- Search Menu
- Advance Articles

Editor's Choice
- Information for authors
- Submission Site
- Open Access Options
- Why publish with the journal
- About DNA Research
- About the Kazusa DNA Research Institute
- Editorial Board
- Advertising and Corporate Services
- Journals Career Network
- Self-Archiving Policy
- Dispatch Dates
- Journals on Oxford Academic
- Books on Oxford Academic

Editor-in-Chief
Satoshi Tabata
About the journal
DNA Research is an internationally peer-reviewed journal which aims at publishing papers of highest quality in broad aspects of DNA and genome-related research …
Latest Articles

High-Impact Research Collection
Explore a collection of freely available high-impact research from 2020 and 2021 published in DNA Research .
Browse the collection

DNA Research is the official journal of Kazusa DNA Research Institute, published by Oxford University Press and supported by funding from Chiba Prefecture, Japan.

Why publish in DNA Research?
Growing Impact Factor, fully open access journal, low open access charges, and more.

Volume 26, Issue 6: TASUKE+: a web-based platform for exploring GWAS results and large-scale resequencing data
Read the Executive Editor’s commentary
Resource Articles: Genomes Explored

Email alerts
Register to receive table of contents email alerts as soon as new issues of DNA Research are published online.

Recommend to your library
Fill out our simple online form to recommend DNA Research to your library.
Recommend now

Committee on Publication Ethics (COPE)
This journal is a member of and subscribes to the principles of the Committee on Publication Ethics (COPE)
publicationethics.org

PubMed Central
This journal enables compliance with the NIH Public Access Policy Read more

- Open access
Open access options for authors.

Accepting high quality papers on broad aspects of DNA and genome-related research.
Related Titles

- Author Guidelines
Affiliations
- Online ISSN 1756-1663
- Copyright © 2024 Kazusa DNA Research Institute
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
Recombinant DNA technology and DNA sequencing
Affiliation.
- 1 Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, United Kingdon.
- PMID: 31652313
- DOI: 10.1042/EBC20180039
DNA present in all our cells acts as a template by which cells are built. The human genome project, reading the code of the DNA within our cells, completed in 2003, is undoubtedly one of the great achievements of modern bioscience. Our ability to achieve this and to further understand and manipulate DNA has been tightly linked to our understanding of the bacterial and viral world. Outside of the science, the ability to understand and manipulate this code has far-reaching implications for society. In this article, we explore some of the basic techniques that enable us to read, copy and manipulate DNA sequences alongside a brief consideration of some of the implications for society.
Keywords: CRISPR; DNA sequencing; biochemical techniques and resources.
© 2019 The Author(s).
Publication types
- Cloning, Molecular / methods
- DNA / genetics*
- DNA / isolation & purification
- DNA, Recombinant / genetics*
- Electrophoresis, Agar Gel / methods
- Genetic Testing*
- Genetic Vectors / genetics
- Plants, Genetically Modified / genetics*
- Polymerase Chain Reaction / methods
- DNA, Recombinant
- Reference Manager
- Simple TEXT file
People also looked at
Mini review article, past, present, and future of dna typing for analyzing human and non-human forensic samples.

