
Want to create or adapt books like this? Learn more about how Pressbooks supports open publishing practices.

Section 5.3: Multiple Regression Explanation, Assumptions, Interpretation, and Write Up
Learning Objectives
At the end of this section you should be able to answer the following questions:
- Explain the difference between Multiple Regression and Simple Regression.
- Explain the assumptions underlying Multiple Regression.
Multiple Regression is a step beyond simple regression. The main difference between simple and multiple regression is that multiple regression includes two or more independent variables – sometimes called predictor variables – in the model, rather than just one.
As such, the purpose of multiple regression is to determine the utility of a set of predictor variables for predicting an outcome, which is generally some important event or behaviour. This outcome can be designated as the outcome variable, the dependent variable, or the criterion variable. For example, you might hypothesise that the need to belong will predict motivations for Facebook use and that self-esteem and meaningful existence will uniquely predict motivations for Facebook use.
Before beginning your analysis, you should consider the following points:
- Regression analyses reveal relationships among variables (relationship between the criterion variable and the linear combination of a set of predictor variables) but do not imply a causal relationship.
- A regression solution – or set of predictor variables – is sensitive to combinations of variables. Whether a predictor is important in a solution depends on the other predictors in the set. If the predictor of interest is the only one that assesses some important facet of the outcome, it will appear important. If a predictor is only one of several predictors that assess the same important facet of the outcome, it will appear less important. For a good set of predictor variables – the smallest set of uncorrelated variables is best.
PowerPoint: Venn Diagrams
Please click on the link labeled “Venn Diagrams” to work through an example.
- Chapter Five – Venn Diagrams
In these Venn Diagrams, you can see why it is best for the predictors to be strongly correlated with the dependent variable but uncorrelated with the other Independent Variables. This reduces the amount of shared variance between the independent variables. The illustration in Slide 2 shows logical relationships between predictors, for two different possible regression models in separate Venn diagrams. On the left, you can see three partially correlated independent variables on a single dependent variable. The three partially correlated independent variables are physical health, mental health, and spiritual health and the dependent variable is life satisfaction. On the right, you have three highly correlated independent variables (e.g., BMI, blood pressure, heart rate) on the dependent variable of life satisfaction. The model on the left would have some use in discovering the associations between those variables, however, the model on the right would not be useful, as all three of the independent variables are basically measuring the same thing and are mostly accounting for the same variability in the dependent variable.
There are two main types of regression with multiple independent variables:
- Standard or Single Step: Where all predictors enter the regression together.
- Sequential or Hierarchical: Where all predictors are entered in blocks. Each block represents one step.
We will now be exploring the single step multiple regression:
All predictors enter the regression equation at once. Each predictor is treated as if it had been analysed in the regression model after all other predictors had been analysed. These predictors are evaluated by the shared variance (i.e., level of prediction) shared between the dependant variable and the individual predictor variable.
Multiple Regression Assumptions
There are a number of assumptions that should be assessed before performing a multiple regression analysis:
- The dependant variable (the variable of interest) needs to be using a continuous scale.
- There are two or more independent variables. These can be measured using either continuous or categorical means.
- The three or more variables of interest should have a linear relationship, which you can check by using a scatterplot.
- The data should have homoscedasticity. In other words, the line of best fit is not dissimilar as the data points move across the line in a positive or negative direction. Homoscedasticity can be checked by producing standardised residual plots against the unstandardized predicted values.
- The data should not have two or more independent variables that are highly correlated. This is called multicollinearity which can be checked using Variance-inflation-factor or VIF values. High VIF indicates that the associated independent variable is highly collinear with the other variables in the model.
- There should be no spurious outliers.
- The residuals (errors) should be approximately normally distributed. This can be checked by a histogram (with a superimposed normal curve) and by plotting the of the standardised residuals using either a P-P Plot, or a Normal Q-Q Plot .
Multiple Regression Interpretation
For our example research question, we will be looking at the combined effect of three predictor variables – perceived life stress, location, and age – on the outcome variable of physical health?
PowerPoint: Standard Regression
Please open the output at the link labeled “Chapter Five – Standard Regression” to view the output.
- Chapter Five – Standard Regression
Slide 1 contains the standard regression analysis output.
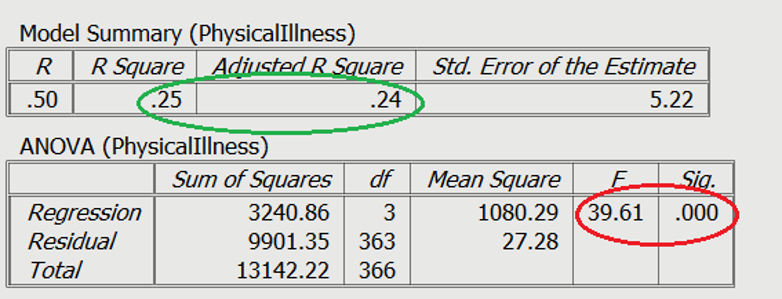
On Slide 2 you can see in the red circle, the test statistics are significant. The F-statistic examines the overall significance of the model, and shows if your predictors as a group provide a better fit to the data than no predictor variables, which they do in this example.
The R 2 values are shown in the green circle. The R 2 value shows the total amount of variance accounted for in the criterion by the predictors, and the adjusted R 2 is the estimated value of R 2 in the population.
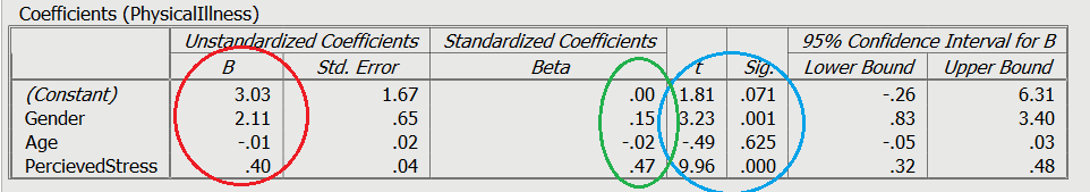
Moving on to the individual variable effects on Slide 3, you can see the significance of the contribution of individual predictors in light blue. The unstandardized slope or the B value is shown in red, which represents the change caused by the variable (e.g., increasing 1 unit of perceived stress will raise physical illness by .40). Finally, you can see the standardised slope value in green, which are also known as beta values. These values are standardised ranging from +/-0 to 1, similar to an r value.
We should also briefly discuss dummy variables:
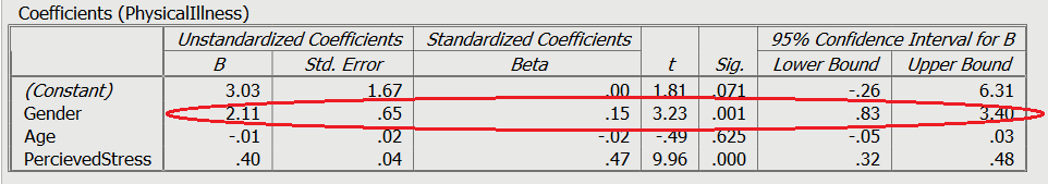
A dummy variable is a variable that is used to represent categorical information relating to the participants in a study. This could include gender, location, race, age groups, and you get the idea. Dummy variables are most often represented as dichotomous variables (they only have two values). When performing a regression, it is easier for interpretation if the values for the dummy variable is set to 0 or 1. 1 usually resents when a characteristic is present. For example, a question asking the participants “Do you have a drivers license” with a forced choice response of yes or no.
In this example on Slide 3 and circled in red, the variable is gender with male = 0, and female = 1. A positive Beta (B) means an association with 1, whereas a negative beta means an association with 0. In this case, being female was associated with greater levels of physical illness.
Multiple Regression Write Up
Here is an example of how to write up the results of a standard multiple regression analysis:
In order to test the research question, a multiple regression was conducted, with age, gender (0 = male, 1 = female), and perceived life stress as the predictors, with levels of physical illness as the dependent variable. Overall, the results showed the utility of the predictive model was significant, F (3,363) = 39.61, R 2 = .25, p < .001. All of the predictors explain a large amount of the variance between the variables (25%). The results showed that perceived stress and gender of participants were significant positive predictors of physical illness ( β =.47, t = 9.96, p < .001, and β =.15, t = 3.23, p = .001, respectively). The results showed that age ( β =-.02, t = -0.49 p = .63) was not a significant predictor of perceived stress.
Statistics for Research Students Copyright © 2022 by University of Southern Queensland is licensed under a Creative Commons Attribution 4.0 International License , except where otherwise noted.
Share This Book

Statistics Made Easy
Introduction to Multiple Linear Regression
When we want to understand the relationship between a single predictor variable and a response variable, we often use simple linear regression .
However, if we’d like to understand the relationship between multiple predictor variables and a response variable then we can instead use multiple linear regression .
If we have p predictor variables, then a multiple linear regression model takes the form:
Y = β 0 + β 1 X 1 + β 2 X 2 + … + β p X p + ε
- Y : The response variable
- X j : The j th predictor variable
- β j : The average effect on Y of a one unit increase in X j , holding all other predictors fixed
- ε : The error term
The values for β 0 , β 1 , B 2 , … , β p are chosen using the least square method , which minimizes the sum of squared residuals (RSS):
RSS = Σ(y i – ŷ i ) 2
- Σ : A greek symbol that means sum
- y i : The actual response value for the i th observation
- ŷ i : The predicted response value based on the multiple linear regression model
The method used to find these coefficient estimates relies on matrix algebra and we will not cover the details here. Fortunately, any statistical software can calculate these coefficients for you.
How to Interpret Multiple Linear Regression Output
Suppose we fit a multiple linear regression model using the predictor variables hours studied and prep exams taken and a response variable exam score .
The following screenshot shows what the multiple linear regression output might look like for this model:
Note: The screenshot below shows multiple linear regression output for Excel , but the numbers shown in the output are typical of the regression output you’ll see using any statistical software.

From the model output, the coefficients allow us to form an estimated multiple linear regression model:
Exam score = 67.67 + 5.56*(hours) – 0.60*(prep exams)
The way to interpret the coefficients are as follows:
- Each additional one unit increase in hours studied is associated with an average increase of 5.56 points in exam score, assuming prep exams is held constant.
- Each additional one unit increase in prep exams taken is associated with an average decrease of 0.60 points in exam score, assuming hours studied is held constant.
We can also use this model to find the expected exam score a student will receive based on their total hours studied and prep exams taken. For example, a student who studies for 4 hours and takes 1 prep exam is expected to score a 89.31 on the exam:
Exam score = 67.67 + 5.56*(4) -0.60*(1) = 89.31
Here is how to interpret the rest of the model output:
- R-Square: This is known as the coefficient of determination. It is the proportion of the variance in the response variable that can be explained by the explanatory variables. In this example, 73.4% of the variation in the exam scores can be explained by the number of hours studied and the number of prep exams taken.
- Standard error: This is the average distance that the observed values fall from the regression line. In this example, the observed values fall an average of 5.366 units from the regression line.
- F: This is the overall F statistic for the regression model, calculated as regression MS / residual MS.
- Significance F: This is the p-value associated with the overall F statistic. It tells us whether or not the regression model as a whole is statistically significant. In other words, it tells us if the two explanatory variables combined have a statistically significant association with the response variable. In this case the p-value is less than 0.05, which indicates that the explanatory variables hours studied and prep exams taken combined have a statistically significant association with exam score.
- Coefficient P-values. The individual p-values tell us whether or not each explanatory variable is statistically significant. We can see that hours studied is statistically significant (p = 0.00) while prep exams taken (p = 0.52) is not statistically significant at α = 0.05. Since prep exams taken is not statistically significant, we may end up deciding to remove it from the model.
How to Assess the Fit of a Multiple Linear Regression Model
There are two numbers that are commonly used to assess how well a multiple linear regression model “fits” a dataset:
1. R-Squared: This is the proportion of the variance in the response variable that can be explained by the predictor variables.
The value for R-squared can range from 0 to 1. A value of 0 indicates that the response variable cannot be explained by the predictor variable at all. A value of 1 indicates that the response variable can be perfectly explained without error by the predictor variable.
The higher the R-squared of a model, the better the model is able to fit the data.
2. Standard Error: This is the average distance that the observed values fall from the regression line. The smaller the standard error, the better a model is able to fit the data.
If we’re interested in making predictions using a regression model, the standard error of the regression can be a more useful metric to know than R-squared because it gives us an idea of how precise our predictions will be in terms of units.
For a complete explanation of the pros and cons of using R-squared vs. Standard Error for assessing model fit, check out the following articles:
- What is a Good R-squared Value?
- Understanding the Standard Error of a Regression Model
Assumptions of Multiple Linear Regression
There are four key assumptions that multiple linear regression makes about the data:
1. Linear relationship: There exists a linear relationship between the independent variable, x, and the dependent variable, y.
2. Independence: The residuals are independent. In particular, there is no correlation between consecutive residuals in time series data.
3. Homoscedasticity: The residuals have constant variance at every level of x.
4. Normality: The residuals of the model are normally distributed.
For a complete explanation of how to test these assumptions, check out this article .
Multiple Linear Regression Using Software
The following tutorials provide step-by-step examples of how to perform multiple linear regression using different statistical software:
How to Perform Multiple Linear Regression in R How to Perform Multiple Linear Regression in Python How to Perform Multiple Linear Regression in Excel How to Perform Multiple Linear Regression in SPSS How to Perform Multiple Linear Regression in Stata How to Perform Linear Regression in Google Sheets
Featured Posts

Hey there. My name is Zach Bobbitt. I have a Masters of Science degree in Applied Statistics and I’ve worked on machine learning algorithms for professional businesses in both healthcare and retail. I’m passionate about statistics, machine learning, and data visualization and I created Statology to be a resource for both students and teachers alike. My goal with this site is to help you learn statistics through using simple terms, plenty of real-world examples, and helpful illustrations.
One Reply to “Introduction to Multiple Linear Regression”
i came here to find the interaction or in terms of marketing ‘synergy effect’ in multiple linear regression and how to handle dummy variable in multiple linear regression. But i found some perspective which is fine.
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Join the Statology Community
Sign up to receive Statology's exclusive study resource: 100 practice problems with step-by-step solutions. Plus, get our latest insights, tutorials, and data analysis tips straight to your inbox!
By subscribing you accept Statology's Privacy Policy.
Multiple Regression Analysis using SPSS Statistics
Introduction.
Multiple regression is an extension of simple linear regression. It is used when we want to predict the value of a variable based on the value of two or more other variables. The variable we want to predict is called the dependent variable (or sometimes, the outcome, target or criterion variable). The variables we are using to predict the value of the dependent variable are called the independent variables (or sometimes, the predictor, explanatory or regressor variables).
For example, you could use multiple regression to understand whether exam performance can be predicted based on revision time, test anxiety, lecture attendance and gender. Alternately, you could use multiple regression to understand whether daily cigarette consumption can be predicted based on smoking duration, age when started smoking, smoker type, income and gender.
Multiple regression also allows you to determine the overall fit (variance explained) of the model and the relative contribution of each of the predictors to the total variance explained. For example, you might want to know how much of the variation in exam performance can be explained by revision time, test anxiety, lecture attendance and gender "as a whole", but also the "relative contribution" of each independent variable in explaining the variance.
This "quick start" guide shows you how to carry out multiple regression using SPSS Statistics, as well as interpret and report the results from this test. However, before we introduce you to this procedure, you need to understand the different assumptions that your data must meet in order for multiple regression to give you a valid result. We discuss these assumptions next.
SPSS Statistics
Assumptions.
When you choose to analyse your data using multiple regression, part of the process involves checking to make sure that the data you want to analyse can actually be analysed using multiple regression. You need to do this because it is only appropriate to use multiple regression if your data "passes" eight assumptions that are required for multiple regression to give you a valid result. In practice, checking for these eight assumptions just adds a little bit more time to your analysis, requiring you to click a few more buttons in SPSS Statistics when performing your analysis, as well as think a little bit more about your data, but it is not a difficult task.
Before we introduce you to these eight assumptions, do not be surprised if, when analysing your own data using SPSS Statistics, one or more of these assumptions is violated (i.e., not met). This is not uncommon when working with real-world data rather than textbook examples, which often only show you how to carry out multiple regression when everything goes well! However, don’t worry. Even when your data fails certain assumptions, there is often a solution to overcome this. First, let's take a look at these eight assumptions:
- Assumption #1: Your dependent variable should be measured on a continuous scale (i.e., it is either an interval or ratio variable). Examples of variables that meet this criterion include revision time (measured in hours), intelligence (measured using IQ score), exam performance (measured from 0 to 100), weight (measured in kg), and so forth. You can learn more about interval and ratio variables in our article: Types of Variable . If your dependent variable was measured on an ordinal scale, you will need to carry out ordinal regression rather than multiple regression. Examples of ordinal variables include Likert items (e.g., a 7-point scale from "strongly agree" through to "strongly disagree"), amongst other ways of ranking categories (e.g., a 3-point scale explaining how much a customer liked a product, ranging from "Not very much" to "Yes, a lot").
- Assumption #2: You have two or more independent variables , which can be either continuous (i.e., an interval or ratio variable) or categorical (i.e., an ordinal or nominal variable). For examples of continuous and ordinal variables , see the bullet above. Examples of nominal variables include gender (e.g., 2 groups: male and female), ethnicity (e.g., 3 groups: Caucasian, African American and Hispanic), physical activity level (e.g., 4 groups: sedentary, low, moderate and high), profession (e.g., 5 groups: surgeon, doctor, nurse, dentist, therapist), and so forth. Again, you can learn more about variables in our article: Types of Variable . If one of your independent variables is dichotomous and considered a moderating variable, you might need to run a Dichotomous moderator analysis .
- Assumption #3: You should have independence of observations (i.e., independence of residuals ), which you can easily check using the Durbin-Watson statistic, which is a simple test to run using SPSS Statistics. We explain how to interpret the result of the Durbin-Watson statistic, as well as showing you the SPSS Statistics procedure required, in our enhanced multiple regression guide.
- Assumption #4: There needs to be a linear relationship between (a) the dependent variable and each of your independent variables, and (b) the dependent variable and the independent variables collectively . Whilst there are a number of ways to check for these linear relationships, we suggest creating scatterplots and partial regression plots using SPSS Statistics, and then visually inspecting these scatterplots and partial regression plots to check for linearity. If the relationship displayed in your scatterplots and partial regression plots are not linear, you will have to either run a non-linear regression analysis or "transform" your data, which you can do using SPSS Statistics. In our enhanced multiple regression guide, we show you how to: (a) create scatterplots and partial regression plots to check for linearity when carrying out multiple regression using SPSS Statistics; (b) interpret different scatterplot and partial regression plot results; and (c) transform your data using SPSS Statistics if you do not have linear relationships between your variables.
- Assumption #5: Your data needs to show homoscedasticity , which is where the variances along the line of best fit remain similar as you move along the line. We explain more about what this means and how to assess the homoscedasticity of your data in our enhanced multiple regression guide. When you analyse your own data, you will need to plot the studentized residuals against the unstandardized predicted values. In our enhanced multiple regression guide, we explain: (a) how to test for homoscedasticity using SPSS Statistics; (b) some of the things you will need to consider when interpreting your data; and (c) possible ways to continue with your analysis if your data fails to meet this assumption.
- Assumption #6: Your data must not show multicollinearity , which occurs when you have two or more independent variables that are highly correlated with each other. This leads to problems with understanding which independent variable contributes to the variance explained in the dependent variable, as well as technical issues in calculating a multiple regression model. Therefore, in our enhanced multiple regression guide, we show you: (a) how to use SPSS Statistics to detect for multicollinearity through an inspection of correlation coefficients and Tolerance/VIF values; and (b) how to interpret these correlation coefficients and Tolerance/VIF values so that you can determine whether your data meets or violates this assumption.
- Assumption #7: There should be no significant outliers , high leverage points or highly influential points . Outliers, leverage and influential points are different terms used to represent observations in your data set that are in some way unusual when you wish to perform a multiple regression analysis. These different classifications of unusual points reflect the different impact they have on the regression line. An observation can be classified as more than one type of unusual point. However, all these points can have a very negative effect on the regression equation that is used to predict the value of the dependent variable based on the independent variables. This can change the output that SPSS Statistics produces and reduce the predictive accuracy of your results as well as the statistical significance. Fortunately, when using SPSS Statistics to run multiple regression on your data, you can detect possible outliers, high leverage points and highly influential points. In our enhanced multiple regression guide, we: (a) show you how to detect outliers using "casewise diagnostics" and "studentized deleted residuals", which you can do using SPSS Statistics, and discuss some of the options you have in order to deal with outliers; (b) check for leverage points using SPSS Statistics and discuss what you should do if you have any; and (c) check for influential points in SPSS Statistics using a measure of influence known as Cook's Distance, before presenting some practical approaches in SPSS Statistics to deal with any influential points you might have.
- Assumption #8: Finally, you need to check that the residuals (errors) are approximately normally distributed (we explain these terms in our enhanced multiple regression guide). Two common methods to check this assumption include using: (a) a histogram (with a superimposed normal curve) and a Normal P-P Plot; or (b) a Normal Q-Q Plot of the studentized residuals. Again, in our enhanced multiple regression guide, we: (a) show you how to check this assumption using SPSS Statistics, whether you use a histogram (with superimposed normal curve) and Normal P-P Plot, or Normal Q-Q Plot; (b) explain how to interpret these diagrams; and (c) provide a possible solution if your data fails to meet this assumption.
You can check assumptions #3, #4, #5, #6, #7 and #8 using SPSS Statistics. Assumptions #1 and #2 should be checked first, before moving onto assumptions #3, #4, #5, #6, #7 and #8. Just remember that if you do not run the statistical tests on these assumptions correctly, the results you get when running multiple regression might not be valid. This is why we dedicate a number of sections of our enhanced multiple regression guide to help you get this right. You can find out about our enhanced content as a whole on our Features: Overview page, or more specifically, learn how we help with testing assumptions on our Features: Assumptions page.
In the section, Procedure , we illustrate the SPSS Statistics procedure to perform a multiple regression assuming that no assumptions have been violated. First, we introduce the example that is used in this guide.
A health researcher wants to be able to predict "VO 2 max", an indicator of fitness and health. Normally, to perform this procedure requires expensive laboratory equipment and necessitates that an individual exercise to their maximum (i.e., until they can longer continue exercising due to physical exhaustion). This can put off those individuals who are not very active/fit and those individuals who might be at higher risk of ill health (e.g., older unfit subjects). For these reasons, it has been desirable to find a way of predicting an individual's VO 2 max based on attributes that can be measured more easily and cheaply. To this end, a researcher recruited 100 participants to perform a maximum VO 2 max test, but also recorded their "age", "weight", "heart rate" and "gender". Heart rate is the average of the last 5 minutes of a 20 minute, much easier, lower workload cycling test. The researcher's goal is to be able to predict VO 2 max based on these four attributes: age, weight, heart rate and gender.
Setup in SPSS Statistics
In SPSS Statistics, we created six variables: (1) VO 2 max , which is the maximal aerobic capacity; (2) age , which is the participant's age; (3) weight , which is the participant's weight (technically, it is their 'mass'); (4) heart_rate , which is the participant's heart rate; (5) gender , which is the participant's gender; and (6) caseno , which is the case number. The caseno variable is used to make it easy for you to eliminate cases (e.g., "significant outliers", "high leverage points" and "highly influential points") that you have identified when checking for assumptions. In our enhanced multiple regression guide, we show you how to correctly enter data in SPSS Statistics to run a multiple regression when you are also checking for assumptions. You can learn about our enhanced data setup content on our Features: Data Setup page. Alternately, see our generic, "quick start" guide: Entering Data in SPSS Statistics .
Test Procedure in SPSS Statistics
The seven steps below show you how to analyse your data using multiple regression in SPSS Statistics when none of the eight assumptions in the previous section, Assumptions , have been violated. At the end of these seven steps, we show you how to interpret the results from your multiple regression. If you are looking for help to make sure your data meets assumptions #3, #4, #5, #6, #7 and #8, which are required when using multiple regression and can be tested using SPSS Statistics, you can learn more in our enhanced guide (see our Features: Overview page to learn more).
Note: The procedure that follows is identical for SPSS Statistics versions 18 to 28 , as well as the subscription version of SPSS Statistics, with version 28 and the subscription version being the latest versions of SPSS Statistics. However, in version 27 and the subscription version , SPSS Statistics introduced a new look to their interface called " SPSS Light ", replacing the previous look for versions 26 and earlier versions , which was called " SPSS Standard ". Therefore, if you have SPSS Statistics versions 27 or 28 (or the subscription version of SPSS Statistics), the images that follow will be light grey rather than blue. However, the procedure is identical .

