EDITORIAL article
Editorial: theoretical advances and practical applications of spiking neural networks.

- 1 Neuromorphic Sensor Signal Processing Lab, Department of Electronic and Electrical Engineering, Centre for Image and Signal Processing, University of Strathclyde, Glasgow, United Kingdom
- 2 Centre for Future Media, School of Computer Science and Engineering, University of Electronic Science and Technology of China, Chengdu, Sichuan, China
- 3 Learning and Intelligent Systems Lab, School of Electrical Engineering and Computer Science, Ohio University, Athens, OH, United States
Editorial on the Research Topic Theoretical advances and practical applications of spiking neural networks

1 Introduction
Neuromorphic engineering has experienced a significant growth in popularity over the last 10 years, going from being a niche academic research area, often confused with deep learning and mostly unknown to the wider industrial community, to being the main focus of many funding calls, significant industrial endeavours, and national and international initiatives. The advent to market of neuromorphic sensors, with a related widening understanding of the event-based sensing paradigm, combined with the development of the first neuromorphic processors, has steered the wider academic community and industry toward the investigation and use of Spiking Neural Networks (SNN). Very often overlooked in favour of the now extremely popular Deep Neural Networks (DNN), SNNs have become a serious alternative to DNNs, in application domains where size, weight and power are key limiting factors to the deployment of AI systems, such in Space applications, Security and Defence, Automotive, and more generally AI at the Edge. Nonetheless, there are many aspects of SNNs that still require significant investigation, as there are many unexplored avenues in this regard. To this aim, the articles accepted in this special topic present novel research works that focus on methodologies for training of SNNs and on the use of SNN in real life applications.
2 About the papers
The articles published in this special topic cover a varied set of application domains, including image processing (such as denoising and segmentation), analysis of biosignals (for seizure detection), activity recognition through wearable devices, and audio processing for the cocktail party effect. Moreover, the topic hosts also two papers on novel learning methods, such as meta-spikepropamine, and the chip-in-loop SNN proxy learning.
In the article “ Efficient and generalizable cross-patient epileptic seizure detection through a spiking neural network ”, Zhang et al. propose an EEG-based spiking neural network (EESNN) with a recurrent spiking convolution structure, leveraging the biological plausibility of SNNs, to take better advantage of temporal and biological characteristics of EEG signals.
Li Y. et al. propose the application of the SNNs to time series of binary spikes generated through wearable devices, in their work entitled “ Efficient human activity recognition with spatio-temporal spiking neural networks ”. The results reported indicate that SNNs achieve competitive accuracy while significantly reducing energy in this application.
In “ Explaining cocktail party effect and McGurk effect with a spiking neural network improved by Motif-topology ” Jia et al. introduce a novel Motif-topology improved SNN (M-SNN) to enhance the network's ability to tackle complex cognitive tasks. The experimental results show a lower computational cost and higher accuracy and a better explanation of some key phenomena of these two effects, such as new concept generation and anti-background noise.
In their article “ Meta-SpikePropamine: learning to learn with synaptic plasticity in spiking neural networks ”, Schmidgall et al. propose a novel bi-level optimization framework that integrates neuroscience principles into SNNs to enhance online learning capabilities. The experimental outcomes underscore the potential of neuroscience-inspired models to advance the field of online learning, marking a promising direction for future research.
Li X. et al. propose a biologically-plausible algorithm named the mixture of personality (MoP) improved spiking actor network (SAN), in their work “ Mixture of personality improved spiking actor network for efficient multi-agent cooperation ”. The experimental results on the benchmark cooperative overcooked task show that the proposed MoP-SAN algorithm could achieve higher performance for the paradigms with (learning) and without (generalization) unseen partners.
In the article “ SPIDEN: Deep Spiking Neural Networks for Efficient Image Denoising ” Castagnetti et al. explore the use of SNNs for image denoising applications, with the goal of reaching the accuracy of conventional Deep Convolutional Neural Networks (DCNNs), while reducing computational costs. The authors present a formal analysis of data flow through Integrate and Fire (IF) spiking neurons, and establish the trade-off between conversion error and activation sparsity in SNNs. Experimental results demonstrate that the design goals are effectively achieved.
Yue et al. propose a novel three-stage SNN training scheme for segmenting human brain images, in the work “ Spiking Neural Networks Fine-Tuning for Brain Image Segmentation ”. Their pipeline begins with fully optimizing an ANN, followed by a quick ANN-to-SNN conversion to initialize the corresponding spiking network. Spike-based backpropagation is then employed to fine-tune the converted SNN. Experimental results show a significant advantage of the proposed scheme over both ANN-to-SNN conversion and direct SNN training solutions in terms of segmentation accuracy and training efficiency.
In “ Chip-In-Loop SNN Proxy Learning: A New Method for Efficient Training of Spiking Neural Networks ” Liu et al. introduce the Chip-In-Loop SNN Proxy Learning (CIL-SPL) method. Their approach applies proxy learning principles and use hardware devices as proxy agents. This combination allows the hardware to exhibit event-driven, asynchronous behavior, while enabling training of the synchronous SNN structure using backward loss gradients. Experiments on the N-MNIST dataset demonstrate that CIL-SPL achieves the best performance on actual hardware chips.
3 Discussion and future trends
As it can be seen, the range and type of applications covered demonstrate the wide potential of SNNs in being applicable and effective in different domains, and not just for event-based visual sensing and processing, which are sometimes confused, by non-experts, almost as synonymous for SNNs. Indeed, event-based cameras are currently the most popular example of event-based sensors. The reason for this is quite likely due to the fact that event-based vision sensors provide an easy to understand approach to spike generation by mimicking the human retina. In fact, the output of an event-based camera is rather straightforward to visualise and interpret. Therefore, it is encouraging to see that the research community is exploring other avenues of application of SNNs, and this should be encouraged further. More specifically, event extraction from other sensing modalities is a very interesting and welcome feature of some of the articles in this topic.
In this respect, new commercially available neuromorphic sensors are welcome. In fact, researchers have indeed proposed novel neuromorphic sensors in other modalities, as for example audio, olfactory, tactile. However, these are still pretty much confined at academic research level. Furthermore, an interesting future direction is the investigation of novel event-based sensing approaches based on conventional, i.e. non natively neuromorphic, sensors and sensing hardware architectures. For example, as Radars and Lidars already handle information in the form of pulses, it would be ideal if existing technology could be used, as is, to generate event-based data to be fed to SNNs directly, in raw format. This would avoid the need for redundant initial transformation of the sensed data into more conventional formats, which may be unnecessary in the context of SNN-based processing.
In another sense, research on SNNs can borrow ideas from DNN approaches, but it should avoid closely mimicking the development of DNNs. As while this can provide useful research directions and hints, it may also steer SNNs toward use that does not fully exploit all the potential that event-based sensing and processing have to offer. More applications which leverage key aspects of SNN should be welcome, such as the sparse nature of event-base data, and its timing and asynchronous processing aspect. Along this line of thought, these two key aspects should be investigated to devise learning methods, that mimic the human brain and its learning, not from a mere dogmatic biological plausibility aspect, but also from a more pragmatic engineering angle.
4 Conclusion
Spiking Neural Networks and Neuromorphic Engineering have come out from their niche and are now known to the wider academic community and to industry, and are now frequently indicated as the key technologies that can successfully achieve AI at the Edge. This is a fundamental step forward, in order to entice further research focusing on the core aspects: sensors, processing algorithms, and hardware architectures. Nonetheless, common misconceptions should be cleared, and there should be a conscious effort to use SNNs and their capabilities for what they are, to ensure that they are used as the right tool for the job, rather than just based on their similarity to other generations of neural networks.
Author contributions
GD: Writing – original draft, Writing – review & editing. JL: Writing – review & editing. MZ: Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Spiking Neural Networks (SNN), Neuromorphic Engineering (NE), event-based sensing, neural networks, artificial intelligence
Citation: Di Caterina G, Zhang M and Liu J (2024) Editorial: Theoretical advances and practical applications of spiking neural networks. Front. Neurosci. 18:1406502. doi: 10.3389/fnins.2024.1406502
Received: 25 March 2024; Accepted: 29 March 2024; Published: 22 April 2024.
Edited and reviewed by: André van Schaik , Western Sydney University, Australia
Copyright © 2024 Di Caterina, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaetano Di Caterina, gaetano.di-caterina@strath.ac.uk
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
An Overview of Spikingneural Networks
Ieee account.
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.
SMT-Based Modeling and Verification of Spiking Neural Networks: A Case Study
- Conference paper
- First Online: 17 January 2023
- Cite this conference paper

- Soham Banerjee 10 ,
- Sumana Ghosh 10 ,
- Ansuman Banerjee 10 &
- Swarup K. Mohalik 11
Part of the book series: Lecture Notes in Computer Science ((LNCS,volume 13881))
Included in the following conference series:
- International Conference on Verification, Model Checking, and Abstract Interpretation
539 Accesses
In this paper, we present a case study on modeling and verification of Spiking Neural Networks (SNN) using Satisfiability Modulo Theory (SMT) solvers. SNN are special neural networks that have great similarity in their architecture and operation with the human brain. These networks have shown similar performance when compared to traditional networks with comparatively lesser energy requirement. We discuss different properties of SNNs and their functioning. We then use Z3, a popular SMT solver to encode the network and its properties. Specifically, we use the theory of Linear Real Arithmetic (LRA). Finally, we present a framework for verification and adversarial robustness analysis and demonstrate it on the Iris and MNIST benchmarks.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Code and Benchmarks. https://github.com/Soham-Banerjee/SMT-Encoding-for-Spiking-Neural-Network
Alur, R.: Timed automata. In: Peled, D. (ed.) CAV 1999. LNCS, vol. 1633, pp. 8–22. Springer, Heidelberg (1999). https://doi.org/10.1007/3-540-48683-6_3
Chapter Google Scholar
Aman, B., Ciobanu, G.: Modelling and verification of weighted spiking neural systems. Theoret. Comput. Sci. 623 , 92–102 (2016)
Article MathSciNet Google Scholar
De Maria, E., Di Giusto, C., Laversa, L.: Spiking neural networks modelled as timed automata with parameter learning (2018)
Google Scholar
De Maria, E., Muzy, A., Gaffé, D., Ressouche, A., Grammont, F.: Verification of temporal properties of neuronal archetypes modeled as synchronous reactive systems. In: Cinquemani, E., Donzé, A. (eds.) HSB 2016. LNCS, vol. 9957, pp. 97–112. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-47151-8_7
Demin, V., Nekhaev, D.: Recurrent spiking neural network learning based on a competitive maximization of neuronal activity. Front. Neuroinf. 12 , 79 (2018)
Article Google Scholar
Deng, L.: The MNIST database of handwritten digit images for machine learning research. IEEE Signal Process. Mag. 29 (6), 141–142 (2012)
Diehl, P.U., Cook, M.: Unsupervised learning of digit recognition using spike-timing-dependent plasticity. Front. Comput. Neurosci. 9 , 99 (2015)
Ding, J., Yu, Z., Tian, Y., Huang, T.: Optimal ANN-SNN conversion for fast and accurate inference in deep spiking neural networks (2021)
Elboher, Y.Y., Gottschlich, J., Katz, G.: An abstraction-based framework for neural network verification. In: Lahiri, S.K., Wang, C. (eds.) CAV 2020. LNCS, vol. 12224, pp. 43–65. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-53288-8_3
Eshraghian, J.K., et al.: Training spiking neural networks using lessons from deep learning (2021)
Fisher, R.: Iris. UCI Machine Learning Repository (1988). https://archive.ics.uci.edu/ml/datasets/Iris
Gokulanathan, S., Feldsher, A., Malca, A., Barrett, C., Katz, G.: Simplifying neural networks using formal verification. In: Lee, R., Jha, S., Mavridou, A., Giannakopoulou, D. (eds.) NFM 2020. LNCS, vol. 12229, pp. 85–93. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-55754-6_5
Goldberger, B., Katz, G., Adi, Y., Keshet, J.: Minimal modifications of deep neural networks using verification. In: LPAR23. LPAR-23: 23rd International Conference on Logic for Programming, Artificial Intelligence and Reasoning, vol. 73, pp. 260–278 (2020)
Guo, W., Fouda, M.E., Eltawil, A.M., Salama, K.N.: Neural coding in spiking neural networks: a comparative study for robust neuromorphic systems. Front. Neurosci. 15 , 638474 (2021)
Katz, G., Barrett, C., Dill, D.L., Julian, K., Kochenderfer, M.J.: Reluplex: a calculus for reasoning about deep neural networks. Formal Methods Syst. Design 1–30 (2021)
Kim, T., et al.: Spiking neural network (SNN) with memristor synapses having non-linear weight update. Front. Comput. Neurosci. 15 , 646125 (2021)
Kuper, L., Katz, G., Gottschlich, J., Julian, K., Barrett, C., Kochenderfer, M.: Toward scalable verification for safety-critical deep networks (2018)
Lahav, O., Katz, G.: Pruning and slicing neural networks using formal verification (2021)
Li, S., Zhang, Z., Mao, R., Xiao, J., Chang, L., Zhou, J.: A fast and energy-efficient SNN processor with adaptive clock/event-driven computation scheme and online learning. IEEE Trans. Circuits Syst. I Regul. Pap. 68 (4), 1543–1552 (2021)
Liu, T.Y., Mahjoubfar, A., Prusinski, D., Stevens, L.: Neuromorphic computing for content-based image retrieval. PLOS One 17 (4), 1–13 (2022). https://doi.org/10.1371/journal.pone.0264364
Malik, N.: Artificial neural networks and their applications (2005)
de Maria, E., Gaffé, D., Ressouche, A., Girard Riboulleau, C.: A model-checking approach to reduce spiking neural networks. In: BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms, pp. 1–8 (2018)
de Moura, L., Bjørner, N.: Z3: an efficient SMT solver. In: Ramakrishnan, C.R., Rehof, J. (eds.) TACAS 2008. LNCS, vol. 4963, pp. 337–340. Springer, Heidelberg (2008). https://doi.org/10.1007/978-3-540-78800-3_24
Stimberg, M., Brette, R., Goodman, D.F.: Brian 2, an intuitive and efficient neural simulator. eLife 8 , e47314 (2019)
Tavanaei, A., Ghodrati, M., Kheradpisheh, S.R., Masquelier, T., Maida, A.S.: Deep learning in spiking neural networks (2018)
Tjeng, V., Xiao, K., Tedrake, R.: Evaluating robustness of neural networks with mixed integer programming (2017)
Yu, Z., Abdulghani, A.M., Zahid, A., Heidari, H., Imran, M.A., Abbasi, Q.H.: An overview of neuromorphic computing for artificial intelligence enabled hardware-based hopfield neural network. IEEE Access 8 , 67085–67099 (2020). https://doi.org/10.1109/ACCESS.2020.2985839
Download references
Author information
Authors and affiliations.
Indian Statistical Institute, Kolkata, India
Soham Banerjee, Sumana Ghosh & Ansuman Banerjee
Ericsson Research, Bangalore, India
Swarup K. Mohalik
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Ansuman Banerjee .
Editor information
Editors and affiliations.
Inria, Amazon Web Services, Courbevoie, France
Cezara Dragoi
Amazon Web Services, Seattle, WA, USA
Michael Emmi
University of Southern California, Los Angeles, CA, USA
Jingbo Wang
Rights and permissions
Reprints and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper.
