Academia Bees

How to Write Acknowledgement for Research Paper (5 Samples)
July 12, 2023
No Comments
By Mohsin Khurshid
Writing acknowledgements is an essential part of crafting a comprehensive research paper. It allows you to express gratitude and recognize the contributions of individuals and institutions who have supported your work. In this article, we will delve into the art of writing acknowledgement for research papers, providing you with valuable insights, practical tips, and five sample acknowledgements to guide you in acknowledging the people and resources that have played a significant role in your research journey.
Table of Contents
- 1 Understanding the Role of Acknowledgements in Research Papers
- 2 Key Elements of an Effective Acknowledgement
- 3 10 Tips for Writing an Acknowledgement for a Research Paper
- 4.1 Sample 1: Acknowledgement for Collaborative Research:
- 4.2 Sample 2: Acknowledgement for Funding Support:
- 4.3 Sample 3: Acknowledgement for Mentorship and Guidance:
- 4.4 Sample 4: Acknowledgement for Institutional Support:
- 4.5 Sample 5: Acknowledgement for Peer Reviewers:
- 6 Conclusion
Understanding the Role of Acknowledgements in Research Papers
Acknowledgements serve as a platform to express appreciation and recognize the collective effort that goes into the completion of a research paper. They provide an opportunity to acknowledge the guidance, support, and assistance received throughout the research process. By including acknowledgements, you can demonstrate your gratitude and give credit to those who have contributed to your success.
Key Elements of an Effective Acknowledgement
Crafting an effective acknowledgement involves considering various elements to ensure its sincerity and clarity. It is crucial to mention specific individuals, institutions, and their contributions, while keeping the acknowledgement concise and relevant. By adhering to ethical considerations and cultural norms, you can create an acknowledgement that reflects your gratitude and professionalism.
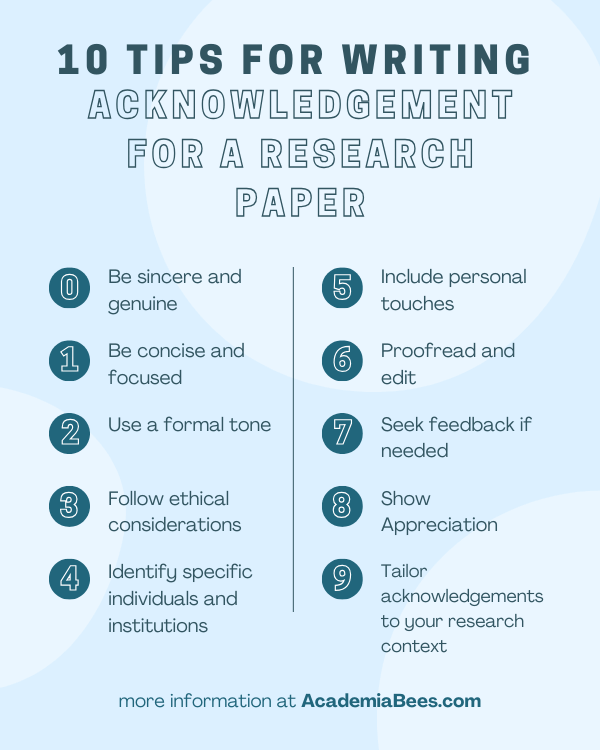
10 Tips for Writing an Acknowledgement for a Research Paper
- Be sincere and genuine : Write your acknowledgements with heartfelt gratitude, expressing sincere appreciation for the support and contributions received during your research.
- Identify specific individuals and institutions: Mention the names of people who have played a significant role in your research, such as mentors, advisors, collaborators, and funding agencies. Also, acknowledge the contributions of institutions that provided resources or facilities.
- Be concise and focused: Keep your acknowledgements concise and to the point. Focus on the key individuals and organizations that have made a substantial impact on your research.
- Use a formal tone: Maintain a professional and formal tone throughout your acknowledgements. Remember that this section is a formal acknowledgment of support, not a casual conversation.
- Follow ethical considerations: Ensure that you acknowledge individuals and organizations according to ethical guidelines and norms. Respect the privacy and confidentiality of individuals who may prefer not to be mentioned.
- Include personal touches: If appropriate, you can include personal anecdotes or specific instances where individuals or organizations made a significant impact on your research.
- Tailor acknowledgements to your research context: Consider the nature of your research and tailor your acknowledgements accordingly. For example, if you conducted interdisciplinary research, acknowledge experts from different fields who provided valuable insights.
- Proofread and edit: Like any other section of your research paper, proofread and edit your acknowledgements for grammar, spelling, and clarity. Ensure that the acknowledgements are well-written and free of errors.
- Seek feedback if needed: If you’re uncertain about whom to acknowledge or how to express your gratitude, seek feedback from your mentors, advisors, or colleagues. They can provide valuable guidance and suggestions.
- Show appreciation beyond formal requirements: While it’s important to acknowledge the required individuals and institutions, also consider extending your appreciation to others who may have supported you indirectly, such as family, friends, or colleagues who provided emotional support during your research journey.
Remember, acknowledgements are an opportunity to express your gratitude and recognize the contributions of those who have helped you along the way. Take the time to craft a thoughtful and sincere acknowledgement section that reflects the support and collaborative spirit of your research endeavor.
5 Samples for Acknowledgment in Research Paper
Explore these 5 carefully crafted acknowledgment samples to effectively express gratitude in your research paper.
Sample 1: Acknowledgement for Collaborative Research:
In this sample, we showcase an acknowledgement that acknowledges the collaborative efforts of research collaborators and team members. It highlights the importance of recognizing the joint contributions made towards the research project.
“I would like to express my deepest appreciation to the members of the research team, [Collaborators’ Names], for their invaluable contributions and collaborative spirit throughout this research project. Our collective efforts and synergistic teamwork have significantly enhanced the quality and depth of this study. Each member’s unique expertise and perspectives have brought forth diverse insights, resulting in a more comprehensive and well-rounded analysis.
I am grateful for the dedication, commitment, and professionalism demonstrated by each team member. The constructive discussions, intellectual debates, and shared enthusiasm have fostered an enriching research environment that has truly pushed the boundaries of our collective knowledge. This research project stands as a testament to the power of collaboration and the collective pursuit of knowledge.”
Sample 2: Acknowledgement for Funding Support:
This sample acknowledgement focuses on acknowledging the financial support received for the research. It emphasizes the significance of recognizing funding agencies or organizations that have provided the necessary resources for the research to take place.
“I would like to extend my sincere gratitude to the funding agencies and organizations that have provided financial support for this research. Their generous contributions have made it possible to conduct this study and have significantly contributed to its successful completion. The financial support has allowed for the procurement of necessary research materials, access to specialized equipment, and the opportunity to engage in valuable research experiences.
I would like to express my appreciation to [Name of Funding Agency/Organization 1] for their generous grant, which has played a crucial role in supporting this research project. Their belief in the significance of this study and their commitment to advancing knowledge in this field have been instrumental in its realization.
Furthermore, I would like to acknowledge the support received from [Name of Funding Agency/Organization 2]. Their funding has been vital in facilitating data collection, analysis, and the dissemination of research findings. Their investment in this project has not only provided financial resources but has also validated the importance and potential impact of this research.”
Sample 3: Acknowledgement for Mentorship and Guidance:
Here, we present a sample acknowledgement that expresses gratitude towards mentors and advisors who have provided guidance and support throughout the research journey. It underscores the critical role of mentorship in academic and research endeavors.
“I am deeply grateful to my mentor, [Mentor’s Name], for their exceptional guidance and unwavering support throughout this research endeavor. Their expertise, insightful feedback, and continuous encouragement have been invaluable in shaping the direction and outcomes of this study. Their unwavering commitment to my academic growth and professional development has been truly inspiring.
I am indebted to [Mentor’s Name] for their generous allocation of time and resources, their willingness to share their wealth of knowledge, and their unwavering dedication to pushing me to new heights. Their mentorship has not only enriched the quality of this research but has also had a profound impact on my personal and intellectual growth. I am truly fortunate to have had the privilege of working under their guidance.”
Sample 4: Acknowledgement for Institutional Support:
In this sample, we illustrate an acknowledgement that acknowledges the support and resources provided by institutions. It emphasizes the institutional backing that has facilitated the research process and contributed to its success.
“I would like to express my heartfelt gratitude to the faculty members and academic advisors who have provided guidance, feedback, and support throughout my academic journey. Their expertise, wisdom, and dedication to teaching and mentoring have been instrumental in shaping my research skills and scholarly pursuits.
I am grateful to [Name of Faculty Member/Advisor 1] for their unwavering support and invaluable insights. Their expertise and guidance have been critical in refining the research design, analyzing data, and interpreting findings. Their constructive feedback and intellectual discussions have truly enriched this study.
I would also like to acknowledge the contributions of [Name of Faculty Member/Advisor 2]. Their mentorship and encouragement have played a pivotal role in the development of my research abilities and have inspired me to reach for new heights. Their belief in my potential has been a constant source of motivation throughout this research journey.”
Sample 5: Acknowledgement for Peer Reviewers:
“I would like to express my deepest gratitude to the anonymous peer reviewers who have dedicated their time and expertise to provide valuable feedback and constructive criticism on this research paper. Their rigorous evaluation, insightful comments, and suggestions for improvement have immensely contributed to the quality and credibility of this work.
The meticulous review process conducted by the peer reviewers has helped shape and refine the content, methodology, and interpretation of this study. Their expertise in the field and their commitment to upholding scholarly standards have been crucial in ensuring the accuracy, validity, and relevance of the research findings.
I am sincerely grateful for the time and effort invested by each reviewer in thoroughly assessing this paper. Their detailed comments and recommendations have not only helped enhance the clarity and coherence of the manuscript but have also encouraged further reflection and refinement of the research.
The contributions of the peer reviewers are invaluable in the advancement of scientific knowledge and the improvement of academic publications. Their commitment to maintaining the rigor and integrity of the research process plays a pivotal role in fostering academic excellence and promoting the dissemination of high-quality research outcomes.”

When writing an acknowledgement in a research paper, begin by expressing gratitude to individuals, institutions, or organizations who have contributed to the research. Provide a sincere and concise acknowledgement, mentioning their specific contributions and the impact they made on the study.
While specific examples may vary depending on the research context, an acknowledgement section in a journal article typically acknowledges the contributions of individuals, funding sources, or institutions involved in the research process. It expresses gratitude for their support, guidance, or resources.
The purpose of the acknowledgement section in a research paper is to recognize and express gratitude to individuals or entities who have supported the research. It acknowledges their contributions, whether through funding, mentorship, technical assistance, data provision, or other forms of support.
When writing acknowledgements for a publication , start by identifying the key individuals or entities that have contributed to the research. Express gratitude for their support, mentioning specific contributions and the value they added to the study. Keep the acknowledgements concise and focused on the research context.
The acknowledgement section of a research paper should include acknowledgements for individuals or entities that have contributed to the research process. This may include mentors, advisors, funding agencies, research collaborators, or others who have provided valuable support, guidance, or resources.
While including an acknowledgement section in a research paper is not mandatory, it is a common practice in academic publishing. It provides an opportunity to acknowledge and appreciate the contributions of individuals or entities who have supported the research.
When writing an acknowledgements section for a literature review, acknowledge individuals or sources that have influenced and contributed to your understanding of the topic. Express gratitude for their insights, guidance, or resources that have shaped your literature review.
The terms “acknowledgement” and “acknowledgment” are both correct and interchangeable. The choice of spelling (with or without the “e”) may depend on regional or personal preferences.
To acknowledge a source in a research paper, use proper citation and referencing techniques according to the specific citation style guidelines. Include in-text citations and a corresponding entry in the reference list or bibliography to give credit to the original source.
Yes, you can acknowledge individuals who provided personal support in the acknowledgement section, such as family, friends, or loved ones. Recognize their emotional support, encouragement, or understanding during the research process.
Writing acknowledgements for a research paper allows you to express gratitude and acknowledge the invaluable contributions of individuals and institutions who have supported your work. By following the tips and utilizing the sample acknowledgements provided in this article, you can effectively and sincerely express your appreciation. Remember, acknowledgements are an opportunity to show your gratitude and give credit where it is due.
Acknowledgement for Paper Publication (10 Samples)
Acknowledgement for internship report: 10 samples and tips, leave a comment cancel reply.
Save my name, email, and website in this browser for the next time I comment.
Thesis Acknowledgements Examples
5 Examples For Your Inspiration
By: Derek Jansen (MBA) | Reviewers: Dr Eunice Rautenbach | May 2024
The acknowledgements section of your thesis or dissertation is an opportunity to say thanks to all the people who helped you along your research journey. In this post, we’ll share five thesis acknowledgement examples to provide you with some inspiration.
Overview: Acknowledgements Examples
- The acknowledgements 101
- Example: Formal and professional
- Example: Warm and personal
- Example: Challenge-centric
- Example: Institution-centric
- Example: Reflective and philosophical
- FREE Acknowledgements template
Acknowledgements 101: The Basics
The acknowledgements section in your thesis or dissertation is where you express gratitude to those who helped bring your project to fruition. This section is typically brief (a page or less) and less formal, but it’s crucial to thank the right individuals in the right order .
As a rule of thumb, you’ll usually begin with academic support : your supervisors, advisors, and faculty members. Next, you’ll acknowledge any funding bodies or sponsors that supported your research. You’ll then follow this with your intellectual contributors , such as colleagues and peers. Lastly, you’ll typically thank your personal support network , including family, friends, and even pets who offered emotional support during your studies.
As you can probably see, this order moves from the most formal acknowledgements to the least . Typically, your supervisor is mentioned first, due to their significant role in guiding and potentially evaluating your work. However, while this structure is recommended, it’s essential to adapt it based on any specific guidelines from your university. So, be sure to always check (and adhere to) any requirements or norms specific to your university.
With that groundwork laid, let’s look at a few dissertation and thesis acknowledgement examples . If you’d like more, check out our collection of dissertation examples here .
Need a helping hand?
Example #1: Formal and Professional
This acknowledgement formally expresses gratitude to academic mentors and peers, emphasising professional support and academic guidance.
I extend my deepest gratitude to my supervisor, Professor Jane Smith, for her unwavering support and insightful critiques throughout my research journey. Her deep commitment to academic excellence and meticulous attention to detail have significantly shaped this dissertation. I am equally thankful to the members of my thesis committee, Dr. John Doe and Dr. Emily White, for their constructive feedback and essential suggestions that enhanced the quality of my work.
My appreciation also goes to the faculty and staff in the Department of Biology at University College London, whose resources and assistance have been invaluable. I would also like to acknowledge my peers for their camaraderie and the stimulating discussions that inspired me throughout my academic journey. Their collective wisdom and encouragement have been a cornerstone of my research experience.
Finally, my sincere thanks to the technical staff whose expertise in managing laboratory equipment was crucial for my experiments. Their patience and readiness to assist at all times have left a profound impact on the completion of my project.
Example #2: Warm and Personal
This acknowledgement warmly credits the emotional and personal support received from family, friends and an approachable mentor during the research process.
This thesis is a reflection of the unwavering support and boundless love I received from my family and friends during this challenging academic pursuit. I owe an immense debt of gratitude to my parents, who nurtured my curiosity and supported my educational endeavours from the very beginning.
To my partner, Alex, your endless patience and understanding, especially during the most demanding phases of this research, have been my anchor. A heartfelt thank you to my supervisor, Dr. Mark Brown, whose mentorship extended beyond the academic realm into personal guidance, providing comfort and motivation during tough times.
I am also thankful for my friends, who provided both distractions when needed and encouragement when it seemed impossible to continue. The discussions and unwavering support from my peers at the university have enriched my research experience, making this journey not only possible but also enjoyable. Their presence and insights have been pivotal in navigating the complexities of my research topic.
Example #3: Inclusive of Challenges
This acknowledgement recognises the wide range of support received during unexpected personal and academic challenges, highlighting resilience and communal support.
The path to completing this dissertation has been fraught with both professional challenges and personal adversities. I am profoundly grateful for the enduring support of my supervisor, Dr. Lisa Green, whose steadfast belief in my capabilities and academic potential encouraged me to persevere even during my lowest moments. Her guidance was a beacon of light in times of doubt.
I must also express my deepest appreciation for my family, who stepped in not only with emotional reassurance, but also with critical financial support, when unexpected personal challenges arose. Their unconditional love and sacrifice have been the foundation of my resilience and success.
I am thankful, too, for the support services at the university, including the counselling centre, whose staff provided me with the tools to manage stress and maintain focus on my academic goals. Each of these individuals and institutions played a crucial role in my journey, reminding me that the pursuit of knowledge is not a solo expedition but a communal effort.
Example #4: Institution-Centric
This acknowledgement succinctly appreciates the financial, academic, and operational support provided by the university and its staff.
This dissertation would not have been possible without the generous financial support from the University of Edinburgh Scholarship Programme, which enabled me to focus fully on my research without financial burden. I am profoundly grateful to my advisor, Professor Richard Miles, for his sage advice, rigorous academic guidance, and the confidence he instilled in me. His expertise in the field of microbiology greatly enriched my work.
I would also like to thank the staff at the university library, whose assistance was indispensable. Their patience and willingness to help locate rare and essential resources facilitated a thorough and comprehensive literature review. Additionally, the administrative and technical staff, who often go unmentioned, provided necessary support that greatly enhanced my research experience. Their dedication and hard work create an environment conducive to academic success.
Lastly, my peers’ encouragement and the intellectual environment at the university have been vital in completing my research.
Example #5: Reflective and philosophical
This acknowledgement reflects on the philosophical growth and personal insights gained through the support of mentors, peers, and family, framing the dissertation as a journey of intellectual discovery.
Completing this thesis has been not only an academic challenge but also a profound journey of personal and philosophical growth.
I am immensely grateful to my mentor, Professor Sarah Johnson, for encouraging me to explore complex ideas and to challenge conventional wisdom. Her guidance helped me navigate the philosophical underpinnings of my research and deepened my analytical skills. My fellow students in the Philosophy Department provided a supportive and intellectually stimulating community. Our discussions extended beyond the classroom, offering new insights and perspectives that were crucial to my thesis.
I am also deeply thankful for my family, whose intellectual curiosity and spirited philosophical debates at home sparked my interest in philosophy from a young age. Their continuous support and belief in my academic pursuits have been instrumental in my success.
This dissertation reflects not only my work but also the collective support of everyone who has touched my life academically and personally. The journey has taught me the value of questioning and the importance of diverse perspectives in enriching our understanding of complex philosophical issues.

FREE Acknowledgements Template
To help fast-track your writing process, we’ve created a free, fully editable template . This template covers all the necessary content for a generic thesis or dissertation acknowledgements section. If you’re interested, you can download a copy here .

Psst... there’s more!
This post was based on one of our popular Research Bootcamps . If you're working on a research project, you'll definitely want to check this out ...
You Might Also Like:

Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly

- Acknowledgements for PhD Thesis and Dissertations – Explained
- Doing a PhD
The Purpose of Acknowledgements
The acknowledgement section of a thesis or dissertation is where you recognise and thank those who supported you during your PhD. This can be but is not limited to individuals, institutions or organisations.
Although your acknowledgements will not be used to evaluate your work, it is still an important section of your thesis. This is because it can have a positive (or negative for that matter) influence the perception of your reader before they even reach the main body of your work.
Who Should I Acknowledge?
Acknowledgements for a PhD thesis will typically fall into one of two categories – professional or personal.
Within these categories, who you thank will ultimately be your decision. However, it’s imperative that you pay special attention to the ‘professional’ group. This is because not thanking someone who has played an important role in your studies, whether it be intentional or accidental, will more often than not be seen as a dismissal of their efforts. Not only would this be unfair if they genuinely helped you, but from a certain political aspect, it could also jeopardise any opportunities for future collaborations .
Professional Acknowledgements
This may include, but is not limited to:
- Funding bodies/sponsorship providers
- Supervisors
- Research group and lab assistants
- Research participants
- Proofreaders
Personal Acknowledgements
- Key family members and friends
- Individuals who inspired you or directly influenced your academic journey
- Anyone else who has provided personal support that you would like to mention
It should be noted that certain universities have policies which state only those who have directly supported your work, such as supervisors and professors, should be included in your acknowledgements. Therefore, we strongly recommend that you read your university guidelines before writing this section of your thesis.
How to Write Acknowledgements for PhD Thesis
When producing this section, your writing style can be more informal compared to the rest of your thesis. This includes writing in first person and using more emotive language. Although in most cases you will have complete freedom in how you write this section of your thesis, it is still highly advisable to keep it professional. As mentioned earlier, this is largely because it will be one of the first things your assessors will read, and so it will help set the tone for the rest of your work.
In terms of its structure, acknowledgements are expected to be ordered in a manner that first recognises the most formal support before moving onto the less formal support. In most cases, this follows the same order that we have outlined in the ‘Who Should I Thank’ section.
When thanking professionals, always write out their full name and provide their title. This is because although you may be on a first-name basis with them, those who read your thesis will not. By providing full names and titles, not only do you help ensure clarity, but it could also indirectly contribute to the credibility of your thesis should the individual you’re thanking be well known within your field.
If you intend to include a list of people from one institution or organisation, it is best to list their names in alphabetical order. The exception to this is when a particular individual has been of significant assistance; here, it would be advisable to list them.
How Long Should My Acknowledgements Be?
Acknowledgements vary considerably in length. Some are a single paragraph whilst some continue for up to three pages. The length of your acknowledgement page will mostly depend on the number of individuals you want to recognise.
As a general rule, try to keep your acknowledgements section to a single page. Although there are no word limits, creating a lengthy acknowledgements section dilutes the gratitude you’re trying to express, especially to those who have supported you the most.
Where Should My Acknowledgements Go?
In the vast majority of cases, your acknowledgements should appear directly after your abstract and before your table of contents.
However, we highly advise you to check your university guidelines as a few universities set out their own specific order which they will expect you to follow.
Phrases to Help You Get Started

We appreciate how difficult it can be to truly show how grateful you are to those who have supported you over the years, especially in words.
To help you get started, we’ve provided you with a few examples of sentences that you can complete or draw ideas from.
- I am deeply grateful to XXX…
- I would like to express my sincere gratitude to XXX…
- I would like to offer my special thanks to XXX…
- I would like to extend my sincere thanks to XXX…
- …for their assistance at every stage of the research project.
- …for their insightful comments and suggestions.
- …for their contribution to XXX.
- …for their unwavering support and belief in me.
Thesis Acknowledgement Examples
Below are three PhD thesis acknowledgment samples from which you can draw inspiration. It should be noted that the following have been extracted from theses which are freely available in the public domain. Irrespective of this, references to any individual, department or university have been removed for the sake of privacy.
First and foremost I am extremely grateful to my supervisors, Prof. XXX and Dr. XXX for their invaluable advice, continuous support, and patience during my PhD study. Their immense knowledge and plentiful experience have encouraged me in all the time of my academic research and daily life. I would also like to thank Dr. XXX and Dr. XXX for their technical support on my study. I would like to thank all the members in the XXX. It is their kind help and support that have made my study and life in the UK a wonderful time. Finally, I would like to express my gratitude to my parents, my wife and my children. Without their tremendous understanding and encouragement in the past few years, it would be impossible for me to complete my study.
I would like to thank my supervisors Dr. XXX and Dr. XXX for all their help and advice with this PhD. I would also like to thank my sisters, whom without this would have not been possible. I also appreciate all the support I received from the rest of my family. Lastly, I would like to thank the XXX for the studentship that allowed me to conduct this thesis.
I would like to thank my esteemed supervisor – Dr. XXX for his invaluable supervision, support and tutelage during the course of my PhD degree. My gratitude extends to the Faculty of XXX for the funding opportunity to undertake my studies at the Department of XXX, University of XXX. Additionally, I would like to express gratitude to Dr. XXX for her treasured support which was really influential in shaping my experiment methods and critiquing my results. I also thank Dr. XXX, Dr. XXX, Dr. XXX for their mentorship. I would like to thank my friends, lab mates, colleagues and research team – XXX, XXX, XXX, XXX for a cherished time spent together in the lab, and in social settings. My appreciation also goes out to my family and friends for their encouragement and support all through my studies.
Browse PhDs Now
Join thousands of students.
Join thousands of other students and stay up to date with the latest PhD programmes, funding opportunities and advice.

The Savvy Scientist
Experiences of a London PhD student and beyond
Thesis acknowledgements: Samples and how to write your own thesis or dissertation acknowledgements

Writing a thesis can be tricky. That’s why I’m starting a new series covering each section of the thesis, from thesis acknowledgements all the way to conclusions. I’ll be guiding you through the whole process, from what to include in your thesis to how to write it, along with examples from defended theses to help you to write your own.
We’ll begin by covering thesis acknowledgments. The acknowledgements section appears at the start of the thesis so it is often one of the first parts that everyone tries to tackle. As this will likely be your first taste of your thesis it can often feel quite intimidating to write!
Thankfully it’s also one of the easiest parts of the thesis to complete, which may help to give you a boost for the rest.
In this post we’ll cover everything to do with thesis acknowledgements: samples, what to include and how to write them. At the end I’ll also outline a 60 minute exercise which will get you preparing a first draft of your own! I’ve also got a similar post to craft your thesis title, which you can check out here .
I’m writing this post with a PhD thesis in mind but it could work just as well if you’re looking for help including acknowledgements in your Master’s or undergraduate thesis/ dissertation.
What is the purpose of the acknowledgements section in a thesis?
The acknowledgements section of your thesis is an opportunity to reflect on the people who have supported and shaped your PhD experience.
Don’t worry, although your examiners will be interested to read your acknowledgements section, you won’t really get judged on it in your PhD viva. This section is for you to share as little, or as much, as you want about everyone involved in your PhD journey.
The acknowledgements are a very personal section of your thesis and each PhD student will have different things they want to include. For example, many people wonder: How do I thank my family in a thesis? And the acknowledgements section is the answer!
Note – You can also use a thesis dedication to thank your family. This is a separate section to your thesis acknowledgements and is entirely optional. It’s usually just a single line, just like you might find at the front of some books. Most people don’t include a separate dedication section but you can if you want to go that extra step.
What to include in your thesis acknowledgements
There are usually no formal requirements dictating what to include in your acknowledgements. However, do double check for any potential rules at your specific institution.
In general the acknowledgements are the section of your thesis where you have some creative liberty and are not bound by rigid research protocols or guidelines.
Many students choose to use the acknowledgements section to thank people (or organisations) who:
- Introduced them to the topic
- Helped with their PhD application
- Funded the project
- Supervisors
- Technicians
- Partners, friends or family
- Or anyone else who made an impression along the way!
But remember, you can include whatever you want! For example in my own PhD acknowledgements, which you’ll read further down this post, I thanked the university for providing a green outdoor space for us.
Acknowledge whoever and whatever influenced your own PhD experience.
You may find it helpful to start by writing a list of everyone you wish to thank.
How do you write an acknowledgements section?
Since there are no guidelines to worry about, it is really up to you how you write your own thesis acknowledgements. You have a lot of freedom for what to include and how to write it.
However you may find the following suggested phases helpful as a starting point.
Who you want to thank…
- “First and foremost, I would like to express my sincere gratitude to…”
- “I must thank…”
- “A special thanks to…”
- “I would like to highlight two truly exceptional people from…”
- “I want to thank…”
- “In addition, I would like to mention”
- “I would also like to extend my thanks to…”
- “I want to give my deepest appreciation to…”
- “Finally, but the most importantly, I would like to thank…”
…then, why you want to thank them
It can be nice to also include why you’re thanking these people, using phrases such as:
- “…for the opportunity to be a part of this project”
- “…for always being there when I needed his support, reviewing my progress constantly, and guiding me through my PhD studies”
- “….for being a great bunch of people in and out of the lab”
- …”for all the guidance, support and outstanding feedback”
- “… who took their time to help teach me…”
- “…for her unlimited support and unconditional guidance during my PhD journey”
- “…were always there for discussions about anything that I was unsure on”
- “…whom has offered invaluable advice that will benefit me throughout my life”
- “…for supporting me since my undergraduate, and for the valuable discussions we had along the road”
- “…for making the past 4 years much more enjoyable and keeping me sane throughout the whole process”
Here is a whole example from an accepted PhD thesis:
Firstly, I want to thank [supervisor’s name(s)] for giving me the opportunity to work on this project, providing valuable guidance and feedback, and challenging me to grow as a scientist. Excerpt from Dr Wane’s thesis acknowledgements, available via this page or use this direct download link .
Some people will choose to use full names and titles for any professional acknowledgements and first names for any personal ones. Again, this is up to you.
To help illustrate the variety of thesis acknowledgement formats, we’ll shortly be coming on to some examples of acknowledgment sections from successfully defended theses.
Before then I want to cover some of the main questions relating to how to write your own thesis acknowledgements section:
How long should you spend writing your thesis acknowledgements?
My suggestion is to spend only an hour or two making a first draft. I suggest doing this well ahead of your final deadline so that you have time to come back to it. Even so, I’d certainly look to spend far less than one day’s work on it in total.
It is a “nice to have” and means a lot to a lot of people, but remember you’re really only writing this section for yourself. I probably spent about two hours writing mine in total, simply because it wasn’t a priority for me.
What order should you write your acknowledgements in?
A typical way to write your acknowledgements is to go from the most formal/academic relationships to the least.
It is normal to start with any funding bodies, then formal people like your PhD supervisors, then move through labmates, friends and family. But again, there are generally no rules!
How long should the acknowledgements section be?
You can include as much or as little as you want. My own PhD acknowledgements section was just under a page long and it consisted of 386 words or 1892 characters (without spaces).
Here is how it was formatted:

But let’s not just look at my thesis. Using Imperial’s publicly accessible database I went through 25 published PhD theses for you.
The average (mean) length of these 25 theses was 365 words and 1793 characters without spaces. Writing an acknowledgements section of length 350-450 words was the most common:

The shortest acknowledgements sections was 122 words(653 characters) long. The longest one consisted of 1022 words and 5082 characters. Hopefully this illustrates that you’re not really bound by any limits. Write as much or as little as you want for this section.
Sample thesis acknowledgements
My own phd thesis acknowledgement.
My own PhD thesis is available here *, the acknowledgements section is on page 5. Here is the complete version of my acknowledgements section:
I would like to acknowledge both EPSRC and the Class of 1964 Scholarship for their financial support. It has been an honour to be the inaugural recipient of the Class of 1964 Scholarship and I am indebted to the donors in providing me complete academic freedom in this research. An immense thank you to my PhD supervisors: Jonathan Jeffers, Ulrich Hansen and Julian Jones. Support and guidance throughout the project from you all has been invaluable. JJ in particular you’ve been a fantastic primary supervisor. Thank you to all the academics who helped me get to this stage. The late Dr Kajal Mallick and his Biomedical Materials course at the University of Warwick was a huge influence and without which I would have never followed this path. My “pre-doc” supervisors in Dr Helen Lee of University of Cambridge and in particular the remarkable Prof Judith Hall OBE of Cardiff University from whom I learned so much. Thanks to Alison Paul and Michael Lim for being so supportive when I was considering applying for PhDs. It has been an amazing experience working between two research groups across different departments, thanks to everyone from the Biomechanics and JRJ groups I’ve worked with and from whom I’ve learned so much. Thank you of course to the Hybrids team I’ve worked so closely on this project with: Fra, Gloria, Agathe, Maria, Silvia, it’s been great fun working with you all! Gloria in particular thanks for you all your help, support and friendship: your inclusivity is appreciated by many. Saman, I’ve been so pleased to have you working on DVC with me and being able to discuss ideas with you really has been invaluable. I am grateful to everyone I’ve collaborated with externally: Farah, Amin and Brett (Natural History Museum) plus Andy and Behzad (Royal Veterinary College), thank you all for your support and input. Thanks also to everyone I’ve met through the Environmental Society at Imperial in particular Chelcie: your friendship and support have added a lot to my life. Thanks to Imperial for providing space for the ESoc garden, taking a break and enjoy nature in this space has certainly improved my work. Thanks of course to my family for their support. Finally, thank you Jo for always being so supportive and helping me every step of the way. My PhD thesis, available here . Acknowledgements are on page 5.
*For me the thesis was a means to an end. I wanted my PhD and didn’t want to spend too long agonising over each page. Therefore, it is possible there are typos in there, if you read any of it: firstly well done, I haven’t looked at it much since submitting the final copy, secondly, please don’t tell me about any typos you find!
Other PhD thesis acknowledgement examples
Below are the other 24 published and openly accessible STEM PhD theses I found for this article.
For each person’s thesis, either follow the first link to be taken to the landing page or follow the second link to directly download their thesis: I gave you a choice in case you don’t want stuff to start downloading automatically from a random text link!

