An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Int J Environ Res Public Health


Worldwide Research Trends on Medicinal Plants
Esther salmerón-manzano.
1 Faculty of Law, Universidad Internacional de La Rioja (UNIR), 26006 Logroño, Spain; [email protected]
Jose Antonio Garrido-Cardenas
2 Department of Biology and Geology, University of Almeria, ceiA3, 04120 Almeria, Spain; se.lau@anedracj
Francisco Manzano-Agugliaro
3 Department of Engineering, University of Almeria, ceiA3, 04120 Almeria, Spain
The use of medicinal plants has been done since ancient times and may even be considered the origin of modern medicine. Compounds of plant origin have been and still are an important source of compounds for drugs. In this study a bibliometric study of all the works indexed in the Scopus database until 2019 has been carried out, analyzing more than 100,000 publications. On the one hand, the main countries, institutions and authors researching this topic have been identified, as well as their evolution over time. On the other hand, the links between the authors, the countries and the topics under research have been analyzed through the detection of communities. The last two periods, from 2009 to 2014 and from 2015 to 2019, have been examined in terms of research topics. It has been observed that the areas of study or clusters have been reduced, those of the last period being those engaged in unclassified drug, traditional medicine, cancer, in vivo study—antidiabetic activity, and animals—anti-inflammatory activity. In summary, it has been observed that the trend in global research is focused more on the search for new medicines or active compounds rather than on the cultivation or domestication of plant species with this demonstrated potential.
1. Introduction
Ten percent of all vascular plants are used as medicinal plants [ 1 ], and there are estimated to be between 350,000 [ 2 ] and almost half a million [ 3 ] species of them. Since ancient times, plants have been used in medicine and are still used today [ 4 ]. In the beginning, the trial and error method was used to treat illnesses or even simply to feel better, and in this way, to distinguish useful plants with beneficial effects [ 5 ]. The use of these plants has been gradually refined over the generations, and this has become known in many contexts as traditional medicine. The official definition of traditional medicine can be considered as “the sum total of the knowledge, skills and practices based on the theories, beliefs and experiences indigenous to different cultures, whether explicable or not, used in the maintenance of health, as well as in the prevention, diagnosis, improvement or treatment of physical and mental illnesses” [ 6 ].
It is a fact that all civilizations have developed this form of medicine [ 7 ] based on the plants in their own habitat [ 8 ]. There are even authors who claim that this transmitted knowledge is the origin of medicine and pharmacy. Even today, hundreds of higher plants are cultivated worldwide to obtain useful substances in medicine and pharmacy [ 9 ]. The therapeutic properties of plants gave rise to medicinal drugs made from certain plants with these benefits [ 10 ].
Until the 18th century, the therapeutic properties of many plants, their effect on the human organism and their method of treatment were known, but the active compound was unknown [ 11 ]. As an example, the Canon of Medicine written by the Persian physician and scientist Avicenna (Ibn Sina) was used until the 18th century [ 12 ].
The origin of modern science, especially in the Renaissance, in particular chemical analysis, and the associated instrumentation such as the microscope, was what made it possible to isolate the active principles of medical plants [ 13 ]. Since then, these active principles have been obtained synthetically in the laboratory to produce the medicines later [ 14 ]. The use of medicines was gradually expanded. Until today, the direct use of medicinal plants is apparently displaced in modern medicine [ 15 ]. Today’s medicine needs the industry producing pharmaceutical medicines, which are largely based on the active principles of plants, and therefore, these are used as raw materials in many cases [ 16 ]. Yet, today, the underdeveloped world does not have access to this modern medicine of synthetic origin, and therefore, large areas of the world continue to use traditional medicine based on the direct use of medicinal plants due to their low cost [ 17 ].
However, it should be noted that the possible trend to return to this type of traditional medicine may have two major drawbacks. The first is the use of medicinal plants without sanitary control, without thinking about the possible harmful aspects for health [ 18 ]. Although many plants do not have side effects like the aromatic plants used in infusions: chamomile, rosemary, mint, or thyme; however, others may have dangerous active principles. To cite an example, Bitter melon ( Momordica charantia L. ) used to cure fever and in cases of malaria [ 19 ], its green seeds are very toxic as they can cause a sharp drop in blood sugar and induce a patient’s coma (hypoglycemic coma) [ 20 ]; this is due to the fact that the components of bitter melon extract appear to have structural similarities to animal insulin [ 21 ]. Secondly, there has been a proliferation of products giving rise to false perspectives, as they are not sufficiently researched [ 22 ].
Examining the specialized literature of reviews and bibliometric studies on medicinal plants, three types of studies are found: those focused on a geographical area, those focused on a specific plant or family, and those focused on some type of medical interest activity. Regarding the studies of geographical areas, for example, there are the studies of Africa. Specifically, in South Africa, the plants that are marketed [ 23 ], as these plants of medical interest have been promoted [ 24 ], or for the treatment of specific diseases such as Alzheimer’s [ 25 ]. In Central Africa, the studies of Cameroon are remarkable, where for general bibliometric studies of its scientific output, the topic of medicinal plants stands out as one of the most important in this country [ 26 ]. Or those of Ghana, regarding frequent diseases in this country such as malaria, HIV/AIDS, hypertension, tuberculosis, or bleeding disorders [ 27 ]. Other countries that have conducted a bibliometric study of their medicinal plants have been Cuba [ 28 ] and China [ 29 ].
The other direction of the bibliometric studies mentioned, those that focus on specific plants, are those of: Artemisia annua L. [ 30 ], Aloe vera [ 31 ], Panax ginseng [ 32 ], Punica grantum L. [ 33 ], Apocynum cannabinum [ 34 ], or Andrographis paniculata [ 35 ]. The third line of the bibliometric research on medicinal plants deals with some kind of specific activity; there are studies for example for the activities of: antibacterial or antifungal [ 36 ], antioxidant [ 37 ], and anticancer [ 38 , 39 , 40 ].
As a common feature of the bibliometric studies published so far, none of them has a worldwide perspective. Furthermore, they are generally based on Web of Science and some of them on other more specific databases such as CAB Abstracts or PlantMedCUBA, but no work based on Scopus has been observed. Therefore, this paper aims to study what types of scientific advances are being developed around medicinal plants, what research trends are being carried out, and by which countries and research institutions. To this purpose, it is proposed to carry out a bibliometric analysis of all the scientific publications on this topic.
2. Materials and Methods
The data analyzed in this work have been obtained through a query in the Scopus database, which has been successfully used in a large number of bibliometric studies [ 41 ]. Due to the large amount of results, it was necessary to use the Scopus API to download the data, whose methodology has been developed in previous works [ 42 , 43 ]. In this study, the query used was: (TITLE-ABS-KEY(“medic* plant*”)). An outline of the methodology used is shown in Figure 1 . The analysis of the scientific communities, both in terms of keywords and the relationship between authors or between countries was done with the SW VosViewer [ 44 ].

Methodology.
3.1. Global Evolution Trend
From 1960 to 2019, more than 110,000 studies related to medicinal plants have been published. Figure 2 shows the trend in research in this field. Overall, it can be said that there was a continuous increase from 1960 to 2001, with just over 1300 published studies. From here, the trend increases faster until 2011, when it reaches a maximum of just over 6200 publications. After this period, publications stabilize at just over 5000 per year. These three periods identified are highlighted in Figure 2 .

Worldwide temporal evolution of medical plants publications.
3.2. Global Subject Category
If the results are analyzed according to the categories in which they have been published (see Figure 3 ), according to the Scopus database, it can be seen that most of them have been carried out in the Pharmacology, Toxicology and Pharmaceutics category with 27.1 % of the total. Other categories with significant relative relevance have been: Medicine (23.8%), Biochemistry, Genetics and Molecular Biology (16.7%), Agricultural and Biological Sciences (11%), Chemistry (8.7%), Immunology and Microbiology (2.5%), Environmental Science (2.1%), and Chemical Engineering (1.5%). All other categories are below 1%, such as: Nursing, Multidisciplinary, or Engineering.

Medicinal plants publications by scientific categories indexed in Scopus.
3.3. Distribution of Publications by Countries
If the results obtained are analyzed by country, a total of 159 countries have published on this topic. Figure 4 shows the countries that have published on the subject and the intensity with which they published has been shown. It is observed that China and India stand out over the rest of the countries with more than 10,000 publications, perhaps influenced by traditional medicine, although their most cited works are related to antioxidant activity, both for China [ 45 ], and for India [ 46 , 47 ], and in this last country also antidiabetic potential [ 4 ]. The third place is the USA followed by Brazil, both with more than 5000 publications. The most frequently cited publications from these countries focus on antioxidant activity [ 48 ], and antimicrobial activity [ 49 ] for the USA and anti-inflammatory activity for Brazil [ 50 , 51 ].

Worldwide research on medical plants.
As mentioned, the list of countries is very long, but those with more than 2000 publications are included: Japan, South Korea, Germany, Iran, United Kingdom, Pakistan, Italy, and France. If the overall results obtained are analyzed in their evolution by years, for this list of countries with more than 2000 publications, Figure 5 is obtained. From this point onwards, three groups of countries can be identified.

Temporal evolution on medical plants publications for Top 12 countries.
The first group is the leaders of this research, China and India, with between 800 and 1100 publications per year. China led the research from 1996 to 2010, and from this year to 2016, the leader was India, after which it returned to China. The second group of five countries is formed in order in the last year of the study: Iran, Brazil, USA, South Korea and Pakistan. This group of countries has a sustained growth over time, with a rate of publications between 200 and 400 per year. It should be noted that Brazil led the third place for a decade, from 2007 to 2016, since then that position is for Iran. The third group of five countries is made up of: Japan, Germany, United Kingdom, Italy, and France. They are keeping the publications around 100 a year, with an upward trend, but at a very slight rate.
If the analysis of the publications by country is made according to the categories in which they publish, Figure 6 is obtained, which shows the relative effort between the different themes or categories is shown. At first look, it might seem that they have a similar distribution. However, in relative terms the category of Pharmacology, Toxicology and Pharmaceutics is led by Brazil with 35% of its own publications followed by India with 33%. For the Medicine category, in relative terms it is led by China with 29 %, followed by Germany with 27 %. The category of Biochemistry, Genetics and Molecular Biology always takes second or third place for this ranking of countries, standing out especially for Japan and South Korea with 23% and for France with 22%. The fourth category for many countries is Agricultural and Biological Sciences, with Pakistan standing out with 20%, followed by Italy with 16%. The category of Chemistry occupies the fourth category for countries such as Japan with 20% or Iran with 14%. The other categories: Chemical Engineering, Immunology and Microbiology, Environmental Science, Multidisciplinary, or Engineering, are below 5 % in all countries.

Distribution by scientific categories according to countries.
According to these results, it can be seen the relative lack of relevance of the category of Agricultural and Biological Sciences for medicinal plants, compared to the categories of Pharmacology, Toxicology and Pharmaceutics, Medicine, or Biochemistry, Genetics and Molecular Biology.
3.4. Institutions (Affiliations)
So far, the distribution by country has been seen, but the research is done in specific research centers (institution or affiliations as are indexed in Scopus) and therefore, it is important to study them. Table 1 shows the 25 institutions with more than 400 publications, of which 13 are from China (including the first 7), 3 from Brazil, 2 from South Korea, and now with 1: Saudi Arabia, Pakistan, Iran, Mexico, Cameroon, France, and Malaysia.
Top 25 affiliations and main keywords.
If the three main keywords of these affiliations are analyzed, it can be seen that there are no great differences, and in fact, they are often the same: Unclassified Drug, Drug Isolation, Drug Structure, Chemistry, Controlled Study, Isolation And Purification, Chemistry, and Plant Extract. They only call attention to “Drugs, Chinese Herbal” which appears in two affiliations: China Academy of Chinese Medical Sciences, and Beijing University of Chinese Medicine, which of course is a very specific issue in this country.
3.5. Authors
The main authors researching this topic are shown in Table 2 , which are those with more than 100 publications on this topic. It is observed that they are authors with a significantly high h-index. On the other hand, it is noteworthy that the first two are not from China or India, which as we have seen were the most productive countries, and also had the most relevant institutions in this area. The lead author is from South Africa, J. Van Staden, and the second from Bangladesh, M. Rahmatullah. The author with the highest h-index is from Germany, T. Efferth.
Main authors in medicinal plants.
If the network of collaboration between authors with more than 40 documents is established, Figure 7 is obtained. Here, there are 33 clusters, where the most important is the red one with 195 authors, where the central author is Huang, L.Q. The second more abundant cluster is the green one, composed of 69 authors. In this cluster, there is no central author, but instead, a collaboration between prominent authors such as Kim, J.S., Lee, K.R. or Park, J.S. The third cluster, in blue, is composed of 64 authors, led by the authors M.I. Choudhary and M. Ahmad.

A collaborative network of authors with more than 40 publications on medicinal plants.
The fourth cluster, of yellow color is composed of 63 authors, the central authors are Y. Li and H-D. Sun. The fifth cluster, in purple, is also composed of 51 authors, the central author is W. Villegas. It should be noted that this cluster is not linked to the whole network, so they must research very specific topics in their field. The sixth cluster is composed of 48 authors and is cyan colored, the central author is Rahmatullah, M. The cluster of the main author of Table 2 , Van Staden, J., is composed of 23 authors, and would be number 17 in order of importance by number of authors, is light brown, and is located next to that of W. Vilegas but without any apparent connection.
3.6. Keywords
3.6.1. global perspective.
The central aspect of bibliometric studies is to study the keywords in the publications and, through the relationships between them, to establish the clusters or scientific communities in which the different topics associated with a field of study can be grouped together. If keywords are extracted from the total number of publications, an overview can be made of the most used keywords in relation to the subject of medicinal plants (see Figure 8 ). As expected, the search terms are the main ones, but then, there are two indexing terms, Human and Nonhuman, and then Unclassified Drug and Plant Extract.

Cloudword of keywords in medical plants publications.
If the keywords are analyzed by country, and we do not take into account the search terms, the results are obtained in Table 3 , where the four main keywords of the main countries that research this topic are shown. It can be seen that the terms: Unclassified Drug, Plant Extract, and Controlled Study, are the ones that dominate without a doubt.
Main keywords by country.
3.6.2. Keywords Related to Plants
If this keyword analysis is done by parts of the plant (see Table 4 ), which shows which parts of the plant have been most investigated. It should be noted that the number of documents is less than the sum of the individual keywords, since a publication contains more than one keyword. It has been obtained that the parts of the plant most studied in order of importance have been the value expressed in relative terms: Leaf-Leaves (33%), Root-Roots (22%), Seed (12%), Stem (10%), Fruit (10%), Bark (7%), and Flower (6%). The table also shows which plant families have been most used for the study of that part of the plant.
Main keywords related to plant parts and plant families studied.
To give an idea of the most studied plant families, see Table 5 . Although the first two are the same family, it has been left separately to indicate the indexing preferences of the two main affiliations that study them. This is also the situation with Compositae that correspond to the family of Asteraceae. This table lists for each plant family the main institution working on its study. However, it is curious that even if a country is a leader in certain studies related to plant families, most often it is found that the institution leading the issue is not from the country leading the study on that plant family. This helps to establish a certain amount of global leadership on the side of the institutions.
Plant families and Institutions.
3.7. Clusters
The analysis of the clusters formed by the keywords allows the classification of the different groups into which the research trends are grouped. A first analysis has been made with the documents published between 2009 and 2019 and in two periods, from 2009 to 2014 and from 2015 to 2019. Figure 9 shows the clusters obtained for the period 2009 to 2014, showing seven clusters, which can be distinguished by color, and in Table 6 its main keywords have been collected.

Network of keywords in medical plants publications: Clusters between 2009–2014.
Main keywords used by the communities detected in the topic in the period 2009–2014.
The first of these clusters, in red (1-1), is linked to traditional medicine. This is reflected in the main keywords associated with this cluster: phytotherapy, herbaceous agent, traditional medicine, ethnobotany. Within this cluster, the most cited publications are related to the antioxidant function of plants. This includes the prevention of hyperglycemia hypertension [ 52 ], and the prevention of cancer. Of the latter, studies suggest that a reduced risk of cancer is associated with high consumption of vegetables and fruits [ 53 ]. Another topic frequently addressed is the antidiabetic properties, as some plants have hypoglycemic properties [ 34 ]. It should be remembered that diabetes mellitus is one of the common metabolic disorders, acquiring around 2.8% of the world’s population and is expected to double by 2025 [ 54 ].
The second cluster, in green (1-2), appears to be the central cluster, and is related to drugs—chemistry. The main keywords are: drug isolation, drug structure, chemistry, drug determination, and molecular structure. Here, the most cited publications are the search for new drugs [ 55 ] or in natural antimicrobials for food preservation [ 56 ].
The third cluster, in purple (1-3), is focused on in vivo study through studies with laboratory animals, as shown by keywords such as mouse and mice. As it is known that in vivo drug trials are initiated in laboratory animals such as mice, in general studies focused on anti-inflammatory effect [ 57 , 58 ].
The fourth cluster, in yellow (1-4), is engaged in the search for drugs. The main keywords in this regard are unclassified drug and drug screening. Within this cluster, the studies of flavonoids stand out [ 59 ]. Flavonoids have been shown to be antioxidant, free radical scavenger, coronary heart disease prevention, hepatoprotective, anti-inflammatory and anticancer, while some flavonoids show possible antiviral activities [ 60 ].
The fifth cluster, in blue (1-5), is focused on the effectiveness of some drugs, and their experimentation on animals. Some of the most cited publications of this cluster over this period are those focused on genus Scutellaria [ 61 ], Epimedium ( Berberidaceae ) [ 62 ] and Vernonia ( Asteraceae ) [ 63 ].
The sixth cluster, in cyan (1-6), is aimed at the effect of extraction solvent/technique on the antioxidant activity. One of the most cited publications in this regard studies the effects on barks of Azadirachta indica , Acacia nilotica , Eugenia jambolana , Terminalia arjuna , leaves and roots of Moringa oleifera , fruit of Ficus religiosa , and leaves of Aloe barbadensis [ 64 ]. Regarding neuroprotection, some publications are the related to genus Peucedanum [ 65 ] or Bacopa monnieri [ 66 ]. This cluster is among the clusters of traditional medicine (1-1) and drug efficacy (1-5).
Finally, the seventh orange cluster (1-7) is of small relative importance within this cluster analysis and is focused on malaria. As it is known, malaria is one of the most lethal diseases in the world every year [ 67 ]. Malaria causes nearly half a million deaths and was estimated at over 200 million cases, 90 per cent of which occurred in African countries [ 68 ]. Of the Plasmodium species affecting humans, Plasmodium falciparum causes the most deaths, although Plasmodium vivax is the most widely spread except in sub-Saharan Africa [ 69 ]. On the other hand, this cluster cites Plasmodium berghei , which mainly affects mice, and is often used as a model for testing medicines or vaccines [ 70 ].
The second period under study, from 2015 to 2019, is shown in Figure 10 , where five clusters have been identified, Table 7 , as opposed to the previous period which was seven. Now, there is no cluster focusing on malaria. In Figure 10 , the colors of the cluster have been unified with those of Figure 9 , when the clusters have the same topic as in the previous period.

Network of keywords in medical plants publications: Clusters between 2015–2019.
Main keywords used by the communities detected in the topic in the period 2015–2019.
The first cluster in order of importance (2-1), the red one in Figure 10 , can be seen to be that of unclassified drug, which has gone from fourth place (1-4) to first in this last period. In this period, research works include one on the therapeutic potential of spirooxindoles as antiviral agents [ 71 ], or the antimicrobial peptides from plants [ 72 ].
The second cluster of this last period (2-2), the one in green in Figure 10 , is the one assigned to traditional medicine, which has now moved up to second place (1-1) in decreasing order of significance. It seems that this cluster of traditional medicine is now the merging with the drug efficacy cluster of the previous period (1-4). This cluster includes research such as oxidative stress and Parkinson’s disease [ 73 ].
The cluster from the previous period that was devoted to animals-in vivo study (1-3), we assume is now divided into three new clusters. The first of these would be the third cluster (2-3), blue in Figure 10 , which can be considered to be dedicated to cancer. One of the works in this cluster is “Anticancer activity of silver nanoparticles from Panax ginseng fresh leaves in human cancer cells” [ 74 ]. Then, the other two are committed to in vivo studies or with animals. The first one seems to be more engaged in vivo study at antidiabetic activity [ 75 , 76 ], would be the cyan-colored cluster 4 (2-4). The other cluster (2-5) involved in testing anti-inflammatory activity, with plants such as Curcumin [ 77 ], Rosmarinus officinalis [ 78 ], would be the purple cluster in Figure 10 .
3.8. Collaboration Network of Countries
Figure 11 shows the collaborative network between countries doing research on medicinal plants. Table 8 lists the countries of each cluster identified and the main country of each cluster. The countries that are most central to this network of collaboration between countries are India, Iran, Indonesia, and the USA. The largest cluster is led by Brazil, which is also not restricted to its own geographical area as it has strong collaborative links with European countries as well as with neighboring countries such as Argentina. The second cluster led by South Africa also presents the same features as the previous one, some collaborations with nearby countries, Tanzania, Congo, or Sudan, but also with European countries such as France, Belgium, or the Netherlands.

