Get science-backed answers as you write with Paperpal's Research feature

What is a Literature Review? How to Write It (with Examples)

A literature review is a critical analysis and synthesis of existing research on a particular topic. It provides an overview of the current state of knowledge, identifies gaps, and highlights key findings in the literature. 1 The purpose of a literature review is to situate your own research within the context of existing scholarship, demonstrating your understanding of the topic and showing how your work contributes to the ongoing conversation in the field. Learning how to write a literature review is a critical tool for successful research. Your ability to summarize and synthesize prior research pertaining to a certain topic demonstrates your grasp on the topic of study, and assists in the learning process.
Table of Contents
- What is the purpose of literature review?
- a. Habitat Loss and Species Extinction:
- b. Range Shifts and Phenological Changes:
- c. Ocean Acidification and Coral Reefs:
- d. Adaptive Strategies and Conservation Efforts:
How to write a good literature review
- Choose a Topic and Define the Research Question:
- Decide on the Scope of Your Review:
- Select Databases for Searches:
- Conduct Searches and Keep Track:
- Review the Literature:
- Organize and Write Your Literature Review:
- How to write a literature review faster with Paperpal?
- Frequently asked questions
What is a literature review?
A well-conducted literature review demonstrates the researcher’s familiarity with the existing literature, establishes the context for their own research, and contributes to scholarly conversations on the topic. One of the purposes of a literature review is also to help researchers avoid duplicating previous work and ensure that their research is informed by and builds upon the existing body of knowledge.

What is the purpose of literature review?
A literature review serves several important purposes within academic and research contexts. Here are some key objectives and functions of a literature review: 2
1. Contextualizing the Research Problem: The literature review provides a background and context for the research problem under investigation. It helps to situate the study within the existing body of knowledge.
2. Identifying Gaps in Knowledge: By identifying gaps, contradictions, or areas requiring further research, the researcher can shape the research question and justify the significance of the study. This is crucial for ensuring that the new research contributes something novel to the field.
Find academic papers related to your research topic faster. Try Research on Paperpal
3. Understanding Theoretical and Conceptual Frameworks: Literature reviews help researchers gain an understanding of the theoretical and conceptual frameworks used in previous studies. This aids in the development of a theoretical framework for the current research.
4. Providing Methodological Insights: Another purpose of literature reviews is that it allows researchers to learn about the methodologies employed in previous studies. This can help in choosing appropriate research methods for the current study and avoiding pitfalls that others may have encountered.
5. Establishing Credibility: A well-conducted literature review demonstrates the researcher’s familiarity with existing scholarship, establishing their credibility and expertise in the field. It also helps in building a solid foundation for the new research.
6. Informing Hypotheses or Research Questions: The literature review guides the formulation of hypotheses or research questions by highlighting relevant findings and areas of uncertainty in existing literature.
Literature review example
Let’s delve deeper with a literature review example: Let’s say your literature review is about the impact of climate change on biodiversity. You might format your literature review into sections such as the effects of climate change on habitat loss and species extinction, phenological changes, and marine biodiversity. Each section would then summarize and analyze relevant studies in those areas, highlighting key findings and identifying gaps in the research. The review would conclude by emphasizing the need for further research on specific aspects of the relationship between climate change and biodiversity. The following literature review template provides a glimpse into the recommended literature review structure and content, demonstrating how research findings are organized around specific themes within a broader topic.
Literature Review on Climate Change Impacts on Biodiversity:
Climate change is a global phenomenon with far-reaching consequences, including significant impacts on biodiversity. This literature review synthesizes key findings from various studies:
a. Habitat Loss and Species Extinction:
Climate change-induced alterations in temperature and precipitation patterns contribute to habitat loss, affecting numerous species (Thomas et al., 2004). The review discusses how these changes increase the risk of extinction, particularly for species with specific habitat requirements.
b. Range Shifts and Phenological Changes:
Observations of range shifts and changes in the timing of biological events (phenology) are documented in response to changing climatic conditions (Parmesan & Yohe, 2003). These shifts affect ecosystems and may lead to mismatches between species and their resources.
c. Ocean Acidification and Coral Reefs:
The review explores the impact of climate change on marine biodiversity, emphasizing ocean acidification’s threat to coral reefs (Hoegh-Guldberg et al., 2007). Changes in pH levels negatively affect coral calcification, disrupting the delicate balance of marine ecosystems.
d. Adaptive Strategies and Conservation Efforts:
Recognizing the urgency of the situation, the literature review discusses various adaptive strategies adopted by species and conservation efforts aimed at mitigating the impacts of climate change on biodiversity (Hannah et al., 2007). It emphasizes the importance of interdisciplinary approaches for effective conservation planning.

Strengthen your literature review with factual insights. Try Research on Paperpal for free!
Writing a literature review involves summarizing and synthesizing existing research on a particular topic. A good literature review format should include the following elements.
Introduction: The introduction sets the stage for your literature review, providing context and introducing the main focus of your review.
- Opening Statement: Begin with a general statement about the broader topic and its significance in the field.
- Scope and Purpose: Clearly define the scope of your literature review. Explain the specific research question or objective you aim to address.
- Organizational Framework: Briefly outline the structure of your literature review, indicating how you will categorize and discuss the existing research.
- Significance of the Study: Highlight why your literature review is important and how it contributes to the understanding of the chosen topic.
- Thesis Statement: Conclude the introduction with a concise thesis statement that outlines the main argument or perspective you will develop in the body of the literature review.
Body: The body of the literature review is where you provide a comprehensive analysis of existing literature, grouping studies based on themes, methodologies, or other relevant criteria.
- Organize by Theme or Concept: Group studies that share common themes, concepts, or methodologies. Discuss each theme or concept in detail, summarizing key findings and identifying gaps or areas of disagreement.
- Critical Analysis: Evaluate the strengths and weaknesses of each study. Discuss the methodologies used, the quality of evidence, and the overall contribution of each work to the understanding of the topic.
- Synthesis of Findings: Synthesize the information from different studies to highlight trends, patterns, or areas of consensus in the literature.
- Identification of Gaps: Discuss any gaps or limitations in the existing research and explain how your review contributes to filling these gaps.
- Transition between Sections: Provide smooth transitions between different themes or concepts to maintain the flow of your literature review.
Write and Cite as you go with Paperpal Research. Start now for free.
Conclusion: The conclusion of your literature review should summarize the main findings, highlight the contributions of the review, and suggest avenues for future research.
- Summary of Key Findings: Recap the main findings from the literature and restate how they contribute to your research question or objective.
- Contributions to the Field: Discuss the overall contribution of your literature review to the existing knowledge in the field.
- Implications and Applications: Explore the practical implications of the findings and suggest how they might impact future research or practice.
- Recommendations for Future Research: Identify areas that require further investigation and propose potential directions for future research in the field.
- Final Thoughts: Conclude with a final reflection on the importance of your literature review and its relevance to the broader academic community.

Conducting a literature review
Conducting a literature review is an essential step in research that involves reviewing and analyzing existing literature on a specific topic. It’s important to know how to do a literature review effectively, so here are the steps to follow: 1
Choose a Topic and Define the Research Question:
- Select a topic that is relevant to your field of study.
- Clearly define your research question or objective. Determine what specific aspect of the topic do you want to explore?
Decide on the Scope of Your Review:
- Determine the timeframe for your literature review. Are you focusing on recent developments, or do you want a historical overview?
- Consider the geographical scope. Is your review global, or are you focusing on a specific region?
- Define the inclusion and exclusion criteria. What types of sources will you include? Are there specific types of studies or publications you will exclude?
Select Databases for Searches:
- Identify relevant databases for your field. Examples include PubMed, IEEE Xplore, Scopus, Web of Science, and Google Scholar.
- Consider searching in library catalogs, institutional repositories, and specialized databases related to your topic.
Conduct Searches and Keep Track:
- Develop a systematic search strategy using keywords, Boolean operators (AND, OR, NOT), and other search techniques.
- Record and document your search strategy for transparency and replicability.
- Keep track of the articles, including publication details, abstracts, and links. Use citation management tools like EndNote, Zotero, or Mendeley to organize your references.
Review the Literature:
- Evaluate the relevance and quality of each source. Consider the methodology, sample size, and results of studies.
- Organize the literature by themes or key concepts. Identify patterns, trends, and gaps in the existing research.
- Summarize key findings and arguments from each source. Compare and contrast different perspectives.
- Identify areas where there is a consensus in the literature and where there are conflicting opinions.
- Provide critical analysis and synthesis of the literature. What are the strengths and weaknesses of existing research?
Organize and Write Your Literature Review:
- Literature review outline should be based on themes, chronological order, or methodological approaches.
- Write a clear and coherent narrative that synthesizes the information gathered.
- Use proper citations for each source and ensure consistency in your citation style (APA, MLA, Chicago, etc.).
- Conclude your literature review by summarizing key findings, identifying gaps, and suggesting areas for future research.
Whether you’re exploring a new research field or finding new angles to develop an existing topic, sifting through hundreds of papers can take more time than you have to spare. But what if you could find science-backed insights with verified citations in seconds? That’s the power of Paperpal’s new Research feature!
How to write a literature review faster with Paperpal?
Paperpal, an AI writing assistant, integrates powerful academic search capabilities within its writing platform. With the Research feature, you get 100% factual insights, with citations backed by 250M+ verified research articles, directly within your writing interface with the option to save relevant references in your Citation Library. By eliminating the need to switch tabs to find answers to all your research questions, Paperpal saves time and helps you stay focused on your writing.
Here’s how to use the Research feature:
- Ask a question: Get started with a new document on paperpal.com. Click on the “Research” feature and type your question in plain English. Paperpal will scour over 250 million research articles, including conference papers and preprints, to provide you with accurate insights and citations.
- Review and Save: Paperpal summarizes the information, while citing sources and listing relevant reads. You can quickly scan the results to identify relevant references and save these directly to your built-in citations library for later access.
- Cite with Confidence: Paperpal makes it easy to incorporate relevant citations and references into your writing, ensuring your arguments are well-supported by credible sources. This translates to a polished, well-researched literature review.
The literature review sample and detailed advice on writing and conducting a review will help you produce a well-structured report. But remember that a good literature review is an ongoing process, and it may be necessary to revisit and update it as your research progresses. By combining effortless research with an easy citation process, Paperpal Research streamlines the literature review process and empowers you to write faster and with more confidence. Try Paperpal Research now and see for yourself.
Frequently asked questions
A literature review is a critical and comprehensive analysis of existing literature (published and unpublished works) on a specific topic or research question and provides a synthesis of the current state of knowledge in a particular field. A well-conducted literature review is crucial for researchers to build upon existing knowledge, avoid duplication of efforts, and contribute to the advancement of their field. It also helps researchers situate their work within a broader context and facilitates the development of a sound theoretical and conceptual framework for their studies.
Literature review is a crucial component of research writing, providing a solid background for a research paper’s investigation. The aim is to keep professionals up to date by providing an understanding of ongoing developments within a specific field, including research methods, and experimental techniques used in that field, and present that knowledge in the form of a written report. Also, the depth and breadth of the literature review emphasizes the credibility of the scholar in his or her field.
Before writing a literature review, it’s essential to undertake several preparatory steps to ensure that your review is well-researched, organized, and focused. This includes choosing a topic of general interest to you and doing exploratory research on that topic, writing an annotated bibliography, and noting major points, especially those that relate to the position you have taken on the topic.
Literature reviews and academic research papers are essential components of scholarly work but serve different purposes within the academic realm. 3 A literature review aims to provide a foundation for understanding the current state of research on a particular topic, identify gaps or controversies, and lay the groundwork for future research. Therefore, it draws heavily from existing academic sources, including books, journal articles, and other scholarly publications. In contrast, an academic research paper aims to present new knowledge, contribute to the academic discourse, and advance the understanding of a specific research question. Therefore, it involves a mix of existing literature (in the introduction and literature review sections) and original data or findings obtained through research methods.
Literature reviews are essential components of academic and research papers, and various strategies can be employed to conduct them effectively. If you want to know how to write a literature review for a research paper, here are four common approaches that are often used by researchers. Chronological Review: This strategy involves organizing the literature based on the chronological order of publication. It helps to trace the development of a topic over time, showing how ideas, theories, and research have evolved. Thematic Review: Thematic reviews focus on identifying and analyzing themes or topics that cut across different studies. Instead of organizing the literature chronologically, it is grouped by key themes or concepts, allowing for a comprehensive exploration of various aspects of the topic. Methodological Review: This strategy involves organizing the literature based on the research methods employed in different studies. It helps to highlight the strengths and weaknesses of various methodologies and allows the reader to evaluate the reliability and validity of the research findings. Theoretical Review: A theoretical review examines the literature based on the theoretical frameworks used in different studies. This approach helps to identify the key theories that have been applied to the topic and assess their contributions to the understanding of the subject. It’s important to note that these strategies are not mutually exclusive, and a literature review may combine elements of more than one approach. The choice of strategy depends on the research question, the nature of the literature available, and the goals of the review. Additionally, other strategies, such as integrative reviews or systematic reviews, may be employed depending on the specific requirements of the research.
The literature review format can vary depending on the specific publication guidelines. However, there are some common elements and structures that are often followed. Here is a general guideline for the format of a literature review: Introduction: Provide an overview of the topic. Define the scope and purpose of the literature review. State the research question or objective. Body: Organize the literature by themes, concepts, or chronology. Critically analyze and evaluate each source. Discuss the strengths and weaknesses of the studies. Highlight any methodological limitations or biases. Identify patterns, connections, or contradictions in the existing research. Conclusion: Summarize the key points discussed in the literature review. Highlight the research gap. Address the research question or objective stated in the introduction. Highlight the contributions of the review and suggest directions for future research.
Both annotated bibliographies and literature reviews involve the examination of scholarly sources. While annotated bibliographies focus on individual sources with brief annotations, literature reviews provide a more in-depth, integrated, and comprehensive analysis of existing literature on a specific topic. The key differences are as follows:
References
- Denney, A. S., & Tewksbury, R. (2013). How to write a literature review. Journal of criminal justice education , 24 (2), 218-234.
- Pan, M. L. (2016). Preparing literature reviews: Qualitative and quantitative approaches . Taylor & Francis.
- Cantero, C. (2019). How to write a literature review. San José State University Writing Center .
Paperpal is an AI writing assistant that help academics write better, faster with real-time suggestions for in-depth language and grammar correction. Trained on millions of research manuscripts enhanced by professional academic editors, Paperpal delivers human precision at machine speed.
Try it for free or upgrade to Paperpal Prime , which unlocks unlimited access to premium features like academic translation, paraphrasing, contextual synonyms, consistency checks and more. It’s like always having a professional academic editor by your side! Go beyond limitations and experience the future of academic writing. Get Paperpal Prime now at just US$19 a month!
Related Reads:
- Empirical Research: A Comprehensive Guide for Academics
- How to Write a Scientific Paper in 10 Steps
- How Long Should a Chapter Be?
- How to Use Paperpal to Generate Emails & Cover Letters?
6 Tips for Post-Doc Researchers to Take Their Career to the Next Level
Self-plagiarism in research: what it is and how to avoid it, you may also like, how to write a high-quality conference paper, how paperpal’s research feature helps you develop and..., how paperpal is enhancing academic productivity and accelerating..., how to write a successful book chapter for..., academic editing: how to self-edit academic text with..., 4 ways paperpal encourages responsible writing with ai, what are scholarly sources and where can you..., how to write a hypothesis types and examples , measuring academic success: definition & strategies for excellence, what is academic writing: tips for students.
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, automatically generate references for free.
- Knowledge Base
- Dissertation
- What is a Literature Review? | Guide, Template, & Examples
What is a Literature Review? | Guide, Template, & Examples
Published on 22 February 2022 by Shona McCombes . Revised on 7 June 2022.
What is a literature review? A literature review is a survey of scholarly sources on a specific topic. It provides an overview of current knowledge, allowing you to identify relevant theories, methods, and gaps in the existing research.
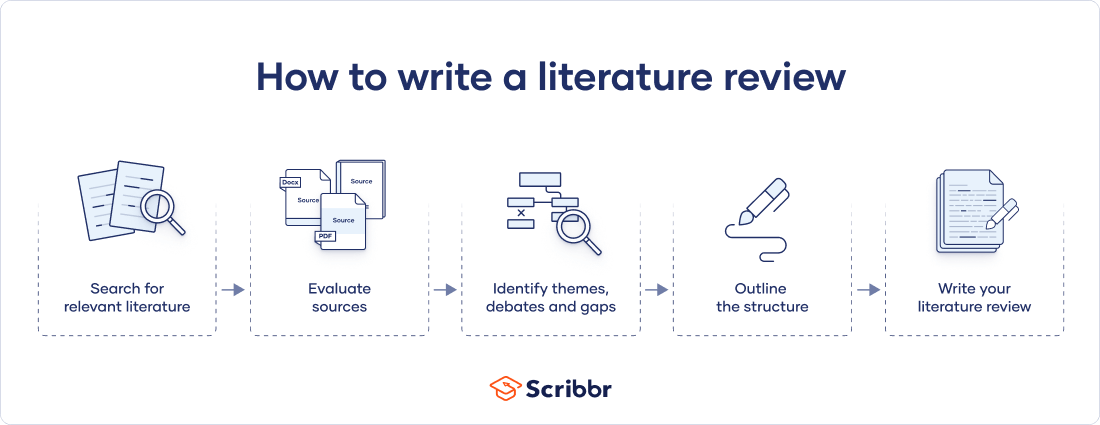
There are five key steps to writing a literature review:
- Search for relevant literature
- Evaluate sources
- Identify themes, debates and gaps
- Outline the structure
- Write your literature review
A good literature review doesn’t just summarise sources – it analyses, synthesises, and critically evaluates to give a clear picture of the state of knowledge on the subject.
Instantly correct all language mistakes in your text
Be assured that you'll submit flawless writing. Upload your document to correct all your mistakes.
Table of contents
Why write a literature review, examples of literature reviews, step 1: search for relevant literature, step 2: evaluate and select sources, step 3: identify themes, debates and gaps, step 4: outline your literature review’s structure, step 5: write your literature review, frequently asked questions about literature reviews, introduction.
- Quick Run-through
- Step 1 & 2
When you write a dissertation or thesis, you will have to conduct a literature review to situate your research within existing knowledge. The literature review gives you a chance to:
- Demonstrate your familiarity with the topic and scholarly context
- Develop a theoretical framework and methodology for your research
- Position yourself in relation to other researchers and theorists
- Show how your dissertation addresses a gap or contributes to a debate
You might also have to write a literature review as a stand-alone assignment. In this case, the purpose is to evaluate the current state of research and demonstrate your knowledge of scholarly debates around a topic.
The content will look slightly different in each case, but the process of conducting a literature review follows the same steps. We’ve written a step-by-step guide that you can follow below.
The only proofreading tool specialized in correcting academic writing
The academic proofreading tool has been trained on 1000s of academic texts and by native English editors. Making it the most accurate and reliable proofreading tool for students.
Correct my document today
Writing literature reviews can be quite challenging! A good starting point could be to look at some examples, depending on what kind of literature review you’d like to write.
- Example literature review #1: “Why Do People Migrate? A Review of the Theoretical Literature” ( Theoretical literature review about the development of economic migration theory from the 1950s to today.)
- Example literature review #2: “Literature review as a research methodology: An overview and guidelines” ( Methodological literature review about interdisciplinary knowledge acquisition and production.)
- Example literature review #3: “The Use of Technology in English Language Learning: A Literature Review” ( Thematic literature review about the effects of technology on language acquisition.)
- Example literature review #4: “Learners’ Listening Comprehension Difficulties in English Language Learning: A Literature Review” ( Chronological literature review about how the concept of listening skills has changed over time.)
You can also check out our templates with literature review examples and sample outlines at the links below.
Download Word doc Download Google doc
Before you begin searching for literature, you need a clearly defined topic .
If you are writing the literature review section of a dissertation or research paper, you will search for literature related to your research objectives and questions .
If you are writing a literature review as a stand-alone assignment, you will have to choose a focus and develop a central question to direct your search. Unlike a dissertation research question, this question has to be answerable without collecting original data. You should be able to answer it based only on a review of existing publications.
Make a list of keywords
Start by creating a list of keywords related to your research topic. Include each of the key concepts or variables you’re interested in, and list any synonyms and related terms. You can add to this list if you discover new keywords in the process of your literature search.
- Social media, Facebook, Instagram, Twitter, Snapchat, TikTok
- Body image, self-perception, self-esteem, mental health
- Generation Z, teenagers, adolescents, youth
Search for relevant sources
Use your keywords to begin searching for sources. Some databases to search for journals and articles include:
- Your university’s library catalogue
- Google Scholar
- Project Muse (humanities and social sciences)
- Medline (life sciences and biomedicine)
- EconLit (economics)
- Inspec (physics, engineering and computer science)
You can use boolean operators to help narrow down your search:
Read the abstract to find out whether an article is relevant to your question. When you find a useful book or article, you can check the bibliography to find other relevant sources.
To identify the most important publications on your topic, take note of recurring citations. If the same authors, books or articles keep appearing in your reading, make sure to seek them out.
You probably won’t be able to read absolutely everything that has been written on the topic – you’ll have to evaluate which sources are most relevant to your questions.
For each publication, ask yourself:
- What question or problem is the author addressing?
- What are the key concepts and how are they defined?
- What are the key theories, models and methods? Does the research use established frameworks or take an innovative approach?
- What are the results and conclusions of the study?
- How does the publication relate to other literature in the field? Does it confirm, add to, or challenge established knowledge?
- How does the publication contribute to your understanding of the topic? What are its key insights and arguments?
- What are the strengths and weaknesses of the research?
Make sure the sources you use are credible, and make sure you read any landmark studies and major theories in your field of research.
You can find out how many times an article has been cited on Google Scholar – a high citation count means the article has been influential in the field, and should certainly be included in your literature review.
The scope of your review will depend on your topic and discipline: in the sciences you usually only review recent literature, but in the humanities you might take a long historical perspective (for example, to trace how a concept has changed in meaning over time).
Remember that you can use our template to summarise and evaluate sources you’re thinking about using!
Take notes and cite your sources
As you read, you should also begin the writing process. Take notes that you can later incorporate into the text of your literature review.
It’s important to keep track of your sources with references to avoid plagiarism . It can be helpful to make an annotated bibliography, where you compile full reference information and write a paragraph of summary and analysis for each source. This helps you remember what you read and saves time later in the process.
You can use our free APA Reference Generator for quick, correct, consistent citations.
To begin organising your literature review’s argument and structure, you need to understand the connections and relationships between the sources you’ve read. Based on your reading and notes, you can look for:
- Trends and patterns (in theory, method or results): do certain approaches become more or less popular over time?
- Themes: what questions or concepts recur across the literature?
- Debates, conflicts and contradictions: where do sources disagree?
- Pivotal publications: are there any influential theories or studies that changed the direction of the field?
- Gaps: what is missing from the literature? Are there weaknesses that need to be addressed?
This step will help you work out the structure of your literature review and (if applicable) show how your own research will contribute to existing knowledge.
- Most research has focused on young women.
- There is an increasing interest in the visual aspects of social media.
- But there is still a lack of robust research on highly-visual platforms like Instagram and Snapchat – this is a gap that you could address in your own research.
There are various approaches to organising the body of a literature review. You should have a rough idea of your strategy before you start writing.
Depending on the length of your literature review, you can combine several of these strategies (for example, your overall structure might be thematic, but each theme is discussed chronologically).
Chronological
The simplest approach is to trace the development of the topic over time. However, if you choose this strategy, be careful to avoid simply listing and summarising sources in order.
Try to analyse patterns, turning points and key debates that have shaped the direction of the field. Give your interpretation of how and why certain developments occurred.
If you have found some recurring central themes, you can organise your literature review into subsections that address different aspects of the topic.
For example, if you are reviewing literature about inequalities in migrant health outcomes, key themes might include healthcare policy, language barriers, cultural attitudes, legal status, and economic access.
Methodological
If you draw your sources from different disciplines or fields that use a variety of research methods , you might want to compare the results and conclusions that emerge from different approaches. For example:
- Look at what results have emerged in qualitative versus quantitative research
- Discuss how the topic has been approached by empirical versus theoretical scholarship
- Divide the literature into sociological, historical, and cultural sources
Theoretical
A literature review is often the foundation for a theoretical framework . You can use it to discuss various theories, models, and definitions of key concepts.
You might argue for the relevance of a specific theoretical approach, or combine various theoretical concepts to create a framework for your research.
Like any other academic text, your literature review should have an introduction , a main body, and a conclusion . What you include in each depends on the objective of your literature review.
The introduction should clearly establish the focus and purpose of the literature review.
If you are writing the literature review as part of your dissertation or thesis, reiterate your central problem or research question and give a brief summary of the scholarly context. You can emphasise the timeliness of the topic (“many recent studies have focused on the problem of x”) or highlight a gap in the literature (“while there has been much research on x, few researchers have taken y into consideration”).
Depending on the length of your literature review, you might want to divide the body into subsections. You can use a subheading for each theme, time period, or methodological approach.
As you write, make sure to follow these tips:
- Summarise and synthesise: give an overview of the main points of each source and combine them into a coherent whole.
- Analyse and interpret: don’t just paraphrase other researchers – add your own interpretations, discussing the significance of findings in relation to the literature as a whole.
- Critically evaluate: mention the strengths and weaknesses of your sources.
- Write in well-structured paragraphs: use transitions and topic sentences to draw connections, comparisons and contrasts.
In the conclusion, you should summarise the key findings you have taken from the literature and emphasise their significance.
If the literature review is part of your dissertation or thesis, reiterate how your research addresses gaps and contributes new knowledge, or discuss how you have drawn on existing theories and methods to build a framework for your research. This can lead directly into your methodology section.
A literature review is a survey of scholarly sources (such as books, journal articles, and theses) related to a specific topic or research question .
It is often written as part of a dissertation , thesis, research paper , or proposal .
There are several reasons to conduct a literature review at the beginning of a research project:
- To familiarise yourself with the current state of knowledge on your topic
- To ensure that you’re not just repeating what others have already done
- To identify gaps in knowledge and unresolved problems that your research can address
- To develop your theoretical framework and methodology
- To provide an overview of the key findings and debates on the topic
Writing the literature review shows your reader how your work relates to existing research and what new insights it will contribute.
The literature review usually comes near the beginning of your dissertation . After the introduction , it grounds your research in a scholarly field and leads directly to your theoretical framework or methodology .
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the ‘Cite this Scribbr article’ button to automatically add the citation to our free Reference Generator.
McCombes, S. (2022, June 07). What is a Literature Review? | Guide, Template, & Examples. Scribbr. Retrieved 14 May 2024, from https://www.scribbr.co.uk/thesis-dissertation/literature-review/
Is this article helpful?
Shona McCombes
Other students also liked, how to write a dissertation proposal | a step-by-step guide, what is a theoretical framework | a step-by-step guide, what is a research methodology | steps & tips.
State-of-the-art literature review methodology: A six-step approach for knowledge synthesis
- Original Article
- Open access
- Published: 05 September 2022
- Volume 11 , pages 281–288, ( 2022 )
Cite this article
You have full access to this open access article

- Erin S. Barry ORCID: orcid.org/0000-0003-0788-7153 1 , 2 ,
- Jerusalem Merkebu ORCID: orcid.org/0000-0003-3707-8920 3 &
- Lara Varpio ORCID: orcid.org/0000-0002-1412-4341 3
27k Accesses
8 Citations
18 Altmetric
Explore all metrics
Introduction
Researchers and practitioners rely on literature reviews to synthesize large bodies of knowledge. Many types of literature reviews have been developed, each targeting a specific purpose. However, these syntheses are hampered if the review type’s paradigmatic roots, methods, and markers of rigor are only vaguely understood. One literature review type whose methodology has yet to be elucidated is the state-of-the-art (SotA) review. If medical educators are to harness SotA reviews to generate knowledge syntheses, we must understand and articulate the paradigmatic roots of, and methods for, conducting SotA reviews.
We reviewed 940 articles published between 2014–2021 labeled as SotA reviews. We (a) identified all SotA methods-related resources, (b) examined the foundational principles and techniques underpinning the reviews, and (c) combined our findings to inductively analyze and articulate the philosophical foundations, process steps, and markers of rigor.
In the 940 articles reviewed, nearly all manuscripts (98%) lacked citations for how to conduct a SotA review. The term “state of the art” was used in 4 different ways. Analysis revealed that SotA articles are grounded in relativism and subjectivism.
This article provides a 6-step approach for conducting SotA reviews. SotA reviews offer an interpretive synthesis that describes: This is where we are now. This is how we got here. This is where we could be going. This chronologically rooted narrative synthesis provides a methodology for reviewing large bodies of literature to explore why and how our current knowledge has developed and to offer new research directions.
Similar content being viewed by others

