Writing an Abstract for Your Research Paper

Definition and Purpose of Abstracts
An abstract is a short summary of your (published or unpublished) research paper, usually about a paragraph (c. 6-7 sentences, 150-250 words) long. A well-written abstract serves multiple purposes:
- an abstract lets readers get the gist or essence of your paper or article quickly, in order to decide whether to read the full paper;
- an abstract prepares readers to follow the detailed information, analyses, and arguments in your full paper;
- and, later, an abstract helps readers remember key points from your paper.
It’s also worth remembering that search engines and bibliographic databases use abstracts, as well as the title, to identify key terms for indexing your published paper. So what you include in your abstract and in your title are crucial for helping other researchers find your paper or article.
If you are writing an abstract for a course paper, your professor may give you specific guidelines for what to include and how to organize your abstract. Similarly, academic journals often have specific requirements for abstracts. So in addition to following the advice on this page, you should be sure to look for and follow any guidelines from the course or journal you’re writing for.
The Contents of an Abstract
Abstracts contain most of the following kinds of information in brief form. The body of your paper will, of course, develop and explain these ideas much more fully. As you will see in the samples below, the proportion of your abstract that you devote to each kind of information—and the sequence of that information—will vary, depending on the nature and genre of the paper that you are summarizing in your abstract. And in some cases, some of this information is implied, rather than stated explicitly. The Publication Manual of the American Psychological Association , which is widely used in the social sciences, gives specific guidelines for what to include in the abstract for different kinds of papers—for empirical studies, literature reviews or meta-analyses, theoretical papers, methodological papers, and case studies.
Here are the typical kinds of information found in most abstracts:
- the context or background information for your research; the general topic under study; the specific topic of your research
- the central questions or statement of the problem your research addresses
- what’s already known about this question, what previous research has done or shown
- the main reason(s) , the exigency, the rationale , the goals for your research—Why is it important to address these questions? Are you, for example, examining a new topic? Why is that topic worth examining? Are you filling a gap in previous research? Applying new methods to take a fresh look at existing ideas or data? Resolving a dispute within the literature in your field? . . .
- your research and/or analytical methods
- your main findings , results , or arguments
- the significance or implications of your findings or arguments.
Your abstract should be intelligible on its own, without a reader’s having to read your entire paper. And in an abstract, you usually do not cite references—most of your abstract will describe what you have studied in your research and what you have found and what you argue in your paper. In the body of your paper, you will cite the specific literature that informs your research.
When to Write Your Abstract
Although you might be tempted to write your abstract first because it will appear as the very first part of your paper, it’s a good idea to wait to write your abstract until after you’ve drafted your full paper, so that you know what you’re summarizing.
What follows are some sample abstracts in published papers or articles, all written by faculty at UW-Madison who come from a variety of disciplines. We have annotated these samples to help you see the work that these authors are doing within their abstracts.
Choosing Verb Tenses within Your Abstract
The social science sample (Sample 1) below uses the present tense to describe general facts and interpretations that have been and are currently true, including the prevailing explanation for the social phenomenon under study. That abstract also uses the present tense to describe the methods, the findings, the arguments, and the implications of the findings from their new research study. The authors use the past tense to describe previous research.
The humanities sample (Sample 2) below uses the past tense to describe completed events in the past (the texts created in the pulp fiction industry in the 1970s and 80s) and uses the present tense to describe what is happening in those texts, to explain the significance or meaning of those texts, and to describe the arguments presented in the article.
The science samples (Samples 3 and 4) below use the past tense to describe what previous research studies have done and the research the authors have conducted, the methods they have followed, and what they have found. In their rationale or justification for their research (what remains to be done), they use the present tense. They also use the present tense to introduce their study (in Sample 3, “Here we report . . .”) and to explain the significance of their study (In Sample 3, This reprogramming . . . “provides a scalable cell source for. . .”).
Sample Abstract 1
From the social sciences.
Reporting new findings about the reasons for increasing economic homogamy among spouses
Gonalons-Pons, Pilar, and Christine R. Schwartz. “Trends in Economic Homogamy: Changes in Assortative Mating or the Division of Labor in Marriage?” Demography , vol. 54, no. 3, 2017, pp. 985-1005.
![what is research abstract brainly “The growing economic resemblance of spouses has contributed to rising inequality by increasing the number of couples in which there are two high- or two low-earning partners. [Annotation for the previous sentence: The first sentence introduces the topic under study (the “economic resemblance of spouses”). This sentence also implies the question underlying this research study: what are the various causes—and the interrelationships among them—for this trend?] The dominant explanation for this trend is increased assortative mating. Previous research has primarily relied on cross-sectional data and thus has been unable to disentangle changes in assortative mating from changes in the division of spouses’ paid labor—a potentially key mechanism given the dramatic rise in wives’ labor supply. [Annotation for the previous two sentences: These next two sentences explain what previous research has demonstrated. By pointing out the limitations in the methods that were used in previous studies, they also provide a rationale for new research.] We use data from the Panel Study of Income Dynamics (PSID) to decompose the increase in the correlation between spouses’ earnings and its contribution to inequality between 1970 and 2013 into parts due to (a) changes in assortative mating, and (b) changes in the division of paid labor. [Annotation for the previous sentence: The data, research and analytical methods used in this new study.] Contrary to what has often been assumed, the rise of economic homogamy and its contribution to inequality is largely attributable to changes in the division of paid labor rather than changes in sorting on earnings or earnings potential. Our findings indicate that the rise of economic homogamy cannot be explained by hypotheses centered on meeting and matching opportunities, and they show where in this process inequality is generated and where it is not.” (p. 985) [Annotation for the previous two sentences: The major findings from and implications and significance of this study.]](https://writing.wisc.edu/wp-content/uploads/sites/535/2019/08/Abstract-1.png)
Sample Abstract 2
From the humanities.
Analyzing underground pulp fiction publications in Tanzania, this article makes an argument about the cultural significance of those publications
Emily Callaci. “Street Textuality: Socialism, Masculinity, and Urban Belonging in Tanzania’s Pulp Fiction Publishing Industry, 1975-1985.” Comparative Studies in Society and History , vol. 59, no. 1, 2017, pp. 183-210.
![what is research abstract brainly “From the mid-1970s through the mid-1980s, a network of young urban migrant men created an underground pulp fiction publishing industry in the city of Dar es Salaam. [Annotation for the previous sentence: The first sentence introduces the context for this research and announces the topic under study.] As texts that were produced in the underground economy of a city whose trajectory was increasingly charted outside of formalized planning and investment, these novellas reveal more than their narrative content alone. These texts were active components in the urban social worlds of the young men who produced them. They reveal a mode of urbanism otherwise obscured by narratives of decolonization, in which urban belonging was constituted less by national citizenship than by the construction of social networks, economic connections, and the crafting of reputations. This article argues that pulp fiction novellas of socialist era Dar es Salaam are artifacts of emergent forms of male sociability and mobility. In printing fictional stories about urban life on pilfered paper and ink, and distributing their texts through informal channels, these writers not only described urban communities, reputations, and networks, but also actually created them.” (p. 210) [Annotation for the previous sentences: The remaining sentences in this abstract interweave other essential information for an abstract for this article. The implied research questions: What do these texts mean? What is their historical and cultural significance, produced at this time, in this location, by these authors? The argument and the significance of this analysis in microcosm: these texts “reveal a mode or urbanism otherwise obscured . . .”; and “This article argues that pulp fiction novellas. . . .” This section also implies what previous historical research has obscured. And through the details in its argumentative claims, this section of the abstract implies the kinds of methods the author has used to interpret the novellas and the concepts under study (e.g., male sociability and mobility, urban communities, reputations, network. . . ).]](https://writing.wisc.edu/wp-content/uploads/sites/535/2019/08/Abstract-2.png)
Sample Abstract/Summary 3
From the sciences.
Reporting a new method for reprogramming adult mouse fibroblasts into induced cardiac progenitor cells
Lalit, Pratik A., Max R. Salick, Daryl O. Nelson, Jayne M. Squirrell, Christina M. Shafer, Neel G. Patel, Imaan Saeed, Eric G. Schmuck, Yogananda S. Markandeya, Rachel Wong, Martin R. Lea, Kevin W. Eliceiri, Timothy A. Hacker, Wendy C. Crone, Michael Kyba, Daniel J. Garry, Ron Stewart, James A. Thomson, Karen M. Downs, Gary E. Lyons, and Timothy J. Kamp. “Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors.” Cell Stem Cell , vol. 18, 2016, pp. 354-367.
![what is research abstract brainly “Several studies have reported reprogramming of fibroblasts into induced cardiomyocytes; however, reprogramming into proliferative induced cardiac progenitor cells (iCPCs) remains to be accomplished. [Annotation for the previous sentence: The first sentence announces the topic under study, summarizes what’s already known or been accomplished in previous research, and signals the rationale and goals are for the new research and the problem that the new research solves: How can researchers reprogram fibroblasts into iCPCs?] Here we report that a combination of 11 or 5 cardiac factors along with canonical Wnt and JAK/STAT signaling reprogrammed adult mouse cardiac, lung, and tail tip fibroblasts into iCPCs. The iCPCs were cardiac mesoderm-restricted progenitors that could be expanded extensively while maintaining multipo-tency to differentiate into cardiomyocytes, smooth muscle cells, and endothelial cells in vitro. Moreover, iCPCs injected into the cardiac crescent of mouse embryos differentiated into cardiomyocytes. iCPCs transplanted into the post-myocardial infarction mouse heart improved survival and differentiated into cardiomyocytes, smooth muscle cells, and endothelial cells. [Annotation for the previous four sentences: The methods the researchers developed to achieve their goal and a description of the results.] Lineage reprogramming of adult somatic cells into iCPCs provides a scalable cell source for drug discovery, disease modeling, and cardiac regenerative therapy.” (p. 354) [Annotation for the previous sentence: The significance or implications—for drug discovery, disease modeling, and therapy—of this reprogramming of adult somatic cells into iCPCs.]](https://writing.wisc.edu/wp-content/uploads/sites/535/2019/08/Abstract-3.png)
Sample Abstract 4, a Structured Abstract
Reporting results about the effectiveness of antibiotic therapy in managing acute bacterial sinusitis, from a rigorously controlled study
Note: This journal requires authors to organize their abstract into four specific sections, with strict word limits. Because the headings for this structured abstract are self-explanatory, we have chosen not to add annotations to this sample abstract.
Wald, Ellen R., David Nash, and Jens Eickhoff. “Effectiveness of Amoxicillin/Clavulanate Potassium in the Treatment of Acute Bacterial Sinusitis in Children.” Pediatrics , vol. 124, no. 1, 2009, pp. 9-15.
“OBJECTIVE: The role of antibiotic therapy in managing acute bacterial sinusitis (ABS) in children is controversial. The purpose of this study was to determine the effectiveness of high-dose amoxicillin/potassium clavulanate in the treatment of children diagnosed with ABS.
METHODS : This was a randomized, double-blind, placebo-controlled study. Children 1 to 10 years of age with a clinical presentation compatible with ABS were eligible for participation. Patients were stratified according to age (<6 or ≥6 years) and clinical severity and randomly assigned to receive either amoxicillin (90 mg/kg) with potassium clavulanate (6.4 mg/kg) or placebo. A symptom survey was performed on days 0, 1, 2, 3, 5, 7, 10, 20, and 30. Patients were examined on day 14. Children’s conditions were rated as cured, improved, or failed according to scoring rules.
RESULTS: Two thousand one hundred thirty-five children with respiratory complaints were screened for enrollment; 139 (6.5%) had ABS. Fifty-eight patients were enrolled, and 56 were randomly assigned. The mean age was 6630 months. Fifty (89%) patients presented with persistent symptoms, and 6 (11%) presented with nonpersistent symptoms. In 24 (43%) children, the illness was classified as mild, whereas in the remaining 32 (57%) children it was severe. Of the 28 children who received the antibiotic, 14 (50%) were cured, 4 (14%) were improved, 4(14%) experienced treatment failure, and 6 (21%) withdrew. Of the 28children who received placebo, 4 (14%) were cured, 5 (18%) improved, and 19 (68%) experienced treatment failure. Children receiving the antibiotic were more likely to be cured (50% vs 14%) and less likely to have treatment failure (14% vs 68%) than children receiving the placebo.
CONCLUSIONS : ABS is a common complication of viral upper respiratory infections. Amoxicillin/potassium clavulanate results in significantly more cures and fewer failures than placebo, according to parental report of time to resolution.” (9)
Some Excellent Advice about Writing Abstracts for Basic Science Research Papers, by Professor Adriano Aguzzi from the Institute of Neuropathology at the University of Zurich:

Academic and Professional Writing
This is an accordion element with a series of buttons that open and close related content panels.
Analysis Papers
Reading Poetry
A Short Guide to Close Reading for Literary Analysis
Using Literary Quotations
Play Reviews
Writing a Rhetorical Précis to Analyze Nonfiction Texts
Incorporating Interview Data
Grant Proposals
Planning and Writing a Grant Proposal: The Basics
Additional Resources for Grants and Proposal Writing
Job Materials and Application Essays
Writing Personal Statements for Ph.D. Programs
- Before you begin: useful tips for writing your essay
- Guided brainstorming exercises
- Get more help with your essay
- Frequently Asked Questions
Resume Writing Tips
CV Writing Tips
Cover Letters
Business Letters
Proposals and Dissertations
Resources for Proposal Writers
Resources for Dissertators
Research Papers
Planning and Writing Research Papers
Quoting and Paraphrasing
Writing Annotated Bibliographies
Creating Poster Presentations
Thank-You Notes
Advice for Students Writing Thank-You Notes to Donors
Reading for a Review
Critical Reviews
Writing a Review of Literature
Scientific Reports
Scientific Report Format
Sample Lab Assignment
Writing for the Web
Writing an Effective Blog Post
Writing for Social Media: A Guide for Academics
- Privacy Policy

Home » Research Paper Abstract – Writing Guide and Examples
Research Paper Abstract – Writing Guide and Examples
Table of Contents

Research Paper Abstract
Research Paper Abstract is a brief summary of a research pape r that describes the study’s purpose, methods, findings, and conclusions . It is often the first section of the paper that readers encounter, and its purpose is to provide a concise and accurate overview of the paper’s content. The typical length of an abstract is usually around 150-250 words, and it should be written in a concise and clear manner.
Research Paper Abstract Structure
The structure of a research paper abstract usually includes the following elements:
- Background or Introduction: Briefly describe the problem or research question that the study addresses.
- Methods : Explain the methodology used to conduct the study, including the participants, materials, and procedures.
- Results : Summarize the main findings of the study, including statistical analyses and key outcomes.
- Conclusions : Discuss the implications of the study’s findings and their significance for the field, as well as any limitations or future directions for research.
- Keywords : List a few keywords that describe the main topics or themes of the research.
How to Write Research Paper Abstract
Here are the steps to follow when writing a research paper abstract:
- Start by reading your paper: Before you write an abstract, you should have a complete understanding of your paper. Read through the paper carefully, making sure you understand the purpose, methods, results, and conclusions.
- Identify the key components : Identify the key components of your paper, such as the research question, methods used, results obtained, and conclusion reached.
- Write a draft: Write a draft of your abstract, using concise and clear language. Make sure to include all the important information, but keep it short and to the point. A good rule of thumb is to keep your abstract between 150-250 words.