- 1 Department of Biological Sciences, Florida International University, Miami, FL, United States
- 2 International Forensic Research Institute, Florida International University, Miami, FL, United States
Forensic DNA analysis has vastly evolved since the first forensic samples were evaluated by restriction fragment length polymorphism (RFLP). Methodologies advanced from gel electrophoresis techniques to capillary electrophoresis and now to next generation sequencing (NGS). Capillary electrophoresis was and still is the standard method used in forensic analysis. However, dependent upon the information needed, there are several different techniques that can be used to type a DNA fragment. Short tandem repeat (STR) fragment analysis, Sanger sequencing, SNapShot, and capillary electrophoresis-single strand conformation polymorphism (CE-SSCP) are a few of the techniques that have been used for the genetic analysis of DNA samples. NGS is the newest and most revolutionary technology and has the potential to be the next standard for genetic analysis. This review briefly encompasses many of the techniques and applications that have been utilized for the analysis of human and nonhuman DNA samples.
Introduction
Forensic genetics applies genetic tools and scientific methodology to solve criminal and civil litigations ( Editorial, 2007 ). Locard’s Exchange Principle states that every contact leaves a trace, making any evidence a key component in forensic analysis. Biological evidence can comprise of cellular material or cell-free DNA from crime scenes, and as technologies improved, genetic methodologies were expanded to include human and non-human forensic analyses. Although these methodologies can be used for any genome, the prevalence of databases and standard guidelines has allowed human DNA typing to become the gold standard. This review will discuss the historical progression of DNA analysis techniques, strengths and limitations, and their possible forensic applications applied to human and non-human genetics.
Methodologies to Detect Genetic Differences in Humans Is the “Gold Standard”
“dna fingerprinting”: the beginning of human forensic dna typing.
“DNA fingerprinting” was serendipitously discovered in 1984 ( Jeffreys, 2013 ). What they found propelled DNA “fingerprinting,” or DNA typing, to the forefront in legal cases to become the “gold standard” for forensic genetics in a court of law. Jeffreys first used restriction enzymes to fragment DNA, a method in which restriction endonucleases (RE) enzymes fragment the genomic DNA, producing restriction fragment length polymorphisms (RFLP) patterns. Since each RE recognizes specific DNA sequences to enzymatically cut the DNA, then inherent differences between gene sequences, due to evolutionary changes, will produce different fragment lengths. If the enzyme site is present in one individual but has changed in a different individual, the fragment lengths, once separated and visualized, will differ. While this technique was useful for some studies, Jeffreys did not find it useful for his particular genetic studies. Subsequently when working with the myoglobin gene in seals, he discovered that a short section of that gene – a minisatellite – was conserved and when isolated and cloned could be used to detect inherited genetic lineages as well as individualize a subject. Fragment length separation by electrophoresis, followed by transfer to Southern blot membranes, hybridized with a specific or non-specific complementary isotopic DNA probe, allowed for DNA fragments visualization ( Jeffreys et al., 1985b ). Upon careful analysis, Jeffreys determined that the fragments represented different combinations of DNA repetitive elements, unique to each individual, and could be used to better identify individuals or kinship lineages ( Jeffreys et al., 1985b ). Jeffreys’ technology was used in several subsequent paternity, immigration, and forensic genetics cases ( Gill et al., 1985 ; Jeffreys et al., 1985a ; Evans, 2007 ). This was just the beginning of a whole new era in DNA typing.
Restriction Fragment Length Polymorphism (RFLP) Analysis: The Past
After Jeffreys’ discoveries, many DNA analyses methods involving electrophoretic fragment separation were discovered. Many were based on RFLP principles ( Botstein et al., 1980 ), e.g., amplified fragment length polymorphism (AFLP) ( Vos et al., 1995 ), and terminal restriction fragment length polymorphism (TRFLP) ( Liu et al., 1997 ). Others like length heterogeneity- polymerase chain reaction (LH-PCR) ( Suzuki et al., 1998 ) were based on intrinsic insertions and deletions of bases within specific genetic markers. Sanger sequencing ( Sanger and Coulson, 1975 ), and single-strand conformational polymorphism (SSCP) analysis ( Orita et al., 1989 ), while separated by electrophoresis, are theoretically based on single base sequence changes rather than insertions, deletions or RE site differences. While Jeffrey’s DNA fingerprinting method provided a very high power of discrimination, the main limitations were it was very time-consuming and required at least 10–25 ng of DNA to be successful ( Wyman and White, 1980 ). With these limitations, RFLP was not always feasible for forensic cases.
Short Tandem Repeat (STR) Analysis: The Present
The polymerase chain reaction (PCR) was discovered by Kary Mullis in 1985 and helped transform all DNA analyses ( Mullis et al., 1986 ). The current standard for human DNA typing is short tandem repeat (STR) analysis ( McCord et al., 2019 ). This method amplifies highly polymorphic, repetitive DNA regions by PCR and separates them by amplicon length using capillary electrophoresis. These inheritable markers are a series of 2–7 bases tandemly repeated at a specific locus, often in non-coding genetic regions. Forensic STRs are commonly tetranucleotide repeats ( Goodwin et al., 2011 ), chosen because of their technical robustness and high variation among individuals ( Kim et al., 2015 ). The combined DNA index system (CODIS) uses 20 core STR loci, expanded in 2017, and several commercial kits are available that contain these STRs ( Oostdik et al., 2014 ; Ludeman et al., 2018 ). After amplification, different fluorochromes on each primer set allow for visualization of STRs after deconvolution, creating a STR profile consisting of a combination of genotypes ( Gill et al., 2015 ). This method has become the gold standard for human forensics. Its greatest strength is the standardization of loci used by all laboratories and an extremely large searchable database of genetic profiles. However, some limitations and challenges are faced when dealing with highly degraded or low template DNA samples. To overcome these technical challenges, standardized mini-STR kits have been developed which use shorter versions of the core STRs and can be used in the same manner for forensic cases ( Butler et al., 2007 ; Constantinescu et al., 2012 ). Keep in mind, DNA typing of humans – a single species – is the gold standard because of (a) the concerted scientific effort to standardize loci to analyze, (b) the development of commercial kits that can produce the same results regardless of instrumentation or laboratory performing the work, (c) a compatible and very large database that provides allelic frequencies for all sub-populations of humans, (d) standardized statistical methods used to report the results and (e) many court cases that have accepted human DNA typing evidence in a court of law – setting the precedent for future cases to use DNA typing results.
Methodologies to Detect Genetic Differences in Non-Humans: Past and Present
Amplified fragment length polymorphism (aflp) analysis.
It was not long before scientists realized that non-human DNA could provide informative genetic evidence in forensic cases. Applications include bioterrorism, wildlife crimes, human identification through skin microorganisms, and so much more ( Arenas et al., 2017 ). Since large quantities of biological materials are frequently not found at crime scenes, successful RFLP analyses were unlikely. Combining restriction enzymes and PCR technology, a process known as AFLP analysis ( Vos et al., 1995 ), became a method for DNA fingerprinting using minute amounts of unknown sourced DNA. REs digest genomic DNA, then ligation of a constructed adapter sequence to the ends of all fragments allows the annealing of primers designed to recognize the adaptor sequences. Subsequent amplification generates many amplicons ranging in length when separated and visualized in an electropherogram or on a gel ( Vos et al., 1995 ; Butler, 2012 ). AFLP markers for plant forensic DNA typing have been used because it provides high discrimination, requires only small amounts of DNA and the method is reproducible, all forensically important characteristics ( Datwyler and Weiblen, 2006 ). For example, since most cannabis is clonally propagated, subsequent generations will have identical genetic profiles as seen with AFLP ( Miller Coyle et al., 2003 ), providing useful intelligence links back to the source population. But there are significant variation between cultivars and within populations, so not having a standard database representing the species’ diversity for statistical comparisons greatly limits the method’s applicability. Another forensic example of its use is differentiating between marijuana and hemp, two morphologically and genetically similar plants, one an illicit drug while the other is not. In this study, three populations of hemp and one population of marijuana were analyzed with AFLP producing 18 bands that were specific to hemp samples. Additionally, 51.9% of molecular variance occurred within populations indicating these polymorphisms were useful for forensic individualization ( Datwyler and Weiblen, 2006 ).
Terminal Restriction Fragment Length Polymorphism (TRFLP) Analysis
As a result of the anthrax letter attacks of 2001, microbial forensics came to the forefront ( Schmedes et al., 2016 ), a discipline that combines multiple scientific specialties – microbiology, genetics, forensic science, and analytical chemistry. One method used to compare microbial communities is TRFLP ( Liu et al., 1997 ; Osborn et al., 2000 ; Butler, 2012 ). With this method, the DNA is amplified using “universal,” highly conserved primer sequences shared across all organisms of interest, i.e., the 16S rRNA genes in bacteria and Archaea, and then uses REs to fragment the PCR products ( Table 1 ). Separated by capillary electrophoresis, only the fluorescently tagged terminal restricted fragments are visualized ( Mrkonjic Fuka et al., 2007 ), reducing the profile complexity and providing high discrimination. TRFLP has been used to characterize complex microbial communities for forensic applications by linking the similarity of the amplicon patterns generated from the intrinsic soil communities to the evidence from a crime scene ( Meyers and Foran, 2008 ; Habtom et al., 2017 ). This method does provide a distinct pattern reflective of the microbial community, useful for forensic genetics but the method does not provide any sequence information. Another limitation is no standardization of which primer pairs or REs are used, making direct comparisons between studies difficult. This lack of standardization also hinders the development of a database for species identification. Additionally, the method is time-consuming due to the additional step of restriction digestion and the possibility of incomplete enzymatic digestion can complicate the interpretation of results ( Osborn et al., 2000 ; Moreno et al., 2006 ).
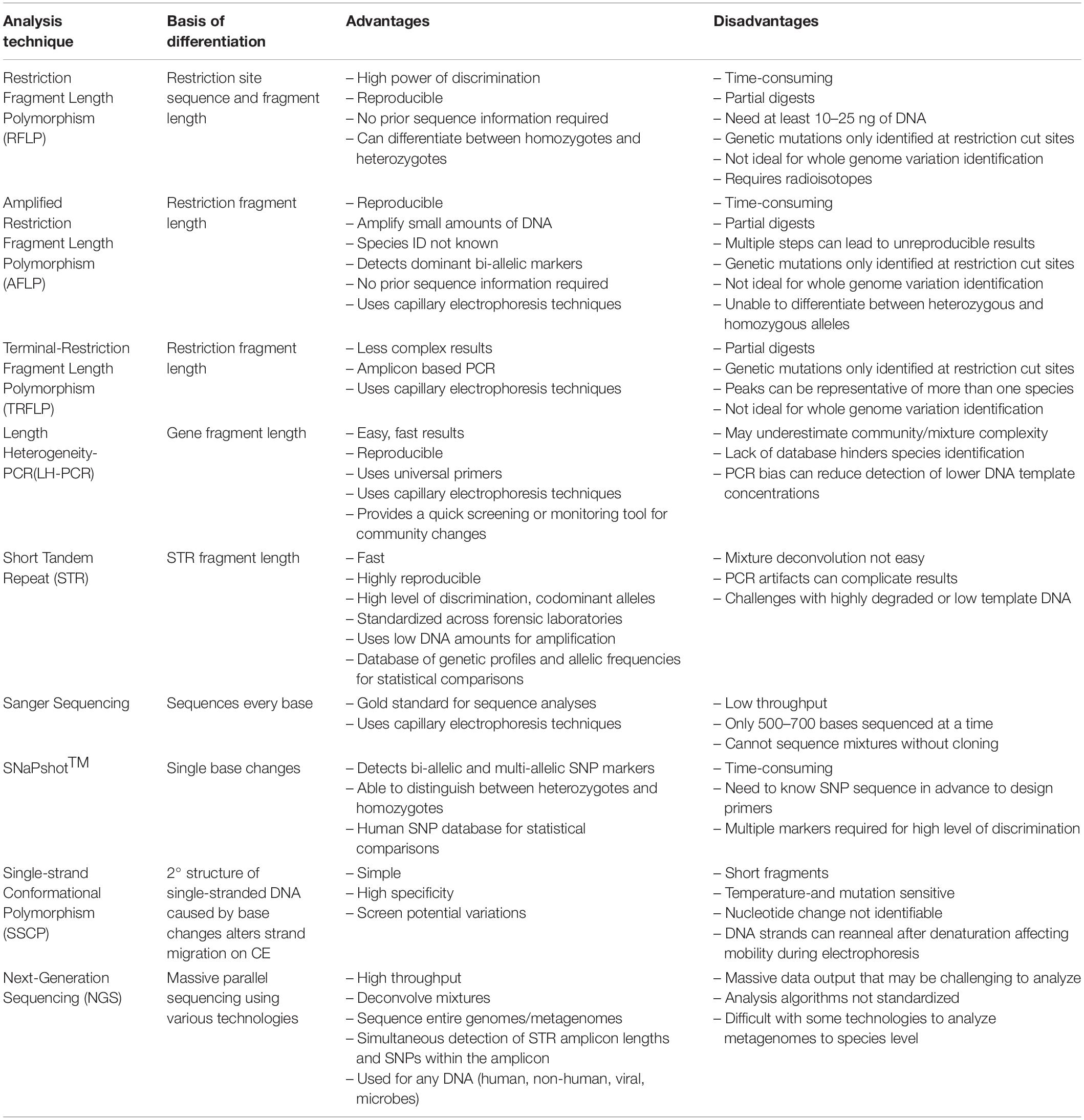
Table 1. The basis of differentiation, advantages, and disadvantages of past and current technologies.
Length Heterogeneity-Polymerase Chain Reaction (LH-PCR)
Another methodology has been used to characterize microbial communities is length heterogeneity- polymerase chain reaction (LH-PCR) ( Suzuki et al., 1998 ). Universal primers complementary to highly conserved domains within genomes are used to amplify hypervariable sequences within specific sequence domains. The 16S/18S rRNA genes, the chloroplast genes or Internal Transcribed Spacer (ITS) regions are commonly used. This technique is based on the natural sequence length variation due to insertions and deletions of bases that occur within a domain ( Moreno et al., 2006 ). It has been used to characterize microbial communities for forensic soil applications where a correlation between geographic location and microbial profiles has proven to be more discriminating than elemental soil analysis ( Moreno et al., 2006 , 2011 ; Damaso et al., 2018 ). With LH-PCR, metagenomic DNA extracted from the soil is amplified using fluorescently labeled universal primers with amplicon peaks within the electropherogram representing the minimum diversity within the community. However, specific sequence information is not known as many peaks of the same size could represent more than one species, thereby masking the community’s actual taxonomic diversity. A recent study showed the intrinsic diversity of a microbial mat, masked by LH-PCR, could be further resolved by the inherent sequence differences using capillary electrophoresis-single strand conformational polymorphism (CE-SSCP) analysis ( Damaso et al., 2014 ) and confirmed by sequencing. The advantage of LH-PCR is it is a fast and reproducible method that can correlate geographical areas to microbial patterns with bioinformatics ( Damaso et al., 2018 ); but a soil database would need to be developed to be useful beyond specific geographical areas.
Methodologies to Detect Intersequence Variation: The Past and Present
Sanger sequencing and single nucleotide polymorphism (snp) variation.
The basis of genomic differentiation is the intrinsic order of base pairs within a region that can be evaluated by sequencing. Sanger sequencing has been the gold standard since the 1970s ( Sanger and Coulson, 1975 ). Sanger sequencing was termed the gold standard because of the ability for single base pair resolution allowing for full sequence information to be determined. Robust and extensive databases are also readily available for comparison, i.e., GenBank, to identify an organism. However, it does have some limitations such as the short length (<500–700 bp) and it cannot sequence mixtures of organisms, for example, without cloning, so it would not be useful for sequencing complex microbial communities without intense time, effort and cost.
Other approaches use the ability to identify intrinsic single base sequence variation using single nucleotide polymorphisms (SNPs) within four forensically relevant SNP classes: identity-testing, ancestry informative, phenotype informative, and lineage informative. SNPs are particularly useful when typing degraded DNA or increasing the amount of genetic information retrieved from a sample ( Budowle and van Daal, 2008 ; Goodwin et al., 2011 ). SNaPshot TM is a commercially available SNP kit that can identify known SNPs using single base extension (SBE) technology ( Daniel et al., 2015 ; Fondevila et al., 2017 ). Wildlife forensics has used SNaPshot TM to identify endangered or trafficked species that are illegally poached to support criminal prosecutions. Elephant species identification from ivory and ivory products ( Kitpipit et al., 2017 ) or differentiating wolf species from dog subspecies ( Jiang et al., 2020 ) are both examples of SNaPshot TM assays developed for wildlife forensics. By using species-specific SNPs, the samples could be identified. But yet again, the limitation becomes the need for species-specific reference databases and the monumental task of developing a robust database for each species. Human SNPs databases with allele frequencies, as seen in dbSNP, however, are available making their forensic application more feasible in some cases.
Next-Generation Sequencing: The Present
Massively parallel sequencing (MPS) or next-generation sequencing (NGS) allows for mixtures of genomes of any species to be sequenced in one analysis ( Ansorge, 2009 ). This technology can sequence thousands of genomic regions simultaneously, allowing for whole-genome, metagenomic sequencing or targeted amplicon sequencing ( Gettings et al., 2016 ). Various NGS technologies are available each using slightly different technologies to sequence DNA ( Heather and Chain, 2016 ). Verogen has developed kits explicitly for human forensic genomics using Illumina’s MiSeq FGx system ( Guo et al., 2017 ; Moreno et al., 2018 ). The FBI recently approved DNA profiles generated by Verogen forensic technology to be uploaded into the National DNA Index System (NDIS) ( SWGDAM, 2019 ), making it the first NGS technology approved for NDIS.
Short tandem repeat mixture deconvolution, degraded, low template samples, and even microbial community samples are just a few of the potential NGS applications for forensic genomics and metagenomics ( Borsting and Morling, 2015 ). In human STR analyses, the greatest challenge is mixture deconvolution. NGS technology presents an increased power of discrimination of STR alleles using the intrinsic SNPs genetic microhaplotypes – a combination of 2–4 closely linked SNPs within an allele ( Kidd et al., 2014 ; Pang et al., 2020 ). However, the acceptance of analyses programs to deconvolve mixtures has not been standardized to the same level as it has for STRs.
Microbes are the first responders to changes in any environment because they are rapidly affected by the availability of nutrients and their intrinsic habitats. This makes them excellent indicators for studies investigating post-mortem interval (PMI) or as an indicator of soil geographical provenance ( Giampaoli et al., 2014 ; Finley et al., 2015 ). In decaying organisms, shifts in epinecrotic communities or the thanatomicrobiome are becoming increasingly critical components in investigating PMI ( Javan et al., 2016 ). Sequencing of the thanatomicrobiome revealed the Clostridium spp. varied during different stages human decomposition, the “Postmortem Clostridium Effect” (PCE), providing a time signature of the thanatomicrobiome, which could only have been uncovered through NGS ( Javan et al., 2017 ). However, the lack of consensus in analyses techniques must be addressed before NGS methodologies can be introduced into the justice system ( Table 1 ).
Future Directions and Concluding Remarks
Forensic DNA typing has progressed quickly within a short timeframe ( Figure 1 ), which can be attributed to the many advancements in molecular biology technologies. As these techniques advance, forensic scientists will analyze more atypical forms of evidence to answer questions deemed unresolvable with traditional DNA analyses. For example, epigenetics and DNA methylation markers have been proposed to estimate age, determine the tissue type, and even differentiate between monozygotic twins ( Vidaki and Kayser, 2018 ). However, since epigenetic patterns are also influenced by environmental factors, they can be dynamic, and a number of confounding factors have the potential to affect predictions and must be taken into account when preparing prediction models (i.e., age estimation). Additionally, phenotype informative SNPs across the genome can infer physical characteristics like eye, hair, and skin color, even age, from an unknown source of DNA retrieved from a crime scene. But this technology could pose an “implicit bias” toward minorities, especially in “societies where racism and xenophobia are now on the rise” ( Schneider et al., 2019 ) if not ethically and judicially implemented. With the increased sensitivity of NGS, low biomass samples from environmental DNA (eDNA) – DNA from soil, water, air – can complement and enhance intelligence gathering or provenance in criminal cases. Pollen and dust are two types of eDNA recently explored for their future forensic potential ( Alotaibi et al., 2020 ; Young and Linacre, 2021 ). However, if used in criminal investigations where the eDNA collected has had interaction with other environments, there must be some protocol or quality control established to account for variability that is likely to occur. This makes the prudent validation of this type of DNA analysis, essential. Limitations also arise due to lack of a database for comparison of samples and statistical analyses to evaluate the strength of a match like in the analysis of human STR profiles.
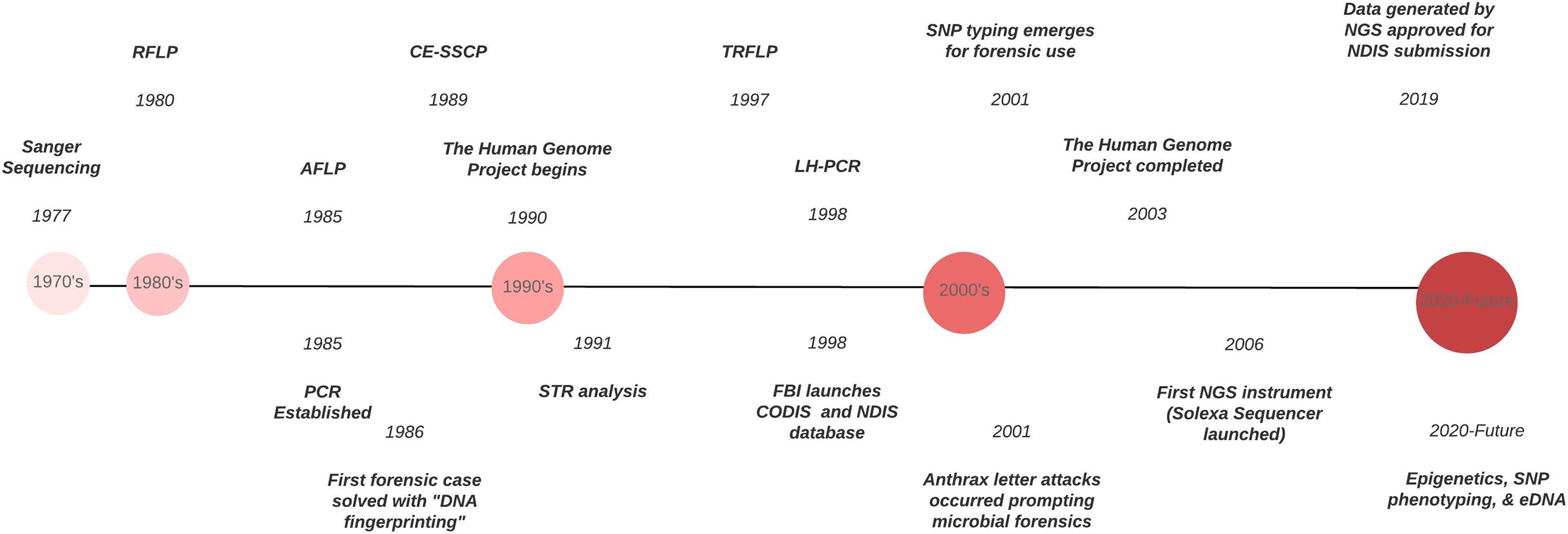
Figure 1. Timeline of the evolution of DNA typing technologies from the 1970’s to the present.
DNA has long been the gold standard in human forensic analysis because of the standardization of DNA markers, databases and statistical analyses. It has laid the foundation for these promising new technologies that will significantly enhance intelligence gathering and species identification – human and non-human – in forensic cases. In order for these methodologies to be useful in criminal investigations, they must adhere to the legal standards such as the Frye or Daubert Standards which determines if an expert testimony or evidence is admissible in court. A method can be deemed acceptable if it follows forensic guidelines set by organizations such as NIST’s Organization Scientific Area Committees (OSAC), Society for Wildlife Forensic Sciences (SWFS), Scientific Working Group on DNA Analysis Methods (SWGDAM), and the International Society for Forensic Genetics (ISFG) ( Linacre et al., 2011 ) just to name a few. These committees provide the guidelines for validation, interpretation, and quality assurance, all necessary components for DNA analysis. The US Fish and Wildlife forensic laboratory has standardized protocols for crimes against federally endangered or threatened species 1 . However, the more common limiting factors in the development of standard guidelines of non-human forensic genetic analyses across different state laboratories are the lack of consensus in methodologies, supporting allelic databases and standardized statistical analyses. Addressing those issues could lay the foundation for non-human analyses to be on par with human analyses.
Author Contributions
DJ designed and wrote the manuscript. DM edited and contributed to the writing of the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge the invitation by the editors to contribute to this special edition. DJ was supported by the Florida Education Fund’s McKnight Doctoral Fellowship.
- ^ https://www.fws.gov/lab/about.php
Alotaibi, S. S., Sayed, S. M., Alosaimi, M., Alharthi, R., Banjar, A., Abdulqader, N., et al. (2020). Pollen molecular biology: Applications in the forensic palynology and future prospects: A review. Saudi J. Biol. Sci. 27, 1185–1190. doi: 10.1016/j.sjbs.2020.02.019
PubMed Abstract | CrossRef Full Text | Google Scholar
Ansorge, W. J. (2009). Next-generation DNA sequencing techniques. N. Biotechnol. 25, 195–203. doi: 10.1016/j.nbt.2008.12.009
Arenas, M., Pereira, F., Oliveira, M., Pinto, N., Lopes, A. M., Gomes, V., et al. (2017). Forensic genetics and genomics: Much more than just a human affair. PLoS Genet. 13:e1006960. doi: 10.1371/journal.pgen.1006960
Borsting, C., and Morling, N. (2015). Next generation sequencing and its applications in forensic genetics. Forensic Sci. Int. Genet. 18, 78–89. doi: 10.1016/j.fsigen.2015.02.002
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314–331.
Google Scholar
Budowle, B., and van Daal, A. (2008). Forensically relevant SNP classes. Biotechniques 60:610. doi: 10.2144/000112806
Butler, J. M. (2012). “Non-human DNA,” in Advanced Topics in Forensic DNA Typing , ed. J. M. Butler (San Diego: Academic Press), 473–495.
Butler, J. M., Coble, M. D., and Vallone, P. M. (2007). STRs vs. SNPs: thoughts on the future of forensic DNA testing. Forensic Sci. Med. Pathol. 3, 200–205. doi: 10.1007/s12024-007-0018-1
Constantinescu, C. M., Barbarii, L. E., Iancu, C. B., Constantinescu, A., Iancu, D., and Girbea, G. (2012). Challenging DNA samples solved with MiniSTR analysis. Brief overview. Rom. J. Leg. Med. 20, 51–56. doi: 10.4323/rjlm.2012.51
CrossRef Full Text | Google Scholar
Damaso, N., Martin, L., Kushwaha, P., and Mills, D. (2014). F-108 polymer and capillary electrophoresis easily resolves complex environmental DNA mixtures and SNPs. Electrophoresis 35, 3208–3211. doi: 10.1002/elps.201400069
Damaso, N., Mendel, J., Mendoza, M., von Wettberg, E. J., Narasimhan, G., and Mills, D. (2018). Bioinformatics Approach to Assess the Biogeographical Patterns of Soil Communities: The Utility for Soil Provenance. J. Forensic. Sci. 63, 1033–1042. doi: 10.1111/1556-4029.13741
Daniel, R., Santos, C., Phillips, C., Fondevila, M., van Oorschot, R. A., Carracedo, A., et al. (2015). A SNaPshot of next generation sequencing for forensic SNP analysis. Forensic Sci. Int. Genet. 14, 50–60. doi: 10.1016/j.fsigen.2014.08.013
Datwyler, S. L., and Weiblen, G. D. (2006). Genetic variation in hemp and marijuana (Cannabis sativa L.) according to amplified fragment length polymorphisms. J. Forensic Sci. 51, 371–375. doi: 10.1111/j.1556-4029.2006.00061.x
Editorial. (2007). Launching Forensic Science International daughter journal in 2007: Forensic Science International: Genetics. Forensic Sci. Int. Genet. 1, 1–2. doi: 10.1016/j.fsigen.2006.10.001
Evans, C. (2007). The Casebook of Forensic Detection: How Science Solved 100 of the World’s Most Baffling Crimes. New York, NY: Berkley Books.
Finley, S. J., Benbow, M. E., and Javan, G. T. (2015). Potential applications of soil microbial ecology and next-generation sequencing in criminal investigations. Appl. Soil. Ecol. 88, 69–78. doi: 10.1016/j.apsoil.2015.01.001
Fondevila, M., Borsting, C., Phillips, C., de la Puente, M., Consortium, E. N., Carracedo, A., et al. (2017). Forensic SNP genotyping with SNaPshot: Technical considerations for the development and optimization of multiplexed SNP assays. Forensic Sci. Rev. 29, 57–76.
Gettings, K. B., Kiesler, K. M., Faith, S. A., Montano, E., Baker, C. H., Young, B. A., et al. (2016). Sequence variation of 22 autosomal STR loci detected by next generation sequencing. Forensic Sci. Int. Genet. 21, 15–21. doi: 10.1016/j.fsigen.2015.11.005
Giampaoli, S., Berti, A., Di Maggio, R. M., Pilli, E., Valentini, A., Valeriani, F., et al. (2014). The environmental biological signature: NGS profiling for forensic comparison of soils. Forensic Sci. Int. 240, 41–47. doi: 10.1016/j.forsciint.2014.02.028
Gill, P., Haned, H., Bleka, O., Hansson, O., Dorum, G., and Egeland, T. (2015). Genotyping and interpretation of STR-DNA: Low-template, mixtures and database matches-Twenty years of research and development. Forensic Sci. Int. Genet. 18, 100–117. doi: 10.1016/j.fsigen.2015.03.014
Gill, P., Jeffreys, A. J., and Werrett, D. J. (1985). Forensic application of DNA ‘fingerprints’. Nature 318, 577–579. doi: 10.1038/318577a0
Goodwin, W., Linacre, A., and Hadi, S. (2011). “An introduction to forensic genetics.” 2nd ed. West Sussex, UK: Wiley-Blackwell, 53–62.
Guo, F., Yu, J., Zhang, L., and Li, J. (2017). Massively parallel sequencing of forensic STRs and SNPs using the Illumina((R)) ForenSeq DNA Signature Prep Kit on the MiSeq FGx Forensic Genomics System. Forensic Sci. Int. Genet. 31, 135–148. doi: 10.1016/j.fsigen.2017.09.003
Habtom, H., Demaneche, S., Dawson, L., Azulay, C., Matan, O., Robe, P., et al. (2017). Soil characterisation by bacterial community analysis for forensic applications: A quantitative comparison of environmental technologies. Forensic Sci. Int. Genet. 26, 21–29. doi: 10.1016/j.fsigen.2016.10.005
Heather, J. M., and Chain, B. (2016). The sequence of sequencers: The history of sequencing DNA. Genomics 107, 1–8. doi: 10.1016/j.ygeno.2015.11.003
Javan, G. T., Finley, S. J., Abidin, Z., and Mulle, J. G. (2016). The Thanatomicrobiome: A Missing Piece of the Microbial Puzzle of Death. Front. Microbiol. 7:225. doi: 10.3389/fmicb.2016.00225
Javan, G. T., Finley, S. J., Smith, T., Miller, J., and Wilkinson, J. E. (2017). Cadaver Thanatomicrobiome Signatures: The Ubiquitous Nature of Clostridium Species in Human Decomposition. Front. Microbiol. 8:2096. doi: 10.3389/fmicb.2017.02096
Jeffreys, A. J. (2013). The man behind the DNA fingerprints: an interview with Professor Sir Alec Jeffreys. Investig. Genet. 4:21. doi: 10.1186/2041-2223-4-21
Jeffreys, A. J., Brookfield, J. F., and Semeonoff, R. (1985a). Positive identification of an immigration test-case using human DNA fingerprints. Nature 317, 818–819. doi: 10.1038/317818a0
Jeffreys, A. J., Wilson, V., and Thein, S. L. (1985b). Hypervariable ‘minisatellite’ regions in human DNA. Nature 314, 67–73. doi: 10.1038/314067a0
Jiang, H. H., Li, B., Ma, Y., Bai, S. Y., Dahmer, T. D., Linacre, A., et al. (2020). Forensic validation of a panel of 12 SNPs for identification of Mongolian wolf and dog. Sci. Rep. 10:13249. doi: 10.1038/s41598-020-70225-5
Kidd, K. K., Pakstis, A. J., Speed, W. C., Lagace, R., Chang, J., Wootton, S., et al. (2014). Current sequencing technology makes microhaplotypes a powerful new type of genetic marker for forensics. Forensic Sci. Int. Genet. 12, 215–224. doi: 10.1016/j.fsigen.2014.06.014
Kim, Y. T., Heo, H. Y., Oh, S. H., Lee, S. H., Kim, D. H., and Seo, T. S. (2015). Microchip-based forensic short tandem repeat genotyping. Electrophoresis 36, 1728–1737. doi: 10.1002/elps.201400477
Kitpipit, T., Thongjued, K., Penchart, K., Ouithavon, K., and Chotigeat, W. (2017). Mini-SNaPshot multiplex assays authenticate elephant ivory and simultaneously identify the species origin. Forensic Sci. Int. Genet. 27, 106–115. doi: 10.1016/j.fsigen.2016.12.007
Linacre, A., Gusmão, L., Hecht, W., Hellmann, A. P., Mayr, W. R., Parson, W., et al. (2011). ISFG: Recommendations regarding the use of non-human (animal) DNA in forensic genetic investigations. Forensic Sci. Int. Genet. 5, 501–505. doi: 10.1016/j.fsigen.2010.10.017
Liu, W. T., Marsh, T. L., Cheng, H., and Forney, L. J. (1997). Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63, 4516–4522. doi: 10.1128/AEM.63.11.4516-4522.1997
Ludeman, M. J., Zhong, C., Mulero, J. J., Lagace, R. E., Hennessy, L. K., Short, M. L., et al. (2018). Developmental validation of GlobalFiler PCR amplification kit: a 6-dye multiplex assay designed for amplification of casework samples. Int. J. Legal. Med. 132, 1555–1573. doi: 10.1007/s00414-018-1817-5
McCord, B. R., Gauthier, Q., Cho, S., Roig, M. N., Gibson-Daw, G. C., Young, B., et al. (2019). Forensic DNA Analysis. Anal. Chem. 91, 673–688. doi: 10.1021/acs.analchem.8b05318
Meyers, M. S., and Foran, D. R. (2008). Spatial and temporal influences on bacterial profiling of forensic soil samples. J. Forensic Sci. 53, 652–660. doi: 10.1111/j.1556-4029.2008.00728.x
Miller Coyle, H., Palmbach, T., Juliano, N., Ladd, C., and Lee, H. C. (2003). An overview of DNA methods for the identification and individualization of marijuana. Croat Med. J. 44, 315–321.
Moreno, L. I., Galusha, M. B., and Just, R. (2018). A closer look at Verogen’s Forenseq DNA Signature Prep kit autosomal and Y-STR data for streamlined analysis of routine reference samples. Electrophoresis 39, 2685–2693. doi: 10.1002/elps.201800087
Moreno, L. I., Mills, D. K., Entry, J., Sautter, R. T., and Mathee, K. (2006). Microbial metagenome profiling using amplicon length heterogeneity-polymerase chain reaction proves more effective than elemental analysis in discriminating soil specimens. J. Forensic Sci. 51, 1315–1322. doi: 10.1111/j.1556-4029.2006.00264.x
Moreno, L. I., Mills, D., Fetscher, J., John-Williams, K., Meadows-Jantz, L., and McCord, B. (2011). The application of amplicon length heterogeneity PCR (LH-PCR) for monitoring the dynamics of soil microbial communities associated with cadaver decomposition. J. Microbiol. Methods 84, 388–393. doi: 10.1016/j.mimet.2010.11.023
Mrkonjic Fuka, M., Gesche Braker, S. H., and Philippot, L. (2007). “Molecular Tools to Assess the Diversity and Density of Denitrifying Bacteria in Their Habitats,” in Biology of the Nitrogen Cycle , eds H. Bothe, S. J. Ferguson, and W. E. Newton (Amsterdam: Elsevier), 313–330.
Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G., and Erlich, H. (1986). Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1, 263–273. doi: 10.1101/sqb.1986.051.01.032
Oostdik, K., Lenz, K., Nye, J., Schelling, K., Yet, D., Bruski, S., et al. (2014). Developmental validation of the PowerPlex((R)) Fusion System for analysis of casework and reference samples: A 24-locus multiplex for new database standards. Forensic Sci. Int. Genet. 12, 69–76. doi: 10.1016/j.fsigen.2014.04.013
Orita, M., Iwahana, H., Kanazawa, H., Hayashi, K., and Sekiya, T. (1989). Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl. Acad. Sci. U S A 86, 2766–2770.
Osborn, A. M., Moore, E. R., and Timmis, K. N. (2000). An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2, 39–50. doi: 10.1046/j.1462-2920.2000.00081.x
Pang, J. B., Rao, M., Chen, Q. F., Ji, A. Q., Zhang, C., Kang, K. L., et al. (2020). A 124-plex Microhaplotype Panel Based on Next-generation Sequencing Developed for Forensic Applications. Sci. Rep. 10:1945. doi: 10.1038/s41598-020-58980-x
Sanger, F., and Coulson, A. R. (1975). A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 94, 441–448. doi: 10.1016/0022-2836(75)90213-2
Schmedes, S. E., Sajantila, A., and Budowle, B. (2016). Expansion of Microbial Forensics. J. Clin. Microbiol. 54, 1964–1974. doi: 10.1128/JCM.00046-16
Schneider, P. M., Prainsack, B., and Kayser, M. (2019). The Use of Forensic DNA Phenotyping in Predicting Appearance and Biogeographic Ancestry. Dtsch Arztebl. Int. 52, 873–880. doi: 10.3238/arztebl.2019.0873
Suzuki, M., Rappe, M. S., and Giovannoni, S. J. (1998). Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64, 4522–4529.
SWGDAM (2019). Addendum to SWGDAM Autosomal Interpretation Guidelines for NGS.Swgdam. Available online at: https://www.swgdam.org/publications
Vidaki, A., and Kayser, M. (2018). Recent progress, methods and perspectives in forensic epigenetics. Forensic Sci. Int. Genet. 37, 180–195. doi: 10.1016/j.fsigen.2018.08.008
Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. doi: 10.1093/nar/23.21.4407
Wyman, A. R., and White, R. (1980). A highly polymorphic locus in human DNA. Proc. Natl. Acad. Sci. U S A 77, 6754–6758. doi: 10.1073/pnas.77.11.6754
Young, J. M., and Linacre, A. (2021). Massively parallel sequencing is unlocking the potential of environmental trace evidence. Forensic Sci. Int. Genet. 50:102393. doi: 10.1016/j.fsigen.2020.102393
Keywords : forensic genetics, DNA typing, metabarcoding, soil, microbes, minisatellites, next-generation sequencing
Citation: Jordan D and Mills D (2021) Past, Present, and Future of DNA Typing for Analyzing Human and Non-Human Forensic Samples. Front. Ecol. Evol. 9:646130. doi: 10.3389/fevo.2021.646130
Received: 25 December 2020; Accepted: 02 March 2021; Published: 22 March 2021.
Reviewed by:
Copyright © 2021 Jordan and Mills. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: DeEtta Mills, [email protected]
This article is part of the Research Topic
Life and Death: New Perspectives and Applications in Forensic Science
recombinant dna technology Recently Published Documents
Total documents.
- Latest Documents
- Most Cited Documents
- Contributed Authors
- Related Sources
- Related Keywords
Pharmacological profile, clinical efficacy, and safety of Follitropin Delta produced by recombinant DNA technology in a human cell line (REKOVELLE® PEN for S.C. Injection 12 μg, 36 μg, 72 μg)
Genetic engineering technique in virus-like particle vaccine construction.
Vaccine becomes a very effective strategy to deal with various infectious diseases even to the point of eradication as in the smalpox virus. At present many conventional vaccines such as inactivated and live-attenuated vaccines. However, these vaccine methods have side effects on the population. Viral-like particle (VLP) is an alternative vaccine based on recombinant DNA technology that is safe with the same immunogenicity as conventional viruses. This vaccine has been shown to induce humoral immune responses mediated by antibodies and cellular immune responses mediated by cytotoxic T cells. With these advantages, currently various types of vaccines have only been developed on a VLP basis. VLP can be produced from a variety of recombinant gene expression systems including bacterial cell expression systems, yeast cells, insect cells, mammalian cells, plant cells, and cell-free systems.
Structural and Functional Characterization of a Novel Recombinant Antimicrobial Peptide from Hermetia illucens
Antibiotics are commonly used to treat pathogenic bacteria, but their prolonged use contributes to the development and spread of drug-resistant microorganisms raising the challenge to find new alternative drugs. Antimicrobial peptides (AMPs) are small/medium molecules ranging 10–100 residues synthesized by all living organisms and playing important roles in the defense systems. These features, together with the inability of microorganisms to develop resistance against the majority of AMPs, suggest that these molecules might represent effective alternatives to classical antibiotics. Because of their high biodiversity, with over one million described species, and their ability to live in hostile environments, insects represent the largest source of these molecules. However, production of insect AMPs in native forms is challenging. In this work we investigate a defensin-like antimicrobial peptide identified in the Hermetia illucens insect through a combination of transcriptomics and bioinformatics approaches. The C-15867 AMP was produced by recombinant DNA technology as a glutathione S-transferase (GST) fusion peptide and purified by affinity chromatography. The free peptide was then obtained by thrombin proteolysis and structurally characterized by mass spectrometry and circular dichroism analyses. The antibacterial activity of the C-15867 peptide was evaluated in vivo by determination of the minimum inhibitory concentration (MIC). Finally, crystal violet assays and SEM analyses suggested disruption of the cell membrane architecture and pore formation with leaking of cytosolic material.
Nursing Skill and Responsibility in Administration of Injection Actilyse
Actilyse can break blood clots that form in the heart, blood arteries, or lungs during a heart attack. This medication is also given to stroke patients to improve recovery and reduce the likelihood of impairment. Recombinant DNA technology was used to create Activase, a tissue plasminogen activator. It is a sterile, purified glycoprotein of 527 amino acids. It is made by combining complementary DNA (cDNA) from a human melanoma cell line with the natural human tissue-type plasminogen activator. After reconstitution with Sterile Water for Injection, USP, Activase is a sterile, white to off-white lyophilized powder for intravenous injection.
Towards the development of a chemical lure to improve the management of invading rat populations
<p>Introduced species, such as Rattus norvegicus and Rattus rattus,have contributed to the extinction of many native animals and plants in New Zealand(NZ). Current strategies exist to monitor, manage and eradicate pest species. However, these haven’t always been completely successful and tools to detect small or invading densities remain to be developed. One possible new method to address this problem is the application of chemical attractants (lures). Recently, a major urinary protein (MUP) has been shown in male miceto act as a sexual attractant. MUPs modulate the release of volatile attractants and have potential to act as attractants themselves. Our aim was to determine if a similar MUP(s) and associated volatiles are present in the urine of rats, with the prospect of creating a chemical lure to use in rat detection and eradication. Using Gas Chromatography/Mass Spectrometry, potential volatiles in rat urine have been identified. Analysis of rat urine by gel electrophoresis has shown MUPs present in both sexes. A 22.4 kDa MUP in Rattus norvegicushas been synthesised and expressed in E.coliusing recombinant DNA technology. Preliminary steps have been made towards the production of a MUP based on ship rat DNA sequence. Future behavioral trials are needed to investigate whether the synthesised protein, in the presence or absence of the urinary-derived volatiles, is a sexual attractant.</p>
If Not Now, When? Nonserotype Pneumococcal Protein Vaccines
Abstract The sudden emergence and global spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have greatly accelerated the adoption of novel vaccine strategies, which otherwise would have likely languished for years. In this light, vaccines for certain other pathogens could certainly benefit from reconsideration. One such pathogen is Streptococcus pneumoniae (pneumococcus), an encapsulated bacterium that can express >100 antigenically distinct serotypes. Current pneumococcal vaccines are based exclusively on capsular polysaccharide—either purified alone or conjugated to protein. Since the introduction of conjugate vaccines, the valence of pneumococcal vaccines has steadily increased, as has the associated complexity and cost of production. There are many pneumococcal proteins invariantly expressed across all serotypes, which have been shown to induce robust immune responses in animal models. These proteins could be readily produced using recombinant DNA technology or by mRNA technology currently used in SARS-CoV-2 vaccines. A door may be opening to new opportunities in affordable and broadly protective vaccines.
A comprehensive comparison between camelid nanobodies and single chain variable fragments
AbstractBy the emergence of recombinant DNA technology, many antibody fragments have been developed devoid of undesired properties of natural immunoglobulins. Among them, camelid heavy-chain variable domains (VHHs) and single-chain variable fragments (scFvs) are the most favored ones. While scFv is used widely in various applications, camelid antibodies (VHHs) can serve as an alternative because of their superior chemical and physical properties such as higher solubility, stability, smaller size, and lower production cost. Here, these two counterparts are compared in structure and properties to identify which one is more suitable for each of their various therapeutic, diagnosis, and research applications.
RISPR-Cas9: a weapon against COVID-19
In current pandemic circumstances, novel coronavirus is a salutary challenge for all over the world and coronavirus used the host cell for replication. Coronavirus usually use the host cellular products to perform their basic functions. Various specific target sites also present in coronavirus proteins for target-specific therapy such as small inhibitor molecule for viral polymerase or prevent the attachment of viruses to the receptor sites for vaccination purpose. The virus attaches to ACE2 receptors and uses enzyme to cleave translated products which encodes for various enzymes like RNA polymerase, helicase etc. The system needs some processes which lead for the disturbance and make the virus unable to replicate. The recombinant DNA technology makes a great advancement in every field of life with a number of importance in agriculture, industries, and clinics. It is used to manipulate the genetic material of living organism for the purpose of producing desirable products such as disease resistant crops, treatment of cancer, genetic disease and viral disease. Thus, for the purpose of antiviral strategies, the specific technique called CRISPR/Cas9 is used, and this technique has the capability to target specific nucleotide sequence inside the genome of coronavirus.
Recombinant DNA Technology
Export citation format, share document.
More From Forbes
Gene therapy, dna's past, rna's future: a new wave of hope.
- Share to Facebook
- Share to Twitter
- Share to Linkedin
Artist representation of gene therapy
This story is part of a series on the current progression in Regenerative Medicine. In 1999, I defined regenerative medicine as the collection of interventions that restore tissues and organs damaged by disease, injured by trauma, or worn by time to normal function. I include a full spectrum of chemical, gene, and protein-based medicines, cell-based therapies, and biomechanical interventions that achieve that goal.
In this subseries, we focus specifically on gene therapies. We explore the current treatments and examine the advances poised to transform healthcare. Each article in this collection delves into a different aspect of gene therapy's role within the larger narrative of Regenerative Medicine.
Gene therapy represents the frontier of hope in combating genetic disorders, promising to rewrite the rulebook on how we address some of the most persistent and challenging diseases known to humankind. Over the last ten years, we have witnessed a transformation from speculative science to a reality where genetic disorders are treated not just in theory but in patients' lives.
A New Wave of Gene Therapy Trials and Approvals
Gene therapy made significant strides throughout the 2010s, culminating in multiple regulatory trial approvals for gene therapy products across 38 countries . The work done in these trials led to formal regulatory approval for the use of gene therapies in a variety of countries, some of the most significant of these were from 2012 to 2017.
Apple Confirms Major iPhone Changes With New App Features Enabled
Aew dynamite results winners and grades as cm punk destroys jack perry, chiefs rashee rice hit with 8 criminal charges in connection to multi car crash.
Number of gene therapy clinical trials approved worldwide 1989-2017
In 2012, Glybera became the first-ever gene therapy approved in the European Union . This therapy treats lipoprotein lipase deficiency by compensating for the missing or ineffective enzyme. Similarly, Strimvelis, approved in 2016 , has been used to treat ADA-SCID. This rare genetic disorder severely compromises the immune system in children. This therapy corrects the gene responsible for the disease.
In 2017, three FDA approvals were game-changers for the medical field. The first was Kymriah , a new way of treating acute lymphoblastic leukemia by genetically modifying the patient's T-cells to attack cancer cells. The second approval was for Luxturna , which treats a genetic form of blindness by directly providing a standard copy of the RPE65 gene to the eye. Lastly, the FDA approved Yescarta , the first CAR T-cell therapy, in 2017 to treat large B-cell lymphoma.
Zolgensma, approved in 2019 , is a gene therapy for spinal muscular atrophy (SMA), which causes muscle wasting. This therapy introduces a new, functional copy of the human SMN gene into a patient's motor neuron cells, providing a one-time treatment for this lifelong disease. Gene therapies like Zolgensma offer hope for people who have genetic disorders that have been difficult or impossible to treat until recently.
While these approvals were happening, CRISPR-Cas9 became synonymous with a new era in gene editing.
The CRISPR Revolution
During the mid-2010s, the genetic research and therapy field experienced a seismic shift with the introduction of the CRISPR-Cas9 system . This tool has dramatically changed gene editing, allowing for precise and efficient modification of genes in living organisms. Unlike traditional gene-editing methods, the CRISPR system enables accurate targeting of specific genes with unprecedented speed and precision .
Diagram of CRISPR for patients with cancer-tested T cells modified to better "see" and kill cancer. ... [+] In this example, CRISPR removed three genes: two that can interfere with the NY-ESO-1 receptor and another that limits the cells’ cancer-killing abilities.
Technology development has significantly reduced the barriers to entry for genetic research and therapy, making it more accessible and affordable. With the capability to customize treatments with unprecedented specificity, new therapies can be tested and developed with greater accuracy and efficiency. This has paved the way for more effective genetic research and treatment, potentially leading to cures for previously thought untreatable diseases.
Every Challenge is an Open Door
While gene therapy has seen notable successes and approvals, there have also been setbacks in recent years. In science, every challenge is an open door, and gene therapy has walked through many of these, with more to open. The biggest challenges in the field have been the complexities in delivering treatments, persistent safety concerns, and daunting immune responses.
Adeno-associated viruses (AAVs) are one of the most commonly used gene therapy vectors . However, high doses of AAVs can cause potential side effects, such as inflammation and liver damage . While these side effects are usually manageable, they can lead to more severe complications.
The immune system may recognize AAVs as foreign particles and launch a full attack against them, resulting in a reduced therapeutic effect or a system-wide immune response. In some cases, this immune response is so severe it has even resulted in death, such was the case in the untimely passing of Terry Horgan, a 27-year-old man with Duchenne muscular dystrophy, following an investigational CRISPR-based treatment.
A Death in a Gene Therapy Trial
Terry Horgan's story is both brave and tragic. In October 2022, he was the only participant in an early-stage safety trial . He was given a high dosage of gene-editing therapy tailored to his unique condition. This trial used an adeno-associated virus as the vector to introduce the CRISPR tool into his body.
In Horgan's case, the viral vector precipitated an unexpected and severe immune response , leading to organ failure and, ultimately, his death. This incident has thrust the use of these vectors into the limelight, prompting intense scrutiny from the medical community and invigorating the discourse on gene therapy safety.
Drawing parallels with the 1999 case of Jesse Gelsinger , the first person publicly identified to have died in a clinical trial for gene therapy, Horgan's experience emerges both familiar and foreboding. Similarities can be seen in the unexpected immune reactions elicited by gene therapy vectors, underlying the need for revamped vigilance in early trials. Yet, the divergence lies in the progress made over the two decades separating these tragedies, a period marked by leaps in genetic understanding and technological advances.
Charting a Course for Transformation
Gene therapy has transitioned from the margins to the epicenter of medical science. The innovations of the early 21st century are now yielding concrete treatments for diseases once considered incurable. Conditions such as inherited blindness, spinal muscular atrophy, and various forms of blood cancer now benefit from FDA-approved gene therapies.
Now is an opportune moment for the medical community and others to delve into the vast potential of gene therapy. After all, we are not just the products of our genes but the architects of our genetic health.
To learn more about regenerative medicine, read more stories at www.williamhaseltine.com
- Editorial Standards
- Reprints & Permissions

