana . Science. 2011;334(6052):86–9.
Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334(6052):83–6.
Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature. 2000;408(6814):796–815.
Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, et al. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana . Cell. 2016;166(2):481–91.
Durvasula A, Fulgione A, Gutaker RM, Alacakaptan SI, Flood PJ, Neto C, et al. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2017;114(20):5213–8.
Frachon L, Mayjonade B, Bartoli C, Hautekèete N-C, Roux F. Adaptation to plant communities across the genome of Arabidopsis thaliana . Mol Biol Evol. 2019;36(7):1442–56.
Fulgione A, Koornneef M, Roux F, Hermisson J, Hancock AM. Madeiran Arabidopsis thaliana reveals ancient long-range colonization and clarifies demography in Eurasia. Mol Biol Evol. 2018;35(3):564–74.
Fulgione A, Neto C, Elfarargi AF, Tergemina E, Ansari S, Göktay M, et al. Parallel reduction in flowering time from de novo mutations enable evolutionary rescue in colonizing lineages. Nat Commun. 2022;13(1):1461.
Kasulin L, Rowan BA, León RJC, Schuenemann VJ, Weigel D, Botto JF. A single haplotype hyposensitive to light and requiring strong vernalization dominates Arabidopsis thaliana populations in Patagonia. Argentina Mol Ecol. 2017;26(13):3389–404.
Picó FX, Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics. 2008;180(2):1009–21.
Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465(7298):627–31.
Flood PJ, Kruijer W, Schnabel SK, van der Schoor R, Jalink H, Snel JFH, et al. Phenomics for photosynthesis, growth and reflectance in Arabidopsis thaliana reveals circadian and long-term fluctuations in heritability. Plant Methods. 2016;12(1):14.
Marchadier E, Hanemian M, Tisné S, Bach L, Bazakos C, Gilbault E, et al. The complex genetic architecture of shoot growth natural variation in Arabidopsis thaliana . PLoS Genet. 2019;15(4):e1007954.
Tisné S, Serrand Y, Bach L, Gilbault E, Ben Ameur R, Balasse H, et al. Phenoscope: an automated large-scale phenotyping platform offering high spatial homogeneity. Plant J. 2013;74(3):534–44.
Tschiersch H, Junker A, Meyer RC, Altmann T. Establishment of integrated protocols for automated high throughput kinetic chlorophyll fluorescence analyses. Plant Methods. 2017;13:54.
Chen X, MacGregor DR, Stefanato FL, Zhang N, Barros-Galvão T, Penfield S. A VEL3 histone deacetylase complex establishes a maternal epigenetic state controlling progeny seed dormancy. Nat Commun. 2023;14(1):2220.
Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–101.
Davey JW, Blaxter ML. RADSeq: next-generation population genetics. Brief Funct Genomics. 2010;9(5–6):416–23.
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6(5):e19379.
MacGregor DR. What makes a weed a weed? How virus-mediated reverse genetics can help to explore the genetics of weediness. Outlooks Pest Manag. 2020;31(5):224–9.
Mellado-Sánchez M, McDiarmid F, Cardoso V, Kanyuka K, MacGregor DR. Virus-mediated transient expression techniques enable gene function studies in blackgrass. Plant Physiol. 2020;183(2):455–9.
Dimaano NG, Yamaguchi T, Fukunishi K, Tominaga T, Iwakami S. Functional characterization of Cytochrome P450 CYP81A subfamily to disclose the pattern of cross-resistance in Echinochloa phyllopogon . Plant Mol Biol. 2020;102(4–5):403–16.
de Figueiredo MRA, Küpper A, Malone JM, Petrovic T, de Figueiredo ABTB, Campagnola G, et al. An in-frame deletion mutation in the degron tail of auxin coreceptor IAA2 confers resistance to the herbicide 2,4-D in Sisymbrium orientale . Proc Natl Acad Sci U S A. 2022;119(9):e2105819119.
Patzoldt WL, Hager AG, McCormick JS, Tranel PJ. A codon deletion confers resistance to herbicides inhibiting protoporphyrinogen oxidase. Proc Natl Acad Sci U S A. 2006;103(33):12329–34.
Zabala-Pardo D, Gaines T, Lamego FP, Avila LA. RNAi as a tool for weed management: challenges and opportunities. Adv Weed Sci. 2022;40(spe1):e020220096.
Fattorini R, Glover BJ. Molecular mechanisms of pollination biology. Annu Rev Plant Biol. 2020;71:487–515.
Rollin O, Benelli G, Benvenuti S, Decourtye A, Wratten SD, Canale A, et al. Weed-insect pollinator networks as bio-indicators of ecological sustainability in agriculture. A review Agron Sustain Dev. 2016;36(1):8.
Irwin RE, Strauss SY. Flower color microevolution in wild radish: evolutionary response to pollinator-mediated selection. Am Nat. 2005;165(2):225–37.
Ma B, Wu J, Shi T-L, Yang Y-Y, Wang W-B, Zheng Y, et al. Lilac ( Syringa oblata ) genome provides insights into its evolution and molecular mechanism of petal color change. Commun Biol. 2022;5(1):686.
Xing A, Wang X, Nazir MF, Zhang X, Wang X, Yang R, et al. Transcriptomic and metabolomic profiling of flavonoid biosynthesis provides novel insights into petals coloration in Asian cotton ( Gossypium arboreum L.). BMC Plant Biol. 2022;22(1):416.
Zheng Y, Chen Y, Liu Z, Wu H, Jiao F, Xin H, et al. Important roles of key genes and transcription factors in flower color differences of Nicotiana alata . Genes (Basel). 2021;12(12):1976.
Krizek BA, Anderson JT. Control of flower size. J Exp Bot. 2013;64(6):1427–37.
Powell AE, Lenhard M. Control of organ size in plants. Curr Biol. 2012;22(9):R360–7.
Spencer V, Kim M. Re"CYC"ling molecular regulators in the evolution and development of flower symmetry. Semin Cell Dev Biol. 2018;79:16–26.
Amrad A, Moser M, Mandel T, de Vries M, Schuurink RC, Freitas L, et al. Gain and loss of floral scent production through changes in structural genes during pollinator-mediated speciation. Curr Biol. 2016;26(24):3303–12.
Delle-Vedove R, Schatz B, Dufay M. Understanding intraspecific variation of floral scent in light of evolutionary ecology. Ann Bot. 2017;120(1):1–20.
Pichersky E, Gershenzon J. The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol. 2002;5(3):237–43.
Ballerini ES, Kramer EM, Hodges SA. Comparative transcriptomics of early petal development across four diverse species of Aquilegia reveal few genes consistently associated with nectar spur development. BMC Genom. 2019;20(1):668.
Corbet SA, Willmer PG, Beament JWL, Unwin DM, Prys-Jones OE. Post-secretory determinants of sugar concentration in nectar. Plant Cell Environ. 1979;2(4):293–308.
Galliot C, Hoballah ME, Kuhlemeier C, Stuurman J. Genetics of flower size and nectar volume in Petunia pollination syndromes. Planta. 2006;225(1):203–12.
Vila-Aiub MM, Neve P, Powles SB. Fitness costs associated with evolved herbicide resistance alleles in plants. New Phytol. 2009;184(4):751–67.
Baucom RS. Evolutionary and ecological insights from herbicide-resistant weeds: what have we learned about plant adaptation, and what is left to uncover? New Phytol. 2019;223(1):68–82.
Bajwa AA, Latif S, Borger C, Iqbal N, Asaduzzaman M, Wu H, et al. The remarkable journey of a weed: biology and management of annual ryegrass ( Lolium rigidum ) in conservation cropping systems of Australia. Plants (Basel). 2021;10(8):1505.
Bitarafan Z, Andreasen C. Fecundity allocation in some european weed species competing with crops. Agronomy. 2022;12(5):1196.
Costea M, Weaver SE, Tardif FJ. The biology of Canadian weeds. 130. Amaranthus retroflexus L., A. powellii , A. powellii S. Watson, and A. hybridus L. Can J Plant Sci. 2004;84(2):631–68.
