Acknowledgement for Assignment
An acknowledgement is an important part of every assignment or project. A project or an assignment is completed by an individual or group with the help and guidance of their teachers, mentors, friends, group members, etc. It is a good practice for the individual or the group to acknowledge others’ contributions towards the completion of the assignment. Thus, while submitting a project, a student or a group should attach an acknowledgement page, wherein a short write-up thanking all the people who played an instrumental role in helping the person complete the project is acknowledged. In this article, we shall guide you in writing an acknowledgement for assignment.


Acknowledgement for Assignment: Guidelines and Tips
- Writing an acknowledgement for an assignment is simply thanking everyone who helped you in completing the assignment.
- You can include teachers, schools, friends family, peers etc. in your acknowledgement.
- If the acknowledgement is for a school project it should be kept simple. So just write a couple of sentences to make a short and sweet acknowledgement section.
- If the acknowledgement is for a college project, you can just thank your teacher, lab assistant and your school/college.
- You may add anyone in the acknowledgement for a school or college project, whom you think has helped you with the project.
- You can explain briefly how their support and guidance have helped you complete the assignment.
- Make sure that the acknowledgement is formal and professional.
Format of Acknowledgement for Assignment
For acknowledgement of any assignment a student can write in the following manner:
Place(name of the city)
Date (dd/mm/yyyy)
Body of the letter
Name of the student
Sample Acknowledgement for Assignment 1 – Individual Assignment
Andheri, Mumbai
In making this project, and completing it successfully, I had to get help and guidance from some respected people. I am grateful that I was given the opportunity to make this project, which has enhanced my knowledge in so many aspects.
I would like to show my gratitude to my course instructor, Mr Bhuvan Sharma and our principal Mrs Dubey for giving me great guidance for this assignment. I wish to extend my special thanks to my friends and family who have always been supportive and guided me throughout this assignment.
Kiran Khatri
Sample Acknowledgement for Assignment 2 – Group Assignment/Project
We as a group would like to thank Mr Srikant Jain, our economics mentor and our principal Dr Ratnakar, of BM College for their help and guidance in completing our project on the topic ‘stock market’. You have always taught us and guided us to understand things that we should know while studying the topic and also in producing good project work.
I would like to take this opportunity to express my heartfelt gratitude to all my group members Sakshi Jain, Kratika Shah, Aarti Sharma and Vartika Gupta. Without their cooperation and understanding of the subject matter, we would not have been able to complete this project.
Kanika Gaur
Explore More Sample Letters
- Leave Letter
- Letter to Uncle Thanking him for Birthday Gift
- Joining Letter After Leave
- Invitation Letter for Chief Guest
- Letter to Editor Format
- Consent Letter
- Complaint Letter Format
- Authorization Letter
- Application for Bank Statement
- Apology Letter Format
- Paternity Leave Application
- Salary Increment Letter
- Permission Letter Format
- Enquiry Letter
- Cheque Book Request Letter
- Application For Character Certificate
- Name Change Request Letter Sample
- Internship Request Letter
- Application For Migration Certificate
- NOC Application Format
- Application For ATM Card
- DD Cancellation Letter
Sample Acknowledgement for Assignment 3 – University Assignment/Project
5 August 2022
I would like to show my special gratitude to Mrs Karishma Khanna, of the biology department, and Mr Darshan Singh, dean of Goodword University for the immense amount of cooperation and contribution they have provided in the completion of my project titled ‘human’s nervous system’.
I would also like to acknowledge Miss Kavya for the time and efforts she provided throughout the semester. Her useful guidance and suggestions were really helpful to me. Last but not least I would like to thank my friends who always provided feedback about the project at all levels.
I would like to acknowledge that this project was completed entirely by me and not someone else.
Sample Acknowledgement for Assignment 4 – PhD. Thesis
2 August 2022
I would like to express my gratitude to my guide, Mr Suresh Prabhakar who gave me the great opportunity to conduct this wonderful thesis on the topic, Coding for Kids which helped me gain a vast extent of knowledge.
I would also like to thank my colleague, Ms Pooja Sharma who made an immense contribution to the completion of this thesis and helped me a lot in finishing this thesis within the limited frame time.
Niharika Verma
FAQs about Acknowledgement for Assignment
Question 1. What is an acknowledgement in a project/assignment paper?
Answer. An acknowledgement section of a project or assignment is to thank all the people/institutions who have helped and contributed during the project and in the completion of the project. They can be teachers, mentors, principals, fellow students, guides, parents etc.
Question 2. How long should an acknowledgement for an assignment be?
Answer. An acknowledgement for an assignment should be professional and formal. Along with that, the length of acknowledgement can be dependent on the type of assignment. Usually, the acknowledgement can be anywhere between 250-400 words but can be extended to 500-1000 words for a more complex assignment.
Question 3. How do you start an acknowledgement for an assignment?
Answer. Some common phrases one can use in the acknowledgement section of the assignment are:
- I wish to show my appreciation
- I would like to thank
- I would like to show my heartfelt gratitude
- I would like to thank the following people for providing me with their guidance throughout the assignment
- The assistance provided by Mr Z was greatly appreciated
Question 4. What should be kept in mind while writing an acknowledgement of an assignment?
Answer. One of the important points to keep in mind while writing an acknowledgement is to mention the title of the project or assignment and also mention the names of the mentors/teachers and principal/HOD.
Customize your course in 30 seconds
Which class are you in.

Letter Writing
- Letter to School Principal from Parent
- ATM Card Missing Letter Format
- Application for Quarter Allotment
- Change of Address Letter to Bank
- Name Change Letter to Bank
- Application for School Teacher Job
- Parents Teacher Meeting Format
- Application to Branch Manager
- Request Letter for School Admission
- No Due Certificate From Bank
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Download the App

The Acknowledgements Section
How to write the acknowledgements for your thesis or dissertation
By: Derek Jansen (MBA) | Reviewers: Dr Eunice Rautenbach | January 2024
Writing the acknowledgements section of your thesis might seem straightforward, but it’s more than just a list of names . In this post, we’ll unpack everything you need to know to write up a rock-solid acknowledgements section for your dissertation or thesis.
Overview: The Acknowledgements
- What (exactly) is the acknowledgements section?
Who should you acknowledge?
- How to write the section
- Practical example
- Free acknowledgements template
- Key takeaways
What is the acknowledgements section?
The acknowledgements section of your thesis or dissertation is where you give thanks to the people who contributed to your project’s success. Generally speaking, this is a relatively brief, less formal section.
With the acknowledgements section, you have the opportunity to show appreciation for the guidance, support, and resources provided by others during your research journey. We’ll unpack the exact contents, order and structure of this section in this post.
Need a helping hand?
Although this is a less “academic” section, acknowledging the right people in the correct order is still important. Typically, you’ll start with the most formal (academic) support received, before moving on to other types of support.
Here’s a suggested order that you can follow when writing up your acknowledgements:
Level 1: Supervisors and academic staff
Start with those who have provided you with academic guidance, including your supervisor, advisors, and other faculty members.
Level 2: Funding bodies or sponsors
If your research was funded, acknowledging these organisations is essential. You don’t need to get into the specifics of the funding, but you should recognise the important role that this made in bringing your project to life.
Level 3: Colleagues and peers
Next you’ll want to mention those who contributed intellectually to your work, including your fellow cohort members and researchers.
Level 4: Family, friends and pets
Last but certainly not least, you should acknowledge your personal (non-academic) support system – those who have provided emotional and moral support. If Fido kept you company during those long nights hunched over the keyboard, you can also thank him here 🙂
As you can see, the order of the acknowledgements goes from the most academic to the least . Importantly, your thesis or dissertation supervisor (sometimes also called an advisor) generally comes first . This is because they are typically the person most involved in shaping your project (or at least, they should be). Plus, they’re oftentimes involved in marking your final work and so a kind word never hurts…
All that said, remember that your acknowledgements section is personal . So, feel free to adjust this order, but do pay close attention to any guidelines or rules provided by your university. If they specify a certain order or set of contents, follow their instructions to the letter.

How to write the acknowledgements section
In terms of style, try to strike a balance between conveying a formal tone and a personal touch . In practical terms, this means that you should use plain, straightforward language (this isn’t the time for heavy academic jargon), but avoid using any slang, nicknames, etc.
As a guide, you’ll typically use some of the following phrases in the acknowledgements section:
I would like to express my appreciation to… for their help with… I’m particularly grateful to… as they provided… I could not have completed this project without… as this allowed me to… Special thanks to… who did… I had the pleasure of working with… who helped me… I’d also like to recognise… who assisted me with…
In terms of positioning, the acknowledgements section is typically in the preliminary matter , most commonly after the abstract and before the table of contents. In terms of length, this section usually spans one to three paragraphs , but there’s no strict word limit (unless your university’s brief states otherwise, of course).
If you’re unsure where to place your acknowledgements or what length to make this section, it’s a good idea to have a look at past dissertations and theses from your university and/or department to get a clearer view of what the norms are.

Practical Example
Alright, let’s look at an example to give you a better idea of what this section looks like in practice.
I would like to express my deepest gratitude to Professor Smith, whose expertise and knowledge were invaluable during this research. My sincere thanks also go to the University Research Fund for their financial support. I am deeply thankful to my colleagues, John and Jane, for their insightful discussions and moral support. Lastly, I must acknowledge my family for their unwavering love and encouragement. Without your support, this project would not have been possible.
As you can see in this example, the section is short and to the point , working from formal support through to personal support.
To simplify the process, we’ve created a free template for the acknowledgements section. If you’re interested, you can download a copy here .

FAQs: Acknowledgements
Can i include some humour in my acknowledgements.
A touch of light humour is okay, but keep it appropriate and professional. Remember that this is still part of an academic document.
Can I acknowledge someone who provided informal or emotional support?
Yes, you can thank anyone who offered emotional support, motivation, or even informal advice that helped you during your studies. This can include friends, family members, or a mentor/coach who provided guidance outside of an academic setting.
Should I mention any challenges or difficulties I faced during my research?
While the acknowledgements section is primarily for expressing gratitude, briefly mentioning significant challenges you overcame can highlight the importance of the support you received. That said, you’ll want to keep the focus on the gratitude aspect and avoid delving too deeply into the challenges themselves.
Can I acknowledge the contribution of participants in my research?
Absolutely. If your research involved participants, especially in fields like social sciences or human studies, acknowledging their contribution is not only courteous but also an ethical practice. It shows respect for their participation and contribution to your research.
How do I acknowledge posthumous gratitude, for someone who passed away during my study period?
Acknowledging a deceased individual who played a significant role in your academic journey can be done respectfully. Mention them in the same way you would a living contributor, perhaps adding a note of remembrance.
For example, “I would like to posthumously acknowledge John McAnders for their invaluable advice and support in the early stages of this research.”.
Is there a limit to the number of people I can acknowledge?
How do i acknowledge a group or organisation.
When thanking a group or organization, mention the entity by name and, if applicable, include specific individuals within the organization who were particularly helpful.
For example, “I extend my thanks to The Speakers Foundation for their support, particularly Mr Joe Wilkins, for their guidance.”
Recap: Key Takeaways
Writing the acknowledgements section of your thesis or dissertation is an opportunity to express gratitude to everyone who helped you along the way.
Remember to:
- Acknowledge those people who significantly contributed to your research journey
- Order your thanks from formal support to personal support
- Maintain a balance between formal and personal tones
- Keep it concise
In a nutshell, use this section to reflect your appreciation in a genuinely and professionally way.

Psst... there’s more!
This post was based on one of our popular Research Bootcamps . If you're working on a research project, you'll definitely want to check this out ...
You Might Also Like:

Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly
- Link to facebook
- Link to linkedin
- Link to twitter
- Link to youtube
- Writing Tips
How to Write Acknowledgements
2-minute read
- 25th March 2015
If you are near the end of your thesis, you can start to think about putting on the finishing touches . One thing you will have to do here is write your acknowledgements. A lot of people worry about this, but there’s really no need. As we are about to show, it’s actually quite simple!
What Are Acknowledgements?
The acknowledgement section of a thesis is where you can thank everyone who has helped you in your research. It is typically located at the beginning of your thesis, right after the contents page, and shouldn’t really be more than one or two pages long. The best thing to do is to keep it concise.
Who Should I Thank?
Think about the people who were of crucial importance during your research. This could include friends, family or professors, or even volunteers who have taken part in your research. It’s also important to acknowledge professional bodies who have given you funding or other help.
How Should I Write It?
There are no strict requirements for the tone of your acknowledgements; it’s essentially the one section where you can be a little bit more informal! Try to make sure the tone fits the person or organization you’re thanking, though, maintaining a formal approach when addressing funding bodies or other official groups.
Find this useful?
Subscribe to our newsletter and get writing tips from our editors straight to your inbox.
Other than trying to be concise, the main thing you should keep in mind is varying your language, if only because starting every sentence with “I would like to thank…” will sound a bit dull.
There are several ways you could word your thanks. A few suggestions are:
- This research would not have been possible without…
- My sincere thanks go to…
- I am grateful to…
- Heartfelt thanks to…
- I would like to express my gratitude to…
- Appreciation is due to…
- I acknowledge the contribution of…
- I am indebted to…
And that’s all there is to it! Relax and enjoy writing your acknowledgements. If you’ve got this far in the thesis-writing process, you’re over the toughest bit now! For more information about writing a dissertation or thesis, read our full dissertation writing guide .
Share this article:
Post A New Comment
Got content that needs a quick turnaround? Let us polish your work. Explore our editorial business services.
3-minute read
How to Insert a Text Box in a Google Doc
Google Docs is a powerful collaborative tool, and mastering its features can significantly enhance your...
How to Cite the CDC in APA
If you’re writing about health issues, you might need to reference the Centers for Disease...
5-minute read
Six Product Description Generator Tools for Your Product Copy
Introduction If you’re involved with ecommerce, you’re likely familiar with the often painstaking process of...
What Is a Content Editor?
Are you interested in learning more about the role of a content editor and the...
4-minute read
The Benefits of Using an Online Proofreading Service
Proofreading is important to ensure your writing is clear and concise for your readers. Whether...
6 Online AI Presentation Maker Tools
Creating presentations can be time-consuming and frustrating. Trying to construct a visually appealing and informative...

Make sure your writing is the best it can be with our expert English proofreading and editing.
Acknowledgement Examples
Examples of Acknowledgement for Project, Thesis and Assignments
Acknowledgement Samples
Acknowledgement is a vital part of our lives. It’s how we show appreciation for the things people do for us, and let them know that we noticed and appreciated their efforts.
But sometimes, it can be hard to know how to properly acknowledge someone. You may not know what words to use, or you may not be sure how to act.
In this blog post, I’ll be discussing six different ways to acknowledge someone. I’ll provide examples of each one, and I’ll also explain when and how to use them. So if you’re looking for some guidance on how to write acknowledgement samples, then this blog post is for you!
Writing Tips For Acknowledgement Samples
When it comes to writing an acknowledgement, it is important to be clear, concise and to the point. The following are some tips that will help you write acknowledgement samples that will properly thank those who have helped you along the way:
- Make sure to list everyone who has helped you. This includes anyone who has given you a hand, whether it be big or small.
- Thank those who have helped you in a timely manner. The sooner you send out your acknowledgements, the sooner those who have helped you will know that their assistance was appreciated.
- Keep your acknowledgements short and sweet. Nobody wants to read a long, drawn-out paragraph thanking everyone who has helped you.
- Be sure to personalize your acknowledgements. Thanking someone for their help is much more meaningful when you take the time to mention how their assistance specifically made a difference.
- Proofread your acknowledgements before sending them out. This is important, as it ensures that your acknowledgements are free of errors and look professional.
Other Acknowledgement Article:
- Acknowledgement for Science Project
- Acknowledgement For Commerce Project
- Acknowledgement For Biology Project
Overall, acknowledgement samples serve as a useful tool for expressing gratitude and recognizing the contributions of others. Whether it’s for a research paper, thesis, or project, these samples provide a clear and concise way to acknowledge the support and guidance of those who have helped along the way. Whether it’s a mentor, colleague, or family member, an acknowledgement is a simple yet powerful way to show appreciation and acknowledge the role others have played in your success.
Leave a Comment Cancel reply
Save my name, email, and website in this browser for the next time I comment.
- Letter Writing
- Formal Letter Writing In English
- Acknowledgement For College Project
Acknowledgement for College Project | How to Write and Samples
It is always a great relief when you are done with something you have been working on for a long time. It is the same when complete your final project. Unlike other assignments, a project is successfully completed with the mental and moral guidance of your mentors, principal, family, friends, etc. While submitting a project, you are supposed to acknowledge all those who helped you directly or indirectly in the completion of your project. Learn how to write an acknowledgement for a college project in this article.
Table of Contents
How to write an acknowledgement for a college project, sample 2 -acknowledgement for group project, acknowledgement sample for university project, frequently asked questions on acknowledgement for college project.
appreciating and acknowledging the help you receive is a good practice. All through your school and college life, you will have to work on multiple assignments/projects. Even though you are the one working on the project, you will have direct and indirect help and support from people around you.
When writing an acknowledgement, make sure you use the very term ‘acknowledgement’ as the title. Use simple and professional language. See to it that you mention all those who have been involved in the process. Structure the paragraphs properly so that it is coherent.
Samples of Acknowledgement for Project and Assignment
An acknowledgement for a college project is similar to an acknowledgement for assignment . Go through the samples given below to have a clear idea of how they are written.
Acknowledgement Sample for College Project
Sample 1 – acknowledgement for individual project.
I would like to convey my heartfelt gratitude to Mrs. Debjani Nanda for her tremendous support and assistance in the completion of my project. I would also like to thank our Principal, Mrs.Jyothi Kumar, for providing me with this wonderful opportunity to work on a project with the topic Food Culture during the Pandemic. The completion of the project would not have been possible without their help and insights.
Anisha Sinha
We would like to thank Mr. Satish Kaipa, our Professor-in-charge and our Principal, Dr.Srinivasan for their support and guidance in completing our project on the topic (topic name). It was a great learning experience.
I would like to take this opportunity to express my gratitude to all of my group members Mitra Jain, Suresh Chandra, Mahesh Shah, Kritika Mittal, and V S Athira. The project would not have been successful without their cooperation and inputs.
A student of a university will have to write the acknowledgement for their project or research paper stating that the submission is done and is not copied. The acknowledgement sample for the university project is mostly attached after the dedication page thanking all the faculty members of the department, HOD, the Dean and the mentor.
I would like to express my profound gratitude to Mr./Mrs.____ (name of the HOD), of ____ (designation and department name) department, and Mr./Mrs. _____ (Dean) of _____ university for their contributions to the completion of my project titled _____.
I would like to express my special thanks to our mentor Mr./Mrs. ______ for his/her time and efforts he/she provided throughout the year. Your useful advice and suggestions were really helpful to me during the project’s completion. In this aspect, I am eternally grateful to you.
I would like to acknowledge that this project was completed entirely by me and not by someone else.
What is an acknowledgement in a project/research paper?
The acknowledgment section of a project or research is written to show gratitude to the persons or institutions who significantly contributed during the project/research and helped you complete your project.
Who should be mentioned in an acknowledgement for a college project report?
While writing an acknowledgement for a college project report, a student can mention the names and designation of everyone who has contributed in his/her learning process. You can thank your teachers, principal, guides, peers, and parents.
How can we write an acknowledgement for a group project?
A group project can be submitted as one whole project or individually by every member. Every child should keep in mind that while writing the acknowledgement for college, either of himself/herself or on behalf of the entire group, should mention the names of all group members.
Leave a Comment Cancel reply
Your Mobile number and Email id will not be published. Required fields are marked *
Request OTP on Voice Call
Post My Comment
- Share Share
Register with BYJU'S & Download Free PDFs
Register with byju's & watch live videos.