Published with written permission from SPSS Statistics, IBM Corporation.
Note: Don't worry that you're selecting A nalyze > R egression > L inear... on the main menu or that the dialogue boxes in the steps that follow have the title, Linear Regression . You have not made a mistake. You are in the correct place to carry out the multiple regression procedure. This is just the title that SPSS Statistics gives, even when running a multiple regression procedure.

Interpreting and Reporting the Output of Multiple Regression Analysis
SPSS Statistics will generate quite a few tables of output for a multiple regression analysis. In this section, we show you only the three main tables required to understand your results from the multiple regression procedure, assuming that no assumptions have been violated. A complete explanation of the output you have to interpret when checking your data for the eight assumptions required to carry out multiple regression is provided in our enhanced guide. This includes relevant scatterplots and partial regression plots, histogram (with superimposed normal curve), Normal P-P Plot and Normal Q-Q Plot, correlation coefficients and Tolerance/VIF values, casewise diagnostics and studentized deleted residuals.
However, in this "quick start" guide, we focus only on the three main tables you need to understand your multiple regression results, assuming that your data has already met the eight assumptions required for multiple regression to give you a valid result:
Determining how well the model fits
The first table of interest is the Model Summary table. This table provides the R , R 2 , adjusted R 2 , and the standard error of the estimate, which can be used to determine how well a regression model fits the data:

The " R " column represents the value of R , the multiple correlation coefficient . R can be considered to be one measure of the quality of the prediction of the dependent variable; in this case, VO 2 max . A value of 0.760, in this example, indicates a good level of prediction. The " R Square " column represents the R 2 value (also called the coefficient of determination), which is the proportion of variance in the dependent variable that can be explained by the independent variables (technically, it is the proportion of variation accounted for by the regression model above and beyond the mean model). You can see from our value of 0.577 that our independent variables explain 57.7% of the variability of our dependent variable, VO 2 max . However, you also need to be able to interpret " Adjusted R Square " ( adj. R 2 ) to accurately report your data. We explain the reasons for this, as well as the output, in our enhanced multiple regression guide.
Statistical significance
The F -ratio in the ANOVA table (see below) tests whether the overall regression model is a good fit for the data. The table shows that the independent variables statistically significantly predict the dependent variable, F (4, 95) = 32.393, p < .0005 (i.e., the regression model is a good fit of the data).

Estimated model coefficients
The general form of the equation to predict VO 2 max from age , weight , heart_rate , gender , is:
predicted VO 2 max = 87.83 – (0.165 x age ) – (0.385 x weight ) – (0.118 x heart_rate ) + (13.208 x gender )
This is obtained from the Coefficients table, as shown below:

Unstandardized coefficients indicate how much the dependent variable varies with an independent variable when all other independent variables are held constant. Consider the effect of age in this example. The unstandardized coefficient, B 1 , for age is equal to -0.165 (see Coefficients table). This means that for each one year increase in age, there is a decrease in VO 2 max of 0.165 ml/min/kg.
Statistical significance of the independent variables
You can test for the statistical significance of each of the independent variables. This tests whether the unstandardized (or standardized) coefficients are equal to 0 (zero) in the population. If p < .05, you can conclude that the coefficients are statistically significantly different to 0 (zero). The t -value and corresponding p -value are located in the " t " and " Sig. " columns, respectively, as highlighted below:

You can see from the " Sig. " column that all independent variable coefficients are statistically significantly different from 0 (zero). Although the intercept, B 0 , is tested for statistical significance, this is rarely an important or interesting finding.
Putting it all together
You could write up the results as follows:
A multiple regression was run to predict VO 2 max from gender, age, weight and heart rate. These variables statistically significantly predicted VO 2 max, F (4, 95) = 32.393, p < .0005, R 2 = .577. All four variables added statistically significantly to the prediction, p < .05.
If you are unsure how to interpret regression equations or how to use them to make predictions, we discuss this in our enhanced multiple regression guide. We also show you how to write up the results from your assumptions tests and multiple regression output if you need to report this in a dissertation/thesis, assignment or research report. We do this using the Harvard and APA styles. You can learn more about our enhanced content on our Features: Overview page.
Lesson 5: Multiple Linear Regression (MLR) Model & Evaluation
Overview of this lesson.
In this lesson, we make our first (and last?!) major jump in the course. We move from the simple linear regression model with one predictor to the multiple linear regression model with two or more predictors. That is, we use the adjective "simple" to denote that our model has only predictor, and we use the adjective "multiple" to indicate that our model has at least two predictors.
In the multiple regression setting, because of the potentially large number of predictors, it is more efficient to use matrices to define the regression model and the subsequent analyses. This lesson considers some of the more important multiple regression formulas in matrix form. If you're unsure about any of this, it may be a good time to take a look at this Matrix Algebra Review .
The good news is that everything you learned about the simple linear regression model extends — with at most minor modification — to the multiple linear regression model. Think about it — you don't have to forget all of that good stuff you learned! In particular:
- The models have similar "LINE" assumptions. The only real difference is that whereas in simple linear regression we think of the distribution of errors at a fixed value of the single predictor, with multiple linear regression we have to think of the distribution of errors at a fixed set of values for all the predictors. All of the model checking procedures we learned earlier are useful in the multiple linear regression framework, although the process becomes more involved since we now have multiple predictors. We'll explore this issue further in Lesson 6.
- The use and interpretation of r 2 (which we'll denote R 2 in the context of multiple linear regression) remains the same. However, with multiple linear regression we can also make use of an "adjusted" R 2 value, which is useful for model building purposes. We'll explore this measure further in Lesson 11.
- With a minor generalization of the degrees of freedom, we use t -tests and t -intervals for the regression slope coefficients to assess whether a predictor is significantly linearly related to the response, after controlling for the effects of all the opther predictors in the model.
- With a minor generalization of the degrees of freedom, we use confidence intervals for estimating the mean response and prediction intervals for predicting an individual response. We'll explore these further in Lesson 6.
For the simple linear regression model, there is only one slope parameter about which one can perform hypothesis tests. For the multiple linear regression model, there are three different hypothesis tests for slopes that one could conduct. They are:
- a hypothesis test for testing that one slope parameter is 0
- a hypothesis test for testing that all of the slope parameters are 0
- a hypothesis test for testing that a subset — more than one, but not all — of the slope parameters are 0
In this lesson, we also learn how to perform each of the above three hypothesis tests.
- 5.1 - Example on IQ and Physical Characteristics
- 5.2 - Example on Underground Air Quality
- 5.3 - The Multiple Linear Regression Model
- 5.4 - A Matrix Formulation of the Multiple Regression Model
- 5.5 - Three Types of MLR Parameter Tests
- 5.6 - The General Linear F-Test
- 5.7 - MLR Parameter Tests
- 5.8 - Partial R-squared
- 5.9 - Further MLR Examples
Start Here!
- Welcome to STAT 462!
- Search Course Materials
- Lesson 1: Statistical Inference Foundations
- Lesson 2: Simple Linear Regression (SLR) Model
- Lesson 3: SLR Evaluation
- Lesson 4: SLR Assumptions, Estimation & Prediction
- 5.9- Further MLR Examples
- Lesson 6: MLR Assumptions, Estimation & Prediction
- Lesson 7: Transformations & Interactions
- Lesson 8: Categorical Predictors
- Lesson 9: Influential Points
- Lesson 10: Regression Pitfalls
- Lesson 11: Model Building
- Lesson 12: Logistic, Poisson & Nonlinear Regression
- Website for Applied Regression Modeling, 2nd edition
- Notation Used in this Course
- R Software Help
- Minitab Software Help

Copyright © 2018 The Pennsylvania State University Privacy and Legal Statements Contact the Department of Statistics Online Programs

- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons
Margin Size
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.

3.3.4: Hypothesis Test for Simple Linear Regression
- Last updated
- Save as PDF
- Page ID 28708
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
We will now describe a hypothesis test to determine if the regression model is meaningful; in other words, does the value of \(X\) in any way help predict the expected value of \(Y\)?
Simple Linear Regression ANOVA Hypothesis Test
Model Assumptions
- The residual errors are random and are normally distributed.
- The standard deviation of the residual error does not depend on \(X\)
- A linear relationship exists between \(X\) and \(Y\)
- The samples are randomly selected
Test Hypotheses
\(H_o\): \(X\) and \(Y\) are not correlated
\(H_a\): \(X\) and \(Y\) are correlated
\(H_o\): \(\beta_1\) (slope) = 0
\(H_a\): \(\beta_1\) (slope) ≠ 0
Test Statistic
\(F=\dfrac{M S_{\text {Regression }}}{M S_{\text {Error }}}\)
\(d f_{\text {num }}=1\)
\(d f_{\text {den }}=n-2\)
Sum of Squares
\(S S_{\text {Total }}=\sum(Y-\bar{Y})^{2}\)
\(S S_{\text {Error }}=\sum(Y-\hat{Y})^{2}\)
\(S S_{\text {Regression }}=S S_{\text {Total }}-S S_{\text {Error }}\)
In simple linear regression, this is equivalent to saying “Are X an Y correlated?”
In reviewing the model, \(Y=\beta_{0}+\beta_{1} X+\varepsilon\), as long as the slope (\(\beta_{1}\)) has any non‐zero value, \(X\) will add value in helping predict the expected value of \(Y\). However, if there is no correlation between X and Y, the value of the slope (\(\beta_{1}\)) will be zero. The model we can use is very similar to One Factor ANOVA.
The Results of the test can be summarized in a special ANOVA table:
Example: Rainfall and sales of sunglasses
Design : Is there a significant correlation between rainfall and sales of sunglasses?
Research Hypothese s:
\(H_o\): Sales and Rainfall are not correlated \(H_o\): 1 (slope) = 0
\(H_a\): Sales and Rainfall are correlated \(H_a\): 1 (slope) ≠ 0
Type I error would be to reject the Null Hypothesis and \(t\) claim that rainfall is correlated with sales of sunglasses, when they are not correlated. The test will be run at a level of significance (\(\alpha\)) of 5%.
The test statistic from the table will be \(\mathrm{F}=\dfrac{\text { MSRegression }}{\text { MSError }}\). The degrees of freedom for the numerator will be 1, and the degrees of freedom for denominator will be 5‐2=3.
Critical Value for \(F\) at \(\alpha\)of 5% with \(df_{num}=1\) and \(df_{den}=3} is 10.13. Reject \(H_o\) if \(F >10.13\). We will also run this test using the \(p\)‐value method with statistical software, such as Minitab.
Data/Results
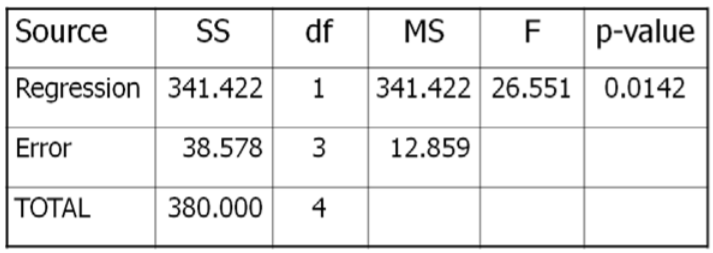
\(F=341.422 / 12.859=26.551\), which is more than the critical value of 10.13, so Reject \(H_o\). Also, the \(p\)‐value = 0.0142 < 0.05 which also supports rejecting \(H_o\).
Sales of Sunglasses and Rainfall are negatively correlated.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 28 May 2024
Gut microbiome remodeling and metabolomic profile improves in response to protein pacing with intermittent fasting versus continuous caloric restriction
- Alex E. Mohr ORCID: orcid.org/0000-0001-5401-3702 1 , 2 ,
- Karen L. Sweazea 1 , 2 , 3 ,
- Devin A. Bowes ORCID: orcid.org/0000-0001-9819-2503 2 ,
- Paniz Jasbi 4 , 5 ,
- Corrie M. Whisner ORCID: orcid.org/0000-0003-3888-6348 1 , 2 ,
- Dorothy D. Sears ORCID: orcid.org/0000-0002-9260-3540 1 ,
- Rosa Krajmalnik-Brown ORCID: orcid.org/0000-0001-6064-3524 2 ,
- Yan Jin 6 ,
- Haiwei Gu 1 , 6 ,
- Judith Klein-Seetharaman ORCID: orcid.org/0000-0002-4892-6828 1 , 4 ,
- Karen M. Arciero 7 ,
- Eric Gumpricht 8 &
- Paul J. Arciero ORCID: orcid.org/0000-0001-7445-6164 7 , 9
Nature Communications volume 15 , Article number: 4155 ( 2024 ) Cite this article
545 Accesses
265 Altmetric
Metrics details
- Metabolomics
- Risk factors
The gut microbiome (GM) modulates body weight/composition and gastrointestinal functioning; therefore, approaches targeting resident gut microbes have attracted considerable interest. Intermittent fasting (IF) and protein pacing (P) regimens are effective in facilitating weight loss (WL) and enhancing body composition. However, the interrelationships between IF- and P-induced WL and the GM are unknown. The current randomized controlled study describes distinct fecal microbial and plasma metabolomic signatures between combined IF-P ( n = 21) versus a heart-healthy, calorie-restricted (CR, n = 20) diet matched for overall energy intake in free-living human participants (women = 27; men = 14) with overweight/obesity for 8 weeks. Gut symptomatology improves and abundance of Christensenellaceae microbes and circulating cytokines and amino acid metabolites favoring fat oxidation increase with IF-P (p < 0.05), whereas metabolites associated with a longevity-related metabolic pathway increase with CR (p < 0.05). Differences indicate GM and metabolomic factors play a role in WL maintenance and body composition. This novel work provides insight into the GM and metabolomic profile of participants following an IF-P or CR diet and highlights important differences in microbial assembly associated with WL and body composition responsiveness. These data may inform future GM-focused precision nutrition recommendations using larger sample sizes of longer duration. Trial registration, March 6, 2020 (ClinicalTrials.gov as NCT04327141), based on a previous randomized intervention trial.
Similar content being viewed by others

Protein supplementation during an energy-restricted diet induces visceral fat loss and gut microbiota amino acid metabolism activation: a randomized trial


Gut microbiota plasticity is correlated with sustained weight loss on a low-carb or low-fat dietary intervention