Dixon A, Comont D, Slavov GT, Neve P. Population genomics of selectively neutral genetic structure and herbicide resistance in UK populations of Alopecurus myosuroides . Pest Manag Sci. 2021;77(3):1520–9.
Kersten S, Chang J, Huber CD, Voichek Y, Lanz C, Hagmaier T, et al. Standing genetic variation fuels rapid evolution of herbicide resistance in blackgrass. Proc Natl Acad Sci U S A. 2023;120(16):e2206808120.
Qiu J, Zhou Y, Mao L, Ye C, Wang W, Zhang J, et al. Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat Commun. 2017;8(1):15323.
Kreiner JM, Caballero A, Wright SI, Stinchcombe JR. Selective ancestral sorting and de novo evolution in the agricultural invasion of Amaranthus tuberculatus . Evolution. 2022;76(1):70–85.
Kreiner JM, Latorre SM, Burbano HA, Stinchcombe JR, Otto SP, Weigel D, et al. Rapid weed adaptation and range expansion in response to agriculture over the past two centuries. Science. 2022;378(6624):1079–85.
Wu D, Shen E, Jiang B, Feng Y, Tang W, Lao S, et al. Genomic insights into the evolution of Echinochloa species as weed and orphan crop. Nat Commun. 2022;13(1):689.
Yeaman S, Hodgins KA, Lotterhos KE, Suren H, Nadeau S, Degner JC, et al. Convergent local adaptation to climate in distantly related conifers. Science. 2016;353(6306):1431–3.
Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 2013;45(8):891–8.
Sackton TB, Grayson P, Cloutier A, Hu Z, Liu JS, Wheeler NE, et al. Convergent regulatory evolution and loss of flight in paleognathous birds. Science. 2019;364(6435):74–8.
Ye CY, Fan L. Orphan crops and their wild relatives in the genomic era. Mol Plant. 2021;14(1):27–39.
Clements DR, Jones VL. Ten ways that weed evolution defies human management efforts amidst a changing climate. Agronomy. 2021;11(2):284.
Article CAS Google Scholar
Weinig C. Rapid evolutionary responses to selection in heterogeneous environments among agricultural and nonagricultural weeds. Int J Plant Sci. 2005;166(4):641–7.
Cousens RD, Fournier-Level A. Herbicide resistance costs: what are we actually measuring and why? Pest Manag Sci. 2018;74(7):1539–46.
Lasky JR, Josephs EB, Morris GP. Genotype–environment associations to reveal the molecular basis of environmental adaptation. Plant Cell. 2023;35(1):125–38.
Lotterhos KE. The effect of neutral recombination variation on genome scans for selection. G3-Genes Genom Genet. 2019;9(6):1851–67.
Lovell JT, MacQueen AH, Mamidi S, Bonnette J, Jenkins J, Napier JD, et al. Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature. 2021;590(7846):438–44.
Todesco M, Owens GL, Bercovich N, Légaré J-S, Soudi S, Burge DO, et al. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature. 2020;584(7822):602–7.
Revolinski SR, Maughan PJ, Coleman CE, Burke IC. Preadapted to adapt: Underpinnings of adaptive plasticity revealed by the downy brome genome. Commun Biol. 2023;6(1):326.
Kuester A, Conner JK, Culley T, Baucom RS. How weeds emerge: a taxonomic and trait-based examination using United States data. New Phytol. 2014;202(3):1055–68.
Arnaud JF, Fénart S, Cordellier M, Cuguen J. Populations of weedy crop-wild hybrid beets show contrasting variation in mating system and population genetic structure. Evol Appl. 2010;3(3):305–18.
Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci U S A. 2000;97(13):7043–50.
Nakabayashi K, Leubner-Metzger G. Seed dormancy and weed emergence: from simulating environmental change to understanding trait plasticity, adaptive evolution, and population fitness. J Exp Bot. 2021;72(12):4181–5.
Busi R, Yu Q, Barrett-Lennard R, Powles S. Long distance pollen-mediated flow of herbicide resistance genes in Lolium rigidum . Theor Appl Genet. 2008;117(8):1281–90.
Délye C, Clément JAJ, Pernin F, Chauvel B, Le Corre V. High gene flow promotes the genetic homogeneity of arable weed populations at the landscape level. Basic Appl Ecol. 2010;11(6):504–12.
Roumet M, Noilhan C, Latreille M, David J, Muller MH. How to escape from crop-to-weed gene flow: phenological variation and isolation-by-time within weedy sunflower populations. New Phytol. 2013;197(2):642–54.
Moghadam SH, Alebrahim MT, Mohebodini M, MacGregor DR. Genetic variation of Amaranthus retroflexus L. and Chenopodium album L. (Amaranthaceae) suggests multiple independent introductions into Iran. Front Plant Sci. 2023;13:1024555.
Muller M-H, Latreille M, Tollon C. The origin and evolution of a recent agricultural weed: population genetic diversity of weedy populations of sunflower ( Helianthus annuus L.) in Spain and France. Evol Appl. 2011;4(3):499–514.
Wesse C, Welk E, Hurka H, Neuffer B. Geographical pattern of genetic diversity in Capsella bursa-pastoris (Brassicaceae) -A global perspective. Ecol Evol. 2021;11(1):199–213.
Fraimout A, Debat V, Fellous S, Hufbauer RA, Foucaud J, Pudlo P, et al. Deciphering the routes of invasion of Drosophila suzukii by means of ABC random forest. Mol Biol Evol. 2017;34(4):980–96.
CAS PubMed PubMed Central Google Scholar
Battlay P, Wilson J, Bieker VC, Lee C, Prapas D, Petersen B, et al. Large haploblocks underlie rapid adaptation in the invasive weed Ambrosia artemisiifolia . Nat Commun. 2023;14(1):1717.
van Boheemen LA, Hodgins KA. Rapid repeatable phenotypic and genomic adaptation following multiple introductions. Mol Ecol. 2020;29(21):4102–17.
Putra A, Hodgins K, Fournier-Level A. Assessing the invasive potential of different source populations of ragweed ( Ambrosia artemisiifolia L.) through genomically-informed species distribution modelling. Authorea. 2023;17(1):e13632.
Google Scholar
Bourguet D, Delmotte F, Franck P, Guillemaud T, Reboud X, Vacher C, et al. Heterogeneity of selection and the evolution of resistance. Trends Ecol Evol. 2013;28(2):110–8.
The International Herbicide-Resistant Weed Database. www.weedscience.org . Accessed 20 June 2023.
Powles S. Herbicide discovery through innovation and diversity. Adv Weed Sci. 2022;40(spe1):e020220074.
Murphy BP, Tranel PJ. Target-site mutations conferring herbicide resistance. Plants (Basel). 2019;8(10):382.
Gaines TA, Duke SO, Morran S, Rigon CAG, Tranel PJ, Küpper A, et al. Mechanisms of evolved herbicide resistance. J Biol Chem. 2020;295(30):10307–30.
Lonhienne T, Cheng Y, Garcia MD, Hu SH, Low YS, Schenk G, et al. Structural basis of resistance to herbicides that target acetohydroxyacid synthase. Nat Commun. 2022;13(1):3368.
Comont D, MacGregor DR, Crook L, Hull R, Nguyen L, Freckleton RP, et al. Dissecting weed adaptation: fitness and trait correlations in herbicide-resistant Alopecurus myosuroides . Pest Manag Sci. 2022;78(7):3039–50.
Neve P. Simulation modelling to understand the evolution and management of glyphosate resistance in weeds. Pest Manag Sci. 2008;64(4):392–401.
Torra J, Alcántara-de la Cruz R. Molecular mechanisms of herbicide resistance in weeds. Genes (Basel). 2022;13(11):2025.
Délye C, Gardin JAC, Boucansaud K, Chauvel B, Petit C. Non-target-site-based resistance should be the centre of attention for herbicide resistance research: Alopecurus myosuroides as an illustration. Weed Res. 2011;51(5):433–7.
Chandra S, Leon RG. Genome-wide evolutionary analysis of putative non-specific herbicide resistance genes and compilation of core promoters between monocots and dicots. Genes (Basel). 2022;13(7):1171.
Margaritopoulou T, Tani E, Chachalis D, Travlos I. Involvement of epigenetic mechanisms in herbicide resistance: the case of Conyza canadensis . Agriculture. 2018;8(1):17.
