- Skip to main content
- Skip to primary sidebar
- Skip to footer
- QuestionPro

- Solutions Industries Gaming Automotive Sports and events Education Government Travel & Hospitality Financial Services Healthcare Cannabis Technology Use Case NPS+ Communities Audience Contactless surveys Mobile LivePolls Member Experience GDPR Positive People Science 360 Feedback Surveys
- Resources Blog eBooks Survey Templates Case Studies Training Help center
Home Market Research

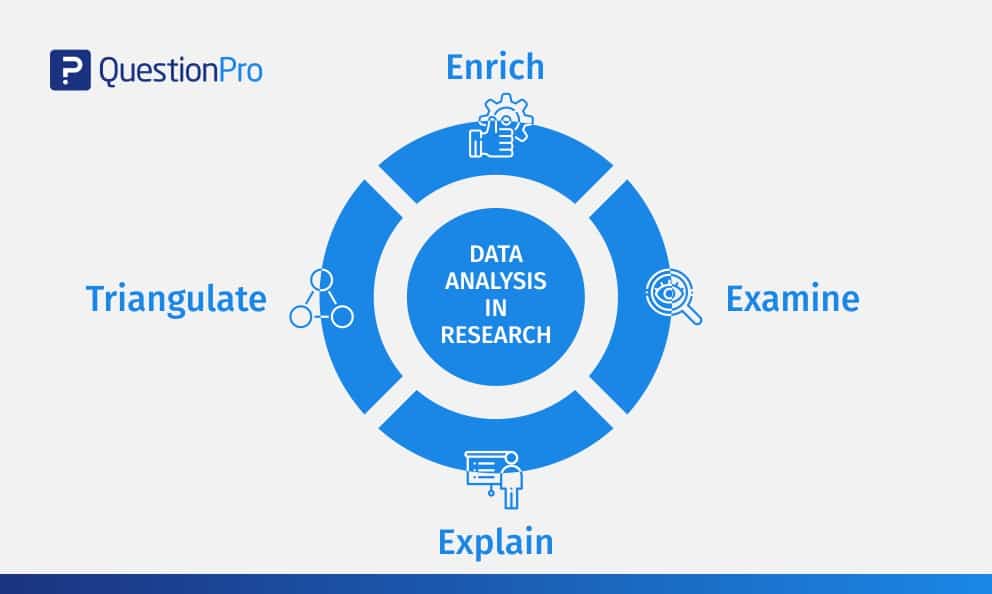
Data Analysis in Research: Types & Methods
Content Index
Why analyze data in research?
Types of data in research, finding patterns in the qualitative data, methods used for data analysis in qualitative research, preparing data for analysis, methods used for data analysis in quantitative research, considerations in research data analysis, what is data analysis in research.
Definition of research in data analysis: According to LeCompte and Schensul, research data analysis is a process used by researchers to reduce data to a story and interpret it to derive insights. The data analysis process helps reduce a large chunk of data into smaller fragments, which makes sense.
Three essential things occur during the data analysis process — the first is data organization . Summarization and categorization together contribute to becoming the second known method used for data reduction. It helps find patterns and themes in the data for easy identification and linking. The third and last way is data analysis – researchers do it in both top-down and bottom-up fashion.
LEARN ABOUT: Research Process Steps
On the other hand, Marshall and Rossman describe data analysis as a messy, ambiguous, and time-consuming but creative and fascinating process through which a mass of collected data is brought to order, structure and meaning.
We can say that “the data analysis and data interpretation is a process representing the application of deductive and inductive logic to the research and data analysis.”
Researchers rely heavily on data as they have a story to tell or research problems to solve. It starts with a question, and data is nothing but an answer to that question. But, what if there is no question to ask? Well! It is possible to explore data even without a problem – we call it ‘Data Mining’, which often reveals some interesting patterns within the data that are worth exploring.
Irrelevant to the type of data researchers explore, their mission and audiences’ vision guide them to find the patterns to shape the story they want to tell. One of the essential things expected from researchers while analyzing data is to stay open and remain unbiased toward unexpected patterns, expressions, and results. Remember, sometimes, data analysis tells the most unforeseen yet exciting stories that were not expected when initiating data analysis. Therefore, rely on the data you have at hand and enjoy the journey of exploratory research.
Create a Free Account
Every kind of data has a rare quality of describing things after assigning a specific value to it. For analysis, you need to organize these values, processed and presented in a given context, to make it useful. Data can be in different forms; here are the primary data types.
- Qualitative data: When the data presented has words and descriptions, then we call it qualitative data . Although you can observe this data, it is subjective and harder to analyze data in research, especially for comparison. Example: Quality data represents everything describing taste, experience, texture, or an opinion that is considered quality data. This type of data is usually collected through focus groups, personal qualitative interviews , qualitative observation or using open-ended questions in surveys.
- Quantitative data: Any data expressed in numbers of numerical figures are called quantitative data . This type of data can be distinguished into categories, grouped, measured, calculated, or ranked. Example: questions such as age, rank, cost, length, weight, scores, etc. everything comes under this type of data. You can present such data in graphical format, charts, or apply statistical analysis methods to this data. The (Outcomes Measurement Systems) OMS questionnaires in surveys are a significant source of collecting numeric data.
- Categorical data: It is data presented in groups. However, an item included in the categorical data cannot belong to more than one group. Example: A person responding to a survey by telling his living style, marital status, smoking habit, or drinking habit comes under the categorical data. A chi-square test is a standard method used to analyze this data.
Learn More : Examples of Qualitative Data in Education
Data analysis in qualitative research
Data analysis and qualitative data research work a little differently from the numerical data as the quality data is made up of words, descriptions, images, objects, and sometimes symbols. Getting insight from such complicated information is a complicated process. Hence it is typically used for exploratory research and data analysis .
Although there are several ways to find patterns in the textual information, a word-based method is the most relied and widely used global technique for research and data analysis. Notably, the data analysis process in qualitative research is manual. Here the researchers usually read the available data and find repetitive or commonly used words.
For example, while studying data collected from African countries to understand the most pressing issues people face, researchers might find “food” and “hunger” are the most commonly used words and will highlight them for further analysis.
LEARN ABOUT: Level of Analysis
The keyword context is another widely used word-based technique. In this method, the researcher tries to understand the concept by analyzing the context in which the participants use a particular keyword.
For example , researchers conducting research and data analysis for studying the concept of ‘diabetes’ amongst respondents might analyze the context of when and how the respondent has used or referred to the word ‘diabetes.’
The scrutiny-based technique is also one of the highly recommended text analysis methods used to identify a quality data pattern. Compare and contrast is the widely used method under this technique to differentiate how a specific text is similar or different from each other.
For example: To find out the “importance of resident doctor in a company,” the collected data is divided into people who think it is necessary to hire a resident doctor and those who think it is unnecessary. Compare and contrast is the best method that can be used to analyze the polls having single-answer questions types .
Metaphors can be used to reduce the data pile and find patterns in it so that it becomes easier to connect data with theory.
Variable Partitioning is another technique used to split variables so that researchers can find more coherent descriptions and explanations from the enormous data.
LEARN ABOUT: Qualitative Research Questions and Questionnaires
There are several techniques to analyze the data in qualitative research, but here are some commonly used methods,
- Content Analysis: It is widely accepted and the most frequently employed technique for data analysis in research methodology. It can be used to analyze the documented information from text, images, and sometimes from the physical items. It depends on the research questions to predict when and where to use this method.
- Narrative Analysis: This method is used to analyze content gathered from various sources such as personal interviews, field observation, and surveys . The majority of times, stories, or opinions shared by people are focused on finding answers to the research questions.
- Discourse Analysis: Similar to narrative analysis, discourse analysis is used to analyze the interactions with people. Nevertheless, this particular method considers the social context under which or within which the communication between the researcher and respondent takes place. In addition to that, discourse analysis also focuses on the lifestyle and day-to-day environment while deriving any conclusion.
- Grounded Theory: When you want to explain why a particular phenomenon happened, then using grounded theory for analyzing quality data is the best resort. Grounded theory is applied to study data about the host of similar cases occurring in different settings. When researchers are using this method, they might alter explanations or produce new ones until they arrive at some conclusion.
LEARN ABOUT: 12 Best Tools for Researchers
Data analysis in quantitative research
The first stage in research and data analysis is to make it for the analysis so that the nominal data can be converted into something meaningful. Data preparation consists of the below phases.
Phase I: Data Validation
Data validation is done to understand if the collected data sample is per the pre-set standards, or it is a biased data sample again divided into four different stages
- Fraud: To ensure an actual human being records each response to the survey or the questionnaire
- Screening: To make sure each participant or respondent is selected or chosen in compliance with the research criteria
- Procedure: To ensure ethical standards were maintained while collecting the data sample
- Completeness: To ensure that the respondent has answered all the questions in an online survey. Else, the interviewer had asked all the questions devised in the questionnaire.
Phase II: Data Editing
More often, an extensive research data sample comes loaded with errors. Respondents sometimes fill in some fields incorrectly or sometimes skip them accidentally. Data editing is a process wherein the researchers have to confirm that the provided data is free of such errors. They need to conduct necessary checks and outlier checks to edit the raw edit and make it ready for analysis.
Phase III: Data Coding
Out of all three, this is the most critical phase of data preparation associated with grouping and assigning values to the survey responses . If a survey is completed with a 1000 sample size, the researcher will create an age bracket to distinguish the respondents based on their age. Thus, it becomes easier to analyze small data buckets rather than deal with the massive data pile.
LEARN ABOUT: Steps in Qualitative Research
After the data is prepared for analysis, researchers are open to using different research and data analysis methods to derive meaningful insights. For sure, statistical analysis plans are the most favored to analyze numerical data. In statistical analysis, distinguishing between categorical data and numerical data is essential, as categorical data involves distinct categories or labels, while numerical data consists of measurable quantities. The method is again classified into two groups. First, ‘Descriptive Statistics’ used to describe data. Second, ‘Inferential statistics’ that helps in comparing the data .
Descriptive statistics
This method is used to describe the basic features of versatile types of data in research. It presents the data in such a meaningful way that pattern in the data starts making sense. Nevertheless, the descriptive analysis does not go beyond making conclusions. The conclusions are again based on the hypothesis researchers have formulated so far. Here are a few major types of descriptive analysis methods.
Measures of Frequency
- Count, Percent, Frequency
- It is used to denote home often a particular event occurs.
- Researchers use it when they want to showcase how often a response is given.
Measures of Central Tendency
- Mean, Median, Mode
- The method is widely used to demonstrate distribution by various points.
- Researchers use this method when they want to showcase the most commonly or averagely indicated response.
Measures of Dispersion or Variation
- Range, Variance, Standard deviation
- Here the field equals high/low points.
- Variance standard deviation = difference between the observed score and mean
- It is used to identify the spread of scores by stating intervals.
- Researchers use this method to showcase data spread out. It helps them identify the depth until which the data is spread out that it directly affects the mean.
Measures of Position
- Percentile ranks, Quartile ranks
- It relies on standardized scores helping researchers to identify the relationship between different scores.
- It is often used when researchers want to compare scores with the average count.
For quantitative research use of descriptive analysis often give absolute numbers, but the in-depth analysis is never sufficient to demonstrate the rationale behind those numbers. Nevertheless, it is necessary to think of the best method for research and data analysis suiting your survey questionnaire and what story researchers want to tell. For example, the mean is the best way to demonstrate the students’ average scores in schools. It is better to rely on the descriptive statistics when the researchers intend to keep the research or outcome limited to the provided sample without generalizing it. For example, when you want to compare average voting done in two different cities, differential statistics are enough.
Descriptive analysis is also called a ‘univariate analysis’ since it is commonly used to analyze a single variable.
Inferential statistics
Inferential statistics are used to make predictions about a larger population after research and data analysis of the representing population’s collected sample. For example, you can ask some odd 100 audiences at a movie theater if they like the movie they are watching. Researchers then use inferential statistics on the collected sample to reason that about 80-90% of people like the movie.
Here are two significant areas of inferential statistics.
- Estimating parameters: It takes statistics from the sample research data and demonstrates something about the population parameter.
- Hypothesis test: I t’s about sampling research data to answer the survey research questions. For example, researchers might be interested to understand if the new shade of lipstick recently launched is good or not, or if the multivitamin capsules help children to perform better at games.
These are sophisticated analysis methods used to showcase the relationship between different variables instead of describing a single variable. It is often used when researchers want something beyond absolute numbers to understand the relationship between variables.
Here are some of the commonly used methods for data analysis in research.
- Correlation: When researchers are not conducting experimental research or quasi-experimental research wherein the researchers are interested to understand the relationship between two or more variables, they opt for correlational research methods.
- Cross-tabulation: Also called contingency tables, cross-tabulation is used to analyze the relationship between multiple variables. Suppose provided data has age and gender categories presented in rows and columns. A two-dimensional cross-tabulation helps for seamless data analysis and research by showing the number of males and females in each age category.
- Regression analysis: For understanding the strong relationship between two variables, researchers do not look beyond the primary and commonly used regression analysis method, which is also a type of predictive analysis used. In this method, you have an essential factor called the dependent variable. You also have multiple independent variables in regression analysis. You undertake efforts to find out the impact of independent variables on the dependent variable. The values of both independent and dependent variables are assumed as being ascertained in an error-free random manner.
- Frequency tables: The statistical procedure is used for testing the degree to which two or more vary or differ in an experiment. A considerable degree of variation means research findings were significant. In many contexts, ANOVA testing and variance analysis are similar.
- Analysis of variance: The statistical procedure is used for testing the degree to which two or more vary or differ in an experiment. A considerable degree of variation means research findings were significant. In many contexts, ANOVA testing and variance analysis are similar.
- Researchers must have the necessary research skills to analyze and manipulation the data , Getting trained to demonstrate a high standard of research practice. Ideally, researchers must possess more than a basic understanding of the rationale of selecting one statistical method over the other to obtain better data insights.
- Usually, research and data analytics projects differ by scientific discipline; therefore, getting statistical advice at the beginning of analysis helps design a survey questionnaire, select data collection methods , and choose samples.
LEARN ABOUT: Best Data Collection Tools
- The primary aim of data research and analysis is to derive ultimate insights that are unbiased. Any mistake in or keeping a biased mind to collect data, selecting an analysis method, or choosing audience sample il to draw a biased inference.
- Irrelevant to the sophistication used in research data and analysis is enough to rectify the poorly defined objective outcome measurements. It does not matter if the design is at fault or intentions are not clear, but lack of clarity might mislead readers, so avoid the practice.
- The motive behind data analysis in research is to present accurate and reliable data. As far as possible, avoid statistical errors, and find a way to deal with everyday challenges like outliers, missing data, data altering, data mining , or developing graphical representation.
LEARN MORE: Descriptive Research vs Correlational Research The sheer amount of data generated daily is frightening. Especially when data analysis has taken center stage. in 2018. In last year, the total data supply amounted to 2.8 trillion gigabytes. Hence, it is clear that the enterprises willing to survive in the hypercompetitive world must possess an excellent capability to analyze complex research data, derive actionable insights, and adapt to the new market needs.
LEARN ABOUT: Average Order Value
QuestionPro is an online survey platform that empowers organizations in data analysis and research and provides them a medium to collect data by creating appealing surveys.
MORE LIKE THIS

Why Multilingual 360 Feedback Surveys Provide Better Insights
Jun 3, 2024

Raked Weighting: A Key Tool for Accurate Survey Results
May 31, 2024

Top 8 Data Trends to Understand the Future of Data
May 30, 2024

Top 12 Interactive Presentation Software to Engage Your User
May 29, 2024
Other categories
- Academic Research
- Artificial Intelligence
- Assessments
- Brand Awareness
- Case Studies
- Communities
- Consumer Insights
- Customer effort score
- Customer Engagement
- Customer Experience
- Customer Loyalty
- Customer Research
- Customer Satisfaction
- Employee Benefits
- Employee Engagement
- Employee Retention
- Friday Five
- General Data Protection Regulation
- Insights Hub
- Life@QuestionPro
- Market Research
- Mobile diaries
- Mobile Surveys
- New Features
- Online Communities
- Question Types
- Questionnaire
- QuestionPro Products
- Release Notes
- Research Tools and Apps
- Revenue at Risk
- Survey Templates
- Training Tips
- Uncategorized
- Video Learning Series
- What’s Coming Up
- Workforce Intelligence
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, automatically generate references for free.
- Knowledge Base
- Methodology
Research Methods | Definition, Types, Examples
Research methods are specific procedures for collecting and analysing data. Developing your research methods is an integral part of your research design . When planning your methods, there are two key decisions you will make.
First, decide how you will collect data . Your methods depend on what type of data you need to answer your research question :
- Qualitative vs quantitative : Will your data take the form of words or numbers?
- Primary vs secondary : Will you collect original data yourself, or will you use data that have already been collected by someone else?
- Descriptive vs experimental : Will you take measurements of something as it is, or will you perform an experiment?
Second, decide how you will analyse the data .
- For quantitative data, you can use statistical analysis methods to test relationships between variables.
- For qualitative data, you can use methods such as thematic analysis to interpret patterns and meanings in the data.
Table of contents
Methods for collecting data, examples of data collection methods, methods for analysing data, examples of data analysis methods, frequently asked questions about methodology.
Data are the information that you collect for the purposes of answering your research question . The type of data you need depends on the aims of your research.
Qualitative vs quantitative data
Your choice of qualitative or quantitative data collection depends on the type of knowledge you want to develop.
For questions about ideas, experiences and meanings, or to study something that can’t be described numerically, collect qualitative data .
If you want to develop a more mechanistic understanding of a topic, or your research involves hypothesis testing , collect quantitative data .
You can also take a mixed methods approach, where you use both qualitative and quantitative research methods.
Primary vs secondary data
Primary data are any original information that you collect for the purposes of answering your research question (e.g. through surveys , observations and experiments ). Secondary data are information that has already been collected by other researchers (e.g. in a government census or previous scientific studies).
If you are exploring a novel research question, you’ll probably need to collect primary data. But if you want to synthesise existing knowledge, analyse historical trends, or identify patterns on a large scale, secondary data might be a better choice.
Descriptive vs experimental data
In descriptive research , you collect data about your study subject without intervening. The validity of your research will depend on your sampling method .
In experimental research , you systematically intervene in a process and measure the outcome. The validity of your research will depend on your experimental design .
To conduct an experiment, you need to be able to vary your independent variable , precisely measure your dependent variable, and control for confounding variables . If it’s practically and ethically possible, this method is the best choice for answering questions about cause and effect.
Prevent plagiarism, run a free check.
Your data analysis methods will depend on the type of data you collect and how you prepare them for analysis.
Data can often be analysed both quantitatively and qualitatively. For example, survey responses could be analysed qualitatively by studying the meanings of responses or quantitatively by studying the frequencies of responses.
Qualitative analysis methods
Qualitative analysis is used to understand words, ideas, and experiences. You can use it to interpret data that were collected:
- From open-ended survey and interview questions, literature reviews, case studies, and other sources that use text rather than numbers.
- Using non-probability sampling methods .
Qualitative analysis tends to be quite flexible and relies on the researcher’s judgement, so you have to reflect carefully on your choices and assumptions.
Quantitative analysis methods
Quantitative analysis uses numbers and statistics to understand frequencies, averages and correlations (in descriptive studies) or cause-and-effect relationships (in experiments).
You can use quantitative analysis to interpret data that were collected either:
- During an experiment.
- Using probability sampling methods .
Because the data are collected and analysed in a statistically valid way, the results of quantitative analysis can be easily standardised and shared among researchers.
Quantitative research deals with numbers and statistics, while qualitative research deals with words and meanings.
Quantitative methods allow you to test a hypothesis by systematically collecting and analysing data, while qualitative methods allow you to explore ideas and experiences in depth.
In mixed methods research , you use both qualitative and quantitative data collection and analysis methods to answer your research question .
A sample is a subset of individuals from a larger population. Sampling means selecting the group that you will actually collect data from in your research.
For example, if you are researching the opinions of students in your university, you could survey a sample of 100 students.
Statistical sampling allows you to test a hypothesis about the characteristics of a population. There are various sampling methods you can use to ensure that your sample is representative of the population as a whole.
The research methods you use depend on the type of data you need to answer your research question .
- If you want to measure something or test a hypothesis , use quantitative methods . If you want to explore ideas, thoughts, and meanings, use qualitative methods .
- If you want to analyse a large amount of readily available data, use secondary data. If you want data specific to your purposes with control over how they are generated, collect primary data.
- If you want to establish cause-and-effect relationships between variables , use experimental methods. If you want to understand the characteristics of a research subject, use descriptive methods.
Methodology refers to the overarching strategy and rationale of your research project . It involves studying the methods used in your field and the theories or principles behind them, in order to develop an approach that matches your objectives.
Methods are the specific tools and procedures you use to collect and analyse data (e.g. experiments, surveys , and statistical tests ).
In shorter scientific papers, where the aim is to report the findings of a specific study, you might simply describe what you did in a methods section .
In a longer or more complex research project, such as a thesis or dissertation , you will probably include a methodology section , where you explain your approach to answering the research questions and cite relevant sources to support your choice of methods.
Is this article helpful?
More interesting articles.
- A Quick Guide to Experimental Design | 5 Steps & Examples
- Between-Subjects Design | Examples, Pros & Cons
- Case Study | Definition, Examples & Methods
- Cluster Sampling | A Simple Step-by-Step Guide with Examples
- Confounding Variables | Definition, Examples & Controls
- Construct Validity | Definition, Types, & Examples
- Content Analysis | A Step-by-Step Guide with Examples
- Control Groups and Treatment Groups | Uses & Examples
- Controlled Experiments | Methods & Examples of Control
- Correlation vs Causation | Differences, Designs & Examples
- Correlational Research | Guide, Design & Examples
- Critical Discourse Analysis | Definition, Guide & Examples
- Cross-Sectional Study | Definitions, Uses & Examples
- Data Cleaning | A Guide with Examples & Steps
- Data Collection Methods | Step-by-Step Guide & Examples
- Descriptive Research Design | Definition, Methods & Examples
- Doing Survey Research | A Step-by-Step Guide & Examples
- Ethical Considerations in Research | Types & Examples
- Explanatory Research | Definition, Guide, & Examples
- Explanatory vs Response Variables | Definitions & Examples
- Exploratory Research | Definition, Guide, & Examples
- External Validity | Types, Threats & Examples
- Extraneous Variables | Examples, Types, Controls
- Face Validity | Guide with Definition & Examples
- How to Do Thematic Analysis | Guide & Examples
- How to Write a Strong Hypothesis | Guide & Examples
- Inclusion and Exclusion Criteria | Examples & Definition
- Independent vs Dependent Variables | Definition & Examples
- Inductive Reasoning | Types, Examples, Explanation
- Inductive vs Deductive Research Approach (with Examples)
- Internal Validity | Definition, Threats & Examples
- Internal vs External Validity | Understanding Differences & Examples
- Longitudinal Study | Definition, Approaches & Examples
- Mediator vs Moderator Variables | Differences & Examples
- Mixed Methods Research | Definition, Guide, & Examples
- Multistage Sampling | An Introductory Guide with Examples
- Naturalistic Observation | Definition, Guide & Examples
- Operationalisation | A Guide with Examples, Pros & Cons
- Population vs Sample | Definitions, Differences & Examples
- Primary Research | Definition, Types, & Examples
- Qualitative vs Quantitative Research | Examples & Methods
- Quasi-Experimental Design | Definition, Types & Examples
- Questionnaire Design | Methods, Question Types & Examples
- Random Assignment in Experiments | Introduction & Examples
- Reliability vs Validity in Research | Differences, Types & Examples
- Reproducibility vs Replicability | Difference & Examples
- Research Design | Step-by-Step Guide with Examples
- Sampling Methods | Types, Techniques, & Examples
- Semi-Structured Interview | Definition, Guide & Examples
- Simple Random Sampling | Definition, Steps & Examples
- Stratified Sampling | A Step-by-Step Guide with Examples
- Structured Interview | Definition, Guide & Examples
- Systematic Review | Definition, Examples & Guide
- Systematic Sampling | A Step-by-Step Guide with Examples
- Textual Analysis | Guide, 3 Approaches & Examples
- The 4 Types of Reliability in Research | Definitions & Examples
- The 4 Types of Validity | Types, Definitions & Examples
- Transcribing an Interview | 5 Steps & Transcription Software
- Triangulation in Research | Guide, Types, Examples
- Types of Interviews in Research | Guide & Examples
- Types of Research Designs Compared | Examples
- Types of Variables in Research | Definitions & Examples
- Unstructured Interview | Definition, Guide & Examples
- What Are Control Variables | Definition & Examples
- What Is a Case-Control Study? | Definition & Examples
- What Is a Cohort Study? | Definition & Examples
- What Is a Conceptual Framework? | Tips & Examples
- What Is a Double-Barrelled Question?
- What Is a Double-Blind Study? | Introduction & Examples
- What Is a Focus Group? | Step-by-Step Guide & Examples
- What Is a Likert Scale? | Guide & Examples
- What is a Literature Review? | Guide, Template, & Examples
- What Is a Prospective Cohort Study? | Definition & Examples
- What Is a Retrospective Cohort Study? | Definition & Examples
- What Is Action Research? | Definition & Examples
- What Is an Observational Study? | Guide & Examples
- What Is Concurrent Validity? | Definition & Examples
- What Is Content Validity? | Definition & Examples
- What Is Convenience Sampling? | Definition & Examples
- What Is Convergent Validity? | Definition & Examples
- What Is Criterion Validity? | Definition & Examples
- What Is Deductive Reasoning? | Explanation & Examples
- What Is Discriminant Validity? | Definition & Example
- What Is Ecological Validity? | Definition & Examples
- What Is Ethnography? | Meaning, Guide & Examples
- What Is Non-Probability Sampling? | Types & Examples
- What Is Participant Observation? | Definition & Examples
- What Is Peer Review? | Types & Examples
- What Is Predictive Validity? | Examples & Definition
- What Is Probability Sampling? | Types & Examples
- What Is Purposive Sampling? | Definition & Examples
- What Is Qualitative Observation? | Definition & Examples
- What Is Qualitative Research? | Methods & Examples
- What Is Quantitative Observation? | Definition & Examples
- What Is Quantitative Research? | Definition & Methods
- What Is Quota Sampling? | Definition & Examples
- What is Secondary Research? | Definition, Types, & Examples
- What Is Snowball Sampling? | Definition & Examples
- Within-Subjects Design | Explanation, Approaches, Examples
Your Modern Business Guide To Data Analysis Methods And Techniques
Table of Contents
1) What Is Data Analysis?
2) Why Is Data Analysis Important?
3) What Is The Data Analysis Process?
4) Types Of Data Analysis Methods
5) Top Data Analysis Techniques To Apply
6) Quality Criteria For Data Analysis
7) Data Analysis Limitations & Barriers
8) Data Analysis Skills
9) Data Analysis In The Big Data Environment
In our data-rich age, understanding how to analyze and extract true meaning from our business’s digital insights is one of the primary drivers of success.
Despite the colossal volume of data we create every day, a mere 0.5% is actually analyzed and used for data discovery , improvement, and intelligence. While that may not seem like much, considering the amount of digital information we have at our fingertips, half a percent still accounts for a vast amount of data.
With so much data and so little time, knowing how to collect, curate, organize, and make sense of all of this potentially business-boosting information can be a minefield – but online data analysis is the solution.
In science, data analysis uses a more complex approach with advanced techniques to explore and experiment with data. On the other hand, in a business context, data is used to make data-driven decisions that will enable the company to improve its overall performance. In this post, we will cover the analysis of data from an organizational point of view while still going through the scientific and statistical foundations that are fundamental to understanding the basics of data analysis.
To put all of that into perspective, we will answer a host of important analytical questions, explore analytical methods and techniques, while demonstrating how to perform analysis in the real world with a 17-step blueprint for success.
What Is Data Analysis?
Data analysis is the process of collecting, modeling, and analyzing data using various statistical and logical methods and techniques. Businesses rely on analytics processes and tools to extract insights that support strategic and operational decision-making.
All these various methods are largely based on two core areas: quantitative and qualitative research.
To explain the key differences between qualitative and quantitative research, here’s a video for your viewing pleasure:
Gaining a better understanding of different techniques and methods in quantitative research as well as qualitative insights will give your analyzing efforts a more clearly defined direction, so it’s worth taking the time to allow this particular knowledge to sink in. Additionally, you will be able to create a comprehensive analytical report that will skyrocket your analysis.
Apart from qualitative and quantitative categories, there are also other types of data that you should be aware of before dividing into complex data analysis processes. These categories include:
- Big data: Refers to massive data sets that need to be analyzed using advanced software to reveal patterns and trends. It is considered to be one of the best analytical assets as it provides larger volumes of data at a faster rate.
- Metadata: Putting it simply, metadata is data that provides insights about other data. It summarizes key information about specific data that makes it easier to find and reuse for later purposes.
- Real time data: As its name suggests, real time data is presented as soon as it is acquired. From an organizational perspective, this is the most valuable data as it can help you make important decisions based on the latest developments. Our guide on real time analytics will tell you more about the topic.
- Machine data: This is more complex data that is generated solely by a machine such as phones, computers, or even websites and embedded systems, without previous human interaction.
Why Is Data Analysis Important?
Before we go into detail about the categories of analysis along with its methods and techniques, you must understand the potential that analyzing data can bring to your organization.
- Informed decision-making : From a management perspective, you can benefit from analyzing your data as it helps you make decisions based on facts and not simple intuition. For instance, you can understand where to invest your capital, detect growth opportunities, predict your income, or tackle uncommon situations before they become problems. Through this, you can extract relevant insights from all areas in your organization, and with the help of dashboard software , present the data in a professional and interactive way to different stakeholders.
- Reduce costs : Another great benefit is to reduce costs. With the help of advanced technologies such as predictive analytics, businesses can spot improvement opportunities, trends, and patterns in their data and plan their strategies accordingly. In time, this will help you save money and resources on implementing the wrong strategies. And not just that, by predicting different scenarios such as sales and demand you can also anticipate production and supply.
- Target customers better : Customers are arguably the most crucial element in any business. By using analytics to get a 360° vision of all aspects related to your customers, you can understand which channels they use to communicate with you, their demographics, interests, habits, purchasing behaviors, and more. In the long run, it will drive success to your marketing strategies, allow you to identify new potential customers, and avoid wasting resources on targeting the wrong people or sending the wrong message. You can also track customer satisfaction by analyzing your client’s reviews or your customer service department’s performance.
What Is The Data Analysis Process?
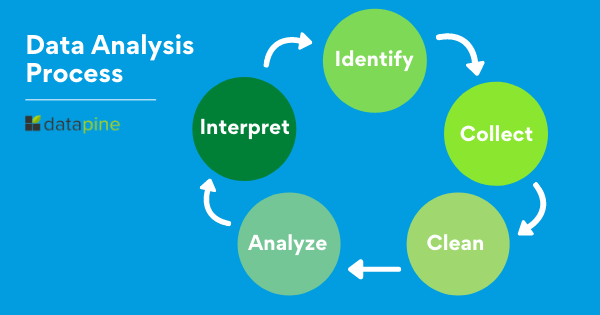
When we talk about analyzing data there is an order to follow in order to extract the needed conclusions. The analysis process consists of 5 key stages. We will cover each of them more in detail later in the post, but to start providing the needed context to understand what is coming next, here is a rundown of the 5 essential steps of data analysis.
- Identify: Before you get your hands dirty with data, you first need to identify why you need it in the first place. The identification is the stage in which you establish the questions you will need to answer. For example, what is the customer's perception of our brand? Or what type of packaging is more engaging to our potential customers? Once the questions are outlined you are ready for the next step.
- Collect: As its name suggests, this is the stage where you start collecting the needed data. Here, you define which sources of data you will use and how you will use them. The collection of data can come in different forms such as internal or external sources, surveys, interviews, questionnaires, and focus groups, among others. An important note here is that the way you collect the data will be different in a quantitative and qualitative scenario.
- Clean: Once you have the necessary data it is time to clean it and leave it ready for analysis. Not all the data you collect will be useful, when collecting big amounts of data in different formats it is very likely that you will find yourself with duplicate or badly formatted data. To avoid this, before you start working with your data you need to make sure to erase any white spaces, duplicate records, or formatting errors. This way you avoid hurting your analysis with bad-quality data.
- Analyze : With the help of various techniques such as statistical analysis, regressions, neural networks, text analysis, and more, you can start analyzing and manipulating your data to extract relevant conclusions. At this stage, you find trends, correlations, variations, and patterns that can help you answer the questions you first thought of in the identify stage. Various technologies in the market assist researchers and average users with the management of their data. Some of them include business intelligence and visualization software, predictive analytics, and data mining, among others.
- Interpret: Last but not least you have one of the most important steps: it is time to interpret your results. This stage is where the researcher comes up with courses of action based on the findings. For example, here you would understand if your clients prefer packaging that is red or green, plastic or paper, etc. Additionally, at this stage, you can also find some limitations and work on them.
Now that you have a basic understanding of the key data analysis steps, let’s look at the top 17 essential methods.
17 Essential Types Of Data Analysis Methods
Before diving into the 17 essential types of methods, it is important that we go over really fast through the main analysis categories. Starting with the category of descriptive up to prescriptive analysis, the complexity and effort of data evaluation increases, but also the added value for the company.
a) Descriptive analysis - What happened.
The descriptive analysis method is the starting point for any analytic reflection, and it aims to answer the question of what happened? It does this by ordering, manipulating, and interpreting raw data from various sources to turn it into valuable insights for your organization.
Performing descriptive analysis is essential, as it enables us to present our insights in a meaningful way. Although it is relevant to mention that this analysis on its own will not allow you to predict future outcomes or tell you the answer to questions like why something happened, it will leave your data organized and ready to conduct further investigations.
b) Exploratory analysis - How to explore data relationships.
As its name suggests, the main aim of the exploratory analysis is to explore. Prior to it, there is still no notion of the relationship between the data and the variables. Once the data is investigated, exploratory analysis helps you to find connections and generate hypotheses and solutions for specific problems. A typical area of application for it is data mining.
c) Diagnostic analysis - Why it happened.
Diagnostic data analytics empowers analysts and executives by helping them gain a firm contextual understanding of why something happened. If you know why something happened as well as how it happened, you will be able to pinpoint the exact ways of tackling the issue or challenge.
Designed to provide direct and actionable answers to specific questions, this is one of the world’s most important methods in research, among its other key organizational functions such as retail analytics , e.g.
c) Predictive analysis - What will happen.
The predictive method allows you to look into the future to answer the question: what will happen? In order to do this, it uses the results of the previously mentioned descriptive, exploratory, and diagnostic analysis, in addition to machine learning (ML) and artificial intelligence (AI). Through this, you can uncover future trends, potential problems or inefficiencies, connections, and casualties in your data.
With predictive analysis, you can unfold and develop initiatives that will not only enhance your various operational processes but also help you gain an all-important edge over the competition. If you understand why a trend, pattern, or event happened through data, you will be able to develop an informed projection of how things may unfold in particular areas of the business.
e) Prescriptive analysis - How will it happen.
Another of the most effective types of analysis methods in research. Prescriptive data techniques cross over from predictive analysis in the way that it revolves around using patterns or trends to develop responsive, practical business strategies.
By drilling down into prescriptive analysis, you will play an active role in the data consumption process by taking well-arranged sets of visual data and using it as a powerful fix to emerging issues in a number of key areas, including marketing, sales, customer experience, HR, fulfillment, finance, logistics analytics , and others.
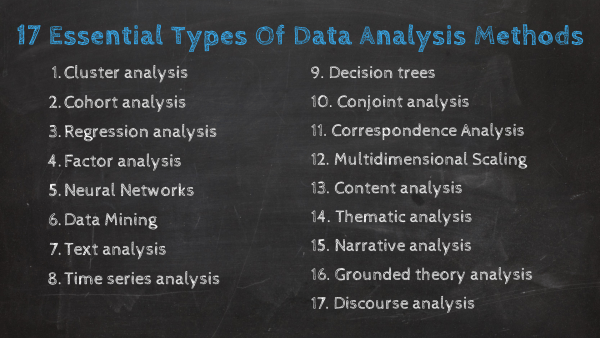
As mentioned at the beginning of the post, data analysis methods can be divided into two big categories: quantitative and qualitative. Each of these categories holds a powerful analytical value that changes depending on the scenario and type of data you are working with. Below, we will discuss 17 methods that are divided into qualitative and quantitative approaches.
Without further ado, here are the 17 essential types of data analysis methods with some use cases in the business world:
A. Quantitative Methods
To put it simply, quantitative analysis refers to all methods that use numerical data or data that can be turned into numbers (e.g. category variables like gender, age, etc.) to extract valuable insights. It is used to extract valuable conclusions about relationships, differences, and test hypotheses. Below we discuss some of the key quantitative methods.
1. Cluster analysis
The action of grouping a set of data elements in a way that said elements are more similar (in a particular sense) to each other than to those in other groups – hence the term ‘cluster.’ Since there is no target variable when clustering, the method is often used to find hidden patterns in the data. The approach is also used to provide additional context to a trend or dataset.
Let's look at it from an organizational perspective. In a perfect world, marketers would be able to analyze each customer separately and give them the best-personalized service, but let's face it, with a large customer base, it is timely impossible to do that. That's where clustering comes in. By grouping customers into clusters based on demographics, purchasing behaviors, monetary value, or any other factor that might be relevant for your company, you will be able to immediately optimize your efforts and give your customers the best experience based on their needs.
2. Cohort analysis
This type of data analysis approach uses historical data to examine and compare a determined segment of users' behavior, which can then be grouped with others with similar characteristics. By using this methodology, it's possible to gain a wealth of insight into consumer needs or a firm understanding of a broader target group.
Cohort analysis can be really useful for performing analysis in marketing as it will allow you to understand the impact of your campaigns on specific groups of customers. To exemplify, imagine you send an email campaign encouraging customers to sign up for your site. For this, you create two versions of the campaign with different designs, CTAs, and ad content. Later on, you can use cohort analysis to track the performance of the campaign for a longer period of time and understand which type of content is driving your customers to sign up, repurchase, or engage in other ways.
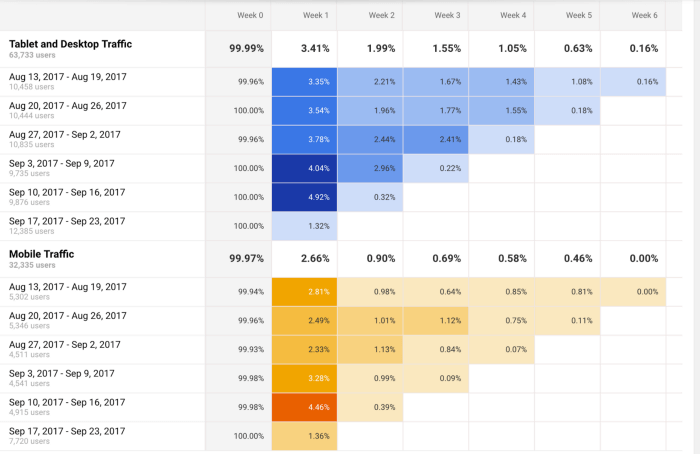
A useful tool to start performing cohort analysis method is Google Analytics. You can learn more about the benefits and limitations of using cohorts in GA in this useful guide . In the bottom image, you see an example of how you visualize a cohort in this tool. The segments (devices traffic) are divided into date cohorts (usage of devices) and then analyzed week by week to extract insights into performance.
3. Regression analysis
Regression uses historical data to understand how a dependent variable's value is affected when one (linear regression) or more independent variables (multiple regression) change or stay the same. By understanding each variable's relationship and how it developed in the past, you can anticipate possible outcomes and make better decisions in the future.
Let's bring it down with an example. Imagine you did a regression analysis of your sales in 2019 and discovered that variables like product quality, store design, customer service, marketing campaigns, and sales channels affected the overall result. Now you want to use regression to analyze which of these variables changed or if any new ones appeared during 2020. For example, you couldn’t sell as much in your physical store due to COVID lockdowns. Therefore, your sales could’ve either dropped in general or increased in your online channels. Through this, you can understand which independent variables affected the overall performance of your dependent variable, annual sales.
If you want to go deeper into this type of analysis, check out this article and learn more about how you can benefit from regression.
4. Neural networks
The neural network forms the basis for the intelligent algorithms of machine learning. It is a form of analytics that attempts, with minimal intervention, to understand how the human brain would generate insights and predict values. Neural networks learn from each and every data transaction, meaning that they evolve and advance over time.
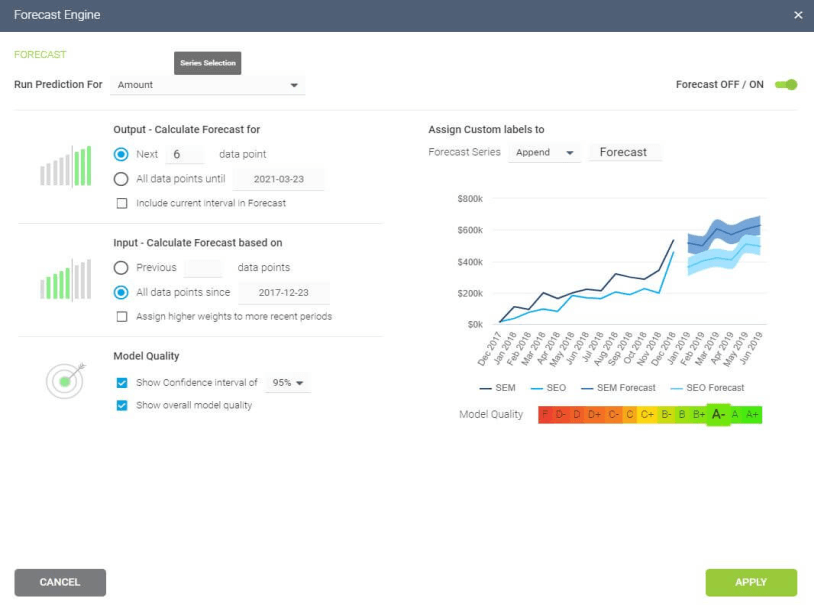
A typical area of application for neural networks is predictive analytics. There are BI reporting tools that have this feature implemented within them, such as the Predictive Analytics Tool from datapine. This tool enables users to quickly and easily generate all kinds of predictions. All you have to do is select the data to be processed based on your KPIs, and the software automatically calculates forecasts based on historical and current data. Thanks to its user-friendly interface, anyone in your organization can manage it; there’s no need to be an advanced scientist.
Here is an example of how you can use the predictive analysis tool from datapine:
**click to enlarge**
5. Factor analysis
The factor analysis also called “dimension reduction” is a type of data analysis used to describe variability among observed, correlated variables in terms of a potentially lower number of unobserved variables called factors. The aim here is to uncover independent latent variables, an ideal method for streamlining specific segments.
A good way to understand this data analysis method is a customer evaluation of a product. The initial assessment is based on different variables like color, shape, wearability, current trends, materials, comfort, the place where they bought the product, and frequency of usage. Like this, the list can be endless, depending on what you want to track. In this case, factor analysis comes into the picture by summarizing all of these variables into homogenous groups, for example, by grouping the variables color, materials, quality, and trends into a brother latent variable of design.
If you want to start analyzing data using factor analysis we recommend you take a look at this practical guide from UCLA.
6. Data mining
A method of data analysis that is the umbrella term for engineering metrics and insights for additional value, direction, and context. By using exploratory statistical evaluation, data mining aims to identify dependencies, relations, patterns, and trends to generate advanced knowledge. When considering how to analyze data, adopting a data mining mindset is essential to success - as such, it’s an area that is worth exploring in greater detail.
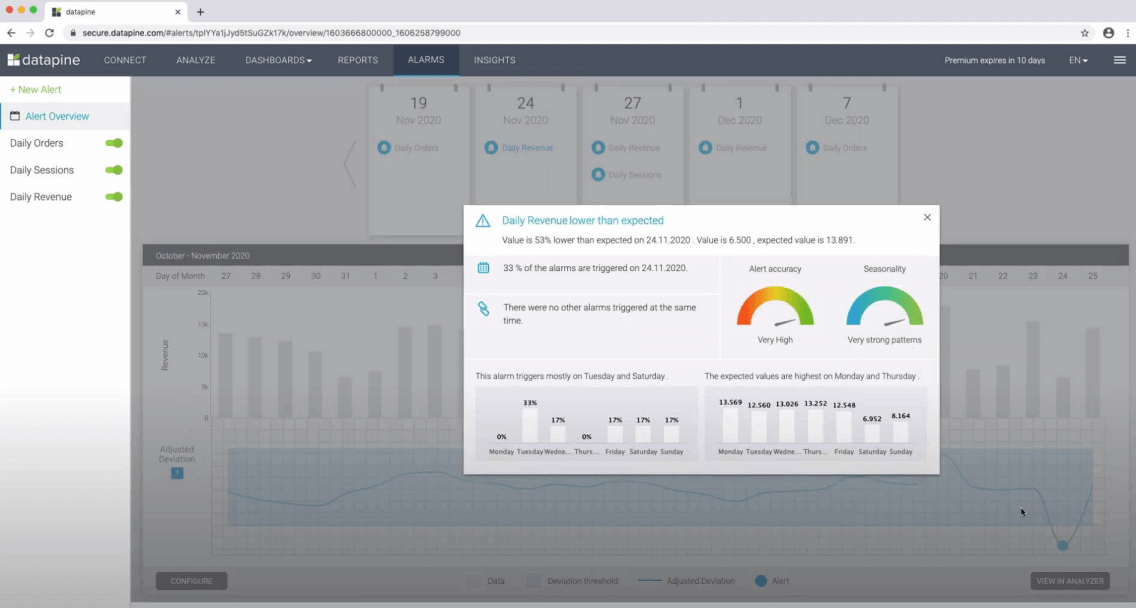
An excellent use case of data mining is datapine intelligent data alerts . With the help of artificial intelligence and machine learning, they provide automated signals based on particular commands or occurrences within a dataset. For example, if you’re monitoring supply chain KPIs , you could set an intelligent alarm to trigger when invalid or low-quality data appears. By doing so, you will be able to drill down deep into the issue and fix it swiftly and effectively.
In the following picture, you can see how the intelligent alarms from datapine work. By setting up ranges on daily orders, sessions, and revenues, the alarms will notify you if the goal was not completed or if it exceeded expectations.
7. Time series analysis
As its name suggests, time series analysis is used to analyze a set of data points collected over a specified period of time. Although analysts use this method to monitor the data points in a specific interval of time rather than just monitoring them intermittently, the time series analysis is not uniquely used for the purpose of collecting data over time. Instead, it allows researchers to understand if variables changed during the duration of the study, how the different variables are dependent, and how did it reach the end result.
In a business context, this method is used to understand the causes of different trends and patterns to extract valuable insights. Another way of using this method is with the help of time series forecasting. Powered by predictive technologies, businesses can analyze various data sets over a period of time and forecast different future events.
A great use case to put time series analysis into perspective is seasonality effects on sales. By using time series forecasting to analyze sales data of a specific product over time, you can understand if sales rise over a specific period of time (e.g. swimwear during summertime, or candy during Halloween). These insights allow you to predict demand and prepare production accordingly.
8. Decision Trees
The decision tree analysis aims to act as a support tool to make smart and strategic decisions. By visually displaying potential outcomes, consequences, and costs in a tree-like model, researchers and company users can easily evaluate all factors involved and choose the best course of action. Decision trees are helpful to analyze quantitative data and they allow for an improved decision-making process by helping you spot improvement opportunities, reduce costs, and enhance operational efficiency and production.
But how does a decision tree actually works? This method works like a flowchart that starts with the main decision that you need to make and branches out based on the different outcomes and consequences of each decision. Each outcome will outline its own consequences, costs, and gains and, at the end of the analysis, you can compare each of them and make the smartest decision.
Businesses can use them to understand which project is more cost-effective and will bring more earnings in the long run. For example, imagine you need to decide if you want to update your software app or build a new app entirely. Here you would compare the total costs, the time needed to be invested, potential revenue, and any other factor that might affect your decision. In the end, you would be able to see which of these two options is more realistic and attainable for your company or research.
9. Conjoint analysis
Last but not least, we have the conjoint analysis. This approach is usually used in surveys to understand how individuals value different attributes of a product or service and it is one of the most effective methods to extract consumer preferences. When it comes to purchasing, some clients might be more price-focused, others more features-focused, and others might have a sustainable focus. Whatever your customer's preferences are, you can find them with conjoint analysis. Through this, companies can define pricing strategies, packaging options, subscription packages, and more.
A great example of conjoint analysis is in marketing and sales. For instance, a cupcake brand might use conjoint analysis and find that its clients prefer gluten-free options and cupcakes with healthier toppings over super sugary ones. Thus, the cupcake brand can turn these insights into advertisements and promotions to increase sales of this particular type of product. And not just that, conjoint analysis can also help businesses segment their customers based on their interests. This allows them to send different messaging that will bring value to each of the segments.
10. Correspondence Analysis
Also known as reciprocal averaging, correspondence analysis is a method used to analyze the relationship between categorical variables presented within a contingency table. A contingency table is a table that displays two (simple correspondence analysis) or more (multiple correspondence analysis) categorical variables across rows and columns that show the distribution of the data, which is usually answers to a survey or questionnaire on a specific topic.
This method starts by calculating an “expected value” which is done by multiplying row and column averages and dividing it by the overall original value of the specific table cell. The “expected value” is then subtracted from the original value resulting in a “residual number” which is what allows you to extract conclusions about relationships and distribution. The results of this analysis are later displayed using a map that represents the relationship between the different values. The closest two values are in the map, the bigger the relationship. Let’s put it into perspective with an example.
Imagine you are carrying out a market research analysis about outdoor clothing brands and how they are perceived by the public. For this analysis, you ask a group of people to match each brand with a certain attribute which can be durability, innovation, quality materials, etc. When calculating the residual numbers, you can see that brand A has a positive residual for innovation but a negative one for durability. This means that brand A is not positioned as a durable brand in the market, something that competitors could take advantage of.
11. Multidimensional Scaling (MDS)
MDS is a method used to observe the similarities or disparities between objects which can be colors, brands, people, geographical coordinates, and more. The objects are plotted using an “MDS map” that positions similar objects together and disparate ones far apart. The (dis) similarities between objects are represented using one or more dimensions that can be observed using a numerical scale. For example, if you want to know how people feel about the COVID-19 vaccine, you can use 1 for “don’t believe in the vaccine at all” and 10 for “firmly believe in the vaccine” and a scale of 2 to 9 for in between responses. When analyzing an MDS map the only thing that matters is the distance between the objects, the orientation of the dimensions is arbitrary and has no meaning at all.
Multidimensional scaling is a valuable technique for market research, especially when it comes to evaluating product or brand positioning. For instance, if a cupcake brand wants to know how they are positioned compared to competitors, it can define 2-3 dimensions such as taste, ingredients, shopping experience, or more, and do a multidimensional scaling analysis to find improvement opportunities as well as areas in which competitors are currently leading.
Another business example is in procurement when deciding on different suppliers. Decision makers can generate an MDS map to see how the different prices, delivery times, technical services, and more of the different suppliers differ and pick the one that suits their needs the best.
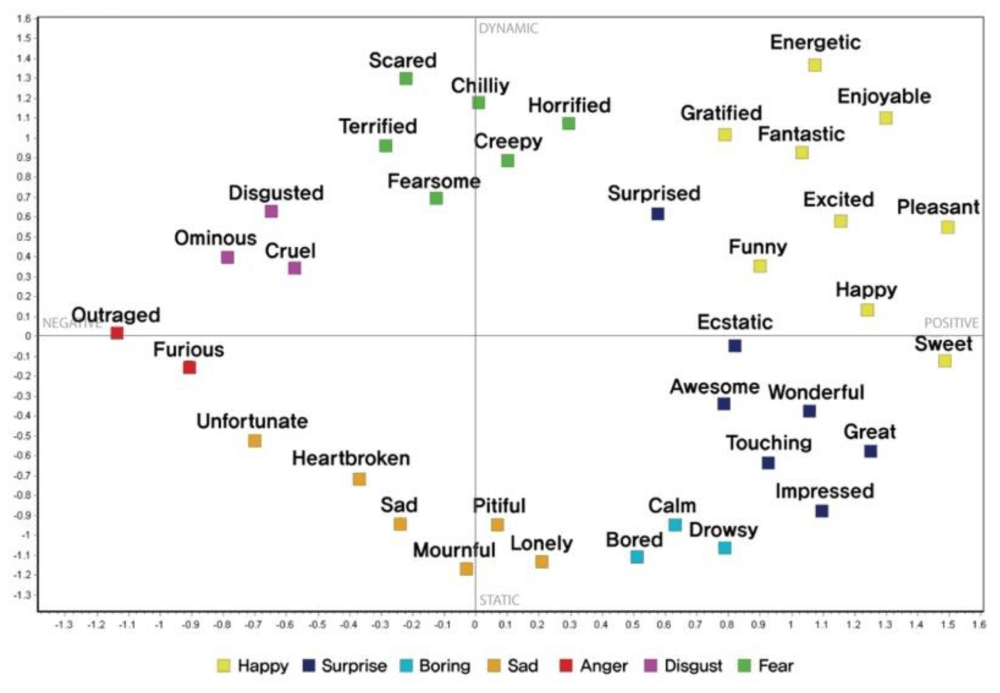
A final example proposed by a research paper on "An Improved Study of Multilevel Semantic Network Visualization for Analyzing Sentiment Word of Movie Review Data". Researchers picked a two-dimensional MDS map to display the distances and relationships between different sentiments in movie reviews. They used 36 sentiment words and distributed them based on their emotional distance as we can see in the image below where the words "outraged" and "sweet" are on opposite sides of the map, marking the distance between the two emotions very clearly.
Aside from being a valuable technique to analyze dissimilarities, MDS also serves as a dimension-reduction technique for large dimensional data.
B. Qualitative Methods
Qualitative data analysis methods are defined as the observation of non-numerical data that is gathered and produced using methods of observation such as interviews, focus groups, questionnaires, and more. As opposed to quantitative methods, qualitative data is more subjective and highly valuable in analyzing customer retention and product development.
12. Text analysis
Text analysis, also known in the industry as text mining, works by taking large sets of textual data and arranging them in a way that makes it easier to manage. By working through this cleansing process in stringent detail, you will be able to extract the data that is truly relevant to your organization and use it to develop actionable insights that will propel you forward.
Modern software accelerate the application of text analytics. Thanks to the combination of machine learning and intelligent algorithms, you can perform advanced analytical processes such as sentiment analysis. This technique allows you to understand the intentions and emotions of a text, for example, if it's positive, negative, or neutral, and then give it a score depending on certain factors and categories that are relevant to your brand. Sentiment analysis is often used to monitor brand and product reputation and to understand how successful your customer experience is. To learn more about the topic check out this insightful article .
By analyzing data from various word-based sources, including product reviews, articles, social media communications, and survey responses, you will gain invaluable insights into your audience, as well as their needs, preferences, and pain points. This will allow you to create campaigns, services, and communications that meet your prospects’ needs on a personal level, growing your audience while boosting customer retention. There are various other “sub-methods” that are an extension of text analysis. Each of them serves a more specific purpose and we will look at them in detail next.
13. Content Analysis
This is a straightforward and very popular method that examines the presence and frequency of certain words, concepts, and subjects in different content formats such as text, image, audio, or video. For example, the number of times the name of a celebrity is mentioned on social media or online tabloids. It does this by coding text data that is later categorized and tabulated in a way that can provide valuable insights, making it the perfect mix of quantitative and qualitative analysis.
There are two types of content analysis. The first one is the conceptual analysis which focuses on explicit data, for instance, the number of times a concept or word is mentioned in a piece of content. The second one is relational analysis, which focuses on the relationship between different concepts or words and how they are connected within a specific context.
Content analysis is often used by marketers to measure brand reputation and customer behavior. For example, by analyzing customer reviews. It can also be used to analyze customer interviews and find directions for new product development. It is also important to note, that in order to extract the maximum potential out of this analysis method, it is necessary to have a clearly defined research question.
14. Thematic Analysis
Very similar to content analysis, thematic analysis also helps in identifying and interpreting patterns in qualitative data with the main difference being that the first one can also be applied to quantitative analysis. The thematic method analyzes large pieces of text data such as focus group transcripts or interviews and groups them into themes or categories that come up frequently within the text. It is a great method when trying to figure out peoples view’s and opinions about a certain topic. For example, if you are a brand that cares about sustainability, you can do a survey of your customers to analyze their views and opinions about sustainability and how they apply it to their lives. You can also analyze customer service calls transcripts to find common issues and improve your service.
Thematic analysis is a very subjective technique that relies on the researcher’s judgment. Therefore, to avoid biases, it has 6 steps that include familiarization, coding, generating themes, reviewing themes, defining and naming themes, and writing up. It is also important to note that, because it is a flexible approach, the data can be interpreted in multiple ways and it can be hard to select what data is more important to emphasize.
15. Narrative Analysis
A bit more complex in nature than the two previous ones, narrative analysis is used to explore the meaning behind the stories that people tell and most importantly, how they tell them. By looking into the words that people use to describe a situation you can extract valuable conclusions about their perspective on a specific topic. Common sources for narrative data include autobiographies, family stories, opinion pieces, and testimonials, among others.
From a business perspective, narrative analysis can be useful to analyze customer behaviors and feelings towards a specific product, service, feature, or others. It provides unique and deep insights that can be extremely valuable. However, it has some drawbacks.
The biggest weakness of this method is that the sample sizes are usually very small due to the complexity and time-consuming nature of the collection of narrative data. Plus, the way a subject tells a story will be significantly influenced by his or her specific experiences, making it very hard to replicate in a subsequent study.
16. Discourse Analysis
Discourse analysis is used to understand the meaning behind any type of written, verbal, or symbolic discourse based on its political, social, or cultural context. It mixes the analysis of languages and situations together. This means that the way the content is constructed and the meaning behind it is significantly influenced by the culture and society it takes place in. For example, if you are analyzing political speeches you need to consider different context elements such as the politician's background, the current political context of the country, the audience to which the speech is directed, and so on.
From a business point of view, discourse analysis is a great market research tool. It allows marketers to understand how the norms and ideas of the specific market work and how their customers relate to those ideas. It can be very useful to build a brand mission or develop a unique tone of voice.
17. Grounded Theory Analysis
Traditionally, researchers decide on a method and hypothesis and start to collect the data to prove that hypothesis. The grounded theory is the only method that doesn’t require an initial research question or hypothesis as its value lies in the generation of new theories. With the grounded theory method, you can go into the analysis process with an open mind and explore the data to generate new theories through tests and revisions. In fact, it is not necessary to collect the data and then start to analyze it. Researchers usually start to find valuable insights as they are gathering the data.
All of these elements make grounded theory a very valuable method as theories are fully backed by data instead of initial assumptions. It is a great technique to analyze poorly researched topics or find the causes behind specific company outcomes. For example, product managers and marketers might use the grounded theory to find the causes of high levels of customer churn and look into customer surveys and reviews to develop new theories about the causes.
How To Analyze Data? Top 17 Data Analysis Techniques To Apply

