- Skip to primary navigation
- Skip to main content
- Skip to primary sidebar
- Skip to footer
- Image & Use Policy
- Translations
UC MUSEUM OF PALEONTOLOGY

Understanding Evolution
Your one-stop source for information on evolution
- ES en Español
Mutations are changes in the information contained in genetic material. For most of life, this means a change in the sequence of DNA, the hereditary material of life. An organism’s DNA affects how it looks, how it behaves, its physiology — all aspects of its life. So a change in an organism’s DNA can cause changes in all aspects of its life.
Mutations are random Mutations can be beneficial, neutral, or harmful for the organism, but mutations do not “try” to supply what the organism “needs.” In this respect, mutations are random — whether a particular mutation happens or not is unrelated to how useful that mutation would be.
Not all mutations matter to evolution Since all cells in our body contain DNA, there are lots of places for mutations to occur; however, not all mutations matter for evolution. Somatic mutations occur in non-reproductive cells and so won’t be passed on to offspring.
For example, the yellow color on half of a petal on this red tulip was caused by a somatic mutation. The seeds of the tulip do not carry the mutation. Cancer is also caused by somatic mutations that cause a particular cell lineage (e.g., in the breast or brain) to multiply out of control. Such mutations affect the individual carrying them but are not passed directly on to offspring.

The only mutations that matter for the evolution of life’s diversity are those that can be passed on to offspring. These occur in reproductive cells like eggs and sperm and are called germline mutations .
- More Details
- Evo Examples
- Teaching Resources
Read more about how mutations are random and the famous Lederberg experiment that demonstrated this. Or read more about how mutations factored into the history of evolutionary thought . Or dig into DNA and mutations in this primer.
Learn more about mutation in context:
- Evolution at the scene of the crime , a news brief with discussion questions.
- A chink in HIV's evolutionary armor , a news brief with discussion questions.
Find lessons, activities, videos, and articles that focus on mutation.
Reviewed and updated June, 2020.
Genetic variation
The effects of mutations
Subscribe to our newsletter
- Teaching resource database
- Correcting misconceptions
- Conceptual framework and NGSS alignment
- Image and use policy
- Evo in the News
- The Tree Room
- Browse learning resources
- Biology Article
- Mutation Genetic Change
Mutation - A Genetic Change
Mutation definition.
“Mutation is the change in our DNA base pair sequence due to various environmental factors such as UV light, or mistakes during DNA replication.”
Table of Contents
Classification & types of mutations, frequently asked questions, what are mutations.
The DNA sequence is specific to each organism. It can sometimes undergo changes in its base-pairs sequence. It is termed as a mutation. A mutation may lead to changes in proteins translated by the DNA. Usually, the cells can recognize any damage caused by mutation and repair it before it becomes permanent.
A mutation is a sudden, heritable modification in an organism’s traits. The term “mutant” refers to a person who exhibits these heritable alterations. Mutations usually produce recessive genes.
Causes of Mutations
The mutation leads to genetic variations among species. Positive mutations are transferred to successive generations.
E.g. Mutation in the gene coding for haemoglobin causes sickle cell anaemia. The R.B.Cs become sickle in shape. However, in the African population, this mutation provides protection against malaria.
A mutation in the gene controlling the cell division leads to cancer.
Let us have an overview of the causes and impacts of mutation.
Also Read: Mutagens
The mutation is caused due to the following reasons:
Internal Causes
Most of the mutations occur when the DNA fails to copy accurately. All these mutations lead to evolution. During cell division , the DNA makes a copy of its own. Sometimes, the copy of the DNA is not perfect and this slight difference from the original DNA is called a mutation.
External Causes
When the DNA is exposed to certain chemicals or radiations, it causes the DNA to break down. The ultraviolet radiations cause the thymine dimers to break resulting in a mutated DNA.
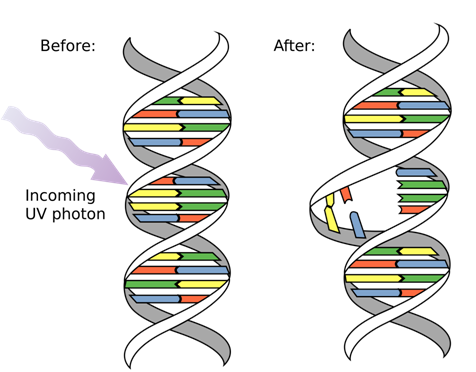
DNA Mutation
Effects of Mutation
There are several mutations that cannot be passed on to the offsprings. Such mutations occur in the somatic cells and are known as somatic mutations.
The germline mutations can be passed on to successive generations and occur in the reproductive cells.
Let us have a look at some of the effects of mutation:
Beneficial Effects of Mutation
- Few mutations result in new versions of proteins and help the organisms to adapt to changes in the environment. Such mutations lead to evolution.
- Mutations in many bacteria result in antibiotic-resistant strains of bacteria that can survive in the presence of antibiotics.
- A unique mutation found in the population of Italy protects them from atherosclerosis, where fatty materials build up in the blood vessels.
Effects of Mutations
- Genetic disorders can be caused by the mutation of one or more genes. Cystic fibrosis is one such genetic disorder caused by the mutation in one or more genes.
- Cancer is another disease caused by the mutation in genes that regulate the cell cycle.
For more information on mutation, its causes and effects, keep visiting BYJU’S website or download BYJU’S app for further reference.
Give the meaning of mutation.
Mutation means an alteration in the genes or chromosomes of a cell. This shift in the gametes may impact the development and structure of the progeny. A mutation in biology is a modification of the nucleic acid sequence of a virus, extrachromosomal DNA, or the genome of an organism. The observable traits of an organism (phenotype) may or may not change as a result of a mutation.
Define gene mutation. Give examples.
Gene or genetic mutations are modifications to the DNA sequence that take place as the cells divide and generate copies of themselves. The DNA provides instructions on how to develop and run the human body. Genetic changes may result in diseases like cancer or, in the long run, enable people to adapt to their surroundings more successfully.
Examples include animals possessing extra body parts after birth, such as four-legged ducks, cyclops kittens, and snakes with two heads. Genetic disorders in humans, like Sickle-cell disease, are frequently brought on by gene mutations or chromosomal aberrations. Angelman syndrome, Canavan disease, colour blindness, cystic fibrosis, cri-du-chat syndrome, Down syndrome, haemophilia, Klinefelter syndrome, Duchenne muscular dystrophy, phenylketonuria, Prader-Willi syndrome, Tay-Sachs disease, and Turner syndrome are additional examples of common mutations in human beings.
What is DNA mutation?
A DNA mutation is a long-lasting alteration to the nucleotide sequence of DNA that can occur during replication and recombination. Most of the time, mutations are benign unless they result in tumour growth or cell death. Base pair substitution, deletion, or insertion can all result in mutations in damaged DNA. Cells have developed processes for repairing damaged DNA owing to the deadly consequences of DNA mutations.
Base substitutions, deletions, and insertions are the three different forms of DNA mutations.
What are the causes of gene mutation?
A gene mutation is a permanent change to a DNA sequence that makes it different from the sequence found in other people. A genetic mutation occurs during cell division when the grow and divide and replicate. Mutations can impact anything from a single DNA base pair (a unit of genetic composition) to a significant portion of a chromosome that contains numerous genes.
How does mutation affect genetic change?
A population can acquire new alleles through mutations, increasing the genetic diversity of the population. For mutations to impact an organism’s offspring, they must: 1) arise in cells that give rise to the succeeding generation; and 2) alter the genetic code. Diversity among organisms is ultimately produced through the interaction of hereditary mutations and environmental stresses.

Put your understanding of this concept to test by answering a few MCQs. Click ‘Start Quiz’ to begin!
Select the correct answer and click on the “Finish” button Check your score and answers at the end of the quiz
Visit BYJU’S for all Biology related queries and study materials
Your result is as below
Request OTP on Voice Call
Leave a Comment Cancel reply
Your Mobile number and Email id will not be published. Required fields are marked *
Post My Comment
Superbb explanation
Register with BYJU'S & Download Free PDFs
Register with byju's & watch live videos.
If you're seeing this message, it means we're having trouble loading external resources on our website.
If you're behind a web filter, please make sure that the domains *.kastatic.org and *.kasandbox.org are unblocked.
To log in and use all the features of Khan Academy, please enable JavaScript in your browser.
Middle school biology
Course: middle school biology > unit 7.
- Apply: mutations
Key points:
- A mutation is any change to the nucleotide sequence of a DNA molecule. Some mutations arise as DNA is copied. Others are due to environmental factors.
- A mutation in a gene can change the structure and function of the protein encoded by that gene. This, in turn, can affect an organism’s traits.
- Harmful mutations have negative effects on an organism’s health and survival. For example, some mutations cause inherited disorders such as sickle cell anemia and cystic fibrosis.
- Beneficial mutations have positive effects on an organism’s health and survival. For example, some people have mutations that lower their risk of developing type 2 diabetes.
- Neutral mutations have no observable effect on an organism’s traits. For example, some gene mutations do not lead to amino acid changes, and so do not affect protein function.
Want to join the conversation?
- Upvote Button navigates to signup page
- Downvote Button navigates to signup page
- Flag Button navigates to signup page


- Previous Article
Cover Image
Introduction, the human genome and variation, chromosome structure and chromosomal disorders, the sex chromosomes, x and y, single-gene disorders, mitochondrial disorders, epigenetics, complex disorders, cancer: mutation and epigenetics, genetic testing in the diagnostic laboratory, diagnosis, management and therapy of genetic disease, challenges in delivering a genetics service, competing interests, author contribution, appendix. glossary of terms, abbreviations, further reading: the human genome and variation, the genetic basis of disease.
- Split-Screen
- Article contents
- Figures & tables
- Supplementary Data
- Peer Review
- Open the PDF for in another window
- Cite Icon Cite
- Get Permissions
Maria Jackson , Leah Marks , Gerhard H.W. May , Joanna B. Wilson; The genetic basis of disease. Essays Biochem 3 December 2018; 62 (5): 643–723. doi: https://doi.org/10.1042/EBC20170053
Download citation file:
- Ris (Zotero)
- Reference Manager
Genetics plays a role, to a greater or lesser extent, in all diseases. Variations in our DNA and differences in how that DNA functions (alone or in combinations), alongside the environment (which encompasses lifestyle), contribute to disease processes. This review explores the genetic basis of human disease, including single gene disorders, chromosomal imbalances, epigenetics, cancer and complex disorders, and considers how our understanding and technological advances can be applied to provision of appropriate diagnosis, management and therapy for patients.
When most people consider the genetic basis of disease, they might think about the rare, single gene disorders, such as cystic fibrosis (CF), phenylketonuria or haemophilia, or perhaps even cancers with a clear heritable component (for example, inherited predisposition to breast cancer). However, although genetic disorders are individually rare, they account for approximately 80% of rare disorders, of which there are several thousand. The sheer number of rare disorders means that, collectively, approximately 1 in 17 individuals are affected by them. Moreover, our genetic constitution plays a role, to a greater or lesser extent, in all disease processes, including common disorders, as a consequence of the multitude of differences in our DNA. Some of these differences, alone or in combinations, might render an individual more susceptible to one disorder (for example, a type of cancer), but could render the same individual less susceptible to develop an unrelated disorder (for example, diabetes). The environment (including lifestyle) plays a significant role in many conditions (for example, diet and exercise in relation to diabetes), but our cellular and bodily responses to the environment may differ according to our DNA. The genetics of the immune system, with enormous variation across the population, determines our response to infection by pathogens. Furthermore, most cancers result from an accumulation of genetic changes that occur through the lifetime of an individual, which may be influenced by environmental factors. Clearly, understanding genetics and the genome as a whole and its variation in the human population, are integral to understanding disease processes and this understanding provides the foundation for curative therapies, beneficial treatments and preventative measures.
With so many genetic disorders, it is impossible to include more than a few examples within this review, to illustrate the principles. For further information on specific conditions, there are a number of searchable internet resources that provide a wealth of reliable detail. These include Genetics Home Reference ( https://ghr.nlm.nih.gov/ ), Gene Reviews ( https://www.ncbi.nlm.nih.gov/books/NBK1116/ ), the ‘Education’ section from the National Human Genome Research Institute ( https://www.genome.gov/education/ ) and Online Mendelian Inheritance in Man ( https://www.omim.org/ ). In this review, an understanding and knowledge of basic principles and techniques in molecular biology, such as the structure of DNA and the PCR will be assumed, but explanations and animations of PCR (and some other processes) are available from the DNA Learning Center ( https://www.dnalc.org/resources/ ). The focus here will be on human disease, although much of the research that defines our understanding comes from the study of animal models that share similar or related genes.
The human genome and the human genome reference sequence
The complete instructions for generating a human are encoded in the DNA present in our cells: the human genome, comprising roughly 3 billion bp of DNA. Scientists from across the world collaborated in the ‘Human Genome Project’ to generate the first DNA sequence of the entire human genome (published in 2001), with many additions and corrections made in the following years. Genome sequence information for humans and many other species is freely accessible through a number of portals, including the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/ ) and Ensembl ( http://www.ensembl.org/ ), which also provide a wealth of related information.
The majority of our DNA is present within the nucleus as chromosomes (the nuclear DNA or nuclear genome), but there is also a small amount of DNA in the mitochondria (the mtDNA or mitochondrial genome). Most individuals possess 23 pairs of chromosomes ( Figure 2 ), therefore much of the DNA content is present in two copies, one from our mother and one from our father.
The human nuclear genome encodes roughly 20000 protein-coding genes, which typically consists of both protein-coding (exon) and non-coding (intron) sequences. Our genome also contains roughly 22000 genes that encode RNA molecules only; some of these RNAs form components of the translation machinery (rRNA, tRNA) but there are many more that perform various roles within the cell, including regulation of expression of other genes. In fact it is now believed that as much as 80% of our genome has biological activity that may influence structure and function. The human genome also contains over 14000 ‘pseudogenes’; these are imperfect copies of protein-coding genes that have lost the ability to code for protein. Although originally considered as evolutionary relics, there is now evidence that some may be involved in regulating their protein-coding relatives, and in fact dysregulation of pseudogene-encoded transcripts has been reported in cancer. Additionally, sequence similarity between a pseudogene and its normal counterpart may promote recombination events which inactivate the normal copy, as seen in some cases of perinatal lethal Gaucher disease. Furthermore, some pseudogenes have the potential to be harnessed in gene therapy to generate functional genes by gene editing approaches. The distribution of genes between chromosomes is not equal: chromosome 19 is particularly gene-dense, while the autosomes for which trisomy is viable (13, 18, 21) are relatively gene-poor ( Table 1 ).
Note that although these numbers seem very precise they should be taken as indicative only, since (i) chromosomes of each individual will vary from the reference sequence, and (ii) the human reference genome sequence is continuously updated with corrections (the data here are from GRCh38.p12, which represents a particular ‘build’ of the human genome). Note that the data for the acrocentric chromosomes 13, 14, 15, 21, 22 does not include the shared ribosomal DNA array repeats present on the p arms (see Figure 2 ). Data from Ensembl, June 2018.
From the very beginning of the Human Genome Project, it was recognised that there was a huge amount of DNA sequence variation between healthy individuals, and therefore there is no such thing as a ‘normal’ human DNA sequence. However, if we are to describe changes to the DNA sequence, we need to describe these changes with respect to some baseline; this baseline is the human reference genome sequence.
Variation versus mutation
A geneticist’s definition of mutation is ‘any heritable change to the DNA sequence’, where heritable refers to both somatic cell division (the proliferation of cells in tissues) and germline inheritance (from parent to child). Such changes to the DNA may have no consequences but sometimes lead to observable differences in the individual (the ‘phenotype’). Consequently, in the past such alterations in the human population, particularly when they were associated with a disease state, were referred to as ‘mutations’. However, for many people this terminology has negative connotations, and brings to mind the ‘mutants’ seen in science fiction and zombie films! Therefore modern practice, particularly for medical genetics within the context of a health service, is to refer to differences from the reference sequence as ‘variants’. Variants may be further classified as benign (not associated with disease) or pathogenic (associated with disease), although there are increasing numbers of human DNA variants identified for which we are still not sure of the effect; these are termed ‘variants of uncertain significance’ or VUS ( Table 2 ).
Although this system was designed for classification of variants in relation to a potential role in cancer predisposition, it can also be used to classify variants in other situations.
Where two (or more) different versions of a DNA sequence exist in the population, these are referred to as ‘alleles’: each allele represents one particular version (or variant) of that sequence. By analysing many human genomes we can calculate the frequency at which a particular variant occurs in the population, often expressed as the ‘minor allele frequency’ or MAF. Where the MAF is at least 1%, a variant can be called a ‘polymorphism’, although this is a fairly arbitrary cut-off.
Single nucleotide variants: The most frequent variants in our genome are substitutions that affect only one base pair (bp), referred to as single nucleotide variants (SNV) or as single nucleotide polymorphisms (SNP) ( Figure 1 ) depending upon the MAF. It has been estimated that there are at least 11 million SNPs in the human genome (averaging approximately 1 per 300 bp). It also seems likely that if we sequenced the genomes of everyone on the planet, for most positions in our genome we would discover at least one individual with an SNV, wherever such variation is compatible with life.
Some types of variants found in human genomes

Variation involving one or a few nucleotides are shown above the chromosome icon, and structural variants below; in each case the variants are depicted in relation to the reference sequence. For depiction of structural variants A, B, C and D represent large segments of DNA; Y and Z represent segments of DNA from a different chromosome. Note that differentiation between CNVs and deletions/insertions depends upon the size of the relevant DNA segment (see text for further details). Abbreviation: CNV, copy number variant. Chromosome ideogram from NCBI Genome Decoration Page.
Insertions and deletions (indels): Insertions or deletions of less than 1000 bp are also relatively common in the human genome, with the smallest indels being the most numerous.
Structural variants: Structural variants are defined as variants affecting segments of DNA greater than 1000 bp (1 kb). They include translocations, inversions, large deletions and copy number variants (CNV). CNVs are segments of our genome that range in size from 1000 to millions of bp, and which, in healthy individuals, may vary in copy number from zero to several copies ( Figure 1 ). By analysis of many human genomes it is apparent that CNV exists for approximately 12% of the human genome sequence. The largest CNVs may contain several entire genes. Where the population frequency of a CNV reaches 1% or more, it may be referred to as a copy number polymorphism (CNP).
Repeat variations: Human genomes contain large numbers of repetitive sequences. These include ‘interspersed repeats’ which constitute approximately 45% of our genome, and represent remnants of mobile DNA elements (transposons). There are also several classes of ‘tandem repeats’, in which the repeated units are side-by-side in a head-to-tail fashion forming arrays of repeats of the same (or very similar) sequence. The number of repeats in each array can vary, generating multiple alleles, so that these loci have high variability within the population, and can be used in identifying individuals (see below). Tandem repeats include minisatellites and microsatellites ( Figure 1 / Table 3 ). Although generally inherited stably (i.e. with the same number of repeats) from parent to child, expansions in some microsatellites are associated with disease.
Note that repeats with unit lengths of 7–9 bp may be classified as micro- or minisatellites depending on their biological behaviour.
Many (but not all) authors include mononucleotide repeats in the category of microsatellites.
Variation between healthy individuals
Given that no two individuals look exactly alike (apart from identical twins) it will come as no surprise that this is reflected in our DNA. What is surprising is the amount of variation between us. Looking at any one human genome, compared with the reference sequence, we would find approximately 3 million SNPs, and approximately 2000 structural variants. The genomes of any two unrelated individuals will differ in approximately 0.5% of their DNA (approximately 15 million bp), and most of this variation can be attributed to CNVs and large deletions. Although much of the variation in our genome lies within the non-coding DNA, we now know that, on average, each individual has several hundred variants that are either known, or predicted, to be damaging to gene function, including roughly 85 variants that lead to truncated (incomplete) protein products. Furthermore, the total number of functional genes per human genome may vary by up to 10% between individuals as a consequence of CNVs, large deletions and loss-of-function variants. Faced with this enormous level of variation you might wonder, not why some individuals are affected by disease due to inherited ‘mutations’, but rather how any of us manage to remain relatively healthy! Clearly there is no requirement for all of our genes to be functional: for many genes only one working copy is required, and in other cases there appears to be a level of redundancy or plasticity built into the system. However it is becoming increasingly apparent that some of the variations in our genomes may lead to higher susceptibility to common diseases.
Variation between populations
The greatest amount of variation is found within populations of African ancestry, which is consistent with initial migration out of Africa, with each group of migrants taking subsets of variants with them. Common variants tend to be shared between all populations, whereas rare variants are more likely to be specific to particular populations or related populations. Some of the differences will be related to environmental adaptation, for example skin pigmentation or enzymes to detoxify dietary plant toxins. These same enzymes are also responsible for the metabolism of many pharmaceutical (and recreational) drugs; genetic variants may lead to some individuals being ultrarapid metabolisers or poor metabolisers, which may translate into poor drug response or adverse side effects. For example, deficiency in dihydropyrimidine dehydrogenase, leading to a toxic response to the cancer treatment 5-fluorouracil, is two to three times more common in African-American populations than in Caucasians.
DNA profiling
In the early 1980s, with the discovery of minisatellites, which are highly variable within the population but inherited stably from parent to child, it became possible to use these in forensic analyses and paternity testing, to generate unique patterns (similar to supermarket barcodes) for each individual, a technique referred to as ‘DNA fingerprinting’. This technology needed large amounts of sample (micrograms of DNA) and tended to be time-consuming (1–2 weeks) in addition to requiring use of radioactive labels. Towards the end of the 1980s, microsatellites were first reported and since these could be analysed with simple and rapid PCR-based assays, needing only approximately 1 nanogram of sample DNA, ‘DNA profiling’ using microsatellites quickly replaced the earlier DNA fingerprinting approach. Forensic DNA profiling in the U.K. currently analyses 16 microsatellites from across the genome, together with a region from the amelogenin gene present on both X and Y chromosomes that is 4 bp different in size between them, allowing gender identification. The process is similar to QF-PCR for prenatal aneuploidy testing, which will be discussed later. Finding a perfect match between the two samples (e.g. from crime scene and suspect) strongly suggests that these came from the same individual – the likelihood of finding a perfect match between samples from two different individuals is estimated at 1 in a billion – unless of course they are identical twins. On the other hand, if the two samples do not match, it can be concluded that the crime scene sample was not from the suspect. Likewise, in paternity testing, DNA profiling can exclude a man as the father of a child, but cannot prove he is the father with absolute certainty. DNA profiling is also useful in helping to identify human remains, for example where decomposition makes physical identification difficult. The fact that certain variants (including microsatellite alleles) are more frequently found in populations of particular ancestry means that the capability already exists to make some inferences on likely ancestral origin based on only a DNA sample and research is underway to establish whether particular features (for example, eye colour, hair colour and even facial characteristics) can be predicted from DNA. Thus the DNA profiling of the future may generate an identikit image of a wanted individual.
De novo mutations and mosaicism
Most of the variants in our genome were inherited from one of our parents. However, our DNA is constantly bombarded with DNA damaging agents and furthermore every time a cell’s DNA is replicated prior to division there is opportunity for errors. Genomic sequencing of trios (child plus both parents) has demonstrated that on average each individual has 74 de novo SNVs that were not present in either parent, in addition to approximately three de novo insertions/deletions. Approximately 1–2% of children will have a de novo CNV greater than 100 kb in size. Microsatellites have a relatively high mutation frequency, with gain or loss of a repeat unit occurring in roughly 1 per 1000 microsatellites per gamete per generation. In contrast with aneuploidy, which is most often a consequence of meiotic error during oocyte generation, new mutations are almost four times more common in the male germline than the female germline, which is likely to relate to the high number of cell divisions during spermatogenesis. For both sexes the new mutation rate increases with age, though again, the increase is more marked in the male germline. Most new mutations will have little or no effect on health, particularly those outside coding sequences, but some are associated with disease.
If a new mutation occurs during embryogenesis or development this can lead to mosaicism, where some cells in the individual have that new variant while others do not. Mosaicism for a new mutation may also be present in the gonads (‘gonadal mosaicism’), such that a new variant may be transmitted to less than 50% of the offspring, depending upon the percentage of gonadal cells in which the new variant is present. New mutations occurring during embryogenesis and development also generate a few differences between the genomes of identical twins.
Very rarely fusion of two embryos will generate a chimera: an individual that has two genetically distinct cell lines present. Where the same sex chromosome constitution is present in both cell lines chimerism might only come to light with the observation of apparent non-maternity or non-paternity amongst offspring (where one cell line predominates in the gonads and the other predominates in blood cells). Fusion of two embryos of different sex can lead to characteristics of both genders being present, and chimerism is found in approximately 13% of cases of hermaphroditism.
The massive amount of variation between individual human genomes can make it very difficult to determine which variants are benign and which might be associated with a disease. Even where a disease-associated variant is present, this will be present within a genomic context of millions of other differences from the ‘reference’ sequence, some of which may impact upon the severity of that disease in the individual. Thus it will become increasingly common to investigate wider genomic influences when considering contribution of variants to disease. Note that several scientific conventions are used when referring to chromosomes, genes, proteins and variants affecting them; these ensure unambiguous communication between scientists and health professionals. International System for Human Cytogenetic Nomenclature (ISCN) is used for describing karyotypes and changes at the chromosomal level. Individual loci and genes, for which there are often multiple different historical names, have now been assigned specific unique names by the HUGO Gene Nomenclature Committee (HGCN) ( https://www.genenames.org/ ). Sequence variants are described according to Human Genome Variation Society (HGVS) guidelines ( http://varnomen.hgvs.org/ ) for both DNA and proteins. Finally, since the same names are applied to genes and the proteins they encode, italics are used to refer to the gene, with standard font used when referring to the protein.
Almost every human cell contains a full diploid genome, consisting of 2 metres of DNA arranged into 46 chromosomes: 22 homologous autosomal pairs, and the sex chromosomes comprising two X chromosomes in females and an X and a Y in males. The exceptions are anucleate cells like erythrocytes (red blood cells), cell fragments (platelets) and haploid germline cells (sperm and eggs) which contain 23 chromosomes. Although mechanisms have evolved which ensure that during cell division, daughter cells will inherit a complete genome, those mechanisms occasionally make mistakes. This can lead to cells with chromosomal abnormalities, which can be categorised as numerical abnormalities, i.e. the resulting daughter cell contains too many or too few chromosomes, or structural abnormalities, where more complex rearrangements of the genome have taken place.
The normal chromosome complement of a species (i.e. the number, size and shape of chromosomes) is called its karyotype. According to the ISCN, the ‘normal’ human karyotype is denoted by either 46,XX (female) or 46,XY (male). Human chromosomes consist of DNA which is wrapped around a core of histone proteins to form chromatin. Most of the time, chromatin exists in a diffuse form within a cell’s nucleus, however, during metaphase of the cell division cycle, the chromosomes condense. It is these condensed chromosomes which can be stained with a variety of chemicals, and which can then be observed under a light microscope, to reveal the characteristic banding patterns. The bands reflect regions of chromatin with different characteristics, and therefore different functional elements. A photographic representation of a person’s metaphase chromosomes, arranged by size, may be referred to as a karyogram or karyotype ( Figure 2 A,B) and a graphical representation is called an ideogram ( Figure 2 C). The available stains for chromosomes differ in their chemical properties and consequently in the resulting banding pattern. The most commonly used stain is called Giemsa after the chemist who developed it in 1904; the resulting banding pattern of chromosomes is referred to as G-banding. The microscopic analysis of stained chromosomes is termed cytogenetics. Depending on the quality of the chromosome preparation, trained cytogeneticists can identify abnormalities with a resolution of approximately 3–4 Mb (millions of bp), however, abnormalities below this resolution threshold cannot be identified using conventional cytogenetics and require alternative, molecular techniques (see section ‘Genetic testing in the diagnostic laboratory’).
Giemsa banding (G-banding) to form a karyogram

( A ) Metaphase spreads like this are obtained from cultured cells arrested in metaphase using colcemid, followed by Giemsa staining to create characteristic light and dark bands. Generally the dark bands represent regions which are AT-rich and gene-poor. ( B ) The chromosomes from the spread are arranged in pairs to view the karyotype, often using specialist software like Cytovision. ( C ) Diagrammatic representations of the G-banding patterns, called ideograms, are used as a reference. The ideograms have been aligned at the centromere (dotted line); blue shaded regions are highly variable – note for example the variation between p arms of chromosome 13, 14 and 15 in (B). In fact the p arms of the acrocentric chromosomes (13, 14, 15, 21, 22) all have very similar content, which includes the nucleolar organiser regions or NORs. Each NOR contains a tandem repeat of ribosomal DNA (rDNA) which encodes the rRNAs. Between all five acrocentrics there are approximately 300–400 rDNA repeats, though the actual number varies between individuals. Chromosome ideograms from NCBI Genome Decoration Page.
When viewing condensed metaphase chromosomes under a microscope, some key features can be identified ( Figure 3 ). All mammalian chromosomes have a centromere, which appears like a narrow waist, here proteins attach for separation of chromosomes during cell division. In humans, the centromere is located between the two arms of the chromosome, the shorter arm is called the ‘p’ arm (for ‘petite’), while the longer arm is called ‘q’ (‘queue’). Depending on the location of the centromere relative to the two arms, human chromosomes are classified as ‘metacentric’, where the centromere is more or less in the middle of the chromosome, ‘submetacentric’, where the centromere is somewhat offset from the centre or ‘acrocentric’, where the centromere is significantly offset from the centre, with only a very short p arm. In some species such as the mouse, the centromere is located at one end of the chromosome, termed as telocentric. In humans, chromosomes 1, 3, 16, 19 and 20 are metacentric, chromosomes 13, 14, 15, 21, 22 and Y are acrocentric, while the remainder are submetacentric. In eukaryotes, the structures at the ends of each linear chromosome are called telomeres and consist of 300–8000 repeats of the sequence TTAGGG, which forms a loop at the end. One function of telomeres is to protect the ends of chromosomes from being recognised as ‘damaged DNA’ and erroneously repaired by the cell’s DNA repair machinery. They also accommodate the loss of sequences during each round of replication, which occurs as a result of the so-called ‘end replication problem’. In cells without the enzyme telomerase (which extends existing telomeres), a short stretch of sequence is lost from the 5′ end of the newly replicated strand with each cell division, which ultimately can lead to cell senescence.
Chromosome structure and band nomenclature
![what is genetic mutation essay This ideogram of the complete chromosome 8 illustrates the general structure of all human chromosomes: short (p) and long (q) arms, joined at the centromere. Each chromosome has a characteristic G-banding pattern, with each band annotated, for example p22 or q23. In chromosomes which are less condensed, more bands are seen as separate entities, while bands may merge together in more condensed chromosomes (for example, q21.1, q21.2 and q21.3 appear as a single band [q21] in a more condensed chromosome 8). The approved way of stating the location q21.1 is q-two-one-point-one (not q-twenty-one-point-one). Telomeres, with a shared structure, are present at both ends of each chromosome. Each telomere is composed of arrays of TTAGGG repeats, followed by a subtelomere, which is formed of repetitive sequences which can be similar between several telomeres. Chromosome ideogram from NCBI Genome Decoration Page.](https://port.silverchair-cdn.com/port/content_public/journal/essaysbiochem/62/5/10.1042_ebc20170053/2/m_ebc-2017-0053ci003.jpeg?Expires=1718722264&Signature=aOY3wo~9tZJbPIuv5pYQ9-8evM46w1t4hfv0GmH8glj3-dCWj9k8IfS7224rjBUGHLiJ2qSrNdoRQZd0pECGzjXzUB9laMc0h1amSa4ofvQxZRnVi~GE003abefrNPbmkTLXfF7yYiYfl9cXIWaieuexx3dqraTRwPru0VnT5~bf2ItsEzkb-ltz1NO3t6GhrLLvG1vWTequNKZmXF0mDymBsmCEwH4isvDLX0HB7XMzDrVhsE2WmBxHfc5gR5ZHASBmw4ye8nKCArpWRnD-Q11n7YiPmevj-jph4WGRFkC3NNkUHc4~9bG2js1Z2fm~Tz1gtTxB8L62QJp~P69zyw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This ideogram of the complete chromosome 8 illustrates the general structure of all human chromosomes: short (p) and long (q) arms, joined at the centromere. Each chromosome has a characteristic G-banding pattern, with each band annotated, for example p22 or q23. In chromosomes which are less condensed, more bands are seen as separate entities, while bands may merge together in more condensed chromosomes (for example, q21.1, q21.2 and q21.3 appear as a single band [q21] in a more condensed chromosome 8). The approved way of stating the location q21.1 is q-two-one-point-one ( not q-twenty-one-point-one). Telomeres, with a shared structure, are present at both ends of each chromosome. Each telomere is composed of arrays of TTAGGG repeats, followed by a subtelomere, which is formed of repetitive sequences which can be similar between several telomeres. Chromosome ideogram from NCBI Genome Decoration Page.
Numerical abnormalities
An abnormality where a cell contains more than two complete sets of the human haploid genome (69 chromosomes or more) is termed as polyploidy. Triploidy (three haploid sets of chromosomes) occurs in 1–3% of pregnancies and usually arises from fertilisation of a single egg with two sperms or sometimes from fertilisation involving a diploid gamete (egg or sperm). Viability of triploid foetuses is usually very low and leads to early spontaneous abortion during pregnancy while tetraploidy (four haploid sets of chromosomes) is even rarer and not compatible with life. However, a situation where the chromosome number is not an exact multiple of the haploid chromosome number is called aneuploidy.
Aneuploidy usually arises because a gamete is formed that contains more or fewer chromosomes than the normal complement. This results from a phenomenon called non-disjunction, where the replicated chromosomes do not separate properly at cell division, and can happen during either meiosis I (non-disjunction of paired chromosomes) or meiosis II (non-disjunction of sister chromatids) ( Figure 4 ). Non-disjunction generates germ cells which either contain an extra copy of one of the chromosomes or lack one chromosome. Fertilisation then leads to the formation of a zygote with an extra chromosome or a missing chromosome respectively ( Figure 5 ). Non-disjunction most commonly occurs during meiosis II of oocyte formation, and is influenced by the mother’s age and other environmental factors. The risk of delivering a trisomic foetus increases from 1.9% in women aged 25–29 years to over 19% in women aged over 39 years. There is also evidence that folic acid deficiency, smoking, obesity and low-dose irradiation with radioactive contaminants increases the risk of non-disjunction.
Principles of meiosis and non-disjunction

For simplicity, only one pair of newly replicated autosomes is shown in two different colours to distinguish the maternal from the paternal chromosome and crossover is not considered. During spermatogenesis, all four meiotic products can form the gametes (sperm), while in oogenesis, only one of the four products will actually become the ovum (egg) as one daughter cell forms a polar body at meiosis I (MI) and another forms a polar body at meiosis II (MII). For clarity, all four potential meiotic products are shown. ( A ) During normal meiosis, four haploid meiotic products are formed. ( B ) If non-disjunction occurs during MI, two daughter cells are formed which completely lack this particular chromosome (nullisomic for this chromosome), while two others contain two copies of the chromosome (disomic). ( C ) If non-disjunction occurs during MII, one nullisomic and one disomic daughter cell is formed, while the remaining two form normally.
Fertilisation outcomes

( A ) Fertilisation of a normal oocyte with a normal sperm cell leads to the formation of a diploid (2n) zygote. ( B ) If a nullisomic oocyte is fertilised, the resulting zygote will be monosomic for one chromosome. ( C , D ) Fertilisation of a disomic oocyte results in trisomic zygotes. Note that in (C), the oocyte has resulted from non-disjunction in meiosis I, and the resulting zygote contains one chromosome (ignoring crossover) from each maternal grandparent as well as the paternal contribution. In (D), the oocyte has resulted from non-disjunction in meiosis II, and the resulting zygote contains two chromosomes (aside from crossover regions) from one grandparent.
Examples of syndromes caused by aneuploidy
Most aneuploidies are lethal. However, those that are viable are listed in Table 4 , together with approximate incidence rates and common symptoms. Foetuses with trisomy 13 or 18 may survive to term, while individuals with trisomy 21 can survive beyond the age of 40. Presence of an extra autosome generally leads to severe developmental abnormalities, and only trisomies of small, gene-poor chromosomes ( Table 1 ) appear to be tolerated. Autosomal monosomies have even more severe consequences, as they invariably lead to miscarriage during the early stages of pregnancy. The developmental consequences of such trisomies and monosomies are a result of an imbalance of the levels of critical gene products encoded on the affected chromosomes. For example, the major features of Down syndrome (DS) are associated with the presence of three copies of a 1.6-Mb region at chromosome location 21q22.2, called the Down Syndrome Critical Region.
Common names are given, where available, together with estimated incidence rates and symptoms frequently associated with the condition.
Having an abnormal number of sex chromosomes generally has milder consequences than abnormal numbers of autosomes and is discussed in more detail in the section ‘The sex chromosomes, X and Y’.
Structural abnormalities
DNA damage, e.g. by radiation or mutagenic chemicals, can lead to chromosome breaks. Complex cell cycle checkpoints prevent cells with unrepaired chromosome breaks, in particular free broken ends (i.e. ends without telomeres), from entering mitosis. DNA repair mechanisms exist which recognise chromosome breaks and attempt to repair them. However, these mechanisms occasionally repair broken chromosomes incorrectly, which can then result in chromosomes with structural abnormalities. Errors during recombination, e.g. between mispaired homologues, may also result in such abnormalities.
If a single chromosome sustains breaks, incorrect repair can lead to material being lost (deletion), inverted or incorporated into a circular structure: a ring chromosome. The resulting structurally abnormal chromosomes can be stably propagated during cell division, as long as they possess a single centromere. Chromosomes without a centromere are eventually lost. Chromosomes with two centromeres are rarely found, in these cases one centromere appears to be suppressed.
If single breaks occur in two separate chromosomes, incorrect joining of the resulting fragments may lead to the exchange of material between chromosomes (translocation). In a balanced reciprocal translocation DNA from two different chromosomes is exchanged without net loss. If both resulting hybrid (or ‘derivative’) chromosomes carry one centromere, they will be replicated and segregated stably. However, during gamete formation, it can happen that only one of the hybrid chromosomes, together with one of the unaltered chromosomes, are segregated into a gamete ( Figure 6 ). Fertilisation of such gametes leads to the formation of a zygote with partial trisomy of genetic material from one of the chromosomes involved in the translocation, and partial monosomy of material from the other participating chromosome. Depending on the location of the breakpoints, and therefore on how much genetic material is present in trisomic or monosomic form, such embryos can be viable, but have a high risk of developmental abnormalities. Nevertheless, approximately 1 in 500 individuals carry a balanced reciprocal translocation. These carriers frequently appear asymptomatic, however, there is an increased miscarriage rate associated with either parent being a translocation carrier and offspring of carriers may present with congenital abnormalities.
Segregation of reciprocal translocations

( A ) A carrier of a reciprocal translocation has one unaltered copy of each chromosome that participates in the translocation, together with two hybrid chromosomes. Only the relevant chromosomes are shown, for illustration each is labelled with a circled number. ( B ) During meiosis, replicated sister chromatids pair up with their homologues. In the case of a translocation carrier, so-called ‘quadrivalents’ can form, in which four instead of two chromosomes pair up. ( C ) Three possible segregation paths are illustrated. During ‘alternate’ segregation, chromosomes 1 and 4, and chromosomes 2 and 3 are segregated into separate gametes. ‘Adjacent 1’ and ‘Adjacent 2’ segregation leads to different combinations as indicated. Note that other segregation patterns can also occur, e.g. where three chromosomes segregate into one gamete, and only one into the other. ( D ) Only alternate segregation leads to gametes which either carry the two unaltered, ‘normal’ chromosomes, or the two hybrid chromosomes. Zygotes formed from these gametes are expected to be phenotypically normal (unless there is a critical gene disruption at the translocation breakpoint). However, in the other two instances, all gametes carry one unaltered and one hybrid chromosome. Fertilisation of these gametes leads to zygotes carrying partial trisomy of one chromosomal segment, and partial monosomy of a different segment.
A second type of translocation is the Robertsonian translocation. Here, two acrocentric chromosomes both break at the centromere, lose their short p arms, and form a single chromosome, containing one centromere and the q arms of both original chromosomes. Carriers of Robertsonian translocations are usually phenotypically normal since only a small amount of genetic material, the nucleolar organiser region (NOR), is present in the short arms of all acrocentric chromosomes (see Figure 2 ). Therefore, the loss of two short arms can be compensated by the remaining acrocentric chromosomes. However, similar to reciprocal translocations, gamete formation and subsequent fertilisation can lead to the formation of zygotes with either monosomy or trisomy of one of the participating acrocentric chromosomes and therefore children with chromosomal imbalances. As is the case with meiosis in carriers of translocations, meiosis in carriers of inversions can also lead to the formation of gametes carrying an unbalanced combination of chromosomes. Therefore, such carriers may also have children with chromosomal imbalances. Carrier frequencies for Robertsonian translocations and inversions which are not considered normal variants are estimated to be 1:1000 and 1:2000 respectively.
Truly balanced translocations and inversions do not lead to the net loss of genetic material, therefore only affect the phenotype of the carrier if either a chromosome break has disrupted an important gene or a break affects the expression of a gene without disrupting its coding region, e.g. by juxtaposing the complete coding region of one gene to the control sequences of a different gene.
Microdeletions, microduplications, CNVs
Molecular genetic analysis of patients with symptoms that cannot be explained using cytogenetics can lead to the identification of the underlying causes, which in many cases are microdeletions, microduplications and other CNVs. Such variations can involve single genes or relatively few genes, which can then allow researchers to determine which particular gene is responsible for specific symptoms. Table 5 shows example of microdeletion and microduplication syndromes, together with key genes, where known, and associated symptoms. Note that in some cases, both microdeletion of a key region as well as microduplication of the same region have been identified as causative for ‘reciprocal’ syndromes. An example is a 3.6-Mb region at 17p11.2, which, when deleted, causes Smith–Magenis syndrome, but when duplicated, causes Potocki–Lupski syndrome.
Where specific genes have been identified as associated with particular features of the syndrome these are noted, but this does not exclude a role for additional genes in the region. Extent of the deletions/duplications often varies between patients but in general larger imbalances are associated with greater severity of symptoms.
Primary sex determination in mammals is chromosomal, meaning that the development of the gonads into male (testes) or female (ovary) is determined by the sex chromosomes. The female caries two X chromosomes (46,XX) and the male has one X and one Y chromosome (46,XY). In some animals, sex determination is in part, or whole, environmentally determined (for example by temperature in most turtles), but in mammals, initiation of sexual fate is entirely driven by the chromosomes. Along with the haploid set of autosomes (22 in humans), each egg of the female has a single X chromosome, while the male can generate a sperm carrying either an X or a Y chromosome. If the egg receives an X chromosome from the sperm, the resulting XX individual will form ovaries through development and be female. If the egg receives a Y chromosome from the sperm, the resulting XY individual will form testes and be male ( Figure 7 ). The Y chromosome is relatively small (57 Mb, with 171 genes and of these only approximately one-third are protein encoding (see Table 1 )), but carries a gene that is crucial for the formation of the testes, encoding a testis-determining factor (TDF), also known as the sex-determining region Y ( SRY ) ( Figure 8 ). All else being wild-type, an individual carrying a normally functioning copy of this gene will develop as a male. Thus, if the Y chromosome is missing (45,X) or if SRY is deleted, female development will ensue, although, two X chromosomes are needed for complete ovarian development. Development of primary sexual characteristics aside from the gonads, that is the reproductive structures (penis, epididymides, seminal vesicles and prostate gland in males; oviducts, vagina, cervix and uterus in females) as well as secondary sexual characteristics (mammary glands in females, along with other sex-specific features such as size, musculature, facial hair and vocal cartilage) are determined by hormones that are secreted by the gonads and this is influenced by many other genetic and environmental factors. Oestrogen, secreted by the ovaries, directs female development, while the newly formed testes secrete anti-Müllerian duct hormone and testosterone which masculinises the foetus.
The X and Y chromosomes determine male or female sexual development

Males produce haploid gametes (sperm) that are either 23,X or 23,Y. Females produce haploid gametes (eggs) that are 23,X. Daughters inherit an X chromosome from their mother and an X chromosome from their father. Sons inherit an X chromosome from their mother and a Y chromosome from their father (paternal chromosomes indicated in blue, maternal chromosomes indicated in green).
Schematic map of the X and Y chromosomes

