- USC Libraries
- Research Guides

Organizing Your Social Sciences Research Paper
- Quantitative Methods
- Purpose of Guide
- Design Flaws to Avoid
- Independent and Dependent Variables
- Glossary of Research Terms
- Reading Research Effectively
- Narrowing a Topic Idea
- Broadening a Topic Idea
- Extending the Timeliness of a Topic Idea
- Academic Writing Style
- Applying Critical Thinking
- Choosing a Title
- Making an Outline
- Paragraph Development
- Research Process Video Series
- Executive Summary
- The C.A.R.S. Model
- Background Information
- The Research Problem/Question
- Theoretical Framework
- Citation Tracking
- Content Alert Services
- Evaluating Sources
- Primary Sources
- Secondary Sources
- Tiertiary Sources
- Scholarly vs. Popular Publications
- Qualitative Methods
- Insiderness
- Using Non-Textual Elements
- Limitations of the Study
- Common Grammar Mistakes
- Writing Concisely
- Avoiding Plagiarism
- Footnotes or Endnotes?
- Further Readings
- Generative AI and Writing
- USC Libraries Tutorials and Other Guides
- Bibliography
Quantitative methods emphasize objective measurements and the statistical, mathematical, or numerical analysis of data collected through polls, questionnaires, and surveys, or by manipulating pre-existing statistical data using computational techniques . Quantitative research focuses on gathering numerical data and generalizing it across groups of people or to explain a particular phenomenon.
Babbie, Earl R. The Practice of Social Research . 12th ed. Belmont, CA: Wadsworth Cengage, 2010; Muijs, Daniel. Doing Quantitative Research in Education with SPSS . 2nd edition. London: SAGE Publications, 2010.
Need Help Locating Statistics?
Resources for locating data and statistics can be found here:
Statistics & Data Research Guide
Characteristics of Quantitative Research
Your goal in conducting quantitative research study is to determine the relationship between one thing [an independent variable] and another [a dependent or outcome variable] within a population. Quantitative research designs are either descriptive [subjects usually measured once] or experimental [subjects measured before and after a treatment]. A descriptive study establishes only associations between variables; an experimental study establishes causality.
Quantitative research deals in numbers, logic, and an objective stance. Quantitative research focuses on numeric and unchanging data and detailed, convergent reasoning rather than divergent reasoning [i.e., the generation of a variety of ideas about a research problem in a spontaneous, free-flowing manner].
Its main characteristics are :
- The data is usually gathered using structured research instruments.
- The results are based on larger sample sizes that are representative of the population.
- The research study can usually be replicated or repeated, given its high reliability.
- Researcher has a clearly defined research question to which objective answers are sought.
- All aspects of the study are carefully designed before data is collected.
- Data are in the form of numbers and statistics, often arranged in tables, charts, figures, or other non-textual forms.
- Project can be used to generalize concepts more widely, predict future results, or investigate causal relationships.
- Researcher uses tools, such as questionnaires or computer software, to collect numerical data.
The overarching aim of a quantitative research study is to classify features, count them, and construct statistical models in an attempt to explain what is observed.
Things to keep in mind when reporting the results of a study using quantitative methods :
- Explain the data collected and their statistical treatment as well as all relevant results in relation to the research problem you are investigating. Interpretation of results is not appropriate in this section.
- Report unanticipated events that occurred during your data collection. Explain how the actual analysis differs from the planned analysis. Explain your handling of missing data and why any missing data does not undermine the validity of your analysis.
- Explain the techniques you used to "clean" your data set.
- Choose a minimally sufficient statistical procedure ; provide a rationale for its use and a reference for it. Specify any computer programs used.
- Describe the assumptions for each procedure and the steps you took to ensure that they were not violated.
- When using inferential statistics , provide the descriptive statistics, confidence intervals, and sample sizes for each variable as well as the value of the test statistic, its direction, the degrees of freedom, and the significance level [report the actual p value].
- Avoid inferring causality , particularly in nonrandomized designs or without further experimentation.
- Use tables to provide exact values ; use figures to convey global effects. Keep figures small in size; include graphic representations of confidence intervals whenever possible.
- Always tell the reader what to look for in tables and figures .
NOTE: When using pre-existing statistical data gathered and made available by anyone other than yourself [e.g., government agency], you still must report on the methods that were used to gather the data and describe any missing data that exists and, if there is any, provide a clear explanation why the missing data does not undermine the validity of your final analysis.
Babbie, Earl R. The Practice of Social Research . 12th ed. Belmont, CA: Wadsworth Cengage, 2010; Brians, Craig Leonard et al. Empirical Political Analysis: Quantitative and Qualitative Research Methods . 8th ed. Boston, MA: Longman, 2011; McNabb, David E. Research Methods in Public Administration and Nonprofit Management: Quantitative and Qualitative Approaches . 2nd ed. Armonk, NY: M.E. Sharpe, 2008; Quantitative Research Methods. Writing@CSU. Colorado State University; Singh, Kultar. Quantitative Social Research Methods . Los Angeles, CA: Sage, 2007.
Basic Research Design for Quantitative Studies
Before designing a quantitative research study, you must decide whether it will be descriptive or experimental because this will dictate how you gather, analyze, and interpret the results. A descriptive study is governed by the following rules: subjects are generally measured once; the intention is to only establish associations between variables; and, the study may include a sample population of hundreds or thousands of subjects to ensure that a valid estimate of a generalized relationship between variables has been obtained. An experimental design includes subjects measured before and after a particular treatment, the sample population may be very small and purposefully chosen, and it is intended to establish causality between variables. Introduction The introduction to a quantitative study is usually written in the present tense and from the third person point of view. It covers the following information:
- Identifies the research problem -- as with any academic study, you must state clearly and concisely the research problem being investigated.
- Reviews the literature -- review scholarship on the topic, synthesizing key themes and, if necessary, noting studies that have used similar methods of inquiry and analysis. Note where key gaps exist and how your study helps to fill these gaps or clarifies existing knowledge.
- Describes the theoretical framework -- provide an outline of the theory or hypothesis underpinning your study. If necessary, define unfamiliar or complex terms, concepts, or ideas and provide the appropriate background information to place the research problem in proper context [e.g., historical, cultural, economic, etc.].
Methodology The methods section of a quantitative study should describe how each objective of your study will be achieved. Be sure to provide enough detail to enable the reader can make an informed assessment of the methods being used to obtain results associated with the research problem. The methods section should be presented in the past tense.
- Study population and sampling -- where did the data come from; how robust is it; note where gaps exist or what was excluded. Note the procedures used for their selection;
- Data collection – describe the tools and methods used to collect information and identify the variables being measured; describe the methods used to obtain the data; and, note if the data was pre-existing [i.e., government data] or you gathered it yourself. If you gathered it yourself, describe what type of instrument you used and why. Note that no data set is perfect--describe any limitations in methods of gathering data.
- Data analysis -- describe the procedures for processing and analyzing the data. If appropriate, describe the specific instruments of analysis used to study each research objective, including mathematical techniques and the type of computer software used to manipulate the data.
Results The finding of your study should be written objectively and in a succinct and precise format. In quantitative studies, it is common to use graphs, tables, charts, and other non-textual elements to help the reader understand the data. Make sure that non-textual elements do not stand in isolation from the text but are being used to supplement the overall description of the results and to help clarify key points being made. Further information about how to effectively present data using charts and graphs can be found here .
- Statistical analysis -- how did you analyze the data? What were the key findings from the data? The findings should be present in a logical, sequential order. Describe but do not interpret these trends or negative results; save that for the discussion section. The results should be presented in the past tense.
Discussion Discussions should be analytic, logical, and comprehensive. The discussion should meld together your findings in relation to those identified in the literature review, and placed within the context of the theoretical framework underpinning the study. The discussion should be presented in the present tense.
- Interpretation of results -- reiterate the research problem being investigated and compare and contrast the findings with the research questions underlying the study. Did they affirm predicted outcomes or did the data refute it?
- Description of trends, comparison of groups, or relationships among variables -- describe any trends that emerged from your analysis and explain all unanticipated and statistical insignificant findings.
- Discussion of implications – what is the meaning of your results? Highlight key findings based on the overall results and note findings that you believe are important. How have the results helped fill gaps in understanding the research problem?
- Limitations -- describe any limitations or unavoidable bias in your study and, if necessary, note why these limitations did not inhibit effective interpretation of the results.
Conclusion End your study by to summarizing the topic and provide a final comment and assessment of the study.
- Summary of findings – synthesize the answers to your research questions. Do not report any statistical data here; just provide a narrative summary of the key findings and describe what was learned that you did not know before conducting the study.
- Recommendations – if appropriate to the aim of the assignment, tie key findings with policy recommendations or actions to be taken in practice.
- Future research – note the need for future research linked to your study’s limitations or to any remaining gaps in the literature that were not addressed in your study.
Black, Thomas R. Doing Quantitative Research in the Social Sciences: An Integrated Approach to Research Design, Measurement and Statistics . London: Sage, 1999; Gay,L. R. and Peter Airasain. Educational Research: Competencies for Analysis and Applications . 7th edition. Upper Saddle River, NJ: Merril Prentice Hall, 2003; Hector, Anestine. An Overview of Quantitative Research in Composition and TESOL . Department of English, Indiana University of Pennsylvania; Hopkins, Will G. “Quantitative Research Design.” Sportscience 4, 1 (2000); "A Strategy for Writing Up Research Results. The Structure, Format, Content, and Style of a Journal-Style Scientific Paper." Department of Biology. Bates College; Nenty, H. Johnson. "Writing a Quantitative Research Thesis." International Journal of Educational Science 1 (2009): 19-32; Ouyang, Ronghua (John). Basic Inquiry of Quantitative Research . Kennesaw State University.
Strengths of Using Quantitative Methods
Quantitative researchers try to recognize and isolate specific variables contained within the study framework, seek correlation, relationships and causality, and attempt to control the environment in which the data is collected to avoid the risk of variables, other than the one being studied, accounting for the relationships identified.
Among the specific strengths of using quantitative methods to study social science research problems:
- Allows for a broader study, involving a greater number of subjects, and enhancing the generalization of the results;
- Allows for greater objectivity and accuracy of results. Generally, quantitative methods are designed to provide summaries of data that support generalizations about the phenomenon under study. In order to accomplish this, quantitative research usually involves few variables and many cases, and employs prescribed procedures to ensure validity and reliability;
- Applying well established standards means that the research can be replicated, and then analyzed and compared with similar studies;
- You can summarize vast sources of information and make comparisons across categories and over time; and,
- Personal bias can be avoided by keeping a 'distance' from participating subjects and using accepted computational techniques .
Babbie, Earl R. The Practice of Social Research . 12th ed. Belmont, CA: Wadsworth Cengage, 2010; Brians, Craig Leonard et al. Empirical Political Analysis: Quantitative and Qualitative Research Methods . 8th ed. Boston, MA: Longman, 2011; McNabb, David E. Research Methods in Public Administration and Nonprofit Management: Quantitative and Qualitative Approaches . 2nd ed. Armonk, NY: M.E. Sharpe, 2008; Singh, Kultar. Quantitative Social Research Methods . Los Angeles, CA: Sage, 2007.
Limitations of Using Quantitative Methods
Quantitative methods presume to have an objective approach to studying research problems, where data is controlled and measured, to address the accumulation of facts, and to determine the causes of behavior. As a consequence, the results of quantitative research may be statistically significant but are often humanly insignificant.
Some specific limitations associated with using quantitative methods to study research problems in the social sciences include:
- Quantitative data is more efficient and able to test hypotheses, but may miss contextual detail;
- Uses a static and rigid approach and so employs an inflexible process of discovery;
- The development of standard questions by researchers can lead to "structural bias" and false representation, where the data actually reflects the view of the researcher instead of the participating subject;
- Results provide less detail on behavior, attitudes, and motivation;
- Researcher may collect a much narrower and sometimes superficial dataset;
- Results are limited as they provide numerical descriptions rather than detailed narrative and generally provide less elaborate accounts of human perception;
- The research is often carried out in an unnatural, artificial environment so that a level of control can be applied to the exercise. This level of control might not normally be in place in the real world thus yielding "laboratory results" as opposed to "real world results"; and,
- Preset answers will not necessarily reflect how people really feel about a subject and, in some cases, might just be the closest match to the preconceived hypothesis.
Research Tip
Finding Examples of How to Apply Different Types of Research Methods
SAGE publications is a major publisher of studies about how to design and conduct research in the social and behavioral sciences. Their SAGE Research Methods Online and Cases database includes contents from books, articles, encyclopedias, handbooks, and videos covering social science research design and methods including the complete Little Green Book Series of Quantitative Applications in the Social Sciences and the Little Blue Book Series of Qualitative Research techniques. The database also includes case studies outlining the research methods used in real research projects. This is an excellent source for finding definitions of key terms and descriptions of research design and practice, techniques of data gathering, analysis, and reporting, and information about theories of research [e.g., grounded theory]. The database covers both qualitative and quantitative research methods as well as mixed methods approaches to conducting research.
SAGE Research Methods Online and Cases
- << Previous: Qualitative Methods
- Next: Insiderness >>
- Last Updated: May 2, 2024 4:39 PM
- URL: https://libguides.usc.edu/writingguide
Academia.edu no longer supports Internet Explorer.
To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your browser .
Enter the email address you signed up with and we'll email you a reset link.
- We're Hiring!
- Help Center

Example of a Quantitative Research Paper for Students & Researchers

This example of a quantitative research paper is designed to help students and other r esearchers who are learning how to write about their work. The reported research obs erves the behaviour of restaurant customers, and example paragraphs are combined with instructions for logical argumentation. Authors are encouraged to observe a traditional structure for organising quantitative research papers, to formulate research que stions, working hypotheses and investigative tools, to report results accurately and thor oughly, and to present thoughtful interpretation and logical discussion of evidence.
Related Papers
Journal of Foodservice
Christina Fjellström
Rohit Taraporewala
Noor Mustafa
FAST FOOD OBESITY 16
Princess Moon Galindez
Journal of Hospitality & Leisure Marketing
Tajulurrus mohammad
Food industry, the world over, is witnessing unprecedented increase in the number of multinational enterprises. These multinational enterprises, when deciding to expand their operations to a new country, have to make a choice between following uniform business strategies as in their home country or modify their strategies to suit the host country socioeconomic and political environment. Given the economic cost of modification of business strategies, the choice has widespread implications for the sustainability of multinational enterprises. The present paper argues that this decision-making is particularly critical in the case of multinational food enterprises because of large scale variability in food habits across countries and even within a country. Drawing from case studies of three multinational food enterprises in India, the paper points out that, in order to operate successfully in their host countries, the multinational food enterprises must adopt Glocalized strategies in marketing, product development, advertisement etc.
Modern China Series,North American Business Press
Robert Tian
Food is an important aspect of social culture and has a close relationship with economic development. The Chinese food culture has the characteristics of inheritability and development, and throughout the history of Chinese food culture, it has maintained its momentum of development since its primitive society. Neither the change of dynasty nor the change of social system has had a profound influence on it, and the philosophy of supplying enough food to people and food being the top priority was very popular. Eating was a top priority for people in China. Long ago, Confucius said that the desire for food and sex is part of human nature. As such, in the Chinese culture food became the priority. Because of the attention to diet, Chinese people would, when they had leisure time or abundant raw materials, work out a variety of food. Chinese cooking is flexible, which is characterized by saying that there is no fixed taste and what is delicious is valued. The beauty of food is one of the important roots of Chinese aesthetics, which inspires people with the stimulation of eating. Triggering art inspiration is the inevitable result of Chinese food culture pursuing complete and beautiful color, fragrance, taste, shape, and utensils. It makes food culture a comprehensive art containing multiple cultural connotations of diet, diet mentality, beautiful utensils and etiquette, food enjoyment and eating. Chinese foods have not only exquisite craftsmanship and rich nutrition, but also elegant and graceful names, which are literary and romantic, poetic and fancy. Food functions to not only satiate people’s hunger; it has also become an integral aspect of life enjoyment, which represents an essential component of food anthropology. Food anthropologists stress that changes in people’s eating habits not only depend on the local food culture, which may be specific to a given region, but also varies with economic development in different regions. Food anthropology, as a sub branch of applied anthropology, adapts anthropological theories and methods to study food industry, food culture, food consumption and food commerce. Seminal work in this regard has been provided by scholars and consultants in the field of food anthropology. This book describes the anthropological studies on Chinese foodways, outlines the Chinese food anthropology basic theories and methods. Anthropology in China is still at its development stage in China, while food anthropology is just at its initial stages of development. Nevertheless, China’s economic and social development, especially in ethnic minority regions in Western China, needs the theoretical guidance of some disciplines, including food anthropology, economic anthropology and business anthropology. At the same time, it has provided opportunities to develop food anthropology with the Chinese characteristics. Therefore, when Chinese scholars are learning and adopting Western food anthropology theories and methodologies, they must innovate and develop the related theories and methodologies with Chinese characteristics, so that they can better serve the well-off of the entire society.
MUHAMMAD IMAD UD DIN
City & Community
Petra Kuppinger
RELATED PAPERS
Golden Arches East: McDonald's in East …
Anuththara Wanaguru
Adrian Paul Padilla
Freya Higgins-Desbiolles , Gayathri Wijesinghe
Jeroen Struben
Harris Solomon
Divina Seming
AIMS Agriculture and Food
Giuseppe Sortino , Pietro Columba
Emmanuel Marillier
Anshul Garg
American Journal of Public Health
Janelle Gunn
Łukasz Korus
Asmaliyana Ghani
Denise Mainville
Dayangku Nurul Asyiqin
The 18 th Annual …
Anil Bilgihan
Celyrah B Castillo
Asian Journal of Tourism Research
Kathleen M Adams
- We're Hiring!
- Help Center
- Find new research papers in:
- Health Sciences
- Earth Sciences
- Cognitive Science
- Mathematics
- Computer Science
- Academia ©2024
- Privacy Policy

Home » Quantitative Research – Methods, Types and Analysis
Quantitative Research – Methods, Types and Analysis
Table of Contents

Quantitative Research
Quantitative research is a type of research that collects and analyzes numerical data to test hypotheses and answer research questions . This research typically involves a large sample size and uses statistical analysis to make inferences about a population based on the data collected. It often involves the use of surveys, experiments, or other structured data collection methods to gather quantitative data.
Quantitative Research Methods

