What is a research repository, and why do you need one?
Last updated
31 January 2024
Reviewed by
Miroslav Damyanov
Without one organized source of truth, research can be left in silos, making it incomplete, redundant, and useless when it comes to gaining actionable insights.
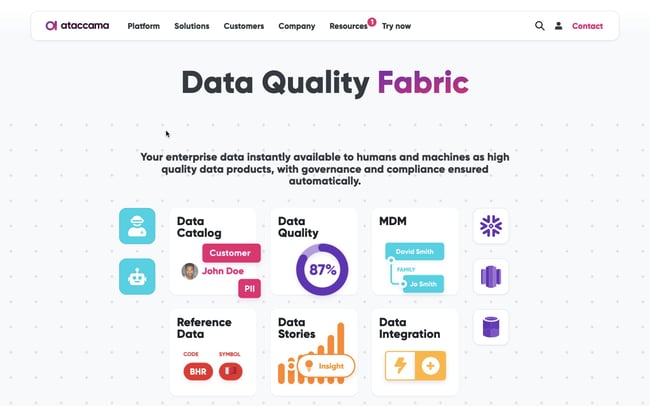
A research repository can act as one cohesive place where teams can collate research in meaningful ways. This helps streamline the research process and ensures the insights gathered make a real difference.
- What is a research repository?
A research repository acts as a centralized database where information is gathered, stored, analyzed, and archived in one organized space.
In this single source of truth, raw data, documents, reports, observations, and insights can be viewed, managed, and analyzed. This allows teams to organize raw data into themes, gather actionable insights , and share those insights with key stakeholders.
Ultimately, the research repository can make the research you gain much more valuable to the wider organization.
- Why do you need a research repository?
Information gathered through the research process can be disparate, challenging to organize, and difficult to obtain actionable insights from.
Some of the most common challenges researchers face include the following:
Information being collected in silos
No single source of truth
Research being conducted multiple times unnecessarily
No seamless way to share research with the wider team
Reports get lost and go unread
Without a way to store information effectively, it can become disparate and inconclusive, lacking utility. This can lead to research being completed by different teams without new insights being gathered.
A research repository can streamline the information gathered to address those key issues, improve processes, and boost efficiency. Among other things, an effective research repository can:
Optimize processes: it can ensure the process of storing, searching, and sharing information is streamlined and optimized across teams.
Minimize redundant research: when all information is stored in one accessible place for all relevant team members, the chances of research being repeated are significantly reduced.
Boost insights: having one source of truth boosts the chances of being able to properly analyze all the research that has been conducted and draw actionable insights from it.
Provide comprehensive data: there’s less risk of gaps in the data when it can be easily viewed and understood. The overall research is also likely to be more comprehensive.
Increase collaboration: given that information can be more easily shared and understood, there’s a higher likelihood of better collaboration and positive actions across the business.
- What to include in a research repository
Including the right things in your research repository from the start can help ensure that it provides maximum benefit for your team.
Here are some of the things that should be included in a research repository:

An overall structure
There are many ways to organize the data you collect. To organize it in a way that’s valuable for your organization, you’ll need an overall structure that aligns with your goals.
You might wish to organize projects by research type, project, department, or when the research was completed. This will help you better understand the research you’re looking at and find it quickly.
Including information about the research—such as authors, titles, keywords, a description, and dates—can make searching through raw data much faster and make the organization process more efficient.
All key data and information
It’s essential to include all of the key data you’ve gathered in the repository, including supplementary materials. This prevents information gaps, and stakeholders can easily stay informed. You’ll need to include the following information, if relevant:
Research and journey maps
Tools and templates (such as discussion guides, email invitations, consent forms, and participant tracking)
Raw data and artifacts (such as videos, CSV files, and transcripts)
Research findings and insights in various formats (including reports, desks, maps, images, and tables)
Version control
It’s important to use a system that has version control. This ensures the changes (including updates and edits) made by various team members can be viewed and reversed if needed.
- What makes a good research repository?
The following key elements make up a good research repository that’s useful for your team:
Access: all key stakeholders should be able to access the repository to ensure there’s an effective flow of information.
Actionable insights: a well-organized research repository should help you get from raw data to actionable insights faster.
Effective searchability : searching through large amounts of research can be very time-consuming. To save time, maximize search and discoverability by clearly labeling and indexing information.
Accuracy: the research in the repository must be accurately completed and organized so that it can be acted on with confidence.
Security: when dealing with data, it’s also important to consider security regulations. For example, any personally identifiable information (PII) must be protected. Depending on the information you gather, you may need password protection, encryption, and access control so that only those who need to read the information can access it.
- How to create a research repository
Getting started with a research repository doesn’t have to be convoluted or complicated. Taking time at the beginning to set up the repository in an organized way can help keep processes simple further down the line.
The following six steps should simplify the process:
1. Define your goals
Before diving in, consider your organization’s goals. All research should align with these business goals, and they can help inform the repository.
As an example, your goal may be to deeply understand your customers and provide a better customer experience . Setting out this goal will help you decide what information should be collated into your research repository and how it should be organized for maximum benefit.
2. Choose a platform
When choosing a platform, consider the following:
Will it offer a single source of truth?
Is it simple to use
Is it relevant to your project?
Does it align with your business’s goals?
3. Choose an organizational method
To ensure you’ll be able to easily search for the documents, studies, and data you need, choose an organizational method that will speed up this process.
Choosing whether to organize your data by project, date, research type, or customer segment will make a big difference later on.
4. Upload all materials
Once you have chosen the platform and organization method, it’s time to upload all the research materials you have gathered. This also means including supplementary materials and any other information that will provide a clear picture of your customers.
Keep in mind that the repository is a single source of truth. All materials that relate to the project at hand should be included.
5. Tag or label materials
Adding metadata to your materials will help ensure you can easily search for the information you need. While this process can take time (and can be tempting to skip), it will pay off in the long run.
The right labeling will help all team members access the materials they need. It will also prevent redundant research, which wastes valuable time and money.
6. Share insights
For research to be impactful, you’ll need to gather actionable insights. It’s simpler to spot trends, see themes, and recognize patterns when using a repository. These insights can be shared with key stakeholders for data-driven decision-making and positive action within the organization.
- Different types of research repositories
There are many different types of research repositories used across organizations. Here are some of them:
Data repositories: these are used to store large datasets to help organizations deeply understand their customers and other information.
Project repositories: data and information related to a specific project may be stored in a project-specific repository. This can help users understand what is and isn’t related to a project.
Government repositories: research funded by governments or public resources may be stored in government repositories. This data is often publicly available to promote transparent information sharing.
Thesis repositories: academic repositories can store information relevant to theses. This allows the information to be made available to the general public.
Institutional repositories: some organizations and institutions, such as universities, hospitals, and other companies, have repositories to store all relevant information related to the organization.
- Build your research repository in Dovetail
With Dovetail, building an insights hub is simple. It functions as a single source of truth where research can be gathered, stored, and analyzed in a streamlined way.
1. Get started with Dovetail
Dovetail is a scalable platform that helps your team easily share the insights you gather for positive actions across the business.
2. Assign a project lead
It’s helpful to have a clear project lead to create the repository. This makes it clear who is responsible and avoids duplication.
3. Create a project
To keep track of data, simply create a project. This is where you’ll upload all the necessary information.
You can create projects based on customer segments, specific products, research methods , or when the research was conducted. The project breakdown will relate back to your overall goals and mission.
4. Upload data and information
Now, you’ll need to upload all of the necessary materials. These might include data from customer interviews , sales calls, product feedback , usability testing , and more. You can also upload supplementary information.
5. Create a taxonomy
Create a taxonomy to organize the data effectively by ensuring that each piece of information will be tagged and organized.
When creating a taxonomy, consider your goals and how they relate to your customers. Ensure those tags are relevant and helpful.
6. Tag key themes
Once the taxonomy is created, tag each piece of information to ensure you can easily filter data, group themes, and spot trends and patterns.
With Dovetail, automatic clustering helps quickly sort through large amounts of information to uncover themes and highlight patterns. Sentiment analysis can also help you track positive and negative themes over time.
7. Share insights
With Dovetail, it’s simple to organize data by themes to uncover patterns and share impactful insights. You can share these insights with the wider team and key stakeholders, who can use them to make customer-informed decisions across the organization.
8. Use Dovetail as a source of truth
Use your Dovetail repository as a source of truth for new and historic data to keep data and information in one streamlined and efficient place. This will help you better understand your customers and, ultimately, deliver a better experience for them.
Should you be using a customer insights hub?
Do you want to discover previous research faster?
Do you share your research findings with others?
Do you analyze research data?
Start for free today, add your research, and get to key insights faster
Editor’s picks
Last updated: 13 April 2023
Last updated: 14 February 2024
Last updated: 27 January 2024
Last updated: 18 April 2023
Last updated: 8 February 2023
Last updated: 23 January 2024
Last updated: 30 January 2024
Last updated: 7 February 2023
Last updated: 7 March 2023
Last updated: 18 May 2023
Last updated: 13 May 2024
Latest articles
Related topics, .css-je19u9{-webkit-align-items:flex-end;-webkit-box-align:flex-end;-ms-flex-align:flex-end;align-items:flex-end;display:-webkit-box;display:-webkit-flex;display:-ms-flexbox;display:flex;-webkit-flex-direction:row;-ms-flex-direction:row;flex-direction:row;-webkit-box-flex-wrap:wrap;-webkit-flex-wrap:wrap;-ms-flex-wrap:wrap;flex-wrap:wrap;-webkit-box-pack:center;-ms-flex-pack:center;-webkit-justify-content:center;justify-content:center;row-gap:0;text-align:center;max-width:671px;}@media (max-width: 1079px){.css-je19u9{max-width:400px;}.css-je19u9>span{white-space:pre;}}@media (max-width: 799px){.css-je19u9{max-width:400px;}.css-je19u9>span{white-space:pre;}} decide what to .css-1kiodld{max-height:56px;display:-webkit-box;display:-webkit-flex;display:-ms-flexbox;display:flex;-webkit-align-items:center;-webkit-box-align:center;-ms-flex-align:center;align-items:center;}@media (max-width: 1079px){.css-1kiodld{display:none;}} build next, decide what to build next.

Users report unexpectedly high data usage, especially during streaming sessions.

Users find it hard to navigate from the home page to relevant playlists in the app.

It would be great to have a sleep timer feature, especially for bedtime listening.

I need better filters to find the songs or artists I’m looking for.
Log in or sign up
Get started for free
- Skip to main content
- Skip to primary sidebar
- Skip to footer
- QuestionPro

- Solutions Industries Gaming Automotive Sports and events Education Government Travel & Hospitality Financial Services Healthcare Cannabis Technology Use Case NPS+ Communities Audience Contactless surveys Mobile LivePolls Member Experience GDPR Positive People Science 360 Feedback Surveys
- Resources Blog eBooks Survey Templates Case Studies Training Help center
Home Market Research Research Tools and Apps
What is a Data Repository? Definition, Types and Examples

Collecting data isn’t that hard, but what’s hard is creating and maintaining a data repository. Even harder is making sense out of a data repository.
The concept of a data repository has grown in popularity to manage and utilize data efficiently. A data repository is a centralized storage site that allows for easy access, data management, and analysis.
Here, we start by defining a data repository, explaining how to create one for research insights, and outlining its benefits.
What is a data repository?
A data repository is a data library or archive. It may refer to large database management systems or several databases that collect, manage, and store sensitive data sets for data analysis , sharing, and reporting.
Authorized users can easily access and retrieve data by using query and search tools, which helps with research and decision-making. It gives a complete and unified view of the data by combining data from different sources, like databases, apps, and external systems.
Data can be collected and stored in different ways, like aggregated data, which is usually collected from multiple sources or segments of a business. Then, it can be stored in a structured or unstructured manner and later tagged with different metadata.
The data repository uses structured organization methods, standardized schemas, and metadata to ensure that the data is always the same and easy to find. It has tools for storing, managing, and protecting data, such as compression, indexing, access controls, encryption, and reporting.
Data repositories generally maintain subscriptions to licensed data resources for their users to access the information.
Data Repository Examples
In the data management industry, various types of data repositories allow users to make the most of the information they have available, each with its own limitations and characteristics.
Security is crucial as more organizations adopt data repositories to manage and store data. Data repositories are generally categorized into four types of data repositories:
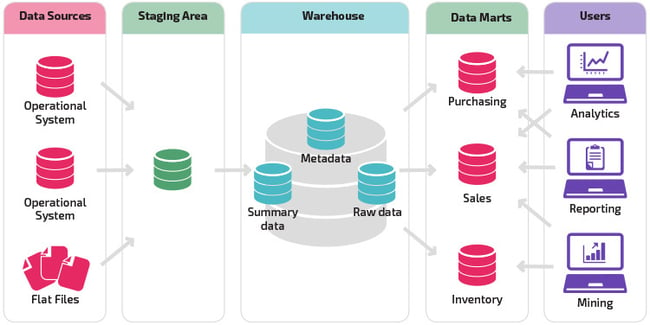
Data warehouse
This is the largest repository type, where data is collected from several business segments or sources. The data stored in this repository is generally used for analysis and reporting, which will help the data users or teams make the correct decisions in their business or project.
In this type of repository, data can be in any form, whether structured, semi-structured, or unstructured. It is a huge storehouse of unstructured data categorized and labeled with metadata.
The main reason for the existence of a data lake is the limitation of the data warehouses. It helps to gain better data governance and data governance framework total control of the data it has in it.
Data marts are often confused with data warehouses. However, they serve different functions.
This subset of the data warehouse is focused on a particular subject, department, or other specific area. Since the data is stored for a specific area, a user can swiftly access the insights without spending much time searching an entire data warehouse, ultimately making users’ lives easy.
This repository contains the most complex data in it. It may be described as the multidimensional extensions of different tables, and they’re generally used to represent data that is too complex to be described by just tables, rows, and columns.
So basically, a data cube can be used when we analyze data available to us and beyond 3-D. Here, we’ll particularly talk about data repositories used in market research.
Benefits of using a research data repository
Using research data repositories has many benefits for both researchers and the scientific community as a whole. Here are some significant benefits:
Greater visibility
Data saved in data repositories can be viewed anytime. Keeping it siloed in Excel sheets or applications not used by a team reduces its visibility and usability, wasting time and resources.
Enhanced discoverability
Saving data in digital format makes it more accessible. Just search for the piece of data you’re looking for, and voila! Also, the metadata added along with the data repository enables others to understand the large context and make more sense of it.
A data repository contains many pieces of data. However, it’s more than just a warehouse. Discrete datasets are joined so that you can derive interesting insights into your area of research and generate various types of reports using the same datasets.
For instance, if you conduct an online survey and collect data from your target audience , you can generate a comparison report to compare responses from various demographic groups. You can also generate trend reports to understand how people’s choices have changed over time. Both of these reports use the same data.
Gain insights from multiple sources of data
Integrating data repositories with other applications lets you see a multi-dimensional view of your data. For instance, you can analyze the historical survey data and the actual sales data to understand the accuracy of insights gained in the past.
How to create a Data Repository using online tools?
Creating data repositories for your research data is simple with the right online tools. If you are conducting your research using surveys , communities, focus groups , or any other method, here are some of the ways to create one.
Create a questionnaire
Many online tools allow you to drag and drop question types . You can create a survey in under 5 minutes! You can also save time by using a ready-to-use survey template. Customize the template per your needs, and you’re ready.
Brand your survey
Customize the header and footer, and add a logo to look more professional. You can also choose a font style and color that suit your brand voice. Branding your surveys increases the chances of getting more responses.
Distribute your survey
Many tools offer different ways to distribute your survey, such as email, embedding data on the website, or sharing it on social media sites. You can also generate a QR code or let your audience answer questions using a mobile app.
Analyze the data
Finally, once you have collected your data, generating the reports is just a matter of time. Use tools that let you create dashboards and generate reports with ease.
How does QuestionPro help implement data repositories?
QuestionPro is a powerful online survey and research platform that collects, analyzes, and manages data. It mostly creates surveys, collects data, and helps establish and maintain data repositories. QuestionPro helps data repository management in several ways:
- Data collection : QuestionPro lets you develop and send surveys to collect data. Surveys can use multiple choice, rating scales, open-ended questions , and more. Your data repositories get important data from this data collection process.
- Data Management : With QuestionPro, you can effectively organize and manage your gathered data. It filters, categorizes, and validates data to ensure accuracy and quality. These management tools help maintain a well-organized and ordered data repository.
- Data Analysis: QuestionPro has built-in tools to help you examine and visualize your data. You can create reports, charts, and graphs based on survey answers to help you find trends, patterns, and insights. The analysis results can be saved in your data repository.
- Real-time Reporting: Real-time reporting lets you view and analyze your data. After collecting replies, you may instantly generate reports to assess trends and progress and make data-driven decisions.
- Data Security: QuestionPro prioritizes data security. It encrypts, transfers, and restricts data access to prevent data breaches. This makes sure that the data in your repository is safe and that users’ privacy is protected.
- Data Integration: QuestionPro integrates with Excel, Google Sheets, and SPSS. This connection lets you import external data or survey responses into your data repositories for analysis and storage.
Data collecting, customer data integration , management, analysis, and security features in QuestionPro can help you manage your repository. It’s useful for data repository management since it centralizes data collection, storage, and analysis.
Learn more about best data collection tools to help you choose the best one.
If you need any help conducting research or creating a data repository, connect with our team of experts. We can guide you through the process and help you make the most of your data.
Frequently Asking Questions (FAQ)
Your data repository should suit your demands. You should choose a repository that is popular and relevant to your research domain. Your data format should be supported by the repository.
Data repositories are managed digital environments that specialize in gathering, characterizing, distributing, and tracking research data. Sharing data in a repository is a best practice that is frequently mandated by federal authorities.
The difference between a database and a data repository lies in its functionality, the former is only a data storage system while a data repository is a data management system.
MORE LIKE THIS

Cannabis Industry Business Intelligence: Impact on Research
May 28, 2024
Top 10 Dynata Alternatives & Competitors
May 27, 2024
What Are My Employees Really Thinking? The Power of Open-ended Survey Analysis
May 24, 2024

I Am Disconnected – Tuesday CX Thoughts
May 21, 2024
Other categories
- Academic Research
- Artificial Intelligence
- Assessments
- Brand Awareness
- Case Studies
- Communities
- Consumer Insights
- Customer effort score
- Customer Engagement
- Customer Experience
- Customer Loyalty
- Customer Research
- Customer Satisfaction
- Employee Benefits
- Employee Engagement
- Employee Retention
- Friday Five
- General Data Protection Regulation
- Insights Hub
- Life@QuestionPro
- Market Research
- Mobile diaries
- Mobile Surveys
- New Features
- Online Communities
- Question Types
- Questionnaire
- QuestionPro Products
- Release Notes
- Research Tools and Apps
- Revenue at Risk
- Survey Templates
- Training Tips
- Uncategorized
- Video Learning Series
- What’s Coming Up
- Workforce Intelligence
Research Data Management
- Data Management Plans
- National Institutes of Health (NIH)
- National Endowment for the Humanities (NEH)
Data Repositories
- Resources on Campus
- File Formats
- Learning Resources
UDSpace - UD's Institutional Repository
University of Delaware original research in digital form. Includes articles, technical reports, working & conference papers, images, data sets, & more.
The UDSpace Institutional Repository collects and disseminates research material from the University of Delaware. The information is organized by Communities, corresponding to administrative entities such as college, school, department, and research center, and then by Collection. Select a Community to browse its collections, or perform a keyword search and then filter by author, subject, or date.
- What are data repositories?
- Where can I find a data repository?
- About Open Data
A data repository is simply a place to store datasets for the long-term. Using a repository or archive to store your data (versus simply hosting it on your website) facilitates its discovery and preservation, ensuring that it will be found by others looking for data and that care will be taken to preserve the data over time.
There are thousands of data repositories, often for a specific subject or type of data, and many organized by universities or scholarly organizations.
Released in May 2022 by OSTP, the report Desirable Characteristics of Data Repositories for Federally Funded Research aims to improve consistency across Federal departments and agencies in the instructions they provide to researchers about selecting repositories for data resulting from Federally funded research. This document contains clearly defined desirable characteristics for two classes of online research data repositories: a general class appropriate for all types of Federally funded data—including free and easy access—and a specific class that has special considerations for the sharing of human data, including additional data security and privacy considerations.
For subject data repositories, please also visit your discipline's research guide or contact your subject librarian directly. Note that some subjects may not have discipline specific repositories.
See the Open Data tab for other repositories
- DataCite Commons Web portal for users to search for works, people, organizations or repositories based on the DataCite metadata catalog
- figshare figshare helps academic institutions store, share and manage all of their research outputs. Upload files up to 5GB. We accept any file format and aim to preview all of them in the browser.
- ICPSR Data Portal An international consortium of more than 750 academic institutions and research organizations, Inter-university Consortium for Political and Social Research (ICPSR) provides leadership and training in data access, curation, and methods of analysis for the social science research community. ICPSR maintains a data archive of more than 250,000 files of research in the social and behavioral sciences. It hosts 21 specialized collections of data in education, aging, criminal justice, substance abuse, terrorism, and other fields.
- re3data.org re3data.org is a global registry of research data repositories that covers research data repositories from different academic disciplines.
- Simmons Data Repositories Listing A listing of various subject oriented data repositories, part of the Open Access Directory.
Open Data is part of the broader Open Access movement, promoting the idea that research, especially publicly funded research, should be made widely available. This allows for more and more rapid development and dissemination of knowledge.
Open Data is data that is permanently and freely available for the world to use, allowing for maximum exposure and benefit from current research.
- Open Access Directory The Open Access Directory (OAD) is a compendium of simple factual lists about open access (OA) to science and scholarship, maintained by the OA community at large.
- Open Data Commons Provides a set of legal tools and licenses to help you publish, provide and use open data
- Harvard Dataverse Repository A free data repository open to all researchers from any discipline, both inside and outside of the Harvard community, where you can share, archive, cite, access, and explore research data.
- Dryad Digital Repository The Dryad Digital Repository is a curated resource that makes research data discoverable, freely reusable, and citable. Dryad provides a general-purpose home for a wide diversity of data types.
- << Previous: National Endowment for the Humanities (NEH)
- Next: Resources on Campus >>
- Last Updated: May 16, 2024 8:35 AM
- URL: https://guides.lib.udel.edu/researchdata