Pan L, Guo Q, Wang J, Shi L, Yang X, Zhou Y, et al. CYP81A68 confers metabolic resistance to ALS and ACCase-inhibiting herbicides and its epigenetic regulation in Echinochloa crus-galli . J Hazard Mater. 2022;428:128225.
Sen MK, Hamouzová K, Košnarová P, Roy A, Soukup J. Herbicide resistance in grass weeds: Epigenetic regulation matters too. Front Plant Sci. 2022;13:1040958.
Han H, Yu Q, Beffa R, González S, Maiwald F, Wang J, et al. Cytochrome P450 CYP81A10v7 in Lolium rigidum confers metabolic resistance to herbicides across at least five modes of action. Plant J. 2021;105(1):79–92.
Kubis GC, Marques RZ, Kitamura RS, Barroso AA, Juneau P, Gomes MP. Antioxidant enzyme and Cytochrome P450 activities are involved in horseweed ( Conyza sumatrensis ) resistance to glyphosate. Stress. 2023;3(1):47–57.
Qiao Y, Zhang N, Liu J, Yang H. Interpretation of ametryn biodegradation in rice based on joint analyses of transcriptome, metabolome and chemo-characterization. J Hazard Mater. 2023;445:130526.
Rouse CE, Roma-Burgos N, Barbosa Martins BA. Physiological assessment of non–target site restistance in multiple-resistant junglerice ( Echinochloa colona ). Weed Sci. 2019;67(6):622–32.
Abou-Khater L, Maalouf F, Jighly A, Alsamman AM, Rubiales D, Rispail N, et al. Genomic regions associated with herbicide tolerance in a worldwide faba bean ( Vicia faba L.) collection. Sci Rep. 2022;12(1):158.
Gupta S, Harkess A, Soble A, Van Etten M, Leebens-Mack J, Baucom RS. Interchromosomal linkage disequilibrium and linked fitness cost loci associated with selection for herbicide resistance. New Phytol. 2023;238(3):1263–77.
Kreiner JM, Tranel PJ, Weigel D, Stinchcombe JR, Wright SI. The genetic architecture and population genomic signatures of glyphosate resistance in Amaranthus tuberculatus . Mol Ecol. 2021;30(21):5373–89.
Parcharidou E, Dücker R, Zöllner P, Ries S, Orru R, Beffa R. Recombinant glutathione transferases from flufenacet-resistant black-grass ( Alopecurus myosuroides Huds.) form different flufenacet metabolites and differ in their interaction with pre- and post-emergence herbicides. Pest Manag Sci. 2023;79(9):3376–86.
Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21(3):181–200.
Acuner Ozbabacan SE, Engin HB, Gursoy A, Keskin O. Transient protein-protein interactions. Protein Eng Des Sel. 2011;24(9):635–48.
Lu H, Zhou Q, He J, Jiang Z, Peng C, Tong R, et al. Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials. Signal Transduct Target Ther. 2020;5(1):213.
Benson CW, Sheltra MR, Maughan PJ, Jellen EN, Robbins MD, Bushman BS, et al. Homoeologous evolution of the allotetraploid genome of Poa annua L. BMC Genom. 2023;24(1):350.
Robbins MD, Bushman BS, Huff DR, Benson CW, Warnke SE, Maughan CA, et al. Chromosome-scale genome assembly and annotation of allotetraploid annual bluegrass ( Poa annua L.). Genome Biol Evol. 2022;15(1):evac180.
Montgomery JS, Giacomini D, Waithaka B, Lanz C, Murphy BP, Campe R, et al. Draft genomes of Amaranthus tuberculatus , Amaranthus hybridus and Amaranthus palmeri . Genome Biol Evol. 2020;12(11):1988–93.
Jeschke MR, Tranel PJ, Rayburn AL. DNA content analysis of smooth pigweed ( Amaranthus hybridus ) and tall waterhemp ( A. tuberculatus ): implications for hybrid detection. Weed Sci. 2003;51(1):1–3.
Rayburn AL, McCloskey R, Tatum TC, Bollero GA, Jeschke MR, Tranel PJ. Genome size analysis of weedy Amaranthus species. Crop Sci. 2005;45(6):2557–62.
Laforest M, Martin SL, Bisaillon K, Soufiane B, Meloche S, Tardif FJ, et al. The ancestral karyotype of the Heliantheae Alliance, herbicide resistance, and human allergens: Insights from the genomes of common and giant ragweed. Plant Genome . 2024;e20442. https://doi.org/10.1002/tpg2.20442 .
Mulligan GA. Chromosome numbers of Canadian weeds. I Canad J Bot. 1957;35(5):779–89.
Meyer L, Causse R, Pernin F, Scalone R, Bailly G, Chauvel B, et al. New gSSR and EST-SSR markers reveal high genetic diversity in the invasive plant Ambrosia artemisiifolia L. and can be transferred to other invasive Ambrosia species. PLoS One. 2017;12(5):e0176197.
Pustahija F, Brown SC, Bogunić F, Bašić N, Muratović E, Ollier S, et al. Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa. Plant Soil. 2013;373(1):427–53.
Kubešová M, Moravcova L, Suda J, Jarošík V, Pyšek P. Naturalized plants have smaller genomes than their non-invading relatives: a flow cytometric analysis of the Czech alien flora. Preslia. 2010;82(1):81–96.
Thébaud C, Abbott RJ. Characterization of invasive Conyza species (Asteraceae) in Europe: quantitative trait and isozyme analysis. Am J Bot. 1995;82(3):360–8.
Garcia S, Hidalgo O, Jakovljević I, Siljak-Yakovlev S, Vigo J, Garnatje T, et al. New data on genome size in 128 Asteraceae species and subspecies, with first assessments for 40 genera, 3 tribes and 2 subfamilies. Plant Biosyst. 2013;147(4):1219–27.
Zhao X, Yi L, Ren Y, Li J, Ren W, Hou Z, et al. Chromosome-scale genome assembly of the yellow nutsedge ( Cyperus esculentus ). Genome Biol Evol. 2023;15(3):evad027.
Bennett MD, Leitch IJ, Hanson L. DNA amounts in two samples of angiosperm weeds. Ann Bot. 1998;82:121–34.
Schulz-Schaeffer J, Gerhardt S. Cytotaxonomic analysis of the Euphorbia spp. (leafy spurge) complex. II: Comparative study of the chromosome morphology. Biol Zentralbl. 1989;108(1):69–76.
Schaeffer JR, Gerhardt S. The impact of introgressive hybridization on the weediness of leafy spurge. Leafy Spurge Symposium. 1989;1989:97–105.
Bai C, Alverson WS, Follansbee A, Waller DM. New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann Bot. 2012;110(8):1623–9.
Aarestrup JR, Karam D, Fernandes GW. Chromosome number and cytogenetics of Euphorbia heterophylla L. Genet Mol Res. 2008;7(1):217–22.
Wang L, Sun X, Peng Y, Chen K, Wu S, Guo Y, et al. Genomic insights into the origin, adaptive evolution, and herbicide resistance of Leptochloa chinensis , a devastating tetraploid weedy grass in rice fields. Mol Plant. 2022;15(6):1045–58.
Paril J, Pandey G, Barnett EM, Rane RV, Court L, Walsh T, et al. Rounding up the annual ryegrass genome: high-quality reference genome of Lolium rigidum . Front Genet. 2022;13:1012694.
Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM. Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am J Bot. 2006;93(1):148–56.
Towers G, Mitchell J, Rodriguez E, Bennett F, Subba Rao P. Biology & chemistry of Parthenium hysterophorus L., a problem weed in India. Biol Rev. 1977;48:65–74.
CAS Google Scholar
Moghe GD, Hufnagel DE, Tang H, Xiao Y, Dworkin I, Town CD, et al. Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish ( Raphanus raphanistrum ) and three other Brassicaceae species. Plant Cell. 2014;26(5):1925–37.
Zhang X, Liu T, Wang J, Wang P, Qiu Y, Zhao W, et al. Pan-genome of Raphanus highlights genetic variation and introgression among domesticated, wild, and weedy radishes. Mol Plant. 2021;14(12):2032–55.