Countries network collaboration.
Countries collaboration in the period 2009–2019.
The third cluster is led by India and has very strong collaboration with Iran, but it could also be considered as the central country in the whole international collaboration network. The cooperation with European countries comprises mainly Eastern countries like Poland, Serbia, or Croatia.
The fourth cluster, led by Germany and Pakistan, includes Middle Eastern countries such as Jordan, Saudi Arabia, and United Arab Emirates, which are quite related to the cluster led by China. The fifth cluster seems to have a geographical consideration within Asia by including countries such as Indonesia, Malaysia, Thailand, and Australia. The sixth cluster includes very technologically advanced countries such as USA, UK, Japan, Canada, or South Korea. The seventh cluster is very small in the number of countries. It is made up of very different countries like some in Africa: Cameroon and Kenya; some of Europe as Denmark, and some from Asia like Nepal. In this sense, most of the research linked to African countries in general and to Cameroon particularly is linked to the most frequent parasitic diseases [ 79 ], such as African trypanosomiasis [ 80 ], diarrhea [ 81 ] or tuberculosis [ 82 ]. Finally, the China cluster is made up of nearby areas of influence such as Taiwan, Singapore, Hong Kong, Macau, or Taiwan.
4. Conclusions
The use of plants as a source of research in the search for active compounds for medicine has been proven to have a significant scientific output. An analysis of the scientific literature indexed in the Scopus database concerning medicinal plants clearly shows that in the last 20 years, progress has been rapid, with a peak in 2010. From this year onwards, publications have stabilized at just over 5000 per year.
The research of products derived from the plants shows great collaboration between the countries of the first world and the countries with a traditional use of these plants from Asia, Africa or Latin America, all this to produce new medicines with scientific tests of safety and effectiveness. Within the analysis of the different clusters of collaboration between countries, there are four from Asia, led by China, India, Indonesia and Pakistan; two from Africa, led by South Africa and Cameroon, and then one from Latin America, led by Brazil and another from North America, led by the USA. It has been proven that there is no cluster of European countries, but that they generally collaborate with countries with which they have a commercial relationship. The research of medicinal plants in Africa is greatly underdeveloped, in contrast with China and India. In fact, there is no African country among the countries that published the most in this field. Among the first 25 institutions there is only one that belongs to the African continent. From this top 25, 13 are from China (including the first 7), 3 from Brazil, 2 from South Korea, and 1 of Saudi Arabia, Pakistan, Iran, Mexico, Cameroon, France, and Malaysia.
The most widely used search terms by the main institutions researching in this field are Unclassified Drug, Plant Extract, and Controlled Study. From the study of the keywords in the period from 2009 to 2014, seven clusters have been found, those dedicated to: Traditional medicine, Drug determination, Animals-in vivo study, Unclassified drug, Drug efficacy, Effect of extraction solvent, and Malaria. Subsequently, from the period 2015 to 2019, the clusters are reduced to five, and those focused on: Unclassified drug, Traditional medicine, Cancer, In vivo study—antidiabetic activity, and Animals—anti-inflammatory activity.
This is proven by the fact that of the total number of publications analyzed, more than 100,000, only 11% are in the Agricultural and Biological Sciences category, while more than 50% are grouped in the Pharmacology, Toxicology and Pharmaceutics category and Medicine. This study highlights the scarce research from the agronomic perspective regarding domestication, production or genetic or biotechnological research on breeding of medicinal plants.
Acknowledgments
The authors would like to thank to the CIAIMBITAL (University of Almeria, CeiA3) for its support.
Author Contributions
E.S.-M., J.A.G.-C. and F.M.-A. conceived the research, designed the search, and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- ADVERTISEMENT FEATURE Advertiser retains sole responsibility for the content of this article
Revitalizing the science of traditional medicinal plants
Produced by

As early as the Qin and Han Dynasty (roughly 221 BCE to 220 CE), Sheng Nong’s Herbal Classic recorded 365 medicines. By the time of the Ming Dynasty (1368–1644), the number of Chinese herbal medicines had grown to close to 2,000. Credit: Marilyna/iStock/Getty Images Plus
Plants can be frustratingly inconsistent. With so much dependent on environmental factors, even clones can produce foliage, roots and fruits of varying quantity and quality. Issues with consistency in plant studies have thwarted attempts to characterize the many botanical extracts used in traditional medicines. But traditional knowledge could be a rich resource for drug discovery, says Timothy Mitchison at Harvard Medical School’s Department of Systems Biology.
For example, pharmaceutical chemist, Youyou Tu, discovered artemisinin, an antimalarial extract from the plant Artemisia after being inspired by an entry in the sixteenth century tome, Compendium of Materia Medica . Used as an ancient remedy for fever, artemisinin was isolated and refined by Tu in the 1970s, and according to the World Health Organization, antimalarials containing artemisinin have saved more than three million lives since 2000. Tu was awarded a Nobel Prize for her work in 2015.
“The long history of human data we have for traditional Chinese medicine could be most valuable thing you can get to help characterize any drug,” says Mitchison. He adds that while traditional Chinese medicine-derived molecules typically exhibit poor pharmacology by the standards expected of a synthetic oral drug, that has implications that are under-explored. He says that short plasma half-lives could suggest these molecules have higher action in the liver or kidney, while low oral bioavailability could be the result of action in the gut, which, he says, might be useful for targeting gut diseases.

1,892: The number of herbs mentioned Compendium of Materia Medica. Credit: Lou-Foto/Alamy Stock Photo
In the case of a plant molecule, colchicine, Mitchison’s long-time study subject, its short half-life corresponds to local action in the liver. “These special features of plant-derived molecules cannot be achieved using standard synthetic drugs, which are systemically adsorbed,” he says. “I would encourage medical researchers to have an open mind regarding different medical traditions.”
In Tu’s lecture after winning the Nobel Prize in Physiology or Medicine, she recalled the difficulties of plant research, ranging from managing extraction and purification technologies, to the variables involved in the study of the six Artemisia species, such as accounting for origin, harvest season, and the use of different plant parts.

7,000: Roughly the number of samples in the traditional Chinese medicine collection at the Royal Botanical Gardens, Kew. Credit: Ileana_bt/Shutterstock
The technical and taxonomic challenges of plant research are a source of fascination for Monique Simmonds, director of the Commercial Innovation Unit at the Royal Botanical Gardens, Kew, in London, one of the world’s largest botanical collections. But increased scrutiny of plant research aimed at pharmaceuticals is crucial, she says.
In 1998, Simmonds helped raise funds to create a 7,000-sample traditional Chinese medicine plant collection at Kew, and she currently leads a 300-strong research team focused on unlocking potential drugs derived from plants.
“Some fellow scientists are rightfully cynical about traditional Chinese medicine − some of the research, unfortunately, hasn’t been done with the level of accuracy that you would need for a medicinal drug,” she explains. “A common mistake would be to study different plant species in the same family, such as mistaking Korean and Chinese ginseng.”

17,810: The number of plant species that have a medicinal use, out of some 30,000 plants for which a use of any kind is documented. Credit: Marilyna/iStock/Getty Images Plus
Improving plant study replication through more controlled global standards is part of Simmonds’ mission as the president of the Good Practice in Traditional Chinese Medicine Research Association. Established in 2012, the association now involves 112 institutions and 24 countries, who work on creating better guidelines.
“For example, we would recommend consultation with taxonomists to help independently verify the plants or plant parts being used in research,” says Simmonds. “While taxonomy has been the backbone of Kew’s scientific research, in the next 10 years accelerating taxonomy with machine learning and trait research − from genomic and chemical to morphological and ecological − will also be vital.”
Speeding up drug discovery
At Kew, drug discovery is also being accelerated by machine learning and high-throughput mass spectrometry that reveals the chemical structures of plant compounds. Kew’s Small Molecule Analysis Laboratory, for example, profiles small molecules produced by plants and fungi to help identify chemical structures that might be useful for drug development.
Kaixian Chen, a professor at the Shanghai University of Traditional Chinese Medicine (SUTCM), points out that these types of resources have radically sped up the shortlisting process for drug candidate study.
Chen was an early user of computer-aided drug design in the 1990s. “One of the biggest technological leaps during my career has been in virtual screening: we pair our small molecule libraries of traditional Chinese medicine bioactive components with protein structures that are most likely to bind to specific drug targets in our database, saving us a lot of research time and money,” he explains.
In 2021, for example, using high-throughput screening of natural product libraries, Chen’s colleagues at SUTCM discovered an agonist to bile acid receptor TGR5 that is a potential target for drugs to treat obesity. The agonist, notoginsenoside Ft1, is derived from Panax notoginseng , a ginseng species used for 2,000 years in traditional Chinese medicine to enhance circulation.
A small, early mouse model study has validated notoginsenoside Ft1’s potential in treating obesity. “But if we are to continue to make the most of accelerating drug screening technologies, we must ensure scientific rigour in traditional Chinese medicine studies,” Chen says.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
EDITORIAL article
Editorial: current trends in medicinal plant research and neurodegenerative disorders.

- 1 Department of Pharmacy, Faculty of Biological Sciences, University of Malakand, Chakdara, Pakistan
- 2 Calgary Prion Research Unit, Department of Comparative Biology and Experimental Medicine, Faculty of Veterinary Medicine, University of Calgary, Calgary, AB, Canada
- 3 Hotchkiss Brain Institute, Cumming School of Medicine, University of Calgary, Calgary, AB, Canada
- 4 Center of Excellence in Genomic Medicine Research, King Abdulaziz University, Jeddah, Saudi Arabia
- 5 Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
Editorial on the Research Topic Current Trends in Medicinal Plant Research and Neurodegenerative Disorders
In natural product research, the ethnopharmacological approach is unique because it requires input from the cultural and social sciences. For the first time in 1967, the term “ethnopharmacology” was used as a book title “ Ethnopharmacological Search for Psychoactive Drugs” ( Efron et al., 1967 ). Ethnopharmacology is the scientific exploration of biologically active agents which are traditionally used or observed by man ( Bruhn and Helmstedt, 1981 ). In many parts of the word, medicinal plants are considered as part of the traditional knowledge of a culture due to their significance in indigenous medical systems ( Ayaz et al., 2019b ). Thus, those studies which focus on the documentation of traditional uses of plants have ethnopharmacological relevance. The uses of medicinal plants have been described by many explorers, merchants, missionaries, and respective knowledgeable experts of healing and traditions which serve as a basis for ethnopharmacology-based drug development. Such knowledge has been widely used as a starting point for the development of drug ( Heinrich, 2007 ).
The medicinal plants used by common people act as a significant part of all medical systems occurring in the world ( Ayaz et al., 2017b ; Ayaz et al., 2019c ). It has been reported that in 17th century, an English housewife used Digitalis purpurea L. [Plantaginaceae] (foxglove) for the treatment of dropsy. After that, it was used by a physician WilliamWithering more systematically and he transformed this knowledge into medicine form that could be used by medical doctors ( Griggs, 1981 ; Heinrich, 2010 ). Some of the ethno-pharmacologically driven natural products, identified during 19th century include morphine, emetine, strychnine, quinine, caffeine, coniine, atropine and capsaicin ( Heinrich, 2010 ). Natural products are one of the most important sources of new drug leads. In past, crude materials isolated from various plants or their extracts were used as medicines for medical treatment and then after the second half of the 19th century due to rapid expansion of pharmaceutical industries the researchers started to develop and characterize various drugs from plant origin ( Ovais et al., 2021 ; Heinrich, 2010 ). Chin et al., reported that among the marketed launched products, more than half of all new chemical entities are natural products or their derivatives ( Sneader, 2005 ).
Since ancient times, natural products (NP) have been used as medicines to cure various illnesses ( Ayaz et al., 2017a ; Ayaz et al., 2020 ). As a source of therapeutic molecules, NP have historically proven their value and still act as an important pool for the recognition of novel drug leads ( Atanasov et al., 2015 ). Galanthamine is a natural product obtained from several members of amaryllidaceae family and is commonly used for the treatment of Alzheimer’s disease (AD). As per the ethnobotanical information, the development of galanthamine as anti-Alzheimer’s drug consists of three main periods, including early development (for the treatment of poliomyelitis), preclinical development (as anti-Alzheimer’s drug in 1980s) and clinical development in 1990s ( Heinrich and Teoh, 2004 ). In 1951, the acetylcholine esterase (AChE) inhibiting properties of galanthamine obtained from Galanthus woronowii Losinsk. [Amaryllidaceae] was proved by M. D. Mashkovsky and R. P. Kruglikojva-Lvov using ex vivo system of rat smooth muscle ( Heinrich, 2010 ). Another example is the leaves extract of Ginkgo biloba L. [Ginkgoaceae], which is not considered to be a medicine in many countries but in other countries it is used to prevent dementia, memory deterioration and to enhances cognitive processes ( Heinrich, 2010 ). Flavonoid glycosides were identified as active constituents in the leaf extracts of G. biloba L. in the mid of 1960 during initial research. The first patent on the complete extraction and standardization was filed in 1971 (in Germany) and 1972 (in France) ( DeFeudis and Drieu, 2000 ). This example highlights the development of a standardized extract on the basis of traditional knowledge into an over-the-counter herbal medicine. In later years, many similar novel phytomedicines were development including Hypericum perforatum L. [Hypericaceae] (used for mild to moderate depression), Harpagophytumprocumbens (Burch.) DC. ex Meisn. [Pedaliaceae] (used for chronic pain), and Piper methysticum G. Forst. [Piperaceae] (used for relieving anxiety) ( Collocott, 1927 ). Drug development for neurological disorders on the basis of ethnopharmacology persists to an exciting opportunity. According to the information available in the libraries of Swiss university, more than 150 plant species in different preparations have the potential for research and development (R&D) to develop new drugs against cognitive disorders ( Adams et al., 2007 ).
Alzheimer’s disease (AD) is a multifactorial and progressive neurodegenerative disease. AD is the major cause of dementia and clinically characterized by loss of cognition and memory functions. Currently, there are more than 50 million AD patients affected across the globe and this number is anticipated to double every 5 years and will increase to higher than 150 million by 2050. Besides the health problem for patients and their families, AD also represents a socioeconomic burden, with estimated global costs of US$1 trillion annually, which will be doubled by 2030 ( Khalil et al., 2018 ; Saleem et al., 2021 ). Neuropathologically, AD is characterized by accumulation of plaques composed of aggregated amyloid-β (Aβ) and intraneuronal neurofibrillary tangles (NFTs) of hyperphosphorylated tau proteins. In early onset familial AD, Aβ generates from the proteolytic cleavage of amyloid precursor protein (APP), by the proteolytic and enzymatic action of β- and γ-secretases, a mechanism called amyloidogenic pathway. The Aβ aggregation and deficits in Aβ clearance led to the most neurotoxic AβO species. The hyperphosphorylation of tau proteins are also associated with amyloidogenic pathway. The hyperphosphorylated tau proteins aggregate intraneuronal and forming NFTs. According to amyloidogenic pathway the elevation of AβO induces hyperphosphorylation of tau proteins, resulting intraneuronal NFTs, resulting to synaptic and neuronal degeneration and subsequently cell death ( Kunkle et al., 2019 ; Mahnashi et al., 2021 ). However, more than 95% of AD cases are sporadic with late onset and very heterogeneous neuropathology. Currently, there is no cure for AD. Hence, a better understanding of the contributing factors leading to neuropathology is essential to explore the underlying causes and mediating factors to cure AD.
The purpose of this editorial is to shed light on the recent development of compounds that could prevent or treat AD. The exact underlying cause of pathological changes in AD is still unknown. However, the therapeutic strategies were applied by targeting several pathological mechanisms including protein misfolding such as aggregation of Aβ and tau proteins, pro-inflammatory mediators (IL-1β, TNF-α, TLRs, NF-kβ) and neuroinflammation, oxidative damage and accumulated reactive oxygen species (ROS) as well as its associated pathways such as heme oxygenase-1 and nuclear factor-erythroid factor 2-related factor 2 (HO-1/Nrf2), aberrant cellular and energy homeostasis signaling (e.g., AMPK, SIRT1, mTOR etc) and signalling related with elevated phosphases and kinases, including MAPK/ERK, JNK, PI3K/Akt/GSK3β, as well as synaptic trafficking and its associated pathologies ( Majd and Power, 2018 ; Yu et al., 2021 ).
Aging is a process that is the reason of many diseases such as cancer, heart diseases, diabetes, and many neurological disorders such as Huntington’s disease (HD), Alzheimer’s disease (AD), and Parkinson’s disease (PD) ( Tong et al., 2020 ). It has been reported in many studies that increased level of Reactive oxygen species (ROS) is reason of many neurodegenerative disease in different age-linked disorders such as diabetes, AD, and PD ( Ovais et al., 2018 ; Saleem et al., 2021 ; Mahnashi et al., 2022 ). The increased ROS activate the destruction of the macromolecules such as lipids, proteins and DNA that is directly involved in the neurodegeneration through the disturbance of physiological activities of the brain ( Ayaz et al., 2019a ). The Research Topic, fifteen papers related to different aspects of neuroprotective drugs from natural sources were published. In the first study, Ahmad et al. reported that D-galactose (D-gal) effects neurological damage by inducing ROS signaling pathway while, Fisetin (natural flavonoid) play a protective potentials role against D-galactose-induced stress, neuroinflammation, and memory loss through adaptable anti-oxidant mechanisms, such as Sirt1/Nrf2 signaling, suppression of activated p-JNK/NF-kB signaling pathway and further downstream targets leading to inflammatory cytokines. Similarly, in another study showed neuroprotective effect of medicinal herb known as Bacopa monnieri (L.) Wettst. [Plantaginaceae], that is used as a brain tonic showed its neuroprotective effect PD when the compound extracted from Wettst extract (BME) in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced mice model. Further, more the BME exerts is effective and showed it neurorescue and neuroprotective and effects against MPTP-induced neurodegeneration of the nigrostriatal dopaminergic neurons. Further, it was also studied that BME help in slow down the disease progression and delay the process of neuronal damage in PD ( Singh et al. ). Bacopa monnieri(L.) Wettst. [Plantaginaceae] (BM) extract and the compounds isolated from it mainly used in many disease animal models. Previous studies revealed that Bacoside A may decrease the level of oxidative stress in the CNS by increasing the activities of superoxide dismutase (SOD), glutathione peroxidase (GPx), glutathione reductase (GSR) and catalase level in brain ( Comens, 1983 ). Furthermore, BM extract was also studies in a Caenorhabditis elegans model of 6-hydroxydopamine (6-OHDA)-induced Parkinson’s disease (PD), and results showed that it may decrease the aggregation of α-synuclein by increasing the expression level of hsp-70 protein ( Chowdhuri et al., 2002 ; Jadiya et al., 2011 ). Yet in another study, Pushparaj et al. , evaluated an innovative tool (Next generation Knowledge discovery NGKD) to evaluate the AD-associated gene expression implicated in abnormal signaling pathways.
Rasool et al. have studied the role of antioxidant s in Schizophrenic patients. The study was carried out on 288 Schizophrenic patients of both sexes and various ages. The study reveals that there is an alteration of liver function, increase of stress marker and decrease in the level of antioxidant in the patients. It was also concluded in the study that in patients with thyroid disorder, the deficiencies of certain vitamin (B6, B9 and B12) can lead to hyperhomocysteinemia which ultimately results in the decline of antioxidants and cause oxidative disorders. Panax ginsengC.A.Mey. [Araliaceae] is a perennial plant which has wide variety of useful applications. The major components of ginseng are ginsenosides and gintonin. Li et al. has compiled a literature review on the anti-Alzheimer effect of ginseng. Their literature conclusion reveals that ginseng has therapeutic effect in neurological disorders like Alzheimer. It was further summarized that it exerts the neuroprotective effect by targeting neuro-inflammation, amyloid plaques, mitochondria and function as an antioxidant. Though there is no clinically effective drug for the management of AD. However, the summary related to the clinical findings of ginseng in the management of AD have also been compiled.
Modern society is highly advanced and has many stressful stimuli in life and these event leads to depression ( Post, 1992 ). Mood disorders due to the stressful life are become a serious problem for health that need serious attention ( Gooren and Giltay, 2014 ). Recently, studies in male animals model with chronic stress showed nonorganic erectile dysfunction, testicular injury, less sexual motivation was reported ( Chen et al., 2019 ). In china, for the control of emotion and to decrease sexual dysfunction a drug name as Bupleurum falcatum L. [Apiaceae] had been widely used. Its main active component is saikosaponin D (SSD) act as antidepressant. One of the study in this Research Topic investigated that SSD exposure help to restore sexual functions after chronically stressed mice and the brain mechanisms involved in these effects ( Wang et al. ). Salidroside (SLDS), a phenolic glycoside compound extracted from Rhodiola rosea L. [Crassulaceae] an old medicinal plant from China has been extensively used for the treatment of multiple inflammatory diseases. Yet in another study, SLDS was showed to exhibit protective against depressive behaviors via microglia activation ( Fan et al. ). The study revealed that SLDS exposure significantly declined microglial immuno-reactivity for both CD68 and Iba-1. Moreover, SLDS reserved microglial activation connecting the suppression of P38 MAPK, ERK1/2, and p65 NF-κB activation and thus decreased the expression level and release of neuroinflammatory cytokines in stress mice as well as in lipopolysaccharide (LPS)-induced primary microglia ( Fan et al. ). Further, it was also observed that SLDS changed morphology of microglial cells by reducing the phagocytic and the decreasing the ability of attachment in LPS-induced primary microglia. The results of the study showed that SLDS exposure may improve the depressive symptoms caused by chronic stress due to the unpredictable conditions and also having the potential therapeutic application of SLDS for the treatment of depression by controlling the microglia related neuroinflammation ( Fan et al. ). The Catha edulis (Vahl) Endl. [Celastraceae] (Khat) is most commonly known as a stimulant. The major constituents of Khat are cathinone and cathine. Abou-Elhamd et al. have evaluated the role of Khat extract in molecular signaling using SKOV3 cells. Their observations were that the extract have significant effect on molecular level using SKOV3 cells, and thus, can cause wide variety of neurological disorders. So, in countries where Khat leaves are chewed to induce excitement and euphorbia will have severe effects on the health. Lai et al. studied effect of carnosic acid on the levodopa (L-dopa)-induced dyskinesia (LID) in rats treated with 6-hydroxydopamine (6-OHDA). They proved that by regulating the D1R signaling, CA improves the development of LID in 6-OHDA-treated rats. This leads to prevention of L-dopa-induced apoptotic cell death through modulating the ERK1/2-c-Jun and inducing the parkin. This indicates beneficial role of CA in delaying development of LID in PD patients.
Wide variety of medicinal plants with its ethnomedicinal background are a big source of drug discovery. The Centella asiatica (L.) Urb. [Apiaceae] have been explored to have neuroprotective and anti-inflammatory properties. The plant exert its effect by protecting the mitochondria and have antioxidant properties ( Wong et al. ). Lee et al. tested herbal extract from Glycyrrhiza uralensis Fisch. ex DC. [Fabaceae], Atractylodes macrocephala Koidz. [Asteraceae], Panax ginseng C.A.Mey. [Araliaceae], Astragalus mongholicus Bunge [Fabaceae] to study the anti-inflammatory in the Muscle and Spinal Cord of an Amyotrophic Lateral Sclerosis Animal Model. They performed behavioral tests, including rotarod test and foot printing, immunohistochemistry, and Western blotting, in hSOD1 G93A mice. Their experiments resulted in improved motor activity and reduced motor neuron loss in hSOD1 G93A mice. They also found that the herbal extract reduced levels of oxidative stress-related proteins (HO1, NQO1, Bax, and ferritin) and inflammatory proteins ((GFAP, CD11b, and TNF-α)) in the skeletal muscles and spinal cord of hSOD1 G93A mice.
Cerebral amyloid angiopathy (CAA) is considered by the accretion of β-amyloid (Aβ) in the walls of cerebral vessels, further causing the complications such as convexity subarachnoid hemorrhage, intracerebral hemorrhage as well as cerebral microinfarcts ( Love et al., 2014 ). Dementia and strokes may develop in the patients with CAA-related intracerebral hemorrhage. Many experimental studies explained and demonstrated the pathology of more than 90% of AD patients have associated with CAA and leading to common pathogenic mechanisms. Possible causes of CAA include impaired Aβ removal from the brain through the system called as intramural periarterial drainage (IPAD) ( Saito et al., 2019 ). Moreover, CAA causes control of IPAD causing the limiting clearance. Early interference in CAA may help in the prevention of AD. In another paper published in this Research Topic, Saito et al. , summarized that Taxifolin (dihydroquercetin) is a plant flavonoid is a safe and effective therapy for CAA. Taxifolin is a flavonoid extracted from plant is widely existing in the supplement product, which has been used to exhibit against anti-inflammatory effects, anti-oxidative effect and used as protective agents against the advanced glycation end products as well as mitochondrial damage. Further the flavonoid also showed that it help to facilitate disassembly and prevent oligomer formation and increase clearance of Aβ in CAA of mouse model. Taxifolin treatment also prevent the spatial reference memory impairment and cerebrovascular reactivity in CAA animal model. Further studied required to prove and explain the exact mechanism of Taxifolin that will help to use this drug with effectiveness and safe for the patients with CAA Saito et al. Corona virus disease (COVID-19) is a pandemic of the current era. The COVID-19 has the symptoms from simple common cold to more complex and even leading to the neuro-COVID complications. Pushparaj et al. has worked on the gene sequencing targeting the neuro-COVID. They were able to embark RNA sequencing and find out that some small organic molecules from natural or synthetic source can be useful in the treatment of neurological disorders related to COVID-19. Neuroprotective and anti-inflammatory effect of Pterostilbene was tested against Cerebral Ischemia/Reperfusion injury via suppression of COX-2 in middle cerebral artery occlusion (MCAO) rodent model by Yan et al. Treatment of Pterostilbene significantly reduced neurological score, infarct volume and brain edema. Hepatic parameters (ALT, AST and ALP), renal parameters (uric acid, creatinine, BUN and urea), lipid parameters (TG, HDL, LDL, TC and VLDL), antioxidant parameters (SOD, CAT, GSH, GPx, MDA), inflammatory cytokines (TNF-α, IL-1β, IL-6, and IL-10), inflammatory mediators (COX-2, PGE 2 , iNOS) AND metalloproteinases (MMP) (MMP-2, and MMP-9) levels were improved. Results of these studies show that Pterostilbene is effective in the treatment of cerebral ischemic stroke and cerebral ischemia reperfusion.
Cerebral hypoperfusion (CH) causes neurological diseases like Alzheimer’s-type dementia and vascular cognitive impairment and dementia. To find plant-based treatment for this problem, Liu et al. carried out experiments to unearth potential of Cucurbitacin E (steroidal tetracyclic terpene) in a rat model of CH. Treatment of the rats with Cucurbitacin E (CuE) for 28 days resulted in reduced CH-Induced neurological, sensorimotor and memory deficits, low lipid peroxidation (TBARS content) and protein carbonyls, increased GSH and catalase and diminished inflammatory cytokines (TNF-α, NF-κB, MPO, MMP-9, and iNOS). LDH, caspase-3, glutamate and acetylcholinesterase activities were decreased in Cu-E treated rats subjected to CH. Viable neuron density in the cortex was increased after treatment with CuE. These findings suggest that CuE is a potential compound against CH-associated disorders.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams, M., Gmünder, F., and Hamburger, M. (2007). Plants Traditionally Used in Age Related Brain Disorders-Aa Survey of Ethnobotanical Literature. J. Ethnopharmacol. 113 (3), 363–381. doi:10.1016/j.jep.2007.07.016
PubMed Abstract | CrossRef Full Text | Google Scholar
Atanasov, A. G., Waltenberger, B., Pferschy-Wenzig, E. M., Linder, T., Wawrosch, C., Uhrin, P., et al. (2015). Discovery and Resupply of Pharmacologically Active Plant-Derived Natural Products: A Review. Biotechnol. Adv. 33 (8), 1582–1614. doi:10.1016/j.biotechadv.2015.08.001
Ayaz, M., Ahmad, I., Sadiq, A., Ullah, F., Ovais, M., Khalil, A. T., et al. (2020). Persicaria Hydropiper (L.) Delarbre: A Review on Traditional Uses, Bioactive Chemical Constituents and Pharmacological and Toxicological Activities. J. Ethnopharmacol. 251, 112516. doi:10.1016/j.jep.2019.112516
Ayaz, M., Junaid, M., Ullah, F., Subhan, F., Sadiq, A., Ali, G., et al. (2017a). Anti-Alzheimer's Studies on β-Sitosterol Isolated from Polygonum Hydropiper L. Front. Pharmacol. 8, 697. doi:10.3389/fphar.2017.00697
Ayaz, M., Sadiq, A., Junaid, M., Ullah, F., Ovais, M., Ullah, I., et al. (2019a). Flavonoids as Prospective Neuroprotectants and Their Therapeutic Propensity in Aging Associated Neurological Disorders. Front. Aging Neurosci. 11, 155. doi:10.3389/fnagi.2019.00155
Ayaz, M., Sadiq, A., Junaid, M., Ullah, F., Subhan, F., and Ahmed, J. (2017b). Neuroprotective and Anti-aging Potentials of Essential Oils from Aromatic and Medicinal Plants. Front. Aging Neurosci. 9, 168. doi:10.3389/fnagi.2017.00168
Ayaz, M., Ullah, F., Sadiq, A., Kim, M. O., and Ali, T. (2019b). Editorial: Natural Products-Based Drugs: Potential Therapeutics against Alzheimer's Disease and Other Neurological Disorders. Front. Pharmacol. 10, 1417. doi:10.3389/fphar.2019.01417
Ayaz, M., Ullah, F., Sadiq, A., Ullah, F., Ovais, M., Ahmed, J., et al. (2019c). Synergistic Interactions of Phytochemicals with Antimicrobial Agents: Potential Strategy to Counteract Drug Resistance. Chemico-biological Interact. 308, 294–303. doi:10.1016/j.cbi.2019.05.050
Bruhn, J. G., and Helmstedt, B. (1981). Ethnopharmacology: Objectives, Principles and Perspectives. Nat. Prod. as Med. agents , 405–430.
Google Scholar
Chen, G., Chen, J., Yang, B., Yu, W., Chen, Y., and Dai, Y. (2019). Dopamine D2 Receptors in the Basolateral Amygdala Modulate Erectile Function in a Rat Model of Nonorganic Erectile Dysfunction. Andrologia 51 (1), e13160. doi:10.1111/and.13160
Chowdhuri, D. K., Parmar, D., Kakkar, P., Shukla, R., Seth, P. K., and Srimal, R. C. (2002). Antistress Effects of Bacosides of Bacopa Monnieri: Modulation of Hsp70 Expression, Superoxide Dismutase and Cytochrome P450 Activity in Rat Brain. Phytother. Res. 16 (7), 639–645. doi:10.1002/ptr.1023
Collocott, E. E. V. (1927). Kava Ceremonial in Tonga. J. Polyn. Soc. 36 (141), 2
Comens, C. (1983). Fixed Drug Eruption. Australas. J. Dermatol 24 (1), 1–8. doi:10.1111/j.1440-0960.1983.tb00240.x
DeFeudis, F. V., and Drieu, K. (2000). Ginkgo Biloba Extract (EGb 761) and CNS Functions: Basic Studies and Clinical Applications. Curr. Drug Targets 1 (1), 25–58. doi:10.2174/1389450003349380
Efron, D. H., Holmstedt, B., and Kline, N. S. (1967). “Ethnopharmacologic Search for Psychoactive Drugs,” in Proceedings of a symposium , San Francisco, CA , January, 1967 . National Institute of Mental Health, Editor C. Chase (Washington, DC: American Association for the Advancement of Science ), 468.
PubMed Abstract | Google Scholar
Gooren, L. J., and Giltay, E. J. (2014). Men and Women, So Different, So Similar: Observations from Cross-Sex Hormone Treatment of Transsexual Subjects. Andrologia 46 (5), 570–575. doi:10.1111/and.12111
Griggs, B. (1981). Green Pharmacy: A History of Herbal Medicine . London: Viking Press .
Heinrich, M., and Lee Teoh, H. (2004). Galanthamine from Snowdrop-Tthe Development of a Modern Drug against Alzheimer's Disease from Local Caucasian Knowledge. J. Ethnopharmacol. 92 (2-3), 147–162. doi:10.1016/j.jep.2004.02.012
Heinrich, M. (2010). Ethnopharmacology and Drug Discovery.” in Comprehensive Natural Products II . Oxford: Elsevier , 351–381. doi:10.1016/b978-008045382-8.00666-3
CrossRef Full Text | Google Scholar
Heinrich, M. (2007). W. Sneader, Drug Discovery: A History, John Wiley and Sons Ltd., Chichester (2005) 468 pp.; Numerous Figures (Mostly Chemical Line Drawings), Foreword by Arthur Hollman, ISBN 13: 978-0-471-89979-2 (HB), £ 85/978-0-471-89980-8 (PB), £ 34.95. J. Ethnopharmacol. 112, 596–597. doi:10.1016/j.jep.2007.04.017
Jadiya, P., Khan, A., Sammi, S. R., Kaur, S., Mir, S. S., and Nazir, A. (2011). Anti-Parkinsonian Effects of Bacopa Monnieri: Insights from Transgenic and Pharmacological Caenorhabditis elegans Models of Parkinson's Disease. Biochem. Biophys. Res. Commun. 413 (4), 605–610. doi:10.1016/j.bbrc.2011.09.010
Khalil, A. T., Ayaz, M., Ovais, M., Wadood, A., Ali, M., Shinwari, Z. K., et al. (2018). In Vitro cholinesterase Enzymes Inhibitory Potential and In Silico Molecular Docking Studies of Biogenic Metal Oxides Nanoparticles. Inorg. Nano-Metal Chem. 48 (9), 441–448. doi:10.1080/24701556.2019.1569686
Kunkle, B. W., Grenier-Boley, B., Sims, R., Bis, J. C., Damotte, V., Naj, A. C., et al. (2019). Genetic Meta-Analysis of Diagnosed Alzheimer's Disease Identifies New Risk Loci and Implicates Aβ, Tau, Immunity and Lipid Processing. Nat. Genet. 51 (3), 414–430. doi:10.1038/s41588-019-0358-2
Love, S., Chalmers, K., Ince, P., Esiri, M., Attems, J., Jellinger, K., et al. (2014). Development, Appraisal, Validation and Implementation of a Consensus Protocol for the Assessment of Cerebral Amyloid Angiopathy in Post-Mortem Brain Tissue. Am. J. Neurodegener. Dis. 3 (1), 19–32.
Mahnashi, M. H., Alqahtani, Y. S., Alyami, B. A., Alqarni, A. O., Alqahl, S. A., Ullah, F., et al. (2022). HPLC-DAD Phenolics Analysis, α-glucosidase, α-amylase Inhibitory, Molecular Docking and Nutritional Profiles of Persicaria Hydropiper L. BMC Complement. Med. Ther. 22 (1), 26–20. doi:10.1186/s12906-022-03510-7
Mahnashi, M. H., Alyami, B. A., Alqahtani, Y. S., Alqarni, A. O., Jan, M. S., Ayaz, M., et al. (2021). Neuroprotective Potentials of Selected Natural Edible Oils Using Enzyme Inhibitory, Kinetic and Simulation Approaches. BMC complementary Med. Ther. 21 (1), 1–14. doi:10.1186/s12906-021-03420-0
Majd, S., and Power, J. H. T. (2018). Oxidative Stress and Decreased Mitochondrial Superoxide Dismutase 2 and Peroxiredoxins 1 and 4 Based Mechanism of Concurrent Activation of AMPK and mTOR in Alzheimer's Disease. Curr. Alzheimer Res. 15 (8), 764–776. doi:10.2174/1567205015666180223093020
Ovais, M., Zia, N., Ahmad, I., Khalil, A. T., Raza, A., Ayaz, M., et al. (2018). Phyto-Therapeutic and Nanomedicinal Approaches to Cure Alzheimer's Disease: Present Status and Future Opportunities. Front. Aging Neurosci. 10, 284. doi:10.3389/fnagi.2018.00284
Ovais, M., Hoque, M. Z., Khalil, A. T., Ayaz, M., Ahmad, I., et al. (2021). “Mechanisms Underlying the Anticancer Applications of Biosynthesized Nanoparticles,” in Biogenic Nanoparticles for Cancer Theranostics (Cambridge, MA: Elsevier ), 229–248.
Post, R. M. (1992). Transduction of Psychosocial Stress into the Neurobiology of Recurrent Affective Disorder. Am. J. psychiatry 149 (8), 999–1010. doi:10.1176/ajp.149.8.999
Saito, S., Yamamoto, Y., and Ihara, M. (2019). Development of a Multicomponent Intervention to Prevent Alzheimer's Disease. Front. Neurol. 10, 490. doi:10.3389/fneur.2019.00490
Saleem, U., Akhtar, R., Anwar, F., Shah, M. A., Chaudary, Z., Ayaz, M., et al. (2021). Neuroprotective Potential of Malva Neglecta Is Mediated via Down-Regulation of Cholinesterase and Modulation of Oxidative Stress Markers. Metab. Brain Dis. 36 (5), 889–900. doi:10.1007/s11011-021-00683-x
Sneader, W. (2005). Drug Discovery: A History . New Jersey: John Wiley & Sons .
Tong, X., Li, X., Ayaz, M., Ullah, F., Sadiq, A., Ovais, M., et al. (2020). Neuroprotective Studies on Polygonum Hydropiper L. Essential Oils Using Transgenic Animal Models. Front. Pharmacol. 11, 580069. doi:10.3389/fphar.2020.580069
Yu, M., Zhang, H., Wang, B., Zhang, Y., Zheng, X., Shao, B., et al. (2021). Key Signaling Pathways in Aging and Potential Interventions for Healthy Aging. Cells 10 (3), 660. doi:10.3390/cells10030660
Keywords: Alzheime´s disease, natural products, neuroprotection, signaling pathways, oxidative stress
Citation: Ayaz M, Ali T, Sadiq A, Ullah F and Naseer MI (2022) Editorial: Current Trends in Medicinal Plant Research and Neurodegenerative Disorders. Front. Pharmacol. 13:922373. doi: 10.3389/fphar.2022.922373
Received: 17 April 2022; Accepted: 13 June 2022; Published: 30 June 2022.
Reviewed by:
Copyright © 2022 Ayaz, Ali, Sadiq, Ullah and Naseer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Ayaz, [email protected]
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
- All Journals
- {{subColumn.name}}
{{lists.name}}
© 2024 Maximum Academic Press, except Open Access articles. MAP is a member of ALPSP, CrossRef and cOAlition S.
Privacy Policy | Site Map | Terms and Conditions
- Share facebook twitter linkedin