Please enable JavaScript or use a Browser that supports JavaScript
- Advanced Search
- Browse by Analyte
- Browse by Pre-analytical Factor
- Search SOPs
- SOP Compendiums
- Suggest a New Paper
- Submit an SOP
- Curator Home
- Back Office
Analysis of quality metrics in comprehensive cancer genomic profiling using a dual DNA-RNA panel.
Author(s): Watanabe K, Kohsaka S, Tatsuno K, Shinozaki-Ushiku A, Isago H, Kage H, Ushiku T, Aburatani H, Mano H, Oda K
Publication: Pract Lab Med , 2024, Vol. 39 , Page e00368
PubMed ID: 38404525 PubMed Review Paper? No
Purpose of Paper
This paper compared two DNA integrity markers (Q-value and ddCq) and one RNA integrity metric (DV200) using DNA and RNA extracted from 585 formalin-fixed, paraffin-embedded (FFPE) blocks that had been stored for <1, 1-2, 2-3, 3-5 y or approximately 5 years before extraction. The authors investigated the predictive performance of these metrics for four next-generation sequencing quality metrics. Potential effects associated with the hospital of collection/processing and cancer type on the DNA/RNA quality metrics evaluated were also examined.
Conclusion of Paper
The duration of FFPE block storage had a deleterious effect on DNA and RNA integrity, with a higher ratio of short to long real-time PCR products (ddCq), a lower ratio of PCR-amplified DNA to double-stranded DNA (Q-value), and a lower percentage of RNA fragments >200 bp (DV200) noted with progressive storage. While ddCq was weakly to modestly negatively correlated with the percentage on-target reads, and mean and target exon coverage, Q-value was weakly to modestly positively correlated with the percentage of on-target reads and coverage uniformity and DV200 were weakly positively correlated with coverage of housekeeping genes. A ddCq ≤5.36 was highly predictive of a mean depth >500 but less predictive of coverage uniformity or target exon coverage. A Q-value ≥0.928 was predictive of a mean depth >500, coverage uniformity ≥99%, and target exon coverage ≥99%. The predictive power of a DV200 >41 for coverage of housekeeping genes that was ≥70% was 92.1%. ddCq and Q-values displayed significant differences among hospitals even after correcting for tumor type, but DV200 was not affected by hospital. Q-values were lowest and DV200 highest in lung tumors.
Study Purpose
This study compared two DNA integrity markers (Q-value and ddCq) and one RNA integrity metric (DV200) using DNA and RNA extracted from 585 formalin-fixed, paraffin-embedded (FFPE) blocks that had been stored for <1, 1-2, 2-3, 3-5 y or approximately 5 years before extraction. The authors investigated the predictive performance of these metrics for four next-generation sequencing quality metrics. Potential effects associated with the hospital of collection/processing and cancer type on the DNA/RNA quality metrics were also examined. A total of 585 tissues that included 114 lung, 90 bowel, 58 ovarian/fallopian tube, 51 uterus, 38 soft tissue, 31 cervix, and 203 specimens of types representing <5% of the total were collected (further details were not provided) and fixed in 10-20% neutral buffered formalin for 24-72 h before paraffin embedding. The specimens were contributed from multiple hospitals, with six hospitals providing >10 specimens. DNA (582 specimens) and RNA (572 specimens) were extracted using the GeneRead DNA FFPE Kit and the RNeasy FFPE Kit, respectively. Double stranded DNA was quantified by Qubit and amplifiable DNA was quantified using the TaqMan Copy Number Reference Assay Human RNase P. The Q value was then calculated as the percentage of dsDNA that was amplifiable. NA quantity was defined as the amount of double-stranded DNA when the Q-value was >1 and as the amount of amplifiable DNA when the Q-Value was ≥1. The ddCq value was calculated based on the ratio of two real-time PCR amplicons in the FFPE DNA QC Assay version 2 Kit. The percentage of RNA fragments >200 bp (DV200) was assessed using a 2200 TapeStation. DNA and the RNA libraries were prepared using a SureSelectXT Custom Kit and the SureSelect RNA Capture Kit, respectively, and sequenced on a Next-seq Instrument.
Summary of Findings:
The duration of FFPE block storage had a deleterious effect on DNA integrity, with significantly higher ddCq values and lower Q-values noted with progressive storage. ddCq values were higher in specimens stored for >1 year compared to those stored for approximately 1 year (P<0.001, all), stored >3 years compared to 1-2 years (P<0.001, both), or stored approximately 5 years compared to those stored 2-3 or 3-5 years (P<0.001, both). Q-values were lower in specimens that were stored for 5 years than 3-5 years (P<0.01), 2-3 years (P<0.001), 1-2 years (P<0.001), or approximately 1 year (P<0.001), or stored for 3-5 years compared to those stored 1-2 years (P<0.01) or approximately 1 year (P<0.01) but were not significantly different in specimens stored <3 years when compared to those stored for approximately 1 year. ddCq was negatively correlated with the percentage on-target reads (r=-0.47, P<0.001), mean depth (r=-0664, P<0.001), and target exon coverage (r=-0.362, P<0.001); Q-value was correlated with the percentage on-target reads (r=0.353, P<0.001) and coverage uniformity (r=0.411, P<0.001). In a generalized linear model, the percentage of on-target reads was associated with ddCq, but mean depth was associated with ddCq and Q-value. The predictive power of a ddCq ≤5.36 for a mean depth >500 was 91.6%, but the same cut-off was less predictive of coverage uniformity ≥99% (55.1%) or target exon coverage ≥99% (60.6%). The predictive power of a Q-value ≥0.928 for a mean depth >500, coverage uniformity ≥99%, and target exon coverage ≥99% was 79.9%, 81.5%, and 83.1% respectively. For specimens with a low ddCq (≤5.36) but unacceptable Q-values, the on-target read percentage and mean depth were still generally acceptable but uniformity and target exon coverage were <99%. ddCq and Q-values displayed significant differences among hospitals even after correcting for tumor type. Q-values were lowest in lung tumors.
Similarly, DNA and RNA integrity were adversely affected by block storage. The percentage of RNA fragments >200 nt (DV200) decreased progressively with FFPE block storage duration, with significantly lower DV200 values from specimens that were stored >2 years than those stored approximately 1 year (P<0.001 all) or those stored for 3-5 years or approximately 5 years compared to those stored approximately 1 year (P<0.01, both). DV200 was weakly positively correlated with the coverage of housekeeping genes (r=0.369, P<0.001). The predictive power of a DV200 >41 for a coverage of housekeeping genes ≥70% was 92.1%. DV200 was not significantly different among hospitals but was significantly higher in lung cancer specimens than in bowel or ovarian/fallopian cancer specimens.
Please enable JavaScript or use a Browser that supports JavaScript for more on this study.
Biospecimens
- Tissue - Colorectal
- Tissue - Lung
- Tissue - Other
- Tissue - Uterus
- Tissue - Cervix
- Tissue - Ovary
Preservative Types
- Neoplastic - Carcinoma
- Not specified
- Neoplastic - Not specified
Pre-analytical Factors:
You recently viewed , news and announcements.
- Most Popular SOPs in March 2024
- New SOPs Available
- Most Downloaded SOPs in January and February of 2024
- U.S. Department of Health and Human Services
- National Institutes of Health
- National Cancer Institute
- Skip to primary navigation
- Skip to main content
- Skip to primary sidebar
- Skip to footer
Don't Miss a Post! Subscribe
- Guest Posts

- Educational AI
- Edtech Tools
- Edtech Apps
- Teacher Resources
- Special Education
- Edtech for Kids
- Buying Guides for Teachers

Educators Technology
Innovative EdTech for teachers, educators, parents, and students
10 Great AI Tools for Researchers
By Med Kharbach, PhD | Last Update: April 8, 2024

Today, I want to talk about some really cool AI tools that are changing the way we do research. This is just a small preview of what I’m putting together in an eBook full of AI tools for researchers like us. If you don’t want to miss out on the full thing, be sure to sign up for our email updates.
There’s a lot of talk about AI in universities, and not everyone agrees about using it. But, like it or not, AI is becoming a big part of research, and it’s here to stay. I believe we should use AI the right way. It’s not about just copying and pasting stuff; that’s not real research. You still have to do the hard work of reading and writing yourself. But, AI can be a huge help, kind of like having an extra assistant who’s always there when you need it. I’m all for using AI to make our research better, as long as we keep doing the important parts ourselves.
AI Tools for Researchers
Here are are some good AI Tools I recommend for student researchers and academics:
Litmaps is a tool for research students that makes finding papers and authors on a topic easy and quick. Instead of spending lots of time reading through hundreds of papers, you can use Litmaps to find the important ones in seconds. It helps you find papers you might miss otherwise and keeps you updated on new research without getting overwhelmed. You can see which papers are connected and important for your work through visual maps, making it simple to keep track of your literature review.
Jenni is an AI-powered writing tool that helps you write, edit, and reference your work easily. It’s like having a helpful friend who’s always there to get you past writer’s block, suggest ways to say things differently, and make sure your citations are in order. More specifically, Jenni can:
- Suggest words and sentences as you write to help you keep going.
- Help you cite sources correctly in styles like APA, MLA, and others, using your own PDFs or research.
- Let you change the wording of any text to match the tone you need.
- Turn your research papers into written content by analyzing and summarizing them.
- Chat with your PDFs to quickly understand and summarize them.
- Import a bunch of sources at once if you have them saved.
- Export your work to LaTeX, Word, or HTML without messing up your formatting.
- Create an outline for your paper just from a prompt you provide.
- Work in multiple languages, including English (US and British), Spanish, German, French, and Chinese.
- Keep all your research organized in one place for easy citing in any document.
- Offer suggestions and help expand notes into full paragraphs, so you’re never stuck staring at a blank page.
3. Paperpa l
Paperpal is a handy tool for anyone who writes academic texts like essays, theses, dissertations, or research papers. It checks your writing for grammar mistakes and makes sure you’re using the right language for academic work. With the help of generative AI, Paperpal can also create outlines, abstracts, and titles for your papers. This tool makes it easy to paraphrase your work for clarity .You can also check your work for plagiarism with detailed reports, get help generating various parts of your academic text, and even translate text from over 25 languages to English.
Related: 6 Best Text to Video AI Tools
4. Unriddle
Unriddle is a cool tool that changes how students and researchers work with documents. It gives you an AI helper for any document you’re looking at, making it super quick to find info, sum up tricky topics, and take notes easily. Unriddle is all about making your research easier and faster, so you don’t have to read every single word to find what you need.
Unriddle also helps you write and reference sources the right way. It can point out the most important sources when you highlight text, so your references are spot-on. It works in over 90 languages and has some cool extra features like a Chrome extension to summarize online articles, settings you can change to fit your needs, and the ability to work with many documents at once.

5. Connected Papers
Connected Papers helps you discover recent important works without needing to maintain extensive lists. It is a visual tool for research students and academics who are diving into a new field or ensuring their research is comprehensive. It starts with a paper you’re interested in and creates a graph showing similar papers in that field. This visual approach helps you understand the trends and main contributors quickly. It’s especially useful in fast-moving fields where new studies are constantly published.
With Connected Papers, you can also build a bibliography for your thesis more efficiently. By starting with a few key references, it finds additional relevant papers, helping you to fill in the gaps. It offers views for finding significant prior works or the latest reviews and state-of-the-art papers following your chosen study.
6. Scite Assistant
Scite Assistant is like a research companion powered by large language models (LLMs), designed to make your research process smoother and more insightful. You can ask scite Assistant any research-related question and you will get insights and explanations for its responses, helping you understand the reasoning behind its conclusions.
scite Assistant offers customizable settings to tailor the tool to your specific research needs. You can control whether you want references included, filter your searches by year, topics, or journals, and even specify the sources the Assistant should use, like your own dashboard collection or preferred journals. This level of customization ensures that the responses and sources are relevant to your specific research questions and preferences, making it an invaluable tool for academics and researchers seeking detailed and reliable information.
7. DocAnalyzer
DocAnalyzer.ai makes talking to your documents easy and smart. You can upload one or many documents and start chatting right away, getting answers to your questions in real time. This tool is great because it understands the context of your documents, making it super helpful for finding exactly what you need without any confusion. What makes docAnalyzer.ai special is how simple and smart it is to use. You can ask your PDFs questions and get back clear, detailed answers quickly. You can even share your document chats with others, making teamwork easier.
8. SciSummary
SciSummary is all about making it easier to get the gist of scientific articles fast. You can email or upload a document, and in minutes, you’ll get a summary sent right back to you. This is perfect for scientists, students, and anyone who’s busy but needs to stay on top of the latest research without reading long articles.
SciSummary uses advanced AI, like a super smart robot that can summarize any scientific article. The AI gets better over time, learning from summaries that experts check. This means you can quickly understand new discoveries and research without spending hours reading. SciSummary offers a free option for summarizing articles, and if you need more, there are affordable plans with more features.
9. Explainpaper
Explainpaper is like having a smart friend that helps you understand research papers quickly. You just upload a paper, highlight the parts you find confusing, and get an explanation. This tool is perfect for diving into complex topics and for speeding up your review process. With Explainpaper, you’re not alone when facing intimidating jargon or complex concepts.
10. SciSpace
SciSpace aims to make finding and understanding research papers a breeze. It’s an all-in-one platform where you can read papers, get straightforward explanations from AI, and explore related research. SciSpace is designed to cut down on the time researchers spend looking for information and dealing with the hassle of formatting papers. With access to metadata for over 200 million papers and more than 50 million full-text PDFs, SciSpace provides tools like a citation generator, AI detector, and paraphraser to make your research process smoother and more productive. It’s a dedicated workspace for researchers, publishers, and institutions to collaborate and discover information effortlessly.
Final thoughts
As I mentioned, these tools are just a part of the bigger picture I’m assembling in the upcoming eBook. The AI tools we explored today are stepping stones towards a more efficient, insightful, and innovative research process. But remember, they’re tools to aid us, not to replace the foundational skills of rigorous research. Embracing AI in our work, when used ethically and wisely, opens up new horizons for discovery and understanding. Stay tuned for the full eBook release, and let’s navigate this promising future of research together.

Join our mailing list
Never miss an EdTech beat! Subscribe now for exclusive insights and resources .

Meet Med Kharbach, PhD
Dr. Med Kharbach is an influential voice in the global educational technology landscape, with an extensive background in educational studies and a decade-long experience as a K-12 teacher. Holding a Ph.D. from Mount Saint Vincent University in Halifax, Canada, he brings a unique perspective to the educational world by integrating his profound academic knowledge with his hands-on teaching experience. Dr. Kharbach's academic pursuits encompass curriculum studies, discourse analysis, language learning/teaching, language and identity, emerging literacies, educational technology, and research methodologies. His work has been presented at numerous national and international conferences and published in various esteemed academic journals.

Join our email list for exclusive EdTech content.
Suggestions or feedback?
MIT News | Massachusetts Institute of Technology
- Machine learning
- Social justice
- Black holes
- Classes and programs
Departments
- Aeronautics and Astronautics
- Brain and Cognitive Sciences
- Architecture
- Political Science
- Mechanical Engineering
Centers, Labs, & Programs
- Abdul Latif Jameel Poverty Action Lab (J-PAL)
- Picower Institute for Learning and Memory
- Lincoln Laboratory
- School of Architecture + Planning
- School of Engineering
- School of Humanities, Arts, and Social Sciences
- Sloan School of Management
- School of Science
- MIT Schwarzman College of Computing
A new way to detect radiation involving cheap ceramics
Press contact :.