Quantitative Research Methods are as follows:
Descriptive Research Design
Descriptive research design is used to describe the characteristics of a population or phenomenon being studied. This research method is used to answer the questions of what, where, when, and how. Descriptive research designs use a variety of methods such as observation, case studies, and surveys to collect data. The data is then analyzed using statistical tools to identify patterns and relationships.
Correlational Research Design
Correlational research design is used to investigate the relationship between two or more variables. Researchers use correlational research to determine whether a relationship exists between variables and to what extent they are related. This research method involves collecting data from a sample and analyzing it using statistical tools such as correlation coefficients.
Quasi-experimental Research Design
Quasi-experimental research design is used to investigate cause-and-effect relationships between variables. This research method is similar to experimental research design, but it lacks full control over the independent variable. Researchers use quasi-experimental research designs when it is not feasible or ethical to manipulate the independent variable.
Experimental Research Design
Experimental research design is used to investigate cause-and-effect relationships between variables. This research method involves manipulating the independent variable and observing the effects on the dependent variable. Researchers use experimental research designs to test hypotheses and establish cause-and-effect relationships.
Survey Research
Survey research involves collecting data from a sample of individuals using a standardized questionnaire. This research method is used to gather information on attitudes, beliefs, and behaviors of individuals. Researchers use survey research to collect data quickly and efficiently from a large sample size. Survey research can be conducted through various methods such as online, phone, mail, or in-person interviews.
Quantitative Research Analysis Methods
Here are some commonly used quantitative research analysis methods:
Statistical Analysis
Statistical analysis is the most common quantitative research analysis method. It involves using statistical tools and techniques to analyze the numerical data collected during the research process. Statistical analysis can be used to identify patterns, trends, and relationships between variables, and to test hypotheses and theories.
Regression Analysis
Regression analysis is a statistical technique used to analyze the relationship between one dependent variable and one or more independent variables. Researchers use regression analysis to identify and quantify the impact of independent variables on the dependent variable.
Factor Analysis
Factor analysis is a statistical technique used to identify underlying factors that explain the correlations among a set of variables. Researchers use factor analysis to reduce a large number of variables to a smaller set of factors that capture the most important information.
Structural Equation Modeling
Structural equation modeling is a statistical technique used to test complex relationships between variables. It involves specifying a model that includes both observed and unobserved variables, and then using statistical methods to test the fit of the model to the data.
Time Series Analysis
Time series analysis is a statistical technique used to analyze data that is collected over time. It involves identifying patterns and trends in the data, as well as any seasonal or cyclical variations.
Multilevel Modeling
Multilevel modeling is a statistical technique used to analyze data that is nested within multiple levels. For example, researchers might use multilevel modeling to analyze data that is collected from individuals who are nested within groups, such as students nested within schools.
Applications of Quantitative Research
Quantitative research has many applications across a wide range of fields. Here are some common examples:
- Market Research : Quantitative research is used extensively in market research to understand consumer behavior, preferences, and trends. Researchers use surveys, experiments, and other quantitative methods to collect data that can inform marketing strategies, product development, and pricing decisions.
- Health Research: Quantitative research is used in health research to study the effectiveness of medical treatments, identify risk factors for diseases, and track health outcomes over time. Researchers use statistical methods to analyze data from clinical trials, surveys, and other sources to inform medical practice and policy.
- Social Science Research: Quantitative research is used in social science research to study human behavior, attitudes, and social structures. Researchers use surveys, experiments, and other quantitative methods to collect data that can inform social policies, educational programs, and community interventions.
- Education Research: Quantitative research is used in education research to study the effectiveness of teaching methods, assess student learning outcomes, and identify factors that influence student success. Researchers use experimental and quasi-experimental designs, as well as surveys and other quantitative methods, to collect and analyze data.
- Environmental Research: Quantitative research is used in environmental research to study the impact of human activities on the environment, assess the effectiveness of conservation strategies, and identify ways to reduce environmental risks. Researchers use statistical methods to analyze data from field studies, experiments, and other sources.
Characteristics of Quantitative Research
Here are some key characteristics of quantitative research:
- Numerical data : Quantitative research involves collecting numerical data through standardized methods such as surveys, experiments, and observational studies. This data is analyzed using statistical methods to identify patterns and relationships.
- Large sample size: Quantitative research often involves collecting data from a large sample of individuals or groups in order to increase the reliability and generalizability of the findings.
- Objective approach: Quantitative research aims to be objective and impartial in its approach, focusing on the collection and analysis of data rather than personal beliefs, opinions, or experiences.
- Control over variables: Quantitative research often involves manipulating variables to test hypotheses and establish cause-and-effect relationships. Researchers aim to control for extraneous variables that may impact the results.
- Replicable : Quantitative research aims to be replicable, meaning that other researchers should be able to conduct similar studies and obtain similar results using the same methods.
- Statistical analysis: Quantitative research involves using statistical tools and techniques to analyze the numerical data collected during the research process. Statistical analysis allows researchers to identify patterns, trends, and relationships between variables, and to test hypotheses and theories.
- Generalizability: Quantitative research aims to produce findings that can be generalized to larger populations beyond the specific sample studied. This is achieved through the use of random sampling methods and statistical inference.
Examples of Quantitative Research
Here are some examples of quantitative research in different fields:
- Market Research: A company conducts a survey of 1000 consumers to determine their brand awareness and preferences. The data is analyzed using statistical methods to identify trends and patterns that can inform marketing strategies.
- Health Research : A researcher conducts a randomized controlled trial to test the effectiveness of a new drug for treating a particular medical condition. The study involves collecting data from a large sample of patients and analyzing the results using statistical methods.
- Social Science Research : A sociologist conducts a survey of 500 people to study attitudes toward immigration in a particular country. The data is analyzed using statistical methods to identify factors that influence these attitudes.
- Education Research: A researcher conducts an experiment to compare the effectiveness of two different teaching methods for improving student learning outcomes. The study involves randomly assigning students to different groups and collecting data on their performance on standardized tests.
- Environmental Research : A team of researchers conduct a study to investigate the impact of climate change on the distribution and abundance of a particular species of plant or animal. The study involves collecting data on environmental factors and population sizes over time and analyzing the results using statistical methods.
- Psychology : A researcher conducts a survey of 500 college students to investigate the relationship between social media use and mental health. The data is analyzed using statistical methods to identify correlations and potential causal relationships.
- Political Science: A team of researchers conducts a study to investigate voter behavior during an election. They use survey methods to collect data on voting patterns, demographics, and political attitudes, and analyze the results using statistical methods.
How to Conduct Quantitative Research
Here is a general overview of how to conduct quantitative research:
- Develop a research question: The first step in conducting quantitative research is to develop a clear and specific research question. This question should be based on a gap in existing knowledge, and should be answerable using quantitative methods.
- Develop a research design: Once you have a research question, you will need to develop a research design. This involves deciding on the appropriate methods to collect data, such as surveys, experiments, or observational studies. You will also need to determine the appropriate sample size, data collection instruments, and data analysis techniques.
- Collect data: The next step is to collect data. This may involve administering surveys or questionnaires, conducting experiments, or gathering data from existing sources. It is important to use standardized methods to ensure that the data is reliable and valid.
- Analyze data : Once the data has been collected, it is time to analyze it. This involves using statistical methods to identify patterns, trends, and relationships between variables. Common statistical techniques include correlation analysis, regression analysis, and hypothesis testing.
- Interpret results: After analyzing the data, you will need to interpret the results. This involves identifying the key findings, determining their significance, and drawing conclusions based on the data.
- Communicate findings: Finally, you will need to communicate your findings. This may involve writing a research report, presenting at a conference, or publishing in a peer-reviewed journal. It is important to clearly communicate the research question, methods, results, and conclusions to ensure that others can understand and replicate your research.
When to use Quantitative Research
Here are some situations when quantitative research can be appropriate:
- To test a hypothesis: Quantitative research is often used to test a hypothesis or a theory. It involves collecting numerical data and using statistical analysis to determine if the data supports or refutes the hypothesis.
- To generalize findings: If you want to generalize the findings of your study to a larger population, quantitative research can be useful. This is because it allows you to collect numerical data from a representative sample of the population and use statistical analysis to make inferences about the population as a whole.
- To measure relationships between variables: If you want to measure the relationship between two or more variables, such as the relationship between age and income, or between education level and job satisfaction, quantitative research can be useful. It allows you to collect numerical data on both variables and use statistical analysis to determine the strength and direction of the relationship.
- To identify patterns or trends: Quantitative research can be useful for identifying patterns or trends in data. For example, you can use quantitative research to identify trends in consumer behavior or to identify patterns in stock market data.
- To quantify attitudes or opinions : If you want to measure attitudes or opinions on a particular topic, quantitative research can be useful. It allows you to collect numerical data using surveys or questionnaires and analyze the data using statistical methods to determine the prevalence of certain attitudes or opinions.
Purpose of Quantitative Research
The purpose of quantitative research is to systematically investigate and measure the relationships between variables or phenomena using numerical data and statistical analysis. The main objectives of quantitative research include:
- Description : To provide a detailed and accurate description of a particular phenomenon or population.
- Explanation : To explain the reasons for the occurrence of a particular phenomenon, such as identifying the factors that influence a behavior or attitude.
- Prediction : To predict future trends or behaviors based on past patterns and relationships between variables.
- Control : To identify the best strategies for controlling or influencing a particular outcome or behavior.
Quantitative research is used in many different fields, including social sciences, business, engineering, and health sciences. It can be used to investigate a wide range of phenomena, from human behavior and attitudes to physical and biological processes. The purpose of quantitative research is to provide reliable and valid data that can be used to inform decision-making and improve understanding of the world around us.
Advantages of Quantitative Research
There are several advantages of quantitative research, including:
- Objectivity : Quantitative research is based on objective data and statistical analysis, which reduces the potential for bias or subjectivity in the research process.
- Reproducibility : Because quantitative research involves standardized methods and measurements, it is more likely to be reproducible and reliable.
- Generalizability : Quantitative research allows for generalizations to be made about a population based on a representative sample, which can inform decision-making and policy development.
- Precision : Quantitative research allows for precise measurement and analysis of data, which can provide a more accurate understanding of phenomena and relationships between variables.
- Efficiency : Quantitative research can be conducted relatively quickly and efficiently, especially when compared to qualitative research, which may involve lengthy data collection and analysis.
- Large sample sizes : Quantitative research can accommodate large sample sizes, which can increase the representativeness and generalizability of the results.
Limitations of Quantitative Research
There are several limitations of quantitative research, including:
- Limited understanding of context: Quantitative research typically focuses on numerical data and statistical analysis, which may not provide a comprehensive understanding of the context or underlying factors that influence a phenomenon.
- Simplification of complex phenomena: Quantitative research often involves simplifying complex phenomena into measurable variables, which may not capture the full complexity of the phenomenon being studied.
- Potential for researcher bias: Although quantitative research aims to be objective, there is still the potential for researcher bias in areas such as sampling, data collection, and data analysis.
- Limited ability to explore new ideas: Quantitative research is often based on pre-determined research questions and hypotheses, which may limit the ability to explore new ideas or unexpected findings.
- Limited ability to capture subjective experiences : Quantitative research is typically focused on objective data and may not capture the subjective experiences of individuals or groups being studied.
- Ethical concerns : Quantitative research may raise ethical concerns, such as invasion of privacy or the potential for harm to participants.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Questionnaire – Definition, Types, and Examples

Case Study – Methods, Examples and Guide

Observational Research – Methods and Guide

Qualitative Research Methods

Explanatory Research – Types, Methods, Guide

Survey Research – Types, Methods, Examples
Quantitative Research
- Reference work entry
- First Online: 13 January 2019
- Cite this reference work entry

- Leigh A. Wilson 2 , 3
4279 Accesses
4 Citations
Quantitative research methods are concerned with the planning, design, and implementation of strategies to collect and analyze data. Descartes, the seventeenth-century philosopher, suggested that how the results are achieved is often more important than the results themselves, as the journey taken along the research path is a journey of discovery. High-quality quantitative research is characterized by the attention given to the methods and the reliability of the tools used to collect the data. The ability to critique research in a systematic way is an essential component of a health professional’s role in order to deliver high quality, evidence-based healthcare. This chapter is intended to provide a simple overview of the way new researchers and health practitioners can understand and employ quantitative methods. The chapter offers practical, realistic guidance in a learner-friendly way and uses a logical sequence to understand the process of hypothesis development, study design, data collection and handling, and finally data analysis and interpretation.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Durable hardcover edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Babbie ER. The practice of social research. 14th ed. Belmont: Wadsworth Cengage; 2016.
Google Scholar
Descartes. Cited in Halverston, W. (1976). In: A concise introduction to philosophy, 3rd ed. New York: Random House; 1637.
Doll R, Hill AB. The mortality of doctors in relation to their smoking habits. BMJ. 1954;328(7455):1529–33. https://doi.org/10.1136/bmj.328.7455.1529 .
Article Google Scholar
Liamputtong P. Research methods in health: foundations for evidence-based practice. 3rd ed. Melbourne: Oxford University Press; 2017.
McNabb DE. Research methods in public administration and nonprofit management: quantitative and qualitative approaches. 2nd ed. New York: Armonk; 2007.
Merriam-Webster. Dictionary. http://www.merriam-webster.com . Accessed 20th December 2017.
Olesen Larsen P, von Ins M. The rate of growth in scientific publication and the decline in coverage provided by Science Citation Index. Scientometrics. 2010;84(3):575–603.
Pannucci CJ, Wilkins EG. Identifying and avoiding bias in research. Plast Reconstr Surg. 2010;126(2):619–25. https://doi.org/10.1097/PRS.0b013e3181de24bc .
Petrie A, Sabin C. Medical statistics at a glance. 2nd ed. London: Blackwell Publishing; 2005.
Portney LG, Watkins MP. Foundations of clinical research: applications to practice. 3rd ed. New Jersey: Pearson Publishing; 2009.
Sheehan J. Aspects of research methodology. Nurse Educ Today. 1986;6:193–203.
Wilson LA, Black DA. Health, science research and research methods. Sydney: McGraw Hill; 2013.
Download references
Author information
Authors and affiliations.
School of Science and Health, Western Sydney University, Penrith, NSW, Australia
Leigh A. Wilson
Faculty of Health Science, Discipline of Behavioural and Social Sciences in Health, University of Sydney, Lidcombe, NSW, Australia
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Leigh A. Wilson .
Editor information
Editors and affiliations.
Pranee Liamputtong
Rights and permissions
Reprints and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this entry
Cite this entry.
Wilson, L.A. (2019). Quantitative Research. In: Liamputtong, P. (eds) Handbook of Research Methods in Health Social Sciences. Springer, Singapore. https://doi.org/10.1007/978-981-10-5251-4_54
Download citation
DOI : https://doi.org/10.1007/978-981-10-5251-4_54
Published : 13 January 2019
Publisher Name : Springer, Singapore
Print ISBN : 978-981-10-5250-7
Online ISBN : 978-981-10-5251-4
eBook Packages : Social Sciences Reference Module Humanities and Social Sciences Reference Module Business, Economics and Social Sciences
Share this entry
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
Log in using your username and password
- Search More Search for this keyword Advanced search
- Latest content
- Current issue
- Write for Us
- BMJ Journals More You are viewing from: Google Indexer
You are here
- Volume 21, Issue 4
- How to appraise quantitative research
- Article Text
- Article info
- Citation Tools
- Rapid Responses
- Article metrics
This article has a correction. Please see:
- Correction: How to appraise quantitative research - April 01, 2019
- Xabi Cathala 1 ,
- Calvin Moorley 2
- 1 Institute of Vocational Learning , School of Health and Social Care, London South Bank University , London , UK
- 2 Nursing Research and Diversity in Care , School of Health and Social Care, London South Bank University , London , UK
- Correspondence to Mr Xabi Cathala, Institute of Vocational Learning, School of Health and Social Care, London South Bank University London UK ; cathalax{at}lsbu.ac.uk and Dr Calvin Moorley, Nursing Research and Diversity in Care, School of Health and Social Care, London South Bank University, London SE1 0AA, UK; Moorleyc{at}lsbu.ac.uk
https://doi.org/10.1136/eb-2018-102996
Statistics from Altmetric.com
Request permissions.
If you wish to reuse any or all of this article please use the link below which will take you to the Copyright Clearance Center’s RightsLink service. You will be able to get a quick price and instant permission to reuse the content in many different ways.
Introduction
Some nurses feel that they lack the necessary skills to read a research paper and to then decide if they should implement the findings into their practice. This is particularly the case when considering the results of quantitative research, which often contains the results of statistical testing. However, nurses have a professional responsibility to critique research to improve their practice, care and patient safety. 1 This article provides a step by step guide on how to critically appraise a quantitative paper.
Title, keywords and the authors
The authors’ names may not mean much, but knowing the following will be helpful:
Their position, for example, academic, researcher or healthcare practitioner.
Their qualification, both professional, for example, a nurse or physiotherapist and academic (eg, degree, masters, doctorate).
This can indicate how the research has been conducted and the authors’ competence on the subject. Basically, do you want to read a paper on quantum physics written by a plumber?
The abstract is a resume of the article and should contain:
Introduction.
Research question/hypothesis.
Methods including sample design, tests used and the statistical analysis (of course! Remember we love numbers).
Main findings.
Conclusion.
The subheadings in the abstract will vary depending on the journal. An abstract should not usually be more than 300 words but this varies depending on specific journal requirements. If the above information is contained in the abstract, it can give you an idea about whether the study is relevant to your area of practice. However, before deciding if the results of a research paper are relevant to your practice, it is important to review the overall quality of the article. This can only be done by reading and critically appraising the entire article.
The introduction
Example: the effect of paracetamol on levels of pain.
My hypothesis is that A has an effect on B, for example, paracetamol has an effect on levels of pain.
My null hypothesis is that A has no effect on B, for example, paracetamol has no effect on pain.
My study will test the null hypothesis and if the null hypothesis is validated then the hypothesis is false (A has no effect on B). This means paracetamol has no effect on the level of pain. If the null hypothesis is rejected then the hypothesis is true (A has an effect on B). This means that paracetamol has an effect on the level of pain.
Background/literature review
The literature review should include reference to recent and relevant research in the area. It should summarise what is already known about the topic and why the research study is needed and state what the study will contribute to new knowledge. 5 The literature review should be up to date, usually 5–8 years, but it will depend on the topic and sometimes it is acceptable to include older (seminal) studies.
Methodology
In quantitative studies, the data analysis varies between studies depending on the type of design used. For example, descriptive, correlative or experimental studies all vary. A descriptive study will describe the pattern of a topic related to one or more variable. 6 A correlational study examines the link (correlation) between two variables 7 and focuses on how a variable will react to a change of another variable. In experimental studies, the researchers manipulate variables looking at outcomes 8 and the sample is commonly assigned into different groups (known as randomisation) to determine the effect (causal) of a condition (independent variable) on a certain outcome. This is a common method used in clinical trials.
There should be sufficient detail provided in the methods section for you to replicate the study (should you want to). To enable you to do this, the following sections are normally included:
Overview and rationale for the methodology.
Participants or sample.
Data collection tools.
Methods of data analysis.
Ethical issues.
Data collection should be clearly explained and the article should discuss how this process was undertaken. Data collection should be systematic, objective, precise, repeatable, valid and reliable. Any tool (eg, a questionnaire) used for data collection should have been piloted (or pretested and/or adjusted) to ensure the quality, validity and reliability of the tool. 9 The participants (the sample) and any randomisation technique used should be identified. The sample size is central in quantitative research, as the findings should be able to be generalised for the wider population. 10 The data analysis can be done manually or more complex analyses performed using computer software sometimes with advice of a statistician. From this analysis, results like mode, mean, median, p value, CI and so on are always presented in a numerical format.
The author(s) should present the results clearly. These may be presented in graphs, charts or tables alongside some text. You should perform your own critique of the data analysis process; just because a paper has been published, it does not mean it is perfect. Your findings may be different from the author’s. Through critical analysis the reader may find an error in the study process that authors have not seen or highlighted. These errors can change the study result or change a study you thought was strong to weak. To help you critique a quantitative research paper, some guidance on understanding statistical terminology is provided in table 1 .
- View inline
Some basic guidance for understanding statistics
Quantitative studies examine the relationship between variables, and the p value illustrates this objectively. 11 If the p value is less than 0.05, the null hypothesis is rejected and the hypothesis is accepted and the study will say there is a significant difference. If the p value is more than 0.05, the null hypothesis is accepted then the hypothesis is rejected. The study will say there is no significant difference. As a general rule, a p value of less than 0.05 means, the hypothesis is accepted and if it is more than 0.05 the hypothesis is rejected.
The CI is a number between 0 and 1 or is written as a per cent, demonstrating the level of confidence the reader can have in the result. 12 The CI is calculated by subtracting the p value to 1 (1–p). If there is a p value of 0.05, the CI will be 1–0.05=0.95=95%. A CI over 95% means, we can be confident the result is statistically significant. A CI below 95% means, the result is not statistically significant. The p values and CI highlight the confidence and robustness of a result.
Discussion, recommendations and conclusion
The final section of the paper is where the authors discuss their results and link them to other literature in the area (some of which may have been included in the literature review at the start of the paper). This reminds the reader of what is already known, what the study has found and what new information it adds. The discussion should demonstrate how the authors interpreted their results and how they contribute to new knowledge in the area. Implications for practice and future research should also be highlighted in this section of the paper.
A few other areas you may find helpful are:
Limitations of the study.
Conflicts of interest.
Table 2 provides a useful tool to help you apply the learning in this paper to the critiquing of quantitative research papers.
Quantitative paper appraisal checklist
- 1. ↵ Nursing and Midwifery Council , 2015 . The code: standard of conduct, performance and ethics for nurses and midwives https://www.nmc.org.uk/globalassets/sitedocuments/nmc-publications/nmc-code.pdf ( accessed 21.8.18 ).
- Gerrish K ,
- Moorley C ,
- Tunariu A , et al
- Shorten A ,
Competing interests None declared.
Patient consent Not required.
Provenance and peer review Commissioned; internally peer reviewed.
Correction notice This article has been updated since its original publication to update p values from 0.5 to 0.05 throughout.
Linked Articles
- Miscellaneous Correction: How to appraise quantitative research BMJ Publishing Group Ltd and RCN Publishing Company Ltd Evidence-Based Nursing 2019; 22 62-62 Published Online First: 31 Jan 2019. doi: 10.1136/eb-2018-102996corr1
Read the full text or download the PDF:
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, automatically generate references for free.
- Knowledge Base
- Methodology
- Qualitative vs Quantitative Research | Examples & Methods
Qualitative vs Quantitative Research | Examples & Methods
Published on 4 April 2022 by Raimo Streefkerk . Revised on 8 May 2023.
When collecting and analysing data, quantitative research deals with numbers and statistics, while qualitative research deals with words and meanings. Both are important for gaining different kinds of knowledge.
Common quantitative methods include experiments, observations recorded as numbers, and surveys with closed-ended questions. Qualitative research Qualitative research is expressed in words . It is used to understand concepts, thoughts or experiences. This type of research enables you to gather in-depth insights on topics that are not well understood.
Table of contents
The differences between quantitative and qualitative research, data collection methods, when to use qualitative vs quantitative research, how to analyse qualitative and quantitative data, frequently asked questions about qualitative and quantitative research.
Quantitative and qualitative research use different research methods to collect and analyse data, and they allow you to answer different kinds of research questions.