The list is formatted as follows:
- [Link to thesis page on repository], [which page the acknowledgements appear on], [direct link to download the thesis]
- Dr Shipman’s thesis , for the acknowledgements go to page 3. Direct download here .
- Longest acknowledgements section of the list at 1022 words.
- Dr Li’s thesis , page 11. Direct download here .
- Dr Podgurschi’s thesis , page 5. Direct download here .
- Dr Medjeral-Thomas’ thesis page 3. Direct download here .
- Dr Sztuc’s thesis , page 5. Direct download here .
- Dr Yap’s thesis , page 5. Direct download here .
- Dr Sukkar’s thesis , page 9. Direct download here .
- Dr Lo’s thesis , page 11. Direct download here .
- Dr Sullivan’s thesis , page 5. Direct download here .
- Dr Tawy’s thesis , page 3. Direct download here .
- Dr Wane’s thesis , page 2. Direct download here .
- Dr Addison’s thesis , page 4. Direct download here .
- Dr Wang’s thesis , page 5. Direct download here .
- Dr Sebest’s thesis , page 3. Direct download here .
- Dr Hopkins’ thesis , page 7. Direct download here .
- Dr Bates’s thesis , page 4. Direct download here .
- Dr Somuyiwa’s thesis , page 6. Direct download here .
- Dr Reynolds’ thesis , page 5. Direct download here .
- My labmate’s thesis, who wrote the acknowledgements in a different style to the rest by using bullet points.
- Shortest acknowledgements section of the list at 122 words.
- Dr Manca’s thesis , acknowledgements on page 5. Direct download here .
- Dr Liu’s thesis , page 5. Direct download here .
- Dr Hotinli’s thesis , page 7. Direct download here .
My top tips for writing your own thesis acknowledgements
- Don’t spend too long on them. The acknowledgements section is really not worth spending too much time on. Even worse, since they appear at the start of your thesis, it is tempting to write your acknowledgements first. This can be fine, or, it can be an opportunity for lots of unnecessary procrastination. Which I why I instead suggest that you…
- Write your acknowledgements at the end of your first draft of the thesis. There is no need to write your thesis in the order it is presented. If you write your acknowledgements at the end you’ll be less likely to spend precious time on a section which really doesn’t warrant too much brain power.
- Don’t stress about it. The acknowledgements are merely for yourself and for anyone close to you that you want to thank. There are far more important sections for you to be particular about!
- Remember: You can make changes after you submit the copy for your viva. As with everything in your thesis, you can make changes after you submit the thesis for your viva. The real “final” copy is when you submit your thesis to the university for archiving. Which is even more reason to not spend too much time writing it the first time around.
Draft your own thesis or dissertation acknowledgements in 60 minutes
Hopefully you now feel inspired to start writing your own thesis acknowledgments!
For the exercise below I’d suggest setting a stop-watch on your phone and move on to the next section when the alarm goes, even if you’ve not fully finished. The aim is to have a rough draft at the end which you can polish off at a later point in time.
- Read a few of the example thesis acknowledgements above to get a feel for the structure ( 15 mins )
- List everyone (or everything!) you wish to thank – including any personal and professional acknowledgements in addition to funding bodies if relevant ( 10 mins )
- Decide on a rough order in which to thank them ( 5 mins )
- Craft some sentences using the phrases mentioned above ( 30 mins )
Congratulations you’re now well on your way to having one section of your PhD thesis completed!
I hope this post has been useful for constructing your own thesis or dissertation acknowledgements. It is the first in a series of posts aiming to help your thesis writing by delving into each section in depth. Be sure to let me know if you have any questions or suggestions for other content which you would find useful.
Subscribe below to stay updated about future posts in the series:
Share this:
- Click to share on Facebook (Opens in new window)
- Click to share on LinkedIn (Opens in new window)
- Click to share on Twitter (Opens in new window)
- Click to share on Reddit (Opens in new window)
Related Posts

Minor Corrections: How To Make Them and Succeed With Your PhD Thesis
2nd June 2024 2nd June 2024

How to Master Data Management in Research
25th April 2024 27th April 2024

Thesis Title: Examples and Suggestions from a PhD Grad
23rd February 2024 23rd February 2024
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Notify me of follow-up comments by email.
This site uses Akismet to reduce spam. Learn how your comment data is processed .
Privacy Overview

How to write acknowledgements in a thesis or dissertation
Navigating the intricate process of writing a thesis or dissertation can be challenging.
One crucial, yet often overlooked part is the thesis acknowledgement. It is also the only bit of my thesis that anyone really reads.
This section allows you to express gratitude to those who contributed to your academic journey. From supervisors and professors to family and friends, the acknowledgement section provides a platform to thank all who played a part in your work.
Whether you’re unsure about how to begin or looking for the best ways to acknowledge your mentors, this blog will provide valuable insights and practical advice to help you create an impactful thesis acknowledgement.
What is your thesis acknowledgement?
A thesis acknowledgement is a section in your thesis where you express gratitude to those who helped and supported you during your research and writing process.

It typically comprises two parts: professional and personal acknowledgements.
- Professional acknowledgements include your supervisor, colleagues, other academics, funding bodies, or institutions that significantly contributed to your work.
- Personal acknowledgements encompass your family and friends who provided emotional support or helped with editing and proofreading.
The acknowledgements section is usually more informal than the rest of your thesis , and it’s acceptable to write in the first person. It’s typically placed at the beginning of your thesis, either before the abstract or the table of contents.
Although the length may vary, it usually doesn’t exceed one page. It’s crucial to plan ahead, listing everyone you wish to thank and consider their specific contribution to your work.
Who to thank in your acknowledgements
In your acknowledgements, you should first thank the members of academia who contributed to your research, including:
- funding bodies,
- supervisors,
- professors,
- proofreaders,
- and research participants.
Mention them using their full names and titles.
If an authoritative figure in your field provided feedback, their acknowledgement adds weight to your research.
Despite the circumstances, a brief thank you to your supervisor is necessary.
Personal acknowledgements can include friends, family members, or even pets who provided inspiration or support during the writing process. Always refer to your university’s guidelines on acknowledgements.
Creating an acknowledgement can be slightly subjective, as the order and individuals to be thanked can vary greatly depending on the circumstances of the work and the author’s preferences.
However, generally, this example follows a common structure:
The order can be customized based on the importance of the roles these individuals played in the author’s journey.
Some may prefer to thank family or significant others first, while others might start with professional relationships such as advisors or collaborators.
It’s also crucial to keep in mind that the way of expressing gratitude can differ significantly between cultures and individuals.
How Long Should My Acknowledgements Be?
The length of an acknowledgement section varies depending on the individual and the nature of the project.
Some people prefer to keep their acknowledgements brief and only thank those individuals who made significant contributions to their work.
Others may choose to include a more extensive list of people, such as mentors, colleagues, and friends, who provided support and encouragement throughout the process.
In general, it is recommended to keep your acknowledgements concise and focused on those who had a direct impact on the project
. Including a heartfelt thank you to these individuals is a meaningful way to show appreciation for their efforts.
However, it is important not to get carried away and turn the acknowledgement page into a long list of names. Remember that the focus should be on quality rather than quantity, as the acknowledgement section should not overshadow the main content of the project.
Where Should My Acknowledgements Go?
The placement of your acknowledgements can vary, but it’s typically located in the first part of your thesis.
Mine is right after the abstract and before the introduction of my PhD thesis.
You can place it right before your dissertation abstract or before the table of contents. However, the exact positioning may depend on the guidelines and requirements provided by your university.
Always ensure to check your university’s formatting requirements to be sure you’ve chosen the correct location for your acknowledgements section.
Thesis acknowledgement examples
Here is my PhD thesis acknowledgement.

Here are some sentence starters that you can use for inspiration:
1. “This thesis acknowledgement is a tribute to all the people who made my academic journey worthwhile.” 2. “I would like to thank my supervisor, whose unwavering support has been instrumental in the completion of this thesis.” 3. “In this acknowledgement section, I extend my deepest gratitude to all who have walked with me on this challenging but fulfilling journey.” 4. “Firstly, I would like to express my sincere thanks to the academic staff who provided their invaluable expertise and guidance.” 5. “My thesis would not have been possible without the endless help and support from my colleagues.” 6. “Special thanks go to my family, whose constant encouragement fueled my perseverance during the completion of this dissertation.” 7. “In the professional acknowledgements, I would like to acknowledge the significant contributions made by my research participants.” 8. “I would also like to thank the funding bodies, whose financial support made this research possible.” 9. “Through this acknowledgment, I express my heartfelt gratitude to my friends who have been my pillars of strength.” 10. “The completion of this thesis or dissertation is the culmination of efforts from various individuals whom I would like to express my sincere appreciation.” 11. “This thesis acknowledgement section is an opportunity to give thanks to those who made this journey less daunting.” 12. “I would like to express my gratitude to my editor, whose meticulous proofreading greatly improved my thesis.” 13. “Without their dedication, this thesis would not have been possible.” 14. “I express my sincere gratitude to all those whose names appear in this acknowledgement for their invaluable input.” 15. “In this acknowledgement for my thesis, I extend my appreciation to all those who have been part of this journey.”
Top tips to write acknowledgements
- Plan Ahead : Make a list of the people you want to acknowledge and their specific contributions to your work.
- Follow University Guidelines : Check your university’s formatting and content guidelines to ensure your acknowledgements adhere to them.
- Use First Person : Unlike the rest of your thesis, the acknowledgements can be written in the first person.
- Keep it Brief : The acknowledgement section should generally not exceed one page. Be concise and precise in expressing your gratitude.
- Maintain Professional-Personal Order : Start with professional acknowledgements (e.g., supervisors, colleagues, funders) before moving on to personal ones (e.g., friends, family).
- Be Specific : Highlight the specific contributions each person or organization made to your thesis.
- Use Full Names and Titles : When acknowledging academic contributors, use their full names and appropriate titles.
- Use Informal Language : Acknowledgements can be written in a more informal style, but avoid colloquial language.
- Proofread : Ensure your acknowledgements are free of spelling and grammar errors.
- Be Genuine and Sincere : The acknowledgements section should sincerely reflect your gratitude to the people who helped you in your academic journey.
Wrapping up – writing your acknowledgements section
As we reach the conclusion of this informative journey into the art of writing acknowledgements for a thesis or dissertation, it’s clear that this often-overlooked section carries significant emotional and professional weight.
A dissertation acknowledgements page is more than just a list of names; it’s a chance to express genuine gratitude and give due credit to all who have contributed to your academic journey.
Remember, writing this section of your thesis isn’t an obligatory chore but a genuine opportunity to thank those who supported you.
From the tireless members of your thesis committee to the friends and family who offered emotional support, it’s a platform to acknowledge all the people who helped.
From mentors who provided expert guidance, colleagues who offered invaluable insights, to the institutions that funded your research – everyone deserves a heartfelt note of thanks.
Sample acknowledgements in a thesis often include both professional acknowledgements first, followed by personal ones, ensuring that all contributors are recognized appropriately. Always remember to use full names and titles for professional acknowledgements, and express your gratitude sincerely.
The acknowledgement page isn’t a place for long tales, jokes or anecdotes; instead, keep your acknowledgements concise, specific, and heartfelt.
As shown in the thesis acknowledgement examples, you should reflect on the people and organizations that significantly contributed to your research or writing, whether in a substantial technical manner or through support and guidance throughout the process.
Studentship that allowed you to pursue your research, faculty who guided your studies, even friends who provided distractions when they were most needed – all these contributors deserve your thanks. Remember, it’s okay to use their first names for those who’ve been part of your personal journey, but for professional acknowledgments, full names and titles are recommended.
As a PhD student, your acknowledgements should reflect your journey – the struggles, the triumphs, and most importantly, the people who have helped you along the way. Whether you include a list of names in alphabetical order, or you decide to group people or organizations, remember to be genuine, concise, and respectful.
Whether it’s a thesis dedication to a mentor, expressing gratitude to your parents, thanking your friends for their love and encouragement, or even including certain political aspects that influenced your research, the acknowledgments section is yours to personalize.
Writing a thesis or dissertation is a monumental task, and the people who support you through it are worth acknowledging. Keep this guide in mind when you write your thesis acknowledgements, and don’t forget to thank those who’ve been there for you – for in the journey of research and writing, no one truly walks alone.
The last sentence may be a heartfelt statement, “I would like to express my gratitude to all those who walked with me throughout my research journey – your support was my strength, and this achievement is as much yours as it is mine.”

Dr Andrew Stapleton has a Masters and PhD in Chemistry from the UK and Australia. He has many years of research experience and has worked as a Postdoctoral Fellow and Associate at a number of Universities. Although having secured funding for his own research, he left academia to help others with his YouTube channel all about the inner workings of academia and how to make it work for you.
Thank you for visiting Academia Insider.
We are here to help you navigate Academia as painlessly as possible. We are supported by our readers and by visiting you are helping us earn a small amount through ads and affiliate revenue - Thank you!

2024 © Academia Insider

- Thesis Action Plan New
- Academic Project Planner
- Literature Navigator
- Thesis Dialogue Blueprint
- Writing Wizard's Template
- Research Proposal Compass
- Why students love us
- Why professors love us
- Why we are different
- All Products
- Coming Soon

Exploring the Implications for Research in Modern Science

Crafting Effective Literature Review Questions for Academic Success

Understanding and Overcoming Test Anxiety: A Comprehensive Thesis PDF

Navigating Anxiety in Your PhD Thesis: Tips and Strategies

Friends and Thesis Writing: Striking the Right Balance for Your Thesis

Mastering the Art of Communication: Effective Strategies for Voice Your Research in Interviews
Navigating ethical dilemmas: the morality of conducting interviews.
Data Anomalies: Strategies for Analyzing and Interpreting Outlier Data
Crafting the perfect thesis acknowledgement: tips and examples.

Crafting the perfect thesis acknowledgement is a respectful and important part of completing your academic thesis. It provides an opportunity to express gratitude to those who have supported you throughout your research journey. This article aims to guide you through the essential elements of thesis acknowledgements and offer practical examples to help you write a sincere and structured acknowledgement section.
Key Takeaways
- Understanding the purpose of acknowledgements in a thesis is crucial; they recognize the support and contributions of individuals and organizations.
- A well-structured acknowledgement should be genuine, concise, and specific, mentioning each contributor by name and the nature of their assistance.
- Providing examples from various categories such as mentors, family, classmates, and funding support can serve as a template for crafting personalized acknowledgements.
Essential Elements of Thesis Acknowledgements
Understanding the purpose.
The acknowledgement section of your thesis is not merely a formality; it is a professional tribute to the individuals and entities that have supported your academic journey . It serves as a genuine expression of gratitude , reflecting the assistance and encouragement you've received. When crafting this section, consider the purpose of each acknowledgment, ensuring that your thanks are directed appropriately and thoughtfully.
To effectively recognize the contributions of others, you may want to:
- Identify key individuals such as advisors, mentors, and family who have provided substantial support.
- Consider the roles of classmates, research participants, and institutional entities in your work.
- Acknowledge any financial support or technical assistance that was crucial to your research.
Remember, the acknowledgements section is yours to personalize . While there are formalities to observe, the essence lies in providing a sincere and specific account of the support you've received. This is your opportunity to convey appreciation in a meaningful way, setting the tone for the professional and academic relationships you've built during your thesis endeavor.
Structuring Your Acknowledgement
When you begin to structure your acknowledgement, start with a courteous introduction, expressing your sincere gratitude. Identify each contributor by name , and succinctly state how they supported your thesis journey. It's important to maintain a balance between brevity and meaningfulness, ensuring that each acknowledgment is both concise and heartfelt.
Consider the following structure for your acknowledgement:
- Formal address or opening
- Thanks to academic advisors and faculty
- Acknowledgement of funding sources
- Gratitude towards research participants
- Appreciation for family and friends
- Mention of technical and editorial assistance
- Closing remarks
Remember to review the Thesis / Dissertation Formatting Manual (2024) for specific guidelines on citing previously published material and obtaining necessary permissions. The structure of your acknowledgement should reflect the unique contributions to your work, as outlined in the ' Acknowledgement for Thesis | Definition & Sample - BachelorPrint'. Lastly, ensure that your acknowledgement aligns with the general recommendations provided in '4. Writing up your Research: Thesis Formatting (MS Word)', which includes a title page, abstract, and table of contents.
Providing Genuine Thanks
In crafting your thesis acknowledgement, the essence of genuine gratitude cannot be overstated. Express your thanks in a personal and meaningful way , ensuring that each contributor feels valued for their unique support . Reflect on the specific ways individuals or organizations have aided your journey, whether through intellectual guidance, emotional support, or logistical assistance.
- Begin with a heartfelt opening that sets the tone of gratitude.
- Mention each contributor by name, highlighting their specific input.
- Conclude with a warm closing that encapsulates your overall appreciation.
Remember, an acknowledgement is more than a mere formality; it's a reflection of your academic integrity and personal character. By being sincere and specific , you honor the collective effort that has shaped your scholarly work.
Reviewing and Finalizing Your Acknowledgement
Once you have expressed your gratitude and acknowledged all the significant contributors to your thesis, it's crucial to review and finalize your acknowledgement. Ensure that your acknowledgement reflects sincerity and professionalism by meticulously checking for any errors or omissions. Here are some steps to consider:
- Reread your acknowledgement to verify that all names are correctly spelled and titles are accurate.
- Reflect on the contributions of each individual or entity, making sure you haven't missed anyone who played a pivotal role in your thesis journey .
- Seek feedback from peers or mentors to gain an outside perspective on the tone and content of your acknowledgement.
- Revise your acknowledgement for clarity, conciseness, and flow, ensuring it reads well and conveys your genuine appreciation.
Remember, the acknowledgement is not just a courtesy, but a meaningful part of your thesis that showcases your professional relationships and gratitude. Before you consider this section complete, take a moment to restate the significance of each contribution, mirroring the care you've invested in your research.
Practical Examples of Thesis Acknowledgements
Acknowledgement for course instructor.
When crafting your thesis acknowledgement, it's essential to express gratitude to those who have played a pivotal role in your academic journey , including your course instructor. Begin with a heartfelt thank you , acknowledging the specific contributions of your instructor. For instance, you might appreciate their insightful lectures , constructive feedback, or the encouragement they provided that spurred your intellectual growth.
Remember to keep your language genuine and concise . A bulleted list can help you structure your thanks effectively:
- Thank your instructor for their dedication to your learning.
- Mention the ways they have enriched your understanding of the subject.
- Acknowledge any personal encouragement or academic advice they have offered.
By following these steps, you ensure that your acknowledgement resonates with sincerity and adequately reflects the importance of your instructor's support.
Acknowledgement for the Mentor
The mentorship you receive during your thesis journey is pivotal to your academic and personal growth. Your mentor's expertise and guidance are the bedrock of your research success. It is essential to acknowledge their contribution with gratitude and respect. Here are some points you might consider including in your acknowledgement for your mentor:
- Expressing appreciation for their insightful feedback and continuous support.
- Recognizing the mentor's role in shaping your academic path and research skills.
- Mentioning specific instances where their guidance was particularly influential.
- Acknowledging the time and effort they invested in your success.
Remember, a heartfelt and personalized acknowledgement will always resonate more than a generic thank you. Take the time to reflect on the unique aspects of your mentorship experience when crafting this section of your thesis.
Acknowledgement for Family Support
When it comes to acknowledging family support in your thesis, it's essential to convey your gratitude in a manner that reflects the personal and emotional support you've received. Your family's unwavering belief in your academic journey deserves a special mention. They've provided a foundation of encouragement and understanding, often without the expectation of anything in return.
Expressing thanks to your family can be structured in a simple list to ensure each member's contribution is recognized:
- To my parents, for their wisdom and sacrifices.
- To my siblings, for their companionship and humor during stressful times.
- To my partner, for patience and love that gave me strength.
Remember, a heartfelt acknowledgment resonates more than a mere listing of names. It's about capturing the essence of their support, which has been as crucial as any academic guidance. As one source puts it, special thanks are due to family for their patience and understanding during the long hours spent in academic pursuit. In finalizing this section, review it to ensure it genuinely reflects your appreciation and the unique role your family has played in your academic success.
Acknowledgement for Classmates
Your journey through the rigorous academic challenge of thesis writing is often shared with a group of individuals who are in the same boat - your classmates. Their camaraderie and mutual support play a pivotal role in your success . It is essential to acknowledge their contribution, as they provide not just academic support, but also a sense of community during this intense period.
Collaborative efforts with classmates can take various forms, from group study sessions to peer reviews of your work. Here are some ways to express gratitude to your classmates in your thesis acknowledgement:
- Thank them for specific instances of support, such as providing feedback on a draft or sharing resources.
- Acknowledge the collective spirit that helped you stay motivated and focused.
- Mention any classmate who went above and beyond to assist you in your research or writing process.
Remember, a heartfelt acknowledgement can strengthen your bonds and show appreciation for the shared academic journey.
Acknowledgement for Research Participants
When acknowledging research participants , it's essential to express your sincere appreciation for their time and insights. These individuals have played a pivotal role in the advancement of your study, offering valuable data that has likely shaped the outcome of your research. Their willingness to share personal experiences and information is the cornerstone of your project's success.
Begin by thanking them collectively, and if appropriate, mention any groups or communities specifically. It's important to respect the privacy and confidentiality agreements made with participants, so avoid disclosing sensitive information. Here's a simple structure you can follow:
- Express gratitude for their participation
- Highlight the importance of their contributions
- Acknowledge the impact of their data on your research
- Reiterate your commitment to confidentiality and ethical standards
Remember, without their involvement, the richness of your research would be diminished. As you draft this section, reflect on the tools and resources that aided you in defining your thesis purpose and maintaining research focus , such as worksheets, templates, and action plans. These tools not only facilitate organization but also underscore the significance of each participant's contribution.
Acknowledgement for Funding Support
When acknowledging funding support , it's crucial to express gratitude in a manner that reflects the significance of the financial contribution to your research. Your benefactors have provided more than just funds; they have invested in your academic potential.
- Begin by naming the specific funding bodies , grants, or scholarships that supported your work. For example, "This research was supported by the XYZ Scholarship Fund."
- Mention how the funding has directly impacted your research, such as enabling fieldwork, purchasing equipment, or allowing for full-time dedication to the study.
- If applicable, outline the broader impact of the funding, such as contributing to the advancement of knowledge in your field or supporting future scholars.
Remember to review the requirements or guidelines provided by your funding bodies as some may have specific stipulations on how their support should be acknowledged. A sincere and thoughtful acknowledgement not only shows your appreciation but also demonstrates your professionalism and attention to detail.
Acknowledgement for Technical Assistance
When acknowledging technical assistance in your thesis, it's important to highlight the specific contributions made by individuals or groups that provided technical support . Your acknowledgement should reflect the significance of their expertise to your research. For instance:
- Mr. Smith's proficiency with analytical tools greatly facilitated the research process.
- The guidance in navigating complex data sets was crucial.
- The dedication to ensuring the accuracy and rigor of findings was invaluable.
Expressing gratitude for technical assistance not only shows your appreciation but also underscores the collaborative nature of academic research. Remember to be specific about the type of assistance received and the impact it had on your work. This can range from data analysis to specialized equipment handling or software troubleshooting.
Incorporate these tips for effective time management to overcome procrastination and make progress on your thesis. Break tasks into smaller chunks, eliminate distractions, set goals, take breaks, and seek support when you're dealing with technical challenges. This proactive approach will help you maintain momentum and express genuine thanks in your acknowledgement.
Acknowledgement for Institutional Support
When it comes to acknowledging institutional support in your thesis, it's important to recognize the broader infrastructure that has facilitated your research. Your institution is not just a backdrop for your academic journey; it is a pivotal enabler of your scholarly pursuits. Expressing gratitude towards your university or college demonstrates an awareness of the collective effort that underpins individual success.
Begin by thanking the institution for specific resources or opportunities provided, such as access to libraries, laboratories, or funding programs. For instance:
- ABC University for the use of their state-of-the-art laboratories
- XYZ College for granting special research fellowships
- The 123 Institute for providing critical data sets
Remember to mention any significant contributions that have directly impacted the success of your research. This acknowledgment seeks to recognize the invaluable support provided by the institution, which often includes administrative staff, facilities management, and academic departments . It's a way to show gratitude and appreciation for the collective support that has been instrumental in reaching this pivotal point in your academic career.
Acknowledgement for Editorial Assistance
When it comes to editorial assistance, your acknowledgement should reflect the meticulous effort and valuable contributions of those who helped refine your thesis. Express your gratitude for their attention to detail and the significant improvement they brought to the quality of your work. Editorial experts not only enhance the clarity and coherence of your manuscript but also ensure adherence to the highest standards of academic integrity .
Remember to mention specific instances where their insights were particularly beneficial. For example:
- Their suggestions on rephrasing complex sentences
- Corrections made to ensure accurate referencing
- Guidance on maintaining a consistent tone throughout the document
By acknowledging the editorial support you received, you highlight the collaborative nature of academic work and the importance of clear communication. It's a gesture that underscores the value of their role in the journey of your scholarly publication .
Acknowledgement of Industry Collaboration
When acknowledging industry collaboration in your thesis, it's important to highlight the practical impact of such partnerships. Your innovative ideas and constructive feedback have enhanced the overall quality of our work. Thank you for being an invaluable partner in this endeavor. The collaboration with industry experts not only brings a wealth of knowledge but also ensures that your research remains relevant and applicable in a real-world context .
In expressing gratitude, consider the following points:
- The specific contributions and resources provided by the industry partner.
- The ways in which the collaboration has shaped your research.
- Any challenges that were overcome through the partnership.
Remember, a well-crafted acknowledgement can serve as a testament to the symbiotic relationship between academia and industry. It's a gesture of appreciation that underscores the mutual benefits of such collaborations. Ensure that your thanks are not just a formality, but a genuine recognition of the value added to your research.
Embarking on the journey of thesis writing can be daunting, but you don't have to do it alone. Our comprehensive Thesis Action Plan is designed to guide you through every step, ensuring a stress-free and successful completion. From practical examples of thesis acknowledgements to step-by-step instructions, we've got you covered. Don't let anxiety and sleepless nights take over. Visit our website now to claim your special offer and start your path to thesis mastery with confidence!
In crafting the perfect thesis acknowledgement, we must remember that it is more than a mere formality; it is a heartfelt expression of gratitude towards those who have contributed to our academic journey. The acknowledgement section is a unique space within our thesis to extend our sincerest thanks to mentors, family, peers, and institutions that have provided support, guidance, and resources. By following the tips and examples provided in this article, students can ensure that their acknowledgements are not only appropriately structured and sincere but also reflective of the collaborative spirit inherent in academic pursuits. As we conclude, let us acknowledge that the success of a thesis is not a solo achievement but the culmination of a collective effort, and it is this spirit of collaboration and support that the acknowledgement section should encapsulate.
Frequently Asked Questions
What is the purpose of a thesis acknowledgement.
The purpose of a thesis acknowledgement is to express gratitude to individuals, groups, or institutions that have contributed to the completion of your thesis. It's an opportunity to thank those who provided support, guidance, resources, or inspiration throughout your research and writing process.
How should I structure my thesis acknowledgement?
Your thesis acknowledgement should be structured with a polite opening, followed by specific thanks to each contributor. Start with those most closely associated with your research, such as advisors and committee members, then acknowledge other supporters such as peers, family, and funding bodies. Keep it concise and sincere.
Can I include personal acknowledgements in my thesis?
Yes, it is common to include personal acknowledgements in your thesis. You can express gratitude to family members, friends, or anyone else who provided personal support or encouragement. However, maintain a professional tone and keep the focus on how they contributed to your thesis journey.

- Rebels Blog
- Blog Articles
- Terms and Conditions
- Payment and Shipping Terms
- Privacy Policy
- Return Policy
© 2024 Research Rebels, All rights reserved.
Your cart is currently empty.

Acknowledgement in Research Paper – A Quick Guide [5 Examples]
The acknowledgement section in your research paper is where you thank those who have helped or supported you throughout your research and writing. It is a short section of 3-5 paragraphs or no more than 300 words you put on a page after the title page.
In this post, we are going to provide you with five examples of acknowlegdement section and a handful of best practices you can make your work look professional.

Saying thank you with style
How to write an acknowledgement: the complete guide for students, why should i include an acknowledgement in my research paper.
Acknowledging assistance and contributions from others can establish your integrity as a researcher. This will eventually make your work more credible.
What should be acknowledged about (aka thankful for)?
In your acknowledgement, you can show gratitude for those who provide you with resources in the following area:
- Technical help may include people who helped you by providing materials and supplies.
- Intellectual help includes academic advice and assistance.
- Mental help can be any kind of verbal support and encouragement.
- Financial support that is obviously related to monetary support
Who should be included in the acknowledgement of a research paper?
You can include everyone who helped you technically, intellectually, or financially (assistance with grants or monetary help) in the process of researching and writing your research paper. Except for your family and friends, you should always include the full names with the title of these individuals:
- Your profession, supervisor, or teacher
- Academic staff (e.g. lab assistant) of your school/college
- Your department, faculty, college, or school
- Classmates, teammates, co-workers, or colleague
- Friends and family members
You can start with your professor or the individuals who supported you the most throughout the research. And then you can continue by thanking your institution and then the reviewer who reviewed your paper. Then you can thank your friends and families and any other individual who helped.
What is the tone of the acknowledgement in a research paper?
You should write your acknowledgement in formal language with complete sentences. It is appropriate to write in the first person (‘I’ for a single author or ‘we’ for two or more).
Note that personal pronouns such as ‘I, my, me …’ are nearly always used in the acknowledgements only. For the rest of the research paper, such personal pronouns are generally avoided.
Writing an acknowledgement for research paper is one of the important parts of your project report. You need to thank everyone for helping you with your paper . Here are some examples of acknowledgement for your research paper.
Acknowledgement in Research Paper: Example 1
Acknowledgement in research paper: example 2, acknowledgement in research paper: example 3, acknowledgement in research paper: example 4, acknowledgement in research paper: example 5.
You can use these or try to create your own version for your project report. Also, you can use our auto acknowledgement generator tool to automatically generate acknowledgement for your project.
Where should I put the acknowledgement section?
The acknowledgements section should appear between your title page and your introduction in your research paper.
How long is an acknowledgement in a research paper?
The acknowledgement section (usualy inserted as a page) of your research paper should consist of 3-5 paragraphs or no more than 300 words you put on a page after the title page.
Should I use the full names of family members in an acknowledgement?
You do not necessarily need to use the full name for your family and friends (it would sound pretty awkward to use the full name of your parent or spouse right?), you should always include the full names with the title for all other individuals in your acknowledgement.
Can I use “first person” in an acknowledgement?
Yes. It is appropriate to write in the first person (‘I’ for a single author or ‘we’ for two or more).
What is an acknowledgement in academic writing?
An acknowledgement is a page is where you show appreciation to people who helped or supported you intellectually, mentally, or financially in your academic writing.
It should be no longer than one page.

More Definitions on Acknowledgement
- Acknowledgement vs Empathy What is the Difference Between Acknowledgement and Empathy?
- Acknowledgement vs Acceptance Does Acknowledgement Mean Acceptance? Lessons From History and the Bible
- Acknowledgement vs Agreement Is Acknowledgement the Same as Agreement?
- Acknowledgement Receipts vs Official Receipts What’s the Difference Between Acknowledgement Receipts and Official Receipts?
“Acknowledgement” vs “Acknowledgment”… …what the hack?

Both “acknowledgement” and “acknowledgment” are used in the English-speaking world. However, acknowledgement with the “e” in the middle is more commonly used. It is up to 24.5 times more popular in the top 5 English-speaking countries in the world.
Other Popular Acknowledgement Examples
For work or business Acknowledgement Receipt of Payment [4 Examples] Acknowledging Receipt of Documents: A Quick Guide with Examples Acknowledgement for Presentation [9 Examples] Acknowledgement for Job Offer [3 Examples] Acknowledgement for Business Plan [4 Examples] Acknowledgement for Work Immersion [5 Examples] Acknowledgement of Receipt of Appraisal [3 Examples] Acknowledegment of Debt [5 Examples] Resignation Acknowledgement for Employers [5 Examples]
Academic Acknowledgement for Research Paper [5 Examples] Acknowledgement for Internship Report [5 Examples] Acknowledgement for Thesis and Dissertation [15 Examples] Acknowledgement for Portfolio [5 Examples] Acknowledgement for Case Study [4 Examples] Acknowledgement for Academic Research Paper [5 Examples] Acknowledgement for College/School Assignment [5 Examples] Acknowledgemet to God in Reports [5 Examples]
Others Acknowledgement to Funeral Attendees [5 Examples] Funeral Acknowledgement Templates (for Newspapers and Websites) Common Website Disclaimers to Protect Your Online Business Notary Acknowledgement [5 Examples]
Acknowledgement Examples for School/College Projects
Most popular Acknowledgement For School/College Projects [7 Examples] Acknowledgement for English Project [5 Examples] Acknowledgement for Project Class 11 and 12 Acknowledgement for Project of Class 8, 9 and 10 By subjects Acknowledgement for Accounting Project [3 Examples] Acknowledgement for Business Studies Project [5 Examples] Acknowledgement for Chemistry Project [5 Examples] Acknowledgement for Computer Project [5 Examples] Acknowledgement for Economics Project [5 Examples] Acknowledgement for English Project [5 Examples] Acknowledgement for Geography Project [5 Examples] Acknowledgement for History Project [5 Examples] Acknowledgement for Maths Project for Students [5 Examples] Acknowledgement for Physics Project [5 Examples] Acknowledgement for Social Science Project [5 Examples] Others Acknowledgement for Group Project [5 Examples] Acknowledgement for Graduation Project [5 Examples] Acknowledgement for Disaster Management Project [3 Examples] Acknowledgement for Yoga Project [3 Samples]
How-to Guides on Academic Writing and Others
Most popular How to Write an Acknowledgement: The Complete Guide for Students How to Write an Acknowledgement for College Project? How to Write a Dedication Page for a Thesis or Dissertation? More on acknowledgements How to Write Acknowledgment for a Dissertation or a Thesis? Is Acknowledgement and Dedication the Same? Thesis or Dissertation How to Write a Master’s Thesis: The Ultimate Guide How to Write a Thesis Proposal? How to Write an Abstract for a Thesis? How to Write a Preface for a Thesis? Others How to Write an Introduction for a Research Paper? 7 Real Research Paper Examples to Get You Started How to Write Cover Letter for an Internship Program? How to Write an Internship Acceptance Letter? How to Write a Leave Application? For Schools and the Workplace How to Write a Resignation Letter?
Introduction to Academic Writing
By O.P. Jindal Global University Duration: 16-hour Cost: FREE Gain an in-depth understanding of reading and writing as essential skills to conduct robust and critical research for your writing.
Writing in English at University
By Lund University Duration: 24-hour Cost: FREE Learn how to structure your text and arguments, quote sources, and incorporate editing and proofreading in your academic writing.
Academic English: Writing Specialization
By the University of California, Irvine Duration: 6 months Cost: Free 7-day trial, USD39 per month The skills taught in this Specialization will empower you to succeed in any college-level course or professional field. You’ll learn to conduct rigorous academic research and to express your ideas clearly in an academic format. Share your Course Certificates in your LinkedIn profile, on printed resumes, CVs, or other documents.

Thank you, your samples really helped me.
This is great! Your samples really helped me in my research. Thank you and more power!
I’m so grateful that you make this kind of blog, I really need this for my research. Thank you so much… God bless you.
Thank you so much for the samples you have provided, it has helped me a lot.
Leave a Comment Cancel Reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.