- Use clear and concise language : Use clear and concise language to explain the purpose of your study, the methods used, the results obtained, and the conclusions drawn.
- Emphasize your findings: Emphasize your findings in the abstract, highlighting the key results and the significance of your study.
- Revise and edit: Once you have a draft, revise and edit it to ensure that it is clear, concise, and free from errors.
- Check the formatting: Finally, check the formatting of your abstract to make sure it meets the requirements of the journal or conference where you plan to submit it.
Research Paper Abstract Examples
Research Paper Abstract Examples could be following:
Title : “The Effectiveness of Cognitive-Behavioral Therapy for Treating Anxiety Disorders: A Meta-Analysis”
Abstract : This meta-analysis examines the effectiveness of cognitive-behavioral therapy (CBT) in treating anxiety disorders. Through the analysis of 20 randomized controlled trials, we found that CBT is a highly effective treatment for anxiety disorders, with large effect sizes across a range of anxiety disorders, including generalized anxiety disorder, panic disorder, and social anxiety disorder. Our findings support the use of CBT as a first-line treatment for anxiety disorders and highlight the importance of further research to identify the mechanisms underlying its effectiveness.
Title : “Exploring the Role of Parental Involvement in Children’s Education: A Qualitative Study”
Abstract : This qualitative study explores the role of parental involvement in children’s education. Through in-depth interviews with 20 parents of children in elementary school, we found that parental involvement takes many forms, including volunteering in the classroom, helping with homework, and communicating with teachers. We also found that parental involvement is influenced by a range of factors, including parent and child characteristics, school culture, and socio-economic status. Our findings suggest that schools and educators should prioritize building strong partnerships with parents to support children’s academic success.
Title : “The Impact of Exercise on Cognitive Function in Older Adults: A Systematic Review and Meta-Analysis”
Abstract : This paper presents a systematic review and meta-analysis of the existing literature on the impact of exercise on cognitive function in older adults. Through the analysis of 25 randomized controlled trials, we found that exercise is associated with significant improvements in cognitive function, particularly in the domains of executive function and attention. Our findings highlight the potential of exercise as a non-pharmacological intervention to support cognitive health in older adults.
When to Write Research Paper Abstract
The abstract of a research paper should typically be written after you have completed the main body of the paper. This is because the abstract is intended to provide a brief summary of the key points and findings of the research, and you can’t do that until you have completed the research and written about it in detail.
Once you have completed your research paper, you can begin writing your abstract. It is important to remember that the abstract should be a concise summary of your research paper, and should be written in a way that is easy to understand for readers who may not have expertise in your specific area of research.
Purpose of Research Paper Abstract
The purpose of a research paper abstract is to provide a concise summary of the key points and findings of a research paper. It is typically a brief paragraph or two that appears at the beginning of the paper, before the introduction, and is intended to give readers a quick overview of the paper’s content.
The abstract should include a brief statement of the research problem, the methods used to investigate the problem, the key results and findings, and the main conclusions and implications of the research. It should be written in a clear and concise manner, avoiding jargon and technical language, and should be understandable to a broad audience.
The abstract serves as a way to quickly and easily communicate the main points of a research paper to potential readers, such as academics, researchers, and students, who may be looking for information on a particular topic. It can also help researchers determine whether a paper is relevant to their own research interests and whether they should read the full paper.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Research Objectives – Types, Examples and...

APA Research Paper Format – Example, Sample and...

Figures in Research Paper – Examples and Guide

Research Design – Types, Methods and Examples

Future Research – Thesis Guide

Ethical Considerations – Types, Examples and...
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Saudi J Anaesth
- v.13(Suppl 1); 2019 Apr
Writing the title and abstract for a research paper: Being concise, precise, and meticulous is the key
Milind s. tullu.
Department of Pediatrics, Seth G.S. Medical College and KEM Hospital, Parel, Mumbai, Maharashtra, India
This article deals with formulating a suitable title and an appropriate abstract for an original research paper. The “title” and the “abstract” are the “initial impressions” of a research article, and hence they need to be drafted correctly, accurately, carefully, and meticulously. Often both of these are drafted after the full manuscript is ready. Most readers read only the title and the abstract of a research paper and very few will go on to read the full paper. The title and the abstract are the most important parts of a research paper and should be pleasant to read. The “title” should be descriptive, direct, accurate, appropriate, interesting, concise, precise, unique, and should not be misleading. The “abstract” needs to be simple, specific, clear, unbiased, honest, concise, precise, stand-alone, complete, scholarly, (preferably) structured, and should not be misrepresentative. The abstract should be consistent with the main text of the paper, especially after a revision is made to the paper and should include the key message prominently. It is very important to include the most important words and terms (the “keywords”) in the title and the abstract for appropriate indexing purpose and for retrieval from the search engines and scientific databases. Such keywords should be listed after the abstract. One must adhere to the instructions laid down by the target journal with regard to the style and number of words permitted for the title and the abstract.
Introduction
This article deals with drafting a suitable “title” and an appropriate “abstract” for an original research paper. Because the “title” and the “abstract” are the “initial impressions” or the “face” of a research article, they need to be drafted correctly, accurately, carefully, meticulously, and consume time and energy.[ 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 ] Often, these are drafted after the complete manuscript draft is ready.[ 2 , 3 , 4 , 5 , 9 , 10 , 11 ] Most readers will read only the title and the abstract of a published research paper, and very few “interested ones” (especially, if the paper is of use to them) will go on to read the full paper.[ 1 , 2 ] One must remember to adhere to the instructions laid down by the “target journal” (the journal for which the author is writing) regarding the style and number of words permitted for the title and the abstract.[ 2 , 4 , 5 , 7 , 8 , 9 , 12 ] Both the title and the abstract are the most important parts of a research paper – for editors (to decide whether to process the paper for further review), for reviewers (to get an initial impression of the paper), and for the readers (as these may be the only parts of the paper available freely and hence, read widely).[ 4 , 8 , 12 ] It may be worth for the novice author to browse through titles and abstracts of several prominent journals (and their target journal as well) to learn more about the wording and styles of the titles and abstracts, as well as the aims and scope of the particular journal.[ 5 , 7 , 9 , 13 ]
The details of the title are discussed under the subheadings of importance, types, drafting, and checklist.
Importance of the title
When a reader browses through the table of contents of a journal issue (hard copy or on website), the title is the “ first detail” or “face” of the paper that is read.[ 2 , 3 , 4 , 5 , 6 , 13 ] Hence, it needs to be simple, direct, accurate, appropriate, specific, functional, interesting, attractive/appealing, concise/brief, precise/focused, unambiguous, memorable, captivating, informative (enough to encourage the reader to read further), unique, catchy, and it should not be misleading.[ 1 , 2 , 3 , 4 , 5 , 6 , 9 , 12 ] It should have “just enough details” to arouse the interest and curiosity of the reader so that the reader then goes ahead with studying the abstract and then (if still interested) the full paper.[ 1 , 2 , 4 , 13 ] Journal websites, electronic databases, and search engines use the words in the title and abstract (the “keywords”) to retrieve a particular paper during a search; hence, the importance of these words in accessing the paper by the readers has been emphasized.[ 3 , 4 , 5 , 6 , 12 , 14 ] Such important words (or keywords) should be arranged in appropriate order of importance as per the context of the paper and should be placed at the beginning of the title (rather than the later part of the title, as some search engines like Google may just display only the first six to seven words of the title).[ 3 , 5 , 12 ] Whimsical, amusing, or clever titles, though initially appealing, may be missed or misread by the busy reader and very short titles may miss the essential scientific words (the “keywords”) used by the indexing agencies to catch and categorize the paper.[ 1 , 3 , 4 , 9 ] Also, amusing or hilarious titles may be taken less seriously by the readers and may be cited less often.[ 4 , 15 ] An excessively long or complicated title may put off the readers.[ 3 , 9 ] It may be a good idea to draft the title after the main body of the text and the abstract are drafted.[ 2 , 3 , 4 , 5 ]
Types of titles
Titles can be descriptive, declarative, or interrogative. They can also be classified as nominal, compound, or full-sentence titles.
Descriptive or neutral title
This has the essential elements of the research theme, that is, the patients/subjects, design, interventions, comparisons/control, and outcome, but does not reveal the main result or the conclusion.[ 3 , 4 , 12 , 16 ] Such a title allows the reader to interpret the findings of the research paper in an impartial manner and with an open mind.[ 3 ] These titles also give complete information about the contents of the article, have several keywords (thus increasing the visibility of the article in search engines), and have increased chances of being read and (then) being cited as well.[ 4 ] Hence, such descriptive titles giving a glimpse of the paper are generally preferred.[ 4 , 16 ]

Declarative title
This title states the main finding of the study in the title itself; it reduces the curiosity of the reader, may point toward a bias on the part of the author, and hence is best avoided.[ 3 , 4 , 12 , 16 ]
Interrogative title
This is the one which has a query or the research question in the title.[ 3 , 4 , 16 ] Though a query in the title has the ability to sensationalize the topic, and has more downloads (but less citations), it can be distracting to the reader and is again best avoided for a research article (but can, at times, be used for a review article).[ 3 , 6 , 16 , 17 ]
From a sentence construct point of view, titles may be nominal (capturing only the main theme of the study), compound (with subtitles to provide additional relevant information such as context, design, location/country, temporal aspect, sample size, importance, and a provocative or a literary; for example, see the title of this review), or full-sentence titles (which are longer and indicate an added degree of certainty of the results).[ 4 , 6 , 9 , 16 ] Any of these constructs may be used depending on the type of article, the key message, and the author's preference or judgement.[ 4 ]
Drafting a suitable title
A stepwise process can be followed to draft the appropriate title. The author should describe the paper in about three sentences, avoiding the results and ensuring that these sentences contain important scientific words/keywords that describe the main contents and subject of the paper.[ 1 , 4 , 6 , 12 ] Then the author should join the sentences to form a single sentence, shorten the length (by removing redundant words or adjectives or phrases), and finally edit the title (thus drafted) to make it more accurate, concise (about 10–15 words), and precise.[ 1 , 3 , 4 , 5 , 9 ] Some journals require that the study design be included in the title, and this may be placed (using a colon) after the primary title.[ 2 , 3 , 4 , 14 ] The title should try to incorporate the Patients, Interventions, Comparisons and Outcome (PICO).[ 3 ] The place of the study may be included in the title (if absolutely necessary), that is, if the patient characteristics (such as study population, socioeconomic conditions, or cultural practices) are expected to vary as per the country (or the place of the study) and have a bearing on the possible outcomes.[ 3 , 6 ] Lengthy titles can be boring and appear unfocused, whereas very short titles may not be representative of the contents of the article; hence, optimum length is required to ensure that the title explains the main theme and content of the manuscript.[ 4 , 5 , 9 ] Abbreviations (except the standard or commonly interpreted ones such as HIV, AIDS, DNA, RNA, CDC, FDA, ECG, and EEG) or acronyms should be avoided in the title, as a reader not familiar with them may skip such an article and nonstandard abbreviations may create problems in indexing the article.[ 3 , 4 , 5 , 6 , 9 , 12 ] Also, too much of technical jargon or chemical formulas in the title may confuse the readers and the article may be skipped by them.[ 4 , 9 ] Numerical values of various parameters (stating study period or sample size) should also be avoided in the titles (unless deemed extremely essential).[ 4 ] It may be worthwhile to take an opinion from a impartial colleague before finalizing the title.[ 4 , 5 , 6 ] Thus, multiple factors (which are, at times, a bit conflicting or contrasting) need to be considered while formulating a title, and hence this should not be done in a hurry.[ 4 , 6 ] Many journals ask the authors to draft a “short title” or “running head” or “running title” for printing in the header or footer of the printed paper.[ 3 , 12 ] This is an abridged version of the main title of up to 40–50 characters, may have standard abbreviations, and helps the reader to navigate through the paper.[ 3 , 12 , 14 ]
Checklist for a good title
Table 1 gives a checklist/useful tips for drafting a good title for a research paper.[ 1 , 2 , 3 , 4 , 5 , 6 , 12 ] Table 2 presents some of the titles used by the author of this article in his earlier research papers, and the appropriateness of the titles has been commented upon. As an individual exercise, the reader may try to improvise upon the titles (further) after reading the corresponding abstract and full paper.
Checklist/useful tips for drafting a good title for a research paper
Some titles used by author of this article in his earlier publications and remark/comment on their appropriateness
The Abstract
The details of the abstract are discussed under the subheadings of importance, types, drafting, and checklist.
Importance of the abstract
The abstract is a summary or synopsis of the full research paper and also needs to have similar characteristics like the title. It needs to be simple, direct, specific, functional, clear, unbiased, honest, concise, precise, self-sufficient, complete, comprehensive, scholarly, balanced, and should not be misleading.[ 1 , 2 , 3 , 7 , 8 , 9 , 10 , 11 , 13 , 17 ] Writing an abstract is to extract and summarize (AB – absolutely, STR – straightforward, ACT – actual data presentation and interpretation).[ 17 ] The title and abstracts are the only sections of the research paper that are often freely available to the readers on the journal websites, search engines, and in many abstracting agencies/databases, whereas the full paper may attract a payment per view or a fee for downloading the pdf copy.[ 1 , 2 , 3 , 7 , 8 , 10 , 11 , 13 , 14 ] The abstract is an independent and stand-alone (that is, well understood without reading the full paper) section of the manuscript and is used by the editor to decide the fate of the article and to choose appropriate reviewers.[ 2 , 7 , 10 , 12 , 13 ] Even the reviewers are initially supplied only with the title and the abstract before they agree to review the full manuscript.[ 7 , 13 ] This is the second most commonly read part of the manuscript, and therefore it should reflect the contents of the main text of the paper accurately and thus act as a “real trailer” of the full article.[ 2 , 7 , 11 ] The readers will go through the full paper only if they find the abstract interesting and relevant to their practice; else they may skip the paper if the abstract is unimpressive.[ 7 , 8 , 9 , 10 , 13 ] The abstract needs to highlight the selling point of the manuscript and succeed in luring the reader to read the complete paper.[ 3 , 7 ] The title and the abstract should be constructed using keywords (key terms/important words) from all the sections of the main text.[ 12 ] Abstracts are also used for submitting research papers to a conference for consideration for presentation (as oral paper or poster).[ 9 , 13 , 17 ] Grammatical and typographic errors reflect poorly on the quality of the abstract, may indicate carelessness/casual attitude on part of the author, and hence should be avoided at all times.[ 9 ]
Types of abstracts
The abstracts can be structured or unstructured. They can also be classified as descriptive or informative abstracts.