Medicinal Plant Biology : A new era for medicinal plant research
- Xiaoya Chen 1 , , ,
- Cathie Martin 2 , , &
- Wansheng Chen 3 , ,
Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, China
John Innes Centre, Norwich NR4 7UH, UK
Shanghai University of Traditional Chinese Medicine, Shanghai 201203, China
- Corresponding authors: [email protected] ; [email protected] ; [email protected]
- Received Date: 06 January 2022 Accepted Date: 10 January 2022 Published Online: 14 January 2022 Medicinal Plant Biology 1 , Article number: 1 (2022) | Cite this article
- Medicinal Plant Biology ,
- Medicinal Plant ,
- Inaugural Editorial
Rights and permissions
About this article, cite this article, article metrics.
Article views( 5304 ) PDF downloads( 854 )
Access History

Other Articles By Authors
- Xiaoya Chen
- Cathie Martin
- Wansheng Chen
- Received Date: 06 January 2022
- Accepted Date: 10 January 2022
- Published Online: 14 January 2022
- Medicinal Plant Biology /
- Medicinal Plant /
Plants are amazing chemical factories, and medicinal plants provide a myriad of pharmaceutically active compounds that have been commonly used as traditional medicines for thousands of years. The practice of traditional medicine in China dates back at least 4,500 years. The Shen Nong Ben Cao Jing ("Shen Nong's Herbal Classic" in 770−475 BC) has been considered the oldest list of medicinal plants. Recent rapid economic development has enabled China to invest substantially in science and technology research. In many ethnic groups worldwide, herbal medicines are, in the same way as traditional Chinese medicines, still commonly used today. A wide array of plant-extract health supplements has become increasingly popular in Western societies. Numerous drugs derived from a broad range of plant species have been discoveried, such as taxol and artemisinin and their derivatives. Investigations of the chemotaxonomy, molecular phylogeny, and pharmacology of these diverse plants and derived compounds through molecular biology and omics-based techniques have led to a new frontier of medicinal plant research, i.e., Medicinal Plant Biology. For example, improvements in sequencing technology—with drastically reduced costs—have offered unpresidented access to genomic, transcriptomic, proteomic and metabolomic information for large numbers of medicinal plants. The massive amount of new data will surely lead to new discoveries in plant-derived medicine. This has been witnessed by a substantial increase in MPB-related research papers. The time is clearly right to initiate an international journal with focus on the biology of these specific groups of plants, and it is our great honor to announce the launch of Medicinal Plant Biology (MPB) with the aims of filling the gap and meeting the need for publications of the highest standards in this field. We aim to build MPB into a flagship journal, publishing leading research, which will have a profound impact on the field of medicinal plants, not only in the advance of science but also in providing a venue for international scholarly exchange. To ensure that MPB attracts high-quality publications, the journal will be guided by a distinguished Advisory Board consisting of preeminent, world-class scholars in the field and will be edited by a distinguished international Editorial Board comprised of outstanding front-line researchers. All papers will be subjected to rigorous peer review, and accepted papers will be published online immediately with free access and global dissemination. We sincerely hope that you will help the journal excel by submitting your excellent research and review articles, by serving as reviewers, and by becoming frequent readers of MPB.
On behalf of the journal's distinguished Editorial Board and the publisher, Maximum Academic Press, we warmly welcome you to visit the MPB webpage ( www.maxapress.com/mpb ) and read the incoming articles of high quality and impact.
The authors declare that they have no conflict of interest.
- Open Access This work is licensed under a Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
- Copyright: 2022 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
Export File