Banerjee, S., Ghosh, S., Banerjee, A., Mohalik, S.K. (2023). SMT-Based Modeling and Verification of Spiking Neural Networks: A Case Study. In: Dragoi, C., Emmi, M., Wang, J. (eds) Verification, Model Checking, and Abstract Interpretation. VMCAI 2023. Lecture Notes in Computer Science, vol 13881. Springer, Cham. https://doi.org/10.1007/978-3-031-24950-1_2
Download citation
DOI : https://doi.org/10.1007/978-3-031-24950-1_2
Published : 17 January 2023
Publisher Name : Springer, Cham
Print ISBN : 978-3-031-24949-5
Online ISBN : 978-3-031-24950-1
eBook Packages : Computer Science Computer Science (R0)
Share this paper
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Front Comput Neurosci
Spiking neural network connectivity and its potential for temporal sensory processing and variable binding
1 Multimedia and Vision Research Group, School of Electronic Engineering and Computer Science, Queen Mary, University of London, London, UK
Cornelius Glackin
2 Adaptive Systems Research Group, Department of Computer Science, University of Hertfordshire, Hatfield, Hertfordshire, UK
The most biologically-inspired artificial neurons are those of the third generation, and are termed spiking neurons, as individual pulses or spikes are the means by which stimuli are communicated. In essence, a spike is a short-term change in electrical potential and is the basis of communication between biological neurons. Unlike previous generations of artificial neurons, spiking neurons operate in the temporal domain, and exploit time as a resource in their computation. In 1952, Alan Lloyd Hodgkin and Andrew Huxley produced the first model of a spiking neuron; their model describes the complex electro-chemical process that enables spikes to propagate through, and hence be communicated by, spiking neurons. Since this time, improvements in experimental procedures in neurobiology, particularly with in vivo experiments, have provided an increasingly more complex understanding of biological neurons. For example, it is now well-understood that the propagation of spikes between neurons requires neurotransmitter, which is typically of limited supply. When the supply is exhausted neurons become unresponsive. The morphology of neurons, number of receptor sites, amongst many other factors, means that neurons consume the supply of neurotransmitter at different rates. This in turn produces variations over time in the responsiveness of neurons, yielding various computational capabilities. Such improvements in the understanding of the biological neuron have culminated in a wide range of different neuron models, ranging from the computationally efficient to the biologically realistic. These models enable the modeling of neural circuits found in the brain.
In recent years, much of the focus in neuron modeling has moved to the study of the connectivity of spiking neural networks. Spiking neural networks provide a vehicle to understand from a computational perspective, aspects of the brain's neural circuitry. This understanding can then be used to tackle some of the historically intractable issues with artificial neurons, such as scalability and lack of variable binding. Current knowledge of feed-forward, lateral, and recurrent connectivity of spiking neurons, and the interplay between excitatory and inhibitory neurons is beginning to shed light on these issues, by improved understanding of the temporal processing capabilities and synchronous behavior of biological neurons. This research topic spans current research on neuron models to spiking neural networks and their application to interesting and current computational problems. The research papers submitted to this topic can be categorized into the following major areas of more efficient neuron modeling; lateral and recurrent spiking neural network connectivity; exploitation of biological neural circuitry by means of spiking neural networks; optimization of spiking neural networks; and spiking neural networks for sensory processing.
Moujahid and d'Anjou ( 2012 ) stimulate the giant squid axon with simulated spikes to develop some new insights into the development of more relevant models of biological neurons. They observed that temperature mediates the efficiency of action potentials by reducing the overlap between sodium and potassium currents in the ion exchange and subsequent energy consumption. The original research article by Dockendorf and Srinivasa ( 2013 ) falls into the area of lateral and recurrent spiking neural network connectivity. It presents a recurrent spiking model capable of learning episodes featuring missing and noisy data. The presented topology provides a means of recalling previously encoded patterns where inhibition is of the high frequency variety aiming to promote stability of the network. Kaplan et al. ( 2013 ) also investigated the use of recurrent spiking connectivity in their work on motion-based prediction and the issue of missing data. Here they address how anisotropic connectivity patterns that consider the tuning properties of neurons efficiently predict the trajectory of a disappearing moving stimulus. They demonstrate and test this by simulating the network response in a moving-dot blanking experiment.
Garrido et al. ( 2013 ) investigate how systematic modifications of synaptic weights can exert close control over the timing of spike transmissions. They demonstrate this using a network of leaky integrate-and-fire spiking neurons to simulate cells of the cerebellar granular layer. Börgers and Walker ( 2013 ) investigate simulations of excitatory pyramidal cells and inhibitory interneurons which interact and exhibit gamma rhythms in the hippocampus and neocortex. They focus on how inhibitory interneurons maintain synchrony using gap junctions. Similarly, Ponulak and Hopfield ( 2013 ) also take inspiration from the neural structure of the hippocampus to hypothesize about the problem of spatial navigation. Their topology encodes the spatial environment through an exploratory phase which utilizes “place” cells to reflect all possible trajectory boundaries and environmental constraints. Subsequently, a wave propagation process maps the trajectory between the target or multiple targets and the current location by altering the synaptic connectivity of the aforementioned “place” cells in a single pass. A novel viewpoint of the state-of-the-art for the exploitation of biological neural circuitry by means of spiking neural networks is provided by Aimone and Weick ( 2013 ). In their paper, a thorough and comprehensive review of modeling cortical damage due to stroke is provided. They argue that a theoretical understanding of the damaged cortical area post-disease is vital while taking into account current thinking of models for adult neurogenesis.
One of the issues with modeling large-scale spiking neural networks is the lack of tools to analyse such a large parameter space, as Buice and Chow ( 2013 ) discuss in their hypothesis and theory article. They propose a possible approach which combines mean field theory with information about spiking correlations; thus reducing the complexity to that of a more comprehensible rate-like description. Demonstrations of spiking neural networks for sensory processing include the work of Srinivasa and Jiang ( 2013 ). Their research consists of the development of spiking neuron models, initially assembled into an unstructured map topology. The authors show how the combination of self-organized and STDP-based continuous learning can provide the initial formation and on-going maintenance of orientation and ocular dominance maps of the kind commonly found in the visual cortex.
It is clear that research on spiking neural networks has expanded beyond computational models of individual neurons and now encompasses large-scale networks which aim to model the behavior of whole neural regions. This has resulted in a diverse and exciting field of research with many perspectives and a multitude of potential applications.
- Aimone J. B., Weick J. P. (2013). Perspectives for computational modeling of cell replacement for neurological disorders . Front. Comput. Neurosci . 7 :150 10.3389/fncom.2013.00150 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Börgers C., Walker B. (2013). Toggling between gamma-frequency activity and suppression of cell assemblies . Front. Comput. Neurosci . 7 :33 10.3389/fncom.2013.00033 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Buice M. A., Chow C. C. (2013). Generalized activity equations for spiking neural network dynamics . Front. Comput. Neurosci . 7 :162 10.3389/fncom.2013.00162 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Dockendorf K., Srinivasa N. (2013). Learning and prospective recall of noisy spike pattern episodes . Front. Comput. Neurosci . 7 :80 10.3389/fncom.2013.00080 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Garrido J. A., Ros E., D'Angelo E. (2013). Spike timing regulation on the millisecond scale by distributed synaptic plasticity at the cerebellum input stage: a simulation study . Front. Comput. Neurosci . 7 :64 10.3389/fncom.2013.00064 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kaplan B. A., Lansner A., Masson G. S., Perrinet L. U. (2013). Anisotropic connectivity implements motion-based prediction in a spiking neural network . Front. Comput. Neurosci . 7 :112 10.3389/fncom.2013.00112 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Moujahid A., d'Anjou A. (2012). Metabolic efficiency with fast spiking in the squid axon . Front. Comput. Neurosci . 6 :95 10.3389/fncom.2012.00095 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ponulak F. J., Hopfield J. J. (2013). Rapid, parallel path planning by propagating wavefronts of spiking neural activity. Front. Comput. Neurosci . 7 :98 10.3389/fncom.2013.00098 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Srinivasa N., Jiang Q. (2013). Stable learning of functional maps in self-organizing spiking neural networks with continuous synaptic plasticity . Front. Comput. Neurosci . 7 :10 10.3389/fncom.2013.00010 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
IM-Loss: Information Maximization Loss for Spiking Neural Networks
Part of Advances in Neural Information Processing Systems 35 (NeurIPS 2022) Main Conference Track
Yufei Guo, Yuanpei Chen, Liwen Zhang, Xiaode Liu, Yinglei Wang, Xuhui Huang, Zhe Ma
Spiking Neural Network (SNN), recognized as a type of biologically plausible architecture, has recently drawn much research attention. It transmits information by $0/1$ spikes. This bio-mimetic mechanism of SNN demonstrates extreme energy efficiency since it avoids any multiplications on neuromorphic hardware. However, the forward-passing $0/1$ spike quantization will cause information loss and accuracy degradation. To deal with this problem, the Information maximization loss (IM-Loss) that aims at maximizing the information flow in the SNN is proposed in the paper. The IM-Loss not only enhances the information expressiveness of an SNN directly but also plays a part of the role of normalization without introducing any additional operations (\textit{e.g.}, bias and scaling) in the inference phase. Additionally, we introduce a novel differentiable spike activity estimation, Evolutionary Surrogate Gradients (ESG) in SNNs. By appointing automatic evolvable surrogate gradients for spike activity function, ESG can ensure sufficient model updates at the beginning and accurate gradients at the end of the training, resulting in both easy convergence and high task performance. Experimental results on both popular non-spiking static and neuromorphic datasets show that the SNN models trained by our method outperform the current state-of-the-art algorithms.
Name Change Policy
Requests for name changes in the electronic proceedings will be accepted with no questions asked. However name changes may cause bibliographic tracking issues. Authors are asked to consider this carefully and discuss it with their co-authors prior to requesting a name change in the electronic proceedings.
Use the "Report an Issue" link to request a name change.
Subscribe to the PwC Newsletter
Join the community, edit social preview.

Add a new code entry for this paper
Remove a code repository from this paper, mark the official implementation from paper authors, add a new evaluation result row, remove a task, add a method, remove a method, edit datasets, temporal spiking neural networks with synaptic delay for graph reasoning.
27 May 2024 · Mingqing Xiao , Yixin Zhu , Di He , Zhouchen Lin · Edit social preview
Spiking neural networks (SNNs) are investigated as biologically inspired models of neural computation, distinguished by their computational capability and energy efficiency due to precise spiking times and sparse spikes with event-driven computation. A significant question is how SNNs can emulate human-like graph-based reasoning of concepts and relations, especially leveraging the temporal domain optimally. This paper reveals that SNNs, when amalgamated with synaptic delay and temporal coding, are proficient in executing (knowledge) graph reasoning. It is elucidated that spiking time can function as an additional dimension to encode relation properties via a neural-generalized path formulation. Empirical results highlight the efficacy of temporal delay in relation processing and showcase exemplary performance in diverse graph reasoning tasks. The spiking model is theoretically estimated to achieve $20\times$ energy savings compared to non-spiking counterparts, deepening insights into the capabilities and potential of biologically inspired SNNs for efficient reasoning. The code is available at https://github.com/pkuxmq/GRSNN.
Code Edit Add Remove Mark official
Tasks edit add remove, datasets edit.

Results from the Paper Edit Add Remove
Methods edit add remove.
Stochastic Spiking Neural Networks with First-to-Spike Coding
Spiking Neural Networks (SNNs), recognized as the third generation of neural networks, are known for their bio-plausibility and energy efficiency, especially when implemented on neuromorphic hardware. However, the majority of existing studies on SNNs have concentrated on deterministic neurons with rate coding, a method that incurs substantial computational overhead due to lengthy information integration times and fails to fully harness the brain’s probabilistic inference capabilities and temporal dynamics. In this work, we explore the merger of novel computing and information encoding schemes in SNN architectures where we integrate stochastic spiking neuron models with temporal coding techniques. Through extensive benchmarking with other deterministic SNNs and rate-based coding, we investigate the tradeoffs of our proposal in terms of accuracy, inference latency, spiking sparsity, energy consumption, and robustness. Our work is the first to extend the scalability of direct training approaches of stochastic SNNs with temporal encoding to VGG architectures and beyond-MNIST datasets.
Index Terms:
I introduction.
Spiking neural networks (SNNs) bridge the gap between artificial and biological neural networks (ANNs), offering insights into neurological processes. In the human neuronal system, most of the information is propagated between neurons using spike-based activation signals over time. Inspired by this property, SNNs use binary spike signals to transmit, encode, and process information. Compared with analog neural networks where neuron and synaptic states are represented by non-binary multi-bit representations, SNNs have demonstrated significant energy and computational power savings, especially when deployed on neuromorphic hardware [ 1 ] .
In SNNs, rate coding is one of the most popular coding methods. Under such a scheme, the information is represented by the rate or frequency of spikes over a defined period of time. However, such coding methods overlook the information of precise spike timings [ 2 ] and are constrained by slow information transmission and large processing latency. On the other hand, temporal coding represents information by the precise timing of individual spikes but often lacks scalability and robustness [ 3 ] . Compared to rate coding, which requires high firing rates to represent the same information, temporal coding can represent complex temporal patterns with relatively few spikes. In particular, First-to-Spike coding is a temporal coding scheme inspired by the rapid information processing observed in certain biological neural systems such as the retina [ 4 ] and auditory system [ 5 ] . In First-to-Spike coding, prediction is made when the first spike is observed on any one of the output neurons, thereby saving the need to operate over a redundant period of time as in rate coding. Therefore, it is often claimed that temporal coding is more computationally efficient than rate coding [ 3 ] .
Nevertheless, it is a challenging task to train an SNN due to the disruptive nature of information representation and processing, especially for frameworks based on temporal coding. Most existing works on scalable SNN training with temporal encoding convert pre-trained ANN to SNN [ 6 , 7 ] . The conversion process typically involves mapping the analog activation values of the ANN’s neurons to the timing of spikes of the SNN’s neurons. SNN training algorithms with conventional architectures and rate encoding [ 8 ] have witnessed rapid development [ 9 , 10 ] in recent years ranging from global spike-driven backpropagation techniques [ 11 ] to more local approaches like Deep Continuous Local Learning (DECOLLE) [ 12 ] , Equilibrium Propagation (EP) [ 13 , 14 ] , Deep Spike Timing Dependent Plasticity [ 15 ] , among others. In stark contrast, literature on direct training of SNNs with temporal encoding remains extremely sparse with demonstrations primarily on toy datasets like MNIST.
Although the vast majority of algorithm development and applications in SNNs employ deterministic neuron models such as the deterministic Spike Response Model (SRM) [ 16 ] , Integrate and Fire (IF), and Leaky Integrate and Fire (LIF) models [ 17 ] , it is important to recognize that biological neurons generate spikes in a stochastic fashion [ 18 ] . Furthermore, deterministic neuron models are discontinuous and non-differentiable, which presents substantial challenges in the application of gradient-based optimization methods. On the other hand, stochastic neuron models smoothen the network model to be continuously differentiable [ 19 ] and therefore have the potential to offer enhanced efficiency and robustness.
Recently, there has been growing interest in exploiting stochastic devices in the neuromorphic hardware community [ 20 ] , [ 21 ] . With the scaling of device dimensions, memristive devices lose their programming resolution and are characterized by increased cycle-to-cycle variation. Work has started in earnest to design stochastic state-compressed SNNs using such scaled neuromorphic devices that exhibit iso-accuracies in comparison to their multi-bit deterministic counterparts enabled by the alternate encoding of information in the probability domain [ 22 , 23 ] . Ref. [ 24 ] proposes noisy spiking neural network (NSNN) which leverages stochastic noise as a resource for training SNNs using rate-based coding. While there is significant progress in the domain of stochastic SNN training algorithms, there remains a noticeable gap in the design of SNN architectures that integrate the benefits of both stochastic neuronal computing and temporal information coding. Most of the current efforts on directly training stochastic SNNs with temporal coding have primarily demonstrated success on simple datasets and shallow network structures [ 25 , 26 , 27 ] . Scaling these networks to deeper architectures and more complex datasets presents significant challenges. Further, as we demonstrate in this work, many of the supposed benefits of temporal encoding, like enhanced spiking sparsity, may not necessarily hold true for deep architectures. This necessitates a co-design approach to identify the relative tradeoffs of stochastic temporally encoded SNNs in terms of accuracy, latency, sparsity, energy cost, and robustness. The specific contributions of this work are summarized below:
(i) Algorithm Development: We present a simple and structured algorithm framework to train stochastic SNNs directly with First-to-Spike coding. We also present training frameworks to train deterministic SNNs with temporal encoding that serve as a comparison baseline for our work to identify the relative merits/demerits of the computing and encoding scheme. We present empirical results to substantiate the scalability of our approach by demonstrating state-of-the-art accuracies on MNIST [ 28 ] and CIFAR-10 [ 29 ] datasets for 2-layer MLP, LeNet5, and VGG15 architectures. Notably, this is the first work to demonstrate direct SNN training employing First-to-Spike coding for VGG architectures on the CIFAR dataset.