Now that we’ve answered the questions “what is data analysis’”, why is it important, and covered the different data analysis types, it’s time to dig deeper into how to perform your analysis by working through these 17 essential techniques.
1. Collaborate your needs
Before you begin analyzing or drilling down into any techniques, it’s crucial to sit down collaboratively with all key stakeholders within your organization, decide on your primary campaign or strategic goals, and gain a fundamental understanding of the types of insights that will best benefit your progress or provide you with the level of vision you need to evolve your organization.
2. Establish your questions
Once you’ve outlined your core objectives, you should consider which questions will need answering to help you achieve your mission. This is one of the most important techniques as it will shape the very foundations of your success.
To help you ask the right things and ensure your data works for you, you have to ask the right data analysis questions .
3. Data democratization
After giving your data analytics methodology some real direction, and knowing which questions need answering to extract optimum value from the information available to your organization, you should continue with democratization.
Data democratization is an action that aims to connect data from various sources efficiently and quickly so that anyone in your organization can access it at any given moment. You can extract data in text, images, videos, numbers, or any other format. And then perform cross-database analysis to achieve more advanced insights to share with the rest of the company interactively.
Once you have decided on your most valuable sources, you need to take all of this into a structured format to start collecting your insights. For this purpose, datapine offers an easy all-in-one data connectors feature to integrate all your internal and external sources and manage them at your will. Additionally, datapine’s end-to-end solution automatically updates your data, allowing you to save time and focus on performing the right analysis to grow your company.
4. Think of governance
When collecting data in a business or research context you always need to think about security and privacy. With data breaches becoming a topic of concern for businesses, the need to protect your client's or subject’s sensitive information becomes critical.
To ensure that all this is taken care of, you need to think of a data governance strategy. According to Gartner , this concept refers to “ the specification of decision rights and an accountability framework to ensure the appropriate behavior in the valuation, creation, consumption, and control of data and analytics .” In simpler words, data governance is a collection of processes, roles, and policies, that ensure the efficient use of data while still achieving the main company goals. It ensures that clear roles are in place for who can access the information and how they can access it. In time, this not only ensures that sensitive information is protected but also allows for an efficient analysis as a whole.
5. Clean your data
After harvesting from so many sources you will be left with a vast amount of information that can be overwhelming to deal with. At the same time, you can be faced with incorrect data that can be misleading to your analysis. The smartest thing you can do to avoid dealing with this in the future is to clean the data. This is fundamental before visualizing it, as it will ensure that the insights you extract from it are correct.
There are many things that you need to look for in the cleaning process. The most important one is to eliminate any duplicate observations; this usually appears when using multiple internal and external sources of information. You can also add any missing codes, fix empty fields, and eliminate incorrectly formatted data.
Another usual form of cleaning is done with text data. As we mentioned earlier, most companies today analyze customer reviews, social media comments, questionnaires, and several other text inputs. In order for algorithms to detect patterns, text data needs to be revised to avoid invalid characters or any syntax or spelling errors.
Most importantly, the aim of cleaning is to prevent you from arriving at false conclusions that can damage your company in the long run. By using clean data, you will also help BI solutions to interact better with your information and create better reports for your organization.
6. Set your KPIs
Once you’ve set your sources, cleaned your data, and established clear-cut questions you want your insights to answer, you need to set a host of key performance indicators (KPIs) that will help you track, measure, and shape your progress in a number of key areas.
KPIs are critical to both qualitative and quantitative analysis research. This is one of the primary methods of data analysis you certainly shouldn’t overlook.
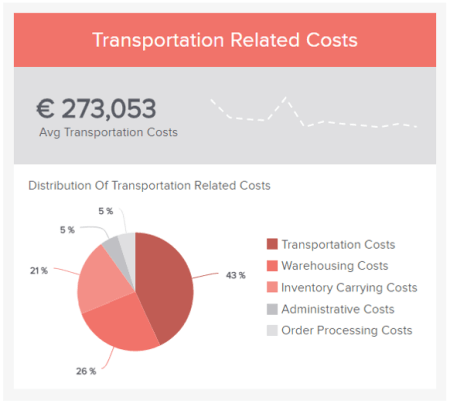
To help you set the best possible KPIs for your initiatives and activities, here is an example of a relevant logistics KPI : transportation-related costs. If you want to see more go explore our collection of key performance indicator examples .
7. Omit useless data
Having bestowed your data analysis tools and techniques with true purpose and defined your mission, you should explore the raw data you’ve collected from all sources and use your KPIs as a reference for chopping out any information you deem to be useless.
Trimming the informational fat is one of the most crucial methods of analysis as it will allow you to focus your analytical efforts and squeeze every drop of value from the remaining ‘lean’ information.
Any stats, facts, figures, or metrics that don’t align with your business goals or fit with your KPI management strategies should be eliminated from the equation.
8. Build a data management roadmap
While, at this point, this particular step is optional (you will have already gained a wealth of insight and formed a fairly sound strategy by now), creating a data governance roadmap will help your data analysis methods and techniques become successful on a more sustainable basis. These roadmaps, if developed properly, are also built so they can be tweaked and scaled over time.
Invest ample time in developing a roadmap that will help you store, manage, and handle your data internally, and you will make your analysis techniques all the more fluid and functional – one of the most powerful types of data analysis methods available today.
9. Integrate technology
There are many ways to analyze data, but one of the most vital aspects of analytical success in a business context is integrating the right decision support software and technology.
Robust analysis platforms will not only allow you to pull critical data from your most valuable sources while working with dynamic KPIs that will offer you actionable insights; it will also present them in a digestible, visual, interactive format from one central, live dashboard . A data methodology you can count on.
By integrating the right technology within your data analysis methodology, you’ll avoid fragmenting your insights, saving you time and effort while allowing you to enjoy the maximum value from your business’s most valuable insights.
For a look at the power of software for the purpose of analysis and to enhance your methods of analyzing, glance over our selection of dashboard examples .
10. Answer your questions
By considering each of the above efforts, working with the right technology, and fostering a cohesive internal culture where everyone buys into the different ways to analyze data as well as the power of digital intelligence, you will swiftly start to answer your most burning business questions. Arguably, the best way to make your data concepts accessible across the organization is through data visualization.
11. Visualize your data
Online data visualization is a powerful tool as it lets you tell a story with your metrics, allowing users across the organization to extract meaningful insights that aid business evolution – and it covers all the different ways to analyze data.
The purpose of analyzing is to make your entire organization more informed and intelligent, and with the right platform or dashboard, this is simpler than you think, as demonstrated by our marketing dashboard .
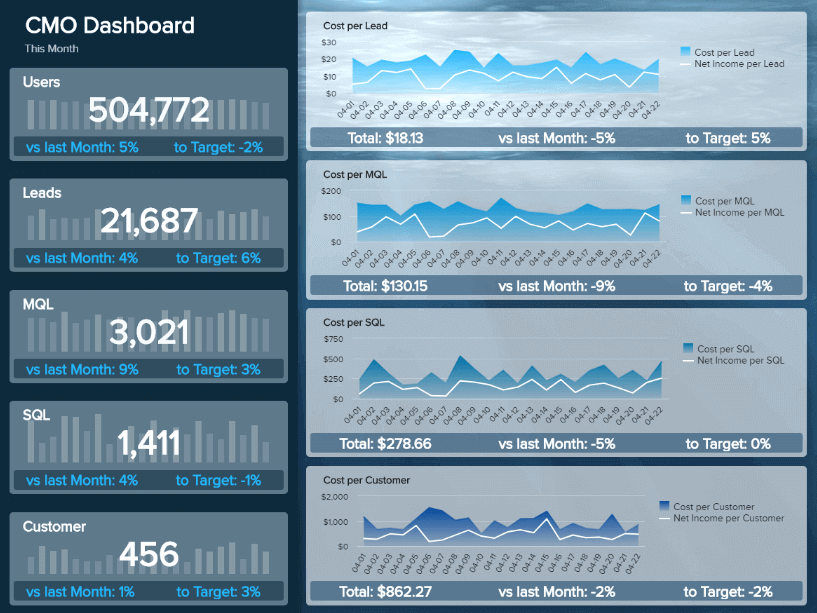
This visual, dynamic, and interactive online dashboard is a data analysis example designed to give Chief Marketing Officers (CMO) an overview of relevant metrics to help them understand if they achieved their monthly goals.
In detail, this example generated with a modern dashboard creator displays interactive charts for monthly revenues, costs, net income, and net income per customer; all of them are compared with the previous month so that you can understand how the data fluctuated. In addition, it shows a detailed summary of the number of users, customers, SQLs, and MQLs per month to visualize the whole picture and extract relevant insights or trends for your marketing reports .
The CMO dashboard is perfect for c-level management as it can help them monitor the strategic outcome of their marketing efforts and make data-driven decisions that can benefit the company exponentially.
12. Be careful with the interpretation
We already dedicated an entire post to data interpretation as it is a fundamental part of the process of data analysis. It gives meaning to the analytical information and aims to drive a concise conclusion from the analysis results. Since most of the time companies are dealing with data from many different sources, the interpretation stage needs to be done carefully and properly in order to avoid misinterpretations.
To help you through the process, here we list three common practices that you need to avoid at all costs when looking at your data:
- Correlation vs. causation: The human brain is formatted to find patterns. This behavior leads to one of the most common mistakes when performing interpretation: confusing correlation with causation. Although these two aspects can exist simultaneously, it is not correct to assume that because two things happened together, one provoked the other. A piece of advice to avoid falling into this mistake is never to trust just intuition, trust the data. If there is no objective evidence of causation, then always stick to correlation.
- Confirmation bias: This phenomenon describes the tendency to select and interpret only the data necessary to prove one hypothesis, often ignoring the elements that might disprove it. Even if it's not done on purpose, confirmation bias can represent a real problem, as excluding relevant information can lead to false conclusions and, therefore, bad business decisions. To avoid it, always try to disprove your hypothesis instead of proving it, share your analysis with other team members, and avoid drawing any conclusions before the entire analytical project is finalized.
- Statistical significance: To put it in short words, statistical significance helps analysts understand if a result is actually accurate or if it happened because of a sampling error or pure chance. The level of statistical significance needed might depend on the sample size and the industry being analyzed. In any case, ignoring the significance of a result when it might influence decision-making can be a huge mistake.
13. Build a narrative
Now, we’re going to look at how you can bring all of these elements together in a way that will benefit your business - starting with a little something called data storytelling.
The human brain responds incredibly well to strong stories or narratives. Once you’ve cleansed, shaped, and visualized your most invaluable data using various BI dashboard tools , you should strive to tell a story - one with a clear-cut beginning, middle, and end.
By doing so, you will make your analytical efforts more accessible, digestible, and universal, empowering more people within your organization to use your discoveries to their actionable advantage.
14. Consider autonomous technology
Autonomous technologies, such as artificial intelligence (AI) and machine learning (ML), play a significant role in the advancement of understanding how to analyze data more effectively.
Gartner predicts that by the end of this year, 80% of emerging technologies will be developed with AI foundations. This is a testament to the ever-growing power and value of autonomous technologies.
At the moment, these technologies are revolutionizing the analysis industry. Some examples that we mentioned earlier are neural networks, intelligent alarms, and sentiment analysis.
15. Share the load
If you work with the right tools and dashboards, you will be able to present your metrics in a digestible, value-driven format, allowing almost everyone in the organization to connect with and use relevant data to their advantage.
Modern dashboards consolidate data from various sources, providing access to a wealth of insights in one centralized location, no matter if you need to monitor recruitment metrics or generate reports that need to be sent across numerous departments. Moreover, these cutting-edge tools offer access to dashboards from a multitude of devices, meaning that everyone within the business can connect with practical insights remotely - and share the load.
Once everyone is able to work with a data-driven mindset, you will catalyze the success of your business in ways you never thought possible. And when it comes to knowing how to analyze data, this kind of collaborative approach is essential.
16. Data analysis tools
In order to perform high-quality analysis of data, it is fundamental to use tools and software that will ensure the best results. Here we leave you a small summary of four fundamental categories of data analysis tools for your organization.
- Business Intelligence: BI tools allow you to process significant amounts of data from several sources in any format. Through this, you can not only analyze and monitor your data to extract relevant insights but also create interactive reports and dashboards to visualize your KPIs and use them for your company's good. datapine is an amazing online BI software that is focused on delivering powerful online analysis features that are accessible to beginner and advanced users. Like this, it offers a full-service solution that includes cutting-edge analysis of data, KPIs visualization, live dashboards, reporting, and artificial intelligence technologies to predict trends and minimize risk.
- Statistical analysis: These tools are usually designed for scientists, statisticians, market researchers, and mathematicians, as they allow them to perform complex statistical analyses with methods like regression analysis, predictive analysis, and statistical modeling. A good tool to perform this type of analysis is R-Studio as it offers a powerful data modeling and hypothesis testing feature that can cover both academic and general data analysis. This tool is one of the favorite ones in the industry, due to its capability for data cleaning, data reduction, and performing advanced analysis with several statistical methods. Another relevant tool to mention is SPSS from IBM. The software offers advanced statistical analysis for users of all skill levels. Thanks to a vast library of machine learning algorithms, text analysis, and a hypothesis testing approach it can help your company find relevant insights to drive better decisions. SPSS also works as a cloud service that enables you to run it anywhere.
- SQL Consoles: SQL is a programming language often used to handle structured data in relational databases. Tools like these are popular among data scientists as they are extremely effective in unlocking these databases' value. Undoubtedly, one of the most used SQL software in the market is MySQL Workbench . This tool offers several features such as a visual tool for database modeling and monitoring, complete SQL optimization, administration tools, and visual performance dashboards to keep track of KPIs.
- Data Visualization: These tools are used to represent your data through charts, graphs, and maps that allow you to find patterns and trends in the data. datapine's already mentioned BI platform also offers a wealth of powerful online data visualization tools with several benefits. Some of them include: delivering compelling data-driven presentations to share with your entire company, the ability to see your data online with any device wherever you are, an interactive dashboard design feature that enables you to showcase your results in an interactive and understandable way, and to perform online self-service reports that can be used simultaneously with several other people to enhance team productivity.
17. Refine your process constantly
Last is a step that might seem obvious to some people, but it can be easily ignored if you think you are done. Once you have extracted the needed results, you should always take a retrospective look at your project and think about what you can improve. As you saw throughout this long list of techniques, data analysis is a complex process that requires constant refinement. For this reason, you should always go one step further and keep improving.
Quality Criteria For Data Analysis
So far we’ve covered a list of methods and techniques that should help you perform efficient data analysis. But how do you measure the quality and validity of your results? This is done with the help of some science quality criteria. Here we will go into a more theoretical area that is critical to understanding the fundamentals of statistical analysis in science. However, you should also be aware of these steps in a business context, as they will allow you to assess the quality of your results in the correct way. Let’s dig in.
- Internal validity: The results of a survey are internally valid if they measure what they are supposed to measure and thus provide credible results. In other words , internal validity measures the trustworthiness of the results and how they can be affected by factors such as the research design, operational definitions, how the variables are measured, and more. For instance, imagine you are doing an interview to ask people if they brush their teeth two times a day. While most of them will answer yes, you can still notice that their answers correspond to what is socially acceptable, which is to brush your teeth at least twice a day. In this case, you can’t be 100% sure if respondents actually brush their teeth twice a day or if they just say that they do, therefore, the internal validity of this interview is very low.
- External validity: Essentially, external validity refers to the extent to which the results of your research can be applied to a broader context. It basically aims to prove that the findings of a study can be applied in the real world. If the research can be applied to other settings, individuals, and times, then the external validity is high.
- Reliability : If your research is reliable, it means that it can be reproduced. If your measurement were repeated under the same conditions, it would produce similar results. This means that your measuring instrument consistently produces reliable results. For example, imagine a doctor building a symptoms questionnaire to detect a specific disease in a patient. Then, various other doctors use this questionnaire but end up diagnosing the same patient with a different condition. This means the questionnaire is not reliable in detecting the initial disease. Another important note here is that in order for your research to be reliable, it also needs to be objective. If the results of a study are the same, independent of who assesses them or interprets them, the study can be considered reliable. Let’s see the objectivity criteria in more detail now.
- Objectivity: In data science, objectivity means that the researcher needs to stay fully objective when it comes to its analysis. The results of a study need to be affected by objective criteria and not by the beliefs, personality, or values of the researcher. Objectivity needs to be ensured when you are gathering the data, for example, when interviewing individuals, the questions need to be asked in a way that doesn't influence the results. Paired with this, objectivity also needs to be thought of when interpreting the data. If different researchers reach the same conclusions, then the study is objective. For this last point, you can set predefined criteria to interpret the results to ensure all researchers follow the same steps.
The discussed quality criteria cover mostly potential influences in a quantitative context. Analysis in qualitative research has by default additional subjective influences that must be controlled in a different way. Therefore, there are other quality criteria for this kind of research such as credibility, transferability, dependability, and confirmability. You can see each of them more in detail on this resource .
Data Analysis Limitations & Barriers
Analyzing data is not an easy task. As you’ve seen throughout this post, there are many steps and techniques that you need to apply in order to extract useful information from your research. While a well-performed analysis can bring various benefits to your organization it doesn't come without limitations. In this section, we will discuss some of the main barriers you might encounter when conducting an analysis. Let’s see them more in detail.
- Lack of clear goals: No matter how good your data or analysis might be if you don’t have clear goals or a hypothesis the process might be worthless. While we mentioned some methods that don’t require a predefined hypothesis, it is always better to enter the analytical process with some clear guidelines of what you are expecting to get out of it, especially in a business context in which data is utilized to support important strategic decisions.
- Objectivity: Arguably one of the biggest barriers when it comes to data analysis in research is to stay objective. When trying to prove a hypothesis, researchers might find themselves, intentionally or unintentionally, directing the results toward an outcome that they want. To avoid this, always question your assumptions and avoid confusing facts with opinions. You can also show your findings to a research partner or external person to confirm that your results are objective.
- Data representation: A fundamental part of the analytical procedure is the way you represent your data. You can use various graphs and charts to represent your findings, but not all of them will work for all purposes. Choosing the wrong visual can not only damage your analysis but can mislead your audience, therefore, it is important to understand when to use each type of data depending on your analytical goals. Our complete guide on the types of graphs and charts lists 20 different visuals with examples of when to use them.
- Flawed correlation : Misleading statistics can significantly damage your research. We’ve already pointed out a few interpretation issues previously in the post, but it is an important barrier that we can't avoid addressing here as well. Flawed correlations occur when two variables appear related to each other but they are not. Confusing correlations with causation can lead to a wrong interpretation of results which can lead to building wrong strategies and loss of resources, therefore, it is very important to identify the different interpretation mistakes and avoid them.
- Sample size: A very common barrier to a reliable and efficient analysis process is the sample size. In order for the results to be trustworthy, the sample size should be representative of what you are analyzing. For example, imagine you have a company of 1000 employees and you ask the question “do you like working here?” to 50 employees of which 49 say yes, which means 95%. Now, imagine you ask the same question to the 1000 employees and 950 say yes, which also means 95%. Saying that 95% of employees like working in the company when the sample size was only 50 is not a representative or trustworthy conclusion. The significance of the results is way more accurate when surveying a bigger sample size.
- Privacy concerns: In some cases, data collection can be subjected to privacy regulations. Businesses gather all kinds of information from their customers from purchasing behaviors to addresses and phone numbers. If this falls into the wrong hands due to a breach, it can affect the security and confidentiality of your clients. To avoid this issue, you need to collect only the data that is needed for your research and, if you are using sensitive facts, make it anonymous so customers are protected. The misuse of customer data can severely damage a business's reputation, so it is important to keep an eye on privacy.
- Lack of communication between teams : When it comes to performing data analysis on a business level, it is very likely that each department and team will have different goals and strategies. However, they are all working for the same common goal of helping the business run smoothly and keep growing. When teams are not connected and communicating with each other, it can directly affect the way general strategies are built. To avoid these issues, tools such as data dashboards enable teams to stay connected through data in a visually appealing way.
- Innumeracy : Businesses are working with data more and more every day. While there are many BI tools available to perform effective analysis, data literacy is still a constant barrier. Not all employees know how to apply analysis techniques or extract insights from them. To prevent this from happening, you can implement different training opportunities that will prepare every relevant user to deal with data.
Key Data Analysis Skills
As you've learned throughout this lengthy guide, analyzing data is a complex task that requires a lot of knowledge and skills. That said, thanks to the rise of self-service tools the process is way more accessible and agile than it once was. Regardless, there are still some key skills that are valuable to have when working with data, we list the most important ones below.
- Critical and statistical thinking: To successfully analyze data you need to be creative and think out of the box. Yes, that might sound like a weird statement considering that data is often tight to facts. However, a great level of critical thinking is required to uncover connections, come up with a valuable hypothesis, and extract conclusions that go a step further from the surface. This, of course, needs to be complemented by statistical thinking and an understanding of numbers.
- Data cleaning: Anyone who has ever worked with data before will tell you that the cleaning and preparation process accounts for 80% of a data analyst's work, therefore, the skill is fundamental. But not just that, not cleaning the data adequately can also significantly damage the analysis which can lead to poor decision-making in a business scenario. While there are multiple tools that automate the cleaning process and eliminate the possibility of human error, it is still a valuable skill to dominate.
- Data visualization: Visuals make the information easier to understand and analyze, not only for professional users but especially for non-technical ones. Having the necessary skills to not only choose the right chart type but know when to apply it correctly is key. This also means being able to design visually compelling charts that make the data exploration process more efficient.
- SQL: The Structured Query Language or SQL is a programming language used to communicate with databases. It is fundamental knowledge as it enables you to update, manipulate, and organize data from relational databases which are the most common databases used by companies. It is fairly easy to learn and one of the most valuable skills when it comes to data analysis.
- Communication skills: This is a skill that is especially valuable in a business environment. Being able to clearly communicate analytical outcomes to colleagues is incredibly important, especially when the information you are trying to convey is complex for non-technical people. This applies to in-person communication as well as written format, for example, when generating a dashboard or report. While this might be considered a “soft” skill compared to the other ones we mentioned, it should not be ignored as you most likely will need to share analytical findings with others no matter the context.
Data Analysis In The Big Data Environment
Big data is invaluable to today’s businesses, and by using different methods for data analysis, it’s possible to view your data in a way that can help you turn insight into positive action.
To inspire your efforts and put the importance of big data into context, here are some insights that you should know:
- By 2026 the industry of big data is expected to be worth approximately $273.4 billion.
- 94% of enterprises say that analyzing data is important for their growth and digital transformation.
- Companies that exploit the full potential of their data can increase their operating margins by 60% .
- We already told you the benefits of Artificial Intelligence through this article. This industry's financial impact is expected to grow up to $40 billion by 2025.
Data analysis concepts may come in many forms, but fundamentally, any solid methodology will help to make your business more streamlined, cohesive, insightful, and successful than ever before.
Key Takeaways From Data Analysis
As we reach the end of our data analysis journey, we leave a small summary of the main methods and techniques to perform excellent analysis and grow your business.
17 Essential Types of Data Analysis Methods:
- Cluster analysis
- Cohort analysis
- Regression analysis
- Factor analysis
- Neural Networks
- Data Mining
- Text analysis
- Time series analysis
- Decision trees
- Conjoint analysis
- Correspondence Analysis
- Multidimensional Scaling
- Content analysis
- Thematic analysis
- Narrative analysis
- Grounded theory analysis
- Discourse analysis
Top 17 Data Analysis Techniques:
- Collaborate your needs
- Establish your questions
- Data democratization
- Think of data governance
- Clean your data
- Set your KPIs
- Omit useless data
- Build a data management roadmap
- Integrate technology
- Answer your questions
- Visualize your data
- Interpretation of data
- Consider autonomous technology
- Build a narrative
- Share the load
- Data Analysis tools
- Refine your process constantly
We’ve pondered the data analysis definition and drilled down into the practical applications of data-centric analytics, and one thing is clear: by taking measures to arrange your data and making your metrics work for you, it’s possible to transform raw information into action - the kind of that will push your business to the next level.
Yes, good data analytics techniques result in enhanced business intelligence (BI). To help you understand this notion in more detail, read our exploration of business intelligence reporting .
And, if you’re ready to perform your own analysis, drill down into your facts and figures while interacting with your data on astonishing visuals, you can try our software for a free, 14-day trial .
News alert: UC Berkeley has announced its next university librarian
Secondary menu
- Log in to your Library account
- Hours and Maps
- Connect from Off Campus
- UC Berkeley Home
Search form
Research methods--quantitative, qualitative, and more: overview.
- Quantitative Research
- Qualitative Research
- Data Science Methods (Machine Learning, AI, Big Data)
- Text Mining and Computational Text Analysis
- Evidence Synthesis/Systematic Reviews
- Get Data, Get Help!
About Research Methods
This guide provides an overview of research methods, how to choose and use them, and supports and resources at UC Berkeley.
As Patten and Newhart note in the book Understanding Research Methods , "Research methods are the building blocks of the scientific enterprise. They are the "how" for building systematic knowledge. The accumulation of knowledge through research is by its nature a collective endeavor. Each well-designed study provides evidence that may support, amend, refute, or deepen the understanding of existing knowledge...Decisions are important throughout the practice of research and are designed to help researchers collect evidence that includes the full spectrum of the phenomenon under study, to maintain logical rules, and to mitigate or account for possible sources of bias. In many ways, learning research methods is learning how to see and make these decisions."
The choice of methods varies by discipline, by the kind of phenomenon being studied and the data being used to study it, by the technology available, and more. This guide is an introduction, but if you don't see what you need here, always contact your subject librarian, and/or take a look to see if there's a library research guide that will answer your question.
Suggestions for changes and additions to this guide are welcome!
START HERE: SAGE Research Methods
Without question, the most comprehensive resource available from the library is SAGE Research Methods. HERE IS THE ONLINE GUIDE to this one-stop shopping collection, and some helpful links are below:
- SAGE Research Methods
- Little Green Books (Quantitative Methods)
- Little Blue Books (Qualitative Methods)
- Dictionaries and Encyclopedias
- Case studies of real research projects
- Sample datasets for hands-on practice
- Streaming video--see methods come to life
- Methodspace- -a community for researchers
- SAGE Research Methods Course Mapping
Library Data Services at UC Berkeley
Library Data Services Program and Digital Scholarship Services
The LDSP offers a variety of services and tools ! From this link, check out pages for each of the following topics: discovering data, managing data, collecting data, GIS data, text data mining, publishing data, digital scholarship, open science, and the Research Data Management Program.
Be sure also to check out the visual guide to where to seek assistance on campus with any research question you may have!
Library GIS Services
Other Data Services at Berkeley
D-Lab Supports Berkeley faculty, staff, and graduate students with research in data intensive social science, including a wide range of training and workshop offerings Dryad Dryad is a simple self-service tool for researchers to use in publishing their datasets. It provides tools for the effective publication of and access to research data. Geospatial Innovation Facility (GIF) Provides leadership and training across a broad array of integrated mapping technologies on campu Research Data Management A UC Berkeley guide and consulting service for research data management issues
General Research Methods Resources
Here are some general resources for assistance:
- Assistance from ICPSR (must create an account to access): Getting Help with Data , and Resources for Students
- Wiley Stats Ref for background information on statistics topics
- Survey Documentation and Analysis (SDA) . Program for easy web-based analysis of survey data.
Consultants
- D-Lab/Data Science Discovery Consultants Request help with your research project from peer consultants.
- Research data (RDM) consulting Meet with RDM consultants before designing the data security, storage, and sharing aspects of your qualitative project.
- Statistics Department Consulting Services A service in which advanced graduate students, under faculty supervision, are available to consult during specified hours in the Fall and Spring semesters.
Related Resourcex
- IRB / CPHS Qualitative research projects with human subjects often require that you go through an ethics review.
- OURS (Office of Undergraduate Research and Scholarships) OURS supports undergraduates who want to embark on research projects and assistantships. In particular, check out their "Getting Started in Research" workshops
- Sponsored Projects Sponsored projects works with researchers applying for major external grants.
- Next: Quantitative Research >>
- Last Updated: Apr 25, 2024 11:09 AM
- URL: https://guides.lib.berkeley.edu/researchmethods
Quantitative Methods
- Living reference work entry
- First Online: 11 June 2021
- Cite this living reference work entry

- Juwel Rana 2 , 3 , 4 ,
- Patricia Luna Gutierrez 5 &
- John C. Oldroyd 6
422 Accesses
1 Citations
Quantitative analysis ; Quantitative research methods ; Study design
Quantitative method is the collection and analysis of numerical data to answer scientific research questions. Quantitative method is used to summarize, average, find patterns, make predictions, and test causal associations as well as generalizing results to wider populations. It allows us to quantify effect sizes, determine the strength of associations, rank priorities, and weigh the strength of evidence of effectiveness.
Introduction
This entry aims to introduce the most common ways to use numbers and statistics to describe variables, establish relationships among variables, and build numerical understanding of a topic. In general, the quantitative research process uses a deductive approach (Neuman 2014 ; Leavy 2017 ), extrapolating from a particular case to the general situation (Babones 2016 ).
In practical ways, quantitative methods are an approach to studying a research topic. In research, the...
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
Institutional subscriptions
Babones S (2016) Interpretive quantitative methods for the social sciences. Sociology. https://doi.org/10.1177/0038038515583637
Balnaves M, Caputi P (2001) Introduction to quantitative research methods: an investigative approach. Sage, London
Book Google Scholar
Brenner PS (2020) Understanding survey methodology: sociological theory and applications. Springer, Boston
Google Scholar
Creswell JW (2014) Research design: qualitative, quantitative, and mixed methods approaches. Sage, London
Leavy P (2017) Research design. The Gilford Press, New York
Mertens W, Pugliese A, Recker J (2018) Quantitative data analysis, research methods: information, systems, and contexts: second edition. https://doi.org/10.1016/B978-0-08-102220-7.00018-2
Neuman LW (2014) Social research methods: qualitative and quantitative approaches. Pearson Education Limited, Edinburgh
Treiman DJ (2009) Quantitative data analysis: doing social research to test ideas. Jossey-Bass, San Francisco
Download references
Author information
Authors and affiliations.
Department of Public Health, School of Health and Life Sciences, North South University, Dhaka, Bangladesh
Department of Biostatistics and Epidemiology, School of Health and Health Sciences, University of Massachusetts Amherst, MA, USA
Department of Research and Innovation, South Asia Institute for Social Transformation (SAIST), Dhaka, Bangladesh
Independent Researcher, Masatepe, Nicaragua
Patricia Luna Gutierrez
School of Behavioral and Health Sciences, Australian Catholic University, Fitzroy, VIC, Australia
John C. Oldroyd
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Juwel Rana .
Editor information
Editors and affiliations.
Florida Atlantic University, Boca Raton, FL, USA
Ali Farazmand
Rights and permissions
Reprints and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this entry
Cite this entry.
Rana, J., Gutierrez, P.L., Oldroyd, J.C. (2021). Quantitative Methods. In: Farazmand, A. (eds) Global Encyclopedia of Public Administration, Public Policy, and Governance. Springer, Cham. https://doi.org/10.1007/978-3-319-31816-5_460-1
Download citation
DOI : https://doi.org/10.1007/978-3-319-31816-5_460-1
Received : 31 January 2021
Accepted : 14 February 2021
Published : 11 June 2021
Publisher Name : Springer, Cham
Print ISBN : 978-3-319-31816-5
Online ISBN : 978-3-319-31816-5
eBook Packages : Springer Reference Economics and Finance Reference Module Humanities and Social Sciences Reference Module Business, Economics and Social Sciences
- Publish with us
Policies and ethics
- Find a journal
- Track your research
Qualitative Data Analysis Methods 101:
The “big 6” methods + examples.
By: Kerryn Warren (PhD) | Reviewed By: Eunice Rautenbach (D.Tech) | May 2020 (Updated April 2023)
Qualitative data analysis methods. Wow, that’s a mouthful.
If you’re new to the world of research, qualitative data analysis can look rather intimidating. So much bulky terminology and so many abstract, fluffy concepts. It certainly can be a minefield!
Don’t worry – in this post, we’ll unpack the most popular analysis methods , one at a time, so that you can approach your analysis with confidence and competence – whether that’s for a dissertation, thesis or really any kind of research project.
What (exactly) is qualitative data analysis?
To understand qualitative data analysis, we need to first understand qualitative data – so let’s step back and ask the question, “what exactly is qualitative data?”.
Qualitative data refers to pretty much any data that’s “not numbers” . In other words, it’s not the stuff you measure using a fixed scale or complex equipment, nor do you analyse it using complex statistics or mathematics.
So, if it’s not numbers, what is it?
Words, you guessed? Well… sometimes , yes. Qualitative data can, and often does, take the form of interview transcripts, documents and open-ended survey responses – but it can also involve the interpretation of images and videos. In other words, qualitative isn’t just limited to text-based data.
So, how’s that different from quantitative data, you ask?
Simply put, qualitative research focuses on words, descriptions, concepts or ideas – while quantitative research focuses on numbers and statistics . Qualitative research investigates the “softer side” of things to explore and describe , while quantitative research focuses on the “hard numbers”, to measure differences between variables and the relationships between them. If you’re keen to learn more about the differences between qual and quant, we’ve got a detailed post over here .
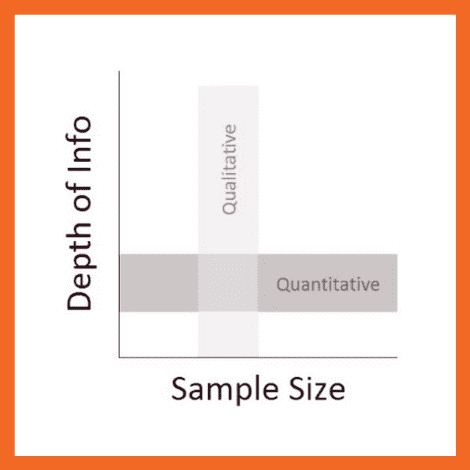
So, qualitative analysis is easier than quantitative, right?
Not quite. In many ways, qualitative data can be challenging and time-consuming to analyse and interpret. At the end of your data collection phase (which itself takes a lot of time), you’ll likely have many pages of text-based data or hours upon hours of audio to work through. You might also have subtle nuances of interactions or discussions that have danced around in your mind, or that you scribbled down in messy field notes. All of this needs to work its way into your analysis.
Making sense of all of this is no small task and you shouldn’t underestimate it. Long story short – qualitative analysis can be a lot of work! Of course, quantitative analysis is no piece of cake either, but it’s important to recognise that qualitative analysis still requires a significant investment in terms of time and effort.
Need a helping hand?
In this post, we’ll explore qualitative data analysis by looking at some of the most common analysis methods we encounter. We’re not going to cover every possible qualitative method and we’re not going to go into heavy detail – we’re just going to give you the big picture. That said, we will of course includes links to loads of extra resources so that you can learn more about whichever analysis method interests you.
Without further delay, let’s get into it.
The “Big 6” Qualitative Analysis Methods
There are many different types of qualitative data analysis, all of which serve different purposes and have unique strengths and weaknesses . We’ll start by outlining the analysis methods and then we’ll dive into the details for each.
The 6 most popular methods (or at least the ones we see at Grad Coach) are:
- Content analysis
- Narrative analysis
- Discourse analysis
- Thematic analysis
- Grounded theory (GT)
- Interpretive phenomenological analysis (IPA)
Let’s take a look at each of them…
QDA Method #1: Qualitative Content Analysis
Content analysis is possibly the most common and straightforward QDA method. At the simplest level, content analysis is used to evaluate patterns within a piece of content (for example, words, phrases or images) or across multiple pieces of content or sources of communication. For example, a collection of newspaper articles or political speeches.
With content analysis, you could, for instance, identify the frequency with which an idea is shared or spoken about – like the number of times a Kardashian is mentioned on Twitter. Or you could identify patterns of deeper underlying interpretations – for instance, by identifying phrases or words in tourist pamphlets that highlight India as an ancient country.
Because content analysis can be used in such a wide variety of ways, it’s important to go into your analysis with a very specific question and goal, or you’ll get lost in the fog. With content analysis, you’ll group large amounts of text into codes , summarise these into categories, and possibly even tabulate the data to calculate the frequency of certain concepts or variables. Because of this, content analysis provides a small splash of quantitative thinking within a qualitative method.
Naturally, while content analysis is widely useful, it’s not without its drawbacks . One of the main issues with content analysis is that it can be very time-consuming , as it requires lots of reading and re-reading of the texts. Also, because of its multidimensional focus on both qualitative and quantitative aspects, it is sometimes accused of losing important nuances in communication.
Content analysis also tends to concentrate on a very specific timeline and doesn’t take into account what happened before or after that timeline. This isn’t necessarily a bad thing though – just something to be aware of. So, keep these factors in mind if you’re considering content analysis. Every analysis method has its limitations , so don’t be put off by these – just be aware of them ! If you’re interested in learning more about content analysis, the video below provides a good starting point.
QDA Method #2: Narrative Analysis
As the name suggests, narrative analysis is all about listening to people telling stories and analysing what that means . Since stories serve a functional purpose of helping us make sense of the world, we can gain insights into the ways that people deal with and make sense of reality by analysing their stories and the ways they’re told.
You could, for example, use narrative analysis to explore whether how something is being said is important. For instance, the narrative of a prisoner trying to justify their crime could provide insight into their view of the world and the justice system. Similarly, analysing the ways entrepreneurs talk about the struggles in their careers or cancer patients telling stories of hope could provide powerful insights into their mindsets and perspectives . Simply put, narrative analysis is about paying attention to the stories that people tell – and more importantly, the way they tell them.
Of course, the narrative approach has its weaknesses , too. Sample sizes are generally quite small due to the time-consuming process of capturing narratives. Because of this, along with the multitude of social and lifestyle factors which can influence a subject, narrative analysis can be quite difficult to reproduce in subsequent research. This means that it’s difficult to test the findings of some of this research.
Similarly, researcher bias can have a strong influence on the results here, so you need to be particularly careful about the potential biases you can bring into your analysis when using this method. Nevertheless, narrative analysis is still a very useful qualitative analysis method – just keep these limitations in mind and be careful not to draw broad conclusions . If you’re keen to learn more about narrative analysis, the video below provides a great introduction to this qualitative analysis method.
QDA Method #3: Discourse Analysis
Discourse is simply a fancy word for written or spoken language or debate . So, discourse analysis is all about analysing language within its social context. In other words, analysing language – such as a conversation, a speech, etc – within the culture and society it takes place. For example, you could analyse how a janitor speaks to a CEO, or how politicians speak about terrorism.
To truly understand these conversations or speeches, the culture and history of those involved in the communication are important factors to consider. For example, a janitor might speak more casually with a CEO in a company that emphasises equality among workers. Similarly, a politician might speak more about terrorism if there was a recent terrorist incident in the country.
So, as you can see, by using discourse analysis, you can identify how culture , history or power dynamics (to name a few) have an effect on the way concepts are spoken about. So, if your research aims and objectives involve understanding culture or power dynamics, discourse analysis can be a powerful method.
Because there are many social influences in terms of how we speak to each other, the potential use of discourse analysis is vast . Of course, this also means it’s important to have a very specific research question (or questions) in mind when analysing your data and looking for patterns and themes, or you might land up going down a winding rabbit hole.
Discourse analysis can also be very time-consuming as you need to sample the data to the point of saturation – in other words, until no new information and insights emerge. But this is, of course, part of what makes discourse analysis such a powerful technique. So, keep these factors in mind when considering this QDA method. Again, if you’re keen to learn more, the video below presents a good starting point.
QDA Method #4: Thematic Analysis
Thematic analysis looks at patterns of meaning in a data set – for example, a set of interviews or focus group transcripts. But what exactly does that… mean? Well, a thematic analysis takes bodies of data (which are often quite large) and groups them according to similarities – in other words, themes . These themes help us make sense of the content and derive meaning from it.
Let’s take a look at an example.
With thematic analysis, you could analyse 100 online reviews of a popular sushi restaurant to find out what patrons think about the place. By reviewing the data, you would then identify the themes that crop up repeatedly within the data – for example, “fresh ingredients” or “friendly wait staff”.
So, as you can see, thematic analysis can be pretty useful for finding out about people’s experiences , views, and opinions . Therefore, if your research aims and objectives involve understanding people’s experience or view of something, thematic analysis can be a great choice.
Since thematic analysis is a bit of an exploratory process, it’s not unusual for your research questions to develop , or even change as you progress through the analysis. While this is somewhat natural in exploratory research, it can also be seen as a disadvantage as it means that data needs to be re-reviewed each time a research question is adjusted. In other words, thematic analysis can be quite time-consuming – but for a good reason. So, keep this in mind if you choose to use thematic analysis for your project and budget extra time for unexpected adjustments.