A worked example of Braun and Clarke’s approach to reflexive thematic analysis

Qualitative Research: Ethical Considerations
Criteria for Good Qualitative Research: A Comprehensive Review
Avoid common mistakes on your manuscript.
Literature reviews play a foundational role in scientific research; they support knowledge advancement by collecting, describing, analyzing, and integrating large bodies of information and data [ 1 , 2 ]. Indeed, as Snyder [ 3 ] argues, all scientific disciplines require literature reviews grounded in a methodology that is accurate and clearly reported. Many types of literature reviews have been developed, each with a unique purpose, distinct methods, and distinguishing characteristics of quality and rigor [ 4 , 5 ].
Each review type offers valuable insights if rigorously conducted [ 3 , 6 ]. Problematically, this is not consistently the case, and the consequences can be dire. Medical education’s policy makers and institutional leaders rely on knowledge syntheses to inform decision making [ 7 ]. Medical education curricula are shaped by these syntheses. Our accreditation standards are informed by these integrations. Our patient care is guided by these knowledge consolidations [ 8 ]. Clearly, it is important for knowledge syntheses to be held to the highest standards of rigor. And yet, that standard is not always maintained. Sometimes scholars fail to meet the review’s specified standards of rigor; other times the markers of rigor have never been explicitly articulated. While we can do little about the former, we can address the latter. One popular literature review type whose methodology has yet to be fully described, vetted, and justified is the state-of-the-art (SotA) review.
While many types of literature reviews amalgamate bodies of literature, SotA reviews offer something unique. By looking across the historical development of a body of knowledge, SotA reviews delves into questions like: Why did our knowledge evolve in this way? What other directions might our investigations have taken? What turning points in our thinking should we revisit to gain new insights? A SotA review—a form of narrative knowledge synthesis [ 5 , 9 ]—acknowledges that history reflects a series of decisions and then asks what different decisions might have been made.
SotA reviews are frequently used in many fields including the biomedical sciences [ 10 , 11 ], medicine [ 12 , 13 , 14 ], and engineering [ 15 , 16 ]. However, SotA reviews are rarely seen in medical education; indeed, a bibliometrics analysis of literature reviews published in 14 core medical education journals between 1999 and 2019 reported only 5 SotA reviews out of the 963 knowledge syntheses identified [ 17 ]. This is not to say that SotA reviews are absent; we suggest that they are often unlabeled. For instance, Schuwirth and van der Vleuten’s article “A history of assessment in medical education” [ 14 ] offers a temporally organized overview of the field’s evolving thinking about assessment. Similarly, McGaghie et al. published a chronologically structured review of simulation-based medical education research that “reviews and critically evaluates historical and contemporary research on simulation-based medical education” [ 18 , p. 50]. SotA reviews certainly have a place in medical education, even if that place is not explicitly signaled.
This lack of labeling is problematic since it conceals the purpose of, and work involved in, the SotA review synthesis. In a SotA review, the author(s) collects and analyzes the historical development of a field’s knowledge about a phenomenon, deconstructs how that understanding evolved, questions why it unfolded in specific ways, and posits new directions for research. Senior medical education scholars use SotA reviews to share their insights based on decades of work on a topic [ 14 , 18 ]; their junior counterparts use them to critique that history and propose new directions [ 19 ]. And yet, SotA reviews are generally not explicitly signaled in medical education. We suggest that at least two factors contribute to this problem. First, it may be that medical education scholars have yet to fully grasp the unique contributions SotA reviews provide. Second, the methodology and methods of SotA reviews are poorly reported making this form of knowledge synthesis appear to lack rigor. Both factors are rooted in the same foundational problem: insufficient clarity about SotA reviews. In this study, we describe SotA review methodology so that medical educators can explicitly use this form of knowledge synthesis to further advance the field.
We developed a four-step research design to meet this goal, illustrated in Fig. 1 .
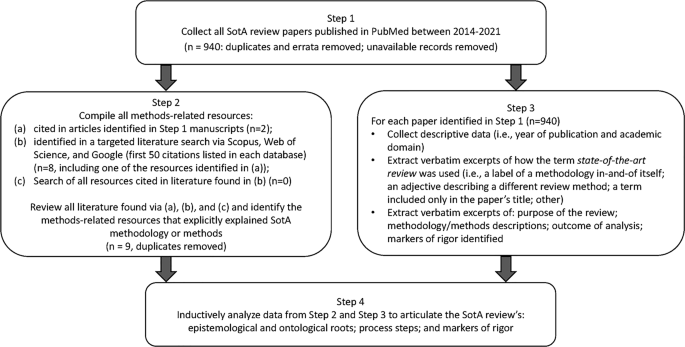
Four-step research design process used for developing a State-of-the-Art literature review methodology
Step 1: Collect SotA articles
To build our initial corpus of articles reporting SotA reviews, we searched PubMed using the strategy (″state of the art review″[ti] OR ″state of the art review*″) and limiting our search to English articles published between 2014 and 2021. We strategically focused on PubMed, which includes MEDLINE, and is considered the National Library of Medicine’s premier database of biomedical literature and indexes health professions education and practice literature [ 20 ]. We limited our search to 2014–2021 to capture modern use of SotA reviews. Of the 960 articles identified, nine were excluded because they were duplicates, erratum, or corrigendum records; full text copies were unavailable for 11 records. All articles identified ( n = 940) constituted the corpus for analysis.
Step 2: Compile all methods-related resources
EB, JM, or LV independently reviewed the 940 full-text articles to identify all references to resources that explained, informed, described, or otherwise supported the methods used for conducting the SotA review. Articles that met our criteria were obtained for analysis.
To ensure comprehensive retrieval, we also searched Scopus and Web of Science. Additionally, to find resources not indexed by these academic databases, we searched Google (see Electronic Supplementary Material [ESM] for the search strategies used for each database). EB also reviewed the first 50 items retrieved from each search looking for additional relevant resources. None were identified. Via these strategies, nine articles were identified and added to the collection of methods-related resources for analysis.
Step 3: Extract data for analysis
In Step 3, we extracted three kinds of information from the 940 articles papers identified in Step 1. First, descriptive data on each article were compiled (i.e., year of publication and the academic domain targeted by the journal). Second, each article was examined and excerpts collected about how the term state-of-the-art review was used (i.e., as a label for a methodology in-and-of itself; as an adjective qualifying another type of literature review; as a term included in the paper’s title only; or in some other way). Finally, we extracted excerpts describing: the purposes and/or aims of the SotA review; the methodology informing and methods processes used to carry out the SotA review; outcomes of analyses; and markers of rigor for the SotA review.
Two researchers (EB and JM) coded 69 articles and an interrater reliability of 94.2% was achieved. Any discrepancies were discussed. Given the high interrater reliability, the two authors split the remaining articles and coded independently.
Step 4: Construct the SotA review methodology
The methods-related resources identified in Step 2 and the data extractions from Step 3 were inductively analyzed by LV and EB to identify statements and research processes that revealed the ontology (i.e., the nature of reality that was reflected) and the epistemology (i.e., the nature of knowledge) underpinning the descriptions of the reviews. These authors studied these data to determine if the synthesis adhered to an objectivist or a subjectivist orientation, and to synthesize the purposes realized in these papers.
To confirm these interpretations, LV and EB compared their ontology, epistemology, and purpose determinations against two expectations commonly required of objectivist synthesis methods (e.g., systematic reviews): an exhaustive search strategy and an appraisal of the quality of the research data. These expectations were considered indicators of a realist ontology and objectivist epistemology [ 21 ] (i.e., that a single correct understanding of the topic can be sought through objective data collection {e.g., systematic reviews [ 22 ]}). Conversely, the inverse of these expectations were considered indicators of a relativist ontology and subjectivist epistemology [ 21 ] (i.e., that no single correct understanding of the topic is available; there are multiple valid understandings that can be generated and so a subjective interpretation of the literature is sought {e.g., narrative reviews [ 9 ]}).
Once these interpretations were confirmed, LV and EB reviewed and consolidated the methods steps described in these data. Markers of rigor were then developed that aligned with the ontology, epistemology, and methods of SotA reviews.
Of the 940 articles identified in Step 1, 98% ( n = 923) lacked citations or other references to resources that explained, informed, or otherwise supported the SotA review process. Of the 17 articles that included supporting information, 16 cited Grant and Booth’s description [ 4 ] consisting of five sentences describing the overall purpose of SotA reviews, three sentences noting perceived strengths, and four sentences articulating perceived weaknesses. This resource provides no guidance on how to conduct a SotA review methodology nor markers of rigor. The one article not referencing Grant and Booth used “an adapted comparative effectiveness research search strategy that was adapted by a health sciences librarian” [ 23 , p. 381]. One website citation was listed in support of this strategy; however, the page was no longer available in summer 2021. We determined that the corpus was uninformed by a cardinal resource or a publicly available methodology description.
In Step 2 we identified nine resources [ 4 , 5 , 24 , 25 , 26 , 27 , 28 ]; none described the methodology and/or processes of carrying out SotA reviews. Nor did they offer explicit descriptions of the ontology or epistemology underpinning SotA reviews. Instead, these resources provided short overview statements (none longer than one paragraph) about the review type [ 4 , 5 , 24 , 25 , 26 , 27 , 28 ]. Thus, we determined that, to date, there are no available methodology papers describing how to conduct a SotA review.
Step 3 revealed that “state of the art” was used in 4 different ways across the 940 articles (see Fig. 2 for the frequency with which each was used). In 71% ( n = 665 articles), the phrase was used only in the title, abstract, and/or purpose statement of the article; the phrase did not appear elsewhere in the paper and no SotA methodology was discussed. Nine percent ( n = 84) used the phrase as an adjective to qualify another literature review type and so relied entirely on the methodology of a different knowledge synthesis approach (e.g., “a state of the art systematic review [ 29 ]”). In 5% ( n = 52) of the articles, the phrase was not used anywhere within the article; instead, “state of the art” was the type of article within a journal. In the remaining 15% ( n = 139), the phrase denoted a specific methodology (see ESM for all methodology articles). Via Step 4’s inductive analysis, the following foundational principles of SotA reviews were developed: (1) the ontology, (2) epistemology, and (3) purpose of SotA reviews.
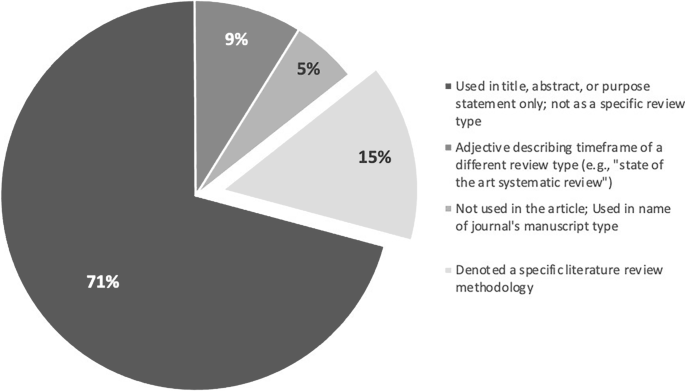
Four ways the term “state of the art” is used in the corpus and how frequently each is used
Ontology of SotA reviews: Relativism
SotA reviews rest on four propositions:
The literature addressing a phenomenon offers multiple perspectives on that topic (i.e., different groups of researchers may hold differing opinions and/or interpretations of data about a phenomenon).
The reality of the phenomenon itself cannot be completely perceived or understood (i.e., due to limitations [e.g., the capabilities of current technologies, a research team’s disciplinary orientation] we can only perceive a limited part of the phenomenon).
The reality of the phenomenon is a subjective and inter-subjective construction (i.e., what we understand about a phenomenon is built by individuals and so their individual subjectivities shape that understanding).
The context in which the review was conducted informs the review (e.g., a SotA review of literature about gender identity and sexual function will be synthesized differently by researchers in the domain of gender studies than by scholars working in sex reassignment surgery).
As these propositions suggest, SotA scholars bring their experiences, expectations, research purposes, and social (including academic) orientations to bear on the synthesis work. In other words, a SotA review synthesizes the literature based on a specific orientation to the topic being addressed. For instance, a SotA review written by senior scholars who are experts in the field of medical education may reflect on the turning points that have shaped the way our field has evolved the modern practices of learner assessment, noting how the nature of the problem of assessment has moved: it was first a measurement problem, then a problem that embraced human judgment but needed assessment expertise, and now a whole system problem that is to be addressed from an integrated—not a reductionist—perspective [ 12 ]. However, if other scholars were to examine this same history from a technological orientation, learner assessment could be framed as historically constricted by the media available through which to conduct assessment, pointing to how artificial intelligence is laying the foundation for the next wave of assessment in medical education [ 30 ].
Given these foundational propositions, SotA reviews are steeped in a relativist ontology—i.e., reality is socially and experientially informed and constructed, and so no single objective truth exists. Researchers’ interpretations reflect their conceptualization of the literature—a conceptualization that could change over time and that could conflict with the understandings of others.
Epistemology of SotA reviews: Subjectivism
SotA reviews embrace subjectivism. The knowledge generated through the review is value-dependent, growing out of the subjective interpretations of the researcher(s) who conducted the synthesis. The SotA review generates an interpretation of the data that is informed by the expertise, experiences, and social contexts of the researcher(s). Furthermore, the knowledge developed through SotA reviews is shaped by the historical point in time when the review was conducted. SotA reviews are thus steeped in the perspective that knowledge is shaped by individuals and their community, and is a synthesis that will change over time.
Purpose of SotA reviews
SotA reviews create a subjectively informed summary of modern thinking about a topic. As a chronologically ordered synthesis, SotA reviews describe the history of turning points in researchers’ understanding of a phenomenon to contextualize a description of modern scientific thinking on the topic. The review presents an argument about how the literature could be interpreted; it is not a definitive statement about how the literature should or must be interpreted. A SotA review explores: the pivotal points shaping the historical development of a topic, the factors that informed those changes in understanding, and the ways of thinking about and studying the topic that could inform the generation of further insights. In other words, the purpose of SotA reviews is to create a three-part argument: This is where we are now in our understanding of this topic. This is how we got here. This is where we could go next.
The SotA methodology
Based on study findings and analyses, we constructed a six-stage SotA review methodology. This six-stage approach is summarized and guiding questions are offered in Tab. 1 .
Stage 1: Determine initial research question and field of inquiry
In Stage 1, the researcher(s) creates an initial description of the topic to be summarized and so must determine what field of knowledge (and/or practice) the search will address. Knowledge developed through the SotA review process is shaped by the context informing it; thus, knowing the domain in which the review will be conducted is part of the review’s foundational work.
Stage 2: Determine timeframe
This stage involves determining the period of time that will be defined as SotA for the topic being summarized. The researcher(s) should engage in a broad-scope overview of the literature, reading across the range of literature available to develop insights into the historical development of knowledge on the topic, including the turning points that shape the current ways of thinking about a topic. Understanding the full body of literature is required to decide the dates or events that demarcate the timeframe of now in the first of the SotA’s three-part argument: where we are now . Stage 2 is complete when the researcher(s) can explicitly justify why a specific year or event is the right moment to mark the beginning of state-of-the-art thinking about the topic being summarized.
Stage 3: Finalize research question(s) to reflect timeframe
Based on the insights developed in Stage 2, the researcher(s) will likely need to revise their initial description of the topic to be summarized. The formal research question(s) framing the SotA review are finalized in Stage 3. The revised description of the topic, the research question(s), and the justification for the timeline start year must be reported in the review article. These are markers of rigor and prerequisites for moving to Stage 4.
Stage 4: Develop search strategy to find relevant articles
In Stage 4, the researcher(s) develops a search strategy to identify the literature that will be included in the SotA review. The researcher(s) needs to determine which literature databases contain articles from the domain of interest. Because the review describes how we got here , the review must include literature that predates the state-of-the-art timeframe, determined in Stage 2, to offer this historical perspective.
Developing the search strategy will be an iterative process of testing and revising the search strategy to enable the researcher(s) to capture the breadth of literature required to meet the SotA review purposes. A librarian should be consulted since their expertise can expedite the search processes and ensure that relevant resources are identified. The search strategy must be reported (e.g., in the manuscript itself or in a supplemental file) so that others may replicate the process if they so choose (e.g., to construct a different SotA review [and possible different interpretations] of the same literature). This too is a marker of rigor for SotA reviews: the search strategies informing the identification of literature must be reported.
Stage 5: Analyses
The literature analysis undertaken will reflect the subjective insights of the researcher(s); however, the foundational premises of inductive research should inform the analysis process. Therefore, the researcher(s) should begin by reading the articles in the corpus to become familiar with the literature. This familiarization work includes: noting similarities across articles, observing ways-of-thinking that have shaped current understandings of the topic, remarking on assumptions underpinning changes in understandings, identifying important decision points in the evolution of understanding, and taking notice of gaps and assumptions in current knowledge.
The researcher(s) can then generate premises for the state-of-the-art understanding of the history that gave rise to modern thinking, of the current body of knowledge, and of potential future directions for research. In this stage of the analysis, the researcher(s) should document the articles that support or contradict their premises, noting any collections of authors or schools of thinking that have dominated the literature, searching for marginalized points of view, and studying the factors that contributed to the dominance of particular ways of thinking. The researcher(s) should also observe historical decision points that could be revisited. Theory can be incorporated at this stage to help shape insights and understandings. It should be highlighted that not all corpus articles will be used in the SotA review; instead, the researcher(s) will sample across the corpus to construct a timeline that represents the seminal moments of the historical development of knowledge.
Next, the researcher(s) should verify the thoroughness and strength of their interpretations. To do this, the researcher(s) can select different articles included in the corpus and examine if those articles reflect the premises the researcher(s) set out. The researcher(s) may also seek out contradictory interpretations in the literature to be sure their summary refutes these positions. The goal of this verification work is not to engage in a triangulation process to ensure objectivity; instead, this process helps the researcher(s) ensure the interpretations made in the SotA review represent the articles being synthesized and respond to the interpretations offered by others. This is another marker of rigor for SotA reviews: the authors should engage in and report how they considered and accounted for differing interpretations of the literature, and how they verified the thoroughness of their interpretations.
Stage 6: Reflexivity
Given the relativist subjectivism of a SotA review, it is important that the manuscript offer insights into the subjectivity of the researcher(s). This reflexivity description should articulate how the subjectivity of the researcher(s) informed interpretations of the data. These reflections will also influence the suggested directions offered in the last part of the SotA three-part argument: where we could go next. This is the last marker of rigor for SotA reviews: researcher reflexivity must be considered and reported.
SotA reviews have much to offer our field since they provide information on the historical progression of medical education’s understanding of a topic, the turning points that guided that understanding, and the potential next directions for future research. Those future directions may question the soundness of turning points and prior decisions, and thereby offer new paths of investigation. Since we were unable to find a description of the SotA review methodology, we inductively developed a description of the methodology—including its paradigmatic roots, the processes to be followed, and the markers of rigor—so that scholars can harness the unique affordances of this type of knowledge synthesis.
Given their chronology- and turning point-based orientation, SotA reviews are inherently different from other types of knowledge synthesis. For example, systematic reviews focus on specific research questions that are narrow in scope [ 32 , 33 ]; in contrast, SotA reviews present a broader historical overview of knowledge development and the decisions that gave rise to our modern understandings. Scoping reviews focus on mapping the present state of knowledge about a phenomenon including, for example, the data that are currently available, the nature of that data, and the gaps in knowledge [ 34 , 35 ]; conversely, SotA reviews offer interpretations of the historical progression of knowledge relating to a phenomenon centered on significant shifts that occurred during that history. SotA reviews focus on the turning points in the history of knowledge development to suggest how different decisions could give rise to new insights. Critical reviews draw on literature outside of the domain of focus to see if external literature can offer new ways of thinking about the phenomenon of interest (e.g., drawing on insights from insects’ swarm intelligence to better understand healthcare team adaptation [ 36 ]). SotA reviews focus on one domain’s body of literature to construct a timeline of knowledge development, demarcating where we are now, demonstrating how this understanding came to be via different turning points, and offering new research directions. Certainly, SotA reviews offer a unique kind of knowledge synthesis.
Our six-stage process for conducting these reviews reflects the subjectivist relativism that underpins the methodology. It aligns with the requirements proposed by others [ 24 , 25 , 26 , 27 ], what has been written about SotA reviews [ 4 , 5 ], and the current body of published SotA reviews. In contrast to existing guidance [ 4 , 5 , 20 , 21 , 22 , 23 ], our description offers a detailed reporting of the ontology, epistemology, and methodology processes for conducting the SotA review.
This explicit methodology description is essential since many academic journals list SotA reviews as an accepted type of literature review. For instance, Educational Research Review [ 24 ], the American Academy of Pediatrics [ 25 ], and Thorax all lists SotA reviews as one of the types of knowledge syntheses they accept [ 27 ]. However, while SotA reviews are valued by academia, guidelines or specific methodology descriptions for researchers to follow when conducting this type of knowledge synthesis are conspicuously absent. If academics in general, and medical education more specifically, are to take advantage of the insights that SotA reviews can offer, we need to rigorously engage in this synthesis work; to do that, we need clear descriptions of the methodology underpinning this review. This article offers such a description. We hope that more medical educators will conduct SotA reviews to generate insights that will contribute to further advancing our field’s research and scholarship.
Cooper HM. Organizing knowledge syntheses: a taxonomy of literature reviews. Knowl Soc. 1988;1:104.
Google Scholar
Badger D, Nursten J, Williams P, Woodward M. Should all literature reviews be systematic? Eval Res Educ. 2000;14:220–30.
Article Google Scholar
Snyder H. Literature review as a research methodology: an overview and guidelines. J Bus Res. 2019;104:333–9.
Grant MJ, Booth A. A typology of reviews: an analysis of 14 review types and associated methodologies. Health Info Libr J. 2009;26:91–108.
Sutton A, Clowes M, Preston L, Booth A. Meeting the review family: exploring review types and associated information retrieval requirements. Health Info Libr J. 2019;36:202–22.
Moher D, Liberati A, Tetzlaff J, Altman DG, Prisma Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
Tricco AC, Langlois E, Straus SE, World Health Organization, Alliance for Health Policy and Systems Research. Rapid reviews to strengthen health policy and systems: a practical guide. Geneva: World Health Organization; 2017.
Jackson R, Feder G. Guidelines for clinical guidelines: a simple, pragmatic strategy for guideline development. Br Med J. 1998;317:427–8.
Greenhalgh T, Thorne S, Malterud K. Time to challenge the spurious hierarchy of systematic over narrative reviews? Eur J Clin Invest. 2018;48:e12931.
Bach QV, Chen WH. Pyrolysis characteristics and kinetics of microalgae via thermogravimetric analysis (TGA): a state-of-the-art review. Bioresour Technol. 2017;246:88–100.
Garofalo C, Milanović V, Cardinali F, Aquilanti L, Clementi F, Osimani A. Current knowledge on the microbiota of edible insects intended for human consumption: a state-of-the-art review. Food Res Int. 2019;125:108527.
Carbone S, Dixon DL, Buckley LF, Abbate A. Glucose-lowering therapies for cardiovascular risk reduction in type 2 diabetes mellitus: state-of-the-art review. Mayo Clin Proc. 2018;93:1629–47.
Hofkens PJ, Verrijcken A, Merveille K, et al. Common pitfalls and tips and tricks to get the most out of your transpulmonary thermodilution device: results of a survey and state-of-the-art review. Anaesthesiol Intensive Ther. 2015;47:89–116.
Schuwirth LW, van der Vleuten CP. A history of assessment in medical education. Adv Health Sci Educ Theory Pract. 2020;25:1045–56.
Arena A, Prete F, Rambaldi E, et al. Nanostructured zirconia-based ceramics and composites in dentistry: a state-of-the-art review. Nanomaterials. 2019;9:1393.
Bahraminasab M, Farahmand F. State of the art review on design and manufacture of hybrid biomedical materials: hip and knee prostheses. Proc Inst Mech Eng H. 2017;231:785–813.
Maggio LA, Costello JA, Norton C, Driessen EW, Artino AR Jr. Knowledge syntheses in medical education: a bibliometric analysis. Perspect Med Educ. 2021;10:79–87.
McGaghie WC, Issenberg SB, Petrusa ER, Scalese RJ. A critical review of simulation-based medical education research: 2003–2009. Med Educ. 2010;44:50–63.
Krishnan DG, Keloth AV, Ubedulla S. Pros and cons of simulation in medical education: a review. Education. 2017;3:84–7.
National Library of Medicine. MEDLINE: overview. 2021. https://www.nlm.nih.gov/medline/medline_overview.html . Accessed 17 Dec 2021.
Bergman E, de Feijter J, Frambach J, et al. AM last page: a guide to research paradigms relevant to medical education. Acad Med. 2012;87:545.
Maggio LA, Samuel A, Stellrecht E. Systematic reviews in medical education. J Grad Med Educ. 2022;14:171–5.
Bandari J, Wessel CB, Jacobs BL. Comparative effectiveness in urology: a state of the art review utilizing a systematic approach. Curr Opin Urol. 2017;27:380–94.
Elsevier. A guide for writing scholarly articles or reviews for the educational research review. 2010. https://www.elsevier.com/__data/promis_misc/edurevReviewPaperWriting.pdf . Accessed 3 Mar 2020.
American Academy of Pediatrics. Pediatrics author guidelines. 2020. https://pediatrics.aappublications.org/page/author-guidelines . Accessed 3 Mar 2020.
Journal of the American College of Cardiology. JACC instructions for authors. 2020. https://www.jacc.org/pb-assets/documents/author-instructions-jacc-1598995793940.pdf . Accessed 3 Mar 2020.
Thorax. Authors. 2020. https://thorax.bmj.com/pages/authors/ . Accessed 3 Mar 2020.
Berven S, Carl A. State of the art review. Spine Deform. 2019;7:381.
Ilardi CR, Chieffi S, Iachini T, Iavarone A. Neuropsychology of posteromedial parietal cortex and conversion factors from mild cognitive impairment to Alzheimer’s disease: systematic search and state-of-the-art review. Aging Clin Exp Res. 2022;34:289–307.
Chan KS, Zary N. Applications and challenges of implementing artificial intelligence in medical education: integrative review. JMIR Med Educ. 2019;5:e13930.
World Health Organization. Framework for action on interprofessional education and collaborative practice. 2010. https://www.who.int/publications/i/item/framework-for-action-on-interprofessional-education-collaborative-practice . Accessed July 1 2021.
Hammersley M. On ‘systematic’ reviews of research literatures: a ‘narrative’ response to Evans & Benefield. Br Educ Res J. 2001;27:543–54.
Chen F, Lui AM, Martinelli SM. A systematic review of the effectiveness of flipped classrooms in medical education. Med Educ. 2017;51:585–97.
Arksey H, O’Malley L. Scoping studies: towards a methodological framework. Int J Soc Res Methodol. 2005;8:19–32.
Matsas B, Goralnick E, Bass M, Barnett E, Nagle B, Sullivan E. Leadership development in US undergraduate medical education: a scoping review of curricular content and competency frameworks. Acad Med. 2022;97:899–908.
Cristancho SM. On collective self-healing and traces: How can swarm intelligence help us think differently about team adaptation? Med Educ. 2021;55:441–7.
Download references
Acknowledgements
We thank Rhonda Allard for her help with the literature review and compiling all available articles. We also want to thank the PME editors who offered excellent development and refinement suggestions that greatly improved this manuscript.
Author information
Authors and affiliations.
Department of Anesthesiology, F. Edward Hébert School of Medicine, Uniformed Services University, Bethesda, MD, USA
Erin S. Barry
School of Health Professions Education (SHE), Maastricht University, Maastricht, The Netherlands
Department of Medicine, F. Edward Hébert School of Medicine, Uniformed Services University, Bethesda, MD, USA
Jerusalem Merkebu & Lara Varpio
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Erin S. Barry .
Ethics declarations
Conflict of interest.
E.S. Barry, J. Merkebu and L. Varpio declare that they have no competing interests.
Additional information
The opinions and assertions contained in this article are solely those of the authors and are not to be construed as reflecting the views of the Uniformed Services University of the Health Sciences, the Department of Defense, or the Henry M. Jackson Foundation for the Advancement of Military Medicine.
Supplementary Information
40037_2022_725_moesm1_esm.docx.
For information regarding the search strategy to develop the corpus and search strategy for confirming capture of any available State of the Art review methodology descriptions. Additionally, a list of the methodology articles found through the search strategy/corpus is included
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Barry, E.S., Merkebu, J. & Varpio, L. State-of-the-art literature review methodology: A six-step approach for knowledge synthesis. Perspect Med Educ 11 , 281–288 (2022). https://doi.org/10.1007/s40037-022-00725-9
Download citation
Received : 03 December 2021
Revised : 25 July 2022
Accepted : 27 July 2022
Published : 05 September 2022
Issue Date : October 2022
DOI : https://doi.org/10.1007/s40037-022-00725-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- State-of-the-art literature review
- Literature review
- Literature review methodology
- Find a journal
- Publish with us
- Track your research
How To Structure Your Literature Review
3 options to help structure your chapter.
By: Amy Rommelspacher (PhD) | Reviewer: Dr Eunice Rautenbach | November 2020 (Updated May 2023)
Writing the literature review chapter can seem pretty daunting when you’re piecing together your dissertation or thesis. As we’ve discussed before , a good literature review needs to achieve a few very important objectives – it should:
- Demonstrate your knowledge of the research topic
- Identify the gaps in the literature and show how your research links to these
- Provide the foundation for your conceptual framework (if you have one)
- Inform your own methodology and research design
To achieve this, your literature review needs a well-thought-out structure . Get the structure of your literature review chapter wrong and you’ll struggle to achieve these objectives. Don’t worry though – in this post, we’ll look at how to structure your literature review for maximum impact (and marks!).

But wait – is this the right time?
Deciding on the structure of your literature review should come towards the end of the literature review process – after you have collected and digested the literature, but before you start writing the chapter.
In other words, you need to first develop a rich understanding of the literature before you even attempt to map out a structure. There’s no use trying to develop a structure before you’ve fully wrapped your head around the existing research.
Equally importantly, you need to have a structure in place before you start writing , or your literature review will most likely end up a rambling, disjointed mess.
Importantly, don’t feel that once you’ve defined a structure you can’t iterate on it. It’s perfectly natural to adjust as you engage in the writing process. As we’ve discussed before , writing is a way of developing your thinking, so it’s quite common for your thinking to change – and therefore, for your chapter structure to change – as you write.
Need a helping hand?
Like any other chapter in your thesis or dissertation, your literature review needs to have a clear, logical structure. At a minimum, it should have three essential components – an introduction , a body and a conclusion .
Let’s take a closer look at each of these.
1: The Introduction Section
Just like any good introduction, the introduction section of your literature review should introduce the purpose and layout (organisation) of the chapter. In other words, your introduction needs to give the reader a taste of what’s to come, and how you’re going to lay that out. Essentially, you should provide the reader with a high-level roadmap of your chapter to give them a taste of the journey that lies ahead.
Here’s an example of the layout visualised in a literature review introduction:
Your introduction should also outline your topic (including any tricky terminology or jargon) and provide an explanation of the scope of your literature review – in other words, what you will and won’t be covering (the delimitations ). This helps ringfence your review and achieve a clear focus . The clearer and narrower your focus, the deeper you can dive into the topic (which is typically where the magic lies).
Depending on the nature of your project, you could also present your stance or point of view at this stage. In other words, after grappling with the literature you’ll have an opinion about what the trends and concerns are in the field as well as what’s lacking. The introduction section can then present these ideas so that it is clear to examiners that you’re aware of how your research connects with existing knowledge .

2: The Body Section
The body of your literature review is the centre of your work. This is where you’ll present, analyse, evaluate and synthesise the existing research. In other words, this is where you’re going to earn (or lose) the most marks. Therefore, it’s important to carefully think about how you will organise your discussion to present it in a clear way.
The body of your literature review should do just as the description of this chapter suggests. It should “review” the literature – in other words, identify, analyse, and synthesise it. So, when thinking about structuring your literature review, you need to think about which structural approach will provide the best “review” for your specific type of research and objectives (we’ll get to this shortly).
There are (broadly speaking) three options for organising your literature review.

Option 1: Chronological (according to date)
Organising the literature chronologically is one of the simplest ways to structure your literature review. You start with what was published first and work your way through the literature until you reach the work published most recently. Pretty straightforward.
The benefit of this option is that it makes it easy to discuss the developments and debates in the field as they emerged over time. Organising your literature chronologically also allows you to highlight how specific articles or pieces of work might have changed the course of the field – in other words, which research has had the most impact . Therefore, this approach is very useful when your research is aimed at understanding how the topic has unfolded over time and is often used by scholars in the field of history. That said, this approach can be utilised by anyone that wants to explore change over time .

For example , if a student of politics is investigating how the understanding of democracy has evolved over time, they could use the chronological approach to provide a narrative that demonstrates how this understanding has changed through the ages.
Here are some questions you can ask yourself to help you structure your literature review chronologically.
- What is the earliest literature published relating to this topic?
- How has the field changed over time? Why?
- What are the most recent discoveries/theories?
In some ways, chronology plays a part whichever way you decide to structure your literature review, because you will always, to a certain extent, be analysing how the literature has developed. However, with the chronological approach, the emphasis is very firmly on how the discussion has evolved over time , as opposed to how all the literature links together (which we’ll discuss next ).
Option 2: Thematic (grouped by theme)
The thematic approach to structuring a literature review means organising your literature by theme or category – for example, by independent variables (i.e. factors that have an impact on a specific outcome).
As you’ve been collecting and synthesising literature , you’ll likely have started seeing some themes or patterns emerging. You can then use these themes or patterns as a structure for your body discussion. The thematic approach is the most common approach and is useful for structuring literature reviews in most fields.
For example, if you were researching which factors contributed towards people trusting an organisation, you might find themes such as consumers’ perceptions of an organisation’s competence, benevolence and integrity. Structuring your literature review thematically would mean structuring your literature review’s body section to discuss each of these themes, one section at a time.

Here are some questions to ask yourself when structuring your literature review by themes:
- Are there any patterns that have come to light in the literature?
- What are the central themes and categories used by the researchers?
- Do I have enough evidence of these themes?
PS – you can see an example of a thematically structured literature review in our literature review sample walkthrough video here.
Option 3: Methodological
The methodological option is a way of structuring your literature review by the research methodologies used . In other words, organising your discussion based on the angle from which each piece of research was approached – for example, qualitative , quantitative or mixed methodologies.
Structuring your literature review by methodology can be useful if you are drawing research from a variety of disciplines and are critiquing different methodologies. The point of this approach is to question how existing research has been conducted, as opposed to what the conclusions and/or findings the research were.

For example, a sociologist might centre their research around critiquing specific fieldwork practices. Their literature review will then be a summary of the fieldwork methodologies used by different studies.
Here are some questions you can ask yourself when structuring your literature review according to methodology:
- Which methodologies have been utilised in this field?
- Which methodology is the most popular (and why)?
- What are the strengths and weaknesses of the various methodologies?
- How can the existing methodologies inform my own methodology?
3: The Conclusion Section
Once you’ve completed the body section of your literature review using one of the structural approaches we discussed above, you’ll need to “wrap up” your literature review and pull all the pieces together to set the direction for the rest of your dissertation or thesis.
The conclusion is where you’ll present the key findings of your literature review. In this section, you should emphasise the research that is especially important to your research questions and highlight the gaps that exist in the literature. Based on this, you need to make it clear what you will add to the literature – in other words, justify your own research by showing how it will help fill one or more of the gaps you just identified.
Last but not least, if it’s your intention to develop a conceptual framework for your dissertation or thesis, the conclusion section is a good place to present this.

Example: Thematically Structured Review
In the video below, we unpack a literature review chapter so that you can see an example of a thematically structure review in practice.
Let’s Recap
In this article, we’ve discussed how to structure your literature review for maximum impact. Here’s a quick recap of what you need to keep in mind when deciding on your literature review structure:
- Just like other chapters, your literature review needs a clear introduction , body and conclusion .
- The introduction section should provide an overview of what you will discuss in your literature review.
- The body section of your literature review can be organised by chronology , theme or methodology . The right structural approach depends on what you’re trying to achieve with your research.
- The conclusion section should draw together the key findings of your literature review and link them to your research questions.
If you’re ready to get started, be sure to download our free literature review template to fast-track your chapter outline.

Psst… there’s more!
This post is an extract from our bestselling short course, Literature Review Bootcamp . If you want to work smart, you don't want to miss this .
You Might Also Like:

27 Comments
Great work. This is exactly what I was looking for and helps a lot together with your previous post on literature review. One last thing is missing: a link to a great literature chapter of an journal article (maybe with comments of the different sections in this review chapter). Do you know any great literature review chapters?
I agree with you Marin… A great piece
I agree with Marin. This would be quite helpful if you annotate a nicely structured literature from previously published research articles.
Awesome article for my research.
I thank you immensely for this wonderful guide
It is indeed thought and supportive work for the futurist researcher and students
Very educative and good time to get guide. Thank you
Great work, very insightful. Thank you.
Thanks for this wonderful presentation. My question is that do I put all the variables into a single conceptual framework or each hypothesis will have it own conceptual framework?
Thank you very much, very helpful
This is very educative and precise . Thank you very much for dropping this kind of write up .
Pheeww, so damn helpful, thank you for this informative piece.
I’m doing a research project topic ; stool analysis for parasitic worm (enteric) worm, how do I structure it, thanks.
comprehensive explanation. Help us by pasting the URL of some good “literature review” for better understanding.
great piece. thanks for the awesome explanation. it is really worth sharing. I have a little question, if anyone can help me out, which of the options in the body of literature can be best fit if you are writing an architectural thesis that deals with design?
I am doing a research on nanofluids how can l structure it?
Beautifully clear.nThank you!
Lucid! Thankyou!
Brilliant work, well understood, many thanks
I like how this was so clear with simple language 😊😊 thank you so much 😊 for these information 😊
Insightful. I was struggling to come up with a sensible literature review but this has been really helpful. Thank you!
You have given thought-provoking information about the review of the literature.
Thank you. It has made my own research better and to impart your work to students I teach
I learnt a lot from this teaching. It’s a great piece.
I am doing research on EFL teacher motivation for his/her job. How Can I structure it? Is there any detailed template, additional to this?
You are so cool! I do not think I’ve read through something like this before. So nice to find somebody with some genuine thoughts on this issue. Seriously.. thank you for starting this up. This site is one thing that is required on the internet, someone with a little originality!
I’m asked to do conceptual, theoretical and empirical literature, and i just don’t know how to structure it
Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly
Purdue Online Writing Lab Purdue OWL® College of Liberal Arts
Writing a Literature Review

Welcome to the Purdue OWL
This page is brought to you by the OWL at Purdue University. When printing this page, you must include the entire legal notice.
Copyright ©1995-2018 by The Writing Lab & The OWL at Purdue and Purdue University. All rights reserved. This material may not be published, reproduced, broadcast, rewritten, or redistributed without permission. Use of this site constitutes acceptance of our terms and conditions of fair use.
A literature review is a document or section of a document that collects key sources on a topic and discusses those sources in conversation with each other (also called synthesis ). The lit review is an important genre in many disciplines, not just literature (i.e., the study of works of literature such as novels and plays). When we say “literature review” or refer to “the literature,” we are talking about the research ( scholarship ) in a given field. You will often see the terms “the research,” “the scholarship,” and “the literature” used mostly interchangeably.
Where, when, and why would I write a lit review?
There are a number of different situations where you might write a literature review, each with slightly different expectations; different disciplines, too, have field-specific expectations for what a literature review is and does. For instance, in the humanities, authors might include more overt argumentation and interpretation of source material in their literature reviews, whereas in the sciences, authors are more likely to report study designs and results in their literature reviews; these differences reflect these disciplines’ purposes and conventions in scholarship. You should always look at examples from your own discipline and talk to professors or mentors in your field to be sure you understand your discipline’s conventions, for literature reviews as well as for any other genre.
A literature review can be a part of a research paper or scholarly article, usually falling after the introduction and before the research methods sections. In these cases, the lit review just needs to cover scholarship that is important to the issue you are writing about; sometimes it will also cover key sources that informed your research methodology.
Lit reviews can also be standalone pieces, either as assignments in a class or as publications. In a class, a lit review may be assigned to help students familiarize themselves with a topic and with scholarship in their field, get an idea of the other researchers working on the topic they’re interested in, find gaps in existing research in order to propose new projects, and/or develop a theoretical framework and methodology for later research. As a publication, a lit review usually is meant to help make other scholars’ lives easier by collecting and summarizing, synthesizing, and analyzing existing research on a topic. This can be especially helpful for students or scholars getting into a new research area, or for directing an entire community of scholars toward questions that have not yet been answered.
What are the parts of a lit review?
Most lit reviews use a basic introduction-body-conclusion structure; if your lit review is part of a larger paper, the introduction and conclusion pieces may be just a few sentences while you focus most of your attention on the body. If your lit review is a standalone piece, the introduction and conclusion take up more space and give you a place to discuss your goals, research methods, and conclusions separately from where you discuss the literature itself.
Introduction:
- An introductory paragraph that explains what your working topic and thesis is
- A forecast of key topics or texts that will appear in the review
- Potentially, a description of how you found sources and how you analyzed them for inclusion and discussion in the review (more often found in published, standalone literature reviews than in lit review sections in an article or research paper)
- Summarize and synthesize: Give an overview of the main points of each source and combine them into a coherent whole
- Analyze and interpret: Don’t just paraphrase other researchers – add your own interpretations where possible, discussing the significance of findings in relation to the literature as a whole
- Critically Evaluate: Mention the strengths and weaknesses of your sources
- Write in well-structured paragraphs: Use transition words and topic sentence to draw connections, comparisons, and contrasts.
Conclusion:
- Summarize the key findings you have taken from the literature and emphasize their significance
- Connect it back to your primary research question
How should I organize my lit review?
Lit reviews can take many different organizational patterns depending on what you are trying to accomplish with the review. Here are some examples:
- Chronological : The simplest approach is to trace the development of the topic over time, which helps familiarize the audience with the topic (for instance if you are introducing something that is not commonly known in your field). If you choose this strategy, be careful to avoid simply listing and summarizing sources in order. Try to analyze the patterns, turning points, and key debates that have shaped the direction of the field. Give your interpretation of how and why certain developments occurred (as mentioned previously, this may not be appropriate in your discipline — check with a teacher or mentor if you’re unsure).
- Thematic : If you have found some recurring central themes that you will continue working with throughout your piece, you can organize your literature review into subsections that address different aspects of the topic. For example, if you are reviewing literature about women and religion, key themes can include the role of women in churches and the religious attitude towards women.
- Qualitative versus quantitative research
- Empirical versus theoretical scholarship
- Divide the research by sociological, historical, or cultural sources
- Theoretical : In many humanities articles, the literature review is the foundation for the theoretical framework. You can use it to discuss various theories, models, and definitions of key concepts. You can argue for the relevance of a specific theoretical approach or combine various theorical concepts to create a framework for your research.
What are some strategies or tips I can use while writing my lit review?
Any lit review is only as good as the research it discusses; make sure your sources are well-chosen and your research is thorough. Don’t be afraid to do more research if you discover a new thread as you’re writing. More info on the research process is available in our "Conducting Research" resources .
As you’re doing your research, create an annotated bibliography ( see our page on the this type of document ). Much of the information used in an annotated bibliography can be used also in a literature review, so you’ll be not only partially drafting your lit review as you research, but also developing your sense of the larger conversation going on among scholars, professionals, and any other stakeholders in your topic.
Usually you will need to synthesize research rather than just summarizing it. This means drawing connections between sources to create a picture of the scholarly conversation on a topic over time. Many student writers struggle to synthesize because they feel they don’t have anything to add to the scholars they are citing; here are some strategies to help you:
- It often helps to remember that the point of these kinds of syntheses is to show your readers how you understand your research, to help them read the rest of your paper.
- Writing teachers often say synthesis is like hosting a dinner party: imagine all your sources are together in a room, discussing your topic. What are they saying to each other?
- Look at the in-text citations in each paragraph. Are you citing just one source for each paragraph? This usually indicates summary only. When you have multiple sources cited in a paragraph, you are more likely to be synthesizing them (not always, but often
- Read more about synthesis here.
The most interesting literature reviews are often written as arguments (again, as mentioned at the beginning of the page, this is discipline-specific and doesn’t work for all situations). Often, the literature review is where you can establish your research as filling a particular gap or as relevant in a particular way. You have some chance to do this in your introduction in an article, but the literature review section gives a more extended opportunity to establish the conversation in the way you would like your readers to see it. You can choose the intellectual lineage you would like to be part of and whose definitions matter most to your thinking (mostly humanities-specific, but this goes for sciences as well). In addressing these points, you argue for your place in the conversation, which tends to make the lit review more compelling than a simple reporting of other sources.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- PLoS Comput Biol
- v.9(7); 2013 Jul

Ten Simple Rules for Writing a Literature Review
Marco pautasso.
1 Centre for Functional and Evolutionary Ecology (CEFE), CNRS, Montpellier, France
2 Centre for Biodiversity Synthesis and Analysis (CESAB), FRB, Aix-en-Provence, France
Literature reviews are in great demand in most scientific fields. Their need stems from the ever-increasing output of scientific publications [1] . For example, compared to 1991, in 2008 three, eight, and forty times more papers were indexed in Web of Science on malaria, obesity, and biodiversity, respectively [2] . Given such mountains of papers, scientists cannot be expected to examine in detail every single new paper relevant to their interests [3] . Thus, it is both advantageous and necessary to rely on regular summaries of the recent literature. Although recognition for scientists mainly comes from primary research, timely literature reviews can lead to new synthetic insights and are often widely read [4] . For such summaries to be useful, however, they need to be compiled in a professional way [5] .
When starting from scratch, reviewing the literature can require a titanic amount of work. That is why researchers who have spent their career working on a certain research issue are in a perfect position to review that literature. Some graduate schools are now offering courses in reviewing the literature, given that most research students start their project by producing an overview of what has already been done on their research issue [6] . However, it is likely that most scientists have not thought in detail about how to approach and carry out a literature review.
Reviewing the literature requires the ability to juggle multiple tasks, from finding and evaluating relevant material to synthesising information from various sources, from critical thinking to paraphrasing, evaluating, and citation skills [7] . In this contribution, I share ten simple rules I learned working on about 25 literature reviews as a PhD and postdoctoral student. Ideas and insights also come from discussions with coauthors and colleagues, as well as feedback from reviewers and editors.
Rule 1: Define a Topic and Audience
How to choose which topic to review? There are so many issues in contemporary science that you could spend a lifetime of attending conferences and reading the literature just pondering what to review. On the one hand, if you take several years to choose, several other people may have had the same idea in the meantime. On the other hand, only a well-considered topic is likely to lead to a brilliant literature review [8] . The topic must at least be:
- interesting to you (ideally, you should have come across a series of recent papers related to your line of work that call for a critical summary),
- an important aspect of the field (so that many readers will be interested in the review and there will be enough material to write it), and
- a well-defined issue (otherwise you could potentially include thousands of publications, which would make the review unhelpful).
Ideas for potential reviews may come from papers providing lists of key research questions to be answered [9] , but also from serendipitous moments during desultory reading and discussions. In addition to choosing your topic, you should also select a target audience. In many cases, the topic (e.g., web services in computational biology) will automatically define an audience (e.g., computational biologists), but that same topic may also be of interest to neighbouring fields (e.g., computer science, biology, etc.).
Rule 2: Search and Re-search the Literature
After having chosen your topic and audience, start by checking the literature and downloading relevant papers. Five pieces of advice here:
- keep track of the search items you use (so that your search can be replicated [10] ),
- keep a list of papers whose pdfs you cannot access immediately (so as to retrieve them later with alternative strategies),
- use a paper management system (e.g., Mendeley, Papers, Qiqqa, Sente),
- define early in the process some criteria for exclusion of irrelevant papers (these criteria can then be described in the review to help define its scope), and
- do not just look for research papers in the area you wish to review, but also seek previous reviews.
The chances are high that someone will already have published a literature review ( Figure 1 ), if not exactly on the issue you are planning to tackle, at least on a related topic. If there are already a few or several reviews of the literature on your issue, my advice is not to give up, but to carry on with your own literature review,