PDF not found!
- Open access
- Published: 25 April 2018
Quantitative study of medicinal plants used by the communities residing in Koh-e-Safaid Range, northern Pakistani-Afghan borders
- Wahid Hussain 1 ,
- Lal Badshah 1 ,
- Manzoor Ullah 2 ,
- Maroof Ali 3 ,
- Asghar Ali 4 &
- Farrukh Hussain 5
Journal of Ethnobiology and Ethnomedicine volume 14 , Article number: 30 ( 2018 ) Cite this article
59k Accesses
40 Citations
1 Altmetric
Metrics details
The residents of remote areas mostly depend on folk knowledge of medicinal plants to cure different ailments. The present study was carried out to document and analyze traditional use regarding the medicinal plants among communities residing in Koh-e-Safaid Range northern Pakistani-Afghan border.
A purposive sampling method was used for the selection of informants, and information regarding the ethnomedicinal use of plants was collected through semi-structured interviews. The collected data was analyzed through quantitative indices viz. relative frequency citation, use value, and family use value. The conservation status of medicinal plants was enumerated with the help of International Union for Conservation of Nature Red List Categories and Criteria (2001). Plant samples were deposited at the Herbarium of Botany Department, University of Peshawar for future reference.
One hundred eight informants including 72 male and 36 female were interviewed. The informants provided information about 92 plants species used in the treatment of 53 ailments. The informant reported maximum number of species used for the treatment of diabetes (16 species), followed by carminatives (12 species), laxatives (11 species), antiseptics (11 species), for cough (10 species), to treat hepatitis (9 species), for curing diarrhea (7 species), and to cure ulcers (7 species), etc. Decoction (37 species, i.e., 40%) was the common method of recipe preparation. Most familiar medicinal plants were Withania coagulans , Caralluma tuberculata , and Artemisia absinthium with relative frequency (0.96), (0.90), and (0.86), respectively. The relative importance of Withania coagulans was highest (1.63) followed by Artemisia absinthium (1.34), Caralluma tuberculata (1.20), Cassia fistula (1.10), Thymus linearis (1.06), etc. This study allows identification of novel uses of plants. Abies pindrow , Artemisia scoparia , Nannorrhops ritchiana , Salvia reflexa , and Vincetoxicum cardiostephanum have not been reported previously for their medicinal importance. The study also highlights many medicinal plants used to treat chronic metabolic conditions in patients with diabetes.
Conclusions
The folk knowledge of medicinal plants species of Koh-e-Safaid Range was unexplored. We, for the first time, conducted this quantitative study in the area to document medicinal plants uses, to preserve traditional knowledge, and also to motivate the local residents against the vanishing wealth of traditional knowledge of medicinal flora. The vast use of medicinal plants reported shows the significance of traditional herbal preparations among tribal people of the area for their health care. Knowledge about the medicinal use of plants is rapidly disappearing in the area as a new generation is unwilling to take interest in medicinal plant use, and the knowledgeable persons keep their knowledge a secret. Thus, the indigenous use of plants needs conservational strategies and further investigation for better utilization of natural resources.
The residents of remote areas mostly depend on folk knowledge of medicinal plants to cure different ailments. Plants not only provide food, shelter, fodder, drugs, timber, and fuel wood, but also provide different other services such as regulating different air gases, water recycling, and control of different soil erosion. Hence, phytodiversity is required to fulfill several human daily livelihood needs. Millions of people in developing countries commonly derive their income from different wild plant products [ 1 ]. Ethnomedicinal plants have been extensively applied in traditional medicine systems to treat various ailments [ 2 ]. This relationship goes back to the Neanderthal man who used plants as a healing agent. In spite of their ancient nature, international community has recognized that many indigenous communities depend on biological resources including medicinal plants [ 3 ]. About 80% of the populations in developing countries rely on medicinal plants to treat diseases, maintaining and improving the lives of their generation [ 4 , 5 ]. The people, in most parts of the world particularly in rural areas, rely on traditional medicinal plants’ remedies due to easy availability, cultural acceptability, and poor economic conditions. Out of the total 422,000 known angiosperms, more than 50,000 are used for medicinal purposes [ 6 ]. Some 75% of the herbal drugs have been developed through research on traditional medicinal plants, and 25% of prescribed drugs belong to higher plants [ 7 ]. Traditional knowledge has a long historical cultural heritage and rich natural resources that have accumulated in the indigenous communities through oral and discipleship practices [ 8 ]. Traditional indigenous knowledge is important in the formulation of herbal remedies and isolates bioactive constituents which are a precursor for semisynthetic drugs. It is the most successful criterion for the development of novelties in drugs [ 9 , 10 , 11 ]. Traditional knowledge can also contribute to conserve and sustain the use of biological diversity. However, traditional knowledge, especially herbal health care system, has declined in remote communities and in younger generations as a result of a shift in attitude and ongoing socio-economic changes [ 12 ]. The human communities are facing health and socio-economic problems due to changing environmental conditions and socio-economic status [ 13 ]. The tribal people have rich unwritten traditional medicinal knowledge. It rests with elders and transfers to younger orally. With rapid economic development and oral transmitted nature of traditional knowledge, there is an urgent need to systematically document traditional medicinal knowledge from these communities confined in rural and tribal areas of the world including Pakistan. The Koh-e-Safaid Range is one of the remote tribal areas of Pakistan having unique and century-old ethnic characteristics. A single hospital with limited insufficient health facilities is out of reach for most inhabitants. Nature has gifted the area with rich diversity of medicinal plants. The current advancement in the use of synthetic medicines has severely affected the indigenous health care system through the use of medicinal traditional practices in the area. The young generation has lost interest in using medicinal plants, and they are reluctant to practice traditional health care system that is one of the causes of the decline in traditional knowledge system. Quantitative approaches can explain and analyze the variables quantitatively. In such approach, authentic information can be used for conservation and development of existing resources. Therefore, the present research was conducted in the area to document medicinal uses of local plants with their relative importance, to record information for future investigation and discovery of novelty in drug use, and to educate the locals about the declining wealth of traditional and medicinal flora from the area.
Ethnographic and socio-economic background of the study area
Koh-e-Safaid Range is a tribal territory banding Pakistan with Afghanistan in Kurram Agency. It lies between 33° 20′ to 34° 10′ N latitudes and 69° 50′ to 70° 50′ E longitudes (Fig. 1 ).This area is federally administered by the Government of Pakistan. The Agency is surrounded on the east by Orakzai and Khyber agencies, in the southeast by Hangu district, and in the south by North Waziristan Agency and Nangarhar and Pukthia of Afghanistan lies on its west. The highest range of Koh-e-Safaid is Sikaram peak with, 4728 m height. The Agency is well-populated with many small fortified villages receiving irrigation water from Kurram River that flows through it. The weather of the Agency is mostly pleasant in summer; however, in winters, freezing temperature is experienced, and sometimes falls to − 10 °C. The weather charts website “Climate-Charts” ranked it as the fourth coldest location in Pakistan. Autumn and winter are usually dry seasons while summer and spring receive much of the precipitation. The total population of the Agency according to the 2017 censuses report is 253,478. Turi, Bangash, Sayed, Maqbal, Mangel, Khushi, Hazara, Kharote, and Jaji are the major tribes in the research area. The joint family system is practiced in the area. Most of the marriages are held within the tribe; however, there is no ban on the marriages outside the tribe. Marriage functions are communal whereby all relatives, friends, and village people participate with songs, music, and dances male and female separately. The death and funeral ceremonies are jointly attended by the friends and relatives. The people of the area follow Jirga to resolve their social and administrative problems. This is one of the most active and strong social institutions in the area. Economically, most people in the area are poor and earning their livelihoods by menial jobs. The professional includes farmers, pastoralists, shopkeepers, horticulturists, local health healers, wood sellers, and government servants. In the adjoining areas of the city, pastorals keep domestic animals and are considered a better source of income.
Map of the study area and area location in Pakistan
Sampling method
The study was conducted through purposive sampling by informants’ selection method. The selection of informants was primary based on the ethnomedicinal plants and their willingness to share the information. The selection criteria include people who prescribe recipes for treatment; people involved in buying, collection, or cultivation of plants; elder members of above 60 years age; and young literate members. The participants were traditional healers, plant collectors, farmers, traders, and selected knowledgeable elders above 60 years age and young ones. The interviews were conducted in local Pashto language in the local dialect. The informants were involved in the gathering of data with a consent of village tribe chieftains called Maliks.
Data collection
Semi-structured open-ended interviews were conducted for the collection of ethnomedicinal information from April 2015 to August 2017. Informants from 19 localities were interviewed including Sultan, Malikhail, Daal, Mali kali, Alam Sher, Kirman, Zeran, Malana, Luqman Khail, Shalozan, Pewar, Teri Mangal, Bughdi, Burki, Kharlachi, Shingak, Nastikot, Karakhila, and Parachinar city (Fig. 1 ). The objectives of this study were thoroughly explained to all the informants before the interview [ 14 ]. Data about medicinal plants and informants including local names of plants, preparation of recipes, storage of plant parts, informant age, occupation, and education were collected during face-to-face interviews. A questionnaire was set with the following information: informant bio-data, medicinal plant use, plant parts used and modes of preparation, and administration of the remedies. Plants were confirmed through repeated group discussion with informants [ 15 , 16 ]. For the identification of plants, informants were requested for transect walks in the field to locate the cited plant for confirmation.
Collection and identification of medicinal plants
The medicinal plants used in traditional treatment of ailments in the study area were collected with the help local knowledgeable persons, traditional healers, and botanists. The plants were pressed, dried, and mounted on herbarium sheet. The field identification was confirmed by a taxonomist in the Herbarium Department of Botany, University of Peshawar. The voucher specimens of all species were numbered and deposited in the Herbarium of Peshawar University (Fig. 2 ).
Landscape of Kurram Valley ( a winter, b summer). c , d Traditional healers selling herbal drugs on footpath. e Trader crushing Artemisia absinthium for marketing. f Principal author in the field during data collection. g , h Plant collectors in subalpine zone. i Lilium polyphyllum rare species distributed in subalpine zone. j Ziziphora tenuior endangered species of subtropical zone
Data analysis
The information about ethnomedicinal uses of plants and informants included in questionnaires such as botanical name, local name, family name, parts used, mode of preparation, use reports, frequency of citation, relative importance, and voucher number were tabulated for all reported plant species. Informants’ use reports for various ailments and frequency of citation were calculated for each species. The relative importance of species was calculated according to use-value formula (UV = UVi/Ni) [ 17 ], where “UVi” is the number of citations for species across all informants and “Ni” the number of informants. The citation probability of each medicinal plant across all informants was equal to avoid researchers’ biasness. Family use value was calculated using the formula FUV = UVs/Ns, where “UVs” represent the sum of use values of species falling within family, and Ns represents the number of species reported for the family. The conservation status of wild medicinal plants species was enumerated by applying International Union for Conservation of Nature (IUCN) criterion (2001) [ 18 ].
Informants’ knowledge about medicinal plants and their demography
A total of 108 including 72 male and 36 female informants were interviewed from 19 locations. The three groups of male respondents were falling in the age groups of 21 to 40, 41 to 60, and 61 to 80 years having the numbers of 19, 19, and 34, respectively. Among the female respondents, 10 aged 21 to 40, 14 aged 41 to 60, and 12 aged 61 to 80 years. Among the informants, 15 males were illiterate, 34 were matriculate, 13 were intermediate, and 10 were graduates. Among the females, 19 were illiterate, 16 were matriculate, and only 1 was graduate (Table 1 ). Informants were shepherd, healers, plant collectors, gardeners, and farmers. Twenty-eight informants of above 60 years age, living a retired life, were also interviewed. It was found that males were more knowledgeable than females. Furthermore, health healers were more knowledgeable.
Diversity of medicinal plants
A total of 92 medicinal species including 91vascular plant species belonging to 50 families and 1 mushroom Morchella of Ascomycetes of family Morchellaceae were reported (Table 2 ) . Asteraceae had eight species followed by seven species of Lamiaceae and Rosaceae. Three species were contributed by each of Moraceae, Asclepiadaceae, Polygonaceae, Brassicaceae, Solanaceae, Cucurbitaceae, and Liliaceae. Of the remaining eight families, namely, Poaceae, Pinaceae, Zingiberaceae, Chenopodiaceae, Plantaginaceae, Apiaceae, Fabaceae, and Zygophyllaceae, each one contributed two species [ 19 , 20 ]. Asteraceae, Lamiaceae, and Rosaceae were also reported with a high number of plants used for medicinal purposes. The reported plants were collected both from the wild (86.9%) and cultivated (13.1%) sources. However, greater percentage of medicinal plants from wild sources indicated higher species’ diversity in the study area. The 62 herbs species, 16 tree, 12 shrubs, and 2 undershrubs species were used in medicinal preparation for remedies.
Plant parts used in preparation of remedies
The plant parts used in the preparation of remedies were root, rhizome, bulbils, stem, branches, leaves, flowers, fruits, seeds, bark, resin, and latex. The relative use of these plant parts is shown in (Fig. 3 ). Fruits were frequently used plant part (26 species), followed by leaves (23 species) and remaining parts (21 species).
Plant parts used in the formulation of remedies
Preparation and mode of administration of remedies
The collection of data for the preparation of remedies from medicinal plants is extremely important. Such information is essential for identification of active ingredients and intake of relevant amount of drug. The present research observed seven methods for preparing recipes. It included decoction, powder, juice, infusions, roast, and ash methods (Fig. 4 ). The 37 species (40%) were most frequently used for the preparation of remedies. A plant part is boiled while infusion is obtained by soaking plant material in cold or hot water overnight. Eleven species (14%) are in powdered form, 11species (14%) in vegetable form, 7 species (9%) in juice form, 7 species (9%) in infusions form, 3 species (4%) in roasted form, and 1 species (2%) in ash form were used. Twenty-seven plant parts were used directly. It included wild fruits that were consumed for their nutritional and medicinal purpose. The most frequently used mode of administration of remedies was oral intake practice of 74 species (79%) followed by both orally and topically practice of 11 species (12%) and topically of 8 species (9%) (Fig. 5 ).
Different modes of drug formulation
Route of administration of drugs
Medicinal plants use categories
The inhabitants used medicinal plants in the treatment of 53 health disorders. The important disorders were cancer, diabetic, diarrhea, dysentery, hepatitis, malaria, and ulcer (Table. 3 ). These disorders were classified into 17 categories. Among the ailments, most plants were used for the treatment of digestive problems mainly as carminative (12 species), diarrhea (11 species), laxative (11 species), ulcer (7 species), appetizer (5 species), colic pain (4 species), and anthelmintic (4 species). Such higher use of plants for the treatment of digestive problems had been reported in ethnobotanical studies conducted in another tribal area of Pakistan [ 21 ]. The other categories (18 species) were used to treat respiratory disorders, followed by endocrine disorders (16 species); antiseptic and anti-inflammatory (15 species); circulatory system disorders (15 species); integumentary problems (15 species); antipyretic, refrigerant, and analgesic (9 species); and hepatic disorders (9 species). However, among the ailments, the highest number of plants were used in the treatment of diabetes (16 species), followed by antiseptic (11 species), cough (10 species), hepatitis (9 species), and ulcer (7 species). Among the remaining species, the informants reported three and two species used against malaria and cancer, respectively (Table 3 ).
Quantitative appraisal of ethnomedicinal use
Based on the quantitative indices, the analyzed data showed that few plants were cited by the majority of the informants for their medicinal value. Seventeen plant species with the highest citation frequency are shown in (Fig. 6 ). The highest citation frequency was calculated for Withania coagulans (0.96), followed by Caralluma tuberculata (0.90), and Artemisia absanthium (0.86). The high values of these species indicated that most of the informants were familiar with their medicinal value. However, the familiarity of these three plants could be linked to their collection for economic purposes [ 22 ]. Withania coagulans (1.63), Artemisia absinthium (1.34), Caralluma tuberculata (1.20), Cassia fistula (1.10), and Thymus linearis (1.06) were reported having the highest used values for medicinal purposes (Fig. 7 ). All these species were used for the cure of three or more diseases. The powdered fruit of Withania coagulans is used for the cure of stomach pain, constipation, diabetes, and ulcer. The next highest use value was calculated for Artemisia absinthium with five medical indications as diabetes, malaria, fever, blood pressure, and urologic problems. Among the remaining three plants, Caralluma tuberculata is used for diabetes, cancer, and stomachic problems, and as blood purifier; Cassia fistula for colic pain and stomach pain and as a carminative agent; and Thymus linearis for cough and as carminative and appetizer. Lowest use value was calculated for Rununculus muricatus (0.04) with next three species having same lowest use value: Abies pindrow (0.05), Lepidium virginicum (0.05), and Oxalis corniculata (0.05). Highest family use value was calculated for Juglandaceae (0.86), followed by Cannabaceae (0.78), Apiaceae (0.75), Asclepiadaceae (0.71), Fumariaceae (0.71), Berberidaceae (0.70), Fabaceae (0.67), Punicaceae (0.65), Solanaceae (0.64), and Asteraceae (0.61). This is the first study that presents a quantitative value of medicinal plants used in the investigated area.
Medicinal plants with highest relative frequency citation
Medicinal plants with highest relative importance
Conservation status of the medicinal flora
Plant preservation means the study of plant declination, their causes, and techniques to protect rare and scarce plants. Plant conservation is a fairly new field that emphasizes the conservation of biodiversity and whole ecosystems as opposed to the conservation of individual species [ 23 ]. The ex situ conservation must be encouraged for the protection of medicinal plants [ 24 ]. In the present case, the area under study is under tremendous anthropogenic pressure as well. Therefore, ex situ conservation of endangered species is recommended. The woody plants, cut down for miscellaneous purposes, are facing conservational problems. Sayer et al. [ 25 ] reported that large investments are being made in the establishment of tree plantation on degraded area in Asia [ 25 ]. Alam and Ali stressed that proper conservation studies are almost negligible in Pakistan [ 26 ]. Same is the case with the study area as no project has been initiated for the conservation of forest or vegetation so far. Anthropogenic activities, small size population, distribution in limited area, and specificity of habitat were observed as the chief threats to endangered species.
According to IUCN Red List Criteria (2001) [ 18 ] conservation status of 80 wild medicinal species have been assessed based on availability, collection status, growth status, and their parts used. The remaining 12 medicinal plants were cultivated species. Of these, 7 (8.7%) species are endangered, 34 (42.5%) species are vulnerable, 29 (36.2%) species are rare, 9 (11.2%) species are infrequent, and only 1 (1.3%) species is dominant. The endangered species were Caralluma tuberculata , Morchella esculenta , Rheum speciforme , Tanacetum artemisioides , Vincetoxicum cardiostephanum , Withania coagulans , and Polygonatum verticillatum.
Traditional medicines are a vital and often underestimated part of health care. Nowadays, it is practiced in almost every country of the world. Its demand is currently increasing rapidly in the form of alternative medicine [ 20 ]. Ethnomedicinal plants have been widely applied in traditional medicine systems to treat various ailments. About 80% of the populations in developing countries rely on medicinal plants to treat diseases, maintaining and improving the lives of their generation [ 19 ]. Traditional knowledge has a long historical cultural heritage and rich natural resources that has accumulated in the indigenous communities through oral and discipleship practices [ 8 ]. Traditional indigenous knowledge is important in the formulation of herbal remedies and isolates bioactive constituents which are a precursor for semisynthetic drugs. It is the most successful criterion for the development of novelties in drugs [ 11 ]. A total of 92 medicinal species including 91 vascular plant species belonging to 50 families and 1 mushroom Morchella of Ascomycetes of family Morchellaceae were reported (Table 2 ) . The current study reveals that the family Asteraceae represents eight species followed by seven species of Lamiaceae and Rosaceae each which showed a higher number of medicinal plants. Three species were contributed by each of Moraceae, Asclepiadaceae, Polygonaceae, Brassicaceae, Solanaceae, Cucurbitaceae, and Amaryllidaceae. While the remaining eight families, namely, Poaceae, Pinaceae, Zingiberaceae, Chenopodiaceae, Plantaginaceae, Apiaceae, Fabaceae, and Zygophylaceae, contributed two species each. Asteraceae, Lamiaceae, and Rosaceae were also reported with a high number of plants used for medicinal purposes. Indigenous use of medicinal plants in the communities residing in Koh-e-Safid Range of Pakistan is evident. Traditional health healers are important to fulfill the basic health needs of the economically poor people of the area. The high dependency on traditional healers is due to limited and inaccessible health facilities. Most people either take recipes from local healers or select wild medicinal plants prescribed by them. Some elders also knew how to preserve medicinal plant parts for future use. Traditional knowledge of medicinal plants is declining in the area due to lack of interest in the young generation to acquire this traditional treasure. Furthermore, most traditional health healers and knowledgeable elders hesitate to disseminate their recipes. Therefore, traditional knowledge in the area is diminishing as aged persons are passing away. Vernacular names of plants are the roots of ethnomedicinal diversity knowledge [ 27 ]. They can clear the ambiguity in the identification of medicinal plants within an area. It also helps in the preservation of indigenous knowledge of medicinal plants. The medicinal plants were mostly reported with one specific vernacular name in the investigated area. While Rosa moschata and Rosa webbiana were known by same single vernacular name as Jangle Gulab. Few species were known by two vernacular names: Curcuma longa as Korkaman or Hildi, Ficus carica as Togh or Anzer, Fumaria indica as Chamtara or Chaptara, Marrubium vulgare as Dorshol or Butaka, Solanum nigrum as Bartang or Kharsobay, Teucrium stocksianum as Harboty or Gulbahar, and Thymus linearis as Paney or Mawory. The informants also mentioned different vernacular names for species even belonging to single genus; Plantago lanceolata as Chamchapan or Ghuyezaba and Plantago major as Ghazaki or Palisepary. Majority of the species commonly had a single name. However, local dialects varied in few species, i. e., Withania coagulans was known by three names: Hapyanaga, Hafyanga, and Shapynga, Caralluma tuberculata as Pamenny or Pawanky, Foeniculum vulgare as Koglany or Khoglany, and Viola canescens was called as Banafsha or Balamsha. The species with high use value need conservation for maintaining biodiversity in the study area. However, in the present case, no project or programs for the conservation of forest or vegetation are operating. Grazing and unsustainable medicinal uses were observed as the chief hazard to highly medicinal plant species. The higher use of herbs can be attributed to their abundance, diversity, and therapeutic potentials as antidiabetic, antimalarial, antipyretic, antiulcerogenic, antipyretic, blood purifier, and emollient and for blood pressure, hepatitis, stomach pain, and itching. Aloe vera , cultivated for ornamental purpose, is used as wound healing agent. Among the plant parts, the higher use of fruit may relate to its nutritional value. The aerial parts of the herbaceous plants were mostly collected in abundance and frequently used for medicinal purposes. In many recipes, more than one part was used. The utilization of roots, rhizomes, and the whole plant is the main threat in the regeneration of the medicinal plants [ 28 ]. In the current study, decoction was found to be the main method of remedy preparation as reported in the ethnopharmacological studies from other parts [ 29 , 30 , 31 ]. Fortunately, we collected important information like preparation of remedies and their mode of administration for all the reported plants. However, the therapeutic potential of few plants are connected to their utilization method. A roasted bulb of Allium cepa is wrapped on the spine-containing wound to release the spine. The leaf of Aloe vera containing viscous juice is scratched and wrapped on a wound. The latex of Calotropis procera is first mixed with flour and then topically applied on the skin for wound healing. Infusion of Cassia fistula fruit’s inner septa is prepared for stomach pain and carminative and colic pain in children. The fruit of Citrullus colocynthis boiled in water is orally taken for the treatment of diabetes. Grains of Hordeum vulgare are kept in water for a day, and its extraction is taken for the treatment of diabetes. The decoction of Seriphidium kurramensis shoots are used as anti-anthelmintic and antimalaria. The leaves of Juglans regia are locally used for cleaning the teeth and to prevent them from decaying. Furthermore, its fruit is used as brain tonic, and its roasted form is useful in the treatment of dysentery. The roots of Pinus wallichiana are cut into small pieces and put into the pot. The cut pieces are boiled, and the extracted liquid is poured into the container. One drop of the extracted liquid is mixed with one glass of milk and taken orally once a day as blood purifier. An infusion of Thymus linearis aerial parts is prepared like hot tea and is drunk for cough and as appetizer and carminative. A decoction of Zingiber officinale rhizome is drunk at night time for relief of cough. Medicinal