Previous image Next image
The radiation detectors used today for applications like inspecting cargo ships for smuggled nuclear materials are expensive and cannot operate in harsh environments, among other disadvantages. Now, in work funded largely by the U.S. Department of Homeland Security with early support from the U.S. Department of Energy, MIT engineers have demonstrated a fundamentally new way to detect radiation that could allow much cheaper detectors and a plethora of new applications.
They are working with Radiation Monitoring Devices , a company in Watertown, Massachusetts, to transfer the research as quickly as possible into detector products.
In a 2022 paper in Nature Materials , many of the same engineers reported for the first time how ultraviolet light can significantly improve the performance of fuel cells and other devices based on the movement of charged atoms, rather than those atoms’ constituent electrons.
In the current work, published recently in Advanced Materials , the team shows that the same concept can be extended to a new application: the detection of gamma rays emitted by the radioactive decay of nuclear materials.
“Our approach involves materials and mechanisms very different than those in presently used detectors, with potentially enormous benefits in terms of reduced cost, ability to operate under harsh conditions, and simplified processing,” says Harry L. Tuller, the R.P. Simmons Professor of Ceramics and Electronic Materials in MIT’s Department of Materials Science and Engineering (DMSE).
Tuller leads the work with key collaborators Jennifer L. M. Rupp, a former associate professor of materials science and engineering at MIT who is now a professor of electrochemical materials at Technical University Munich in Germany, and Ju Li, the Battelle Energy Alliance Professor in Nuclear Engineering and a professor of materials science and engineering. All are also affiliated with MIT’s Materials Research Laboratory
“After learning the Nature Materials work, I realized the same underlying principle should work for gamma-ray detection — in fact, may work even better than [UV] light because gamma rays are more penetrating — and proposed some experiments to Harry and Jennifer,” says Li.
Says Rupp, “Employing shorter-range gamma rays enable [us] to extend the opto-ionic to a radio-ionic effect by modulating ionic carriers and defects at material interfaces by photogenerated electronic ones.”
Other authors of the Advanced Materials paper are first author Thomas Defferriere, a DMSE postdoc, and Ahmed Sami Helal, a postdoc in MIT’s Department of Nuclear Science and Engineering.
Modifying barriers
Charge can be carried through a material in different ways. We are most familiar with the charge that is carried by the electrons that help make up an atom. Common applications include solar cells. But there are many devices — like fuel cells and lithium batteries — that depend on the motion of the charged atoms, or ions, themselves rather than just their electrons.
The materials behind applications based on the movement of ions, known as solid electrolytes, are ceramics. Ceramics, in turn, are composed of tiny crystallite grains that are compacted and fired at high temperatures to form a dense structure. The problem is that ions traveling through the material are often stymied at the boundaries between the grains.
In their 2022 paper, the MIT team showed that ultraviolet (UV) light shone on a solid electrolyte essentially causes electronic perturbations at the grain boundaries that ultimately lower the barrier that ions encounter at those boundaries. The result: “We were able to enhance the flow of the ions by a factor of three,” says Tuller, making for a much more efficient system.
Vast potential
At the time, the team was excited about the potential of applying what they’d found to different systems. In the 2022 work, the team used UV light, which is quickly absorbed very near the surface of a material. As a result, that specific technique is only effective in thin films of materials. (Fortunately, many applications of solid electrolytes involve thin films.)
Light can be thought of as particles — photons — with different wavelengths and energies. These range from very low-energy radio waves to the very high-energy gamma rays emitted by the radioactive decay of nuclear materials. Visible light — and UV light — are of intermediate energies, and fit between the two extremes.
The MIT technique reported in 2022 worked with UV light. Would it work with other wavelengths of light, potentially opening up new applications? Yes, the team found. In the current paper they show that gamma rays also modify the grain boundaries resulting in a faster flow of ions that, in turn, can be easily detected. And because the high-energy gamma rays penetrate much more deeply than UV light, “this extends the work to inexpensive bulk ceramics in addition to thin films,” says Tuller. It also allows a new application: an alternative approach to detecting nuclear materials.
Today’s state-of-the-art radiation detectors depend on a completely different mechanism than the one identified in the MIT work. They rely on signals derived from electrons and their counterparts, holes, rather than ions. But these electronic charge carriers must move comparatively great distances to the electrodes that “capture” them to create a signal. And along the way, they can be easily lost as they, for example, hit imperfections in a material. That’s why today’s detectors are made with extremely pure single crystals of material that allow an unimpeded path. They can be made with only certain materials and are difficult to process, making them expensive and hard to scale into large devices.
Using imperfections
In contrast, the new technique works because of the imperfections — grains — in the material. “The difference is that we rely on ionic currents being modulated at grain boundaries versus the state-of-the-art that relies on collecting electronic carriers from long distances,” Defferriere says.
Says Rupp, “It is remarkable that the bulk ‘grains’ of the ceramic materials tested revealed high stabilities of the chemistry and structure towards gamma rays, and solely the grain boundary regions reacted in charge redistribution of majority and minority carriers and defects.”
Comments Li, “This radiation-ionic effect is distinct from the conventional mechanisms for radiation detection where electrons or photons are collected. Here, the ionic current is being collected.”
Igor Lubomirsky, a professor in the Department of Materials and Interfaces at the Weizmann Institute of Science, Israel, who was not involved in the current work, says, “I found the approach followed by the MIT group in utilizing polycrystalline oxygen ion conductors very fruitful given the [materials’] promise for providing reliable operation under irradiation under the harsh conditions expected in nuclear reactors where such detectors often suffer from fatigue and aging. [They also] benefit from much-reduced fabrication costs.”
As a result, the MIT engineers are hopeful that their work could result in new, less expensive detectors. For example, they envision trucks loaded with cargo from container ships driving through a structure that has detectors on both sides as they leave a port. “Ideally, you’d have either an array of detectors or a very large detector, and that’s where [today’s detectors] really don’t scale very well,” Tuller says.
Another potential application involves accessing geothermal energy, or the extreme heat below our feet that is being explored as a carbon-free alternative to fossil fuels. Ceramic sensors at the ends of drill bits could detect pockets of heat — radiation — to drill toward. Ceramics can easily withstand extreme temperatures of more than 800 degrees Fahrenheit and the extreme pressures found deep below the Earth’s surface.
The team is excited about additional applications for their work. “This was a demonstration of principle with just one material,” says Tuller, “but there are thousands of other materials good at conducting ions.”
Concludes Defferriere: “It’s the start of a journey on the development of the technology, so there’s a lot to do and a lot to discover.”
This work is currently supported by the U.S. Department of Homeland Security, Countering Weapons of Mass Destruction Office. This support does not constitute an express or implied endorsement on the part of the government. It was also funded by the U.S. Defense Threat Reduction Agency.
Share this news article on:
Related links.
- Harry Tuller
- Tuller Research Group
- Materials Research Laboratory
Related Topics
- Nuclear security and policy
- Materials science and engineering
- Nuclear science and engineering
- Department of Energy (DoE)

Related Articles

A simple way to significantly increase lifetimes of fuel cells and other devices

Harry Tuller honored for career advancing solid-state chemistry and electrochemistry

Light could boost performance of fuel cells, lithium batteries, and other devices
Previous item Next item
More MIT News

A biomedical engineer pivots from human movement to women’s health
Read full story →

MIT tops among single-campus universities in US patents granted

A crossroads for computing at MIT

Growing our donated organ supply
New AI method captures uncertainty in medical images

Improving drug development with a vast map of the immune system
- More news on MIT News homepage →
Massachusetts Institute of Technology 77 Massachusetts Avenue, Cambridge, MA, USA
- Map (opens in new window)
- Events (opens in new window)
- People (opens in new window)
- Careers (opens in new window)
- Accessibility
- Social Media Hub
- MIT on Facebook
- MIT on YouTube
- MIT on Instagram
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
DNA computing articles from across Nature Portfolio
DNA computing is a branch of biomolecular computing concerned with the use of DNA as a carrier of information to make arithmetic and logic operations.
Latest Research and Reviews

Efficient DNA-based data storage using shortmer combinatorial encoding
- Inbal Preuss
- Michael Rosenberg
Pattern recognition in the nucleation kinetics of non-equilibrium self-assembly
Examination of nucleation during self-assembly of multicomponent structures illustrates how ubiquitous molecular phenomena inherently classify high-dimensional patterns of concentrations in a manner similar to neural network computation.
- Constantine Glen Evans
- Jackson O’Brien
- Arvind Murugan
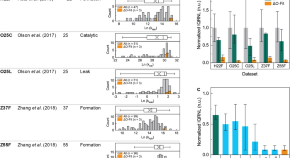
Generation of DNA oligomers with similar chemical kinetics via in-silico optimization
Networks of interacting DNA oligomers have various applications in molecular biology, chemistry and materials science, however, kinetic dispersions during DNA hybridization can be problematic for some applications. Here, the authors reveal that limiting unnecessary duplexes using in-silico optimization can reduce in-vitro kinetic dispersions by as much as 96%.
- Michael Tobiason
- Bernard Yurke
- William L. Hughes
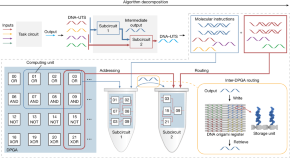
DNA-based programmable gate arrays for general-purpose DNA computing
Generic single-stranded oligonucleotides used as a uniform transmission signal can reliably integrate large-scale DNA integrated circuits with minimal leakage and high fidelity for general-purpose computing.
- Chunhai Fan
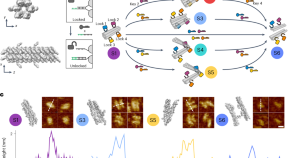
A temporally resolved DNA framework state machine in living cells
The heterogeneous and compartmentalized environments within living cells make it difficult to deploy theranostic agents with precise spatiotemporal accuracy. Zhao et al. demonstrate a DNA framework state machine that can switch among multiple structural states according to the temporal sequence of molecular cues, enabling temporally controlled CRISPR–Cas9 targeting in living mammalian cells.
- Shuting Cao
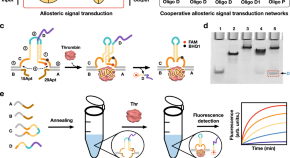
Programming conformational cooperativity to regulate allosteric protein-oligonucleotide signal transduction
Conformational cooperativity is a universal molecular effect mechanism and plays a critical role in signalling pathways. Here the authors present a programmable conformational cooperativity strategy to construct the oligo-protein signal transduction platform for logic operations and gene regulations which can be cooperatively regulated by conformational signals.
- Cheng Zhang
News and Comment
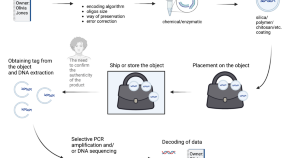
Unlocking the potential of DNA-based tagging: current market solutions and expanding horizons
The commercialization of DNA tagging is a growing trend that demonstrates the increasing practicality of this novel approach. This interdisciplinary technology is based on the distinctive characteristics of DNA as a molecule that can remain stable in varying environmental conditions and store data following appropriate preparation. Moreover, newly developed technologies could simplify DNA synthesis and the encoding of data within DNA. The implementation of DNA tagging presents distinctive benefits in comparison to conventional labelling techniques, including universal product code (UPC) barcoding, radio-frequency identification (RFID), quick response (QR) codes, and Bluetooth technologies, by surmounting the limitations encountered by these systems. The discourse pertains to extant DNA-tagging mechanisms along with prospective implementations in a wide range of domains, including but not limited to art, the metaverse, forensics, wildlife monitoring, and the military. The potential of DNA labelling in various contexts underscores the importance of continued research and development in this rapidly evolving field.
- Adam Kuzdraliński
- Marek Miśkiewicz
- Bogdan Księżopolski
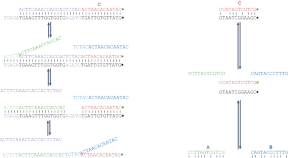
Non-complementary computation
Molecular computing programmed with complementary nucleic acid strands allows the construction of sophisticated biomolecular circuits. Now, systems with partially complementary strands have been shown to enable more compact and faster molecular circuits, and may illuminate biological processes.
- Philip Petersen
- Grigory Tikhomirov
In vitro convolutional neural networks
Neural networks can be implemented by using purified DNA molecules that interact in a test tube. Convolutional neural networks to classify high-dimensional data have now been realized in vitro, in one of the most complex demonstrations of molecular programming so far.
- William Poole
Spatially localized DNA domino
Fast and scalable molecular logic circuits have been constructed by spatially localizing DNA hairpins on DNA origami scaffolds.
- André Estevez-Torres
- Yannick Rondelez
Programmed communication
Autocatalytic nanoparticles activated and deactivated by DNA 'programs' can trigger through-space molecular communication and give rise to collective particle behaviours.
- Kristiana Kandere-Grzybowska
- Bartosz A. Grzybowski
Nothing more than DNA
- Bryden Le Bailly
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Share full article

There’s an Explosion of Plastic Waste. Big Companies Say ‘We’ve Got This.’
Big brands like Procter & Gamble and Nestlé say a new generation of plants will help them meet environmental goals, but the technology is struggling to deliver.
Recycled polypropylene pellets at a PureCycle Technologies plant in Ironton, Ohio. Credit... Maddie McGarvey for The New York Times
Supported by

By Hiroko Tabuchi
- Published April 5, 2024 Updated April 8, 2024
By 2025, Nestle promises not to use any plastic in its products that isn’t recyclable. By that same year, L’Oreal says all of its packaging will be “refillable, reusable, recyclable or compostable.”
And by 2030, Procter & Gamble pledges that it will halve its use of virgin plastic resin made from petroleum.
To get there, these companies and others are promoting a new generation of recycling plants, called “advanced” or “chemical” recycling, that promise to recycle many more products than can be recycled today.
So far, advanced recycling is struggling to deliver on its promise. Nevertheless, the new technology is being hailed by the plastics industry as a solution to an exploding global waste problem.
The traditional approach to recycling is to simply grind up and melt plastic waste. The new, advanced-recycling operators say they can break down the plastic much further, into more basic molecular building blocks, and transform it into new plastic.
PureCycle Technologies, a company that features prominently in Nestlé, L’Oréal, and Procter & Gamble’s plastics commitments, runs one such facility, a $500 million plant in Ironton, Ohio. The plant was originally to start operating in 2020 , with the capacity to process as much as 182 tons of discarded polypropylene, a hard-to-recycle plastic used widely in single-use cups, yogurt tubs, coffee pods and clothing fibers, every day.