University Libraries
- Ohio University Libraries
- Library Guides
Research Data Repositories: Finding and Storing Data
- Business & Economics
- Crime, Government, & Law
- Sociology, Anthropology, & Archaeology
- Biological Sciences
- Engineering and Computer Science
- Health, Medicine, and Psychology
- Physics and Astronomy
- Chemistry and Biochemistry
- General Humanities
- Literature, Linguistics, & Languages
- Contact/Need Help?
We have three guides about data: Which one do you need?
- Research Data Literacy 101 This guide covers research data generally, what data is, the difference between data and statistics, understanding open data, library databases that offer statistics and data, and other overview topics.
- Research Data Repositories: Finding and Storing Data This guide is a annotated list of data repositories by subject where a researcher can deposit their data per gov standards and find data sets from research done by others. Note: For library databases that offer statistics and data, see Research Data Literacy 101: Find Data and Statistics.
- Research Data Management This guide covers how to create a Data Management Plan, including funders, metadata, and other important aspects. Includes help using DMPTool.
Often, the place to find and store data are the very same. Researchers will place the data they collect into general or disciplinary repositories. While other researchers can search those repositories for data and datasets on their topic. Some repositories are costly while others are considered "open" and offer data freely for anyone to download.
Data and Statistics Are Not Equivalent
Although both terms are commonly used synonymously, they are, in fact, very different. Before you start searching for either, think about which one best applies to your needs.
- Data: are collected raw numbers or bits of information that have not been analyzed or organized.
- Statistics: are the product of collected data after it has been analyzed or organized that will help derive meaning from the data.
The National Library of Medicine has a great resource full of other data-related definitions.
What to Consider When Choosing a Data Repository
A data repository is a storage space for researchers to deposit data sets associated with their research. And if you’re an author seeking to comply with a journal or funder data sharing policy, you’ll need to identify a suitable repository for your data.
An open access data repository openly stores data in a way that allows immediate user access to anyone. There are no limitations to the repository access.
When choosing a repository for your data, keep in mind the following:
- It is likely that your funder or journal will have specific guidelines for sharing your data
- Ensure the repository issues a persistent identifier (like a DOI) or you can link to your ORCID account
- Repository has a preservation plan in perpetuity
- Does the repository have a cost to store your data? There may also be a cost to access datasets.
- Is the repository certified or indexed?
- Is the repository completely open or are there restrictions to access?
- Consider FAIR data Principles - Data should be Findable, Accessible, Interoperable, and Re-usable
NIH guidelines for selecting a data repository
3 Ways to use Google to find Data
Google has a Dataset Search! Here is a video tutorial on how to use this search tool .
You can search for specific file types in Google, for example CSV files for datasets. By typing into Google filetype:csv in the search bar you are "telling" Google to only search for things that have that specific file type. For example: (poverty AND ohio) filetype:xls will result in XLS (Excel) files mentioning Poverty in Ohio.
Limit search results by web domain by typing into Google: site:.gov (YOUR TOPIC HERE) . This will limit datasets, files, etc. from specific websites. You could even do .org for professional organizations.
- Next: Social Sciences >>
Research Data Management
- Data Repositories
A key aspect of data management involves not only making articles available, but also the data, code, and materials used to conduct that research. Data repositories are a centralized place to hold data, make data available for use, and organize data in a logical manner.
Which Repository Should I Use?
The best repository for sharing your data depends on your discipline. If there is a national or subject-level repository in your discipline, that would be your first choice. To determine if such repositories exist, you can search the registry of repositories re3data , or check the Open Access Directory of Data Repositories at Simmons University .
It is also possible that your funding agency or your state has a repository you can use. In the absence of these, there are a number of general subject repositories which can take your data. The NNLM Data Repository Finder is a tool that was developed to help locate NIH-supported repositories for sharing research data
- << Previous: Unique Identifiers
- Data Management Plans
- Data Policies & Compliance
- Directory Structures
- File Naming Conventions
- Roles & Responsibilities
- Collaborative Tools & Software
- Electronic Lab Notebooks
- Documentation & Metadata
- Reproducibility
- Analysis Ready Datasets
- Image Management
- Version Control
- Data Storage
- Data & Safety Monitoring
- Data Privacy & Confidentiality
- Retention & Preservation
- Data Destruction
- Data Sharing
- Public Access
- Data Transfer Agreements
- Intellectual Property & Copyright
- Unique Identifiers
Research Support
Request a data management services consultation.
Email [email protected] to schedule a consultation related to the organization, storage, preservation, and sharing of data.
- Last Updated: May 23, 2024 10:37 AM
- URL: https://guides.library.ucdavis.edu/data-management
When you choose to publish with PLOS, your research makes an impact. Make your work accessible to all, without restrictions, and accelerate scientific discovery with options like preprints and published peer review that make your work more Open.
- PLOS Biology
- PLOS Climate
- PLOS Complex Systems
- PLOS Computational Biology
- PLOS Digital Health
- PLOS Genetics
- PLOS Global Public Health
- PLOS Medicine
- PLOS Mental Health
- PLOS Neglected Tropical Diseases
- PLOS Pathogens
- PLOS Sustainability and Transformation
- PLOS Collections
- About This Blog
- Official PLOS Blog
- EveryONE Blog
- Speaking of Medicine
- PLOS Biologue
- Absolutely Maybe
- DNA Science
- PLOS ECR Community
- All Models Are Wrong
- About PLOS Blogs
What a difference a data repository makes: Six ways depositing data maximizes the impact of your science

Data is key to verification, replication, reuse, and enhanced understanding of research conclusions. When your data is in a repository—instead of an old hard drive, say, or even a Supporting Information file—its impact and its relevance are magnified. Here are six ways that putting your data in a public repository can help your research go further.
1. You can’t lose data that’s in a public data repository
Have you ever lost track of a dataset? Maybe you’ve upgraded your computer or moved to a new institution. Maybe you deleted a file by mistake, or simply can’t remember the name of the file you’re looking for. No matter the cause, lost data can be embarrassing and time consuming. You’re unable to supply requested information to journals during the submission process or to readers after publication. Future meta analyses or systematic reviews are impossible. And you may end up redoing experiments in order to move forward with your line of inquiry. With data securely deposited in a repository with a unique DOI for tracking, archival standards to prevent loss, and metadata and readme materials to make sure your data is used correctly, fulfilling journal requests or revisiting past work is easy.
2. Public data repositories support understanding, reanalysis and reuse
Transparently posting raw data to a public repository supports trustworthy, reproducible scientific research. Insight into the data and analysis gives readers a deeper understanding of published research articles. Offering the opportunity for others to interpret results demonstrates integrity and opens new avenues for discussion and collaboration. Machine-readable data formatting allows the work to be incorporated into future systematic reviews or meta analyses, expanding its usefulness.
3. Public data repositories facilitate discovery
Even the best data can’t be used unless it can be found. Detailed metadata, database indexing, and bidirectional linking to and from related articles helps to make data in public repositories easily searchable—so that it reaches the readers who need it most, maximizing the impact and influence of the study as a whole.
4. Public data repositories reflect the true value of data
Data shouldn’t be treated like an ancillary bi-product of a research article. Data is research . And researchers deserve academic credit for collecting, capturing and curating the data they generate through their work. Public repositories help to illustrate the true importance and lasting relevance of datasets by assigning them their own unique DOI, distinct from that of related research articles—so that datasets can accumulate citations in their own right.
5. Public data demonstrates rigor
There’s no better way to illustrate the rigor of your results than explaining exactly how you achieved them. Sharing data lets you demonstrate your credibility and inspires confidence in readers by contextualizing results and facilitating reproducibility.
6. Research with data in public data repositories attracts more citations
A 2020 study of more than 500,000 published research articles found articles that link to data in a public repository have a 25% higher citation rate on average than articles where data is available on request or as Supporting Information. The precise reasons for the association remain unclear. Are researchers who deposit carefully curated data in a repository also more likely to produce rigorous, citation-worthy research? Are researchers with the time and resources to devote to data curation and deposition more established in their careers, and therefore more highly cited? Are readers more likely to cite research when they trust that they can verify the conclusions with data? Perhaps some combination?
What do you see as the most important reason for posting data in a repository?
Access to raw scientific data enhances understanding, enables replication and reanalysis, and increases trust in published research. The vitality and utility of…
The latest quarterly update to the Open Science Indicators (OSIs) dataset was released in December, marking the one year anniversary of OSIs…
For PLOS, increasing data-sharing rates—and especially increasing the amount of data shared in a repository—is a high priority. Research data is a…
Home / Blogs / Data Repository: Definition, Types, and Benefits with Best Practices
The Automated, No-Code Data Stack
Learn how Astera Data Stack can simplify and streamline your enterprise’s data management.

Data Repository: Definition, Types, and Benefits with Best Practices
With time, data is becoming more significant to business decision-making. This means you need solutions to gather, store, and analyze data. A data repository is a virtual storage entity that can help you consolidate and manage critical enterprise data.
In this blog, we’ll give a brief overview of a data repository, its common examples, and critical benefits.
What is a Data Repository?
A data repository , often called a data archive or library, is a generic terminology that refers to a segmented data set used for reporting or analysis.
A data repository serves as a centralized storage facility for managing and storing various datasets . It encompasses:
- Large database management systems: These systems efficiently collect, organize, and store extensive datasets.
- Data archives: These archives securely preserve sensitive data sets for analysis, sharing, and reporting purposes.
Data repositories facilitate data management, ensuring accessibility, security, and efficiency in handling diverse datasets.
It’s a vast database infrastructure that gathers, manages, and stores varying data sets for analysis, distribution, and reporting.
Types of Data Repositories
Some common types of data repositories include:
Data Warehouse
A data warehouse is a large central data repository that gathers data from several sources or business segments. The stored data is generally used for reporting and analysis to help users make critical business decisions.
In a broader perspective, a data warehouse offers a consolidated view of either a physical or logical data repository gathered from numerous systems. The main objective of a data warehouse is to establish a connection between data from current systems, such as product catalog data stored in one system and procurement orders for a client stored in another one.
A data lake is a unified data repository that allows you to store structured, semi-structured, and unstructured enterprise data at any scale. Data can be in raw form and used for different tasks like reporting, visualizations, advanced analytics, and machine learning.
A data mart is a subject-oriented data repository often a segregated section of a data warehouse. It holds a subset of data usually aligned with a specific business department, such as marketing, finance, or support.
Due to its smaller size, a data mart can fast-track business procedures as you can easily access relevant data within days instead of months. As it only includes the data pertinent to a specific area, a data mart is an economical way to acquire actionable insights swiftly.
Metadata Repositories
While metadata incorporates information about the structures that include the actual data, metadata repositories contain information about the data model that store and share this data. They describe where the data source is, how it was collected, and what it signifies. It may define the arrangement of any data or subject deposited in any format.
For businesses, metadata repositories are essential in helping people understand administrative changes, as they contain detailed information about the data.
Data cubes are data lists with multidimensions (usually three or more dimensions) stored as a table. They are used to describe the time sequence of an image’s data and help assess gathered data from a range of standpoints.
Each dimension of a data cube signifies specific database characteristics such as day-to-day, monthly or annual sales. The data within a data cube allows you to analyze all the information for almost any client, sales representative, products, and more. Consequently, a data cube can help you identify trends and scrutinize business performance.
Why Do You Need A Data Repository?
A data repository can help businesses fast-track decision-making by offering a consolidated space to store data critical to your operations. This segmentation enables easier data access and troubleshooting and streamlines reporting and analysis.
For instance, if you want to find out which of your workplaces incur the most cost, you can create an information repository for leases, energy expenses, amenities, security, and utilities, excluding employees or business function information. Storing this data in one place can make it easier for you to come to a decision.
Challenges Associated with a Data Repository
Although an information repository offers many benefits, it also includes several challenges that you must manage efficiently to alleviate possible data security risks.
Some challenges in maintaining data repositories include:
- An increase in data sets can reduce your system’s speed. To rectify this problem, ensure that the database management system can scale with data expansion.
- In case a system crashes, it can negatively impact your data. It’s best to maintain a backup of all the databases and restrict access to control the system risk.
- Unauthorized operators can access sensitive data more quickly if stored in a single location than if it’s dispersed across numerous sources. On the contrary, implementing security protocols on a single data storage location is more accessible than multiple ones.
Best Practices to Create and Manage Data Repositories
When creating and maintaining software repositories, you have to make several hardware and software decisions. Therefore, it is best to involve all stakeholders during the development and usage phase of the data repositories. For example, in case of building a clinical data repository architecture, it is a good idea to involve doctors, data experts, analysts and data pipeline engineers in the initial planning stages.
Here are some of the best practices to help you make the most of this storage solution:
1. Select the Right Tool
Using ETL tools to create a data repository and transfer data can help ensure data quality is maintained during the process. But keep in mind that different data repository tools offer additional features to create, maintain, and control the repository. So, find a tool that provides the features that support your business requirements.
2. Limit the Scope Initially
It’s best to narrow down the scope of your information repository in the initial days. Accumulate smaller data sets and limit the number of subject areas. Gradually increase the complexity as the data operators get familiar with the system.
3. Automate as Much as Possible
Automating the process for loading and maintaining the data repository saves the user from manual efforts and reduces the chances of errors.
4. Prioritize Flexibility
The data repository should be scalable enough to accommodate evolving data types and increase volumes. So, make flexible plans that make allowance for alterations in technology.
As more and more businesses adopt data repositories to store and administer their ever-increasing data, a secure approach becomes imperative for your company’s overall security. Creating comprehensive access rules to permit only authorized operators to access, change, or transfer data will help secure your enterprise data.
Astera Centerprise is an automated data integration tool that helps in data management with features such as data cleansing, profiling, and transformation all in one solution. Contact our team for a personalized demo .
You MAY ALSO LIKE
What is olap (online analytical processing).
What is Online Analytical Processing (OLAP)? Online Analytical Processing (OLAP) is a computing technology through which users can extract and...
The Best Data Ingestion Tools in 2024
Data ingestion is important in collecting and transferring data from various sources to storage or processing systems. In this blog,...
Data Ingestion vs. ETL: Understanding the Difference
Working with large volumes of data requires effective data management practices and tools, and two of the frequently used processes...
Considering Astera For Your Data Management Needs?
Establish code-free connectivity with your enterprise applications, databases, and cloud applications to integrate all your data.
This article provides an overview of the current state of research data repositories, what they are, why your library needs one, and pragmatic steps toward actualization. It surveys the present changing data repository landscape and uses practical examples from a large, current Texas consortial effort to create a research data repository for universities across the Lone Star State. Libraries in general need to think seriously about data repositories to partner with state, national, and global efforts to begin providing the next generation of information services and infrastructures.
Data Research Repositories: Definitions
Online research data repositories are large database infrastructures set up to manage, share, access, and archive researchers’ datasets. Repositories may be specialized and relegated to aggregating disciplinary data or more general, collecting over larger knowledge areas, such as the sciences or social sciences. Online repositories may also aggregate experts’ data globally or locally, collecting a university or consortium of universities researcher’s data for mutual benefit. The simple idea is that sharing data improves results and drives research and discovery forward. A repository allows examination, proof, review, transparency, and validation of a researcher’s results by other experts beyond the published refereed academic article. Placing research data online allows instantaneous access by a globally dispersed group of researchers to share, understand, and synthesize results. This aggregation and synthesis provide an opportunity for insight, progress, and that uniquely human quest for larger understanding. Data repositories also allow for the publication of previously hidden negative data, essentially experiments that didn’t work. This enables other researchers to avoid previous dead ends of those who have tried a path before them to better find their way toward more fertile territory. A global community of experts benefits from online sharing and the aggregation of research data.
The Nuts and Bolts
Data repositories allow long-term archiving and preservation of data by the ingestion/uploading of various data types. This includes simple Excel files, SPSS, and more exotic disciplinary formats (i.e., GIS shapefiles and Genome data-specific formats). Usually, a repository will also provide a permalink strategy for online citation and instant access so that researchers may offer a direct link to their data and ancillary files in the later published article or conference paper. This is usually provided through a digital object identifier (DOI) or universal numerical fingerprint (UNF), which allows later linking of data and possibilities for interoperability and mashing up of data archives. Within data archives, para-textual research material is also stored for later archiving and sharing. Users include social scientists and hard scientists. Data files include spreadsheets, field notes, lab methodology recipes, multimedia, and specific software programs for analyzing and working with the accompanying datasets.
The data repository infrastructure trajectory moves through a lifecycle. It begins with the experiment or research project and initial data capture and progresses to uploading, cataloging, adding disciplinary metadata schema, and assigning DOI and/or UNF (see above). Repositories will typically allow instant searching, retrievability, linking, and downloading of data. As data repositories progress, they will allow synthesis of datasets and data fields to facilitate insight, discovery, and verification. In an online global networked environment, this is accomplished through data harvesting and the possibilities of linked data and data visualization with current applications such as Tableau.
Why a Repository Now?
Besides being a good thing for the sharing and verification of data-driven research results, data research repositories are now necessary for university campuses. Placing one’s research data online has become mandatory for any researcher wishing to receive grants from any public U.S. agency. This includes the National Institutes of Health (NIH), National Science Foundation (NSF), U.S. Department of Agriculture (USDA), and National Endowment for the Humanities (NEH). The rational is that if a researcher is drawing from the public taxpayers’ trough, the research must be publicly accessible through both the article and original data. Sharing this data helps keep the wider economy vital, facilitating healthy competition toward commercialization and dissemination of discovery. If researchers do not have data management plans in place, their chance of obtaining a grant decreases. Currently, a majority of grant-funded researchers do not share data. With recent mandated changes, this situation is rapidly changing. Ivy League institutions have already capitalized on it—sharing data leverages and enhances faculty, departmental, and a university’s global research standing.
The Current State of Affairs
Among research-intensive institutions and academic research libraries, about 74% provide data archiving services (see figure below). Of this group, only 13% provide data-specific repositories. Another 13% use more general digital repositories, and 74% use temporary stopgaps—text-centric repositories such as DSpace—to accommodate current grant stipulations until new data-centered applications can be put in place. The vast majority of academic libraries lag behind this cohort. In terms of Roger’s technological adoption curve, phases of innovators and early adopters of data repositories are complete; we are entering the early majority and primary adoption phases (see the bottom figure). It is a great time to be thinking about a data repository.
Research Data Repository Software
There are currently several possibilities with regard to research data repository software, some specifically created for data (i.e., Dataverse, HUBzero, and Chronopolis), others cobbled together from previous text-based institutional repository/digital library sources (i.e., DSpace, Fedora, and Hydra). The software may be hosted or installed on university servers. Different infrastructures also contain various ranges of data management and data collaboration options. There are both well-established open source software (notably, Dataverse and HUBzero) and proprietary/commercial sources (Interuniversity Consortium for Political and Social Resource [ICSPR], figshare, and Digital Commons).
Repositories, Institutions, and Consortiums
Currently, at Texas State University, we are part of a Texas-wide university effort championed by the Texas Digital Library (TDL) to implement a statewide consortial data repository based on Harvard University’s open source solution, Dataverse. Dataverse is a software framework that enables institutions to host research data repositories and has its roots in the social sciences and Harvard’s Institute for Quantitative Social Sciences (IQSS).
Because of Dataverse’s largely customizable metadata schema abilities and open source flexibilities, TDL is using it as a data archiving infrastructure for the state (officially scheduled for launch in summer/fall 2016). The software allows data sharing, persistent data citation, data publishing, and various administrative management functions. The architecture also allows customization for a consortial effort for future systemwide sharing and interoperability of datasets for a stronger data research network (see diagram directly above).
If an institution is looking seriously at open source data repositories, other software also worth considering is Purdue’s data repository system—HUBzero ( hubzero.org )—and a customized instance, Purdue University Research Repository (PURR; purr.purdue.edu ). Different from Dataverse’s social science antecedents, HUBzero originally began as a platform for hard scientific collaboration (nanoHUB; nanohub.org). In re-creating or customizing one’s institution’s or consortium’s data repository, PURR’s interface is particularly user-friendly. It is worth looking at how different data repositories step their researchers through the data management process via various online examples (see PURR example below).
Other more proprietary data repositories such as figshare, bepress’s Digital Commons, or ICPSR are worth looking at, depending on an institution’s size, needs, and present infrastructure. As the landscape is changing quickly, an environmental scan is a good idea. A good example scan recently conducted by TDL prior to choosing Harvard’s Dataverse is available here: tinyurl.com/h36w93v .
Data Size Matters
Beyond the specific data repository an institution chooses, another factor that needs to be considered is size of datasets. To generalize, researcher, project, and data storage needs come in all different shapes and sizes. Preliminarily thinking about these factors will be important as an institution moves from implementation and customization to setting policy and data storage requirements.
Research data projects may be divided into size categories of 1) small/medium, 2) large, and 3) very large. For small-to-medium datasets, these are data projects that can be stored on a researcher’s current desktop hard drive, typically sets of Excel or other specialized disciplinary data files. These may be uploaded by a researcher, emailed, or transferred through university network drives to a server or the cloud and/or uploaded by a data archivist into a repository. Many of the current data-specific repositories allow researchers’ self-uploading processes to begin or facilitate this process.
For medium-to-large projects, data may require special back-end storage systems or relationships engendered with core university IT to set up larger storage options (i.e., dedicated network space allocation and RAID). Typically, these types of datasets can still be linked online, but there is a larger weight toward data curation, adding robust metadata for access points and considering logical divisions of datasets/fields in consultation with researchers.
For very large projects, relationships may be required to be engendered with consortial, national, or proprietary data preservation and archiving efforts. For example, TDL partners with both state and national organizations—the Texas Advanced Computing Center (TACC) and the Digital Preservation Network (DPN)—and proprietary solutions, DuraCloud and Amazon Web Services (Amazon Glacier and Amazon S3). Funds become a factor here.
Typically, a university will have a spectrum of researchers with low to very high data storage requirements. Infrastructure bridges should be set up to accommodate the range of possibilities that will arise. Longer-term storage needs that a university or consortial environment anticipates and will require should also be factored in here.
Data Management Planning: The Wide Angle
Data management repositories are an important, but single, piece in any researcher’s larger data management plans. Other infrastructure bridges will necessarily involve offices of sponsored research, university core IT, and library personnel working together to build these new paradigms. Fortunately, several good planning tools have been created. A good starting place for planning considerations is the California Digital Library’s DMPTool ( dmp.cdlib.org ). This will help researchers, libraries, and other infrastructure personnel begin thinking and stepping through the multi-tiered process of managing their data.
With an institutional or consortial data repository initially in place and a few key staff members to help researchers navigate this new world, a data repository infrastructure may be enabled. This article has given a whirlwind tour of the fast-changing and now required area of data repositories. A larger presentation with more detailed links and references for further exploration and research is available here: tinyurl.com/jljmmcz .
The Sheridan Libraries
- Data Management and Sharing
- Sheridan Libraries
- How to Find a Data Repository
- Write A Proposal
- Write a Data Management and Sharing Plan
- Human Participant Research Considerations
- JHU Policies for Data Management and Sharing
- Access and Collect Data
- Manage Data
- Analyze and Visualize Data
- Document Data
- Store and Preserve Data
- Overview of Data Sharing
- Prepare Your Data For Sharing
Find a Repository
Repository-specific data submission guidelines.
- Share Code or Software
- Conditions for Access and Reuse
When sharing your research data code, and documentation, there are many repositories to choose from. Some repositories are domain specific, and focus on a very narrow type of research. Other repositories, known as generalists, accept data from multiple disciplines and in various file formats. Another aspect of repositories is the ease with which data is accessible (controlled-access or public).
Qualities to Look For in a Repository
- Data Services selecting a repository for data deposit : Tips and set of questions researchers can use in determining whether a particular research data repository will work for their circumstances.
- NIH guidance on selecting a repository : List of desirable characteristics to look for when choosing a repository to manage and share data resulting from Federally funded research.
- Nature Data Repository guidance: Because Nature does not host data, this is guide for researchers who are publishing with them on qualities to look for when determining where to share their data.
- Data Sharing Tiers for Broad Sharing of JHM Clinically Derived Data : Written by JHU Data Trust , a guide to determining the types of repositories you can use to share clinical data. Requires JHU affiliation JHED to login .
- General JHU IRB Expectations for Sharing of Individual Level Research Data : Written by JHU IRBs. The table outlines considerations researchers should be aware of when developing plans for data sharing when the data to be shared is from human research participants.
Data Repository Registries/Lists
- re3data.org : A global registry of data repositories that covers research data repositories from different academic disciplines and with varying access controls.
- NIH-supported Scientific Data Repositories : A browsable list of NIH-supported repositories including a description and data submission instructions for each repository.
- Welch Medical Library Research Data Repositories & Databases guide : Description of medically-related databases and repositories to find datasets for secondary analysis. Many of those listed are also options for data submission.
- Welch Medical Library Data Catalogs & Search Engines : A list of repositories that you can find secondary data and publish your own data in.
- FAIRsharing Database Registry : A registry of knowledgebases and repositories of data and other digital assets.
- Johns Hopkins Research Data Repository
- Johns Hopkins Research Data Repository : Open access repository for the long-term management and preservation of research data. Johns Hopkins researchers can use the JHU Data Archive to meet their funder and/or journal data sharing requirements.
- Description of the Repository : an overview of the benefits of using the JHU Data Archive and the procedures for getting started
- FAQs about the Johns Hopkins Research Data Repository
Inter-university Consortium for Political and Social Research (ICPSR)
ICPSR has one of the largest archives of social science datasets in the world and is free to deposit your data into. It is designed for both public-use and restricted-use data.
- Information on how to deposit
- FAQs for depositing
Prior to depositing into a repository you should review the submission guidelines as they likely will require you to format your data a particular way and include specific metadata with your submission.
- Submitting data to the Gene Expression Omnibus
- dbGaP Study Submission Guide
- Data Sharing for Next-Generation Sequencing : an online course created by Welch medical library that discusses preparing genomic data for sharing including into dbGaP.
- SRA Submission Quick Start
- << Previous: Prepare Your Data For Sharing
- Next: Share Code or Software >>
- Last Updated: May 10, 2024 9:46 AM
- URL: https://guides.library.jhu.edu/dataservices
What Is a Data Repository? [+ Examples and Tools]
Published: April 19, 2022
Businesses are collecting, storing, and using more data than ever before. This data is being used to improve the customer experience, support marketing and advertising efforts, and drive decision making. But more data means more challenges.