Prevent plagiarism, run a free check.
Quantitative and qualitative data can be collected using various methods. It is important to use a data collection method that will help answer your research question(s).
Many data collection methods can be either qualitative or quantitative. For example, in surveys, observations or case studies , your data can be represented as numbers (e.g. using rating scales or counting frequencies) or as words (e.g. with open-ended questions or descriptions of what you observe).
However, some methods are more commonly used in one type or the other.
Quantitative data collection methods
- Surveys : List of closed or multiple choice questions that is distributed to a sample (online, in person, or over the phone).
- Experiments : Situation in which variables are controlled and manipulated to establish cause-and-effect relationships.
- Observations: Observing subjects in a natural environment where variables can’t be controlled.
Qualitative data collection methods
- Interviews : Asking open-ended questions verbally to respondents.
- Focus groups: Discussion among a group of people about a topic to gather opinions that can be used for further research.
- Ethnography : Participating in a community or organisation for an extended period of time to closely observe culture and behavior.
- Literature review : Survey of published works by other authors.
A rule of thumb for deciding whether to use qualitative or quantitative data is:
- Use quantitative research if you want to confirm or test something (a theory or hypothesis)
- Use qualitative research if you want to understand something (concepts, thoughts, experiences)
For most research topics you can choose a qualitative, quantitative or mixed methods approach . Which type you choose depends on, among other things, whether you’re taking an inductive vs deductive research approach ; your research question(s) ; whether you’re doing experimental , correlational , or descriptive research ; and practical considerations such as time, money, availability of data, and access to respondents.
Quantitative research approach
You survey 300 students at your university and ask them questions such as: ‘on a scale from 1-5, how satisfied are your with your professors?’
You can perform statistical analysis on the data and draw conclusions such as: ‘on average students rated their professors 4.4’.
Qualitative research approach
You conduct in-depth interviews with 15 students and ask them open-ended questions such as: ‘How satisfied are you with your studies?’, ‘What is the most positive aspect of your study program?’ and ‘What can be done to improve the study program?’
Based on the answers you get you can ask follow-up questions to clarify things. You transcribe all interviews using transcription software and try to find commonalities and patterns.
Mixed methods approach
You conduct interviews to find out how satisfied students are with their studies. Through open-ended questions you learn things you never thought about before and gain new insights. Later, you use a survey to test these insights on a larger scale.
It’s also possible to start with a survey to find out the overall trends, followed by interviews to better understand the reasons behind the trends.
Qualitative or quantitative data by itself can’t prove or demonstrate anything, but has to be analysed to show its meaning in relation to the research questions. The method of analysis differs for each type of data.
Analysing quantitative data
Quantitative data is based on numbers. Simple maths or more advanced statistical analysis is used to discover commonalities or patterns in the data. The results are often reported in graphs and tables.
Applications such as Excel, SPSS, or R can be used to calculate things like:
- Average scores
- The number of times a particular answer was given
- The correlation or causation between two or more variables
- The reliability and validity of the results
Analysing qualitative data
Qualitative data is more difficult to analyse than quantitative data. It consists of text, images or videos instead of numbers.
Some common approaches to analysing qualitative data include:
- Qualitative content analysis : Tracking the occurrence, position and meaning of words or phrases
- Thematic analysis : Closely examining the data to identify the main themes and patterns
- Discourse analysis : Studying how communication works in social contexts
Quantitative research deals with numbers and statistics, while qualitative research deals with words and meanings.
Quantitative methods allow you to test a hypothesis by systematically collecting and analysing data, while qualitative methods allow you to explore ideas and experiences in depth.
In mixed methods research , you use both qualitative and quantitative data collection and analysis methods to answer your research question .
The research methods you use depend on the type of data you need to answer your research question .
- If you want to measure something or test a hypothesis , use quantitative methods . If you want to explore ideas, thoughts, and meanings, use qualitative methods .
- If you want to analyse a large amount of readily available data, use secondary data. If you want data specific to your purposes with control over how they are generated, collect primary data.
- If you want to establish cause-and-effect relationships between variables , use experimental methods. If you want to understand the characteristics of a research subject, use descriptive methods.
Data collection is the systematic process by which observations or measurements are gathered in research. It is used in many different contexts by academics, governments, businesses, and other organisations.
There are various approaches to qualitative data analysis , but they all share five steps in common:
- Prepare and organise your data.
- Review and explore your data.
- Develop a data coding system.
- Assign codes to the data.
- Identify recurring themes.
The specifics of each step depend on the focus of the analysis. Some common approaches include textual analysis , thematic analysis , and discourse analysis .
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the ‘Cite this Scribbr article’ button to automatically add the citation to our free Reference Generator.
Streefkerk, R. (2023, May 08). Qualitative vs Quantitative Research | Examples & Methods. Scribbr. Retrieved 29 April 2024, from https://www.scribbr.co.uk/research-methods/quantitative-qualitative-research/
Is this article helpful?
Raimo Streefkerk
- Discoveries
- Right Journal
- Journal Metrics
- Journal Fit
- Abbreviation
- In-Text Citations
- Bibliographies
- Writing an Article
- Peer Review Types
- Acknowledgements
- Withdrawing a Paper
- Form Letter
- ISO, ANSI, CFR
- Google Scholar
- Journal Manuscript Editing
- Research Manuscript Editing
Book Editing
- Manuscript Editing Services
Medical Editing
- Bioscience Editing
- Physical Science Editing
- PhD Thesis Editing Services
- PhD Editing
- Master’s Proofreading
- Bachelor’s Editing
- Dissertation Proofreading Services
- Best Dissertation Proofreaders
- Masters Dissertation Proofreading
- PhD Proofreaders
- Proofreading PhD Thesis Price
- Journal Article Editing
- Book Editing Service
- Editing and Proofreading Services
- Research Paper Editing
- Medical Manuscript Editing
- Academic Editing
- Social Sciences Editing
- Academic Proofreading
- PhD Theses Editing
- Dissertation Proofreading
- Proofreading Rates UK
- Medical Proofreading
- PhD Proofreading Services UK
- Academic Proofreading Services UK
Medical Editing Services
- Life Science Editing
- Biomedical Editing
- Environmental Science Editing
- Pharmaceutical Science Editing
- Economics Editing
- Psychology Editing
- Sociology Editing
- Archaeology Editing
- History Paper Editing
- Anthropology Editing
- Law Paper Editing
- Engineering Paper Editing
- Technical Paper Editing
- Philosophy Editing
- PhD Dissertation Proofreading
- Lektorat Englisch
- Akademisches Lektorat
- Lektorat Englisch Preise
- Wissenschaftliches Lektorat
- Lektorat Doktorarbeit
PhD Thesis Editing
- Thesis Proofreading Services
- PhD Thesis Proofreading
- Proofreading Thesis Cost
- Proofreading Thesis
- Thesis Editing Services
- Professional Thesis Editing
- Thesis Editing Cost
- Proofreading Dissertation
- Dissertation Proofreading Cost
- Dissertation Proofreader
- Correção de Artigos Científicos
- Correção de Trabalhos Academicos
- Serviços de Correção de Inglês
- Correção de Dissertação
- Correção de Textos Precos
- 定額 ネイティブチェック
- Copy Editing
- FREE Courses
- Revision en Ingles
- Revision de Textos en Ingles
- Revision de Tesis
- Revision Medica en Ingles
- Revision de Tesis Precio
- Revisão de Artigos Científicos
- Revisão de Trabalhos Academicos
- Serviços de Revisão de Inglês
- Revisão de Dissertação
- Revisão de Textos Precos
- Corrección de Textos en Ingles
- Corrección de Tesis
- Corrección de Tesis Precio
- Corrección Medica en Ingles
- Corrector ingles
Select Page
Example of a Quantitative Research Paper
Posted by Rene Tetzner | Sep 4, 2021 | How To Get Published | 0 |

Example of a Quantitative Research Paper for Students & Researchers This example of a quantitative research paper is designed to help students and other researchers who are learning how to write about their work. The reported research observes the behaviour of restaurant customers, and example paragraphs are combined with instructions for logical argumentation. Authors are encouraged to observe a traditional structure for organising quantitative research papers, to formulate research questions, working hypotheses and investigative tools, to report results accurately and thoroughly, and to present thoughtful interpretation and logical discussion of evidence.
The structure of the example and the nature of its contents follow the recommendations of the Publication Manual of the American Psychological Association . This APA style calls for parenthetical author–date citations in the paper’s main text (with page numbers when material is quoted) and a final list of complete references for all sources cited, so I have given a few sample references here. Content has been kept as simple as possible to focus attention on the way in which the paper presents the research process and its results. As is the case in many research projects, the more the author learns and thinks about the topic, the more complex the issues become, and here the researcher discusses a hypothesis that proved incorrect. An APA research paper would normally include additional elements such as an abstract, keywords and perhaps tables, figures and appendices similar to those referred to in the example. These elements have been eliminated for brevity here, so do be sure to check the APA Manual (or any other guidelines you are following) for the necessary instructions.

Surprises at a Local “Family” Restaurant: Example Quantitative Research Paper
A quantitative research paper with that title might start with a paragraph like this:
Quaintville, located just off the main highway only five miles from the university campus, may normally be a sleepy community, but recent plans to close the only fast-food restaurant ever to grace its main street have been met with something of a public outcry. Regular clients argue that Pudgy’s Burgers fills a vital function and will be sorely missed. As the editor of the Quaintville Times would have it, “good old Pudgy’s is the only restaurant in Quaintville where a working family can still get a decent meal for a fair buck, and a comfortable place to eat it too, out of the winter wind where the kids can run about and play a bit” (Chapton, 2017, p. A3). On the other hand, the most outspoken of Quaintville residents in favour of the planned closure look forward to the eradication of a local eyesore and tend to consider the restaurant more of “a hazard than a benefit to the health of some of our poorest families” (“Local dive,” 2017, p. 1).
Following this opening a brief introduction to published scholarship and other issues associated with the problem would be appropriate, so here the researcher might add a paragraph or two discussing:
• A selection of recently published studies that investigate the effect of inexpensive fast-food restaurants on the health of low-income families, especially their children (Shunts, 2013; Whinner, 2015). • Fast-food restaurants that have responded to criticism about the quality of their food by offering healthy menu items. This could be enhanced with evidence that when such choices are available, they are rare purchases for many families (Parkson, 2016), particularly in small towns and rural areas (Shemble, 2017). • The interesting trend in several independent studies suggesting that families form a much smaller portion of the clientele of fast-food restaurants than anticipated.

Explaining how the current research is related to the published scholarship as well as the specific problem is vital. Here, for instance, the author might be thinking that Pudgy’s, which has healthy menu items as well as the support of so many long-term residents, will prove an exception to the trends revealed by other studies. Research questions and hypotheses should be constructed to articulate and explore that idea. Research questions, for instance, could be developed from that claim in the Quaintville Times as well as from the published scholarship:
• Do families constitute the majority of Pudgy’s regular clientele? • Does the restaurant offer a decent family meal for a fair price? • Do families linger in the restaurant’s comfort and warmth?> • Do children use the indoor play area provided by the restaurant?
Working hypotheses can be constructed by anticipating answers to these questions. The example paper assumes a simple hypothesis something along the lines of “Families do indeed constitute the majority of Pudgy’s clientele.” The exact opposite supposition would work as well – “Families do not constitute the majority of Pudgy’s clientele” – and so would hypotheses exploring and combining other aspects of the situation, such as “Pudgy’s healthy menu options and indoor play area are positive and appealing considerations for families” or “The comfortable atmosphere of Pudgy’s with its play area makes it much more than a restaurant for local families.”

The exact wording of your questions and hypotheses will ultimately depend on your focus and aims, but certain terms, concepts and categories may require definition to ensure precision in communicating your ideas to readers. Here, for instance, exactly what is meant by ‘a family,’ ‘a decent meal,’ ‘a fair price’ and even ‘comfortable’ could be briefly but carefully defined. A general statement about your understanding of how the current research will explore the problem, answer your questions and test your hypotheses is usually required as well, setting the stage for the more detailed Method section that follows. This statement might be something as simple as “I intend to observe the restaurant’s customers over a two-month period with the objective of learning about Pudgy’s clientele and measuring the use and value of the establishment for local families.” On the other hand, outlining your research might require a paragraph or two of introductory discussion.
Method Whether a brief general statement or a longer explanation of how the research will proceed appears among your introductory material, it is in the Method section that you should report exactly what you did to conduct your investigation, explain the conditions and controls you applied to increase the reliability and value of your research, and reveal any difficulties you encountered. For example:
My observations took place at Pudgy’s Burgers in January and February of 2018. Each session was approximately four hours long, and I aimed to obtain an equivalent number of observations for all opening hours of the week (the restaurant’s hours are listed in Table 1), but course requirements made this difficult. Tuesday and Thursday afternoons are therefore underrepresented, and observations from 1:00 pm to 5:00 pm on two consecutive Tuesdays (6 and 13 February) are the work of my classmate, Jake Jenkins. Without his assistance, I could not have met my objective of gathering observations for every opening hour of the week at least twice (Table 2 outlines the overall pattern of observation sessions). Serving staff at the restaurant assure me that I have now “seen ‘em all,” so I believe my observations have resulted in a representative sampling of local customers over two months when that “winter wind” has been especially busy about its work.
To avoid detection by the customers I was observing and the possibility of altering their behaviour, I obtained permission from Pudgy’s manager, Mr Jobson, to sit at the staff table in a dark and quiet corner of the restaurant where clients never go. This table is labelled in the plan of Pudgy’s Burgers and its grounds that I have included as Figure 1. From there I could see the customers both at the service counter and at their tables, but they could not see me, at least not clearly, and if they did, they paid me no more attention than they did the restaurant employees. From the staff table I could also see the row of indoor park-style children’s toys running down the north wall of windows, as well as the take out lane and the people waiting in their cars.
A Method section often features subheadings to separate and present particularly important aspects of the research methodology, such as the Customer Fact Sheet developed and used by the author of this study.
The Customer Fact Sheet Recording thorough and equivalent information about every Pudgy’s customer I observed was crucial for quantifying and analysing the results of my study. I therefore prepared a Customer Fact Sheet (included as Appendix I at the end of this paper) for gathering key pieces of information and recording observations about each individual, couple or group who purchased food or beverages. This sheet ensured that vital details such as date, weather conditions, time of arrival, eat in or take out order, number in party, approximate age of individuals, food purchased, food consumed, healthy choices, amount spent, who paid, dessert or extra beverage, children playing, interaction with other children and families, time of departure and other important details were recorded in every case. The Customer Fact Sheet proved particularly helpful when my classmate performed observations for me and was invaluable for evaluating the data I collected. I initially hoped to complete at least 500 of these Customer Fact Sheets and was pleased to increase that number by 100 for a total of 600 or an average of just over 10 per day over the 59 days of the study.
Notice in the three example paragraphs for the Method section that clear references to Tables 1 & 2, Figure 1 and Appendix I are provided to let readers know when and why these extra elements are relevant and helpful. Be sure also to include in your description of methods any additional approaches or sources of information that should be considered part of your research procedures, such as:
• Receipt information about customer purchases provided by the restaurant manager. • Conversations with restaurant servers who might confirm family relationships and estimated ages or tell you what was eaten and what was not by particular customer groups. • The analysis you performed to make sense of your results, such as counting customers, meals and behaviours and working out percentages and averages overall as well as for certain categories in order to answer the research questions.
Results The Results section is where you report what you discovered during your research, including the findings that do not support your hypothesis (or hypotheses) as well as those that do. Returning to your research questions to indicate exactly how the data you gathered answers them is an excellent way to stay focused and enable the selectivity that may be necessary to meet length requirements or maintain a clear line of argumentation. A Results section for the Pudgy’s research project might start like this:
The results of my investigation were both surprising and more complex than I had anticipated. I asked whether families constituted the majority of Pudgy’s clientele and assumed they did, but my research shows that they do not (see Figure 2 for information on customer categories). Even when the loosest definition of family as explained in my introduction is applied, only slightly over 25% (152) of the 600 Customer Fact Sheets record family visits to the restaurant. Among them fathers alone with their children are the most frequent patrons (68 Customer Fact Sheets or nearly 45% of the family category). The only day of the week on which families approach 50% of the restaurant’s customers is Sunday, particularly in the afternoon, when family groups account for 48% of the total customers averaged over the eight Sundays of observation. On all other days of the week, individual customers are the most frequent patrons, with their numbers hovering around 50% on most days. Single men visit the restaurant more often than any other customers and constitute as much as 61% of the clientele on a few weekday evenings.
The report of results might then continue by providing information about other categories of customer, what different types of customers ate and did, and any additional results that help answer the other research questions posed in the introductory paragraphs. Major trends revealed by the data should be reported, and both content and writing style should be clear and factual. Interpretation and discussion are best saved for the Discussion section except in those rare instances when guidelines indicate that research results and discussion should be combined in a single section. Although you will need to inform readers about any mathematical or statistical analysis of your raw data if you have not already done so in the Method section, the raw data itself is usually not appropriate for a short research paper. Selecting the most convincing and relevant evidence as the focus is, however, and the raw data can usually be made available via a university’s website or a journal’s online archives for expert readers and future researchers.
Discussion The Discussion section of a quantitative paper is where you interpret your research results and discuss their implications. Here the hypotheses as well as the research questions established in the introductory material are important. Were your primary suppositions confirmed by your results or not? Be precise and concise as you discuss your findings, but keep in mind that matters need not be quite as black and white or as strictly factual as they were in the Results section. Your ideas and argument should be soundly based on the data you collected, of course, but the Discussion is the place for describing complexities and expressing uncertainties as well as offering interpretations and explanations. The following opening briefly restates primary findings, picks up other important threads from the Results section and sets the stage for discussing the complexities involved in assessing the true value of Pudgy’s to the Quaintville community:
Although I had anticipated that families constitute the majority of Pudgy’s clientele, the evidence gathered over two months of observation does not support this supposition. In fact, individuals are the most frequent customers, with groups of teenagers running a close second. These teenagers are often in the restaurant when families are and they sometimes sit on the indoor toys instead of at the plastic tables and chairs, which I can confirm as extremely uncomfortable. On a few occasions the presence of teenagers appeared to intimidate the children and prevent them from playing on the facilities intended for them. In accordance with Parkson (2016) and Shemble (2017), my research also showed that most families who eat at Pudgy’s do not choose the healthier low-fat menu items, with the limited number and extremely high prices of these items offering little incentive. The few parents who make healthy choices for themselves and their children often do not insist upon the children eating those items, adding waste (of both food and money) to the problem. Furthermore, although Pudgy’s prices for their more traditional fast-food items are the lowest in town, at least two of the restaurants in Quaintville offer equivalent meals for similar prices and far healthier ones for just a little more.
The claim, then, in the Quaintville Times that “good old Pudgy’s is the only restaurant in Quaintville where a working family can still get a decent meal for a fair buck, and a comfortable place to eat it too, out of the winter wind where the kids can run about and play a bit” (Chapton, 2017, p.A3) is revealed as more sentiment than fact. It would be equally erroneous, however, to insist that Pudgy’s Burgers has no value for the local community or to call it more of “a hazard…to the health of some of our poorest families” (“Local dive,” 2017, p.1) than any other restaurants serving burgers and chips in Quaintville. Indeed, I suspect those “poorest families” very rarely visit local restaurants at all, but my observations have revealed a great deal about who does eat at Pudgy’s, what they do when they are there and what kind of value the establishment actually has for Quaintville residents.
The discussion could then continue with information about the customers, behaviours and other issues that render the findings more complex and the restaurant more valuable to the community than the primary results noted above may indicate:
• Perhaps the restaurant serves a vital function as a social gathering place for all those single customers. Do they usually remain alone or do they meet up with others to linger and talk over coffee or lunch? • Do the teenagers who gather at Pudgy’s have an alternative place to meet out of the cold? In towns without recreation centres or other facilities for teens, restaurants with informal, open-door policies can be vital. Where might those teenagers go or what might they be doing were Pudgy’s not there? • Even though the evidence showed that families are not the most frequent customers, you may want to consider the value the restaurant has for the families who do use it. Those single fathers are certainly worthy of some attention, for instance, and perhaps family groups occasionally met up with other families, ate together and then lingered for dessert and talk as their children enjoyed the toys. This would be worth discussing too. • Less measurable considerations viewed through a qualitative research lens may be helpful as well, but the data collected through observations should support such discussions. Remember as you analyse your data, reflect on your findings, determine their meaning and develop your argument that it is important to keep the limitations of your methodology and thus of your results and their implications clearly in mind.
Offering recommendations is also standard in the Discussion section of a quantitative research paper, and here recommendations might be particularly useful if the franchise had not yet finalised its decision about closing Pudgy’s and was actively seeking community feedback. The researcher might suggest that Pudgy’s could better serve families by increasing the number of healthy food items on the menu, offering these for more affordable prices and making an effort to keep the teenagers off the children’s toys. Finally, the last part of a Discussion usually provides concluding comments, so summarising your key points and clearly articulating the main messages you want your readers to take away with them are essential. In some organisational templates, Conclusions are offered in a separate final section of the paper instead of at the end of the Discussion, so always check the guidelines.
References These references follow APA style, but since special fonts may not display properly in all online situations, please note that the titles of books and the names and volume numbers of journals are (and should be) in italic font. The list represents a sample only; a paper the length of the one posited in this example would almost certainly mention, discuss and list more than half a dozen studies and sources.
Chapton, D. (2017, September 29). Will Quaintville lose its favourite family restaurant? Quaintville Times , pp. A1, A3. Local dive sees last days. (2017, Autumn). Quaintville Community Newsletter , pp. 1–2. Shemble, M. (2017). Is anyone really eating healthy fast food in rural towns? Country Food & Families , 14 , 12–23. Shunts, P. (2013). The true cost of high-fat fast food for low-income families. Journal of Family Health & Diet , 37 , 3–19. Parkson, L. (2016). Family diets, fast foods and unhealthy choices. In S. Smith & J. Jones (eds.), Modern diets and family health (pp. 277–294). Philadelphia, PA: The Family Press. Whinner, N. (2015). Healthy families take time: The impact of fatty fast foods on child health. Journal of Family Health & Diet , 39 , 31–43.
You might be interested in Services offered by Proof-Reading-Service.com
Journal editing.
Journal article editing services
PhD thesis editing services
Scientific Editing
Manuscript editing.
Manuscript editing services
Expert Editing
Expert editing for all papers

Research Editing
Research paper editing services
Professional book editing services
Example of a Quantitative Research Paper This example is organised into introductory material, method, results & discussion.
Related Posts

Choosing the Right Journal
September 10, 2021

What Is a Good H-Index Required for an Academic Position?
September 3, 2021

Acknowledgements Example for an Academic Research Paper
September 1, 2021

Free Sample Letters for Withdrawing a Manuscript
August 31, 2021
Our Recent Posts