But PureCycle’s recent months have instead been filled with setbacks: technical issues at the plant, shareholder lawsuits, questions over the technology and a startling report from contrarian investors who make money when a stock price falls. They said that they had flown a drone over the facility that showed that the plant was far from being able to make much new plastic.
PureCycle, based in Orlando, Fla., said it remained on track. “We’re ramping up production,” its chief executive, Dustin Olson, said during a recent tour of the plant, a constellation of pipes, storage tanks and cooling towers in Ironton, near the Ohio River. “We believe in this technology. We’ve seen it work,” he said. “We’re making leaps and bounds.”
Nestlé, Procter & Gamble and L’Oréal have also expressed confidence in PureCycle. L’Oréal said PureCycle was one of many partners developing a range of recycling technologies. P.&G. said it hoped to use the recycled plastic for “numerous packaging applications as they scale up production.” Nestlé didn’t respond to requests for comment, but has said it is collaborating with PureCycle on “groundbreaking recycling technologies.”
PureCycle’s woes are emblematic of broad trouble faced by a new generation of recycling plants that have struggled to keep up with the growing tide of global plastic production, which scientists say could almost quadruple by midcentury .
A chemical-recycling facility in Tigard, Ore., a joint venture between Agilyx and Americas Styrenics, is in the process of shutting down after millions of dollars in losses. A plant in Ashley, Ind., that had aimed to recycle 100,000 tons of plastic a year by 2021 had processed only 2,000 tons in total as of late 2023, after fires, oil spills and worker safety complaints.
At the same time, many of the new generation of recycling facilities are turning plastic into fuel, something the Environmental Protection Agency doesn’t consider to be recycling, though industry groups say some of that fuel can be turned into new plastic .
Overall, the advanced recycling plants are struggling to make a dent in the roughly 36 million tons of plastic Americans discard each year, which is more than any other country. Even if the 10 remaining chemical-recycling plants in America were to operate at full capacity, they would together process some 456,000 tons of plastic waste, according to a recent tally by Beyond Plastics , a nonprofit group that advocates stricter controls on plastics production. That’s perhaps enough to raise the plastic recycling rate — which has languished below 10 percent for decades — by a single percentage point.
For households, that has meant that much of the plastic they put out for recycling doesn’t get recycled at all, but ends up in landfills. Figuring out which plastics are recyclable and which aren’t has turned into, essentially, a guessing game . That confusion has led to a stream of non-recyclable trash contaminating the recycling process, gumming up the system.
“The industry is trying to say they have a solution,” said Terrence J. Collins, a professor of chemistry and sustainability science at Carnegie Mellon University. “It’s a non-solution.”
‘Molecular washing machine’
It was a long-awaited day last June at PureCycle’s Ironton facility: The company had just produced its first batch of what it describes as “ultra-pure” recycled polypropylene pellets.
That milestone came several years late and with more than $350 million in cost overruns. Still, the company appeared to have finally made it. “Nobody else can do this,” Jeff Kramer, the plant manager, told a local news crew .
PureCycle had done it by licensing a game-changing method — developed by Procter & Gamble researchers in the mid-2010s, but unproven at scale — that uses solvent to dissolve and purify the plastic to make it new again. “It’s like a molecular washing machine,” Mr. Olson said.
There’s a reason Procter & Gamble, Nestlé and L’Oréal, some of the world’s biggest users of plastic, are excited about the technology. Many of their products are made from polypropylene, a plastic that they transform into a plethora of products using dyes and fillers. P.&G. has said it uses more polypropylene than any other plastic, more than a half-million tons a year.
But those additives make recycling polypropylene more difficult.
The E.P.A. estimates that 2.7 percent of polypropylene packaging is reprocessed. But PureCycle was promising to take any polypropylene — disposable beer cups, car bumpers, even campaign signs — and remove the colors, odors, and contaminants to transform it into new plastic.
Soon after the June milestone, trouble hit.
On Sept. 13, PureCycle disclosed that its plant had suffered a power failure the previous month that had halted operations and caused a vital seal to fail. That meant the company would be unable to meet key milestones, it told lenders.
Then in November, Bleecker Street Research — a New York-based short-seller, an investment strategy that involves betting that a company’s stock price will fall — published a report asserting that the white pellets that had rolled off PureCycle’s line in June weren’t recycled from plastic waste. The short-sellers instead claimed that the company had simply run virgin polypropylene through the system as part of a demonstration run.
Mr. Olson said PureCycle hadn’t used consumer waste in the June 2023 run, but it hadn’t used virgin plastic, either. Instead it had used scrap known as “post industrial,” which is what’s left over from the manufacturing process and would otherwise go to a landfill, he said.
Bleecker Street also said it had flown heat-sensing drones over the facility and said it found few signs of commercial-scale activity. The firm also raised questions about the solvent PureCycle was using to break down the plastic, calling it “a nightmare concoction” that was difficult to manage.
PureCycle is now being sued by other investors who accuse the company of making false statements and misleading investors about its setbacks.
Mr. Olson declined to describe the solvent. Regulatory filings reviewed by The New York Times indicate that it is butane, a highly flammable gas, stored under pressure. The company’s filing described the risks of explosion, citing a “worst case scenario” that could cause second-degree burns a half-mile away, and said that to mitigate the risk the plant was equipped with sprinklers, gas detectors and alarms.
Chasing the ‘circular economy’
It isn’t unusual, of course, for any new technology or facility to experience hiccups. The plastics industry says these projects, once they get going, will bring the world closer to a “circular” economy, where things are reused again and again.
Plastics-industry lobbying groups are promoting chemical recycling. At a hearing in New York late last year, industry lobbyists pointed to the promise of advanced recycling in opposing a packaging-reduction bill that would eventually mandate a 50 percent reduction in plastic packaging. And at negotiations for a global plastics treaty , lobby groups are urging nations to consider expanding chemical recycling instead of taking steps like restricting plastic production or banning plastic bags.
A spokeswoman for the American Chemistry Council, which represents plastics makers as well as oil and gas companies that produce the building blocks of plastic, said that chemical recycling potentially “complements mechanical recycling, taking the harder-to-recycle plastics that mechanical often cannot.”
Environmental groups say the companies are using a timeworn strategy of promoting recycling as a way to justify selling more plastic, even though the new recycling technology isn’t ready for prime time. Meanwhile, they say, plastic waste chokes rivers and streams, piles up in landfills or is exported .
“These large consumer brand companies, they’re out over their skis,” said Judith Enck, the president of Beyond Plastics and a former regional E.P.A. administrator. “Look behind the curtain, and these facilities aren’t operating at scale, and they aren’t environmentally sustainable,” she said.
The better solution, she said, would be, “We need to make less plastic.”
Touring the plant
Mr. Olson recently strolled through a cavernous warehouse at PureCycle’s Ironton site, built at a former Dow Chemical plant. Since January, he said, PureCycle has been processing mainly consumer plastic waste and has produced about 1.3 million pounds of recycled polypropylene, or about 1 percent of its annual production target.
“This is a bag that would hold dog food,” he said, pointing to a bale of woven plastic bags. “And these are fruit carts that you’d see in street markets. We can recycle all of that, which is pretty cool.”
The plant was dealing with a faulty valve discovered the day before, so no pellets were rolling off the line. Mr. Olson pulled out a cellphone to show a photo of a valve with a dark line ringing its interior. “It’s not supposed to look like that,” he said.
The company later sent video of Mr. Olson next to white pellets once again streaming out of its production line.
PureCycle says every kilogram of polypropylene it recycles emits about 1.54 kilograms of planet-warming carbon dioxide. That’s on par with a commonly used industry measure of emissions for virgin polypropylene. PureCycle said that it was improving on that measure.
Nestlé, L’Oréal and Procter & Gamble continue to say they’re optimistic about the technology. In November, Nestlé said it had invested in a British company that would more easily separate out polypropylene from other plastic waste.
It was “just one of the many steps we are taking on our journey to ensure our packaging doesn’t end up as waste,” the company said.
Hiroko Tabuchi covers the intersection of business and climate for The Times. She has been a journalist for more than 20 years in Tokyo and New York. More about Hiroko Tabuchi
Learn More About Climate Change
Have questions about climate change? Our F.A.Q. will tackle your climate questions, big and small .
“Buying Time,” a new series from The New York Times, looks at the risky ways humans are starting to manipulate nature to fight climate change.
Big brands like Procter & Gamble and Nestlé say a new generation of recycling plants will help them meet environmental goals, but the technology is struggling to deliver .
The Italian energy giant Eni sees future profits from collecting carbon dioxide and pumping it into natural gas fields that have been exhausted.
New satellite-based research reveals how land along the East Coast is slumping into the ocean, compounding the danger from global sea level rise . A major culprit: the overpumping of groundwater.
Did you know the ♻ symbol doesn’t mean something is actually recyclable ? Read on about how we got here, and what can be done.
Advertisement
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Forensic Sci Int Synerg
Recent advances in forensic biology and forensic DNA typing: INTERPOL review 2019–2022
Associated data.
This review paper covers the forensic-relevant literature in biological sciences from 2019 to 2022 as a part of the 20th INTERPOL International Forensic Science Managers Symposium. Topics reviewed include rapid DNA testing, using law enforcement DNA databases plus investigative genetic genealogy DNA databases along with privacy/ethical issues, forensic biology and body fluid identification, DNA extraction and typing methods, mixture interpretation involving probabilistic genotyping software (PGS), DNA transfer and activity-level evaluations, next-generation sequencing (NGS), DNA phenotyping, lineage markers (Y-chromosome, mitochondrial DNA, X-chromosome), new markers and approaches (microhaplotypes, proteomics, and microbial DNA), kinship analysis and human identification with disaster victim identification (DVI), and non-human DNA testing including wildlife forensics. Available books and review articles are summarized as well as 70 guidance documents to assist in quality control that were published in the past three years by various groups within the United States and around the world.
1. Introduction
This review explores developments in forensic biology and forensic DNA analysis of biological evidence during the years 2019–2022. In some cases, there may be overlap with 2019 articles mentioned in the previous INTERPOL review covering 2016 to 2019 [ 1 ]. This review includes books and review articles, published guidance documents to assist in quality control, rapid DNA testing, using law enforcement DNA databases plus investigative genetic genealogy DNA databases along with privacy/ethical issues, forensic biology and body fluid identification, DNA extraction and typing methods, mixture interpretation involving probabilistic genotyping software (PGS), DNA transfer and activity level evaluations, next-generation sequencing (NGS), DNA phenotyping, lineage markers (Y-chromosome, mitochondrial DNA, X-chromosome), new markers and approaches (microhaplotypes, proteomics, and microbial DNA), kinship analysis and human identification with disaster victim identification (DVI), and non-human DNA testing including wildlife forensics.
Multiple searches, using the Scopus (Elsevier) and Web of Science (Clarivate) databases, were conducted in the first half of 2022 with “forensic” and “DNA” or “biology” and “2019 to 2022” as search options. Over 4000 articles were returned with these searches. Through visual examination of titles and authors, duplicates were removed, and articles sorted into 32 subcategories to arrive at a list of almost 2000 publications that were supplemented throughout the remainder of the year as this review was being prepared. The tables of contents for non-indexed journals, such as WIRES Forensic Science , Journal of Forensic Identification , and Forensic Genomics were also examined to locate potentially relevant articles.
For example, a Scopus search conducted on June 13, 2022, using “forensic DNA” and “2019 to 2022” found a total of 3059 documents. Table 1 lists the top ten journals from this search. The Forensic Science International: Genetics Supplement Series (see row #4 in Table 1 ) provides the proceedings of the International Society for Forensic Genetics (ISFG) meeting held in Prague in September 2019. This volume contains 914 pages with 347 articles (although only 172 showed up in the Scopus search) that are freely available at https://www.fsigeneticssup.com /[ 2 ]. Thus, searches conducted with one or even multiple databases (e.g., Scopus and Web of Science) may not be comprehensive or exhaustive.
Top ten journals with forensic DNA articles published from 2019 to 2022 based on a Scopus search on June 13, 2022.
1.1. Books, special issues, and review articles of note
Books published during the period of this review relating to forensic biology and forensic DNA include Essential Forensic Biology, Third Edition [ 3 ], Principles and Practices of DNA Analysis: A Laboratory Manual for Forensic DNA Typing [ 4 ], Forensic DNA Profiling: A Practical Guide to Assigning Likelihood Ratios [ 5 ], Forensic Practitioner's Guide to the Interpretation of Complex DNA Profiles [ 6 ], Silent Witness: Forensic DNA Evidence in Criminal Investigations and Humanitarian Disasters [ 7 ], Mass Identifications: Statistical Methods in Forensic Genetics [ 8 ], Probability and Forensic Evidence: Theory, Philosophy, and Applications [ 9 ], Interpreting Complex Forensic DNA Evidence [ 10 ], Understanding DNA Ancestry [ 11 ], Understanding Forensic DNA [ 12 ], and Handbook of DNA Profiling [ 13 ]. The 2022 Handbook of DNA Profiling spans two volumes and 1206 pages with 54 chapters from 115 contributors representing 17 countries.
Over the past three years, several special issues on topics related to forensic biology were published in Forensic Science International: Genetics and Genes . These special issues were typically collated virtually rather than physically as invited articles were published online over some period of time and then bundled together virtually as a special issue. Some of these review articles or a set of special issue articles are open access (i.e., the authors paid a publication fee so that the article would be available online for free to readers).
During the time frame of this INTERPOL DNA review, FSI Genetics published two special issues: (1) “Trends and Perspectives in Forensic Genetics” (editor: Manfred Kayser) 1 with nine review and two original research articles published between September 2018 and January 2019, and (2) “Forensic Genetics – Unde venisti et quo vadis?” [Latin for “where did you come from and where are you going?”] (editor: Manfred Kayser) with nine articles published in 2021 and early 2022 and likely two more before the end of 2022. Topics for review articles in these special issues include DNA transfer [ 14 ], probabilistic genotyping software [ 15 ], microhaplotypes in forensic genetics [ 16 ], investigative genetic genealogy [ 17 ], forensic proteomics [ 18 ], distinguishing male monozygotic twins [ 19 ], and using the human microbiome for estimating post-mortem intervals and identifying individuals, tissues, or body fluids [ 20 , 21 ]. All of these topics will be discussed later in this article.
A Genes special issue “Forensic Genetics and Genomics” (editors: Emiliano Giardina and Michele Ragazzo) 2 published 11 online articles plus an editorial from April 2020 to January 2021 while another Genes special issue “Forensic Mitochondrial Genomics” (editors: Mitch Holland and Charla Marshall) 3 compiled 11 articles from February 2020 to April 2021. An “Advances in Forensic Genetics” Genes special issue (editor: Niels Morling) 4 included 25 articles shared between April 2021 and May 2022. In July 2022, the Advances in Forensic Genetics articles were compiled as a 518-page book. 5 Other Genes special issues in development or forthcoming covering aspects of forensic DNA and requesting potential manuscripts by late 2022 or early 2023 include “State-of-the-Art in Forensic Genetics” (editor: Chiara Turchi), 6 “Trends in Population Genetics and Identification—Impact on Anthropology (editors: Antonio Amorim, Veronica Gomes, Luisa Azevedo), 7 “Identification of Human Remains for Forensic and Humanitarian Purposes: From Molecular to Physical Methods” (editors: Elena Pilli, Cristina Cattaneo), 8 “Improved Methods in Forensic and DNA Analysis” (editor: Marie Allen), 9 “Forensic DNA Mixture Interpretation and Probabilistic Genotyping” (editor: Michael Coble) 10 , and “Advances in Forensic Molecular Genetics” (editors: Erin Hanson and Claire Glynn). 11 There has been a proliferation of review articles and special issues in this field in the past several years!
A new journal Forensic Science International: Reports was launched in November 2019. As of June 2022, it has published 89 articles involving DNA, most of which are descriptions of population genetic data. Likewise, a June 27, 2022, PubMed search with “forensic DNA” and the journal “Genes” found 88 articles – many of which are part of the previously mentioned special issues.
1.2. Guidance documents
Numerous documentary standards and guidance documents related to forensic DNA have been published by various organizations around the world. Table 2 lists 70 such documents released in the past three years (2019–2022) in the United States, UK, Australia, and the European Union.
Guidance documents related to forensic DNA published from 2019 to 2022. The titles are hyperlinked to available documents. Abbreviations: FBI (Federal Bureau of Investigation), CODIS (Combined DNA Index System), SWGDAM (Scientific Working Group on DNA Analysis Methods), NGS (next generation sequencing), US DOJ (United States Department of Justice), ULTR (Uniform Language for Testimony and Reports), AABB (Association for the Advancement of Blood and Biotherapies), ASB (Academy Standards Board), OSAC (Organization of Scientific Area Committees for Forensic Science), UKFSR (United Kingdom Forensic Science Regulator), ENFSI (European Network of Forensic Science Institutes), NIFS (National Institute of Forensic Science), ISFG (International Society for Forensic Genetics).
1.2.1. SWGDAM, FBI, and other US DOJ activities
The Federal Bureau of Investigation (FBI) Laboratory funds the Scientific Working Group on DNA Analysis Methods (SWGDAM) 12 to serve as a forum for discussing, sharing, and evaluating forensic biology methods, protocols, training, and research. In addition to creating guidelines on various topics, SWGDAM, which meets semiannually in January and July, provides recommendations to the FBI Director on the Quality Assurance Standards (QAS) used to assess U.S. forensic DNA laboratories involved in the National DNA Index System (NDIS) that perform DNA databasing and forensic casework. New versions of the QAS became effective July 1, 2020.
SWGDAM work products from the timeframe of 2019–2022 (see Table 2 ) include QAS audit and guidance documents, mitochondrial DNA analysis and short tandem repeat (STR) interpretation guideline revisions related to next-generation sequencing (NGS), training and Y-chromosome interpretation guidelines, a Y-chromosome Haplotype Reference Database (YHRD) update for U.S. laboratories, and reports on investigative genetic genealogy and Y-screening of sexual assault evidence kits. These documents are all accessible online. 13
In January 2022, the FBI produced a 13-page guide 14 on rapid DNA testing describing booking station applications and their vision for future integration of crime scene sample analysis and the Combined DNA Index System (CODIS), which builds on a joint position statement published in July 2020 by leaders of U.S. and European groups [ 22 ]. In addition, the FBI has shared guidance on their website for non-CODIS use of rapid DNA testing with law enforcement applications 15 and considerations for court. 16
United States Department of Justice (US DOJ) Uniform Language for Testimony and Reports (ULTRs), 17 contain three ULTRs for the forensic DNA discipline that became effective in March 2019: autosomal DNA with probabilistic genotyping, mitochondrial DNA, and Y-STR DNA. USDOJ also released an interim policy on investigative genetic genealogy in November 2019 [ 23 ] along with an opinion piece in the journal Science calling for responsible genetic genealogy [ 24 ].
Other agencies within US DOJ, namely the Bureau of Justice Assistance (BJA) and the National Institute of Justice (NIJ), published a guide for prosecutors on triaging forensic evidence [ 25 ] and best practices for improving DNA laboratory process efficiency [ 26 ]. A 200-page report to Congress on the needs assessment of forensic laboratories and medical examiner/coroner offices was released in December 2019 calling for $640 million annually in additional funding to support U.S. forensic efforts [ 27 ].
In September 2021, the Forensic Technology Center of Excellence (FTCOE), which is funded by NIJ, published a 29-page implementation strategy on next-generation sequencing for DNA analysis that was written by the NIJ Forensic Laboratory Needs Technology Working Group (FLN-TWG) [ 28 ]. In May 2022, FTCOE released a 50-page landscape study examining technologies and automation for differential extraction and sperm separation used in sexual assault investigations [ 29 ]. An introduction to forensic genetic genealogy was released in September 2022 [ 30 ].
The FTCOE also published a human factors forensic science sourcebook 18 in March 2022 through open access articles in the journal Forensic Science International: Synergy . This sourcebook, which has general applicability rather than being specific to forensic DNA analysts, includes an overview article [ 31 ] along with articles on personnel selection and assessment [ 32 ], the benefits of committing errors during training [ 33 ], how characteristics of human reasoning and certain situations can contribute to errors [ 34 ], stressors that impact performance [ 35 ], and the impact of communication between forensic analysts and detectives using a new metaphor [ 36 ].
1.2.2. OSAC and ASB activities
The Organization of Scientific Area Committees for Forensic Science (OSAC) 19 is congressionally-funded and administered by the Special Programs Office within the National Institute of Standards and Technology (NIST). OSAC consists of a governing board and over 600 members and associates organized into seven scientific area committees (SACs) and 22 subcommittees. The Biology SAC is divided into human and wildlife forensic biology activities. The Human Forensic Biology Subcommittee 20 focuses on standards and guidelines related to training, method development and validation, data analysis, interpretation, and statistical analysis as well as reporting and testimony for human forensic serological and DNA testing. The Wildlife Forensics Subcommittee 21 works on standards and guidelines related to taxonomic identification, individualization, and geographic origin of non-human biological evidence based on morphological and genetic analyses.
The Academy Standards Board (ASB) 22 is a wholly owned subsidiary of the American Academy of Forensic Sciences (AAFS) and was established as a standards developing organization (SDO). In 2015, ASB was accredited as an SDO by the American National Standards Institute (ANSI). The ASB DNA Consensus Body, with a membership consisting of practitioners, researchers, and lawyers, develops standards and guidelines related to the use of DNA in legal proceedings. Many of the documents developed by ASB were originally proposed OSAC standards or guidelines.
The OSAC Registry 23 is a repository of high-quality and technically-sound standards (both published and proposed) that are intended for implementation in forensic science laboratories. As of July 2022, the OSAC Registry contains 11 standards published by ASB as well as two (2) proposed OSAC standards or best practice recommendations related to human forensic biology. Another four ASB standards and two proposed OSAC standards related to wildlife forensic biology are on the OSAC Registry. The ASB standards issued in the past three years related to human forensic biology cover interpretation and comparison protocols, training in various parts of the process, and validation of forensic serological and DNA analysis methods as well as probabilistic genotyping systems (see Table 2 for names of these documents). A number of other documents 24 related to serological testing methods, assigning propositions for likelihood ratios in forensic DNA interpretations, validation of forensic DNA methods and software, familial DNA searching, management and use of quality assurance DNA elimination databases, setting thresholds, evaluative forensic DNA testimony, and training in use of statistics are in development within OSAC and ASB.
Additional work products of OSAC include (1) a lexicon 25 with 3282 records (although multiple records may exist for the same word, e.g., there are five definitions provided for “validation” from various sources), (2) a 35-page technical guidance document 26 on human factors in validation and performance testing that describes key issues in designing, conducting, and reporting validation research, (3) a listing of research and development needs in forensic science 27 including 18 identified by the OSAC Human Forensic Biology Subcommittee during their deliberations ( Table 3 ), and (4) process maps for several forensic disciplines including a 42-page depiction of current practices and decisions in human forensic DNA analysis released in May 2022 [ 37 ]. As a visual representation of critical steps and decision points, a process map is intended to help improve efficiencies and reduce errors, and highlight gaps where further research or standardization would be beneficial. Process maps can assist with training new examiners and enable development of specific laboratory policies or help identify best practices for the field.
Research and development needs in forensic biology as identified by the OSAC Human Forensic Biology Subcommittee (as of July 2022, see https://www.nist.gov/osac/osac-research-and-development-needs ).
1.2.3. UK Forensic Science Regulator
The UK Forensic Science Regulator (UKFSR) oversees forensic science efforts in England, Wales, and Northern Ireland. In March 2021, the Regulator released the seventh issue 28 of the Codes of Practice and Conduct for forensic science providers and practitioners in the criminal justice system. This 114-page document, which has been updated every few years, provides the overall framework for forensic science activities in the UK with other supporting guidance documents on specific areas like DNA analysis or general tasks like validation. In September 2020, a number of the Regulator documents were revised and reissued. As noted in Table 2 (see rows with documents containing “Issue 1” in the title), new guidance documents were also released in the past few years on sexual assault examinations, development of evaluative opinions, proficiency testing for DNA mixture interpretation, Y-STR profiling, DNA relationship testing, and methods employing rapid DNA testing devices. Table 2 lists 20 guidance documents pertinent to forensic biology from the UKFSR.
1.2.4. European Union and Australia
The European Network of Forensic Science Institutes (ENFSI) DNA Working Group published two documents in the past three years: one on DNA database management and the other on training of staff in forensic DNA laboratories (see Table 2 ). A best practice manual for human forensic biology and DNA profiling is also under development.
The Australian National Institute of Forensic Science (NIFS) published three documents of relevance to forensic biology on case record review, empirical study design, and transitioning technology from the laboratory to the field (see Table 2 ).
1.2.5. Other international efforts
The Association for the Advancement of Blood and Biotherapies (AABB) 29 published the 15th edition of their Standard for Relationship Testing Laboratories, which became effective on January 1, 2022. This documentary standard was developed by the AABB Relationship Testing Standards Committee and applies to laboratories accredited for paternity testing and other forms of genetic relationship assessment.
The International Society for Forensic Genetics (ISFG) DNA Commission 30 published two articles during the timeframe of this INTERPOL review (see Table 2 ). In 2020, guidelines and considerations were published on evaluating DNA results under activity level propositions [ 38 ]. In addition, the state of the field regarding interpretation of Y-STR results was examined along with different approaches for haplotype frequency estimation using population data – with the Discrete Laplace approach being recommended [ 39 ]. Future ISFG DNA Commission efforts will address STR allele sequence nomenclature and phenotyping.
2. Advancements in current practices
This section (Section 2 ) is intended to be law enforcement and practitioner-focused through examination of advances in current practices. The following section (Section 3 ) is intended to be researcher-focused through emphasis on emerging technologies and new developments. In this section, topics specifically covered include rapid DNA analysis, use of DNA databases to aid investigations (including familial searching, investigative genetic genealogy, genetic privacy and ethical concerns, and sexual assault kit testing), body fluid identification, DNA extraction and typing methods, and DNA interpretation at the sub-source and activity level.
2.1. Rapid DNA analysis
Rapid DNA instruments that provide integrated “swab-in-profile-out” results in 90 min or less can be used in police booking station environments and assist investigations outside of a traditional laboratory environment. These instruments were initially designed for analysis of buccal swabs to help speed processing of reference samples associated with criminal cases. Such samples are expected to contain relatively large quantities of DNA from a single contributor. Some attempts to extend the range of sample types to low quantities of DNA or mixtures have been published with various levels of success (see Table 4 ). Researcher and practitioners from Australia [ [40] , [41] , [42] ], Canada [ 43 ], China [ 44 ], Italy [ 45 ], Japan [ 46 , 47 ], and the United States [ [48] , [49] , [50] , [51] , [52] , [53] , [54] , [55] , [56] , [57] ] have contributed to an increased understanding of rapid DNA testing capabilities and limitations.
Summary of 20 rapid DNA instrument validation and evaluation studies published from 2019 to 2022. Abbreviations: A-Chip (arrestee cartridge, designed for high-quantity DNA samples), I-Chip (investigative cartridge, designed for low-quantity DNA samples), ACE (arrestee cartridge with GlobalFiler STR markers), RapidINTEL (uses 32 rather than 28 PCR cycles to increase success with low-quantity DNA samples). A-Chip and I-Chip amplify the FlexPlex set of 23 autosomal STRs, three Y-STRs, and amelogenin [ 51 ]. ACE and RapidINTEL utilize the GlobalFiler set of 21 autosomal STRs, one Y-STR, one Y-chromosome InDel, and amelogenin.
The Accelerated Nuclear DNA Equipment (ANDE) 6C (ANDE, Longmont, CO, USA) and the RapidHIT ID (Thermo Fisher Scientific, Waltham, MA, USA) are the current 31 commercially available rapid DNA systems. Each system consists of a swab for introducing the sample, a cartridge or biochip with pre-packed reagents, the instrument, and analysis software with an expert system for automated STR allele calling. Different sample cartridges can be run on each system depending on the sample type and expected quantity of DNA.
For ANDE, the arrestee cartridge (A-Chip), can accommodate up to five samples and is intended for relatively high quantities of DNA typically collected from reference buccal swabs, while the investigative cartridge (I-Chip), can process up to four samples and is intended for lower quantities of DNA that might be present in casework or disaster victim identification samples. Both ANDE cartridges use the FlexPlex27 STR assay that tests 23 autosomal STR loci, three Y-chromosome STRs, and amelogenin to generate data compatible with DNA databases around the world [ 51 ]. The RapidHIT ID ACE cartridge and RapidINTEL cartridge serve similar purposes as the ANDE A-Chip and I-Chip using GlobalFiler Express kit markers (21 autosomal STRs, DYS391, a Y-chromosome insertion/deletion marker, and amelogenin) instead of the FlexPlex assay. The ACE sample cartridge uses buccal swabs while the EXT sample cartridge processes DNA extracts [ 56 ]. Sensitivity is enhanced in the RapidINTEL cartridge by increasing the number of PCR cycles from 28 to 32 and decreasing the lysis buffer volume from 500 μL to 300 μL compared to the ACE cartridge parameters [ 46 ].
With rapid DNA testing's swab-in and answer-out integrated configuration, limited options exist for testing conditions (e.g., either A-Chip or I-Chip with ANDE). Therefore, users should evaluate performance for the sample types they desired to routinely test in their specific environment. Table 4 summarizes recently published studies containing rapid DNA assessments.
National DNA Index System (NDIS) approval has been provided by the FBI Laboratory for accredited forensic DNA laboratories to use either the ANDE 6C or RapidHIT ID Systems (A-Chip and ACE cartridges only) 32 with eligible reference mouth swabs. As noted in Table 2 , the FBI.gov website contains three documents related to rapid DNA testing: “Non-CODIS Rapid DNA Considerations and Best Practices for Law Enforcement Use” (7-pages), “Rapid DNA Testing for Non-CODIS Uses: Considerations for Court” (5-pages), and “A Guide to All Things Rapid DNA” (13-pages) in January 2022 to provide information on the topic to law enforcement agencies.
The ENFSI DNA Working Group, SWGDAM, and an FBI Rapid DNA Crime Scene Technology Advancement Task Group co-published a position statement on the use of rapid DNA testing from crime scene samples [ 22 ]. These groups emphasized the need to have future rapid DNA systems with (1) methods to identify low quantity, degradation, and inhibition as well as meeting the human quantification requirements shared by SWGDAM and others, (2) the ability to export analyzable raw data for analysis or reanalysis by trained and qualified forensic DNA analysts, (3) an on-board fully automated expert system to accurately flag single-source or mixture DNA profiles requiring analyst evaluation, (4) improved peak height ratio balance (per locus and across loci) for low-quality and mixture samples “through enhancements in extraction efficiencies, changes in cycling parameters, and/or changes in STR kit chemistries,” and (5) published developmental validation studies on a wide variety of forensic evidence type samples with “data-supported recommendations regarding types of forensic evidence that are suitable and unsuitable for use with Rapid DNA technology” [ 22 ].
With a likely increase in the capabilities and the availability of rapid DNA systems, investigators will need to decide whether to use this capability onsite in specific situations or to send collected samples to a conventional forensic laboratory for processing at a later time. A group in the Netherlands collaborated with the New York City Police Department Crime Scene Unit and Evidence Collection Team to explore a decision support system [ 60 ]. In this study, participants were informed that rapid DNA testing was less sensitive compared to laboratory analysis and that the sample would be consumed, but that results from rapid DNA testing could identify a suspect within 2 h as opposed to waiting an average of 45 days for the laboratory results [presumably due to sample backlogs]. They were also told that a DNA profile obtained with rapid DNA would be acceptable in court. In the end, “>90% of the participants (85 out of 91) saw added value for using a Rapid DNA device in their investigative process …” with “a systematic approach, which consists of weighing all possible outcomes before deciding to use a Rapid DNA analysis device” [ 60 ]. The authors note that for such an approach to be successful “knowledge on DNA success rates [with various evidence types] is necessary in making evidence-based decisions for Rapid DNA analysis” [ 60 ].
A group in Australia performed a cost-benefit analysis of a decentralized rapid DNA workflow that might exist in the future with instruments placed at police stations around their country [ 61 ]. A virtual assessment considered all reference DNA samples collected during a two-month time period at 10 participating police stations in five regions of Australia. Processing times at the corresponding DNA analysis laboratories were calculated based on when the sample was received compared to the day when a DNA profile was obtained for that sample. From the survey conducted, it was estimated that up to 80,000 reference DNA samples are currently processed each year in forensic DNA laboratories across Australia [ 61 ].
Consumable costs for conventional DNA testing reagents in Australia were found to range from $17 to $35 whereas the rapid DNA consumable costs were estimated to be $100 per sample along with an anticipated $100,000 instrument cost per police station. Of course, the rate of use is expected to vary based on the number of reference samples collected in that jurisdiction. Since rapid DNA instruments utilize consumable cartridges with expiration dates, it was estimated that a police station would need to process six DNA samples per week to avoid having to discard an expired cartridge and thus increase the overall cost of their rapid DNA testing efforts. The authors of this study conclude “that routine laboratory DNA analysis meets the current needs for the majority of cases … It is anticipated that while the cost discrepancy between laboratory and rapid DNA processing remains high, the uptake of the technology in Australia will be limited [at least for a police booking station scenario]” [ 61 ].
