
One size does not fit all

Safety Notification

Deliver powerful randomization methods

Fast and accurate clinical coding

- Drug and Device Logistics
Mitigate risk and create closer connections

Essential document collection

Accelerate your trial and ensure data integrity

Meet patients where they are

Faster, cleaner and more reliable data

- Patient Poll
Quick insights from patients

We are hiring!

Capture better data, faster

Get up and running in no time

- PV Management
Identify and handle your safety management needs

- Data Management
Make more informed business decisions

Managed Service Model for MDR
Migration, Consulting and Customization for your MDR

- Submission and Data Mapping
Move and consolidate data

- eTechnology Strategy
Integrate and capitalize on critical technologies

- Protocol Development
Turning your research question into a study

- Observational Studies
Digital, cost effective and time efficient

- Primary Market Research
Direct to patient, unique, and quality data

- IRB and EC Management
End to end regulatory management

- PV and GDPR Compliance
Safety, consent and data protection

- Scientific Communications
Results dissemination

- Digital Communications
Increase patient awareness

- Data Linkage
Generate, combine and analyze data
Press Releases
Success Stories

What are the main clinical software? EDC, RTSM, CTMS, eTMF, ePRO and others.
Home » From Controlled Experiments to Real-Life Insights: How Clinical Trials and Real-World Evidence Shape Healthcare » What is a Clinical Trial? Definition, Phases, Compliance & Methodology » What are the main clinical software? EDC, RTSM, CTMS, eTMF, ePRO and others.
Table of Contents

Clinical trials are the backbone of medical research, providing essential evidence on the safety and effectiveness of medical interventions. In today’s digital era, software solutions play a pivotal role in streamlining trial processes, ensuring data accuracy, and enhancing patient safety . This article explores the various software applications used in clinical trials and their significance in driving research progress. From electronic data capture (EDC) systems to electronic patient-reported outcomes (ePRO) tools, electronic trial master files (eTMF), clinical trial management systems (CTMS), randomization and trial supply management (RTSM), electronic informed consent (eConsent), medical coding software, and data management and analysis tools, each software application contributes to the efficient management and analysis of trial data. Additionally, we will delve into emerging technologies and future trends that are shaping the landscape of clinical trial software. By leveraging these software solutions, researchers can optimize trial processes, generate reliable evidence, and ultimately improve patient care and outcomes .
Electronic Data Capture (EDC) Systems
Electronic data capture (EDC) systems have revolutionized data collection and management in clinical trials. These systems replace traditional paper-based data collection with digital platforms, streamlining data entry, validation, and query management. EDC systems offer numerous advantages, including improved data accuracy, real-time data access, and efficient data cleaning processes.
With EDC systems, researchers can design electronic case report forms (eCRFs) that align with the trial protocol. These forms are customized to capture specific data elements and ensure standardization across trial sites. By eliminating manual data entry, EDC systems reduce transcription errors and enhance data accuracy. Real-time data access allows researchers and sponsors to monitor trial progress, identify data discrepancies, and address queries promptly.
Data cleaning processes are also streamlined with EDC systems. These systems incorporate data validation rules and edit checks to ensure data consistency and integrity . Automated queries can be generated to resolve data discrepancies, accelerating the query resolution process. Additionally, EDC systems enable remote data monitoring, allowing for timely identification of data anomalies and ensuring data quality throughout the trial.
Overall, EDC systems improve data accuracy, data management efficiency, and data accessibility, facilitating evidence-based decision-making and speeding up the trial timeline.
Electronic Trial Master Files (eTMF)
Clinical trial documentation is vast and complex, requiring meticulous organization and management. Electronic trial master file (eTMF) systems digitize the management of trial-related documents, providing a centralized repository for essential trial documents and ensuring compliance with regulatory requirements.
eTMF systems streamline document management processes by replacing paper-based files with electronic counterparts. Trial documents, including study protocols, investigator brochures, informed consent forms, and monitoring reports, are securely stored and readily accessible in a digital format. This improves document version control, facilitates document sharing among trial stakeholders, and improves overall document traceability.
eTMF offer features such as document categorization, metadata tagging, and document versioning, enabling efficient organization and retrieval of trial documents. Automatic notifications and reminders help ensure document completeness and facilitate document review and approval workflows.
Moreover, eTMF software enhance collaboration among trial stakeholders by providing controlled access to trial documents based on user roles and permissions. This enables effective collaboration between sponsors, investigators, monitors, and regulatory authorities.
Compliance with regulatory requirements is a critical aspect of clinical trial documentation, and eTMF systems simplify this process. These systems support the implementation of standardized document structures and naming conventions , ensuring consistency and compliance. They also enable the generation of comprehensive reports on document status, completeness, and compliance, aiding in preparation for regulatory inspections.
Overall, eTMF systems streamline document management, enhance collaboration, and ensure regulatory compliance in clinical trials.
Clinical Trial Management Systems (CTMS)
Clinical trial management systems are an integral part of clinical trial software, facilitating the planning, execution, and monitoring of trials. CTMS platforms provide a comprehensive suite of tools and functionalities that enable efficient trial management and oversight.
CTMS systems assist in the management of trial timelines, budgets, resources, and site performance . Researchers can track the progress of study milestones, monitor enrollment rates, and manage study budgets and expenses. CTMS platforms often integrate with other software applications such as EDC systems and eTMF systems, enabling seamless data exchange and providing a unified view of trial data.
CTMS systems support site management activities, including site selection, site initiation, and site monitoring. These systems streamline the communication between study sites and sponsors, facilitating the exchange of essential trial documents and enabling remote monitoring of site activities. They often include features such as visit tracking, monitoring visit reports, and site performance metrics .
CTMS systems also offer reporting and analytics capabilities, providing real-time insights into trial progress, patient recruitment, and data quality. These systems generate comprehensive reports on key performance indicators (KPIs), enabling stakeholders to identify bottlenecks, make data-driven decisions, and take proactive measures to ensure trial success.
By leveraging CTMS systems, researchers can optimize trial management, improve site performance, and ensure efficient trial execution. These systems contribute to improved operational efficiency, data quality, and overall trial success.
Randomization and Trial Supply Management (RTSM)
RTSM systems play a crucial role in clinical trials, facilitating randomization of participants and efficient management of trial supplies . These systems have revolutionized the randomization process by replacing manual methods with automated, web-based platforms. RTSM systems offer numerous advantages, including enhanced randomization algorithms, real-time allocation of treatment arms, and improved inventory management .
With RTSM systems, researchers can implement sophisticated randomization algorithms that ensure unbiased and stratified assignment of participants to different treatment groups. This helps in balancing participant characteristics and minimizing potential confounding factors , enhancing the reliability and validity of trial results. Real-time treatment arm allocation allows for prompt decision-making and optimization of enrollment, ensuring efficient trial progression.
Moreover, RTSM systems enable seamless management of trial supplies, including study medications, placebos, and ancillary materials . These systems provide real-time visibility into inventory levels, enabling proactive planning and replenishment. Automated alerts and notifications help prevent stockouts or expired supplies, reducing the risk of disruption to the trial. Additionally, RTSM systems integrate with other clinical trial software applications, such as EDC and eTMF, enabling smooth data exchange and comprehensive trial management.
Overall, RTSM systems streamline the randomization process, improve inventory management, and enhance trial efficiency . By leveraging these systems, researchers can ensure unbiased participant assignment, optimize resource utilization, and ultimately accelerate the pace of clinical research.
Electronic Patient-Reported Outcomes (ePRO) Tools
Patient-reported outcomes (PROs) provide valuable insights into the patient’s perspective on treatment efficacy, symptoms, and quality of life. ePRO tools, which are electronic platforms for collecting PRO data, have transformed the way PROs are captured and analyzed in clinical trials.
ePRO tools enable patients to directly report their symptoms, treatment experiences, and quality of life using digital devices such as smartphones or tablets . These tools enhance data accuracy by minimizing recall bias and ensuring real-time data capture. Patients can conveniently report their PROs from their own environment, improving compliance and reducing the burden of frequent clinic visits.
ePRO platforms offer various advantages, including automated data capture, data standardization, and customizable assessments . The data captured through ePRO tools can be automatically integrated with the trial database, eliminating manual data entry and reducing the potential for data transcription errors. Standardized assessments ensure consistency in data collection across different trial sites and allow for direct comparisons between treatment groups.
Customizable assessments enable researchers to tailor PRO questionnaires to specific trial objectives and capture disease-specific outcomes. ePRO tools also facilitate the administration of validated PRO questionnaires and can incorporate skip patterns to optimize the patient’s survey experience.
By leveraging ePRO tools, researchers can capture high-quality PRO data, gain insights into the patient experience, and enhance patient-centered outcomes assessment in clinical trials.
Electronic Informed Consent (eConsent)
Informed consent is a fundamental aspect of ethical clinical research . Traditional paper-based informed consent processes have limitations in terms of comprehension and documentation management. eConsent platforms digitize and streamline the consent process, providing interactive and multimedia-rich content. This enhances participant understanding, engagement, and documentation management.
eConsent platforms present trial information to participants in an interactive and engaging manner. Multimedia elements such as videos, graphics, and interactive modules improve participants’ understanding of trial procedures, potential risks, benefits, and their rights . Participants can navigate through the consent document at their own pace, seeking clarification on specific sections or terms.
Real-time updates enable researchers to communicate protocol amendments, study updates, and safety information promptly. Consent forms can be dynamically updated, ensuring that participants have the most up-to-date information throughout the trial. Efficient consent documentation management allows researchers to track and manage consent forms electronically, simplifying document version control and ensuring compliance with regulatory requirements.
eConsent platforms also facilitate remote consent processes, enabling participants to review and provide consent remotely . This feature improves participant convenience, particularly for trials conducted across multiple sites or geographically dispersed locations. Remote consent enhances trial accessibility and increases participation rates, improving trial diversity and generalizability of results.
By leveraging eConsent platforms, researchers can elevate participant comprehension, streamline consent documentation management, and uphold ethical standards in clinical trials.
Medical Coding and Terminology Management Software
Accurate and standardized representation of medical concepts is crucial in clinical trials. Medical coding software, commonly known as “ Coder ,” assists in assigning appropriate codes to medical terms, facilitating data analysis, reporting, and regulatory compliance . Coder software improves data accuracy, supports interoperability, and streamlines the coding process.
Coding software automates the process of assigning standardized codes to medical terms based on coding dictionaries such as MedDRA (Medical Dictionary for Regulatory Activities) or SNOMED CT (Systematized Nomenclature of Medicine – Clinical Terms). This automation improves coding accuracy and consistency, reducing the risk of human error and ensuring standardized representation of medical concepts across trials.
Moreover, coding software facilitates efficient data analysis by enabling researchers to group and aggregate coded data. By categorizing adverse events, medical procedures, and other medical concepts, researchers can analyze safety profiles, identify common patterns, and assess treatment effects accurately.
Standardized medical coding also ensures interoperability and data exchange across different systems and healthcare organizations. Consistent coding facilitates data sharing, collaboration, and comparative analyses between trials and real-world data sources.
Furthermore, coding software aids in regulatory compliance by ensuring that trial data is accurately classified and reported according to regulatory requirements. Consistent and standardized coding improves the quality and reliability of safety and efficacy data submitted to regulatory agencies .
Overall, medical coding software improves data accuracy, supports interoperability, and ensures regulatory compliance in clinical trials.
Software Solutions: Advancing Clinical Trials and Healthcare
Software solutions have revolutionized the landscape of clinical trials, streamlining processes, optimizing data management, and driving research progress . From electronic data capture systems to electronic patient-reported outcomes tools, electronic trial master files, electronic informed consent platforms, and medical coding software, each software application contributes to the efficient management and analysis of trial data. These software solutions enhance data accuracy, improve participant engagement, ensure regulatory compliance, and expedite the trial timeline . As technology continues to evolve, embracing emerging technologies such as artificial intelligence, machine learning, and blockchain will further enhance the capabilities of clinical trial software. By leveraging software solutions, researchers can optimize trial processes, generate reliable evidence, and ultimately improve patient care and outcomes . Embracing these advancements in software technology is essential for driving the future of clinical research and transforming healthcare.
Last updated:
- October 24, 2023
Understanding Health Technology Assessment (HTA): Process, Steps, and Real-World Examples
RTSM (Randomization and Trial Supply Management): Enhancing Efficiency in Clinical
Exploring eConsent: Revolutionizing Informed Consent in Clinical Trials
What is an eTMF (electronic Trial Master File)?
Streamlining Clinical Trials: The Power of CTMS Software
What is an Electronic Data Capture (EDC) software?
EvidentIQ Group GmbH Rathausmarkt 5, 20095 Hamburg, Germany.
- Tel: +49 89 4522775-000
- Email: [email protected]
Quick Links
Whitepapers
Subscribe for monthly newsletter
Copyright 2024 EvidentIQ
All Rights Reserved
Commercial Register: Hamburg HRB 147319
Sales Tax Identification Number: DE312852940
- Privacy Policy
- Legal Notice
Managing Directors
Andreas Weber, Manuel Neukum, Axel Jansen, Lars Kloppsteck, Helge Hofmeister
Commercial Register: Hamburg, HRB 147319
Sales tax identification number: DE312852940
Subscribe for monthly newsletters
- Terms & Conditions
Copyright 2023 EvidentIQ. All Rights Reserved
REQUEST FOR PROPOSAL
Schedule a meeting, request a demo.