The X and Y chromosomes are depicted, showing the short (p) and long (q) arms and centromeres (black circle). The pseudoautosomal regions (PAR) 1 and 2 are highlighted in red. The sex-determining genes SRY and DAX1 are indicated. The location of the X inactivation centre (XIC) and the XIST gene is shown. Locations of other genes specifically mentioned in the text are indicated.
The X chromosome is relatively large (156 Mb) and incorporates approximately 1500 genes more than half of which are protein encoding (see Table 1 ), the vast majority of these have nothing to do with sex determination and are needed by both males and females. As such, the imbalance between males and females with respect to the number of X chromosomes in the genome (and therefore potential gene expression levels) needs to be rectified and this is accomplished by different mechanisms in different animal species. This balancing phenomenon, referred to as ‘dosage compensation’ is achieved in mammals by a process termed X chromosome inactivation. Early in development, in every cell in the female embryo, one of the two X chromosomes becomes inactivated, such that the majority, but not all, of the genes are not expressed from the inactive chromosome (Xi). As a consequence, the levels of expression of these genes on the active X chromosome (Xa) in female cells are equivalent to levels in male cells that only have one X chromosome. With respect to which of the two X chromosomes are inactivated, this occurs randomly from one cell to another, but then persists through subsequent cell divisions. This means that as development continues, female tissues become a patchwork, an expression-mosaic, with one of the two X chromosomes activated in some patches and the other X chromosome activated in adjacent patches. The result of this process is visible in female tortoiseshell cats, which are heterozygous for X-linked black and orange coat colour genes; the consequence of X inactivation is evident as random black and orange patches of fur in the adult. As a stochastic process, the proportion of cells that have inactivated the paternally inherited X chromosome, versus the maternally inherited X chromosome, will average 50% each. However, from one individual to another and even from one tissue to another within an individual, there can be considerable skewing from the 50% mean. Think of the tortoiseshell cat, most show roughly equivalent areas of orange and black fur, but some are more black than orange, while others are more orange than black. All female mammals are effectively expression-mosaics with respect to their X chromosomes.
One consequence for geneticists of the male-specific Y chromosome and X-inactivation in females is that the terminology of recessive and dominant allele variants becomes complicated. In addition, as described below, pathogenic allele variants on the X-chromosome can show hugely variable disease penetrance in females.
The pseudoautosomal regions
During human oogenesis, the two X chromosomes synapse in meiosis I and engage in crossover, exactly as the autosomes do. In male spermatogenesis, despite the X and Y chromosomes being of very different sizes and different genetic make-up, the chromosomes do pair (and undergo recombination) in meiosis, at short regions of homology at the ends of each chromosome. These regions are termed pseudoautosomal regions (PAR) 1 and 2 ( Figure 8 ), because they are present on both the X and Y chromosomes; most of the genes in these regions are not subject to X inactivation in females and they behave like autosomal sequences in terms of inheritance patterns. All genes tested within the larger PAR1 escape inactivation in female cells, thus both alleles are expressed in both male and female cells. The smaller PAR2 region has been a recent acquisition in evolutionary terms, there is no equivalent in the mouse (and even some primates). PAR2 genes behave differently. The two most telomeric genes of PAR2, IL9R and CXYorf1 , escape inactivation and are expressed from the Xi as well as Xa in female cells. However, two other genes of PAR2, SYBL1 and HSPRY3 , do become inactivated on Xi. To compensate for this in male cells, the Y chromosome alleles of these two genes are hypermethylated and not expressed, thus in both female and male cells, only one allele is expressed.
In addition to genes within the PARs, there are several homologous gene pairs (or gametologues) present on the X and Y chromosomes, that are located in the X and Y-specific regions, that do not undergo recombination. Consequently these gene pairs have diverged from one another through evolution and often have quite different sequence from each other, although may retain a similar function. An example is the RPS4X and RPS4Y pair, which encode ribosomal proteins of essentially the same function, but differ in 19 of the 263 encoded amino acids. RPS4X escapes inactivation, therefore both alleles are expressed in female cells, while male cells express both single alleles RPS4X and RPS4Y .
SRY, DAX1 and sex determination
The SRY gene is located on the short arm of the Y chromosome, just 5 kb away from the PAR1 boundary ( Figure 8 ). It encodes a transcription factor and is a trigger for driving male sexual development. In 46,XY individuals where the gene is dysfunctional or deleted, female development ensues. Furthermore, in approximately 80% of 46,XX male cases, the SRY gene is found translocated to an X chromosome.
In the developing embryo, a long and narrow structure called the genital ridge is the precursor to gonad formation in both sexes. The somatic cells of the genital ridge differentiate into either Sertoli cells, which promote the testicular differentiation programme or into granulosa cells, which promote ovarian differentiation. Expression of SRY in the genital ridge induces the start of Sertoli cell differentiation. While expression of SRY is brief, it initiates a cascade of events that will lead to male development. The next gene in the cascade is SOX9 , an autosomal gene (located on chromosome 17) which also encodes a transcription factor and is essential for testes development. Expression of SOX9 in the genital ridge acts to induce the expression of several other genes required for testicular development, and also anti-Müllerian duct hormone which suppresses ovarian development. Thus the reverse is the case during ovarian development in XX individuals, where SOX9 is repressed. Some rare 46,XX individuals who have an extra copy of the SOX9 gene, develop as males (despite the absence of an SRY gene). Conversely, individuals who have a non-functional variant of SOX9 or gene deletion, develop a syndrome called campomelic dysplasia (which involves multiple organ systems) and 75% of 46,XY patients with this syndrome develop as phenotypic females or hermaphrodites.
It was initially thought that female development was the default state in the absence of SRY, however, this does not accurately reflect the situation that female sexual development is an active, genetically controlled process. A gene on the X chromosome, DAX1 (aka NROB1 ), which encodes a hormone receptor/transcriptional regulator, is required for female sexual development. DAX1 is expressed in the genital ridge shortly after SRY and antagonises the function of SRY by interfering with the induction of SOX9 , in a dose-dependent manner. Normally, in 46,XY genital ridge cells, DAX1 is expressed from the X chromosome and SRY from the Y chromosome, in a ratio that leads to testes development. In 46,XX genital ridge cells, one copy of DAX1 is expressed (the other is inactivated on Xi), in the absence of SRY, to produce ovaries. However, if there are two active copies of DAX1 (for example, through gene duplication on Xa), along with SRY in 46,XY individuals, this leads to poorly formed gonads that produce neither anti-Müllerian duct hormone nor testosterone and individuals appear phenotypically female.
Following initiation of the female developmental pathway, several genes play a key role in female sexual development. The WNT4A gene (chromosome 1) encodes a secreted factor that is essential for the growth of ovarian follicle cells and is down-regulated by SOX9. The gene NR5A1/SF1 (on chromosome 9) encodes another transcriptional regulator important in both male and female pathways (as well as in the adrenal glands). Along with SRY, SF1 co-regulates SOX9 expression and is therefore critical in male sex determination. However, this multifunctional protein also plays a role later in ovarian follicular development. Thus, mutations in this gene can lead to disorders of sex development (DSD), including sex reversal, in both XX and XY individuals. Thus the SRY and DAX1 genes (present on Y and X chromosomes respectively) determine sex, as they act to flip the switch between male and female sexual fates. However, in each case, there are critical downstream regulators that promote one programme and/or inhibit the other programme and variant or mutated forms of these genes can cause DSD.
The human Y chromosome has 63 protein encoding genes, and aside from genes within the PARs and the gametologues, the majority are expressed in the testis and are involved in male fertility. The Y chromosome also has a large number (392 at last count) of pseudogenes. These have resulted from the fact that the Y chromosome (outside the PARs) has no recombination partner. As a consequence, through evolution, harmful gene mutations cannot uncouple (and thereby be selected against) from necessary genes and therefore such deleterious mutations, now in the form of pseudogenes, hitchhike along with the necessary genes.
The process of X inactivation
Early in female development (the early blastocyst), one X chromosome in each 46,XX cell becomes inactivated. This is initiated from a region called the X-inactivation centre (XIC) at Xq13 and in humans occurs at random, it can be either the X chromosome inherited from the mother, or the one inherited from the father. Inactivation starts with the expression of a long, non-coding RNA located within the XIC, called the X-inactive specific transcript ( XIST ), from the chromosome that will be silenced ( Figure 9 ). XIST RNA coats the chromosome from which it is expressed, spreading from the XIC outwards in both directions along the entire length of the chromosome. This then leads to several epigenetic changes along the coated chromosome, including depletion of RNA polymerase II, loss of histone acetylation and an increase in histone ubiquitination and repressive methylation to silence gene expression on Xi. The Xi chromosome becomes condensed and can be seen microscopically as a dense area to the side of the nucleus referred to as the Barr body (as it was first described by the cytogeneticist Murray Barr). The inactivation is stable through subsequent cell divisions so that the same Xi is maintained in each cell lineage throughout development and adult life, with the exception of the germline. In germ cells X inactivation is reversed, so that all oocytes contain an active X.

A simplified view of genes located at the XIC (not to scale), which maps at Xq13.2 on the human X chromosome and spans over 1 Mb. XIST (red) is surrounded by a number of other non-coding RNA genes (blue), which have an effect upon XIST regulation, including TSIX . Additionally, there are protein encoding genes (yellow), within the XIC region and recently RLIM has been shown to regulate XIST expression. Deletions across this region affect the process of X chromosome inactivation, but the function of all the genes and sequence regions located at XIC are yet to be fully understood.
At the XIC another non-coding transcript is expressed, partially overlapping and in the opposite (antisense) direction to XIST , the TSIX transcript is specifically expressed from the active X chromosome Xa ( Figure 9 ). TSIX acts locally as a negative regulator of XIST expression and protects Xa against inactivation. In conditions of aneuploidy, with more than two X chromosomes, only one X remains activated, which reveals that there is a counting mechanism at play. For each autosome set, one X chromosome remains active, although how this occurs is currently poorly understood. Deletion of XIC sequences (including the XIST and TSIX genes) from one X chromosome still allows inactivation of the other, wild-type X chromosome, in 46,XX individuals. Furthermore, in transgenic mice, introduction of an XIC into an autosome, renders the autosome subject to silencing. These studies show that the XIC (and XIST ) is required for initiation of chromosome inactivation in cis , but that the counting mechanism must involve factors and regions outside XIST and TSIX . It is thought that X chromosome inactivation (and the counting mechanism) must be regulated by X-encoded activators and autosomally encoded suppressors which control XIST . Recently, an X-linked gene, RLIM (or RNF12 ), located 500 kb upstream of XIST , has been identified as an important regulator of XIST expression, as the RLIM protein works to degrade an inhibitor of XIST transcription. However, the complexities of this process are yet far from being fully understood.
Approximately one-fifth of the genes on the X chromosome escape inactivation on Xi. These either have a Y chromosome homologue (for example, located in a PAR, as described above), or those that do not have a Y homologue tend to lie in clusters (mostly on the short arm, Xp) and apparently the 2:1 dosage (expression in female:male cells) is not problematic. It is thought that these genes are surrounded by a DNA sequence that binds a protein factor (termed CTCF) that can insulate the genes from the actions of XIST .
Due to the low gene count of the Y chromosome and the process of X inactivation, having an abnormal number of sex chromosomes has milder consequences than abnormal numbers of autosomes. Females with Triple X syndrome (47,XXX) or males with 47,XYY tend to be taller than average, but usually show few other physical differences ( Table 4 ) and have normal fertility, thus can go undiagnosed. X inactivation in 47,XXX cells will lead to the inactivation of two X chromosomes, so despite the presence of the trisomy, the vast majority of the X-linked genes will be expressed from just the single Xa in each cell. However, overexpression of genes that escape X inactivation (as described above) gives rise to the syndrome. In 47,XYY men, there is double the Y chromosome gene dose. Therefore, there will be double the levels of Y-specific gene expression and an extra dose of products of the genes that have an X chromosome homologue (one dose from X and two doses from Y). Males with 47,XYY, as well as the above noted features, tend to have an increased risk of behavioural, emotional and social difficulties.
Men with Klinefelter syndrome (47,XXY) carry an extra X chromosome and tend to be sterile, however symptoms are frequently very subtle ( Table 4 ) and only noticed at puberty. Again, one of the two X chromosome will be inactivated (Xi) in each cell, therefore, 47,XXY cells will only show overexpression of genes (compared with 46,XY cells) that escape X inactivation (in 47,XXX individuals, this extra expression is in the context of female development, while in 47,XXY individuals it is in the context of male development, so can have different consequences). Men with 48,XXXY display a more severe syndrome, resulting from the extra overexpression of genes that escape X inactivation, as well as the expression of Y-specific genes.
Complete loss of the X chromosome, 45,Y is early embryonic lethal. However, females with monosomy of chromosome X (45,X) or partial loss of an X chromosome, develop Turner syndrome ( Table 4 ). In 45,X cases where an entire sex chromosome (X or Y) is lost, the remaining X chromosome does not undergo inactivation, however, this leads to half the normal expression levels of genes that do not undergo X chromosome inactivation. As such, the loss of dosage of several of these genes contributes to the syndrome. In cases where there is only partial loss of the X chromosome, such individuals will display some features of the syndrome. A gene that lies within PAR1, with alleles on both X and Y chromosomes, SHOX ( Figure 8 ), is responsible for approximately two-thirds of the height deficit seen in Turner syndrome individuals. With loss of this region of the X chromosome, SHOX is expressed from the other allele, therefore at only half the usual levels and this is not sufficient (haploinsufficiency) to fully achieve its required growth-related function. Heterozygous loss-of-function mutations in this gene alone in both males and females causes Leri–Weill dyschondrosteosis which is characterised by skeletal dysplasia and short stature. Loss of function in both alleles causes Langer mesomelic dysplasia, which is associated with severe limb aplasia and severe height deficit. Conversely, duplication of the SHOX gene is associated with tall stature.
Pathogenic single gene variants
Haemophilia a, an example of x-linked recessive inheritance.
Haemophilia A is a condition where blood clotting is defective, due to deficiency in the activity of one of the blood clotting factors, factor VIII. Symptoms can vary considerably, from mild cases, where patients only bleed excessively after major trauma or surgery, to severe cases, where patients suffer up to 30 annual episodes of spontaneous or excessive bleeding, even after minor trauma. Factor VIII is encoded by the F8 gene, located on chromosome Xq28 ( Figure 8 ) and the gene is subject to X inactivation. There is no Y homologue, therefore if males inherit a pathogenic variant allele on the maternal X chromosome (or in 30% cases have a de novo mutation), they will be affected and disease severity will reflect the type of mutation (ranging from expression of a dysfunctional protein to complete absence of the factor). Females who are heterozygous, carrying one copy of a pathogenic variant allele, are generally asymptomatic and thus haemophilia shows a typical X-linked recessive inheritance pattern (see section on ‘Single-gene disorders’). However, as a result of X inactivation, some cells will express the wild-type F8 allele (from Xa), while other cells will express the pathogenic variant allele and the overall expression ratio between the two alleles can be 50:50 or skewed to one or the other. No cell will express both alleles. As a consequence, most female carriers produce enough factor VIII (between 30 and 70% of normal circulating levels, the variation depending both upon the nature of the variant allele and the degree to which it is Xi silenced) to appear largely unaffected. However, a significant proportion of female carriers do show some bleeding problems (for example, heavy menstrual bleeding) and in some cases (carriers who have less than 30% of normal factor VIII levels) can show mild haemophilia symptoms. In rare cases, where females inherit two variant alleles, they are more severely affected, as in males.
Rett syndrome, an example of X-linked dominant inheritance
Rett syndrome is an X-linked, dominant, neurodevelopmental disorder seen predominantly in females and becomes apparent in babies between the age of 6 and 18 months. After an initial phase of apparently normal development, individuals develop severe mental and physical disabilities, displaying coordination problems, slower growth, repetitive movements, seizures, scoliosis and other problems. The age at which symptoms first appear and the severity, varies considerably from one individual to another. Rett syndrome affects approximately 1 in 10000 females and is a single gene disorder involving the X-linked MECP2 gene. Due to the severity of symptoms, this usually arises as a de novo mutation. Boys with a similar mutation have a more severe phenotype, for example congenital encephalopathy, and die shortly after birth. MECP2 encodes a protein that binds to methylated DNA and has an important epigenetic function as a repressor of gene expression. Although the gene is normally expressed throughout the body, its function is essential in mature nerve cells and the phenotypic consequences of loss-of-function mutations (for example, loss of expression mutations or mutations that give rise to a non-functional protein) are most profound in the brain. The nature of the mutation and the extent to which the allele has lost function dictates one variable seen in disease severity. Additionally, the MECP2 gene is subject to X chromosome inactivation, this therefore also contributes to the variation seen in disease severity. If the mutant allele shows skewed inactivation (such that this allele is more frequently inactivated on Xi than the wild-type allele), this can result in considerably milder symptoms. It has been proposed that following X inactivation during development, cells which inactivated the chromosome carrying the wild-type MECP2 allele and therefore express the mutant MECP2 allele, may be selected against, resulting in a skewed X inactivation body pattern. In addition other genetic factors may exacerbate or alleviate the disease pathogenicity to contribute to the variation observed. The levels of MECP2 are critical and both too little and too much are deleterious. Therefore, mutations that result in overexpression of this gene give rise to a different syndrome ( MECP2 duplication syndrome). In males MECP2 duplication leads to severe intellectual disability and epilepsy; similar duplications in females lead to a more variable condition depending upon the proportion of cells that inactivate the X chromosome containing the duplication.
In conclusion, the presentation and severity of syndromes and diseases resulting from variants of the sex chromosomes are not only influenced by the nature of the variant itself, but also by the sex-linked ploidy of these chromosomes and the consequences of X chromosome inactivation.
Many conditions and diseases depend on the genotype at a single locus (or gene), with inheritance following Mendel’s laws of segregation, independent assortment and dominance. Therefore, these diseases are often called ‘Mendelian’ although not all inherited disorders follow Mendel’s laws (e.g. triplet-repeat diseases and imprinting disorders). To date, over 6000 phenotypes have been identified for which the molecular basis is known, these phenotypes and the associated genes are collected in the database OMIM (‘Online Mendelian Inheritance in Man’, https://www.omim.org/ ). Some well-characterised examples are shown in Table 6 .
Diseases are shown together with their inheritance patterns, the affected gene, the most commonly found types of mutation, and estimated incidence rates. Note, some diseases, for example osteogenesis imperfecta (of which there are several forms), can be caused by pathogenic variants in one of a number of different genes.
Modes of inheritance and examples
Mendelian diseases can be recognised by their characteristic patterns of inheritance in family trees or pedigrees. Pedigrees can also reveal if the locus in question resides on an autosome or a sex chromosome and if a genetic variant is dominant or recessive. To understand the concepts of dominant and recessive variants, it is important to recall that each diploid human cell carries two copies (called alleles) of each autosomal gene, one inherited from the mother and one from the father. Frequently, these alleles are not identical. A person carrying two identical copies of the same allele on both autosomes is homozygous for this allele, while a person carrying two different alleles is heterozygous for the locus. A dominant allele is one which leads to a particular phenotype (e.g. a genetic disorder) no matter if the second allele is ‘normal’ or not. This is because the second, ‘normal’ allele cannot compensate for the effect of the dominant allele. A recessive allele, on the other hand, does not lead to a phenotype on its own, here, the ‘normal’ allele is sufficient or can compensate. However, if a person inherits recessive alleles of a gene from both parents, then the corresponding phenotype is displayed because no ‘normal’ allele is present to compensate. Consequently, five distinct types of Mendelian inheritance patterns can be distinguished.
Autosomal dominant
A person with an autosomal dominant disorder usually has at least one similarly affected parent and on average 50% of their children will have the disorder. Such conditions are revealed in pedigrees ( Figure 10 ) because the disease occurs in each generation, affects both males and females, and transmission can occur from either parent to offspring of either sex. Frequently, but not always, autosomal dominant disorders are caused by genetic variants which convey a novel function to a gene product (termed gain-of-function). In this case, the presence of a ‘normal’, non-pathogenic allele of the gene on the homologous autosome cannot compensate for the altered function of the mutated gene. Note that autosomal dominant disorders can also be caused by loss-of-function alleles, if 50% of normal gene expression from the normal allele is not sufficient, a phenomenon termed haploinsufficiency.
Autosomal dominant inheritance and pedigree

( A ) Inheritance pattern of an autosomal dominant variant (red). Only the relevant chromosomes are shown. ( B ) Pedigree of a family with an autosomal dominant condition. ( C ) Key for pedigree symbols.
Autosomal recessive
Autosomal recessive conditions are caused by loss-of-function pathogenic variants which on their own do not lead to a recognisable phenotype. Here, the presence of a second, functional allele of the gene in question on the homologous autosome is sufficient to compensate. Consequently, such conditions only manifest themselves in individuals who carry pathogenic variants at both the homologous loci (either two identical or two different recessive variants). Usually, such individuals have two unaffected parents, who are both non-symptomatic heterozygous carriers of a single pathogenic allele ( Figure 11 ). The children of two carrier parents have a 25% chance of inheriting both pathogenic variants, while the children of affected individuals are obligate carriers of a pathogenic variant. Disease incidence is frequently increased in families where parents are consanguineous (related by descent). In families with multiple affected generations, autosomal recessive diseases often skip one or more generations.
Autosomal recessive inheritance and pedigree

( A ) Inheritance pattern of an autosomal recessive variant (red). Only the relevant chromosomes are shown. ( B ) Pedigree of a family with an autosomal recessive condition. See Figure 10 C for key to symbols.
X-linked recessive
Diseases which are caused by recessive variants in loci located on the X chromosome affect females and males differently. Males have a single X chromosome, therefore, if they carry a pathogenic variant, they have no second allele to compensate for its effect, and will be affected by the disease. All their daughters will inherit their X chromosome, therefore will be carriers, while their sons will be unaffected ( Figure 12 ). Since females carry two X chromosomes, they will typically only be affected by the disease if they inherited one pathogenic variant of the relevant gene from their (affected) father and a second pathogenic variant from their mother, who could be an unaffected carrier or a homozygous, affected individual. However, due to the phenomenon of X chromosome inactivation in females, such variants are often not completely recessive and can show some aspects of the phenotype in female carriers (as explained in the section ‘The sex chromosomes, X and Y’).
X-linked recessive inheritance and pedigree

( A ) Inheritance pattern of an X-linked recessive variant (red). Only the relevant chromosomes are shown. ( B ) Pedigree of a family with an X-linked recessive condition. See Figure 10 C for key to symbols.
X-linked dominant
A dominant pathogenic variant on the X chromosome will typically affect both males and females (but this is also complicated by X-inactivation). All daughters of an affected male will inherit the condition, while all of his sons will be unaffected. In the case of an affected female, if she has a single pathogenic variant, her children will have a 50% chance of being affected. However, if she has inherited a pathogenic variant from each of her parents, both her parents will typically have been affected, and all her children will also be affected. The actual situation may be more complicated, for example in late-onset conditions if an apparently unaffected parent died before they developed the disorder or if one of the pathogenic alleles is non-penetrant due to non-random X-inactivation. X-linked dominant disorders are rare, but examples are shown in Table 6 .
Since the Y chromosome is very small and only contains comparatively few genes, Y-linked single-gene disorders are even rarer than X-linked dominant ones. As much of the Y chromosome exists in a hemizygous state (with the exception of genes with homologues on the X chromosome), recessive and dominant definitions do not apply; as such, the phenotype of Y chromosome variants will be manifest. Consequently, affected males also have affected fathers, unless a de novo mutation has occurred, and all their sons will be affected. Since daughters of affected males will inherit their father’s normal X chromosome, and not the affected Y chromosome, they will be unaffected, and their offspring will also be unaffected. An example of a Y-linked condition is nonobstructive spermatogenic failure, which leads to fertility problems in males which may be addressed by assisted reproductive methods such as in vitro fertilisation (IVF).
Types of variants and their effects
Mendelian disorders are caused by pathogenic variants at single loci (in single genes), therefore, it is relevant to briefly discuss what kinds of mutations are involved and what their consequences upon gene function are. This depends on where within the sequence of a gene the change has occurred (e.g. within the coding region, in a control region or in a region involved in post-transcriptional modification).
Mutations can be categorised into those where nucleotides are exchanged against different ones where the total number of nucleotides do not change, and those where nucleotides are deleted, inserted or a combination thereof, with a concomitant change in the overall number of nucleotides. Where only a few nucleotides are involved, this is referred to as a microlesion, if only a single nucleotide is involved, this is referred to as a point mutation.
The following section will briefly describe mutations in coding regions, but microlesions outside the coding region of a gene can still have severe consequences. Mutations that change the regulation of a gene’s expression (e.g. promoter mutations) can lead to the production of too much or too little of the resulting protein, which may lead to a noticeable phenotype. Mutations can also occur in the conserved intronic sequences directly adjacent to intron–exon boundaries. Such mutations can then lead to aberrant splicing of the resulting transcript, with subsequent consequences on the encoded protein. In addition, mutations in non-coding RNAs can have profound effects, for example in one of the numerous miRNAs, which act to control the expression of other genes.
Point mutations
A point mutation is one which changes one nucleotide by substitution (one base pair is replaced by another), deletion or addition. If a substitution point mutation occurs within the coding region of a gene, various outcomes are possible: silent or synonymous mutations lead to the exchange of one codon for a different codon which still encodes the same amino acid. For example, the codons ATT, ATC and ATA all code for the amino acid isoleucine and if a mutation changed ATT to ATC, this would not lead to a change in the encoded protein sequence, therefore, such mutations are not expected to change the function of the encoded proteins. Missense mutations lead to a change in the codon such that it encodes a different amino acid. For example, a change from GAG to GTG will lead to the incorporation of valine into the resulting protein instead of glutamic acid. Such changes could lead to the complete loss-of-function of the resulting protein, or a dramatic change in function, or may have only a small effect that can be tolerated. This depends on the context of the amino acid within the mature protein, its function and on the chemical characteristics of the amino acids that are exchanged. Nonsense mutations lead to a codon for an amino acid being exchanged for one of the three stop codons. For example, a change from TAC to TAG leads to a change from the codon for tyrosine to a stop codon. Translation of the resulting mutated coding sequence leads to the formation of a prematurely truncated polypeptide, and most such truncations lead to non-functional proteins, although if they occur towards the C-terminal end of the protein they may have less effect.
Insertions, deletions and indels
The term ‘indel’ refers to mutations that change the total number of nucleotides in a genome, by in sertion, del etion or a combination of both. Indels generally refer to small changes from a point mutation, up to 1 kb. Indels occurring in sequences which control levels of gene expression or transcript splicing will lead to aberrant gene expression or splicing, but the effect of indels in coding sequences depends on the actual number of nucleotides inserted or deleted.
Deletion of three (or a multiple of three) nucleotides from a coding sequence corresponds to the deletion of one (or more) codons and will lead to the expression of a protein where one (or more) amino acids are deleted, without changes to the remaining amino acid sequence ( Figure 13 ). Such polypeptides may still be functional. However, if a number of nucleotides which is not divisible by three is deleted from a coding region, all subsequent codons will be altered, a phenomenon called a ‘frameshift’ ( Figure 13 ). A ribosome translates mRNA molecules one triplet (codon) at a time, before moving to the next triplet. If a single nucleotide is deleted, the ribosome will still translate one triplet at a time, but from the deletion point onwards, each triplet will now differ from those originally present. This will lead to the formation of a polypeptide whose sequence will differ completely from that originally present. Frequently, such frame shifts lead to stop codons being brought into the reading frame soon after the deletion point, thereby truncating the protein.
Insertions and deletions

( 1 ) A wild-type sequence consisting of three-letter words. ( 2 ) Insertion of three letters inserts another three-letter word, and keeps the remainder of the sequence unchanged. ( 3 , 4 ) Deletion of a number of letters which is a multiple of three removes a number of words, but leaves the remainder of the sequence unchanged. Note that the resulting sentence still makes (more or less) sense. Deletion ( 5 ) or insertion ( 6 ) of a number of letters which is not a multiple of three changes all three-letter words after the point of deletion/insertion. The resulting sentence does not make sense any more. These two cases are examples of frameshift mutations.
Insertion of nucleotides follows the same principle. Insertion of three (or a multiple of three) nucleotides leads to the insertion of one (or more) amino acids into the translated protein, while insertion of a number of nucleotides not divisible by three will lead to a frameshift mutation.
Examples of single-gene disorders
A special case of insertion mutations are the nucleotide repeat expansion disorders, typically triplet-repeat disorders ( Table 7 ). Trinucleotide repeats are repetitive sequences where triplets of nucleotides are repeated in tandem multiple times; in some cases these repeats are located in coding sequences and are translated as stretches of polypeptide consisting of a repeat of the same amino acid. Other trinucleotide repeats are located in non-coding sequences. It is possible that the presence of expanded repeats within a transcript is toxic for the RNA processing machinery in the nucleus. The nucleotide sequence of trinucleotide repeats is often (CAG) n or (CTG) n , where n is the number of repeat units, but other trinucleotide repeats are also known. One unusual aspect of these repeats is that they can become unstable and expand, such that the number of repeats increases dramatically from one generation to the next, a phenomenon termed ‘anticipation’. In addition, the repeats (over a certain number) can be unstable through somatic cell division, such that cells of some tissues show hugely varying numbers of repeats in the expanded allele. In all known cases of trinucleotide repeat expansion disorders, individuals carrying a number of repeats up to a threshold do not show any clinical symptoms, while individuals carrying longer repeats show progressively severe symptoms.
Four genes subject to repeat expansions are shown, with the gene affected, repeat sequence, normal and pathogenic range of repeat, main disease features and estimated incidence.
Huntington disease
Huntington disease (HD) is one of the trinucleotide repeat expansion disorders where the CAG repeat encodes a polyglutamine tract within the coding region of the huntingtin gene HTT on chromosome 4p16. It is a progressive neurodegenerative disorder with patients suffering from progressive neural cell loss and atrophy. Symptoms start with personality and mood changes, followed by a steady deterioration of physical and mental abilities. The function of the huntingtin protein is unclear, but it is essential for development. Inheritance follows an autosomal dominant pattern, caused by a gain-of-function associated with the repeat expansion. Unaffected individuals carry between 9 and 35 CAG repeats, incomplete penetrance occurs in carriers of 36–39 repeats, while the disease is fully penetrant when 40 or more repeats are present. Alleles containing 250 and more repeats have been reported.
While repeat alleles of 9–30 are almost always transmitted without change to the next generation, larger alleles show instability, both in somatic tissues and in the germline, with a tendency towards expansion from one generation to the next. There is a correlation between the number of repeats and the severity of disease and also an inverse correlation between the number of repeats and the age of disease onset. The degree of repeat instability is also largely proportional to the number of repeats, and is also affected by the sex of the transmitting parent, with larger expansions occurring in male transmission. This leads to ‘anticipation’ where an apparently healthy individual might have a child with late onset HD and a grandchild with more severe symptoms and an earlier onset, and so on.
Achondroplasia
Achondroplasia (ACH) is the most common form of dwarfism in humans and is inherited in an autosomal dominant fashion with 100% penetrance. Individuals with ACH have shortened limbs, a large head, and a trunk of relatively normal size.
ACH is caused by specific variants in FGFR3 , the gene for fibroblast growth factor (FGF) receptor 3 (FGFR3), on chromosome 4p16. Almost all individuals with ACH are heterozygous for a variant which leads to a substitution of arginine for glycine at position 380 (p.Gly380Arg) in the mature protein. Eighty percent of ACH cases are due to spontaneous, de novo mutations, often occurring during spermatogenesis. FGFR3 is a transmembrane receptor protein which binds to FGF ligands and triggers intracellular signalling processes. One of these processes is the inhibition of chondrocyte proliferation in the growth plate of long bones. The p.Gly380Arg variant in FGFR3 generates a constitutively active version of the receptor which can be further activated by binding of FGF. Therefore, this variant acts as a gain-of-function mutation. Consequently, chondrocyte proliferation in growth plates is constitutively inhibited. While one such variant allele (in the heterozygous state) leads to ACH, homozygosity is lethal before birth or perinatally. Interestingly, loss-of-function variants in FGFR3 have also been described which cause a different condition, camptodactyly, tall stature and hearing loss (CATSHL) syndrome. This is an example where different variants of the same gene result in different phenotypes, so-called ‘allelic disorders’.
Cystic fibrosis
Cystic fibrosis (CF) mostly affects the lungs (resulting in breathing difficulty and frequent lung infections) and the pancreas (with disruption of the exocrine function), but the liver, kidney, intestines and male reproductive system are also frequently affected. It is the most common lethal genetic disease among Caucasians, and is inherited in an autosomal recessive pattern.
CF is caused by pathogenic variants in the CFTR gene, which encodes the CF transmembrane conductance regulator, a transmembrane protein which functions as a selective chloride channel. If the CFTR protein does not function properly, the chloride balance between the inside and outside of cells becomes disrupted, leading to the build-up of mucus in narrow passages in affected organs such as the lungs. Such blockages lead to damage and reduced function in these organs. The CFTR gene is located on chromosome 7q31 and encodes a protein of 1480 amino acids. The gene is approximately 250000 nts long and over 2000 pathogenic variants have been identified in its sequence. These variants fall into different classes (e.g. those where protein synthesis is defective, those where reduced amounts of normal protein is made, those where the synthesised protein is not processed properly and does not reach the membrane and others). As long as an individual carries one functional allele of CFTR , they may show no or only very mild symptoms ( Figure 14 ), but an individual carrying two pathogenic variants will display symptoms that depend on the amount of functional protein generated. The most common pathogenic variant, representing approximately 70% of Caucasian CF alleles, is a deletion of the codon for a phenylalanine at position 508 (p.Phe508del) in the mature protein. This particular variant leads to the synthesis of a protein which does not fold properly into its 3D shape, and is degraded by the cell before it can reach the membrane, therefore representing a loss of function.
CF symptoms depend on residual CFTR activity

The left-hand side shows a scale of residual CFTR protein activity, between 0 and 100%. The right-hand side shows corresponding symptoms. Note that heterozygotes, carrying one pathogenic CF allele, still retain 50% of CFTR activity, therefore may be asymptomatic or show only mild symptoms. Only patients with 5% or less residual CFTR activity show full CF symptoms. Figure adapted from Davis 2001 .
Mitochondria – structure and function
Mitochondria are cellular organelles containing a genome which is independent of the nuclear genome, and are thought to have arisen from a symbiotic relationship between a primitive bacterial cell and a precursor of a eukaryotic cell. Mitochondria are the ‘powerhouse’ of the cell and are responsible for generating energy in the form of ATP. In addition, these organelles are key structures in controlling the process of programmed cell death or apoptosis. As such, they embody the life and death of the cell. They are capsule-like in structure ( Figure 15 ) with two membranes, an outer and an inner, and an intermembrane space in between the two. The inner membrane folds on itself forming cristae which increase the overall surface area and contain a series of protein complexes and transport chains involved in the production of ATP.
Mitochondrial structure

The mitochondrion has two membranes; the inner membrane folds into series of cristae on which are the electron carriers and ATP synthase, responsible for the generation of ATP. The matrix of the mitochondrion contains multiple copies of the circular mtDNA (image by Mariana Ruiz Villarreal, reproduced from Wikimedia Commons: Public Domain).
The generation of energy by the mitochondria involves several stages which are collectively known as cellular respiration. Firstly, a sugar (usually glucose) taken in by a cell is broken down by glycolysis in the cytoplasm, to generate two molecules of pyruvate, NADH and ATP. The pyruvate produced in glycolysis is converted into acetyl CoA which then enters the Krebs cycle – both the reactions occur in the mitochondrial matrix. The product of the Krebs cycle is NADH, which undergoes oxidative phosphorylation in the last stage of cellular respiration. In oxidative phosphorylation, which occurs on the cristae of the inner membrane, electrons are transferred from NADH or FADH 2 to O 2 by a series of electron carriers ( Figure 16 ). As a result of this process, ATP is formed.
The electron transport chain

On the inner membrane of the mitochondria, electrons from NADH and FADH 2 pass through the electron transport chain to oxygen which then undergoes a reduction reaction, producing water. The chain comprises five complexes and a series of electron transporters and electrons are passed from donors to acceptors down the chain, releasing energy which is utilised to generate a proton gradient across the mitochondrial membrane (image reproduced from Wikimedia Commons: Public Domain).
Control of some aspects of apoptosis or programmed cell death (so called because the survival or death of the cell is determined by a balance of cellular components) is another important mitochondrial function. In the very early stages of apoptosis, mitochondria release cytochrome c , an essential component of the electron transport chain, which initiates a pathway of procaspase activation, central to apoptosis. Key in mediating the mitochondria to release cytochrome c are proteins from the BCL family which reside in the outer mitochondrial membrane.
The mitochondrial genome
Located in the mitochondrial matrix, the mitochondrial genome exists as a circular, dsDNA molecule ( Figure 17 ), of approximately 16500 bp encoding 37 genes, each of which is vital to the function of mitochondria. However, numerous nuclear DNA encoded proteins also contribute to the formation and function of the mitochondria. Each cell contains thousands of mitochondria and each mitochondrion contains multiple copies of the mitochondrial genome. Thirteen of the 37 mitochondrial genes encode subunits of the four respiratory complexes involved in oxidative phosphorylation, which are situated in the inner membrane, while the rest code for 22 tRNAs and 2 rRNAs. The majority of components of the respiratory complexes are expressed from the nuclear genome and transported into the mitochondria.

Circular and double stranded, with no introns, the mitochondrial genome comprises 37 genes which code for components of the respiratory complexes involved in oxidative phosphorylation as well as for tRNA and rRNA. Many of the components of the respiratory complexes are coded for by the nuclear DNA (image reproduced from Wikimedia Commons: Public Domain).
The mitochondrial genome is unique in several ways. Almost 93% of mtDNA is coding, compared with the nuclear genome of which only 3% is coding, and mtDNA is free from introns, histones and epigenetic marks. MtDNA is subject to a mutation rate 100-times higher than that of the nuclear genome, since the mtDNA repair systems are less robust (more error prone) than nuclear DNA repair systems and because the internal environment of the organelle has more reactive molecules that can damage DNA (from the products of the respiratory transport chain). Importantly, mtDNA is only inherited maternally – a female will pass on mitochondria to all her children while a male will not normally pass on any of his mitochondria ( Figure 18 ).
Mitochondrial inheritance

In the case of a mitochondrial mutation (see following section), an affected mother ( A ) will pass the mutation on to all of her children since mitochondria are maternally inherited. On the other hand paternal mitochondrial mutations ( B ) will not be transmitted to his children.
Heteroplasmy is an important feature of the mitochondrial genome – this means that not all copies of the mitochondrial genome within a cell are the same ( Figure 19 ). If a particular variant is present in all copies of the mitochondrial genome in a cell, the cell is said to be homoplasmic for the mutation. The situation where some mitochondria contain the mutation while others which do not are referred to as heteroplasmy. Obviously this is an important phenomenon when considering the inheritance of mitochondrial mutations (discussed below) as offspring may inherit a high or low proportion of mutant mitochondria from a carrier mother.
Heteroplasmy

A primordial germ cell containing a mixture of mutant and normal mtDNA may give rise to oocytes which have high, intermediate or low mutation loads, dependent on the region of cytoplasm which contributes to it.
Mitochondria and disease
A number of conditions, from age-related hearing loss to specific forms of epilepsy and diabetes, are associated with mutations in the mitochondrial genome, with the phenotypes and severity varying widely. Often affecting tissues with high energy requirements such as muscle and nerve tissue, there is significant overlap between phenotypes caused by different mutations. As an example, two phenotypically distinct conditions caused by mitochondrial mutations are described below.
Leber hereditary optic neuropathy (LHON) is a mitochondrially inherited disorder of vision, with a reported incidence of approximately 1/30000 to 1/50000 births in European populations and symptoms which typically appear in the teens or twenties. The central area of vision tends to be most severely affected, with blurring and cloudy vision progressing to loss of sharpness and colour vision in the later stages. The pathogenesis of the condition results from cell death in the optic nerve. Mutations in a number of mitochondrial genes, including those encoding several NADH dehydrogenases (MT-ND 1, 4 and 6 ), are known to cause LHON.
The precise link between these mutations and the optic nerve cell death is unknown, and further complicated by the fact that up to 85% of individuals harbouring these mutations never develop visual problems. In some families, additional complications are present, including cardiac conduction and mild neurological problems. Although many different mutations have been associated with LHON, one of three missense mutations is thought to be present in 90% of affected families. Genetic diagnosis of the mutations causing mitochondrial disease such as LHON can be carried out using PCR-based techniques, such as allele-specific PCR.
Leigh syndrome, in contrast with LHON, typically appears within the first year of life. It is a severe and progressive neurological condition, with progressive loss of both mental and motor skills. Affected children usually die within 2 or 3 years of first showing symptoms, generally because of respiratory failure. With an overall similar frequency to LHON, Leigh syndrome is found in some populations (for example in Quebec, Canada) at a much higher frequency. Lesions in the basal ganglia, cerebellum and brainstem are seen on MRI scans in affected individuals. Both nuclear and mtDNA mutations have been observed in this condition, affecting complexes II, III, IV as well as ATP synthase, also known as complex V.
Therapy for mitochondrial disease
A number of traditional approaches have been used to combat the symptoms of mitochondrial disease, mainly in the form of dietary supplements, with little success. More recently, several strategies, including gene transfer using adeno-associated viral vectors have been trialled, in an attempt to replace the mutated gene.
However, as there is currently no effective strategy for treating mitochondrial disease, the approach of preventing rather than curing the conditions seems very attractive. Women who carry a mitochondrial mutation have the option of using a donor egg or adoption in order to avoid the possibility of having an affected child, but the possibility of having their own healthy biological child is more attractive to many. Thus mitochondrial replacement therapy or the ‘three parent baby’, in which the nucleus is removed from an egg containing mutated mitochondria and is placed into an enucleated egg from a healthy donor, has gained increasing interest in recent years.
There are two main approaches to achieve this – pronuclear transfer and spindle nuclear transfer ( Figure 20 ). With pronuclear transfer, which is currently the technique approved in the U.K., the mother’s egg as well as a donor egg are both fertilised with the father’s sperm. Subsequently the nucleus from each fertilised egg is removed and the donor egg’s nucleus is then replaced with that of the mother. In the alternative technique (spindle transfer), the nucleus is removed from an unfertilised mother’s egg and this is then inserted into a donor egg which has had its nucleus removed. Fertilisation with the father’s sperm then takes place. Although the U.K. approved pronuclear transfer in 2016, the first baby to be born using this technique was a boy born to Jordanian parents at a clinic in Mexico, to avoid the transmission of a mitochondrial mutation causing Leigh syndrome. It is expected that more babies will be conceived using this technique.
Mitochondrial replacement therapy (‘three-parent baby’)

Eggs are harvested ( 1 ) from the mother-to-be, whose mitochondria are affected by a pathogenic DNA variant. The nucleus is extracted from the egg ( 2 ). Eggs are also harvested ( 3 ) from a donor female who has healthy mitochondria, and the nucleus is removed from the egg, leaving only the cytoplasm ( 4 ). Now the nucleus (2) is injected into the enucleated egg (4) to generate an egg (5) that has the nuclear DNA of the mother-to-be, and healthy mitochondria. This can now be fertilised in vitro using sperm from the father (6), and allowed to develop for a few days before implantation into the mother-to-be.
The nucleotide sequence of the human genome contains within it a vast amount of complex information that is required for a healthy human, and changes to the DNA sequence may lead to disease states as described in previous sections. However, there is another layer of information that is superimposed upon the nucleotide sequence information, and study of this additional information is referred to as ‘epigenetics’, a term derived from the Greek and literally meaning ‘above the genetics’. This epigenetic information takes the form of chemical groups (for example, methyl groups) attached to the DNA or attached to the histone proteins around which the DNA is wrapped in chromatin. A key concept is that epigenetic information can be inherited between cell generations.
DNA methylation
In humans (as well as other mammals) cytosines within the dinucleotide sequence CG may become methylated ( Figure 21 ). Note the distinction between a C–G base pair (where the C and G are on opposite DNA strands) and a CG dinucleotide, which is a CG sequence along one strand of the DNA read in the 5′ to 3′direction (consequently, this is often referred to as CpG, to indicate the phosphate linkage on the DNA backbone). However, due to the complementarity of DNA there will also be a CG sequence in the 5′ to 3′ direction on the other strand of DNA ( Figure 22 ). The methylated cytosines are recognised by specific proteins, which bind to the DNA at these locations, and often lead to the recruitment of further proteins; these protein complexes can lead to alterations in chromatin structure and thereby activity of genes, typically by affecting the modification of histones.
Methylation of cytosine and consequences of deamination of methyl-C

( A ) Cytosine can be methylated to generate 5-methylcytosine; this may undergo spontaneous deamination to generate thymine. The arrow indicates position of attachment to the deoxyribose ring. ( B ) CG dinucleotides (indicated by vertical lines) are relatively rare in the genome overall compared with the expected frequency. This is believed to be an evolutionary consequence of deamination of methylcytosine leading to conversion of many C–G base pairs within CG dinucleotides into T–A. However due to the importance of DNA methylation in transcription regulation (see Figure 25 ), CG dinucleotides are present at higher frequency around promoter regions (transcriptional start sites) of genes, generating so-called ‘CG islands’ (also known as CpG islands).
Methylation occurs on the C of the sequence CG and facilitates binding of specific regulatory proteins to the DNA

Note that the sequence CG is palindromic, in the sense that the same sequence occurs in the 5′ to 3′ direction on both strands of the DNA. Abbreviation: N, any nucleotide.
The cell has several different enzymes which are responsible for DNA methylation, the DNA methyltransferases (DNMTs). Two of these, DNMT3A and DNMT3B, appear to specialise in de novo methylation, while the major role for DNMT1 appears to be in maintenance methylation ( Figure 23 ). The importance of maintenance methylation is evident when DNA replication is considered ( Figure 24 ). When methylated DNA is replicated by the action of DNA polymerase, the new strand will be generated by incorporation of standard nucleotide triphosphates, and thus no methyl groups will be present. If this DNA is replicated again, the newly synthesised DNA would have no methylation. However, hemimethylated DNA is recognised by DNMT1, which will methylate the cytosine on the complementary strand. This ensures that DNA methylation patterns are heritable between cell generations.
Methylation and demethylation

