Get science-backed answers as you write with Paperpal's Research feature

What is Research Methodology? Definition, Types, and Examples

Research methodology 1,2 is a structured and scientific approach used to collect, analyze, and interpret quantitative or qualitative data to answer research questions or test hypotheses. A research methodology is like a plan for carrying out research and helps keep researchers on track by limiting the scope of the research. Several aspects must be considered before selecting an appropriate research methodology, such as research limitations and ethical concerns that may affect your research.
The research methodology section in a scientific paper describes the different methodological choices made, such as the data collection and analysis methods, and why these choices were selected. The reasons should explain why the methods chosen are the most appropriate to answer the research question. A good research methodology also helps ensure the reliability and validity of the research findings. There are three types of research methodology—quantitative, qualitative, and mixed-method, which can be chosen based on the research objectives.
What is research methodology ?
A research methodology describes the techniques and procedures used to identify and analyze information regarding a specific research topic. It is a process by which researchers design their study so that they can achieve their objectives using the selected research instruments. It includes all the important aspects of research, including research design, data collection methods, data analysis methods, and the overall framework within which the research is conducted. While these points can help you understand what is research methodology, you also need to know why it is important to pick the right methodology.
Why is research methodology important?
Having a good research methodology in place has the following advantages: 3
- Helps other researchers who may want to replicate your research; the explanations will be of benefit to them.
- You can easily answer any questions about your research if they arise at a later stage.
- A research methodology provides a framework and guidelines for researchers to clearly define research questions, hypotheses, and objectives.
- It helps researchers identify the most appropriate research design, sampling technique, and data collection and analysis methods.
- A sound research methodology helps researchers ensure that their findings are valid and reliable and free from biases and errors.
- It also helps ensure that ethical guidelines are followed while conducting research.
- A good research methodology helps researchers in planning their research efficiently, by ensuring optimum usage of their time and resources.
Writing the methods section of a research paper? Let Paperpal help you achieve perfection
Types of research methodology.
There are three types of research methodology based on the type of research and the data required. 1
- Quantitative research methodology focuses on measuring and testing numerical data. This approach is good for reaching a large number of people in a short amount of time. This type of research helps in testing the causal relationships between variables, making predictions, and generalizing results to wider populations.
- Qualitative research methodology examines the opinions, behaviors, and experiences of people. It collects and analyzes words and textual data. This research methodology requires fewer participants but is still more time consuming because the time spent per participant is quite large. This method is used in exploratory research where the research problem being investigated is not clearly defined.
- Mixed-method research methodology uses the characteristics of both quantitative and qualitative research methodologies in the same study. This method allows researchers to validate their findings, verify if the results observed using both methods are complementary, and explain any unexpected results obtained from one method by using the other method.
What are the types of sampling designs in research methodology?
Sampling 4 is an important part of a research methodology and involves selecting a representative sample of the population to conduct the study, making statistical inferences about them, and estimating the characteristics of the whole population based on these inferences. There are two types of sampling designs in research methodology—probability and nonprobability.
- Probability sampling
In this type of sampling design, a sample is chosen from a larger population using some form of random selection, that is, every member of the population has an equal chance of being selected. The different types of probability sampling are:
- Systematic —sample members are chosen at regular intervals. It requires selecting a starting point for the sample and sample size determination that can be repeated at regular intervals. This type of sampling method has a predefined range; hence, it is the least time consuming.
- Stratified —researchers divide the population into smaller groups that don’t overlap but represent the entire population. While sampling, these groups can be organized, and then a sample can be drawn from each group separately.
- Cluster —the population is divided into clusters based on demographic parameters like age, sex, location, etc.
- Convenience —selects participants who are most easily accessible to researchers due to geographical proximity, availability at a particular time, etc.
- Purposive —participants are selected at the researcher’s discretion. Researchers consider the purpose of the study and the understanding of the target audience.
- Snowball —already selected participants use their social networks to refer the researcher to other potential participants.
- Quota —while designing the study, the researchers decide how many people with which characteristics to include as participants. The characteristics help in choosing people most likely to provide insights into the subject.
What are data collection methods?
During research, data are collected using various methods depending on the research methodology being followed and the research methods being undertaken. Both qualitative and quantitative research have different data collection methods, as listed below.
Qualitative research 5
- One-on-one interviews: Helps the interviewers understand a respondent’s subjective opinion and experience pertaining to a specific topic or event
- Document study/literature review/record keeping: Researchers’ review of already existing written materials such as archives, annual reports, research articles, guidelines, policy documents, etc.
- Focus groups: Constructive discussions that usually include a small sample of about 6-10 people and a moderator, to understand the participants’ opinion on a given topic.
- Qualitative observation : Researchers collect data using their five senses (sight, smell, touch, taste, and hearing).
Quantitative research 6
- Sampling: The most common type is probability sampling.
- Interviews: Commonly telephonic or done in-person.
- Observations: Structured observations are most commonly used in quantitative research. In this method, researchers make observations about specific behaviors of individuals in a structured setting.
- Document review: Reviewing existing research or documents to collect evidence for supporting the research.
- Surveys and questionnaires. Surveys can be administered both online and offline depending on the requirement and sample size.
Let Paperpal help you write the perfect research methods section. Start now!
What are data analysis methods.
The data collected using the various methods for qualitative and quantitative research need to be analyzed to generate meaningful conclusions. These data analysis methods 7 also differ between quantitative and qualitative research.
Quantitative research involves a deductive method for data analysis where hypotheses are developed at the beginning of the research and precise measurement is required. The methods include statistical analysis applications to analyze numerical data and are grouped into two categories—descriptive and inferential.
Descriptive analysis is used to describe the basic features of different types of data to present it in a way that ensures the patterns become meaningful. The different types of descriptive analysis methods are:
- Measures of frequency (count, percent, frequency)
- Measures of central tendency (mean, median, mode)
- Measures of dispersion or variation (range, variance, standard deviation)
- Measure of position (percentile ranks, quartile ranks)
Inferential analysis is used to make predictions about a larger population based on the analysis of the data collected from a smaller population. This analysis is used to study the relationships between different variables. Some commonly used inferential data analysis methods are:
- Correlation: To understand the relationship between two or more variables.
- Cross-tabulation: Analyze the relationship between multiple variables.
- Regression analysis: Study the impact of independent variables on the dependent variable.
- Frequency tables: To understand the frequency of data.
- Analysis of variance: To test the degree to which two or more variables differ in an experiment.
Qualitative research involves an inductive method for data analysis where hypotheses are developed after data collection. The methods include:
- Content analysis: For analyzing documented information from text and images by determining the presence of certain words or concepts in texts.
- Narrative analysis: For analyzing content obtained from sources such as interviews, field observations, and surveys. The stories and opinions shared by people are used to answer research questions.
- Discourse analysis: For analyzing interactions with people considering the social context, that is, the lifestyle and environment, under which the interaction occurs.
- Grounded theory: Involves hypothesis creation by data collection and analysis to explain why a phenomenon occurred.
- Thematic analysis: To identify important themes or patterns in data and use these to address an issue.
How to choose a research methodology?
Here are some important factors to consider when choosing a research methodology: 8
- Research objectives, aims, and questions —these would help structure the research design.
- Review existing literature to identify any gaps in knowledge.
- Check the statistical requirements —if data-driven or statistical results are needed then quantitative research is the best. If the research questions can be answered based on people’s opinions and perceptions, then qualitative research is most suitable.
- Sample size —sample size can often determine the feasibility of a research methodology. For a large sample, less effort- and time-intensive methods are appropriate.
- Constraints —constraints of time, geography, and resources can help define the appropriate methodology.
Got writer’s block? Kickstart your research paper writing with Paperpal now!
How to write a research methodology .
A research methodology should include the following components: 3,9
- Research design —should be selected based on the research question and the data required. Common research designs include experimental, quasi-experimental, correlational, descriptive, and exploratory.
- Research method —this can be quantitative, qualitative, or mixed-method.
- Reason for selecting a specific methodology —explain why this methodology is the most suitable to answer your research problem.
- Research instruments —explain the research instruments you plan to use, mainly referring to the data collection methods such as interviews, surveys, etc. Here as well, a reason should be mentioned for selecting the particular instrument.
- Sampling —this involves selecting a representative subset of the population being studied.
- Data collection —involves gathering data using several data collection methods, such as surveys, interviews, etc.
- Data analysis —describe the data analysis methods you will use once you’ve collected the data.
- Research limitations —mention any limitations you foresee while conducting your research.
- Validity and reliability —validity helps identify the accuracy and truthfulness of the findings; reliability refers to the consistency and stability of the results over time and across different conditions.
- Ethical considerations —research should be conducted ethically. The considerations include obtaining consent from participants, maintaining confidentiality, and addressing conflicts of interest.
Streamline Your Research Paper Writing Process with Paperpal
The methods section is a critical part of the research papers, allowing researchers to use this to understand your findings and replicate your work when pursuing their own research. However, it is usually also the most difficult section to write. This is where Paperpal can help you overcome the writer’s block and create the first draft in minutes with Paperpal Copilot, its secure generative AI feature suite.
With Paperpal you can get research advice, write and refine your work, rephrase and verify the writing, and ensure submission readiness, all in one place. Here’s how you can use Paperpal to develop the first draft of your methods section.
- Generate an outline: Input some details about your research to instantly generate an outline for your methods section
- Develop the section: Use the outline and suggested sentence templates to expand your ideas and develop the first draft.
- P araph ras e and trim : Get clear, concise academic text with paraphrasing that conveys your work effectively and word reduction to fix redundancies.
- Choose the right words: Enhance text by choosing contextual synonyms based on how the words have been used in previously published work.
- Check and verify text : Make sure the generated text showcases your methods correctly, has all the right citations, and is original and authentic. .
You can repeat this process to develop each section of your research manuscript, including the title, abstract and keywords. Ready to write your research papers faster, better, and without the stress? Sign up for Paperpal and start writing today!
Frequently Asked Questions
Q1. What are the key components of research methodology?
A1. A good research methodology has the following key components:
- Research design
- Data collection procedures
- Data analysis methods
- Ethical considerations
Q2. Why is ethical consideration important in research methodology?
A2. Ethical consideration is important in research methodology to ensure the readers of the reliability and validity of the study. Researchers must clearly mention the ethical norms and standards followed during the conduct of the research and also mention if the research has been cleared by any institutional board. The following 10 points are the important principles related to ethical considerations: 10
- Participants should not be subjected to harm.
- Respect for the dignity of participants should be prioritized.
- Full consent should be obtained from participants before the study.
- Participants’ privacy should be ensured.
- Confidentiality of the research data should be ensured.
- Anonymity of individuals and organizations participating in the research should be maintained.
- The aims and objectives of the research should not be exaggerated.
- Affiliations, sources of funding, and any possible conflicts of interest should be declared.
- Communication in relation to the research should be honest and transparent.
- Misleading information and biased representation of primary data findings should be avoided.
Q3. What is the difference between methodology and method?
A3. Research methodology is different from a research method, although both terms are often confused. Research methods are the tools used to gather data, while the research methodology provides a framework for how research is planned, conducted, and analyzed. The latter guides researchers in making decisions about the most appropriate methods for their research. Research methods refer to the specific techniques, procedures, and tools used by researchers to collect, analyze, and interpret data, for instance surveys, questionnaires, interviews, etc.
Research methodology is, thus, an integral part of a research study. It helps ensure that you stay on track to meet your research objectives and answer your research questions using the most appropriate data collection and analysis tools based on your research design.
Accelerate your research paper writing with Paperpal. Try for free now!
- Research methodologies. Pfeiffer Library website. Accessed August 15, 2023. https://library.tiffin.edu/researchmethodologies/whatareresearchmethodologies
- Types of research methodology. Eduvoice website. Accessed August 16, 2023. https://eduvoice.in/types-research-methodology/
- The basics of research methodology: A key to quality research. Voxco. Accessed August 16, 2023. https://www.voxco.com/blog/what-is-research-methodology/
- Sampling methods: Types with examples. QuestionPro website. Accessed August 16, 2023. https://www.questionpro.com/blog/types-of-sampling-for-social-research/
- What is qualitative research? Methods, types, approaches, examples. Researcher.Life blog. Accessed August 15, 2023. https://researcher.life/blog/article/what-is-qualitative-research-methods-types-examples/
- What is quantitative research? Definition, methods, types, and examples. Researcher.Life blog. Accessed August 15, 2023. https://researcher.life/blog/article/what-is-quantitative-research-types-and-examples/
- Data analysis in research: Types & methods. QuestionPro website. Accessed August 16, 2023. https://www.questionpro.com/blog/data-analysis-in-research/#Data_analysis_in_qualitative_research
- Factors to consider while choosing the right research methodology. PhD Monster website. Accessed August 17, 2023. https://www.phdmonster.com/factors-to-consider-while-choosing-the-right-research-methodology/
- What is research methodology? Research and writing guides. Accessed August 14, 2023. https://paperpile.com/g/what-is-research-methodology/
- Ethical considerations. Business research methodology website. Accessed August 17, 2023. https://research-methodology.net/research-methodology/ethical-considerations/
Paperpal is a comprehensive AI writing toolkit that helps students and researchers achieve 2x the writing in half the time. It leverages 21+ years of STM experience and insights from millions of research articles to provide in-depth academic writing, language editing, and submission readiness support to help you write better, faster.
Get accurate academic translations, rewriting support, grammar checks, vocabulary suggestions, and generative AI assistance that delivers human precision at machine speed. Try for free or upgrade to Paperpal Prime starting at US$19 a month to access premium features, including consistency, plagiarism, and 30+ submission readiness checks to help you succeed.
Experience the future of academic writing – Sign up to Paperpal and start writing for free!
Related Reads:
- Dangling Modifiers and How to Avoid Them in Your Writing
- Webinar: How to Use Generative AI Tools Ethically in Your Academic Writing
- Research Outlines: How to Write An Introduction Section in Minutes with Paperpal Copilot
- How to Paraphrase Research Papers Effectively
Language and Grammar Rules for Academic Writing
Climatic vs. climactic: difference and examples, you may also like, how to write a high-quality conference paper, how paperpal is enhancing academic productivity and accelerating..., academic editing: how to self-edit academic text with..., 4 ways paperpal encourages responsible writing with ai, what are scholarly sources and where can you..., how to write a hypothesis types and examples , what is academic writing: tips for students, what is hedging in academic writing , how to use ai to enhance your college..., how to use paperpal to generate emails &....
What Is Research Methodology? A Plain-Language Explanation & Definition (With Examples)
By Derek Jansen (MBA) and Kerryn Warren (PhD) | June 2020 (Last updated April 2023)
If you’re new to formal academic research, it’s quite likely that you’re feeling a little overwhelmed by all the technical lingo that gets thrown around. And who could blame you – “research methodology”, “research methods”, “sampling strategies”… it all seems never-ending!
In this post, we’ll demystify the landscape with plain-language explanations and loads of examples (including easy-to-follow videos), so that you can approach your dissertation, thesis or research project with confidence. Let’s get started.
Research Methodology 101
- What exactly research methodology means
- What qualitative , quantitative and mixed methods are
- What sampling strategy is
- What data collection methods are
- What data analysis methods are
- How to choose your research methodology
- Example of a research methodology

What is research methodology?
Research methodology simply refers to the practical “how” of a research study. More specifically, it’s about how a researcher systematically designs a study to ensure valid and reliable results that address the research aims, objectives and research questions . Specifically, how the researcher went about deciding:
- What type of data to collect (e.g., qualitative or quantitative data )
- Who to collect it from (i.e., the sampling strategy )
- How to collect it (i.e., the data collection method )
- How to analyse it (i.e., the data analysis methods )
Within any formal piece of academic research (be it a dissertation, thesis or journal article), you’ll find a research methodology chapter or section which covers the aspects mentioned above. Importantly, a good methodology chapter explains not just what methodological choices were made, but also explains why they were made. In other words, the methodology chapter should justify the design choices, by showing that the chosen methods and techniques are the best fit for the research aims, objectives and research questions.
So, it’s the same as research design?
Not quite. As we mentioned, research methodology refers to the collection of practical decisions regarding what data you’ll collect, from who, how you’ll collect it and how you’ll analyse it. Research design, on the other hand, is more about the overall strategy you’ll adopt in your study. For example, whether you’ll use an experimental design in which you manipulate one variable while controlling others. You can learn more about research design and the various design types here .
Need a helping hand?
What are qualitative, quantitative and mixed-methods?
Qualitative, quantitative and mixed-methods are different types of methodological approaches, distinguished by their focus on words , numbers or both . This is a bit of an oversimplification, but its a good starting point for understanding.
Let’s take a closer look.
Qualitative research refers to research which focuses on collecting and analysing words (written or spoken) and textual or visual data, whereas quantitative research focuses on measurement and testing using numerical data . Qualitative analysis can also focus on other “softer” data points, such as body language or visual elements.
It’s quite common for a qualitative methodology to be used when the research aims and research questions are exploratory in nature. For example, a qualitative methodology might be used to understand peoples’ perceptions about an event that took place, or a political candidate running for president.
Contrasted to this, a quantitative methodology is typically used when the research aims and research questions are confirmatory in nature. For example, a quantitative methodology might be used to measure the relationship between two variables (e.g. personality type and likelihood to commit a crime) or to test a set of hypotheses .
As you’ve probably guessed, the mixed-method methodology attempts to combine the best of both qualitative and quantitative methodologies to integrate perspectives and create a rich picture. If you’d like to learn more about these three methodological approaches, be sure to watch our explainer video below.
What is sampling strategy?
Simply put, sampling is about deciding who (or where) you’re going to collect your data from . Why does this matter? Well, generally it’s not possible to collect data from every single person in your group of interest (this is called the “population”), so you’ll need to engage a smaller portion of that group that’s accessible and manageable (this is called the “sample”).
How you go about selecting the sample (i.e., your sampling strategy) will have a major impact on your study. There are many different sampling methods you can choose from, but the two overarching categories are probability sampling and non-probability sampling .
Probability sampling involves using a completely random sample from the group of people you’re interested in. This is comparable to throwing the names all potential participants into a hat, shaking it up, and picking out the “winners”. By using a completely random sample, you’ll minimise the risk of selection bias and the results of your study will be more generalisable to the entire population.
Non-probability sampling , on the other hand, doesn’t use a random sample . For example, it might involve using a convenience sample, which means you’d only interview or survey people that you have access to (perhaps your friends, family or work colleagues), rather than a truly random sample. With non-probability sampling, the results are typically not generalisable .
To learn more about sampling methods, be sure to check out the video below.
What are data collection methods?
As the name suggests, data collection methods simply refers to the way in which you go about collecting the data for your study. Some of the most common data collection methods include:
- Interviews (which can be unstructured, semi-structured or structured)
- Focus groups and group interviews
- Surveys (online or physical surveys)
- Observations (watching and recording activities)
- Biophysical measurements (e.g., blood pressure, heart rate, etc.)
- Documents and records (e.g., financial reports, court records, etc.)
The choice of which data collection method to use depends on your overall research aims and research questions , as well as practicalities and resource constraints. For example, if your research is exploratory in nature, qualitative methods such as interviews and focus groups would likely be a good fit. Conversely, if your research aims to measure specific variables or test hypotheses, large-scale surveys that produce large volumes of numerical data would likely be a better fit.
What are data analysis methods?
Data analysis methods refer to the methods and techniques that you’ll use to make sense of your data. These can be grouped according to whether the research is qualitative (words-based) or quantitative (numbers-based).
Popular data analysis methods in qualitative research include:
- Qualitative content analysis
- Thematic analysis
- Discourse analysis
- Narrative analysis
- Interpretative phenomenological analysis (IPA)
- Visual analysis (of photographs, videos, art, etc.)
Qualitative data analysis all begins with data coding , after which an analysis method is applied. In some cases, more than one analysis method is used, depending on the research aims and research questions . In the video below, we explore some common qualitative analysis methods, along with practical examples.
Moving on to the quantitative side of things, popular data analysis methods in this type of research include:
- Descriptive statistics (e.g. means, medians, modes )
- Inferential statistics (e.g. correlation, regression, structural equation modelling)
Again, the choice of which data collection method to use depends on your overall research aims and objectives , as well as practicalities and resource constraints. In the video below, we explain some core concepts central to quantitative analysis.
How do I choose a research methodology?
As you’ve probably picked up by now, your research aims and objectives have a major influence on the research methodology . So, the starting point for developing your research methodology is to take a step back and look at the big picture of your research, before you make methodology decisions. The first question you need to ask yourself is whether your research is exploratory or confirmatory in nature.
If your research aims and objectives are primarily exploratory in nature, your research will likely be qualitative and therefore you might consider qualitative data collection methods (e.g. interviews) and analysis methods (e.g. qualitative content analysis).
Conversely, if your research aims and objective are looking to measure or test something (i.e. they’re confirmatory), then your research will quite likely be quantitative in nature, and you might consider quantitative data collection methods (e.g. surveys) and analyses (e.g. statistical analysis).
Designing your research and working out your methodology is a large topic, which we cover extensively on the blog . For now, however, the key takeaway is that you should always start with your research aims, objectives and research questions (the golden thread). Every methodological choice you make needs align with those three components.
Example of a research methodology chapter
In the video below, we provide a detailed walkthrough of a research methodology from an actual dissertation, as well as an overview of our free methodology template .

Psst... there’s more!
This post was based on one of our popular Research Bootcamps . If you're working on a research project, you'll definitely want to check this out ...
You Might Also Like:

199 Comments
Thank you for this simple yet comprehensive and easy to digest presentation. God Bless!
You’re most welcome, Leo. Best of luck with your research!
I found it very useful. many thanks
This is really directional. A make-easy research knowledge.
Thank you for this, I think will help my research proposal
Thanks for good interpretation,well understood.
Good morning sorry I want to the search topic
Thank u more
Thank you, your explanation is simple and very helpful.
Very educative a.nd exciting platform. A bigger thank you and I’ll like to always be with you
That’s the best analysis
So simple yet so insightful. Thank you.
This really easy to read as it is self-explanatory. Very much appreciated…
Thanks for this. It’s so helpful and explicit. For those elements highlighted in orange, they were good sources of referrals for concepts I didn’t understand. A million thanks for this.
Good morning, I have been reading your research lessons through out a period of times. They are important, impressive and clear. Want to subscribe and be and be active with you.
Thankyou So much Sir Derek…
Good morning thanks so much for the on line lectures am a student of university of Makeni.select a research topic and deliberate on it so that we’ll continue to understand more.sorry that’s a suggestion.
Beautiful presentation. I love it.
please provide a research mehodology example for zoology
It’s very educative and well explained
Thanks for the concise and informative data.
This is really good for students to be safe and well understand that research is all about
Thank you so much Derek sir🖤🙏🤗
Very simple and reliable
This is really helpful. Thanks alot. God bless you.
very useful, Thank you very much..
thanks a lot its really useful
in a nutshell..thank you!
Thanks for updating my understanding on this aspect of my Thesis writing.
thank you so much my through this video am competently going to do a good job my thesis
Thanks a lot. Very simple to understand. I appreciate 🙏
Very simple but yet insightful Thank you
This has been an eye opening experience. Thank you grad coach team.
Very useful message for research scholars
Really very helpful thank you
yes you are right and i’m left
Research methodology with a simplest way i have never seen before this article.
wow thank u so much
Good morning thanks so much for the on line lectures am a student of university of Makeni.select a research topic and deliberate on is so that we will continue to understand more.sorry that’s a suggestion.
Very precise and informative.
Thanks for simplifying these terms for us, really appreciate it.
Thanks this has really helped me. It is very easy to understand.
I found the notes and the presentation assisting and opening my understanding on research methodology
Good presentation
Im so glad you clarified my misconceptions. Im now ready to fry my onions. Thank you so much. God bless
Thank you a lot.
thanks for the easy way of learning and desirable presentation.
Thanks a lot. I am inspired
Well written
I am writing a APA Format paper . I using questionnaire with 120 STDs teacher for my participant. Can you write me mthology for this research. Send it through email sent. Just need a sample as an example please. My topic is ” impacts of overcrowding on students learning
Thanks for your comment.
We can’t write your methodology for you. If you’re looking for samples, you should be able to find some sample methodologies on Google. Alternatively, you can download some previous dissertations from a dissertation directory and have a look at the methodology chapters therein.
All the best with your research.
Thank you so much for this!! God Bless
Thank you. Explicit explanation
Thank you, Derek and Kerryn, for making this simple to understand. I’m currently at the inception stage of my research.
Thnks a lot , this was very usefull on my assignment
excellent explanation
I’m currently working on my master’s thesis, thanks for this! I’m certain that I will use Qualitative methodology.
Thanks a lot for this concise piece, it was quite relieving and helpful. God bless you BIG…
I am currently doing my dissertation proposal and I am sure that I will do quantitative research. Thank you very much it was extremely helpful.
Very interesting and informative yet I would like to know about examples of Research Questions as well, if possible.
I’m about to submit a research presentation, I have come to understand from your simplification on understanding research methodology. My research will be mixed methodology, qualitative as well as quantitative. So aim and objective of mixed method would be both exploratory and confirmatory. Thanks you very much for your guidance.
OMG thanks for that, you’re a life saver. You covered all the points I needed. Thank you so much ❤️ ❤️ ❤️
Thank you immensely for this simple, easy to comprehend explanation of data collection methods. I have been stuck here for months 😩. Glad I found your piece. Super insightful.
I’m going to write synopsis which will be quantitative research method and I don’t know how to frame my topic, can I kindly get some ideas..
Thanks for this, I was really struggling.
This was really informative I was struggling but this helped me.
Thanks a lot for this information, simple and straightforward. I’m a last year student from the University of South Africa UNISA South Africa.
its very much informative and understandable. I have enlightened.
An interesting nice exploration of a topic.
Thank you. Accurate and simple🥰
This article was really helpful, it helped me understanding the basic concepts of the topic Research Methodology. The examples were very clear, and easy to understand. I would like to visit this website again. Thank you so much for such a great explanation of the subject.
Thanks dude
Thank you Doctor Derek for this wonderful piece, please help to provide your details for reference purpose. God bless.
Many compliments to you
Great work , thank you very much for the simple explanation
Thank you. I had to give a presentation on this topic. I have looked everywhere on the internet but this is the best and simple explanation.
thank you, its very informative.
Well explained. Now I know my research methodology will be qualitative and exploratory. Thank you so much, keep up the good work
Well explained, thank you very much.
This is good explanation, I have understood the different methods of research. Thanks a lot.
Great work…very well explanation
Thanks Derek. Kerryn was just fantastic!
Great to hear that, Hyacinth. Best of luck with your research!
Its a good templates very attractive and important to PhD students and lectuter
Thanks for the feedback, Matobela. Good luck with your research methodology.
Thank you. This is really helpful.
You’re very welcome, Elie. Good luck with your research methodology.
Well explained thanks
This is a very helpful site especially for young researchers at college. It provides sufficient information to guide students and equip them with the necessary foundation to ask any other questions aimed at deepening their understanding.
Thanks for the kind words, Edward. Good luck with your research!
Thank you. I have learned a lot.
Great to hear that, Ngwisa. Good luck with your research methodology!
Thank you for keeping your presentation simples and short and covering key information for research methodology. My key takeaway: Start with defining your research objective the other will depend on the aims of your research question.
My name is Zanele I would like to be assisted with my research , and the topic is shortage of nursing staff globally want are the causes , effects on health, patients and community and also globally
Thanks for making it simple and clear. It greatly helped in understanding research methodology. Regards.
This is well simplified and straight to the point
Thank you Dr
I was given an assignment to research 2 publications and describe their research methodology? I don’t know how to start this task can someone help me?
Sure. You’re welcome to book an initial consultation with one of our Research Coaches to discuss how we can assist – https://gradcoach.com/book/new/ .
Thanks a lot I am relieved of a heavy burden.keep up with the good work
I’m very much grateful Dr Derek. I’m planning to pursue one of the careers that really needs one to be very much eager to know. There’s a lot of research to do and everything, but since I’ve gotten this information I will use it to the best of my potential.
Thank you so much, words are not enough to explain how helpful this session has been for me!
Thanks this has thought me alot.
Very concise and helpful. Thanks a lot
Thank Derek. This is very helpful. Your step by step explanation has made it easier for me to understand different concepts. Now i can get on with my research.
I wish i had come across this sooner. So simple but yet insightful
really nice explanation thank you so much
I’m so grateful finding this site, it’s really helpful…….every term well explained and provide accurate understanding especially to student going into an in-depth research for the very first time, even though my lecturer already explained this topic to the class, I think I got the clear and efficient explanation here, much thanks to the author.
It is very helpful material
I would like to be assisted with my research topic : Literature Review and research methodologies. My topic is : what is the relationship between unemployment and economic growth?
Its really nice and good for us.
THANKS SO MUCH FOR EXPLANATION, ITS VERY CLEAR TO ME WHAT I WILL BE DOING FROM NOW .GREAT READS.
Short but sweet.Thank you
Informative article. Thanks for your detailed information.
I’m currently working on my Ph.D. thesis. Thanks a lot, Derek and Kerryn, Well-organized sequences, facilitate the readers’ following.
great article for someone who does not have any background can even understand
I am a bit confused about research design and methodology. Are they the same? If not, what are the differences and how are they related?
Thanks in advance.
concise and informative.
Thank you very much
How can we site this article is Harvard style?
Very well written piece that afforded better understanding of the concept. Thank you!
Am a new researcher trying to learn how best to write a research proposal. I find your article spot on and want to download the free template but finding difficulties. Can u kindly send it to my email, the free download entitled, “Free Download: Research Proposal Template (with Examples)”.
Thank too much
Thank you very much for your comprehensive explanation about research methodology so I like to thank you again for giving us such great things.
Good very well explained.Thanks for sharing it.
Thank u sir, it is really a good guideline.
so helpful thank you very much.
Thanks for the video it was very explanatory and detailed, easy to comprehend and follow up. please, keep it up the good work
It was very helpful, a well-written document with precise information.
how do i reference this?
MLA Jansen, Derek, and Kerryn Warren. “What (Exactly) Is Research Methodology?” Grad Coach, June 2021, gradcoach.com/what-is-research-methodology/.
APA Jansen, D., & Warren, K. (2021, June). What (Exactly) Is Research Methodology? Grad Coach. https://gradcoach.com/what-is-research-methodology/
Your explanation is easily understood. Thank you
Very help article. Now I can go my methodology chapter in my thesis with ease
I feel guided ,Thank you
This simplification is very helpful. It is simple but very educative, thanks ever so much
The write up is informative and educative. It is an academic intellectual representation that every good researcher can find useful. Thanks
Wow, this is wonderful long live.
Nice initiative
thank you the video was helpful to me.
Thank you very much for your simple and clear explanations I’m really satisfied by the way you did it By now, I think I can realize a very good article by following your fastidious indications May God bless you
Thanks very much, it was very concise and informational for a beginner like me to gain an insight into what i am about to undertake. I really appreciate.
very informative sir, it is amazing to understand the meaning of question hidden behind that, and simple language is used other than legislature to understand easily. stay happy.
This one is really amazing. All content in your youtube channel is a very helpful guide for doing research. Thanks, GradCoach.
research methodologies
Please send me more information concerning dissertation research.
Nice piece of knowledge shared….. #Thump_UP
This is amazing, it has said it all. Thanks to Gradcoach
This is wonderful,very elaborate and clear.I hope to reach out for your assistance in my research very soon.
This is the answer I am searching about…
realy thanks a lot
Thank you very much for this awesome, to the point and inclusive article.
Thank you very much I need validity and reliability explanation I have exams
Thank you for a well explained piece. This will help me going forward.
Very simple and well detailed Many thanks
This is so very simple yet so very effective and comprehensive. An Excellent piece of work.
I wish I saw this earlier on! Great insights for a beginner(researcher) like me. Thanks a mil!
Thank you very much, for such a simplified, clear and practical step by step both for academic students and general research work. Holistic, effective to use and easy to read step by step. One can easily apply the steps in practical terms and produce a quality document/up-to standard
Thanks for simplifying these terms for us, really appreciated.
Thanks for a great work. well understood .
This was very helpful. It was simple but profound and very easy to understand. Thank you so much!
Great and amazing research guidelines. Best site for learning research
hello sir/ma’am, i didn’t find yet that what type of research methodology i am using. because i am writing my report on CSR and collect all my data from websites and articles so which type of methodology i should write in dissertation report. please help me. i am from India.
how does this really work?
perfect content, thanks a lot
As a researcher, I commend you for the detailed and simplified information on the topic in question. I would like to remain in touch for the sharing of research ideas on other topics. Thank you
Impressive. Thank you, Grad Coach 😍
Thank you Grad Coach for this piece of information. I have at least learned about the different types of research methodologies.
Very useful content with easy way
Thank you very much for the presentation. I am an MPH student with the Adventist University of Africa. I have successfully completed my theory and starting on my research this July. My topic is “Factors associated with Dental Caries in (one District) in Botswana. I need help on how to go about this quantitative research
I am so grateful to run across something that was sooo helpful. I have been on my doctorate journey for quite some time. Your breakdown on methodology helped me to refresh my intent. Thank you.
thanks so much for this good lecture. student from university of science and technology, Wudil. Kano Nigeria.
It’s profound easy to understand I appreciate
Thanks a lot for sharing superb information in a detailed but concise manner. It was really helpful and helped a lot in getting into my own research methodology.
Comment * thanks very much
This was sooo helpful for me thank you so much i didn’t even know what i had to write thank you!
You’re most welcome 🙂
Simple and good. Very much helpful. Thank you so much.
This is very good work. I have benefited.
Thank you so much for sharing
This is powerful thank you so much guys
I am nkasa lizwi doing my research proposal on honors with the university of Walter Sisulu Komani I m on part 3 now can you assist me.my topic is: transitional challenges faced by educators in intermediate phase in the Alfred Nzo District.
Appreciate the presentation. Very useful step-by-step guidelines to follow.
I appreciate sir
wow! This is super insightful for me. Thank you!
Indeed this material is very helpful! Kudos writers/authors.
I want to say thank you very much, I got a lot of info and knowledge. Be blessed.
I want present a seminar paper on Optimisation of Deep learning-based models on vulnerability detection in digital transactions.
Need assistance
Dear Sir, I want to be assisted on my research on Sanitation and Water management in emergencies areas.
I am deeply grateful for the knowledge gained. I will be getting in touch shortly as I want to be assisted in my ongoing research.
The information shared is informative, crisp and clear. Kudos Team! And thanks a lot!
hello i want to study
Hello!! Grad coach teams. I am extremely happy in your tutorial or consultation. i am really benefited all material and briefing. Thank you very much for your generous helps. Please keep it up. If you add in your briefing, references for further reading, it will be very nice.
All I have to say is, thank u gyz.
Good, l thanks
thank you, it is very useful
Trackbacks/Pingbacks
- What Is A Literature Review (In A Dissertation Or Thesis) - Grad Coach - […] the literature review is to inform the choice of methodology for your own research. As we’ve discussed on the Grad Coach blog,…
- Free Download: Research Proposal Template (With Examples) - Grad Coach - […] Research design (methodology) […]
- Dissertation vs Thesis: What's the difference? - Grad Coach - […] and thesis writing on a daily basis – everything from how to find a good research topic to which…
Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly
- USC Libraries
- Research Guides
Organizing Your Social Sciences Research Paper
- 6. The Methodology
- Purpose of Guide
- Design Flaws to Avoid
- Independent and Dependent Variables
- Glossary of Research Terms
- Reading Research Effectively
- Narrowing a Topic Idea
- Broadening a Topic Idea
- Extending the Timeliness of a Topic Idea
- Academic Writing Style
- Applying Critical Thinking
- Choosing a Title
- Making an Outline
- Paragraph Development
- Research Process Video Series
- Executive Summary
- The C.A.R.S. Model
- Background Information
- The Research Problem/Question
- Theoretical Framework
- Citation Tracking
- Content Alert Services
- Evaluating Sources
- Primary Sources
- Secondary Sources
- Tiertiary Sources
- Scholarly vs. Popular Publications
- Qualitative Methods
- Quantitative Methods
- Insiderness
- Using Non-Textual Elements
- Limitations of the Study
- Common Grammar Mistakes
- Writing Concisely
- Avoiding Plagiarism
- Footnotes or Endnotes?
- Further Readings
- Generative AI and Writing
- USC Libraries Tutorials and Other Guides
- Bibliography
The methods section describes actions taken to investigate a research problem and the rationale for the application of specific procedures or techniques used to identify, select, process, and analyze information applied to understanding the problem, thereby, allowing the reader to critically evaluate a study’s overall validity and reliability. The methodology section of a research paper answers two main questions: How was the data collected or generated? And, how was it analyzed? The writing should be direct and precise and always written in the past tense.
Kallet, Richard H. "How to Write the Methods Section of a Research Paper." Respiratory Care 49 (October 2004): 1229-1232.
Importance of a Good Methodology Section
You must explain how you obtained and analyzed your results for the following reasons:
- Readers need to know how the data was obtained because the method you chose affects the results and, by extension, how you interpreted their significance in the discussion section of your paper.
- Methodology is crucial for any branch of scholarship because an unreliable method produces unreliable results and, as a consequence, undermines the value of your analysis of the findings.
- In most cases, there are a variety of different methods you can choose to investigate a research problem. The methodology section of your paper should clearly articulate the reasons why you have chosen a particular procedure or technique.
- The reader wants to know that the data was collected or generated in a way that is consistent with accepted practice in the field of study. For example, if you are using a multiple choice questionnaire, readers need to know that it offered your respondents a reasonable range of answers to choose from.
- The method must be appropriate to fulfilling the overall aims of the study. For example, you need to ensure that you have a large enough sample size to be able to generalize and make recommendations based upon the findings.
- The methodology should discuss the problems that were anticipated and the steps you took to prevent them from occurring. For any problems that do arise, you must describe the ways in which they were minimized or why these problems do not impact in any meaningful way your interpretation of the findings.
- In the social and behavioral sciences, it is important to always provide sufficient information to allow other researchers to adopt or replicate your methodology. This information is particularly important when a new method has been developed or an innovative use of an existing method is utilized.
Bem, Daryl J. Writing the Empirical Journal Article. Psychology Writing Center. University of Washington; Denscombe, Martyn. The Good Research Guide: For Small-Scale Social Research Projects . 5th edition. Buckingham, UK: Open University Press, 2014; Lunenburg, Frederick C. Writing a Successful Thesis or Dissertation: Tips and Strategies for Students in the Social and Behavioral Sciences . Thousand Oaks, CA: Corwin Press, 2008.
Structure and Writing Style
I. Groups of Research Methods
There are two main groups of research methods in the social sciences:
- The e mpirical-analytical group approaches the study of social sciences in a similar manner that researchers study the natural sciences . This type of research focuses on objective knowledge, research questions that can be answered yes or no, and operational definitions of variables to be measured. The empirical-analytical group employs deductive reasoning that uses existing theory as a foundation for formulating hypotheses that need to be tested. This approach is focused on explanation.
- The i nterpretative group of methods is focused on understanding phenomenon in a comprehensive, holistic way . Interpretive methods focus on analytically disclosing the meaning-making practices of human subjects [the why, how, or by what means people do what they do], while showing how those practices arrange so that it can be used to generate observable outcomes. Interpretive methods allow you to recognize your connection to the phenomena under investigation. However, the interpretative group requires careful examination of variables because it focuses more on subjective knowledge.
II. Content
The introduction to your methodology section should begin by restating the research problem and underlying assumptions underpinning your study. This is followed by situating the methods you used to gather, analyze, and process information within the overall “tradition” of your field of study and within the particular research design you have chosen to study the problem. If the method you choose lies outside of the tradition of your field [i.e., your review of the literature demonstrates that the method is not commonly used], provide a justification for how your choice of methods specifically addresses the research problem in ways that have not been utilized in prior studies.
The remainder of your methodology section should describe the following:
- Decisions made in selecting the data you have analyzed or, in the case of qualitative research, the subjects and research setting you have examined,
- Tools and methods used to identify and collect information, and how you identified relevant variables,
- The ways in which you processed the data and the procedures you used to analyze that data, and
- The specific research tools or strategies that you utilized to study the underlying hypothesis and research questions.
In addition, an effectively written methodology section should:
- Introduce the overall methodological approach for investigating your research problem . Is your study qualitative or quantitative or a combination of both (mixed method)? Are you going to take a special approach, such as action research, or a more neutral stance?
- Indicate how the approach fits the overall research design . Your methods for gathering data should have a clear connection to your research problem. In other words, make sure that your methods will actually address the problem. One of the most common deficiencies found in research papers is that the proposed methodology is not suitable to achieving the stated objective of your paper.
- Describe the specific methods of data collection you are going to use , such as, surveys, interviews, questionnaires, observation, archival research. If you are analyzing existing data, such as a data set or archival documents, describe how it was originally created or gathered and by whom. Also be sure to explain how older data is still relevant to investigating the current research problem.
- Explain how you intend to analyze your results . Will you use statistical analysis? Will you use specific theoretical perspectives to help you analyze a text or explain observed behaviors? Describe how you plan to obtain an accurate assessment of relationships, patterns, trends, distributions, and possible contradictions found in the data.
- Provide background and a rationale for methodologies that are unfamiliar for your readers . Very often in the social sciences, research problems and the methods for investigating them require more explanation/rationale than widely accepted rules governing the natural and physical sciences. Be clear and concise in your explanation.
- Provide a justification for subject selection and sampling procedure . For instance, if you propose to conduct interviews, how do you intend to select the sample population? If you are analyzing texts, which texts have you chosen, and why? If you are using statistics, why is this set of data being used? If other data sources exist, explain why the data you chose is most appropriate to addressing the research problem.
- Provide a justification for case study selection . A common method of analyzing research problems in the social sciences is to analyze specific cases. These can be a person, place, event, phenomenon, or other type of subject of analysis that are either examined as a singular topic of in-depth investigation or multiple topics of investigation studied for the purpose of comparing or contrasting findings. In either method, you should explain why a case or cases were chosen and how they specifically relate to the research problem.
- Describe potential limitations . Are there any practical limitations that could affect your data collection? How will you attempt to control for potential confounding variables and errors? If your methodology may lead to problems you can anticipate, state this openly and show why pursuing this methodology outweighs the risk of these problems cropping up.
NOTE : Once you have written all of the elements of the methods section, subsequent revisions should focus on how to present those elements as clearly and as logically as possibly. The description of how you prepared to study the research problem, how you gathered the data, and the protocol for analyzing the data should be organized chronologically. For clarity, when a large amount of detail must be presented, information should be presented in sub-sections according to topic. If necessary, consider using appendices for raw data.
ANOTHER NOTE : If you are conducting a qualitative analysis of a research problem , the methodology section generally requires a more elaborate description of the methods used as well as an explanation of the processes applied to gathering and analyzing of data than is generally required for studies using quantitative methods. Because you are the primary instrument for generating the data [e.g., through interviews or observations], the process for collecting that data has a significantly greater impact on producing the findings. Therefore, qualitative research requires a more detailed description of the methods used.
YET ANOTHER NOTE : If your study involves interviews, observations, or other qualitative techniques involving human subjects , you may be required to obtain approval from the university's Office for the Protection of Research Subjects before beginning your research. This is not a common procedure for most undergraduate level student research assignments. However, i f your professor states you need approval, you must include a statement in your methods section that you received official endorsement and adequate informed consent from the office and that there was a clear assessment and minimization of risks to participants and to the university. This statement informs the reader that your study was conducted in an ethical and responsible manner. In some cases, the approval notice is included as an appendix to your paper.
III. Problems to Avoid
Irrelevant Detail The methodology section of your paper should be thorough but concise. Do not provide any background information that does not directly help the reader understand why a particular method was chosen, how the data was gathered or obtained, and how the data was analyzed in relation to the research problem [note: analyzed, not interpreted! Save how you interpreted the findings for the discussion section]. With this in mind, the page length of your methods section will generally be less than any other section of your paper except the conclusion.
Unnecessary Explanation of Basic Procedures Remember that you are not writing a how-to guide about a particular method. You should make the assumption that readers possess a basic understanding of how to investigate the research problem on their own and, therefore, you do not have to go into great detail about specific methodological procedures. The focus should be on how you applied a method , not on the mechanics of doing a method. An exception to this rule is if you select an unconventional methodological approach; if this is the case, be sure to explain why this approach was chosen and how it enhances the overall process of discovery.
Problem Blindness It is almost a given that you will encounter problems when collecting or generating your data, or, gaps will exist in existing data or archival materials. Do not ignore these problems or pretend they did not occur. Often, documenting how you overcame obstacles can form an interesting part of the methodology. It demonstrates to the reader that you can provide a cogent rationale for the decisions you made to minimize the impact of any problems that arose.
Literature Review Just as the literature review section of your paper provides an overview of sources you have examined while researching a particular topic, the methodology section should cite any sources that informed your choice and application of a particular method [i.e., the choice of a survey should include any citations to the works you used to help construct the survey].
It’s More than Sources of Information! A description of a research study's method should not be confused with a description of the sources of information. Such a list of sources is useful in and of itself, especially if it is accompanied by an explanation about the selection and use of the sources. The description of the project's methodology complements a list of sources in that it sets forth the organization and interpretation of information emanating from those sources.
Azevedo, L.F. et al. "How to Write a Scientific Paper: Writing the Methods Section." Revista Portuguesa de Pneumologia 17 (2011): 232-238; Blair Lorrie. “Choosing a Methodology.” In Writing a Graduate Thesis or Dissertation , Teaching Writing Series. (Rotterdam: Sense Publishers 2016), pp. 49-72; Butin, Dan W. The Education Dissertation A Guide for Practitioner Scholars . Thousand Oaks, CA: Corwin, 2010; Carter, Susan. Structuring Your Research Thesis . New York: Palgrave Macmillan, 2012; Kallet, Richard H. “How to Write the Methods Section of a Research Paper.” Respiratory Care 49 (October 2004):1229-1232; Lunenburg, Frederick C. Writing a Successful Thesis or Dissertation: Tips and Strategies for Students in the Social and Behavioral Sciences . Thousand Oaks, CA: Corwin Press, 2008. Methods Section. The Writer’s Handbook. Writing Center. University of Wisconsin, Madison; Rudestam, Kjell Erik and Rae R. Newton. “The Method Chapter: Describing Your Research Plan.” In Surviving Your Dissertation: A Comprehensive Guide to Content and Process . (Thousand Oaks, Sage Publications, 2015), pp. 87-115; What is Interpretive Research. Institute of Public and International Affairs, University of Utah; Writing the Experimental Report: Methods, Results, and Discussion. The Writing Lab and The OWL. Purdue University; Methods and Materials. The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. Department of Biology. Bates College.
Writing Tip
Statistical Designs and Tests? Do Not Fear Them!
Don't avoid using a quantitative approach to analyzing your research problem just because you fear the idea of applying statistical designs and tests. A qualitative approach, such as conducting interviews or content analysis of archival texts, can yield exciting new insights about a research problem, but it should not be undertaken simply because you have a disdain for running a simple regression. A well designed quantitative research study can often be accomplished in very clear and direct ways, whereas, a similar study of a qualitative nature usually requires considerable time to analyze large volumes of data and a tremendous burden to create new paths for analysis where previously no path associated with your research problem had existed.
To locate data and statistics, GO HERE .
Another Writing Tip
Knowing the Relationship Between Theories and Methods
There can be multiple meaning associated with the term "theories" and the term "methods" in social sciences research. A helpful way to delineate between them is to understand "theories" as representing different ways of characterizing the social world when you research it and "methods" as representing different ways of generating and analyzing data about that social world. Framed in this way, all empirical social sciences research involves theories and methods, whether they are stated explicitly or not. However, while theories and methods are often related, it is important that, as a researcher, you deliberately separate them in order to avoid your theories playing a disproportionate role in shaping what outcomes your chosen methods produce.
Introspectively engage in an ongoing dialectic between the application of theories and methods to help enable you to use the outcomes from your methods to interrogate and develop new theories, or ways of framing conceptually the research problem. This is how scholarship grows and branches out into new intellectual territory.
Reynolds, R. Larry. Ways of Knowing. Alternative Microeconomics . Part 1, Chapter 3. Boise State University; The Theory-Method Relationship. S-Cool Revision. United Kingdom.
Yet Another Writing Tip
Methods and the Methodology
Do not confuse the terms "methods" and "methodology." As Schneider notes, a method refers to the technical steps taken to do research . Descriptions of methods usually include defining and stating why you have chosen specific techniques to investigate a research problem, followed by an outline of the procedures you used to systematically select, gather, and process the data [remember to always save the interpretation of data for the discussion section of your paper].
The methodology refers to a discussion of the underlying reasoning why particular methods were used . This discussion includes describing the theoretical concepts that inform the choice of methods to be applied, placing the choice of methods within the more general nature of academic work, and reviewing its relevance to examining the research problem. The methodology section also includes a thorough review of the methods other scholars have used to study the topic.
Bryman, Alan. "Of Methods and Methodology." Qualitative Research in Organizations and Management: An International Journal 3 (2008): 159-168; Schneider, Florian. “What's in a Methodology: The Difference between Method, Methodology, and Theory…and How to Get the Balance Right?” PoliticsEastAsia.com. Chinese Department, University of Leiden, Netherlands.
- << Previous: Scholarly vs. Popular Publications
- Next: Qualitative Methods >>
- Last Updated: May 20, 2024 9:47 AM
- URL: https://libguides.usc.edu/writingguide

Thesis, major paper, and major project proposals
- Definitions
- Introductory section
- Literature review
Methodology
- Schedule/work plan
- Other potential elements
- Proposal references
- Ask for help

The methodology section can include (but isn't limited to):
- A description of the research design and methods
- A description of data-gathering instruments
- Methods of data collection
- Ethical considerations
- Analysis strategies and techniques
- Potential participants
- Rationale for your choice of methodological choices
- How the methodology is appropriate for the organization or participants
- The advantages and disadvantages of the methodology
- References to scholarly literature that support the chosen research design and methods
If you are unsure if including the methodology is required in your thesis, major project, or research paper proposal, please consult with your instructor or advisor.
This information regarding the methodology section of a proposal was gathered from RRU thesis and major project handbooks, current in 2020, from programs in the Faculty of Social and Applied Sciences, the Faculty of Management, and the College of Interdisciplinary Studies. If the details here differ from the information provided in the handbook for your project, please follow the handbook's directions.
Image credit: Image by Gerd Altmann from Pixabay

- In SAGE Research Methods Project Planner ; access via this link requires a RRU username and password.
Data Collection
How Do I Write About Theory?
- In SAGE Research Methods: Writing Up ; look for the How Do I Write About Theory? drop down option. Access via this link requires a RRU username and password.
How Do I Write My Methodology Section?
- In SAGE Research Methods: Writing Up ; look for the How Do I Write My Methodology Section? drop down option. Access via this link requires a RRU username and password.
Research Ethics
Image credit: Image by Mohamed Assan from Pixabay
- << Previous: Literature review
- Next: Schedule/work plan >>
- Last Updated: Jan 8, 2024 12:29 PM
- URL: https://libguides.royalroads.ca/proposals
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Indian J Anaesth
- v.60(9); 2016 Sep
How to write a research proposal?
Department of Anaesthesiology, Bangalore Medical College and Research Institute, Bengaluru, Karnataka, India
Devika Rani Duggappa
Writing the proposal of a research work in the present era is a challenging task due to the constantly evolving trends in the qualitative research design and the need to incorporate medical advances into the methodology. The proposal is a detailed plan or ‘blueprint’ for the intended study, and once it is completed, the research project should flow smoothly. Even today, many of the proposals at post-graduate evaluation committees and application proposals for funding are substandard. A search was conducted with keywords such as research proposal, writing proposal and qualitative using search engines, namely, PubMed and Google Scholar, and an attempt has been made to provide broad guidelines for writing a scientifically appropriate research proposal.
INTRODUCTION
A clean, well-thought-out proposal forms the backbone for the research itself and hence becomes the most important step in the process of conduct of research.[ 1 ] The objective of preparing a research proposal would be to obtain approvals from various committees including ethics committee [details under ‘Research methodology II’ section [ Table 1 ] in this issue of IJA) and to request for grants. However, there are very few universally accepted guidelines for preparation of a good quality research proposal. A search was performed with keywords such as research proposal, funding, qualitative and writing proposals using search engines, namely, PubMed, Google Scholar and Scopus.
Five ‘C’s while writing a literature review