In a survey on customer experience (CX) among businesses in the United States , 49.8% identified the lack of reliability and integrity of available data as the main challenge affecting data analysis capability for CX. Data security, data privacy, and too many data sources were also identified as challenges.
To help you overcome these issues and get the most out of your data, you can store it in a data repository. Let’s take a close look at this term, then walk through some examples, benefits, and tools that can help you store and manage your data .
![define research data repository Download Now: Introduction to Data Analytics [Free Guide]](https://no-cache.hubspot.com/cta/default/53/8982b6b5-d870-4c07-9ccd-49a31e661036.png)
What is a data repository?
A data repository is a data storage entity in which data has been isolated for analytical or reporting purposes. Since it provides long-term storage and access to data, it is a type of sustainable information infrastructure.
While commonly used for scientific research, a data repository can also be used to manage business data. Let’s take a look at some challenges and benefits below.
What are the challenges of a data repository?
The challenges of a data repository all revolve around management. For example, data repositories can slow down enterprise systems as they grow so it’s important you have a software or mechanism in place to scale your repository. You also need to ensure your repository is backed up and secure. That’s because a system crash or attack could compromise all your data since it’s stored in one place instead of distributed across multiple locations.
These challenges can be addressed by a solid data management strategy that addresses data quality, privacy, and other data trends .
To create your own, check out our guide Everything You Need to Know About Data Management .
What are the benefits of a data repository?
Having data from multiple sources in one place makes it easier to manage, analyze, and report on. A data repository makes it faster and easier to analyze and report data because it’s stored in one place and compartmentalized. It also improves the quality of data since it’s aggregated and preserved. Without a single repository, you’ll likely deal with duplicate data, missing data, and other issues that affect the quality of your analysis.
Now that we understand both the challenges and benefits of a data repository, let’s look at some examples.
Data Repository Examples
Data repository is a general term. There are several more specific terms or subtypes. Let’s take a look at some of these examples below.
Data Warehouse
A data warehouse is a centralized repository that stores large volumes of data from multiple sources in order to more efficiently organize, analyze, and report on it. Unlike a data mart and lake, it covers multiple subjects and is already filtered, cleaned, and defined for a specific use.
We’ll take a closer look at the difference between a data repository and warehouse below (jump link).
Data Repository Software
Choosing a data repository software comes down to a few key factors, including sustainability, usability, and flexibility. Here are some questions to ask when evaluating different software:
- Is the repository supported by a company or community?
- What does the user interface look like?
- Is the documentation clear and comprehensive?
- What data formats does it support?
Answering these and other questions will help you pick the software that best meets your needs. Let’s take a look at some popular data repository software options below.
1. Ataccama
Best for: Multinational corporations and mid-sized businesses
Don't forget to share this post!
Related articles.
Materialized View: What You Need to Know [+Best Practices]

What Is Data Hygiene?: Why You Need It & How to Do It Right
API Management: What Is It & Why Does It Matter?

5 Best Data Governance Tools

How to Create a Data Quality Management Plan

Single Source of Truth: Benefits, Challenges, & Examples

Data Governance (DG): A Straightforward Guide


What Is Event-Driven Architecture? Everything You Need to Know
Data Stream: Use Cases, Benefits, & Examples

ETL vs. ELT: What's the Difference & Which Is Better?
Unlock the power of data and transform your business with HubSpot's comprehensive guide to data analytics.
Marketing software that helps you drive revenue, save time and resources, and measure and optimize your investments — all on one easy-to-use platform
.png)
How to build a research repository: a step-by-step guide to getting started
Research repositories have the potential to be incredibly powerful assets for any research-driven organisation. But when it comes to building one, it can be difficult to know where to start. In this post, we provide some practical tips to define a clear vision and strategy for your repository.

Done right, research repositories have the potential to be incredibly powerful assets for any research-driven organisation. But when it comes to building one, it can be difficult to know where to start.
As a result, we see tons of teams jumping in without clearly defining upfront what they actually hope to achieve with the repository, and ending up disappointed when it doesn't deliver the results.
Aside from being frustrating and demoralising for everyone involved, building an unused repository is a waste of money, time, and opportunity.
So how can you avoid this?
In this post, we provide some practical tips to define a clear vision and strategy for your repository in order to help you maximise your chances of success.
🚀 This post is also available as a free, interactive Miro template that you can use to work through each exercise outlined below - available for download here .
Defining the end goal for your repository
To start, you need to define your vision.
Only by setting a clear vision, can you start to map out the road towards realising it.
Your vision provides something you can hold yourself accountable to - acting as a north star. As you move forward with the development and roll out of your repository, this will help guide you through important decisions like what tool to use, and who to engage with along the way.
The reality is that building a research repository should be approached like any other product - aiming for progress, over perfection with each iteration of the solution.
Starting with a very simple question like "what do we hope to accomplish with our research repository within the first 12 months?" is a great starting point.
You need to be clear on the problems that you’re looking to solve - and the desired outcomes from building your repository - before deciding on the best approach.
Building a repository is an investment, so it’s important to consider not just what you want to achieve in the next few weeks or months, but also in the longer term to ensure your repository is scalable.
Whatever the ultimate goal (or goals), capturing the answer to this question will help you to focus on outcomes over output .
🔎 How to do this in practice…
1. complete some upfront discovery.
In a previous post we discussed how to conduct some upfront discovery to help with understanding today’s biggest challenges when it comes to accessing and leveraging research insights.
⏰ You should aim to complete your upfront discovery within a couple of hours, spending 20-30 mins interviewing each stakeholder (we recommend talking with at least 5 people, both researchers and non-researchers).
2. Prioritise the problems you want to solve
Start by spending some time reviewing the current challenges your team and organisation are facing when it comes to leveraging research and insights.
You can run a simple affinity mapping exercise to highlight the common themes from your discovery and prioritise the top 1-3 problems that you’d like to solve using your repository.

💡 Example challenges might include:
Struggling to understand what research has already been conducted to-date, leading to teams repeating previous research
Looking for better ways to capture and analyse raw data e.g. user interviews
Spending lots of time packaging up research findings for wider stakeholders
Drowning in research reports and artefacts, and in need of a better way to access and leverage existing insights
Lacking engagement in research from key decision makers across the organisation
⏰ You should aim to confirm what you want to focus on solving with your repository within 45-60 mins (based on a group of up to 6 people).
3. Consider what future success looks like
Next you want to take some time to think about what success looks like one year from now, casting your mind to the future and capturing what you’d like to achieve with your repository in this time.
A helpful exercise is to imagine the headline quotes for an internal company-wide newsletter talking about the impact that your new research repository has had across the business.
The ‘ Jobs to be done ’ framework provides a helpful way to format the outputs for this activity, helping you to empathise with what the end users of your repository might expect to experience by way of outcomes.

💡 Example headlines might include:
“When starting a new research project, people are clear on the research that’s already been conducted, so that we’re not repeating previous research” Research Manager
“During a study, we’re able to quickly identify and share the key insights from our user interviews to help increase confidence around what our customers are currently struggling with” Researcher
“Our designers are able to leverage key insights when designing the solution for a new user journey or product feature, helping us to derisk our most critical design decisions” Product Design Director
“Our product roadmap is driven by customer insights, and building new features based on opinion is now a thing of the past” Head of Product
“We’ve been able to use the key research findings from our research team to help us better articulate the benefits of our product and increase the number of new deals” Sales Lead
“Our research is being referenced regularly by C-level leadership at our quarterly townhall meetings, which has helped to raise the profile of our team and the research we’re conducting” Head of Research
Ask yourself what these headlines might read and add these to the front page of a newspaper image.

You then want to discuss each of these headlines across the group and fold these into a concise vision statement for your research repository - something memorable and inspirational that you can work towards achieving.
💡Example vision statements:
‘Our research repository makes it easy for anyone at our company to access the key learnings from our research, so that key decisions across the organisation are driven by insight’
‘Our research repository acts as a single source of truth for all of our research findings, so that we’re able to query all of our existing insights from one central place’
‘Our research repository helps researchers to analyse and synthesise the data captured from user interviews, so that we’re able to accelerate the discovery of actionable insights’
‘Our research repository is used to drive collaborative research across researchers and teams, helping to eliminate data silos, foster innovation and advance knowledge across disciplines’
‘Our research repository empowers people to make a meaningful impact with their research by providing a platform that enables the translation of research findings into remarkable products for our customers’
⏰ You should aim to agree the vision for your repository within 45-60 mins (based on a group of up to 6 people).
Creating a plan to realise your vision
Having a vision alone isn't going to make your repository a success. You also need to establish a set of short-term objectives, which you can use to plan a series of activities to help you make progress towards this.
Focus your thinking around the more immediate future, and what you want to achieve within the first 3 months of building your repository.
Alongside the short-term objectives you’re going to work towards, it’s also important to consider how you’ll measure your progress, so that you can understand what’s working well, and what might require further attention.
Agreeing a set of success metrics is key to holding yourself accountable to making a positive impact with each new iteration. This also helps you to demonstrate progress to others from as early on in the process as possible.
1. Establish 1-3 short term objectives
Take your vision statement and consider the first 1-3 results that you want to achieve within the first 3 months of working towards this.
These objectives need to be realistic and achievable given the 3 month timeframe, so that you’re able to build some momentum and set yourself up for success from the very start of the process.
💡Example objectives:
Improve how insights are defined and captured by the research team
Revisit our existing research to identify what data we want to add to our new research repository
Improve how our research findings are organised, considering how our repository might be utilised by researchers and wider teams
Initial group of champions bought-in and actively using our research repository
Improve the level of engagement with our research from wider teams and stakeholders
Capture your 3 month objectives underneath your vision, leaving space to consider the activities that you need to complete in order to realise each of these.

2. Identify how to achieve each objective
Each activity that you commit to should be something that an individual or small group of people can comfortably achieve within the first 3 months of building your repository.
Come up with some ideas for each objective and then prioritise completing the activities that will result in the biggest impact, with the least effort first.
💡Example activities:
Agree a definition for strategic and tactical insights to help with identifying the previous data that we want to add to our new research repository
Revisit the past 6 months of research and capture the data we want to add to our repository as an initial body of knowledge
Create the first draft taxonomy for our research repository, testing this with a small group of wider stakeholders
Launch the repository with an initial body of knowledge to a group of wider repository champions
Start distributing a regular round up of key insights stored in the repository
You can add your activities to a simple kanban board , ordering your ‘To do’ column with the most impactful tasks up top, and using this to track your progress and make visible who’s working on which tasks throughout the initial build of your repository.

This is something you can come back to a revisit as you move throughout the wider roll out of your repository - adding any new activities into the board and moving these through to ‘Done’ as they’re completed.
⚠️ At this stage it’s also important to call out any risks or dependencies that could derail your progress towards completing each activity, such as capacity, or requiring support from other individuals or teams.
3. Agree how you’ll measure success
Lastly, you’ll need a way to measure success as you work on the activities you’ve associated with each of your short term objectives.
We recommend choosing 1-3 metrics that you can measure and track as you move forward with everything, considering ways to capture and review the data for each of these.
⚠️ Instead of thinking of these metrics as targets, we recommend using them to measure your progress - helping you to identify any activities that aren’t going so well and might require further attention.
💡Example success metrics:
Usage metrics - Number of insights captured, Active users of the repository, Number of searches performed, Number of insights viewed and shared
User feedback - Usability feedback for your repository, User satisfaction ( CSAT ), NPS aka how likely someone is to recommend using your repository
Research impact - Number of stakeholder requests for research, Time spent responding to requests, Level of confidence, Repeatable value of research, Amount of duplicated research, Time spent onboarding new joiners
Wider impact - Mentions of your research (and repository) internally, Links to your research findings from other initiatives e.g. discovery projects, product roadmaps, Customers praising solutions that were fuelled by your research
Think about how often you want to capture and communicate this information to the rest of the team, to help motivate everyone to keep making progress.
By establishing key metrics, you can track your progress and determine whether your repository is achieving its intended goals.
⏰ You should aim to create a measurable action plan for your repository within 60-90 mins (based on a group of up to 6 people).
🚀 Why not use our free, downloadable Miro template to start putting all of this into action today - available for download here .
To summarise
As with the development of any product, the cost of investing time upfront to ensure you’re building the right thing for your end users, is far lower than the cost of building the wrong thing - repositories are no different!
A well-executed research repository can be an extremely valuable asset for your organisation, but building one requires consideration and planning - and defining a clear vision and strategy upfront will help to maximise your chances of success.
It’s important to not feel pressured to nail every objective that you set in the first few weeks or months. Like any product, the further you progress, the more your strategy will evolve and shift. The most important thing is getting started with the right foundations in place, and starting to drive some real impact.
We hope this practical guide will help you to get started on building an effective research repository for your organisation. Thanks and happy researching!