plants are still practiced in tribal and rural areas as they are considered as main therapeutic agents in maintaining better health. Such practices have been described in the ethnobotanical studies conducted across Pakistan. The current study reveals several plant species with more than one medical use including Artemisia absanthium , Cichorium intybus , Fumaria indica , Punica granatum , Tanacetum artemisioides , Teucrium stocksianum , and Withania coagulans . Their medicinal importance can be validated from indigenous studies conducted in various parts of the country. Amaranthus viridis leaf extract is an emollient and is used for curing cough and asthma as well [ 32 ]. Artemisia absanthium is used for the treatment of malaria and diabetes [ 33 , 34 , 35 , 36 ]. Cichorium intybus is used against diabetes, malaria, and gastric ulcer, and it is also used as digestive and laxative agent [ 28 , 37 , 38 , 39 , 40 , 41 ]. Leaves of Cannabis sativa are used as bandage for wound healing; powdered leaves as anodyne, sedative, tonic, and narcotic; and juice added with milk and nuts as a cold drink [ 42 ]. Whole plant of Fumaria indica [ 36 ] and Tanacetum artemisioides [ 43 ] is used for treating constipation and diabetes, respectively. Dried rind powder and fruit extract of Punica granatum are taken orally for the treatment of anemia, diarrhea, dysentery, and diabetes [ 44 , 45 , 46 , 47 ]. A decoction of aerial parts of Teucrium stocksianum is used for curing diabetes [ 29 , 48 ]. Withania coagulans is known worldwide [ 38 , 49 ] as a medicinal plant, whose fruit decoction is best remedy for skin diseases and diabetes. Its seeds are used against digestive problems, gastritis, diabetes, and constipation [ 21 , 28 , 50 ]. Our results are in line with the traditional uses of plants in the neighboring counties [ 8 ]. For example, Fumaria indica is used as blood purifier, and Hordeum vulgare grains decoction for diabetes; Juglans regia bark for toothaches and scouring teeth; Mangifera indica seed decoction for diarrhea; Solanum nigrum extract for jaundice; and Solanum surattense fruit decoction for cough have been documented in the study (40) . Such agreements strengthen our results and provide good opportunity to evaluate therapeutic potential of the reported plants. Three plants species Adiantum capillus-veneris , Malva parviflora , and Peganum harmala have been documented for their medicinal use in the ethnobotanical study [ 51 ]. According to this, the decoction of the aerial parts of Adiantum capillus-veneris is used for the treatment of asthma and dyspnea. Malva parviflora root and flower are used for stomach ulcers. Peganum harmala fruit powder and decoction are used for toothache, gynecological infections, and menstruation. The dried leaves of Artemisia absanthium is used to cure stomach pain and intestinal worm while an inflorescence paste prepared from its fresh leaves is used as wound healing agent and antidiabetic [ 52 , 53 ]. The bulb of Allium sativum is used in rheumatism while its seed vessel mixed with hot milk is useful for the prevention of tuberculosis and high blood pressure. The fruit bark of Punica granatum is used in herbal mixture for intestinal problems [ 54 ]. Avena sativa decoction is used for skin diseases including eczema, wounds, irritation, inflammation, erythema, burns, itching, and sunburn [ 55 ]. Foeniculum vulgare and Lepidium sativum are used for the treatment of diabetes and renal diseases [ 53 ]. Verbascum thapsus leaves and flowers can be used to reduce mucous formation and stimulate the coughing up of phlegm. Externally, it is used as a good emollient and wound healer. Leaves of Thymus linearis are effective against whooping cough, asthma, and round worms and are an antiseptic agent [ 21 ]. Berberis lycium wood decoction with sugar is the best treatment for jaundice. Chenopodium album has anthelmintic, diuretic, and laxative properties, and its root decoction is effective against jaundice. The whole plant decoction of Fumaria indica is used for blood purification. Dried leaves and flowers of Mentha longifolia are used as a remedy for jaundice, fever, asthma, and high blood pressure [ 36 ]. Morus alba fruit is used to treat constipation and cough [ 42 ]. Oxalis corniculata roots are anthelmintic, and powder of Chenopodium album is used for headache and seminal weakness [ 47 ]. Boiled leaves of Cichorium intybus are used for stomachic pain and laxative while boiled leaves of Plantago major are used against gastralgia [ 56 ]. Viola canescens flower is used as a purgative [ 32 ]. The above ethnomedicinal information confirms the therapeutic importance of the reported plants. The reported plant species show biological activities which suggest their therapeutic uses. The aqueous extract of Allium sativum has been studied for its lipid lowering ability and was found to be effective at the amount of 200 mg/kg of body weight. It also has significant antioxidant effect and normalizes the activities of superoxide dismutase, catalase, glutathione peroxidase, and glutathione reductase in the liver [ 57 ]. An extract of Artemisia absanthium antinociception in mice has been found and was linked to cholinergic, serotonergic, dopaminergic, and opioidergic system [ 58 ]. The ethanolic extract of Artemisia absanthium at a dose of 500 and 1000 mg/kg body weight has reduced blood glucose to significant level [ 59 ]. The hepatoprotective activity of crude extract of aerial parts of Artemisia scoparia was investigated against experimentally produced hepatic damage through carbon tetrachloride. The experimental data showed that crude extract of Artemisia scoparia is hepatoprotective [ 60 ]. Ethanolic and aqueous extracts from Asparagus exhibited strong hypolipidemic and hepatoprotective action when administered at a daily dose of 200 mg/kg for 8 weeks in hyperlipidemic mice [ 61 , 62 ]. The extract of Calotropis procera was evaluated for the antiulcerogenic activity by using different in vivo ulcer in pyloric-ligated rats, and significant protection was observed in histamine-induced duodenal ulcers in guinea pigs [ 63 ]. Cannabidiol of Cannabis sativa was found as anxiolytic, antipsychotic, and schizophrenic agent [ 64 ]. Caralluma tuberculata methanolic extract of aerial parts (500 mg/kg) in fasting blood glucose level in hyperglycemic condition decreased up to 54% at fourth week with concomitant increase in plasma insulin by 206.8% [ 65 ]. The aqueous and methanol crude extract of Celtis australis , traditionally used in Indian system of medicine, was screened for its antibacterial activity [ 66 ]. Cichorium intybus L. whole plant 80% ethanolic extract a percent change in serum glucose has been observed after 30 min in rats administrated with vehicle, 125, 250, and 500 mg notified as 52.1, 25.2, 39, and 30.9%, respectively [ 67 ]. Citrullus colocynthis fruit, pulp, leaves, and root have significantly decreased blood glucose level and restored beta cells [ 30 , 68 , 69 , 70 ]. The two new aromatic esters horizontoates A and B and one new sphingolipid C were isolated from Cotoneaster horizontalis . The compounds A and B showed significant inhibitory effects on acetylcholinesterase and butylcholinesterase in a dose-dependent manner [ 71 ]. The alkaloids found in Datura stramonium are organic esters used clinically as anticholinergic agents [ 72 ]. The methanolic extract of Momordica charantia fruits on gastric and duodenal ulcers was evaluated in pylorus-ligated rats; the extract showed significant decrease in ulcer index [ 73 ]. Antifungal activity of Nannorrhops ritchiana was investigated against fungal strains Aspergillus flavus , Trichophyton longifusis , Trichophyton mentagrophytes , Aspergillus flavus , and Microsporum canis were found susceptible to the extracts with percentage inhibition of 70–80% [ 74 ]. The inhibitory effects of Olea ferruginea crude leaf extract on bacterial and fungal pathogens have been evaluated [ 75 ]. The aqueous extract of Plantago lanceolata showed that higher doses provide an overall better protection against gastro-duodenal ulcers [ 76 ]. The oral and intraperitoneal management of extracts reduced the gastric acidity in pylorus-ligated mice [ 77 ]. The antiulcer effect of Solanum nigrum fruit extract on cold restraint stress, indomethacin, pyloric ligation, and ethanol-induced gastric ulcer models and ulcer healing activity on acetic acid-induced ulcer model in rats [ 78 , 79 ]. The antifungal activity (17.62 mm) of Viola canescens acetone extract 1000 mg/ml against Fusarium oxysporum has been observed [ 80 ]. Leaf methanolic extract of Xanthium strumarium has inhibited eight pathogenic bacteria at a concentration of 50 and 100 mg/ml [ 81 ]. Aqueous extract of the fruits of Withania coagulans in streptozotocin-induced rats at dose of 1 g/kg for 7 days has shown significant decrease ( p < 0.01) in the blood glucose level (52%), triglyceride, total cholesterol, and low density lipoprotein and very significant increase ( p < 0.01) in high density lipoprotein level [ 31 ]. This shows that further investigation on the reported ethnomedicinal plants can lead to the discovery of novel agents with therapeutic properties.
In the current study, conservation status of 80 medicinal species was reported which was growing wild in the area. The information was collected and recorded for different conservation attributes by following International Union for Conservation and Nature (2001) [ 18 ]. It was reported that seven species (8.7%) were endangered due to the much collection, anthropogenic activities, adverse climatic conditions, small size population and distribution in limited area, specificity of habitat, and over grazing in the research area. However, the below-mentioned species were found to be endangered: Caralluma tuberculata , Morchella esculenta , Rheum speciforme , Tanacetum artemisioides , Vincetoxicum cardiostephanum , Withania coagulans , and Polygonatum verticillatum . Unsustainable use and lack of suitable habitat have affected their regeneration and pushed them to endangered category. Traditional knowledge can also contribute to conservation and sustainable use of biological diversity [ 19 , 20 ].
Novelty and future prospects
Ethnomedicinal literature research indicated that five plant species, Abies pindrow , Artemisia scoparia , Nannorrhops ritchiana , Salvia reflexa , and Vincetoxicum cardiostephanum , have not been reported previously for their medicinal importance from this area. The newly documented uses of these plants were Abies pindrow and Salvia reflexa (antidiabetic), Artemisia scoparia (anticancer), Nannorrhops ritchiana (laxative), and Vincetoxicum cardiostephanum (chest problems). Adiantum capillus-veneris is reported for the first time for its use in the treatment of skin problems. These plant species can be further screened for therapeutic agents and their pharmacological activities in search of novel drugs. The study also highlights 16 species of antidiabetic plants Caralluma tuberculata , Momordica charantia , Marrubium vulgare , Artemisia scoparia , Melia azedarach , Salvia reflexa , Citrullus colocynthis , Tanacetum artemisioides , Quercus baloot , Olea ferruginea , Cichorium intybus , Artemisia absinthium , Hordeum vulgare , Teucrium stocksianum , Withania coagulans , and Abies pindrow . Except sole paper from District Attack, Pakistan [ 28 ], such a high number of antidiabetic plants have not been reported previously from any part of Pakistan in the ethnobotanical studies.
Traditional knowledge about medicinal plants and preparation of plant-based remedies is still common in tribal area of Koh-e-Safaid Range. People due to closeness to medicinal plants and inaccessible health facilities still rely on indigenous traditional knowledge of plants. The role of traditional healers in the area is observable in primary health care. The locals used medicinal plants in treatment of important disorders such as cancer, diabetes, hepatitis, malaria, and ulcer. The analyzed data may provide opportunities for extraction of new bioactive constituents and to develop herbal remedies. The study also confirmed that the communities residing in the area have not struggled for conservation of this traditional treasure of indigenous knowledge and medicinal plants. Medicinal plant diversity in the remote and backward area of Koh-e-Safaid Range has great role in maintaining better health conditions of local communities. Therefore, conservation strategies should be adopted for the protection of medicinal plants and traditional knowledge in the study area to sustain them in the future.
Abbreviations
Family use value
International Union for Conservation of Nature
The number of informants
Represent the number of species reported for the family
The number of citations for a species across all informants
Represent sum of use values of species falling within family
Maroyi A. Diversity of use and local knowledge of wild and cultivated plants in the Eastern Cape province. South Africa J Ethnobiol Ethnomed. 2017;13:43.
Article PubMed Google Scholar
El-Seedi HR, Burman R, Mansour A, Turki Z, Boulos L, Gullbo J. The traditional medical uses and cytotoxic activities of sixty-one Egyptian plants: discovery of an active cardiac glycoside from Urginea maritima. J Ethnopharmacol. 2013;145:746–57.
Akerele O. Nature’s medicinal bounty: don’t throw it away. 1993.
Google Scholar
Calixto JB. Twenty-five years of research on medicinal plants in Latin America: a personal view. J Ethnopharmacol. 2005;100:131–4.
Health WHOR. Managing complications in pregnancy and childbirth: a guide for midwives and doctors: World Health Organization; 2003.
Hamilton AC. Medicinal plants, conservation and livelihoods. Biodivers Conserv. 2004;13:1477–517.
Article Google Scholar
Wang M-Y, West BJ, Jensen CJ, Nowicki D, Su C, Palu AK. Morinda citrifolia (Noni): a literature review and recent advances in noni research. Acta Pharmacol Sin. 2002;23:1127–41.
CAS PubMed Google Scholar
Ouelbani R, Bensari S, Mouas TN, Khelifi D. Ethnobotanical investigations on plants used in folk medicine in the regions of Constantine and Mila (north-east of Algeria). J Ethnopharmacol. 2016;194:196–218.
Farnsworth NR. The role of ethnopharmacology in drug development. Bioact Compd from plants. 1990;154:2–21.
CAS Google Scholar
Cox PA, Balick MJ. The ethnobotanical approach to drug discovery. Sci Am. 1994;6:82–7.
Fabricant DS, Farnsworth NR. The value of plants used in traditional medicine for drug discovery. Environ Health Perspect. 2001;109:69.
Article CAS PubMed PubMed Central Google Scholar
Kala CP. Current status of medicinal plants used by traditional Vaidyas in Uttaranchal state of India. 2005.
Abbas Z, Khan SM, Alam J, Khan SW, Abbasi AM. Medicinal plants used by inhabitants of the Shigar Valley, Baltistan region of Karakorum range-Pakistan. J Ethnobiol Ethnomed. 2017;13:53.
Article PubMed PubMed Central Google Scholar
Cunningham AB. Applied ethnobotany: people, wild plant use and conservation. London: Earthscan. Ersity and sustaining local livelihood. Annu Rev Environ Resour. 2001;30:219–52.
Martin GJ. Ethnobotany: a people and plants conservation manual. London: Chapman and Hall CrossRef Google Scholar; 1995.
Book Google Scholar
Maundu P. Methodology for collecting and sharing indigenous knowledge: a case study. Indig Knowl Dev Monit. 1995;3:3–5.
Phillips O, Gentry AH, Reynel C, Wilkin P, Galvez DB. Quantitative ethnobotany and Amazonian conservation. Conserv Biol. 1994;8:225–48.
Anonymous. IUCN Red List Categories and Criteria: version 3.1. IUCN species survival commission IUCN, Gland, witzerland and Cambridge, U.K. 2001.
Tuasha N, Petros B, Asfaw Z. Medicinal plants used by traditional healers to treat malignancies and other human ailments in Dalle District, Sidama Zone, Ethiopia. J Ethnobiol Ethnomed. 2018;14(1):15.
Aziz MA, Khan AH, Adnan M, Ullah H. Traditional uses of medicinal plants used by indigenous communities for veterinary practices at Bajaur Agency, Pakistan. J Ethnobiol Ethnomed. 2018;14(1):11.
Ullah M, Khan MU, Mahmood A, Malik RN, Hussain M, Wazir SM. An ethnobotanical survey of indigenous medicinal plants in Wana district south Waziristan agency. Pakistan J Ethnopharmacol. 2013;3:150–8.
Hussain W, Hussain J, Ali R, Khan I, Shinwari ZK, Nascimento IA. Tradable and conservation status of medicinal plants of KurramValley. Parachinar, Pakistan. 2012;2:66–70.
Soulé ME. What is conservation biology? Bioscience. 1985;35:727–34.
Heywood VH, Iriondo JM. Plant conservation: old problems, new perspectives. Biol Conserv. 2003;113(3):321–35.
Sayer J, Chokkalingam U, Poulsen J. The restoration of forest biodiversity and ecological values. For Ecol Manag. 2004;201(1):3–11.
Alam J, Ali SI. Conservation status of Androsace Russellii Y. Nasir: a critically endangered species in Gilgit District, Pakistan. Pak J Bot. 2010;42(3):1381–93.
Khasbagan S. Indigenous knowledge for plant species diversity: a case study of wild plants’ folk names used by the Mongolians in Ejina desert area, Inner Mongolia. PR China J Ethnobiol Ethnomed. 2008;4:2.
Article CAS PubMed Google Scholar
Ahmad M, Qureshi R, Arshad M, Khan MA, Zafar M. Traditional herbal remedies used for the treatment of diabetes from district Attock (Pakistan). Pak J Bot. 2009;41(6):2777–82.
Ullah R, Iqbal ZHZ, Hussain J, Khan FU, Khan N, Muhammad Z. Traditional uses of medicinal plants in Darra Adam Khel NWFP Pakistan. J Med Plants Res. 2010;4(17):1815–21.
Gurudeeban S, Ramanathan T. Antidiabetic effect of Citrullus colocynthis in alloxon-induced diabetic rats. Inven Rapid Ethno Pharmacol. 2010;1:112.
Hoda Q, Ahmad S, Akhtar M, Najmi AK, Pillai KK, Ahmad SJ. Antihyperglycaemic and antihyperlipidaemic effect of poly-constituents, in aqueous and chloroform extracts, of Withania coagulans Dunal in experimental type 2 diabetes mellitus in rats. Hum Exp Toxicol. 2010;29(8):653–8.
Shinwari MI, Khan MA. Folk use of medicinal herbs of Margalla hills national park. Islamabad J Ethnopharmacol. 2000;69(1):45–56.
Abbas G, Abbas Q, Khan SW, Hussain I, Najumal-ul-Hassan S. Medicinal plants diversity and their utilization in Gilgit region. Northern Pakistan.
Ashraf M, Hayat MQ, Jabeen S, Shaheen N, Khan MA, Yasmin G, Artemisia L. Species recognized by the local community of the northern areas of Pakistan as folk therapeutic plants. J Med Plants Res. 2010;4(2):112–9.
Murad W, Ahmad A, Ishaq G, Saleem KM, Muhammad KA, Ullah I. Ethnobotanical studies on plant resources of Hazar Nao forest, district Malakand, Pakistan. Pakistan J Weed Sci Res. 2012;18(4):509–27.
Khan SW, Khatoon S. Ethnobotanical studies on some useful herbs of Haramosh and Bugrote valleys in Gilgit, northern areas of Pakistan. Pakistan J Bot. 2008;40(1):43.
Mohammad I, Rahmatullah Q, Shinwari ZK, Muhammad A, Mirza SN. Some ethnoecological aspects of the plants of Qalagai hills, Kabal valley, swat. Pakistan Int J Agric Biol. 2013;15(5):801–10.
Jabeen N, Ajaib M, Siddiqui MF, Ulfat M, Khan B. A survey of ethnobotanically important plants of District Ghizer. Gilgit-Baltistan FUUAST J Biol. 2015;5(1):153–60.
Jan G, Khan MA, Jan F. Medicinal value of the Asteraceae of Dir Kohistan Valley, NWFP, Pakistan. Ethnobot Leafl. 2009;13:1205–15.
Ali H, Sannai J, Sher H, Rashid A. Ethnobotanical profile of some plant resources in Malam Jabba valley of Swat. Pakistan J Med Plants Res. 2011;5(18):4676–87.
Jan G, Khan MA, Farhatullah JFG, Ahmad M, Jan M, Zafar M. Ethnobotanical studies on some useful plants of Dir Kohistan valleys, KPK. Pakistan. Pak J Bot. 2011;43(4):1849–52.
Akhtar N, Rashid A, Murad W, Bergmeier E. Diversity and use of ethno-medicinal plants in the region of Swat, north Pakistan. J Ethnobiol Ethnomed. 2013;9(1):25.
Marwat SK. Ethnophytomedicines for treatment of various diseases in DI Khan district. Sarhad J Agric. 2008;24(2):305–15.
Ijaz F, Iqbal Z, Alam J, Khan SM, Afzal A, Rahman IU. Ethno medicinal study upon folk recipes against various human diseases in Sarban Hills, Abbottabad. Pakistan World J Zool. 2015;10(1):41–6.
Abbasi AM, Khan MA, Khan N, Shah MH. Ethnobotanical survey of medicinally important wild edible fruits species used by tribal communities of lesser Himalayas-Pakistan. J Ethnopharmacol. 2013;148(2):528–36.
Haq F, Ahmad H, Alam M. Traditional uses of medicinal plants of Nandiar Khuwarr catchment (district Battagram). Pakistan. J Med Plants Res. 2011;5(1):39–48.
Devi U, Seth MK, Sharma P, Rana JC. Study on ethnomedicinal plants of Kibber wildlife sanctuary: a cold desert in trans Himalaya. India J Med Plants Res. 2013;7(47):3400–19.
Alamgeer TA, Rashid M, Malik MNH, Mushtaq MN. Ethnomedicinal survey of plants of Valley Alladand Dehri, Tehsil Batkhela, District Malakand, Pakistan. Int J Basic Med Sci Pharm. 2013;3(1):23–32.
Khan B, Abdukadir A, Qureshi R, Mustafa G. Medicinal uses of plants by the inhabitants of Khunjerab National Park, Gilgit, Pakistan. Pak J Bot. 2011;43(5):2301–10.
Shah A, Marwat SK, Gohar F, Khan A, Bhatti KH, Amin M. Ethnobotanical study of medicinal plants of semi-tribal area of Makerwal & Gulla Khel (lying between Khyber Pakhtunkhwa and Punjab provinces), Pakistan. Am J Plant Sci. 2013;4(1):98.
Mosaddegh M, Naghibi F, Moazzeni H, Pirani A, Esmaeili S. Ethnobotanical survey of herbal remedies traditionally used in Kohghiluyeh va Boyer Ahmad province of Iran. J Ethnopharmacol. 2012;141(1):80–95.
Malik AH, Khuroo AA, Dar GH, Khan ZS. Ethnomedicinal uses of some plants in the Kashmir Himalaya. 2011.
Jouad H, Haloui M, Rhiouani H, El Hilaly J, Eddouks M. Ethnobotanical survey of medicinal plants used for the treatment of diabetes, cardiac and renal diseases in the north centre region of Morocco (Fez–Boulemane). J Ethnopharmacol. 2001;77(2–3):175–82.
Tumpa SI, Hossain MI, Ishika T. Ethnomedicinal uses of herbs by indigenous medicine practitioners of Jhenaidah district, Bangladesh. J Pharmacogn Phytochem. 2014;3(2):509–27.
Zari ST, Zari TA. A review of four common medicinal plants used to treat eczema. J Med Plants Res. 2015;9(24):702–11.
Dogan Y, Ugulu I. Medicinal plants used for gastrointestinal disorders in some districts of Izmir province, Turkey. Study Ethno-Medicine. 2013;7(3):149–61.
Shrivastava A, Chaturvedi U, Singh SV, Saxena JK, Bhatia G. A mechanism based pharmacological evaluation of efficacy of Allium sativum in regulation of dyslipidemia and oxidative stress in hyperlipidemic rats. Asian J Pharm Clin Res. 2012;5:123–6.
Zeraati F, Esna-Ashari F, Araghchian M, Emam AH, Rad MV, Seif S. Evaluation of topical antinociceptive effect of Artemisia absinthium extract in mice and possible mechanisms. African J Pharm Pharmacol. 2014;8(19):492–6.
Daradka HM, Abas MM, Mohammad MAM, Jaffar MM. Antidiabetic effect of Artemisia absinthium extracts on alloxan-induced diabetic rats. Comp Clin Path. 2014;23(6):1733–42.
Article CAS Google Scholar
Gliani AH, Janbaz KH. Hepatoprotective effects of Artemisia scoparia against carbon tetrachloride: an environmental contaminant. Journal-Pakistan Med Assoc. 1994;44:65.
Zhu X, Zhang W, Zhao J, Wang J, Qu W. Hypolipidaemic and hepatoprotective effects of ethanolic and aqueous extracts from Asparagus officinalis L. by-products in mice fed a high-fat diet. J Sci Food Agric. 2010;90(7):1129–35.
Mathur R, Gupta SK, Mathur SR, Velpandian T. Anti-tumor studies with extracts of Calotropis procera (Ait.) R. Br. Root employing Hep2 cells and their possible mechanism of action. 2009.
Basu A, Sen T, Pal S, Mascolo N, Capasso F, Nag Chaudhuri AK. Studies on the antiulcer activity of the chloroform fraction of Calotropis procera root extract. Phyther Res. 1997;11(2):163–5.
Zuardi AW, Crippa JAS, Hallak JEC, Moreira FA, Guimaraes FS. Cannabidiol, a Cannabis sativa constituent, as an antipsychotic drug. Brazilian J Med Biol Res. 2006;39(4):421–9.
Abdel-Sattar E, Harraz FM, Ghareib SA, Elberry AA, Gabr S, Suliaman MI. Antihyperglycaemic and hypolipidaemic effects of the methanolic extract of Caralluma tuberculata in streptozotocin-induced diabetic rats. Nat Prod Res. 2011;25(12):1171–9.
Ahmad S, Sharma R, Mahajan S, Gupta A. Antibacterial activity of Celtis australis by invitro study. 2012.
Pushparaj PN, Low HK, Manikandan J, Tan BKH, Tan CH. Anti-diabetic effects of Cichorium intybus in streptozotocin-induced diabetic rats. J Ethnopharmacol. 2007;111(2):430–4.
Vinaykumar T, Eswarkumar K, Roy H. Evaluation of antihyperglycemic activity of Citrullus colocynthis fruit pulp in streptozotocin induced diabetic rats.
Abdel-Hassan IA, Abdel-Barry JA, Mohammeda ST. The hypoglycaemic and antihyperglycaemic effect of Citrullus colocynthis fruit aqueous extract in normal and alloxan diabetic rabbits. J Ethnopharmacol. 2000;71(1–2):325–30.
Sebbagh N, Cruciani GC, Ouali F, Berthault MF, Rouch C, Sari DC. Comparative effects of Citrullus colocynthis, sunflower and olive oil-enriched diet in streptozotocin-induced diabetes in rats. Diabetes Metab. 2009;35(3):178–84.
Khan S, Wang Z, Wang R, Zhang L. Horizontoates AC. New cholinesterase inhibitors from Cotoneaster horizontalis. Phytochem Lett 2014;10:204–208.
Soni P, Siddiqui AA, Dwivedi J, Soni V. Pharmacological properties of Datura stramonium L. as a potential medicinal tree: an overview. Asian Pac J Trop Biomed. 2012;2(12):1002–8.
Alam S, Asad M, Asdaq SMB, Prasad VS. Antiulcer activity of methanolic extract of Momordica charantia L. in rats. J Ethnopharmacol. 2009;123(3):464–9.
Rashid R, Mukhtar F, Khan A. Antifungal and cytotoxic activities of Nannorrhops ritchiana roots extract. Acta Pol Pharm. 2014;71(5):789.
PubMed Google Scholar
Amin A, Khan MA, Shah S, Ahmad M, Zafar M, Hameed A. Inhibitory effects of Olea ferruginea crude leaves extract against some bacterial and fungal pathogen. Pak J Pharm Sci. 2013;26(2):251–54.
Melese E, Asres K, Asad M, Engidawork E. Evaluation of the antipeptic ulcer activity of the leaf extract of Plantago lanceolata L. in rodents. Phyther Res. 2011;25(8):1174–80.
Karimi G, Hosseinzadeh H, Ettehad N. Evaluation of the gastric antiulcerogenic effects of Portulaca oleracea L. extracts in mice. Phyther Res. 2004;18(6):484–7.
Jainu M, Devi CSS. Antiulcerogenic and ulcer healing effects of Solanum nigrum (L.) on experimental ulcer models: possible mechanism for the inhibition of acid formation. J Ethnopharmacol. 2006;104(1–2):156–63.
Rashid M, Mushtaq MN, Malik MNH, Ghumman SA, Numan M, Khan AQ. Pharmacological evaluation of antidiabetic effect of ethyl acetate extract of Teucrium stocksianum Boiss in alloxan-induced diabetic rabbits. JAPS, J Anim Plant Sci. 2013;23(2):436–9.
Rawal P, Adhikari R, Tiwari A. Antifungal activity of Viola canescens against fusarium oxysporum f. sp. lycopersici. Int J Curr Microbiol App Sci. 2015;4(5):1025–32.
PSV R. Phytochemical screening and in vitro antimicrobial investigation of the methanolic extract of Xanthium strumarium leaf. Int J Drug Dev Res. 2011;3(4):286–93.
Download references
Acknowledgements
This work is part of the Doctoral research work of the principal (first) author. The authors also acknowledge the participants for sharing their valuable information.
Authors’ contribution
WH conducted the collection of field data and wrote the initial draft of the manuscript. LB supervised the project. MU and MA helped in the field survey, sampling, and identification of taxon. AA and FH helped in the data analysis and revision of the manuscript. All the authors approved the final manuscript after revision.
Availability of data and materials
All the supporting data is available in Additional files 1 and 2 .
Author information
Authors and affiliations.
Department of Botany, University of Peshawar, Peshawar, 25000, Pakistan
Wahid Hussain & Lal Badshah
Department of Botany, University of Science and Technology, Bannu, Pakistan
Manzoor Ullah
Department of Plant Science, Quaid-i-Azam University, Islamabad, Pakistan
Dr. Khan Shaheed Govt. Degree College Kabal, Swat, Pakistan
Institute of Biological Sciences, Sarhad University of Science and Information Technology, Peshawar, Pakistan
Farrukh Hussain
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Maroof Ali .
Ethics declarations
Ethics approval and consent to participate.
Letters of permission were taken from Peshawar University and local administration office prior to the data collections. Oral agreements were also got from the local informants about the aims and objectives of the study prior to the interviews, and all the field data were collected through their oral consents. No further ethics approval was required.
Competing interests
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:.
Field data of the research project Quantitative study of medicinal plants used by the communities residing in Koh-e-Safaid Range northern Pakistani-Afghan border. (XLSX 167 kb)
Additional file 2:
Annexures. (DOCX 27 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated.
Reprints and permissions
About this article
Cite this article.
Hussain, W., Badshah, L., Ullah, M. et al. Quantitative study of medicinal plants used by the communities residing in Koh-e-Safaid Range, northern Pakistani-Afghan borders. J Ethnobiology Ethnomedicine 14 , 30 (2018). https://doi.org/10.1186/s13002-018-0229-4
Download citation
Received : 23 November 2017
Accepted : 05 April 2018
Published : 25 April 2018
DOI : https://doi.org/10.1186/s13002-018-0229-4

Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Quantitative study
- Medicinal plants
- Traditional knowledge
- Koh-e-Safaid Range
Journal of Ethnobiology and Ethnomedicine
ISSN: 1746-4269
- General enquiries: [email protected]

Medicinal and Aromatic Plants
Current Research Status, Value-Addition to Their Waste, and Agro-Industrial Potential (Vol I)
- Lakhan Kumar 0 ,
- Navneeta Bharadvaja 1 ,
- Ram Singh 2 ,
- Raksha Anand 3
Department of Biotechnology, Delhi Technological University, New Delhi, India
You can also search for this editor in PubMed Google Scholar
Department of Applied Chemistry, Delhi Technological University, New Delhi, India
- Compiles knowledge on recent progress, challenges, and way forward in medicinal plant research
- Presents qualitative and quantitative analysis of phytocompounds of industrial importance and their extraction
- Addresses the issue of management of waste plant parts post extraction as well post-harvest zero waste approach
Part of the book series: Sustainable Landscape Planning and Natural Resources Management (SLPNRM)
Buy print copy
- Durable hardcover edition
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
About this book
Due to complex phytochemical components and associated beneficial properties, numerous medicinal and aromatic plants, in whole or parts, have been used for nutritional purposes or the treatment of various diseases and disorders in humans and animals. Essential oils from medicinal and aromatic plants (MAPs) have been exploited for product formulations of pharmaceuticals, cosmetics, food and beverage, colorants, biopesticides, and several other utility chemicals of industrial importance. There is scientific evidence of many medicinal plant extracts possessing immunomodulatory, immunostimulatory, antidiabetic, anticarcinogenic, antimicrobial, and antioxidant properties, thus demonstrating their traditional use in popular medicine. With the advent of modern technology, the exploitation of natural resources has exponentially increased in order to fulfill the demand of an increased human population with improved quality of life. The traditional agriculture and production-based supply of commodities is inadequate to meet the current demand. Biotechnological approaches are gaining importance to bridge the gaps in demand and supply. In the proposed book, medicinal and aromatic plant-based secondary metabolites have been discussed in terms of their therapeutic potential and industrial relevance. To discuss the qualitative and quantitative analysis of a range of medicinal and aromatic plants-based secondary metabolites (SMs), bioprocess development for their extraction and bioseparation, a brief overview of their industrial relevance, various tissue culturing strategies, biotechnological approaches to enhance production, scale-up strategies, management of residual biomass post extraction of target SMs is central to the idea of the proposed book. A section will explore the verticals mentioned above. In the next section, the book addresses the approaches for conserving and improving medicinal and aromatic plant genetic resources. In the third section, approaches to managing the post-harvest crop residue and secondary metabolites extracted plant biomass will be thoroughly discussed. The recent integration of artificial intelligence to improve medicinal and aromatic plant research at several levels, including the development and employment of computational approaches to enhance secondary metabolite production, tissue culture, drug design and discovery, and disease treatment, will be included in the fourth section. The book summarizes current research status, gaps in knowledge, agro-industrial potential, waste or residual plant biomass management, conservation strategies, and computational approaches in the area of medicinal and aromatic plants with an aim to translate biotechnological interventions into reality.
- Biotechnological approaches
- Bioprocess development
- Computational approaches on MAPs
- Conservation of genetic resources
- Medicinal and aromatic plants
- Secondary metabolites
- Translational research
- Zero-waste approach
Editors and Affiliations
Lakhan Kumar, Navneeta Bharadvaja, Raksha Anand
About the editors
Lakhan Kumar works toward Environmental Sustainability. He completed his B.Tech. in Biotechnology from the National Institute of Technology, Jalandhar, and his M.Tech. in Industrial Biotechnology from Delhi Technological University, Delhi. He obtained his Ph.D. in Biotechnology from Delhi Technological University, Delhi, India. His areas of interest include bioenergy, bioprocess engineering, algal biorefinery, plant biotechnology, and remediation of environmental pollutants.
Navneeta Bharadvaja is working as an Assistant Professor at the Department of Biotechnology, Delhi Technological University, Delhi, India-110042. She is an accomplished plant biotechnologist. She has more than 16 years of Research and Teaching experience. She has guided 5 Ph.D. students and more than 100 B.Tech./M.Tech./M.Sc Students. She has published more than 60 peer-reviewed scientific articles in the fields of Medicinal and Aromatic Plants, Algal Biotechnology, Bioremediation, and Biofuels.
Ram Singh is currently working as a Professor at the Department of Applied Chemistry, Delhi Technological University, Delhi, India-110042. He has extensive experience in organic synthesis, plants, natural product chemistry, biomimetic chemistry, and chemical biology. He has published over 100 research papers in peer-reviewed journals, authored eight books, 20 book chapters, and 31 Modules for ePG-Pathshala, and contributed to more than 100 conferences. He has supervised 6 Ph.D. and 10 M.Tech students. His research has been funded by DST, CSIR, and DRDO, and he has carried out several projects in the area of natural product chemistry. He is on the Editorial Advisory Board of various journals of repute and is a Life Member of various societies.
Raksha Anand works on the development of algal nutraceuticals and waste and biomass valorization. She completed her B.Sc. (Hons.) degree in Biotechnology from the School of Basic Sciences and Research (SBSR), Sharda University, and her Master’s degree in Biotechnology from Delhi Technological University, Delhi. Her areas of interest include Nutraceuticals and Lifestyle Disease Management, Algal Biorefinery, Plant Biotechnology, and Environmental remediation. She has published several peer-reviewed articles and book chapters majorly in Nutraceuticals, Wastewater Treatment, Microbial Fuel Cells, and Bioremediation. She is editing a contributed book on algal-derived nutraceuticals.
Bibliographic Information
Book Title : Medicinal and Aromatic Plants
Book Subtitle : Current Research Status, Value-Addition to Their Waste, and Agro-Industrial Potential (Vol I)
Editors : Lakhan Kumar, Navneeta Bharadvaja, Ram Singh, Raksha Anand
Series Title : Sustainable Landscape Planning and Natural Resources Management
Publisher : Springer Cham
eBook Packages : Earth and Environmental Science , Earth and Environmental Science (R0)
Copyright Information : The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Switzerland AG 2024
Hardcover ISBN : 978-3-031-60116-3 Due: 23 July 2024
Softcover ISBN : 978-3-031-60119-4 Due: 23 July 2024
eBook ISBN : 978-3-031-60117-0 Due: 23 July 2024
Series ISSN : 2948-1910
Series E-ISSN : 2948-1929
Edition Number : 1
Number of Pages : XV, 407
Number of Illustrations : 88 b/w illustrations, 12 illustrations in colour
- Publish with us
Policies and ethics
- Find a journal
- Track your research
- Open access
- Published: 17 May 2024
The chromosome-level genome and functional database accelerate research about biosynthesis of secondary metabolites in Rosa roxburghii
- Jiaotong Yang 1 na1 ,
- Jingjie Zhang 1 na1 ,
- Hengyu Yan 2 ,
- Yahua Liu 1 ,
- Mian Zhang 1 ,
- Jun Li 1 &
- Qiaoqiao Xiao 1
BMC Plant Biology volume 24 , Article number: 410 ( 2024 ) Cite this article
36 Accesses
Metrics details
Rosa roxburghii Tratt, a valuable plant in China with long history, is famous for its fruit. It possesses various secondary metabolites, such as L-ascorbic acid (vitamin C), alkaloids and poly saccharides, which make it a high nutritional and medicinal value. Here we characterized the chromosome-level genome sequence of R. roxburghii , comprising seven pseudo-chromosomes with a total size of 531 Mb and a heterozygosity of 0.25%. We also annotated 45,226 coding gene loci after masking repeat elements. Orthologs for 90.1% of the Complete Single-Copy BUSCOs were found in the R. roxburghii annotation. By aligning with protein sequences from public platform, we annotated 85.89% genes from R. roxburghii . Comparative genomic analysis revealed that R. roxburghii diverged from Rosa chinensis approximately 5.58 to 13.17 million years ago, and no whole-genome duplication event occurred after the divergence from eudicots. To fully utilize this genomic resource, we constructed a genomic database RroFGD with various analysis tools. Otherwise, 69 enzyme genes involved in L-ascorbate biosynthesis were identified and a key enzyme in the biosynthesis of vitamin C, GDH (L-Gal-1-dehydrogenase), is used as an example to introduce the functions of the database. This genome and database will facilitate the future investigations into gene function and molecular breeding in R. roxburghii .
Peer Review reports
Introduction
Rosa roxburghii Tratt, which belongs to Rosaceae family, is a wild deciduous, perennial shrub. It is also named Cili in China because its golden fruit covered with tiny prickles. The fruit of R. roxburghii has a faint aroma with a bit sour and astringent taste, but it is famous for its nutritious and medical function [ 1 ]. R. roxburghii is wildly distributed in the alpine and hilly areas of southwest China, especially in Guizhou province with large temperature difference [ 2 ]. In China, in addition to wild R. roxburghii , several germplasm resources are cultured artificially, such as ‘Guinong 5’ (Rr-5) [ 3 ]. Since the medicinal and commercial value are discovered, the research on R. roxburghii has been more and more popular. R. roxburghii fruits contain various nutrients, such as carbohydrates, amino acids, vitamins, proteins, minerals and dietary fibers, which are benefit to our health [ 2 , 4 ]. Otherwise, active ingredients including superoxide dismutase (SOD), organic acids, polysaccharides, flavonoids, polyphenols, triterpenoids, glycosides etc., play important roles in medical function [ 2 ]. Several research have demonstrated that R. roxburghii poses the function of antioxidant, anti-tumor, anti-inflammatory, anti-radiation, anti-diabetes, anti-radiation and anti-aging [ 5 , 6 , 7 , 8 , 9 ]. All these functions depend on the active ingredients in the plant. Furthermore, R. roxburghii is widely used in the food industry, such as herbal tea, jam, vinegar, yoghurt and moon cake, which take R. roxburghii as raw materials to enhance the flavor of food [ 10 ]. Wide application of R. roxburghii in various field makes it a promising crop with broad market prospects.
Recently, long-read sequencing technology and high-throughput chromosome conformation capture technology make it possible to obtain the genome sequences of various plants [ 10 , 11 ]. Pu et al. assembled the chromosome level genome of Lonicera japonica [ 12 ]. Combination of genome and transcriptome analysis, they elucidated the molecular mechanism of dynamic flower color, which provided valuable genetic resources for molecular breeding and important clues for the evolution of L. japonica family [ 12 ]. Tu et al. constructed a high-quality Tripterygium wilfordii genome and found that cytochrome P450 participated in the metabolism of triptolide [ 13 ]. This important study contributed to elucidation the pathway of triptolide biosynthesis and further laid the foundation for the heterologous bioproduction of triptolide [ 13 ]. Zhang et al. completed whole genome assembly of Dendrobium chrysotoxum and comparative genomic analysis revealed molecular regulatory mechanisms such as medicinal components, flower color, flowering duration and stress tolerance, providing genetic basis for the medicinal and horticultural development of this plant [ 14 ]. Jiang et al. deciphered haplotype-resolved genome of Bletilla striata and revealed the mechanism of B. striata polysaccharide (BSP) biosynthesis [ 15 ]. Comparative genomic analysis further indicated that the expansion of B. striata gene family may played an important role in secondary metabolite biosynthesis and environmental adaptation [ 15 ]. These chromosome-scale genome information provided the basis for the heredity and evolution of species, and also laid the foundation for the study of gene function. However, there is a lack of a functional genomic database and its associated applications based on a high-quality genome assembly of R.roxburghii .
In our study, we assembled a chromosome-scale genome of R. roxburghii . Combined with genome and transcription data, we constructed a gene functional database, named RroFGD ( http://www.gzybioinformatics.cn/RroFGD ). In addition, we integrated basic local alignment search tool (Blast), extract sequence, gene set enrichment analysis (GSEA), heatmap and JBrowse analysis tools into the database for mining gene function. Using the database, we identified 69 key enzyme genes involved in ascorbate biosynthesis based on KEGG annotation information. We also took a vitamin C synthetase RrGDH as an example to introduce the functions of the database. The analysis results indicated that RrGDH gene might play an important role in the biosynthesis of L-ascorbate. The genome and database revealed here will provide reference not just for comprehension of R. roxburghii evolution but also for mining the gene function.
Materials and methods
Plant materials.
Plant materials of R. roxburghii seedling were collected in the wild of Hongfeng Lake Scenic Area, Qingzhen City, Guizhou, China (Fig. 1 A). The voucher specimen has been identified as R. roxburghii Tratt by Professor Chenggang Hu at Guizhou University of Traditional Chinese Medicine and is deposited at the Miao Medicine Museum of Guizhou University of Traditional Chinese Medicine (Collection Number: GZTM0220111).