BASIC REQUIREMENTS OF A RESEARCH PROPOSAL
A proposal needs to show how your work fits into what is already known about the topic and what new paradigm will it add to the literature, while specifying the question that the research will answer, establishing its significance, and the implications of the answer.[ 2 ] The proposal must be capable of convincing the evaluation committee about the credibility, achievability, practicality and reproducibility (repeatability) of the research design.[ 3 ] Four categories of audience with different expectations may be present in the evaluation committees, namely academic colleagues, policy-makers, practitioners and lay audiences who evaluate the research proposal. Tips for preparation of a good research proposal include; ‘be practical, be persuasive, make broader links, aim for crystal clarity and plan before you write’. A researcher must be balanced, with a realistic understanding of what can be achieved. Being persuasive implies that researcher must be able to convince other researchers, research funding agencies, educational institutions and supervisors that the research is worth getting approval. The aim of the researcher should be clearly stated in simple language that describes the research in a way that non-specialists can comprehend, without use of jargons. The proposal must not only demonstrate that it is based on an intelligent understanding of the existing literature but also show that the writer has thought about the time needed to conduct each stage of the research.[ 4 , 5 ]
CONTENTS OF A RESEARCH PROPOSAL
The contents or formats of a research proposal vary depending on the requirements of evaluation committee and are generally provided by the evaluation committee or the institution.
In general, a cover page should contain the (i) title of the proposal, (ii) name and affiliation of the researcher (principal investigator) and co-investigators, (iii) institutional affiliation (degree of the investigator and the name of institution where the study will be performed), details of contact such as phone numbers, E-mail id's and lines for signatures of investigators.
The main contents of the proposal may be presented under the following headings: (i) introduction, (ii) review of literature, (iii) aims and objectives, (iv) research design and methods, (v) ethical considerations, (vi) budget, (vii) appendices and (viii) citations.[ 4 ]
Introduction
It is also sometimes termed as ‘need for study’ or ‘abstract’. Introduction is an initial pitch of an idea; it sets the scene and puts the research in context.[ 6 ] The introduction should be designed to create interest in the reader about the topic and proposal. It should convey to the reader, what you want to do, what necessitates the study and your passion for the topic.[ 7 ] Some questions that can be used to assess the significance of the study are: (i) Who has an interest in the domain of inquiry? (ii) What do we already know about the topic? (iii) What has not been answered adequately in previous research and practice? (iv) How will this research add to knowledge, practice and policy in this area? Some of the evaluation committees, expect the last two questions, elaborated under a separate heading of ‘background and significance’.[ 8 ] Introduction should also contain the hypothesis behind the research design. If hypothesis cannot be constructed, the line of inquiry to be used in the research must be indicated.
Review of literature
It refers to all sources of scientific evidence pertaining to the topic in interest. In the present era of digitalisation and easy accessibility, there is an enormous amount of relevant data available, making it a challenge for the researcher to include all of it in his/her review.[ 9 ] It is crucial to structure this section intelligently so that the reader can grasp the argument related to your study in relation to that of other researchers, while still demonstrating to your readers that your work is original and innovative. It is preferable to summarise each article in a paragraph, highlighting the details pertinent to the topic of interest. The progression of review can move from the more general to the more focused studies, or a historical progression can be used to develop the story, without making it exhaustive.[ 1 ] Literature should include supporting data, disagreements and controversies. Five ‘C's may be kept in mind while writing a literature review[ 10 ] [ Table 1 ].
Aims and objectives
The research purpose (or goal or aim) gives a broad indication of what the researcher wishes to achieve in the research. The hypothesis to be tested can be the aim of the study. The objectives related to parameters or tools used to achieve the aim are generally categorised as primary and secondary objectives.
Research design and method
The objective here is to convince the reader that the overall research design and methods of analysis will correctly address the research problem and to impress upon the reader that the methodology/sources chosen are appropriate for the specific topic. It should be unmistakably tied to the specific aims of your study.
In this section, the methods and sources used to conduct the research must be discussed, including specific references to sites, databases, key texts or authors that will be indispensable to the project. There should be specific mention about the methodological approaches to be undertaken to gather information, about the techniques to be used to analyse it and about the tests of external validity to which researcher is committed.[ 10 , 11 ]
The components of this section include the following:[ 4 ]
Population and sample
Population refers to all the elements (individuals, objects or substances) that meet certain criteria for inclusion in a given universe,[ 12 ] and sample refers to subset of population which meets the inclusion criteria for enrolment into the study. The inclusion and exclusion criteria should be clearly defined. The details pertaining to sample size are discussed in the article “Sample size calculation: Basic priniciples” published in this issue of IJA.
Data collection
The researcher is expected to give a detailed account of the methodology adopted for collection of data, which include the time frame required for the research. The methodology should be tested for its validity and ensure that, in pursuit of achieving the results, the participant's life is not jeopardised. The author should anticipate and acknowledge any potential barrier and pitfall in carrying out the research design and explain plans to address them, thereby avoiding lacunae due to incomplete data collection. If the researcher is planning to acquire data through interviews or questionnaires, copy of the questions used for the same should be attached as an annexure with the proposal.
Rigor (soundness of the research)
This addresses the strength of the research with respect to its neutrality, consistency and applicability. Rigor must be reflected throughout the proposal.
It refers to the robustness of a research method against bias. The author should convey the measures taken to avoid bias, viz. blinding and randomisation, in an elaborate way, thus ensuring that the result obtained from the adopted method is purely as chance and not influenced by other confounding variables.
Consistency
Consistency considers whether the findings will be consistent if the inquiry was replicated with the same participants and in a similar context. This can be achieved by adopting standard and universally accepted methods and scales.
Applicability
Applicability refers to the degree to which the findings can be applied to different contexts and groups.[ 13 ]
Data analysis
This section deals with the reduction and reconstruction of data and its analysis including sample size calculation. The researcher is expected to explain the steps adopted for coding and sorting the data obtained. Various tests to be used to analyse the data for its robustness, significance should be clearly stated. Author should also mention the names of statistician and suitable software which will be used in due course of data analysis and their contribution to data analysis and sample calculation.[ 9 ]
Ethical considerations
Medical research introduces special moral and ethical problems that are not usually encountered by other researchers during data collection, and hence, the researcher should take special care in ensuring that ethical standards are met. Ethical considerations refer to the protection of the participants' rights (right to self-determination, right to privacy, right to autonomy and confidentiality, right to fair treatment and right to protection from discomfort and harm), obtaining informed consent and the institutional review process (ethical approval). The researcher needs to provide adequate information on each of these aspects.
Informed consent needs to be obtained from the participants (details discussed in further chapters), as well as the research site and the relevant authorities.
When the researcher prepares a research budget, he/she should predict and cost all aspects of the research and then add an additional allowance for unpredictable disasters, delays and rising costs. All items in the budget should be justified.
Appendices are documents that support the proposal and application. The appendices will be specific for each proposal but documents that are usually required include informed consent form, supporting documents, questionnaires, measurement tools and patient information of the study in layman's language.
As with any scholarly research paper, you must cite the sources you used in composing your proposal. Although the words ‘references and bibliography’ are different, they are used interchangeably. It refers to all references cited in the research proposal.
Successful, qualitative research proposals should communicate the researcher's knowledge of the field and method and convey the emergent nature of the qualitative design. The proposal should follow a discernible logic from the introduction to presentation of the appendices.
Financial support and sponsorship
Conflicts of interest.
There are no conflicts of interest.
- SUGGESTED TOPICS
- The Magazine
- Newsletters
- Managing Yourself
- Managing Teams
- Work-life Balance
- The Big Idea
- Data & Visuals
- Reading Lists
- Case Selections
- HBR Learning
- Topic Feeds
- Account Settings
- Email Preferences
How to Present to an Audience That Knows More Than You
- Deborah Grayson Riegel

Lean into being a facilitator — not an expert.
What happens when you have to give a presentation to an audience that might have some professionals who have more expertise on the topic than you do? While it can be intimidating, it can also be an opportunity to leverage their deep and diverse expertise in service of the group’s learning. And it’s an opportunity to exercise some intellectual humility, which includes having respect for other viewpoints, not being intellectually overconfident, separating your ego from your intellect, and being willing to revise your own viewpoint — especially in the face of new information. This article offers several tips for how you might approach a roomful of experts, including how to invite them into the discussion without allowing them to completely take over, as well as how to pivot on the proposed topic when necessary.
I was five years into my executive coaching practice when I was invited to lead a workshop on “Coaching Skills for Human Resource Leaders” at a global conference. As the room filled up with participants, I identified a few colleagues who had already been coaching professionally for more than a decade. I felt self-doubt start to kick in: Why were they even here? What did they come to learn? Why do they want to hear from me?
- Deborah Grayson Riegel is a professional speaker and facilitator, as well as a communication and presentation skills coach. She teaches leadership communication at Duke University’s Fuqua School of Business and has taught for Wharton Business School, Columbia Business School’s Women in Leadership Program, and Peking University’s International MBA Program. She is the author of Overcoming Overthinking: 36 Ways to Tame Anxiety for Work, School, and Life and the best-selling Go To Help: 31 Strategies to Offer, Ask for, and Accept Help .
Partner Center
Company Filings | More Search Options
Company Filings More Search Options -->