- SpringerLink shop
Acknowledgments and References
Acknowledgments.
This usually follows the Discussion and Conclusions sections. Its purpose is to thank all of the people who helped with the research but did not qualify for authorship (check the target journal’s Instructions for Authors for authorship guidelines). Acknowledge anyone who provided intellectual assistance, technical help (including with writing and editing), or special equipment or materials.
TIP: The International Committee of Medical Journal Editors has detailed guidelines on who to list as an author and who to include in the Acknowledgments that are useful for scientists in all fields.
Some journals request that you use this section to provide information about funding by including specific grant numbers and titles. Check your target journal’s instruction for authors for specific instructions. If you need to include funding information, list the name(s) of the funding organization(s) in full, and identify which authors received funding for what.
As references have an important role in many parts of a manuscript, failure to sufficiently cite other work can reduce your chances of being published. Every statement of fact or description of previous findings requires a supporting reference.
TIP: Be sure to cite publications whose results disagree with yours. Not citing conflicting work will make readers wonder whether you are really familiar with the research literature. Citing conflicting work is also a chance to explain why you think your results are different.
It is also important to be concise. You need to meet all the above needs without overwhelming the reader with too many references—only the most relevant and recent articles need to be cited. There is no correct number of references for a manuscript, but be sure to check the journal’s guidelines to see whether it has limits on numbers of references.
TIP: Never cite a publication based on what you have read in a different publication (such as a review), or based only on the publication’s abstract. These may mislead you and readers. Read the publication itself before you cite it, and then check the accuracy of the citation again before submitting your manuscript.
You should reference other work to:
- Establish the origin of ideas
When you refer to an idea or theory, it is important to let your readers know which researcher(s) came up with the idea. By citing publications that have influenced your own work, you give credit to the authors and help others evaluate the importance of particular publications. Acknowledging others’ contributions is also an important ethical principle.
- Justify claims
In a scientific manuscript, all statements must be supported with evidence. This evidence can come from the results of the current research, common knowledge, or from previous publications. A citation after a claim makes it clear which previous study supports the claim.
- Provide a context for your work
By highlighting related works, citations help show how a manuscript fits into the bigger picture of scientific research. When readers understand what previous studies found and what puzzles or controversies your study relates to, they will better understand the meaning of your work.
- Show there is interest your field of research
Citations show that other researchers are performing work similar to your own. Having current citations will help journal editors see that there is a potential audience for your manuscript.
Back │ Next
- Discoveries
- Right Journal
- Journal Metrics
- Journal Fit
- Abbreviation
- In-Text Citations
- Bibliographies
- Writing an Article
- Peer Review Types
- Acknowledgements
- Withdrawing a Paper
- Form Letter
- ISO, ANSI, CFR
- Google Scholar
- Journal Manuscript Editing
- Research Manuscript Editing
Book Editing
- Manuscript Editing Services
Medical Editing
- Bioscience Editing
- Physical Science Editing
- PhD Thesis Editing Services
- PhD Editing
- Master’s Proofreading
- Bachelor’s Editing
- Dissertation Proofreading Services
- Best Dissertation Proofreaders
- Masters Dissertation Proofreading
- PhD Proofreaders
- Proofreading PhD Thesis Price
- Journal Article Editing
- Book Editing Service
- Editing and Proofreading Services
- Research Paper Editing
- Medical Manuscript Editing
- Academic Editing
- Social Sciences Editing
- Academic Proofreading
- PhD Theses Editing
- Dissertation Proofreading
- Proofreading Rates UK
- Medical Proofreading
- PhD Proofreading Services UK
- Academic Proofreading Services UK
Medical Editing Services
- Life Science Editing
- Biomedical Editing
- Environmental Science Editing
- Pharmaceutical Science Editing
- Economics Editing
- Psychology Editing
- Sociology Editing
- Archaeology Editing
- History Paper Editing
- Anthropology Editing
- Law Paper Editing
- Engineering Paper Editing
- Technical Paper Editing
- Philosophy Editing
- PhD Dissertation Proofreading
- Lektorat Englisch
- Akademisches Lektorat
- Lektorat Englisch Preise
- Wissenschaftliches Lektorat
- Lektorat Doktorarbeit
PhD Thesis Editing
- Thesis Proofreading Services
- PhD Thesis Proofreading
- Proofreading Thesis Cost
- Proofreading Thesis
- Thesis Editing Services
- Professional Thesis Editing
- Thesis Editing Cost
- Proofreading Dissertation
- Dissertation Proofreading Cost
- Dissertation Proofreader
- Correção de Artigos Científicos
- Correção de Trabalhos Academicos
- Serviços de Correção de Inglês
- Correção de Dissertação
- Correção de Textos Precos
- 定額 ネイティブチェック
- Copy Editing
- FREE Courses
- Revision en Ingles
- Revision de Textos en Ingles
- Revision de Tesis
- Revision Medica en Ingles
- Revision de Tesis Precio
- Revisão de Artigos Científicos
- Revisão de Trabalhos Academicos
- Serviços de Revisão de Inglês
- Revisão de Dissertação
- Revisão de Textos Precos
- Corrección de Textos en Ingles
- Corrección de Tesis
- Corrección de Tesis Precio
- Corrección Medica en Ingles
- Corrector ingles
Select Page
Acknowledgements Example for an Academic Research Paper
Posted by Rene Tetzner | Sep 1, 2021 | How To Get Published | 0 |

Acknowledgements Example for an Academic or Scientific Research Paper This example of acknowledgements for a research paper is designed to demonstrate how intellectual, financial and other research contributions should be formally acknowledged in academic and scientific writing. As brief acknowledgements for a research paper, the example gathers contributions of different kinds – intellectual assistance, financial support, image credits etc. – into a single Acknowledgements section. Do note, however, that the formats preferred by some scholarly journals require the separation of certain contributions such as financial support of research into their own sections.

Although authors often write acknowledgements hastily, the Acknowledgements section is an important part of a research paper. Acknowledging assistance and contributions establishes your integrity as a researcher as well as your connections and collaborations. It can also help your readers with their own research, affect the influence and impact of the researchers and other professionals you thank, and demonstrate the value and purpose of the agencies that fund your work. The contents of the example I have prepared here are appropriate for a research paper intended for publication in a peer-reviewed journal, but the author, the research project, the manuscript studied, the journal publishing the paper and all those to whom gratitude is extended are entirely fictional. They were created for the purpose of demonstrating the following key concerns when writing the acknowledgements for a formal research paper:

• Writing in the first person (‘I’ for a single author or ‘we’ for two or more) to offer concise but sincere acknowledgements of specific contributions to your research. • Maintaining formal language, complete sentences and a professional tone to give specific and thorough information about contributions and convey collegial gratitude. • Expressing respect and appreciation in an appropriate fashion for each and every contribution and avoiding artificial or excessive flattery. • Using the complete names and preferred name formats for individuals, funding agencies, libraries, businesses and other organisations. Here, for example, I posit that the library holding the relevant manuscript has indicated that the name of the collection (lengthy though it is) should not be abbreviated. • Acknowledging contributions to your research and paper in the order that best represents the nature and importance of those contributions. The assistance of the author’s mentor comes first here, for instance, whereas the language editor is acknowledged much further down the list. • Meeting the requirements for acknowledgements set by the journal or other publisher of the research paper. For the example below, the goal is to record all relevant contributions to the research and paper in a single brief Acknowledgements section of 500 words or less – a set of parameters that would suit the acknowledgement requirements or expectations of many academic and scientific journals and even fit into a footnote or endnote if necessary.

Example Acknowledgements for an Academic Research Paper This paper and the research behind it would not have been possible without the exceptional support of my supervisor, Lawrence Magister. His enthusiasm, knowledge and exacting attention to detail have been an inspiration and kept my work on track from my first encounter with the log books of British Naval Ships MS VII.2.77 to the final draft of this paper. Margaret Kempis and Matthew Brown, my colleagues at Western University, have also looked over my transcriptions and answered with unfailing patience numerous questions about the language and hands of British Naval Ships MS VII.2.77. Samantha McKenzie, head librarian of the Southern Region Central Collegiate Library Special Collections and Microfilms Department where British Naval Ships MS VII.2.77 currently resides, not only provided colour images of the manuscript overnight, but unexpectedly shared the invaluable information on the book that she has been gathering for almost twenty years. I am also grateful for the insightful comments offered by the anonymous peer reviewers at Books & Texts. The generosity and expertise of one and all have improved this study in innumerable ways and saved me from many errors; those that inevitably remain are entirely my own responsibility.
Studying British Naval Ships MS VII.2.77 has proved extremely costly and I am most thankful for the Western University Doctoral Fellowship that has provided financial support for the larger project from which this paper grew. A travel grant from the Literary Society of the Southern Region turned the hope of working in person with British Naval Ships MS VII.2.77 into a reality, and the generous offer of free accommodation from Ms McKay (Samantha McKenzie’s aunt) allowed me to continue my research with the book much longer than I could have hoped. The final design of the complicated transcription tables in Appendices I–III is the creative and technical work of Sam Stone at A+AcaSciTables.com, and the language and format of the paper have benefited enormously from the academic editing services of Veronica Perfect. Finally, it is with true pleasure that I acknowledge the contributions of my amazing partner, Kendric James, who has given up many a Friday evening and Sunday afternoon to read every version of this paper and the responses it has generated with a combination of compassion and criticism that only he could muster for what he fondly calls ‘my odd obsession with books about the sea.’
You might be interested in Services offered by Proof-Reading-Service.com
Journal editing.
Journal article editing services
PhD thesis editing services
Scientific Editing
Manuscript editing.
Manuscript editing services
Expert Editing
Expert editing for all papers
Research Editing
Research paper editing services
Professional book editing services
Related Posts

Choosing the Right Journal
September 10, 2021

Example of a Quantitative Research Paper
September 4, 2021

What Is a Good H-Index Required for an Academic Position?
September 3, 2021

Free Sample Letters for Withdrawing a Manuscript
August 31, 2021
Our Recent Posts

Our review ratings
- Examples of Research Paper Topics in Different Study Areas Score: 98%
- Dealing with Language Problems – Journal Editor’s Feedback Score: 95%
- Making Good Use of a Professional Proofreader Score: 92%
- How To Format Your Journal Paper Using Published Articles Score: 95%
- Journal Rejection as Inspiration for a New Perspective Score: 95%
Explore our Categories
- Abbreviation in Academic Writing (4)
- Career Advice for Academics (5)
- Dealing with Paper Rejection (11)
- Grammar in Academic Writing (5)
- Help with Peer Review (7)
- How To Get Published (146)
- Paper Writing Advice (17)
- Referencing & Bibliographies (16)
We use cookies on this site to enhance your experience
By clicking any link on this page you are giving your consent for us to set cookies.
A link to reset your password has been sent to your email.
Back to login
We need additional information from you. Please complete your profile first before placing your order.
Thank you. payment completed., you will receive an email from us to confirm your registration, please click the link in the email to activate your account., there was error during payment, orcid profile found in public registry, download history, what to include in your acknowledgments section.
- Charlesworth Author Services
- 02 June, 2018
- Academic Writing Skills
Most academic papers have many people who have helped in some way in the preparation of the written version or the research itself. This could be someone from a sponsoring institution, a funding body, other researchers, or even family, friends or colleagues who have helped in the preparation. These people need to be mentioned in the Acknowledgments section of the paper.
Acknowledgments section in different academic documents
The Acknowledgments section is present in both a paper and an academic thesis . For papers, the Acknowledgments section is usually presented at the back, whereas in a thesis, this section is located towards the front of the manuscript and is commonly placed somewhere between the abstract and Introduction . However, the exact location varies between each university , as each establishment possesses its own style guide for theses and student submissions. So, it is always worthwhile consulting your university’s academic style guide before writing a manuscript for undergraduate/postgraduate submission.
Acknowledgments section in theses
For academic theses, there is no right or wrong way to acknowledge people, and who you want to acknowledge is down to personal preference. However, the common types of people authors acknowledge in their academic theses include:
- Supervisor’s contributions
- Research group (especially if the thesis in question is a master’s and the work is helped along by a PhD student)
- Support staff (laboratory technicians, etc.)
- Any students who undertook side projects with them (e.g. final year undergraduates, summer students, master’s students)
- Administrative staff (there can be a lot of bureaucracy for thesis submissions)
- Referees that got them onto the course (postgraduate only)
- Funding bodies
- Any collaboration with industry and the people they worked with at said establishment(s)
Acknowledgments section in journal papers
Now, whilst university manuscripts can include any combination of the above (including all and none in some cases), academic publications in journals more commonly acknowledge the same kind of people/organizations, but again it is up to the author(s) what they feel should be acknowledged; not every piece of help needs to be acknowledged, just the most useful/prevalent help. Also, acknowledgments should be written in the first person .
Examples of whom and what should be acknowledged in a journal publication are listed below:
- Direct technical help (e.g. supply of animal subjects, cells, equipment setup, methods , statistics/data manipulation, samples, chemicals/reagents, analytical/spectroscopy techniques)
- Indirect assistance (topical and intellectual discussions about the research which can lead to generation of new ideas)
- Affiliated institutions
- Grant numbers
- Who received the funding (if not the author, e.g. a supervisor)
- Any associated fellowships
Whom to acknowledge - and whom not to acknowledge
- Other authors/contributors : It is not common practice for the lead paper writer (i.e. the person writing and publishing the manuscript) to acknowledge the other authors/direct contributors to the paper. Only those who are not recognized as authors may be thanked and acknowledged.
- Reviewers : Authors are also not allowed to thank reviewers personally, or those who inspire them but cannot directly receive their appreciation – although reviewers can be thanked if they are kept anonymous .
- Friends and family : Unlike university manuscripts, journal manuscripts should not include help and guidance from family and friends.
Other acknowledgments
- Titles and institutions : Titles such as Mr, Mrs, Miss, etc. are not commonly included, but honorary titles such as Dr, Professor, etc. are. The institutions of the acknowledged people are usually mentioned.
For example, the following would not be acceptable:
We dedicate this work to the deceased Prof. Bloggs.
However, the following would be acceptable:
We acknowledge Prof. Bloggs for discovering the secret of anonymity.
Additional pointers for writing the Acknowledgments section
- The tone of the section should be in an active voice.
- Do not use pronouns indicating possession (i.e. his, her, their, etc.).
- Terms associated with specific companies should be written out in full, e.g. Limited, Corporation, etc.
- If the results have been published elsewhere, then this should also be acknowledged.
- Any abbreviations should be expanded unless the abbreviation appears in the main body of the text.
Below are examples of the Acknowledgments sections taken from a couple of papers from Nature Communications :

Duan L., Hope J., Ong Q., Lou H-Y., Kim N., McCarthy C., Acero V., Lin M., Cui B., Understanding CRY2 interactions for optical control of intracellular signalling, Nature Communications, 2017, 8:547
Xu Q., Jensen K., Boltyanskiy R., Safarti R., Style R., Dufresne E., Direct measurement of strain-dependent solid surface stress, Nature Communications, 2017, 8:555
Many people think that the Acknowledgments section of a manuscript is a trivial and unimportant component. However, it constitutes a vital means to ensure that all affiliated support for the paper can be duly and transparently mentioned. By acknowledging people for their efforts and contributions, you demonstrate your integrity as an academic researcher. In addition, crediting other people for their help can also increase their presence in the academic world and possibly help to boost their career as well as your own.
Maximise your publication success with Charlesworth Author Services.
Charlesworth Author Services, a trusted brand supporting the world’s leading academic publishers, institutions and authors since 1928.
To know more about our services, visit: Our Services
Share with your colleagues
Related articles.

How to write an Introduction to an academic article
Charlesworth Author Services 17/08/2020 00:00:00
Writing an Abstract: Purpose and Tips
Related webinars.

Bitesize Webinar: How to write and structure your academic article for publication- Module 3: Understand the structure of an academic paper
Charlesworth Author Services 04/03/2021 00:00:00

Bitesize Webinar: How to write and structure your academic article for publication: Module 6: Choose great titles and write strong abstracts
Charlesworth Author Services 05/03/2021 00:00:00

Bitesize Webinar: How to write and structure your academic article for publication: Module 11: Know when your article is ready for submission

Bitesize Webinar: How to write and structure your academic article for publication - Module 14: Increase your chances for publication
Charlesworth Author Services 20/04/2021 00:00:00

Authorship of academic papers
Charlesworth Author Services 11/07/2017 00:00:00

Ethics in academic publishing: Understanding ‘gift’ authorships
Charlesworth Author Services 21/11/2019 00:00:00

Who gets the CRediT? Authorship issues and fairness in academic writing
Charlesworth Author Services 15/07/2019 00:00:00
.webp)
How to Write an Acknowledgement for a Research Paper

Hey guys, Phill Collins here! Today, I will teach you how to write an acknowledgment section in a research paper. Let’s do this!
Acknowledging contributions is a crucial aspect of creating a thorough research paper. It provides an opportunity to convey appreciation and acknowledge the support from individuals and institutions throughout your work. This piece will explore the intricacies of crafting acknowledgments for papers, offering valuable insights, practical advice, and sample acknowledgments. It aims to assist you in expressing gratitude to those who have played a substantial role in your research journey. As usual, I recommend those of you who struggle with your writings to pay for a research paper to save time and have a stress-free evening.
What Is Acknowledgement in a Research Paper
Acknowledgment in a research paper is a section dedicated to expressing gratitude and recognizing the individuals, institutions, or resources that have contributed to the completion of the research. This section is an opportunity for the author to appreciate the support, guidance, or assistance received during the research process. Acknowledgments go beyond the academic content of the paper and serve as a personal and professional gesture of recognition for those who played a significant role in the research endeavor.
In this section, researchers typically acknowledge the contributions of mentors, advisors, colleagues, or peers who provided valuable insights, feedback, or assistance in shaping the research project. Additionally, institutions, funding agencies, or organizations that supported the research financially or through resources may be acknowledged. The acknowledgment section reflects the collaborative and communal nature of academic work, highlighting the interconnected web of individuals and entities that contribute to the scholarly pursuit.
While there is no strict format for writing acknowledgments, it is important to balance professionalism and sincerity. Authors can use this space to express genuine gratitude, share personal reflections on the collaborative process, and convey the impact of the support received. The acknowledgment section adds a human touch to the paper, recognizing the collective effort that goes into the creation of academic knowledge.

The Role of an Acknowledgment in a Research Papers
The acknowledgment section in a paper plays a vital role in recognizing and appreciating the various contributors and influences that have shaped the research journey. Beyond the academic rigor captured in the main body of the paper, acknowledgments offer a space to express gratitude for the support and guidance received during the research process. This section often serves as a heartfelt acknowledgment of the collaborative effort to bring a research project to fruition.
Essentials of an Acknowledgement in Research Paper
An essential component of a complex paper, the acknowledgment section serves as a heartfelt expression of gratitude towards individuals and entities who have contributed significantly to the research process. In this section, authors typically recognize mentors, advisors, colleagues, and peers who provided valuable insights, guidance, or support. Additionally, institutions, funding sources, or organizations that played a role in the research project are acknowledged. The acknowledgment is a personal touch within the scholarly document, acknowledging the collaborative nature of academic work and underscoring the importance of communal support in the research journey. It adds a human element to the paper, recognizing the interconnected network of individuals and resources that contribute to the scholarly endeavor.
How to Write an Acknowledgement for Research Paper Using 8 Simple Tips
Keep in mind that acknowledgments offer a chance to express gratitude and acknowledge the valuable contributions of those who assisted you. Dedicate time to creating a genuine and thoughtful acknowledgment section that mirrors the collaborative and supportive nature of your research endeavor.
Sincerity and Authenticity
Write your acknowledgments with heartfelt gratitude, conveying genuine appreciation for the support and contributions you received throughout your research journey.
Specific Individuals and Institutions
Identify key figures and entities that played a substantial role in your research, including mentors, advisors, collaborators, and funding agencies. Acknowledge institutions that provided resources or facilities.
Conciseness and Focus
Keep your acknowledgments brief and to the point. Concentrate on highlighting the pivotal individuals and organizations that significantly influenced your research.
Formal Tone
Maintain a professional and formal tone throughout your acknowledgments. Remember that this section serves as a formal recognition of support, not a casual conversation.
Ethical Considerations
Adhere to ethical guidelines and norms when acknowledging individuals and organizations. Respect the privacy and confidentiality of those who may prefer not to be mentioned.
Personal Touches
If appropriate, include personal anecdotes or specific instances where individuals or organizations made a noteworthy impact on your research.
Tailor to Research Context
Consider the nature of your research and customize your acknowledgments accordingly. For instance, if your research is interdisciplinary, recognize experts from various fields who provide valuable insights.
Appreciation Beyond Formal Requirements
While an acknowledgement in research paper has to list individuals and institutions, extend your appreciation to others who indirectly supported you. This may include family, friends, or colleagues who provided emotional support during your research journey.
Example of Acknowledgement in Research Paper
.webp)
An Additional Example of Acknowledgement in Research Paper
.webp)
Final Words
In my opinion, acknowledgements in a research paper provide an avenue to convey appreciation and recognize the indispensable contributions of individuals and institutions that have bolstered your work. In this article, I did my best to offer tips and a sample acknowledgment to assist you in authentically expressing your gratitude. Keep in mind that acknowledgments are a chance to genuinely convey appreciation and attribute credit where it is rightfully due.

How to write acknowledgement in research paper?
Begin your acknowledgment section with a formal salutation, expressing gratitude to those who contributed to your research. Use a sincere and appreciative tone, mentioning specific individuals and institutions, and keep it concise.
What is the purpose of the acknowledgement section in a research paper?
The acknowledgment section serves to express gratitude and recognize the contributions of individuals and institutions who supported the research. It reflects the researcher's appreciation for guidance, resources, and collaboration during the project.
What should the acknowledgement section of a research paper include?
An acknowledgement for research paper includes thanks to advisors, committee members, funding agencies, collaborators, institutions providing resources, and anyone who significantly contributed. Maintain a formal tone, adhere to ethical considerations, and, if appropriate, add a personal touch or anecdotes.
How to acknowledge someone in a research paper?
Acknowledge individuals by mentioning their names, roles, and specific contributions. Follow a formal and respectful tone, adhering to ethical guidelines. If applicable, express personal appreciation and consider tailoring acknowledgments to the nature of the research and relationships involved.
Frequently asked questions
She was flawless! first time using a website like this, I've ordered article review and i totally adored it! grammar punctuation, content - everything was on point
This writer is my go to, because whenever I need someone who I can trust my task to - I hire Joy. She wrote almost every paper for me for the last 2 years
Term paper done up to a highest standard, no revisions, perfect communication. 10s across the board!!!!!!!
I send him instructions and that's it. my paper was done 10 hours later, no stupid questions, he nailed it.
Sometimes I wonder if Michael is secretly a professor because he literally knows everything. HE DID SO WELL THAT MY PROF SHOWED MY PAPER AS AN EXAMPLE. unbelievable, many thanks
You Might Also Like

New Posts to Your Inbox!
Stay in touch
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, automatically generate references for free.
- Knowledge Base
- Dissertation
- Thesis & Dissertation Acknowledgements | Tips & Examples
Thesis & Dissertation Acknowledgements | Tips & Examples
Published on 4 May 2022 by Tegan George . Revised on 4 November 2022.
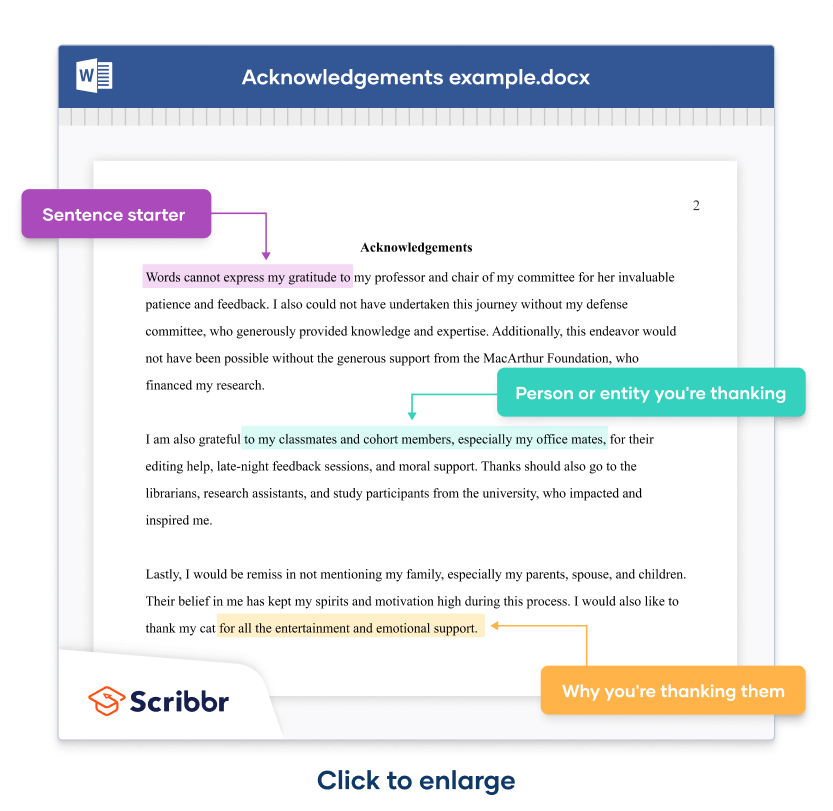
The acknowledgements section is your opportunity to thank those who have helped and supported you personally and professionally during your thesis or dissertation process.
Thesis or dissertation acknowledgements appear between your title page and abstract and should be no longer than one page.
In your acknowledgements, it’s okay to use a more informal style than is usually permitted in academic writing , as well as first-person pronouns . Acknowledgements are not considered part of the academic work itself, but rather your chance to write something more personal.
To get started, download our step-by-step template in the format of your choice below. We’ve also included sample sentence starters to help you construct your acknowledgments section from scratch.
Download Word doc Download Google doc
Instantly correct all language mistakes in your text
Be assured that you'll submit flawless writing. Upload your document to correct all your mistakes.
Table of contents
Who to thank in your acknowledgements, how to write acknowledgements, acknowledgements section example, acknowledgements dos and don’ts, frequently asked questions.
Generally, there are two main categories of acknowledgements: professional and personal .
A good first step is to check your university’s guidelines, as they may have rules or preferences about the order, phrasing, or layout of acknowledgements. Some institutions prefer that you keep your acknowledgements strictly professional.
Regardless, it’s usually a good idea to place professional acknowledgements first, followed by any personal ones. You can then proceed by ranking who you’d like to thank from most formal to least.
- Chairs, supervisors, or defence committees
- Funding bodies
- Other academics (e.g., colleagues or cohort members)
- Editors or proofreaders
- Librarians, research/laboratory assistants, or study participants
- Family, friends, or pets
Typically, it’s only necessary to mention people who directly supported you during your thesis or dissertation. However, if you feel that someone like a secondary school physics teacher was a great inspiration on the path to your current research, feel free to include them as well.
Professional acknowledgements
It is crucial to avoid overlooking anyone who helped you professionally as you completed your thesis or dissertation. As a rule of thumb, anyone who directly contributed to your research should be mentioned.
A few things to keep in mind include:
- Even if you feel your chair didn’t help you very much, you should still thank them first to avoid looking like you’re snubbing them.
- Be sure to follow academic conventions, using full names with titles where appropriate.
- If several members of a group or organisation assisted you, mention the collective name only.
- Remember the ethical considerations around anonymised data. If you wish to protect someone’s privacy, use only their first name or a generic identifier (such as ‘the interviewees’).
Personal acknowledgements
There is no need to mention every member of your family or friend group. However, if someone was particularly inspiring or supportive, you may wish to mention them specifically. Many people choose to thank parents, partners, children, friends, and even pets, but you can mention anyone who offered moral support or encouragement, or helped you in a tangible or intangible way.
Some students may wish to dedicate their dissertation to a deceased influential person in their personal life. In this case, it’s okay to mention them first, before any professional acknowledgements.
Prevent plagiarism, run a free check.
After you’ve compiled a list of who you’d like to thank, you can then sort your list into rank order. Separate everyone you listed into ‘major thanks’, ‘big thanks’, and ‘minor thanks’ categories.
- ‘Major thanks’ are given to people who your project would be impossible without. These are often predominantly professional acknowledgements, such as your advisor , chair, and committee, as well as any funders.
- ‘Big thanks’ are an in-between, for those who helped you along the way or helped you grow intellectually, such as classmates, peers, or librarians.
- ‘Minor thanks’ can be a catch-all for everyone else, especially those who offered moral support or encouragement. This can include personal acknowledgements, such as parents, partners, children, friends, or even pets.
How to phrase your acknowledgements
To avoid acknowledgements that sound repetitive or dull, consider changing up your phrasing. Here are some examples of common sentence starters you can use for each category.
Note that you do not need to write any sort of conclusion or summary at the end. You can simply end the acknowledgements with your last thank-you.
Here’s an example of how you can combine the different sentences to write your acknowledgements.
A simple construction consists of a sentence starter (in purple highlight ), followed by the person or entity mentioned (in green highlight ), followed by what you’re thanking them for (in yellow highlight .)
Acknowledgements
Words cannot express my gratitude to my professor and chair of my committee for her invaluable patience and feedback. I also could not have undertaken this journey without my defense committee, who generously provided knowledge and expertise. Additionally, this endeavor would not have been possible without the generous support from the MacArthur Foundation, who financed my research .
I am also grateful to my classmates and cohort members, especially my office mates, for their editing help, late-night feedback sessions, and moral support. Thanks should also go to the librarians, research assistants, and study participants from the university, who impacted and inspired me.
Lastly, I would be remiss in not mentioning my family, especially my parents, spouse, and children. Their belief in me has kept my spirits and motivation high during this process. I would also like to thank my cat for all the entertainment and emotional support.
- Write in first-person, professional language
- Thank your professional contacts first
- Include full names, titles, and roles of professional acknowledgements
- Include personal or intangible supporters, like friends, family, or even pets
- Mention funding bodies and what they funded
- Appropriately anonymise or group research participants or non-individual acknowledgments
Don’t:
- Use informal language or slang
- Go over one page in length
- Mention people who had only a peripheral or minor impact on your work
You may acknowledge God in your thesis or dissertation acknowledgements , but be sure to follow academic convention by also thanking the relevant members of academia, as well as family, colleagues, and friends who helped you.
Yes, it’s important to thank your supervisor(s) in the acknowledgements section of your thesis or dissertation .
Even if you feel your supervisor did not contribute greatly to the final product, you still should acknowledge them, if only for a very brief thank you. If you do not include your supervisor, it may be seen as a snub.
In the acknowledgements of your thesis or dissertation, you should first thank those who helped you academically or professionally, such as your supervisor, funders, and other academics.
Then you can include personal thanks to friends, family members, or anyone else who supported you during the process.
The acknowledgements are generally included at the very beginning of your thesis or dissertation, directly after the title page and before the abstract .
In a thesis or dissertation, the acknowledgements should usually be no longer than one page. There is no minimum length.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the ‘Cite this Scribbr article’ button to automatically add the citation to our free Reference Generator.
George, T. (2022, November 04). Thesis & Dissertation Acknowledgements | Tips & Examples. Scribbr. Retrieved 3 June 2024, from https://www.scribbr.co.uk/thesis-dissertation/acknowledgements/
Is this article helpful?
Tegan George
Other students also liked, dissertation title page, how to write an abstract | steps & examples, dissertation table of contents in word | instructions & examples.
- Utility Menu
FAS Research Administration Services
Guidelines on authorship and acknowledgement.
Disagreements may arise regarding who should be named as an author or contributor to intellectual work and the order in which individuals should be listed. These Guidelines are meant to serve as a set of standards that are shared by the academic community as a whole in order to help facilitate open communication through the adherence to common principles. These principles apply to all intellectual products, whether published or prepared for internal use or for broad dissemination. For a printable pdf of these guidelines, please click here .
Applicability
These Guidelines apply to all faculty, students postdoctoral researchers, and staff. Ownership of research data and materials resulting from Harvard University (“University”) research activities rests with the University (see Research Data Ownership Policy ).
Designing an ethical and transparent approach to authorship and publication of research, whether in a peer-reviewed journal or in an open access e-print or pre-print repository (e.g., arXiv, PsyArXiv), is a shared responsibility of all research team members but is primarily the responsibility of the Principal Investigator. The University recognizes that there are different standards across disciplines regarding authorship and the order in which authors are listed or acknowledged. Additionally, journals often specify their requirements in their guidance for authors and require attestations regarding individual authors intellectual contributions to the work. As a result, each laboratory, department, and/or school should engage in conversations regarding their own discipline-specific standards of authorship and, if needed, are encouraged to supplement the Guidelines herein with a description of these respective discipline-specific processes for deciding who should be an author and the order in which authors will be listed.
Note that these Guidelines are not intended for allegations related to research misconduct, defined as fabrication or falsification of data or plagiarism, which are subject to the Procedures for Responding to Allegations of Misconduct in Research and reviewed by the Committee on Professional Conduct (CPC).
Criteria for Authorship
FAS and SEAS recommend that authorship consider the following criteria [1] ;
- Each author is expected to have made substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data; or the creation of new software used in the work; or have drafted the work or substantively revised it; AND
- To have approved the submitted version (and any substantially modified version that involves the author’s contribution to the study); AND
- To have agreed both to be personally accountable for the author’s own contributions and to help ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated and resolved..
Some diversity exists across academic disciplines regarding acceptable standards for substantive contributions that would lead to attribution of authorship. Many journals have adopted discipline-specific standards. The University expects that researchers will act in accordance with accepted practice of the relevant research community. This Guidance is intended to allow for such variation of best practices within a specific discipline, while ensuring authorship is not inappropriately assigned.
Acknowledgment Versus Authorship
Financial sponsorship or donation of gift funding does not constitute criteria for authorship. Individuals who do not meet the recommended requirements for authorship, but have provided a valuable contribution to the work, should be acknowledged for their contributing role as appropriate to the publication. Authorship should not be conferred on those who have not made intellectual contributions to the work, or whose intellectual contributions are limited.
Implementation
Implementation of these Guidelines should include a commitment to collegiality, open communication, and expectation-setting throughout the research and scholarly process as well as the following considerations (see Authorship Best Practices Guidance (Addendum A) and Authorship Discussion Tool (Addendum B):
- Research groups should discuss authorship credit/criteria, presentation of joint work, and future direction of the research as early as practical, frequently during the course of their work, and as research team members begin or end their involvement. The Principal Investigator should initiate these discussions; however, any collaborator should feel free to raise questions or seek clarity throughout the course of the collaboration. Each lab or group may consider having a written document in place as guidance.
- All members of the research team are expected to adhere to good laboratory practices including maintaining an accurate laboratory notebook and annotating electronic files, as these practices will aide in identifying and clarifying individuals’ contributions to a project.
- Disposition of collaborative data and research materials should be mutually agreed upon among collaborators as early as practical and in accordance with any data-sharing and retention requirements.
- Laboratories, departments, centers, and programs supporting scholarly work should have available these Guidelines and a description of their discipline-specific processes of determining who should be an author, and the order in which authors are listed. These Guidelines should be included in the orientation of new research team members.
Authorship Disputes and Resolution
Disputes over authorship are best settled by the authors themselves; however, conflicts related to authorship may arise at any time during the research or scholarly process, resulting from differing perceptions of one’s contributions and resulting attribution of credit. It is expected that the resolution of disputes among collaborators will occur through open and collegial discourse, and mutual agreement is strongly encouraged. To facilitate this process, any prior decisions or discussions among authors, including verbal or written agreements between coauthors, should be reviewed and considered. These Guidelines and any documented customary practices in the relevant discipline should be applied, as appropriate. The authors should utilize the Authorship Discussion Tool (see Addendum B) in order to guide authors through a robust series of questions that can be jointly discussed by the authors in an effort to resolve the dispute. Extending an invitation to a mutually agreed-upon party outside the group who is familiar with publication norms in the field to informally serve as a neutral facilitator may ensure that all viewpoints are considered and objectively applied. It is expected that most disputes will be resolved collegially among collaborators. Should an authorship dispute arise that includes a question of the veracity of underlying data supporting a manuscript or the misappropriation of the work of others , consultation with the Research Integrity Officer may be helpful to support resolution.
If the dispute cannot be resolved at the local level, it is the responsibility of the FAS Department Chair or SEAS Area Chair or their designee to take the lead in effecting a resolution of the dispute, assuming that the FAS Department Chair or SEAS Area Chair is not a direct party to the dispute and does not have a conflict of interest.
If strenuous, good faith efforts to resolve the dispute utilizing the Authorship Discussion Tool (see Addendum B) are unsuccessful, one or more of the parties may then contact their FAS Divisional Dean(s)/SEAS Area Dean, sharing the completed Addendum B, which records the nature of the dispute and the efforts undertaken, and requesting further consideration. The FAS Divisional Dean(s)/SEAS Area Dean will review the submitted information and determine whether or not to appoint a committee to examine the case. As necessary, the Dean(s) will appoint a committee (and designate a committee chair), in consultation with the relevant FAS department(s)/SEAS area(s). The committee will consist of the following:
- A[n additional] faculty member from the field or fields relevant to the dispute
- Two faculty members from an adjacent field/department/area
FAS/SEAS Research Integrity Officer
- If the case involves a graduate student, an appropriate (non-student) representative from the Graduate School of Arts and Sciences
- If the case involves a postdoctoral researcher, an appropriate (non-postdoctoral) representative from the FAS Office of Postdoctoral Affairs
The committee will review the case and develop a recommendation to make to the authors. The committee chair will first inform the FAS Divisional Dean(s)/SEAS Area Dean of this recommendation and then inform the authors.
Related Resources
University Statement of Policy in Regard to Intellectual Property (IP Policy)
Graduate School of Arts and Sciences Office of Student Affairs
Harvard Ombuds Office
Committee on Publication Ethics (COPE) Authorship Resources
FAS/SEAS Procedures for Responding to Allegations of Research Misconduct
Harvard Medical School Authorship Guidelines
[1] As published in McNutt et al., Transparency in authors’ contributions and responsibilities to promote integrity in scientific publication. Proceedings of the National Academy of Sciences of the United States of America (PNAS) March 13, 2018 115 (11) 2557-2560. These criteria were adapted from the International Committee for Medical Journal Editors (ICMJE) framework for broader applicability across scientific fields.
Filter by Policy Area
- Faculty Research Policies
- Effort Policies
- Research Compliance Policies
- Research Finances Policies
- Proposal Submission Policies
Filter by Policy Type
- FAS Policies
- University Policies

The Plagiarism Checker Online For Your Academic Work
Start Plagiarism Check
Editing & Proofreading for Your Research Paper
Get it proofread now
Online Printing & Binding with Free Express Delivery
Configure binding now
- Academic essay overview
- The writing process
- Structuring academic essays
- Types of academic essays
- Academic writing overview
- Sentence structure
- Academic writing process
- Improving your academic writing
- Titles and headings
- APA style overview
- APA citation & referencing
- APA structure & sections
- Citation & referencing
- Structure and sections
- APA examples overview
- Commonly used citations
- Other examples
- British English vs. American English
- Chicago style overview
- Chicago citation & referencing
- Chicago structure & sections
- Chicago style examples
- Citing sources overview
- Citation format
- Citation examples
- College essay overview
- Application
- How to write a college essay
- Types of college essays
- Commonly confused words
- Definitions
- Dissertation overview
- Dissertation structure & sections
- Dissertation writing process
- Graduate school overview
- Application & admission
- Study abroad
- Master degree
- Harvard referencing overview
- Language rules overview
- Grammatical rules & structures
- Parts of speech
- Punctuation
- Methodology overview
- Analyzing data
- Experiments
- Observations
- Inductive vs. Deductive
- Qualitative vs. Quantitative
- Types of validity
- Types of reliability
- Sampling methods
- Theories & Concepts
- Types of research studies
- Types of variables
- MLA style overview
- MLA examples
- MLA citation & referencing
- MLA structure & sections
- Plagiarism overview
- Plagiarism checker
- Types of plagiarism
- Printing production overview
- Research bias overview
- Types of research bias
- Example sections
- Types of research papers
- Research process overview
- Problem statement
- Research proposal
- Research topic
- Statistics overview
- Levels of measurment
- Frequency distribution
- Measures of central tendency
- Measures of variability
- Hypothesis testing
- Parameters & test statistics
- Types of distributions
- Correlation
- Effect size
- Hypothesis testing assumptions
- Types of ANOVAs
- Types of chi-square
- Statistical data
- Statistical models
- Spelling mistakes
- Tips overview
- Academic writing tips
- Dissertation tips
- Sources tips
- Working with sources overview
- Evaluating sources
- Finding sources
- Including sources
- Types of sources
Your Step to Success
Plagiarism Check within 10min
Printing & Binding with 3D Live Preview
Acknowledgement for Thesis
How do you like this article cancel reply.
Save my name, email, and website in this browser for the next time I comment.