Our review ratings
- Examples of Research Paper Topics in Different Study Areas Score: 98%
- Dealing with Language Problems – Journal Editor’s Feedback Score: 95%
- Making Good Use of a Professional Proofreader Score: 92%
- How To Format Your Journal Paper Using Published Articles Score: 95%
- Journal Rejection as Inspiration for a New Perspective Score: 95%
Explore our Categories
- Abbreviation in Academic Writing (4)
- Career Advice for Academics (5)
- Dealing with Paper Rejection (11)
- Grammar in Academic Writing (5)
- Help with Peer Review (7)
- How To Get Published (146)
- Paper Writing Advice (17)
- Referencing & Bibliographies (16)
- Open access
- Published: 18 April 2024
Research ethics and artificial intelligence for global health: perspectives from the global forum on bioethics in research
- James Shaw 1 , 13 ,
- Joseph Ali 2 , 3 ,
- Caesar A. Atuire 4 , 5 ,
- Phaik Yeong Cheah 6 ,
- Armando Guio Español 7 ,
- Judy Wawira Gichoya 8 ,
- Adrienne Hunt 9 ,
- Daudi Jjingo 10 ,
- Katherine Littler 9 ,
- Daniela Paolotti 11 &
- Effy Vayena 12
BMC Medical Ethics volume 25 , Article number: 46 ( 2024 ) Cite this article
1214 Accesses
6 Altmetric
Metrics details
The ethical governance of Artificial Intelligence (AI) in health care and public health continues to be an urgent issue for attention in policy, research, and practice. In this paper we report on central themes related to challenges and strategies for promoting ethics in research involving AI in global health, arising from the Global Forum on Bioethics in Research (GFBR), held in Cape Town, South Africa in November 2022.
The GFBR is an annual meeting organized by the World Health Organization and supported by the Wellcome Trust, the US National Institutes of Health, the UK Medical Research Council (MRC) and the South African MRC. The forum aims to bring together ethicists, researchers, policymakers, research ethics committee members and other actors to engage with challenges and opportunities specifically related to research ethics. In 2022 the focus of the GFBR was “Ethics of AI in Global Health Research”. The forum consisted of 6 case study presentations, 16 governance presentations, and a series of small group and large group discussions. A total of 87 participants attended the forum from 31 countries around the world, representing disciplines of bioethics, AI, health policy, health professional practice, research funding, and bioinformatics. In this paper, we highlight central insights arising from GFBR 2022.
We describe the significance of four thematic insights arising from the forum: (1) Appropriateness of building AI, (2) Transferability of AI systems, (3) Accountability for AI decision-making and outcomes, and (4) Individual consent. We then describe eight recommendations for governance leaders to enhance the ethical governance of AI in global health research, addressing issues such as AI impact assessments, environmental values, and fair partnerships.
Conclusions
The 2022 Global Forum on Bioethics in Research illustrated several innovations in ethical governance of AI for global health research, as well as several areas in need of urgent attention internationally. This summary is intended to inform international and domestic efforts to strengthen research ethics and support the evolution of governance leadership to meet the demands of AI in global health research.
Peer Review reports
Introduction
The ethical governance of Artificial Intelligence (AI) in health care and public health continues to be an urgent issue for attention in policy, research, and practice [ 1 , 2 , 3 ]. Beyond the growing number of AI applications being implemented in health care, capabilities of AI models such as Large Language Models (LLMs) expand the potential reach and significance of AI technologies across health-related fields [ 4 , 5 ]. Discussion about effective, ethical governance of AI technologies has spanned a range of governance approaches, including government regulation, organizational decision-making, professional self-regulation, and research ethics review [ 6 , 7 , 8 ]. In this paper, we report on central themes related to challenges and strategies for promoting ethics in research involving AI in global health research, arising from the Global Forum on Bioethics in Research (GFBR), held in Cape Town, South Africa in November 2022. Although applications of AI for research, health care, and public health are diverse and advancing rapidly, the insights generated at the forum remain highly relevant from a global health perspective. After summarizing important context for work in this domain, we highlight categories of ethical issues emphasized at the forum for attention from a research ethics perspective internationally. We then outline strategies proposed for research, innovation, and governance to support more ethical AI for global health.
In this paper, we adopt the definition of AI systems provided by the Organization for Economic Cooperation and Development (OECD) as our starting point. Their definition states that an AI system is “a machine-based system that can, for a given set of human-defined objectives, make predictions, recommendations, or decisions influencing real or virtual environments. AI systems are designed to operate with varying levels of autonomy” [ 9 ]. The conceptualization of an algorithm as helping to constitute an AI system, along with hardware, other elements of software, and a particular context of use, illustrates the wide variety of ways in which AI can be applied. We have found it useful to differentiate applications of AI in research as those classified as “AI systems for discovery” and “AI systems for intervention”. An AI system for discovery is one that is intended to generate new knowledge, for example in drug discovery or public health research in which researchers are seeking potential targets for intervention, innovation, or further research. An AI system for intervention is one that directly contributes to enacting an intervention in a particular context, for example informing decision-making at the point of care or assisting with accuracy in a surgical procedure.
The mandate of the GFBR is to take a broad view of what constitutes research and its regulation in global health, with special attention to bioethics in Low- and Middle- Income Countries. AI as a group of technologies demands such a broad view. AI development for health occurs in a variety of environments, including universities and academic health sciences centers where research ethics review remains an important element of the governance of science and innovation internationally [ 10 , 11 ]. In these settings, research ethics committees (RECs; also known by different names such as Institutional Review Boards or IRBs) make decisions about the ethical appropriateness of projects proposed by researchers and other institutional members, ultimately determining whether a given project is allowed to proceed on ethical grounds [ 12 ].
However, research involving AI for health also takes place in large corporations and smaller scale start-ups, which in some jurisdictions fall outside the scope of research ethics regulation. In the domain of AI, the question of what constitutes research also becomes blurred. For example, is the development of an algorithm itself considered a part of the research process? Or only when that algorithm is tested under the formal constraints of a systematic research methodology? In this paper we take an inclusive view, in which AI development is included in the definition of research activity and within scope for our inquiry, regardless of the setting in which it takes place. This broad perspective characterizes the approach to “research ethics” we take in this paper, extending beyond the work of RECs to include the ethical analysis of the wide range of activities that constitute research as the generation of new knowledge and intervention in the world.
Ethical governance of AI in global health
The ethical governance of AI for global health has been widely discussed in recent years. The World Health Organization (WHO) released its guidelines on ethics and governance of AI for health in 2021, endorsing a set of six ethical principles and exploring the relevance of those principles through a variety of use cases. The WHO guidelines also provided an overview of AI governance, defining governance as covering “a range of steering and rule-making functions of governments and other decision-makers, including international health agencies, for the achievement of national health policy objectives conducive to universal health coverage.” (p. 81) The report usefully provided a series of recommendations related to governance of seven domains pertaining to AI for health: data, benefit sharing, the private sector, the public sector, regulation, policy observatories/model legislation, and global governance. The report acknowledges that much work is yet to be done to advance international cooperation on AI governance, especially related to prioritizing voices from Low- and Middle-Income Countries (LMICs) in global dialogue.
One important point emphasized in the WHO report that reinforces the broader literature on global governance of AI is the distribution of responsibility across a wide range of actors in the AI ecosystem. This is especially important to highlight when focused on research for global health, which is specifically about work that transcends national borders. Alami et al. (2020) discussed the unique risks raised by AI research in global health, ranging from the unavailability of data in many LMICs required to train locally relevant AI models to the capacity of health systems to absorb new AI technologies that demand the use of resources from elsewhere in the system. These observations illustrate the need to identify the unique issues posed by AI research for global health specifically, and the strategies that can be employed by all those implicated in AI governance to promote ethically responsible use of AI in global health research.
RECs and the regulation of research involving AI
RECs represent an important element of the governance of AI for global health research, and thus warrant further commentary as background to our paper. Despite the importance of RECs, foundational questions have been raised about their capabilities to accurately understand and address ethical issues raised by studies involving AI. Rahimzadeh et al. (2023) outlined how RECs in the United States are under-prepared to align with recent federal policy requiring that RECs review data sharing and management plans with attention to the unique ethical issues raised in AI research for health [ 13 ]. Similar research in South Africa identified variability in understanding of existing regulations and ethical issues associated with health-related big data sharing and management among research ethics committee members [ 14 , 15 ]. The effort to address harms accruing to groups or communities as opposed to individuals whose data are included in AI research has also been identified as a unique challenge for RECs [ 16 , 17 ]. Doerr and Meeder (2022) suggested that current regulatory frameworks for research ethics might actually prevent RECs from adequately addressing such issues, as they are deemed out of scope of REC review [ 16 ]. Furthermore, research in the United Kingdom and Canada has suggested that researchers using AI methods for health tend to distinguish between ethical issues and social impact of their research, adopting an overly narrow view of what constitutes ethical issues in their work [ 18 ].
The challenges for RECs in adequately addressing ethical issues in AI research for health care and public health exceed a straightforward survey of ethical considerations. As Ferretti et al. (2021) contend, some capabilities of RECs adequately cover certain issues in AI-based health research, such as the common occurrence of conflicts of interest where researchers who accept funds from commercial technology providers are implicitly incentivized to produce results that align with commercial interests [ 12 ]. However, some features of REC review require reform to adequately meet ethical needs. Ferretti et al. outlined weaknesses of RECs that are longstanding and those that are novel to AI-related projects, proposing a series of directions for development that are regulatory, procedural, and complementary to REC functionality. The work required on a global scale to update the REC function in response to the demands of research involving AI is substantial.
These issues take greater urgency in the context of global health [ 19 ]. Teixeira da Silva (2022) described the global practice of “ethics dumping”, where researchers from high income countries bring ethically contentious practices to RECs in low-income countries as a strategy to gain approval and move projects forward [ 20 ]. Although not yet systematically documented in AI research for health, risk of ethics dumping in AI research is high. Evidence is already emerging of practices of “health data colonialism”, in which AI researchers and developers from large organizations in high-income countries acquire data to build algorithms in LMICs to avoid stricter regulations [ 21 ]. This specific practice is part of a larger collection of practices that characterize health data colonialism, involving the broader exploitation of data and the populations they represent primarily for commercial gain [ 21 , 22 ]. As an additional complication, AI algorithms trained on data from high-income contexts are unlikely to apply in straightforward ways to LMIC settings [ 21 , 23 ]. In the context of global health, there is widespread acknowledgement about the need to not only enhance the knowledge base of REC members about AI-based methods internationally, but to acknowledge the broader shifts required to encourage their capabilities to more fully address these and other ethical issues associated with AI research for health [ 8 ].
Although RECs are an important part of the story of the ethical governance of AI for global health research, they are not the only part. The responsibilities of supra-national entities such as the World Health Organization, national governments, organizational leaders, commercial AI technology providers, health care professionals, and other groups continue to be worked out internationally. In this context of ongoing work, examining issues that demand attention and strategies to address them remains an urgent and valuable task.
The GFBR is an annual meeting organized by the World Health Organization and supported by the Wellcome Trust, the US National Institutes of Health, the UK Medical Research Council (MRC) and the South African MRC. The forum aims to bring together ethicists, researchers, policymakers, REC members and other actors to engage with challenges and opportunities specifically related to research ethics. Each year the GFBR meeting includes a series of case studies and keynotes presented in plenary format to an audience of approximately 100 people who have applied and been competitively selected to attend, along with small-group breakout discussions to advance thinking on related issues. The specific topic of the forum changes each year, with past topics including ethical issues in research with people living with mental health conditions (2021), genome editing (2019), and biobanking/data sharing (2018). The forum is intended to remain grounded in the practical challenges of engaging in research ethics, with special interest in low resource settings from a global health perspective. A post-meeting fellowship scheme is open to all LMIC participants, providing a unique opportunity to apply for funding to further explore and address the ethical challenges that are identified during the meeting.
In 2022, the focus of the GFBR was “Ethics of AI in Global Health Research”. The forum consisted of 6 case study presentations (both short and long form) reporting on specific initiatives related to research ethics and AI for health, and 16 governance presentations (both short and long form) reporting on actual approaches to governing AI in different country settings. A keynote presentation from Professor Effy Vayena addressed the topic of the broader context for AI ethics in a rapidly evolving field. A total of 87 participants attended the forum from 31 countries around the world, representing disciplines of bioethics, AI, health policy, health professional practice, research funding, and bioinformatics. The 2-day forum addressed a wide range of themes. The conference report provides a detailed overview of each of the specific topics addressed while a policy paper outlines the cross-cutting themes (both documents are available at the GFBR website: https://www.gfbr.global/past-meetings/16th-forum-cape-town-south-africa-29-30-november-2022/ ). As opposed to providing a detailed summary in this paper, we aim to briefly highlight central issues raised, solutions proposed, and the challenges facing the research ethics community in the years to come.
In this way, our primary aim in this paper is to present a synthesis of the challenges and opportunities raised at the GFBR meeting and in the planning process, followed by our reflections as a group of authors on their significance for governance leaders in the coming years. We acknowledge that the views represented at the meeting and in our results are a partial representation of the universe of views on this topic; however, the GFBR leadership invested a great deal of resources in convening a deeply diverse and thoughtful group of researchers and practitioners working on themes of bioethics related to AI for global health including those based in LMICs. We contend that it remains rare to convene such a strong group for an extended time and believe that many of the challenges and opportunities raised demand attention for more ethical futures of AI for health. Nonetheless, our results are primarily descriptive and are thus not explicitly grounded in a normative argument. We make effort in the Discussion section to contextualize our results by describing their significance and connecting them to broader efforts to reform global health research and practice.
Uniquely important ethical issues for AI in global health research
Presentations and group dialogue over the course of the forum raised several issues for consideration, and here we describe four overarching themes for the ethical governance of AI in global health research. Brief descriptions of each issue can be found in Table 1 . Reports referred to throughout the paper are available at the GFBR website provided above.
The first overarching thematic issue relates to the appropriateness of building AI technologies in response to health-related challenges in the first place. Case study presentations referred to initiatives where AI technologies were highly appropriate, such as in ear shape biometric identification to more accurately link electronic health care records to individual patients in Zambia (Alinani Simukanga). Although important ethical issues were raised with respect to privacy, trust, and community engagement in this initiative, the AI-based solution was appropriately matched to the challenge of accurately linking electronic records to specific patient identities. In contrast, forum participants raised questions about the appropriateness of an initiative using AI to improve the quality of handwashing practices in an acute care hospital in India (Niyoshi Shah), which led to gaming the algorithm. Overall, participants acknowledged the dangers of techno-solutionism, in which AI researchers and developers treat AI technologies as the most obvious solutions to problems that in actuality demand much more complex strategies to address [ 24 ]. However, forum participants agreed that RECs in different contexts have differing degrees of power to raise issues of the appropriateness of an AI-based intervention.
The second overarching thematic issue related to whether and how AI-based systems transfer from one national health context to another. One central issue raised by a number of case study presentations related to the challenges of validating an algorithm with data collected in a local environment. For example, one case study presentation described a project that would involve the collection of personally identifiable data for sensitive group identities, such as tribe, clan, or religion, in the jurisdictions involved (South Africa, Nigeria, Tanzania, Uganda and the US; Gakii Masunga). Doing so would enable the team to ensure that those groups were adequately represented in the dataset to ensure the resulting algorithm was not biased against specific community groups when deployed in that context. However, some members of these communities might desire to be represented in the dataset, whereas others might not, illustrating the need to balance autonomy and inclusivity. It was also widely recognized that collecting these data is an immense challenge, particularly when historically oppressive practices have led to a low-trust environment for international organizations and the technologies they produce. It is important to note that in some countries such as South Africa and Rwanda, it is illegal to collect information such as race and tribal identities, re-emphasizing the importance for cultural awareness and avoiding “one size fits all” solutions.
The third overarching thematic issue is related to understanding accountabilities for both the impacts of AI technologies and governance decision-making regarding their use. Where global health research involving AI leads to longer-term harms that might fall outside the usual scope of issues considered by a REC, who is to be held accountable, and how? This question was raised as one that requires much further attention, with law being mixed internationally regarding the mechanisms available to hold researchers, innovators, and their institutions accountable over the longer term. However, it was recognized in breakout group discussion that many jurisdictions are developing strong data protection regimes related specifically to international collaboration for research involving health data. For example, Kenya’s Data Protection Act requires that any internationally funded projects have a local principal investigator who will hold accountability for how data are shared and used [ 25 ]. The issue of research partnerships with commercial entities was raised by many participants in the context of accountability, pointing toward the urgent need for clear principles related to strategies for engagement with commercial technology companies in global health research.
The fourth and final overarching thematic issue raised here is that of consent. The issue of consent was framed by the widely shared recognition that models of individual, explicit consent might not produce a supportive environment for AI innovation that relies on the secondary uses of health-related datasets to build AI algorithms. Given this recognition, approaches such as community oversight of health data uses were suggested as a potential solution. However, the details of implementing such community oversight mechanisms require much further attention, particularly given the unique perspectives on health data in different country settings in global health research. Furthermore, some uses of health data do continue to require consent. One case study of South Africa, Nigeria, Kenya, Ethiopia and Uganda suggested that when health data are shared across borders, individual consent remains necessary when data is transferred from certain countries (Nezerith Cengiz). Broader clarity is necessary to support the ethical governance of health data uses for AI in global health research.
Recommendations for ethical governance of AI in global health research
Dialogue at the forum led to a range of suggestions for promoting ethical conduct of AI research for global health, related to the various roles of actors involved in the governance of AI research broadly defined. The strategies are written for actors we refer to as “governance leaders”, those people distributed throughout the AI for global health research ecosystem who are responsible for ensuring the ethical and socially responsible conduct of global health research involving AI (including researchers themselves). These include RECs, government regulators, health care leaders, health professionals, corporate social accountability officers, and others. Enacting these strategies would bolster the ethical governance of AI for global health more generally, enabling multiple actors to fulfill their roles related to governing research and development activities carried out across multiple organizations, including universities, academic health sciences centers, start-ups, and technology corporations. Specific suggestions are summarized in Table 2 .
First, forum participants suggested that governance leaders including RECs, should remain up to date on recent advances in the regulation of AI for health. Regulation of AI for health advances rapidly and takes on different forms in jurisdictions around the world. RECs play an important role in governance, but only a partial role; it was deemed important for RECs to acknowledge how they fit within a broader governance ecosystem in order to more effectively address the issues within their scope. Not only RECs but organizational leaders responsible for procurement, researchers, and commercial actors should all commit to efforts to remain up to date about the relevant approaches to regulating AI for health care and public health in jurisdictions internationally. In this way, governance can more adequately remain up to date with advances in regulation.
Second, forum participants suggested that governance leaders should focus on ethical governance of health data as a basis for ethical global health AI research. Health data are considered the foundation of AI development, being used to train AI algorithms for various uses [ 26 ]. By focusing on ethical governance of health data generation, sharing, and use, multiple actors will help to build an ethical foundation for AI development among global health researchers.
Third, forum participants believed that governance processes should incorporate AI impact assessments where appropriate. An AI impact assessment is the process of evaluating the potential effects, both positive and negative, of implementing an AI algorithm on individuals, society, and various stakeholders, generally over time frames specified in advance of implementation [ 27 ]. Although not all types of AI research in global health would warrant an AI impact assessment, this is especially relevant for those studies aiming to implement an AI system for intervention into health care or public health. Organizations such as RECs can use AI impact assessments to boost understanding of potential harms at the outset of a research project, encouraging researchers to more deeply consider potential harms in the development of their study.
Fourth, forum participants suggested that governance decisions should incorporate the use of environmental impact assessments, or at least the incorporation of environment values when assessing the potential impact of an AI system. An environmental impact assessment involves evaluating and anticipating the potential environmental effects of a proposed project to inform ethical decision-making that supports sustainability [ 28 ]. Although a relatively new consideration in research ethics conversations [ 29 ], the environmental impact of building technologies is a crucial consideration for the public health commitment to environmental sustainability. Governance leaders can use environmental impact assessments to boost understanding of potential environmental harms linked to AI research projects in global health over both the shorter and longer terms.
Fifth, forum participants suggested that governance leaders should require stronger transparency in the development of AI algorithms in global health research. Transparency was considered essential in the design and development of AI algorithms for global health to ensure ethical and accountable decision-making throughout the process. Furthermore, whether and how researchers have considered the unique contexts into which such algorithms may be deployed can be surfaced through stronger transparency, for example in describing what primary considerations were made at the outset of the project and which stakeholders were consulted along the way. Sharing information about data provenance and methods used in AI development will also enhance the trustworthiness of the AI-based research process.
Sixth, forum participants suggested that governance leaders can encourage or require community engagement at various points throughout an AI project. It was considered that engaging patients and communities is crucial in AI algorithm development to ensure that the technology aligns with community needs and values. However, participants acknowledged that this is not a straightforward process. Effective community engagement requires lengthy commitments to meeting with and hearing from diverse communities in a given setting, and demands a particular set of skills in communication and dialogue that are not possessed by all researchers. Encouraging AI researchers to begin this process early and build long-term partnerships with community members is a promising strategy to deepen community engagement in AI research for global health. One notable recommendation was that research funders have an opportunity to incentivize and enable community engagement with funds dedicated to these activities in AI research in global health.
Seventh, forum participants suggested that governance leaders can encourage researchers to build strong, fair partnerships between institutions and individuals across country settings. In a context of longstanding imbalances in geopolitical and economic power, fair partnerships in global health demand a priori commitments to share benefits related to advances in medical technologies, knowledge, and financial gains. Although enforcement of this point might be beyond the remit of RECs, commentary will encourage researchers to consider stronger, fairer partnerships in global health in the longer term.
Eighth, it became evident that it is necessary to explore new forms of regulatory experimentation given the complexity of regulating a technology of this nature. In addition, the health sector has a series of particularities that make it especially complicated to generate rules that have not been previously tested. Several participants highlighted the desire to promote spaces for experimentation such as regulatory sandboxes or innovation hubs in health. These spaces can have several benefits for addressing issues surrounding the regulation of AI in the health sector, such as: (i) increasing the capacities and knowledge of health authorities about this technology; (ii) identifying the major problems surrounding AI regulation in the health sector; (iii) establishing possibilities for exchange and learning with other authorities; (iv) promoting innovation and entrepreneurship in AI in health; and (vi) identifying the need to regulate AI in this sector and update other existing regulations.
Ninth and finally, forum participants believed that the capabilities of governance leaders need to evolve to better incorporate expertise related to AI in ways that make sense within a given jurisdiction. With respect to RECs, for example, it might not make sense for every REC to recruit a member with expertise in AI methods. Rather, it will make more sense in some jurisdictions to consult with members of the scientific community with expertise in AI when research protocols are submitted that demand such expertise. Furthermore, RECs and other approaches to research governance in jurisdictions around the world will need to evolve in order to adopt the suggestions outlined above, developing processes that apply specifically to the ethical governance of research using AI methods in global health.
Research involving the development and implementation of AI technologies continues to grow in global health, posing important challenges for ethical governance of AI in global health research around the world. In this paper we have summarized insights from the 2022 GFBR, focused specifically on issues in research ethics related to AI for global health research. We summarized four thematic challenges for governance related to AI in global health research and nine suggestions arising from presentations and dialogue at the forum. In this brief discussion section, we present an overarching observation about power imbalances that frames efforts to evolve the role of governance in global health research, and then outline two important opportunity areas as the field develops to meet the challenges of AI in global health research.
Dialogue about power is not unfamiliar in global health, especially given recent contributions exploring what it would mean to de-colonize global health research, funding, and practice [ 30 , 31 ]. Discussions of research ethics applied to AI research in global health contexts are deeply infused with power imbalances. The existing context of global health is one in which high-income countries primarily located in the “Global North” charitably invest in projects taking place primarily in the “Global South” while recouping knowledge, financial, and reputational benefits [ 32 ]. With respect to AI development in particular, recent examples of digital colonialism frame dialogue about global partnerships, raising attention to the role of large commercial entities and global financial capitalism in global health research [ 21 , 22 ]. Furthermore, the power of governance organizations such as RECs to intervene in the process of AI research in global health varies widely around the world, depending on the authorities assigned to them by domestic research governance policies. These observations frame the challenges outlined in our paper, highlighting the difficulties associated with making meaningful change in this field.
Despite these overarching challenges of the global health research context, there are clear strategies for progress in this domain. Firstly, AI innovation is rapidly evolving, which means approaches to the governance of AI for health are rapidly evolving too. Such rapid evolution presents an important opportunity for governance leaders to clarify their vision and influence over AI innovation in global health research, boosting the expertise, structure, and functionality required to meet the demands of research involving AI. Secondly, the research ethics community has strong international ties, linked to a global scholarly community that is committed to sharing insights and best practices around the world. This global community can be leveraged to coordinate efforts to produce advances in the capabilities and authorities of governance leaders to meaningfully govern AI research for global health given the challenges summarized in our paper.
Limitations
Our paper includes two specific limitations that we address explicitly here. First, it is still early in the lifetime of the development of applications of AI for use in global health, and as such, the global community has had limited opportunity to learn from experience. For example, there were many fewer case studies, which detail experiences with the actual implementation of an AI technology, submitted to GFBR 2022 for consideration than was expected. In contrast, there were many more governance reports submitted, which detail the processes and outputs of governance processes that anticipate the development and dissemination of AI technologies. This observation represents both a success and a challenge. It is a success that so many groups are engaging in anticipatory governance of AI technologies, exploring evidence of their likely impacts and governing technologies in novel and well-designed ways. It is a challenge that there is little experience to build upon of the successful implementation of AI technologies in ways that have limited harms while promoting innovation. Further experience with AI technologies in global health will contribute to revising and enhancing the challenges and recommendations we have outlined in our paper.
Second, global trends in the politics and economics of AI technologies are evolving rapidly. Although some nations are advancing detailed policy approaches to regulating AI more generally, including for uses in health care and public health, the impacts of corporate investments in AI and political responses related to governance remain to be seen. The excitement around large language models (LLMs) and large multimodal models (LMMs) has drawn deeper attention to the challenges of regulating AI in any general sense, opening dialogue about health sector-specific regulations. The direction of this global dialogue, strongly linked to high-profile corporate actors and multi-national governance institutions, will strongly influence the development of boundaries around what is possible for the ethical governance of AI for global health. We have written this paper at a point when these developments are proceeding rapidly, and as such, we acknowledge that our recommendations will need updating as the broader field evolves.
Ultimately, coordination and collaboration between many stakeholders in the research ethics ecosystem will be necessary to strengthen the ethical governance of AI in global health research. The 2022 GFBR illustrated several innovations in ethical governance of AI for global health research, as well as several areas in need of urgent attention internationally. This summary is intended to inform international and domestic efforts to strengthen research ethics and support the evolution of governance leadership to meet the demands of AI in global health research.
Data availability
All data and materials analyzed to produce this paper are available on the GFBR website: https://www.gfbr.global/past-meetings/16th-forum-cape-town-south-africa-29-30-november-2022/ .
Clark P, Kim J, Aphinyanaphongs Y, Marketing, Food US. Drug Administration Clearance of Artificial Intelligence and Machine Learning Enabled Software in and as Medical devices: a systematic review. JAMA Netw Open. 2023;6(7):e2321792–2321792.
Article Google Scholar
Potnis KC, Ross JS, Aneja S, Gross CP, Richman IB. Artificial intelligence in breast cancer screening: evaluation of FDA device regulation and future recommendations. JAMA Intern Med. 2022;182(12):1306–12.
Siala H, Wang Y. SHIFTing artificial intelligence to be responsible in healthcare: a systematic review. Soc Sci Med. 2022;296:114782.
Yang X, Chen A, PourNejatian N, Shin HC, Smith KE, Parisien C, et al. A large language model for electronic health records. NPJ Digit Med. 2022;5(1):194.
Meskó B, Topol EJ. The imperative for regulatory oversight of large language models (or generative AI) in healthcare. NPJ Digit Med. 2023;6(1):120.
Jobin A, Ienca M, Vayena E. The global landscape of AI ethics guidelines. Nat Mach Intell. 2019;1(9):389–99.
Minssen T, Vayena E, Cohen IG. The challenges for Regulating Medical Use of ChatGPT and other large Language models. JAMA. 2023.
Ho CWL, Malpani R. Scaling up the research ethics framework for healthcare machine learning as global health ethics and governance. Am J Bioeth. 2022;22(5):36–8.
Yeung K. Recommendation of the council on artificial intelligence (OECD). Int Leg Mater. 2020;59(1):27–34.
Maddox TM, Rumsfeld JS, Payne PR. Questions for artificial intelligence in health care. JAMA. 2019;321(1):31–2.
Dzau VJ, Balatbat CA, Ellaissi WF. Revisiting academic health sciences systems a decade later: discovery to health to population to society. Lancet. 2021;398(10318):2300–4.
Ferretti A, Ienca M, Sheehan M, Blasimme A, Dove ES, Farsides B, et al. Ethics review of big data research: what should stay and what should be reformed? BMC Med Ethics. 2021;22(1):1–13.
Rahimzadeh V, Serpico K, Gelinas L. Institutional review boards need new skills to review data sharing and management plans. Nat Med. 2023;1–3.
Kling S, Singh S, Burgess TL, Nair G. The role of an ethics advisory committee in data science research in sub-saharan Africa. South Afr J Sci. 2023;119(5–6):1–3.
Google Scholar
Cengiz N, Kabanda SM, Esterhuizen TM, Moodley K. Exploring perspectives of research ethics committee members on the governance of big data in sub-saharan Africa. South Afr J Sci. 2023;119(5–6):1–9.
Doerr M, Meeder S. Big health data research and group harm: the scope of IRB review. Ethics Hum Res. 2022;44(4):34–8.
Ballantyne A, Stewart C. Big data and public-private partnerships in healthcare and research: the application of an ethics framework for big data in health and research. Asian Bioeth Rev. 2019;11(3):315–26.
Samuel G, Chubb J, Derrick G. Boundaries between research ethics and ethical research use in artificial intelligence health research. J Empir Res Hum Res Ethics. 2021;16(3):325–37.
Murphy K, Di Ruggiero E, Upshur R, Willison DJ, Malhotra N, Cai JC, et al. Artificial intelligence for good health: a scoping review of the ethics literature. BMC Med Ethics. 2021;22(1):1–17.
Teixeira da Silva JA. Handling ethics dumping and neo-colonial research: from the laboratory to the academic literature. J Bioethical Inq. 2022;19(3):433–43.
Ferryman K. The dangers of data colonialism in precision public health. Glob Policy. 2021;12:90–2.
Couldry N, Mejias UA. Data colonialism: rethinking big data’s relation to the contemporary subject. Telev New Media. 2019;20(4):336–49.
Organization WH. Ethics and governance of artificial intelligence for health: WHO guidance. 2021.
Metcalf J, Moss E. Owning ethics: corporate logics, silicon valley, and the institutionalization of ethics. Soc Res Int Q. 2019;86(2):449–76.
Data Protection Act - OFFICE OF THE DATA PROTECTION COMMISSIONER KENYA [Internet]. 2021 [cited 2023 Sep 30]. https://www.odpc.go.ke/dpa-act/ .
Sharon T, Lucivero F. Introduction to the special theme: the expansion of the health data ecosystem–rethinking data ethics and governance. Big Data & Society. Volume 6. London, England: SAGE Publications Sage UK; 2019. p. 2053951719852969.
Reisman D, Schultz J, Crawford K, Whittaker M. Algorithmic impact assessments: a practical Framework for Public Agency. AI Now. 2018.
Morgan RK. Environmental impact assessment: the state of the art. Impact Assess Proj Apprais. 2012;30(1):5–14.
Samuel G, Richie C. Reimagining research ethics to include environmental sustainability: a principled approach, including a case study of data-driven health research. J Med Ethics. 2023;49(6):428–33.
Kwete X, Tang K, Chen L, Ren R, Chen Q, Wu Z, et al. Decolonizing global health: what should be the target of this movement and where does it lead us? Glob Health Res Policy. 2022;7(1):3.
Abimbola S, Asthana S, Montenegro C, Guinto RR, Jumbam DT, Louskieter L, et al. Addressing power asymmetries in global health: imperatives in the wake of the COVID-19 pandemic. PLoS Med. 2021;18(4):e1003604.
Benatar S. Politics, power, poverty and global health: systems and frames. Int J Health Policy Manag. 2016;5(10):599.
Download references
Acknowledgements
We would like to acknowledge the outstanding contributions of the attendees of GFBR 2022 in Cape Town, South Africa. This paper is authored by members of the GFBR 2022 Planning Committee. We would like to acknowledge additional members Tamra Lysaght, National University of Singapore, and Niresh Bhagwandin, South African Medical Research Council, for their input during the planning stages and as reviewers of the applications to attend the Forum.
This work was supported by Wellcome [222525/Z/21/Z], the US National Institutes of Health, the UK Medical Research Council (part of UK Research and Innovation), and the South African Medical Research Council through funding to the Global Forum on Bioethics in Research.
Author information
Authors and affiliations.
Department of Physical Therapy, Temerty Faculty of Medicine, University of Toronto, Toronto, Canada
Berman Institute of Bioethics, Johns Hopkins University, Baltimore, MD, USA
Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, USA
Department of Philosophy and Classics, University of Ghana, Legon-Accra, Ghana
Caesar A. Atuire
Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, UK
Mahidol Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand
Phaik Yeong Cheah
Berkman Klein Center, Harvard University, Bogotá, Colombia
Armando Guio Español
Department of Radiology and Informatics, Emory University School of Medicine, Atlanta, GA, USA
Judy Wawira Gichoya
Health Ethics & Governance Unit, Research for Health Department, Science Division, World Health Organization, Geneva, Switzerland
Adrienne Hunt & Katherine Littler
African Center of Excellence in Bioinformatics and Data Intensive Science, Infectious Diseases Institute, Makerere University, Kampala, Uganda
Daudi Jjingo
ISI Foundation, Turin, Italy
Daniela Paolotti
Department of Health Sciences and Technology, ETH Zurich, Zürich, Switzerland
Effy Vayena
Joint Centre for Bioethics, Dalla Lana School of Public Health, University of Toronto, Toronto, Canada
You can also search for this author in PubMed Google Scholar
Contributions
JS led the writing, contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. JA contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. CA contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. PYC contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. AE contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. JWG contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. AH contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. DJ contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. KL contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. DP contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper. EV contributed to conceptualization and analysis, critically reviewed and provided feedback on drafts of this paper, and provided final approval of the paper.
Corresponding author
Correspondence to James Shaw .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Shaw, J., Ali, J., Atuire, C.A. et al. Research ethics and artificial intelligence for global health: perspectives from the global forum on bioethics in research. BMC Med Ethics 25 , 46 (2024). https://doi.org/10.1186/s12910-024-01044-w
Download citation
Received : 31 October 2023
Accepted : 01 April 2024
Published : 18 April 2024
DOI : https://doi.org/10.1186/s12910-024-01044-w
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Artificial intelligence
- Machine learning
- Research ethics
- Global health
BMC Medical Ethics
ISSN: 1472-6939
- General enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 30 April 2024
Microbiome confounders and quantitative profiling challenge predicted microbial targets in colorectal cancer development
- Raúl Y. Tito ORCID: orcid.org/0000-0001-9660-7621 1 , 2 na1 ,
- Sara Verbandt 3 na1 ,
- Marta Aguirre Vazquez 3 ,
- Leo Lahti ORCID: orcid.org/0000-0001-5537-637X 1 , 4 ,
- Chloe Verspecht 1 , 2 ,
- Verónica Lloréns-Rico 1 , 2 , 5 ,
- Sara Vieira-Silva ORCID: orcid.org/0000-0002-4616-7602 1 , 6 , 7 ,
- Janine Arts 8 ,
- Gwen Falony 1 , 2 , 6 ,
- Evelien Dekker 9 ,
- Joke Reumers ORCID: orcid.org/0000-0001-5434-6515 10 ,
- Sabine Tejpar ORCID: orcid.org/0000-0003-3281-8643 3 na1 &
- Jeroen Raes ORCID: orcid.org/0000-0002-1337-041X 1 , 2 na1
Nature Medicine ( 2024 ) Cite this article
3100 Accesses
178 Altmetric
Metrics details
- Colon cancer
- Diagnostic markers
Despite substantial progress in cancer microbiome research, recognized confounders and advances in absolute microbiome quantification remain underused; this raises concerns regarding potential spurious associations. Here we study the fecal microbiota of 589 patients at different colorectal cancer (CRC) stages and compare observations with up to 15 published studies (4,439 patients and controls total). Using quantitative microbiome profiling based on 16S ribosomal RNA amplicon sequencing, combined with rigorous confounder control, we identified transit time, fecal calprotectin (intestinal inflammation) and body mass index as primary microbial covariates, superseding variance explained by CRC diagnostic groups. Well-established microbiome CRC targets, such as Fusobacterium nucleatum , did not significantly associate with CRC diagnostic groups (healthy, adenoma and carcinoma) when controlling for these covariates. In contrast, the associations of Anaerococcus vaginalis , Dialister pneumosintes , Parvimonas micra , Peptostreptococcus anaerobius , Porphyromonas asaccharolytica and Prevotella intermedia remained robust, highlighting their future target potential. Finally, control individuals (age 22–80 years, mean 57.7 years, standard deviation 11.3) meeting criteria for colonoscopy (for example, through a positive fecal immunochemical test) but without colonic lesions are enriched for the dysbiotic Bacteroides2 enterotype, emphasizing uncertainties in defining healthy controls in cancer microbiome research. Together, these results indicate the importance of quantitative microbiome profiling and covariate control for biomarker identification in CRC microbiome studies.
Similar content being viewed by others