- Commissioners
- Reports and Publications
- Securities Laws
- Commission Votes
- Corporation Finance
- Enforcement
- Investment Management
- Economic and Risk Analysis
- Trading and Markets
- Office of Administrative Law Judges
- Examinations
- Litigation Releases
- Administrative Proceedings
- Opinions and Adjudicatory Orders
- Accounting and Auditing
- Trading Suspensions
- How Investigations Work
- Receiverships
- Information for Harmed Investors
- Rulemaking Activity
- Proposed Rules
- Final Rules
- Interim Final Temporary Rules
- Other Orders and Notices
- Self-Regulatory Organizations
- Staff Interpretations
- Investor Education
- Small Business Capital Raising
- EDGAR – Search & Access
- EDGAR – Information for Filers
- Company Filing Search
- How to Search EDGAR
- About EDGAR
- Press Releases
- Speeches and Statements
- Securities Topics
- Upcoming Events
- Media Gallery
- Divisions & Offices
- Public Statements
Notice of Filing of Proposed Rule Change by The Options Clearing Corporation Concerning Amendments to Its Rules and Comprehensive Stress Testing & Clearing Fund Methodology, and Liquidity Risk Management Description
Public comments.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 20 May 2024
A novel anti-loosening bolt looseness diagnosis of bolt connections using a vision-based technique
- Jun Luo 1 ,
- Kaili Li 1 ,
- ChengQian Xie 1 ,
- Zhitao Yan 1 ,
- Feng Li 2 ,
- Xiaogang Jia 2 &
- Yuanlai Wang 2
Scientific Reports volume 14 , Article number: 11441 ( 2024 ) Cite this article
Metrics details
- Civil engineering
- Energy infrastructure
Bolt looseness detection is a common problem in engineering. Most vision-based detection techniques focus on diagnosing ordinary bolt looseness, i.e., the methods used for diagnosis are based only on the sidelines of nuts. These methods cannot be used for anti-loosening bolt looseness diagnosis because of the simultaneous rotation of screws and nuts. Therefore, a novel anti-loosening bolt looseness diagnosis method based on a vision-based technique is proposed in this paper. First, a regular hexagonal cap was installed on the screw, which can be used as a reference for the nut. Then, to automatically distinguish the hexagonal borders of the screw cap and nut, a new hexagonal border reconstruction algorithm is proposed. Furthermore, the relative rotation angles of the screw cap and nut hexagons can be determined using the sidelines of the reconstructed hexagonal borders of the screw cap and nut. Finally, a novel anti-loosening bolt looseness diagnosis method is established by using the relative rotation angle of the regular hexagonal borders of the screw cap and nut under initial status and loose status. A prototype flange node of the transmission tower was used for experimental verification. The results show that the proposed method can effectively detect the loosening angle of anti-loosening bolts.
Introduction
Bolted connections have been widely used in engineering structures due to their advantages of easy disassembly and maintenance, strong adaptability, and flexibility and reliability. However, because of vibration, impact, and dynamic and periodic loads, the bolts in bolted connections may loosen, which will reduce the reliability of bolted connections in structures and seriously affect their safety. Therefore, the detection of bolt looseness is highly important. First, bolt looseness detection was achieved manually by observing the surface texture of the bolt, tapping the bolt with a hammer, or measuring the bolt torque using a torque wrench 1 , 2 . However, these methods are inefficient, carry a high risk of personnel injury and are unable to achieve real-time detection.
To establish more efficient and nonmanual detection methods, several sensors have been introduced to detect bolt looseness, and several sensor-based methods for detecting bolt looseness have been proposed, such as the guided wave method 3 , 4 and the piezoelectric impedance method 5 , 6 , 7 . In the guided wave method, the attenuation of ultrasonic waves in the elastic bolt connection structure is monitored, which can be used to diagnose bolt loosening. In the piezoelectric impedance method, based on the electromechanical coupling characteristics of the piezoelectric material and structure, changes in the amplitude or characteristic frequency of the piezoelectric impedance spectrum can be used to identify bolt loosening. These sensor-based methods for detecting bolt looseness can improve the accuracy of bolt loosening detection and help achieve online health monitoring. However, the cost of these sensors is high, and regular maintenance is needed. Moreover, these sensors are relatively sensitive and susceptible to external interference. Therefore, sensor-based bolt looseness detection methods are difficult to deploy and apply in large engineering structures.
Currently, with the improvement of image processing technology, vision-based techniques have been introduced for bolt looseness detection because of their lower cost, greater intelligence and greater accuracy. Based on the Canny edge detection algorithm 8 and Hough line detection algorithm 9 in image processing technology, the edge lines of nuts can be identified. The angles between these edge lines and the horizontal coordinate axes are related to the loosening angle of the mainstream regular hexagonal bolts 10 , 11 , which can be used to quantify the loosening angle. The vision-based bolt loosening detection method is stable and has high detection accuracy 12 . With the rapid development of neural networks, deep learning and image processing technology have been combined to improve the efficiency of bolt location identification 13 , detect the integrity of bolts, and classify loose and nonloose bolts 14 , 15 , 16 , 17 . Breakthroughs in deep learning technology have greatly improved the accuracy and efficiency of bolt looseness detection methods based on vision technology, greatly increasing their application value.
However, there are still certain shortcomings in most vision-based detection methods, i.e., anti-loosening bolt looseness cannot be detected quantitatively. The reason is that the current quantitative detection methods for bolt looseness using a vision-based technique are almost exclusively based on the sideline angles of nuts. As shown in Fig. 1 , under loose state 1, only the nut of the bolt rotates clockwise, and the rotation angle is 30 degrees. Therefore, using the existing quantitative detection method of bolt looseness based on vision technology, the rotation of the nut can be detected, and the conclusion that the bolt is loosened by 30 degrees can be reached. However, for anti-loosening bolts, the screw and nut may rotate together. For example, under loose state 2 in Fig. 1 , the nut and screw rotate 30 degrees synchronously, and the bolt is actually not loose because there is no relative rotation between the screw and nut. Based on the existing method, the same conclusion as loose state 1 will be given, which is an incorrect conclusion. Hence, the relative rotation between the screw and nut will have a negative impact on the identification of the loosening angle.
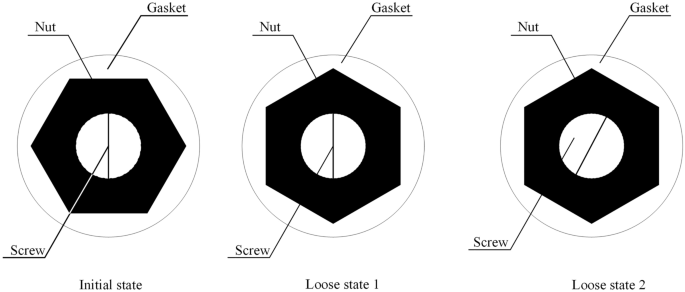
Two bolt looseness states.
To this end, a novel anti-loosening bolt looseness diagnosis method is proposed to quantitatively detect anti-loosening bolt looseness. First, a regular hexagonal cap was installed on the screw, which can be used as a reference for the nut. Then, to automatically distinguish the hexagonal borders of the screw cap and nut, a new hexagonal border reconstruction algorithm is proposed. Furthermore, the relative rotation angles of the screw cap and nut hexagons can be determined using the sidelines of the reconstructed hexagonal borders of the screw cap and nut. Finally, a novel anti-loosening bolt looseness diagnosis method is established by using the relative rotation angle of the regular hexagonal borders of the screw cap and nut under initial status and loose status.
The main contributions of this paper are summarized as follows:
A novel anti-loosening bolt looseness diagnosis method is proposed for anti-loosening bolt looseness detection. The proposed method can quantitatively detect bolt loosening with high detection accuracy and low cost.
A new hexagonal border reconstruction algorithm is proposed for automatically distinguishing the hexagonal borders of the screw cap and nut. The centroid of the hexagonal cap is used as the reference point, and the distances from the endpoints on the edge lines of the hexagonal borders of the screw cap and nut to the reference point are used as the clustering constraints. Then, the intra-class variance algorithm is introduced to automatically distinguish the edge lines of the hexagonal borders of the screw cap and nut, and the hexagonal borders of the screw cap and nut can be reconstructed.
The proposed method was verified to be effective, and the potential influencing factors were also analyzed. The results show that the proposed method in this paper can more comprehensively detect anti-loosening bolt looseness with high accuracy and good stability.
The remainder of this paper is arranged as follows. Related works on bolt loosening detection are presented in “Related Work”. The “Proposed Method” section mainly introduces the theory of the proposed novel anti-loosening bolt looseness diagnosis method. The “Experiments” section mainly verifies the feasibility of the proposed method, including method validation and analysis of the influencing factors. The “ Conclusion ” section contains the conclusion.
Related work
Bolt loosening diagnosis based on hardware sensors.
Due to the inefficiency of traditional manual maintenance of bolt loosening, the detection accuracy is inadequate. In recent years, bolt looseness detection methods based on hardware sensors have attracted widespread attention from researchers. An et al. 18 proposed a bolt loose detection method based on integrated impedance and guided waves, which utilizes piezoelectric transducers to obtain impedance and guided wave signals for bolt loose detection. Yin et al. 6 proposed a bolt looseness detection method based on the piezoelectric active sensing method, which determines the actual contact area of bolts through ultrasonic energy transmission, thereby determining the looseness of bolts. Zhao et al. 19 proposed a detection method for timber structural bolts based on wavelet analysis that combines the pasting of lead zirconate titanate with the time-recursive method. Zhang et al. 20 proposed a bolt looseness detection method based on audio classification using a support vector machine, which mainly records and extracts the hammer sound produced by the varying looseness of bolt connections. Wang et al. 21 proposed a vibroacoustic method for detecting bolt looseness. The above methods can achieve unmanned detection and sufficient detection accuracy. However, some of these sensors are relatively expensive.
Bolt loosening diagnosis based on visual technology
Currently, with the improvement of image processing technology, vision-based techniques have been introduced for bolt looseness detection because of their lower cost, greater intelligence and greater accuracy. Ramana et al. 22 proposed a fully automatic bolt looseness detection method based on the Viola–Jones algorithm and a support vector machine. The Viola–Jones algorithm was used to determine the bolt location, and a support vector machine was used to classify the loose bolts. Park et al. 10 and Nguyen et al. 11 proposed a bolt connection looseness detection technology-based Canny edge detection algorithm 8 and Hough transform 9 , which identifies bolt loosening by comparing the sideline angles before and after bolt loosening. Kong et al. 12 proposed a visual noncontact bolt looseness detection method by judging whether the bolt alignment process has rotated between two bolt images at different times. Cha et al. 23 proposed a loose bolt detection method using the Hough transform and support vector machine, which can automatically calculate the damage sensitive features of bolts to train the support vector machine and construct a robust classifier to distinguish tight bolts and loose bolts. Luo et al. 24 proposed a new bolt image correction method based on a square gasket for flange connection node bolts.
Additionally, with the rapid development of deep learning, vision-based techniques have become more intelligent and efficient. The main methods include deep learning only and the combination of deep learning and image processing technology 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 . Huynh et al. 13 used convolutional neural networks (CNNs) 25 to identify bolts and segment bolt images, and the nut edge line angles from the initial and current states were used to diagnose bolt looseness based on image processing technology. Zhang et al. 15 used the Faster-RCNN 26 , 27 deep learning algorithm for autonomous bolt looseness detection based on different screw heights, and the experimental results showed that the average accuracy of looseness detection reached 0.9503. Yang et al. 16 comparatively tested the detection performance of three YOLO target detection algorithms 28 for bolt loosening identification, namely, YOLO v3, YOLO v4 and YOLO v4-Tiny. Yuan et al. 29 proposed an automatic bolt loosening identification method based on Mask-RCNN 30 . Gong et al. 31 proposed a quantitative bolt loosening identification method by calculating the exposed length of the screw using vision-based deep learning and geometric imaging theory, and the results showed that the average measurement error was limited to within 0.61 mm. However, the relationship between the loosening angle and the exposed length of the screw is still unclear. Pan et al. 32 , 33 proposed a bolt looseness detection method based on 3D vision. The bolts in the 3D model are located and extracted based on the CNN convolutional neural network and point cloud processing algorithm, and the height of the screw is used to determine whether the screw is loose.
The above studies mainly focused on the quantitative nut rotation and the exposed height of the screw. However, less research has focused on identifying loosening of anti-loosening bolts. Therefore, in this paper, a new anti-loosening bolt loosening detection method is proposed.
Proposed method
Bolts with regular hexagonal caps.
The vertical and top views of the proposed anti-loosening bolt construction in this paper are shown in Figs. 2 and 3 , respectively. An additional regular hexagonal cap was installed at the end of the screw. The distance between the screw cap and nut hexagons was 7.5 mm, as shown in Fig. 2 . Except for the regular hexagonal cap, the other bolts are the same as the normal anti-loosening bolt, including the screw, nut, circular washer, and connected components. The regular hexagonal cap is white, and the nut is black, which can maximize the gray gradient difference between the screw cap and nut edge 34 and improve the stability of edge detection.
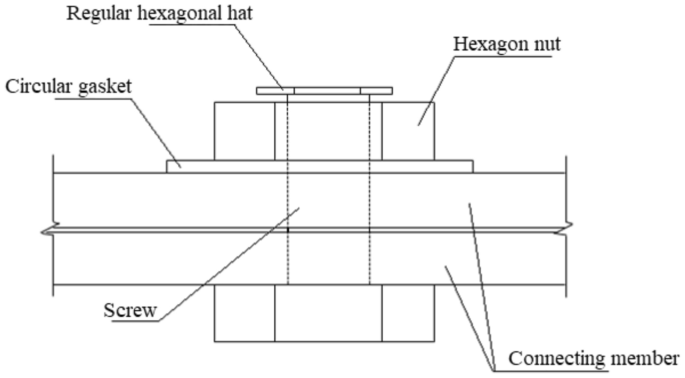
Vertical view of the constructed bolt.
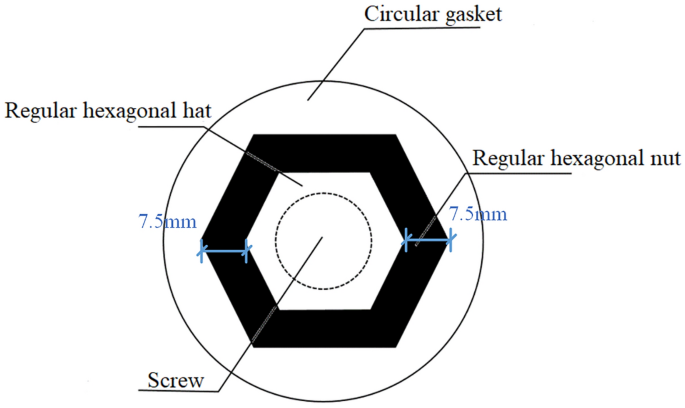
Top view of the constructed bolt.
The proposed hexagonal border reconstruction algorithm
When photos of the anti-loosening bolt are obtained using a camera at the top, the hexagonal borders of the screw cap and nut can be determined using the Canny edge detection method, as shown in Fig. 4 . However, the relative rotation angle between the screw cap and nut is the desired quantity. Therefore, a new hexagonal border reconstruction algorithm must be proposed to automatically distinguish the hexagonal borders of the screw cap and nut.
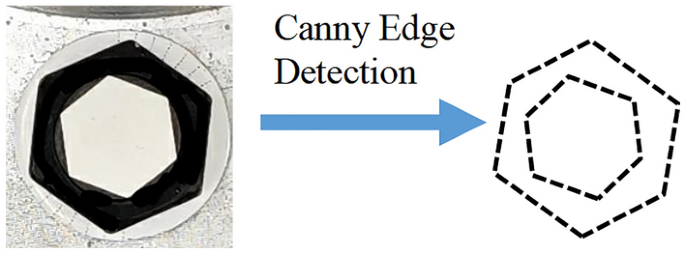
Bolt picture and the hexagonal borders.
The proposed hexagonal border reconstruction algorithm is shown in Fig. 5 . First, the initial image is transformed into grayscale, denoised by a Wiener adaptive filter 35 , and binarized by the Otsu method 36 , 37 . Second, connected domain processing and morphological operations are used, and the morphological operations usually include erosion, expansion, deletion of interfering binary image areas, calculation of binary image centroid coordinates, etc. Then, the interference areas are deleted, and the centroid coordinates of the regular hexagonal cap can be obtained. Third, the edge lines of the nut and regular hexagonal cap can be segmented. The Canny algorithm 8 and Hough algorithm 9 are used to detect edge lines on the initial bolt image, and the endpoint coordinates of the straight edge lines can be extracted. Then, the distances between all endpoint coordinates and the identified centroid point of the regular hexagonal cap can be calculated, and the intra-class variance method 38 , 39 , 40 is used to adaptively calculate the optimal segmentation distance, which can be used to segment the edge lines of the nut and the regular hexagonal cap. Finally, the hexagonal borders of the nut and regular hexagonal cap can be reconstructed using individual edge lines. Since the Otsu threshold segmentation method can maximize the difference between the front and rear backgrounds and is not affected by image brightness, the Canny algorithm is not sensitive to image brightness. Therefore, the program in this section is not affected by image brightness and has high stability.
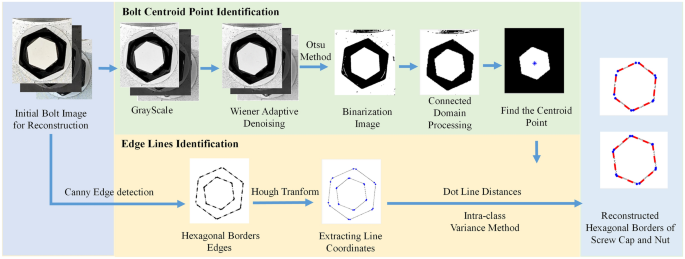
Process of the proposed hexagonal border reconstruction algorithm.
Relative rotation angle of the screw cap and nut hexagons
After reconstructing the hexagonal border of the screw cap or nut, the rotation angle of the screw cap or nut hexagon can be determined. As shown in Fig. 6 , a regular hexagonal border has six edges parallel to each other, so there are only three angles on the x-o-y coordinate system, namely, θ 1 , θ 2 , and θ 3 . θ 1 , θ 2 , and θ 3 have the following angular relationships:
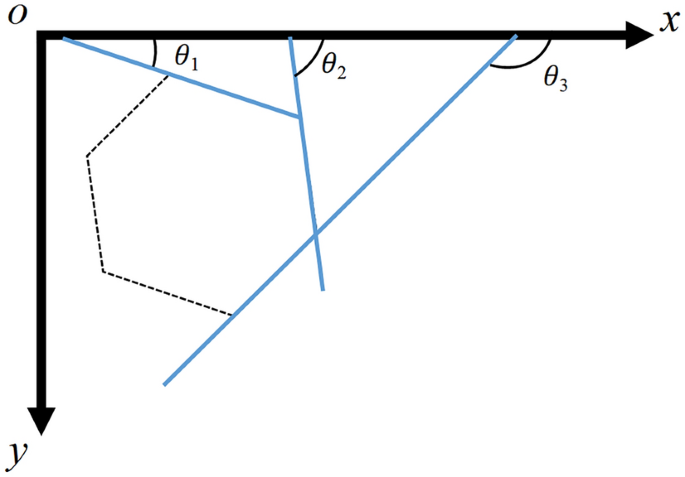
Three angles of a regular hexagonal border.
Therefore, when the hexagonal border starts to rotate, the angles θ 1 , θ 2 and θ 3 will undergo changes. Thus, based on the relationships among these three angles, the rotation angle of the hexagonal border can be calculated using the angles θ 1 , θ 2 and θ 3 as follows:
where θ j is the angle between the j th edge line and the x-axis , n is the number of identified sidelines, and rem ( . ) is the operator used to determine the remainder after division.
Based on Eqs. ( 1 ) and ( 2 ), the rotation angle of the hexagonal border of the screw cap can be calculated and defined as θ inner . Similarly, the rotation angle of the hexagonal border of the nut can be calculated and defined as θ out . Then, the relative rotation angle of the screw cap and nut hexagons can be defined as
The proposed anti-loosening bolt looseness diagnosis
When the bolt is not loose, the relative rotation angle of the hexagonal borders of the screw cap and nut can be determined and defined as \(\hat{\theta }_{u}\) . Meanwhile, when the bolt is loose, the relative rotation angle of the hexagonal borders of the screw cap and nut can be determined and defined as \(\hat{\theta }_{d}\) . Then, the bolt looseness angle of the anti-loosening bolt can be expressed as:
In other words, the proposed method in this paper involves the installation of a regular hexagonal screw cap, the reconstruction of hexagonal borders, the identification of the relative rotation angle of the screw cap and nut, and the comparison of the relative rotation angles under the initial status and loose status. The process of the proposed anti-loosening bolt looseness diagnosis method is shown in Fig. 7 .
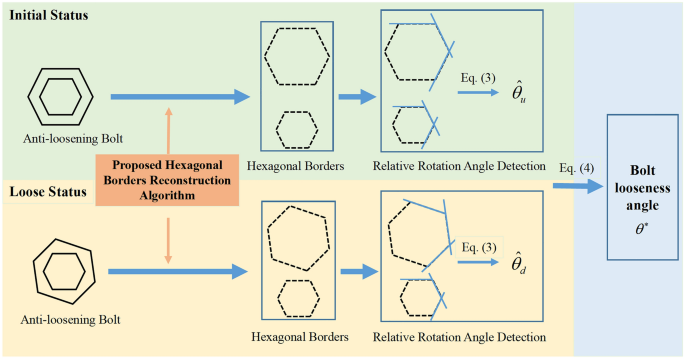
The process of the proposed anti-loosening bolt looseness diagnosis method.
Experimental
Experimental overview.
To verify the feasibility of the proposed method, a laboratory experiment is implemented. A prototype flange node of the transmission tower was used, as shown in Fig. 8 . The hexagonal cap and the screw were connected by an adhesive. One of the bolts was selected for study, and images were captured using an iPhone 12 in the laboratory.
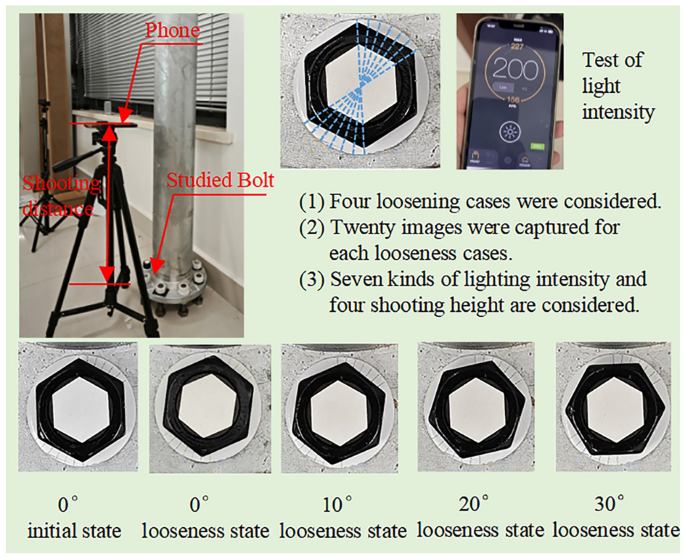
Experimental overview.
In the experiment, one initial bolt status is considered. Four bolt looseness cases were considered, i.e., the relative rotation angles of the screw cap and nut hexagons were 0°, 10°, 20°, and 30°. For the 0° bolt looseness case, the screw and nut rotate synchronously, and the location of the screw cap and nut is not the same as that for the 0° initial status, as shown in Fig. 7 . For the 10°, 20°, and 30° bolt looseness cases, only the nut rotation and looseness angles are simulated by using the change in the relative rotation angle between the screw cap and nut, as shown in Fig. 7 . To set the angles accurately, seven adjacent 10-degree scale lines were drawn on the gasket, as shown in Fig. 7 .
The influences of lighting intensity and shooting distance were also studied. Seven lighting intensities, four shooting heights and twelve shooting perspective angles are considered and are discussed in the “Analysis of influencing factors” section.
The validation experiment of the proposed method
Images obtained under a lighting intensity of 200 lx and a shooting height of 50 cm were used to verify the proposed method. Under the initial status, one image was captured to calculate the quantity \(\hat{\theta }_{u}\) . Under each looseness case, twenty images were captured to study the stability of the identification results. Twenty images under each looseness case were processed, and twenty relative rotation angles \(\hat{\theta }_{d}\) between the screw cap and nut were obtained. Examples of edge line segmentation results under different bolt states are shown in Fig. 9 .
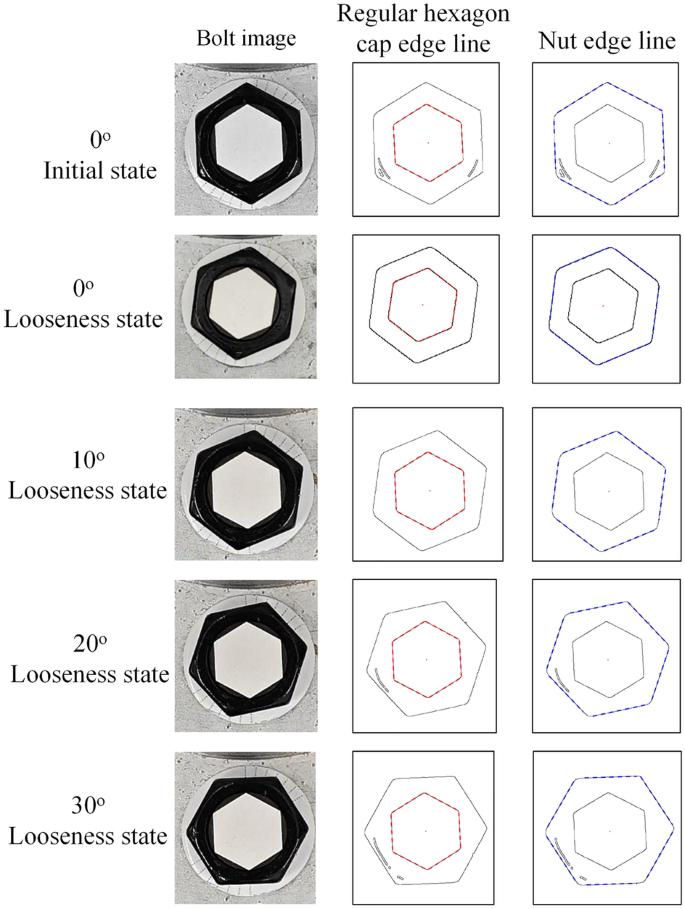
Experimental edge line segmentation results.
Then, based on Eq. ( 4 ), twenty looseness angles θ* can be calculated for each looseness case, as shown in Fig. 10 . Ave represents the average value of 20 test results per group. Std represents the standard deviation of 20 test results per group. The results in Fig. 10 show that the mean value deviations are small, with a maximum of only 0.62° under the 30° looseness case. The standard deviations do not exceed 0.54° under the 30° looseness case. The proposed method has good effectiveness and feasibility and high accuracy.
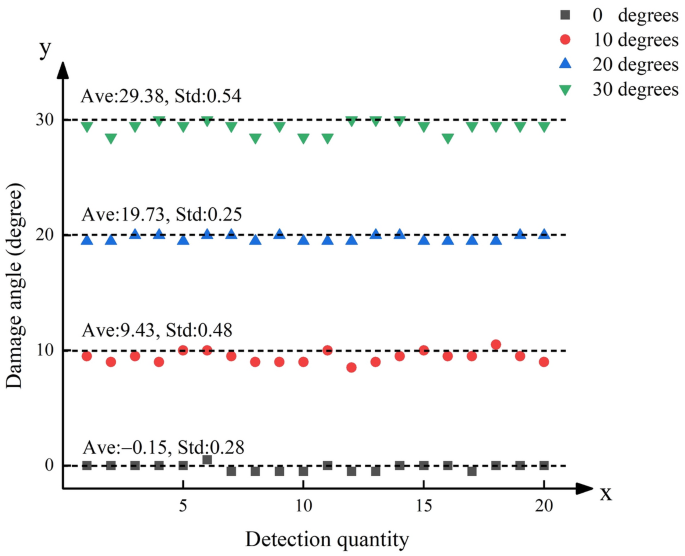
Test results for the 20 bolt looseness cases.
Comparison of the proposed method and existing methods
One of the images under the 0° initial state and 0° looseness state are used to compare the proposed method and existing bolt looseness quantitative identification methods 13 . To make the calculation process more intuitive, the calculation results obtained using the two methods are shown in Fig. 11 . Based on the existing method, the results show that the bolt is already loose and that the rotation angle is 10.5°. In contrast, based on the proposed method, the results show that there is no loose bolt. The method proposed in this article can effectively avoid the impact of the synchronous rotation of the nut and screw on bolt looseness detection.
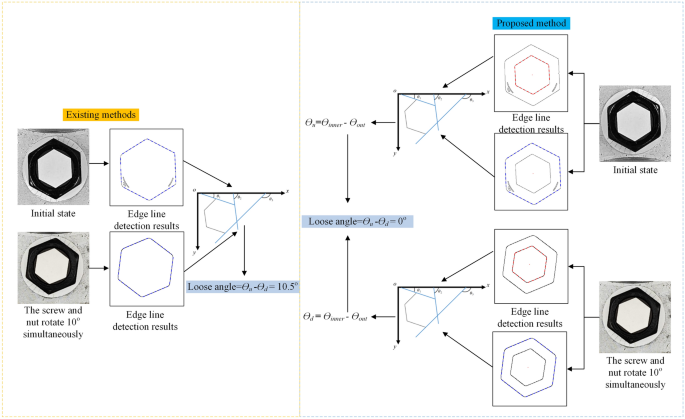
Comparison of the proposed method and existing methods.
Analysis of the influencing factors
To study the influence of camera height, light intensity and shooting perspective angle, bolt looseness detection tests under different test conditions were implemented. Seven lighting intensities, four shooting heights and twelve shooting perspective angles are considered.
Effect test and result analysis of different shooting heights
Four shooting heights are considered, i.e., 50 cm, 60 cm, 80 cm and 100 cm. The minimum shooting height is limited by the integrity of the bolt images and is set to 50 cm. The looseness angles are set to 10° and 30°. The bolt images with different shooting heights under 10° looseness state are shown in Fig. 12 . The indoor lighting intensity was measured as 200 lx. The image acquisition and calculation of the looseness angles θ* are consistent with those in the previous section.
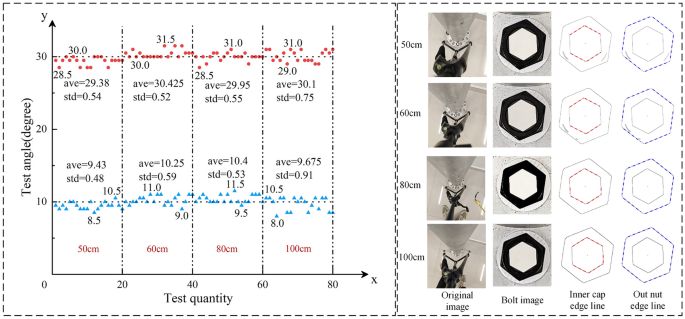
Results for different shooting heights.
Based on the proposed method, the looseness angles under different shooting heights can be calculated, as shown in Fig. 12 . The results show that the impact of shooting height is very limited. Generally, for all the shooting heights, the maximum mean deviation is 0.62° under the 30° looseness case, and the maximum standard deviation is 0.91° under the 10° looseness case, which are still within an acceptable range. However, it is not recommended to shoot bolts at a high height. The reason is that the lower the camera height is, the more details the camera captures, the better the edge identification, and the greater the stability of the bolt looseness detection.
Effect test and result analysis of different lighting intensities
Seven light intensities are considered, i.e., 22,686 lx, 14,142 lx, 5172 lx, 350 lx, 200 lx, 14 lx and 7 lx. The lighting intensities correspond to outdoor strong sunshine weather, outdoor sunshine weather, outdoor bright cloudy weather, outdoor cloudy weather, indoor weather, outdoor evening, and outdoor night, respectively. The shooting height is set as 50 cm. The bolt images with different light intensities under 10° looseness state are shown in Fig. 13 . Under each light intensity, 20 bolt images have been obtained. In order to better showcase its fluctuations under different light intensities, the first image under each light intensity is used as the initial state. Therefore, the looseness angle is actually set to 0°. The image acquisition and calculation of the looseness angles θ* are consistent with those in the previous section.
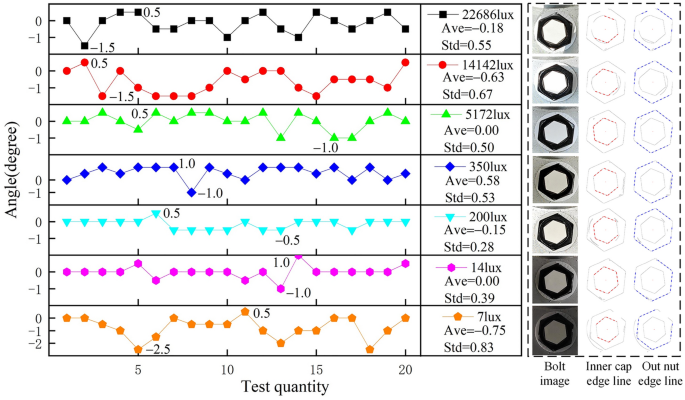
Results under different lighting intensities.
Based on the proposed method, the looseness angles under different lighting intensities can be calculated, as shown in Fig. 13 . Figure 13 shows that there is not much difference between the mean value deviations under different light intensities, and the impact of light intensity is very limited. The mean deviation is limited to between -0.75° and 0.58°, which is within an acceptable range. However, the standard deviations under strong or dark light intensities are generally greater than those under normal light intensities, which means that there is greater fluctuation in the identified results under strong or dark light intensities. Under a strong lighting environment, the luminescence on the bolt interferes with imaging, reducing the stability of the proposed method. In a dark lighting environment, the number of noise points in the image will increase. Hence, it is not recommended to shoot images under extreme lighting environments, such as strong or dark lighting environments. In a strong lighting environment, the luminescence on the bolt interferes with imaging. In a dark lighting environment, the number of noise points in the image will increase.
Effect test and result analysis of different shooting perspective angles
The considered shooting perspective angles include horizontal perspective, vertical perspective, and horizontal-vertical perspective angles. The horizontal and vertical perspective angles are set to 10°, 20°, 30° and 45°. The horizontal-vertical perspective angles are set to 10°–10°, 10°–30°, 30°–10° and 45°–45°. The looseness angles are set to 0°, 10° and 30°. The camera shooting height was set to 50 cm within an indoor environment. A series of 20 images were captured under each perspective angle and each looseness state. Figure 14 displays some bolt images with various perspective angles under a 10° looseness state.
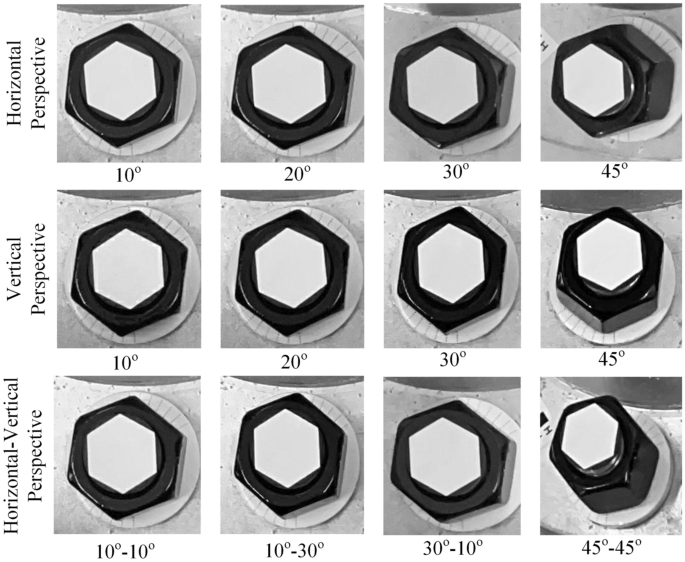
Schematic diagram of bolt images from different perspective angles under a 10° looseness state.
Then, the collected images first undergo distortion correction, and the distortion correction method based on the homography matrix is used 41 . Finally, based on the corrected images and the proposed method in this paper, the looseness angles can be calculated, as shown in Figs. 15 , 16 and 17 . The average and standard deviation values of the results are shown in Fig. 18 . The results show the following:
The perspective angle has a significant impact on the bolt looseness angle. The identification results using bolt images without perspective angle correction fluctuate more significantly. The maximum fluctuation is from the 0° looseness state and 45° horizontal perspective angle, where the maximum identification error reaches 6.5°. Furthermore, based on the standard deviation results, when the perspective angle is greater than 20°, the fluctuations in the identification results increase when using uncorrected bolt images.
The distortion correction method based on the homography matrix is effective. Using the corrected images, the fluctuation of the results is small. The maximum fluctuation is from the 0° looseness state and 45°–45° bidirectional perspective angle, where the maximum identification average value error is 1.1°.
In summary, the perspective angle has a significant impact on loosening angle identification. However, the distortion correction method based on the homography matrix is effective, and the impact of the perspective angle on the identification of loosening of the anti-loosening bolt can be effectively reduced.
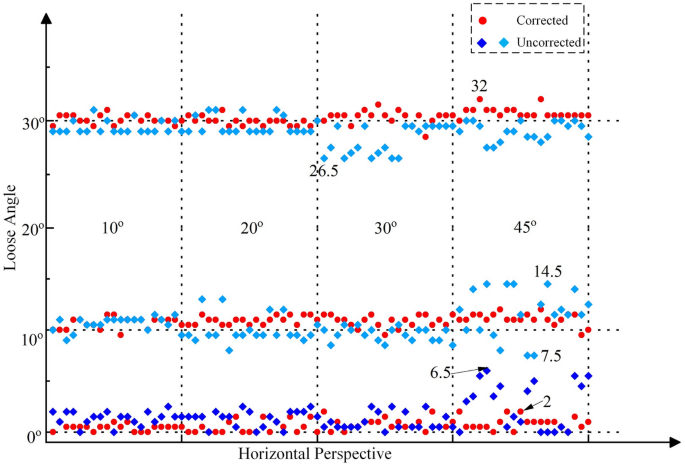
Calculated bolt looseness angles from a horizontal perspective under different looseness states.
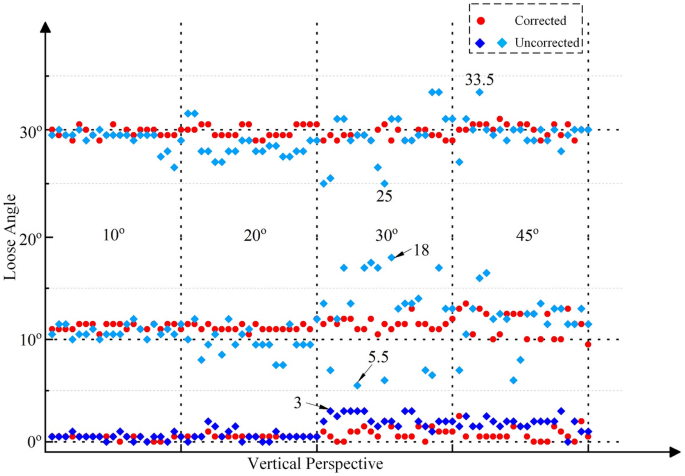
Calculated bolt looseness angle from the vertical perspective under the different looseness states.
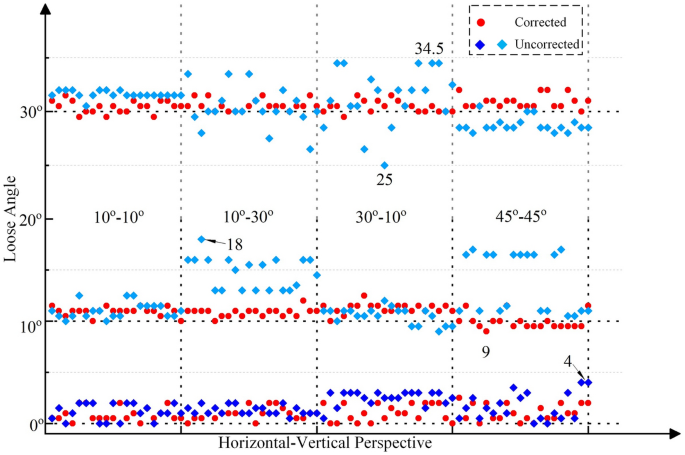
Calculated bolt looseness angle from the horizontal-vertical perspective under the different looseness states.
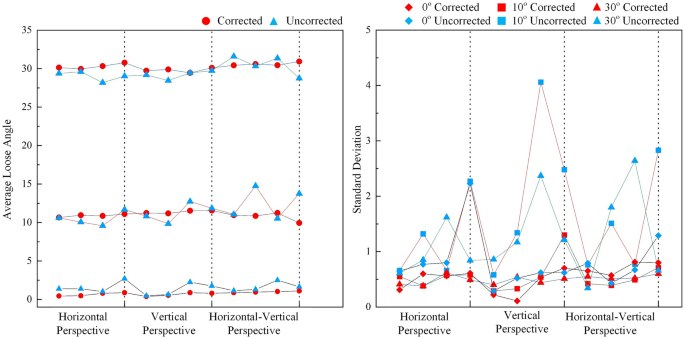
Average and standard deviation values of the anti-loosening bolt loosening diagnostic results for different perspective angles.
This paper proposes a novel anti-loosening bolt looseness diagnosis method using a vision-based technique. A regular hexagonal cap was installed on the screw. Then, a new hexagonal border reconstruction algorithm is proposed to automatically distinguish the hexagonal borders of the screw cap and nut. Finally, a novel anti-loosening bolt looseness diagnosis method is established by using the relative rotation angle of the regular hexagonal borders of the screw cap and nut under initial status and loose status. A prototype flange node is used to verify the proposed method. Four bolt looseness cases, four shooting heights, seven lighting intensities and twelve shooting perspective angles were considered in the experiment. The results show that the proposed method can effectively detect the looseness angle of anti-loosening bolts. Meanwhile, it should be noted that only the standard hexagonal bolts are concerned in this paper and the applicability of the method to other bolt feature needs further discussion.
Data availability
All data generated or analysed during this study are included in this published article.
Motosh, N. Development of design charts for bolts preloaded up to the plastic range. J. Eng. Ind. 98 , 849–851 (1976).
Article Google Scholar
Wang, Z., Zhang, Z. & Cui, C. Measurement and control method for miniature fastener tension in precision instrument. In Proceedings of the 7th CIRP Conference on Assembly Technologies and Systems, Tianjin, China, 10–12 May 2018 , Vol. 76, 110–114.
Yang, J. & Chang, F. K. Detection of bolt loosening in C–C composite thermal protection panels: I. Diagnostic principle. Smart Mater. Struct. 15 (2), 581 (2006).
Article ADS Google Scholar
Chaki, S., Corneloup, G., Lillamand, I. & Walaszek, H. Combination of longitudinal and transverse ultrasonic waves for in situ control of the tightening of bolts. J. Press. Vessel Technol. 129 , 383–390 (2007).
Wang, T. et al. Proof-of-concept study of monitoring bolt connection status using a piezoelectric based active sensing method. Smart Mater. Struct. 22 (8), 087001 (2013).
Yin, H. et al. A smart washer for bolt looseness monitoring based on piezoelectric active sensing method. Appl. Sci. 6 (11), 320 (2016).
Min, J., Park, S. & Yun, C. B. Impedance-based structural health monitoring using neural networks for autonomous frequency range selection. Smart Mater. Struct. 19 (12), 125011 (2010).
Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 6 , 679–698 (1986).
Rahmdel, P., Comley, R., Shi, D., et al. A review of hough transform and line segment detection approaches. In Proceedings of the 10th International Conference on Computer Vision Theory and Applications-Volume 2: VISAPP. 82w90 , 411–418 (2015).
Park, J., Kim, T. & Kim, J. Image-based bolt-loosening detection technique of bolt joint in steel bridges. In 6th International Conference on Advances in Experimental Structural Engineering 11th International Workshop on Advanced Smart Materials and Smart Structures Technology 1–2 (2015).
Nguyen, T. C., Huynh, T. C., Ryu, J. Y., et al. Bolt-loosening identification of bolt connections by vision image-based technique. In Nondestructive Characterization and Monitoring of Advanced Materials, Aerospace, and Civil Infrastructure 2016. SPIE , Vol. 9804, 227–243 (2016).
Kong, X. & Li, J. Image registration-based bolt loosening detection of steel joints. Sensors 18 (4), 1000 (2018).
Article ADS PubMed PubMed Central Google Scholar
Huynh, T. C. et al. Quasi-autonomous bolt-loosening detection method using vision-based deep learning and image processing. Autom. Constr. 105 , 102844 (2019).
Zhao, X., Zhang, Y. & Wang, N. Bolt loosening angle detection technology using deep learning. Struct. Control Health Monit. 26 (1), e2292 (2019).
Zhang, Y. et al. Autonomous bolt loosening detection using deep learning. Struct. Health Monit. 19 (1), 105–122 (2020).
Article CAS Google Scholar
Yang, X. et al. Deep learning-based bolt loosening detection for wind turbine towers. Struct. Control Health Monit. 29 (6), e2943 (2022).
Yuan, C. et al. Automated structural bolt looseness detection using deep learning-based prediction model. Struct. Control Health Monit. 29 (3), e2899 (2022).
An, Y. K. & Sohn, H. Integrated impedance and guided wave based damage detection. Mech. Syst. Signal Process. 28 , 50–62 (2012).
Zhao, Z. et al. Health monitoring of bolt looseness in timber structures using PZT-enabled time-reversal method. J. Sens. 2019 , 1–8 (2019).
CAS Google Scholar
Zhang, Y. et al. Bolt loosening detection based on audio classification. Adv. Struct. Eng. 22 (13), 2882–2891 (2019).
Wang, F. & Song, G. Monitoring of multi-bolt connection looseness using a novel vibro-acoustic method. Nonlinear Dyn. 100 (1), 243–254 (2020).
Ramana, L., Choi, W. & Cha, Y. J. Fully automated vision-based loosened bolt detection using the Viola-Jones algorithm. Struct. Health Monit. 18 (2), 422–434 (2019).
Cha, Y. J., You, K. & Choi, W. Vision-based detection of loosened bolts using the Hough transform and support vector machines. Autom. Constr. 71 , 181–188 (2016).
Luo, J. et al. Image-based bolt-loosening detection using an improved homography-based perspective rectification method. Civ. Struct. Health Monit. 14 , 513–526 (2023).
Zeiler, M. D. & Fergus, R. Visualizing and understanding convolutional networks. In Computer Vision—ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6–12, 2014, Proceedings, Part I 13 818–833 (Springer, 2014).
Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision 1440–1448 (2015).
Ren, S., He, K., Girshick, R., et al. Faster r-cnn: Towards real-time object detection with region proposal networks. In Advances in Neural Information Processing Systems , Vol. 28 (2015).
Jiang, P. et al. A review of Yolo algorithm developments. Procedia Comput. Sci. 199 , 1066–1073 (2022).
Yuan, C. et al. Near real-time bolt-loosening detection using mask and region-based convolutional neural network. Struct. Control Health Monit. 28 (7), e2741 (2021).
He, K., Gkioxari, G., Dollár, P., et al. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision 2961–2969 (2017).
Gong, H. et al. Quantitative loosening detection of threaded fasteners using vision-based deep learning and geometric imaging theory. Autom. Constr. 133 , 104009 (2022).
Pan, X., Tavasoli, S. & Yang, T. Y. Autonomous 3D vision-based bolt loosening assessment using micro aerial vehicles. Comput. Aid. Civ. Infrastruct. Eng. 38 , 2443–2454 (2023).
Pan, X. & Yang, T. Y. 3D vision-based bolt loosening assessment using photogrammetry, deep neural networks, and 3D point-cloud processing. J. Build. Eng. 70 , 106326 (2023).
Luo, J. et al. Bolt-loosening detection using vision technique based on a gray gradient enhancement method. Adv. Struct. Eng. 26 (4), 668–678 (2023).
Jin, F., Fieguth, P., Winger, L., et al. Adaptive Wiener filtering of noisy images and image sequences. In Proceedings 2003 International Conference on Image Processing (Cat. No. 03CH37429). IEEE , Vol. 3, III-349 (2003).
Huang, M., Yu, W. & Zhu, D. An improved image segmentation algorithm based on the Otsu method. In 2012 13th ACIS International Conference on Software Engineering, Artificial Intelligence, Networking and Parallel/Distributed Computing. IEEE 135–139 (2012).
Wu, B. et al. An ameliorated teaching–learning-based optimization algorithm based study of image segmentation for multilevel thresholding using Kapur’s entropy and Otsu’s between class variance. Inf. Sci. 533 , 72–107 (2020).
Article MathSciNet Google Scholar
Xing, J., Yang, P. & Qingge, L. Automatic thresholding using a modified valley emphasis. IET Image Process. 14 (3), 536–544 (2020).
Pare, S. et al. Image segmentation using multilevel thresholding: a research review. Iran. J. Sci. Technol. Trans. Electr. Eng. 44 , 1–29 (2020).
Luo, J. et al. UAV-based operational modal analysis method using improved homography-based perspective rectification method. J. Vib. Control 30 (7–8), 1829–1840 (2024).
Download references
Acknowledgements
This work was funded by the Natural Science Foundation of Chongqing [Grant Number CSTB2023NSCQ-LZX0051]; the Special Funding Project of Chongqing Postdoctoral Research Project [Grant Number 2022CQBSHTB3071]; the Chongqing Urban Investment Infrastructure Construction Co., Ltd. Program [Grant Number CQCT-JS-SC-GC-2022-0079]; and the Research Foundation of Chongqing University of Science and Technology [Grant Number ckrc2019033].