Rapid DNA technology can be used in a variety of contexts including some that extend beyond traditional law enforcement. Seven distinct use contexts for rapid DNA capabilities have been described [ 62 ]: (1) evidence processing at or near crime scenes to generate leads for confirmation by a forensic laboratory, (2) booking or detection stations to compare an individual's DNA profile to a forensic database while the individual is still in custody, (3) disaster victim identification to permit rapid DNA processing of a victim's family members during their visit to family assistance centers when filing missing persons reports, (4) missing persons investigations to quickly process unidentified human remains and/or family reference samples to generate leads for confirmation by a forensic laboratory, (5) border security to develop DNA data from detainees for comparison to indices of prior border crossers while the individual is still in custody, (6) human trafficking and immigration fraud detection to permit immigration officials to verify family relationship claims, and (7) migrant family reunification to allow immigration officials to verify parentage claims and reunite family members separated at the border. Social and ethical considerations have been proposed for each of these use contexts in terms of data collection, data access and storage, and oversight and data protection [ 62 ].
One study [ 47 ] evaluating buccal swabs and mock disaster victim identification samples drew an important conclusion worth repeating here: “The Rapid DNA system provides robust and automated analysis of forensic samples without human review. Sample analysis failure can happen by chance in both the Rapid DNA system and conventional laboratory STR testing. While re-injection of PCR product is easily possible in the conventional method, this is not an option with the Rapid DNA system. Accordingly, the Rapid DNA system is a suitable choice but should be limited to samples that can easily be collected again if necessary or to samples that are of sufficient amount for repeated analysis. Application of this system to valuable samples such as those related to casework need to be considered carefully before analysis.”
2.2. Using DNA databases to aid investigations (national databases, familial searching, investigative genetic genealogy, genetic privacy & ethical concerns, sexual assault kit testing)
Forensic DNA databases can aid investigations by demonstrating connections between crime scenes, linking a previously enrolled DNA profile from an arrestee or convicted offender to biological material recovered from a crime scene, or aiding identification of missing persons through association of remains with biological relatives. Establishment of these databases requires significant investments over time to enroll data from crime scenes and potential serial offenders or unidentified human remains and relatives of missing persons. This section explores issues around national DNA databases, familial searching, investigative genetic genealogy, and genetic privacy and ethical concerns.
A systematic review regarding the effectiveness of forensic DNA databases looked at 19 articles published between 1985 and 2018 and found most studies support the assumption that DNA databases are an effective tool for the police, society, and forensic scientists [ 63 ]. Recommendations have been proposed to make cross-border exchange of DNA data more transparent and accountable with the Prüm system that enables information sharing across the European Union [ 64 ]. An analysis of news articles discussing the use of DNA testing in family reunification with migrants separated at the U.S.-Mexico border has been performed [ 65 ], and a standalone humanitarian DNA identification database has been proposed [ 66 ]. Aspects of international DNA kinship matching were explored to aid missing persons investigations and disaster victim identification processes [ 67 ]. A business case was presented for expanded DNA indirect matching using additional genetic markers, such as Y-chromosome STRs, mitochondrial DNA, and X-chromosome STRs, to reveal previously undetected familial relationships [ 68 ].
Approaches to transnational exchange of DNA data include (1) creation of an international DNA database, (2) linked or networked national DNA databases, (3) request-based exchange of data, and (4) a combination of these [ 69 ]. For example, the INTERPOL DNA database 33 contains more than 247,000 profiles contributed by 84 member countries. The I-Familia global database assists with missing persons identification based on international DNA kinship matching. 34
2.2.1. National DNA databases
Since the United Kingdom launched the first national DNA database in 1995, national DNA databases continue to be added in many countries including Brazil [ 70 , 71 ], India [ 72 ], Pakistan [ 73 , 74 ], Portugal [ 75 ], and Serbia [ 76 ]. A survey of 15 Latin American countries found that 13 of them had some kind of DNA database [ 77 ]. The opinions of 210 prisoners and prison officials in three Spanish penitentiary centers were also collected regarding DNA databases [ 78 ].
The effectiveness of databases has been debated over the years. Seven key indicators were used in a 2019 examination of the effectiveness of the UK national DNA database. These indicators included (1) implementation cost – the financial input required to implement the database system, (2) crime-solving capability – the ability of the database to assist criminal justice officials in case resolution, (3) incapacitation effect – the ability of the database to reduce crime through the incapacitation of offenders, (4) deterrence effect – the preventative potential of the database through deterrence of individuals from committing crime, (5) privacy protection – protection of the privacy or civil liberty rights of individuals, (6) legitimacy – compliance of the databasing system to the principle of proportionality, and (7) implementation efficiency – the time and non-monetary resource required to implement the database system [ 79 ].
A follow-up article concluded: “Available evidence shows that while DNA analysis has contributed to successful investigations in many individual cases, its aggregate value to the resolution of all crime is low” [ 80 ]. The systematic review of 19 articles on DNA databases cited previously noted “the expansion of DNA databases would only have positive effects on detection and clearance if the offender were already included in the database” [ 63 ]. When previous offenders are not already in a law enforcement DNA database to provide a hit to a crime scene profile, efforts are increasingly turning to familial searching and investigative genetic genealogy as described in the following sections.
2.2.2. Familial DNA searching
Familial DNA searching (FDS) extends the traditional direct matching of STR profiles within law enforcement databases to search for potential close family relationships, such as a parent or sibling, of a profile in the database. 35 FDS typically uses Y-STR lineage testing to narrow the set of candidate possibilities along with other case information such as geographic details of the crime and age of the person(s) of interest. For example, FDS helped solve murder cases in Romania [ 81 ] and China [ 82 ] by locating the perpetrator through a relative in the DNA database. A survey of 103 crime laboratories in the United States found that 11 states use FDS while laboratories in 24 states use a similar but distinct practice of partial matching [ 83 ].
The expansion of the number of STRs from 15 to 20 or 21 helps distinguish between true and false matches during a DNA database search by reducing the number of FDS adventitious matches [ 84 ]. Another study noted that the choice of allele frequencies affects the rate at which non-relatives are erroneously classified as relatives and found that using ancestry inference on the query profile can reduce false positive rates [ 85 ]. New Y-STR kits have been developed to assist with familial searching [ 86 , 87 ]. FDS of law enforcement databases differs from investigative genetic genealogy in two important ways – the genetic markers and the databases used for searching [ 88 , 89 ].
2.2.3. Investigative genetic genealogy
In recent years when national DNA databases fail to generate a lead to a potential person of interest, law enforcement agencies have started to utilize the capabilities of investigative genetic genealogy (IGG), also called forensic genetic genealogy (FGG) or forensic investigative genetic genealogy (FIGG), as an approach to locate potential persons of interest in criminal or missing persons cases. For example, a pilot case study in Sweden used IGG to locate the perpetrator of a double murder from 2004 who had evaded detection despite 15 years of various investigation efforts including more than 9000 interrogations and mass DNA screenings of more than 6000 men [ 90 ]. Hardly a week goes by without mention in the global media of another cold case being solved with IGG. Since the arrest of Joseph DeAngelo in April 2018 identified as the infamous Golden State Killer using IGG, hundreds of cold criminal and unidentified human remains cases have been resolved [ 91 ].
IGG involves examination of about 600,000 single nucleotide polymorphisms (SNPs), rather than the 20 or so STRs used in conventional forensic DNA testing, to enable associations of relatives as distant as third or fourth cousins [ 17 ]. IGG relies on a combination of publicly accessible records and the consent of individuals who have uploaded their genetic genealogy DNA profiles to genetic genealogy databases [ 92 ]. Multiple reviews and research articles have been published describing current IGG methods, knowledge, and practice along with the effectiveness and operational limits of the technique [ 17 , 30 , [93] , [94] , [95] , [96] , [97] ]. IGG works best with high-quality, single-source DNA samples. A case study involving whole genome sequencing of human remains from a 2003 murder victim found that it was possible to perform IGG for identification of the victim in this situation [ 98 ].
The four main direct-to-consumer (DTC) genetic genealogy companies, 23andMe (Mountain View, CA), Ancestry (Salt Lake City, UT), FamilyTree DNA (Houston, TX), and My Heritage (Lehi, UT), have DNA data from over 41 million individuals 36 as of July 2022 [ 97 ]. Individuals can upload their DTC data to GEDmatch, which is a DNA comparison and analysis website launched in 2010 and purchased in 2019 by Verogen (San Diego, CA). Law enforcement IGG searches are currently permitted with DTC data for individuals who opt into the GEDmatch database or do not opt out of the FamilyTree DNA database [ 99 , 100 ]. Currently most DTC genetic genealogy data comes from the United States and individuals of European origin. A UK study found that 4 of 10 volunteer donors could be identified with IGG including someone of Indian heritage demonstrating that under the right circumstances individuals of non-European origin can be identified [ 101 ].
As noted previously in Section 1.2.1 , the U.S. Department of Justice released an interim policy guide to forensic genetic genealogical DNA analysis and searching [ 23 ], and the FBI Laboratory's chief biometric scientist published an editorial in Science calling for responsible genetic genealogy [ 24 ]. SWGDAM has provided an overview of IGG that emphasizes the approach being used only after a regular STR profile search of a law enforcement DNA database fails to produce any investigative leads [ 102 ]. Policy and practical implications of IGG have been explored in Australia [ 103 ] and within the UK as part of probing the perceptions of 45 professional and public stakeholders [ 104 , 105 ].
Four misconceptions about IGG were examined by several members of the SWGDAM group: (1) when law enforcement conducts IGG in a genetic genealogy database, they are given special access to participants' SNP profiles, (2) law enforcement will arrest a genetic genealogy database participant's relatives based on the genetic information the participant provided to the database, (3) IGG necessarily involves collecting and testing DNA samples from a larger number of innocent persons than would be the case if IGG were not used in the investigation, and (4) IGG is or soon will be ubiquitous because there are no barriers to IGG that limit the cases in which it can be conducted [ 106 ].
In May 2021, the state of Maryland passed the first law in the United States and in the world that regulates law enforcement's use of DTC genetic data to investigate crimes. A policy forum article in Science explained how this new law provides a model for others in this area [ 107 ]. Six important features were described: (1) requiring judicial authorization for the initiation of an IGG search, (2) affirming individual control over the investigative use of one's genetic data, (3) establishing strong protections for third parties who are not suspects in the case, (4) ensuring that IGG is available to prove either guilt or innocence, (5) imposing consequences and fines for violations, and (6) requiring annual public reporting and review to enable informed oversight of IGG methods. However, as of September 2022, these regulations have not been implemented apparently due to lack of resources with these unfunded requirements. 37
Efforts have been made to raise awareness among defense attorneys about how IGG searches can potentially invade people's privacy in unique ways [ 108 ]. Important perspectives on ethical, legal, and social issues have been offered along with directions for future research [ 109 ]. These concerns about data privacy, public trust, proficiency and agency trust, and accountability have led to a call for standards and certification of IGG to address issues raised by privacy scholars, law enforcement agencies, and traditional genealogists [ 110 , 111 ] and for an ethical and privacy assessment framework covering transparency, access criteria, quality assurance, and proportionality [ 112 ].
2.2.4. Genetic privacy and ethical concerns
Two important topics are considered in this section: (1) do the genetic markers used in traditional forensic DNA typing reveal more than identity and therefore potentially impact privacy of the individuals tested? and (2) are samples collected and tested according to ethical principles?
Forensic DNA databases utilize STR markers that were intentionally selected to avoid phenotypic associations. An extensive review of the literature examined 107 articles associating a forensic STR with some genetic trait and found “no demonstration of forensic STR variants directly causing or predicting disease” [ 113 ]. A study of the potential association of 15 STRs and 3 facial characteristics on 721 unrelated Han Chinese individuals also found “scarcely any association between [the] STRs with studied facial characteristics” [ 114 ].
In 2021, the American Type Culture Collection (ATCC) published a standard for authentication of human cell lines using DNA profiling with the 13 CODIS STR markers [ 115 ]. This use of forensic STR markers for biospecimen authentication led a bioethicist and a law professor to write a policy forum article in Science titled “Get law enforcement out of biospecimen authentication” [ 116 ]. The authors of this policy forum believe that using the same genetic markers could potentially: (1) undermine efforts to recruit research participants from historically marginalized and excluded groups that are underrepresented in research, (2) risk drawing law enforcement interest in gaining access to these research data, and (3) impose additional potential harms on already vulnerable populations, particularly children. Instead they advocate for using non-CODIS STRs or a new SNP assay to distinguish biospecimens in repositories, something done recently at the Coriell Institute for Medical Research with six new STR markers [ 117 ]. A responsive letter to the editor regarding this policy forum article expressed that “their proposal could potentially create artificial silos between genomic data in the justice system and in biomedical research, making it inefficient and ultimately counterproductive” [ 118 ]. The authors of the original article responded that “the risk of attracting law enforcement interest to research data increases when the data are available in a recognizable way” [ 119 ].
Modern scientific research seeks to protect the dignity, rights, and welfare of research participants by following ethical requirements. Six forensic science journals over the time period of 2010–2019 were examined for their reporting of ethical approval and informed consent in original research using human or animal subjects [ 120 ]. These journals were Forensic Science International: Genetics , Science & Justice , Journal of Forensic and Legal Medicine , the Australian Journal of Forensic Sciences , Forensic Science International , and the International Journal of Legal Medicine . A total of 3010 studies that described research on human or animal subjects and/or samples were selected from these journals with only 1079 articles (36%) reporting that they had obtained ethical approval and 527 articles (18%) stating that informed consent was sought either by written or verbal agreement. The authors of this study noted that reported compliance with ethical guidelines in forensic science research and publication was below what is considered minimal reporting rates in biomedical research and encouraged widespread adoption of the 2020 guidelines described below [ 120 ].
Guidelines and recommendations for ethnical research on genetics and genomics of biological material were jointly adopted and published in Forensic Science International: Genetics [ 121 ] and Forensic Science International: Reports [ 122 ]. These guidelines utilize the following principles as prerequisites for publication in these two journals as well as the Forensic Science International: Genetics Supplement Series : (1) general ethics principles that are regulated by national boards and represent widely signed international agreements, (2) universal declarations that require implementations in state members, such as the World Medical Association Declaration of Helsinki biomedical research on human subjects, and (3) universal declarations and principles drafted by independent organizations that have been widely adopted by the scientific community. This includes the U.S. Federal Policy for the Protection of Human Subjects (“Common Rule”) that was revised in 2017 (with a compliance date delayed to January 21, 2019). 38
Submitted manuscripts must provide the following supporting documentation to demonstrate compliance with the publication guidelines: (1) ethical approval in the country of [sample] collection by the appropriate local ethical committee or institutional review board, (2) ethical approval in the country of experimental work according to local legislation; if material collection and experimentation are conducted in different countries, both (1) and (2) are required, (3) template of consent forms in the case of human material as approved by the relevant ethical committee, and (4) approved export/import permits as applicable. Authors must declare in their submitted manuscript that these guidelines have been strictly followed [ 121 , 122 ].
Forensic genetic frequency databases, such as the Y-chromosome Haplotype Reference Database (YHRD), have been challenged over the ethics of DNA holdings, specifically of samples originating from the minority Muslim Uyghur population in western China [ 123 , 124 ]. A survey of U.S. state policies on potential law enforcement access to newborn screening samples found that nearly one-third of states permit these samples or their related data to be disclosed to or used by law enforcement and more than 25% of states have no discernible policy in place regarding law enforcement access [ 125 ].
A framework for ethical conduct of forensic scientists as “lived practice” has been proposed, and three case studies were discussed in terms of decision-making processes involving forensic DNA phenotyping and biographical ancestry testing, investigative genetic genealogy, and forensic epigenetics [ 126 ]. An ethos for forensic genetics involving the values of integrity, trustworthiness, and effectiveness has likewise been described [ 127 ].
2.2.5. Sexual assault kit testing
Unsubmitted or untested sexual assault kits (SAKs) may exist in police or laboratory evidence lockers for many years leading to rape kit backlogs that can spark community outrage when discovered. A number of articles have been published in the past three years describing success rates with examining SAKs and the policies surrounding them. For example, an evaluation of 3422 unsubmitted SAKs in Michigan found 1239 that produced a DNA profile eligible for upload into CODIS with 585 yielding a CODIS hit [ 128 ]. In addition, results from a groping and sexual assault case were presented to support the expansion of touch DNA evidence in these types of cases [ 129 ].
To assess success rates in their jurisdiction, the Houston Police Department randomly selected 491 cases of over 6500 previously unsubmitted sexual assault kits [ 130 ]. Of these, 336 cases (68%; 336/491) screened positive for biological evidence; a DNA profile was developed in 270 cases (55%; 270/491) with 213 (43%; 213/491) uploaded to CODIS; and 104 (21% total; 104/491 or 49% of uploaded profiles; 104/213) resulted in a CODIS hit. The statute of limitation had expired in 44% of these CODIS-hit cases, which prohibited arrests and prosecution. Victims were unwilling to participate in a follow-up investigation in another 25% of these cases. When the data were compiled for the publication, charges had been filed in only one CODIS-hit case [ 130 ].
Sexual assault cases can be difficult to prosecute as victims may be re-traumatized when a cold case is reopened. The authors of one study shared: “A key to successful pursuit of cold case sexual assaults is to have a well-crafted victim-notification plan and a victim advocate as part of the investigative team” [ 131 ]. Interviews with eight assistant district attorneys provided important prosecutors’ perspectives on SAK cases, the development of narratives to explain the evidence in a case, and the decision on whether a case should be pursued or what further investigative activities may be needed [ 132 ]. The authors concluded: “Our findings suggest that forensic evidence does not magically lead to criminal justice outcomes by itself, but must be used thoughtfully in conjunction with other evidence as part of a well-considered strategy of investigation and prosecution” [ 132 ].
Discussing a data set from Denver, Colorado where 1200 sexual assault cold cases with testable DNA samples were examined and 600 cases were processed through the laboratory resulting in 97 CODIS hits, 55 arrests and court filings, and 48 convictions, the authors conclude that the cost of the Denver cold case sexual assault program was worth the investment [ 131 ].
From December 2015 to July 2018, the Palm Beach County Sheriff's Office (Florida, USA) researched more than 5500 cases and evaluated evidence from previously untested sexual assault kits spanning a 43-year period at a cost of over $1 million. Of the 1558 sexual assaults examined, there were 686 cases (44%; 686/1558) with CODIS-eligible profiles, 261 CODIS hits, and 5 arrests when the article was written in mid-2019 [ 133 ]. The Palm Beach County Sheriff's Office also helped develop a backlog reduction effort through creating a biological processing laboratory within the Boca Raton Police Services Department [ 134 ]. With this joint effort from 2016 to 2018, the total average turnaround time decreased from 30 days to under 20 days with the 3489 DNA profiles entered into CODIS resulting in 1254 associations and 965 investigations aided. Important takeaway lessons include the value of (1) engaging legal counsel early to outline necessary legal procedures and the timeline, (2) bringing all stakeholders “to the table” early to discuss expectations, as well as legal and operational responsibilities, and (3) creating a realistic timeline with a comprehensive memorandum of understanding so all parties have agreed to their roles and responsibilities [ 134 ].
From 275 previously untested sexual assault kits submitted for DNA testing in one region of Central Brazil, a total of 176 profiles were uploaded to their DNA database resulting in 60 matches (34%; 60/176) and 32 assisted investigations (18%; 32/176) with information about the suspect identity or the connection of serial sexual assaults assigned to the same individual [ 135 ]. Another study from the same region of Brazil examined 2165 cases and noted that 13% (286/2165) had information regarding the victim-offender relationship with 63% (179/286) being stranger-perpetrated rapes and 37% (107/286) being non-stranger [ 136 ]. The authors then summarize: “Hits were detected only with stranger-perpetrated assaults ( n = 41), which reinforces that DNA databases are fundamental to investigate sexual crimes. Without DNA typing and DNA databases, probably these cases would never be solved” [ 136 ].
Given that laboratories have limited resources and need to prioritize their efforts, some business analytics have been applied to SAK testing. An analysis of the potential societal return on investment (ROI) for processing backlogged, untested SAKs reported a range of 10%–65% ROI depending on the volume of activity for the laboratory conducting the analysis [ 137 ]. An evaluation of data from 868 SAKs tested by the San Francisco Policy Department Criminalistics Laboratory during 2017–2019 found that machine learning algorithms outperformed forensic examiners in flagging potentially probative samples [ 138 ].
An examination of 5165 SAKs collected in Cuyahoga County (Ohio, USA) from 1993 through 2011 found 3099 with DNA of which 2127 produced a CODIS hit, with 803 investigations leading to an indictment and eventually 78 to trial along with 330 pleas [ 139 ]. The authors report a “cost savings to the community of $26.48 million after the inclusion of tangible and intangible costs of future sexual assaults averted through convictions” and advocate for “the cost-effectiveness of investigating no CODIS hit cases and support an ‘investigate all’ approach” [ 139 ]. Likewise an assessment of 900 previously-untested SAKs from Detroit (Michigan, USA) found that “few of the tested variables were significant predictors of CODIS hit rate” and “testing all previously-unsubmitted kits may generate information that is useful to the criminal justice system, while also potentially addressing the institutional betrayal victims experienced when their kits were ignored” [ 140 ].
A group in the Philippines described an integrated system to improve their SAK processing [ 141 ]. With an optimized workflow in Montreal, Canada, SAK processing median turnaround time decreased from 140 days to 45 days with a foreign DNA profile being obtained in 44% of cases [ 142 ]. In addition, this group examined casework data to guide resource allocation through identifying the likelihood of specific types of cases and samples yielding foreign biological material [ 142 ]. Decision trees and logistic regression models were also used to try and predict whether or not SAKs will yield a CODIS-eligible DNA profile [ 143 ]. Finally, direct PCR and rapid DNA approaches to streamline SAK testing were reviewed [ 144 ].
2.3. Forensic biology and body fluid identification
The basic workflow for biological samples in forensic examinations typically involves a visual examination of the evidence, a presumptive and/or confirmatory test for a suspected body fluid (e.g., the amylase assay for saliva), and DNA analysis and interpretation [ 145 ]. Body fluid identification (BFID), in particular with blood, saliva, semen, or vaginal fluid stains, provides valuable evidence in many investigations that can aid in the resolution of a crime [ 146 ]. Many of these BFID tests are presumptive and not nearly as sensitive as modern DNA tests meaning that “obtaining a DNA profile without being able to associate [it] with a body fluid is an increasingly regular occurrence” and “it is necessary and important, especially in the eyes of the law, to be able to say which body fluid that the DNA profile was obtained from” [ 147 ].
A number of approaches are being taken to improve the sensitivity and specificity of BFID in recent years including DNA methylation [ [148] , [149] , [150] , [151] , [152] , [153] , [154] , [155] , [156] , [157] , [158] , [159] , [160] , [161] ], messenger RNA (mRNA) [ [162] , [163] , [164] , [165] , [166] ], microRNA (miRNA) [ 167 ], protein mass spectrometry for seminal fluid detection [ 168 ], and microbiome analysis [ 169 , 170 ]. Although many new techniques are being described in the scientific literature, traditional methods for semen identification are still widely used in regular forensic casework [ 171 ].
When using RNA assays, DNA and RNA are co-extracted from examined samples [ 172 , 173 ]. Some tests may only distinguish between two possible body fluids, such as saliva and vaginal fluid [ 174 ], while other tests may attempt to distinguish six forensically relevant body fluids – vaginal fluid, seminal fluids, sperm cells, saliva, menstrual blood, and peripheral blood – although not always as clearly as desired [ 175 ]. BFID assays must also cope with mixed body fluids [ 176 ].
2.4. DNA collection and extraction
The process of obtaining a DNA profile begins with collecting a biological sample and extracting DNA from it. A review of recent trends and developments in forensic DNA extraction focused on isolating male DNA in sexual assault cases, using portable rapid DNA testing instruments, recovering DNA from difficult samples such as human remains, and bypassing DNA extraction altogether with direct PCR methods [ 177 ].
2.4.1. Touch evidence and fingerprint processing methods
Various studies have explored the compatibility of common fingerprint processing methods with DNA typing results [ [178] , [179] , [180] , [181] , [182] , [183] , [184] , [185] , [186] , [187] , [188] ]. For example, DNA recovery was explored after various steps in three different latent fingerprint processing methods – and fewer treatments were judged preferable with a 1,2-indanedione-zinc (IND/Zn) method appearing least harmful to downstream DNA analysis [ 187 ]. A different study found improved recovery of DNA from cigarette butts following latent fingerprint processing with 1,8-diazafluoren-9-one (DFO) compared to IND/Zn [ 179 ].
DNA losses were quantified with mock fingerprints deposited on four different surfaces to better understand DNA collection and extraction method performance [ 189 ]. The application of Diamond Dye has been shown to enable visualization of cells deposited on surfaces without interfering with subsequent PCR amplification and DNA typing [ [190] , [191] , [192] ].
It was possible to recover DNA profiles from clothing that someone touched for as little as 2 s [ 193 ]. DNA sampling success rates from car seats and steering wheels were studied [ 194 ] and recovery of DNA from vehicle surfaces using different swabs was explored [ 195 ]. In addition, the double-swab technique, where a wipe using a wet swab is followed by a wipe with a dry one, was revisited with an observation that for non-absorbing surfaces, the first web swab yielded 16 times more DNA than the second dry swab [ 196 ]. Swabs of cotton, flocked nylon, and foam reportedly provided equivalent DNA recoveries for smooth/non-absorbing surfaces, and an optimized swabbing technique involving the application of a 60-degree angle and rotating the swab during sampling improved DNA yields for cotton swabs [ 197 ].
2.4.2. Results from unfired and fired cartridge cases
Ammunition needs to be handled to load a weapon and thus DNA from the handler may be deposited onto the ammunition via touch [ 198 ]. Important progress has been made in recovering DNA from ammunition such as unfired cartridges or fired cartridge cases (FCCs) that may remain at a crime scene after a weapon has been fired. Trace quantities of DNA recovered from firearm or FCC surfaces has been used to try and link results to gun-related crimes.
A 2019 review of the literature regarding obtaining successful DNA results from ammunition examined collection techniques, extraction methodologies, and various amplification kits and conditions [ 199 ]. A direct PCR approach detected more STR alleles than methods using DNA extraction, and the authors noted that mixtures are commonly observed from gun surfaces, bullets, and cartridges in both controlled experimental conditions and from actual casework evidence and they encourage careful interpretation of these results [ 200 ]. The development of a crime scene FCC collector was combined with a new DNA recovery method that uses a rinse-and-swab technique [ 201 ].
Research studies and review articles have considered factors affecting DNA recovery from cartridge cases and the impact of metal surfaces on DNA recovery [ [202] , [203] , [204] , [205] , [206] , [207] , [208] , [209] ]. Recovery of mtDNA from unfired ammunition components has been assessed for sequence quality [ 210 ].
2.5. DNA typing
Following collection of DNA evidence and its extraction from biological samples, the typical typing process involves DNA quantitation, PCR amplification of STR markers, and STR typing using capillary electrophoresis. Direct PCR avoids the DNA extraction and quantitation steps, which can improve recovery of trace amounts of DNA [ 211 , 212 ]. Whole genome amplification prior to STR analysis has also been examined to aid recovery of degraded DNA [ 213 ] and to enable profiling of single sperm cells [ 214 ].
PCR amplification using STR typing kits can sometimes produce artifacts that impact DNA interpretation including missing (null) alleles [ 215 ], false tri-allelic patterns [ 216 ] or extra peaks when amplified in the presence of microbial DNA [ [217] , [218] , [219] ].
Applied Biosystems Genetic Analyzers have been the primary means of performing multi-colored capillary electrophoresis for many years [ 4 ]. First experiences with Promega's new Spectrum Compact CE System have recently been reported [ 220 ]. A number of new research and commercial STR kits have been introduced in recent years along with the publication of at least 24 validation studies ( Table 5 ). These validation studies typically follow guidelines outlined by the ENFSI DNA Working Group, 39 SWGDAM 40 , or a 2009 Chinese National Standard. 41
STR kits assessed with 24 published validation studies during 2019–2022.
A report on the first two years of submissions to the STRidER 42 (STRs for Identity ENFSI Reference) database for online allele frequencies revealed that 96% of the submitted 165 autosomal STR datasets generated by CE contained errors, showing the value of centralized quality control and data curation [ 245 ].
2.6. DNA interpretation at the source or sub-source level
The designation of STR alleles and genotypes of contributors in DNA mixtures are key aspects of DNA interpretation [ 246 , 247 ]. Electropherograms generated by CE instruments exhibit both STR alleles and artifacts that complicate data interpretation. Efforts are underway to understand and model instrumental artifacts [ [248] , [249] , [250] , [251] ] as well as biological artifacts of the PCR amplification process such as STR stutter products [ 252 , 253 ]. Machine learning approaches are being applied to classify artifacts versus alleles with the goal to eventually replace manual data interpretation with computer algorithms [ [254] , [255] , [256] , [257] ]. One such program, FaSTR DNA, enables potential artifact peaks from stutter, pull-up, and spikes to be filtered or flagged, and a developmental validation has been published examining 3403 profiles generated with seven different STR kits [ 258 ].
2.6.1. DNA mixture interpretation
Forensic evidence routinely contains contributions from multiple donors, which result in DNA mixtures. A number of approaches have been taken and advances made in DNA mixture interpretation [ 259 ]. These include probabilistic genotyping software [ 15 ], using genetic markers beyond traditional autosomal STR typing [ 260 ], or separating contributor cells and performing single-cell analysis [ [261] , [262] , [263] , [264] , [265] , [266] ].
In June 2021, the National Institute of Standards and Technology (NIST) released a draft report regarding the scientific foundations of DNA mixture interpretation [ 267 ]. This 250-page document described 16 principles that underpin DNA mixture interpretation, provided 25 key takeaways, and cited 528 references. NIST also began a Human Factors Expert Working Group on DNA Interpretation in February 2020 and plans to release a report with recommendations in 2023.
Assessment of the number of contributors (NoC) is a critical element of accurate DNA mixture interpretation. For example, the LRs relating to minor contributors can be reduced when the incorrect number of contributors is assumed [ 268 ]. Allele sharing among contributors to a mixture and masking of alleles due to STR stutter artifacts can lead to inaccurate NoC estimates based on simply counting the number of alleles at a locus. Different approaches and software programs have been used for NoC estimation [ [269] , [270] , [271] , [272] , [273] , [274] , [275] ]. Total allele count (TAC) distribution via TAC curves showed an improvement in manually estimating the number of contributors with complex mixtures [ 276 ]. Sequence analysis of STR loci expands the number of possible alleles compared to CE-based length measurements and thus can improve NoC estimates [ 277 ].
In the past three years, validation studies have been performed with a number of probabilistic genotyping software (PGS) systems including EuroForMix [ 278 ], DNAStatistX [ 279 , 280 ], TrueAllele [ 281 ], STRmix [ 282 ], Statistefix [ 283 ], Mixture Solution [ 284 ], Kongoh [ 285 ], and MaSTR [ 286 , 287 ]. Developers of EuroForMix, DNAStatistX, and STRmix provided a review of these systems [ 288 ]. Multi-laboratory assessments have been described [ 289 , 290 ] and likelihood ratios obtained from EuroForMix and STRmix compared [ [291] , [292] , [293] , [294] ]. With a growing literature in this area, there are many other articles that could have been cited.
2.7. DNA interpretation at the activity level
DNA interpretation at the source or sub-source level helps to answer the question of who deposited the cell material, whether attribution for the result can be made to a specific cell type (i.e., source level) or simply to the DNA if no attribution can be made to a specific cell type (i.e., sub-source level). Activity-level propositions seek to answer the question of how did an individual's cell material get there. Interpretation at the activity level is sometimes referred to as evaluative reporting [ 295 , 296 ].
In 2020, the ISFG DNA Commission [ 38 ] discussed the why, when, and how to carry out evaluative reporting given activity level propositions through providing examples of formulating these propositions. These Commission recommendations emphasize that reports using a likelihood ratio based on case-specific propositions and relevant conditioning information should highlight the assumptions being made and that “it is not valid to carry over a likelihood ratio from a low level, such as sub-source, to a higher level such as source or activity propositions … because the LRs given sub-source level propositions are often very high and LRs given activity level propositions will often be many orders of magnitude lower” [ 38 ]. Another recommendation specifies that “scientists must not give their opinion on what is the ‘most likely way of transfer’ (direct or indirect), as this would amount to giving an opinion on the activities and result in a prosecutor's fallacy (i.e., give the probability that X is true). The scientists' role is to assess the value of the results if each proposition is true in accordance with the likelihood ratio framework (the probability of the results if X is true and if Y is true)” [ 38 ] (emphasis in the original). This DNA Commission provided 11 recommendations and 4 considerations that should be studied carefully by those who implement activity-level DNA interpretation.