Structured and unstructured abstracts
Structured abstracts are followed by most journals, are more informative, and include specific subheadings/subsections under which the abstract needs to be composed.[ 1 , 7 , 8 , 9 , 10 , 11 , 13 , 17 , 18 ] These subheadings usually include context/background, objectives, design, setting, participants, interventions, main outcome measures, results, and conclusions.[ 1 ] Some journals stick to the standard IMRAD format for the structure of the abstracts, and the subheadings would include Introduction/Background, Methods, Results, And (instead of Discussion) the Conclusion/s.[ 1 , 2 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 17 , 18 ] Structured abstracts are more elaborate, informative, easy to read, recall, and peer-review, and hence are preferred; however, they consume more space and can have same limitations as an unstructured abstract.[ 7 , 9 , 18 ] The structured abstracts are (possibly) better understood by the reviewers and readers. Anyway, the choice of the type of the abstract and the subheadings of a structured abstract depend on the particular journal style and is not left to the author's wish.[ 7 , 10 , 12 ] Separate subheadings may be necessary for reporting meta-analysis, educational research, quality improvement work, review, or case study.[ 1 ] Clinical trial abstracts need to include the essential items mentioned in the CONSORT (Consolidated Standards Of Reporting Trials) guidelines.[ 7 , 9 , 14 , 19 ] Similar guidelines exist for various other types of studies, including observational studies and for studies of diagnostic accuracy.[ 20 , 21 ] A useful resource for the above guidelines is available at www.equator-network.org (Enhancing the QUAlity and Transparency Of health Research). Unstructured (or non-structured) abstracts are free-flowing, do not have predefined subheadings, and are commonly used for papers that (usually) do not describe original research.[ 1 , 7 , 9 , 10 ]
The four-point structured abstract: This has the following elements which need to be properly balanced with regard to the content/matter under each subheading:[ 9 ]
Background and/or Objectives: This states why the work was undertaken and is usually written in just a couple of sentences.[ 3 , 7 , 8 , 9 , 10 , 12 , 13 ] The hypothesis/study question and the major objectives are also stated under this subheading.[ 3 , 7 , 8 , 9 , 10 , 12 , 13 ]
Methods: This subsection is the longest, states what was done, and gives essential details of the study design, setting, participants, blinding, sample size, sampling method, intervention/s, duration and follow-up, research instruments, main outcome measures, parameters evaluated, and how the outcomes were assessed or analyzed.[ 3 , 7 , 8 , 9 , 10 , 12 , 13 , 14 , 17 ]
Results/Observations/Findings: This subheading states what was found, is longer, is difficult to draft, and needs to mention important details including the number of study participants, results of analysis (of primary and secondary objectives), and include actual data (numbers, mean, median, standard deviation, “P” values, 95% confidence intervals, effect sizes, relative risks, odds ratio, etc.).[ 3 , 7 , 8 , 9 , 10 , 12 , 13 , 14 , 17 ]
Conclusions: The take-home message (the “so what” of the paper) and other significant/important findings should be stated here, considering the interpretation of the research question/hypothesis and results put together (without overinterpreting the findings) and may also include the author's views on the implications of the study.[ 3 , 7 , 8 , 9 , 10 , 12 , 13 , 14 , 17 ]
The eight-point structured abstract: This has the following eight subheadings – Objectives, Study Design, Study Setting, Participants/Patients, Methods/Intervention, Outcome Measures, Results, and Conclusions.[ 3 , 9 , 18 ] The instructions to authors given by the particular journal state whether they use the four- or eight-point abstract or variants thereof.[ 3 , 14 ]
Descriptive and Informative abstracts
Descriptive abstracts are short (75–150 words), only portray what the paper contains without providing any more details; the reader has to read the full paper to know about its contents and are rarely used for original research papers.[ 7 , 10 ] These are used for case reports, reviews, opinions, and so on.[ 7 , 10 ] Informative abstracts (which may be structured or unstructured as described above) give a complete detailed summary of the article contents and truly reflect the actual research done.[ 7 , 10 ]
Drafting a suitable abstract
It is important to religiously stick to the instructions to authors (format, word limit, font size/style, and subheadings) provided by the journal for which the abstract and the paper are being written.[ 7 , 8 , 9 , 10 , 13 ] Most journals allow 200–300 words for formulating the abstract and it is wise to restrict oneself to this word limit.[ 1 , 2 , 3 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 22 ] Though some authors prefer to draft the abstract initially, followed by the main text of the paper, it is recommended to draft the abstract in the end to maintain accuracy and conformity with the main text of the paper (thus maintaining an easy linkage/alignment with title, on one hand, and the introduction section of the main text, on the other hand).[ 2 , 7 , 9 , 10 , 11 ] The authors should check the subheadings (of the structured abstract) permitted by the target journal, use phrases rather than sentences to draft the content of the abstract, and avoid passive voice.[ 1 , 7 , 9 , 12 ] Next, the authors need to get rid of redundant words and edit the abstract (extensively) to the correct word count permitted (every word in the abstract “counts”!).[ 7 , 8 , 9 , 10 , 13 ] It is important to ensure that the key message, focus, and novelty of the paper are not compromised; the rationale of the study and the basis of the conclusions are clear; and that the abstract is consistent with the main text of the paper.[ 1 , 2 , 3 , 7 , 9 , 11 , 12 , 13 , 14 , 17 , 22 ] This is especially important while submitting a revision of the paper (modified after addressing the reviewer's comments), as the changes made in the main (revised) text of the paper need to be reflected in the (revised) abstract as well.[ 2 , 10 , 12 , 14 , 22 ] Abbreviations should be avoided in an abstract, unless they are conventionally accepted or standard; references, tables, or figures should not be cited in the abstract.[ 7 , 9 , 10 , 11 , 13 ] It may be worthwhile not to rush with the abstract and to get an opinion by an impartial colleague on the content of the abstract; and if possible, the full paper (an “informal” peer-review).[ 1 , 7 , 8 , 9 , 11 , 17 ] Appropriate “Keywords” (three to ten words or phrases) should follow the abstract and should be preferably chosen from the Medical Subject Headings (MeSH) list of the U.S. National Library of Medicine ( https://meshb.nlm.nih.gov/search ) and are used for indexing purposes.[ 2 , 3 , 11 , 12 ] These keywords need to be different from the words in the main title (the title words are automatically used for indexing the article) and can be variants of the terms/phrases used in the title, or words from the abstract and the main text.[ 3 , 12 ] The ICMJE (International Committee of Medical Journal Editors; http://www.icmje.org/ ) also recommends publishing the clinical trial registration number at the end of the abstract.[ 7 , 14 ]
Checklist for a good abstract
Table 3 gives a checklist/useful tips for formulating a good abstract for a research paper.[ 1 , 2 , 3 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 17 , 22 ]
Checklist/useful tips for formulating a good abstract for a research paper
Concluding Remarks
This review article has given a detailed account of the importance and types of titles and abstracts. It has also attempted to give useful hints for drafting an appropriate title and a complete abstract for a research paper. It is hoped that this review will help the authors in their career in medical writing.
Financial support and sponsorship
Conflicts of interest.
There are no conflicts of interest.
Acknowledgement
The author thanks Dr. Hemant Deshmukh - Dean, Seth G.S. Medical College & KEM Hospital, for granting permission to publish this manuscript.
Research Skills
An abstract is a summary. In academic articles, the abstract is a one-paragraph synopsis of the content of the article that follows it. The abstract, therefore, is not an introduction. While it does appear first, it is meant to stand apart from the article as a whole and to enable researchers to quickly decide whether the article discusses an applicable problem, subject, and/or methodology. An abstract is not terribly detailed, and it doesn’t usually divulge an article’s ultimate claims. Rather, it provides a brief glimpse of the topics covered, the research methods used, and the general findings discovered. The abstract is useful for helping you decide, relatively quickly, whether or not the article overlaps with your research question(s). This short paragraph is sometimes followed by a list of author-supplied keywords that, if used as search terms, could reasonably be expected to pull up the article in question. An abstract also pretty clearly identifies a piece of writing as an academic research article instead of something that is less formal, such as an informative piece. In other words, if you aren’t sure whether what you have found is in fact an academic research article, ask yourself, “Does it have an abstract?” If the answer is, “No,” it may not be an academic research article, in which case it would be unsuitable for our purposes.
- Parts of An Article. Authored by : Kerry Bowers. Provided by : University of Mississippi. Project : WRIT 250 Committee OER Project. License : CC BY-SA: Attribution-ShareAlike

Privacy Policy
- Open access
- Published: 27 May 2024
Current status of community resources and priorities for weed genomics research
- Jacob Montgomery 1 ,
- Sarah Morran 1 ,
- Dana R. MacGregor ORCID: orcid.org/0000-0003-0543-0408 2 ,
- J. Scott McElroy ORCID: orcid.org/0000-0003-0331-3697 3 ,
- Paul Neve ORCID: orcid.org/0000-0002-3136-5286 4 ,
- Célia Neto ORCID: orcid.org/0000-0003-3256-5228 4 ,
- Martin M. Vila-Aiub ORCID: orcid.org/0000-0003-2118-290X 5 ,
- Maria Victoria Sandoval 5 ,
- Analia I. Menéndez ORCID: orcid.org/0000-0002-9681-0280 6 ,
- Julia M. Kreiner ORCID: orcid.org/0000-0002-8593-1394 7 ,
- Longjiang Fan ORCID: orcid.org/0000-0003-4846-0500 8 ,
- Ana L. Caicedo ORCID: orcid.org/0000-0002-0378-6374 9 ,
- Peter J. Maughan 10 ,
- Bianca Assis Barbosa Martins 11 ,
- Jagoda Mika 11 ,
- Alberto Collavo 11 ,
- Aldo Merotto Jr. ORCID: orcid.org/0000-0002-1581-0669 12 ,
- Nithya K. Subramanian ORCID: orcid.org/0000-0002-1659-7396 13 ,
- Muthukumar V. Bagavathiannan ORCID: orcid.org/0000-0002-1107-7148 13 ,
- Luan Cutti ORCID: orcid.org/0000-0002-2867-7158 14 ,
- Md. Mazharul Islam 15 ,
- Bikram S. Gill ORCID: orcid.org/0000-0003-4510-9459 16 ,
- Robert Cicchillo 17 ,
- Roger Gast 17 ,
- Neeta Soni ORCID: orcid.org/0000-0002-4647-8355 17 ,
- Terry R. Wright ORCID: orcid.org/0000-0002-3969-2812 18 ,
- Gina Zastrow-Hayes 18 ,
- Gregory May 18 ,
- Jenna M. Malone ORCID: orcid.org/0000-0002-9637-2073 19 ,
- Deepmala Sehgal ORCID: orcid.org/0000-0002-4141-1784 20 ,
- Shiv Shankhar Kaundun ORCID: orcid.org/0000-0002-7249-2046 20 ,
- Richard P. Dale 20 ,
- Barend Juan Vorster ORCID: orcid.org/0000-0003-3518-3508 21 ,
- Bodo Peters 11 ,
- Jens Lerchl ORCID: orcid.org/0000-0002-9633-2653 22 ,
- Patrick J. Tranel ORCID: orcid.org/0000-0003-0666-4564 23 ,
- Roland Beffa ORCID: orcid.org/0000-0003-3109-388X 24 ,
- Alexandre Fournier-Level ORCID: orcid.org/0000-0002-6047-7164 25 ,
- Mithila Jugulam ORCID: orcid.org/0000-0003-2065-9067 15 ,
- Kevin Fengler 18 ,
- Victor Llaca ORCID: orcid.org/0000-0003-4822-2924 18 ,
- Eric L. Patterson ORCID: orcid.org/0000-0001-7111-6287 14 &
- Todd A. Gaines ORCID: orcid.org/0000-0003-1485-7665 1
Genome Biology volume 25 , Article number: 139 ( 2024 ) Cite this article
653 Accesses
9 Altmetric
Metrics details
Weeds are attractive models for basic and applied research due to their impacts on agricultural systems and capacity to swiftly adapt in response to anthropogenic selection pressures. Currently, a lack of genomic information precludes research to elucidate the genetic basis of rapid adaptation for important traits like herbicide resistance and stress tolerance and the effect of evolutionary mechanisms on wild populations. The International Weed Genomics Consortium is a collaborative group of scientists focused on developing genomic resources to impact research into sustainable, effective weed control methods and to provide insights about stress tolerance and adaptation to assist crop breeding.
Each year globally, agricultural producers and landscape managers spend billions of US dollars [ 1 , 2 ] and countless hours attempting to control weedy plants and reduce their adverse effects. These management methods range from low-tech (e.g., pulling plants from the soil by hand) to extremely high-tech (e.g., computer vision-controlled spraying of herbicides). Regardless of technology level, effective control methods serve as strong selection pressures on weedy plants and often result in rapid evolution of weed populations resistant to such methods [ 3 , 4 , 5 , 6 , 7 ]. Thus, humans and weeds have been locked in an arms race, where humans develop new or improved control methods and weeds adapt and evolve to circumvent such methods.
Applying genomics to weed science offers a unique opportunity to study rapid adaptation, epigenetic responses, and examples of evolutionary rescue of diverse weedy species in the face of widespread and powerful selective pressures. Furthermore, lessons learned from these studies may also help to develop more sustainable control methods and to improve crop breeding efforts in the face of our ever-changing climate. While other research fields have used genetics and genomics to uncover the basis of many biological traits [ 8 , 9 , 10 , 11 ] and to understand how ecological factors affect evolution [ 12 , 13 ], the field of weed science has lagged behind in the development of genomic tools essential for such studies [ 14 ]. As research in human and crop genetics pushes into the era of pangenomics (i.e., multiple chromosome scale genome assemblies for a single species [ 15 , 16 ]), publicly available genomic information is still lacking or severely limited for the majority of weed species. Recent reviews of current weed genomes identified 26 [ 17 ] and 32 weed species with sequenced genomes [ 18 ]—many assembled to a sub-chromosome level.
Here, we summarize the current state of weed genomics, highlighting cases where genomics approaches have successfully provided insights on topics such as population genetic dynamics, genome evolution, and the genetic basis of herbicide resistance, rapid adaptation, and crop dedomestication. These highlighted investigations all relied upon genomic resources that are relatively rare for weedy species. Throughout, we identify additional resources that would advance the field of weed science and enable further progress in weed genomics. We then introduce the International Weed Genomics Consortium (IWGC), an open collaboration among researchers, and describe current efforts to generate these additional resources.
Evolution of weediness: potential research utilizing weed genomics tools
Weeds can evolve from non-weed progenitors through wild colonization, crop de-domestication, or crop-wild hybridization [ 19 ]. Because the time span in which weeds have evolved is necessarily limited by the origins of agriculture, these non-weed relatives often still exist and can be leveraged through population genomic and comparative genomic approaches to identify the adaptive changes that have driven the evolution of weediness. The ability to rapidly adapt, persist, and spread in agroecosystems are defining features of weedy plants, leading many to advocate agricultural weeds as ideal candidates for studying rapid plant adaptation [ 20 , 21 , 22 , 23 ]. The insights gained from applying plant ecological approaches to the study of rapid weed adaptation will move us towards the ultimate goals of mitigating such adaptation and increasing the efficacy of crop breeding and biotechnology [ 14 ].
Biology and ecological genomics of weeds
The impressive community effort to create and maintain resources for Arabidopsis thaliana ecological genomics provides a motivating example for the emerging study of weed genomics [ 24 , 25 , 26 , 27 ]. Arabidopsis thaliana was the first flowering plant species to have its genome fully sequenced [ 28 ] and rapidly became a model organism for plant molecular biology. As weedy genomes become available, collection, maintenance, and resequencing of globally distributed accessions of these species will help to replicate the success found in ecological studies of A. thaliana [ 29 , 30 , 31 , 32 , 33 , 34 , 35 ]. Evaluation of these accessions for traits of interest to produce large phenomics data sets (as in [ 36 , 37 , 38 , 39 , 40 ]) enables genome-wide association studies and population genomics analyses aimed at dissecting the genetic basis of variation in such traits [ 41 ]. Increasingly, these resources (e.g. the 1001 genomes project [ 29 ]) have enabled A. thaliana to be utilized as a model species to explore the eco-evolutionary basis of plant adaptation in a more realistic ecological context. Weedy species should supplement lessons in eco-evolutionary genomics learned from these experiments in A. thaliana .