The bottom-right situation (many literature reviews but few research papers) is not just a theoretical situation; it applies, for example, to the study of the impacts of climate change on plant diseases, where there appear to be more literature reviews than research studies [33] .
- discussing in your review the approaches, limitations, and conclusions of past reviews,
- trying to find a new angle that has not been covered adequately in the previous reviews, and
- incorporating new material that has inevitably accumulated since their appearance.
When searching the literature for pertinent papers and reviews, the usual rules apply:
- be thorough,
- use different keywords and database sources (e.g., DBLP, Google Scholar, ISI Proceedings, JSTOR Search, Medline, Scopus, Web of Science), and
- look at who has cited past relevant papers and book chapters.
Rule 3: Take Notes While Reading
If you read the papers first, and only afterwards start writing the review, you will need a very good memory to remember who wrote what, and what your impressions and associations were while reading each single paper. My advice is, while reading, to start writing down interesting pieces of information, insights about how to organize the review, and thoughts on what to write. This way, by the time you have read the literature you selected, you will already have a rough draft of the review.
Of course, this draft will still need much rewriting, restructuring, and rethinking to obtain a text with a coherent argument [11] , but you will have avoided the danger posed by staring at a blank document. Be careful when taking notes to use quotation marks if you are provisionally copying verbatim from the literature. It is advisable then to reformulate such quotes with your own words in the final draft. It is important to be careful in noting the references already at this stage, so as to avoid misattributions. Using referencing software from the very beginning of your endeavour will save you time.
Rule 4: Choose the Type of Review You Wish to Write
After having taken notes while reading the literature, you will have a rough idea of the amount of material available for the review. This is probably a good time to decide whether to go for a mini- or a full review. Some journals are now favouring the publication of rather short reviews focusing on the last few years, with a limit on the number of words and citations. A mini-review is not necessarily a minor review: it may well attract more attention from busy readers, although it will inevitably simplify some issues and leave out some relevant material due to space limitations. A full review will have the advantage of more freedom to cover in detail the complexities of a particular scientific development, but may then be left in the pile of the very important papers “to be read” by readers with little time to spare for major monographs.
There is probably a continuum between mini- and full reviews. The same point applies to the dichotomy of descriptive vs. integrative reviews. While descriptive reviews focus on the methodology, findings, and interpretation of each reviewed study, integrative reviews attempt to find common ideas and concepts from the reviewed material [12] . A similar distinction exists between narrative and systematic reviews: while narrative reviews are qualitative, systematic reviews attempt to test a hypothesis based on the published evidence, which is gathered using a predefined protocol to reduce bias [13] , [14] . When systematic reviews analyse quantitative results in a quantitative way, they become meta-analyses. The choice between different review types will have to be made on a case-by-case basis, depending not just on the nature of the material found and the preferences of the target journal(s), but also on the time available to write the review and the number of coauthors [15] .
Rule 5: Keep the Review Focused, but Make It of Broad Interest
Whether your plan is to write a mini- or a full review, it is good advice to keep it focused 16 , 17 . Including material just for the sake of it can easily lead to reviews that are trying to do too many things at once. The need to keep a review focused can be problematic for interdisciplinary reviews, where the aim is to bridge the gap between fields [18] . If you are writing a review on, for example, how epidemiological approaches are used in modelling the spread of ideas, you may be inclined to include material from both parent fields, epidemiology and the study of cultural diffusion. This may be necessary to some extent, but in this case a focused review would only deal in detail with those studies at the interface between epidemiology and the spread of ideas.
While focus is an important feature of a successful review, this requirement has to be balanced with the need to make the review relevant to a broad audience. This square may be circled by discussing the wider implications of the reviewed topic for other disciplines.
Rule 6: Be Critical and Consistent
Reviewing the literature is not stamp collecting. A good review does not just summarize the literature, but discusses it critically, identifies methodological problems, and points out research gaps [19] . After having read a review of the literature, a reader should have a rough idea of:
- the major achievements in the reviewed field,
- the main areas of debate, and
- the outstanding research questions.
It is challenging to achieve a successful review on all these fronts. A solution can be to involve a set of complementary coauthors: some people are excellent at mapping what has been achieved, some others are very good at identifying dark clouds on the horizon, and some have instead a knack at predicting where solutions are going to come from. If your journal club has exactly this sort of team, then you should definitely write a review of the literature! In addition to critical thinking, a literature review needs consistency, for example in the choice of passive vs. active voice and present vs. past tense.
Rule 7: Find a Logical Structure
Like a well-baked cake, a good review has a number of telling features: it is worth the reader's time, timely, systematic, well written, focused, and critical. It also needs a good structure. With reviews, the usual subdivision of research papers into introduction, methods, results, and discussion does not work or is rarely used. However, a general introduction of the context and, toward the end, a recapitulation of the main points covered and take-home messages make sense also in the case of reviews. For systematic reviews, there is a trend towards including information about how the literature was searched (database, keywords, time limits) [20] .
How can you organize the flow of the main body of the review so that the reader will be drawn into and guided through it? It is generally helpful to draw a conceptual scheme of the review, e.g., with mind-mapping techniques. Such diagrams can help recognize a logical way to order and link the various sections of a review [21] . This is the case not just at the writing stage, but also for readers if the diagram is included in the review as a figure. A careful selection of diagrams and figures relevant to the reviewed topic can be very helpful to structure the text too [22] .
Rule 8: Make Use of Feedback
Reviews of the literature are normally peer-reviewed in the same way as research papers, and rightly so [23] . As a rule, incorporating feedback from reviewers greatly helps improve a review draft. Having read the review with a fresh mind, reviewers may spot inaccuracies, inconsistencies, and ambiguities that had not been noticed by the writers due to rereading the typescript too many times. It is however advisable to reread the draft one more time before submission, as a last-minute correction of typos, leaps, and muddled sentences may enable the reviewers to focus on providing advice on the content rather than the form.
Feedback is vital to writing a good review, and should be sought from a variety of colleagues, so as to obtain a diversity of views on the draft. This may lead in some cases to conflicting views on the merits of the paper, and on how to improve it, but such a situation is better than the absence of feedback. A diversity of feedback perspectives on a literature review can help identify where the consensus view stands in the landscape of the current scientific understanding of an issue [24] .
Rule 9: Include Your Own Relevant Research, but Be Objective
In many cases, reviewers of the literature will have published studies relevant to the review they are writing. This could create a conflict of interest: how can reviewers report objectively on their own work [25] ? Some scientists may be overly enthusiastic about what they have published, and thus risk giving too much importance to their own findings in the review. However, bias could also occur in the other direction: some scientists may be unduly dismissive of their own achievements, so that they will tend to downplay their contribution (if any) to a field when reviewing it.
In general, a review of the literature should neither be a public relations brochure nor an exercise in competitive self-denial. If a reviewer is up to the job of producing a well-organized and methodical review, which flows well and provides a service to the readership, then it should be possible to be objective in reviewing one's own relevant findings. In reviews written by multiple authors, this may be achieved by assigning the review of the results of a coauthor to different coauthors.
Rule 10: Be Up-to-Date, but Do Not Forget Older Studies
Given the progressive acceleration in the publication of scientific papers, today's reviews of the literature need awareness not just of the overall direction and achievements of a field of inquiry, but also of the latest studies, so as not to become out-of-date before they have been published. Ideally, a literature review should not identify as a major research gap an issue that has just been addressed in a series of papers in press (the same applies, of course, to older, overlooked studies (“sleeping beauties” [26] )). This implies that literature reviewers would do well to keep an eye on electronic lists of papers in press, given that it can take months before these appear in scientific databases. Some reviews declare that they have scanned the literature up to a certain point in time, but given that peer review can be a rather lengthy process, a full search for newly appeared literature at the revision stage may be worthwhile. Assessing the contribution of papers that have just appeared is particularly challenging, because there is little perspective with which to gauge their significance and impact on further research and society.
Inevitably, new papers on the reviewed topic (including independently written literature reviews) will appear from all quarters after the review has been published, so that there may soon be the need for an updated review. But this is the nature of science [27] – [32] . I wish everybody good luck with writing a review of the literature.
Acknowledgments
Many thanks to M. Barbosa, K. Dehnen-Schmutz, T. Döring, D. Fontaneto, M. Garbelotto, O. Holdenrieder, M. Jeger, D. Lonsdale, A. MacLeod, P. Mills, M. Moslonka-Lefebvre, G. Stancanelli, P. Weisberg, and X. Xu for insights and discussions, and to P. Bourne, T. Matoni, and D. Smith for helpful comments on a previous draft.
Funding Statement
This work was funded by the French Foundation for Research on Biodiversity (FRB) through its Centre for Synthesis and Analysis of Biodiversity data (CESAB), as part of the NETSEED research project. The funders had no role in the preparation of the manuscript.
- UWF Libraries
Literature Review: Conducting & Writing
- Sample Literature Reviews
- Steps for Conducting a Lit Review
- Finding "The Literature"
- Organizing/Writing
- APA Style This link opens in a new window
- Chicago: Notes Bibliography This link opens in a new window
- MLA Style This link opens in a new window
Sample Lit Reviews from Communication Arts
Have an exemplary literature review.
- Literature Review Sample 1
- Literature Review Sample 2
- Literature Review Sample 3
Have you written a stellar literature review you care to share for teaching purposes?
Are you an instructor who has received an exemplary literature review and have permission from the student to post?
Please contact Britt McGowan at [email protected] for inclusion in this guide. All disciplines welcome and encouraged.
- << Previous: MLA Style
- Next: Get Help! >>
- Last Updated: Mar 22, 2024 9:37 AM
- URL: https://libguides.uwf.edu/litreview
Harvey Cushing/John Hay Whitney Medical Library
- Collections
- Research Help
YSN Doctoral Programs: Steps in Conducting a Literature Review
- Biomedical Databases
- Global (Public Health) Databases
- Soc. Sci., History, and Law Databases
- Grey Literature
- Trials Registers
- Data and Statistics
- Public Policy
- Google Tips
- Recommended Books
- Steps in Conducting a Literature Review
What is a literature review?
A literature review is an integrated analysis -- not just a summary-- of scholarly writings and other relevant evidence related directly to your research question. That is, it represents a synthesis of the evidence that provides background information on your topic and shows a association between the evidence and your research question.
A literature review may be a stand alone work or the introduction to a larger research paper, depending on the assignment. Rely heavily on the guidelines your instructor has given you.
Why is it important?
A literature review is important because it:
- Explains the background of research on a topic.
- Demonstrates why a topic is significant to a subject area.
- Discovers relationships between research studies/ideas.
- Identifies major themes, concepts, and researchers on a topic.
- Identifies critical gaps and points of disagreement.
- Discusses further research questions that logically come out of the previous studies.
APA7 Style resources
APA Style Blog - for those harder to find answers
1. Choose a topic. Define your research question.
Your literature review should be guided by your central research question. The literature represents background and research developments related to a specific research question, interpreted and analyzed by you in a synthesized way.
- Make sure your research question is not too broad or too narrow. Is it manageable?
- Begin writing down terms that are related to your question. These will be useful for searches later.
- If you have the opportunity, discuss your topic with your professor and your class mates.
2. Decide on the scope of your review
How many studies do you need to look at? How comprehensive should it be? How many years should it cover?
- This may depend on your assignment. How many sources does the assignment require?
3. Select the databases you will use to conduct your searches.
Make a list of the databases you will search.
Where to find databases:
- use the tabs on this guide
- Find other databases in the Nursing Information Resources web page
- More on the Medical Library web page
- ... and more on the Yale University Library web page
4. Conduct your searches to find the evidence. Keep track of your searches.
- Use the key words in your question, as well as synonyms for those words, as terms in your search. Use the database tutorials for help.
- Save the searches in the databases. This saves time when you want to redo, or modify, the searches. It is also helpful to use as a guide is the searches are not finding any useful results.
- Review the abstracts of research studies carefully. This will save you time.
- Use the bibliographies and references of research studies you find to locate others.
- Check with your professor, or a subject expert in the field, if you are missing any key works in the field.
- Ask your librarian for help at any time.
- Use a citation manager, such as EndNote as the repository for your citations. See the EndNote tutorials for help.

Review the literature
Some questions to help you analyze the research:
- What was the research question of the study you are reviewing? What were the authors trying to discover?
- Was the research funded by a source that could influence the findings?
- What were the research methodologies? Analyze its literature review, the samples and variables used, the results, and the conclusions.
- Does the research seem to be complete? Could it have been conducted more soundly? What further questions does it raise?
- If there are conflicting studies, why do you think that is?
- How are the authors viewed in the field? Has this study been cited? If so, how has it been analyzed?
Tips:
- Review the abstracts carefully.
- Keep careful notes so that you may track your thought processes during the research process.
- Create a matrix of the studies for easy analysis, and synthesis, across all of the studies.
- << Previous: Recommended Books
- Last Updated: Jan 4, 2024 10:52 AM
- URL: https://guides.library.yale.edu/YSNDoctoral
Covidence website will be inaccessible as we upgrading our platform on Monday 23rd August at 10am AEST, / 2am CEST/1am BST (Sunday, 15th August 8pm EDT/5pm PDT)
How to write the methods section of a systematic review
Home | Blog | How To | How to write the methods section of a systematic review
Covidence breaks down how to write a methods section
The methods section of your systematic review describes what you did, how you did it, and why. Readers need this information to interpret the results and conclusions of the review. Often, a lot of information needs to be distilled into just a few paragraphs. This can be a challenging task, but good preparation and the right tools will help you to set off in the right direction 🗺️🧭.
Systematic reviews are so-called because they are conducted in a way that is rigorous and replicable. So it’s important that these methods are reported in a way that is thorough, clear, and easy to navigate for the reader – whether that’s a patient, a healthcare worker, or a researcher.
Like most things in a systematic review, the methods should be planned upfront and ideally described in detail in a project plan or protocol. Reviews of healthcare interventions follow the PRISMA guidelines for the minimum set of items to report in the methods section. But what else should be included? It’s a good idea to consider what readers will want to know about the review methods and whether the journal you’re planning to submit the work to has expectations on the reporting of methods. Finding out in advance will help you to plan what to include.
Describe what happened
While the research plan sets out what you intend to do, the methods section is a write-up of what actually happened. It’s not a simple case of rewriting the plan in the past tense – you will also need to discuss and justify deviations from the plan and describe the handling of issues that were unforeseen at the time the plan was written. For this reason, it is useful to make detailed notes before, during, and after the review is completed. Relying on memory alone risks losing valuable information and trawling through emails when the deadline is looming can be frustrating and time consuming!
Keep it brief
The methods section should be succinct but include all the noteworthy information. This can be a difficult balance to achieve. A useful strategy is to aim for a brief description that signposts the reader to a separate section or sections of supporting information. This could include datasets, a flowchart to show what happened to the excluded studies, a collection of search strategies, and tables containing detailed information about the studies.This separation keeps the review short and simple while enabling the reader to drill down to the detail as needed. And if the methods follow a well-known or standard process, it might suffice to say so and give a reference, rather than describe the process at length.
Follow a structure
A clear structure provides focus. Use of descriptive headings keeps the writing on track and helps the reader get to key information quickly. What should the structure of the methods section look like? As always, a lot depends on the type of review but it will certainly contain information relating to the following areas:
- Selection criteria ⭕
- Data collection and analysis 👩💻
- Study quality and risk of bias ⚖️
Let’s look at each of these in turn.
1. Selection criteria ⭕
The criteria for including and excluding studies are listed here. This includes detail about the types of studies, the types of participants, the types of interventions and the types of outcomes and how they were measured.
2. Search 🕵🏾♀️
Comprehensive reporting of the search is important because this means it can be evaluated and replicated. The search strategies are included in the review, along with details of the databases searched. It’s also important to list any restrictions on the search (for example, language), describe how resources other than electronic databases were searched (for example, non-indexed journals), and give the date that the searches were run. The PRISMA-S extension provides guidance on reporting literature searches.
Systematic reviewer pro-tip:
Copy and paste the search strategy to avoid introducing typos
3. Data collection and analysis 👩💻
This section describes:
- how studies were selected for inclusion in the review
- how study data were extracted from the study reports
- how study data were combined for analysis and synthesis
To describe how studies were selected for inclusion , review teams outline the screening process. Covidence uses reviewers’ decision data to automatically populate a PRISMA flow diagram for this purpose. Covidence can also calculate Cohen’s kappa to enable review teams to report the level of agreement among individual reviewers during screening.
To describe how study data were extracted from the study reports , reviewers outline the form that was used, any pilot-testing that was done, and the items that were extracted from the included studies. An important piece of information to include here is the process used to resolve conflict among the reviewers. Covidence’s data extraction tool saves reviewers’ comments and notes in the system as they work. This keeps the information in one place for easy retrieval ⚡.
To describe how study data were combined for analysis and synthesis, reviewers outline the type of synthesis (narrative or quantitative, for example), the methods for grouping data, the challenges that came up, and how these were dealt with. If the review includes a meta-analysis, it will detail how this was performed and how the treatment effects were measured.
4. Study quality and risk of bias ⚖️
Because the results of systematic reviews can be affected by many types of bias, reviewers make every effort to minimise it and to show the reader that the methods they used were appropriate. This section describes the methods used to assess study quality and an assessment of the risk of bias across a range of domains.
Steps to assess the risk of bias in studies include looking at how study participants were assigned to treatment groups and whether patients and/or study assessors were blinded to the treatment given. Reviewers also report their assessment of the risk of bias due to missing outcome data, whether that is due to participant drop-out or non-reporting of the outcomes by the study authors.
Covidence’s default template for assessing study quality is Cochrane’s risk of bias tool but it is also possible to start from scratch and build a tool with a set of custom domains if you prefer.
Careful planning, clear writing, and a structured approach are key to a good methods section. A methodologist will be able to refer review teams to examples of good methods reporting in the literature. Covidence helps reviewers to screen references, extract data and complete risk of bias tables quickly and efficiently. Sign up for a free trial today!

Laura Mellor. Portsmouth, UK
Perhaps you'd also like....

Top 5 Tips for High-Quality Systematic Review Data Extraction
Data extraction can be a complex step in the systematic review process. Here are 5 top tips from our experts to help prepare and achieve high quality data extraction.

How to get through study quality assessment Systematic Review
Find out 5 tops tips to conducting quality assessment and why it’s an important step in the systematic review process.