QDA Method #5: Grounded theory (GT)
Grounded theory is a powerful qualitative analysis method where the intention is to create a new theory (or theories) using the data at hand, through a series of “ tests ” and “ revisions ”. Strictly speaking, GT is more a research design type than an analysis method, but we’ve included it here as it’s often referred to as a method.
What’s most important with grounded theory is that you go into the analysis with an open mind and let the data speak for itself – rather than dragging existing hypotheses or theories into your analysis. In other words, your analysis must develop from the ground up (hence the name).
Let’s look at an example of GT in action.
Assume you’re interested in developing a theory about what factors influence students to watch a YouTube video about qualitative analysis. Using Grounded theory , you’d start with this general overarching question about the given population (i.e., graduate students). First, you’d approach a small sample – for example, five graduate students in a department at a university. Ideally, this sample would be reasonably representative of the broader population. You’d interview these students to identify what factors lead them to watch the video.
After analysing the interview data, a general pattern could emerge. For example, you might notice that graduate students are more likely to read a post about qualitative methods if they are just starting on their dissertation journey, or if they have an upcoming test about research methods.
From here, you’ll look for another small sample – for example, five more graduate students in a different department – and see whether this pattern holds true for them. If not, you’ll look for commonalities and adapt your theory accordingly. As this process continues, the theory would develop . As we mentioned earlier, what’s important with grounded theory is that the theory develops from the data – not from some preconceived idea.
So, what are the drawbacks of grounded theory? Well, some argue that there’s a tricky circularity to grounded theory. For it to work, in principle, you should know as little as possible regarding the research question and population, so that you reduce the bias in your interpretation. However, in many circumstances, it’s also thought to be unwise to approach a research question without knowledge of the current literature . In other words, it’s a bit of a “chicken or the egg” situation.
Regardless, grounded theory remains a popular (and powerful) option. Naturally, it’s a very useful method when you’re researching a topic that is completely new or has very little existing research about it, as it allows you to start from scratch and work your way from the ground up .

QDA Method #6: Interpretive Phenomenological Analysis (IPA)
Interpretive. Phenomenological. Analysis. IPA . Try saying that three times fast…
Let’s just stick with IPA, okay?
IPA is designed to help you understand the personal experiences of a subject (for example, a person or group of people) concerning a major life event, an experience or a situation . This event or experience is the “phenomenon” that makes up the “P” in IPA. Such phenomena may range from relatively common events – such as motherhood, or being involved in a car accident – to those which are extremely rare – for example, someone’s personal experience in a refugee camp. So, IPA is a great choice if your research involves analysing people’s personal experiences of something that happened to them.
It’s important to remember that IPA is subject – centred . In other words, it’s focused on the experiencer . This means that, while you’ll likely use a coding system to identify commonalities, it’s important not to lose the depth of experience or meaning by trying to reduce everything to codes. Also, keep in mind that since your sample size will generally be very small with IPA, you often won’t be able to draw broad conclusions about the generalisability of your findings. But that’s okay as long as it aligns with your research aims and objectives.
Another thing to be aware of with IPA is personal bias . While researcher bias can creep into all forms of research, self-awareness is critically important with IPA, as it can have a major impact on the results. For example, a researcher who was a victim of a crime himself could insert his own feelings of frustration and anger into the way he interprets the experience of someone who was kidnapped. So, if you’re going to undertake IPA, you need to be very self-aware or you could muddy the analysis.

How to choose the right analysis method
In light of all of the qualitative analysis methods we’ve covered so far, you’re probably asking yourself the question, “ How do I choose the right one? ”
Much like all the other methodological decisions you’ll need to make, selecting the right qualitative analysis method largely depends on your research aims, objectives and questions . In other words, the best tool for the job depends on what you’re trying to build. For example:
- Perhaps your research aims to analyse the use of words and what they reveal about the intention of the storyteller and the cultural context of the time.
- Perhaps your research aims to develop an understanding of the unique personal experiences of people that have experienced a certain event, or
- Perhaps your research aims to develop insight regarding the influence of a certain culture on its members.
As you can probably see, each of these research aims are distinctly different , and therefore different analysis methods would be suitable for each one. For example, narrative analysis would likely be a good option for the first aim, while grounded theory wouldn’t be as relevant.
It’s also important to remember that each method has its own set of strengths, weaknesses and general limitations. No single analysis method is perfect . So, depending on the nature of your research, it may make sense to adopt more than one method (this is called triangulation ). Keep in mind though that this will of course be quite time-consuming.
As we’ve seen, all of the qualitative analysis methods we’ve discussed make use of coding and theme-generating techniques, but the intent and approach of each analysis method differ quite substantially. So, it’s very important to come into your research with a clear intention before you decide which analysis method (or methods) to use.
Start by reviewing your research aims , objectives and research questions to assess what exactly you’re trying to find out – then select a qualitative analysis method that fits. Never pick a method just because you like it or have experience using it – your analysis method (or methods) must align with your broader research aims and objectives.

Let’s recap on QDA methods…
In this post, we looked at six popular qualitative data analysis methods:
- First, we looked at content analysis , a straightforward method that blends a little bit of quant into a primarily qualitative analysis.
- Then we looked at narrative analysis , which is about analysing how stories are told.
- Next up was discourse analysis – which is about analysing conversations and interactions.
- Then we moved on to thematic analysis – which is about identifying themes and patterns.
- From there, we went south with grounded theory – which is about starting from scratch with a specific question and using the data alone to build a theory in response to that question.
- And finally, we looked at IPA – which is about understanding people’s unique experiences of a phenomenon.
Of course, these aren’t the only options when it comes to qualitative data analysis, but they’re a great starting point if you’re dipping your toes into qualitative research for the first time.
If you’re still feeling a bit confused, consider our private coaching service , where we hold your hand through the research process to help you develop your best work.

Psst... there’s more!
This post was based on one of our popular Research Bootcamps . If you're working on a research project, you'll definitely want to check this out ...
You Might Also Like:
85 Comments
This has been very helpful. Thank you.
Thank you madam,
Thank you so much for this information
I wonder it so clear for understand and good for me. can I ask additional query?
Very insightful and useful
Good work done with clear explanations. Thank you.
Thanks so much for the write-up, it’s really good.
Thanks madam . It is very important .
thank you very good
Great presentation
This has been very well explained in simple language . It is useful even for a new researcher.
Great to hear that. Good luck with your qualitative data analysis, Pramod!
This is very useful information. And it was very a clear language structured presentation. Thanks a lot.
Thank you so much.
very informative sequential presentation
Precise explanation of method.
Hi, may we use 2 data analysis methods in our qualitative research?
Thanks for your comment. Most commonly, one would use one type of analysis method, but it depends on your research aims and objectives.
You explained it in very simple language, everyone can understand it. Thanks so much.
Thank you very much, this is very helpful. It has been explained in a very simple manner that even a layman understands
Thank nicely explained can I ask is Qualitative content analysis the same as thematic analysis?
Thanks for your comment. No, QCA and thematic are two different types of analysis. This article might help clarify – https://onlinelibrary.wiley.com/doi/10.1111/nhs.12048
This is my first time to come across a well explained data analysis. so helpful.
I have thoroughly enjoyed your explanation of the six qualitative analysis methods. This is very helpful. Thank you!
Thank you very much, this is well explained and useful
i need a citation of your book.
Thanks a lot , remarkable indeed, enlighting to the best
Hi Derek, What other theories/methods would you recommend when the data is a whole speech?
Keep writing useful artikel.
It is important concept about QDA and also the way to express is easily understandable, so thanks for all.
Thank you, this is well explained and very useful.
Very helpful .Thanks.
Hi there! Very well explained. Simple but very useful style of writing. Please provide the citation of the text. warm regards
The session was very helpful and insightful. Thank you
This was very helpful and insightful. Easy to read and understand
As a professional academic writer, this has been so informative and educative. Keep up the good work Grad Coach you are unmatched with quality content for sure.
Keep up the good work Grad Coach you are unmatched with quality content for sure.
Its Great and help me the most. A Million Thanks you Dr.
It is a very nice work
Very insightful. Please, which of this approach could be used for a research that one is trying to elicit students’ misconceptions in a particular concept ?
This is Amazing and well explained, thanks
great overview
What do we call a research data analysis method that one use to advise or determining the best accounting tool or techniques that should be adopted in a company.
Informative video, explained in a clear and simple way. Kudos
Waoo! I have chosen method wrong for my data analysis. But I can revise my work according to this guide. Thank you so much for this helpful lecture.
This has been very helpful. It gave me a good view of my research objectives and how to choose the best method. Thematic analysis it is.
Very helpful indeed. Thanku so much for the insight.
This was incredibly helpful.
Very helpful.
very educative
Nicely written especially for novice academic researchers like me! Thank you.
choosing a right method for a paper is always a hard job for a student, this is a useful information, but it would be more useful personally for me, if the author provide me with a little bit more information about the data analysis techniques in type of explanatory research. Can we use qualitative content analysis technique for explanatory research ? or what is the suitable data analysis method for explanatory research in social studies?
that was very helpful for me. because these details are so important to my research. thank you very much
I learnt a lot. Thank you
Relevant and Informative, thanks !
Well-planned and organized, thanks much! 🙂
I have reviewed qualitative data analysis in a simplest way possible. The content will highly be useful for developing my book on qualitative data analysis methods. Cheers!
Clear explanation on qualitative and how about Case study
This was helpful. Thank you
This was really of great assistance, it was just the right information needed. Explanation very clear and follow.
Wow, Thanks for making my life easy
This was helpful thanks .
Very helpful…. clear and written in an easily understandable manner. Thank you.
This was so helpful as it was easy to understand. I’m a new to research thank you so much.
so educative…. but Ijust want to know which method is coding of the qualitative or tallying done?
Thank you for the great content, I have learnt a lot. So helpful
precise and clear presentation with simple language and thank you for that.
very informative content, thank you.
You guys are amazing on YouTube on this platform. Your teachings are great, educative, and informative. kudos!
Brilliant Delivery. You made a complex subject seem so easy. Well done.
Beautifully explained.
Thanks a lot
Is there a video the captures the practical process of coding using automated applications?
Thanks for the comment. We don’t recommend using automated applications for coding, as they are not sufficiently accurate in our experience.
content analysis can be qualitative research?
THANK YOU VERY MUCH.
Thank you very much for such a wonderful content
do you have any material on Data collection
What a powerful explanation of the QDA methods. Thank you.
Great explanation both written and Video. i have been using of it on a day to day working of my thesis project in accounting and finance. Thank you very much for your support.
very helpful, thank you so much
Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly

NCRM delivers training and resources at core and advanced levels, covering quantitative, qualitative, digital, creative, visual, mixed and multimodal methods
Join our mailing list
Receive monthly updates about our latest courses and resources

Short courses
Browse our calendar of training courses and events

Featured course
12-14 June 2024

Our resources
NCRM hosts a huge range of online resources, including video tutorials and podcasts, plus an extensive publications catalogue.

Online tutorials
Access more than 80 free research methods tutorials

Resources for trainers
Browse our materials for teachers of research methods
Methods News

What is agent-based modelling and how can it be useful for public health research?

Learn about UK education datasets at free webinar

Dad-shaped holes and opportunities in data about UK children
- Privacy Policy

Home » Qualitative Research – Methods, Analysis Types and Guide
Qualitative Research – Methods, Analysis Types and Guide
Table of Contents

Qualitative Research
Qualitative research is a type of research methodology that focuses on exploring and understanding people’s beliefs, attitudes, behaviors, and experiences through the collection and analysis of non-numerical data. It seeks to answer research questions through the examination of subjective data, such as interviews, focus groups, observations, and textual analysis.
Qualitative research aims to uncover the meaning and significance of social phenomena, and it typically involves a more flexible and iterative approach to data collection and analysis compared to quantitative research. Qualitative research is often used in fields such as sociology, anthropology, psychology, and education.
Qualitative Research Methods

Qualitative Research Methods are as follows:
One-to-One Interview
This method involves conducting an interview with a single participant to gain a detailed understanding of their experiences, attitudes, and beliefs. One-to-one interviews can be conducted in-person, over the phone, or through video conferencing. The interviewer typically uses open-ended questions to encourage the participant to share their thoughts and feelings. One-to-one interviews are useful for gaining detailed insights into individual experiences.
Focus Groups
This method involves bringing together a group of people to discuss a specific topic in a structured setting. The focus group is led by a moderator who guides the discussion and encourages participants to share their thoughts and opinions. Focus groups are useful for generating ideas and insights, exploring social norms and attitudes, and understanding group dynamics.
Ethnographic Studies
This method involves immersing oneself in a culture or community to gain a deep understanding of its norms, beliefs, and practices. Ethnographic studies typically involve long-term fieldwork and observation, as well as interviews and document analysis. Ethnographic studies are useful for understanding the cultural context of social phenomena and for gaining a holistic understanding of complex social processes.
Text Analysis
This method involves analyzing written or spoken language to identify patterns and themes. Text analysis can be quantitative or qualitative. Qualitative text analysis involves close reading and interpretation of texts to identify recurring themes, concepts, and patterns. Text analysis is useful for understanding media messages, public discourse, and cultural trends.
This method involves an in-depth examination of a single person, group, or event to gain an understanding of complex phenomena. Case studies typically involve a combination of data collection methods, such as interviews, observations, and document analysis, to provide a comprehensive understanding of the case. Case studies are useful for exploring unique or rare cases, and for generating hypotheses for further research.
Process of Observation
This method involves systematically observing and recording behaviors and interactions in natural settings. The observer may take notes, use audio or video recordings, or use other methods to document what they see. Process of observation is useful for understanding social interactions, cultural practices, and the context in which behaviors occur.
Record Keeping
This method involves keeping detailed records of observations, interviews, and other data collected during the research process. Record keeping is essential for ensuring the accuracy and reliability of the data, and for providing a basis for analysis and interpretation.
This method involves collecting data from a large sample of participants through a structured questionnaire. Surveys can be conducted in person, over the phone, through mail, or online. Surveys are useful for collecting data on attitudes, beliefs, and behaviors, and for identifying patterns and trends in a population.
Qualitative data analysis is a process of turning unstructured data into meaningful insights. It involves extracting and organizing information from sources like interviews, focus groups, and surveys. The goal is to understand people’s attitudes, behaviors, and motivations
Qualitative Research Analysis Methods
Qualitative Research analysis methods involve a systematic approach to interpreting and making sense of the data collected in qualitative research. Here are some common qualitative data analysis methods:
Thematic Analysis
This method involves identifying patterns or themes in the data that are relevant to the research question. The researcher reviews the data, identifies keywords or phrases, and groups them into categories or themes. Thematic analysis is useful for identifying patterns across multiple data sources and for generating new insights into the research topic.
Content Analysis
This method involves analyzing the content of written or spoken language to identify key themes or concepts. Content analysis can be quantitative or qualitative. Qualitative content analysis involves close reading and interpretation of texts to identify recurring themes, concepts, and patterns. Content analysis is useful for identifying patterns in media messages, public discourse, and cultural trends.
Discourse Analysis
This method involves analyzing language to understand how it constructs meaning and shapes social interactions. Discourse analysis can involve a variety of methods, such as conversation analysis, critical discourse analysis, and narrative analysis. Discourse analysis is useful for understanding how language shapes social interactions, cultural norms, and power relationships.
Grounded Theory Analysis
This method involves developing a theory or explanation based on the data collected. Grounded theory analysis starts with the data and uses an iterative process of coding and analysis to identify patterns and themes in the data. The theory or explanation that emerges is grounded in the data, rather than preconceived hypotheses. Grounded theory analysis is useful for understanding complex social phenomena and for generating new theoretical insights.
Narrative Analysis
This method involves analyzing the stories or narratives that participants share to gain insights into their experiences, attitudes, and beliefs. Narrative analysis can involve a variety of methods, such as structural analysis, thematic analysis, and discourse analysis. Narrative analysis is useful for understanding how individuals construct their identities, make sense of their experiences, and communicate their values and beliefs.
Phenomenological Analysis
This method involves analyzing how individuals make sense of their experiences and the meanings they attach to them. Phenomenological analysis typically involves in-depth interviews with participants to explore their experiences in detail. Phenomenological analysis is useful for understanding subjective experiences and for developing a rich understanding of human consciousness.
Comparative Analysis
This method involves comparing and contrasting data across different cases or groups to identify similarities and differences. Comparative analysis can be used to identify patterns or themes that are common across multiple cases, as well as to identify unique or distinctive features of individual cases. Comparative analysis is useful for understanding how social phenomena vary across different contexts and groups.
Applications of Qualitative Research
Qualitative research has many applications across different fields and industries. Here are some examples of how qualitative research is used:
- Market Research: Qualitative research is often used in market research to understand consumer attitudes, behaviors, and preferences. Researchers conduct focus groups and one-on-one interviews with consumers to gather insights into their experiences and perceptions of products and services.
- Health Care: Qualitative research is used in health care to explore patient experiences and perspectives on health and illness. Researchers conduct in-depth interviews with patients and their families to gather information on their experiences with different health care providers and treatments.
- Education: Qualitative research is used in education to understand student experiences and to develop effective teaching strategies. Researchers conduct classroom observations and interviews with students and teachers to gather insights into classroom dynamics and instructional practices.
- Social Work : Qualitative research is used in social work to explore social problems and to develop interventions to address them. Researchers conduct in-depth interviews with individuals and families to understand their experiences with poverty, discrimination, and other social problems.
- Anthropology : Qualitative research is used in anthropology to understand different cultures and societies. Researchers conduct ethnographic studies and observe and interview members of different cultural groups to gain insights into their beliefs, practices, and social structures.
- Psychology : Qualitative research is used in psychology to understand human behavior and mental processes. Researchers conduct in-depth interviews with individuals to explore their thoughts, feelings, and experiences.
- Public Policy : Qualitative research is used in public policy to explore public attitudes and to inform policy decisions. Researchers conduct focus groups and one-on-one interviews with members of the public to gather insights into their perspectives on different policy issues.

How to Conduct Qualitative Research
Here are some general steps for conducting qualitative research:
- Identify your research question: Qualitative research starts with a research question or set of questions that you want to explore. This question should be focused and specific, but also broad enough to allow for exploration and discovery.
- Select your research design: There are different types of qualitative research designs, including ethnography, case study, grounded theory, and phenomenology. You should select a design that aligns with your research question and that will allow you to gather the data you need to answer your research question.
- Recruit participants: Once you have your research question and design, you need to recruit participants. The number of participants you need will depend on your research design and the scope of your research. You can recruit participants through advertisements, social media, or through personal networks.
- Collect data: There are different methods for collecting qualitative data, including interviews, focus groups, observation, and document analysis. You should select the method or methods that align with your research design and that will allow you to gather the data you need to answer your research question.
- Analyze data: Once you have collected your data, you need to analyze it. This involves reviewing your data, identifying patterns and themes, and developing codes to organize your data. You can use different software programs to help you analyze your data, or you can do it manually.
- Interpret data: Once you have analyzed your data, you need to interpret it. This involves making sense of the patterns and themes you have identified, and developing insights and conclusions that answer your research question. You should be guided by your research question and use your data to support your conclusions.
- Communicate results: Once you have interpreted your data, you need to communicate your results. This can be done through academic papers, presentations, or reports. You should be clear and concise in your communication, and use examples and quotes from your data to support your findings.
Examples of Qualitative Research
Here are some real-time examples of qualitative research:
- Customer Feedback: A company may conduct qualitative research to understand the feedback and experiences of its customers. This may involve conducting focus groups or one-on-one interviews with customers to gather insights into their attitudes, behaviors, and preferences.
- Healthcare : A healthcare provider may conduct qualitative research to explore patient experiences and perspectives on health and illness. This may involve conducting in-depth interviews with patients and their families to gather information on their experiences with different health care providers and treatments.
- Education : An educational institution may conduct qualitative research to understand student experiences and to develop effective teaching strategies. This may involve conducting classroom observations and interviews with students and teachers to gather insights into classroom dynamics and instructional practices.
- Social Work: A social worker may conduct qualitative research to explore social problems and to develop interventions to address them. This may involve conducting in-depth interviews with individuals and families to understand their experiences with poverty, discrimination, and other social problems.
- Anthropology : An anthropologist may conduct qualitative research to understand different cultures and societies. This may involve conducting ethnographic studies and observing and interviewing members of different cultural groups to gain insights into their beliefs, practices, and social structures.
- Psychology : A psychologist may conduct qualitative research to understand human behavior and mental processes. This may involve conducting in-depth interviews with individuals to explore their thoughts, feelings, and experiences.
- Public Policy: A government agency or non-profit organization may conduct qualitative research to explore public attitudes and to inform policy decisions. This may involve conducting focus groups and one-on-one interviews with members of the public to gather insights into their perspectives on different policy issues.
Purpose of Qualitative Research
The purpose of qualitative research is to explore and understand the subjective experiences, behaviors, and perspectives of individuals or groups in a particular context. Unlike quantitative research, which focuses on numerical data and statistical analysis, qualitative research aims to provide in-depth, descriptive information that can help researchers develop insights and theories about complex social phenomena.
Qualitative research can serve multiple purposes, including:
- Exploring new or emerging phenomena : Qualitative research can be useful for exploring new or emerging phenomena, such as new technologies or social trends. This type of research can help researchers develop a deeper understanding of these phenomena and identify potential areas for further study.
- Understanding complex social phenomena : Qualitative research can be useful for exploring complex social phenomena, such as cultural beliefs, social norms, or political processes. This type of research can help researchers develop a more nuanced understanding of these phenomena and identify factors that may influence them.
- Generating new theories or hypotheses: Qualitative research can be useful for generating new theories or hypotheses about social phenomena. By gathering rich, detailed data about individuals’ experiences and perspectives, researchers can develop insights that may challenge existing theories or lead to new lines of inquiry.
- Providing context for quantitative data: Qualitative research can be useful for providing context for quantitative data. By gathering qualitative data alongside quantitative data, researchers can develop a more complete understanding of complex social phenomena and identify potential explanations for quantitative findings.
When to use Qualitative Research
Here are some situations where qualitative research may be appropriate:
- Exploring a new area: If little is known about a particular topic, qualitative research can help to identify key issues, generate hypotheses, and develop new theories.
- Understanding complex phenomena: Qualitative research can be used to investigate complex social, cultural, or organizational phenomena that are difficult to measure quantitatively.
- Investigating subjective experiences: Qualitative research is particularly useful for investigating the subjective experiences of individuals or groups, such as their attitudes, beliefs, values, or emotions.
- Conducting formative research: Qualitative research can be used in the early stages of a research project to develop research questions, identify potential research participants, and refine research methods.
- Evaluating interventions or programs: Qualitative research can be used to evaluate the effectiveness of interventions or programs by collecting data on participants’ experiences, attitudes, and behaviors.
Characteristics of Qualitative Research
Qualitative research is characterized by several key features, including:
- Focus on subjective experience: Qualitative research is concerned with understanding the subjective experiences, beliefs, and perspectives of individuals or groups in a particular context. Researchers aim to explore the meanings that people attach to their experiences and to understand the social and cultural factors that shape these meanings.
- Use of open-ended questions: Qualitative research relies on open-ended questions that allow participants to provide detailed, in-depth responses. Researchers seek to elicit rich, descriptive data that can provide insights into participants’ experiences and perspectives.
- Sampling-based on purpose and diversity: Qualitative research often involves purposive sampling, in which participants are selected based on specific criteria related to the research question. Researchers may also seek to include participants with diverse experiences and perspectives to capture a range of viewpoints.
- Data collection through multiple methods: Qualitative research typically involves the use of multiple data collection methods, such as in-depth interviews, focus groups, and observation. This allows researchers to gather rich, detailed data from multiple sources, which can provide a more complete picture of participants’ experiences and perspectives.
- Inductive data analysis: Qualitative research relies on inductive data analysis, in which researchers develop theories and insights based on the data rather than testing pre-existing hypotheses. Researchers use coding and thematic analysis to identify patterns and themes in the data and to develop theories and explanations based on these patterns.
- Emphasis on researcher reflexivity: Qualitative research recognizes the importance of the researcher’s role in shaping the research process and outcomes. Researchers are encouraged to reflect on their own biases and assumptions and to be transparent about their role in the research process.
Advantages of Qualitative Research
Qualitative research offers several advantages over other research methods, including:
- Depth and detail: Qualitative research allows researchers to gather rich, detailed data that provides a deeper understanding of complex social phenomena. Through in-depth interviews, focus groups, and observation, researchers can gather detailed information about participants’ experiences and perspectives that may be missed by other research methods.
- Flexibility : Qualitative research is a flexible approach that allows researchers to adapt their methods to the research question and context. Researchers can adjust their research methods in real-time to gather more information or explore unexpected findings.
- Contextual understanding: Qualitative research is well-suited to exploring the social and cultural context in which individuals or groups are situated. Researchers can gather information about cultural norms, social structures, and historical events that may influence participants’ experiences and perspectives.
- Participant perspective : Qualitative research prioritizes the perspective of participants, allowing researchers to explore subjective experiences and understand the meanings that participants attach to their experiences.
- Theory development: Qualitative research can contribute to the development of new theories and insights about complex social phenomena. By gathering rich, detailed data and using inductive data analysis, researchers can develop new theories and explanations that may challenge existing understandings.
- Validity : Qualitative research can offer high validity by using multiple data collection methods, purposive and diverse sampling, and researcher reflexivity. This can help ensure that findings are credible and trustworthy.
Limitations of Qualitative Research
Qualitative research also has some limitations, including:
- Subjectivity : Qualitative research relies on the subjective interpretation of researchers, which can introduce bias into the research process. The researcher’s perspective, beliefs, and experiences can influence the way data is collected, analyzed, and interpreted.
- Limited generalizability: Qualitative research typically involves small, purposive samples that may not be representative of larger populations. This limits the generalizability of findings to other contexts or populations.
- Time-consuming: Qualitative research can be a time-consuming process, requiring significant resources for data collection, analysis, and interpretation.
- Resource-intensive: Qualitative research may require more resources than other research methods, including specialized training for researchers, specialized software for data analysis, and transcription services.
- Limited reliability: Qualitative research may be less reliable than quantitative research, as it relies on the subjective interpretation of researchers. This can make it difficult to replicate findings or compare results across different studies.
- Ethics and confidentiality: Qualitative research involves collecting sensitive information from participants, which raises ethical concerns about confidentiality and informed consent. Researchers must take care to protect the privacy and confidentiality of participants and obtain informed consent.
Also see Research Methods
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Survey Research – Types, Methods, Examples

Phenomenology – Methods, Examples and Guide

One-to-One Interview – Methods and Guide

Qualitative Research Methods

Observational Research – Methods and Guide

Basic Research – Types, Methods and Examples
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Korean J Anesthesiol
- v.71(2); 2018 Apr
Introduction to systematic review and meta-analysis
1 Department of Anesthesiology and Pain Medicine, Inje University Seoul Paik Hospital, Seoul, Korea
2 Department of Anesthesiology and Pain Medicine, Chung-Ang University College of Medicine, Seoul, Korea
Systematic reviews and meta-analyses present results by combining and analyzing data from different studies conducted on similar research topics. In recent years, systematic reviews and meta-analyses have been actively performed in various fields including anesthesiology. These research methods are powerful tools that can overcome the difficulties in performing large-scale randomized controlled trials. However, the inclusion of studies with any biases or improperly assessed quality of evidence in systematic reviews and meta-analyses could yield misleading results. Therefore, various guidelines have been suggested for conducting systematic reviews and meta-analyses to help standardize them and improve their quality. Nonetheless, accepting the conclusions of many studies without understanding the meta-analysis can be dangerous. Therefore, this article provides an easy introduction to clinicians on performing and understanding meta-analyses.
Introduction
A systematic review collects all possible studies related to a given topic and design, and reviews and analyzes their results [ 1 ]. During the systematic review process, the quality of studies is evaluated, and a statistical meta-analysis of the study results is conducted on the basis of their quality. A meta-analysis is a valid, objective, and scientific method of analyzing and combining different results. Usually, in order to obtain more reliable results, a meta-analysis is mainly conducted on randomized controlled trials (RCTs), which have a high level of evidence [ 2 ] ( Fig. 1 ). Since 1999, various papers have presented guidelines for reporting meta-analyses of RCTs. Following the Quality of Reporting of Meta-analyses (QUORUM) statement [ 3 ], and the appearance of registers such as Cochrane Library’s Methodology Register, a large number of systematic literature reviews have been registered. In 2009, the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [ 4 ] was published, and it greatly helped standardize and improve the quality of systematic reviews and meta-analyses [ 5 ].

Levels of evidence.
In anesthesiology, the importance of systematic reviews and meta-analyses has been highlighted, and they provide diagnostic and therapeutic value to various areas, including not only perioperative management but also intensive care and outpatient anesthesia [6–13]. Systematic reviews and meta-analyses include various topics, such as comparing various treatments of postoperative nausea and vomiting [ 14 , 15 ], comparing general anesthesia and regional anesthesia [ 16 – 18 ], comparing airway maintenance devices [ 8 , 19 ], comparing various methods of postoperative pain control (e.g., patient-controlled analgesia pumps, nerve block, or analgesics) [ 20 – 23 ], comparing the precision of various monitoring instruments [ 7 ], and meta-analysis of dose-response in various drugs [ 12 ].
Thus, literature reviews and meta-analyses are being conducted in diverse medical fields, and the aim of highlighting their importance is to help better extract accurate, good quality data from the flood of data being produced. However, a lack of understanding about systematic reviews and meta-analyses can lead to incorrect outcomes being derived from the review and analysis processes. If readers indiscriminately accept the results of the many meta-analyses that are published, incorrect data may be obtained. Therefore, in this review, we aim to describe the contents and methods used in systematic reviews and meta-analyses in a way that is easy to understand for future authors and readers of systematic review and meta-analysis.
Study Planning
It is easy to confuse systematic reviews and meta-analyses. A systematic review is an objective, reproducible method to find answers to a certain research question, by collecting all available studies related to that question and reviewing and analyzing their results. A meta-analysis differs from a systematic review in that it uses statistical methods on estimates from two or more different studies to form a pooled estimate [ 1 ]. Following a systematic review, if it is not possible to form a pooled estimate, it can be published as is without progressing to a meta-analysis; however, if it is possible to form a pooled estimate from the extracted data, a meta-analysis can be attempted. Systematic reviews and meta-analyses usually proceed according to the flowchart presented in Fig. 2 . We explain each of the stages below.

Flowchart illustrating a systematic review.
Formulating research questions
A systematic review attempts to gather all available empirical research by using clearly defined, systematic methods to obtain answers to a specific question. A meta-analysis is the statistical process of analyzing and combining results from several similar studies. Here, the definition of the word “similar” is not made clear, but when selecting a topic for the meta-analysis, it is essential to ensure that the different studies present data that can be combined. If the studies contain data on the same topic that can be combined, a meta-analysis can even be performed using data from only two studies. However, study selection via a systematic review is a precondition for performing a meta-analysis, and it is important to clearly define the Population, Intervention, Comparison, Outcomes (PICO) parameters that are central to evidence-based research. In addition, selection of the research topic is based on logical evidence, and it is important to select a topic that is familiar to readers without clearly confirmed the evidence [ 24 ].
Protocols and registration
In systematic reviews, prior registration of a detailed research plan is very important. In order to make the research process transparent, primary/secondary outcomes and methods are set in advance, and in the event of changes to the method, other researchers and readers are informed when, how, and why. Many studies are registered with an organization like PROSPERO ( http://www.crd.york.ac.uk/PROSPERO/ ), and the registration number is recorded when reporting the study, in order to share the protocol at the time of planning.
Defining inclusion and exclusion criteria
Information is included on the study design, patient characteristics, publication status (published or unpublished), language used, and research period. If there is a discrepancy between the number of patients included in the study and the number of patients included in the analysis, this needs to be clearly explained while describing the patient characteristics, to avoid confusing the reader.
Literature search and study selection
In order to secure proper basis for evidence-based research, it is essential to perform a broad search that includes as many studies as possible that meet the inclusion and exclusion criteria. Typically, the three bibliographic databases Medline, Embase, and Cochrane Central Register of Controlled Trials (CENTRAL) are used. In domestic studies, the Korean databases KoreaMed, KMBASE, and RISS4U may be included. Effort is required to identify not only published studies but also abstracts, ongoing studies, and studies awaiting publication. Among the studies retrieved in the search, the researchers remove duplicate studies, select studies that meet the inclusion/exclusion criteria based on the abstracts, and then make the final selection of studies based on their full text. In order to maintain transparency and objectivity throughout this process, study selection is conducted independently by at least two investigators. When there is a inconsistency in opinions, intervention is required via debate or by a third reviewer. The methods for this process also need to be planned in advance. It is essential to ensure the reproducibility of the literature selection process [ 25 ].
Quality of evidence
However, well planned the systematic review or meta-analysis is, if the quality of evidence in the studies is low, the quality of the meta-analysis decreases and incorrect results can be obtained [ 26 ]. Even when using randomized studies with a high quality of evidence, evaluating the quality of evidence precisely helps determine the strength of recommendations in the meta-analysis. One method of evaluating the quality of evidence in non-randomized studies is the Newcastle-Ottawa Scale, provided by the Ottawa Hospital Research Institute 1) . However, we are mostly focusing on meta-analyses that use randomized studies.
If the Grading of Recommendations, Assessment, Development and Evaluations (GRADE) system ( http://www.gradeworkinggroup.org/ ) is used, the quality of evidence is evaluated on the basis of the study limitations, inaccuracies, incompleteness of outcome data, indirectness of evidence, and risk of publication bias, and this is used to determine the strength of recommendations [ 27 ]. As shown in Table 1 , the study limitations are evaluated using the “risk of bias” method proposed by Cochrane 2) . This method classifies bias in randomized studies as “low,” “high,” or “unclear” on the basis of the presence or absence of six processes (random sequence generation, allocation concealment, blinding participants or investigators, incomplete outcome data, selective reporting, and other biases) [ 28 ].
The Cochrane Collaboration’s Tool for Assessing the Risk of Bias [ 28 ]
Data extraction
Two different investigators extract data based on the objectives and form of the study; thereafter, the extracted data are reviewed. Since the size and format of each variable are different, the size and format of the outcomes are also different, and slight changes may be required when combining the data [ 29 ]. If there are differences in the size and format of the outcome variables that cause difficulties combining the data, such as the use of different evaluation instruments or different evaluation timepoints, the analysis may be limited to a systematic review. The investigators resolve differences of opinion by debate, and if they fail to reach a consensus, a third-reviewer is consulted.
Data Analysis
The aim of a meta-analysis is to derive a conclusion with increased power and accuracy than what could not be able to achieve in individual studies. Therefore, before analysis, it is crucial to evaluate the direction of effect, size of effect, homogeneity of effects among studies, and strength of evidence [ 30 ]. Thereafter, the data are reviewed qualitatively and quantitatively. If it is determined that the different research outcomes cannot be combined, all the results and characteristics of the individual studies are displayed in a table or in a descriptive form; this is referred to as a qualitative review. A meta-analysis is a quantitative review, in which the clinical effectiveness is evaluated by calculating the weighted pooled estimate for the interventions in at least two separate studies.
The pooled estimate is the outcome of the meta-analysis, and is typically explained using a forest plot ( Figs. 3 and and4). 4 ). The black squares in the forest plot are the odds ratios (ORs) and 95% confidence intervals in each study. The area of the squares represents the weight reflected in the meta-analysis. The black diamond represents the OR and 95% confidence interval calculated across all the included studies. The bold vertical line represents a lack of therapeutic effect (OR = 1); if the confidence interval includes OR = 1, it means no significant difference was found between the treatment and control groups.

Forest plot analyzed by two different models using the same data. (A) Fixed-effect model. (B) Random-effect model. The figure depicts individual trials as filled squares with the relative sample size and the solid line as the 95% confidence interval of the difference. The diamond shape indicates the pooled estimate and uncertainty for the combined effect. The vertical line indicates the treatment group shows no effect (OR = 1). Moreover, if the confidence interval includes 1, then the result shows no evidence of difference between the treatment and control groups.

Forest plot representing homogeneous data.
Dichotomous variables and continuous variables
In data analysis, outcome variables can be considered broadly in terms of dichotomous variables and continuous variables. When combining data from continuous variables, the mean difference (MD) and standardized mean difference (SMD) are used ( Table 2 ).
Summary of Meta-analysis Methods Available in RevMan [ 28 ]
The MD is the absolute difference in mean values between the groups, and the SMD is the mean difference between groups divided by the standard deviation. When results are presented in the same units, the MD can be used, but when results are presented in different units, the SMD should be used. When the MD is used, the combined units must be shown. A value of “0” for the MD or SMD indicates that the effects of the new treatment method and the existing treatment method are the same. A value lower than “0” means the new treatment method is less effective than the existing method, and a value greater than “0” means the new treatment is more effective than the existing method.
When combining data for dichotomous variables, the OR, risk ratio (RR), or risk difference (RD) can be used. The RR and RD can be used for RCTs, quasi-experimental studies, or cohort studies, and the OR can be used for other case-control studies or cross-sectional studies. However, because the OR is difficult to interpret, using the RR and RD, if possible, is recommended. If the outcome variable is a dichotomous variable, it can be presented as the number needed to treat (NNT), which is the minimum number of patients who need to be treated in the intervention group, compared to the control group, for a given event to occur in at least one patient. Based on Table 3 , in an RCT, if x is the probability of the event occurring in the control group and y is the probability of the event occurring in the intervention group, then x = c/(c + d), y = a/(a + b), and the absolute risk reduction (ARR) = x − y. NNT can be obtained as the reciprocal, 1/ARR.
Calculation of the Number Needed to Treat in the Dichotomous table
Fixed-effect models and random-effect models
In order to analyze effect size, two types of models can be used: a fixed-effect model or a random-effect model. A fixed-effect model assumes that the effect of treatment is the same, and that variation between results in different studies is due to random error. Thus, a fixed-effect model can be used when the studies are considered to have the same design and methodology, or when the variability in results within a study is small, and the variance is thought to be due to random error. Three common methods are used for weighted estimation in a fixed-effect model: 1) inverse variance-weighted estimation 3) , 2) Mantel-Haenszel estimation 4) , and 3) Peto estimation 5) .
A random-effect model assumes heterogeneity between the studies being combined, and these models are used when the studies are assumed different, even if a heterogeneity test does not show a significant result. Unlike a fixed-effect model, a random-effect model assumes that the size of the effect of treatment differs among studies. Thus, differences in variation among studies are thought to be due to not only random error but also between-study variability in results. Therefore, weight does not decrease greatly for studies with a small number of patients. Among methods for weighted estimation in a random-effect model, the DerSimonian and Laird method 6) is mostly used for dichotomous variables, as the simplest method, while inverse variance-weighted estimation is used for continuous variables, as with fixed-effect models. These four methods are all used in Review Manager software (The Cochrane Collaboration, UK), and are described in a study by Deeks et al. [ 31 ] ( Table 2 ). However, when the number of studies included in the analysis is less than 10, the Hartung-Knapp-Sidik-Jonkman method 7) can better reduce the risk of type 1 error than does the DerSimonian and Laird method [ 32 ].
Fig. 3 shows the results of analyzing outcome data using a fixed-effect model (A) and a random-effect model (B). As shown in Fig. 3 , while the results from large studies are weighted more heavily in the fixed-effect model, studies are given relatively similar weights irrespective of study size in the random-effect model. Although identical data were being analyzed, as shown in Fig. 3 , the significant result in the fixed-effect model was no longer significant in the random-effect model. One representative example of the small study effect in a random-effect model is the meta-analysis by Li et al. [ 33 ]. In a large-scale study, intravenous injection of magnesium was unrelated to acute myocardial infarction, but in the random-effect model, which included numerous small studies, the small study effect resulted in an association being found between intravenous injection of magnesium and myocardial infarction. This small study effect can be controlled for by using a sensitivity analysis, which is performed to examine the contribution of each of the included studies to the final meta-analysis result. In particular, when heterogeneity is suspected in the study methods or results, by changing certain data or analytical methods, this method makes it possible to verify whether the changes affect the robustness of the results, and to examine the causes of such effects [ 34 ].
Heterogeneity
Homogeneity test is a method whether the degree of heterogeneity is greater than would be expected to occur naturally when the effect size calculated from several studies is higher than the sampling error. This makes it possible to test whether the effect size calculated from several studies is the same. Three types of homogeneity tests can be used: 1) forest plot, 2) Cochrane’s Q test (chi-squared), and 3) Higgins I 2 statistics. In the forest plot, as shown in Fig. 4 , greater overlap between the confidence intervals indicates greater homogeneity. For the Q statistic, when the P value of the chi-squared test, calculated from the forest plot in Fig. 4 , is less than 0.1, it is considered to show statistical heterogeneity and a random-effect can be used. Finally, I 2 can be used [ 35 ].
I 2 , calculated as shown above, returns a value between 0 and 100%. A value less than 25% is considered to show strong homogeneity, a value of 50% is average, and a value greater than 75% indicates strong heterogeneity.
Even when the data cannot be shown to be homogeneous, a fixed-effect model can be used, ignoring the heterogeneity, and all the study results can be presented individually, without combining them. However, in many cases, a random-effect model is applied, as described above, and a subgroup analysis or meta-regression analysis is performed to explain the heterogeneity. In a subgroup analysis, the data are divided into subgroups that are expected to be homogeneous, and these subgroups are analyzed. This needs to be planned in the predetermined protocol before starting the meta-analysis. A meta-regression analysis is similar to a normal regression analysis, except that the heterogeneity between studies is modeled. This process involves performing a regression analysis of the pooled estimate for covariance at the study level, and so it is usually not considered when the number of studies is less than 10. Here, univariate and multivariate regression analyses can both be considered.
Publication bias
Publication bias is the most common type of reporting bias in meta-analyses. This refers to the distortion of meta-analysis outcomes due to the higher likelihood of publication of statistically significant studies rather than non-significant studies. In order to test the presence or absence of publication bias, first, a funnel plot can be used ( Fig. 5 ). Studies are plotted on a scatter plot with effect size on the x-axis and precision or total sample size on the y-axis. If the points form an upside-down funnel shape, with a broad base that narrows towards the top of the plot, this indicates the absence of a publication bias ( Fig. 5A ) [ 29 , 36 ]. On the other hand, if the plot shows an asymmetric shape, with no points on one side of the graph, then publication bias can be suspected ( Fig. 5B ). Second, to test publication bias statistically, Begg and Mazumdar’s rank correlation test 8) [ 37 ] or Egger’s test 9) [ 29 ] can be used. If publication bias is detected, the trim-and-fill method 10) can be used to correct the bias [ 38 ]. Fig. 6 displays results that show publication bias in Egger’s test, which has then been corrected using the trim-and-fill method using Comprehensive Meta-Analysis software (Biostat, USA).