Author information
Authors and affiliations.
School of Civil Engineering and Architecture, Chongqing University of Science and Technology, No. 20, East University Town Road, Shapingba District, Chongqing, 401331, China
Jun Luo, Kaili Li, ChengQian Xie & Zhitao Yan
Chongqing Urban Investment Infrastructure Construction Co., Ltd, Chongqing, China
Feng Li, Xiaogang Jia & Yuanlai Wang
You can also search for this author in PubMed Google Scholar
Contributions
Conceptualization, Z.Y.; methodology, J.L. and C.X.; data curation, Y.W.; validation, F.L. and X.J.; writing—original draft, J.L. and K.L.
Corresponding author
Correspondence to Zhitao Yan .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Luo, J., Li, K., Xie, C. et al. A novel anti-loosening bolt looseness diagnosis of bolt connections using a vision-based technique. Sci Rep 14 , 11441 (2024). https://doi.org/10.1038/s41598-024-62560-8
Download citation
Received : 10 March 2024
Accepted : 18 May 2024
Published : 20 May 2024
DOI : https://doi.org/10.1038/s41598-024-62560-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Bolt connections
- Anti-loosening bolts
- Looseness detection
- Vision-based technique
- Hexagonal border reconstruction
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- Open access
- Published: 13 May 2024
SCIPAC: quantitative estimation of cell-phenotype associations
- Dailin Gan 1 ,
- Yini Zhu 2 ,
- Xin Lu 2 , 3 &
- Jun Li ORCID: orcid.org/0000-0003-4353-5761 1
Genome Biology volume 25 , Article number: 119 ( 2024 ) Cite this article
402 Accesses
2 Altmetric
Metrics details
Numerous algorithms have been proposed to identify cell types in single-cell RNA sequencing data, yet a fundamental problem remains: determining associations between cells and phenotypes such as cancer. We develop SCIPAC, the first algorithm that quantitatively estimates the association between each cell in single-cell data and a phenotype. SCIPAC also provides a p -value for each association and applies to data with virtually any type of phenotype. We demonstrate SCIPAC’s accuracy in simulated data. On four real cancerous or noncancerous datasets, insights from SCIPAC help interpret the data and generate new hypotheses. SCIPAC requires minimum tuning and is computationally very fast.
Single-cell RNA sequencing (scRNA-seq) technologies are revolutionizing biomedical research by providing comprehensive characterizations of diverse cell populations in heterogeneous tissues [ 1 , 2 ]. Unlike bulk RNA sequencing (RNA-seq), which measures the average expression profile of the whole tissue, scRNA-seq gives the expression profiles of thousands of individual cells in the tissue [ 3 , 4 , 5 , 6 , 7 ]. Based on this rich data, cell types may be discovered/determined in an unsupervised (e.g., [ 8 , 9 ]), semi-supervised (e.g., [ 10 , 11 , 12 , 13 ]), or supervised manner (e.g., [ 14 , 15 , 16 ]). Despite the fast development, there are still limitations with scRNA-seq technologies. Notably, the cost for each scRNA-seq experiment is still high; as a result, most scRNA-seq data are from a single or a few biological samples/tissues. Very few datasets consist of large numbers of samples with different phenotypes, e.g., cancer vs. normal. This places great difficulties in determining how a cell type contributes to a phenotype based on single-cell studies (especially if the cell type is discovered in a completely unsupervised manner or if people have limited knowledge of this cell type). For example, without having single-cell data from multiple cancer patients and multiple normal controls, it could be hard to computationally infer whether a cell type may promote or inhibit cancer development. However, such association can be critical for cancer research [ 17 ], disease diagnosis [ 18 ], cell-type targeted therapy development [ 19 ], etc.
Fortunately, this difficulty may be overcome by borrowing information from bulk RNA-seq data. Over the past decade, a considerable amount of bulk RNA-seq data from a large number of samples with different phenotypes have been accumulated and made available through databases like The Cancer Genome Atlas (TCGA) [ 20 ] and cBioPortal [ 21 , 22 ]. Data in these databases often contain comprehensive patient phenotype information, such as cancer status, cancer stages, survival status and time, and tumor metastasis. Combining single-cell data from a single or a few individuals and bulk data from a relatively large number of individuals regarding a particular phenotype can be a cost-effective way to determine how a cell type contributes to the phenotype. A recent method Scissor [ 23 ] took an essential step in this direction. It uses single-cell and bulk RNA-seq data with phenotype information to classify the cells into three discrete categories: Scissor+, Scissor−, and null cells, corresponding to cells that are positively associated, negatively associated, and not associated with the phenotype.
Here, we present a method that takes another big step in this direction, which is called Single-Cell and bulk data-based Identifier for Phenotype Associated Cells or SCIPAC for short. SCIPAC enables quantitative estimation of the strength of association between each cell in a scRNA-seq data and a phenotype, with the help of bulk RNA-seq data with phenotype information. Moreover, SCIPAC also enables the estimation of the statistical significance of the association. That is, it gives a p -value for the association between each cell and the phenotype. Furthermore, SCIPAC enables the estimation of association between cells and an ordinal phenotype (e.g., different stages of cancer), which could be informative as people may not only be interested in the emergence/existence of cancer (cancer vs. healthy, a binary problem) but also in the progression of cancer (different stages of cancer, an ordinal problem).
To study the performance of SCIPAC, we first apply SCIPAC to simulated data under three schemes. SCIPAC shows high accuracy with low false positive rates. We further show the broad applicability of SCIPAC on real datasets across various diseases, including prostate cancer, breast cancer, lung cancer, and muscular dystrophy. The association inferred by SCIPAC is highly informative. In real datasets, some cell types have definite and well-studied functions, while others are less well-understood: their functions may be disease-dependent or tissue-dependent, and they may contain different sub-types with distinct functions. In the former case, SCIPAC’s results agree with current biological knowledge. In the latter case, SCIPAC’s discoveries inspire the generation of new hypotheses regarding the roles and functions of cells under different conditions.
An overview of the SCIPAC algorithm
SCIPAC is a computational method that identifies cells in single-cell data that are associated with a given phenotype. This phenotype can be binary (e.g., cancer vs. normal), ordinal (e.g., cancer stage), continuous (e.g., quantitative traits), or survival (i.e., survival time and status). SCIPAC uses input data consisting of three parts: single-cell RNA-seq data that measures the expression of p genes in m cells, bulk RNA-seq data that measures the expression of the same set of p genes in n samples/tissues, and the statuses/values of the phenotype of the n bulk samples/tissues. The output of SCIPAC is the strength and the p -value of the association between each cell and the phenotype.
SCIPAC proposes the following definition of “association” between a cell and a phenotype: A group of cells that are likely to play a similar role in the phenotype (such as cells of a specific cell type or sub-type, cells in a particular state, cells in a cluster, cells with similar expression profiles, or cells with similar functions) is considered to be positively/negatively associated with a phenotype if an increase in their proportion within the tissue likely indicates an increased/decreased probability of the phenotype’s presence. SCIPAC assigns the same association to all cells within such a group. Taking cancer as the phenotype as an example, if increasing the proportion of a cell type indicates a higher chance of having cancer (binary), having a higher cancer stage (ordinal), or a higher hazard rate (survival), all cells in this cell type is positively associated with cancer.
The algorithm of SCIPAC follows the following four steps. First, the cells in the single-cell data are grouped into clusters according to their expression profiles. The Louvain algorithm from the Seurat package [ 24 , 25 ] is used as the default clustering algorithm, but the user may choose any clustering algorithm they prefer. Or if information of the cell types or other groupings of cells is available a priori, it may be supplied to SCIPAC as the cell clusters, and this clustering step can be skipped. In the second step, a regression model is learned from bulk gene expression data with the phenotype. Depending on the type of the phenotype, this model can be logistic regression, ordinary linear regression, proportional odds model, or Cox proportional hazards model. To achieve a higher prediction power with less variance, by default, the elastic net (a blender of Lasso and ridge regression [ 26 ]) is used to fit the model. In the third step, SCIPAC computes the association strength \(\Lambda\) between each cell cluster and the phenotype based on a mathematical formula that we derive. Finally, the p -values are computed. The association strength and its p -value between a cell cluster and the phenotype are given to all cells in the cluster.
SCIPAC requires minimum tuning. When the cell-type information is given in step 1, SCIPAC does not have any (hyper)parameter. Otherwise, the Louvain algorithm used in step 1 has a “resolution” parameter that controls the number of cell clusters: a larger resolution results in more clusters. SCIPAC inherits this parameter as its only parameter. Since SCIPAC gives the same association strength and p -value to cells from the same cluster, this parameter also determines the resolution of results provided by SCIPAC. Thus, we still call it “resolution” in SCIPAC. Because of its meaning, we recommend setting it so that the number of cell clusters given by the clustering algorithm is comparable to, or reasonably larger than, the number of cell types (or sub-types) in the data. We will see that the performance of SCIPAC is insensitive to this resolution parameter, and the default value 2.0 typically works well.
The details of the SCIPAC algorithm are given in the “ Methods ” section.
Performance in simulated data
We assess the performance of SCIPAC in simulated data under three different schemes. The first scheme is simple and consists of only three cell types. The second scheme is more complicated and consists of seven cell types, which better imitates actual scRNA-seq data. In the third scheme, we simulate cells under different cell development stages to test the performance of SCIPAC under an ordinal phenotype. Details of the simulation are given in Additional file 1.
Simulation scheme I
Under this scheme, the single-cell data consists of three cell types: one is positively associated with the phenotype, one is negatively associated, and the third is not associated (we call it “null”). Figure 1 a gives the UMAP [ 27 ] plot of the three cell types, and Fig. 1 b gives the true associations of these three cell types with the phenotype, with red, blue, and light gray denoting positive, negative, and null associations.
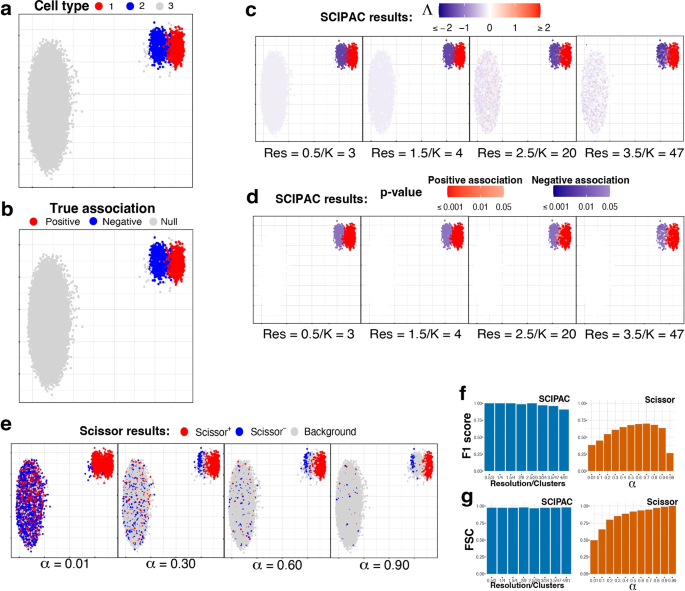
UMAP visualization and numeric measures of the simulated data under scheme I. All the plots in a–e are scatterplots of the two dimensional single-cell data given by UMAP. The x and y axes represent the two dimensions, and their scales are not shown as their specific values are not directly relevant. Points in the plots represents single cells, and they are colored differently in each subplot to reflect different information/results. a Cell types. b True associations. The association between cell types 1, 2, and 3 and the phenotype are positive, negative, and null, respectively. c Association strengths \(\Lambda\) given by SCIPAC under different resolutions. Red/blue represents the sign of \(\Lambda\) , and the shade gives the absolute value of \(\Lambda\) . Every cell is colored red or blue since no \(\Lambda\) is exactly zero. Below each subplot, Res stands for resolution, and K stands for the number of cell clusters given by this resolution. d p -values given by SCIPAC. Only cells with p -value \(< 0.05\) are colored red (positive association) or blue (negative association); others are colored white. e Results given by Scissor under different \(\alpha\) values. Red, blue, and light gray stands for Scissor+, Scissor−, and background (i.e., null) cells. f F1 scores and g FSC for SCIPAC and Scissor under different parameter values. For SCIPAC, each bar is the value under a resolution/number of clusters. For Scissor, each bar is the value under an \(\alpha\)
We apply SCIPAC to the simulated data. For the resolution parameter (see the “ Methods ” section), values 0.5, 1.0, and 1.5 give 3, 4, and 4 clusters, respectively, close to the actual number of cell types. They are good choices based on the guidance for choosing this parameter. To show how SCIPAC behaves under parameter misspecification, we also set the resolution up to 4.0, which gives a whopping 61 clusters. Figure 1 c and d give the association strengths \(\Lambda\) and the p -values given by four different resolutions (results under other resolutions are provided in Additional file 1: Fig. S1 and S2). In Fig. 1 c, red and blue denote positive and negative associations, respectively, and the shade of the color represents the strength of the association, i.e., the absolute value of \(\Lambda\) . Every cell is colored blue or red, as none of \(\Lambda\) is exactly zero. In Fig. 1 d, red and blue denote positive and negative associations that are statistically significant ( p -value \(< 0.05\) ). Cells whose associations are not statistically significant ( p -value \(\ge 0.05\) ) are shown in white. To avoid confusion, it is worth repeating that cells that are colored in red/blue in Fig. 1 c are shown in red/blue in Fig. 1 d only if they are statistically significant; otherwise, they are colored white in Fig. 1 d.
From Fig. 1 c, d (as well as Additional file 1: Fig. S1 and S2), it is clear that the results of SCIPAC are highly consistent under different resolution values, including both the estimated association strengths and the p -values. It is also clear that SCIPAC is highly accurate: most truly associated cells are identified as significant, and most, if not all, truly null cells are identified as null.
As the first algorithm that quantitatively estimates the association strength and the first algorithm that gives the p -value of the association, SCIPAC does not have a real competitor. A previous algorithm, Scissor, is able to classify cells into three discrete categories according to their associations with the phenotype. So, we compare SCIPAC with Scissor in respect of the ability to differentiate positively associated, negatively associated, and null cells.
Running Scissor requires tuning a parameter called \(\alpha\) , which is a number between 0 and 1 that balances the amount of regularization for the smoothness and for the sparsity of the associations. The Scissor R package does not provide a default value for this \(\alpha\) or a function to help select this value. The Scissor paper suggests the following criterion: “the number of Scissor-selected cells should not exceed a certain percentage of total cells (default 20%) in the single-cell data. In each experiment, a search on the above searching list is performed from the smallest to the largest until a value of \(\alpha\) meets the above criteria.” In practice, we have found that this criterion does not often work properly, as the truly associated cells may not compose 20% of all cells in actual data. Therefore, instead of setting \(\alpha\) to any particular value, we set \(\alpha\) values that span the whole range of \(\alpha\) to see the best possible performance of Scissor.
The performance of Scissor in our simulation data under four different \(\alpha\) values are shown in Fig. 1 e, and results under more \(\alpha\) values are shown in Additional file 1: Fig. S3. In the figures, red, blue, and light gray denote Scissor+, Scissor−, and null (called “background” in Scissor) cells, respectively. The results of Scissor have several characteristics different from SCIPAC. First, Scissor does not give the strength or statistical significance of the association, and thus the colors of the cells in the figures do not have different shades. Second, different \(\alpha\) values give very different results. Greater \(\alpha\) values generally give fewer Scissor+ and Scissor− cells, but there are additional complexities. One complexity is that the Scissor+ (or Scissor−) cells under a greater \(\alpha\) value are not a strict subset of Scissor+ (or Scissor−) cells under a smaller \(\alpha\) value. For example, the number of truly negatively associated cells detected as Scissor− increases when \(\alpha\) increases from 0.01 to 0.30. Another complexity is that the direction of the association may flip as \(\alpha\) increases. For example, most cells of cell type 2 are identified as Scissor+ under \(\alpha =0.01\) , but many of them are identified as Scissor− under larger \(\alpha\) values. Third, Scissor does not achieve high power and low false-positive rate at the same time under any \(\alpha\) . No matter what the \(\alpha\) value is, there is only a small proportion of cells from cell type 2 that are correctly identified as negatively associated, and there is always a non-negligible proportion of null cells (i.e., cells from cell type 3) that are incorrectly identified as positively or negatively associated. Fourth, Scissor+ and Scissor− cells can be close to each other in the figure, even under a large \(\alpha\) value. This means that cells with nearly identical expression profiles are detected to be associated with the phenotype in opposite directions, which can place difficulties in interpreting the results.
SCIPAC overcomes the difficulties of Scissor and gives results that are more informative (quantitative strengths with p -values), more accurate (both high power and low false-positive rate), less sensitive to the tuning parameter, and easier to interpret (cells with similar expression typically have similar associations to the phenotype).
SCIPAC’s higher accuracy in differentiating positively associated, negatively associated, and null cells than Scissors can also be measured numerically using the F1 score and the fraction of sign correctness (FSC). F1, which is the harmonic mean of precision and recall, is a commonly used measure of calling accuracy. Note that precision and recall are only defined for two-class problems, which try to classify desired signals/discoveries (so-called “positives”) against noises/trivial results (so-called “negatives”). Our case, on the other hand, is a three-class problem: positive association, negative association, and null. To compute F1, we combine the positive and negative associations and treat them as “positives,” and treat null as “negatives.” This F1 score ignores the direction of the association; thus, it alone is not enough to describe the performance of an association-detection algorithm. For example, an algorithm may have a perfect F1 score even if it incorrectly calls all negative associations positive. To measure an algorithm’s ability to determine the direction of the association, we propose a statistic called FSC, defined as the fraction of true discoveries that also have the correct direction of the association. The F1 score and FSC are numbers between 0 and 1, and higher values are preferred. A mathematical definition of these two measures is given in Additional file 1.
Figure 1 f, g show the F1 score and FSC of SCIPAC and Scissor under different values of tuning parameters. The F1 score of Scissor is between 0.2 and 0.7 under different \(\alpha\) ’s. The FSC of Scissor increases from around 0.5 to nearly 1 as \(\alpha\) increases, but Scissor does not achieve high F1 and FSC scores at the same time under any \(\alpha\) . On the other hand, the F1 score of SCIPAC is close to perfection when the resolution parameter is properly set, and it is still above 0.90 even if the resolution parameter is set too large. The FSC of SCIPAC is always above 0.96 under different resolutions. That is, SCIPAC achieves high F1 and FSC scores simultaneously under a wide range of resolutions, representing a much higher accuracy than Scissor.
Simulation scheme II
This more complicated simulation scheme has seven cell types, which are shown in Fig. 2 a. As shown in Fig. 2 b, cell types 1 and 3 are negatively associated (colored blue), 2 and 4 are positively associated (colored red), and 5, 6, and 7 are not associated (colored light gray).
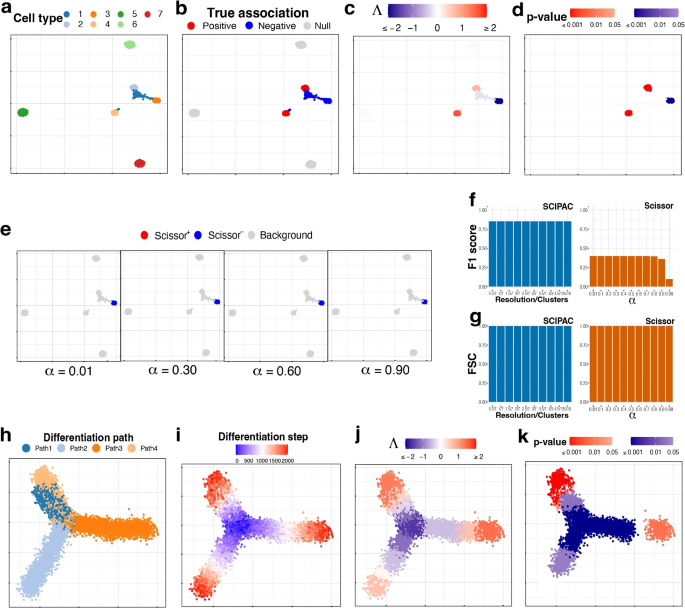
UMAP visualization of the simulated data under a–g scheme II and h–k scheme III. a Cell types. b True associations. c , d Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution. e Results given by Scissor under different \(\alpha\) values. f F1 scores and g FSC for SCIPAC and Scissor under different parameter values. h Cell differentiation paths. The four paths have the same starting location, which is in the center, but different ending locations. This can be considered as a progenitor cell type differentiating into four specialized cell types. i Cell differentiation steps. These steps are used to create four stages, each containing 500 steps. Thus, this plot of differentiation steps can also be viewed as the plot of true association strengths. j , k Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution
The association strengths and p -values given by SCIPAC under the default resolution are illustrated in Fig. 2 c, d, respectively. Results under several other resolutions are given in Additional file 1: Fig. S4 and S5. Again, we find that SCIPAC gives highly consistent results under different resolutions. SCIPAC successfully identifies three out of the four truly associated cell types. For the other truly associated cell type, cell type 1, SCIPAC correctly recognizes its association with the phenotype as negative, although the p -values are not significant enough. The F1 score is 0.85, and the FSC is greater than 0.99, as shown in Fig. 2 f, g.
The results of Scissor under four different \(\alpha\) values are given in Fig. 2 e. (More shown in Additional file 1: Fig. S6.) Under this highly challenging simulation scheme, Scissor can only identify one out of four truly associated cell types. Its F1 score is below 0.4.
Simulation scheme III
This simulation scheme is to assess the performance of SCIPAC for ordinal phenotypes. We simulate cells along four cell-differentiation paths with the same starting location but different ending locations, as shown in Fig. 2 h. These cells can be considered as a progenitor cell population differentiating into four specialized cell types. In Fig. 2 i, the “step” reflects their position in the differentiation path, with step 0 meaning the start and step 2000 meaning the end of the differentiation. Then, the “stage” is generated according to the step: cells in steps 0 \(\sim\) 500, 501 \(\sim\) 1000, 1001 \(\sim\) 1500, and 1501 \(\sim\) 2000 are assigned to stages I, II, III, and IV, respectively. This stage is treated as the ordinal phenotype. Under this simulation scheme, Fig. 2 i also gives the actual associations, and all cells are associated with the phenotype.
The results of SCIPAC under the default resolution are shown in Fig. 2 j, k. Clearly, the associations SCIPAC identifies are highly consistent with the truth. Particularly, it successfully identifies the cells in the center as early-stage cells and most cells at the end of branches as last-stage cells. The results of SCIPAC under other resolutions are given in Additional file 1: Fig. S7 and S8, which are highly consistent. Scissor does not work with ordinal phenotypes; thus, no results are reported here.
Performance in real data
We consider four real datasets: a prostate cancer dataset, a breast cancer dataset, a lung cancer dataset, and a muscular dystrophy dataset. The bulk RNA-seq data of the three cancer datasets are obtained from the TCGA database, and that of the muscular dystrophy dataset is obtained from a published paper [ 28 ]. A detailed description of these datasets is given in Additional file 1. We will use these datasets to assess the performance of SCIPAC on different types of phenotypes. The cell type information (i.e., which cell belongs to which cell type) is available for the first three datasets, but we ignore this information so that we can make a fair comparison with Scissor, which cannot utilize this information.
Prostate cancer data with a binary phenotype
We use the single-cell expression of 8,700 cells from prostate-cancer tumors sequenced by [ 29 ]. The cell types of these cells are known and given in Fig. 3 a. The bulk data comprises 550 TCGA-PRAD (prostate adenocarcinoma) samples with phenotype (cancer vs. normal) information. Here the phenotype is cancer, and it is binary: present or absent.
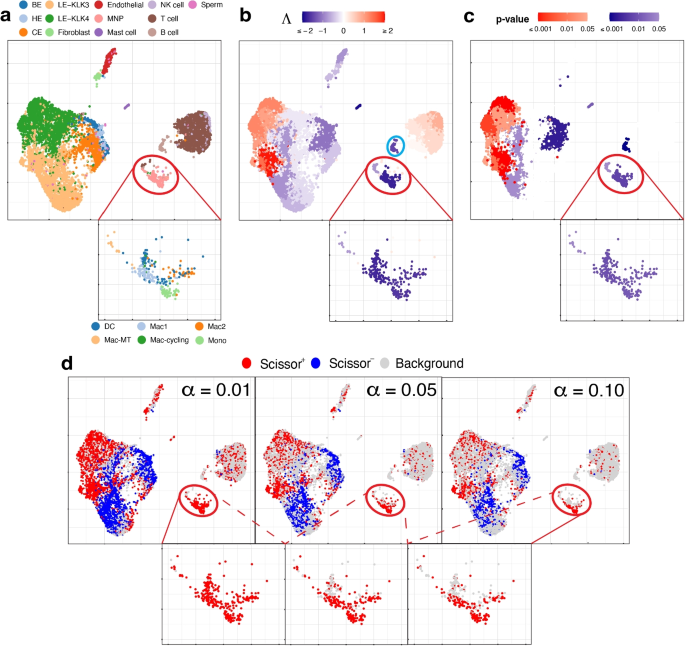
UMAP visualization of the prostate cancer data, with a zoom-in view for the red-circled region (cell type MNP). a True cell types. BE, HE, and CE stand for basal, hillock, club epithelial cells, LE-KLK3 and LE-KLK4 stand for luminal epithelial cells with high levels of kallikrein related peptidase 3 and 4, and MNP stands for mononuclear phagocytes. In the zoom-in view, the sub-types of MNP cells are given. b Association strengths \(\Lambda\) given by SCIPAC under the default resolution. The cyan-circled cells are B cells, which are estimated by SCIPAC as negatively associated with cancer but estimated by Scissor as Scissor+ or null. c p -values given by SCIPAC. The MNP cell type, which is red-circled in the plot, is estimated by SCIPAC to be strongly negatively associated with cancer but estimated by Scissor to be positively associated with cancer. d Results given by Scissor under different \(\alpha\) values
Results from SCIPAC with the default resolution are shown in Fig. 3 b, c (results with other resolutions, given in Additional file 1: Fig. S9 and S10, are highly consistent with results here.) Compared with results from Scissor, shown in Fig. 3 d, results from SCIPAC again show three advantages. First, results from SCIPAC are richer and more comprehensive. SCIPAC gives estimated associations and the corresponding p -values, and the estimated associations are quantitative (shown in Fig. 3 b as different shades to the red or blue color) instead of discrete (shown in Fig. 3 d as a uniform shade to the red, blue, or light gray color). Second, SCIPAC’s results can be easier to interpret as the red and blue colors are more block-wise instead of scattered. Third, unlike Scissor, which produces multiple sets of results varying based on the parameter \(\alpha\) —a parameter without a default value or tuning guidance—typically, a single set of results from SCIPAC under its default settings suffices.
Comparing the results from our SCIPAC method with those from Scissor is non-trivial, as the latter’s outcomes are scattered and include multiple sets. We propose the following solutions to summarize the inferred association of a known cell type with the phenotype using a specific method (Scissor under a specific \(\alpha\) value, or SCIPAC with the default setting). We first calculate the proportion of cells in this cell type identified as Scissor+ (by Scissor at a specific \(\alpha\) value) or as significantly positively associated (by SCIPAC), denoted by \(p_{+}\) . We also calculate the proportion of all cells, encompassing any cell type, which are identified as Scissor+ or significantly positively associated, serving as the average background strength, denoted by \(p_{a}\) . Then, we compute the log odds ratio for this cell type to be positively associated with the phenotype compared to the background, represented as:
Similarly, the log odds ratio for the cell type to be negatively associated with the phenotype, \(\rho _-\) , is computed in a parallel manner.
For SCIPAC, a cell type is summarized as positively associated with the phenotype if \(\rho _+ \ge 1\) and \(\rho _- < 1\) and negatively associated if \(\rho _- \ge 1\) and \(\rho _+ < 1\) . If neither condition is met, the association is inconclusive. For Scissor, we apply it under six different \(\alpha\) values: 0.01, 0.05, 0.10, 0.15, 0.20, and 0.25. A cell type is summarized as positively associated with the phenotype if \(\rho _+ \ge 1\) and \(\rho _- < 1\) in at least four of these \(\alpha\) values and negatively associated if \(\rho _- \ge 1\) and \(\rho _+ < 1\) in at least four \(\alpha\) values. If these criteria are not met, the association is deemed inconclusive. The above computation of log odds ratios and the determination of associations are performed only on cell types that each compose at least 1% of the cell population, to ensure adequate power.
For the prostate cancer data, the log odds ratios for each cell type using each method are presented in Tables S1 and S2. The final associations determined for each cell type are summarized in Table S3. In the last column of this table, we also indicate whether the conclusions drawn from SCIPAC and Scissor are consistent or not.
We find that SCIPAC’s results agree with Scissor on most cell types. However, there are three exceptions: mononuclear phagocytes (MNPs), B cells, and LE-KLK4.
MNPs are red-circled and zoomed in in each sub-figure of Fig. 3 . Most cells in this cell type are colored red in Fig. 3 d but colored dark blue in Fig. 3 b. In other words, while Scissor determines that this cell type is Scissor+, SCIPAC makes the opposite inference. Moreover, SCIPAC is confident about its judgment by giving small p -values, as shown in Fig. 3 c. To see which inference is closer to the biological fact is not easy, as biologically MNPs contain a number of sub-types that each have different functions [ 30 , 31 ]. Fortunately, this cell population has been studied in detail in the original paper that generated this dataset [ 29 ], and the sub-type information of each cell is provided there: this MNP population contains six sub-types, which are dendritic cells (DC), M1 macrophages (Mac1), metallothionein-expressing macrophages (Mac-MT), M2 macrophages (Mac2), proliferating macrophages (Mac-cycling), and monocytes (Mono), as shown in the zoom-in view of Fig. 3 a. Among these six sub-types, DC, Mac1, and Mac-MT are believed to inhibit cancer development and can serve as targets in cancer immunotherapy [ 29 ]; they compose more than 60% of all MNP cells in this dataset. SCIPAC makes the correct inference on this majority of MNP cells. Another cell type, Mac2, is reported to promote tumor development [ 32 ], but it only composes less than \(15\%\) of the MNPs. How the other two cell types, Mac-cycling and Mono, are associated with cancer is less studied. Overall, the results given by SCIPAC are more consistent with the current biological knowledge.
B cells are cyan-circled in Fig. 3 b. B cells are generally believed to have anti-tumor activity by producing tumor-reactive antibodies and forming tertiary lymphoid structures [ 29 , 33 ]. This means that B cells are likely to be negatively associated with cancer. SCIPAC successfully identifies this negative association, while Scissor fails.
LE-KLK4, a subtype of cancer cells, is thought to be positively associated with the tumor phenotype [ 29 ]. SCIPAC successfully identified this positive association, in contrast to Scissor, which failed to do so (in the figure, a proportion of LE-KLK4 cells are identified as Scissor+, especially under the smallest \(\alpha\) value; however, this proportion is not significantly higher than the background Scissor+ level under the majority of \(\alpha\) values).
In summary, across all three cell types, the results from SCIPAC appear to be more consistent with current biological knowledge. For more discussions regarding this dataset, refer to Additional file 1.
Breast cancer data with an ordinal phenotype
The scRNA-seq data for breast cancer are from [ 34 ], and we use the 19,311 cells from the five HER2+ tumor tissues. The true cell types are shown in Fig. 4 a. The bulk data include 1215 TCGA-BRCA samples with information on the cancer stage (I, II, III, or IV), which is treated as an ordinal phenotype.
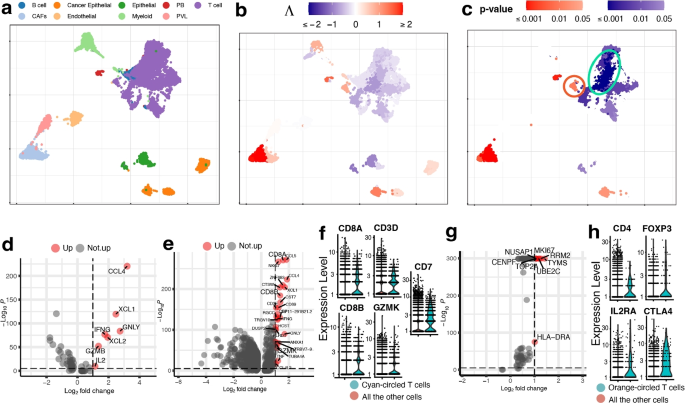
UMAP visualization of the breast cancer data. a True cell types. CAFs stand for cancer-associated fibroblasts, PB stands for plasmablasts and PVL stands for perivascular-like cells. b , c Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution. Cyan-circled are a group of T cells that are estimated by SCIPAC to be most significantly associated with the cancer stage in the negative direction, and orange-circled are a group of T cells that are estimated by SCIPAC to be significantly positively associated with the cancer stage. d DE analysis of the cyan-circled T cells vs. all the other T cells. e DE analysis of the cyan-circled T cells vs. all the other cells. f Expression of CD8+ T cell marker genes in the cyan-circled cells and all the other cells. g DE analysis of the orange-circled T cells vs. all the other cells. h Expression of regulatory T cell marker genes in the orange-circled cells and all the other cells
Association strengths and p -values given by SCIPAC under the default resolution are shown in Fig. 4 b, c. Results under other resolutions are given in Additional file 1: Fig. S11 and S12, and again they are highly consistent with results under the default resolution. We do not present the results from Scissor, as Scissor does not take ordinal phenotypes.
In the SCIPAC results, cells that are most strongly and statistically significantly associated with the phenotype in the positive direction are the cancer-associated fibroblasts (CAFs). This finding agrees with the literature: CAFs contribute to therapy resistance and metastasis of cancer cells via the production of secreted factors and direct interaction with cancer cells [ 35 ], and they are also active players in breast cancer initiation and progression [ 36 , 37 , 38 , 39 ]. Another large group of cells identified as positively associated with the phenotype is the cancer epithelial cells. They are malignant cells in breast cancer tissues and are thus expected to be associated with severe cancer stages.
Of the cells identified as negatively associated with severe cancer stages, a large portion of T cells is the most noticeable. Biologically, T cells contain many sub-types, including CD4+, CD8+, regulatory T cells, and more, and their functions are diverse in the tumor microenvironment [ 40 ]. To explore SCIPAC’s discoveries, we compare T cells that are identified as most statistically significant, with p -values \(< 10^{-6}\) and circled in Fig. 4 d, with the other T cells. Differential expression (DE) analysis (details about DE analysis and other analyses are given in Additional file 1) identifies seven genes upregulated in these most significant T cells. Of these seven genes, at least five are supported by the literature: CCL4, XCL1, IFNG, and GZMB are associated with CD8+ T cell infiltration; they have been shown to have anti-tumor functions and are involved in cancer immunotherapy [ 41 , 42 , 43 ]. Also, IL2 has been shown to serve an important role in combination therapies for autoimmunity and cancer [ 44 ]. We also perform an enrichment analysis [ 45 ], in which a pathway called Myc stands out with a \(\textit{p}\text{-value}<10^{-7}\) , much smaller than all other pathways. Myc is downregulated in the T cells that are identified as most negatively associated with cancer stage progress. This agrees with current biological knowledge about this pathway: Myc is known to contribute to malignant cell transformation and tumor metastasis [ 46 , 47 , 48 ].
On the above, we have compared T cells that are most significantly associated with cancer stages in the negative direction with the other T cells using DE and pathway analysis, and the results could suggest that these cells are tumor-infiltrated CD8+ T cells with tumor-inhibition functions. To check this hypothesis, we perform DE analysis of these cells against all other cells (i.e., the other T cells and all the other cell types). The DE genes are shown in Fig. 4 e. It can be noted that CD8+ T cell marker genes such as CD8A, CD8B, and GZMK are upregulated. We further obtain CD8+ T cell marker genes from CellMarker [ 49 ] and check their expression, as illustrated in Fig. 4 f. Marker genes CD8A, CD8B, CD3D, GZMK, and CD7 show significantly higher expression in these T cells. This again supports our hypothesis that these cells are tumor-infiltrated CD8+ T cells that have anti-tumor functions.
Interestingly, not all T cells are identified as negatively associated with severe cancer stages; a group of T cells is identified as positively associated, as circled in Fig. 4 c. To explore the function of this group of T cells, we perform DE analysis of these T cells against the other T cells. The DE genes are shown in Fig. 4 g. Based on the literature, six out of eight over-expressed genes are associated with cancer development. The high expression of NUSAP1 gene is associated with poor patient overall survival, and this gene also serves as a prognostic factor in breast cancer [ 50 , 51 , 52 ]. Gene MKI67 has been treated as a candidate prognostic prediction for cancer proliferation [ 53 , 54 ]. The over-expression of RRM2 has been linked to higher proliferation and invasiveness of malignant cells [ 55 , 56 ], and the upregulation of RRM2 in breast cancer suggests it to be a possible prognostic indicator [ 57 , 58 , 59 , 60 , 61 , 62 ]. The high expression of UBE2C gene always occurs in cancers with a high degree of malignancy, low differentiation, and high metastatic tendency [ 63 ]. For gene TOP2A, it has been proposed that the HER2 amplification in HER2 breast cancers may be a direct result of the frequent co-amplification of TOP2A [ 64 , 65 , 66 ], and there is a high correlation between the high expressions of TOP2A and the oncogene HER2 [ 67 , 68 ]. Gene CENPF is a cell cycle-associated gene, and it has been identified as a marker of cell proliferation in breast cancers [ 69 ]. The over-expression of these genes strongly supports the correctness of the association identified by SCIPAC. To further validate this positive association, we perform DE analysis of these cells against all the other cells. We find that the top marker genes obtained from CellMarker [ 49 ] for the regulatory T cells, which are known to be immunosuppressive and promote cancer progression [ 70 ], are over-expressed with statistical significance, as shown in Fig. 4 h. This finding again provides strong evidence that the positive association identified by SCIPAC for this group of T cells is correct.
Lung cancer data with survival information
The scRNA-seq data for lung cancer are from [ 71 ], and we use two lung adenocarcinoma (LUAD) patients’ data with 29,888 cells. The true cell types are shown in Fig. 5 a. The bulk data consist of 576 TCGA-LUAD samples with survival status and time.
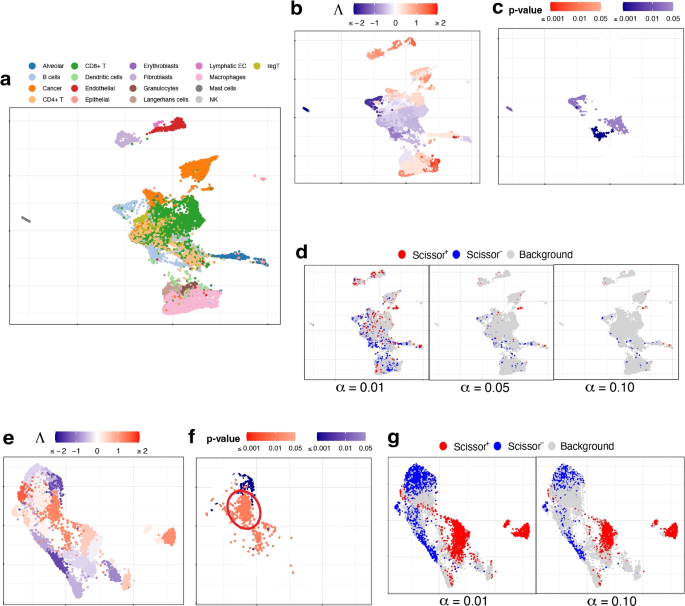
UMAP visualization of a–d the lung cancer data and e–g the muscular dystrophy data. a True cell types. b , c Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution. d Results given by Scissor under different \(\alpha\) values. e , f Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution. Circled are a group of cells that are identified by SCIPAC as significantly positively associated with the disease but identified by Scissor as null. g Results given by Scissor under different \(\alpha\) values
Association strengths and p -values given by SCIPAC are given in Fig. 5 b, c (results under other resolutions are given in Additional file 1: Fig. S13 and S14). In Fig. 5 c, most cells with statistically significant associations are CD4+ T cells or B cells. These associations are negative, meaning that the abundance of these cells is associated with a reduced death rate, i.e., longer survival time. This agrees with the literature: CD4+ T cells primarily mediate anti-tumor immunity and are associated with favorable prognosis in lung cancer patients [ 72 , 73 , 74 ]; B cells also show anti-tumor functions in all stages of human lung cancer development and play an essential role in anti-tumor responses [ 75 , 76 ].
The results by Scissor under different \(\alpha\) values are shown in Fig. 5 d. The highly scattered Scissor+ and Scissor− cells make identifying and interpreting meaningful phenotype-associated cell groups difficult.
Muscular dystrophy data with a binary phenotype
This dataset contains cells from four facioscapulohumeral muscular dystrophy (FSHD) samples and two control samples [ 77 ]. We pool all the 7047 cells from these six samples together. The true cell types of these cells are unknown. The bulk data consists of 27 FSHD patients and eight controls from [ 28 ]. Here the phenotype is FSHD, and it is binary: present or absent.
The results of SCIPAC with the default resolution are given in Fig. 5 e, f. Results under other resolutions are highly similar (shown in Additional file 1: Fig. S15 and S16). For comparison, results given by Scissor under different \(\alpha\) values are presented in Fig. 5 g. The agreements between the results of SCIPAC and Scissor are clear. For example, both methods identify cells located at the top and lower left part of UMAP plots to be negatively associated with FSHD, and cells located at the center and right parts of UMAP plots to be positively associated. However, the discrepancies in their results are also evident. The most pronounced one is a large group of cells (circled in Fig. 5 f) that are identified by SCIPAC as significantly positively associated but are completely ignored by Scissor. Checking into this group of cells, we find that over 90% (424 out of 469) come from the FSHD patients, and less than 10% come from the control samples. However, cells from FSHD patients only compose 73% (5133) of all the 7047 cells. This statistically significant ( p -value \(<10^{-15}\) , Fisher’s exact test) over-representation (odds ratio = 3.51) suggests that the positive association identified SCIPAC is likely to be correct.
SCIPAC is computationally highly efficient. On an 8-core machine with 2.50 GHz CPU and 16 GB RAM, SCIPAC takes 7, 24, and 2 s to finish all the computation and give the estimated association strengths and p -values on the prostate cancer, lung cancer, and muscular dystrophy datasets, respectively. As a reference, Scissor takes 314, 539, and 171 seconds, respectively.
SCIPAC works with various phenotype types, including binary, continuous, survival, and ordinal. It can easily accommodate other types by using a proper regression model with a systematic component in the form of Eq. 3 (see the “ Methods ” section). For example, a Poisson or negative binomial log-linear model can be used if the phenotype is a count (i.e., non-negative integer).
In SCIPAC’s definition of association, a cell type is associated with the phenotype if increasing the proportion of this cell type leads to a change of probability of the phenotype occurring. The strength of association represents the extent of the increase or decrease in this probability. In the case of binary-response, this change is measured by the log odds ratio. For example, if the association strength of cell type A is twice that of cell type B, increasing cell type A by a certain proportion leads to twice the amount of change in the log odds ratio of having the phenotype compared to increasing cell type B by the same proportion. The association strength under other types of phenotypes can be interpreted similarly, with the major difference lying in the measure of change in probability. For quantitative, ordinal, and survival outcomes, the difference in the quantitative outcome, log odds ratio of the right-tail probability, and log hazard ratio respectively are used. Despite the differences in the exact form of the association strength under different types of phenotypes, the underlying concept remains the same: a larger (absolute value of) association strength indicates that the same increase/decrease in a cell type leads to a larger change in the occurrence of the phenotype.
As SCIPAC utilizes both bulk RNA-seq data with phenotype and single-cell RNA-seq data, the estimated associations for the cells are influenced by the choice of the bulk data. Although different bulk data can yield varying estimations of the association for the same single cells, the estimated associations appear to be reasonably robust even when minor changes are made to the bulk data. See Additional file 1 for further discussions.
When using the Louvain algorithm in the Seurat package to cluster cells, SCIPAC’s default resolution is 2.0, larger than the default setting of Seurat. This allows for the identification of potential subtypes within the major cell type and enables the estimation of individual association strengths. Consequently, a more detailed and comprehensive description of the association between single cells and the phenotype can be obtained by SCIPAC.
When applying SCIPAC to real datasets, we made a deliberate choice to disregard the cell annotation provided by the original publications and instead relied on the inferred cell clusters produced by the Louvain algorithm. We made this decision for several reasons. Firstly, we aimed to ensure a fair comparison with Scissor, as it does not utilize cell-type annotations. Secondly, the original annotation might not be sufficiently comprehensive or detailed. Presumed cell types could potentially encompass multiple subtypes, each of which may exhibit distinct associations with the phenotype under investigation. In such cases, employing the Louvain algorithm with a relatively high resolution, which is the default setting in SCIPAC, enables us to differentiate between these subtypes and allows SCIPAC to assign varying association strengths to each subtype.
SCIPAC fits the regression model using the elastic net, a machine-learning algorithm that maximizes a penalized version of the likelihood. The elastic net can be replaced by other penalized estimates of regression models, such as SCAD [ 78 ], without altering the rest of the SCIPAC algorithm. The combination of a regression model and a penalized estimation algorithm such as the elastic net has shown comparable or higher prediction power than other sophisticated methods such as random forests, boosting, or neural networks in numerous applications, especially for gene expression data [ 79 ]. However, there can still be datasets where other models have higher prediction power. It will be future work to incorporate these models into SCIPAC.
The use of metacells is becoming an efficient way to handle large single-cell datasets [ 80 , 81 , 82 , 83 ]. Conceptually, SCIPAC can incorporate metacells and their representatives as an alternative to its default setting of using cell clusters/types and their centroids. We have explored this aspect using metacells provided by SEACells [ 81 ]. Details are given in Additional file 1. Our comparative analysis reveals that combining SCIPAC with SEACells results in significantly reduced performance compared to using SCIPAC directly on original single-cell data. The primary reason for this appears to be the subpar performance of SEACells in cell grouping, especially when contrasted with the Louvain algorithm. Given these findings, we do not suggest using metacells provided by SEACells for SCIPAC applications in the current stage.
Conclusions
SCIPAC is a novel algorithm for studying the associations between cells and phenotypes. Compared to the previous algorithm, SCIPAC gives a much more detailed and comprehensive description of the associations by enabling a quantitative estimation of the association strength and by providing a quality control—the p -value. Underlying SCIPAC are a general statistical model that accommodates virtually all types of phenotypes, including ordinal (and potentially count) phenotypes that have never been considered before, and a concise and closed-form mathematical formula that quantifies the association, which minimizes the computational load. The mathematical conciseness also largely frees SCIPAC from parameter tuning. The only parameter (i.e., the resolution) barely changes the results given by SCIPAC. Overall, compared with its predecessor, SCIPAC represents a substantially more capable software by being much more informative, versatile, robust, and user-friendly.
The improvement in accuracy is also remarkable. In simulated data, SCIPAC achieves high power and low false positives, which is evident from the UMAP plot, F1 score, and FSC score. In real data, SCIPAC gives results that are consistent with current biological knowledge for cell types whose functions are well understood. For cell types whose functions are less studied or more multifaceted, SCIPAC gives support to certain biological hypotheses or helps identify/discover cell sub-types.
SCIPAC’s identification of cell-phenotype associations closely follows its definition of association: when increasing the fraction of a cell type increases (or decreases) the probability for a phenotype to be present, this cell type is positively (or negatively) associated with the phenotype.
The increase of the fraction of a cell type
For a bulk sample, let vector \(\varvec{G} \in \mathbb {R}^p\) be its expression profile, that is, its expression on the p genes. Suppose there are K cell types in the tissue, and let \(\varvec{g}_{k}\) be the representative expression of the k ’th cell type. Usually, people assume that \(\varvec{G}\) can be decomposed by
where \(\gamma _{k}\) is the proportion of cell type k in the bulk tissue, with \(\sum _{k = 1}^{K}\gamma _{k} = 1\) . This equation links the bulk and single-cell expression data.
Now consider increasing cells from cell type k by \(\Delta \gamma\) proportion of the original number of cells. Then, the new proportion of cell type k becomes \(\frac{\gamma _{k} + \Delta \gamma }{1 + \Delta \gamma }\) , and the new proportion of cell type \(j \ne k\) becomes \(\frac{\gamma _{j}}{1 + \Delta \gamma }\) (note that the new proportions of all cell types should still add up to 1). Thus, the bulk expression profile with the increase of cell type k becomes
Plugging Eq. 1 , we get
Interestingly, this expression of \(\varvec{G}^*\) does not include \(\gamma _{1}, \ldots , \gamma _{K}\) . This means that there is no need actually to compute \(\gamma _{1}, \ldots , \gamma _{K}\) in Eq. 1 , which could otherwise be done using a cell-type-decomposition software, but an accurate and robust decomposition is non-trivial [ 84 , 85 , 86 ]. See Additional file 1 for a more in-depth discussion on the connections of SCIPAC with decomposition/deconvolution.
The change in chance of a phenotype
In this section, we consider how the increase in the fraction of a cell type will change the chance for a binary phenotype such as cancer to occur. Other types of phenotypes will be considered in the next section.
Let \(\pi (\varvec{G})\) be the chance of an individual with gene expression profile \(\varvec{G}\) for this phenotype to occur. We assume a logistic regression model to describe the relationship between \(\pi (\varvec{G})\) and \(\varvec{G}\) :
here the left-hand side is the log odds of \(\pi (\varvec{G})\) , \(\beta _{0}\) is the intercept, and \(\varvec{\beta }\) is a length- p vector of coefficients. In the section after the next, we will describe how we obtain \(\beta _{0}\) and \(\varvec{\beta }\) from the data.
When increasing cells from cell type k by \(\Delta \gamma\) , \(\varvec{G}\) becomes \(\varvec{G}^*\) in Eq. 3 . Plugging Eq. 2 , we get
We further take the difference between Eqs. 4 and 3 and get
The left-hand side of this equation is the log odds ratio (i.e., the change of log odds). On the right-hand side, \(\frac{\Delta \gamma }{1 + \Delta \gamma }\) is an increasing function with respect to \(\Delta \gamma\) , and \(\varvec{\beta }^T(\varvec{g}_{k} - \varvec{G})\) is independent of \(\Delta \gamma\) . This indicates that given any specific \(\Delta \gamma\) , the log odds ratio under over-representation of cell type k is proportional to
\(\lambda _k\) describes the strength of the effect of increasing cell type k to a bulk sample with expression profile \(\varvec{G}\) . Given the presence of numerous bulk samples, employing multiple \(\lambda _k\) ’s could be cumbersome and obscure the overall effect of a particular cell type. To concisely summarize the association of cell type k , we propose averaging their effects. The average effect on all bulk samples can be obtained by
where \(\bar{\varvec{G}}\) is the average expression profile of all bulk samples.
\(\Lambda _k\) gives an overall impression of how strong the effect is when cell type k over-represents to the probability for the phenotype to be present. Its sign represents the direction of the change: a positive value means an increase in probability, and a negative value means a decrease in probability. Its absolute value represents the strength of the effect. In SCIPAC, we call \(\Lambda _k\) the association strength of cell type k and the phenotype.
Note that this derivation does not involve likelihood, although the computation of \(\varvec{\beta }\) does. Here, it serves more as a definitional approach.
Definition of the association strength for other types of phenotype
Our definition of \(\Lambda _k\) relies on vector \(\varvec{\beta }\) . In the case of a binary phenotype, \(\varvec{\beta }\) are the coefficients of a logistic regression that describes a linear relationship between the expression profile and the log odds of having the phenotype, as shown in Eq. 3 . For other types of phenotype, \(\varvec{\beta }\) can be defined/computed similarly.
For a quantitative (i.e., continuous) phenotype, an ordinary linear regression can be used, and the left-hand side of Eq. 3 is changed to the quantitative value of the phenotype.
For a survival phenotype, a Cox proportional hazards model can be used, and the left-hand side of Eq. 3 is changed to the log hazard ratio.
For an ordinal phenotype, we use a proportional odds model
where \(j \in \{1, 2, ..., (J - 1)\}\) and J is the number of ordinal levels. It should be noted that here we use the right-tail probability \(\Pr (Y_{i} \ge j + 1 | X)\) instead of the commonly used cumulative probability (left-tail probability) \(\Pr (Y_{i} \le j | X)\) . Such a change makes the interpretation consistent with other types of phenotypes: in our model, a larger value on the right-hand side indicates a larger chance for \(Y_{i}\) to have a higher level, which in turn guarantees that the sign of the association strength defined according to this \(\varvec{\beta }\) has the usual meaning: a positive \(\Lambda _k\) value means a positive association with the phenotype-using the cancer stage as an example. A positive \(\Lambda _k\) means the over-representation of cell type k increases the chance of a higher cancer stage. In contrast, using the commonly used cumulative probability leads to a counter-intuitive, reversed interpretation.
Computation of the association strength in practice
In practice, \(\varvec{\beta }\) in Eq. 3 needs to be learned from the bulk data. By default, SCIPAC uses the elastic net, a popular and powerful penalized regression method:
In this model, \(l(\beta _{0}, \varvec{\beta })\) is a log-likelihood of the linear model (i.e., logistic regression for a binary phenotype, ordinary linear regression for a quantitative phenotype, Cox proportional odds model for a survival phenotype, and proportional odds model for an ordinal phenotype). \(\alpha\) is a number between 0 and 1, denoting a combination of \(\ell _1\) and \(\ell _2\) penalties, and \(\lambda\) is the penalty strength. SCIPAC fixes \(\alpha\) to be 0.4 (see Additional file 1 for discussions on this choice) and uses 10-fold cross-validation to decide \(\lambda\) automatically. This way, they do not become hyperparameters.
In SCIPAC, the fitting and cross-validation of the elastic net are done by calling the ordinalNet [ 87 ] R package for the ordinal phenotype and by calling the glmnet R package [ 88 , 89 , 90 , 91 ] for other types of phenotypes.
The computation of the association strength, as defined by Eq. 7 , does not only require \(\varvec{\beta }\) , but also \(\varvec{g}_k\) and \(\bar{\varvec{G}}\) . \(\bar{\varvec{G}}\) is simply the average expression profile of all bulk samples. On the other hand, \(\varvec{g}_k\) requires knowing the cell type of each cell. By default, SCIPAC does not assume this information to be given, and it uses the Louvain clustering implemented in the Seurat [ 24 , 25 ] R package to infer it. This clustering algorithm has one tuning parameter called “resolution.” SCIPAC sets its default value as 2.0, and the user can use other values. With the inferred or given cell types, \(\varvec{g}_k\) is computed as the centroid (i.e., the mean expression profile) of cells in cluster k .
Given \(\varvec{\beta }\) , \(\bar{\varvec{G}}\) , and \(\varvec{g}_k\) , the association strength can be computed using Eq. 7 . Knowing the association strength for each cell type and the cell-type label for each cell, we also know the association strength for every single cell. In practice, we standardize the association strengths for all cells. That is, we compute the mean and standard deviation of the association strengths of all cells and use them to centralize and scale the association strength, respectively. We have found such standardization makes SCIPAC more robust to the possible unbalance in sample size of bulk data in different phenotype groups.
Computation of the p -value
SCIPAC uses non-parametric bootstrap [ 92 ] to compute the standard deviation and hence the p -value of the association. Fifty bootstrap samples, which are believed to be enough to compute the standard error of most statistics [ 93 ], are generated for the bulk expression data, and each is used to compute (standardized) \(\Lambda\) values for all the cells. For cell i , let its original \(\Lambda\) values be \(\Lambda _i\) , and the bootstrapped values be \(\Lambda _i^{(1)}, \ldots , \Lambda _i^{(50)}\) . A z -score is then computed using
and then the p -value is computed according to the cumulative distribution function of the standard Gaussian distribution. See Additional file 1 for more discussions on the calculation of p -value.
Availability of data and materials
The simulated datasets [ 94 ] under three schemes are available at Zenodo with DOI 10.5281/zenodo.11013320 [ 95 ]. The SCIPAC package is available at GitHub website https://github.com/RavenGan/SCIPAC under the MIT license [ 96 ]. The source code of SCIPAC is also deposited at Zenodo with DOI 10.5281/zenodo.11013696 [ 97 ]. A vignette of the R package is available on the GitHub page and in the Additional file 2. The prostate cancer scRNA-seq data is obtained from the Prostate Cell Atlas https://www.prostatecellatlas.org [ 29 ]; the scRNA-seq data for the breast cancer are from the Gene Expression Omnibus (GEO) under accession number GSE176078 [ 34 , 98 ]; the scRNA-seq data for the lung cancer are from E-MTAB-6149 [ 99 ] and E-MTAB-6653 [ 71 , 100 ]; the scRNA-seq data for facioscapulohumeral muscular dystrophy data are from the GEO under accession number GSE122873 [ 101 ]. The bulk RNA-seq data are obtained from the TCGA database via TCGAbiolinks (ver. 2.25.2) R package [ 102 ]. More details about the simulated and real scRNA-seq and bulk RNA-seq data can be found in the Additional file 1.
Yofe I, Dahan R, Amit I. Single-cell genomic approaches for developing the next generation of immunotherapies. Nat Med. 2020;26(2):171–7.
Article CAS PubMed Google Scholar
Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179(4):829–45.
Fan J, Slowikowski K, Zhang F. Single-cell transcriptomics in cancer: computational challenges and opportunities. Exp Mol Med. 2020;52(9):1452–65.
Article CAS PubMed PubMed Central Google Scholar
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–201.
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–14.
Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360(6385):176–82.
Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8(1):1–12.
Article Google Scholar
Abdelaal T, Michielsen L, Cats D, Hoogduin D, Mei H, Reinders MJ, et al. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 2019;20(1):1–19.
Article CAS Google Scholar
Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15(6):e8746.
Article PubMed PubMed Central Google Scholar
Guo H, Li J. scSorter: assigning cells to known cell types according to marker genes. Genome Biol. 2021;22(1):1–18.
Pliner HA, Shendure J, Trapnell C. Supervised classification enables rapid annotation of cell atlases. Nat Methods. 2019;16(10):983–6.
Zhang AW, O’Flanagan C, Chavez EA, Lim JL, Ceglia N, McPherson A, et al. Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling. Nat Methods. 2019;16(10):1007–15.
Zhang Z, Luo D, Zhong X, Choi JH, Ma Y, Wang S, et al. SCINA: a semi-supervised subtyping algorithm of single cells and bulk samples. Genes. 2019;10(7):531.
Johnson TS, Wang T, Huang Z, Yu CY, Wu Y, Han Y, et al. LAmbDA: label ambiguous domain adaptation dataset integration reduces batch effects and improves subtype detection. Bioinformatics. 2019;35(22):4696–706.
Ma F, Pellegrini M. ACTINN: automated identification of cell types in single cell RNA sequencing. Bioinformatics. 2020;36(2):533–8.
Tan Y, Cahan P. SingleCellNet: a computational tool to classify single cell RNA-Seq data across platforms and across species. Cell Syst. 2019;9(2):207–13.
Salcher S, Sturm G, Horvath L, Untergasser G, Kuempers C, Fotakis G, et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell. 2022;40(12):1503–20.
Good Z, Sarno J, Jager A, Samusik N, Aghaeepour N, Simonds EF, et al. Single-cell developmental classification of B cell precursor acute lymphoblastic leukemia at diagnosis reveals predictors of relapse. Nat Med. 2018;24(4):474–83.
Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell. 2019;177(5):1330–45.
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–20.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Disc. 2012;2(5):401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):1.
Sun D, Guan X, Moran AE, Wu LY, Qian DZ, Schedin P, et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat Biotechnol. 2022;40(4):527–38.
Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008;2008(10):P10008.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–902.
Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B Stat Methodol. 2005;67(2):301–20.
McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. 2018. arXiv preprint arXiv:1802.03426 .
Wong CJ, Wang LH, Friedman SD, Shaw D, Campbell AE, Budech CB, et al. Longitudinal measures of RNA expression and disease activity in FSHD muscle biopsies. Hum Mol Genet. 2020;29(6):1030–43.
Tuong ZK, Loudon KW, Berry B, Richoz N, Jones J, Tan X, et al. Resolving the immune landscape of human prostate at a single-cell level in health and cancer. Cell Rep. 2021;37(12):110132.
Hume DA. The mononuclear phagocyte system. Curr Opin Immunol. 2006;18(1):49–53.
Hume DA, Ross IL, Himes SR, Sasmono RT, Wells CA, Ravasi T. The mononuclear phagocyte system revisited. J Leukoc Biol. 2002;72(4):621–7.
Raggi F, Bosco MC. Targeting mononuclear phagocyte receptors in cancer immunotherapy: new perspectives of the triggering receptor expressed on myeloid cells (TREM-1). Cancers. 2020;12(5):1337.
Largeot A, Pagano G, Gonder S, Moussay E, Paggetti J. The B-side of cancer immunity: the underrated tune. Cells. 2019;8(5):449.
Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53(9):1334–47.
Fernández-Nogueira P, Fuster G, Gutierrez-Uzquiza Á, Gascón P, Carbó N, Bragado P. Cancer-associated fibroblasts in breast cancer treatment response and metastasis. Cancers. 2021;13(13):3146.
Ao Z, Shah SH, Machlin LM, Parajuli R, Miller PC, Rawal S, et al. Identification of cancer-associated fibroblasts in circulating blood from patients with metastatic breast cancer. Identification of cCAFs from metastatic cancer patients. Cancer Res. 2015;75(22):4681–7.
Arcucci A, Ruocco MR, Granato G, Sacco AM, Montagnani S. Cancer: an oxidative crosstalk between solid tumor cells and cancer associated fibroblasts. BioMed Res Int. 2016;2016. https://pubmed.ncbi.nlm.nih.gov/27595103/ .
Buchsbaum RJ, Oh SY. Breast cancer-associated fibroblasts: where we are and where we need to go. Cancers. 2016;8(2):19.
Ruocco MR, Avagliano A, Granato G, Imparato V, Masone S, Masullo M, et al. Involvement of breast cancer-associated fibroblasts in tumor development, therapy resistance and evaluation of potential therapeutic strategies. Curr Med Chem. 2018;25(29):3414–34.
Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24(7):986–93.
Bassez A, Vos H, Van Dyck L, Floris G, Arijs I, Desmedt C, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med. 2021;27(5):820–32.
Romero JM, Grünwald B, Jang GH, Bavi PP, Jhaveri A, Masoomian M, et al. A four-chemokine signature is associated with a T-cell-inflamed phenotype in primary and metastatic pancreatic cancer. Chemokines in Pancreatic Cancer. Clin Cancer Res. 2020;26(8):1997–2010.
Tamura R, Yoshihara K, Nakaoka H, Yachida N, Yamaguchi M, Suda K, et al. XCL1 expression correlates with CD8-positive T cells infiltration and PD-L1 expression in squamous cell carcinoma arising from mature cystic teratoma of the ovary. Oncogene. 2020;39(17):3541–54.
Hernandez R, Põder J, LaPorte KM, Malek TR. Engineering IL-2 for immunotherapy of autoimmunity and cancer. Nat Rev Immunol. 2022:22:1–15. https://pubmed.ncbi.nlm.nih.gov/35217787/ .
Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. BioRxiv. 2016:060012. https://www.biorxiv.org/content/10.1101/060012v3.abstract .
Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35.
Gnanaprakasam JR, Wang R. MYC in regulating immunity: metabolism and beyond. Genes. 2017;8(3):88.
Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, et al. G2M cell cycle pathway score as a prognostic biomarker of metastasis in estrogen receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21(8):2921.
Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng C, et al. Cell Marker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–8.
Chen L, Yang L, Qiao F, Hu X, Li S, Yao L, et al. High levels of nucleolar spindle-associated protein and reduced levels of BRCA1 expression predict poor prognosis in triple-negative breast cancer. PLoS ONE. 2015;10(10):e0140572.
Li M, Yang B. Prognostic value of NUSAP1 and its correlation with immune infiltrates in human breast cancer. Crit Rev TM Eukaryot Gene Expr. 2022;32(3). https://pubmed.ncbi.nlm.nih.gov/35695609/ .
Zhang X, Pan Y, Fu H, Zhang J. Nucleolar and spindle associated protein 1 (NUSAP1) inhibits cell proliferation and enhances susceptibility to epirubicin in invasive breast cancer cells by regulating cyclin D kinase (CDK1) and DLGAP5 expression. Med Sci Monit: Int Med J Exp Clin Res. 2018;24:8553.
Geyer FC, Rodrigues DN, Weigelt B, Reis-Filho JS. Molecular classification of estrogen receptor-positive/luminal breast cancers. Adv Anat Pathol. 2012;19(1):39–53.
Karamitopoulou E, Perentes E, Tolnay M, Probst A. Prognostic significance of MIB-1, p53, and bcl-2 immunoreactivity in meningiomas. Hum Pathol. 1998;29(2):140–5.
Duxbury MS, Whang EE. RRM2 induces NF- \(\kappa\) B-dependent MMP-9 activation and enhances cellular invasiveness. Biochem Biophys Res Commun. 2007;354(1):190–6.
Zhou BS, Tsai P, Ker R, Tsai J, Ho R, Yu J, et al. Overexpression of transfected human ribonucleotide reductase M2 subunit in human cancer cells enhances their invasive potential. Clin Exp Metastasis. 1998;16(1):43–9.
Zhang H, Liu X, Warden CD, Huang Y, Loera S, Xue L, et al. Prognostic and therapeutic significance of ribonucleotide reductase small subunit M2 in estrogen-negative breast cancers. BMC Cancer. 2014;14(1):1–16.
Putluri N, Maity S, Kommagani R, Creighton CJ, Putluri V, Chen F, et al. Pathway-centric integrative analysis identifies RRM2 as a prognostic marker in breast cancer associated with poor survival and tamoxifen resistance. Neoplasia. 2014;16(5):390–402.
Koleck TA, Conley YP. Identification and prioritization of candidate genes for symptom variability in breast cancer survivors based on disease characteristics at the cellular level. Breast Cancer Targets Ther. 2016;8:29.
Li Jp, Zhang Xm, Zhang Z, Zheng Lh, Jindal S, Liu Yj. Association of p53 expression with poor prognosis in patients with triple-negative breast invasive ductal carcinoma. Medicine. 2019;98(18). https://pubmed.ncbi.nlm.nih.gov/31045815/ .
Gong MT, Ye SD, Lv WW, He K, Li WX. Comprehensive integrated analysis of gene expression datasets identifies key anti-cancer targets in different stages of breast cancer. Exp Ther Med. 2018;16(2):802–10.
PubMed PubMed Central Google Scholar
Chen Wx, Yang Lg, Xu Ly, Cheng L, Qian Q, Sun L, et al. Bioinformatics analysis revealing prognostic significance of RRM2 gene in breast cancer. Biosci Rep. 2019;39(4). https://pubmed.ncbi.nlm.nih.gov/30898978/ .
Hao Z, Zhang H, Cowell J. Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker. Tumor Biol. 2012;33(3):723–30.
Arriola E, Rodriguez-Pinilla SM, Lambros MB, Jones RL, James M, Savage K, et al. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res Treat. 2007;106(2):181–9.
Knoop AS, Knudsen H, Balslev E, Rasmussen BB, Overgaard J, Nielsen KV, et al. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol. 2005;23(30):7483–90.
Tanner M, Isola J, Wiklund T, Erikstein B, Kellokumpu-Lehtinen P, Malmstrom P, et al. Topoisomerase II \(\alpha\) gene amplification predicts favorable treatment response to tailored and dose-escalated anthracycline-based adjuvant chemotherapy in HER-2/neu-amplified breast cancer: Scandinavian Breast Group Trial 9401. J Clin Oncol. 2006;24(16):2428–36.
Arriola E, Moreno A, Varela M, Serra JM, Falo C, Benito E, et al. Predictive value of HER-2 and topoisomerase II \(\alpha\) in response to primary doxorubicin in breast cancer. Eur J Cancer. 2006;42(17):2954–60.
Järvinen TA, Tanner M, Bärlund M, Borg Å, Isola J. Characterization of topoisomerase II \(\alpha\) gene amplification and deletion in breast cancer. Gene Chromosome Cancer. 1999;26(2):142–50.
Landberg G, Erlanson M, Roos G, Tan EM, Casiano CA. Nuclear autoantigen p330d/CENP-F: a marker for cell proliferation in human malignancies. Cytom J Int Soc Anal Cytol. 1996;25(1):90–8.
CAS Google Scholar
Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441(7090):235–8.
Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24(8):1277–89.
Bremnes RM, Busund LT, Kilvær TL, Andersen S, Richardsen E, Paulsen EE, et al. The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non-small cell lung cancer. J Thorac Oncol. 2016;11(6):789–800.
Article PubMed Google Scholar
Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN, et al. Objective measurement and clinical significance of TILs in non–small cell lung cancer. J Natl Cancer Inst. 2015;107(3):dju435.
Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4+ T cells in cancer immunotherapy—new insights into old paradigms. Cancer Gene Ther. 2021;28(1):5–17.
Dieu-Nosjean MC, Goc J, Giraldo NA, Sautès-Fridman C, Fridman WH. Tertiary lymphoid structures in cancer and beyond. Trends Immunol. 2014;35(11):571–80.
Wang Ss, Liu W, Ly D, Xu H, Qu L, Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol. 2019;16(1):6–18.
van den Heuvel A, Mahfouz A, Kloet SL, Balog J, van Engelen BG, Tawil R, et al. Single-cell RNA sequencing in facioscapulohumeral muscular dystrophy disease etiology and development. Hum Mol Genet. 2019;28(7):1064–75.
Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. J Am Stat Assoc. 2001;96(456):1348–60.
Hastie T, Tibshirani R, Friedman JH, Friedman JH. The elements of statistical learning: data mining, inference, and prediction, vol. 2. New York: Springer; 2009.
Book Google Scholar
Baran Y, Bercovich A, Sebe-Pedros A, Lubling Y, Giladi A, Chomsky E, et al. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 2019;20(1):1–19.
Persad S, Choo ZN, Dien C, Sohail N, Masilionis I, Chaligné R, et al. SEACells infers transcriptional and epigenomic cellular states from single-cell genomics data. Nat Biotechnol. 2023;41:1–12. https://pubmed.ncbi.nlm.nih.gov/36973557/ .
Ben-Kiki O, Bercovich A, Lifshitz A, Tanay A. Metacell-2: a divide-and-conquer metacell algorithm for scalable scRNA-seq analysis. Genome Biol. 2022;23(1):100.
Bilous M, Tran L, Cianciaruso C, Gabriel A, Michel H, Carmona SJ, et al. Metacells untangle large and complex single-cell transcriptome networks. BMC Bioinformatics. 2022;23(1):336.
Avila Cobos F, Alquicira-Hernandez J, Powell JE, Mestdagh P, De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat Commun. 2020;11(1):1–14.
Jin H, Liu Z. A benchmark for RNA-seq deconvolution analysis under dynamic testing environments. Genome Biol. 2021;22(1):1–23.
Wang X, Park J, Susztak K, Zhang NR, Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun. 2019;10(1):380.
Wurm MJ, Rathouz PJ, Hanlon BM. Regularized ordinal regression and the ordinalNet R package. 2017. arXiv preprint arXiv:1706.05003 .
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1.
Simon N, Friedman J, Hastie T. A blockwise descent algorithm for group-penalized multiresponse and multinomial regression. 2013. arXiv preprint arXiv:1311.6529 .
Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39(5):1.
Tibshirani R, Bien J, Friedman J, Hastie T, Simon N, Taylor J, et al. Strong rules for discarding predictors in lasso-type problems. J R Stat Soc Ser B Stat Methodol. 2012;74(2):245–66.
Efron B. Bootstrap methods: another look at the jackknife. In: Breakthroughs in statistics. New York: Springer; 1992. pp. 569–593.
Efron B, Tibshirani RJ. An introduction to the bootstrap. London: CRC Press; 1994.
Zappia L, Phipson B, Oshlack A. Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 2017;18(1):174.
Gan D, Zhu Y, Lu X, Li J. Simulated datasets used in SCIPAC analysis. Zenodo. 2024. https://doi.org/10.5281/zenodo.11013320 .
Gan D, Zhu Y, Lu X, Li J. SCIPAC R package. GitHub. 2024. https://github.com/RavenGan/SCIPAC . Accessed 24 Apr 2024.
Gan D, Zhu Y, Lu X, Li J. SCIPAC source code. Zenodo. 2024. https://doi.org/10.5281/zenodo.11013696 .
Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single-cell and spatially resolved atlas of human breast cancers. Datasets. 2021. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE176078 . Gene Expression Omnibus. Accessed 1 Oct 2022.
Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Datasets. 2018. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-6149 . ArrayExpress. Accessed 24 July 2022.
Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Datasets. 2018. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-6653 . ArrayExpress. Accessed 24 July 2022.
van den Heuvel A, Mahfouz A, Kloet SL, Balog J, van Engelen BG, Tawil R, et al. Single-cell RNA sequencing in facioscapulohumeral muscular dystrophy disease etiology and development. Datasets. 2019. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122873 . Gene Expression Omnibus. Accessed 13 Aug 2022.
Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71.
Download references
Review history
The review history is available as Additional file 3.
Peer review information
Veronique van den Berghe was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
This work is supported by the National Institutes of Health (R01CA280097 to X.L. and J.L, R01CA252878 to J.L.) and the DOD BCRP Breakthrough Award, Level 2 (W81XWH2110432 to J.L.).
Author information
Authors and affiliations.
Department of Applied and Computational Mathematics and Statistics, University of Notre Dame, Notre Dame, 46556, IN, USA
Dailin Gan & Jun Li
Department of Biological Sciences, Boler-Parseghian Center for Rare and Neglected Diseases, Harper Cancer Research Institute, Integrated Biomedical Sciences Graduate Program, University of Notre Dame, Notre Dame, 46556, IN, USA
Yini Zhu & Xin Lu
Tumor Microenvironment and Metastasis Program, Indiana University Melvin and Bren Simon Comprehensive Cancer Center, Indianapolis, 46202, IN, USA
You can also search for this author in PubMed Google Scholar
Contributions
J.L. conceived and supervised the study. J.L. and D.G. proposed the methods. D.G. implemented the methods and analyzed the data. D.G. and J.L. drafted the paper. D.G., Y.Z., X.L., and J.L. interpreted the results and revised the paper.
Corresponding author
Correspondence to Jun Li .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1. supplementary materials that include additional results and plots., additional file 2. a vignette of the scipac package., additional file 3. review history., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Gan, D., Zhu, Y., Lu, X. et al. SCIPAC: quantitative estimation of cell-phenotype associations. Genome Biol 25 , 119 (2024). https://doi.org/10.1186/s13059-024-03263-1
Download citation
Received : 30 January 2023
Accepted : 30 April 2024
Published : 13 May 2024
DOI : https://doi.org/10.1186/s13059-024-03263-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Phenotype association
- Single cell
- RNA sequencing
- Cancer research
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]