Chytrý M, Danihelka J, Kaplan Z, Wild J, Holubová D, Novotný P, et al. Pladias database of the Czech flora and vegetation. Preslia. 2021;93(1):1–87.
Patterson EL, Pettinga DJ, Ravet K, Neve P, Gaines TA. Glyphosate resistance and EPSPS gene duplication: Convergent evolution in multiple plant species. J Hered. 2018;109(2):117–25.
Jugulam M, Niehues K, Godar AS, Koo DH, Danilova T, Friebe B, et al. Tandem amplification of a chromosomal segment harboring 5-enolpyruvylshikimate-3-phosphate synthase locus confers glyphosate resistance in Kochia scoparia . Plant Physiol. 2014;166(3):1200–7.
Patterson EL, Saski CA, Sloan DB, Tranel PJ, Westra P, Gaines TA. The draft genome of Kochia scoparia and the mechanism of glyphosate resistance via transposon-mediated EPSPS tandem gene duplication. Genome Biol Evol. 2019;11(10):2927–40.
Zhang C, Johnson N, Hall N, Tian X, Yu Q, Patterson E. Subtelomeric 5-enolpyruvylshikimate-3-phosphate synthase ( EPSPS ) copy number variation confers glyphosate resistance in Eleusine indica . Nat Commun. 2023;14:4865.
Koo D-H, Molin WT, Saski CA, Jiang J, Putta K, Jugulam M, et al. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri . Proc Natl Acad Sci U S A. 2018;115(13):3332–7.
Molin WT, Yaguchi A, Blenner M, Saski CA. The eccDNA Replicon: A heritable, extranuclear vehicle that enables gene amplification and glyphosate resistance in Amaranthus palmeri . Plant Cell. 2020;32(7):2132–40.
Jugulam M. Can non-Mendelian inheritance of extrachromosomal circular DNA-mediated EPSPS gene amplification provide an opportunity to reverse resistance to glyphosate? Weed Res. 2021;61(2):100–5.
Kreiner JM, Giacomini DA, Bemm F, Waithaka B, Regalado J, Lanz C, et al. Multiple modes of convergent adaptation in the spread of glyphosate-resistant Amaranthus tuberculatus . Proc Natl Acad Sci U S A. 2019;116(42):21076–84.
Cai L, Comont D, MacGregor D, Lowe C, Beffa R, Neve P, et al. The blackgrass genome reveals patterns of non-parallel evolution of polygenic herbicide resistance. New Phytol. 2023;237(5):1891–907.
Chen K, Yang H, Peng Y, Liu D, Zhang J, Zhao Z, et al. Genomic analyses provide insights into the polyploidization-driven herbicide adaptation in Leptochloa weeds. Plant Biotechnol J. 2023;21(8):1642–58.
Ohadi S, Hodnett G, Rooney W, Bagavathiannan M. Gene flow and its consequences in Sorghum spp. Crit Rev Plant Sci. 2017;36(5–6):367–85.
Renzi JP, Coyne CJ, Berger J, von Wettberg E, Nelson M, Ureta S, et al. How could the use of crop wild relatives in breeding increase the adaptation of crops to marginal environments? Front Plant Sci. 2022;13:886162.
Ward SM, Cousens RD, Bagavathiannan MV, Barney JN, Beckie HJ, Busi R, et al. Agricultural weed research: a critique and two proposals. Weed Sci. 2014;62(4):672–8.
Evans JA, Tranel PJ, Hager AG, Schutte B, Wu C, Chatham LA, et al. Managing the evolution of herbicide resistance. Pest Manag Sci. 2016;72(1):74–80.
International Weed Genomics Consortium Website. https://www.weedgenomics.org . Accessed 20 June 2023.
WeedPedia Database. https://weedpedia.weedgenomics.org/ . Accessed 20 June 2023.
Hall N, Chen J, Matzrafi M, Saski CA, Westra P, Gaines TA, et al. FHY3/FAR1 transposable elements generate adaptive genetic variation in the Bassia scoparia genome. bioRxiv . 2023; DOI: https://doi.org/10.1101/2023.05.26.542497 .
Jarvis DE, Sproul JS, Navarro-Domínguez B, Krak K, Jaggi K, Huang Y-F, et al. Chromosome-scale genome assembly of the hexaploid Taiwanese goosefoot “Djulis” ( Chenopodium formosanum ). Genome Biol Evol. 2022;14(8):evac120.
Ferreira LAI, de Oliveira RS, Jr., Constantin J, Brunharo C. Evolution of ACCase-inhibitor resistance in Chloris virgata is conferred by a Trp2027Cys mutation in the herbicide target site. Pest Manag Sci. 2023;79(12):5220–9.
Laforest M, Martin SL, Bisaillon K, Soufiane B, Meloche S, Page E. A chromosome-scale draft sequence of the Canada fleabane genome. Pest Manag Sci. 2020;76(6):2158–69.
Guo L, Qiu J, Ye C, Jin G, Mao L, Zhang H, et al. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat Commun. 2017;8(1):1031.
Sato MP, Iwakami S, Fukunishi K, Sugiura K, Yasuda K, Isobe S, et al. Telomere-to-telomere genome assembly of an allotetraploid pernicious weed, Echinochloa phyllopogon . DNA Res. 2023;30(5):dsad023.
Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza . Nat Genet. 2018;50(2):285–96.
Wu D, Xie L, Sun Y, Huang Y, Jia L, Dong C, et al. A syntelog-based pan-genome provides insights into rice domestication and de-domestication. Genome Biol. 2023;24(1):179.
Wang Z, Huang S, Yang Z, Lai J, Gao X, Shi J. A high-quality, phased genome assembly of broomcorn millet reveals the features of its subgenome evolution and 3D chromatin organization. Plant Commun. 2023;4(3):100557.
Mao Q, Huff DR. The evolutionary origin of Poa annua L. Crop Sci. 2012;52(4):1910–22.
Benson CW, Sheltra MR, Maughan JP, Jellen EN, Robbins MD, Bushman BS, et al. Homoeologous evolution of the allotetraploid genome of Poa annua L. Res Sq. 2023. https://doi.org/10.21203/rs.3.rs-2729084/v1 .
Brunharo C, Benson CW, Huff DR, Lasky JR. Chromosome-scale genome assembly of Poa trivialis and population genomics reveal widespread gene flow in a cool-season grass seed production system. Plant Direct. 2024;8(3):e575.
Mo C, Wu Z, Shang X, Shi P, Wei M, Wang H, et al. Chromosome-level and graphic genomes provide insights into metabolism of bioactive metabolites and cold-adaption of Pueraria lobata var. montana . DNA Research. 2022;29(5):dsac030.
Thielen PM, Pendleton AL, Player RA, Bowden KV, Lawton TJ, Wisecaver JH. Reference genome for the highly transformable Setaria viridis ME034V. G3 (Bethesda, Md). 2020;10(10):3467–78.
Yoshida S, Kim S, Wafula EK, Tanskanen J, Kim Y-M, Honaas L, et al. Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Curr Biol. 2019;29(18):3041–52.
Qiu S, Bradley JM, Zhang P, Chaudhuri R, Blaxter M, Butlin RK, et al. Genome-enabled discovery of candidate virulence loci in Striga hermonthica , a devastating parasite of African cereal crops. New Phytol. 2022;236(2):622–38.
Nunn A, Rodríguez-Arévalo I, Tandukar Z, Frels K, Contreras-Garrido A, Carbonell-Bejerano P, et al. Chromosome-level Thlaspi arvense genome provides new tools for translational research and for a newly domesticated cash cover crop of the cooler climates. Plant Biotechnol J. 2022;20(5):944–63.
USDA-ARS Germplasm Resources Information Network (GRIN). https://www.ars-grin.gov/ . Accessed 20 June 2023.
Buck M, Hamilton C. The Nagoya Protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their utilization to the convention on biological diversity. RECIEL. 2011;20(1):47–61.
Chauhan BS, Matloob A, Mahajan G, Aslam F, Florentine SK, Jha P. Emerging challenges and opportunities for education and research in weed science. Front Plant Sci. 2017;8:1537.