Acknowledgement in Research Paper – A Quick Guide [5 Examples]
The acknowledgement section in your research paper is where you thank those who have helped or supported you throughout your research and writing. It is a short section of 3-5 paragraphs or no more than 300 words you put on a page after the title page.
In this post, we are going to provide you with five examples of acknowlegdement section and a handful of best practices you can make your work look professional.

Saying thank you with style
How to write an acknowledgement: the complete guide for students, why should i include an acknowledgement in my research paper.
Acknowledging assistance and contributions from others can establish your integrity as a researcher. This will eventually make your work more credible.
What should be acknowledged about (aka thankful for)?
In your acknowledgement, you can show gratitude for those who provide you with resources in the following area:
- Technical help may include people who helped you by providing materials and supplies.
- Intellectual help includes academic advice and assistance.
- Mental help can be any kind of verbal support and encouragement.
- Financial support that is obviously related to monetary support
Who should be included in the acknowledgement of a research paper?
You can include everyone who helped you technically, intellectually, or financially (assistance with grants or monetary help) in the process of researching and writing your research paper. Except for your family and friends, you should always include the full names with the title of these individuals:
- Your profession, supervisor, or teacher
- Academic staff (e.g. lab assistant) of your school/college
- Your department, faculty, college, or school
- Classmates, teammates, co-workers, or colleague
- Friends and family members
You can start with your professor or the individuals who supported you the most throughout the research. And then you can continue by thanking your institution and then the reviewer who reviewed your paper. Then you can thank your friends and families and any other individual who helped.
What is the tone of the acknowledgement in a research paper?
You should write your acknowledgement in formal language with complete sentences. It is appropriate to write in the first person (‘I’ for a single author or ‘we’ for two or more).
Note that personal pronouns such as ‘I, my, me …’ are nearly always used in the acknowledgements only. For the rest of the research paper, such personal pronouns are generally avoided.
Writing an acknowledgement for research paper is one of the important parts of your project report. You need to thank everyone for helping you with your paper . Here are some examples of acknowledgement for your research paper.
Acknowledgement in Research Paper: Example 1
Acknowledgement in research paper: example 2, acknowledgement in research paper: example 3, acknowledgement in research paper: example 4, acknowledgement in research paper: example 5.
You can use these or try to create your own version for your project report. Also, you can use our auto acknowledgement generator tool to automatically generate acknowledgement for your project.
Where should I put the acknowledgement section?
The acknowledgements section should appear between your title page and your introduction in your research paper.
How long is an acknowledgement in a research paper?
The acknowledgement section (usualy inserted as a page) of your research paper should consist of 3-5 paragraphs or no more than 300 words you put on a page after the title page.
Should I use the full names of family members in an acknowledgement?
You do not necessarily need to use the full name for your family and friends (it would sound pretty awkward to use the full name of your parent or spouse right?), you should always include the full names with the title for all other individuals in your acknowledgement.
Can I use “first person” in an acknowledgement?
Yes. It is appropriate to write in the first person (‘I’ for a single author or ‘we’ for two or more).
What is an acknowledgement in academic writing?
An acknowledgement is a page is where you show appreciation to people who helped or supported you intellectually, mentally, or financially in your academic writing.
It should be no longer than one page.

More Definitions on Acknowledgement
- Acknowledgement vs Empathy What is the Difference Between Acknowledgement and Empathy?
- Acknowledgement vs Acceptance Does Acknowledgement Mean Acceptance? Lessons From History and the Bible
- Acknowledgement vs Agreement Is Acknowledgement the Same as Agreement?
- Acknowledgement Receipts vs Official Receipts What’s the Difference Between Acknowledgement Receipts and Official Receipts?
“Acknowledgement” vs “Acknowledgment”… …what the hack?

Both “acknowledgement” and “acknowledgment” are used in the English-speaking world. However, acknowledgement with the “e” in the middle is more commonly used. It is up to 24.5 times more popular in the top 5 English-speaking countries in the world.
Other Popular Acknowledgement Examples
For work or business Acknowledgement Receipt of Payment [4 Examples] Acknowledging Receipt of Documents: A Quick Guide with Examples Acknowledgement for Presentation [9 Examples] Acknowledgement for Job Offer [3 Examples] Acknowledgement for Business Plan [4 Examples] Acknowledgement for Work Immersion [5 Examples] Acknowledgement of Receipt of Appraisal [3 Examples] Acknowledegment of Debt [5 Examples] Resignation Acknowledgement for Employers [5 Examples]
Academic Acknowledgement for Research Paper [5 Examples] Acknowledgement for Internship Report [5 Examples] Acknowledgement for Thesis and Dissertation [15 Examples] Acknowledgement for Portfolio [5 Examples] Acknowledgement for Case Study [4 Examples] Acknowledgement for Academic Research Paper [5 Examples] Acknowledgement for College/School Assignment [5 Examples] Acknowledgemet to God in Reports [5 Examples]
Others Acknowledgement to Funeral Attendees [5 Examples] Funeral Acknowledgement Templates (for Newspapers and Websites) Common Website Disclaimers to Protect Your Online Business Notary Acknowledgement [5 Examples]
Acknowledgement Examples for School/College Projects
Most popular Acknowledgement For School/College Projects [7 Examples] Acknowledgement for English Project [5 Examples] Acknowledgement for Project Class 11 and 12 Acknowledgement for Project of Class 8, 9 and 10 By subjects Acknowledgement for Accounting Project [3 Examples] Acknowledgement for Business Studies Project [5 Examples] Acknowledgement for Chemistry Project [5 Examples] Acknowledgement for Computer Project [5 Examples] Acknowledgement for Economics Project [5 Examples] Acknowledgement for English Project [5 Examples] Acknowledgement for Geography Project [5 Examples] Acknowledgement for History Project [5 Examples] Acknowledgement for Maths Project for Students [5 Examples] Acknowledgement for Physics Project [5 Examples] Acknowledgement for Social Science Project [5 Examples] Others Acknowledgement for Group Project [5 Examples] Acknowledgement for Graduation Project [5 Examples] Acknowledgement for Disaster Management Project [3 Examples] Acknowledgement for Yoga Project [3 Samples]
How-to Guides on Academic Writing and Others
Most popular How to Write an Acknowledgement: The Complete Guide for Students How to Write an Acknowledgement for College Project? How to Write a Dedication Page for a Thesis or Dissertation? More on acknowledgements How to Write Acknowledgment for a Dissertation or a Thesis? Is Acknowledgement and Dedication the Same? Thesis or Dissertation How to Write a Master’s Thesis: The Ultimate Guide How to Write a Thesis Proposal? How to Write an Abstract for a Thesis? How to Write a Preface for a Thesis? Others How to Write an Introduction for a Research Paper? 7 Real Research Paper Examples to Get You Started How to Write Cover Letter for an Internship Program? How to Write an Internship Acceptance Letter? How to Write a Leave Application? For Schools and the Workplace How to Write a Resignation Letter?
Introduction to Academic Writing
By O.P. Jindal Global University Duration: 16-hour Cost: FREE Gain an in-depth understanding of reading and writing as essential skills to conduct robust and critical research for your writing.
Writing in English at University
By Lund University Duration: 24-hour Cost: FREE Learn how to structure your text and arguments, quote sources, and incorporate editing and proofreading in your academic writing.
Academic English: Writing Specialization
By the University of California, Irvine Duration: 6 months Cost: Free 7-day trial, USD39 per month The skills taught in this Specialization will empower you to succeed in any college-level course or professional field. You’ll learn to conduct rigorous academic research and to express your ideas clearly in an academic format. Share your Course Certificates in your LinkedIn profile, on printed resumes, CVs, or other documents.

Thank you, your samples really helped me.
This is great! Your samples really helped me in my research. Thank you and more power!
I’m so grateful that you make this kind of blog, I really need this for my research. Thank you so much… God bless you.
Thank you so much for the samples you have provided, it has helped me a lot.
Leave a Comment Cancel Reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.

- Acknowledgements for PhD Thesis and Dissertations – Explained
- Doing a PhD
The Purpose of Acknowledgements
The acknowledgement section of a thesis or dissertation is where you recognise and thank those who supported you during your PhD. This can be but is not limited to individuals, institutions or organisations.
Although your acknowledgements will not be used to evaluate your work, it is still an important section of your thesis. This is because it can have a positive (or negative for that matter) influence the perception of your reader before they even reach the main body of your work.
Who Should I Acknowledge?
Acknowledgements for a PhD thesis will typically fall into one of two categories – professional or personal.
Within these categories, who you thank will ultimately be your decision. However, it’s imperative that you pay special attention to the ‘professional’ group. This is because not thanking someone who has played an important role in your studies, whether it be intentional or accidental, will more often than not be seen as a dismissal of their efforts. Not only would this be unfair if they genuinely helped you, but from a certain political aspect, it could also jeopardise any opportunities for future collaborations .
Professional Acknowledgements
This may include, but is not limited to:
- Funding bodies/sponsorship providers
- Supervisors
- Research group and lab assistants
- Research participants
- Proofreaders
Personal Acknowledgements
- Key family members and friends
- Individuals who inspired you or directly influenced your academic journey
- Anyone else who has provided personal support that you would like to mention
It should be noted that certain universities have policies which state only those who have directly supported your work, such as supervisors and professors, should be included in your acknowledgements. Therefore, we strongly recommend that you read your university guidelines before writing this section of your thesis.
How to Write Acknowledgements for PhD Thesis
When producing this section, your writing style can be more informal compared to the rest of your thesis. This includes writing in first person and using more emotive language. Although in most cases you will have complete freedom in how you write this section of your thesis, it is still highly advisable to keep it professional. As mentioned earlier, this is largely because it will be one of the first things your assessors will read, and so it will help set the tone for the rest of your work.
In terms of its structure, acknowledgements are expected to be ordered in a manner that first recognises the most formal support before moving onto the less formal support. In most cases, this follows the same order that we have outlined in the ‘Who Should I Thank’ section.
When thanking professionals, always write out their full name and provide their title. This is because although you may be on a first-name basis with them, those who read your thesis will not. By providing full names and titles, not only do you help ensure clarity, but it could also indirectly contribute to the credibility of your thesis should the individual you’re thanking be well known within your field.
If you intend to include a list of people from one institution or organisation, it is best to list their names in alphabetical order. The exception to this is when a particular individual has been of significant assistance; here, it would be advisable to list them.
How Long Should My Acknowledgements Be?
Acknowledgements vary considerably in length. Some are a single paragraph whilst some continue for up to three pages. The length of your acknowledgement page will mostly depend on the number of individuals you want to recognise.
As a general rule, try to keep your acknowledgements section to a single page. Although there are no word limits, creating a lengthy acknowledgements section dilutes the gratitude you’re trying to express, especially to those who have supported you the most.
Where Should My Acknowledgements Go?
In the vast majority of cases, your acknowledgements should appear directly after your abstract and before your table of contents.
However, we highly advise you to check your university guidelines as a few universities set out their own specific order which they will expect you to follow.
Phrases to Help You Get Started

We appreciate how difficult it can be to truly show how grateful you are to those who have supported you over the years, especially in words.
To help you get started, we’ve provided you with a few examples of sentences that you can complete or draw ideas from.
- I am deeply grateful to XXX…
- I would like to express my sincere gratitude to XXX…
- I would like to offer my special thanks to XXX…
- I would like to extend my sincere thanks to XXX…
- …for their assistance at every stage of the research project.
- …for their insightful comments and suggestions.
- …for their contribution to XXX.
- …for their unwavering support and belief in me.
Thesis Acknowledgement Examples
Below are three PhD thesis acknowledgment samples from which you can draw inspiration. It should be noted that the following have been extracted from theses which are freely available in the public domain. Irrespective of this, references to any individual, department or university have been removed for the sake of privacy.
First and foremost I am extremely grateful to my supervisors, Prof. XXX and Dr. XXX for their invaluable advice, continuous support, and patience during my PhD study. Their immense knowledge and plentiful experience have encouraged me in all the time of my academic research and daily life. I would also like to thank Dr. XXX and Dr. XXX for their technical support on my study. I would like to thank all the members in the XXX. It is their kind help and support that have made my study and life in the UK a wonderful time. Finally, I would like to express my gratitude to my parents, my wife and my children. Without their tremendous understanding and encouragement in the past few years, it would be impossible for me to complete my study.
I would like to thank my supervisors Dr. XXX and Dr. XXX for all their help and advice with this PhD. I would also like to thank my sisters, whom without this would have not been possible. I also appreciate all the support I received from the rest of my family. Lastly, I would like to thank the XXX for the studentship that allowed me to conduct this thesis.
I would like to thank my esteemed supervisor – Dr. XXX for his invaluable supervision, support and tutelage during the course of my PhD degree. My gratitude extends to the Faculty of XXX for the funding opportunity to undertake my studies at the Department of XXX, University of XXX. Additionally, I would like to express gratitude to Dr. XXX for her treasured support which was really influential in shaping my experiment methods and critiquing my results. I also thank Dr. XXX, Dr. XXX, Dr. XXX for their mentorship. I would like to thank my friends, lab mates, colleagues and research team – XXX, XXX, XXX, XXX for a cherished time spent together in the lab, and in social settings. My appreciation also goes out to my family and friends for their encouragement and support all through my studies.
Browse PhDs Now
Join thousands of students.
Join thousands of other students and stay up to date with the latest PhD programmes, funding opportunities and advice.

How To Write Acknowledgement for Project [Sample and Best Practices]
An acknowledgement is a statement of gratitude or appreciation for support, effort, or inspiration from others towards a specific task or endeavor.
In simple words, an acknowledgement expresses thankfulness and recognizes the positive contribution of people who have helped in carrying out a project or activity.
Acknowledgement in project is a way for the author to express gratitude to individuals or organizations that have provided support, guidance, or resources for the project. This section is commonly found at the beginning of a research paper, thesis, project, assignment or a book.
It is a thoughtful gesture to recognize the contributions of professors, colleagues, research assistants, funding agencies, and anyone else who has helped in the development and completion of the project work. Acknowledgement can also extend to friends and family members who have provided emotional support during the research process.
Acknowledgement Synonyms
Appreciation, recognition, gratitude, thanks, indebtedness, beholdenness, credit, shout-out, homage, praise, commendation, acclaim, favor, regard, compliments
What is the Purpose and Importance of Acknowledgement
Acknowledgement serves a crucial purpose in various settings such as in school, college, or within a project. It is a way to express gratitude and appreciation towards individuals or organizations who have contributed to the success of a particular endeavor.
Acknowledgement is important because it serves as a form of recognition and appreciation for the contributions of others. In a school or college project, acknowledging the sources and individuals who have helped in the research or development of the project shows integrity and respect for the work of others. It also helps to build a sense of community and collaboration.
Sample Acknowledgement for Project
Here is an Acknowledgement example for School Project File:
First and foremost, I would like to express my deepest gratitude to my class teacher, [Teacher’s Name], for invaluable guidance, feedback, and constant encouragement throughout the duration of this project. Her/his passion for physics was contagious and her/his insights greatly facilitated the progress of my work.
I am extremely grateful to the school principal, [Principal’s Name], for granting me access to the physics lab and permitting me to use the necessary equipment and resources. Having access to quality infrastructure was vital for the experimental aspect of my project.
In addition, I would like to acknowledge my classmates [You can mention names but not necessary] for their camaraderie and assistance during the project. Their help, advice and company made this journey enjoyable.
Finally, I must thank my parents for their unwavering support and encouragement needed to take on this challenging but enriching project. Their practical assistance at every step was invaluable.
I could not have completed this project successfully without everyone who has contributed. I will forever remain grateful to them.