Upstate businessowner concerned over proposed sanitation station
G REENVILLE COUNTY, S.C. (FOX Carolina) - The future of a new trash site in Greenville County now sits in the hands of the Department of Health and Environmental Control (DHEC).
Even though the time to submit public input ended May 15, a local business owner is using his voice to express concerns over the proposed waste transfer station along Old Easley Highway.
“So much has been done to protect our rivers and natural resources in the area, so it seems like a step backwards to put a transfer station within 100 feet of wetlands, Cliff Carden, the owner of Saluda Outdoor River Company and Saluda Outdoor Center, said.
The team at Saluda Outdoor River Company is gearing up for the start of summer and tubing season, which officially begins Memorial Day Weekend on Friday at 10 a.m.
“We are so excited to have everybody out. We’re seven minutes from Downtown Greenville, and we are so excited to be here and get you out on the river,” operations manager Evie Buisch said.
“We wanted to have a place for people from Greenville to just come and get out in nature....I had a vision of doing something that families could enjoy,” Carden continued.
But that vision could soon be in jeopardy with Greater Greenville Sanitation’s proposal of a new waste transfer station less than a mile away from the tubing center.
Carden is concerned about potential ecological impacts.
“When you have a trash bag and things bust...that runoff water runs downhill to those creeks and rivers which go into the Saluda River,” he said.
Greater Greenville Sanitation’s permit proposal offers some proposals for runoff. One part of the proposal states:
“Surface water impact will be minimized using erosion control methods and stormwater runoff treatment BMPs.” Greater Greenville Sanitation Transfer Station Proposal
Greater Greenville Sanitation is also a special purpose district, which state law defines as an entity that “provides any governmental power or function including, but not limited to, fire protection, sewerage treatment, water or natural gas distribution or recreation.”
This means it is also supported by taxes.
“If there is something that happens to the river or [if] there’s problems with that property, guess who gets to pay for the fix? That’s the taxpayer,” Carden said.
Odor is another concern.
“It’s summertime when we run. So you got hot trash right here, and the least bit of wind will blow right down to our people who are coming just to enjoy being out in nature. So that is a concern for us as business owners,” Carden said.
DHEC said they are considering public comments and do not have a timeline as to when they will make their final decision.
FOX Carolina also reached out to Greater Greenville Sanitation to provide a comment. We will be sure to update this story as more information comes in.
PREVIOUS COVERAGE: Neighbors say “no” to proposed trash site