(ii) Co-Design Analysis: We present a comprehensive quantitative analysis of previously unexplored trade-offs for stochastic SNNs with temporal encoding in terms of neuromorphic compute specific metrics like accuracy, latency, sparsity, energy efficiency, and robustness.
The rest of the paper is organized as follows. Section II describes related works. Section III introduces the training frameworks of SNNs with First-to-Spike coding for both deterministic and stochastic computing architectures. Section IV presents the experimental results and Section V provides conclusions and future outlook.
II Related Works
Temporal Coding: Temporal coding is characterized by its emphasis on the timing of spikes rather than the frequency in rate coding. Time-to-First-Spike (TTFS) coding [ 30 ] is a popular temporal coding scheme that is demonstrated to have rapid and low power processing [ 31 , 32 , 33 , 34 ] since it typically imposes a limitation that each neuron should only generate at most one spike. This limitation lacks biological plausibility and it cannot handle the complex temporal structure of sequences of real-world events [ 35 ] . Although latency is significantly reduced compared to rate coding, it still suffers from high latency, particularly when processing complex datasets [ 33 , 36 , 37 ] . Therefore, we focus on First-to-Spike coding based temporal coding strategy that can further reduce the latency in comparison to TTFS coding. First-to-Spike coding is distinct from other approaches since it does not primarily rely on the precise timing of each spike. Instead, this coding strategy focuses on the order of the first spike of all the output neurons. The efficacy and potential applications of the First-to-Spike coding mechanism have been extensively explored in recent literature [ 35 , 38 ] . Nevertheless, the process of generating a spike in SNNs is non-differentiable. To tackle this problem, there are several common methods for developing and training SNNs with temporal coding, which will be introduced next.
ANN-SNN Conversion Approaches for Temporal Coding: ANN-SNN Conversion is a widely adopted method for converting pre-trained ANNs to SNNs [ 39 , 40 , 41 ] . The neurons with continuous activation functions, such as sigmoid or ReLU, need to be mapped to spiking neurons like IF/LIF neurons. Algorithmic approaches usually aim to reduce information loss caused during the conversion process. Proposal by [ 6 ] designed an exact mapping from an ANN with ReLUs to a corresponding SNN with TTFS coding. The key achievement of this mapping is that it maintains the network accuracy after conversion with minimal drop. However, the conversion process involves complex steps which can make the process difficult to implement and optimize. Additionally, the necessity to use different conversion strategies for different types of layers further adds to the complexity. Also, it is important to note that the ANNs are trained without any temporal information, which typically results in high latency when converted to SNNs [ 11 ] . Hence, it is critical to explore direct training strategies for SNNs with temporal encoding.
Direct SNN Training Approaches for Temporal Coding: In the domain of TTFS coding, a convolutional-like coding method [ 42 , 43 ] was proposed to directly train an SNN, which uses a temporal kernel to integrate temporal and spatial information. It can significantly reduce the model size and transform the spatial localities into temporal localities which can improve efficiency and accuracy. Some recent works use the surrogate gradient [ 35 , 44 ] or surrogate model [ 37 ] to solve the non-differentiable backpropagation issue in deterministic neurons with temporal coding. Another technique is to directly train SNNs with stochastic neurons. The smoothing effect of stochastic neurons is crucial for enabling gradient-based optimization methods in SNNs [ 19 ] by solving the non-linear, non-differentiable aspects of the spiking mechanism. Research by [ 25 , 26 ] introduced a stochastic neuron model for directly training SNNs. This model uses the generalized linear model (GLM) [ 45 ] and first-to-spike coding. The GLM consists of a set of linear filters to process the incoming spikes, followed by a nonlinear function that computes the neuron’s firing probability based on the filtered inputs. Subsequently, this model employs a stochastic process such as the Poisson process to generate spike trains. However, the computational complexity of adapting a GLM for large-scale SNN training can be quite high. Other recent works have also explored stochastic SNNs with TTFS coding where the stochastic neuron is implemented by the intrinsic physics of spin devices [ 27 ] . However, existing research primarily focuses on shallow networks and MNIST-level datasets and lacks quantification of benefits offered by stochastic computing and temporal encoding in SNNs at scale.
III Methods
In this section, we introduce the methodology to train deterministic and stochastic SNNs with First-to-Spike coding where the key idea is to find the neuron that generates the first spike signal, thereby terminating the inference process. Associated loss function design and weight gradient calculations are also elaborated considering discontinuity issues observed in spiking neurons.
III-A Deterministic SNN
The Leaky Integrate-and-Fire (LIF) neuron model is one of the most recognized spiking neuron models in SNNs, primarily chosen for its balance between simplicity and biological plausibility [ 46 ] . The LIF model simulates the behavior of neurons by accumulating input signals (voltage) until they reach the threshold. During this period, the accumulated voltage decays over time, which simulates the electrical resistance seen in real neuronal membranes. However, the process to generate a spike in LIF models is non-differentiable which makes it challenging for traditional gradient-based methods. Defining a surrogate gradient (SG) as a continuous relaxation of the real gradients is one of the common ways to tackle the discontinuous spiking nonlinearity [ 19 ] . The deterministic LIF neuron model used in our network can be summarized as:
The output spike train o i t superscript subscript 𝑜 𝑖 𝑡 o_{i}^{t} italic_o start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT start_POSTSUPERSCRIPT italic_t end_POSTSUPERSCRIPT is generated by following this equation:
The temporal cross-entropy loss function [ 47 ] , which integrates the principles of First-to-Spike coding, is formalized as follows: For each neuron i 𝑖 i italic_i , the estimated activation probability is computed using the equation:
where, t i subscript 𝑡 𝑖 t_{i} italic_t start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT is the time of the first spike of neuron i 𝑖 i italic_i and n 𝑛 n italic_n is the number of output neurons. The loss function is given by the following equation:
where, y i ∈ { 0 , 1 } subscript 𝑦 𝑖 0 1 y_{i}\in\{0,1\} italic_y start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT ∈ { 0 , 1 } is a one-hot target vector and n 𝑛 n italic_n is the number of output neurons. In the context of First-to-Spike coding, the goal is to minimize the time of the first spike of the correct neuron, which leads to maximizing its corresponding probability, as indicated by the cross-entropy loss function.
The gradient of the weights corresponding to the deterministic LIF neuron model is given by the following equation:
superscript subscript 𝑉 𝑖 𝑡 {\partial o_{i}^{t}}/{\partial V_{i}^{t}} ∂ italic_o start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT start_POSTSUPERSCRIPT italic_t end_POSTSUPERSCRIPT / ∂ italic_V start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT start_POSTSUPERSCRIPT italic_t end_POSTSUPERSCRIPT , we need a surrogate gradient to solve the discontinuous spiking nonlinearity. In this paper, the A r c t a n 𝐴 𝑟 𝑐 𝑡 𝑎 𝑛 Arctan italic_A italic_r italic_c italic_t italic_a italic_n surrogate [ 48 ] is used. After employing the A r c t a n 𝐴 𝑟 𝑐 𝑡 𝑎 𝑛 Arctan italic_A italic_r italic_c italic_t italic_a italic_n surrogate, Eqn. 2 can be written as:
superscript subscript 𝑜 𝑖 𝑡 {\partial t_{i}}/{\partial o_{i}^{t}} ∂ italic_t start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT / ∂ italic_o start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT start_POSTSUPERSCRIPT italic_t end_POSTSUPERSCRIPT can be expressed as:
This allows the network to be trained using variants of backpropagation. In this paper, Backpropagation through time (BPTT) [ 49 ] is used where the network is unrolled across timesteps for backpropagation.
III-B Stochastic SNN
Contrary to the usage of SG for deterministic neuron models, another way to solve the discontinuous spiking nonlinearity is by using stochastic neuron models. Inspired by [ 25 ] , we integrate stochastic LIF neurons with First-to-Spike coding. The membrane potential is computed by using the following equation:
where, k i subscript 𝑘 𝑖 k_{i} italic_k start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT is a scaling factor of the membrane potential of the neuron. Subsequently, the sigmoid activation function is used to calculate p i t superscript subscript 𝑝 𝑖 𝑡 p_{i}^{t} italic_p start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT start_POSTSUPERSCRIPT italic_t end_POSTSUPERSCRIPT , which is the probability of neuron i 𝑖 i italic_i generating a spike at time t 𝑡 t italic_t . The probability p i t superscript subscript 𝑝 𝑖 𝑡 p_{i}^{t} italic_p start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT start_POSTSUPERSCRIPT italic_t end_POSTSUPERSCRIPT is used to generate an independent and identically distributed (i.i.d.) Bernoulli value, which represents the discrete spike train generated by the neuron. Due to the non-differentiable nature of the Bernoulli function, it poses a problem for backpropagation techniques which rely on gradient-based optimization. To address this issue, we use the Straight-Through (ST) estimator [ 50 ] , which passes the gradient received from the deeper layer directly to the preceding layer without any modification in the backward phase. In the output layer, we compute the probability P t subscript 𝑃 𝑡 P_{t} italic_P start_POSTSUBSCRIPT italic_t end_POSTSUBSCRIPT of the correct neuron to generate the earliest spike at time t 𝑡 t italic_t by the following equation:
where, p c t superscript subscript 𝑝 𝑐 𝑡 p_{c}^{t} italic_p start_POSTSUBSCRIPT italic_c end_POSTSUBSCRIPT start_POSTSUPERSCRIPT italic_t end_POSTSUPERSCRIPT is the probability of correct neuron c generating a spike at time t 𝑡 t italic_t . This equation represents the probability that no wrong neurons generate a spike before the correct neuron produces a spike at time t 𝑡 t italic_t . We use the same ML (Maximum Likelihood) criterion used in [ 25 ] by maximizing the sum of all P t subscript 𝑃 𝑡 P_{t} italic_P start_POSTSUBSCRIPT italic_t end_POSTSUBSCRIPT s through the following equation:
As the timestep increases, P t subscript 𝑃 𝑡 P_{t} italic_P start_POSTSUBSCRIPT italic_t end_POSTSUBSCRIPT reduces, and the contributions to the overall losses diminish progressively which encourages neurons to fire earlier but not before the correct neuron, resulting in reduced latency. Furthermore, the BPTT algorithm is employed in a similar fashion as the deterministic SNN model, unfolding the network across timesteps and calculating the gradients of Eqn. 10 with respect to the weights at each timestep.
In this section, we evaluate the accuracy, latency, sparsity, energy cost, and noise sensitivity of different types of models to evaluate the influence of information encoding, computing scheme and training methods independently: ANN-SNN conversion utilizing deterministic neurons and rate coding (D-R-CONV) [ 39 ] , BPTT trained models with deterministic neurons utilizing rate coding (D-R-BPTT) [ 11 ] , deterministic neural networks trained by BPTT utilizing First-to-Spike coding (D-F-BPTT) and the stochastic neural networks trained by BPTT utilizing First-to-Spike coding (S-F-BPTT). Acronyms are used to simplify the naming that reflects its key features: The first part indicates the type of neurons: deterministic (D) and stochastic (S), the second part denotes the coding method: rate coding (R) and First-to-Spike coding (F), and the third part represents the training method: ANN-SNN conversion (CONV) and Backpropagation Through Time (BPTT). We will use these acronyms throughout the remainder of the paper for brevity. We conduct experiments for three neural network architectures, ranging from shallow to deep: 2-layer MLP, LeNet5, and VGG15.
IV-A Datasets
In this paper, we use the MNIST [ 28 ] and CIFAR-10 [ 29 ] datasets for our experiments. In the preprocessing stage for the MNIST dataset, we adjust the pixel intensities from their original range of 0-255 to a normalized range of 0-1. For the CIFAR-10 dataset, we use data augmentation to effectively increase the diversity of the training data and reduce overfitting. In our case, the random horizontal flipping is applied with a probability of 0.5, and the image is rotated at an angle randomly selected from a range of -15 to 15 degrees [ 51 ] . Our preprocessing also includes random cropping of images, with a padding of 4 pixels [ 52 ] . To further augment the dataset, a random affine transformation is applied to the image. This includes shear-based transformations, where the degree of shear is precisely set to 10, effectively introducing a specific level of distortion to the images. Additionally, scaling adjustments are applied, altering the image size to fluctuate between 80% and 120% of the original size. The image attributes such as brightness, contrast, and saturation are adjusted [ 53 ] , each by a factor of 0.2, to enhance model robustness against varying lighting and color conditions. Furthermore, normalization of the input image data is employed based on the mean and standard deviation for each color channel in the CIFAR-10 dataset.
IV-B Model Training
In the training of all architectures, the Adam optimizer [ 54 ] is utilized, accompanied by a learning rate scheduler. For deterministic LIF neurons, the parameter α 𝛼 \alpha italic_α is set to 2 in Eqn 7 . The detailed hyperparameter settings are listed in Table I . Additionally, a critical aspect of SNN-specific optimization involves layerwise tuning of the neuron’s firing threshold V t h subscript 𝑉 𝑡 ℎ V_{th} italic_V start_POSTSUBSCRIPT italic_t italic_h end_POSTSUBSCRIPT in the D-F-BPTT model and the scaling factor k i subscript 𝑘 𝑖 k_{i} italic_k start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT in the S-F-BPTT model (see Section III). For this purpose, we used a Neuroevolutionary optimized hybrid SNN training approach [ 55 ] where the trained model was subsequently optimized using the gradient-free differential evolution (DE) algorithm [ 56 , 57 ] to achieve the best accuracy-latency tradeoff. Prior work has demonstrated that such a hybrid framework significantly outperforms approaches that combine such hyperparameter tuning during the BPTT training process itself [ 55 ] .
IV-C Quantitative Analysis
Accuracy: The performance of each network is summarized in Table II . The accuracy is determined by calculating the mean value across ten independent runs. Transitioning from rate coding to temporal coding actually does not reduce the accuracy and even increases the accuracy in some cases. However, the introduction of stochasticity to the model causes a consistent increase in the network accuracy on the more complex CIFAR-10 dataset . For complex datasets, the variability introduced by stochasticity could act as a form of data augmentation, presenting the network with a wider range of data during the training. This can prevent the model from overfitting, leading to better generalization.
Inference Latency: In SNNs, reducing latency without sacrificing accuracy can be a critical goal, allowing for faster and more energy-efficient computation. In particular, for the D-R-CONV and D-R-BPTT models, the optimal number of timesteps is determined by identifying the saturation point on a plot of timesteps versus accuracy, where further increase in timesteps no longer significantly improves model accuracy. The differences in SNN inference latency in terms of timesteps are noted in Table II . Compared to other temporal coding models, First-to-Spike coding models show a significantly lower latency. In the First-to-Spike coding scheme, the result is based on which neuron in the output layer is the first to generate a spike. This approach requires only a single spike in the output layer to ascertain the result, reducing the number of timesteps significantly. On the other hand, rate coding relies on the frequency of spikes over time, and therefore the network needs a longer observation window to establish an accurate spike rate. This phenomenon is magnified when the dataset becomes complex. On the CIFAR-10 dataset, the rate coding approaches require a substantially higher number of timesteps to achieve the same level of accuracy compared to the models employing First-to-Spike coding. Interestingly, we find that the S-F-BPTT model reduces the latency even further in comparison to the D-F-BPTT model. The stochastic nature of spike generation in S-F-BPTT models may cause an output spike generation even when the input stimulus is not too strong or the membrane potential is low, allowing for a faster response to the input.