Work with our team of experts
At Dualo we help teams to define a clear vision and strategy for their research repository as part of the ‘Discover, plan and set goals’ module facilitated by our Dualo Academy team. If you’re interested in learning more about how we work with teams, book a short call with us to discuss how we can support you with the development of your research repository and knowledge management process.
Nick Russell
I'm one of the Co-Founders of Dualo, passionate about research, design, product, and AI. Always open to chatting with others about these topics.
Insights to your inbox
Join our growing community and be the first to see fresh content.
Repo Ops ideas worth stealing
Interviews with leaders

Related Articles

How top 1% researchers build UXR case studies

Navigating generative AI in UX Research: a deep dive into data privacy

Welcoming a new age of knowledge management for user research

Building a research repository? Avoid these common pitfalls

Unlocking hidden insights: why research teams must conduct meta-analysis

Unlocking the exponential power of insights – an interview with Zachary Heinemann
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- HHS Author Manuscripts

The Clinical Research Data Repository of the US National Institutes of Health
The US National Institutes of Health (NIH) includes 27 institutes and centers, many of which conduct clinical research. Previously, data collected in research trials has existed in multiple, disparate databases. This paper describes the design, implementation and experience to date with the Biomedical Translational Research Information System (BTRIS), being developed at NIH to consolidate clinical research data. BTRIS is intended to simplify data access and analysis of data from active clinical trials and to facilitate reuse of existing data to answer new questions. Unique aspects of the system includes a Research Entities Dictionary that unifies all controlled terminologies used by source systems and a hybrid data model that unifies parts of the source data models and include other data in entity-attribute-value tables. BTRIS currently includes over 300 million rows of data, from three institutes, ranging from 1976 to present. Users are able to retrieve data on their own research subjects in identified form as well as deidentified data on all subjects.
Introduction
The United States National Institutes of Health (NIH) consists of 27 institutes and centers (ICs) dedicated to biomedical research for improving the health of the public. Most ICs are located wholly or in part at the main NIH campus in Bethesda, Maryland, just north of Washington, DC. All clinical research on the Bethesda campus is coordinated through the Clinical Center (CC), the 242-bed, 90-day-station hospital of the NIH.
Much of the information collected on human subjects at the CC exists in the Clinical Research Information System (CRIS). Many researchers also collect data in other locations, including IC systems, laboratory systems within the ICs, and even individual researchers’ computers and notebooks. This data distribution causes two problems for researchers. First, CRIS is primarily an electronic medical record system, concerned with the tasks involved in patient care. Although it can support tasks related to research protocols, it is not designed to support research data analysis (e.g, queries for data across subjects in a clinical trial). Second, distribution of data across multiple sources complicates the ability of researchers to use the data to answer their research questions.
The clinical research data at NIH are also of interest to researchers besides those who are collecting them in the course of active trials. The US government mandates the sharing of clinical data that have been collected with federal funding, yet there is no mechanism at NIH to share the data that have been collected here for over half a century.
This paper describes the new NIH Biomedical Translational Research Information System (BTRIS), which has been providing investigators with access to clinical research data since July of 2009. Although still in evolution, BTRIS contains a substantial database and makes use of unique data models and terminology management techniques to merge data from multiple, disparate sources, to support active clinical trials and reuse of data.
NIH Efforts to Consolidate and Reuse Research Data
NIH initiatives have recognized the need for a clinical research data repository to support reuse and sharing of data for clinical research, including the NIH Roadmap NECTAR project,[ 1 ] the CABIG project through the National Cancer Institute[ 2 ] and the current Clinical and Translational Science Awards (CTSA) program.[ 3 ] Based on researcher requirements as well as a business case, the NIH endorsed the concept of a clinical research data repository for aggregation and re-use of data collected at the NIH itself (as opposed to data collected by NIH-funded projects at other institutions). Initial funding for the project was received in 2007 and the development of BTRIS began in earnest in 2008.
The Columbia University Clinical Data Repository
The initial design of BTRIS has been based on experience with the creation of the Clinical Data Repository (CDR) at the Columbia University Medical Center in New York.[ 4 ] That system has accrued patient care data since 1988 from many different sources, including laboratories, pharmacies, radiology departments, order entry, and clinician documentation. Over the years, the repository has supported a number of systems for clinical care[ 5 ] and clinical research.[ 6 ] All data in the Columbia CDR have been merged using a single, common relational data model that simplifies representation of disparate data while maintaining important distinctions and details.[ 7 ] The model makes extensive use of the “Entity-Attribute-Value” (EAV) approach, which allows specifications about the meanings of data to be stored with the data themselves, rather than being modeled as tables or columns in the data model. This method provides great flexibility for accommodating changes in data sources.[ 8 ]
The data in the Columbia CDR are represented with a single coding system, called the Medical Entities Dictionary (MED),[ 9 ] that unifies terminologies from all the sources providing data to the CDR. The MED provides a one-to-one mapping of individual concepts from each source and organizes them into a multiple-hierarchy ontology that provides definitional information about the concepts and supports data aggregation and inferencing functions.
Design Considerations
System architecture.
The first issue to be addressed in the BTRIS design was whether we should attempt to create a single, centralized repository (as was done at Columbia) or seek to create a federated system in which individual sources systems could be queried to provide data on demand. Although there are potential advantages to the federated model, [ 10 ] we quickly realized that most of the sources we would be dealing with (including archived repositories from defunct systems) would be incapable of participating in a federated design. We therefore proceeded to design a centralized repository.
In designing the BTRIS data model, we considered the various advantages and disadvantages of traditional modeling approaches and the EAV modeling approach. We chose to take a hybrid approach in which data from disparate sources (for example laboratory test results from CRIS, from archives of the system that preceded CRIS, and from various IC systems) are analyzed and commonalities (such as the fact that all laboratory tests have primary times and results) are represented with columns in tables, while distinct source-specific differences are captured in EAV tables.
In addition to data collected from research subjects, we recognized that we would need a repository of information about the subjects themselves, including the protocols with which they are affiliated and the dates of those affiliations. While some of this information is available from CRIS, much is missing and some individual data may be “tagged” with protocol affiliations that do not match the CRIS database.
Data Acquisition, Extraction, Translation and Loading
The approach to adding data to the BTRIS database is a fairly typical extraction, translation and loading (ETL) process. Acquired data are dissected into their component elements and converted into a form compatible with the BTRIS database. They are then stored in the appropriate tables, rows and columns, according to a set of mapping rules. Sources include archived files, copies of active databases, and collections of transaction messages (typically in HL7 format). Sources may provide data on a one-time basis (from archives) or on a periodic basis (typically, daily or weekly). Mapping rules are created manually for each source, based on careful analysis of the source systems’ documentation and the actual data provided (which do not always match the documentation).
Data Coding
Early in the project, we established a repository of controlled terminologies used by source systems to represent their data (for example, laboratory codes for tests and unique names for medications). This “Research Entities Dictionary” (RED) is based on the experience with Columbia’s MED: each source term corresponds to a unique concept in the dictionary, with additional knowledge about the terms represented in hierarchical and non-hierarchical semantic relationships between concepts. The ETL process maps individual data elements to their corresponding entries in the RED so that the RED Codes can be stored along with the original data. Although the source systems do not use standard terminologies, concepts in the RED are being mapped to international standards to facilitate data sharing, including those contained in the US National Library of Medicine’s Unified Medical Language System (UMLS).[ 11 ]
Data Reporting
Another key early decision in the system architecture was to determine that BTRIS users would perform retrievals themselves, using predetermined queries that could be tailored by the users for their specific needs. Given that a number of mature commercial “business intelligence” tools currently exist to support such capabilities, we evaluated several options and ultimate chose one to be our user interface to the database. System developers create query templates with general retrieval strategies (for example, to obtain demographic information or laboratory test results) and search filters (e.g., an age range, date range, type of laboratory test, or type of medication). Users provide values for the search filters when running the query to limit retrieval to specific subsets of data.
Queries were developed in response to a variety of perceived information needs. Some of these were identified in the original requirements gathering process for CRIS (see above), while others were developed through interactions with a BTRIS user group composed of interested NIH investigators.
Progress to Date
The BTRIS project officially began in January of 2008, with assembly of the development team in March, acquisition of sample data from several systems in May, and demonstration of a proof-of-concept prototype in July. The initial prototype used SQL Server (Microsoft, Redmond, WA) as the database management system, Terminology Development Editor (Apelon, Mountainview CA) for the RED, and Business Objects (SAP, Newton Square, PA) as the reporting tool.
Experience with the BTRIS prototype informed a number of changes in database design and user requirements, which led to the selection of Cognos (IBM, Armonk, NY) as the reporting tool. Based on the performance and user feedback with the prototype, approval for the project was secured in October. The first version of the actual BTRIS system was released on July 30, 2009 to PIs with active clinical protocols.
The Research Entities Dictionary
Each data source incorporated into BTRIS has one or more controlled terminologies that have been added to the RED. For example, the radiology system has a list of codes for procedures, while the laboratory has codes for tests, panels, organisms, antibiotics, specimens, and results. The RED currently contains 120,636 concepts with 155,321 hierarchical relationships (i.e., each concept has, on average, 1.3 parents).
Database Design
The BTRIS database contains five general sections, with information about investigators, protocols, subjects, the RED, and subject data. Investigator, protocol, and subject tables are related to each other in a typical manner.
Subject data are considered measurable (for data with normal ranges, such as laboratory tests), substance (for data with routes of administration, such as medications), and general (everything else). Data stored in event tables (for “things that happen”, such as orders and procedures) and observations (for “things that report something”, such as results and dosages given). Most events are associated with one or more observations. Each table has an associated EAV table ( Figure 1 ); thus, there are a total of 12 tables for subject data (three event tables, three observation tables, and an EAV for each).

Simplified view of part of the BTRIS data model. * = primary keys, [F]=foreign keys, [R]=elements coded in RED, [R+]=multiple column elements
The six main tables include information common to all data from all the sources, such as subject ID, source, name, etc. These tables also include data elements that have been judged to be similar enough across sources that they form part of the merged data model. For example, each source provides one or more times (including date) for observations; for each source, we choose one of these to be the primary time. For radiology procedures, it is the time the procedures were performed. For laboratory tests, it may be the specimen collection time, or it might be specimen analysis time. Other times for each source are retained in the EAV tables.
Another example of the merged data model can be found in the way results of observations are treated. In the BTRIS model, results of observations that have normal ranges (such as laboratory test results and vital sign measurements) are all stored in the same table. Separate columns are used to store the result (or parts of the result) that are numeric, text, controlled terms, and comments. Controlled term results are stored as they appear in the data and as the corresponding RED Codes. Observations without normal ranges, whether nurse’s notes, radiology reports, or discharge summaries, are all included as text results in the general observations table.
The RED is represented in two particularly interesting tables. One table relates every concept in the RED to one or more data sources, such that an identifier (such as a laboratory test code from the laboratory system or a medication name from the order entry system) can be uniquely mapped to a particular RED Code. This information is managed in the RED (see below) and exported to this table for use by the ETL process.
Another important RED table is the ancestor-descendant table. This table is derived from the RED hierarchy and supports class-based queries of the subject data. Figure 2 shows a simplified example of the use of the ancestor-descendant table for class-based queries. As of this writing, there are 1,214,646 ancestor-descendant relationships (that is, each concept subsumes, on average, ten concepts including itself).

Class-based query for “Anti-Platelet Drugs” using ancestor-descendant table.
Database Content
As of this writing, data have been accrued from three ICs: the Clinical Center (including CRIS and archived tapes of CRIS’s predecessor), the National Institute of Allergy and Infectious Diseases (NIAID), and the National Institute of Alcohol Abuse and Alcoholism (NIAAA). Data types include demographics, vital signs, laboratory test results, medication orders, medication administrations, medication lists, and problem lists. Additional data to be added in the near-term include clinical documents (e.g., progress notes and discharge summaries) and procedure notes (radiology, pathology, etc.). Next steps include obtaining radiology image data and gene sequence and expression data from the National Cancer Institute (NCI). Thus far, there are over 86 million rows in event tables, 180 million rows in observation tables, and 855 million rows in EAV tables. Data are derived from 436,422 subjects, 196,036 of whom have been affiliated with one or more of the 9,055 protocols involving 3180 investigators, for a total of 393,447 protocol-subject affiliations.
Thus far, we have created three types of reports for identifiable data: summary reports (enrollment inclusion report for the Institutional Review Boards (IRBs)), detailed data reports (demographics, vital signs, laboratory test results, and medication data) and “list” reports (which create lists of patients, tests and medications that can be used as filters for other reports). Summary reports and detailed reports for all of these data have been created for de-identified access as well. When running reports, users interact with the RED in one of two ways. A text-based search produces a list of terms from which users may select terms ( Figure 3 ). The users may also browse the RED hierarchy to select terms ( Figure 4 ).

Example of text-based terminology searches in BTRIS. Note that each of the selected terms will be used to query against the Ancestor-Descendant table.

Example of tree-based terminology search in BTRIS. A subsequent query will use the ancestor-descendant table to select all data with any of the 12 amiodarone medications.
User Experience
Currently, BTRIS has been providing access to identified data for 30 weeks; 111 users have logged on to date. Of the 4,349 reports run thus far, 3,015 have been to retrieve laboratory test results, 303 to retrieve medication information, 275 to retrieve vital signs, and 310 to create summary reports for IRBs. De-identified data have recently been made available. User feedback been extremely positive. Additional reports have been requested; the current BTRIS model appears to be capable of supporting these requests.
BTRIS is intended to encompass all clinical research data collected on subjects at the NIH. We began with some initial assumptions about design requirements and desired functionality and have proceeded rapidly through design, construction, and deployment. Our hybridization between column-oriented and EAV data models has allowed us to accommodate diverse data from disparate source in a way that supports aggregation across multiple data sources. The use of the RED and the EAV tables allows us to maintain the distinct aspects of data that are unique to their sources. The combination of the hybrid data modeling and the rich terminology representation provides a novel approach to the creation of a multi-purpose clinical data repository.
Thus far, BTRIS appears to be meeting the needs for researchers to obtain identified data on active clinical protocols. BTRIS is poised to provide access to de-identified NIH data, across protocols, to analyze old data in new ways and ask new questions. We do not yet know how our researchers will make use of such functionality, but we believe that it will be in creative, unforeseen ways. BTRIS is designed to be flexible enough to meet a wide variety of such needs.
In particular, coding data with the RED supports queries that can aggregate or distinguish data as needed for the users’ purposes. For example, instances of the administration of a 325mg aspirin tablet will be retrieved when a user requests that specific information, or all instances of the administration of any aspirin, any analgesic, any antipyretic, any platelet inhibitor, or simply any drug of any kind.
Elements of different data sources that have been stored in common columns in our six main tables have been carefully chosen to support what we believe will be the kinds of data aggregation that researchers are likely to want. For example, a user interested in the use of aspirin in a set of research subjects can request all instances of aspirin administration from the CRIS system, all instances of aspirin orders from the CRIS system, all instances of aspirin on a subject’s medication list from the NIAID system, or a combination of any of these. Together with the flexible class-based queries supported by the RED, users have a range of ways to retrieve desired data.
The commercial reporting tool we have chosen (Cognos), allows us to create a variety of reports that appear to meet many of the users’ needs, while giving them the power to tailor their queries and immediately obtain results. However, we fully expect that there will be information needs that will not be easily met with this approach. For example, a user may require a complex query that makes use of data from several main tables and EAV tables. In these cases, we may create specialized reports within Cognos, or we may perform retrievals, directly against the database, on the user’s behalf.
In addition to the technical challenges, the development of BTRIS has required addressing a variety of policy issues that are beyond the scope of this paper. We have been successful at overcoming these issues in ways that address human subjects protection, privacy, data ownership, data access, and data sharing concerns. Solutions have required combinations of administrative and technical methods.
As with any system, BTRIS is faced with a number of potential limitations, particularly with regard to scaling (as we add image and genomic data), scope (as we add new data from institutes) and performance. Thus far, however, we have been able to address these issues and are not yet close to reaching capacity. BTRIS is still very much in development, as we add new reports for identified data, explore creative ways to reuse de-identified data, and expand to include new sources and types of data. The NIH has demonstrated deep commitment to creating a repository that serves all of the NIH community and eventually the research community at large, for the betterment of the health of humankind.
BTRIS addresses a long-standing need to consolidate clinical research data across the NIH for a variety of purposes. Our design includes a combination of novel approaches, development has been rapid, and it is already successfully addressing the information needs of NIH researchers.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH.

Research Data Management
- Research data lifecycle
- Searching for research data
- NTU Research Data Policy
Defining Research data
- Classification, Storage & Handling
- Anonymisation This link opens in a new window
- NTU DMP Template v3 (18 Jun 2020 - )
- NTU DMP Template v2 (15 Jan 2018 - 17 Jun 2020)
- Where to share data?
- How to share data?
- Current Workshops This link opens in a new window
- Singapore Open Research Conference 2024 This link opens in a new window
- Singapore Open Research Conference 2022 This link opens in a new window
- DR-NTU (Data) This link opens in a new window
- Open Science This link opens in a new window
- Blog This link opens in a new window
An Introduction to the Basics of Research Data
An animated video that explains the basics of research data.
(Source: Created by Louise Patterton of the Council for Scientific and Industrial Research in South Africa)
Giving an authoritative definition to “research data” is challenging. Different disciplines usually develop their own preferred definition which is appropriate and suitable for their domain.
Some higher education institutions around the world have developed their definition in developing their policy on research data.
Definition by Nanyang Technological University
“Research data are data in whatever formats or form collected, observed, generated, created and obtained during the entire course of a research project. This would include numerical, descriptive, aural, visual or physical forms recorded by the researcher, generated by equipment and derived from models, simulations.”
(Source: NTU Research Data Policy )
Definition by National Medical Research Council
The National Medical Research Council (NMRC) from Ministry of Health (MOH) in Singapore has defined it in its ' Research Data Governance and Sharing ' briefing slides presented in March 2015 data sharing roadshows as:
"Recorded factual material commonly accepted in the scientific community as necessary to document and support research findings."
(Source : National Medical Research Council )
Further readings:
- What is Research Data? (Source: Yale University)
- Service Standards and Policies/EPSRC Policy Framework on Research Data/ Scope and Benefits (Source: Engineering and Physical Sciences Research Council [EPSRC])
- Defining Research Data (Source: University of Oregon)
- Management of Research Data and Records Policy (Source: University of Melbourne)
- Research Data Management: Staff, Adjuncts and Visitors Procedures (Source: Monash University)
- Management of Research Data (Source: Queensland University of Technology)
- Research Data in Context (Source: MANTRA)
- << Previous: NTU Research Data Policy
- Next: Classification, Storage & Handling >>
- Last Updated: May 24, 2024 3:27 PM
- URL: https://libguides.ntu.edu.sg/rdm
You are expected to comply with University policies and guidelines namely, Appropriate Use of Information Resources Policy , IT Usage Policy and Social Media Policy . Users will be personally liable for any infringement of Copyright and Licensing laws. Unless otherwise stated, all guide content is licensed by CC BY-NC 4.0 .
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 22 May 2024
From Planning Stage Towards FAIR Data: A Practical Metadatasheet For Biomedical Scientists
- Lea Seep ORCID: orcid.org/0000-0002-0399-1896 1 ,
- Stephan Grein 1 ,
- Iva Splichalova 2 ,
- Danli Ran 3 ,
- Mickel Mikhael 3 ,
- Staffan Hildebrand 3 ,
- Mario Lauterbach 4 ,
- Karsten Hiller ORCID: orcid.org/0000-0001-9322-5820 4 ,
- Dalila Juliana Silva Ribeiro 5 ,
- Katharina Sieckmann ORCID: orcid.org/0009-0008-0336-8835 5 ,
- Ronja Kardinal 5 ,
- Hao Huang ORCID: orcid.org/0000-0003-3878-3947 2 ,
- Jiangyan Yu 1 , 6 ,
- Sebastian Kallabis 7 ,
- Janina Behrens 8 ,
- Andreas Till 9 ,
- Viktoriya Peeva 9 ,
- Akim Strohmeyer 10 ,
- Johanna Bruder 10 ,
- Tobias Blum 11 ,
- Ana Soriano-Arroquia 3 ,
- Dominik Tischer 3 ,
- Katharina Kuellmer 10 ,
- Yuanfang Li 12 ,
- Marc Beyer ORCID: orcid.org/0000-0001-9704-148X 12 , 13 ,
- Anne-Kathrin Gellner 14 , 15 ,
- Tobias Fromme 10 ,
- Henning Wackerhage 16 ,
- Martin Klingenspor 10 , 17 , 18 ,
- Wiebke K. Fenske 9 , 19 ,
- Ludger Scheja 8 ,
- Felix Meissner 7 , 20 ,
- Andreas Schlitzer 6 ,
- Elvira Mass ORCID: orcid.org/0000-0003-2318-2356 2 ,
- Dagmar Wachten ORCID: orcid.org/0000-0003-4800-6332 5 ,
- Eicke Latz 5 ,
- Alexander Pfeifer 3 , 21 &
- Jan Hasenauer 1 , 22
Scientific Data volume 11 , Article number: 524 ( 2024 ) Cite this article
3 Altmetric
Metrics details
- Data acquisition
- Research management
Datasets consist of measurement data and metadata. Metadata provides context, essential for understanding and (re-)using data. Various metadata standards exist for different methods, systems and contexts. However, relevant information resides at differing stages across the data-lifecycle. Often, this information is defined and standardized only at publication stage, which can lead to data loss and workload increase. In this study, we developed Metadatasheet, a metadata standard based on interviews with members of two biomedical consortia and systematic screening of data repositories. It aligns with the data-lifecycle allowing synchronous metadata recording within Microsoft Excel, a widespread data recording software. Additionally, we provide an implementation, the Metadata Workbook, that offers user-friendly features like automation, dynamic adaption, metadata integrity checks, and export options for various metadata standards. By design and due to its extensive documentation, the proposed metadata standard simplifies recording and structuring of metadata for biomedical scientists, promoting practicality and convenience in data management. This framework can accelerate scientific progress by enhancing collaboration and knowledge transfer throughout the intermediate steps of data creation.
Similar content being viewed by others
Machine actionable metadata models