Intermittent fasting modulates the intestinal microbiota and improves obesity and host energy metabolism
Introduction.
As a principal modulator of the gut microbiome (GM) and weight status, nutritional input holds great therapeutic promise for addressing a wide range of metabolic dysregulation 1 . Dependent on the host for nutrients and fluid, one of the main processes by which the GM affects host physiology is producing bioactive metabolites from the gastrointestinal (GI) contents. Nutrient composition, feeding frequency, and meal timing impact this dependency 2 , 3 . To maintain a stable community and ecosystem, the GM must regulate its growth rate and diversity in response to nutrient availability and population density 4 . Such maintenance is affected by caloric restriction (CR) coupled with periods of feeding and intermittent fasting (IF) 5 . Moreover, we’ve recently shown the nutritional composition and meal frequency during these periods alter the metabolizable energy for the host 6 . The current study incorporates protein pacing (P), defined as four meals/day consumed evenly spaced every 4 h, consisting of 25–50 g of protein/meal 7 , 8 , 9 . Indeed, we have previously characterized a dietary approach of calorie-restricted IF-P combined and P alone 7 , 8 . These studies included nutrient-dense meal replacement shakes, along with whole foods, to quantitatively examine beneficial changes in body composition and cardiometabolic, inflammatory, and toxin-related outcomes in healthy and overweight individuals 7 , 8 , 10 , 11 , 12 . Further, recent preclinical work in mice has identified dietary protein as having anti-obesity effects after CR that are partially modulated through the GM 13 . Thus, the need to examine this in humans is warranted.
In this current work, we compare the effects of two low-calorie dietary interventions matched for weekly energy intake and expenditure; continuous caloric restriction on a heart-healthy diet (CR) aligned with current United States (US) dietary recommendations 14 versus our calorie-restricted IF-P diet 8 , 15 , in forty-one individuals with overweight or obesity, over an 8-week intervention. We hypothesize an IF-P diet may favorably influence the GM and metabolome to a greater extent than a calorie-matched CR alone. This exploratory investigation utilizes data and samples from a randomized controlled trial (NCT04327141) that compares the effects of the CR versus IF-P diet on anthropometric and cardiometabolic outcomes, as previously published 15 . As an additional analysis, we select “high” and “low” responders based on relative weight loss (WL) for a subgroup examination of the IF-P diet to better elucidate potential differential responses to intermittent fasting and protein pacing. Of special note, one individual lost 15% of their initial body weight over the 8-week intervention; this individual is followed longitudinally for a year to explore the dynamics of their GM and fecal metabolome. Novel findings from the current study shows an IF-P regimen results in improved gut symptomatology, a more pronounced community shift, and greater divergence of the gut microbiome, including microbial families and genera, such as Christensenellaceae , Rikenellaceae , and Marvinbryantia , associated with favorable metabolic profiles, compared to CR. Furthermore, IF-P significantly increases cytokines linked to lipolysis, weight loss, inflammation, and immune response. These findings shed light on the differential effects of IF-P as a promising dietary intervention for obesity management and microbiotic and metabolic health.
Intermittent fasting - protein pacing (IF-P) significantly influences gut microbiome (GM) dynamics compared to calorie restriction (CR)
We compared an IF-P vs. a CR per-protocol dietary intervention (matched for total energy intake and expenditure) over eight weeks to compare changes in weight, cardiometabolic outcomes, and the GM in men and women with overweight/obesity (IF-P: n = 21; CR: n = 20). One participant in each group were lost to follow-up due to non-compliance with dietary intervention (Fig. 1a ; CONSORT flow diagram: Supplementary Fig. S1a ). The primary outcomes of dietary intake, body weight and composition responses, cardiometabolic outcomes, and hunger ratings after both dietary interventions are provided in our companion paper 15 . Briefly, after a one-week run-in period consuming their usual dietary intake (baseline diet), with no differences between groups at baseline for any dietary intake variable 15 , both dietary interventions significantly reduced total fat, carbohydrate, sodium, sugar, and energy intake by approximately 40% (~1000 kcals/day) from baseline levels (Fig. 1b ; Supplementary Data 1 ). By design, IF-P increased protein intake greater than CR during the intervention. The IF-P regimen consisted of 35% carbohydrate, 30% fat, and 35% protein for five to six days per week and a weekly extended modified fasting period (36–60 h) consisting of 350–550 kcals per day using randomization, as detailed previously 7 , 8 , 9 , 10 , 15 . In comparison, the CR regimen consisted of 41% carbohydrate, 38% fat, and 21% protein in accordance with current US dietary recommendations (Supplementary Table S1 ) 14 , 16 . Using two-way factorial mixed model analysis of variance (ANOVA), significant macronutrient decreases drove energy reduction from dietary fat and carbohydrate ( p < 0.001), with increased protein in the IF-P compared to CR ( p < 0.001; Supplementary Fig. S1b ; Supplementary Data 1 ). Regarding GI functioning and GM modulation, IF-P significantly decreased sugar and increased dietary fiber relative to CR (IF-P; pre, 20 ± 2 vs. post, 26 ± 2: CR; pre, 24 ± 3 vs. 24 ± 2 g/day; p < 0.05). Despite similar average weekly energy intake (~9000 kcals/week) and physical activity energy expenditure (~350 kcals/day; p = 0.260) during the intervention, participants following the IF-P regimen lost significantly more body weight (−8.81 ± 0.71% vs. −5.40 ± 0.67%; p = 0.003; Fig. 1c ; Supplementary Data 1 ) and total, abdominal, and visceral fat mass and increased fat-free mass percentage (~2×; p ≤ 0.030), as previously reported 15 . In addition, within-group analyses revealed a significant decrease in the reported frequency of total and lower-moderate GI symptoms (GI symptom rating score [GSRS] ≥4) over time for both IF-P and CR participants. However, when comparing the two dietary interventions at each time point, a more substantial reduction was observed in IF-P participants compared to CR participants (i.e., −9.3% vs. −5.4% and −13.2% vs. −3.9%, respectively; Table 1 ). The increased protein and lower sugar intake in IF-P compared to CR may have favorably mediated the GM and symptomatology.
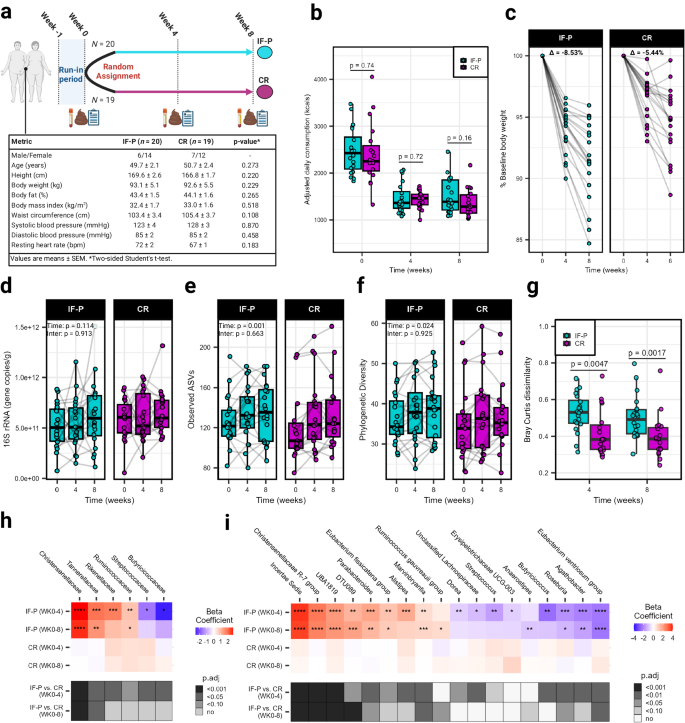
a Study design with baseline participant characteristics. A registered dietitian counseled individuals from both groups each week. Time points with data collection are shown for both IF-P and CR participants. Icons created using BioRender.com. b Total daily caloric intake at each time point was not significantly different between IF-P and CR diet groups (two-sided Student’s t -test, p < 0.05). Adjusted values are displayed by dividing total weekly intake by seven, to account for the fasting periods of IF-P. c IF-P participants lost significantly more weight over time versus CR participants. Points connected by line represent percent of weight compared to baseline weight for each participant. d Overall gut microbial colonization, as demonstrated by qPCR-based quantification of 16S rRNA gene copies per gram wet weight was unaffected by time or intervention (linear-mixed effects [LME] model, two-sided p > 0.05). Alpha diversity metrics, e observed amplicon sequence variants (ASVs), and f Phylogenetic diversity at the ASV level significantly increased over time, independent of the intervention. g Intra-individual changes in GM community structure from baseline to weeks four and eight in IF-P participants shifted significantly throughout the IF-P intervention compared to CR as measured by the Bray-Curtis dissimilarity index (two-sided Wilcoxon rank-sum test). All box and whiskers plots display the box ranging from the first to the third quartile, and the center the median value, while the whiskers extend from each quartile to the minimum or maximum values. Heatmap of significant changes in h family- and i genus-level bacter i a by intervention. Colors indicate the within-group change beta coefficients over time for each cell, and asterisks denote significance. Black-white annotations on the bottom denote the significance of between-group change difference (by MaAsLin2 group × time interactions; p -values were corrected to produce adjusted values [ p .adj] using the Benjamini–Hochberg method). For all panels, IF-P: n = 20, CR: n = 19. Source data are provided as a Source Data file.
The substantial reduction in calorie intake of both groups (~40% from baseline) led us to investigate its potential impact on transient microbial colonization in the gut, as estimated by 16S rRNA gene copies (linear-mixed effects model [LME] time effect, p = 0.114; Fig. 1d ; Supplementary Data 2 ). While it might be expected that a significant reduction in calorie intake could influence gut microbial colonization, our findings indicate that this reduction did not reach statistical significance within the timeframe of our study. This result contrasts with previous research that imposed more substantial energy restriction, such as a four-week regimen of ~800 kcal/day in participants with overweight/obesity, where overall gut microbial colonization notably decreased 4 . In addition to assessing microbial colonization, we also investigated whether the calorie reduction significantly influenced principal stool characteristics, including wet stool weight, Bristol stool scale (BSS), and fecal pH ( p ≥ 0.066; Table 1 ). However, we did not observe statistically significant changes in these parameters over the course of the study. Moreover, there were no significant differences between the two dietary intervention groups over time (interaction effect, p ≥ 0.051). In contrast, there were significant time effects for observed amplicon sequence variants (ASVs) and phylogenetic diversity (LME time effect, p ≤ 0.023; Fig. 1e, f ; Supplementary Data 2 ), with values increasing at weeks four and eight compared to baseline (pairwise comparisons, p ≤ 0.048); however, no interaction was observed for either alpha diversity metric (group × time effect, p ≥ 0.925). To rule out the potential confounding effects of GI transit time 17 , BSS (as a surrogate marker) and stool pH were not significantly correlated with alpha diversity (Spearman correlations, p ≥ 0.210). In relation to community composition, much of the intervention variance could be attributed to individual response upon testing nested permutational analysis of variance (PERMANOVA; R 2 = 0.749, p = 0.001; Supplementary Table S2 ), showcasing the highly individualistic landscape of the human GM in response to dietary intervention. However, a significant 1.8% of the variance was accounted for by the group × time interaction ( p = 0.001). Moreover, individual responses over time showed variance between the two dietary interventions (PERMANOVA, R 2 = 0.123, p = 0.003). This variability was apparent by assessing intra-individual differences, where a pronounced increase in Bray-Curtis dissimilarity was observed in the IF-P compared to the CR group after four (median Bray-Curtis dissimilarity, 0.53 [IQR: 0.47–0.61] vs. 0.38 [IQR: 0.33–0.47]) and eight weeks (0.50 [IQR: 0.41–0.55] vs. 0.39 [IQR: 0.33–0.45]; Fig. 1g ; Wilcoxon rank-sum test, p ≤ 0.005).
To understand the taxa driving this GM variation from baseline to weeks four and eight between the two dietary interventions, we constructed MaAsLin2 linear-mixed models with the individual participant as a random factor 18 . We observed differential abundance patterns at the family and genus level in response to the IF-P but not the CR intervention. Of the 28 family and 69 genus-level features captured after filtering, a respective total of six and 18 taxa displayed significant interaction effects, with all significant time effects occurring from IF-P ( p .adj ≤ 0.10; Fig. 1h, i ; Supplementary Data 3 , 4 ). Notably, the changes observed at the four-week mark were more pronounced compared to those at eight weeks. These early alterations may signify an initial adaptation phase during which microbial populations respond to the modified substrate availability and nutrient composition, suggesting a degree of community resilience 19 . Increases were sustained to the third fecal collection for the family Christensenellaceae and the genera Incertae Sedis ( Ruminococcaceae family), Christensenellaceae R-7 group , and UBA1819 ( Ruminococcaceae family) (effect size > 2.0). Christensenellaceae is well regarded as a marker of a lean (anti-obesity) phenotype 20 and is associated with higher protein intake 21 . Other notable increases included Rikenellaceae , which, like Christensenellaceae , has been linked to reduced visceral adipose tissue and healthy metabolic profiles 22 , and Marvinbryantia , a candidate marker for predicting long-term weight loss success in individuals with obesity 23 . In addition, IF-P increased Ruminococcaceae , which has been noted to have an increased proteolytic and lipolytic capacity 24 . This shift in IF-P participants likely represents a change in GM substrate fermentation preferences as the diet regimen (relative protein and carbohydrate) and energy restriction is expected to increase the proteolytic: saccharolytic potential ratio 25 . In contrast, all taxa that decreased in IF-P participants were butyrate producers. These included the family Butyricicoccaceae and several genera such as Butyricicoccus (week four), Eubacterium ventriosum group (weeks four and eight), and Agathobacter (week four) (effect size < −2.0). When comparing monozygotic twin pairs, Eubacterium ventriosum group and another reduced genus, Roseburia , were more abundant in the higher body mass index (BMI) siblings 26 . Others, such as the mucosa-associated Butyricicoccus and Erysipelotricaceae UCG-003, have been positively correlated with insulin resistance and speculated to contribute to impaired glycolipid metabolism 27 .
Despite these changes in GM composition and increased fiber intake (+30% vs. baseline) of the IF-P participants 15 , we did not detect a significant shift in the abundance of the principal fecal short-chain fatty acids (SCFAs), acetate, propionate, butyrate, or valerate, as assessed by gas chromatography-mass spectrometry (GC–MS) (LME, p ≥ 0.470; Supplementary Fig. S1c ; Supplementary Data 5 ). Several factors likely contribute to this finding. For example, the distinct physical-chemical properties of fiber sources between IF-P and CR are inherently different. Participants adhering to the IF-P diet consumed most of their dietary fiber as liquid meal replacements (shakes) that are rich in non-digestible, oligosaccharide dietary-resistant starch 5 (RS5). In contrast, subjects on the CR regimen consumed their fiber from whole food sources such as vegetables, whole grains, and legumes. These fiber sources provided a mixture of soluble and insoluble fibers and a more complex fiber profile than IF-P participants. Moreover, even similar fiber profiles may function differently due to differences in food matrices and/or food preparation (cooking, raw consumption, etc.). Also of relevance is the timing of their fiber consumption. IF-P participants’ fiber intake was concentrated in fiber-rich shakes, offering immediate availability of fiber to the GI tract. In contrast, CR participants consumed fiber through whole foods, leading to a slower digestion and absorption process influenced by individual digestive transit times and enzymatic profiles. Interestingly, our results parallel recent work where participants more than doubled their fiber intake without affecting fecal SCFAs 28 . The disparate findings may be due to the type of dietary-resistant starch (RS) as a component of the nutrition regimen. In the current study, RS5 was included in the meal replacement shakes (eight grams/shake, two shakes/day, 16 g/day total). Prior research supports resistant starch intakes of >20 g/day favorably modulate SCFA production, primarily butyrate, over four to 12-week interventions 29 , 30 . Moreover, this lack of response in fecal SCFAs in both groups may have been further compounded by the significant reduction in energy intake in both groups, where the epithelia of the GI tract may have absorbed any potential increase in SCFAs from the dietary shift. It is worth noting that stool analysis may not be the most reliable biological surrogate for capturing SCFA flux over time 28 . Nevertheless, the changes in nutrient quality, timing, ratios, and the observed shift toward proteolytic activity suggest that the luminal matrix of digesta in the IF-P group impacted substrate availability for GM. This effect appears to be an influencing force in driving the observed beneficial shifts in microbial communities, such as Christensenellaceae and Incertae Sedis , as well as improvements in GI symptomatology in IF-P compared to CR. These results underscore the complexity of dietary influences on GM and highlight the need for further research to explore the impact of liquid meal replacements versus whole food sources on GM changes and SCFA status.
IF-P modulates circulating cytokines and gut microbiome taxa compared to CR
Caloric restriction and WL have been well known to positively influence inflammatory cytokine expression, with GM now emerging as an important modulator 31 . Surveying a panel of 14 plasma cytokines, we noted significant interaction (group × time) effects for IL-4, IL-6, IL-8, and IL-13 (LME, p ≤ 0.034; Fig. 2a–d ; Supplementary Table S3 ; Supplementary Data 6 ). These cytokines exhibited increases at weeks four and/or eight compared to baseline exclusively in the IF-P group (pairwise comparisons, p .adj ≤ 0.098), while no significant changes were observed in the CR group ( p .adj ≥ 0.562). Notably, IL-4 has been reported to display lipolytic effects 32 , and IL-8 has been positively associated with weight loss and maintenance 33 . Regarded as a proinflammatory myokine, IL-6 can acutely increase lipid mobilization in adipose tissue under fasting or exercise conditions 34 , 35 , 36 . IL-13 may be important for gut mucosal immune responses and is a stimulator of mucus production from goblet cells 37 , which has been recently reported to be influenced during a two-day-a-week fasting regimen in mice 38 . These results were of note considering the significant total body weight, fat, and visceral fat loss in the IF-P compared to the CR group. Surprisingly, correlational analysis with change (post – pre) in anthropometric and select plasma biomarker values with the cytokine profile did not reveal any significant associations after correcting for multiple testing effects ( p .adj ≥ 0.476; Supplementary Data 7 ). Plasma cytokines were, however, correlated with microbial composition for samples collected in the IF-P group during the intervention period (weeks four and eight) using graph-guided fused least absolute shrinkage and selection operator (GFLASSO) regression, revealing associations between cytokine-taxa pairs (Supplementary Fig. S2a ). Of the four cytokines that increased in IF-P participants, we identified multiple significant correlations: Colidextribacter (rho = −0.55, p .adj = 0.015), Ruminococcus gauvreauii group (rho = 0.50, p .adj = 0.036), and Intestinibacter (rho = 0.45, p .adj = 0.086) with IL-4 (Supplementary Fig. S2b ) and an unclassified genus from Oscillospiraceae (rho = −0.53, p .adj = 0.019), Colidextribacter (rho = −0.52, p .adj = 0.019), and Ruminoccus gauvreauii group (rho = 0.51, p .adj = 0.019) with IL-13 (Supplementary Fig. S2c ).
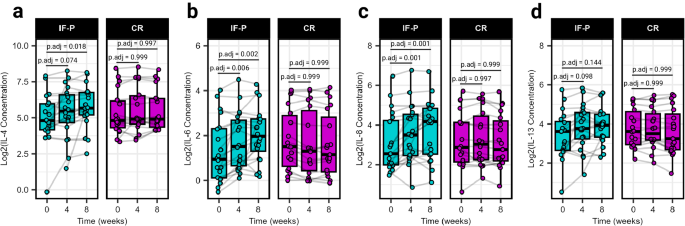
a IL-4, b IL-6, c IL-8, and d IL-13: Each panel shows the cytokine concentration levels. Significant time effects and interaction effects (group × time) were detected using linear-mixed effects models (LME, two-sided p < 0.05), indicating differential changes over the intervention period. IF-P participants exhibited significant increases in cytokine levels compared to baseline, as evidenced by pairwise comparisons adjusted for multiple testing using the Benjamini–Hochberg method (two-sided p .adj < 0.10). All box and whiskers plots display the box ranging from the first to the third quartile, and the center the median value, while the whiskers extend from each quartile to the minimum or maximum values. For all panels, IF-P: n = 20, CR: n = 19. Source data are provided as a Source Data file.
Displaying negative correlations for IL-4 and IL-13, Colidextribacter has been shown to be positively correlated to fat accumulation, insulin, and triglyceride levels in mice fed a high-fat diet 39 and positively correlated with products of lipid peroxidation, suggesting its potential role in promoting oxidative stress 40 . Conversely, Ruminoccus gauvreauii group was positively correlated with IL-4 and IL-13. Although limited information is available regarding the host interactions of this microbe, this genus is considered a commensal part of the core human GM and able to convert complex polysaccharides into a variety of nutrients for their hosts 41 . While these findings highlight the potential interplay between specific microbes and cytokine profiles, the directional influence—whether microbial changes drive cytokine alterations or vice versa—cannot be determined in this study setting. Furthermore, despite the change in cytokine profiles in the IF-P group, we did not detect any significant time or group × time effects when measuring lipopolysaccharide-binding protein (LBP; Δ pre/post, IF-P: 0.24 ± 0.31 vs CR: −0.93 ± 0.49 μg/mL; p ≥ 0.254), a surrogate marker for gut permeability 42 . While the GM plays a crucial role in modulating the gut-immune axis, the observed cytokine fluctuations and microbial associations might also involve other factors. These include the production of specific metabolites due to shifts in microbial composition as well as the influence of the dietary regimen itself, which may have a central role in shaping these interactions.
IF-P and CR yield distinct circulating metabolite signatures and convergence of multiple metabolic pathways