Sparsity: Spiking sparsity in SNNs is an important metric for evaluating the efficiency and functionality of models. The average spiking rate of a particular layer, defined as the average number of spikes that a neuron generates over a fixed time interval, is utilized to quantitatively represent sparsity. In this context, a higher average spike rate indicates lower sparsity, and vice versa. The average spiking rate of each model across various layers for the LeNet5 architecture trained on the MNIST dataset and VGG15 architecture trained on the CIFAR-10 dataset is shown in Fig. 1 . Contrary to the common assumption of higher sparsity in temporal coding models than in rate coding models, the figure shows that first-to-spike models do show higher sparsity in the final layer, with the hidden layers presenting a contrary trend. The main reason is the necessity to encode the same information with reduced latency for temporally encoded models, which demands an increased spike count. It also explains why the S-F-BPTT model has the highest spiking rate with the lowest latency. Moreover, to achieve a reliable and consistent output in the presence of stochasticity, the stochastic SNN model needs to increase its spiking rate. This compensates for the unpredictability of individual spikes, ensuring that the overall signal transmission between neurons remains stable and correct.

Energy Cost: The total number of SNN computations, that serves as a proxy metric for the resultant energy consumption of the model when deployed on neuromorphic hardware [ 39 ] , is also a crucial factor in designing SNN models. The total “energy cost” E 𝐸 E italic_E of each model, defined as the ratio of the number of computations performed in the SNN relative to that of an iso-architecture ANN, can be estimated as:
where, L 𝐿 L italic_L is the total number of layers, S i − 1 subscript 𝑆 𝑖 1 S_{i-1} italic_S start_POSTSUBSCRIPT italic_i - 1 end_POSTSUBSCRIPT is the average spiking rate of the ( i − 1 ) t h superscript 𝑖 1 𝑡 ℎ (i-1)^{th} ( italic_i - 1 ) start_POSTSUPERSCRIPT italic_t italic_h end_POSTSUPERSCRIPT layer, T 𝑇 T italic_T is the number of timesteps used for inference, and O P i 𝑂 subscript 𝑃 𝑖 {OP}_{i} italic_O italic_P start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT is the number of operations in the i t h superscript 𝑖 𝑡 ℎ i^{th} italic_i start_POSTSUPERSCRIPT italic_t italic_h end_POSTSUPERSCRIPT layer. Following [ 39 ] , O P i 𝑂 subscript 𝑃 𝑖 {OP}_{i} italic_O italic_P start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT of convolutional layers and linear layers can be summarized as:
where, l i subscript 𝑙 𝑖 l_{i} italic_l start_POSTSUBSCRIPT italic_i end_POSTSUBSCRIPT is the i t h superscript 𝑖 𝑡 ℎ i^{th} italic_i start_POSTSUPERSCRIPT italic_t italic_h end_POSTSUPERSCRIPT layer, C I subscript 𝐶 𝐼 C_{I} italic_C start_POSTSUBSCRIPT italic_I end_POSTSUBSCRIPT and C O subscript 𝐶 𝑂 C_{O} italic_C start_POSTSUBSCRIPT italic_O end_POSTSUBSCRIPT are the number of input and output channels, K H subscript 𝐾 𝐻 K_{H} italic_K start_POSTSUBSCRIPT italic_H end_POSTSUBSCRIPT and K W subscript 𝐾 𝑊 K_{W} italic_K start_POSTSUBSCRIPT italic_W end_POSTSUBSCRIPT are the height and width of the kernel, O H subscript 𝑂 𝐻 O_{H} italic_O start_POSTSUBSCRIPT italic_H end_POSTSUBSCRIPT and O W subscript 𝑂 𝑊 O_{W} italic_O start_POSTSUBSCRIPT italic_W end_POSTSUBSCRIPT are the height and width of the output, and I F subscript 𝐼 𝐹 I_{F} italic_I start_POSTSUBSCRIPT italic_F end_POSTSUBSCRIPT and O F subscript 𝑂 𝐹 O_{F} italic_O start_POSTSUBSCRIPT italic_F end_POSTSUBSCRIPT are the number of input and output features. The results depicted in Table II demonstrate the energy cost of the four models: D-R-BPTT, D-R-CONV, S-F-BPTT, and D-F-BPTT. The D-R-BPTT model has the highest energy requirement, followed by the D-R-CONV, S-F-BPTT, and D-F-BPTT models, in descending order. The results demonstrate that the models using First-to-Spike coding are more energy efficient than rate-coded models. Furthermore, it is observed that the benefit of lower latency for the S-F-BPTT model is outweighed by its significantly higher spiking rate in contrast to the D-F-BPTT model, ultimately resulting in comparable or increased energy expenditure. However, on the CIFAR-10 dataset, this difference is less pronounced, as the D-F-BPTT model requires almost twice the number of timesteps compared to the S-F-BPTT model, resulting in only a slight difference in the total energy cost.

Noise Sensitivity: A key aspect of ML model design is to ensure robustness to noise. A model’s noise sensitivity can be measured by adding different levels of noise to the input and observing the impact on the network’s accuracy. In this paper, Gaussian noise is used to assess how well the model can maintain its performance under noisy conditions. The variance of Gaussian noise is adjusted from 0 to 1. Fig. 2 shows the relationship between accuracy degradation and the magnitude of applied noise. It can be observed that the D-R-BPTT model demonstrates a higher tolerance to noise, maintaining higher accuracy as the noise intensity increases. Since it uses rate coding, which encodes information by the frequency of multiple spikes over time, individual perturbations caused by noise have less impact on the overall information conveyed. Temporal coding models (D-F-BPTT and S-F-BPTT) are more sensitive to noise because they rely on the precise timing of spikes to encode information. However, the stochastic model has better performance than the deterministic model at high noise levels. This can be attributed to the stochasticity which is incorporated during the training process itself and therefore can provide more resilience to noise (through more tolerance towards the precision of individual spikes).
V Conclusions
In summary, our research explores the interplay of deterministic/stochastic computing with First-to-Spike information coding in SNNs. This integration bridges a gap in current research, demonstrating scalable direct training of SNNs with temporal encoding on large-scale datasets and deep architectures. We showcase that First-to-Spike coding has significant performance benefits for SNN architectures in contrast to traditional rate-based models with regard to various metrics, including latency, sparsity, and energy efficiency. We also underscore notable trade-offs between the stochastic and deterministic SNN models in temporal encoding scenarios. Stochastic models reduce latency and provide enhanced noise robustness, which is important for real-time confidence-critical applications. However, this advantage is offered at the expense of a slight decrement in sparsity, which consequentially results in higher energy costs compared to deterministic SNNs employing First-to-Spike coding. In terms of accuracy, stochastic SNNs have the potential to aid in better generalization, especially for complex datasets. Although our results are promising, scaling this method to ImageNet level vision tasks as well as beyond vision applications could be a future research direction. Energy and sparsity aware training techniques can be also considered for stochastic SNN models with temporal encoding to further enhance its applicability for resource-constrained edge devices.
Acknowledgments
This material is based upon work supported in part by the U.S. Department of Energy, Office of Science, Office of Advanced Scientific Computing Research, under Award Number #DE-SC0021562, the U.S. National Science Foundation under award No. CCSS #2333881, CCF #1955815, CAREER #2337646 and EFRI BRAID #2318101 and by Oracle Cloud credits and related resources provided by the Oracle for Research program.
- [1] A. Javanshir, T. T. Nguyen, M. A. P. Mahmud, and A. Z. Kouzani, “Advancements in Algorithms and Neuromorphic Hardware for Spiking Neural Networks,” Neural Computation , vol. 34, no. 6, pp. 1289–1328, 05 2022.
- [2] W. Maas, “Networks of spiking neurons: The third generation of neural network models,” Trans. Soc. Comput. Simul. Int. , vol. 14, no. 4, p. 1659–1671, dec 1997.
- [3] W. Guo, M. E. Fouda, A. M. Eltawil, and K. N. Salama, “Neural coding in spiking neural networks: A comparative study for robust neuromorphic systems,” Frontiers in Neuroscience , vol. 15, 2021.
- [4] T. Gollisch and M. Meister, “Rapid neural coding in the retina with relative spike latencies,” Science , vol. 319, no. 5866, pp. 1108–1111, 2008.
- [5] P. Heil, “First-spike latency of auditory neurons revisited,” Current Opinion in Neurobiology , vol. 14, no. 4, pp. 461–467, 2004.
- [6] A. Stanojevic, S. Woźniak, G. Bellec, G. Cherubini, A. Pantazi, and W. Gerstner, “An exact mapping from relu networks to spiking neural networks,” 2022.
- [7] B. Rueckauer and S.-C. Liu, “Conversion of analog to spiking neural networks using sparse temporal coding,” in 2018 IEEE International Symposium on Circuits and Systems (ISCAS) , 2018, pp. 1–5.
- [8] J. Lin, S. Lu, M. Bal, and A. Sengupta, “Benchmarking spiking neural network learning methods with varying locality,” arXiv preprint arXiv:2402.01782 , 2024.
- [9] M. Bal and A. Sengupta, “Spikingbert: Distilling bert to train spiking language models using implicit differentiation,” in Proceedings of the AAAI Conference on Artificial Intelligence , vol. 38, no. 10, 2024, pp. 10 998–11 006.
- [10] R.-J. Zhu, Q. Zhao, G. Li, and J. K. Eshraghian, “Spikegpt: Generative pre-trained language model with spiking neural networks,” arXiv preprint arXiv:2302.13939 , 2023.
- [11] N. Rathi, G. Srinivasan, P. Panda, and K. Roy, “Enabling deep spiking neural networks with hybrid conversion and spike timing dependent backpropagation,” in International Conference on Learning Representations , 2020.
- [12] J. Kaiser, H. Mostafa, and E. Neftci, “Synaptic plasticity dynamics for deep continuous local learning (decolle),” Frontiers in Neuroscience , vol. 14, May 2020. [Online]. Available: http://dx.doi.org/10.3389/fnins.2020.00424
- [13] B. Scellier and Y. Bengio, “Equilibrium propagation: Bridging the gap between energy-based models and backpropagation,” 2017.
- [14] M. Bal and A. Sengupta, “Sequence learning using equilibrium propagation,” arXiv preprint arXiv:2209.09626 , 2022.
- [15] S. Lu and A. Sengupta, “Deep Unsupervised Learning Using Spike-Timing-Dependent Plasticity,” Neuromorphic Computing and Engineering , 2023.
- [16] W. Gerstner, R. Ritz, and J. L. van Hemmen, “Why spikes? hebbian learning and retrieval of time-resolved excitation patterns,” Biol. Cybern. , vol. 69, no. 5-6, pp. 503–515, Sep. 1993.
- [17] L. Lapicque, “Recherches quantitatives sur l’excitation électrique des nerfs traitée comme une polarisation,” J Physiol Paris , vol. 9, pp. 620–635, 1907.
- [18] W. Maass, “To spike or not to spike: That is the question,” Proceedings of the IEEE , vol. 103, no. 12, pp. 2219–2224, 2015.
- [19] E. O. Neftci, H. Mostafa, and F. Zenke, “Surrogate gradient learning in spiking neural networks,” 2019.
- [20] A. Sengupta, M. Parsa, B. Han, and K. Roy, “Probabilistic deep spiking neural systems enabled by magnetic tunnel junction,” IEEE Transactions on Electron Devices , vol. 63, no. 7, p. 2963–2970, Jul. 2016.
- [21] K. Yang and A. Sengupta, “Stochastic magnetoelectric neuron for temporal information encoding,” Applied Physics Letters , vol. 116, no. 4, Jan. 2020.
- [22] A. Islam, K. Yang, A. K. Shukla, P. Khanal, B. Zhou, W.-G. Wang, and A. Sengupta, “Hardware in loop learning with spin stochastic neurons,” arXiv preprint arXiv:2305.03235 , 2023.
- [23] A. Islam, A. Saha, Z. Jiang, K. Ni, and A. Sengupta, “Hybrid stochastic synapses enabled by scaled ferroelectric field-effect transistors,” Applied Physics Letters , vol. 122, no. 12, 2023.
- [24] Ma, G., Yan, R. & Tang, H. Exploiting noise as a resource for computation and learning in spiking neural networks. Patterns , vol. 4, no. 10, pp. 100831, 2023.
- [25] A. Bagheri, O. Simeone, and B. Rajendran, “Training probabilistic spiking neural networks with first-to-spike decoding,” 2018.
- [26] B. Rosenfeld, O. Simeone, and B. Rajendran, “Learning first-to-spike policies for neuromorphic control using policy gradients,” in 2019 IEEE 20th International Workshop on Signal Processing Advances in Wireless Communications (SPAWC) , 2019, pp. 1–5.
- [27] K. Yang, D. P. Gm, and A. Sengupta, “Leveraging probabilistic switching in superparamagnets for temporal information encoding in neuromorphic systems,” IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems , 2023.
- [28] Y. LeCun and C. Cortes, “MNIST handwritten digit database,” 2010.
- [29] A. Krizhevsky, V. Nair, and G. Hinton, “Cifar-10 (canadian institute for advanced research).”
- [30] W. Gerstner and W. M. Kistler, Spiking Neuron Models: Single Neurons, Populations, Plasticity . Cambridge University Press, 2002.
- [31] J. Göltz, L. Kriener, A. Baumbach, S. Billaudelle, O. Breitwieser, B. Cramer, D. Dold, A. F. Kungl, W. Senn, J. Schemmel, K. Meier, and M. A. Petrovici, “Fast and energy-efficient neuromorphic deep learning with first-spike times,” Nature Machine Intelligence , vol. 3, no. 9, p. 823–835, Sep. 2021.
- [32] S. Oh, D. Kwon, G. Yeom, W.-M. Kang, S. Lee, S. Y. Woo, J. Kim, and J.-H. Lee, “Neuron circuits for low-power spiking neural networks using time-to-first-spike encoding,” IEEE Access , vol. 10, pp. 24 444–24 455, 2022.
- [33] S. Park, S. Kim, B. Na, and S. Yoon, “T2fsnn: Deep spiking neural networks with time-to-first-spike coding,” 2020.
- [34] Y. Kim, A. Kahana, R. Yin, Y. Li, P. Stinis, G. E. Karniadakis, and P. Panda, “Rethinking skip connections in spiking neural networks with time-to-first-spike coding,” Frontiers in Neuroscience , vol. 18, 2024. [Online]. Available: https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2024.1346805
- [35] S. Liu, V. C. H. Leung, and P. L. Dragotti, “First-spike coding promotes accurate and efficient spiking neural networks for discrete events with rich temporal structures,” Front. Neurosci. , vol. 17, p. 1266003, Oct. 2023.
- [36] W. Wei, M. Zhang, H. Qu, A. Belatreche, J. Zhang, and H. Chen, “Temporal-coded spiking neural networks with dynamic firing threshold: Learning with event-driven backpropagation,” in Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) , October 2023, pp. 10 552–10 562.
- [37] S. Park and S. Yoon, “Training energy-efficient deep spiking neural networks with time-to-first-spike coding,” 2021.
- [38] M. Mozafari, S. R. Kheradpisheh, T. Masquelier, A. Nowzari-Dalini, and M. Ganjtabesh, “First-spike-based visual categorization using reward-modulated stdp,” IEEE Transactions on Neural Networks and Learning Systems , vol. 29, no. 12, p. 6178–6190, Dec. 2018.
- [39] S. Lu and A. Sengupta, “Exploring the connection between binary and spiking neural networks,” Frontiers in Neuroscience , vol. 14, jun 2020.
- [40] A. Sengupta, Y. Ye, R. Wang, C. Liu, and K. Roy, “Going deeper in spiking neural networks: Vgg and residual architectures,” 2019.
- [41] Y. Li, S. Deng, X. Dong, R. Gong, and S. Gu, “A free lunch from ann: Towards efficient, accurate spiking neural networks calibration,” 2021.
- [42] T. Liu, Z. Liu, F. Lin, Y. Jin, G. Quan, and W. Wen, “Mt-spike: A multilayer time-based spiking neuromorphic architecture with temporal error backpropagation,” 2018.
- [43] T. Liu, L. Jiang, Y. Jin, G. Quan, and W. Wen, “Pt-spike: A precise-time-dependent single spike neuromorphic architecture with efficient supervised learning,” in 2018 23rd Asia and South Pacific Design Automation Conference (ASP-DAC) , 2018, pp. 568–573.