Funnel plot showing the effect size on the x-axis and sample size on the y-axis as a scatter plot. (A) Funnel plot without publication bias. The individual plots are broader at the bottom and narrower at the top. (B) Funnel plot with publication bias. The individual plots are located asymmetrically.

Funnel plot adjusted using the trim-and-fill method. White circles: comparisons included. Black circles: inputted comparisons using the trim-and-fill method. White diamond: pooled observed log risk ratio. Black diamond: pooled inputted log risk ratio.
Result Presentation
When reporting the results of a systematic review or meta-analysis, the analytical content and methods should be described in detail. First, a flowchart is displayed with the literature search and selection process according to the inclusion/exclusion criteria. Second, a table is shown with the characteristics of the included studies. A table should also be included with information related to the quality of evidence, such as GRADE ( Table 4 ). Third, the results of data analysis are shown in a forest plot and funnel plot. Fourth, if the results use dichotomous data, the NNT values can be reported, as described above.
The GRADE Evidence Quality for Each Outcome
N: number of studies, ROB: risk of bias, PON: postoperative nausea, POV: postoperative vomiting, PONV: postoperative nausea and vomiting, CI: confidence interval, RR: risk ratio, AR: absolute risk.
When Review Manager software (The Cochrane Collaboration, UK) is used for the analysis, two types of P values are given. The first is the P value from the z-test, which tests the null hypothesis that the intervention has no effect. The second P value is from the chi-squared test, which tests the null hypothesis for a lack of heterogeneity. The statistical result for the intervention effect, which is generally considered the most important result in meta-analyses, is the z-test P value.
A common mistake when reporting results is, given a z-test P value greater than 0.05, to say there was “no statistical significance” or “no difference.” When evaluating statistical significance in a meta-analysis, a P value lower than 0.05 can be explained as “a significant difference in the effects of the two treatment methods.” However, the P value may appear non-significant whether or not there is a difference between the two treatment methods. In such a situation, it is better to announce “there was no strong evidence for an effect,” and to present the P value and confidence intervals. Another common mistake is to think that a smaller P value is indicative of a more significant effect. In meta-analyses of large-scale studies, the P value is more greatly affected by the number of studies and patients included, rather than by the significance of the results; therefore, care should be taken when interpreting the results of a meta-analysis.
When performing a systematic literature review or meta-analysis, if the quality of studies is not properly evaluated or if proper methodology is not strictly applied, the results can be biased and the outcomes can be incorrect. However, when systematic reviews and meta-analyses are properly implemented, they can yield powerful results that could usually only be achieved using large-scale RCTs, which are difficult to perform in individual studies. As our understanding of evidence-based medicine increases and its importance is better appreciated, the number of systematic reviews and meta-analyses will keep increasing. However, indiscriminate acceptance of the results of all these meta-analyses can be dangerous, and hence, we recommend that their results be received critically on the basis of a more accurate understanding.
1) http://www.ohri.ca .
2) http://methods.cochrane.org/bias/assessing-risk-bias-included-studies .
3) The inverse variance-weighted estimation method is useful if the number of studies is small with large sample sizes.
4) The Mantel-Haenszel estimation method is useful if the number of studies is large with small sample sizes.
5) The Peto estimation method is useful if the event rate is low or one of the two groups shows zero incidence.
6) The most popular and simplest statistical method used in Review Manager and Comprehensive Meta-analysis software.
7) Alternative random-effect model meta-analysis that has more adequate error rates than does the common DerSimonian and Laird method, especially when the number of studies is small. However, even with the Hartung-Knapp-Sidik-Jonkman method, when there are less than five studies with very unequal sizes, extra caution is needed.
8) The Begg and Mazumdar rank correlation test uses the correlation between the ranks of effect sizes and the ranks of their variances [ 37 ].
9) The degree of funnel plot asymmetry as measured by the intercept from the regression of standard normal deviates against precision [ 29 ].
10) If there are more small studies on one side, we expect the suppression of studies on the other side. Trimming yields the adjusted effect size and reduces the variance of the effects by adding the original studies back into the analysis as a mirror image of each study.
2.2 Research Methods
Learning objectives.
By the end of this section, you should be able to:
- Recall the 6 Steps of the Scientific Method
- Differentiate between four kinds of research methods: surveys, field research, experiments, and secondary data analysis.
- Explain the appropriateness of specific research approaches for specific topics.
Sociologists examine the social world, see a problem or interesting pattern, and set out to study it. They use research methods to design a study. Planning the research design is a key step in any sociological study. Sociologists generally choose from widely used methods of social investigation: primary source data collection such as survey, participant observation, ethnography, case study, unobtrusive observations, experiment, and secondary data analysis , or use of existing sources. Every research method comes with plusses and minuses, and the topic of study strongly influences which method or methods are put to use. When you are conducting research think about the best way to gather or obtain knowledge about your topic, think of yourself as an architect. An architect needs a blueprint to build a house, as a sociologist your blueprint is your research design including your data collection method.
When entering a particular social environment, a researcher must be careful. There are times to remain anonymous and times to be overt. There are times to conduct interviews and times to simply observe. Some participants need to be thoroughly informed; others should not know they are being observed. A researcher wouldn’t stroll into a crime-ridden neighborhood at midnight, calling out, “Any gang members around?”
Making sociologists’ presence invisible is not always realistic for other reasons. That option is not available to a researcher studying prison behaviors, early education, or the Ku Klux Klan. Researchers can’t just stroll into prisons, kindergarten classrooms, or Klan meetings and unobtrusively observe behaviors or attract attention. In situations like these, other methods are needed. Researchers choose methods that best suit their study topics, protect research participants or subjects, and that fit with their overall approaches to research.
As a research method, a survey collects data from subjects who respond to a series of questions about behaviors and opinions, often in the form of a questionnaire or an interview. The survey is one of the most widely used scientific research methods. The standard survey format allows individuals a level of anonymity in which they can express personal ideas.
At some point, most people in the United States respond to some type of survey. The 2020 U.S. Census is an excellent example of a large-scale survey intended to gather sociological data. Since 1790, United States has conducted a survey consisting of six questions to received demographical data pertaining to residents. The questions pertain to the demographics of the residents who live in the United States. Currently, the Census is received by residents in the United Stated and five territories and consists of 12 questions.
Not all surveys are considered sociological research, however, and many surveys people commonly encounter focus on identifying marketing needs and strategies rather than testing a hypothesis or contributing to social science knowledge. Questions such as, “How many hot dogs do you eat in a month?” or “Were the staff helpful?” are not usually designed as scientific research. The Nielsen Ratings determine the popularity of television programming through scientific market research. However, polls conducted by television programs such as American Idol or So You Think You Can Dance cannot be generalized, because they are administered to an unrepresentative population, a specific show’s audience. You might receive polls through your cell phones or emails, from grocery stores, restaurants, and retail stores. They often provide you incentives for completing the survey.
Sociologists conduct surveys under controlled conditions for specific purposes. Surveys gather different types of information from people. While surveys are not great at capturing the ways people really behave in social situations, they are a great method for discovering how people feel, think, and act—or at least how they say they feel, think, and act. Surveys can track preferences for presidential candidates or reported individual behaviors (such as sleeping, driving, or texting habits) or information such as employment status, income, and education levels.
A survey targets a specific population , people who are the focus of a study, such as college athletes, international students, or teenagers living with type 1 (juvenile-onset) diabetes. Most researchers choose to survey a small sector of the population, or a sample , a manageable number of subjects who represent a larger population. The success of a study depends on how well a population is represented by the sample. In a random sample , every person in a population has the same chance of being chosen for the study. As a result, a Gallup Poll, if conducted as a nationwide random sampling, should be able to provide an accurate estimate of public opinion whether it contacts 2,000 or 10,000 people.
After selecting subjects, the researcher develops a specific plan to ask questions and record responses. It is important to inform subjects of the nature and purpose of the survey up front. If they agree to participate, researchers thank subjects and offer them a chance to see the results of the study if they are interested. The researcher presents the subjects with an instrument, which is a means of gathering the information.
A common instrument is a questionnaire. Subjects often answer a series of closed-ended questions . The researcher might ask yes-or-no or multiple-choice questions, allowing subjects to choose possible responses to each question. This kind of questionnaire collects quantitative data —data in numerical form that can be counted and statistically analyzed. Just count up the number of “yes” and “no” responses or correct answers, and chart them into percentages.
Questionnaires can also ask more complex questions with more complex answers—beyond “yes,” “no,” or checkbox options. These types of inquiries use open-ended questions that require short essay responses. Participants willing to take the time to write those answers might convey personal religious beliefs, political views, goals, or morals. The answers are subjective and vary from person to person. How do you plan to use your college education?
Some topics that investigate internal thought processes are impossible to observe directly and are difficult to discuss honestly in a public forum. People are more likely to share honest answers if they can respond to questions anonymously. This type of personal explanation is qualitative data —conveyed through words. Qualitative information is harder to organize and tabulate. The researcher will end up with a wide range of responses, some of which may be surprising. The benefit of written opinions, though, is the wealth of in-depth material that they provide.
An interview is a one-on-one conversation between the researcher and the subject, and it is a way of conducting surveys on a topic. However, participants are free to respond as they wish, without being limited by predetermined choices. In the back-and-forth conversation of an interview, a researcher can ask for clarification, spend more time on a subtopic, or ask additional questions. In an interview, a subject will ideally feel free to open up and answer questions that are often complex. There are no right or wrong answers. The subject might not even know how to answer the questions honestly.
Questions such as “How does society’s view of alcohol consumption influence your decision whether or not to take your first sip of alcohol?” or “Did you feel that the divorce of your parents would put a social stigma on your family?” involve so many factors that the answers are difficult to categorize. A researcher needs to avoid steering or prompting the subject to respond in a specific way; otherwise, the results will prove to be unreliable. The researcher will also benefit from gaining a subject’s trust, from empathizing or commiserating with a subject, and from listening without judgment.
Surveys often collect both quantitative and qualitative data. For example, a researcher interviewing people who are incarcerated might receive quantitative data, such as demographics – race, age, sex, that can be analyzed statistically. For example, the researcher might discover that 20 percent of incarcerated people are above the age of 50. The researcher might also collect qualitative data, such as why people take advantage of educational opportunities during their sentence and other explanatory information.
The survey can be carried out online, over the phone, by mail, or face-to-face. When researchers collect data outside a laboratory, library, or workplace setting, they are conducting field research, which is our next topic.
Field Research
The work of sociology rarely happens in limited, confined spaces. Rather, sociologists go out into the world. They meet subjects where they live, work, and play. Field research refers to gathering primary data from a natural environment. To conduct field research, the sociologist must be willing to step into new environments and observe, participate, or experience those worlds. In field work, the sociologists, rather than the subjects, are the ones out of their element.
The researcher interacts with or observes people and gathers data along the way. The key point in field research is that it takes place in the subject’s natural environment, whether it’s a coffee shop or tribal village, a homeless shelter or the DMV, a hospital, airport, mall, or beach resort.
While field research often begins in a specific setting , the study’s purpose is to observe specific behaviors in that setting. Field work is optimal for observing how people think and behave. It seeks to understand why they behave that way. However, researchers may struggle to narrow down cause and effect when there are so many variables floating around in a natural environment. And while field research looks for correlation, its small sample size does not allow for establishing a causal relationship between two variables. Indeed, much of the data gathered in sociology do not identify a cause and effect but a correlation .
Sociology in the Real World
Beyoncé and lady gaga as sociological subjects.
Sociologists have studied Lady Gaga and Beyoncé and their impact on music, movies, social media, fan participation, and social equality. In their studies, researchers have used several research methods including secondary analysis, participant observation, and surveys from concert participants.
In their study, Click, Lee & Holiday (2013) interviewed 45 Lady Gaga fans who utilized social media to communicate with the artist. These fans viewed Lady Gaga as a mirror of themselves and a source of inspiration. Like her, they embrace not being a part of mainstream culture. Many of Lady Gaga’s fans are members of the LGBTQ community. They see the “song “Born This Way” as a rallying cry and answer her calls for “Paws Up” with a physical expression of solidarity—outstretched arms and fingers bent and curled to resemble monster claws.”
Sascha Buchanan (2019) made use of participant observation to study the relationship between two fan groups, that of Beyoncé and that of Rihanna. She observed award shows sponsored by iHeartRadio, MTV EMA, and BET that pit one group against another as they competed for Best Fan Army, Biggest Fans, and FANdemonium. Buchanan argues that the media thus sustains a myth of rivalry between the two most commercially successful Black women vocal artists.
Participant Observation
In 2000, a comic writer named Rodney Rothman wanted an insider’s view of white-collar work. He slipped into the sterile, high-rise offices of a New York “dot com” agency. Every day for two weeks, he pretended to work there. His main purpose was simply to see whether anyone would notice him or challenge his presence. No one did. The receptionist greeted him. The employees smiled and said good morning. Rothman was accepted as part of the team. He even went so far as to claim a desk, inform the receptionist of his whereabouts, and attend a meeting. He published an article about his experience in The New Yorker called “My Fake Job” (2000). Later, he was discredited for allegedly fabricating some details of the story and The New Yorker issued an apology. However, Rothman’s entertaining article still offered fascinating descriptions of the inside workings of a “dot com” company and exemplified the lengths to which a writer, or a sociologist, will go to uncover material.
Rothman had conducted a form of study called participant observation , in which researchers join people and participate in a group’s routine activities for the purpose of observing them within that context. This method lets researchers experience a specific aspect of social life. A researcher might go to great lengths to get a firsthand look into a trend, institution, or behavior. A researcher might work as a waitress in a diner, experience homelessness for several weeks, or ride along with police officers as they patrol their regular beat. Often, these researchers try to blend in seamlessly with the population they study, and they may not disclose their true identity or purpose if they feel it would compromise the results of their research.
At the beginning of a field study, researchers might have a question: “What really goes on in the kitchen of the most popular diner on campus?” or “What is it like to be homeless?” Participant observation is a useful method if the researcher wants to explore a certain environment from the inside.
Field researchers simply want to observe and learn. In such a setting, the researcher will be alert and open minded to whatever happens, recording all observations accurately. Soon, as patterns emerge, questions will become more specific, observations will lead to hypotheses, and hypotheses will guide the researcher in analyzing data and generating results.
In a study of small towns in the United States conducted by sociological researchers John S. Lynd and Helen Merrell Lynd, the team altered their purpose as they gathered data. They initially planned to focus their study on the role of religion in U.S. towns. As they gathered observations, they realized that the effect of industrialization and urbanization was the more relevant topic of this social group. The Lynds did not change their methods, but they revised the purpose of their study.
This shaped the structure of Middletown: A Study in Modern American Culture , their published results (Lynd & Lynd, 1929).
The Lynds were upfront about their mission. The townspeople of Muncie, Indiana, knew why the researchers were in their midst. But some sociologists prefer not to alert people to their presence. The main advantage of covert participant observation is that it allows the researcher access to authentic, natural behaviors of a group’s members. The challenge, however, is gaining access to a setting without disrupting the pattern of others’ behavior. Becoming an inside member of a group, organization, or subculture takes time and effort. Researchers must pretend to be something they are not. The process could involve role playing, making contacts, networking, or applying for a job.
Once inside a group, some researchers spend months or even years pretending to be one of the people they are observing. However, as observers, they cannot get too involved. They must keep their purpose in mind and apply the sociological perspective. That way, they illuminate social patterns that are often unrecognized. Because information gathered during participant observation is mostly qualitative, rather than quantitative, the end results are often descriptive or interpretive. The researcher might present findings in an article or book and describe what he or she witnessed and experienced.
This type of research is what journalist Barbara Ehrenreich conducted for her book Nickel and Dimed . One day over lunch with her editor, Ehrenreich mentioned an idea. How can people exist on minimum-wage work? How do low-income workers get by? she wondered. Someone should do a study . To her surprise, her editor responded, Why don’t you do it?
That’s how Ehrenreich found herself joining the ranks of the working class. For several months, she left her comfortable home and lived and worked among people who lacked, for the most part, higher education and marketable job skills. Undercover, she applied for and worked minimum wage jobs as a waitress, a cleaning woman, a nursing home aide, and a retail chain employee. During her participant observation, she used only her income from those jobs to pay for food, clothing, transportation, and shelter.
She discovered the obvious, that it’s almost impossible to get by on minimum wage work. She also experienced and observed attitudes many middle and upper-class people never think about. She witnessed firsthand the treatment of working class employees. She saw the extreme measures people take to make ends meet and to survive. She described fellow employees who held two or three jobs, worked seven days a week, lived in cars, could not pay to treat chronic health conditions, got randomly fired, submitted to drug tests, and moved in and out of homeless shelters. She brought aspects of that life to light, describing difficult working conditions and the poor treatment that low-wage workers suffer.
The book she wrote upon her return to her real life as a well-paid writer, has been widely read and used in many college classrooms.
Ethnography
Ethnography is the immersion of the researcher in the natural setting of an entire social community to observe and experience their everyday life and culture. The heart of an ethnographic study focuses on how subjects view their own social standing and how they understand themselves in relation to a social group.
An ethnographic study might observe, for example, a small U.S. fishing town, an Inuit community, a village in Thailand, a Buddhist monastery, a private boarding school, or an amusement park. These places all have borders. People live, work, study, or vacation within those borders. People are there for a certain reason and therefore behave in certain ways and respect certain cultural norms. An ethnographer would commit to spending a determined amount of time studying every aspect of the chosen place, taking in as much as possible.
A sociologist studying a tribe in the Amazon might watch the way villagers go about their daily lives and then write a paper about it. To observe a spiritual retreat center, an ethnographer might sign up for a retreat and attend as a guest for an extended stay, observe and record data, and collate the material into results.
Institutional Ethnography
Institutional ethnography is an extension of basic ethnographic research principles that focuses intentionally on everyday concrete social relationships. Developed by Canadian sociologist Dorothy E. Smith (1990), institutional ethnography is often considered a feminist-inspired approach to social analysis and primarily considers women’s experiences within male- dominated societies and power structures. Smith’s work is seen to challenge sociology’s exclusion of women, both academically and in the study of women’s lives (Fenstermaker, n.d.).
Historically, social science research tended to objectify women and ignore their experiences except as viewed from the male perspective. Modern feminists note that describing women, and other marginalized groups, as subordinates helps those in authority maintain their own dominant positions (Social Sciences and Humanities Research Council of Canada n.d.). Smith’s three major works explored what she called “the conceptual practices of power” and are still considered seminal works in feminist theory and ethnography (Fensternmaker n.d.).
Sociological Research
The making of middletown: a study in modern u.s. culture.
In 1924, a young married couple named Robert and Helen Lynd undertook an unprecedented ethnography: to apply sociological methods to the study of one U.S. city in order to discover what “ordinary” people in the United States did and believed. Choosing Muncie, Indiana (population about 30,000) as their subject, they moved to the small town and lived there for eighteen months.
Ethnographers had been examining other cultures for decades—groups considered minorities or outsiders—like gangs, immigrants, and the poor. But no one had studied the so-called average American.
Recording interviews and using surveys to gather data, the Lynds objectively described what they observed. Researching existing sources, they compared Muncie in 1890 to the Muncie they observed in 1924. Most Muncie adults, they found, had grown up on farms but now lived in homes inside the city. As a result, the Lynds focused their study on the impact of industrialization and urbanization.
They observed that Muncie was divided into business and working class groups. They defined business class as dealing with abstract concepts and symbols, while working class people used tools to create concrete objects. The two classes led different lives with different goals and hopes. However, the Lynds observed, mass production offered both classes the same amenities. Like wealthy families, the working class was now able to own radios, cars, washing machines, telephones, vacuum cleaners, and refrigerators. This was an emerging material reality of the 1920s.
As the Lynds worked, they divided their manuscript into six chapters: Getting a Living, Making a Home, Training the Young, Using Leisure, Engaging in Religious Practices, and Engaging in Community Activities.
When the study was completed, the Lynds encountered a big problem. The Rockefeller Foundation, which had commissioned the book, claimed it was useless and refused to publish it. The Lynds asked if they could seek a publisher themselves.
Middletown: A Study in Modern American Culture was not only published in 1929 but also became an instant bestseller, a status unheard of for a sociological study. The book sold out six printings in its first year of publication, and has never gone out of print (Caplow, Hicks, & Wattenberg. 2000).
Nothing like it had ever been done before. Middletown was reviewed on the front page of the New York Times. Readers in the 1920s and 1930s identified with the citizens of Muncie, Indiana, but they were equally fascinated by the sociological methods and the use of scientific data to define ordinary people in the United States. The book was proof that social data was important—and interesting—to the U.S. public.
Sometimes a researcher wants to study one specific person or event. A case study is an in-depth analysis of a single event, situation, or individual. To conduct a case study, a researcher examines existing sources like documents and archival records, conducts interviews, engages in direct observation and even participant observation, if possible.
Researchers might use this method to study a single case of a foster child, drug lord, cancer patient, criminal, or rape victim. However, a major criticism of the case study as a method is that while offering depth on a topic, it does not provide enough evidence to form a generalized conclusion. In other words, it is difficult to make universal claims based on just one person, since one person does not verify a pattern. This is why most sociologists do not use case studies as a primary research method.
However, case studies are useful when the single case is unique. In these instances, a single case study can contribute tremendous insight. For example, a feral child, also called “wild child,” is one who grows up isolated from human beings. Feral children grow up without social contact and language, which are elements crucial to a “civilized” child’s development. These children mimic the behaviors and movements of animals, and often invent their own language. There are only about one hundred cases of “feral children” in the world.
As you may imagine, a feral child is a subject of great interest to researchers. Feral children provide unique information about child development because they have grown up outside of the parameters of “normal” growth and nurturing. And since there are very few feral children, the case study is the most appropriate method for researchers to use in studying the subject.
At age three, a Ukranian girl named Oxana Malaya suffered severe parental neglect. She lived in a shed with dogs, and she ate raw meat and scraps. Five years later, a neighbor called authorities and reported seeing a girl who ran on all fours, barking. Officials brought Oxana into society, where she was cared for and taught some human behaviors, but she never became fully socialized. She has been designated as unable to support herself and now lives in a mental institution (Grice 2011). Case studies like this offer a way for sociologists to collect data that may not be obtained by any other method.
Experiments
You have probably tested some of your own personal social theories. “If I study at night and review in the morning, I’ll improve my retention skills.” Or, “If I stop drinking soda, I’ll feel better.” Cause and effect. If this, then that. When you test the theory, your results either prove or disprove your hypothesis.
One way researchers test social theories is by conducting an experiment , meaning they investigate relationships to test a hypothesis—a scientific approach.
There are two main types of experiments: lab-based experiments and natural or field experiments. In a lab setting, the research can be controlled so that more data can be recorded in a limited amount of time. In a natural or field- based experiment, the time it takes to gather the data cannot be controlled but the information might be considered more accurate since it was collected without interference or intervention by the researcher.
As a research method, either type of sociological experiment is useful for testing if-then statements: if a particular thing happens (cause), then another particular thing will result (effect). To set up a lab-based experiment, sociologists create artificial situations that allow them to manipulate variables.
Classically, the sociologist selects a set of people with similar characteristics, such as age, class, race, or education. Those people are divided into two groups. One is the experimental group and the other is the control group. The experimental group is exposed to the independent variable(s) and the control group is not. To test the benefits of tutoring, for example, the sociologist might provide tutoring to the experimental group of students but not to the control group. Then both groups would be tested for differences in performance to see if tutoring had an effect on the experimental group of students. As you can imagine, in a case like this, the researcher would not want to jeopardize the accomplishments of either group of students, so the setting would be somewhat artificial. The test would not be for a grade reflected on their permanent record of a student, for example.
And if a researcher told the students they would be observed as part of a study on measuring the effectiveness of tutoring, the students might not behave naturally. This is called the Hawthorne effect —which occurs when people change their behavior because they know they are being watched as part of a study. The Hawthorne effect is unavoidable in some research studies because sociologists have to make the purpose of the study known. Subjects must be aware that they are being observed, and a certain amount of artificiality may result (Sonnenfeld 1985).
A real-life example will help illustrate the process. In 1971, Frances Heussenstamm, a sociology professor at California State University at Los Angeles, had a theory about police prejudice. To test her theory, she conducted research. She chose fifteen students from three ethnic backgrounds: Black, White, and Hispanic. She chose students who routinely drove to and from campus along Los Angeles freeway routes, and who had had perfect driving records for longer than a year.
Next, she placed a Black Panther bumper sticker on each car. That sticker, a representation of a social value, was the independent variable. In the 1970s, the Black Panthers were a revolutionary group actively fighting racism. Heussenstamm asked the students to follow their normal driving patterns. She wanted to see whether seeming support for the Black Panthers would change how these good drivers were treated by the police patrolling the highways. The dependent variable would be the number of traffic stops/citations.
The first arrest, for an incorrect lane change, was made two hours after the experiment began. One participant was pulled over three times in three days. He quit the study. After seventeen days, the fifteen drivers had collected a total of thirty-three traffic citations. The research was halted. The funding to pay traffic fines had run out, and so had the enthusiasm of the participants (Heussenstamm, 1971).
Secondary Data Analysis
While sociologists often engage in original research studies, they also contribute knowledge to the discipline through secondary data analysis . Secondary data does not result from firsthand research collected from primary sources, but are the already completed work of other researchers or data collected by an agency or organization. Sociologists might study works written by historians, economists, teachers, or early sociologists. They might search through periodicals, newspapers, or magazines, or organizational data from any period in history.
Using available information not only saves time and money but can also add depth to a study. Sociologists often interpret findings in a new way, a way that was not part of an author’s original purpose or intention. To study how women were encouraged to act and behave in the 1960s, for example, a researcher might watch movies, televisions shows, and situation comedies from that period. Or to research changes in behavior and attitudes due to the emergence of television in the late 1950s and early 1960s, a sociologist would rely on new interpretations of secondary data. Decades from now, researchers will most likely conduct similar studies on the advent of mobile phones, the Internet, or social media.
Social scientists also learn by analyzing the research of a variety of agencies. Governmental departments and global groups, like the U.S. Bureau of Labor Statistics or the World Health Organization (WHO), publish studies with findings that are useful to sociologists. A public statistic like the foreclosure rate might be useful for studying the effects of a recession. A racial demographic profile might be compared with data on education funding to examine the resources accessible by different groups.
One of the advantages of secondary data like old movies or WHO statistics is that it is nonreactive research (or unobtrusive research), meaning that it does not involve direct contact with subjects and will not alter or influence people’s behaviors. Unlike studies requiring direct contact with people, using previously published data does not require entering a population and the investment and risks inherent in that research process.
Using available data does have its challenges. Public records are not always easy to access. A researcher will need to do some legwork to track them down and gain access to records. To guide the search through a vast library of materials and avoid wasting time reading unrelated sources, sociologists employ content analysis , applying a systematic approach to record and value information gleaned from secondary data as they relate to the study at hand.
Also, in some cases, there is no way to verify the accuracy of existing data. It is easy to count how many drunk drivers, for example, are pulled over by the police. But how many are not? While it’s possible to discover the percentage of teenage students who drop out of high school, it might be more challenging to determine the number who return to school or get their GED later.
Another problem arises when data are unavailable in the exact form needed or do not survey the topic from the precise angle the researcher seeks. For example, the average salaries paid to professors at a public school is public record. But these figures do not necessarily reveal how long it took each professor to reach the salary range, what their educational backgrounds are, or how long they’ve been teaching.
When conducting content analysis, it is important to consider the date of publication of an existing source and to take into account attitudes and common cultural ideals that may have influenced the research. For example, when Robert S. Lynd and Helen Merrell Lynd gathered research in the 1920s, attitudes and cultural norms were vastly different then than they are now. Beliefs about gender roles, race, education, and work have changed significantly since then. At the time, the study’s purpose was to reveal insights about small U.S. communities. Today, it is an illustration of 1920s attitudes and values.
As an Amazon Associate we earn from qualifying purchases.
This book may not be used in the training of large language models or otherwise be ingested into large language models or generative AI offerings without OpenStax's permission.
Want to cite, share, or modify this book? This book uses the Creative Commons Attribution License and you must attribute OpenStax.
Access for free at https://openstax.org/books/introduction-sociology-3e/pages/1-introduction
- Authors: Tonja R. Conerly, Kathleen Holmes, Asha Lal Tamang
- Publisher/website: OpenStax
- Book title: Introduction to Sociology 3e
- Publication date: Jun 3, 2021
- Location: Houston, Texas
- Book URL: https://openstax.org/books/introduction-sociology-3e/pages/1-introduction
- Section URL: https://openstax.org/books/introduction-sociology-3e/pages/2-2-research-methods
© Jan 18, 2024 OpenStax. Textbook content produced by OpenStax is licensed under a Creative Commons Attribution License . The OpenStax name, OpenStax logo, OpenStax book covers, OpenStax CNX name, and OpenStax CNX logo are not subject to the Creative Commons license and may not be reproduced without the prior and express written consent of Rice University.
Research Methods In Psychology
Saul Mcleod, PhD
Editor-in-Chief for Simply Psychology
BSc (Hons) Psychology, MRes, PhD, University of Manchester
Saul Mcleod, PhD., is a qualified psychology teacher with over 18 years of experience in further and higher education. He has been published in peer-reviewed journals, including the Journal of Clinical Psychology.
Learn about our Editorial Process
Olivia Guy-Evans, MSc
Associate Editor for Simply Psychology
BSc (Hons) Psychology, MSc Psychology of Education
Olivia Guy-Evans is a writer and associate editor for Simply Psychology. She has previously worked in healthcare and educational sectors.
Research methods in psychology are systematic procedures used to observe, describe, predict, and explain behavior and mental processes. They include experiments, surveys, case studies, and naturalistic observations, ensuring data collection is objective and reliable to understand and explain psychological phenomena.

Hypotheses are statements about the prediction of the results, that can be verified or disproved by some investigation.
There are four types of hypotheses :
- Null Hypotheses (H0 ) – these predict that no difference will be found in the results between the conditions. Typically these are written ‘There will be no difference…’
- Alternative Hypotheses (Ha or H1) – these predict that there will be a significant difference in the results between the two conditions. This is also known as the experimental hypothesis.
- One-tailed (directional) hypotheses – these state the specific direction the researcher expects the results to move in, e.g. higher, lower, more, less. In a correlation study, the predicted direction of the correlation can be either positive or negative.
- Two-tailed (non-directional) hypotheses – these state that a difference will be found between the conditions of the independent variable but does not state the direction of a difference or relationship. Typically these are always written ‘There will be a difference ….’
All research has an alternative hypothesis (either a one-tailed or two-tailed) and a corresponding null hypothesis.
Once the research is conducted and results are found, psychologists must accept one hypothesis and reject the other.
So, if a difference is found, the Psychologist would accept the alternative hypothesis and reject the null. The opposite applies if no difference is found.
Sampling techniques
Sampling is the process of selecting a representative group from the population under study.
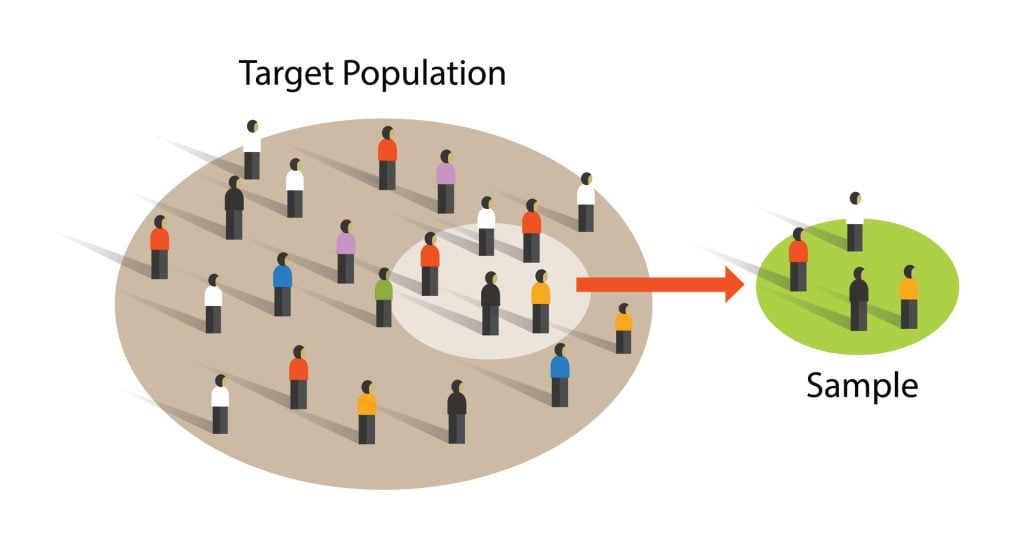
A sample is the participants you select from a target population (the group you are interested in) to make generalizations about.
Representative means the extent to which a sample mirrors a researcher’s target population and reflects its characteristics.
Generalisability means the extent to which their findings can be applied to the larger population of which their sample was a part.
- Volunteer sample : where participants pick themselves through newspaper adverts, noticeboards or online.
- Opportunity sampling : also known as convenience sampling , uses people who are available at the time the study is carried out and willing to take part. It is based on convenience.
- Random sampling : when every person in the target population has an equal chance of being selected. An example of random sampling would be picking names out of a hat.
- Systematic sampling : when a system is used to select participants. Picking every Nth person from all possible participants. N = the number of people in the research population / the number of people needed for the sample.
- Stratified sampling : when you identify the subgroups and select participants in proportion to their occurrences.
- Snowball sampling : when researchers find a few participants, and then ask them to find participants themselves and so on.
- Quota sampling : when researchers will be told to ensure the sample fits certain quotas, for example they might be told to find 90 participants, with 30 of them being unemployed.
Experiments always have an independent and dependent variable .
- The independent variable is the one the experimenter manipulates (the thing that changes between the conditions the participants are placed into). It is assumed to have a direct effect on the dependent variable.
- The dependent variable is the thing being measured, or the results of the experiment.
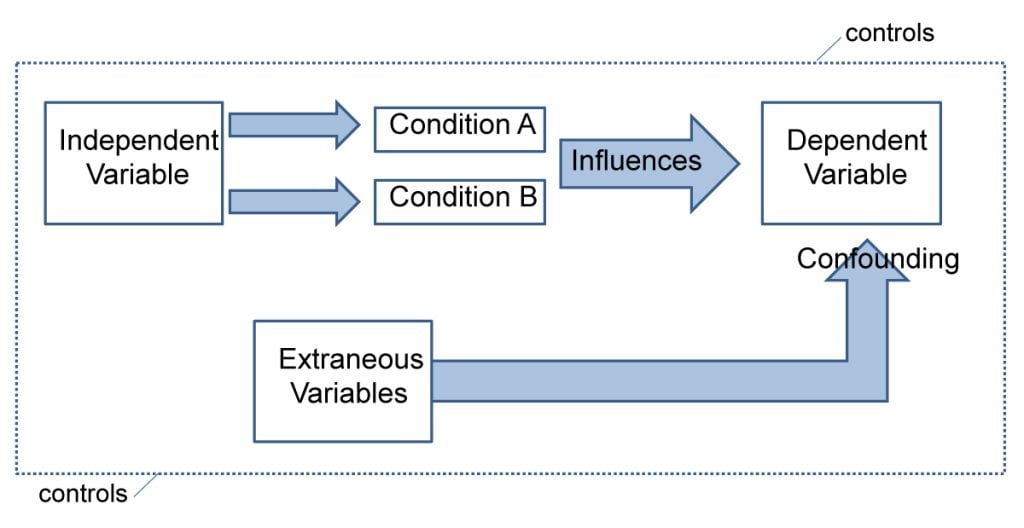
Operationalization of variables means making them measurable/quantifiable. We must use operationalization to ensure that variables are in a form that can be easily tested.
For instance, we can’t really measure ‘happiness’, but we can measure how many times a person smiles within a two-hour period.
By operationalizing variables, we make it easy for someone else to replicate our research. Remember, this is important because we can check if our findings are reliable.
Extraneous variables are all variables which are not independent variable but could affect the results of the experiment.
It can be a natural characteristic of the participant, such as intelligence levels, gender, or age for example, or it could be a situational feature of the environment such as lighting or noise.
Demand characteristics are a type of extraneous variable that occurs if the participants work out the aims of the research study, they may begin to behave in a certain way.
For example, in Milgram’s research , critics argued that participants worked out that the shocks were not real and they administered them as they thought this was what was required of them.
Extraneous variables must be controlled so that they do not affect (confound) the results.
Randomly allocating participants to their conditions or using a matched pairs experimental design can help to reduce participant variables.
Situational variables are controlled by using standardized procedures, ensuring every participant in a given condition is treated in the same way
Experimental Design
Experimental design refers to how participants are allocated to each condition of the independent variable, such as a control or experimental group.
- Independent design ( between-groups design ): each participant is selected for only one group. With the independent design, the most common way of deciding which participants go into which group is by means of randomization.
- Matched participants design : each participant is selected for only one group, but the participants in the two groups are matched for some relevant factor or factors (e.g. ability; sex; age).
- Repeated measures design ( within groups) : each participant appears in both groups, so that there are exactly the same participants in each group.
- The main problem with the repeated measures design is that there may well be order effects. Their experiences during the experiment may change the participants in various ways.
- They may perform better when they appear in the second group because they have gained useful information about the experiment or about the task. On the other hand, they may perform less well on the second occasion because of tiredness or boredom.
- Counterbalancing is the best way of preventing order effects from disrupting the findings of an experiment, and involves ensuring that each condition is equally likely to be used first and second by the participants.
If we wish to compare two groups with respect to a given independent variable, it is essential to make sure that the two groups do not differ in any other important way.
Experimental Methods
All experimental methods involve an iv (independent variable) and dv (dependent variable)..
- Field experiments are conducted in the everyday (natural) environment of the participants. The experimenter still manipulates the IV, but in a real-life setting. It may be possible to control extraneous variables, though such control is more difficult than in a lab experiment.
- Natural experiments are when a naturally occurring IV is investigated that isn’t deliberately manipulated, it exists anyway. Participants are not randomly allocated, and the natural event may only occur rarely.
Case studies are in-depth investigations of a person, group, event, or community. It uses information from a range of sources, such as from the person concerned and also from their family and friends.
Many techniques may be used such as interviews, psychological tests, observations and experiments. Case studies are generally longitudinal: in other words, they follow the individual or group over an extended period of time.
Case studies are widely used in psychology and among the best-known ones carried out were by Sigmund Freud . He conducted very detailed investigations into the private lives of his patients in an attempt to both understand and help them overcome their illnesses.
Case studies provide rich qualitative data and have high levels of ecological validity. However, it is difficult to generalize from individual cases as each one has unique characteristics.
Correlational Studies
Correlation means association; it is a measure of the extent to which two variables are related. One of the variables can be regarded as the predictor variable with the other one as the outcome variable.
Correlational studies typically involve obtaining two different measures from a group of participants, and then assessing the degree of association between the measures.
The predictor variable can be seen as occurring before the outcome variable in some sense. It is called the predictor variable, because it forms the basis for predicting the value of the outcome variable.
Relationships between variables can be displayed on a graph or as a numerical score called a correlation coefficient.
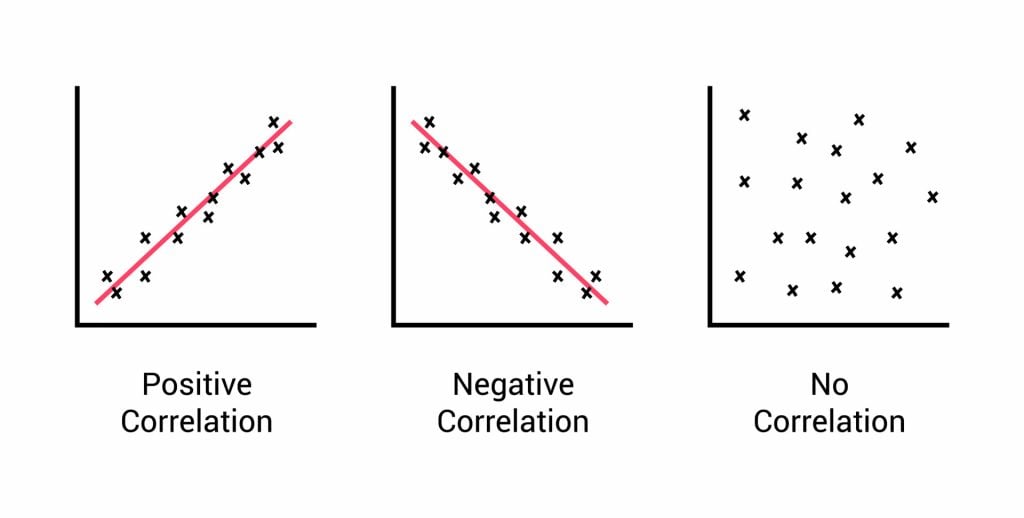
- If an increase in one variable tends to be associated with an increase in the other, then this is known as a positive correlation .
- If an increase in one variable tends to be associated with a decrease in the other, then this is known as a negative correlation .
- A zero correlation occurs when there is no relationship between variables.
After looking at the scattergraph, if we want to be sure that a significant relationship does exist between the two variables, a statistical test of correlation can be conducted, such as Spearman’s rho.
The test will give us a score, called a correlation coefficient . This is a value between 0 and 1, and the closer to 1 the score is, the stronger the relationship between the variables. This value can be both positive e.g. 0.63, or negative -0.63.
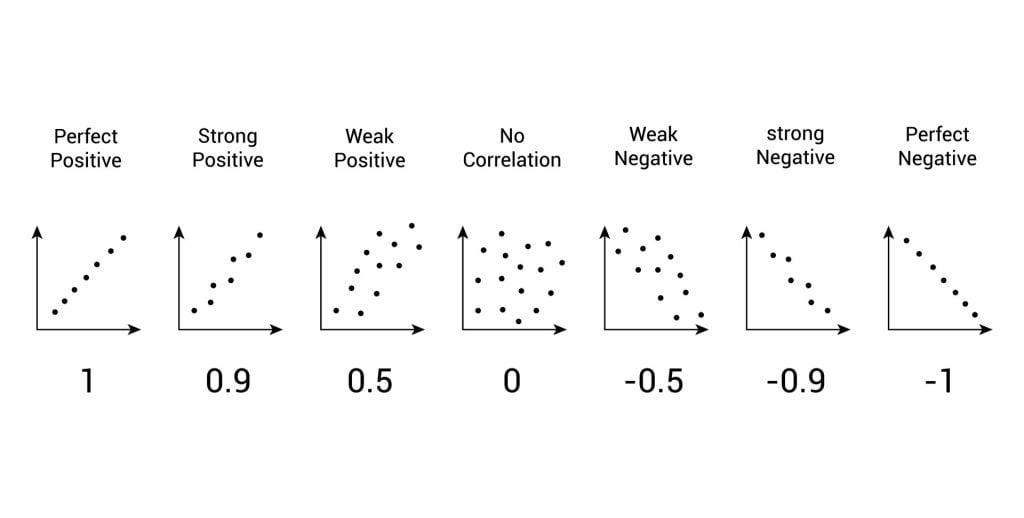
A correlation between variables, however, does not automatically mean that the change in one variable is the cause of the change in the values of the other variable. A correlation only shows if there is a relationship between variables.
Correlation does not always prove causation, as a third variable may be involved.
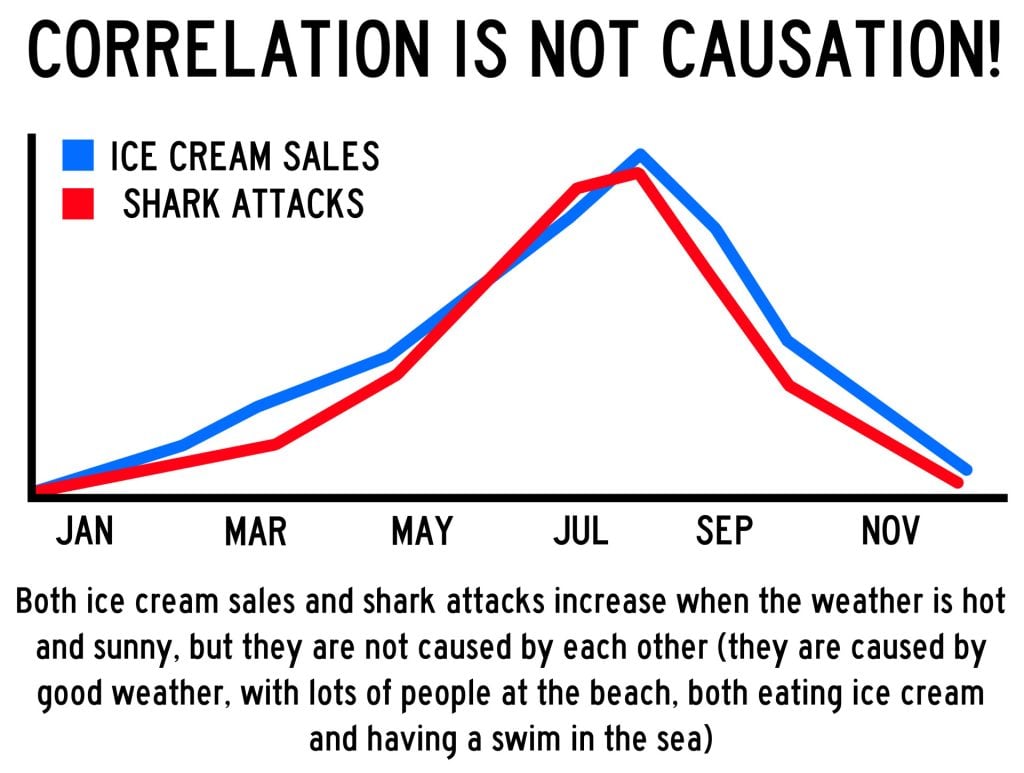
Interview Methods
Interviews are commonly divided into two types: structured and unstructured.
A fixed, predetermined set of questions is put to every participant in the same order and in the same way.
Responses are recorded on a questionnaire, and the researcher presets the order and wording of questions, and sometimes the range of alternative answers.
The interviewer stays within their role and maintains social distance from the interviewee.
There are no set questions, and the participant can raise whatever topics he/she feels are relevant and ask them in their own way. Questions are posed about participants’ answers to the subject
Unstructured interviews are most useful in qualitative research to analyze attitudes and values.
Though they rarely provide a valid basis for generalization, their main advantage is that they enable the researcher to probe social actors’ subjective point of view.
Questionnaire Method
Questionnaires can be thought of as a kind of written interview. They can be carried out face to face, by telephone, or post.
The choice of questions is important because of the need to avoid bias or ambiguity in the questions, ‘leading’ the respondent or causing offense.
- Open questions are designed to encourage a full, meaningful answer using the subject’s own knowledge and feelings. They provide insights into feelings, opinions, and understanding. Example: “How do you feel about that situation?”
- Closed questions can be answered with a simple “yes” or “no” or specific information, limiting the depth of response. They are useful for gathering specific facts or confirming details. Example: “Do you feel anxious in crowds?”
Its other practical advantages are that it is cheaper than face-to-face interviews and can be used to contact many respondents scattered over a wide area relatively quickly.
Observations
There are different types of observation methods :
- Covert observation is where the researcher doesn’t tell the participants they are being observed until after the study is complete. There could be ethical problems or deception and consent with this particular observation method.
- Overt observation is where a researcher tells the participants they are being observed and what they are being observed for.
- Controlled : behavior is observed under controlled laboratory conditions (e.g., Bandura’s Bobo doll study).
- Natural : Here, spontaneous behavior is recorded in a natural setting.
- Participant : Here, the observer has direct contact with the group of people they are observing. The researcher becomes a member of the group they are researching.
- Non-participant (aka “fly on the wall): The researcher does not have direct contact with the people being observed. The observation of participants’ behavior is from a distance
Pilot Study
A pilot study is a small scale preliminary study conducted in order to evaluate the feasibility of the key s teps in a future, full-scale project.
A pilot study is an initial run-through of the procedures to be used in an investigation; it involves selecting a few people and trying out the study on them. It is possible to save time, and in some cases, money, by identifying any flaws in the procedures designed by the researcher.
A pilot study can help the researcher spot any ambiguities (i.e. unusual things) or confusion in the information given to participants or problems with the task devised.
Sometimes the task is too hard, and the researcher may get a floor effect, because none of the participants can score at all or can complete the task – all performances are low.
The opposite effect is a ceiling effect, when the task is so easy that all achieve virtually full marks or top performances and are “hitting the ceiling”.
Research Design
In cross-sectional research , a researcher compares multiple segments of the population at the same time
Sometimes, we want to see how people change over time, as in studies of human development and lifespan. Longitudinal research is a research design in which data-gathering is administered repeatedly over an extended period of time.
In cohort studies , the participants must share a common factor or characteristic such as age, demographic, or occupation. A cohort study is a type of longitudinal study in which researchers monitor and observe a chosen population over an extended period.
Triangulation means using more than one research method to improve the study’s validity.
Reliability
Reliability is a measure of consistency, if a particular measurement is repeated and the same result is obtained then it is described as being reliable.
- Test-retest reliability : assessing the same person on two different occasions which shows the extent to which the test produces the same answers.
- Inter-observer reliability : the extent to which there is an agreement between two or more observers.
Meta-Analysis
A meta-analysis is a systematic review that involves identifying an aim and then searching for research studies that have addressed similar aims/hypotheses.
This is done by looking through various databases, and then decisions are made about what studies are to be included/excluded.
Strengths: Increases the conclusions’ validity as they’re based on a wider range.
Weaknesses: Research designs in studies can vary, so they are not truly comparable.
Peer Review
A researcher submits an article to a journal. The choice of the journal may be determined by the journal’s audience or prestige.
The journal selects two or more appropriate experts (psychologists working in a similar field) to peer review the article without payment. The peer reviewers assess: the methods and designs used, originality of the findings, the validity of the original research findings and its content, structure and language.
Feedback from the reviewer determines whether the article is accepted. The article may be: Accepted as it is, accepted with revisions, sent back to the author to revise and re-submit or rejected without the possibility of submission.
The editor makes the final decision whether to accept or reject the research report based on the reviewers comments/ recommendations.
Peer review is important because it prevent faulty data from entering the public domain, it provides a way of checking the validity of findings and the quality of the methodology and is used to assess the research rating of university departments.
Peer reviews may be an ideal, whereas in practice there are lots of problems. For example, it slows publication down and may prevent unusual, new work being published. Some reviewers might use it as an opportunity to prevent competing researchers from publishing work.
Some people doubt whether peer review can really prevent the publication of fraudulent research.
The advent of the internet means that a lot of research and academic comment is being published without official peer reviews than before, though systems are evolving on the internet where everyone really has a chance to offer their opinions and police the quality of research.
Types of Data
- Quantitative data is numerical data e.g. reaction time or number of mistakes. It represents how much or how long, how many there are of something. A tally of behavioral categories and closed questions in a questionnaire collect quantitative data.
- Qualitative data is virtually any type of information that can be observed and recorded that is not numerical in nature and can be in the form of written or verbal communication. Open questions in questionnaires and accounts from observational studies collect qualitative data.
- Primary data is first-hand data collected for the purpose of the investigation.
- Secondary data is information that has been collected by someone other than the person who is conducting the research e.g. taken from journals, books or articles.
Validity means how well a piece of research actually measures what it sets out to, or how well it reflects the reality it claims to represent.
Validity is whether the observed effect is genuine and represents what is actually out there in the world.
- Concurrent validity is the extent to which a psychological measure relates to an existing similar measure and obtains close results. For example, a new intelligence test compared to an established test.
- Face validity : does the test measure what it’s supposed to measure ‘on the face of it’. This is done by ‘eyeballing’ the measuring or by passing it to an expert to check.
- Ecological validit y is the extent to which findings from a research study can be generalized to other settings / real life.
- Temporal validity is the extent to which findings from a research study can be generalized to other historical times.
Features of Science
- Paradigm – A set of shared assumptions and agreed methods within a scientific discipline.
- Paradigm shift – The result of the scientific revolution: a significant change in the dominant unifying theory within a scientific discipline.
- Objectivity – When all sources of personal bias are minimised so not to distort or influence the research process.
- Empirical method – Scientific approaches that are based on the gathering of evidence through direct observation and experience.
- Replicability – The extent to which scientific procedures and findings can be repeated by other researchers.
- Falsifiability – The principle that a theory cannot be considered scientific unless it admits the possibility of being proved untrue.
Statistical Testing
A significant result is one where there is a low probability that chance factors were responsible for any observed difference, correlation, or association in the variables tested.
If our test is significant, we can reject our null hypothesis and accept our alternative hypothesis.
If our test is not significant, we can accept our null hypothesis and reject our alternative hypothesis. A null hypothesis is a statement of no effect.
In Psychology, we use p < 0.05 (as it strikes a balance between making a type I and II error) but p < 0.01 is used in tests that could cause harm like introducing a new drug.
A type I error is when the null hypothesis is rejected when it should have been accepted (happens when a lenient significance level is used, an error of optimism).
A type II error is when the null hypothesis is accepted when it should have been rejected (happens when a stringent significance level is used, an error of pessimism).
Ethical Issues
- Informed consent is when participants are able to make an informed judgment about whether to take part. It causes them to guess the aims of the study and change their behavior.
- To deal with it, we can gain presumptive consent or ask them to formally indicate their agreement to participate but it may invalidate the purpose of the study and it is not guaranteed that the participants would understand.
- Deception should only be used when it is approved by an ethics committee, as it involves deliberately misleading or withholding information. Participants should be fully debriefed after the study but debriefing can’t turn the clock back.
- All participants should be informed at the beginning that they have the right to withdraw if they ever feel distressed or uncomfortable.
- It causes bias as the ones that stayed are obedient and some may not withdraw as they may have been given incentives or feel like they’re spoiling the study. Researchers can offer the right to withdraw data after participation.
- Participants should all have protection from harm . The researcher should avoid risks greater than those experienced in everyday life and they should stop the study if any harm is suspected. However, the harm may not be apparent at the time of the study.
- Confidentiality concerns the communication of personal information. The researchers should not record any names but use numbers or false names though it may not be possible as it is sometimes possible to work out who the researchers were.
Related Articles
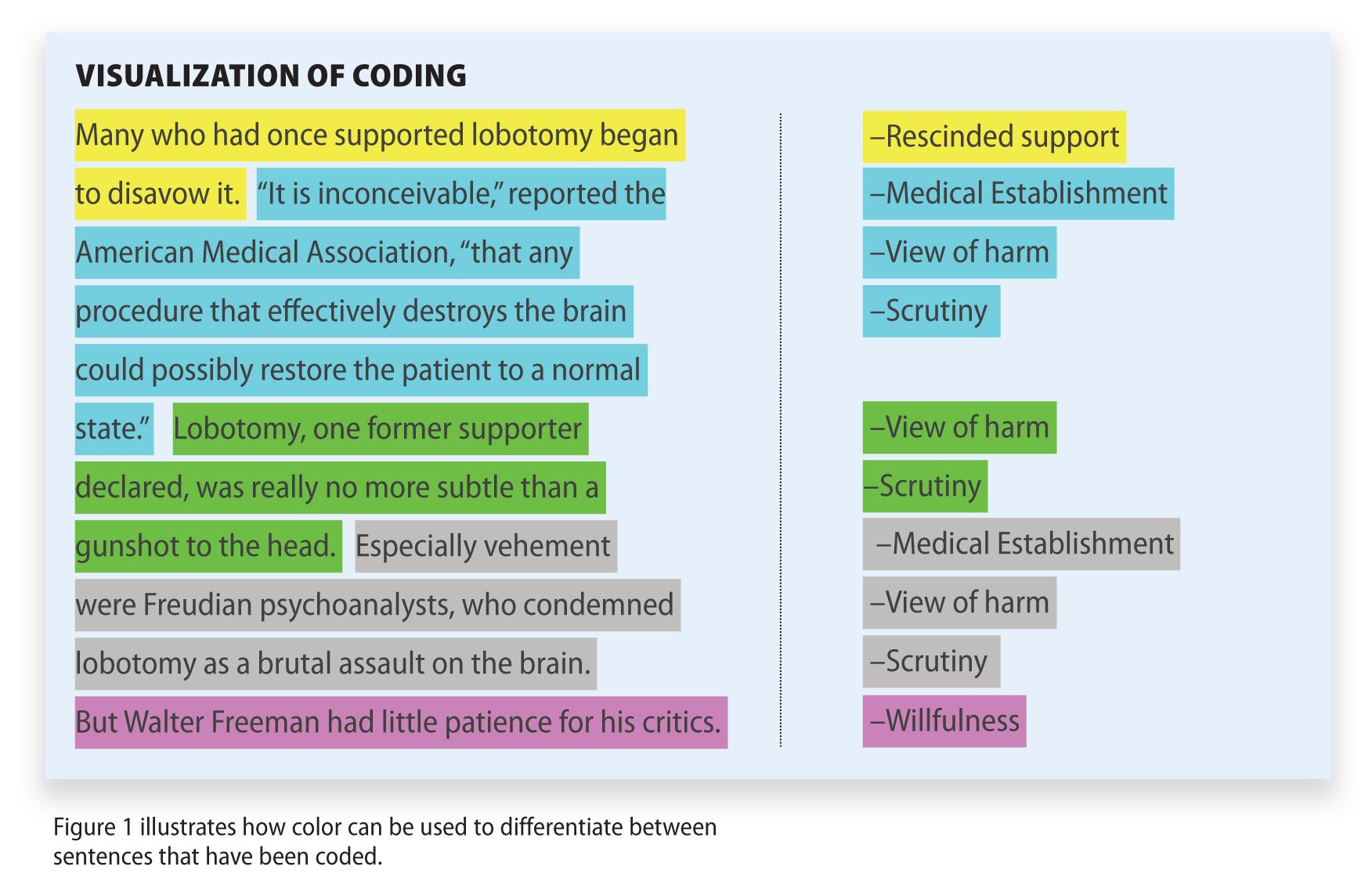
Research Methodology
Qualitative Data Coding