Also Check: Acknowledgement for Project All Subjects Sample with PDF
How to Write Acknowledgement for Project
The acknowledgement section in a project provides an opportunity to recognize and thank everyone who contributed to making the project possible.
- Start by listing the names of those who provided significant assistance, guidance or support for the project.
- Mention specifically how they helped – for example, by providing expertise, tools, data access, funding, mentorship, facilities, or other forms of support.
- The people and groups who contributed should be properly credited for their specific roles.
- Phrase the acknowledgement statements clearly and concisely.
- Keep them simple, sincere and specific without overstating contributions.
- Make sure to get prior consent from everyone being acknowledged.
- Review the list thoroughly to ensure no significant contributors are missed.
- A thoughtfully crafted acknowledgement section is important for properly recognizing those whose efforts were crucial in enabling the project to be undertaken and completed successfully.
Key Elements of a Well-Crafted Acknowledgement
- Specifically mention each person or organization and their contribution
- Be clear, concise and sincere
- Obtain prior consent from everyone being acknowledged
- Use proper titles and spell names correctly
- Place in proper position (after table of contents and before body of work)
- Thank financial contributors, advisors, institutional support
- Credit sources of any permissions granted
- Mention groups, labs, institutions that provided resources/support
- Note grants/funding that made the work possible
- Do not exaggerate contributions or imply endorsement
Tone and Style in Acknowledgement
- Be gracious, humble and appreciative in tone
- Use sincerity – avoid sounding artificial or over-the-top
- Maintain professional yet personal warmth
- Keep it simple – avoid flowery overly complex language
- Use clear, concrete language – avoid ambiguity
- Make statements specific yet concise
- Remain brief but do not leave out important contributors
- Check for consistent tone aligned with piece as a whole
- Read aloud to ensure tone conveys genuine gratitude
- Be thoughtful and diplomatic in phrasing
How to write Acknowledgement for School Project
When writing acknowledgement for school project , it is important to recognize those who provided assistance and support specifically within the school context.
Some unique considerations for school project acknowledgement:
- Thank your teacher and advisor for guidance and direction on the project. Recognize their time spent supervising and providing feedback.
- Mention the principal and any other school administrators who enabled the project by approving resources or providing facilities.
- Credit classmates who directly collaborated or provided input on the project.
- Acknowledge school staff like librarians or IT support who may have assisted with research or technical aspects.
- Note access to any special school equipment, labs, or rooms used for the project.
- If relevant, recognize a particular school department/program or curriculum that contributed to the foundations for the project.
Thoughtfully acknowledging school contributors reinforces the value of the guidance, resources and environment the school provided to make the project possible. This shows appreciation for the unique role the school community played in the project journey.
Sample Acknowledgement for School Project
I would first like to thank my science teacher, Mrs. Pathak, for her invaluable advice and guidance throughout this project. Thank you for your unwavering encouragement, feedback on my ideas, and for pushing me to make this project the best it could be.
I would also like to express my gratitude to my school principal, Mr. J.K. Verma, for enabling me to use the school laboratory and providing access to needed equipment and resources. This project would not have been possible without that support.
In addition, I must thank my family for listening to my ideas, calming my worries, and cheering me on through every step. Also, thanks to my classmates who offered me advice, collaborated on experiments, and commiserated through the challenges. I appreciate your insights and camaraderie.
How to write Acknowledgement for College Project
When writing acknowledgement for a college project, it is important to keep in mind you are addressing an academic audience. The tone should remain formal rather than too casual or conversational. Supporters and contributors should be acknowledged professionally using appropriate titles.
Technical/field-specific language can be used more liberally compared to a high school audience, but avoid overly complex jargon. The acknowledgement still require sincerity, clarity and brevity like any well-crafted acknowledgement, but tailored for a college setting.
Crafting Effective Acknowledgement for College Project
- Thank your professor/advisor first for guidance provided throughout the project. Specifically mention forms of support like providing direction on the methodology, lending equipment, reviewing drafts, etc.
- Recognize assistance from other professors who allowed you access to labs, gave feedback, or provided academic resources.
- Acknowledge any graduate students, teaching assistants or peers who collaborated, offered insights or provided technical help.
- Thank external industry partners or academic institutions that may have provided facilities, data access, funding or other project support.
- Recognize relevant campus departments or university libraries that enabled literature reviews, lab work, computing resources, etc.
- Mention grants, scholarships or programs that provided financial support to undertake the project.
- Use appropriate academic terminology to accurately describe contributions to college-level audiences.
- Maintain a professional tone, but inject some warmth into the acknowledgement.
Sample Acknowledgement for College Project
I would like to express my deepest gratitude to my mechanics professors, Mr. Akash Singh for heis invaluable guidance and support throughout this project. Thank you for pushing me to refine my methodology, providing constructive feedback on multiple drafts, and supplying the equipment I needed from your lab. This project would not have been possible without your dedication as my mentor.
I would also like to thank my branch mate Santosh for all of the long hours we collaborated in the campus together brainstorming ideas and troubleshooting issues side-by-side. Your insights greatly enriched this project.
In addition, I received important assistance from other professors in the mechanical Department and the library staff who allowed me access to essential research materials.
To everyone mentioned here – thank you again for all of your guidance and support throughout my journey with this project. I am tremendously appreciative.
How to write Acknowledgement for University Project
When writing acknowledgement for a university project, it is important to adopt a sophisticated tone suited for an academic audience. Use appropriate professional titles and terminology. While acknowledgement require formality, inject modest warmth and remain personable.
The language should be sophisticated without being overly ornate or verbose. Demonstrate your technical knowledge when describing contributions, but avoid excessive jargon. Articulate how support furthered your academic enrichment within the field.
Balancing Formality and Appreciation
- Thank your university advisor and highlight specific guidance provided throughout your journey with grace and formality.
- Recognize research partners, collaborators and assistants with sincerity while maintaining professional decorum.
- Use appropriate, field-specific language to accurately describe technical, theoretical, conceptual or other forms of support.
- Note any facilities provided by external institutions or industry partners using proper, sophisticated phrasing.
- Acknowledge funding sources formally using their complete official names and grant numbers.
- Adopt a refined but accessible style that displays gratitude without over-familiarity.
- Proofread acknowledgement to ensure tone is consistently formal yet warm.
Sample Acknowledgement for University Project
I would like to extend my deepest gratitude to my research supervisor, Dr. P.K. Sahu, for the invaluable mentorship provided to me over the past two years. Your patience, encouragement, and insightful feedback challenged me to grow and enriched my academic experience immensely. I sincerely thank you for guiding me through both the triumphs and obstacles of this research endeavor.
I must also recognize my fellow lab members Sumit Joshi and Ankita Gandhi for their collaborative spirit and willingness to lend their expertise whenever I encountered difficulties in my methodology and analysis. Thank you all for the community we built together.
In addition, I am grateful to the University Library staff for their assistance in locating numerous critical resources to build the foundations of my literature review.
Furthermore, I wish to acknowledge the GSK Emerging Researchers in Oncology Grant for providing the funding that enabled this ambitious project to come to fruition. I am tremendously appreciative of this opportunity.
To all those recognized here, I extend my wholehearted gratitude. Your contributions strengthened this project and my growth as a budding researcher tremendously.
Analyzing Well-Written Acknowledgements
You are skilled at crafting project acknowledgments. Let’s examine acknowledgments from top universities’ projects to gather inspiration and ideas.
This is the acknowledgement of a B.Tech project report of IIT Delhi.
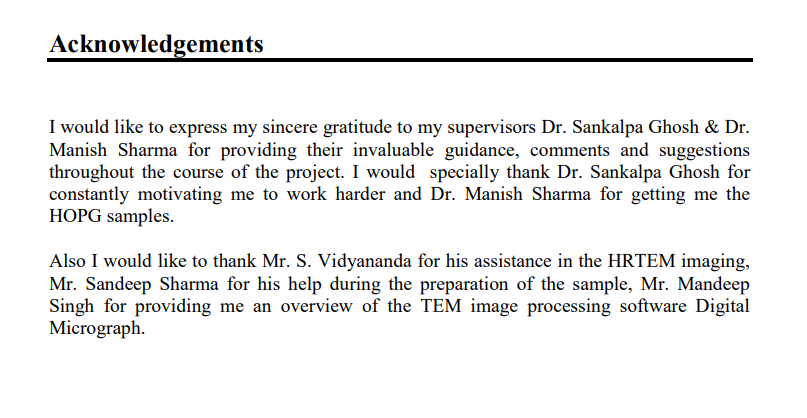
This is the acknowledgement section of an M.Tech project report of IIT Hyderabad.

Lessons Learned from Notable Acknowledgements
- Specifically mention each person and their contribution. Both acknowledgements call out individuals by name and identify how they helped.
- Use titles and honorifics properly. They use “Dr.” and “Mr.” where appropriate to show respect.
- Note different types of support like guidance, mentorship, resources, encouragement, opportunity, advice, etc. They highlight diverse forms of assistance.
- Mention various contributors – supervisors, co-advisors, team members, lab staff, classmates, family. They cover the full range.
- Express gratitude sincerely using words like “profound,” “immense,” “privilege,” “sincerely,” etc. The gratitude comes across as authentic.
- Show appreciation for intangibles too like motivation, enthusiasm and support. They acknowledge not just technical help but also emotional/moral support.
- Maintain an academic tone but also inject warmth and care. The tone balances professionalism with heartfelt appreciation.
- Keep sentences focused. Each acknowledgment statement is concise while still being specific.
- Organize coherently grouping similar types of contributors together. There is logical flow.
- Proofread carefully with proper names, titles and no typos. Attention to detail is evident.
The lessons and examples provided have equipped you with the key skills to write strong acknowledgement. You now have all the knowledge needed to acknowledge everyone who made your work possible.
This will let contributors know their efforts mattered and will stand as a permanent note of appreciation. With grace and care, you can write acknowledgements that convey genuine gratitude to recognizes those who paved the way for your success. You’ve got this!
- Resources Home 🏠
- Try SciSpace Copilot
- Search research papers
- Add Copilot Extension
- Try AI Detector
- Try Paraphraser
- Try Citation Generator
- April Papers
- June Papers
- July Papers

How to Write Acknowledgement in Research Paper

Table of Contents
Writing an acknowledgement in a research paper is an integral part of the process. It is a formal way of expressing gratitude to the individuals and institutions that contributed to the completion of your research.
This section, though not mandatory, holds significant value as it acknowledges the efforts of those who assisted you in the successful completion of your project. In this comprehensive guide, we will delve into the intricacies of writing an effective acknowledgement for your research paper.
Introduction
Acknowledgements serve a crucial role in research papers . They not only express gratitude but also provide a sense of credibility to your work. Acknowledging the contributions of others shows that your research is a collective effort, which can enhance the perceived validity of your findings.
Moreover, acknowledgements can also serve as a platform for you to demonstrate your professional courtesy and respect for the individuals and institutions that have supported your research. This can help in fostering positive relationships, which can be beneficial for your future research endeavors.
Who to acknowledge in your research paper ?
Deciding who to acknowledge in your research paper can be a challenging task. It is important to ensure that you do not overlook anyone who has contributed to your research. Here are some categories of individuals and institutions that you might consider acknowledging:
Academic advisors and supervisors:
Your academic advisors and supervisors are likely to be your first point of contact for guidance and support during your research. They provide valuable insights, feedback, and direction, which can significantly influence the outcome of your research.
Therefore, acknowledging them in your research paper is a way of expressing your gratitude for their assistance and guidance. It also shows your respect for their expertise and dedication to your research.
Research participants and collaborators
Research participants and collaborators play a crucial role in the success of your research. They provide the data or information necessary for your research, making their contribution invaluable.
Acknowledging them in your research paper is a way of showing your appreciation for their time and effort. It also symbolizes your respect for their contribution to your research.
Funding bodies and institutions:
Funding bodies and institutions provide the financial support necessary for conducting your research. Without their support, it might be challenging to carry out your research effectively.
Therefore, acknowledging them in your research paper is a way of expressing your gratitude for their financial support. It also shows your appreciation for their trust in your research capabilities.
How to write acknowledgements for your research paper?
Writing acknowledgements for your research paper involves more than simply listing names. It requires a thoughtful and sincere expression of gratitude. Here are some steps to guide you in writing effective acknowledgements:
Start with the most significant contributions:
Begin your acknowledgements by expressing gratitude to those who have made the most significant contributions to your research. This could be your academic advisors, supervisors, or funding bodies. Starting with the most significant contributions helps to set the tone for the rest of your acknowledgements.
Ensure that you express your gratitude sincerely and professionally. Avoid using overly emotional or informal language as this can undermine the professionalism of your acknowledgements.
Acknowledge other contributors:
After acknowledging the most significant contributors, proceed to acknowledge other individuals and institutions that have supported your research. This could include research participants, collaborators, and other supportive individuals or institutions.
When acknowledging these contributors, be sure to express your gratitude sincerely and professionally. Also, ensure that you acknowledge each contributor individually to show your appreciation for their unique contribution.
Use appropriate language and tone:
The language and tone you use in your acknowledgements can significantly influence how they are perceived. Therefore, it is important to use appropriate language and maintain a professional tone throughout your acknowledgements.
Use formal language and avoid using jargon or colloquial expressions. Also, maintain a consistent tone throughout your acknowledgements to ensure that they are coherent and easy to read.

Examples of acknowledgements in research papers
Here are a few examples that demonstrate how to acknowledge different contributors effectively:
"I would like to express my deepest gratitude to my advisor, Professor ABC, for his invaluable guidance and support throughout this research. His expertise and dedication have been a source of inspiration and motivation."
Research participants and collaborators:
"I am deeply grateful to all the participants who generously shared their time and experiences for this research. Their contributions have been instrumental in the success of this study."
"This research was made possible by the generous funding from ABC Foundation. I am profoundly grateful for their support and trust in my research capabilities."
Writing acknowledgements in a research paper is a thoughtful process that requires careful consideration of who to acknowledge and how to express gratitude. By following the guidelines and examples provided in this article, you can write effective acknowledgements that reflect your appreciation and respect for the contributions of others to your research.
Remember, acknowledgements are more than just a formality. They are an opportunity to express your gratitude and respect for the individuals and institutions that have supported your research journey. So, take the time to write acknowledgements that are sincere, professional, and reflective of your gratitude.
Frequently Asked Questions
In a research paper, the acknowledgment section is where the author shows appreciation to those who helped with the research. It's usually found at the start of the paper, before the main text begins. However, the exact location varies depending on the university guidelines.
Citations are formal ways to acknowledge the sources you used, while acknowledgments are more personal and can serve as a confidential way to thank someone for their help or contribution.
The purpose of including acknowledgement is to express gratitude to everyone who assisted with the research but didn't meet the criteria for being listed as an author.
There is no minimum length for writing an acknowledgement in a research paper but it should not be more than one page.
If you conducted the research entirely by yourself and received no assistance or support from others, it's not necessary to include acknowledgements. However, if you received any form of support or assistance, even if minimal, it's appropriate to acknowledge it.
Yes, it's appropriate to acknowledge funding sources in your research.
You might also like