- [44] M. Zhang, J. Wang, B. Amornpaisannon, Z. Zhang, V. Miriyala, A. Belatreche, H. Qu, J. Wu, Y. Chua, T. E. Carlson, and H. Li, “Rectified linear postsynaptic potential function for backpropagation in deep spiking neural networks,” 2020.
- [45] J. W. Pillow, L. Paninski, V. J. Uzzell, E. P. Simoncelli, and E. J. Chichilnisky, “Prediction and decoding of retinal ganglion cell responses with a probabilistic spiking model,” J. Neurosci. , vol. 25, no. 47, pp. 11 003–11 013, Nov. 2005.
- [46] E. Izhikevich, “Which model to use for cortical spiking neurons?” IEEE Transactions on Neural Networks , vol. 15, no. 5, pp. 1063–1070, 2004.
- [47] J. K. Eshraghian, M. Ward, E. Neftci, X. Wang, G. Lenz, G. Dwivedi, M. Bennamoun, D. S. Jeong, and W. D. Lu, “Training spiking neural networks using lessons from deep learning,” Proceedings of the IEEE , vol. 111, no. 9, pp. 1016–1054, 2023.
- [48] W. Fang, Z. Yu, Y. Chen, T. Masquelier, T. Huang, and Y. Tian, “Incorporating learnable membrane time constant to enhance learning of spiking neural networks,” 2021.
- [49] Y. Wu, L. Deng, G. Li, J. Zhu, and L. Shi, “Spatio-temporal backpropagation for training high-performance spiking neural networks,” Frontiers in Neuroscience , vol. 12, May 2018.
- [50] Y. Bengio, N. Léonard, and A. C. Courville, “Estimating or propagating gradients through stochastic neurons for conditional computation,” CoRR , vol. abs/1308.3432, 2013.
- [51] C. Shorten and T. M. Khoshgoftaar, “A survey on image data augmentation for deep learning,” J. Big Data , vol. 6, no. 1, Dec. 2019.
- [52] C.-Y. Lee, S. Xie, P. Gallagher, Z. Zhang, and Z. Tu, “Deeply-supervised nets,” 2014.
- [53] E. D. Cubuk, B. Zoph, D. Mane, V. Vasudevan, and Q. V. Le, “Autoaugment: Learning augmentation policies from data,” 2019.
- [54] D. P. Kingma and J. Ba, “Adam: A method for stochastic optimization,” 2017.
- [55] S. Lu and A. Sengupta, “Neuroevolution guided hybrid spiking neural network training,” Front. Neurosci. , vol. 16, p. 838523, Apr. 2022.
- [56] R. Storn and K. Price, “Differential evolution - a simple and efficient heuristic for global optimization over continuous spaces,” Journal of Global Optimization , vol. 11, pp. 341–359, 01 1997.
- [57] P. Virtanen, R. Gommers, T. E. Oliphant, M. Haberland, T. Reddy, D. Cournapeau, E. Burovski, P. Peterson, W. Weckesser, J. Bright et al. , “SciPy 1.0: fundamental algorithms for scientific computing in python,” Nature methods , vol. 17, no. 3, pp. 261–272, 2020.
- [58] Y. Sakemi, K. Morino, T. Morie, and K. Aihara, “A supervised learning algorithm for multilayer spiking neural networks based on temporal coding toward energy-efficient vlsi processor design,” IEEE Transactions on Neural Networks and Learning Systems , vol. 34, no. 1, pp. 394–408, 2023.
Academia.edu no longer supports Internet Explorer.
To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your browser .
- We're Hiring!
- Help Center
SPIKING NEURAL NETWORK
- Most Cited Papers
- Most Downloaded Papers
- Newest Papers
- Save to Library
- Last »
- Spiking Neurons Follow Following
- Optic Nerve Follow Following
- Spiking neuron models Follow Following
- Neuronal Activity Follow Following
- Spiking Neural Networks Follow Following
- Temporal Data Mining Follow Following
- Computer and Communication Systems Engineering Follow Following
- MASTERS Follow Following
- Character Recognition Follow Following
- Microarray Data Analysis Follow Following
Enter the email address you signed up with and we'll email you a reset link.
- Academia.edu Publishing
- We're Hiring!
- Help Center
- Find new research papers in:
- Health Sciences
- Earth Sciences
- Cognitive Science
- Mathematics
- Computer Science
- Academia ©2024
- Help & FAQ
Training spiking neural networks with metaheuristic algorithms
Research output : Contribution to journal › Article › Research › peer-review
Taking inspiration from the brain, spiking neural networks (SNNs) have been proposed to understand and diminish the gap between machine learning and neuromorphic computing. Supervised learning is the most commonly used learning algorithm in traditional ANNs. However, directly training SNNs with backpropagation-based supervised learning methods is challenging due to the discontinuous and non-differentiable nature of the spiking neuron. To overcome these problems, this paper proposes a novel metaheuristic-based supervised learning method for SNNs by adapting the temporal error function. We investigated seven well-known metaheuristic algorithms called Harmony Search (HS), Cuckoo Search (CS), Differential Evolution (DE), Particle Swarm Optimization (PSO), Genetic Algorithm (GA), Artificial Bee Colony (ABC), and Grammatical Evolution (GE) as search methods for carrying out network training. Relative target firing times were used instead of fixed and predetermined ones, making the computation of the error function simpler. The performance of our proposed approach was evaluated using five benchmark databases collected in the UCI Machine Learning Repository. The experimental results showed that the proposed algorithm had a competitive advantage in solving the four classification benchmark datasets compared to the other experimental algorithms, with accuracy levels of 0.9858, 0.9768, 0.7752, and 0.6871 for iris, cancer, diabetes, and liver datasets, respectively. Among the seven metaheuristic algorithms, CS reported the best performance.
- classification
- metaheuristic
- spiking neural network

Access to Document
- 10.3390/app13084809
- 594976585-oa Final published version, 3.71 MB
Other files and links
- Link to publication in Scopus
T1 - Training spiking neural networks with metaheuristic algorithms
AU - Javanshir, Amirhossein
AU - Nguyen, Thanh Thi
AU - Mahmud, M. A.Parvez
AU - Kouzani, Abbas Z.
N1 - Publisher Copyright: © 2023 by the authors.
PY - 2023/4/11
Y1 - 2023/4/11
N2 - Taking inspiration from the brain, spiking neural networks (SNNs) have been proposed to understand and diminish the gap between machine learning and neuromorphic computing. Supervised learning is the most commonly used learning algorithm in traditional ANNs. However, directly training SNNs with backpropagation-based supervised learning methods is challenging due to the discontinuous and non-differentiable nature of the spiking neuron. To overcome these problems, this paper proposes a novel metaheuristic-based supervised learning method for SNNs by adapting the temporal error function. We investigated seven well-known metaheuristic algorithms called Harmony Search (HS), Cuckoo Search (CS), Differential Evolution (DE), Particle Swarm Optimization (PSO), Genetic Algorithm (GA), Artificial Bee Colony (ABC), and Grammatical Evolution (GE) as search methods for carrying out network training. Relative target firing times were used instead of fixed and predetermined ones, making the computation of the error function simpler. The performance of our proposed approach was evaluated using five benchmark databases collected in the UCI Machine Learning Repository. The experimental results showed that the proposed algorithm had a competitive advantage in solving the four classification benchmark datasets compared to the other experimental algorithms, with accuracy levels of 0.9858, 0.9768, 0.7752, and 0.6871 for iris, cancer, diabetes, and liver datasets, respectively. Among the seven metaheuristic algorithms, CS reported the best performance.
AB - Taking inspiration from the brain, spiking neural networks (SNNs) have been proposed to understand and diminish the gap between machine learning and neuromorphic computing. Supervised learning is the most commonly used learning algorithm in traditional ANNs. However, directly training SNNs with backpropagation-based supervised learning methods is challenging due to the discontinuous and non-differentiable nature of the spiking neuron. To overcome these problems, this paper proposes a novel metaheuristic-based supervised learning method for SNNs by adapting the temporal error function. We investigated seven well-known metaheuristic algorithms called Harmony Search (HS), Cuckoo Search (CS), Differential Evolution (DE), Particle Swarm Optimization (PSO), Genetic Algorithm (GA), Artificial Bee Colony (ABC), and Grammatical Evolution (GE) as search methods for carrying out network training. Relative target firing times were used instead of fixed and predetermined ones, making the computation of the error function simpler. The performance of our proposed approach was evaluated using five benchmark databases collected in the UCI Machine Learning Repository. The experimental results showed that the proposed algorithm had a competitive advantage in solving the four classification benchmark datasets compared to the other experimental algorithms, with accuracy levels of 0.9858, 0.9768, 0.7752, and 0.6871 for iris, cancer, diabetes, and liver datasets, respectively. Among the seven metaheuristic algorithms, CS reported the best performance.
KW - classification
KW - metaheuristic
KW - spiking neural network
UR - http://www.scopus.com/inward/record.url?scp=85156169307&partnerID=8YFLogxK
U2 - 10.3390/app13084809
DO - 10.3390/app13084809
M3 - Article
AN - SCOPUS:85156169307
SN - 2076-3417
JO - Applied Sciences
JF - Applied Sciences
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 28 August 2023
Neural encoding with unsupervised spiking convolutional neural network
- Chong Wang 1 , 2 , 3 ,
- Hongmei Yan ORCID: orcid.org/0000-0002-9629-1396 2 , 3 ,
- Wei Huang 2 , 3 ,
- Wei Sheng 2 , 3 ,
- Yuting Wang 2 , 3 ,
- Yun-Shuang Fan 2 , 3 ,
- Tao Liu 2 ,
- Ting Zou 2 ,
- Rong Li ORCID: orcid.org/0000-0001-7266-0241 2 , 3 &
- Huafu Chen ORCID: orcid.org/0000-0002-4062-4753 1 , 2 , 3
Communications Biology volume 6 , Article number: 880 ( 2023 ) Cite this article
1 Citations
6 Altmetric
Metrics details
- Neural encoding
- Striate cortex
Accurately predicting the brain responses to various stimuli poses a significant challenge in neuroscience. Despite recent breakthroughs in neural encoding using convolutional neural networks (CNNs) in fMRI studies, there remain critical gaps between the computational rules of traditional artificial neurons and real biological neurons. To address this issue, a spiking CNN (SCNN)-based framework is presented in this study to achieve neural encoding in a more biologically plausible manner. The framework utilizes unsupervised SCNN to extract visual features of image stimuli and employs a receptive field-based regression algorithm to predict fMRI responses from the SCNN features. Experimental results on handwritten characters, handwritten digits and natural images demonstrate that the proposed approach can achieve remarkably good encoding performance and can be utilized for “brain reading” tasks such as image reconstruction and identification. This work suggests that SNN can serve as a promising tool for neural encoding.
Similar content being viewed by others

Limits to visual representational correspondence between convolutional neural networks and the human brain
Emergence of Visual Center-Periphery Spatial Organization in Deep Convolutional Neural Networks
Optimized spiking neurons can classify images with high accuracy through temporal coding with two spikes
Introduction.
The objective of neural encoding is to predict the brain’s response to external stimuli, providing an effective means to explore the brain’s mechanism for processing sensory information and serving as the foundation for brain–computer interface (BCI) systems. Visual perception, being one of the primary ways in which we receive external information, has been a major focus of neural encoding research. With the advancement of non-invasive brain imaging techniques, such as functional magnetic resonance imaging (fMRI), scientists have made remarkable progress in vision-based neural encoding 1 , 2 , 3 , 4 over the past two decades, making it a hot topic in neuroscience.
The process of vision-based encoding typically involves two main steps: feature extraction and response prediction 5 . Feature extraction aims to produce visual features of the stimuli by stimulating the visual cortex. An accurate feature extractor that approximates real visual mechanisms is crucial for successful encoding. Response prediction aims to predict voxel-wise fMRI responses based on the extracted visual features. Linear regression 6 is commonly used for this step, as the relationship between the features and responses should be as simple as possible. Previous studies have shown that the early visual cortex processes information in a manner similar to Gabor wavelets 7 , 8 , 9 . Building on this finding, Gabor filter-based encoding models have been proposed and successfully applied in tasks such as image identification and movie reconstruction 1 , 3 . In recent years, convolutional neural networks (CNNs) have garnered significant attention due to their impressive accomplishments in the field of computer vision. Several studies 10 , 11 have utilized representational similarity analysis 12 to compare the dissimilarity patterns of CNN and fMRI representations, revealing that the human visual cortex shares similar hierarchical representations to CNNs. As a result, CNN-based encoding models have become widely used and have demonstrated excellent performance 2 , 4 , 13 , 14 . However, it is important to note that despite the success of CNNs in encoding applications, the differences between CNNs and the brain in processing visual information cannot be overlooked 15 .
In terms of computational mechanisms, a fundamental distinction exists between the artificial neurons in CNNs and the biological neurons, whereby the former propagate continuous digital values, while the latter propagate action potentials (spikes). The introduction of spiking neural networks (SNNs), considered the third generation of neural networks 16 , has significantly reduced this difference. Unlike traditional artificial neural networks (ANNs), SNNs transmit information through spike timing. In SNNs, each neuron integrates spikes from the previous layer and emits spikes to the next layer when its internal voltage surpasses the threshold. The spike-timing-dependent plasticity (STDP) 17 , 18 algorithm, which is an unsupervised method for weight update and has been discovered in mammalian visual cortex 19 , 20 , 21 , is the most commonly used learning algorithm for SNNs. Recent studies have applied STDP-based SNNs to object recognition and achieved considerable performance 22 , 23 , 24 . The biological plausibility of SNNs provides them with an advantage in neural encoding.
In this paper, a spiking convolutional neural network (SCNN)-based encoding framework was proposed to bridge the gap between CNNs and the realistic visual system. The encoding procedure comprised three steps. Firstly, a SCNN was trained using the STDP algorithm to extract the visual features of the images. Secondly, the coordinates of each voxel’s receptive field in the SNN feature maps were annotated based on the retinal topological properties of the visual cortex, where each voxel receives visual input from only one fixed location of the feature map. Thirdly, linear regression models were built for each voxel to predict their responses from corresponding SNN features. The framework was evaluated using four publicly available image-fMRI datasets, including handwritten character 25 , handwritten digit 26 , grayscale natural image 1 , and colorful natural image datasets 27 . Additionally, two downstream decoding tasks, namely image reconstruction and image identification, were performed based on the encoding models. The encoding and decoding performance of the proposed method was compared with that of previous methods.
Encoding performance on handwritten character dataset
We built SCNN-based encoding models (see Fig. 1a ) on four image-fMRI datasets and realized image reconstruction and image identification tasks based on the pre-trained encoding models (see Fig. 1b, c ). Table 1 provides the basic information about these datasets, and details can be found in Methods. To predict the fMRI responses evoked by handwritten characters, the SCNN was first constructed using the images in the TICH dataset (with the exclusion of images in the test set and the inclusion of 14,854 images for the 6 characters). This was done to maximize the representation ability of the SCNN. Subsequently, voxel-wise linear regression models were trained with the fMRI data in the train set for each participant. The encoding performance was measured using Pearson’s correlation coefficients (PCC) between the predicted and measured responses to the test set images. Moreover, the proposed model was compared with a CNN-based encoding model, where the network architecture of CNN was constrained to be consistent with that of the SCNN (Supplementary Table 1 ). The CNN was trained using the Adam optimizer 28 with a learning rate of 0.0001 for 50 epochs on the TICH dataset, achieving a classification accuracy of 99% on the test set images. The subsequent encoding procedures for CNN were identical to those for SCNN. To eliminate the influence of noise (unrelated to the visual task) voxels on the result, 500 voxels with the highest encoding performance for each subject were selected for comparison. Figure 2a displays the prediction accuracies for SCNN and CNN-based encoding models. The results indicate that the accuracies of SCNN on all three subjects were significantly higher than those of CNN ( p < \({10}^{-18}\) , one-tailed, two-sample t -test). This finding suggests that the SCNN has greater potential than CNN for encoding tasks.
a The illustration of the encoding model. The proposed model uses a two-layer SCNN to extract visual features of the input images and uses linear regression models to predict the fMRI responses for each voxel. b The diagram for the image reconstruction task, which aims to reconstruct the perceived images from the brain activity. The handwritten character images are adapted from the TICH character dataset 47 with permission. c The diagram for the image identification task, which aims to identify which image is perceived based on the fMRI responses. The grayscale natural images are reproduced with permission from Kay et al. 1 .
a The encoding accuracies ( n = 500) of different subjects in the handwritten character dataset. b The mean stimulus intensities in the train set of the handwritten character dataset. c The receptive field locations of the 100 most predictable voxels of the handwritten character dataset. A smaller transparency represents a larger number of voxels. d The encoding accuracies ( n = 500) of the handwritten digit dataset. e The encoding accuracies ( n = 200) of different visual areas in the grayscale natural image dataset. f The encoding accuracies ( n = 500) and noise ceilings (mean ± standard deviation) of different subjects in the colorful natural image dataset. The bar charts represent the mean ± SEM (standard error of the mean) of the encoding accuracies, and * represents p < \({10}^{-12}\) for a one-tailed two-sample t -test.
