An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- J Cell Biol
- v.216(5); 2017 May 1


The signal hypothesis matures with age
Secretory proteins are targeted for secretion by sequences in their mature domains, as well as by N-terminal signal peptides.

Focal Point (left to right) Katerina Chatzi, Marios Sardis, Lily Karamanou, Tassos Economou, Alexandra Tsirigotaki, and colleagues reveal that bacterial secretory proteins, such as PhoA (pictured), are targeted for secretion by multiple hydrophobic patches (orange) in their mature domains, as well as by their N-terminal signal peptides (green). These mature targeting signals are buried in the fully folded form of the protein (left), but are exposed in the unstructured, secretion-competent form (right), allowing them to bind to hydrophobic patches on the translocase subunit SecA. Once bound to the translocase, the mature targeting signals are subsequently required for the protein’s translocation across the cell membrane. Photos courtesy of the authors.
For over 40 years, researchers have known that the majority of membrane and secretory proteins are targeted for secretion by an N-terminal signal peptide that is subsequently cleaved off to generate the mature form of the protein. But Chatzi et al. now reveal that sequences in the mature regions of secretory proteins are also crucial for directing them to the translocation machinery and mediating their passage from the cytoplasm to the extracellular space ( 1 ).
Günter Blobel won a Nobel Prize for his “signaling hypothesis” explaining how signal peptides target secretory proteins to a translocase channel embedded in the membrane of the endoplasmic reticulum ( 2 ). Once the proteins have passed through this channel into the lumen of the ER, the signal peptide is removed by proteases and the mature protein is secreted from the cell. A similar process takes place in bacteria, where cleavable signal peptides direct secretory proteins to the SecA–SecYEG translocase complex embedded in the bacterial plasma membrane ( 3 ).
The signal peptide is generally considered to be sufficient for secretory protein targeting. “Undoubtedly, the signal peptide is really important,” says Tassos Economou, from Katholieke Universiteit Leuven in Belgium. “But several lines of evidence suggest that additional targeting information may reside in the mature domains of secretory proteins.”
Economou’s group has provided some of this evidence themselves. In 2009, they demonstrated that bacterial secretory proteins lacking their signal peptides could still bind to the SecA–SecYEG complex with high affinity ( 4 ). Though the signal peptide further enhances this affinity, its essential role is to activate the translocase complex to transport the secretory protein across the membrane. In fact, signal peptides can even play this role in trans, stimulating the translocation of secretory protein mature domains to which they are not covalently linked ( 4 ).
“The signal peptide isn’t enough for secretion.”
Economou and colleagues, led by graduate students Katerina Chatzi and Marios Sardis and senior scientist Lily Karamanou, set out to identify targeting signals in the mature domains of bacterial secretory proteins ( 1 ). The researchers realized that the mature domains of proteins such as the alkaline phosphatase PhoA contain multiple stretches of hydrophobic amino acids that can bind to the SecA component of the bacterial translocase. One or two of these hydrophobic patches were sufficient to target the proteins to the translocase, but deleting or mutating all of them greatly reduced the proteins’ affinity for SecA, particularly if they also lacked an N-terminal signal peptide.
These hydrophobic residues would generally be buried on the inside of fully folded secretory proteins, but Chatzi et al. found that proteins targeting the bacterial translocase are largely unstructured. The buried hydrophobic stretches therefore become exposed on the surface of the secretory proteins, where they can interact with conserved hydrophobic patches on the surface of the SecA receptor, including one patch that lies next to SecA’s previously identified signal peptide-binding cleft ( 5 ).
Surprisingly, the mature domain targeting signals don’t just mediate binding to SecA; they are also required for the protein’s subsequent translocation across the bacterial membrane. Secretory proteins lacking their mature targeting signals could still bind to SecA through their signal peptides but they weren’t secreted in vivo or in vitro. Mutating the hydrophobic patches on SecA similarly impaired protein secretion.
“So, the signal peptide isn’t enough for secretion, and neither are the targeting sequences in the mature domain,” Economou says. “You need both for the translocase to be activated and secretion to occur.” This “dual key mechanism” allows the translocase to distinguish genuine secretory proteins from cytoplasmic proteins with exposed hydrophobic residues. The researchers now want to investigate how the mature targeting signals contribute to protein translocation.