How to extract study data for your systematic review
Learn the basic process and some tips to build data extraction forms for your systematic review with Covidence.
Better systematic review management
Head office, working for an institution or organisation.
Find out why over 350 of the world’s leading institutions are seeing a surge in publications since using Covidence!
Request a consultation with one of our team members and start empowering your researchers:
By using our site you consent to our use of cookies to measure and improve our site’s performance. Please see our Privacy Policy for more information.
- Open access
- Published: 17 August 2023
Data visualisation in scoping reviews and evidence maps on health topics: a cross-sectional analysis
- Emily South ORCID: orcid.org/0000-0003-2187-4762 1 &
- Mark Rodgers 1
Systematic Reviews volume 12 , Article number: 142 ( 2023 ) Cite this article
3658 Accesses
13 Altmetric
Metrics details
Scoping reviews and evidence maps are forms of evidence synthesis that aim to map the available literature on a topic and are well-suited to visual presentation of results. A range of data visualisation methods and interactive data visualisation tools exist that may make scoping reviews more useful to knowledge users. The aim of this study was to explore the use of data visualisation in a sample of recent scoping reviews and evidence maps on health topics, with a particular focus on interactive data visualisation.
Ovid MEDLINE ALL was searched for recent scoping reviews and evidence maps (June 2020-May 2021), and a sample of 300 papers that met basic selection criteria was taken. Data were extracted on the aim of each review and the use of data visualisation, including types of data visualisation used, variables presented and the use of interactivity. Descriptive data analysis was undertaken of the 238 reviews that aimed to map evidence.
Of the 238 scoping reviews or evidence maps in our analysis, around one-third (37.8%) included some form of data visualisation. Thirty-five different types of data visualisation were used across this sample, although most data visualisations identified were simple bar charts (standard, stacked or multi-set), pie charts or cross-tabulations (60.8%). Most data visualisations presented a single variable (64.4%) or two variables (26.1%). Almost a third of the reviews that used data visualisation did not use any colour (28.9%). Only two reviews presented interactive data visualisation, and few reported the software used to create visualisations.
Conclusions
Data visualisation is currently underused by scoping review authors. In particular, there is potential for much greater use of more innovative forms of data visualisation and interactive data visualisation. Where more innovative data visualisation is used, scoping reviews have made use of a wide range of different methods. Increased use of these more engaging visualisations may make scoping reviews more useful for a range of stakeholders.
Peer Review reports
Scoping reviews are “a type of evidence synthesis that aims to systematically identify and map the breadth of evidence available on a particular topic, field, concept, or issue” ([ 1 ], p. 950). While they include some of the same steps as a systematic review, such as systematic searches and the use of predetermined eligibility criteria, scoping reviews often address broader research questions and do not typically involve the quality appraisal of studies or synthesis of data [ 2 ]. Reasons for conducting a scoping review include the following: to map types of evidence available, to explore research design and conduct, to clarify concepts or definitions and to map characteristics or factors related to a concept [ 3 ]. Scoping reviews can also be undertaken to inform a future systematic review (e.g. to assure authors there will be adequate studies) or to identify knowledge gaps [ 3 ]. Other evidence synthesis approaches with similar aims have been described as evidence maps, mapping reviews or systematic maps [ 4 ]. While this terminology is used inconsistently, evidence maps can be used to identify evidence gaps and present them in a user-friendly (and often visual) way [ 5 ].
Scoping reviews are often targeted to an audience of healthcare professionals or policy-makers [ 6 ], suggesting that it is important to present results in a user-friendly and informative way. Until recently, there was little guidance on how to present the findings of scoping reviews. In recent literature, there has been some discussion of the importance of clearly presenting data for the intended audience of a scoping review, with creative and innovative use of visual methods if appropriate [ 7 , 8 , 9 ]. Lockwood et al. suggest that innovative visual presentation should be considered over dense sections of text or long tables in many cases [ 8 ]. Khalil et al. suggest that inspiration could be drawn from the field of data visualisation [ 7 ]. JBI guidance on scoping reviews recommends that reviewers carefully consider the best format for presenting data at the protocol development stage and provides a number of examples of possible methods [ 10 ].
Interactive resources are another option for presentation in scoping reviews [ 9 ]. Researchers without the relevant programming skills can now use several online platforms (such as Tableau [ 11 ] and Flourish [ 12 ]) to create interactive data visualisations. The benefits of using interactive visualisation in research include the ability to easily present more than two variables [ 13 ] and increased engagement of users [ 14 ]. Unlike static graphs, interactive visualisations can allow users to view hierarchical data at different levels, exploring both the “big picture” and looking in more detail ([ 15 ], p. 291). Interactive visualizations are often targeted at practitioners and decision-makers [ 13 ], and there is some evidence from qualitative research that they are valued by policy-makers [ 16 , 17 , 18 ].
Given their focus on mapping evidence, we believe that scoping reviews are particularly well-suited to visually presenting data and the use of interactive data visualisation tools. However, it is unknown how many recent scoping reviews visually map data or which types of data visualisation are used. The aim of this study was to explore the use of data visualisation methods in a large sample of recent scoping reviews and evidence maps on health topics. In particular, we were interested in the extent to which these forms of synthesis use any form of interactive data visualisation.
This study was a cross-sectional analysis of studies labelled as scoping reviews or evidence maps (or synonyms of these terms) in the title or abstract.
The search strategy was developed with help from an information specialist. Ovid MEDLINE® ALL was searched in June 2021 for studies added to the database in the previous 12 months. The search was limited to English language studies only.
The search strategy was as follows:
Ovid MEDLINE(R) ALL
(scoping review or evidence map or systematic map or mapping review or scoping study or scoping project or scoping exercise or literature mapping or evidence mapping or systematic mapping or literature scoping or evidence gap map).ab,ti.
limit 1 to english language
(202006* or 202007* or 202008* or 202009* or 202010* or 202011* or 202012* or 202101* or 202102* or 202103* or 202104* or 202105*).dt.
The search returned 3686 records. Records were de-duplicated in EndNote 20 software, leaving 3627 unique records.
A sample of these reviews was taken by screening the search results against basic selection criteria (Table 1 ). These criteria were piloted and refined after discussion between the two researchers. A single researcher (E.S.) screened the records in EPPI-Reviewer Web software using the machine-learning priority screening function. Where a second opinion was needed, decisions were checked by a second researcher (M.R.).
Our initial plan for sampling, informed by pilot searching, was to screen and data extract records in batches of 50 included reviews at a time. We planned to stop screening when a batch of 50 reviews had been extracted that included no new types of data visualisation or after screening time had reached 2 days. However, once data extraction was underway, we found the sample to be richer in terms of data visualisation than anticipated. After the inclusion of 300 reviews, we took the decision to end screening in order to ensure the study was manageable.
Data extraction
A data extraction form was developed in EPPI-Reviewer Web, piloted on 50 reviews and refined. Data were extracted by one researcher (E. S. or M. R.), with a second researcher (M. R. or E. S.) providing a second opinion when needed. The data items extracted were as follows: type of review (term used by authors), aim of review (mapping evidence vs. answering specific question vs. borderline), number of visualisations (if any), types of data visualisation used, variables/domains presented by each visualisation type, interactivity, use of colour and any software requirements.
When categorising review aims, we considered “mapping evidence” to incorporate all of the six purposes for conducting a scoping review proposed by Munn et al. [ 3 ]. Reviews were categorised as “answering a specific question” if they aimed to synthesise study findings to answer a particular question, for example on effectiveness of an intervention. We were inclusive with our definition of “mapping evidence” and included reviews with mixed aims in this category. However, some reviews were difficult to categorise (for example where aims were unclear or the stated aims did not match the actual focus of the paper) and were considered to be “borderline”. It became clear that a proportion of identified records that described themselves as “scoping” or “mapping” reviews were in fact pseudo-systematic reviews that failed to undertake key systematic review processes. Such reviews attempted to integrate the findings of included studies rather than map the evidence, and so reviews categorised as “answering a specific question” were excluded from the main analysis. Data visualisation methods for meta-analyses have been explored previously [ 19 ]. Figure 1 shows the flow of records from search results to final analysis sample.
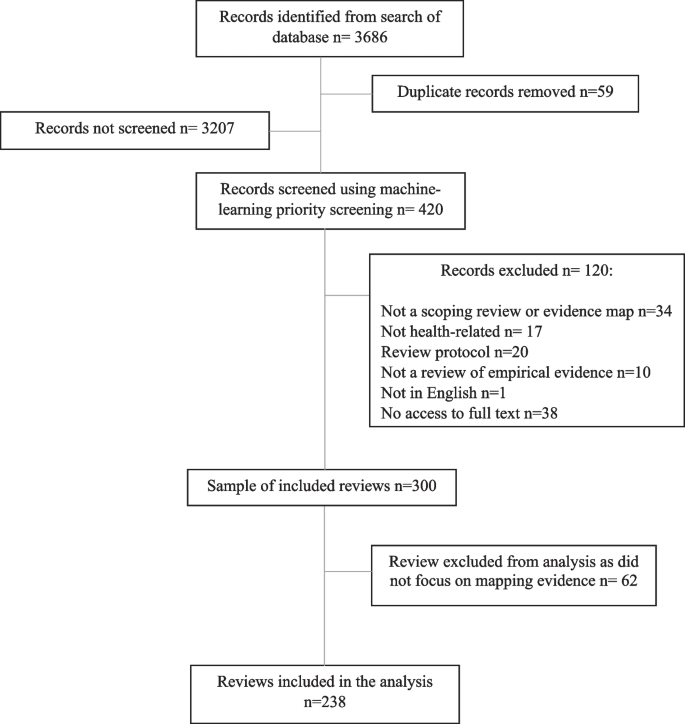
Flow diagram of the sampling process
Data visualisation was defined as any graph or diagram that presented results data, including tables with a visual mapping element, such as cross-tabulations and heat maps. However, tables which displayed data at a study level (e.g. tables summarising key characteristics of each included study) were not included, even if they used symbols, shading or colour. Flow diagrams showing the study selection process were also excluded. Data visualisations in appendices or supplementary information were included, as well as any in publicly available dissemination products (e.g. visualisations hosted online) if mentioned in papers.
The typology used to categorise data visualisation methods was based on an existing online catalogue [ 20 ]. Specific types of data visualisation were categorised in five broad categories: graphs, diagrams, tables, maps/geographical and other. If a data visualisation appeared in our sample that did not feature in the original catalogue, we checked a second online catalogue [ 21 ] for an appropriate term, followed by wider Internet searches. These additional visualisation methods were added to the appropriate section of the typology. The final typology can be found in Additional file 1 .
We conducted descriptive data analysis in Microsoft Excel 2019 and present frequencies and percentages. Where appropriate, data are presented using graphs or other data visualisations created using Flourish. We also link to interactive versions of some of these visualisations.
Almost all of the 300 reviews in the total sample were labelled by review authors as “scoping reviews” ( n = 293, 97.7%). There were also four “mapping reviews”, one “scoping study”, one “evidence mapping” and one that was described as a “scoping review and evidence map”. Included reviews were all published in 2020 or 2021, with the exception of one review published in 2018. Just over one-third of these reviews ( n = 105, 35.0%) included some form of data visualisation. However, we excluded 62 reviews that did not focus on mapping evidence from the following analysis (see “ Methods ” section). Of the 238 remaining reviews (that either clearly aimed to map evidence or were judged to be “borderline”), 90 reviews (37.8%) included at least one data visualisation. The references for these reviews can be found in Additional file 2 .
Number of visualisations
Thirty-six (40.0%) of these 90 reviews included just one example of data visualisation (Fig. 2 ). Less than a third ( n = 28, 31.1%) included three or more visualisations. The greatest number of data visualisations in one review was 17 (all bar or pie charts). In total, 222 individual data visualisations were identified across the sample of 238 reviews.
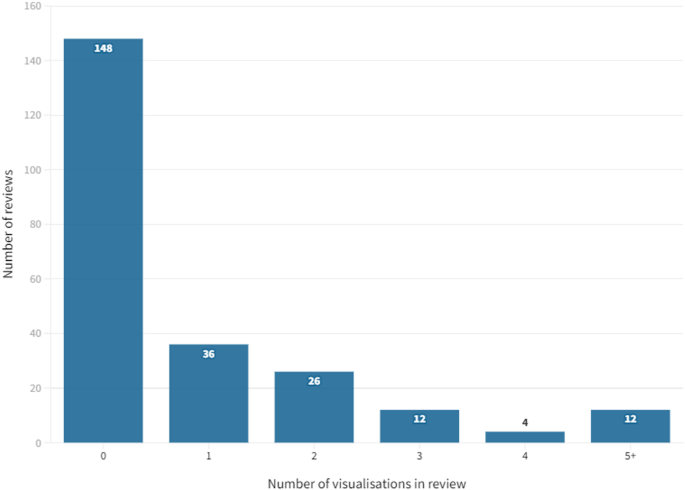
Number of data visualisations per review
Categories of data visualisation
Graphs were the most frequently used category of data visualisation in the sample. Over half of the reviews with data visualisation included at least one graph ( n = 59, 65.6%). The least frequently used category was maps, with 15.6% ( n = 14) of these reviews including a map.
Of the total number of 222 individual data visualisations, 102 were graphs (45.9%), 34 were tables (15.3%), 23 were diagrams (10.4%), 15 were maps (6.8%) and 48 were classified as “other” in the typology (21.6%).
Types of data visualisation
All of the types of data visualisation identified in our sample are reported in Table 2 . In total, 35 different types were used across the sample of reviews.
The most frequently used data visualisation type was a bar chart. Of 222 total data visualisations, 78 (35.1%) were a variation on a bar chart (either standard bar chart, stacked bar chart or multi-set bar chart). There were also 33 pie charts (14.9% of data visualisations) and 24 cross-tabulations (10.8% of data visualisations). In total, these five types of data visualisation accounted for 60.8% ( n = 135) of all data visualisations. Figure 3 shows the frequency of each data visualisation category and type; an interactive online version of this treemap is also available ( https://public.flourish.studio/visualisation/9396133/ ). Figure 4 shows how users can further explore the data using the interactive treemap.
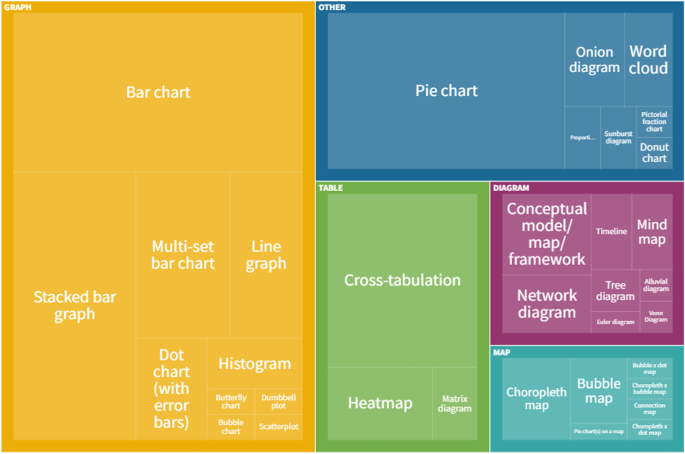
Data visualisation categories and types. An interactive version of this treemap is available online: https://public.flourish.studio/visualisation/9396133/ . Through the interactive version, users can further explore the data (see Fig. 4 ). The unit of this treemap is the individual data visualisation, so multiple data visualisations within the same scoping review are represented in this map. Created with flourish.studio ( https://flourish.studio )
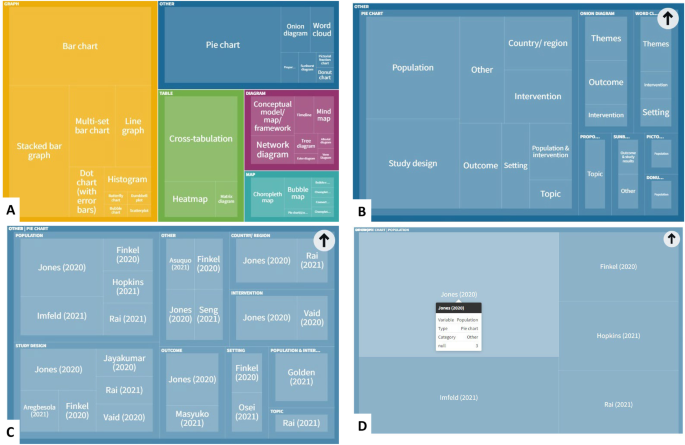
Screenshots showing how users of the interactive treemap can explore the data further. Users can explore each level of the hierarchical treemap ( A Visualisation category > B Visualisation subcategory > C Variables presented in visualisation > D Individual references reporting this category/subcategory/variable permutation). Created with flourish.studio ( https://flourish.studio )
Data presented
Around two-thirds of data visualisations in the sample presented a single variable ( n = 143, 64.4%). The most frequently presented single variables were themes ( n = 22, 9.9% of data visualisations), population ( n = 21, 9.5%), country or region ( n = 21, 9.5%) and year ( n = 20, 9.0%). There were 58 visualisations (26.1%) that presented two different variables. The remaining 21 data visualisations (9.5%) presented three or more variables. Figure 5 shows the variables presented by each different type of data visualisation (an interactive version of this figure is available online).
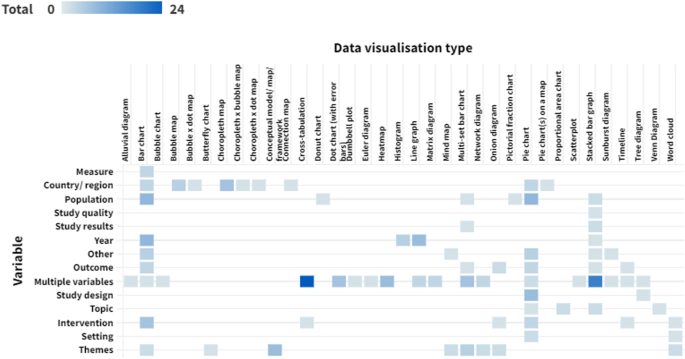
Variables presented by each data visualisation type. Darker cells indicate a larger number of reviews. An interactive version of this heat map is available online: https://public.flourish.studio/visualisation/10632665/ . Users can hover over each cell to see the number of data visualisations for that combination of data visualisation type and variable. The unit of this heat map is the individual data visualisation, so multiple data visualisations within a single scoping review are represented in this map. Created with flourish.studio ( https://flourish.studio )
Most reviews presented at least one data visualisation in colour ( n = 64, 71.1%). However, almost a third ( n = 26, 28.9%) used only black and white or greyscale.
Interactivity
Only two of the reviews included data visualisations with any level of interactivity. One scoping review on music and serious mental illness [ 22 ] linked to an interactive bubble chart hosted online on Tableau. Functionality included the ability to filter the studies displayed by various attributes.
The other review was an example of evidence mapping from the environmental health field [ 23 ]. All four of the data visualisations included in the paper were available in an interactive format hosted either by the review management software or on Tableau. The interactive versions linked to the relevant references so users could directly explore the evidence base. This was the only review that provided this feature.
Software requirements
Nine reviews clearly reported the software used to create data visualisations. Three reviews used Tableau (one of them also used review management software as discussed above) [ 22 , 23 , 24 ]. Two reviews generated maps using ArcGIS [ 25 ] or ArcMap [ 26 ]. One review used Leximancer for a lexical analysis [ 27 ]. One review undertook a bibliometric analysis using VOSviewer [ 28 ], and another explored citation patterns using CitNetExplorer [ 29 ]. Other reviews used Excel [ 30 ] or R [ 26 ].
To our knowledge, this is the first systematic and in-depth exploration of the use of data visualisation techniques in scoping reviews. Our findings suggest that the majority of scoping reviews do not use any data visualisation at all, and, in particular, more innovative examples of data visualisation are rare. Around 60% of data visualisations in our sample were simple bar charts, pie charts or cross-tabulations. There appears to be very limited use of interactive online visualisation, despite the potential this has for communicating results to a range of stakeholders. While it is not always appropriate to use data visualisation (or a simple bar chart may be the most user-friendly way of presenting the data), these findings suggest that data visualisation is being underused in scoping reviews. In a large minority of reviews, visualisations were not published in colour, potentially limiting how user-friendly and attractive papers are to decision-makers and other stakeholders. Also, very few reviews clearly reported the software used to create data visualisations. However, 35 different types of data visualisation were used across the sample, highlighting the wide range of methods that are potentially available to scoping review authors.
Our results build on the limited research that has previously been undertaken in this area. Two previous publications also found limited use of graphs in scoping reviews. Results were “mapped graphically” in 29% of scoping reviews in any field in one 2014 publication [ 31 ] and 17% of healthcare scoping reviews in a 2016 article [ 6 ]. Our results suggest that the use of data visualisation has increased somewhat since these reviews were conducted. Scoping review methods have also evolved in the last 10 years; formal guidance on scoping review conduct was published in 2014 [ 32 ], and an extension of the PRISMA checklist for scoping reviews was published in 2018 [ 33 ]. It is possible that an overall increase in use of data visualisation reflects increased quality of published scoping reviews. There is also some literature supporting our findings on the wide range of data visualisation methods that are used in evidence synthesis. An investigation of methods to identify, prioritise or display health research gaps (25/139 included studies were scoping reviews; 6/139 were evidence maps) identified 14 different methods used to display gaps or priorities, with half being “more advanced” (e.g. treemaps, radial bar plots) ([ 34 ], p. 107). A review of data visualisation methods used in papers reporting meta-analyses found over 200 different ways of displaying data [ 19 ].
Only two reviews in our sample used interactive data visualisation, and one of these was an example of systematic evidence mapping from the environmental health field rather than a scoping review (in environmental health, systematic evidence mapping explicitly involves producing a searchable database [ 35 ]). A scoping review of papers on the use of interactive data visualisation in population health or health services research found a range of examples but still limited use overall [ 13 ]. For example, the authors noted the currently underdeveloped potential for using interactive visualisation in research on health inequalities. It is possible that the use of interactive data visualisation in academic papers is restricted by academic publishing requirements; for example, it is currently difficult to incorporate an interactive figure into a journal article without linking to an external host or platform. However, we believe that there is a lot of potential to add value to future scoping reviews by using interactive data visualisation software. Few reviews in our sample presented three or more variables in a single visualisation, something which can easily be achieved using interactive data visualisation tools. We have previously used EPPI-Mapper [ 36 ] to present results of a scoping review of systematic reviews on behaviour change in disadvantaged groups, with links to the maps provided in the paper [ 37 ]. These interactive maps allowed policy-makers to explore the evidence on different behaviours and disadvantaged groups and access full publications of the included studies directly from the map.
We acknowledge there are barriers to use for some of the data visualisation software available. EPPI-Mapper and some of the software used by reviews in our sample incur a cost. Some software requires a certain level of knowledge and skill in its use. However numerous online free data visualisation tools and resources exist. We have used Flourish to present data for this review, a basic version of which is currently freely available and easy to use. Previous health research has been found to have used a range of different interactive data visualisation software, much of which does not required advanced knowledge or skills to use [ 13 ].
There are likely to be other barriers to the use of data visualisation in scoping reviews. Journal guidelines and policies may present barriers for using innovative data visualisation. For example, some journals charge a fee for publication of figures in colour. As previously mentioned, there are limited options for incorporating interactive data visualisation into journal articles. Authors may also be unaware of the data visualisation methods and tools that are available. Producing data visualisations can be time-consuming, particularly if authors lack experience and skills in this. It is possible that many authors prioritise speed of publication over spending time producing innovative data visualisations, particularly in a context where there is pressure to achieve publications.
Limitations
A limitation of this study was that we did not assess how appropriate the use of data visualisation was in our sample as this would have been highly subjective. Simple descriptive or tabular presentation of results may be the most appropriate approach for some scoping review objectives [ 7 , 8 , 10 ], and the scoping review literature cautions against “over-using” different visual presentation methods [ 7 , 8 ]. It cannot be assumed that all of the reviews that did not include data visualisation should have done so. Likewise, we do not know how many reviews used methods of data visualisation that were not well suited to their data.
We initially relied on authors’ own use of the term “scoping review” (or equivalent) to sample reviews but identified a relatively large number of papers labelled as scoping reviews that did not meet the basic definition, despite the availability of guidance and reporting guidelines [ 10 , 33 ]. It has previously been noted that scoping reviews may be undertaken inappropriately because they are seen as “easier” to conduct than a systematic review ([ 3 ], p.6), and that reviews are often labelled as “scoping reviews” while not appearing to follow any established framework or guidance [ 2 ]. We therefore took the decision to remove these reviews from our main analysis. However, decisions on how to classify review aims were subjective, and we did include some reviews that were of borderline relevance.
A further limitation is that this was a sample of published reviews, rather than a comprehensive systematic scoping review as have previously been undertaken [ 6 , 31 ]. The number of scoping reviews that are published has increased rapidly, and this would now be difficult to undertake. As this was a sample, not all relevant scoping reviews or evidence maps that would have met our criteria were included. We used machine learning to screen our search results for pragmatic reasons (to reduce screening time), but we do not see any reason that our sample would not be broadly reflective of the wider literature.
Data visualisation, and in particular more innovative examples of it, is currently underused in published scoping reviews on health topics. The examples that we have found highlight the wide range of methods that scoping review authors could draw upon to present their data in an engaging way. In particular, we believe that interactive data visualisation has significant potential for mapping the available literature on a topic. Appropriate use of data visualisation may increase the usefulness, and thus uptake, of scoping reviews as a way of identifying existing evidence or research gaps by decision-makers, researchers and commissioners of research. We recommend that scoping review authors explore the extensive free resources and online tools available for data visualisation. However, we also think that it would be useful for publishers to explore allowing easier integration of interactive tools into academic publishing, given the fact that papers are now predominantly accessed online. Future research may be helpful to explore which methods are particularly useful to scoping review users.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
Organisation formerly known as Joanna Briggs Institute
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
Munn Z, Pollock D, Khalil H, Alexander L, McLnerney P, Godfrey CM, Peters M, Tricco AC. What are scoping reviews? Providing a formal definition of scoping reviews as a type of evidence synthesis. JBI Evid Synth. 2022;20:950–952.
Peters MDJ, Marnie C, Colquhoun H, Garritty CM, Hempel S, Horsley T, Langlois EV, Lillie E, O’Brien KK, Tunçalp Ӧ, et al. Scoping reviews: reinforcing and advancing the methodology and application. Syst Rev. 2021;10:263.
Article PubMed PubMed Central Google Scholar
Munn Z, Peters MDJ, Stern C, Tufanaru C, McArthur A, Aromataris E. Systematic review or scoping review? Guidance for authors when choosing between a systematic or scoping review approach. BMC Med Res Methodol. 2018;18:143.
Sutton A, Clowes M, Preston L, Booth A. Meeting the review family: exploring review types and associated information retrieval requirements. Health Info Libr J. 2019;36:202–22.
Article PubMed Google Scholar
Miake-Lye IM, Hempel S, Shanman R, Shekelle PG. What is an evidence map? A systematic review of published evidence maps and their definitions, methods, and products. Syst Rev. 2016;5:28.
Tricco AC, Lillie E, Zarin W, O’Brien K, Colquhoun H, Kastner M, Levac D, Ng C, Sharpe JP, Wilson K, et al. A scoping review on the conduct and reporting of scoping reviews. BMC Med Res Methodol. 2016;16:15.
Khalil H, Peters MDJ, Tricco AC, Pollock D, Alexander L, McInerney P, Godfrey CM, Munn Z. Conducting high quality scoping reviews-challenges and solutions. J Clin Epidemiol. 2021;130:156–60.
Lockwood C, dos Santos KB, Pap R. Practical guidance for knowledge synthesis: scoping review methods. Asian Nurs Res. 2019;13:287–94.
Article Google Scholar
Pollock D, Peters MDJ, Khalil H, McInerney P, Alexander L, Tricco AC, Evans C, de Moraes ÉB, Godfrey CM, Pieper D, et al. Recommendations for the extraction, analysis, and presentation of results in scoping reviews. JBI Evidence Synthesis. 2022;10:11124.
Google Scholar
Peters MDJ GC, McInerney P, Munn Z, Tricco AC, Khalil, H. Chapter 11: Scoping reviews (2020 version). In: Aromataris E MZ, editor. JBI Manual for Evidence Synthesis. JBI; 2020. Available from https://synthesismanual.jbi.global . Accessed 1 Feb 2023.
Tableau Public. https://www.tableau.com/en-gb/products/public . Accessed 24 January 2023.
flourish.studio. https://flourish.studio/ . Accessed 24 January 2023.
Chishtie J, Bielska IA, Barrera A, Marchand J-S, Imran M, Tirmizi SFA, Turcotte LA, Munce S, Shepherd J, Senthinathan A, et al. Interactive visualization applications in population health and health services research: systematic scoping review. J Med Internet Res. 2022;24: e27534.
Isett KR, Hicks DM. Providing public servants what they need: revealing the “unseen” through data visualization. Public Adm Rev. 2018;78:479–85.
Carroll LN, Au AP, Detwiler LT, Fu T-c, Painter IS, Abernethy NF. Visualization and analytics tools for infectious disease epidemiology: a systematic review. J Biomed Inform. 2014;51:287–298.
Lundkvist A, El-Khatib Z, Kalra N, Pantoja T, Leach-Kemon K, Gapp C, Kuchenmüller T. Policy-makers’ views on translating burden of disease estimates in health policies: bridging the gap through data visualization. Arch Public Health. 2021;79:17.
Zakkar M, Sedig K. Interactive visualization of public health indicators to support policymaking: an exploratory study. Online J Public Health Inform. 2017;9:e190–e190.
Park S, Bekemeier B, Flaxman AD. Understanding data use and preference of data visualization for public health professionals: a qualitative study. Public Health Nurs. 2021;38:531–41.
Kossmeier M, Tran US, Voracek M. Charting the landscape of graphical displays for meta-analysis and systematic reviews: a comprehensive review, taxonomy, and feature analysis. BMC Med Res Methodol. 2020;20:26.
Ribecca, S. The Data Visualisation Catalogue. https://datavizcatalogue.com/index.html . Accessed 23 November 2021.
Ferdio. Data Viz Project. https://datavizproject.com/ . Accessed 23 November 2021.
Golden TL, Springs S, Kimmel HJ, Gupta S, Tiedemann A, Sandu CC, Magsamen S. The use of music in the treatment and management of serious mental illness: a global scoping review of the literature. Front Psychol. 2021;12: 649840.
Keshava C, Davis JA, Stanek J, Thayer KA, Galizia A, Keshava N, Gift J, Vulimiri SV, Woodall G, Gigot C, et al. Application of systematic evidence mapping to assess the impact of new research when updating health reference values: a case example using acrolein. Environ Int. 2020;143: 105956.
Article CAS PubMed PubMed Central Google Scholar
Jayakumar P, Lin E, Galea V, Mathew AJ, Panda N, Vetter I, Haynes AB. Digital phenotyping and patient-generated health data for outcome measurement in surgical care: a scoping review. J Pers Med. 2020;10:282.
Qu LG, Perera M, Lawrentschuk N, Umbas R, Klotz L. Scoping review: hotspots for COVID-19 urological research: what is being published and from where? World J Urol. 2021;39:3151–60.
Article CAS PubMed Google Scholar
Rossa-Roccor V, Acheson ES, Andrade-Rivas F, Coombe M, Ogura S, Super L, Hong A. Scoping review and bibliometric analysis of the term “planetary health” in the peer-reviewed literature. Front Public Health. 2020;8:343.
Hewitt L, Dahlen HG, Hartz DL, Dadich A. Leadership and management in midwifery-led continuity of care models: a thematic and lexical analysis of a scoping review. Midwifery. 2021;98: 102986.
Xia H, Tan S, Huang S, Gan P, Zhong C, Lu M, Peng Y, Zhou X, Tang X. Scoping review and bibliometric analysis of the most influential publications in achalasia research from 1995 to 2020. Biomed Res Int. 2021;2021:8836395.
Vigliotti V, Taggart T, Walker M, Kusmastuti S, Ransome Y. Religion, faith, and spirituality influences on HIV prevention activities: a scoping review. PLoS ONE. 2020;15: e0234720.
van Heemskerken P, Broekhuizen H, Gajewski J, Brugha R, Bijlmakers L. Barriers to surgery performed by non-physician clinicians in sub-Saharan Africa-a scoping review. Hum Resour Health. 2020;18:51.
Pham MT, Rajić A, Greig JD, Sargeant JM, Papadopoulos A, McEwen SA. A scoping review of scoping reviews: advancing the approach and enhancing the consistency. Res Synth Methods. 2014;5:371–85.
Peters MDJ, Marnie C, Tricco AC, Pollock D, Munn Z, Alexander L, McInerney P, Godfrey CM, Khalil H. Updated methodological guidance for the conduct of scoping reviews. JBI Evid Synth. 2020;18:2119–26.
Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, Moher D, Peters MDJ, Horsley T, Weeks L, et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. 2018;169:467–73.
Nyanchoka L, Tudur-Smith C, Thu VN, Iversen V, Tricco AC, Porcher R. A scoping review describes methods used to identify, prioritize and display gaps in health research. J Clin Epidemiol. 2019;109:99–110.
Wolffe TAM, Whaley P, Halsall C, Rooney AA, Walker VR. Systematic evidence maps as a novel tool to support evidence-based decision-making in chemicals policy and risk management. Environ Int. 2019;130:104871.
Digital Solution Foundry and EPPI-Centre. EPPI-Mapper, Version 2.0.1. EPPI-Centre, UCL Social Research Institute, University College London. 2020. https://eppi.ioe.ac.uk/cms/Default.aspx?tabid=3790 .
South E, Rodgers M, Wright K, Whitehead M, Sowden A. Reducing lifestyle risk behaviours in disadvantaged groups in high-income countries: a scoping review of systematic reviews. Prev Med. 2022;154: 106916.
Download references
Acknowledgements
We would like to thank Melissa Harden, Senior Information Specialist, Centre for Reviews and Dissemination, for advice on developing the search strategy.
This work received no external funding.
Author information
Authors and affiliations.
Centre for Reviews and Dissemination, University of York, York, YO10 5DD, UK
Emily South & Mark Rodgers
You can also search for this author in PubMed Google Scholar
Contributions
Both authors conceptualised and designed the study and contributed to screening, data extraction and the interpretation of results. ES undertook the literature searches, analysed data, produced the data visualisations and drafted the manuscript. MR contributed to revising the manuscript, and both authors read and approved the final version.
Corresponding author
Correspondence to Emily South .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1..
Typology of data visualisation methods.
Additional file 2.
References of scoping reviews included in main dataset.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
South, E., Rodgers, M. Data visualisation in scoping reviews and evidence maps on health topics: a cross-sectional analysis. Syst Rev 12 , 142 (2023). https://doi.org/10.1186/s13643-023-02309-y
Download citation
Received : 21 February 2023
Accepted : 07 August 2023
Published : 17 August 2023
DOI : https://doi.org/10.1186/s13643-023-02309-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Scoping review
- Evidence map
- Data visualisation
Systematic Reviews
ISSN: 2046-4053
- Submission enquiries: Access here and click Contact Us
- General enquiries: [email protected]
- Open access
- Published: 13 May 2024
Neighborhood based computational approaches for the prediction of lncRNA-disease associations
- Mariella Bonomo 1 &
- Simona E. Rombo 1 , 2
BMC Bioinformatics volume 25 , Article number: 187 ( 2024 ) Cite this article
46 Accesses
Metrics details
Long non-coding RNAs (lncRNAs) are a class of molecules involved in important biological processes. Extensive efforts have been provided to get deeper understanding of disease mechanisms at the lncRNA level, guiding towards the detection of biomarkers for disease diagnosis, treatment, prognosis and prevention. Unfortunately, due to costs and time complexity, the number of possible disease-related lncRNAs verified by traditional biological experiments is very limited. Computational approaches for the prediction of disease-lncRNA associations allow to identify the most promising candidates to be verified in laboratory, reducing costs and time consuming.
We propose novel approaches for the prediction of lncRNA-disease associations, all sharing the idea of exploring associations among lncRNAs, other intermediate molecules (e.g., miRNAs) and diseases, suitably represented by tripartite graphs. Indeed, while only a few lncRNA-disease associations are still known, plenty of interactions between lncRNAs and other molecules, as well as associations of the latters with diseases, are available. A first approach presented here, NGH, relies on neighborhood analysis performed on a tripartite graph, built upon lncRNAs, miRNAs and diseases. A second approach (CF) relies on collaborative filtering; a third approach (NGH-CF) is obtained boosting NGH by collaborative filtering. The proposed approaches have been validated on both synthetic and real data, and compared against other methods from the literature. It results that neighborhood analysis allows to outperform competitors, and when it is combined with collaborative filtering the prediction accuracy further improves, scoring a value of AUC equal to 0966.
Availability
Source code and sample datasets are available at: https://github.com/marybonomo/LDAsPredictionApproaches.git
Peer Review reports
Introduction
More than \(98\%\) of the human genome consists of non-coding regions, considered in the past as “junk” DNA. However, in the last decades evidence has been shown that non-coding genome elements often play an important role in regulating various critical biological processes [ 1 ]. An important class of non-coding molecules which have started to receive great attention in the last few years is represented by long non-coding RNAs (lncRNAs), that is, RNAs not translated into functional proteins, and longer than 200 nucleotides.
LncRNAs have been found to interplay with other molecules in order to perform important biological tasks, such as modulating chromatin function, regulating the assembly and function of membraneless nuclear bodies, interfering with signalling pathways [ 2 , 3 ]. Many of these functions ultimately affect gene expression in diverse biological and physiopathological contexts, such as in neuronal disorders, immune responses and cancer. Therefore, the alteration and dysregulation of lncRNAs have been associated with the occurrence and progress of many complex diseases [ 4 ].
The discovery of novel lncRNA-disease associations (LDAs) may provide valuable input to the understanding of disease mechanisms at lncRNA level, as well as to the detection of disease biomarkers for disease diagnosis, treatment, prognosis and prevention. Unfortunately, verifying that a specific lncRNA may have a role in the occurrence/progress of a given disease is an expensive process, therefore the number of disease-related lncRNAs verified by traditional biological experiments is yet very limited. Computational approaches for the prediction of potential LDAs can effectively decrease the time and cost of biological experiments, allowing for the identification of the most promising lncRNA-disease pairs to be further verified in laboratory (see [ 5 ] for a comprehensive review on the topic). Such approaches often train predictive models on the basis of the known and experimentally validated lncRNA-disease pairs (e.g., [ 6 , 7 , 8 , 9 ]). In other cases, they rely on the analysis of lncRNAs related information stored in public databases, such as their interaction with other types of molecules (e.g., [ 10 , 11 , 12 , 13 , 14 , 15 ]). As an example, large amounts of lncRNA-miRNA interactions have been collected in public databases, and plenty of experimentally confirmed miRNA-disease associations are available as well. However, although non-coding RNA function and its association with human complex diseases have been widely studied in the literature (see [ 16 , 17 , 18 ]), how to provide biologists with more accurate and ready-to-use software tools for LDAs prediction is yet an open challenge, due to the specific characteristics of lncRNAs (e.g., they are much less characterized than other non-coding RNAs.)
We propose three novel computational approaches for the prediction of LDAs, relying on the use of known lncRNA-miRNA interactions (LMIs) and miRNA-disease associations (MDAs). In particular, we model the problem of LDAs prediction as a neighborhood analysis performed on tripartite graphs, where the three sets of vertices represent lncRNAs, miRNAs and diseases, respectively, and vertices are linked according to LMIs and MDAs. Based on the assumption that similar lncRNAs interact with similar diseases [ 12 ], the first approach proposed here (NGH) aims at identifying novel LDAs by analyzing the behaviour of lncRNAs which are neighbors , in terms of their intermediate relationships with miRNAs. The main idea here is that neighborhood analysis automatically guides towards the detection of similar behaviours, and without the need of using a-priory known LDAs for training. Therefore, differently than other approaches from the literature, those proposed here do not involve verified LDAs in the prediction step, thus avoiding possible biases due to the fact that the number and variety of verified LDAs is yet very limited. The second presented approach (CF) relies on collaborative filtering, applied on the basis of common miRNAs shared by different lncRNAs. We have also explored the combination of neighborhood analysis with collaborative filtering, showing that this notably improves the LDAs prediction accuracy. Indeed, the third approach we have designed (NGH-CF) boosts NGH with collaborative filtering, and it is the best performing one, although also NGH and CF have been able to reach high accuracy values across all the different considered validation tests. In particular, Fig. 1 summarizes the research flowchart explained above.
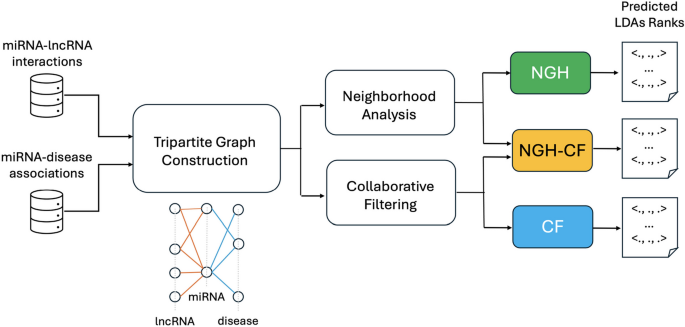
Flowchart of the research pipeline. The miRNA-lncRNA interactions and miRNA-disease associations are exploited for the construction of the tripartite graph. The tripartite graph, in its turn, is at the basis of both neighborhood analysis and collaborative filtering steps, from which the three proposed approaches are obtained: NGH from neighborhood analysis, CF from collaborative filtering, NGH-CF from the combination of the two ones. Each prediction approach returns in output a LDAs rank
The proposed approaches have been exhaustively validated on both synthetic and real datasets, and the result is that they outperform (also significantly) the other methods from the literature. The experimental analysis shows that the improvement in accuracy achieved by the methods proposed here is due to their ability in capturing specific situations neglected by competitors. Examples of that are represented by true LDAs, detected by our approaches and not by the other approaches in the literature, where the involved lncRNA does not present intermediate molecules in common with the associated disease, although its neighbor lncRNAs share a large number of miRNAs with that disease. Moreover, it is shown that our approaches are robust to noise obtained by perturbing a controlled percentage of lncRNA-miRNA interactions and miRNA-disease associations, with NGH-CF the best one also for robustness. The obtained experimental results show that the prediction methods proposed here may effectively support biologists in selecting significant associations to be further verified in laboratory.
Novel putative LDAs coming from the consensus of the three proposed methods, and not yet registered in the available databases as experimentally verified, are provided. Interestingly, the core of novel LDAs returned with highest score by all three approaches finds evidence in the recent literature, while many other high scored predicted LDAs involve less studied lncRNAs, thus providing useful insights for their better characterization.
A first group of approaches aim at using existing true validated cases to train the prediction system, in order to make it able to correctly detect novel cases.
In [ 19 ] a Laplacian Regularized Least Squares is proposed to infer candidates LDAs ( LRLSLDA ) by applying a semi-supervised learning framework. LRLSLDA assumes that similar diseases tend to correlate with functionally similar lncRNAs, and vice versa. Thus, known LDAs and lncRNA expression profiles are combined to prioritize disease-associated lncRNA candidates by LRLSLDA, which does not require negative samples (i.e., confirmed uncorrelated LDAs). In [ 20 ] the method SKF-LDA is proposed that constructs a lncRNA-disease correlation matrix, based on the known LDAs. Then, it calculates the similarity between lncRNAs and that between diseases, according to specific metrics, and integrates such data. Finally, a predicted LDA matrix is obtained by the Laplacian Regularized Least Squares method. The method ENCFLDA [ 6 ] combines matrix decomposition and collaborative filtering. It uses matrix factorization combined with elastic networks and a collaborative filtering algorithm, making the prediction model more stable and eliminating the problem of data over-fitting. HGNNLDA recently proposed in [ 21 ] is based on hypergraph neural network, where the associations are modeled as a lncRNA-drug bipartite graph to build lncRNA hypergraph and drug hypergraph. Hypergraph convolution is then used to learn correlation of higher-order neighbors from the lncRNA and drug hypergraphs. LDAI-ISPS proposed in [ 22 ] is a LDAs inference approach based on space projections of integrated networks, recostructing the disease (lncRNA) integrated similarities network via integrating multiple information, such as disease semantic similarities, lncRNA functional similarities, and known LDAs. A space projection score is finally obtained via vector projections of the weighted networks. In [ 7 ] a consensual prediction approach called HOPEXGB is presented, to identify disease-related miRNAs and lncRNAs by high-order proximity preserved embedding and extreme gradient boosting. The authors build a heterogeneous disease-miRNA-lncRNA (DML) information network by linking lncRNA, miRNA, and disease nodes based on their correlation, and generate a negative dataset based on the similarities between unknown and known associations, in order to reduce the false negative rate in the data set for model construction. The method MAGCNSE proposed in [ 23 ] builds multiple feature matrices based on semantic similarity and disease Gaussian interaction profile kernel similarity of both lncRNAs and diseases. MAGCNSE adaptively assigns weights to the different feature matrices built upon the lncRNAs and diseases similarities. Then, it uses a convolutional neural network to further extract features from multi-channel feature matrices, in order to obtain the final representations of lncRNAs and diseases that is used for the LDAs prediction task.