The degree of involvement of a voxel in the visual task is a determining factor in its predictability. Specifically, if a voxel receives a substantial amount of stimulus information, its fMRI activities will be more predictable, and vice versa. To validate this hypothesis, we visualized the distributions of stimulus intensities and voxel receptive fields. By annotating the receptive field for each voxel through threefold cross-validation on the train set data, the top 100 voxels with the highest \({R}^{2}\) of each participant were selected for analysis. The mean stimulus intensities of the train set and the receptive fields of the selected voxels are shown in Fig. 2b, c . Their spatial distribution patterns, which approximately followed Gaussian distributions along the x -axis and uniform distributions along the y -axis, were found to be quite similar. This suggests that the receptive fields of these informative voxels tended to be located in areas with higher stimulus intensity. This finding provides further evidence of the efficacy of the receptive field-based feature selection algorithm employed in this study.
Encoding performance on handwritten digit dataset
To verify the encoding performance of the proposed approach on handwritten digit stimuli, we trained the SCNN using 2000 prior images that were not utilized in the fMRI experiment. Voxel-wise encoding models were then constructed on the train set of this dataset. Similarly, CNN-based encoding models were built on the handwritten digit dataset, and the top 500 voxels with the highest encoding performance were selected for comparison. The encoding results are presented in Fig. 2d , and the results indicate that the encoding accuracies of SCNN were significantly higher than those of CNN ( \({{{{{\rm{p}}}}}}=6.78\times {10}^{-18}\) , one-tailed two-sample t -test).
Encoding performance on natural image datasets
In comparison to handwritten characters and digit images, natural images are more intricate and closely resemble our everyday visual experiences. To assess the feasibility of the proposed approach for encoding natural image stimuli, we trained and tested the encoding model on grayscale and colorful natural image datasets. The SCNNs utilized for encoding were trained on the train set images of these datasets.
For the grayscale natural image dataset, the utilization of task-optimized CNN-based encoding models is not feasible due to the absence of category labels in the visual stimuli. A comparison was conducted between our approach and the Gabor wavelet pyramid (GWP) model proposed by Kay et al. 1 , as well as the brain-optimized CNN (GNet) 13 , 29 . Instead of classifying the input images, the CNN in GNet was trained to predict the fMRI responses in an end-to-end fashion. The architecture of GNet can be found in Supplementary Table 2 . Independently, we trained the GNet for each visual area in each subject (a total of 6 models were trained). Regions of interest (ROI)-level analysis was performed on this dataset, and for each visual area, 200 voxels with the highest encoding performance (100 for each subject) were selected for comparison. The encoding results are presented in Fig. 2e . It was observed that the encoding accuracies of V3 were lower than those of V1 and V2, which may be attributed to its lower signal-to-noise ratio 1 . Significant differences were observed between the accuracies of SCNN and GWP ( p < \({10}^{-24}\) , one-tailed two-sample t -test) for all visual areas, with no significant difference between SCNN and GNet ( p > 0.12, two-tailed two-sample t -test) for V2 and V3. For the colorful natural image dataset, we compared the encoding performance of SCNN with CNN and GWP and selected 500 voxels with the highest encoding performance for each subject for comparison. As depicted in Fig. 2f , the accuracies of SCNN were significantly higher than those of CNN ( p < \({10}^{-36}\) , one-tailed two-sample t -test) for all subjects. Moreover, SCNN demonstrated comparable results to GNet for subject1 (SCNN higher than GNet, \({{{{{\rm{p}}}}}}=1.58\times {10}^{-19}\) , one-tailed two sample t-test) and subject4 (no significant difference, p = 0.725, two-tailed two-sample t -test).
In general, the encoding results of the natural image datasets suggest that the unsupervised SCNN-based encoding model outperforms traditional GWP and CNN-based models and can even achieve comparable performance with neural networks optimized with brain response as the target.
Image reconstruction result
The image reconstruction task aims to reconstruct the images perceived by the participant from the fMRI responses. Based on the pre-trained encoding models, we accomplished this task on handwritten characters, handwritten digits, and colorful natural image datasets. The prior image set for the handwritten character dataset consisted of the images of six characters in the TICH dataset (excluding the test set images). For the handwritten digit dataset, the prior image set comprised 2000 prior handwritten 6 and 9 images. The images in the validation set of ImageNet were used as the prior image set for the colorful natural image dataset. It is noteworthy that only 200 voxels selected from the train set data were utilized for this task. To reconstruct each image in the test set, the top 15 images of the prior image set with the highest likelihood with observed responses were averaged, resulting in the reconstructed image.
The reconstruction results of the handwritten character dataset demonstrated that our reconstructions can effectively distinguish different characters and can reconstruct images that belong to the same character with different writing styles (see Fig. 3a, b ). Similarly, our approach yielded promising reconstruction results on the handwritten digit dataset (see Fig. 3c ). The reconstruction results of the colorful natural image dataset are presented in Fig. 3d . Although our model can only deal with grayscale images, which resulted in the loss of color information in the reconstruction results, the reconstructions retained the structural information, such as shape and position, of the original stimuli. Additionally, we observed that the prior images with the highest likelihood exhibited high structural similarities to the real stimuli (see Fig. 3e ). The reconstruction results were quantitatively evaluated using PCC and Structural Similarity Index (SSIM) 30 and were compared with other benchmark methods, including CNN, GNet, SMLR 31 , DCCAE 32 , DGMM+ 33 , and Denoiser GAN 34 . As presented in Table 2 , our approach achieved competitive or superior performance compared to these methods.
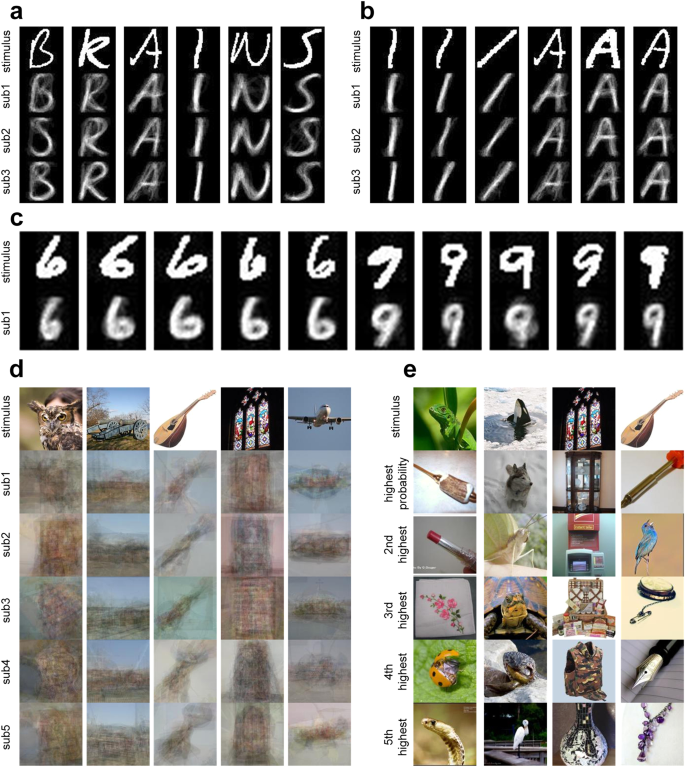
a The reconstructions of different handwritten characters (B, R, A, I, N, and S). The images in the first row are the presented images (ground truth), and the images in the second to fourth rows are the reconstruction results of the 3 subjects. b The reconstructions of the same character with different writing styles. c The reconstructions of handwritten digits. The handwritten digit images are adapted from the MNIST database ( http://yann.lecun.com/exdb/mnist/ ) with permission. d The reconstructions of natural images. e Examples of prior images with the highest likelihoods of the colorful natural image datasets. The colorful natural images in d and e are adapted from the ImageNet database 52 with permission.
Image identification result
The image identification task aims to identify the image seen by the participant from the fMRI responses, and this task was accomplished on the grayscale natural image dataset. The encoding model was utilized to generate predicted fMRI responses for all images in the test set. The images perceived by the participants were identified by matching the measured responses to the predicted responses. As per a previous study 1 , 500 voxels with the highest predictive power were employed for this task. Our approach achieved identification accuracies of 96.67% (116/120) and 90.83% (109/120) for the two participants, respectively, which were higher than those of the GWP model (92% and 72%) and GNet (90% and 73.33%). The correlation maps between measured and predicted responses for the two participants are presented in Fig. 4 . For most of the rows in the correlation maps, the elements on the diagonal were significantly larger than the others, indicating that our approach exhibited excellent identification ability.
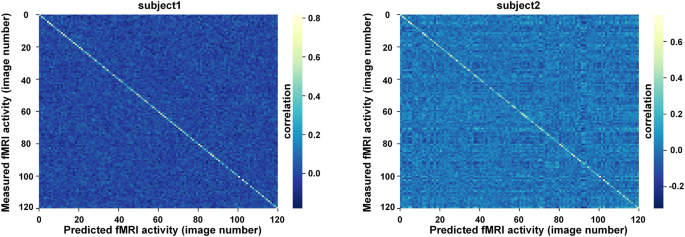
The correlation maps of the measured and predicted fMRI responses to test set images for the two participants. The element in the \({m}_{{th}}\) column and \({n}_{{th}}\) row represents the correlation between the measured fMRI response for the \({m}_{{th}}\) image and the predicted fMRI response for the \({n}_{{th}}\) image.
Effect of hyperparameters on decoding tasks
The selection of hyper-parameters directly affects the performance of downstream decoding tasks. To evaluate the impact of hyper-parameters on the image reconstruction task, we investigated the reconstruction performance with two hyper-parameters: the number of selected voxels and the number of averaged images. Specifically, we examined the reconstruction performance using 50, 100, 200, and 500 voxels and 1, 5, 10, 15, 20, 25, and 30 images on the handwritten character dataset. As illustrated in Fig. 5a , the PCC index increased with the number of images and reached its peak at the voxel number of 200. Conversely, the SSIM index decreased with the increase in the number of images and reached its peak at the voxel number of 200 and 500. A larger number of voxels contained more stimulus information but also introduced more noise. Similarly, a larger number of images made the reconstruction more realistic but also blurred the reconstruction. To evaluate the impact of hyper-parameters on the image identification task, we investigated the identification accuracies with 100, 500, 1000, and 2000 voxels. As depicted in Fig. 5b , our approach achieved the highest accuracies when 500 voxels were utilized.
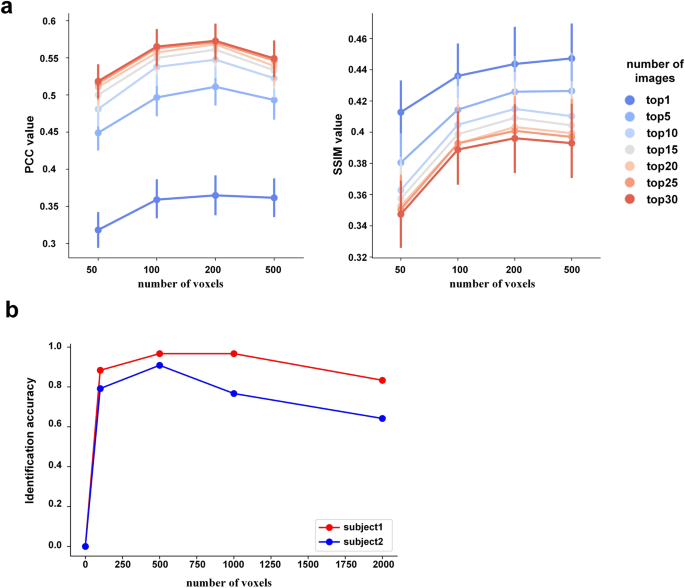
a The reconstruction performance (PCC and SSIM) of different hyper-parameters (number of selected voxels and number of averaged images) on the handwritten character dataset, the dots represent mean values, and the error bars represent 95% confidence intervals. b The identification accuracies with different numbers of voxels for the two subjects in the grayscale natural image dataset.
Reproducibility analysis
In the proposed encoding model, the unsupervised SCNN was utilized to extract features of the visual stimuli, and the training process of SCNN was influenced by its initial values. To investigate the impact of initial values on the encoding performance, we trained another SCNN with different initial values on the grayscale natural image dataset and compared its encoding performance with the original one. For each subject, the top 500 voxels with the highest encoding performance were selected for comparison, and no significant differences were observed between the two encoding results (subject1: p = 0.1, subject2: p = 0.47, two-tailed two-sample t -test).
In this work, a visual perception encoding model based on SCNN was proposed, comprising the SCNN feature extractor and voxel-wise response predictors. Unlike conventional Gabor and CNN-based methods that employ real-value computation, the proposed model utilized spike-driven SCNN to process visual information in a more biologically plausible manner. The model demonstrated remarkable success in predicting brain activity evoked by handwritten characters, handwritten digits, and natural images, using a simple two-layer unsupervised SCNN and four publicly available datasets as the test bed. Moreover, promising results were obtained in image reconstruction and identification tasks using our encoding models, suggesting the potential of the model in addressing practical brain-reading problems.
Neural encoding can bridge artificial intelligence models and the human brain. By establishing a linear mapping from model features to brain activity, the similarity of information processing between the model and the brain can be quantitatively evaluated. Therefore, it is reasonable to assume that a model with high biological plausibility is more likely to achieve superior encoding performance. In light of this, we developed an SCNN-based encoding model to predict brain responses elicited by various visual inputs. The SCNN architecture combines the network structure of CNN, which has been shown to be effective for neural encoding 2 , 4 , 13 , 14 , with the computational rules of SNN that are more biologically realistic. To extract meaningful visual features, we employed an SCNN consisting of a DoG layer and a convolutional layer, which simulate information processing in the retina and visual cortex, respectively. Our model outperformed other benchmark methods (Gabor and CNN-based encoding models), in terms of encoding performance on experimental data, highlighting the superiority of SCNN in visual perception encoding.
Despite its biological plausibility, SCNN simulates information processing at the level of individual neurons, while fMRI measures large-scale brain activity, with each voxel’s signal representing the joint activity of a large number of neurons. Therefore, regression models are crucial for voxel-level encoding, as they map the activations of multiple SCNN neurons to the responses of single voxels. Previous studies have demonstrated the neuronal population receptive field properties 35 , 36 of fMRI data, indicating that each voxel in the visual cortex (especially in V1–3) only receives visual input from a fixed range of the visual field. Based on this theory, we employed a feature selection algorithm that matched the receptive field location for each voxel, which was more consistent with the real visual mechanism and reduced the risk of overfitting.
The question of whether the brain operates under supervised or unsupervised conditions has been a topic of debate. In lieu of utilizing supervised CNNs, we employed an unsupervised SCNN trained via STDP in our model. The findings of this study suggest that the early visual areas of the visual cortex are more inclined to acquire visual representations in an unsupervised manner. Additionally, the STDP-based SCNN offers several advantages in terms of neural encoding. Firstly, it is biologically plausible due to the bioinspired nature of STDP as a learning rule. Secondly, it is capable of handling both labeled and unlabeled data. Lastly, it is particularly well-suited for small sample datasets, such as those obtained via fMRI.