Untargeted genomic approaches for understanding the evolutionary trajectories of populations and the genetic basis of traits as described above rely on the collection of genotypic information from across the genome of many individuals. While whole-genome resequencing accomplishes this requirement and requires no custom methodology, this approach provides more information than is necessary and is prohibitively expensive in species with large genomes. Development and optimization of genotype-by-sequencing methods for capturing reduced representations of newly sequence genomes like those described by [ 42 , 43 , 44 ] will reduce the cost and computational requirements of genetic mapping and population genetic experiments. Most major weed species do not currently have protocols for stable transformation, a key development in the popularity of A. thaliana as a model organism and a requirement for many functional genomic approaches. Functional validation of genes/variants believed to be responsible for traits of interest in weeds has thus far relied on transiently manipulating endogenous gene expression [ 45 , 46 ] or ectopic expression of a transgene in a model system [ 47 , 48 , 49 ]. While these methods have been successful, few weed species have well-studied viral vectors to adapt for use in virus induced gene silencing. Spray induced gene silencing is another potential option for functional investigation of candidate genes in weeds, but more research is needed to establish reliable delivery and gene knockdown [ 50 ]. Furthermore, traits with complex genetic architecture divergent between the researched and model species may not be amenable to functional genomic approaches using transgenesis techniques in model systems. Developing protocols for reduced representation sequencing, stable transformation, and gene editing/silencing in weeds will allow for more thorough characterization of candidate genetic variants underlying traits of interest.
Beyond rapid adaptation, some weedy species offer an opportunity to better understand co-evolution, like that between plants and pollinators and how their interaction leads to the spread of weedy alleles (Additional File 1 : Table S1). A suite of plant–insect traits has co-evolved to maximize the attraction of the insect pollinator community and the efficiency of pollen deposition between flowers ensuring fruit and seed production in many weeds [ 51 , 52 ]. Genetic mapping experiments have identified genes and genetic variants responsible for many floral traits affecting pollinator interaction including petal color [ 53 , 54 , 55 , 56 ], flower symmetry and size [ 57 , 58 , 59 ], and production of volatile organic compounds [ 60 , 61 , 62 ] and nectar [ 63 , 64 , 65 ]. While these studies reveal candidate genes for selection under co-evolution, herbicide resistance alleles may also have pleiotropic effects on the ecology of weeds [ 66 ], altering plant-pollinator interactions [ 67 ]. Discovery of genes and genetic variants involved in weed-pollinator interaction and their molecular and environmental control may create opportunities for better management of weeds with insect-mediated pollination. For example, if management can disrupt pollinator attraction/interaction with these weeds, the efficiency of reproduction may be reduced.
A more complete understanding of weed ecological genomics will undoubtedly elucidate many unresolved questions regarding the genetic basis of various aspects of weediness. For instance, when comparing populations of a species from agricultural and non-agricultural environments, is there evidence for contemporary evolution of weedy traits selected by agricultural management or were “natural” populations pre-adapted to agroecosystems? Where there is differentiation between weedy and natural populations, which traits are under selection and what is the genetic basis of variation in those traits? When comparing between weedy populations, is there evidence for parallel versus non-parallel evolution of weediness at the phenotypic and genotypic levels? Such studies may uncover fundamental truths about weediness. For example, is there a common phenotypic and/or genotypic basis for aspects of weediness among diverse weed species? The availability of characterized accessions and reference genomes for species of interest are required for such studies but only a few weedy species have these resources developed.
Population genomics
Weed species are certainly fierce competitors, able to outcompete crops and endemic species in their native environment, but they are also remarkable colonizers of perturbed habitats. Weeds achieve this through high fecundity, often producing tens of thousands of seeds per individual plant [ 68 , 69 , 70 ]. These large numbers in terms of demographic population size often combine with outcrossing reproduction to generate high levels of diversity with local effective population sizes in the hundreds of thousands [ 71 , 72 ]. This has two important consequences: weed populations retain standing genetic variation and generate many new mutations, supporting weed success in the face of harsh control. The generation of genomic tools to monitor weed populations at the molecular level is a game-changer to understanding weed dynamics and precisely testing the effect of artificial selection (i.e., management) and other evolutionary mechanisms on the genetic make-up of populations.
Population genomic data, without any environmental or phenotypic information, can be used to scan the genomes of weed and non-weed relatives to identify selective sweeps, pointing at loci supporting weed adaptation on micro- or macro-evolutionary scales. Two recent within-species examples include weedy rice, where population differentiation between weedy and domesticated populations was used to identify the genetic basis of weedy de-domestication [ 73 ], and common waterhemp, where consistent allelic differences among natural and agricultural collections resolved a complex set of agriculturally adaptive alleles [ 74 , 75 ]. A recent comparative population genomic study of weedy barnyardgrass and crop millet species has demonstrated how inter-specific investigations can resolve the signatures of crop and weed evolution [ 76 ] (also see [ 77 ] for a non-weed climate adaptation example). Multiple sequence alignments across numerous species provide complementary insight into adaptive convergence over deeper timescales, even with just one genomic sample per species (e.g., [ 78 , 79 ]). Thus, newly sequenced weed genomes combined with genomes available for closely related crops (outlined by [ 14 , 80 ]) and an effort to identify other non-weed wild relatives will be invaluable in characterizing the genetic architecture of weed adaptation and evolution across diverse species.
Weeds experience high levels of genetic selection, both artificial in response to agricultural practices and particularly herbicides, and natural in response to the environmental conditions they encounter [ 81 , 82 ]. Using genomic analysis to identify loci that are the targets of selection, whether natural or artificial, would point at vulnerabilities that could be leveraged against weeds to develop new and more sustainable management strategies [ 83 ]. This is a key motivation to develop genotype-by-environment association (GEA) and selective sweep scan approaches, which allow researchers to resolve the molecular basis of multi-dimensional adaptation [ 84 , 85 ]. GEA approaches, in particular, have been widely used on landscape-wide resequencing collections to determine the genetic basis of climate adaptation (e.g., [ 27 , 86 , 87 ]), but have yet to be fully exploited to diagnose the genetic basis of the various aspects of weediness [ 88 ]. Armed with data on environmental dimensions of agricultural settings, such as focal crop, soil quality, herbicide use, and climate, GEA approaches can help disentangle how discrete farming practices have influenced the evolution of weediness and resolve broader patterns of local adaptation across a weed’s range. Although non-weedy relatives are not technically required for GEA analyses, inclusion of environmental and genomic data from weed progenitors can further distinguish genetic variants underpinning weed origins from those involved in local adaptation.
New weeds emerge frequently [ 89 ], either through hybridization between species as documented for sea beet ( Beta vulgaris ssp. maritima) hybridizing with crop beet to produce progeny that are well adapted to agricultural conditions [ 90 , 91 , 92 ], or through the invasion of alien species that find a new range to colonize. Biosecurity measures are often in place to stop the introduction of new weeds; however, the vast scale of global agricultural commodity trade precludes the possibility of total control. Population genomic analysis is now able to measure gene flow between populations [ 74 , 93 , 94 , 95 ] and identify populations of origin for invasive species including weeds [ 96 , 97 , 98 ]. For example, the invasion route of the pest fruitfly Drosophila suzukii from Eastern Asia to North America and Europe through Hawaii was deciphered using Approximate Bayesian Computation on high-throughput sequencing data from a global sample of multiple populations [ 99 ]. Genomics can also be leveraged to predict invasion rather than explain it. The resequencing of a global sample of common ragweed ( Ambrosia artemisiifolia L.) elucidated a complex invasion route whereby Europe was invaded by multiple introductions of American ragweed that hybridized in Europe prior to a subsequent introduction to Australia [ 100 , 101 ]. In this context, the use of genomically informed species distribution models helps assess the risk associated with different source populations, which in the case of common ragweed, suggests that a source population from Florida would allow ragweed to invade most of northern Australia [ 102 ]. Globally coordinated research efforts to understand potential distribution models could support the transformation of biosecurity from perspective analysis towards predictive risk assessment.
Herbicide resistance and weed management
Herbicide resistance is among the numerous weedy traits that can evolve in plant populations exposed to agricultural selection pressures. Over-reliance on herbicides to control weeds, along with low diversity and lack of redundancy in weed management strategies, has resulted in globally widespread herbicide resistance [ 103 ]. To date, 272 herbicide-resistant weed species have been reported worldwide, and at least one resistance case exists for 21 of the 31 existing herbicide sites of action [ 104 ]—significantly limiting chemical weed control options available to agriculturalists. This limitation of control options is exacerbated by the recent lack of discovery of herbicides with new sites of action [ 105 ].
Herbicide resistance may result from several different physiological mechanisms. Such mechanisms have been classified into two main groups, target-site resistance (TSR) [ 4 , 106 ] and non-target-site resistance (NTSR) [ 4 , 107 ]. The first group encompasses changes that reduce binding affinity between a herbicide and its target [ 108 ]. These changes may provide resistance to multiple herbicides that have a common biochemical target [ 109 ] and can be effectively managed through mixture and/or rotation of herbicides targeting different sites of action [ 110 ]. The second group (NTSR), includes alterations in herbicide absorption, translocation, sequestration, and/or metabolism that may lead to unpredictable pleotropic cross-resistance profiles where structurally and functionally diverse herbicides are rendered ineffective by one or more genetic variant(s) [ 47 ]. This mechanism of resistance threatens not only the efficacy of existing herbicidal chemistries, but also ones yet to be discovered. While TSR is well understood because of the ease of identification and molecular characterization of target site variants, NTSR mechanisms are significantly more challenging to research because they are often polygenic, and the resistance causing element(s) are not well understood [ 111 ].
Improving the current understanding of metabolic NTSR mechanisms is not an easy task, since genes of diverse biochemical functions are involved, many of which exist as extensive gene families [ 109 , 112 ]. Expression changes of NTSR genes have been implicated in several resistance cases where the protein products of the genes are functionally equivalent across sensitive and resistant plants, but their relative abundance leads to resistance. Thus, regulatory elements of NTSR genes have been scrutinized to understand their role in NTSR mechanisms [ 113 ]. Similarly, epigenetic modifications have been hypothesized to play a role in NTSR, with much remaining to be explored [ 114 , 115 , 116 ]. Untargeted approaches such as genome-wide association, selective sweep scans, linkage mapping, RNA-sequencing, and metabolomic profiling have proven helpful to complement more specific biochemical- and chemo-characterization studies towards the elucidation of NTSR mechanisms as well as their regulation and evolution [ 47 , 117 , 118 , 119 , 120 , 121 , 122 , 123 , 124 ]. Even in cases where resistance has been attributed to TSR, genetic mapping approaches can detect other NTSR loci contributing to resistance (as shown by [ 123 ]) and provide further evidence for the role of TSR mutations across populations. Knowledge of the genetic basis of NTSR will aid the rational design of herbicides by screening new compounds for interaction with newly discovered NTSR proteins during early research phases and by identifying conserved chemical structures that interact with these proteins that should be avoided in small molecule design.
Genomic resources can also be used to predict the protein structure for novel herbicide target site and metabolism genes. This will allow for prediction of efficacy and selectivity for new candidate herbicides in silico to increase herbicide discovery throughput as well as aid in the design and development of next-generation technologies for sustainable weed management. Proteolysis targeting chimeras (PROTACs) have the potential to bind desired targets with great selectivity and degrade proteins by utilizing natural protein ubiquitination and degradation pathways within plants [ 125 ]. Spray-induced gene silencing in weeds using oligonucleotides has potential as a new, innovative, and sustainable method for weed management, but improved methods for design and delivery of oligonucleotides are needed to make this technique a viable management option [ 50 ]. Additionally, success in the field of pharmaceutical drug discovery in the development of molecules modulating protein–protein interactions offers another potential avenue towards the development of herbicides with novel targets [ 126 , 127 ]. High-quality reference genomes allow for the design of new weed management technologies like the ones listed here that are specific to—and effective across—weed species but have a null effect on non-target organisms.
Comparative genomics and genome biology
The genomes of weed species are as diverse as weed species themselves. Weeds are found across highly diverged plant families and often have no phylogenetically close model or crop species relatives for comparison. On all measurable metrics, weed genomes run the gamut. Some have smaller genomes like Cyperus spp. (~ 0.26 Gb) while others are larger, such as Avena fatua (~ 11.1 Gb) (Table 1 ). Some have high heterozygosity in terms of single-nucleotide polymorphisms, such as the Amaranthus spp., while others are primarily self-pollinated and quite homozygous, such as Poa annua [ 128 , 129 ]. Some are diploid such as Conyza canadensis and Echinochloa haploclada while others are polyploid such as C. sumetrensis , E. crus-galli , and E. colona [ 76 ]. The availability of genomic resources in these diverse, unexplored branches of the tree of life allows us to identify consistencies and anomalies in the field of genome biology.
The weed genomes published so far have focused mainly on weeds of agronomic crops, and studies have revolved around their ability to resist key herbicides. For example, genomic resources were vital in the elucidation of herbicide resistance cases involving target site gene copy number variants (CNVs). Gene CNVs of 5-enolpyruvylshikimate-3-phosphate synthase ( EPSPS ) have been found to confer resistance to the herbicide glyphosate in diverse weed species. To date, nine species have independently evolved EPSPS CNVs, and species achieve increased EPSPS copy number via different mechanisms [ 153 ]. For instance, the EPSPS CNV in Bassia scoparia is caused by tandem duplication, which is accredited to transposable element insertions flanking EPSPS and subsequent unequal crossing over events [ 154 , 155 ]. In Eleusine indica , a EPSPS CNV was caused by translocation of the EPSPS locus into the subtelomere followed by telomeric sequence exchange [ 156 ]. One of the most fascinating genome biology discoveries in weed science has been that of extra-chromosomal circular DNAs (eccDNAs) that harbor the EPSPS gene in the weed species Amaranthus palmeri [ 157 , 158 ]. In this case, the eccDNAs autonomously replicate separately from the nuclear genome and do not reintegrate into chromosomes, which has implications for inheritance, fitness, and genome structure [ 159 ]. These discoveries would not have been possible without reference assemblies of weed genomes, next-generation sequencing, and collaboration with experts in plant genomics and bioinformatics.
Another question that is often explored with weedy genomes is the nature and composition of gene families that are associated with NTSR. Gene families under consideration often include cytochrome P450s (CYPs), glutathione- S -transferases (GSTs), ABC transporters, etc. Some questions commonly considered with new weed genomes include how many genes are in each of these gene families, where are they located, and which weed accessions and species have an over-abundance of them that might explain their ability to evolve resistance so rapidly [ 76 , 146 , 160 , 161 ]? Weed genome resources are necessary to answer questions about gene family expansion or contraction during the evolution of weediness, including the role of polyploidy in NTSR gene family expansion as explored by [ 162 ].