Microbiota in health and diseases

A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche

A sustainable approach to universal metabolic cancer diagnosis
Colorectal cancer (CRC) incidence is steadily increasing 1 , especially in people under 50 years 2 . It is estimated that approximately 16 and approximately 14 individuals per 100,000 people in the United States and Belgium, respectively, die every year from CRC 3 . As medical interventions can effectively reduce CRC progression and associated mortality, it is imperative to identify individuals at increased risk.
Colonoscopies with polypectomy of adenomas reduce up to 90% of CRC risk 4 . Early identification of individuals with polyps would reduce the global burden of CRC. Yet, ascertainment of patients at an increased risk remains challenging, highlighting the need for population-wide screening.
Microbiota shifts have been associated with a wide array of disease phenotypes 5 . Some bacterial markers, such as Fusobacterium , have been reported enriched in lesions and stools of patients with CRC 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 across developing and developed countries 15 , suggesting a potential role for microbiome-based diagnostics and/or prognostics.
Although microbiome profiles are affected by multiple variables that may confound or compound biological phenomena, covariate control is far from standard. For example, moisture content, a proxy for transit time, remains uncontrolled despite showing the biggest explanatory power for overall gut microbiota variation in multiple cohorts 16 , 17 . Intestinal inflammation, measured as fecal calprotectin 18 , 19 that reflects increased neutrophil shedding into the intestinal lumen 20 , is more sensitive than fecal occult blood for identifying patients with CRC 21 , thus a potential untapped target for molecular stool CRC-screening 19 .
Relative microbiome profiling (RMP, taxon abundances are expressed in percentages) remains the dominant approach in microbiome research. However, given issues with compositionality 22 and interpretation of relative profiles 23 , the use of experimental and quantitative approaches is increasingly recommended 23 , 24 , 25 . This reduces both false-positive and false-negative rates in downstream analyses, thereby lowering the risk of erroneous interpretation of microbiome associations, and allows focusing clinical programs on biologically relevant targets 25 . Although quantitative microbiome profiling (QMP) facilitates normalized comparisons across different samples or conditions 24 , 25 , so far, no QMP CRC microbiota studies were performed.
In this Article, we address these two gaps in CRC microbiota studies: (1) to quantitively characterize the microbiota profile associated with malignant colonic transformation and (2) to identify microbiota covariates that may obscure biological phenomena behind microbiota-CRC associations. To this end, we examined the microbial profiles of 589 Belgian patients from Universitair Ziekenhuis Leuven (UZL) who warranted colonoscopies based on clinical presentations, including patients with CRC, and compared these to existing published datasets (total n = 4,439 patients and controls). To the best of our knowledge, this is the first large scale study of the gut microbiota across colonic cancer developmental stages that combines QMP analysis with extensive analysis of microbiota covariates to disentangle disease-associated from confounder-based signals to identify taxa specifically associated with CRC.
Intestinal inflammation is higher in patients with colorectal tumors
We recruited 650 volunteers referred for colonoscopy and colonic resections at UZL between 2017 and 2018 who provided a stool sample before the colonic procedure. Most participants were from the Flemish region of Belgium. For this study, cancer developmental stages were defined as diagnosis groups, and we classified participants into three groups according to a thorough colonoscopy and clinical assessment: (1) patients without evidence of colonic lesions (CTLs, n = 205), (2) patients with polyps (considering polyps as a precancerous lesion; n < 10 and size between 6 and 10 mm) (ADE, n = 337) and (3) patients with CRC ( n = 47; 2 (4%) stage 0, 14 (30%) stage I, 13 (28%) stage II, 11 (23%) stage III, 3 (6%) stage IV and 4 (9%) of undetermined stage). We excluded patients outside these criteria, as well as those with insufficient clinical and molecular data. The final Leuven CRC Progression Microbiome (LCPM) study cohort consisted of 589 patients. The most frequent indications for colonoscopy were either a positive fecal immunochemical test (FIT) or adenoma surveillance. Other indications included familial risk, abdominal symptoms and change in bowel habits (Fig. 1a and Supplementary Table 1 ). The study was registered at clinicaltrials.gov (NCT02947607).
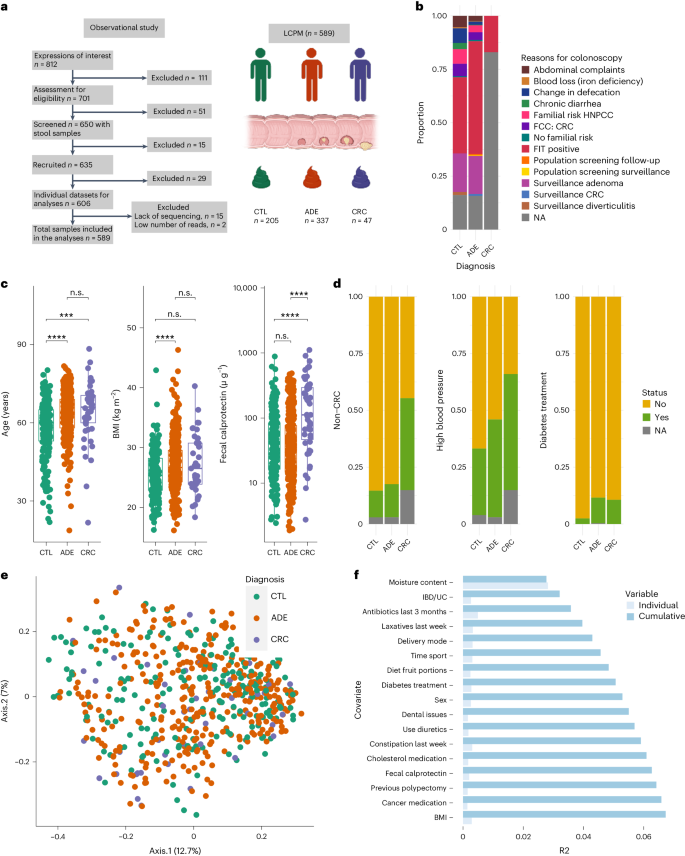
a , STROBE flowchart and cohort size. CTL represents patients without colonic lesions, ADE denotes patients with colonic polyps and CRC refers to patients with colorectal tumors (generated in BioRender.com ). b , Colonoscopy referral reasons for patients of the LCPM cohort: positive FIT, adenoma surveillance, familial risk cancer (FCC), hereditary nonpolyposis CRC (HNPCC) and changes in defecation. NA, denotes the proportion of patients without information. c , Age, BMI and calprotectin are associated with diagnosis groups. The patients without lesions were younger ( n = 589, two-sided KW test χ 2 = 35.77, adjusted P = 2.6 × 10 −7 ; phD tests) and had lower BMI ( n = 553, two-sided KW test χ 2 = 15.73, adjusted P = 1.9 × 10 −3 ; phD tests), while patients with tumors had higher fecal calprotectin levels ( n = 583, two-sided KW test χ 2 = 29.43, adjusted P = 3.0 × 10 −6 ; phD tests, adjusted *** P <0.001, ** P <0.01, * P <0.05 and n.s., non-significant P > 0.05; Supplementary Table 3 ). The box plot center represents the median value whiskers extend from the quartiles to the last data point within 1.5 times of the interquartile range, with outliers beyond. d , Previous non-CRC cancer, high blood pressure and diabetes treatment are associated with the distribution of diagnosis groups. The patients with CRC have a higher proportion of previous cancer (47.5% versus 15.0 % and 12.1%, two-sided CS test, CV effect size of 0.24, χ 2 = 31.65, d.f. of 2, adjusted P = 1.98 × 10 −2 ) and high blood pressure (60.0% versus 44.3% and 30.5%, CV of 0.17, two-sided CS test, χ 2 = 16.55, d.f. of 2, adjusted P = 1.98 × 10 −2 ) while the CTL group has the lowest proportion of patients with diabetes treatment (2.4% versus 10.3 and 10.6, two-sided CV effect size of 0.15, CS test, χ 2 = 13.79, d.f. of 2, adjusted P = 1.98 × 10 −2 ). e , PCoA on BCD representing QMP species-level microbiota variation in the LCPM cohort ( n = 589), PCoA1 (Axis.1) and PCoA2 (Axis.2) respectively explained 12.7% and 7% of the variance. Each dot represents one sample, colored by assigned diagnosis group. f , Cumulative effect sizes of significant covariates on microbiota community variation (cumulative bars; stepwise dbRDA on BCD) as compared to individual effect sizes (R 2 ) assuming covariate independence in the LCPM cohort ( n = 589; Supplementary Table 5 ). UC, ulcerative colitis.
Source data
We collected an extensive set of 165 universal metadata variables (nonspecific for any of the three groups) from each participant. After curation, we excluded variables that were colinear (if Pearson | r | > 0.8, we kept the variable with fewer missing data) or had incomplete data collection (variables missing more than 20% of the values). The final set consisted of 95 high-quality variables (Supplementary Table 2 ).
To identify metadata variables associated with diagnosis groups, we applied two statistical approaches: (1) nonparametric Kruskal–Wallis (KW) test and its η 2 effect size (Supplementary Table 3 ) for all numerical variables and (2) chi-square (CS) tests and Cramer’s V effect size (CV) (Supplementary Table 4 ) for categorical variables, followed by the Benjamini–Hochberg method for multiple testing correction (adjusted P ). We found eight variables associated with diagnosis groups (false discovery rate <5%), namely: age, body mass index (BMI), calprotectin, reported hours of sleep, previous cancer (including CRC), dental status (complete, partial and so on), diabetes treatment and high blood pressure (Supplementary Tables 3 and 4 ). The CTL patients were younger ( n = 589, KW test, η 2 = 0.058, χ 2 = 35.77, adjusted P = 2.6 × 10 −7 ; post hoc Dunn (phD) tests, adjusted P < 0.05 for CTL versus ADE or CRC groups), had a lower BMI ( n = 553, KW test, η 2 = 0.023, χ 2 = 15.73, adjusted P = 1.9 × 10 −3 ; phD tests, adjusted P < 0.05 for CTL versus ADE) and reported fewer hours of sleep than participants from the other two diagnosis groups ( n = 557, KW test, η 2 = 0.019, χ 2 = 13.41, adjusted P = 4.6 × 10 −3 ; phD tests, adjusted P < 0.05 for CTL versus ADE; Fig. 1 ; see Supplementary Table 3 for full results). Moisture content, an important microbiota covariate 16 , was not significant across diagnosis groups ( n = 589, KW test, η 2 = −0.001, χ 2 = 1.32, adjusted P = 7.0 × 10 −1 ).
The calprotectin levels were positively associated with malignant transformation. The patients with CRC showed higher intestinal inflammation, measured by fecal calprotectin 18 , 26 (Fig. 1a and Supplementary Table 3 ). Specifically, CRC exhibited higher levels (219.42 µg g −1 , range 2.74–1,114.42, n = 47) compared to ADE (70.24 µg g −1 , range 1.87–487.21, n = 337) or CTL (73.25 µg g −1 , range 2.42–884.82, n = 202) (Fig. 1a , N = 583, KW test, η 2 = 0.047, χ 2 = 29.43, adjusted P = 3.0 × 10 −6 ; phD tests, adjusted P < 0.05 for CRC versus CTL and CRC versus ADE). We also observed increased fecal calprotectin in patients reporting previous cancers (primarily breast and prostate cancer) (Wilcoxon ranksum (WR) test, W = 11,067, adjusted P = 4.1 × 10 −3 ), consumption of cancer medication (WR test, W = 3,671, adjusted P < 0.05), heartburn complaints (WR test, W = 11,067, adjusted P = 1.0 × 10 −10 ) and lower dietary fiber (WR test, W = 20,964, adjusted P = 3.3 × 10 −2 ).
The history of chronic diseases was distinct across diagnosis groups. The patients with CRC showed higher proportions of previous non-CRC cancer (47.5% versus 15.0 % and 12.1%, CS test, CV of 0.24, χ 2 = 31.65, d.f. of 2, adjusted P = 1.98 × 10 −2 ) and high blood pressure (60.0% versus 44.3% and 30.5%, CS test, CV of 0.17, χ 2 = 16.55, d.f. of 2, adjusted P = 1.98 × 10 −2 ) (Fig. 1b and Supplementary Table 4 ). The CTL group had the lowest diabetes treatment (2.4% versus 10.3% and 10.6%, CS test, CV of 0.15, χ 2 = 13.79, d.f. of 2, adjusted P = 1.98 × 10 −2 ) (Fig. 1b and Supplementary Table 4 ) and mostly complete dental sets (53.3% versus 35.2% and 32.5%, CS test, CV of 0.03, χ 2 = 30.78, d.f. of 10, adjusted P = 1.98 × 10 −2 ) (Supplementary Table 4 ).
Known confounders, not diagnosis groups, explain overall microbiota variation across CRC developmental stages
The influence of microbiota covariates and the quantitative amplitude of observed microbiota shifts are understudied in CRC. We combined sequencing data with flow cytometry measurements of fecal microbial load 23 to generate QMP data from our study cohort. 23 We studied the QMP variation in the context of the 94 potential covariates mentioned above (the 95th being microbial load) using established procedures 17 .
A principal coordinate analysis (PCoA; Fig. 1c ) on a species-level Bray–Curtis dissimilarity (BCD) matrix revealed no significant separation between diagnosis groups. Furthermore, no difference in total microbial load was found between groups ( n = 589, KW test, χ 2 = 0.68, adjusted P = 8.2 × 10 −1 ). Distance-based redundancy analysis (dbRDA) revealed 24 microbiota covariates associated with microbial variation in this cohort (Fig. 1d and Supplementary Table 5 ). We identified 17 nonredundant covariates that jointly explained 6.7% of microbiota compositional variation (Supplementary Table 5 ).
Consistent with previous reports 16 , 17 , moisture content exhibited the highest explanatory value (2.8%) of all covariates ( n = 589, stepwise dbRDA, R 2 = 2.8%, adjusted P = 2 × 10 −3 ). Intestinal bowel disease/ulcerative colitis (IBD/UC) status, a CRC-risk factor, possibly associated with its microbial dysbiotic community and intestinal inflammation 27 , was the second largest covariate. IBD/UC explained 0.4% of the microbiota variation ( n = 569, stepwise dbRDA, R 2 = 0.4%, adjusted P = 2 × 10 −3 ). Other top microbiota covariates included antibiotics and laxatives use (Fig. 1d ). Delivery mode (cesarean or natural birth) explained 0.3% variation ( n = 533, stepwise dbRDA, R 2 = 0.3%, adjusted P = 2 ×10 −3 ), although it is probably confounded by diet in this cohort (proportion of dietary vegetables; CS test, χ 2 = 33.09, d.f. of 14, P = 2.8 × 10 −3 , adjusted P < 0.05). Intestinal inflammation (fecal calprotectin) explained 0.2% ( n = 583, stepwise dbRDA, R 2 = 0.2%, adjusted P = 2.6 × 10 −2 ). In contrast with our previous study in the Flemish population (Flemish Gut Flora Project, FGFP) 17 , age did not explain microbiota variation ( n = 589, univariate dbRDA, R 2 = 0.2%, adjusted P = 5.9 × 10 −2 ). Surprisingly, the cancer diagnosis group (CTL, ADE and CRC), as a covariate, was not associated with microbial variation ( n = 589, univariate dbRDA, R 2 = 0.2%, adjusted P = 0.22; Supplementary Table 5 ).
Fusobacterium association with CRC stages disappears when controlling for confounders or when using QMP
Microbiota signals can be specific to taxonomic groups and, thus, not reflected in broad community shifts. While a multitude of microbial associations have been reported in CRC studies using RMP 6 , 7 , 8 , 13 , we used QMP to identify species whose absolute abundance associated with diagnosis groups. The comparisons were limited to the 138 species with a prevalence of greater than 5% in at least one of the diagnosis groups of the LCPM cohort (Supplementary Table 6 ). Only eight species showed significant differential abundance (absolute or relative) among diagnosis groups: Anaerococcus vaginalis ( Anaerococcus obesiensis ), Alistipes onderdonkii , Dialister pneumosintes , Fusobacterium nucleatum , Parvimonas micra , Peptostreptococcus anaerobius , Porphyromonas asaccharolytica and Prevotella intermedia (KW test, adjusted P < 0.05; Fig. 2a,b and Supplementary Table 7 ). While Fusobacterium nucleatum has been consistently associated with colorectal lesions across cohorts of diverse backgrounds 13 , 14 , in the LCPM cohort, Fusobacterium nucleatum absolute abundance was positively correlated with high fecal calprotectin levels (Spearman’s rank and Kendall’s tau correlations, adjusted P < 0.05; Fig. 2c , Extended Data Fig. 1 and Supplementary Table 8 ) and cancer progression (diagnosis groups) (KW test, η 2 = 0.010, adjusted P = 1.84 × 10 −5 ; phD test adjusted P = 8.80 × 10 −1 for CTL versus ADE, adjusted P = 3.84 × 10 −7 for CTL versus CRC and adjusted P = 3.84 × 10 −7 for ADE versus CRC; Fig. 2c and Supplementary Table 7 ). However, after deconfounding for calprotectin only or combined BMI, moisture content and calprotectin, and neither absolute nor relative Fusobacterium nucleatum abundance were associated with diagnosis (generalized linear model analysis of variance (ANOVA), n = 547, P > 0.05; Extended Data Fig. 2 ).
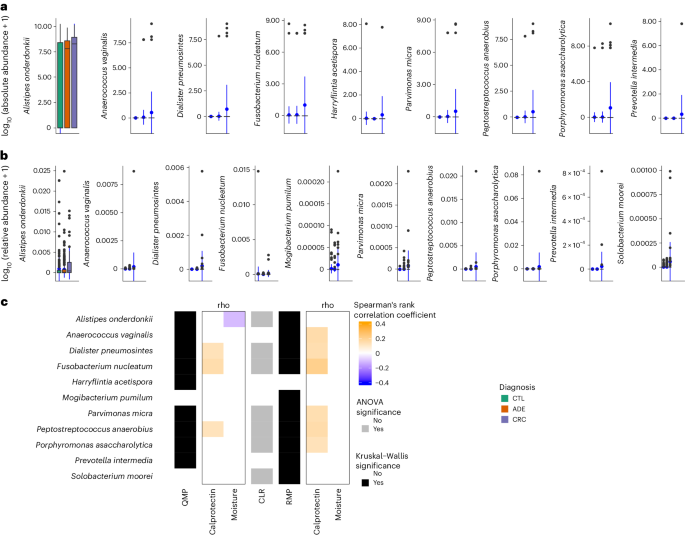
a , Nine species were identified with differential absolute abundance across diagnosis groups ( n = 589, KW test, adjusted P < 0.05; Supplementary Table 7 ). b , Ten species were identified with differential relative abundance across diagnosis groups ( n = 589, KW test, adjusted P < 0.05; Supplementary Table 7 ). The center of the box plot represents the median value of the data, and the whiskers extend from the quartiles to the last data point within 1.5 times of the interquartile range, with outliers beyond. The blue circles represent the mean. c , Biomarkers associations and their confounders. Species Spearman’s rank correlation with calprotectin levels and moisture proportions using QMP (first rho column panel) and RMP (second rho column panel) data. The effect size of the associations between species and calprotectin, moisture and diagnosis variables for QMP and RMP ( n = 589, Spearman’s rank correlation comparison, adjusted P < 0.05). Significant associations were tested using two-sided KW tests for QMP and RMP data and ANOVA for CLR data. The associations for Harryflintia acetispora , Parvimonas micra and Prevotella intermedia are sensitive to bias by the extreme values (absolute abundance) in the higher range. Removing these values leads to loss of significance. As rank-based approaches were used, it is not clear if this loss is due to the strength of the signal or the loss of power.
Multiple established CRC microbial markers are associated with transit time, intestinal inflammation and body mass index but not with CRC stages
The association of Fusobacterium abundance with fecal calprotectin urged us to investigate the influence of this confounder on previously reported CRC-associated genera, adding moisture content since it is the top microbiome covariate, and BMI, which showed differences among diagnosis groups.
To this end, we compiled a list of 89 CRC species-level markers from ten published cohorts 6 , 9 , 11 , 13 , 14 , 28 , 29 , 30 , 31 (including 1,633 samples) and 67 genera-level markers from 15 cohorts 6 , 7 , 8 , 9 , 11 , 12 , 13 , 14 , 15 , 28 , 29 , 30 , 31 , 32 (representing 4,439 samples). We used this compiled list of taxa as a criterion to test whether the CRC association of these taxa in our cohort is influenced by the target covariates. To reduce the impact of distinct statistical treatments, we downloaded the microbial profiles of nine out of ten studies at species level from the curated MetagenomicData 33 resource and analyzed them using the statistical component of our pipeline.
Spearman correlation between taxa abundances and the three focus covariates revealed strong associations between microbial targets and these confounders at the species (Extended Data Fig. 3a ) and genus level (Fig. 3b ). Most of these associations were replicated in an independent population cohort (FGFP), suggesting these associations are robust and not specifically linked to CRC (Extended Data Fig. 3 ). Moisture content, the known major covariate in microbiome studies 17 , is unsurprisingly associated with many taxa validated in both cohorts.