It remains to be seen whether eukaryotic secretory proteins also carry targeting information in their mature domains. Economou suspects that this may be the case, at least for proteins that are translocated into the ER posttranslationally.
Advertisement
Lost in translation : the signal hypothesis
- Standard View
- Article contents
- Figures & tables
- Supplementary Data
- Peer Review
- Open the PDF for in another window
- Get Permissions
- Cite Icon Cite
- Search Site
Mitch Leslie; Lost in translation : the signal hypothesis . J Cell Biol 1 August 2005; 170 (3): 338. doi: https://doi.org/10.1083/jcb1703fta1
Download citation file:
- Ris (Zotero)
- Reference Manager
It was cell biology's version of the ship in the bottle. How do proteins a cell intends to secrete end up in the endoplasmic reticulum? Winkling out the details of the translocation mechanism that spirits these proteins into the ER required more than 20 years and earned Günter Blobel of Rockefeller University the 1999 Nobel Prize in Physiology or Medicine.
Another Rockefeller laureate, George Palade, had demonstrated that ribosomes free in the cytoplasm manufactured nonsecreted proteins, whereas ribosomes stuck to the ER made proteins for export. Cell biologists searched in vain for distinctions between free and attached ribosomes that might explain their contrasting behavior. A new assistant professor at Rockefeller and Palade's protege, Blobel suspected that the difference must lie in the proteins themselves. He and colleague David Sabatini conjectured that secretory proteins might carry a short segment near the NH 2 terminus (Blobel and Sabatini, 1971). Once this sequence protruded from the ribosome during translation, a “binding factor” would hook onto the protein and guide it and the ribosome to the ER membrane. Continued translation would then thread the elongating protein into the ER's interior. “It was a beautiful idea,” says Blobel. It was also, he admits, “pure speculation.”

The signal hypothesis in 1975, with the signal peptide as a dotted line.
Unaware of Blobel and Sabatini's hypothesis, Cesar Milstein of Cambridge University proposed a similar idea based on his team's cell-free system. It also pumped out an overweight light chain, but when the researchers checked the output of microsomes (ER fragments), they found only the normal-sized protein (Milstein et al., 1972). Milstein speculated that the extra amino acids help direct the growing protein to the ER.
Despite this suggestive data, detractors argued that the protein's extra heft was an artifact of in vitro translation or isolation errors, Blobel recalls. To answer their complaints, he crafted a protein-synthesizing system with help from post-doc Bernhard Dobberstein (now at the University of Heidelberg). Using detergent, they dislodged ribosomes from rough microsomes, and then slipped the particles—which carried unfinished light chains—into a solution that allowed protein making to resume. Because the researchers also added a compound that blocks new translation, the ribosomes could only complete chains they had started.
At first, only the smaller, processed chain appeared (Blobel and Dobberstein, 1975a). These proteins came from ribosomes that were well into translation when they parted from microsomes, the researchers concluded, and the chains they held had already undergone pruning to remove the signal sequence. After a few minutes, however, the synthesis mixture started producing longer chains as well. The bulkier proteins emerged from ribosomes that had just started translating when isolated from microsomes. At the time, they bore stubby chains that hadn't yet shed their signal sequence. When translation restarted, these short chains didn't lose the sequence—evidence that the processing enzyme that removes the signal is part of the ER membrane.
In another key experiment, Blobel and Dobberstein let rough microsomes—which carry ribosomes and some associated mRNA—produce proteins. The scientists detected only the shorter version. Adding the protein-dissolving enzymes trypsin and chymotrypsin (which rarely enter the microsomes) did not digest most of the chains, confirming that the trimmed protein ends up tucked away within the microsomes, as the signal hypothesis predicted.

Ribosomes severed from microsomes make first a smaller, processed protein (left) and later a longer form with signal sequence intact (upper band on right).
Blobel, G., and B. Dobberstein. 1975 a. J. Cell Biol. 67 : 835 –851.
Blobel, G., and B. Dobberstein. 1975 b. J. Cell Biol. 67 : 852 –862.