Inhaltsverzeichnis
- 1 Definition
- 3 Sample Acknowledgement for Thesis
- 4 Writing an Acknowledgement
- 5 Do’s and Don`ts
- 6 In a Nutshell
The acknowledgement for thesis is the section where you thank all people, institutions, and companies that helped you complete the project successfully. It is similar to a dedication, except for the fact that it is formal. Also, you don’t need to mention every single person who helped you with the research- just those who were most important to your research. For example, you don’t need to thank your boyfriend for making you dinner as you worked on the project.
What is an acknowledgement for thesis/for research?
In a research paper, an acknowledgement refers to the section at the beginning of your thesis formatting where you show your appreciation for the people who contributed to your project. It is up to you to determine who you are most grateful to for helping you with the research.
How do you write the thesis/dissertation acknowledgements?
Unlike acknowledgements in a book, a thesis or dissertation acknowledgement has to be formal. You should avoid showing strong emotions in the acknowledgement for thesis and should simply show your appreciation for their input.
Helpful: If you’re having trouble with phrasing your acknowledgement for thesis, transition words will help to enhance the flow of your writing.
Where do you put acknowledgements in a thesis?
The acknowledgement for thesis section is included right at the beginning in your thesis formatting . It is placed immediately after the table of contents, before the body of the thesis. The acknowledgement for thesis section is relatively brief.
Who should you thank in the acknowledgement for thesis section?
You should thank anyone who helped you with the project. Some people who are commonly included in the acknowledgement for thesis include your primary supervisor, other academic staff in your department, anyone who financed the research, and family and friends. Perhaps they helped you with your research proposal right at the beginning, or maybe they helped with editing your thesis. Regardless, you can recognise them in your acknowledgement for thesis. Although the first person is not used in academic research projects, you can use it in the acknowledgement for thesis section.
Can you use the first person in the acknowledgement?
Although the first person is not used in academic research projects, or most academic writing , you can use it in the acknowledgement for thesis section. The acknowledgement for thesis section is informal compared to the rest of your paper. However, you should still refrain from including strong emotional words in your acknowledgements.
Sample Acknowledgement for Thesis
The acknowledgement for thesis should be brief and should not include personal details. Here are some good sample thesis acknowledgements:
- I would like to express my gratitude to my primary supervisor, Michael Brown, who guided me throughout this project. I would also like to thank my friends and family who supported me and offered deep insight into the study.
- I wish to acknowledge the help provided by the technical and support staff in the Economics department of the University of London. I would also like to show my deep appreciation to my supervisors who helped me finalize my project.
Some common phrases you can use in the acknowledgement section of your project include:
- I wish to show my appreciation
- I would like to thank
- The assistance provided by Mr X was greatly appreciated
- I wish to extend my special thanks to
- I would like to thank the following people for helping me finalize the project
- Mr. X offered valuable data and statistics which I used in my project
Writing an Acknowledgement
The acknowledgement for thesis is typically written in the first person, singular or plural. You will have to avoid getting too personal as this section is not meant to be a dedication.You will typically start with the person who was most important in your study. This could be your professor, your supervisor, the staff, or even your family and friends. The last people you should acknowledge are those who played a smaller role in your research.
Acknowledgements don’t have a standard length. It could be just a few paragraphs, or it could run for a few pages. The length will primarily depend on the number of people you want to thank and acknowledge. It is advisable to keep the length of your acknowledgement for thesis as short as possible. If it gets very long, it could easily become meaningless. You can limit longer acknowledgements to the few people who had a significant impact on the study.
In the acknowledgement for thesis, you should try to be very specific. Mention the names of the people you are acknowledging, and not just their titles. Some people have trouble remembering the names of people and how they helped them with the research projects. If you have such tendencies, you can consider writing down the names of the people as they offer their help with the research. You should not miss out any party that played a major role in the study.
Do’s and Don`ts

- State the full names of the people you are acknowledging
- Use the first person singular or plural
- Write in formal language
- Identify the role played by each of the acknowledged parties
- List the funding organizations along with the parts of the projects that they funded

- Use personal or informal language
- Acknowledge every single person who had even the smallest impact on your research. For example, you don’t need to acknowledge the waitress at the restaurant where you used to relax after working on the project.
- Include people who qualified for authorship in the research project. A person will qualify for authorship if they make a substantial contribution to the project, if they draft and revise the work for intellectual content, and if they agree to be accountable for the content of the work.
In a Nutshell
- The acknowledgement for thesis section shows all the people who played an important role in the creation of the final paper, except for those who qualified for authorship.
- You should use the first person to show gratitude to the people who contributed to the project.
- You should show the specific role played by each person and party in the research project.
- The thesis acknowledgement should first show the people who contributed the most to the research and should end with the people who contributed the least.
- The thesis acknowledgement should be added after the table of contents in your research paper.
We use cookies on our website. Some of them are essential, while others help us to improve this website and your experience.
- External Media
Individual Privacy Preferences
Cookie Details Privacy Policy Imprint
Here you will find an overview of all cookies used. You can give your consent to whole categories or display further information and select certain cookies.
Accept all Save
Essential cookies enable basic functions and are necessary for the proper function of the website.
Show Cookie Information Hide Cookie Information
Statistics cookies collect information anonymously. This information helps us to understand how our visitors use our website.
Content from video platforms and social media platforms is blocked by default. If External Media cookies are accepted, access to those contents no longer requires manual consent.
Privacy Policy Imprint
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List

Acknowledgements are not just thank you notes: A qualitative analysis of acknowledgements content in scientific articles and reviews published in 2015
Adèle paul-hus.
École de bibliothéconomie et des sciences de l'information, Université de Montréal, Downtown Station, Montreal, Quebec, Canada
Nadine Desrochers
Associated data.
Restrictions apply to the availability of the acknowledgement data, which is used under license from Clarivate Analytics. Readers can contact Clarivate Analytics at the following URL: http://clarivate.com/scientific-and-academic-research/research-discovery/web-of-science/ . References for the acknowledgement excerpts used are available in S1 Table .
Acknowledgements in scientific articles can be described as miscellaneous, their content ranging from pre-formulated financial disclosure statements to personal testimonies of gratitude. To improve understanding of the context and various uses of expressions found in acknowledgements, this study analyses their content qualitatively. The most frequent noun phrases from a Web of Science acknowledgements corpus were analysed to generate 13 categories. When 3,754 acknowledgement sentences were manually coded into the categories, three distinct axes emerged: the contributions, the disclaimers, and the authorial voice. Acknowledgements constitute a space where authors can detail the division of labour within collaborators of a research project. Results also show the importance of disclaimers as part of the current scholarly communication apparatus, an aspect which was not highlighted by previous analyses and typologies of acknowledgements. Alongside formal disclaimers and acknowledgements of various contributions, there seems to remain a need for a more personal space where the authors can speak for themselves, in their own name, on matters they judge worth mentioning.
Introduction
The idea of using acknowledgements as a source for bibliometric indicators has been surrounding their study since the 1990s. In 1991, Cronin was already asking, “why are acknowledgement counts excluded from formal assessments of individual merit or influence, such as tenure review?” ([ 1 ]: p. 236). In 1995, Cronin and Weaver were encouraging the development of an Acknowledgement Index, based on the model of the Science Citation Index [ 2 ]. Almost two decades later, Costas and van Leeuwen [ 3 ] suggested that it was perhaps time “to employ this sort of tool to facilitate development of the so-called ‘influmetrics’” ([ 3 ]: p. 1659). For their part, Díaz-Faes and Bordons [ 4 ] highlighted that the inclusion of acknowledgement information in the Web of Science (WoS) was offering new avenues to study collaboration in science, going beyond traditional bibliometric indicators. McCain [ 5 ] went further and assessed the feasibility of a formal Personal Acknowledgements Index. And yet, despite decades of studies positioning acknowledgements alongside citations and authorship in what Cronin called the “reward triangle” [ 6 ], the consideration of acknowledgements as an indicator of scientific credit has not materialized and, at best, remains a proposal at the exploratory stage, or even simply a rhetorical idea (see [ 7 ] for a meta-synthesis of this literature).
At the same time, many studies have used funding-related indicators based on acknowledgement data (e.g. [ 8 – 11 ]). In fact, acknowledgement studies can no longer be separated from the financial aspect of scientific research. In 2008, WoS started to collect and index funding sources found in the acknowledgements of scientific papers. These new data were added by WoS in response to many funding bodies’ requirement to acknowledge the sources supporting research. Since then, large-scale acknowledgement data have been used as a bibliometric tool to follow the money trail of research and funding-related analyses have become a dominant trend in recent acknowledgement literature [ 7 ]. To this day, acknowledgements have been more closely related to funding indicators than to any other kind of scientific credit indicators.
The literature also underlines the elusive nature of acknowledgements, pointing to their form and tone, which have been described as sometimes flowery, personal, and even manipulative:
- Acknowledgements are permeated by hyperbole, effusiveness, overstatement, and exaggeration. ([ 12 ]: p. 64)
- Acknowledgements have been discussed as a form of patronage in scholarly communication, where the reality of the past may be purposefully glossed over and where the author could be looking toward the possibility of receiving future favours. ([ 13 ]: p. 4)
Furthermore, several studies mention the lack of standardization of acknowledgements as one important limitation hindering their analyses:
- The format of acknowledgement varies from field to field and from journal to journal. As noted, persons and institutional sources may be listed in the methods and materials section of an article or explicitly thanked in an acknowledgement section. ([ 14 ]: p. 506)
- Since there are no established formats for acknowledgements in papers, as there are for citations, expressions of gratitude vary greatly and sometimes it was difficult to identify the correct type of support, and even more difficult, the correct funding organization. ([ 15 ]: p. 238)
- The first source of simple error may arise through the misspelling of the names of funding bodies and potentially the names of grants and grant codes […]. A second difficulty will be that researchers will not correctly remember the funding bodies and grants that they used to support the research. ([ 16 ]: p. 368–369)
Acknowledgements may thus contain formally required statements of gratitude but have also been used as personal spaces of authorial expression, and as such, acknowledgement texts have been analysed as a genre per se. Several discourse and linguistic analyses have studied acknowledgements found in dissertations, theses, monographies, and research articles (e.g. [ 17 – 19 ]).
Acknowledgements analyses have also led to numerous typologies or classifications of the contributions acknowledged in scientific publications. In 1972, Mackintosh [ 20 ] proposed the first qualitative content analysis of acknowledgements based on a typology of the three main types of “services” acknowledged in scientific papers: facilities , access to data , and help of individuals . Twenty years later, McCain [ 14 ] offered a finer typology of acknowledgements, using five categories: access to research-related information , access to unpublished results and data , peer interactive communication , technical assistance , and manuscript preparation . The same year, Cronin introduced his first version of a six-part typology of acknowledgements ( paymaster , moral support , dogsbody , technical , prime mover , and trusted assessor ) which was created before encountering Mackintosh’s 1972 and McCain’s 1991 work [ 1 , 21 ]. Subsequent versions of this typology—developed with different collaborators through the years (namely McKenzie, Rubio and Weaver(-Wozniak))—include the peer interactive communication category borrowed from McCain [ 14 ] alongside moral support , access (to resources, materials and infrastructure), clerical support , technical support , and financial support [ 2 , 22 – 24 ]. Cronin’s model has since been adopted, adapted, and augmented in several studies (e.g. [ 25 – 30 ].
More recently, Giles and Councill [ 31 ] used natural language processing to extract named entities from more than 180,000 acknowledgements published in computer science research papers. In their content analysis, the most frequently acknowledged entities are classified into four categories: funding agencies , corporations , universities and individuals . Other studies have analysed the content of acknowledgements focusing on funding bodies and classifying them by sectors and subsectors (e.g. [ 10 , 32 – 35 ]).
Finally, linguistic studies have also used classifications of acknowledgements, focusing on the structure and patterns of dissertation acknowledgement texts (e.g. [ 18 , 36 – 40 ]) and on the socio-pragmatic construction of acknowledgements found in research articles and academic books [ 19 , 41 – 43 ].
Typologies and classifications aim to describe and categorize the content of acknowledgements in a synthetic manner. However, these taxonomies are based on small-scale samples of acknowledgements, the only exception being the work of Giles and Councill [ 31 ] which focused solely on named entities. More recently, a large-scale multidisciplinary analysis of acknowledgement texts was published by the authors and collaborators in PLOS One [ 44 ]. This analysis of acknowledgements from more than one million articles and reviews published in 2015, highlighted important variations in the practices of acknowledging. Focusing on the 214 most frequent noun phrases of that corpus, the study showed that acknowledgement practices truly do vary across disciplines. Noun phrases referring to technical support appeared more frequently in natural sciences while noun phrases related to peers (colleagues, editors and reviewers) were more frequent in earth and space, professional fields, and social sciences. Noun phrases referring to logistics and fieldwork-related tasks appeared prominently in biology. Pre-formulated statements used in the context of conflict of interest or responsibility disclosures were more frequently found in acknowledgements from clinical medicine, health, and psychology. However, this analysis also led to further questions concerning the interpretation of these noun phrases in their original context. Findings from this study showed that acknowledgements are not limited to credit attribution and that the numerous taxonomies and classifications found in the literature do not account for the current acknowledgement practices where pre-formulated statements of financial assistance and conflict of interest disclosures appear to be frequent [ 44 ]. Conclusions from this study raise further questions because these pre-formulated statements could have an influence on large-scale analyses that use automated linguistic methods, thus calling for a qualitative analysis of acknowledgements in the context of their use.
Objective and research questions
To improve understanding of the context and various uses of expressions found in acknowledgements, this study proposes to analyse their content qualitatively. More specifically, this study aims at answering the following research questions:
- In which contexts are specific expressions used?
- Do the contexts and meanings vary by discipline?
- What does a qualitative analysis reveal in terms of offering avenues for a more contextualized use of acknowledgements in large-scale studies?
Data and methods
Data for this study were retrieved from WoS’s Science Citation Index Expanded (SCI-E) and Social Sciences Citation Index (SSCI), which both include funding acknowledgement data. It bears repeating that acknowledgments are collected and indexed by WoS only if they include funding source information [ 45 ]. Access to WoS data in a relational database format was provided by the Observatoire des sciences et des technologies ( http://www.ost.uqam.ca ). The full text of acknowledgements from all 2015 articles and reviews indexed in the SCI-E and the SSCI were extracted. The original corpus includes a total of 1,009,411 acknowledgements for as many papers.
In a previous analysis, we identified the 214 most frequent noun phrases of that corpus of acknowledgement using natural language processing [ 44 ]. For the purpose the present qualitative analysis, these 214 noun phrases were reduced to single words (e.g. “technical assistance” was reduced to “technical” and “assistance”) and redundant words were excluded, for a final corpus of 154 single words. Each single word could therefore be found in context, no matter its proximity to other single words; this offered us the possibility to code various types of occurrences of each word, whether it was part of a noun phrase or not.
The coding was done in two steps. First, an initial codebook was established inductively by one researcher to classify each of the 154 words and revised by a second researcher. All words were then coded by both researchers and their work was reconciled through “negotiated agreement” ([ 46 ]: p. 305, see also [ 47 , 48 ]). Second, 20 words were selected from the corpus of 154 words by purposeful sampling, where cases for study are selected because “they offer useful manifestations of the phenomenon of interest” ([ 49 ]: p. 40). Selection of the words included in the final sample was based on the quantitative analysis findings [ 44 ], which highlighted the potential importance of pre-formulated statements such as “The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript” (ut 000367510900041). Special attention was given to the words frequently used in those statements (e.g. analysis, collection, design, preparation). Sampling decisions were also oriented towards potential polysemous words which could lead to different contextual meanings (e.g. “assistance”). The 20 words of the final sample were coded within the context of their original sentences, extracted from acknowledgements. Words were thus used as a seed to refer back to full acknowledgement sentences.
The coding process entails data reduction where the many meanings of a sentence must be reduced or summarized under one main category [ 50 ] in order to reflect a practice or a phenomenon on a humanly manageable scale. The principles of saturation and qualitative sampling, whereby the sample is “conceptually representative of the set of all possible units” ([ 51 ]: p. 84), ensures that the phenomenon is reflected in its full complexity. Therefore, acknowledgements were stratified by discipline to reflect potentially different disciplinary uses of a word. Coding was then performed on this sample of 20 words within their original acknowledgement contexts, using the sentence as the unit of analysis and adapting the codebook in an iterative manner as finer meanings emerged.
The final codebook is composed of 13 categories, presented in Table 1 . The coding was done by one researcher and guided by the question, “in which context is this word used?” One category was selected for each sentence coded, aiming at qualifying the context in which a word is used. Each word of the sample was coded in a minimum of 15 original sentences per discipline, for all 12 disciplines, resulting in a total of 3,754 sentences coded. Results are reported in “thick description” using sufficient descriptions and quotations to allow “thick interpretation”, which means connecting individual cases to the larger context without going into trivial details ([ 49 ]: p. 503).
The results of the coding process are summarized in Table 2 which presents, for each word of the sample, the percentage of all the occurrences attributed to a specific category. The analysis reveals the importance of three distinct axes: the contributions, the disclaimers, and the authorial voice. Moreover, disciplinary patterns bring another layer of analysis as divergent uses of the coded words emerge.
Words are presented in the table in descending order of their frequency in the corpus.
* “Other” regroups the following categories: Supervision and Management, Combination, and Vague or other.
Contributions
Acknowledgements constitute a space where authors can detail “who has done what” during the research process. Most often, authors use this space to thank colleagues that contributed to the research, as in the following example: “The authors thank Colleen Dalton and four anonymous reviewers for their helpful comments that improved the manuscript. We thank Fan-Chi Lin for providing FTAN measurements for comparison, and Anna Foster, Jiayi Xie and Goran Ekstrom for informative discussion.” (ut 000355321800013; earth and space). However, in some cases acknowledgements can also include contributorship statements from the authors in order to reflect the distribution of labour: “A.P., V.M. and V.P were involved in writing the manuscript. A.B.G and Y.A.K. were responsible for conception of the idea” (ut 000365808000014; clinical medicine).
The categories peer communication, investigation and analysis, materials and resources, and writing refer to specific types of contribution to research. These categories, taken together, represent half (50%) of the sample coded, confirming the importance of the contributions axis within the acknowledgements’ context. Moreover, some words are used most often to refer to specific categories of contribution, such as “access” which is used mainly in the category materials and resources (70% of the occurrences coded), “discussion” which is almost exclusively associated to the peer communication category (98% of the occurrences coded), and “assistance”, “experiment”, “help”, and “measurement”, which are all mainly associated to the category investigation and analysis (more than 60% of the occurrences coded).
Disclaimers
Acknowledgements are not necessarily thank-you notes or recognition of responsibility. Financial disclosure, conflict of interest, disclaimer, and ethics account for more than 40% of the sample coded. In fact, the categories financial disclosure and disclaimer are among the most frequent in the sample, accounting respectively for 22% and 18% of all occurrences coded. The words “analysis”, “collection”, “decision”, “design”, “interpretation”, “preparation”, and “writing”, which could all seemingly refer to types of contributions, were in fact used in the context of responsibility statements in a substantial share of the cases analysed. Moreover, the words “decision”, “design” and “interpretation” also are mostly found in those kinds of responsibility disclaimers (in respectively 65%, 55% and 61% of the occurrences coded for these specific words).
Non-responsibility statements of funding bodies are the most frequent disclaimers. The following example presents a typical statement: “The funding source had no role in the design of the study, the analysis and interpretation of the data or the writing of, nor the decision to publish the manuscript.” (ut 000352854700010). However, we found declarations of non-responsibility for other types of contributors regarding some part of a research project, as in the following sentence: “The data collectors have no responsibility over the analysis and interpretations presented in this study.” (ut 000349266800011). Furthermore, disclaimers are not always non-responsibility statements and can, on the contrary, disclose the specific responsibility of an organization, such as: “This study was funded by Xi'an Janssen Pharmaceutical Ltd (Beijing, People's Republic of China) who was responsible for study design and data collection, analysis, and interpretation.” (ut 000356594900001).
Contributions and disclaimers crossovers
In many cases, the disciplinary stratification provided a further level of analysis. The words “analysis”, “assistance”, and “code” present clear disciplinary patterns where the coding highlights the distinction between the two main contextual uses: the contributions axis and the disclaimers axis. For instance, the word “analysis” is used primarily in the sample to describe an investigation and analysis type of contribution: “We are grateful to Nahoko Adachi for her help in conducting the statistical analysis” (ut 000353959400005; psychology). However, for biomedical research, clinical medicine, and health, “analysis” is used mainly within the category disclaimer (example: “The funding agencies did not have any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript” [ut 000346498800018; clinical medicine]). Mathematics is a divergent discipline, where the dominant category for “analysis” is financial disclosure, as exemplified by the following sentence: “This work was supported by the International Max-Planck Research School, 'Analysis, Design and Optimization in Chemical and Bio-chemical Process Engineering', Otto-von-Guericke-Universitat Magdeburg” (ut 000362588800005; mathematics).
Similarly, the word “assistance” is generally used across disciplines to describe a contribution pertaining to the category investigation and analysis (example: “The authors thank S. Watmough and K. Finder for assistance with field sampling at Dorset, and A. McDonough for assistance with the classification of plant species” [ut 000347756900044; earth and space]), except in engineering and technology and in mathematics where “assistance” is used to disclose financial help (financial disclosure) in the majority of the cases examined, as in this sentence: “The financial assistance of the National Research Foundation (NRF grant: Unlocking the future- FA2007043000003) towards this research is hereby acknowledged” (ut 000350024900008; mathematics).
Two distinct contextual uses emerge for the word “code”: it is found most often within the disclaimers axis (financial disclosure category) in biology, biomedical research, chemistry, health, psychology and social sciences (example: “The research (project code: TSY-11-3820) was supported by the Research Fund of Erciyes University” [ut 000363704000011; biology]) while it is used to describe a specific contribution (investigation and analysis category) in the majority of the cases studied in earth and space, engineering and technology, mathematics, physics and professional fields (example: “We thank Prof. D. Karaboga and Dr. B. Basturk for providing their excellent ABC MATLAB codes to implement this research” [ut 000361400900022; earth and space]).
In the case of the word “review”, the coding process also highlights two dominant uses, varying with the discipline: in biology, biomedical research, earth and space, mathematics, physics, and in the professional fields, “review” is used primarily to describe some part of the peer communication process (peer communication category), as in the following example: “We would like to express our gratitude to the anonymous referee for his or her careful review and insightful comments, in particular, for pointing out a simple proof of Lemma 1.8.” (ut 000347714700003; engineering and technology). However, in clinical medicine, a different use is made of the word “review,” mainly to refer to the document per se (dissemination category), as in this example: “We are grateful to Dr. Mozzetta for critically reading the manuscript and all members of the lab for stimulating discussions during the preparation of this review” (ut 000352374400001; clinical medicine). For all the remaining disciplines (chemistry, health, psychology, and social sciences), both categories (peer communication and dissemination) appear frequently.
The word “data” also presents distinct disciplinary patterns in the sample coded. “Data” is used mainly within the contributions axis (materials and resources category) in biology, clinical medicine, earth and space, engineering and technology, and social sciences (example: “The authors thank Chesapeake Energy for providing access to the VSP data we used” [ut 000364362900035; earth and space]). Moreover, the word “data” refers to a task within the investigation and analysis category in an important share of the cases coded in chemistry, physics, professional fields, and psychology (example: “We thank all graduate research assistants who helped with data collection” [ut 000348882900009; psychology]). However, “data” is mainly found within the disclaimers axis in clinical medicine and health (disclaimer category) as in the following example: “The funding agencies had no role in the study design, data collection and analysis, the decision to publish or preparation of the manuscript” [ut 000345586900003; clinical medicine].
Authorial voice
Although details of contributions and various disclaimers represent a substantive share of their content, acknowledgements also constitute a space for personal testimony. Notwithstanding the expectations of funders and ethical considerations, acknowledgements remain the subjective presentation of researchers’ practices and of research contexts. The authors are the voice of the acknowledgements and as such, the word “author” is one of the most frequent with more than 339,000 occurrences in our dataset. Moreover, even when the word “author” is absent, the concept is not. In fact, the authorial voice cannot be reduced to a single category, because it pervades the acknowledgements whether the authors speak in the first or third persons:
- “ I would like to thank Iliana Flores, Amy Harrison, and Shannon Kahlden for their help with data collection.” (ut 000361977300090)
- “ We would also like to thank two anonymous reviewers for the contributions to this manuscript.” (ut 000364777400031)
- “Also, our thanks go to Mr Vit Hanousek who designed an original computer tool suitable for making all the above-discussed measurements.” (ut 000346267600010)
- “The authors declare that they have no competing interests.” (ut 000369908800022)
- “The authors wish to express their appreciation to the National Iranian Copper Industry Company (NICICO) for funding this work.” (ut 000344595900005)
- “Schuster is profoundly grateful to all the families who hosted her but especially Hasidullah, his wife, son and grandson who were unfailingly patient and kind with the strange cuckoo in their nest and to the Leverhulme Trust for funding her time in Afghanistan.” (ut 000350285300006)
- “This review is dedicated to the memory of my father who was a source of inspiration.” (ut 000349637500005)
Furthermore, as exemplified by the cases presented above, the varied nature of the testimonies found in acknowledgements underlines a need for a “free space” within research publications. Alongside formal disclaimers and acknowledgements of various contributions, authors seem to require a more personal space where they can speak for themselves, in their own name, on matters they judge worth mentioning.
Discussion and conclusion
In the last decades, acknowledgements have become a “constitutive element of academic writing” ([ 52 ]: p. 160). However, the acknowledgement section is not a mandatory part of a scientific article and its content could certainly be described as miscellaneous, ranging from pre-formulated financial disclosure statements to personal testimonies of gratitude. Moreover, acknowledgements’ content and practices have evolved over time, just as citations and authorship attribution practices have changed following the transformations that are affecting the whole reward system of science [ 53 ].
Typologies and classifications of acknowledgements have been a consistent topic in the acknowledgement literature [ 7 ]. Most of these typologies and classifications revolve around the contributions axis of acknowledgements, focusing on “who gets thanked for what” and “what types of contributions are acknowledged”. This qualitative analysis of acknowledgement content confirms the importance of the contributions axis: acknowledgements are indeed still a space where authors can detail the division of labour within all collaborators of a research project. Our findings also reveal the importance of disclaimers as part of the current scholarly communication apparatus, an aspect which was not highlighted by previous analyses and typologies.
It should be noted that our analysis was restricted to a corpus of single words, sampled from noun phrases identified by correspondence analysis [ 44 ]. Further research could now seek to recombine those single words into noun phrases that present variations in meaning around a common concept, such as “assistance” (e.g. “technical assistance” and “financial assistance”). Furthermore, our coding of acknowledgement sentences was done using mutually exclusive categories, an epistemological choice. Given the fact that sentences can perform more than one kind of action, another avenue would be to use open coding and place occurrences in non-exclusive, mutually complementary categories.
Our qualitative results show that caution should be used when working with acknowledgement data. Large-scale acknowledgement data are limited to funded research, given that in the two main bibliographic databases, Web of Science and Scopus, acknowledgements are collected with the intended objective of identifying funding sponsors and tracking funded research [ 54 , 55 ]. The indexation of acknowledgements are thus limited to acknowledgements that contain some kind of funding information; this could in turn induce a potential bias toward funding-related aspects within acknowledgements’ content [ 45 ]. This indexation bias could then, at least in part, explain the importance of funding disclosures in the dataset analysed here, but also elsewhere in large-scale studies.
Yet, our findings show that acknowledgements cannot be described as having one single and homogeneous purpose; they can include expected, if not imposed, acknowledgement of financial resources as well as infrastructure alongside very personal testimonies of gratitude, all at the same time, as the following excerpt exemplifies: “Data presented herein were obtained at the W. M. Keck Observatory, which is operated as a scientific partnership among the California Institute of Technology, the University of California, and the National Aeronautics and Space Administration. […]. The authors wish to extend special thanks to those of Hawaiian ancestry, on whose sacred mountain we are privileged to be guests. Without their generous hospitality, the observations would not have been possible” (ut 000363471600015). On rare occasions, personal matters discussed in the acknowledgements become the center of attention, such as when an author proposed to his girlfriend in the acknowledgement of a paper: “C.M.B. would specifically like to highlight the ongoing and unwavering support of Lorna O’Brien. Lorna, will you marry me?” [ 56 ]. This particular paper was covered by many news outlets and online media sites when it was published, ranking in the 20 th position of the Altmetrics Top100 ranking for the year 2015. Such a case highlights the potential unexpected effect an acknowledgement can have on the visibility of a paper.
Clearly delimited and dedicated spaces for funding information, conflict of interest disclosures and contributorship statements are already implemented in some scientific journals (e.g. PLOS One , The Lancet , Science ). Nonetheless, such examples are far from the norm at the moment. In light of our findings, if an effort of standardization of acknowledgements is to be made, acknowledgements should at least include three main sections: ethics of research (financial disclosure, conflict of interest and responsibility disclaimers), contributions made to research, and personal testimony. These three indexation fields would, in turn, allow large-scale analysis of acknowledgements without the equivocality that currently characterizes these texts, yet without narrowing the space left for the authorial voice. The question remains as to whether there is a real wish within the scientific community to delineate such acknowledgement sections; if not, acknowledgement data are likely destined to remain simple tracking devices for science funding, the contributions and the authorial voices lost in large-scale analyses of scientific credit.
Supporting information
References are presented in order of in-text appearance.
Acknowledgments
The authors would like to thank Vincent Larivière for his comments and the three anonymous reviewers for their insightful suggestions and careful reading of the manuscript. This research was supported by the Social Sciences and Humanities Research Council of Canada: Joseph-Armand Bombardier CGS Doctoral Scholarships (Paul-Hus) and, Insight Development [grant number 430-2014-0617] (Desrochers).
Funding Statement
APH was supported by the Social Sciences and Humanities Research Council of Canada ( http://www.sshrc-crsh.gc.ca/ ): Joseph-Armand Bombardier CGS Doctoral Scholarships. ND was supported the Social Sciences and Humanities Research Council of Canada ( http://www.sshrc-crsh.gc.ca/ ): Insight Development [grant number 430-2014-0617]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
Publisher's Note: The article involves the independent analysis of data from publications in PLOS ONE. PLOS ONE staff had no knowledge or involvement in the study design, funding, execution or manuscript preparation. The evaluation and editorial decision for this manuscript have been managed by an Academic Editor independent of PLOS ONE staff, per our standard editorial process. The findings and conclusions reported in this article are strictly those of the author(s).
Phone Numbers
Routine and emergency care.
Companion Animal Hospital in Ithaca, NY for cats, dogs, exotics, and wildlife
Equine and Nemo Farm Animal Hospitals in Ithaca, NY for horses and farm animals
Cornell Ruffian Equine Specialists, on Long Island for every horse
Ambulatory and Production Medicine for service on farms within 30 miles of Ithaca, NY
Animal Health Diagnostic Center New York State Veterinary Diagnostic Laboratory
General Information
Cornell University College of Veterinary Medicine Ithaca, New York 14853-6401