a , b , Species ( a ) and genera ( b ) previously reported in association with CRC (blue and green represent enrichment or depletion; the squares indicate reported in corresponding publications, while circles represent our reanalysis of the MetaPhlAn 3.0 profiles generated from the curatedMetagenomicData 33 of these cohorts using the statistical part of our pipeline). Graphic representation of Spearman’s rank correlation of pairwise analysis of fecal calprotectin, BMI, and moisture values against absolute species abundance (QMP) and RMP from the LCPM ( N = 589) and FGFP ( N = 1,045) cohorts (adjusted P < 0.05, Supplementary Table 8 ). The species enriched or depleted in relation to CRC diagnosis groups were tested using QMP, CLR and RMP data before ( n = 589, two-sided KW test and Spearman’s rank correlation comparison, adjusted P < 0.05) and after controlling for microbiota covariates (before adjustment for BMI, calprotectin and moisture; generalized linear model ANOVA, adjusted P < 0.05).
As we compiled the CRC-associated taxa from non-QMP studies, we conducted analyses using both RMP and QMP to assess whether confounder associations influence quantitative association of biomarkers or targets to diagnosis groups in LCPM. We found only 8% (6 out of 89) and 10% (9 out of 89) of species previously associated with CRC using QMP and RMP replicating after confounder control. Anaerococcus vaginalis , Dialister pneumosintes , Parvimonas micra , Peptostreptococcus anaerobius , Prevotella intermeia and Porphyromonas asaccharolytica , were identified by controlled QMP and RMP. Controlled QMP excluded Fusobacterium nucleatum and Alistipes onderdonkii , suggesting previous associations of these two species may be spurious (Fig. 3a ).
We identified eight species previously linked to CRC (that is, using QMP and/or RMP), including Fusobacterium nucleatum and Peptostreptococcus anaerobius , to be associated with inflammation (Fig. 3 and Supplementary Tables 8 and 9 ). This association was previously reported for only three out of the eight taxa above ( Escherichia , Fusobacterium and Streptococcus ) 24 . Further validation of this association was conducted using the FGFP (Extended Data Fig. 3 and Supplementary Tables 8 and 9 ).
Recognizing that inflammation is a risk factor, not a requirement, for CRC progression, we further investigated markers associated with diagnosis groups in relation to inflammatory status. To this end, we focused on a subset of 340 samples, which, regardless of their CRC status, exhibited normal levels of calprotectin (fecal calprotectin under 50 μg g −1 (ref. 34 )), indicating no evidence of local inflammation (112 CTL, 216 ADE and 12 CRC). Assessment of the 89 CRC species-level markers mentioned above confirmed that the association of three of the six replicating species ( Anaerococcus vaginalis , Prevotella intermedia and Porphyromonas asaccharolytica) is independent of intestinal inflammation (Supplementary Table 10 ).
Colonoscopy patients, with or without CRC, exhibit an excess of the Bacteroides2 enterotype
To study the LCPM cohort in a population context, we enterotyped participants using Dirichlet multinomial mixtures (DMM) on a genus matrix against the background of microbial variation as observed in the FGFP samples ( n = 1,045) 17 . Consistent with previous description of the Flemish population 23 , we identified four community types based on selecting the optimal number of clusters using the Bayesian Information Criterion (Fig. 4a,b and Extended Data Fig. 4 ), ‘Bacteroides1’ (Bact1), ‘Bacteroides2’ (Bact2), ‘Prevotella’ (Prev) and ‘Ruminococcaceae’ (Rum). The enterotype distribution was different between LCPM and FGFP (CS test, χ 2 = 34.3, d.f. of 3, adjusted P = 1.7 × 10 −7 ), but no differences were observed among diagnosis groups within the LCPM cohort (pairwise CS tests, adjusted P > 0.1). Pairwise comparisons of the prevalence of the dysbiotic Bact2 enterotype in the LCPM cohort diagnosis groups revealed that compared to the FGFP population, this enterotype was enriched in all CRC diagnosis groups (test of equal or given proportions, FGFP versus CTL: χ 2 = 15.09, d.f. of 1, adjusted P = 1.1 × 10 −4 ; FGFP versus ADE: χ 2 = 18.93, d.f. of 1, adjusted P = 2.4 × 10 −5 ; and FGFP versus CRC: χ 2 = 4.34, d.f. of 1, adjusted P = 3.4 × 10 −2 ). Although dysbiosis and CRC development were previously linked 13 , 35 , the high prevalence of this enterotype in the LCPM, even in samples from patients free of lesions, is unexpected. Consistent with previous reports 24 , 25 , the Bact2 enterotype in this group exhibited all hallmarks of dysbiosis: low cell count, low richness, higher calprotectin values, reduced butyrate producers and increased proinflammatory bacteria.
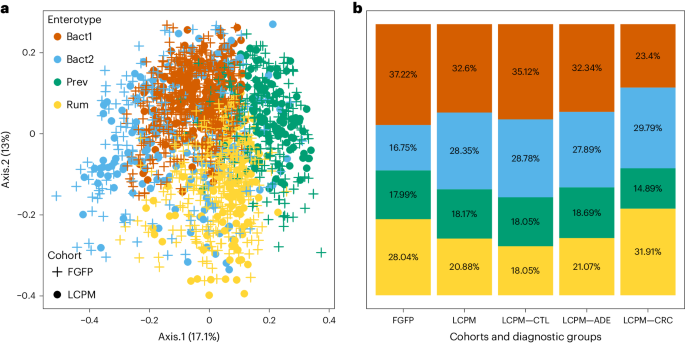
a , PCoA of interindividual differences (BCD) in relative microbiota profiles of the LCPM cohort ( n = 589 samples) using a cross-section of the Flemish population ( n = 1,045 samples) as a background dataset. PCoA1 (Axis.1) and PCoA2 (Axis.2) respectively explained 13% and 17.1% of the variance of microbiota at the genus level. b , Enterotype distribution across the FGFP, LCPM and LCPM diagnosis groups (CTL, ADE and CRC), increased prevalence of the Bact2 enterotype in the three groups from the LCPM cohort ( n = 589) as compared to FGFP samples ( n = 1,045); pairwise two-sided test of equal or given proportions ( P < 0.05).
Additional categorical variables appeared associated with the Bact2 enterotype. They included antibiotic consumption (CS test, χ 2 = 30.78, d.f. of 3, adjusted P = 2.1 × 10 −2 ), current treatment with anti-inflammatory medications (CS test, χ 2 = 30.78, d.f. of 3, adjusted P = 2.1 × 10 −2 ), diabetes treatment (CS test, χ 2 = 30.78, d.f. of 3, adjusted P = 3.3 × 10 −2 ), recent diarrhea (last week) (CS test, χ 2 = 30.78, d.f. of 3, adjusted P = 2.1 × 10 −2 ), history of gallstones (CS test, χ 2 = 30.78, d.f. of 3, adjusted P = 4.7 × 10 −2 ) and recent use of laxatives (last week) ( χ 2 = 30.78, d.f. of 3, adjusted P = 4.2 × 10 −2 ) (Supplementary Table 11 ).
While associations between the gut microbiota and CRC have been extensive, this is the first study using QMP and extensive metadata collection to systematically investigate microbiota covariates that potentially are masking or creating spurious associations between specific taxa and malignant transformation.
At first glance, this study yielded a gut microbial profile partially consistent with previous reports of CRC-associated taxa. Further analysis, however, suggested that many of the previously reported associations, including those of prominent biomarkers, such as Fusobacterium (nucleatum), are confounded by microbiota covariates. A total of 17 of 94 variables explained 6.7% of the observed variation. Of those, the moisture content had highest explanatory power (2.7%), greater than eight times that of the next covariate (IBD status). The explanatory power of fecal calprotectin was lower (0.2%) but significant; age and, most importantly, diagnosis groups were not.
Some associations were complex in nature. For example, BMI, consistent with previous reports, showed an association with both microbial composition 17 , 25 and cancer progression 36 , while others, such as age, suggested to modify the BMI-association with cancer progression 37 , were not significant in this cohort.
Inflammation is a known risk factor for CRC 38 , but its effect size in shaping the cancer-associated microbiota is yet to be described. Fecal calprotectin is a well-documented marker of intestinal local inflammation 39 , 40 and has been associated with cancer progression, probably having an effect on tumor development rather than on tumor initiation 41 . We observed participants with normal and elevated fecal calprotectin levels within each diagnosis group and covariate-controlled analysis of the LCPM cohort revealed that 8 and 19 CRC-associated markers, at the species and genus levels, respectively, associated with fecal calprotectin rather than with the diagnosis group. We replicated these observations in an independent cohort of apparently healthy individuals (FGFP).
High levels of fecal calprotectin have been associated with intestinal inflammatory pathologies 19 . However, when removing patients with IBD from our analysis, CRC diagnosis groups remained not significant, and the significance of Fusobacterium nucleatum , among other six species, was unaltered after differential abundance analysis. In patients with CRC, increased levels of fecal calprotectin (>50 µg g −1 stool 18 , 26 ) are directly associated with tumor presence, as the level decreases after tumor resection 42 . Here, fecal calprotectin was increased in CRC, consistent with previous associations between malignant transformation, local inflammation 43 and advanced tumor stages (T3 and T4) 42 . No difference in calprotectin levels was observed between CTL and ADE (mean 73.25 versus 70.24 µg g −1 ), suggesting that although no lesions are visible in the colon of the CTL group, they have a detectable level of local inflammation. The potential effect of local inflammation in shaping the colonic microbiota in the context of malignant transformation, or its potential confounding effect, remains largely obscure, as most studies surveying the association between gut microbiota and CRC, including meta-analysis 13 , 14 , do not control for local inflammation.
We argue that strict control of covariates is a must in any microbiota analysis assessing potential clinical associations, as for example, three of the species with repeated CRC association 11 , 13 , 14 , 28 , 29 , 30 , 32 , Escherichia coli , Fusobacterium nucleatun and Parvimonas micra , exhibit association with local inflammation, unfortunately uncontrolled for in previous studies, that may or may not be associated with cancer progression.
Fusobacterium nucleatum is one of the species that attracts more attention as there is a substantial body of work linking it to CRC 44 . In this study, Fusobacterium was enriched in patients with CRC. However, this apparent association disappears when the analysis is covariate controlled. Our study suggests that the association of Fusobacterium nucleatum to cancer may be driven by its association to intestinal inflammatory conditions; there are no differences in the abundance of Fusobacterium nucleatum across diagnostic groups once calprotectin is controlled for. These results suggest reassessment of the diagnostic utility of this marker. At the same time, our results do not mean that Fusobacterium nucleatum is not linked to CRC; they rather suggest that the reasons behind this association might be less straightforward than originally considered. They, thus, present a cautionary tale of the importance to control for covariates as the microbiome field moves forward. Given that inflammation is a risk factor for CRC but not a requirement 41 , potential use of Fusobacterium nucleatum as a marker of CRC development could fail to identify those cases of inflammation-independent cancer progression. While not yet commercialized, there are already publications proposing the use of microbial markers, including Fusobacterium nucleatum , for CRC screening 7 , 45 , which, in light of our results, raises concerns as uncontrolled variables may be obscuring actual biological mechanisms. We present evidence that purported CRC biomarkers, even those replicated in multiple studies, may suffer from the compounding or confounding effect of covariates, which in addition to the use of nonquantitative signals, may result in misleading conclusions on what diagnostic signals really mean—complicating the path towards potential clinical applications.
BMI, in combination or independent of inflammation, has been independently associated with changes in the gut microbiota 46 , which in turn are associated with increased risk of CRC 47 . Yet, microbial dysbiosis by itself does not explain the higher risk of colon cancer observed in the obese population 48 , indicating that the underlying process that associates obesity and CRC is more complex and demands further investigation.
Among four described gut enterotypes, the Bact2 enterotype is defined as a dysbiotic microbial profile 24 , 25 . Bact2 enrichment is observed in obesity 25 and in conditions such as PSC (Primary sclerosing cholangitis) and IBD 24 , further supporting the potential disease association of this enterotype. The analysis of the LCPM cohort revealed an excess of the Bact2 enterotype across all diagnosis subgroups, regardless of BMI.
Increased Bact2 prevalence in the no-lesions group compared to FGFP is particularly striking. While patients in the CTL group have no observable lesions, they may be considered at increased risk for colorectal perturbations based on clinical referrals (blood loss in the stool, familiar risk to colonic lesion and so on) that warranted colonoscopies—something that might also be reflected by their Bact2 enterotype. Of importance, ‘healthy’ biopsies included in CRC microbiome studies are often selected using colonoscopies with a negative result as the main criterium, posing a potential problem, as no other markers of colonic health are considered to qualify these healthy individuals. The reasons for the appearance of Bact2 in the no-lesion group are multifold, but these findings suggest that such individuals, while representing a useful category for biomarker discovery, may harbor an unhealthy gut ecosystem, from a microbial point of view.
There is a plethora of variables identified as modifiers of the gut microbiota. Yet, covariate control is far from standard and notably absent from most association studies. As intestinal microbial taxa are being nominated as potential biomarkers of malignant transformation, it is imperative to explore the influence of microbiota covariates as potential confounders or compounders of observed associations. Rather than denying previous associations, our analysis emphasizes the need for covariate-controlled analysis for any microbiota study aiming to establish clinical associations, as these covariates by themselves may explain most of the stool microbiota variation, independent of CRC status.
Out of the multiple taxa previously associated with CRC, six species remain significant after strict control of covariates in this quantitative cohort. Without denying other potential biomarkers, further studies are warranted on Anaerococcus vaginalis , Dialister pneumosintes , Parvimonas micra , Peptostreptococcus anaerobius , Prevotella intermedia and Porphyromonas asaccharolytica , as their reported association to CRC 6 , 7 is robust enough to remain independent of the method. Our data present a strong argument in favor of revisiting potential microbial associations with clinical phenotypes to ensure that the purported associations are not driven by uncontrolled covariates warranting further follow up of the mechanisms underlying these associations. Refining the approaches to discover microbial biomarkers will undoubtedly impact the microbiota field, facilitating the path towards the much-coveted clinical applications.
Limitations
We aim to identify taxa associated with malignant colonic transformation. While our cohort includes a set of participants without lesions, we make no claim that these are healthy controls, as there is an apparent increased incidence of gut dysbiosis in this group. Considering that all participants in this study had a medical need for a colonoscopy, there is an implicit increased risk to CRC. Thus, the present study cannot rule out that the group without polyps is undergoing potential molecular or cellular changes that are not detectable via colonoscopy. In addition, as this is a cross-sectional study, the term cancer progression is an extrapolation of what is seen at cancer development stages (operationalized here as diagnosis groups). We cannot rule out potential particularities of our cohort that may be contributing to our observations, as most studies do not report sufficient metadata for us to compare across cohorts. It is important to consider that certain taxonomic groups may not even be represented in current databases, and specific microbial species may require longer hypervariable regions or alternative sequencing approaches to achieve accurate species-level identification. Nonetheless, the V4 region for our cohort seems to be able to resolve species taxonomy of the biomarkers previously associated with CRC, as we show for the case of Fusobacterium .
Furthermore, it has been proposed that the potential diagnostic value of colonic microbial profiles goes beyond bacteria, as fungal and viral species have been proposed as CRC biomarkers 49 . We recognize that multidomain approaches to discover CRC biomarkers and longitudinal prospective studies to better study the dynamics of cancer progression are warranted to comprehensively inform cancer detection and treatment.
Participant recruitment
The LCPM project was an observational cross-sectional survey for which procedures were approved by the medical ethics committee of the UZL (ethical approval number S57084). Between 2017 and 2018, we recruited patients through the study nurse following a standardized procedure. Briefly, we invited patients scheduled for lower gastrointestinal endoscopy or abdominal surgery for CRC removal at the UZL were invited. After explaining the research project and if they expressed their agreement, participants signed an informed consent, and no compensation was offered. A set of stool sample collection material was provided.
Each patient completed an extensive questionnaire containing information about the date of sample collection, the consistency of the stool, diet, antibiotics usage, clinical symptoms or disease among other variables 17 , as well as an extensive medical and clinical questionnaire using the Websurvey service of KU Leuven.
As a validation cohort we included the FGFP 17 , a population-wide microbiota monitoring effort, representing one of the largest and best characterized fecal microbiota database currently available. Its extensive metadata including health and lifestyle allowed the identification of 69 factors associated with microbiota variation (microbiota covariates). The QMP transformation was conducted in parallel, with the same protocol, for both the FGFP and the LCPM cohorts.
CRC status classification
We invited patients referred for colonoscopy or colectomy to participate in the study. Those that consented were instructed to collect a stool sample at home, which was kept frozen using a sample kit provided by the research team. Upon completion of the medically necessary procedures (colonoscopy or colon resection), we stratified study participants into three diagnosis groups according to their clinical phenotype: (1) patients without evidence of lesions, (2) patients with polyps ( n < 10 and size between 6 and 10 mm) (ADE) and (3) patients with CRC. Patients whose clinical presentation did not fit any of these three groups were excluded from the study. Once the participants were included in the corresponding groups, extensive metadata was collected from their medical records as stated in the informed consent.
Sample collection
The stool samples of patients from UZL were collected as part of the LCPM project using aliquot ready mat without any buffer or preservative (Supplementary Fig. 1 ). The samples were kept at −20 °C freezers at the patients’ homes and brought to our laboratory on icepacks. Upon arrival, samples were stored in the Raes’ Lab at −80 °C until further analysis. Each stool sample had a temperature logger to make sure that, during the storage at home or transport to the laboratory, low stable temperature was maintained.
Stool sample analyses
Microbial load measurement by flow cytometry.
We determined microbial loads of stool samples of LCPM patients following published procedures 23 . We performed cell counting for all other samples in triplicate. Briefly, we dissolved 0.2 g frozen (−80 °C) aliquots in physiological solution to a total volume of 100 ml (8.5 g l −1 NaCl; VWR International). Subsequently, the slurry was diluted 1,000 times. The samples were filtered using a sterile syringe filter (pore size of 5 μm; Sartorius Stedim Biotech). Next, we stained 1 ml of the microbial cell suspension obtained with 1 μl SYBR Green I (1:100 dilution in dimethylsulfoxide; shaded for 15 min of incubation at 37 °C; 10,000 concentrate, Thermo Fisher Scientific) and monitored fluorescence events using the FL1 533/530 nm and FL3 >670 nm optical detectors of the C6 Accuri flow cytometer (BD Biosciences). In addition, forward and sideward scattered light was collected. The BD Accuri CFlow (v.1.0.264.21) software was used to gate and separate the microbial fluorescence events on the FL1/FL3 density plot from background events Supplementary Fig. 2 . A threshold value of 2,000 was applied on the FL1 channel. We evaluated the gated fluorescence events on the forward and sideward density plot, as to exclude remaining background events. We kept instrument and gating settings identical for all samples as described previously 24 . Based on the exact weight of the aliquots analyzed, we converted cell counts to microbial loads per gram of fecal material.
Fecal moisture content
We determined moisture content as the percentage of mass loss after lyophilization from 0.2 g frozen aliquots of nonhomogenized fecal material (−80 °C) as described previously 24 .
Fecal calprotectin measurement
We quantified fecal calprotectin concentrations using the fCAL ELISA Kit (Buhlmann). For patients and FGFP participants, we conducted analyses on frozen fecal material (−80 °C) as described previously 24 .
Microbiota phylogenetic profiling
Dna extraction and sequencing data preprocessing.
The fecal microbiota profile of the FGFP cohort was described previously 17 . For fecal DNA extraction and microbiota profiling of the new cohort, we followed the same protocols 17 .
The bacterial profiling was carried out as described previously 50 . Briefly, we extracted nucleic acids from frozen fecal aliquots using the MagAttract PowerMicrobiome DNA/RNA kit (Qiagen). We modified the manufacturer’s protocol by the addition of a heating step at 90 °C for 10 min after vortexing and excluding the steps where DNA is removed. For bacterial and archaeal characterization, we used 16S ribosomal RNA primers 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) targeting the V4 region. These primers were modified to contain a barcode sequence between each primer and the Illumina adapter sequences to produce dual-barcoded libraries from the extracted DNA (dilution 1:10) in triplicate. Deep sequencing was performed on a MiSeq platform (2 × 250 paired end (PE) reads, Illumina). We randomized all samples and negative controls (polymerase chain reaction (PCR) and extraction controls) taken along for sequencing. After demultiplexing with sdm as part of the LotuS pipeline (v. 1.60) 51 without allowing for mismatches, we further analyzed fastq sequences per sample using DADA2 pipeline (v. 1.6) 52 . Briefly, we removed the primer sequences and the first ten nucleotides after the primer. After merging paired sequences and removing chimeras, we assigned taxonomy using formatted Silva set ‘SLV_nr99_v138.1’. We performed taxonomic assignments at the domain, class, order, family, genus and species levels were performed using the ‘assignTaxonomy’ function from the DADA2 R library, by a naive Bayesian classifier method with a minimum bootstrap confidence of 50, using the ‘silva_nr99_v138.1_wSpecies_train_set.fa.gz’ training database (Extended Data Fig. 5 ). Deep sequencing was performed on a MiSeq platform from the DADA2 R library with the formatted Silva SSU database ‘silva_species_assignment_v138.1.fa.gz’ to obtain species assignments for the amplicon sequence variants (ASVs). We labeled any unassigned ASVs at any taxonomic level, with the prefix ‘uc’ along with the assigned taxonomic level (not species level) to avoid the lack of labels.
Before the analyses, we removed sequences annotated to the class Chloroplast, family mitochondria or unknown archaea and bacteria from eukaryotic origin. phyloseq (v. 1.36.0) 53 and MicroViz (v. 0.11.0) 54 libraries were used for data curation and figure generation.
For the relative microbiome matrix, we transformed ASV counts to relative abundances. In other words, we divided ASV counts by the total counts of ASV per sample. We agglomerated ASV to species level using the phyloseq (v. 1.36.0) 53 function ‘tax_glom’.
We agglomerated ASV to the species level, and the abundance matrix was centered log-ratio (CLR)-transformed using ‘codaSeq.clr ’ in the CoDaSeq (v. 0.99.6) 55 using the minimum proportional abundance detected for each taxon for the imputation of zeros.
Workflow Assessment
We conducted a workflow assessment using (1) a commercial mock community, ZymoBIOMICS Gut, and (2) two Fusobacterium species: Fusobacterium hwasookii (THCT14E2) and Fusobacterium nucleatum (DSM 20482T). The assessment followed our standard methods, involving the amplification, sequencing and analysis of the extracted DNA. This evaluation aimed to assess the performance of our full methodology, as depicted in Extended Data Fig. 6 .
Quality control assessment for amplicon sequencing data (16S rRNA) using RMP
In short, we sequenced all samples in six MiSeq runs (Extended Data Fig. 7a ). Per each run, we used a set of internal controls to identify: 1) Technical variation within and between runs 1) Contamination events during the DNA extraction, 2) Contamination events during the amplification and sequencing procedures and, 3) Carry-over contamination at the sequencing facility and barcode crosstalk.
We amplified all samples, including biological material (stool samples), positive controls (DNA from a stool sample previously profiled and RS: nonhuman gut bacteria strain ‘ Runella slithyformis’ ), negative controls (negative control of extraction (NCE) and negative control during PCR (NCP)) in triplicate using a unique barcode combination, while omitting several barcode combinations to control for primer synthesis cross contamination. We used Runella slithyformis in duplicate within each sequencing library to detect barcode crosstalk during the sequencing procedure (Extended Data Fig. 7b ). This genus is not detected in human gut samples; therefore, we expected no Runella slithyformis reads in any of the stool samples analyzed. We determined technical variation based on the BCD of positive control samples (Extended Data Fig. 7c ). Finally, we included NCEs along the whole process from extraction to bioinformatic analysis. For amplification and sequencing contamination 56 , we used NCP and NCE (Extended Data Fig. 7d and Supplementary Table 12 ), and for carry-over contamination events, we used a different set of barcode combinations in consecutive MiSeq runs 56 .
We built the QMP matrix as described previously 23 . In brief, we downsized samples to even sampling depth, defined as the ratio between sampling size (16S rRNA gene copy number-corrected sequencing depth) and microbial load (the average total cell counts per gram of frozen fecal material; Supplementary Table 2 ). We imputed 16S rRNA genome copies (GC) numbers using RasperGade16S (v. 0.0.1) 57 , a new tool that utilizes a heterogeneous pulsed evolution model for predicting 16S rRNA GC. It not only predicts the GC but also provides confidence estimates for the predictions 57 . We used a minimum rarefied read count of less than 150 for QMP analyses. We converted rarefied ASV abundances into numbers of cells per gram. The QMP matrices had a final size of 589 samples for the study cohort and 1,045 samples for the FGFP validation cohort 17 . We agglomerate the QMP matrix at ASV level to species level using the phyloseq (v. 1.36.0) 53 function ‘tax_glom’. We used the resulting species QMP matrix for the main analysis.
Statistical analysis
We performed all statistical analyses with R (Version 4.2.1, RStudio v.2022.12.0 + 353, 86_64-apple-darwin17.0 (64-bit)) and packages phyloseq (v. 1.36.0) 53 , vegan (v. 2.6.2) 58 , coin(v. 1.4.2) 59 , effectsize (v. 0.8.3), vcd(1.4.11) 60 , DirichletMultinomial(v. 1.34.0) 61 , pairwiseAdonis (v. 0.4.1) and microbiome (v. 1.14.0) 62 . We used nonparametric statistical tests for robust comparisons among unbalanced groups. For multiple testing, we corrected all P values using the Benjamini–Hochberg method (reported as adjusted P ) as appropriate on lists ( n > 1) of features (for example, taxa–metadata or metadata–metadata associations) and also when performing multiple pairwise group ( n > 2) comparisons (for example, KW test with phD test).