LDAFGAN [ 8 ] is a model designed for predicting associations between long non-coding RNAs (lncRNAs) and diseases. This method is based on a generative and a discriminative networks, typically implemented as multilayer fully connected neural networks, which generate synthetic data based on some underlying distribution. The generative and discriminative networks are trained together in an adversarial manner. The generative network tries to generate realistic representations of lncRNA-disease associations, while the discriminative network tries to distinguish between real and fake associations. This adversarial training process helps the generative network learn to generate more realistic associations. Once the model is trained, it can predict associations between new lncRNAs and diseases without requiring associated data for those specific lncRNAs. The model captures the data distribution during training, which enables it to make predictions even for unseen lncRNAs. The approach GCNFORMER [ 9 ] is based on graph convolutional network and transformer. First, it integrates the intraclass similarity and interclass connections between miRNAs, lncRNAs and diseases, building a graph adjacency matrix. Then, the method extracts the features between various nodes, by a graph convolutional network. To obtain the global dependencies between inputs and outputs, a transformer encoder with a multiheaded attention mechanism to forecast lncRNA-disease associations is finally applied.
As for the approaches summarized above, it is worth to point out that they may suffer of the fact that the experimentally verified LDAs are still very limited, therefore the training set may be rather incomplete and not enough diversified. For this reason, when such approaches are applied for de novo LDAs prediction, their performance may drastically go down [ 12 ].
Other approaches from the literature use intermediate molecules (e.g., miRNA) to infer novel LDAs. Such approaches are the most related to those we propose here.
The author in [ 11 ] proposes HGLDA , relying on HyperGeometric distribution for LDAs inference, that integrates MDAs and LMIs information. HGLDA has been successfully applied to predict Breast Cancer, Lung Cancer and Colorectal Cancer-related lncRNAs. NcPred [ 10 ] is a resource propagation technique, using a tripartite network where the edges associate each lncRNA with a disease through its targets. The algorithm proposed in [ 10 ] is based on a multilevel resource transfer technique, which computes the weights between each lncRNA-disease pair and, at each step, considers the resource transferred from the previous step. The approach in [ 24 ], referred to as LDA-TG for short in the following, is the antecedent of the approaches proposed here. It relies on the construction of a tripartite graph, built upon MDAs and LMIs. A score is assigned to each possible LDA ( l , d ) by considering both their respective interactions with common miRNAs, and the interactions with miRNAs shared by the considered disease d and other lncRNAs in the neighborhood of l on the tripartite graph. The approaches proposed here differ from LDA-TG for two main reasons. First, the score of LDA-TG is different from the one we introduce here, that allows to reach a better accuracy. Second, a further step based on collaborative filtering is considered here, which also improves the accuracy performance. A method for LDAs prediction relying on a matrix completion technique inspired by recommender systems is presented in [ 14 ]. A two-layer multi-weighted nearest-neighbor prediction model is adopted, using a method similar to memory-based collaborative filtering. Weights are assigned to neighbors for reassigning values to the target matrix, that is an adjacency matrix consisting of lncRNAs, diseases and miRNA. SSMF-BLNP [ 25 ] is based on the combination of selective similarity matrix fusion (SSMF) and bidirectional linear neighborhood label propagation (BLNP). In SSMF, self-similarity networks of lncRNAs and diseases are obtained by selective preprocessing and nonlinear iterative fusion. In BLNP, the initial LDAs are employed in both lncRNA and disease directions as label information for linear neighborhood label propagation.
A third category includes approaches based on integrative frameworks, proposed to take into account different types of information related to lncRNAs, such as their interactions with other molecules, their involvement in disorders and diseases, their similarities. This may improve the prediction step, taking into account simultaneously independent factors.
IntNetLncSim [ 26 ] relies on the construction of an integrated network that comprises lncRNA regulatory data, miRNA-mRNA and mRNA-mRNA interactions. The method computes a similarity score for all pairs of lncRNAs in the integrated network, then analyzes the information flow based on random walk with damping. This allows to infer novel LDAs by exploring the function of lncRNAs. SIMCLDA [ 12 ] identifies LDAs by using inductive matrix completion, based on the integration of known LDAs, disease-gene interactions and gene-gene interactions. The main idea in [ 12 ] is to extract feature vectors of lncRNAs and diseases by principal component analysis, and to calculate the interaction profile for a new lncRNA by the interaction profiles. MFLDA [ 27 ] is a Matrix Factorization based LDAs prediction model that first encodes directly (or indirectly) relevant data sources related to lncRNAs or diseases in individual relational data matrices, and presets weights for these matrices. Then, it simultaneously optimizes the weights and low-rank matrix tri-factorization of each relational data matrix. RWSF-BLP , proposed in [ 28 ], applies a random walk-based multi-similarity fusion method to integrate different similarity matrices, mainly based on semantic and expression data, and bidirectional label propagation. The framework LRWRHLDA is proposed in [ 15 ] based on the construction of a global multi-layer network for LDAs prediction. First, four isomorphic networks including a lncRNA similarity network, a disease similarity network, a gene similarity network and a miRNA similarity network are constructed. Then, six heterogeneous networks involving known lncRNA-disease, lncRNA-gene, lncRNA-miRNA, disease-gene, disease-miRNA, and gene-miRNA associations are built to design the multi-layer network. In [ 29 ] the LDAP-WMPS LDA prediction model is proposed, based on weight matrix and projection score. LDAP-WMPS consists on three steps: the first one computes the disease projection score; the second step calculates the lncRNA projection score; the third step fuses the disease projection score and the lncRNA projection score proportionally, then it normalizes them to get the prediction score matrix.
For most of the approaches summarized above, the performance is evaluated using the LOOCV framework, such that each known LDA is left out in turn as a test sample, and how well this test sample is ranked relative to the candidate samples (all the LDAs without the evidence to confirm their relationships) is computed.
The main goal of the research presented here is to provide more accurate computational methods for the prediction of novel LDAs, candidate for experimental validation in laboratory. To this aim, external information on both molecular interactions (e.g., lncRNA-miRNA interactions) and genotype-phenotype associations (e.g., miRNA-disease associations) is assumed to be available. Indeed, while only a restricted number of validated LDAs is yet available, large amounts of interactions between lncRNAs and other molecules (e.g., miRNAs, genes, proteins), as well as associations between these other molecules and diseases, are known and annotated in curated databases.
A commonly recognized assumption is that lncRNAs with similar behaviour in terms of their molecular interactions with other molecules, may also reflect such a similarity for their involvement in the occurrence and progress of disorders and diseases [ 12 ]. This is even more effective if the correlation with diseases is “mediated” by the molecules they interact with. Based on this observation, we have designed three novel prediction methods that all consider the notion of lncRNA “neighbors”, intended as lncRNAs which share common mediators among the molecules they physically interact with. Here, we focus on miRNAs as mediator molecules. However, the proposed approaches are general enough to allow also the inclusion of other different molecules. Relationships among lncRNAs, mediators and diseases are modeled through tripartite graphs in all the proposed approaches (see Fig. 1 that illustrates the flowchart of the presented research pipeline).
Problem statement Let \({\mathcal {L}}=\{l_1, l_2, \ldots , l_h\}\) be a set of lncRNAs and \({\mathcal {D}}=\{d_1, d_2, \ldots , d_k\}\) be a set of diseases. The goal is to return an ordered set of triplets \({\mathcal {R}}=\{\langle l_x, d_y, s_{xy}\rangle \}\) (with \(x\in [1,h]\) , and \(y\in [1,k]\) ), ranked according to the score \(s_{xy}\) .
The top triplets in \({\mathcal {R}}\) correspond to those pairs \((l_x, d_y)\) with most chances to represent putative LDAs which may be considered for further analysis in laboratory, while the triplets in the bottom correspond to lncRNAs and diseases which are unlikely to be related each other. A key aspect for the solution of the problem defined above is the score computation, that is the main aim of the approaches introduced in the following.
NGH: neighborhood based approach
A model of tripartite graph is adopted here to take into account that lncRNAs interacting with common mediators may be involved in common diseases.
Let \(T_{LMD}=\langle I, A \rangle\) be a tripartite graph defined on the three sets of disjoint vertexes L , M and D , such that \((l,m) \in I\) are edges between vertexes \(l \in L\) and \(m \in M\) , \((m,d) \in A\) are edges between vertexes \(m \in M\) and \(d \in D\) , respectively. In particular, L is associated to a set of lncRNAs, M to a set of miRNA and D to a set of diseases. Moreover, edges of the type ( l , m ) represent molecular interactions between lncRNAs and miRNA, experimentally validated in laboratory; edges of the type ( m , d ) correspond to known miRNA-disease associations, according to the existing literature. In both cases, interactions and associations annotated and stored in public databases may be taken into account.
The following definitions hold.
Definition 1
(Neighbors) Two lncRNAs \(l_h, l_k \in L\) are neighbors in \(T_{LMD}=\langle I, A \rangle\) if there exists at least a \(m_x \in M\) such that \((l_h, m_x) \in I\) and \((l_k, m_x) \in I\) .
Definition 2
(Prediction Score) The Prediction Score for the pair \((l_i,d_j)\) such that \(l_i \in L\) and \(d_j \in D\) is defined as:
\(M_{l_i}\) is the set of annotated miRNA interacting with \(l_i\) ,
\(M_{d_j}\) is the set of miRNA found to be associated to \(d_j\) ,
\(M_{l_x}\) is the set of miRNA interacting with the neighbor \(l_x\) of \(l_i\) (for each neighbor of \(l_i\) ),
\(\alpha\) is a real value in [0, 1] used to balance the two terms of the formula.
Definition 3
(Normalized prediction score) The Normalized Prediction Score for the pair \((l_i,d_j)\) such that \(l_i \in L\) , \(d_j \in D\) and \(s_{ij}\) is the Prediction Score for \((l_i,d_j)\) , is defined as:
NGH-CF: NGH extended with collaborative filtering
We remark that the main idea here is trying to infer the behaviour of a lncRNA, from that of its neighbors. Moreover, it is worth to point out that the notion of neighbor is related to the presence of miRNAs interacting with the same lncRNAs. However, not all the miRNA-lncRNA interactions have already been discovered, and miRNA-disease associations as well. This intuitively reminds to a typical context of data incompleteness where Collaborative Filtering may be successful in supporting the prediction process [ 30 ].
In more detail, what to be encoded by the Collaborative Filter is that lncRNAs presenting similar behaviours in terms of interactions with miRNAs, should reflect such a similarity also in their involvement with the occurrence and progress of diseases, mediated by those miRNAs. To this aim, a matrix R is considered here such that each element \(r_{ij}\) represents if (or to what extent) the lncRNA i and the disease j may be considered related. We call R relationship matrix (it is also known as rating matrix in other contexts, such as for example in the prediction of user-item associations). How to obtain \(r_{ij}\) is at the basis of the two variants of the approach presented in this section.
Due to the fact that R is usually a very sparse matrix, it can be factored into other two matrices L and D such that R \(\approx\) \(L\) \(^T\) \(D\) . In particular, matrix factorization models map both lncRNAs and diseases to a joint latent factor space F of dimensionality f , such that each lncRNA i is associated with a vector \(l_i \in F\) , each disease j with a vector \(d_j \in F\) , and their relationships are modeled as inner products in that space. Indeed, for each lncRNA i , the elements of \(l_i\) measure the extent to which it possesses those latent factors, and the same holds for each disease j and the corresponding elements of \(d_j\) . The resulting dot product in the factor space captures the affinity between lncRNA i and disease j , with reference to the considered latent factors. To this aim, there are two important tasks to be solved:
Mapping lncRNAs and diseases into the corresponding latent factors vectors.
Fill the matrix R , that is, the training set.
To learn the factor vectors \(l_i\) and \(d_j\) , a possible choice is to minimize the regularized squared error on the set of known relationships:
where \(\chi\) is the set of ( i , j ) pairs for which \(r_{ij}\) is not equal to zero in the matrix R . To this aim, we apply the ALS technique [ 31 ], which rotates between fixing the \(l_i\) ’s and fixing the \(d_j\) ’s. When all \(l_i\) ’s are fixed, the system recomputes the \(d_j\) ’s by solving a least-squares problem, and vice versa.
Filling the matrix R is performed according to two different criteria, resulting in the two different variants of the approach presented in this section, namely, CF and NGH-CF, respectively. According to the first criteria (CF), \(r_{ij}\) is set equal to 1 if the lncRNA i and the disease j share at least one miRNA in common, to 0 otherwise. The second variant (NGH-CF) works instead as a booster to improve the accuracy of NGH. In this latter case, the matrix R is filled by the normalized score ( 2 ). For both variants, the considered score to rank the predicted LDAs is given by the final value returned by the ALS technique applied on the corresponding matrix R .
Validation methodologies
We remark that the proposed approaches for LDAs prediction return a rank of LDAs, sorted according to the score that is characteristic of the considered approach, such that top triplets may be assumed as the most promising putative LDAs for further analysis in laboratory. As in other contexts [ 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 ], the performance of a prediction tool may be evaluated using suitable external criteria . Here, an external criterion relies on the existence of LDAs that are known to be true from the literature or, even better, from public repositories, where associations already verified in laboratory are annotated. A gold standard is constructed, containing only such true LDAs. The putative LDAs returned by the prediction method can thus be compared against those in the gold standard. In order to work properly, this validation methodology requires the gold standard information to be independent on that considered, in its turn, from the method under evaluation during its prediction task. This is satisfied in our case, due to the fact that all three approaches introduced in the previous sections do not exploit any type of knowledge referred to known LDAs during prediction, relying instead on known miRNA-lncRNA interactions and miRNA-disease associations, which come from independent sources.
According to the above mentioned validation methodology, the proposed approaches can be validated with references to the Receiver Operating Characteristics (ROC) analysis [ 34 ]. In particular, each predicted LDA is associated to a label, that is true if that association is contained in the considered gold standard, and false otherwise.
By varying the threshold value, it is possible to compute the true positive rate (TPR) and the false positive rate (FPR), by refferring to the percentage of the true/false predictions whose ranking is higher/below than the considered threshold value. ROC curve can be drawn by plotting TPR versus FPR at different threshold values. The Area Under ROC Curve (ROC-AUC) is further calculated to evaluate the performance of the tested methods. ROC-AUC equal to 1 indicates perfect performance, ROC-AUC equal to 0.5 random performance.
Similarly to the ROC curve, the Precision-Recall (PR) curve can be drawn as well, combining the positive predictive value (PPV, Precision), i.e., the fraction of predicted LDAs which are true in the gold standard, and the TPR (Recall), in a single visualization, at the threshold varying. The higher on y-axis the obtained curve is, the better the prediction method performance. The Area Under PR curve (AUPR) is more sensitive than AUC to the improvements for the positive class prediction [ 35 ], that is important for the case studied here. Indeed, only true LDAs are known, therefore no negative samples are included in the gold standard.
Another important measure useful to evaluate the prediction accuracy of a method and that can be considered here is the F1-score, defined as the harmonic mean of Precision and Recall to symmetrically represent both metrics in a single one.
We have validated the proposed approaches on both syntetic and real datasets, as explained below.
Synthetic data
A synthetic dataset has been built with 15 lncRNAs, 35 miRNA and 10 diseases, such that three different sets of LDAs may be identified, as follows (see also Table 1 , where the characteristics of each LDA are summarized).
Set 1: 26 LDAs, such that each lncRNA has from 3 to 4 miRNAs shared with the same disease (strongly linked lncRNAs) .
Set 2: 16 LDAs, each lncRNA having only one miRNA shared with a disease, and from 2 to 5 neighbors that are strongly linked with that same disease (directly linked lncRNAs and strong neighborhood) .
Set 3: 12 LDAs involving lncRNAs without any miRNA in common with a certain disease, and a number between 2 and 5 neighbors that are strongly linked with that same disease (only strong neighborhood) .
Experimentally verified data downloaded from starBase [ 36 ] and from HMDD [ 37 ] have been considered for the lncRNA-miRNA interactions and for the miRNA-disease associations, respectively. In particular, the latest version of HMDD, updated at 2019, has been used. Overall, \(1,\!114\) lncRNAs, \(1,\!058\) miRNAs, 885 diseases, \(10,\!112\) lncRNA-miRNA interactions and \(16,\!904\) miRNA-disease associations have been included in the analysis.
In order to evaluate the prediction accuracy of the approaches proposed here against those from the literature, three different gold standards have been considered. A first gold standard dataset GS1 has been obtained from the LncRNA-Disease database [ 38 ], resulting in 183 known and verified LDAs. A second, more restrictive, gold standard GS2 with 157 LDAs has been built by the intersection of data from [ 38 ] and [ 39 ]. Finally, also a larger gold standard dataset GS3 has been included in the analysis, by extracting LDAs from MNDRv2.0 database [ 40 ], where associations both experimentally verified and retrieved from manual literature curation are stored, resulting in 408 known LDAs.
Comparison on real data
The approaches proposed here have been compared against other approaches from the literature, over the three different gold standards described in the previous Section. In particular, all approaches considered from the literature have been run according to the default setting of their parameters, reported on the corresponding scientific publications and/or on their manual instructions.
Our approaches have been compared at first on GS1 against those approaches taking exactly the same input than ours, that are HGLDA [ 11 ], ncPred [ 10 ] and LDA-TG [ 24 ]. In particular, we have implemented HGLDA and used the corresponding p-value score, corrected by FDR as suggested by [ 11 ], for the ROC analysis. Moreover, we have normalized also the scores returned by ncPred and LDA-TG for the predicted LDAs, according to the formula in Definition 3 . Indeed, we have observed experimentally that such a normalization improves the accuracy of both methods from the literature, resulting in a better AUC. As for the novel approaches proposed here, the Normalized Prediction Score has been considered for NGH, while the approximated rating score resulting from ALS [ 31 ] is used for both CF and NGH-CF. Figure 2 shows the AUC scored by each method on GS1, while in Fig. 3 the different ROC curves are plotted. In particular, NGH scores a value of AUC equal to 0.914, thus outperforming the other three methods previously presented in the literature, i.e., HGLDA, ncPred and LDA-TG, that reach 0.876, 0.886 and 0.866, respectively (we remark also that performance of both ncPred and LDA-TG has been slightly improved with respect to their original one, by normalizing their scores). As for the novel approaches based on collaborative filtering, they both present a better accuracy than the others, with CF having AUC equal to 0.957 and NGH-CF to 0.966, respectively. Therefore, these results confirm that taking into account the collaborative effects of lncRNAs and miRNAs is useful to improve LDAs prediction, and the most successful approach is NGH-CF, that is, the neighborhood based approach boosted by collaborative filtering.
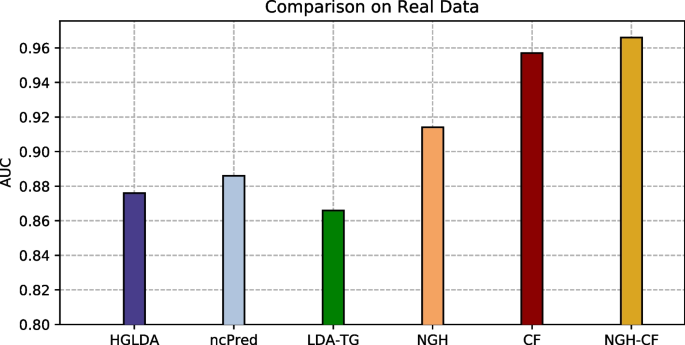
Comparison of the scored AUC on GS1
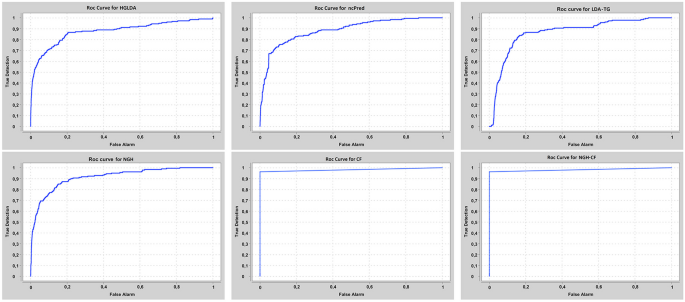
ROC curves for the compared methods on GS1
Another interesting issue is represented by the “agreement” between the different methods taking the same input, in terms of the returned best scoring LDAs. Table 2 shows the Jaccard Index computed between the proposed approaches and those receiving the same input, on the top \(5\%\) LDAs in the corresponding ranks, sorted from the best to the worst score values for each method. It emerges that results by HGLDA and ncPred have a small match with the other approaches (at most 0.23), while NGH-CF has high agreement with CF (0.74), as well as with NGH and LDA-TG (both 0.70). LDA-TG and CF present a sufficient match in their best predictions (0.59). This latter comparison based on agreement shows that approaches based on neighborhood analysis share a larger set of LDAs, in the top part of their ranks.
The proposed approaches have been compared also against other two recent methods from the literature, i.e., SIMCLDA and HGNNLDA, which receive in input different data than ours, including mRNA and drugs. For this reason, the more restrictive gold standard GS2 has been exploited for the comparison, where only lncRNAs and diseases having some correspondences with the additional input data of SIMCLDA and HGNNLDA are included. Figure 4 shows the comparison of the scored AUC on GS2, while Fig. 5 the corresponding ROC curves. In particular, the behaviour of all approaches previously tested does not change significantly on this other gold standard, moreover all the other approaches overcome SIMCLDA. On the other hand, HGNNLDA has a better performance than HGLDA, NcPred and LDA-TG, although it has a worse accuracy than NGH, CF and NGH-CF. The former confirms its superiority with regards to all considered approaches.
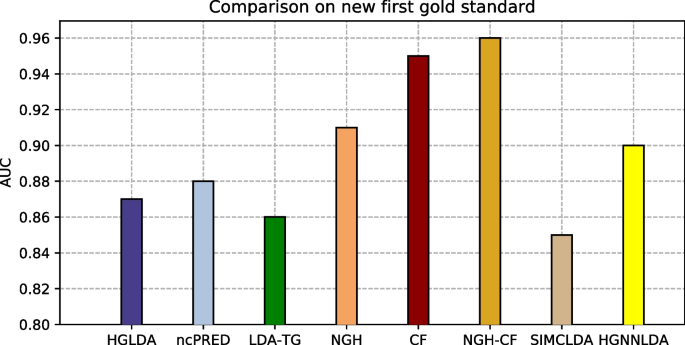
Comparison of the scored AUC on GS2
ROC curves for the compared methods on GS2
The proposed approaches have been compared also against LDAP-WMPS on GS3. Figure 6 shows the AUC values scored by all compared approaches on GS3, while Fig. 7 the corresponding ROC curves. In particular, the behaviour of all approaches previously tested does not change on this other gold standard, and LDAP-WMPS has better performance than the other approaches except for NGH, CF, NGH-CF and HGNNLDA.
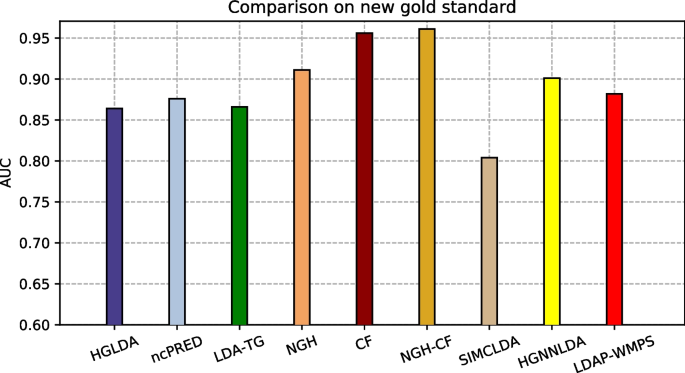
Comparison of the scored AUC on GS3
ROC curves for the compared methods on GS3
The AUPR values scored by the compared methods on GS1, GS2, and GS3 are shown in Fig. 8 , while the corresponding PR-curves are plotted in Fig. 9 . In particular, for GS1 results are analogous to the ROC analysis, with NGH-CF the best performing one, followed by CF and NGH, while HGLDA is the worst. On GS2, NGH-CF and CF keep their superiority, followed by SMCLDA and NGH, while HGLDA is yet the worst one. On GS3, NGH-CF is the first, Cf the second and both HGNNLDA and LDAP-WMPS outperform NGH, while HGLDA in this case slightly outperforms LDA-TG, ncPred and SMCLDA, which results to be the worst one.
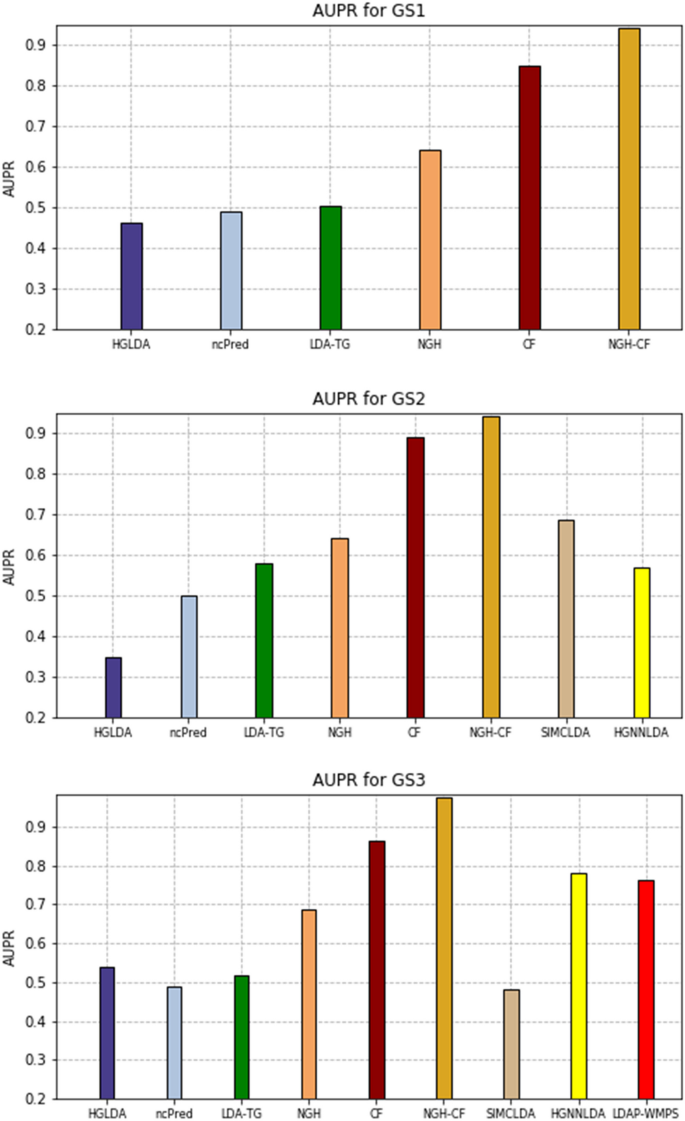
AUPR hystogram for the compared methods on GS1, GS2, GS3
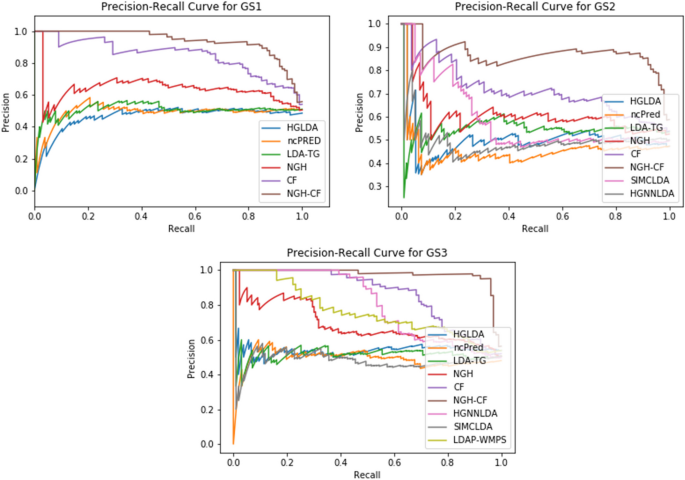
Precision-recall curves for the compared methods on GS1,GS2,GS3
Figures 10 , 11 and 12 show the F1-score values obtained, for all methods compared on GS1, GS2 and GS3, respectively, at the varying of a threshold fixed on the method score. In Tables 3 , 4 and 5 it is shown, for each gold standard, the highest value of F1-score obtained by each considered method, as well as the corresponding Precision and Recall values, and the minimum threshold value for which the highest F1-score value has been reached. On GS1 and GS2, the three best performing approaches are NGH-CF, CF and NGH, in this order. On GS3 the order is the same, and LDAP-WMPS performs equally to NGH.
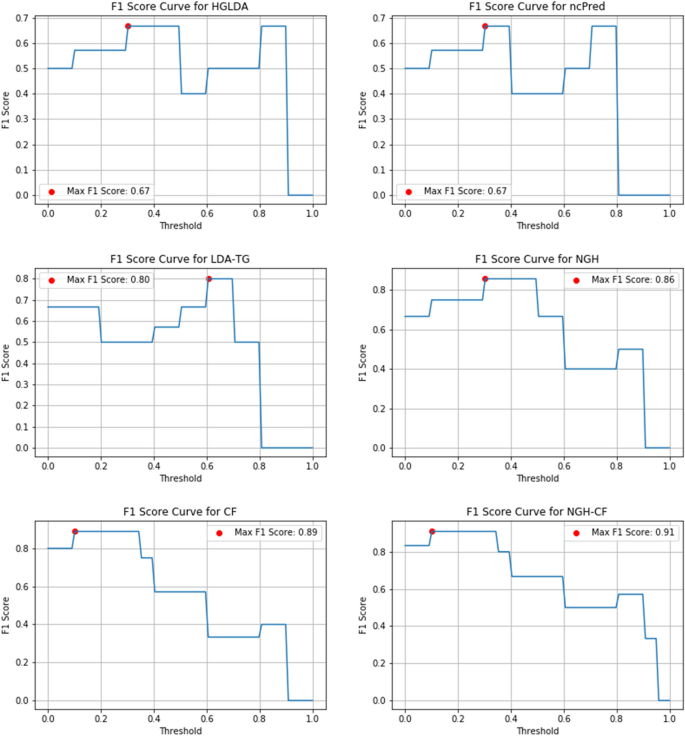
F1-score for the compared methods on GS1
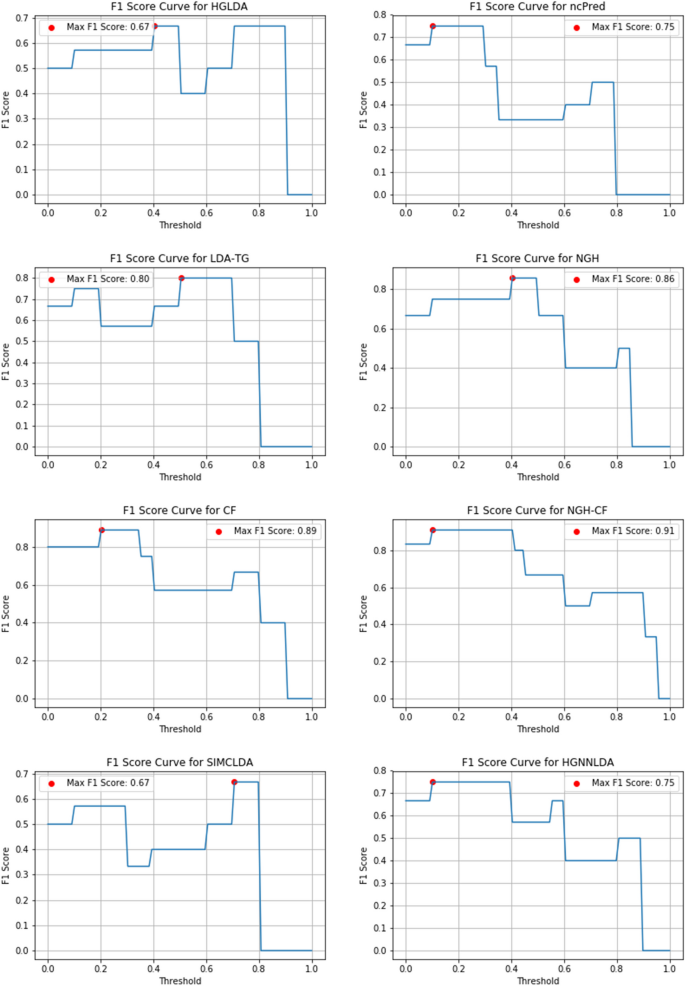
F1-Score for the compared methods on GS2
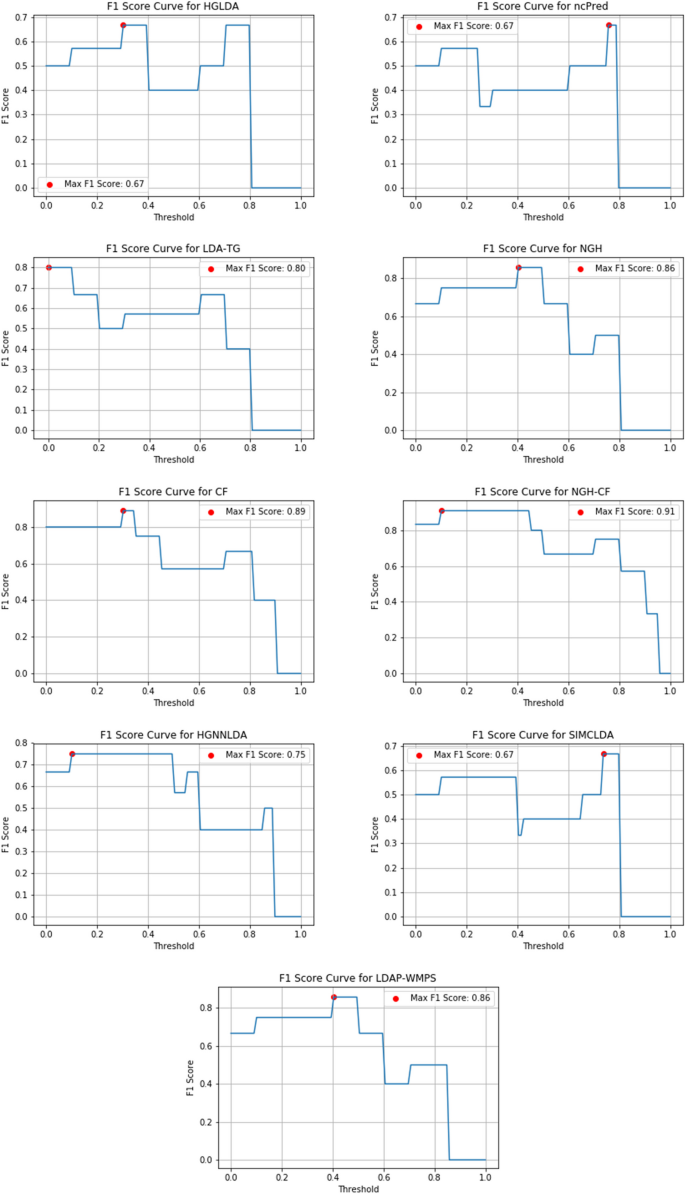
F1-Score for the compared methods on GS3
Robustness analysis
The main aim of the analysis discussed here is to measure to what extent the proposed methods are able to correctly recognize verified LDAs, even if part of the existing associations are missed, i.e., the sets of known and verified lncRNA-miRNA interactions and miRNA-disease associations are not complete. This is important to verify that the proposed approaches can provide reliable predictions also in presence of data incompleteness, that is often the case when lncRNAs are involved. Therefore, the robustness of each proposed method has been evaluated by performing progressive alterations of the input associations coming from the real datasets, according to the following three different criteria.
Progressively eliminate the \(5\%\) , \(10\%\) , \(15\%\) and \(20\%\) of lncRNA-miRNA interactions from the input data.
Progressively eliminate the \(5\%\) , \(10\%\) , \(15\%\) and \(20\%\) of miRNA-disease associations from the input data.
Progressively eliminate the \(5\%\) , \(10\%\) , \(15\%\) and \(20\%\) of both lncRNA-miRNA interactions and miRNA-disease associations (half and half), from the input data.
Tests summarized above have been performed for 20 times each. Tables 6 , 7 and 8 show the mean of the AUC values for NGH, CF and NGH-CF, respectively, over the 20 tests. In particular, all methods perform well on the three test typologies at \(5\%\) , the worst being NGH-CF, which however presents an average AUC equal to 0.84 for case 1), that is still a high value. NGH-CF is also the method that presents the best robustness on case 3), keeping the value of 0.92 also at \(20\%\) , while CF is the worst performing in case 3), indeed its average AUC decreases from 0.95 at \(5\%\) to 0.63 already at \(10\%\) , and then to 0.50 at \(20\%\) . This behaviour in case 3), where both lncRNA-miRNA interactions and miRNA-disease associations are progressively eliminated, deserves some observations. Indeed, results show that the combination of neighborhood analysis and collaborative filtering is the most robust one with regards to this perturbation, while collaborative filtering alone is the worst performing. On the other hand, CF results to be the most robust in case 1), where only lncRNA-miRNA interactions are eliminated, and this is due to the fact that CF does not take into account how many miRNAs are shared by pairs of lncRNAs. As for case 2), performance of all methods is comparable and generally good, possibly in consideration of the fact that a large number of miRNA-disease associations are available, therefore discarding small percentages of them does not affect largely the final prediction.
Comparison on specific situations
In this section further experimental tests are described, showing how well the considered methods perform in detecting specific situations, depicted through the synthetic dataset first, and then searched for in the real data. In particular, the basic observation here is that prediction approaches from the literature usually fail in detecting true LDAs, when the involved lncRNAs and diseases do not have a large number of shared miRNAs (referring to those approaches taking the same input than ours). The novel approaches we propose are particularly effective in managing the situation depicted above, through neighborhood analysis and collaborative filtering, allowing to detect similar behaviours shared by different lncRNAs, depending on the miRNAs they interact with.
For each set of LDAs defined in the synthetic data (i.e., set 1, set 2, and set 3), and for each tested method (i.e., HGLDA, NCPRED, NHG, CF, NGH-CF), Table 9 shows the percentage of LDAs in that set which is recognized at the top \(10\%\) , \(20\%\) , \(30\%\) , \(50\%\) of the rank of all LDAs, sorted by the score returned by the considered method. As an example, for HGLDA the \(32\%\) of LDAs of set 1 are located in the top \(10\%\) of its rank, where instead none LDAs in set 2 or 3 find place.
Looking at these results some interesting considerations come out. First of all, for the methods HGLDA, NCPRED, NHG and CF most associations of the set 1 are located in the top \(50\%\) of their corresponding ranks, while NGH-CF has a different behaviour. Indeed, it locates a lower number of such LDAs in the highest part of its rank than the other approaches, possibly due to the fact that it leaves room for a larger number of associations in the other two sets in the top ranked positions. As for LDAs in the set 2, all methods recognize some of them already in the top \(10\%\) , except for HGLDA, as alredy highlighted. The approaches able to recognize the larger percentages of these associations at the top \(50\%\) of their rank are NGH and NGH-CF. LDAs in the set 3 are the most difficult to recognize, due to the fact that the lncRNA and the disease do not share any miRNA in common. Indeed, the worst performing methods in this case are HGLDA, which is able to locate some of these associations only at the top \(50\%\) (according to the percentages we considered here), and NCPRED, which performs slightly better although it reaches the same percentage of located associations than HGLDA at \(50\%\) (the \(28\%\) ). As expected, approaches based on neighborhood analysis and collaborative filtering perform better, with the best one resulting to be NGH-CF.
In the previous section we have shown that all methods proposed here are able to detect specific situations, characterized by the fact that a lncRNA may have very few (or none) common miRNAs with a disease, and its neighbors share instead a large set of miRNAs with that disease. We have checked if this case occurs among the verified LDAs that our approaches find and their competitors do not. Table 10 shows, only by meaning of example, 10 experimentally verified LDAs, included in GS1, that are top ranked for the novel approaches proposed here, whereas they are in the bottom rank of the other approaches from the literature compared on GS1. Six out of such LDAs do not present any common miRNAs between the lncRNA and the disease, while four share only one miRNA. All involved lncRNAs present neighbors with a large number of miRNAs in common with the disease in that LDA, in accordance with the hypothesis that the ability in capturing this situation allows to obtain a better accuracy.
Survival analysis has been also performed by one of the TCGA Computational Tools, that is, TANRIC [ 41 ], on four of the pairs in Table 10 . In particular, those lncRNAs and diseases available in TANRIC have been chosen. Results are reported in Figures 13 , 14 , 15 and 16 , showing that the over-expression of the considered lncRNA determines a lower survival probability over the time, for all four considered cases.
Survival analysis related to SNHG16 and bladder neoplasm
Survival analysis related to CBR3-AS1 and prostate neoplasm
Survival analysis related to MALAT1 and bladder neoplasm
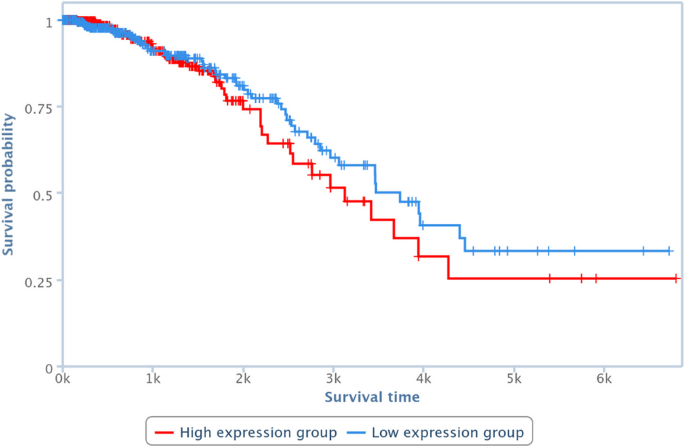
Survival analysis related to MEG3 and breast neoplasm
In the previous sections the effectiveness and robustness of the proposed approaches have been illustrated, showing that all three are able to return reliable predictions, as well as to detect specific situations which may occur in true predictions and are missed by competitors. Here we provide a discussion on some novel LDAs predicted by NGH, CF and NGH-CF.
Table 11 shows seven LDAs which are not present in the considered gold standards, and that have been returned by all three methods proposed here, with highest score. The first of these associations is between CDKN2B-AS1 and LEUKEMIA, confirmed by recent literature [ 42 , 43 ]. Indeed, CDKN2B-AS1 was found to be highly expressed in pediatric T-ALL peripheral blood mononuclear cells [ 42 ], moreover genome-wide association studies show that it is associated to Chronic Lymphocytic Leukaemia risk in Europeans [ 43 ]. As for the second association between DLEU2 and LEUKEMIA, DLEU2 is a long non-coding transcript with several splice variants, which has been identified by [ 44 ] through a comprehensive sequencing of a commonly deleted region in leukemia (i.e., the 13q14 region). Different investigations reported up regulation of this lncRNA in several types of cancers. The lncRNA H19 regulates GLIOMA angiogenesis [ 45 , 46 ], while MAP3K14 is one of the well-recognized biomarkers in the prognosis of renal cancer, which is reminiscent of the pancreatic metastasis from renal cell carcinoma [ 47 ]. MEG3 has been recently found to be important for the prediction of LEUKEMIA risk [ 48 ]. Multiple studies have shown that MIR155HG is highly expressed in diffuse large B-cell (DLBC) lymphoma and primary mediastinal B-cell lymphoma, and in chronic lymphocytic leukemia. The transcription factor MYB activates MIR155HG activity, which causes the epigenetic state of MIR155HG to be dysregulated and causes an abnormal increase in MIR155 [ 49 ]. Also the last top-ranked association in Table 11 between TUG1 and NON-SMALL CELL LUNG CARCINOMA has found evidence in the literature [ 50 , 51 , 52 ].
Tables 12 , 13 , and 14 show the top 100 (sorted by the scores returned by each method) novel LDA predictions that NGH and CF, NGH and NGH-CF, CF and NGH-CF have in common, respectively. Many of the lncRNAs involved in such top-ranked LDAs are not yet characterized in the literature, therefore results presented here may be considered a first attempt to provide novel knowledge about them, through their inferred association with known diseases.
We have explored the application of neighborhood analysis, combined with collaborative filtering, for the improvement of LDAs prediction accuracy. The three approaches proposed here have been evaluated and compared first against their direct competitors from the literature, i.e., the other methods which also use lncRNA-miRNA interactions and miRNA-disease associations, without exploiting a priori known LDAs. It results that all methods proposed here are able to outperform direct competitors, the best one (NGH-CF) also significantly (AUC equal to 0.966 against the 0.886 by NCPRED). In particular, it has been shown that the improvement in accuracy is due to the fact that our approaches capture specific situations neglected by competitors, relying on similar lncRNAs behaviour in terms of their interactions with the considered intermediate molecules (i.e., miRNAs). The proposed approaches have been then compared also against other recent methods, taking different inputs (e.g., integrative approaches), and the experimental evaluation shows that they are able to outperform them as well.
It is worth pointing out the importance of providing reliable data in input to the LDAs prediction approaches. As discussed in this manuscript, information on the lncRNAs relationships with other molecules, and between intermediate molecules and diseases, is provided in input to the proposed approaches. Reliable datasets have been used to perform the experimental analysis provided here. However, as the user may provide also different input datasets, it is important to point out that the reliability of the obtained predictions strictly depends on that of input information.
As neighborhood analysis has resulted to be effective in characterizing lncRNAs with regards to their association with known diseases, we plan to apply it also for predicting possible common functions among lncRNAs, for example by clustering them according to their interactions, which has shown to be successful for other types of molecules [ 53 ]. Moreover, due to the success of integrative approaches on the analysis of biological data [ 54 ], we expect that including other types of intermediate molecules, such as for example genes and proteins, in the main pipeline of the proposed approaches may further improve their accuracy.
In conclusion, the use of reliable input data and the integration of different types of information coming from molecular interactions seem to be the most promising future directions for LDAs prediction.
Availability of data and materials
The source code is available at: https://github.com/marybonomo/LDAsPredictionApproaches.git In particular, executable software for NGH, CF, and NGH-CF are provided, as well as syntetic and real input datasets used here; the three different gold standard datasets GS1, GS2, GS3; the final obtained results.
Medico-Salsench E, et al. The non-coding genome in genetic brain disorders: New targets for therapy? Essays Biochem. 2021;65(4):671–83.
Article CAS PubMed PubMed Central Google Scholar
Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118.
Article CAS PubMed Google Scholar
Zhao H, Shi J, Zhang Y, et al. LncTarD: a manually-curated database of experimentally-supported functional lncRNA–target regulations in human diseases. Nucl Acids Res. 2019;48(D1):D118–D126. ISSN: 0305-1048.
Liao Q, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co- expression network. Nuc Acids Res. 2011;39:3864–78.
Article CAS Google Scholar
Chen X, et al. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinf. 2017;18(4):558–76.
CAS Google Scholar
Wang B, et al. lncRNA-disease association prediction based on matrix decomposition of elastic network and collaborative filtering. Sci Rep. 2022;12:7.
Google Scholar
He J, et al. HOPEXGB: a consensual model for predicting miRNA/lncRNA-disease associations using a heterogeneous disease-miRNA-lncRNA information network. J Chem Inf Model 2023
Zhong H, et al. Association filtering and generative adversarial networks for predicting lncRNA-associated disease. BMC Bioinf. 2023;24(1):234.
Dengju Y, et al. GCNFORMER: graph convolutional network and transformer for predicting lncRNA-disease associations. BMC Bioinf. 2024;25(1):5.
Article Google Scholar
Alaimo S, Giugno R, Pulvirenti A. ncPred: ncRNA-disease association prediction through Tripartite network-based inference. Front Bioeng Biot. 2014;2:71.
Chen X. Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci Rep. 2015;5:13186.
Lu C, et al. Prediction of lncRNA-disease associations based on inductive matrix completion. Bioinformatics. 2018;34(19):3357–64.
Xuan Z, Li J, Yu X, Feng J, et al. A probabilistic matrix factorization method for identifying lncRNA-disease associations. Genes 2019;10(2)
Du X, et al. lncRNA-disease association prediction method based on the nearest neighbor matrix completion model. Sci Rep. 2022;12(1):21653.
Wang L, et al. Prediction of lncRNA-disease association based on a Laplace normalized random walk with restart algorithm on heterogeneous networks. BMC Bioinf. 2022;23(1):1–20.
Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: taxonomy, trends and challenges of computational models. Brief Bioinf. 2022;23(5):bbac358.
Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: experimental results, databases, webservers and data fusion. Brief Bioinf. 2022;23(6):bbac397.
Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: towards systematic evaluation of computational models. Brief Bioinf. 2022;23(6):bbac407.
Chen X, Yan G. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29(20):2617–24.
Xie G, et al. SKF-LDA: similarity kernel fusion for predicting lncRNA-disease association. Mol Therapy-Nucleic Acids. 2019;18:45–55.
Liu D, et al. HGNNLDA: predicting lncRNA-drug sensitivity associations via a dual channel hypergraph neural network. IEEE/ACM transactions on computational biology and bioinformatics, 2023;1–11.
Zhang Y, et al. LDAI-ISPS: lncRNA-disease associations inference based on integrated space projection scores. Int J Molecular Sci. 2020;21(4):1508.
Liang Y, et al. MAGCNSE: predicting lncRNA-disease associations using multi-view attention graph convolutional network and stacking ensemble model. BMC Bioinf. 2022;23(1):189.
Bonomo M, La Placa A, Rombo SE. Prediction of lncRNA-disease associations from tripartite graphs. In: Heterogeneous data management, polystores, and analytics for healthcare - VLDB workshops, poly 2020 and DMAH 2020, virtual event, August 31 and September 4, 2020, Revised Selected Papers. Springer, Berlin, 2020;205–210. ISSN: 978-3-030-71054-5
Xie G, et al. Predicting lncRNA-disease associations based on combining selective similarity matrix fusion and bidirectional linear neighborhood label propagation. Brief Bioinform. 2023;24(1):bbac595.
Article PubMed Google Scholar
Cheng L, et al. ntNetLncSim: an integrative network analysis method to infer human lncRNA functional similarity. Oncotarget. 2016;7(30):47864–74.
Article PubMed PubMed Central Google Scholar
Guangyuan F, et al. Matrix factorization-based data fusion for the prediction of lncRNA-disease associations. Bioinformatics. 2018;34:1529–37.
Xie G, et al. RWSF-BLP: a novel lncRNA-disease association prediction model using random walk-based multi-similarity fusion and bidirectional label propagation. Mol Genet Genom. 2021;296:473–83.
Wang B, et al. lncRNA-disease association prediction based on the weight matrix and projection score. PLOS One. 2023;18(1): e0278817.
Duan R, Jiang C, Jain HK. Combining review-based collaborative filtering and matrix factorization: a solution to rating’s sparsity problem”. Decis Support Syst 2022;156:113748. ISSN: 0167–9236.
Koren Y, Bell R, Volinsky C. Matrix factorization techniques for recommender systems. Computer. 2009;42(8):30–7.
Parida L, Pizzi C, Rombo SE. Irredundant tandem motifs. Theoret Comput Sci. 2014;525:89–102.
Bonomo M, et al. Topological ranks reveal functional knowledge encoded in biological networks: a comparative analysis. Brief Bioinform. 2022;23(3):bbac101.
Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27(8):861–74.
Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLOS One. 2015;10(3): e0118432.
Li J, et al. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013;42:D92–7.
Li Y, et al. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42:D1070–4.
Chen G, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–6.
Gao Y, et al. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021;49(D1):D1251–8.
Cui T, et al. MNDR v2. 0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018;46(D1):D371–4.
CAS PubMed Google Scholar
Li J, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75(18):3728–37.
Chen L, et al. lncRNA CDKN2B-AS1 contributes to tumorigenesis and chemoresistance in pediatric T-cell acute lymphoblastic leukemia through miR-335-3p/TRAF5 axis. In: Anti-cancer drugs, Wolters Kluwer Health, Inc. (2020)
Song C, et al. CDKN2B-AS1: an indispensable long non-coding RNA in multiple diseases. Current Pharm Des. 2020;26(41):5335–46.
Ghafouri-Fard S, et al. Deleted in lymphocytic leukemia 2 (DLEU2): an lncRNA with dissimilar roles in different cancers. Biomed Pharmacother. 2021;133: 111093.
Jia P, et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381(2):359–69.
Liu Z, et al. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1 α /VEGFaxis. Neoplasma. 2020;67(1):111–8.
Zhou S, et al. A novel immune-related gene prognostic Index (IRGPI) in pancreatic adenocarcinoma (PAAD) and its implications in the tumor microenvironment. Cancers. 2022;14(22):5652.
Pei J, et al. Novel contribution of long non-coding RNA MEG3 genotype to prediction of childhood leukemia risk. Cancer Genom Proteom. 2022;19(1):27–34.
Peng L, et al. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019;8(17):7161–73.
Zhang E, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5(5):e1243–e1243.
Lin P, et al. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer. 2016;16:1–10.
Niu Y, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16(1):1–13.
Pizzuti C, Rombo SE. An evolutionary restricted neighborhood search clustering approach for PPI networks. Neurocomputing. 2014;145:53–61.
Rombo SE, Ursino D (2021) Integrative bioinformatics and omics data source interoperability in the next-generation sequencing era
Download references
Acknowledgements
The authors are grateful to the Anonymous Reviewers, for the constructive and useful suggestions that allowed to significantly improve the quality of this manuscript. Some of the results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga .
PRIN “multicriteria data structures and algorithms: from compressed to learned indexes, and beyond”, Grant No. 2017WR7SHH, funded by MIUR (closed). “Modelling and analysis of big knowledge graphs for web and medical problem solving” (CUP: E55F22000270001), “Computational Approaches for Decision Support in Precision Medicine” (CUP:E53C22001930001), and “Knowledge graphs e altre rappresentazioni compatte della conoscenza per l’analisi di big data” (CUP: E53C23001670001), funded by INdAM GNCS 2022, 2023, 2024 projects, respectively. “Models and Algorithms relying on knowledge Graphs for sustainable Development goals monitoring and Accomplishment - MAGDA” (CUP: B77G24000050001), funded by the European Union under the PNRR program related to “Future Artificial Intelligence - FAIR”.
Author information
Authors and affiliations.
Kazaam Lab s.r.l., Palermo, Italy
Mariella Bonomo & Simona E. Rombo
Department of Mathematics and Computer Science, University of Palermo, Palermo, Italy
Simona E. Rombo
You can also search for this author in PubMed Google Scholar
Contributions
MB and SER equally contributed to the research presented in this manuscript. MB implemented and run the software, SER performed the analysis of results. Both authors wrote and reviewed the entire manuscript.
Corresponding author
Correspondence to Mariella Bonomo .
Ethics declarations
Ethics approval and consent to participate.
Not Applicable
Consent for publication
Competing interests.
SER is editor of BMC Bionformatics. MB has no Conflict of interest.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Bonomo, M., Rombo, S.E. Neighborhood based computational approaches for the prediction of lncRNA-disease associations. BMC Bioinformatics 25 , 187 (2024). https://doi.org/10.1186/s12859-024-05777-8
Download citation
Received : 13 December 2023
Accepted : 11 April 2024
Published : 13 May 2024
DOI : https://doi.org/10.1186/s12859-024-05777-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- LncRNA-disease associations
- Molecular interactions
- Bioinformatics
- Long non-coding RNA
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]
- Reference Manager
- Simple TEXT file
People also looked at
Review article, sonication protocols and their contributions to the microbiological diagnosis of implant-associated infections: a review of the current scenario.