Baker Institute for Animal Health
Dedicated to the study of veterinary infectious diseases, immunology, cancer, reproduction, genomics and epigenomics, canine embryonic atlas, acknowledgements.
- Vicki N. Meyers-Wallen, VMD, PhD, Dipl. ACT, Baker Institute for Animal Health and Dept. of Biomedical Sciences, College of Veterinary Medicine, Cornell University, Ithaca, NY. Author of the Canine Embryonic Atlas, sample collection, determination of gestational age from P4 and LH data, determination of canine developmental stages (CfS 14 to >23), text descriptions, photomicrographs of histologic sections, comparative staging of canine embryo samples compared to published information on human and mouse embryos, and photographs of histologic sections.
- Linda Mizer, DVM, MS, PhD, Dept. of Biomedical Sciences, College of Veterinary Medicine, Cornell University, Ithaca, NY. Special acknowledgment for assistance in curating anatomical details and terminology.
- John C. Fyfe 1,2 , DVM, PhD, Brian Schutte 1,3 , PhD, Katharina Freiberger 1 , Shelby Hemker 1 , 1 Microbiology & Molecular Genetics, 2 Small Animal Clinical Sciences, and 3 Pediatrics & Development, Michigan State University, College of Veterinary Medicine, East Lansing, MI. Preparation of palate histologic sections, photography and annotation.
- Alexander J. Travis 1,2 , VMD, PhD, Chinatsu Mukai 1 , PhD, YeunHee Kim 1 , PhD, Jacque Harrington Nelson 1 , BS, 1 Baker Institute for Animal Health and 2 Dept. of Biomedical Sciences, College of Veterinary Medicine, Cornell University, Ithaca, NY. Sample collection, determination of gestational age from P4 and LH data for CfS 1-4, photomicrographs, and text descriptions.
- Lynne Anguish, MS, Baker Institute for Animal Health, College of Veterinary Medicine, Cornell University, Ithaca, NY. Assistance in sample collection, preservation, and photomicrographs of histologic sections.
- Julie A. Jordan, AS, Veterinary BioBank, and Mary Lou Norman, Clinical Programs, College of Veterinary Medicine, Cornell University, Ithaca, NY. Preparation of histologic sections.
- Aimee Decker, Nazrin Tingstrom, Anthony Adinolfi, Custom Development, Cornell Information Technologies, and Starr Todd, Baker Institute for Animal Health, College of Veterinary Medicine, Cornell University, Ithaca, NY. Website design and production.
- Shannon Kellogg, Julie Reynolds, Kevin Draiss, Jackie Wright, and Scott Soprano for animal husbandry. Mary Martin, DVM, and Cheryl Brown, LVT, Cornell Center for Animal Resources and Education for Veterinary Medical Care, College of Veterinary Medicine, Cornell University, Ithaca, NY.
- Cornelia Farnum, Professor Emerita, Dept. of Biomedical Sciences, College of Veterinary Medicine, Cornell University, Ithaca, NY. Special acknowledgment for helpful advice and encouragement.
- Canine embryonic samples were collected during studies of canine gonadal development that were supported in part by the National Institutes of Child Health and Human Development (R03HD077119, R01HD19393), the Collaborative Research Program, College of Veterinary Medicine (M-W), and the Baker Institute for Animal Health Pursuit of Excellence Fund (M-W), Cornell University, Ithaca, NY USA. The funders had no role in study design, data collection or website preparation or content.
The compilation and production of the Canine Atlas was made possible by support from the Baker Institute for Animal Health, College of Veterinary Medicine, Cornell University, Ithaca, NY, and in part, from the Herndon Canine Disease Research Fund.
Connect with the Baker Institute on Twitter and Facebook:
- Letter to the Editor
- Open access
- Published: 03 June 2024
Mapping the global research landscape on molecular mimicry: a visualization and bibliometric study
- Sa’ed H. Zyoud ORCID: orcid.org/0000-0002-7369-2058 1 , 2
Journal of Translational Medicine volume 22 , Article number: 531 ( 2024 ) Cite this article
Metrics details
To the Editor,
This letter represents my interest in participating in the recently released Journal of Translational Medicine Collection on Molecular Mimicry in Human diseases. A better understanding of many human diseases is possible, and new therapeutic approaches based on molecular mimicry have been developed. Damian first described “molecular mimicry” in 1964. This theory suggests that pathogenic microbes may escape the immune system by expressing antigens that are closely related to those found in human hosts [ 1 ]. Since then, growing data from experimental and epidemiological studies have supported the connection between autoimmune and infectious diseases. This suggests that molecular mimicry and the resulting cross-reactivity are very important [ 2 ]. A better understanding of molecular mimicry in this environment will greatly influence the diagnosis, prevention, and treatment of these diseases [ 3 ].
Numerous disciplines have employed bibliometric analysis to identify and emphasize the most significant countries, institutions, journals, and citations [ 4 , 5 ]. The application of mathematical and statistical techniques to quantitatively analyze and characterize published literature, provide evidence to support the formation of future research hotspots, master the discipline’s development trend, track the frontier of scientific research, improve the efficiency of scientific research, and propose research directions is known as bibliometrics [ 6 ]. It also summarizes the state of affairs and highlights hotspots in particular research fields. On the other hand, no bibliometric analysis of molecular mimicry has been performed. Therefore, the objective of this study was to conduct an in-depth review of the scientific advances in molecular mimicry. Consequently, the purpose of this bibliometric analysis was to investigate molecular mimicry research trends and to pinpoint potential future research hotspots. Furthermore, by offering references and concepts for further research on molecular mimicry pathogenicity, autoimmunity, and clinical applications, this study contributes significantly to the body of knowledge.
The current study uses the Scopus database, which is widely regarded among researchers for the purposes of high-quality bibliometric studies [ 7 ], although a significant number of databases are used for evaluative research at the global level [ 8 ]. The largest abstract and citation database of peer-reviewed research literature in the world, Scopus, is a trusted source for locating biomedical research, including MEDLINE documents. Because I am more interested in molecular mimicry as a novel idea in study than in related subjects, I used the terms “molecular mimicry” and its synonyms as my key words. The MeSH terms in PubMed and the findings of earlier studies on molecular mimicry were used to determine the relevant keywords [ 3 , 9 , 10 , 11 ]. The date of data mining was May 14, 2024. The main focus was on journal articles that used “molecular mimicry” to identify objects based on searches conducted in the fields of titles and abstracts.
To understand research trends, a quantitative analysis of the obtained articles was carried out. A comprehensive picture of global research production was obtained by analyzing important indicators such as publication rates, the distribution of articles in journals, and institutional and geographical origins. The VOSviewer software (version 1.6.20) was used to create visual maps based on the most common terms in the title of the article and the abstract, further clarifying research areas and emerging themes [ 12 ]. This visualization approach offers important new perspectives on current hot topics of research and suggests possible paths for molecular mimicry.
Our search strategy identified 3,391 articles on molecular mimicry published between 1965 and 2023. Most of the research articles were of original type (63.78%, n = 2117), followed by reviews (30.94%, n = 1027) and other types (7.44%, n = 247), such as letters and editorials. As shown in Fig. 1 , there was a strong positive correlation ( r = 0.876, p < 0.001) between the publication year and the number of articles on molecular mimicry. Initially (1965–1990), there were few publications (annual average of approximately 7). However, from 1991 onward, there was a significant increase in publications (annual average of approximately 97), reaching a peak of 182 articles in 2023.

Annual number of publications related to molecular mimicry from 1965 to 2023
A worldwide review of publications on molecular mimicry research indicates a concentration in the US. With 1186 articles (34.97%) among the 94 contributing nations, the USA took the lead, followed by the UK (316, 9.32%), Italy (310, 9.14%), and other countries (Table 1 ). The National Institutes of Health (NIH) published 73 articles (2.15%), followed by Tel Aviv University ( n = 56; 1.65%). American institutions dominate the field. King’s College London is another notable contributor ( n = 53; 1.56%). The Journal of Autoimmunity (70 articles, 2.06%), Journal of Immunology (70 articles, 2.06%), Frontiers in Immunology (52 articles, 1.53%), and Autoimmunity Reviews (50 articles, 1.47%) are the most active journals that publish research on molecular mimicry.
The most cited article by Wucherpfennig and Strominger [ 13 ], published in Cell in 1995, investigated the role of molecular mimicry in T-cell-mediated autoimmunity. This study tested 129 peptides on seven myelin basic protein-specific T-cell clones from multiple sclerosis patients. Although only one peptide was identified as a molecular mimic, seven viral peptides and one bacterial peptide activated three of the clones. This finding suggested that a single T-cell receptor can recognize distinct but structurally related peptides from multiple pathogens, potentially contributing to the development of autoimmunity.
Three main areas of molecular mimicry research were identified by a thematic analysis of the literature (Fig. 2 A):
Mapping of terms used in research on molecular mimicry. A : Research topics clustered by mapping the cooccurrences of terms for publications on molecular mimicry. Of the 59,752 terms, 305 terms occurred at least 50 times. B : Overlay visualization map of the time sequence of frequently used terms in molecular mimicry (1965–2023). The yellow terms represent the most recent research
This group investigates how the immune system can become confused by pathogen molecules that have structures similar to those of our own cells. Rheumatoid arthritis and Guillain–Barré syndrome are examples of autoimmune diseases that can arise when the immune system targets these ‘mimicked’ self-cells. The purpose of this research was to discover possible targets for treatment and elucidate the role that molecular mimicry plays in autoimmunity.
This group looks into the possibility of creating vaccines through molecular mimicry. Vaccines have the potential to induce a potent immune response by mimicking particular pathogen molecules without actually triggering the disease. These studies were aimed to develop safe and efficient mimicry-based vaccines as well as optimal mimicry targets.
This group addresses the difficulty of attaining accurate targeting in molecular mimicry research. Important aspects include creating computational tools to detect highly specific mimics and ensuring that these mimics do not cause unwanted immune responses. Here, research has attempted to improve targeting tactics and make use of computing power to speed up the detection of mimics. Researchers are looking for ways to improve the specificity and design therapies that target specific mimics involved in the disease.
To illustrate the distribution of keyword frequencies across publications, I classified keywords by publication year and assigned them the corresponding colors, as shown in Fig. 2 B. The blue keywords represent research areas emphasized in earlier studies (before 2012), while the yellow keywords highlight topics addressed in more recent publications (post 2012). Notably, keywords related to “targeting, mimicry discovery through specificity, and computational tools” emerged as a prominent theme after 2012, suggesting their growing importance for future research. On the contrary, research prior to 2012 appears to have focused more on “molecular mimicry and vaccine development” and “molecular mimicry’s function in autoimmune diseases”.
In conclusion, molecular mimicry has enormous potential for improving our understanding and ability to treat a wide range of human diseases. There has been a significant increase in publications on molecular mimicry since 1991, indicating a growing interest and activity in this area of research. After 2012, keyword analysis revealed a shift in research focus toward specificity and computational tools, indicating a growing emphasis on improving therapeutic and targeting approaches. In addition to their academic value, the findings of this study emphasize the importance of molecular mimicry in understanding disease mechanisms and developing therapeutic interventions, with implications for future research directions and practical applications.
Data availability
All the data generated or analyzed during this study are included in this published article. In addition, other datasets used during the current study are available from the author upon reasonable request ([email protected]).
Damian RT. Molecular mimicry: antigen sharing by parasite and host and its consequences. Am Nat. 1964;98(900):129–49.
Article Google Scholar
Rojas M, Restrepo-Jimenez P, Monsalve DM, Pacheco Y, Acosta-Ampudia Y, Ramirez-Santana C, Leung PSC, Ansari AA, Gershwin ME, Anaya JM. Molecular mimicry and autoimmunity. J Autoimmun. 2018;95:100–23.
Article CAS PubMed Google Scholar
Suliman BA. Potential clinical implications of molecular mimicry-induced autoimmunity. Immun Inflamm Dis. 2024;12(2):e1178.
Article CAS PubMed PubMed Central Google Scholar
Ellegaard O, Wallin JA. The bibliometric analysis of scholarly production: how great is the impact? Scientometrics 2015, 105(3):1809–31.
Ninkov A, Frank JR, Maggio LA. Bibliometrics: methods for studying academic publishing. Perspect Med Educ. 2022;11(3):173–6.
Article PubMed Google Scholar
Hou A, Yang L, Lü J, Yang L, Kuang H, Jiang H. A bibliometrics visualization analysis and hotspots prediction for natural product on osteoporosis research from 2000 to 2021. J Future Foods. 2022;2(4):326–37.
Baas J, Schotten M, Plume A, Côté G, Karimi R. Scopus as a curated, high-quality bibliometric data source for academic research in quantitative science studies. Quant Sci Stud. 2020;1(1):377–86.
AlRyalat SAS, Malkawi LW, Momani SM. Comparing Bibliometric Analysis using PubMed, Scopus, and web of Science Databases. J Vis Exp 2019(152).
Garg A, Kumari B, Singhal N, Kumar M. Using molecular-mimicry-inducing pathways of pathogens as novel drug targets. Drug Discov Today. 2019;24(9):1943–52.
Miraglia F, Colla E. Microbiome, Parkinson’s Disease and Molecular Mimicry. Cells 2019, 8(3).
Xiao S, Wang Y, Shan S, Zhou Z. The interplay between viral molecular mimicry and host chromatin dynamics. Nucleus. 2023;14(1):2216560.
Article PubMed PubMed Central Google Scholar
van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2010;84(2):523–38.
Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80(5):695–705.
Download references
Acknowledgements
The author thanks An-Najah National University for all its administrative assistance during the implementation of the project. The English language of some sentences in this manuscript was edited by American Journal Experts (AJE) AI for digital editing.
No support was received for conducting this study.
Author information
Authors and affiliations.
Department of Clinical and Community Pharmacy, College of Medicine and Health Sciences, An-Najah National University, Nablus, 44839, Palestine
Sa’ed H. Zyoud
Clinical Research Centre, An-Najah National University Hospital, Nablus, 44839, Palestine
You can also search for this author in PubMed Google Scholar
Contributions
S.Z., the sole author, read and approved the final manuscript.
Corresponding author
Correspondence to Sa’ed H. Zyoud .
Ethics declarations
Ethics approval and consent to participate.
Given that this was a bibliometric study without human involvement, there was no need for ethical approval.
Consent for publication
Not applicable.
Competing interests
The author declares that he has no competing interest.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Zyoud, S.H. Mapping the global research landscape on molecular mimicry: a visualization and bibliometric study. J Transl Med 22 , 531 (2024). https://doi.org/10.1186/s12967-024-05357-7
Download citation
Received : 18 May 2024
Accepted : 29 May 2024
Published : 03 June 2024
DOI : https://doi.org/10.1186/s12967-024-05357-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Journal of Translational Medicine
ISSN: 1479-5876
- Submission enquiries: Access here and click Contact Us
- General enquiries: [email protected]
Rosacea and autoimmune disease: a case-control study in the all of us database
- Research Letter
- Published: 25 May 2024
- Volume 316 , article number 252 , ( 2024 )
Cite this article