Introducing SciSpace’s Citation Booster To Increase Research Visibility

AI for Meta-Analysis — A Comprehensive Guide

How To Write An Argumentative Essay
Acknowledgement For Project [5 Examples]
This article aims to emphasize the importance of acknowledgment in project , going beyond mere formality.
In the realm of project completion, expressing gratitude through acknowledgments holds significant importance. Beyond being a courteous gesture, acknowledging those who have contributed to your project serves several essential purposes.
Let’s delve into why it’s crucial and explore effective ways to craft acknowledgements for various project scenarios.It explains why saying thank you matters, from acknowledging efforts and building professional relationships to boosting team morale. Offering practical advice with bold examples, the article guides readers on crafting sincere acknowledgements for different scenarios.
Table of Contents
Why acknowledge contributors?
Recognition of efforts: .
Acknowledging contributors is a way to recognize and appreciate the efforts they’ve invested in your project. This recognition fosters a positive atmosphere and motivates individuals to continue their valuable contributions.
Building Professional Relationships:
Acknowledging collaborators helps in building strong professional relationships. It establishes a sense of reciprocity and mutual respect, laying the foundation for potential future collaborations.
Team Morale Boost:
Publicly acknowledging team members can significantly boost morale. Feeling appreciated enhances job satisfaction by increasing productivity and commitment to future endeavors.
How to Write an Effective Acknowledgement?
Crafting an effective acknowledgment requires thoughtfulness and consideration. Here’s a guide to help you express your gratitude sincerely.
Personalize Your Message:
Tailor your acknowledgement to each contributor. Mention specific contributions, skills, or support they provided. Personalization adds a genuine touch to your gratitude.
Scenario: “Dear Justin Taylor, your unparalleled coding skills brought our project to life. Your dedication to perfection was evident in every line of code, and I am truly grateful for your exceptional contribution.”
Be Specific and Concise:
Clearly state how each individual or group contributed to the project. Be concise in your expression, avoiding overly elaborate language.
Scenario : “James Wilson, your meticulous data analysis greatly contributed to the project’s accuracy. Your ability to distill complex information into actionable insights made a significant impact on our success.”
Use a Formal Tone:
Maintain a formal tone in your acknowledgement, especially if it’s for academic or professional projects. This ensures the recognition is respectful and professional.
Scenario: “Dear Justin Taylor, your professionalism and attention to detail were crucial to the success of our project. Your commitment to excellence set a standard that inspired the entire team.”
Include Relevant Details:
If appropriate, include details such as the project timeline, challenges faced, and the significance of contributions. This adds context to the acknowledgment.
Scenario : “In the midst of tight deadlines, Justin Taylor’s strategic problem-solving skills proved invaluable. His quick thinking during project challenges played a pivotal role in overcoming obstacles.”
Samples of Acknowledgement For a Project
Acknowledgement for project (general).
Dear Justin Taylor,
I would like to express my sincere gratitude to you for your invaluable contributions to Project Symphony. Your expertise and dedication played a pivotal role in its successful completion. Your insights were truly instrumental, and I appreciate the time and effort you invested in this project.
Best Regards, James Wilson
Acknowledgement Sample For School Project
To Justin Taylor:
I extend my heartfelt thanks for your support and guidance throughout the Quantum Physics Project. Your encouragement and valuable suggestions greatly enhanced the project’s quality. It’s truly a pleasure to have had the opportunity to work under your mentorship.
Warm Regards, James Wilson
Acknowledgement for a Research Project for the School
I am immensely grateful for your guidance and mentorship during the Genetic Research Project. Your expertise and constructive feedback significantly contributed to the project’s success. Your commitment to academic excellence has been truly inspiring.
Sincerely, James Wilson
Acknowledgement for College Project Samples
Acknowledgement for college project (individual project).
I want to express my heartfelt appreciation for your unwavering support and encouragement throughout the Environmental Science Project. Your insightful feedback and guidance were instrumental in shaping the project’s outcomes. Thank you for being a source of inspiration.
Warmest Regards, James Wilson
Read More: Acknowledgement For English Project
Acknowledgement for College Project (Group Project)
To Justin Taylor and Team,
Our sincere thanks to each member of the team for the collective efforts that led to the success of the Marketing Strategy Group Project. Your collaboration, dedication, and unique skills brought diversity and excellence to our work. It was truly a collaborative triumph.
Acknowledgement for University Project
My sincere gratitude for your support and mentorship throughout the International Relations Project. Your guidance, both academically and professionally, has been invaluable. I am fortunate to have had the opportunity to work under your tutelage.
Kind Regards, James Wilson
Acknowledgement for a Research Project
I wish to express my deepest appreciation for your guidance and support during the Neuroscience Research Project. Your expertise and constructive criticism were pivotal in refining the project. Your mentorship has left an indelible mark on my academic journey.
Yours Gratefully, James Wilson
Various Acknowledgment Samples for Different Projects, Subjects, and Topics
Dos and don’ts of writing acknowledgements.
- Express Genuine Gratitude: Ensure your acknowledgment reflects genuine appreciation for the contributions received.
- Be Specific: Clearly mention the roles and contributions of each individual or group.
- Use Formal Language: Maintain a formal and respectful tone, especially in academic or professional settings.
- Proofread: Check for grammatical errors and ensure clarity in your expressions.
- Generic Statements: Avoid generic acknowledgments. Be specific about how each person contributed.
- Overly Complex Language: Keep the language clear and straightforward. Avoid unnecessary complexity.
- Oversights: Double-check to ensure you haven’t overlooked anyone who made a significant contribution.
As Oscar Wilde aptly said, “ I have simple tastes, and I am easily satisfied with the best.” Expressing gratitude through acknowledgments is a simple yet powerful way to appreciate the best in others.
What is an acknowledgment page?
An acknowledgment page is a section in a document or project where the author expresses gratitude and recognition to individuals or groups who have contributed to its completion. It is a way of giving credit to those who played a role in the project’s success.
What is an example of acknowledgment for a group assignment?
An example of acknowledgment for a group assignment could be: “We would like to express our gratitude to each member of the group—Sarah, David , and Emily —for their dedication and collaborative efforts in completing this assignment. Each contribution was vital to our collective success.”
What is a sample acknowledgment letter?
A sample acknowledgment letter is a written document expressing thanks or recognition for someone’s help, support, or contribution . For example, “Dear Mr. Johnson, I am writing to express my sincere gratitude for your invaluable support during the marketing campaign project.”
What is an acknowledgment description and example?
Acknowledgment is the act of recognizing and appreciating someone’s efforts or contributions . For instance, in a project report, an acknowledgement may read, “I would like to extend my appreciation to my colleagues, Mark and Jessica , for their significant contributions to this project.”
What is an example of an acknowledgment in a thesis?
In a thesis, an acknowledgment might be: “I would like to express my heartfelt gratitude to my supervisor, Dr. Anderson , for their unwavering support and guidance throughout the research process.”
How do you write an acknowledgement for a project supervisor?
An acknowledgment for a project supervisor can be written like this: “I want to extend my sincere thanks to Professor Davis for their continuous support and valuable guidance during the entire duration of this project.”
How do I write an acknowledgment PDF?
To write an acknowledgment in a PDF, create a new page or section in your document. Use a formal tone, mention specific contributions , and express gratitude . Save or export the document as a PDF for sharing or printing.
What is an acknowledgment message?
An acknowledgment message is a brief note expressing thanks or confirming the receipt of something. For instance, “Thank you for your email. We have received the documents and will review them promptly.”
How do you write an acknowledgement of receipt?
To acknowledge receipt, send a simple message like, “Thank you for the documents received. We confirm their safe arrival and will proceed with the necessary actions.” can be used.
Final thoughts
In conclusion, acknowledging the contributions of those who supported a project is not just a courtesy but a powerful way to celebrate collaboration and express gratitude. By recognizing the efforts of individuals, fostering professional relationships, and boosting team morale, acknowledgments become a vital aspect of successful project completion. Through the practical guidance provided in this article, individuals are encouraged to craft sincere and personalized acknowledgments, fostering a positive atmosphere that enhances both project outcomes and professional connections. As we embrace the power of gratitude, let us remember that a simple “thank you” has the potential to leave a lasting impact on individuals and the collaborative spirit of future endeavors. Acknowledgments are more than a formality—they are a genuine expression of gratitude . Whether it’s a school project, research endeavor, or professional collaboration, taking the time to acknowledge those who supported you enhances both the project’s quality and your professional relationships.
I’m Matthew Porter , the creative mind behind “ Acknowledgment Templates .” I’ve had a blast creating templates that capture the essence of gratitude in acknowledgment sections. At Acknowledgment Templates, we’re all about turning appreciation into a well-crafted art. Let’s make your acknowledgment section a masterpiece—join me in the creative process at Acknowledgment Templates!
1 thought on “Acknowledgement For Project [5 Examples]”
Good very helpful
Leave a Comment Cancel reply
Save my name, email, and website in this browser for the next time I comment.
I'm Dariel Campbell , the expert helping you navigate acknowledgment sections at " Acknowledgment Templates ."
Playing with words and expressions is my thing. At Acknowledgment Templates, we're here to make your acknowledgment stand out and shine.
Let's make your appreciation heartfelt and memorable—come and join the fun at Acknowledgment Templates!
Recent Posts

Acknowledgement For Art Integrated Project |Best Examples

Acknowledgement For Dissertation|12+Best Samples

Acknowledgement Sample For Undergraduate Thesis|12 Best Samples

Acknowledgement Letter Sample For A Great Customer Service|9+ Samples

Acknowledgement Letter Sample For Order Cancellation|12 Best Samples

Sample Of Acknowledgement For The Ngo Project Report|10+ Samples

Order Acknowledgement| How to write full guide

What To Write In Acknowledgement?|Best Examples
Make your acknowledgment stand out and shine
Privacy Policy
Latest Articles

March 23, 2024

© 2024 Acknowledgement Templates
Acknowledgement Letter
Get All Types Of Acknowledgement Samples.
Acknowledgement For Project
Post by Ruben Patel Leave a Comment
Completing a project is a rewarding experience, but it’s never done without the help and support of others. That’s why it’s important to include an acknowledgement section in your project report. This is a chance to express your gratitude to everyone who helped you along the way, whether it was a teacher, mentor, colleague, friend, or family member, there’s always someone who goes above and beyond to help us achieve our goals. Acknowledgements are typically included in project reports, research papers, and other formal documents.
Writing an effective acknowledgement is not difficult, but it does require some thought and care. When writing an acknowledgement, it is important to be specific and to thank people by name. It is also important to be sincere and to express your gratitude in a meaningful way.

If you’re not sure who to thank, or what to say, don’t worry. I’ve got you covered. In this blog post, I’ll share 100+ Samples of acknowledgements for projects , so you can find the perfect inspiration for your own. Whether you are writing an acknowledgement for a college project, a research project, a group project, or a professional project , these examples will help you get started.
Table of Contents
Why is it important to acknowledge people who have helped you with your project?
Acknowledging people who helped you with your project is a gracious and essential way to show your appreciation for their contributions and to build strong relationships. It shows that you value their time, effort, and expertise and that you recognize that your success is not solely your own.
When you acknowledge someone for their help, you are also sending a message to others that you are a team player and that you are supportive of others’ success. This can create a more positive and productive work environment for everyone involved. It also gives credit where credit is due, and it is an opportunity to build relationships and showcase your professionalism.
Here are some tips for acknowledging people who helped you with your project:
- Be specific. Mention the specific things that each person did to help you, and how their contributions made a difference.
- Be sincere. Let people know that you are truly grateful for their help.
- Be timely. Acknowledge people as soon as possible after you have completed your project.
- Be creative. There are many ways to acknowledge people, such as writing a thank-you note, giving them a small gift, or simply saying thank you in person. Choose a way that is meaningful to the person and the situation.
By acknowledging the people who helped you with your project, you are showing your appreciation for their contributions and building stronger relationships. You are also sending a message that you are a team player and that you value the success of others.
How to write an effective acknowledgement for a project
An acknowledgement is a way to recognize and appreciate the contributions of others to your project. It is a chance to say thank you to those who have helped you in any way, big or small.
A well-written acknowledgement can show your gratitude, build relationships, and reflect well on you and your work. When crafting your acknowledgment, be specific, mentioning names and detailing their impact.
To write an effective acknowledgement, start by identifying the specific things that each person or group of people did to help you. Then, write a brief statement expressing your gratitude for their contributions. Keep your acknowledgement to the point, and avoid including unnecessary details. Be sure to include the person’s name or title, and avoid using generic language.
Some tips for writing a good acknowledgement –
- Use Names and Roles: Say thanks by mentioning people’s names and what they did. It’s like giving each person a special shout-out for their unique contribution.
- Be Honest and Real: Don’t use fancy words. Just say what you really feel. People can tell if you’re being genuine.
- Keep it Proper: Use proper grammar and spelling, and avoid using slang or informal language.
- Thank Everyone: Don’t forget anyone! Say thanks to the main helpers and even those who worked behind the scenes. Everyone deserves a shout-out!
Samples of Acknowledgement For a Project
Sample 1 – acknowledgement for project (general).
I want to express my gratitude to my teacher, _____ [Teacher’s Name], for their encouragement, insightful suggestions, and mentorship. I’d also like to extend my thanks to my principal, _______ [Principal Name], for granting me this wonderful opportunity to be part of this project.
Also, I would like to express my appreciation to all those who have supported and contributed to the completion of this project. Your assistance, guidance, and encouragement have been invaluable. Thank you for being a part of this project.
Lastly, I want to thank my family & friends for their understanding and support during this project.
Sample 2 – Acknowledgement Sample For School Project
I would like to acknowledge my advisor, Dr. Ranjan, for his guidance and support throughout the project. His insights and feedback were invaluable in helping me to develop my research questions and methodology. I would also like to thank my collaborators, Shyam Roy and Sumit Sen, for their contributions to the data collection and analysis. Their expertise and hard work were essential in completing the project on time. Finally, I would like to thank my research assistant, Sima Gupta, for her help with the literature review and bibliography. Her attention to detail and organizational skills were essential in helping me to keep track of my research.
Sample 3 – Acknowledgement for a Research Project for the School
I would like to express my deepest gratitude to my project supervisor, ______ [Your Supervisor’s Name], for their invaluable guidance and support throughout the research process. Their expertise and dedication were instrumental in shaping this project. I would also like to thank my classmates for their constructive feedback and encouragement. Additionally, I extend my appreciation to the library staff for their assistance in accessing research materials.
Lastly, I would like to express my gratitude to my _____ [family name] for their love and support during this project. This project truly reflects the collective effort of many, and I’m deeply appreciative of their contributions.
Acknowledgement for College Project Samples
Acknowledgement for college project (individual project).
I want to convey my heartfelt gratitude to my professor, ______ [Professor’s Name], for their support and encouragement during the research and writing of this history project. Their expertise in the subject matter greatly contributed to the depth and quality of the project.
Also, I would like to express my sincere gratitude to our Principal, _______ [Principal Name], for his unwavering support and encouragement throughout this project. I am grateful for the opportunity to have worked on this project under her guidance, and I am confident that my learning and personal growth have been enriched as a result.
I would also like to thank my friends, for their feedback and support. Their input was invaluable in helping me to develop and refine my ideas. Lastly, my family’s support and encouragement were essential throughout the completion of this project.
Acknowledgement for College Project (Group Project)
We would like to extend our heartfelt appreciation to everyone who contributed to the successful completion of this project. Our sincere thanks go to our project team members for their dedication and collaboration throughout the project. Each member played a significant role in shaping the outcome. Special thanks to our professor, ______ [Professor’s Name], for their guidance and valuable feedback, which enriched our work.
I’d like to express my heartfelt gratitude to The University of _______ [University Name] for affording us the opportunity to collaborate on the _______ [Project Name]. Lastly, we want to thank our families and friends for their patience and encouragement during this project. Their belief in me helped me to stay motivated and to persevere through difficult times.
Acknowledgement for University Project
I would like to express my sincere gratitude to my mentor, Dr. _______ [mentor’s name], for their guidance and support throughout this project. Dr. ______ [mentor’s name] was always available to answer my questions and provide me with feedback. They were also instrumental in helping me to develop my research skills and to write my thesis. I am grateful for their patience and encouragement.
I would also like to thank my classmates, [classmates’ names], for their collaboration and support. We worked together to overcome challenges and to achieve our goals. We shared resources and ideas, and we helped each other to stay on track. I am grateful for their friendship and support.
Finally, I would like to thank my family and friends for their love and support. They encouraged me to pursue my goals and to never give up. I could not have completed this project without them.
Acknowledgement for a Research Project
I would like to express my special thanks of gratitude to my supervisor, [Supervisor’s Name], whose encouragement, guidance and support from the initial to the final level enabled me to develop an understanding of the subject. This research wouldn’t have been possible without the funding and resources provided by [Funding Organization/Institution]. I am also grateful to my research team members, [team members’ names], for their hard work and dedication. We worked together to collect data, analyze results, and write the research paper.
Various Acknowledgment Samples for Different Projects, Subjects, and Topics (100+ Samples)
Acknowledgment Samples On Different Subjects
Acknowledgment Samples On Different Topics

Dos and Don’ts of Writing Acknowledgements
Faqs on acknowledgement for project.
1) What is an acknowledgment for a project?
An acknowledgement for a project is a section of the project report where you express your gratitude to the people and organizations who helped you complete the project. It’s a way to recognize and appreciate their involvement.
2) What should I include in my acknowledgment?
Include the names of individuals or organizations who played a significant role in your project. You should also briefly mention their specific contributions. For example, you could thank your mentor for providing guidance and support, or thank your team members for their hard work and dedication. Express genuine gratitude and keep the tone respectful and appreciative.
3) How can I write an acknowledgement for school?
Start by thanking teachers, classmates, or anyone who supported your academic journey. Be specific about their contributions, whether it’s guidance, assistance, or inspiration. Mention any school resources or facilities that played a role, and keep the tone sincere and respectful. You can also express your gratitude for the opportunity to learn and grow.
4) How can we write an acknowledgment for a group project?
Start by thanking the team collectively for their contributions. Ensure that you individually thank each group member for their specific contributions, specifying their roles in the project. For example, you could thank someone for their research, writing, editing, or design skills.
5) Is acknowledgement compulsory for project?
No, acknowledgement is not compulsory for a project, but it is a good practice to acknowledge the people who helped you with your project. This shows that you are grateful for their contributions and that you value their work. It can also help to build relationships and trust with the people you have worked with.
6) What if I don’t know who to thank?
If you are not sure who to thank, It’s better to acknowledge a few extras than miss out on someone who played a part. Acknowledge anyone who made a significant contribution, including t eam members, mentors, advisors, funders , or those who provided resources and support. You can also thank your department, institution, or other organizations that provided you with support.
7) How lengthy should an acknowledgement be?
It depends on your project’s size and how many people or groups you want to thank. But a good idea is to keep it short—around one page or 100 to 200 words.
8) What is the difference between acknowledgment and acknowledgement?
There is no difference between acknowledgement and acknowledgment. Both spellings are correct, but “ acknowledgment ” is more commonly used in American English, while “ acknowledgement ” is preferred in British English. Choose the one that fits the spelling conventions of your intended audience or follow the style guide provided for your project.
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.