The methylation process is initiated by addition of a methyl group to one strand of the DNA by DNMT3A or DNMT3B. The resultant hemimethylated DNA becomes fully methylated by the action of DNMT1. Removal of methyl groups from DNA may be active (involving DNA demethylases) or passive (in the absence of maintenance – see also Figure 24 ). Abbreviation: m, methyl (CH 3 ) group.
DNA methylation status is heritable but requires maintenance

( A ) The Cs within CG dinucleotides (blue) represent potential methylation sites. In this piece of DNA sites 1 and 2 are fully methylated (i.e. on both strands of the DNA), but site 3 is not methylated. ( B ) Following replication the new strands of DNA (green) are unmethylated; further rounds of replication of this hemimethylated DNA would lead to some unmethylated DNA. However, hemimethylated DNA is a substrate for the maintenance methylase, DNMT1. ( C ) The daughter DNA molecules are now fully methylated at the originally methylated positions (1 and 2), but remain unmethylated at position 3, which was unmethylated in the original DNA template. Abbreviation: m, methyl (CH 3 ) group.
Methylation patterns in DNA are dynamic and can be altered in response to stimuli or at particular stages of development. To achieve this, it is necessary to have enzymes which can remove methyl groups from DNA: the DNA demethylases.
Histone modifications
For packaging into chromatin in eukaryotes, DNA becomes wrapped around octamers of histones; each octamer includes two molecules each of H2A, H2B, H3 and H4. The histone octamer forms a disc-like shape, with the N-terminal tails of each of the individual histones protruding from this disc. The histone proteins are subject to a variety of post-translational modifications, which include acetylation, methylation, phosphorylation and ubiquitination. Like DNA methylation, these modifications may be altered in response to changing circumstances, and influence the compaction of the chromatin and its accessibility by transcription factors and other proteins ( Figure 25 ). The best understood histone modifications are those which occur on the N-terminal tails. For example, acetylation of lysine residues in the N-terminal tails renders chromatin less compact and thus facilitates transcription. Acetyl groups can be added by the histone acetyl transferase (HAT) group of enzymes, and removed by histone deacetylases (HDACs); thus the activity of HATs will facilitate transcription, while the activity of HDACs will tend to inhibit transcription.
Chromatin status is influenced by DNA methylation and histone acetylation

In active chromatin the N-terminal tails of histone proteins are acetylated (by HATs); the additional positive charges encourage a looser packing of nucleosomes that makes the DNA more accessible for other proteins and thus facilitates transcription. Addition of methyl groups to the CG dinucleotides (by DNMTs) and removal of the acetyl groups from the histones (by HDACs) provokes a more compact chromatin structure, which is not easily accessible by transcription factors, generating inactive chromatin. Reactivation of inactive chromatin is facilitated by DNA demethylases (which remove the methyl groups) and HATs.
DNA methylation status can influence histone modification and vice versa. For example, methyl-C binding proteins can recruit HDACs to sites of DNA methylation, and histone methylation may either facilitate or inhibit DNA methylation, depending upon which amino acid has been methylated within the histones. One consequence of all this modification is that, although all our cells may contain the same genome and therefore the same genes, the pattern of which genes may or may not be active in a given cell or tissue type is dependent upon the modifications present in the DNA and on the histone proteins: the epigenetics.
Chromosomal imprinting
Several regions of the human genome, involving roughly 100 genes, are rendered inactive by epigenetic mechanisms depending upon parent of origin. For some genes only the paternal allele is active, while the maternal copy is epigenetically silenced throughout the life of the individual. For other genes it is the maternal copy that is active and the paternal copy which becomes epigenetically silenced. The process of epigenetic silencing in a parent-of-origin specific manner is termed as ‘chromosomal imprinting’, and involves DNA methylation, histone modification and additional protein and RNA factors. During gametogenesis all previous imprints are removed from the DNA, and new imprints are established: female imprints during oogenesis and male imprints during spermatogenesis ( Figure 26 ), thus each embryo should receive chromosomes with a complementary set of imprints and therefore one active copy of each imprinted gene.
Imprints are erased and reset during gametogenesis

For each imprinted chromosome region, healthy adult humans ( 1 ) will have, in their somatic cells, one maternal and one paternal imprint. During gametogenesis, the imprints present ( 2 ) must first be erased ( 3 ), and then the imprints are reset ( 4 ) according to the sex of the parent. Therefore during oogenesis all relevant regions are given a female imprint, and during spermatogenesis all relevant regions are given a male imprint. Thus at fertilisation ( 5 ) the union of egg and sperm generates a baby ( 6 ) with one maternal and one paternal imprint for each imprinted region. Chromosomes are shown in green, with male and female imprints indicated by blue and pink respectively.
What is the purpose of imprinting? In general, paternal imprints lead to gene expression patterns associated with increased growth, and maternal imprints lead to gene expression patterns associated with decreased growth. Some theories regarding the origin of imprinting relate to adaptive co-evolution. Other theories are based on conflict relating to strategies for reproductive success: the mother has to make a greater investment of energy in the child than the father, and too much maternal investment in one child may be detrimental to her ability to reproduce again; however the father’s reproductive success may be facilitated by larger babies that are better equipped to survive. There is also a potential conflict between the needs of the developing foetus and the continued health of the mother. Imprinting is observed in all mammals and in some other species; there is much debate about its origins, but correct imprinting is critical for normal development.
For imprinted genes, with one copy epigenetically silenced throughout the lifetime of the individual, it is clearly vital that a functional copy of that gene is inherited from the other parent. Where this is not the case, for example due to gene mutations or errors affecting the resetting of imprints during gametogenesis, imprinting disorders will be seen.
Imprinting disorders and uniparental disomy
There are a number of genetic conditions which are related to imprinting, including Beckwith–Wiedemann syndrome, Silver–Russell syndrome, Angelman syndrome (AS) and Prader–Willi syndrome (PWS). One of the best characterised imprinted regions is located close to the centromere on the long arm of chromosome 15 (15q11.2). The genes in this region include SNRPN , two clusters of genes for small nucleolar RNAs (snoRNA), and UBE3A . The products of SNRPN and snoRNA genes appear to play roles in RNA processing within the nucleus, while the UBE3A protein functions in targeting proteins for degradation by the proteasome. Thus the products of genes in this region clearly have wide-ranging effects within the cell. SNRPN and the two snoRNA clusters are active only on the paternal chromosome 15, while UBE3A is active on the maternal copy ( Figure 27 ). Absence of the paternally expressed genes leads to PWS ( Table 8 ). In contrast, if there is no maternal copy of 15q11.2, and thus no active UBE3A gene, then the individual will be affected by AS. While the majority of PWS and AS cases are a consequence of microdeletions, there are other mechanisms ( Table 8 ), including uniparental disomy (UPD).
Imprinting on chromosome 15

A cluster of genes near the centromere of chromosome 15 (at band 15q11.2) is subject to imprinting in a parent-of-origin specific manner (indicated by blue and pink shading): a number of genes including SNRPN and many snoRNA genes are expressed exclusively from the paternal chromosome 15, and are silenced on the maternal 15. Conversely, the UBE3A gene is silenced on the paternal copy and active on the maternal copy. Thus loss of function of these genes has different consequences depending upon the parental origin: loss of UBE3A on the maternal 15 leads to Angelman syndrome ( Table 8 ) whereas loss of UBE3A on the paternal 15 is without consequence since the gene is inactive anyway on the paternal copy. The mechanism involves DNA methylation (indicated by hatching) of the SNRPN region of the maternal chromosome 15.
UPD is a rare phenomenon in which both homologues of a chromosome pair are derived from the same parent. If both chromosomes 15 are maternal in origin then the child will be affected by PWS, and if both 15s are paternal in origin the child will be affected by AS. There are essentially three mechanisms which can lead to UPD, each requiring two errors affecting meiosis/mitosis. Monosomy rescue is a rare, sporadic event which can allow a monosomic zygote to survive by duplication of the monosomic chromosome; the mechanism permitting this is not entirely clear, but it will always result in UPD. Trisomy rescue is, likewise, a rare, sporadic event, in which trisomic cells lose one chromosome, for example by anaphase lag, in which one chromosome does not get incorporated into a daughter nucleus during mitosis. Depending upon which of the three chromosomes is lost, trisomy rescue leads to UPD in one third of cases. A final possibility is for a nullisomic gamete to combine with a disomic gamete (in other words, meiotic errors in both parents), which appears statistically unlikely. UPD of some chromosomes is without consequence (for example 1, 5, 9, 10, 13, 21, 22), but for chromosomes that harbour imprinted genes (which include 6, 11, 14, 15, 20), the relevant imprinting disorder will ensue.
UPD can be in the form of heterodisomy (both homologues from one parent are present) or isodisomy (two copies of one of the parental homologues). In isodisomy, because the two chromosomes are identical there will be homozygosity for all alleles and therefore in this case UPD may also unmask a recessive disorder.
Although most cases of imprinting disorders occur as a consequence of de novo mutations, there are also cases in which mutations have been transmitted through families. For example, if a new UBE3A mutation is present in a spermatozoon that fertilises an egg, the resultant child would not show any phenotype, since the paternal copy of UBE3A is epigenetically silenced anyway. If the child is a male, he could also transmit this same mutation to offspring without consequence to their health. However, if the child is female, and she transmits this mutation to her offspring, they would be affected by AS.
Interestingly, imprinting of UBE3A appears to be maintained only in the brain; other tissues have biallelic expression. Furthermore, overexpression of UBE3A , for example as a consequence of gene duplication on the maternal chromosome 15, is associated with autism.
Epigenetic contributions to other disorders
During development, epigenetic modifications at many genetic loci change as part of the differentiation process, generating tissue types with specific patterns of gene expression. Epigenetic changes may also occur in response to environmental stimuli. Deficits affecting components of epigenetic mechanisms can lead to disease states. A well-characterised example involves the MECP2 gene which encodes a methyl-C binding protein. This protein recognises and binds to methylated cytosines in DNA, acting to recruit other complexes like HDAC which can alter the transcriptional status of the DNA. Pathogenic loss-of-function variants in MECP2 lead to Rett syndrome, as discussed earlier. Another example is the rare autosomal recessive disorder: immunodeficiency, centromeric instability and facial anomalies (ICF) syndrome. Roughly half of ICF cases are a consequence of biallelic pathogenic variants in the DNMT3B gene. From mouse studies, complete loss of function of DNMT3B appears to be incompatible with life, and thus the variants that lead to ICF are generally missense, with each patient having at least one missense variant which retains some DNMT3B function. Note that epigenetic changes can also involve non-coding RNAs, for which X-inactivation provides a good example.
It is becoming increasingly apparent that changes in epigenetic programming play a role in many conditions, including cancer, mental illness, autoimmune disease, diabetes and obesity. Identical twins have different patterns of DNA methylation and histone modification that may contribute to different disease susceptibility between them. Because epigenetic marks like DNA methylation can be faithfully copied between cell generations (in other words they are heritable), epigenetics can also help explain how the foetal or childhood environment can influence later susceptibility to disease. There is also evidence that epigenetic marks, including those on imprinted genes, may be perturbed during IVF, and that this may lead to health problems, for example imprinting disorders, in some children conceived by IVF.
The clear inheritance patterns associated with single gene (monogenic) disorders have facilitated the identification of the causative genetic changes for these conditions. However, increasing attention is now focused on genetic contributions to complex multifactorial disorders like diabetes, heart disease and schizophrenia, where disease is the outcome of a complex interplay of multiple genetic and environmental influences. The impact of an individual variant in one gene may be very small, but when present together with multiple variants in other genes, in the context of a particular environment, may lead to an increased risk of disease. The same is true for many traits (for example, height) and behaviours (for example, aggression or novelty seeking).
Type 2 diabetes (T2D) exemplifies complex disorders and is one that is increasing in incidence across the world. The major environmental contributions to T2D relate to diet and exercise: high-calorie diets in the context of a less active lifestyle, often involving long periods spent in front of a computer or television. This lifestyle presents a stark contrast with the environment that our ancestors had to survive, and traits that might once have been advantageous (like an energy-saving metabolism) have become a disadvantage. Identifying the genetic factors underlying T2D is challenging because of the small effects of individual contributions.
Whereas type 1 diabetes (T1D) is characterised by loss of insulin production as a consequence of autoimmune destruction of pancreatic islet cells, T2D is most commonly associated with insulin resistance – the body is no longer able to respond appropriately to insulin. T2D typically has a late onset (>35 years) and is associated with obesity, though onset at younger ages is increasing.
Family studies
Initial clues that genetics plays a role in complex disorders like T2D tend to come from family studies, in particular, the study of twins. If one of a pair of monozygotic (identical) twins is affected by a monogenic disorder like CF it is practically certain that the other twin also has CF, in other words, is concordant, because they have virtually identical DNA. However, for dizygotic (non-identical) twins, who share, on average, 50% of their DNA, the chance of being concordant for CF is 50%. For a disorder that is completely environmental in origin there would be expected to be little difference in concordance when comparing dizygotic with monozygotic twin pairs. For T2D concordance is approximately 70% for monozygotic twins, but only approximately 25% for dizygotic twins, therefore indicating significant genetic involvement. Furthermore, the risk of T2D is higher for any individual if a parent is also affected (higher still if both are affected). The relative contribution of genetic factors to a disease phenotype is known as ‘heritability’, and for T2D estimates of the heritability vary between 25 and 80%.
Identifying genetic loci associated with complex disorders
One way of identifying the loci involved is a ‘candidate gene’ approach. For T2D potential candidates might be genes involved in glucose metabolism or the response to insulin, or in predisposition to obesity. Investigation of the PPARG gene, based on its known role in adipocyte differentiation and glucose homoeostasis, identified a common polymorphism which was protective for T2D ( Table 9 ). However, due to the complexity of regulatory and metabolic networks in the cell, the number of potential candidate genes for T2D is immense, and fully investigating all possible candidates would require massive investment of time and money. Furthermore, by looking only at genes that are expected to play a role based on current understanding some key players might be missed.
To facilitate the identification of susceptibility loci, approaches based on genetic linkage or ‘association’ have been used ( Figure 28 ). Such approaches rely upon the observation that genetic recombination does not occur randomly across our genome, but instead tends to occur at ‘hotspots’ spread throughout chromosomes. The consequence is that particular segments of the genome tend to remain together through many generations. This means that we can use ‘genetic markers’, most often SNPs, as tags for the genome segments, and then look within populations to test whether particular markers associate with the disease. This approach is typified by the ‘genome-wide association study’ or GWAS ( Figure 29 ).
The principles of genetic association

SNP1 (with alleles C and T) and SNP2 (with alleles A and G) are two polymorphic sites present on one chromosome. Within one of the group of ancestors, a new mutation (yellow star) has occurred very close to the position of SNP1; this is a new pathogenic variant which contributes to a particular disease condition (yellow individual). Because SNP1 is so close to the pathogenic variant, there will be little or no recombination between these sites down the generations, whereas recombination is likely to occur between SNP2 and the pathogenic variant. Thus when the descendants are genotyped, SNP2 has identical allele distribution in both healthy and affected individuals (no association of SNP2 with the disease). However, for SNP1 there is an excess of the T allele (and a corresponding deficit in the C allele) in the affected population, in other words SNP1 shows association with the disease. In reality, for complex conditions where there may be many different predisposing variants in several different genes, the scale of association would be less extreme, requiring analysis of thousands of individuals.
Genome wide association study for T2D-related loci

Appropriate study groups are selected from the general population ( 1 ). For example, a group of individuals who have T2D ( 2 ) and a group of individuals who are healthy to act as controls ( 3 ). These groups must be matched as far as possible in terms of their constitution to avoid confounding effects – for example, matched for gender, ethnicity, smoking, socioeconomic status, education and so on. For each individual, thousands of SNPs across the genome are genotyped ( 4 ), and then the overall allele frequencies are compared between the two groups for SNPs across each chromosome ( 5 ). Each spot on the graph represents one SNP at a known location on a particular chromosome. The expectation is that, for the vast majority of SNPs, the MAF will be similar between the two groups. However, where there is a difference, either a significantly higher ( 6 ) or significantly lower ( 7 ) MAF in the affected group, this identifies a specific chromosome location which may play a role in T2D. Note that the actual variants which either predispose to obesity or protect from T2D may be close to the relevant SNPs (i.e. genetic linkage) rather than the SNPs themselves being causative or protective.
One problem with GWASs is that even with high statistical stringency, reproducibility is not guaranteed and results from different studies can appear contradictory, although this may reflect that there are too many other variables between the study groups. Large meta-analyses attempt to pull results from multiple GWAS together to determine significance on a large scale. It is important to note that association of a SNP with a particular trait or condition does not necessarily indicate causation, instead it is quite likely that the SNP, through close linkage, has been inherited along with the change which contributes to the trait ( Figure 28 ). Once a locus has been identified by GWAS the next step is to look at the genomic region to see which genes in that region are likely to be relevant to the observed phenotype, and to see if the association can be confirmed by other studies, which will need to include functional studies in cells and/or animal models. Over 120 genetic loci have been identified by GWAS for T2D; some of these are shown in Table 9 .
GWAS can be an effective way of linking common variants with associated diseases, but rare variants may also play a significant role within some families and subpopulations, for example the p.Glu508Lys allele of HNF1A in Native Americans ( Table 9 ). Identification of additional rare variants is likely to come from approaches involving genome sequencing. It should be noted that variants might either increase or decrease disease risk, depending upon their functional effect (see KCNJ11 and SLC30A8 in Table 9 ). The mechanism by which variants might contribute to complex disorders is often not obvious; for some loci implicated in T2D, there is a clear link to pancreatic function/glucose homoeostasis or obesity, but the significance of other loci identified by GWAS is less clear. At first glance, the function of the CDKAL1 product (tRNA modification) has no relationship to diabetes, but a potential impact emerges when the effect of lysine mistranslation on proinsulin cleavage is considered. Another locus, CDKN2A/2B , encodes products involved in cell cycle/cell proliferation, and it is feasible that there might be a link to the overall number of islet cells in the mature pancreas (having more islet cells may equate to better ability to sustain insulin production).
Despite the large number of T2D-associated loci that have been identified by GWAS, these are not able to explain all of the heritability of T2D. However, it is becoming increasingly clear that epigenetics also plays a significant role in complex diseases like T2D. Imprinting may be involved: the risk that offspring will be affected by T2D is greater when the mother is affected than when the father is affected. Interestingly, this is the reverse of the situation for T1D, for which the risks for a child are higher if the father is affected, than if the mother is affected. A parent-of-origin effect has been observed for some alleles of KCNQ1 which only increase T2D risk when transmitted maternally.
The impact of epigenetics appears to begin preconceptionally: mouse experiments have demonstrated effects on health and also in DNA methylation patterns for offspring following paternal exposure to high-fat diet or low nutrition. Early foetal life is also a critical time. Key observations have come from the study of individuals who were exposed to an adverse intrauterine environment, particularly during first trimester, as a consequence of severe famine during the ‘Dutch hunger winter’ of 1944/45. These individuals had normal birth weight (since the famine had ended by the later stages of the pregnancy), but had significantly increased rates of obesity and T2D as adults, suggesting that a form of foetal programming had occurred. This is supported by the observation of altered methylation patterns 60 years later when comparing the DNA of these individuals with that of their siblings.
Lifestyle choices and environmental exposures also lead to epigenetic change. Clearly, sedentary lifestyles combined with high calorie obesogenic diets contribute directly to T2D risk, but there are also many studies that demonstrate epigenetic changes associated with different foods. Tobacco smoking leads to decreased methylation in several genes associated with T2D, including KCNQ1 , and exercise has been shown to promote methylation changes in T2D-associated genes as well as altering histone deacetylase expression. Many epigenetic marks in the genome can change throughout the life of an individual as a result of changing lifestyle ( Figure 30 ), and may provide a useful target for management of T2D risk. The epigenetic marks present in the DNA of monozygotic twins have been shown to diverge as they age, and, when comparing individuals with/without particular complex disorders, differential epigenetic marks can be demonstrated in key genes. Epigenome-wide association studies (EWAS), which operate on similar principles to GWAS, may help to elucidate the epigenetic basis of complex diseases.
Many factors, environmental, genetic and epigenetic interact together in the overall risk for T2D

Epigenetic factors can alter throughout the life of an individual, and may be affected by parental environment preconception, and maternal environment during pregnancy. A multitude of inherited variants (some of which may be protective) combine with epigenetic status to generate an overall genetic risk for T2D, but the disease state will generally only be manifested in the presence of environmental triggers. Although inherited genetic variants are hard to change, it is apparent that environmental change (improved diet and more exercise) may impact on disease severity not only directly, but also indirectly by epigenetic changes. Pictograms from PictArts.
There is still a long way to go in understanding genetic contributions in complex diseases. T2D has only reached epidemic proportions over the last few decades, but the majority of genetic variants implicated as risk factors for T2D have been around for much longer in the human gene pool. Therefore, it is the environmental context of high calorie intake plus sedentary lifestyle which potentiates the effect of the genetic risk variants in T2D. Furthermore, it is important to recognise that variants which appear to be ‘bad’ as risk factors for one disease may in fact be protective against another (see HNF1B in Table 9 ), which underlines the fact that there is no such thing as a ‘perfect’ human genome!
Cancer affects approximately 1 in 4 people worldwide, with an estimated 14.1 million new cases in 2012 and a prediction that there will be 23.6 million new cases per year by 2030. In the U.K. there are nearly 990 new cases diagnosed every day, which equates to a new diagnosis approximately every 2 min. In the U.S.A., there are over 4600 new cases every day (a new case every 19 s). These rates are rising, due both to the increase in population size and increasing longevity. More than one-third of cancer cases in the U.K. are diagnosed in people aged 75 and older.
There are many different types of cancer and these types reflect the tissue and cell type from which the cancer originated. The most common types in the U.K. are cancer of the breast, prostate, lung and bowel, which together comprise over 53% of all cancer cases. Overall, half of the people diagnosed with cancer survive their disease for more than 10 years and this is improving, from only 24%, 40 years ago. However, each type of cancer shows a very different mortality rate, for example 98% of people diagnosed with testicular cancer survive for more than 10 years after diagnosis, while the figure is only 1% for pancreatic cancer.
Although the different types of cancer show different disease patterns and survival rates, the commonality of all cancers is that the cells have lost the normal controls over growth and movement. Cancer cells proliferate in an uncontrolled way and this can lead to the formation of a lump, or tumour. When the growth of the cells within the mass is limited and the cells retain certain normal features, do not invade adjacent tissues, nor spread to other parts of the body, the tumour is called ‘benign’. However, if the growth becomes more uncontrolled, such that the cells divide indefinitely and spread to other parts of the body, the cancer is termed malignant and the tumours that form at secondary sites are called metastases and it is metastatic disease that is the primary cause of cancer mortality.
There are several contributing factors that enable cancer cells to grow in this uncontrolled way and Hanahan and Weinberg described these in a seminal review in 2011 as the ‘hallmarks of cancer’. These features of cancer cells can be summarised as follows: (i) they have an inherent ability to divide; (ii) the cells do not respond to factors in the body that might inhibit their growth; (iii) they develop ways to avoid being destroyed by the immune system; (iv) they overcome the in-built clock that limits normal somatic cell division; (v) the cells release factors that in turn promote the surrounding normal cells to release other factors that will support the growth of the cancer cells, in other words, cancer cells can promote a permissive environment for their growth; (vi) they acquire the ability to move and invade other tissues; (vii) they promote the growth of a blood supply to the tumours to provide the oxygen and nutrients the cancer cells need to grow; (viii) the cancer cell genome becomes more prone to mutation; (ix) they become resistant to the normal mechanisms of cell death and (x) the cells adjust their metabolic pathways to better support rapid cell proliferation. All these changes from a normal cell are brought about by different somatic mutations in the cancer cell genome, and/or by epigenetic modifications. As such, cancer is essentially a disease of mutation.
The genome of a cancer cell is full of somatic mutations. Indeed, in some cancers, such as lung cancer and melanoma, there may be hundreds of thousands of mutations ( Figure 31 ). Many such mutations are observable at the level of the karyotype, including whole and part chromosome duplications, deletions, inversions and translocations. In addition, cancer cells typically carry large numbers of point and micromutations.
Different cancer types typically display different numbers of mutations in the cell genome

These can range from less than one hundred observed in some retinoblastomas to hundreds of thousands in lung cancer.
Many of these mutations will have been involved in the disease process to some degree (called causative or driver mutations), however, the cell also accumulates mutations that are not causative, i.e. ‘passenger’ mutations (as many as 99.9% of the mutations present) and one challenge is to distinguish between the two (see next section on ‘Genomics’). While there are a number of genes known to be powerful oncogenes when mutated or critical tumour suppressor genes, cancer cells never have only one causative mutation. This is because for the cancer cell to acquire all of the changes (as described above), mutations in numerous genes are needed to overcome normal growth regulatory processes. This is also reflected by the incidence of cancer. The statistics show that the biggest risk factor for cancer is age. The older an individual is, the more likely they are to develop cancer, indeed most cancer types are quite rare in young people ( Figure 32 ). The very shape of the incidence curve reveals that cancer is caused by multiple changes to the cellular genome. Mutations occur and accumulate in the cells of the body throughout life. The vast majority of these will be harmless and may not affect the phenotype of the cell at all. However, over time, as mutations accrue, there is a risk, a statistical chance, that eventually a cell undergoes enough causative mutations that it begins to develop cancerous properties and with time, this may progress to a cancerous state. Agents that speed up the mutation rate in cells will also increase the risk of cancer, so for example, overexposure to sunlight (a component of which is mutagenic UV rays), will increase the risk of melanoma, a type of skin cancer. Similarly, cigarette smoke contains multiple mutagens and smoking tobacco increases the risk of lung cancer.
Cancer increases with age

Theoretical cancer incidence curves are shown reflecting a set mutation rate. If only one mutation could cause a normal cell to become cancerous, the cancer incidence rate would be linear (blue line). The actual incidence increase with age (red curve) reflecting the accumulation of cancer causing changes that act together over time.
Oncogenes are genes whose activation contributes to the development of cancer. Oncogenes are usually mutated versions or pathogenic variants of normal cellular genes (the unmutated genes are often called proto-oncogenes for this reason), and this reflects that the normal function of the gene is involved in the control of cell growth in some way. Thus genes that promote or are involved in cell division (mitosis) or inhibit programmed cell death (apoptosis), differentiation, quiescence or senescence are genes that when mutated, could become oncogenic. In addition, some pathogens carry oncogenes, for example a small number of viruses can lead to a higher risk of specific cancers, because they encode genes that promote cell proliferation or survival. It is estimated that approximately 15% of cancers have developed with the involvement of an infectious agent, for example some human papillomaviruses can increase the risk of developing cervical cancer.
Many proteins expressed by potential oncogenes act in a process called signal transduction, or are transcription factors that are activated by this mechanism. Signal transduction is the method by which a cell converts a signal, typically received from the outside of the cell, into a change in gene expression which will lead to a response ( Figure 33 ). For example, if a growth factor acts upon a cell, this triggers a signal that passes through the cytoplasm, to induce the expression of genes required to initiate cell division. This is usually (but not exclusively) achieved by the growth factor interacting with a receptor at the cell surface. The interaction activates the receptor, which leads to a cascade of changes in the state of other factors that are located in the cytoplasm. Transcription factors are often activated by this process through phosphorylation upon specific serine or threonine residues, resulting in their translocation to the nucleus to regulate gene expression ( Figure 34 ). Importantly, once the signal has concluded, all components are deactivated. Mutations in genes involved in this process that lead to overactivation of the protein, or just too much of it, can result in excess signalling, instructing the cell to divide continuously.
Signal transduction

The cell receives signals from contact with other cells, from the extracellular matrix and from soluble molecules, including secreted proteins. The information received is integrated and transduced to the nucleus. The cytoplasmic signalling pathways and networks that are activated will depend upon the cell’s status and which genes are currently expressed. The combined signalling input can result in an altered programme of gene expression to achieve one of several possible responses.
A simplified, typical signal transduction pathway

Many growth factors (secreted proteins) interact with a receptor at the cell surface and both ligand and receptor can act in the form of a monomer or in a complex. The interaction causes the receptor to become active, and this will lead to a signalling cascade (depicted by the dashed red lines), passed from one molecule to the next. The signal may culminate in the activation of a transcription factor with a consequent change in gene expression, or influence mitochondrial membrane integrity and cell survival, or have an alternative destination to elicit a cellular response. The activation signal may be transmitted in several ways, the most common method is through the action of kinases, enzymes which phosphorylate their substrates (depicted by P), thereby activating the substrate for the next step. While a simple pathway is depicted, the reality is that a vast network of complex interactions occur, integrating numerous signals allowing for an extensive array of subtly different responses.
Essentially all types of mutation have been observed in the conversion of a proto-oncogene into an oncogene, including gene duplication, point mutation, partial deletion or rearrangement and chromosomal translocation. Oncogenic mutations tend to be gain-of-function and thus are usually dominant. Such mutations generally lead to overexpression of the gene or overactivation.
Oncogene activation by overexpression
There are three main mutational mechanisms by which gene overexpression is achieved ( Figure 35 ). These are: amplification of the gene, with increased expression due to the increased copy number, novel juxtaposition of sequences that enhance expression (for example, through chromosomal translocation) and mutations in the gene expression control sequences that either prevent gene silencing or directly enhance expression. In addition, epigenetic modification of the gene promoter sequences can act to increase or suppress expression.
Oncogenic mutations

Examples of oncogene activating mutations are depicted. ( A ) Gene amplification leading to increased expression of the product. ( B ) Reciprocal chromosomal translocation leading to enhanced expression of a gene at the breakpoint, as observed in follicular lymphoma and the 18:14 translocation involving the BCL2 gene and the IGH locus (above); or as observed in chronic myeloid leukaemia and the 9:22 translocation leading to a BCR-ABL fusion protein (fused in frame with respect to the ORF). ( C ) Loss of a protein regulatory region by small deletion. ( D ) Activating change in the coding sequence brought about by a point mutation (exemplified by RAS genes). In each case, the gene region is depicted by a coloured box and expression indicated by a bent arrow.
An example of mutation leading to overexpression is seen with the gene encoding the receptor protein HER2 (aka ERBB2, a member of the epidermal growth factor receptor (EGFR) family of tyrosine kinases), which is frequently mutated in breast cancer. One type of HER2 mutation found in cancer cells is amplification. The whole gene is duplicated resulting in more than one copy, sometimes several copies. This leads to excess production of the protein within the cell. As a consequence, the cell sends more signals to the nucleus to initiate mitosis, which contributes to the cancerous state by increasing cell proliferation. This knowledge led to the development of a drug called Herceptin, which blocks the action of HER2 and is an effective co-treatment for those breast cancers that overexpress HER2.
Another example of mutation causing gene overexpression is exemplified by the BCL2 gene in follicular lymphoma, a type of B-cell cancer. The BCL2 protein sits at the surface of the mitochondria and inhibits a form of programmed cell death called apoptosis. If a cell is destined to die (and there are several instances where this is normal), overexpression of BCL2 can inhibit cell death. Follicular lymphoma cells display a typical chromosomal translocation, juxtaposing part of chromosome 18, the location of the BCL2 gene, to chromosome 14, the location of an antibody gene (the immunoglobulin heavy chain ( IGH ) gene) ( Figure 35 ). The result is that BCL2 comes under the control of an enhancer sequence which would normally drive the expression of the IGH gene in B cells, but in the mutant cells, leads to overexpression of BCL2 . Thus the B cells are resistant to apoptosis and this contributes to the development of B-cell lymphoma.
Oncogene activation through increased activity
There are several mutational mechanisms by which the activity of an encoded protein can be increased or rendered constitutive. These include point (or small) mutations at critical regulatory residues, for example loss of negative regulatory phosphorylation sites, deletion of negative regulatory domains (which can occur by simple deletion or result from larger rearrangements such as chromosomal translocation), or activating mutations in catalytic domains or interaction domains.
Proteins that relay growth signals in cells have to be exquisitely regulated. These proteins generally exist in an inactive state, are briefly activated by signal transduction and then return to an inactive state, thereby tightly controlling cell proliferation. Mutations that lead to increased or constitutive activity can be oncogenic. A classic example of this occurs with the RAS genes ( H-RAS, N-RAS and K-RAS ). These three related genes encode small proteins that are pivotal in multiple cell signalling pathways and in the development of numerous cancers, and therefore RAS has been referred to as a ‘molecular switch’. The RAS proteins bind to either GDP or GTP (guanosine di- or tri-phosphate). When bound to GDP, the protein is inactive. As a consequence of signalling from receptors, RAS switches to bind to GTP and in doing so, changes conformation and becomes active and thereby can interact with the next protein in the chain to pass on the activation signal ( Figure 36 ). In the absence of an activating signal, RAS proteins are rapidly deactivated via an intrinsic GTPase activity (which hydrolyses the bound GTP to GDP). At just a few key points along the gene, mutation changing a single amino acid (typically codons 12, 13 or 61), can prevent GTP hydrolysis, thus locking RAS into the active, GTP-bound state and lead to constitutive signalling.
RAS activation

( A ) RAS is bound to GDP in the inactive state. Signal transduction can lead to the activation of RAS, via a GEF (GDP/GTP exchange factor), which displaces GDP from RAS, allowing the binding of GTP. This causes a change in conformation of RAS that enables interaction with effector proteins, thereby passing the activation signal onwards. Deactivation of RAS occurs by hydrolysis of GTP to GDP, assisted by GTPase-activating proteins (GAP). ( B ) Proteins of the RAF family are among several effectors of RAS. RAF proteins are serine/threonine kinases. Activated RAS binds to RAF, leading to a change in conformation of the latter and activating its kinase activity. RAF then phosphorylates its substrate MEK (which is also a kinase) thus activating it, and so the signal proceeds. This describes part of well-known signal transduction pathway, the mitogen activated protein kinase (MAPK) pathway.
Another example of protein activation is observed with the C-SRC gene, which encodes a tyrosine protein kinase. The C-SRC protein locates to the inside surface of the plasma membrane and passes on the mitogenic signals from several growth factor receptors. The kinase activity of the protein is controlled by a regulatory site at the C-terminal end, the location of a critical tyrosine residue (Tyr 527 ). The phosphorylation status of Tyr 527 is determined by other protein tyrosine kinases and phosphatases and when Tyr 527 is phosphorylated, this inhibits the kinase activity of C-SRC. Mutations that delete this residue result in a protein that is constitutively active and constantly relaying growth signals to the nucleus. The mutation can be as small as a point mutation, such that the tyrosine residue is lost or replaced by an alternative amino acid. Such mutations are frequently found in cancer of the colon, lung, liver, breast and pancreas. A further example of mutation leading to loss of a regulatory domain is seen with a chromosomal translocation that leads to the expression of a fusion protein (derived from two genes), called the BCR-ABL fusion. This particular chromosomal translocation (between chromosomes 9 and 22), called the Philadelphia chromosome, is a characteristic of chronic myeloid leukaemia and leads to the loss of a regulatory domain from the ABL1 gene, which encodes a tyrosine kinase. The fusion protein has constitutive kinase activity which promotes cell division.
Tumour suppressor genes
Tumour suppressor genes (TSG) are genes whose action inhibits the growth of tumour cells, therefore their inactivation is advantageous to a cancer cell. Consequently, the function of several TSGs is lost from all forms of cancer. The loss of function can be achieved by mutation affecting a critical region of the protein or by loss of expression, the latter is frequently brought about by deletion mutations (deleting part or all of the gene) or alternatively, by epigenetic modification of the gene regulatory sequences to suppress expression. Several TSGs function to inhibit the progression of the cell cycle, each acting at different points of the cycle, or in different tissues, or under different circumstances. The cell cycle is directed by a complex of proteins, including cyclins and their partners, the cyclin-dependent kinases (CDK). The CDKs, in turn through the cycle, phosphorylate and activate a plethora of proteins that orchestrate the cycle moving forward, from the initial growth phase (G 1 ), through DNA synthesis (S), the completion of DNA synthesis and continued growth (G 2 ) and mitosis (M) ( Figure 37 ). One of the central players in the cell cycle is a TSG called the retinoblastoma ( RB ) gene. In the unphosphorylated state, the RB protein inhibits both entry into S phase and the progress of the cell cycle by obstructing crucial cell cycle progression factors. When a cell receives signals to undergo division, this activates the cyclin/CDK complexes and one of the substrates is RB. As RB becomes phosphorylated, it releases the cell cycle progression factors to allow the cell cycle to move forward. If RB is lost from the cell, this critical inhibitory mechanism is lost, rendering the cell subject to uncontrolled proliferation. Many cancer cells show complete loss of RB expression. This means that both alleles must be affected, either by mutation (typically deletion) or epigenetic suppression. At the level of the cell phenotype, loss of RB is recessive, with one functional copy sufficient to control the cell cycle, however, as discussed below under heritable cancers, the dominant/recessive issue is more complex.
The cell cycle and RB

The cell cycle is depicted, showing the phases (divided by chevrons): growth or gap 1 (G 1 ), DNA synthesis (S), growth or gap 2 (G 2 ) and mitosis (M) in a circle, with exit from cycle represented as G 0 . Cell cycle check points are depicted as bars, G 1 /S check point (red: a DNA damage check), S-phase check point (blue: a DNA damage and replication fork check), G 2 /M check point (green: a DNA damage and completion of replication check) and spindle check point (yellow: ensuring correct alignment of the chromosomes upon the spindle, ready for division). The cyclin and CDK complexes relevant to each phase are shown. Central to cell cycle control is the TSG RB. RB becomes increasingly phosphorylated by the activated CDKs through G 1 (as depicted by increasing P). As the cycle progresses, it becomes hyperphosphorylated and this allows entry into S phase and further progression. Un-phosphorylated RB blocks cell cycle progression. During G 1 , the cycle can be initiated via mitogenic signalling (purple arrows). Once past ‘start’ the cell is committed to cycle.
Several other TSG products also act to inhibit cell cycle components to tightly control proliferation, such that this proceeds only when required and when all conditions are optimal, acting directly to inhibit the kinase activity of CDKs. A number of these (when first characterised), were given the unimaginative name that simply refers to the apparent size of the protein (in kiloDaltons), including p21, p16, p27 etc., with a superscript to distinguish between it and other proteins of the same size. The TSG encoded protein p21 Waf1 (expressed from the CDKN1A gene), inhibits CDK2 and therefore blocks entry into S phase and progression through G 2 , while p16 Ink4a (expressed from the CDKN2A gene) inactivates the CDK4 or CDK6 complex bound to cyclin D and therefore plays a key role in the passage of cells into G 0 (exiting cell cycle) and entering quiescence (reversible) or senescence (irreversible).
Numerous other TSG products function in processes that are not directly related to the cell cycle, instead affecting the growth conditions of the tumour and its environment. For example, if a solid tumour does not acquire a blood supply to provide sufficient oxygen and glucose to the dividing cells, it can grow little more than a few millimetres in diameter. The TSG termed von Hippel–Lindau ( VHL ) encodes an enzyme required in the processes of protein degradation; a ubiquitin ligase, its primary target, HIF (hypoxia-inducible factor), is a key protein that controls the growth of blood vessels from existing vessels (angiogenesis). Normally angiogenesis occurs when metabolic activity is high and oxygen availability low (like in a growing muscle in response to exercise), but otherwise is kept repressed. Mutations in VHL that result in the loss of the ability to cause HIF degradation, permit the persistent activation of HIF, thence the unrestrained angiogenesis necessary for tumour growth.
Genome integrity
As somatic mutation is a driving force in cancer, loss or aberrant function of genes involved in the processes of repair of damaged DNA can be tumorigenic. DNA damage can be caused by exogenous mutagenic agents, but the vast majority of mutations that are present in a cancer cell have occurred as a result of errors during DNA replication or have been caused by endogenous biochemical processes. As such, replication proofreading and repair proteins work continuously to maintain the integrity of the genome. Not all DNA repair genes are TSGs or proto-oncogenes, many are essential and therefore their mutation or loss will lead to cell death. However, if mutation of such a gene permits cell survival, but increases the chance of incorrect repair (i.e. mutation), this will increase the cancer risk.
The most commonly mutated TSG in human cancer is a gene termed TP53 . The encoded TP53 (or simply p53) protein is a DNA-binding transcription factor. Normally present at low levels in the cell, it becomes stabilised and activated when the cellular genome is at risk, particularly following DNA damage ( Figure 38 ). When DNA damage is detected by a complex of proteins, the information is transduced by either the ATM kinase or the ATR kinase to activate effectors, CHK1, CHK2 and p53 ( Figure 38 ). P53 then acts to induce the expression of other genes that will halt the cell cycle, including p21 Waf1 . As such, there is an effective ‘check point’ at the G 1 /S boundary of the cell cycle ( Figure 37 ). If DNA is damaged, activated p53 induces p21 Waf1 expression, CDK2 is then inhibited and the cycle does not proceed. P53 is also involved in DNA repair processes, therefore, following DNA damage, the repair machinery can work while the cell cycle is delayed. Additionally, p53 can induce the expression of genes that will lead to cell death (counteracting the survival function of BCL2) if the cell is beyond repair, thus the cell can be neatly removed. The most common mutations in TP53 in cancer cells occur in the DNA-binding region of the protein and prevent p53 from binding to DNA and therefore disable its function as a transcription factor. As a consequence, cells with damaged DNA can progress through the cell cycle check points with a high risk of mutation. For this reason, p53 has been referred to as ‘the guardian of the genome’.
Sensing and responding to damaged DNA