- Austin J. Piontkowski 1 ,
- Daniela Mikhaylov 1 ,
- Omar Alani 1 ,
- Jeremy Orloff 1 ,
- Camille Powers 1 ,
- Nicholas Gulati 1 &
- Benjamin Ungar 1
52 Accesses
Explore all metrics
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Data availability
The data that support these findings are publicly available to registered and verified researchers via the All of Us Researcher Workbench.
Egeberg A, Hansen RP, Gislason HG, Thyssen PJ (2016) Clustering of autoimmune diseases in patients with rosacea. J Am Acad Dermatol 74(4):667–72e1
Article PubMed Google Scholar
Rainer MB, Kang S, Chien LA (2017) Rosacea: Epidemiology, pathogenesis, and treatment. Dermato-Endocrinology 9(1):e1361574
Article PubMed PubMed Central Google Scholar
Damsky W, King AB (2017) JAK inhibitors in dermatology: the promise of a new drug class. J Am Acad Dermatol 76(4):736–744
Article CAS PubMed PubMed Central Google Scholar
Ren M, Yang X, Teng Y, Lu W, Ding Y, Tao X (2023) Successful Treatment of Granulomatous Rosacea by JAK Inhibitor Abrocitinib: A Case Report. Clinical, Cosmetic and Investigational Dermatology 16:3369-74
Sun YH, Man XY, Xuan XY, Huang CZ, Shen Y, Lao LM (2022) Tofacitinib for the treatment of erythematotelangiectatic and papulopustular rosacea: a retrospective case series. Dermatol Ther 35(11)
Download references
Acknowledgements
The All of Us Research Program is supported by the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549; 1 OT2 OD026554; 1 OT2 OD026557; 1 OT2 OD026556; 1 OT2 OD026550; 1 OT2 OD 026552; 1 OT2 OD026553; 1 OT2 OD026548; 1 OT2 OD026551; 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205; 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277; 3 OT2 OD025315; 1 OT2 OD025337; 1 OT2 OD025276. In addition, the All of Us Research Program would not be possible without the partnership of its participants.
The authors have not received any funding for this project.
Author information
Authors and affiliations.
Department of Dermatology, Icahn School of Medicine at Mount Sinai, 5 East 98th St, 5th Floor, New York, NY, 10029, USA
Austin J. Piontkowski, Daniela Mikhaylov, Omar Alani, Jeremy Orloff, Camille Powers, Nicholas Gulati & Benjamin Ungar
You can also search for this author in PubMed Google Scholar
Contributions
AP contributed to conceptualization, data curation, formal analysis, investigation, methodology, and writing – original draft. DM and OA contributed to writing – original draft. JO and CP contributed to writing – review & editing. NG contributed to writing – review & editing and supervision. BU contributed to conceptualization, project administration, methodology, supervision, and writing – review & editing. All authors reviewed the manuscript.
Corresponding author
Correspondence to Benjamin Ungar .
Ethics declarations
Conflict of interest.
BU is an employee of Mount Sinai and has received research funds (grants paid to the institution) from: Incyte, Rapt Therapeutics, and Pfizer. He is also a consultant for Arcutis Biotherapeutics, Bristol Myers Squibb, Castle Biosciences, Fresenius Kabi, Galderma, Janssen, Lilly, Pfizer, Primus Pharmaceuticals, Sanofi, and UCB. The other authors have no conflicts of interest.
IRB approval status
Reviewed and deemed exempt by Icahn School of Medicine at Mount Sinai Institutional Review Board; STUDY-22-00884. The data that support these findings are publicly available and de-identified.
Patient consent
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Reprints and permissions
About this article
Piontkowski, A.J., Mikhaylov, D., Alani, O. et al. Rosacea and autoimmune disease: a case-control study in the all of us database. Arch Dermatol Res 316 , 252 (2024). https://doi.org/10.1007/s00403-024-02993-3
Download citation
Received : 11 April 2024
Revised : 11 April 2024
Accepted : 26 April 2024
Published : 25 May 2024
DOI : https://doi.org/10.1007/s00403-024-02993-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Rheumatoid arthritis
- General dermatology
- Medical dermatology
- Clinical research
- Find a journal
- Publish with us
- Track your research
- Open access
- Published: 27 May 2024
Discovery of novel RNA viruses through analysis of fungi-associated next-generation sequencing data
- Xiang Lu 1 , 2 na1 ,
- Ziyuan Dai 3 na1 ,
- Jiaxin Xue 2 na1 ,
- Wang Li 4 ,
- Ping Ni 4 ,
- Juan Xu 4 ,
- Chenglin Zhou 4 &
- Wen Zhang 1 , 2 , 4
BMC Genomics volume 25 , Article number: 517 ( 2024 ) Cite this article
350 Accesses
1 Altmetric
Metrics details
Like all other species, fungi are susceptible to infection by viruses. The diversity of fungal viruses has been rapidly expanding in recent years due to the availability of advanced sequencing technologies. However, compared to other virome studies, the research on fungi-associated viruses remains limited.
In this study, we downloaded and analyzed over 200 public datasets from approximately 40 different Bioprojects to explore potential fungal-associated viral dark matter. A total of 12 novel viral sequences were identified, all of which are RNA viruses, with lengths ranging from 1,769 to 9,516 nucleotides. The amino acid sequence identity of all these viruses with any known virus is below 70%. Through phylogenetic analysis, these RNA viruses were classified into different orders or families, such as Mitoviridae , Benyviridae , Botourmiaviridae , Deltaflexiviridae , Mymonaviridae , Bunyavirales , and Partitiviridae . It is possible that these sequences represent new taxa at the level of family, genus, or species. Furthermore, a co-evolution analysis indicated that the evolutionary history of these viruses within their groups is largely driven by cross-species transmission events.
Conclusions
These findings are of significant importance for understanding the diversity, evolution, and relationships between genome structure and function of fungal viruses. However, further investigation is needed to study their interactions.
Peer Review reports
Introduction
Viruses are among the most abundant and diverse biological entities on Earth; they are ubiquitous in the natural environment but difficult to culture and detect [ 1 , 2 , 3 ]. In recent decades, the significant advancements in omics have transformed the field of virology and enabled researchers to detect potential viruses in a variety of environmental samples, helping us to expand the known diversity of viruses and explore the “dark matter” of viruses that may exist in vast quantities [ 4 ]. In most cases, the hosts of these newly discovered viruses exhibit only asymptomatic infections [ 5 , 6 ], and they even play an important role in maintaining the balance, stability, and sustainable development of the biosphere [ 7 ]. But some viruses may be involved in the emergence and development of animal or plant diseases. For example, the tobacco mosaic virus (TMV) causes poor growth in tobacco plants, while norovirus is known to cause diarrhea in mammals [ 8 , 9 ]. In the field of fungal research, viral infections have significantly reduced the yield of edible fungi, thereby attracting increasing attention to fungal diseases caused by viruses [ 10 ]. However, due to their apparent relevance to health [ 11 ], fungal-associated viruses have been understudied compared to viruses affecting humans, animals, or plants.
Mycoviruses (also known as fungal viruses) are widely distributed in various fungi and fungal-like organisms [ 12 ]. The first mycoviruses were discovered in the 1960s by Hollings M in the basidiomycete Agaricus bisporus , an edible cultivated mushroom [ 13 ]. Shortly thereafter, Ellis LF et al. reported mycoviruses in the ascomycete Penicillium stoloniferum , confirming that viral dsRNA is responsible for interferon stimulation in mammals [ 13 , 14 , 15 ]. In recent years, the diversity of known mycoviruses has rapidly increased with the development and widespread application of sequencing technologies [ 16 , 17 , 18 , 19 , 20 ]. According to the classification principles of the International Committee for the Taxonomy of Viruses (ICTV), mycoviruses are currently classified into 24 taxa, consisting of 23 families and 1 genus ( Botybirnavirus ) [ 21 ]. Most mycoviruses belong to double-stranded (ds) RNA viruses, such as families Totiviridae , Partitiviridae , Reoviridae , Chrysoviridae , Megabirnaviridae , Quadriviridae , and genus Botybirnavirus , or positive-sense single-stranded (+ ss) RNA viruses, such as families Alphaflexiviridae , Gammaflexiviridae , Barnaviridae , Hypoviridae , Endornaviridae , Metaviridae and Pseudoviridae . However, negative-sense single-stranded (-ss) RNA viruses (family Mymonaviridae ) and single-stranded (ss) DNA viruses (family Genomoviridae ) have also been described [ 22 ]. The taxonomy of mycoviruses is continually refined as novel mycoviruses that cannot be classified into any established taxon are identified. While the vast majority of fungi-infecting viruses do not show infection characteristics and have no significant impact on their hosts, some mycoviruses have inhibitory effects on the phenotype of the host, leading to hypovirulence in phytopathogenic fungi [ 23 ]. The use of environmentally friendly, low-virulence-related mycoviruses such as Chryphonectria hypovirus 1 (CHV-1) for biological control has been considered a viable alternative to chemical fungicides [ 24 ]. With the deepening of research, an increasing number of mycoviruses that can cause fungal phenotypic changes have been identified [ 3 , 23 , 25 ]. Therefore, understanding the distribution of these viruses and their effects on hosts will allow us to determine whether their infections can be prevented and treated.
To explore the viral dark matter hidden within fungi, this study collected over 200 available fungal-associated libraries from approximately 40 Bioprojects in the Sequence Read Archive (SRA) database, uncovering novel RNA viruses within them. We further elucidated the genetic relationships between known viruses and these newfound ones, thereby expanding our understanding of fungal-associated viruses and providing assistance to viral taxonomy.
Materials and methods
Genome assembly.
To discover novel fungal-associated viruses, we downloaded 236 available libraries from the SRA database, corresponding to 32 fungal species (Supplementary Table 1). Pfastq-dump v0.1.6 ( https://github.com/inutano/pfastq-dump ) was used to convert SRA format files to fastq format files. Subsequently, Bowtie2 v2.4.5 [ 26 ] was employed to remove host sequences. Primer sequences of raw reads underwent trimming using Trim Galore v0.6.5 ( https://www.bioinformatics.babraham.ac.uk/projects/trim_galore ), and the resulting files underwent quality control with the options ‘–phred33 –length 20 –stringency 3 –fastqc’. Duplicated reads were marked using PRINSEQ-lite v0.20.4 (-derep 1). All SRA datasets were then assembled in-house pipeline. Paired-end reads were assembled using SPAdes v3.15.5 [ 27 ] with the option ‘-meta’, while single-end reads were assembled with MEGAHIT v1.2.9 [ 28 ], both using default parameters. The results were then imported into Geneious Prime v2022.0.1 ( https://www.geneious.com ) for sorting and manual confirmation. To reduce false negatives during sequence assembly, further semi-automatic assembly of unmapped contigs and singlets with a sequence length < 500 nt was performed. Contigs with a sequence length > 1,500 nt after reassembly were retained. Individual contigs were then used as references for mapping to the raw data using the Low Sensitivity/Fastest parameter in Geneious Prime. In addition, mixed assembly was performed using MEGAHIT in combination with BWA v0.7.17 [ 29 ] to search for unused reads that might correspond to low-abundance contigs.
Searching for novel viruses in fungal libraries
We identified novel viral sequences present in fungal libraries through a series of steps. To start, we established a local viral database, consisting of the non-redundant protein (nr) database downloaded in August 2023, along with IMG/VR v3 [ 30 ], for screening assembled contigs. The contigs labeled as “viruses” and exhibiting less than 70% amino acid (aa) sequence identity with the best match in the database were imported into Geneious Prime for manual mapping. Putative open reading frames (ORFs) were predicted by Geneious Prime using built-in parameters (Minimum size: 100) and were subsequently verified by comparison to related viruses. The annotations of these ORFs were based on comparisons to the Conserved Domain Database (CDD). The sequences after manual examination were subjected to genome clustering using MMseqs2 (-k 0 -e 0.001 –min-seq-id 0.95 -c 0.9 –cluster-mode 0) [ 31 ]. After excluding viruses with high aa sequence identity (> 70%) to known viruses, a dataset containing a total of 12 RNA viral sequences was obtained. The non-redundant fungal virus dataset was compared against the local database using the BLASTx program built in DIAMOND v2.0.15 [ 32 ], and significant sequences with a cut-off E-value of < 10 –5 were selected. The coverage of each sequence in all libraries was calculated using the pileup tool in BBMap. Taxonomic identification was conducted using TaxonKit [ 33 ] software, along with the rma2info program integrated into MEGAN6 [ 34 ]. The RNA secondary structure prediction of the novel viruses was conducted using RNA Folding Form V2.3 ( http://www.unafold.org/mfold/applications/rna-folding-form-v2.php ).
Phylogenetic analysis
To infer phylogenetic relationships, nucleotide and their encoded protein sequences of reference strains belonging to different groups of corresponding viruses were downloaded from the NCBI GenBank database, along with sequences of proposed species pending ratification. Related sequences were aligned using the alignment program within the CLC Genomics Workbench 10.0, and the resulting alignment was further optimized using MUSCLE in MEGA-X [ 35 ]. Sites containing more than 50% gaps were temporarily removed from the alignments. Maximum-likelihood (ML) trees were then constructed using IQ-TREE v1.6.12 [ 36 ]. All phylogenetic trees were created using IQ-TREE with 1,000 bootstrap replicates (-bb 1000) and the ModelFinder function (-m MFP). Interactive Tree Of Life (iTOL) was used for visualizing and editing phylogenetic trees [ 37 ]. Colorcoded distance matrix analysis between novel viruses and other known viruses were performed with Sequence Demarcation Tool v1.2 [ 38 ].
To illustrate cross-species transmission and co-divergence between viruses and their hosts across different virus groups, we reconciled the co-phylogenetic relationships between these viruses and their hosts. The evolutionary tree and topologies of the hosts involved in this study were obtained from the TimeTree [ 39 ] website by inputting their Latin names. The viruses in the phylogenetic tree for which the host cannot be recognized through published literature or information provided by the authors are disregarded. The co-phylogenetic plots (or ‘tanglegram’) generated using the R package phytools [ 40 ] visually represent the correspondence between host and virus trees, with lines connecting hosts and their respective viruses. The event-based program eMPRess [ 41 ] was employed to determine whether the pairs of virus groups and their hosts undergo coevolution. This tool reconciles pairs of phylogenetic trees according to the Duplication-Transfer-Loss (DTL) model [ 42 ], employing a maximum parsimony formulation to calculate the cost of each coevolution event. The cost of duplication, host-jumping (transfer), and extinction (loss) event types were set to 1.0, while host-virus co-divergence was set to zero, as it was considered the null event.
Data availability
The data reported in this paper have been deposited in the GenBase in National Genomics Data Center [ 43 ], Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, under accession numbers C_AA066339.1-C_AA066350.1 that are publicly accessible at https://ngdc.cncb.ac.cn/genbase . Please refer to Table 1 for details.
Twelve novel RNA viruses associated with fungi
We investigated fungi-associated novel viruses by mining publicly available metagenomic and transcriptomic fungal datasets. In total, we collected 236 datasets, which were categorized into four fungal phyla: Ascomycota (159), Basidiomycota (47), Chytridiomycota (15), and Zoopagomycota (15). These phyla corresponded to 20, 8, 2, and 2 different fungal genera, respectively (Supplementary Table 1). A total of 12 sequences containing complete coding DNA sequences (CDS) for RNA-dependent RNA polymerase (RdRp) have been identified, ranging in length from 1,769 nt to 9,516 nt. All of these sequences have less than 70% aa identity with RdRp sequences from any currently known virus (ranging from 32.97% to 60.43%), potentially representing novel families, genera, or species (Table 1 ). Some of the identified sequences were shorter than the reference genomes of RNA viruses, suggesting that these viral sequences represented partial sequences of viral genomes. To exclude the possibility of transient viral infections in hosts or de novo assembly artefacts in co-infection detection, we extracted the nucleotide sequences of the coding regions of these 12 sequences and mapped them to all collected libraries to compute coverage (Supplementary Table 2). The results revealed varying degrees of read matches for these viral genomes across different libraries, spanning different fungal species. Although we only analyzed sequences longer than 1,500 nt, it is worth noting that we also discovered other viral reads in many libraries. However, we were unable to assemble them into sufficiently long contigs, possibly due to library construction strategies or sequencing depth. In any case, this preliminary finding reveals a greater diversity of fungal-associated viruses than previously considered.
Positive-sense single-stranded RNA viruses
(i) mitoviridae.
Members of the family Mitoviridae (order Cryppavirales ) are monopartite, linear, positive-sense ( +) single-stranded (ss) RNA viruses with genome size of approximately 2.5–2.9 kb [ 44 ], carrying a single long open reading frame (ORF) which encodes a putative RdRp. Mitoviruses have no true virions and no structural proteins, virus genome is transmitted horizontally through mating or vertically from mother to daughter cells [ 45 ]. They use mitochondria as their sites of replication and have typical 5' and 3' untranslated regions (UTRs) of varying sizes, which are responsible for viral translation and replicase recognition [ 46 ]. According to the taxonomic principles of ICTV, the viruses belonging to the family Mitoviridae are divided into four genera, namely Duamitovirus , Kvaramitovirus , Triamitovirus and Unuamitovirus . In this study, two novel viruses belonging to the family Mitoviridae were identified in the same library (SRR12744489; Species: Thielaviopsis ethacetica ), named Thielaviopsis ethacetica mitovirus 1 (TeMV01) and Thielaviopsis ethacetica mitovirus 2 (TeMV02), respectively (Fig. 1 A). The genome sequence of TeMV01 spans 2,689 nucleotides in length with a GC content of 32.2%. Its 5' and 3' UTRs comprise 406 nt and 36 nt, respectively. Similarly, the genome sequence of TeMV02 extends 3,087 nucleotides in length with a GC content of 32.6%. Its 5' and 3' UTRs consist of 553 and 272 nt, respectively. The 5' and 3' ends of both genomes are predicted to have typical stem-loop structures (Fig. 1 B). In order to determine the evolutionary relationship between these two mitoviruses and other known mitoviruses, phylogenetic analysis based on RdRp showed that viral strains were divided into 2 genetic lineages in the genera Duamitovirus and Unuamitovirus (Fig. 1 C). In the genus Unuamitovirus , TeMV01 was clustered with Ophiostoma mitovirus 4, exhibiting the highest aa identity of 51.47%, while in the genus Duamitovirus , TeMV02 was clustered with a strain isolated from Plasmopara viticola , showing the highest aa identity of 42.82%. According to the guidelines from the ICTV regarding the taxonomy of the family Mitoviridae , a species demarcation cutoff of < 70% aa sequence identity is established [ 47 ]. Drawing on this recommendation and phylogenetic inferences, these two viral strains could be presumed to be novel viral species [ 48 ].
Identification of novel positive-sense single-stranded RNA viruses in fungal sequencing libraries. A Genome organization of two novel mitoviruses; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. B Predicted RNA secondary structures of the 5'- and 3'-terminal regions. C ML phylogenetic tree of members of the family Mitoviridae . The best-fit model (LG + F + R6) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified viruses represented in red font. D The genome organization of GtBeV is depicted at the top; in the middle is the ML phylogenetic tree of members of the family Benyviridae . The best-fit model (VT + F + R5) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font. At the bottom is the distance matrix analysis of GeBeV identified in Gaeumannomyces tritici . Pairwise sequence comparison produced with the RdRp amino acid sequences within the ML tree. E The genome organization of CrBV is depicted at the top; in the middle is the ML phylogenetic tree of members of the family Botourmiaviridae . The best-fit model (VT + F + R5) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font. At the bottom is the distance matrix analysis of CrBV identified in Clonostachys rosea . Pairwise sequence comparison produced with the RdRp amino acid sequences within the ML tree
(ii) Benyviridae
The family Benyviridae is comprised of multipartite plant viruses that are rod-shaped, approximately 85–390 nm in length and 20 nm in diameter. Within this family, there is a single genus, Benyvirus [ 49 ]. It is reported that one species within this genus,Beet necrotic yellow vein virus, can cause widespread and highly destructive soil-borne ‘rhizomania’ disease of sugar beet [ 50 ]. A full-length RNA1 sequence related to Benyviridae has been detected from Gaeumannomyces tritici (ERR3486062), with a length of 6,479 nt. It possesses a poly(A) tail at the 3' end and is temporarily designated as Gaeumannomyces tritici benyvirus (GtBeV). BLASTx results indicate a 34.68% aa sequence identity with the best match found (Fig. 1 D). The non-structural polyprotein CDS of RNA1 encodes a large replication-associated protein of 1,688 amino acids with a molecular mass of 190 kDa. Four domains were predicted in this polyprotein corresponding to representative species within the family Benyviridae . The viral methyltransferase (Mtr) domain spans from nucleotide position 386 to 1411, while the RNA helicase (Hel) domain occupies positions 2113 to 2995 nt. Additionally, the protease (Pro) domain is located between positions 3142 and 3410 nt, and the RdRp domain is located at 4227 to 4796 nt. A phylogenetic analysis was conducted by integrating RdRp sequences of viruses closely related to GtBeV. The result revealed that GtBeV clustered within the family Benyviridae , exhibiting substantial evolutionary divergence from any other sequences. Consequently, this virus likely represents a novel species in the family Benyviridae .
(iii) Botourmiaviridae
The family Botourmiaviridae comprises viruses infecting plants and filamentous fungi, which may possess mono- or multi-segmented genomes [ 51 ]. Recent research has led to a rapid expansion in the number of viruses within the family Botourmiaviridae , increasing from the confirmed 4 genera in 2020 to a total of 12 genera. A contig identified from Clonostachys rosea (ERR5928658) using the BLASTx method exhibited similarity to viruses in the family Botourmiaviridae . After manual mapping, a 2,903 nt-long genome was obtained, tentatively named Clonostachys rosea botourmiavirus (CrBV), which includes a complete RdRP region (Fig. 1 E). Based on phylogenetic analysis using RdRp, CrBV clustered with members of the genus Magoulivirus , sharing 56.58% aa identity with a strain identified from Eclipta prostrata . However, puzzlingly, according to the ICTV's Genus/Species demarcation criteria, members of different genera/species within the family Botourmiaviridae share less than 70%/90% identity in their complete RdRP amino acid sequences. Furthermore, the RdRp sequences with accession numbers NC_055143 and NC_076766, both considered to be members of the genus Magoulivirus , exhibited only 39.05% aa identity to each other. Therefore, CrBV should at least be considered as a new species within the family Botourmiaviridae .
(iv) Deltaflexiviridae
An assembled sequence of 3,425 nucleotides in length Lepista sordida deltaflexivirus (LsDV), derived from Lepista sordida (DRR252167) and showing homology to Deltaflexiviridae within the order Tymovirales , was obtained. The Tymovirales comprises five recognized families: Alphaflexiviridae , Betaflexiviridae , Deltaflexiviridae , Gammaflexiviridae , and Tymoviridae [ 52 ]. The Deltaflexiviridae currently only includes one genus, the fungal-associated deltaflexivirus; they are mostly identified in fungi or plants pathogens [ 53 ]. LsDV was predicted to have a single large ORF, VP1, which starts with an AUG codon at nt 163–165 and ends with a UAG codon at nt 3,418–3,420. This ORF encodes a putative polyprotein of 1,086 aa with a calculated molecular mass of 119 kDa. Two conserved domains within the VP1 protein were identified: Hel and RdRp (Fig. 2 A). However, the Mtr was missing, indicating that the 5' end of this polyprotein is incomplete. According to the phylogenetic analysis of RdRp, LsDV was closely related to viruses of the family Deltaflexiviridae and shared 46.61% aa identity with a strain (UUW06602) isolated from Macrotermes carbonarius . Despite this, according to the species demarcation criteria proposed by ICTV, because we couldn't recover the entire replication-associated polyprotein, LsDV cannot be regarded as a novel species at present.
Identification of novel members of family Deltaflexiviridae and Toga-like virus in fungal sequencing libraries. A On the right side of the image is the genome organization of LsDV; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. ML phylogenetic tree of members of the family Deltaflexiviridae . The best-fit model (VT + F + R6) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font. B The genome organization of GtTlV is depicted at the top; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. ML phylogenetic tree of members of the order Martellivirales . The best-fit model (LG + R7) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font
(v) Toga-like virus
Members of the family Togaviridae are primarily transmitted by arthropods and can infect a wide range of vertebrates, including mammals, birds, reptiles, amphibians, and fish [ 54 ]. Currently, this family only contains a single confirmed genus, Alphavirus . A contig was discovered in Gaeumannomyces tritici (ERR3486058), it is 7,588 nt in length with a complete ORF encoding a putative protein of 1,928 aa, which had 60.43% identity to Fusarium sacchari alphavirus-like virus 1 (QIQ28421) with 97% coverage. Phylogenetic analysis showed that it did not cluster with classical alphavirus members such as VEE, WEE, EEE, SF complex [ 54 ], but rather with several sequences annotated as Toga-like that were available (Fig. 2 B). It was provisionally named Gaeumannomyces tritici toga-like virus (GtTIV). However, we remain cautious about the accuracy of these so-called Toga-like sequences, as they show little significant correlation with members of the order Martellivirales .
Negative-sense single-stranded RNA viruses
(i) mymonaviridae.
Mymonaviridae is a family of linear, enveloped, negative-stranded RNA genomes in the order Mononegavirales , which infect fungi. They are approximately 10 kb in size and encode six proteins [ 55 ]. The famliy Mymonaviridae was established to accommodate Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV-1), a novel virus discovered in a hypovirulent strain of Sclerotinia sclerotiorum [ 56 ]. According to the ICTV, the family Mymonaviridae currently includes 9 genera, namely Auricularimonavirus , Botrytimonavirus , Hubramonavirus , Lentimonavirus , Penicillimonavirus , Phyllomonavirus , Plasmopamonavirus , Rhizomonavirus and Sclerotimonavirus . Two sequences originating from Gaeumannomyces tritici (ERR3486068) and Aspergillus puulaauensis (DRR266546), respectively, and associated with the family Mymonaviridae , have been identified and provisionally named Gaeumannomyces tritici mymonavirus (GtMV) and Aspergillus puulaauensis mymonavirus (ApMV). GtMV is 9,339 nt long with a GC content of 52.8%. It was predicted to contain 5 discontinuous ORFs, with the largest one encoding RdRp. Additionally, a nucleoprotein and three hypothetical proteins with unknown function were also predicted. A multiple alignment of nucleotide sequences among these ORFs identified a semi-conserved sequence, 5'-UAAAA-CUAGGAGC-3', located downstream of each ORF (Fig. 3 A). These regions are likely gene-junction regions in the GtMV genome, a characteristic feature shared by mononegaviruses [ 57 , 58 ]. For ApMV, a complete RdRp CDS with a length of 1,978 aa was predicted. The BLASTx searches showed that GtMV shared 45.22% identity with the RdRp of Soybean leaf-associated negative-stranded RNA virus 2 (YP_010784557), while ApMV shared 55.90% identity with the RdRp of Erysiphe necator associated negative-stranded RNA virus 23 (YP_010802816). The representative members of the family Mymonaviridae were included in the phylogenetic analysis. The results showed that GtMV and ApMV clustered closely with members of the genera Sclerotimonavirus and Plasmopamonavirus , respectively (Fig. 3 B). Members of the genus Plasmopamonavirus are about 6 kb in size and encode for a single protein. Therefore, GtMV and ApMV should be considered as representing new species within their respective genera.
Identification of two new members in the family Mymonaviridae . A At the top is the nucleotide multiple sequence alignment result of GtMV with the reference genomes. the putative ORF for the viral RdRp is depicted by a green box, the predicted nucleoprotein is displayed in a yellow box, and three hypothetical proteins are displayed in gray boxes. The comparison of putative semi-conserved regions between ORFs in GtMV is displayed in the 5' to 3' orientation, with conserved sequences are highlighted. At the bottom is the genome organization of AmPV; the putative ORF for the viral RdRp is depicted by a green box. B ML phylogenetic tree of members of the family Mymonaviridae . The best-fit model (LG + F + R6) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified viruses represented in red font
(ii) Bunyavirales
The Bunyavirales (the only order in the class Ellioviricetes ) is one of the largest groups of segmented negative-sense single-stranded RNA viruses with mainly tripartite genomes [ 59 ], which includes many pathogenic strains that infect arthropods(such as mosquitoes, ticks, sand flies), plants, protozoans, and vertebrates, and even cause severe human diseases. Order Bunyavirales consists of 14 viral families, including Arenaviridae , Cruliviridae , Discoviridae , Fimoviridae , Hantaviridae , Leishbuviridae , Mypoviridae , Nairoviridae , Peribunyaviridae , Phasmaviridae , Phenuiviridae , Tospoviridae , Tulasviridae and Wupedeviridae . In this study, three complete or near complete RNA1 sequences related to bunyaviruses were identified and named according to their respective hosts: CoBV ( Conidiobolus obscurus bunyavirus; SRR6181013; 7,277 nt), GtBV ( Gaeumannomyces tritici bunyavirus; ERR3486069; 7,364 nt), and TaBV ( Thielaviopsis aethacetica bunyavirus; SRR12744489; 9,516 nt) (Fig. 4 A). The 5' and 3' terminal RNA segments of GtBV and TaBV complement each other, allowing the formation of a panhandle structure [ 60 ], which plays an essential role as promoters of genome transcription and replication [ 61 ], except for CoBV, as the 3' terminal of CoBV has not been fully obtained (Fig. 4 B). BLASTx results indicated that these three viruses had identities ranging from 32.97% to 54.20% to the best matches in the GenBank database. Phylogenetic analysis indicated that CoBV was classified into the family Phasmaviridae , with distant relationships to any of its genera; GtBV clustered well with members of the genus Entovirus of family Phenuiviridae ; while TaBV did not cluster with any known members of families within Bunyavirales , hence provisionally placed within the Bunya-like group (Fig. 4 C). Therefore, these three sequences should be considered as potential new family, genus, or species within the order Bunyavirales .
Identification of three new members in the order Bunyavirales . A The genome organization of CoBV, GtBV, and TaBV; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. B The complementary structures formed at the 5' and 3' ends of GtBV and TaBV. C ML phylogenetic tree of members of the order Bunyavirales . The best-fit model (VT + F + R8) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified viruses represented in red font
Double-stranded RNA viruses
Partitiviridae.
The Partitiviridae is a family of small, non-enveloped viruses, approximately 35–40 nm in diameter, with bisegmented double-stranded (ds) RNA genomes. Each segment is about 1.4–3.0 kb in size, resulting in a total size about 4 kb [ 62 ]. The family Partitiviridae is now divided into five genera: Alphapartitivirus , Betapartiivirus , Cryspovirus , Deltapartitivirus and Gammapartitivirus . Each genus has characteristic hosts: plants or fungi for Alphapartitivirus and Betapartitivirus , fungi for Gammapartitivirus , plants for Deltapartitivirus , and protozoa for Cryspovirus [ 62 ]. A complete dsRNA1 sequence Neocallimastix californiae partitivirus (NcPV) retrieved from Neocallimastix californiae (SRR15362281) has been identified as being associated with the family Partitiviridae . The BLASTp result indicated that it shared the highest aa identity of 41.5% with members of the genus Gammapartitivirus . According to the phylogenetic tree constructed based on RdRp, NcPV was confirmed to fall within the genus Gammapartitivirus (Fig. 5 ). Typical members of the genus Gammapartitivirus have two segments in their complete genome, namely dsRNA1 and dsRNA2, encoding RdRp and coat protein, respectively [ 62 ]. The larger dsRNA1 segment of NcPV measures 1,769 nt in length, with a GC content of 35.8%. It contains a single ORF encoding a 561 aa RdRp. A CDD search revealed that the RdRp of NcPV harbors a catalytic region spanning from 119 to 427aa. Regrettably, only the complete dsRNA1 segment was obtained. According to the classification principles of ICTV, due to the lack of information regarding dsRNA2, we are unable to propose it as a new species. It is worth noting that according to the Genus demarcation criteria ( https://ictv.global/report/chapter/partitiviridae/partitiviridae ), members of the genus Gammapartitivirus should have a dsRNA1 length ranging from 1645 to 1787 nt, and the RdRp length should fall between 519 and 539 aa. However, the length of dsRNA1 in NcPV is 1,769 nt, with RdRp being 561 aa, challenging this classification criterion. In fact, multiple strains have already exceeded this criterion, such as GenBank accession numbers: WBW48344, UDL14336, QKK35392, among others.
Identification of a new member in the family Partitiviridae . The genome organization of NcPV is depicted at the top; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. At the bottom is the ML phylogenetic tree of members of the family Partitiviridae . The best-fit model (VT + F + R4) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font
Long-term evolutionary relationships between fungal-associated viruses and hosts