Exploring The 22 Best Clinical Trial Management Software Of 2024
The Medical Practice is reader-supported. We may earn a commission when you click through links on our site—read our affiliate disclosure to learn more about how we aim to stay transparent.
12 Best Clinical Trial Management Software Shortlist
As an expert, I've scrutinized numerous tools to deliver the 12 best clinical trial management software:
- Viedoc - Best for robust data management and trial design
- Vial - Best for user-friendly interfaces and trial execution
- Clario - Best for comprehensive trial enablement and management
- Medidata Rave CTMS - Best for optimized clinical operations and study management
- RealTime Software Solutions - Best for real-time tracking of clinical trials and participant management
- Medrio - Best for decentralized clinical trials and data collection
- Advarra - Best for multi-site trials and compliance management
- Phlexglobal - Best for electronic trial master file (eTMF) management
- Citeline - Best for global clinical trial solutions and comprehensive market analysis
- ResearchManager - Best for collaborative research and workflow management
- Calyx - Best for integrated trial management and regulatory compliance
- Florence eTMF - Best for document management and streamlined trial collaboration
Navigating the complex world of clinical research can feel like a Herculean task. Clinical Trial Management Software (CTMS) serves as an essential tool, simplifying this intricate process by offering an end-to-end, web-based solution.
It's a valuable asset to any research organization, pharma startup, or biotech company looking to streamline its clinical data management and trial supply management. The built-in API also provides flexibility for custom integrations, enhancing interactions among stakeholders, and improving contract research organizations' output. Let's dive into some remarkable options that are primed to help you manage your clinical conductor duties with efficiency and compliance at the forefront.
What Is a Clinical Trial Management Software?
Clinical trial management software is a powerful tool that aids researchers, clinicians, and other healthcare professionals in planning, executing, and managing clinical trials. Often used by pharmaceutical and biotechnology companies, research institutions, and medical device manufacturers, these systems streamline the complex processes associated with clinical trials.
Using a CTMS, you can leverage its cloud-based system to store and manage patient data, monitor enrollment, and coordinate activities across different research sites. Particularly valuable features include eCRF and eCOA, as well as eConsent and eSource functionalities that provide a comprehensive audit trail. With features like templates, financial management tools, messaging, notifications, and mobile app accessibility, it offers you an efficient way to manage your clinical study.
With features like data collection and analysis, regulatory compliance tracking, participant management, and more, they serve to improve efficiency, enhance collaboration, and ensure the integrity and accuracy of trials. The end goal is a more structured, organized, and reliable approach to conducting and managing clinical research, driving innovation in healthcare and medical fields.
Overviews of the 12 Best Clinical Trial Management Software
1. viedoc - best for robust data management and trial design.
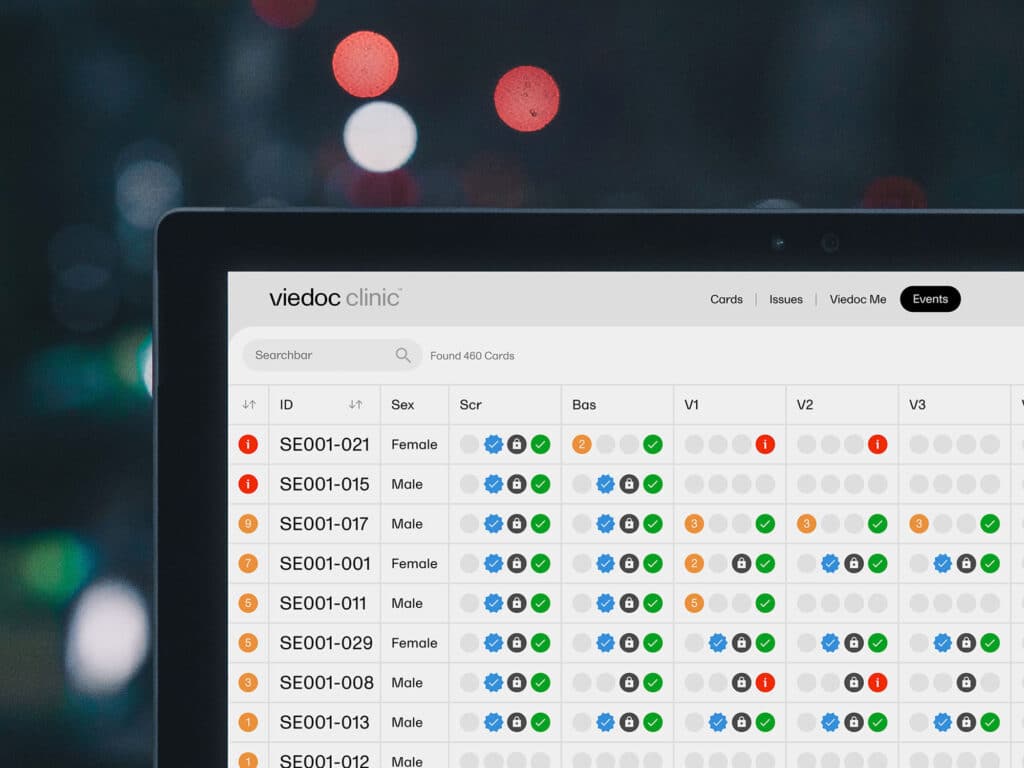
Viedoc stands as a powerful software dedicated to streamlining and strengthening data management and trial design for clinical research. Its robust suite of features and user-friendly interface make it an optimal choice for researchers and institutions needing an intuitive yet robust solution for their clinical trials.
Why I Picked Viedoc:
I selected Viedoc due to its unique ability to integrate complex data management systems with a comprehensive trial design toolkit. Its capacity to handle large volumes of data with precision sets it apart from its peers. Viedoc's superior functionality makes it ideal for robust data management and intricate trial design, two critical factors in successful clinical research.
Standout Features & Integrations:
Viedoc includes data collection, patient engagement, trial design, and electronic data capture features. Its analytics capabilities ensure an efficient and accurate understanding of trial data. Notable integrations include connections to medical imaging systems, laboratory systems, and electronic medical record systems, further enhancing its utility.
From $150/user/month (min 5 seats)
- Comprehensive data management and trial design functionalities
- High-volume data handling capabilities
- User-friendly interface with intuitive navigation
- Starting price may be higher than some competitors
- Minimum of five seats required for the basic package
- May have more features than smaller trials require
2. Vial - Best for user-friendly interfaces and trial execution
As a software solution for clinical trial management, Vial offers a blend of simplicity and robust functionality. Its design caters to user-friendliness, making it a valuable tool for individuals and teams who prioritize ease of use and effective trial execution.
Why I Picked Vial:
In my evaluation process, Vial stood out due to its intuitive interface and efficient execution of trial processes. Its attention to user experience, paired with its solid feature set, sets it apart in the crowded field of clinical trial software. Its strengths in user-friendly interface design and trial execution made it a clear choice for this list.
Vial boasts a variety of important features, such as participant tracking, data capture, and analysis, as well as comprehensive trial execution tools. Noteworthy integrations include connections to laboratory systems, electronic health records, and other trial-related software, which further enhance its capabilities.
From $120/user/month (billed annually)
- User-friendly interface for easy navigation
- Comprehensive trial execution tools
- Integration with a variety of trial-related systems
- Annual billing may limit flexibility for some users
- Interface may be too simplistic for users desiring complex features
- Pricing may be steep for smaller research teams
3. Clario - Best for comprehensive trial enablement and management
Clario is a comprehensive clinical trial management software that offers a powerful suite of features for trial enablement and management. It provides a streamlined, organized approach to clinical research, making it an optimal choice for those who require full control and oversight of their trials.
Why I Picked Clario:
I chose Clario for its comprehensive approach to trial enablement and management. Its robust feature set, coupled with its ability to manage all aspects of a clinical trial, make it a unique choice among other software options.
Clario's strength in comprehensive trial enablement and management makes it best suited for this role.
Clario comes packed with essential features including robust data collection and analysis, regulatory compliance tracking, and participant management. As for integrations, it provides compatibility with major electronic health records systems, laboratory systems, and other clinical trial software, enabling a more interconnected trial process.
From $130/user/month (min 5 seats)
- Comprehensive suite of features for trial enablement and management
- Robust data collection and analysis
- Good integration with other clinical systems
- Might be overwhelming for smaller teams due to its extensive features
- A minimum of five seats required for the basic package
- Slightly higher starting price compared to some other options
4. Medidata Rave CTMS - Best for optimized clinical operations and study management
Medidata Rave CTMS is a trailblazer in providing specialized features for optimized clinical operations and comprehensive study management. Its smart approach to trial oversight ensures researchers have every resource at their fingertips to carry out efficient trials.
Why I Picked Medidata Rave CTMS:
I picked Medidata Rave CTMS for its focus on enhancing the efficiency of clinical operations. Its comprehensive set of features targeted at study management set it apart from the crowd. This strong focus makes Medidata Rave CTMS the best for optimizing clinical operations and managing studies effectively.
Medidata Rave CTMS offers a rich suite of features like real-time data capture, study planning and tracking, and regulatory compliance management. As for integrations, it offers a broad range of interoperability with various data capture systems, electronic health record systems, and other CTMS platforms.
From $100/user/month (billed annually)
- Comprehensive set of features for clinical operations and study management
- Robust integrations with other healthcare and clinical trial systems
- Real-time data capture for instantaneous insights
- Pricing may be high for smaller research teams
- Only offers annual billing, not monthly
- Could be complex for teams new to CTMS software
5. RealTime Software Solutions - Best for real-time tracking of clinical trials and participant management

RealTime Software Solutions brings a fresh perspective to the clinical trials management space with its emphasis on real-time tracking and participant management. The software's capabilities extend from patient recruitment to data management, all in real-time.
Why I Picked RealTime Software Solutions:
I chose RealTime Software Solutions based on its emphasis on real-time monitoring and participant management. This focus allows the software to provide an immediate snapshot of trial progress and participant status, an aspect not all competitors offer.
Given this, RealTime Software Solutions stands as the best for real-time tracking and participant management in clinical trials.
RealTime Software Solutions excels in offering features such as instant trial monitoring, patient recruitment, and management, and streamlined data management. When it comes to integrations, the platform offers significant compatibility with EDC systems, patient recruitment tools, and data analytics solutions, among others.
From $50/user/month (billed annually)
- Real-time monitoring capabilities enhance trial oversight
- Strong focus on participant management
- Diverse integrations with other healthcare and trial systems
- May be complex for small teams or beginners in clinical trial management
- Pricing might be steep for some budgets
6. Medrio - Best for decentralized clinical trials and data collection
Medrio is a clinical trials management tool that specializes in decentralized trials and data collection. The platform supports data collection from multiple sources, and its decentralization capabilities enable remote management of clinical trials, which is key in an era when remote work is increasingly common.
Why I Picked Medrio:
I selected Medrio for its focus on decentralized trials and robust data collection. These two areas of focus are becoming more important in clinical trial management due to increasing trends toward remote work and data-driven decision-making. Thus, Medrio shines in this area and is, in my view, the best tool for decentralized clinical trials and data collection.
Key features of Medrio include its comprehensive data collection tools, support for decentralized clinical trials, and an intuitive user interface that simplifies complex processes. Medrio integrates seamlessly with a variety of data analysis tools, and EDC systems, and also supports direct data imports from various sources.
Pricing upon request
- Emphasis on decentralized clinical trials makes it ideal for remote teams
- Robust data collection and management capabilities
- Seamless integrations with data analysis tools
- Pricing is not publicly available, which could deter some users
- May have a learning curve for those not familiar with decentralized trials
- Its strong focus on decentralization may not suit all trial management styles
7. Advarra - Best for multi-site trials and compliance management

Advarra is a platform built to aid clinical trial management across multiple sites. It also includes advanced tools for regulatory compliance, making it a sound choice for organizations seeking to manage complex, multi-site clinical trials while adhering to all necessary rules and regulations.
Why I Picked Advarra:
I chose Advarra because of its emphasis on multi-site trial management and compliance. When compared to other clinical trial management tools, Advarra stood out due to its robust features and specific focus on managing trials across multiple sites while maintaining compliance. Therefore, I determined it to be the best tool for this specific use case.
Advarra provides features like comprehensive multi-site trial management, regulatory compliance tools, and robust reporting functionalities. It integrates smoothly with many EDC systems, data analysis tools, and document management platforms, enabling a connected and streamlined workflow.
- Comprehensive features for multi-site trial management
- Strong emphasis on regulatory compliance
- Seamless integrations with other clinical trial tools
- Pricing information is not easily accessible
- Could be complex for smaller trials or single-site management
- The user interface may have a steep learning curve for some users
8. Phlexglobal - Best for electronic trial master file (eTMF) management
Phlexglobal is a solution specifically designed for managing electronic trial master files (eTMFs). By providing an easy-to-use interface and robust tools, it simplifies the organization, storage, and retrieval of eTMF data. This specialization makes Phlexglobal an ideal fit for organizations looking to improve their eTMF management.
Why I Picked Phlexglobal:
I picked Phlexglobal due to its unique focus on electronic trial master file management. Comparing different platforms, Phlexglobal differentiated itself with its specialized tools for eTMF organization, making it an excellent choice for those specific needs. It is why I've deemed it as "best for eTMF management."
Phlexglobal features a comprehensive eTMF management system, advanced search functionalities, and a variety of reporting tools. It also integrates well with various other clinical trial software, which makes it easier to import and export data.
- Dedicated eTMF management system
- Advanced search and reporting capabilities
- Seamless integrations with various other clinical trial software
- Lack of clear pricing information
- May be over-specialized for organizations that need a broader range of tools
- Requires training to effectively use all the functionalities
9. Citeline - Best for global clinical trial solutions and comprehensive market analysis