The proteins that initiate DNA repair after damage or replication blockage can be divided into sensors, transducers and effectors. The sensors: a complex of proteins recognise the broken ends of DNA double strand breaks (DSB) and complexes of different composition recognise stalled DNA replication forks (pink and yellow bubbles). These complexes attract two key kinases, ATM and ATR, the transducers, which function to phosphorylate, and thereby activate, two further kinases, CHK1 and CHK2. TP53 is stabilised both directly via phosphorylation by ATM and by phosphorylation by CHK1 and CHK2. Following on, the effectors lead to one of several responses, as indicated.
BRCA1 and BRCA2 (breast cancer 1 and 2) are also TSGs involved in DNA repair. The encoded proteins act in a complex to repair DNA double strand breaks (DSB). Mutations that lead to loss of function of the proteins or loss of expression result in a high chance of inaccurate repair of DNA DSB and thereby markedly increase the mutation rate of the cells and therefore also the cancer risk.
Some oncogene products act to down-regulate DNA repair, either as one of several functions, or in some cases, their primary function. The mitogenic protein HER2 (described above), when activated, down-regulates several DNA repair factors and this contributes to its oncogenic properties. In the last few decades, the importance of small non-coding RNAs in controlling gene expression has become apparent. Numerous tiny RNAs (called miRNAs) are expressed by cells and these function by down-regulating the expression of specific target genes. The miRNA-182 (miR-182) specifically down-regulates the expression of BRCA1 and a few other TSGs and miR-182 has been found overexpressed in several cancer cells (including breast cancer), often as a result of gene duplication. It can therefore be viewed as a distinct type of oncogene.
Cancer types show characteristic mutation profiles
Multiple powerful oncogenes have been known for several decades and historically, many were identified by virtue of the action of retroviruses in animal tumour model systems. Many of these are mutated in particular types of cancer and with different frequencies, for example RAS gene mutations are found in roughly 60% of pancreatic cancers, 50% of colon cancers, 20% of lung cancers, but rarely (1%) in kidney cancer. Several critical TSGs were discovered as a result of determining the pathogenic variant in familial cancer syndromes, such as heritable retinoblastoma. Furthermore, it has long been known that certain types of mutations are typical of particular cancer types, for example follicular lymphoma cells carry a chromosomal translocation involving the BCL2 locus and Burkitt’s lymphoma cells always harbour a translocation involving the C-MYC gene. However, the vast majority of oncogenes and TSGs make just a small contribution to the development of disease and are therefore harder to discern. Tackling this problem has been revolutionised by the huge advances in genome sequencing technologies over the last few decades. As will be described in the next section on ‘Genomics’, large sequencing projects comparing the mutated genome of a tumour with that derived from the normal tissue of the same individual has enabled the identification of causative mutations in that cancer. Taking this approach, not only have many more mutations (and therefore genes) which contribute to the cancer process been identified, but it has also been found that cancer types (and even subtypes) show characteristic somatic mutation profiles and this can then inform treatment options.
Epigenetic modifiers
In addition to mutation, overexpression of oncogenes and loss or reduced expression of TSGs are found in cancer cells through epigenetic modification of the expression control sequences. The genes whose products are epigenetic modifiers, are themselves frequently mutated in cancer cells. The mutation of these genes, (or their abnormal expression as a consequence of epigenetic modification of their control sequences) leads to aberrant chromatin modelling and the misexpression (under or over) of multiple genes. Epigenetic silencing of TSGs has been found to play a significant role in the genesis of cancer. Moreover, mutation in epigenetic modifier genes can have widespread effects and in some cases, they can be viewed as TSGs. For example, loss-of-function mutation in the DNA methyltransferase 3A ( DNMT3A ) gene is frequently found in blood cell tumours (lymphoid and myeloid malignancies). The loss of DNMT3A appears to increase the proliferative capacity of the cells and inhibit differentiation. Conversely, other genetic modifier genes have gain-of-function mutations in cancer cells. The histone-methyltransferase encoding gene EZH2 has been found to be activated by point mutation or is overexpressed through gene amplification in a variety of tumours. However, this gene cannot be simply classified as an oncogene, as its loss is observed in other tumours indicative of a TSG role in some cell types. Thus, altered epigenetic modification of genes is widespread in cancer cells, however its control is highly complex.
Heritable cancer and predisposition
Three factors essentially determine whether a cell becomes cancerous: the environment (including lifestyle), chance and the genotype. As described above, environmental mutagens (such as sunlight and tobacco smoke) clearly increase the risk of cancer and, although some are highly controversial, dietary factors (such as alcohol) may also influence risk. Statistical chance reflects that only a tiny proportion of all possible somatic mutations will be causative in cancer processes and this is why carcinogens are described as increasing the ‘risk’ of cancer. The genotype is all-important. Heritable loss-of-function variants in some TSGs increase the risk of cancer so substantially, that these present as dominant pathogenic variants, however, they contribute to a relatively small proportion of cancers overall ( Table 10 ). This is exemplified by heritable loss of function variants in TP53, RB, BRCA1 and BRCA2 . Breast cancer is a common cancer, and approximately 1 in 8 women in the U.K. will develop it during their lifetime, while only 3% of these cases are caused by inherited pathogenic variants in BRCA1, BRCA2 or TP53 . Conversely, retinoblastoma is a very rare childhood cancer, with approximately 45 children diagnosed per year in the U.K. Of these, approximately 40% carry inherited pathogenic variants in the RB gene. Germline loss of function variants in TP53 cause Li–Fraumeni syndrome (affecting approximately 1–4 individuals per 20000), which is characterised by early-onset cancer of several different types. The risk of developing cancer in such an individual is approximately 50% by the age of 30, rising to 90% by the age 70. The apparent paradox, that loss of function of these TSGs is recessive in the cellular phenotype, yet dominant in the individual, is explained by Knudson’s ‘two hit hypothesis’, first put forward to account for this phenomenon with respect to the RB gene and familial retinoblastoma (from which the gene was discovered). The function of one allele is heritably inactivated and the other allele is inactivated through somatic mutation or epigenetic silencing, such that tumours arising in these individuals show loss of function of both RB alleles. The same applies to other TSGs and tumour types, including heritable loss of TP53 function and the development of Li–Fraumeni syndrome tumours and heritable loss-of-function variants of BRCA1 and BRCA2 in breast cancer.
The population frequency % given under organ affected, indicates the proportion of individuals in the U.K. that are likely to develop this cancer at some point in their life. Numbers indicate new cases diagnosed in the U.K. Statistics were derived from the Cancer Research UK website.
Some heritable pathogenic allele variants increase the cancer risk by a small but significant degree, for example patients with a hyperactivated variant of the PIK3CD gene (a proto-oncogene which functions in signal transduction) suffer from the dominant disorder activated PI3Kδ syndrome (APDS) but also have an increased risk of B-cell lymphoma.
Aside from the clear cancer-associated, high risk, inherited gene variants, the genetic profile of an individual has a profound effect upon the predisposition to cancer. Thousands of allele polymorphisms or variants may increase or decrease the cancer risk (relative to each other) by a tiny degree, or under specific circumstances. Combinations of many such medium or low risk alleles together may have a compound effect upon risk. These low, medium and condition-specific risk alleles are being identified and explored using GWAS approaches and mass sequencing endeavours like the 100,000 Genomes Project (see next section). As such, although the statistics currently give this impression, there is really no clear distinction between ‘heritable’ and ‘spontaneous’ cancers, it is better described as a sliding scale of inherited risk. The large proportion of cancers currently not classified as heritable, nevertheless arose against a genomic background with a certain risk value. The massive GWAS and sequencing studies currently underway hold enormous promise for the future; there will come a time when the genotype of an individual can be used to determine the lifetime risk of certain diseases, particularly cancer and this can then be used to suggest preventative lifestyle measures or treatments.
Whereas genetic studies have traditionally focused on the effects of variants in individual genes, there is a shift towards consideration of the impact of the whole genome in health and medicine. Many conditions with a strong genetic basis, like T2D, epilepsy, hypertrophic cardiomyopathy and intellectual disability are associated, not with pathogenic variants in single genes, but rather, with variants in any one of a growing number of genes. There are (in 2018) 84 genes in which variants are reported to be associated with epilepsy as a core symptom, and several hundred other genes in which variants lead to conditions with epilepsy appearing as part of a wider spectrum of symptoms. There are over 600 genes in which pathogenic variants have been reported to be associated with intellectual disability, and the list is expanding. This diversity underlies the fact that for many individuals and families affected by genetic disease, there has been a ‘diagnostic odyssey’, with a series of misdiagnoses over many years, and perhaps a series of genetic tests, which may or may not have culminated in a definitive outcome.
As explained earlier, cancer results from an accumulation of somatic mutations which lead to disruption of the pathways which normally regulate processes including cell proliferation, cell death and cell motility. These pathways collectively involve input from the products of hundreds of genes (over 1% of our genome), and it is a challenge to identify all the genes in which mutation can contribute to cancer (the causative mutations). In addition to this, the inherited genome of all individuals influences the risk of developing certain types of cancers, upon which the somatic mutation profile builds.
Whole genome approaches, both GWAS and full sequencing are now being used by collaborative groups of scientists and consortiums to investigate the genetic predisposition to disease and the contribution of somatic mutation. To help address the gaps in our understanding of genes involved in rare diseases and complex disorders and genes associated with cancer, as well as to enhance understanding of our genome as a whole, the 100,000 Genomes Project was initiated in 2012 by Genomics England (owned and funded by the U.K. Department of Health).
The 100,000 Genomes Project and Scottish Genomes Partnership
The 100,000 Genomes Project set out with two targets. Firstly, to fully sequence entire ‘normal’ and ‘tumour’ genomes from approximately 50000 cancer patients, in order to be able to identify all the alterations in the cancer genomes; and secondly to fully sequence the genomes of approximately 17000 rare disease patients together with two very close relatives (ideally mother and father) ( Table 11 ). Analysis of parent-child trios facilitates identification of de novo mutations that may contribute to disease, as well as recognition of variants that may be benign (present in a healthy parent). The Scottish Genomes Partnership (SGP) plans to sequence genomes of at least 3000 individuals, with goals that are similar to those of the 100,000 Genomes Project.
In a few cases, it may be another close relative.
Each genome sequenced generates roughly 200 GB of data, which represents huge challenges for data storage and analysis, as well as data security. Nevertheless, the benefits should include not only a molecular diagnosis for thousands of rare disease patients, but also a huge amount of data that will contribute to our understanding of the role of genomic variations and mutations in disease, and to the development of better diagnostic approaches and improved therapies that are targeted to key alterations.
Genetic modifiers
The complexity of, and interactions between, biochemical pathways and physiological processes within the body mean that it is important to consider not only pathogenic variants in a single gene, but to also take into account the potential effects of other genetic variants elsewhere in the genome. This is exemplified by long QT syndrome (LQTS; named for an alteration seen on electrocardiogram traces of the heart’s rhythm, where the ‘QT interval’ is very prolonged) which presents as a defect in the electrical activity of the heart affecting approximately 1 in 2000 individuals, and can result in sudden death. The commonest cause of autosomal dominant LQTS is a loss-of-function variant in one copy of the KCNQ1 gene. Some common variants in another gene, NOS1AP , have been demonstrated to have a small but significant effect on QT interval, even in healthy individuals. These variants in NOS1AP can cause further prolongation of the QT interval that is associated with an increased risk of sudden cardiac death in carriers of a pathogenic KCNQ1 variant.
Variants in several genes have been identified as potential modifiers of lung disease phenotypes in CF, and it is likely that multiple genetic modifiers exist for a major proportion of genetic diseases. Identification and understanding of genetic modifiers and the mechanisms by which they operate will extend the range of potential therapeutic targets, and also allow better prognostic information and risk stratification. It is hoped that initiatives like the 100,000 Genomes Project and SGP will contribute to the identification of such genetic modifiers.
Investigating cancer predisposition
There are many examples in which the identical pathogenic variant leads to different severity of disease in different individuals. For many variants the penetrance (the proportion of individuals with the variant who exhibit the phenotype) is less than 100%, and the expression (pattern of effects observed in individuals with the variant) can also vary. For example, inherited pathogenic BRCA1 variants are associated with predisposition to breast and ovarian cancer. However, some women inherit a clearly pathogenic BRCA1 variant but are not affected during their lifetime. Women who inherited the same pathogenic BRCA1 variant may be affected by breast cancer only (unilateral or bilateral), by ovarian cancer, or by both breast and ovarian cancer, and in fact other cancer types may also be seen as a consequence of the inherited BRCA1 variant. Differences in penetrance and expression are likely to depend to a great extent upon a combination of environmental and genetic modifiers. Thus, while such individuals carry a high risk pathogenic variant, multiple other genes contribute to the overall risk of disease.
The majority of cancers arise in individuals who do not carry a high risk pathogenic variant (such as the BRCA1/2 variants). Nevertheless, the genetic make-up of the individual influences the chance of cancer occurring, affording protection against risk, or increasing the predisposition. By comparing the genotypes of individuals who have suffered from one type of cancer with those who have not, it is possible to begin to build information about the relative risk that certain gene variants might hold, also, to relate the risk to particular conditions. Such studies typically involve GWAS approaches (see Figures 28 and 29 ), but can also be revealed by full genome sequence analysis. For example, a specific SNP might be linked with some protection for the individual from the damaging effects of cigarette smoke in the lung, but might have little effect on lung cancer incidence in a non-smoker. Similarly, another variant might increase the cancer risk, but only in a smoker. Risk of lung cancer attributed to such variants therefore would be conditional upon the smoking status. Similarly, risk alleles might reflect diet or other environmental factors, or may show a disease association that is not dependent upon any known environmental condition. In relation to cancer, such studies hold great promise for future cancer prevention, as building a risk profile for individuals would facilitate informed lifestyle choices and preventative treatment measures (as is currently the case with prophylactic surgery for BRCA1 and BRCA2 pathogenic variant carriers).
Identifying causative or driver mutations in cancer
Cancer cells harbour hundreds, even thousands of somatic mutations, but only a small proportion of these are causative in the cancer process; the vast majority are generally ‘passenger’ mutations. By comparing the genomic sequence of the cancer cells with that of the unaffected genome from the same individual (from non-cancer tissue, usually the blood), all of the somatic mutations that have occurred in the cancer cell can be identified. Then by comparing the genes that have been subject to mutation, between cancers arising in hundreds of different people, it is possible to establish which genes are commonly mutated in a given cancer type. On the basis that mutation occurs largely at random, the chance that a passenger mutation in a given gene will be observed in multiple cancer samples is low. Conversely, if a causative mutation occurs, this will give the cell a growth advantage and it will be selected during the ‘evolution’ of the cancer. Therefore, genes that are found to be mutated in multiple different cancer samples constitute candidate driver mutations. The gene’s properties and function can then be researched to assess if it can indeed contribute to cancerous processes.
Understanding the contribution of risk alleles and the role of all the cancer-associated mutations is crucial to enable the design and successful delivery of novel therapies which target particular mutations present in a given cancer. Furthermore, it appears likely that future genetic diagnostics will be aiming to identify not only the primary causative variants in rare and complex diseases, but also the genetic modifier variants that may influence disease severity and progression: a genomic rather than genetic approach.
Advances in technology, particularly in DNA sequencing technologies, have led to identification of even more genes for which loss or changes in function can lead to disease. This in turn increases the need for genetic testing, to confirm potential diagnoses, to provide information on recurrence risk (for example, in future pregnancies) and to facilitate testing in relatives where appropriate. The range of tests available in genetic diagnostic laboratories has changed dramatically in recent years, with time-consuming tests like Southern blot being replaced by approaches with much faster turn-around times.
Karyotyping
Genetic testing has its roots in being able to visualise whole chromosomes in a process known as karyotyping (see Figure 2 ). This process usually requires culture of cells (can take 1–2 weeks) and arrests cells in metaphase, the stage of the cell cycle in which the chromosomes are in their most condensed form and thus easiest to visualise. In early karyotyping analyses, the imaged chromosomes were simply arranged, in order of their size and position of the centromere. Advances in this procedure came in the 1970s when various banding techniques were developed, including G-banding, currently the most widely used technique in the U.K. and U.S.A. Following trypsin digestion, Giemsa dye is used to stain the chromosomes, with AT-rich, gene-poor regions staining more readily than the gene-rich regions. The resultant pattern allows the chromosomes to be distinguished and thus permits the presence of large abnormalities such as deletions, duplications, inversions and translocations to be assessed. One drawback of karyotyping is the lack of resolution; in general changes of less than 3–4 Mb are virtually impossible to detect.
Fluorescence in situ hybridisation
A later development, in the 1980s, was fluorescence in situ hybridisation (FISH), which uses labelled DNA probes to assess the presence, copy number and location of the complementary sequence of DNA in an individual’s genome by hybridisation. The sample under investigation can comprise interphase cells or cultured cells arrested in metaphase, and is immobilised on a surface, usually a glass slide. After incubation with the fluorescently tagged probe followed by washing to remove unbound probe, binding of the probe is viewed using a fluorescence microscope ( Figure 39 ). An example of a condition where FISH might be useful is DiGeorge syndrome (DGS), a genetic condition usually caused by microdeletions of 3 kb or less, including the TBX1 gene, at 22q11.2. Conventional karyotyping does not have sufficient resolution to identify such deletions. However, using FISH probes for the TBX1 gene, the deletion can be easily identified. Various types of FISH probes can be used, including single locus probes (like the TBX1 probe), centromeric probes (which recognise centromeres of specific chromosomes), subtelomeric probes (which target unique sequences close to the telomere) and chromosome paints (which can target the non-repetitive content of an entire chromosome). Chromosome paints can facilitate the identification of the chromosomal origin of constituent parts of rearranged chromosomes. A drawback of FISH is the cost of the probes (£ 50–100) plus the time taken for the analysis, so that there is a limit to the number of loci that it would be economically feasible to test for in one patient. Thus the subsequent development of microarrays has superseded the use of FISH, particularly where the clinical features do not narrow the region of the genome that is likely involved.

( A ) One or more fluorescently labelled probes are required for the targets that are to be detected. The patient sample containing chromosomal DNA (which may be cultured cells for metaphase FISH or uncultured cells for interphase FISH) is immobilised on a microscope slide. Probes and chromosomes are denatured and allowed to hybridise together, thus localising the fluorescent probes to the regions where complementary chromosomal DNA is present. ( B ) Probes for detection of DiGeorge syndrome. One probe (red) targets the DiGeorge locus at 22q11.2 (including the TBX1 gene), and a control probe (green) targeting genes at 22q13 ( ARSA and SHANK3 ) helps to identify both copies of chromosome 22. Note that FISH probes are very long, typically hundreds of kb, in order to render the target detectable; this means that a small deletion may be missed. ( C ) A fluorescence microscope is used to visualise the results; the image here includes an interphase nucleus as well as a metaphase spread. The chromosomal material has been counterstained blue by DAPI. The normal chromosome 22 is indicated by ‘ n ’ in the metaphase spread and in the inset; both red and green probes hybridise (the presence of two separate spots for each probe is due to the presence of sister chromatids each possessing a copy of the relevant locus). The chromosome 22 with deletion of the DiGeorge locus (indicated by ‘d’ in the metaphase and inset) shows hybridisation only to the control probe. Note that the interphase nucleus shows only the number of loci present (two green control loci and one red DiGeorge locus), not their location with respect to each other. Note also that in interphase nuclei the chromosomes are very extended so that even loci on the same chromosome become widely separated. FISH image courtesy of West of Scotland Genetics Service. Chromosome ideogram from NCBI Genome Decoration Page.
Mutation specific assays
Although karyotyping and FISH offer a large scale view of chromosomes, there are often times where resolution at the nucleotide level is required. Conditions where the underlying genetic change is known allows the design of specific assays which can be used in a clinical context. Using CF as an example, it is known that by testing for a defined set of 31 pathogenic variants, it is possible to detect almost 90% of CF cases in a Caucasian population. One of the approaches used is known as allele-specific PCR (or amplification refractory mutation system, ARMS).
This allele-specific PCR is based on the principle that complementarity at the 3′ binding site of the primer is crucial for amplification to occur. Although base mismatches at other points in the primer may be tolerated, mismatches at the 3′ end prevent amplification in the extension phase of the PCR. Thus it is possible to design two sets of primers – one which matches the ‘normal’ allele and one set where the 3′ end of one primer is complementary to the pathogenic allele ( Figure 40 ). By determining which set of primers generate a PCR product, it can be determined whether normal, pathogenic or both alleles are present in the individual. This process is repeated for multiple variants, often carried out simultaneously in the same reaction.
Allele-specific PCR by positioning the variant at the 3′ end of one primer

( A ) As with all PCRs, both forward and reverse primers are required, one of which will be a common primer (here the forward primer) and one of which will have specific versions for each allele (here the reverse primer). The specificity is generated by the sequence at the 3′ end of the primer. For assay of a SNP with two alleles, two PCR amplifications are set up, both containing the common primer, but containing the alternate versions of the allele-specific primer. ( B ) An assay for a T/C SNP. ( 1 ) One version of the reverse primer has an A at the 3′ end; this matches the T allele, and extension can occur from the primer when the T allele is present, so PCR products are obtained from homozygotes or heterozygotes for T (TT or TC). ( 2 ) The reverse primer with A at the 3′ end does not allow extension, so PCR would fail if only the C allele were present (CC homozygotes). ( 3 ) Reverse primer ending in G does not allow PCR amplification when the template contains only the T allele (TT homozygotes). ( 4 ) Reverse primer ending in G allows extension if the C allele is present (CC or TC).
Several other techniques, including reverse dot blot and the oligonucleotide ligation assay are alternatives to consider when detecting pathogenic variants that affect one or a few nucleotides. For conditions where pathogenic change is more likely to be a deletion or duplication, for example DGS or Duchenne muscular dystrophy (DMD), a technique such as multiplex ligation-dependent probe amplification (MLPA) may be used ( Figure 41 ). Although FISH ( Figure 39 ) can also detect deletions such as those found in DGS, MLPA, utilising multiple smaller probes across a larger genomic region, can provide a higher resolution view of the extent of any deletion. However FISH is able to provide information on chromosomal position of the target sequence(s), whereas MLPA only informs on copy number.
The MLPA assay and application to diagnosis of DGS

( A ) Short regions of DNA, roughly 50–70 bp, are selected as targets, indicated by S and T ( 1 ). Single-stranded oligonucleotide probe pairs are designed for each target ( 2 ), with half of the target sequence present in the ‘left’ probe and the other half in the ‘right’ probe. Each left probe contains an additional sequence ‘F’ and each right probe contains an additional sequence ‘R’. The right probes also contain a ‘stuffer’ sequence which is a different length for each target to be detected. Genomic DNA is denatured and probes allowed to anneal ( 3 ). Annealed probe pairs will lie precisely adjacent to each other on the target DNA ( 4 ) allowing DNA ligase to join left and right probes together into a single molecule ( 5 ). The ligated probes are denatured away from the target ( 6 ), and then PCR amplification is carried out ( 7 ) using the same primer pair for all ligated probes (fluorescently-labelled F and the complement of R). The final quantity of each amplified product is dependent upon the copy number of that target sequence within the sample genome. ( B ) For each target the amount of fluorescent product from the test sample is compared with the amount of product from a control genome, and the ratio is plotted. This plot depicts typical MLPA results for 29 target sequences across the 22q11 region for a patient suspected of having DGS. The gene targeted by each probe is indicated below the plot; distance along the X-axis indicates distance along the chromosome. A ratio of approximately 1.0 indicates normal copy number (blue squares). However the results for 14 targets (including two for the TBX1 gene) give a ratio of only 0.5, indicating a halving of the copy number (one copy instead of the two expected from two complete copies of chromosome 22). This result confirms a diagnosis of DGS. ( C ) The region of chromosome 22 which is targeted by MLPA probes from the ‘P250-B2 DiGeorge’ kit from MRC-Holland. Cat eye syndrome (CES) is caused by duplication of the region indicated by the green bar; duplications can be identified by amplification ratios of 1.5 compared with control. Roughly 90% of DGS cases result from a 3-Mb deletion, indicated by the blue bar, and including approximately 60 genes, while approximately 8% of cases have a 1.5-Mb deletion, indicated by the purple bar, involving 28 genes. A small number of cases have atypical deletions which may be larger than 3 Mb. While FISH using the TBX1 probe (red) can identify a deletion in the region, MLPA can provide a more accurate picture of the extent of any imbalance due to the greater number of probes used. The P250-B2 DiGeorge kit also contains probes that target relevant regions on chromosomes 4q, 8p, 9q, 10p and 17p, in which copy imbalance generates phenotypes which overlap with DGS; thus a single MLPA assay can assess multiple target regions. Chromosome ideogram from NCBI Genome Decoration Page.
Sanger sequencing
The techniques described above are useful in detecting conditions where there is already some knowledge of where in the gene the pathogenic variant is likely to be, for example it is known that p.Phe508del accounts for approximately 70% of CF alleles worldwide, which can be easily detected using allele-specific PCR. However, many conditions do not have a subset of common mutations, a good example being the inherited predisposition to breast cancer, where pathogenic variants are spread throughout the relevant genes, which include BRCA1 and BRCA2 . In order to detect these types of changes it is often necessary to determine the sequence of the whole gene(s) in a patient and to then compare this with a reference genome to identify changes in the patient. For many years this has been done using Sanger sequencing, a technique which was first developed in the 1970s, but which has undergone many developments to generate the currently used automated method ( Figure 42 ).
Automated Sanger sequencing

( A ) PCR products ( 1 ) are generated from the region to be sequenced so that there are billions of template molecules for the sequencing reaction. A sequencing primer is annealed to the denatured PCR products ( 2 ). A mixture of deoxynucleotide triphosphates (dNTPs) and dideoxynucleotide triphosphates (ddNTPs) is added ( 3 ), which are used by DNA polymerase ( 4 ) to synthesise a new strand. The four ddNTPs are each labelled with a different fluorescent dye: ddATP green, ddCTP blue, ddGTP yellow and ddTTP red. When a dNTP has been added, DNA synthesis can continue in the normal way. However, ddNTPs are chain terminators, so that once a ddNTP is added, synthesis will terminate; the colour of fluorescence of the terminated chain will indicate which nucleotide is present at that position (here the red fluorescence indicates a T). Because there are billions of templates, and because ddNTPs are added randomly, terminating different chains at different positions, billions of terminated chains are generated, with many chains terminated at each nucleotide position. ( B ) The products of the sequencing reaction are denatured away from the template and electrophoresed to separate them by size; the shortest chains will move fastest. A laser-based detector registers the colours of fluorescence emitted as each of the sequencing products travels past. ( C ) The data from the detector is processed to generate an ‘electropherogram’, in which successive peaks represent products which are each one nucleotide longer than the previous one, allowing the sequence to be read by using the fluorescent colour to identify the nucleotide(s) at that position in the DNA. Here the control sequence is TTACAGC, while the patient sample shows a heterozygous substitution of T in the middle of this sequence: since two different alleles are present in the patient, both are represented in the sequence trace at this position, indicated by the pink star.
Aneuploidy testing by quantificative fluorescence PCR
Karyotyping was the traditional approach to diagnose aneuploidy, but the complete process took approximately 1–2 weeks due to the time taken to culture cells. To provide a rapid result, for example for testing during pregnancy, interphase FISH has also been used, but more recently quantitative fluorescence PCR (QF-PCR) has become the technique of choice for initial aneuploidy testing. This involves the analysis of microsatellites with high variability in the population. The copy number of each chromosome tested is deduced from a combination of the number of different microsatellite alleles present, and their ratios ( Figure 43 ). For each chromosome to be tested (usually 13, 18, 21 and sometimes including sex chromosomes) four or five loci are analysed along the length of the chromosome. This approach is generally effective in rapidly (next day) identifying trisomy, and a positive finding can be confirmed by karyotyping or another test if desired. Sometimes one or more of the loci used are homozygous and therefore non-informative, but results from the other loci on that chromosome are generally informative, allowing a result to be reported. Consanguinity is likely to lead to multiple uninformative microsatellites, in which case it is possible to use additional loci, or if that also fails, to use FISH/karyotyping.

Several microsatellite loci (here R, S and T) on the relevant chromosome(s) are analysed by PCR using an appropriate pair of flanking primers, one of which is fluorescently labelled, so that all products will be fluorescent, and quantity of product is measurable by amount of fluorescent product generated. The particular loci are selected on the basis of high variability (many different alleles) in the population. ( A ) Where there are two copies of the chromosome, there should be two copies of each microsatellite, which ideally have different repeat numbers. Thus at locus R one chromosome has 12 repeats, whilst the other has 14 repeats. Following PCR and electrophoretic analysis (see Figure 42 ), two product peaks are generated, one for each allele, at a 1:1 ratio. The same pattern is observed for loci S and T. ( B ) Trisomy should result in three copies of each microsatellite. Where there are three different alleles present (as for locus R) three peaks, at a ratio of 1:1:1, will be generated by the analysis. If two of the chromosomes share the same allele while te other is different (as for loci S and T) there will be two peaks with a ratio of 1:2 or 2:1. Thus disomy can be differentiated from trisomy by number of peaks generated and the ratios between them. Note that if all chromosomes present share the same allele at a particular locus, then only one peak is generated and that locus is therefore uninformative.
New approaches to diagnostics
Where there is clear suspicion relating to a particular gene or group of genes, it may be that specific assays, as discussed above, are the most cost-effective diagnostic approach ( Figure 44 ). But it is not always possible to pinpoint the relevant genes or genomic region from the patient symptoms. In such circumstances whole genome approaches are frequently used as the first-line strategy. Developmental delay and learning difficulties, for example can be associated with not only full aneuploidy, but also with gains and/or losses involving large segments of DNA which might be the consequence of microdeletions, microduplications or unbalanced rearrangements of parental translocations/inversions or specific gene variants.
Selecting an appropriate test

Where the pathogenic variant in a family is known then testing will generally use a specific assay for that variant. For new diagnoses the most appropriate and cost-effective test must be selected according to technology and expertise available to the laboratory as well as the clinical features of the patient. Definitive findings (blue, underlined) can be reported in cases where a specific family variant was being tested for, or where a clearly pathogenic variant that is causative of the patient phenotype is detected. Note that use of some techniques, such as karyotyping, is declining with the advent of NGS- and array-based approaches. *It is possible NGS gene panels may be used in future even where the disease gene is known. Abbreviations: ASP, allele-specific PCR; NGS, next generation sequencing; SS, Sanger sequencing; WES, whole exome sequencing; WGS, whole genome sequencing.
Microarrays
Microarrays allow simultaneous analysis of hundreds of thousands of individual targets across the genome. For whole genome analysis, SNP genotyping by array has largely replaced the earlier array comparative genomic hybridisation (aCGH) approach, so only SNP arrays will be described here. There are several different approaches (or ‘platforms’) for SNP array, but all are based on oligonucleotide ‘probes’ for each of the targets to be assayed, that are immobilised on a solid surface ( Figure 45 ).
Basic microarray

The microarray ( 1 ) is a grid of hundreds of thousands of microscopic ‘spots’; each spot ( 2 ) contains billions of copies of an oligonucleotide ‘probe’ that represents a specific target. These single-stranded probes will be able to hybridise to, and therefore capture ( 3 ), complementary ssDNA from a denatured sample – for example DNA from a patient (purple strands). Fluorescent label (yellow starbursts) can be attached to the patient DNA prior to hybridisation to facilitate detection – the presence and amount of label at each spot on the array is determined by scanning of the entire array.
DNA from patient samples can be fragmented, denatured and hybridised to the microarray; each spot on the microarray will capture its complementary sequence from the patient sample, provided that the particular sequence is present in the patient’s genome. The quantity of patient DNA that is captured on each spot will be proportional to the amount of that sequence present in the sample. The hybridisation process is optimised such that only perfectly complementary matches can occur. Thus the hybridisation can be sensitive to a single nucleotide difference, allowing genotyping to determine which allele of a particular SNP is present, for example by having one spot on the array for each of the two alleles. A typical SNP array can be used to genotype hundreds of thousands of SNPs from across the genome of a patient, and the interpretation of allele ratios plus total fluorescence ( Figure 46 ) can reveal multiple chromosomal imbalances ( Figure 47 ).
SNP array data can reveal copy number imbalance and homozygosity associated with consanguinity and UPD

For ease of display and interpretation in SNP arrays the two alleles of each SNP are conventionally designated A and B, thus, for a T/C SNP, T would become the ‘A’ allele and C would become the ‘B’ allele. Each SNP is genotyped and the result plotted (as a green spot) according to position along the chromosome in terms of ‘B allele ratio’, which will be 0% for AA, 50% for AB and 100% for BB. Trisomy is revealed by B allele frequencies of 33 and 66%, and by an overall gain in fluorescence. In monosomy there is only one allele for each SNP so there will be no heterozygosity, and the total fluorescence will be only half the expected value. Long runs of homozygosity resulting from UPD or consanguinity will generate normal fluorescence levels. Other states can also be diagnosed, for example mosaicism (mixtures of two or more cell lines) will lead to skewed B allele ratios.
SNP array demonstrates areas of loss and gain across the genome at high resolution

( A ) ‘LogR’ plot for all 22 autosomes and the sex chromosomes demonstrates balanced copy number for most chromosomes. The pink star indicates a duplication (upward shift of LogR) affecting the chromosome 2p terminus, which is expanded in ( i ). The blue star indicates a deletion affecting the 15q terminus, which is expanded in ( ii ). The green star indicates apparent loss affecting the X chromosome but this reflects the presence of only one X chromosome in a male. ( B ) The B allele frequency plot confirms the 2p duplication ( i ) and 15q deletion ( ii ); each spot represents the result for a single SNP – there are 843551 SNPs represented in this array. ( C , D ) The sample in this case came from the child of a balanced reciprocal translocation carrier. The chromosomes 2 and 15 of the parent are depicted in (C), with arrows indicating the breakpoints. The array result indicates that the child received an unbalanced arrangement from this parent: a normal chromosome 2 together with the translocation 15 (D). Note that the array result (A,B) also demonstrates a duplication of chromosome 15 material (yellow arrow) associated with the translocation breakpoint. This would have been below the resolution of a standard karyotype, but is clear from the array result. Small gains and losses can be seen in other chromosomes on close inspection; these may represent CNVs. Array images courtesy of West of Scotland Genetics Service using Illumina CytoSNP 850K Beadchip; chromosome images generated using CyDAS ( www.cydas.org ).
SNP arrays are more effective than karyotyping at revealing chromosomal abnormalities, since the resolution is higher: typically 50 kb, with a resolution of 10 kb in critical regions for diagnostic SNP arrays, compared with approximately 3–4 Mb for karyotyping.
Next-generation sequencing
While microarrays provide whole genome coverage at high resolution, many genetic conditions result from much smaller alterations, typically SNVs or small indels. Sanger sequencing provides accurate data but, even when automated, only one or two genes can generally be analysed at a time. A number of newer technologies, collectively referred to as next-generation sequencing (NGS), overcome the limitations of Sanger sequencing and can provide an entire human genome sequence by ‘massively parallel sequencing’. DNA from a patient sample can be fragmented and then the fragments can be sequenced as part of massive arrays generating millions or even billions of short sequence ‘reads’ per run. Bioinformatics is then used to map all these reads to the reference genome, which can then be assembled into a whole genome sequence for that individual ( Figure 48 ). Differences from the reference sequence are identified by the software, and this is where the limitations of whole genome approaches start to become apparent. Because of the sheer amount of variation present in each human genome, the task of filtering the information to identify relevant variants is enormous. In fact, in 2010 Elaine Mardis described the whole genome approach as ‘the $1,000 genome, the $100,000 analysis’ in recognition of the fact that while obtaining the full genome sequence is now relatively cheap, the subsequent analysis requires huge input of time and effort. Development of improved bioinformatics approaches will undoubtedly decrease the cost of analysis, however whole-genome sequencing (WGS) is currently beyond the scope of routine diagnostics in a healthcare context.
NGS data analysis

The screenshots cover one exon from the analysis of a patient sample with an epilepsy gene panel that examines exon sequences and flanking regions from 104 genes. ( A ) The top of the screenshot provides chromosomal context (in this case 14q32). The ‘read depth’ window provides an indication of the number of times each nucleotide was represented among all the reads. Individual reads are shown as blue (forward strand read) or green (reverse strand read) bars in the ‘pile-up’. Each read is approximately 100 nucleotides in length, and represents the output from one of the millions of massively parallel sequencing reactions. The reads are aligned to the matching segment of the genome reference sequence, to generate the ‘pile-up’ view. Where the sequence of a read differs from the reference sequence that nucleotide is highlighted in a different colour. Differences seen in only one or two of the reads are likely sequencing errors (for example, those indicated by orange arrows), while the pink arrow indicates a position at which approximately 50% of the reads differ from the reference, which indicates a heterozygous variant. At the bottom is shown the intron/exon structure: the data are for exon 65 plus flanking sequence of the DYNC1N1 gene. ( B ) The view is zoomed in to the heterozygous variant detected in the first nucleotide of exon 65 (the second base of a codon); this is a characterised variant known as rs138428684, present in 1 per 1000 European alleles, changing the coded amino acid from threonine to arginine at position 3981 in the encoded protein. Readers can explore this variant in databases like Ensembl genome browser ( www.ensembl.org ) by inserting the variant name (rs138428684) into the search box. Images courtesy of West of Scotland Genetics Service using SeqVar software.
An alternative to WGS is whole-exome sequencing (WES), in which either exon sequences are specifically captured/amplified from the sample for sequencing, or bioinformatics tools remove non-coding sequences from the subsequent analysis. A typical WGS will generate 3–4 million variants per genome, whereas WES will generate only 30000–60000. In cases where a particular variant leads to loss-of-function (e.g. nonsense, frameshift) in a gene that has been previously associated with the patient’s condition, or where the particular variant has been reported as causative for that condition, then an unambiguous diagnosis can be provided. However, large numbers of VUS are identified during NGS, in particular missense variants, or variants that might affect splicing. These can be analysed in silico , for example based on properties of the new amino acid compared with the original, or conservation of that amino acid between species, or potential to create or destroy a splice recognition site in the RNA.
The scale and difficulties associated with variant analysis mean that a preferred approach for many genetic conditions is to use a gene panel representing the specific set of genes that are known to be associated with the particular condition, for example epilepsy, cardiomyopathy, inherited cancer predisposition or even broader panels, such as for recessive paediatric-onset conditions. Even with a reduced sequencing target the problem of VUS is still significant, and many patients will still not receive a diagnosis. As knowledge improves, for example with functional studies by research laboratories, some VUS will be reclassified as either benign or pathogenic. However, the current position is that NGS technology surpasses our ability to effectively utilise the information generated for the benefit of patients.
Variants which are expected to have a severe effect on an encoded protein (for example nonsense or frameshift) are easy to classify as pathogenic. However, variants which might affect the quantity or sequence of the mRNA (for example, variants affecting splicing or promoter activity) are harder to interpret using the DNA sequence alone. Although in silico analysis can help, the optimal approach is to investigate the transcripts generated, by isolating RNA from a patient sample and conversion into complementary DNA (cDNA) for analysis. Of course, transcription and splicing patterns of many genes are tissue-specific, so there may be a requirement to use a tissue biopsy rather than a blood sample. RNA-based analysis is currently uncommon in diagnostic laboratories, with the notable exception of cancer diagnostics (see below). Nevertheless, NGS technology can also be applied to the analysis of cDNA, and this ‘RNA-seq’ approach, currently used extensively in research, has enormous potential in clinical diagnostics.
The role of molecular pathology in cancer diagnostics and management
Cancer cells accumulate large numbers of mutations, many of which are passengers, but some of which are drivers of the cancer phenotype. Identification of the driver mutations present in the cancer of a particular patient can help direct therapy. A relatively new application of genetic diagnostics is in molecular pathology, which is fast becoming a core discipline within cancer management. Immunohistochemistry is frequently used to detect levels of particular proteins in tissue sections from cancers, for example the detection of HER2 overexpression to inform decisions about use of the drug Herceptin. However, many of the genetic diagnostic approaches used for inherited disorders can also be applied to investigation of cancers. The presence of genome rearrangements that are associated with particular diagnoses and/or success of a particular therapy ( Figure 35 ) can be identified by use of specific combinations of FISH probes applied to interphase cells or tissue sections. Likewise, techniques including allele-specific PCR and DNA sequencing can be used to identify mutations of diagnostic or therapeutic significance, and MLPA can be used to identify gene copy number changes. The presence of cancer-associated transcripts, particularly fusion transcripts such as those from BCR-ABL gene fusions, can be assessed by PCR amplification of cDNA. Such PCR-based approaches have much higher sensitivity than FISH; for example interphase FISH can detect one leukaemia cell per 200 normal cells, whereas PCR using cDNA can detect one leukaemia cell per million cells. This level of sensitivity is critical in monitoring response to therapy, and in early detection of relapse (by reappearance of fusion transcripts in blood samples). In fact, the sensitivity of PCR is being harnessed in the development of ‘liquid biopsy’, which targets circulating cancer DNA present in the blood, and has the potential to replace invasive tissue biopsies.
As with all genetic diagnostics, NGS-based approaches are starting to play a role in molecular pathology, which is not surprising, given the large numbers of mutations in each individual cancer. Gene panels can be applied to the analysis of hundreds of targets within cancer DNA for alterations that provide information relevant to diagnosis, prognosis and response to therapy. Whereas the PCR-based approaches mentioned above are effective in identifying fusion transcripts, the number of targets that can be assessed in one assay is very limited, and therefore some fusions will be missed. RNA-seq represents a powerful approach to screen for hundreds of potential fusion targets in a single assay, and will be particularly useful in initial identification of relevant gene fusions in individual patients. Following identification of the fusion, standard PCR-based approaches can be used to monitor that patient in relation to the identified fusion.
Clearly health service genetics laboratories have many different approaches available for the detection of pathogenic variants, either inherited or in relation to cancer tissue. One of the key roles of a clinical scientist is therefore to ensure that the most appropriate and most cost-effective approach is used for each patient sample. This can also require that the clinician who is referring the patient for genetic analysis provides a sufficiently clear and thorough view of the clinical features, to guide appropriate genetic analysis. However, the increasing use of NGS-based approaches will facilitate a more comprehensive genetic analysis that will improve diagnostic success rates.
When considering genetic diseases, it is always vital to remember the impact they can have on the life of an individual, family and society. Diagnosis, management and therapy are all important aspects of genetic disease and these issues are discussed briefly in this section. Obtaining a clear diagnosis is often important for several reasons, potentially directing therapeutic intervention, management or informing reproductive decisions. A diagnosis can also be useful in terms of gaining access to relevant support, whether this be financial, social or practical. In addition, many individuals for whom obtaining a diagnosis has been problematic, report great relief when a diagnosis is finally made. Genetic testing can be carried out at any time during the life of an individual who is symptomatic or has a high risk. However, in some situations it is desirable to be proactive in testing whole populations in order for timely intervention to occur. This is typified by genetic screening programmes.