Understanding the co-divergence history between viruses and hosts helps reveal patterns of virus transmission and infection and influences the biodiversity and stability of ecosystems. To explore the frequency of cross-species transmission and co-divergence among fungi-associated viruses, we constructed tanglegrams illustrating the interconnected evolutionary histories of viral families and their respective hosts through phylogenetic trees (Fig. 6 A). The results indicated that cross-species transmission (Host-jumping) consistently emerged as the most frequent evolutionary event among all groups of RNA viruses examined in this study (median, 66.79%; range, 60.00% to 79.07%) (Fig. 6 B). This finding is highly consistent with the evolutionary patterns of RNA viruses recently identified by Mifsud et al. in their extensive transcriptome survey of plants [ 63 ]. Members of the families Botourmiaviridae (79.07%) and Deltaflexiviridae (72.41%) were most frequently involved in cross-species transmission. The frequencies of co-divergence (median, 20.19%; range, 6.98% to 27.78%), duplication (median, 10.60%; range, 0% to 22.45%), and extinction (median, 2.42%; range, 0% to 5.56%) events involved in the evolution of fungi-associated viruses gradually decrease. Specifically, members of the family Benyviridae exhibited the highest frequency of co-divergence events, which also supports the findings reported by Mifsud et al.; certain studies propose that members of Benyviridae are transmitted via zoospores of plasmodiophorid protist [ 64 ]. It's speculated that the ancestor of these viruses underwent interkingdom horizontal transfer between plants and protists over evolutionary timelines [ 65 ]. Members of the family Mitoviridae showed the highest frequency of duplication events; and members of the families Benyviridae and Partitiviridae demonstrated the highest frequency of extinction events. Not surprisingly, this result is influenced by the current limited understanding of virus-host relationships. On one hand, viruses whose hosts cannot be recognized through published literature or information provided by authors have been overlooked. On the other hand, the number of viruses recorded in reference databases represents just the tip of the iceberg within the entire virosphere. The involvement of a more extensive sample size in the future should change this evolutionary landscape.
Co-evolutionary analysis of virus and host. A Tanglegram of phylogenetic trees for virus orders/families and their hosts. Lines and branches are color-coded to indicate host clades. The cophylo function in phytools was employed to enhance congruence between the host (left) and virus (right) phylogenies. B Reconciliation analysis of virus groups. The bar chart illustrates the proportional range of possible evolutionary events, with the frequency of each event displayed at the top of its respective column
Our understanding of the interactions between fungi and their associated viruses has long been constrained by insufficient sampling of fungal species. Advances in metagenomics in recent decades have led to a rapid expansion of the known viral sequence space, but it is far from saturated. The diversity of hosts, the instability of the viral structures (especially RNA viruses), and the propensity to exchange genetic material with other host viruses all contribute to the unparalleled diversity of viral genomes [ 66 ]. Fungi are diverse and widely distributed in nature and are closely related to humans. A few fungi can parasitize immunocompromised humans, but their adverse effects are limited. As decomposers in the biological chain, fungi can decompose the remains of plants and animals and maintain the material cycle in the biological world [ 67 ]. In agricultural production, many fungi are plant pathogens, and about 80% of plant diseases are caused by fungi. However, little is currently known about the diversity of mycoviruses and how these viruses affect fungal phenotypes, fungal-host interactions, and virus evolution, and the sequencing depth of fungal libraries in most public databases only meets the needs of studying bacterial genomes. Sampling viruses from a larger diversity of fungal hosts should lead to new and improved evolutionary scenarios.
RNA viruses are widespread in deep-sea sediments [ 68 ], freshwater [ 69 ], sewage [ 70 ], and rhizosphere soils [ 71 ]. Compared to DNA viruses, RNA viruses are less conserved, prone to mutation, and can transfer between different hosts, potentially forming highly differentiated and unrecognized novel viruses. This characteristic increases the difficulty of monitoring these viruses. Previously, all discovered mycoviruses were RNA viruses. Until 2010, Yu et al. reported the discovery of a DNA virus, namely SsHADV-1, in fungi for the first time [ 72 ]. Subsequently, new fungal-related DNA viruses are continually being identified [ 73 , 74 , 75 ]. Currently, viruses have been found in all major groups of fungi, and approximately 100 types of fungi can be infected by viruses, instances exist where one virus can infect multiple fungi, or one fungus can be infected by several viruses simultaneously. The transmission of mycoviruses differs from that of animal and plant viruses and is mainly categorized into vertical and horizontal transmission [ 76 ]. Vertical transmission refers to the spread of the mycovirus to the next generation through the sexual or asexual spores of the fungus, while horizontal transmission refers to the spread of the mycovirus from one strain to another through fusion between hyphae. In the phylum Ascomycota , mycoviruses generally exhibit a low ability to transmit vertically through ascospores, but they are commonly transmitted vertically to progeny strains through asexual spores [ 77 ].
In this study, we identified two novel species belonging to different genera within the family Mitoviridae . Interestingly, they both simultaneously infect the same fungus— Thielaviopsis ethacetica , the causal agent of pineapple sett rot disease in sugarcane [ 78 ]. Previously, a report identified three different mitoviruses in Fusarium circinatum [ 79 ]. These findings suggest that there may be a certain level of adaptability or symbiotic relationship among members of the family Mitoviridae . Benyviruses are typically considered to infect plants, but recent evidence suggests that they can also infect fungi, such as Agaricus bisporus [ 80 ], further reinforced by the virus we discovered in Gaeumannomyces tritici . Moreover, members of the family Botourmiaviridae commonly exhibit a broad host range, with viruses closely related to CrBV capable of infecting members of Eukaryota , Viridiplantae , and Metazoa , in addition to fungi (Supplementary Fig. 1). The LsDV identified in this study shared the closest phylogenetic relationship with a virus identified from Macrotermes carbonarius in southern Vietnam (17_N1 + N237) [ 81 ]. M. carbonarius is an open-air foraging species that collects plant litter and wood debris to cultivate fungi in fungal gardens [ 82 ], termites may act as vectors, transmitting deltaflexivirus to other fungi. Furthermore, the viruses we identified, typically associated with fungi, also deepen their connections with species from other kingdoms on the tanglegram tree. For example, while Partitiviridae are naturally associated with fungi and plants, NcPV also shows close connections with Metazoa . In fact, based largely on phylogenetic predictions, various eukaryotic viruses have been found to undergo horizontal transfer between organisms of plants, fungi, and animals [ 83 ]. The rice dwarf virus was demonstrated to infect both plant and insect vectors [ 84 ]; moreover, plant-infecting rhabdoviruses, tospoviruses, and tenuiviruses are now known to replicate and spread in vector insects and shuttle between plants and animals [ 85 ]. Furthermore, Bian et al. demonstrated that plant virus infection in plants enables Cryphonectria hypovirus 1 to undergo horizontal transfer from fungi to plants and other heterologous fungal species [ 86 ].
Recent studies have greatly expanded the diversity of mycoviruses [ 87 , 88 ]. Gilbert et al. [ 20 ] investigated publicly available fungal transcriptomes from the subphylum Pezizomycotina, resulting in the detection of 52 novel mycoviruses; Myers et al. [ 18 ] employed both culture-based and transcriptome-mining approaches to identify 85 unique RNA viruses across 333 fungi; Ruiz-Padilla et al. identified 62 new mycoviral species from 248 Botrytis cinerea field isolates; Zhou et al. identified 20 novel viruses from 90 fungal strains (across four different macrofungi species) [ 89 ]. However, compared to these studies, our work identified fewer novel viruses, possibly due to the following reasons: 1) The libraries from the same Bioproject are usually from the same strains (or isolates). Therefore, there is a certain degree of redundancy in the datasets collected for this study. 2) Contigs shorter than 1,500 nt were discarded, potentially resulting in the oversight of short viral molecules. 3) Establishing a threshold of 70% aa sequence identity may also lead to the exclusion of certain viruses. 4) Some poly(A)-enriched RNA-seq libraries are likely to miss non-polyadenylated RNA viral genomes.
Taxonomy is a dynamic science, evolving with improvements in analytical methods and the emergence of new data. Identifying and rectifying incorrect classifications when new information becomes available is an ongoing and inevitable process in today's rapidly expanding field of virology. For instance, in 1975, members of the genera Rubivirus and Alphavirus were initially grouped under the family Togaviridae ; however, in 2019, Rubivirus was reclassified into the family Matonaviridae due to recognized differences in transmission modes and virion structures [ 90 ]. Additionally, the conflicts between certain members of the genera Magoulivirus and Gammapartitivirus mentioned here and their current demarcation criteria (e.g., amino acid identity, nucleotide length thresholds) need to be reconsidered.
Taken together, these findings reveal the potential diversity and novelty within fungal-associated viral communities and discuss the genetic similarities among different fungal-associated viruses. These findings advance our understanding of fungal-associated viruses and suggest the importance of subsequent in-depth investigations into the interactions between fungi and viruses, which will shed light on the important roles of these viruses in the global fungal kingdom.
Availability of data and materials
The data reported in this paper have been deposited in the GenBase in National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, under accession numbers C_AA066339.1-C_AA066350.1 that are publicly accessible at https://ngdc.cncb.ac.cn/genbase . Please refer to Table 1 for details.
Leigh DM, Peranic K, Prospero S, Cornejo C, Curkovic-Perica M, Kupper Q, et al. Long-read sequencing reveals the evolutionary drivers of intra-host diversity across natural RNA mycovirus infections. Virus Evol. 2021;7(2):veab101. https://doi.org/10.1093/ve/veab101 . Epub 2022/03/19 PubMed PMID: 35299787; PubMed Central PMCID: PMCPMC8923234.
Article PubMed PubMed Central Google Scholar
Ghabrial SA, Suzuki N. Viruses of plant pathogenic fungi. Annu Rev Phytopathol. 2009;47:353–84. https://doi.org/10.1146/annurev-phyto-080508-081932 . Epub 2009/04/30 PubMed PMID: 19400634.
Article CAS PubMed Google Scholar
Ghabrial SA, Caston JR, Jiang D, Nibert ML, Suzuki N. 50-plus years of fungal viruses. Virology. 2015;479–480:356–68. https://doi.org/10.1016/j.virol.2015.02.034 . Epub 2015/03/17 PubMed PMID: 25771805.
Chen YM, Sadiq S, Tian JH, Chen X, Lin XD, Shen JJ, et al. RNA viromes from terrestrial sites across China expand environmental viral diversity. Nat Microbiol. 2022;7(8):1312–23. https://doi.org/10.1038/s41564-022-01180-2 . Epub 2022/07/29 PubMed PMID: 35902778.
Pearson MN, Beever RE, Boine B, Arthur K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol. 2009;10(1):115–28. https://doi.org/10.1111/j.1364-3703.2008.00503.x . Epub 2009/01/24 PubMed PMID: 19161358; PubMed Central PMCID: PMCPMC6640375.
Santiago-Rodriguez TM, Hollister EB. Unraveling the viral dark matter through viral metagenomics. Front Immunol. 2022;13:1005107. https://doi.org/10.3389/fimmu.2022.1005107 . Epub 2022/10/04 PubMed PMID: 36189246; PubMed Central PMCID: PMCPMC9523745.
Article CAS PubMed PubMed Central Google Scholar
Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol. 2008;159(5):349–57. https://doi.org/10.1016/j.resmic.2008.04.010 . Epub 2008/06/21 PubMed PMID: 18565737.
Guo W, Yan H, Ren X, Tang R, Sun Y, Wang Y, et al. Berberine induces resistance against tobacco mosaic virus in tobacco. Pest Manag Sci. 2020;76(5):1804–13. https://doi.org/10.1002/ps.5709 . Epub 2019/12/10 PubMed PMID: 31814252.
Villabruna N, Izquierdo-Lara RW, Schapendonk CME, de Bruin E, Chandler F, Thao TTN, et al. Profiling of humoral immune responses to norovirus in children across Europe. Sci Rep. 2022;12(1):14275. https://doi.org/10.1038/s41598-022-18383-6 . Epub 2022/08/23 PubMed PMID: 35995986.
Zhang Y, Gao J, Li Y. Diversity of mycoviruses in edible fungi. Virus Genes. 2022;58(5):377–91. https://doi.org/10.1007/s11262-022-01908-6 . Epub 2022/06/07 PubMed PMID: 35668282.
Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 2019;26(4):527–41. https://doi.org/10.1016/j.chom.2019.09.009 . Epub 2019/10/11 PubMed PMID: 31600503.
Botella L, Janousek J, Maia C, Jung MH, Raco M, Jung T. Marine Oomycetes of the Genus Halophytophthora harbor viruses related to Bunyaviruses. Front Microbiol. 2020;11:1467. https://doi.org/10.3389/fmicb.2020.01467 . Epub 2020/08/08 PubMed PMID: 32760358; PubMed Central PMCID: PMCPMC7375090.
Kotta-Loizou I. Mycoviruses and their role in fungal pathogenesis. Curr Opin Microbiol. 2021;63:10–8. https://doi.org/10.1016/j.mib.2021.05.007 . Epub 2021/06/09 PubMed PMID: 34102567.
Ellis LF, Kleinschmidt WJ. Virus-like particles of a fraction of statolon, a mould product. Nature. 1967;215(5101):649–50. https://doi.org/10.1038/215649a0 . Epub 1967/08/05 PubMed PMID: 6050227.
Banks GT, Buck KW, Chain EB, Himmelweit F, Marks JE, Tyler JM, et al. Viruses in fungi and interferon stimulation. Nature. 1968;218(5141):542–5. https://doi.org/10.1038/218542a0 . Epub 1968/05/11 PubMed PMID: 4967851.
Jia J, Fu Y, Jiang D, Mu F, Cheng J, Lin Y, et al. Interannual dynamics, diversity and evolution of the virome in Sclerotinia sclerotiorum from a single crop field. Virus Evol. 2021;7(1):veab032. https://doi.org/10.1093/ve/veab032 .
Mu F, Li B, Cheng S, Jia J, Jiang D, Fu Y, et al. Nine viruses from eight lineages exhibiting new evolutionary modes that co-infect a hypovirulent phytopathogenic fungus. Plos Pathog. 2021;17(8):e1009823. https://doi.org/10.1371/journal.ppat.1009823 . Epub 2021/08/25 PubMed PMID: 34428260; PubMed Central PMCID: PMCPMC8415603.
Myers JM, Bonds AE, Clemons RA, Thapa NA, Simmons DR, Carter-House D, et al. Survey of early-diverging lineages of fungi reveals abundant and diverse Mycoviruses. mBio. 2020;11(5):e02027. https://doi.org/10.1128/mBio.02027-20 . Epub 2020/09/10 PubMed PMID: 32900807; PubMed Central PMCID: PMCPMC7482067.
Ruiz-Padilla A, Rodriguez-Romero J, Gomez-Cid I, Pacifico D, Ayllon MA. Novel Mycoviruses discovered in the Mycovirome of a Necrotrophic fungus. MBio. 2021;12(3):e03705. https://doi.org/10.1128/mBio.03705-20 . Epub 2021/05/13 PubMed PMID: 33975945; PubMed Central PMCID: PMCPMC8262958.
Gilbert KB, Holcomb EE, Allscheid RL, Carrington JC. Hiding in plain sight: new virus genomes discovered via a systematic analysis of fungal public transcriptomes. Plos One. 2019;14(7):e0219207. https://doi.org/10.1371/journal.pone.0219207 . Epub 2019/07/25 PubMed PMID: 31339899; PubMed Central PMCID: PMCPMC6655640.
Khan HA, Telengech P, Kondo H, Bhatti MF, Suzuki N. Mycovirus hunting revealed the presence of diverse viruses in a single isolate of the Phytopathogenic fungus diplodia seriata from Pakistan. Front Cell Infect Microbiol. 2022;12:913619. https://doi.org/10.3389/fcimb.2022.913619 . Epub 2022/07/19 PubMed PMID: 35846770; PubMed Central PMCID: PMCPMC9277117.
Kotta-Loizou I, Coutts RHA. Mycoviruses in Aspergilli: a comprehensive review. Front Microbiol. 2017;8:1699. https://doi.org/10.3389/fmicb.2017.01699 . Epub 2017/09/22 PubMed PMID: 28932216; PubMed Central PMCID: PMCPMC5592211.
Garcia-Pedrajas MD, Canizares MC, Sarmiento-Villamil JL, Jacquat AG, Dambolena JS. Mycoviruses in biological control: from basic research to field implementation. Phytopathology. 2019;109(11):1828–39. https://doi.org/10.1094/PHYTO-05-19-0166-RVW . Epub 2019/08/10 PubMed PMID: 31398087.
Rigling D, Prospero S. Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Mol Plant Pathol. 2018;19(1):7–20. https://doi.org/10.1111/mpp.12542 . Epub 2017/02/01 PubMed PMID: 28142223; PubMed Central PMCID: PMCPMC6638123.
Okada R, Ichinose S, Takeshita K, Urayama SI, Fukuhara T, Komatsu K, et al. Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: down-regulation of host growth and up-regulation of host plant pathogenicity. Virology. 2018;519:23–32. https://doi.org/10.1016/j.virol.2018.03.027 . Epub 2018/04/10 PubMed PMID: 29631173.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. https://doi.org/10.1038/nmeth.1923 . Epub 2012/03/06 PubMed PMID: 22388286; PubMed Central PMCID: PMCPMC3322381.
Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes De Novo assembler. Curr Protoc Bioinform. 2020;70(1):e102. https://doi.org/10.1002/cpbi.102 . Epub 2020/06/20 PubMed PMID: 32559359.
Article CAS Google Scholar
Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, et al. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. https://doi.org/10.1016/j.ymeth.2016.02.020 . Epub 2016/03/26 PubMed PMID: 27012178.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. https://doi.org/10.1093/bioinformatics/btp698 . Epub 2010/01/19 PubMed PMID: 20080505; PubMed Central PMCID: PMCPMC2828108.
Roux S, Paez-Espino D, Chen IA, Palaniappan K, Ratner A, Chu K, et al. IMG/VR v3: an integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 2021;49(D1):D764–75. https://doi.org/10.1093/nar/gkaa946 . Epub 2020/11/03 PubMed PMID: 33137183; PubMed Central PMCID: PMCPMC7778971.
Mirdita M, Steinegger M, Soding J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics. 2019;35(16):2856–8. https://doi.org/10.1093/bioinformatics/bty1057 . Epub 2019/01/08 PubMed PMID: 30615063; PubMed Central PMCID: PMCPMC6691333.
Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18(4):366–8. https://doi.org/10.1038/s41592-021-01101-x . Epub 2021/04/09 PubMed PMID: 33828273; PubMed Central PMCID: PMCPMC8026399.
Shen W, Ren H. TaxonKit: A practical and efficient NCBI taxonomy toolkit. J Genet Genomics. 2021;48(9):844–50. https://doi.org/10.1016/j.jgg.2021.03.006 . Epub 2021/05/19 PubMed PMID: 34001434.
Article PubMed Google Scholar
Gautam A, Felderhoff H, Bagci C, Huson DH. Using AnnoTree to get more assignments, faster, in DIAMOND+MEGAN microbiome analysis. mSystems. 2022;7(1):e0140821. https://doi.org/10.1128/msystems.01408-21 . Epub 2022/02/23 PubMed PMID: 35191776; PubMed Central PMCID: PMCPMC8862659.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. https://doi.org/10.1093/molbev/msy096 . Epub 2018/05/04 PubMed PMID: 29722887; PubMed Central PMCID: PMCPMC5967553.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–4. https://doi.org/10.1093/molbev/msaa015 . Epub 2020/02/06 PubMed PMID: 32011700; PubMed Central PMCID: PMCPMC7182206.
Letunic I, Bork P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res (2024). https://doi.org/10.1093/nar/gkae268
Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. Plos One. 2014;9(9):e108277. https://doi.org/10.1371/journal.pone.0108277 . Epub 2014/09/27 PubMed PMID: 25259891; PubMed Central PMCID: PMCPMC4178126.
Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, et al. TimeTree 5: an expanded resource for species divergence times. Mol Biol Evol. 2022;39(8):msac174. https://doi.org/10.1093/molbev/msac174 . Epub 2022/08/07 PubMed PMID: 35932227; PubMed Central PMCID: PMCPMC9400175.
Revell LJ. phytools 2.0: an updated R ecosystem for phylogenetic comparative methods (and other things). PeerJ. 2024;12:e16505. https://doi.org/10.7717/peerj.16505 . Epub 2024/01/09 PubMed PMID: 38192598; PubMed Central PMCID: PMCPMC10773453.
Santichaivekin S, Yang Q, Liu J, Mawhorter R, Jiang J, Wesley T, et al. eMPRess: a systematic cophylogeny reconciliation tool. Bioinformatics. 2021;37(16):2481–2. https://doi.org/10.1093/bioinformatics/btaa978 . Epub 2020/11/21 PubMed PMID: 33216126.
Ma W, Smirnov D, Libeskind-Hadas R. DTL reconciliation repair. BMC Bioinformatics. 2017;18(Suppl 3):76. https://doi.org/10.1186/s12859-017-1463-9 . Epub 2017/04/01 PubMed PMID: 28361686; PubMed Central PMCID: PMCPMC5374596.
Members C-N, Partners. Database resources of the national genomics data center, China national center for bioinformation in 2024. Nucleic Acids Res. 2024;52(D1):D18–32. https://doi.org/10.1093/nar/gkad1078 . Epub 2023/11/29 PubMed PMID: 38018256; PubMed Central PMCID: PMCPMC10767964.
Article Google Scholar
Shafik K, Umer M, You H, Aboushedida H, Wang Z, Ni D, et al. Characterization of a Novel Mitovirus infecting Melanconiella theae isolated from tea plants. Front Microbiol. 2021;12: 757556. https://doi.org/10.3389/fmicb.2021.757556 . Epub 2021/12/07 PubMed PMID: 34867881; PubMed Central PMCID: PMCPMC8635788
Kamaruzzaman M, He G, Wu M, Zhang J, Yang L, Chen W, et al. A novel Partitivirus in the Hypovirulent isolate QT5–19 of the plant pathogenic fungus Botrytis cinerea. Viruses. 2019;11(1):24. https://doi.org/10.3390/v11010024 . Epub 2019/01/06 PubMed PMID: 30609795; PubMed Central PMCID: PMCPMC6356794.
Akata I, Keskin E, Sahin E. Molecular characterization of a new mitovirus hosted by the ectomycorrhizal fungus Albatrellopsis flettii. Arch Virol. 2021;166(12):3449–54. https://doi.org/10.1007/s00705-021-05250-4 . Epub 2021/09/24 PubMed PMID: 34554305.
Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, et al. Recent changes to virus taxonomy ratified by the international committee on taxonomy of viruses (2022). Arch Virol. 2022;167(11):2429–40. https://doi.org/10.1007/s00705-022-05516-5 . Epub 2022/08/24 PubMed PMID: 35999326; PubMed Central PMCID: PMCPMC10088433.
Alvarez-Quinto R, Grinstead S, Jones R, Mollov D. Complete genome sequence of a new mitovirus associated with walking iris (Trimezia northiana). Arch Virol. 2023;168(11):273. https://doi.org/10.1007/s00705-023-05901-8 . Epub 2023/10/17 PubMed PMID: 37845386.
Gilmer D, Ratti C, Ictv RC. ICTV Virus taxonomy profile: Benyviridae. J Gen Virol. 2017;98(7):1571–2. https://doi.org/10.1099/jgv.0.000864 . Epub 2017/07/18 PubMed PMID: 28714846; PubMed Central PMCID: PMCPMC5656776.
Wetzel V, Willlems G, Darracq A, Galein Y, Liebe S, Varrelmann M. The Beta vulgaris-derived resistance gene Rz2 confers broad-spectrum resistance against soilborne sugar beet-infecting viruses from different families by recognizing triple gene block protein 1. Mol Plant Pathol. 2021;22(7):829–42. https://doi.org/10.1111/mpp.13066 . Epub 2021/05/06 PubMed PMID: 33951264; PubMed Central PMCID: PMCPMC8232027.
Ayllon MA, Turina M, Xie J, Nerva L, Marzano SL, Donaire L, et al. ICTV Virus taxonomy profile: Botourmiaviridae. J Gen Virol. 2020;101(5):454–5. https://doi.org/10.1099/jgv.0.001409 . Epub 2020/05/08 PubMed PMID: 32375992; PubMed Central PMCID: PMCPMC7414452.
Xiao J, Wang X, Zheng Z, Wu Y, Wang Z, Li H, et al. Molecular characterization of a novel deltaflexivirus infecting the edible fungus Pleurotus ostreatus. Arch Virol. 2023;168(6):162. https://doi.org/10.1007/s00705-023-05789-4 . Epub 2023/05/17 PubMed PMID: 37195309.
Canuti M, Rodrigues B, Lang AS, Dufour SC, Verhoeven JTP. Novel divergent members of the Kitrinoviricota discovered through metagenomics in the intestinal contents of red-backed voles (Clethrionomys gapperi). Int J Mol Sci. 2022;24(1):131. https://doi.org/10.3390/ijms24010131 . Epub 2023/01/09 PubMed PMID: 36613573; PubMed Central PMCID: PMCPMC9820622.
Hermanns K, Zirkel F, Kopp A, Marklewitz M, Rwego IB, Estrada A, et al. Discovery of a novel alphavirus related to Eilat virus. J Gen Virol. 2017;98(1):43–9. https://doi.org/10.1099/jgv.0.000694 . Epub 2017/02/17 PubMed PMID: 28206905.
Jiang D, Ayllon MA, Marzano SL, Ictv RC. ICTV Virus taxonomy profile: Mymonaviridae. J Gen Virol. 2019;100(10):1343–4. https://doi.org/10.1099/jgv.0.001301 . Epub 2019/09/04 PubMed PMID: 31478828.
Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, et al. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci U S A. 2014;111(33):12205–10. https://doi.org/10.1073/pnas.1401786111 . Epub 2014/08/06 PubMed PMID: 25092337; PubMed Central PMCID: PMCPMC4143027.
Zhong J, Li P, Gao BD, Zhong SY, Li XG, Hu Z, et al. Novel and diverse mycoviruses co-infecting a single strain of the phytopathogenic fungus Alternaria dianthicola. Front Cell Infect Microbiol. 2022;12:980970. https://doi.org/10.3389/fcimb.2022.980970 . Epub 2022/10/15 PubMed PMID: 36237429; PubMed Central PMCID: PMCPMC9552818.
Wang W, Wang X, Tu C, Yang M, Xiang J, Wang L, et al. Novel Mycoviruses discovered from a Metatranscriptomics survey of the Phytopathogenic Alternaria Fungus. Viruses. 2022;14(11):2552. https://doi.org/10.3390/v14112552 . Epub 2022/11/25 PubMed PMID: 36423161; PubMed Central PMCID: PMCPMC9693364.
Sun Y, Li J, Gao GF, Tien P, Liu W. Bunyavirales ribonucleoproteins: the viral replication and transcription machinery. Crit Rev Microbiol. 2018;44(5):522–40. https://doi.org/10.1080/1040841X.2018.1446901 . Epub 2018/03/09 PubMed PMID: 29516765.
Li P, Bhattacharjee P, Gagkaeva T, Wang S, Guo L. A novel bipartite negative-stranded RNA mycovirus of the order Bunyavirales isolated from the phytopathogenic fungus Fusarium sibiricum. Arch Virol. 2023;169(1):13. https://doi.org/10.1007/s00705-023-05942-z . Epub 2023/12/29 PubMed PMID: 38155262.
Ferron F, Weber F, de la Torre JC, Reguera J. Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res. 2017;234:118–34. https://doi.org/10.1016/j.virusres.2017.01.018 . Epub 2017/02/01 PubMed PMID: 28137457; PubMed Central PMCID: PMCPMC7114536.
Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S, et al. ICTV Virus taxonomy profile: Partitiviridae. J Gen Virol. 2018;99(1):17–8. https://doi.org/10.1099/jgv.0.000985 . Epub 2017/12/08 PubMed PMID: 29214972; PubMed Central PMCID: PMCPMC5882087.
Mifsud JCO, Gallagher RV, Holmes EC, Geoghegan JL. Transcriptome mining expands knowledge of RNA viruses across the plant Kingdom. J Virol. 2022;96(24):e0026022. https://doi.org/10.1128/jvi.00260-22 . Epub 2022/06/01 PubMed PMID: 35638822; PubMed Central PMCID: PMCPMC9769393.
Tamada T, Kondo H. Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors. J Gen Plant Pathol. 2013;79:307–20.
Dolja VV, Krupovic M, Koonin EV. Deep roots and splendid boughs of the global plant virome. Annu Rev Phytopathol. 2020;58:23–53.
Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, et al. Global organization and proposed Megataxonomy of the virus world. Microbiol Mol Biol Rev. 2020;84(2):e00061. https://doi.org/10.1128/MMBR.00061-19 . Epub 2020/03/07 PubMed PMID: 32132243; PubMed Central PMCID: PMCPMC7062200.
Osono T. Role of phyllosphere fungi of forest trees in the development of decomposer fungal communities and decomposition processes of leaf litter. Can J Microbiol. 2006;52(8):701–16. https://doi.org/10.1139/w06-023 . Epub 2006/08/19 PubMed PMID: 16917528.
Li Z, Pan D, Wei G, Pi W, Zhang C, Wang JH, et al. Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J. 2021;15(8):2366–78. https://doi.org/10.1038/s41396-021-00932-y . Epub 2021/03/03 PubMed PMID: 33649554; PubMed Central PMCID: PMCPMC8319345.
Hierweger MM, Koch MC, Rupp M, Maes P, Di Paola N, Bruggmann R, et al. Novel Filoviruses, Hantavirus, and Rhabdovirus in freshwater fish, Switzerland, 2017. Emerg Infect Dis. 2021;27(12):3082–91. https://doi.org/10.3201/eid2712.210491 . Epub 2021/11/23 PubMed PMID: 34808081; PubMed Central PMCID: PMCPMC8632185.
La Rosa G, Iaconelli M, Mancini P, Bonanno Ferraro G, Veneri C, Bonadonna L, et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ. 2020;736:139652. https://doi.org/10.1016/j.scitotenv.2020.139652 . Epub 2020/05/29 PubMed PMID: 32464333; PubMed Central PMCID: PMCPMC7245320.
Sutela S, Poimala A, Vainio EJ. Viruses of fungi and oomycetes in the soil environment. FEMS Microbiol Ecol. 2019;95(9):fiz119. https://doi.org/10.1093/femsec/fiz119 . Epub 2019/08/01 PubMed PMID: 31365065.
Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci U S A. 2010;107(18):8387–92. https://doi.org/10.1073/pnas.0913535107 . Epub 2010/04/21 PubMed PMID: 20404139; PubMed Central PMCID: PMCPMC2889581.
Li P, Wang S, Zhang L, Qiu D, Zhou X, Guo L. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci Adv. 2020;6(14):eaay9634. https://doi.org/10.1126/sciadv.aay9634 . Epub 2020/04/15 PubMed PMID: 32284975; PubMed Central PMCID: PMCPMC7138691.
Khalifa ME, MacDiarmid RM. A mechanically transmitted DNA Mycovirus is targeted by the defence machinery of its host, Botrytis cinerea. Viruses. 2021;13(7):1315. https://doi.org/10.3390/v13071315 . Epub 2021/08/11 PubMed PMID: 34372522; PubMed Central PMCID: PMCPMC8309985.
Yu X, Li B, Fu Y, Xie J, Cheng J, Ghabrial SA, et al. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc Natl Acad Sci U S A. 2013;110(4):1452–7. https://doi.org/10.1073/pnas.1213755110 . Epub 2013/01/09 PubMed PMID: 23297222; PubMed Central PMCID: PMCPMC3557086.
Nuss DL. Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol. 2005;3(8):632–42. https://doi.org/10.1038/nrmicro1206 . Epub 2005/08/03 PubMed PMID: 16064055.
Coenen A, Kevei F, Hoekstra RF. Factors affecting the spread of double-stranded RNA viruses in Aspergillus nidulans. Genet Res. 1997;69(1):1–10. https://doi.org/10.1017/s001667239600256x . Epub 1997/02/01 PubMed PMID: 9164170.
Freitas CSA, Maciel LF, Dos Correa Santos RA, Costa O, Maia FCB, Rabelo RS, et al. Bacterial volatile organic compounds induce adverse ultrastructural changes and DNA damage to the sugarcane pathogenic fungus Thielaviopsis ethacetica. Environ Microbiol. 2022;24(3):1430–53. https://doi.org/10.1111/1462-2920.15876 . Epub 2022/01/08 PubMed PMID: 34995419.
Martinez-Alvarez P, Vainio EJ, Botella L, Hantula J, Diez JJ. Three mitovirus strains infecting a single isolate of Fusarium circinatum are the first putative members of the family Narnaviridae detected in a fungus of the genus Fusarium. Arch Virol. 2014;159(8):2153–5. https://doi.org/10.1007/s00705-014-2012-8 . Epub 2014/02/13 PubMed PMID: 24519462.
Deakin G, Dobbs E, Bennett JM, Jones IM, Grogan HM, Burton KS. Multiple viral infections in Agaricus bisporus - characterisation of 18 unique RNA viruses and 8 ORFans identified by deep sequencing. Sci Rep. 2017;7(1):2469. https://doi.org/10.1038/s41598-017-01592-9 . Epub 2017/05/28 PubMed PMID: 28550284; PubMed Central PMCID: PMCPMC5446422.
Litov AG, Zueva AI, Tiunov AV, Van Thinh N, Belyaeva NV, Karganova GG. Virome of three termite species from Southern Vietnam. Viruses. 2022;14(5):860. https://doi.org/10.3390/v14050860 . Epub 2022/05/29 PubMed PMID: 35632601; PubMed Central PMCID: PMCPMC9143207.
Hu J, Neoh KB, Appel AG, Lee CY. Subterranean termite open-air foraging and tolerance to desiccation: Comparative water relation of two sympatric Macrotermes spp. (Blattodea: Termitidae). Comp Biochem Physiol A Mol Integr Physiol. 2012;161(2):201–7. https://doi.org/10.1016/j.cbpa.2011.10.028 . Epub 2011/11/17 PubMed PMID: 22085890.
Kondo H, Botella L, Suzuki N. Mycovirus diversity and evolution revealed/inferred from recent studies. Annu Rev Phytopathol. 2022;60:307–36. https://doi.org/10.1146/annurev-phyto-021621-122122 . Epub 2022/05/25 PubMed PMID: 35609970.
Fukushi T. Relationships between propagative rice viruses and their vectors. 1969.
Google Scholar
Sun L, Kondo H, Bagus AI. Cross-kingdom virus infection. Encyclopedia of Virology: Volume 1–5. 4th Ed. Elsevier; 2020. pp. 443–9. https://doi.org/10.1016/B978-0-12-809633-8.21320-4 .
Bian R, Andika IB, Pang T, Lian Z, Wei S, Niu E, et al. Facilitative and synergistic interactions between fungal and plant viruses. Proc Natl Acad Sci U S A. 2020;117(7):3779–88. https://doi.org/10.1073/pnas.1915996117 . Epub 2020/02/06 PubMed PMID: 32015104; PubMed Central PMCID: PMCPMC7035501.
Chiapello M, Rodriguez-Romero J, Ayllon MA, Turina M. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol. 2020;6(2):veaa058. https://doi.org/10.1093/ve/veaa058 . Epub 2020/12/17 PubMed PMID: 33324489; PubMed Central PMCID: PMCPMC7724247.
Sutela S, Forgia M, Vainio EJ, Chiapello M, Daghino S, Vallino M, et al. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol. 2020;6(2):veaa076. https://doi.org/10.1093/ve/veaa076 . Epub 2020/12/17 PubMed PMID: 33324490; PubMed Central PMCID: PMCPMC7724248.
Zhou K, Zhang F, Deng Y. Comparative analysis of viromes identified in multiple macrofungi. Viruses. 2024;16(4):597. https://doi.org/10.3390/v16040597 . Epub 2024/04/27 PubMed PMID: 38675938; PubMed Central PMCID: PMCPMC11054281.
Siddell SG, Smith DB, Adriaenssens E, Alfenas-Zerbini P, Dutilh BE, Garcia ML, et al. Virus taxonomy and the role of the International Committee on Taxonomy of Viruses (ICTV). J Gen Virol. 2023;104(5):001840. https://doi.org/10.1099/jgv.0.001840 . Epub 2023/05/04 PubMed PMID: 37141106; PubMed Central PMCID: PMCPMC10227694.
Download references
Acknowledgements
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; WZ and CZ contributed to the conception and design; XL, ZD, JXU, WL and PN contributed to the collection and assembly of data; XL, ZD and JXE contributed to the data analysis and interpretation.
This research was supported by National Key Research and Development Programs of China [No.2023YFD1801301 and 2022YFC2603801] and the National Natural Science Foundation of China [No.82341106].
Author information
Xiang Lu, Ziyuan Dai and Jiaxin Xue are equally contributed to this works.
Authors and Affiliations
Institute of Critical Care Medicine, The Affiliated People’s Hospital, Jiangsu University, Zhenjiang, 212002, China
Xiang Lu & Wen Zhang
Department of Microbiology, School of Medicine, Jiangsu University, Zhenjiang, 212013, China
Xiang Lu, Jiaxin Xue & Wen Zhang
Department of Clinical Laboratory, Affiliated Hospital 6 of Nantong University, Yancheng Third People’s Hospital, Yancheng, Jiangsu, China
Clinical Laboratory Center, The Affiliated Taizhou People’s Hospital of Nanjing Medical University, Taizhou, 225300, China
Wang Li, Ping Ni, Juan Xu, Chenglin Zhou & Wen Zhang
You can also search for this author in PubMed Google Scholar
Contributions
Corresponding authors.
Correspondence to Juan Xu , Chenglin Zhou or Wen Zhang .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary material 1., supplementary material 2., supplementary material 3., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Lu, X., Dai, Z., Xue, J. et al. Discovery of novel RNA viruses through analysis of fungi-associated next-generation sequencing data. BMC Genomics 25 , 517 (2024). https://doi.org/10.1186/s12864-024-10432-w
Download citation
Received : 19 March 2024
Accepted : 20 May 2024
Published : 27 May 2024
DOI : https://doi.org/10.1186/s12864-024-10432-w
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
BMC Genomics
ISSN: 1471-2164
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Published: 27 May 2024
A single infusion of engineered long-lived and multifunctional T cells confers durable remission of asthma in mice
- Gang Jin 1 , 2 , 3 na1 ,
- Yanyan Liu 1 , 2 , 3 na1 ,
- Lixia Wang ORCID: orcid.org/0000-0001-7991-4013 1 , 2 , 3 ,
- Zihao He 1 , 2 , 3 ,
- Xiaocui Zhao 1 , 2 , 3 ,
- Yuying Ma 1 , 2 , 3 ,
- Yuting Jia 2 ,
- Zhuoyang Li ORCID: orcid.org/0009-0006-1379-8279 1 , 2 , 3 ,
- Na Yin 1 , 2 , 3 &
- Min Peng ORCID: orcid.org/0000-0002-5885-0660 1 , 2 , 3
Nature Immunology volume 25 , pages 1059–1072 ( 2024 ) Cite this article
3783 Accesses
1 Citations
274 Altmetric
Metrics details
- Immunotherapy
- Inflammatory diseases
Asthma, the most prevalent respiratory disease, affects more than 300 million people and causes more than 250,000 deaths annually. Type 2-high asthma is characterized by interleukin (IL)-5-driven eosinophilia, along with airway inflammation and remodeling caused by IL-4 and IL-13. Here we utilize IL-5 as the targeting domain and deplete BCOR and ZC3H12A to engineer long-lived chimeric antigen receptor (CAR) T cells that can eradicate eosinophils. We call these cells immortal-like and functional IL-5 CAR T cells (5T IF ) cells. 5T IF cells were further modified to secrete an IL-4 mutein that blocks IL-4 and IL-13 signaling, designated as 5T IF 4 cells. In asthma models, a single infusion of 5T IF 4 cells in fully immunocompetent mice, without any conditioning regimen, led to sustained repression of lung inflammation and alleviation of asthmatic symptoms. These data show that asthma, a common chronic disease, can be pushed into long-term remission with a single dose of long-lived CAR T cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
195,33 € per year
only 16,28 € per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Selective haematological cancer eradication with preserved haematopoiesis