To understand the potential differential impact of IF-P versus CR on the host, we surveyed the plasma metabolome, reliably detecting 136 plasma metabolites across 117 samples (i.e., QC CV < 20% and relative abundance > 1000 in 80% of samples). Based on outlier examination (random forest [RF] and principal component analysis [PCA]), no samples were categorized as outliers, and all data were retained for subsequent analysis. Metabolomic profile shifts were observed in both IF-P and CR groups compared with baseline (Canberra distance), however, these did not differ significantly by group or time (weeks four and eight; Wilcoxon rank-sum test, p ≥ 0.087; Supplementary Fig. S3a ). We prepared a general linear model (GLM) with age, sex, and time as covariates and corrected for false discovery rate (FDR). When controlling for these relevant covariates, we observed significant differences between IF-P and CR for 15 metabolites (Fig. 3a , Supplementary Table S4 ): 2,3-dihydroxybenzoic acid, malonic acid, choline, agmatine, protocatechuic acid, myoinositol, oxaloacetic acid, xylitol, dulcitol, asparagine, n-acetylglutamine, sorbitol, cytidine, acetylcarnitine, and urate ( p .adj ≤ 0.089). To estimate the univariate classification performance of the 15 significant metabolites, we performed a receiver operating characteristic (ROC) analysis. Ten metabolites demonstrated a moderate area under the curve (AUC) (0.718–0.819), while five metabolites had an AUC < 0.70. Therefore, to improve classification performance, we constructed a supervised PLS-DA model using levels of the 15 significant metabolites ( p .adj ≤ 0.089) and analyzed variable importance in projection (VIP) scores (Supplementary Fig. S3b ). Five metabolites with a VIP > 1.0 (2,3-dihydroxybenzoic acid, malonic acid, protocatechuic acid, agmatine, and myoinositol) were retained to construct an enhanced orthogonal projection to latent structures discriminant analysis (OPLS-DA) model. In contrast, the model fit was assessed with 100-fold leave-one-out cross-validation (LOOCV; see “Methods” section). Permutation testing showed the refined OPLS-DA model to have an acceptable fit to data ( Q 2 = 0.460, p < 0.001), with appreciable explanatory capacity ( R 2 = 0.506, p < 0.001; Supplementary Fig. S3c ). The ROC analysis produced an area under the curve (AUC) of 0.929 (95% CI: 0.868–0.973, sensitivity = 0.8, specificity = 0.9; Supplementary Fig. S3d ) between the CR and IF-P groups showing good accuracy of the GLM and providing strong support for the differential expression of these 15 metabolites between groups.
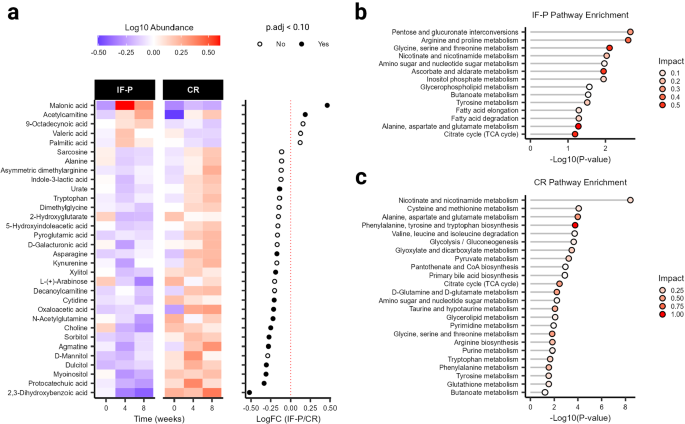
a Abundance and log fold-change of significant plasma metabolites between IF-P and CR groups as determined by a general linear model (GLM) adjusted for age, sex, and time. All GLM analyses utilized two-sided p -values, with multiple testing corrections applied using the Benjamini–Hochberg method ( p .adj). Metabolome pathway analysis was conducted for b IF-P and c CR using all reliably detected metabolites showing significantly altered pathways ( p .adj < 0.10) with moderate and above impact (>0.10). Impact scores were calculated using a hypergeometric test, while significance was assessed via a test of relative betweenness centrality, emphasizing the changes in metabolic network connectivity. For all panels, IF-P: n = 20, CR: n = 19. Source data are provided as a Source Data file.
Two metabolites, malonic acid, and acetylcarnitine, increased compared to the CR intervention. Several other investigators have noted the increase in acetylcarnitine via fasting protocols 43 , 44 . This increase is consistent with free fatty acid mobilization and increased transportation of these fatty acids via carnitine acylation into the mitochondria for fatty acid oxidation. These results would also be consistent with the expected ketogenesis, although not documented in our study, but noted by similar fasting interventions 44 . Relatedly, malonic acid, a naturally occurring organic acid, is a key regulatory molecule in fatty acid synthesis via its conversion to acetoacetate; hence, our results may reflect this increased synthesis in response to the mobilization and oxidation of fatty acids occurring during fasting. Other metabolites that decreased with IF-P include several sugar alcohols (myoinositol, dulcitol, and xylitol). Dulcitol (galactitol) is a sugar alcohol derived from galactose. It is possible that during fasting, levels of dulcitol decrease as glucose (initially) and free fatty acids (after 24–36 h of fasting) are preferentially utilized as energy substrates. One amino acid (asparagine) and one amino acid analog (N-acetylglutamine, associated with consumption of a Mediterranean diet 45 ) also decreased with IF-P relative to CR. Finally, 2,3-dihydroxybenzoic acid significantly decreased with IF-P. This metabolite is formed during the metabolism of flavonoids, as it is found abundantly in fruits, vegetables, and some spices. At the cellular level, this hydroxybenzoic acid functions as a cell signaling agent and has been speculated as a potentially protective molecule in various cancers 46 . It is unclear whether this metabolite decreased due to either dietary intake or metabolic processes related to high-protein intake or the fasting protocol. Collectively, the metabolic responses to these dietary regimens reflect the interrelationships of several anabolic and catabolic physiologic responses to three key components of these interventions: (a) the WL process itself, (b) changes in amount (and type) of macronutrient distribution (i.e., meal replacement shakes vs. whole food diet approach; higher vs. normal protein intakes), and (c) the adherence to fasting (IF-P only).
To determine the significantly impacted pathways of the dietary interventions, we grouped participant samples according to baseline or intervention period (weeks four and eight), with IF-P and CR assessed separately. A total of 14 pathways were significant in the IF-P group ( p .adj < 0.10; Fig. 3b ), with three displaying large impact coefficients (>0.5): (1) Glycine, serine, and threonine metabolism, (2) alanine, aspartate, and glutamate metabolism, and (3) ascorbate and aldarate metabolism. In comparison, 24 pathways were significant for the CR group (Fig. 3c ), with four showing large impact coefficients (>0.5): (1) Phenylalanine, tyrosine, and tryptophan biosynthesis, (2) alanine, aspartate, and glutamate metabolism, (3) citrate cycle (TCA cycle), and (4) glycine, serine and threonine metabolism. Notably, the glycine, serine, and threonine pathway has recently been found in preclinical models to play a pivotal role in longevity and related life-sustaining mechanisms independent of diet, though heavily impacted by fasting time and caloric restriction 47 . This may be partially related to the ability of glycine to increase tissue glutathione 48 , 49 and protect against oxidative stress 50 . In our analysis, this pathway was significant in both diet groups and is biochemically and topologically related to the additionally captured amino acid pathway, alanine, aspartate, and glutamate metabolism, as well as the energy-releasing pathway, the citrate cycle (TCA cycle). Notably, in the CR group, phenylalanine, tyrosine, and tryptophan biosynthesis, are important for neurotransmitter production and reported to be suppressed (tryptophan) in obesity 51 . This representation may have also been attributed to the differences in protein intake 52 or differences in dietary diversity 53 , yet to be determined. Regardless, we noted similar representations of pathway impact between IF-P and CR, with metabolic response centered on utilization of amino acids in addition to lipid turnover and energy pathways.
Gut microbiome and plasma metabolome latent factors indicate differential multi-omic signatures between IF-P and CR regimens
As the plasma metabolome has been suggested as a bidirectional mediator of GM influence on the host 54 , we performed a multi-omics factor analysis (MOFA) 55 to identify potential patterns of covariation and co-occurrence between the microbiome and circulating metabolites. Operating in a probabilistic Bayesian framework, MOFA simultaneously performs unsupervised matrix factorization to obtain overall sources of variability via a limited number of inferred factors and identifies shared versus exclusive variation across multiple omic data sets 55 . Eight latent factors were identified (minimum explained variance ≥2%; see “Methods” section), with the plasma metabolome and GM explaining 37.12% and 17.49% of the overall sample variability, respectively (Fig. 4a ). Based on significance and the proportion of total variance explained by individual factors for each omic assay, Factors 1 ( R 2 = 11.98) and 6 ( R 2 = 5.28) captured the greatest covariation between the two omic layers (Fig. 4a ; Supplementary Table S5 ). In contrast, Factors 2 and 5 were nearly exclusive to the metabolome, and factors 3 and 4 to the GM. Interestingly, Factor 1 was significantly negatively correlated to dietary protein intake (Spearman rho = −0.270, p.adj = 0.021; Fig. 4b ) and captured the variation associated with the CR diet (Wilcoxon rank-sum test, p .adj = 3.2e-04; Fig. 4c ). Factor 6 had the greatest number of significant correlations, including negative associations with visceral adipose tissue, waist circumference, body weight, BMI, fat mass, android fat, subcutaneous adipose tissue, dietary sodium, carbohydrate, fat, energy intake (kcal), and sugar (Spearman rho ≤ −0.220, p .adj ≤ 0.075) and captured the variation associated with IF-P (Wilcoxon rank-sum test, p .adj = 0.007).
a The cumulative proportion of total variance explained ( R 2 ) and proportion of total variance explained by eight individual latent factors for each omic layer. b Spearman correlation matrix of the eight latent factors and clinical anthropometric and dietary covariates. Each circle represents a separate association, with the size indicating the significance (-log10 ( p -values)) and the color representing the effect size (hue) with its direction (red: positive; blue: negative). All correlations are calculated using two-sided tests. Asterisks within a circle denote significance after adjustment with the Benjamini–Hochberg method. c Scatter plot of Factors 1 and 6, with each dot representing a sample colored by intervention. Box and whisker plots illustrate significant differences between groups after adjusting for multiple testing using the Benjamini–Hochberg method (Wilcoxon rank-sum test; top = Factor 1, p .adj = 3.2e-04; right = Factor 6, p .adj = 0.007). The plots show boxes ranging from the first to the third quartile and the median at the center, with whiskers extending to the minimum and maximum values. d Factor 1 and 6 loadings of genera and metabolites with the largest weights annotated. Symbols: * p .adj < 0.10, ** p .adj < 0.01, *** p .adj < 0.001, **** p .adj < 1.0e-04. For all panels, IF-P: n = 20, CR: n = 19. Source data are provided as a Source Data file.
Assessing the positive weights (feature importance) of Factor 1 revealed a microbial and metabolomic signature linked with CR, including the taxa Faecalibacterium , Romboutsia , and Roseburia , and the plasma metabolites myoinositol, agmatine, N-acetylglutamine, erythrose, and mucic acid (Fig. 4d ). Previous dietary restriction studies have reported co-occurrence of gut microbial taxa and plasma metabolites that span a wide variety of applications and investigations 56 . The specific co-occurrences observed in Factor 1 exhibited an abundance of butyrate-producing bacterial taxa that utilize carbohydrates as their predominant substrate and plasma metabolites that are generally involved in carbohydrate metabolism, such as erythrose, an intermediate in the pentose phosphate pathway (PPP), and mucic acid which is derived from galactose and/or galactose-containing compounds (i.e., lactose). These co-occurrence patterns biologically cohere considering the nutritional profile of the CR group and the large contribution of fiber-rich, unrefined carbohydrates and reduction in sugar (~50% kcal from sugar). Indeed, these nutritional changes may have influenced the GM to accommodate changes in dietary substrate more efficiently. One interesting co-occurrence was the genus Romboutsia and metabolite N-acetylglutamine. Romboutsia has been shown to produce several SCFAs and ferment certain amino acids, including glutamate 57 . N-acetylglutamine is biosynthesized from glutamate; thus, its co-occurrence with the abundance of Romboutsia encourages further exploration into this interaction 58 .
Factor 6 captured the signature associated with IF-P, with positive contributions from the taxa Incertae Sedis ( Ruminococcaceae family), Erysipelatoclostridium , Christensenellaceae R-7 group , Oscillospiraceae UCG-002, and Alistipes , and the plasma metabolites malonic acid, adipic acid, succinate, methylmalonic acid, and mucic acid (Fig. 4d ). Prior work has established that Alistipes increases from diets rich in protein and fat, and contributes to the highest number of putrefaction pathways (i.e., fermentation of undigested proteins in the GI tract) over the other commensals 59 . This could explain the co-occurrence of plasma metabolites from protein catabolism, such as 2-aminoadipid acid, adipic acid, and glutamic acid 22 , 59 . Oscillospiraceae has recently been viewed with next-generation probiotic potential, harboring positive regulatory effects in areas related to obesity and chronic inflammation 60 . Mentioned prior, recent studies have reported on the role of Christensenellaceae on human health, participating in host amino acid and lipid metabolism as well as fiber fermentation 20 , with Christensenellaceae R-7 group notably evidenced to correlate with visceral adipose tissue reduction 22 . As such, the elevated abundance of microbes in the GM of IF-P participants observed in this study in tandem with the co-occurrence of metabolites indicative of protein degradation and mobilization and oxidation of fatty acids, such as methylmalonic acid, malonic acid, and succinate, presents a nascent multi-omic signature of IF-P. In addition, and more pronounced in the IF-P vs CR group, participants decreased sugar intake by ~75% (kcals) compared to baseline levels. Considering the other regimental components of IF-P, the differences in multi-omic signatures likely display the selective pressures of these two interventions.
Gut microbiome (GM) composition is associated with weight loss (WL) responsiveness to IF-P diet
The IF-P intervention produced a microbiome and metabolomic response; however, the loss in body weight and fat across individuals varied (Fig. 5a ). To provide deeper characterization and explore differential features of WL responsiveness, we performed a GM-focused subgroup analysis by employing shotgun metagenomic and untargeted fecal metabolomic surveys in 10 individuals that either achieved ≥10% loss in body weight or bordered on clinically important WL (i.e., >5% BW; herein, ‘High’ and ‘Low’ responders) 61 . Importantly, baseline characteristics between WL responder classification did not differ significantly (baseline body weight: High, 108.9 ± 30.8 vs. Low, 81.9 ± 18.1 kg, p = 0.117; Supplementary Table S6 ). Assessing the GM at the fundamental taxonomic rank, species composition showed significant separation by weight loss response evaluated by Bray-Curtis dissimilarity (group × time: R 2 = 0.114, p = 0.001; Fig. 5b ; Supplementary Table S7 ), with most of the variation explained by the individual ( R 2 = 0.711, p = 0.001). In comparison, species level alpha diversity did not differ significantly between classifications (group × time: p ≥ 0.674; Fig. 5c, d ). Identifying 212 species after filtering, we noted significant differences in bacterial abundances between groups over time (Fig. 5e ; Supplementary Data 8 ). A total of 10 features increased in the High-responder group relative to the Low-response group over the eight-week study period, including Collinsella SGB14861 , Clostridium leptum , Blautia hydrogenotrophica , and less typified species; GGB74510 SGB47635 (unclassified Firmicutes), GGB3511 SGB4688 (unclassified Firmicutes), Faecalicatena contorta , Lachnospiraceae bacterium NSJ-29 , Phascolarctobacterium SGB4573 , GGB38744 SGB14842 (unclassified Oscillospiraceae ), and Massiliimalia timonensis (effect size ≥ 1.163, p .adj ≤ 0.092). The increase in Collinsella , a less characterized anaerobic pathobiont that produces lactate and has been associated with low-fiber intakes 62 , 63 and lipid metabolism 64 , may have been related to the periods of CR and IF, in conjunction with the greater influx of host-released fatty acids in the High-responder group. Relatedly, Clostridium leptum growth has been linked with increases in monounsaturated fat intake, reductions in blood cholesterol 65 , and stimulation of Treg induction (i.e., anti-inflammatory) 66 . The latter association is relevant to the SCFA-promoting (primarily butyrate) qualities of Clostridium leptum 67 . Blautia hydrogenotrophica , an acetogen with bidirectional metabolic cross-feeding properties (e.g., transfer of hydrogen and acetate), is also important for butyrate formation 68 . Taxa that decreased relative to the Low-responder group; Eubacterium ventriosum , Streptococcus salivarius , Eubacterium rectale , Anaerostipes hadrus , Roseburia inulinivorans , Mediterraneibacter glycyrrhizinilyticus , and Blautia massiliensis (effect size ≤ −1.690, p .adj ≤ 0.078), included butyrate producers, Eubacterium ventriosum , Eubacterium rectale , Roseburia inulinivorans , and others, such as Streptococcus salivarius , a nuclear factor kappa B (NF-κB) activity repressor 69 and Peroxisome proliferator-activated receptor gamma (PPARγ) inhibitor potentially influencing lipid and glucose metabolism 70 . Investigating monozygotic (MZ) twin pairs, Eubacterium ventriosum was more abundant in the higher BMI siblings 26 , with enhanced scavenging fermentation capabilities 71 . Roseburia inulinivorans is a mobile firmicute (flagella) that harbors a wide-ranging enzymatic repertoire able to act on various dietary polysaccharide substrates suggestive of the ability to respond to the availability of alternative dietary substrates 72 . While we noted a more variable shift in fecal total SCFAs, acetate, propionate, butyrate, or valerate (via targeted GC–MS), in the Low weight loss responders, there was no significant difference when compared to High weight loss responders (Wilcoxon rank-sum test, p ≥ 0.210; Supplementaryl Fig. S4a ; Supplementary Data 9 ).
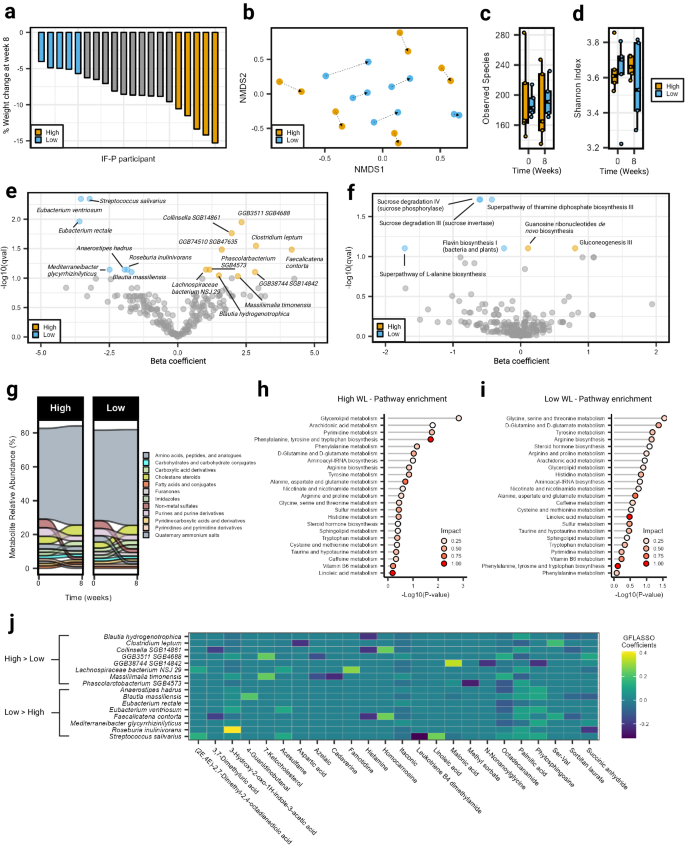
a Relative weight loss over the eight-week intervention for each participant in the IF-P group. b NMDS ordination showed the personalized trajectories of participants’ microbiomes over time. Dotted lines connect the same individual and point toward the final sample collection. No significant time or group × time interaction effects for alpha diversity metrics, c observed species, and d the Shannon index. Box and whiskers plots display the box ranging from the first to the third quartile, and the center the median value, while the whiskers extend from each quartile to the minimum or maximum values. Volcano plots displaying differential abundance between High and Low weight loss responders for e microbial species and f functional pathways. Significant features were more enriched in High and Low weight loss responders colored orange and light blue, respectively. g Alluvial plot displaying the fecal metabolite profile at the subclass level (Human Microbiome Database). Most abundant metabolite subclasses displayed (i.e., ≥1%). Metabolome pathway analysis for h High and i Low weight loss responders using all reliably detected fecal metabolites showing altered pathways with moderate and above impact (>0.10). Impact was calculated using a hypergeometric test, while significance was determined using a test of relative betweenness centrality. j Grid-fused least absolute shrinkage and selection operator (GFLASSO) regression of species from differential abundance analysis displayed correlative relationships with fecal metabolites. Species with greater abundance in High (High > Low) and Low (Low > High) weight loss responders are separate‘. For all panels, High: n = 5, Low: n = 5. Source data are provided as a Source Data file.
Less affected compared to taxonomic features were the 275 microbial-affiliated metabolic pathways identified after filtering, of which gluconeogenesis III and guanosine ribonucleotides de novo biosynthesis were increased (effect size ≥ 0.108, p .adj = 0.079), while super pathway of L-alanine biosynthesis, sucrose degradation IV (sucrose phosphorylase), sucrose degradation III (sucrose invertase), super pathway of thiamine diphosphate biosynthesis III, and flavin biosynthesis I (bacteria and plants) were decreased in the High relative to the Low weight loss responder group (effect size ≤ −0.247, p .adj ≤ 0.079; Fig. 5f ; Supplementary Data 10 )
As the difference in microbial shifts versus function is well established, we also tracked the fecal metabolome to better understand metabolic modification/production and identify potential microbial metabolic targets for future weight loss interventions. Overall, we reliably detected (QC relative standard deviation > 20% and mean intensity value > 1000 in 80% of samples) and annotated 607 (Human Metabolome Database) compounds across fecal samples. Notably, we found the fecal metabolite profile of both subgroups abundant in amino acids, peptides, and analogs, with decreases in sulfates, furanones, and quaternary ammonium salts and increases in cholestane steroids, carboxylic acid derivatives, and imidazoles (Fig. 5g ). Assessing metabolite changes between groups did not yield significance when comparing logFC values (Wilcoxon rank-sum test, p .adj > 0.10; Supplementary Fig. S4b ). Pathway analysis of High weight loss responders revealed prominent metabolic signatures relevant to lipid metabolism (glycerolipid and arachidonic metabolism), nucleotide turnover (pyrimidine metabolism), and aromatic amino acid formation (phenylalanine, tyrosine, and tryptophan biosynthesis; Fig. 5h , Supplementary Data 11 ). In comparison, the more prominent enriched pathways for Low weight loss responders included those related to amino acid and peptide metabolism (glycine, serine, and threonine, d-glutamine and d-glutamate, and tyrosine metabolism and arginine biosynthesis; Fig. 5i , Supplementary Data 12 ).
Finally, species captured by our differential abundance analysis were channeled into a GFLASSO model with the fecal metabolome library to select metabolically relevant compounds best predicted by microbial abundances. Restricting taxa and metabolites displaying stronger co-occurrence signals (GFLASSO coefficients > 0.02), we noted several patterns (Fig. 5j ). This included positive associations between GGB3511 SGB4688 (unclassified Firmicute) and malonic acid (important to fatty acid metabolism), as well as Roseburia inulinivorans and 3-Hydroxy-2-oxo-1H-indole-3-acetic acid. Negative associations included Phascolarctobacterium SGB4573 with the fatty acid ester, methyl sorbate, and Streptococcus salivarius (anti-inflammatory) with leukotriene B4 dimethylamide.
Differences detected in our subgroup analysis suggest that the GM composition plays a role in WL responsiveness during IF-P interventions. Notable differences in taxa and fecal metabolites suggest differing substrate utilization capabilities and nutrient-acquiring pathways between High and Low responders, despite being on the same dietary regimen. Although differences between High and Low responders were statistically significant for the microbiome data, the magnitude of differences varied, suggesting further research is needed to clarify these differences.
Long-term IF-P remodels the gut microbiome after substantial weight loss – A case study