Blobel, G., and D.D. Sabatini. 1971. In Biomembranes. L.A. Manson, ed. 2:193–195.
Milstein, C., et al. 1972 . Nat. New Biol. 239 : 117 –120.
Schechter, I. 1973 . Proc. Natl. Acad. Sci. USA. 70 : 2256 –2260.
Swan, D., et al. 1972 . Proc. Natl. Acad. Sci. USA. 69 : 1967 –1971.
Tonegawa, S., and I. Baldi. 1973 . Biochem. Biophys. Res. Commun. 51 : 81 –87.
Data & Figures
- Previous Article
- Next Article
Supplements
Related & metrics, suggested content, email alerts, affiliations.
- Newest Articles
- Current Issue
- Email Alerts
- Submit a Manuscript
- Instructions for Authors
- For Librarians
- Editors & Staff
- Policies & Permissions
- Accessibility Statement
- Privacy Policy
- Online ISSN 1540-8140
- Print ISSN 0021-9525
This Feature Is Available To Subscribers Only
Sign In or Create an Account
Signal Sequence
- Reference work entry
- First Online: 01 January 2017
- Cite this reference work entry

588 Accesses
Signal peptide
Is a N -terminal, short (18–26) amino acid sequence that acts like a zip code in nascent polypeptides and determines their subcellular targeting. A signal sequence is a protein region with which a protein can be directed to the appropriate cellular compartment within a cell; they initiate co-translational transfer through the membrane of the endoplasmic reticulum (ER). Proteins are often synthesized in an immature version (pre-protein) that is larger than the mature functional form. This is due to the presence of N-terminal amino acid stretches, referred to as leader sequences. The pre-protein is a transient precursor, since the leader sequence is cleaved off during protein processing. This signal sequence is a short stretch of 15–30 amino acids that mediate the transfer of any attached polypeptide to the endoplasmic reticulum. It provides the means for the ribosomes to attach to the ER membrane (ER regions with associated ribosomes are called “rough...
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Editor information
Editors and affiliations.
University Professor of Genetics, German Cancer Research Center - DKFZ, Heidelberg, Germany
Manfred Schwab
Rights and permissions
Reprints and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this entry
Cite this entry.
(2011). Signal Sequence. In: Schwab, M. (eds) Encyclopedia of Cancer. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-16483-5_5297
Download citation
DOI : https://doi.org/10.1007/978-3-642-16483-5_5297
Published : 10 March 2017
Publisher Name : Springer, Berlin, Heidelberg
Print ISBN : 978-3-642-16482-8
Online ISBN : 978-3-642-16483-5
eBook Packages : Biomedical and Life Sciences Reference Module Biomedical and Life Sciences
Share this entry
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
Spatial expression of the genome: the signal hypothesis at forty
Affiliation.
- 1 Department of Surgery, The University of Chicago, 5841 South Maryland Avenue, MC 5032, SBRI J557, Chicago, Illinois 60637-1470, USA. [email protected].
- PMID: 21487438
- DOI: 10.1038/nrm3105
The signal hypothesis, formulated by Günter Blobel and David Sabatini in 1971, and elaborated by Blobel and his colleagues between 1975 and 1980, fundamentally expanded our view of cells by introducing the concept of topogenic signals. Cells were no longer just morphological entities with compartmentalized biochemical functions; they were now active participants in the creation and perpetuation of their own form and identity, the decoders of linear genetic information into three dimensions.
Publication types
- Research Support, N.I.H., Extramural
- Endoplasmic Reticulum / metabolism*
- Models, Genetic
- Protein Biosynthesis / genetics*
- Protein Sorting Signals / genetics*
- Ribosomes / metabolism*
- Signal Transduction / genetics
- Protein Sorting Signals
Grants and funding
- G13 LM009860/LM/NLM NIH HHS/United States
- G13 LM009860-02/LM/NLM NIH HHS/United States
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Published: November 1999

A Nobel Prize for cell biology
Nature Cell Biology volume 1 , page E169 ( 1999 ) Cite this article
15k Accesses
4 Citations
10 Altmetric
Metrics details
This year’s Nobel Prize in Physiology or Medicine has been awarded to Günter Blobel, of the Rockefeller University, for ‘‘the discovery that proteins have intrinsic signals that govern their transport and localisation in the cell’’. Angus Lamond of the University of Dundee says that the prize is “a landmark in its recognition of the fundamental importance of basic research in molecular cell biology for modern medicine and the fight against disease”.