Developing a standardized but extendable framework to increase the findability of infectious disease datasets
Modeling community standards for metadata as templates makes data FAIR
Introduction.
Collaboration along with the open exchange of techniques, protocols and data is the backbone of modern biomedical research 1 . Data usage and retrieval requires the structured collection of information, such as study design, experimental conditions, sample preparation and sample processing, on the performed measurements. This information is generally referred to as metadata, which grows along the research data-lifecycle (Fig. 1A ), from planning to its final storage alongside publication 2 , 3 , 4 , 5 , 6 . There is a growing consensus among researchers, journals and funding agencies that data should adhere to the principles of being findable, accessible, inter-operable and reusable (FAIR). The adherence to these FAIR data principles 7 requires metadata 8 , 9 .
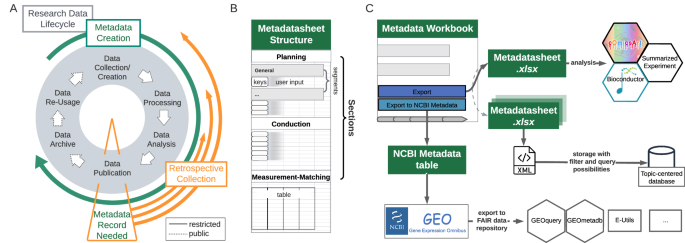
Alignment of Metadata Lifecycle with the Research Data-Lifecycle. ( A ) Metadata is created alongside the research data creation, however, often only gathered at the point of publication when it is requested from, e.g., repositories, marking a clear decisive point before open accessibility of produced data. ( B ) The structure of the proposed Metadatasheet is defined by its sections, which further encompass segments. Within each segment user input is required, which can be of different forms, e.g., values to keys or table entries. ( C ) Upon complete records, the Metadata Workbook can export either to a plain xlsx file or to the requested NCBI GEO metadata format. Deposited data can be accessed by a plethora of tools (examples given). Outside the workbook a single xlsx file can be converted to a SummarizedExperiment object for data analysis, multiple Metadatasheets can be transformed to xml files using the provided ontology to build the input for a topic-centred database.
Metadata for an experiment exists in different formats and locations including handwritten notes (in classical labbooks), electronic Notebooks (e.g., RSpace 10 or Signals 11 ) and various (more-or-less) standardized electronic formats (e.g. automatic measurement machine output for experimental systems). The choice of recording systems often depends on the individual scientist conducting the experiment or his/her research group 12 . Recording supporting tools can be the open source ISA-tool suite 13 or commercial solutions such as Laboratory-Information management systems (commonly referred to as LIMS). Successful management can yield in high quality data deposited on trustworthy digital repositories. Trustworthiness is marked by Transparency, Responsibility, User focus, Sustainability and Technology (TRUST) 14 .
Repositories are subdivided into cross-discipline and domain-specific categories. Cross-discipline repositories intentionally do not impose any requirements on format or size to allow sharing without boundaries. Domain-specific repositories in the field of biomedicine impose requirements during submission in form of data and metadata standards. Example biomedical domain repositories are BioSample and GEO 15 , maintained by the National Center for Biotechnology Information (NCBI), or PRIDE 16 and BioModels 17 , 18 , maintained by European Bioinformatics Institute (EBI).
Standards often make use of controlled vocabularies and ontologies to ensure consistency and comparability. Controlled vocabularies, consisting of standardized terms, describe requested characteristics and keys 5 , while ontologies, such as the Gene Ontology (GO) 19 , establish structured frameworks for depicting relationships between entities, fostering comprehensive and searchable knowledge structures. Current metadata standards can be divided into two categories. First, comprehensive high-level documents that are often tailored to specific use cases. These documents primarily consist of lists of requested terms or guidelines, often interconnected with corresponding ontologies. For instance, ARRIVE (Animal Research: Reporting of In Vivo Experiments) provides a checklist of information to include in publications of in vivo experiments 20 or MIRIAM (minimum information requested in the annotation of biochemical models) 21 standardizes the curation of biochemical models including their annotations. Second, there are structured metadata standards supplied and requested by respective repositories. Irrespective of the suitable metadata standard, it is common to adhere to requested standards at the stage of data publication evoking a retrospective collection (Fig. 1A ). Necessary information resides at all stages of the data-lifecycle and may involve different responsible individuals, thereby rendering the retrospective metadata collection resource-intensive. Furthermore, data scientists or third parties, not involved in data acquisition, dedicate most of their time to cleaning and comprehending the data 22 . This task becomes particularly challenging when lacking explicit experimental knowledge. On a large scale, data curation companies might be involved.
Despite the existence of various metadata standards in biomedical sciences and widespread recognition of the relevance of metadata, a practical issue persists: the absence of a dedicated metadata standard that effectively and with low burden directs researchers in capturing metadata along the data-lifecycle without loss of information, towards FAIRness during and after the experiment (Fig. S1 ). Standardized metadata capture lowers the researcher’s efforts and enhances the suitability and turn over of data and metadata for repositories and therefore availability for third parties 23 . Thus, we propose a metadata standard tailored for wet-lab scientists mirroring the phases of the biomedical research lifecycle, offering transferability across distinct stages and among diverse stakeholders.
The proposed standard, further referred to as Metadatasheet, is embedded in a macro-enabled Excel workbook, further referred to as Metadata Workbook. The Metadata Workbook offers various usability features, such as automation, integrity checks, extensive documentation, usage of templates, and a set of export functionalities to other metadata standards. By design, the proposed Metadatasheet, accompanied by the Metadata Workbook, naturally allows stage-by-stage collection, embodying a paradigm shift in metadata collection strategies, promoting the efficient use of knowledge in the pre-publication phase and its turn-over to the community.
The Metadatasheet is based on comprehensive interview of biomedical researchers
Metadata information consists of a set of characteristics, attributes, herein named keys, that intend to provide a common understanding of the data. Example keys are experimental system, tissue type, or measurement type. Accordingly, the Metadatasheet is built upon requested keys gathered from comprehensive interviews of research groups and systematical collection from public repositories. In the initial phase, more than 30 experimental researchers from the biomedical sciences participated, who were from two consortia focusing on metaflammation ( https://www.sfb1454-metaflammation.de/ ) and metabolism of brown adipose tissue ( https://www.trr333.uni-bonn.de/ ). The participating researchers reported common general keys as well as diverse experimental designs covering five major experimental systems and 15 common measurement techniques, each accompanied by their specific set of keys. To refine and enhance the set of metadata keys, we engaged in iterative consultations with biomedical researchers. In parallel, we systematically collected relevant keys from three popular public repositories, namely NCBI’s GEO 15 , the Metabolomics Workbench 24 and the PRIDE 16 database. Moreover, expected input, summarized under the term ‘controlled vocabulary’, for all keys needed to be specified. From second iteration on, specifications of the controlled vocabulary, as well as the set of keys, were improved based on researchers’ feedback. The comprehensive key and controlled vocabulary collection process revealed the dynamic, unique and growing requirements of different projects, in terms of values within the controlled vocabulary and performed measurements. Those requirements lead to the choice of allowing customisation and expansion of key sets and controlled vocabulary as an integral part of the Metadatasheet. To handle the dynamic and adaptable nature of the Metadatasheet, it was embedded within a reactive framework with additional functionalities, the Metadata Workbook.
In the following, the overall concept and design of the Metadatasheet is introduced, afterwards key aspects of the Metadata Workbook are highlighted. The results section concludes with an example Metadatasheet generated by the Metadata Workbook.
The Metadatasheet design follows and allows metadata recording along the data-lifecycle
The proposed Metadatasheet is organized into three main sections: ‘planning’, ‘conduction’ and ‘measurement-matching’ section. These sections mirror the stages of the data-lifecycle and align with the general experimental timeline (Fig. 1B ). The analogous top-to-bottom structure allows sequential metadata recording, acknowledging the continuous growth of metadata. Each section further subdivides into segments, which hold the keys, that need to be specified by the user through values. The segmentation aims to group keys into logical units, that are likely provided by a single individual. This grouping enables the assignment of responsible persons, resulting in a clear emergent order for data entry if multiple persons are involved. Moreover, within a section the segments are independent of each other, allowing also parallel data entry.
Metadatasheet keys can be categorized based on the form of the expected input. First, providing a single value (key:value pair), e.g. the analysed ‘tissue’ (key) originates from the ‘liver’ (value). Second, filling tables, whereby the row names can be interpreted as keys, but multiple values need to be provided (one per column). Third, changing a key:value entry to a table entry by the keyword ‘CHANGES’. If the keyword is supplied as a value, the respective target key changes from key:value pair to a table entry. The switch of form allows data entries to be minimal if sufficient or exhaustively detailed if needed. This flexible data entry minimizes the need for repetition, gaining easier readability but allows recording fine-grained information whenever needed.
Required values can be entered in form of controlled vocabulary items, date-format, free text including numbers or filenames. Filenames are a special type of free text and specify additional resources, where corresponding files are either expected within the same directory as the Metadatasheet itself or given as relative path. Suitable form of values is naturally determined by the key, e.g., ‘Date’ is of date format, ‘weight’ is of number format and ‘tissue’ of discrete nature to be selected from the controlled vocabulary. The format choice is constraining the allowed values. Providing such input constraints to each key, allows harmonization of metadata. Harmonization enables machine readability, which is a starting point for further applications.
A single Metadatasheet captures the combination of an experimental design and a measurement type, as those come with a distinct set of keys, also referred to as dependent keys. An experimental design is here defined as a specific experimental system exposed to a contrasting setting. Within the Metadatasheet five contrasting settings, herein named comparison groups, are set: ‘diet’, ‘treatment’, ‘genotype’, ‘age’, ‘temperature’ and ‘other’ (non-specific). Experimental designs exhibit a range of complexities, they can span multiple comparison groups such as different treatments exposed to different genotypes, while each group can have multiple instances such as LPS-treatment and control-treatment.
The varying complexity in experimental designs is reflected in the Metadatasheet structure. This reflection is achieved through hierarchies, organized into up to three levels. The top-level keys are mandatory, while the inclusion of other-level keys depends on the design’s complexity. Present hierarchies within the samples are also important to consider for statistical analysis. Hierarchies emerge, if the sample is divided into subsamples prior to the measurement. For instance, if the experimental system involves a mouse with two extracted organs for measurement, the relation to the sample should be specified. Moreover, subsamples are also present when measurements were conducted on technical replicates of the extracted sample. The Metadatasheet accommodates up to two levels of sample partitioning. By leveraging a hierarchical structure, details are displayed only when necessary, avoiding unnecessary intricacies. Moreover, relationships of the measured samples can be recorded, enhancing clarity.
To ensure coherence between a sample’s actual measurement data and recorded metadata, it is crucial to link them accurately by a unique personal ID. To guide through matching and prevent mismatches, we have designed the Measurement-Matching section to summarize essential information and focusing on differences between samples. This information includes their association with an instance of a comparison group, the number of replicate, and the presence or absence of subsamples. If subsamples are present, they are organized in a separate table, referencing their higher, preceding sample. Careful recording also involves specified covariates. They are expected at the lowest level, the measurement level, and must be carefully matched to the correct ID within the set of replicates within a comparison group instance.
The inherent innovative force within the research community risks hitting boundaries of anything predefined, here, particularly evident in controlled vocabulary and dependent keys. Those predefined sets come as additional tables, associated with the Metadatasheet. Subsequently, the resources of the Metadatasheet require an ongoing commitment to be extended and further developed. Ontology terms can be integrated into every controlled vocabulary set. If necessary, users can search for the appropriate terms outside the Metadata Workbook using services such as the Ontology Lookup service 25 or OntoBee 26 . The separation of the Metadatasheet and its resources also allows the creation of group-specific subsets of controlled vocabulary. This feature proves helpful when a group wants a more constrained set of controlled vocabulary, e.g., using specific ontologies and respective value specifications. The ontology terms intended for use are incorporated into the controlled vocabulary set, ensuring that users only have access to those terms. The group-specific validation should be a subset of the overall validation.
The Metadatasheet design aligns with the data-lifecycle to allow analogous metadata recording. The presented design choices allow to adapt to various settings biomedical researchers are confronted with and thereby provide a high degree of flexibility.
The implementation of the metadatasheet, the metadata workbook, enhances user experience by automation, integrity checks, customisation and export to other formats
Gathering the diverse resources, specifically the Metadatasheet, the validation and dependent fields resources, we created an Excel Workbook including all of those sheets. To promote usage through user-friendliness, dynamic adaption and automation, we further introduced Excel macros (a set of custom functions) resolving to a macro enabled Excel workbook, called the Metadata Workbook. This Metadata Workbook is designed to guide the Metadatasheet application while providing automation whenever possible. Advancements through the implementation include specifically the ability to automatically insert dependent keys, enhance user experience and updating the controlled vocabulary. Additionally, there are options to use templates, automatic input validation and export functions that enable long-term storage. Crucial advancements are explained in more detail in the following.
The Metadata Workbook creates tailored Metadatasheets for common biomedical experimental systems and measurement techniques. Those segments come with their unique set of dependent keys and therefore change between individual Metadatasheets. Static sheets result therefore in a high amount of sheets. The Metadata Workbook provides a dynamic solution, reducing different requirements to a single Metadata Workbook that needs to be handled. The dependent, inserted keys, can be extended, but not changed, by adding values to the respective column within the dependent field sheet. The new addition is automatically added to the validation sheet, holding the controlled vocabulary. For new additions, the key’s input constraints can be changed. These features enable flexibility through expansion, allowing to match current and future research contexts.
The Metadata Workbook employs various features to enhance user experience and convenience while facilitating to capture simple to advanced setups of an experiment: sections of the sheet collapse, such as second levels of hierarchical segments, if not applicable; DropDown menus based on the provided controlled vocabulary enrich value fields, facilitating ease of selection. Furthermore, visual cues notify users in several situations: any segment where the structure deviates from the typical key:value format to adapt to a tabular arrangement is highlighted automatically; text-highlighting is used to mark mistakes, e.g., if input values for key fields do not align with the controlled vocabulary. Altogether, Metadata Workbook provides a user-friendly environment to guide users to record metadata.
Disruptive redundancy across and within the proposed Metadatasheet is tackled within the Metadata Workbook. Redundancy across Metadatasheets occurs if multiple studies are conducted in the same context, with similar designs, systems or experimental techniques. To reduce redundancy and prevent mistakes from copying and pasting, existing Metadatasheets can serve as templates. All information from the first two sections (planning and conduction) are exported from an uploaded Metadatasheet. Upon upload, users only need to update the ID information in the Measurement-Matching section for the new setting. This exception prevents not updating these crucial IDs. Redundancy within a single Metadatasheet occurs while providing the ‘final groups’ as well as the table within the Measurement-Matching section at the beginning of section two and three, respectively. The Metadata Workbook provides ‘generate’ buttons to produce both those tables automatically. Hence, the first ‘generate’ button creates all possible combinations based on the Planning section, while the measurement-matching table is generated based on the Conduction section. To maintain structural integrity, the Metadata Workbook requires a sequential input of the sections. The generate buttons prevent violations by evoking an error if input in the preceding section is invalid. The ‘generate’ functionalities remove through automation the need for copy paste actions and redundant actions for the user.
Upon the completion of the Metadata Workbook, it can be exported to various formats serving different objectives. Current supported formats are xlsx, the NCBI GEO metadata format, SummarizedExperiment (an R object specification from the Bioconductor family 27 , 28 ) and xml. Through export functionality, users gain several benefits, such as compatibility with open-source software, long-term storage through TRUST repositories and minimization of work by don’t repeat yourself (DRY) principles 29 . Compatibility of the Metadata Workbook with open-source software, like LibreOffice, is facilitated by the export option to a simple Excel ( xlsx file type) file while simultaneously removing any associated functionalities. Notably, a unique identifier is automatically assigned upon export. Providing metadata represents a critical prerequisite before uploading data to repositories or publication. Repositories normally adhere to their distinct metadata standards. Some offer submission tools featuring user interfaces, e.g. MetabolomicsWorkbench. Conversely, others like GEO or NCBI require the manual completion of an Excel table. For both repositories, export capabilities have been added to transform the Metadata Workbook compliant with the repositories’ requirements. The proposed structure covers all mandatory fields from the major repositories. These export functionalities reduce the hours spend on reformatting to meet different requirements and are a crucial step towards DRY principles within the metadata annotation procedure. Further, a converter is provided that turns the proposed structure, given as an exported xlsx file, to an object, commonly used as input to data analysis. The converter, applicable to omics-data and associated metadata, returns an R object called SummarizedExperiment 30 . The SummarizedExperiment object can be easily shared and lays the foundation for a plethora of standardized bioinformatic analyses within R. The object contains all available metadata from previous data-lifecycle stages limiting issues due to missing information, like unmentioned covariates.
In essence, the introduced implementation results in a macro-enhanced Excel Workbook, the Metadata Workbook, with advanced functionalities that choose the appropriate keys, enhances user experience with colour cues and automation while maintaining data integrity.
Showcase and application of the metadatasheet demonstrate its use in recording metadata and subsequent data analysis
To assess the suitability and adaptability of the designed Metadatasheet, we asked researchers from 40 different groups to gather and transfer their metadata in this format. The initiation of capturing standardized metadata alongside the data generation process has made a range of practical applications possible, yielding multiple advantages within the consortia. The versatility of the proposed structure is demonstrated by a curated collection of sheets (Table 1 ), each accompanied by a concise description of the study’s setting. The provided selection encompasses various measurement types and differing experimental systems. The experimental designs within this selection range from straightforward setups to nested designs, as well as two-way comparisons. For all complete Metadatasheets, see Supplementary Material. As the Metadatasheet records metadata from the start of the data-lifecycle, some measurement data in certain showcases is not included here due to its non-disclosure status before publication.
In the following, a single Metadatasheet from the showcase collection is highlighted, which has been created with the Metadata Workbook. The picked Metadatasheet for demonstration encompasses one of the datasets associated with the study of developmental programming of Kupffer cells by maternal obesity 31 . The associated data is deposited on GEO and are accessible through GEO Series accession number GSE237408 .
Example planning section
The Metadatasheet starts with the Planning section which captures all information already available during the conceptualization of an experiment. The section is subdivided into the segments ‘General’, ‘Experimental System’ and ‘Comparison groups’ (Fig. 2 ). The requested information in ‘General’ (Fig. 2A ) includes personal information, the title of the project as well as the specification whether the sheet is part of a collection of multiple related Metadatasheets. Collections allow users to link individual Metadatasheets from the same project to spread awareness of such connections, in this example linking multiple datasets associated with the same project. ‘Experimental System’ segment provides automatically predefined keys (dependent fields sheet) after the selection within the Metadata Workbook, for example, ‘line’ and ‘genotype’ information will be needed upon selecting ‘mouse’ (Fig. 2B ). To illustrate the incorporation of ontology terms, note the BRENDA Tissue Ontology (BTO) term for tissue type.
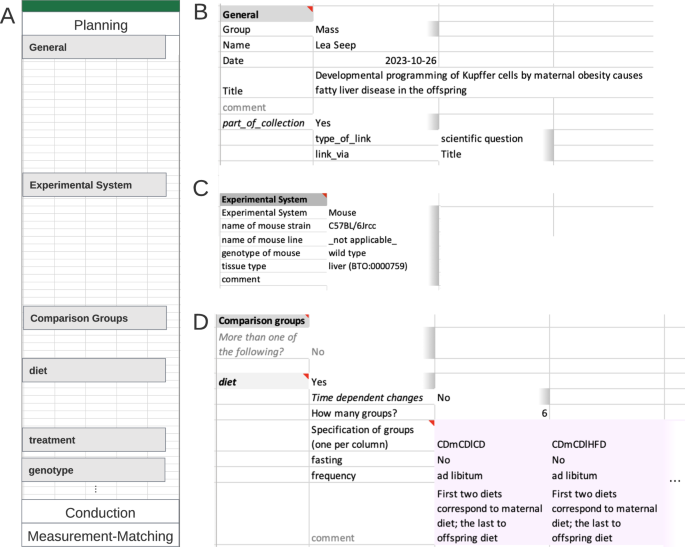
Example of an instance of the Planning section. ( A ) Overview Planning section. ( B ) General segment contains contact information and general project information in form of key:value pairs; on its second level, linked Metadatasheets can be specified. ( C ) The experimental system segment is requesting keys dependent on the value given to key ‘Experimental System’. For tissue type, the controlled vocabulary encompasses ontology terms taken from BRENDA Tissue Ontology (BTO). ( D ) Comparison group segment; here the only comparison group is ‘diet’. defined through diet (other comparison group options as treatment etc. not shown). As six groups are requested by the user a table is present with six columns (only two shown). Information per specified group is expected column-wise. Note that the full Metadatasheet of this example can be found in Supplementary Material.
The ‘Comparison groups’ segment (Fig. 2C ) specifies the experimental design linked to the current research question. The experiment design for each comparison group involves two levels: broader comparison group, here ‘diet’ and details for each instance within the broader comparison group. Users are not restricted to a single comparison group. At the second level, details for each chosen comparison group are entered. Here, 6 different groups with varying diet schemes were studied. The established feeding scheme is unique within the consortia, those special requirements were easily added to the controlled vocabulary for ‘diet’ with the Metadata Workbook, leveraging on its adaptability.
Example conduction section
The Conduction section is divided into six segments and captures all information created during the experimental/ wet-lab phase. The section starts with the specification of the ‘final groups’ resulting from previously specified comparison groups. As diet is the only comparison group with six instances, the final groups resolve to those types (Fig. 3A ). If multiple groups are planned, for example, if six diet groups and two genotype groups, 12 final groups would be present due to all combination possibilities. Within the Metadata Workbook those final groups are generated automatically, the user then defines the respective replicates.
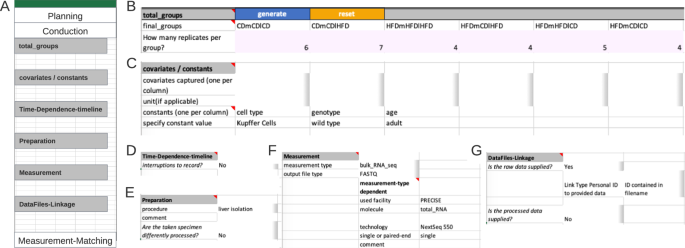
Example of an instance of the Conduction section. ( A ) Overview Conduction section. ( B ) The ‘total_groups’ segment expects all possible combinations of the comparison groups defined in the Planning section. Number of replicates belongs underneath each group. In the Metadatasheet implementation, ‘final_groups’ are generated; pink colour marks an expected table. ( C ) The segment covariates/constants requests respective specification including units. For constants, the value is expected in place, whereas covariates values are expected within the measurement-matching table. ( D ) Time-Dependence-timeline segment collapses completely if not required. ( E ) Preparation segment expects the procedure that is required before the actual measurement. Here, the reference to either a fixed protocol, chosen from the controlled vocabulary or a filename is expected. The specified file is expected to be on the same level as the Metadatasheet in the filesystem. ( F ) The Measurement segment is requesting keys depending on the value given to key measurement type. ( G ) The DataFiles-Linkage segment specifies how to identify the correct measurement file given the subsequent (within the measurement matching section) specified personal ID. If there is no clear pattern, one can choose keyword ‘CHANGES’ to promote filename specification to the measurement matching section. Note that the full Metadatasheet of this example can be found in Supplementary Material.
The segment ‘Covariates/Constants’, expects each constant or covariate to fill a single column with the respective suitable unit (table form). For clarification, a covariate refers to any additional variable or factor, beyond the main variables of interest (comparison groups), that is considered or observed in the experimental design. This could include factors such as age, gender, environmental conditions but also unusual colour of serum or day of preparation. Here, no covariate but the constants ‘cell type’ and ‘genotype’ were recorded, respective values, ‘Kupffer Cells’ and ‘wild type’ occupying a single column each (Fig. 3B ).
The next segment ‘Time-Dependence-Timeline’ is organized hierarchically. On the first level, one decides whether this segment is applicable, by answering if interruptions are present. The presence of an interrupted timeline is given, when the designated comparison group is to be augmented with temporal details that occurred during the experimental period. The second level distinguishes between two types of an interrupted timeline: ‘continued’ and ‘discontinued’. A ‘continued’ timeline is identified when temporal details are annotated. On the other hand, if the temporal details describe a change, such as a modification in treatment, it falls under the ‘discontinued’ type. For example, an interrupted timeline is present when a mouse undergoes several glucose tolerance tests during a contrasting diet setting (interrupted timeline type continued), or when a treatment consists of administering agent A for 24 hours followed by agent B for the next 24 hours (interrupted timeline type discontinued) before the actual measurement. While not present in the example at hand, both types of interrupted timelines would require further details (Fig. 4A ).
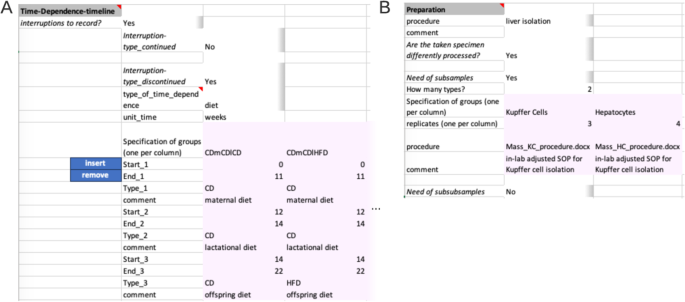
Advanced example of segments within the Conduction section. ( A ) Within the Time-Dependence Timeline segment, given comparison groups can be enriched with time dependent information on the second hierarchy level. One specifies which of the comparison groups is to be enriched with timeline information and the unit of time. Then, time-steps can be specified. Pink colour marks the table, which needs to be filled. ( B ) Within the Preparation segment, one can supply up to two divisions of the original experimental system sample. Here, from the liver of mice, two cell types are isolated. The liver isolation has the same protocol, while cell type isolation has differing protocols. The respective files are expected to be on the same level as the Metadatasheet in the filesystem.
The next two segments ‘Preparation’ (Fig. 3D ) and ‘Measurement’ (Fig. 3E ) capture the information for sample preparation approaches and measurement techniques, respectively. The ‘Preparation’ segment holds the information about the process of the experimental system to the specimen that gets measured. The respective protocol can be selected from a predefined set of terms, such as common workflows or entering a filename in the designated comment field, as shown here. When there are subsamples present (Fig. 4B ), information at segments’ secondary level is necessary, such as the number of subsamples per sample, their instances, replicates, and preparation information must be provided in a tabular format. The ‘Measurement’ segment requests details depending on the respective choice of the measurement technique (Fig. 3E ). Note, that ‘used facility’ was an additional dependent key added upon the process of filling the Metadatasheet. The user can easily add further keys by entering the wanted key in both dependent fields sheet in respective column of Measurement type: ‘bulk_RNA_seq’ and specify its type of constraints, e.g., free-text, date or controlled vocabulary, within the ‘Validation’ sheet.
The final segment ‘DataFiles-Linkage’ (Fig. 3F ), connects the measurement results with metadata. On the first level, one specifies whether raw or processed data is available. Raw data denotes the original machine-generated output, untouched by any processing, here the raw data are the fastq files. At secondary levels, users would provide more details about their file naming system. Three options are provided: ‘ID contained in filename’, ‘single file for all’, and ‘CHANGES’. The options ‘ID contained in filename’ and ‘single file for all’ require the data to be positioned at the same level as the metadata document within a file system, whereby relative paths can be given. The option of ‘CHANGES’ (switching key:value pair to tabular form) allows the user to define their unique naming system in the Measurement-Matching section. For processed data the procedure is required, and to be provided like the preparation protocol.
Example measurement-matching section
The last but the most important step for Metadatasheet is the ‘measurement-matching’ section, which links the recorded metadata to the measurement data. This section involves an ID-specific metadata table to facilitate matching (Fig. 5 ). Here, the measurement for each replicate within a group requires a unique measurement ID. Given this ID and the group name (defined at the top of Metadatasheet), one must be able to identify the respective measurement. If there are subgroups or further subdivisions of samples, a table per division is expected. By design, the actual measurement happens at the last division stage, hence the measurement ID belongs to the last stage, as well. If available, further personal IDs can be given on sample level, too.
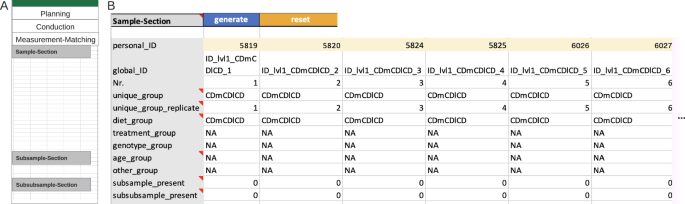
Example of an instance of the Measurement-Matching section. ( A ) Overview Measurement-Matching section. ( B ) An ID-specific metadata table example with the minimal number of required rows. The yellow marked cells hold measurement IDs (‘personal_ID’) required for the matching of metadata column with the respective measured data. ‘NA’ indicates non-available information (‘Diet’ is the only comparison group specified). The last two rows indicate that neither subsamples nor subsubsamples are needed in this instance. The table is column cropped; based on previous final groups and given replicates, a total of 30 columns are expected in the full table. Note that the full Metadatasheet of this example can be found in Supplementary Material.
The automatically generated ID-specific metadata table summarizes the preceding input of the user to ease the measurement to metadata matching. Hence, besides the default rows, the ID-specific metadata table will expand depending on inputs from the Conduction section. Expansion includes previously mentioned covariates and constants, along with any keys where the ‘CHANGES’ value was applied. The Measurement-Matching section overall ensures the flexibility tailored to capture information individually for each measured sample or division of such. Moreover, the arrangement of subsamples and subsubsamples clearly reveals any nested design, which is important for choosing appropriate statistics.
Hence, the application example showcases the Metadatasheet in differing context.
Additional examples of metadata management in practice are available in the supplementary materials, which include distribution and update-handling of the Metadata Workbook and its associated resources, along with example workflows of different users within a research group. The use of Metadatasheets benefit individual users and the scientific community by streamlining data management and enabling program development.
Applications of completed metadatasheets within and beyond the metadata workbook
The availability of standardized Metadatasheets offers advantages to individual users, the associated scientific community, ranging from the respective group to large-scale consortia, as well as not involved third parties.
The individual’s benefits from utilizing the Metadatasheet as a live document or central hub guides their data management for conducted or planned experiments. This approach simplifies the process of handing or taking over projects, as documentation follows a streamlined format, as opposed to each person maintaining individual data management methods. Furthermore, standardization plays a pivotal role in enabling the development of programs for analysis and processing, thanks to uniform input formats. A notable example is the provided conversion program that parses the Metadatasheet involving bulk-omics measurements to an R object. This SummarizedExperiment object 30 itself is the standardized input for many Bioconductor based analysis 27 , 28 .
A group or consortia introducing the Metadatasheet will have access to multiple Metadatasheets. This in turn evokes the possibility for creation of a comprehensive database. Within this database, numerous sheets can be easily searched for specific information. To support this application, we have developed a dedicated, publicly accessible ontology for seamless integration of data into a custom database. The provided ontology is specific for the proposed Metadatasheet and incorporated terms. Essentially, this database functions as a centralized knowledge hub, enabling swift access to available data, available specimen and planned experiments across groups. A database facilitates meta-analyses and aids in identifying gaps in the current local research landscape, potentially discovering collaboration opportunities.
Ensuring both human and machine readability of the Metadatasheet is essential for facilitating seamless interactions with the data it represents. By accommodating both, the Metadatasheet enables users to query and access data more efficiently, from a single sheet up to a large collection. By a careful design and through a hierarchical structuring approach for the metadata sheet, additionally accompanied by instant help texts (mouse-over) and available training resources, input metadata remains human-readable and allows for a quick and efficient look up of, e.g., single sets of interest. Machine readability is given through the provided ontology and export functionality into OWL/XML or RDF/XML formats. The Metadata Workbook offers the export functionality for derived metadata formats required e.g. for upload to the NCBI Geo repository. Upon the upload of data and metadata to repositories, research employing methods capable of reading and processing data from these repositories will benefit. Example for such methods are GeoQuery 32 , GEOmetadb 33 or E-Utils provided by NCBI directly 34 . The Metadatasheet captures a broad range of measurement techniques and experimental systems, which may pose challenges in finding a suitable domain-specific repository, especially if datasets are linked. In such cases, the Metadatasheet offers a solution through the creation of topic-centered databases using its machine-readable format. These topic-centered databases can transition from restricted to public access upon publication. The use of Metadatasheets benefit individual users, the associated scientific community as well as third parties through enabled program development, export to repositories and creation of topic-centered databases if suitable.
The developed metadata standard facilitates comprehensive recording of all relevant metadata for a broad spectrum of biomedical applications throughout the data-lifecycle. The standard’s implementation ensures efficient documentation of metadata and with a user-friendly design. The provided Metadata Workbook enriched with custom, open-source functionalities can be extended on various levels to adjust to additional setups.
The presented framework, encompasses two parts. The first part involved the iterative collection and organisation of keys, while the second part focused on the implementation of the user experience within the Metadata Workbook. During the collection phase, it became apparent that the specific set of keys varies enormously depending on the research groups, while multiple keys are found repeatedly across the assessed repositories. To address the high variability, we made adaptability of the Metadatasheet a priority. While the set of comparisons (‘comparison groups’) is tailored to our context, e.g. diet or temperature, the implementation is designed to be extensible ad-hoc. This means the Metadatasheet can be customized by specifying requested keys and adding experimental groups and measurement types, as well as expanding the controlled vocabulary. Moreover, a versatile comparison group labelled as ‘Others’ has been introduced. This ‘Others’ group adapts to any comparison scenario, not covered. Adding another ‘comparison group’ to the structure is also possible when adhering to the segment’s structural characteristics, only requiring additions to the provided Metadatasheet ontology. For version tracking and other ontology management means, tools such as CENTree 35 or OntoBrowser 36 could be employed.
To follow the DRY principle, the Metadatasheet key collection aims for comprehensiveness, capturing metadata required in other contexts. The adaptability of the Metadatasheet allows for the introduction of additional formal means, although not strictly enforced.
The Metadatasheet has been implemented within a macro-enabled Microsoft Excel workbook. Despite the fact that Excel is not open-source, nor free, it has several severe advantages. Its widespread availability, familiarity and standard-use within the biomedical research community makes it a valuable choice, especially when compared to custom standalone applications. Furthermore, most users are experienced Excel user, allowing for seamless integration of our proposed sheet into existing workflows. This immediate integration would not be as straightforward with open-source spreadsheet software like LibreOffice, also lacking required automation aspects. An online, browser-based, operating system independent approach such as GoogleSheets, besides being accessible for everyone, violates the needs of sensitive data, particularly in cases involving unpublished studies. If data sensitivity isn’t an issue, a browser approach might be preferable to the proposed solution. However, our solution within Excel suits all data protection levels. Addtionally, given Excel’s wide spread, some electronic lab books readily offer Excel integrations. It’s important to note that the Metadata Workbook offers a user-friendly solution for completing and expanding the Metadatasheet, whereby the Metadatasheet itself is a standalone solution for metadata recording. The complete Metadatasheet can be converted into machine-readable XML files and SummarizedExperiment objects, using provided tools. Recently, Microsoft has introduced Excel365, a browser-based software. However, our provided Metadata Workbook, requires adjustments to function within the Excel365 framework, as the used automation languages differ.
Metadata labels provide meaning to data, especially if keys and values are not only comprehensive but also interconnected, enabling cross-study comparisons. Providing metadata labels is commonly referred to as semantic interoperability, and it is considered a pivotal aspect of data management 37 . In order to attain semantic interoperability, there are domain-specific ontologies that establish meaningful connections between the labels of metadata. However, it is important to note that there is no single ontology that can comprehensively address the diverse requirements, even within a relatively homogeneous domain of investigation within a single consortium in the field of biomedical sciences. In fact, the choice of the appropriate ontology is far from straightforward and can vary for the same keys depending on the context. Pending ontology decisions might delay the recording of metadata, which in turn can lead to data loss. Involvement of inexperienced users, due to common high fluctuations of early-stage researchers, can further exacerbate the delay. Therefore, we have made the conscious choice, following our adaptability priority, to employ an extendable controlled vocabulary. This decision empowers biomedical researchers to directly and effortlessly record metadata without the need to immediately handle ontologies and their unavoidable complexities. While this decision will require additional retrospective annotation efforts to adhere to appropriate ontologies, it is manageable in contrast to retrospectively recovering metadata information that was never recorded. To support the handling of introduced expansions, we also offer a merge Workbook to unite differently extended controlled vocabularies. This serves as an initial aid in managing retrospective individual metadata items.
The presented framework enables and directs researchers to document FAIR data. However, for the process to be completed, researchers must undertake final steps, such as selecting appropriate ontologies and exporting and depositing data in repositories like NCBI GEO. Our strategy prioritizes ease of initial data recording and acknowledges the practical challenges associated with ontology selection and application.
Ontologies enrich any set of collected metadata, therefore, we do not aim to discourage the use of ontologies. Integration of ontologies into the workflow could be facilitated by Metadata Annotation Services, such as RightField 8 , Ontology LookUp service (OLS) 25 or OntoBee 26 . RightField is a standalone tool populating cells within a spreadsheet with ontology-based controlled vocabulary. OntoBee and OLS are linked data servers and can be used to query suitable ontologies and IDs given a keyword. Groups can enforce the partial or complete usage of ontology for keys in the Metadatasheet by leveraging on the option of group-specific validation and creating a tailored validation sheet. The supplementary material includes a table that lists potentially suitable ontologies for the keys, offering guidance for users (Table S1 ).
We anticipate our proposed Metadatasheet accompanied by its implementation, the Metadata Workbook, being used for more than just data recording. Even in a partially filled state and at the start of a research cycle, the findability, accessibility, and interoperability provided by standardized Metadatasheets can speed up experiment preparation between groups, encourage effective specimen usage, and foster collaborations. Beyond individual and group benefits, these platforms can serve as the foundation for topic-centered public databases. This offers an alternative solution for managing interconnected and diverse datasets, potentially linked with an Application Programming Interface (API) to facilitate computational access through queries. However, researchers still need to assess suitable domain-specific repositories, potentially sharing datasets across multiple resources, thereby enhancing their findability. Given that many datasets are often deposited as supplementary material 38 , likely due to the challenges of adhering to metadata standards, our aim is to enhance both the structure of supplementary material using the Metadatasheet and facilitate the transition to repositories through automatic export. We envision the Metadata Workbook to lower the burden associated with adhering to metadata standards, thereby encouraging more frequent submissions to repositories initially. Ultimately, this process aims to foster the generation of more FAIR data.
A tool for facilitating FAIR data recording is valuable and effective only when it is maintained and actively utilized. However, small to medium-sized academic labs often lack dedicated personnel solely responsible for such tasks. Therefore, we have designed our proposed solution, integrated into the Metadata Workbook, to be easily adaptable and extendable without requiring any programming skills or other domain-specific knowledge, thus enhancing its sustainability. Detailed documentation outlines the processes involved thoroughly. Our open source solution is built upon basic VBA code, avoiding complex functionalities, which is the most likely to stay functional. Consequently, the maintenance of the framework can be decentralized, promoting low-cost while having enough flexibility to extensively adapt.
We are currently developing analysis tools that facilitate seamless integration, including integration with custom databases, to promote usage by delivering numerous and immediate advantages. By establishing local hubs of uniformly structured data through these efforts, it becomes significantly easier for data management entities, now prevalent throughout academia, to undertake the, e.g., mapping process.
Planned development of the Metadatasheet and the Metadata Workbook includes adding export options, a database for Standard Operation Protocols, analysing sets of collected metadata, and providing project monitoring tools. Additionally, we aim to further automate the filling of the Metadatasheet to further close the gap between good documentation need and associated effort for the scientist 39 . Automation extensions are auto-completion upon typing, transferring information from in-place LIMS resources, as well as other metadata locations. Furthermore, we aim to establish the option to assign specific sections of the Metadatasheet to responsible individuals, allowing for proper crediting of their work and acknowledgment of the numerous scientists involved throughout the recording process.
In conclusion, the framework leverages the widespread use of Excel, enabling comprehensive metadata documentation and improving the efficiency of data deposit on repositories. Our practical solution offers a user-friendly and sequential approach to manage metadata, thereby addressing the need for FAIR data in the field of biomedical science at intermediate stages during the data life cycle up to publication. We expect this to be of high relevance for a broad spectrum of biomedical researchers, and think that it can also be easily adapted to adjacent fields.
Metadata workbook structure
The proposed Metadatasheet is implemented within Microsoft Excel macro-enabled workbook, which consists out of multiple sheets with macros modules. The input sheet resembles the Metadatasheet. The other sheets hold the validation resources, the dependent fields for the differing experimental systems and measurement types, a plain Metadatasheet for reset, the repositories’ metadata standards, and additional resources for user guidance, such as a glossary. Input, validation, dependent fields and user guidance sheets are visible to the user, whereby only the input sheet is extensively editable by the user. Within validation and dependent fields sheets, only blank cells can be filled.
The structure of the individual sheets ensures their functionality. An example is the validation sheet, which holds per column the controlled vocabulary for a respective key. Each column starts with the three rows where the type of validation - freetext, date, DropDown or DropDown_M (multiple selection possible) - any specification in form of help text and the respective key is specified. The ‘dependentFields’ sheet is constructed in a similar manner. Here, the first two rows for each column determine the general category - measurement type or experimental system - as well as the specification from the controlled vocabulary set, e.g. of mouse. After those specifications, the dependent keys are enumerated.
The input sheet and attached functionalities utilize different font faces as well as colour cues for structuring, and segment specific automatised processes. All grey cells with bold font content signal different segments of each section. This provides a fine-grid structure. Italic font characterize boolean validation requests, hence expecting ‘yes’ or ‘no’. This does not only help for structure but also is done for performance reasons as just by checking font, actions can be precisely called.
Custom add-on functionalities
The Workbook including VBA based macros was developed using Excel Version 16.77. The implementation is tested for use on both macOS (Ventura 13.5) and Windows (Windows 11) and respective variations of Microsoft Excel Version 16. The differences in Excel functionality between Windows and macOS influenced our implementation, such as bypassing ‘ActiveX-controls’ being not available on MacOS platforms.
The Metadata Workbook incorporates various functionalities organized into VBA modules. Users invoke actions by either actively pressing a button or upon input, which is a change of a cell within the input sheet. The latter allows for reactive updates. Reactivity functionality is directly attached to the input sheet, unlike VBA modules. The Metadata Workbook key functionalities include a validation function, an insertion-of-dependent-keys function, and a reset-import function, which are further discussed in the following. Furthermore, the reactivity procedure evoked upon cell change is outlined.
The custom validation function leverages the Excels Data-Validation feature. The feature checks predefined conditions for a given cell upon the user’s input, e.g. if the input value lies within a range of allowed values. If those values are of discrete nature, one can display all possible values as a DropDown to the user. Our custom validation function populates Excels Data-Validation feature automatically, passing the appropriate data constraints to determine a valid input. An exception exists for all keys that allow multiple selections, marked in the validation sheet as type DropDown_M. To allow the selection of multiple items, reactive functionalities had to be included. Any user values that fail validation are marked. To simplify searching within the DropDown list, the allowed values are automatically sorted alphabetically.
In the case of extensive controlled vocabulary or the wish to tight constraints, users have the option to subset the main validation sheet. The subset sheet must be named ‘Validation_[Group]’, whereby ‘[Group]’ is to be replaced by the respective value to the requested key group. The structure of the subset sheet is expected to be the same as within the validation sheet. To use this predefined subset, one has to choose ‘yes’ for ‘group specific?’ on top of the sheet.
The insertion functionalities handle the automatic dependent key insertion, inserting necessary keys dependent on the user’s choice of the experimental system and measurement type. Here, the subroutines conduct a search for a match with the user’s input within the ‘dependentFields’ sheet, retrieving the corresponding column with associated keys for insertion in the Metadatasheet. Note that dependent key sets can be extended by adding keys to the list, whereby additional keys subsequently need to be added to the validation sheet to provide constraints.
The reset/import function allows users to reset the sheet to its initial state or to a chosen template state. Two options are available upon pressing the ‘Reset’ button and displayed to the user with a pop-up window. The first option resets to a blank input sheet. The function deletes the current input sheet, copies a ‘ResetSheet’ and renames it to ‘Input’. The ‘ResetSheet’ has the same VBA-code as the ‘Input’ Sheet attached. The second option resets to a user chosen template. A template may be a previous complete Metadatasheet or a partially filled Metadatasheet. The inputs from the template sheet are copied upon a duplication of the ‘ResetSheet’ to retain reactivity-functionality. The duplication with the template’s input is renamed to ‘Input’. The original ‘ResetSheet’ is always hidden to prevent accidental deletion.
Metadatasheet ontology creation
Our custom ontology was modelled by following a top-down approach using established tools in the realm of semantic web (cf. Protégé 40 and accompanying tools), giving rise to a consistent contextual data model, logical data model and physical data model eventually leading to an integration of individuals (metadata samples) into a semantic database.
Conversion program creation
The conversion program uses a completed Metadatasheet as input and checks for suitability of conversion based on the measurement type. If the type is one of ‘bulk-metabolomics’,‘bulk-transcriptomics’ or ‘bulk-lipidomics’, the conversion starts. The Measurement-Matching section will be saved within ‘colData’-slot. The actual data matrix is identified, guided by the Data File Linkage information. Given the personal ID and the given file measurement data is identified. Note, the location of the input Metadatasheet is seen as root and given filenames are expected as relative paths. If ‘single file for all’ is selected, the filename given in the comment section is directly searched for. If nothing is found, measurement data is searched for by the given extension in processed data and returned to the user asking for clarification. The program is written in R.
Data availability
The ontology needed to create a database upon a set of Metadatasheets (version 1.8.0) is available under the following link on Github https://github.com/stephanmg/metadata_ontology .
Code availability
The Metadata Workbook and related content is freely available on Zenodo 41 ( https://zenodo.org/records/10278069 ) and GitHub ( https://github.com/LeaSeep/MetaDataFormat ). The repository contains the macro-embedded Metadata Workbook, the isolated VBA scripts, the macro-embedded Merge Workbook, as well as the converter to turn a Metadatasheet to a SummarizedExperiment Object. The repository includes a pre-commit hook that extracts the associated VBA scripts automatically, facilitating easy evaluation of code changes directly within GitHub.
Morillo, F., Bordons, M. & Gómez, I. Interdisciplinarity in science: A tentative typology of disciplines and research areas. Journal of the American Society for Information Science and Technology 54 , 1237–1249, https://doi.org/10.1002/asi.10326 (2003).
Article Google Scholar
Cioffi, M., Goldman, J. & Marchese, S. Harvard biomedical research data lifecycle. Zenodo https://doi.org/10.5281/zenodo.8076168 (2023).
Habermann, T. Metadata life cycles, use cases and hierarchies. Geosciences 8 , https://doi.org/10.3390/geosciences8050179 (2018).
Stevens, I. et al . Ten simple rules for annotating sequencing experiments. PLOS Computational Biology 16 , 1–7, https://doi.org/10.1371/journal.pcbi.1008260 (2020).
Article CAS Google Scholar
Shaw, F. et al . Copo: a metadata platform for brokering fair data in the life sciences. F1000Research 9 , 495, https://doi.org/10.12688/f1000research.23889.1 (2020).
Ulrich, H. et al . Understanding the nature of metadata: Systematic review. J Med Internet Res 24 , e25440, https://doi.org/10.2196/25440 (2022).
Article PubMed PubMed Central Google Scholar
Wilkinson, M. D. et al . Comment: The fair guiding principles for scientific data management and stewardship. Scientific Data 3 , https://doi.org/10.1038/sdata.2016.18 (2016).
Wolstencroft, K. et al . Rightfield: Embedding ontology annotation in spreadsheets. Bioinformatics 27 , 2021–2022, https://doi.org/10.1093/bioinformatics/btr312 (2011).
Article CAS PubMed Google Scholar
Leipzig, J., Nüst, D., Hoyt, C. T., Ram, K. & Greenberg, J. The role of metadata in reproducible computational research. Patterns 2 , https://doi.org/10.1016/j.patter.2021.100322 (2021).
Researchspace. https://www.researchspace.com/ . Accessed: 12th March 2024 (2024).
Revvity signals notebook eln. https://revvitysignals.com/products/research/signals-notebook-eln . Accessed: 12th March 2024 (2024).
Kowalczyk, S. T. Before the repository: Defining the preservation threats to research data in the lab. In Proceedings of the 15th ACM/IEEE-CS Joint Conference on Digital Libraries , JCDL ‘15, 215–222, https://doi.org/10.1145/2756406.2756909 (Association for Computing Machinery, New York, NY, USA, 2015).
Rocca-Serra, P. et al . ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics 26 , 2354–2356, https://doi.org/10.1093/bioinformatics/btq415 (2010).
Article CAS PubMed PubMed Central Google Scholar
Lin, D. et al . The trust principles for digital repositories. Scientific Data 7 , 144, https://doi.org/10.1038/s41597-020-0486-7 (2020).
Article ADS PubMed PubMed Central Google Scholar
Barrett, T. et al . NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research 41 , D991–D995, https://doi.org/10.1093/nar/gks1193 (2012).
VizcaÃno, J. A. et al . 2016 update of the PRIDE database and its related tools. Nucleic Acids Research 44 , D447–D456, https://doi.org/10.1093/nar/gkv1145 (2015).
Malik-Sheriff, R. S. et al . BioModels—15 years of sharing computational models in life science. Nucleic Acids Research 48 , D407–D415, https://doi.org/10.1093/nar/gkz1055 (2019).
Article CAS PubMed Central Google Scholar
Glont, M. et al . BioModels: expanding horizons to include more modelling approaches and formats. Nucleic Acids Research 46 , D1248–D1253, https://doi.org/10.1093/nar/gkx1023 (2017).
Consortium, T. G. O. et al . The Gene Ontology knowledgebase in 2023. Genetics 224 , iyad031, https://doi.org/10.1093/genetics/iyad031 (2023).
Percie du Sert, N. et al . The arrive guidelines 2.0: Updated guidelines for reporting animal research. PLOS Biology 18 , 1–12, https://doi.org/10.1371/journal.pbio.3000410 (2020).
Novère, N. L. et al . Minimum information requested in the annotation of biochemical models (miriam. Nature Biotechnology 23 , 1509–1515, https://doi.org/10.1038/nbt1156 (2005).
Gil Press. Cleaning big data: Most time-consuming, least enjoyable data science task, survey says. https://www.forbes.com/sites/gilpress/2016/03/23/data-preparation-most-time-consuming-least-enjoyable-data-science-task-survey-says/?sh=27709ef76f63 . Accessed: 2024-4-3 (2016).
Hughes, L. D. et al . Addressing barriers in fair data practices for biomedical data. Scientific Data 10 , 98, https://doi.org/10.1038/s41597-023-01969-8 (2023).
The metabolomics workbench, https://www.metabolomicsworkbench.org/ .
EMBL. Ontology lookup service, https://www.ebi.ac.uk/ols4 .
Xiang, Z., Mungall, C. J., Ruttenberg, A. & He, Y. O. Ontobee: A linked data server and browser for ontology terms. In International Conference on Biomedical Ontology (2011).
Huber, W. et al . Orchestrating high-throughput genomic analysis with bioconductor. Nature Methods 12 , 115–121, https://doi.org/10.1038/nmeth.3252 (2015).
Gentleman, R. C. et al . Bioconductor: open software development for computational biology and bioinformatics. Genome Biology 5 , R80, https://doi.org/10.1186/gb-2004-5-10-r80 (2004).
Hunt, A. & Thomas, D. The pragmatic programmer: From journeyman to master . (Addison Wesley, Boston, MA, 1999).
Google Scholar
Morgan, M., Obenchain, V., Hester, J. & Pages, H. Summarizedexperiment: Summarizedexperiment container. Bioconductor (2003).
Mass, E. et al . Developmental programming of kupffer cells by maternal obesity causes fatty liver disease in the offspring. Research Square Platform LLC https://doi.org/10.21203/rs.3.rs-3242837/v1 (2023).
Davis, S. & Meltzer, P. S. Geoquery: a bridge between the gene expression omnibus (geo) and bioconductor. Bioinformatics 23 , 1846–1847, https://doi.org/10.1093/bioinformatics/btm254 (2007).
Zhu, Y., Davis, S., Stephens, R., Meltzer, P. S. & Chen, Y. Geometadb: powerful alternative search engine for the gene expression omnibus. Bioinformatics 24 , 2798–2800, https://doi.org/10.1093/bioinformatics/btn520 (2008).
National Center for Biotechnology Information (US). Entrez programming utilities help. Internet. Accessed on 02.04.2024 (2010).
SciBite, CENtree, https://scibite.com/platform/centree-ontology-management-platform/
Ravagli, C., Pognan, F. & Marc, P. Ontobrowser: a collaborative tool for curation of ontologies by subject matter experts. Bioinformatics 33 , 148–149, https://doi.org/10.1093/bioinformatics/btw579 (2016).
Sasse, J., Darms, J. & Fluck, J. Semantic metadata annotation services in the biomedical domain—a literature review. Applied Sciences (Switzerland) 12 , https://doi.org/10.3390/app12020796 (2022).
Tedersoo, L. et al . Data sharing practices and data availability upon request differ across scientific disciplines. Scientific Data 8 , 192, https://doi.org/10.1038/s41597-021-00981-0 (2021).
Menzel, J. & Weil, P. Metadata capture in an electronic notebook: How to make it as simple as possible? Metadatenerfassung in einem elektronischen laborbuch: Wie macht man es so einfach wie möglich? GMS Medizinische Informatik, Biometrie Epidemiologie 5 , 11, https://doi.org/10.3205/mibe000162 (2015).
Musen, M. A. The protégé project: A look back and a look forward. AI Matters 1 , 4–12, https://doi.org/10.1145/2757001.2757003 (2015).
Seep, L. METADATASHEET - Showcases, Zenodo , https://doi.org/10.5281/zenodo.10278069 (2023).
Download references
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (project IDs 390685813 - EXC 2047 and 390873048 - EXC 2151) and through Metaflammation, project ID 432325352 – SFB 1454 (L.Se., I.S., H.H., D.Ri., J.Y., T.B., K.S., R.K., S.K., E.M., D.W., E.L., F.M., A.Sch., J.H), BATenergy, project ID 450149205 - TRR 333 (S.G., A.S.A., S.H., M.M., D.Ra., J.Be., D.W., A.T., V.P., K.K., A.P., H.W., L.Sch., T.F., W. K. F., M.K., J.H), the Research Unit “Deciphering the role of primary ciliary dynamics in tissue organisation and function”, Project-ID 503306912 - FOR5547 (D.W., E.M.), and SEPAN, project ID 458597554 (L.Se.), and by the University of Bonn via the Schlegel professorship to J.H. E.M. is supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant Agreement No. 851257). W.K.F. is further supported by the DFG (FE 1159/6-1, FE 1159/5-1, DFG FE 1159/2-1), by the European Research Council (ERC, under the European Union’s Horizon Europe research and innovation program; Grant Agreement No. 101080302) and by grants from the Gabriele Hedwig Danielewski foundation and the Else Kroener Fresenius Foundation. A.T. is supported by the Gabriele Hedwig Danielewski foundation. A.K.G. is supported by Medical Faculty, University of Bonn, BONFOR grants 2018-1A-05, 2019-2-07, 2020-5-01. We thank all members, including associated, of the SFB Metaflammation and TRR BATenergy for the iterative discussions and their input throughout.
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and affiliations.
Computational Biology, Life & Medical Sciences (LIMES) Institute, University of Bonn, Bonn, Germany
Lea Seep, Stephan Grein, Jiangyan Yu & Jan Hasenauer
Developmental Biology of the Immune System, Life & Medical Sciences (LIMES) Institute, University of Bonn, Bonn, Germany
Iva Splichalova, Hao Huang & Elvira Mass
Institute of Pharmacology and Toxicology, University Hospital, University of Bonn, Bonn, Germany
Danli Ran, Mickel Mikhael, Staffan Hildebrand, Ana Soriano-Arroquia, Dominik Tischer & Alexander Pfeifer
Department of Bioinformatics and Biochemistry, Technical University Braunschweig, Braunschweig, Germany
Mario Lauterbach & Karsten Hiller
Institute of Innate Immunity, University Hospital Bonn, University of Bonn, Bonn, Germany
Dalila Juliana Silva Ribeiro, Katharina Sieckmann, Ronja Kardinal, Dagmar Wachten & Eicke Latz
Quantitative Systems Biology, Life & Medical Sciences (LIMES) Institute, University of Bonn, Bonn, Germany
Jiangyan Yu & Andreas Schlitzer
Systems Immunology and Proteomics, Institute of Innate Immunity, Medical Faculty, University of Bonn, Bonn, Germany
Sebastian Kallabis & Felix Meissner
Department of Biochemistry and Molecular Cell Biology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
Janina Behrens & Ludger Scheja
Department of Internal Medicine I, Division of Endocrinology, Diabetes and Metabolism, University Medical Center Bonn, Bonn, Germany
Andreas Till, Viktoriya Peeva & Wiebke K. Fenske
Chair of Molecular Nutritional Medicine, TUM School of Life Sciences, Technical University of Munich, Freising, Germany
Akim Strohmeyer, Johanna Bruder, Katharina Kuellmer, Tobias Fromme & Martin Klingenspor
Immunology and Environment, Life & Medical Sciences (LIMES) Institute, University of Bonn, Bonn, Germany
Tobias Blum
Immunogenomics & Neurodegeneration, German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany
Yuanfang Li & Marc Beyer
PRECISE, Platform for Single Cell Genomics and Epigenomics at the German Center for Neurodegenerative Diseases and the University of Bonn, Bonn, Germany
Department of Psychiatry and Psychotherapy, University Hospital Bonn, Bonn, Germany
Anne-Kathrin Gellner
Institute of Physiology II, Medical Faculty, University of Bonn, Bonn, Germany
School for Medicine and Health, Faculty of Sport and Health Sciences, Technical University of Munich, Munich, Germany
Henning Wackerhage
EKFZ—Else Kröner-Fresenius Center for Nutritional Medicine, Technical University of Munich, Freising, Germany
Martin Klingenspor
ZIEL Institute for Food & Health, Technical University of Munich, Freising, Germany
Department of Internal Medicine I - Endocrinology, Diabetology and Metabolism, Gastroenterology and Hepatology, University Hospital Bergmannsheil, Bochum, Germany
Wiebke K. Fenske
Experimental Systems Immunology, Max Planck Institute of Biochemistry, Martinsried, Germany
Felix Meissner
PharmaCenter Bonn, University of Bonn, Bonn, Germany
Alexander Pfeifer
Helmholtz Center Munich, German Research Center for Environmental Health, Computational Health Center, Munich, Germany
Jan Hasenauer
You can also search for this author in PubMed Google Scholar
Contributions
J.H. and S.G. conceived the concept. L.Se. implemented and extended the Metadatasheet and created the Metadata Workbook. T.B., M.K., J.Br., A.St. tested and provided feedback on initial version of the Metadatasheet. I.S., D.Ra., M.M., S.H., M.L., K.H., D.Ri, K.S., R.K., H.H., J.Y., S.K., J.Be., A.T., V.P., A.S.A., D.T., K.K., Y.L., M.B., A.K.G., T.F., H.W., M.K., W.K.F., L.Sch., F.M., A.Sch., E.M., D.W. provided in-depth feedback to the Metadatasheet and the Metadata Workbook and contributed to the showcases. E.L. and A.P. lead the discussion rounds as representatives of the consortia. L.Se. and J.H. wrote the first draft of the manuscript. All authors reviewed the manuscript.
Corresponding author
Correspondence to Jan Hasenauer .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Seep, L., Grein, S., Splichalova, I. et al. From Planning Stage Towards FAIR Data: A Practical Metadatasheet For Biomedical Scientists. Sci Data 11 , 524 (2024). https://doi.org/10.1038/s41597-024-03349-2
Download citation
Received : 07 December 2023
Accepted : 08 May 2024
Published : 22 May 2024
DOI : https://doi.org/10.1038/s41597-024-03349-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