Citeline provides solutions for global clinical trials and in-depth market analysis. The platform aggregates and analyzes data from a myriad of sources, enabling users to keep track of clinical trials worldwide and conduct comprehensive market research.
These abilities make it an exceptional choice for businesses operating or observing trials on a global scale.
Why I Picked Citeline:
In selecting Citeline, its strength in providing a broad view of global clinical trials and offering comprehensive market analysis stood out. Judging by its comprehensive dataset and analytical capabilities, Citeline provides unmatched insights into global clinical trials.
This specificity and focus make it the "Best for global clinical trial solutions and comprehensive market analysis."
Citeline offers a comprehensive data set of global clinical trials and a suite of tools to analyze and visualize that data. Additionally, it integrates well with various business intelligence tools, extending the power of data analysis and reporting capabilities.
- Provides data on global clinical trials
- Comprehensive market analysis tools
- Effective integrations with business intelligence tools
- Absence of transparent pricing information
- Can be complex for new users
- Focus on global data may not be useful for localized operations
10. ResearchManager - Best for collaborative research and workflow management
ResearchManager is a dynamic platform designed to streamline the research process and facilitate collaboration. It allows users to manage their research workflows effectively while promoting teamwork among researchers, thereby making it suitable for collaborative research and workflow management.
Why I Picked ResearchManager:
I chose ResearchManager for its unique capability to blend workflow management with a collaboration-friendly environment. In making this selection, I observed that ResearchManager stands out for its focus on enabling smooth cooperation between research team members while keeping workflows organized and efficient.
This uniqueness solidifies its position as "Best for collaborative research and workflow management."
ResearchManager provides a suite of tools for managing workflows, data collection, and storage. The platform also boasts of a strong collaborative feature set that includes real-time data sharing and multi-user project management. It integrates effectively with standard data analysis and project management tools, enhancing its usability.
- Emphasizes collaborative work and workflow management
- Real-time data sharing
- Effective integrations with analysis and project management tools
- Steep learning curve for beginners
- Collaboration features may be overkill for small-scale or solo research projects
11. Calyx - Best for integrated trial management and regulatory compliance
Calyx provides a comprehensive clinical research platform that combines trial management and regulatory compliance functionality. This makes it a versatile tool for ensuring organized, compliant research practices and makes it a top choice for integrated trial management and regulatory compliance.
Why I Picked Calyx:
In deciding on this list, Calyx emerged as a preferred option because of its unique integration of trial management with regulatory compliance mechanisms. It's distinctive in its approach, providing a holistic solution to trial management that considers regulatory needs.
This blend of functionality made me see Calyx as "Best for integrated trial management and regulatory compliance."
Key features of Calyx include its extensive trial management tools and robust compliance tracking capabilities. Furthermore, it provides an integrated electronic data capture system for seamless data management. It offers important integrations with data management and visualization tools that enhance its trial management capabilities.
- Integrated trial management and regulatory compliance
- Extensive management tools
- Seamless data management through integrated electronic data capture
- Lack of transparent pricing
- Might be complex for users new to clinical trial management
- Certain features may exceed the needs of small-scale trials
12. Florence eTMF - Best for document management and streamlined trial collaboration
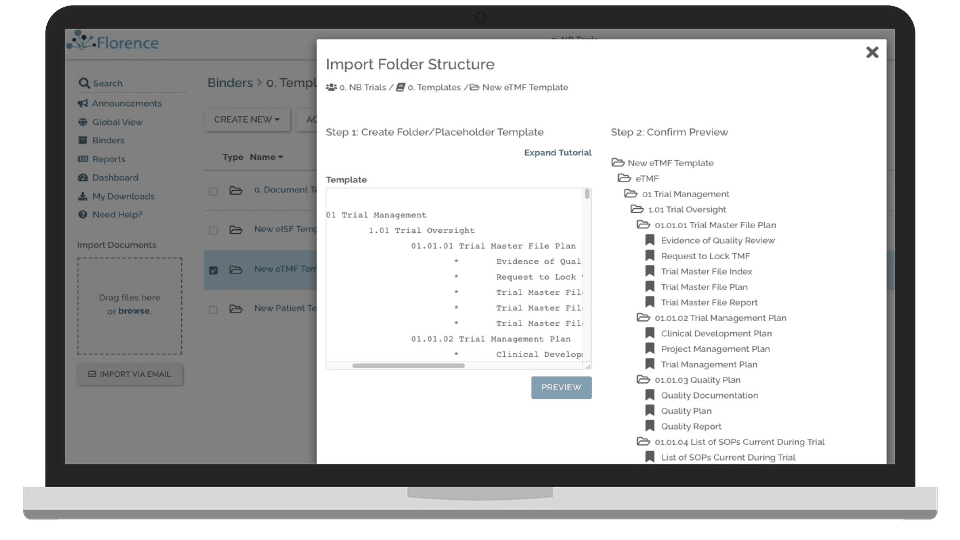
Florence eTMF is a dedicated tool that focuses on facilitating efficient document management and fostering improved collaboration within clinical trials. Its robust document-handling capabilities and collaborative features make it an ideal solution for document management and streamlined trial collaboration.
Why I Picked Florence eTMF:
Florence eTMF earned its spot on this list primarily because of its emphasis on simplifying document management and enhancing collaboration within clinical trials. It stands out with its dedicated tools for managing documents and facilitating communication between trial participants.
Because of these reasons, I consider Florence eTMF to be the "Best for document management and streamlined trial collaboration."
Florence eTMF shines with features like its centralized document repository, which provides an organized, easily accessible hub for all trial documents. It also offers robust collaboration tools designed to streamline communication and coordination between team members.
Integration-wise, Florence eTMF integrates well with a range of commonly used software in the clinical trial space, including data management, project management, and communication tools.
- Centralized document repository for efficient management
- Robust collaboration tools for improved trial coordination
- Effective integrations with other clinical trial tools
- Lack of transparent pricing information
- May be too specialized for some users, lacking broad trial management features
- Learning curve may be steep for less tech-savvy users
Other Clinical Trial Management Software
- Oracle Clinical - Good for decentralizing clinical trials
- Veeva Vault CTMS - Good for medical technology clinical trials
- ClinCapture - Good for user-friendly electronic data capture
- OpenClinica - Good for open-source clinical trial solutions
- Datatrak - Good for enterprise-level clinical trial management
- TrialKit - Good for mobile data capture in clinical trials
- Fusion eClinical Suite - Good for integrating multiple clinical trial systems
- Octalsoft - Good for scalable clinical trial management systems
- Clinion - Good for paperless clinical trial management
- BS CTMS - Good for comprehensive trial management solution
Selection Criteria For Clinical Trial Management Software
When it comes to clinical trial management software, the nuances of each platform can significantly impact a team's efficacy and the overall success of a study. I've extensively tested and researched a multitude of these tools, and I've been particularly vigilant about certain key aspects that can enhance or hinder a research team's work.
Core Functionality
A competent clinical trial management software should enable users to:
- Conduct data collection and management efficiently
- Facilitate patient recruitment and retention
- Coordinate site management
- Streamline trial monitoring
- Oversee inventory management
Key Features
For this niche, the following features matter the most:
- Electronic Data Capture (EDC) : A tool that allows the collection of clinical trial data in an electronic format is a must for real-time, accurate data monitoring.
- Randomization : This feature helps balance patient allocation across different trial arms, ensuring the validity of the study.
- Electronic Patient-Reported Outcomes (ePRO) : An essential feature for collecting patient responses or feedback directly.
- Regulatory Compliance Management : Tools with this feature ensure that all data and processes comply with necessary regulations like HIPAA, GCP, and GDPR.
- Study Designing : A tool should allow users to design their study layout according to their specific requirements.
When it comes to usability, the following factors are significant:
- Intuitive Interface : Clinical trial software needs to have a clear, easy-to-navigate layout that allows users to quickly find and access different modules of the platform.
- Scalability : The ability to handle increasing amounts of work and accommodate growth is crucial, especially for larger trials or expanding organizations.
- Role-Based Access Control : A system that allows admins to limit access based on user roles is key for maintaining data integrity and confidentiality.
- Training and Support : Given the complexity of clinical trials, having readily available and responsive customer support, along with comprehensive training resources, is essential.
People Also Ask
What are the benefits of using clinical trial management software.
Clinical trial management software offers multiple benefits, including:
- Efficiency : By automating various tasks, these tools reduce manual labor, save time, and streamline workflow.
- Data Accuracy : Digital data collection minimizes errors associated with manual data entry, thus ensuring data accuracy.
- Collaboration : These platforms offer a central hub for all trial-related activities, fostering better communication and collaboration among teams.
- Regulatory Compliance : Built-in compliance features help maintain adherence to regulatory standards like HIPAA, GCP, and GDPR.
- Real-Time Monitoring : The software allows real-time monitoring of trial progress, facilitating timely intervention and decision-making.
How much does clinical trial management software cost?
The cost of clinical trial management software varies significantly based on the size of the clinical trial, the number of users, and the specific functionalities required. Some providers offer tiered pricing, where you pay per user per month, while others may charge a flat rate.
What are the pricing models for these tools?
Typically, vendors offer several pricing models:
- Per User/Month : You pay a certain amount for each user every month.
- Per Study : You pay a one-time fee for each study conducted using the software.
- Flat Rate : You pay a flat monthly or annual fee for unlimited access to the software.
What is the typical range of pricing?
Prices can range from $25/user/month for basic packages to several thousand dollars per month for enterprise-level solutions that offer advanced features.
What is the cheapest and most expensive software?
The cheapest paid software can start as low as $25/user/month, such as TrialKit, while the most expensive ones like Oracle Clinical can cost several thousand dollars per month, especially for larger clinical trials or for more complex features.
Are there any free clinical trial management software options?
Yes, there are a few free options available like OpenClinica, which offers a free version of their software. However, these free options often have limitations on features and user capacity and are best suited for small-scale trials or for initial exploratory use before transitioning to a paid solution.
Other Clinical Management Software Reviews
- Medical Lab Software
- Patient Engagement Software
- EMR Systems
- Patient Case Management Software
In conclusion, selecting the ideal clinical trial management software relies heavily on understanding your unique needs and trial requirements. Such software offers substantial benefits like streamlined workflows, data accuracy, enhanced collaboration, regulatory compliance, and real-time monitoring of trial progress. However, the perfect tool for one organization may not be the same for another.
Key Takeaways
- Define Your Requirements : Clarify what specific functionalities, features, and compliance standards your trial necessitates. Do you need document management capabilities? Are integrations with existing systems critical? Answering these questions helps narrow down suitable options.
- Consider User Experience : Software usability significantly affects team productivity and software adoption rates. Seek software with a user-friendly interface, easy onboarding, and good customer support. Role-based access, an intuitive dashboard, and simple navigation are crucial aspects of clinical trial management software.
- Budget Appropriately : Pricing varies widely among these tools. Understand the pricing model that aligns best with your budget and the scale of your trials. While affordability is essential, ensure not to compromise on crucial features for the sake of cost savings. Some vendors offer free versions or trials, which are worth exploring before committing to a paid plan.
Navigating the landscape of clinical trial management software may seem daunting, but armed with the right knowledge and clear objectives, you can make an informed decision that will significantly improve your clinical trial processes.
What Do You Think?
Of course, this guide is not exhaustive, and the world of clinical trial management software continues to evolve with new tools and solutions entering the market. If you've come across an excellent tool that wasn't included in this list or have personal experiences with the tools I've mentioned, I invite you to share your insights. Your contributions could help others make more informed decisions in their quest for the perfect clinical trial management software.
Clinical Trial Management Software
Find the best Clinical Trial Management Software
Popular comparisons, buyers guide, filter products, company size.
- Self-Employed

Pricing Options
- # of User Reviews
- Average Rating
- Alphabetically (A-Z)
Compare Products
Showing 1 - 20 of 84 products

Health Cloud
Salesforce Health Cloud is a patient management platform designed to help medium to large healthcare organizations manage inquiries, track issues, deliver post-acute care processes and more. Key features include clinical data mana... Read more about Health Cloud
4.6 ( 5 reviews )

Designed for hospitals, medical centers, trial sites and more, Complion is a cloud-based clinical trial management solution that helps handle manual processes on a centralized interface. The software automates investigator regulat... Read more about Complion
4.5 ( 2 reviews )

RealTime-CTMS
RealTime-CTMS is a web-based clinical trial management system. It is designed for sponsors, CROs, research sites, university medical centers, plus other research-based organizations. To help with overall study operations, from set... Read more about RealTime-CTMS
4.9 ( 66 reviews )

ClinCapture
Captivate EDC is a cloud-based electronic data capture solution that assists medical researchers and practitioners with data capture and forms management. Key features include WYSIWYG form editor, risk-based monitoring, activity d... Read more about ClinCapture
4.8 ( 15 reviews )

Visual Planning
Visual Planning is a hybrid resource management and scheduling platform that helps businesses to manage their assets and day-to-day operations. The solution can be deployed on-premise or hosted in the cloud. Visual Planning o... Read more about Visual Planning
4.7 ( 39 reviews )

eTMF Master
ClinPlus CTMS by Anju Software is a clinical trial management system (CTMS) designed to help sponsors and contract research organizations manage clinical trials. This solution offers workflow tools to help reduce outdated processe... Read more about eTMF Master
4.3 ( 4 reviews )

Florence eTMF
Get set up fast with the top-rated platform in ease-of-use, ease-of-setup, and customer support. Plus, connect directly to your sites with the only eTMF integrated with the industry-standard electronic investigator site file netwo... Read more about Florence eTMF
4.6 ( 18 reviews )

Florence SiteLink
SiteLink facilitates automated workflows like electronic logs, placeholders, eSignatures and quality assurance workflows, reduces start-up times by 40% for most of our customers and accelerates overall study timelines to reduce ti... Read more about Florence SiteLink
4.7 ( 3 reviews )

Florence eBinders
Florence is a clinical trial workflow platform that is easy to use and set up and provides excellent customer support. Even the most paper-loving PI will enjoy moving to Florence eBinders and the industry-leading implementation an... Read more about Florence eBinders
4.5 ( 74 reviews )

RegDocs365 is an electronic regulatory document management (EDM) system for life sciences. It caters to biotech, pharmaceutical, and medical device companies. The platform offers core features for managing regulatory documents. It... Read more about RegDocs365
3.8 ( 14 reviews )

LeadSquared
LeadSquared is a cloud-based marketing automation and customer relationship management (CRM) solution for businesses of all sizes. It serves clients in industries such as finance, e-commerce, education, health and wellness, market... Read more about LeadSquared
4.3 ( 159 reviews )

PatienTrials
PatienTrials is a clinical trial software that helps businesses engage and inform patients regarding trials. The platform enables managers to connect with teams, patients, and other stakeholders in real-time to share updates, disc... Read more about PatienTrials
No reviews yet

QuesGen Platform
QG-Case is a cloud-based comprehensive case management solution built specifically for the needs of programs focused on CVI (5t). It supports the program efforts of providing targeted counseling and support to those who have been ... Read more about QuesGen Platform

Datacubed Health
Designed for virtual, hybrid, or traditional clinical trial settings, our Datacubed Health App is proven to deliver 85%+ compliance for your clinical studies. Offline data capture allows patients to remain engaged even in the ab... Read more about Datacubed Health
4.0 ( 1 reviews )