The idea that proteins contain targeting zip codes originated in 1971, when Blobel and his colleague David Sabatini postulated that the information needed to direct a nascent peptide to the membrane of the endoplasmic reticulum is contained within the peptide itself. A year later, César Milstein and colleagues provided experimental evidence for a transient signal sequence at the amino-terminal end of a secretory protein. Building on this important discovery and work from his own laboratory, Blobel, together with Bernhard Dobberstein, formulated the ‘signal hypothesis’. In a 1975 paper — which has since become a citation classic — they predicted that newly synthesized proteins have a built-in signal which directs them to the endoplasmic reticulum and through a channel in the membrane. This principle has turned out to be a common mechanism, operating in a variety of intracellular pathways in all organisms, from bacteria to yeast to man. According to Peter Walter of the University of San Francisco, ‘‘Günter’s accomplishment is not defined by a single paper or even a set of papers, but rather is the fruit of his casting a wide net over a fundamentally important problem in biology. Many firsts came out of his lab, but it’s the sum of the individual pieces that adds up to the big picture’’.

As well as his proposal of the signal hypothesis, it is Blobel’s imaginative approach towards the in vitro reconstitution of protein transport that has revolutionized the field of cell biology. ‘‘The early realisation that many, perhaps most, reactions that occur in the cell could ultimately be recapitulated in vitro ,’’ says Hidde Ploegh of Harvard University, ‘‘is in no small measure due to Blobel’s work in this area’’. According to Randy Schekman of the University of California, Berkeley, it was the first time that ‘‘a cell-free system was developed to dissect the process of intracellular transport using the tools of the enzymologist. Before this landmark event, the field had relied essentially exclusively on morphology and cell fractionation, techniques that by themselves had little prospect of elucidating the machinery required to move proteins from one place to another in the cell’’. Blobel’s in vitro experimental system inspired other cell biologists and, according to Graham Warren of Yale University, it ‘‘made people believe that any cellular function, no matter how complicated, can be mimicked in the test tube’’.
Founder of a distinguished scientific lineage
Like any high-profile award, this Nobel Prize has sparked some controversy and speculation. It is, however, undeniable that Blobel has played a pivotal part in training an impressive group of cell biologists, including Bernhard Dobberstein, Peter Walter, Reid Gilmore, Larry Gerace, Vishwanath Lingappa, Gerry Waters, David Anderson, Sandy Simon and Mary Moore, to name just a few. These scientists are now shaping the field of molecular cell biology. Blobel himself was trained by the 1974 Nobel laureate George Palade, who fondly remembers him as a ‘‘young, impetuous and highly promising postdoctoral fellow’’. Palade, who is well known for his dedication to his students and postdoctoral fellows, no doubt provided an excellent role model in this regard. Waters, who was a graduate student with Blobel, says that he ‘‘was always available to discuss science, and was an extraordinarily supportive and enthusiastic graduate advisor’’. Lingappa ‘‘is, and always has been, immensely proud to be one of his scientific offsprings’’. It seems that Blobel’s extraordinary track record as a mentor reflects his ability to bring out the best in people by making them believe in themselves. Perhaps it is this, and not the Nobel Prize, that is his greatest achievement.
Rights and permissions
Reprints and permissions
About this article
Cite this article.
A Nobel Prize for cell biology. Nat Cell Biol 1 , E169 (1999). https://doi.org/10.1038/15602
Download citation
Issue Date : November 1999
DOI : https://doi.org/10.1038/15602
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Blobel and sabatini’s “beautiful idea”: visual representations of the conception and refinement of the signal hypothesis.