Newborn screening
Newborn screening for genetic conditions has been used for several decades in order to detect and treat genetic conditions and potentially avert serious outcomes. Diagnosis and treatment of the autosomal recessive condition phenylketonuria (PKU) (see Table 6 ), is one such success story. In this situation both parents are carriers and phenotypically unaffected but with a one-fourth chance of having an affected child. Affected individuals lack the enzyme phenylalanine hydroxylase which converts phenylalanine into tyrosine and this results in the toxic build-up of phenylalanine in the body. This has a devastating effect on the developing brain in particular, and causes irreversible damage.
Newborn screening for PKU in the U.K. began in 1969 and identifies affected babies by measuring the phenylalanine levels in a bloodspot taken from the heel between 5 and 8 days after birth. Once PKU is diagnosed, a strict phenylalanine-free diet is implemented which ideally should be continued throughout life (although adherence is less essential during adulthood with the exception of pregnancy). If this diet is implemented prior to day 23 of life, the individual will not suffer from brain damage and is likely to lead a normal life. Testing for other conditions such as CF and sickle cell disease is now also routinely offered and new conditions are added to the programme as part of regular review processes. Mass Spectrometry with its ability to give insight into the metabolome has already proven significant in these developments and will likely contribute to further expansion.
Pre-marital genetic testing for thalassemia
While newborn screening attempts to identify affected infants very early in life, other strategies are aimed at preventing the conception of affected individuals. Specifically, pre-marital or pre-conception testing identifies couples where both are carriers of the same condition so that they can make informed choices. Examples of conditions in which pre-marital testing has been successfully used are the thalassemias and sickle cell anaemia as well as Tay–Sachs disease. The widely reported pre-marital screening programme for thalassaemia in Cyprus began in 1973, with couples being required to produce a certificate confirming they had been tested for thalassemia carrier status before marriage could legally take place. In the majority of countries where this is practiced, marriage between two carrier individuals is not forbidden, however the social structure in many communities means that it is discouraged. In other settings, religious leaders, for example in some Jewish communities, have used the genetic information (with full consent from the individuals) as part of their marriage arrangement considerations. This was deemed to be both acceptable and indeed welcomed by the community, who for many years had suffered the effects of a high incidence of Tay–Sachs disease, a fatal inborn error of metabolism.
Prenatal diagnosis
Prenatal diagnosis (PND), the testing of an unborn foetus for a specific condition, is also offered by genetics services. An initial requirement of any PND is that the sample is obtained from the foetus. This can be achieved in a number of ways, primarily chorionic villus sampling (CVS) between 10 and 14 weeks of gestation, and amniotic fluid sampling, usually between 14 and 20 weeks of gestation ( Figure 49 ). As both have an intrinsic risk of miscarriage (1–2% and 0.5–1% respectively), they tend to only be offered where there is a substantial risk of disease. Later in pregnancy, taking a cord blood sample is also a possibility, with a similar risk of miscarriage to CVS. Amniocentesis and cordocentesis have the advantage that they represent foetal rather than placental tissue – although both are derived from the embryo, the possibility exists that mosaicism may be present, resulting in the placenta and the foetus having different genotypes and thus a small risk of misdiagnosis using a CVS.
Amniocentesis procedure

Under ultrasound guidance, a thin needle is passed through the abdominal wall into the amniotic sac. A small amount of amniotic fluid is then removed and subjected to analysis. The amniotic fluid contains a mixture of cells, a proportion of which will be foetal. By BruceBlaus, CC BY-SA 4.0, from Wikimedia Commons.
Once a sample has been obtained through CVS or amniocentesis, testing can be carried out. The genetic test used depends upon the reason for the PND; QF-PCR ( Figure 43 ) for aneuploidies is currently a routine step, and additional testing can be either directed (for a particular condition or chromosomal imbalance that the foetus is at-risk for) or a whole genome approach like SNP array ( Figure 47 ), where abnormalities of unknown aetiology have been detected on ultrasound. Individuals undergoing these procedures most often opt for termination of pregnancy if a genetic problem is found, while others use the knowledge to enable them to prepare for the birth of an affected child.
Pregnancy screening and non-invasive prenatal diagnosis
Traditionally, women have been offered maternal blood screening tests in early pregnancy, the results of which place them in either high or low risk categories for DS other trisomies. In the maternal blood sample, the level of a number of proteins is quantified and these results are combined with ultrasound measurements and factors such as age to assess risk. Women in the high risk category are then offered PND using CVS or amniocentesis to obtain a sample.
However, the relatively low sensitivity and specificity of the traditional maternal blood screening tests result in both false positives (unaffected pregnancies placed in the high risk category and therefore undergoing invasive diagnostic tests, with inherent miscarriage risk) as well as false negatives (affected pregnancies where the mother is falsely reassured by being placed in the low risk category).
Therefore, more recently, non-invasive prenatal diagnosis (NIPD) has been offered in pilot schemes in the U.K., in an attempt to reduce the number of healthy pregnancies lost due to diagnostic testing and to offer women greater choice. In NIPD, a sample of maternal blood is taken from approximately 7 weeks of gestation onwards and analysed directly, using cell-free foetal DNA in the maternal circulation ( Figure 50 ). The amount of DNA present can be quantified and a risk for aneuploidies given with a very high sensitivity and specificity (approximately 99% for DS). This screening can be followed up by diagnostic testing if a high risk result is obtained. NIPD can also be applied to some inherited disorders, for example by testing for foetal sex by presence of Y chromosome (for X-linked recessive disorders) or looking for paternal pathogenic variants.
NIPD for trisomy 21

( A ) Foetal cell-free DNA (cfDNA) from the foetal circulation crosses the placenta into the maternal circulation, which thus contains both maternal and foetal cfDNA. ( B ) cfDNA is collected from maternal blood samples. The maternal cfDNA tends to be longer fragments than foetal cfDNA so that separation is possible but not straightforward. In general, however, there is no separation stage. In cases where the foetus is affected by trisomy 21, there will be more foetal cfDNA fragments derived from chromosome 21 (coloured red and indicated by asterisks) in comparison with a case where the foetus is unaffected. ( C ) The total cfDNA is analysed by DNA sequencing using NGS, allowing a count of how many reads have been obtained from each chromosome. If there was an over-representation of chromosome 21 fragments in the cfDNA sample then there will be increased representation of NGS sequence reads that match chromosome 21 (asterisked). The analysis is often quantified by calculating ratios of (for example) chromosome 21 reads to chromosome 1 reads. If only foetal cfDNA was present the chr 21:chr 1ratio would be expected to be 1:1 from an unaffected foetus, and 1.5:1 from an affected foetus, but the additional presence of disomic maternal cfDNA in the sample means that the ratio will be lower.
Preimplantation genetic diagnosis and three parent babies
As an alternative to prenatal diagnosis and possible termination of an affected pregnancy, some couples prefer to prevent the implantation of an affected embryo. Pre-implantation genetic diagnosis (PGD) is an approach which combines IVF and genetic technology to ensure only embryos unaffected by a specific genetic condition are implanted in the uterus ( Figure 51 ). After IVF, and growth to approximately the 8-cell stage, 1 or 2 cells are removed from the developing blastocyst and, depending on the condition being tested for, undergo FISH or PCR-based analysis, looking for specific variants or imbalances. Only unaffected embryos are then implanted. Follow-up PND is recommended during the pregnancy as occasionally technical limitations may lead to false results at the genetic analysis stage. PGD has been successfully used for a number of conditions, including both single gene disorders and chromosomal disorders.
Pre-implantation genetic diagnosis by biopsy of early embryos

Traditional IVF procedures ( 1 ) are used to generate fertilised embryos ( 2 ), which are allowed to develop to the 8-celled stage ( 3 ). One or two cells are then removed for genetic analysis ( 4 ) and only embryos without the condition tested for are implanted into the mother’s uterus ( 5 ).
Furthermore, as discussed earlier, in a family with a mitochondrial disorder, transmission to the next generation can be prevented using the new approach known as mitochondrial replacement therapy in which a maternal and donor egg are combined, either prior or subsequent to fertilisation.
Personalised medicine
In addition to provision of diagnostic information for genetic disease, treatment is also an important area to address. While, general therapies have been used for centuries, the concept of personalised medicine has long been seen as the ‘holy grail’ of genetics. Encompassed in the idea of giving the right treatment to the right patient at the right time, personalised medicine relies on the ability to specifically diagnose the underlying molecular and genetic aspects of the condition.
With some cancers, a personalised approach is, to some extent, mainstream. In the treatment of breast cancer, only patients whose cancer expresses specific hormone receptors on their cell surface will be offered hormonal treatments such as tamoxifen for those whose tumours overexpress the oestrogen receptor. Similar approaches in leukaemia (using the drug imatinib to target cancer cells with BCR-ABL translocations) have been used for several years and are now beginning to be used in lung cancers (using erlotonib to target cancer cells which have EGFR mutations) and a variety of others ( Figures 52 and 53 ).
EGFR mutation status determines the outcome of erlotinib as a therapy

( A ) EGFR is a transmembrane receptor that has a tyrosine kinase (TK) domain. In the absence of EGF the normal receptor is in an inactive state. ( B ) When EGF binds, the TK domain undergoes a conformational change and becomes activated, generating signals for cell proliferation. ( C ) Cancer associated mutations in EGFR lead to hyperactivation of EGFR that represents a key driver of tumorigenesis. ( D ) The TK inhibitor erlotinib binds to EGFR and prevents downstream signalling. Therefore in cases where EGFR mutations are driving tumorigenesis, erlotinib can block this process. ( E ) The acquisition of a second mutation that prevents erlotinib binding leads to resistance to this inhibitor.
EGFR mutation status must be determined in order to ensure that erlotinib is only used in those cases where it will provide benefit

The EGFR activating and TKI resistance mutations are clustered in the tyrosine kinase domain (see Figure 52 ) between amino acids 688 and 875 of the EGFR protein, so that mutation analysis can be focussed on the coding sequences for this region. Abbreviation: NSCLC, non-small cell lung cancer.
More recently, personalised approaches to single gene disorders, for example CF are beginning to be used. For CF, drugs have been developed which target the specific protein defects caused by particular mutations, for example some mutations which cause the protein to form channels that cannot open and close properly. A drug called KALYDECO (ivacaftor) can be used in these patients to help the channel to stay open allowing normal passage of ions through the open channel.
Gene therapy/gene editing
While personalised treatments in cancer and in CF target the affected protein, therapies which address the underlying genetic aspects, to ensure that a functional protein can be generated, also hold great appeal. Broadly speaking, gene therapy aims to replace the defective gene by delivering a new working copy to the cell, while gene editing aims to correct the defective gene.
One condition in which these approaches have been studied is DMD, an X-linked recessive disorder which causes progressive muscle degeneration and weakness. DMD is mainly caused by large deletions and duplications (and less commonly nonsense mutations) in the dystrophin gene and affected individuals produce virtually no dystrophin, with resultant muscle cell damage. Males with the condition commonly use a wheelchair by 12 years old and the average lifespan for an affected individual is approximately 30 years.
In a condition such as DMD, where the underlying mutation is known, a variety of approaches have been trialled ( Figure 54 ). It is hoped that to induce exon skipping, whereby the RNA processing machinery excludes the affected exon, can result in a milder form of the condition, similar to Becker muscular dstrophy (BMD). Replacing the dystrophin gene through delivery by adenovirus particles has also been attempted, although not without difficulty due to both the size of the gene and the immune reaction to the viral particles.
Potential therapeutic approaches for DMD

( A ) In healthy muscle the dystrophin protein functions as a link between the actin fibres in the cell and the dystroglycan complex (DGC) in the cell membrane, preventing damage to the cell during muscle contraction. ( B ) In the absence of dystrophin, the muscle cell sustains damage during contractions, eventually leading to muscle cell death. A number of therapeutic approaches are possible (those in pink boxes have been trialled in DMD patients, with some success reported; the relevant drug names are in green font). ( 1 ) Stem cell therapy, by injecting either healthy, tissue matched muscle stem cells from a donor, or stem cells from the patient which have been isolated, cultured in vitro , and then subjected to genome modification (see 6 , 7 ) to correct the defect prior to injection back into the patient. ( 2 ) The utrophin protein has a similar structure and function to dystrophin, but is not normally expressed in sufficient quantity to substitute for dystrophin; by up-regulating utrophin gene expression the function of dystrophin can be replaced ( C ); this approach has been effective in mouse models. ( 3 ) A significant proportion (approximately 15%) of DMD is due to nonsense mutations which lead to premature translation termination, and would therefore generate non-functional protein fragments. By use of a nonsense suppressor drug, which influences the ribosome to read through nonsense codons by incorporating an amino acid and continuing translation, full length dystrophin protein can be generated ( D ). ( 4 ) Roughly 70% of DMD is due to deletion or duplication of exons, leading to frameshift; a proportion of microlesions also lead to frameshift. By use of molecules that target the splicing process, and cause selected exons to be skipped (removed during splicing), the reading frame can be restored, generating a shorter version of the dystrophin protein, which is, nevertheless, still able to form a link between actin and DGC ( E ). Although not the same as normal dystrophin, these shorter dystrophin proteins are associated with much milder symptoms, i.e. Becker muscular dystrophy (BMD). Exon skipping could also be used to skip exons harbouring nonsense mutations. ( 5 ) The full-length dystrophin gene is too large to be accommodated in current gene therapy vectors, but because shorter versions of dystrophin are effective in restoring function, gene therapy with minigenes is a possibility. (6) Genome editing using strategies like CRISPR-Cas may be utilised to correct the pathogenic change within the genome, either by in vitro targeting of stem cells removed from the patient prior to injection back into the patient, or by delivering the CRISPR-Cas system directly to the muscle cells. (7) Genome editing (exon snipping) to remove particular exons from the genome is an alternative approach to generating an in-frame gene that is free of pathogenic variants.
Clinical trials using gene therapy have had a notoriously difficult path, with death of a patient affected by ornithine transcarbamoylase (OTC) deficiency (Jesse Gelsinger) in one trial and development of leukaemia in others. Even in conditions which seem naturally amenable to gene therapy (e.g. CF with its well-understood pathophysiology and relative ease of access to affected tissues), achieving stable and sustained expression of replacement genes has proven difficult.
Recent success in treating the neuromuscular disorder spinal muscular atrophy (SMA) using a treatment known as antisense oligonucleotide therapy has aroused much interest. With the condition being caused by loss of a gene called SMN1 , research has focused on attempting to restore the function of a homologue, the SMN2 gene. Under normal conditions SMN2 is largely non-functional due to a point mutation which results in exon 7 being spliced out. Antisense oligonucleotide therapy employs an oligonucleotide which binds to the SMN2 mRNA and changes how it is spliced, allowing the SMN2 protein to substitute for the absent SMN1. Trials have so far been very successful, with every indication that this is a remarkable breakthrough in treating this severe and life-limiting condition.
Great excitement has also surrounded the advent of gene editing technology, in particular the use of the CRISPR Cas9 (clustered regularly interspaced short palindromic repeats (CRISPR) associated nuclease 9) system, a genome editing tool which acts as a pair of ‘molecular scissors’ to cut a specific piece of DNA. Made from two components and originating as part of bacterial immune defences, Cas9 is a nuclease, guided to its target by a single guide RNA which binds to its cDNA sequence. The Cas9–RNA complex creates specific DSB non-homologous end joining or by homology directed repair if a suitable donor DNA is present. This allows precise sequence modification to edit out the genetic defect and replace it with a desired sequence. Recent studies, for example in DMD mouse models show promise and there are hopes that the technique may also be applicable in humans ( Figure 55 ), although difficulties such as specificity and efficiency of the repair still have to be fully addressed, as does the issue of immunogenicity. Nevertheless this is an exciting area, holding great promise for the future.
The use of CRISPR/Cas9 in DMD

In stem cells from the patient CRISPR/Cas9 can be used to remove the section of the dystrophin gene which harbours the mutation. The cells will then repair the DNA, creating a gene which, when expressed, will result in a shorter but functional form of the protein. When the cells are then grown and caused to differentiate into skeletal and heart muscle cells which can then be transplanted into the patient resulting in a milder phenotype. This has already been achieved in mice and the same treatment could potentially be applicable to humans.
As options available to patients, healthcare workers and scientists continue to expand, we face an array of issues and dilemmas in genetics.
The first full human genome was sequenced at a cost of $1 billion over a period of 13 years. Today, a full genome can be sequenced in 1 h costing approximately $ 1000 ( Figure 56 ). As we understand the genetic basis of increasing numbers of conditions, and as full genome sequencing becomes ever more available, it seems inevitable that there will be a clash between cost effectiveness and the patient’s ‘right to know’. Additionally, although it is relatively straightforward to sequence the human genome, the interpretation of these data are an altogether more complex issue, highlighted by the issues surrounding VUS.
Cost per genome over time

As a comparison, expected fall in cost as predicted by Moore’s law, a commonly used model for tracking technological development, was surpassed beginning approximately 2008. Image courtesy of National Human Genome Research Institute https://www.genome.gov
VUS pose increasing dilemmas for the healthcare sector because, while the capability exists to detect virtually all variants in the human genome, the ability to understand this information has yet to catch up. Simply put, although it is possible to identify that there has been, for example a single base change in a particular gene, it is much more difficult to determine what the resultant effect of that change is at a protein or even cellular level. Variants are investigated using a range of computer algorithms, a lengthy process which examines aspects such as conservation across species, amino acid similarity, protein domain structure etc., but which will not always yield a definitive result (see Table 2 ). As explained earlier, each individual harbours approximately 3 million variations in their genome, the vast majority of which are harmless. Deciding which, if any, of all the variants found is responsible for, or may predispose to a specific condition is a potential minefield. Similarly challenging is the issue of which of these variants should be disclosed to patients, and with what explanation. As our ability to catalogue and share information on variants expands, their significance will likely become clearer. Ensuring that patients are kept abreast of significant discoveries relevant to their healthcare is likely to be challenging. Finally, if interpreting VUS in post-natal, child and adult genetic services is complex, the challenge is even greater in prenatal genetics, where the lack of phenotype information compounds uncertainty. Additionally, the issue of data storage is an increasingly important one, with the concern that health service systems may soon struggle to keep pace with the increasing quantity of data being generated.
Direct to consumer testing
While individuals affected with genetic conditions have typically been seen by genetics hospital services, in the past decade the availability of ‘Direct to Consumer’ (DTC) genetic testing has risen exponentially. DTC testing is ‘sold’ directly to the customer with no healthcare service/provider involvement and through companies such as ‘23andMe’ and ‘Living DNA’, individuals can be tested for both ancestry and health information. As an example, ‘23andMe’ (at the time of writing) offers to screen customers for their carrier status for 40 diseases and their susceptibility to an additional ten conditions (including Alzheimer’s, Parkinson’s and Celiac disease), as well as offering insights into ‘traits’ such as earlobe type, sweet taste preference and hair curliness. Samples, generally saliva or cheek swaps are sent through the post with results being communicated online after a period of approximately 3 months.
The testing carried out by these companies is generally SNP array-based and ranges from testing for established mutations (e.g. CF carrier screening tests for 29 mutations in the CFTR gene) to the highly controversial susceptibility testing. For example in the test used to determine an individual’s risk of developing Parkinson’s disease, two SNPs in the LRRK2 and GBA genes are used to provide an estimate of the individual’s risk of developing the condition. Evidence surrounding these SNPs is reasonably convincing, however they are only two of several such SNPs that have been reported. The U.S. Food and Drug Administration withdrew ‘23andMe’s’ licence to provide heath information in 2013 due to concerns regarding ‘medical guidance reasons and the accuracy of the data gathered’, however this has since been reinstated for a limited number of conditions. Uncoupling of genetic testing from professional genetic counselling has raised concerns and several studies have suggested that many consumers are not able to fully understand the implications of their test results. This in turn may lead to increased pressure on GPs, with whom patients are likely to consult for help interpreting this information.
Whose information is it?
As the amount of genetic information generated grows, the question of data ownership becomes increasingly pertinent. Both within and out-with individual families, it is often not clear who has a right to know certain genetic information. Consider the situation where a child has been diagnosed with DS, and more specifically with DS resulting from a translocation present in one of the parents (accounting for approximately 4% of cases). Other members of the family, (for instance, the carrier parent’s siblings) may well be at risk of having similarly affected children. The question of communicating the results to other family members is generally left to the discretion of the family themselves, however this may not be information they want to share. Emotions such as guilt and blame, as well as existing family dynamics all come into play when such personal information is disclosed. Thus at risk relatives may be left uninformed. Similarly, a young adult receiving an ‘affected’ predictive test result for a condition such as HD has simultaneously uncovered a parent’s result because it is almost certain that they inherited this affected allele from one of their parents. This is irrespective of whether the parent wished to be tested or not. In such cases, patients are advised during pre-test counselling that they may wish not to disclose to family members that they are considering testing, on the premise that once a result is known it is very difficult to conceal the news.
Perhaps arousing more debate is the question of who, if anyone, outside a family has the right to access genetic test results. Clearly such results may be of considerable use to certain public services, for example the police, however, the ethics of release of information remains unclear. Regulatory bodies such as the U.K. Driver and Vehicle Licensing Agency also have a vested interest in genetic test information – for example from patients whose genetic condition means they may not be safe to continue holding a driving licence.
Insurance is also an important issue with the moratorium on the use of genetic test results by insurers in the U.K. until 2019. Some would argue that the current health questionnaires required to gain insurance already screen for some of the information, e.g. a family history of HD, and that by including genetic testing results those not at risk would not be unfairly penalised. However, requiring genetic results to be disclosed on application could prevent some from having useful genetic testing carried out, or may prevent other vulnerable people from obtaining relevant insurance. Thus while the understanding and interpretation of genetic data are challenges for the scientific community and healthcare providers, the encompassing issues require considerable ethical debate and supporting legislation.
Concluding remarks
During recent decades the role of genetics in medicine has changed dramatically, beginning as a speciality dealing with conditions that were relatively rare in the population, and becoming a discipline that underpins developments in wider areas of patient care. Better understanding of the contributions of genetic predisposition and epigenetic changes to common diseases, and of the role of genetic alterations in response to therapy, means that genetics will be relevant to everyone in the population. Education, both for health professionals and for their patients, will play a key part in ensuring that new developments in genetics and genomics continue to be successfully translated and implemented into clinical practice. There will be an increasing need for clinical scientists, genetic technologists and bioinformaticians to provide the laboratory services, as well as an increasing need for clinicians, genetic counsellors and nurses who are conversant with the new genetic and genomic technologies. Additionally, there will be a great need for further generations of research scientists to address the huge remaining gaps in our understanding and to generate innovative approaches in the application of our knowledge to the delivery of cost-effective healthcare. For anyone exploring career options, genetics and genomics should provide many interesting and rewarding possibilities for the future!
We thank the West of Scotland Genetics Service for providing images of diagnostic results.
The authors declare that there are no competing interests associated with the manuscript.
The main sections of this article were authored as follows:
- The human genome and variation by Maria Jackson
- Chromosome structure and chromosomal disorders by Gerhard H.W. May
- Single‐gene disorders by Gerhard H.W. May
- The sex chromosomes, X and Y by Joanna B. Wilson
- Mitochondrial disorders by Leah Marks
- Epigenetics by Maria Jackson
- Complex disorders by Maria Jackson and Leah Marks
- Cancer: mutation and epigenetics by Joanna B. Wilson
- Genomics by Maria Jackson and Joanna B. Wilson
- Genetic testing in the diagnostic laboratory by Maria Jackson and Leah Marks
- Diagnosis, Management and therapy of genetic disease by Leah Marks
- Challenges in delivering a genetics service by Leah Marks
Acetylation/acetyltransferase The process of acetylation involves the addition of acetyl group (O=C–CH 3 ) to the target molecule, for example proteins, by the action of the appropriate acetyltransferase enzymes.
Allele A particular form of a given gene, usually one of several versions that differ in sequence and may differ in phenotype. Multiple different alleles (gene versions that differ in sequence) can exist within a population (some quite common), giving rise to phenotypic variation. Germ line differences from the sequence database for the human reference genome are called polymorphisms, or those that are more rare, called variants. Polymorphisms and variants arose in the germ line gene pool by mutation. Therefore the distinction between what is called a polymorphic or variant allele and what is called a mutant allele by research geneticists, can be blurred. In general, the frequency of the allele in the population and the extent to which it is disease causing or not, determines what it is called. Thus, the currently accepted definitions used by medical geneticists, in relation to the human reference genome are: pathogenic variant, likely pathogenic variant, VUS, likely benign variant and benign variant.
Allele-specific PCR A PCR-based method in which particular allele(s) are amplified, facilitating genotyping of the locus, typically by placing the variant nucleotide at the 3′ end of either the forward or the reverse PCR primer.
Aneuploidy An abnormal number of chromosomes in a cell, with one or more extra chromosomes or chromosomes missing.
Angiogenesis The process of ‘growing’ new blood vessels from pre-existing vessels.
Anticipation Anticipation can occur in some genetic conditions as a pathogenic variant is passed from one generation to the next. In those cases, the age of onset of the symptoms decreases from one generation to the next, and often the severity of symptoms increases as well.
Antisense oligonucleotide Short deoxynucleotide which is complementary to a ‘sense’ sequence of DNA.
Apoptosis This is a form of ‘programmed cell death’. The phenomenon results when a cell is genetically determined to die or receives internal and/or external signals to self-destruct. The cell dismantles into component parts that are neatly disposed of or recycled and this does not cause inflammation. A classic example of normal apoptosis occurs during the formation of the fingers and toes in development. During mammalian embryogenesis, as an ‘evolutionary throwback’, the digits form, linked by webbing. The cells that make up the webbing are programmed to die and as development proceeds, they die by apoptosis, separating the digits.
Association The occurrence of a specific polymorphism together with a particular trait more often than would be expected by change.
Autosome A chromosome that is not a sex chromosome.
Benign growth/tumour This results from limited new growth of a cell such that a small (occasionally large) lump forms within a tissue, but does not progress on to become invasive. A classic example of this is a mole or nevus on the skin. Benign tumours are mostly harmless, but some may acquire further mutations to become cancerous.
Benign variant A variant allele which is believed to have no effect on health either in the heterozygous or homozygous state.
Biallelic Relating to both alleles of a gene; for example biallelic expression means that products are generated from both copies of the gene.
Carcinogen Any agent that acts to increase the risk of cancer. Not all carcinogens are mutagens, but many are.
Candidate gene A gene thought to have a high chance of being involved in a particular phenotype, often due to pathways it is known to be involved in.
Cell cycle The process by which a cell divides into two cells. The cycle usually follows the four stages: G 1 (gap or growth 1), S (synthesis of DNA), G 2 (gap or growth 2), finally mitosis (note in meiosis, the cell cycle follows a different pattern, as described below). G 1 , S and G 2 together make up ‘interphase’.
Cell-free DNA (cfDNA) DNA which is not contained within a cell and is found in small amounts in the circulation or other fluids, e.g. urine.
Cell proliferation The term used when cells divide by mitosis, resulting in an increase in cell number.
Chromatin Describes the way the human genome is organised/packaged within a cell. Usually, DNA is wrapped around a core of histone proteins, forming nucleosomes.
Centromere The waist-like constriction of a chromosome which separates the short from the long arm. During cell division, spindle fibres attach at the centromere to pull replicated chromatids apart.
Chimera An organism consisting of cells which are genetically different, which can be generated by fusion of early embryos.
Codon Three consecutive nucleotides which instruct a ribosome to incorporate a specific amino acid into a growing polypeptide, or to stop translation. Note that strictly it makes only sense to speak of codons in mRNA sequences, but codons are also usually referred to when describing nucleotide triplets in the coding sequences of genomic DNA.
Complementary DNA ( cDNA ) This is generated by use of the enzyme reverse transcriptase to make a DNA copy (a ‘complementary’ copy) from RNA that has been isolated from a sample (for example, blood or tissue). This cDNA can be used to analyse splicing patterns and relative transcription levels.
Compound heterozygote An individual with (usually) pathogenic variants in both copies of a gene, where the variants are different from each other, for example a CF patient with p.Phe508del in one copy of the CFTR gene and p.Gly542X affecting the other copy.
Consanguineous Refers to families where both parents share at least one recent common ancestor.
Copy number variant (CNV)/copy number polymorphism (CNP) Segments of our genome that range in size from 1000 to millions of bp, and which, in healthy individuals, may vary in copy number from zero to several copies. Where the population frequency reaches 1% or more it may be referred to as a copy number polymorphism.
Cytogenetics The study of chromosomes.
De novo Latin for ‘anew; starting from the beginning’. Used to describe newly arisen mutations, as opposed to variants which have been inherited from a parent.
Dideoxynucleotide Used in Sanger DNA sequencing, the deoxyribose moiety of these nucleotides lacks the 3′ hydroxy group so that, while dideoxynucleotides can be incorporated into a growing DNA chain, they do not allow the addition of further nucleotides, so that the chain is terminated.
Differentiation The process by which cells and tissues acquire specialized characteristics, for example during embryonic development.
Diploid Having two copies of each autosome, and two sex chromosomes. This is the normal state of most human somatic cells.
Disorders of sex development (DSD) A diverse group of conditions that affect the development of the gonads and/or sexual differentiation and include partial or complete sex reversal in relation to the XX or XY genotype.
Dominant An allele or mutant gene version that leads to a phenotype when in a heterozygous state (for example the other allele is wild-type) is referred to as dominant, is also often used to describe a condition.
Dosage compensation The mechanism by which an imbalance in gene dose (gene copy number) is compensated for by differential gene expression. This is particularly relevant to genes residing on the X chromosome that have no Y chromosome homologue. In mammals, the process of X chromosome inactivation results in only one copy of the two alleles in female cells being available for expression, balancing with the fact that male cells only have one allele.
Dysgenesis Defective or abnormal development of an organ, for example of the gonads.
Electropherogram This is a visualisation of the results of electrophoretic separation of molecules; in the case of genetic analysis, DNA molecules can be separated based on size, and detected by the use of fluorescent labels previously attached to the DNA.
Enhancer A specific DNA sequence often adjacent to the coding region of a gene, which functions in gene regulation, for example by binding transcription factors.
Epigenetic modification This refers to modification marks that do not change a DNA sequence, but can effect gene expression and includes methylation of DNA bases (usually cytosine in mammals), and methylation, phosphorylation and acetylation of the proteins that DNA is wrapped around, the histones.
Epigenetics The study of changes in gene function that are heritable mitotically and/or meiotically and that are not a consequence of change in the DNA sequence.
Exome The portion of the genome that codes for proteins – the complete collection of all the exons.
Exon A section of a protein-coding gene that encodes part of the protein sequence; within the gene exons are separated by intervening sequences (introns) and in order to generate a functional mRNA the relevant exons must be spliced together to generate an uninterrupted coding sequence.
Gene expression represents the processes including transcription and translation which lead to the production of products (for example, proteins) from genes
Expression is also used to describe physical traits (or phenotypes) generated as a consequence of variants; the term expressivity is the degree to which the traits observed differ between individuals who have the same genotype.
Frameshift Ribosomes translate mRNA molecules one triplet codon at a time, in a continuous ‘reading frame’. Any mutation that leads to the insertion or deletion of a number of nucleotides into the mRNA, which is not a multiple of three, leads to a shift in this reading frame. This usually leads to premature truncation of the resulting polypeptide.
Gametologue A gene that has homologues on both X and Y chromosomes that are not subject to crossover in meiosis. These are not termed alleles due to the fact that they do not recombine and therefore evolve independently on the two chromosomes.
Gene amplification The duplication of a gene, often at the site of the original gene, leading to multiple copies. The duplicated gene may be wild-type or mutant and duplication usually results in its overexpression.
Genome The complete set of genetic information (usually DNA) of an organism, including all genes plus all other sequences, and in humans includes both nuclear and mtDNA.
Genomics In contrast with genetics, which often focuses on single genes, genomics represents the study of large groups of genes, often the entire genome of one or more organisms.
Genotype The genetic make-up of a cell or organism, relating to the sequence of the genes and genome as a whole. Often the genotype(s) at one or a few loci only are considered. Genotyping is the process of determining which alleles are present at one or more loci.
Germ cells Cells that will form the gametes, to become the haploid oocytes or sperm cells upon differentiation.
Gonadal mosaic Having cells of different genotypes within one or both gonads, often as a consequence of somatic mutation, with the consequence that an apparently de novo mutation, not present in the parent, may be transmitted to more than one child.
Haploid Having only a single copy of each autosome, and one sex chromosome. The usual state of gametes.
Haploinsufficiency This occurs when one allele of the homologous pair of genes in a diploid organism is lost or not expressed and this results in an abnormal phenotype. The remaining allele is expressed, but can only provide half the normal level of gene product and this is not sufficient to fully conduct the required function. Such loss-of-function mutations are dominant, as they give rise to a phenotype.
Hemizygous Having only one locus/allele within the cell.
Heteroplasmy The presence of differing mitochondrial genomes within a cell.
Heterozygous Having two different alleles at one locus.
Histone acetyl transferase (HAT)/histone deacetylase (HDAC) These enzymes add or remove acetyl groups (O=C–CH 3 ) from histone proteins leading to changes in chromatin structure that affect function of the DNA.
This is a genetical term referring to genes that are related by evolutionary descent, that is, homologues have evolved from the same gene in an ancient organism. There are also subdivisions of the term homologue. Paralogue: indicating descent within a single species, for example the human RAS genes ( HRAS, NRAS and KRAS ) are paralogues of each other. In the ancestral organism, the RAS gene underwent duplications and three of these remain in humans (and many other mammals) today that have evolved to take on slightly different roles. Orthologue: indicating the same gene in different species, for example human HRAS and mouse HRas are orthologues.
In a diploid organism, each autosome and the X chromosome in females (and therefore also each gene on these chromosomes), has a homologue, thus comprising the homologous pairs of chromosomes present in the nucleus of the diploid cell.
Homoplasmy The presence of only identical mitochondrial genomes within a cell.
Homozygous Having two identical alleles at one locus.
Ideogram A graphical representation of a cell’s or organism’s karyogram.
Imprinting (chromosomal or genomic) The process by which epigenetic marks are attached to particular loci in a parent-of-origin specific manner, leading to differential expression of maternally and paternally derived genes.
Indel A term describing any variant which represents the insertion or deletion (or a combination of both) of nucleotides at a specific position, compared with a reference genome.
Inflammation This describes an immune response, usually wounding or infection, but can be of unknown cause. Immune cells enter the damaged or infected tissue and release factors that are designed to repair the wound and combat infection. Short-lived inflammation, for example in response to a small wound, is called acute inflammation. Prolonged inflammation, sometimes of unknown cause, is called chronic inflammation and can be damaging to tissues if it continues unabated.
In silico Performed using a computer; for example the application of software or algorithms that use existing information to predict the effect of DNA variants.
Intron A segment of a gene, between two coding segments (exons), in other words an intervening sequence, which is transcribed into RNA and then removed by splicing during generation of the final mRNA.
Karyogram A (usually photographic) representation of the chromosomes of a cell, arranged in pairs.
Karyotype The number and appearance of the chromosomes in the nucleus.
Kinase Kinases are enzymes that add a phosphate group onto their substrates. Protein kinases phosphorylate (by transfer of the phosphate group onto the oxygen atom of the amino acid side chain) their protein substrates upon serine, threonine or tyrosine residues.
Locus Genetical term referring to a specific location in a genome, usually defining the position of a gene or DNA sequence of interest. Plural: loci.
Metaphase One of the phases of the mitotic cell division cycle, during which chromosomes become condensed and visible under a light microscope.
Metastasis (plural metastases)/metastatic disease Cancer cells that are dividing out of control and have spread (from the primary tumour site) to other tissues or organ sites around the body.
Meiosis The final stages of germ cell division to produce four haploid gametes, each of which is genetically distinct. A germ cell undergoes DNA replication and then, before separation of the replicated ‘sister chromatids’, homologous chromosomes pair and undergo recombination, such that DNA is swapped or ‘crossed over’. Then two rounds of cell division occur: Meiosis I and Meiosis II. In Meiosis I, the chromosome pairs separate into daughter cells, in Meiosis II, the sister chromatids separate into daughter cells. In some organisms and in the production of mammalian sperm, all four meiotic products form gametes. In female mammals, only one cell develops into the ovum or oocyte, with asymmetric division of the cytoplasm, the other three meiotic nuclei are extruded as polar bodies.
Methylation/methyltransferase The process of methylation involves the addition of methyl (CH 3 ) groups to target molecules, for example DNA or proteins, by the action of the appropriate methyltransferase enzymes.
Microarray A set of targets, most often DNA probes, arranged in a grid to facilitate testing. DNA microarrays, including SNP arrays, usually contain hundreds of thousands of probes to which sample DNA or RNA can be hybridised.
Microdeletion/microduplication A deletion/duplication that is generally defined as below the resolution of karyotyping and therefore less than 4–5 Mb, but larger than 1 kb. However, precise definitions may vary between authors.
Microsatellite A variable number tandem repeat in which the repeating unit is generally between 2 and 6 bp in length, and under some definitions includes mononucleotide repeats.
Minisatellite A variable number tandem repeat in which the repeating unit is generally between 10 and 100 bp in length.
Minor allele frequency (MAF) The frequency at which the second most common allele occurs in a population.
Missense An alteration (mutation), often affecting a single nucleotide, which leads to the change of a codon for one specific amino acid into one for a different amino acid.
Mitosis The process of somatic cell division, as part of the cell cycle after DNA replication, whereby a diploid cell goes through the stages: prophase, metaphase, anaphase and telophase; to separate the chromosomes into two nuclei. This is followed by cytokinesis, dividing the cytoplasm and organelles to produce two diploid daughter cells.
Mitochondrial replacement therapy A modification of IVF in which the mitochondria of the embryo are obtained from someone other than the mother or father of the child.
Modifier gene A gene in which variation can alter the severity or phenotype of disease caused by a pathogenic variant at another locus.
Molecular pathology The study and diagnosis of disease by analysis of molecules such as nucleic acids and proteins within tissues or body fluids.
Mosaic A condition in which cells of distinct genotypes or distinct karyotypes are present in one individual, often as a consequence of somatic mutation or mitotic non-disjunction.
Mutagen Any agent that can lead to DNA mutation. Therefore, by definition, all mutagens are potential carcinogens.
Mutant see Wild-type.
Mutation Any heritable (through somatic cell division or germline) change in the DNA sequence. It does not have to result in a phenotypic change or a change to an encoded protein sequence (which would be a silent mutation). See also the definition of wild-type (and polymorphic, variant and mutant alleles).
Non-invasive prenatal diagnosis (NIPD) A form of PND in which foetal DNA is obtained from the maternal blood rather than from a CVS or amniotic fluid sample.
Nonsense An alteration (mutation) affecting a single nucleotide which leads to the change of a codon for one specific amino acid into one of the three stop codons.
Oncogene A gene whose product can positively contribute to the cancerous process. Often an oncogene is a mutated form of a normal cellular gene.
Pathogenic variant A variant which is associated with disease.
Penetrance The extent to which a pathogenic variant leads to observable clinical symptoms, that is, the proportion of individuals with the variant who exhibit the disease phenotype. A variant with 100% penetrance would affect all carriers, whereas for a variant with 80% penetrance the particular phenotype would not be seen in 20% of carriers.
Phenotype The appearance, properties and behaviour of a cell or organism that are a direct consequence of its genotype.
Point mutation: The mutational change of one base pair, either to any of the other three possibilities, or deletion of the base pair, or addition of a new base pair in the sequence.
Polygenic The situation where a trait or phenotype is controlled by multiple genes.
Polymorphism Any variant allele that is present in the population at a frequency of at least 1% of all alleles.
Polyploidy State of a cell or organism which contains more than two complete sets of all chromosomes. Triploidy indicates the presence of three sets of chromosomes, tetraploidy of four.
Primer A short sequence of DNA or RNA which can act as a starting place for a new DNA strand to be synthesised.
Programmed cell death see Apoptosis.
Pronuclear transfer A technique in which the mother’s egg as well as a donor egg are both fertilised with the father’s sperm. Subsequently the nucleus from each fertilised egg is removed and the donor egg’s nucleus is then replaced with that of the mother.
Pseudogene A locus that resembles a protein-coding gene, and likely arose via an ancient gene-duplication event, but which, as a consequence of mutation, is not able to generate the original protein. Pseudogenes may, however, have regulatory roles in expression of other genes.
Quiescence Describes the state when cells are not in cell cycle (reversible G 0 ).
Recessive An allele or mutant gene version that leads only to a phenotype when in a homozygous state (or heterozygous with another pathogenic allele) is referred to as recessive. Also used to describe a condition.
Reference sequence A genomic sequence that represents a baseline from which to compare individual human sequences; the reference sequence is intended not as a ‘norm’, but only as a reference point for describing human variation.
Senescence Describes the state when cells can no longer divide, they are irreversibly in G 0 . They may still be able to function.
Sex chromosome Chromosomes determining the genetic gender of an organism. Human sex chromosomes are X and Y, male somatic cells carry one of each, female somatic cells carry two X chromosomes.
Signal transduction The process by which a cell converts an external signal into a responding action. For example, when a growth factor binds to its receptor on the cell surface, this sends a cascade of signals inside the cell, to the nucleus (or other target, such as mitochondria), which can result in a change in gene expression resulting in the cell following a new action (such as initiating the cell cycle).
Silent mutation A mutation that results in no observable phenotypic effect.
Somatic cells Comprise all cells of the body, other than those that contribute to the germ line.
Spindle transfer A technique in which the nucleus is removed from an unfertilised mother’s egg and this is then inserted into a donor egg which has had its nucleus removed. Fertilisation with the father’s sperm then takes place.
Synonymous mutation A mutation in a coding region which, despite changing the DNA sequence does not change the sequence of the resulting polypeptide (due to the redundancy of the genetic code).
Tetraploidy Having four complete sets of chromosomes – i.e. four times the haploid number.
Telomere Specific, repetitive, sequences present at the ends of linear chromosomes that protect the chromosome ends and play a key role in chromosome maintenance and stability.
Translocation This describes a large rearrangement, often visible by karyotypic analysis, where a large chunk of one chromosome has moved and joined another chromosome. Reciprocal translocation is when two chromosomes have effectively exchanged large portions. In a Robertsonian translocation, two acrocentric chromosomes lose their short arms and fuse at the centromere.
Transcription factor A protein that, typically, binds to DNA (often as part of a complex of proteins) and regulates (up or down) the expression of a gene that is usually located at, or near, the site of binding.
Triploidy Having three complete sets of chromosomes – i.e. three times the haploid number.
Trisomy Three copies of a specific chromosome, e.g. three copies of chromosome 21 in Down syndrome.
Tumour suppressor gene (TSG) A gene whose product can inhibit tumour cell growth, often through inhibiting cell cycle progression. One or more TSGs are frequently silenced in cancer cells.
Ubiquitin A small protein of 76 amino acids, that can be covalently attached to other proteins, by ubiquitin ligases, resulting in a variety of potential regulatory effects that include targeting for degradation, and changes in localisation or activity of the ubiquitinated protein.
Uniparental disomy (UPD) A situation in which both homologues of a chromosome are from the same parent.
Variant Any DNA sequence that differs from the reference genome sequence.
Variant of uncertain significance (VUS) A classification applied to variants for which the effect on the phenotype is not clear, and there is insufficient evidence to support either benign or pathogenic classification.
Wild-type allele A term used by research geneticists to indicate the allele that is most commonly found in the population. Different alleles commonly found in the population are referred to as polymorphisms, or more rare alleles, variants. Medical geneticists tend not to use the term wild-type and instead refer to the human reference genome (see also Allele). Mutant alleles are those that have undergone a sequence change as a result of somatic mutation. Constitutional pathogenic variants are often referred to by research geneticists as ‘mutant’ alleles (and this term will be found in the scientific literature), but this terminology is out of favour with medical geneticists.
X chromosome inactivation The almost complete expressional silencing of one of the two X chromosomes in every mammalian female cell, with the exception of the gametes. The process is initiated in early embryonic development and persists throughout adult life.
achondroplasia
Angelman syndrome
Becker muscular dystrophy
breast cancer susceptibility
cyclin-dependent kinase
complementary DNA
cystic fibrosis
cell-free DNA
CF transmembrane conductance regulator
copy number polymorphism
copy number variant
chorionic villus sampling
Duchenne muscular dystrophy
DNA methyl transferase
Down syndrome
double strand break
disorder of sex development
direct to consumer (genetic testing)
fluorescence in situ hybridisation
genome-wide association study
histone acetyl transferase
histone deacetylase
hypoxia-inducible factor
Immunodeficiency, centromeric instability, and facial anomalies syndrome
International System for Human Cytogenetic Nomenclature
in vitro fertilisation
kilobase pair (1000 bp)
Leber hereditary optic neuropathy
long QT syndrome
minor allele frequency
multiplex ligation dependent probe amplification
next-generation sequencing
non-invasive prenatal diagnosis
nucleolar organiser region
pseudoautosomal region
programmed cell death
pre-implantation genetic diagnosis
pre-natal diagnosis
Prader–Willi syndrome
quantificative fluorescence PCR
ribosomal DNA
Scottish Genomes Partnership
spinal muscular atrophy
single nucleotide polymorphism
single nucleotide variant
sex-determining region Y
simple sequence repeat
short tandem repeat
type 1 diabetes
type 2 diabetes
testis determining factor
tyrosine kinase
tyrosine kinase inhibitor
tumour suppressor gene
uniparental disomy
variable number tandem repeat
variant of unknown significance
whole exome sequencing
whole genome sequencing
active X chromosome
inactive X chromosome
X inactivation centre
Author notes
Authors in alphabetical order.
This article is a reviewed, revised and updated version of the 1999 Biochemistry Across the School Curriculum (BASC) booklet The Genetic Basis of Human Disease by G. Wallis. For further information or to provide feedback on this or other Biochemical Society education resources, contact [email protected] .
Get Email Alerts
- Online ISSN 1744-1358
- Print ISSN 0071-1365
- Submit Your Work
- Language-editing services
- Recommend to Your Librarian
- Accessibility
- Sign up for alerts
- Sign up to our mailing list
- Biochemical Society Membership
- About Biochemical Society
- Publishing Life Cycle
- Biochemical Society Events
- Sponsored award winners
- About Portland Press
- Portland Press Tel
- +44 (0)20 3880 2795
- Portland Press Company no. 02453983
- Biochemical Society Tel
- +44 (0)20 3880 2793
- Email: [email protected]
- Biochemical Society Company no. 00892796
- Registered Charity no. 253894
- VAT no. GB 523 2392 69
- Privacy and cookies
- © Copyright 2024 Portland Press
This Feature Is Available To Subscribers Only
Sign In or Create an Account

- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons
Margin Size
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.

4.10: Mutation Effects
- Last updated
- Save as PDF
- Page ID 6519
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
DNA Repair Pathway. This flow chart shows one way that damaged DNA is repaired in E. coli bacteria.
Beneficial Mutations
Some mutations have a positive effect on the organism in which they occur. They are called beneficial mutations . They lead to new versions of proteins that help organisms adapt to changes in their environment. Beneficial mutations are essential for evolution to occur. They increase an organism’s changes of surviving or reproducing, so they are likely to become more common over time. There are several well-known examples of beneficial mutations. Here are just two:
- Mutations in many bacteria that allow them to survive in the presence of antibiotic drugs. The mutations lead to antibiotic-resistant strains of bacteria.
- A unique mutation is found in people in a small town in Italy. The mutation protects them from developing atherosclerosis, which is the dangerous buildup of fatty materials in blood vessels. The individual in which the mutation first appeared has even been identified.
Harmful Mutations
Imagine making a random change in a complicated machine such as a car engine. The chance that the random change would improve the functioning of the car is very small. The change is far more likely to result in a car that does not run well or perhaps does not run at all. By the same token, any random change in a gene's DNA is likely to result in a protein that does not function normally or may not function at all. Such mutations are likely to be harmful. Harmful mutations may cause genetic disorders or cancer .
- A genetic disorder is a disease caused by a mutation in one or a few genes. A human example is cystic fibrosis. A mutation in a single gene causes the body to produce thick, sticky mucus that clogs the lungs and blocks ducts in digestive organs. You can watch a video about cystic fibrosis and other genetic disorders at this link: http://www.youtube.com/watch?v=8s4he3wLgkM (9:31).
- Cancer is a disease in which cells grow out of control and form abnormal masses of cells. It is generally caused by mutations in genes that regulate the cell cycle. Because of the mutations, cells with damaged DNA are allowed to divide without limits. Cancer genes can be inherited. You can learn more about hereditary cancer by watching the video at the following link: http://www.youtube.com/watch?v=LWk5FplsKwM (4:29)
Albino Redwoods, Ghosts of the Forest
What happens if a plant does not have chlorophyll? They would lack the part of the leaf that makes them green. So these plants could be referred to as albino. This would have to result from a genetic mutation. Do these plants die because they cannot photosynthesize? Not necessarily. What can these plants tell us about the biochemistry, genetics and physiology of plants?
See Science on the SPOT: Albino Redwoods, Ghosts of the Forest at http://science.kqed.org/quest/video/science-on-the-spot-albino-redwoods-ghosts-of-the-forest/ , Science on the SPOT: Revisiting Albino Redwoods, Biological Mystery at http://science.kqed.org/quest/video/science-on-the-spot-revisiting-albino-redwoods-biological-mystery/ , and Science on the SPOT: Revisiting Albino Redwoods, Cracking the Code at http://science.kqed.org/quest/video/science-on-the-spot-revisiting-albino-redwoods-cracking-the-code/ for more information.
- Mutations are essential for evolution to occur because they increase genetic variation and the potential for individuals to differ.
- The majority of mutations are neutral in their effects on the organisms in which they occur.
- Beneficial mutations may become more common through natural selection.
- Harmful mutations may cause genetic disorders or cancer .
Making Connections
Explore more, explore more i.
Use these resources to answer the questions that follow.
- http://www.hippocampus.org/Biology → Non-Majors Biology → Search: Genetic Disorders
- Define genetic disorders.
- What are the two primary types of genetic aberrations?
- What is a carrier?
Explore More II
- http://www.hippocampus.org/Biology → Non-Majors Biology → Search: Gene Defects
- What are the results of a mutation or defect in a single gene?
- Describe the causes and effects of cystic fibrosis, Huntington's Disease, and hemophilia.
Explore More III
- http://www.hippocampus.org/Biology → Non-Majors Biology → Search: Chromosomal Abnormalities
- What is a chromosomal disorder?
- When and how do chromosomal errors occur?
- Describe an inversion and translocation.
- Describe the causes of Cri-du-chat Syndrome and Down Syndrome.
Explore More IV
- Test Neurofibromin Activity in a Cell athttp://learn.genetics.utah.edu/con...neurofibromin/ .
- Why are mutations essential for evolution to occur?
- What is a genetic disorder?
- What is cancer? What usually causes cancer?
Frontiers for Young Minds
- Download PDF
What Is Genetic Diversity and Why Does it Matter?