Acknowledgement Sample

The Power of Acknowledgements in your school project: Building Professional Relationships through Gratitude
Learn how acknowledging those who have helped with your school project can lead to building strong professional relationships and potentially opening doors for future career opportunities. Discover the benefits of expressing gratitude and maintaining those connections over time.
Sample acknowledgement for a social science project
We have been asked by many of you to provide some samples of acknowledgement for a social science project. In the most of the schools
Sample acknowledgement for computer project
How to write acknowledgement for computer science project? Sample acknowledgement for computer science project file.
Acknowledgement sample for maths project
How to write acknowledgement for maths project? Sample acknowledgement of maths project. What to include in mathematics project acknowledgement?
SAMPLE ACKNOWLEDGEMENT FOR ACCOUNTS PROJECT
We have been asked by many of you to provide some samples of acknowledgement for accounts project. In the most of the schools now, it

Latest Posts
We have been asked by many of you to provide some samples of acknowledgement for a social science project. In the most of the schools now, it is required to include acknowledgement with project paper,
We have been asked by many of you to provide some samples of acknowledgement for accounts project. In the most of the schools now, it is required to include acknowledgement with project paper, and it
What is acknowledgement in research paper ?
In the majority of research papers acknowledgement relates to the part of research project where author shows gratitude to the people who contributed to the research project. An acknowledgement in research paper is actually a
Sample Acknowledgement for physics project
How to write acknowledgement for physics project? Sample acknowledgement of physics project. What to include in physics project acknowledgement?
How to write acknowledgement for history project?
How to write acknowledgement for history project? Sample acknowledgement of history project. What to include in history project acknowledgement?
Sample acknowledgement for economics project
Many of you have asked us to provide them samples of acknowledgement for an economics project. In the most of the schools now, it is required to include acknowledgement within project paper, and it became
Sample acknowledgement for Chemistry Project
How to write acknowledgement for for chemistry project. Sample acknowledgement for chemistry investigatory project. What to include in chemistry project acknowledgement.
How To Write Your Book Acknowledgments