What is a Document Repository? A Complete Overview
Discover what a document repository is and how it can streamline your document management process in our comprehensive overview. This resource is essential for professionals in engineering and manufacturing seeking to enhance organization and accessibility of their documentation.
Consider an architectural firm working across multiple international projects at the same time. Team members are scattered across the globe, but still need access to countless of the same documents daily. With a document repository, they have access to a centralized digital archive that meticulously organizes, manages, and secures every piece of documentation.
These document management systems are beneficial across numerous industries – such as manufacturing, oil & gas, and the pharmaceutical world. By providing an easy way to retrieve and work on documents, workers spend less time on repetitive manual tasks and more time invested in higher levels of thinking. Eliminating inefficiency around data is the first step on a path toward overall organizational success.
What is a Document Repository?
A document repository is a centralized digital archive where organizations store, manage, and track their documents. By systematically organizing files, these repositories make sure documents are easily accessible and secure. This caters to various operational needs across an organization.
Document repositories typically handle multiple document formats and types. This can range from text files and spreadsheets to presentations and technical drawings. Their versatility makes them particularly useful tools in corporate, scientific, and technical environments where document integrity and availability are of the utmost importance.
The primary function of a document repository is to provide a secure environment where documents can be stored, accessed, and managed. Features include document version control , access permissions, and audit trails.
The Importance of a Document Repository
Document repositories streamline the management of files and markedly reduce the time and effort spent on locating and verifying documents. It fosters a culture of collaboration across teams by providing all members with centralized access to necessary documents. The use of document control systems leads to more efficient teamwork and project management.
Furthermore, these systems help tremendously with compliance management. By maintaining rigorous standards of document integrity and security, document repositories help organizations meet legal and regulatory requirements. This is particularly helpful in industries where adherence to compliance is mandatory and closely monitored.
Industries in Need of Document Repositories
Multiple industries can benefit from document repositories, but they are important for engineering project teams, maintenance crews, and compliance groups. They are especially beneficial in sectors like manufacturing, engineering, and pharmaceuticals.
These industries face unique challenges – such as high standards of quality control, strict regulatory requirements, and managing extensive and complex records. It is about simplifying compliance with safety standards.