Fecal microbiota derived features and visualization
We visualized microbiota interindividual variation by PCoA using BCD on the species QMP matrix 24 , 25 . All the rest of the microbiota derived features were calculated based on QMP. We determined the contribution of metadata variables to microbiota community variation (effect size) of each of 94 variables by dbRDA on a species-level BCD with the capscale function in the vegan package 58 . We visualized absolute abundance species as log10 (abundance +1). This was the same for relative abundance.
Microbiota and physiological features associations
We excluded from analyzes any taxa unclassified at the species level or present in less than 5% of samples per each diagnosis group (Supplementary Table 6 ). We used Spearman correlations for rank–order correlations, between continuous variables complemented by Kendall’s tau correlation, including species abundances, calprotectin values and moisture content. We used the Mann–Whitney U -test to test median differences of continuous variables between two different groups. For more than two groups, for example, for differential abundance analysis for QMP and RMP taxa versus diagnosis groups, we used the KW test with phD test. For differential abundance analysis among diagnosis groups and bacteria species abundances from CLR transformed data, we performed an ANOVA test.
We evaluated statistical differences in the proportions of categorical variables (enterotypes) between patient groups using pairwise CS tests. We tested for deconfounded microbiota contributions to the diagnosis groups variable by using a nested model comparison (ANOVA) of generalized linear models as follows:
[alternative model] glm1 = rank(abundance) + rank(calprotectin) + rank(moisture) + rank(BMI) + diagnosis, where the diagnosis groups were recoded as 1, 2 and 3 for patients without evidence of CTLs, patients with polyps and patients with CRC, respectively. We treated this variable as a continuous variable, translating the directional increase in disease progression, from healthy to lesions, in the colonic mucosa. For the nested model comparison, we used taxa abundances (quantitative or relative) as explanatory variables, the diagnosis groups variable as response variable and BMI, fecal calprotectin and moisture as covariates. Additionally, we employed rank-transformed modeling to perform nonparametric testing on data that is not normally distributed, such as species abundances.
Previous reported CRC microbial markers
To compile a list of published CRC markers that would define taxa that should be tested against covariates in our data set, we conducted a PubMed search query using the keywords ‘CRC AND microbiome AND stool AND human AND biomarkers’. We found ten studies that met our inclusion criteria, namely: (1) a sample size minimum of 60 and (2) the CRC biomarker described at the species level, with statistical significance, in the main text of the publication. We included this list of published biomarkers in our correlation analysis between taxa and the three main covariates (fecal calprotectin, BMI and moisture) within the LCPM cohort. A similar procedure was followed at the genus level, which included 15 studies found in our PubMed search.
CRC microbial markers identification
We performed differential abundance analyzes on nine different CRC shotgun datasets as part of ‘curatedMetagenomicData’ 33 using MetaPhlAn 3.0 profiles to compare the results while controlling for potential differences arising from the classification tools and statistical methods used in each independent study. The results of the meta-analysis are presented in Extended Data Fig. 8 and Supplementary Table 13 .
Enterotyping and visualization
Using the genus matrix (agglomerated and downsized to 10,000 reads), we enterotyped and calculated observed genus richness 53 , as already reported for previous studies 24 , 25 . For enterotyping (or community typing) based on the DMM approach we used R as described previously 61 . We performed enterotyping on a combined genus-level abundance RMP matrix including LCPM samples compiled with 1,045 samples originating from the FGFP 17 . The optimal number of Dirichlet components based on the Bayesian information criterion was four. The four clusters were named ‘Bact1’, ‘Bact2’, ‘Prev’ and ‘Rum’, as described previously 23 .
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw amplicon sequencing data and metadata reported in this study have been deposited in European Nucleotide Archive with accession code EGAS00001007413 . FGFP 16S rRNA gene sequencing data and metadata are available at the European Genome-phenome Archive ( EGAS00001003296 ). The diagnosis metadata and processed microbiome data required for the reanalysis are provided as Supplementary Tables 1 and 14 , respectively. Formatted Silva set ‘SLV_nr99_v138.1’ files were downloaded from Zenodo via https://zenodo.org/records/4587955/files/silva_nr99_v138.1_wSpecies_train_set.fa.gz?download=1 (silva_nr99_v138.1_wSpecies_train_set.fa.gz) 63 and https://zenodo.org/records/4587955/files/silva_species_assignment_v138.1.fa.gz?download=1 (silva_species_assignment_v138.1.fa.gz) 63 . The nine CRC cohort MetaPhlAn 3.0 profiles were collected from curatedMetagenomicData, study names: FengQ_2015, HanniganGD_2017, ThomasAM_2018a, ThomasAM_2018b, VogtmannE_2016, WirbelJ_2018, YachidaS_2019 and YuJ_2015, ZellerG_2014 ( https://doi.org/10.18129/B9.bioc.curatedMetagenomicData ). Source data are provided with this paper.
Code availability
Analysis codes are available via Github at https://github.com/raeslab/QMP-Microbiome-CRC-confounders .
Yang, L. et al. Changes in colorectal cancer incidence by site and age from 1973 to 2015: a SEER database analysis. Aging Clin. Exp. Res. 33 , 1–10 (2020).
CAS Google Scholar
Keum, N. & Giovannucci, E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 16 , 713–732 (2019).
Article PubMed Google Scholar
Araghi, M. et al. Global trends in colorectal cancer mortality: projections to the year 2035. Int. J. Cancer https://doi.org/10.1002/ijc.32055 (2018).
Rex, D. K. & Eid, E. Considerations regarding the present and future roles of colonoscopy in colorectal cancer prevention. Clin. Gastroenterol. Hepatol. 6 , 506–514 (2008).
Gupta, V. K. et al. A predictive index for health status using species-level gut microbiome profiling. Nat. Commun. 11 , 4635 (2020).
Article CAS PubMed PubMed Central Google Scholar
Yachida, S. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25 , 968–976 (2019).
Young, C. et al. Microbiome analysis of more than 2,000 NHSbowel cancer screening programme samples shows the potential to improve screening accuracy. Clin. Cancer Res. 27 , 2246–2254 (2021).
Clos-Garcia, M. et al. Integrative analysis of fecal metagenomics and metabolomics in colorectal cancer. Cancers https://doi.org/10.3390/cancers12051142 (2020).
Article PubMed PubMed Central Google Scholar
Yu, Y. N. et al. Berberine may rescue Fusobacterium nucleatum- induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget 6 , 32013–32026 (2015).
Yu, T. C. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170 , 548–563.e16 (2017).
He, T., Cheng, X. & Xing, C. The gut microbial diversity of colon cancer patients and the clinical significance. Bioengineered 12 , 7046–7060 (2021).
Kasai, C. et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol. Rep. 35 , 325–333 (2016).
Article CAS PubMed Google Scholar
Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. https://doi.org/10.1038/s41591-019-0405-7 (2019).
Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. https://doi.org/10.1038/s41591-019-0406-6 (2019).
Young, C. et al. The colorectal cancer-associated faecal microbiome of developing countries resembles that of developed countries. Genome Med. 13 , 1–13 (2021).
Article Google Scholar
Vandeputte, D. et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65 , 57–62 (2016).
Falony, G. et al. Population-level analysis of gut microbiome variation. Science 352 , 560–564 (2016).
Poullis, A., Foster, R., Shetty, A., Fagerhol, M. K. & Mendall, M. A. Bowel inflammation as measured by fecal calprotectin: a link between lifestyle factors and colorectal cancer risk. Cancer Epidemiol. Biomarkers Prev. https://doi.org/10.1158/1055-9965.EPI-03-0160 (2004).
Högberg, C., Karling, P., Rutegård, J. & Lilja, M. Diagnosing colorectal cancer and inflammatory bowel disease in primary care: the usefulness of tests for faecal haemoglobin, faecal calprotectin, anaemia and iron deficiency. A prospective study. Scand. J. Gastroenterol. 52 , 69–75 (2017).
Schreuders, E. H., Grobbee, E. J., Spaander, M. C. W. & Kuipers, E. J. Advances in fecal tests for colorectal cancer screening. Curr. Treat. Options Gastroenterol. 14 , 152–162 (2016).
Røseth, A. G. et al. Faecal calprotectin: a novel test for the diagnosis of colorectal cancer? Scand. J. Gastroenterol. 28 , 1073–1076 (1993).
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V. & Egozcue, J. J. Microbiomedatasets are compositional: and this is not optional. Front. Microbiol . 8 , 2224 (2017).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551 , 507–511 (2017).
Vieira-Silva, S. et al. Quantitative microbiome profiling disentangles inflammation-and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat. Microbiol . 4 , 1826–1831(2019).
Vieira-Silva, S. et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature https://doi.org/10.1038/s41586-020-2269-x (2020).
Tibble, J. A. & Bjarnason, I. Fecal calprotectin as an index of intestinal inflammation. Drugs Today https://doi.org/10.1358/dot.2001.37.2.614846 (2001).
Quaglio, A. E. V., Grillo, T. G., De Oliveira, E. C. S., Di Stasi, L. C. & Sassaki, L. Y. Gut microbiota, inflammatory bowel disease and colorectal cancer. World J. Gastroenterol. 28 , 4053–4060 (2022).
Zeller, G. et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10 , 766 (2014).
Feng, Q. et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat. Commun. 6 , 6528 (2015).
Vogtmann, E. et al. Colorectal cancer and the human gut microbiome: reproducibility with whole-genome shotgun sequencing. PLoS ONE 11 , e0155362 (2016).
Hannigan, G. D., Duhaime, M. B., Ruffin, M. T., Koumpouras, C. C. & Schloss, P. D. Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio 9 , e02248-18 (2018).
Yu, J. et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 66 , 70–78 (2017).
Pasolli, E. et al. Accessible, curated metagenomic data through ExperimentHub. Nat. Methods 14 , 1023–1024 (2017).
Bjarnason, I. The use of fecal calprotectin in inflammatory bowel disease. Gastroenterol. Hepatol. 13 , 53–56 (2017).
Google Scholar
Dai, Z. et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome https://doi.org/10.1186/s40168-018-0451-2 (2018).
Zheng, R. et al. Body mass index (BMI) trajectories and risk of colorectal cancer in the PLCO cohort. Br. J. Cancer 119 , 130–132 (2018).
Carr, P. R. et al. Association of BMI and major molecular pathological markers of colorectal cancer in men and women. Am. J. Clin. Nutr. https://doi.org/10.1093/ajcn/nqz315 (2020).
Rutter, M. et al. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology 126 , 451–459 (2004).
Costa, F. et al. Role of faecal calprotectin as non-invasive marker of intestinal inflammation. Digest. Liver Dis. 35 , 642–647 (2003).
Article CAS Google Scholar
Konikoff, M. R. & Denson, L. A. Role of fecal calprotectin as a biomarker of intestinal inflammation in inflammatory bowel disease. Inflamm. Bowel Dis. https://doi.org/10.1097/00054725-200606000-00013 (2006).
Terzić, J., Grivennikov, S., Karin, E. & Karin, M. Inflammation and colon cancer. Gastroenterology 138 , 2101–2114 (2010).
Lehmann, F. S. et al. Clinical and histopathological correlations of fecal calprotectin release in colorectal carcinoma. World J. Gastroenterol. https://doi.org/10.3748/wjg.v20.i17.4994 (2014).
Pathirana, W. G. W., Chubb, S. P., Gillett, M. J., & Vasikaran, S. D. Faecal calprotectin. Clin. Biochem. Rev. https://doi.org/10.1097/mpg.0000000000001847 (2018).
Bullman, S. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358 , 1443–1448 (2017).
Osman, M. A. et al. Parvimonas micra , Peptostreptococcus stomatis , Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Sci. Rep. 11 , 1–12 (2021).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457 , 480–484 (2009).
Moghaddam, A. A., Woodward, M. & Huxley, R. Obesity and risk of colorectal cancer: a meta-analysis of 31 studies with 70,000 events. Cancer Epidemiol. Biomarkers Prev. 16 , 2533–2547 (2007).
Greathouse, K. L. et al. Gut microbiome meta-analysis reveals dysbiosis is independent of body mass index in predicting risk of obesity-associated CRC. BMJ Open Gastroenterol. https://doi.org/10.1136/bmjgast-2018-000247 (2019).
Liu, N. N. et al. Multi-kingdom microbiota analyses identify bacterial–fungal interactions and biomarkers of colorectal cancer across cohorts. Nat. Microbiol. 7 , 238–250 (2022).
Tito, R. Y. et al. Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut https://doi.org/10.1136/gutjnl-2018-316106 (2018).
Hildebrand, F., Tadeo, R., Voigt, A. Y., Bork, P. & Raes, J. LotuS: an efficient and user-friendly OTU processing pipeline. Microbiome 2 , 30 (2014).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13 , 581–583 (2016).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8 , e61217 (2013).
Barnett, D., Arts, I. & Penders, J. microViz: an R package for microbiome data visualization and statistics. J. Open Source Softw. 6 , 3201 (2021).
Gloor, G. B., Wu, J. R., Pawlowsky-Glahn, V. & Egozcue, J. J. It’s all relative: analyzing microbiome data as compositions. Ann. Epidemiol. 26 , 322–329 (2016).
Seitz, V. et al. A new method to prevent carry-over contaminations in two-step PCR NGS library preparations. Nucleic Acids Res. https://doi.org/10.1093/nar/gkv694 (2015).
Gao, Y. & Wu, M. Accounting for 16S rRNA copy number prediction uncertainty and its implications in bacterial diversity analyses. ISME Commun. 3 , 59–67 (2023).
Oksanen, F. J. et al. Vegan: Community Ecology Package. R package Version 2.4-3 https://CRAN.R-project.org/package=vegan (2017).
Hothorn, T., Hornik, K., Van De Wiel, M. A. & Zeileis, A. A Lego system for conditional inference. Am. Stat. https://doi.org/10.1198/000313006×118430 (2006).
Friendly, M. & Institute, S. A. S. Visualizing Categorical Data (SAS Institute, 2000).
Holmes, I., Harris, K. & Quince, C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS ONE 7 , e30126 (2012).
Shetty, S. A. & Lahti, L. Microbiome data science. J. Biosci. 44 , 1–6 (2019).
McLaren, M. R. & Callahan, B. J. Silva 138.1 prokaryotic SSU taxonomic training data formatted for DADA2. Zenodo https://doi.org/10.5281/zenodo.4587955 (2021).
Download references
Acknowledgements
We thank all study participants and the different staff members involved in the recruitment and execution of this project. We acknowledge L. Rymenans for her contribution to sample analysis. R.Y.T., S.V. and V.L.R. are funded by postdoctoral fellowships from the Research Fund–Flanders (1234321N, 12R6119N and 12V9421N, respectively). This work was funded by the Innovatie door Wetenschap en Technologie project ‘CRC_µBiome: characterization of human and microbial genetic components in premalignant adenoma and colorectal cancer’. The Raes lab is supported by Vlaams Instituut voor Biotechnologie (VIB), KU Leuven and the Rega Institute for Medical Research. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
These authors contributed equally: Raúl Y. Tito, Sara Verbandt, Sabine Tejpar, Jeroen Raes.
Authors and Affiliations
Laboratory of Molecular Bacteriology, Department of Microbiology and Immunology, Rega Institute, Katholieke Universiteit Leuven, Leuven, Belgium
Raúl Y. Tito, Leo Lahti, Chloe Verspecht, Verónica Lloréns-Rico, Sara Vieira-Silva, Gwen Falony & Jeroen Raes
Center for Microbiology, Vlaams Instituut voor Biotechnologie, Leuven, Belgium
Raúl Y. Tito, Chloe Verspecht, Verónica Lloréns-Rico, Gwen Falony & Jeroen Raes
Digestive Oncology, Department of Oncology, Katholieke Universiteit Leuven, Leuven, Belgium
Sara Verbandt, Marta Aguirre Vazquez & Sabine Tejpar
Department of Computing, University of Turku, Turku, Finland
Systems Biology of Host–Microbiome Interactions Laboratory, Principe Felipe Research Center (CIPF), Valencia, Spain
Verónica Lloréns-Rico
Institute of Medical Microbiology and Hygiene and Research Center for Immunotherapy, University Medical Center of the Johannes Gutenberg-University Mainz, Mainz, Germany
Sara Vieira-Silva & Gwen Falony
Institute of Molecular Biology, Mainz, Germany
Sara Vieira-Silva
Oncology, Janssen Pharmaceutica NV, Beerse, Belgium
Janine Arts
Department of Gastroenterology and Hepatology, Amsterdam University Medical Centers, Amsterdam, the Netherlands
Evelien Dekker
Therapeutics Discovery, Janssen Pharmaceutica NV, Beerse, Belgium
Joke Reumers
You can also search for this author in PubMed Google Scholar
Contributions
This study was conceived by J.A., S.T., J. Reumers and J. Raes. The experiments were designed by R.Y.T. and J. Raes. The data were collected and curated by S.V., M.A.V., L.L., J. Reumers, V.L.R., S.V.S., G.F. and S.T. The molecular data were generated by C.V. and R.Y.T. The statistical analyses were planned and executed by R.Y.T. and J. Raes R.Y.T. and J. Raes drafted the manuscript. All authors revised the article and approved the final version for publication.
Corresponding author
Correspondence to Jeroen Raes .
Ethics declarations
Competing interests.
J.A. and J. Reumers are employees of Janssen Pharmaceutica NV. J. Raes and R.T. are inventors on the patent application WO2017109059A1 in the name of VIB VZW, Katholieke Universiteit Leuven, KU Leuven R&D and Universiteit Gent covering methods for detecting the presence or assessing the risk of development of inflammatory arthritis disease. J. Raes, S.V.S. and G.F. are inventors on the patent application PCT/EP2018/084920 in the name of VIB VZW, KAtholieke Universiteit Leuven, KU Leuven Research and Development and Vrije Universiteit Brussel covering microbiome features associated with inflammation described in Vieira-Silva et al. Nature Microbiology 2019. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Medicine thanks Ruixin Zhu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling editor: Alison Farrell, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended data fig. 1 association of intestinal inflammation with fusobacterium nucleatum ..
Intestinal calprotectin levels associate Fusobacterium nucleatum absolute ( a) and relative ( b ) abundance in the LCMP. Two-sided Spearman rank correlation (adjP <0.05) and ‘x’ axes are log 10 transformed just for plotting. To rule out that the observed association is driven by a few samples with high abundance of Fusobacterium nucleatum, panel a has an insert of the plot removing samples with Fusobacterium nucleatum values above 1E8 cells per gram of stool. Best-fitting regression line in blue and 95% confidence interval shown in grey shading.
Extended Data Fig. 2 Fusobacterium nucleatum abundances before and after correction for intestinal calprotectin across diagnosis groups.
Absolute abundance of Fusobacterium nucleatum before ( a ) and after ( b ) correcting for intestinal calprotectin. Relative abundance of Fusobacterium nucleatum before ( c ) and after ( d ) correcting for intestinal calprotectin. The whiskers extend from the quartiles to the last data point within 1.5× of the interquartile range, with outliers beyond. The ‘y’ axes for (a) are log 10 transformed values (absolute abundance +1). The whiskers extend from the quartiles to the last data point within 1.5× of the interquartile range, with outliers beyond.
Extended Data Fig. 3 Spearman correlation between species abundance and microbiota covariates in the LCPM and FGFP cohorts.
Two-sided Spearman’s rank correlation comparison between absolute species abundance (QMP) and relative abundance (RMP) from the LCPM (N = 589 samples) and FGFP (N = 1045 samples) cohorts and a, BMI b, faecal calprotectin and c, moisture content values. Spearman correlation adjP < 0.05 (QMP and RMP, Supplementary Table 8 ).
Extended Data Fig. 4 Enterotype stratification by DMM community typing.
a , Identification of optimal number of clusters (Dirichlet components) in the LCPM cohort (n = 589) complemented with 1045 samples from the FGFP cohort, based on the Bayesian Information Criterion (BIC). b , Barplot representation of the average relative abundance of a few representative genera split into the four enterotypes identified by DMM community typing on the combined LCPM and FGFP cohorts (n = 1634).
Extended Data Fig. 5 Taxa assignation performance of the V4 amplicon marker in the LCPM.
a , Bootstrap values distribution across different ranks, b , Proportion of ASVs assigned from species to phylum, c , Proportion of ASVs assigned from species to phylum to each sample. The whiskers extend from the quartiles to the last data point within 1.5× of the interquartile range, with outliers beyond. The figure below (Panel a) illustrates our taxa assignation performance, showing that more than half of the ASVs were assigned to species level with bootstrap values above 80. Panel b shows the ASV assignation proportions from phylum (100%) to species level (50%). A comparison of proportions of ASVs assigned from each sample at different taxonomic levels revealed no significant differences in the distributions of assigned ASVs per sample across diagnosis groups, as indicated in panel c (KW test, p-values > 0.05). The center of the box plot represents the median value of the data, and the whiskers extend from the quartiles to the last data point within 1.5× of the interquartile range, with outliers beyond.
Extended Data Fig. 6 Performance of our methodology in small communities and isolated microorganisms.
a , Species composition of the ZymoBIOMICS gut controls, ten successfully identified species and b , two Fusobacterium species: Fusobacterium hwasookii (THCT14E2) and Fusobacterium nucleatum (DSM 20482T) were successfully identified using our methodology.
Extended Data Fig. 7 Quality control assessment for amplicon sequencing data (V4 16S rRNA gene).
a , The obtained reads for each sample are shown after processing with DADA2 (red and orange dashed lines represent 10, 000 and 1,000 reads, respectively; NCP: PCR negative control, NCE: DNA extraction Negative control, PC: positive control, and RS: Runella slithyformis control). b , Sequencing controls reveal the absence of barcode crosstalk. RS sequences serve as a marker for barcode crosstalk during sequencing. The absence of RS sequences in the samples without RS (no_RS) ruled out barcode crosstalk during the sequencing or PCR setup procedures. c , BCD among technical replicates demonstrating reproducibility. Pairwise comparisons between PC samples within and among MiSeq runs showed values under 0.2 (depicted by the pointed blue line). The center of the box plot represents the median value of the data, and the whiskers extend from the quartiles to the last data point within 1.5× of the interquartile range, with outliers beyond. d , Species composition of negative controls is presented, indicating the relative abundance and prevalence of the top 20 species. None of the species detected with differential abundance using QMP, RMP or CLR were found as background contaminants. Non-significant differences in bacteria composition were observed among DNA sequencing runs (Padj > 0.05, pairwiseAdonis test). A full list of detected species is available in Supplementary Table 12 . Of note, DI18R24 is not shown as the negative controls (NCE and NCP) did not produce reads.
Extended Data Fig. 8 Species and genera associated with CRC on a subset of the curatedMetagenomicData.
After performing our differential abundance procedure on the MataPhalAn 3.0 profiles downloaded from the curatedMetagenomicData, 108 species ( a ) and 63 genera ( b ) were identified across the 9 metagenomics datasets.
Supplementary information
Supplementary information.
Supplementary Figs. 1 and 2 and Tables 1–14.
Reporting Summary
Supplementary tables 1–14.
Supplementary Table 1. Reasons for the colonoscopy referral of the LCPM cohort. Supplementary Table 2. LCMP cohort variable names, 95 variables plus enterotypes. Supplementary Table 3. Associations between continuous variables and cancer progression (KW test with phD tests. N is specified for each test, and statistical significance was derived from two-sided testing and adjusted for multiple testing (adjusted P , Benjamini–Hochberg method)). Supplementary Table 4. Associations between categorical variables and cancer progression (two-sided CS test; statistical significance was derived from two-sided testing and adjusted for multiple testing (adjusted P , Benjamini–Hochberg method)). Supplementary Table 5. Microbiome variation in the LCMP cohort. Independent and cumulative contribution of metadata variables to species-level microbiome variation (dbRDA and stepwise dbRDA; false discovery rate by Benjamini–Hochberg). Cumulative explanatory power and significance level of the included variables are reported. Supplementary Table 6. List of species excluded and included from the analysis. Supplementary Table 7. Differences in absolute (QMP) and relative (RMP) species abundances over diagnostic groups LCMP cohort ( n = 589, KW, phD test; statistical significance was derived from two-sided testing and adjusted for multiple testing (adjusted P , Benjamini–Hochberg method)). Supplementary Table 8. Associations between species abundances (QMP and RMP) and BMI, intestinal calprotectin and moisture in the LCPM cohort ( n = 589, Spearman and Kendall’s tau; statistical significance was derived from two-sided testing and adjusted for multiple testing (adjusted P , Benjamini–Hochberg method)). Supplementary Table 9. Associations between species abundances (QMP and RMP) and BMI, intestinal calprotectin and moisture in the FGFP cohort ( n = 1,045, Spearman; statistical significance was derived from two-sided testing and adjusted for multiple testing (adjusted P , Benjamini–Hochberg method)). Supplementary Table 10. Differences in absolute (QMP) and relative (RMP) species abundances over diagnostic groups in the LCMP cohort subset with normal levels of fecal calprotectin ( n = 340 (112 PWoL, 216 PWP and 12 PWT, KW and adjusted for multiple testing (adjusted P , Benjamini–Hochberg method)). Supplementary Table 11. Associations between categorical variables and enterotype distribution (two-sided CS test; statistical significance was derived from two-sided testing and adjusted for multiple testing (adjusted P , Benjamini–Hochberg method)). Supplementary Table 12. Full list of the species detected in the negative controls (NCE and NCP). Supplementary Table 13. Differences in relative abundances of species profiles from MetaPhlAn 3.0 between CRC and controls from nine published CRC cohorts from the curatedMetagenomicData ( n = 1,254, two-sided Wilcoxon signed-rank test and adjusted for multiple testing (adjusted P , Benjamini–Hochberg method)). Supplementary Table 14. Absolute taxonomic abundances at species level in the LCMP cohort ( n = 589).
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Source data fig. 3, source data fig. 4, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Tito, R.Y., Verbandt, S., Aguirre Vazquez, M. et al. Microbiome confounders and quantitative profiling challenge predicted microbial targets in colorectal cancer development. Nat Med (2024). https://doi.org/10.1038/s41591-024-02963-2
Download citation
Received : 18 November 2022
Accepted : 29 March 2024
Published : 30 April 2024
DOI : https://doi.org/10.1038/s41591-024-02963-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
By continuing to browse the site you are agreeing to our use of cookies and similar tracking technologies described in our privacy policy .
Resource Library
The AAEP develops numerous resources to assist veterinarians and the broader equine industry with issues affecting the horse and connection to those with common goals.
Search & Filter
Resource Type
Audience Type
Snake Bite Vaccination Guidelines
Tetanus Vaccination Guidelines
Tetanus toxoid is a core equine vaccine and should be included in equine immunization programs for every horse annually.
Venezuelan Equine Encephalomyelitis Vaccination Guidelines
Equine Viral Arteritis (EVA) Vaccination Guidelines
Equine Influenza Vaccination Guidelines
West Nile Virus Vaccination Guidelines
- Next »
Join Us Today!
Our community of horse doctors connects you to more than 9,000 veterinarians and veterinary students who make a difference every day in horse health, just like you!