ResearchManager
ResearchManager offers a sophisticated, flexible, and cost-effective eClinical platform encompassing Clinical Data Management Tools (EDC, ePRO, RTSM, eConsent) and Clinical Operations (CTMS, eTMF, LIMS, RIMS). Backed by a decade ... Read more about ResearchManager
4.2 ( 109 reviews )

Track.health
Track.health helps businesses in the healthcare industry streamline data acquisition for clinical study or trials. It captures EDC and eCOA data on a centralized platform. Track.health allows professionals to implement remote... Read more about Track.health

ClinicDr is a cloud-based medical practice management software which helps chiropractors handle treatment documentation, billing services, credentialing, claim scrubbing, scheduling, appointment reminders, telemedicine and more. K... Read more about ClinicDr
4.8 ( 10 reviews )

iMednet is a cloud-based electronic data capture platform designed to help the healthcare industry capture, clean, adjudicate and manage data across clinical trials. The application enables research teams to build and implement st... Read more about iMednet
5.0 ( 2 reviews )

Complex eClinical Solution for EDC/ERT/eCOA that significantly speeds up the start of clinical trials (up to 5 days), saves budget (up to 80% for monitoring), increases the capitalization and attractiveness of your company (noted ... Read more about MainEDC
4.8 ( 5 reviews )

Prelude EDC
Designed for clinical research, Prelude EDC is a CFR, Annex 11, and HIPAA compliant EDC solution that streamlines the data collection process, automates data cleaning efforts, and monitors the state of electronic case reports form... Read more about Prelude EDC
5.0 ( 12 reviews )

Google Cloud

Clinical research organizations (CROs) face several challenges, such as conducting multiple trials simultaneously, adhering to regulatory compliance, and ensuring transparency in financial transactions. To deal with these challenges, CROs need to be proactive, and clinical trial management software helps them do that. The software assists CROs and other healthcare organizations in protecting patients’ health and financial data to avoid compliance issues.
There are various clinical trial management software applications available on the market. However, choosing software with features that are right for your organization can be time-consuming. To help you, we've created this buyers guide with the information you need to know when making a purchase decision.
Here's what we'll cover:
What is clinical trial management software?
Common features of clinical trial management software, what type of buyer are you, benefits of clinical trial management software, key considerations when selecting clinical trial management software.
Market trends to understand
Clinical trial management software is used to manage clinical trials done in clinical research. It serves as a single, centralized repository to store all the clinical research studies conducted in a CRO.
The software helps CROs streamline daily tasks, such as team collaboration, performance management, tracking of deadlines, and regulatory document management. It also assists in planning, reporting, and benchmarking clinical research processes.
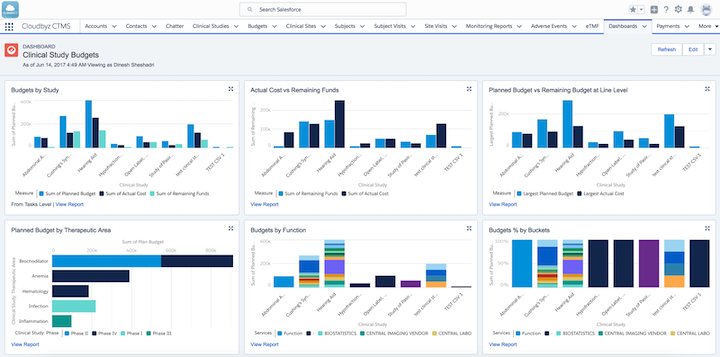
Clinical study budget analysis in Cloudbyz clinical trial management software ( Source )
Different clinical trial management software vendors' features offerings will vary. Selecting software with the right features for you is easier when you know what the most common features are, and what they do.
Before you start evaluating software options, you'll want to know which buyer category you belong to, so you can understand what to look for in a solution. Most buyers fall into one of the below categories.
Small and midsize CROs (5-100 employees): These CROs are usually founded by individuals or groups of physicians/scientists with expertise in a specific field. They have fewer employees, focus on a niche area of research, and usually operate out of a single location. Such buyers should consider software with features that are aligned with their area of clinical research or specialization. Some of the common software features they should consider are enrollment management, document management, patient database, and patient scheduling.
Large CROs (more than 100 employees): These CROs have a larger number of employees and several clinical research trials and studies on their annual agendas. They have to manage large volumes of data—documents, contracts, and financial transactions—for all the patients involved in trials. They also have to take care of legal and compliance requirements for clients, mostly big pharma companies. Such buyers should opt for a complete clinical trial management software suite that offers features such as compliance management, financial management, reporting, and third-party integration.
While some of the benefits of clinical trial management software may already be evident, we've listed the most notable ones in this section.
Ensuring compliance: Large volumes of data (patient data, billing information, study designs, deadlines, etc.) are processed during clinical trials. The software helps streamline the clinical audit process by tracking, managing, and consolidating all data into a single, centralized system. It also consolidates data from previous internal and external audits, compliance certificates, and other related processes. Further, it tracks any process deviations in daily protocols.
Tracking financial data: Clinical trial management software ensures accurate invoicing for all stakeholders and timely payment for all research trials and studies. It also lets CROs monitor their bottom lines to help control costs and filter out trials that aren’t bringing in revenue.
Managing data: The software offers a single and secure location for data collection, storage, and retrieval. Information—such as patient data or clinical study-related documents—is stored in a centralized repository and accessed by all authorized personnel through a searchable library.
Listed below are some important points to consider before you purchase clinical trial management software.
Mobile app availability: The software you’re planning to purchase should be available as a mobile app on iOS and Android smartphones or tablets. Mobile-ready software will allow you to perform tasks such as enrolling patients, updating contact details, and signing documents digitally, even when you’re on the move.
Customer support: Check out the type of support offered by your shortlisted vendors: 24/7, 24/5, or only during business hours. Also, ask about the available support channels, such as email, phone, or live chat. Support services will help you fix software issues that can hamper productivity or result in downtime.
Market trend to understand
Virtual clinical trials: New technologies, such as wearable sensors, smartphone apps, and mobile health (mHealth), are paving the way for virtual clinical trials. In virtual clinical trials, all steps, including patient enrollment, data collection, and follow-ups, are executed through video or audio calls. Such trials are likely to help reduce patient enrollment time and result in a more diverse pool of participants. Pharma companies across the globe are increasingly collaborating with major players in the clinical care space to conduct virtual trials.
Note: The application selected in this article is an example to show a feature in context and isn’t intended as an endorsement or recommendation. It has been obtained from sources believed to be reliable at the time of publication.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 23 August 2023
Making Biomedical Research Software FAIR: Actionable Step-by-step Guidelines with a User-support Tool
- Bhavesh Patel ORCID: orcid.org/0000-0002-0307-262X 1 ,
- Sanjay Soundarajan 1 ,
- Hervé Ménager ORCID: orcid.org/0000-0002-7552-1009 2 &
- Zicheng Hu ORCID: orcid.org/0000-0002-4168-1725 3
Scientific Data volume 10 , Article number: 557 ( 2023 ) Cite this article
1925 Accesses
1 Citations
11 Altmetric
Metrics details
- Data publication and archiving
Findable, Accessible, Interoperable, and Reusable (FAIR) guiding principles tailored for research software have been proposed by the FAIR for Research Software (FAIR4RS) Working Group. They provide a foundation for optimizing the reuse of research software. The FAIR4RS principles are, however, aspirational and do not provide practical instructions to the researchers. To fill this gap, we propose in this work the first actionable step-by-step guidelines for biomedical researchers to make their research software compliant with the FAIR4RS principles. We designate them as the FAIR Biomedical Research Software (FAIR-BioRS) guidelines. Our process for developing these guidelines, presented here, is based on an in-depth study of the FAIR4RS principles and a thorough review of current practices in the field. To support researchers, we have also developed a workflow that streamlines the process of implementing these guidelines. This workflow is incorporated in FAIRshare, a free and open-source software application aimed at simplifying the curation and sharing of FAIR biomedical data and software through user-friendly interfaces and automation. Details about this tool are also presented.
Similar content being viewed by others
FAIR in action - a flexible framework to guide FAIRification

The FAIR Cookbook - the essential resource for and by FAIR doers
Perceptions and behavior of clinical researchers and research support staff regarding data FAIRification
Introduction.
Research software (including scripts, computational models, notebooks, code libraries, etc.) is becoming increasingly vital in scientific research. A survey by the Software Sustainability Institute in the UK revealed that 92% of academics use research software, 69% say that their research would not be practical without it, and 56% develop their own software 1 . Other surveys have similarly emphasized the importance of research software in scientific research 2 , 3 , 4 . Research software plays a fundamental role not only in collecting, analyzing, and processing data but has also become the centerpiece of many scientific endeavors aimed at developing computational models to understand and predict various physical phenomena. In line with this general trend in scientific research, research software has also become an essential part of biomedical research over the last decade, especially with the emergence of machine learning and artificial intelligence in the field. The growing number of new biomedical-related software repositories created on GitHub every year (c.f. Fig. 1 ) provide an overview of this shift.

Evolution of the number of new biomedical-related software repositories created on GitHub in a given year (rounded to the hundredth when >1,000). The search consisted of looking for new repositories created in a given year with “biomedical” included in their name, tags, README, or description. To exclude repositories with data only (e.g., CSV files or markdown text), the search was limited to repositories with a major coding language being one of the popular software programming languages. This list of popular software programming languages was established based on GitHub’s list of popular programming languages to which we added a couple of languages that we deemed relevant for biomedical research software. For more details, see the code associated with this manuscript (c.f. Code Availability section).
Research software has consequently become an essential asset of scientific research and, therefore, it has become critical to preserve, share, and make it reusable. The Findable, Accessible, Interoperable, and Reusable (FAIR) guiding principles provide a foundation for achieving that 5 . Published in 2016, these principles are aimed at optimizing data reuse by humans and machines. While postulated for all digital research objects, several research groups have shown that the FAIR principles as written do not directly apply to software because they do not capture the specific traits of research software 6 , 7 . Lamprecht et al . were the first in 2019 to work on this shortcoming and published in 2020 reformulated FAIR principles that are tailored to research software 6 . Noticing yet a need for more software-specific principles, the FAIR for Research Software (FAIR4RS) Working Group, jointly convened as a Research Data Alliance (RDA) Working Group, FORCE11 Working Group, and Research Software Alliance (ReSA) Task Force, initiated a large-scale effort shortly after in mid-2020 to tackle this need. A subgroup of this working group took a fresh look at the FAIR principles and their applicability to research software, and suggested a set of reformulated FAIR principles tailored for research software that were published as a preprint in January 2021 8 and then as a peer-reviewed article in March 2021 9 . Based on that effort and the efforts of other subgroups, the FAIR4RS working group published in June 2021 a draft for formal community review of their new principles called the FAIR4RS principles (v0.3) 10 . After revising the FAIR4RS principles based on community feedback, they published the final version (v1.0) in May 2022 11 . This version was introduced in a peer-reviewed article in September 2022 12 .
Just like the original FAIR principles, the FAIR4RS principles are aspirational and aimed at providing a general framework. The “From FAIR research data toward FAIR and open research software” study provides some actionable guidelines for making research software FAIR but it does not include a clear process and is based on the original FAIR principles rather than the FAIR4RS principles 13 . The “Five recommendations for FAIR software” publication also provides some actionable guidelines for making research software FAIR but it is not aimed at complying with each principle and is also based on the original FAIR principles 14 . The absence of actionable guidelines, which researchers can adhere to in order to comply with each of the FAIR4RS principles, constitutes a hindrance to the widespread adoption of FAIR practices in research software development. Particularly in biomedical research, the COVID-19 pandemic has emphasized the need for such guidelines 15 . As noted by the RDA COVID-19 Working Group: “Whilst preprints and papers are increasingly openly shared to accelerate COVID-19 responses, the software and/or source code for these papers is often not cited and hard to find, making reproducibility of this research challenging, if not impossible” 15 .
To fill this gap, we proposed in this work the first minimal and actionable step-by-step guidelines for researchers to make their biomedical research software compliant with the FAIR4RS principles. Our focus was on biomedical research software given that this effort was initiated as part of a larger project aimed at supporting the curation and sharing of COVID-19 related research data and software. We designate these guidelines as the FAIR Biomedical Research Software (FAIR-BioRS) guidelines. Numerous challenges have been reported in formulating such actionable guidelines, including the absence of consensus within the research community concerning metadata and identifiers 8 . Rather than waiting for these challenges to be overcome, our approach consisted of deriving the best actionable guidelines possible around these challenges such that researchers can already start making their research software FAIR. This manuscript presents the FAIR-BioRS guidelines and details the approach we used to develop them.
As funding to support the reusability of software is lacking, the main responsibility of making research software FAIR falls on the researchers developing the software since they may receive only limited support to assist them in this effort. Our proposed guidelines, while crucial for enhancing software reusability, may impose an additional burden on these researchers who already grapple with significant challenges during the software development process 16 , 17 , 18 . To address this concern and ensure the FAIR-BioRS guidelines are easily accessible and widely embraced by researchers, we have developed a workflow to streamline the process of implementing the FAIR-BioRS guidelines. This workflow is incorporated into FAIRshare, a free and open-source desktop software application aimed at simplifying the processes for making biomedical data and software FAIR. FAIRshare takes the user’s software repository as input and guides them through step-by-step implementation of the FAIR-BioRS guidelines. FAIRshare provides an intuitive graphical user interface at each step where the user can easily provide required inputs for items that cannot be automatically assessed (e.g., selecting a desired license for the software), and includes automation in the backend to take over complex and/or time-consuming tasks that can be automated (e.g., creating a LICENSE file with standard license terms once the license is selected). More details about this tool are provided in this manuscript.
The work in this manuscript is based on the FAIR4RS principles v1.0 11 given that they are the most thorough to date and have garnered substantial community support. Our work was initiated in December 2021 and was initially based on the formal draft version of the FAIR4RS principles (v0.3) 10 . We then reiterated on our work between November 2022 and June 2023 to align with the final version (v1.0) of the FAIR4RS principles as well as incorporate community feedback we received along the way. Our overall process for establishing the FAIR-BioRS guidelines is illustrated in Fig. 2 . More details are provided in the Methods section.