Considering the microbiomic and metabolic importance of sustained WL, we additionally performed a longitudinal, exploratory case study analysis on the participant who lost the most body weight during the eight-week WL period (−15.3% BW, −24.9 kg). Under rigorous clinical supervision, this individual was guided through and comprehensively tracked over 52 weeks, strictly adhering to an IF-P regimen, including WL (0–16 weeks) and maintenance (16–52 weeks) periods, which included adjusting the calorie intake to maintain energy balance. Microbial richness and evenness at the species level displayed a general inverse trend with body weight reduction, although they converged at 52 weeks (Fig. 6a, b ). Species dissimilarity peaked at weeks four and 16, after which it plateaued, but remained consistently higher in comparison to baseline over the 52-week period (Fig. 6c ). Examining positive linear coefficients of a PERMANOVA model, constructed to detect variation between community compositions over time, dominant influences included several species within the Lachnospiraceae family such as Fusicatenibacter saccharivorans , Blautia wexlerae , Blautia massillensis , Anaerostipes hadrus , and Coprococcus comes and others like Akkermansia muciniphila (Fig. 6d ). Negative contributions included species from the Oscillospiraceae family, such as Ruminococcus bromii and Ruminococcus torques . Indeed, visualizing community composition over the sampling time points suggested specific GM remodeling (Fig. 6e ; Supplementary Data 13 ). Many keystone taxa prominent over time in the microbiome are highly relevant to the significant reduction in body weight and metabolic improvement of the case-study participant. For example, Blautia wexlerae , a commensal bacterium recently reported to confer anti-adipogenesis and anti-inflammatory properties to adipocytes 73 became visually more prominent over time. This association was also the case for the health-associated microbe, Anaerostipes hadrus , which converts inositol stereoisomers (including myoinositol) to propionate and acetate, apt to improve insulin sensitivity and reduce serum triglyceride levels 74 , translating to reduced host metabolic disease risk 75 . Other elevated taxa, like the mucin-degrading Akkermansia muciniphila and Bacteroides faecis , are negatively correlated with markers for insulin resistance 76 . There was also a notable bloom of Collinsella SGB14861 (anaerobic pathobiont producing lactate) 63 and suppression of Eubacterium rectale , Ruminococcus torques (associated with circadian rhythm disruption in mice) 77 , and Ruminococcus bromii (an exceptional starch degrader) 78 .
Change in alpha diversity metrics a observed species and b Shannon index with percentage of baseline body weight. c Bray-Curtis dissimilarity at the species level with d top PERMANOVA model coefficients (analysis: species~time). e Alluvial plot displaying the variation in abundance of the 20 most prevalent bacteria over time. For visual clarity, the less abundant taxa are not displayed. f Canberra distance of fecal metabolome with g top PERMANOVA model coefficients (analysis: pathway~time). h Pathway analysis of fecal metabolites comparing baseline to subsequent sample collections. Data are plotted as -log10(p) versus pathway impact. Node size corresponds to the proportion of metabolites captured in each pathway set, while node color signifies significance. Impact was calculated using a hypergeometric test, while significance was determined using a test of relative betweenness centrality. No p -value adjustments were made. Source data are provided as a Source Data file.
Compared to the more pronounced shifts in the GM, an inspection of Bray-Curtis dissimilarity at the microbial metabolic pathway level was much less affected (Supplementary Fig. S5a ). Though positive contributions in multiple biosynthesis pathways were noted, as well as reductions in the superpathway of UDP-glucose-derived O-antigen building blocks biosynthesis and glucose and glucose-1-phosphate degradation (Supplementary Fig. S5b ; Supplementary Data 14 ). We also tracked the fecal metabolome concordance with the GM to corroborate potential metabolic output. Shifts in metabolites captured by calculating the Canberra distance were prominent (Fig. 6f ), with positive influences from agrocybin (possessing antifungal activity 79 ), nicotinic acid (nicotinamide adenine dinucleotide precursor), and sulfate, and reductions in cadaverine (involved in the inhibition of intestinal motility 80 ), maltitol, acetohydroxamic acid (a urease inhibitor), and hypoxanthine, after removing the dominant amino acid subclass (Fig. 6g ; Supplementary Fig. S5c ). At the chemical class level, we observed apparent shifts in chemical subclasses; cholestane steroids, amines, purines, and purine derivatives, and amino acids, peptides, and analogs (Supplementary Fig. S5d ). Given our case-study approach, we performed a pathway analysis using all reliably detected fecal metabolites at each collection point over 52 weeks. Pathway analysis (Fig. 6h ) identified primary bile acid biosynthesis ( p = 0.014) and cysteine and methionine metabolism ( p = 0.096) as having the greatest significance, while the greatest impact (I) was observed in phenylalanine, tyrosine, and tryptophan biosynthesis and linoleic acid metabolism ( I = 1.0). Alanine, aspartate, and glutamate metabolism ( I = 0.756), vitamin B6 metabolism ( I = 0.647), sulfur metabolism ( I = 0.532), phenylalanine metabolism (I = 0.357), and nicotinate and nicotinamide metabolism ( I = 0.194) also displayed marked pathway impacts (Supplementary Fig. S5e ; Supplementary Data 15 ). Together, these integrated findings from the group comparisons (IF-P vs. CR), high vs. low responders, and the case study, suggest that the remodeling of the gut microbiome through sustained weight loss on an IF-P regimen not only alters the microbial composition but also influences key metabolic pathways and output, reflective of fat mobilization and metabolic improvement.
Our study demonstrates distinct effects of IF-P on gut symptomatology and microbiome, as well as circulating metabolites compared to continuous CR. We observed significant changes in the GM response to both interventions; however, the IF-P group exhibited a more pronounced community shift and greater divergence from baseline (i.e., intra-individual Bray-Curtis dissimilarities). This shift was characterized by increased specific microbial families and genera, such as Christensenellaceae , Rikenellaceae , and Marvinbryantia , associated with favorable metabolic profiles. Furthermore, IF-P significantly increased circulating cytokine concentrations of IL-4, IL-6, IL-8, and IL-13. These cytokines have been linked to lipolysis, WL, inflammation, and immune response. The plasma metabolome analysis revealed distinct metabolite signatures in IF-P and CR groups, with the convergence of multiple metabolic pathways. These findings shed light on the differential effects of IF regimens, including IF-P as a promising dietary intervention for obesity management and microbiotic and metabolic health.
While acknowledging individual contributions of WL, protein pacing, and IF, we propose that the beneficial shifts observed may be best characterized as the culmination of features inherent in our IF-P approach. For example, it is possible that microbial competition is leveraged during reduced and intermittent nutritional input periods, emphasizing nutrient composition and food matrix type (combination of whole food and meal replacements vs. primarily whole food), affecting available substrates for gut microbes. IF-P participants’ fiber intake was concentrated in fiber-rich (RS5 type) shakes, offering immediate availability of fiber to the GI tract. In contrast, CR participants consumed fiber through whole foods, leading to a slower digestion and absorption process influenced by individual digestive transit times and enzymatic profiles. This nutritional environment may create ecological niches that support symbiont microbial communities. In this investigation, we provide support of such remodeling, with intentional fasting and increased relative protein (protein pacing) consumption well-validated to improve body composition and metabolism during weight loss 7 , 8 , 15 . Our results align with previous studies on CR, where greater relative protein intake was associated with an increased abundance of Christensenella 81 . This increase is likely a result of increased amino acid-derived metabolites 21 . We also observed increased signatures of amino acid metabolism in the GM of IF-P participants, which may be attributed to increased nitrogen availability, prompting de novo amino acid biosynthesis. The liquid format of two of the daily meals and precise timing of high-quality protein consumption (Protein Pacing) in the IF-P regimen may have influenced these results, as amino acids play essential roles in microbial communities, acting as energy and nitrogen sources and essential nutrients for amino acid auxotrophs.
In addition to the differences in nutrient composition, the IF-P group exhibited a profound reduction (33%) in visceral fat 15 . This reduction is significant because visceral fat is highly correlated with GM. While the specific influence of GM on fat depots in our study remains unclear, the shift in cytokine profile and metabolic pathways suggests an interaction between GM and fat metabolism. Regarding GM-host interaction, we did not detect changes in gut permeability assaying LBP. However, correlations were found with cytokines IL-4 and IL-13 and microbes Colidextribacter (negative association) and Ruminoccus gauveauii group (positive association). These associations may reflect the direct impact of the dietary intervention, yet they also hint at a deeper crosstalk within the gut-immune axis. This crosstalk is known to play a pivotal role in modulating host inflammation and influencing adipose tissue signaling pathways 42 . Furthermore, the observed microbial shifts, including changes in populations of Christensenella , suggest a nuanced role for certain microbes in regulating metabolic health. Notably, certain strains of Christensenella have been implicated in the regulation of key metabolic markers, such as glycemia and leptin levels, and in promoting hepatic fat oxidation 82 .
Our findings also underscore that GM composition plays a role in WL responsiveness during IF-P interventions. Subgroup analysis based on WL responsiveness revealed significant differences in species composition at the taxonomic level. The High-responder group showed an increased abundance of certain bacteria associated with metabolic benefits and anti-inflammatory effects. In contrast, the Low-responder group exhibited an increased abundance of butyrate-producing and nutritionally adaptive species (e.g., Eubacterium ventriosum 71 and Roseburia inulinivorans 72 ). Fecal metabolome analysis further highlighted differences between the two subgroups, with distinct metabolic signatures and enrichment in specific metabolic pathways. Notably, the High WL responders displayed enrichment of fecal metabolites involved in lipid metabolism. In contrast, Low responders were more prominent in pathways related to the metabolism of amino acids and peptides, including glycine, serine, and threonine, d-glutamine, and d-glutamate, as well as tyrosine metabolism and arginine biosynthesis. The latter metabolic signature has been reported in individuals with severe obesity undergoing high-protein, low-calorie diets 83 . As both High and Low WL responders were consuming the same diet, our results suggest differences in GM composition and metabolism, which could play a role in determining the success of an IF-P regimen. Though, as these enrichment analyses were performed in an exploratory manner, we acknowledge the need for a more systematic approach to validate these findings.
Finally, we provide evidence of long-term GM stabilization from these changes by following one individual over 12 months. Dietary restriction is widely used to reduce fat mass and weight in individuals with or without obesity; however, weight regain after such periods presents a critical challenge, and the underlying homeostatic mechanisms remain largely elusive. Notably, keystone taxa that became more prominent over time were associated with anti-adipogenesis, improved insulin sensitivity, and reduced metabolic disease risk. The microbial shifts were accompanied by noticeable changes in the fecal metabolome, with shifts in various metabolites and chemical subclasses. Pathway analysis identified impacts on primary bile acid biosynthesis, cysteine and methionine metabolism, and other fat mobilization and metabolic improvement pathways. These shifts were accompanied by noticeable changes in the fecal metabolome, particularly in metabolites and chemical subclasses related to lipid metabolism, nucleotide turnover, and aromatic amino acid formation.
Despite the valuable insights from our study on the complex interactions between intermittent fasting, higher protein intake using protein pacing, the GM, and circulating metabolites in obese individuals, several limitations should be acknowledged. First, our reliance on fecal samples to represent the GM may have overlooked potential microbial populations in the upper GI tract. Including samples from proximal regions in future studies would provide a more comprehensive understanding of the gut microbiome’s response to IF-P and CR. In addition, the sample size for our study was determined based on the primary outcomes related to body weight and composition from the parent study 15 . This sample size may have reduced statistical power and potentially amplified individual variability among participants. However, it is important to note that the smaller RCT design allowed for more precise control over diet and lifestyle factors, minimizing potential confounding influences on the study outcomes. Furthermore, the study’s duration was limited to eight weeks, which prevented potential insights into the differential long-term effects between the two interventions. However, we were able to extend the follow-up duration and conduct periodic assessments for a year in our case-study participant, offering a more comprehensive understanding of the sustainability of the observed changes and the potential for weight regain for IF-P. The current study compared a combination of whole food and supplements (shakes and bars; IF-P) versus primarily whole food (CR), which together with variations in protein and fiber content and type may have influenced the gut symptomatology and nutrient absorption between groups. Additionally, study participants self-reported dietary intake daily, although there was close monitoring of intake through the return of empty food packaging/containers of consumed food and daily monitoring by investigators and weekly meetings with a registered dietitian. Overall, knowledge gaps are present in this research, including how the microbiome is rebuilt after food reintroduction and how overall caloric restriction and specific macronutrients contribute to this process. However, considering the multifactorial nature of weight loss and metabolic health, our work represents an important precedent for future work. Future investigators should consider integrating these factors to provide a more comprehensive understanding of the underlying mechanisms. Additional research is warranted to characterize the metabolic signature of IF-P, the time relationship between these fasting periods, and the analysis of these metabolic changes. A strength of our High-Low-responder and case-study analyses is the hypothesis-driving nature of the findings, from which targeted microbiome and/or precision nutrition interventions can be designed and tested.
In conclusion, our study provides valuable insights into the complex interactions among intermittent fasting and protein pacing, the GM, and circulating metabolites in individuals with obesity. Specifically, intermittent fasting - protein pacing significantly reduces gut symptomatology and increases gut microbes associated with a lean phenotype ( Christensenella ) and circulating cytokines mediating total body weight and fat loss. These findings highlight the importance of personalized approaches in tailoring dietary interventions for optimal weight management and metabolic health outcomes. Further research is necessary to elucidate the underlying mechanisms driving these associations and to explore the therapeutic implications for developing personalized strategies in obesity management. Additionally, future studies should consider investigating microbial populations in upper GI sections and potential intestinal tissue remodeling to gain a more comprehensive understanding of the gut microbiome’s role in these interventions.
Study design and participants
The protocol of the clinical trial was registered on March 6, 2020 (Clinicaltrials.gov; NCT04327141), and the results of the primary analysis have been published previously 15 . Briefly, participants were recruited from Saratoga Springs, NY, and were provided informed written consent in accordance with the Skidmore College Human Subjects Institutional Review Board before participation (IRB#: 1911-859), including consent for the use of samples and data from the current study. Each procedure performed was in adherence with New York state regulations and the Federal Wide Assurance, which follows the National Commission for the Protection of Human Subjects of Biomedical and Behavioral Research, and in agreement with the Helsinki Declaration (revised in 1983). Their physicians performed a comprehensive medical examination/history assessment to rule out any current cardiovascular or metabolic disease. For at least six months before the start of the study, all eligible participants were either sedentary or lightly active (<30 min, two days/week of organized physical activity), with overweight or obesity (BMI > 27.5 kg/m2; % body fat > 30%), weight stable (±2 kg), and middle-aged (30–65 years). In addition, participants taking antibiotics, antifungals, or probiotics within the previous two months were excluded. Enrolled participants were matched for body weight, BMI, and body fat and randomly assigned to one of two groups: (a) IF-P ( n = 21; 14 women; 7 men) or (b) CR ( n = 20; 12 women; 8 men) for eight weeks. During a one-week run-in period, subjects maintained a stable body weight by consuming a similar caloric intake as their pre-enrollment caloric intake while maintaining their sedentary lifestyle. This was confirmed by matching their pre-enrollment dietary intake to the one-week run-in diet period 15 . Following baseline testing, participants were provided detailed instructions on their weight loss dietary regimen (Supplementary Table S1 ) and received weekly dietary counseling and compliance/adherence monitoring from the research team via daily food records, and weekly registered dietitian meetings, along with weekly visits to the Human Nutrition and Metabolism laboratory at Skidmore College (Saratoga Springs, NY) for meal distribution and empty packet/container returns. All outcome variables were assessed pre (week 0), mid (week 4), and post (week 8). All participants were compensated $100 for successful completion of the study and received an additional monthly stipend of $75 for groceries (CR group only) or up to two meals per day of food supplements and meal replacements (IF-P only).
IF days consisted of ~350–550 kcals per day, in which participants were provided a variety of supplements and snacks. Protein pacing (P) days for IF-P consisted of four and five meals/day for women and men, respectively, two of which (breakfast and one other meal) were liquid meal replacement shakes with added whole foods (Whole Blend IsaLean® Shakes, 350/400 kcals, 30/36 g of protein/meal, 9 g of fiber); a whole food evening dinner meal (450/500 kcals men), an afternoon snack (200 kcals, men only), and an evening protein snack (IsaLean® or IsaPro® Shake or IsaLean Whole Blend® Bar; 200–250 kcals). This dietary regimen provided 1350–1500 and 1700–1850 kcals/day for women and men, respectively, and a macronutrient distribution targeting 35% protein, 35% carbohydrate, 20–30 g/day of fiber, and 30% fat. Isagenix International, LLC (Gilbert, AZ, USA) provided all meal replacement shakes, bars, beverages, and supplements. In comparison, participants assigned to the CR diet followed specific guidelines of the National Cholesterol Education Program Therapeutics Lifestyle Changes (TLC) diet of the American Heart Association with a strong Mediterranean diet influence of a variety of fresh vegetables, fruits, nuts, and legumes. The specific macronutrient distribution recommended was <35% of kcal as fat; 50%–60% of kcal as carbohydrates; 15% kcal as protein; <200 mg/dL of dietary cholesterol; and 20–30 g/day of fiber. The total calorie intake was 1200 and 1500 calories per day for women and men, respectively, during the 8-week weight loss intervention. In addition to weekly meetings with the registered dietitian and daily contact with research team members, subjects were provided detailed written instructions for their meal plans. They were closely monitored through daily participant-researcher communication (e.g., email, text, and mobile phone), two-day food diary analysis, weekly dietary intake journal inspections, weekly meal/supplement container distribution, and returning empty packets and containers.
Gastrointestinal (GI) symptom rating scale
Participants completed the 15-question GI symptom rating scale (GSRS) 84 at baseline, week four, and week eight. Briefly, each question is rated on a 7-point Likert scale (1 = absent; 2 = minor; 3 = mild; 4 = moderate; 5 = moderately severe; 6 = severe and 7 = very severe) and recalled from the previous week. Questions include symptoms related to upper abdominal pain, heartburn, regurgitation (acid reflux), empty feeling in the stomach, nausea, abdominal rumbling, bloating, belching, flatulence, and questions on defecation. The GSRS questionnaire provides explanations of each symptom, is understandable, and has reproducibility for measuring the presence of GI symptoms 85 . In our analysis, a score of ≥2 (minor) was defined as symptom presence, and a score ≥ 4 (moderate) was defined as moderate symptom presence. Furthermore, to better categorize symptom location, bloating, flatulence, constipation, diarrhea, stool consistency, defecation urgency, and sensation of not completely emptying bowels were classified as lower GI symptoms, and nausea, heartburn, regurgitation, upper abdominal pain, empty feeling in the stomach, stomach rumbling, and belching was classified as upper GI symptoms. Total scores were also generated for overall symptom and moderate symptom presence.
Fecal sample collection and DNA extraction