All living things on Earth contain a unique code within them, called DNA. DNA is organised into genes, similar to the way letters are organised into words. Genes give our bodies instructions on how to function. However, the exact DNA code is different even between individuals within the same species. We call this genetic diversity. Genetic diversity causes differences in the shape of bird beaks, in the flavours of tomatoes, and even in the colour of your hair! Genetic diversity is important because it gives species a better chance of survival. However, genetic diversity can be lost when populations get smaller and isolated, which decreases a species’ ability to adapt and survive. In this article, we explore the importance of genetic diversity, discuss how it is formed and maintained in wild populations, how it is lost and why that is dangerous, and what we can do to conserve it.
Why is Everything and Everyone A Little Bit Different?
Earth contains millions of different species that all look different from one another. While some species look more similar to each other than others, like lions and tigers, they will still have differences between them. Even within each species, individuals look similar to each other but they are not identical. These differences and similarities are because of many small differences between individuals’ genes . All organisms have DNA and each individual’s DNA is organised into genes. These contain the instructions to build our bodies. This is similar to the way that letters are combined to make words that then make a story. DNA can be seen as the letters, genes the words, and their instructions are the story. Small differences in DNA might change blue eyes to green, or a butterfly’s wings from black to white, like how a word can change when you replace a letter.
The combined differences in the DNA of all individuals in a species make up the genetic diversity of that species. Genetic diversity causes individuals to have different characteristics, which we can see even in our groceries. Although all tomatoes belong to the same species, the tomatoes we eat are hugely diverse, ranging from giant beefeater tomatoes to tiny cherry tomatoes. There are also hundreds of apple varieties ( Figure 1 ), that range from red to green, tart to sweet, and some apples even have pink flesh inside! Genetic diversity is what makes these types of tomatoes and apples look so different [ 1 ]. Genetic diversity is also seen in animals. For example, dogs can be large enough to pull sledges or small enough to sit nicely on your lap. All dogs are from the same species, but they look different because of genetic diversity! Though often more difficult to see, genetic diversity is also extremely important in wild animals and plants.

- Figure 1 - An example of genetic diversity in the food we eat.
- All these apples are one species. Different alleles of the genes that control their colour cause the apples to be green, yellow, red, or almost purple. Differences in the alleles that control flavour make each type taste different.
How is Genetic Diversity Generated?
Changes to an individual’s DNA are called mutations ( Figure 2 ). Mutations can arise when mistakes are made while cells are copying DNA, like making a spelling mistake when copying a word. These mutations make up a species’ genetic diversity. Over generations, more and more mistakes are made, leading to more mutations. Most mutations are either harmful or have no impact at all, but sometimes these mutations can cause changes that are helpful for a species. The individuals that have these helpful mutations might have greater chances of survival, and have more babies as a result [ 2 ]. This is adaptation . When a mother and a father have babies, the DNA of their baby is a mix of the parents’ DNA. Babies have two copies of every gene in their DNA, one from each parent. Copies of the same gene with different mutations are called alleles . When parents make a sperm or an egg, alleles in each parent are shuffled and recombined, and only one allele of a gene ends up in each sperm or egg cell. When the reshuffled alleles from a mother and a father are combined when sperm and eggs join, new mixes of alleles are created in the babies [ 2 , 3 ]. The mixing of alleles allows for new combinations of mutations and characteristics, adding to a species’ genetic diversity ( Figure 2 ).
- Figure 2 - (A) Genetic diversity is generated when mutations create new alleles over time.
- Mixing alleles from parents creates new combinations of alleles in their babies. Organisms that can clone themselves, like bacteria, can pass alleles to each other. Each coloured dot represents a different allele. (B) Genetic diversity can be lost when habitat loss divides populations or when buildings or highways isolate populations. (C) Creating protected areas where individuals from different populations can migrate and spread their genes can help a species to maintain its genetic diversity.
Not all species need a mother and a father to make a baby. Bacteria can clone themselves ( Figure 2 ) and directly pass their alleles from a parent to its identical clone [ 3 ]. Any mistakes in the parent’s DNA will be passed on to the clone. Amazingly, bacteria can also give alleles to each other, even if they are not related! This is a unique way simple species like bacteria can increase their genetic diversity, without relying on the mixing of alleles between a mother and a father [ 4 ].
Why is Genetic Diversity Important?
When a species has a lot of differences in its DNA, we say that genetic diversity is high [ 2 ]. In species with high genetic diversity, there are lots of mutations in the DNA, which cause differences in the way individuals look as well as differences in important traits that we cannot see [ 2 ]. This is called adaptation . For example, some types of apples can grow better in hotter environments, thanks to their genes. The variety of characteristics in species with high genetic diversity means they are more likely to successfully cope with changes in their environment. A great example of this is seen in the peppered moths during the industrial revolution [ 4 ]. Natural genetic diversity in peppered moths produced different wing colours, ranging from light to dark. Before the Industrial Revolution, peppered moths with light wings were more common because they had the best camouflage on white tree trunks. The Industrial Revolution caused a lot of air pollution that started to cover tree trunks, making them black. Light-winged moths were no longer camouflaged and were easy prey for birds. But dark-winged individuals were now hidden! This meant that dark moths had an advantage and were more likely to live long enough to have babies. The babies of dark moths were also dark because of the alleles they inherited from their parents, so they were also more likely to survive. The dark moths had higher fitness and became more common as a result [ 4 ].
What Happens When Genetic Diversity is Low?
When few mutations are found in the DNA of a species, genetic diversity is said to be low [ 2 ]. Low genetic diversity means that there is a limited variety of alleles for genes within that species and so there are not many differences between individuals. This can mean that there are fewer opportunities to adapt to environmental changes. Low genetic diversity often occurs due to habitat loss. For example, when a species’ habitat is destroyed or broken up into small pieces, populations become small. Small, fragmented populations can lead to loss of genetic diversity because fewer individuals can survive in the remaining habitat so fewer individuals breed to pass on their alleles. In small populations, the choice of mates is also limited. Over time, individuals will all become related and will be forced to mate with relatives. This is inbreeding . Inbred animals often have two identical alleles for their genes because the same gene was passed on from both parents. If this allele has harmful mutations, an inbred baby can be unhealthy. This is called inbreeding depression [ 2 ].
If genetic diversity gets too low, species can go extinct and be lost forever. This is due to the combined effects of inbreeding depression and failure to adapt to change. In such cases, the introduction of new alleles can save a population. This is called genetic rescue [ 2 ]. In the 1990s conservation scientists had to use genetic rescue to save the Florida panther, which was threatened by extinction due to low genetic diversity ( Figure 3 ) [ 5 ]. Very few Florida panthers remained and their genetic diversity was extremely low. Many Florida panther babies were sick because of inbreeding depression. A closely related panther with high genetic diversity was present in Texas. Texan panthers were moved to Florida to have babies with the Florida panthers. This increased genetic diversity because of the mixing of alleles we spoke about before. Soon after the Texan panthers arrived, many healthy kittens were born [ 5 ].
- Figure 3 - (A) The Florida panther was once widespread, with high genetic diversity.
- (B) Hunting and habitat loss reduced population size and resulted in very low genetic diversity and inbreeding. (C) Eight female panthers from Texas were moved to Florida to breed with Florida panthers. (D) When the Texas and Florida panthers bred, new alleles were introduced into the population, helping the Florida panther population become bigger and healthier over time.
What’s Happening to Genetic Diversity Around the World?
We hear a lot about the loss of species in the world, but we are also seeing a loss of genetic diversity within species. The increasing number of people on Earth and our increasing use of natural resources has reduced space and resources for wild species. Over time, many wild animal and plant populations have become smaller or more isolated. Many species have also gone through local extinctions. This has led to a global loss of genetic diversity. Scientists think that the genetic diversity within species may have declined by as much as 6% globally since the Industrial Revolution [ 6 ]. This means that many species are less able to adapt when facing new challenges, like climate change, pollution, and new diseases. If too much genetic diversity is lost, more and more species could become unhealthy and in need of conservation actions similar to the Florida panther. However, there are steps we can take to conserve and restore genetic diversity across many species.
How Do We Stop Genetic Diversity Loss?
We must preserve and protect genetic diversity. This can be done through the conservation of our remaining wild populations [ 2 ]. We can use nature reserves and wildlife bridges to reconnect wild populations that have become separated by our cities and highways. We can also restore habitats, because this will allow wild populations to get bigger. Sometimes we can even remove harmful stressors and pests so that populations can naturally regrow. We can also reintroduce species that have been lost from habitats they used to live in. Taken together, these strategies can help stop genetic diversity loss. It is important to protect genetic diversity because it is the foundation for healthy species. Healthy species are necessary for human health and for the health of the whole planet!
Gene : ↑ A section of DNA that contains the instructions for a trait.
Genetic Diversity : ↑ The overall diversity in the DNA between the individuals of a species.
Mutation : ↑ A change in an organism’s DNA. This can be a change of a single letter or a much bigger change of hundreds of letters at once.
Adaptation : ↑ The process of a species changing in order to better survive in its environment.
Alleles : ↑ Different variations of a gene caused by mutations. Many species have two alleles for every gene, one copy from each parent.
Inbreeding : ↑ Breeding between closely related individuals. Inbreeding often happens when populations are small and there are few options for mating. Inbred individuals are usually less healthy.
Inbreeding Depression : ↑ Inbred individuals share ancestors and are more likely to have identical copies of genes. If these genes contain harmful mutations, they will be expressed and cause lower health of inbred individuals.
Genetic Rescue : ↑ A conservation strategy, new individuals are moved into a population to increase genetic diversity and improve population health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
[1] ↑ Meyer, R., and Purugganan, M. 2013. Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14:840–52. doi: 10.1038/nrg3605
[2] ↑ Frankham, R., Ballou, J. D., and Briscoe, D. A. 2002. Introduction to Conservation Genetics. Cambridge: Cambridge University Press. p. 617.
[3] ↑ Emamalipour M., Seidi K., Zununi V. S., Jahanban-Esfahlan A., Jaymand M., Majdi H., et al. 2020. Horizontal gene transfer: from evolutionary flexibility to disease progression. Front. Cell. Dev. Biol. 8:229. doi: 10.3389/fcell.2020.00229
[4] ↑ Cook, L. M., and Saccheri, I. J. 2013. The peppered moth and industrial melanism: evolution of a natural selection case study. Heredity 110:207–12. doi: 10.1038/hdy.2012.92
[5] ↑ Johnson, W. E., Onorato, D. P., Roelke, M. E., Land, E. D., Cunningham, M., Belden, R. C., et al. 2010. Genetic restoration of the Florida panther. Science . 329:1641–5. doi: 10.1126/science.1192891
[6] ↑ Leigh, D. M., Hendry, A. P., Vázquez-Domínguez, E., and Friesen, V. L. 2019. Estimated six per cent loss of genetic variation in wild populations since the industrial revolution. Evol. Appl. 12:1505–12. doi: 10.1111/eva.12810
Loading metrics
Open Access
What is mutation? A chapter in the series: How microbes “jeopardize” the modern synthesis
Affiliations Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, United States of America, Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, Texas, United States of America, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, Texas, United States of America, The Dan L Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, Texas, United States of America
* E-mail: [email protected]
- Devon M. Fitzgerald,
- Susan M. Rosenberg

Published: April 1, 2019
- https://doi.org/10.1371/journal.pgen.1007995
- Reader Comments
Mutations drive evolution and were assumed to occur by chance: constantly, gradually, roughly uniformly in genomes, and without regard to environmental inputs, but this view is being revised by discoveries of molecular mechanisms of mutation in bacteria, now translated across the tree of life. These mechanisms reveal a picture of highly regulated mutagenesis, up-regulated temporally by stress responses and activated when cells/organisms are maladapted to their environments—when stressed—potentially accelerating adaptation. Mutation is also nonrandom in genomic space, with multiple simultaneous mutations falling in local clusters, which may allow concerted evolution—the multiple changes needed to adapt protein functions and protein machines encoded by linked genes. Molecular mechanisms of stress-inducible mutation change ideas about evolution and suggest different ways to model and address cancer development, infectious disease, and evolution generally.
Citation: Fitzgerald DM, Rosenberg SM (2019) What is mutation? A chapter in the series: How microbes “jeopardize” the modern synthesis. PLoS Genet 15(4): e1007995. https://doi.org/10.1371/journal.pgen.1007995
Editor: W. Ford Doolittle, Dalhousie University, CANADA
Copyright: © 2019 Fitzgerald, Rosenberg. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding: This work was supported by the American Cancer Society Postdoctoral Fellowship 132206-PF-18-035-01-DMC (DMF) and NIH grant R35-GM122598. The funders had no role in the preparation of the article.
Competing interests: The authors have declared that no competing interests exist.
Introduction
Mutation is any change in the sequence of an organism’s genome or the process by which the changes occur. Mutations range from single-basepair alterations to megabasepair deletions, insertions, duplications, and inversions. Though seemingly simple, ideas about mutation became entangled with the initially simplifying assumptions of both Darwin himself and the “Modern Synthesis”—the geneticists who embraced Darwin in the pre-DNA early 20th century, beginning evolutionary biology. The assumptions of purely “chance” mutations that occur constantly, gradually, and uniformly in genomes have underpinned biology for almost a century but began as a “wait-and-see”–based acknowledgment by early evolutionary biologists that they did not know the chemical nature of genes or how mutations in genes might occur.
Darwin considered generation of variation by chance to be a simplifying assumption, given that the origins of variation (and genes!) were unknown in his time, but he appears to have thought chance variation to be unlikely: “I have hitherto sometimes spoken as if the variations—so common and multiform in organic beings under domestication, and in a lesser degree in those in a state of nature—had been due to chance. This, of course, is a wholly incorrect expression, but it serves to acknowledge plainly our ignorance of the cause of particular variation [Chapter 5, 1].”
He also described multiple instances in which the degree and types of observable variation change in response to environmental exposures, thus seeming open to the possibility that the generation of variation might be environmentally responsive [ 1 ]. However, even once mutations were described on a molecular level, many continued to treat spontaneous mutations as necessarily chance occurrences—typically as mistakes occurring during DNA replication or repair. Darwinian evolution, however, requires only two things: heritable variation (usually genetic changes) and selection imposed by the environment. Any of many possible modes of mutation—purely “chance” or highly biased, regulated mechanisms—are compatible with evolution by variation and selection.
Here, we review some of the wealth of evidence, much of which originated in microbes, that reframes mutagenesis as dynamic and highly regulated processes. Mutation is regulated temporally by stress responses, occurring when organisms are poorly adapted to their environments, and occurs nonrandomly in genomes. Both biases may accelerate adaptation.
Bacteria teach biologists about evolution
Microbes were initially held as proof of the independence of mutational processes and selective environments. The Luria–Delbruck experiment (1943) demonstrated that bacterial mutations to phage resistance can occur prior to phage exposure [ 2 ], and the Lederbergs showed similar results for resistance to many antibiotics [ 3 ]. However, discovery of the SOS DNA-damage response and its accompanying mutagenesis [ 4 – 7 ] in the post-DNA world of molecular genetics began to erode the random-mutation zeitgeist. Harrison Echols thought that the SOS response conferred “inducible evolution” [ 8 ], echoing Barbara McClintock’s similar SOS-inspired suggestion of adaptation by regulated bursts of genome instability [ 9 ]. But SOS mutagenesis might be an unavoidable byproduct of DNA repair, and high-fidelity repair might be difficult to evolve, many argued. John Cairns’ later proposal of “directed” or “adaptive” mutagenesis in starvation-stressed Escherichia coli [ 10 , 11 ] reframed the supposed randomness of mutation as an exciting problem not yet solved. The mutagenesis they studied under the nonlethal environment of starvation is now known to reflect stress-induced mutagenesis—mutation up-regulated by stress responses. Its molecular mechanism(s), reviewed here, demonstrate regulation of mutagenesis. Similar mechanisms are now described from bacteria to humans, suggesting that regulated mutagenesis may be the rule, not the exception (discussed here and reviewed more extensively, [ 12 ]).
Stress-induced mutagenic DNA break repair in E . coli
DNA double-strand breaks (DSBs) occur spontaneously in approximately 1% of proliferating E . coli [ 13 , 14 ]. In unstressed E . coli , DSB repair by homologous recombination (HR) is relatively high fidelity. However, activation of the general stress response, for example, by starvation, flips a switch, causing DSB repair to become mutagenic [ 15 , 16 ]. This process of mutagenic break repair (MBR) causes mutations preferentially when cells are poorly adapted to their environment—when stressed—and, as modeling indicates [ 17 – 20 ], may accelerate adaptation.
At least three stress responses cooperate to increase mutagenesis in starving E . coli . The membrane stress response contributes to DSB formation at some loci [ 21 ]; the SOS response up-regulates error-prone DNA polymerases used in one of two MBR mechanisms [ 22 – 24 ]; and the general stress response licenses the use of, or persistence of errors made by, those DNA polymerases in DSB repair [ 15 , 16 ]. The requirement for multiple stress responses indicates that cells check a few environmental conditions before flipping the switch to mutation [ 25 ]. E . coli MBR is a model of general principles in mutation from bacteria to human: the regulation of mutation in time, by stress responses, and its restriction in genomic space, limited to small genomic regions, in the case of MBR, near DNA breaks. We look at MBR, then other mutation mechanisms in microbes and multicellular organisms, which share these common features.
MBR mechanisms
Two distinct but related MBR mechanisms occur in starving E . coli , and both require activation of the general/starvation response. Moreover, both occur without the starvation stress if the general stress response is artificially up-regulated [ 15 , 16 ], indicating that the stress response itself without actual stress is sufficient. Homologous-recombinational (homology-directed) MBR (HR-MBR) generates base substitutions and small indels via DNA-polymerase errors during DSB-repair synthesis ( Fig 1A–1F ). Microhomologous MBR causes amplifications and other gross chromosomal rearrangements (GCRs) [ 26 – 28 ], most probably by microhomology-mediated break-induced replication (MMBIR) [ 28 , 29 ] ( Fig 1A–1C , 1G and 1H ). Both MBR pathways challenge traditional assumptions about the "chance" nature of mutations.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
(a–c) RecBCD nuclease loads RecA HR protein onto ssDNA, similarly to human BRCA2 loading RAD51; basepairing with a strand of identical duplex DNA (gray, e.g., a sister chromosome). Parallel lines, basepaired DNA strands. Repair synthesis (dashed lines) is switched to a mutagenic mode by the general stress response (sigma S). DNA polymerase errors (d, purple X) generate indels (e, purple XX) and base substitutions (f, purple XX). Microhomologous MBR requires DNA Pol I for template switching to regions containing microhomology (g), of as little as a few basepairs, and initiates replication, creating genome rearrangements; (h) a duplicated chromosome segment (blue arrows) is shown here. Circled numbers and shading indicate the three main events in HR-MBR: ① a DSB and its repair by HR, ② the SOS response (pink), and ③ the general stress response (blue). Note that HR-MBR (d–f, purple) requires both the SOS response (②, pink, which up-regulates error-prone DNA Pol IV, necessary for HR-MBR) and general stress response (③, blue), but microhomologous MBR (g–h, blue) requires the general stress response but not SOS (③, blue). Figure modified from [ 12 ]. HR, homologous recombination; MBR, mutagenic break repair; ssDNA, single-stranded DNA.
https://doi.org/10.1371/journal.pgen.1007995.g001
Both MBR mechanisms are initiated by a DSB and require HR DSB-repair proteins ( Fig 1 , ①) [ 15 , 28 , 30 – 33 ]. The first steps mirror standard HR DSB repair: RecBCD nuclease processes DSB ends and loads RecA HR protein ( Fig 1A and 1B ). Next, the RecA–DNA nucleoprotein filament can activate the SOS response ( Fig 1 , ② pink), which is required for HR-MBR but not microhomologous MBR. RecA also facilitates strand invasion—the initial contact between the broken DNA molecule and an identical sister chromosome from which repair is templated ( Fig 1C ). In unstressed cells, this intermediate leads to high-fidelity HR repair; however, if the general stress response is activated, repair proceeds via one of two mutagenic pathways ( Fig 1D–1H , ③). In HR-DSB repair, errors generated by error-prone SOS-up-regulated DNA polymerases IV (DinB), V (UmuDC), and II (PolB) accumulate in the tracts of repair synthesis during HR repair ( Fig 1D ) [ 22 , 23 , 34 ]. Activation of the general stress response licenses the use of these polymerases and/or prevents the removal of errors they generate: base substitutions and small indels ( Fig 1E, 1F ) [ 35 , 36 ] that are located mostly in clusters/hotspots of about 100 kb around the original DSB location [ 30 ]. Microhomologous MBR requires DNA Pol I, which is proposed to promote microhomology-dependent template switching during repair synthesis to generate GCRs ( Fig 1G and 1H ) [ 28 ]. Similar MMBIR mechanisms are proposed to underlie many DSB-driven GCRs in human genetic diseases and cancers [ 28 , 29 , 37 ].
Stress response regulation of E . coli MBR
Environmentally responsive and temporally regulated MBR mechanisms challenge long-held assumptions about the constant, gradual nature of mutagenesis and its blindness to an organism’s environmental suitability, or the lack of it, showing that mutagenesis is regulated tightly via environmental inputs. The general stress response controls the switch between high-fidelity or mutagenic DSB repair [ 15 , 16 ]. This stress response, controlled by the alternative sigma factor σ S , is activated by starvation, cold, acid, antibiotic, oxidative, and osmotic stresses, among others. During a general stress response, the σ S transcriptional activator increases the transcription of hundreds of genes (approximately 10% of all E . coli genes) that provide a range of protective functions (reviewed, [ 38 ]). We do not know exactly how the general stress response promotes mutagenesis. Two possibilities are as follows. First, the general stress response modestly up-regulates error-prone Pol IV above SOS-induced levels [ 39 ]. This might be the rate-limiting step. Also, the general stress response down-regulates mismatch repair (MMR) enzymes MutS and MutH [ 40 , 41 ]. The HR-MBR mutation spectrum is similar to that of unstressed MMR-deficient strains [ 35 , 36 , 42 ], suggesting that MMR becomes limiting transiently during HR-MBR [ 36 , 43 , 44 ]. Other σ S targets are also plausible, including down-regulation of the high-fidelity replicative DNA Pol III. Together, these observations suggest a model in which the general stress response enables error-prone polymerases to participate in DSB repair and/or allows the errors introduced by these polymerases to escape mismatch repair.
At least two other stress responses also contribute to one or both MBR mechanisms. The SOS DNA-damage response is required for HR-MBR [ 45 ] but not microhomologous MBR [ 22 ]. The SOS response is detected in about 25% of cells with a reparable DSB [ 13 ] and so comes automatically with the DSB that initiates MBR. (The 75% without SOS may repair fast enough to avoid SOS [ 13 ].) The SOS response halts cell division and activates DNA-damage tolerance and repair pathways. The primary role of the SOS response in HR-MBR is the upregulation of the error-prone DNA polymerases IV and V and possibly II. In some assays, production of Pol IV completely restores mutagenesis in SOS-defective cells [ 23 ]. In others, Pols II and V also contribute to mutagenesis [ 16 , 34 , 46 ]. Finally, the membrane stress response, regulated by σ E , promotes MBR at some loci by playing a role in spontaneous DSB formation through an unknown mechanism (see “Localization of MBR-dependent mutations”) [ 21 ]. The membrane stress response is triggered by an accumulation of unfolded envelope proteins caused by heat and other stressors [ 47 ] and therefore appears to couple these stressors to mutagenesis.
A genome-wide screen revealed a network of 93 genes required for starvation stress–induced MBR [ 25 ]. Strikingly, over half participate in sensing or signaling various types of stress and act upstream of activation of the key stress response regulators, which are hubs in the MBR network. During starvation stress, at least 31 genes function upstream of (in activation of) the general stress response. Most encode proteins used in electron transfer and other metabolic pathways, suggesting that these may be the primary sensors of starvation stress. Additionally, at least six genes are required for activation of the SOS response during MBR, and at least 33 MBR-network genes are required for activation of the membrane stress response. The 93 MBR genes form a highly connected network based on protein–protein interactions with the three stress response regulators (σ S , RecA/LexA, and σ E ) as nonredundant network hubs [ 25 ]. The MBR network highlights the importance of tight, combinatorial stress response regulation of mutagenesis in response to multiple inputs.
Generality of general stress response–promoted mutation
In E . coli , σ S -dependent mutagenesis has a mutational signature that is distinct from that seen in low-stress mutation accumulation (MA) studies and generation-dependent mutagenesis [ 34 , 35 , 42 , 48 ]. Importantly, the nucleotide diversity in genomes of extant E . coli and other bacteria is described better by the σ S -dependent signature than the signature seen in MA studies [ 48 ]. Specifically, both σ S -dependent mutations and those seen in extant species have much higher ratios of transitions to transversions than is seen in MA experiments or expected by chance. This suggests that a significant portion of adaptive mutations in bacteria arise from σ S -dependent stress-induced mutation mechanisms such as MBR [ 48 ]. Furthermore, mathematical modeling suggests that stress response–regulated mutagenesis, such as MBR, promotes adaptation in changing environments [ 17 – 20 ]. Organisms that encode regulated mutagenesis mechanisms may have an increased ability to evolve, which would promote the evolution and maintenance of such mechanisms by second-order selection [ 17 , 19 , 20 ].
Localization of MBR-dependent mutations
MBR generates mutations in hotspots close to the site of the instigating DSB, not at random locations in the genome [ 30 , 49 ]. Hotspotting near DSBs is best described for HR-MBR initiated by engineered DSBs at various sites in the bacterial chromosome [ 30 ]. Mutations are most frequent within the first kilobase (kb) pair on either side of the DSB, and then fall off to near background levels approximately 60 kb from the break, with a weak long-distance hot zone of around 1 MB from the DSB site. This pattern of mutations supports the model that most MBR-dependent mutations arise from DNA polymerase errors during HR repair synthesis, and the remainder arise during more processive error-prone break-induced replication. The observation that mutations occur near DSBs does not, in itself, suggest that mutations are more likely to occur in certain genomic regions or in locations related to an organism’s adaptive “need.” However, it does suggest that the distribution of mutations is likely to mirror the distribution of DSBs, and the following lines of evidence suggest that DSB distributions may be nonrandom and reflect potential utility of genes in particular environments.
The sources and distributions of spontaneous DSBs are poorly understood in all organisms (reviewed, [ 14 ]), but we have some clues about the origins of DSBs that lead to MBR. First, transcriptional RNA–DNA hybrids (R-loops) are one source of MBR-promoting DSBs [ 50 ]. R-loops have been implicated in DSB formation in many experimental systems, although the exact mechanism(s) of DNA breakage is unresolved (reviewed, [ 51 ]). Though the distribution of R-loops has not been thoroughly assessed in starving E . coli , R-loops tend to be biased toward highly transcribed genes, promoters, and noncoding-RNA genes [ 52 – 54 ] and might, therefore, target DSBs and mutations to those sites. Also, activation of the σ E membrane stress response is required for DSB formation in some assays and might target DSBs in genomic space [ 21 ]. The mechanism by which the σ E stress response causes DSBs is unknown, but one possibility is that σ E -activated transcription causes DSBs directly (rather than via gene products’ up- or down-regulation), via an R-loop–dependent or other transcription-dependent mechanism. R-loops and the σ E stress response might direct DSBs, and thus mutations, to regions of the genome with more adaptive potential for a given environment: transcribed genes and regulatory elements (promoters and regulatory small RNAs).
Additionally, MBR-dependent mutations can occur in clusters [ 55 ]. When a MBR-induced mutation occurs, the probability of finding another mutation at neighboring sites 10 kb away is approximately 10 3 times higher than if the first mutation did not occur [ 55 ], and this is not true for a distant unlinked site in the genome [ 43 ], indicating that nearby mutations are not independent events. That is, linked mutations appear to occur simultaneously, in single MBR events. Such clusters are predicted to promote concerted evolution by simultaneously introducing changes to multiple domains of a protein or subunits of a complex protein machine [ 15 , 20 , 55 ]. Because multiple mutations are often needed for new functions to emerge, and often, the intermediate mutated states are less fit and counter selected, how complex protein machines evolve has been a long-standing problem [ 56 ]. Similar clusters have been identified in many organisms [ 57 ] and in cancer genomes, in which mutation clusters are called kataegis , Greek for (mutation) storms [ 58 – 60 ]. The mechanisms of mutation localization and co-occurrence revealed by MBR in E . coli have guided more mechanistic understanding of how mutation clusters occur across the tree of life.
Analyses of E . coli mutation accumulation lines and natural isolates indicate that local mutation rates vary by about one order of magnitude on the scale of approximately 10–100 kb [ 61 , 62 ]. It is possible, even likely, that the DSB-dependent mutation localization and co-occurring mutation clusters characteristic of MBR are important contributors to this nonuniformity in mutation rate. Similar degrees of variation in local mutation rates have been reported for other bacteria [ 63 ], yeast [ 64 ], and mammals (mouse, human, and other primates [ 65 , 66 ]) and could also result from MBR-like mutation mechanisms. Further analysis of natural isolates, with a specific focus on identifying clusters of cosegregating single-nucleotide variants, could indicate how frequent MBR-dependent mutation clusters are and how they shape genomes.
The molecular mechanisms of MBR reveal many ways by which mutations do not occur uniformly or independently from one another in genomic space. More work is needed to assess fully whether the MBR mechanism or genomes themselves have evolved to bias mutations to locations where they are most likely to be beneficial, such as genes actively transcribed in response to the experienced stressor.
Other regulated mutagenesis mechanisms in microbes
In addition to starvation-induced MBR in E . coli , diverse bacteria and single-celled eukaryotes display examples of stress response–up-regulated mutagenesis. Some of these mutation mechanisms provide additional insight into how mutation rates vary across genomes in ways that may accelerate adaptive evolution. Many share characteristics with E . coli MBR but differ enough to suggest that regulated mutagenesis has evolved independently multiple times, thus highlighting the importance of regulated mutagenesis to evolution-driven problems, such as combatting infectious disease and antimicrobial resistance. Potential strategies to counteract pathogen evolution require understanding of how genetic variation is generated in these organisms. Continued study of regulated-mutagenesis mechanisms may reveal potential new drug targets to block mutagenesis and thus evolution [ 12 , 25 , 67 ].
Other mechanisms of starvation stress–induced mutagenesis in bacteria
Diverse wild E . coli isolates show increased mutation rates during extended incubation on solid medium compared with vegetative growth, known as mutagenesis in aging colonies (MAC) [ 68 ]. In the one isolate tested for genetic requirements, MAC required σ S , decreased MMR capacity and error-prone Pol II but not DSB-repair proteins or SOS activation [ 68 ]—like, but not identical to, MBR in E . coli . Bacillus subtilis undergoes starvation-induced mutagenesis that is up-regulated by the ComK starvation-stress response and requires the SOS-induced Pol IV homolog YqjH but does not require DSB repair [ 69 , 70 ]. In B . subtilis , starvation-induced mutation of reporter genes increases with increased levels of transcription of those genes, dependently on the transcription-coupled repair factor Mfd [ 71 ], similarly to E . coli MBR [ 50 ]. This suggests that transcription directs starvation-induced mutations to transcribed regions of the B . subtilis genome, where they are more likely to be adaptive. This is similar to the hypothesized targeting of E . coli MBR but occurs through a DSB-independent mechanism.
Antibiotic-induced mutagenesis in bacteria
Many antibiotics, especially at subinhibitory concentrations, increase mutation rate and generate de novo resistance and cross-resistance in a variety of bacteria, including important pathogens. The β-lactam antibiotic ampicillin induces mutagenesis in E . coli , Pseudomonas aeruginosa , and Vibrio cholera via a mechanism requiring σ S , Pol IV, and limiting mismatch repair [ 41 ]. Whether DSBs are involved remains untested. The topoisomerase-inhibiting antibiotic ciprofloxacin (cipro) induces cipro resistance rapidly in E . coli , requiring HR proteins, SOS induction, and error-prone Pols II, IV, and V [ 72 ]. A requirement for σ S has only very recently been demonstrated, along with the demonstration that cipro-induced mutagenesis is σ S -dependent MBR, similar to that induced by starvation[ 73 ]. In fact, diverse antibiotics both create DSBs [ 74 ] and activate the general stress response in E . coli [ 41 ], suggesting that these antibiotics may increase mutagenesis both by increasing DNA damage and triggering a switch to low-fidelity repair of that damage.
Stress response regulation of mobile DNA elements in bacteria
Environmental stress up-regulates the activity of mobile DNA elements in many organisms, and this inducible genome instability is likely to be an important driver of evolution (reviewed, [ 75 ]). Although the mechanisms of regulation are poorly understood, stress response regulators have been implicated in a few cases. The general stress response promotes excision of an E . coli transposable prophage [ 76 ] and a Pseudomonas transposon [ 77 ]. Starvation increases the retromobility of Lactobacillus lactis LtrB group II intron through signaling by the small molecule regulators guanine pentaphosphate (ppGpp) and cyclic adenosine monophosphate (cAMP) [ 78 ]. Mobility of an E . coli transposon is increased by metabolic disruptions and negatively regulated by the σ E membrane stress response [ 79 ]. Also, stress can directly regulate mobile element activity without an intervening stress response: movement of the T4 td intron becomes promiscuous during oxidative stress through ROS-induced oxidation of an amino acid in the intron-encoded homing endonuclease, which makes it a transposase [ 80 ].
Regulated mutagenesis in eukaryotic microbes
Many examples of stress-associated mutagenesis and MBR have been reported in yeast, but stress response regulation has been demonstrated in only two cases. First, in the budding yeast Saccharomyces cerevisiae , the proteotoxic drug canavanine induces mutagenesis dependently on the MSN environmental stress response [ 81 ]. MSN-dependent mutagenesis requires the nonhomologous end-joining (NHEJ) protein Ku and two error-prone polymerases, Rev1 and Pol zeta (ζ) [ 81 ]. NHEJ is a relatively genome-destabilizing DSB-repair pathway, so MSN-dependent mutagenesis represents a stress-induced switch to MBR. NHEJ proteins are required for starvation-induced mutations in yeast as well [ 82 ]. Others have reported yeast MBR dependent on the error-prone DNA polymerase Rev3 [ 83 ] and spontaneous mutations dependent on error-prone polymerases Rev1 and Pol ζ [ 84 ]. Yeast also form mutation clusters by MBR [ 85 ] and undergo MMBIR similar to E . coli microhomologous MBR [ 86 ]. It is unknown whether these observations represent one or more mechanisms of mutation and whether MSN or other stress responses regulate mutagenesis in these cases. In all cases of yeast MBR, mutations are likely to occur near DSBs and, therefore, may be localized within genomes, as discussed for E . coli MBR.
Second, a heat shock response, activated by heat shock or protein denaturation, induces aneuploidy in S . cerevisiae by titration of the chaperone heat shock protein 90 (HSP90) [ 87 ]. Inhibitors of HSP90, such as radicicol, also induce aneuploidy. HSP90 is required for proper folding of kinetochore proteins in unstressed cells, so HSP90 titration or inhibition probably triggers aneuploidy through the disruption of kinetochore assembly [ 87 ]. The resulting yeast cell populations show high karyotypic and phenotypic variation and harbor cells resistant to radicicol and other drugs [ 87 ]. Aneuploidy in the form of extra chromosome copies may also facilitate adaptive evolution by providing a larger mutational target. Extra chromosomes may also buffer otherwise deleterious mutations through the sharing of gene products. Similar heat- and other stress-induced aneuploidy has been reported in Candida albicans and other yeast species, and can cause resistance to a variety of compounds, including clinically relevant antifungal drugs (reviewed, [ 88 ]). Some of these examples are likely to result from HSP90 titration, but other stress responses may be involved also.
Regulated mutagenesis in multicellular organisms
Although microbes led the way in revealing mechanisms of stress response–up-regulated mutagenesis, many microbial mutation mechanisms are mirrored throughout the tree of life, including in multicellular organisms. Stress response–up-regulated mutation mechanisms have been discovered in plants, flies, and human cells (reviewed, [ 12 ]). The potential adaptive roles of these mutation mechanisms are less clear in multicellular organisms than in microbes. Do these mechanisms contribute to germline variation (and thus organismal evolution), mosaicism and somatic cell evolution, or both? Or are they simply biproducts of other required cellular functions or stress-induced dysfunctions?
In the Drosophila germline, the HSP90 heat shock response increases transposon-mediated mutagenesis and can drive organismal adaptation [ 89 ]. Most other regulated mutation mechanisms characterized to date have been in somatic cells, in which they might contribute to mosaicism. Somatic diversity may be important during development and contribute to organismal fitness, as is the case with antibody diversification during B-cell maturation. For example, neural development might require genetic complexity and plasticity as organisms get differently “wired” during development, based on their experiences. However, up-regulated mutagenesis is also likely to drive pathogenic somatic evolution, such as during cancer development. For example, hypoxic stress responses trigger down-regulation of mismatch repair and down-regulate HR DSB-repair proteins RAD51 and BRCA1, leaving only chromosome-rearranging nonhomologous or microhomologous DSB-repair mechanisms (reviewed, [ 90 ]). Hypoxic stress response–induced mutagenesis occurs in mouse and human, suggesting an adaptive function in addition to its probable relevance to tumor biology. Tumors become hypoxic and induce hypoxic stress responses, which promote angiogenesis. Hypoxic stress responses may also promote tumor evolution via mutagenesis. The tumor growth factor β (TGF-β) signaling pathway also induces genome rearrangement by reduction of HR DSB repair in human cancer cell lines, leading to increased copy number alterations and resistance to multiple chemotherapeutic drugs [ 91 , 92 ]. Stress-induced and localized mutagenesis in multicellular organisms and the relevance of these mechanisms to cancer are reviewed in more detail elsewhere [ 12 ].
Evolution and applications of stress-induced mutation
Mutations provide the raw material for evolution but can also decrease the fitness of an organism. Therefore, mutation rates have, presumably, been finely tuned, apparently through second-order selection. Constitutively high mutation rates are advantageous in rapidly changing environments but decrease fitness in more stable (or periodically changing) environments. By biasing mutation to times of stress and to particular genomic regions, perhaps such regions relevant to a specific stress, stress-induced mutagenesis mechanisms provide the benefits of high mutation rate, while mitigating the risks. The ubiquity of these mechanisms throughout the tree of life supports their crucial role in evolution.
Stress-induced mutation mechanisms, first discovered in bacteria, challenge historical assumptions about the constancy and uniformity of mutation but do not violate strict interpretations of the Modern Synthesis. Mutation is still viewed as probabilistic, not deterministic, but we argue that regulated mutagenesis mechanisms greatly increase the probability that the useful mutations will occur at the right time, thus increasing an organism’s ability to evolve and, possibly, in the right places. Assumptions about the constant, gradual, clock-like, and environmentally blind nature of mutation are ready for retirement.
Stress-induced mutation mechanisms are likely to play important roles in human disease by promoting pathogen and tumor evolution and may drive evolution more generally. Mutation mechanisms may also be attractive drug targets for combatting infectious disease, cancer, and drug-resistance evolution in both [ 73 ]. Although many mechanisms of stress-inducible mutation have been identified in the past two decades [ 12 ], these are likely to be the tip of the iceberg. Some current pressing questions are highlighted below.
Open questions in mutation research
- What fraction of total “spontaneous” mutagenesis results from mutagenesis up-regulated by stress responses? Do stress response–regulated mutation programs drive much of adaptive evolution in microbes? Multicellular organisms?
- Are DSBs and the mutations they cause randomly distributed in genomic space? Or is DSB formation regulated, biased, or directed? By what mechanisms? Is this targeting adaptive?
- Can stress response–regulated mutation mechanisms be targeted by anti-evolvability drugs that limit the generation of heritable diversity? Can these drugs prevent pathogens and cancers from out-evolving host responses and drugs?
Acknowledgments
We thank P.J. Hastings for comments on the manuscript and our colleagues in this bundle for extreme patience.
- 1. Darwin CR. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray; 1859.
- View Article
- Google Scholar
- PubMed/NCBI
- 6. Radman M. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli : SOS hypothesis. In: Prokash L, Sherman F, Miller M, Lawrence C, Tabor H, editors. Molecular and Environmental Aspects of Mutagenesis. Springfield, Illinois: Charles C. Thomas; 1974. p. 128–42.
- 7. Radman M. SOS Repair Hypothesis: Phenomenology of an Inducible DNA Repair Which Is Accomplanied by Mutagenesis. In: Hanawalt P, editor. Molecular Mechanisms for Repair of DNA. New York: Plenum Press; 1975.
- 73. Pribis JP, García-Villada L, Zhai Y, Lewin-Epstein O, Wang A, Liu J, et al. Gamblers: an antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen-induced general stress response. Molecular cell. 2019;74 (in press). https://doi.org/10.1016/j.molcel.2019.02.037
An official website of the United States government
The .gov means it's official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you're on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- Browse Titles
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Institute of Medicine (US) Committee on Assessing Interactions Among Social, Behavioral, and Genetic Factors in Health; Hernandez LM, Blazer DG, editors. Genes, Behavior, and the Social Environment: Moving Beyond the Nature/Nurture Debate. Washington (DC): National Academies Press (US); 2006.