Shah S, Lonhienne T, Murray CE, Chen Y, Dougan KE, Low YS, et al. Genome-guided analysis of seven weed species reveals conserved sequence and structural features of key gene targets for herbicide development. Front Plant Sci. 2022;13:909073.
International Weed Genomics Consortium Training Resources. https://www.weedgenomics.org/training-resources/ . Accessed 20 June 2023.
Blackford S. Harnessing the power of communities: career networking strategies for bioscience PhD students and postdoctoral researchers. FEMS Microbiol Lett. 2018;365(8):fny033.
Pender M, Marcotte DE, Sto Domingo MR, Maton KI. The STEM pipeline: The role of summer research experience in minority students’ Ph.D. aspirations. Educ Policy Anal Arch. 2010;18(30):1–36.
PubMed PubMed Central Google Scholar
Burke A, Okrent A, Hale K. The state of U.S. science and engineering 2022. Foundation NS. https://ncses.nsf.gov/pubs/nsb20221 . 2022.
Wu J-Y, Liao C-H, Cheng T, Nian M-W. Using data analytics to investigate attendees’ behaviors and psychological states in a virtual academic conference. Educ Technol Soc. 2021;24(1):75–91.
Download references
Peer review information
Wenjing She was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
The International Weed Genomics Consortium is supported by BASF SE, Bayer AG, Syngenta Ltd, Corteva Agriscience, CropLife International (Global Herbicide Resistance Action Committee), the Foundation for Food and Agriculture Research (Award DSnew-0000000024), and two conference grants from USDA-NIFA (Award numbers 2021–67013-33570 and 2023-67013-38785).
Author information
Authors and affiliations.
Department of Agricultural Biology, Colorado State University, 1177 Campus Delivery, Fort Collins, CO, 80523, USA
Jacob Montgomery, Sarah Morran & Todd A. Gaines
Protecting Crops and the Environment, Rothamsted Research, Harpenden, Hertfordshire, UK
Dana R. MacGregor
Department of Crop, Soil, and Environmental Sciences, Auburn University, Auburn, AL, USA
J. Scott McElroy
Department of Plant and Environmental Sciences, University of Copenhagen, Taastrup, Denmark
Paul Neve & Célia Neto
IFEVA-Conicet-Department of Ecology, University of Buenos Aires, Buenos Aires, Argentina
Martin M. Vila-Aiub & Maria Victoria Sandoval
Department of Ecology, Faculty of Agronomy, University of Buenos Aires, Buenos Aires, Argentina
Analia I. Menéndez
Department of Botany, The University of British Columbia, Vancouver, BC, Canada
Julia M. Kreiner
Institute of Crop Sciences, Zhejiang University, Hangzhou, China
Longjiang Fan
Department of Biology, University of Massachusetts Amherst, Amherst, MA, USA
Ana L. Caicedo
Department of Plant and Wildlife Sciences, Brigham Young University, Provo, UT, USA
Peter J. Maughan
Bayer AG, Weed Control Research, Frankfurt, Germany
Bianca Assis Barbosa Martins, Jagoda Mika, Alberto Collavo & Bodo Peters
Department of Crop Sciences, Federal University of Rio Grande Do Sul, Porto Alegre, Rio Grande Do Sul, Brazil
Aldo Merotto Jr.
Department of Soil and Crop Sciences, Texas A&M University, College Station, TX, USA
Nithya K. Subramanian & Muthukumar V. Bagavathiannan
Department of Plant, Soil and Microbial Sciences, Michigan State University, East Lansing, MI, USA
Luan Cutti & Eric L. Patterson
Department of Agronomy, Kansas State University, Manhattan, KS, USA
Md. Mazharul Islam & Mithila Jugulam
Department of Plant Pathology, Kansas State University, Manhattan, KS, USA
Bikram S. Gill
Crop Protection Discovery and Development, Corteva Agriscience, Indianapolis, IN, USA
Robert Cicchillo, Roger Gast & Neeta Soni
Genome Center of Excellence, Corteva Agriscience, Johnston, IA, USA
Terry R. Wright, Gina Zastrow-Hayes, Gregory May, Kevin Fengler & Victor Llaca
School of Agriculture, Food and Wine, University of Adelaide, Glen Osmond, South Australia, Australia
Jenna M. Malone
Jealott’s Hill International Research Centre, Syngenta Ltd, Bracknell, Berkshire, UK
Deepmala Sehgal, Shiv Shankhar Kaundun & Richard P. Dale
Department of Plant and Soil Sciences, University of Pretoria, Pretoria, South Africa
Barend Juan Vorster
BASF SE, Ludwigshafen Am Rhein, Germany
Jens Lerchl
Department of Crop Sciences, University of Illinois, Urbana, IL, USA
Patrick J. Tranel
Senior Scientist Consultant, Herbicide Resistance Action Committee / CropLife International, Liederbach, Germany
Roland Beffa
School of BioSciences, University of Melbourne, Parkville, VIC, Australia
Alexandre Fournier-Level
You can also search for this author in PubMed Google Scholar
Contributions
JMo and TG conceived and outlined the article. TG, DM, EP, RB, JSM, PJT, MJ wrote grants to obtain funding. MMI, BSG, and MJ performed mitotic chromosome visualization. VL performed sequencing. VL and KF assembled the genomes. LC and ELP annotated the genomes. JMo, SM, DRM, JSM, PN, CN, MV, MVS, AIM, JMK, LF, ALC, PJM, BABM, JMi, AC, MVB, LC, AFL, and ELP wrote the first draft of the article. All authors edited the article and improved the final version.
Corresponding author
Correspondence to Todd A. Gaines .
Ethics declarations
Ethics approval and consent to participate.
Ethical approval is not applicable for this article.
Competing interests
Some authors work for commercial agricultural companies (BASF, Bayer, Corteva Agriscience, or Syngenta) that develop and sell weed control products.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
13059_2024_3274_moesm1_esm.docx.
Additional file 1. List of completed and in-progress genome assemblies of weed species pollinated by insects (Table S1).
13059_2024_3274_MOESM2_ESM.docx
Additional file 2. Methods and results for visualizing and counting the metaphase chromosomes of hexaploid Avena fatua (Fig S1); diploid Lolium rigidum (Fig S2); tetraploid Phalaris minor (Fig S3); and tetraploid Salsola tragus (Fig S4).
Additional file 3. Review history.
Rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Montgomery, J., Morran, S., MacGregor, D.R. et al. Current status of community resources and priorities for weed genomics research. Genome Biol 25 , 139 (2024). https://doi.org/10.1186/s13059-024-03274-y
Download citation
Received : 11 July 2023
Accepted : 13 May 2024
Published : 27 May 2024
DOI : https://doi.org/10.1186/s13059-024-03274-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Weed science
- Reference genomes
- Rapid adaptation
- Herbicide resistance
- Public resources
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Help | Advanced Search
Computer Science > Robotics
Title: robust perception and navigation of autonomous surface vehicles in challenging environments.
Abstract: Research on coastal regions traditionally involves methods like manual sampling, monitoring buoys, and remote sensing, but these methods face challenges in spatially and temporally diverse regions of interest. Autonomous surface vehicles (ASVs) with artificial intelligence (AI) are being explored, and recognized by the International Maritime Organization (IMO) as vital for future ecosystem understanding. However, there is not yet a mature technology for autonomous environmental monitoring due to typically complex coastal situations: (1) many static (e.g., buoy, dock) and dynamic (e.g., boats) obstacles not compliant with the rules of the road (COLREGs); (2) uncharted or uncertain information (e.g., non-updated nautical chart); and (3) high-cost ASVs not accessible to the community and citizen science while resulting in technology illiteracy. To address the above challenges, my research involves both system and algorithmic development: (1) a robotic boat system for stable and reliable in-water monitoring, (2) maritime perception to detect and track obstacles (such as buoys, and boats), and (3) navigational decision-making with multiple-obstacle avoidance and multi-objective optimization.
Submission history
Access paper:.
- Other Formats
References & Citations
- Google Scholar
- Semantic Scholar
BibTeX formatted citation
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .

Generative AI and the Future of Social Research: Opportunities, Challenges and Implications
This one-day workshop will explore the methodological implications of generative AI tools for social inquiry. It will take place in Edinburgh on 1 October 2024 and is free to attend.
Attendees will learn about the exciting opportunities and profound challenges that generative AI presents for social research in the digital age. The event is presented by NCRM and the British Sociological Association's Digital Sociology Study Group .
This workshop will bring together leading scholars and practitioners to discuss the challenges and opportunites presented by AI and chart a path forward for social research in an era of generative AI.
Through a series of panel discussions, the event will explore the methodological innovations, ethical challenges and theoretical frameworks necessary to navigate this new frontier of social inquiry.
The workshop will take place between 09:00 and 17:00 on Tuesday, 1 October 2024.
View the full programme
Apply to attend
If you would like to attend this event, please complete our online application form. The workshop is free of charge. However, places are limited.
Complete the application form
Please note, we are unfortunately unable to cover travel costs.
Application process
- Application process opens: Monday, 3 June 2024
- Closing date: Wednesday, 31 July 2024
- Outcome of application: week commencing Monday, 19 August 2024
- Registration deadline: Monday, 2 September 2024 (17:00 BST)
The workshop will take place at the Radisson Blu Hotel, 80 High Street, The Royal Mile, Edinburgh, EH1 1TH. The address is:
Radisson Blu Hotel, 80 High Street, The Royal Mile, Edinburgh, EH1 1TH
Find out more about the venue
More information about AI and social research
The rapid advancement of generative AI presents both exciting opportunities and profound challenges for social research in the digital age. This seminar aims to explore the methodological implications of generative AI tools for social inquiry.
Generative AI opens up novel avenues for research, enabling the creation and analysis of vast amounts of synthetic data, simulating and modelling complex social systems. Natural language processing techniques powered by generative AI provide us with powerful tools for the analysis of large volumes of data, facilitating the exploration of complex social phenomena and enabling researchers to identify trends and correlations not readily visible when using traditional methods.
However, these tools come with significant practical and ethical challenges. How can we ensure the reproducibility of data? How transparent are the algorithms used in AI-driven research, and how can we ensure researchers and participants understand the impact of these algorithms? How can we navigate the ethical complexities of working with AI-generated content? Given the well documented issues of bias in training datasets for large language models (LLMs) how can we ensure that we represent and include diverse populations rather than perpetuating structural inequalities?
Concerns around the usefulness of the work produced by LLMs continues to be a notable issue, with the idea of LLMs as "stochastic parrots" suggesting that while AI may produce plausible enough responses, these responses can be devoid of meaning.
In the realm of digital social research, generative AI poses fundamental questions about the nature of online content and its relationship to human agency. The rise of computational propaganda has already begun to test the assumed link between online content and human actors, but generative AI threatens to sever this connection entirely. As AI-generated content becomes increasingly pervasive, researchers must grapple with the epistemological implications of studying a digital landscape shaped by artificial intelligence.
How this workshop will address these issues
Combined, the issues outlined above pose concerns for what role AI can meaningfully play in sociological research. Join us as we confront the profound implications of generative AI for social research and work towards a future in which these powerful tools are harnessed for the advancement of knowledge and the betterment of society.
Key questions will include:
- How might AI affect research design, sampling, data collection and analysis?
- What ethical challenges emerge when using AI in social research?
- What role do human researchers play in overseeing AI-driven processes?
- How can we ensure that AI-driven research is accessible and inclusive to researchers with varying levels of technical expertise and resources?
- What are the implications of AI on power dynamics within social research, and how can we mitigate potential biases and avoid reinforcing existing inequalities?
- What impact might AI have on research practices, disciplinary norms and understandings of knowledge production?
- How can we foster interdisciplinary collaborations between social scientists, computer scientists, ethicists, and other stakeholders to address the challenges and enrich the development and application of AI in social research?
- How might our research practices be configured to consider how we meet the emerging research agendas of our students as they move into their academic and industrial careers? How do we best equip them for the research questions they will address in years to come?
Further information
If you have any questions, please contact Laura Marshall: [email protected]
Landslide prediction with severity analysis using efficient computer vision and soft computing algorithms
- Published: 30 May 2024
Cite this article

- Payal Varangaonkar 1 &
- S. V. Rode 2
Since the preceding decade, there has been a great deal of interest in forecasting landslides using remote-sensing images. Early detection of possible landslide zones will help to save lives and money. However, this approach presents several obstacles. Computer vision systems must be carefully built since normal image processing does not apply to images obtained by remote sensing (RS). This research proposes a novel landslide prediction method with a severity analysis model based on real-time hyperspectral RS images. The proposed model consists of phases of pre-processing, dynamic segmentation, hybrid feature extraction, landslide prediction, and landslide severity detection. The pre-processing step performs the geometric correction of input RS images to suppress the built-up regions, water, and vegetation using the Normal Difference Vegetation Index (NDVI). The pre-processing stage encompasses many steps, including atmospheric adjustments, geometric corrections, and the elimination of superfluous regions by denoising techniques such as 2D median filtering. Dynamic segmentation is employed to segment the pre-processed picture for Region of Interest (ROI) localization. The ROI image is utilized to extract manually designed features that accurately depict spatial and temporal variations within the input RS image. For each input RS image, the hybrid feature vector is normalized. We trained ANN and SVM to predict landslides. If the input image predicts a landslide, its severity is identified. For the performance analysis, we collected real-time RS images of the western region of India (Goa and Maharashtra). Simulation results show the efficiency of the proposed model.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
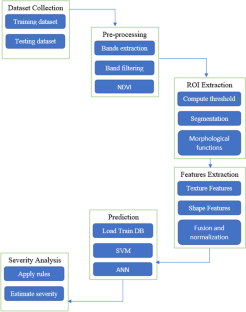
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Guzzetti F, Reichenbach P, Cardinali M, Galli M, Ardizzone F (2005) Probablistic landslide hazard assessment at the basin scale. Geomorphology 72:272–299. https://doi.org/10.1016/j.geomorph.2005.06.002
Article Google Scholar
Perkins S (2012) Death toll from landslides vastly underestimated. http://www.nature.com/ , http://www.emdat.be/database . Accessed 15 Jan 2022
Nguyen TL (2008) Landslide susceptibility mapping of the mountainous area in a Luoi District. Thua Thien Hue Province, Vietnam Vrije Universiteit Brusse
de Listo FLR, Carvalho Vieira B (2012) Mapping of risk and susceptibility of shallow-landslide in the city of São Paulo, Brazil. Geomorphology 169–170:30–44. https://doi.org/10.1016/j.geomorph.2012.01.010
Pradhan B (2010) Landslide susceptibility mapping of a catchment area using frequency ratio, fuzzy logic and multivariate logistic regression approaches. J Indian Soc Remote Sens 38(2):301–320. https://doi.org/10.1007/s12524-010-0020-z