What Is The Acknowledgments Section?
The Acknowledgments section is where you recognize and thank everyone who helped you with your book. It’s a way to display your appreciation to them in a public and permanent forum.
Who Should You Thank In The Acknowledgments?
This is entirely up to you. Recognize whoever you feel contributed enough to your book that they deserve it. For example, common groups of people that Authors thank include:
- Family members (spouse, children, parents)
- Editors/people who worked on the book production
- Coworkers/assistants
- Agents/managers
- Contributors/advisers/sources of information
- Teachers/mentors/bosses
- Inspirations
Do You Need One?
No. Most books do have them, but by no means are they required or mandatory.
How To Write Your Acknowledgments Section
1. Remember: people will read this
People will read the Acknowledgments section, and it will impact them—especially the people who are in it. This section is about those people you are naming, not about you, so approach this as you should your entire book: make it good for the people you are naming, who will read it.
2. Start with a list of who will go in (by full name)
This method has worked well in many situations: write out all the people you want to thank BEFORE you start writing this section. Doing this allows you to see them all together in a list and helps ensure that everyone is on the list who should be there.
Note: a great way to make sure you are not missing anyone is to group people by category so that you are more likely to remember them. When you put your family members together, you are less likely to leave any out.
3. Be specific for the important people
For the most important people, the more specific you can be in your thanks, the better.
For example, this is not specific:
“I want to thank my wife, Veronica. Thank you.”
This is specific:
“I have to start by thanking my awesome wife, Veronica. From reading early drafts to giving me advice on the cover to keeping the munchkins out of my hair so I could edit, she was as important to this book getting done as I was. Thank you so much, dear.”
Being specific in thanks is all about making them feel special. The more detailed you can be in your thanks, the more you’re showing that you recognized and appreciated their help. It is rewarding when someone thanks you for a particular thing you did, as opposed to just thanking you overall.
As you go further into your list, you can group people. But again, be specific in your thanks, even to groups. For example, this is not specific:
“Thanks to everyone on my publishing team.”
“Thanks to everyone on the Scribe team who helped me so much. Special thanks to Ellie, the ever-patient Publishing Manager; Meghan, my amazing Scribe; and Erin, the greatest cover designer I could ever imagine.”
4. Be sincere in your thanks
The worst thing you can do in an Acknowledgments section is say things you don’t believe. If you aren’t willing to be sincere, then you are better off not doing one at all.
Sincerity means honestly and deeply thanking the people who helped you (mentioning the specific ways they helped, as noted above) and remembering the way that they sacrificed for you.
At the same time, don’t feel the need to go overboard. You’re not accepting an Oscar, so don’t go on and on or say things just to make yourself look good. Make it meaningful and sincere.
5. Don’t worry about length
You might see some people recommend the Acknowledgments section be only one page. Ignore that.
This is the only section I will tell you that you can go long if you want. You may only write one book in your life, and if that is the case, then take all the time and space you need to thank everyone who helped you.
At Scribe, we put the Acknowledgments section in the back so it doesn’t interfere with the average reader’s experience. Readers can skip this section if they are bored, but you can never go back and re-thank the people you left out because of some arbitrary “rule.”
Examples of Acknowledgments Sections
You can pull almost any book off your shelf and read the Acknowledgments section for examples. Here are some examples of different Acknowledgments sections for books we have done.
Bill Hicks, in The Leadership Manifesto , starts his acknowledgments off with a blanket acknowledgment of leaders everywhere, before naming a handful of them by name. He then thanks his book publishing team and closes with a paragraph acknowledging his CEO. This is a good example of an acknowledgment from a business executive:
The world is a better place thanks to people who want to develop and lead others. What makes it even better are people who share the gift of their time to mentor future leaders. Thank you to everyone who strives to grow and help others grow. It is the business version of The Lion King song, “Circle of Life.”
To all the individuals I have had the opportunity to lead, be led by, or watch their leadership from afar, I want to say thank you for being the inspiration and foundation for The Leadership Manifesto .
Without the experiences and support from my peers and team at Ultimate Software, this book would not exist. You have given me the opportunity to lead a great group of individuals—to be a leader of great leaders is a blessed place to be. Thank you to Chad, Dan, Dave, Gretchen, JC, Laura, Patrick, Scott, and Susan.
Having an idea and turning it into a book is as hard as it sounds. The experience is both internally challenging and rewarding. I especially want to thank the individuals that helped make this happen. Complete thanks to Joanie, Randy Walton, Patrick O’Neill, Barbara Boyd, Carol Raphael, and Dan Bernitt.
Scott Scherr, thank you for being a leader I trust, honor, and respect. I will always welcome the chance to represent you. “Au Au Au!”
Tiffany Haddish, a well-known comedian, continues her “simple yet emotionally powerful” style from her book The Last Black Unicorn into the acknowledgments. She thanks her close family and then closes with a joke in line with the subject matter of the book and a blanket acknowledgment to the untold masses who have encouraged her.
I want to thank:
My Grandma.
My Aunties.
My Daddy for donating the sperm that made me.
All my brothers and sisters.
My best friends Selena, Shermona, Aiko, Shana, Richea.
My old agent, my current agents and managers, and Tucker Max.
Department of Children Services and the court system for taking care of me when no one else would.
I want to thank EVERYONE who ever said anything positive to me or taught me something. I heard it all, and it meant something.
All the dudes I ever slept with, I appreciate the experiences, but I ain’t naming none of you!
I want to thank God most of all, because without God I wouldn’t be able to do any of this.
The Scribe Crew
Read this next.
Book Ghostwriters for Hire: Find the Perfect Writer
How to Use AI When Writing a Book
Why Work With a Memoir Ghostwriter?
- Open access
- Published: 26 April 2024
hadge: a comprehensive pipeline for donor deconvolution in single-cell studies
- Fabiola Curion 1 , 2 na1 ,
- Xichen Wu 1 , 2 na1 ,
- Lukas Heumos 1 , 4 , 6 na1 ,
- Mylene Mariana Gonzales André 1 , 2 ,
- Lennard Halle 1 ,
- Matiss Ozols 7 , 8 ,
- Melissa Grant-Peters 3 ,
- Charlotte Rich-Griffin 3 ,
- Hing-Yuen Yeung 3 ,
- Calliope A. Dendrou 3 , 5 ,
- Herbert B. Schiller 10 , 4 , 9 &
- Fabian J. Theis ORCID: orcid.org/0000-0002-2419-1943 1 , 2 , 6
Genome Biology volume 25 , Article number: 109 ( 2024 ) Cite this article
8 Altmetric
Metrics details
Single-cell multiplexing techniques (cell hashing and genetic multiplexing) combine multiple samples, optimizing sample processing and reducing costs. Cell hashing conjugates antibody-tags or chemical-oligonucleotides to cell membranes, while genetic multiplexing allows to mix genetically diverse samples and relies on aggregation of RNA reads at known genomic coordinates. We develop hadge (hashing deconvolution combined with genotype information), a Nextflow pipeline that combines 12 methods to perform both hashing- and genotype-based deconvolution. We propose a joint deconvolution strategy combining best-performing methods and demonstrate how this approach leads to the recovery of previously discarded cells in a nuclei hashing of fresh-frozen brain tissue.
Single-cell RNA sequencing (scRNA–seq) technologies have unlocked unprecedented resolution to discover complex mechanisms of health and disease in human biology [ 1 ]. Droplet-based methods, which encapsulate aqueous cells into oil constituting a micro-chamber for lysis and retrotranscription of the RNA of individual cells, have made single-cell sequencing more accessible and dramatically increased the throughput of single cells from individual samples [ 2 ]. The cDNA produced in these reactions is uniquely barcoded for each droplet, such that the retrieval of these barcodes enables the association of sequencing readouts to individual cells. Despite considerable strides made in cellular profiling methods, the application of scRNA-seq to biomedical studies and clinical applications, which often require complex multi-sample and multi-condition experiments, has been limited by sample throughput, cost, and susceptibility to technical variability [ 2 ]. When samples cannot be acquired fresh or immediately processed after the acquisition, as may be the case for biobank specimens, fixation techniques that allow to preserve the biological material and optimize single-cell and nuclei profiling via multiplexing are a viable option [ 3 , 4 , 5 , 6 ]. In recent years, methods have emerged that allow the pooling of single cells and nuclei from individual samples [ 7 ], often relying on multiplexing techniques [ 8 ]. These methods have found wide applicability [ 8 ] and are now routinely used to carry out population-scale studies with single-cell sequencing protocols [ 9 , 10 ].
To date, there are two major protocols to generate a mixture of cells from multi-sample studies: “cell hashing” and “genotype-based multiplexing.” Cell hashing is a sample processing technique that tags the membrane or nuclei of cells in individual cell-suspension samples with unique oligonucleotide barcodes. One option is to stain the individual samples with oligonucleotide-labeled antibodies that target proteins ubiquitously expressed on the cell or nucleus surface [ 11 ]. Another option is to chemically conjugate oligonucleotides directly to the membrane constituents, for example by hybridization of a lipid-modified oligonucleotide (LMO) to the hydrophobic cell membrane, a technique called “lipid tagging” [ 12 ], or by chemical ligation of the oligonucleotide to exposed N-Hydroxysuccinimide-reactive amines, a technique called “chemical barcoding” [ 11 , 13 , 14 ]. After staining or tagging, cells undergo a washing or quenching process, allowing for the safe combination of different samples into a single mixture in one tube. From this mixture, two separate sequencing libraries are created: one for single-cell RNA (scRNA) and one for hashing oligos (HTO). These libraries are independently sequenced to yield two distinct single-cell count matrices, corresponding respectively to scRNA and HTO data. To deconvolve the cell’s source sample, the HTO counts are processed to discover cell barcodes positive for at least one hashtag, using cell-hashing deconvolution methods [ 11 , 12 , 15 , 16 ]. Cell barcodes are classified into “Singlets,” if they are positive for one tag; “Doublets,” if positive for two or more; and “Negatives,” when only a low background-noise signal is detectable (Fig. 1 A). Cell-tagging approaches suffer from constraints such as low starting cell numbers, as these methods require washing steps that may result in cell-number loss. Furthermore, different issues can impair the quality of a hashing experiment and therefore decrease the final number of uniquely identified cells. Antibodies or free oligonucleotides can persist in suspensions if an adequate number of washes is not performed, or can attach to debris from membrane lysis in fixed samples [ 15 ].
Overview of donor deconvolution and the hadge pipeline. A Schematic example of the cellular components leveraged by single-cell multiplexing experiments. Hashing cell counts and scRNAseq reads with SNP calling by cell are the input to the hadge deconvolution pipeline. B hadge implements 12 methods across two sub-workflows of which seven are hashing-based and five are genotype-based deconvolution that can be run independently, in parallel or jointly, in rescue mode. In rescue mode, the pipeline offers the option to refine hashing results with genotype-based deconvolution methods to rescue failed hashing experiments in the donor-matching process. It compares the concordance in donor identification between hashing and genotype-based methods and identifies the best pair of two strategies based on the calculated Phi score
Genotype-based multiplexing allows the mixing of samples with unique genetic composition, where natural genetic variants serve as inherent cell barcodes [ 17 ]. Users can harness these genetic barcodes to determine the identity of each cell in the mixture. Provided a genotype reference, the scRNA reads are scanned for single nucleotide polymorphisms (SNPs) in the reference, and a table of SNP assignment to cells is produced to computationally infer the donors (Fig. 1 A). Cell barcodes with a genetic composition matching one donor are called “Singlets,” cell-barcodes with a mixture of at least two donors genotypes are deemed “Doublets,” and cells where the read coverage is insufficient to identify their genetic composition are “Negatives.” One limitation of this approach is the need to rely on additional data to correctly assign the cell mixtures. Users can genotype the individual samples through SNP arrays or bulk RNA-seq followed by variant calling and then aggregate the expression values at these genomic positions for deconvolution. The same process can be conducted without genotype of origin or “genotype-free,” by piling up the mixture of scRNA onto an unrelated genomic reference of genotypes such as that provided by the “1000 Genome Project.” However, this approach can only deconvolve the cell mixture in the form of anonymous donors and additional processing is needed to match them to the sample of origin.
The limitations of each of these protocols can be mitigated when combining demultiplexing approaches. Experiments, where the hashing libraries are of low quality, can be rescued and successfully demultiplexed using the natural genetic variation of their RNA libraries. The combination of hashing and genetic deconvolution methods represents a viable option for combinatorial experimental design and can result in increased cell recovery rate and calling accuracy [ 18 ]. Moreover, joint demultiplexing can be a cost-effective deconvolution strategy as it further avoids having to produce sample-specific genotyping data in the form of SNP arrays or bulk sequencing methods for variant calling [ 18 ]. To date, at least nine hashing and five genotype-based deconvolution methods have been developed, each with unique strengths and weaknesses [ 12 , 15 , 17 , 19 , 20 , 21 , 22 ]. However, investigations on joint demultiplexing strategies have been limited to the combination of two specific tools instead of computationally testing the best combination of demultiplexing methods, therefore neglecting the utility of other widely used tools [ 18 ]. Although single workflows for hashing-based deconvolution and genotype-based deconvolution exist [ 23 , 24 ], no study has combined all the tools for both approaches in a single comprehensive and efficient pipeline, such that both hashing and genotype deconvolution pipelines can be run in parallel on multiple samples, providing a score to discover the best methods across the board, with the final goal of maximizing the number of rescued cells and increasing the confidence of deconvolution.
Therefore, there is a critical need for a unified pipeline that integrates the strengths of multiple donor deconvolution tools. Here we present the hadge (hashing deconvolution combined with genetic information) pipeline. Our Nextflow-based pipeline [ 25 ] enables deconvolving samples of both hashing and genetic multiplexing experiments either independently or simultaneously. hadge allows for the automatic determination of the best combination of hashing and SNP-based donor deconvolution tools. Moreover, hadge provides a rescue mode to run both genetic and hashing approaches jointly to rescue problematic hashing experiments in cases where donors are genetically distinct. We demonstrate our pipeline using a single nuclei hashing experiment of fresh frozen multiple sclerosis (MS) brain tissue and show that joint deconvolution allows us to rescue high-quality cells that would have been otherwise discarded. Finally, we benchmark our pipeline with the state-of-the-art tools and a large-scale scRNA dataset [ 9 ].
The hadge pipeline
Hadge offers a user-friendly, zero-config solution for analyzing multiplexed single-cell data at scale (Fig. 1 B). Our pipeline takes advantage of Nextflow’s cloud-computing capabilities, enabling efficient use of cloud resources to accelerate the analysis of large datasets. Furthermore, Nextflow’s built-in containerization functionality simplifies deployment, providing a more reliable and reproducible analysis environment. The hadge pipeline consists of 12 deconvolution tools, including five genetics-based tools (Demuxlet [ 17 ], Freemuxlet [ 26 ], Vireo [ 22 ], scSplit [ 20 ], and Souporcell [ 21 ]), seven hashing-based tools (HTODemux [ 27 ], Multiseq [ 12 ], HashedDrops [ 28 ], Demuxem [ 15 ], gmm-demux [ 29 ], BFF [ 24 ], and Hashsolo [ 30 ]), and one doublet-detection method (Solo [ 30 ]). All of these tools have been benchmarked in independent publications and are widely used by the scientific community [ 14 , 23 , 31 , 32 ]. Furthermore, for methods that require additional preprocessing such as normalization of the HTO counts or variant calling, the hadge pipeline includes a preprocessing step before the genotype-based deconvolution algorithm is applied.
The hadge pipeline has three modes: “genetic,” “hashing,” and “rescue.” In the genetic or hashing mode, the pipeline runs either the genotype- or hashing-based deconvolution workflow allowing for choice of methods and customization of input parameters. Each of these pipelines can be run in parallel across multiple samples, reducing the time and effort required for deconvolution. Finally, in the rescue mode, hadge allows jointly deconvolving hashing experiments with genotype-based deconvolution tools, with the option to recover cells from failed hashing. Lacking prior individual genetic profiles that associate SNPs to explicit donors, genotype-based deconvolution tools assign cells to anonymous donors. Hadge de-anonymizes the donors by calculating the best match between a hashing and a genetic demultiplexing method. After the conversion of the cell deconvolution assignments into a binary matrix with rows representing cell barcodes and columns representing the assigned donors or hashtags, donor genotypes are matched with hashtags by measuring the concordance of two methods in assigning the droplets, computing pairwise Pearson correlation to determine the optimal match, hereby termed “Phi score” (see the “ Methods ” section). hadge then generates a new assignment of the cells based on this optimal match between hashing and genotype-based deconvolution to uncover the true donor identity of the cells effectively rescuing cells from failed hashing with a valid genotyped-based deconvolution assignment. Finally, hadge outputs the results of the donor deconvolution for all combinations of methods and hyperparameters tested, both as a separate tabular format and as cell metadata in either Anndata [ 33 ] or MuData [ 34 ] objects, depending on the users’ choice.
Hashing-based methods’ performance greatly varies with noisy HTO libraries
We applied the hadge pipeline to a hashing dataset of single nuclei sequencing collected from post-mortem brain tissue from multiple sclerosis donors [ 35 ]. The hashing count matrix of this dataset presented a high background noise from non-specific antibody binding, which originally resulted in a high number of doublets and negative cells (Fig. 2 A, B). We ran both hashing and genetic deconvolution workflows to assess the performance of the two types of approaches. We observed inconsistent hashtag counts (Fig. 2 A, B and Additional file 1 : Fig. S1). Specifically, hashtag 453 showed a high overall expression, while hashtags 454 and 455 were expressed in relatively low levels (Fig. 2 A and Additional file 1 : Fig. S1, S6). Due to the variable readout of the hashing oligos, the sample assignment of the hashing-based methods was not consistent. The number of detected singlets varied greatly between different methods (Fig. 2 C and Additional file 1 : Fig. S1-2, S7). While Hashsolo classified almost every droplet as a singlet, HashedDrops detected only 32 singlets among 4048 non-empty droplets. Notably, DemuxEM and Multiseq exhibited nearly identical performance (Additional file 1 : Fig. S1-2,S4), both assigning nearly 1800 singlets, (Additional file 2 : Table S1) with Multiseq identifying slightly more singlets and being considerably faster than DemuxEM. (Additional file 3 : Table S1). Despite the noisy HTO readouts, the RNA profiles of these cells are still of good quality, allowing demultiplexing to be performed from the RNA library. Since the donor-specific reference genotypes are not available for this experiment, we run all genetic deconvolution tools in reference genotype-free mode. Compared to hashing, genotype-based deconvolution methods performed more consistently and identified significantly more singlets (Fig. 2 D and Additional file 1 : Fig. S3, S8-9). Each tool classified over 90% of the droplets as singlets, and there was consistent agreement between all tools for 3914 singlets (Fig. 3 D). However, scSplit identified 296 droplets as doublets, which were consistently identified as singlets by three other methods. Due to the high consistency among Vireo, Freemuxlet, and Souporcell, and available benchmarks showcasing its favorable performance compared to the other tools [ 23 ], we decided to use Vireo as a baseline for genotype-based deconvolution methods.
Comparison of the performance of donor deconvolution methods. A The violin plot of raw HTO counts shows high count levels of Hashtag 453 in cells with noisy or undetectable expression of the other HTOs. B UMAP visualization of normalized HTO counts colored by HTODemux assignment shows poor separation of the cells based on hashtags, with most droplets assigned to Hashtag 453. C Hashing-based deconvolution methods show the inconsistent assignment of cells, reported as the different proportions of cells identified as one of either singlet, negative, or doublet. D Genetic deconvolution tools show a more consistent assignment of the cell mixture to singlets, doublets, and negatives
Joint deconvolution recovers high-quality cells. A Overview of the steps to extract cell variants from common SNPs in the population based on the assignment of Multiseq and Vireo. B Heatmap summarizing the donor matching result shows that DemuxEM and Multiseq are in high concordance with all genotype-based deconvolution methods, where all the donors are matched with a high matching score. C Correlation heatmap of donor identification between Vireo and Multiseq. D Sankey plot summarizing the percentages of cells deconvoluted by hashing (Multiseq) and after the joint deconvolution step (Vireo). E Number of donor-specific variants used as input for the refinement step. F Sankey plot summarizing the percentages of cells deconvoluted by hashing (Multiseq) and after the refinement step
Joint deconvolution recovers cells with low-quality hashing data
Beyond determining the optimal combination of hashing- and genotype-based deconvolution methods, hadge aims to rescue cells whose hashing quality was low or whose hashes were missing (Fig. 3 A). Hadge performs joint deconvolution with both hashing and genetic deconvolution tools to rescue high-confidence singlets. Only cells that can be confidently genetically deconvoluted are eligible to be rescued. After having demultiplexed the experiment in genotype-free mode [ 20 , 22 ], the anonymous donors need to be matched to their original sample to be identified. Here, we rely on the hashing deconvolution to provide the known correspondence between the antibody hashtags and the original sample. Cells that are jointly deconvolved provide the key to de-anonymize the genetically rescued cells.
We first define the hashing method that matches the genetic demultiplexing method by calculating the Phi score (see the “ Methods ” section). For each pair of hashing and genetic deconvolution outputs, we compute the pairwise Pearson correlation on the binarized cell classification vectors, thereby matching donors where a high correlation is observed. We then compute the matching score by summing the non-negative correlations and dividing by the number of expected donors, obtaining the degree of consistency in donor identification between any two methods. Based on the observed high matching score and the successful matching of all anonymous donors (Additional file 1 : Fig. S4-5, S10), two hashing demultiplexing methods performed best compared to Vireo, namely Multiseq and Demuxem, both recovering identical matches between genetic and hashing donors (Fig. 3 B, Additional file 1 : Fig S4-5 and Additional file 2 : Table S1). When the optimal match is identified, the identities recovered using the cells in the intersection between genetic and hashing can be propagated to the cells that are identified by genetic deconvolution alone. Here, we decided to use the joint demultiplexing of Multiseq and Vireo to showcase the rescue mode because of Multiseq’s reduced runtime. For every anonymous donor recovered by Vireo, there was only one hashtag with a high correlation, with scores ranging from 0.53 to 0.89 (Fig. 3 C). Using the cells that are jointly deconvoluted into singlets by hashing and genetic demultiplexing, we extended the classification to those cells whose hashing was undetectable (negatives). We identify 90% of the cells as singlets, rescuing 89.7% of the original negatives (Fig. 3 D), and double the number of recovered singlets for the hashes with the lowest detection rate (Hash454-456, Fig. 2 A). Vireo is implemented to rely on cellSNP, which outputs the recovered genetic variants in each cell. We implemented an optional process in hadge to refine and confirm the quality of the deconvolution, by extracting cell-variants to reconstruct minimal donor genotypes from the common SNPs in the populations. Variants with low coverage (allele depth < 10) or a low frequency of the overrepresented allele (frequency < 0.1) were excluded, revealing 7866 variants that were unique to each donor (Fig. 3 E). Since only a small fraction of the hashing-recovered singlets is sufficient to de-anonymize the genetic-singlets, we can use these reconstructed genotypes to run an additional genetic demultiplexing. Therefore, this final refinement step allows to effectively demultiplex cell mixtures without having to generate new SNP references. Using this refining approach, we identify 75% of the cells as singlets, with the number of rescued negatives decreasing to 69.7% (Fig. 3 F). Nevertheless, we obtain 97.6% consistent donor assignment between the rescued and the refined assignments (Additional file 1 : Table S1), suggesting that these variants were crucial in distinguishing a donor cluster from others during deconvolution.