Overall process followed to establish the current version of the FAIR-BioRS guidelines.
High-level categories and instructions to fulfill the FAIR4RS principles
For each of the FAIR4RS principles, we derived concise high-level instructions to fulfill that principle. These instructions were derived from pertinent information found in the FAIR4RS v1.0 publication 11 and the corresponding introduction manuscript 12 . These instructions are available in the “fair4rsv1.0Instructions” tab of the “data.xlsx” file associated with this manuscript (c.f. Data Availability section). We identified that the instructions for the different principles had overlapping themes and therefore organized the instructions into five action-based categories to help in our process of deriving actionable guidelines:
Category 1: Develop software following standards and best practices
Category 2: include metadata, category 3: provide a license, category 4: share software in a repository.
Category 5: Register in a registry
In Supplementary Table 1 , we present the instructions related to each category, along with the respective FAIR4RS principles they encompass. Then, for each category, we identified outstanding questions we needed to answer for deriving actionable guidelines that allow us to fulfill the high-level instructions from that category. These questions are also provided in Supplementary Table 1 . To answer these questions, we combined findings from a comprehensive review of relevant resources, our own understanding of research software development, and external suggestions we received from various communities (c.f. Discussion section) to derive relevant recommendations.
Review of current practices and resulting recommendations
Reviewed studies.
Our methodology for conducting the review is outlined in the Methods section. A list of the reviewed resources is available in the “resourcesList” tab of the “data.xlsx” file included in the dataset associated with this manuscript (c.f. Data Availability section). During the review, we sought actionable recommendations that addressed the questions listed in Supplementary Table 1 . A total of 39 resources were deemed relevant to these questions and included in the analysis presented next. The information collected from each resource can be found in the “resourcesReview” tab of the “data.xlsx” file included in the dataset associated with this manuscript (c.f. Data Availability section). We summarize the key findings below for each of the categories and provide our resulting recommendations.
None of the reviewed resources provided actionable items for following standard development practices, except one that mentions the PEP 8 Style Guide for Python Code ( http://peps.python.org/pep-0008 ). This is understandable since such standards vary depending on many factors such as the domain of research and coding language.
Similarly, there are no clear standards provided for the format of the data that software read, write, and exchange. Few of the reviewed resources 10 , 11 , 19 refers to the FAIRsharing Registry ( https://fairsharing.org ) for a curated list of community standards 20 .
Suggestions for best development practices are provided in 21 of the reviewed resources. Some of the reviewed resources are fully dedicated to best practices, including general best practices for scientific software development 21 and specific best practices for biomedical software 22 , 23 . Overall, developing with a version control system is commonly suggested 8 , 12 , 13 , 14 , 15 , 19 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 . GitHub, BitBucket, and GitLab are commonly mentioned ready-to-use version control system platforms. Using container technologies such as Docker and Singularity 13 , 18 , 19 , 22 , 24 , 29 , having code level documentation (in code comments, description in the headers) 19 , 21 , 22 , 25 , 30 , and recording dependencies 11 , 12 , 19 , 21 are other best practices mentioned in the reviewed resources.
There is clearly a lack of community-agreed standards and best practices. Therefore, we used findings from this review, combined with our own assessment and external suggestions we received when consensus was lacking, to establish our recommendations for fulfilling the instructions from Category 1. We recommend working from a version control system platform (e.g., GitHub, Bitbucket, GitLab) as this seems essential for complying with several of the FAIR4RS principles. We also recommend having code-level documentation (e.g., in code comments, description in the headers) when deemed necessary for code reusability. Additionally, we recommend recording dependencies as per standard practices for the coding language (e.g., in a requirement.txt file for Python code, in a package.json file for Node projects, or in a DESCRIPTION file for R packages). Some of the dependencies can also be recorded in the software documentation such as in the README file (see documentation discussion in Category 2 below). Following language-specific standards and best practices, which depend on the development stack used, is also recommended. For instance, we suggest following the PEP 8 Style Guide for Python Code or Google’s R Style Guide for R code ( https://google.github.io/styleguide/Rguide ). Finally, we recommend ensuring that inputs/outputs of the software follow any applicable community standards (e.g., General Feature Format (GFF) for genomic annotation files) as this is essential for interoperability. We refer to FAIRsharing Registry for finding relevant standards. We leave out recommendations on using container technologies as this is a complex topic that we deem out of the scope of the minimal guidelines we are aiming for here.
From the reviewed resources, 24 have made a recommendation of metadata files and ontologies to use for research software. A majority of them 6 , 8 , 10 , 12 , 13 , 14 , 17 , 18 , 19 , 24 , 25 , 31 , 32 , 33 , 34 , 35 , 36 suggest following the guidelines of the CodeMeta Project for including metadata in research software. The CodeMeta Project is an academic-led community initiative that was the result of the FORCE11 Software Citation working group. It aims to formalize the metadata fields included in typical software metadata records and introduces important fields that did not have clear equivalents. The CodeMeta vocabulary resulting from this effort is built over the schema.org classes SoftwareApplication and SoftwareSourceCode, which links the data for semantic web discovery. Some additional terms necessary to describe software that are not part of schema.org have been included in the CodeMeta schema. There is a continued effort to propose these terms to schema.org so that the CodeMeta schema completely aligns with schema.org rather than becoming a separate standard. Metadata information conformant to the CodeMeta vocabulary is to be included in JSON-LD format in a file named codemeta.json and stored in the root directory of the software. The Software Heritage group has developed a CodeMeta generator ( https://codemeta.github.io/codemeta-generator ) that can be used to create a codemeta.json file or edit an existing one. Similar tools are also available to include a codemeta.json file in R packages 37 .
Including a citation file following the Citation File Format (CFF) is another popular suggestion amongst the reviewed resources 13 , 14 , 18 , 19 , 25 , 26 , 27 , 28 , 30 , 34 , 35 , 36 . The CFF was developed by a group of academics assembled under the group “Development and implementation of a standard format for CITATION files” 38 . The goal of the CFF is to provide an all-purpose citation format and specifically provide optimized means for the citation of software via the provision of software-specific reference keys and types. CFF files must be named CITATION.cff, implemented in YAML 1.2, which is a machine-readable format that optimizes human readability and must be stored in the root directory of the software. The CFF file initializer ( https://citation-file-format.github.io/cff-initializer-javascript ) is mentioned by several reviewed resources as a tool for easily generating a CITATION.cff file. Note that the CITATION.cff file integrates with the GitHub citation feature such that if a CITATION.cff file is included in the root folder of a GitHub repository, a “Cite this repository” option is automatically displayed on the repository landing page making it easier to cite a software repository on GitHub.
The codemeta.json and CITATION.cff are two general metadata files that can be used for any research software repository. One of the reviewed resources 6 suggests including language-specific metadata for instance based on the DESCRIPTION file for R packages or the PEP 566 for Python packages. Other reviewed resources 6 , 24 , 25 also suggested preparing metadata using bio-specific ontologies such as EDAM 19 , 39 , 40 , biotoolsSchema 41 , and Bioschemas 42 . EDAM is a comprehensive ontology of well-established, familiar concepts that are prevalent within bioinformatics and computational biology. The biotoolsSchema is a formalized schema (XSD) that also includes EDAM ontology and is used by the Tools & Data Services Registry bio.tools 43 . Bioschemas is a community project built on top of schema.org, aiming to improve interoperability in Life Sciences so resources can better communicate and work together by using a common markup on their websites. Bioschemas has overlap with CodeMeta. These schemas and ontologies do not have formal file formats for inclusion in research software and are rather suited for repositories and registries hosting software metadata.
While the recommendations above are tailored toward machine-friendly metadata files, software metadata in a human-friendly format, i.e. documentation, is also required to comply with the FAIR4RS principles. Suggestions for such documentation are provided in 9 of the reviewed resources with one study fully dedicated to best practices for documenting scientific software 30 . Including a README file (called README, README.txt, or README.md) is the method suggested by all of the resources for maintaining documentation 14 , 19 , 21 , 24 , 28 , 30 , 33 , 44 . For more visibility, one of the reviewed resources 24 also suggests maintaining a website using GitHub pages ( https://pages.github.com ) or Read the Docs ( https://readthedocs.org ). Several resources are suggested in the reviewed literature to help with preparing documentation for research software: Write the Docs, Doxygen, Sphinx3, Javadoc (Java code), Roxygen (R code). Documenting changes between versions of software in a CHANGELOG file is also suggested 19 .
There is clearly a lack of community agreement on metadata files and ontologies to use for documenting research software. As a result, we formulated our recommendations for fulfilling the instructions in Category 2 by combining the findings from this review with our internal assessment and external suggestions received when consensus was lacking. We recommend including both a codemeta.json and a CITATION.cff metadata files in the software code’s root directory. Despite some overlapping fields, both files are recommended since codemeta.json offers a machine-oriented format, while CITATION.cff provides a more human-readable format. Moreover, they both fulfill the requirement of using controlled vocabulary and include typically suggested metadata fields in the FAIR4RS principles. The CodeMeta generator and CFF file initializer are available to help prepare both. We suggest providing all available fields in both files. To align with the FAIR4RS principles, we recommend providing at least the following fields in the codemeta.json file:
Software name (“name”)
Software description/abstract (“description”)
Unique identifier (“identifier”)
Authors (“givenName”, “familyName”) with their organization name (“affiliation”)
Keywords (“keywords”)
Programming Language (“programmingLanguage”)
First and current release date (“dataPublished” and “dateModified”)
License used (“license”)
Similarly, we recommend providing at least the following fields in the CITATION.cff file:
Authors (“given-names”, “family-names”) with their organization name (“affiliation”)
Software description/abstract (“abstract”)
Unique identifier (“identifiers”)
License (“license”)
Release date (“date-released”)
In addition, our suggestion is to maintain human-friendly documentation at the very least in a README.md or README.txt file located in the root directory of the software. Mature/complex software may require additional, more sophisticated documentation that can be developed e.g. using tools such as GitHub pages or Read the Docs. To comply with the FAIR4RS principles, the following aspects must be documented: overall description of the software (e.g., in an “About” section), high-level dependencies of the software (e.g., Node or Python version), inputs and outputs of the software, parameters and data required to run the software, the standards followed, how to contribute to the software, and how to cite the software. We also recommend following any community-agreed standard documentation approach when available (e.g., the Common Workflow Language (CWL) 45 for describing command line tools). Finally, we recommend documenting changes between different versions of the software in a file called “CHANGELOG” using plain text or markdown syntax. Locate it in the root directory of the software. We suggest following the “Keep a changelog” ( https://keepachangelog.com ) conventions for the content of the CHANGELOG file and using the Semantic Versioning v2.0.0 ( https://semver.org ) for software version numbers.
From the reviewed resources, 18 have made suggestions about a suitable license for research software 6 , 10 , 12 , 13 , 14 , 15 , 18 , 21 , 22 , 23 , 24 , 26 , 31 , 33 , 36 , 46 , 47 . All agree that it is preferable to use an open-source license to make the software as reusable as possible. Since there are a large variety of open-source licenses available, it should be possible to find one that fits everyone’s needs 13 . Therefore, if an open-source license is not used, that decision is expected to be properly motivated 31 . Most of the reviewed resources don’t explicitly suggest a specific license, but typically encourage choosing a license approved by the Open Source Initiative (OSI) ( https://opensource.org/licenses ) since they are well known and understood thus making reuse easier 18 , 24 . Some of the reviewed resources do recommend using permissive licenses since they have very few restrictions making them optimal for reuse 6 , 21 . Others explicitly encourage the use of the permissive MIT and Apache 2.0 licenses 12 , 14 , 18 , 21 , 26 , 47 . The Software Package Data Exchange (SPDX) ( https://spdx.dev ), Choose a License ( https://choosealicense.com ), and the lesson on license from the 4 Simple Recommendations for Open Source Software ( https://softdev4research.github.io/4OSS-lesson/03-use-license/index.html ) are some suggested resources for getting help with selecting a suitable license. It is typically suggested to include the license terms in a LICENSE.txt or LICENSE.md file stored in the root directory of the software 21 , 22 , 24 , 28 .
After analyzing these findings along with our own assessment and external suggestions, we recommend fulfilling the instructions from Category 3 by including the license term in a LICENSE.txt or LICENSE.md file placed in the software’s root directory. While the FAIR4RS principles do not require research software to be open-source, we highly recommend using a license approved by the OSI. Amongst those licenses, we encourage the use of the permissive MIT or Apache 2.0 licenses. Resources such as Choose a License and/or the SPDX License List can be used to help with selecting a license and including standard terms in the LICENSE file. It is desirable to select the license at the beginning of the development of the software so that it is easier to ensure that the software dependencies are compatible with the license.
From the reviewed resources, 9 have made a recommendation on the files to share for software. Sharing the source code is recommended by most of them to optimize reusability 8 , 9 , 11 , 23 , 29 , 48 . Providing an executable 9 and some input/result data 19 , 23 , 48 when available is also recommended.
From the reviewed resources, 27 made a suggestion about repositories to use for sharing research software. Three types of repositories are typically mentioned: archival, deployment, and domain-specific. The Registry of Research Data Repositories ( https://www.re3data.org ) is often mentioned as a tool for finding a suitable repository.
Zenodo ( https://zenodo.org ) is the most commonly suggested archival repository across all reviewed studies 6 , 8 , 12 , 13 , 14 , 15 , 17 , 18 , 19 , 21 , 25 , 26 , 27 , 28 , 31 , 32 , 34 , 36 , 47 , 48 , 49 , 50 . Funded by the European Commission and developed and hosted by the Centre Européen pour la Recherche Nucléaire (CERN), Zenodo allows any digital resources, including research software, to be shared and preserved in line with the FAIR principles ( https://about.zenodo.org/principles ). Zenodo assigns a separate Digital Object Identifier (DOI) for each released version of a software and also creates a high-level DOI that refers to all versions 51 . Of the 50,000 plus DOIs registered for software, more than 80% were registered via Zenodo showing its popularity amongst the research software community 32 , 52 . The integration between GitHub and Zenodo facilitates the automatic archiving on Zenodo for each GitHub release of the software. Figshare ( https://figshare.com ) is also a highly recommended archival repository 6 , 14 , 15 , 18 , 21 , 23 , 27 , 48 , 49 , 50 . Figshare is a repository supported by Digital Science where researchers can preserve and share any research outputs, including research software, in line with the FAIR principles 53 . Archiving GitHub repositories on Figshare is also made easy through a tool developed in collaboration with the Mozilla Science Lab ( https://mozillascience.github.io/code-research-object ). Software Heritage ( https://www.softwareheritage.org ) is a commonly suggested archival repository as well 6 , 8 , 12 , 13 , 14 , 17 , 19 , 25 , 31 , 32 , 50 , 54 . Software Heritage is an international initiative to provide a universal archive and reference system for all software. It automatically and regularly harvests source code from version control system platforms such as GitHub and also allows anyone to initiate the submission of their source code for archival. Software Heritage assigns intrinsic identifiers that follow a standardized format called SoftWare Heritage persistent IDentifiers (SWHIDs). Contrary to Zenodo and Figshare, unique identifiers are assigned to all artifacts over all levels of granularity such as project status, project release, state of source code, and code fragment, which can be useful to unambiguously refer to any component of a software.
Some of the reviewed resources 6 , 17 , 18 , 25 , 31 , 50 mention using deployment repositories which are typically language-specific such as PyPI ( https://pypi.org ) and Conda ( https://conda.io ) for Python packages, CRAN ( https://cran.r-project.org ) for R packages 55 , and Dockstore ( https://dockstore.org ) for Docker-based tools. Only two biomedical-specific repositories were mentioned in the reviewed studies 6 , 48 : ModelDB for computational neuroscience models 56 and Bioconductor for R-packages aimed at the analysis of genomics data 57 , 58 .
After considering these findings, our internal assessment, and the external suggestions we received, we have identified three recommendations for meeting the instructions in Category 4. If applicable, we suggest sharing each version of a software on a deployment repository (e.g., PyPI or Conda for Python packages, CRAN for R package, Dockstore for Docker-based tools). While such repositories do not provide unique and persistent identifiers as required in the FAIR4RS, this is still useful for increasing the findability and reusability of the software. In addition, we suggest always archiving each version of a software on Zenodo or Figshare as they both allow software to be archived in line with the FAIR4RS principles. Zenodo is preferable due to its popularity for archiving research software. The source code of the software with all the above-mentioned metadata files must be archived. Executables and sample input and out data, if available, must be archived as well. Finally, we suggest archiving the software on Software Heritage directly from your version control system platform as it will automatically assign a unique identifier for all levels of granularity. Note that there is no community agreement on the identifier (e.g., DOI or SWHID) to use for software, which is a gap that will need to be addressed by the community.
Category 5: Register on a registry
From the reviewed resources, 8 made a suggestion about registries to use for research software 6 , 8 , 12 , 17 , 24 , 25 , 34 , 59 . The repositories Zenodo, Figshare, Software Heritage, CRAN, PyPI, and Conda are also mentioned as registries in the reviewed literature since they require specific metadata that is used for indexing. bio.tools ( https://bio.tools ) 43 is the most commonly suggested registry. It is supported by ELIXIR, an intergovernmental organization that brings together life science resources from across Europe and whose goal is to coordinate these resources so that they form a single infrastructure. bio.tools contains more than 28,000 registered tools (as of June 2023). As explained in the metadata files section above, bio.tools uses the biotoolsSchema, including EDAM ontology, to describe registered bioinformatics software. A unique ID called biotoolsID is assigned to each software. Another possibility, not found in the reviewed resources but known to the authors, is to register the software on the Research Resource Identifiers (RRIDs) portal ( https://www.rrids.org ) and obtain an RRID. RRIDs are unique numbers assigned to help researchers cite key resources, including software projects, in the biomedical literature to improve the transparency of research methods 60 . Note that an RRID identifies a software as a whole, but not its different versions. Moreover, there exists a collaboration between bio.tools and the RRID portal to share software metadata and cross-link entries.
Based on these findings, combined with our own assessment and external suggestions we received, our recommendation for fulfilling instructions from Category 5 is to register the software on bio.tools. Even if Zenodo and Figshare already act as registries, it is still suggested to register on bio.tools to increase findability and create additional rich metadata about the software following biotoolsSchema. The software can optionally be registered on the RRIDs portal as well. Registering on bio.tools and the RRID portal is only required once but the registry-specific metadata must be updated as needed for each new software version.
FAIR Biomedical research software guidelines
To make the above-mentioned recommendations easy to implement, we organized them into step-by-step guidelines that align with the typical software development process. This led to the FAIR Biomedical Research Software (FAIR-BioRS) guidelines. The first versions of the guidelines (v1.0.0 and v1.0.1) were based on the FAIR4RS principles v0.3. They then evolved as we aligned our work with the FAIR4RS principles v1.0 as presented in the manuscript to the FAIR-BioRS guidelines v2.0.0 presented in Table 1 . Implementing these guidelines will ensure compliance with the FAIR4RS principles as shown in the crosswalk presented in Supplementary Table 2 . We have established a “FAIR-BioRS” organization on GitHub to maintain all the related resources ( http://github.com/FAIR-BioRS ). These guidelines and the crosswalk are maintained in a GitHub repository called “Guidelines” within that organization. It documents all the versions and is also archived on Zenodo every time a new version is released 61 . This repository will serve as a tool to collect community feedback on the FAIR-BioRS guidelines. New versions of the guidelines, which will be established based on community feedback or the evolution of current standards, will be maintained there. We recommend readers consult it to access the latest versions of the guidelines.
Guidelines typically have little effect and are very unlikely to be adopted without proper tools to support them. Tools and resources are available to implement some steps of the FAIR-BioRS guidelines as mentioned in our recommendations above, but they are dispersed and not logically connected to streamline the process. Therefore, we developed and integrated a workflow to implement the FAIR-BioRS guidelines in our software application called FAIRshare. FAIRshare is aimed at streamlining the processes for making biomedical research data and software FAIR. Specifically, FAIRshare combines intuitive user interfaces, automation, and user support resources into a single application to guide and assist the researchers through a suitable workflow for making their research data and software FAIR and sharing it on an adequate repository. FAIRshare is free to use and developed under the permissible open-source MIT license to encourage community contributions for updating existing workflows and including new ones. Details about the development of FAIRshare are provided in the Methods section. The current version of FAIRshare (v2.1.0) and its documentation (v5.0.0) can be accessed through their respective GitHub repositories and their Zenodo archive (c.f. Code Availability section).
Typically, a user will use FAIRshare to comply with the FAIR-BioRS guidelines when they are ready to publish a new version of their software. The workflow to make research software FAIR with FAIRshare guides users through the steps of the FAIR-BioRS guidelines in a slightly different order to optimize the workflow, as illustrated in Fig. 3 . In step 1, FAIRshare asks the user to select the location of their software directory. FAIRshare provides users with the option to select a directory from their computer or select one of their GitHub repositories. If the user selects GitHub, FAIRshare provides an interface to connect FAIRshare to their GitHub account. In step 2, FAIRshare asks a series of “Yes/No” questions to verify that the user has followed applicable standards and best practices during the development and documentation of the software as dictated by Steps 2 and 3 of the FAIR-BioRS guidelines. Links to the relevant resources on standards and best practices suggested by the FAIR-BioRS guidelines are provided in the interface. In step 3, FAIRshare provides a convenient form-type interface so the user can provide information about their software. The fields of the form correspond to fields from the codemeta.json and CITATION.cff metadata files and the user’s entries are used to automatically generate and include these metadata files in the user’s root software directory. If a codemeta.json file is found at the location selected by the user in step 1, information from that file is automatically pulled by FAIRshare to pre-populate the form. If no codemeda.json file is found and the user files are located in a GitHub repository, FAIRshare will automatically pull available information from the repository metadata (authors, keywords, etc.) and pre-populate related fields of the form. Throughout the forms, tooltips have been included to help users enter the required information. Suggestions are also made for some of the fields to help the user provide inputs rapidly (e.g., “National Institutes of Health” is suggested for the “Funder” field, “Scientific” is suggested for the “Software type” field, etc.). If any of the fields require an entry following a controlled vocabulary for the codemeta.json or CITATION.cff metadata files, FAIRshare typically provides a dropdown list of standardized options to select from to ensure metadata files are generated without error. FAIRshare also makes it mandatory to provide inputs for the metadata fields that were deemed essential by the FAIR-BioRS guidelines. In step 4, the user is prompted to select a license for their software if a LICENSE file is not found in their software directory. A dropdown list is provided to select any one of the licenses recommended by the OSI. The “MIT” and “Apache 2.0” licenses are suggested in the user interface since they have been deemed optimal for making research software FAIR. The user can read and edit the terms of a selected license directly in the app and can request FAIRshare to include a LICENSE file with associated terms in their software. In step 5, the user is prompted to select an archival repository to share their software files. In the current release, FAIRshare supports sharing on both Zenodo and Figshare. A convenient interface is provided to connect FAIRshare to the user’s Zenodo or Figshare account using a token. Upon login, the user is prompted to provide information about the software that is required by the selected repository. FAIRshare provides a form-type interface such that the user can easily input that information. The interface closely mimics the interface from the selected repository platform to maintain familiarity. Since most of the information required by Zenodo and Figshare is similar to that required in the codemeta.json file, FAIRshare pre-populates the form based on user inputs during step 3. Step 6 depends on the location of the user’s software files as specified during step 1:
If the source files are located on the user’s computer, a summary of the user’s software files, including files to be generated by FAIRshare, is provided to the user in step 6. Once the user hits the “Start upload” button, FAIRshare creates a draft deposition on the selected archival repository to reserve a DOI, generates the codemeta.json and CITATION metadata files (with the reserved DOI included) and LICENSE file in the software folder, zips the folder, and uploads it in the draft deposition to create a draft software dataset.
If the files are located on a GitHub repository, FAIRshare provides an interface to include any additional files in their archive if desired besides those already on their GitHub repository (e.g., executables). Then, a summary of the user’s software files, including files to be generated by FAIRshare, is shown to the user in step 6. Once the user hits the “Start upload” button, FAIRshare creates a draft deposition on the selected archival repository to reserve a DOI, generate and push the codemeta.json and CITATION metadata files (with the reserved DOI included) and LICENSE file on their GitHub repository. The GitHub repository content is then downloaded, and any additional files specified by the user are included in the downloaded folder. FAIRshare then zips that software folder and uploads it into the draft deposition to create a draft software dataset.