Cancer therapy with antibodies
Engineered CD47 protects T cells for enhanced antitumour immunity
Data availability.
RNA-seq and scRNA-seq data were deposited in the Gene Expression Omnibus under accession numbers GSE262359 and GSE262360 , respectively. All other data are available in the article and Supplementary Information or from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
The code utilized in this study was crafted using openly accessible R (v4.1.2) packages, such as DESeq2, ggplot2, pheatmap, clusterProfiler and Seurat. No custom software was generated for the purpose of this research. Upon request, the analysis scripts will be made available.
Hinrichs, C. S. & Rosenberg, S. A. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol. Rev. 257 , 56–71 (2014).
Article CAS PubMed PubMed Central Google Scholar
Neelapu, S. S. et al. Chimeric antigen receptor T-cell therapy—assessment and management of toxicities. Nat. Rev. Clin. Oncol. 15 , 47–62 (2018).
Article CAS PubMed Google Scholar
Luke, J. J., Flaherty, K. T., Ribas, A. & Long, G. V. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 14 , 463–482 (2017).
Kochenderfer, J. N. et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 116 , 4099–4102 (2010).
Israel, E. & Reddel, H. K. Severe and difficult-to-treat asthma in adults. N. Engl. J. Med. 377 , 965–976 (2017).
Papi, A., Brightling, C., Pedersen, S. E. & Reddel, H. K. Asthma. Lancet 391 , 783–800 (2018).
Article PubMed Google Scholar
Hammad, H. & Lambrecht, B. N. The basic immunology of asthma. Cell 184 , 2521–2522 (2021).
Brusselle, G. G. & Koppelman, G. H. Biologic therapies for severe asthma. N. Engl. J. Med. 386 , 157–171 (2022).
Hearn, A. P., Kent, B. D. & Jackson, D. J. Biologic treatment options for severe asthma. Curr. Opin. Immunol. 66 , 151–160 (2020).
Jacobsen, E. A. et al. Eosinophil knockout humans: uncovering the role of eosinophils through eosinophil-directed biological therapies. Annu. Rev. Immunol. 39 , 719–757 (2021).
Bleecker, E. R. et al. Efficacy and safety of benralizumab for patients with severe asthma uncontrolled with high-dosage inhaled corticosteroids and long-acting beta(2)-agonists (SIROCCO): a randomised, multicentre, placebo-controlled phase 3 trial. Lancet 388 , 2115–2127 (2016).
FitzGerald, J. M. et al. Benralizumab, an anti-interleukin-5 receptor alpha monoclonal antibody, as add-on treatment for patients with severe, uncontrolled, eosinophilic asthma (CALIMA): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet 388 , 2128–2141 (2016).
Wang, L. et al. Induction of immortal-like and functional CAR T cells by defined factors. J. Exp. Med. 221 , e20232368 (2024).
Cornelis, S. et al. Detailed analysis of the IL-5-IL-5R alpha interaction: characterization of crucial residues on the ligand and the receptor. EMBO J. 14 , 3395–3402 (1995).
Dudley, M. E. et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 298 , 850–854 (2002).
Murad, J. P. et al. Pre-conditioning modifies the TME to enhance solid tumor CAR T cell efficacy and endogenous protective immunity. Mol. Ther. 29 , 2335–2349 (2021).
Amor, C. et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature 583 , 127–132 (2020).
Mackensen, A. et al. Anti-CD19 CAR T cell therapy for refractory systemic lupus erythematosus. Nat. Med. 28 , 2124–2132 (2022).
Rosenberg, H. F., Dyer, K. D. & Foster, P. S. Eosinophils: changing perspectives in health and disease. Nat. Rev. Immunol. 13 , 9–22 (2013).
Wenzel, S., Wilbraham, D., Fuller, R., Getz, E. B. & Longphre, M. Effect of an interleukin-4 variant on late phase asthmatic response to allergen challenge in asthmatic patients: results of two phase 2a studies. Lancet 370 , 1422–1431 (2007).
Grunewald, S. M. et al. A murine interleukin-4 antagonistic mutant protein completely inhibits interleukin-4-induced cell proliferation, differentiation, and signal transduction. J. Biol. Chem. 272 , 1480–1483 (1997).
Grunewald, S. M. et al. Antagonistic IL-4 mutant prevents type I allergy in the mouse: Inhibition of the IL-4/IL-13 receptor system completely abrogates humoral immune response to allergen and development of allergic symptoms in vivo. J. Immunol. 160 , 4004–4009 (1998).
Kouro, T., Ikutani, M., Kariyone, A. & Takatsu, K. Expression of IL-5Rα on B-1 cell progenitors in mouse fetal liver and involvement of Bruton’s tyrosine kinase in their development. Immunol. Lett. 123 , 169–178 (2009).
Dubucquoi, S. et al. Interleukin 5 synthesis by eosinophils: association with granules and immunoglobulin-dependent secretion. J. Exp. Med. 179 , 703–708 (1994).
Davoine, F. & Lacy, P. Eosinophil cytokines, chemokines, and growth factors: emerging roles in immunity. Front Immunol. 5 , 570 (2014).
Article PubMed PubMed Central Google Scholar
Turner, D. L. et al. Biased generation and in situ activation of lung tissue-resident memory CD4 T cells in the pathogenesis of allergic asthma. J. Immunol. 200 , 1561–1569 (2018).
Ulrich, B. J. et al. Allergic airway recall responses require IL-9 from resident memory CD4 + T cells. Sci. Immunol. 7 , eabg9296 (2022).
McLane, L. M., Abdel-Hakeem, M. S. & Wherry, E. J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37 , 457–495 (2019).
Matsushita, K. et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458 , 1185–1190 (2009).
Kotov, J. A. et al. BCL6 corepressor contributes to Th17 cell formation by inhibiting Th17 fate suppressors. J. Exp. Med. 216 , 1450–1464 (2019).
Kishimoto, T. & Kang, S. IL-6 revisited: from rheumatoid arthritis to CAR T cell therapy and COVID-19. Annu. Rev. Immunol. 40 , 323–348 (2022).
Anderson, K. G. et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9 , 209–222 (2014).
Mueller, S. N. & Mackay, L. K. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16 , 79–89 (2016).
Wong, T. W., Doyle, A. D., Lee, J. J. & Jelinek, D. F. Eosinophils regulate peripheral B cell numbers in both mice and humans. J. Immunol. 192 , 3548–3558 (2014).
Berek, C. Eosinophils: important players in humoral immunity. Clin. Exp. Immunol. 183 , 57–64 (2016).
Yang, G. et al. Anti-IL-13 monoclonal antibody inhibits airway hyperresponsiveness, inflammation and airway remodeling. Cytokine 28 , 224–232 (2004).
Aegerter, H. & Lambrecht, B. N. The pathology of asthma: what is obstructing our view?. Annu. Rev. Pathol. 18 , 387–409 (2023).
Gause, W. C., Rothlin, C. & Loke, P. Heterogeneity in the initiation, development and function of type 2 immunity. Nat. Rev. Immunol. 20 , 603–614 (2020).
Chen, S. et al. Treatment of allergic eosinophilic asthma through engineered IL-5-anchored chimeric antigen receptor T cells. Cell Discov. 8 , 80 (2022).
Hitoshi, Y. et al. IL-5 receptor positive B cells, but not eosinophils, are functionally and numerically influenced in mice carrying the X-linked immune defect. Int. Immunol. 5 , 1183–1190 (1993).
Agache, I. et al. Efficacy and safety of treatment with biologicals (benralizumab, dupilumab, mepolizumab, omalizumab and reslizumab) for severe eosinophilic asthma. A systematic review for the EAACI Guidelines - recommendations on the use of biologicals in severe asthma. Allergy 75 , 1023–1042 (2020).
Xing, X. & Hu, X. Risk factors of cytokine release syndrome: stress, catecholamines, and beyond. Trends Immunol. 44 , 93–100 (2023).
Rafiq, S. et al. Targeted delivery of a PD-1-blocking scFv by CAR-T cells enhances anti-tumor efficacy in vivo. Nat. Biotechnol. 36 , 847–856 (2018).
Allen, G. M. et al. Synthetic cytokine circuits that drive T cells into immune-excluded tumors. Science 378 , eaba1624 (2022).
Debeuf, N., Haspeslagh, E., van Helden, M., Hammad, H. & Lambrecht, B. N. Mouse models of asthma. Curr. Protoc. Mouse Biol. 6 , 169–184 (2016).
Grunewald, S. M. et al. An antagonistic IL-4 mutant prevents type I allergy in the mouse: inhibition of the IL-4/IL-13 receptor system completely abrogates humoral immune response to allergen and development of allergic symptoms in vivo. J. Immunol. 160 , 4004–4009 (1998).
Tomkinson, A. et al. A murine IL-4 receptor antagonist that inhibits IL-4- and IL-13-induced responses prevents antigen-induced airway eosinophilia and airway hyperresponsiveness. J. Immunol. 166 , 5792–5800 (2001).
Myou, S. et al. Blockade of inflammation and airway hyperresponsiveness in immune-sensitized mice by dominant-negative phosphoinositide 3-kinase-TAT. J. Exp. Med. 198 , 1573–1582 (2003).
Tanaka, H. et al. The effect of allergen-induced airway inflammation on airway remodeling in a murine model of allergic asthma. Inflamm. Res 50 , 616–624 (2001).
Download references
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (82350108, to M.P.), Vanke Special Fund for Public Health and Health Discipline Development Tsinghua University (2022Z82WKJ013, to M.P.), Tsinghua University DUSHI Program (52302102323, to M.P.), Tsinghua-Peking Center for Life Sciences (to M.P.) and SXMU-Tsinghua Collaborative Innovation Center for Frontier Medicine (to M.P.).
Author information
These authors contributed equally: Gang Jin, Yanyan Liu.
Authors and Affiliations
State Key Laboratory of Molecular Oncology, Institute for Immunology, Beijing Key Laboratory for Immunological Research on Chronic Diseases, School of Basic Medical Sciences, Tsinghua University, Beijing, China
Gang Jin, Yanyan Liu, Lixia Wang, Zihao He, Xiaocui Zhao, Yuying Ma, Zhuoyang Li, Na Yin & Min Peng
SXMU-Tsinghua Collaborative Innovation Center for Frontier Medicine, Shanxi Medical University, Taiyuan, China
Gang Jin, Yanyan Liu, Lixia Wang, Zihao He, Xiaocui Zhao, Yuying Ma, Yuting Jia, Zhuoyang Li, Na Yin & Min Peng
Tsinghua-Peking Center for Life Sciences, Beijing, China
You can also search for this author in PubMed Google Scholar
Contributions
G.J. and Y.L. performed experiments and analyzed the data; L.W., Z.H., X.Z., Y.M., Y.J., Z.L. and N.Y. provided technical help; N.Y. supervised the project; M.P. conceived and supervised the project, analyzed and interpreted data, and wrote the paper with inputs from all authors.
Corresponding author
Correspondence to Min Peng .
Ethics declarations
Competing interests.
A patent application based on the findings described in this study has been filed by Tsinghua University with M.P. listed as an inventor. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Immunology thanks Hirohito Kita, Rod Rahimi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: N. Bernard, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended data fig. 1 il-5 car t cells do not expand nor kill eosinophils in vivo..
a , Experimental design. Activated CD8 + T cells were transduced with IL-5 CAR (Thy1.1 + ), and one million Thy1.1 + CD8 + cells were transferred into B6 mice without conditioning. IL-5 CAR T cells (5T) and eosinophils (Siglec-F + SSC hi ) in peripheral blood were examined 7 days post-transfer. b-d , Representative plots ( b , c ) and statistics ( d ) of 5T and eosinophils in peripheral blood are shown (n = 4). e-j , Activated CD8 + T cells were transduced with IL-5 CAR (Thy1.1 + ), and 1, 3, or 5 million of Thy1.1 + CD8 + cells were transferred into B6 mice without conditioning. 5T and eosinophils in blood were examined 7- and 28-days post-transfer. On 28 days post-transfer, 5T and eosinophils in spleen (SPL) and bone marrow (BM) were examined. Representative FACS plots ( e ) and statistics ( f , g ) of 5T and eosinophils in blood are shown (n = 4). Representative FACS plots ( h ) and statistics ( i , j ) of 5T and eosinophils in SPL and BM are shown (n = 4). Data represent mean ± SEM from one of two independent experiments. n = number of mice. Exact p values were determined by two-tailed unpaired Student’s t-test in ( d ), one-way ANOVA for Tukey’s multiple-comparisons test in ( f , g , i , j ); ns, not significant.
Source data
Extended data fig. 2 the generation, phenotype and safety of 5t if..
a , A one-vector system for expressing IL-5 CAR and sgRNA. b , The editing of Bcor and Zc3h12a was validated by genomic PCR (left), Sanger sequencing (middle), and quantitative PCR (right, n = 3 biologically independent samples). Representative data from three independent experiments. c − f , Representative FACS plots ( c , e ) and statistics ( d , f ) of the expression of Thy1.1 and IL-5 CAR on the plasma membrane of CD8 + T cells co-expressing indicated sgRNAs (n = 4 or 3 biologically independent samples). g , h , Representative FACS plots ( g ) and statistics ( h ) of eosinophils (Siglec-F + SSC hi ) in spleen of indicated mice (3 months post-transfer) are shown (n = 4 in PBS and 5T IF (response) group, n = 3 in 5T IF (relapse) group). i , j , Representative FACS plots ( i ) and statistics ( j ) of IL5Rα expression on eosinophils from indicated mice are shown (n = 4). k , l , B6 mice were transferred with PBS or one million 5T IF at 6-week-old age, then monitored for health status and analyzed 3 months post-transfer. Representative t-SNE plots derived from FACS data ( k ) and statistics ( l ) are shown (n = 4). EOS, eosinophils (CD11b + CD11c - Siglec-F + ); NEU, neutrophils (CD11b + Ly6G + ); MON, monocytes (CD11b + CD11c - Siglec-F - Ly6G − ); T, T cells (CD3 + ); B, B cells (B220 + ). m , n , FACS analysis of the activation status of endogenous T cells from spleen. Representative FACS plots ( m ) and statistics ( n ) are shown (n = 4). Data represent mean ± SEM from one of two independent experiments. n = number of mice. Exact p values were determined by two-tailed unpaired Student’s t-test in ( j , l ), one-way ANOVA for Tukey’s multiple-comparisons test in ( f , h ); ns, not significant.
Extended Data Fig. 3 5T IF 4 maintain their specificity and cytotoxicity against target cells.
a , The construct for expression of IL-4 mutein. b , Co-transduction strategy for 5T IF 4 production. c , d , FACS analysis of Thy1.1 and IL-5 CAR expression on the plasma membrane of CD8 + T cells. Representative FACS plots ( c ) and statistics ( d ) are shown (n = 3). e , In vitro killing of IL-5Rα + and IL-5Rα - MC38 tumor cells by indicated CAR T cells at the effector vs target (E: T) ratio of 3:1 (n = 4). f , In vitro killing of mouse eosinophils by indicated IL-5 CAR T cells (5T) and non-transduced activated T cells (control) at the E:T ratio of 3:1 (n = 4). g , h , FACS analysis of IL5Rα expression on the plasma membrane of indicated cells. Representative FACS plots ( g ) and statistics ( h ) are shown (n = 4). i , In vitro killing of mouse resting non-activated B cells and LPS-activated B cells by indicated CAR T cells at the E:T ratio of 3:1 (n = 4). j , k , LPS-activated B cells were transduced with retrovirus overexpressing (OE) mIL-5Rα, and the expression of mIL-5Rα on transduced B cells was examined by FACS. Representative plots ( j ) and statistics ( k ) are shown (n = 4). l , In vitro killing of indicated B cells by 5T at the E:T ratio of 3:1 (n = 4). m , Cytotoxicity of indicated CAR T cells against activated B cells overexpressing mIL-5Rα (n = 4). n - t , Cytotoxic effects of indicated CAR T cells on plasma cells. n , In vitro co-culture experiment strategy. o , p , FACS analysis of plasma cells (induced by LPS and IL-4 in vitro) after co-culture with indicated CAR T cells. Representative FACS plots ( o ) and statistics ( p ) are shown (n = 4). q , r , FACS analysis of IgG1 + B cells after co-culture with indicated CAR T cells. Representative FACS plots ( q ) and statistics ( r ) are shown (n = 4). s , t , FACS analysis of IgE + B cells after co-culture with indicated CAR T cells. Representative FACS plots ( s ) and statistics ( t ) are shown (n = 4). Data represent mean ± SEM from one of two independent experiments. n = number of biologically independent samples. Exact p values were determined by two-tailed unpaired Student’s t-test in ( d , e , f ), one-way ANOVA for Tukey’s multiple-comparisons test in ( h, k , l ); ns, not significant.
Extended Data Fig. 4 5T IF 4 suppress memory Th2 cells in the lung.
a , Experimental design for OVA-induced memory Th2 cells. b - d , FACS analysis of IL-4 + , IL-5 + , and IL-13 + production by endogenous CD4 + T cells isolated from lung (n = 4). Data represent mean ± SEM from one of two independent experiments. n = number of mice. Exact p values were determined by one-way ANOVA for Tukey’s multiple-comparisons test in ( b, c , d ); ns, not significant.
Extended Data Fig. 5 Characterization of 5T IF and 5T IF 4.
a - c , FACS analysis of IL-5 CAR T cells (5T) and eosinophils in peripheral blood 2 weeks post-infusion. Representative plots ( a , b ) and statistics ( c ) are shown (n = 4). d - o , Cellular phenotypes of 5T IF and 5T IF 4 were examined 2 months post-transfer. d , e , FACS gating of 5T from spleen of mice. Representative plots ( d ) and statistics ( e ) are shown (n = 4). f , g , FACS gating of CD44 + CD62L + cells of endogenous CD8 + T cells from PBS group and Thy1.1 + 5T from indicated groups. Representative plots ( f ) and statistics ( g ) are shown (n = 4). h , i , FACS analysis of the expression of indicated proteins by endogenous CD8 + T cells and Thy1.1 + 5T. Representative plots ( h ) and statistical analysis of mean fluorescence intensity (MFI) ( i ) are shown (n = 4). j , k , FACS gating of Ly108 + CD62L + cells of endogenous CD8 + T cells from PBS group and Thy1.1 + 5T from indicated groups. Representative plots ( j ) and statistics ( k ) are shown (n = 4). l , m , FACS gating of PD-1 + TIM-3 + cells and PD-1 + TIM-3 - cells of endogenous CD8 + T cells from PBS group and Thy1.1 + 5T from indicated groups. Representative plots ( l ) and statistics ( m ) are shown (n = 4). n , o , FACS gating of Ly108 + CX3XR1 − cells and Ly108 − CX3XR1 + cells of endogenous CD8 + T cells from PBS group and Thy1.1 + 5T from indicated groups. Representative plots ( n ) and statistics ( o ) are shown (n = 4). Data represent mean ± SEM from one of two independent experiments. n = number of mice. Exact p values were determined by one-way ANOVA for Tukey’s multiple-comparisons test in ( c , e , g , i , k , m , o ); ns, not significant.
Extended Data Fig. 6 Transcriptional analysis of 5T IF and 5T IF 4.
B6 mice were transferred with one million 5T IF or 5T IF 4. CD8 + Thy1.1 + cells were FACS sorted from spleen 4 weeks post-transfer for mRNA extraction and RNA-seq analysis. Endogenous CD8 + T cells were used as control since wild-type IL-5 CAR T cells do not expand in mice. a , A volcano plot of differentially expressed genes (DEGs) between endogenous CD8 + T cells and 5T IF . b , A volcano plot of DEGs between 5T IF and 5T IF 4. c , Heatmaps showing the expression of selected genes. d , GSEA of 5T IF 4 using indicated gene sets. NES, normalized enrichment score; P , nominal P value. The nominal P value is calculated by comparing the tails of the observed and null distributions for the NES in ( d ). Two-sided Wald test in Deseq2 in ( a , b ).
Extended Data Fig. 7 Tissue distribution of 5T IF and 5T IF 4, and their impact on cytokines.
a , Experimental design for monitoring cytokine production in the serum. b , c , ELISA analysis of indicated cytokines in the serum of mice transferred with PBS, 5T IF , or 5T IF 4 at 1 week or 4 weeks post-transfer (n = 4). d - i , FACS analysis of 5T IF or 5T IF 4 in peripheral lymph nodes (pLN), spleen, liver, and lung of indicated mice one month post-transfer. Representative plots ( d , f ) and statistics ( e , g ) are shown (n = 4). h - j , Extravascular distribution of 5T IF and 5T IF 4 in the liver and lung. h , Experimental design of intravenous injection of αCD45-APC-Cy7 antibodies. i , j , FACS analysis of endogenous CD8 + T cells, 5T IF , and 5T IF 4 in liver and lung of indicated mice 4 weeks post-transfer. Representative plots ( i ) and statistics ( j ) are shown (n = 4). k , l , FACS analysis of CD69 and CD103 expression on indicated extravascular T cells in the lung. Representative plots ( k ) and statistics ( l ) are shown (n = 4). Data represent mean ± SEM from one of two independent experiments. n = number of mice. Exact p values were determined by one-way ANOVA for Tukey’s multiple-comparisons test in ( b , e , g , j , l ); ns, not significant.
Extended Data Fig. 8 Long-term safety of 5T IF and 5T IF 4.
a , Experimental design. Eight-week-old male B6 mice were transferred with PBS, one million 5T IF , or 5T IF 4, then monitored for health status and analyzed at 12 months post-transfer. b , Survival curve of mice from indicated groups (n = 8). c , Representative images of mice from indicated groups. d , Body weight of mice from indicated groups (n = 8). e , Images of spleen from indicated groups. f , g , FACS analysis of immune cells from spleen. Representative t-SNE plots derived from FACS data ( f ) and statistics ( g ) are shown (n = 4). EOS, eosinophils (CD11b + CD11c − Siglec − F + ); NEU, neutrophils (CD11b + Ly6G + ); MON, monocytes (CD11b + CD11c - Siglec-F - Ly6G − ); T, T cells (CD3 + ); B, B cells (B220 + ). h , i , FACS analysis of activation status of endogenous T cells from spleen. Representative FACS plots ( h ) and statistics ( i ) are shown (n = 4). j , H&E staining of slices of heart, kidney, liver, and lung. Representative sections are shown. Data represent mean ± SEM from one of two independent experiments. n = number of mice. Exact p values were determined by one-way ANOVA for Tukey’s multiple-comparisons test in ( g ).
Extended Data Fig. 9 Impact of 5T IF or 5T IF 4 on the health of mice.
a , Experimental design. b , c , FACS analysis of plasma cells in spleen of mice from indicated mice. Representative plots ( b ) and statistics ( c ) are shown (n = 4). d , ELISA analysis of IgM, IgG, IgE, IgG1, and IgG2a in serum of indicated mice 5 months post-transfer (n = 4). e , Experimental design of LCMV Armstrong virus (LCMV-Arm) infection. Mice transferred with PBS or 5T IF 4 for 4 weeks were infected with LCMV-Arm. f - h , FACS analysis of GP33-tetramer + CD44 + cells in endogenous CD8 + T cells. Representative plots ( f ) and statistics ( g , h ) are shown (n = 6). i , j , FACS analysis of IFNγ + cells in endogenous CD8 + T cells. Representative plots ( i ) and statistics ( j ) are shown (n = 6). k , Experimental design for fertility test. l , The number of pups and litters. Data represent mean ± SEM from one of two independent experiments. n = number of mice. Exact p values were determined by two-tailed unpaired Student’s t-test in ( g , h , j ), one-way ANOVA for Tukey’s multiple-comparisons test in ( c , d ); ns, not significant.
Extended Data Fig. 10 Induction and characterization of human 5T IF 4.
a , A lentiviral construct for the expression of IL-5 CAR in human T cells. b , The expression of GFP and IL-5 CAR on the plasma membrane of human T cells (gated on CD8 + T cells). A representative FACS plot is shown. c , In vitro killing of IL-5Rα + and IL-5Rα - 143B tumor cells by indicated CAR T cells at the E:T ratio of 1:1 (n = 4). d , Dose response of IL-5 CAR T cells (5T) killing of IL-5Rα + 143B tumor cells at indicated E:T ratios (n = 3). e , Lentiviral vectors for the expression of IL-5 CAR, sgRNA, and IL-4 mutein in human T cells. f , The expression of IL-5 CAR (GFP + IL-5 + ) and IL-4 mutein (GFP + IL-5 - ) by human T cells. A representative FACS plot is shown. g , IL-4 mutein in supernatants of 5T and h5T IF 4 examined by ELISA (n = 4). h , The editing of BCOR and ZC3H12A loci in h5T IF 4 was examined by sequencing. Representative tracks are shown. i , Clustered heatmap of representative marker genes across C0 to C8. j , UMAP plots of representative marker genes across all clusters. k , Violin plots of inflammation-related gene expression among 9 clusters. Data represent mean ± SEM from one of two independent experiments. n = number of biologically independent samples. Exact p values were determined by two-tailed unpaired Student’s t-test in ( c , g ); ns, not significant.
Supplementary information
Supplementary information.
Supplementary Figs. 1–5.
Reporting Summary
Supplementary data.
Source data for Supplementary Figs. 1–5.
Source Data Figs. 1–8 and Extended Data Figs. 1–10
Statistical source data for Figs. 1–8 and Extended Data Figs. 1–10.
Source Data Fig. 4 and Extended Data Fig. 2
Unprocessed western blots and/or gels for Fig. 4b and Extended Data Fig. 2b.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Cite this article.
Jin, G., Liu, Y., Wang, L. et al. A single infusion of engineered long-lived and multifunctional T cells confers durable remission of asthma in mice. Nat Immunol 25 , 1059–1072 (2024). https://doi.org/10.1038/s41590-024-01834-9
Download citation
Received : 16 July 2023
Accepted : 06 April 2024
Published : 27 May 2024
Issue Date : June 2024
DOI : https://doi.org/10.1038/s41590-024-01834-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Car t cells put the brakes on asthma.
- Bart N. Lambrecht
- Hamida Hammad
Nature Immunology (2024)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- Open access
- Published: 04 June 2024
Single-cell RNA sequencing opens a new era for cotton genomic research and gene functional analysis
- Pan Xiaoping 1 ,
- Peng Renhai 2 &
- Zhang Baohong ORCID: orcid.org/0000-0002-9308-4340 1
Journal of Cotton Research volume 7 , Article number: 17 ( 2024 ) Cite this article
Metrics details
Single-cell RNA sequencing (scRNA-seq) is one of the most advanced sequencing technologies for studying transcriptome landscape at the single-cell revolution. It provides numerous advantages over traditional RNA-seq. Since it was first used to profile single-cell transcriptome in plants in 2019, it has been extensively employed to perform different research in plants. Recently, scRNA-seq was also quickly adopted by the cotton research community to solve lots of scientific questions which have been never solved. In this comment, we highlighted the significant progress in employing scRNA-seq to cotton genetic and genomic study and its future potential applications.
Introduction
With the quick development of next generation sequencing technology, including RNA sequencing (RNA-seq), in the past two decades, great progress has been made in the cotton genome and transcriptome landscape (Peng et al., 2021 ; Wen et al., 2023 ). The expression profiles have been sequenced in different tissue, organs, and cells under different conditions, which enhance our understanding of individual genes, including both protein-coding and non-coding genes (such as microRNAs), in cotton growth and development as well as response to different environmental stresses, which has fueled lots of discovery and innovation in cotton gene functional study in recent years (Peng et al., 2021 ; Wen et al., 2023 ). However, as we know, traditional RNA-seq generally performes gene expression on samples with mixed cell populations that contents at least thousands of different cells, this limited and even hindered us from understanding the functions of an individual gene in cell-type heterogeneity of biological samples at a single cell revolution level. The quick development of single-cell RNA sequencing technology revolutionized gene expression profiling analysis. Since it was first employed to profile the transcriptome in different Arabidopsis root cells and others in 2019 (Denyer et al., 2019 ; Horvath et al., 2019 ; Jean-Baptiste et al., 2019 ; Nelms et al., 2019 ; Ryu et al., 2019 ; Shulse et al., 2019 ; Zhang et al., 2019 ), scRNA-seq has been attracting more and more attention from both scientific and industrial communities, and the number of publications on scRNA-seq has been dramatically increased. Right now, the studied plant species not only include model plant species, such as Arabidopsis and rice, but also many agriculturally important crops, such as corn (Satterlee et al., 2020 ; Xu et al., 2021 ), peanut (Liu et al., 2021 ; Deng et al., 2024 ), and tobacco (Feng et al., 2023 ). The investigated field includes plant tissue and organ development and response to various environmental factors, including plant nutrient deficiency.
scRNA-seq is also revolutionizing cotton transcriptome and gene functional analysis
Due to its great potential to distinguish transcriptomes in different types of cells even adjacent cells, scRNA-seq has also quickly attracted lots of attention from cotton research communities (Table 1 ). In 2022, Qin et al. ( 2022 ) reported, for the first time, scRNA-seq was employed to study gene expression in cotton. They identified a total of 14 535 cells from cotton ovule cells, and these cells were divided into three major cell types, including fiber cells. Furthermore, they found that several core transcription factors, such as MYB25-like and HOX3, play a key role in regulating cotton fiber differentiation and tip-biased diffuse growth (Qin et al., 2022 ). Since then, in only about one year, at least 8 papers have been published utilizing scRNA-seq to study different biological processes in cotton (Table 1 ). Due to the fiber is the major product of cotton, there are always interest in investigating the molecular mechanisms underlying cotton fiber initiation and development. Following the research performed by Qin et al. ( 2022 ), Wang et al. ( 2023 ) also employed scRNA-seq to investigate the cell-specific clock-controlled gene expression landscape regulating rhythmic fiber cell growth. Their study used a well known cotton fibreless (fuzzless/lintless) mutant Xuzhou142fl and its wiltype Xue 142. Through scRNA-seq analysis, they identified five cell populations and successfully constructed the development trajectory for cotton finer lineage cells (Wang et al., 2023 ). Based on their study, they also found the circadian rhythmic process may regulate the primary growth of fiber cells, in which both small peptide RALF1 and certain cis -regulatory elements (such as CRE and TCPs) may play an important role (Wang et al., 2023 ).
scRNA-seq was also employed to study gland development and metabolite biosynthesis. Gossypol and its associated pigment gland are important for cotton evolution, which plays an important role for cotton plants against certain insects and diseases. Sun et al. ( 2023 ) employed scRNA-seq to study molecular mechanisms underlying gland morphogenesis using a unique wild cotton species, G. bickii , whose seeds are glandless but there are glands in other tissues. They found that both light and plant hormone gibberellin promoted the formation of pigment glands and three new genes, including ETHYLENE RESPONSE FACTOR 114 (ERF114), ZINC FINGER OF ARABIDOPSIS THALIANA 11 (ZAT11), and NAC TRANSCRIPTION FACTOR-LIKE 9 (NTL9) affected gland formation (Sun et al., 2023 ). Long et al. ( 2023 ) also employed scRNA-seq to study the pigment gland morphogenesis, in which the authors used two near-isogenic lines (NIL) (Gland cotton cultivar CCRI 12 and glandless cotton cultivar CCRI12gl); based on their results, several transcription factors, including PGF, ERF12, MYB14, and JUB1, serve as regulators for pigment gland morphogenesis in cotton. Recently, Lin et al. ( 2023 ) identified a hierarchical transcriptional regulatory network for terpenoid biosynthesis in cotton secretory glandular cells; they also found that two transcription factors, HSF4a and NAC42, directly affected terpenoid biosynthesis by targeting the expression of terpenoid biosynthetic genes.
Both transgenic and genome editing have become powerful tools for studying gene function and crop improvement (Zhang et al., 2021 ). However, the advanced techniques are majorly based on plant tissue culture and regeneration. Currently, compared with other plant species, it is hard to obtain transgenic or genome-edited regenerated plants because of the long period of culture from callus induction to plant regeneration and the genotype recalcitrance, only a few genotypes can be used for plant regeneration in cotton. Thus, studying the potential mechanisms underlying plant regeneration is extremely important. Recently, Zhu et al. ( 2023 ) employed scRNA-seq to compare the transcriptome profile between an easily regenerable cotton genotype Jin668, and the recalcitrant genotype, also cotton genetic standard, TM-1. Based on the single-cell levels of mRNA profiling, they identified nine putative cell clusters and 23 cluster-specific marker genes for these two different cotton genotypes with different regeneration capacities. Combining with transgenic and genome editing, they also found that certain genes, such as PLT3 , LOX3 , and LAX1/2 , are involved in reprogramming the fate of cotton cells and further control plant regeneration in cotton (Zhu et al., 2023 ). Guo et al. ( 2024 ) also employed scRNA-seq to analyze the transcriptome atlas during somatic embryogenesis in cotton; based on their scRNA-seq results, cotton calli were partitioned into four broad populations with six distinct cell clusters.
scRNA-seq was also used to study the transcriptome atlas and specific genes during cotton anther development, and plant response to environmental stress (Li et al., 2024a; 2024b). scRNA-seq shows that both high temperature and salinity stress induced certain gene expression aberrance in specific cell types in anther (Li et al., 2024b ) and root (Li et al., 2024a ), respectively (Table 1 ).
Summary and future prospects
Since scRNA-seq can be used to directly compare the gene expression profiling of an individual cell, it can assess transcriptional similarities and differences within a population of cells. Thus, it is possible for us for the first time to elucidate the cell heterogeneity and regulatory mechanism of each individual cell. Due to these unique features, scRNA-seq has been quickly adopted by the cotton research community to elucidate the molecular mechanism underlying many biological processes in cotton, such as fiber initiation and development, somatic embryogenesis, and response to different environmental stresses.
In scRNA-seq studies across various plant species, the initial and critical step of scRNA-seq is to isolate viable single cells from the targeted tissues. Because all plant cells have cell wall that not only provide support and protection to the cells but also allow the cells link together (Zhang et al., 2020 ), thus, for plant scRNA-seq, the first step generally is to isolate plant protoplast by using different cell wall breakdown methods and then followed by cell sorting using different methods, such as microfluidics. To achieve the high quality of cotton cell protoplast, there are a couple of explored methods for isolating protoplasts for cotton scRNA-seq (Liu et al., 2022 ; Zhang et al., 2023 ). Currently, most scRNA-seq studies in cotton employ protoplast isolation for harvesting single cell. However, enzymatic digestion-based protoplast preparation for scRNA-seq suffers one major weakness: no matter how good to isolate the protoplast, it needs a process to remove the cell walls which the process of protoplast preparation may be considered as cellular stress that will affect the gene expression levels. This suggests that some scRNA-seq results may pose false positives. Thus, better cell isolation methods may be considered to avoid these negative effects.
With the mature of scRNA-seq technology, more and more researchers will adopt this technique to explore new applications in cotton genomic research and crop improvement. Particularly as newly emerging technologies are created, such as the more advanced scRNA-seq, single cell stereo-sequencing technology (Stereo-seq) (Bawa et al., 2024 ), this technology will become more powerful for constructing spatially resolved developmental trajectories and the molecular regulatory mechanisms underlying cotton growth and development as well as yield and quality under different biotic and abiotic stresses. Combining scRNA-seq with the advanced clustered regularly interspaced short palindromic repeats (CRISPR) genome editing technology, the key genes controlling cotton fiber yield and quality as well as tolerance to stress will be identified. These findings will further be used for precision cotton molecular breed for creating new cultivars for sustainable cotton development.
Availability of data and materials
Not applicable.
Bawa G, Liu Z, Yu X, et al. Introducing single cell stereo-sequencing technology to transform the plant transcriptome landscape. Trends Plant Sci. 2024;29(2):249–65. https://doi.org/10.1016/j.tplants.2023.10.002 .
Article CAS PubMed Google Scholar
Deng Q, Du P, Gangurde SS, et al. scRNA-Seq reveals dark- and light-induced differentially expressed gene atlases of seedling leaves in Arachis hypogaea L. Plant Biotechnol J. 2024. https://doi.org/10.1111/pbi.14306 . Accessed 6 Mar 2024.
Article PubMed Google Scholar
Denyer T, Ma X, Klesen S, et al. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev Cell. 2019;48(6):840–52. https://doi.org/10.1016/j.devcel.2019.02.022 .
Feng Y, Zhao Y, Ma Y, et al. Single-cell transcriptome analyses reveal cellular and molecular responses to low nitrogen in burley tobacco leaves. Physiol Plant. 2023;175(6):e14118. https://doi.org/10.1111/ppl.14118 .
Guo H, Zhang L, Guo H, et al. Single-cell transcriptome atlas reveals somatic cell embryogenic differentiation features during regeneration. Plant Physiol. 2024:kiae107. https://doi.org/10.1093/plphys/kiae107 . Accessed 6 Mar 2024.
Horvath R, Laenen B, Takuno S, et al. Single-cell expression noise and gene-body methylation in Arabidopsis thaliana . Heredity (Edinb). 2019;123(2):81–91. https://doi.org/10.1038/s41437-018-0181-z .
Jean-Baptiste K, Mcfaline-Figueroa JL, Alexandre CM, et al. Dynamics of gene expression in single root cells of Arabidopsis thaliana . Plant Cell. 2019;31(5):993–1011. https://doi.org/10.1105/tpc.18.00785 .
Article CAS PubMed PubMed Central Google Scholar
Li P, Liu Q, Wei Y, et al. Transcriptional landscape of cotton roots in response to salt stress at single-cell resolution. Plant Commun. 2024a;5(2):100740. https://doi.org/10.1016/j.xplc.2023.100740 .
Li Y, Ma H, Wu Y, et al. Single-cell transcriptome atlas and regulatory dynamics in developing cotton anthers. Adv Sci (Weinh). 2024b;11(3):e2304017. https://doi.org/10.1002/advs.202304017 .
Lin JL, Chen L, Wu WK, et al. Single-cell RNA sequencing reveals a hierarchical transcriptional regulatory network of terpenoid biosynthesis in cotton secretory glandular cells. Mol Plant. 2023;16(12):1990–2003. https://doi.org/10.1016/j.molp.2023.10.008 .
Liu H, Hu D, Du P, et al. Single-cell RNA-seq describes the transcriptome landscape and identifies critical transcription factors in the leaf blade of the allotetraploid peanut ( Arachis hypogaea L.). Plant Biotechnol J. 2021;19(11):2261–76. https://doi.org/10.1111/pbi.13656 .
Liu Q, Li P, Cheng S, et al. Protoplast dissociation and transcriptome analysis provides insights to salt stress response in cotton. Int J Mol Sci. 2022;23(5):2845. https://doi.org/10.3390/ijms23052845 .
Long L, Xu FC, Wang CH, et al. Single-cell transcriptome atlas identified novel regulators for pigment gland morphogenesis in cotton. Plant Biotechnol J. 2023;21(6):1100–2. https://doi.org/10.1111/pbi.14035 .
Nelms B, Walbot V. Defining the developmental program leading to meiosis in maize. Science. 2019;364(6435):52–6. https://doi.org/10.1126/science.aav6428 .
Peng R, Jones DC, Liu F, et al. From sequencing to genome editing for cotton improvement. Trends Biotechnol. 2021;39(3):221–4. https://doi.org/10.1016/j.tibtech.2020.09.001 .
Qin Y, Sun M, Li W, et al. Single-cell RNA-seq reveals fate determination control of an individual fibre cell initiation in cotton ( Gossypium hirsutum ). Plant Biotechnol J. 2022;20(12):2372–88. https://doi.org/10.1111/pbi.13918 .
Ryu KH, Huang L, Kang HM, et al. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 2019;179(4):1444–56. https://doi.org/10.1104/pp.18.01482 .
Satterlee JW, Strable J, Scanlon MJ. Plant stem-cell organization and differentiation at single-cell resolution. Proc Natl Acad Sci U S A. 2020;117(52):33689–99. https://doi.org/10.1073/pnas.2018788117 .
Shulse CN, Cole BJ, Ciobanu D, et al. High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 2019;27(7):2241–7.e4. https://doi.org/10.1016/j.celrep.2019.04.054 .
Sun Y, Han Y, Sheng K, et al. Single-cell transcriptomic analysis reveals the developmental trajectory and transcriptional regulatory networks of pigment glands in Gossypium bickii. Mol Plant. 2023;16(4):694–708. https://doi.org/10.1016/j.molp.2023.02.005 .
Wang D, Hu X, Ye H, et al. Cell-specific clock-controlled gene expression program regulates rhythmic fiber cell growth in cotton. Genome Biol. 2023;24(1):49. https://doi.org/10.1186/s13059-023-02886-0 .
Wen X, Chen Z, Yang Z, et al. A comprehensive overview of cotton genomics, biotechnology and molecular biological studies. Sci China Life Sci. 2023;66(10):2214–56. https://doi.org/10.1007/s11427-022-2278-0 .
Xu X, Crow M, Rice BR, et al. Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev Cell. 2021;56(4):557–68.e6. https://doi.org/10.1016/j.devcel.2020.12.015 .
Zhang D, Zhang B. Pectin drives cell wall morphogenesis without turgor pressure. Trends Plant Sci. 2020;25(8):719–22. https://doi.org/10.1016/j.tplants.2020.05.007 .
Zhang TQ, Xu ZG, Shang GD, et al. A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol Plant. 2019;12(5):648–60. https://doi.org/10.1016/j.molp.2019.04.004 .
Zhang D, Zhang Z, Unver T, et al. CRISPR/Cas: a powerful tool for gene function study and crop improvement. J Adv Res. 2021;29:207–21. https://doi.org/10.1016/j.jare.2020.10.003 .
Zhang K, Liu S, Fu Y, et al. Establishment of an efficient cotton root protoplast isolation protocol suitable for single-cell RNA sequencing and transient gene expression analysis. Plant Methods. 2023;19(1):5. https://doi.org/10.1186/s13007-023-00983-6 .
Zhu X, Xu Z, Wang G, et al. Single-cell resolution analysis reveals the preparation for reprogramming the fate of stem cell niche in cotton lateral meristem. Genome Biol. 2023;24(1):194. https://doi.org/10.1186/s13059-023-03032-6 .
Download references
Acknowledgements
Author information, authors and affiliations.
Department of Biology, East Carolina University, Greenville, NC, 27858, USA
Pan Xiaoping & Zhang Baohong
College of Biology and Food Engineering, Anyang Institute of Technology, Anyang, Henan, 455000, China
Peng Renhai
You can also search for this author in PubMed Google Scholar
Contributions
Pan XP, Peng RH, and Zhang BH wrote the comment. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Zhang Baohong .
Ethics declarations
Ethics approval and consent to participate, consent for publication, competing interests, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Pan, X., Peng, R. & Zhang, B. Single-cell RNA sequencing opens a new era for cotton genomic research and gene functional analysis. J Cotton Res 7 , 17 (2024). https://doi.org/10.1186/s42397-024-00181-2
Download citation
Received : 25 March 2024
Accepted : 28 April 2024
Published : 04 June 2024
DOI : https://doi.org/10.1186/s42397-024-00181-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Single-cell RNA sequencing
- Transcriptome
Journal of Cotton Research
ISSN: 2523-3254
- Submission enquiries: [email protected]
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
3 10 Tips for Writing an Acknowledgement for a Research Paper. 4 5 Samples for Acknowledgment in Research Paper. 4.1 Sample 1: Acknowledgement for Collaborative Research: 4.2 Sample 2: Acknowledgement for Funding Support: 4.3 Sample 3: Acknowledgement for Mentorship and Guidance: 4.4 Sample 4: Acknowledgement for Institutional Support:
The acknowledgements section is your opportunity to thank those who have helped and supported you personally and professionally during your thesis or dissertation process. ... research assistants, and study participants from the university, who impacted and inspired me. Lastly, I would be remiss in not mentioning my family, especially my ...
Writing the acknowledgements section of your thesis or dissertation is an opportunity to express gratitude to everyone who helped you along the way. Remember to: Acknowledge those people who significantly contributed to your research journey. Order your thanks from formal support to personal support. Maintain a balance between formal and ...
Acknowledgements 101: The Basics. The acknowledgements section in your thesis or dissertation is where you express gratitude to those who helped bring your project to fruition. This section is typically brief (a page or less) and less formal, but it's crucial to thank the right individuals in the right order.. As a rule of thumb, you'll usually begin with academic support: your supervisors ...
Formatting the acknowledgements. As a rule of thumb, the acknowledgement section should be a single short paragraph of say half a dozen lines. Examine the target journal for the format: whether the heading appears on a separate line or run on (that is, the text follows the heading on the same line). Check also whether the heading is in bold or ...
Example 1. First and foremost I am extremely grateful to my supervisors, Prof. XXX and Dr. XXX for their invaluable advice, continuous support, and patience during my PhD study. Their immense knowledge and plentiful experience have encouraged me in all the time of my academic research and daily life.
Begin your acknowledgements by expressing gratitude to those who have made the most significant contributions to your research. This could be your academic advisors, supervisors, or funding bodies. Starting with the most significant contributions helps to set the tone for the rest of your acknowledgements. Ensure that you express your gratitude ...
My labmate's thesis, who wrote the acknowledgements in a different style to the rest by using bullet points. Dr Wei's thesis, acknowledgements on page 6. Direct download here. Shortest acknowledgements section of the list at 122 words. Dr Manca's thesis, acknowledgements on page 5. Direct download here. Dr Liu's thesis, page 5.
10. "The completion of this thesis or dissertation is the culmination of efforts from various individuals whom I would like to express my sincere appreciation.". 11. "This thesis acknowledgement section is an opportunity to give thanks to those who made this journey less daunting.". 12.
Acknowledgement for Research Participants. When acknowledging research participants, it's essential to express your sincere appreciation for their time and insights. These individuals have played a pivotal role in the advancement of your study, offering valuable data that has likely shaped the outcome of your research.
Finally, I wish to thank my parents for their support and encouragement throughout my study. Acknowledgement in Research Paper: Example 5 I'd like to thank Professor X and Professor Y, my research supervisors, for their patient instruction, passionate support, and constructive criticisms of this study effort. I'd also want to thank Dr.
Acknowledging others' contributions is also an important ethical principle. In a scientific manuscript, all statements must be supported with evidence. This evidence can come from the results of the current research, common knowledge, or from previous publications. A citation after a claim makes it clear which previous study supports the claim.
help you write your Acknowledgements section of your dissertation. According to one source, the Acknowledgements section of a Ph.D. dissertation is the most widely read section. Whether you believe this or not, many individuals who helped you in the process of writing may check to see if, indeed, they have meant something to you.
As brief acknowledgements for a research paper, the example gathers contributions of different kinds - intellectual assistance, financial support, image credits etc. - into a single Acknowledgements section. ... The generosity and expertise of one and all have improved this study in innumerable ways and saved me from many errors; those that ...
Dedications: In addition, work dedicated to people directly, such as those who are deceased, may be included in the Acknowledgments section, but this must be done in a certain way and the appreciation put into an open dialogue. For example, the following would not be acceptable: We dedicate this work to the deceased Prof. Bloggs.
Acknowledgment in a research paper is a section dedicated to expressing gratitude and recognizing the individuals, institutions, or resources that have contributed to the completion of the research. This section is an opportunity for the author to appreciate the support, guidance, or assistance received during the research process.
The acknowledgements section is your opportunity to thank those who have helped and supported you personally and professionally during your thesis or dissertation process. ... research assistants, and study participants from the university, who impacted and inspired me. Lastly, I would be remiss in not mentioning my family, especially my ...
Guidelines on Authorship and Acknowledgement. Determining authorship is an important component of upholding the integrity of the research and scholarly enterprise and serves as an explicit way of assigning responsibility and giving credit for intellectual work. Only by honestly reflecting the contribution of all members of the research team can ...
The acknowledgement for thesis is the section where you thank all people, institutions, and companies that helped you complete the project successfully. It is similar to a dedication, except for the fact that it is formal. Also, you don't need to mention every single person who helped you with the research- just those who were most important ...
In fact, acknowledgement studies can no longer be separated from the financial aspect of scientific research. In 2008, WoS started to collect and index funding sources found in the acknowledgements of scientific papers. These new data were added by WoS in response to many funding bodies' requirement to acknowledge the sources supporting research.
This is an acknowledgement for your practical research acknowledgement (pr) first and foremost, praises and thanks to god, the almighty, for his showers of. Skip to document. University; High School; ... Sir Kobe Brynt C. Patungan for providing us invaluable supervision, support, and tutelage during the course of our research study. We would ...
Canine embryonic samples were collected during studies of canine gonadal development that were supported in part by the National Institutes of Child Health and Human Development (R03HD077119, R01HD19393), the Collaborative Research Program, College of Veterinary Medicine (M-W), and the Baker Institute for Animal Health Pursuit of Excellence ...
The current study uses the Scopus database, which is widely regarded among researchers for the purposes of high-quality bibliometric studies [], although a significant number of databases are used for evaluative research at the global level [].The largest abstract and citation database of peer-reviewed research literature in the world, Scopus, is a trusted source for locating biomedical ...
It is important to note the majority of patients included in this study were white and female, limiting the generalizability of our findings. However, our data are in line with existing research suggesting higher rates of diagnosis among women and patients with lighter skin tones [], potentially reflecting gender disparities in healthcare seeking behavior and underdiagnosis in darker skin types.
Background Like all other species, fungi are susceptible to infection by viruses. The diversity of fungal viruses has been rapidly expanding in recent years due to the availability of advanced sequencing technologies. However, compared to other virome studies, the research on fungi-associated viruses remains limited. Results In this study, we downloaded and analyzed over 200 public datasets ...
Acknowledgements. The authors would like to thank the anonymous reviewers for their valuable feedback which enriched this piece further. ... Andrew Silke, "Contemporary Terrorism Studies: Issues in Research," in Critical Terrorism Studies: A New Research Agenda, ed. Richard Jackson, Marie Breen Smyth, and Jeroen Gunning (London: Routledge ...
In a recent study, we demonstrated that a single infusion of CAR19T IF (CD19 CAR T cells devoid of BCOR and ZC3H12A, termed immortal-like and functional T (T IF) cells) into immunocompetent mice ...
Single-cell RNA sequencing (scRNA-seq) is one of the most advanced sequencing technologies for studying transcriptome landscape at the single-cell revolution. It provides numerous advantages over traditional RNA-seq. Since it was first used to profile single-cell transcriptome in plants in 2019, it has been extensively employed to perform different research in plants. Recently, scRNA-seq was ...