What Is a Focus Group?
Cross-Cultural Research Methodology In Psychology

A-Level Psychology
A-level Psychology AQA Revision Notes

What Is Internal Validity In Research?

Research Methodology , Statistics
What Is Face Validity In Research? Importance & How To Measure
- Python For Data Analysis
- Data Science
- Data Analysis with R
- Data Analysis with Python
- Data Visualization with Python
- Data Analysis Examples
- Math for Data Analysis
- Data Analysis Interview questions
- Artificial Intelligence
- Data Analysis Projects
- Machine Learning
- Deep Learning
- Computer Vision
- Types of Research - Methods Explained with Examples
- GRE Data Analysis | Methods for Presenting Data
- Financial Analysis: Objectives, Methods, and Process
- Financial Analysis: Need, Types, and Limitations
- Methods of Marketing Research
- Top 10 SQL Projects For Data Analysis
- What is Statistical Analysis in Data Science?
- 10 Data Analytics Project Ideas
- Predictive Analysis in Data Mining
- How to Become a Research Analyst?
- Data Analytics and its type
- Types of Social Networks Analysis
- What is Data Analysis?
- Six Steps of Data Analysis Process
- Multidimensional data analysis in Python
- Attributes and its Types in Data Analytics
- Exploratory Data Analysis (EDA) - Types and Tools
- Data Analyst Jobs in Pune
Data Analysis in Research: Types & Methods
Data analysis is a crucial step in the research process, transforming raw data into meaningful insights that drive informed decisions and advance knowledge. This article explores the various types and methods of data analysis in research, providing a comprehensive guide for researchers across disciplines.

Data Analysis in Research
Overview of Data analysis in research
Data analysis in research is the systematic use of statistical and analytical tools to describe, summarize, and draw conclusions from datasets. This process involves organizing, analyzing, modeling, and transforming data to identify trends, establish connections, and inform decision-making. The main goals include describing data through visualization and statistics, making inferences about a broader population, predicting future events using historical data, and providing data-driven recommendations. The stages of data analysis involve collecting relevant data, preprocessing to clean and format it, conducting exploratory data analysis to identify patterns, building and testing models, interpreting results, and effectively reporting findings.
- Main Goals : Describe data, make inferences, predict future events, and provide data-driven recommendations.
- Stages of Data Analysis : Data collection, preprocessing, exploratory data analysis, model building and testing, interpretation, and reporting.
Types of Data Analysis
1. descriptive analysis.
Descriptive analysis focuses on summarizing and describing the features of a dataset. It provides a snapshot of the data, highlighting central tendencies, dispersion, and overall patterns.
- Central Tendency Measures : Mean, median, and mode are used to identify the central point of the dataset.
- Dispersion Measures : Range, variance, and standard deviation help in understanding the spread of the data.
- Frequency Distribution : This shows how often each value in a dataset occurs.
2. Inferential Analysis
Inferential analysis allows researchers to make predictions or inferences about a population based on a sample of data. It is used to test hypotheses and determine the relationships between variables.
- Hypothesis Testing : Techniques like t-tests, chi-square tests, and ANOVA are used to test assumptions about a population.
- Regression Analysis : This method examines the relationship between dependent and independent variables.
- Confidence Intervals : These provide a range of values within which the true population parameter is expected to lie.
3. Exploratory Data Analysis (EDA)
EDA is an approach to analyzing data sets to summarize their main characteristics, often with visual methods. It helps in discovering patterns, spotting anomalies, and checking assumptions with the help of graphical representations.
- Visual Techniques : Histograms, box plots, scatter plots, and bar charts are commonly used in EDA.
- Summary Statistics : Basic statistical measures are used to describe the dataset.
4. Predictive Analysis
Predictive analysis uses statistical techniques and machine learning algorithms to predict future outcomes based on historical data.
- Machine Learning Models : Algorithms like linear regression, decision trees, and neural networks are employed to make predictions.
- Time Series Analysis : This method analyzes data points collected or recorded at specific time intervals to forecast future trends.
5. Causal Analysis
Causal analysis aims to identify cause-and-effect relationships between variables. It helps in understanding the impact of one variable on another.
- Experiments : Controlled experiments are designed to test the causality.
- Quasi-Experimental Designs : These are used when controlled experiments are not feasible.
6. Mechanistic Analysis
Mechanistic analysis seeks to understand the underlying mechanisms or processes that drive observed phenomena. It is common in fields like biology and engineering.
Methods of Data Analysis
1. quantitative methods.
Quantitative methods involve numerical data and statistical analysis to uncover patterns, relationships, and trends.
- Statistical Analysis : Includes various statistical tests and measures.
- Mathematical Modeling : Uses mathematical equations to represent relationships among variables.
- Simulation : Computer-based models simulate real-world processes to predict outcomes.
2. Qualitative Methods
Qualitative methods focus on non-numerical data, such as text, images, and audio, to understand concepts, opinions, or experiences.
- Content Analysis : Systematic coding and categorizing of textual information.
- Thematic Analysis : Identifying themes and patterns within qualitative data.
- Narrative Analysis : Examining the stories or accounts shared by participants.
3. Mixed Methods
Mixed methods combine both quantitative and qualitative approaches to provide a more comprehensive analysis.
- Sequential Explanatory Design : Quantitative data is collected and analyzed first, followed by qualitative data to explain the quantitative results.
- Concurrent Triangulation Design : Both qualitative and quantitative data are collected simultaneously but analyzed separately to compare results.
4. Data Mining
Data mining involves exploring large datasets to discover patterns and relationships.
- Clustering : Grouping data points with similar characteristics.
- Association Rule Learning : Identifying interesting relations between variables in large databases.
- Classification : Assigning items to predefined categories based on their attributes.
5. Big Data Analytics
Big data analytics involves analyzing vast amounts of data to uncover hidden patterns, correlations, and other insights.
- Hadoop and Spark : Frameworks for processing and analyzing large datasets.
- NoSQL Databases : Designed to handle unstructured data.
- Machine Learning Algorithms : Used to analyze and predict complex patterns in big data.
Applications and Case Studies
Numerous fields and industries use data analysis methods, which provide insightful information and facilitate data-driven decision-making. The following case studies demonstrate the effectiveness of data analysis in research:
Medical Care:
- Predicting Patient Readmissions: By using data analysis to create predictive models, healthcare facilities may better identify patients who are at high risk of readmission and implement focused interventions to enhance patient care.
- Disease Outbreak Analysis: Researchers can monitor and forecast disease outbreaks by examining both historical and current data. This information aids public health authorities in putting preventative and control measures in place.
- Fraud Detection: To safeguard clients and lessen financial losses, financial institutions use data analysis tools to identify fraudulent transactions and activities.
- investing Strategies: By using data analysis, quantitative investing models that detect trends in stock prices may be created, assisting investors in optimizing their portfolios and making well-informed choices.
- Customer Segmentation: Businesses may divide up their client base into discrete groups using data analysis, which makes it possible to launch focused marketing efforts and provide individualized services.
- Social Media Analytics: By tracking brand sentiment, identifying influencers, and understanding consumer preferences, marketers may develop more successful marketing strategies by analyzing social media data.
- Predicting Student Performance: By using data analysis tools, educators may identify at-risk children and forecast their performance. This allows them to give individualized learning plans and timely interventions.
- Education Policy Analysis: Data may be used by researchers to assess the efficacy of policies, initiatives, and programs in education, offering insights for evidence-based decision-making.
Social Science Fields:
- Opinion mining in politics: By examining public opinion data from news stories and social media platforms, academics and policymakers may get insight into prevailing political opinions and better understand how the public feels about certain topics or candidates.
- Crime Analysis: Researchers may spot trends, anticipate high-risk locations, and help law enforcement use resources wisely in order to deter and lessen crime by studying crime data.
Data analysis is a crucial step in the research process because it enables companies and researchers to glean insightful information from data. By using diverse analytical methodologies and approaches, scholars may reveal latent patterns, arrive at well-informed conclusions, and tackle intricate research inquiries. Numerous statistical, machine learning, and visualization approaches are among the many data analysis tools available, offering a comprehensive toolbox for addressing a broad variety of research problems.
Data Analysis in Research FAQs:
What are the main phases in the process of analyzing data.
In general, the steps involved in data analysis include gathering data, preparing it, doing exploratory data analysis, constructing and testing models, interpreting the results, and reporting the results. Every stage is essential to guaranteeing the analysis’s efficacy and correctness.
What are the differences between the examination of qualitative and quantitative data?
In order to comprehend and analyze non-numerical data, such text, pictures, or observations, qualitative data analysis often employs content analysis, grounded theory, or ethnography. Comparatively, quantitative data analysis works with numerical data and makes use of statistical methods to identify, deduce, and forecast trends in the data.
What are a few popular statistical methods for analyzing data?
In data analysis, predictive modeling, inferential statistics, and descriptive statistics are often used. While inferential statistics establish assumptions and draw inferences about a wider population, descriptive statistics highlight the fundamental characteristics of the data. To predict unknown values or future events, predictive modeling is used.
In what ways might data analysis methods be used in the healthcare industry?
In the healthcare industry, data analysis may be used to optimize treatment regimens, monitor disease outbreaks, forecast patient readmissions, and enhance patient care. It is also essential for medication development, clinical research, and the creation of healthcare policies.
What difficulties may one encounter while analyzing data?
Answer: Typical problems with data quality include missing values, outliers, and biased samples, all of which may affect how accurate the analysis is. Furthermore, it might be computationally demanding to analyze big and complicated datasets, necessitating certain tools and knowledge. It’s also critical to handle ethical issues, such as data security and privacy.
Please Login to comment...
Similar reads.
- Data Science Blogathon 2024
- AI-ML-DS Blogs
- Data Analysis
Improve your Coding Skills with Practice
What kind of Experience do you want to share?
SYSTEMATIC REVIEW article
This article is part of the research topic.
Reviews in Gastroenterology 2023
Electrogastrography Measurement Systems and Analysis Methods Used in Clinical Practice and Research: Comprehensive Review Provisionally Accepted

- 1 VSB-Technical University of Ostrava, Czechia
The final, formatted version of the article will be published soon.
Electrogastrography (EGG) is a non-invasive method with high diagnostic potential for the prevention of gastroenterological pathologies in clinical practice. In this paper, a review of the measurement systems, procedures, and methods of analysis used in electrogastrography is presented. A critical review of historical and current literature is conducted, focusing on electrode placement, measurement apparatus, measurement procedures, and time-frequency domain methods of filtration and analysis of the non-invasively measured electrical activity of the stomach.As a result a total of 129 relevant articles with primary aim on experimental diet were reviewed in this study. Scopus, PubMed and Web of Science databases were used to search for articles in English language, according to the specific query and using PRISMA method. The research topic of electrogastrography has been continuously growing in popularity since the first measurement by professor Alvarez 100 years ago and there are many researchers and companies interested in EGG nowadays. Measurement apparatus and procedures are still being developed in both commercial and research settings. There are plenty variable electrode layouts, ranging from minimal numbers of electrodes for ambulatory measurements to very high numbers of electrodes for spatial measurements. Most authors used in their research anatomically approximated layout with 2 active electrodes in bipolar connection and commercial electrogastrograph with sampling rate of 2 or 4 Hz. Test subjects were usually healthy adults and diet was controlled. However, evaluation methods are being developed at a slower pace and usually the signals are classified only based on dominant frequency. The main review contributions include the overview of spectrum of measurement systems and procedures for electrogastrography developed by many authors, but a firm medical standard has not yet been defined. Therefore, it is not possible to use this method in clinical practice for objective diagnosis.
Keywords: electrogastrography, non-invasive method, Measurement systems, Electrode placement, Measurement apparatus, Signal processing
Received: 19 Jan 2024; Accepted: 03 Jun 2024.
Copyright: © 2024 Oczka, Augustynek, Penhaker and Kubicek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
* Correspondence: Dr. Jan Kubicek, VSB-Technical University of Ostrava, Ostrava, 708 33, Moravian-Silesian Region, Czechia
People also looked at
Numbers, Facts and Trends Shaping Your World
Read our research on:
Full Topic List
Regions & Countries
- Publications
- Our Methods
- Short Reads
- Tools & Resources
Read Our Research On:
Table of Contents
Which social media platforms are most common, who uses each social media platform, find out more, social media fact sheet.
Many Americans use social media to connect with one another, engage with news content, share information and entertain themselves. Explore the patterns and trends shaping the social media landscape.
To better understand Americans’ social media use, Pew Research Center surveyed 5,733 U.S. adults from May 19 to Sept. 5, 2023. Ipsos conducted this National Public Opinion Reference Survey (NPORS) for the Center using address-based sampling and a multimode protocol that included both web and mail. This way nearly all U.S. adults have a chance of selection. The survey is weighted to be representative of the U.S. adult population by gender, race and ethnicity, education and other categories.
Polls from 2000 to 2021 were conducted via phone. For more on this mode shift, read our Q&A.
Here are the questions used for this analysis , along with responses, and its methodology .
A note on terminology: Our May-September 2023 survey was already in the field when Twitter changed its name to “X.” The terms Twitter and X are both used in this report to refer to the same platform.

YouTube and Facebook are the most-widely used online platforms. About half of U.S. adults say they use Instagram, and smaller shares use sites or apps such as TikTok, LinkedIn, Twitter (X) and BeReal.
Note: The vertical line indicates a change in mode. Polls from 2012-2021 were conducted via phone. In 2023, the poll was conducted via web and mail. For more details on this shift, please read our Q&A . Refer to the topline for more information on how question wording varied over the years. Pre-2018 data is not available for YouTube, Snapchat or WhatsApp; pre-2019 data is not available for Reddit; pre-2021 data is not available for TikTok; pre-2023 data is not available for BeReal. Respondents who did not give an answer are not shown.
Source: Surveys of U.S. adults conducted 2012-2023.

Usage of the major online platforms varies by factors such as age, gender and level of formal education.
% of U.S. adults who say they ever use __ by …
- RACE & ETHNICITY
- POLITICAL AFFILIATION