2.7.1. DNA transfer and persistence studies
To evaluate DNA findings given activity-level propositions it is important to understand the factors and variables that may impact DNA transfer, persistence, prevalence, and recovery (DNA-TPPR). These factors include history of contacting surfaces, biological material type, quantity and quality of DNA, dryness of biological material, manner and duration of contact, number and order of contacts, substrate type(s), time lapses and environment, and methods and thresholds used in the forensic DNA laboratory to generate the available data [ 297 ].
Three valuable review articles were published on this topic in 2019 [ 14 , 28 , 299 ]. Following a comprehensive January 2019 review that cited [ 298 ] references on DNA-TPPR [ 14 ], the same authors provided an update in November 2021 on recent progress towards meeting challenges and a synopsis of 144 relevant articles published between January 2018 and March 2021 [ 297 ]. While few studies provide the information needed to help assign probabilities of obtaining DNA results given specific sets of circumstances, progress includes use of Bayesian Networks [ 300 ] to identify variables for complex transfer scenarios [ 38 , [301] , [302] , [303] , [304] , [305] ] as well as development of an online database DNA-TrAC 43 for relevant research articles [ 299 ] and a structured knowledge base 44 with information to help practitioners interpret general transfer events at an activity level [ 306 ].
Forensic DNA pioneer Peter Gill emphasized that awareness of the limitations of DNA evidence is important for users of this data given that an increased sensitivity of modern DNA methods means that DNA may be recovered that is irrelevant to the crime under investigation [ 307 ]. An ISFG DNA Commission (see Section 1.2.5 ) emphasized that the strength of evidence associated with a DNA match at the sub-source level cannot be carried over to activity level propositions [ 38 ]. Structuring case details into propositions, assumptions, and undisputed case information has been encouraged [ 308 ].
Factors affecting variability of DNA recovery on firearms were studied with four realistic, casework-relevant handling scenarios along with results obtained including DNA quantities, number of contributors, and relative profile contributions for known and unknown contributors [ 309 ]. These studies found that sampling several smaller surfaces on a firearm and including the sampling location in the evaluation process can be helpful in assessing results given alternative activity-level propositions in gun-related crimes. The authors recommend that “further extensive, detailed and systematic DNA transfer studies are needed to acquire the knowledge required for reliable activity-level evaluations” [ 309 ].
Other recent studies on DNA-TPPR include examining prevalence and persistence of DNA or saliva from car drivers and passengers [ [310] , [311] , [312] ], evaluation of DNA from regularly-used knives after a brief use by someone else [ 313 ], studying the accumulation of endogenous and exogenous DNA on hands [ 314 ] and non-self-DNA on the neck [ 315 ], considering the potential of DNA transfer via work gloves [ 316 , 317 ] or during lock picking [ 318 ], and investigating whether DNA can be recovered from illicit drug capsules [ 319 , 320 ] or packaging [ 321 ] to identify those individuals preparing or handling the drugs.
Efforts have been made to estimate the quantity of DNA transferred in primary versus secondary transfer scenarios [ 322 ]. As quantities of DNA transferred can be highly variable and thought to be dependent on the so-called “shedder status” – how much DNA an individual exudes, several studies explored this topic [ [323] , [324] , [325] , [326] , [327] ]. Studies have also considered the level of DNA an individual transfers to untouched items in their immediate surroundings [ 328 ], the position and level of DNA transferred during digital sexual assault [ 329 ] or during various activities with worn upper garments [ 330 , 331 ], and the DNA composition on the surface of evidence bags pre- and post-exhibit examination [ 332 ]. Studies assessing background levels of male DNA on underpants worn by females [ 333 ] and background levels of DNA on flooring within houses [ 334 ] are providing important knowledge about the possibilities and probabilities of DNA transfer and persistence.
The authors of one study summarize some key points that could be extended to many other studies as words of caution: “From a wider trace DNA point of view, this study has demonstrated that the person who most recently handled an item may not be the major contributor and someone who handled an item for longer may still not be the major contributor if they remove more DNA than they deposit. The amount of DNA transferred and retained on an item is highly variable between individuals and even within the same individual between replicates” [ 320 ].
3. Emerging technologies, research studies, and other topics
New technologies to aid forensic DNA typing are constantly under development. This section explores recent activities with next-generation DNA sequencing, DNA phenotyping for estimating a sample donor's age, ancestry, and appearance, lineage markers, other markers and approaches, and non-human DNA and wildlife forensics, and is expected to be of value to researchers and those practitioners looking to future directions in the field.
3.1. Next-generation sequencing
Next-generation sequencing (NGS), also known as massively parallel sequencing (MPS) in the forensic DNA community, expands the measurement capabilities and information content of a DNA sample beyond the traditional length-based results with STR markers obtained with capillary electrophoresis (CE) methods. Additional genetic markers, such as single nucleotide polymorphisms (SNPs), microhaplotypes, and mitochondrial genome (mtGenome) sequence, may be analyzed along with the full sequence of STR alleles. This higher information content per sample opens up new potential applications such as phenotyping of externally visible characteristics and biogeographical ancestry as described in review articles [ 335 , 336 ].
As mentioned in Section 1.2.1 , the NIJ Forensic Laboratory Needs Technology Working Group (FLN-TWG) published a 29-page implementation strategy on next-generation sequencing for DNA analysis in September 2021 [ 28 ]. This guide discusses how NGS works and its advantages and disadvantages, the various instrument platforms and commercial kits available with approximate costs, items to consider regarding facilities, data storage, and personnel training, and resources for implementing NGS technology. A total of 73% of 105 forensic DNA laboratories surveyed from 32 European countries already own an MPS platform or plan to acquire one in the next year or two and one-third of the survey participants already conduct MPS-based STR sequencing, identity, or ancestry SNP typing [ 337 ].
Validation studies have been described with the ForenSeq DNA Signature Prep kit and the MiSeq FGx system [ [338] , [339] , [340] ], with the Verogen ForenSeq Primer Mix B for phenotyping and biogeographical ancestry predictions [ 341 , 342 ], and for resizing reaction volumes with the ForenSeq DNA Signature Prep kit library preparation [ 343 ]. MPS sequence data showed excellent allele concordance with CE results for 31 autosomal STRs in the Precision ID GlobalFiler NGS STR Panel from 496 Spanish individuals [ 344 ] and from 22 autosomal STR loci in the PowerSeq 46GY panel with 247 Austrians [ 345 ].
STR flanking region sequence variation has been explored [ 346 ] and reports of population data and sequence variation were published for samples from India [ 347 ], France [ 348 ], China [ 349 , 350 ], Korea [ 351 ], Brazil [ 352 ], Tibet [ 353 ], and the United States [ 354 ].
In April 2019 the STRAND ( S hort T andem R epeat: A lign, N ame, D efine) Working Group was formalized [ 355 ] to consider several possible approaches to sequence-based STR nomenclature that have been proposed [ 356 , 357 ]. An overview of software options has been provided for analysis of forensic sequencing data [ 358 ]. Some recent published options include STRinNGS [ 359 ], STRait Razor [ 360 ], ArmedXpert tools MixtureAce and Mixture Interpretation to analyze MPS-STR data [ 361 ], and STRsearch for targeted profiling of STRs in MPS data [ 362 ]. To aid interpretation of MPS-STR data, sensitivity studies were performed with single-source samples and sequence data analyzed by DNA quantity and method used [ 363 ]. A procedure has been described to address calculation of match probabilities when results are generated using MPS kits with different trim sites than those present in the relevant population frequency database [ 364 ]. Performance of different MPS kits, markers, or methods can be compared for accuracy and precision using the Levenshtein distance metric [ 365 ].
Novel MPS STR and SNP panels developed in recent years include IdPrism [ 366 ], a QIAGEN 140-locus SNP panel [ 367 ], the 21plex monSTR identity panel [ 368 ], a 42plex STR NGS panel to assist with kinship analysis [ 369 ], the 5422 marker FORCE (FORensic Capture Enrichment) panel [ 370 ], a forensic panel with 186 SNPs and 123 STRs [ 371 ], the SifaMPS panel for targeting 87 STRs and 294 SNPs [ 372 ], a 1245 SNP panel [ 373 ], 90 STRs and 100 SNPs for application with kinship cases [ 374 ], an adaption of the SNPforID 52plex panel to MPS [ 375 ], 448plex SNP panel [ 376 ], a 133plex panel with 52 autosomal and 81 Y-chromosome STRs [ 377 ], and a forensic identification multiplex with 1270 tri-allelic SNPs involving 1241 autosomal and 29 X-chromosome markers [ 378 ]. The 124 SNPs in the Precision ID Identity Panel were examined in a central Indian population [ 379 ] and human leukocyte antigen (HLA) alleles used in the early 1990s were revisited with MPS capability [ [380] , [381] , [382] ].
MPS methods have demonstrated utility with compromised samples [ [383] , [384] , [385] , [386] , [387] , [388] ] and mixture interpretation [ [389] , [390] , [391] , [392] , [393] , [394] , [395] ]. Microhaplotype assays have also been developed to assist with DNA mixture deconvolution [ 396 , 397 ]. Collaborative studies have explored variability with laboratory performance using MPS methods [ 398 , 399 ]. Population structure [ 400 ] and linkage and linkage disequilibrium [ 401 ] were examined among the markers in forensic MPS panels.
A review of transcriptome analysis using MPS discussed efforts with body fluid and tissue identification, determination of the time since deposition of stains and the age of donors, the estimation of post-mortem interval, and assistance to post-mortem death investigations [ 402 ]. The potential for MPS methods to assist with environmental trace analysis was reviewed in terms of forensic soil analysis, forensic botany, and human identification utilizing the skin microbiome [ 403 ]. The possibility of non-invasive prenatal paternity testing using cell-free fetal DNA from maternal plasma was explored with the Precision ID Identity Panel [ 404 ] and the ForenSeq DNA Signature Prep Kit [ 405 ]. Pairwise kinship analysis was also examined using the ForenSeq DNA Signature Prep Kit and multi-generational family pedigrees [ 406 , 407 ]. Nanopore sequencing has also been explored for sequencing STR and SNP markers [ [408] , [409] , [410] , [411] , [412] , [413] , [414] , [415] , [416] ].
3.2. DNA phenotyping (ancestry, appearance, age)
Continuing research into the genetic components of biogeographic ancestry, appearance, and age predictions have improved forensic DNA phenotyping capabilities [ 417 ]. These forensic innovations may sometimes impact public expectations [ 418 ]. The investigation in a murder case was assisted using information from forensic DNA phenotyping that predicted eye, hair, and skin color of an unknown suspect with the HIrisPlex-S system involving targeted massively parallel sequencing [ 419 ].
The VISAGE ( Vis ible A ttributes Through Ge nomics) Consortium, which consists of 13 partners from academic, police, and justice institutions in 8 European countries, has established new scientific knowledge and developed and tested prototype tools for DNA analysis and statistical interpretation as well as conducted education for stakeholders. In the 2019 to 2022 time window of this review, this concerted effort produced 45 one review article [ 417 ], 22 original research publications [ 337 , [420] , [421] , [422] , [423] , [424] , [425] , [426] , [427] , [428] , [429] , [430] , [431] , [432] , [433] , [434] , [435] , [436] , [437] , [438] , [439] , [440] ], and three reports [ [441] , [442] , [443] ].
DNA phenotyping is currently an active area of research, and numerous activities and publications exist beyond the VISAGE articles noted here. Another 137 articles have appeared in the literature in the past three years on biogeographical ancestry, appearance (primarily hair color, eye color, and skin color), and biological age predictions (typically utilizing DNA methylation) (see Supplemental File ).
3.3. Lineage markers (Y-chromosome, mtDNA, X-chromosome)
Lineage markers consist of Y-chromosome, mitochondrial DNA, and X-chromosome genetic information that may be inherited from just one parent without the regular recombination that occurs with autosomal DNA markers. Research in terms of new markers, assays, and population studies continue to be published for these lineage markers.
3.3.1. Y-chromosome
Several recent review articles were published on forensic applications of Y-chromosome testing [ [444] , [445] , [446] ]. As discussed previously in Section 1.2 , an ISFG DNA Commission summarized the state of the field with Y-STR interpretation [ 39 ]. Rapidly mutating Y-STR loci can be used to differentiate closely related males [ [447] , [448] , [449] ]. New statistical approaches to assessing evidence with Y-chromosome information have been described [ 450 , 451 ]. Four commercial Y-STR multiplexes were compared with the NIST 1032 U S. population sample set and the allele and haplotype diversities explored with length-based versus sequence-based information [ 452 ].
A number of Y-STR typing systems have been described along with validation studies, such as a 36plex [ 453 ], a 41plex [ 454 ], a 29plex [ 455 ], a 17plex [ 456 ], a 24plex [ 457 ], the Microreader 40Y ID System [ 458 ], the 24 Y-STRs in the AGCU Y SUPP STR kit [ 459 ], the DNATyper Y26 PCR amplification kit [ 460 ], a multiplex with 12 multicopy Y-STR loci [ 461 ], the Yfiler Platinum PCR Amplification Kit [ 462 ], a 45plex [ 463 ], the Microreader 29Y Prime ID system [ 464 ], an assay with 30 slow and moderate mutation Y-STR markers [ 465 ], the 17plex Microreader RM-Y ID System [ 466 ], and a 26plex for rapidly mutating Y-STRs [ 467 ]. A machine learning program predicted Y haplogroups using two Y-STR multiplexes with 32 Y-STRs [ 468 ].
Deletions and duplications with 42 Y-STR were reported in a sample of 1420 unrelated males and 1160 father-son pairs from a Chinese Han population [ 469 ]. Using Y-STR allele sequences has enabled locating parallel mutations in deep-rooting family pedigrees [ 470 ]. The surname match frequency with Y-chromosome haplotypes was explored using 2401 males genotyped for 46 Y-STRs and 183 Y-SNPs [ 471 ]. In the Y-chromosome's role as a valuable kinship indicator to assist in genetic genealogy and forensic research, models to improve prediction of the time to the most recent common paternal ancestor have been studied with 46 Y-STRs and 1120 biologically related genealogical pairs [ 472 ]. A massively parallel sequencing tool was developed to analyze 859 Y-SNPs to infer 640 Y haplogroups [ 473 ]. Another MPS tool, the CSYseq panel, targeted 15,611 Y-SNPs to categorize 1443 Y-sub-haplogroup lineages worldwide along with 202 Y-STRs including 81 slow, 68 moderate, 27 fast, and 26 rapidly mutating Y-STRs to individualize close paternal relatives [ 474 ].
3.3.2. Mitochondrial DNA
Mitochondrial DNA (mtDNA), which is maternally inherited with a high copy number per cell, can aid human identification, missing persons investigations, and challenging forensic specimens containing low quantities of nuclear DNA such as hair shafts [ [475] , [476] , [477] ]. Validation studies have been published using traditional Sanger sequencing [ 478 ] and next-generation sequencing [ [479] , [480] , [481] ]. Illumina and Thermo Fisher now provide mtDNA whole genome NGS assays [ [482] , [483] , [484] , [485] ]. Many mtDNA population data sets were published in the past three years including high-quality data from U.S. populations [ 486 ]. The suitability of current mtDNA interpretation guidelines for whole mtDNA genome (mtGenome) comparisons has been evaluated [ 487 ].
NGS methods have increased sensitivity of mtDNA heteroplasmy detection [ 488 , 489 ], which can influence the ability to connect buccal reference samples and rootless hairs from the same individual [ 490 , 491 ]. Twelve polymerases were compared in terms of mtDNA amplification yields from challenging hairs – with KAPA HiFi HotStart and PrimeSTR HS outperforming AmpliTaq Gold DNA polymerase that is widely used in forensic laboratories [ 492 ]. Multiple studies and review articles have discussed distinguishing mtDNA from nuclear DNA elements of mtDNA (NUMTs) that have been inserted into our nuclear DNA [ [493] , [494] , [495] , [496] ].
NGS sequencing of the mtGenome has permitted improved resolution of the most common West Eurasian mtDNA control region haplotype [ 497 ]. Phylogenetic alignment and haplogroup classification have continued to be refined with new sequence information [ 498 ], and new assays have been developed to aid haplogroup classification [ 499 ]. Concerns over potential paternal inheritance of mtDNA have also been addressed [ 500 , 501 ].
3.3.3. X-chromosome
A 20-year review of X-chromosome use in forensic genetics examined the number and types of markers available, an overview of worldwide population data, the use of X-chromosome markers in complex kinship testing, mutation studies, current weaknesses, and future prospects [ 502 ]. One example of the forensic application of X-chromosome markers include use in relationship testing cases involving suspicion of incest or paternity without a maternal sample for comparison [ 503 ]. Four new X-STR multiplex assays were described along with validation studies including a 19plex [ 504 ], a 16plex [ 505 ], another 19plex – the Microreader 19X Direct ID System [ 506 ], and an 18plex named TYPER-X19 multiplex assay [ 507 ]. A collaborative study examined paternal and maternal mutations in X-STR markers [ 508 ]. A software program for performing population statistics on X-STR data was introduced [ 509 ] and sequence-based U.S. population data described for 7 X-STR loci [ 510 ].
3.4. New markers and approaches (microhaplotypes, InDels, proteomics, human microbiome)
In this section on new markers and approaches, publications related to microhaplotypes and insertion/deletion (InDel, or DIP for deletion insertion polymorphisms) markers are reviewed along with proteomic and microbiome approaches to supplement standard human DNA typing methods.
3.4.1. Microhaplotypes
Microhaplotype (MH) markers consist of multiple SNPs in close proximity (e.g., typically <200 bp or <300 bp) that can be simultaneously genotyped with each DNA sequence read using NGS. Two or more linked SNPs will define three or more haplotypes. Compared to STR markers, MHs do not have stutter artifacts (which complicate mixture interpretation), can be designed with shorter amplicon lengths in some cases (which benefits recovery of genetic information from degraded DNA samples), possess a higher degree of polymorphism compared to single SNP loci (which benefits discrimination power), and exhibit low mutation rates (which enables relationship testing and biogeographical ancestry inference). Thus, MH markers bring advantages to human identification, ancestry inference, kinship analysis, and mixture deconvolution to potentially assist missing person investigations, relationship testing, and forensic casework as discussed in several recent reviews [ 16 , 511 ]. A new database, MicroHapDB, has compiled information on over 400 published MH markers and frequency data from 26 global population groups [ 512 ].
A number of MH panels have been described [ [513] , [514] , [515] , [516] , [517] , [518] , [519] ]. Population data has been collected from a number of sources around the world including four U.S. population groups examined with a 74plex assay with 74 MH loci and 230 SNPs [ 520 ]. Various MH panels have been evaluated for effectiveness with kinship analysis [ [521] , [522] , [523] ]. Likewise the ability to detect minor contributors in DNA mixtures has been assessed [ [524] , [525] , [526] ].
3.4.2. InDel markers
InDel markers can be detected using a CE-based length analysis, and thus use instrumentation that forensic DNA laboratories already have. InDels can also be designed to amplify short DNA fragments (e.g., <125 bp) to help improve amplification success rates with low DNA quantity and/or quality. However, with only two possible alleles like SNPs, InDels are not as polymorphic as STRs and thus require more markers to obtain similar powers of discrimination as multi-allelic STR markers and do not work as well with mixed DNA samples. InDels possess a lower mutation rate than STRs and can be used as ancestry informative markers (AIMs) since allele frequencies may differ among geographically separated population groups.
Two commercial InDel kit exist: (1) Investigator DIPlex (QIAGEN, Hilden, Germany) with 30 InDels [ [527] , [528] , [529] , [530] , [531] ] and (2) InnoTyper 21 (InnoGenomics, New Orleans, Louisiana, USA) with 21 autosomal insertion-null (INNUL) markers [ [532] , [533] , [534] , [535] ]. In addition, a number of InDel assays have been published including a 32plex [ 536 ], a 35plex [ 537 ], a 38plex [ 538 ], a 39plex with AIMs [ 539 ], a 43plex [ 540 ], a 57plex [ 541 ], a 60plex with 57 autosomal InDels, 2 Y-chromosome InDels, and amelogenin [ 542 ], a 32plex with X-chromosome InDels [ 543 ], and a 21plex with AIMs [ 544 ].
A multi-InDel marker is a specific DNA fragment with more than one InDel marker located tightly in the physical position that provides a microhaplotype [ 545 ]. Several multi-InDel assays have been published include a 12plex [ 546 ] and an 18plex [ 547 ].
3.4.3. Proteomics
Protein analysis, often through immunological assays, has traditionally been used to identify body fluids and tissues. With improvements in protein mass spectrometry in recent years, genetic variation can be observed in hair shafts via single amino acid polymorphisms. Detection of these genetically variant peptides (GVPs) can infer the presence of corresponding SNP alleles in the genome of the individual who is the source of the protein sample. A thorough review of forensic proteomics in 2021 cited 375 references [ 18 ]. Recent efforts in this area have focused on using GVPs to differentiate individuals through their human skin cells [ [548] , [549] , [550] ] or hair samples [ [551] , [552] , [553] , [554] , [555] , [556] , [557] , [558] , [559] ]. An algorithm has been proposed for calculating random match probabilities with GVP information [ 560 ].
3.4.4. Human microbiome
Microorganisms live in and on the human body, and efforts are underway to utilize the human microbiome for a variety of potential forensic applications [ 21 , [561] , [562] , [563] ]. There are also active efforts with analysis of microbiomes in the environment (e.g., soil or water samples), which could be classified under non-human DNA testing. Forensic microbiome research covers at least six areas: (1) individual identification, (2) tissue/body fluid identification, (3) geolocation, (4) time since stain deposition estimation, (5) forensic medicine, and (6) post-mortem interval (PMI) estimation. Biological, technical, and data issues have been raised and potential solutions explored in a recent review article [ 21 ]. For example, microbes on deceased individuals are being studied to estimate the postmortem interval [ 20 ] and postmortem skin microbiomes were found to be stable during repeated sampling up to 60 h postmortem [ 564 ].
Sequence analysis of 16S rRNA using NGS provides information on the microbiome community present in a tested sample [ 565 ]. The Forensic Microbiome Database 46 correlates publicly available 16S rRNA sequence data as a community resource. If the skin microbiome is extremely diverse among individuals, then the potential exists to associate the bacterial communities on an individual's skin with objects touched by this individual assuming that the bacteria originating from the donor's skin are deposited (i.e., transfer to and persist on the surface) and can be detected and interpreted.
Specific aspects of the microbiome (e.g., the bacterial community) may be able to provide details about the donor through bacterial profiling. For example, in one study correlations were observed between the bacterial profile and gender, ethnicity, diet type, and hand sanitizer used [ 566 ]. Another study with 30 individuals found that each person left behind microbial signatures that could be used to track interaction with various surfaces within a building, but the authors concluded “we believe the human microbiome, while having some potential value as a trace evidence marker for forensic analysis, is currently under-developed and unable to provide the level of security, specificity and accuracy required for a forensic tool” [ 565 ].
Direct and indirect transfer of microbiomes between individuals has been studied [ 567 , 568 ] along with identifying background microbiomes [ 569 ] and the possibility of transfer of microbiomes within a forensic laboratory setting [ 570 ]. Changes in four bacterial species in saliva stains were charted, showing that it was possible to correctly predict deposition time within one week in 80% of the stains [ 571 ]. The ability to detect sexual contact has been explored through using the microbiome of the pubic region [ [572] , [573] , [574] ]. The microbiomes on skin, saliva, vaginal fluid, and stool samples have been compared [ 575 ]. The stability, diversity, and individualization of the human skin virome was explored with 59 viral biomarkers being found that differed across the 42 individuals studied [ 576 ]. It will be interesting to see what the future holds and what other findings come from this active area of research.
3.5. Kinship analysis, human identification, and disaster victim identification
Kinship analysis, which uses genetic markers and statistics to evaluate the potential for specific biological relationships, is important for parentage testing, disaster victim identification (DVI), and human identification of remains that may be recovered in missing person cases. New open-source software programs have been described that can assist with kinship analysis [ 577 , 578 ].
A potential biological relationship is commonly evaluated using a likelihood ratio (LR) by comparing the likelihoods of observing the genetic data given two alternative hypotheses, such as (1) an individual is related to another individual in a defined relationship versus (2) the two individuals not related. Higher LR values indicate stronger support with the genetic data if the proposed relationship is true. Multiple factors influence LR kinship calculations including the specific hypotheses, the genetic markers examined, the allele frequencies of the relevant population(s), the co-ancestry coefficient applied, and approaches to address potential mutations. STR genotypes were reported for 11 population groups used by the FBI Laboratory [ 579 ]. The status quo has been challenged in recent articles regarding how hypotheses are commonly established [ 580 ] and whether race-specific U.S. population databases should be used for allele frequency calculations [ 581 ].
Depending on the relationship being explored, information can be optimized through genetic information from additional known relatives or through collecting results at more loci [ 582 ]. Potential error rates have been modeled with the observation that false negatives, which occur when related individuals are misinterpreted as being unrelated, are more common than false positives, where unrelated people are interpreted as being related [ 583 ]. While LRs are generally reliable in detecting or confirming parent/child pairs, limitations of kinship determinations exist (e.g., distinguishing siblings from half-siblings) when using STR data [ 584 ].
Pairwise comparisons have been studied in forensic kinship analysis [ [585] , [586] , [587] ]. The effectiveness of 40 STRs plus 91 SNPs was shown to be better than 27 STRs and 91 SNPs or 40 STRs alone [ 588 ]. Only a minor increase in LRs was observed when taking NGS-generated allele sequence variation rather than fragment length allele variation [ 589 ]. The statistical power of exclusion and inclusion can be used to prioritize family members selected for testing in resolving missing person cases [ 590 ]. A strategy for making decisions when facing low statistical power in missing person and DVI cases was published [ 591 ].
The most challenging kinship cases involve efforts to separate pairs of individuals who are typically thought to be genetically indistinguishable (i.e., monozygotic twins) or distant relatives (e.g., fourth cousins) where there is an increased uncertainty in the possible relationship. In some situations, somatic mutations may permit distinguishing monozygotic twins following whole genome sequencing – and this approach was successful in four of six cases reported recently [ 19 ]. The probative value of NGS data for distinguishing monozygotic twins was explored [ 592 ]. A unique case of heteropaternal twinning was reported where opposite-sex twins apparently had different fathers [ 593 ]. An impressive effort in kinship analysis using direct-to-consumer genetic genealogy information from 56 living descendants of multiple genealogical lineages helped resolve a contested paternity case from over a century and a half ago to identify the biological father of Josephine Lyon [ 594 ].
Techniques for identification of human remains continue to improve particularly with the capabilities of NGS and hybridization capture [ 595 ] and ancient DNA extraction protocols [ 596 , 597 ]. Studies have reported variation in skeletal DNA preservation [ 598 ] and retrospectively considered success rates with compromised human remains [ 599 ].
A simulated airplane crash enabled six forensic laboratories in Switzerland to gain valuable DVI experience with kinship cases of varying complexity [ 600 ]. The ISFG Spanish-Portuguese Speaking Working Group likewise conducted a DVI collaborative exercise with a simulated airplane crash to explore fragment re-associations, victim identification through kinship analysis, coping with related victims, handling mutations or insufficient number of family references, working in a Bayesian framework, and the correct use of DVI software [ 601 ]. Other groups have explored the capability of a particular software tool [ 602 ] or implemented rapid DNA analysis to accelerate victim identification [ 603 ]. The International Commission on Missing Persons (ICMP) has gained considerable experience with DNA extraction and STR amplification from degraded skeletal remains and kinship matching procedures in large databases [ 604 ]. To supplement the INTERPOL DVI Guide, 47 some lessons learned and experienced-based recommendations for DVI operations have recently been provided [ 605 ].
3.6. Non-human DNA testing and wildlife forensics
Non-human biological evidence may inform criminal investigations when animals or plants are victims or perpetrators of crime or the presence of specific material, such as cat or dog hair, may contribute to reconstructing events at a crime scene. Non-human DNA testing includes wildlife forensics and domestic animal species as well as forensic botany and has many commonalities and some important differences compared to human DNA testing [ [606] , [607] , [608] , [609] , [610] ]. Pollen analysis can assist criminal investigations [ 611 , 612 ]. The potential for and the barriers associated with the wider application of forensic botany in civil proceedings and criminal cases have been examined [ 613 , 614 ].
Mammalian species identification can assist in determining the origins of non-human biological material found at crime scenes through narrowing the range of possibilities [ 615 ]. New sequencing methods have been developed to assist species identification [ 616 ]. A multiplex PCR assay was developed to simultaneously identify 22 mammalian species (alpaca, Asiatic black bear, Bactrian camel, brown rat, cat, cow, common raccoon, dog, European rabbit, goat, horse, house mouse, human, Japanese badger, Japanese wild boar, masked palm civet, pig, raccoon dog, red fox, sheep, Siberian weasel, and sika deer) and four poultry species (chicken, domestic turkey, Japanese quail, and mallard) [ 617 ]. A number of other species identification assays have also been reported [ [618] , [619] , [620] ].
An important effort for harmonizing canine DNA analysis is an ISFG working group known as the Canine DNA Profiling Group, or CaDNAP. 48 The CaDNAP group published an analysis of 13 STR markers in 1184 dogs from Germany, Austria, and Switzerland [ 621 ]. Six traits for predicting visible characteristics in dogs, namely coat color, coat pattern, coat structure, body size, ear shape, and tail length, were explored with 15 SNPs and six InDel markers [ 622 ]. Canine breed classification and skeletal phenotype prediction has been explored using various genetic markers [ 623 ]. A novel assay using a feline leukemia virus was developed to demonstrate that a contested bobcat was not a domestic cat hybrid [ 624 ] and a core panel of 101 SNP markers was selected for domestic cat parentage verification and identification [ 625 ].
DNA tests have been developed to assist with illegal trafficking investigations involving elephant ivory seizures [ 626 ], falcons [ 627 ], and precious coral material [ 628 ]. Accuracy in animal forensic genetic testing was explored with interlaboratory assessments performed in 2016 and 2018 [ 629 ]. A collaborative exercise conducted in 2020 and 2021 by the ISFG Italian Speaking Working Group examined performance across 21 laboratories with a 13-locus STR marker test for Cannabis sativa [ 630 ]. A molecular approach was explored to distinguish drug-type versus fiber-type hemp varieties [ 631 ].
Acknowledgments and disclaimer
I am grateful to Dominique Saint-Dizier from the French National Scientific Police for the invitation and opportunity to conduct this review and for the support of my supervisor, Shyam Sunder, for granting the time to work on this extensive review. Input and suggestions on this manuscript by Todd Bille, Thomas Callaghan, Kevin Kiesler, François-Xavier Laurent, Robert Ramotowski, Kathy Sharpless, and Robert Thompson are greatly appreciated. Certain commercial entities, equipment, or materials may be identified in this document in order to describe an experimental procedure or concept adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the entities, materials, or equipment are necessarily the best available for the purpose.
1 https://www.sciencedirect.com/journal/forensic-science-international-genetics/special-issue/10TSDS4360H .
2 https://www.mdpi.com/journal/genes/special_issues/Forensic_Genetic .
3 https://www.mdpi.com/journal/genes/special_issues/forensic_mitochondrial_genomics .
4 https://www.mdpi.com/journal/genes/special_issues/Advances_Forensic_Genetics .
5 https://www.mdpi.com/books/pdfdownload/book/5798 .
6 https://www.mdpi.com/journal/genes/special_issues/Bioinformatics_Forensic_Genetics .
7 https://www.mdpi.com/journal/genes/special_issues/genetics_anthropology .
8 https://www.mdpi.com/journal/genes/special_issues/Identification_of_Human_Remains .
9 https://www.mdpi.com/journal/genes/special_issues/Forensic_DNA_analysis .
10 https://www.mdpi.com/journal/genes/special_issues/Forensic_DNA_Mixture .
11 https://www.mdpi.com/journal/genes/special_issues/28FBA0G4DH .
12 See https://www.swgdam.org/ .
13 https://www.swgdam.org/publications .
14 https://www.fbi.gov/file-repository/rapid-dna-guide-january-2022.pdf/view .
15 https://www.fbi.gov/file-repository/non-codis-rapid-dna-best-practices-092419.pdf/view .
16 https://www.fbi.gov/file-repository/rapid-dna-testing-for-non-codis-uses-considerations-for-court-073120.pdf/view .
17 https://www.justice.gov/olp/uniform-language-testimony-and-reports .
18 https://forensiccoe.org/human_factors_forensic_science_sourcebook/ .
19 https://www.nist.gov/organization-scientific-area-committees-forensic-science .
20 https://www.nist.gov/organization-scientific-area-committees-forensic-science/human-forensic-biology-subcommittee .
21 https://www.nist.gov/topics/organization-scientific-area-committees-forensic-science/wildlife-forensics-subcommittee .
22 https://www.aafs.org/academy-standards-board .
23 https://www.nist.gov/organization-scientific-area-committees-forensic-science/osac-registry .
24 See https://www.nist.gov/organization-scientific-area-committees-forensic-science/human-forensic-biology-subcommittee .
25 https://lexicon.forensicosac.org/ .
26 https://www.nist.gov/osac/human-factors-validation-and-performance-testing-forensic-science .
27 https://www.nist.gov/organization-scientific-area-committees-forensic-science/osac-research-and-development-needs .
28 https://www.gov.uk/government/publications/forensic-science-providers-codes-of-practice-and-conduct-2021-issue-7 .
29 https://www.aabb.org/standards-accreditation/standards/relationship-testing-laboratories .
30 https://www.isfg.org/DNA+Commission .
31 Previously available rapid DNA systems included the RapidHIT 200 from IntegenX and MiDAS (Miniaturized integrated DNA Analysis System) from the Center for Applied NanoBioscience at the University of Arizona.
32 See https://le.fbi.gov/science-and-lab-resources/biometrics-and-fingerprints/codis/rapid-dna .
33 See https://www.interpol.int/How-we-work/Forensics/DNA .
34 See https://www.interpol.int/How-we-work/Forensics/I-Familia .
35 See https://le.fbi.gov/science-and-lab-resources/biometrics-and-fingerprints/codis#Familial-Searching .
36 See https://isogg.org/wiki/Autosomal_DNA_testing_comparison_chart .
37 See https://www.wmar2news.com/infocus/maryland-quietly-shelves-parts-of-genealogy-privacy-law .
38 See https://www.hhs.gov/ohrp/regulations-and-policy/regulations/finalized-revisions-common-rule/index.html .
39 See https://enfsi.eu/about-enfsi/structure/working-groups/dna/ .
40 See https://www.swgdam.org/publications .
41 See https://www.chinesestandard.net/PDF/English.aspx/GAT815-2009 .
42 See https://strider.online/ .
43 See https://bit.ly/2R4bFgL (DNA-TrAC).
44 See https://cieqfmweb.uqtr.ca/fmi/webd/OD_CIEQ_CRIMINALISTIQUE (Transfer Traces Activity DataBase).
45 See https://www.visage-h2020.eu/index.html#publications .
46 See http://fmd.jcvi.org/ .
47 See https://www.interpol.int/en/How-we-work/Forensics/Disaster-Victim-Identification-DVI .
48 See https://www.isfg.org/Working+Groups/CaDNAP .
Appendix A Supplementary data to this article can be found online at https://doi.org/10.1016/j.fsisyn.2022.100311 .
Appendix A. Supplementary data
The following is the supplementary data to this article:

IMAGES
VIDEO
COMMENTS
3. Current Research Progress. Recombinant DNA technology is a fast growing field and researchers around the globe are developing new approaches, devices, and engineered products for application in different sectors including agriculture, health, and environment.
Biotechnology which is synonymous with genetic engineering or recombinant DNA (rDNA) is an industrial process that uses the scientific research on DNA for practical applications. rDNA is a form of ...
Genomic research has evolved from seeking to understand the fundamentals of the human ... genomic technology advanced dramatically. DNA-sequencing throughput increased from 1000 base pairs per day ...
Effective DNA synthesis is therefore vital to close the gap between the ability to read and write DNA. Fig. 1: The state of the art in DNA synthesis. a, Productivity of DNA reading and DNA writing ...
DNA (deoxyribonucleic acid) is the nucleic acid polymer that forms the genetic code for a cell or virus. Most DNA molecules consist of two polymers (double-stranded) of four nucleotides that each ...
Conference Paper PDF Available. ... scientific research on DNA for practical applications. rDNA is a form . ... the shift to molecular genetics and recombinant DNA technology (1970 to die present ...
We could observe how a broken DNA fragment can scout the nucleus to identify a similar sequence and use it as a template for repair. News & Views 23 Aug 2023 Nature Structural & Molecular Biology ...
Chromosomal DNA sequences of the Pacific saury genome: versatile resources for fishery science and comparative biology. Mana Sato and others. Pacific saury ( Cololabis saira ) is a commercially important small pelagic fish species in Asia. In this study, we conducted the first-ever whole genome sequencing of this species, with single molecule ...
Based on my ethnographic research, I analysed the mutual relationships among scientists' bodywork, assorted bio-objects, their specific disembodied ontology and the DNA recombinant technology. This showed the significance of ideas of future life or immortal life and their mobilizations within contemporary editing genome technologies and ...
DNA present in all our cells acts as a template by which cells are built. The human genome project, reading the code of the DNA within our cells, completed in 2003, is undoubtedly one of the great achievements of modern bioscience. ... Recombinant DNA technology and DNA sequencing Essays Biochem. 2019 Oct 16;63(4):457-468. doi: 10.1042 ...
Forensic DNA analysis has vastly evolved since the first forensic samples were evaluated by restriction fragment length polymorphism (RFLP). Methodologies advanced from gel electrophoresis techniques to capillary electrophoresis and now to next generation sequencing (NGS). Capillary electrophoresis was and still is the standard method used in forensic analysis. However, dependent upon the ...
Step 4: Amp lification of gene of interest using PCR: Polymerase Chain Reaction (PCR) is. best defined as the in -vitro DNA replication. This is the step of cloning genes by Polymerase. Chain ...
The advancements in microscope and fluorescence imaging technology for biomolecules enabled the direct observation of DNA and proteins at the single-molecule level. ... , (iii) the stretching of DNA molecules by DNA molecule absorption using filter paper ... The development of DNA-origami research has dramatically improved the complexity of DNA ...
Biotechnology which is synonymous with genetic engineering or recombinant DNA (rDNA) is an industrial process that uses the scientific research on DNA for practical applications. rDNA is a form of artificial DNA that is made through the combination or insertion of one or more DNA strands, therefore combining DNA sequences, within different species, that is, DNA sequences that would not ...
A paper by Mauro ... subcellular localization, PTMs, and protein extraction and purification methodologies. DNA technology and genetic transformation methodologies have also involved to a great extent, with substantial improvements. ... In future, thorough research is required, giving attention to the integration of various bioprocessing steps ...
Nanopore design. The concept of nanopore sequencing emerged in the 1980s and was realized through a series of technical advances in both the nanopore and the associated motor protein 1,4,5,6,7,8 ...
Recombinant DNA technology was used to create Activase, a tissue plasminogen activator. It is a sterile, purified glycoprotein of 527 amino acids. It is made by combining complementary DNA (cDNA) from a human melanoma cell line with the natural human tissue-type plasminogen activator.
Technology development has significantly reduced the barriers to entry for genetic research and therapy, making it more accessible and affordable. With the capability to customize treatments with ...
DISCOVERY OF THE DNA FINGERPRINT. Historically, identity testing in the forensic field started with the analysis of the ABO blood group system. Later, new markers for identity and paternity identification were based on variations of serum proteins and red blood cell enzymes; eventually the human leukocyte antigen system was used ().It was not until 20 years ago that Sir Alec Jeffreys ...
The Q value was then calculated as the percentage of dsDNA that was amplifiable. NA quantity was defined as the amount of double-stranded DNA when the Q-value was >1 and as the amount of amplifiable DNA when the Q-Value was ≥1. The ddCq value was calculated based on the ratio of two real-time PCR amplicons in the FFPE DNA QC Assay version 2 Kit.
Abstract: - DNA chips are tiny devices tha t work by. allowing DNA to stick to its matching DNA in a very. specific way. This process is called hybridization. Scientists use DNA chips in medical ...
The Hill reporter Sharon Udasin writes that MIT researchers have developed a new solar-powered desalination device that "could last several years and generate water at a rate and price that is less expensive than tap water." The researchers estimated that "if their model was scaled up to the size of a small suitcase, it could produce about 4 to 6 liters of drinking water per hour ...
Two new papers in Nature Biotechnology report methods for targeted sequencing of complex DNA samples, achieved in real time during nanopore sequencing runs. Darren J. Burgess Research Highlight ...
Here are are some good AI Tools I recommend for student researchers and academics: 1. Litmaps. Litmaps is a tool for research students that makes finding papers and authors on a topic easy and quick. Instead of spending lots of time reading through hundreds of papers, you can use Litmaps to find the important ones in seconds.
In the current paper they show that gamma rays also modify the grain boundaries resulting in a faster flow of ions that, in turn, can be easily detected. And because the high-energy gamma rays penetrate much more deeply than UV light, "this extends the work to inexpensive bulk ceramics in addition to thin films," says Tuller.
The period in the 1990s was the golden research age of DNA fingerprinting succeeded by two decades of engineering, implementation, and high-throughput application. From the Foreword of Alec Jeffreys in Fingerprint News, Issue 1, January 1989: 'Dear Colleagues, […] I hope that Fingerprint News will cover all aspects of hypervariable DNA and ...
DNA computing is a branch of biomolecular computing concerned with the use of DNA as a carrier of information to make arithmetic and logic operations. Latest Research and Reviews
Since January, he said, PureCycle has been processing mainly consumer plastic waste and has produced about 1.3 million pounds of recycled polypropylene, or about 1 percent of its annual production ...
This review paper covers the forensic-relevant literature in biological sciences from 2019 to 2022 as a part of the 20th INTERPOL International Forensic Science Managers Symposium. Topics reviewed include rapid DNA testing, using law enforcement DNA databases plus investigative genetic genealogy DNA databases along with privacy/ethical issues ...