Remondo J, Bonachea J, Cendrero A (2008) Quantitative landslide risk assessment and mapping on the basis of recent occurrences. Geomorphology 94(3–4):496–507. https://doi.org/10.1016/j.geomorph.2006.10.041
Mahajan HB, Uke N, Pise P et al (2022) Automatic robot Manoeuvres detection using computer vision and deep learning techniques: a perspective of internet of robotics things (IoRT). Multimed Tools Appl. https://doi.org/10.1007/s11042-022-14253-5
Aksoy B, Ercanoglu M (2012) Landslide identification and classification by object-based image analysis and fuzzy logic: An example from the Azdavay region (Kastamonu, Turkey). Computers, Geosciences 38(1):87–98. https://doi.org/10.1016/j.cageo.2011.05.010
Tien Bui D (2012) Modeling of rainfall-induced landslide hazard for the Hoa Binh, province of Vietnam. Norwegian University of Life Sciences. Ph.D Thesis
Pradhan B, Lee S, Buchroithner M (2010) A GIS-based back-propagation neural network model and its cross-application and validation for landslide susceptibility analyses. Comput Environ Urban Syst 34:216–235. https://doi.org/10.1016/j.compenvurbsys.2009.12.004
Arabameri A, Saha S, Roy J, Chen W, Blaschke T, Tien Bui D (2020) Landslide susceptibility evaluation and management using different machine learning methods in the Gallicash River Watershed, Iran. Remote Sens 12(3):475. https://doi.org/10.3390/rs1203047
Sezer E, Pradhan B, Gokceoglu C (2011) Manifestation of an adaptive neuro-fuzzy model on landslide susceptibility mapping: Klang Valley, Malaysia. Expert Syst Appl 38:8208–8219. https://doi.org/10.1016/j.eswa.2010.12.167
Khanlari GR, Heidari M, Momeni AA, Abdilor Y (2012) Prediction of shear strength parameters of soils using artificial neural networks and multivariate regression methods. Eng Geol 131–132:11. https://doi.org/10.1016/j.enggeo.2011.12.006
Lee S, Hwang J, Park I (2013) Application of data-driven evidential belief functions to landslide susceptibility mapping in Jinbu, Korea. Catena 100:15–30. https://doi.org/10.1016/j.catena.2012.07.014
Lee S, Min K (2001) Statistical analysis of landslide susceptibility at Yongin, Korea. Environ Geol 40(9):1095–1113. https://doi.org/10.1007/s002540100310
Pradhan B (2013) A comparative study on the predictive ability of the decision tree, support vector machine and neuro-fuzzy models in landslide susceptibility mapping using GIS. Comput Geosci 51:350–365. https://doi.org/10.1016/j.cageo.2012.08.023
San BT (2014) An evaluation of SVM using polygon-based random sampling in landslide susceptibility mapping: the Candir catchment area (western Antalya, Turkey). Int J Appl Earth Obs Geoinf 26:399–412. https://doi.org/10.1016/j.jag.2013.09.010
Hwang S, Guevarra IF, Yu B (2009) Slope failure prediction using a decision tree: a case of engineered slopes in South Korea. Eng Geol 104(1–2):126–134. https://doi.org/10.1016/j.enggeo.2008.09.004
Marjanović M, Kovačević M, Bajat B, Voženílek V (2011) Landslide susceptibility assessment using SVM machine learning algorithm. Eng Geol 123(3):225–234. https://doi.org/10.1016/j.enggeo.2011.09.006
Mohan A, Kumar B, Dwivedi R (2021) Review on remote sensing methods for landslide detection using machine and deep learning. Trans Emerg TelecommunTechnol 32. https://doi.org/10.1002/ett.3998
Lacroix P, Zavala B, Berthier E, Audin L (2013) Supervised method of landslide inventory using Panchromatic SPOT5 images and application to the earthquake-triggered landslides of Pisco (Peru, 2007, Mw8.0). Remote Sens 5(6):2590–2616. https://doi.org/10.3390/rs5062590
Rai PK, Mohan K, Kumra VK (2014) Landslide hazard and its mapping using remote sensing and GIS. J Sci Res 58:1–133333
Google Scholar
Chaturvedi P, Dutt V, Jaiswal B, Tyagi N, Sharma S, Mishra Sp, Dhar S, Joglekar P (2014) Remote sensing based regional landslide risk assessment. Int J Emerg Trends Electr Electron 2320–9569(10):135–140
Dou J, Chang K-T, Chen S, Yunus A, Liu J-K, Xia H, Zhu Z (2015) Automatic case-based reasoning approach for landslide detection: Integration of object-oriented image analysis and a genetic algorithm. Remote Sens 7(4):4318–4342. https://doi.org/10.3390/rs70404318
Li X, Cheng X, Chen W, Chen G, Liu S (2015) Identification of forested landslides using LiDar data, object-based image analysis, and machine learning algorithms. Remote Sensing 7(8):9705–9726. https://doi.org/10.3390/rs70809705
Golovko D, Roessner S, Behling R, Wetzel H-U, Kleinschmit B (2017) Evaluation of remote-sensing-based landslide inventories for hazard assessment in Southern Kyrgyzstan. Remote Sens 9(9):943. https://doi.org/10.3390/rs9090943
Chen Z, Zhang Y, Ouyang C, Zhang F, Ma J (2018) Automated landslides detection for Mountain cities using Multi-temporal Remote sensing imagery. Sensors 18(3):821. https://doi.org/10.3390/s18030821
Konishi T, Suga Y (2018) Landslide detection using COSMO-SkyMed images: a case study of a landslide event on Kii Peninsula, Japan. Eur J Remote Sens 51(1):205–221. https://doi.org/10.1080/22797254.2017.1418185
Si A, Zhang J, Tong S, Lai Q, Wang R, Li N, Bao Y (2018) Regional landslide identification based on susceptibility analysis and change detection. ISPRS Int J Geo-Information 7(10):394. https://doi.org/10.3390/ijgi7100394
Tavakkoli Piralilou S, Shahabi H, Jarihani B, Ghorbanzadeh O, Blaschke T, Gholamnia K, Aryal J (2019) Landslide detection using multi-scale image segmentation and different machine learning models in the higher himalayas. Remote Sens 11(21):2575. https://doi.org/10.3390/rs11212575
Pawłuszek K, Marczak S, Borkowski A, Tarolli P (2019) Multi-aspect analysis of object-oriented landslide detection based on an Extended Set of LiDAR-Derived Terrain features. ISPRS Int J Geo-Information 8(8):321. https://doi.org/10.3390/ijgi8080321
Wang H, Zhang L, Yin K, Luo H, Li J (2020) Landslide identification using machine learning. Geosci Front. https://doi.org/10.1016/j.gsf.2020.02.012
Li L, Cheng S, Wen Z (2021) Landslide prediction based on improved principal component analysis and mixed kernel function least squares support vector regression model. J Mt Sci 18:2130–2142. https://doi.org/10.1007/s11629-020-6396-5
Ye C, Wei R, Ge Y et al (2022) GIS-based spatial prediction of landslide using road factors and random forest for Sichuan-Tibet Highway. J Mt Sci 19:461–476. https://doi.org/10.1007/s11629-021-6848-6
Meena SR, Soares LP, Grohmann CH et al (2022) Landslide detection in the Himalayas using machine learning algorithms and U-Net. Landslides. https://doi.org/10.1007/s10346-022-01861-3
Sajadi P, Sang Y-F, Gholamnia M, Bonafoni S (2022) Evaluation of the landslide susceptibility and its spatial difference in the whole Qinghai-Tibetan Plateau region by five learning algorithms. Geosci Lett 9. https://doi.org/10.1186/s40562-022-00218-x
https://bhuvan.nrsc.gov.in/home/index.php . Accessed 21 Dec 2021
https://www.usgs.gov/centers/eros/science/usgs-eros-archive-isro-resourcesat-1-and-resourcesat-2-liss-3 . Accessed 4 Dec 2021
Ma S, Chen J, Wu S, Li Y (2023) Landslide susceptibility prediction using machine learning methods: a case study of landslides in the Yinghu Lake Basin in Shaanxi. Sustainability 15(22):15836. https://doi.org/10.3390/su152215836
Hussain MA, Chen Z, Zheng Y, Zhou Y, Daud H (2023) Deep learning and machine learning models for landslide susceptibility mapping with remote sensing data. Remote Sens 15(19):4703. https://doi.org/10.3390/rs15194703
Sun D, Chen D, Zhang J, Mi C, Gu Q, Wen H (2023) Landslide susceptibility mapping based on interpretable machine learning from the perspective of geomorphological differentiation. Land 12(5):1018. https://doi.org/10.3390/land12051018
Varangaonkar P, Rode SV (2023) Lightweight deep learning model for automatic landslide prediction and localization. Multimed Tools Appl 82:33245–33266. https://doi.org/10.1007/s11042-023-15049-x
Mahajan HB, Junnarkar AA (2023) Smart healthcare system using integrated and lightweight ECC with private blockchain for multimedia medical data processing. Multimed Tools Appl. https://doi.org/10.1007/s11042-023-15204-4
Kadam MV, Mahajan HB, Uke NJ, Futane PR (2023) Cybersecurity threats mitigation in internet of vehicles communication system using reliable clustering and routing. Microprocess Microsyst 102:104926. https://doi.org/10.1016/j.micpro.2023.104926
Mahajan H, Reddy KTV (2023) Secure gene profile data processing using lightweight cryptography and blockchain. Cluster Comput. https://doi.org/10.1007/s10586-023-04123-6
Download references
Author information
Authors and affiliations.