Recovered cells recapitulate known cell types
To investigate whether the cells that are rescued are of good quality and biologically relevant, we reanalyzed the MS samples, including the recovered cells. We first merged the already existing annotation of the cells with the deconvolution information obtained from the hadge pipeline. We then applied quality filtering, removing cells based on gene content, mitochondrial percentage, and doublet rates (Additional file 1 : Fig S12) (see the “ Methods ” section), reproducing the quality control performed in the original study but with a more stringent doublet detection threshold. With this approach, we retained 3208 cells, rescuing 952 cells that were excluded in the original study. We then embedded the cells using UMAP and calculated Leiden clustering. Most of the rescued cells were distributed across existing clusters, with comparable marker expression between the old and new cells (Fig. 4 A, B, D, Additional file 1 : Fig. S11). Intriguingly though, the percentage of rescued cells per cluster varied. While most of the clusters consisted predominantly of previously annotated cells mixing with a smaller part of rescued cells, two clusters were composed of more than half or even 100% rescued cells (Fig. 4 E). While the smaller one of these, consisting solely of rescued cells, had an almost exclusively high expression of the marker HTR2C , we found the gene marker expression of, e.g., SYT1 , SLC17A7 , and low GAD2 to be consistent with a neuronal profile with excitatory and non-inhibitory properties in both clusters. Reassuringly, the latter marker expression was in accordance with that of known neuronal clusters [ 36 , 37 ] (Fig. 4 C, D, Additional file 1 : Fig. S11).
Recovered cells recapitulate known cell types. A UMAP of the single-cell gene expression data with old and rescued cells. B Leiden clustering of the dataset with old and rescued cells. C SYT1 expression defines rescued cells as a new cluster of neurons. D Dotplot of a selection of marker genes shows concordant expression of markers in old and rescued cells. E Barplot showing the cluster composition in old and rescued cells, with two neuronal clusters enriched for rescued cells. Colors on top of the barplot identify the cell annotation from A
Benchmarking hadge’s runtime and robustness
To demonstrate the robustness and superior runtime of our proposed pipeline, we benchmarked its performance against two existing pipelines, demuxafy and cellHashR (Table 1 ). We submitted each pipeline on a Linux server with 32 CPU cores and 160 GB of RAM memory. In all benchmarks, hadge showed superior performance with respect to the optimization of computational resources and runtime (Fig. 5 A). Both hadge-genetic and demuxafy successfully executed all methods for the two mpxMS samples and an additional dataset. However, in the hashing deconvolution of the mpxMS data, some methods ( bff_cluster, bff_raw ) ran but failed to deconvolve the cells in both hadge-hashing and cellhashR . Additionally, one method ( demuxmix ) failed to initialize in both pipelines and as a standalone method. Hence, we excluded demuxmix from hadge. Notably, despite successfully running gmm_demux within hadge-hashing or when called outside the pipeline, we were not able to run cellhashR’s gmm-demux module.
Benchmarking performance. A Hadge genetic and hashing demultiplexing pipelines were benchmarked against demuxafy and cellhashR. The benchmark was performed on three samples for each pairwise comparison, for a total of four samples (mpxMS:gx12, mpxMS:gx38, demuxafy dataset, CR-438–21 dataset). B Results of hadge genetic on the onek1k cohort; each boxplot represents the distribution of percentage singlets identified across 75 pools by each genetic deconvolution tool. C Percentage of correctly matched singlets for each tool; each boxplot represents the distribution across 75 pools. D Dotplot showing the effect of the number of cells per pool on the percentage of recovered and matched singlets. The regression line represents the fit of a linear model on the percentage of singlets identified by Freemuxlet ( R . 2 0.35, p.adj < 0.0001)
Next, we leveraged hadge’s fast multi-sample, multi-process handling to investigate how the input number of cells affects the performance of genetic and hashing demultiplexing. We ran hadge with default parameters on the onek1k dataset, which comprises 75 pools of cell mixtures from 9 to 15 donors each [ 9 ] and has ground truth donor-genotypes available. All tools detected a proportion of singlets per pool between 75 and 98% (Fig. 5 B). However, when looking at matched donors within the singlets, the performance of the tools varied substantially, with scSplit having the lowest concordance with the original donor identities, while Demuxlet had the best performance in terms of recovered singlets and Vireo recovering the most matched singlets (Fig. 5 C). We investigated if these two metrics were associated with the number of cells in each pool. Only Freemuxlet’s singlets percentage had a significant association with the number of cells per pool ( R 2 = 0.35, p.adj < 0.0001) (Fig. 5 D), but all methods were significantly affected by the number of donors in the pools (Additional file 1 : Fig. S13A, Additional file 3 : Table S2), with the highest concordant calls reached on pools with 14 multiplexed donors (Additional file 1 : Fig. S13B). Across all the pools, Vireo, Demuxlet, Freemuxlet, and Souporcell were mostly consistent, confirming these tools’ superior performance, and as observed on the MS dataset (Figs. 2 and 3 ). Additionally, we ran downsampling on two hashing and one genetic multiplexing experiments and used hadge to obtain the percentages of correctly assigned singlets. The hashing methods showed an overall similar trend with the percentage of matched singlets decreasing with the number of cells, except for HTODemux and Demuxem (Additional file 1 : Fig. S14). All the hashing demultiplexing tools were able to correctly assign at least 90% of the cells after downsampling. The performance of the genetic demultiplexing tools was in line with what we observed on the onek1k dataset, and consistent for each tool across the different subsamples, with Vireo having the best performance across the board (~ 99% recovered matching singlets) (Additional file 1 : Fig. S14). Hadge allowed us to efficiently benchmark the demultiplexing performances of all the methods across the two workflows. Collectively, these results indicate that the number of donors and cells in the cell mixtures can significantly affect the number of recovered cells and the quality of the deconvolution for both families of demultiplexing methods.
Single-cell multiplexing techniques enhance sample throughput, reduce costs, minimize technical variation, and improve cell type identification in single-cell genomics studies by increasing the number of samples and therefore reducing the gene expression variation associated with single-cell RNA sequencing. Some of the techniques for generating multiplexed single-cell mixtures require additional processing steps, which can introduce technical noise and result in a low yield of usable data. Furthermore, computational donor deconvolution errors can occur due to technical noise or experimental artifacts, leading to the misidentification of cells or barcodes.
We developed hadge, a comprehensive pipeline for donor deconvolution experiments generated with both genetic and hashing multiplexing methods. hadge is the only pipeline capable of processing both types of data inputs allowing for fine tuning of deconvolution experiments and performs favorably compared to the state-of-the-art pipelines. We leveraged the optimized multi-sample handling implemented in hadge to investigate the demultiplexing performance of the 12 demultiplexing methods included with varying numbers of cells and donors in the input cell mixtures. We showed that the different numbers of input cells and donors can significantly affect the performance of the tools, and users may need to take this into account when designing their experiment and interpreting the deconvolution results. As different tools rely on varying hyperparameters, it is possible to tune them to investigate the effect on cell deconvolution. To ensure confident identification of cell mixtures, hadge enables complete customization of input hyperparameters and selection of methods and offers a host of diagnostic plots and statistics to compare results between methods. Additionally, hadge performs joint genotype- and hashing-based deconvolution of cell mixtures generated from genetically diverse inputs to enable users to retrieve only confidently assigned singlets. This functionality is particularly relevant for experiments where hashing data quality may be compromised by technical noise, tissue-specific variability, or variability in reagent performance. In these experiments, genotype-free deconvolution followed by donor matching can increase the number of good-quality singlets which can be further investigated for biological signatures. Another recent work [ 18 ] proposed joint deconvolution to increase the confidence in called singlets, but offers limited options to customize the selection of tools or parameters to run the joint deconvolution step (Table 1 ). Given the importance of retaining only correctly assigned cells for downstream tasks, such as cell annotation and differential expression between multiplexed conditions, joint deconvolution is a necessary step for experiments threatened by suboptimal hashing libraries. Existing strategies generally only retain the union of singlets called by two methods [ 35 ]. Instead, hadge allows both automated matching of the best hashing and genotype-based deconvolution tools based on the optimal concordance between methods, or the selection of individual methods for each workflow, ensuring an additional level of control over the joint deconvolution step. To guarantee that the joint deconvolution retains only confidently donor-assigned singlets, we developed an additional refinement component that allows the generation of donor genotypes from recovered single-cell variants, which are then used as input for a new round of deconvolution. One limitation of this approach is that, by reducing the number of input variants to include only donor-specific variants, the read coverage in the already shallow-depth single-cell data may decrease at individual genetic variants, resulting in a higher number of cells discarded as negatives. Nevertheless, in the data presented here, only 15 (0.03%) of the total cells are misclassified into a different donor at this step, suggesting the relevance of the selected genetic variants. Furthermore, applying the joint demultiplexing approach can reduce the cost of multi-sample, multi-condition experiments, when the same donors are challenged with multiple perturbations. In such cases, staining only one condition provides enough data to generate donor-specific genotypes, removing the need for additional genotyping and reducing the costs of the staining procedure.
Other pipelines have been proposed to benchmark either genotype-based [ 23 ] or hashing-based deconvolution [ 16 , 24 ] individually (Table 1 ). However, some deconvolution tools do not integrate well with downstream analysis pipelines, making it difficult to perform integrated analyses across multiple samples or experiments. hadge seamlessly integrates within the scverse [ 38 ] ecosystem, and its outputs can be processed with existing pipelines for automated single-cell analysis [ 39 ], minimizing the friction between preprocessing and data analysis steps and ensuring quality and reproducibility of results.
Conclusions
In conclusion, hadge is a powerful and flexible pipeline that addresses the challenges associated with all commercially available single-cell multiplexing techniques in genomics studies. By allowing customization of input parameters, selection of methods, and joint deconvolution, hadge ensures confident identification of cell mixtures and retrieval of high-quality singlets. Its integration with existing analysis pipelines and compatibility with the scverse ecosystem further streamlines the data processing and analysis workflow, promoting reproducibility and enabling integrated analyses across multiple samples and experiments. With its comprehensive features and robust performance, hadge is poised to greatly enhance the accuracy and efficiency of single-cell genomics research.
Implementation of the hadge pipeline
The hadge pipeline, implemented in Nextflow, provides hashing- and genotype-based deconvolution workflows. Both workflows support the execution of multiple methods simultaneously.
The genotype-based deconvolution workflow includes five deconvolution methods: Demuxlet [ 17 ], Freemuxlet [ 26 ], Vireo [ 22 ], scSplit [ 20 ], and Souporcell [ 21 ].
The hashing-based deconvolution workflow includes seven hashing deconvolution algorithms: HTODemux [ 27 ], Multiseq [ 12 ], HashedDrops [ 28 ], Demuxem [ 15 ], gmm-demux [ 29 ], BFF [ 24 ], and Hashsolo [ 30 ], and one doublet-detection method (Solo [ 30 ]).
In addition to the two multiplexing workflows, the hadge pipeline includes a doublet detection method, Solo, which is based on a semi-supervised deep learning approach. Since Solo only identifies singlets without revealing the true donor identity of the droplets, we only use it as a supplementary method.
As genotype-based deconvolution techniques rely on SNPs to distinguish samples in the pools, the workflow also includes a preprocessing component with samtools, Freebayes [ 40 ] and cellsnp-lite [ 41 ] as two separate processes for variant calling. The Freebayes process is designed as per the instruction of scSplit ( https://github.com/jon-xu/scSplit ) to find variants in pooled samples. To optimize runtime, the process is carried out separately for each chromosome. With an additional filtering step, SNPs with a minimum allele count of 2, minimum base quality of 1 and quality scores greater than 30 from each chromosome are retained and merged. As suggested by Vireo, the Mode 1a of cellsnp-lite is called in the cellsnp-lite process to genotype single cells against candidate SNPs. Two allele count matrices for each given SNP are generated, one for the reference and another one for the alternative allele, which can be then fed into Vireo.
The hashing-based deconvolution workflow also has a pre-processing step to prepare the input data for both HTODemux and Multiseq based on the Seurat vignette ( https://satijalab.org/seurat/articles/hashing_vignette.html ). A Seurat object is initialized with the cell containing barcodes for the RNA matrix and HTO raw count matrix. Only the cell barcodes that are at the intersection between RNA and HTO counts are retained. The HTO data is added as an independent assay and normalized using centered log-ratio transformation (CLR).
The hadge pipeline features three distinct modes: genetic, hashing , and rescue mode. The hashing and genetic mode are two independent workflows, and the rescue mode allows joint demultiplexing by combining the outputs of the two workflows. Different inputs are required for the two workflows, specifically:
For the hashing workflow, raw and filtered HTO and RNA counts are the minimum required input. Each of these outputs is normally generated by the cellranger pipeline, which outputs the required HTO and RNA counts in the unfiltered (raw) and filtered feature-barcode matrices in two file formats: the Market Exchange Format (MEX), and Hierarchical Data Format (HDF5). Hadge accepts the files in the MEX format.
For the genetic workflow, the minimal requirements are as follows: the indexed sequence alignment file in BAM format along with its index (.BAI format), the barcodes of the cell-containing droplets in a TSV file, the number of expected donors in a mixture, and the reference genotypes and the variants present in the pooled sample, both in VCF format. The VCF of the reference genotype can be an unrelated genomic reference to run methods in “genotype-free” mode. Optionally, if the pooled sample’s VCF is not available, we include two processes for variant calling (cellsnp and freebayes). Users can provide the mixed FASTA file to be used as input to generate the VCF file with freebayes, which is the default preprocessing for the scSplit method. All of the inputs, except for the reference VCF files, are commonly generated by the cellranger pipeline. Following deconvolution in each workflow, the output files are passed to the summary process to generate summary files. Within this module, three CSV files are produced per tool as output, with each column representing a trial conducted during a single run of the pipeline. These output files provide a comprehensive summary of three aspects, including the specified parameters for each trial, the classification of individual droplets as singlets, doublets, or negative droplets, and the assignment of cell barcodes to their respective donors. As multiple tools are executed within a single run, additional CSV files are generated to merge the classification and assignment results from different tools into unified data frames.
In the rescue mode, hashing and genotype-based deconvolution workflows work jointly with the aim (i) to recover the droplets where the classification is discordant between the two approaches and (ii) optionally to extract donor-specific variants from the droplets with coherent classification and to reconstruct donor genotypes for mixed samples, which can then be used to rerun genotyped-based deconvolution as a sanity check to prove whether the result is reliable. The pipeline first runs the two workflows in parallel and saves the results of all methods in a single CSV file. Next, the file is passed to the “donor matching” process which computes a score (Phi score) to associate an identity to the anonymous donors using the droplets where the concordance between one genetic donor and one hashtag is maximized.
The process converts the assignment of two tools into a matrix of binary values, with rows representing cell barcodes and columns representing donors or hashtags. The value is set to 1 if the cell is assigned to the donor or hashtag, and 0 otherwise. The similarity between two matrices is calculated column-by-column using Pearson correlation, and hashtags and donors are matched if they have the highest mutual Pearson correlations. If every donor is paired with a hashtag, the pipeline generates a new assignment of the tools with mapped donors and a heat map to visualize the correlation between the donors and hashtags. If Vireo is the optimal genotype-based deconvolution method in donor matching, the process has the option to extract informative variants from donor genotypes estimated by Vireo. Using the input of cellsnp-lite, genotyped SNPs are first filtered based on the SNPs (read depth > 10) among cells with consistent assignment between Vireo and the hashing tool with which it is compared. Only variants with an overrepresented allele are retained, i.e., the frequency of the alternative or reference allele in the group of cells must be greater than a specified threshold (frequency > 90%). The pipeline compares the genotypes of these variants in cells that have been inconsistently deconvolved and keeps only the SNPs that have the same overrepresented allele in cells with and without consistent assignment. These are candidate variants used to distinguish cells from different donors. The process is performed separately on cells from different donors to retrieve donor-specific informative variants. Finally, BCFtools sorts and indexes the donor genotype from Vireo and filters the donor-specific variants. The samples are renamed by the matching hashtags.
Demuxlet/Freemuxlet
Dsc-pileup, Demuxlet, and Freemuxlet implemented in popscle (v0.1) were performed one after another. Using the BAM file and filtered barcode file produced by cellranger [ 42 ] as input, dsc-pileup aggregated reads around common SNPs in the human population, which in the case of Freemuxlet are derived from the 1000 Genomes Project and filtered by cellsnp-lite with minor allele frequency (MAF) > 0.05 as reference variant sites ( https://sourceforge.net/projects/cellsnp/files/SNPlist/ ). Demuxlet/Freemuxlet then uses the pileup files from dsc-pileup to deconvolve cells. We ran these methods in default mode.
Cell genotypes were generated at common SNPs from the 1000 Genomes Project ( https://sourceforge.net/projects/cellsnp/files/SNPlist/ ) using cellsnp-lite (v1.2.2) with default parameters before performing Vireo. Subsequently, the output of cellsnp-lite was processed by Vireosnp (v0.5.6) to perform the deconvolution with default parameters.
Souporcell (v2.0) was run on the BAM file and filtered barcode file produced by cellranger and the human reference ( http://cf.10xgenomics.com/supp/cell-exp/refdata-cellranger-GRCh38-3.0.0.tar.gz ). We also used common SNPs from the 1000 Genomes Project [ 43 ] with a minor allele frequency of 2% (provided by https://github.com/wheaton5/souporcell ) as input to skip repeated and memory-intensive steps, remapping, and variant-calling.
scSplit was executed only after the pre-processing and variant calling modules were completed. The input BAM file was pre-processed by SAMtools (v1.15.1) and UMI-tools (v1.1.2). In the variant calling module, freebayes (v1.2) was performed on the pre-processed BAM file to call variants from mixed samples. Taking the pre-processed BAM file and called variants, scSplit (v1.0.8.2) deconvolved the cell mixture in three steps. The count command of scSplit constructed two count matrices for the reference and alternative alleles. To increase the accuracy of donor identification, a list of common SNPs provided by scSplit ( https://melbourne.figshare.com/articles/dataset/Common_SNVS_hg38/17032163 ) was used to filter the count matrices. The run command identified the cells in the pool to the clusters according to the allele matrices, with doublets being assigned to a separate cluster. Finally, the genotype command predicted individual genotypes for every cluster.
HTODemux begins with loading the Seurat object, which was created during the pre-processing module using the Seurat R package (v4.3.0). HTODemux (also included in Seurat R package v4.3.0) was then called with default parameters to deconvolve cells based on clr-normalized HTO counts.
Similar to HTODemux, MULTIseqDemux (included in Seurat R package v4.3.0) function was performed on the pre-processed Seurat object, with default parameters allowing for automatic determination of the optimal quantile to use in a range from 0.1 to 0.9 by a step of 0.05.
We used Demuxem with default settings. The raw RNA and HTO count data were loaded as a MultimodalData object (pegasuspy Python package v1.7.1). Demuxem then deconvolved cells with at least 100 expressed genes and 100 UMIs in two main steps. The antibody background was first determined based on empty barcodes using the KMeans algorithm. The signal hashtag counts were then calculated using background information, and cells with a minimum signal of 10 were assigned to their signal hashtag.
The process expects to start from the raw HTO counts in hdf5 file format into an Anndata object (Scanpy v1.9.1) (solo-sc v1.3). We ran Hashsolo with default parameters, setting the priors for the hypothesis of negative droplets, singlets, and doublets each to 1/3.
HashedDrops
This process requires as input both RNA and HTO raw counts. First, emptyDrops finds cell-containing droplets, this list of barcodes is then used as input to the HashedDrops call (both algorithms are included in DropletUtils R package v1.18.0). We used HashedDrops with default settings.
BFF accepts raw or preprocessed HTO data, while offering a preprocessing step (ProcessCountMatrix), included in the CellHashR pipeline (CellHashR v.1.14.0). Two different alternatives of BFF are available, “BFF raw” and “BFF cluster,” which apply different processing on the HTO raw counts. Both methods can be run in parallel and the tool will generate a consensus call between the two. We ran both alternatives for the benchmark.
GMM-demux (GMM-demux Python package v.0.2.1.3) expects the HTO raw counts as csv or tsv files and the names of the expected cell hashtags. We ran GMM-demux using tsv files under default parameters.
Benchmarking
Mpxms-dataset.