Translational research and communication with weed management stakeholders
Whereas genomics of model plants is typically aimed at addressing fundamental questions in plant biology, and genomics of crop species has the obvious goal of crop improvement, goals of genomics of weedy plants also include the development of more effective and sustainable strategies for their management. Weed genomic resources assist with these objectives by providing novel molecular ecological and evolutionary insights from the context of intensive anthropogenic management (which is lacking in model plants), and offer knowledge and resources for trait discovery for crop improvement, especially given that many wild crop relatives are also important agronomic weeds (e.g., [ 163 ]). For instance, crop-wild relatives are valuable for improving crop breeding for marginal environments [ 164 ]. Thus, weed genomics presents unique opportunities and challenges relative to plant genomics more broadly. It should also be noted that although weed science at its core is an applied discipline, it draws broadly from many scientific disciplines such as, plant physiology, chemistry, ecology, and evolutionary biology, to name a few. The successful integration of weed-management strategies, therefore, requires extensive collaboration among individuals collectively possessing the necessary expertise [ 165 ].
With the growing complexity of herbicide resistance management, practitioners are beginning to recognize the importance of understanding resistance mechanisms to inform appropriate management tactics [ 14 ]. Although weed science practitioners do not need to understand the technical details of weed genomics, their appreciation of the power of weed genomics—together with their unique insights from field observations—will yield novel opportunities for applications of weed genomics to weed management. In particular, combining field management history with information on weed resistance mechanisms is expected to provide novel insights into evolutionary trajectories (e.g. [ 6 , 166 ]), which can be utilized for disrupting evolutionary adaptation. It can be difficult to obtain field history information from practitioners, but developing an understanding among them of the importance of such information can be invaluable.
Development of weed genomics resources by the IWGC
Weed genomics is a fast-growing field of research with many recent breakthroughs and many unexplored areas of study. The International Weed Genomics Consortium (IWGC) started in 2021 to address the roadblocks listed above and to promote the study of weedy plants. The IWGC is an open collaboration among academic, government, and industry researchers focused on producing genomic tools for weedy species from around the world. Through this collaboration, our initial aim is to provide chromosome-level reference genome assemblies for at least 50 important weedy species from across the globe that are chosen based on member input, economic impact, and global prevalence (Fig. 1 ). Each genome will include annotation of gene models and repetitive elements and will be freely available through public databases with no intellectual property restrictions. Additionally, future funding of the IWGC will focus on improving gene annotations and supplementing these reference genomes with tools that increase their utility.
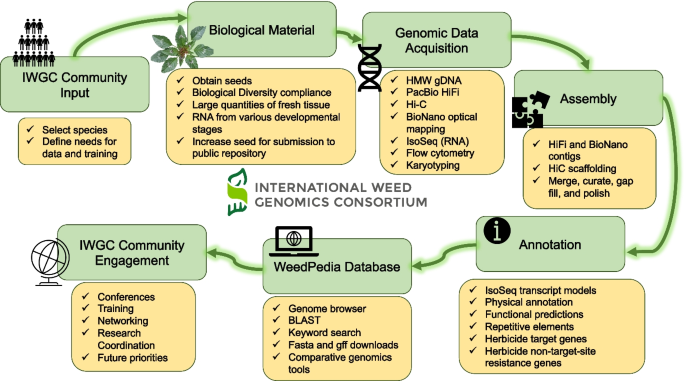
The International Weed Genomics Consortium (IWGC) collected input from the weed genomics community to develop plans for weed genome sequencing, annotation, user-friendly genome analysis tools, and community engagement
Reference genomes and data analysis tools
The first objective of the IWGC is to provide high-quality genomic resources for agriculturally important weeds. The IWGC therefore created two main resources for information about, access to, or analysis of weed genomic data (Fig. 1 ). The IWGC website (available at [ 167 ]) communicates the status and results of genome sequencing projects, information on training and funding opportunities, upcoming events, and news in weed genomics. It also contains details of all sequenced species including genome size, ploidy, chromosome number, herbicide resistance status, and reference genome assembly statistics. The IWGC either compiles existing data on genome size, ploidy, and chromosome number, or obtains the data using flow cytometry and cytogenetics (Fig. 1 ; Additional File 2 : Fig S1-S4). Through this website, users can request an account to access our second main resource, an online genome database called WeedPedia (accessible at [ 168 ]), with an account that is created within 3–5 working days of an account request submission. WeedPedia hosts IWGC-generated and other relevant publicly accessible genomic data as well as a suite of bioinformatic tools. Unlike what is available for other fields, weed science did not have a centralized hub for genomics information, data, and analysis prior to the IWGC. Our intention in creating WeedPedia is to encourage collaboration and equity of access to information across the research community. Importantly, all genome assemblies and annotations from the IWGC (Table 1 ), along with the raw data used to produce them, will be made available through NCBI GenBank. Upon completion of a 1-year sponsoring member data confidentiality period for each species (dates listed in Table 1 ), scientific teams within the IWGC produce the first genome-wide investigation to submit for publication including whole genome level analyses on genes, gene families, and repetitive sequences as well as comparative analysis with other species. Genome assemblies and data will be publicly available through NCBI as part of these initial publications for each species.
WeedPedia is a cloud-based omics database management platform built from the software “CropPedia” and licensed from KeyGene (Wageningen, The Netherlands). The interface allows users to access, visualize, and download genome assemblies along with structural and functional annotation. The platform includes a genome browser, comparative map viewer, pangenome tools, RNA-sequencing data visualization tools, genetic mapping and marker analysis tools, and alignment capabilities that allow searches by keyword or sequence. Additionally, genes encoding known target sites of herbicides have been specially annotated, allowing users to quickly identify and compare these genes of interest. The platform is flexible, making it compatible with future integration of other data types such as epigenetic or proteomic information. As an online platform with a graphical user interface, WeedPedia provides user-friendly, intuitive tools that encourage users to integrate genomics into their research while also allowing more advanced users to download genomic data to be used in custom analysis pipelines. We aspire for WeedPedia to mimic the success of other public genomic databases such as NCBI, CoGe, Phytozome, InsectBase, and Mycocosm to name a few. WeedPedia currently hosts reference genomes for 40 species (some of which are currently in their 1-year confidentiality period) with additional genomes in the pipeline to reach a currently planned total of 55 species (Table 1 ). These genomes include both de novo reference genomes generated or in progress by the IWGC (31 species; Table 1 ), and publicly available genome assemblies of 24 weedy or related species that were generated by independent research groups (Table 2 ). As of May 2024, WeedPedia has over 370 registered users from more than 27 countries spread across 6 continents.
The IWGC reference genomes are generated in partnership with the Corteva Agriscience Genome Center of Excellence (Johnston, Iowa) using a combination of single-molecule long-read sequencing, optical genome maps, and chromosome conformation mapping. This strategy has already yielded highly contiguous, phased, chromosome-level assemblies for 26 weed species, with additional assemblies currently in progress (Table 1 ). The IWGC assemblies have been completed as single or haplotype-resolved double-haplotype pseudomolecules in inbreeding and outbreeding species, respectively, with multiple genomes being near gapless. For example, the de novo assemblies of the allohexaploids Conyza sumatrensis and Chenopodium album have all chromosomes captured in single scaffolds and most chromosomes being gapless from telomere to telomere. Complementary full-length isoform (IsoSeq) sequencing of RNA collected from diverse tissue types and developmental stages assists in the development of gene models during annotation.
As with accessibility of data, a core objective of the IWGC is to facilitate open access to sequenced germplasm when possible for featured species. Historically, the weed science community has rarely shared or adopted standard germplasm (e.g., specific weed accessions). The IWGC has selected a specific accession of each species for reference genome assembly (typically susceptible to herbicides). In collaboration with a parallel effort by the Herbicide Resistant Plants committee of the Weed Science Society of America, seeds of the sequenced weed accessions will be deposited in the United States Department of Agriculture Germplasm Resources Information Network [ 186 ] for broad access by the scientific community and their accession numbers will be listed on the IWGC website. In some cases, it is not possible to generate enough seed to deposit into a public repository (e.g., plants that typically reproduce vegetatively, that are self-incompatible, or that produce very few seeds from a single individual). In these cases, the location of collection for sequenced accessions will at least inform the community where the sequenced individual came from and where they may expect to collect individuals with similar genotypes. The IWGC ensures that sequenced accessions are collected and documented to comply with the Nagoya Protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their utilization under the Convention on Biological Diversity and related Access and Benefit Sharing Legislation [ 187 ]. As additional accessions of weed species are sequenced (e.g., pangenomes are obtained), the IWGC will facilitate germplasm sharing protocols to support collaboration. Further, to simplify the investigation of herbicide resistance, the IWGC will link WeedPedia with the International Herbicide-Resistant Weed Database [ 104 ], an already widely known and utilized database for weed scientists.
Training and collaboration in weed genomics
Beyond producing genomic tools and resources, a priority of the IWGC is to enable the utilization of these resources across a wide range of stakeholders. A holistic approach to training is required for weed science generally [ 188 ], and we would argue even more so for weed genomics. To accomplish our training goals, the IWGC is developing and delivering programs aimed at the full range of IWGC stakeholders and covering a breadth of relevant topics. We have taken care to ensure our approaches are diverse as to provide training to researchers with all levels of existing experience and differing reasons for engaging with these tools. Throughout, the focus is on ensuring that our training and outreach result in impacts that benefit a wide range of stakeholders.
Although recently developed tools are incredibly enabling and have great potential to replace antiquated methodology [ 189 ] and to solve pressing weed science problems [ 14 ], specialized computational skills are required to fully explore and unlock meaning from these highly complex datasets. Collaboration with, or training of, computational biologists equipped with these skills and resources developed by the IWGC will enable weed scientists to expand research programs and better understand the genetic underpinnings of weed evolution and herbicide resistance. To fill existing skill gaps, the IWGC is developing summer bootcamps and online modules directed specifically at weed scientists that will provide training on computational skills (Fig. 1 ). Because successful utilization of the IWGC resources requires more than general computational skills, we have created three targeted workshops that teach practical skills related to genomics databases, molecular biology, and population genomics (available at [ 190 ]). The IWGC has also hosted two official conference meetings, one in September of 2021 and one in January of 2023, with more conferences planned. These conferences have included invited speakers to present successful implementations of weed genomics, educational workshops to build computational skills, and networking opportunities for research to connect and collaborate.
Engagement opportunities during undergraduate degrees have been shown to improve academic outcomes [ 191 , 192 ]. As one activity to help achieve this goal, the IWGC has sponsored opportunities for US undergraduates to undertake a 10-week research experience, which includes an introduction to bioinformatics, a plant genomics research project that results in a presentation, and access to career building opportunities in diverse workplace environments. To increase equitable access to conferences and professional communities, we supported early career researchers to attend the first two IWGC conferences in the USA as well as workshops and bootcamps in Europe, South America, and Australia. These hybrid or in-person travel grants are intentionally designed to remove barriers and increase participation of individuals from backgrounds and experiences currently underrepresented within weed/plant science or genomics [ 193 ]. Recipients of these travel awards gave presentations and gained the measurable benefits that come from either virtual or in-person participation in conferences [ 194 ]. Moving forward, weed scientists must amass skills associated with genomic analyses and collaborate with other area experts to fully leverage resources developed by the IWGC.
The tools generated through the IWGC will enable many new research projects with diverse objectives like those listed above. In summary, contiguous genome assemblies and complete annotation information will allow weed scientists to join plant breeders in the use of genetic mapping for many traits including stress tolerance, plant architecture, and herbicide resistance (especially important for cases of NTSR). These assemblies will also allow for investigations of population structure, gene flow, and responses to evolutionary mechanisms like genetic bottlenecking and artificial selection. Understanding gene sequences across diverse weed species will be vital in modeling new herbicide target site proteins and designing novel effective herbicides with minimal off-target effects. The IWGC website will improve accessibility to weed genomics data by providing a single hub for reference genomes as well as phenotypic and genotypic information for accessions shared with the IWGC. Deposition of sequenced germplasm into public repositories will ensure that researchers are able to access and utilize these accessions in their own research to make the field more standardized and equitable. WeedPedia allows users of all backgrounds to quickly access information of interest such as herbicide target site gene sequence or subcellular localization of protein products for different genes. Users can also utilize server-based tools such as BLAST and genome browsing similar to other public genomic databases. Finally, the IWGC is committed to training and connecting weed genomicists through hosting trainings, workshops, and conferences.
Conclusions
Weeds are unique and fascinating plants, having significant impacts on agriculture and ecosystems; and yet, aspects of their biology, ecology, and genetics remain poorly understood. Weeds represent a unique area within plant biology, given their repeated rapid adaptation to sudden and severe shifts in the selective landscape of anthropogenic management practices. The production of a public genomics database with reference genomes and annotations for over 50 weed species represents a substantial step forward towards research goals that improve our understanding of the biology and evolution of weeds. Future work is needed to improve annotations, particularly for complex gene families involved in herbicide detoxification, structural variants, and mobile genetic elements. As reference genome assemblies become available; standard, affordable methods for gathering genotype information will allow for the identification of genetic variants underlying traits of interest. Further, methods for functional validation and hypothesis testing are needed in weeds to validate the effect of genetic variants detected through such experiments, including systems for transformation, gene editing, and transient gene silencing and expression. Future research should focus on utilizing weed genomes to investigate questions about evolutionary biology, ecology, genetics of weedy traits, and weed population dynamics. The IWGC plans to continue the public–private partnership model to host the WeedPedia database over time, integrate new datasets such as genome resequencing and transcriptomes, conduct trainings, and serve as a research coordination network to ensure that advances in weed science from around the world are shared across the research community (Fig. 1 ). Bridging basic plant genomics with translational applications in weeds is needed to deliver on the potential of weed genomics to improve weed management and crop breeding.
Availability of data and materials
All genome assemblies and related sequencing data produced by the IWGC will be available through NCBI as part of publications reporting the first genome-wide analysis for each species.
Gianessi LP, Nathan PR. The value of herbicides in U.S. crop production. Weed Technol. 2007;21(2):559–66.
Article Google Scholar
Pimentel D, Lach L, Zuniga R, Morrison D. Environmental and economic costs of nonindigenous species in the United States. Bioscience. 2000;50(1):53–65.
Barrett SH. Crop mimicry in weeds. Econ Bot. 1983;37(3):255–82.
Powles SB, Yu Q. Evolution in action: plants resistant to herbicides. Annu Rev Plant Biol. 2010;61:317–47.
Article CAS PubMed Google Scholar
Thurber CS, Reagon M, Gross BL, Olsen KM, Jia Y, Caicedo AL. Molecular evolution of shattering loci in U.S. weedy rice. Mol Ecol. 2010;19(16):3271–84.
Article PubMed PubMed Central Google Scholar
Comont D, Lowe C, Hull R, Crook L, Hicks HL, Onkokesung N, et al. Evolution of generalist resistance to herbicide mixtures reveals a trade-off in resistance management. Nat Commun. 2020;11(1):3086.
Article CAS PubMed PubMed Central Google Scholar
Ashworth MB, Walsh MJ, Flower KC, Vila-Aiub MM, Powles SB. Directional selection for flowering time leads to adaptive evolution in Raphanus raphanistrum (wild radish). Evol Appl. 2016;9(4):619–29.
Chan EK, Rowe HC, Kliebenstein DJ. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics. 2010;185(3):991–1007.
Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94.
Harkess A, Zhou J, Xu C, Bowers JE, Van der Hulst R, Ayyampalayam S, et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun. 2017;8(1):1279.
Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L, et al. The gene Sr33 , an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science. 2013;341(6147):786–8.
Ågren J, Oakley CG, McKay JK, Lovell JT, Schemske DW. Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2013;110(52):21077–82.
Article PubMed Central Google Scholar
Schartl M, Walter RB, Shen Y, Garcia T, Catchen J, Amores A, et al. The genome of the platyfish, Xiphophorus maculatus , provides insights into evolutionary adaptation and several complex traits. Nat Genet. 2013;45(5):567–72.
Ravet K, Patterson EL, Krähmer H, Hamouzová K, Fan L, Jasieniuk M, et al. The power and potential of genomics in weed biology and management. Pest Manag Sci. 2018;74(10):2216–25.
Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou S, Liu J, et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science. 2021;373(6555):655–62.
Liao W-W, Asri M, Ebler J, Doerr D, Haukness M, Hickey G, et al. A draft human pangenome reference. Nature. 2023;617(7960):312–24.
Huang Y, Wu D, Huang Z, Li X, Merotto A, Bai L, et al. Weed genomics: yielding insights into the genetics of weedy traits for crop improvement. aBIOTECH. 2023;4:20–30.
Chen K, Yang H, Wu D, Peng Y, Lian L, Bai L, et al. Weed biology and management in the multi-omics era: progress and perspectives. Plant Commun. 2024;5(4):100816.
De Wet JMJ, Harlan JR. Weeds and domesticates: evolution in the man-made habitat. Econ Bot. 1975;29(2):99–108.
Mahaut L, Cheptou PO, Fried G, Munoz F, Storkey J, Vasseur F, et al. Weeds: against the rules? Trends Plant Sci. 2020;25(11):1107–16.
Neve P, Vila-Aiub M, Roux F. Evolutionary-thinking in agricultural weed management. New Phytol. 2009;184(4):783–93.
Article PubMed Google Scholar
Sharma G, Barney JN, Westwood JH, Haak DC. Into the weeds: new insights in plant stress. Trends Plant Sci. 2021;26(10):1050–60.
Vigueira CC, Olsen KM, Caicedo AL. The red queen in the corn: agricultural weeds as models of rapid adaptive evolution. Heredity (Edinb). 2013;110(4):303–11.
Donohue K, Dorn L, Griffith C, Kim E, Aguilera A, Polisetty CR, et al. Niche construction through germination cueing: life-history responses to timing of germination in Arabidopsis thaliana . Evolution. 2005;59(4):771–85.
PubMed Google Scholar
Exposito-Alonso M. Seasonal timing adaptation across the geographic range of Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2020;117(18):9665–7.
Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. A map of local adaptation in Arabidopsis thaliana . Science. 2011;334(6052):86–9.
Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334(6052):83–6.
Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature. 2000;408(6814):796–815.
Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, et al. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana . Cell. 2016;166(2):481–91.
Durvasula A, Fulgione A, Gutaker RM, Alacakaptan SI, Flood PJ, Neto C, et al. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2017;114(20):5213–8.
Frachon L, Mayjonade B, Bartoli C, Hautekèete N-C, Roux F. Adaptation to plant communities across the genome of Arabidopsis thaliana . Mol Biol Evol. 2019;36(7):1442–56.
Fulgione A, Koornneef M, Roux F, Hermisson J, Hancock AM. Madeiran Arabidopsis thaliana reveals ancient long-range colonization and clarifies demography in Eurasia. Mol Biol Evol. 2018;35(3):564–74.
Fulgione A, Neto C, Elfarargi AF, Tergemina E, Ansari S, Göktay M, et al. Parallel reduction in flowering time from de novo mutations enable evolutionary rescue in colonizing lineages. Nat Commun. 2022;13(1):1461.
Kasulin L, Rowan BA, León RJC, Schuenemann VJ, Weigel D, Botto JF. A single haplotype hyposensitive to light and requiring strong vernalization dominates Arabidopsis thaliana populations in Patagonia. Argentina Mol Ecol. 2017;26(13):3389–404.
Picó FX, Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics. 2008;180(2):1009–21.
Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465(7298):627–31.
Flood PJ, Kruijer W, Schnabel SK, van der Schoor R, Jalink H, Snel JFH, et al. Phenomics for photosynthesis, growth and reflectance in Arabidopsis thaliana reveals circadian and long-term fluctuations in heritability. Plant Methods. 2016;12(1):14.
Marchadier E, Hanemian M, Tisné S, Bach L, Bazakos C, Gilbault E, et al. The complex genetic architecture of shoot growth natural variation in Arabidopsis thaliana . PLoS Genet. 2019;15(4):e1007954.
Tisné S, Serrand Y, Bach L, Gilbault E, Ben Ameur R, Balasse H, et al. Phenoscope: an automated large-scale phenotyping platform offering high spatial homogeneity. Plant J. 2013;74(3):534–44.
Tschiersch H, Junker A, Meyer RC, Altmann T. Establishment of integrated protocols for automated high throughput kinetic chlorophyll fluorescence analyses. Plant Methods. 2017;13:54.
Chen X, MacGregor DR, Stefanato FL, Zhang N, Barros-Galvão T, Penfield S. A VEL3 histone deacetylase complex establishes a maternal epigenetic state controlling progeny seed dormancy. Nat Commun. 2023;14(1):2220.
Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–101.
Davey JW, Blaxter ML. RADSeq: next-generation population genetics. Brief Funct Genomics. 2010;9(5–6):416–23.
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6(5):e19379.
MacGregor DR. What makes a weed a weed? How virus-mediated reverse genetics can help to explore the genetics of weediness. Outlooks Pest Manag. 2020;31(5):224–9.
Mellado-Sánchez M, McDiarmid F, Cardoso V, Kanyuka K, MacGregor DR. Virus-mediated transient expression techniques enable gene function studies in blackgrass. Plant Physiol. 2020;183(2):455–9.
Dimaano NG, Yamaguchi T, Fukunishi K, Tominaga T, Iwakami S. Functional characterization of Cytochrome P450 CYP81A subfamily to disclose the pattern of cross-resistance in Echinochloa phyllopogon . Plant Mol Biol. 2020;102(4–5):403–16.
de Figueiredo MRA, Küpper A, Malone JM, Petrovic T, de Figueiredo ABTB, Campagnola G, et al. An in-frame deletion mutation in the degron tail of auxin coreceptor IAA2 confers resistance to the herbicide 2,4-D in Sisymbrium orientale . Proc Natl Acad Sci U S A. 2022;119(9):e2105819119.
Patzoldt WL, Hager AG, McCormick JS, Tranel PJ. A codon deletion confers resistance to herbicides inhibiting protoporphyrinogen oxidase. Proc Natl Acad Sci U S A. 2006;103(33):12329–34.
Zabala-Pardo D, Gaines T, Lamego FP, Avila LA. RNAi as a tool for weed management: challenges and opportunities. Adv Weed Sci. 2022;40(spe1):e020220096.
Fattorini R, Glover BJ. Molecular mechanisms of pollination biology. Annu Rev Plant Biol. 2020;71:487–515.
Rollin O, Benelli G, Benvenuti S, Decourtye A, Wratten SD, Canale A, et al. Weed-insect pollinator networks as bio-indicators of ecological sustainability in agriculture. A review Agron Sustain Dev. 2016;36(1):8.
Irwin RE, Strauss SY. Flower color microevolution in wild radish: evolutionary response to pollinator-mediated selection. Am Nat. 2005;165(2):225–37.
Ma B, Wu J, Shi T-L, Yang Y-Y, Wang W-B, Zheng Y, et al. Lilac ( Syringa oblata ) genome provides insights into its evolution and molecular mechanism of petal color change. Commun Biol. 2022;5(1):686.
Xing A, Wang X, Nazir MF, Zhang X, Wang X, Yang R, et al. Transcriptomic and metabolomic profiling of flavonoid biosynthesis provides novel insights into petals coloration in Asian cotton ( Gossypium arboreum L.). BMC Plant Biol. 2022;22(1):416.
Zheng Y, Chen Y, Liu Z, Wu H, Jiao F, Xin H, et al. Important roles of key genes and transcription factors in flower color differences of Nicotiana alata . Genes (Basel). 2021;12(12):1976.
Krizek BA, Anderson JT. Control of flower size. J Exp Bot. 2013;64(6):1427–37.
Powell AE, Lenhard M. Control of organ size in plants. Curr Biol. 2012;22(9):R360–7.
Spencer V, Kim M. Re"CYC"ling molecular regulators in the evolution and development of flower symmetry. Semin Cell Dev Biol. 2018;79:16–26.
Amrad A, Moser M, Mandel T, de Vries M, Schuurink RC, Freitas L, et al. Gain and loss of floral scent production through changes in structural genes during pollinator-mediated speciation. Curr Biol. 2016;26(24):3303–12.
Delle-Vedove R, Schatz B, Dufay M. Understanding intraspecific variation of floral scent in light of evolutionary ecology. Ann Bot. 2017;120(1):1–20.
Pichersky E, Gershenzon J. The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol. 2002;5(3):237–43.
Ballerini ES, Kramer EM, Hodges SA. Comparative transcriptomics of early petal development across four diverse species of Aquilegia reveal few genes consistently associated with nectar spur development. BMC Genom. 2019;20(1):668.
Corbet SA, Willmer PG, Beament JWL, Unwin DM, Prys-Jones OE. Post-secretory determinants of sugar concentration in nectar. Plant Cell Environ. 1979;2(4):293–308.
Galliot C, Hoballah ME, Kuhlemeier C, Stuurman J. Genetics of flower size and nectar volume in Petunia pollination syndromes. Planta. 2006;225(1):203–12.
Vila-Aiub MM, Neve P, Powles SB. Fitness costs associated with evolved herbicide resistance alleles in plants. New Phytol. 2009;184(4):751–67.
Baucom RS. Evolutionary and ecological insights from herbicide-resistant weeds: what have we learned about plant adaptation, and what is left to uncover? New Phytol. 2019;223(1):68–82.
Bajwa AA, Latif S, Borger C, Iqbal N, Asaduzzaman M, Wu H, et al. The remarkable journey of a weed: biology and management of annual ryegrass ( Lolium rigidum ) in conservation cropping systems of Australia. Plants (Basel). 2021;10(8):1505.
Bitarafan Z, Andreasen C. Fecundity allocation in some european weed species competing with crops. Agronomy. 2022;12(5):1196.
Costea M, Weaver SE, Tardif FJ. The biology of Canadian weeds. 130. Amaranthus retroflexus L., A. powellii , A. powellii S. Watson, and A. hybridus L. Can J Plant Sci. 2004;84(2):631–68.
Dixon A, Comont D, Slavov GT, Neve P. Population genomics of selectively neutral genetic structure and herbicide resistance in UK populations of Alopecurus myosuroides . Pest Manag Sci. 2021;77(3):1520–9.
Kersten S, Chang J, Huber CD, Voichek Y, Lanz C, Hagmaier T, et al. Standing genetic variation fuels rapid evolution of herbicide resistance in blackgrass. Proc Natl Acad Sci U S A. 2023;120(16):e2206808120.
Qiu J, Zhou Y, Mao L, Ye C, Wang W, Zhang J, et al. Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat Commun. 2017;8(1):15323.
Kreiner JM, Caballero A, Wright SI, Stinchcombe JR. Selective ancestral sorting and de novo evolution in the agricultural invasion of Amaranthus tuberculatus . Evolution. 2022;76(1):70–85.
Kreiner JM, Latorre SM, Burbano HA, Stinchcombe JR, Otto SP, Weigel D, et al. Rapid weed adaptation and range expansion in response to agriculture over the past two centuries. Science. 2022;378(6624):1079–85.
Wu D, Shen E, Jiang B, Feng Y, Tang W, Lao S, et al. Genomic insights into the evolution of Echinochloa species as weed and orphan crop. Nat Commun. 2022;13(1):689.
Yeaman S, Hodgins KA, Lotterhos KE, Suren H, Nadeau S, Degner JC, et al. Convergent local adaptation to climate in distantly related conifers. Science. 2016;353(6306):1431–3.
Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 2013;45(8):891–8.
Sackton TB, Grayson P, Cloutier A, Hu Z, Liu JS, Wheeler NE, et al. Convergent regulatory evolution and loss of flight in paleognathous birds. Science. 2019;364(6435):74–8.
Ye CY, Fan L. Orphan crops and their wild relatives in the genomic era. Mol Plant. 2021;14(1):27–39.
Clements DR, Jones VL. Ten ways that weed evolution defies human management efforts amidst a changing climate. Agronomy. 2021;11(2):284.
Article CAS Google Scholar
Weinig C. Rapid evolutionary responses to selection in heterogeneous environments among agricultural and nonagricultural weeds. Int J Plant Sci. 2005;166(4):641–7.
Cousens RD, Fournier-Level A. Herbicide resistance costs: what are we actually measuring and why? Pest Manag Sci. 2018;74(7):1539–46.
Lasky JR, Josephs EB, Morris GP. Genotype–environment associations to reveal the molecular basis of environmental adaptation. Plant Cell. 2023;35(1):125–38.
Lotterhos KE. The effect of neutral recombination variation on genome scans for selection. G3-Genes Genom Genet. 2019;9(6):1851–67.
Lovell JT, MacQueen AH, Mamidi S, Bonnette J, Jenkins J, Napier JD, et al. Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature. 2021;590(7846):438–44.
Todesco M, Owens GL, Bercovich N, Légaré J-S, Soudi S, Burge DO, et al. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature. 2020;584(7822):602–7.
Revolinski SR, Maughan PJ, Coleman CE, Burke IC. Preadapted to adapt: Underpinnings of adaptive plasticity revealed by the downy brome genome. Commun Biol. 2023;6(1):326.
Kuester A, Conner JK, Culley T, Baucom RS. How weeds emerge: a taxonomic and trait-based examination using United States data. New Phytol. 2014;202(3):1055–68.
Arnaud JF, Fénart S, Cordellier M, Cuguen J. Populations of weedy crop-wild hybrid beets show contrasting variation in mating system and population genetic structure. Evol Appl. 2010;3(3):305–18.
Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci U S A. 2000;97(13):7043–50.
Nakabayashi K, Leubner-Metzger G. Seed dormancy and weed emergence: from simulating environmental change to understanding trait plasticity, adaptive evolution, and population fitness. J Exp Bot. 2021;72(12):4181–5.
Busi R, Yu Q, Barrett-Lennard R, Powles S. Long distance pollen-mediated flow of herbicide resistance genes in Lolium rigidum . Theor Appl Genet. 2008;117(8):1281–90.
Délye C, Clément JAJ, Pernin F, Chauvel B, Le Corre V. High gene flow promotes the genetic homogeneity of arable weed populations at the landscape level. Basic Appl Ecol. 2010;11(6):504–12.
Roumet M, Noilhan C, Latreille M, David J, Muller MH. How to escape from crop-to-weed gene flow: phenological variation and isolation-by-time within weedy sunflower populations. New Phytol. 2013;197(2):642–54.
Moghadam SH, Alebrahim MT, Mohebodini M, MacGregor DR. Genetic variation of Amaranthus retroflexus L. and Chenopodium album L. (Amaranthaceae) suggests multiple independent introductions into Iran. Front Plant Sci. 2023;13:1024555.