- Michelle Lynne LaBonte
Journal of the History of Biology (2017)
Controlling Protein Compartmentalization to Overcome Disease
- James R. Davis
- Mudit Kakar
- Carol S. Lim
Pharmaceutical Research (2006)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- Subscriber Services
- For Authors
- Publications
- Archaeology
- Art & Architecture
- Bilingual dictionaries
- Classical studies
- Encyclopedias
- English Dictionaries and Thesauri
- Language reference
- Linguistics
- Media studies
- Medicine and health
- Names studies
- Performing arts
- Science and technology
- Social sciences
- Society and culture
- Overview Pages
- Subject Reference
- English Dictionaries
- Bilingual Dictionaries
Recently viewed (0)
- Save Search

Edited by: Robert Hine
- Find at OUP.com
- Google Preview
- Share This Facebook LinkedIn Twitter
- Publishing Information
- The List of Entries by Subject
- Simplified phylogenetic tree of the animal kingdom
- Simplified phylogenetic tree for plants
- Geological time scale
- Navigating the body
- Model organisms and their genomes
- Major mass extinction of species
- Nobel prizewinning contributions to biology
- Useful websites
- Previous Version
signal hypothesis
A hypothesis to explain how ribosomes become attached to membranes within cells in order to deliver the appropriate proteins to cell organelles, such as mitochondria and chloroplasts, or transport proteins outside the cell membrane. It proposes that the leading end of the nascent polypeptide chain consists of a ... ...
Access to the complete content on Oxford Reference requires a subscription or purchase. Public users are able to search the site and view the abstracts and keywords for each book and chapter without a subscription.
Please subscribe or login to access full text content.
If you have purchased a print title that contains an access token, please see the token for information about how to register your code.
For questions on access or troubleshooting, please check our FAQs , and if you can''t find the answer there, please contact us .
- Oxford University Press
PRINTED FROM OXFORD REFERENCE (www.oxfordreference.com). (c) Copyright Oxford University Press, 2023. All Rights Reserved. Under the terms of the licence agreement, an individual user may print out a PDF of a single entry from a reference work in OR for personal use (for details see Privacy Policy and Legal Notice ).
date: 16 May 2024
- Cookie Policy
- Privacy Policy
- Legal Notice
- Accessibility
- [66.249.64.20|185.80.149.115]
- 185.80.149.115
Character limit 500 /500

- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons
Margin Size
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.

7.4: Propagation of the signal
- Last updated
- Save as PDF
- Page ID 69040
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
Once a ligand binds to a receptor, the signal is transmitted through the membrane and into the cytoplasm. Continuation of a signal in this manner is called signal transduction . Signal transduction only occurs with cell-surface receptors because internal receptors are able to interact directly with DNA in the nucleus to initiate protein synthesis.
Binding Initiates a Signaling Pathway
After the ligand binds to the cell-surface receptor, the activation of the receptor’s intracellular components sets off a chain of events that is called a signaling pathway or a signaling cascade . Signaling pathways can get very complicated very quickly because most cellular proteins can affect different downstream events, depending on the conditions within the cell. A single pathway can branch off toward different endpoints based on the interplay between two or more signaling pathways, and the same ligands are often used to initiate different signals in different cell types. This variation in response is due to differences in protein expression in different cell types. Another complicating element is signal integration of the pathways, in which signals from two or more different cell-surface receptors merge to activate the same response in the cell. This process can ensure that multiple external requirements are met before a cell commits to a specific response.
The effects of extracellular signals can also be amplified by enzymatic cascades. At the initiation of the signal, a single ligand binds to a single receptor. However, activation of a receptor-linked enzyme can activate many copies of a component of the signaling cascade, which amplifies the signal.