Genes, Behavior, and the Social Environment: Moving Beyond the Nature/Nurture Debate.
- Hardcopy Version at National Academies Press
3 Genetics and Health
Although there are many possible causes of human disease, family history is often one of the strongest risk factors for common disease complexes such as cancer, cardiovascular disease (CVD), diabetes, autoimmune disorders, and psychiatric illnesses. A person inherits a complete set of genes from each parent, as well as a vast array of cultural and socioeconomic experiences from his/her family. Family history is thought to be a good predictor of an individual’s disease risk because family members most closely represent the unique genomic and environmental interactions that an individual experiences ( Kardia et al., 2003 ). Inherited genetic variation within families clearly contributes both directly and indirectly to the pathogenesis of disease. This chapter focuses on what is known or theorized about the direct link between genes and health and what still must be explored in order to understand the environmental interactions and relative roles among genes that contribute to health and illness.
- GENETIC SUSCEPTIBILITY
For more than 100 years, human geneticists have been studying how variations in genes contribute to variations in disease risk. These studies have taken two approaches. The first approach focuses on identifying the individual genes with variations that give rise to simple Mendelian patterns of disease inheritance (e.g., autosomal dominant, autosomal recessive, and X-linked) (see Table 3-1 ; Mendelian Inheritance in Man). The second approach seeks to understand the genetic susceptibility to disease as the con sequence of the joint effects of many genes. Each of these approaches will be discussed below.
Online Mendelian Inheritance in Man (OMIM) Statistics (as of May 15, 2006), Number of Entries.
In general, diseases with simple Mendelian patterns of inheritance tend to be relatively uncommon or frequently rare, with early ages of onset, such as phenylketonuria, sickle cell anemia, Tay-Sachs disease, and cystic fibrosis. In addition, some of these genes have been associated with extreme forms of common diseases, such as familial hypercholesterolemia, which is caused by mutations in the low-density lipoprotein (LDL) receptor that predispose individuals to early onset of heart disease ( Brown and Goldstein, 1981 ).
Another example of Mendelian inheritance is familial forms of breast cancer associated with mutations in the BRCA1 and BRCA2 genes that predispose women to early onset breast cancer and often ovarian cancer. The genes identified have mutations that often are highly penetrant—that is, the probability of developing the disease in someone carrying the disease susceptibility genotype is relatively high (greater than 50 percent). These genetic diseases often exhibit a genetic phenomenon known as allelic heterogeneity, in which multiple mutations within the same gene (i.e., alleles) are found to be associated with the same disease. This allelic heterogeneity often is population specific and can represent the unique demographic and mutational history of the population.
In some cases, genetic diseases also are associated with locus heterogeneity , meaning that a deleterious mutation in any one of several genes can give rise to an increased risk of the disease. This is a finding common to many human diseases including Alzheimer’s disease and polycystic kidney disease. Both allelic heterogeneity and locus heterogeneity are sources of variation in these disease phenotypes since they can have varying effects on the disease initiation, progression, and clinical severity.
Environmental factors also vary across individuals and the combined effect of environmental and genetic heterogeneity is etiologic heterogeneity. Etiologic heterogeneity refers to a phenomenon that occurs in the general population when multiple groups of disease cases, such as breast cancer clusters, exhibit similar clinical features, but are in fact the result of differing events or exposures. Insight into the etiology of specific diseases as well as identification of possible causative agents is facilitated by discovery and examination of disease cases demonstrating etiologic heterogeneity. The results of these studies may also highlight possible gene-gene interactions and gene-environment interactions important in the disease process. Identifying etiologic heterogeneity can be an important step toward analysis of diseases using molecular epidemiology techniques and may eventually lead to improved disease prevention strategies ( Rebbeck et al., 1997 ).
As opposed to the Mendelian approach, the second approach to studying how variations in genes contribute to variations in disease risk focuses on understanding the genetic susceptibility to diseases as the consequence of the joint effects of many genes, each with small to moderate effects (i.e., polygenic models of disease) and often interacting among themselves and with the environment to give rise to the distribution of disease risk seen in a population (i.e., multifactorial models of disease). This approach has been used primarily for understanding the genetics of birth defects and common diseases and their risk factors. As described below, several steps are involved in developing such an understanding.
As a first step, study participants are asked to provide a detailed family history to assess the presence of familial aggregation. If individuals with the disease in question have more relatives affected by the disease than individuals without the disease, familial aggregation is identified. While familial aggregation may be accounted for through genetic etiology, it may also represent an exposure (e.g., pesticides, contaminated drinking water, or diet) common to all family members due to the likelihood of shared environment.
When there is evidence of familial aggregation, the second step is to focus research studies on estimating the heritability of the disease and/or its risk factors. Heritability is defined as the proportion of variation in disease risk in a population that is attributable to unmeasured genetic variations inferred through familial patterns of disease. It is a broad population-based measure of genetic influence that is used to determine whether further genetic studies are warranted, since it allows investigators to test the overarching null hypothesis that no genes are involved in determining disease risk. Twin studies and family studies are frequently used in the study of heritability.
Twin studies comparing the disease and risk factor variability of monozygotic and dizygotic twins have been a common study design used to easily estimate both genetic and cultural inheritance. Studies of monozygotic twins reared together versus those reared apart also have been important in estimating both genetic and environmental contributions to patterns of inheritance. The modeling of the sources of phenotypic variation using family studies has become quite sophisticated, allowing the inclusion of model parameters to represent the additive genetic component (i.e., polygenes), the nonadditive genetic component (i.e., genetic dominance, as well as gene-environment and gene-gene interactions), shared family environment, and individual environments. The contributions of these factors have been shown to vary by age and population.
When significant evidence of genetic involvement is established, the next step is to identify the responsible genes and the mutations that are associated with increased or decreased risk, using either genetic linkage analysis or genetic association studies. For example, in the study of birth defects, this often involves the search for chromosomal deletions, insertions, duplications, or translocations.
- GENETIC LINKAGE ANALYSIS AND GENETIC ASSOCIATION STUDIES
The human genome is made up of tens of thousands of genes. With approximately 30,000 genes to choose from, assigning a specific gene or group of genes to a corresponding human disease demands a methodical approach consisting of many steps. Traditionally, the process of gene discovery begins with a linkage analysis that assesses disease within families. Linkage analyses are typically followed by genetic association studies that assess disease across families or across unrelated individuals.
Genetic Linkage Analysis
The term linkage refers to the tendency of genes proximally located on the same chromosome to be inherited together. Linkage analysis is one step in the search for a disease susceptibility gene. The goal of this analysis is to approximate the location of the disease gene in relation to a known genetic marker, applying an understanding of the patterns of linkage. Traditional linkage analysis that traces patterns of heredity of both the disease phenotype and genetic markers in large, high-risk families have been used to locate disease-causing gene mutations such as the breast cancer gene (BRCA1) on chromosome 17 ( Hall et al., 1990 ).
Because the mode of inheritance is often not clear for common diseases, an alternative approach to classic linkage analysis was developed to capitalize on the basic genetic principle that siblings share half of their alleles on average. By investigating the degree of allelic sharing across their genomes, pairs of affected siblings (i.e., two or more siblings with the same disease) can be used to identify chromosomal regions that may contain genes whose variations are related to the disease being studied. If numerous sibling pairs affected by the disease of interest exhibit a greater than expected sharing of the known alleles of the polymorphic genetic marker being used, then the genetic marker is likely to be linked (that is, within close proximity along the chromosome) to the susceptibility gene responsible for the disease being studied. To find chromosomal regions that show evidence for linkage using this affected sibling pair method typically requires typing numerous affected sibships with hundreds of highly polymorphic markers uniformly positioned along the human genome ( Mathew, 2001 ).
This approach has been widely used to identify regions of the genome thought to contribute to common chronic diseases. However, results of linkage analyses have not been consistently replicated. The inability to successfully replicate linkage findings may be a result of insufficient statistical power (that is, including an inadequate number of sibling pairs with the disease of interest) or results that included false positives in the original study. An alternate explanation could be that different populations are affected by different susceptibility genes than those that were studied originally ( Mathew, 2001 ). Without consistent replication of results it is premature to draw conclusions about the contribution of a gene locus to a specific disease.
Upon the confirmation of a linkage, researchers can begin to search the region for the candidate susceptibility gene. The search for a single susceptibility gene for common diseases often involves examination of very large linkage regions, containing 20 to 30 million base pairs and potentially hundreds of genes ( Mathew, 2001 ). It is also important to note, however, that while linkage mapping is a powerful tool for finding Mendelian disease genes, it often produces weak and sometimes inconsistent signals in studies of complex diseases that may be multifactorial. Linkage studies perform best when there is a single susceptibility allele at any given disease locus and generally performs poorly when there is substantial genetic heterogeneity.
Genetic Association Studies
Technological advances in high-throughput genotyping have allowed the direct examination of specific genetic differences among sizable numbers of people. Genetic association techniques are often the most efficient approach for assessing how specific genetic variation can affect disease risk. Genetic association studies, which have been used for decades, have perpetually progressed in terms of the development of new study designs (such as case-only and family-based association designs), new genotyping systems (such as array-based genotyping and multiplexing assays), and new methods used for addressing biases such as population ( Haines and Pericak-Vance, 1998 ).
Analysis of the effects of genetic variation typically involves first the discovery of single nucleotide polymorphisms (SNPs) 1 and then the analysis of these variations in samples from populations. SNPs occur on average approximately every 500 to 2,000 bases in the human genome. The most common approach to SNP discovery is to sequence the gene of interest in a representative sample of individuals. Currently, sequencing of entire genes on small numbers of individuals (~25 to 50) can detect polymorphisms occurring in 1 to 3 percent of the population with approximately 95 percent confidence. The Human DNA Polymorphism Discovery Program of the National Institute of Environmental Health Sciences’ Environmental Genome Project is one example of the application of automated DNA sequencing technologies to identify SNPs in human genes that may be associated with disease susceptibility and response to environment ( Livingston et al., 2004 ). The National Heart, Lung, and Blood Institute’s Programs in Genomic Applications also has led to important increases in our knowledge about the distribution of SNPs in key genes thought to be already biologically implicated in disease risk (i.e., biological candidate genes 2 ).
Impressive and rapid advances in SNP analysis technology are rapidly redefining the scope of SNP discovery, mapping, and genotyping. New array-based genotyping technology enables “whole genome association” analyses of SNPs between individuals or between strains of laboratory animal species ( Syvanen, 2005 ). Arrays used for these analyses can represent hundreds of thousands of SNPs mapped across a genome ( Klein et al., 2005 ; Hinds et al., 2005 ; Gunderson et al., 2005 ). This approach allows rapid identification of SNPs associated with disease and susceptibility to environmental factors. The strength of this technology is the massive amount of easily measurable genetic variation it puts in the hands of researchers in a cost-effective manner ($500 to $1,000 per chip). The criteria for the selection of SNPs to be included on these arrays are a critical consideration, since they affect the inferences that can be drawn from using these platforms. Of course, the ultimate tool for SNP discovery and genotyping is individual whole genome sequencing. Although not currently feasible, the rapid advancement of technology now being stimulated by the National Human Genome Research Institute’s “$1,000 genome” project likely will make this approach the optimal one for SNP discovery and genotyping in the future.
With the ability to examine large quantities of genetic variations, researchers are moving from investigations of single genes, one at a time, to consideration of entire pathways or physiological systems that include information from genomic, transcriptomic, proteomic, and metabonomic levels that are all subject to different environmental factors ( Haines and Pericak-Vance, 1998 ). However, these genome- and pathway-driven study designs and analytic techniques are still in the early stages of development and will require the joint efforts of multiple disciplines, ranging from molecular biologists to clinicians to social scientists to bioinformaticians, in order to make the most effective use of these vast amounts of data.
- GENE-ENVIRONMENT AND GENE-GENE INTERACTIONS
The study of gene-environment and gene-gene interactions represents a broad class of genetic association studies focused on understanding how human genetic variability is associated with differential responses to environmental exposures and with differential effects depending on variations in other genes. To illustrate the concept of gene-environment interactions, recent studies that identify genetic mutations that appear to be associated with differential response to cigarette smoke and its association with lung cancer are reviewed below. Tobacco smoke contains a broad array of chemical carcinogens that may cause DNA damage. There are several DNA repair pathways that operate to repair this damage, and the genes within this pathway are prime biological candidates for understanding why some smokers develop lung cancers but others do not. In a study by Zhou et al. (2003) , variations in two genes responsible for DNA repair were examined for their potential interaction with the level of cigarette smoking and concomitant association with lung cancer. Briefly, one putatively functional mutation in the XRCC1 (X-ray cross-complementing group 1) gene and two putatively functional mutations in the ERCC2 (excision repair cross-complementing group 2) gene were genotyped in 1,091 lung cancer cases and 1,240 controls. When the cases and controls were stratified into heavy smokers versus nonsmokers, Zhou et al. (2003) found that nonsmokers with the mutant XRCCI genotype had a 2.4 times greater risk of lung cancer than nonsmokers with the normal genotype. In contrast, heavy smokers with the mutant XRCCI genotype had a 50 percent reduction in lung cancer risk compared to their counterparts with the more frequent normal genotype. When the three mutations from these two genes were examined together in the extreme genotype combination (individual with five or six mutations present in his/her genotype) there was a 5.2 time greater risk of lung cancer in nonsmokers and a 70 percent reduction of risk in the heavy smokers compared to individuals with no mutations. The protective effect of these genetic variations in heavy smokers may be caused by the differential increase in the activity of these protective genes stimulated by heavy smoking. Similar types of gene-smoking interactions also have been found for other genes in this pathway, such as ERCC1. These studies illustrate the importance of identifying the genetic variations that are associated with the differential risk of disease related to human behaviors. Note that this type of research also raises many different kinds of ethical and social issues, since it identifies susceptible subgroups and protected subgroups of subjects by both genetic and human behavior strata (see Chapter 10 ).
The study by Zhou et al. (2003) also demonstrates the increased information provided by jointly examining the effects of multiple mutations on toxicity-related disease. Other studies of mutations in genes involved in the Phase II metabolism (GSTM1, GSTT1, GSTP1) also have demonstrated the importance of investigating the joint effects of mutations ( Miller et al., 2002 ) on cancer risk. Although these two studies focused on the additive effects of multiple genes, gene-gene interactions are another important component to develop a better understanding of human susceptibility to disease and to interactions with the environment.
To adequately understand the continuum of genomic susceptibility to environmental agents that influences the public’s health, more studies of the joint effects of multiple mutations need to be conducted. Advances in bioinformatics can play a key role in this endeavor. For example, methods to screen SNP databases for mutations in transcriptional regulatory regions can be used for both discovery and functional validation of polymorphic regulatory elements, such as the antioxidant regulatory element found in the promoter regions of many genes encoding antioxidative and Phase II detoxification enzymes ( Wang et al., 2005 ). Comparative sequence analysis methods also are becoming increasingly valuable to human genetic studies, because they provide a means to rank order SNPs in terms of their potential deleterious effects on protein function or gene regulation ( Wang et al., 2004 ). Methods of performing large-scale analysis of nonsynonymous SNPs to predict whether a particular mutation impairs protein function ( Clifford et al., 2004 ) can help in SNP selection for genetic epidemiological studies and can be used to streamline functional analysis of mutations that are found to be statistically associated with differential response to environmental factors such as diet, stress, and socioeconomic factors.
- MECHANISMS OF GENE EXPRESSION
Identifying genes whose variations are associated with disease is just the first step in linking genetics and health. Understanding the mechanisms by which the gene is expressed and how it is influenced by other genes, proteins, and the environment is becoming increasingly important to the development of preventive, diagnostic, and therapeutic strategies.
When genes are expressed, the chromosomal DNA must be transcribed into RNA and the RNA is then processed and transported to be translated into protein. Regulating the expression of genes is a vital process in the cell and involves the organization of the chromosomal DNA into an appropriate higher-order chromatin structure. It also involves the action of a host of specific protein factors (to either encourage or suppress gene expression), which can act at different steps in the gene expression pathway.
In all organisms, networks of biochemical reactions and feedback signals organize developmental pathways, cellular metabolism, and progression through the cell cycle. Overall coordination of the cell cycle and cellular metabolism results from feed-forward and feedback controls arising from sets of dependent pathways in which the initiation of events is dependent on earlier events. Within these networks, gene expression is controlled by molecular signals that regulate when, where, and how often a given gene is transcribed. These signals often are stimulated by environmental influences or by signals from other cells that affect the gene expression of many genes through a single regulatory pathway. Since a regulatory gene can act in combination with other signals to control many other genes, complex branching networks of interactions are possible ( McAdams and Arkin, 1997 ).
Gene regulation is critical because by switching genes on or off when needed, cells can be responsive to changes in environment (e.g., changes in diet or activity) and can prevent resources from being wasted. Variation in the DNA sequences associated with the regulation of a gene’s expression are therefore likely candidates for understanding gene-environment interactions at the molecular level, since these variations will affect whether an environmental signal transduced to the nucleus will successfully bind to the promoter sequence in the gene and stimulate or repress gene expression. Combining genomic technologies for SNP genotyping with high-density gene expression arrays in human studies has only recently elucidated the extent to which this type of molecular gene-environment interaction may be occurring.
Cells also regulate gene expression by post-transcriptional modification; by allowing only a subset of the mRNAs to go on to translation; or by restricting translation of specific mRNAs to only when and where the product is needed. The genetic factors that influence post-transcriptional control are much more difficult to study because they often involve multiprotein complexes not easily retrieved or assayed from cells. At other levels, cells regulate gene expression through epigenetic mechanisms, including DNA folding, histone acetylation, and methylation (i.e., chemical modification) of the nucleotide bases. These mechanisms are likely to be influenced by genetic variations in the target genes as well as variations manifested in translated cellular regulatory proteins. Gene regulation occurs throughout life at all levels of organismal development and aging.
A classic example of developmental control of gene expression is the differential expression of embryonic, fetal, and adult hemoglobin genes (see Box 3-1 ). The regulation of the epsilon, delta, gamma, alpha, and beta genes occurs through DNA methylation that is tightly controlled through developmental signals. During development a large number of genes are turned on and off through epigenetic regulation. One of the fastest growing fields in genetics is the study of the developmental consequences of environmental exposures on gene expression patterns and the impact of genetic variations on these developmental trajectories.
Gene Expression and Globin. The production of hemoglobin is regulated by a number of transcriptional controls, such as switching, that dictate the expression of a different set of globin genes in different parts of the body throughout the various stages (more...)
An Example of a Single-Gene Disorder with Significant Clinical Variability: Sickle Cell Disease 3
Sickle cell disease refers to an autosomal recessive blood disorder caused by a variant of the β-globin gene called sickle hemoglobin (Hb S). A single nucleotide substitution (T→A) in the sixth codon of the β-globin gene results in the substitution of valine for glutamic acid (GTG→GAG), which can cause Hb S to polymerize (form long chains) when deoxygenated ( Stuart and Nagel, 2004 ). An individual inheriting two copies of Hb S (Hb SS) is considered to have sickle cell anemia , while an individual inheriting one copy of Hb S plus another deleterious β-globin variant (e.g., Hb C or Hb β-thalassemia) is considered to have sickle cell disease . An individual is considered to be a carrier of the sickle cell trait if he/she has one copy of the normal β-globin gene and one copy of the sickle variant (Hb AS) ( Ashley-Koch et al., 2000 ).
Four major β-globin gene haplotypes have been identified. Three are named for the regions in Africa where the mutations first appeared: BEN (Benin), SEN (Senegal), and CAR (Central African Republic). The fourth haplotype, Arabic-India, occurs in India and the Arabic peninsula ( Quinn and Miller, 2004 ).
Disease severity is associated with several genetic factors ( Ashley-Koch et al., 2000 ). The highest degree of severity is associated with Hb SS, followed by Hb s/β0-thalassemia, and Hb SC. Hb S/β + -thalassemia is associated with a more benign course of the disease ( Ashley-Koch et al., 2000 ). Disease severity also is related to β-globin haplotypes, probably due to variations in hemoglobin level and fetal hemoglobin concentrations. The Senegal haplotype is the most benign form, followed by the Benin, and the Central African Republic haplotype is the most severe form ( Ashley-Koch et al., 2000 ).
Thus, although sickle cell disease is a monogenetic disorder, its phenotypic expression is multigenic (see Appendix D ). There are two cardinal pathophysiologic features of sickle cell disease—chronic hemolytic anemia and vasoocclusion. Two primary consequences of hypoxia secondary to vasoocclusive crisis are pain and damage to organ systems. The organs at greatest risk are those in which blood flow is slow, such as the spleen and bone marrow, or those that have a limited terminal arterial blood supply, including the eye, the head of the femur and the humerus, and the lung as the recipient of deoxygenated sickle cells that escape the spleen or bone marrow. Major clinical manifestations of sickle cell disease include painful events, acute chest syndrome, splenic dysfunction, and cerebrovascular accidents.
Efforts to enhance clinical care are focusing on increasing our understanding of the pathophysiology of sickle cell disease in order to facilitate a precise prognosis and individualized treatment. Required is knowledge about which genes are associated with the hemolytic and vascular complications of sickle cell disease and how variants of these genes interact among themselves and with their environment ( Steinberg, 2005 ).
- ASPECTS OF HEALTH INFLUENCED BY GENETICS
Because every cell in the body, with rare exception, carries an entire genome full of variation as the template for the development of its protein machinery, it can be argued that genetic variation impacts all cellular, biochemical, physiological, and morphological aspects of a human being. How that genetic variation is associated with particular disease risk is the focus of much current research. For common diseases such as CVD, hypertension, cancer, diabetes, and many mental illnesses, there is a growing appreciation that different genes and different genetic variations can be involved in different aspects of their natural history. For example, there are likely to be genes whose variations are associated with a predisposition toward the initiation of disease and other genes or gene variations that are involved in the progression of a disease to a clinically defined endpoint. Furthermore, an entirely different set of genes may be involved in how an individual responds to pharmaceutical treatments for that disease. There also are likely to be genes whose variability controls how much or how little a person is likely to be responsive to the environmental risk factors that are associated with disease risk. Finally, there are thought to be genes that affect a person’s overall longevity that may counteract or interact with genes that may otherwise predispose that person to a particular disease outcome and thus may have an additional impact on survivorship.
In many ways, we are only at the beginning the process of developing a true understanding of how genomic variations give rise to disease susceptibility. Indeed many would argue that, without incorporating the equally important role of the environment, we will never fully understand the role of genetics in health. As progress is made through utilizing the new technologies for measuring biological variation in the genome, transcriptome, proteome, and metabonome, we are likely to have to make large shifts in our conceptual frameworks about the roles of genes in disease. Global patterns of genomic susceptibility are likely to emerge only when we consider the influence of the many interacting components working simultaneously that are dependent on contexts such as age, sex, diet, and physical activity that modify the relationship with risk. For the most part, we are still at the stage of documenting the complexity, finding examples and types of genetic susceptibility genes, understanding disease heterogeneity, and postulating ways to develop models of risk that use the totality of what we know about human biology, from our genomes to our ecologies to model risk.
Cardiovascular Disease (CVD)
The study of CVD can be used to illustrate the issues that are encountered in using genetic information in order to understand the etiology of the most common chronic diseases as well as in identifying those at highest risk of developing these diseases. The majority of CVD cases have a complex multifactorial etiology, and even full knowledge of an individual’s genetic makeup cannot predict with certainty the onset, progression, or severity of disease ( Sing et al., 2003 ). Disease develops as a consequence of interactions between a person’s genotype and exposures to environmental agents, which influence cardiovascular phenotypes beginning at conception and continuing throughout adulthood. CVD research has found many high-risk environmental agents and hundreds of genes, each with many variations that are thought to influence disease risk. As the number of interacting agents involved increases, a smaller number of cases of disease will be found to have the same etiology and be associated with a particular genotype ( Sing et al., 2003 ). The many feedback mechanisms and interactions of agents from the genome through intermediate biochemical and physiological subsystems with exposure to environmental agents contribute to the emergence of a given individual’s clinical phenotype. In attempting to sort out the relative contributions of genes and environment to CVD, a large array of factors must be considered, from the influence of genes on cholesterol (e.g., LDL levels) to psychosocial factors such as stress and anger. Although hundreds of genes have been implicated in the initiation, progression, and clinical manifestation of CVD, relatively little is known about how a person’s environment interacts with these genes to tip the balance between the atherogenic and anti-atherogenic processes that result in clinically manifested CVD. Please see Chapters 4 and 6 for further discussion of effects of social environment on CVD.
It is well known that many social and behavioral factors ranging from socioeconomic status, job stress, and depression, to smoking, exercise, and diet affect cardiovascular disease risk (see Chapters 2 , 3 , and 6 for more detailed discussion of these factors). As more studies of gene-environment interaction consider these factors as part of the “environment,” which are examined in conjunction with genetic variations, multiple intellectual and methodological challenges arise. First, how are the social factors embodied such that an interaction with a particular genotype can be associated with differential risk? Second, how can we handle complex interactions to address questions, such as how does an individual’s genotype influence his/her behavior? For example, one’s genetic susceptibility to nicotine addiction is actually a risk factor for CVD and its effect on CVD risk may be contingent on interactions with other genetic factors.
Pharmacogenetics
It has been well established that individuals often respond differently to the same drug therapy. The drug disposition process is a complex set of physiological reactions that begin immediately upon administration. The drug is absorbed and distributed to the targeted areas of the body where it interacts with cellular components, such as receptors and enzymes, that further metabolize the drug, and ultimately the drug is excreted from the body ( Weinshilboum, 2003 ). At any point during this process, genetic variation may alter the therapeutic response of an individual and cause an adverse drug reaction (ADR) ( Evans and McLeod, 2003 ). It has been estimated that 20 to 95 percent of variations in drug disposition, such as ADRs, can be attributed to genetic variation ( Kalow et al., 1998 ; Evans and McLeod, 2003 ).
Sensitivity to both dose-dependent and dose-independent ADRs can have roots in genetic variation. Polymorphisms in kinetic and dynamic factors, such as cytochrome P450 and specific drug targets can cause these individuals susceptibilities to ADRs. While the characteristics of the ADR dictate the true significance of these factors, in most cases, multiple genes are involved ( Pirmohamed and Park, 2001 ). Future analyses using genome-wide SNP profiling could provide a technique for assessing several genetic susceptibility factors for ADRs and ascertaining their joint effects. One of the challenges to the study of the relationship between genetic variation and ADRs is an inadequate number of patient samples. To remedy this problem, Pirmohamed and Park (2001) have proposed that prospective randomized controlled clinical trials become a part of standardized practice to ultimately prove the clinical utility of genotyping all patients as a measure to prevent ADRs.
Here we review some of the current work in pharmacogenetics as an example of what might be expected to arise from rigorous study of the interaction between social, behavioral, and genetic factors. Researchers have provided a few well-established examples of differences in individual drug response that have been ascribed to genetic variations in a variety of cellular drug disposition machinery, such as drug transporters or enzymes responsible for drug metabolism ( Evans and McLeod, 2003 ). For example:
- With the knowledge that the HER2 gene is overexpressed in approximately one fourth of breast cancer cases, researchers developed a humanized monoclonal antibody against the HER2 receptor in hopes of inhibiting the tumor growth associated with the receptor. Genotyping advanced breast cancer patients to identify those with tumors that overexpress the HER2 receptor has produced promising results in improving the clinical outcomes for these breast cancer patients ( Cobleigh et al., 1999 ).
- A therapeutic class of drugs called thiopurines is used as part of the treatment regimen for childhood acute lymphoblastic leukemia. One in 300 Caucasians has a genetic variation that results in low or nonexistent levels of thiopurine methyltransferase (TPMT), an enzyme that is responsible for the metabolism of the thiopurine drugs. If patients with this genetic variation are given thiopurines, the drug accumulates to toxic levels in their body causing life-threatening myelosuppression. Assessing the TPMT phenotype and genotype of the patient can be used to determine the individualized dosage of the drug ( Armstrong et al., 2004 ).
- The family of liver enzymes called cytochrome P450s plays a major role in the metabolism of as many as 40 different types of drugs. Genetic variants in these enzymes may diminish their ability to effectively break down certain drugs, thus creating the potential for overdose in patients with less active or inactive forms of the cytochrome P450 enzyme. Varying levels of reduced cytochrome P450 activity is also a concern for patients taking multiple drugs that may interact if they are not properly metabolized by well-functioning enzymes. Strategies to evaluate the activity level of cytochrome P450 enzymes have been devised and are valuable in planning and monitoring successful drug therapy. Some pharmaceutical drug trials are now incorporating early tests that evaluate the ability of differing forms of cytochrome P450 to metabolize the new drug compound ( Obach et al., 2006 ).
Some pharmacogenetics research has focused on the treatment of psychiatric disorders. With the introduction of a class of drugs known as selective serotonin re-uptake inhibitors (SSRIs), pharmacological treatment of many psychiatric disorders changed drastically. SSRIs offer significant improvements over the previous generation of treatments, including improved efficacy and tolerance for many patients. However, not all patients respond positively to SSRI treatment and many experience ADRs. New pharmacogenetic studies have indicated that these ADRs may be the result of genetic variations in serotonin transporter genes and cytochrome P450 genes. Further study and replication of these findings are necessary. If the characterization of the genetic variations is completed and is fully understood it would be possible to screen and monitor patients using genotyping techniques to create individualized drug therapies similar to those discussed above ( Mancama and Kerwin, 2003 ).
A significant challenge to the development of individualized drug therapies is the often polygenic or multifactorial inherited component of drug responses. Isolating the polygenic determinants of the drug responses is a sizable task. A good understanding of the drug’s mechanism of action and metabolic and disposition pathways should be the basis of all investigations. This knowledge can aid in directing genome-wide searches for gene variations associated with drug effects and subsequent candidate-gene approaches of investigation. Additionally, proteomic and gene-expression profiling studies are also important ways to substantiate and understand the pathways by which the gene of interest operates to affect the individual’s response to the drug ( Evans and McLeod, 2003 ). It is not enough to show an association; characterization of the underlying biological mechanisms is an essential component of moving genetic findings into the area of risk reduction. Another key component of utilizing genetics to improve prevention and reduce disease is an understanding of the distribution of the genetic variations in the populations being served.
- GENETICS OF POPULATONS AS RELATED TO HEALTH AND DISEASE
Human populations differ in their distribution of genetic variations. This is a consequence of their historical patterns of mutation, migration, reproduction, mating, selection, and genetic drift. Inherited mutations typically occur during gametogenesis within a single individual and then can be passed on to offspring for many generations. Whether that mutation goes on to become a prevalent polymorphism (i.e., a mutation with a population frequency of greater than 1 percent) is determined by both evolutionary forces and chance events. For example, it depends on whether the original child who inherited the mutation survives to adulthood and reproduces and whether that child’s children survive to reproduce, and so on. The number of children in a family also influences the prevalence of the mutation, and this is often tied to environmental factors that impact fertility and mating patterns that influence the speed with which a private mutation becomes a public polymorphism. There are well-known examples of what are called founder mutations in which this trajectory can be documented. For example, one particular district in what is Quebec (Canada) today was originally founded by only a few families from a particular French province. One of the founding fathers carried a 10kb deletion in his LDL receptor (LDL-R) gene that was passed down through the generations quickly and today is carried by 1 in 154 French Canadians in northeastern Quebec. This mutation is associated with familial hypercholesterolemia, and French Ca nadians have one of the highest prevalences of this disease in the world because of the small founding populations followed by population expansion ( Moorjani et al., 1989 ).
There are also a number of examples where mutations that arise in an individual become more prevalent because of the selective advantage they impart on their carriers. The best known example is the mutation associated with sickle cell anemia. The geographical pattern of this mutation strongly mirrors the geographical pattern of malarial infection. It has been molecularly demonstrated that individuals carrying the sickle cell mutation have a resistance to malarial infection. Because many of the selection pressures that may have given rise to the current distribution of mutations in particular populations are in our evolutionary past, it is difficult to assess how much variation within or among populations is due to these types of selection forces.
Another major force in determining the distribution of genetic variations within and among human populations is their migration and reproductive isolation. According to our best knowledge, one of the most important periods in human evolution occurred approximately 100,000 years ago, when some humans migrated to other continents from the African basin and established new communities with relative reproductive isolation. Genetic differences among people in different geographical areas have been associated with the concept of race for hundreds of years. Although race is still used as a label, the original concept of race as genetically distinct subspecies of humans has been rejected through modern genetic information. For numerous reasons, discussed in the section below, it is more appropriate to reconceptualize the old genetics of race into a more accurate genetics of ancestry.
In addition to distant evolutionary patterns of migration, more modern migration patterns also have had a profound effect on the genetics of populations. For example, the current population of the United States and much of North America is very diverse genetically as a consequence of the mixing of many people from many different countries and continents.
A central reason for studying the origins and nature of human genetic variation is that the similarities and differences in the type and frequencies of genetic variations within and among populations can have a profound impact on studies that attempt to understand the influence of genes on disease risk. For example, some genetic variations, such as the apolipoprotein E protein polymorphisms, are found in every population and have very similar genotype frequencies around the world ( Wu et al., 2002 ; Deniz Naranjo et al., 2004 ). The variation’s association with increased heart disease and Alzheimer’s disease could be and has been tested in many of the world’s populations. Other mutations such as the 10kb deletion in the LDL-R gene described above are more population-specific variations.
Furthermore, from a statistical point of view, the effect of a genetic variation on the continuum of risk found in any population is correlated with its frequency. For example, common genetic polymorphisms with frequencies near 50 percent cannot be associated with large phenotypic effects within a population because the genotype classes each represent a large fraction of the population and, since most risk is normally distributed, the average risk for a highly prevalent genotype class cannot deviate from the overall risk of the population to any large degree. This correlation between genotype frequency and effect does not mean that common variations cannot be significant in their effects. The statistical significance of an association between a genetic variant and a disease is a joint function of sample size and the size of the effect. In addition, genetic research among populations that differ in their genotype frequencies can differ in their inferences about which polymorphisms have significant effects even if the absolute phenotypic effect is the same. See Cheverud and Routman (1995) for a more formal statistical explanation of this phenomenon and its impact on assessing gene-gene interactions.
Another key consideration in understanding the relationship between genetic variations and measures of disease risk is the population differences in the correlations between genotype frequencies at different SNP locations. There are two common reasons why the frequency of an allele or genotype at a particular SNP could be correlated with the frequency of an allele or genotype for a different SNP. First, a phenomenon known as linkage disequilibrium creates correlations among SNPs as a consequence of the mutation’s history. When mutations arise, they occur on a particular genetic background, which creates a correlation with the other SNPs on the chromosome. Second, the mixing of populations known as admixture that occurs typically through migration means that SNPs with population-specific frequencies will be correlated in a larger mixed sample. In this case, population stratification is the cause of the correlation, and there has been much genetic epidemiological research on this phenomenon and how to control for it. Population stratification is thought to be a possible source of spurious genetic associations with disease (see Box 3-2 ).
Population Stratification (Confounding). When the risk of disease varies between two ethnic groups, any genetic or environmental factor that also varies between the groups will appear to be related to disease. This phenomenon is called “population (more...)
In large part, the twentieth century was dominated by studies of human health and disease that focused on identifying single genetic and environmental agents that could explain variation in disease susceptibility. This new century has been characterized by huge advances in our understanding of Mendelian disorders with severe clinical outcomes. However, the Men delian paradigm has failed to elucidate the genetic contribution to susceptibility to most common chronic diseases, which researchers know have a substantial genetic component because of their familial aggregation and studies that demonstrate significant heritabilities for these diseases. Likewise, environmental and social epidemiological studies have been wildly successful in illuminating the role of many environmental factors such as diet, exercise, and stress on disease risk. However, these environmental factors still do not, by themselves, fully explain the variance in the prevalence of several diseases in different populations. Researchers are only now beginning to study in earnest the potential interactions between the genetic and environmental factors that are likely to be contributing to a large fraction of disease in most populations. There is much that can be done to incorporate measures of social environment into genetic studies and to also incorporate genetic measures into social epidemiological studies.
Over the last two decades, progress in identifying specific genes and mutations that explain genetic susceptibility to common conditions has been relatively slow, for a variety of reasons. First, the diseases being studied tend to be complex in their etiology, meaning that different people in a population will develop disease for different genetic and/or environmental reasons. Any single genetic or environmental factor is expected to explain only a very small fraction of disease risk in a population. Moreover, these factors are expected to interact, and other biological processes (e.g., epigenetic modifications) are likely to be contributors to the complex puzzle of susceptibility. An accurate phenotypic definition of disease and its subtypes is crucial to identifying and understanding the complexities of disease-specific genetic and environmental causes.
Second, geneticists only recently have developed the knowledge base or methods needed to measure genetic variations and their metabolic consequences with sufficient ease and cost-effectiveness so that the large number of genes thought to be involved can be studied. With the completion of the Human Genome Project in 2003, many different scientific entities (e.g., the Environmental Genome Project and the International HapMap Consortium) have been working to identify the mutational spectra in human populations, and genetic epidemiologists are just now beginning to understand the extensive nature of common variations (>1 percent population frequency) within the human genome that could be affecting people’s risk of disease. The SNP data generated by these initiatives are now centrally located in a number of public databases, including the National Center for Biotechnology Information’s dbSNPs database, the National Cancer Institute’s CGAP Genetic Annotation Initiative SNP Database, and the Karolinska Institute Human Genic Bi-Allelic Sequences Database. At present, the largest dataset on human variation is being generated by the International HapMap Project, 4 which is genotyping millions of SNPs on 270 individuals from 4 geographically separated sites from around the world. The International HapMap Project has greatly increased the number of validated SNPs available to the research community to be used to study human variation and is producing a map of genomic haplotypes in four populations with ancestry from parts of Africa, Asia, and Europe. In addition, high-throughput methods of genotyping large numbers of SNPs (thousands) in large epidemiological cohorts are only now becoming available (see above). Unfortunately, high-throughput methods of measuring the environment have not kept a similar pace. For many studies of common disease, a rate-limiting step to increasing our understanding will continue to be the difficult and costly measurement of environmental factors.
Finally, progress also has been hampered because of a lack of adequate investment in developing new methods of analysis that can incorporate the high-dimensional biological reality that we can now measure. The complex genetic and environmental architecture of multifactorial diseases is not easily detected or deciphered using the traditional statistical modeling methods that are focused on the estimation of a single overall model of disease for a population. For example, using traditional logistic regression methods it would be simply impossible to enter all the hundreds of genetic variations that are thought to be involved in CVD risk or in any of the other common disease complexes currently being studied. Beyond the obvious issues of power and overdetermination in such a large-scale model, we also do not know how to model or interpret interactions among many factors simultaneously or how to incorporate the rare, large effects of some genes relative to the common, small effects of others. New modeling strategies that take advantage of advances in pattern recognition, machine learning, and systems analysis (e.g., scale-free networks, Bayesian belief networks, random forest methods) are going to be needed in order to build more comprehensive, predictive models of these etiologically heterogeneous diseases.
The field of human genetics, like many other disciplines, is in transition, and there is much to be gained by joining forces with a wide range of other disciplines that are focused on improving prevention and reducing the disease burden in our populations.
- Altshuler D, Kruglyak L, Lander E. Genetic polymorphisms and disease. New England Journal of Medicine. 1998; 338 (22):1626. [ PubMed : 9606122 ]
- Ardlie KG, Lunetta KL, Seielstad M. Testing for population subdivision and association in four case-control studies. American Journal of Human Genetics. 2002; 71 (2):304–311. [ PMC free article : PMC379163 ] [ PubMed : 12096349 ]
- Armstrong VW, Shipkova M, von Ahsen N, Oellerich M. Analytic aspects of monitoring therapy with thiopurine medications. Therapeutic Drug Monitoring. 2004; 26 (2):220–226. [ PubMed : 15228169 ]
- Ashley-Koch A, Yang Q, Olney R. Sickle hemoglobin (Hb S) allele and sickle cell disease: A HuGE review. American Journal of Epidemiology. 2000; 151 (9):839–845. [ PubMed : 10791557 ]
- Bridges K. Hemoglobinopathies (Hemoglobin Disorders). 2002. [accessed May 15, 2006]. [Online]. Available: sickle .bwh.harvard.edu/hemoglobinopathy .html .
- Brown MS, Goldstein JL. Lowering plasma cholesterol by raising LDL receptors. New England Journal of Medicine. 1981; 305 (9):515–517. [ PubMed : 6265781 ]
- Cardon LR, Bell JI. Association study designs for complex diseases. Nature Reviews Genetics. 2001; 2 (2):91–99. [ PubMed : 11253062 ]
- Cheverud JM, Routman EJ. Epistasis and its contribution to genetic variance components. Genetics. 1995; 139 (3):1455–1461. [ PMC free article : PMC1206471 ] [ PubMed : 7768453 ]
- Clifford RJ, Edmonson MN, Nguyen C, Buetow KH. Large-scale analysis of non-synonymous coding region single nucleotide polymorphisms. Bioinformatics. 2004; 20 (7):1006–1014. [ PubMed : 14751981 ]
- Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman G, Slamon DJ. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. Journal of Clinical Oncology. 1999; 17 (9):2639–2648. [ PubMed : 10561337 ]
- Deniz Naranjo MC, Munoz Fernandez C, Alemany Rodriguez MJ, Perez Vieitez MC, Irurita Latasa J, Suarez Armas R, Suarez Valentin MP, Sanchez Garcia F. Gender has a strong modulating effect on the risk of Alzheimer’s disease conferred by the apolipoprotein E gene in the population of the Canary Islands, Spain. Revista de Neurologia. 2004; 38 (7):615–618. [ PubMed : 15098180 ]
- Evans WE, McLeod HL. Pharmacogenomics—drug disposition, drug targets, and side effects. New England Journal of Medicine. 2003; 348 (6):538–549. [ PubMed : 12571262 ]
- Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nature Genetics. 2005; 37 (5):549–554. [ PubMed : 15838508 ]
- Haines JL, Pericak-Vance MA. Approaches to Gene Mapping in Complex Human Diseases. New York: Wiley-Liss; 1998.
- Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990; 250 (4988):1684–1689. [ PubMed : 2270482 ]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005; 307 (5712):1072–1079. [ PubMed : 15718463 ]
- IOM (Institute of Medicine). Implications of Genomics for Public Health. Washington, DC: The National Academies Press; 2005. [ PubMed : 22379649 ]
- Kalow W, Tang BK, Endrenyi L. Hypothesis: Comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics. 1998; 8 (4):283–289. [ PubMed : 9731714 ]
- Kardia SL, Modell SM, Peyser PA. Family-centered approaches to understanding and preventing coronary heart disease. American Journal of Preventive Medicine. 2003; 24 (2):143–151. [ PubMed : 12568820 ]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005; 308 (5720):385–389. [ PMC free article : PMC1512523 ] [ PubMed : 15761122 ]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994; 265 (5181):2037–2048. [ PubMed : 8091226 ]
- Livingston RJ, von Niederhausern A, Jegga AG, Crawford DC, Carlson CS, Rieder MJ, Gowrisankar S, Aronow BJ, Weiss RB, Nickerson DA. Pattern of sequence variation across 213 environmental response genes. Genome Research. 2004; 14 (10A):1821–1831. [ PMC free article : PMC524406 ] [ PubMed : 15364900 ]
- Mancama D, Kerwin RW. Role of pharmacogenomics in individualising treatment with SSRIs. CNS Drugs. 2003; 17 (3):143–151. [ PubMed : 12617694 ]
- Mathew C. Science medicine and the future—postgenomic technologies: Hunting the genes for common disorders. British Medical Journal. 2001; 322 (7293):1031–1034. [ PMC free article : PMC1120184 ] [ PubMed : 11325769 ]
- McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1997; 94 (3):814–819. [ PMC free article : PMC19596 ] [ PubMed : 9023339 ]
- Miller DP, Liu G, De Vivo I, Lynch TJ, Wain JC, Su L, Christiani DC. Combinations of the variant genotypes of GSTP1, GSTM1, and p53 are associated with an increased lung cancer risk. Cancer Research. 2002; 62 (10):2819–2823. [ PubMed : 12019159 ]
- Moorjani S, Roy M, Gagne C, Davignon J, Brun D, Toussaint M, Lambert M, Campeau L, Blaichman S, Lupien P. Homozygous familial hypercholesterolemia among French Canadians in Quebec Province. Arteriosclerosis. 1989; 9 (2):211–216. [ PubMed : 2923577 ]
- Obach RS, Walsky RL, Venkatakrishnan K, Gaman EA, Houston JB, Tremaine LM. The utility of in vitro cytochrome P450 inhibition data in the prediction of drug-drug interactions. Journal of Pharmacology and Experimental Therapeutics. 2006; 316 (1):336–348. [ PubMed : 16192315 ]
- Pirmohamed M, Park BK. Genetic susceptibility to adverse drug reactions. Trends in Pharmacological Sciences. 2001; 22 (6):298–305. [ PubMed : 11395158 ]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. American Journal of Human Genetics. 1999; 65 (1):220–228. [ PMC free article : PMC1378093 ] [ PubMed : 10364535 ]
- Quinn CT, Miller ST. Risk factors and prediction of outcomes in children and adolescents who have sickle cell anemia. Hematology/Oncology Clinics of North America. 2004; 18 (6 SPEC.ISS):1339–1354. [ PubMed : 15511619 ]
- Rebbeck TR, Walker AH, Phelan CM, Godwin AK, Buetow KH, Garber JE, Narod SA, Weber BL. Defining etiologic heterogeneity in breast cancer using genetic biomarkers. Progress in Clinical and Biological Research. 1997; 396 :53–61. [ PubMed : 9108589 ]
- Rimoin DL, Connor JM, Pyeritz RE, Korf BR, editors. Emery and Rimoin’s Principles and Practice of Medical Genetics. 4th edition. Vol. 2. New York: Churchill Livingstone; 2002.
- Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000; 405 (6788):847–856. [ PubMed : 10866211 ]
- Satten GA, Flanders WD, Yang Q. Accounting for unmeasured population substructure in case-control studies of genetic association using a novel latent-class model. American Journal of Human Genetics. 2001; 68 (2):466–477. [ PMC free article : PMC1235279 ] [ PubMed : 11170894 ]
- Sing CF, Stengard JH, Kardia SLR. Genes, environment, and cardiovascular disease. Arteriosclerosis, Thrombosis, and Vascular Biology. 2003; 23 :1190–1196. [ PubMed : 12730090 ]
- Smith G. The Genomics Age: How DNA Technology Is Transforming the Way We Live and Who We Are. New York: AMACOM; 2005.
- Steinberg MH. Predicting clinical severity in sickle cell anaemia. British Journal of Haematology. 2005; 129 (4):465–481. [ PubMed : 15877729 ]
- Stuart MJ, Nagel RL. Sickle-cell disease. Lancet. 2004; 364 (9442):1343–1360. [ PubMed : 15474138 ]
- Syvanen AC. Toward genome-wide SNP genotyping. Nature Genetics. 2005;(37 Suppl):S5–S10. [ PubMed : 15920530 ]
- Thompson MW, McInnes RR, Willard, editors. Thompson & Thompson Genetics in Medicine. 5th edition. Philadelphia, PA: W.B. Saunders Company; 1991.
- Wacholder S, Rothman N, Caporaso N. Population stratification in epidemiologic studies of common genetic variants and cancer: Quantification of bias. Journal of the National Cancer Institute. 2000; 92 (14):1151–1158. [ PubMed : 10904088 ]
- Wang X, Tomso DJ, Liu X, Bell DA. Single nucleotide polymorphism in transcriptional regulatory regions and expression of environmentally responsive genes. Toxicology and Applied Pharmacology. 2005; 207 (2 Suppl):84–90. [ PubMed : 16002116 ]
- Wang Z, Fan H, Yang HH, Hu Y, Buetow KH, Lee MP. Comparative sequence analysis of imprinted genes between human and mouse to reveal imprinting signatures. Genomics. 2004; 83 (3):395–401. [ PubMed : 14962665 ]
- Weinshilboum R. Inheritance and drug response. New England Journal of Medicine. 2003; 348 (6):529–537. [ PubMed : 12571261 ]
- Wu JH, Lo SK, Wen MS, Kao JT. Characterization of apolipoprotein E genetic variations in Taiwanese association with coronary heart disease and plasma lipid levels. Human Biology. 2002; 74 (1):25–31. [ PubMed : 11931577 ]
- Zhou W, Liu G, Miller DP, Thurston SW, Xu LL, Wain JC, Lynch TJ, Su L, Christiani DC. Polymorphisms in the DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk. Cancer Epidemiology, Biomarkers and Prevention. 2003; 12 (4):359–365. [ PubMed : 12692111 ]
An SNP is the DNA sequence variation that occurs when a single nucleotide (A, T, C, or G) in the genome sequence is altered ( Smith, 2005 ).
A candidate gene is a gene whose protein product is involved in the metabolic or physiological pathways associated with a particular disease ( IOM, 2005 ).
The sickle cell example is abstracted from a commissioned paper prepared by Robert J. Thompson, Jr., Ph.D. ( Appendix D ).
See www .hapmap.org .
- Cite this Page Institute of Medicine (US) Committee on Assessing Interactions Among Social, Behavioral, and Genetic Factors in Health; Hernandez LM, Blazer DG, editors. Genes, Behavior, and the Social Environment: Moving Beyond the Nature/Nurture Debate. Washington (DC): National Academies Press (US); 2006. 3, Genetics and Health.
- PDF version of this title (2.1M)
In this Page
Other titles in this collection.
- The National Academies Collection: Reports funded by National Institutes of Health
Related information
- PMC PubMed Central citations
- PubMed Links to PubMed
Recent Activity
- Genetics and Health - Genes, Behavior, and the Social Environment Genetics and Health - Genes, Behavior, and the Social Environment
Your browsing activity is empty.
Activity recording is turned off.
Turn recording back on
Connect with NLM
National Library of Medicine 8600 Rockville Pike Bethesda, MD 20894
Web Policies FOIA HHS Vulnerability Disclosure
Help Accessibility Careers
ENCYCLOPEDIC ENTRY
Genetic variation.
Genetic variation is the presence of differences in sequences of genes between individual organisms of a species. It enables natural selection, one of the primary forces driving the evolution of life.
Biology, Genetics
genetic variation
In many species, special genetic variations give animals a camouflaged appearance to blend in with their environment, like this Catalpa Sphinx moth (Ceratomia catalpae) which uses its textured wings to blend in with a tree's bark.
Photograph by J.S. Houser

Genetic variation refers to differences among the genomes of members of the same species. A genome is all the hereditary information—all the genes —of an organism . For instance, the human genome contains somewhere between twenty and twenty-five thousand genes .
Genes are units of hereditary information, and they carry instructions for building proteins. The genes that are encoded within these proteins are what enable cells to function. Most organisms that reproduce sexually have two copies of each gene , because each parent cell or organism donates a single copy of its genes to its offspring. Additionally, genes can exist in slightly different forms, called alleles , which further adds to genetic variation.
The combination of alleles of a gene that an individual receives from both parents determines what biologists call the genotype for a particular trait, such as hair texture. The genotype that an individual possesses for a trait, in turn, determines the phenotype —the observable characteristics—such as whether that individual actually ends up with straight, wavy, or curly hair.
Genetic variation within a species can result from a few different sources. Mutations , the changes in the sequences of genes in DNA , are one source of genetic variation. Another source is gene flow , or the movement of genes between different groups of organisms . Finally, genetic variation can be a result of sexual reproduction , which leads to the creation of new combinations of genes .
Genetic variation in a group of organisms enables some organisms to survive better than others in the environment in which they live. Organisms of even a small population can differ strikingly in terms of how well suited they are for life in a certain environment . An example would be moths of the same species with different color wings. Moths with wings similar to the color of tree bark are better able to camouflage themselves than moths of a different color. As a result, the tree-colored moths are more likely to survive, reproduce, and pass on their genes . This process is called natural selection , and it is the main force that drives evolution .
Media Credits
The audio, illustrations, photos, and videos are credited beneath the media asset, except for promotional images, which generally link to another page that contains the media credit. The Rights Holder for media is the person or group credited.
Production Managers
Program specialists, last updated.
October 19, 2023
User Permissions
For information on user permissions, please read our Terms of Service. If you have questions about how to cite anything on our website in your project or classroom presentation, please contact your teacher. They will best know the preferred format. When you reach out to them, you will need the page title, URL, and the date you accessed the resource.
If a media asset is downloadable, a download button appears in the corner of the media viewer. If no button appears, you cannot download or save the media.
Text on this page is printable and can be used according to our Terms of Service .
Interactives
Any interactives on this page can only be played while you are visiting our website. You cannot download interactives.
Related Resources
Research Snapshot: Understanding protein mutations that affect gene expression
Marissa Shapiro
May 23, 2024, 11:52 AM
Graduate student Hillary Layden studies transcriptional control of cancer in the lab of Scott Hiebert , Hortense B. Ingram Chair in Cancer Research and professor of biochemistry.