Numbers, Facts and Trends Shaping Your World
Read our research on:
Full Topic List
Regions & Countries
- Publications
- Our Methods
- Short Reads
- Tools & Resources
Read Our Research On:
Broad Public Support for Legal Abortion Persists 2 Years After Dobbs
Methodology, table of contents.
- Other abortion attitudes
- Overall attitudes about abortion
- Americans’ views on medication abortion in their states
- How statements about abortion resonate with Americans
- Acknowledgments
- The American Trends Panel survey methodology
The American Trends Panel (ATP), created by Pew Research Center, is a nationally representative panel of randomly selected U.S. adults. Panelists participate via self-administered web surveys. Panelists who do not have internet access at home are provided with a tablet and wireless internet connection. Interviews are conducted in both English and Spanish. The panel is being managed by Ipsos.
Data in this report is drawn from ATP Wave 146, conducted from April 8 to 14, 2024. It includes oversamples of non-Hispanic Asian adults, non-Hispanic Black adults, Hispanic adults and adults ages 18 to 29 in order to provide more precise estimates of the opinions and experiences of these smaller demographic subgroups. It also included an oversample of validated 2016 and 2020 “vote switchers” who voted for Donald Trump in 2020 but not in 2016 or who voted for Joe Biden in 2020 but not for Hillary Clinton in 2016. These oversampled groups are weighted back to reflect their correct proportions in the population. A total of 8,709 panelists responded out of 9,527 who were sampled, for a response rate of 91%. The cumulative response rate accounting for nonresponse to the recruitment surveys and attrition is 3%. The break-off rate among panelists who logged on to the survey and completed at least one item is less than 1%. The margin of sampling error for the full sample of 8,709 respondents is plus or minus 1.5 percentage points.
Panel recruitment
The ATP was created in 2014, with the first cohort of panelists invited to join the panel at the end of a large, national, landline and cellphone random-digit-dial survey that was conducted in both English and Spanish. Two additional recruitments were conducted using the same method in 2015 and 2017, respectively. Across these three surveys, a total of 19,718 adults were invited to join the ATP, of whom 9,942 (50%) agreed to participate.
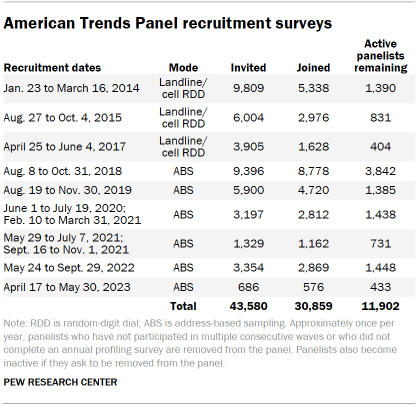
In August 2018, the ATP switched from telephone to address-based sampling (ABS) recruitment. A study cover letter and a pre-incentive are mailed to a stratified, random sample of households selected from the U.S. Postal Service’s Delivery Sequence File. This Postal Service file has been estimated to cover as much as 98% of the population, although some studies suggest that the coverage could be in the low 90% range. 1 Within each sampled household, the adult with the next birthday is asked to participate. Other details of the ABS recruitment protocol have changed over time but are available upon request. 2
We have recruited a national sample of U.S. adults to the ATP approximately once per year since 2014. In some years, the recruitment has included additional efforts (known as an “oversample”) to boost sample size with underrepresented groups. For example, Hispanic, Black and Asian adults were oversampled in 2019, 2022 and 2023, respectively.
Across the six address-based recruitments, a total of 23,862 adults were invited to join the ATP, of whom 20,917 agreed to join the panel and completed an initial profile survey. Of the 30,859 individuals who have ever joined the ATP, 11,902 remained active panelists and continued to receive survey invitations at the time this survey was conducted.
The American Trends Panel never uses breakout routers or chains that direct respondents to additional surveys.
Sample design
The overall target population for this survey was noninstitutionalized persons ages 18 and older living in the U.S., including Alaska and Hawaii. It featured a stratified random sample from the ATP in which the following groups were selected with certainty: non-Hispanic Asian adults; non-Hispanic Black adults; Hispanic adults; adults ages 18 to 29; validated 2016 and 2020 voters who voted for Donald Trump in 2020 but voted for a different candidate in 2016; and validated 2016 and 2020 voters who voted for Joe Biden in 2020 but did not vote for Hillary Clinton in 2016. 3
The remaining panelists were sampled at rates designed to ensure that the share of respondents in each stratum is proportional to its share of the U.S. adult population to the greatest extent possible. Respondent weights are adjusted to account for differential probabilities of selection as described in the Weighting section below.
Questionnaire development and testing
The questionnaire was developed by Pew Research Center in consultation with Ipsos. The web program was rigorously tested on both PC and mobile devices by the Ipsos project management team and Center researchers. The Ipsos project management team also populated test data that was analyzed in SPSS to ensure the logic and randomizations were working as intended before launching the survey.
All respondents were offered a post-paid incentive for their participation. Respondents could choose to receive the post-paid incentive in the form of a check or a gift code to Amazon.com or could choose to decline the incentive. Incentive amounts ranged from $5 to $20 depending on whether the respondent belongs to a part of the population that is harder or easier to reach. Differential incentive amounts were designed to increase panel survey participation among groups that traditionally have low survey response propensities.
Data collection protocol
The data collection field period for this survey was April 8 to 14, 2024. Postcard notifications were mailed to a subset of ATP panelists with a known residential address on April 8. 4
Invitations were sent out in two separate launches: soft launch and full launch. Sixty panelists were included in the soft launch, which began with an initial invitation sent on April 8. The ATP panelists chosen for the initial soft launch were known responders who had completed previous ATP surveys within one day of receiving their invitation. All remaining English- and Spanish-speaking sampled panelists were included in the full launch and were sent an invitation on April 9.
All panelists with an email address received an email invitation and up to two email reminders if they did not respond to the survey. All ATP panelists who consented to SMS messages received an SMS invitation and up to two SMS reminders.
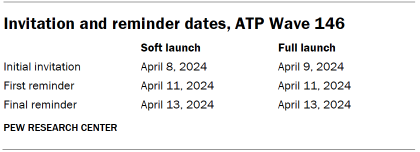
Data quality checks
To ensure high-quality data, the Center’s researchers performed data quality checks to identify any respondents showing clear patterns of satisficing. This includes checking for whether respondents left questions blank at very high rates or always selected the first or last answer presented. As a result of this checking, three ATP respondents were removed from the survey dataset prior to weighting and analysis.
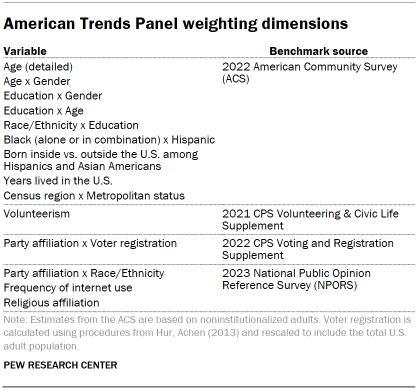
The ATP data is weighted in a multistep process that accounts for multiple stages of sampling and nonresponse that occur at different points in the survey process. First, each panelist begins with a base weight that reflects their probability of selection for their initial recruitment survey. These weights are then rescaled and adjusted to account for changes in the design of ATP recruitment surveys from year to year. Finally, the weights are calibrated to align with the population benchmarks in the accompanying table to correct for nonresponse to recruitment surveys and panel attrition. If only a subsample of panelists was invited to participate in the wave, this weight is adjusted to account for any differential probabilities of selection.
Among the panelists who completed the survey, this weight is then calibrated again to align with the population benchmarks identified in the accompanying table and trimmed at the 2nd and 98th percentiles to reduce the loss in precision stemming from variance in the weights. This trimming is performed separately among non-Hispanic Black, non-Hispanic Asian, Hispanic and all other respondents. Sampling errors and tests of statistical significance take into account the effect of weighting.
The following table shows the unweighted sample sizes and the error attributable to sampling that would be expected at the 95% level of confidence for different groups in the survey.
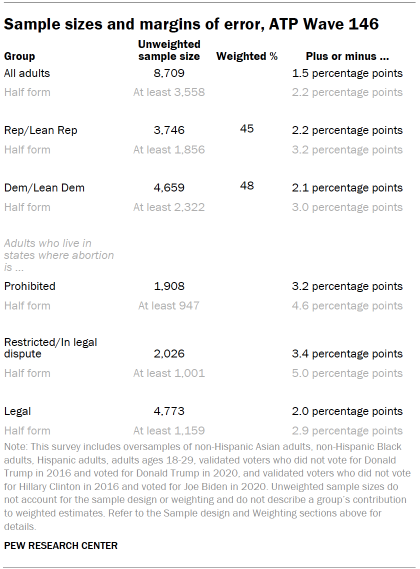
Sample sizes and sampling errors for other subgroups are available upon request. In addition to sampling error, one should bear in mind that question wording and practical difficulties in conducting surveys can introduce error or bias into the findings of opinion polls.
Dispositions and response rates
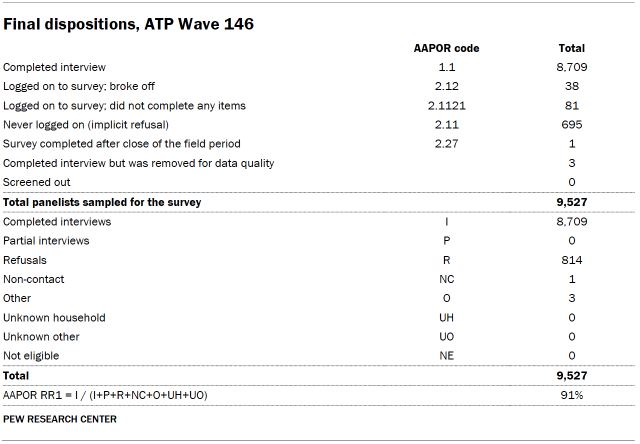
- AAPOR Task Force on Address-based Sampling. 2016. “AAPOR Report: Address-based Sampling.” ↩
- Email [email protected] . ↩
- A validated voter is a citizen who told us that they voted in an election and have a record for voting in that election in a commercial voter file. A voter file is a list of adults that includes information such as which elections they have voted in. Federal law requires states to maintain voter files, and businesses assemble these files to create a nationwide list of adults along with their voter information. ↩
- Postcard notifications are sent to 1) panelists who have been provided with a tablet to take ATP surveys, 2) panelists who were recruited within the last two years, and 3) panelists recruited prior to the last two years who opt to continue receiving postcard notifications. ↩
Sign up for our weekly newsletter
Fresh data delivery Saturday mornings
Sign up for The Briefing
Weekly updates on the world of news & information
- Partisanship & Issues
Support for legal abortion is widespread in many places, especially in Europe
Public opinion on abortion, americans overwhelmingly say access to ivf is a good thing, what the data says about abortion in the u.s., nearly a year after roe’s demise, americans’ views of abortion access increasingly vary by where they live, most popular, report materials.
1615 L St. NW, Suite 800 Washington, DC 20036 USA (+1) 202-419-4300 | Main (+1) 202-857-8562 | Fax (+1) 202-419-4372 | Media Inquiries
Research Topics
- Age & Generations
- Coronavirus (COVID-19)
- Economy & Work
- Family & Relationships
- Gender & LGBTQ
- Immigration & Migration
- International Affairs
- Internet & Technology
- Methodological Research
- News Habits & Media
- Non-U.S. Governments
- Other Topics
- Politics & Policy
- Race & Ethnicity
- Email Newsletters
ABOUT PEW RESEARCH CENTER Pew Research Center is a nonpartisan fact tank that informs the public about the issues, attitudes and trends shaping the world. It conducts public opinion polling, demographic research, media content analysis and other empirical social science research. Pew Research Center does not take policy positions. It is a subsidiary of The Pew Charitable Trusts .
Copyright 2024 Pew Research Center
- Privacy Policy