This fact sheet was compiled by Research Assistant Olivia Sidoti , with help from Research Analyst Risa Gelles-Watnick , Research Analyst Michelle Faverio , Digital Producer Sara Atske , Associate Information Graphics Designer Kaitlyn Radde and Temporary Researcher Eugenie Park .
Follow these links for more in-depth analysis of the impact of social media on American life.
- Americans’ Social Media Use Jan. 31, 2024
- Americans’ Use of Mobile Technology and Home Broadband Jan. 31 2024
- Q&A: How and why we’re changing the way we study tech adoption Jan. 31, 2024
Find more reports and blog posts related to internet and technology .
1615 L St. NW, Suite 800 Washington, DC 20036 USA (+1) 202-419-4300 | Main (+1) 202-857-8562 | Fax (+1) 202-419-4372 | Media Inquiries
Research Topics
- Email Newsletters
ABOUT PEW RESEARCH CENTER Pew Research Center is a nonpartisan fact tank that informs the public about the issues, attitudes and trends shaping the world. It conducts public opinion polling, demographic research, media content analysis and other empirical social science research. Pew Research Center does not take policy positions. It is a subsidiary of The Pew Charitable Trusts .
© 2024 Pew Research Center
- Open access
- Published: 27 May 2024
Discovery of novel RNA viruses through analysis of fungi-associated next-generation sequencing data
- Xiang Lu 1 , 2 na1 ,
- Ziyuan Dai 3 na1 ,
- Jiaxin Xue 2 na1 ,
- Wang Li 4 ,
- Ping Ni 4 ,
- Juan Xu 4 ,
- Chenglin Zhou 4 &
- Wen Zhang 1 , 2 , 4
BMC Genomics volume 25 , Article number: 517 ( 2024 ) Cite this article
371 Accesses
1 Altmetric
Metrics details
Like all other species, fungi are susceptible to infection by viruses. The diversity of fungal viruses has been rapidly expanding in recent years due to the availability of advanced sequencing technologies. However, compared to other virome studies, the research on fungi-associated viruses remains limited.
In this study, we downloaded and analyzed over 200 public datasets from approximately 40 different Bioprojects to explore potential fungal-associated viral dark matter. A total of 12 novel viral sequences were identified, all of which are RNA viruses, with lengths ranging from 1,769 to 9,516 nucleotides. The amino acid sequence identity of all these viruses with any known virus is below 70%. Through phylogenetic analysis, these RNA viruses were classified into different orders or families, such as Mitoviridae , Benyviridae , Botourmiaviridae , Deltaflexiviridae , Mymonaviridae , Bunyavirales , and Partitiviridae . It is possible that these sequences represent new taxa at the level of family, genus, or species. Furthermore, a co-evolution analysis indicated that the evolutionary history of these viruses within their groups is largely driven by cross-species transmission events.
Conclusions
These findings are of significant importance for understanding the diversity, evolution, and relationships between genome structure and function of fungal viruses. However, further investigation is needed to study their interactions.
Peer Review reports
Introduction
Viruses are among the most abundant and diverse biological entities on Earth; they are ubiquitous in the natural environment but difficult to culture and detect [ 1 , 2 , 3 ]. In recent decades, the significant advancements in omics have transformed the field of virology and enabled researchers to detect potential viruses in a variety of environmental samples, helping us to expand the known diversity of viruses and explore the “dark matter” of viruses that may exist in vast quantities [ 4 ]. In most cases, the hosts of these newly discovered viruses exhibit only asymptomatic infections [ 5 , 6 ], and they even play an important role in maintaining the balance, stability, and sustainable development of the biosphere [ 7 ]. But some viruses may be involved in the emergence and development of animal or plant diseases. For example, the tobacco mosaic virus (TMV) causes poor growth in tobacco plants, while norovirus is known to cause diarrhea in mammals [ 8 , 9 ]. In the field of fungal research, viral infections have significantly reduced the yield of edible fungi, thereby attracting increasing attention to fungal diseases caused by viruses [ 10 ]. However, due to their apparent relevance to health [ 11 ], fungal-associated viruses have been understudied compared to viruses affecting humans, animals, or plants.
Mycoviruses (also known as fungal viruses) are widely distributed in various fungi and fungal-like organisms [ 12 ]. The first mycoviruses were discovered in the 1960s by Hollings M in the basidiomycete Agaricus bisporus , an edible cultivated mushroom [ 13 ]. Shortly thereafter, Ellis LF et al. reported mycoviruses in the ascomycete Penicillium stoloniferum , confirming that viral dsRNA is responsible for interferon stimulation in mammals [ 13 , 14 , 15 ]. In recent years, the diversity of known mycoviruses has rapidly increased with the development and widespread application of sequencing technologies [ 16 , 17 , 18 , 19 , 20 ]. According to the classification principles of the International Committee for the Taxonomy of Viruses (ICTV), mycoviruses are currently classified into 24 taxa, consisting of 23 families and 1 genus ( Botybirnavirus ) [ 21 ]. Most mycoviruses belong to double-stranded (ds) RNA viruses, such as families Totiviridae , Partitiviridae , Reoviridae , Chrysoviridae , Megabirnaviridae , Quadriviridae , and genus Botybirnavirus , or positive-sense single-stranded (+ ss) RNA viruses, such as families Alphaflexiviridae , Gammaflexiviridae , Barnaviridae , Hypoviridae , Endornaviridae , Metaviridae and Pseudoviridae . However, negative-sense single-stranded (-ss) RNA viruses (family Mymonaviridae ) and single-stranded (ss) DNA viruses (family Genomoviridae ) have also been described [ 22 ]. The taxonomy of mycoviruses is continually refined as novel mycoviruses that cannot be classified into any established taxon are identified. While the vast majority of fungi-infecting viruses do not show infection characteristics and have no significant impact on their hosts, some mycoviruses have inhibitory effects on the phenotype of the host, leading to hypovirulence in phytopathogenic fungi [ 23 ]. The use of environmentally friendly, low-virulence-related mycoviruses such as Chryphonectria hypovirus 1 (CHV-1) for biological control has been considered a viable alternative to chemical fungicides [ 24 ]. With the deepening of research, an increasing number of mycoviruses that can cause fungal phenotypic changes have been identified [ 3 , 23 , 25 ]. Therefore, understanding the distribution of these viruses and their effects on hosts will allow us to determine whether their infections can be prevented and treated.
To explore the viral dark matter hidden within fungi, this study collected over 200 available fungal-associated libraries from approximately 40 Bioprojects in the Sequence Read Archive (SRA) database, uncovering novel RNA viruses within them. We further elucidated the genetic relationships between known viruses and these newfound ones, thereby expanding our understanding of fungal-associated viruses and providing assistance to viral taxonomy.
Materials and methods
Genome assembly.
To discover novel fungal-associated viruses, we downloaded 236 available libraries from the SRA database, corresponding to 32 fungal species (Supplementary Table 1). Pfastq-dump v0.1.6 ( https://github.com/inutano/pfastq-dump ) was used to convert SRA format files to fastq format files. Subsequently, Bowtie2 v2.4.5 [ 26 ] was employed to remove host sequences. Primer sequences of raw reads underwent trimming using Trim Galore v0.6.5 ( https://www.bioinformatics.babraham.ac.uk/projects/trim_galore ), and the resulting files underwent quality control with the options ‘–phred33 –length 20 –stringency 3 –fastqc’. Duplicated reads were marked using PRINSEQ-lite v0.20.4 (-derep 1). All SRA datasets were then assembled in-house pipeline. Paired-end reads were assembled using SPAdes v3.15.5 [ 27 ] with the option ‘-meta’, while single-end reads were assembled with MEGAHIT v1.2.9 [ 28 ], both using default parameters. The results were then imported into Geneious Prime v2022.0.1 ( https://www.geneious.com ) for sorting and manual confirmation. To reduce false negatives during sequence assembly, further semi-automatic assembly of unmapped contigs and singlets with a sequence length < 500 nt was performed. Contigs with a sequence length > 1,500 nt after reassembly were retained. Individual contigs were then used as references for mapping to the raw data using the Low Sensitivity/Fastest parameter in Geneious Prime. In addition, mixed assembly was performed using MEGAHIT in combination with BWA v0.7.17 [ 29 ] to search for unused reads that might correspond to low-abundance contigs.
Searching for novel viruses in fungal libraries
We identified novel viral sequences present in fungal libraries through a series of steps. To start, we established a local viral database, consisting of the non-redundant protein (nr) database downloaded in August 2023, along with IMG/VR v3 [ 30 ], for screening assembled contigs. The contigs labeled as “viruses” and exhibiting less than 70% amino acid (aa) sequence identity with the best match in the database were imported into Geneious Prime for manual mapping. Putative open reading frames (ORFs) were predicted by Geneious Prime using built-in parameters (Minimum size: 100) and were subsequently verified by comparison to related viruses. The annotations of these ORFs were based on comparisons to the Conserved Domain Database (CDD). The sequences after manual examination were subjected to genome clustering using MMseqs2 (-k 0 -e 0.001 –min-seq-id 0.95 -c 0.9 –cluster-mode 0) [ 31 ]. After excluding viruses with high aa sequence identity (> 70%) to known viruses, a dataset containing a total of 12 RNA viral sequences was obtained. The non-redundant fungal virus dataset was compared against the local database using the BLASTx program built in DIAMOND v2.0.15 [ 32 ], and significant sequences with a cut-off E-value of < 10 –5 were selected. The coverage of each sequence in all libraries was calculated using the pileup tool in BBMap. Taxonomic identification was conducted using TaxonKit [ 33 ] software, along with the rma2info program integrated into MEGAN6 [ 34 ]. The RNA secondary structure prediction of the novel viruses was conducted using RNA Folding Form V2.3 ( http://www.unafold.org/mfold/applications/rna-folding-form-v2.php ).
Phylogenetic analysis
To infer phylogenetic relationships, nucleotide and their encoded protein sequences of reference strains belonging to different groups of corresponding viruses were downloaded from the NCBI GenBank database, along with sequences of proposed species pending ratification. Related sequences were aligned using the alignment program within the CLC Genomics Workbench 10.0, and the resulting alignment was further optimized using MUSCLE in MEGA-X [ 35 ]. Sites containing more than 50% gaps were temporarily removed from the alignments. Maximum-likelihood (ML) trees were then constructed using IQ-TREE v1.6.12 [ 36 ]. All phylogenetic trees were created using IQ-TREE with 1,000 bootstrap replicates (-bb 1000) and the ModelFinder function (-m MFP). Interactive Tree Of Life (iTOL) was used for visualizing and editing phylogenetic trees [ 37 ]. Colorcoded distance matrix analysis between novel viruses and other known viruses were performed with Sequence Demarcation Tool v1.2 [ 38 ].
To illustrate cross-species transmission and co-divergence between viruses and their hosts across different virus groups, we reconciled the co-phylogenetic relationships between these viruses and their hosts. The evolutionary tree and topologies of the hosts involved in this study were obtained from the TimeTree [ 39 ] website by inputting their Latin names. The viruses in the phylogenetic tree for which the host cannot be recognized through published literature or information provided by the authors are disregarded. The co-phylogenetic plots (or ‘tanglegram’) generated using the R package phytools [ 40 ] visually represent the correspondence between host and virus trees, with lines connecting hosts and their respective viruses. The event-based program eMPRess [ 41 ] was employed to determine whether the pairs of virus groups and their hosts undergo coevolution. This tool reconciles pairs of phylogenetic trees according to the Duplication-Transfer-Loss (DTL) model [ 42 ], employing a maximum parsimony formulation to calculate the cost of each coevolution event. The cost of duplication, host-jumping (transfer), and extinction (loss) event types were set to 1.0, while host-virus co-divergence was set to zero, as it was considered the null event.
Data availability
The data reported in this paper have been deposited in the GenBase in National Genomics Data Center [ 43 ], Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, under accession numbers C_AA066339.1-C_AA066350.1 that are publicly accessible at https://ngdc.cncb.ac.cn/genbase . Please refer to Table 1 for details.
Twelve novel RNA viruses associated with fungi
We investigated fungi-associated novel viruses by mining publicly available metagenomic and transcriptomic fungal datasets. In total, we collected 236 datasets, which were categorized into four fungal phyla: Ascomycota (159), Basidiomycota (47), Chytridiomycota (15), and Zoopagomycota (15). These phyla corresponded to 20, 8, 2, and 2 different fungal genera, respectively (Supplementary Table 1). A total of 12 sequences containing complete coding DNA sequences (CDS) for RNA-dependent RNA polymerase (RdRp) have been identified, ranging in length from 1,769 nt to 9,516 nt. All of these sequences have less than 70% aa identity with RdRp sequences from any currently known virus (ranging from 32.97% to 60.43%), potentially representing novel families, genera, or species (Table 1 ). Some of the identified sequences were shorter than the reference genomes of RNA viruses, suggesting that these viral sequences represented partial sequences of viral genomes. To exclude the possibility of transient viral infections in hosts or de novo assembly artefacts in co-infection detection, we extracted the nucleotide sequences of the coding regions of these 12 sequences and mapped them to all collected libraries to compute coverage (Supplementary Table 2). The results revealed varying degrees of read matches for these viral genomes across different libraries, spanning different fungal species. Although we only analyzed sequences longer than 1,500 nt, it is worth noting that we also discovered other viral reads in many libraries. However, we were unable to assemble them into sufficiently long contigs, possibly due to library construction strategies or sequencing depth. In any case, this preliminary finding reveals a greater diversity of fungal-associated viruses than previously considered.
Positive-sense single-stranded RNA viruses
(i) mitoviridae.
Members of the family Mitoviridae (order Cryppavirales ) are monopartite, linear, positive-sense ( +) single-stranded (ss) RNA viruses with genome size of approximately 2.5–2.9 kb [ 44 ], carrying a single long open reading frame (ORF) which encodes a putative RdRp. Mitoviruses have no true virions and no structural proteins, virus genome is transmitted horizontally through mating or vertically from mother to daughter cells [ 45 ]. They use mitochondria as their sites of replication and have typical 5' and 3' untranslated regions (UTRs) of varying sizes, which are responsible for viral translation and replicase recognition [ 46 ]. According to the taxonomic principles of ICTV, the viruses belonging to the family Mitoviridae are divided into four genera, namely Duamitovirus , Kvaramitovirus , Triamitovirus and Unuamitovirus . In this study, two novel viruses belonging to the family Mitoviridae were identified in the same library (SRR12744489; Species: Thielaviopsis ethacetica ), named Thielaviopsis ethacetica mitovirus 1 (TeMV01) and Thielaviopsis ethacetica mitovirus 2 (TeMV02), respectively (Fig. 1 A). The genome sequence of TeMV01 spans 2,689 nucleotides in length with a GC content of 32.2%. Its 5' and 3' UTRs comprise 406 nt and 36 nt, respectively. Similarly, the genome sequence of TeMV02 extends 3,087 nucleotides in length with a GC content of 32.6%. Its 5' and 3' UTRs consist of 553 and 272 nt, respectively. The 5' and 3' ends of both genomes are predicted to have typical stem-loop structures (Fig. 1 B). In order to determine the evolutionary relationship between these two mitoviruses and other known mitoviruses, phylogenetic analysis based on RdRp showed that viral strains were divided into 2 genetic lineages in the genera Duamitovirus and Unuamitovirus (Fig. 1 C). In the genus Unuamitovirus , TeMV01 was clustered with Ophiostoma mitovirus 4, exhibiting the highest aa identity of 51.47%, while in the genus Duamitovirus , TeMV02 was clustered with a strain isolated from Plasmopara viticola , showing the highest aa identity of 42.82%. According to the guidelines from the ICTV regarding the taxonomy of the family Mitoviridae , a species demarcation cutoff of < 70% aa sequence identity is established [ 47 ]. Drawing on this recommendation and phylogenetic inferences, these two viral strains could be presumed to be novel viral species [ 48 ].
Identification of novel positive-sense single-stranded RNA viruses in fungal sequencing libraries. A Genome organization of two novel mitoviruses; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. B Predicted RNA secondary structures of the 5'- and 3'-terminal regions. C ML phylogenetic tree of members of the family Mitoviridae . The best-fit model (LG + F + R6) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified viruses represented in red font. D The genome organization of GtBeV is depicted at the top; in the middle is the ML phylogenetic tree of members of the family Benyviridae . The best-fit model (VT + F + R5) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font. At the bottom is the distance matrix analysis of GeBeV identified in Gaeumannomyces tritici . Pairwise sequence comparison produced with the RdRp amino acid sequences within the ML tree. E The genome organization of CrBV is depicted at the top; in the middle is the ML phylogenetic tree of members of the family Botourmiaviridae . The best-fit model (VT + F + R5) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font. At the bottom is the distance matrix analysis of CrBV identified in Clonostachys rosea . Pairwise sequence comparison produced with the RdRp amino acid sequences within the ML tree
(ii) Benyviridae
The family Benyviridae is comprised of multipartite plant viruses that are rod-shaped, approximately 85–390 nm in length and 20 nm in diameter. Within this family, there is a single genus, Benyvirus [ 49 ]. It is reported that one species within this genus,Beet necrotic yellow vein virus, can cause widespread and highly destructive soil-borne ‘rhizomania’ disease of sugar beet [ 50 ]. A full-length RNA1 sequence related to Benyviridae has been detected from Gaeumannomyces tritici (ERR3486062), with a length of 6,479 nt. It possesses a poly(A) tail at the 3' end and is temporarily designated as Gaeumannomyces tritici benyvirus (GtBeV). BLASTx results indicate a 34.68% aa sequence identity with the best match found (Fig. 1 D). The non-structural polyprotein CDS of RNA1 encodes a large replication-associated protein of 1,688 amino acids with a molecular mass of 190 kDa. Four domains were predicted in this polyprotein corresponding to representative species within the family Benyviridae . The viral methyltransferase (Mtr) domain spans from nucleotide position 386 to 1411, while the RNA helicase (Hel) domain occupies positions 2113 to 2995 nt. Additionally, the protease (Pro) domain is located between positions 3142 and 3410 nt, and the RdRp domain is located at 4227 to 4796 nt. A phylogenetic analysis was conducted by integrating RdRp sequences of viruses closely related to GtBeV. The result revealed that GtBeV clustered within the family Benyviridae , exhibiting substantial evolutionary divergence from any other sequences. Consequently, this virus likely represents a novel species in the family Benyviridae .
(iii) Botourmiaviridae
The family Botourmiaviridae comprises viruses infecting plants and filamentous fungi, which may possess mono- or multi-segmented genomes [ 51 ]. Recent research has led to a rapid expansion in the number of viruses within the family Botourmiaviridae , increasing from the confirmed 4 genera in 2020 to a total of 12 genera. A contig identified from Clonostachys rosea (ERR5928658) using the BLASTx method exhibited similarity to viruses in the family Botourmiaviridae . After manual mapping, a 2,903 nt-long genome was obtained, tentatively named Clonostachys rosea botourmiavirus (CrBV), which includes a complete RdRP region (Fig. 1 E). Based on phylogenetic analysis using RdRp, CrBV clustered with members of the genus Magoulivirus , sharing 56.58% aa identity with a strain identified from Eclipta prostrata . However, puzzlingly, according to the ICTV's Genus/Species demarcation criteria, members of different genera/species within the family Botourmiaviridae share less than 70%/90% identity in their complete RdRP amino acid sequences. Furthermore, the RdRp sequences with accession numbers NC_055143 and NC_076766, both considered to be members of the genus Magoulivirus , exhibited only 39.05% aa identity to each other. Therefore, CrBV should at least be considered as a new species within the family Botourmiaviridae .
(iv) Deltaflexiviridae
An assembled sequence of 3,425 nucleotides in length Lepista sordida deltaflexivirus (LsDV), derived from Lepista sordida (DRR252167) and showing homology to Deltaflexiviridae within the order Tymovirales , was obtained. The Tymovirales comprises five recognized families: Alphaflexiviridae , Betaflexiviridae , Deltaflexiviridae , Gammaflexiviridae , and Tymoviridae [ 52 ]. The Deltaflexiviridae currently only includes one genus, the fungal-associated deltaflexivirus; they are mostly identified in fungi or plants pathogens [ 53 ]. LsDV was predicted to have a single large ORF, VP1, which starts with an AUG codon at nt 163–165 and ends with a UAG codon at nt 3,418–3,420. This ORF encodes a putative polyprotein of 1,086 aa with a calculated molecular mass of 119 kDa. Two conserved domains within the VP1 protein were identified: Hel and RdRp (Fig. 2 A). However, the Mtr was missing, indicating that the 5' end of this polyprotein is incomplete. According to the phylogenetic analysis of RdRp, LsDV was closely related to viruses of the family Deltaflexiviridae and shared 46.61% aa identity with a strain (UUW06602) isolated from Macrotermes carbonarius . Despite this, according to the species demarcation criteria proposed by ICTV, because we couldn't recover the entire replication-associated polyprotein, LsDV cannot be regarded as a novel species at present.
Identification of novel members of family Deltaflexiviridae and Toga-like virus in fungal sequencing libraries. A On the right side of the image is the genome organization of LsDV; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. ML phylogenetic tree of members of the family Deltaflexiviridae . The best-fit model (VT + F + R6) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font. B The genome organization of GtTlV is depicted at the top; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. ML phylogenetic tree of members of the order Martellivirales . The best-fit model (LG + R7) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font
(v) Toga-like virus
Members of the family Togaviridae are primarily transmitted by arthropods and can infect a wide range of vertebrates, including mammals, birds, reptiles, amphibians, and fish [ 54 ]. Currently, this family only contains a single confirmed genus, Alphavirus . A contig was discovered in Gaeumannomyces tritici (ERR3486058), it is 7,588 nt in length with a complete ORF encoding a putative protein of 1,928 aa, which had 60.43% identity to Fusarium sacchari alphavirus-like virus 1 (QIQ28421) with 97% coverage. Phylogenetic analysis showed that it did not cluster with classical alphavirus members such as VEE, WEE, EEE, SF complex [ 54 ], but rather with several sequences annotated as Toga-like that were available (Fig. 2 B). It was provisionally named Gaeumannomyces tritici toga-like virus (GtTIV). However, we remain cautious about the accuracy of these so-called Toga-like sequences, as they show little significant correlation with members of the order Martellivirales .
Negative-sense single-stranded RNA viruses
(i) mymonaviridae.
Mymonaviridae is a family of linear, enveloped, negative-stranded RNA genomes in the order Mononegavirales , which infect fungi. They are approximately 10 kb in size and encode six proteins [ 55 ]. The famliy Mymonaviridae was established to accommodate Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV-1), a novel virus discovered in a hypovirulent strain of Sclerotinia sclerotiorum [ 56 ]. According to the ICTV, the family Mymonaviridae currently includes 9 genera, namely Auricularimonavirus , Botrytimonavirus , Hubramonavirus , Lentimonavirus , Penicillimonavirus , Phyllomonavirus , Plasmopamonavirus , Rhizomonavirus and Sclerotimonavirus . Two sequences originating from Gaeumannomyces tritici (ERR3486068) and Aspergillus puulaauensis (DRR266546), respectively, and associated with the family Mymonaviridae , have been identified and provisionally named Gaeumannomyces tritici mymonavirus (GtMV) and Aspergillus puulaauensis mymonavirus (ApMV). GtMV is 9,339 nt long with a GC content of 52.8%. It was predicted to contain 5 discontinuous ORFs, with the largest one encoding RdRp. Additionally, a nucleoprotein and three hypothetical proteins with unknown function were also predicted. A multiple alignment of nucleotide sequences among these ORFs identified a semi-conserved sequence, 5'-UAAAA-CUAGGAGC-3', located downstream of each ORF (Fig. 3 A). These regions are likely gene-junction regions in the GtMV genome, a characteristic feature shared by mononegaviruses [ 57 , 58 ]. For ApMV, a complete RdRp CDS with a length of 1,978 aa was predicted. The BLASTx searches showed that GtMV shared 45.22% identity with the RdRp of Soybean leaf-associated negative-stranded RNA virus 2 (YP_010784557), while ApMV shared 55.90% identity with the RdRp of Erysiphe necator associated negative-stranded RNA virus 23 (YP_010802816). The representative members of the family Mymonaviridae were included in the phylogenetic analysis. The results showed that GtMV and ApMV clustered closely with members of the genera Sclerotimonavirus and Plasmopamonavirus , respectively (Fig. 3 B). Members of the genus Plasmopamonavirus are about 6 kb in size and encode for a single protein. Therefore, GtMV and ApMV should be considered as representing new species within their respective genera.
Identification of two new members in the family Mymonaviridae . A At the top is the nucleotide multiple sequence alignment result of GtMV with the reference genomes. the putative ORF for the viral RdRp is depicted by a green box, the predicted nucleoprotein is displayed in a yellow box, and three hypothetical proteins are displayed in gray boxes. The comparison of putative semi-conserved regions between ORFs in GtMV is displayed in the 5' to 3' orientation, with conserved sequences are highlighted. At the bottom is the genome organization of AmPV; the putative ORF for the viral RdRp is depicted by a green box. B ML phylogenetic tree of members of the family Mymonaviridae . The best-fit model (LG + F + R6) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified viruses represented in red font
(ii) Bunyavirales
The Bunyavirales (the only order in the class Ellioviricetes ) is one of the largest groups of segmented negative-sense single-stranded RNA viruses with mainly tripartite genomes [ 59 ], which includes many pathogenic strains that infect arthropods(such as mosquitoes, ticks, sand flies), plants, protozoans, and vertebrates, and even cause severe human diseases. Order Bunyavirales consists of 14 viral families, including Arenaviridae , Cruliviridae , Discoviridae , Fimoviridae , Hantaviridae , Leishbuviridae , Mypoviridae , Nairoviridae , Peribunyaviridae , Phasmaviridae , Phenuiviridae , Tospoviridae , Tulasviridae and Wupedeviridae . In this study, three complete or near complete RNA1 sequences related to bunyaviruses were identified and named according to their respective hosts: CoBV ( Conidiobolus obscurus bunyavirus; SRR6181013; 7,277 nt), GtBV ( Gaeumannomyces tritici bunyavirus; ERR3486069; 7,364 nt), and TaBV ( Thielaviopsis aethacetica bunyavirus; SRR12744489; 9,516 nt) (Fig. 4 A). The 5' and 3' terminal RNA segments of GtBV and TaBV complement each other, allowing the formation of a panhandle structure [ 60 ], which plays an essential role as promoters of genome transcription and replication [ 61 ], except for CoBV, as the 3' terminal of CoBV has not been fully obtained (Fig. 4 B). BLASTx results indicated that these three viruses had identities ranging from 32.97% to 54.20% to the best matches in the GenBank database. Phylogenetic analysis indicated that CoBV was classified into the family Phasmaviridae , with distant relationships to any of its genera; GtBV clustered well with members of the genus Entovirus of family Phenuiviridae ; while TaBV did not cluster with any known members of families within Bunyavirales , hence provisionally placed within the Bunya-like group (Fig. 4 C). Therefore, these three sequences should be considered as potential new family, genus, or species within the order Bunyavirales .
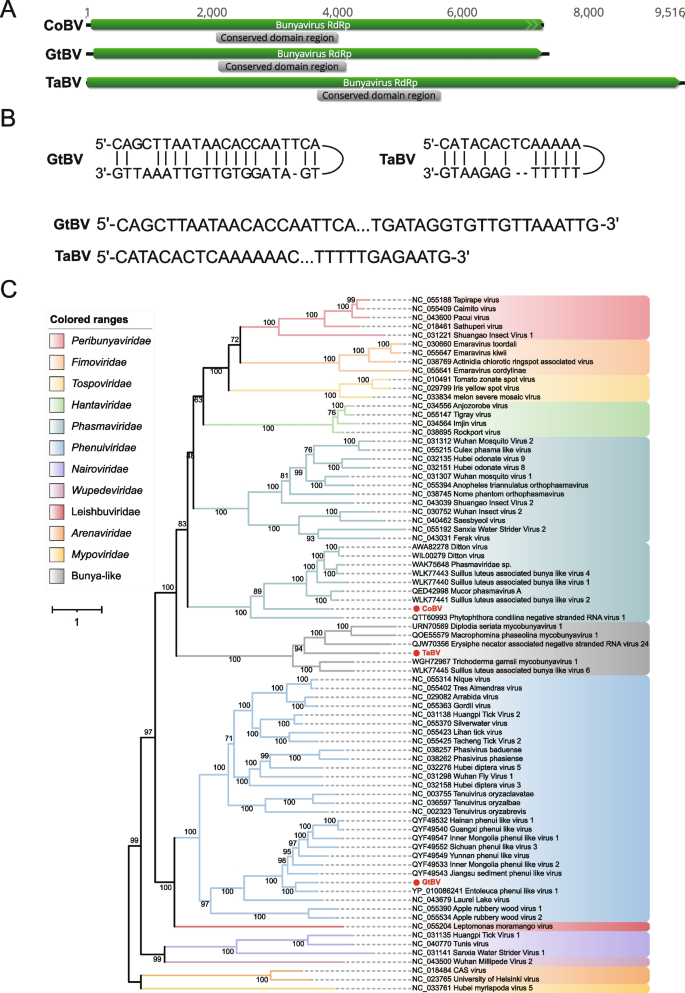
Identification of three new members in the order Bunyavirales . A The genome organization of CoBV, GtBV, and TaBV; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. B The complementary structures formed at the 5' and 3' ends of GtBV and TaBV. C ML phylogenetic tree of members of the order Bunyavirales . The best-fit model (VT + F + R8) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified viruses represented in red font
Double-stranded RNA viruses
Partitiviridae.
The Partitiviridae is a family of small, non-enveloped viruses, approximately 35–40 nm in diameter, with bisegmented double-stranded (ds) RNA genomes. Each segment is about 1.4–3.0 kb in size, resulting in a total size about 4 kb [ 62 ]. The family Partitiviridae is now divided into five genera: Alphapartitivirus , Betapartiivirus , Cryspovirus , Deltapartitivirus and Gammapartitivirus . Each genus has characteristic hosts: plants or fungi for Alphapartitivirus and Betapartitivirus , fungi for Gammapartitivirus , plants for Deltapartitivirus , and protozoa for Cryspovirus [ 62 ]. A complete dsRNA1 sequence Neocallimastix californiae partitivirus (NcPV) retrieved from Neocallimastix californiae (SRR15362281) has been identified as being associated with the family Partitiviridae . The BLASTp result indicated that it shared the highest aa identity of 41.5% with members of the genus Gammapartitivirus . According to the phylogenetic tree constructed based on RdRp, NcPV was confirmed to fall within the genus Gammapartitivirus (Fig. 5 ). Typical members of the genus Gammapartitivirus have two segments in their complete genome, namely dsRNA1 and dsRNA2, encoding RdRp and coat protein, respectively [ 62 ]. The larger dsRNA1 segment of NcPV measures 1,769 nt in length, with a GC content of 35.8%. It contains a single ORF encoding a 561 aa RdRp. A CDD search revealed that the RdRp of NcPV harbors a catalytic region spanning from 119 to 427aa. Regrettably, only the complete dsRNA1 segment was obtained. According to the classification principles of ICTV, due to the lack of information regarding dsRNA2, we are unable to propose it as a new species. It is worth noting that according to the Genus demarcation criteria ( https://ictv.global/report/chapter/partitiviridae/partitiviridae ), members of the genus Gammapartitivirus should have a dsRNA1 length ranging from 1645 to 1787 nt, and the RdRp length should fall between 519 and 539 aa. However, the length of dsRNA1 in NcPV is 1,769 nt, with RdRp being 561 aa, challenging this classification criterion. In fact, multiple strains have already exceeded this criterion, such as GenBank accession numbers: WBW48344, UDL14336, QKK35392, among others.
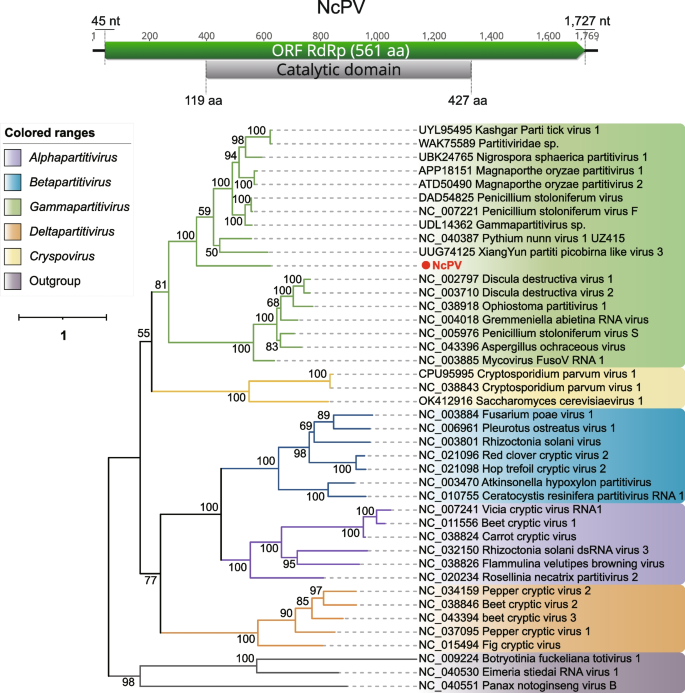
Identification of a new member in the family Partitiviridae . The genome organization of NcPV is depicted at the top; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. At the bottom is the ML phylogenetic tree of members of the family Partitiviridae . The best-fit model (VT + F + R4) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font
Long-term evolutionary relationships between fungal-associated viruses and hosts
Understanding the co-divergence history between viruses and hosts helps reveal patterns of virus transmission and infection and influences the biodiversity and stability of ecosystems. To explore the frequency of cross-species transmission and co-divergence among fungi-associated viruses, we constructed tanglegrams illustrating the interconnected evolutionary histories of viral families and their respective hosts through phylogenetic trees (Fig. 6 A). The results indicated that cross-species transmission (Host-jumping) consistently emerged as the most frequent evolutionary event among all groups of RNA viruses examined in this study (median, 66.79%; range, 60.00% to 79.07%) (Fig. 6 B). This finding is highly consistent with the evolutionary patterns of RNA viruses recently identified by Mifsud et al. in their extensive transcriptome survey of plants [ 63 ]. Members of the families Botourmiaviridae (79.07%) and Deltaflexiviridae (72.41%) were most frequently involved in cross-species transmission. The frequencies of co-divergence (median, 20.19%; range, 6.98% to 27.78%), duplication (median, 10.60%; range, 0% to 22.45%), and extinction (median, 2.42%; range, 0% to 5.56%) events involved in the evolution of fungi-associated viruses gradually decrease. Specifically, members of the family Benyviridae exhibited the highest frequency of co-divergence events, which also supports the findings reported by Mifsud et al.; certain studies propose that members of Benyviridae are transmitted via zoospores of plasmodiophorid protist [ 64 ]. It's speculated that the ancestor of these viruses underwent interkingdom horizontal transfer between plants and protists over evolutionary timelines [ 65 ]. Members of the family Mitoviridae showed the highest frequency of duplication events; and members of the families Benyviridae and Partitiviridae demonstrated the highest frequency of extinction events. Not surprisingly, this result is influenced by the current limited understanding of virus-host relationships. On one hand, viruses whose hosts cannot be recognized through published literature or information provided by authors have been overlooked. On the other hand, the number of viruses recorded in reference databases represents just the tip of the iceberg within the entire virosphere. The involvement of a more extensive sample size in the future should change this evolutionary landscape.
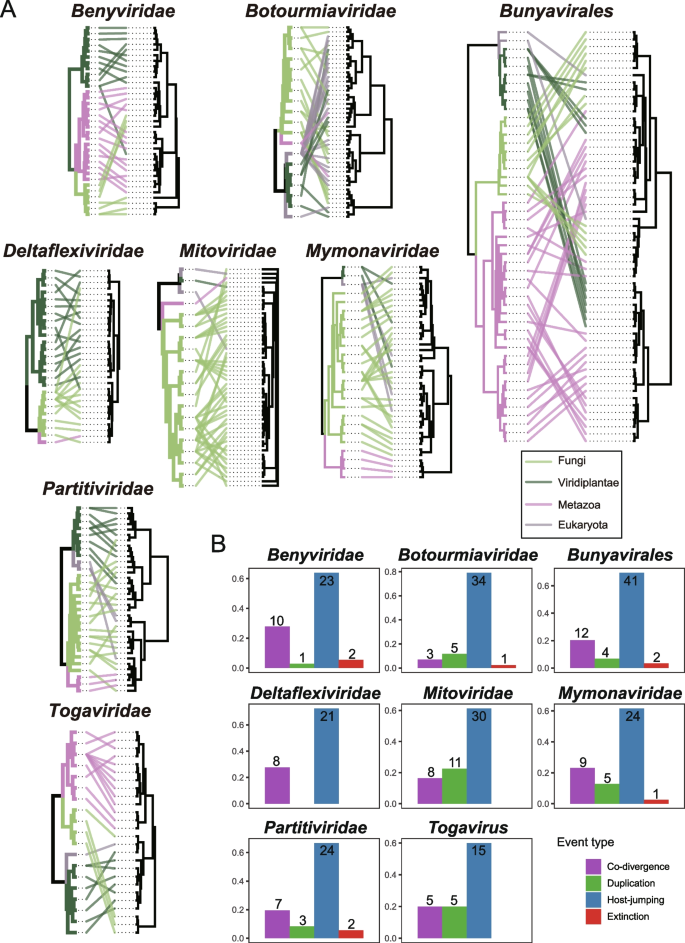
Co-evolutionary analysis of virus and host. A Tanglegram of phylogenetic trees for virus orders/families and their hosts. Lines and branches are color-coded to indicate host clades. The cophylo function in phytools was employed to enhance congruence between the host (left) and virus (right) phylogenies. B Reconciliation analysis of virus groups. The bar chart illustrates the proportional range of possible evolutionary events, with the frequency of each event displayed at the top of its respective column
Our understanding of the interactions between fungi and their associated viruses has long been constrained by insufficient sampling of fungal species. Advances in metagenomics in recent decades have led to a rapid expansion of the known viral sequence space, but it is far from saturated. The diversity of hosts, the instability of the viral structures (especially RNA viruses), and the propensity to exchange genetic material with other host viruses all contribute to the unparalleled diversity of viral genomes [ 66 ]. Fungi are diverse and widely distributed in nature and are closely related to humans. A few fungi can parasitize immunocompromised humans, but their adverse effects are limited. As decomposers in the biological chain, fungi can decompose the remains of plants and animals and maintain the material cycle in the biological world [ 67 ]. In agricultural production, many fungi are plant pathogens, and about 80% of plant diseases are caused by fungi. However, little is currently known about the diversity of mycoviruses and how these viruses affect fungal phenotypes, fungal-host interactions, and virus evolution, and the sequencing depth of fungal libraries in most public databases only meets the needs of studying bacterial genomes. Sampling viruses from a larger diversity of fungal hosts should lead to new and improved evolutionary scenarios.
RNA viruses are widespread in deep-sea sediments [ 68 ], freshwater [ 69 ], sewage [ 70 ], and rhizosphere soils [ 71 ]. Compared to DNA viruses, RNA viruses are less conserved, prone to mutation, and can transfer between different hosts, potentially forming highly differentiated and unrecognized novel viruses. This characteristic increases the difficulty of monitoring these viruses. Previously, all discovered mycoviruses were RNA viruses. Until 2010, Yu et al. reported the discovery of a DNA virus, namely SsHADV-1, in fungi for the first time [ 72 ]. Subsequently, new fungal-related DNA viruses are continually being identified [ 73 , 74 , 75 ]. Currently, viruses have been found in all major groups of fungi, and approximately 100 types of fungi can be infected by viruses, instances exist where one virus can infect multiple fungi, or one fungus can be infected by several viruses simultaneously. The transmission of mycoviruses differs from that of animal and plant viruses and is mainly categorized into vertical and horizontal transmission [ 76 ]. Vertical transmission refers to the spread of the mycovirus to the next generation through the sexual or asexual spores of the fungus, while horizontal transmission refers to the spread of the mycovirus from one strain to another through fusion between hyphae. In the phylum Ascomycota , mycoviruses generally exhibit a low ability to transmit vertically through ascospores, but they are commonly transmitted vertically to progeny strains through asexual spores [ 77 ].
In this study, we identified two novel species belonging to different genera within the family Mitoviridae . Interestingly, they both simultaneously infect the same fungus— Thielaviopsis ethacetica , the causal agent of pineapple sett rot disease in sugarcane [ 78 ]. Previously, a report identified three different mitoviruses in Fusarium circinatum [ 79 ]. These findings suggest that there may be a certain level of adaptability or symbiotic relationship among members of the family Mitoviridae . Benyviruses are typically considered to infect plants, but recent evidence suggests that they can also infect fungi, such as Agaricus bisporus [ 80 ], further reinforced by the virus we discovered in Gaeumannomyces tritici . Moreover, members of the family Botourmiaviridae commonly exhibit a broad host range, with viruses closely related to CrBV capable of infecting members of Eukaryota , Viridiplantae , and Metazoa , in addition to fungi (Supplementary Fig. 1). The LsDV identified in this study shared the closest phylogenetic relationship with a virus identified from Macrotermes carbonarius in southern Vietnam (17_N1 + N237) [ 81 ]. M. carbonarius is an open-air foraging species that collects plant litter and wood debris to cultivate fungi in fungal gardens [ 82 ], termites may act as vectors, transmitting deltaflexivirus to other fungi. Furthermore, the viruses we identified, typically associated with fungi, also deepen their connections with species from other kingdoms on the tanglegram tree. For example, while Partitiviridae are naturally associated with fungi and plants, NcPV also shows close connections with Metazoa . In fact, based largely on phylogenetic predictions, various eukaryotic viruses have been found to undergo horizontal transfer between organisms of plants, fungi, and animals [ 83 ]. The rice dwarf virus was demonstrated to infect both plant and insect vectors [ 84 ]; moreover, plant-infecting rhabdoviruses, tospoviruses, and tenuiviruses are now known to replicate and spread in vector insects and shuttle between plants and animals [ 85 ]. Furthermore, Bian et al. demonstrated that plant virus infection in plants enables Cryphonectria hypovirus 1 to undergo horizontal transfer from fungi to plants and other heterologous fungal species [ 86 ].
Recent studies have greatly expanded the diversity of mycoviruses [ 87 , 88 ]. Gilbert et al. [ 20 ] investigated publicly available fungal transcriptomes from the subphylum Pezizomycotina, resulting in the detection of 52 novel mycoviruses; Myers et al. [ 18 ] employed both culture-based and transcriptome-mining approaches to identify 85 unique RNA viruses across 333 fungi; Ruiz-Padilla et al. identified 62 new mycoviral species from 248 Botrytis cinerea field isolates; Zhou et al. identified 20 novel viruses from 90 fungal strains (across four different macrofungi species) [ 89 ]. However, compared to these studies, our work identified fewer novel viruses, possibly due to the following reasons: 1) The libraries from the same Bioproject are usually from the same strains (or isolates). Therefore, there is a certain degree of redundancy in the datasets collected for this study. 2) Contigs shorter than 1,500 nt were discarded, potentially resulting in the oversight of short viral molecules. 3) Establishing a threshold of 70% aa sequence identity may also lead to the exclusion of certain viruses. 4) Some poly(A)-enriched RNA-seq libraries are likely to miss non-polyadenylated RNA viral genomes.
Taxonomy is a dynamic science, evolving with improvements in analytical methods and the emergence of new data. Identifying and rectifying incorrect classifications when new information becomes available is an ongoing and inevitable process in today's rapidly expanding field of virology. For instance, in 1975, members of the genera Rubivirus and Alphavirus were initially grouped under the family Togaviridae ; however, in 2019, Rubivirus was reclassified into the family Matonaviridae due to recognized differences in transmission modes and virion structures [ 90 ]. Additionally, the conflicts between certain members of the genera Magoulivirus and Gammapartitivirus mentioned here and their current demarcation criteria (e.g., amino acid identity, nucleotide length thresholds) need to be reconsidered.
Taken together, these findings reveal the potential diversity and novelty within fungal-associated viral communities and discuss the genetic similarities among different fungal-associated viruses. These findings advance our understanding of fungal-associated viruses and suggest the importance of subsequent in-depth investigations into the interactions between fungi and viruses, which will shed light on the important roles of these viruses in the global fungal kingdom.
Availability of data and materials
The data reported in this paper have been deposited in the GenBase in National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, under accession numbers C_AA066339.1-C_AA066350.1 that are publicly accessible at https://ngdc.cncb.ac.cn/genbase . Please refer to Table 1 for details.
Leigh DM, Peranic K, Prospero S, Cornejo C, Curkovic-Perica M, Kupper Q, et al. Long-read sequencing reveals the evolutionary drivers of intra-host diversity across natural RNA mycovirus infections. Virus Evol. 2021;7(2):veab101. https://doi.org/10.1093/ve/veab101 . Epub 2022/03/19 PubMed PMID: 35299787; PubMed Central PMCID: PMCPMC8923234.
Article PubMed PubMed Central Google Scholar
Ghabrial SA, Suzuki N. Viruses of plant pathogenic fungi. Annu Rev Phytopathol. 2009;47:353–84. https://doi.org/10.1146/annurev-phyto-080508-081932 . Epub 2009/04/30 PubMed PMID: 19400634.
Article CAS PubMed Google Scholar
Ghabrial SA, Caston JR, Jiang D, Nibert ML, Suzuki N. 50-plus years of fungal viruses. Virology. 2015;479–480:356–68. https://doi.org/10.1016/j.virol.2015.02.034 . Epub 2015/03/17 PubMed PMID: 25771805.
Chen YM, Sadiq S, Tian JH, Chen X, Lin XD, Shen JJ, et al. RNA viromes from terrestrial sites across China expand environmental viral diversity. Nat Microbiol. 2022;7(8):1312–23. https://doi.org/10.1038/s41564-022-01180-2 . Epub 2022/07/29 PubMed PMID: 35902778.
Pearson MN, Beever RE, Boine B, Arthur K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol. 2009;10(1):115–28. https://doi.org/10.1111/j.1364-3703.2008.00503.x . Epub 2009/01/24 PubMed PMID: 19161358; PubMed Central PMCID: PMCPMC6640375.
Santiago-Rodriguez TM, Hollister EB. Unraveling the viral dark matter through viral metagenomics. Front Immunol. 2022;13:1005107. https://doi.org/10.3389/fimmu.2022.1005107 . Epub 2022/10/04 PubMed PMID: 36189246; PubMed Central PMCID: PMCPMC9523745.
Article CAS PubMed PubMed Central Google Scholar
Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol. 2008;159(5):349–57. https://doi.org/10.1016/j.resmic.2008.04.010 . Epub 2008/06/21 PubMed PMID: 18565737.
Guo W, Yan H, Ren X, Tang R, Sun Y, Wang Y, et al. Berberine induces resistance against tobacco mosaic virus in tobacco. Pest Manag Sci. 2020;76(5):1804–13. https://doi.org/10.1002/ps.5709 . Epub 2019/12/10 PubMed PMID: 31814252.
Villabruna N, Izquierdo-Lara RW, Schapendonk CME, de Bruin E, Chandler F, Thao TTN, et al. Profiling of humoral immune responses to norovirus in children across Europe. Sci Rep. 2022;12(1):14275. https://doi.org/10.1038/s41598-022-18383-6 . Epub 2022/08/23 PubMed PMID: 35995986.
Zhang Y, Gao J, Li Y. Diversity of mycoviruses in edible fungi. Virus Genes. 2022;58(5):377–91. https://doi.org/10.1007/s11262-022-01908-6 . Epub 2022/06/07 PubMed PMID: 35668282.
Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 2019;26(4):527–41. https://doi.org/10.1016/j.chom.2019.09.009 . Epub 2019/10/11 PubMed PMID: 31600503.
Botella L, Janousek J, Maia C, Jung MH, Raco M, Jung T. Marine Oomycetes of the Genus Halophytophthora harbor viruses related to Bunyaviruses. Front Microbiol. 2020;11:1467. https://doi.org/10.3389/fmicb.2020.01467 . Epub 2020/08/08 PubMed PMID: 32760358; PubMed Central PMCID: PMCPMC7375090.
Kotta-Loizou I. Mycoviruses and their role in fungal pathogenesis. Curr Opin Microbiol. 2021;63:10–8. https://doi.org/10.1016/j.mib.2021.05.007 . Epub 2021/06/09 PubMed PMID: 34102567.
Ellis LF, Kleinschmidt WJ. Virus-like particles of a fraction of statolon, a mould product. Nature. 1967;215(5101):649–50. https://doi.org/10.1038/215649a0 . Epub 1967/08/05 PubMed PMID: 6050227.
Banks GT, Buck KW, Chain EB, Himmelweit F, Marks JE, Tyler JM, et al. Viruses in fungi and interferon stimulation. Nature. 1968;218(5141):542–5. https://doi.org/10.1038/218542a0 . Epub 1968/05/11 PubMed PMID: 4967851.
Jia J, Fu Y, Jiang D, Mu F, Cheng J, Lin Y, et al. Interannual dynamics, diversity and evolution of the virome in Sclerotinia sclerotiorum from a single crop field. Virus Evol. 2021;7(1):veab032. https://doi.org/10.1093/ve/veab032 .
Mu F, Li B, Cheng S, Jia J, Jiang D, Fu Y, et al. Nine viruses from eight lineages exhibiting new evolutionary modes that co-infect a hypovirulent phytopathogenic fungus. Plos Pathog. 2021;17(8):e1009823. https://doi.org/10.1371/journal.ppat.1009823 . Epub 2021/08/25 PubMed PMID: 34428260; PubMed Central PMCID: PMCPMC8415603.
Myers JM, Bonds AE, Clemons RA, Thapa NA, Simmons DR, Carter-House D, et al. Survey of early-diverging lineages of fungi reveals abundant and diverse Mycoviruses. mBio. 2020;11(5):e02027. https://doi.org/10.1128/mBio.02027-20 . Epub 2020/09/10 PubMed PMID: 32900807; PubMed Central PMCID: PMCPMC7482067.
Ruiz-Padilla A, Rodriguez-Romero J, Gomez-Cid I, Pacifico D, Ayllon MA. Novel Mycoviruses discovered in the Mycovirome of a Necrotrophic fungus. MBio. 2021;12(3):e03705. https://doi.org/10.1128/mBio.03705-20 . Epub 2021/05/13 PubMed PMID: 33975945; PubMed Central PMCID: PMCPMC8262958.
Gilbert KB, Holcomb EE, Allscheid RL, Carrington JC. Hiding in plain sight: new virus genomes discovered via a systematic analysis of fungal public transcriptomes. Plos One. 2019;14(7):e0219207. https://doi.org/10.1371/journal.pone.0219207 . Epub 2019/07/25 PubMed PMID: 31339899; PubMed Central PMCID: PMCPMC6655640.
Khan HA, Telengech P, Kondo H, Bhatti MF, Suzuki N. Mycovirus hunting revealed the presence of diverse viruses in a single isolate of the Phytopathogenic fungus diplodia seriata from Pakistan. Front Cell Infect Microbiol. 2022;12:913619. https://doi.org/10.3389/fcimb.2022.913619 . Epub 2022/07/19 PubMed PMID: 35846770; PubMed Central PMCID: PMCPMC9277117.
Kotta-Loizou I, Coutts RHA. Mycoviruses in Aspergilli: a comprehensive review. Front Microbiol. 2017;8:1699. https://doi.org/10.3389/fmicb.2017.01699 . Epub 2017/09/22 PubMed PMID: 28932216; PubMed Central PMCID: PMCPMC5592211.
Garcia-Pedrajas MD, Canizares MC, Sarmiento-Villamil JL, Jacquat AG, Dambolena JS. Mycoviruses in biological control: from basic research to field implementation. Phytopathology. 2019;109(11):1828–39. https://doi.org/10.1094/PHYTO-05-19-0166-RVW . Epub 2019/08/10 PubMed PMID: 31398087.
Rigling D, Prospero S. Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Mol Plant Pathol. 2018;19(1):7–20. https://doi.org/10.1111/mpp.12542 . Epub 2017/02/01 PubMed PMID: 28142223; PubMed Central PMCID: PMCPMC6638123.
Okada R, Ichinose S, Takeshita K, Urayama SI, Fukuhara T, Komatsu K, et al. Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: down-regulation of host growth and up-regulation of host plant pathogenicity. Virology. 2018;519:23–32. https://doi.org/10.1016/j.virol.2018.03.027 . Epub 2018/04/10 PubMed PMID: 29631173.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. https://doi.org/10.1038/nmeth.1923 . Epub 2012/03/06 PubMed PMID: 22388286; PubMed Central PMCID: PMCPMC3322381.
Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes De Novo assembler. Curr Protoc Bioinform. 2020;70(1):e102. https://doi.org/10.1002/cpbi.102 . Epub 2020/06/20 PubMed PMID: 32559359.
Article CAS Google Scholar
Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, et al. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. https://doi.org/10.1016/j.ymeth.2016.02.020 . Epub 2016/03/26 PubMed PMID: 27012178.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. https://doi.org/10.1093/bioinformatics/btp698 . Epub 2010/01/19 PubMed PMID: 20080505; PubMed Central PMCID: PMCPMC2828108.
Roux S, Paez-Espino D, Chen IA, Palaniappan K, Ratner A, Chu K, et al. IMG/VR v3: an integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 2021;49(D1):D764–75. https://doi.org/10.1093/nar/gkaa946 . Epub 2020/11/03 PubMed PMID: 33137183; PubMed Central PMCID: PMCPMC7778971.
Mirdita M, Steinegger M, Soding J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics. 2019;35(16):2856–8. https://doi.org/10.1093/bioinformatics/bty1057 . Epub 2019/01/08 PubMed PMID: 30615063; PubMed Central PMCID: PMCPMC6691333.
Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18(4):366–8. https://doi.org/10.1038/s41592-021-01101-x . Epub 2021/04/09 PubMed PMID: 33828273; PubMed Central PMCID: PMCPMC8026399.
Shen W, Ren H. TaxonKit: A practical and efficient NCBI taxonomy toolkit. J Genet Genomics. 2021;48(9):844–50. https://doi.org/10.1016/j.jgg.2021.03.006 . Epub 2021/05/19 PubMed PMID: 34001434.
Article PubMed Google Scholar
Gautam A, Felderhoff H, Bagci C, Huson DH. Using AnnoTree to get more assignments, faster, in DIAMOND+MEGAN microbiome analysis. mSystems. 2022;7(1):e0140821. https://doi.org/10.1128/msystems.01408-21 . Epub 2022/02/23 PubMed PMID: 35191776; PubMed Central PMCID: PMCPMC8862659.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. https://doi.org/10.1093/molbev/msy096 . Epub 2018/05/04 PubMed PMID: 29722887; PubMed Central PMCID: PMCPMC5967553.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–4. https://doi.org/10.1093/molbev/msaa015 . Epub 2020/02/06 PubMed PMID: 32011700; PubMed Central PMCID: PMCPMC7182206.
Letunic I, Bork P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res (2024). https://doi.org/10.1093/nar/gkae268
Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. Plos One. 2014;9(9):e108277. https://doi.org/10.1371/journal.pone.0108277 . Epub 2014/09/27 PubMed PMID: 25259891; PubMed Central PMCID: PMCPMC4178126.
Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, et al. TimeTree 5: an expanded resource for species divergence times. Mol Biol Evol. 2022;39(8):msac174. https://doi.org/10.1093/molbev/msac174 . Epub 2022/08/07 PubMed PMID: 35932227; PubMed Central PMCID: PMCPMC9400175.
Revell LJ. phytools 2.0: an updated R ecosystem for phylogenetic comparative methods (and other things). PeerJ. 2024;12:e16505. https://doi.org/10.7717/peerj.16505 . Epub 2024/01/09 PubMed PMID: 38192598; PubMed Central PMCID: PMCPMC10773453.