COMMENTS
The objective of this thesis is to quantify the impact of social media reviews on brand perception. Specifically, this thesis focuses on two diverse media platforms commonly used for sharing opinions about products or services by publishing audio-visual or textual reviews: YouTube and Yelp.
The answer is written in length in the discussion section of the paper. Thus, the research question gives a preview of the different parts and variables of the study meant to address the problem posed in the research question.1 An excellent research ... Definitions and examples of quantitative research questions. Quantitative research questions;
friends on social media. Earlier research conducted by Gross (2004) reflects similar results. In his survey of 261 students in grades 7-10, he found that students spend an average of 40 minutes texting per day. Likewise, research by Kowalski and Limber (2007) reflected comparable results of 3,767
Revised on June 22, 2023. Quantitative research is the process of collecting and analyzing numerical data. It can be used to find patterns and averages, make predictions, test causal relationships, and generalize results to wider populations. Quantitative research is the opposite of qualitative research, which involves collecting and analyzing ...
Quantitative methods emphasize objective measurements and the statistical, mathematical, or numerical analysis of data collected through polls, questionnaires, and surveys, or by manipulating pre-existing statistical data using computational techniques.Quantitative research focuses on gathering numerical data and generalizing it across groups of people or to explain a particular phenomenon.
Quantitative research is the process of collecting and analyzing numerical data to describe, predict, or control variables of interest. This type of research helps in testing the causal relationships between variables, making predictions, and generalizing results to wider populations. The purpose of quantitative research is to test a predefined ...
Research papers in the social and natural sciences often follow APA style. This article focuses on reporting quantitative research methods. In your APA methods section, you should report enough information to understand and replicate your study, including detailed information on the sample, measures, and procedures used.
10 Research Question Examples to Guide your Research Project. Published on October 30, 2022 by Shona McCombes.Revised on October 19, 2023. The research question is one of the most important parts of your research paper, thesis or dissertation.It's important to spend some time assessing and refining your question before you get started.
An Outline for Quantitative Research Pa pers. Rui Pedro Paiva. CISUC - Centre for Informatics and Systems of the University of Coimbra, Portugal. [email protected]. August 2013. Abstract. About ...
Mixed-methods research is a flexible approach, where the research design is determined by what we want to find out rather than by any predetermined epistemological position. In mixed-methods research, qualitative or quantitative components can predominate, or both can have equal status. 1.4. Units and variables.
Summarizing quantitative data and its effective presentation and discussion can be challenging for students and researchers. This chapter provides a framework for adequately reporting findings from quantitative analysis in a research study for those contemplating to write a research paper. The rationale underpinning the reporting methods to ...
Surprises at a Local "Family" Restaurant: Example Quantitative Research Paper A quantitative research paper with that title might start with a paragraph like this: Quaintville, located just off the main highway only five miles from the university campus, may normally be a sleepy community, but recent plans to close the only fast-food ...
Quantitative Research. Quantitative research is a type of research that collects and analyzes numerical data to test hypotheses and answer research questions.This research typically involves a large sample size and uses statistical analysis to make inferences about a population based on the data collected.
These sample papers demonstrate APA Style formatting standards for different student paper types. Students may write the same types of papers as professional authors (e.g., quantitative studies, literature reviews) or other types of papers for course assignments (e.g., reaction or response papers, discussion posts), dissertations, and theses.
Quantitative research methods are concerned with the planning, design, and implementation of strategies to collect and analyze data. Descartes, the seventeenth-century philosopher, suggested that how the results are achieved is often more important than the results themselves, as the journey taken along the research path is a journey of discovery. . High-quality quantitative research is ...
Quantitative research appeared around 1250, and was driven by investigators with the need to quantify data. Since then quantitative research has dominated the western cultural as the research method to create new knowledge. This method was originally developed in the natural sciences to study natural phenomena [Williams, 2007]. In
This section delves into research quantitative design. The quantitative research design procedures employed in the social sciences, natural sciences, and many other domains for gathering and ...
Any quantitative research paper can have aspects of ... The study adopted a cross-sectional research design and quantitative research approach using a sample of 300 respondents from the six public ...
Title, keywords and the authors. The title of a paper should be clear and give a good idea of the subject area. The title should not normally exceed 15 words 2 and should attract the attention of the reader. 3 The next step is to review the key words. These should provide information on both the ideas or concepts discussed in the paper and the ...
This type of research can be used to establish generalisable facts about a topic. Common quantitative methods include experiments, observations recorded as numbers, and surveys with closed-ended questions. Qualitative research. Qualitative research is expressed in words. It is used to understand concepts, thoughts or experiences.
Your SEO optimized title. Sep 4, 2021 How To Get Published 0. Score 94% Score 94%. Example of a Quantitative Research Paper for Students & Researchers. This example of a quantitative research paper is designed to help students and other researchers who are learning how to write about their work. The reported research observes the behaviour of ...
When collecting and analyzing data, quantitative research deals with numbers and statistics, while qualitative research deals with words and meanings. Both are important for gaining different kinds of knowledge. Quantitative research. Quantitative research is expressed in numbers and graphs. It is used to test or confirm theories and assumptions.
COVID-19 is an infectious disease caused by a new strain of coronavirus that attacks the respiratory system (World Health Organization, 2020). As of January 2021, COVID-19 has infected 94 million people and has caused 2 million deaths in 191 countries and territories (John Hopkins University, 2021).
The ethical governance of Artificial Intelligence (AI) in health care and public health continues to be an urgent issue for attention in policy, research, and practice. In this paper we report on central themes related to challenges and strategies for promoting ethics in research involving AI in global health, arising from the Global Forum on Bioethics in Research (GFBR), held in Cape Town ...
Despite substantial progress in cancer microbiome research, recognized confounders and advances in absolute microbiome quantification remain underused; this raises concerns regarding potential ...
A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications. Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the ...
Our community of horse doctors connects you to more than 9,000 veterinarians and veterinary students who make a difference every day in horse health, just like you!