Illustration of the step-by-step guided workflow in FAIRshare for implementing the FAIR-BioRS guidelines and making biomedical research software FAIR. Users of FAIRshare can follow this workflow when they are ready to publish a new version of their biomedical research software.
Upon completion, the user is prompted in step 7 to publish their draft dataset on the archival repository, such that it becomes publicly accessible, and is also given the option to create a GitHub release of their software application. A message is shown to the user after completing step 7 to encourage them to archive their software on Software Heritage and register it on bio.tools. If the user selects to register on their software on bio.tools, an interface is provided in step 8 where the user can connect FAIRshare to their bio.tools account, enter basic metadata required by bio.tools (which is prepopulated based on information already available from the codemeta.json file), and register their software.
Besides combining all required resources for making software FAIR into a single interface, FAIRshare also provides some advantages over existing tools. For instance, the Zenodo/GitHub integration allows users to automatically create a Zenodo deposit every time a GitHub release is created. However, it does not allow to get the DOI of the deposit before it is published and therefore it cannot be included in the codemeta.json and CITATION.cff metadata files or in the software documentation, which violates principles F3 that prescribes for metadata to “include the identifier of the software they describe”. This is addressed in FAIRshare as it creates a draft deposit of the software first on Zenodo and Figshare to reserve a DOI and include it in the metadata before the software is published on either archival repository.
While research software constitutes the backbone of biomedical research, the amount of effort dedicated to ensuring software reusability and long-term sustainability is nowhere near the effort dedicated to data. The Findable, Accessible, Interoperable, and Reusable (FAIR) guiding principles published provide a foundation for managing digital research objects, including software, such that their reusability by humans and machines is optimized. However, they fall short to capture the specific traits of software such as dependencies and versioning. The Research Data Alliance (RDA) FAIR for Research Software (FAIR4RS) Working Group derived much-needed reformulated FAIR principles to address this shortcoming. Similar to the original FAIR guiding principles, the FAIR4RS principles remain aspirational in nature, intending to provide a general framework for all research software. As such, they do not offer specific, practical instructions or actionable instructions to researchers. To bridge this gap, we introduced in this work the first minimal and actionable step-by-step guidelines for biomedical researchers to make their research software compliant with the FAIR4RS principles. Our process for deriving these guidelines started with establishing high-level instructions to fulfill each of the FAIR4RS principles based on FAIR4RS v1.0 publications. Noticing common themes across the instructions, we organized them into five high-level categories. For each category, we identified outstanding questions that needed answers to fulfill the associated instructions effectively. Subsequently, we conducted a thorough literature review to explore existing practices that address these questions. Combining our findings and our own evaluation, we ultimately established the FAIR-BioRS guidelines, which are the first minimal and actional guidelines for making research software FAIR by fulfilling all the requirements of the FAIR4RS principles.
The reviewed resources included the best practices suggested by the NIH for sharing research software ( https://datascience.nih.gov/tools-and-analytics/best-practices-for-sharing-research-software-faq ). Therefore, the FAIR-BioRS guidelines align with the recommendations of the NIH for sharing research software. Additionally, the current version 2.0.0 of the FAIR-BioRS guidelines incorporates suggestions from various communities. Indeed, after establishing v1.0.1 of the guidelines, we initiated a significant effort to raise awareness about the guidelines and get community feedback. The guidelines were presented 62 at the Bioinformatics Open Source Conference (BOSC) 2022 organized by the Open Bioinformatics Foundation (OBF). We also introduced the FAIR-BioRS guidelines as a topic of collective work during the CollaborationFest that followed BOSC, which allowed for in-depth discussions with OBF members. The guidelines were also presented to SciCodes, the consortium of scientific software registries and repositories. In addition, the guidelines are being implemented as part of the Software Development Best Practices of the AI-READI project 63 , a large-scale human-data collection and tools development project funded by the NIH Bridge2AI Program. Finally, the guidelines were also presented through various webinars. Overall, we received valuable suggestions on v1.0.1 of the FAIR-BioRS guidelines during those events which helped greatly in shaping v2.0.0 presented in this manuscript. Our community outreach effort is still ongoing. Our abstract on the FAIR-BioRS guidelines v2.0.0 has been selected for presentation at BOSC 2023. We are in discussion with the RDA Software Source Code Interest Group, the maintenance home for the FAIR4RS principles, to present the FAIR-BioRS guidelines. We are similarly planning a presentation for the Elixir Tools team. We have also initiated discussions with the NIH to recommend the FAIR-BioRS guidelines in addition to or in lieu of their current guidelines for sharing research software that does not fully align with the FAIR4RS principles. We are hoping such an effort will help with raising awareness about the FAIR-BioRS guidelines and lead to their adoption by various biomedical communities.
To ensure the FAIR-BioRS guidelines are easily accessible and widely embraced by researchers, we have developed a workflow to streamline the process of implementing the FAIR-BioRS guidelines. This workflow is incorporated in FAIRshare, a software application aimed at simplifying the processes for making biomedical research data and software FAIR. FAIRshare takes the user’s software-related dataset as input when they are ready to publish a new software version and walks them step-by-step into implementing the FAIR-BioRS guidelines through an intuitive graphical user interface and automation. This way, researchers can implement the FAIR-BioRS guidelines even without prior knowledge of them and in fact learn about these guidelines along the way. FAIRshare is free and open-source to encourage community contributions to keep up with the ever-evolving FAIR guidelines.
While establishing the FAIR-BioRS guidelines, we identified several practical gaps and needs for complying with the FAIR4RS principles. Using our own understanding of research software and external feedback, we established the best possible guidelines to comply with each of the FAIR4RS principles to remedy the gaps. Mainly, there is a lack of community agreement on standards and best practices for developing and documenting software. In addition, there is a lack of community agreement on metadata files and ontologies to use for describing biomedical research software. There is a need to consolidate the various developments (CodeMeta, CFF, Bioschemas, etc.) so developers of biomedical research software can easily navigate through them and ideally use a single metadata file in their software. Moreover, there is a lack of community agreements on standard file formats to use for different biomedical data types, which prevent standardization of the data that software read, write, and exchange. While FAIRsharing is a useful registry, it is still very overwhelming to navigate through a large number of existing standards, and consolidation is required here as well. There is also a lack of agreement on a suitable identifier to use for software (DOI, SWHID, bio.tools ID, RRID, etc.). We hope that our work will help bring awareness about these needs to the relevant communities. Our work and choices made in the FAIR-BioRS guidelines may even guide them in their efforts to address them.
We expect the FAIR-BioRS guidelines and related resources to evolve over time based on several factors. Given the gaps mentioned above, we made several decisions in the guidelines. These decisions may evolve as we receive more suggestions and feedback from the research software community, for instance, to remove some guidelines as they may be above what is expected by the FAIR4RS principles or to add more to cover missing areas (such as linting guidelines, using codes of conduct, community governance, etc). Moreover, the guidelines can evolve as new community agreements are established to address the above-mentioned gaps. We have thus established a dedicated FAIR-BioRS GitHub organization to maintain all related resources. It includes a repository where new versions of the FAIR-BioRS guidelines will be maintained and published. This repository will also be used to receive community feedback and suggestions through the GitHub issues. We plan next to open ownership of the FAIR-BioRS GitHub organization to other members of the biomedical research software community such that this eventually becomes a community-driven effort that is perpetuated over time.
Next, we expect to establish more specific guidelines for different types of software (e.g., establish the FAIR-BioRS Python package guidelines or the FAIR-BioRS Jupyter notebook guidelines) such that it becomes even easier for developers of biomedical research software to comply with the FAIR4RS principles. In parallel, we plan to integrate additional resources in FAIRshare such as sharing on Software Heritage, registering on the RRID portal, or working from other version control system platforms than GitHub such as Bitbucket and GitLab. We also plan to include additional automation to further streamline the FAIRshare process for making research software FAIR. These include for instance automatically validating the implementation of language-specific standards or prefilling keywords and descriptions using Natural Language Processing. A major limitation of FAIRshare is that it is intended to help only when a user is ready to publish a new version of their software, when it might be inconvenient for the user to go back and make changes to their source code, e.g. for aligning with language specific standards or adding in code comments. We are therefore planning the development of a tool that integrates directly with version control system platforms and assists users in complying with the FAIR-BioRS guidelines right from the beginning and throughout the development of their software. As the FAIR-BioRS guidelines, FAIRshare, and other resources related to the FAIR-BioRS guidelines are expected to evolve over time, we have established a dedicated GitHub repository called “Hub” in the FAIR-BioRS organization ( https://github.com/FAIR-BioRS/Hub ). We refer to that repository to access the latest versions of all resources related to the FAIR-BioRS guidelines and track the interaction between their different versions.
By making the FAIR-BioRS guidelines an integral part of their software development practices, we aspire for the biomedical research software community to wholeheartedly embrace FAIR practices. The adoption of these guidelines will enhance the reusability of research software, thereby accelerating the pace of discoveries and innovations within the field.
Definition of research software
The FAIR4RS Working Group was composed of four subgroups, each focusing on different aspects of their effort. Group 3 focused on defining research software through a thorough review of the literature. In the outcome of their effort, they provided the following short and concise definition: “Research Software includes source code files, algorithms, scripts, computational workflows and executables that were created during the research process or for a research purpose. Software components (e.g., operating systems, libraries, dependencies, packages, scripts, etc.) that are used for research but were not created during or with a clear research intent should be considered software in research and not Research Software” 64 . The actionable guidelines developed in this work are for research software based on this definition (although they may be applicable beyond). This includes code, scripts, models, notebooks, libraries, executables, and other forms as long as it fits the previously cited definition. Given that our work was born with the aim of making COVID-19-related data and research software FAIR, our focus was on deriving guidelines for biomedical research software, although some of the findings may be applicable to other fields of research.
Deriving concise instructions for fulfilling the FAIR4RS principles
The FAIR4RS principles v1.0 contain 17 guiding principles for FAIR research software (vs 15 in the original FAIR principles), including 6 for Findability, 4 for Accessibility, 2 for Interoperability, and 5 for Reusability. We analyzed details about each principle provided in the FAIR4RS v1.0 write-up 11 and its introduction manuscript 12 . Based on these details, we derived concise instructions to be followed for fulfilling each of the principles (provided in the “fair4rsv1.0Instructions” tab of the “data.xlsx” file, c.f. Data Availability section), i.e., instructions that need to be followed for implementing each principle. We identified that these instructions had overlapping themes and were fitting overall into five actionable categories: 1) Develop software following standards and best practices, 2) Include metadata, 3) Provide a license, 4) Share software on a repository, 5) Register on a registry. The action-based instructions were thus regrouped into these categories into one instruction paragraph per category as shown in Supplementary Table 1 . Looking at the instructions for each category, we realized that there were several outstanding questions that needed to be answered for deriving actionable guidelines to fulfill the instructions. These questions are provided in Supplementary Table 1 .
Literature review for practical implementation of FAIR4RS principles
We conducted a literature review to help with answering these questions and identifying actionable recommendations for fulfilling the instructions from each category. Since discussion around FAIR for research software is very recent and not widespread in the literature, we reviewed literature not only related to FAIR research software but also related to making research software reusable without necessarily a reference to the FAIR principles. Our review was also not restricted to biomedical-related articles for the same reason. Our overall review strategy consisted of including resources in the English language from the following groups:
Group 1: We started by reviewing the six published resources on FAIR principles for research software 6 , 8 , 9 , 10 , 11 , 12 .
Group 2: Subsequently, we reviewed the resources referenced in studies from group 1 that were deemed relevant based on their title and abstract.
Group 3: We also reviewed resources listed as of February 2023 in the FAIR4Software reading materials ( https://www.rd-alliance.org/group/software-source-code-ig/wiki/fair4software-reading-materials ), FAIR4RS Subgroup 4 reading list of new research 65 , and the Zenodo community page of the FAIR4RS Working Group ( https://zenodo.org/communities/fair4rs ) that were deemed relevant based on their title and abstract and were not already encountered in the previous groups.
Group 4: Given our focus on biomedical research software, we also searched literature related to FAIR for research software on PubMed. The start period was set to January 2015 since the concept of FAIR data was introduced then, and the end period was February 2023. We read resources that were deemed relevant based on their title and abstract and were not already encountered in the previous groups.
Group 5: Finally, we included additional studies available as of February 2023 from the authors’ knowledge that were not already read in the previous groups.
Details about the review process are provided in the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) diagram in Fig. 4 . More details about the review strategy, including the PubMed search, are included in the “reviewStrategy” sheet of the “data.xlsx” file associated with this manuscript (c.f. Data Availability section). A list of all the resources encountered during our review is provided in the “resourcesList” sheet of the same file. Initially, we copied the relevant information word-for-word as written in the reviewed resources. This is available in the “resourcesReview” sheet of the “data.xlsx” file associated with this dataset (c.f. Data Availability section). Subsequently, we only retained keywords such as the name of the suggested metadata file, repository, etc. to facilitate further analysis. Information from any of the topics that applied to only specific fields of research other than biomedical were left out. This is available in the “resourcesReviewKeywords” sheet of the same file.

PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) diagram providing an overview of our literature review process.
We developed a Jupyter notebook-based script (c.f. Code Availability section) that used scientific packages such as pandas 66 , 67 , Matplotlib 68 , 69 , and seaborn 70 to analyze our review data. We used findings from the reviewed resources, combined with our own assessment and external suggestions (c.f. Discussion section) when consensus was lacking in the literature, to derive recommendations for fulfilling the instructions from each category. Finally, we organized the recommendations into step-by-step guidelines that follow the typical software development process to make them easier to implement. This led to the first minimal and actionable step-by-step guidelines for making biomedical research software FAIR such that all the requirements of the FAIR4RS principles are met, i.e., the FAIR-BioRS guidelines.
User support tool: FAIRshare
FAIRshare is inspired by the software called SODA for SPARC, which has been developed by our team to assist researchers funded by the NIH SPARC (Stimulating Peripheral Activity to Relieve Conditions) program in making their data FAIR according to the SPARC data standards 71 , 72 , 73 . Specifically, FAIRshare combines intuitive user interfaces and automation to guide and assist the researchers through the suitable processes for making their research data and software FAIR and sharing it in an adequate repository. FAIRshare is a cross-platform desktop software application, allowing researcher’s data to remain on their computer or a storage medium of their choice until they are ready to share it on a suitable archival repository. It is built using Electron, GitHub’s framework for building cross-platform desktop applications using web technologies (HTML, CSS, JavaScript, NodeJS). Vue.js is used as the frontend framework to build intuitive, interactive, and responsive user interfaces. The backend is built using Flask, a microweb framework written in Python. The use of Python for the backend of the application was motivated by the popularity of Python in the biomedical research field and the availability of relevant existing packages and APIs for curating research data and software. More details are available in the dedicated GitHub repository for FAIRshare (c.f. Code Availability section).
Data availability
The data associated with this manuscript consists of a file named ‘data.xlsx‘ that contains details about the data collected and processed during the review of relevant resources. Since no FAIR guidelines were found for structuring review data, we structured the dataset according to the SPARC Data Structure (SDS), which provides a broad data and metadata structure to organize biomedical research data according to the FAIR principles 72 . The SPARC data curation software SODA for SPARC 71 , 72 , 73 was used to organize the data and prepare the metadata files. The dataset is maintained in a GitHub repository called “Data” in the FAIR-BioRS GitHub organization and the latest version associated with this manuscript (v3.0.0) is also archived on Zenodo 74 under the permissible Creative Commons Attribution 4.0 International (CC-BY) license.
Code availability
The code associated with this manuscript consists of a “main.ipynb” Jupyter notebook, the source code of FAIRshare, and the source code of the FAIRshare documentation. The “main.ipynb” Jupyter notebook contains the code used to analyze the findings from the review and to conduct other analyses presented in this manuscript (e.g., generate Fig. 1 ). This notebook is available in a GitHub repository called “Code” (also maintained in the FAIR-BioRS GitHub organization). The dataset associated with this notebook was made FAIR according to the FAIR-BioRS guidelines using FAIRshare v2.1.0 75 , and shared under the permissible MIT license. The latest version associated with this manuscript (v3.0.0) is archived on Zenodo 76 . The source code for FAIRshare is hosted on GitHub ( https://github.com/fairdataihub/FAIRshare ). The current version of FAIRshare (v2.1.0) discussed in this manuscript was made FAIR using FAIRshare itself, and shared on Zenodo 75 under the permissible MIT license. The source code for the FAIRshare documentation is maintained on GitHub as well ( https://github.com/fairdataihub/FAIRshare-Docs ) and the current version (5.0.0) was shared under the permissible MIT license on Zenodo 77 .
Hettrick, S. softwaresaved/software_in_research_survey_2014: Software in research survey. Zenodo https://doi.org/10.5281/zenodo.1183562 (2018).
Nangia, U. & Katz, D. S. Track 1 Paper: Surveying the U.S. National Postdoctoral Association Regarding Software Use and Training in Research. Figshare https://doi.org/10.6084/m9.figshare.5328442.v1 (2017).
Hannay, J. E. et al . How do scientists develop and use scientific software? in 2009 ICSE Workshop on Software Engineering for Computational Science and Engineering 1–8 (2009).
Prabhu, P. et al . A survey of the practice of computational science. in SC ’11: State of the Practice Reports 1–12 (IEEE, 2011).
Wilkinson, M. D. et al . The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3 , 160018 (2016).
Article PubMed PubMed Central Google Scholar
Lamprecht, A.-L. et al . Towards FAIR principles for research software. Data sci. 3 , 37–59 (2020).
Article Google Scholar
Katz, D. S. et al . Software vs. data in the context of citation. PeerJ Preprints Preprint at https://doi.org/10.7287/peerj.preprints.2630v1 (2016).
Katz, D. S. et al . A Fresh Look at FAIR for Research Software. arXiv Preprint at http://arxiv.org/abs/2101.10883 (2021).
Katz, D. S., Gruenpeter, M. & Honeyman, T. Taking a fresh look at FAIR for research software. Patterns 2 , 100222 (2021).
Chue Hong, N. P. et al . FAIR Principles for Research Software (FAIR4RS Principles). Research Data Alliance https://doi.org/10.15497/RDA00065 (2021).
Chue Hong, N. P. et al . FAIR Principles for Research Software (FAIR4RS Principles) (1.0) . https://doi.org/10.15497/RDA00068 (2022).
Barker, M. et al . Introducing the FAIR Principles for research software. Sci Data 9 , 622 (2022).
Hasselbring, W., Carr, L., Hettrick, S., Packer, H. & Tiropanis, T. From FAIR research data toward FAIR and open research software. it - Information Technology 62 , 39–47 (2020).
Martinez-Ortiz, C., Kuzak, M., Spaaks, J. H., Maassen, J. & Bakker, T. Five recommendations for ‘FAIR software’ (1.0). Zenodo https://doi.org/10.5281/zenodo.4310217 (2020).
RDA COVID-19 Working Group. RDA COVID-19 Recommendations and Guidelines on Data Sharing. Research Data Alliance https://doi.org/10.15497/rda00052 (2020).
Peer, L. et al . Challenges of Curating for Reproducible and FAIR Research Output. Research Data Alliance https://doi.org/10.15497/RDA00063 (2021).
Gruenpeter, M. et al . M2.15 Assessment report on ‘FAIRness of software’ (1.1). Zenodo https://doi.org/10.5281/zenodo.4095092 (2020).
Anzt, H. et al . An environment for sustainable research software in Germany and beyond: current state, open challenges, and call for action. F1000Res. 9 , 295 (2021).
Article PubMed Central Google Scholar
Alves, R. et al . ELIXIR Software Management Plan for Life Sciences. BioHackrXiv Preprint at https://doi.org/10.37044/osf.io/k8znb (2021).
Sansone, S.-A. et al . FAIRsharing as a community approach to standards, repositories and policies. Nat. Biotechnol. 37 , 358–367 (2019).
Article CAS PubMed PubMed Central Google Scholar
Wilson, G. et al . Good enough practices in scientific computing. PLoS Comput. Biol. 13 , e1005510 (2017).
Silva, L. B., Jimenez, R. C., Blomberg, N. & Oliveira, J. L. General guidelines for biomedical software development. F1000Research 6 , 273 (2017).
Leprevost, F. et al . On best practices in the development of bioinformatics software. Front. Genet. 5 , 199 (2014).
Jiménez, R. C. et al . Four simple recommendations to encourage best practices in research software. F1000Res . 6 (2017).
Erdmann, C. et al . Top 10 FAIR Data & Software Things. Zenodo https://doi.org/10.5281/zenodo.2555498 (2019).
Martinez-Ortiz, C. et al . FAIR4RS: Adoption support. Zenodo https://doi.org/10.5281/zenodo.6258366 (2022).
The Software Sustainability Institute. Checklist for a Software Management Plan. Zenodo https://doi.org/10.5281/zenodo.2159713 (2018).
The Turing Way Community. The Turing Way: A handbook for reproducible, ethical and collaborative research. Zenodo https://doi.org/10.5281/zenodo.7625728 (2022).
Madduri, R. et al . Reproducible big data science: A case study in continuous FAIRness. PLoS One 14 , e0213013 (2019).
Lee, B. D. Ten simple rules for documenting scientific software. PLoS Comput. Biol. 14 , e1006561 (2018).
Article ADS PubMed PubMed Central Google Scholar
European Commission, Directorate-General for Research and Innovation. Scholarly Infrastructures for Research Software: Report from the EOSC Executive Board Working Group (WG) Architecture Task Force (TF) SIRS. Publications Office https://doi.org/10.2777/28598 (2020).
Ferguson, C. et al . D3.1 Survey of Current PID Services Landscape. Zenodo https://doi.org/10.5281/zenodo.1324296 (2018).
Di Cosmo, R. et al . Curated archiving of research software artifacts: lessons learned from the French open archive (HAL). in IDCC 2020-International Digital Curation Conference , https://doi.org/10.2218/ijdc.v15i1.698 (2020).
Katz, D. S. et al . Software Citation Implementation Challenges . arXiv Preprint at http://arxiv.org/abs/1905.08674 (2019).
Struck, A. Research Software Discovery: An Overview. in 2018 IEEE 14th International Conference on e-Science (e-Science) 33–37 (2018).
Erdmann, C. & Stall, S. Software Citation Checklist. Zenodo https://doi.org/10.5281/zenodo.4706164 (2021).
Boettiger, C. et al . ropensci/codemetar: codemetar 0.3.0. Zenodo https://doi.org/10.5281/zenodo.4748266 (2021).
Druskat, S. et al . Citation File Format. Zenodo https://doi.org/10.5281/zenodo.5171937 (2021).
Ison, J. et al . EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics 29 , 1325–1332 (2013).
Ison, J. et al . edamontology/edamontology: EDAM 1.25. Zenodo https://doi.org/10.5281/zenodo.3899895 (2020).
Ison, J. et al . biotoolsSchema: a formalized schema for bioinformatics software description. Gigascience 10 , (2021).
Castro, L. J. et al . Data validation and schema interoperability . Preprint at https://biohackrxiv.org/8qdse/ .
Ison, J. et al . The bio.tools registry of software tools and data resources for the life sciences. Genome Biol. 20 , 164 (2019).
Bach, F. et al . Model Policy on sustainable software at the Helmholtz centers. Helmholtz Open Science Office https://doi.org/10.48440/OS.HELMHOLTZ.041 (2019).
Crusoe, M. R. et al . Methods included: standardizing computational reuse and portability with the Common Workflow Language. Commun. ACM 65 , 54–63 (2022).
Katz, D. S. et al . Recognizing the value of software: a software citation guide. F1000Res. 9 , 1257 (2020).
Article PubMed Google Scholar
Bazuine, M. T. U. Delft Guidelines on Research Software: Licensing, Registration and Commercialisation. Zenodo https://doi.org/10.5281/zenodo.4629635 (2021).
Benureau, F. C. Y. & Rougier, N. P. Re-run, Repeat, Reproduce, Reuse, Replicate: Transforming Code into Scientific Contributions. Front. Neuroinform. 11 , 69 (2017).
Smith, A. M., Katz, D. S. & Niemeyer, K. E. Software citation principles. PeerJ Comput. Sci. 2 , e86 (2016).
Jackson, M. Software Deposit: Where to deposit software. Zenodo https://doi.org/10.5281/zenodo.1327329 (2018).
Rix, K. Expert evidence: Frequently asked questions. J. Forensic Leg. Med. 77 , 102106 (2021).
Fenner, M., Katz, D. S., Nielsen, L. H. & Smith, A. DOI Registrations for Software. Datacite Blog https://doi.org/10.5438/1NMY-9902 (2018).
Splawa-Neyman, P. Figshare and the FAIR data principles. Figshare https://doi.org/10.6084/m9.figshare.7476428.v1 (2018).
Gruenpeter, M. Software as a first class output in a FAIR ecosystem. Zenodo https://doi.org/10.5281/zenodo.5563028 (2021).
Hornik, K. The comprehensive R archive network. Wiley Interdiscip. Rev. Comput. Stat. 4 , 394–398 (2012).
McDougal, R. A. et al . Twenty years of ModelDB and beyond: building essential modeling tools for the future of neuroscience. J. Comput. Neurosci. 42 , 1–10 (2017).
Huber, W. et al . Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12 , 115–121 (2015).
Gentleman, R. C. et al . Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5 , R80 (2004).
Chue Hong, N. FAIR4RS Software (FAIR4RS). Zenodo https://doi.org/10.5281/zenodo.6374314 (2022).
Bandrowski, A. et al . The Resource Identification Initiative: A Cultural Shift in Publishing. Neuroinformatics 14 , 169–182 (2016).
Patel, B., Soundarajan, S., Ménager, H. & Hu, Z. FAIR Biomedical Research Software (FAIR-BioRS) guidelines. Zenodo https://doi.org/10.5281/zenodo.8115012 (2023).
Patel, B. & Soundarajan, S. Making biomedical research software findable, accessible, interoperable, reusable (FAIR) with FAIRshare. F1000Res . 11 , (2022).
Patel, B., Soundarajan, S., McWeeney, S., Cordier, B. A. & Benton, E. S. Software Development Best Practices of the AI-READI Project. Zenodo https://doi.org/10.5281/zenodo.7363102 (2022).
Gruenpeter, M. et al . Defining Research Software: a controversial discussion. Zenodo https://doi.org/10.5281/zenodo.5504016 (2021).
FAIR4RS Working Group. FAIR4RS Subgroup 4 - reading list of new research. Zenodo https://doi.org/10.5281/zenodo.4555865 (2021).
McKinney, W. Data Structures for Statistical Computing in Python. in Proceedings of the 9th Python in Science Conference . https://doi.org/10.25080/majora-92bf1922-00a (SciPy, 2010).
The pandas development team. pandas-dev/pandas: Pandas 1.4.2. Zenodo , https://doi.org/10.5281/zenodo.6408044 (2022).
Hunter, J. D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 9 , 90–95 (2007).
Caswell, T. A. et al . matplotlib/matplotlib: REL: v3.5.2. Zenodo https://doi.org/10.5281/zenodo.6513224 (2022).
Waskom, M. seaborn: statistical data visualization. J. Open Source Softw. 6 , 3021 (2021).
Article ADS Google Scholar
Patel, B., Srivastava, H., Aghasafari, P. & Helmer, K. SPARC: SODA, an interactive software for curating SPARC datasets. FASEB J. 34 , 1–1 (2020).
Google Scholar
Bandrowski, A. et al . SPARC Data Structure: Rationale and Design of a FAIR Standard for Biomedical Research Data. bioRxiv 2021.02.10.430563, https://doi.org/10.1101/2021.02.10.430563 (2021).
Patel, B. et al . SODA (Software to Organize Data Automatically) for SPARC v12.0.2. Zenodo https://doi.org/10.5281/zenodo.8111588 (2023).
Patel, B., Soundarajan, S., Ménager, H. & Hu, Z. Dataset: FAIR Biomedical Research Software (FAIR-BioRS) manuscript v3.0.0. Zenodo https://doi.org/10.5281/zenodo.8112100 (2023).
Soundarajan, S. & Patel, B. FAIRshare: FAIR data and software sharing made easy (v2.1.0). Zenodo https://doi.org/10.5281/zenodo.8112716 (2023).
Patel, B. Code: FAIR Biomedical Research Software (FAIR-BioRS) manuscript v3.0.0. Zenodo https://doi.org/10.5281/zenodo.8112631 (2023).
Soundarajan, S. & Patel, B. FAIRshare docs v5.0.0. Zenodo https://doi.org/10.5281/zenodo.8111725 (2023).
Download references
Acknowledgements
This work was supported by grants NIH U01AI150741 and NIH SPARC OT2OD030213. We thank Michael Crusoe, Hilmar Lapp, and Morane Gruenpeter for their valuable feedback that helped shape the FAIR-BioRS guidelines. We also thank the BOSC Community for their tremendous support in establishing and improving the FAIR-BioRS guidelines.
Author information
Authors and affiliations.
FAIR Data Innovations Hub, California Medical Innovations Institute, San Diego, CA, 92121, USA
Bhavesh Patel & Sanjay Soundarajan
Institut Pasteur, Université Paris Cité, Bioinformatics and Biostatistics Hub, 75015, Paris, France
Hervé Ménager
Computational Health Science, University of California San Francisco, San Francisco, CA, 94158, USA
You can also search for this author in PubMed Google Scholar
Contributions
B. Patel led the conception and design of the FAIR-BioRS guidelines, managed the acquisition/analysis/interpretation of data from the reviewed studies, supervised the development of FAIRshare, and contributed to drafting/revising the manuscript. S. Soundarajan led the development of FAIRshare and contributed to revising the manuscript. Z. Hu and H. Ménager contributed to the conception and design of the FAIR-BioRS guidelines, and revising the manuscript.
Corresponding author
Correspondence to Bhavesh Patel .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary tables, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Patel, B., Soundarajan, S., Ménager, H. et al. Making Biomedical Research Software FAIR: Actionable Step-by-step Guidelines with a User-support Tool. Sci Data 10 , 557 (2023). https://doi.org/10.1038/s41597-023-02463-x
Download citation
Received : 09 June 2022
Accepted : 10 August 2023
Published : 23 August 2023
DOI : https://doi.org/10.1038/s41597-023-02463-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: AI and Robotics newsletter — what matters in AI and robotics research, free to your inbox weekly.