Participants were instructed to provide stool samples at baseline, week four, and week eight of the intervention. The case-study participant additionally provided samples at weeks 12, 16, 32, and 52. The entire bowel movement was collected and transported within 24 h of defecation to the Skidmore College Human Nutrition and Metabolism (Saratoga Springs, NY) laboratory using a cooler and ice packs and frozen at −80 °C. Samples were then sent to ASU (Phoenix, AZ) overnight on dry ice for analysis, where they were thawed at 4 °C and processed. Wet weight was recorded to the nearest 0.01 g after subtracting the weight of fecal collection materials. Stool samples were then rated according to the BSS 86 , homogenized in a stomacher bag, and the pH was measured (Symphony SB70P, VWR International, LLC., Radnor, PA, USA). Next, the extraction of DNA was performed using the DNeasy PowerSoil Pro Kit (Cat. No. 47016, Qiagen, Germantown, MD) per the manufacturer’s instructions. DNA concentration and quality were quantified using the NanoDrop™ OneC Microvolume UV-Vis Spectrophotometer (Thermo Scientific™, Waltham, MA) according to manufacturer instructions. The OD 260 /OD 280 ratio of all samples was ≥1.80 (demonstrating DNA purity).
Quantification of bacterial 16S rRNA genes
To estimate total bacterial biomass per sample (16S rRNA gene copies per gram of wet stool), DNA extracted from the fecal collections was assessed via quantitative polymerase chain reaction (qPCR) based on previously published methods 87 , 88 . Briefly, all 20 μL qPCR reactions contained 10 uL of 2X SYBR Premix Ex Taq ™ (Tli RNase H Plus) (Takara Bio USA, Inc., San Jose, CA, USA), 0.3 μM (0.6 μL) of each primer (926 F: AAACTCAAAKGAATTGACGG; 1062 R: CTCACRRCACGAGCTGAC), 2 μL DNA template (or PCR-grade water as negative control), and 6.8 μL nuclease-free water (Thermo Fisher Scientific, Waltham, MA, USA). PCR thermal cycling conditions were as follows: 95 °C for 5 min, followed by 35 cycles of 95 °C for 15 s, 61.5 °C for 15 s, and 72 °C for 20 s, then hold at 72 °C for 5 min, along with a melt curve of 95 °C for 15 s, 60 °C for 1 min, then 95 °C for 1 s. Quantification was performed using a QuantStudio3™ Real-Time PCR System by Applied Biosystems with QuantStudio Design and Analysis Software 1.2 from Thermo Fisher Scientific (Waltham, MA, USA). All samples were analyzed in technical replicates. For quality assurance and quality control, molecular negative template controls (NTC) consisting of PCR-grade water (Invitrogen, Waltham, MA, USA) and positive controls created by linearized plasmids were run on every qPCR plate. Standard curves were run-in triplicate and used for sample quantification, ranging from 10 7 to 10 1 copies/μL with a cycle threshold (CT) detection limit cutoff of 33. Reaction efficiency was approximately 101%, with a slope of −3.29 and R 2 ≥ 0.99.
Fecal microbiome analysis
Amplification of the 16S rRNA gene sequence was completed in triplicate PCRs using 96-well plates. Barcoded universal forward 515 F primers and 806 R reverse primers containing Illumina adapter sequences, which target the highly conserved V4 region, were used to amplify microbial DNA 89 , 90 . PCR, amplicon cleaning, and quantification were performed as previously outlined 90 . Equimolar ratios of amplicons from individual samples were pooled together before sequencing on the Illumina platform (Illumina MiSeq instrument, Illumina, Inc., San Diego, CA). Raw Illumina microbial data were cleaned by removing short and long sequences, sequences with primer mismatches, uncorrectable barcodes, and ambiguous bases using the Quantitative Insights into Microbial Ecology 2 (QIIME2) software, version 2021.8 91 .
16S rRNA sequencing produced 7,366,128 reads with a median of 53,776 per sample (range: 9512–470,848). Paired-end, demultiplexed data were imported and analyzed using QIIME2 software. Upon examination of sequence quality plots, base pairs were trimmed at position 20 and truncated at position 240 and were run through DADA2 to remove low-quality regions and construct a feature table using ASVs. Next, the ASV feature table was passed through the feature-classifier plugin 92 , which was implemented using a naive Bayes machine-learning classifier, pre-trained to discern taxonomy mapped to the latest version of the rRNA database SILVA (138.1; 99% ASVs from 515 F/806 R region of sequences) 93 . Based on an assessment of alpha rarefaction, a threshold of 6500 sequences/sample was established, retaining all samples for downstream analysis. A phylogenic tree was then constructed using the fragment-insertion plugin with SILVA at a p-sampling depth of the rarefaction threshold to impute high-quality reads and normalize for uneven sequencing depth between samples 94 . Alpha diversity (intra-community diversity) was measured using observed ASVs and the Phylogenetic diversity index. Additionally, the Shannon index was calculated for the subgroup and case-study analyses to capture richness and evenness at the species level. Beta diversity (inter-community diversity) was measured using Bray-Curtis dissimilarity.
For shotgun metagenomics, DNA was sequenced on the Illumina NextSeq 500 platform (Illumina, CA, USA) to generate 2 × 150 bp paired-end reads at greater sequencing depth with a minimum of 10 million reads. Raw Illumina sequencing reads underwent standard quality control with FastQC. Adapters were trimmed using TrimGalore. DNA sequences were aligned to Hg38 using bowtie2 95 . DNA sequences were then analyzed via the bio bakery pipeline 96 for taxonomic composition and potential functional content with MetaPhlAn4 and HUMAnN 3.0 (UniRef90 gene-families and MetaCyc metabolic pathways), using standard parameters. Functional profiling resulted in 8528 distinct Kyoto Encyclopedia of Genes and Genomes Orthology (KO) groups and 511 metabolic pathways, which align with previous human gut microbiome studies 96 .
Blood sample collection and biochemical analyses
All participants were tested between the hours of 6:00 a.m. and 9:00 a.m., after an overnight fast for body composition assessments (height, body weight, and total body composition) at weeks 0, 4, and 8. 12-h fasted venous blood samples (~20 mL) were collected into EDTA-coated vacutainer tubes and centrifuged (Hettich Rotina 46R5) for 15 min at 4000 × g at −4 °C. After separation, plasma was stored at −80 °C until analyzed. Undiluted plasma samples were sent to Eve Technologies (Calgary, Alberta, Canada) for assessment of inflammatory cytokines [Granulocyte-macrophage colony-stimulating factor [GM-CSF], interferon-γ (IFNγ), interleukin (IL)-β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, IL-13, IL-17A, IL-23, and Tumor necrosis factor-α (TNFα)] using a high human sensitivity 14-plex cytokine assay (Millipore, Burlington, MA). Circulating LBP concentrations were quantified in duplicate using 1000x diluted plasma samples. A commercially available kit was used per the manufacturer’s protocol (Cat No. EH297RB, Thermo Fisher Scientific, Inc, Waltham, MA; intra-assay coefficient variation [CV] <10%).
Targeted plasma metabolomic analysis
For the plasma metabolomic analysis, a 12-h fasted venous blood sample (~20 mL) was collected into EDTA-coated vacutainer tubes and centrifuged (Hettich Rotina 46R5) for 15 min at 4000 × g at 4 °C. After separation, 2 mL of plasma was aliquoted and stored at −80 °C at the Biochemistry Laboratory at Skidmore College (Saratoga Springs, NY, USA). Samples were then sent to the Arizona Metabolomics Laboratory at ASU (Phoenix, AZ, USA) overnight on dry ice for analysis, where they were thawed at 4 °C and processed. Briefly, 50 μL of plasma from each sample was processed to precipitate proteins and extract metabolites by adding 500 μL MeOH and 50 μL internal standard solution (containing 1810.5 μM 13 C 3 -lactate and 142 μM 13 C 5 -glutamic acid). The mixture was vortexed (10 s) and stored for 30 min at –20 °C, then centrifuged at 224,000 × g for 10 min at 4 °C. Supernatants (450 μL) were extracted, transferred to new Eppendorf vials, and dried (CentriVap Concentrator; Labconco, Fort Scott, KS, USA). Samples were then reconstituted in 150 μL of 40% phosphate-buffered saline (PBS)/60% acetonitrile (ACN) and centrifuged again at 22,000 × g at 4 °C for 10 min. Supernatants (100 µL) were transferred to an LC autosampler vial for subsequent analysis. Quality control (QC) was performed by creating a pooled sample from all plasma samples and injecting once every ten experimental samples to monitor system performance.
The highly-reproducible targeted LC–MS/MS method used in the current investigation was modeled after previous studies 97 , 98 , 99 . The specific metabolites included in our targeted detection panel are representative of more than 35 biological pathways most essential to biological metabolism and have been successfully leveraged for the sensitive and broad detection of effects related to diet 100 , diseases 101 , drug treatment 102 , environmental contamination 103 , and lifestyle factors 104 . Briefly, LC–MS/MS experiments were performed on an Agilent 1290 UPLC-6490 QQQ-MS system (Santa Clara, CA, USA). Each sample was injected twice for analysis, 10 µL using negative and 4 µL using positive ionization modes. Chromatographic separations were performed in hydrophilic interaction chromatography (HILIC) mode on a Waters Xbridge BEH Amide column (150 × 2.1 mm, 2.5 µm particle size, Waters Corporation, Milford, MA, USA). The flow rate was 0.3 mL/min, the autosampler temperature was maintained at 4 °C, and the column compartment was set at 40 °C. The mobile phase system was composed of Solvents A (10 mM ammonium acetate, 10 mM ammonium hydroxide in 95% H 2 O/5% ACN) and B (10 mM ammonium acetate, 10 mM ammonium hydroxide in 95% ACN/5% H 2 O). After the initial 1 min isocratic elution of 90% Solvent B, the percentage of Solvent B decreased to 40% at t = 11 min. The composition of Solvent B was maintained at 40% for 4 min ( t = 15 min).
The mass spectrometer was equipped with an electrospray ionization (ESI) source. Targeted data acquisition was performed in multiple-reaction monitoring (MRM) mode. The LC–MS system was controlled by Agilent MassHunter Workstation software (Santa Clara, CA, USA), and extracted MRM peaks were integrated using Agilent MassHunter Quantitative Data Analysis software (Santa Clara, CA, USA).
GC–MS fecal short-chain fatty acid analysis
Before GC–MS analysis of SCFAs, frozen fecal samples were first thawed overnight under 4 °C. Then, 20 mg of each sample was homogenized with 5 μL hexanoic acid—6,6,6-d 3 (internal standard; 200 µM in H 2 O), 15 μL sodium hydroxide (NaOH [0.5 M]), and 500 μL MeOH. Samples were stored at −20 °C for 20 min and centrifuged at 22,000 × g for 10 min afterward. Next, 450 μL of supernatant was collected, and the sample pH was adjusted to 10 by adding 30 μL of NaOH:H 2 O (1:4, v-v). Samples were then dried, and the residues were initially derivatized with 40 µL of 20 mg/mL MeOX solution in pyridine under 60 °C for 90 min. Subsequently, 60 µL of MTBSTFA containing d 27 -mysristic acid was added, and the mixture was incubated at 60 °C for 30 min. The samples were then vortexed for 30 s and centrifuged at 22,000 × g for 10 min. Finally, 70 µL of supernatant was collected from each sample and injected into new glass vials for GC–MS analysis.
GC–MS conditions used here were adopted from a previously published protocol 105 . Briefly, GC–MS experiments were performed on an Agilent 7820 A GC-5977B MSD system (Santa Clara, CA); all samples were analyzed by injecting 1 µL of prepared samples. Helium was the carrier gas with a constant flow rate of 1.2 mL/min. Separation of metabolites was achieved using an Agilent HP-5 ms capillary column (30 m × 250 µm × 0.25 µm). Ramping parameters were as follows: column temperature was maintained at 60 °C for 1 min, increased at a rate of 10 °C/min to 325 °C, and then held at this temperature for 10 min. Mass spectral signals were recorded at an m/z range of 50–600, and data extraction was performed using Agilent Quantitative Analysis software. Following peak integration, metabolites were filtered for reliability. Only those with QC CV < 20% and a relative abundance of 1000 in > 80% of samples were retained for statistical analysis.
Untargeted fecal metabolomic analysis
Briefly, each fecal sample (~20 mg) was homogenized in 200 µL MeOH:PBS (4:1, v-v, containing 1810.5 μM 13 C 3 -lactate and 142 μM 13 C 5 -glutamic Acid) in an Eppendorf tube using a Bullet Blender homogenizer (Next Advance, Averill Park, NY). Then 800 µL MeOH:PBS (4:1, v-v, containing 1810.5 μM 13 C 3 -lactate and 142 μM 13 C 5 -glutamic Acid) was added, and after vortexing for 10 s, the samples were stored at −20 °C for 30 min. The samples were then sonicated in an ice bath for 30 min. The samples were centrifuged at 22,000 × g for 10 min (4 °C), and 800 µL supernatant was transferred to a new Eppendorf tube. The samples were then dried under vacuum using a CentriVap Concentrator (Labconco, Fort Scott, KS). Prior to MS analysis, the obtained residue was reconstituted in 150 μL 40% PBS/60% ACN. A quality control (QC) sample was pooled from all the study samples.
The untargeted LC–MS metabolomics method used here was modeled after that developed and used in a growing number of studies 106 , 107 , 108 . Briefly, all LC–MS experiments were performed on a Thermo Vanquish UPLC-Exploris 240 Orbitrap MS instrument (Waltham, MA). Each sample was injected twice, 10 µL for analysis using negative ionization mode and 4 µL for analysis using positive ionization mode. Both chromatographic separations were performed in hydrophilic interaction chromatography (HILIC) mode on a Waters XBridge BEH Amide column (150 × 2.1 mm, 2.5 µm particle size, Waters Corporation, Milford, MA). The flow rate was 0.3 mL/min, autosampler temperature was kept at 4 °C, and the column compartment was set at 40 °C. The mobile phase was composed of Solvents A (10 mM ammonium acetate, 10 mM ammonium hydroxide in 95% H 2 O/5% ACN) and B (10 mM ammonium acetate, 10 mM ammonium hydroxide in 95% ACN/5% H 2 O). After the initial 1 min isocratic elution of 90% B, the percentage of Solvent B decreased to 40% at t = 11 min. The composition of Solvent B maintained at 40% for 4 min ( t = 15 min), and then the percentage of B gradually went back to 90%, to prepare for the next injection. Using mass spectrometer equipped with an electrospray ionization (ESI) source, we collected untargeted data from 70 to 1050 m/z.
To identify peaks from the MS spectra, we made extensive use of the in-house chemical standards (~600 aqueous metabolites), and in addition, we searched the resulting MS spectra against the HMDB library, Lipidmap database, METLIN database, as well as commercial databases including mzCloud, Metabolika, and ChemSpider. The absolute intensity threshold for the MS data extraction was 1000, and the mass accuracy limit was set to 5 ppm. Identifications and annotations used available data for retention time (RT), exact mass (MS), MS/MS fragmentation pattern, and isotopic pattern. We used the Thermo Compound Discoverer 3.3 software for aqueous metabolomics data processing. The untargeted data were processed by the software for peak picking, alignment, and normalization. To improve rigor, only the signals/peaks with CV < 20% across quality control (QC) pools, and the signals showing up in >80% of all the samples were included for further analysis. To ensure the robustness of our model validation, we employed an enhanced validation approach by repeating the LOOCV process 100 times. Each iteration involves excluding one sample from the dataset to serve as the test set, with the model being trained on the remaining samples. This approach, referred to as ‘repeated LOOCV’, was adopted to mitigate bias and provide a thorough validation of our model’s predictive capability. The method signifies the number of repetitions of the LOOCV process, rather than splitting the dataset into 100 equal parts.
Multi-omics data analysis
For MOFA, bacterial 16S rRNA ASVs and plasma metabolites were integrated using the MOFA2 package 55 . Before integration, ASV sequences were filtered (minimum of 5 ASV in greater than 10% of all samples), collapsed to the genus level, and scaled using a centralized-log-ratio, as described previously 109 . Plasma metabolites were scaled and normalized as described in the metabolome analysis. The inputs for MOFA model training comprised 53 taxa and 138 metabolites. The latent factors and feature loadings were extracted from the best-trained model with the built-in functions of MOFA2. After model fitting, the number of factors was estimated by requiring a minimum of 2% variance explained across all microbiome modalities.
Integrating microbial taxa with the same filtration as stated above (at the genus level from 16S amplicon sequencing and species level from metagenomic sequencing) and cytokine data and fecal metabolomic data, respectively, was conducted with GFLASSO (R package: GFLASSO, v0.0.0.9000). This correlation-based network solution can handle multiple response variables for a given set of predictors (in this case: 1. cytokine abundances predicted by microbial taxa response; and 2. fecal metabolite response predicted by microbial taxa). Solution parsimony was determined by an unweighted (i.e., presence or absence of association by imposing a correlation threshold) network structure. The regularization and fusion parameters were determined from the smallest root mean squared error (RMSE) estimate via cross-validation, accounting for interdependencies among microbial features. The tested parameters encompassed all combinations between λ and γ with values ranging from 0 to 1 (inclusive) in step increments of 0.1. GFLASSO coefficient matrices were constructed using a threshold coefficient of >0.02 to discern the strongest associative signals.
Statistical analysis
Gastrointestinal symptom scores were on the low end of the GSRS scale and not normally distributed; therefore, nonparametric statistical tests were applied. Symptom prevalence (number of scores ≥ 2) and moderate symptom prevalence (≥4) for total, upper, and lower GI GSRS clusters were analyzed using contingency tables. Specifically, differences between IF-P and CR GI symptoms at baseline were compared using a Fisher’s Exact test, whereas baseline vs. weeks four and eight values were compared with McNemar’s test. Stool weight, BSS, fecal pH, plasma cytokines and LBP, and SCFAs were assessed for normality with Q-Q plots and Shapiro-Wilk tests and log-transformed where appropriate. These were then tested for time and interaction (group × time) effects using linear-mixed effect (LME) models, with each participant included as a random effect.
For analysis and visualization of the microbiome data, artifacts generated in QIIME2 were imported into the R environment (v4.2.2) using the phyloseq package (v1.42.0) 110 . Before conducting downstream analyses, sequences were filtered to remove all non-bacterial sequences, including archaea, mitochondria, and chloroplasts. After assessing normality (Shapiro-Wilk’s tests), LME models were used to test the effect of time and the interaction of group and time with the covariates of age and sex with each participant included as a random effect on the alpha diversity metrics using the nLME package (v3.1.160). For beta diversity, a nested permutational analysis of variance (PERMANOVA) was conducted on Bray-Curtis dissimilarities using the Adonis test in the vegan package (v2.6.2) with 999 permutations. The PERMANOVA model incorporated the factors of time, individual, interaction (group × time), and participant (nested factor). A permutation test for homogeneity in multivariate dispersion (PERMDISP) was conducted using the ‘betadisper’ function in the vegan package to compare dispersion. To support the Adonis analysis, intra-individual differences were also compared between groups, as previously described 111 , by calculating the within-subject distance for paired samples (baseline vs. weeks four and eight) and testing for group distances (Wilcoxon rank-sum test). Differential abundance analysis was performed using MaAsLin2 (v1.12.0) 18 . To detect changes in microbial features between groups over time, we built linear-mixed models that include group, time, and their interaction, with age and sex as covariates and the participant as a random factor. Before analysis, raw counts from the ASV table were filtered for any sequence not present five times in at least 30% of all samples. A significant p-value for the product term indicates that changes in microbial features differed over time between groups. The Benjamini–Hochberg (BH) procedure was used to correct for multiple testing at ≤0.10. To assess the correlation between changes in specific taxa and biomarkers over the eight-week intervention, Spearman correlation tests were performed.
Univariate and multivariate analyses of plasma metabolites and metabolic ontology analysis were performed, and results were visualized using the MetaboAnalystR 5.0 112 . Human metabolomic data were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) human pathway library to analyze predicted states 113 . The data were log 10 -transformed, and Pareto scaled to approximate normality before all analyses. A GLM was constructed with age, sex, and time as covariates to determine significantly affected metabolites by group intervention. Levene’s test was performed to detect significant homogeneity. The BH procedure was used to correct for multiple testing at ≤0.10. Fecal metabolomic analysis for the subgroup comparison was performed by assessing logFC values between groups with a Wilcoxon rank-sum test with BH adjustment. For pathway analysis, the impact was calculated using a hypergeometric test, while significance was determined using a test of relative betweenness centrality. Importantly, the BH procedure was not applied to pathway and enzyme enrichment analyses for the subgroup assessment since these analyses involve testing the significance of multiple related hypotheses rather than independent hypotheses, which is too conservative, resulting in false negative results.
For MOFA, latent factors explaining ≥2.0% of model variance from the plasma metabolomic and amplicon microbiome data were used to perform Spearman correlations on anthropometric and nutritional data and compared between IF-P and CR groups using Wilcoxon rank-sum tests. The highest beta coefficients (>0.3) detected from GFLASSO models were further assessed by performing Spearman correlations of select microbial features with the response variables (i.e., cytokines and fecal metabolites). All statistical tests were performed with a significance level of p < 0.05 and BH correction of p .adj < 0.10. In addition, we present data in this study in accordance with the ‘Strengthening The Organization and Reporting of Microbiome Studies’ (STORMS) guidelines for human microbiome research 114 .
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The microbiome sequencing data generated in this study have been deposited in the BioProject Database of National Centre for Biotechnology Information database under accession code PRJNA847971 . The metadata data linking the microbiome sequences with the appropriate sample ID and intervention in this study are provided in Supplementary Data 1 . The processed data are available at https://github.com/Alex-E-Mohr/GM-Remodeling-IF-ProteinPacing-vs-CaloricRestriction . Source data are provided with this paper.