Muller M-H, Latreille M, Tollon C. The origin and evolution of a recent agricultural weed: population genetic diversity of weedy populations of sunflower ( Helianthus annuus L.) in Spain and France. Evol Appl. 2011;4(3):499–514.
Wesse C, Welk E, Hurka H, Neuffer B. Geographical pattern of genetic diversity in Capsella bursa-pastoris (Brassicaceae) -A global perspective. Ecol Evol. 2021;11(1):199–213.
Fraimout A, Debat V, Fellous S, Hufbauer RA, Foucaud J, Pudlo P, et al. Deciphering the routes of invasion of Drosophila suzukii by means of ABC random forest. Mol Biol Evol. 2017;34(4):980–96.
CAS PubMed PubMed Central Google Scholar
Battlay P, Wilson J, Bieker VC, Lee C, Prapas D, Petersen B, et al. Large haploblocks underlie rapid adaptation in the invasive weed Ambrosia artemisiifolia . Nat Commun. 2023;14(1):1717.
van Boheemen LA, Hodgins KA. Rapid repeatable phenotypic and genomic adaptation following multiple introductions. Mol Ecol. 2020;29(21):4102–17.
Putra A, Hodgins K, Fournier-Level A. Assessing the invasive potential of different source populations of ragweed ( Ambrosia artemisiifolia L.) through genomically-informed species distribution modelling. Authorea. 2023;17(1):e13632.
Google Scholar
Bourguet D, Delmotte F, Franck P, Guillemaud T, Reboud X, Vacher C, et al. Heterogeneity of selection and the evolution of resistance. Trends Ecol Evol. 2013;28(2):110–8.
The International Herbicide-Resistant Weed Database. www.weedscience.org . Accessed 20 June 2023.
Powles S. Herbicide discovery through innovation and diversity. Adv Weed Sci. 2022;40(spe1):e020220074.
Murphy BP, Tranel PJ. Target-site mutations conferring herbicide resistance. Plants (Basel). 2019;8(10):382.
Gaines TA, Duke SO, Morran S, Rigon CAG, Tranel PJ, Küpper A, et al. Mechanisms of evolved herbicide resistance. J Biol Chem. 2020;295(30):10307–30.
Lonhienne T, Cheng Y, Garcia MD, Hu SH, Low YS, Schenk G, et al. Structural basis of resistance to herbicides that target acetohydroxyacid synthase. Nat Commun. 2022;13(1):3368.
Comont D, MacGregor DR, Crook L, Hull R, Nguyen L, Freckleton RP, et al. Dissecting weed adaptation: fitness and trait correlations in herbicide-resistant Alopecurus myosuroides . Pest Manag Sci. 2022;78(7):3039–50.
Neve P. Simulation modelling to understand the evolution and management of glyphosate resistance in weeds. Pest Manag Sci. 2008;64(4):392–401.
Torra J, Alcántara-de la Cruz R. Molecular mechanisms of herbicide resistance in weeds. Genes (Basel). 2022;13(11):2025.
Délye C, Gardin JAC, Boucansaud K, Chauvel B, Petit C. Non-target-site-based resistance should be the centre of attention for herbicide resistance research: Alopecurus myosuroides as an illustration. Weed Res. 2011;51(5):433–7.
Chandra S, Leon RG. Genome-wide evolutionary analysis of putative non-specific herbicide resistance genes and compilation of core promoters between monocots and dicots. Genes (Basel). 2022;13(7):1171.
Margaritopoulou T, Tani E, Chachalis D, Travlos I. Involvement of epigenetic mechanisms in herbicide resistance: the case of Conyza canadensis . Agriculture. 2018;8(1):17.
Pan L, Guo Q, Wang J, Shi L, Yang X, Zhou Y, et al. CYP81A68 confers metabolic resistance to ALS and ACCase-inhibiting herbicides and its epigenetic regulation in Echinochloa crus-galli . J Hazard Mater. 2022;428:128225.
Sen MK, Hamouzová K, Košnarová P, Roy A, Soukup J. Herbicide resistance in grass weeds: Epigenetic regulation matters too. Front Plant Sci. 2022;13:1040958.
Han H, Yu Q, Beffa R, González S, Maiwald F, Wang J, et al. Cytochrome P450 CYP81A10v7 in Lolium rigidum confers metabolic resistance to herbicides across at least five modes of action. Plant J. 2021;105(1):79–92.
Kubis GC, Marques RZ, Kitamura RS, Barroso AA, Juneau P, Gomes MP. Antioxidant enzyme and Cytochrome P450 activities are involved in horseweed ( Conyza sumatrensis ) resistance to glyphosate. Stress. 2023;3(1):47–57.
Qiao Y, Zhang N, Liu J, Yang H. Interpretation of ametryn biodegradation in rice based on joint analyses of transcriptome, metabolome and chemo-characterization. J Hazard Mater. 2023;445:130526.
Rouse CE, Roma-Burgos N, Barbosa Martins BA. Physiological assessment of non–target site restistance in multiple-resistant junglerice ( Echinochloa colona ). Weed Sci. 2019;67(6):622–32.
Abou-Khater L, Maalouf F, Jighly A, Alsamman AM, Rubiales D, Rispail N, et al. Genomic regions associated with herbicide tolerance in a worldwide faba bean ( Vicia faba L.) collection. Sci Rep. 2022;12(1):158.
Gupta S, Harkess A, Soble A, Van Etten M, Leebens-Mack J, Baucom RS. Interchromosomal linkage disequilibrium and linked fitness cost loci associated with selection for herbicide resistance. New Phytol. 2023;238(3):1263–77.
Kreiner JM, Tranel PJ, Weigel D, Stinchcombe JR, Wright SI. The genetic architecture and population genomic signatures of glyphosate resistance in Amaranthus tuberculatus . Mol Ecol. 2021;30(21):5373–89.
Parcharidou E, Dücker R, Zöllner P, Ries S, Orru R, Beffa R. Recombinant glutathione transferases from flufenacet-resistant black-grass ( Alopecurus myosuroides Huds.) form different flufenacet metabolites and differ in their interaction with pre- and post-emergence herbicides. Pest Manag Sci. 2023;79(9):3376–86.
Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21(3):181–200.
Acuner Ozbabacan SE, Engin HB, Gursoy A, Keskin O. Transient protein-protein interactions. Protein Eng Des Sel. 2011;24(9):635–48.
Lu H, Zhou Q, He J, Jiang Z, Peng C, Tong R, et al. Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials. Signal Transduct Target Ther. 2020;5(1):213.
Benson CW, Sheltra MR, Maughan PJ, Jellen EN, Robbins MD, Bushman BS, et al. Homoeologous evolution of the allotetraploid genome of Poa annua L. BMC Genom. 2023;24(1):350.
Robbins MD, Bushman BS, Huff DR, Benson CW, Warnke SE, Maughan CA, et al. Chromosome-scale genome assembly and annotation of allotetraploid annual bluegrass ( Poa annua L.). Genome Biol Evol. 2022;15(1):evac180.
Montgomery JS, Giacomini D, Waithaka B, Lanz C, Murphy BP, Campe R, et al. Draft genomes of Amaranthus tuberculatus , Amaranthus hybridus and Amaranthus palmeri . Genome Biol Evol. 2020;12(11):1988–93.
Jeschke MR, Tranel PJ, Rayburn AL. DNA content analysis of smooth pigweed ( Amaranthus hybridus ) and tall waterhemp ( A. tuberculatus ): implications for hybrid detection. Weed Sci. 2003;51(1):1–3.
Rayburn AL, McCloskey R, Tatum TC, Bollero GA, Jeschke MR, Tranel PJ. Genome size analysis of weedy Amaranthus species. Crop Sci. 2005;45(6):2557–62.
Laforest M, Martin SL, Bisaillon K, Soufiane B, Meloche S, Tardif FJ, et al. The ancestral karyotype of the Heliantheae Alliance, herbicide resistance, and human allergens: Insights from the genomes of common and giant ragweed. Plant Genome . 2024;e20442. https://doi.org/10.1002/tpg2.20442 .
Mulligan GA. Chromosome numbers of Canadian weeds. I Canad J Bot. 1957;35(5):779–89.
Meyer L, Causse R, Pernin F, Scalone R, Bailly G, Chauvel B, et al. New gSSR and EST-SSR markers reveal high genetic diversity in the invasive plant Ambrosia artemisiifolia L. and can be transferred to other invasive Ambrosia species. PLoS One. 2017;12(5):e0176197.
Pustahija F, Brown SC, Bogunić F, Bašić N, Muratović E, Ollier S, et al. Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa. Plant Soil. 2013;373(1):427–53.
Kubešová M, Moravcova L, Suda J, Jarošík V, Pyšek P. Naturalized plants have smaller genomes than their non-invading relatives: a flow cytometric analysis of the Czech alien flora. Preslia. 2010;82(1):81–96.
Thébaud C, Abbott RJ. Characterization of invasive Conyza species (Asteraceae) in Europe: quantitative trait and isozyme analysis. Am J Bot. 1995;82(3):360–8.
Garcia S, Hidalgo O, Jakovljević I, Siljak-Yakovlev S, Vigo J, Garnatje T, et al. New data on genome size in 128 Asteraceae species and subspecies, with first assessments for 40 genera, 3 tribes and 2 subfamilies. Plant Biosyst. 2013;147(4):1219–27.
Zhao X, Yi L, Ren Y, Li J, Ren W, Hou Z, et al. Chromosome-scale genome assembly of the yellow nutsedge ( Cyperus esculentus ). Genome Biol Evol. 2023;15(3):evad027.
Bennett MD, Leitch IJ, Hanson L. DNA amounts in two samples of angiosperm weeds. Ann Bot. 1998;82:121–34.
Schulz-Schaeffer J, Gerhardt S. Cytotaxonomic analysis of the Euphorbia spp. (leafy spurge) complex. II: Comparative study of the chromosome morphology. Biol Zentralbl. 1989;108(1):69–76.
Schaeffer JR, Gerhardt S. The impact of introgressive hybridization on the weediness of leafy spurge. Leafy Spurge Symposium. 1989;1989:97–105.
Bai C, Alverson WS, Follansbee A, Waller DM. New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann Bot. 2012;110(8):1623–9.
Aarestrup JR, Karam D, Fernandes GW. Chromosome number and cytogenetics of Euphorbia heterophylla L. Genet Mol Res. 2008;7(1):217–22.
Wang L, Sun X, Peng Y, Chen K, Wu S, Guo Y, et al. Genomic insights into the origin, adaptive evolution, and herbicide resistance of Leptochloa chinensis , a devastating tetraploid weedy grass in rice fields. Mol Plant. 2022;15(6):1045–58.
Paril J, Pandey G, Barnett EM, Rane RV, Court L, Walsh T, et al. Rounding up the annual ryegrass genome: high-quality reference genome of Lolium rigidum . Front Genet. 2022;13:1012694.
Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM. Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am J Bot. 2006;93(1):148–56.
Towers G, Mitchell J, Rodriguez E, Bennett F, Subba Rao P. Biology & chemistry of Parthenium hysterophorus L., a problem weed in India. Biol Rev. 1977;48:65–74.
CAS Google Scholar
Moghe GD, Hufnagel DE, Tang H, Xiao Y, Dworkin I, Town CD, et al. Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish ( Raphanus raphanistrum ) and three other Brassicaceae species. Plant Cell. 2014;26(5):1925–37.
Zhang X, Liu T, Wang J, Wang P, Qiu Y, Zhao W, et al. Pan-genome of Raphanus highlights genetic variation and introgression among domesticated, wild, and weedy radishes. Mol Plant. 2021;14(12):2032–55.
Chytrý M, Danihelka J, Kaplan Z, Wild J, Holubová D, Novotný P, et al. Pladias database of the Czech flora and vegetation. Preslia. 2021;93(1):1–87.
Patterson EL, Pettinga DJ, Ravet K, Neve P, Gaines TA. Glyphosate resistance and EPSPS gene duplication: Convergent evolution in multiple plant species. J Hered. 2018;109(2):117–25.
Jugulam M, Niehues K, Godar AS, Koo DH, Danilova T, Friebe B, et al. Tandem amplification of a chromosomal segment harboring 5-enolpyruvylshikimate-3-phosphate synthase locus confers glyphosate resistance in Kochia scoparia . Plant Physiol. 2014;166(3):1200–7.
Patterson EL, Saski CA, Sloan DB, Tranel PJ, Westra P, Gaines TA. The draft genome of Kochia scoparia and the mechanism of glyphosate resistance via transposon-mediated EPSPS tandem gene duplication. Genome Biol Evol. 2019;11(10):2927–40.
Zhang C, Johnson N, Hall N, Tian X, Yu Q, Patterson E. Subtelomeric 5-enolpyruvylshikimate-3-phosphate synthase ( EPSPS ) copy number variation confers glyphosate resistance in Eleusine indica . Nat Commun. 2023;14:4865.
Koo D-H, Molin WT, Saski CA, Jiang J, Putta K, Jugulam M, et al. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri . Proc Natl Acad Sci U S A. 2018;115(13):3332–7.
Molin WT, Yaguchi A, Blenner M, Saski CA. The eccDNA Replicon: A heritable, extranuclear vehicle that enables gene amplification and glyphosate resistance in Amaranthus palmeri . Plant Cell. 2020;32(7):2132–40.
Jugulam M. Can non-Mendelian inheritance of extrachromosomal circular DNA-mediated EPSPS gene amplification provide an opportunity to reverse resistance to glyphosate? Weed Res. 2021;61(2):100–5.
Kreiner JM, Giacomini DA, Bemm F, Waithaka B, Regalado J, Lanz C, et al. Multiple modes of convergent adaptation in the spread of glyphosate-resistant Amaranthus tuberculatus . Proc Natl Acad Sci U S A. 2019;116(42):21076–84.
Cai L, Comont D, MacGregor D, Lowe C, Beffa R, Neve P, et al. The blackgrass genome reveals patterns of non-parallel evolution of polygenic herbicide resistance. New Phytol. 2023;237(5):1891–907.
Chen K, Yang H, Peng Y, Liu D, Zhang J, Zhao Z, et al. Genomic analyses provide insights into the polyploidization-driven herbicide adaptation in Leptochloa weeds. Plant Biotechnol J. 2023;21(8):1642–58.
Ohadi S, Hodnett G, Rooney W, Bagavathiannan M. Gene flow and its consequences in Sorghum spp. Crit Rev Plant Sci. 2017;36(5–6):367–85.
Renzi JP, Coyne CJ, Berger J, von Wettberg E, Nelson M, Ureta S, et al. How could the use of crop wild relatives in breeding increase the adaptation of crops to marginal environments? Front Plant Sci. 2022;13:886162.
Ward SM, Cousens RD, Bagavathiannan MV, Barney JN, Beckie HJ, Busi R, et al. Agricultural weed research: a critique and two proposals. Weed Sci. 2014;62(4):672–8.
Evans JA, Tranel PJ, Hager AG, Schutte B, Wu C, Chatham LA, et al. Managing the evolution of herbicide resistance. Pest Manag Sci. 2016;72(1):74–80.
International Weed Genomics Consortium Website. https://www.weedgenomics.org . Accessed 20 June 2023.
WeedPedia Database. https://weedpedia.weedgenomics.org/ . Accessed 20 June 2023.