Santichaivekin S, Yang Q, Liu J, Mawhorter R, Jiang J, Wesley T, et al. eMPRess: a systematic cophylogeny reconciliation tool. Bioinformatics. 2021;37(16):2481–2. https://doi.org/10.1093/bioinformatics/btaa978 . Epub 2020/11/21 PubMed PMID: 33216126.
Ma W, Smirnov D, Libeskind-Hadas R. DTL reconciliation repair. BMC Bioinformatics. 2017;18(Suppl 3):76. https://doi.org/10.1186/s12859-017-1463-9 . Epub 2017/04/01 PubMed PMID: 28361686; PubMed Central PMCID: PMCPMC5374596.
Members C-N, Partners. Database resources of the national genomics data center, China national center for bioinformation in 2024. Nucleic Acids Res. 2024;52(D1):D18–32. https://doi.org/10.1093/nar/gkad1078 . Epub 2023/11/29 PubMed PMID: 38018256; PubMed Central PMCID: PMCPMC10767964.
Article Google Scholar
Shafik K, Umer M, You H, Aboushedida H, Wang Z, Ni D, et al. Characterization of a Novel Mitovirus infecting Melanconiella theae isolated from tea plants. Front Microbiol. 2021;12: 757556. https://doi.org/10.3389/fmicb.2021.757556 . Epub 2021/12/07 PubMed PMID: 34867881; PubMed Central PMCID: PMCPMC8635788
Kamaruzzaman M, He G, Wu M, Zhang J, Yang L, Chen W, et al. A novel Partitivirus in the Hypovirulent isolate QT5–19 of the plant pathogenic fungus Botrytis cinerea. Viruses. 2019;11(1):24. https://doi.org/10.3390/v11010024 . Epub 2019/01/06 PubMed PMID: 30609795; PubMed Central PMCID: PMCPMC6356794.
Akata I, Keskin E, Sahin E. Molecular characterization of a new mitovirus hosted by the ectomycorrhizal fungus Albatrellopsis flettii. Arch Virol. 2021;166(12):3449–54. https://doi.org/10.1007/s00705-021-05250-4 . Epub 2021/09/24 PubMed PMID: 34554305.
Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, et al. Recent changes to virus taxonomy ratified by the international committee on taxonomy of viruses (2022). Arch Virol. 2022;167(11):2429–40. https://doi.org/10.1007/s00705-022-05516-5 . Epub 2022/08/24 PubMed PMID: 35999326; PubMed Central PMCID: PMCPMC10088433.
Alvarez-Quinto R, Grinstead S, Jones R, Mollov D. Complete genome sequence of a new mitovirus associated with walking iris (Trimezia northiana). Arch Virol. 2023;168(11):273. https://doi.org/10.1007/s00705-023-05901-8 . Epub 2023/10/17 PubMed PMID: 37845386.
Gilmer D, Ratti C, Ictv RC. ICTV Virus taxonomy profile: Benyviridae. J Gen Virol. 2017;98(7):1571–2. https://doi.org/10.1099/jgv.0.000864 . Epub 2017/07/18 PubMed PMID: 28714846; PubMed Central PMCID: PMCPMC5656776.
Wetzel V, Willlems G, Darracq A, Galein Y, Liebe S, Varrelmann M. The Beta vulgaris-derived resistance gene Rz2 confers broad-spectrum resistance against soilborne sugar beet-infecting viruses from different families by recognizing triple gene block protein 1. Mol Plant Pathol. 2021;22(7):829–42. https://doi.org/10.1111/mpp.13066 . Epub 2021/05/06 PubMed PMID: 33951264; PubMed Central PMCID: PMCPMC8232027.
Ayllon MA, Turina M, Xie J, Nerva L, Marzano SL, Donaire L, et al. ICTV Virus taxonomy profile: Botourmiaviridae. J Gen Virol. 2020;101(5):454–5. https://doi.org/10.1099/jgv.0.001409 . Epub 2020/05/08 PubMed PMID: 32375992; PubMed Central PMCID: PMCPMC7414452.
Xiao J, Wang X, Zheng Z, Wu Y, Wang Z, Li H, et al. Molecular characterization of a novel deltaflexivirus infecting the edible fungus Pleurotus ostreatus. Arch Virol. 2023;168(6):162. https://doi.org/10.1007/s00705-023-05789-4 . Epub 2023/05/17 PubMed PMID: 37195309.
Canuti M, Rodrigues B, Lang AS, Dufour SC, Verhoeven JTP. Novel divergent members of the Kitrinoviricota discovered through metagenomics in the intestinal contents of red-backed voles (Clethrionomys gapperi). Int J Mol Sci. 2022;24(1):131. https://doi.org/10.3390/ijms24010131 . Epub 2023/01/09 PubMed PMID: 36613573; PubMed Central PMCID: PMCPMC9820622.
Hermanns K, Zirkel F, Kopp A, Marklewitz M, Rwego IB, Estrada A, et al. Discovery of a novel alphavirus related to Eilat virus. J Gen Virol. 2017;98(1):43–9. https://doi.org/10.1099/jgv.0.000694 . Epub 2017/02/17 PubMed PMID: 28206905.
Jiang D, Ayllon MA, Marzano SL, Ictv RC. ICTV Virus taxonomy profile: Mymonaviridae. J Gen Virol. 2019;100(10):1343–4. https://doi.org/10.1099/jgv.0.001301 . Epub 2019/09/04 PubMed PMID: 31478828.
Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, et al. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci U S A. 2014;111(33):12205–10. https://doi.org/10.1073/pnas.1401786111 . Epub 2014/08/06 PubMed PMID: 25092337; PubMed Central PMCID: PMCPMC4143027.
Zhong J, Li P, Gao BD, Zhong SY, Li XG, Hu Z, et al. Novel and diverse mycoviruses co-infecting a single strain of the phytopathogenic fungus Alternaria dianthicola. Front Cell Infect Microbiol. 2022;12:980970. https://doi.org/10.3389/fcimb.2022.980970 . Epub 2022/10/15 PubMed PMID: 36237429; PubMed Central PMCID: PMCPMC9552818.
Wang W, Wang X, Tu C, Yang M, Xiang J, Wang L, et al. Novel Mycoviruses discovered from a Metatranscriptomics survey of the Phytopathogenic Alternaria Fungus. Viruses. 2022;14(11):2552. https://doi.org/10.3390/v14112552 . Epub 2022/11/25 PubMed PMID: 36423161; PubMed Central PMCID: PMCPMC9693364.
Sun Y, Li J, Gao GF, Tien P, Liu W. Bunyavirales ribonucleoproteins: the viral replication and transcription machinery. Crit Rev Microbiol. 2018;44(5):522–40. https://doi.org/10.1080/1040841X.2018.1446901 . Epub 2018/03/09 PubMed PMID: 29516765.
Li P, Bhattacharjee P, Gagkaeva T, Wang S, Guo L. A novel bipartite negative-stranded RNA mycovirus of the order Bunyavirales isolated from the phytopathogenic fungus Fusarium sibiricum. Arch Virol. 2023;169(1):13. https://doi.org/10.1007/s00705-023-05942-z . Epub 2023/12/29 PubMed PMID: 38155262.
Ferron F, Weber F, de la Torre JC, Reguera J. Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res. 2017;234:118–34. https://doi.org/10.1016/j.virusres.2017.01.018 . Epub 2017/02/01 PubMed PMID: 28137457; PubMed Central PMCID: PMCPMC7114536.
Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S, et al. ICTV Virus taxonomy profile: Partitiviridae. J Gen Virol. 2018;99(1):17–8. https://doi.org/10.1099/jgv.0.000985 . Epub 2017/12/08 PubMed PMID: 29214972; PubMed Central PMCID: PMCPMC5882087.
Mifsud JCO, Gallagher RV, Holmes EC, Geoghegan JL. Transcriptome mining expands knowledge of RNA viruses across the plant Kingdom. J Virol. 2022;96(24):e0026022. https://doi.org/10.1128/jvi.00260-22 . Epub 2022/06/01 PubMed PMID: 35638822; PubMed Central PMCID: PMCPMC9769393.
Tamada T, Kondo H. Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors. J Gen Plant Pathol. 2013;79:307–20.
Dolja VV, Krupovic M, Koonin EV. Deep roots and splendid boughs of the global plant virome. Annu Rev Phytopathol. 2020;58:23–53.
Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, et al. Global organization and proposed Megataxonomy of the virus world. Microbiol Mol Biol Rev. 2020;84(2):e00061. https://doi.org/10.1128/MMBR.00061-19 . Epub 2020/03/07 PubMed PMID: 32132243; PubMed Central PMCID: PMCPMC7062200.
Osono T. Role of phyllosphere fungi of forest trees in the development of decomposer fungal communities and decomposition processes of leaf litter. Can J Microbiol. 2006;52(8):701–16. https://doi.org/10.1139/w06-023 . Epub 2006/08/19 PubMed PMID: 16917528.
Li Z, Pan D, Wei G, Pi W, Zhang C, Wang JH, et al. Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J. 2021;15(8):2366–78. https://doi.org/10.1038/s41396-021-00932-y . Epub 2021/03/03 PubMed PMID: 33649554; PubMed Central PMCID: PMCPMC8319345.
Hierweger MM, Koch MC, Rupp M, Maes P, Di Paola N, Bruggmann R, et al. Novel Filoviruses, Hantavirus, and Rhabdovirus in freshwater fish, Switzerland, 2017. Emerg Infect Dis. 2021;27(12):3082–91. https://doi.org/10.3201/eid2712.210491 . Epub 2021/11/23 PubMed PMID: 34808081; PubMed Central PMCID: PMCPMC8632185.
La Rosa G, Iaconelli M, Mancini P, Bonanno Ferraro G, Veneri C, Bonadonna L, et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ. 2020;736:139652. https://doi.org/10.1016/j.scitotenv.2020.139652 . Epub 2020/05/29 PubMed PMID: 32464333; PubMed Central PMCID: PMCPMC7245320.
Sutela S, Poimala A, Vainio EJ. Viruses of fungi and oomycetes in the soil environment. FEMS Microbiol Ecol. 2019;95(9):fiz119. https://doi.org/10.1093/femsec/fiz119 . Epub 2019/08/01 PubMed PMID: 31365065.
Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci U S A. 2010;107(18):8387–92. https://doi.org/10.1073/pnas.0913535107 . Epub 2010/04/21 PubMed PMID: 20404139; PubMed Central PMCID: PMCPMC2889581.
Li P, Wang S, Zhang L, Qiu D, Zhou X, Guo L. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci Adv. 2020;6(14):eaay9634. https://doi.org/10.1126/sciadv.aay9634 . Epub 2020/04/15 PubMed PMID: 32284975; PubMed Central PMCID: PMCPMC7138691.
Khalifa ME, MacDiarmid RM. A mechanically transmitted DNA Mycovirus is targeted by the defence machinery of its host, Botrytis cinerea. Viruses. 2021;13(7):1315. https://doi.org/10.3390/v13071315 . Epub 2021/08/11 PubMed PMID: 34372522; PubMed Central PMCID: PMCPMC8309985.
Yu X, Li B, Fu Y, Xie J, Cheng J, Ghabrial SA, et al. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc Natl Acad Sci U S A. 2013;110(4):1452–7. https://doi.org/10.1073/pnas.1213755110 . Epub 2013/01/09 PubMed PMID: 23297222; PubMed Central PMCID: PMCPMC3557086.
Nuss DL. Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol. 2005;3(8):632–42. https://doi.org/10.1038/nrmicro1206 . Epub 2005/08/03 PubMed PMID: 16064055.
Coenen A, Kevei F, Hoekstra RF. Factors affecting the spread of double-stranded RNA viruses in Aspergillus nidulans. Genet Res. 1997;69(1):1–10. https://doi.org/10.1017/s001667239600256x . Epub 1997/02/01 PubMed PMID: 9164170.
Freitas CSA, Maciel LF, Dos Correa Santos RA, Costa O, Maia FCB, Rabelo RS, et al. Bacterial volatile organic compounds induce adverse ultrastructural changes and DNA damage to the sugarcane pathogenic fungus Thielaviopsis ethacetica. Environ Microbiol. 2022;24(3):1430–53. https://doi.org/10.1111/1462-2920.15876 . Epub 2022/01/08 PubMed PMID: 34995419.
Martinez-Alvarez P, Vainio EJ, Botella L, Hantula J, Diez JJ. Three mitovirus strains infecting a single isolate of Fusarium circinatum are the first putative members of the family Narnaviridae detected in a fungus of the genus Fusarium. Arch Virol. 2014;159(8):2153–5. https://doi.org/10.1007/s00705-014-2012-8 . Epub 2014/02/13 PubMed PMID: 24519462.
Deakin G, Dobbs E, Bennett JM, Jones IM, Grogan HM, Burton KS. Multiple viral infections in Agaricus bisporus - characterisation of 18 unique RNA viruses and 8 ORFans identified by deep sequencing. Sci Rep. 2017;7(1):2469. https://doi.org/10.1038/s41598-017-01592-9 . Epub 2017/05/28 PubMed PMID: 28550284; PubMed Central PMCID: PMCPMC5446422.
Litov AG, Zueva AI, Tiunov AV, Van Thinh N, Belyaeva NV, Karganova GG. Virome of three termite species from Southern Vietnam. Viruses. 2022;14(5):860. https://doi.org/10.3390/v14050860 . Epub 2022/05/29 PubMed PMID: 35632601; PubMed Central PMCID: PMCPMC9143207.
Hu J, Neoh KB, Appel AG, Lee CY. Subterranean termite open-air foraging and tolerance to desiccation: Comparative water relation of two sympatric Macrotermes spp. (Blattodea: Termitidae). Comp Biochem Physiol A Mol Integr Physiol. 2012;161(2):201–7. https://doi.org/10.1016/j.cbpa.2011.10.028 . Epub 2011/11/17 PubMed PMID: 22085890.
Kondo H, Botella L, Suzuki N. Mycovirus diversity and evolution revealed/inferred from recent studies. Annu Rev Phytopathol. 2022;60:307–36. https://doi.org/10.1146/annurev-phyto-021621-122122 . Epub 2022/05/25 PubMed PMID: 35609970.
Fukushi T. Relationships between propagative rice viruses and their vectors. 1969.
Google Scholar
Sun L, Kondo H, Bagus AI. Cross-kingdom virus infection. Encyclopedia of Virology: Volume 1–5. 4th Ed. Elsevier; 2020. pp. 443–9. https://doi.org/10.1016/B978-0-12-809633-8.21320-4 .
Bian R, Andika IB, Pang T, Lian Z, Wei S, Niu E, et al. Facilitative and synergistic interactions between fungal and plant viruses. Proc Natl Acad Sci U S A. 2020;117(7):3779–88. https://doi.org/10.1073/pnas.1915996117 . Epub 2020/02/06 PubMed PMID: 32015104; PubMed Central PMCID: PMCPMC7035501.
Chiapello M, Rodriguez-Romero J, Ayllon MA, Turina M. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol. 2020;6(2):veaa058. https://doi.org/10.1093/ve/veaa058 . Epub 2020/12/17 PubMed PMID: 33324489; PubMed Central PMCID: PMCPMC7724247.
Sutela S, Forgia M, Vainio EJ, Chiapello M, Daghino S, Vallino M, et al. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol. 2020;6(2):veaa076. https://doi.org/10.1093/ve/veaa076 . Epub 2020/12/17 PubMed PMID: 33324490; PubMed Central PMCID: PMCPMC7724248.
Zhou K, Zhang F, Deng Y. Comparative analysis of viromes identified in multiple macrofungi. Viruses. 2024;16(4):597. https://doi.org/10.3390/v16040597 . Epub 2024/04/27 PubMed PMID: 38675938; PubMed Central PMCID: PMCPMC11054281.
Siddell SG, Smith DB, Adriaenssens E, Alfenas-Zerbini P, Dutilh BE, Garcia ML, et al. Virus taxonomy and the role of the International Committee on Taxonomy of Viruses (ICTV). J Gen Virol. 2023;104(5):001840. https://doi.org/10.1099/jgv.0.001840 . Epub 2023/05/04 PubMed PMID: 37141106; PubMed Central PMCID: PMCPMC10227694.
Download references
Acknowledgements
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; WZ and CZ contributed to the conception and design; XL, ZD, JXU, WL and PN contributed to the collection and assembly of data; XL, ZD and JXE contributed to the data analysis and interpretation.
This research was supported by National Key Research and Development Programs of China [No.2023YFD1801301 and 2022YFC2603801] and the National Natural Science Foundation of China [No.82341106].
Author information
Xiang Lu, Ziyuan Dai and Jiaxin Xue are equally contributed to this works.
Authors and Affiliations
Institute of Critical Care Medicine, The Affiliated People’s Hospital, Jiangsu University, Zhenjiang, 212002, China
Xiang Lu & Wen Zhang
Department of Microbiology, School of Medicine, Jiangsu University, Zhenjiang, 212013, China
Xiang Lu, Jiaxin Xue & Wen Zhang
Department of Clinical Laboratory, Affiliated Hospital 6 of Nantong University, Yancheng Third People’s Hospital, Yancheng, Jiangsu, China
Clinical Laboratory Center, The Affiliated Taizhou People’s Hospital of Nanjing Medical University, Taizhou, 225300, China
Wang Li, Ping Ni, Juan Xu, Chenglin Zhou & Wen Zhang
You can also search for this author in PubMed Google Scholar
Contributions
Corresponding authors.
Correspondence to Juan Xu , Chenglin Zhou or Wen Zhang .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary material 1., supplementary material 2., supplementary material 3., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Lu, X., Dai, Z., Xue, J. et al. Discovery of novel RNA viruses through analysis of fungi-associated next-generation sequencing data. BMC Genomics 25 , 517 (2024). https://doi.org/10.1186/s12864-024-10432-w
Download citation
Received : 19 March 2024
Accepted : 20 May 2024
Published : 27 May 2024
DOI : https://doi.org/10.1186/s12864-024-10432-w
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
BMC Genomics
ISSN: 1471-2164
- Submission enquiries: [email protected]
- General enquiries: [email protected]
- Open access
- Published: 27 May 2024
Current status of community resources and priorities for weed genomics research
- Jacob Montgomery 1 ,
- Sarah Morran 1 ,
- Dana R. MacGregor ORCID: orcid.org/0000-0003-0543-0408 2 ,
- J. Scott McElroy ORCID: orcid.org/0000-0003-0331-3697 3 ,
- Paul Neve ORCID: orcid.org/0000-0002-3136-5286 4 ,
- Célia Neto ORCID: orcid.org/0000-0003-3256-5228 4 ,
- Martin M. Vila-Aiub ORCID: orcid.org/0000-0003-2118-290X 5 ,
- Maria Victoria Sandoval 5 ,
- Analia I. Menéndez ORCID: orcid.org/0000-0002-9681-0280 6 ,
- Julia M. Kreiner ORCID: orcid.org/0000-0002-8593-1394 7 ,
- Longjiang Fan ORCID: orcid.org/0000-0003-4846-0500 8 ,
- Ana L. Caicedo ORCID: orcid.org/0000-0002-0378-6374 9 ,
- Peter J. Maughan 10 ,
- Bianca Assis Barbosa Martins 11 ,
- Jagoda Mika 11 ,
- Alberto Collavo 11 ,
- Aldo Merotto Jr. ORCID: orcid.org/0000-0002-1581-0669 12 ,
- Nithya K. Subramanian ORCID: orcid.org/0000-0002-1659-7396 13 ,
- Muthukumar V. Bagavathiannan ORCID: orcid.org/0000-0002-1107-7148 13 ,
- Luan Cutti ORCID: orcid.org/0000-0002-2867-7158 14 ,
- Md. Mazharul Islam 15 ,
- Bikram S. Gill ORCID: orcid.org/0000-0003-4510-9459 16 ,
- Robert Cicchillo 17 ,
- Roger Gast 17 ,
- Neeta Soni ORCID: orcid.org/0000-0002-4647-8355 17 ,
- Terry R. Wright ORCID: orcid.org/0000-0002-3969-2812 18 ,
- Gina Zastrow-Hayes 18 ,
- Gregory May 18 ,
- Jenna M. Malone ORCID: orcid.org/0000-0002-9637-2073 19 ,
- Deepmala Sehgal ORCID: orcid.org/0000-0002-4141-1784 20 ,
- Shiv Shankhar Kaundun ORCID: orcid.org/0000-0002-7249-2046 20 ,
- Richard P. Dale 20 ,
- Barend Juan Vorster ORCID: orcid.org/0000-0003-3518-3508 21 ,
- Bodo Peters 11 ,
- Jens Lerchl ORCID: orcid.org/0000-0002-9633-2653 22 ,
- Patrick J. Tranel ORCID: orcid.org/0000-0003-0666-4564 23 ,
- Roland Beffa ORCID: orcid.org/0000-0003-3109-388X 24 ,
- Alexandre Fournier-Level ORCID: orcid.org/0000-0002-6047-7164 25 ,
- Mithila Jugulam ORCID: orcid.org/0000-0003-2065-9067 15 ,
- Kevin Fengler 18 ,
- Victor Llaca ORCID: orcid.org/0000-0003-4822-2924 18 ,
- Eric L. Patterson ORCID: orcid.org/0000-0001-7111-6287 14 &
- Todd A. Gaines ORCID: orcid.org/0000-0003-1485-7665 1
Genome Biology volume 25 , Article number: 139 ( 2024 ) Cite this article
781 Accesses
11 Altmetric
Metrics details
Weeds are attractive models for basic and applied research due to their impacts on agricultural systems and capacity to swiftly adapt in response to anthropogenic selection pressures. Currently, a lack of genomic information precludes research to elucidate the genetic basis of rapid adaptation for important traits like herbicide resistance and stress tolerance and the effect of evolutionary mechanisms on wild populations. The International Weed Genomics Consortium is a collaborative group of scientists focused on developing genomic resources to impact research into sustainable, effective weed control methods and to provide insights about stress tolerance and adaptation to assist crop breeding.
Each year globally, agricultural producers and landscape managers spend billions of US dollars [ 1 , 2 ] and countless hours attempting to control weedy plants and reduce their adverse effects. These management methods range from low-tech (e.g., pulling plants from the soil by hand) to extremely high-tech (e.g., computer vision-controlled spraying of herbicides). Regardless of technology level, effective control methods serve as strong selection pressures on weedy plants and often result in rapid evolution of weed populations resistant to such methods [ 3 , 4 , 5 , 6 , 7 ]. Thus, humans and weeds have been locked in an arms race, where humans develop new or improved control methods and weeds adapt and evolve to circumvent such methods.
Applying genomics to weed science offers a unique opportunity to study rapid adaptation, epigenetic responses, and examples of evolutionary rescue of diverse weedy species in the face of widespread and powerful selective pressures. Furthermore, lessons learned from these studies may also help to develop more sustainable control methods and to improve crop breeding efforts in the face of our ever-changing climate. While other research fields have used genetics and genomics to uncover the basis of many biological traits [ 8 , 9 , 10 , 11 ] and to understand how ecological factors affect evolution [ 12 , 13 ], the field of weed science has lagged behind in the development of genomic tools essential for such studies [ 14 ]. As research in human and crop genetics pushes into the era of pangenomics (i.e., multiple chromosome scale genome assemblies for a single species [ 15 , 16 ]), publicly available genomic information is still lacking or severely limited for the majority of weed species. Recent reviews of current weed genomes identified 26 [ 17 ] and 32 weed species with sequenced genomes [ 18 ]—many assembled to a sub-chromosome level.
Here, we summarize the current state of weed genomics, highlighting cases where genomics approaches have successfully provided insights on topics such as population genetic dynamics, genome evolution, and the genetic basis of herbicide resistance, rapid adaptation, and crop dedomestication. These highlighted investigations all relied upon genomic resources that are relatively rare for weedy species. Throughout, we identify additional resources that would advance the field of weed science and enable further progress in weed genomics. We then introduce the International Weed Genomics Consortium (IWGC), an open collaboration among researchers, and describe current efforts to generate these additional resources.
Evolution of weediness: potential research utilizing weed genomics tools
Weeds can evolve from non-weed progenitors through wild colonization, crop de-domestication, or crop-wild hybridization [ 19 ]. Because the time span in which weeds have evolved is necessarily limited by the origins of agriculture, these non-weed relatives often still exist and can be leveraged through population genomic and comparative genomic approaches to identify the adaptive changes that have driven the evolution of weediness. The ability to rapidly adapt, persist, and spread in agroecosystems are defining features of weedy plants, leading many to advocate agricultural weeds as ideal candidates for studying rapid plant adaptation [ 20 , 21 , 22 , 23 ]. The insights gained from applying plant ecological approaches to the study of rapid weed adaptation will move us towards the ultimate goals of mitigating such adaptation and increasing the efficacy of crop breeding and biotechnology [ 14 ].
Biology and ecological genomics of weeds
The impressive community effort to create and maintain resources for Arabidopsis thaliana ecological genomics provides a motivating example for the emerging study of weed genomics [ 24 , 25 , 26 , 27 ]. Arabidopsis thaliana was the first flowering plant species to have its genome fully sequenced [ 28 ] and rapidly became a model organism for plant molecular biology. As weedy genomes become available, collection, maintenance, and resequencing of globally distributed accessions of these species will help to replicate the success found in ecological studies of A. thaliana [ 29 , 30 , 31 , 32 , 33 , 34 , 35 ]. Evaluation of these accessions for traits of interest to produce large phenomics data sets (as in [ 36 , 37 , 38 , 39 , 40 ]) enables genome-wide association studies and population genomics analyses aimed at dissecting the genetic basis of variation in such traits [ 41 ]. Increasingly, these resources (e.g. the 1001 genomes project [ 29 ]) have enabled A. thaliana to be utilized as a model species to explore the eco-evolutionary basis of plant adaptation in a more realistic ecological context. Weedy species should supplement lessons in eco-evolutionary genomics learned from these experiments in A. thaliana .
Untargeted genomic approaches for understanding the evolutionary trajectories of populations and the genetic basis of traits as described above rely on the collection of genotypic information from across the genome of many individuals. While whole-genome resequencing accomplishes this requirement and requires no custom methodology, this approach provides more information than is necessary and is prohibitively expensive in species with large genomes. Development and optimization of genotype-by-sequencing methods for capturing reduced representations of newly sequence genomes like those described by [ 42 , 43 , 44 ] will reduce the cost and computational requirements of genetic mapping and population genetic experiments. Most major weed species do not currently have protocols for stable transformation, a key development in the popularity of A. thaliana as a model organism and a requirement for many functional genomic approaches. Functional validation of genes/variants believed to be responsible for traits of interest in weeds has thus far relied on transiently manipulating endogenous gene expression [ 45 , 46 ] or ectopic expression of a transgene in a model system [ 47 , 48 , 49 ]. While these methods have been successful, few weed species have well-studied viral vectors to adapt for use in virus induced gene silencing. Spray induced gene silencing is another potential option for functional investigation of candidate genes in weeds, but more research is needed to establish reliable delivery and gene knockdown [ 50 ]. Furthermore, traits with complex genetic architecture divergent between the researched and model species may not be amenable to functional genomic approaches using transgenesis techniques in model systems. Developing protocols for reduced representation sequencing, stable transformation, and gene editing/silencing in weeds will allow for more thorough characterization of candidate genetic variants underlying traits of interest.
Beyond rapid adaptation, some weedy species offer an opportunity to better understand co-evolution, like that between plants and pollinators and how their interaction leads to the spread of weedy alleles (Additional File 1 : Table S1). A suite of plant–insect traits has co-evolved to maximize the attraction of the insect pollinator community and the efficiency of pollen deposition between flowers ensuring fruit and seed production in many weeds [ 51 , 52 ]. Genetic mapping experiments have identified genes and genetic variants responsible for many floral traits affecting pollinator interaction including petal color [ 53 , 54 , 55 , 56 ], flower symmetry and size [ 57 , 58 , 59 ], and production of volatile organic compounds [ 60 , 61 , 62 ] and nectar [ 63 , 64 , 65 ]. While these studies reveal candidate genes for selection under co-evolution, herbicide resistance alleles may also have pleiotropic effects on the ecology of weeds [ 66 ], altering plant-pollinator interactions [ 67 ]. Discovery of genes and genetic variants involved in weed-pollinator interaction and their molecular and environmental control may create opportunities for better management of weeds with insect-mediated pollination. For example, if management can disrupt pollinator attraction/interaction with these weeds, the efficiency of reproduction may be reduced.
A more complete understanding of weed ecological genomics will undoubtedly elucidate many unresolved questions regarding the genetic basis of various aspects of weediness. For instance, when comparing populations of a species from agricultural and non-agricultural environments, is there evidence for contemporary evolution of weedy traits selected by agricultural management or were “natural” populations pre-adapted to agroecosystems? Where there is differentiation between weedy and natural populations, which traits are under selection and what is the genetic basis of variation in those traits? When comparing between weedy populations, is there evidence for parallel versus non-parallel evolution of weediness at the phenotypic and genotypic levels? Such studies may uncover fundamental truths about weediness. For example, is there a common phenotypic and/or genotypic basis for aspects of weediness among diverse weed species? The availability of characterized accessions and reference genomes for species of interest are required for such studies but only a few weedy species have these resources developed.
Population genomics
Weed species are certainly fierce competitors, able to outcompete crops and endemic species in their native environment, but they are also remarkable colonizers of perturbed habitats. Weeds achieve this through high fecundity, often producing tens of thousands of seeds per individual plant [ 68 , 69 , 70 ]. These large numbers in terms of demographic population size often combine with outcrossing reproduction to generate high levels of diversity with local effective population sizes in the hundreds of thousands [ 71 , 72 ]. This has two important consequences: weed populations retain standing genetic variation and generate many new mutations, supporting weed success in the face of harsh control. The generation of genomic tools to monitor weed populations at the molecular level is a game-changer to understanding weed dynamics and precisely testing the effect of artificial selection (i.e., management) and other evolutionary mechanisms on the genetic make-up of populations.
Population genomic data, without any environmental or phenotypic information, can be used to scan the genomes of weed and non-weed relatives to identify selective sweeps, pointing at loci supporting weed adaptation on micro- or macro-evolutionary scales. Two recent within-species examples include weedy rice, where population differentiation between weedy and domesticated populations was used to identify the genetic basis of weedy de-domestication [ 73 ], and common waterhemp, where consistent allelic differences among natural and agricultural collections resolved a complex set of agriculturally adaptive alleles [ 74 , 75 ]. A recent comparative population genomic study of weedy barnyardgrass and crop millet species has demonstrated how inter-specific investigations can resolve the signatures of crop and weed evolution [ 76 ] (also see [ 77 ] for a non-weed climate adaptation example). Multiple sequence alignments across numerous species provide complementary insight into adaptive convergence over deeper timescales, even with just one genomic sample per species (e.g., [ 78 , 79 ]). Thus, newly sequenced weed genomes combined with genomes available for closely related crops (outlined by [ 14 , 80 ]) and an effort to identify other non-weed wild relatives will be invaluable in characterizing the genetic architecture of weed adaptation and evolution across diverse species.
Weeds experience high levels of genetic selection, both artificial in response to agricultural practices and particularly herbicides, and natural in response to the environmental conditions they encounter [ 81 , 82 ]. Using genomic analysis to identify loci that are the targets of selection, whether natural or artificial, would point at vulnerabilities that could be leveraged against weeds to develop new and more sustainable management strategies [ 83 ]. This is a key motivation to develop genotype-by-environment association (GEA) and selective sweep scan approaches, which allow researchers to resolve the molecular basis of multi-dimensional adaptation [ 84 , 85 ]. GEA approaches, in particular, have been widely used on landscape-wide resequencing collections to determine the genetic basis of climate adaptation (e.g., [ 27 , 86 , 87 ]), but have yet to be fully exploited to diagnose the genetic basis of the various aspects of weediness [ 88 ]. Armed with data on environmental dimensions of agricultural settings, such as focal crop, soil quality, herbicide use, and climate, GEA approaches can help disentangle how discrete farming practices have influenced the evolution of weediness and resolve broader patterns of local adaptation across a weed’s range. Although non-weedy relatives are not technically required for GEA analyses, inclusion of environmental and genomic data from weed progenitors can further distinguish genetic variants underpinning weed origins from those involved in local adaptation.
New weeds emerge frequently [ 89 ], either through hybridization between species as documented for sea beet ( Beta vulgaris ssp. maritima) hybridizing with crop beet to produce progeny that are well adapted to agricultural conditions [ 90 , 91 , 92 ], or through the invasion of alien species that find a new range to colonize. Biosecurity measures are often in place to stop the introduction of new weeds; however, the vast scale of global agricultural commodity trade precludes the possibility of total control. Population genomic analysis is now able to measure gene flow between populations [ 74 , 93 , 94 , 95 ] and identify populations of origin for invasive species including weeds [ 96 , 97 , 98 ]. For example, the invasion route of the pest fruitfly Drosophila suzukii from Eastern Asia to North America and Europe through Hawaii was deciphered using Approximate Bayesian Computation on high-throughput sequencing data from a global sample of multiple populations [ 99 ]. Genomics can also be leveraged to predict invasion rather than explain it. The resequencing of a global sample of common ragweed ( Ambrosia artemisiifolia L.) elucidated a complex invasion route whereby Europe was invaded by multiple introductions of American ragweed that hybridized in Europe prior to a subsequent introduction to Australia [ 100 , 101 ]. In this context, the use of genomically informed species distribution models helps assess the risk associated with different source populations, which in the case of common ragweed, suggests that a source population from Florida would allow ragweed to invade most of northern Australia [ 102 ]. Globally coordinated research efforts to understand potential distribution models could support the transformation of biosecurity from perspective analysis towards predictive risk assessment.
Herbicide resistance and weed management
Herbicide resistance is among the numerous weedy traits that can evolve in plant populations exposed to agricultural selection pressures. Over-reliance on herbicides to control weeds, along with low diversity and lack of redundancy in weed management strategies, has resulted in globally widespread herbicide resistance [ 103 ]. To date, 272 herbicide-resistant weed species have been reported worldwide, and at least one resistance case exists for 21 of the 31 existing herbicide sites of action [ 104 ]—significantly limiting chemical weed control options available to agriculturalists. This limitation of control options is exacerbated by the recent lack of discovery of herbicides with new sites of action [ 105 ].
Herbicide resistance may result from several different physiological mechanisms. Such mechanisms have been classified into two main groups, target-site resistance (TSR) [ 4 , 106 ] and non-target-site resistance (NTSR) [ 4 , 107 ]. The first group encompasses changes that reduce binding affinity between a herbicide and its target [ 108 ]. These changes may provide resistance to multiple herbicides that have a common biochemical target [ 109 ] and can be effectively managed through mixture and/or rotation of herbicides targeting different sites of action [ 110 ]. The second group (NTSR), includes alterations in herbicide absorption, translocation, sequestration, and/or metabolism that may lead to unpredictable pleotropic cross-resistance profiles where structurally and functionally diverse herbicides are rendered ineffective by one or more genetic variant(s) [ 47 ]. This mechanism of resistance threatens not only the efficacy of existing herbicidal chemistries, but also ones yet to be discovered. While TSR is well understood because of the ease of identification and molecular characterization of target site variants, NTSR mechanisms are significantly more challenging to research because they are often polygenic, and the resistance causing element(s) are not well understood [ 111 ].
Improving the current understanding of metabolic NTSR mechanisms is not an easy task, since genes of diverse biochemical functions are involved, many of which exist as extensive gene families [ 109 , 112 ]. Expression changes of NTSR genes have been implicated in several resistance cases where the protein products of the genes are functionally equivalent across sensitive and resistant plants, but their relative abundance leads to resistance. Thus, regulatory elements of NTSR genes have been scrutinized to understand their role in NTSR mechanisms [ 113 ]. Similarly, epigenetic modifications have been hypothesized to play a role in NTSR, with much remaining to be explored [ 114 , 115 , 116 ]. Untargeted approaches such as genome-wide association, selective sweep scans, linkage mapping, RNA-sequencing, and metabolomic profiling have proven helpful to complement more specific biochemical- and chemo-characterization studies towards the elucidation of NTSR mechanisms as well as their regulation and evolution [ 47 , 117 , 118 , 119 , 120 , 121 , 122 , 123 , 124 ]. Even in cases where resistance has been attributed to TSR, genetic mapping approaches can detect other NTSR loci contributing to resistance (as shown by [ 123 ]) and provide further evidence for the role of TSR mutations across populations. Knowledge of the genetic basis of NTSR will aid the rational design of herbicides by screening new compounds for interaction with newly discovered NTSR proteins during early research phases and by identifying conserved chemical structures that interact with these proteins that should be avoided in small molecule design.
Genomic resources can also be used to predict the protein structure for novel herbicide target site and metabolism genes. This will allow for prediction of efficacy and selectivity for new candidate herbicides in silico to increase herbicide discovery throughput as well as aid in the design and development of next-generation technologies for sustainable weed management. Proteolysis targeting chimeras (PROTACs) have the potential to bind desired targets with great selectivity and degrade proteins by utilizing natural protein ubiquitination and degradation pathways within plants [ 125 ]. Spray-induced gene silencing in weeds using oligonucleotides has potential as a new, innovative, and sustainable method for weed management, but improved methods for design and delivery of oligonucleotides are needed to make this technique a viable management option [ 50 ]. Additionally, success in the field of pharmaceutical drug discovery in the development of molecules modulating protein–protein interactions offers another potential avenue towards the development of herbicides with novel targets [ 126 , 127 ]. High-quality reference genomes allow for the design of new weed management technologies like the ones listed here that are specific to—and effective across—weed species but have a null effect on non-target organisms.
Comparative genomics and genome biology
The genomes of weed species are as diverse as weed species themselves. Weeds are found across highly diverged plant families and often have no phylogenetically close model or crop species relatives for comparison. On all measurable metrics, weed genomes run the gamut. Some have smaller genomes like Cyperus spp. (~ 0.26 Gb) while others are larger, such as Avena fatua (~ 11.1 Gb) (Table 1 ). Some have high heterozygosity in terms of single-nucleotide polymorphisms, such as the Amaranthus spp., while others are primarily self-pollinated and quite homozygous, such as Poa annua [ 128 , 129 ]. Some are diploid such as Conyza canadensis and Echinochloa haploclada while others are polyploid such as C. sumetrensis , E. crus-galli , and E. colona [ 76 ]. The availability of genomic resources in these diverse, unexplored branches of the tree of life allows us to identify consistencies and anomalies in the field of genome biology.
The weed genomes published so far have focused mainly on weeds of agronomic crops, and studies have revolved around their ability to resist key herbicides. For example, genomic resources were vital in the elucidation of herbicide resistance cases involving target site gene copy number variants (CNVs). Gene CNVs of 5-enolpyruvylshikimate-3-phosphate synthase ( EPSPS ) have been found to confer resistance to the herbicide glyphosate in diverse weed species. To date, nine species have independently evolved EPSPS CNVs, and species achieve increased EPSPS copy number via different mechanisms [ 153 ]. For instance, the EPSPS CNV in Bassia scoparia is caused by tandem duplication, which is accredited to transposable element insertions flanking EPSPS and subsequent unequal crossing over events [ 154 , 155 ]. In Eleusine indica , a EPSPS CNV was caused by translocation of the EPSPS locus into the subtelomere followed by telomeric sequence exchange [ 156 ]. One of the most fascinating genome biology discoveries in weed science has been that of extra-chromosomal circular DNAs (eccDNAs) that harbor the EPSPS gene in the weed species Amaranthus palmeri [ 157 , 158 ]. In this case, the eccDNAs autonomously replicate separately from the nuclear genome and do not reintegrate into chromosomes, which has implications for inheritance, fitness, and genome structure [ 159 ]. These discoveries would not have been possible without reference assemblies of weed genomes, next-generation sequencing, and collaboration with experts in plant genomics and bioinformatics.
Another question that is often explored with weedy genomes is the nature and composition of gene families that are associated with NTSR. Gene families under consideration often include cytochrome P450s (CYPs), glutathione- S -transferases (GSTs), ABC transporters, etc. Some questions commonly considered with new weed genomes include how many genes are in each of these gene families, where are they located, and which weed accessions and species have an over-abundance of them that might explain their ability to evolve resistance so rapidly [ 76 , 146 , 160 , 161 ]? Weed genome resources are necessary to answer questions about gene family expansion or contraction during the evolution of weediness, including the role of polyploidy in NTSR gene family expansion as explored by [ 162 ].
Translational research and communication with weed management stakeholders
Whereas genomics of model plants is typically aimed at addressing fundamental questions in plant biology, and genomics of crop species has the obvious goal of crop improvement, goals of genomics of weedy plants also include the development of more effective and sustainable strategies for their management. Weed genomic resources assist with these objectives by providing novel molecular ecological and evolutionary insights from the context of intensive anthropogenic management (which is lacking in model plants), and offer knowledge and resources for trait discovery for crop improvement, especially given that many wild crop relatives are also important agronomic weeds (e.g., [ 163 ]). For instance, crop-wild relatives are valuable for improving crop breeding for marginal environments [ 164 ]. Thus, weed genomics presents unique opportunities and challenges relative to plant genomics more broadly. It should also be noted that although weed science at its core is an applied discipline, it draws broadly from many scientific disciplines such as, plant physiology, chemistry, ecology, and evolutionary biology, to name a few. The successful integration of weed-management strategies, therefore, requires extensive collaboration among individuals collectively possessing the necessary expertise [ 165 ].
With the growing complexity of herbicide resistance management, practitioners are beginning to recognize the importance of understanding resistance mechanisms to inform appropriate management tactics [ 14 ]. Although weed science practitioners do not need to understand the technical details of weed genomics, their appreciation of the power of weed genomics—together with their unique insights from field observations—will yield novel opportunities for applications of weed genomics to weed management. In particular, combining field management history with information on weed resistance mechanisms is expected to provide novel insights into evolutionary trajectories (e.g. [ 6 , 166 ]), which can be utilized for disrupting evolutionary adaptation. It can be difficult to obtain field history information from practitioners, but developing an understanding among them of the importance of such information can be invaluable.
Development of weed genomics resources by the IWGC
Weed genomics is a fast-growing field of research with many recent breakthroughs and many unexplored areas of study. The International Weed Genomics Consortium (IWGC) started in 2021 to address the roadblocks listed above and to promote the study of weedy plants. The IWGC is an open collaboration among academic, government, and industry researchers focused on producing genomic tools for weedy species from around the world. Through this collaboration, our initial aim is to provide chromosome-level reference genome assemblies for at least 50 important weedy species from across the globe that are chosen based on member input, economic impact, and global prevalence (Fig. 1 ). Each genome will include annotation of gene models and repetitive elements and will be freely available through public databases with no intellectual property restrictions. Additionally, future funding of the IWGC will focus on improving gene annotations and supplementing these reference genomes with tools that increase their utility.
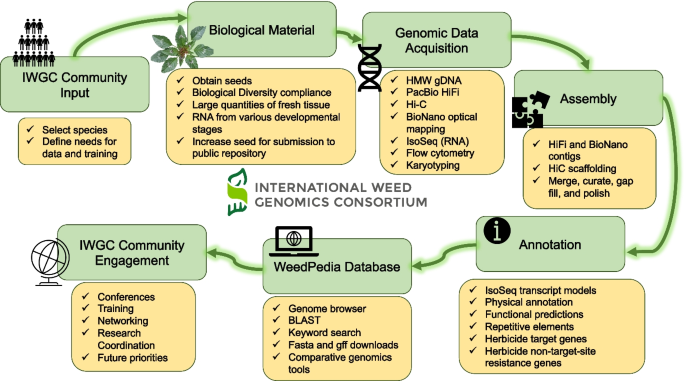
The International Weed Genomics Consortium (IWGC) collected input from the weed genomics community to develop plans for weed genome sequencing, annotation, user-friendly genome analysis tools, and community engagement
Reference genomes and data analysis tools
The first objective of the IWGC is to provide high-quality genomic resources for agriculturally important weeds. The IWGC therefore created two main resources for information about, access to, or analysis of weed genomic data (Fig. 1 ). The IWGC website (available at [ 167 ]) communicates the status and results of genome sequencing projects, information on training and funding opportunities, upcoming events, and news in weed genomics. It also contains details of all sequenced species including genome size, ploidy, chromosome number, herbicide resistance status, and reference genome assembly statistics. The IWGC either compiles existing data on genome size, ploidy, and chromosome number, or obtains the data using flow cytometry and cytogenetics (Fig. 1 ; Additional File 2 : Fig S1-S4). Through this website, users can request an account to access our second main resource, an online genome database called WeedPedia (accessible at [ 168 ]), with an account that is created within 3–5 working days of an account request submission. WeedPedia hosts IWGC-generated and other relevant publicly accessible genomic data as well as a suite of bioinformatic tools. Unlike what is available for other fields, weed science did not have a centralized hub for genomics information, data, and analysis prior to the IWGC. Our intention in creating WeedPedia is to encourage collaboration and equity of access to information across the research community. Importantly, all genome assemblies and annotations from the IWGC (Table 1 ), along with the raw data used to produce them, will be made available through NCBI GenBank. Upon completion of a 1-year sponsoring member data confidentiality period for each species (dates listed in Table 1 ), scientific teams within the IWGC produce the first genome-wide investigation to submit for publication including whole genome level analyses on genes, gene families, and repetitive sequences as well as comparative analysis with other species. Genome assemblies and data will be publicly available through NCBI as part of these initial publications for each species.
WeedPedia is a cloud-based omics database management platform built from the software “CropPedia” and licensed from KeyGene (Wageningen, The Netherlands). The interface allows users to access, visualize, and download genome assemblies along with structural and functional annotation. The platform includes a genome browser, comparative map viewer, pangenome tools, RNA-sequencing data visualization tools, genetic mapping and marker analysis tools, and alignment capabilities that allow searches by keyword or sequence. Additionally, genes encoding known target sites of herbicides have been specially annotated, allowing users to quickly identify and compare these genes of interest. The platform is flexible, making it compatible with future integration of other data types such as epigenetic or proteomic information. As an online platform with a graphical user interface, WeedPedia provides user-friendly, intuitive tools that encourage users to integrate genomics into their research while also allowing more advanced users to download genomic data to be used in custom analysis pipelines. We aspire for WeedPedia to mimic the success of other public genomic databases such as NCBI, CoGe, Phytozome, InsectBase, and Mycocosm to name a few. WeedPedia currently hosts reference genomes for 40 species (some of which are currently in their 1-year confidentiality period) with additional genomes in the pipeline to reach a currently planned total of 55 species (Table 1 ). These genomes include both de novo reference genomes generated or in progress by the IWGC (31 species; Table 1 ), and publicly available genome assemblies of 24 weedy or related species that were generated by independent research groups (Table 2 ). As of May 2024, WeedPedia has over 370 registered users from more than 27 countries spread across 6 continents.
The IWGC reference genomes are generated in partnership with the Corteva Agriscience Genome Center of Excellence (Johnston, Iowa) using a combination of single-molecule long-read sequencing, optical genome maps, and chromosome conformation mapping. This strategy has already yielded highly contiguous, phased, chromosome-level assemblies for 26 weed species, with additional assemblies currently in progress (Table 1 ). The IWGC assemblies have been completed as single or haplotype-resolved double-haplotype pseudomolecules in inbreeding and outbreeding species, respectively, with multiple genomes being near gapless. For example, the de novo assemblies of the allohexaploids Conyza sumatrensis and Chenopodium album have all chromosomes captured in single scaffolds and most chromosomes being gapless from telomere to telomere. Complementary full-length isoform (IsoSeq) sequencing of RNA collected from diverse tissue types and developmental stages assists in the development of gene models during annotation.
As with accessibility of data, a core objective of the IWGC is to facilitate open access to sequenced germplasm when possible for featured species. Historically, the weed science community has rarely shared or adopted standard germplasm (e.g., specific weed accessions). The IWGC has selected a specific accession of each species for reference genome assembly (typically susceptible to herbicides). In collaboration with a parallel effort by the Herbicide Resistant Plants committee of the Weed Science Society of America, seeds of the sequenced weed accessions will be deposited in the United States Department of Agriculture Germplasm Resources Information Network [ 186 ] for broad access by the scientific community and their accession numbers will be listed on the IWGC website. In some cases, it is not possible to generate enough seed to deposit into a public repository (e.g., plants that typically reproduce vegetatively, that are self-incompatible, or that produce very few seeds from a single individual). In these cases, the location of collection for sequenced accessions will at least inform the community where the sequenced individual came from and where they may expect to collect individuals with similar genotypes. The IWGC ensures that sequenced accessions are collected and documented to comply with the Nagoya Protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their utilization under the Convention on Biological Diversity and related Access and Benefit Sharing Legislation [ 187 ]. As additional accessions of weed species are sequenced (e.g., pangenomes are obtained), the IWGC will facilitate germplasm sharing protocols to support collaboration. Further, to simplify the investigation of herbicide resistance, the IWGC will link WeedPedia with the International Herbicide-Resistant Weed Database [ 104 ], an already widely known and utilized database for weed scientists.
Training and collaboration in weed genomics
Beyond producing genomic tools and resources, a priority of the IWGC is to enable the utilization of these resources across a wide range of stakeholders. A holistic approach to training is required for weed science generally [ 188 ], and we would argue even more so for weed genomics. To accomplish our training goals, the IWGC is developing and delivering programs aimed at the full range of IWGC stakeholders and covering a breadth of relevant topics. We have taken care to ensure our approaches are diverse as to provide training to researchers with all levels of existing experience and differing reasons for engaging with these tools. Throughout, the focus is on ensuring that our training and outreach result in impacts that benefit a wide range of stakeholders.
Although recently developed tools are incredibly enabling and have great potential to replace antiquated methodology [ 189 ] and to solve pressing weed science problems [ 14 ], specialized computational skills are required to fully explore and unlock meaning from these highly complex datasets. Collaboration with, or training of, computational biologists equipped with these skills and resources developed by the IWGC will enable weed scientists to expand research programs and better understand the genetic underpinnings of weed evolution and herbicide resistance. To fill existing skill gaps, the IWGC is developing summer bootcamps and online modules directed specifically at weed scientists that will provide training on computational skills (Fig. 1 ). Because successful utilization of the IWGC resources requires more than general computational skills, we have created three targeted workshops that teach practical skills related to genomics databases, molecular biology, and population genomics (available at [ 190 ]). The IWGC has also hosted two official conference meetings, one in September of 2021 and one in January of 2023, with more conferences planned. These conferences have included invited speakers to present successful implementations of weed genomics, educational workshops to build computational skills, and networking opportunities for research to connect and collaborate.