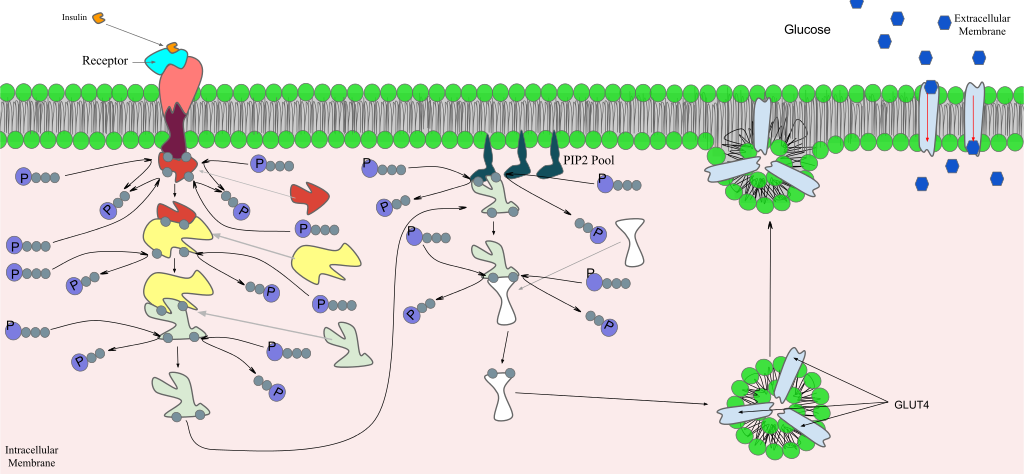
Methods of Intracellular Signaling
The activation of a signaling pathway depends on the modification of a cellular component by an enzyme. There are numerous types of enzymatic modifications that can occur, and they are recognized in turn by the next component downstream. The following are some of the more common events in intracellular signaling.
Phosphorylation
One of the most common chemical modifications that occurs in signaling pathways is the addition of a phosphate group (PO 4 –3 ) to a molecule such as a protein in a process called phosphorylation. The transfer of the phosphate is catalyzed by an enzyme called a kinase. Various kinases are named for the substrate they phosphorylate. Phosphorylation can create a binding site that interacts with downstream components in the signaling cascade. Phosphorylation may activate or inactivate enzymes, and the reversal of phosphorylation, dephosphorylation by a phosphatase, will reverse the effect.
Second Messengers
Second messengers are small molecules that help to spread a signal through the cytoplasm after a ligand binds to a receptor. They do this by altering the behavior of certain cellular proteins. Some examples of second messengers are cAMP (a modified version of AMP, which is related to ATP but only contains one phosphate) and calcium ions.
OpenStax , Biology. OpenStax CNX. October 13, 2017. https://cnx.org/contents/[email protected]

IMAGES
VIDEO
COMMENTS
The signal hypothesis proposes that proteins destined for secretion, which involves the movement of the protein across a biological membrane, are originally manufactured with an initial sequence of amino acids that may or may not present in the mature protein. Work by Blobel and others over two decades established the validity of the proposal.
A remarkable aspect of the work that established the signal hypothesis was its continuity with previous studies carried out in the same Laboratory of Cell Biology at the Rockefeller Institute ...
The signal hypothesis was originally proposed by Günter Blobel and David Sabatini in 1971, and demonstrated by Blobel and colleagues in 1975. The signal hypothesis showed that cytoplasmically synthesised proteins targeted to the ER use a signal sequence to direct them to the ER membrane. A signal sequence is a short peptide that is part of the ...
The signal hypothesis also laid the foundations of modern biotechnology. Proteins such as human insulin and human growth hormone could be tagged for mass production in bacteria, secreted and ...
Günter Blobel won a Nobel Prize for his "signaling hypothesis" explaining how signal peptides target secretory proteins to a translocase channel embedded in the membrane of the endoplasmic reticulum ( 2 ). Once the proteins have passed through this channel into the lumen of the ER, the signal peptide is removed by proteases and the mature ...
Günter Blobel won a Nobel Prize for his "signaling hypothesis" explaining how signal peptides target secretory proteins to a translocase channel embedded in the membrane of the endoplasmic reticulum ( 2 ). Once the proteins have passed through this channel into the lumen of the ER, the signal peptide is removed by proteases and the mature ...
At the time, they bore stubby chains that hadn't yet shed their signal sequence. When translation restarted, these short chains didn't lose the sequence—evidence that the processing enzyme that removes the signal is part of the ER membrane. In another key experiment, Blobel and Dobberstein let rough microsomes—which carry ribosomes and some ...