Document repositories address these challenges by providing a streamlined, secure, and systematic approach to managing extensive documentation. For example, in the pharmaceutical industry, document repositories help maintain detailed records of clinical trials and formulae. It is essential for both compliance and product development.
Key Features of Document Repositories
Document repositories are designed with advanced features to improve the overall document control process. These features help organizations maintain control, security, and efficiency. Here is an in-depth look at some of the key functionalities:
1. Version Control
Version control is fundamental in document management. It tracks all changes made to documents and allows users to revert to previous versions if needed. This helps maintain document integrity over time. The tracking of revisions ensures that all modifications are recorded and therefore reversible.
2. Access Control
Access control mechanisms allow administrators to set specific permissions for viewing, editing, and deleting documents. This feature helps protect sensitive information on various organizational levels. Only authorized personnel have access to certain documents. Additionally, it helps maintain data privacy and security.
3. Search and Retrieval
With advanced search functionalities, users can quickly locate specific documents based on keywords, metadata, or content. This feature reduces the time spent searching for documents. That means employees do not waste their time on manual tasks and have space to think about more innovative and strategic matters.
4. Document Organization
A well-organized document repository allows users to categorize documents into folders, categories, or tags for easy navigation. This simplified organization is a key to easily managing large volumes of documents. It makes it simple for users to find the information they need without unnecessary delays.
5. Security & Compliance
Document repositories provide thick and innovative security features to protect sensitive documents from unauthorized access or data breaches. This takes form in encryption, secure access protocols, and regular security audits. It helps automate compliance with legal and regulatory standards while protecting company assets.
6. Audit Trail
An audit trail records all user actions and document history. It provides a detailed log that can be used for accountability and compliance purposes. This feature is especially important for industries where tracking document handling and access is required by regulatory standards.
7. Scalability
As organizations grow and change, so do their document management needs. Document repositories are designed to scale and accommodate growing or shrinking volumes of documents without compromising performance or security. This flexibility means businesses do not need to worry too much about their document storage and management throughout the natural flow of business.
Main Benefits of a Document Repository
A document repository simplifies the management of records while improving general operations across various organizational levels. Here are four key benefits that these systems provide:
1. Maximize Productivity
Tap the well of potential. Document repositories streamline the storage, retrieval, and sharing of documents. Through this, they significantly reduce the time employees spend searching for information they need. This efficiency boosts productivity by minimizing downtime and accelerating the completion of tasks.
2. Collaborative Edge
Gain a leg up on competitors. These systems foster a culture of real-time collaboration among team members, regardless of their locations. By providing centralized access to all documents, team members can work simultaneously on projects. That means sharing insights instantly and making decisions quickly. That leads to an increase in efficiency and helps organizations stay ahead.
3. Robust Security
Protect what matters. Document repositories provide another layer of data security through comprehensive access controls and robust encryption. These features protect sensitive information from unauthorized access and data breaches. It also leads to compliance with industry-specific regulatory requirements.
4. Informed Decision Making
Make the right choices based on actual information. By maintaining accurate and up-to-date documents, document repositories encourage informed decision-making at all levels of the organization. Access to current and reliable information means managers and executives can make strategic decisions that are based on the latest data. It is about data, not instinct.
Accruent’s Meridian: Best Document Repository Software
Accruent’s Meridian is a comprehensive document repository solution designed to optimize the management, security, and accessibility of documents throughout their lifecycle. The robust features focus on security, convenience, and compliance. With benefits such as streamlined workflows to improved data protection, Meridian is a choice for numerous industry leaders. It provides the tools necessary to manage vast volumes of documents.