Hall N, Chen J, Matzrafi M, Saski CA, Westra P, Gaines TA, et al. FHY3/FAR1 transposable elements generate adaptive genetic variation in the Bassia scoparia genome. bioRxiv . 2023; DOI: https://doi.org/10.1101/2023.05.26.542497 .
Jarvis DE, Sproul JS, Navarro-Domínguez B, Krak K, Jaggi K, Huang Y-F, et al. Chromosome-scale genome assembly of the hexaploid Taiwanese goosefoot “Djulis” ( Chenopodium formosanum ). Genome Biol Evol. 2022;14(8):evac120.
Ferreira LAI, de Oliveira RS, Jr., Constantin J, Brunharo C. Evolution of ACCase-inhibitor resistance in Chloris virgata is conferred by a Trp2027Cys mutation in the herbicide target site. Pest Manag Sci. 2023;79(12):5220–9.
Laforest M, Martin SL, Bisaillon K, Soufiane B, Meloche S, Page E. A chromosome-scale draft sequence of the Canada fleabane genome. Pest Manag Sci. 2020;76(6):2158–69.
Guo L, Qiu J, Ye C, Jin G, Mao L, Zhang H, et al. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat Commun. 2017;8(1):1031.
Sato MP, Iwakami S, Fukunishi K, Sugiura K, Yasuda K, Isobe S, et al. Telomere-to-telomere genome assembly of an allotetraploid pernicious weed, Echinochloa phyllopogon . DNA Res. 2023;30(5):dsad023.
Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza . Nat Genet. 2018;50(2):285–96.
Wu D, Xie L, Sun Y, Huang Y, Jia L, Dong C, et al. A syntelog-based pan-genome provides insights into rice domestication and de-domestication. Genome Biol. 2023;24(1):179.
Wang Z, Huang S, Yang Z, Lai J, Gao X, Shi J. A high-quality, phased genome assembly of broomcorn millet reveals the features of its subgenome evolution and 3D chromatin organization. Plant Commun. 2023;4(3):100557.
Mao Q, Huff DR. The evolutionary origin of Poa annua L. Crop Sci. 2012;52(4):1910–22.
Benson CW, Sheltra MR, Maughan JP, Jellen EN, Robbins MD, Bushman BS, et al. Homoeologous evolution of the allotetraploid genome of Poa annua L. Res Sq. 2023. https://doi.org/10.21203/rs.3.rs-2729084/v1 .
Brunharo C, Benson CW, Huff DR, Lasky JR. Chromosome-scale genome assembly of Poa trivialis and population genomics reveal widespread gene flow in a cool-season grass seed production system. Plant Direct. 2024;8(3):e575.
Mo C, Wu Z, Shang X, Shi P, Wei M, Wang H, et al. Chromosome-level and graphic genomes provide insights into metabolism of bioactive metabolites and cold-adaption of Pueraria lobata var. montana . DNA Research. 2022;29(5):dsac030.
Thielen PM, Pendleton AL, Player RA, Bowden KV, Lawton TJ, Wisecaver JH. Reference genome for the highly transformable Setaria viridis ME034V. G3 (Bethesda, Md). 2020;10(10):3467–78.
Yoshida S, Kim S, Wafula EK, Tanskanen J, Kim Y-M, Honaas L, et al. Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Curr Biol. 2019;29(18):3041–52.
Qiu S, Bradley JM, Zhang P, Chaudhuri R, Blaxter M, Butlin RK, et al. Genome-enabled discovery of candidate virulence loci in Striga hermonthica , a devastating parasite of African cereal crops. New Phytol. 2022;236(2):622–38.
Nunn A, Rodríguez-Arévalo I, Tandukar Z, Frels K, Contreras-Garrido A, Carbonell-Bejerano P, et al. Chromosome-level Thlaspi arvense genome provides new tools for translational research and for a newly domesticated cash cover crop of the cooler climates. Plant Biotechnol J. 2022;20(5):944–63.
USDA-ARS Germplasm Resources Information Network (GRIN). https://www.ars-grin.gov/ . Accessed 20 June 2023.
Buck M, Hamilton C. The Nagoya Protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their utilization to the convention on biological diversity. RECIEL. 2011;20(1):47–61.
Chauhan BS, Matloob A, Mahajan G, Aslam F, Florentine SK, Jha P. Emerging challenges and opportunities for education and research in weed science. Front Plant Sci. 2017;8:1537.
Shah S, Lonhienne T, Murray CE, Chen Y, Dougan KE, Low YS, et al. Genome-guided analysis of seven weed species reveals conserved sequence and structural features of key gene targets for herbicide development. Front Plant Sci. 2022;13:909073.
International Weed Genomics Consortium Training Resources. https://www.weedgenomics.org/training-resources/ . Accessed 20 June 2023.
Blackford S. Harnessing the power of communities: career networking strategies for bioscience PhD students and postdoctoral researchers. FEMS Microbiol Lett. 2018;365(8):fny033.
Pender M, Marcotte DE, Sto Domingo MR, Maton KI. The STEM pipeline: The role of summer research experience in minority students’ Ph.D. aspirations. Educ Policy Anal Arch. 2010;18(30):1–36.
PubMed PubMed Central Google Scholar
Burke A, Okrent A, Hale K. The state of U.S. science and engineering 2022. Foundation NS. https://ncses.nsf.gov/pubs/nsb20221 . 2022.
Wu J-Y, Liao C-H, Cheng T, Nian M-W. Using data analytics to investigate attendees’ behaviors and psychological states in a virtual academic conference. Educ Technol Soc. 2021;24(1):75–91.
Download references
Peer review information
Wenjing She was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
The International Weed Genomics Consortium is supported by BASF SE, Bayer AG, Syngenta Ltd, Corteva Agriscience, CropLife International (Global Herbicide Resistance Action Committee), the Foundation for Food and Agriculture Research (Award DSnew-0000000024), and two conference grants from USDA-NIFA (Award numbers 2021–67013-33570 and 2023-67013-38785).
Author information
Authors and affiliations.
Department of Agricultural Biology, Colorado State University, 1177 Campus Delivery, Fort Collins, CO, 80523, USA
Jacob Montgomery, Sarah Morran & Todd A. Gaines
Protecting Crops and the Environment, Rothamsted Research, Harpenden, Hertfordshire, UK
Dana R. MacGregor
Department of Crop, Soil, and Environmental Sciences, Auburn University, Auburn, AL, USA
J. Scott McElroy
Department of Plant and Environmental Sciences, University of Copenhagen, Taastrup, Denmark
Paul Neve & Célia Neto
IFEVA-Conicet-Department of Ecology, University of Buenos Aires, Buenos Aires, Argentina
Martin M. Vila-Aiub & Maria Victoria Sandoval
Department of Ecology, Faculty of Agronomy, University of Buenos Aires, Buenos Aires, Argentina
Analia I. Menéndez
Department of Botany, The University of British Columbia, Vancouver, BC, Canada
Julia M. Kreiner
Institute of Crop Sciences, Zhejiang University, Hangzhou, China
Longjiang Fan
Department of Biology, University of Massachusetts Amherst, Amherst, MA, USA
Ana L. Caicedo
Department of Plant and Wildlife Sciences, Brigham Young University, Provo, UT, USA
Peter J. Maughan
Bayer AG, Weed Control Research, Frankfurt, Germany
Bianca Assis Barbosa Martins, Jagoda Mika, Alberto Collavo & Bodo Peters
Department of Crop Sciences, Federal University of Rio Grande Do Sul, Porto Alegre, Rio Grande Do Sul, Brazil
Aldo Merotto Jr.
Department of Soil and Crop Sciences, Texas A&M University, College Station, TX, USA
Nithya K. Subramanian & Muthukumar V. Bagavathiannan
Department of Plant, Soil and Microbial Sciences, Michigan State University, East Lansing, MI, USA
Luan Cutti & Eric L. Patterson
Department of Agronomy, Kansas State University, Manhattan, KS, USA
Md. Mazharul Islam & Mithila Jugulam
Department of Plant Pathology, Kansas State University, Manhattan, KS, USA
Bikram S. Gill
Crop Protection Discovery and Development, Corteva Agriscience, Indianapolis, IN, USA
Robert Cicchillo, Roger Gast & Neeta Soni
Genome Center of Excellence, Corteva Agriscience, Johnston, IA, USA
Terry R. Wright, Gina Zastrow-Hayes, Gregory May, Kevin Fengler & Victor Llaca
School of Agriculture, Food and Wine, University of Adelaide, Glen Osmond, South Australia, Australia
Jenna M. Malone
Jealott’s Hill International Research Centre, Syngenta Ltd, Bracknell, Berkshire, UK
Deepmala Sehgal, Shiv Shankhar Kaundun & Richard P. Dale
Department of Plant and Soil Sciences, University of Pretoria, Pretoria, South Africa
Barend Juan Vorster
BASF SE, Ludwigshafen Am Rhein, Germany
Jens Lerchl
Department of Crop Sciences, University of Illinois, Urbana, IL, USA
Patrick J. Tranel
Senior Scientist Consultant, Herbicide Resistance Action Committee / CropLife International, Liederbach, Germany
Roland Beffa
School of BioSciences, University of Melbourne, Parkville, VIC, Australia
Alexandre Fournier-Level
You can also search for this author in PubMed Google Scholar
Contributions
JMo and TG conceived and outlined the article. TG, DM, EP, RB, JSM, PJT, MJ wrote grants to obtain funding. MMI, BSG, and MJ performed mitotic chromosome visualization. VL performed sequencing. VL and KF assembled the genomes. LC and ELP annotated the genomes. JMo, SM, DRM, JSM, PN, CN, MV, MVS, AIM, JMK, LF, ALC, PJM, BABM, JMi, AC, MVB, LC, AFL, and ELP wrote the first draft of the article. All authors edited the article and improved the final version.
Corresponding author
Correspondence to Todd A. Gaines .
Ethics declarations
Ethics approval and consent to participate.
Ethical approval is not applicable for this article.
Competing interests
Some authors work for commercial agricultural companies (BASF, Bayer, Corteva Agriscience, or Syngenta) that develop and sell weed control products.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
13059_2024_3274_moesm1_esm.docx.
Additional file 1. List of completed and in-progress genome assemblies of weed species pollinated by insects (Table S1).
13059_2024_3274_MOESM2_ESM.docx
Additional file 2. Methods and results for visualizing and counting the metaphase chromosomes of hexaploid Avena fatua (Fig S1); diploid Lolium rigidum (Fig S2); tetraploid Phalaris minor (Fig S3); and tetraploid Salsola tragus (Fig S4).
Additional file 3. Review history.
Rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Montgomery, J., Morran, S., MacGregor, D.R. et al. Current status of community resources and priorities for weed genomics research. Genome Biol 25 , 139 (2024). https://doi.org/10.1186/s13059-024-03274-y
Download citation
Received : 11 July 2023
Accepted : 13 May 2024
Published : 27 May 2024
DOI : https://doi.org/10.1186/s13059-024-03274-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Weed science
- Reference genomes
- Rapid adaptation
- Herbicide resistance
- Public resources
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
Definition and Purpose of Abstracts An abstract is a short summary of your (published or unpublished) research paper, usually about a paragraph (c. 6-7 sentences, 150-250 words) long. A well-written abstract serves multiple purposes: an abstract lets readers get the gist or essence of your paper or article quickly, in order to decide whether to…
An abstract is a short summary of a longer work (such as a thesis , dissertation or research paper ). The abstract concisely reports the aims and outcomes of your research, so that readers know exactly what your paper is about.
An abstract is a brief summary, found at the start of scientific papers, that outlines the paper's research and conclusions. It differs from an annotation, which evaluates the paper in addition to providing a summary. An abstract allows readers to understand the key aspects of a paper without reading the whole document.
Research Paper Abstract is a brief summary of a research pape r that describes the study's purpose, methods, findings, and conclusions. It is often the first section of the paper that readers encounter, and its purpose is to provide a concise and accurate overview of the paper's content.
This article deals with formulating a suitable title and an appropriate abstract for an original research paper. The "title" and the "abstract" are the "initial impressions" of a research article, and hence ...
The abstract is the opening section of a scientific paper, which provides a concise summary of the research and conclusions. Scientific papers often follow the IMRaD format (Introduction, Materials and Methods, Results, and Discussion).
In science, an abstract refers to a summary of a scientific research paper or study. It is typically a concise and condensed overview of the key points and findings of the study. The purpose of an abstract is to provide a brief glimpse into the research to help readers determine if the study is relevant to their interests or research.
Reading and understanding abstracts will help you enter the world of academic research, make informed decisions about what research is worth your time and effort to read in full, and prepare you to write your own abstracts. Abstract credit:
An abstract is a short summary of your completed research. It is intended to describe your work without going into great detail. Abstracts should be self-‐contained and concise, explaining your work as briefly and clearly as possible. Different disciplines call for slightly different approaches to abstracts, as will be illustrated by the examples below, so it would be wise to study some ...
An abstract is a summary. In academic articles, the abstract is a one-paragraph synopsis of the content of the article that follows it. The abstract, therefore, is not an introduction. While it does appear first, it is meant to stand apart from the article as a whole and to enable researchers to quickly decide whether the article discusses an ...
Abstract (summary) An abstract is a brief summary of a research article, thesis, review, conference proceeding, or any in-depth analysis of a particular subject and is often used to help the reader quickly ascertain the paper's purpose. [1] When used, an abstract always appears at the beginning of a manuscript or typescript, acting as the point ...
An abstract is a short summary of your completed research. It is intended to describe your work without going into great detail. Abstracts should be self-contained and concise, explaining your work as briefly and clearly as possible What is a research abstract? - 13322274
An abstract is a short summary of a longer work (such as a dissertation or research paper). The abstract concisely reports the aims and outcomes of your research so that readers know exactly what the paper is about.
In a scientific or academic context, an abstract is often a brief summary that provides important highlights of what is going to be discussed in the paper or experiment. This abstract is often found in the initial part of a journal article, for example, and provides a summary of the research and findings.
The statement details the general structure of an abstract in an academic or research paper which provides a concise summary of the study including its purpose, methods, finding, and conclusion along with important keywords.
Weeds are attractive models for basic and applied research due to their impacts on agricultural systems and capacity to swiftly adapt in response to anthropogenic selection pressures. Currently, a lack of genomic information precludes research to elucidate the genetic basis of rapid adaptation for important traits like herbicide resistance and stress tolerance and the effect of evolutionary ...
An abstract is a summary of a research article. It provides a concise overview of the main points, methods, and findings of the article. The purpose of an abstract is to help readers quickly understand the content of the research without having to read the entire article. Here are some key features of an abstract: 1. Concise summary: An abstract should be brief and focused, typically ranging ...
A randomized trial demonstrated consumption of peanut from infancy to age 5 years prevented the development of peanut allergy. An extension of that trial demonstrated the effect persisted after 1 year of peanut avoidance. This follow-up trial examined the durability of peanut tolerance at age 144 months after years of ad libitum peanut consumption.
Research is a tool by which they can test their own, and each others' theories, by using this antagonism to find an answer and advance knowledge. The purpose of research is really an ongoing process of correcting and refining hypotheses, which should lead to the acceptance of certain scientific truths.
The hypothesis of a research paper is usually found in the introduction and is part of the overall structure that includes the abstract, literature review, methodology, results, and conclusion/discussion, each serving a distinct purpose for communicating research findings.