The signal hypothesis proposes that the N-terminus of a secreted protein has a signal sequence whose presence marks it for membrane insertion. Once the protein chain is well inserted into the membrane, the signal sequence is cleaved off by a protease within the membrane and the protein can then enter or even pass through the membrane.
version of the signal hypothesis is proposed 2,3 Signal sequences from pancreatic secretory proteins are shown to be hydrophobic and to resemble the signal sequence of the immunoglobulin light ...
c. Translation (and translocation) are completed, translocon closes. (step 6) e. Signal Peptidase (usually) cuts off signal peptide at arrow. (step 7) Protein is released into lumen of ER. Ribosome and mRNA are released. Now try problem 3-4, especially part D. (If some of the parts are not obvious, wait until later.) B.
The signal hypothesis, formulated by Günter Blobel and David Sabatini in 1971, and elaborated by Blobel and his colleagues between 1975 and 1980, fundamentally expanded our view of cells by introducing the concept of topogenic signals. Cells were no longer just morphological entities with compartmentalized biochemical functions; they were now ...
The signal hypothesis was found to be applicable to protein translocation in all eukaryote and prokaryotes and the concept of signal-mediated targeting of proteins was expanded into a generalised hypothesis of protein topogenesis by Gunter Blobel in 1980. Abstract The signal hypothesis, which describes how secretory and membrane proteins are targeted to the endoplasmic reticulum, was proposed ...
The signal hypothesis, formulated by Günter Blobel and David Sabatini in 1971, and elaborated by Blobel and his colleagues between 1975 and 1980, fundamentally expanded our view of cells by ...
Signal hypothesis was proposed in the early 1971 by Gunter Blobel and David Sabatini. Their work received Nobel prize in Physiology or Medicine in 1999. Signal hypothesis describes how secretory and membrane proteins are targeted to E.R . Although the process of protein synthesis takes place in the cytosol through the interaction of ribosomes ...
The signal hypothesis was originally proposed by Günter Blobel and David Sabatini in 1971, and demonstrated by Blobel and colleagues in 1975. The signal hypothesis showed that cytoplasmically synthesised proteins targeted to the ER use a signal sequence to direct them to the ER membrane. A signal sequence is a short peptide that is part of the ...
Signalling theory. By stotting (also called pronking), a springbok ( Antidorcas marsupialis) signals honestly to predators that it is young, fit, and not worth chasing. Within evolutionary biology, signalling theory is a body of theoretical work examining communication between individuals, both within species and across species.
As well as his proposal of the signal hypothesis, it is Blobel's imaginative approach towards the in vitro reconstitution of protein transport that has revolutionized the field of cell biology ...
signal hypothesis. Definition: Search for: Biology Glossary search by EverythingBio.com. The major mechanism whereby proteins that insert into or cross a membrane are synthesized by a membrane-bound ribosome. The first thirteen to thirty-six amino acids synthesized, termed a signal peptide, are recognized by a signal recognition particle that ...
signal hypothesis Source: A Dictionary of Biology Author(s): Robert HineRobert Hine. A hypothesis to explain how ribosomes become attached to membranes within cells in order to deliver the appropriate proteins to cell organelles, such as mitochondria and chloroplasts, or transport proteins outside the cell membrane.
Abstract. For nascent secretory and membrane proteins to be inserted into the rough endoplasmic reticulum they must first be singled out from the proteins which are destined to remain in the cytoplasm. Moreover, they must contact the rough endoplasmic reticulum. The latest results elucidate these first steps, pointing out the role of specific ...
Synthesis of Membrane Proteins and the "Signal Hypothesis": Principally as a result of the work of G. Blobel, D. D. Sabatini, C. M. Redman, C. Milstein, J. E. Rothman, J. Lenard, and H. F. Lodish, the mechanism that routes newly synthesized proteins to their proper destinations in the cell has gradually unfolded.
Handicap principle. The handicap principle is a disputed hypothesis proposed by the Israeli biologist Amotz Zahavi in 1975. It is meant to explain how "signal selection" during mate choice may lead to "honest" or reliable signalling between male and female animals which have an obvious motivation to bluff or deceive each other.
At the initiation of the signal, a single ligand binds to a single receptor. However, activation of a receptor-linked enzyme can activate many copies of a component of the signaling cascade, which amplifies the signal. Figure 7.4.1 7.4. 1: Example of a signal transduction cascade. In this example, insulin serves as the ligand and activates a ...