Sipna College of Engineering and Technology, Amravati, India
Payal Varangaonkar
Electronics and Telecommunication Department, Sipna College of Engineering & Technology, Amravati, India
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Payal Varangaonkar .
Ethics declarations
Ethical approval.
This article does not contain any studies with human participants performed by any of the authors.
Conflict of interest
All authors declares that they has no conflict of interest.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Varangaonkar, P., Rode, S.V. Landslide prediction with severity analysis using efficient computer vision and soft computing algorithms. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-19454-8
Download citation
Received : 26 September 2023
Revised : 21 March 2024
Accepted : 15 May 2024
Published : 30 May 2024
DOI : https://doi.org/10.1007/s11042-024-19454-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Computer vision methods
- Classifications
- Landslide detection
- Normal digital vegetation index
- Segmentation
- Severity analysis
- Western region
- Find a journal
- Publish with us
- Track your research

IMAGES
VIDEO
COMMENTS
Research techniques refer to the various methods, processes, and tools used to collect, analyze, and interpret data for the purpose of answering research questions or testing hypotheses. ... Mixed-methods research: This is a research method that combines quantitative and qualitative research techniques to provide a more comprehensive ...
Research methods are specific procedures for collecting and analyzing data. Developing your research methods is an integral part of your research design. When planning your methods, there are two key decisions you will make. ... Methods are the specific tools and procedures you use to collect and analyze data (for example, experiments, surveys, ...
(v) Research demands accurate observation and description. (vi) Research involves gathering new data from primary or first-hand sources or using existing data for a new purpose. (vii) Research is characterized by carefully designed procedures that apply rigorous analysis. (viii) Research involves the quest for answers to un-solved problems.
About Research Methods. This guide provides an overview of research methods, how to choose and use them, and supports and resources at UC Berkeley. As Patten and Newhart note in the book Understanding Research Methods, "Research methods are the building blocks of the scientific enterprise. They are the "how" for building systematic knowledge.
Research methods are specific procedures for collecting and analysing data. Developing your research methods is an integral part of your research design. When planning your methods, there are two key decisions you will make. ... Methods are the specific tools and procedures you use to collect and analyse data (e.g. experiments, surveys, ...
About methods, techniques, and research tools. A research method is defined as a way of scientifically studying a phenomenon. It consists of specific activities within the research procedure, supplemented by a set of tools used to collect and analyze data. The intentional, planned, and conscious choice of a research method is a guarantee of the ...
Qualitative research methods. Each of the research approaches involve using one or more data collection methods.These are some of the most common qualitative methods: Observations: recording what you have seen, heard, or encountered in detailed field notes. Interviews: personally asking people questions in one-on-one conversations. Focus groups: asking questions and generating discussion among ...
Data Collection | Definition, Methods & Examples. Published on June 5, 2020 by Pritha Bhandari.Revised on June 21, 2023. Data collection is a systematic process of gathering observations or measurements. Whether you are performing research for business, governmental or academic purposes, data collection allows you to gain first-hand knowledge and original insights into your research problem.
Research methods relate to how researchers gather information and data. For example, surveys, focus groups, customer interviews, and A/B testing are research methods that gather information. On the other hand, tools assist areas of research. Researchers may use tools to more efficiently gather data, store data securely, or uncover insights.
Abstract. This paper aims to provide an overview of the use and assessment of qualitative research methods in the health sciences. Qualitative research can be defined as the study of the nature of phenomena and is especially appropriate for answering questions of why something is (not) observed, assessing complex multi-component interventions ...
The research design is determined prior to the start of data collection and is not flexible. The research process, interventions and data collection tools (e.g. questionnaires) are standardized to minimize or control possible bias. Table 8 provides an overview of quantitative data collection strategies. Qualitative research techniques and tools
research-tools--cancer-genome-atlas.jpg. The Cancer Genome Atlas (TCGA) is a landmark NIH cancer genomics program that transformed our understanding of cancer by analyzing tumors from 11,000 patients with 33 different cancer types. Findings from TCGA identified new ways to prevent, diagnose, and treat cancers, such as gliomas and stomach cancer.
Types of Data Analysis Techniques in Research. Data analysis techniques in research are categorized into qualitative and quantitative methods, each with its specific approaches and tools. These techniques are instrumental in extracting meaningful insights, patterns, and relationships from data to support informed decision-making, validate ...
Statistical methods involved in carrying out a study include planning, designing, collecting data, analysing, drawing meaningful interpretation and reporting of the research findings. The statistical analysis gives meaning to the meaningless numbers, thereby breathing life into a lifeless data. The results and inferences are precise only if ...
Intended for students in research methods courses and for readers seeking a guide to conducting behavioral studies, this book provides an accessible introduction to the techniques and tools of behavioral research used in psychology, sociology, and anthropology, as well as in business, education, design, and program evaluation.
It outlines the techniques, procedures, and tools that researchers use to plan, design, execute, and analyze their studies. A well-defined research m ethodology is crucial
Step 1: Explain your methodological approach. Step 2: Describe your data collection methods. Step 3: Describe your analysis method. Step 4: Evaluate and justify the methodological choices you made. Tips for writing a strong methodology chapter. Other interesting articles.
Research tools refer to a wide range of resources, methods, instruments, software, or techniques that researchers use to collect, analyze, interpret, and communicate data and information during the research process. These tools are designed to facilitate and enhance various aspects of research, such as data collection, organization, analysis ...
Techniques for finding information. Library Search help. Search techniques. Subject guides. Referencing style guides. EndNote referencing software. Digital Essentials. Health Information and Research Essential. Digital Researcher Lab.
A method for developing intra-epithelial innate lymphoid cells (tissue-resident NK cells) for immunotherapy. Uriel Moreno. John Sunwoo. Joshua Tay. Saumyaa Saumyaa. June Ho Shin. Nina Horowitz. This collection contains molecular, cellular, and antibody tools and methods, recombinant technologies, biosensing and analytics technologies ...
Case Studies, Checklists, Interviews, Observation sometimes, and Surveys or Questionnaires are all tools used to collect data. It is important to decide on the tools for data collection because research is carried out in different ways and for different purposes. The objective behind data collection is to capture quality evidence that allows ...
Data integration is considered a classic research field and a pressing need within the information science community. Ontologies play a critical role in such processes by providing well-consolidated support to link and semantically integrate datasets via interoperability. This paper approaches data integration from an application perspective by looking at ontology matching techniques.
Natural language processing (NLP) is a subset of artificial intelligence, computer science, and linguistics focused on making human communication, such as speech and text, comprehensible to computers. NLP is used in a wide variety of everyday products and services. Some of the most common ways NLP is used are through voice-activated digital ...
Weeds are attractive models for basic and applied research due to their impacts on agricultural systems and capacity to swiftly adapt in response to anthropogenic selection pressures. Currently, a lack of genomic information precludes research to elucidate the genetic basis of rapid adaptation for important traits like herbicide resistance and stress tolerance and the effect of evolutionary ...
Revised on June 22, 2023. Quantitative research is the process of collecting and analyzing numerical data. It can be used to find patterns and averages, make predictions, test causal relationships, and generalize results to wider populations. Quantitative research is the opposite of qualitative research, which involves collecting and analyzing ...
Research on coastal regions traditionally involves methods like manual sampling, monitoring buoys, and remote sensing, but these methods face challenges in spatially and temporally diverse regions of interest. Autonomous surface vehicles (ASVs) with artificial intelligence (AI) are being explored, and recognized by the International Maritime Organization (IMO) as vital for future ecosystem ...
Natural language processing techniques powered by generative AI provide us with powerful tools for the analysis of large volumes of data, facilitating the exploration of complex social phenomena and enabling researchers to identify trends and correlations not readily visible when using traditional methods. However, these tools come with ...
Multimedia Tools and Applications - Since the preceding decade, there has been a great deal of interest in forecasting landslides using remote-sensing images. ... This research proposes a novel landslide prediction method with a severity analysis model based on real-time hyperspectral RS images. The proposed model consists of phases of pre ...