- 1 Departamento de Medicina Tropical - Universidade Federal de Pernambuco – UFPE, Recife, Brazil
- 2 Departamento de Microbiologia - Instituto Aggeu Magalhães – Fiocruz, Recife, Brazil
- 3 Dipartimento di Sanità Pubblica e Malattie Infettive - Sapienza University of Rome, Rome, Italy
Addressing the existing problem in the microbiological diagnosis of infections associated with implants and the current debate about the real power of precision of sonicated fluid culture (SFC), the objective of this review is to describe the methodology and analyze and compare the results obtained in current studies on the subject. Furthermore, the present study also discusses and suggests the best parameters for performing sonication. A search was carried out for recent studies in the literature (2019-2023) that addressed this research topic. As a result, different sonication protocols were adopted in the studies analyzed, as expected, and consequently, there was significant variability between the results obtained regarding the sensitivity and specificity of the technique in relation to the traditional culture method (periprosthetic tissue culture – PTC). Coagulase-negative Staphylococcus (CoNS) and Staphylococcus aureus were identified as the main etiological agents by SFC and PTC, with SFC being important for the identification of pathogens of low virulence that are difficult to detect. Compared to chemical biofilm displacement methods, EDTA and DTT, SFC also produced variable results. In this context, this review provided an overview of the most current scenarios on the topic and theoretical support to improve sonication performance, especially with regard to sensitivity and specificity, by scoring the best parameters from various aspects, including sample collection, storage conditions, cultivation methods, microorganism identification techniques (both phenotypic and molecular) and the cutoff point for colony forming unit (CFU) counts. This study demonstrated the need for standardization of the technique and provided a theoretical basis for a sonication protocol that aims to achieve the highest levels of sensitivity and specificity for the reliable microbiological diagnosis of infections associated with implants and prosthetic devices, such as prosthetic joint infections (PJIs). However, practical application and additional complementary studies are still needed.
1 Introduction
Joint replacement surgeries, known as arthroplasties, are increasingly frequent and widely used procedures with the aim of replacing, remodeling or realigning a joint ( Torres et al., 2015 ; Filho et al., 2020 ). Taking into account projections on certain orthopedic procedures, for example, by 2030 in the United States, a significant increase in the number of primary hip (174%) and knee (673%) arthroplasties is expected; for the same period, the United Kingdom expects a 400% increase in demand for arthroplasty ( Torres et al., 2015 ; Ahmed and Haddad, 2019 ; Filho et al., 2020 ).
This increasing use of implantable technology has also increased the risk of deep surgical site infections (SSIs) ( Torres et al., 2015 ). In this context, prosthetic joint infections (PJIs) occur in the joint area up to two years after surgery and are generally acquired during the implant procedure ( Filho et al., 2020 ). They are classified according to the time interval between surgery and the onset of symptoms, which can be classified as follows: early, if it occurs within a time interval of < 3 months after the placement of the prosthesis; early late, if it occurs within a time interval of 3 to 12 months; and chronic delay, if it occurs within a time interval of >12 months. This classification also involves the way the disease is presented, whether it is acute or chronic ( Beam and Osmon, 2018 ; Zardi and Franceschi, 2020 ).
Among the most common pathogens associated with PJI are Staphylococcus coagulase-negative and Staphylococcus aureus , two of which are the most common etiological agents of the disease, followed by Streptococcus sp., Enterococcus sp., gram-negative bacilli, anaerobes and yeasts. These agents are also known as good biofilm formers and are bacterial structures that are favored in PJI because of the abiotic surface of the implant and the lack of a local immunological response, resulting in persistent and progressive infection during treatment ( Karbysheva et al., 2020 ; Zardi and Franceschi, 2020 ).
PJI is still considered the second most common complication, second only to aseptic loosening, and is the most important complication in arthroplasty. It may be responsible for loosening, chronic pain and instability of the prosthesis and is thus associated with a high rate of morbidity, in addition to the risk of death and the need for complex treatment strategies that involve surgical interventions and prolonged antibiotic therapy ( Flurin et al., 2021 ; Zhang et al., 2021b ). The long-term impacts on patients’ quality of life are negative; even following successful clearance of the infection, failure to control the disease can lead to the need for joint fusion and even amputation ( Xu et al., 2023 ).
In addition to causing serious problems for the physical and mental health of patients, PJI also causes relevant economic problems. Hospital fees are generally significantly greater for the treatment of infected joints than for the treatment of noninfected joints. In the United States, the average total cost for revision knee arthroplasty is estimated at US$75,028.07, without considering the costs of prolonged antibiotic therapy at home. Similar patterns have been reported in other developed countries ( Zardi and Franceschi, 2020 ; Xu et al., 2023 ).
According to statistics from the National Healthcare Safety Network (NHSN), which was released in 2017, joint infections are responsible for 1.9% of all surgical site infections (SSIs) worldwide ( Moore et al., 2015 ). However, despite the widespread use of well-established infection prevention measures, these data on the occurrence of PJI may be underestimated due to one of the greatest challenges of this infection: diagnosis. Since there is no single test or finding for safe and accurate diagnosis, a combination of clinical findings, laboratory results of peripheral blood and synovial fluid, histological evaluations, imaging and molecular studies is performed, in addition to the important and necessary microbiological findings. In this scenario, several standardized diagnostic criteria for PJI have been proposed by different groups and societies, such as the Musculoskeletal Infection Society (MSIS), the Infectious Diseases Society of America (IDSA), the International Consensus Meeting (ICM), and the European Bone and Joint Infection Society (EBJIS), each of which adopts different definitions and cutoff points for the same infection ( Trebse and Roskar, 2021 ).
In the process of diagnosing PJI, periprosthetic tissue culture (PTC) is considered the gold standard diagnostic technique because it allows the identification of infectious pathogen(s) and the determination of antimicrobial susceptibility, and this method can be used to determine the best and most targeted therapeutic approach ( Tande and Patel, 2014 ; Salar et al., 2021 ). However, the sensitivity of tissue cultures varies from 65 to 94% and presents high false-negative rates, possibly due to the biofilm formation characteristic of this infection, which makes it difficult to obtain viable loose bacteria (planktonic) for cultivation, especially in chronic and low-grade infections preventing an accurate diagnosis from being made, causing treatment failures and prolonging the patient’s suffering ( Moore et al., 2015 ; Shen et al., 2015 ).
Therefore, Trampuz et al. (2007) ( Trampuz et al., 2007 ) popularized the use of the sonication technique to process removed knee and hip prostheses ( Trampuz et al., 2007 ; Shen et al., 2015 ). Since then, sonication has been suggested as a useful method for sample processing, aiming to physically displace biofilms prior to standard culture. Organizations such as the Swiss Orthopedics and Swiss Society for Infectious Diseases (SOSSID) and EBJIS have supported its use based on studies that reported greater sensitivity and specificity of sonicated fluid culture (SFC) from explanted prostheses compared to standard culture ( Shen et al., 2015 ; Bellova et al., 2019 ).
However, despite most studies in the literature indicating superior results with sonication, several studies observed a variable effect on the physical displacement of the biofilm ( Bellova et al., 2019 ), and some even showed greater sensitivity of PTC ( Oliva et al., 2016 ). Consequently, these discrepancies raise doubts about the reliability of the sonication technique for more accurate diagnosis of PJI ( Oliva et al., 2016 ; Bellova et al., 2019 ). These variations can be attributed to the different protocols used for sonication ( Oliva et al., 2016 ; Bellova et al., 2019 ).
Therefore, this report proposes an analysis of the literature on the subject in a similar way to other recent studies that reviewed the diagnostic methods available for infections associated with implants and their advances, including an overview of sonication ( Birlutiu et al., 2017 ; Portillo and Sancho, 2023 ; Yilmaz et al., 2023 ; Azad and Patel, 2024 ). However, this review sought to analyze and describe the sonication protocols used in studies published in the last five years, with emphasis on the sensitivity and specificity rates achieved by these methods in comparison with PTC. Furthermore, this review also aimed to identify, in depth, the best parameters that should be considered for potential standardization of sonication protocols based on the most recent published studies.
1.1 Literature search
For the literature search, the following terms were used: “prosthetic joint infection,” “sonication,” “tissue culture,” “biofilm,” “sensitivity,” “specificity,” “diagnosis” and combinations of these terms. The search was conducted in the National Center for Biotechnology Information (NCBI) search engine, PubMed ® . To comprehensively examine recent literature, the inclusion criteria for this analysis were original articles that were available electronically, published within the last five years (2019-2023) and written in English. Exclusion criteria included research such as case reports, letters, editorials and books. Furthermore, studies that addressed the microbiological diagnosis of infections other than PJI were excluded.
2 Sonication method
The sonication technique is performed using a device called a sonicator. This device emits sound waves in the ultrasound spectrum, creating high-intensity pressure waves in a liquid medium and causing the formation and collapse of tiny bubbles. When these bubbles collapse, they release energy capable of disrupting intercellular connections on the device’s surface, dislodging the bacteria. Additionally, sonication causes the deagglomeration and lysis of cell adhesion proteins, disrupting the physical structure of the biofilm ( Oliva et al., 2016 ).
Due to these characteristics, sonication has been increasingly utilized to increase the yield of bacterial cultures by releasing organisms embedded in biofilms associated with implants and prostheses, particularly in joints. The sonication technique, apart from dislodging bacteria from the biofilm structure, can also lead to the lysis of bacterial cells. However, this outcome depends on various protocol factors, such as the acoustic frequency, energy, temperature, duration of exposure to ultrasound, and shape of the bacteria ( Oliva et al., 2021 ).
2.1 Review/search results
We identified a total of 11 studies that met the established inclusion criteria, and these studies are described in Table 1 .
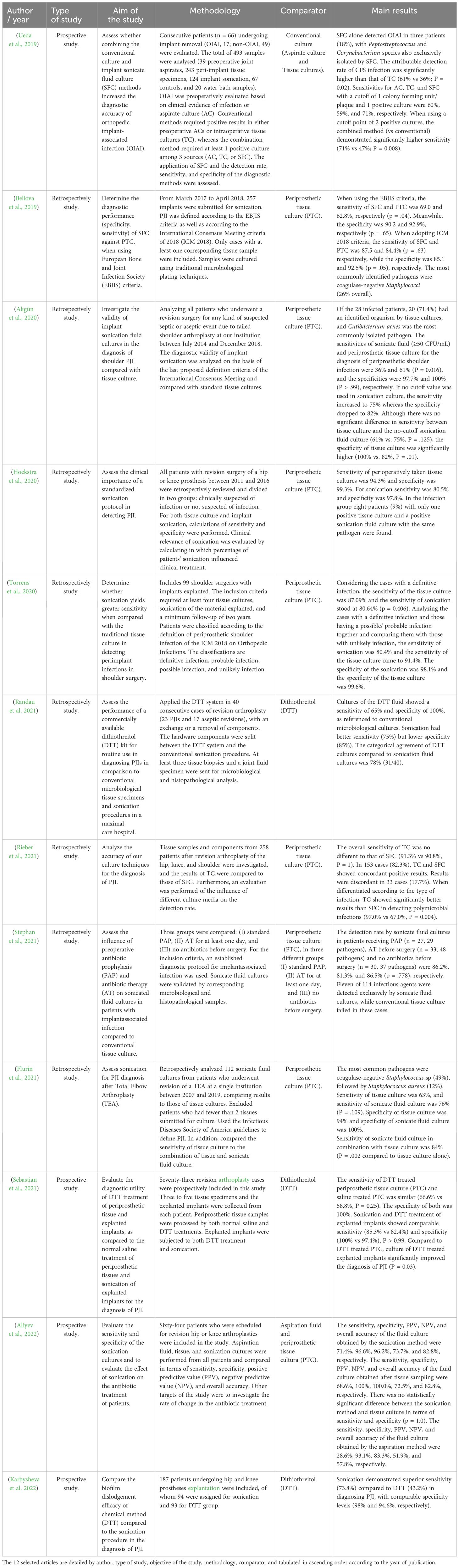
Table 1 Details of selected studies.
After conducting an exploratory reading of the material obtained, the following points were discussed: 1- the sonication protocol used and the results obtained regarding the sensitivity and specificity compared with those of periprosthetic tissue culture; 2- the main microorganisms isolated; and 3- the ability of sonication protocols to displace biofilm structure compared to other displacement techniques.
3 The sensitivity and specificity of sonication protocols are greater than those of periprosthetic tissue culture
Differences in the parameters of the sonication protocols adopted in the selected studies were observed. These differences include the use and duration of vortexing, the use of centrifugation as a method for determining sample concentration after vortex agitation, and variations in the sonication bath concerning frequency, power density, and time. Additionally, cutoff values for microbial count to define infection differed among the studies ( Table 2 ).
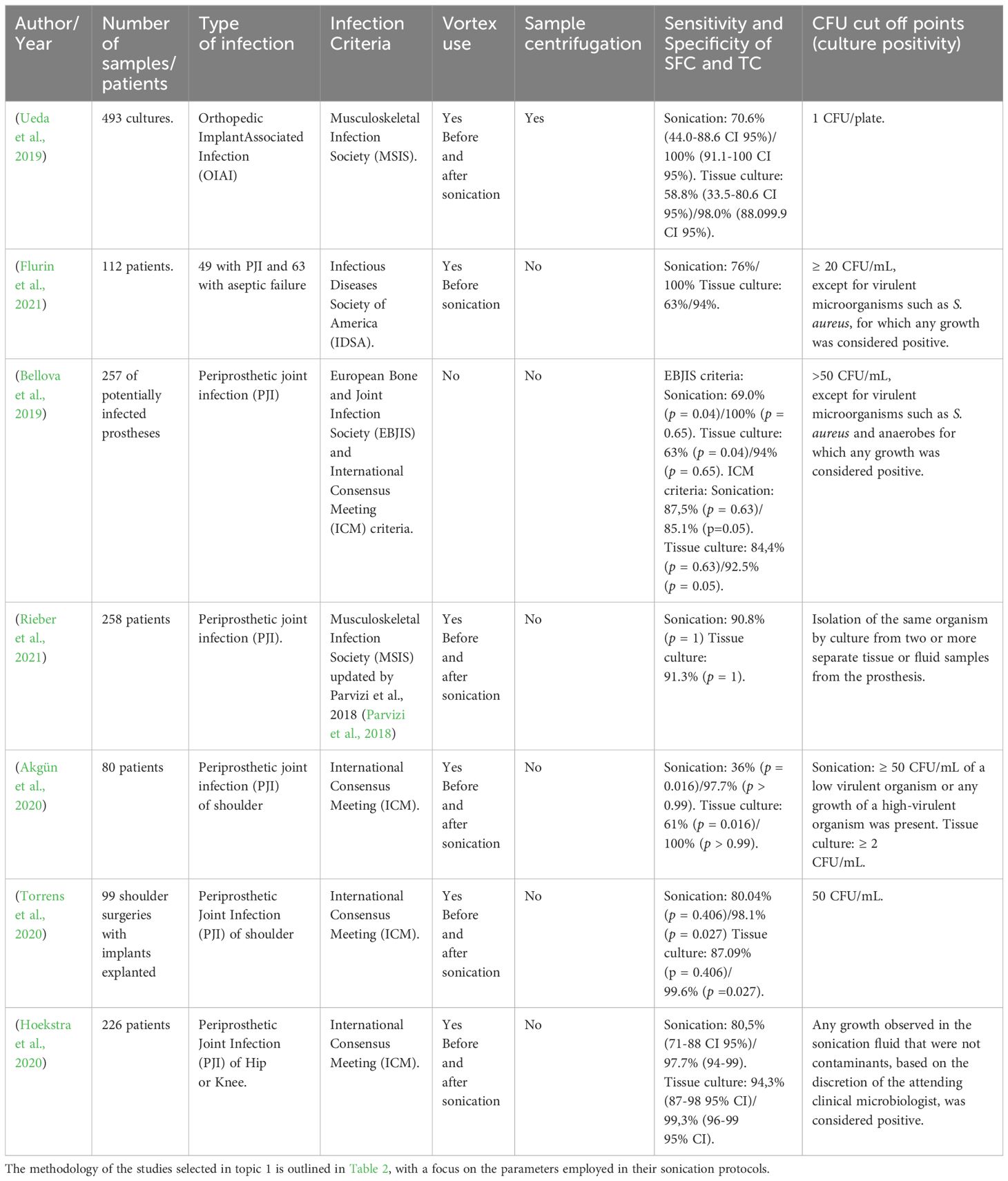
Table 2 Characterization of the studies employing sonication.
The studies analyzed also calculated the sensitivity and specificity percentages of their sonication protocols and standard cultures. Sensitivity is defined as the ability of the diagnostic test to detect individuals who are truly positive and is calculated according to the number of true positives divided by the number of true positives added to the number of false negatives (TP/(TP+FN)), using the gold standard test as a reference. Specificity is defined as the ability of the diagnostic test to detect true negatives and is calculated according to the number of true negatives divided by the number of true negatives plus the number of false positives (TN/(TN+FP)) using the gold standard test as a reference ( Ueda et al., 2019 ).
The first analyzed sonication protocol included vortex mixing of the container with the implant immersed in sterile saline solution for 30 s, an ultrasound bath at a frequency of 40 ± 2 kHz, and 0.22 ± 0.04 W/cm 2 for 1 min, followed by vortexing for another 30 s. Then, 50 mL of sonicated fluid was centrifuged at 2600 rpm for 15 minutes and cultured. The cutoff for a positive result was ≥1 CFU/plate, calculated as CFU/mL based on CFU/plate. For statistical tests, 2x2 contingency tables were constructed consisting of true-positive (TP), false-positive (FP), false-negative (FN) and true-negative (TN) results, taking positive results for the disease as a reference according to MSIS criteria. Ninety-five percent confidence intervals were calculated as exact binomial confidence intervals. The sensitivity and specificity of the different diagnostic culture methods were compared by McNemar’s test of paired proportions. All testing was conducted using SPSS v22.0 software (SPSS, Inc., Chicago, IL), with a p value < 0.05 (in 2-sided testing) considered to indicate statistical significance ( Ueda et al., 2019 ).
The reported sensitivity for SFC was 71%, 95% CI (44.0-88.6), while PTC achieved a sensitivity of 59%, 95% CI (33.5-80.6) at a cutoff point of 1 colony-forming unit/plate and 1 positive culture. Furthermore, the detection rate of orthopedic implant-associated infection (OIAI) attributed to sonicated fluid culture was significantly greater than that attributed to tissue culture (61% vs. 36%; p = 0.02). Using the cutoff point of 2 positive culture, the combination of the two methods (PTC and SFC) showed better sensitivity than the conventional method (94%, 95% CI (69.2-99.7) vs. 82%, 95% CI (55.8-95.3); p = 0.25) ( Ueda et al., 2019 ).
A second study used the sonication protocol proposed by Trampuz et al., 2007 ( Trampuz et al., 2007 ). The container with the prosthetic components was filled with Ringer’s solution (an isotonic solution containing sodium, chloride, potassium, calcium and sodium lactate used to prevent osmotic shock in bacteria in procedures intended for the preparation of suspensions), vortexed (30 s), sonicated at a frequency of 40 ± 2 kHz and 0.22 ± 0.04 W/cm 2 (Aquasonic Model 750T - VWR Scientific Products) for 5 min, vortexed for an additional 30 s, and then cultured. Sensitivities and specificities were also calculated using a 2×2 contingency table for both methods, as well as their 95% confidence intervals. To compare the sensitivities and specificities of the different tests, the McNemar test was used to compare paired proportions ( p value < 0.05) ( Flurin et al., 2021 ).
For sonicate fluids, was considered a culture positive if there was growth of greater than 20 CFU/10 mL of sonicate fluid, with the exception of virulent microorganisms such as S. aureus , for which any growth was considered positive. The sensitivity and specificity of SFC were 76%, 95% CI (62-85), and 100%, 95% IC (94-100), respectively, while for PTC, these values decreased by 63%, 95% CI (49-75), and 94%, 95% IC (85-98), respectively. The sensitivity of both tests combined (84%, 95% CI 71-91) was significantly greater than the sensitivity of tissue culture alone (63%, p = 0.002) ( Flurin et al., 2021 ).
In another study, the explanted prostheses were immersed in Ampuwa ® solution (a highly pure hypotonic water that does not contain dissolved substances) followed by an ultrasonic bath for 1 min at 80% power (P=160 W) (BactoSonic; Bandelin, Berlim, Alemanha), and no vortex was used before culture. Sensitivity and specificity were determined (2x2 contingency tables for SFC and PTC and their proportions calculated using the McNemar test, using SPSS software - IBM Corporation; Armonk, NY, United States) for the diagnosis of PJI defined according to the EBJIS criteria, which considers a count of > 50 CFU/mL of any organism as positive for PJI, and in accordance with the criteria of the ICM 2018, which considers two positive cultures of the same organism as the major criterion for the diagnosis of PJI and a single positive culture as the minor criterion ( Bellova et al., 2019 ).
Based on the EBJIS infection criteria, there was a statistically significant difference in sensitivity between sonication fluid culture and tissue culture ( p = 0.04). SFC exhibited a sensitivity of 69.0% (100/145 patients were accurately identified by the SFC as positive) and a specificity of 90.2% (101/112 patients were accurately identified by the SFC as negative), while PTC demonstrated a sensitivity of 62.8% (91/145) and a specificity of 92.9% (104/112). However, when the ICM 2018 criteria were adopted, the sensitivities of SFC and PTC were 87.5% (84/96) and 84.4% (81/96) ( p = 0.63), respectively, while the specificities were 85.1% (137/161) and 92.5% (149/161) ( p = 0.05), respectively ( Bellova et al., 2019 ).
On the other hand, even when using a standardized protocol ( Trampuz et al., 2007 ) that has already shown positive results ( Flurin et al., 2021 ), the general sensitivity obtained was not significantly different between PTC and SFC (91.3%, 170/186 vs 90.8%, 169/186; p = 1), considering isolation of the same organism by culture from two or more separate tissue or fluid samples from the prosthesis as positive. However, examining the results based on infection type, PTC demonstrated better performance in detecting polymicrobial infections than did SFC (97.0%, 32/33 vs 67.0%, 22/33; p = 0.004), as determined by the two-proportion Z test using RStudio software (version 1.2.5042) ( p < 0.05) ( Rieber et al., 2021 ).
Better results for PTC were also observed using a proposed sonication protocol ( Renz et al., 2018 ) that included the addition of saline solution to the container with the prosthesis to cover most of the implant, then an initial vortex shaking (30 s) of the container, followed by a sonication bath for 1 min at 40 kHz (BactoSonic; Bandelin Electronic, Berlin, Germany), and another 30 seconds of vortex mixing. A cutoff point of ≥ 50 CFU/mL was used to determine positivity by sonication of the fluid and isolation of the same organism from 2 or more tissue samples to determine positivity by standard culture (ICM 2018) ( Akgün et al., 2020 ).
Using a 2x2 contingency table, McNemar comparison test, chi-square test and Fisher’s exact test to determine significant differences between categorical variables ( p < 0.05) (SPSS version 20 - IBM, Armonk, NY, USA) resulted in a sensitivity of 36% for SFC (≥ 50 CFU/mL), while it was 61% for PTC (≥ 2 tissue samples with the same organism) ( p = 0.016). The specificity was 97.7% for SFC and 100% for PTC ( p > 0.99). However, when the cutoff value was eliminated in sonication culture, the sensitivity increased to 75%. Nevertheless, this increase in sensitivity came at the expense of decreased specificity, which decreased to 82%. These changes did not result in a statistically significant difference in the diagnostic benefits of SFC compared to PTC ( Akgün et al., 2020 ).
Similar findings were reported when assessing the sensitivity of SFC compared to that of PTC in detecting peri-implant infections in shoulder surgery using a similar reported protocol ( Akgün et al., 2020 ). After the removed components were transported in a polyethylene container with approximately 200–400 ml of sterile saline, the container was first vortexed for 30 seconds and then sonicated for one minute at a frequency of 40 ± 5 kHz in a Bransonic ® SM25E-MT ultrasound bath (Branson Ultrasonics Corporation, Geneva, Switzerland) after vortexing again for 30 seconds. Sonication was considered positive if at least 50 CFU/mL was detected (ICM 2018). PTC achieved 87.09% sensitivity, and SFC reached 80.64% ( p = 0.406). The specificity of PTC was 99.6%, and that of SFC was 98.1% ( p = 0.175) ( p < 0.05) (sensitivity, specificity, ROC area and Delong comparison test calculated in STATA 15.1). No statistically significant difference was found between the results obtained by the two methods ( Torrens et al., 2020 ).
The sonication procedure, which involved immersing the prosthetic container (90%) in Ringer’s solution, followed by 30 seconds of shaking, a 1-minute sonication bath at 100% power (200 W, 0.22 W/cm 2 ) (Bandelin Bactosonic), and another 30 seconds of shaking before fluid culture, also yielded favorable results for PTC compared to SFC. Using this protocol and ICM 2018 criteria, the PTC sensitivity was 94.3%, 95% CI (87-98), and the specificity was 99.3%, 95% CI (96-99). The sensitivity for SFC was 81%, 95% CI (71-88), and the specificity was 97.8%, 95% CI (94-99), which were considerably lower than the results observed for PTC (2x2 contingency table using SPSS version 22.0). Although the sensitivity and specificity of SFC were lower than those of PTC, it is worth noting that 8 patients (9% of the total) suspected of having a periprosthetic joint infection could be definitively diagnosed based on a positive result from SFC ( Hoekstra et al., 2020 ).
Sonication has shown variable diagnostic accuracy in these studies. It was possible to observe that the best sensitivity and specificity indices, compared to those of PTC, were achieved by sonication protocols that used sterile saline solution for immersion of the prosthesis, low frequencies of ultrasound waves (40 kHz) for a period of 1 or 5 minutes, the use of a vortex (before and after sonication) and centrifugation and even lower cutoff points (≥1 CFU/mL) than the 50 CFU/mL recommended by some consensuses.
In general, the results obtained with SFC were better for diagnosing PJI. Even in some studies in which its sensitivity and specificity were lower than those of PTC, it was possible to observe a small significant difference or even no significant difference. This superiority is magnified when we analyze the cost-benefit of the technique, which presents potential improvement in culture results, in a simple technique with lower recurrence rates (associated with the diagnostic inaccuracy of traditional tissue culture) and costs. Therefore, it is easily applicable in clinical practice from surgical and microbiological points of view ( Flurin et al., 2021 ). However, considering the positive contribution that PTC can add to the diagnosis ( Rieber et al., 2021 ), it is still possible to perform a combination of SFC and PTC ( Ueda et al., 2019 ).
4 Microorganism detection capacities of SFC and PTC and the main microorganisms isolated
Among the predominant microorganisms identified in the OIAI, coagulase-negative Staphylococcus species (CoNS) were the primary causative agents of infections in 24 isolates detected by both the SFC and PTC methods. However, 18% of the positive diagnoses were exclusively identified using sonication. In these cases, less virulent species, such as Streptococcus of the viridans group, Peptostreptococcus , and Corynebacterium spp., were also isolated. Peptostreptococcus and Corynebacterium spp. were isolated by SFC only. Among patients who had prior antibiotic therapy, 67% of those who received SFC had infections ( Ueda et al., 2019 ).
In a recent study, the most common pathogen isolated from periprosthetic elbow infection using both methods was CoNS (49%), followed by Staphylococcus aureus (12%), gram-negative Enterobacter cloacae (3%) and Klebsiella pneumoniae (2%). The authors observed that among the positive cultures, 78% exhibited monomicrobial cultures, while 22% had polymicrobial cultures. The SFC method played a crucial role in identifying the majority of polymicrobial infections, leading to treatment modifications in 4 out of 5 patients. However, it is worth noting that SFC failed to detect Corynebacterium amycolatum , a species that was only identified through tissue culture, but it did not have any impact on the choice of antibiotic regimen. In 10 patients, only sonicate cultures were positive: 7 for S. epidermidis , 1 for coagulase-negative Staphylococcus sp., 1 for S. aureus and 1 for Parvimonas micra ( Flurin et al., 2021 ).
In cases of PJI, the primary pathogens were also CoNS. The SFC method successfully isolated 38 of these microorganisms, whereas the PTC method yielded only 30 isolates. The second most frequently isolated microorganism was S. aureus , which was further classified as methicillin-susceptible Staphylococcus aureus (MSSA) or methicillin-resistant Staphylococcus aureus (MRSA). There were more SFC isolates than PTC isolates for both MSSA (14 vs. 12) and MRSA (5 vs. 4). They also observed that in early PJI detected using sonication, 48.9% of the cases were attributed to high-virulence pathogens, while 51.1% were associated with low-virulence pathogens. A similar pattern was observed for delayed and late infections combined, with 35.7% classified as having high virulence and 64.3% as having low virulence. Significantly, 7.8% of delayed or late infections detected using SFC were positive for anaerobes, with Cutibacterium acnes identified as the predominant species. In comparison, only 2.6% of infections were detected through PTC ( Bellova et al., 2019 ).
In addition, for the microbiological diagnosis of PJI, 220 microorganisms were isolated from the PTC and SFC methods, and concordant positive results were obtained for 153 out of 186 patients (82.3%). The CoNS (n= 60) were also the main group of bacteria isolated, with Staphylococcus epidermidis (n = 43) as the main species. The second most prevalent pathogen was S. aureus (n= 55), followed by anaerobic bacteria (n=43), and members of the Enterobacterales family (n= 27), Streptococcus spp. (n= 17) and Enterococcus spp. (n= 14) were also isolated in this study ( Rieber et al., 2021 ).
However, in periprosthetic elbow infections, the most frequently isolated pathogen by tissue culture was C. acnes , accounting for 46% of the cases. The second most isolated pathogen was CoNS, accounting for 17.9% of the cases, followed by S. aureus , accounting for 7%. Other bacteria isolated included Finegoldia magna (3.6%), Streptococcus agalactiae (13.6%), Enterococcus faecalis (3.6%), and Peptoniphilus asaccharolyticus (3.6%). In 75% of patients, at least one organism was successfully isolated by sonication. However, it is noteworthy that there were discordant results between SFC and PTC in 32% of the patients ( Akgün et al., 2020 ).
This microbial identification was also observed in patients with confirmed infection in hip or knee prostheses. Among these patients, only eight individuals (9%) had positive cultures for the same pathogen using both the SFC and PTC methods. Certain pathogens could only be identified using the SFC method. These pathogens are typically low in virulence and are known to produce biofilms, making them particularly difficult to detect. Examples of such pathogens include Streptococcus mitis , S. epidermidis , Aggregatibacter species, C. acnes , and Corynebacterium striatum ( Hoekstra et al., 2020 ).
Considering that peri-implant infections in the shoulder were definitively diagnosed, S. epidermidis was present in 42% of the patients, followed by C. acnes in 22.5%. Among these cases, 22.6% were classified as polymicrobial infections, with C. acnes being involved in most of these cases (71%) ( Torrens et al., 2020 ).
When evaluating the influence of preoperative antibiotic prophylaxis (PAP) and antibiotic therapy (AT) on SFC in patients with implant-associated infections, 114 important infectious agents were detected, 11 of which were detected exclusively after the use of SFC. The main microorganisms isolated included CoNS, S. aureus , Streptococcus spp., and Enterococcus spp. Microorganisms were identified despite prior antibiotic therapy; therefore, they do not recommend omitting antibiotic prophylaxis in patients with implant‐associated infections ( Stephan et al., 2021 ).
Moreover, although the SFC technique did not enhance the sensitivity of microbiological diagnosis for PJIs in this study, it did demonstrate the ability to identify distinct microorganisms compared to other methods. This finding contributed to changes in the strategy of antibiotic therapy for infected patients, as it relies on antimicrobial sensitivities derived from microbiological culture results ( Aliyev et al., 2022 ).
Sonicated fluid culture plays an important role in the detection of particular microorganisms, such as Peptostreptococcus, Streptococcus mitis, S. epidermidis, Aggregatibacter species, Corynebacterium spp. and Cutibacterium acnes . C. acnes is responsible for chronic and low-grade infections that represent an additional challenge to the diagnosis of PJI, with an emphasis on C. acnes , which decreases the sensitivity of traditional diagnostic tests for infections associated with orthopedic implants ( Renz et al., 2018 ; Hoekstra et al., 2020 ). In addition to the aforementioned bacteria, fungal PJI, although rare (1% to 2%), can be difficult to control and identify because the isolation of organisms by traditional culture can take a long time, resulting in false negatives ( Chisari et al., 2022 ). It is believed that fungi and mycobacteria are responsible for more than 85% of cases of negative cultures in PJI (7%-15%). In this context, sonication is a low-cost method capable of increasing the chances of identifying the causative agent ( Palan et al., 2019 ).
The ability of CFS to identify diverse and especially low-virulence microorganisms, even in the face of preoperative antibiotic prophylaxis, can affect the antibiotic therapy strategy adopted ( Aliyev et al., 2022 ). This characteristic has the potential to increase the effectiveness of treatment, reduce costs associated with prolonged use of antibiotics and longer hospital stays, often requiring multiple surgical procedures, thus reducing unnecessary exposure to antibiotics; consequently, bacterial resistance has increased dramatically in the last ten years, probably due to the excessive and often inappropriate use of antibiotics ( Drago and De Vecchi, 2017 ).
This ability may further permit the use of an antibiotic-loaded bone cement spacer sonication fluid culture technique to confirm the eradication of infection or two-stage revision reinfection prior to reimplantation of new prostheses. It can be used accurately as a complement to evaluate the therapeutic effect of IAP ( Zhang et al., 2021a ).
5 The ability of sonication protocols to dislodge biofilm structure compared to that of other displacement methods
Dislodging of bacterial cells from the biofilm structure can be achieved by mechanical means or chemical or physical methods. Mechanical methods such as scraping the prosthesis or vortices have rarely been evaluated; when studied, they have shown low performance ( Bjerkan et al., 2009 ; Drago and De Vecchi, 2017 ), and in the current literature, they are scarce ( Drago and De Vecchi, 2017 ). The use of chemical substances in explanted implants and periprosthetic tissues is suggested as a possible biofilm dislodgement method with possible applicability in the diagnosis of infections associated with implants. Among the proposed agents are the metal chelator ethylenediaminetetraacetic acid (EDTA) and the strong reducing agent dithiothreitol (DTT) ( Karbysheva et al., 2020 ).
It was suggested that the activity of EDTA against biofilm cells occurs through the chelation of magnesium, calcium, and iron, enhancing the detachment of cells from the biofilm matrix ( Banin et al., 2006 ). Additional observations included that the mean colony count (logCFU/mL) after DTT treatment was comparable to that achieved after sonication or physical methods and greater than the count obtained using the scraping technique ( Drago et al., 2012 ).
Compared with DTT treatments for the diagnosis of PJI, the explanted implants were immersed in 0.1% w/v TDT (Promega, Madison, WI, USA) in sterile saline and kept in an incubator with shaking for 15 minutes at room temperature, followed by centrifugation at 3,000 rpm for 10 minutes before standard cultivation. With a sonication protocol for explanted implants, 90% of the prosthesis was immersed in sterile saline solution, followed by vigorous manual shaking for 30 seconds before and after the sonicated bath, which was programmed for 7 min at 40 kHz in BactoSonic (Bandelin GmbH, Berlin, Germany). The procedure was finished with centrifugation at 4,000 rpm for 20 minutes. Using the MSIS definition of IAP, both methods, SFC and DTT, demonstrated similar sensitivity rates of 85.3% (29/34) and 82.4% (28/34) ( p > 0.05) (analysis performed with STATA software version 14.2 - Stata Corp LLC; Texas, USA), respectively. Although not statistically significant, the specificity was greater when using the SFC technique (100%, 39/39 vs. 97.4%, 38/39 p > 0.99) ( Sebastian et al., 2021 ).
In another study, DTT treatment involved the use of a MicroDTTect closed system for biofilm processing via this chemical method. Explant samples were collected from the sterile system itself, and then, in the laboratory, the chamber valve containing TDT (150 mL, 0.1% p/v) was broken, allowing DTT to flow into the explant. The device was subsequently mechanically stirred for 15 minutes at room temperature, after which standard cultivation continued. For the sonication protocol, the explanted implants were collected in sterile plastic bags that were subsequently filled with sterile saline solution, vortexed for 30 seconds and sonicated in a Bactosonic 14.2 device (Bactosonic, Bandelin, Berlin, Germany) for 5 minutes with a frequency of 40 ± 2 kHz and a power density of 0.22 ± 0.04 W/cm2 followed by 30 seconds of vortexing ( Randau et al., 2021 ).
Using the MSIS criteria to define PJI, the authors found a sensitivity of 65% (13/20) and specificity of 100% (20/20) for DTT fluid culture compared to conventional microbiological cultures. Sonication had better sensitivity (75%, 15/20) but lower specificity (85%, 17/20) than conventional microbiological culture ( p > 0.05) (statistical significance between groups was assessed by the Mann-Whitney test). Fisher’s exact test was used for contingency, sensitivity and specificity analysis. The analysis was performed using GraphPad Prism 8.0.2 (GraphPad Software, La Jolla, CA, USA). The categorical concordance of DTT cultures with that of SFC cultures was 78% (31/40) ( Randau et al., 2021 ).
Based on these results, sonication has been shown to be the main assay for biofilm detection in the microbiological diagnosis of implant-associated infection ( Karbysheva et al., 2020 ). Even in one study that showed a loss of specificity, sonication provided a more reliable diagnosis of PJI, as it identified more pathogens than DTT treatment ( Karbysheva et al., 2022 ). However, given the positive impacts that chemical methods can have on the diagnosis of these infections, especially on culture specificity, DTT treatment could be used as a biofilm displacement technique in situations where sonication is not viable or possible ( Sebastian et al., 2021 ). It is also possible to evaluate the potential additive effect of chemical shift on sonication ( Karbysheva et al., 2020 ).
6 Proposal for a standardized sonication protocol
To propose the best parameters for establishing a sonication protocol, studies by Cieslinski et al., 2021, Trampuz et al., 2006, Oliva et al., 2021, Rosa et al., 2019, Ueda et al., 2019, Dudek et al., 2020, Li et al., 2018, Ribeiro et al., 2022, Beguiristain et al., 2023, Borens et al., 2013 and Morgenstern et al., 2020 ( Trampuz et al., 2006 ; Borens et al., 2013 ; Rosa et al., 2019 ; Ueda et al., 2019 ; Dudek et al., 2020 ; Morgenstern et al., 2020 ; Cieslinski et al., 2021 ; Oliva et al., 2021 ; Ribeiro et al., 2022 ; Beguiristain et al., 2023 ), were also reviewed in addition to the abovementioned studies ( Figure 1 ).
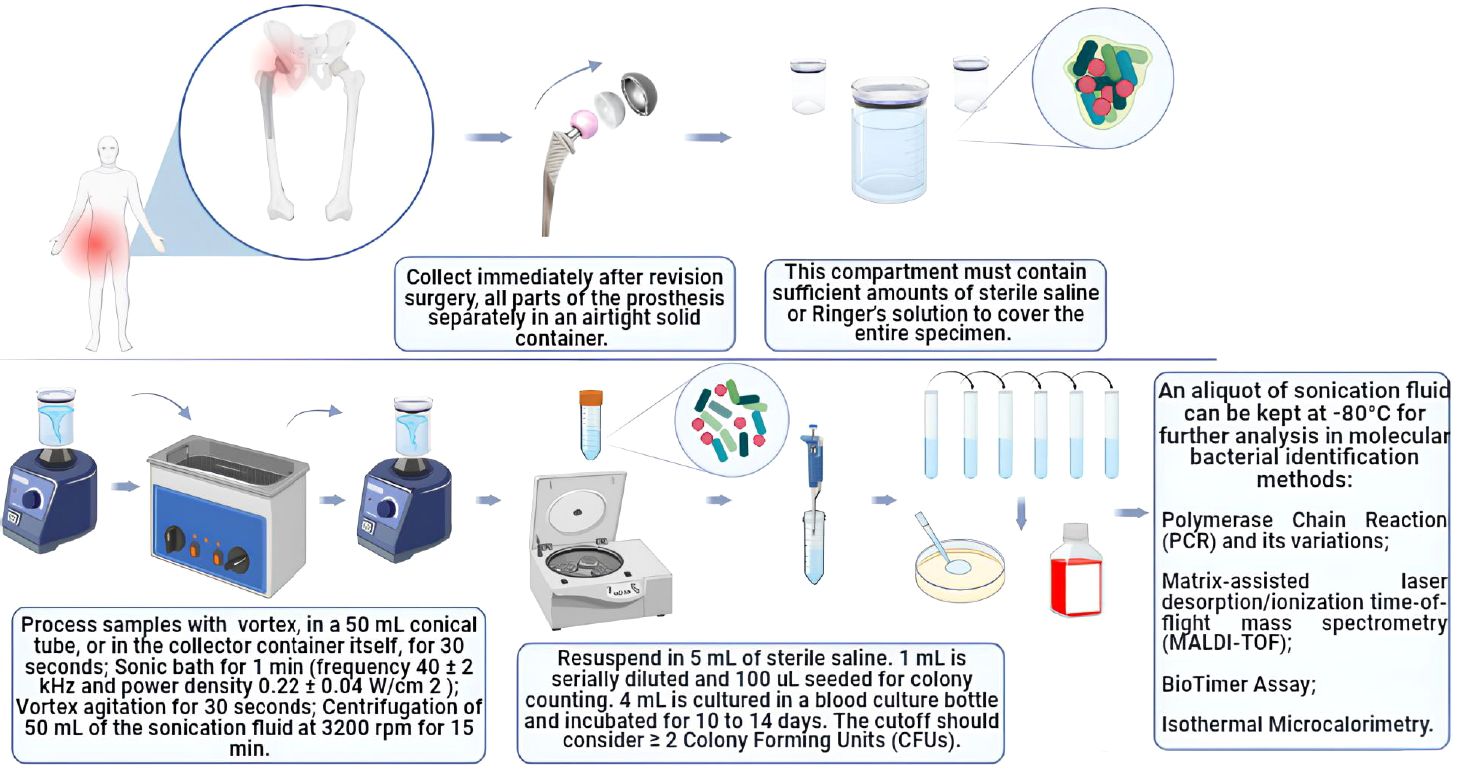
Figure 1 Diagram of the sonication protocol according to the parameters reviewed in this study.
6.1 Material collection
After revision surgery, it is recommended that all prosthetic components, periprosthetic cement or osteosynthesis devices, including polyethylene (PE) materials and metal and polymethylmethacrylate (PMMA) components, be carefully removed to avoid direct contact with the patient’s skin. These items should be collected separately. To ensure the integrity of the collected samples and preserve the viability of microorganisms, it is important to use airtight containers. It has already been demonstrated that storing samples in plastic bags can significantly reduce colony-forming unit (CFU) counts and is associated with the risk of contamination ( Rosa et al., 2019 ; Cieslinski et al., 2021 ).