IMAGES
VIDEO
COMMENTS
Top Clinical Research Software. Choose the right Clinical Research Software using real-time, up-to-date product reviews from 1543 verified user reviews.
Clinical trial management software helps clinical research sites, hospitals, and other pharmaceutical/healthcare companies manage all aspects of clinical trials. These software systems maintain and manage patient data, scheduling, finances, reporting, analysis, and research data.
Pharmaceutical companies, medical research institutes, and research centers managed by hospitals utilize clinical trial management software. This type of software is used by CROs, medical researchers, and trial administrators or sponsors to plan, manage, and monitor the entire lifecycle of clinical trials.
Clinical trials are the backbone of medical research, providing essential evidence on the safety and effectiveness of medical interventions. In today’s digital era, software solutions play a pivotal role in streamlining trial processes, ensuring data accuracy, and enhancing patient safety.
Phlexglobal - Best for electronic trial master file (eTMF) management. Citeline - Best for global clinical trial solutions and comprehensive market analysis. ResearchManager - Best for collaborative research and workflow management. Calyx - Best for integrated trial management and regulatory compliance.
Designed for hospitals, medical centers, trial sites and more, Complion is a cloud-based clinical trial management solution that helps handle manual processes on a centralized interface. The software automates investigator regulat... 4.5 ( 2 reviews) Visit Website.
Founded in a basement in 1979, Epic develops software to help people get well, help people stay well, and help future generations be healthier.
Hervé Ménager & Zicheng Hu. Scientific Data 10, Article number: 557 ( 2023 ) Cite this article. 1922 Accesses. 1 Citations. 10 Altmetric. Metrics. Abstract. Findable, Accessible, Interoperable, and...