Below, Layden shares the results from her research in which she used a deep genomic analysis to determine how protein mutations influence gene expression to promote cancer progression.
What issue does your research address?
Mutations in proteins that control gene expression are common across human cancers, but we don’t understand how these mutations affect gene expression, contribute to disease, or impact response to treatments. In this study, we focused on identifying the direct gene targets of the transcription factor FOXO1 and how mutations in FOXO1 found in B-cell lymphoma patients affect the expression of these genes.
Our approach took advantage of several recently developed technologies, including degron tags and nascent transcript analysis, to analyze transcription immediately after the loss of FOXO1.
What were your findings?
We found that wild-type FOXO1 activates the transcription of genes that are key regulators of B-cell identity and are known oncogenes, maintaining chromatin accessibility at enhancer elements. Mutant FOXO1 controls transcription in the same way but is insensitive to signaling events that inactivate wild-type FOXO1. This allows mutant FOXO1 to maintain the expression of genes that contribute to lymphoma cell growth in cellular conditions that wild-type FOXO1 cannot.
TL;DR: Mutations in FOXO1 affect an on/off switch, locking FOXO1 in the “on” position. When FOXO1 is “on,” it increases the expression of target genes, which are critical for cell growth and survival.
What do you hope will be the outcomes of this study?
Patients with mutations in FOXO1 are more likely to relapse on the current standard of care therapy. We hope that a better understanding of how these mutations affect the function of FOXO1 can be translated into new treatments that better serve this patient population in the long term.
In the short term, this work provides several target genes that can be used as proxies for FOXO1 transcriptional function in B-cells. These targets can be used to identify drugs that affect FOXO1 transcriptional activity or other proteins required for FOXO1 transcriptional function. This work also provides much-needed clarity on how lymphoma-associated mutations in FOXO1 affect the transcription of target genes.
What core facilities were used to conduct the research?
This work made use of the Vanderbilt Computational Structural and Chemical Biology Core Facility, the VUMC Flow Cytometry Shared Resource, Vanderbilt Technologies for Advanced Genomics Analysis and Research Design (VANTAGE), and the VUMC Molecular Cell Biology Resource.
The article “ Mutant FOXO1 controls an oncogenic network via enhancer accessibility ” was published in Cell Genomics in April 2024.
The research was supported by the T. J. Martell Foundation, the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, the National Center for Research Resources, the National Center for Advancing Translational Sciences, and the National Institutes of Health. Core services were performed through grants from the Vanderbilt Digestive Disease Research Center and the Vanderbilt-Ingram Cancer Center.
Explore Story Topics
- Research, News & Discoveries
- Biochemistry
- Molecular Cell Biology Resource
- National Institutes of Health
- School of Medicine Basic Sciences
- Scott Hiebert
- Vanderbilt Computational Structural and Chemical Biology Core Facility
- Vanderbilt Digestive Disease Research Center
- Vanderbilt Technologies for Advanced Genomics Analysis and Research Design
- Vanderbilt-Ingram Cancer Center
- VUMC Flow Cytometry Shared Resource
Evolution and Natural Selection: Distinctions and Connections
This essay is about the concepts of evolution and natural selection, explaining their distinctions and connections. Evolution refers to the changes in the genetic makeup of populations over time, while natural selection is a mechanism that drives this process by favoring traits that enhance survival and reproduction. The essay discusses how mutations, genetic drift, and gene flow contribute to evolution, with natural selection being the most visible mechanism. Examples such as antibiotic resistance and the peppered moth illustrate these concepts. The essay underscores the importance of understanding these mechanisms to appreciate the complexity and dynamic nature of biological change.
How it works
The principles of metamorphosis and innate selection serve as pivotal tenets within the realm of biology, elucidating the astonishing spectrum of life that currently thrives upon our terrestrial sphere. Although frequently mentioned in tandem, they represent distinct facets of biological phenomenon. Metamorphosis delineates the expansive process through which species undergo transformation over successive epochs. Conversely, innate selection emerges as a principal mechanism propelling this transformative odyssey. To fully comprehend the nuances of life’s evolution, an appreciation of the intricate interplay between these concepts becomes imperative.
In delving into the realm of metamorphosis, we encounter the vicissitudes in the hereditary blueprint of populations across successive generations. These fluctuations can engender the genesis of novel species while simultaneously sounding the death knell for others. It’s a sweeping concept encompassing myriad mechanisms such as genetic alteration, gene dispersion, genetic meandering, and innate selection. Each of these elements exerts a distinct influence upon the evolutionary tapestry, yet innate selection stands forth as particularly conspicuous due to its direct correlation with the adaptive responses of organisms to their surroundings.
The advent of innate selection was first elucidated by the erudite Charles Darwin. In its essence, innate selection denotes the mechanism wherein entities bearing traits that afford them a superior fit within their environment stand a higher chance of survival and procreation. These advantageous traits then perpetuate down successive generations. Envision a populace of beetles wherein a fraction is hued green while the remainder dons a brown hue. Should avian predators find it easier to discern the green-hued beetles amidst the verdant forest floor, the green-hued ones are liable to succumb to predation. Consequently, the brown-hued beetles, being better concealed, endure longer and beget more offspring. Over time, the populace shall witness a preponderance of brown-hued beetles. This exemplifies the workings of innate selection.
Viewed metaphorically, metamorphosis unfolds akin to a meandering journey while innate selection emerges as one of the traversed routes. Metamorphosis denotes the overarching process of evolution and development over time, whereas innate selection represents a specific trajectory sculpted by environmental exigencies that steer the course of transformation. This analogy serves to elucidate why innate selection is frequently equated with metamorphosis despite being a mere component of the broader phenomenon.
Mutations represent another pivotal facet of metamorphosis. These alterations in the DNA sequence hold the potential to infuse novel genetic permutations into a populace. Mutations may prove beneficial, detrimental, or neutral in their impact. Beneficial mutations confer upon organisms a survival edge. For instance, a mutation might confer upon a plant resistance to a specific ailment. Should this mutation furnish a notable advantage, the plant stands a greater chance of surviving and reproducing, disseminating the mutation throughout the populace. Subsequently, innate selection may act upon this mutation, augmenting its prevalence across successive generations.
However, not all evolutionary shifts are dictated by innate selection. Genetic meandering, for instance, pertains to stochastic alterations in the prevalence of alleles (distinct variants of a gene) within a populace. This phenomenon can exert significant ramifications, particularly within diminutive populations. Contemplate a populace of lagomorphs wherein a handful by happenstance harbor a rare allele. Were a sudden tempest to decimate a majority of the populace indiscriminately, the frequencies of alleles within the survivors might undergo a drastic upheaval, not on account of any intrinsic advantage but sheer happenstance. This embodies the workings of genetic meandering.
Gene dispersion represents yet another mechanism of metamorphosis and encompasses the transference of genetic material amidst distinct populations. This may introduce novel alleles into a populace, augmenting genetic diversity and potentially reshaping evolutionary trajectories. Should individuals from one populace of avians emigrate to another and crossbreed, they impart an amalgam of genetic material. This can culminate in novel amalgams of traits within the recipient populace, upon which innate selection may then operate. This phenomenon of genetic flux between populations serves to sustain diversity within species and may precipitate novel evolutionary trajectories.
A comprehension of these disparate mechanisms underscores the labyrinthine nature of metamorphosis. Innate selection frequently occupies the forefront owing to its manifest association with survival and procreative success. However, the confluence of mutation, genetic meandering, and gene dispersion profoundly shape the metamorphic process. This interaction underscores the non-linear and heterogeneous nature of metamorphosis.
The dichotomy between metamorphosis and innate selection assumes paramount significance in comprehending the entirety of biological transmutation. Whilst innate selection expounds upon the ascendancy of certain traits owing to environmental pressures, metamorphosis encapsulates the broad spectrum of genetic alterations across time. This expansive purview proves indispensable in deciphering phenomena such as bacterial resistance to antibiotics, the emergence of novel species, and the acclimatization of organisms to shifting environs.
Consider, for instance, the paradigm of bacterial antibiotic resistance. Upon exposure to antibiotics, a bacterial populace may witness the annihilation of the majority, albeit a fraction may harbor stochastic mutations rendering them impervious to the drug. These resistant bacteria survive and propagate, transmitting the resistance trait to ensuing generations. Over time, the populace metamorphoses to harbor augmented resistance to the antibiotic. This constitutes a palpable instance of innate selection steering metamorphic change, as the environmental impetus of antibiotics selects for resistant entities.
Speciation, the genesis of novel species, emerges as another riveting facet of metamorphosis. It frequently transpires when distinct populations of a species become geographically segregated, impeding gene dispersion between them. Across time, the genetic disparities between the isolated populations may burgeon to such an extent that interbreeding becomes untenable, notwithstanding subsequent recontact. This process may be propelled by myriad factors including innate selection, genetic meandering, and mutations. The upshot is the advent of novel species, each tailored to its distinct milieu.
The adaptation to shifting environs constitutes an ongoing process within metamorphosis. As habitats undergo metamorphoses, whether by natural vicissitudes or human interventions, species must adapt to endure. Such adaptation may entail alterations in behavior, physiology, or morphology. For instance, polar bears have evolved dense fur and adipose deposits to weather the frigid climes of the Arctic. Should climate shifts engender pronounced warming in the Arctic, polar bears must adapt to the altered milieu or risk extirpation. Innate selection would favor entities harboring traits better suited to the fresh milieu, thereby precipitating metamorphic change within the populace.
The saga of the peppered moth amid the Industrial Revolution in England offers a quintessential illustration of adaptation via innate selection. Ere industrialization, the majority of peppered moths sported pale hues, affording them seamless camouflage amidst lichen-clad trees. Nonetheless, as industrial effluvia decimated lichens and sooted the bark, moths donning darker hues enjoyed a survival advantage owing to enhanced concealment against predators. The populace of peppered moths transitioned from predominantly pale to overwhelmingly dark-hued in response to the altered environment, thus illustrating the workings of innate selection.
In summation, whilst metamorphosis and innate selection stand inextricably linked, they epitomize distinct facets of the overarching phenomenon. Metamorphosis embodies the sweeping pattern of genetic metamorphosis within populations across time, enshrining diverse mechanisms. Innate selection, conversely, constitutes a specific mechanism impelling metamorphic change by favoring entities endowed with advantageous traits. Discerning this dichotomy fosters a profound comprehension of life’s evolution, adaptation, and diversification. Through the lens of these concepts, we glean insight into the labyrinthine and dynamic nature of the natural world.
By assimilating these mechanisms, we unravel the annals of life’s past and present whilst prognosticating its future trajectory. The scrutiny of metamorphosis and innate selection not only unveils life’s historical tapestry but also informs conservation endeavors, biomedical inquiries, and our comprehension of life’s prospective trajectory amidst ongoing environmental exigencies. The ceaseless interplay of genetic variation, environmental constraints, and the inexorable passage of time continues to sculpt the living fabric in manners both subtle and profound.
Cite this page
Evolution and Natural Selection: Distinctions and Connections. (2024, Jun 01). Retrieved from https://papersowl.com/examples/evolution-and-natural-selection-distinctions-and-connections/
"Evolution and Natural Selection: Distinctions and Connections." PapersOwl.com , 1 Jun 2024, https://papersowl.com/examples/evolution-and-natural-selection-distinctions-and-connections/
PapersOwl.com. (2024). Evolution and Natural Selection: Distinctions and Connections . [Online]. Available at: https://papersowl.com/examples/evolution-and-natural-selection-distinctions-and-connections/ [Accessed: 2 Jun. 2024]
"Evolution and Natural Selection: Distinctions and Connections." PapersOwl.com, Jun 01, 2024. Accessed June 2, 2024. https://papersowl.com/examples/evolution-and-natural-selection-distinctions-and-connections/
"Evolution and Natural Selection: Distinctions and Connections," PapersOwl.com , 01-Jun-2024. [Online]. Available: https://papersowl.com/examples/evolution-and-natural-selection-distinctions-and-connections/. [Accessed: 2-Jun-2024]
PapersOwl.com. (2024). Evolution and Natural Selection: Distinctions and Connections . [Online]. Available at: https://papersowl.com/examples/evolution-and-natural-selection-distinctions-and-connections/ [Accessed: 2-Jun-2024]
Don't let plagiarism ruin your grade
Hire a writer to get a unique paper crafted to your needs.

Our writers will help you fix any mistakes and get an A+!
Please check your inbox.
You can order an original essay written according to your instructions.
Trusted by over 1 million students worldwide
1. Tell Us Your Requirements
2. Pick your perfect writer
3. Get Your Paper and Pay
Hi! I'm Amy, your personal assistant!
Don't know where to start? Give me your paper requirements and I connect you to an academic expert.
short deadlines
100% Plagiarism-Free
Certified writers
- Mobile Site
- Staff Directory
- Advertise with Ars
Filter by topic
- Biz & IT
- Gaming & Culture
Front page layout
Splice of life —
Mutations in a non-coding gene associated with intellectual disability, a gene that only makes an rna is linked to neurodevelopmental problems..
Diana Gitig - May 31, 2024 5:30 pm UTC
Almost 1,500 genes have been implicated in intellectual disabilities; yet for most people with such disabilities, genetic causes remain unknown. Perhaps this is in part because geneticists have been focusing on the wrong stretches of DNA when they go searching. To rectify this, Ernest Turro—a biostatistician who focuses on genetics, genomics, and molecular diagnostics—used whole genome sequencing data from the 100,000 Genomes Project to search for areas associated with intellectual disabilities.
His lab found a genetic association that is the most common one yet to be associated with neurodevelopmental abnormality. And the gene they identified doesn’t even make a protein.
Trouble with the spliceosome
Most genes include instructions for how to make proteins. That’s true. And yet human genes are not arranged linearly—or rather, they are arranged linearly, but not contiguously. A gene containing the instructions for which amino acids to string together to make a particular protein—hemoglobin, insulin, serotonin, albumin, estrogen, whatever protein you like—is modular. It contains part of the amino acid sequence, then it has a chunk of DNA that is largely irrelevant to that sequence, then a bit more of the protein’s sequence, then another chunk of random DNA, back and forth until the end of the protein. It’s as if each of these prose paragraphs were separated by a string of unrelated letters (but not a meaningful paragraph from a different article).
In order to read this piece through coherently, you’d have to take out the letters interspersed between its paragraphs. And that’s exactly what happens with genes. In order to read the gene through coherently, the cell has machinery that splices out the intervening sequences and links up the protein-making instructions into a continuous whole. (This doesn’t happen in the DNA itself; it happens to an RNA copy of the gene.) The cell’s machinery is obviously called the spliceosome.
There are about a hundred proteins that comprise the spliceosome. But the gene just found to be so strongly associated with neurodevelopmental disorders doesn’t encode any of them. Rather, it encodes one of five RNA molecules that are also part of the spliceosome complex and interact with the RNAs that are being spliced. Mutations in this gene were found to be associated with a syndrome with symptoms that include intellectual disability, seizures, short stature, neurodevelopmental delay, drooling, motor delay, hypotonia (low muscle tone), and microcephaly (having a small head).
Supporting data
The researchers buttressed their finding by examining three other databases; in all of them, they found more people with the syndrome who had mutations in this same gene. The mutations occur in a remarkably conserved region of the genome, suggesting that it is very important. Most of the mutations were new in the affected people—i.e. not inherited from their parents—but there was one case of one particular mutation in the gene that was inherited. Based on this, the researchers concluded that this particular variant may cause a less severe disorder than the other mutations.
Many studies that look for genes associated with diseases have focused on searching catalogs of protein coding genes. These results suggest that we could have been missing important mutations because of this focus.
Nature Medicine, 2024. DOI: 10.1038/s41591-024-03085-5
reader comments
Channel ars technica.
- Alzheimer's disease & dementia
- Arthritis & Rheumatism
- Attention deficit disorders
- Autism spectrum disorders
- Biomedical technology
- Diseases, Conditions, Syndromes
- Endocrinology & Metabolism
- Gastroenterology
- Gerontology & Geriatrics
- Health informatics
- Inflammatory disorders
- Medical economics
- Medical research
- Medications
- Neuroscience
- Obstetrics & gynaecology
- Oncology & Cancer
- Ophthalmology
- Overweight & Obesity
- Parkinson's & Movement disorders
- Psychology & Psychiatry
- Radiology & Imaging
- Sleep disorders
- Sports medicine & Kinesiology
- Vaccination
- Breast cancer
- Cardiovascular disease
- Chronic obstructive pulmonary disease
- Colon cancer
- Coronary artery disease
- Heart attack
- Heart disease
- High blood pressure
- Kidney disease
- Lung cancer
- Multiple sclerosis
- Myocardial infarction
- Ovarian cancer
- Post traumatic stress disorder
- Rheumatoid arthritis
- Schizophrenia
- Skin cancer
- Type 2 diabetes
- Full List »
share this!
May 30, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
Q&A: Understanding protein mutations that affect gene expression to promote cancer progression
by Marissa Shapiro, Vanderbilt University
Graduate student Hillary Layden studies transcriptional control of cancer in the lab of Scott Hiebert, Hortense B. Ingram Chair in Cancer Research and professor of biochemistry.
Below, Layden shares the results from her research in which she used a deep genomic analysis to determine how protein mutations influence gene expression to promote cancer progression.
The study is published in the journal Cell Genomics .
What issue does your research address?
Mutations in proteins that control gene expression are common across human cancers , but we don't understand how these mutations affect gene expression, contribute to disease, or impact response to treatments. In this study, we focused on identifying the direct gene targets of the transcription factor FOXO1 and how mutations in FOXO1 found in B-cell lymphoma patients affect the expression of these genes.
Our approach took advantage of several recently developed technologies, including degron tags and nascent transcript analysis, to analyze transcription immediately after the loss of FOXO1.
What were your findings?
We found that wild-type FOXO1 activates the transcription of genes that are key regulators of B-cell identity and are known oncogenes, maintaining chromatin accessibility at enhancer elements. Mutant FOXO1 controls transcription in the same way but is insensitive to signaling events that inactivate wild-type FOXO1. This allows mutant FOXO1 to maintain the expression of genes that contribute to lymphoma cell growth in cellular conditions that wild-type FOXO1 cannot.
In other words, mutations in FOXO1 affect an on/off switch, locking FOXO1 in the "on" position. When FOXO1 is "on," it increases the expression of target genes, which are critical for cell growth and survival.
What do you hope will be the outcomes of this study?
Patients with mutations in FOXO1 are more likely to relapse on the current standard of care therapy. We hope that a better understanding of how these mutations affect the function of FOXO1 can be translated into new treatments that better serve this patient population in the long term.
In the short term, this work provides several target genes that can be used as proxies for FOXO1 transcriptional function in B-cells. These targets can be used to identify drugs that affect FOXO1 transcriptional activity or other proteins required for FOXO1 transcriptional function. This work also provides much-needed clarity on how lymphoma-associated mutations in FOXO1 affect the transcription of target genes .
Explore further
Feedback to editors

Almost 1 in 3 Americans know someone who's died from a drug overdose
12 hours ago

Antibody discovery promises new hope in influenza B battle, paves way for universal vaccine
16 hours ago

Eye-tracking techniques could help primary care providers diagnose autism sooner, more accurately
May 31, 2024
This self-powered sensor could make MRIs more efficient

Not eating can hinder weight loss, study in fruit flies suggests

Neuroscience research suggests ketones can enhance cognitive function and protect brain networks
New research finds antidepressants may help deliver other drugs into the brain

Mediterranean diet tied to one-fifth lower risk of death in women

Scientists find new method to enhance efficacy of bispecific antibodies for solid tumors

Cardiomyocytes study discovers new way to regenerate damaged heart cells
Related stories.
Researchers identify protein that controls CAR T cell longevity
Apr 10, 2024

Uncovering the role of FOXO1 in vascular development and transcriptional dynamics in endothelial cells
Mar 8, 2024

Researchers stimulate gene that enhances CAR T-cell treatments for solid tumors
Apr 11, 2024

Scientists identify key factor regulating abnormal heart growth
Jul 9, 2020

New study uncovers profound impact of diet and genetically induced obesity on ovarian microenvironment
Oct 18, 2023

Zebrafish expose tumor pathway in childhood muscle cancer
Jun 5, 2018
Recommended for you
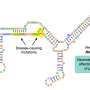
Researchers identify a genetic cause of intellectual disability affecting tens of thousands
Prenatal testing offers a window for finding a mother's cancer risk

Team finds new potential causes of rare and lethal bone cancer

Cancer can be caused by reversible molecular changes, study shows

Gene variants foretell the biology of future breast cancers, study finds
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Medical Xpress in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
Researchers identify a genetic cause of intellectual disability affecting tens of thousands
Novel study links genetic changes in a non-coding gene called rnu4-2 to neurodevelopmental disorders.
Researchers at the Icahn School of Medicine at Mount Sinai and others have identified a neurodevelopmental disorder, caused by mutations in a single gene, that affects tens of thousands of people worldwide. The work, published in the May 31 online issue of Nature Medicine, was done in collaboration with colleagues at the University of Bristol, UK; KU Leuven, Belgium; and the NIHR BioResource, currently based at the University of Cambridge, UK.
The findings will improve clinical diagnostic services for patients with neurodevelopmental disorders.
Through rigorous genetic analysis, the researchers discovered that mutations in a small non-coding gene called RNU4-2 cause a collection of developmental symptoms that had not previously been tied to a distinct genetic disorder. Non-coding genes are parts of DNA that do not produce proteins. The investigators used whole-genome sequencing data in the United Kingdom's National Genomic Research Library to compare the burden of rare genetic variants in 41,132 non-coding genes between 5,529 unrelated cases with intellectual disability and 46,401 unrelated controls.
The discovery is significant, as it represents one of the most common single-gene genetic causes of such disorders, ranking second only to Rett syndrome among patients sequenced by the United Kingdom's Genomic Medicine Service. Notably, these mutations are typically spontaneous and not inherited, providing important insights into the nature of the condition.
"We performed a large genetic association analysis to identify rare variants in non-coding genes that might be responsible for neurodevelopmental disorders," says the study's first author Daniel Greene, PhD, Assistant Professor of Genetics and Genomics Sciences at Icahn Mount Sinai and a Visitor at the University of Cambridge. "Nowadays, finding a single gene that harbors genetic variants responsible for tens of thousands of patients with a rare disease is exceptionally unusual. Our discovery eluded researchers for years due to various sequencing and analytical challenges."
More than 99 percent of genes known to harbor mutations that cause neurodevelopmental disorders encode proteins. The researchers hypothesized that non-coding genes, which don't produce proteins, could also host mutations leading to intellectual disability. Neurodevelopmental disorders, which often appear before grade school, involve developmental deficits affecting personal, social, academic, or occupational functioning. Intellectual disability specifically includes significant limitations in intellectual functioning (e.g., learning, reasoning, problem-solving) and adaptive behavior (e.g., social and practical skills).
"The genetic changes we found affect a very short gene, only 141 units long, but this gene plays a crucial role in a basic biological function of cells, called gene splicing, which is present in all animals, plants and fungi," says senior study author Ernest Turro, PhD, Associate Professor of Genetics and Genomic Sciences at Icahn Mount Sinai and a Visitor at the University of Cambridge. "Most people with a neurodevelopmental disorder do not receive a molecular diagnosis following genetic testing. Thanks to this study, tens of thousands of families will now be able to obtain a molecular diagnosis for their affected family members, bringing many diagnostic odysseys to a close."
Next, the researchers plan to explore the molecular mechanisms underlying this syndrome experimentally. This deeper understanding aims to provide biological insights that could one day lead to targeted interventions.
"What I found remarkable is how such a common cause of a neurodevelopmental disorder has been missed in the field because we've been focusing on coding genes," says Heather Mefford, MD, PhD, of the Center for Pediatric Neurological Disease Research at St. Jude Children's Research Hospital who was not involved with the research. "This study's discovery of mutations in non-coding genes, especially RNU4-2, highlights a significant and previously overlooked cause. It underscores the need to look beyond coding regions, which could reveal many other genetic causes, opening new diagnostic possibilities and research opportunities."
The paper is titled "Mutations in the U4 snRNA gene RNU4-2 cause one of the most prevalent monogenic neurodevelopmental disorders."
The remaining authors of the paper are Chantal Thys (KU Leuven, Belgium); Ian R. Berry, MD (University of Bristol, UK); Joanna Jarvis, MD (Birmingham Womens' Hospital, UK); Els Ortibus, MD, PhD (KU Leuven, Belgium); Andrew D. Mumford, MD (University of Bristol, UK); and Kathleen Freson, PhD (KU Leuven, Belgium).
The work was supported, in part, by NIH awards R01HL161365 and R03HD111492.
- Personalized Medicine
- Gene Therapy
- Diseases and Conditions
- Learning Disorders
- Huntington's Disease
- Social Psychology
- Alzheimer's disease
- Bipolar disorder
- Psychopathology
- Narcissistic personality disorder
- Rett syndrome
- Mental illness
Story Source:
Materials provided by The Mount Sinai Hospital / Mount Sinai School of Medicine . Note: Content may be edited for style and length.
Journal Reference :
- Daniel Greene, Chantal Thys, Ian R. Berry, Joanna Jarvis, Els Ortibus, Andrew D. Mumford, Kathleen Freson, Ernest Turro. Mutations in the U4 snRNA gene RNU4-2 cause one of the most prevalent monogenic neurodevelopmental disorders . Nature Medicine , 2024; DOI: 10.1038/s41591-024-03085-5
Cite This Page :
Explore More
- How Statin Therapy May Prevent Cancer
- Origins of 'Welsh Dragons' Exposed
- Resting Brain: Neurons Rehearse for Future
- Observing Single Molecules
- A Greener, More Effective Way to Kill Termites
- One Bright Spot Among Melting Glaciers
- Martian Meteorites Inform Red Planet's Structure
- Volcanic Events On Jupiter's Moon Io: High Res
- What Negative Adjectives Mean to Your Brain
- 'Living Bioelectronics' Can Sense and Heal Skin
Trending Topics
Strange & offbeat.
Scientists find a likely cause of many unexplained cases of intellectual disability: A genetic disorder
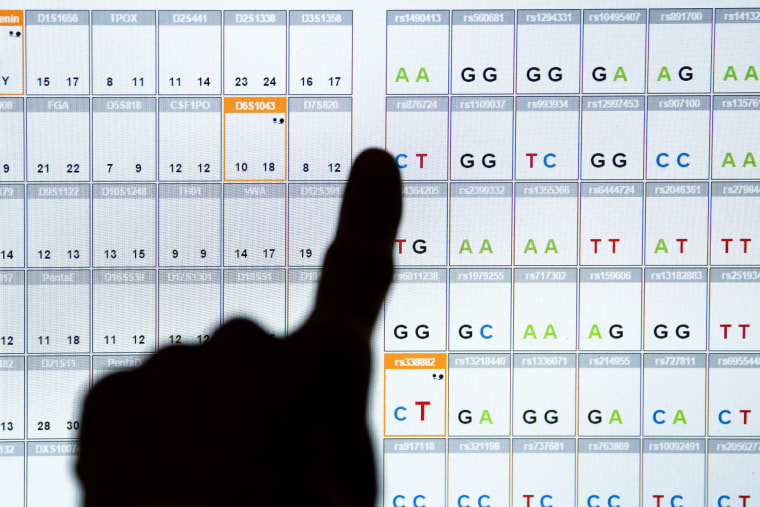
A newly identified neurodevelopmental disorder may explain tens of thousands of cases of intellectual disability whose cause was previously unknown, according to a new study.
The research, published Friday in the journal Nature Medicine, investigates the effects of mutations in the gene RNU4-2, which is found in all animals, plants and fungi.
The gene plays an important role in gene splicing — the process of cutting out portions of genetic material and stitching others together. Ernest Turro, the new study’s senior author and an associate professor of genetics and genomic science at the Icahn School of Medicine at Mount Sinai, said that in theory, mutations in the RNU4-2 gene could disrupt that splicing process, ultimately leading to abnormal brain development and intellectual disability.
This type of disability is characterized by significant limitations to a person’s ability to learn, reason, problem-solve, communicate or socialize, and it is often indicated by a low IQ. People with the disorder might also have seizures, motor delays, small heads, short stature or low muscle mass, according to the research.
The researchers hope that genetic tests for intellectual disabilities in children can quickly be updated to screen for the mutations.
“A considerable number of families will finally be able to have a genetic diagnosis,” Turro said.
Dr. Hakon Hakonarson, director of the Center for Applied Genomics at Children’s Hospital of Philadelphia, who was not involved in the study, said that because most cases of intellectual disability don’t have a known cause, the findings could “explain a good number of cases that are currently unexplained.”
The study estimates that up to 1 in 20,000 young people might have the condition. Researchers don’t know about the life expectancy associated with the disorder, so they have not estimated its prevalence among older adults, but Turro said some people with the genetic mutation have lived into adulthood.
The estimate suggests that the condition is slightly less common than Rett syndrome, a genetic disorder that causes babies to rapidly lose coordination, speech and mobility and affects about 1 in 10,000 female infants.
But Dr. Jeffrey Gruen, a professor of pediatrics and genetics at Yale School of Medicine who was not part of the research, said mutations in the RNU4-2 gene may turn out to be less common than the study suggests. He also questioned whether everyone with the mutations would have obvious learning or developmental issues.
“There are probably tens of thousands of people around the world that carry this, but does it cause intellectual disability in those tens of thousands? I don’t know,” he said. Gruen added, however, that the discovery is significant.
Hakonarson said the mutations probably cause at least some symptoms.
“The likelihood that this is disease-causing with these variants — which are not seen, by the way, in healthy people — is almost 100%,” he said.
The findings are based on data from the National Genomic Research Library, which contains information about the genomes — the entirety of a person’s genetic code — of people in the U.K. The study looked at the genomes of more than 77,000 participants.
Historically, studies of neurodevelopmental disorders have only looked at a small portion of the genome — specifically, so-called coding genes that are involved in the production of proteins. Of the 1,427 genes linked to intellectual disability, all but nine are coding genes.
Instead, Turro and his research team looked at noncoding genes — which don’t produce proteins — in about 5,500 people with intellectual disabilities. Mutations in the RNU4-2 gene were strongly associated with that group, compared with around 46,000 people who did not have intellectual disabilities.
“There’s no question this paper is going to provoke a lot of studies now,” Hakonarson said. “People are going to go hunting for additional genes, because there’s a lot of noncoding RNA genes.”
The mutations in the RNU4-2 gene seem to occur at random, so they most likely can’t be passed from parent to child. For that reason, getting a diagnosis could be a comfort to parents who want to have more children, Turro said.
The researchers said it will be quite some time before they figure out whether the disorder can be treated with drugs or gene therapy.
“These are an extremely tough group of disorders to tackle therapeutically,” Andrew Mumford, a co-author of the study and research director of the South West England NHS Genomic Medicine Service, said on a call with reporters.
But even without an available treatment, he added, families often benefit from having a diagnosis.
“It helps them come to terms with the impact,” he said. “Being able to tell someone, ‘Yes, we have found the cause of development disorder in your child’ is incredibly powerful.”
Gruen said the discovery could also help connect families whose children have the same genetic condition so they can share stories and offer support.
“You could get some idea of what the future holds for them,” Gruen said. “Is this something that could be remediated? Can we expect there to be language? Can we expect there to be motor issues? That’s also very, very important to know.”
Aria Bendix is the breaking health reporter for NBC News Digital.

‘Salty licorice’ cat pattern is the result of a genetic mutation, study reveals
Sign up for CNN’s Wonder Theory science newsletter. Explore the universe with news on fascinating discoveries, scientific advancements and more .
The village of Petäjävesi in central Finland is home to farmland, lakes, an 18th century wood-log church and a group of unusually striking cats. With their white chests, these creatures look similar to other tuxedo cats, but they possess a distinctive bit of flair: ombré strands of fur that start dark at the root and fade to white.
Geneticist Hannes Lohi and his collaborators at the University of Helsinki wanted to know how these cats got their look, so they studied the animals’ DNA. The team’s report, published May 9 in the journal Animal Genetics , found that a novel gene mutation gives rise to the exceptional fur pattern, which they dubbed salmiak, or “salty licorice,” after a popular Finnish treat.
The project was a collaboration between the university, which houses a biobank of 5,000 blood samples from more than 40 feline breeds, a pet care company that makes genetic tests, and cat owners and breeders who offered their companions’ DNA for research.
“Our research approach is community science,” Lohi, who was corresponding author on the study, told CNN. “Often, the research ideas also come from cat owners and breeders having found something interesting in their pets.”
In this case, people first observed the unusual white coloring among Petäjävesi’s cats in 2007. Lohi and his team collected samples from five of them and found that none exhibited the gene variations that typically give rise to white coloration.
To home in on the genetic cause, the researchers sequenced the full genome of two of the cats and discovered a previously unknown mutation that affects a particular gene called KIT.
The team called the gene variant w-sal, for salmiak — black licorice with a speckling of white salt. The researchers tested the salty colored cats and 178 normal-colored samples from the biobank for the new gene variant. Each of the five salmiak cats had two copies of the recessive gene. A few of the other cats had one copy (not resulting in the unique color), and the rest had none.
“One of the fascinating aspects of the study is that it uncovers a really sophisticated way in which this very important KIT gene is normally regulated,” said Greg Barsh, a professor of genetics at Stanford University, who was not involved in the research.
In addition to controlling hair color, the KIT gene encodes for proteins in red blood cells and the cells that become sperm and eggs. Sometimes gene variants that give cats (and dogs) white fur can also cause deafness, though that doesn’t seem to be the case with w-salmiak. It’s one of many ways in which gene mutations that affect hair color can cause issues in other parts of the body.
The larger goal of this and other biobank work, Lohi said, “is to understand the molecular and environmental causes of feline disorders.”
The University of Helsinki has several ongoing projects with Wisdom Panel, which makes pet DNA tests, to look at the genetics of different diseases. And since genes are often similar across mammal species, what Lohi and his team learn could help not just cats but also humans with related medical conditions. None of the study coauthors have a financial stake in Wisdom Panel, Lohi confirmed.
Now that salty licorice cats are officially a thing, could they become the next designer breed?
“It is possible that breeders will choose to develop a population of salty licorice cats,” Lohi said. “However, the health of the salty licorice cats should be followed in more detail to confirm the absence of any color-related health issues.” Tailored genetic testing could be used to ensure the cats are bred without passing on dangerous genes.
“If there are enough people out there that think, ‘Oh, this is really rare and they’re really cute,’ then it could become very popular,” Barsh said. “That is really more of a question having to do with the relationship of humans to companion animals than the science itself.”
Amanda Schupak is a science and health journalist in New York City.
For more CNN news and newsletters create an account at CNN.com


IMAGES
VIDEO
COMMENTS
A genetic mutation is a change to a gene's DNA sequence to produce something different. It creates a permanent change to that gene's DNA sequence. Genetic variations are important for humans to evolve, which is the process of change over generations. A sporadic genetic mutation occurs in one person.
The genome is composed of one to several long molecules of DNA, and mutation can occur potentially anywhere on these molecules at any time. The most serious changes take place in the functional units of DNA, the genes.A mutated form of a gene is called a mutant allele.A gene is typically composed of a regulatory region, which is responsible for turning the gene's transcription on and off at ...
Essay # 2. Gene Mutation: As we come across a really mutated gene only in this type of mutation, sometimes the word mutation is applied to gene mutation or point mutation alone. In nature, genes are constantly duplicating themselves. Every cell division means the formation of new genes—daughter genes from mother genes. In this duplication the ...
Mutations are changes in the information contained in genetic material. For most of life, this means a change in the sequence of DNA, the hereditary material of life. An organism's DNA affects how it looks, how it behaves, its physiology — all aspects of its life. So a change in an organism's DNA can cause changes in all aspects of its life.
Neomorphic mutations are a part of the gain-of-function mutations and are characterized by the control of new protein product synthesis. The newly synthesized gene normally contains a novel gene expression or molecular function. The result of the neomorphic mutation is the gene where the mutation occurs has a complete change in function.
A gene mutation is a permanent change to a DNA sequence that makes it different from the sequence found in other people. A genetic mutation occurs during cell division when the grow and divide and replicate. Mutations can impact anything from a single DNA base pair (a unit of genetic composition) to a significant portion of a chromosome that ...
A mutation is any change to the nucleotide sequence of a DNA molecule. Some mutations arise as DNA is copied. Others are due to environmental factors. A mutation in a gene can change the structure and function of the protein encoded by that gene. This, in turn, can affect an organism's traits.
A mutation is a change in the structure of a gene, the unit of heredity. Genes are made of deoxyribonucleic acid (DNA), a long molecule composed of building blocks called nucleotides.Each . nucleotide is built around one of four different subunits called bases.. These bases are known as guanine, cytosine, adenine, and thymine. A gene carries information in the sequence of its nucleotides, just ...
Definition. A mutation is a change in the DNA sequence of an organism. Mutations can result from errors in DNA replication during cell division, exposure to mutagens or a viral infection. Germline mutations (that occur in eggs and sperm) can be passed on to offspring, while somatic mutations (that occur in body cells) are not passed on.
Mutations that change the regulation of a gene's expression (e.g. promoter mutations) can lead to the production of too much or too little of the resulting protein, which may lead to a noticeable phenotype. Mutations can also occur in the conserved intronic sequences directly adjacent to intron-exon boundaries.
Abstract. Mutation is the engine of evolution in that it generates the genetic variation on which the evolutionary process depends. To understand the evolutionary process we must therefore characterize the rates and patterns of mutation. Starting with the seminal Luria and Delbruck fluctuation experiments in 1943, studies utilizing a variety of ...
Abstract. Population genetics is fundamental to our understanding of evolution, and mutations are essential raw materials for evolution. In this introduction to more detailed papers that follow, we aim to provide an oversight of the field. We review current knowledge on mutation rates and their harmful and beneficial effects on fitness and then ...
Such mutations are likely to be harmful. Harmful mutations may cause genetic disorders or cancer. A genetic disorder is a disease caused by a mutation in one or a few genes. A human example is cystic fibrosis. A mutation in a single gene causes the body to produce thick, sticky mucus that clogs the lungs and blocks ducts in digestive organs.
Genetic testing can also look for changes, or mutations, in a person's DNA that may put a person at risk for a specific disease. The results can help healthcare professionals diagnose conditions.
Gene: ↑ A section of DNA that contains the instructions for a trait. Genetic Diversity: ↑ The overall diversity in the DNA between the individuals of a species. Mutation: ↑ A change in an organism's DNA. This can be a change of a single letter or a much bigger change of hundreds of letters at once.
Mutations drive evolution and were assumed to occur by chance: constantly, gradually, roughly uniformly in genomes, and without regard to environmental inputs, but this view is being revised by discoveries of molecular mechanisms of mutation in bacteria, now translated across the tree of life. These mechanisms reveal a picture of highly regulated mutagenesis, up-regulated temporally by stress ...
A mutation is a change in the nucleotide sequence of a. short region of a genome [1] ( Figure 1 ). Mutation, (a. term coined by Hugo de Yeries in 1900, a rediscover. of Mendels principle s) is ...
evolution. mutation theory, idea that new species are formed from the sudden and unexpected emergence of alterations in their defining traits. Advanced at the beginning of the 20th century by Dutch botanist and geneticist Hugo de Vries in his Die Mutationstheorie (1901-03; The Mutation Theory ), mutation theory joined two seemingly opposed ...
3 A gene drive is a genetic engineering technology—adding, deleting, disrupting, or modifying genes—to rapidly spread a particular genetic trait to an entire offspring population.
These genetic diseases often exhibit a genetic phenomenon known as allelic heterogeneity, in which multiple mutations within the same gene (i.e., alleles) are found to be associated with the same disease. This allelic heterogeneity often is population specific and can represent the unique demographic and mutational history of the population.
Genetic variation refers to differences among the genomes of members of the same species. A genome is all the hereditary information—all the genes —of an organism. For instance, the human genome contains somewhere between twenty and twenty-five thousand genes. Genes are units of hereditary information, and they carry instructions for ...
Genetic disorders occur when a mutation affects your genes or chromosomes. Some disorders cause symptoms at birth, while others develop over time. Genetic testing can help you learn more about the likelihood of experiencing a genetic disorder. If you or a loved one have a genetic disorder, it's important to seek care from an experienced ...
Definition. Mutations in DNA can result in genetic disorders. Genetic disorders are conditions that occur as a result of changes to or mutations in DNA within the body's cells. Most cells in the ...
Mutations in proteins that control gene expression are common across human cancers, but we don't understand how these mutations affect gene expression, contribute to disease, or impact response to treatments. In this study, we focused on identifying the direct gene targets of the transcription factor FOXO1 and how mutations in FOXO1 found in ...
The essay discusses how mutations, genetic drift, and gene flow contribute to evolution, with natural selection being the most visible mechanism. Examples such as antibiotic resistance and the peppered moth illustrate these concepts. The essay underscores the importance of understanding these mechanisms to appreciate the complexity and dynamic ...
Mutations in this gene were found to be associated with a syndrome with symptoms that include intellectual disability, seizures, short stature, neurodevelopmental delay, drooling, motor delay ...
Mutations in proteins that control gene expression are common across human cancers, but we don't understand how these mutations affect gene expression, contribute to disease, or impact response to ...
Researchers have identified a neurodevelopmental disorder, caused by mutations in a single gene, that affects tens of thousands of people worldwide. The findings will improve clinical diagnostic ...
Scientists find a likely cause of many unexplained cases of intellectual disability: A genetic disorder. Many neurodevelopmental issues have no known cause. But new research points to mutations in ...
To home in on the genetic cause, the researchers sequenced the full genome of two of the cats and discovered a previously unknown mutation that affects a particular gene called KIT. The team ...