Engagement opportunities during undergraduate degrees have been shown to improve academic outcomes [ 191 , 192 ]. As one activity to help achieve this goal, the IWGC has sponsored opportunities for US undergraduates to undertake a 10-week research experience, which includes an introduction to bioinformatics, a plant genomics research project that results in a presentation, and access to career building opportunities in diverse workplace environments. To increase equitable access to conferences and professional communities, we supported early career researchers to attend the first two IWGC conferences in the USA as well as workshops and bootcamps in Europe, South America, and Australia. These hybrid or in-person travel grants are intentionally designed to remove barriers and increase participation of individuals from backgrounds and experiences currently underrepresented within weed/plant science or genomics [ 193 ]. Recipients of these travel awards gave presentations and gained the measurable benefits that come from either virtual or in-person participation in conferences [ 194 ]. Moving forward, weed scientists must amass skills associated with genomic analyses and collaborate with other area experts to fully leverage resources developed by the IWGC.
The tools generated through the IWGC will enable many new research projects with diverse objectives like those listed above. In summary, contiguous genome assemblies and complete annotation information will allow weed scientists to join plant breeders in the use of genetic mapping for many traits including stress tolerance, plant architecture, and herbicide resistance (especially important for cases of NTSR). These assemblies will also allow for investigations of population structure, gene flow, and responses to evolutionary mechanisms like genetic bottlenecking and artificial selection. Understanding gene sequences across diverse weed species will be vital in modeling new herbicide target site proteins and designing novel effective herbicides with minimal off-target effects. The IWGC website will improve accessibility to weed genomics data by providing a single hub for reference genomes as well as phenotypic and genotypic information for accessions shared with the IWGC. Deposition of sequenced germplasm into public repositories will ensure that researchers are able to access and utilize these accessions in their own research to make the field more standardized and equitable. WeedPedia allows users of all backgrounds to quickly access information of interest such as herbicide target site gene sequence or subcellular localization of protein products for different genes. Users can also utilize server-based tools such as BLAST and genome browsing similar to other public genomic databases. Finally, the IWGC is committed to training and connecting weed genomicists through hosting trainings, workshops, and conferences.
Conclusions
Weeds are unique and fascinating plants, having significant impacts on agriculture and ecosystems; and yet, aspects of their biology, ecology, and genetics remain poorly understood. Weeds represent a unique area within plant biology, given their repeated rapid adaptation to sudden and severe shifts in the selective landscape of anthropogenic management practices. The production of a public genomics database with reference genomes and annotations for over 50 weed species represents a substantial step forward towards research goals that improve our understanding of the biology and evolution of weeds. Future work is needed to improve annotations, particularly for complex gene families involved in herbicide detoxification, structural variants, and mobile genetic elements. As reference genome assemblies become available; standard, affordable methods for gathering genotype information will allow for the identification of genetic variants underlying traits of interest. Further, methods for functional validation and hypothesis testing are needed in weeds to validate the effect of genetic variants detected through such experiments, including systems for transformation, gene editing, and transient gene silencing and expression. Future research should focus on utilizing weed genomes to investigate questions about evolutionary biology, ecology, genetics of weedy traits, and weed population dynamics. The IWGC plans to continue the public–private partnership model to host the WeedPedia database over time, integrate new datasets such as genome resequencing and transcriptomes, conduct trainings, and serve as a research coordination network to ensure that advances in weed science from around the world are shared across the research community (Fig. 1 ). Bridging basic plant genomics with translational applications in weeds is needed to deliver on the potential of weed genomics to improve weed management and crop breeding.
Availability of data and materials
All genome assemblies and related sequencing data produced by the IWGC will be available through NCBI as part of publications reporting the first genome-wide analysis for each species.
Gianessi LP, Nathan PR. The value of herbicides in U.S. crop production. Weed Technol. 2007;21(2):559–66.
Article Google Scholar
Pimentel D, Lach L, Zuniga R, Morrison D. Environmental and economic costs of nonindigenous species in the United States. Bioscience. 2000;50(1):53–65.
Barrett SH. Crop mimicry in weeds. Econ Bot. 1983;37(3):255–82.
Powles SB, Yu Q. Evolution in action: plants resistant to herbicides. Annu Rev Plant Biol. 2010;61:317–47.
Article CAS PubMed Google Scholar
Thurber CS, Reagon M, Gross BL, Olsen KM, Jia Y, Caicedo AL. Molecular evolution of shattering loci in U.S. weedy rice. Mol Ecol. 2010;19(16):3271–84.
Article PubMed PubMed Central Google Scholar
Comont D, Lowe C, Hull R, Crook L, Hicks HL, Onkokesung N, et al. Evolution of generalist resistance to herbicide mixtures reveals a trade-off in resistance management. Nat Commun. 2020;11(1):3086.
Article CAS PubMed PubMed Central Google Scholar
Ashworth MB, Walsh MJ, Flower KC, Vila-Aiub MM, Powles SB. Directional selection for flowering time leads to adaptive evolution in Raphanus raphanistrum (wild radish). Evol Appl. 2016;9(4):619–29.
Chan EK, Rowe HC, Kliebenstein DJ. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics. 2010;185(3):991–1007.
Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94.
Harkess A, Zhou J, Xu C, Bowers JE, Van der Hulst R, Ayyampalayam S, et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun. 2017;8(1):1279.
Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L, et al. The gene Sr33 , an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science. 2013;341(6147):786–8.
Ågren J, Oakley CG, McKay JK, Lovell JT, Schemske DW. Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2013;110(52):21077–82.
Article PubMed Central Google Scholar
Schartl M, Walter RB, Shen Y, Garcia T, Catchen J, Amores A, et al. The genome of the platyfish, Xiphophorus maculatus , provides insights into evolutionary adaptation and several complex traits. Nat Genet. 2013;45(5):567–72.
Ravet K, Patterson EL, Krähmer H, Hamouzová K, Fan L, Jasieniuk M, et al. The power and potential of genomics in weed biology and management. Pest Manag Sci. 2018;74(10):2216–25.
Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou S, Liu J, et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science. 2021;373(6555):655–62.
Liao W-W, Asri M, Ebler J, Doerr D, Haukness M, Hickey G, et al. A draft human pangenome reference. Nature. 2023;617(7960):312–24.
Huang Y, Wu D, Huang Z, Li X, Merotto A, Bai L, et al. Weed genomics: yielding insights into the genetics of weedy traits for crop improvement. aBIOTECH. 2023;4:20–30.
Chen K, Yang H, Wu D, Peng Y, Lian L, Bai L, et al. Weed biology and management in the multi-omics era: progress and perspectives. Plant Commun. 2024;5(4):100816.
De Wet JMJ, Harlan JR. Weeds and domesticates: evolution in the man-made habitat. Econ Bot. 1975;29(2):99–108.
Mahaut L, Cheptou PO, Fried G, Munoz F, Storkey J, Vasseur F, et al. Weeds: against the rules? Trends Plant Sci. 2020;25(11):1107–16.
Neve P, Vila-Aiub M, Roux F. Evolutionary-thinking in agricultural weed management. New Phytol. 2009;184(4):783–93.
Article PubMed Google Scholar
Sharma G, Barney JN, Westwood JH, Haak DC. Into the weeds: new insights in plant stress. Trends Plant Sci. 2021;26(10):1050–60.
Vigueira CC, Olsen KM, Caicedo AL. The red queen in the corn: agricultural weeds as models of rapid adaptive evolution. Heredity (Edinb). 2013;110(4):303–11.
Donohue K, Dorn L, Griffith C, Kim E, Aguilera A, Polisetty CR, et al. Niche construction through germination cueing: life-history responses to timing of germination in Arabidopsis thaliana . Evolution. 2005;59(4):771–85.
PubMed Google Scholar
Exposito-Alonso M. Seasonal timing adaptation across the geographic range of Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2020;117(18):9665–7.
Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. A map of local adaptation in Arabidopsis thaliana . Science. 2011;334(6052):86–9.
Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334(6052):83–6.
Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature. 2000;408(6814):796–815.
Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, et al. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana . Cell. 2016;166(2):481–91.
Durvasula A, Fulgione A, Gutaker RM, Alacakaptan SI, Flood PJ, Neto C, et al. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2017;114(20):5213–8.
Frachon L, Mayjonade B, Bartoli C, Hautekèete N-C, Roux F. Adaptation to plant communities across the genome of Arabidopsis thaliana . Mol Biol Evol. 2019;36(7):1442–56.
Fulgione A, Koornneef M, Roux F, Hermisson J, Hancock AM. Madeiran Arabidopsis thaliana reveals ancient long-range colonization and clarifies demography in Eurasia. Mol Biol Evol. 2018;35(3):564–74.
Fulgione A, Neto C, Elfarargi AF, Tergemina E, Ansari S, Göktay M, et al. Parallel reduction in flowering time from de novo mutations enable evolutionary rescue in colonizing lineages. Nat Commun. 2022;13(1):1461.
Kasulin L, Rowan BA, León RJC, Schuenemann VJ, Weigel D, Botto JF. A single haplotype hyposensitive to light and requiring strong vernalization dominates Arabidopsis thaliana populations in Patagonia. Argentina Mol Ecol. 2017;26(13):3389–404.
Picó FX, Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics. 2008;180(2):1009–21.
Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465(7298):627–31.
Flood PJ, Kruijer W, Schnabel SK, van der Schoor R, Jalink H, Snel JFH, et al. Phenomics for photosynthesis, growth and reflectance in Arabidopsis thaliana reveals circadian and long-term fluctuations in heritability. Plant Methods. 2016;12(1):14.
Marchadier E, Hanemian M, Tisné S, Bach L, Bazakos C, Gilbault E, et al. The complex genetic architecture of shoot growth natural variation in Arabidopsis thaliana . PLoS Genet. 2019;15(4):e1007954.
Tisné S, Serrand Y, Bach L, Gilbault E, Ben Ameur R, Balasse H, et al. Phenoscope: an automated large-scale phenotyping platform offering high spatial homogeneity. Plant J. 2013;74(3):534–44.
Tschiersch H, Junker A, Meyer RC, Altmann T. Establishment of integrated protocols for automated high throughput kinetic chlorophyll fluorescence analyses. Plant Methods. 2017;13:54.
Chen X, MacGregor DR, Stefanato FL, Zhang N, Barros-Galvão T, Penfield S. A VEL3 histone deacetylase complex establishes a maternal epigenetic state controlling progeny seed dormancy. Nat Commun. 2023;14(1):2220.
Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–101.
Davey JW, Blaxter ML. RADSeq: next-generation population genetics. Brief Funct Genomics. 2010;9(5–6):416–23.
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6(5):e19379.
MacGregor DR. What makes a weed a weed? How virus-mediated reverse genetics can help to explore the genetics of weediness. Outlooks Pest Manag. 2020;31(5):224–9.
Mellado-Sánchez M, McDiarmid F, Cardoso V, Kanyuka K, MacGregor DR. Virus-mediated transient expression techniques enable gene function studies in blackgrass. Plant Physiol. 2020;183(2):455–9.
Dimaano NG, Yamaguchi T, Fukunishi K, Tominaga T, Iwakami S. Functional characterization of Cytochrome P450 CYP81A subfamily to disclose the pattern of cross-resistance in Echinochloa phyllopogon . Plant Mol Biol. 2020;102(4–5):403–16.
de Figueiredo MRA, Küpper A, Malone JM, Petrovic T, de Figueiredo ABTB, Campagnola G, et al. An in-frame deletion mutation in the degron tail of auxin coreceptor IAA2 confers resistance to the herbicide 2,4-D in Sisymbrium orientale . Proc Natl Acad Sci U S A. 2022;119(9):e2105819119.
Patzoldt WL, Hager AG, McCormick JS, Tranel PJ. A codon deletion confers resistance to herbicides inhibiting protoporphyrinogen oxidase. Proc Natl Acad Sci U S A. 2006;103(33):12329–34.
Zabala-Pardo D, Gaines T, Lamego FP, Avila LA. RNAi as a tool for weed management: challenges and opportunities. Adv Weed Sci. 2022;40(spe1):e020220096.
Fattorini R, Glover BJ. Molecular mechanisms of pollination biology. Annu Rev Plant Biol. 2020;71:487–515.
Rollin O, Benelli G, Benvenuti S, Decourtye A, Wratten SD, Canale A, et al. Weed-insect pollinator networks as bio-indicators of ecological sustainability in agriculture. A review Agron Sustain Dev. 2016;36(1):8.
Irwin RE, Strauss SY. Flower color microevolution in wild radish: evolutionary response to pollinator-mediated selection. Am Nat. 2005;165(2):225–37.
Ma B, Wu J, Shi T-L, Yang Y-Y, Wang W-B, Zheng Y, et al. Lilac ( Syringa oblata ) genome provides insights into its evolution and molecular mechanism of petal color change. Commun Biol. 2022;5(1):686.
Xing A, Wang X, Nazir MF, Zhang X, Wang X, Yang R, et al. Transcriptomic and metabolomic profiling of flavonoid biosynthesis provides novel insights into petals coloration in Asian cotton ( Gossypium arboreum L.). BMC Plant Biol. 2022;22(1):416.
Zheng Y, Chen Y, Liu Z, Wu H, Jiao F, Xin H, et al. Important roles of key genes and transcription factors in flower color differences of Nicotiana alata . Genes (Basel). 2021;12(12):1976.
Krizek BA, Anderson JT. Control of flower size. J Exp Bot. 2013;64(6):1427–37.
Powell AE, Lenhard M. Control of organ size in plants. Curr Biol. 2012;22(9):R360–7.
Spencer V, Kim M. Re"CYC"ling molecular regulators in the evolution and development of flower symmetry. Semin Cell Dev Biol. 2018;79:16–26.
Amrad A, Moser M, Mandel T, de Vries M, Schuurink RC, Freitas L, et al. Gain and loss of floral scent production through changes in structural genes during pollinator-mediated speciation. Curr Biol. 2016;26(24):3303–12.
Delle-Vedove R, Schatz B, Dufay M. Understanding intraspecific variation of floral scent in light of evolutionary ecology. Ann Bot. 2017;120(1):1–20.
Pichersky E, Gershenzon J. The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol. 2002;5(3):237–43.
Ballerini ES, Kramer EM, Hodges SA. Comparative transcriptomics of early petal development across four diverse species of Aquilegia reveal few genes consistently associated with nectar spur development. BMC Genom. 2019;20(1):668.
Corbet SA, Willmer PG, Beament JWL, Unwin DM, Prys-Jones OE. Post-secretory determinants of sugar concentration in nectar. Plant Cell Environ. 1979;2(4):293–308.
Galliot C, Hoballah ME, Kuhlemeier C, Stuurman J. Genetics of flower size and nectar volume in Petunia pollination syndromes. Planta. 2006;225(1):203–12.
Vila-Aiub MM, Neve P, Powles SB. Fitness costs associated with evolved herbicide resistance alleles in plants. New Phytol. 2009;184(4):751–67.
Baucom RS. Evolutionary and ecological insights from herbicide-resistant weeds: what have we learned about plant adaptation, and what is left to uncover? New Phytol. 2019;223(1):68–82.
Bajwa AA, Latif S, Borger C, Iqbal N, Asaduzzaman M, Wu H, et al. The remarkable journey of a weed: biology and management of annual ryegrass ( Lolium rigidum ) in conservation cropping systems of Australia. Plants (Basel). 2021;10(8):1505.
Bitarafan Z, Andreasen C. Fecundity allocation in some european weed species competing with crops. Agronomy. 2022;12(5):1196.
Costea M, Weaver SE, Tardif FJ. The biology of Canadian weeds. 130. Amaranthus retroflexus L., A. powellii , A. powellii S. Watson, and A. hybridus L. Can J Plant Sci. 2004;84(2):631–68.
Dixon A, Comont D, Slavov GT, Neve P. Population genomics of selectively neutral genetic structure and herbicide resistance in UK populations of Alopecurus myosuroides . Pest Manag Sci. 2021;77(3):1520–9.
Kersten S, Chang J, Huber CD, Voichek Y, Lanz C, Hagmaier T, et al. Standing genetic variation fuels rapid evolution of herbicide resistance in blackgrass. Proc Natl Acad Sci U S A. 2023;120(16):e2206808120.
Qiu J, Zhou Y, Mao L, Ye C, Wang W, Zhang J, et al. Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat Commun. 2017;8(1):15323.
Kreiner JM, Caballero A, Wright SI, Stinchcombe JR. Selective ancestral sorting and de novo evolution in the agricultural invasion of Amaranthus tuberculatus . Evolution. 2022;76(1):70–85.
Kreiner JM, Latorre SM, Burbano HA, Stinchcombe JR, Otto SP, Weigel D, et al. Rapid weed adaptation and range expansion in response to agriculture over the past two centuries. Science. 2022;378(6624):1079–85.
Wu D, Shen E, Jiang B, Feng Y, Tang W, Lao S, et al. Genomic insights into the evolution of Echinochloa species as weed and orphan crop. Nat Commun. 2022;13(1):689.
Yeaman S, Hodgins KA, Lotterhos KE, Suren H, Nadeau S, Degner JC, et al. Convergent local adaptation to climate in distantly related conifers. Science. 2016;353(6306):1431–3.
Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 2013;45(8):891–8.
Sackton TB, Grayson P, Cloutier A, Hu Z, Liu JS, Wheeler NE, et al. Convergent regulatory evolution and loss of flight in paleognathous birds. Science. 2019;364(6435):74–8.
Ye CY, Fan L. Orphan crops and their wild relatives in the genomic era. Mol Plant. 2021;14(1):27–39.
Clements DR, Jones VL. Ten ways that weed evolution defies human management efforts amidst a changing climate. Agronomy. 2021;11(2):284.
Article CAS Google Scholar
Weinig C. Rapid evolutionary responses to selection in heterogeneous environments among agricultural and nonagricultural weeds. Int J Plant Sci. 2005;166(4):641–7.
Cousens RD, Fournier-Level A. Herbicide resistance costs: what are we actually measuring and why? Pest Manag Sci. 2018;74(7):1539–46.
Lasky JR, Josephs EB, Morris GP. Genotype–environment associations to reveal the molecular basis of environmental adaptation. Plant Cell. 2023;35(1):125–38.
Lotterhos KE. The effect of neutral recombination variation on genome scans for selection. G3-Genes Genom Genet. 2019;9(6):1851–67.
Lovell JT, MacQueen AH, Mamidi S, Bonnette J, Jenkins J, Napier JD, et al. Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature. 2021;590(7846):438–44.
Todesco M, Owens GL, Bercovich N, Légaré J-S, Soudi S, Burge DO, et al. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature. 2020;584(7822):602–7.
Revolinski SR, Maughan PJ, Coleman CE, Burke IC. Preadapted to adapt: Underpinnings of adaptive plasticity revealed by the downy brome genome. Commun Biol. 2023;6(1):326.
Kuester A, Conner JK, Culley T, Baucom RS. How weeds emerge: a taxonomic and trait-based examination using United States data. New Phytol. 2014;202(3):1055–68.
Arnaud JF, Fénart S, Cordellier M, Cuguen J. Populations of weedy crop-wild hybrid beets show contrasting variation in mating system and population genetic structure. Evol Appl. 2010;3(3):305–18.
Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci U S A. 2000;97(13):7043–50.
Nakabayashi K, Leubner-Metzger G. Seed dormancy and weed emergence: from simulating environmental change to understanding trait plasticity, adaptive evolution, and population fitness. J Exp Bot. 2021;72(12):4181–5.
Busi R, Yu Q, Barrett-Lennard R, Powles S. Long distance pollen-mediated flow of herbicide resistance genes in Lolium rigidum . Theor Appl Genet. 2008;117(8):1281–90.
Délye C, Clément JAJ, Pernin F, Chauvel B, Le Corre V. High gene flow promotes the genetic homogeneity of arable weed populations at the landscape level. Basic Appl Ecol. 2010;11(6):504–12.
Roumet M, Noilhan C, Latreille M, David J, Muller MH. How to escape from crop-to-weed gene flow: phenological variation and isolation-by-time within weedy sunflower populations. New Phytol. 2013;197(2):642–54.
Moghadam SH, Alebrahim MT, Mohebodini M, MacGregor DR. Genetic variation of Amaranthus retroflexus L. and Chenopodium album L. (Amaranthaceae) suggests multiple independent introductions into Iran. Front Plant Sci. 2023;13:1024555.
Muller M-H, Latreille M, Tollon C. The origin and evolution of a recent agricultural weed: population genetic diversity of weedy populations of sunflower ( Helianthus annuus L.) in Spain and France. Evol Appl. 2011;4(3):499–514.
Wesse C, Welk E, Hurka H, Neuffer B. Geographical pattern of genetic diversity in Capsella bursa-pastoris (Brassicaceae) -A global perspective. Ecol Evol. 2021;11(1):199–213.
Fraimout A, Debat V, Fellous S, Hufbauer RA, Foucaud J, Pudlo P, et al. Deciphering the routes of invasion of Drosophila suzukii by means of ABC random forest. Mol Biol Evol. 2017;34(4):980–96.
CAS PubMed PubMed Central Google Scholar
Battlay P, Wilson J, Bieker VC, Lee C, Prapas D, Petersen B, et al. Large haploblocks underlie rapid adaptation in the invasive weed Ambrosia artemisiifolia . Nat Commun. 2023;14(1):1717.
van Boheemen LA, Hodgins KA. Rapid repeatable phenotypic and genomic adaptation following multiple introductions. Mol Ecol. 2020;29(21):4102–17.
Putra A, Hodgins K, Fournier-Level A. Assessing the invasive potential of different source populations of ragweed ( Ambrosia artemisiifolia L.) through genomically-informed species distribution modelling. Authorea. 2023;17(1):e13632.
Google Scholar
Bourguet D, Delmotte F, Franck P, Guillemaud T, Reboud X, Vacher C, et al. Heterogeneity of selection and the evolution of resistance. Trends Ecol Evol. 2013;28(2):110–8.
The International Herbicide-Resistant Weed Database. www.weedscience.org . Accessed 20 June 2023.
Powles S. Herbicide discovery through innovation and diversity. Adv Weed Sci. 2022;40(spe1):e020220074.
Murphy BP, Tranel PJ. Target-site mutations conferring herbicide resistance. Plants (Basel). 2019;8(10):382.
Gaines TA, Duke SO, Morran S, Rigon CAG, Tranel PJ, Küpper A, et al. Mechanisms of evolved herbicide resistance. J Biol Chem. 2020;295(30):10307–30.
Lonhienne T, Cheng Y, Garcia MD, Hu SH, Low YS, Schenk G, et al. Structural basis of resistance to herbicides that target acetohydroxyacid synthase. Nat Commun. 2022;13(1):3368.
Comont D, MacGregor DR, Crook L, Hull R, Nguyen L, Freckleton RP, et al. Dissecting weed adaptation: fitness and trait correlations in herbicide-resistant Alopecurus myosuroides . Pest Manag Sci. 2022;78(7):3039–50.
Neve P. Simulation modelling to understand the evolution and management of glyphosate resistance in weeds. Pest Manag Sci. 2008;64(4):392–401.
Torra J, Alcántara-de la Cruz R. Molecular mechanisms of herbicide resistance in weeds. Genes (Basel). 2022;13(11):2025.
Délye C, Gardin JAC, Boucansaud K, Chauvel B, Petit C. Non-target-site-based resistance should be the centre of attention for herbicide resistance research: Alopecurus myosuroides as an illustration. Weed Res. 2011;51(5):433–7.
Chandra S, Leon RG. Genome-wide evolutionary analysis of putative non-specific herbicide resistance genes and compilation of core promoters between monocots and dicots. Genes (Basel). 2022;13(7):1171.
Margaritopoulou T, Tani E, Chachalis D, Travlos I. Involvement of epigenetic mechanisms in herbicide resistance: the case of Conyza canadensis . Agriculture. 2018;8(1):17.
Pan L, Guo Q, Wang J, Shi L, Yang X, Zhou Y, et al. CYP81A68 confers metabolic resistance to ALS and ACCase-inhibiting herbicides and its epigenetic regulation in Echinochloa crus-galli . J Hazard Mater. 2022;428:128225.
Sen MK, Hamouzová K, Košnarová P, Roy A, Soukup J. Herbicide resistance in grass weeds: Epigenetic regulation matters too. Front Plant Sci. 2022;13:1040958.
Han H, Yu Q, Beffa R, González S, Maiwald F, Wang J, et al. Cytochrome P450 CYP81A10v7 in Lolium rigidum confers metabolic resistance to herbicides across at least five modes of action. Plant J. 2021;105(1):79–92.
Kubis GC, Marques RZ, Kitamura RS, Barroso AA, Juneau P, Gomes MP. Antioxidant enzyme and Cytochrome P450 activities are involved in horseweed ( Conyza sumatrensis ) resistance to glyphosate. Stress. 2023;3(1):47–57.
Qiao Y, Zhang N, Liu J, Yang H. Interpretation of ametryn biodegradation in rice based on joint analyses of transcriptome, metabolome and chemo-characterization. J Hazard Mater. 2023;445:130526.
Rouse CE, Roma-Burgos N, Barbosa Martins BA. Physiological assessment of non–target site restistance in multiple-resistant junglerice ( Echinochloa colona ). Weed Sci. 2019;67(6):622–32.
Abou-Khater L, Maalouf F, Jighly A, Alsamman AM, Rubiales D, Rispail N, et al. Genomic regions associated with herbicide tolerance in a worldwide faba bean ( Vicia faba L.) collection. Sci Rep. 2022;12(1):158.
Gupta S, Harkess A, Soble A, Van Etten M, Leebens-Mack J, Baucom RS. Interchromosomal linkage disequilibrium and linked fitness cost loci associated with selection for herbicide resistance. New Phytol. 2023;238(3):1263–77.
Kreiner JM, Tranel PJ, Weigel D, Stinchcombe JR, Wright SI. The genetic architecture and population genomic signatures of glyphosate resistance in Amaranthus tuberculatus . Mol Ecol. 2021;30(21):5373–89.
Parcharidou E, Dücker R, Zöllner P, Ries S, Orru R, Beffa R. Recombinant glutathione transferases from flufenacet-resistant black-grass ( Alopecurus myosuroides Huds.) form different flufenacet metabolites and differ in their interaction with pre- and post-emergence herbicides. Pest Manag Sci. 2023;79(9):3376–86.
Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21(3):181–200.
Acuner Ozbabacan SE, Engin HB, Gursoy A, Keskin O. Transient protein-protein interactions. Protein Eng Des Sel. 2011;24(9):635–48.
Lu H, Zhou Q, He J, Jiang Z, Peng C, Tong R, et al. Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials. Signal Transduct Target Ther. 2020;5(1):213.
Benson CW, Sheltra MR, Maughan PJ, Jellen EN, Robbins MD, Bushman BS, et al. Homoeologous evolution of the allotetraploid genome of Poa annua L. BMC Genom. 2023;24(1):350.
Robbins MD, Bushman BS, Huff DR, Benson CW, Warnke SE, Maughan CA, et al. Chromosome-scale genome assembly and annotation of allotetraploid annual bluegrass ( Poa annua L.). Genome Biol Evol. 2022;15(1):evac180.
Montgomery JS, Giacomini D, Waithaka B, Lanz C, Murphy BP, Campe R, et al. Draft genomes of Amaranthus tuberculatus , Amaranthus hybridus and Amaranthus palmeri . Genome Biol Evol. 2020;12(11):1988–93.
Jeschke MR, Tranel PJ, Rayburn AL. DNA content analysis of smooth pigweed ( Amaranthus hybridus ) and tall waterhemp ( A. tuberculatus ): implications for hybrid detection. Weed Sci. 2003;51(1):1–3.
Rayburn AL, McCloskey R, Tatum TC, Bollero GA, Jeschke MR, Tranel PJ. Genome size analysis of weedy Amaranthus species. Crop Sci. 2005;45(6):2557–62.
Laforest M, Martin SL, Bisaillon K, Soufiane B, Meloche S, Tardif FJ, et al. The ancestral karyotype of the Heliantheae Alliance, herbicide resistance, and human allergens: Insights from the genomes of common and giant ragweed. Plant Genome . 2024;e20442. https://doi.org/10.1002/tpg2.20442 .
Mulligan GA. Chromosome numbers of Canadian weeds. I Canad J Bot. 1957;35(5):779–89.
Meyer L, Causse R, Pernin F, Scalone R, Bailly G, Chauvel B, et al. New gSSR and EST-SSR markers reveal high genetic diversity in the invasive plant Ambrosia artemisiifolia L. and can be transferred to other invasive Ambrosia species. PLoS One. 2017;12(5):e0176197.
Pustahija F, Brown SC, Bogunić F, Bašić N, Muratović E, Ollier S, et al. Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa. Plant Soil. 2013;373(1):427–53.
Kubešová M, Moravcova L, Suda J, Jarošík V, Pyšek P. Naturalized plants have smaller genomes than their non-invading relatives: a flow cytometric analysis of the Czech alien flora. Preslia. 2010;82(1):81–96.
Thébaud C, Abbott RJ. Characterization of invasive Conyza species (Asteraceae) in Europe: quantitative trait and isozyme analysis. Am J Bot. 1995;82(3):360–8.
Garcia S, Hidalgo O, Jakovljević I, Siljak-Yakovlev S, Vigo J, Garnatje T, et al. New data on genome size in 128 Asteraceae species and subspecies, with first assessments for 40 genera, 3 tribes and 2 subfamilies. Plant Biosyst. 2013;147(4):1219–27.
Zhao X, Yi L, Ren Y, Li J, Ren W, Hou Z, et al. Chromosome-scale genome assembly of the yellow nutsedge ( Cyperus esculentus ). Genome Biol Evol. 2023;15(3):evad027.
Bennett MD, Leitch IJ, Hanson L. DNA amounts in two samples of angiosperm weeds. Ann Bot. 1998;82:121–34.
Schulz-Schaeffer J, Gerhardt S. Cytotaxonomic analysis of the Euphorbia spp. (leafy spurge) complex. II: Comparative study of the chromosome morphology. Biol Zentralbl. 1989;108(1):69–76.
Schaeffer JR, Gerhardt S. The impact of introgressive hybridization on the weediness of leafy spurge. Leafy Spurge Symposium. 1989;1989:97–105.
Bai C, Alverson WS, Follansbee A, Waller DM. New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann Bot. 2012;110(8):1623–9.
Aarestrup JR, Karam D, Fernandes GW. Chromosome number and cytogenetics of Euphorbia heterophylla L. Genet Mol Res. 2008;7(1):217–22.
Wang L, Sun X, Peng Y, Chen K, Wu S, Guo Y, et al. Genomic insights into the origin, adaptive evolution, and herbicide resistance of Leptochloa chinensis , a devastating tetraploid weedy grass in rice fields. Mol Plant. 2022;15(6):1045–58.
Paril J, Pandey G, Barnett EM, Rane RV, Court L, Walsh T, et al. Rounding up the annual ryegrass genome: high-quality reference genome of Lolium rigidum . Front Genet. 2022;13:1012694.
Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM. Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am J Bot. 2006;93(1):148–56.
Towers G, Mitchell J, Rodriguez E, Bennett F, Subba Rao P. Biology & chemistry of Parthenium hysterophorus L., a problem weed in India. Biol Rev. 1977;48:65–74.
CAS Google Scholar
Moghe GD, Hufnagel DE, Tang H, Xiao Y, Dworkin I, Town CD, et al. Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish ( Raphanus raphanistrum ) and three other Brassicaceae species. Plant Cell. 2014;26(5):1925–37.
Zhang X, Liu T, Wang J, Wang P, Qiu Y, Zhao W, et al. Pan-genome of Raphanus highlights genetic variation and introgression among domesticated, wild, and weedy radishes. Mol Plant. 2021;14(12):2032–55.
Chytrý M, Danihelka J, Kaplan Z, Wild J, Holubová D, Novotný P, et al. Pladias database of the Czech flora and vegetation. Preslia. 2021;93(1):1–87.
Patterson EL, Pettinga DJ, Ravet K, Neve P, Gaines TA. Glyphosate resistance and EPSPS gene duplication: Convergent evolution in multiple plant species. J Hered. 2018;109(2):117–25.
Jugulam M, Niehues K, Godar AS, Koo DH, Danilova T, Friebe B, et al. Tandem amplification of a chromosomal segment harboring 5-enolpyruvylshikimate-3-phosphate synthase locus confers glyphosate resistance in Kochia scoparia . Plant Physiol. 2014;166(3):1200–7.
Patterson EL, Saski CA, Sloan DB, Tranel PJ, Westra P, Gaines TA. The draft genome of Kochia scoparia and the mechanism of glyphosate resistance via transposon-mediated EPSPS tandem gene duplication. Genome Biol Evol. 2019;11(10):2927–40.
Zhang C, Johnson N, Hall N, Tian X, Yu Q, Patterson E. Subtelomeric 5-enolpyruvylshikimate-3-phosphate synthase ( EPSPS ) copy number variation confers glyphosate resistance in Eleusine indica . Nat Commun. 2023;14:4865.
Koo D-H, Molin WT, Saski CA, Jiang J, Putta K, Jugulam M, et al. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri . Proc Natl Acad Sci U S A. 2018;115(13):3332–7.
Molin WT, Yaguchi A, Blenner M, Saski CA. The eccDNA Replicon: A heritable, extranuclear vehicle that enables gene amplification and glyphosate resistance in Amaranthus palmeri . Plant Cell. 2020;32(7):2132–40.
Jugulam M. Can non-Mendelian inheritance of extrachromosomal circular DNA-mediated EPSPS gene amplification provide an opportunity to reverse resistance to glyphosate? Weed Res. 2021;61(2):100–5.
Kreiner JM, Giacomini DA, Bemm F, Waithaka B, Regalado J, Lanz C, et al. Multiple modes of convergent adaptation in the spread of glyphosate-resistant Amaranthus tuberculatus . Proc Natl Acad Sci U S A. 2019;116(42):21076–84.
Cai L, Comont D, MacGregor D, Lowe C, Beffa R, Neve P, et al. The blackgrass genome reveals patterns of non-parallel evolution of polygenic herbicide resistance. New Phytol. 2023;237(5):1891–907.
Chen K, Yang H, Peng Y, Liu D, Zhang J, Zhao Z, et al. Genomic analyses provide insights into the polyploidization-driven herbicide adaptation in Leptochloa weeds. Plant Biotechnol J. 2023;21(8):1642–58.
Ohadi S, Hodnett G, Rooney W, Bagavathiannan M. Gene flow and its consequences in Sorghum spp. Crit Rev Plant Sci. 2017;36(5–6):367–85.
Renzi JP, Coyne CJ, Berger J, von Wettberg E, Nelson M, Ureta S, et al. How could the use of crop wild relatives in breeding increase the adaptation of crops to marginal environments? Front Plant Sci. 2022;13:886162.
Ward SM, Cousens RD, Bagavathiannan MV, Barney JN, Beckie HJ, Busi R, et al. Agricultural weed research: a critique and two proposals. Weed Sci. 2014;62(4):672–8.
Evans JA, Tranel PJ, Hager AG, Schutte B, Wu C, Chatham LA, et al. Managing the evolution of herbicide resistance. Pest Manag Sci. 2016;72(1):74–80.
International Weed Genomics Consortium Website. https://www.weedgenomics.org . Accessed 20 June 2023.
WeedPedia Database. https://weedpedia.weedgenomics.org/ . Accessed 20 June 2023.
Hall N, Chen J, Matzrafi M, Saski CA, Westra P, Gaines TA, et al. FHY3/FAR1 transposable elements generate adaptive genetic variation in the Bassia scoparia genome. bioRxiv . 2023; DOI: https://doi.org/10.1101/2023.05.26.542497 .
Jarvis DE, Sproul JS, Navarro-Domínguez B, Krak K, Jaggi K, Huang Y-F, et al. Chromosome-scale genome assembly of the hexaploid Taiwanese goosefoot “Djulis” ( Chenopodium formosanum ). Genome Biol Evol. 2022;14(8):evac120.
Ferreira LAI, de Oliveira RS, Jr., Constantin J, Brunharo C. Evolution of ACCase-inhibitor resistance in Chloris virgata is conferred by a Trp2027Cys mutation in the herbicide target site. Pest Manag Sci. 2023;79(12):5220–9.
Laforest M, Martin SL, Bisaillon K, Soufiane B, Meloche S, Page E. A chromosome-scale draft sequence of the Canada fleabane genome. Pest Manag Sci. 2020;76(6):2158–69.
Guo L, Qiu J, Ye C, Jin G, Mao L, Zhang H, et al. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat Commun. 2017;8(1):1031.
Sato MP, Iwakami S, Fukunishi K, Sugiura K, Yasuda K, Isobe S, et al. Telomere-to-telomere genome assembly of an allotetraploid pernicious weed, Echinochloa phyllopogon . DNA Res. 2023;30(5):dsad023.
Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza . Nat Genet. 2018;50(2):285–96.
Wu D, Xie L, Sun Y, Huang Y, Jia L, Dong C, et al. A syntelog-based pan-genome provides insights into rice domestication and de-domestication. Genome Biol. 2023;24(1):179.
Wang Z, Huang S, Yang Z, Lai J, Gao X, Shi J. A high-quality, phased genome assembly of broomcorn millet reveals the features of its subgenome evolution and 3D chromatin organization. Plant Commun. 2023;4(3):100557.
Mao Q, Huff DR. The evolutionary origin of Poa annua L. Crop Sci. 2012;52(4):1910–22.
Benson CW, Sheltra MR, Maughan JP, Jellen EN, Robbins MD, Bushman BS, et al. Homoeologous evolution of the allotetraploid genome of Poa annua L. Res Sq. 2023. https://doi.org/10.21203/rs.3.rs-2729084/v1 .
Brunharo C, Benson CW, Huff DR, Lasky JR. Chromosome-scale genome assembly of Poa trivialis and population genomics reveal widespread gene flow in a cool-season grass seed production system. Plant Direct. 2024;8(3):e575.
Mo C, Wu Z, Shang X, Shi P, Wei M, Wang H, et al. Chromosome-level and graphic genomes provide insights into metabolism of bioactive metabolites and cold-adaption of Pueraria lobata var. montana . DNA Research. 2022;29(5):dsac030.
Thielen PM, Pendleton AL, Player RA, Bowden KV, Lawton TJ, Wisecaver JH. Reference genome for the highly transformable Setaria viridis ME034V. G3 (Bethesda, Md). 2020;10(10):3467–78.
Yoshida S, Kim S, Wafula EK, Tanskanen J, Kim Y-M, Honaas L, et al. Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Curr Biol. 2019;29(18):3041–52.
Qiu S, Bradley JM, Zhang P, Chaudhuri R, Blaxter M, Butlin RK, et al. Genome-enabled discovery of candidate virulence loci in Striga hermonthica , a devastating parasite of African cereal crops. New Phytol. 2022;236(2):622–38.
Nunn A, Rodríguez-Arévalo I, Tandukar Z, Frels K, Contreras-Garrido A, Carbonell-Bejerano P, et al. Chromosome-level Thlaspi arvense genome provides new tools for translational research and for a newly domesticated cash cover crop of the cooler climates. Plant Biotechnol J. 2022;20(5):944–63.
USDA-ARS Germplasm Resources Information Network (GRIN). https://www.ars-grin.gov/ . Accessed 20 June 2023.
Buck M, Hamilton C. The Nagoya Protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their utilization to the convention on biological diversity. RECIEL. 2011;20(1):47–61.
Chauhan BS, Matloob A, Mahajan G, Aslam F, Florentine SK, Jha P. Emerging challenges and opportunities for education and research in weed science. Front Plant Sci. 2017;8:1537.
Shah S, Lonhienne T, Murray CE, Chen Y, Dougan KE, Low YS, et al. Genome-guided analysis of seven weed species reveals conserved sequence and structural features of key gene targets for herbicide development. Front Plant Sci. 2022;13:909073.
International Weed Genomics Consortium Training Resources. https://www.weedgenomics.org/training-resources/ . Accessed 20 June 2023.
Blackford S. Harnessing the power of communities: career networking strategies for bioscience PhD students and postdoctoral researchers. FEMS Microbiol Lett. 2018;365(8):fny033.
Pender M, Marcotte DE, Sto Domingo MR, Maton KI. The STEM pipeline: The role of summer research experience in minority students’ Ph.D. aspirations. Educ Policy Anal Arch. 2010;18(30):1–36.
PubMed PubMed Central Google Scholar
Burke A, Okrent A, Hale K. The state of U.S. science and engineering 2022. Foundation NS. https://ncses.nsf.gov/pubs/nsb20221 . 2022.
Wu J-Y, Liao C-H, Cheng T, Nian M-W. Using data analytics to investigate attendees’ behaviors and psychological states in a virtual academic conference. Educ Technol Soc. 2021;24(1):75–91.
Download references
Peer review information
Wenjing She was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
The International Weed Genomics Consortium is supported by BASF SE, Bayer AG, Syngenta Ltd, Corteva Agriscience, CropLife International (Global Herbicide Resistance Action Committee), the Foundation for Food and Agriculture Research (Award DSnew-0000000024), and two conference grants from USDA-NIFA (Award numbers 2021–67013-33570 and 2023-67013-38785).
Author information
Authors and affiliations.
Department of Agricultural Biology, Colorado State University, 1177 Campus Delivery, Fort Collins, CO, 80523, USA
Jacob Montgomery, Sarah Morran & Todd A. Gaines
Protecting Crops and the Environment, Rothamsted Research, Harpenden, Hertfordshire, UK
Dana R. MacGregor
Department of Crop, Soil, and Environmental Sciences, Auburn University, Auburn, AL, USA
J. Scott McElroy
Department of Plant and Environmental Sciences, University of Copenhagen, Taastrup, Denmark
Paul Neve & Célia Neto
IFEVA-Conicet-Department of Ecology, University of Buenos Aires, Buenos Aires, Argentina
Martin M. Vila-Aiub & Maria Victoria Sandoval
Department of Ecology, Faculty of Agronomy, University of Buenos Aires, Buenos Aires, Argentina
Analia I. Menéndez
Department of Botany, The University of British Columbia, Vancouver, BC, Canada
Julia M. Kreiner
Institute of Crop Sciences, Zhejiang University, Hangzhou, China
Longjiang Fan
Department of Biology, University of Massachusetts Amherst, Amherst, MA, USA
Ana L. Caicedo
Department of Plant and Wildlife Sciences, Brigham Young University, Provo, UT, USA
Peter J. Maughan
Bayer AG, Weed Control Research, Frankfurt, Germany
Bianca Assis Barbosa Martins, Jagoda Mika, Alberto Collavo & Bodo Peters
Department of Crop Sciences, Federal University of Rio Grande Do Sul, Porto Alegre, Rio Grande Do Sul, Brazil
Aldo Merotto Jr.
Department of Soil and Crop Sciences, Texas A&M University, College Station, TX, USA
Nithya K. Subramanian & Muthukumar V. Bagavathiannan
Department of Plant, Soil and Microbial Sciences, Michigan State University, East Lansing, MI, USA
Luan Cutti & Eric L. Patterson
Department of Agronomy, Kansas State University, Manhattan, KS, USA
Md. Mazharul Islam & Mithila Jugulam
Department of Plant Pathology, Kansas State University, Manhattan, KS, USA
Bikram S. Gill
Crop Protection Discovery and Development, Corteva Agriscience, Indianapolis, IN, USA
Robert Cicchillo, Roger Gast & Neeta Soni
Genome Center of Excellence, Corteva Agriscience, Johnston, IA, USA
Terry R. Wright, Gina Zastrow-Hayes, Gregory May, Kevin Fengler & Victor Llaca
School of Agriculture, Food and Wine, University of Adelaide, Glen Osmond, South Australia, Australia
Jenna M. Malone
Jealott’s Hill International Research Centre, Syngenta Ltd, Bracknell, Berkshire, UK
Deepmala Sehgal, Shiv Shankhar Kaundun & Richard P. Dale
Department of Plant and Soil Sciences, University of Pretoria, Pretoria, South Africa
Barend Juan Vorster
BASF SE, Ludwigshafen Am Rhein, Germany
Jens Lerchl
Department of Crop Sciences, University of Illinois, Urbana, IL, USA
Patrick J. Tranel
Senior Scientist Consultant, Herbicide Resistance Action Committee / CropLife International, Liederbach, Germany
Roland Beffa
School of BioSciences, University of Melbourne, Parkville, VIC, Australia
Alexandre Fournier-Level
You can also search for this author in PubMed Google Scholar
Contributions
JMo and TG conceived and outlined the article. TG, DM, EP, RB, JSM, PJT, MJ wrote grants to obtain funding. MMI, BSG, and MJ performed mitotic chromosome visualization. VL performed sequencing. VL and KF assembled the genomes. LC and ELP annotated the genomes. JMo, SM, DRM, JSM, PN, CN, MV, MVS, AIM, JMK, LF, ALC, PJM, BABM, JMi, AC, MVB, LC, AFL, and ELP wrote the first draft of the article. All authors edited the article and improved the final version.
Corresponding author
Correspondence to Todd A. Gaines .
Ethics declarations
Ethics approval and consent to participate.
Ethical approval is not applicable for this article.
Competing interests
Some authors work for commercial agricultural companies (BASF, Bayer, Corteva Agriscience, or Syngenta) that develop and sell weed control products.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
13059_2024_3274_moesm1_esm.docx.
Additional file 1. List of completed and in-progress genome assemblies of weed species pollinated by insects (Table S1).
13059_2024_3274_MOESM2_ESM.docx
Additional file 2. Methods and results for visualizing and counting the metaphase chromosomes of hexaploid Avena fatua (Fig S1); diploid Lolium rigidum (Fig S2); tetraploid Phalaris minor (Fig S3); and tetraploid Salsola tragus (Fig S4).
Additional file 3. Review history.
Rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Montgomery, J., Morran, S., MacGregor, D.R. et al. Current status of community resources and priorities for weed genomics research. Genome Biol 25 , 139 (2024). https://doi.org/10.1186/s13059-024-03274-y
Download citation
Received : 11 July 2023
Accepted : 13 May 2024
Published : 27 May 2024
DOI : https://doi.org/10.1186/s13059-024-03274-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Weed science
- Reference genomes
- Rapid adaptation
- Herbicide resistance
- Public resources
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
Qualitative analysis tends to be quite flexible and relies on the researcher's judgement, so you have to reflect carefully on your choices and assumptions and be careful to avoid research bias. Quantitative analysis methods. Quantitative analysis uses numbers and statistics to understand frequencies, averages and correlations (in descriptive ...
Quantitative research methods are used to collect and analyze numerical data. This type of research is useful when the objective is to test a hypothesis, determine cause-and-effect relationships, and measure the prevalence of certain phenomena. Quantitative research methods include surveys, experiments, and secondary data analysis.
Definition of research in data analysis: According to LeCompte and Schensul, research data analysis is a process used by researchers to reduce data to a story and interpret it to derive insights. The data analysis process helps reduce a large chunk of data into smaller fragments, which makes sense. Three essential things occur during the data ...
To analyse data collected in a statistically valid manner (e.g. from experiments, surveys, and observations). Meta-analysis. Quantitative. To statistically analyse the results of a large collection of studies. Can only be applied to studies that collected data in a statistically valid manner. Thematic analysis.
A method of data analysis that is the umbrella term for engineering metrics and insights for additional value, direction, and context. By using exploratory statistical evaluation, data mining aims to identify dependencies, relations, patterns, and trends to generate advanced knowledge.
About Research Methods. This guide provides an overview of research methods, how to choose and use them, and supports and resources at UC Berkeley. As Patten and Newhart note in the book Understanding Research Methods, "Research methods are the building blocks of the scientific enterprise. They are the "how" for building systematic knowledge.
Qualitative data analysis all begins with data coding, after which an analysis method is applied. In some cases, more than one analysis method is used, depending on the research aims and research questions. In the video below, we explore some common qualitative analysis methods, along with practical examples.
data analysis, the process of systematically collecting, cleaning, transforming, describing, modeling, and interpreting data, generally employing statistical techniques. Data analysis is an important part of both scientific research and business, where demand has grown in recent years for data-driven decision making.Data analysis techniques are used to gain useful insights from datasets, which ...
How to conduct qualitative research? Given that qualitative research is characterised by flexibility, openness and responsivity to context, the steps of data collection and analysis are not as separate and consecutive as they tend to be in quantitative research [13, 14].As Fossey puts it: "sampling, data collection, analysis and interpretation are related to each other in a cyclical ...
Introduction. Statistical analysis is necessary for any research project seeking to make quantitative conclusions. The following is a primer for research-based statistical analysis. It is intended to be a high-level overview of appropriate statistical testing, while not diving too deep into any specific methodology.
2.1 Step 1: defining the research question. The first step in conducting a meta-analysis, as with any other empirical study, is the definition of the research question. Most importantly, the research question determines the realm of constructs to be considered or the type of interventions whose effects shall be analyzed.
This research method is used to gather information on attitudes, beliefs, and behaviors of individuals. Researchers use survey research to collect data quickly and efficiently from a large sample size. Survey research can be conducted through various methods such as online, phone, mail, or in-person interviews. Quantitative Research Analysis ...
Quantitative method is the collection and analysis of numerical data to answer scientific research questions. Quantitative method is used to summarize, average, find patterns, make predictions, and test causal associations as well as generalizing results to wider populations.
QDA Method #3: Discourse Analysis. Discourse is simply a fancy word for written or spoken language or debate. So, discourse analysis is all about analysing language within its social context. In other words, analysing language - such as a conversation, a speech, etc - within the culture and society it takes place.
comprehensive training in research methods. NCRM delivers training and resources at core and advanced levels, covering quantitative, qualitative, digital, creative, visual, mixed and multimodal methods. Join our mailing list. Receive monthly updates about our latest courses and resources ...
Qualitative Research. Qualitative research is a type of research methodology that focuses on exploring and understanding people's beliefs, attitudes, behaviors, and experiences through the collection and analysis of non-numerical data. It seeks to answer research questions through the examination of subjective data, such as interviews, focus ...
Qualitative research methods. Each of the research approaches involve using one or more data collection methods.These are some of the most common qualitative methods: Observations: recording what you have seen, heard, or encountered in detailed field notes. Interviews: personally asking people questions in one-on-one conversations. Focus groups: asking questions and generating discussion among ...
It is easy to confuse systematic reviews and meta-analyses. A systematic review is an objective, reproducible method to find answers to a certain research question, by collecting all available studies related to that question and reviewing and analyzing their results. A meta-analysis differs from a systematic review in that it uses statistical ...
Learning Objectives. By the end of this section, you should be able to: Recall the 6 Steps of the Scientific Method; Differentiate between four kinds of research methods: surveys, field research, experiments, and secondary data analysis.
Research methods in psychology are systematic procedures used to observe, describe, predict, and explain behavior and mental processes. They include experiments, surveys, case studies, and naturalistic observations, ensuring data collection is objective and reliable to understand and explain psychological phenomena. ... A meta-analysis is a ...
Data analysis is a crucial step in the research process, transforming raw data into meaningful insights that drive informed decisions and advance knowledge. This article explores the various types and methods of data analysis in research, providing a comprehensive guide for researchers across disciplines.
Priority is a dimension of research design in a mixed method study that provides a signpost to signify how a study's principal findings were derived. ... In In Onwuegbuzie A. J., Johnson R. B. (Eds.), The Routledge reviewer's guide to mixed methods analysis (pp. 259-276). Sage. Crossref. Google Scholar. Greene J. C. (2007). Mixed methods in ...
Electrogastrography (EGG) is a non-invasive method with high diagnostic potential for the prevention of gastroenterological pathologies in clinical practice. In this paper, a review of the measurement systems, procedures, and methods of analysis used in electrogastrography is presented. A critical review of historical and current literature is conducted, focusing on electrode placement ...
ABOUT PEW RESEARCH CENTER Pew Research Center is a nonpartisan fact tank that informs the public about the issues, attitudes and trends shaping the world. It conducts public opinion polling, demographic research, media content analysis and other empirical social science research. Pew Research Center does not take policy positions.
Additionally, this research can help predict the future impacts of climate change on the local environment, providing references for planning response strategies. ... method to find water bodies. NDWI is calculated using the formula: NDWI = (Green - NIR) / (Green + NIR). This method uses the strong absorption of near-infrared light by water and ...
Textual analysis is a broad term for various research methods used to describe, interpret and understand texts. All kinds of information can be gleaned from a text - from its literal meaning to the subtext, symbolism, assumptions, and values it reveals. The methods used to conduct textual analysis depend on the field and the aims of the ...
Background Like all other species, fungi are susceptible to infection by viruses. The diversity of fungal viruses has been rapidly expanding in recent years due to the availability of advanced sequencing technologies. However, compared to other virome studies, the research on fungi-associated viruses remains limited. Results In this study, we downloaded and analyzed over 200 public datasets ...
Weeds are attractive models for basic and applied research due to their impacts on agricultural systems and capacity to swiftly adapt in response to anthropogenic selection pressures. Currently, a lack of genomic information precludes research to elucidate the genetic basis of rapid adaptation for important traits like herbicide resistance and stress tolerance and the effect of evolutionary ...