Below are the key features that position Meridian as a top choice:
- Advanced Version Control : All document changes are tracked which allows for easy reversion to previous versions and historical integrity.
- Access Restrictions: Detailed control over who can view, edit, or delete documents.
- Powerful Search Capabilities: Quick retrieval of documents through smart search tools that can locate files based on keywords, metadata, or content.
- Multiple File Types: Accommodates a wide range of document formats which makes it versatile for various industry needs.
- Seamless System Integration and Scalability: Integrates easily with existing systems while scaling to meet growing document management needs or demands.
Meridian excels in certain industries. For the manufacturing sector, it improves project management through seamless integration with industry-standard tools. It is easier to maintain high production standards of operations.
In oil and gas, Meridian details site plans, safety procedures, and compliance records. Further, for pharmaceutical companies, document management software helps navigate tricky regulatory compliance with clinical trials, research findings, and drug formulations.
For a real-life example , the global biopharmaceutical company AbbVie faced a restructuring challenge of engineering IT application use after splitting from its parent company. They needed an engineering document management system that could be rolled out quickly and globally while being standardized. They chose Meridian, and now over 625 AbbVie employees across departments use Meridian to manage more than 300,000 CAD (Computer Aided Design) drawings and other files.
Meridian, Your Partner in Document Repository Solutions
Propel the business forward with confidence. Successful document management can and will be fundamentally transformative for your future success. With Meridian’s sophisticated document repository solutions, organizations can increase productivity. It is about streamlining processes, ensuring compliance, and improving overall efficiency.
Learn how Meridian’s features power the pathway toward tomorrow. Schedule your demo today .
Document Repository FAQs
What is the purpose of a document repository.
A document repository is designed to store, manage, and track documents efficiently. It provides a secure platform for organizations to preserve document integrity, ensure easy access, and facilitate collaboration by keeping all documents centralized and organized. For large-scale organizations, the right document repository can be a transformative tool.
What is a centralized document repository?
A centralized document repository collects all organizational documents in one secure, accessible location. The setup helps unify and improve data consistency while reducing redundancies. It can improve access efficiency across departments, making it easier for team members to find and use documents as needed, regardless of their physical location.
What is an example of a document repository?
An example of a document repository is Accruent’s Meridian software. It serves industries like manufacturing, oil & gas, and pharmaceuticals by providing robust document management capabilities such as version control, security measures, and compliance tools. By using software like Meridian, a company can simplify and streamline the potential challenges that come with document management.
- EMS Software
- Maintenance Connection
- Customer Support
- Professional Services
- Customer Support FAQs
- Accruent Academy
- Food & Beverage Manufacturing
- Higher Education
- Manufacturing
- Industries Overview
- Press Releases
- Supplier Diversity
- Data Insights
- ServiceChannel
- Vx Suite/Verisae
- Schedule a Discovery Call
- Interactive Demos
- Pricing Calculators
- Product Walkthrough
- My Accruent

- Learning & Development
- Mobile training
Why your company’s knowledge repository has to be mobile-friendly in 2024
- Posted by by Athena Marousis
- April 10, 2024
- 6 minute read
In an era where information is the cornerstone of innovation, the concept of a knowledge repository has transcended from a buzzword to a fundamental asset for business.
A repository of knowledge serves as a digital sanctuary where information is stored, meticulously managed, and shared. With this, a culture of continuous learning and informed decision making is nurtured.
In today’s global economy, 80% of the working population is made up of deskless workers . As the workforce becomes increasingly decentralized, the ability to access crucial information on the go is imperative. Integrating a knowledge repository with a mobile learning platform makes this possible.
Making a knowledge management repository accessible anywhere and anytime connects people with information. Knowledge is kept current, relevant, and actionable.
Here, we’ll explore the significance of establishing a knowledge repository, its core elements, and strategies to maximize its impact.
What is a knowledge repository?
The core elements of a knowledge repository, why is a knowledge repository important, maximizing the impact of your knowledge repository, strategies for implementing a knowledge repository, leveraging a mobile learning platform for your knowledge repository, key takeaways.
A knowledge repository is a digital database where information is collected, organized, and stored for ongoing access. It serves as a central hub for essential information for businesses and organizations. By having a centralized system that captures and organizes knowledge assets, it becomes easier for staff to access and use what they need.
A knowledge repository is much more than just a digital archive. It’s the heartbeat of your organization’s knowledge ecosystem. By curating your repository of knowledge, you ensure that all information stored is accessible and up-to-date.
A knowledge repository core purpose is to facilitate seamless sharing and management of organizational knowledge . A business’s collective intelligence is safeguarded and made available to every individual, regardless of their role or location. Inside could be a deep dive into past project insights, research findings, procedures, or even tribal knowledge collected over the years. A knowledge repository has it all.
By answering the question, “What is a knowledge repository?” we uncover an indispensable tool for today’s global workforce. It’s a strategic asset that keeps information flowing. As businesses evolve, the amount of knowledge they create increases. Having a well structured knowledge repository is critical, as it connects people with the information they need, exactly when they need it . As a result, knowledge becomes both accessible and actionable.
A knowledge repository is an essential tool with several key components. Understanding these enhances the way information is used.
Content coverage
The foundation of your knowledge repository lies in its content. All topics must have relevant and comprehensive information. This ensures all users can find what they’re looking for.
Technological function
Technological capabilities form the backbone of a knowledge repository. Users must be able to easily access any information they need.
This includes:
- A robust knowledge base for data storage
- Intuitive search functionality for easy information retrieval
- A user-friendly interface
- Accessibility
To make it effective, your knowledge repository must be accessible and well-promoted to its intended users. Ease of access helps with buy-in and regular use.
- Content management
Ongoing management is critical to your repository’s success and reliability. Regularly updating, editing, and maintaining content ensures it will be valuable to its users.
- Analytics and feedback
Tracking usage and collecting user feedback helps with continuous improvement. Understanding user behaviors can guide adjustments to better serve their needs.
- Collaborative features
Tools that aid in knowledge sharing and collaboration are critical for a mobile workforce. These features in your knowledge repository will enrich insights and collective learning.
Investing in a knowledge repository not only supports the operational needs of an organization, but also positions it for future growth.
A knowledge repository is crucial for several reasons:
1. Facilitates knowledge sharing
It breaks down silos by enabling sharing of insights and information across the organization.
2. Supports decision making
Easy access to vital information means informed decisions can be made while reducing errors.
3. Enhances learning
Acting as a central hub for organizational learning, a knowledge repository allows employees to quickly access the information they need.
4. Drives innovation
Making knowledge accessible and actionable fosters an environment of continuous improvement and innovation.
5. Boosts productivity
Streamlining access to information significantly improves efficiency and productivity.
Creating a knowledge repository is an important first step. However, its true value only comes out when it’s actively managed and integrated into daily operations. Making your knowledge repository a valuable resource requires continuous effort and strategic planning.
Maintaining high-quality, up-to-date information is up first. Regularly review and update all information to keep your knowledge repository relevant and accurate. Implementing a structured process works best. Incorporate user feedback and allow for real-time updates. Always keep the content fresh and engaging.
Encourage employee engagement to maximize use and contribution. Consider implementing incentives that reward active participation. Integrate your repository into daily workflow by making it accessible by mobile devices.
Leverage analytics to increase your knowledge repository’s value. Identify how it’s used, which content is most popular, trends, and user engagement levels. This invaluable data helps identify the high-impact areas and where they can be improved.
Enhance accessibility by integrating your knowledge management repository with other tools. Linking it to project management and communication tools along with a mobile learning platform enhances it. These integrations make it a central part of your organization’s ecosystem. Seamless access to information when and where it’s needed most makes it an invaluable asset.
Starting your own knowledge repository might seem like a daunting task. Luckily, it’s completely doable, with a huge payoff. Here’s how to get started:
1. Gather information
Start by collecting all relevant information from across your organization. This ensures a comprehensive collection of existing knowledge, insights, and data spread across departments.
2. Categorize information
Now that you’ve gathered information, the next step is to organize it into categories. The goal is to make it easy and intuitive to navigate so that users can quickly find what they need.
3. Choose the right platform
Evaluate your needs and match them with a knowledge repository platform. Consider search capabilities, user interface, and integration.
4. Engage stakeholders
Involve key stakeholders from various departments. Gather input and promote buy-in to meet your organization’s needs.
5. Develop standards
Establish norms for content creation and review. Define roles and responsibilities for managing the repository.
6. Train users
Provide training and support to ensure your staff gets the most out of your knowledge repository. Highlight best practices for things like searches and adding content.
7. Implement feedback loop
Create a way for users to provide feedback on the content and usability. Use the feedback to make continuous improvements.
8. Monitor and evaluate
Review analytics regularly to identify areas for improvement. Adjust strategies based on user patterns, feedback, and needs.
Critical information cannot be confined to office walls or computers. Keeping knowledge at the fingertips of employees, accessible from anywhere at any time, is a must. Continuous learning and information sharing should always be possible, regardless of physical location.
Integrating a mobile learning platform with your knowledge repository is a strategic move in today’s increasingly digital world. How you manage and disseminate knowledge is more critical than ever. TalentCards , a leading mobile learning platform, exemplifies how this integration can transform knowledge sharing.
Advantages of using a mobile knowledge repository:
Increased engagement.
Mobile platforms encourage higher engagement rates. TalentCards leverages this advantage by presenting info in a microlearning format . This makes info quick to complete and easy to digest, allowing it to easily fit into employees’ busy schedules. This strategy leads to increased user engagement.
Flexible learning
With a mobile learning platform, your staff can learn anytime, and anywhere. Learning at their own pace and on their own terms means they can engage with content in a more meaningful way.
Real-time updates
One of the greatest advantages of using a mobile learning platform with your knowledge repository is the ability to update and push out information in real time. All of the latest information and best practices are always available to your workforce, allowing your repository to simultaneously function as work instruction software . This capability can mean the difference between leading, and lagging behind.
Make information easily accessible from anywhere with a powerful mobile app
What do you mean by knowledge repository?
A knowledge repository is a central place that stores, manages, and shares collective knowledge. It’s designed for easy access to information so people have the resources they need at all times.
What is an organizational knowledge repository?
An organizational knowledge repository is a digital system that captures, stores, and shares knowledge. Unlike broader knowledge repositories, it focuses on the organization’s unique needs.
It acts as the foundation of a culture of continuous learning and innovation within an organization. Knowledge is not just accessible, but actionable for everyone.
What are the elements of a knowledge repository?
The key components of a knowledge repository include:
- Comprehensive content coverage
- Functionality
- Knowledge base
Investing in a knowledge repository boosts operational efficiency, aids in making informed decisions, and encourages innovation across the organization. It cultivates a continuous learning and knowledge-sharing culture, increasing productivity and fostering organizational growth.
Empowering forward-thinking organizations
Establishing a knowledge repository goes beyond an organizational task. It’s a critical investment for your business. Standing at the core of fostering a knowledge driven culture, it enhances decision making and sparks innovation.
In today’s dynamic and often distributed workforces , integrating knowledge repositories with mobile learning platforms is a forward-thinking solution to location challenges. This combination guards valuable information, while keeping it at the fingertips of every employee. Employees can engage with digestible formats conducive to modern learning habits.
Organizations that will succeed in the information age are those that recognize the value of their knowledge assets. They commit to preserving and making them universally accessible and actionable. Integrating robust knowledge repositories with intuitive mobile training platforms represents a powerful, forward-thinking organization.
- A knowledge repository makes organizational knowledge centralized while promoting continuous learning.
- Regular updates keep knowledge repositories relevant and valuable.
- Linking a knowledge repository to a mobile training platform ensures information is accessible to your mobile workforce.
- Mobile access boosts employee engagement.
- Properly managed repositories drive innovation.
Make your company’s knowledge repository accessible on the go with TalentCards

You might also like

- CPA NEW SYLLABUS 2021
- KCSE MARKING SCHEMES
- ACTS OF PARLIAMENT
- UNIVERSITY RESOURCES PDF
- CPA Study Notes
- INTERNATIONAL STANDARDS IN AUDITING (ISA)
- Teach Yourself Computers
KNEC / TVET CDACC STUDY MATERIALS, REVISION KITS AND PAST PAPERS
Quality and Updated
KNEC, TVET CDACC NOTES AND PAST PAPERS
DIPLOMA MATERIALS
- KNEC NOTES – Click to download
- TVET CDACC PAST PAPERS – Click to download
CERTIFICATE MATERIALS
- KNEC CERTIFICATE NOTES – Click to download

- Repository Statistics
Tradition and innovation: six centuries of pottery production, use and disposal in late Saxon to late medieval East Anglia

Anderson, Sue (2023) Tradition and innovation: six centuries of pottery production, use and disposal in late Saxon to late medieval East Anglia. Doctoral thesis, University of East Anglia.
Built upon almost three decades of recording medieval pottery assemblages in East Anglia, this thesis utilises data from over 1800 sites and more than two million sherds – a ‘big data’ approach – to consider distributions of pottery fabrics and forms across the region between the mid-9th and mid-16th centuries. The pottery types in use in the three main period divisions across this date range are described and defined with regard to their fabrics and forms, and related to known and putative production sites. GIS-based study of the distribution of major fabrics and forms provides an insight into the type of pottery used across the region, how the various types related to each other and how these patterns changed over the centuries. Based on this evidence, themes relevant to each period are discussed in order to place the results within the historical and archaeological context of each period. Questions relating to differences between rural and urban pottery manufacture consumption, the role of immigrants, the growth of markets and their impact on pottery manufacture, and changes in patterns of waste disposal are considered. Case studies of the two main manufacturing areas – Grimston in north-west Norfolk and the Sandlings coastal strip of Suffolk – use new data to provide insights into the medieval pottery industry in the region, and the people who bought and used the potters’ products. The study concludes by bringing these themes together to provide an overview of pottery studies in East Anglia in the early 21st century, and provides some key research objectives for the future.
Actions (login required)

COMMENTS
A data repository is also known as a data library or data archive. This is a general term to refer to a data set isolated to be mined for data reporting and analysis. The data repository is a large database infrastructure — several databases — that collect, manage, and store data sets for data analysis, sharing and reporting.
Data Repositories. A key aspect of data sharing involves not only posting or publishing research articles on preprint servers or in scientific journals, but also making public the data, code, and materials that support the research. Data repositories are a centralized place to hold data, share data publicly, and organize data in a logical manner.
A research repository acts as a centralized database where information is gathered, stored, analyzed, and archived in one organized space. In this single source of truth, raw data, documents, reports, observations, and insights can be viewed, managed, and analyzed. This allows teams to organize raw data into themes, gather actionable insights ...
A data repository is a data library or archive. It may refer to large database management systems or several databases that collect, manage, and store sensitive data sets for data analysis, sharing, and reporting. Authorized users can easily access and retrieve data by using query and search tools, which helps with research and decision-making.
A data repository is a storage space for researchers to deposit data sets associated with their research. And if you're an author seeking to comply with a journal data sharing policy, you'll need to identify a suitable repository for your data. An open access data repository openly stores data in a way that allows immediate user access to ...
This document contains clearly defined desirable characteristics for two classes of online research data repositories: a general class appropriate for all types of Federally funded data—including free and easy access—and a specific class that has special considerations for the sharing of human data, including additional data security and ...
A data repository is a storage space for researchers to deposit data sets associated with their research. And if you're an author seeking to comply with a journal or funder data sharing policy, you'll need to identify a suitable repository for your data. An open access data repository openly stores data in a way that allows immediate user ...
Data Repositories. A key aspect of data management involves not only making articles available, but also the data, code, and materials used to conduct that research. Data repositories are a centralized place to hold data, make data available for use, and organize data in a logical manner.
Data is key to verification, replication, reuse, and enhanced understanding of research conclusions. When your data is in a repository—instead of an old hard drive, say, or even a Supporting Information file—its impact and its relevance are magnified. Here are six ways that putting your data in a public data repository can help your research go further.
A data repository, often called a data archive or library, is a generic terminology that refers to a segmented data set used for reporting or analysis. A data repository serves as a centralized storage facility for managing and storing various datasets. It encompasses:
The repository enables authorized users to easily and quickly access, retrieve and manage data using search, query, and other tools. Consequently, users and businesses can perform analysis, research, sharing, and reporting. And this enables them to streamline operations and make better data-driven decisions.
Data Research Repositories: Definitions. Online research data repositories are large database infrastructures set up to manage, share, access, and archive researchers' datasets. Repositories may be specialized and relegated to aggregating disciplinary data or more general, collecting over larger knowledge areas, such as the sciences or social ...
A research repository is a tool that professional user experience (UX) designers use to organize research across multiple professionals. A research repository handles two functions within an organization: growing the awareness of how user experience is important to leadership, product owners and organizations and supporting designers through ...
Open Data is a strategy for incorporating research data into the permanent scientific record by releasing it under an Open Access license. Whether data is deposited in a purpose-built repository or published as Supporting Information alongside a research article, Open Data practices ensure that data remains accessible and discoverable.
Find a Repository. When sharing your research data code, and documentation, there are many repositories to choose from. Some repositories are domain specific, and focus on a very narrow type of research. Other repositories, known as generalists, accept data from multiple disciplines and in various file formats.
A data repository is a data storage entity in which data has been isolated for analytical or reporting purposes. Since it provides long-term storage and access to data, it is a type of sustainable information infrastructure. While commonly used for scientific research, a data repository can also be used to manage business data.
Various studies have found that transparency is associated with trust of digital repositories 9. For example, for users of video data, "transparency of repository practices, and especially data ...
Revisit the past 6 months of research and capture the data we want to add to our repository as an initial body of knowledgeCreate the first draft taxonomy for our research repository, testing this with a small group of wider stakeholdersLaunch the repository with an initial body of knowledge to a group of wider repository champions
Repository traits and types. Descriptions of the sources will refer to repository traits (Table 1) that make them more or less useful and available for research. The first two traits are quantitative ones that we use later (Figure 1) to compare all the repository types. The first trait is the number of patients or research subjects observed.
A data library, data archive, or data repository is a collection of numeric and/or geospatial data sets for secondary use in research. A data library is normally part of a larger institution (academic, corporate, scientific, medical, governmental, etc.). established for research data archiving and to serve the data users of that organisation.
re3data.org is a global registry of research data repositories from all academic disciplines. It provides an overview of existing research data repositories in order to help researchers to identify a suitable repository for their data and thus comply with requirements set out in data policies. [1] [2] The registry went live in autumn 2012.
The Columbia University Clinical Data Repository. The initial design of BTRIS has been based on experience with the creation of the Clinical Data Repository (CDR) at the Columbia University Medical Center in New York.[] That system has accrued patient care data since 1988 from many different sources, including laboratories, pharmacies, radiology departments, order entry, and clinician ...
Definition by Nanyang Technological University. "Research data are data in whatever formats or form collected, observed, generated, created and obtained during the entire course of a research project. This would include numerical, descriptive, aural, visual or physical forms recorded by the researcher, generated by equipment and derived from ...
In May 2005, 16 major Dutch universities cooperatively launched DAREnet, the Digital Academic Repositories, making over 47,000 research papers available. ... US federal agencies must require all results (papers, documents and data) produced as a result of US government-funded research to be available to the public immediately upon publication.
Datasets consist of measurement data and metadata. Metadata provides context, essential for understanding and (re-)using data. Various metadata standards exist for different methods, systems and ...
A document repository simplifies the management of records while improving general operations across various organizational levels. Here are four key benefits that these systems provide: 1. Maximize Productivity . Tap the well of potential. Document repositories streamline the storage, retrieval, and sharing of documents.
A robust knowledge base for data storage; Intuitive search functionality for easy information retrieval; A user-friendly interface; Accessibility. To make it effective, your knowledge repository must be accessible and well-promoted to its intended users. Ease of access helps with buy-in and regular use. Content management
We're a team of professionals who have taught in several schools and been involved in marking of KCPE and KCSE exams. This website has curated content by giving you the best simplified study materials and revision papers for your exams
Built upon almost three decades of recording medieval pottery assemblages in East Anglia, this thesis utilises data from over 1800 sites and more than two million sherds - a 'big data' approach - to consider distributions of pottery fabrics and forms across the region between the mid-9th and mid-16th centuries. The pottery types in use in the three main period divisions across this ...