Code availability
The R code used for analysis and figure generation for reproducibility purposes are available at: https://github.com/Alex-E-Mohr/GM-Remodeling-IF-ProteinPacing-vs-CaloricRestriction . 115
Asnicar, F. et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat. Med. 27 , 321–332 (2021).
Article CAS PubMed PubMed Central Google Scholar
Li, M. et al. Gut microbiota-bile acid crosstalk contributes to the rebound weight gain after calorie restriction in mice. Nat. Commun. 13 , 2060 (2022).
Article ADS CAS PubMed PubMed Central Google Scholar
Machado, A. C. D. et al. Diet and feeding pattern modulate diurnal dynamics of the ileal microbiome and transcriptome. Cell Rep. 40 , 111008 (2022).
Article PubMed Central Google Scholar
von Schwartzenberg R.J. et al. Caloric restriction disrupts the microbiota and colonization resistance. Nature 595 , 272–277 (2021).
Maifeld, A. et al. Fasting alters the gut microbiome reducing blood pressure and body weight in metabolic syndrome patients. Nat. Commun. 12 , 1970 (2021).
Corbin, K. D. et al. Host-diet-gut microbiome interactions influence human energy balance: a randomized clinical trial. Nat. Commun. 14 , 3161 (2023).
Arciero, P. J. et al. Increased protein intake and meal frequency reduces abdominal fat during energy balance and energy deficit. Obesity 21 , 1357–1366 (2013).
Article CAS PubMed Google Scholar
Arciero, P. J. et al. Protein-pacing caloric-restriction enhances body composition similarly in obese men and women during weight loss and sustains efficacy during long-term weight maintenance. Nutrients 8 , 476 (2016).
Article PubMed PubMed Central Google Scholar
Mohr, A. E. et al. Exploratory analysis of one versus two-day intermittent fasting protocols on the gut microbiome and plasma metabolome in adults with overweight/obesity. Front. Nutr. 9 , 1036080 (2022).
He, F., Zuo, L., Ward, E. & Arciero, P. J. Serum polychlorinated biphenyls increase and oxidative stress decreases with a protein-pacing caloric restriction diet in obese men and women. Int. J. Environ. Res. Public Health 14 , 59 (2017).
Ives, S. J. et al. Multi-modal exercise training and protein-pacing enhances physical performance adaptations independent of growth hormone and BDNF but may be dependent on IGF-1 in exercise-trained men. Growth Horm. IGF Res. 32 , 60–70 (2017).
Zuo, L. et al. Comparison of high-protein, intermittent fasting low-calorie diet and heart healthy diet for vascular health of the obese. Front. Physiol. 7 , 350 (2016).
Article ADS PubMed PubMed Central Google Scholar
Zhong, W. et al. High-protein diet prevents fat mass increase after dieting by counteracting Lactobacillus-enhanced lipid absorption. Nat. Metab. 4 , 1713–1731 (2022).
U.S. Department of Agriculture and U.S. Department of Health and Human Services. Dietary Guidelines for Americans, 2020-2025 , 9th edn DietaryGuidelines.gov (2020).
Arciero, P. J. et al. Intermittent fasting and protein pacing are superior to caloric restriction for weight and visceral fat loss. Obesity 31 , 139–149 (2023).
Lichtenstein, A. H. et al. 2021 Dietary guidance to improve cardiovascular health: a scientific statement from the american heart association. Circulation 144 , e472–e487 (2021).
Article PubMed Google Scholar
Falony, G. et al. Population-level analysis of gut microbiome variation. Science 352 , 560–564 (2016).
Article ADS CAS PubMed Google Scholar
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLos Comput. Biol. 17 , e1009442 (2021).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489 , 220–230 (2012).
Waters, J. L. & Ley, R. E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 17 , 83 (2019).
Beaumont, M. et al. Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: a randomized, parallel, double-blind trial in overweight humans. Am. J. Clin. Nutr. 106 , 1005–1019 (2017).
Tavella, T. et al. Elevated gut microbiome abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae is associated with reduced visceral adipose tissue and healthier metabolic profile in Italian elderly. Gut Microbes 13 , 1880221 (2021).
Bischoff, S. C. et al. Gut microbiota patterns predicting long-term weight loss success in individuals with obesity undergoing nonsurgical therapy. Nutrients 14 , 3182 (2022).
Vieira-Silva, S. et al. Species–function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 1 , 16088 (2016).
Cummings, J. H. & Macfarlane, G. T. The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70 , 443–459 (1991).
Tims, S. et al. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 7 , 707–717 (2013).
Atzeni, A. et al. Taxonomic and functional fecal microbiota signatures associated with insulin resistance in non-diabetic subjects with overweight/obesity within the frame of the PREDIMED-plus study. Front. Endocrinol. 13 , 804455 (2022).
Article Google Scholar
Oliver, A. et al. High-fiber, whole-food dietary intervention alters the human gut microbiome but not fecal short-chain fatty acids. Msystems 6 , e00115–e00121 (2021).
McOrist, A. L. et al. Fecal butyrate levels vary widely among individuals but are usually increased by a diet high in resistant starch. J. Nutr. 141 , 883–889 (2011).
Bendiks, Z. A., Knudsen, K. E. B., Keenan, M. J. & Marco, M. L. Conserved and variable responses of the gut microbiome to resistant starch type 2. Nutr. Res. 77 , 12–28 (2020).
Boulangé, C. L., Neves, A. L., Chilloux, J., Nicholson, J. K. & Dumas, M.-E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med 8 , 42 (2016).
Shiau, M.-Y. et al. Mechanism of interleukin-4 reducing lipid deposit by regulating hormone-sensitive lipase. Sci. Rep. 9 , 11974 (2019).
Bruun, J. M., Pedersen, S. B., Kristensen, K. & Richelsen, B. Opposite regulation of interleukin‐8 and tumor necrosis factor‐α by weight loss. Obes. Res. 10 , 499–506 (2002).
Steensberg, A. et al. Production of interleukin‐6 in contracting human skeletal muscles can account for the exercise‐induced increase in plasma interleukin‐6. J. Physiol. 529 , 237–242 (2000).
Hall et al. Interleukin-6 stimulates lipolysis and fat oxidation in humans. J. Clin. Endocrinol. Metab. 88 , 3005–3010 (2003).
Wueest, S. et al. Interleukin-6 contributes to early fasting-induced free fatty acid mobilization in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 306 , R861–R867 (2014).
Wu, D. et al. Interleukin-13 (IL-13)/IL-13 receptor α1 (IL-13Rα1) signaling regulates intestinal epithelial cystic fibrosis transmembrane conductance regulator channel-dependent Cl− secretion. J. Biol. Chem. 286 , 13357–13369 (2011).
Gamage, H. K. A. H. et al. Intermittent fasting has a diet-specific impact on the gut microbiota and colonic mucin O-glycosylation of mice. Preprint at bioRxiv https://doi.org/10.1101/2022.09.15.508181 (2022).
Song, E.-J., Shin, N. R., Jeon, S., Nam, Y.-D. & Kim, H. Lorcaserin and phentermine exert anti-obesity effects with modulation of the gut microbiota. Front. Microbiol. 13 , 1109651 (2023).
Wang, M. et al. Olive fruit extracts supplement improve antioxidant capacity via altering colonic microbiota composition in mice. Front. Nutr. 8 , 645099 (2021).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464 , 59–65 (2010).
Mohr, A. E., Crawford, M., Jasbi, P., Fessler, S. & Sweazea, K. L. Lipopolysaccharide and the gut microbiota: considering structural variation. FEBS Lett. 596 , 849–875 (2022).
Teruya, T., Chaleckis, R., Takada, J., Yanagida, M. & Kondoh, H. Diverse metabolic reactions activated during 58-hr fasting are revealed by non-targeted metabolomic analysis of human blood. Sci. Rep. 9 , 854 (2019).
Collet, T.-H. et al. A metabolomic signature of acute caloric restriction. J. Clin. Endocrinol. Metab. 102 , 4486–4495 (2017).
Vázquez-Fresno, R. et al. Metabolomic pattern analysis after mediterranean diet intervention in a nondiabetic population: a 1- and 3-year follow-up in the PREDIMED study. J. Proteome Res. 14 , 531–540 (2015).
Sankaranarayanan, R., Kumar, D. R., Patel, J. & Bhat, G. J. Do aspirin and flavonoids prevent cancer through a common mechanism involving hydroxybenzoic acids?—The metabolite hypothesis. Molecules 25 , 2243 (2020).
Aon, M. A. et al. Untangling determinants of enhanced health and lifespan through a multi-omics approach in mice. Cell Metab. 32 , 100–116.e4 (2020).
Sekhar, R. V. et al. Glutathione synthesis is diminished in patients with uncontrolled diabetes and restored by dietary supplementation with cysteine and glycine. Diabetes Care 34 , 162–167 (2011).
Nguyen, D., Hsu, J. W., Jahoor, F. & Sekhar, R. V. Effect of increasing glutathione with cysteine and glycine supplementation on mitochondrial fuel oxidation, insulin sensitivity, and body composition in older HIV-infected patients. J. Clin. Endocrinol. Metab. 99 , 169–177 (2014).
Chen, L. et al. Glycine transporter-1 and glycine receptor mediate the antioxidant effect of glycine in diabetic rat islets and INS-1 cells. Free Radic. Biol. Med. 123 , 53–61 (2018).
Breum, L., Rasmussen, M. H., Hilsted, J. & Fernstrom, J. D. Twenty-four–hour plasma tryptophan concentrations and ratios are below normal in obese subjects and are not normalized by substantial weight reduction 1, 2, 3. Am. J. Clin. Nutr. 77 , 1112–1118 (2003).
Fernstrom, J. D. et al. Diurnal variations in plasma concentrations of tryptophan, tryosine, and other neutral amino acids: effect of dietary protein intake. Am. J. Clin. Nutr. 32 , 1912–1922 (1979).
Chen, L. et al. Influence of the microbiome, diet and genetics on inter-individual variation in the human plasma metabolome. Nat. Med. 28 , 2333–2343 (2022).
Fan, Y. & Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19 , 55–71 (2021).
Argelaguet, R. et al. Multi‐Omics Factor Analysis—a framework for unsupervised integration of multi‐omics data sets. Mol. Syst. Biol. 14 , e8124 (2018).
Anderson, E. M. et al. Temporal dynamics of the intestinal microbiome following short-term dietary restriction. Nutrients 14 , 2785 (2022).
Gerritsen, J. et al. A comparative and functional genomics analysis of the genus Romboutsia provides insight into adaptation to an intestinal lifestyle. Preprint at bioRxiv https://doi.org/10.1101/845511 (2019).
Caldovic, L. et al. N-acetylglutamate synthase: structure, function and defects. Mol. Genet. Metab. 100 , S13–S19 (2010).
Parker, B. J., Wearsch, P. A., Veloo, A. C. M. & Rodriguez-Palacios, A. The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 11 , 906 (2020).
Yang, J. et al. Oscillospira - a candidate for the next-generation probiotics. Gut Microbes 13 , 1987783 (2021).
Jensen, M. D. et al. 2013 AHA/ACC/TOS guideline for the management of overweight and obesity in adults. Circulation 129 , S102–S138 (2014).
Gomez-Arango, L. F. et al. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes 9 , 189–201 (2018).
Chen, J. et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 8 , 43 (2016).
Astbury, S. et al. Lower gut microbiome diversity and higher abundance of proinflammatory genus Collinsella are associated with biopsy-proven nonalcoholic steatohepatitis. Gut Microbes 11 , 569–580 (2020).
Lim, R. R. X. et al. Gut microbiome responses to dietary intervention with hypocholesterolemic vegetable oils. npj Biofilms Microbiomes 8 , 24 (2022).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500 , 232–236 (2013).
Louis, P. & Flint, H. J. Diversity, metabolism and microbial ecology of butyrate‐producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294 , 1–8 (2009).
Bui, T. P. N. et al. Mutual metabolic interactions in co-cultures of the intestinal Anaerostipes rhamnosivorans with an acetogen, methanogen, or pectin-degrader affecting butyrate production. Front. Microbiol. 10 , 2449 (2019).
Kaci, G. et al. Anti-inflammatory properties of streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl. Environ. Microbiol. 80 , 928–934 (2014).
Couvigny, B. et al. Commensal streptococcus salivarius modulates PPARγ transcriptional activity in human intestinal epithelial cells. PLoS ONE 10 , e0125371 (2015).
Barcenilla, A. et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66 , 1654–1661 (2000).
Scott, K. P. et al. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proc. Natl Acad. Sci. USA 108 , 4672–4679 (2011).
Hosomi, K. et al. Oral administration of Blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat. Commun. 13 , 4477 (2022).
Bui, T. P. N. et al. Conversion of dietary inositol into propionate and acetate by commensal Anaerostipes associates with host health. Nat. Commun. 12 , 4798 (2021).
Zeevi, D. et al. Structural variation in the gut microbiome associates with host health. Nature 568 , 43–48 (2019).
Brahe, L. K. et al. Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr. Diabetes 5 , e159–e159 (2015).
Deaver, J. A., Eum, S. Y. & Toborek, M. Circadian disruption changes gut microbiome taxa and functional gene composition. Front Microbiol 09 , 737 (2018).
Ze, X., Duncan, S. H., Louis, P. & Flint, H. J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6 , 1535–1543 (2012).
Ngai, P. H. K., Zhao, Z. & Ng, T. B. Agrocybin, an antifungal peptide from the edible mushroom Agrocybe cylindracea. Peptides 26 , 191–196 (2005).
Sánchez, M. et al. Modulatory effect of intestinal polyamines and trace amines on the spontaneous phasic contractions of the isolated ileum and colon rings of mice. Food Nutr. Res. 61 , 1321948 (2017).
Lassen, P. B. et al. Protein supplementation during an energy-restricted diet induces visceral fat loss and gut microbiota amino acid metabolism activation: a randomized trial. Sci. Rep. 11 , 15620 (2021).
Mazier, W. et al. A new strain of christensenella minuta as a potential biotherapy for obesity and associated metabolic diseases. Cells 10 , 823 (2021).
Salazar, N. et al. Fecal metabolome and bacterial composition in severe obesity: impact of diet and bariatric surgery. Gut Microbes 14 , 2106102 (2022).
Svedlund, J., Sjödin, I. & Dotevall, G. GSRS—clinical rating scale for gastrointestinal symptoms in patients with irritable bowel syndrome and peptic ulcer disease. Dig. Dis. Sci. 33 , 129–134 (1988).
Kulich, K. R. et al. Reliability and validity of the Gastrointestinal Symptom Rating Scale (GSRS) and Quality of Life in Reflux and Dyspepsia (QOLRAD) questionnaire in dyspepsia: a six-country study. Health Qual. Life Out. 6 , 12 (2008).
Riegler, G. & Esposito, I. Bristol scale stool form. A still valid help in medical practice and clinical research. Tech. Coloproctol. 5 , 163–164 (2001).
Yang, Y.-W. et al. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl. Environ. Microbiol. 81 , 6749–6756 (2015).
Gregoris, T. B. D., Aldred, N., Clare, A. S. & Burgess, J. G. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J. Microbiol. Methods 86 , 351–356 (2011).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108 , 4516–4522 (2010).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6 , 1621–1624 (2012).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 , 852–857 (2019).
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6 , 90 (2018).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 , D590–D596 (2012).
McKnight, D. T. et al. Methods for normalizing microbiome data: an ecological perspective. Methods Ecol. Evol. 10 , 389–400 (2019).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 , 357–359 (2012).
Beghini, F. et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10 , e65088 (2021).
Bapat, A. et al. Hypoxia promotes erythroid differentiation through the development of progenitors and proerythroblasts. Exp. Hematol. 97 , 32–46.e35 (2021).
Jasbi, P. et al. Metabolic profiling of neocortical tissue discriminates Alzheimer’s disease from mild cognitive impairment, high pathology controls, and normal controls. J. Proteome Res. 20 , 4303–4317 (2021).
Jasbi, P. et al. Microbiome and metabolome profiles of high screen time in a cohort of healthy college students. Sci. Rep. 12 , 3452 (2022).
Basile, A. J. et al. A four-week white bread diet does not alter plasma glucose concentrations, metabolic or vascular physiology in mourning doves, Zenaida macroura. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 247 , 110718 (2020).
Article CAS Google Scholar
Johnston, C. S. et al. Daily vinegar ingestion improves depression scores and alters the metabolome in healthy adults: a randomized controlled trial. Nutrients 13 , 4020 (2021).
Gu, H., Shi, X., Jasbi, P. & Patterson, J. Viruses as therapeutics, methods and protocols. Methods Mol. Biol. 2225 , 179–197 (2020).
He, H. et al. An integrative cellular metabolomic study reveals downregulated tricarboxylic acid cycle and potential biomarkers induced by tetrabromobisphenol A in human lung A549 cells. Environ. Toxicol. 38 , 7–16 (2023).
Mohr, A. E. et al. Association of food insecurity on gut microbiome and metabolome profiles in a diverse college-based sample. Sci. Rep. 12 , 14358 (2022).
Gu, H., Jasbi, P., Patterson, J. & Jin, Y. Enhanced detection of short‐chain fatty acids using gas chromatography mass spectrometry. Curr. Protoc. 1 , e177 (2021).
Qi, Y. et al. Metabolomics study of resina draconis on myocardial ischemia rats using ultraperformance liquid chromatography/quadrupole time-of-flight mass spectrometry combined with pattern recognition methods and metabolic pathway analysis. Évid. Based Complement. Altern. Med. 2013 , 438680 (2013).
Google Scholar
Yao, W. et al. Integrated plasma and urine metabolomics coupled with HPLC/QTOF-MS and chemometric analysis on potential biomarkers in liver injury and hepatoprotective effects of Er-Zhi-Wan. Anal. Bioanal. Chem. 406 , 7367–7378 (2014).
Wei, Y. et al. Early breast cancer detection using untargeted and targeted metabolomics. J. Proteome Res. 20 , 3124–3133 (2021).
Haak, B. W. et al. Integrative transkingdom analysis of the gut microbiome in antibiotic perturbation and critical illness. mSystems 6 , e01148–20 (2021).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLos One 8 , e61217 (2013).
Bokulich, N. A. et al. q2-longitudinal: longitudinal and paired-sample analyses of microbiome data. Msystems 3 , e00219–18 (2018).
Pang, Z. et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49 , W388–W396 (2021).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 , 27–30 (2000).
Mirzayi, C. et al. Reporting guidelines for human microbiome research: the STORMS checklist. Nat. Med. 27 , 1885–1892 (2021).
Mohr, A. E. et al. Gut microbiome remodeling and metabolomic profile in response to protein pacing with intermittent fasting versus continuous caloric restriction https://doi.org/10.5281/zenodo.10971120 (2024).
Download references
Acknowledgements
We thank the trial volunteers for their dedication and commitment to the study protocol. We are grateful for the research assistants from Skidmore College who provided valuable assistance with study protocol design, scheduling, recruitment, data testing, collection, entry, and statistical analysis, and preparation of manuscripts: Molly Boyce, Jenny Zhang, Melissa Haas, Olivia Furlong, Emma Valdez, Jessica Centore, Annika Smith, Kaitlyn Judd, Aaliyah Yarde, Katy Ehnstrom, Dakembay Hoyte, Sheriden Beard, Heather Mak, and Monique Dudar. We are grateful for the extensive guidance and counseling provided by the registered dietitian Jaime Martin. We thank research coordinator Michelle Poe for her superior dedication to all aspects of the study. This study was primarily funded by an unrestricted grant from Isagenix International LLC to P.J.A. (grant #:1911-859), with secondary funding provided to K.L.S.
Author information
Authors and affiliations.
College of Health Solutions, Arizona State University, Phoenix, AZ, USA
Alex E. Mohr, Karen L. Sweazea, Corrie M. Whisner, Dorothy D. Sears, Haiwei Gu & Judith Klein-Seetharaman
Biodesign Institute Center for Health Through Microbiomes, Arizona State University, Tempe, AZ, USA
Alex E. Mohr, Karen L. Sweazea, Devin A. Bowes, Corrie M. Whisner & Rosa Krajmalnik-Brown
Center for Evolution and Medicine, College of Liberal Arts and Sciences, Arizona State University, Tempe, AZ, USA
Karen L. Sweazea
School of Molecular Sciences, Arizona State University, Tempe, AZ, USA
Paniz Jasbi & Judith Klein-Seetharaman
Systems Precision Engineering and Advanced Research (SPEAR), Theriome Inc., Phoenix, AZ, USA
Paniz Jasbi
Center of Translational Science, Florida International University, Port St. Lucie, FL, USA
Yan Jin & Haiwei Gu
Human Nutrition and Metabolism Laboratory, Department of Health and Human Physiological Sciences, Skidmore College, Saratoga Springs, NY, USA
Karen M. Arciero & Paul J. Arciero
Isagenix International, LLC, Gilbert, AZ, USA
Eric Gumpricht
School of Health and Rehabilitation Sciences, Department of Sports Medicine and Nutrition, University of Pittsburgh, Pittsburgh, PA, USA
Paul J. Arciero
You can also search for this author in PubMed Google Scholar
Contributions
Study conceived and designed: P.J.A. Manuscript preparation with input from all authors: A.E.M., K.L.S., D.A.B., P.J., C.M.W., D.D.S., R.K.-B., H.G., J.K.-S., K.M.A., E.G., and P.J.A. Randomized study design and execution: K.M.A., and P.J.A. Microbiome analysis: A.E.M., D.A.B., C.M.W., and R.K.-B. Blood analyte analysis: A.E.M., K.L.S., and P.J.A. Metabolomic analysis: A.E.M., Y.J., H.G., and P.J. Statistical analysis and data presentation: A.E.M., C.M.W., D.D.S., R.K.-B., and P.J.A. Supervision and funding: K.L.S., E.G., and P.J.A.
Corresponding author
Correspondence to Paul J. Arciero .
Ethics declarations
Competing interests.
P.J.A. is a consultant for Isagenix International LLC, the study’s sponsor, he is an advisory board member of the International Protein Board (iPB), and he receives financial compensation for books and keynote presentations on protein pacing ( www.paularciero.com ). Eric Gumpricht is employed by Isagenix International, LLC, the funding source for this research. Isagenix International, LLC had no role in the study design, data collection, analysis, or decision to publish. No authors have financial interests regarding the outcomes of this investigation. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Communications thanks Gertrude Ecklu-Mensah, Saar Shoer, and Levi Teigen for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, reporting summary, source data, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Mohr, A.E., Sweazea, K.L., Bowes, D.A. et al. Gut microbiome remodeling and metabolomic profile improves in response to protein pacing with intermittent fasting versus continuous caloric restriction. Nat Commun 15 , 4155 (2024). https://doi.org/10.1038/s41467-024-48355-5
Download citation
Received : 13 September 2023
Accepted : 26 April 2024
Published : 28 May 2024
DOI : https://doi.org/10.1038/s41467-024-48355-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.
Page not found
The page you requested cannot be found. You may have used an outdated link or may have typed the address (URL) incorrectly. If you entered the URL manually, please check your spelling and try again.
The following resources may help you locate the website you're looking for:
- Stanford Search
- A to Z index of Stanford websites
- Stanford University homepage
If you think you have reached this page due to a server error, please contact HelpSU .
- Maps & Directions
- Search Stanford
- Terms of Use
- Copyright Complaints
© Stanford University , Stanford , California 94305