R. roxburghii plant used for sequencing, assembled genome features, and synteny information. ( A ) R. roxburghii plant. ( B ) a represents chromosome number and relative length, b represents gene density in different chromosomes, c represents GC density in the genome, d represents repeat element density, and e represents synteny blocks of paralogous sequences
Illumina and ONT sequencing
We collected all the samples from an individual R. roxburghii. The sequencing data comprised four main components. The first part was second-generation DNA sequencing data, utilized for genome size assessment, which were sequenced using the illumina platform on leaf DNA. The second part was HIC data, employed for aiding genome scaffolding, also were sequenced using the illumina platform on leaf DNA. The third part was third-generation DNA sequencing data, utilized for genome assembly, which were sequenced using the ONT platform on high-quality leaf DNA. The fourth part was third-generation full-length transcriptome data, utilized for assisting gene structure annotation, which were sequenced using the ONT platform on high-quality RNA from mixed tissues including roots, stems, and leaves. All sequencing was performed in the Wuhan Bebagene Technology Co., Ltd (Wuhan, China).
Genome survey and assembly
After obtaining the illumina paired-end (2 × 150 bp) sequencing data, we measured the k-mer profile (k = 19) according to clean reads by Jellyfish software [ 16 ]. Genome size and heterozygosity were predicted basing on the k-mer profile by gce software (v1.0.2) [ 17 ]. ONT reads were directly assembled using necat software [ 18 ]. The assembled contigs were polished four times by nextPolish software (v1.3.1) [ 19 ], using illumina short reads.
Chromosomal genome assembly and chromatin interactions through Hi-C technology
First, R. roxburghii leaves were crosslinked using 1% formaldehyde. Then, cells were lysed using Dounce homogenizer method to release chromosomes and DNA. DNA was digested using DPNII, generating sticky ends. These ends were filled and labeled with biotin using DNA polymerase. The filled DNA ends were then ligated to form linked products. The biotin labeled Hi-C linked products were pulled down to generate a library suitable for sequencing. Finally, the library was sequenced using illumina sequencing technology. The contigs were clustered and arranged on pre-chromosomes based on the predicted intrachromosomal interaction information using 3d-DNA software [ 20 ]. Then we manually corrected the assembly using the Juicer-box software and recalculated interaction matrix of HIC by HiC-Pro [ 21 ]. Finally, the chromatin interaction heat map was drawn by HiCpotter [ 22 ]. The completeness of the genome assembly was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO) with the Embryophya odb 10 dataset (BUSCO; v5.2.2) [ 23 ].
Genome annotation
RepeatModeler (v2.0.1) [ 24 ] and RepeatMasker (v4.1.0) [ 25 ] were used to detect and annotate repeated elements within the genome of R. roxburghii . Next, we used second-generation, third-generation, homologous alignment evidence, and de novo prediction methods to annotate the gene structure of genome. Specific steps are as follows: (1) We firstly used second-generation transcriptome data (SRA number SRP167314) including leaves, stems, flowers and fruits for gene structure prediction. Then the transcriptome was aligned to the genome using the software hisat2 (version 2.1.0) [ 26 ], and the resulting transcript was reconstructed by stringtie (version 2.1.4) [ 27 ]. We further used the software TransDecoder (version: v5.1.0) to predict coding box of the predicted transcript region, and obtained the coding genes of the second-generation transcriptome prediction. (2) For the full-length transcriptome annotation, we used the software NanoFilt (version: 2.8.0) to conduct data filtering and the software Pychopper (version: v2.7.2) for full-length sequence identification. The resulting full-length sequence was self-corrected using racon (version: v1.4.21) [ 28 ] based on the original reads. We compared the corrected full-length sequence with the genome using minimap2 (version 2.17-r941) [ 29 ] and reconstructed transcript by stringtie (version 2.1.4). The software TransDecoder is used to predict the coding box of the predicted transcript region, and finally the predicted coding genes were obtained. (3) The homologous protein sequences were compared to the genome using the software tblastn [ 30 ], and then Exonerate (version v2.4.0) [ 31 ] was used to predict the transcript and coding region based on the comparison results. (4) Gene structures predicted by the second-generation transcriptome were performed model training and de novo prediction by Augustus (version: 3.3.2). (5) MAKER (v2.31.9) [ 32 ] was used to integrate the genetic annotation results predicted by various software.
For gene functional annotation, we aligned R. roxburghii protein sequence against protein databases such as Nr, Uniprot, Swissprot, and TAIR by diamond blastp software [ 33 ]. The Uniprot database recorded the protein families and their corresponding Gene Ontology (GO) annotations. Using the annotation information from the Uniprot database, GO annotations of the corresponding genes were extracted. The sequences were aligned against the KEGG database by diamond blastp. Then, KOBAS [ 34 ] was used to combined the sequences with KEGG Orthology and Pathway information based on ID mapping. The completeness of the genome annotation was also assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO) with the Embryophya odb 10 dataset (BUSCO; v5.2.2) [ 23 ].
Genome evolution analysis
Orthfinder [ 35 ] was used to cluster the amino acid sequences of R. roxburghii and 13 other angiosperms into orthologous groups. Using the RAxML package (v 8.1.13) [ 36 ], a maximum likelihood phylogenetic tree was constructed according to single-copy genes obtained from R. roxburghii and the 13 other angiosperms. Divergence time were estimated by the mcmctree program in the PAML software package [ 37 ]. The CAFÉ software (v5) [ 38 ] was used to detect gene family expansions and contractions. WGD software [ 39 ] was used to calculate the synonymous substitutions per synonymous site (ks) values between R. roxburghii and R. roxburghii , R. roxburghii and A.thinana , R. roxburghii and Glycine max, R. roxburghii and Malus domestica, R. roxburghii and Fragatia vesca . Syntenic blocks between R. roxburghi and R. roxburghii, R. roxburghii and Rose rugosa, R. roxburghii and Rose Chinensis were identified using MCScanX software [ 40 ]. Furthermore, we constructed a Circos map by Circos v0.52 [ 41 ] to display the genome information of R. roxburghii .
Construction of co-expression network
We downloaded transcriptome data samples from the SRA database and used the Hisat2 software [ 26 ] to map the downloaded transcriptome data to the reference genome of R. roxburghii . Then, we used stringtie software [ 42 ] to obtain the TPM values of each transcriptome sample and constructed an expression matrix. We calculated the correlation between gene expressions for every pair of genes using the PCC algorithm. After that, we ranked the gene correlations using the MR algorithm. Finally, we evaluated the network using ROC curves and selected an appropriate threshold to construct a co-expression network. The formula is as follows:
‘n’ represents the total number of samples present in the RNA-seq data. The variables ‘x’ and ‘y’ represent the TPM values. The term ‘Rank’ denotes the position of PCC values. Specifically, ‘AB’ signifies the ranking of gene A among all genes when compared to gene B, while ‘BA’ indicates the reverse ranking, i.e., the position of gene B when compared to gene A.
For assessment the reliability of network and establishment specific threshold values of both PCC and MR metrics, we identified Gene Ontology (GO) terms related to biological processes, specifically focusing on those with gene counts ranging from 4 to 20, which were designated as prior gene sets. Additionally, we selected co-expressed genes under the defined threshold to form other gene sets. Whether GO could be accurately in co-expression network gene pairs was used as input for a specific binary classifier and calculated the true positive rate (TPR) and false positive rate (FPR), then plotted the ROC curve. By comparing the sizes under the ROC curve (AUC) at various thresholds, we determined MR values that yielded the maximum AUC, representing the optimal cutoff to define the co-expression network.
Protein-protein interaction (PPI) network
The OrthoFinder software [ 35 ] was used to predict orthologous relationships between Arabidopsis and R. roxburghii . Subsequently, PPI network was mapped from Arabidopsis to R. roxburghii , establishing the PPI network in R. roxburghii .
Gene family identification
OrthoFinder [ 35 ] was used to predict the orthologous relationship of proteins between Arabidopsis and R. roxburghii . Subsequently, CAZy and TP were identified based on this orthologous relationship. The iTAK software [ 43 ] was utilized to identify and classify transcription factors, transcription regulators and protein kinases in R. roxburghii . A hidden Markov model obtained from iUUCD 2.0 [ 44 ] was used to identify ubiquitin families successfully in R. roxburghii . Functional annotation of CYP450 genes was performed based on the KEGG annotations.
Construction of RroFGD
Based on the LAMP (Linux, Apache, MySQL, PHP) technical stack, R. roxburghii functional genomics database was built. A MySQL database was created by importing various results and data analyses, such as gene structure annotations, gene functional annotations, co-expression network, PPI network, and gene family classification. To enhance data visualization and analysis, dynamic websites were developed using HTML, PHP, JavaScript, and CSS languages.
Toolkit for gene function analysis
We integrated Gene Set Enrichment Analysis (GSEA) [ 45 ] as previous descriptions [ 46 , 47 , 48 ]. ViroBlast [ 49 ] was used to achieve the sequence alignment function on line. We also incorporated JBrowse software [ 50 ] to display gene structure and RNA-seq mapping states. Furthermore, we introduced a sequence extraction tool using a Perl script and implemented a Heatmap analysis tool based on Highchart Javascript. These additions expanded the capabilities of the platform and improved the visualization and analysis of data.
Identification and analysis of genes involved in L-ascorbate biosynthesis
The L-ascorbate biosynthesis pathway involved 18 gene families, including HK, PGI, PMI, PMM, GMP, GME, GGP, GPP, GDH, GLDH, MDHAR, DHA, APX, GalUR, GulLO, ALase, GulDH and MIOX genes. To identify candidate genes related to the L-ascorbate biosynthesis pathways in R. roxburghii genome, we screened the data based on functional annotation information from the KEGG. According to co-expression network analysis, we identified transcription factors that showed co-expression relationships with the key enzyme genes. In addition, we selected apple, which belonged to the Rosaceae family as the input species and analyzed the binding sites of key enzyme gene promoters using the online tool PlantRegMap [ 51 ].
Genome sequencing and assembly of R. Roxburghii
The genome size of R. roxburghii was estimated to be 497.8 Mb by 19 k-mer distribution analysis, with a heterozygosity of 0.25% (Figure S1 ). Using the Oxford Nanopore PromethION Sequencer, we obtained 50.79Gb of ONT reads with an N50 of 34.8 kb by sequencing and the genome coverage was about 100×. ONT reads were subjected to assemble using necat software [ 18 ], the assembled contigs underwent four rounds of polishing using illumina short reads via nextPolish (v1.3.1) [ 19 ], leading to the construction of a scaffold assembly with an N50 of 13 Mb. The global mapping rate of the DNA illumina sequencing reads to the assembled reference genome was 93.53%. During the construction of the Hi-C sequencing, a total of 380,119,286 raw paired-end reads were generated, which facilitated the anchoring of 96.24% of the draft genome into 7 pseudo-chromosomes (2n = 14) using the 3d-DNA software. The strong intra-chromosomal interaction signal indicated that the Hi-C assembly was of high quality, as shown in Figure S2 . The final genome assembly of R. roxburghii was 531 Mb, which was at the chromosome level. Orthologs for 91.7% of the Complete Single-Copy BUSCOs were found in the R. roxburghii genome assembly.
Genome annotation and whole genome duplication
Transposable elements (TEs) accounted for approximately 61.41% of the R. roxburghii genome, and 38.88% of these TEs were long terminal repeat (LTR) elements (Fig. 1 B, Table S1 ). The R. roxburghii genome was predicted to contain 45,226 coding gene locis after masking repeat elements (Fig. 1 B, Table S2 ). Orthologs for 90.1% of the Complete Single-Copy BUSCOs were found in the R. roxburghii annotation (Table S3 ), indicating that the annotated genome was largely complete. By aligning protein sequences with the Nr, Uniprot, KEGG, swissprot, TAIR and KOG databases obtained the annotation of 38,768, 37,887, 9,344, 28,264, 24,200 and 197 genes with best matches, respectively. A total of 7,417 genes could be mapped to KEGG pathways. The corresponding Gene Ontology (GO) annotation information for 27,451 genes was predicted based on the protein family information recorded in the UniProt database. Additionally, the InterProScan software was used to predict the Pfam and InterPro domain information for 22,056 and 37,057 genes, respectively (Table S4 ).
Orthologous protein groups from 14 angiosperms were delineated, yielding a total of 37,126 orthologous groups that included 453,632 genes. By analyzing the divergence time between R. roxburghii and other species, we found that it diverged from Rose chinensis approximately 5.58 \(\sim\) 13.17 million years ago (mya) (Fig. 2 A). Our analysis indicated that there were 1,837 gene families expanded and 520 gene families contracted in the R. roxburghii lineage (Fig. 2 B). GO enrichment analysis of the expanded gene families revealed significant enrichment of terms related to diterpenoid biosynthetic process, defense response, enzyme activities in multiple function (Table S5 ). Similarly, KEGG enrichment analysis of the expanded gene families showed significant enrichment of terms associated with sesquiterpenoid and triterpenoid biosynthesis, steroid hormone biosynthesis, toluene degradation etc. (Table S6 ). Ks analysis indicated that R. roxburghii shared the eudicot-specific WGT event with any species during its evolutionary process, but no recently WGD occurred (Fig. 3 A). Previous studies had shown that Rosa rugosa and Rosa chinensis did not undergo species-specific whole-genome duplication events [ 52 ]. We conducted macro-collinearity analysis between R. roxburghii and Rosa chinensis , and the results indicated a conserved syntenic relationship between their chromosomes (Fig. 3 B), further supporting the absence of recently WGD in R. roxburghii .

R. roxburghii phylogeny and gene family variation. ( A ) Phylogenetic tree based on single copy genes from 14 plant species showed divergence time. The numbers represented estimated divergence times. ( B ) Variation in the number of gene families relative to the ancestral node. In the pie chart, the red color represented the number of expanded gene families, the green color represented the number of contracted gene families, and the blue color represented the number of gene families with no change
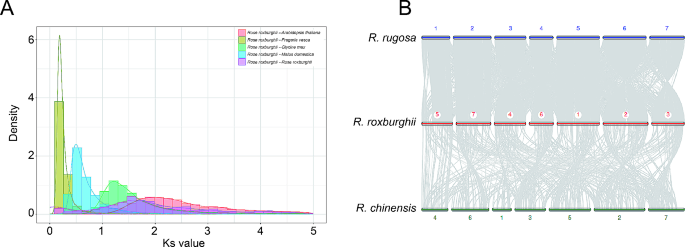
The whole genome duplication event and macro-collinearity analysis. ( A ) The synonymous substitutions per synonymous site (KS) distributions of orthologous and paralogous genes among A. thaliana , F. vesca , G. max, M. domestica and R . roxburghii . ( B ) Macro-collinearity analysis among R. roxburghii , R. rugosa and R. chinensis . The numbers in the chart represented chromosome identifiers
Transcriptome data including 33 samples was obtained from the SRA database in NCBI. These samples included two sets of project data (The SRA numbers are SRP448410 and SRP167314). The SRP448410 was used to study 18 samples of calcium absorption of R. roxburghii seedlings. The SRP167314 was 15 transcriptome samples from different tissues. These RNA-seq datasets were mapped to the reference genome, resulting in a mapping ratio exceeding 80% (Table S7 ). We examined the distribution of Pearson correlation coefficient (PCC) values derived from the expression profiles. Most gene pairs exhibited either no correlation or weak correlation in terms of their expression patterns (Fig. 4 A). To identify gene pairs with strong proximity within every two gene networks, we employed the MR (Matural Rank) approach based on their PCC ranking values.

Network construction and Gene family classification. ( A ) The relationship between Pearson correlation coefficient (PCC) and the number of edges in the co-expression network. ( B ) Statistical analysis of nodes and edges in the positive co-expression network, negative co-expression network, and ( C ) Protein-Protein Interaction (PPI) network. ( D ) Gene family classification information available
Positive co-expressed genes sharing common expression patterns likely contribute to analogous biological processes. Assessment of these shared biological processes can be achieved by GO annotations. The co-expression network’s reliability is heightened with greater similarity in the GO annotation of gene pairs. To bolster the trustworthiness of our constructed network, we integrated a pre-existing gene set grounded in Gene Ontology (GO) terms, specifically selecting 636 GO terms characterized by gene counts ranging from 4 to 20. We conducted mutual predictions of GO between co-expressed gene pairs. We used whether GO could be accurately predicted as input for a specific binary classifier and calculated the true positive rate (TPR) and false positive rate (FPR), then plotted the ROC curve. The larger the Area Under the Curve (AUC) values in the ROC curve, the higher the true positive rate (TPR) for accurately predicting GO, and the more reliable the co-expression network. Comparing AUC values for different MR (Mutual Rank) values with PCC (Pearson Correlation Coefficient) > 0.7, we observed the maximum AUC when MR < 40, prompting the establishment of a network threshold of PCC > 0.7 and MR < 40 for the positive co-expression network (Figure S3 ). The thresholds for the negative co-expression network were set at PCC<-0.7 and MR < 40. The co-expression network of R. roxburghii consisted of 673,980 gene pairs in the positive co-expression network and 357,935 gene pairs in the negative co-expression network (Fig. 4 B).
Protein–protein interaction network
By predicting the orthologous genes between Arabidopsi s and R. roxburghii , we mapped the protein-protein interaction (PPI) network of Arabidopsis onto R. roxburghii , and 31,571 pairs of PPI relationships, involving a total of 5,508 genes were identified (Fig. 4 C).
DEGs in different transcriptome
In order to incorporate gene co-expression and protein-protein interaction (PPI) networks with gene expression data, we performed differential expression analysis on the transcriptome data. SRP448410 descripted that seedlings were transferred to a 2 mmol-L-1 Ca (CH3COO)2 absorbent solution for cultivation. Throughout the calcium starvation period (0 min), rapid calcium uptake period (5 min), and calcium saturation period (6 h), the roots and leaves of R. roxburghii seedlings were sampled for transcriptome sequencing. We compared the transcriptomes between 15 groups at different time points and identified differentially expressed genes. SRP167314 includes transcriptome samples of root, leave, flower, young fruit (YF), and mature fruit. We compared 10 different transcriptomes and identified differentially expressed genes. Finally, we obtained 25 differentially expressed gene (DEG) groups (Table S8 ).
Gene family classification
We utilized the iTAK software to conduct analysis of transcription factors (TFs), transcription regulators (TRs), and protein kinases (PKs) in R. roxburghii . This analysis identified a total of 1,631 potential TFs, 396 TRs, and 1,440 PKs. Next, we employed a hidden Markov model (HMM) derived from the ubiquitin-proteasome dataset in the iUUCD v2.0 database to predict 1,217 genes participated in ubiquitin-proteasome system. Furthermore, based on the orthologous relationship between gene family specific database, we successfully identified 973 transprot protein coding genes, and 959 CAZy family genes. In addition, by KEGG annotation, we predicted 161 cytochrome P450 genes (Fig. 4 D). These analysis provided valuable insights into the transcriptional regulation, protein kinase activity, ubiquitin-proteasome system in R. roxburghii .
Database content
To enhance gene functional analysis in R. roxburghii , a comprehensive database called RroFGD has been developed ( http://www.gzybioinformatics.cn/RroFGD ). RroFGD consists of seven sections, namely Home, Network, Pathway, Tools, Gene Family, Download, and Help, each designed to improve usability and provide valuable insights for researchers. The Network section allowed access to both PPI and co-expression networks, enabling a deeper understanding of the intricate molecular interactions within R. roxburghii . To visualize the integration of these networks and DEGs (differentially expressed genes), a joint display node had been created. Within the network display, up-regulated DEGs were highlighted in red, while down-regulated DEGs were indicated in blue. This color-coded representation allowed for a clear distinction between the different expression patterns exhibited by the DEGs within the network.
Pathway section primarily consisted of gene annotations from the KEGG database. By clicking on the corresponding pathway, users could obtain the coding genes of all key enzymes in corresponding pathway. The Gene Family section encompassed various protein families, including CYP450, TF, TR, PK, TP, Ubiquitin and GAZy. These sections provided researchers with a comprehensive suite of tools for efficient gene functional analysis.
The Search tool allowed users to obtain interesting genes by using keywords, precise genes, transcript, or protein accession numbers. The Blast tool facilitated the screening of nucleic acid or protein sequences, identifying similarities within our database. GSEA (Gene Set Enrichment Analysis) provided an inclusive approach to gene set enrichment analysis. The Extract Sequence tool allowed for quick retrieval of gene sequences based on accession numbers and locations. Additionally, the Heatmap Analysis tool visually presented gene expression data, facilitating the interpretation of candidate gene lists. The integration of JBrowse provided an intuitive visualization of genomic and transcriptomic features, enhancing overall data exploration.
Download section provided convenient access to relevant information, ensuring easy retrieval of necessary resources. Furthermore, the help section offered a comprehensive user manual, guiding researchers how to use the RroFGD effectively.
Identification and analysis of key enzyme genes in ascorbate biosynthesis
First, we identified key enzyme genes related to ascorbate biosynthesis by previous search [ 53 , 54 ], such as PGI and PMI . Subsequently, we filtered these genes based on KEGG annotation information of R. roxburghii and further confirmed their relevance according to functional annotation information. We identified 69 key enzyme genes involved in ascorbate biosynthesis (Table S9 ), including 4 HK , 4 PGI , 4 PMI , 5 PMM , 3 GMP , 2 GME , 3 GGP , 1 GPP , 3 GDH , 2 GLDH , 9 MDHAR , 1 DHAR , 15 APX , 1 GMD , 2 GalUR , 3 GulLO , and 7 MIOX (Fig. 5 , Table S9 ). Additionally, transcriptomic data analysis based on different tissues (leaves, stems, flowers, immature fruits, mature fruits) revealed the expression patterns in different tissues. Previous study showed that the mature fruits of R. roxburghii had the highest content of vitamin C [ 54 ]. Transcriptomic data analysis revealed that some key enzyme genes were significantly higher expression in mature fruits compared to other tissues, including 2 HK , 2 PGI , and 1 GGP (Fig. 5 ).

L-ascorbate biosynthesis and expression heatmap of key enzyme genes in R. roxburghii . The red cross represented key enzymes that were not annotated in the current genome based on the corresponding gene, the red question mark represented key enzymes on the current pathway that were still unclear, and the red star represented key enzymes that were significantly higher expression in mature fruits compared to other tissues
By co-expression network analysis, we analyzed the regulatory genes of key enzyme genes involved in L-ascorbate biosynthesis and found that these key enzyme genes exhibited co-expression relationships with transcription factors, including AP2/ERF-ERF, SRS, C2H2, bZIP, HB-KNOX, C3H, SNF2, GRAS, CAMTA, PHD, B3, DDT, WRKY, TRAF, Trihelix, NAC, mTERF, ARID, HB-other, LOB, and so on (Fig. 6 A). Analysis of transcription factor binding sites in the 1 kb region of these key enzyme promoters demonstrated many binding sites for transcription factors, including 11 co-expressed transcription factor families. The analysis results were partially consistent with the co-expression analysis results (Fig. 6 B).

Gene co-expression network and cis-element analysis for key enzyme in L- ascorbate biosynthesis. ( A ) Gene co-expression network analysis of key enzyme genes in L-ascorbate biosynthesis. The dots in the inner circle represented key enzymes, while the dots in the outer circle represented transcription factors. ( B ) The number of transcription factor binding sites presented in the 1 kb promoter region of key enzymes coding genes. The horizontal axis represented transcription factors, and the vertical axis represented the number of transcription factor binding sites
Functional analysis of the RrGDH gene
GDH (Glutamate dehydrogenase) is one of the key enzymes involved in the synthesis of L-ascorbate [ 55 , 56 ]. A search based on RroFGD could provide detailed gene information for GDH. The gene Rroxscaffold_7G00214970 in R. roxburghii was identified as a member of L-galactose dehydrogenase (Fig. 7 A), located on chromosome 7 from 65,607,324 to 65,612,196 bp with transcript sequence (Fig. 7 B). Network links were also provided (Fig. 7 C). It was found that Aldo/keto reductase family domain was located at protein sequence of GDH (Fig. 7 D). KEGG annotation suggested that enzymes involved in L-ascorbate and aldarate metabolism and biosynthesis of secondary metabolites (Fig. 7 E and F). Studies had also shown that mature R. roxburghii fruits had the highest content of vitamin C (VC) [ 54 ]. According to expression profiling analysis, we found that the expression level of GDH was higher in mature fruit compared to leaves (Fig. 7 G). The display of reads mapping by JBrowse also revealed higher expression in leaves and stem (Fig. 8 A). The expression of GDH showed a similar trend to the biosynthesis and accumulation of active compounds. Therefore, the analysis results suggested that GDH might be involved in the accumulation of L-ascorbate biosynthesis.
Gene details of RrGDH in RroFGD. ( A ) Functional annotations. ( B ) Location and transcript sequences. ( C ) Links for network. ( D ) Protein structure. ( E ) KEGG pathway. ( F ) GO annotation and ( G ) Expression pattern of RrGDH