The use of plastic bags for storage promotes the desiccation of microorganisms, which can lead to changes in their biophysical properties, such as surface tension. This desiccation can disrupt physiological processes, including the growth of microorganisms ( Cieslinski et al., 2021 ).
Thus, physical containers with thicker, completely sealed (hermetic) surfaces can serve as a protective measure by preventing water loss and helping to maintain microbial viability while reducing the risk of contamination. It is important for the entire sample to be covered, so these compartments should contain an adequate amount of sterile saline or Ringer’s solution. By ensuring a sealed and moist environment, physical containers can help preserve the viability of microorganisms and maintain their physiological state during storage. This is crucial for accurate microbiological analysis and for reducing the potential for false-negative results or alterations in microbial characteristics.
6.2 Sample storage
The samples should be processed soon after removal, ideally within 2 to 4 hours. However, if immediate processing is not feasible, the samples can be stored at -4°C without liquid. Refrigerated samples can be stored for 7 days, and although there may be a minor decrease in the bacterial load over time, this decrease is unlikely to have a significant impact on the culture’s positivity. This is particularly true when molecular methods are employed for bacterial identification of sonicated fluid ( Cieslinski et al., 2021 ).
6.3 Vortex-sonication-vortex method
In this proposed protocol, the processing of the samples involves the following steps:
1. Sample vortex: The sample is vortexed for 30 seconds in the collection container, ensuring that the prosthesis is completely submerged in a sterile saline solution (the amount of solution depends on the sample size).
2. Sonication bath: The sample was immersed in a sonication bath for 1 min at a frequency of approximately 40 ± 2 kHz and a power density of 0.22 ± 0.04 W/cm 2 .
3. Vortex Agitation: After sonication, the sample was vortexed again for 30 seconds.
4. Centrifugation: Approximately 15 mL of sonication fluid (without the prosthesis) was centrifuged at 3200 rpm for 15 minutes to concentrate the sample (bacterial cells, probably present in the sonication fluid) for later culturing.
As a standard procedure to minimize the risk of contamination in subsequent protocol steps, it is important to change the water in the ultrasonic bath after each round of sonication.
The use of vortex stirring and centrifugation has been shown to contribute to enhancing specificity ( Rosa et al., 2019 ; Oliva et al., 2021 ). With respect to the duration of sonication, a 1-minute duration produced good results. This short period of sonication helps to avoid potential bactericidal effects of the procedure ( Ueda et al., 2019 ).
The ideal ultrasound frequencies for sonication to identify etiological agents of implant-associated infections, particularly PJI caused by pathogens such as S. aureus , P. aeruginosa , and E. coli , are 35 kHz and 40 kHz ( Dudek et al., 2020 ). These frequencies were effective in displacing bacteria from biofilms, and they had a significant impact on the survival of bacteria, particularly those in a planktonic state.
The majority of studies describing the use of the sonication technique for explant prostheses use a sonication bath, often due to sample size constraints. However, it is worth noting that recently, an article demonstrated greater sensitivity with direct intraoperative sonication culture of implants and soft tissues than with conventional synovial fluid culture utilizing a portable probe sonicator (Shanghai Weimi Ultrasonic Co., Ltd.) ( Ji et al., 2023 ).
6.4 Microbiological analysis
1- The sediment obtained from the centrifuged solution must be resuspended in 5 mL of sterile saline solution. 2- To determine the microbial cell count and viability, 1 mL of the resuspension was serially diluted 10 times at a ratio of 1:10, resulting in a total volume of 1 mL. Between 3-100 μL of the last three dilutions were plated on Mueller Hinton agar plates and incubated for 18-24 hours at 37°C to facilitate bacterial growth. 4- The remaining 4 mL of the resuspension was inoculated into blood culture bottles. The bottles are subsequently incubated for 10 to 14 days to facilitate the detection of slow-growing or fastidious microorganisms that might be present in the sample. In cases where there is clinical evidence of infection but standard microbiological cultures yield negative results, it is advisable to conduct additional fungal and mycobacterial cultures.
The advantages of inoculation into blood culture bottles have recently been highlighted. Among them are the increased sensitivity and specificity of culture, even in patients who received prior antibiotic therapy, due to the presence of antimicrobial removal systems and lytic agents in blood bottles that further promote the release of intracellular microorganisms. In this context, the use of blood culture bottles to inoculate joint fluids directly at the patient’s bedside can be valuable. Additionally, the application of an innovative version of the sonication culture method involves direct sonication of the retrieved implant and soft tissue, without a sonication tube, intraoperatively. This method utilizes a BACT/ALERT 3D blood culture system, contributing to the increased effectiveness of microbiological diagnosis for PJI ( Drago et al., 2019 ; Ji et al., 2023 ).
The incubation period should be extended to up to two weeks to enhance the likelihood of identifying causative agents comprehensively. For instance, species such as Staphylococcus spp. are more likely to emerge during the initial week of incubation, while Cutibacterium spp. are typically detected during the second week ( Oliva et al., 2021 ). Once the etiological agent is accurately identified, it becomes possible to prescribe the most suitable treatment. In regard to antibiotics, selecting the appropriate type and dosage is crucial. Notably, the cure rate for patients with culture-negative PJI is generally low ( Li et al., 2018 ).
6.5 Bacterial phenotypic identification and quantification from sonication fluid
A positive culture result for sonication fluid was determined by a bacterial concentration ≥ 2 CFU/mL in ≥ 2 cultures. In this context, a monomicrobial PJI is considered if only one bacterial species grows above the cutoff in sonicated fluid cultures. Conversely, polymicrobial PJI is diagnosed if more than one species is isolated following the same criteria. Following a positive blood culture, phenotypic identification should be conducted using culture media supporting the growth of both aerobic and anaerobic bacteria. It is important to note that for diagnosis, the identified species are more relevant than the CFU count.
Some studies used cutoff points ≥ 1 and ≥ 5 CFU, as the sensitivity of sonicated liquid cultures can be significantly reduced, especially in patients who have received previous antibiotic therapy or still have chronic, low-grade infection. It is worth mentioning that the cutoff point of 50 CFU/mL, which is defended by most medical societies and widely used in clinical practice, may not be ideal in these cases, despite its ability to distinguish effective infections ( Ueda et al., 2019 ; Oliva et al., 2021 ).
The growth of any virulent microorganism responsible for high-grade acute infections, such as Staphylococcus aureus and gram-negative bacilli, will also be considered. However, the growth of low-virulence microorganisms responsible for chronic and low-grade infections, such as coagulase-negative Staphylococcus, Enterococcus spp., Corynebacterium spp. and Cutibacterium acnes , in a single sample must be evaluated in conjunction with the patient’s clinical context ( Drago et al., 2019 ; Romanò et al., 2019 ; Ueda et al., 2019 ; Oliva et al., 2021 ). This is where sonication presents one of its greatest advantages, which is its ability to more efficiently identify bacteria responsible for chronic, low-grade and difficult-to-detect infections, as previously mentioned, helping to improve the poor performance of conventional microbiological methods for identifying these pathogens ( Renz et al., 2018 ; Hoekstra et al., 2020 ).
According to our suggested protocol, any growth detected in the sonicated fluid culture from patients who received antibiotics within two weeks prior to sample collection should be regarded as a positive result ( Ueda et al., 2019 ; Oliva et al., 2021 ).
6.6 Bacterial identification by molecular methods
The sonication protocol, involving the use of a vortex-sonication vortex followed by CFU counts, may have several limitations. These include the inability to dislodge all adherent microorganisms in the biofilm and the potential for sonication to affect microbial viability, leading to inaccurate CFU counts. Molecular methods can be employed to address these limitations and contribute to the identification of difficult-to-cultivate bacteria, anaerobes, and noncultivable bacteria ( Rosa et al., 2019 ).
These molecular methods include polymerase chain reaction (PCR), such as bacterial identification based on the amplification of 16S ribosomal RNA, and various methods, such as multiplex PCR (mPCR), which can amplify the genetic material of different targets in a single process, thus allowing bacterial identification, as performed in commercial panels of multiplex PCR for IAP ( Schoenmakers et al., 2023 ). It exhibits good sensitivity and requires less sample material and time than culture-based methods. Broad-range PCR can identify the predominant bacterial strain at infection sites of various cultural origins, even in patients undergoing antibiotic therapy. The main limitations of PCR-based diagnosis include the inability to discriminate between live and dead bacteria and DNA contamination. However, when used in conjunction with sonication, it has great diagnostic value for PJI, especially for routine clinical practice when used in panels, as already mentioned ( Liu et al., 2018 ; Schoenmakers et al., 2023 ; Tsikopoulos and Meroni, 2023 ).
Another method is identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF), which allows direct identification of aerobic and anaerobic bacteria from positive blood cultures. MALDI-TOF has been successfully employed for detecting microorganisms in biological samples, whether from colonies or fluids. Several studies support the feasibility of using this technique for bacterial identification in sonicated fluid as well as direct identification in blood culture bottles. This approach facilitates early and reliable identification, serving as an alternative to culture methods ( Cieslinski et al., 2021 ; Ribeiro et al., 2022 ; Beguiristain et al., 2023 ).
As additional techniques, we can also perform fluorescence in situ hybridization (FISH), in which fluorescent probes bind to complementary nucleic acid sequences to identify the presence or absence of these target sequences. The ability to identify bacteria in negative cultures reduces false positives through better identification of environmental contamination and the ability to exclude dead bacteria with a viability stain. Another technique that can help with pathogen identification is DNA microarrays, where microarrays allow the simultaneous measurement of large numbers of genes involving thousands of microscopic DNA sequences (probes) complementary to specific gene fragments of the microorganisms studied. However, both of these methods have the disadvantages of high cost, the need for specialized equipment, the potential for contamination and a lack of probes relevant for diagnosing PJI ( Shoji and Chen, 2020 ).
We can also mention the use of identification methods based on specific bacteriophages for the pathogens studied, where DNA detection by qPCR and adenosine triphosphate (ATP) detection are performed after bacteriophage lysis. This technique aims to contribute to the development of a faster, more sensitive, specific and, at the same time, economical and practical system to establish an accurate diagnosis of PJI, with applicability in sonicated fluid ( Šuster and Cör, 2022 ).
6.7 Alternative identification methods
Among the alternative methods of bacterial identification that have been suggested for the diagnosis of infections associated with implants, the BioTimer Assay (BTA), which indirectly identifies microorganisms through the detection of microbial metabolic products, uses an original reagent containing red phenol or resazurin as indicators. Phenol red changes from red to yellow, indicating the presence of fermenting microorganisms, while resazurin changes from violet to pink, indicating the presence of nonfermenting microorganisms ( Rosa et al., 2019 ).
Another method is isothermal microcalorimetry, which is considered a new method for real-time detection of heat production related to the growth of reproductive microorganisms in biological fluid. This detection method has proven to be highly sensitive and rapid in synovial fluid samples for the diagnosis of septic arthritis. Likewise, sonication fluid microcalorimetry was useful for diagnosing PJI with a considerably faster detection time than conventional microbial culture ( Borens et al., 2013 ; Morgenstern et al., 2020 ).
These methods have the advantages of easy execution and accessibility for the identification process. However, they present important limitations compared to molecular methods, which perform more precise identification. BTA is incapable of identifying microbial genera and species, a problem that can be remedied with sonication, as BTA has good sensitivity for microbial analysis ( Rosa et al., 2019 ).. Microcalorimetry has reduced sensitivity due to the same challenge that culture faces with the presence of biofilms for diagnosing PJI but could complement cultures and support rapid, real-time decisions in orthopedic device-related infections ( Morgenstern et al., 2020 ).
Additionally, imaging techniques to visualize biofilms can also be applied. Confocal laser scanning microscopy and scanning electron microscopy provide imaging of the biofilm without compromising the biofilm structure; in some cases, confocal laser microscopy makes it possible to visualize viable biofilm bacteria in joint fluid, wound tissue and bone cement. Scanning electron microscopy can be used to visualize the coaggregation of microbial cells, but the preparation often results in the loss of the biofilm matrix. The cost and training requirements for obtaining the best images limit the use of these techniques (56).
7 Conclusion
One of the primary challenges in the management of implant-associated infections is microbiological diagnosis. To ensure a reliable diagnosis and successful treatment, complete removal of the implant and dislodgment of the microorganisms causing the infection, which are predominantly present in biofilm structures, are necessary. For this purpose, the sonication technique was successfully proposed, although its diagnostic accuracy is still questioned in the current literature.
When reviewing the literature, it was possible to observe the adoption of different protocols, as expected, and consequently different results regarding the sensitivity and specificity of sonicated fluid cultures compared to those of periprosthetic tissue cultures. It was possible to observe an even greater prevalence of coagulase-negative Staphylococcus species, followed by Staphylococcus aureus , identified as etiological agents of infections associated with implants, by both culture methods, but sonication proved to be important for the identification of low-virulence pathogens that produce biofilms, which are notoriously difficult to detect, such as the species Peptostreptococcus and Corynebacterium spp.
In the analysis of the studies that compared SFC and chemical methods of biofilm displacement, EDTA and DTT, it was observed that the results varied between the superior sensitivity and specificity of SFC, and there was no significant difference between the SFC and chemical methods.
In this context, we conducted an analysis of various aspects, including sample collection, storage conditions, cultivation methods, microorganism identification techniques (both phenotypic and molecular), and the cutoff point for CFU counts. Additionally, we propose optimal parameters for programming the sonication bath and sample processing.
In conclusion, based on our analysis and review of the current literature, we have established a theoretical foundation for standardizing sonication protocols. The aim of this study was to achieve the highest sensitivity and specificity indices for the reliable microbiological diagnosis of infections associated with implants and prosthetic devices, such as PJIs. However, practical application and further complementary studies are still necessary.
Author contributions
ND: Data curation, Methodology, Project administration, Writing – original draft, Writing – review & editing. BS: Data curation, Methodology, Writing – original draft, Writing – review & editing. AO: Formal Analysis, Supervision, Validation, Writing – review & editing. PR: Formal Analysis, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The financial resources to carry out this research were provided through Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (CNPq/MCTI No. 10/2023 – UNIVERSAL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmed, S. S., Haddad, F. S. (2019). Prosthetic joint infection. Bone Joint Res. 8, 570–572. doi: 10.1302/2046-3758.812.BJR-2019-0340
PubMed Abstract | CrossRef Full Text | Google Scholar
Akgün, D., Maziak, N., Plachel, F., Siegert, P., Minkus, M., Thiele, K., et al. (2020). The role of implant sonication in the diagnosis of periprosthetic shoulder infection. J. Shoulder. Elbow. Surg. 29, e222–e228. doi: 10.1016/j.jse.2019.10.011
Aliyev, O., Yıldız, F., Kaya, H. B., Aghazada, A., Sümbül, B., Citak, M., et al. (2022). Sonication of explants enhances the diagnostic accuracy of synovial fluid and tissue cultures and can help determine the appropriate antibiotic therapy for prosthetic joint infections. Int. Orthop. 46, 415–422. doi: 10.1007/s00264-021-05286-w
Azad, M. A., Patel, R. (2024). Practical Guidance for Clinical Microbiology Laboratories: Microbiologic diagnosis of implant-associated infections. Clin. Microbiol. Rev. 20, e0010423. doi: 10.1128/cmr.00104-23
CrossRef Full Text | Google Scholar
Banin, E., Brady, K. M., Greenberg, E. P. (2006). Chelator-Induced Dispersal and Killing of Pseudomonas aeruginosa cells in a biofilm. Society 72, 2064–2069. doi: 10.1128/AEM.72.3.2064
Beam, E., Osmon, D. (2018). Prosthetic joint infection update. Infect. Dis. Clin. North Am. 32, 843–859. doi: 10.1016/j.idc.2018.06.005
Beguiristain, I., Henriquez, L., Sancho, I., Martin, C., Hidalgo-Ovejero, A., Ezpeleta, C., et al. (2023). Direct prosthetic joint infection diagnosis from sonication fluid inoculated in blood culture bottles by direct MALDI-TOF mass spectrometry. Diagnostics 13. doi: 10.3390/diagnostics13050942
Bellova, P., Knop-Hammad, V., Königshausen, M., Mempel, E., Frieler, S., Gessmann, J., et al. (2019). Sonication of retrieved implants improves sensitivity in the diagnosis of periprosthetic joint infection. BMC Musculoskelet. Disord. 20, 1–9. doi: 10.1186/s12891-019-3006-1
Birlutiu, R. M., Birlutiu, V., Mihalache, M., Mihalache, C., Cismasiu, R. S. (2017). Diagnosis and management of orthopedic implant-associated infection: a comprehensive review of the literature. Biomed. Research-tokyo. 28, 5063–5073.
Google Scholar
Bjerkan, G., Witsø, E., Bergh, K. (2009). Sonication is superior to scraping for retrieval of bacteria in biofilm on titanium and steel surfaces in vitro. Acta Orthop. 80, 245–250. doi: 10.3109/17453670902947457
Borens, O., Yusuf, E., Steinrücken, J., Trampuz, A. (2013). Accurate and early diagnosis of orthopedic device-related infection by microbial heat production and sonication. J. Orthopedic. Res. 31, 1700–1703. doi: 10.1002/jor.22419
Chisari, E., Lin, F., Fei, J., Parvizi, J. (2022). Fungal periprosthetic joint infection: Rare but challenging problem. Chin. J. Traumatol. 25, 63–66. doi: 10.1016/j.cjtee.2021.12.006
Cieslinski, J., Ribeiro, V. S. T., Kraft, L., Suss, P. H., Rosa, E., Morello, L. G., et al. (2021). Direct detection of microorganisms in sonicated orthopedic devices after in vitro biofilm production and different processing conditions. Eur. J. Orthopedic. Surg. Traumato. 31, 1113–1120. doi: 10.1007/s00590-020-02856-3
Drago, L., Clerici, P., Morelli, I., Ashok, J., Benzakour, T., Bozhkova, S., et al. (2019). The world association against infection in orthopedics and trauma (WAIOT) procedures for microbiological sampling and processing for periprosthetic joint infections (PJIs) and other implant-related infections. J. Clin. Med. 1, 8. doi: 10.3390/jcm8070933
Drago, L., De Vecchi, E. (2017). Microbiological Diagnosis of Implant-Related Infections: Scientific Evidence and Cost/Benefit Analysis of Routine Antibiofilm Processing [published correction appears in Adv Exp Med Biol. 2017; 971:113]. Adv. Exp. Med. Biol. 971, 51–67. doi: 10.1007/5584_2016_154
Drago, L., Romanò, C. L., Mattina, R., Signori, V., De Vecchi, E. (2012). Does dithiothreitol improve bacterial detection from infected prostheses? A pilot study infection. Clin. Orthop. Relat. Res. 470, 2915–2925. doi: 10.1007/s11999-012-2415-3
Dudek, P., Grajek, A., Kowalczewski, J., Madycki, G., Marczak, D. (2020). Ultrasound frequency of sonication applied in microbiological diagnostics has a major impact on viability of bacteria causing periprosthetic joint infection. Int. J. Infect. Dis. 100, 158–163. doi: 10.1016/j.ijid.2020.08.038
Filho, C. A. M., Aragão, M. T., Santos, R. S. (2020). Clinical and epidemiological profile of infections related to joint prostheses. Arch. Health 1, 7–16. doi: 10.46919/archv1n1-002
Flurin, L., Greenwood-Quaintance, K. E., Esper, R. N., Sanchez-Sotelo, J., Patel, R. (2021). Sonication improves microbiologic diagnosis of periprosthetic elbow infection. J. Shoulder. Elbow. Surg. 30, 1741–1749. doi: 10.1016/j.jse.2021.01.023
Hoekstra, M., Veltman, E. S., Nurmohamed, RFRHA, van Dijk, B., Rentenaar, R. J., Vogely, H. C., et al. (2020). Sonication leads to clinically relevant changes in treatment of periprosthetic hip or knee joint infection. J. Bone Jt. Infect. 5, 128–132. doi: 10.7150/jbji.45006
Ji, B., Aimaiti, A., Wang, F., Maimaitiyiming, A., Zhang, X., Li, G., et al. (2023). Intraoperative direct sonication of implants and soft tissue for the diagnosis of periprosthetic joint infection. J. Bone Joint Surg. 105, 855–874. doi: 10.2106/JBJS.22.00446
Karbysheva, S., Cabric, S., Koliszak, A., Bervar, M., Kirschbaum, S., Hardt, S., et al. (2022). Clinical evaluation of dithiothreitol in comparison with sonication for biofilm dislodgement in the microbiological diagnosis of periprosthetic joint infection. Diagn. Microbiol. Infect. Dis. 103, 115679. doi: 10.1016/j.arth.2017.11.049
Karbysheva, S., Di Luca, M., Butini, M. E., Winkler, T., Schütz, M., Trampuz, A. (2020). Comparison of sonication with chemical biofilm dislodgement methods using chelating and reducing agents: Implications for the microbiological diagnosis of implant associated infection. PloS One 15, 1–15. doi: 10.1371/journal.pone.0231389
Li, C., Renz, N., Thies, C. O., Trampuz, A. (2018). Meta-analysis of sonicate fluid in blood culture bottles for diagnosing periprosthetic joint infection. J. Bone Jt. Infect. 3, 273–279. doi: 10.7150/jbji.29731
Liu, K., Fu, J., Yu, B., Sun, W., Chen, J., Hao, L. (2018). Meta-analysis of sonication prosthetic fluid PCR for diagnosing periprosthetic joint infection. PloS One 1, 13. doi: 10.1371/journal.pone.0196418
Moore, A. J., Blom, A. W., Whitehouse, M. R., Gooberman-Hill, R. (2015). Deep prosthetic joint infection: A qualitative study of the impact on patients and their experiences of revision surgery. BMJ Open 5, 1–13. doi: 10.1136/bmjopen-2015-009495
Morgenstern, C., Renz, N., Cabric, S., Maiolo, E., Perka, C., Trampuz, A. (2020). Thermogenic diagnosis of periprosthetic joint infection by microcalorimetry of synovial fluid. BMC Musculoskelet. Disord. 21, 1–7. doi: 10.1186/s12891-020-03366-3
Oliva, A., Miele, M. C., Al Ismail, D., Di Timoteo, F., De Angelis, M., Rosa, L., et al. (2021). Challenges in the microbiological diagnosis of implant-associated infections: A summary of the current knowledge. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.750460
Oliva, A., Pavone, P., D’Abramo, A., Iannetta, M., Mastroianni, C. M., Vullo, V. (2016). Role of sonication in the microbiological diagnosis of implant-associated infections: Beyond the orthopedic prosthesis. Adv. Exp. Med. Biol. 897, 85–102. doi: 10.1007/5584_2015_5007
Palan, J., Nolan, C., Sarantos, K., Westerman, R., King, R., Foguet, P. (2019). Culture-negative periprosthetic joint infections. EFORT. Open Rev. 4, 585–594. doi: 10.1302/2058-5241.4.180067
Parvizi, J., Tan, T. L., Goswami, K., Higuera, C., Della Valle, C., Chen, A. F., et al. (2018). The 2018 definition of periprosthetic hip and knee infection: an evidence-based and validated criteria. J. Arthroplasty. 33, 1309–1314.e2. doi: 10.1016/j.arth.2018.02.078
Portillo, M. E., Sancho, I. (2023). Advances in the microbiological diagnosis of prosthetic joint infections. Diagn. (Basel). 13, 809. doi: 10.3390/diagnostics13040809
Randau, T. M., Molitor, E., Fröschen, F. S., Hörauf, A., Kohlhof, H., Scheidt, S., et al. (2021). The performance of a dithiothreitol-based diagnostic system in diagnosing periprosthetic joint infection compared to sonication fluid cultures and tissue biopsies. Z. Orthop. Unfall. 159, 447–453. doi: 10.1055/a-1150-8396
Renz, N., Mudrovcic, S., Perka, C., Trampuz, A. (2018). Orthopedic implant-associated infections caused by Cutibacterium spp. – A remaining diagnostic challenge. PloS One 13. doi: 10.1371/journal.pone.0202639
Ribeiro, V. S. T., Cieslinski, J., Bertol, J., Schumacher, A. L., Telles, J. P., Tuon, F. F. (2022). Detection of microorganisms in clinical sonicated orthopedic devices using conventional culture and qPCR. Rev. Bras. Ortop. (Sao. Paulo). 57, 689–696. doi: 10.1055/s-0041-1732386
Rieber, H., Frontzek, A., Heinrich, S., Breil-Wirth, A., Messler, J., Hegermann, S., et al. (2021). Microbiological diagnosis of polymicrobial periprosthetic joint infection revealed superiority of investigated tissue samples compared to sonicate fluid generated from the implant surface. Int. J. Infect. Dis. 106, 302–307. doi: 10.1016/j.ijid.2021.03.085
Romanò, C. L., Al Khawashki, H., Benzakour, T., Bozhkova, S., del Sel, H., Hafez, M., et al. (2019). The W.A.I.O.T. definition of high-grade and low-grade peri-prosthetic joint infection. J. Clin. Med. 1, 8. doi: 10.3390/jcm8050650
Rosa, L., Lepanto, M. S., Cutone, A., Berlutti, F., De Angelis, M., Vullo, V., et al. (2019). BioTimer assay as complementary method to vortex-sonication-vortex technique for the microbiological diagnosis of implant associated infections. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-01944045-1
Salar, O., Phillips, J., Porter, R. (2021). Diagnosis of knee prosthetic joint infection; aspiration and biopsy. Knee 30, 249–253. doi: 10.1016/j.knee.2020.12.023
Schoenmakers, J. W. A., de Boer, R., Gard, L., Kampinga, G. A., van Oosten, M., van Dijl, J. M., et al. (2023). First evaluation of a commercial multiplex PCR panel for rapid detection of pathogens associated with acute joint infections. J. Bone Jt. Infect. 8, 45–50. doi: 10.5194/jbji-8-45-2023
Sebastian, S., Malhotra, R., Sreenivas, V., Kapil, A., Dhawan, B. (2021). The utility of dithiothreitol treatment of periprosthetic tissues and explanted implants in the diagnosis of prosthetic joint infection. Indian J. Med. Microbiol. 39, 179–183. doi: 10.1016/j.ijmmb.2020.12.004
Shen, H., Tang, J., Wang, Q., Jiang, Y., Zhang, X. (2015). Sonication of explanted prosthesis combined with incubation in BD Bactec bottles for pathogen-based diagnosis of prosthetic joint infection. J. Clin. Microbiol. 53, 777–781. doi: 10.1128/JCM.02863-14
Shoji, M. M., Chen, A. F. (2020). Biofilms in periprosthetic joint infections: A review of diagnostic modalities, current treatments, and future directions. J. Knee. Surg. 33, 119–131. doi: 10.1055/s-0040-1701214
Stephan, A., Thürmer, A., Glauche, I., Nowotny, J., Zwingenberger, S., Stiehler, M. (2021). Does preoperative antibiotic prophylaxis affect sonication-based diagnosis in implant-associated infection? J. Orthopedic. Res. 39, 2646–2652. doi: 10.1002/jor.25015
Šuster, K., Cör, A. (2022). Fast and specific detection of staphylococcal PJI with bacteriophage-based methods within 104 sonicate fluid samples. J. Orthop. Res. 40, 1358–1364. doi: 10.1002/jor.25167
Tande, A. J., Patel, R. (2014). Prosthetic joint infection. Clin. Microbiol. Rev. 27, 302–345. doi: 10.1128/CMR.00111-13
Torrens, C., Fraile, A., Santana, F., Puig, L., Alier, A. (2020). Sonication in shoulder surgery: is it necessary? Int. Orthop. 44, 1755–1759. doi: 10.1007/s00264-020-04543-8
Torres, L. M., Turrini, N. R., Merighi, A. M. B., Cruz, A. G. (2015). Readmissão por infecção do sítio cirúrgico ortopédico: uma revisão integrativa Readmission from orthopedic surgical site infections: an integrative review Reingreso por infección del sitio quirúrgico ortopédico: una revisión integradora. Rev. Esc. Enferm. USP. 49, 1004–1011. doi: 10.1590/S0080-623420150000600018
Trampuz, A., Piper, K. E., Hanssen, A. D., Osmon, D. R., Cockerill, F. R., Steckelberg, J. M., et al. (2006). Sonication of explanted prosthetic components in bags for diagnosis of prosthetic joint infection is associated with risk of contamination. J. Clin. Microbiol. 44, 628–631. doi: 10.1128/JCM.44.2.628-631.2006
Trampuz, A., Piper, K. E., Jacobson, M. J., Hanssen, A. D., Unni, K. K., Osmon, D. R., et al. (2007). Sonication of removed hip and knee prostheses for diagnosis of infection. New Engl. J. Med. 357, 654–663. doi: 10.1056/nejmoa061588
Trebse, R., Roskar, S. (2021). Evaluation and interpretation of prosthetic joint infection diagnostic investigations. Int. Orthop. 45, 847–855. doi: 10.1007/s00264-021-04958-x
Tsikopoulos, K., Meroni, G. (2023). Periprosthetic joint infection diagnosis: A narrative review. Antibiotics 1, 12. doi: 10.3390/antibiotics12101485
Ueda, N., Oe, K., Nakamura, T., Tsuta, K., Iida, H., Saito, T. (2019). Sonication of extracted implants improves microbial detection in patients with orthopedic implant-associated infections. J. Arthroplasty. 34, 1189–1196. doi: 10.1016/j.arth.2019.02.020
Xu, Y., Huang, T. B., Schuetz, M. A., Choong, P. F. M. (2023). Mortality, patient-reported outcome measures, and the health economic burden of prosthetic joint infection. EFORT. Open Rev. 8, 690–697. doi: 10.1530/EOR-23-0078
Yilmaz, M. K., Abbaszadeh, A., Tarabichi, S., Azboy, I., Parvizi, J. (2023). Diagnosis of periprosthetic joint infection: the utility of biomarkers in 2023. Antibiotics. (Basel). 12, 1054. doi: 10.3390/antibiotics12061054
Zardi, E. M., Franceschi, F. (2020). Prosthetic joint infection. A relevant public health issue. J. Infect. Public Health 13, 1888–1891. doi: 10.1016/j.jiph.2020.09.006
Zhang, Q., Ding, B., Wu, J., Dong, J., Liu, F. (2021a). Sonication fluid culture of antibiotic-loaded bone cement spacer has high accuracy to confirm eradication of infection before reimplantation of new prostheses. J. Orthop. Surg. Res. 16, 377. doi: 10.1186/s13018-021-02520-4
Zhang, Q., Xi, Y., Li, D., Yuan, Z., Dong, J. (2021b). The yield of sonication fluid culture for presumed aseptic loosening of orthopedic devices: a meta-analysis. Ann. Palliat. Med. 10, 1792–1808. doi: 10.21037/apm-20-1228
Keywords: infections, review, sonication, diagnostic, microbiology
Citation: Silva NS, De Melo BST, Oliva A and de Araújo PSR (2024) Sonication protocols and their contributions to the microbiological diagnosis of implant-associated infections: a review of the current scenario. Front. Cell. Infect. Microbiol. 14:1398461. doi: 10.3389/fcimb.2024.1398461
Received: 09 March 2024; Accepted: 29 April 2024; Published: 13 May 2024.
Reviewed by:
Copyright © 2024 Silva, De Melo, Oliva and de Araújo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natally Dos Santos Silva, [email protected]
† ORCID : Natally Dos Santos Silva, orcid.org/0000-0001-5019-8869
This article is part of the Research Topic
Value of a multidisciplinary approach for modern diagnosis of infectious diseases
- Open access
- Published: 09 May 2024
Exploring age and gender variations in root canal morphology of maxillary premolars in Saudi sub population: a cross-sectional CBCT study
- Mohmed Isaqali Karobari 1 , 2 ,
- Azhar Iqbal 3 ,
- Rumesa Batul 1 ,
- Abdul Habeeb Adil 1 ,
- Jamaluddin Syed 4 , 5 ,
- Hmoud Ali Algarni 3 ,
- Meshal Aber Alonazi 3 &
- Tahir Yusuf Noorani 6 , 7
BMC Oral Health volume 24 , Article number: 543 ( 2024 ) Cite this article
106 Accesses
Metrics details
In complex teeth like maxillary premolars, endodontic treatment success depends on a complete comprehension of root canal anatomy. The research on mandibular premolars’ root canal anatomy has been extensive and well-documented in existing literature. However, there appears to be a notable gap in available data concerning the root canal anatomy of maxillary premolars. This study aimed to explore the root canal morphology of maxillary premolars using cone-beam computed tomography (CBCT) imaging, considering age and gender variations.
From 500 patient CBCT scans, 787 maxillary premolar teeth were evaluated. The sample was divided by gender and age (10–20, 21–30, 31–40, 41–50, 51–60, and 61 years and older). Ahmed et al. classification system was used to record root canal morphology.
The most frequent classifications for right maxillary 1st premolars were 2 MPM 1 B 1 L 1 (39.03%) and 1 MPM 1 (2.81%), while the most frequent classifications for right maxillary 2nd premolars were 2 MPM 1 B 1 L 1 (39.08%) and 1 MPM 1 (17.85%). Most of the premolars typically had two roots (left maxillary first premolars: 81.5%, left maxillary second premolars: 82.7%, right maxillary first premolars: 74.4%, right maxillary second premolars: 75.7%). Left and right maxillary 1st premolars for classes 1 MPM 1 and 1 MPM 1–2−1 showed significant gender differences. For classifications 1 MPM 1 and 1 MPM 1–2−1 , age-related changes were seen in the left and right maxillary first premolars.
This study provides novel insights into the root canal anatomy of maxillary premolars within the Saudi population, addressing a notable gap in the literature specific to this demographic. Through CBCT imaging and analysis of large sample sizes, the complex and diverse nature of root canal morphology in these teeth among Saudi individuals is elucidated. The findings underscore the importance of CBCT imaging in precise treatment planning and decision-making tailored to the Saudi population. Consideration of age and gender-related variations further enhances understanding and aids in personalized endodontic interventions within this demographic.
Peer Review reports
Introduction
The morphology and variability of root canal systems play a crucial role in the success of endodontic treatment [ 1 , 2 ]. Understanding the intricacies of root canal anatomy is essential for effective diagnosis, treatment planning, and applying appropriate techniques. The research on mandibular premolars’ root canal anatomy has been extensive and well-documented in existing literature [ 3 , 4 ]. However, there appears to be a notable gap in available data concerning the root canal anatomy of maxillary premolars [ 5 , 6 , 7 , 8 , 9 ].
Maxillary premolars present unique challenges due to their anatomical complexity, including multiple canals, isthmuses, and accessory canals [ 10 , 11 ]. Accurately identifying and classifying root canal systems in maxillary premolars is crucial for diagnosis and achieving optimal treatment outcomes [ 12 ].
Despite the importance of understanding root canal morphology, there remains a gap in knowledge concerning maxillary premolars. This lack of comprehensive information on the root canal morphology of maxillary premolars hinders endodontic practitioners’ ability to deliver precise and successful treatments [ 13 ]. This study aims to fill this gap by conducting an investigation using cone-beam computed tomography (CBCT) imaging. CBCT, as a non-invasive and highly accurate imaging technique, offers the advantage of providing detailed three-dimensional representations of root canal systems, which were previously not easily achievable through conventional radiographs [ 14 ]. The high-resolution images obtained through CBCT will provide valuable data to enhance the knowledge and clinical management of root canal anatomy in these teeth, leading to better-informed treatment decisions and reduced complications [ 4 , 15 ].
By analyzing a large sample size of CBCT images, we aim to comprehensively understand the root canal configuration in maxillary premolars, considering factors such as age and gender [ 16 ]. The findings of this study will contribute to enhancing the knowledge and clinical management of root canal anatomy in maxillary premolars, improving treatment success rates, and reducing complications.
By elucidating the variations and complexities of root canal morphology in maxillary premolars, this study will aid dental professionals in making informed decisions regarding treatment approaches, instrument selection, and the application of advanced endodontic techniques [ 17 , 18 ]. Furthermore, the results will provide valuable insights for dental educators, researchers, and students, facilitating the development of standardized protocols and guidelines for managing root canal systems in maxillary premolars.
Methodology
Study design.
This study employed a retrospective cross-sectional design to comprehensively investigate the root canal morphology of maxillary premolars using cone-beam computed tomography (CBCT) imaging. This design allows for the examination of a large sample size and facilitates the analysis of root canal anatomy variations among different age groups and genders. By retrospectively analyzing CBCT images, the study aimed to elucidate the complex root canal anatomy of maxillary premolars and identify potential factors influencing their variability.