The realization of neural decoding tasks serves as the foundation for numerous brain-reading applications, such as BCI 37 . Two types of decoding models exist: those derived from encoding models and those constructed directly in an end-to-end manner. The former offers voxel-level functional descriptions while completing decoding tasks 5 . However, recent breakthroughs in decoding have primarily been achieved using the latter models 33 , 38 , 39 . In this study, we successfully completed downstream decoding tasks, including image reconstruction and identification, based on the encoding model. The results demonstrate that our approach outperformed other end-to-end models in both decoding tasks. This finding further confirms the effectiveness of our encoding model and suggests that encoding-based approaches hold significant potential for solving decoding tasks.
Despite the progress made in neural encoding using SCNN, there remain several limitations. First, the architectures of SNNs are typically shallower than those of deep-learning networks, which restricts their ability to extract complex and hierarchical visual features. Recent studies have attempted to address this issue and have made some headway 23 , 24 , 40 . The incorporation of a deeper SCNN into our model would further enhance encoding performance and enable investigation of the hierarchical structure of the visual cortex. Second, the Integrate-and-Fire neuron utilized in our study is a simplification of biological neurons. The use of more realistic neurons, such as leaky Integrate-and-Fire and Hodgkin-Huxley neurons 41 , would further enhance the biological plausibility of our encoding model. Third, the parameters of STDP and network architecture were selected from previous works 23 , 24 , and the impact of different parameters on encoding performance requires further exploration.
In conclusion, this work presents a powerful tool for neural encoding. On the one hand, we combined the structure of CNNs and the calculation rules of SNNs to model the visual system and constructed voxel-wise encoding models based on the receptive field mechanism. On the other hand, we demonstrated that our model can be utilized to perform practical decoding tasks, such as image reconstruction and identification. We anticipate that SCNN-based encoding models will provide valuable insights into the visual mechanism and contribute to the resolution of BCI and computer vision tasks. Furthermore, we plan to extend the use of SNNs to encoding tasks of other cognitive functions (e.g., imagination and memory) in the future.
SCNN-based encoding model
An SCNN-based encoding model was proposed in this study to predict fMRI activities that are elicited by input visual stimuli. The encoding model was comprised of voxel-wise regression models and a SCNN feature extractor. Initially, the unsupervised SCNN was utilized to extract the stimulus features for each input image. Subsequently, linear regression models were constructed to project the SCNN features into fMRI responses. The architecture of the encoding model is depicted in Fig. 1a .
SCNN feature extractor
To extract stimuli features, a simple two-layer SCNN was employed in this study. The first layer, known as the Difference of Gaussians (DoG) layer, was designed to emulate neural processing in retinal ganglion cells 42 , 43 . The parameter settings for this layer were based on previous research 23 , 24 . For both handwritten characters and natural images, each input image underwent convolution with six DoG filters with zero padding. ON- and OFF-center DoG filters with sizes of \(3\times 3\) , \(7\times 7\) , and \(13\times 13\) , and standard deviations of \((3/9,\,6/9)\) , \((7/9,\,14/9)\) , and \((13/9,\,26/9)\) were utilized. The padding size was set to 6 for this study. For handwritten digits, each input image underwent convolution with two DoG filters with zero padding. ON- and OFF-center DoG filters with a size of \(7\times 7\) and standard deviations of \((1,\,2)\) were utilized. The padding size was set to 3. Subsequently, DoG features were transformed into spike waves using intensity-to-latency encoding 44 with a length of 30. Specifically, DoG feature values greater than 50 were sorted in descending order and equally distributed into 30 bins to generate the spike waves. Prior to being passed to the next layer, the output spikes underwent max pooling with a window size of \(2\times 2\) and a stride of 2.
The second layer of the SCNN corresponds to the convolutional layer, which was designed to emulate the information integration mechanism of the visual cortex. In this layer, 64 convolutional kernels comprised of Integrate-and-Fire (IF) neurons were utilized to process the input spikes. The window size of the convolutional kernels was 5×5, and the padding size was 2. Each IF neuron gathered input spikes from its receptive field and emitted a spike when its voltage reached the threshold. This can be expressed mathematically as follows:
where \({v}_{i}\left(t\right)\) represents the voltage of the \({i}_{{th}}\) IF neuron at time step t, while \({w}_{{ij}}\) signifies the synaptic weight between the \({i}_{{th}}\) neuron and the \({j}_{{th}}\) input spikes within the neuron’s receptive field. The firing threshold, denoted by \({v}_{{th}}\) , is set at 10. For each image, neurons are permitted to fire a maximum of once. The inhibition mechanism is employed in the convolutional layer, allowing only the neuron with the earliest spike time to fire at each position in the feature maps. Synaptic weights are updated through Spike-Timing-Dependent Plasticity (STDP), which can be expressed as:
where \(\Delta {w}_{{ij}}\) denotes the weight modification, \({a}^{+}\) and \({a}^{-}\) represent the learning rates (set at 0.004 and −0.003, respectively) 23 , and \({t}_{i}\) and \({t}_{j}\) indicate the spike times of the \({i}_{{th}}\) neuron and \({j}_{{th}}\) input spikes, respectively. The learning convergence, as defined by Kheradpisheh et al. 23 , is calculated using the following equation:
where N represents the total number of synaptic weights. The training of the convolutional layer ceases when C is below 0.01. The SCNN implementation is based on the SpykeTorch platform 45 . After training the SCNN, the firing threshold \({v}_{{th}}\) is set to infinity, and the voltage value at the final time step in each neuron is measured as the SCNN feature of the visual stimuli. As the voltages in the convolutional neurons accumulate over time and are never reset when \({v}_{{th}}\) is infinite, the final voltage values (SCNN feature) reflect the SCNN’s activation in response to the visual stimuli.
Responses prediction algorithm
With the obtained SCNN feature \({{{{{\rm{F}}}}}}\in {{{{{{\mathscr{R}}}}}}}^{64\times h\times w}\) , a linear regression model is constructed for each voxel to predict the fMRI response Y. To avoid the overfitting problem, the receptive field mechanism is introduced into the regression models, where each voxel only receives the input at a specific location of the SCNN feature map. To identify the optimal receptive field location for each voxel (different voxels can have the same preferred receptive field), all locations on the SCNN feature maps are examined to fit the regression model, and threefold cross-validation is performed on the training data. The regression model’s expression and objective function are defined as:
where \({y}_{v}\) represents the fMRI response of voxel v , w denotes the weight parameters in the regression model and \({f}_{{ij}}\in {{{{{{\mathscr{R}}}}}}}^{64\times 1}\,(i={{{{\mathrm{1,2}}}}},\ldots ,h,{j}={{{{\mathrm{1,2}}}}},\ldots ,w)\) signifies the feature vector at location \((i,j)\) of the SCNN feature maps. The regression accuracy is quantified using the coefficient of determination ( \({R}^{2}\) ) of the predicted and observed responses, and the feature location with the highest \({R}^{2}\) is chosen as the receptive field location for each voxel. Lastly, the regression model for each voxel is retrained on the entire training data based on the determined receptive field location.
Downstream decoding tasks
Two downstream decoding tasks were performed based on the encoding models, namely image reconstruction and image identification. The objective of the image reconstruction task is to reconstruct the perceived image from the observed fMRI response, while the image identification task aims to determine the image that was viewed. The specific methodologies employed for these tasks are expounded upon as follows.
Image reconstruction
As depicted in Fig. 1b , the image reconstruction task was executed by utilizing an extensive prior image set. Initially, the encoding model was employed to generate the anticipated fMRI responses for all images in the prior image set. Subsequently, the likelihood of the observed fMRI response r given the prior image s was estimated, which can be mathematically represented as a multivariate Gaussian distribution:
Where \(\hat {{{{{\rm{r}}}}}}({{\mbox{s}}})\) represents the predicted fMRI response of \({{{{{\rm{s}}}}}}\) , and Σ signifies the noise covariance matrix for train samples. Finally, the prior images that elicited the highest likelihood of evoking the observed fMRI response were averaged to derive the reconstruction result.
Image identification
Figure 1c illustrates the methodology employed for the image identification task. The test set images were fed into the encoding model to generate the predicted fMRI responses. Subsequently, the Pearson’s correlation coefficients (PCCs) between the predicted fMRI responses and the observed fMRI response were computed. The image that exhibited the highest correlation between its predicted fMRI response and the observed response was deemed to be the image viewed by the subject.
fMRI datasets
To validate the encoding model, four publicly available datasets that have been extensively utilized in prior research 1 , 25 , 26 , 27 , 33 , 38 , 46 were utilized, namely the handwritten character, handwritten digits, grayscale natural image, and colorful natural image datasets. The fundamental characteristics of these datasets are presented in Table 1 , and a brief overview of each dataset is provided below.
Handwritten character dataset
This dataset comprises fMRI data obtained from three participants as they viewed handwritten character images. A total of 360 images depicting 6 characters (B, R, A, I, N, and S) with the size of \(56\times 56\) were presented to each participant, sourced from the TICH character dataset 47 . A white square was added to each image as a fixation point. During the experiment, each image was displayed for 1 s (flashed at 2.5 Hz), followed by a 3-s black background, and 3 T fMRI data were simultaneously collected (TR = 1.74 s, voxel size = \(2\times 2\times 2\,{{{{{{\rm{mm}}}}}}}^{3}\) ). The voxel-level fMRI responses of visual areas V1 and V2 for each visual stimulus were estimated using general linear models 48 . The same train/test set split as the original work 25 was adopted, which comprised 270 and 90 class-balanced examples, respectively.
Handwritten digit dataset
This dataset comprises fMRI data obtained from one participant while viewing handwritten digit images 26 . During the experiment, 100 handwritten 6 and 9 images with the size of \(28\times 28\) were presented to the participant, with each image displayed for 12.5 s and flashed at 6 Hz. The fMRI responses of V1, V2, and V3 were captured using a Siemens 3 T MRI system (TR = 2.5 s, voxel size = \(2\times 2\times 2\,{{{{{{\rm{mm}}}}}}}^{3}\) ). The train and test sets comprised 90 and 10 examples, respectively. Additionally, this dataset provided 2000 prior handwritten 6 and 9 images that were not utilized in the fMRI experiment for the image reconstruction task.
Grayscale natural image dataset
This dataset comprises fMRI data obtained from two participants as they viewed grayscale natural images 1 . The experiment was divided into train and test stages. During the training stage, the participants were presented with 1750 images, each of which was displayed for a duration of 1 s (flashed at 2 Hz), followed by a 3 s gray background. In the test stage, the participants were shown 120 images that were distinct from the ones used in the training stage. The fMRI data was acquired simultaneously in both stages of the experiment using a 3 T scanner (TR = 1 s, voxel size = \(2\times 2\times 2.5\,{{{{{{\rm{mm}}}}}}}^{3}\) ). The voxel-level fMRI responses of visual areas V1–V3 were estimated for each visual stimulus. To mitigate computational complexity, the natural images were downsampled from \(500\times 500\) to \(128\times 128\) pixels.
Colorful natural image dataset
This dataset comprises fMRI data obtained from five participants as they viewed colorful natural images 27 . The experiment consisted of two sessions, namely the training image session and the test image session. During the training image session, each participant was presented with 1200 images from 150 categories, with each image being displayed only once (flashed at 2 Hz for 9 s). In the test image session, each participant was shown 50 images from 50 categories, with each image being presented 35 times. The fMRI responses of multiple visual areas on the ventral visual pathway were collected using a 3 T Siemens scanner (TR = 3 s, voxel size = \(3\times 3\times 3\,{{{{{{\rm{mm}}}}}}}^{3}\) ), and V1, V2, and V3 were selected as regions of interest for this study. Prior to being fed into the SCNN, the natural images were converted from RGB format to grayscale format and downsampled from \(500\times 500\) to \(128\times 128\) pixels.
Noise ceiling estimation
The encoding accuracies of the colorful natural image dataset were compared with noise ceilings, which represent the upper limit of the accuracies in the presence of noise. To calculate the noise ceiling for each voxel, we employed a method that has been commonly used in previous studies 13 , 49 , 50 , 51 . This method assumes that the noise follows a Gaussian distribution with a mean of zero and that the observed fMRI signal is equal to the response plus noise. Initially, we estimated the standard deviation of the noise \({\hat{\sigma }}_{N}\) using the following formula:
Where \({\sigma }_{R}^{2}\) represents the variance of the responses across 35 repeated sessions of each test image. Subsequently, we calculated the variance of the response by subtracting the variance of the noise from the variance of the mean response:
Where \({\mu }_{R}\) represents the mean responses across the repeated sessions of each test image. Finally, we drew samples from the response and noise distributions to obtain their simulations and generated the simulated signal by summing the simulated response and noise. We conducted 1000 simulations and calculated the PCC between the simulated signal and response in each simulation. The mean PCC value was taken as the noise ceiling.
Statistics and reproducibility
In Fig. 2 , we performed a one-tailed two-sample t -test to compare the encoding accuracies of different methods on each dataset, and the sample sizes were described in figure captions. In reproducibility analysis, we conducted a two-tailed two-sample t -test to estimate whether the encoding accuracies ( n = 500) between the SCNNs with different initial values exhibited any significant statistical differences; the corresponding p -values were reported in the “Results” section.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The handwritten character dataset is publicly available at http://sciencesanne.com/research/ , the handwritten digit dataset is publicly available at http://hdl.handle.net/11633/di.dcc.DSC_2018.00112_485 , the grayscale natural image dataset is publicly available at https://crcns.org/datasets/vc/vim-1 , the colorful natural image dataset is publicly available at https://github.com/KamitaniLab/GenericObjectDecoding . The source data underlying Figs. 2 , 4 , and 5 can be found in Supplementary Data 1 , 2, 3 .
Code availability
The code that supports the findings of this study is available from https://github.com/wang1239435478/Neural-encoding-with-unsupervised-spiking-convolutional-spiking-neural-networks .
Kay, K. N., Naselaris, T., Prenger, R. J. & Gallant, J. L. Identifying natural images from human brain activity. Nature 452 , 352–355 (2008).
CAS PubMed PubMed Central Google Scholar
Güçlü, U. & van Gerven, M. A. Deep neural networks reveal a gradient in the complexity of neural representations across the ventral stream. J. Neurosci. 35 , 10005–10014 (2015).
PubMed PubMed Central Google Scholar
Nishimoto, S. et al. Reconstructing visual experiences from brain activity evoked by natural movies. Curr. Biol. 21 , 1641–1646 (2011).
Wen, H. et al. Neural Encoding and Decoding with Deep Learning for Dynamic Natural Vision. Cereb. Cortex 28 , 4136–4160 (2018).
PubMed Google Scholar
Naselaris, T., Kay, K. N., Nishimoto, S. & Gallant, J. L. Encoding and decoding in fMRI. NeuroImage 56 , 400–410 (2011).
Wu, M. C. K., David, S. V. & Gallant, J. L. Complete functional characterization of sensory neurons by system identification. Annu. Rev. Neurosci. 29 , 477–505 (2006).
CAS PubMed Google Scholar
Adelson, E. H. & Bergen, J. R. Spatiotemporal energy models for the perception of motion. J. Opt. Soc. Am. A 2 , 284–299 (1985).
Jones, J. P. & Palmer, L. A. An evaluation of the two-dimensional Gabor filter model of simple receptive fields in cat striate cortex. J. Neurophysiol. 58 , 1233–1258 (1987).
Carandini, M. et al. Do we know what the early visual system does? J. Neurosci. 25 , 10577–10597 (2005).
Khaligh-Razavi, S. M. & Kriegeskorte, N. Deep supervised, but not unsupervised, models may explain IT cortical representation. PLoS Comput. Biol. 10 , e1003915 (2014).
Cichy, R. M., Khosla, A., Pantazis, D., Torralba, A. & Oliva, A. Comparison of deep neural networks to spatio-temporal cortical dynamics of human visual object recognition reveals hierarchical correspondence. Sci. Rep. 6 , 27755 (2016).
Kriegeskorte, N. & Kievit, R. A. Representational geometry: integrating cognition, computation, and the brain. Trends Cogn. Sci. 17 , 401–412 (2013).