Expression and co-expression network analysis of RrGDH . ( A ) Expression level of RrGDH gene expression in leaf and mature fruit showing by JBrowse. ( B ) Positive co-expression gene network of RrGDH . The largest elliptical point represented the RrGDH gene. ( C ) Analysis of gene differential expression in the positive co-expression network when comparing mature fruit and leave transcriptomes. ( D ) Analysis of gene differential expression in the positive co-expression network when comparing mature fruit and stem transcriptomes. ( E ) Comparative analysis of RrGDH co-expressed genes expression in different transcriptome samples using heatmap analysis tool. The horizontal axis represented different tissues, while the vertical axis represents co-expressed genes of RrGDH
Furthermore, we conducted a co-expression analysis of RrGDH with its expression profiles. Network analysis revealed 35 genes that showed positive co-expression with RrGDH (Fig. 8 B). Additionally, many genes in the co-expression network were significantly upregulated in the mature fruit when compared with leaves or stems (Fig. 8 C, D and E). Therefore, our analysis suggested that the RrGDH gene played an important role in regulating biosynthesis of L-ascorbate.
R. roxburghii is a promising plant endemicity to the southern of China with medicinal and nutritional value. In recent years, the function of R. roxburghii has received wide attention and investigation [ 2 , 10 ]. However, the genome information is still unclear. In our study, we constructed a high-quality R. roxburghii genome and then clustered the contigs into seven pseudochromosomes according to Hi-C data. A total of 90.1% genes were examined by BUSCO, which indicated that genome annotation had relatively high integrality. This is of great significance for the further study of gene function and genetic variation of R. roxburghii . At the same time, this study also provides an important resource for analysis of the evolutionary history, genome structure and identification of functional genes. However, despite the remarkable progress, there are still some challenges, such as how to better deal with heterozygous genomes and improve the precision and continuity of assembly. Future work will continue to focus on this aspect to better interpret the genomic characteristics of R. roxburghii and its relationship to plant morphology and biological function. R. roxburghii belongs to the Rosaceae family, and its genome information provides genetic resources for the future comparative genomic analysis of the Rosaceae family. Similar to other Rosaceae plants, the R. roxburghii we reported has not undergone species-specific whole-genome duplication (WGD) events. Therefore, like grapes, R. roxburghii can be used as a comparative subject to determine whether other species have undergone whole-genome duplication.
R. roxburghii contains numerous active components, especially a high content of L-ascorbate [ 54 ]. Based on the known biosynthetic pathway of L-ascorbate, we analyzed the synthesis pathway in R. roxburghii and identified 69 candidate genes, which might participate in L-ascorbate biosynthesis. The analysis of public expression profile data indicated that certain genes, including 2 HK (Rroxscaffold_2G00149570, Rroxscaffold_7G00165330), 2 PGI (Rroxscaffold_2G00114450, Rroxscaffold_7G00167340) and 1 GGP (Rroxscaffold_1G00060490), exhibited significantly higher expression in mature fruits compared to other tissues. Due to the high content of L-ascorbate in mature fruits, these genes could serve as candidate genes in L-ascorbate biosynthesis for further research. According to the current version of the genome, only the L-galactose biosynthetic pathway was complete. While the Galacturonate, L-gulose, and myo-inositol pathways lacked the corresponding key enzymes, such as ALase for the Galacturonate and myo-inositol pathways, and GulDH for the L-gulose pathway. Additionally, there was no significant expansion of key enzymes in L-ascorbate biosynthesis. Therefore, the reason for the significantly higher L-ascorbate content in R. roxburghii remained to be further explored.
To fully utilize the genome resources of R. roxburghii , we developed a functional genomics database named RroFGD, aiming to provide researchers with a wide range of resources and tools to gain deeper insights into the functional genes and related biological processes. To further elucidate gene functions of R. roxburghii , our database offered various analysis tools. These tools contained gene set enrichment analysis (GSEA), local alignment search tool (Blast), extract sequence, heatmap and JBrowse. Users could use these tools according to their research needs to uncover the biological significance within the co-expression network. To facilitate effective utilization of the database, we provided the detailed usage example that demonstrated how to analyze functional genes in RroFGD.
The establishment of co-expression network and protein interaction network not only contributed to understand the interaction between genes, but also lay an important foundation for further study of the biological function and regulatory mechanism of R. roxburghii . By integrating with co-expression networks and offering various analysis tools along with detailed usage examples, we are committed to R. roxburghii research and provide valuable resources for researchers in related fields. Currently, the database relies on existing gene expression datasets, and how to ensure data quality and coverage remains a challenge. In the future, we plan to expand the scale and diversity of the dataset to provide more comprehensive and accurate analysis results. Additionally, we aim to refine and expand the analysis tools and functionalities of the database, involving continuous I mprovement and updating the latest advancements in the field of R. roxburghii .
The identification of differentially expressed genes helps us to understand the gene expression changes of R. roxburghii in different growth stages and different tissues. Especially in transcriptome samples from calcium absorption experiments, our analysis revealed that many gene expressions changed significantly, which might reflect the plant’s response to changes in calcium content in the environment. In transcriptome analysis of different tissues, we found differences in gene expression between roots, leaves, flowers, young and mature fruits. This gave us clues to further understand the functional differences between the different organs of R. roxburghii . The identification of the differentially expressed genes laid an important foundation for us to further explore the biological characteristics, functional genes and genes related to environmental response and adaptation of R. roxburghii . In addition, our study conducted a detailed classification and identification of the R. roxburghii gene family, and the results of these analyses provided important clues for us to further understand the genomic function and regulatory mechanism. The identification of transcription factors, transcription regulators and protein kinases contributed to understanding of the transcriptional regulatory network of genes. The prediction of ubiquitin-proteasome system provided important information for understanding protein degradation and regulation. The identified gene family and cytochrome P450 gene also provided important references for understanding the metabolic pathway and biosynthesis process of active ingredients in R. roxburghii .
Overall, in our research, we exhibited a high-quality genome of R. roxburghii . We further identified 69 candidate genes for the biosynthesis pathway of active compound of L-ascorbate. In order to facilitate usage, we constructed a functional database RroFGD for researchers to mine more gene function of R. roxburghii . The genomic resources and RroFGD will provide a solid foundation for the future research about R. roxburghii .
Data availability
The raw data of our project have been deposited in the Sequence Read Archive (SRA) in National Center for Biotechnology Information (NCBI) (Project ID: PRJNA1025299). The genome assembly and gff3 file reported in this paper have been deposited in the Genome Warehouse [57] in National Genomics Data Center [58], Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, under accession number GWHEROQ00000000.
Su J, Zhang B, Fu X, Huang Q, Li C, Liu G, Hai Liu R. Recent advances in polysaccharides from Rose Roxburghii Tratt fruits: isolation, structural characterization, and bioactivities. Food Funct. 2022;13(24):12561–71.
Article CAS PubMed Google Scholar
Wang L, Wei T, Zheng L, Jiang F, Ma W, Lu M, Wu X, An H. Recent advances on main active ingredients, pharmacological activities of Rosa Roxbughii and its development and utilization. Foods 2023, 12(5).
Jiang L, Lu M, Rao T, Liu Z, Wu X, An H. Comparative analysis of Fruit Metabolome using widely targeted Metabolomics reveals nutritional characteristics of different Rosa roxburghii genotypes. Foods 2022, 11(6).
Xu J, Vidyarthi SK, Bai W, Pan ZJJFF. Nutritional constituents, health benefits and processing of Rosa Roxburghii: A review. 2019, 60:103456.
Shen C, Wang Y, Zhang H, Li W, Chen W, Kuang M, Song Y, Zhong Z. Exploring the active components and potential mechanisms of Rosa roxburghii Tratt in treating type 2 diabetes mellitus based on UPLC-Q-exactive Orbitrap/MS and network pharmacology. Chin Med. 2023;18(1):12.
Article CAS PubMed PubMed Central Google Scholar
Chen Y, Liu ZJ, Liu J, Liu LK, Zhang ES, Li WL. Inhibition of metastasis and invasion of ovarian cancer cells by crude polysaccharides from rosa roxburghii tratt in vitro. Asian Pac J Cancer Prev. 2014;15(23):10351–4.
Article PubMed Google Scholar
van der Westhuizen FH, van Rensburg CS, Rautenbach GS, Marnewick JL, Loots du T, Huysamen C, Louw R, Pretorius PJ, Erasmus E. In vitro antioxidant, antimutagenic and genoprotective activity of Rosa roxburghii fruit extract. Phytother Res. 2008;22(3):376–83.
Wang L, Zhang B, Xiao J, Huang Q, Li C, Fu X. Physicochemical, functional, and biological properties of water-soluble polysaccharides from Rosa roxburghii Tratt fruit. Food Chem. 2018;249:127–35.
Wang L, Zhang P, Li C, Xu F, Chen J. A polysaccharide from Rosa roxburghii Tratt fruit attenuates high-fat diet-induced intestinal barrier dysfunction and inflammation in mice by modulating the gut microbiota. Food Funct. 2022;13(2):530–47.
Wang LT, Lv MJ, An JY, Fan XH, Dong MZ, Zhang SD, Wang JD, Wang YQ, Cai ZH, Fu YJ. Botanical characteristics, phytochemistry and related biological activities of Rosa roxburghii Tratt fruit, and its potential use in functional foods: a review. Food Funct. 2021;12(4):1432–51.
Lafontaine DL, Yang L, Dekker J, Gibcus JH. Hi-C 3.0: Improved Protocol for genome-wide chromosome conformation capture. Curr Protoc. 2021;1(7):e198.
Pu X, Li Z, Tian Y, Gao R, Hao L, Hu Y, He C, Sun W, Xu M, Peters RJ, et al. The honeysuckle genome provides insight into the molecular mechanism of carotenoid metabolism underlying dynamic flower coloration. New Phytol. 2020;227(3):930–43.
Tu L, Su P, Zhang Z, Gao L, Wang J, Hu T, Zhou J, Zhang Y, Zhao Y, Liu Y, et al. Genome of Tripterygium Wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nat Commun. 2020;11(1):971.
Zhang Y, Zhang GQ, Zhang D, Liu XD, Xu XY, Sun WH, Yu X, Zhu X, Wang ZW, Zhao X, et al. Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic Res. 2021;8(1):183.
Jiang L, Lin M, Wang H, Song H, Zhang L, Huang Q, Chen R, Song C, Li G, Cao Y. Haplotype-resolved genome assembly of Bletilla striata (Thunb.) Reichb.f. To elucidate medicinal value. Plant J. 2022;111(5):1340–53.
Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–70.
Liu B, Shi Y, Yuan J, Hu X, Zhang H, Li N, Li Z, Chen Y, Mu D, Fan WJQB. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. 2013, 35(s 1–3):62–67.
Chen Y, Nie F, Xie SQ, Zheng YF, Dai Q, Bray T, Wang YX, Xing JF, Huang ZJ, Wang DP, et al. Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat Commun. 2021;12(1):60.
Hu J, Fan J, Sun Z, Liu S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2020;36(7):2253–5.
Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356(6333):92–5.
Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259.
Article PubMed PubMed Central Google Scholar
Akdemir KC, Chin L. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 2015;16(1):198.
Manni M, Berkeley MR, Seppey M, Simao FA, Zdobnov EM. BUSCO Update: Novel and Streamlined Workflows along with broader and deeper phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and viral genomes. Mol Biol Evol. 2021;38(10):4647–54.
Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 2020;117(17):9451–7.
Tempel S. Using and understanding RepeatMasker. Methods Mol Biol. 2012;859:29–51.
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–15.
Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019;20(1):278.
Vaser R, Sovic I, Nagarajan N, Sikic M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27(5):737–46.
Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016;32(14):2103–10.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421.
Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31.
Huang X, Yan H, Zhai L, Yang Z, Yi Y. Characterization of the Rosa roxburghii Tratt transcriptome and analysis of MYB genes. PLoS ONE. 2019;14(3):e0203014.
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60.
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–322.
Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20(1):238.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3.
Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91.
De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–71.
Zwaenepoel A, Van de Peer Y. Wgd-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics. 2019;35(12):2153–5.
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45.
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–5.
Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ, et al. iTAK: a program for genome-wide prediction and Classification of Plant Transcription Factors, transcriptional regulators, and Protein Kinases. Mol Plant. 2016;9(12):1667–70.
Zhou J, Xu Y, Lin S, Guo Y, Deng W, Zhang Y, Guo A, Xue Y. iUUCD 2.0: an update with rich annotations for ubiquitin and ubiquitin-like conjugations. Nucleic Acids Res. 2018;46(D1):D447–53.
Yi X, Du Z, Su Z. PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 2013;41(Web Server issue):W98–103.
Yang J, Yan H, Liu Y, Da L, Xiao Q, Xu W, Su Z. GURFAP: a platform for gene function analysis in Glycyrrhiza Uralensis. Front Genet. 2022;13:823966.
Yu J, Zhang Z, Wei J, Ling Y, Xu W, Su Z. SFGD: a comprehensive platform for mining functional information from soybean transcriptome data and its use in identifying acyl-lipid metabolism pathways. BMC Genomics. 2014;15:271.
Yang J, Li P, Li Y, Xiao Q. GelFAP v2.0: an improved platform for Gene functional analysis in Gastrodia Elata. BMC Genomics. 2023;24(1):164.
Deng W, Nickle DC, Learn GH, Maust B, Mullins JI. ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics. 2007;23(17):2334–6.
Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L, et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016;17:66.
Tian F, Yang DC, Meng YQ, Jin J, Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48(D1):D1104–13.
CAS PubMed Google Scholar
Chen F, Su L, Hu S, Xue JY, Liu H, Liu G, Jiang Y, Du J, Qiao Y, Fan Y, et al. A chromosome-level genome assembly of rugged rose (Rosa rugosa) provides insights into its evolution, ecology, and floral characteristics. Hortic Res. 2021;8(1):141.
Lorence A, Chevone BI, Mendes P, Nessler CL. Myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004;134(3):1200–5.
Huang M, Xu Q, Deng XX. L-Ascorbic acid metabolism during fruit development in an ascorbate-rich fruit crop chestnut rose (Rosa roxburghii Tratt). J Plant Physiol. 2014;171(14):1205–16.
Vargas JA, Leonardo DA, D’Muniz Pereira H, Lopes AR, Rodriguez HN, Cobos M, Marapara JL, Castro JC, Garratt RC. Structural characterization of L-Galactose dehydrogenase: an Essential Enzyme for Vitamin C Biosynthesis. Plant Cell Physiol. 2022;63(8):1140–55.
Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393(6683):365–9.
Chen M, Ma Y, Wu S, Zheng X, Kang H, Sang J, Xu X, Hao L, Li Z, Gong Z, et al. Genome warehouse: a public Repository Housing genome-scale data. Genomics Proteom Bioinf. 2021;19(4):584–9.
Article Google Scholar
Members C-N, Partners. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2024. Nucleic Acids Res. 2024;52(D1):D18–32.
Download references
Acknowledgements
We would like to express our gratitude to Professor Chenggang Hu for his invaluable assistance in species identification.
This work was supported by the Guizhou University of Traditional Chinese Medicine Graduate Education Innovation Program (YCXJYS2023033), National Natural Science Foundation of China (NO.32160139 and NO.32260140), the University Science and Technology Innovation Team of the Guizhou Provincial Department of Education ([2023]071), the Guizhou Provincial Science and Technology Projects (ZK[2022]505), the National and Provincial Scientific and Technological Innovation Talent Team of the Guizhou University of Traditional Chinese Medicine (GZYTDHZ[2022]003), and Guizhou Provincial Basic Research Program (Natural Science) under Grant number Qianke He Foundation - ZK[2022] General 506.
Author information
Jiaotong Yang and Jingjie Zhang contributed equally to this work.
Authors and Affiliations
Resource Institute for Chinese and Ethnic Materia Medica, Guizhou University of Traditional Chinese Medicine, Guizhou, 550025, China
Jiaotong Yang, Jingjie Zhang, Qi Pan, Yahua Liu, Mian Zhang, Jun Li & Qiaoqiao Xiao
College of Agronomy, Qingdao Agricultural University, Qingdao, 266109, China
State Key Laboratory of Plant Diversity and Specialty Crops, Institute of Botany, The Chinese Academy of Sciences, Beijing, China
You can also search for this author in PubMed Google Scholar
Contributions
The study’s conception and design were conducted by Jiaotong Yang and Qiaoqiao Xiao. Jingjie Zhang completed the construction of the database. Yahua Liu, Mian Zhang, Jun Li sampled the materials for sequencing and helped in the preparation of the manuscript. Genome assembly and analysis were carried out by Jiaotong Yang, Hengyu Yan, Qi Pan and Xin Yi. The initial draft of the manuscript was authored by Jiaotong Yang, with input and feedback provided by all the authors on earlier versions of the paper. All authors have reviewed and endorsed the final version of the manuscript.
Corresponding authors
Correspondence to Jiaotong Yang or Qiaoqiao Xiao .
Ethics declarations
Ethics approval and consent to participate.
This study did not involve the use of human or animal tissue materials, as such, it did not necessitate ethical approval. The R. roxburghii plant in this research were sourced from Hongfeng Lake Scenic Area, Qingzhen City, Guizhou, China. The materials belong to private ownership, and we have obtained the necessary permissions for their collection. We declared that experimental material collection, steps and methods were in compliance with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary material 2, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Yang, J., Zhang, J., Yan, H. et al. The chromosome-level genome and functional database accelerate research about biosynthesis of secondary metabolites in Rosa roxburghii . BMC Plant Biol 24 , 410 (2024). https://doi.org/10.1186/s12870-024-05109-1
Download citation
Received : 20 November 2023
Accepted : 05 May 2024
Published : 17 May 2024
DOI : https://doi.org/10.1186/s12870-024-05109-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- R. Roxburghii
- Functional annotation
- Metabolites biosynthesis
BMC Plant Biology
ISSN: 1471-2229
- General enquiries: [email protected]

How To Grow Medicinal Plants At Home: In Pots, Terraces, And Backyard
G rowing medicinal plants at home can be a rewarding and sustainable way to have a readily available supply of natural remedies. Whether you have limited space and opt for pots, have a terrace where you can create a small garden, or enjoy a backyard with ample room, here's a guide on how to grow medicinal plants successfully:
1. Choose Suitable Medicinal Plants:
Research and select medicinal plants that are well-suited to your climate and growing conditions. Some popular options include aloe vera, peppermint, chamomile, lavender, rosemary, and calendula. Consider factors such as sunlight requirements, water needs, and available space when making your choices.
2. Select the Right Containers:
If space is limited, grow medicinal plants in pots or containers. Choose containers with proper drainage holes to prevent waterlogging. Opt for lightweight containers made of terracotta, plastic, or fabric. Ensure the containers are large enough to accommodate the plant's root system and allow room for growth.
3. Prepare the Potting Mix:
Create a well-draining potting mix by combining garden soil, compost, coco coir, and perlite or vermiculite. This mixture provides adequate nutrition and good drainage. Avoid using heavy garden soil alone, as it can hinder plant growth.
4. Provide Adequate Sunlight:
Most medicinal plants thrive with at least 6 hours of direct sunlight each day. Place your pots or containers in a location that receives ample sunlight, such as a sunny window, balcony, or terrace. If your available space has limited sunlight, consider using grow lights to supplement the light requirements.
5. Watering and Drainage:
Proper watering is crucial for medicinal plants. Water them when the top inch of soil feels dry, ensuring thorough watering to moisten the entire root system. Avoid overwatering, as it can lead to root rot. Check the drainage holes to prevent water from accumulating in the containers.
6. Prune and Harvest Regularly:
Regular pruning promotes bushier growth and improves air circulation around your medicinal plants. Harvest leaves, flowers, or roots when they are mature and at their peak potency. Follow proper harvesting techniques to ensure plant health and continuous production.
7. Organic Fertilization:
Nourish your medicinal plants with organic fertilizers. Apply compost, worm castings, or diluted organic liquid fertilizers according to the specific requirements of each plant. Be careful not to over-fertilize, as it can negatively impact plant growth and medicinal properties.
8. Pest and Disease Management:
Monitor your plants regularly for pests and diseases. Practice preventive measures such as maintaining good plant hygiene, providing adequate airflow, and spacing plants appropriately. Use organic pest control methods like neem oil, insecticidal soaps, or companion planting to manage pests naturally.
9. Understand Harvesting and Drying Techniques:
Learn about the proper harvesting and drying techniques for each medicinal plant. Some plants require specific methods to preserve their medicinal properties effectively. Harvest plant parts in the morning when essential oils are at their peak, and dry them in a well-ventilated area away from direct sunlight.
10. Research and Educate Yourself:
Continuously educate yourself about the specific medicinal properties and uses of each plant you grow. Research their optimal growing conditions, soil pH requirements, and potential interactions with other plants or medications. Stay updated with reliable sources and consult herbalists or experts for guidance.
Remember to exercise caution and consult a healthcare professional before using any medicinal plants for therapeutic purposes. Some plants may have contraindications or interactions with certain medical conditions or medications.
Growing medicinal plants at home not only provides you with a sustainable source of natural remedies but also offers a fulfilling gardening experience. Embrace the art of herbalism, foster a connection with nature, and enjoy the benefits of cultivating your own healing garden.


IMAGES
VIDEO
COMMENTS
Figure 5. Temporal evolution on medical plants publications for Top 12 countries. The first group is the leaders of this research, China and India, with between 800 and 1100 publications per year. China led the research from 1996 to 2010, and from this year to 2016, the leader was India, after which it returned to China.
Briefly, the research was done using medicinal plants in "article title, abstract, keywords" and then 175,818 documents were retrieved. The second step was the limitation of the results to ...
The Divine Farmer's Materia Medica, the earliest medical text in traditional Chinese medicine, written around ad 200, describes the medicinal and toxicological properties of 365 entries, most of ...
Medicinal plants have a promising future as there are about half a million plants worldwide and most of them have not yet been studied for their medical activities and their hidden potential for medical activities may be important in current and future study treatments (Singh 2015).Medical plants may have had a significant part in the evolution of human civilization, including in religion and ...
In 1998, Simmonds helped raise funds to create a 7,000-sample traditional Chinese medicine plant collection at Kew, and she currently leads a 300-strong research team focused on unlocking ...
Apart from being a founding member of American Council for Medicinally Active Plants (ACMAP), Dr. Parajuli is also a member of AACR and AAI. His research focuses on studying the molecular mechanisms of tumor-immune interaction, neuroinflammation as well as anti-tumor and immune modulatory activities of various medicinal plants.
Resources and conservation of medicinal plants. 2. Biosynthesis and metabolic engineering of medicinal plants. 3. Tissue culture, propagation and bioreactor technology of medicinal plants. 4. Phytochemical research on medicinal plants. 5. Herbal medicines and plant-derived agents in cancer prevention and therapy.
Current Trends in Medicinal Plant Research and Neurodegenerative Disorders. In natural product research, the ethnopharmacological approach is unique because it requires input from the cultural and social sciences. For the first time in 1967, the term "ethnopharmacology" was used as a book title " Ethnopharmacological Search for ...
The path-breaking development in research methodology and interdisciplinary scientific approaches in medicinal plant research is giving rise to innovative standpoints. Therefore, it becomes domineering to keep pace with the development of research and progress in the field of medicinal herbs. ... For planned papers, a title and short abstract ...
Investigations of the chemotaxonomy, molecular phylogeny, and pharmacology of these diverse plants and derived compounds through molecular biology and omics-based techniques have led to a new frontier of medicinal plant research, i.e., Medicinal Plant Biology. For example, improvements in sequencing technology—with drastically reduced costs ...
The residents of remote areas mostly depend on folk knowledge of medicinal plants to cure different ailments. The present study was carried out to document and analyze traditional use regarding the medicinal plants among communities residing in Koh-e-Safaid Range northern Pakistani-Afghan border. A purposive sampling method was used for the selection of informants, and information regarding ...
The use of medicinal plants has been done since ancient times and may even be considered the origin of modern medicine. Compounds of plant origin have been and still are an important source of ...
Medicinal plants. The bark of willow trees contains salicylic acid, the active metabolite of aspirin, and has been used for millennia to relieve pain and reduce fever. [1] Medicinal plants, also called medicinal herbs, have been discovered and used in traditional medicine practices since prehistoric times. Plants synthesize hundreds of chemical ...
Around the world, medicinal and aromatic plants (MAPs) play a fundamental role in the economic, social, cultural, and ecological ambits of local communities. Today, the most important uses of MAPs are their applications in the pharmaceutical, perfume, cosmetics, toothpaste, soap, beverage, and food industries. At the same time, the expression MAPs is often used with a plurality of meanings ...
The use of medicinal plants has been done since ancient times and may even be considered the origin of modern medicine. Compounds of plant origin have been and still are an important source of compounds for drugs. In this study a bibliometric study of all the works indexed in the Scopus database until 2019 has been carried out, analyzing more than 100,000 publications. On the one hand, the ...
Department of Biology for financial support to carry out the research. My special thanks also go to my field guide, Abebe Fikadu, who sacrificed his time and energy to ... Medicinal Plants of the Study Area ..... 22 4.3.1. Medicinal plants used to treat human ailments ..... 24 4.3.1.1. Sources of medicinal plants used to treat human ailments ...
@article{Fadeyi2024UtilisationOM, title={Utilisation of medicinal plants for antifertility activities: A bibliometric analysis of research endeavours from 1968 to 2023}, author={Opeyemi J. Fadeyi and Nneka A. Akwu and Makhotso Lekhooa and Rose Hayeshi and Adeyemi O. Aremu}, journal={Phytomedicine Plus}, year={2024}, url={https://api ...
The book summarizes current research status, gaps in knowledge, agro-industrial potential, waste or residual plant biomass management, conservation strategies, and computational approaches in the area of medicinal and aromatic plants with an aim to translate biotechnological interventions into reality.
Rosa roxburghii Tratt, a valuable plant in China with long history, is famous for its fruit. It possesses various secondary metabolites, such as L-ascorbic acid (vitamin C), alkaloids and poly saccharides, which make it a high nutritional and medicinal value. Here we characterized the chromosome-level genome sequence of R. roxburghii, comprising seven pseudo-chromosomes with a total size of ...
In recent years, there has been a growing interest in the use of medicinal plants and phytochemicals as potential treatments for acne vulgaris. This condition, characterized by chronic inflammation, predominantly affects adolescents and young adults. Conventional treatment typically targets the key factors contributing to its development: the proliferation of Cutibacterium acnes and the ...
The Suaq Balimbing research area spans 350 hectares (865 acres) of mainly peat swamp forest, and has since 1994 served as a site for noninvasive, almost exclusively observational research on ...
2) In 2021, researchers looked at how well an ointment formulation containing Lamium album extract healed wounds; the results were published in the Journal of Medicinal Plants. When tested on rats ...
Allium species are known for their culinary, medicinal, and ornamental purposes. Fusarium basal rot is one of the most damaging soilborne fungal diseases of Allium species and poses a significant threat to yield, quality, and storage life worldwide. Various species of Fusarium have been identified as causal agents for Fusarium basal rot, depending on the Allium species involved. Diverse ...
5. Watering and Drainage: Proper watering is crucial for medicinal plants. Water them when the top inch of soil feels dry, ensuring thorough watering to moisten the entire root system. Avoid ...