IMAGES
VIDEO
COMMENTS
Your null hypothesis in completely fair. You did it the right way. When you have a factor variable as predictor, you omit one of the levels as a reference category (the default is usually the first one, but you also can change that). Then all your other levels' coefficients are tested for a significant difference compared to the omitted category.
As in simple linear regression, under the null hypothesis t 0 = βˆ j seˆ(βˆ j) ∼ t n−p−1. We reject H 0 if |t 0| > t n−p−1,1−α/2. This is a partial test because βˆ j depends on all of the other predictors x i, i 6= j that are in the model. Thus, this is a test of the contribution of x j given the other predictors in the model.
xi: The value of the predictor variable xi. Multiple linear regression uses the following null and alternative hypotheses: H0: β1 = β2 = … = βk = 0. HA: β1 = β2 = … = βk ≠ 0. The null hypothesis states that all coefficients in the model are equal to zero. In other words, none of the predictor variables have a statistically ...
The formula for a multiple linear regression is: = the predicted value of the dependent variable. = the y-intercept (value of y when all other parameters are set to 0) = the regression coefficient () of the first independent variable () (a.k.a. the effect that increasing the value of the independent variable has on the predicted y value ...
Organized by textbook: https://learncheme.com/ See Part 2: https://www.youtube.com/watch?v=ziGbG0dRlsAMade by faculty at the University of Colorado Boulder, ...
This video is an introduction to multiple regression analysis, with a focus on conducting a hypothesis test. If I look tired in the video, it's because I've ...
A population model for a multiple linear regression model that relates a y -variable to p -1 x -variables is written as. y i = β 0 + β 1 x i, 1 + β 2 x i, 2 + … + β p − 1 x i, p − 1 + ϵ i. We assume that the ϵ i have a normal distribution with mean 0 and constant variance σ 2. These are the same assumptions that we used in simple ...
Linear regression has an additive assumption: $ sales = β 0 + β 1 × tv + β 2 × radio + ε $. i.e. An increase of 100 USD dollars in TV ads causes a fixed increase of 100 β 2 USD in sales on average, regardless of how much you spend on radio ads. We saw that in Fig 3.5 above.
12 R2 For+example,+suppose+y is+the+sale+price+of+a+house.+Then+ sensible+predictorsinclude x 1 =the+interior+size+of+the+house, x 2 =the+size+of+the+lot+on+which+the ...
5.3 - The Multiple Linear Regression Model. Notation for the Population Model. A population model for a multiple linear regression model that relates a y -variable to k x -variables is written as. yi = β0 +β1xi,1 +β2xi,2 + … +βkxi,k +ϵi. Here we're using " k " for the number of predictor variables, which means we have k +1 regression ...
The two test statistic formulas are algebraically equal; however, the formulas are different and we use a different parameter in the hypotheses. The formula for the t-test statistic is t = b1 (MSE SSxx)√ t = b 1 ( M S E S S x x) Use the t-distribution with degrees of freedom equal to n − p − 1 n − p − 1.
5.2 - Writing Hypotheses. The first step in conducting a hypothesis test is to write the hypothesis statements that are going to be tested. For each test you will have a null hypothesis (\ (H_0\)) and an alternative hypothesis (\ (H_a\)). Null Hypothesis. The statement that there is not a difference in the population (s), denoted as \ (H_0\)
Photo by Ferdinand Stöhr on Unsplash. Multiple linear regression is one of the most fundamental statistical models due to its simplicity and interpretability of results. For prediction purposes, linear models can sometimes outperform fancier nonlinear models, especially in situations with small numbers of training cases, low signal-to-noise ratio, or sparse data (Hastie et al., 2009).
Testing that individual coefficients take a specific value such as zero or some other value is done in exactly the same way as with the simple two variable regression model. Now suppose we wish to test that a number of coefficients or combinations of coefficients take some particular value. In this case we will use the so called "F-test".
Here, Y is the output variable, and X terms are the corresponding input variables. Notice that this equation is just an extension of Simple Linear Regression, and each predictor has a corresponding slope coefficient (β).The first β term (βo) is the intercept constant and is the value of Y in absence of all predictors (i.e when all X terms are 0). It may or may or may not hold any ...
Multiple Regression Write Up. Here is an example of how to write up the results of a standard multiple regression analysis: In order to test the research question, a multiple regression was conducted, with age, gender (0 = male, 1 = female), and perceived life stress as the predictors, with levels of physical illness as the dependent variable.
Assumptions of Multiple Linear Regression. There are four key assumptions that multiple linear regression makes about the data: 1. Linear relationship: There exists a linear relationship between the independent variable, x, and the dependent variable, y. 2. Independence: The residuals are independent.
The hypothesis in Multiple Linear Regression revolves around the significance of the regression coefficients. Each coefficient corresponds to a specific predictor variable, and the hypothesis tests whether each predictor has a significant impact on the dependent variable. ... (APA) guidelines is crucial for scholarly and professional writing ...
Multiple regression is an extension of simple linear regression. It is used when we want to predict the value of a variable based on the value of two or more other variables. The variable we want to predict is called the dependent variable (or sometimes, the outcome, target or criterion variable). The variables we are using to predict the value ...
a hypothesis test for testing that a subset — more than one, but not all — of the slope parameters are 0. In this lesson, we also learn how to perform each of the above three hypothesis tests. Key Learning Goals for this Lesson: Be able to interpret the coefficients of a multiple regression model. Understand what the scope of the model is ...
Regression allows you to estimate how a dependent variable changes as the independent variable (s) change. Simple linear regression example. You are a social researcher interested in the relationship between income and happiness. You survey 500 people whose incomes range from 15k to 75k and ask them to rank their happiness on a scale from 1 to ...
In simple linear regression, this is equivalent to saying "Are X an Y correlated?". In reviewing the model, Y = β0 +β1X + ε Y = β 0 + β 1 X + ε, as long as the slope ( β1 β 1) has any non‐zero value, X X will add value in helping predict the expected value of Y Y. However, if there is no correlation between X and Y, the value of ...
Linear, Multiple, and Polynomial Regression; Model Selection and Cross-Validation; Bias, Variance, and Hyperparameters; Classification and Logistic Regression; Bootstrap, Confidence Intervals, and Hypothesis Testing; There are many things you would learn from this course. Learn it well; the foundation would be important for the next course.
Multiple regression analyses were used to test 287whether the relationships between each of the pairs of variables remained significant when 288controlling for the third variable. Significance was tested at the α = .05 and α = .01 levels for 289the Pearson correlations and the multiple regression analyses. Following Armstrong's(60)
First, we analyzed several single-stage models instead of the preferred two-stage Heckman models. Specifically, we utilized single-stage OLS, general linear, and fractional logit models and find fully consistent results. Moreover, we tested alternative operationalizations of our dependent variable, extent of activist demand withdrawal.
Here, in a follow-up of a clinical study, the authors show that protein pacing and intermittent fasting improves gut symptomatology and microbial diversity, as well as reduces visceral fat ...
When could this happen in real life: Time series: Each sample corresponds to a different point in time. The errors for samples that are close in time are correlated. Spatial data: Each sample corresponds to a different location in space. Grouped data: Imagine a study on predicting height from weight at birth. If some of the subjects in the study are in the same family, their shared environment ...