We were granted early access to a dataset generated in a study of progressive multiple sclerosis (Calliope Dendrou, University of Oxford) [ 35 ]. In brief, this dataset includes a multiplexed 3′ single nuclear RNA sequencing dataset of brain tissue from 5 controls and 5 cases of progressive multiple sclerosis (mpxMSdataset). The mpxMS-dataset is divided into two sequencing batches (gx12 and gx38) of 6 donors each, with the individual donors hashed with one of six unique TotalSeq™-A anti-nuclear pore complex antibodies. We obtained the count data generated with Cellranger v3.1.0: 6,794,833 barcodes and 6,794,880 barcodes were detected in the raw data of gx12 and gx38, respectively. The number of cells detected in each experiment before deconvolution was 4889 for gx12 and 13,184 for gx38.
The pipeline was applied to the mpxMS-dataset in the rescue mode. In the genotyped-based deconvolution workflow, Freemuxlet, Vireo, Souporcell, and scSplit were used in the absence of reference donor genotypes. To run the algorithm, the number of samples was set to six. All hashing-based deconvolution methods were called to deconvolute the data. All output files were gathered and passed to the corresponding summary component (R v4.2.2). The results of Vireo and Multiseq were used to map donor identities to hashtags in the donor matching component. Donor genotypes estimated by Vireo were then processed by BCFtools (v1.8). The donor-specific variants were extracted from the donor genotypes, where the cell variants were filtered by a minimal cell count of 10 and the overrepresented allele at a given SNP was then determined by a 90% cut-off.
Data analysis was performed with scanpy (v1.9.3) and scrublet (v0.2.3). Plots were generated with scanpy (v1.9.3), seaborn (v0.12.2), and matplotlib (v3.7.1).
We generally followed the recommendations given by the developers of the package ( https://scanpy.readthedocs.io/en/stable/index.html ) and have in part adjusted for this dataset and in accordance with analysis best practices [ 44 ].
For analysis, log-transformation and normalization were achieved with scanpy’s log1p() and normalize_total() function. After this, 50 PCs were generated by principal component analysis (PCA) and dimensionality reduction by UMAP was performed using scanpy’s pca() and umap() functions respectively. Cluster identification was performed using the Leiden algorithm and differential expression of the different clusters was generated using scanpy’s rank_genes_groups() function.
OneK1k-dataset
Raw oneK1K data for scRNA-seq and microarray-based genotype were retrieved from the GEO database (accession numbers GSE196735, GSE196829). Ground truth cell barcode assignment was extracted from the deposited single-cell data ( https://cellxgene.cziscience.com/collections/dde06e0f-ab3b-46be-96a2-a8082383c4a1 ). The demultiplexing was carried out by specifying the original number of donors in each pool ( https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196830 ) and using the donor-genotypes VCFs extracted from the pools bam files using cellsnp, matched against the whole population genotypes. Since the deposited single-cell data contains less cells and donors than the full demultiplexing results (981 as opposed to 1015 donors), the analysis on the percent matched donors was carried out on the 981 donors present in the demultiplexing results. Genotype imputation was performed using the TOPMED-r2 Minimac4 1.7.4 imputation tool. For scRNA-seq data, alignment was conducted using Cellranger version 6.1.1. Hadge genetic demultiplexing was applied under default settings in both genotype aware and genotype absent modes and to ensure tool comparability and data consistency; default cellsnp common variants were used. ( https://sourceforge.net/projects/cellsnp/files/SNPlist/ ).
Cell number downsampling experiments
We performed consecutive downsampling of two publicly available hashing datasets, a PBMC multiplexed sample with 4 hashtags and a total of 15,843 cells ( https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152981 ) and the test data used in the cellHashR pipeline, hereby called CR-438–21, for a total of 11,090 cells ( https://www.github.com/BimberLab/cellhashR/tree/master/tests/testdata/438-21-GEX ) which also provides raw rna data to enable running demuxem. The same dataset was also used for the run time benchmarks (see “Run time benchmarks”). For each sample, we applied random downsampling to 30–50-70% of the cell barcodes in the HTO matrix, using five different seeds to control for performance variations. The downsampled counts were then fed to the hadge hashing workflow and each method was run under default conditions. For the PBMC dataset, the ground truth labels were obtained running gmm-demux on the full sample as described in the original publication [ 29 ]. For the CR-438–21, the ground truth labels were obtained using the instructions provided on the cellhashR repository. The scripts used for downsampling are available at https://github.com/theislab/hadge-reproducibility .
Downsampling of the genetic data was performed on the first batch (gx12) of mpxMS-dataset, using as ground truth the joint deconvolution results between vireo and multiseq demonstrated in Figs. 3 and 4 . We performed random downsampling to 30% and 50% of the cell barcodes associated with each donor, using three different seeds, and evaluated the percentage matching singlets recovered after removing 30 or 50% of the cells for a particular donor across the different tested seeds. To reduce the run time, bam files were pre-processed to contain only reads that overlap with known common SNPs from the 1000 Genomes Project ( https://sourceforge.net/projects/cellsnp/files/SNPlist/ ). The downsampled reads were then used as input for hadge genetic workflow and each method was run under default conditions.
Run time benchmarks
We benchmarked the performance of hadge against demuxafy and cellhashR using four samples with different cell numbers. Each pipeline is developed in different frameworks and requires different configurations. In the demuxafy pipeline, genotype-based deconvolution methods were called sequentially within the singularity container. The benchmark was run on the mpxMS-dataset batch gx12 and gx38, the cellhashR dataset as well as a reduced test dataset provided by demuxafy, using the same parameters as hadge. Since demuxafy does not provide preprocessing functions, we used hadge’s preprocessing module ( which includes freebayes , samtools , and cellsnp ) to provide the same input data to hadge-genetic and demuxafy so the benchmarking starts from the same inputs. For the cellhashR pipeline, we created a conda environment with all the required dependencies as described in the cellhashr GitHub repository [ 45 ]. In the hadge-hashing pipeline, each deconvolution method was called in its own Conda environment separately. For each pipeline run, we allowed 160-GB RAM memory and 32 CPU cores.
Availability of data and materials
The hadge source code is available at https://github.com/theislab/hadge [ 46 ] under the MIT license. We also deposited the version that we used for this manuscript to Zenodo [ 47 ]. Further documentation, tutorials and examples are available at https://hadge.readthedocs.io/en/latest .
Jupyter notebooks to reproduce our analysis and figures including Conda environments that specify all versions are available at https://github.com/theislab/hadge-reproducibility and also deposited to Zenodo [ 47 ].
The mpsMS-dataset applied in this study is an unpublished dataset obtained directly from the authors [ 35 ].
The onek1k data is publicly accessible via the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number SRX14182577 [ 48 ].
The PBMC hashing dataset is publicly available on GEO with accession number GSE152981 [ 49 ].
The CR-438–21 sample is publicly available ( https://www.github.com/BimberLab/cellhashR/tree/master/tests/testdata/438-21-GEX ).
Rood JE, Maartens A, Hupalowska A, Teichmann SA, Regev A. Impact of the Human Cell Atlas on medicine. Nat Med. 2022;28:2486–96.
Article CAS PubMed Google Scholar
Mereu E, et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat Biotechnol. 2020;38:747–55.
Van Phan H, et al. High-throughput RNA sequencing of paraformaldehyde-fixed single cells. Nat Commun. 2021;12:5636.
Article CAS PubMed PubMed Central Google Scholar
Datlinger P, Rendeiro AF, Boenke T, Senekowitsch M. Ultra-high-throughput single-cell RNA sequencing and perturbation screening with combinatorial fluidic indexing. Nat Methods. 2021;18(6):635–42.
Xu Z, et al. High-throughput single nucleus total RNA sequencing of formalin-fixed paraffin-embedded tissues by snRandom-seq. Nat Commun. 2023;14:2734.
Brown DV, et al. A risk-reward examination of sample multiplexing reagents for single cell RNA-Seq. Genomics. 2024;116:110793.
Slyper M, et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med. 2020;26:792–802.
Cheng J, Liao J, Shao X, Lu X, Fan X. Multiplexing methods for simultaneous large-scale transcriptomic profiling of samples at single-cell resolution. Adv Sci. 2021;8:e2101229.
Article Google Scholar
Yazar S, et al. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science. 2022;376:eabf3041.
Perez RK, et al. Single-cell RNA-seq reveals cell type-specific molecular and genetic associations to lupus. Science. 2022;376:eabf1970.
Stoeckius M, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224.
McGinnis CS, et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods. 2019;16:619–26.
Gehring J, Hwee Park J, Chen S, Thomson M, Pachter L. Highly multiplexed single-cell RNA-seq by DNA oligonucleotide tagging of cellular proteins. Nat Biotechnol. 2020;38:35–8.
Mylka V, et al. Comparative analysis of antibody- and lipid-based multiplexing methods for single-cell RNA-seq. Genome Biol. 2022;23:55.
Gaublomme JT, et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat Commun. 2019;10:2907.
Article PubMed PubMed Central Google Scholar
Howitt, G. et al. Benchmarking single-cell hashtag oligo demultiplexing methods. bioRxiv 2022.12.20.521313 (2022) doi: https://doi.org/10.1101/2022.12.20.521313 .
Kang HM, et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018;36:89–94.
Li, L. et al. A hybrid single cell demultiplexing strategy that increases both cell recovery rate and calling accuracy. bioRxiv (2023) doi: https://doi.org/10.1101/2023.04.02.535299 .
Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573-3587.e29.
Xu J, et al. Genotype-free demultiplexing of pooled single-cell RNA-seq. Genome Biol. 2019;20:290.
Heaton H, et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat Methods. 2020;17:615–20.
Huang Y, McCarthy DJ, Stegle O. Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol. 2019;20:273.
Neavin, D. et al. Demuxafy: Improvement in droplet assignment by integrating multiple single-cell demultiplexing and doublet detection methods. bioRxiv 2022.03.07.483367 (2022) doi: https://doi.org/10.1101/2022.03.07.483367 .
Boggy GJ, et al. BFF and cellhashR: analysis tools for accurate demultiplexing of cell hashing data. Bioinformatics. 2022;38:2791–801.
Di Tommaso P, et al. Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35:316–9.
Article PubMed Google Scholar
Zhang, F. Leveraging genetic variants for rapid and robust upstream analysis of massive sequence data. (2019). [Doctoral Dissertation, University of Michigan] https://deepblue.lib.umich.edu/handle/2027.42/151524 .
Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–8.
Lun ATL, et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20:63.
Xin H, et al. GMM-Demux: sample demultiplexing, multiplet detection, experiment planning, and novel cell-type verification in single cell sequencing. Genome Biol. 2020;21:188.
Bernstein NJ, et al. Solo: doublet identification in single-cell RNA-seq via semi-supervised deep learning. Cell Syst. 2020;11:95-101.e5.
Howitt G, et al. Benchmarking single-cell hashtag oligo demultiplexing methods. NAR Genom Bioinform. 2023;5(4):lqad086.
Cardiello JF, et al. Evaluation of genetic demultiplexing of single-cell sequencing data from model species. Life Sci Alliance. 2023;6(8):e202301979.
Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15.
Bredikhin D, Kats I, Stegle O. MUON: multimodal omics analysis framework. Genome Biol. 2022;23:42.
Grant-Peters, M. et al. Biochemical and metabolic maladaption defines pathological niches in progressive multiple sclerosis. bioRxiv 2022.09.26.509462 (2022) doi: https://doi.org/10.1101/2022.09.26.509462 .
Jäkel S, et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature. 2019;566:543–7.
Schirmer L, et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature. 2019;573:75–82.
Virshup I, et al. The scverse project provides a computational ecosystem for single-cell omics data analysis. Nat Biotechnol. 2023. https://doi.org/10.1038/s41587-023-01733-8 .
Rich-Griffin, C. et al. Panpipes: a pipeline for multiomic single-cell data analysis. bioRxiv 2023.03.11.532085 (2023) doi: https://doi.org/10.1101/2023.03.11.532085 .
Garrison, E., & Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907.
Huang X, Huang Y. Cellsnp-lite: an efficient tool for genotyping single cells. Bioinformatics. 2021;37:4569–71.
Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049.
1000 Genomes Project Consortium, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 2023;24:550-72.
cellhashR: an R package designed to demultiplex cell hashing data. Github. 2024. https://www.github.com/BimberLab/cellhashR .
Curion, F. et al. 2024. hadge: a comprehensive pipeline for donor deconvolution in single cell studies Github. 2024. https://www.github.com/theislab/hadge .
Curion, F. et al. hadge: a comprehensive pipeline for donor deconvolution in single cell studies. Zenodo. 2024. https://www.zenodo.org/records/10891138 .
Yazar, S. et al. Single-cell eQTL mapping identifies cell type–specific genetic control of autoimmune disease. Datasets. Sequence Read Archive. 2024. https://www.identifiers.org/ncbi/insdc.sra:SRX14182577 .
Xin, H. et al. GMM-Demux: sample demultiplexing, multiplet detection, experiment planning and novel cell type verification in single cell sequencing. Dataset. Gene Expression Omnibus. 2020. https://www.identifiers.org/geo:GSE152981 .
Download references
Acknowledgements
We thank Luke Zappia for constructive discussion on the design of the pipeline and the overarching project. We are grateful for Lisa Sikkema’s input on the figure design.
A subset of figure panels was created using Biorender.
Review history
The review history is available as Additional file 4 .
Peer review information
Veronique van den Berghe was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Open Access funding enabled and organized by Projekt DEAL. FC acknowledges funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) –SFB- TRR 338/1 2021 –452881907. HBS acknowledges support from the German Center for Lung Research (DZL), the Helmholtz Association (CoViPa—lessons to get prepared for future pandemics), the European Union’s Horizon 2020 research and innovation program (grant agreement 874656—project discovair), and the Chan Zuckerberg Initiative (CZF2019-002438, project Lung Atlas 1.0). MG-P was supported by the Interdisciplinary Bioscience DTP (BBSRC). CAD was supported by the Wellcome Trust and Royal Society (204290/Z/16/Z). Tissue samples and associated clinical and neuropathological data were supplied by the Multiple Sclerosis Society Tissue Bank, funded by the Multiple Sclerosis Society of Great Britain and Northern Ireland, registered charity 207495. MO is supported by the Wellcome Trust grant [206194].
Author information
Fabiola Curion, Xichen Wu, and Lukas Heumos contributed equally to this work.
Authors and Affiliations
Institute of Computational Biology, Computational Health Center, Helmholtz Munich, Neuherberg, Germany
Fabiola Curion, Xichen Wu, Lukas Heumos, Mylene Mariana Gonzales André, Lennard Halle & Fabian J. Theis
Department of Mathematics, School of Computation, Information and Technology, Technical University of Munich, Garching, Germany
Fabiola Curion, Xichen Wu, Mylene Mariana Gonzales André & Fabian J. Theis
Nuffield Department of Medicine, Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK
Melissa Grant-Peters, Charlotte Rich-Griffin, Hing-Yuen Yeung & Calliope A. Dendrou
Comprehensive Pneumology Center, German Center for Lung Research (DZL), Munich, Germany
Lukas Heumos & Herbert B. Schiller
Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, The Kennedy Institute of Rheumatology, University of Oxford, Oxford, UK
Calliope A. Dendrou
TUM School of Life Sciences Weihenstephan, Technical University of Munich, Freising, Germany
Lukas Heumos & Fabian J. Theis
Wellcome Sanger Institute, Hinxton, UK
Matiss Ozols
School of Cell Matrix and Regenerative Medicine, The University of Manchester, Manchester, UK
Research Unit Precision Regenerative Medicine, Helmholtz Munich, Neuherberg, Germany
Herbert B. Schiller
Institute of Experimental Pneumology, LMU University Hospital, Ludwig-Maximilians University, Munich, Germany
You can also search for this author in PubMed Google Scholar
Contributions
FC, XW, and LH contributed equally and have the right to name themselves first in their CV. FC and LH conceived the study. FC, XW, MG, and LH implemented the hadge pipeline. XW, FC, MO, and MG conducted the benchmarking of the tools and the downsampling analyses. FC, XW, and LeH conducted the analysis of the data. HBS and FJT supervised the work. All authors read, corrected, and approved the final manuscript.
Corresponding author
Correspondence to Fabian J. Theis .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
L.H. is an employee of Lamin Labs. F.J.T. consults for Immunai Inc., Singularity Bio B.V., CytoReason Ltd, and Omniscope Ltd and has ownership interest in Dermagnostix GmbH and Cellarity.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:.
Supplementary figures.
Additional file 2:
Supplementary table containing the donor assignments per dataset and tool.
Additional file 3:
Supplementary tables.
Additional file 4:
Review History.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Curion, F., Wu, X., Heumos, L. et al. hadge: a comprehensive pipeline for donor deconvolution in single-cell studies. Genome Biol 25 , 109 (2024). https://doi.org/10.1186/s13059-024-03249-z
Download citation
Received : 08 August 2023
Accepted : 16 April 2024
Published : 26 April 2024
DOI : https://doi.org/10.1186/s13059-024-03249-z
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Single-cell
- Donor deconvolution
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
Acknowledgement Sample For Assignment. I wish to express my deepest gratitude to Mr./Ms ______ (Professor name), for providing me the chance to work on this assignment and for having faith in me. Thanks to their invaluable feedback and their unwavering encouragement, I was able to stay motivated and produce my best work.
Acknowledgement for Assignment Example - Individual School Assignment. First of all, I would like to thank God, who has always been my guidance and support all these years. I would like to thank my Class Teacher, Mrs. Sindhu Krishna, for giving me the opportunity to work on this assignment and for believing in me.
Example 1: Acknowledgement. I would like to express my sincere gratitude to my supervisor [Name], for her invaluable guidance, support and encouragement throughout the course of this assignment. Her insights and expertise greatly enhanced the quality of my work. I would also like to thank [Name], [Name], and [Name] for their assistance and ...
Acknowledgement is a pivotal aspect of any assignment, showcasing gratitude towards those who have contributed to its completion. Acknowledgements are often overlooked or deemed unimportant, but they play a significant role in giving credit where it is due. Whether it be mentors, classmates, or even friends and family, everyone deserves ...
Sample Acknowledgement for Assignment 1 - Individual Assignment. Andheri, Mumbai. 8/06/2022. In making this project, and completing it successfully, I had to get help and guidance from some respected people. I am grateful that I was given the opportunity to make this project, which has enhanced my knowledge in so many aspects.
41 Best Acknowledgement Samples & Examples. Not all professionals do their work by themselves. Although they can be as prolific or as adept in their respective fields, they will still need assistance one way or another. For instance, writing a body of work takes a lot of research. They often depend on their assistants or subordinates to gather ...
The acknowledgements section is your opportunity to thank those who have helped and supported you personally and professionally during your thesis or dissertation process. Thesis or dissertation acknowledgements appear between your title page and abstract and should be no longer than one page. In your acknowledgements, it's okay to use a more ...
Writing the acknowledgements section of your thesis or dissertation is an opportunity to express gratitude to everyone who helped you along the way. Remember to: Acknowledge those people who significantly contributed to your research journey. Order your thanks from formal support to personal support. Maintain a balance between formal and ...
An acknowledgement is a page is where you show appreciation to people who helped or supported you intellectually, mentally, or financially in your academic writing. ... Acknowledgement for Yoga Project [3 Samples] ... [5 Examples] Acknowledgement for College/School Assignment [5 Examples] Acknowledgemet to God in Reports [5 Examples] ...
There are several ways you could word your thanks. A few suggestions are: This research would not have been possible without…. My sincere thanks go to…. I am grateful to…. Heartfelt thanks to…. I would like to express my gratitude to…. Appreciation is due to…. I acknowledge the contribution of….
20 Best Acknowledgement Samples and Templates. Most published works include an Acknowledgement page that thanks to the people who helped the author put the book, thesis, or presentation together. Similarly, it is vital to acknowledge such people when something you write is published or recognized publicly. Doing this will go a long way in ...
Writing Tips For Acknowledgement Samples. When it comes to writing an acknowledgement, it is important to be clear, concise and to the point. The following are some tips that will help you write acknowledgement samples that will properly thank those who have helped you along the way: Make sure to list everyone who has helped you.
How to Write an Acknowledgement for College. When you want to write an acknowledgment page, it is essential to take the following steps to keep things narrowed down: Step 1: Use an appropriate tone. Depending on whether you want to thank a college professor in a typical term paper or a dissertation, your writing form may be different.
Sample 1 - Acknowledgement for Individual Project. I would like to convey my heartfelt gratitude to Mrs. Debjani Nanda for her tremendous support and assistance in the completion of my project. I would also like to thank our Principal, Mrs.Jyothi Kumar, for providing me with this wonderful opportunity to work on a project with the topic Food ...
help you write your Acknowledgements section of your dissertation. According to one source, the Acknowledgements section of a Ph.D. dissertation is the most widely read section. Whether you believe this or not, many individuals who helped you in the process of writing may check to see if, indeed, they have meant something to you.
An acknowledgement is a page is where you show appreciation to people who helped or supported you intellectually, mentally, or financially in your academic writing. ... [5 Examples] Acknowledgement for College/School Assignment [5 Examples] Acknowledgemet to God in Reports [5 Examples] ... Thank you so much for the samples you have provided, it ...
Example 2. I would like to thank my supervisors Dr. XXX and Dr. XXX for all their help and advice with this PhD. I would also like to thank my sisters, whom without this would have not been possible. I also appreciate all the support I received from the rest of my family. Lastly, I would like to thank the XXX for the studentship that allowed me ...
The people and groups who contributed should be properly credited for their specific roles. Phrase the acknowledgement statements clearly and concisely. Keep them simple, sincere and specific without overstating contributions. Make sure to get prior consent from everyone being acknowledged.
Begin your acknowledgements by expressing gratitude to those who have made the most significant contributions to your research. This could be your academic advisors, supervisors, or funding bodies. Starting with the most significant contributions helps to set the tone for the rest of your acknowledgements. Ensure that you express your gratitude ...
An example of acknowledgment for a group assignment could be: "We would like to express our gratitude to each member of the group—Sarah, David, and Emily —for their dedication and collaborative efforts in completing this assignment. Each contribution was vital to our collective success.".
Sample For Assignment; Sample For Thesis; Acknowledgement For Project. ... I've got you covered. In this blog post, I'll share 100+ Samples of acknowledgements for projects, so you can find the perfect inspiration for your own. Whether you are writing an acknowledgement for a college project, a research project, a group project, ...
Sample acknowledgement for a social science project. February 14, 2023 expert Acknowledgement sample for assignment, Acknowledgement sample for project. We have been asked by many of you to provide some samples of acknowledgement for a social science project. In the most of the schools now, it is required to include acknowledgement with project ...
How To Write Your Acknowledgments Section. 1. Remember: people will read this. People will read the Acknowledgments section, and it will impact them—especially the people who are in it. This section is about those people you are naming, not about you, so approach this as you should your entire book: make it good for the people you are naming ...
Comparison of the performance of donor deconvolution methods. A The violin plot of raw HTO counts shows high count levels of Hashtag 453 in cells with noisy or undetectable expression of the other HTOs.B UMAP visualization of normalized HTO counts colored by HTODemux assignment shows poor separation of the cells based on hashtags, with most droplets assigned to Hashtag 453.