Ethical consideration
Ethical approval was obtained from the Local Committee of Bioethics for Research at the Dentistry College, King Abdul-Aziz University (Ethical Approval No. 025-02-22). Informed consent was obtained from the Committee of Bioethics for Research, College of Dentistry, King Abdul-Aziz University, Jeddah, Saudi Arabia, considering the retrospective nature of the study. This ensured that the study adhered to ethical standards and protected the rights and confidentiality of the participants. Additionally, the study complied with all relevant regulations and guidelines regarding the use of patient data for research purposes.
Sample size determination
The sample size for this study was determined using G Power 3.1.9.4 software, considering a chi-square test for goodness-of-fit, statistical power analysis, and an a priori approach. A comprehensive sample of 500 patient records was obtained, resulting in the evaluation of 787 maxillary premolar teeth. This large sample size enhances the statistical power of the study and allows for robust analysis of root canal morphology variations. It also increases the generalizability of the findings to the target population.
Inclusion and exclusion criteria
Inclusion criteria were carefully defined to ensure the selection of appropriate teeth for analysis. Healthy maxillary premolars with small carious or restorative crowns, fully formed root apex, and defect-free radiographic images were included in the study. Exclusion criteria were applied to eliminate potential confounding factors, including root canal-treated teeth, fractured upper and lower posterior teeth, post and core restorations, calcification, resorption defects, and anomalies of crown and root. These criteria helped ensure the homogeneity of the study sample and the validity of the results.
Imaging technique
CBCT images were acquired using the iCAT scanner system (Imaging Sciences International, Hatfield, PA, USA), a widely recognized and reliable imaging device in dentistry. Standardized imaging parameters (120 KVp, 5–7 mA) were employed to ensure consistent image quality across all scans. The use of CBCT allowed for the acquisition of detailed three-dimensional representations of root canal anatomy, enabling precise analysis and classification. High-resolution images obtained through CBCT provided valuable data for evaluating root canal morphology.
Calibration and reliability
Prior to data collection, calibration was conducted involving an expert endodontist and an observer. The observer underwent rigorous training to accurately identify and classify root canal morphology. Calibration involved the examination of 50 CBCT images, with discrepancies resolved through discussion to achieve consensus. The kappa test was utilized to determine the level of agreement between observers, and intra- and interobserver reliability was assessed. Furthermore, specimens were assessed independently by observers following calibration to minimize bias and ensure consistency in the evaluations. A high kappa value (0.8) was obtained, indicating substantial to almost perfect reliability, thereby ensuring the validity of the data collected. This rigorous calibration process helped minimize observer bias and enhance the reliability of the study findings.
Root and canal analysis
Root canal morphology was recorded and classified according to the classification system proposed by Ahmed et al. in 2017. This classification system provides a standardized framework for describing root canal configurations, facilitating comparisons across studies. The obtained CBCT images were meticulously analyzed, with root canal morphology recorded for each maxillary premolar (Fig. 1 ). The images were divided into age groups (10 to 20, 21 to 30, 31 to 40, 41 to 50, 51 to 60, and 61 years above) and categorized by gender (males and females) to explore variations in root canal anatomy. Detailed analysis of each image was conducted to identify the number of roots, canals, and any anatomical variations present.
New classification system for root canal morphology of maxillary left second premolar classified using the new classification system, described as code 1 25 1 . The code consists of three components, the tooth number – Yellow color arrow, number of roots – blue color arrow and the root canal configuration – green color arrow. The number of roots is added as a superscript before the tooth number, so it is single root and tooth number (25). Description of root canal configuration is written as superscript after the tooth number on the course of the root canal starting from the orifices [O], passing through the canal [C], ending by the foramen [F], so it is single canal
Statistical analysis
Statistical analysis was performed using SPSS version 26 software. Descriptive statistics, including mean frequency and standard deviation, were calculated to summarize the data. The association between root canal morphology and age/gender was analyzed using the chi-square test or Fisher exact test, depending on the distribution of the data. Significance levels were set at p ≤ 0.05 to determine the statistical significance of the findings. Additionally, subgroup analyses were conducted to explore potential interactions between age, gender, and root canal morphology.
The distribution of maxillary premolars according to Ahmed’s classification was examined. Table 1 presents the distribution of premolars based on the classification categories. For right maxillary 1st premolars, the majority belonged to 2 MPM 1 B 1 L 1 (39.03%) and 1 MPM 1 (2.81%) categories. Similarly, for right maxillary 2nd premolars, 2 MPM 1 B 1 L 1 (39.08%) and 1 MPM 1 (17.85%) were the most prevalent categories.
Table 2 displays the distribution of maxillary premolars based on the number of roots. The majority of premolars had two roots (73.33% for left maxillary 1st premolars, 24.45% for left maxillary 2nd premolars, 74.03% for right maxillary 1st premolars, and 24.32% for right maxillary 2nd premolars) (Figs. 2 , 3 and 4 ).
CBCT View (Sagittal and axial) of left maxillary second premolar showing the code 1 MPM 1
CBCT View (Sagittal and axial) maxillary first and second premolars showing the canal variations
CBCT View (Sagittal and axial) maxillary first and second premolars showing the canal variations in more than one root
Tables 3 and 4 present the distribution of left and right maxillary 1st and 2nd premolars, respectively, based on gender. In Table 3 , significant gender differences were observed for the classification 1 MPM 1 ( p = 0.515) and 1 MPM 1–2−1 ( p = 0.010*) for both left maxillary 1st and 2nd premolars. The number of males and females for MPM 1 in left maxillary 1st premolars was 121 and 88, respectively, while for 1 MPM 1 in left maxillary 2nd premolars, it was 111 and 72, respectively. Similarly, for 1 MPM 1–2−1 in left maxillary 1st premolars, the number of males and females was 3 and 3, respectively, whereas for left maxillary 2nd premolars, it was 30 and 21, respectively.
Table 4 indicates significant gender differences for the classification MPM 1 ( p = 0.032*) and 1 MPM 1–2−1 ( p = 0.003*) in the right maxillary 1st premolars. The number of males and females for 1 MPM 1 in the right maxillary 1st premolars was 122 and 84, respectively, while for 1 MPM 1 in the right maxillary 2nd premolars, it was 115 and 70, respectively. Additionally, the number of males and females for 1 MPM 1–2−1 in right maxillary 1st premolars was 10 and 11, respectively, whereas, for right maxillary 2nd premolars, it was 33 and 18, respectively.
Tables 5 and 6 demonstrate the distribution of left and right maxillary 1st and 2nd premolars, respectively, based on age groups. In Table 5 , significant differences were observed for the classification 1 MPM 1 ( p = 0.053) and 1 MPM 1–2−1 ( p = 0.002*) in left maxillary 1st premolars. The number of premolars in each age group for 1 MPM 1 in left maxillary 1st premolars ranged from 1 to 7, whereas for 1 MPM 1–2−1, it ranged from 0 to 3. For left maxillary 2nd premolars, significant differences were observed for the classification 1 MPM 1 ( p = 0.002*) and 1 MPM 1–2−1 ( p = 0.002*). The number of premolars in each age group for 1 MPM 1 in left maxillary 2nd premolars ranged from 6 to 38, whereas for 1 MPM 1–2−1, it ranged from 4 to 23.
In Table 6 , significant differences were observed for the classification 1 MPM 1 ( p = 0.055) and MPM 1 ( p = 0.002*) in the right maxillary 1st and 2nd premolars, respectively. The number of premolars in each age group for 1 MPM 1 in the right maxillary 1st premolars ranged from 1 to 6, whereas for 1 MPM 1–2−1, it ranged from 0 to 15. For right maxillary 2nd premolars, significant differences were observed for the classification 1 MPM 1 ( p = 0.002*) and 1 MPM 1–2−1 ( p = 0.002*). The number of premolars in each age group for 1 MPM 1 in the right maxillary 2nd premolars ranged from 6 to 36, whereas for 1 MPM 1–2−1 , it ranged from 3 to 15.
The present study aimed to investigate the root canal morphology of maxillary premolars using cone-beam computed tomography (CBCT) imaging. By analyzing a large sample size of CBCT images, we sought to provide a comprehensive understanding of the complex and variable root canal configuration in maxillary premolars, considering factors such as gender and age.
As mentioned in the literature [ 11 , 19 ], our findings revealed a diverse range of root canal configurations in maxillary premolars. Multiple canals, isthmuses, and accessory canals in these teeth pose a challenge to endodontic treatment, as it necessitates thorough exploration, disinfection, and meticulous instrumentation [ 20 ]. Recognizing such complex anatomy underscores the importance of employing advanced imaging techniques, such as CBCT, to accurately visualize and assess root canal morphology [ 21 , 22 ].
In our study, age emerged as a significant factor influencing the root canal morphology of maxillary premolars. The categorization into different age groups allowed for a nuanced exploration of these variations, corroborating previous research [ 23 , 24 , 25 ]. The age-specific analysis revealed noteworthy trends in the prevalence of certain root canal configurations. For instance, in left maxillary 1st premolars, the marginal significance ( p = 0.053) for 1MPM1 suggests a potential shift in root canal anatomy with increasing age. This finding prompts further investigation into the underlying reasons for such variations across age groups. Similarly, the significant difference ( p = 0.002*) observed in 1MPM1-2-1 in both left and right maxillary 1st premolars indicates distinct patterns in root canal morphology among different age brackets. This finding raises questions about whether these differences are attributed to developmental changes, wear and tear, or other factors associated with aging. These age-related changes can be attributed to factors such as dentin deposition and secondary dentin formation, which may alter the shape and complexity of the root canal system over time. Therefore, endodontists should consider these age-related variations when planning and performing root canal procedures, particularly in older patients [ 26 ]. Younger age groups may exhibit features associated with incomplete root development and open apices, while older age groups may show signs of maturation, closure of apices, and increased calcification [ 27 ]. The correlations between age-related changes in root canal morphology and systemic conditions enhance the clinical context. Systemic factors, such as hormonal changes, metabolic disorders, or medication use, may influence dental development and impact root canal anatomy differently across age groups [ 28 ]. Practitioners should consider these age-related nuances during treatment planning and execution, adjusting their approaches to accommodate the potential variations in root canal anatomy. For example, younger patients may exhibit different anatomical features compared to older individuals, influencing decisions related to instrumentation and obturation techniques.
Furthermore, our study identified gender-based differences in root canal morphology. This finding aligns with Ahmed et al. [ 19 ], who reported similar gender differences in maxillary premolars. Their study revealed a higher prevalence of multiple canals in males than females, which supports our observations of significant gender variations in root canal morphology. However, it is worth noting that Ahmed et al. did not mention the specific classification code 1 MPM 1–2−1 in their study, making a direct comparison somewhat limited.
Likewise, Cleghorn et al. [ 11 ] found that the prevalence of multiple canals in maxillary first premolars ranged from 30 to 73%, a range consistent with our findings. Shi et al., while studying the Chinese population [ 23 ], also noted significant differences in the number of roots and gender in both maxillary first and second premolars.
In a study conducted by Mashyakhy et al. [ 29 ] in a Saudi population, highly statistically significant differences in canal configurations were observed between genders in maxillary teeth. Similarly, Martins et al. [ 30 ] reported a gender difference in the root canal morphology of the Portuguese population. However, it is essential to mention that some contrasting results were found in specific subpopulations. For instance, no significant difference in root canal morphology was noted in the Malaysian subpopulation [ 31 ] and the German subpopulation [ 32 ].
In summary, our study adds to the existing body of literature by providing further evidence of gender-related variations in root canal morphology, and it is in line with previous research in this field.
This study’s utilization of CBCT imaging provided valuable insights into the three-dimensional morphology of maxillary premolars. CBCT has emerged as a powerful diagnostic tool in endodontics, enabling the visualization of intricate root canal anatomy [ 33 ]. Accurately assessing root canal morphology facilitates precise treatment planning, guiding clinicians in determining the appropriate access, instrumentation, and obturation techniques [ 34 ]. The present study has several advantages, reinforcing its conclusions’ reliability and veracity. First and foremost, a large sample size was used in the study, with 500 cone-beam computed tomography (CBCT) images in total, 1230 maxillary premolars included. This large sample size improves the study’s statistical power and broadens the applicability of the results to the intended population.
The study employed qualified endodontists and observers calibrated to evaluate root canal morphology to achieve precise and reliable analysis. To determine the classification of root canal morphology, 50 CBCT images were examined as part of the calibration process. The research boosted the consistency and accuracy of the results by creating a smooth decision-making process that reduced the possibility of observer bias.
In the present study, a standardized classification scheme was used. This classification system offers a reliable and standardized method for classifying root canal morphology. The study’s findings may be easily compared and integrated with those of other research utilizing the same approach because it used a recognized classification system. Understanding root canal morphology in maxillary premolars is ultimately enhanced by this, making it easier for future research and enabling meta-analyses.
Additionally, the study compared its findings to pertinent literature, enabling a thorough interpretation of the data in light of earlier research. The study offers important insights into the heterogeneity of root canal morphology in maxillary premolars by comparing the consistency or divergence of results across different populations and studies. The scientific knowledge base is expanded, and this topic is better understood thanks to the comparative method.
Strengths of our study
One of the key strengths of our study is the large sample size, which enhances the statistical power and generalizability of our findings. Additionally, the utilization of cone-beam computed tomography (CBCT) imaging allowed for detailed three-dimensional analysis of root canal morphology, providing valuable insights into the complexity of maxillary premolars. Our rigorous calibration process, involving expert endodontists and observers, ensured the reliability and accuracy of our data collection and analysis. Furthermore, by considering age and gender variations, we were able to explore the influence of demographic factors on root canal anatomy, contributing to a more nuanced understanding of this topic.
Limitations
Despite these strengths, our study also has several limitations that warrant consideration. Firstly, the retrospective nature of the study may introduce selection bias and limit the generalizability of the findings. Additionally, the study focused on a specific population, which may limit its applicability to other ethnic groups or regions. Furthermore, the reliance on CBCT imaging, while providing detailed anatomical information, is subject to radiation exposure and cost constraints. Moreover, the inclusion and exclusion criteria applied in the study may have inadvertently excluded certain teeth or patient populations, potentially affecting the representativeness of the sample.
Future research endeavors should explore the relationship between root canal morphology and treatment outcomes in maxillary premolars to enhance our knowledge further. Long-term follow-up studies can provide valuable insights into the success rates and potential complications associated with different root canal configurations. Furthermore, advancements in imaging modalities and treatment techniques, such as guided endodontics and regenerative approaches, hold promise for overcoming the challenges posed by complex root canal anatomy.
This study provides novel insights into the root canal anatomy of maxillary premolars within the Saudi population, addressing a notable gap in the literature specific to this demographic. Through CBCT imaging and analysis of large sample sizes, the complex and diverse nature of root canal morphology in these teeth among Saudi individuals is elucidated. The findings underscore the importance of CBCT imaging in precise treatment planning and decision-making tailored to the Saudi population. Consideration of age and gender-related variations further enhances understanding and aids in personalized endodontic interventions within this demographic. Moving forward, these findings inform clinical practice within the Saudi community, emphasizing the need for customized approaches to optimize treatment outcomes.
Data availability
All data supporting the findings of this study are available from the corresponding author upon reasonable request.
Schilder H. Cleaning and shaping the root canal. Dental Clin N Am. 1974;18(2):269–96.
Article CAS Google Scholar
Karobari MI, et al. Application of two systems to classify the root and canal morphology in the human dentition: a national survey in India. Journal of Dental Education; 2023.
Vertucci FJ. Root canal anatomy of the human permanent teeth Oral surgery, oral medicine, oral pathology, 1984. 58(5): pp. 589–599.
Karobari MI, et al. Evaluation of root and canal morphology of mandibular premolar amongst Saudi subpopulation using the new system of classification: a CBCT study. BMC Oral Health. 2023;23(1):1–11.
Article Google Scholar
Martins JN, et al. Worldwide Assessment of the Root and Root Canal characteristics of Maxillary Premolars–A Multi-center Cone-Beam Computed Tomography cross-sectional study with Meta-analysis. J Endod. 2024;50(1):31–54.
Article PubMed Google Scholar
Ahmed HMA. A critical analysis of laboratory and clinical research methods to study root and canal anatomy. Int Endod J. 2022;55:229–80.
Mashyakhy M. Anatomical evaluation of maxillary premolars in a Saudi population: an in vivo cone-beam computed tomography study. J Contemp Dent Pract. 2021;22(3):284–9.
Merhej M-J et al. Root and Root Canal Morphology of Premolars in a Sample of the Lebanese Population: Clinical Considerations 2022.
Karobari MI et al. Assessment of Root Canal Morphology of Maxillary Premolars: A CBCT Study Exploring Age and Gender Variations 2023.
Pécora JD, et al. Internal anatomy, direction and number of roots and size of human mandibular canines. Braz Dent J. 1993;4(1):53–7.
PubMed Google Scholar
Cleghorn BM, et al. Root and root canal morphology of the human permanent maxillary first molar: a literature review. J Endod. 2006;32(9):813–21.
Karobari MI, et al. Root and root canal morphology classification systems. Int J Dent. 2021;2021:1–6.
Google Scholar
Tian YY, et al. Root and canal morphology of maxillary first premolars in a Chinese subpopulation evaluated using cone-beam computed tomography. Int Endod J. 2012;45(11):996–1003.
Karobari MI, et al. Roots and root canals characterization of permanent mandibular premolars analyzed using the cone beam and micro computed tomography—a systematic review and metanalysis. J Clin Med. 2023;12(6):2183.
Article PubMed PubMed Central Google Scholar
Iqbal A, et al. Evaluation of root canal morphology in permanent maxillary and mandibular anterior teeth in Saudi subpopulation using two classification systems: a CBCT study. BMC Oral Health. 2022;22(1):171.
Karobari MI, et al. Root and canal morphology of the anterior permanent dentition in Malaysian population using two classification systems: a CBCT clinical study. Australian Endodontic J. 2021;47(2):202–16.
Versiani MA, et al. Root and root canal morphology of four-rooted maxillary second molars: a micro–computed tomography study. J Endod. 2012;38(7):977–82.
Neelakantan P, et al. Cone-beam computed tomography study of root and canal morphology of maxillary first and second molars in an Indian population. J Endod. 2010;36(10):1622–7.
Ahmad IA, et al. Root and root canal morphology of maxillary first premolars: a literature review and clinical considerations. J Endod. 2016;42(6):861–72.
Dastgerdi AC, et al. Isthmuses, accessory canals, and the direction of root curvature in permanent mandibular first molars: an in vivo computed tomography study. Volume 45. Restorative Dentistry & Endodontics; 2020. 1.
Scarfe WC et al. Use of cone beam computed tomography in endodontics International journal of dentistry, 2009. 2009.
Karobari MI, et al. Root and root canal configuration characterization using microcomputed tomography: a systematic review. J Clin Med. 2022;11(9):2287.
Article CAS PubMed PubMed Central Google Scholar
Shi Z-Y, et al. Root canal morphology of maxillary premolars among the elderly. Chin Med J. 2017;130(24):2999–3000.
Hu R, et al. Aging changes of the root canal morphology in maxillary first premolars observed by cone-beam computerized tomography. Zhonghua Kou Qiang Yi Xue Za Zhi = Zhonghua Kouqiang Yixue Zazhi = Chin J Stomatology. 2016;51(4):224–9.
CAS Google Scholar
Thomas R, et al. Root canal morphology of maxillary permanent first molar teeth at various ages. Int Endod J. 1993;26(5):257–67.
Article CAS PubMed Google Scholar
Mashyakhy M, et al. Root and root canal morphology differences between genders: a comprehensive in-vivo CBCT study in a Saudi population. Acta Stomatol Croatica. 2019;53(3):213.
Nuni E, et al. Endodontic Treatment for Young Permanent Teeth , in contemporary endodontics for children and adolescents . Springer; 2023. pp. 281–321.
Gilroy FG. Perceptions of general health and root canal treatment in New Zealand general dental practice. University of Otago; 2020.
Martins JN, et al. Gender influence on the number of roots and root canal system configuration in human permanent teeth of a Portuguese subpopulation. Quintessence Int. 2018;49(2):103–11.
Pan JYY, et al. Root canal morphology of permanent teeth in a Malaysian subpopulation using cone-beam computed tomography. BMC Oral Health. 2019;19(1):1–15.
Bürklein S, et al. Evaluation of the root canal anatomy of maxillary and mandibular premolars in a selected German population using cone-beam computed tomographic data. J Endod. 2017;43(9):1448–52.
Yoza T, et al. Cone-beam computed tomography observation of maxillary first premolar canal shapes. Anat Cell Biology. 2021;54(4):424–30.
Kulinkovych-Levchuk K, et al. Guided endodontics: a Literature Review. Int J Environ Res Public Health. 2022;19(21):13900.
Karobari MI, et al. Micro computed tomography (Micro-CT) characterization of root and root canal morphology of mandibular first premolars: a systematic review and meta-analysis. BMC Oral Health. 2024;24(1):1.
Download references
Acknowledgements
Not applicable.
The current paper did not receive any external funding.
Author information
Authors and affiliations.
Department of Dental Research, Center for Global Health Research, Saveetha Medical College, and Hospitals, Saveetha Institute of Medical and Technical Sciences, Saveetha University, Chennai, Tamil Nadu, India
Mohmed Isaqali Karobari, Rumesa Batul & Abdul Habeeb Adil
Department of Restorative Dentistry & Endodontics, Faculty of Dentistry, University of Puthisastra, Phnom Penh, 12211, Cambodia
Mohmed Isaqali Karobari
Department of Restorative Dentistry, College of Dentistry, Jouf University, Sakaka, 72345, Saudi Arabia
Azhar Iqbal, Hmoud Ali Algarni & Meshal Aber Alonazi
Director Research & Development, OWA Medical and Research Center, Sugarland, TX, USA
Jamaluddin Syed
Oral Basic and Clinical Sciences, Faculty of Dentistry, King Abdulaziz University, p.o box, Jeddah, 80209, Saudi Arabia
Conservative Dentistry Unit, School of Dental Sciences, Universiti Sains Malaysia, Health Campus, Kubang Kerian, 16150, Kota Bharu, Kelantan, Malaysia
Tahir Yusuf Noorani
Saveetha Dental College, Saveetha Institute of Medical and Technical Sciences (SIMATS), Chennai, Tamil Nadu, India
You can also search for this author in PubMed Google Scholar
Contributions
Conception and design of the study: MIK, and TYN. Acquisition of data: AZ and SJ. Analysis and interpretation of data: RB and AHA. Drafting the article: MIK, RB, AHA and SJ. Revising it critically for important intellectual content: MIK, AZ, HAA, MAA and TYN. All authors approved the final submitted version.
Corresponding author
Correspondence to Mohmed Isaqali Karobari .
Ethics declarations
Consent for publication, competing interests.
The authors declare no competing interests.
Ethics approval
Ethical approval for this retrospective study was obtained from the Local Committee of Bioethics for Research at the Dentistry College, King Abdul-Aziz University, with Ethical Approval No. 025-02-22. Informed consent was obtained from the Committee of Bioethics for Research, College of Dentistry, King Abdul-Aziz University, Jeddah, Saudi Arabia, considering the study’s retrospective nature. Before any investigation or treatment, the patients signed a general consent form, allowing the use of findings in future studies and publications without revealing personal information. The informed consent was obtained from all subjects and/or their legal guardian(s).
Conflict of interest
All the authors declare that they have no known conflicts of interest in terms of competing financial interests or personal relationships that could have an influence or are relevant to the work reported in this paper.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Karobari, M.I., Iqbal, A., Batul, R. et al. Exploring age and gender variations in root canal morphology of maxillary premolars in Saudi sub population: a cross-sectional CBCT study. BMC Oral Health 24 , 543 (2024). https://doi.org/10.1186/s12903-024-04310-w
Download citation
Received : 18 November 2023
Accepted : 29 April 2024
Published : 09 May 2024
DOI : https://doi.org/10.1186/s12903-024-04310-w
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Cone beam computed tomography
- Dental anatomy
- Dental diagnostic imaging
- Dental pulp
- Endodontics
BMC Oral Health
ISSN: 1472-6831
- Submission enquiries: [email protected]
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
Examples of literature reviews. Step 1 - Search for relevant literature. Step 2 - Evaluate and select sources. Step 3 - Identify themes, debates, and gaps. Step 4 - Outline your literature review's structure. Step 5 - Write your literature review.
As mentioned previously, there are a number of existing guidelines for literature reviews. Depending on the methodology needed to achieve the purpose of the review, all types can be helpful and appropriate to reach a specific goal (for examples, please see Table 1).These approaches can be qualitative, quantitative, or have a mixed design depending on the phase of the review.
In this chapter, we have discussed ways that the literature review represents a data collection tool, a method, a mixed research study, and, most of all, a methodology. Further, because oftentimes a methodology can be an abstract process, a methodology needs some type of mechanism, or process, to bring it to fruition.
A literature review is a critical analysis and synthesis of existing research on a particular topic. It provides an overview of the current state of knowledge, identifies gaps, and highlights key findings in the literature. 1 The purpose of a literature review is to situate your own research within the context of existing scholarship ...
1. Outline and identify the purpose of a literature review. As a first step on how to write a literature review, you must know what the research question or topic is and what shape you want your literature review to take. Ensure you understand the research topic inside out, or else seek clarifications.
A literature review is defined as "a critical analysis of a segment of a published body of knowledge through summary, classification, and comparison of prior research studies, reviews of literature, and theoretical articles." (The Writing Center University of Winconsin-Madison 2022) A literature review is an integrated analysis, not just a summary of scholarly work on a specific topic.
9.3. Types of Review Articles and Brief Illustrations. EHealth researchers have at their disposal a number of approaches and methods for making sense out of existing literature, all with the purpose of casting current research findings into historical contexts or explaining contradictions that might exist among a set of primary research studies conducted on a particular topic.
A literature review is a survey of scholarly sources on a specific topic. It provides an overview of current knowledge, allowing you to identify relevant theories, methods, and gaps in the existing research. There are five key steps to writing a literature review: Search for relevant literature. Evaluate sources. Identify themes, debates and gaps.
Okay - with the why out the way, let's move on to the how. As mentioned above, writing your literature review is a process, which I'll break down into three steps: Finding the most suitable literature. Understanding, distilling and organising the literature. Planning and writing up your literature review chapter.
The literature review opening/introduction section; The theoretical framework (or foundation of theory) The empirical research; The research gap; The closing section; We then progress to the sample literature review (from an A-grade Master's-level dissertation) to show how these concepts are applied in the literature review chapter. You can ...
begin by clearing up some misconceptions about what a literature review is and what it is not. Then, I will break the process down into a series of simple steps, looking at examples along the way. In the end, I hope you will have a simple, practical strategy to write an effective literature review.
This article is organized as follows: The next section presents the methodology adopted by this research, followed by a section that discusses the typology of literature reviews and provides empirical examples; the subsequent section summarizes the process of literature review; and the last section concludes the paper with suggestions on how to improve the quality and rigor of literature ...
Introduction Researchers and practitioners rely on literature reviews to synthesize large bodies of knowledge. Many types of literature reviews have been developed, each targeting a specific purpose. However, these syntheses are hampered if the review type's paradigmatic roots, methods, and markers of rigor are only vaguely understood. One literature review type whose methodology has yet to ...
Demonstrate your knowledge of the research topic. Identify the gaps in the literature and show how your research links to these. Provide the foundation for your conceptual framework (if you have one) Inform your own methodology and research design. To achieve this, your literature review needs a well-thought-out structure.
Literature Reviews that are organized methodologically consist of paragraphs/sections that are based on the methods used in the literature found.This approach is most appropriate when you are using new methods on a research question that has already been explored.Since literature review structures are not mutually exclusive, you can organize the use of these methods in chronological order.
A literature review is a document or section of a document that collects key sources on a topic and discusses those sources in conversation with each other (also called synthesis ). The lit review is an important genre in many disciplines, not just literature (i.e., the study of works of literature such as novels and plays).
Literature reviews are in great demand in most scientific fields. Their need stems from the ever-increasing output of scientific publications .For example, compared to 1991, in 2008 three, eight, and forty times more papers were indexed in Web of Science on malaria, obesity, and biodiversity, respectively .Given such mountains of papers, scientists cannot be expected to examine in detail every ...
Literature reviews allow scientists to argue that they are expanding current. expertise - improving on what already exists and filling the gaps that remain. This paper demonstrates the literatu ...
Steps for Conducting a Lit Review; Finding "The Literature" Organizing/Writing; APA Style This link opens in a new window; Chicago: Notes Bibliography This link opens in a new window; MLA Style This link opens in a new window; Sample Literature Reviews. Sample Lit Reviews from Communication Arts; Have an exemplary literature review? Get Help!
This. paper discusses literature review as a methodology for conducting research and o ffers an overview of different. types of reviews, as well as some guidelines to how to both conduct and ...
A literature review is an integrated analysis-- not just a summary-- of scholarly writings and other relevant evidence related directly to your research question.That is, it represents a synthesis of the evidence that provides background information on your topic and shows a association between the evidence and your research question.
Keep it brief. The methods section should be succinct but include all the noteworthy information. This can be a difficult balance to achieve. A useful strategy is to aim for a brief description that signposts the reader to a separate section or sections of supporting information. This could include datasets, a flowchart to show what happened to ...
Scoping reviews and evidence maps are forms of evidence synthesis that aim to map the available literature on a topic and are well-suited to visual presentation of results. A range of data visualisation methods and interactive data visualisation tools exist that may make scoping reviews more useful to knowledge users. The aim of this study was to explore the use of data visualisation in a ...
This review aimed to synthesise existing literature on the efficacy of personalised or precision nutrition (PPN) interventions, including medical nutrition therapy (MNT), in improving outcomes related to glycaemic control (HbA1c, post-prandial glucose [PPG], and fasting blood glucose), anthropometry (weight, BMI, and waist circumference [WC]), blood lipids, blood pressure (BP), and dietary ...
First, the methodology of this literature-review-based paper is clarified. Second, a review of empirical findings of political, social, and economic macro contextual factors and micro firm-level characteristics that influence CSRD is presented. ... Using a sample of 220 reports of Australian and French firms published in 2009, they examined CSR ...
Section "Literature Review and Research Hypotheses" examines the links among the variables being studied and the proposed model. Material and methods are presented in Section "Material and Methods." ... In contrast to these examples, literature has also provided evidence of the positive effect that perceived risk may have on the ...
The proposed approaches have been validated on both synthetic and real data, and compared against other methods from the literature. It results that neighborhood analysis allows to outperform competitors, and when it is combined with collaborative filtering the prediction accuracy further improves, scoring a value of AUC equal to 0966.
Addressing the existing problem in the microbiological diagnosis of infections associated with implants and the current debate about the real power of precision of sonicated fluid culture (SFC), the objective of this review is to describe the methodology and analyze and compare the results obtained in current studies on the subject. Furthermore, the present study also discusses and suggests ...
Background In complex teeth like maxillary premolars, endodontic treatment success depends on a complete comprehension of root canal anatomy. The research on mandibular premolars' root canal anatomy has been extensive and well-documented in existing literature. However, there appears to be a notable gap in available data concerning the root canal anatomy of maxillary premolars. This study ...