Allen, E. J. et al. A massive 7T fMRI dataset to bridge cognitive neuroscience and artificial intelligence. Nat. Neurosci. 25 , 116–126 (2022).
Khosla, M., Ngo, G. H., Jamison, K., Kuceyeski, A. & Sabuncu, M. R. Cortical response to naturalistic stimuli is largely predictable with deep neural networks. Sci. Adv. 7 , eabe7547 (2021).
Xu, Y. & Vaziri-Pashkam, M. Limits to visual representational correspondence between convolutional neural networks and the human brain. Nat. Commun. 12 , 2065 (2021).
Maass, W. Networks of spiking neurons: the third generation of neural network models. Neural Netw. 10 , 1659–1671 (1997).
Google Scholar
Gerstner, W., Kempter, R., van Hemmen, J. L. & Wagner, H. A neuronal learning rule for sub-millisecond temporal coding. Nature 383 , 76–78 (1996).
Bi, G.-Q. & Poo, M.-M. Synaptic modifications in cultured hippocampal neurons: dependence on spike timing, synaptic strength, and postsynaptic cell type. J. Neurosci. 18 , 10464 (1998).
Huang, S. et al. Associative Hebbian synaptic plasticity in primate visual cortex. J. Neurosci. 34 , 7575–7579 (2014).
McMahon, DavidB. T. & Leopold, DavidA. Stimulus timing-dependent plasticity in high-level vision. Curr. Biol. 22 , 332–337 (2012).
Meliza, C. D. & Dan, Y. Receptive-field modification in rat visual cortex induced by paired visual stimulation and single-cell spiking. Neuron 49 , 183–189 (2006).
Diehl, P. & Cook, M. Unsupervised learning of digit recognition using spike-timing-dependent plasticity. Front. Comput. Neurosci. https://doi.org/10.3389/fncom.2015.00099 (2015).
Kheradpisheh, S. R., Ganjtabesh, M., Thorpe, S. J. & Masquelier, T. STDP-based spiking deep convolutional neural networks for object recognition. Neural Netw. 99 , 56–67 (2018).
Mozafari, M., Ganjtabesh, M., Nowzari-Dalini, A., Thorpe, S. J. & Masquelier, T. Bio-inspired digit recognition using reward-modulated spike-timing-dependent plasticity in deep convolutional networks. Pattern Recognit. 94 , 87–95 (2019).
Schoenmakers, S., Barth, M., Heskes, T. & van Gerven, M. Linear reconstruction of perceived images from human brain activity. Neuroimage 83 , 951–961 (2013).
Van Gerven, M. A., De Lange, F. P. & Heskes, T. Neural decoding with hierarchical generative models. Neural Comput. 22 , 3127–3142 (2010).
Horikawa, T. & Kamitani, Y. Generic decoding of seen and imagined objects using hierarchical visual features. Nat. Commun. 8 , 15037 (2017).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. International Conference on Learning Representations . https://doi.org/10.48550/arXiv.1412.6980 (2015).
Seeliger, K. et al. End-to-end neural system identification with neural information flow. PLoS Comput. Biol. 17 , e1008558 (2021).
Zhou, W., Bovik, A. C., Sheikh, H. R. & Simoncelli, E. P. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13 , 600–612 (2004).
Miyawaki, Y. et al. Visual image reconstruction from human brain activity using a combination of multiscale local image decoders. Neuron 60 , 915–929 (2008).
Wang, W., Arora, R., Livescu, K. & Bilmes, J. On deep multi-view representation learning. Proc. 32nd Int. Conf. Mach. Learn. 37 , 1083–1092 (2015).
Du, C., Du, C., Huang, L. & He, H. Reconstructing perceived images from human brain activities with bayesian deep multiview learning. IEEE Trans. Neural Netw. Learn. Syst. 30 , 2310–2323 (2019).
Seeliger, K., Güçlü, U., Ambrogioni, L., Güçlütürk, Y. & van Gerven, M. A. J. Generative adversarial networks for reconstructing natural images from brain activity. NeuroImage 181 , 775–785 (2018).
Victor, J. D., Purpura, K., Katz, E. & Mao, B. Population encoding of spatial frequency, orientation, and color in macaque V1. J. Neurophysiol. 72 , 2151–2166 (1994).
Dumoulin, S. O. & Wandell, B. A. Population receptive field estimates in human visual cortex. NeuroImage 39 , 647–660 (2008).
Gao, X., Wang, Y., Chen, X. & Gao, S. Interface, interaction, and intelligence in generalized brain-computer interfaces. Trends Cogn. Sci. 25 , 671–684 (2021).
Ren, Z. et al. Reconstructing seen image from brain activity by visually-guided cognitive representation and adversarial learning. NeuroImage 228 , 117602 (2021).
Wang, C. et al. Reconstructing rapid natural vision with fMRI-conditional video generative adversarial network. Cerebral Cortex https://doi.org/10.1093/cercor/bhab498 (2022).
Wu, Y., Deng, L., Li, G., Zhu, J. & Shi, L. Spatio-temporal backpropagation for training high-performance spiking neural networks. Front. Neurosci. 12 , 331 (2018).
Izhikevich, E. M. Simple model of spiking neurons. IEEE Trans. Neural Netw. 14 , 1569–1572 (2003).
Enroth-Cugell, C. & Robson, J. G. The contrast sensitivity of retinal ganglion cells of the cat. J. Physiol. 187 , 517–552 (1966).
McMahon, M. J., Packer, O. S. & Dacey, D. M. The classical receptive field surround of primate parasol ganglion cells is mediated primarily by a non-GABAergic pathway. J. Neurosci. 24 , 3736–3745 (2004).
Gautrais, J. & Thorpe, S. Rate coding versus temporal order coding: a theoretical approach. Biosystems 48 , 57–65 (1998).
Mozafari, M., Ganjtabesh, M., Nowzari-Dalini, A. & Masquelier, T. SpykeTorch: efficient simulation of convolutional spiking neural networks with at most one spike per neuron. Front. Neurosci. https://doi.org/10.3389/fnins.2019.00625 (2019).
Du, C., Du, C., Huang, L. & He, H. Conditional generative neural decoding with structured CNN feature prediction. Proc. AAAI Conf. Artif. Intell. 34 , 2629–2636 (2020).
Van der Maaten, L. A new benchmark dataset for handwritten character recognition. Tilburg Univ. 2–5 (2009).
Friston, K. J. et al. Statistical parametric maps in functional imaging: a general linear approach. Hum. Brain Mapp. 2 , 189–210 (1994).
Han, K. et al. Variational autoencoder: an unsupervised model for encoding and decoding fMRI activity in visual cortex. NeuroImage 198 , 125–136 (2019).
Kay, K. N., Winawer, J., Mezer, A. & Wandell, B. A. Compressive spatial summation in human visual cortex. J. Neurophysiol. 110 , 481–494 (2013).
Lage-Castellanos, A., Valente, G., Formisano, E. & De Martino, F. Methods for computing the maximum performance of computational models of fMRI responses. PLoS Comput. Biol. 15 , e1006397 (2019).
Deng, J. et al. Imagenet: a large-scale hierarchical image database. IEEE Conf. Comput. Vis. Pattern Recognit. https://doi.org/10.1109/CVPR.2009.5206848 (2009).
Download references
Acknowledgements
This work was supported by the STI 2030-Major Projects 2022ZD0208900 and the National Natural Science Foundation of China (Nos. 82121003, 62036003, 62276051, and 82072006), Medical-Engineering Cooperation Funds from University of Electronic Science and Technology of China (ZYGX2021YGLH201), Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (No. ZYYCXTD-D-202003).
Author information
Authors and affiliations.
The Center of Psychosomatic Medicine, Sichuan Provincial Center for Mental Health, Sichuan Provincial People’s Hospital, University of Electronic Science and Technology of China, Chengdu, 611731, China
Chong Wang & Huafu Chen
School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu, 610054, China
Chong Wang, Hongmei Yan, Wei Huang, Wei Sheng, Yuting Wang, Yun-Shuang Fan, Tao Liu, Ting Zou, Rong Li & Huafu Chen
MOE Key Lab for Neuroinformation; High-Field Magnetic Resonance Brain Imaging Key Laboratory of Sichuan Province, University of Electronic Science and Technology of China, Chengdu, 610054, China
Chong Wang, Hongmei Yan, Wei Huang, Wei Sheng, Yuting Wang, Yun-Shuang Fan, Rong Li & Huafu Chen
You can also search for this author in PubMed Google Scholar
Contributions
Chong Wang designed the project and wrote the paper; Yuting Wang, Yun-Shuang Fan, and Ting Zou prepared the data; Wei Huang, Wei Sheng, and Tao Liu analyzed data and built models; Hongmei Yan, Rong Li, and Huafu Chen supervised the project and revised the paper.
Corresponding authors
Correspondence to Hongmei Yan , Rong Li or Huafu Chen .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Peer review
Peer review information.
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Joao Valente. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Peer review file, supplementary information, description of additional supplementary files, supplementary data 1, supplementary data 2, supplementary data 3, reporting summary, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Wang, C., Yan, H., Huang, W. et al. Neural encoding with unsupervised spiking convolutional neural network. Commun Biol 6 , 880 (2023). https://doi.org/10.1038/s42003-023-05257-4
Download citation
Received : 06 February 2023
Accepted : 18 August 2023
Published : 28 August 2023
DOI : https://doi.org/10.1038/s42003-023-05257-4
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
VIDEO
COMMENTS
10. Conclusions and Future Perspectives. In this paper, we present a review of the fundamentals of spiking neural networks (SNNs) and provide a survey of the literature on the use of SNNs in computer vision and robotics applications, which demonstrates the great potential of SNNs in the research community.
Biological neural networks continue to inspire breakthroughs in neural network performance. And yet, one key area of neural computation that has been under-appreciated and under-investigated is biologically plausible, energy-efficient spiking neural networks, whose potential is especially attractive for low-power, mobile, or otherwise hardware-constrained settings. We present a literature ...
This paper describes the main research areas in SNNs, including neuronal models, coding methods, network architectures, and learning algorithms, and discusses in detail each perspective's current state of the arts, and reviews future research directions in the field. ... Spiking neural networks (SNNs), known as third-generation neural networks ...
We show that neurons with SFA endow networks of spiking neurons with the capability to learn very fast, even without synaptic plasticity. We focus on two characteristic aspects of the resulting ...
This paper reviews recent developments in the still-off-the-mainstream information and data processing area of spiking neural networks (SNN)—the third generation of artificial neural networks. We provide background information about the functioning of biological neurons, discussing the most important and commonly used mathematical neural models. Most relevant information processing ...
1The final/complete version of this paper has been published in the Neural Networks journal. Please cite as: Tavanaei, A., Ghodrati, M., Kheradpisheh, S. R., Masquelier, T., and Maida, A. ... observation that leads to the realm of spiking neural networks (SNNs). In the brain, the communication be- ... research is based on the fact that coding ...
This paper presents a novel brain-inspired spiking neural network model for the incremental learning of spike sequences. The proposed BI-SNN is a generic architecture that can be applied for the ...
This paper reviews recent developments in the still-off-the-mainstream information and data processing area of spiking neural networks (SNN)—the third generation of artificial neural networks ...
Spiking Neural Networks (SNNs) are distributed trainable systems whose computing elements, or neurons, are characterized by internal analog dynamics and by digital and sparse synaptic communications. The sparsity of the synaptic spiking inputs and the corresponding event-driven nature of neural processing can be leveraged by hardware implementations that have demonstrated significant energy ...
To address the above research questions, we base our methods on the third generation of neural networks, spiking neural networks (SNN), because SNN have the ability to capture dynamic spatio ...
Abstract. Spiking neural networks (SNNs) are a promising energy-efficient alternative to artificial neural networks (ANNs) due to their rich dynamics, capability to process spatiotemporal patterns, and low-power consumption. The complex intrinsic properties of SNNs give rise to a diversity of their learning rules which are essential to ...
Conclusions and Future Perspectives. In this paper, we present a review of the fundamentals of spiking neural networks (SNNs) and provide a survey of the literature on the use of SNNs in computer vision and robotics applications, which demonstrates the great potential of SNNs in the research community.
2 About the papers. ... In the article "Efficient and generalizable cross-patient epileptic seizure detection through a spiking neural network", Zhang et al. propose an EEG-based spiking neural network ... In another sense, research on SNNs can borrow ideas from DNN approaches, but it should avoid closely mimicking the development of DNNs. ...
Spiking neural network (SNN) is a new generation of artificial neural networks (ANNs), which is more analogous with the brain. It has been widely considered with neural computing and brain-like intelligence. SNN is a sparse trigger event-driven model, and it has the characteristics of hardware friendliness and energy saving. SNN is more suitable for hardware implementation and rapid ...
In this context, there is a growing interest in efficient DL algorithms, with Spiking Neural Networks (SNNs) being one of the most promising paradigms. Due to the inherent asynchrony and sparseness of spike trains, these types of networks have the potential to reduce power consumption while maintaining relatively good performance.
Although Spiking neural network has many limitations in accuracy and training difficulty, it has stimulated the research enthusiasm of many researchers. Spiking neural networks has developed rapidly, and many training methods can achieve the same or even higher accuracy than Artificial neural networks.
2 Ericsson Research, Bangalore, India. Abstract. In this paper, we present a case study on modeling and ver-ification of Spiking Neural Networks (SNN) using Satisfiability Mod-ulo Theory (SMT) solvers. SNN are special neural networks that have great similarity in their architecture and operation with the human brain.
Spiking neural networks. The seminal paper from Maass 9 categorizes neural networks into three generations based on their underlying neuronal functionality. The first generation, referred to as ...
Deep neural networks with rate-based neurons have exhibited tremendous progress in the last decade. However, the same level of progress has not been observed in research on spiking neural networks (SNN), despite their capability to handle temporal data, energy-efficiency and low latency. This could be because the benchmarking techniques for SNNs are based on the methods used for evaluating ...
Spiking neural networks play an important role in brain-like neuromorphic computations and in studying working mechanisms of neural circuits. One drawback of training a large scale spiking neural network is that an expensive cost of updating all weights is required. Furthermore, after training, all information related to the computational task is hidden into the weight matrix, prohibiting us ...
The research papers submitted to this topic can be categorized into the following major areas of more efficient neuron modeling; lateral and recurrent spiking neural network connectivity; exploitation of biological neural circuitry by means of spiking neural networks; optimization of spiking neural networks; and spiking neural networks for ...
Spiking Neural Network (SNN), recognized as a type of biologically plausible architecture, has recently drawn much research attention. It transmits information by $0/1$ spikes. This bio-mimetic mechanism of SNN demonstrates extreme energy efficiency since it avoids any multiplications on neuromorphic hardware.
This paper reveals that SNNs, when amalgamated with synaptic delay and temporal coding, are proficient in executing (knowledge) graph reasoning. It is elucidated that spiking time can function as an additional dimension to encode relation properties via a neural-generalized path formulation.
Spiking Neural Networks (SNNs), recognized as the third generation of neural networks, are known for their bio-plausibility and energy efficiency, especially when implemented on neuromorphic hardware. ... by solving the non-linear, non-differentiable aspects of the spiking mechanism. Research by [25, 26] introduced a stochastic neuron model for ...
In this sense, neural assembly computing (NAC) can be seen as a new class of spiking neural network machines. NAC can explain the following points: 1) how neuron groups represent things and states; 2) how they retain binary states in memories that do not require any plasticity mechanism; and 3) how branching, disbanding, and interaction among ...
N2 - Taking inspiration from the brain, spiking neural networks (SNNs) have been proposed to understand and diminish the gap between machine learning and neuromorphic computing. Supervised learning is the most commonly used learning algorithm in traditional ANNs.
In this paper, a spiking convolutional neural network (SCNN)-based encoding framework was proposed to bridge the gap between CNNs and the realistic visual system. The encoding procedure comprised ...
Conversely, Spiking Neural Networks (SNNs), which mimic the neural activity of the brain more closely through impulse-based processing, have not seen widespread adoption. The challenge lies primarily in the complexity of their training methodologies. ... Feature papers represent the most advanced research with significant potential for high ...