Home » How To Write A Proposal – Step By Step Guide [With Template]
How To Write A Proposal – Step By Step Guide [With Template]
Table of Contents

How To Write A Proposal
Writing a Proposal involves several key steps to effectively communicate your ideas and intentions to a target audience. Here’s a detailed breakdown of each step:
Identify the Purpose and Audience
- Clearly define the purpose of your proposal: What problem are you addressing, what solution are you proposing, or what goal are you aiming to achieve?
- Identify your target audience: Who will be reading your proposal? Consider their background, interests, and any specific requirements they may have.
Conduct Research
- Gather relevant information: Conduct thorough research to support your proposal. This may involve studying existing literature, analyzing data, or conducting surveys/interviews to gather necessary facts and evidence.
- Understand the context: Familiarize yourself with the current situation or problem you’re addressing. Identify any relevant trends, challenges, or opportunities that may impact your proposal.
Develop an Outline
- Create a clear and logical structure: Divide your proposal into sections or headings that will guide your readers through the content.
- Introduction: Provide a concise overview of the problem, its significance, and the proposed solution.
- Background/Context: Offer relevant background information and context to help the readers understand the situation.
- Objectives/Goals: Clearly state the objectives or goals of your proposal.
- Methodology/Approach: Describe the approach or methodology you will use to address the problem.
- Timeline/Schedule: Present a detailed timeline or schedule outlining the key milestones or activities.
- Budget/Resources: Specify the financial and other resources required to implement your proposal.
- Evaluation/Success Metrics: Explain how you will measure the success or effectiveness of your proposal.
- Conclusion: Summarize the main points and restate the benefits of your proposal.
Write the Proposal
- Grab attention: Start with a compelling opening statement or a brief story that hooks the reader.
- Clearly state the problem: Clearly define the problem or issue you are addressing and explain its significance.
- Present your proposal: Introduce your proposed solution, project, or idea and explain why it is the best approach.
- State the objectives/goals: Clearly articulate the specific objectives or goals your proposal aims to achieve.
- Provide supporting information: Present evidence, data, or examples to support your claims and justify your proposal.
- Explain the methodology: Describe in detail the approach, methods, or strategies you will use to implement your proposal.
- Address potential concerns: Anticipate and address any potential objections or challenges the readers may have and provide counterarguments or mitigation strategies.
- Recap the main points: Summarize the key points you’ve discussed in the proposal.
- Reinforce the benefits: Emphasize the positive outcomes, benefits, or impact your proposal will have.
- Call to action: Clearly state what action you want the readers to take, such as approving the proposal, providing funding, or collaborating with you.
Review and Revise
- Proofread for clarity and coherence: Check for grammar, spelling, and punctuation errors.
- Ensure a logical flow: Read through your proposal to ensure the ideas are presented in a logical order and are easy to follow.
- Revise and refine: Fine-tune your proposal to make it concise, persuasive, and compelling.
Add Supplementary Materials
- Attach relevant documents: Include any supporting materials that strengthen your proposal, such as research findings, charts, graphs, or testimonials.
- Appendices: Add any additional information that might be useful but not essential to the main body of the proposal.
Formatting and Presentation
- Follow the guidelines: Adhere to any specific formatting guidelines provided by the organization or institution to which you are submitting the proposal.
- Use a professional tone and language: Ensure that your proposal is written in a clear, concise, and professional manner.
- Use headings and subheadings: Organize your proposal with clear headings and subheadings to improve readability.
- Pay attention to design: Use appropriate fonts, font sizes, and formatting styles to make your proposal visually appealing.
- Include a cover page: Create a cover page that includes the title of your proposal, your name or organization, the date, and any other required information.
Seek Feedback
- Share your proposal with trusted colleagues or mentors and ask for their feedback. Consider their suggestions for improvement and incorporate them into your proposal if necessary.
Finalize and Submit
- Make any final revisions based on the feedback received.
- Ensure that all required sections, attachments, and documentation are included.
- Double-check for any formatting, grammar, or spelling errors.
- Submit your proposal within the designated deadline and according to the submission guidelines provided.
Proposal Format
The format of a proposal can vary depending on the specific requirements of the organization or institution you are submitting it to. However, here is a general proposal format that you can follow:
1. Title Page:
- Include the title of your proposal, your name or organization’s name, the date, and any other relevant information specified by the guidelines.
2. Executive Summary:
- Provide a concise overview of your proposal, highlighting the key points and objectives.
- Summarize the problem, proposed solution, and anticipated benefits.
- Keep it brief and engaging, as this section is often read first and should capture the reader’s attention.
3. Introduction:
- State the problem or issue you are addressing and its significance.
- Provide background information to help the reader understand the context and importance of the problem.
- Clearly state the purpose and objectives of your proposal.
4. Problem Statement:
- Describe the problem in detail, highlighting its impact and consequences.
- Use data, statistics, or examples to support your claims and demonstrate the need for a solution.
5. Proposed Solution or Project Description:
- Explain your proposed solution or project in a clear and detailed manner.
- Describe how your solution addresses the problem and why it is the most effective approach.
- Include information on the methods, strategies, or activities you will undertake to implement your solution.
- Highlight any unique features, innovations, or advantages of your proposal.
6. Methodology:
- Provide a step-by-step explanation of the methodology or approach you will use to implement your proposal.
- Include a timeline or schedule that outlines the key milestones, tasks, and deliverables.
- Clearly describe the resources, personnel, or expertise required for each phase of the project.
7. Evaluation and Success Metrics:
- Explain how you will measure the success or effectiveness of your proposal.
- Identify specific metrics, indicators, or evaluation methods that will be used.
- Describe how you will track progress, gather feedback, and make adjustments as needed.
- Present a detailed budget that outlines the financial resources required for your proposal.
- Include all relevant costs, such as personnel, materials, equipment, and any other expenses.
- Provide a justification for each item in the budget.
9. Conclusion:
- Summarize the main points of your proposal.
- Reiterate the benefits and positive outcomes of implementing your proposal.
- Emphasize the value and impact it will have on the organization or community.
10. Appendices:
- Include any additional supporting materials, such as research findings, charts, graphs, or testimonials.
- Attach any relevant documents that provide further information but are not essential to the main body of the proposal.
Proposal Template
Here’s a basic proposal template that you can use as a starting point for creating your own proposal:
Dear [Recipient’s Name],
I am writing to submit a proposal for [briefly state the purpose of the proposal and its significance]. This proposal outlines a comprehensive solution to address [describe the problem or issue] and presents an actionable plan to achieve the desired objectives.
Thank you for considering this proposal. I believe that implementing this solution will significantly contribute to [organization’s or community’s goals]. I am available to discuss the proposal in more detail at your convenience. Please feel free to contact me at [your email address or phone number].
Yours sincerely,
Note: This template is a starting point and should be customized to meet the specific requirements and guidelines provided by the organization or institution to which you are submitting the proposal.
Proposal Sample
Here’s a sample proposal to give you an idea of how it could be structured and written:
Subject : Proposal for Implementation of Environmental Education Program
I am pleased to submit this proposal for your consideration, outlining a comprehensive plan for the implementation of an Environmental Education Program. This program aims to address the critical need for environmental awareness and education among the community, with the objective of fostering a sense of responsibility and sustainability.
Executive Summary: Our proposed Environmental Education Program is designed to provide engaging and interactive educational opportunities for individuals of all ages. By combining classroom learning, hands-on activities, and community engagement, we aim to create a long-lasting impact on environmental conservation practices and attitudes.
Introduction: The state of our environment is facing significant challenges, including climate change, habitat loss, and pollution. It is essential to equip individuals with the knowledge and skills to understand these issues and take action. This proposal seeks to bridge the gap in environmental education and inspire a sense of environmental stewardship among the community.
Problem Statement: The lack of environmental education programs has resulted in limited awareness and understanding of environmental issues. As a result, individuals are less likely to adopt sustainable practices or actively contribute to conservation efforts. Our program aims to address this gap and empower individuals to become environmentally conscious and responsible citizens.
Proposed Solution or Project Description: Our Environmental Education Program will comprise a range of activities, including workshops, field trips, and community initiatives. We will collaborate with local schools, community centers, and environmental organizations to ensure broad participation and maximum impact. By incorporating interactive learning experiences, such as nature walks, recycling drives, and eco-craft sessions, we aim to make environmental education engaging and enjoyable.
Methodology: Our program will be structured into modules that cover key environmental themes, such as biodiversity, climate change, waste management, and sustainable living. Each module will include a mix of classroom sessions, hands-on activities, and practical field experiences. We will also leverage technology, such as educational apps and online resources, to enhance learning outcomes.
Evaluation and Success Metrics: We will employ a combination of quantitative and qualitative measures to evaluate the effectiveness of the program. Pre- and post-assessments will gauge knowledge gain, while surveys and feedback forms will assess participant satisfaction and behavior change. We will also track the number of community engagement activities and the adoption of sustainable practices as indicators of success.
Budget: Please find attached a detailed budget breakdown for the implementation of the Environmental Education Program. The budget covers personnel costs, materials and supplies, transportation, and outreach expenses. We have ensured cost-effectiveness while maintaining the quality and impact of the program.
Conclusion: By implementing this Environmental Education Program, we have the opportunity to make a significant difference in our community’s environmental consciousness and practices. We are confident that this program will foster a generation of individuals who are passionate about protecting our environment and taking sustainable actions. We look forward to discussing the proposal further and working together to make a positive impact.
Thank you for your time and consideration. Should you have any questions or require additional information, please do not hesitate to contact me at [your email address or phone number].
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Grant Proposal – Example, Template and Guide

How To Write A Business Proposal – Step-by-Step...

Business Proposal – Templates, Examples and Guide

How To Write A Research Proposal – Step-by-Step...

Proposal – Types, Examples, and Writing Guide

How to choose an Appropriate Method for Research?

IMAGES
VIDEO
COMMENTS
Step 1: Explain your methodological approach. Step 2: Describe your data collection methods. Step 3: Describe your analysis method. Step 4: Evaluate and justify the methodological choices you made. Tips for writing a strong methodology chapter. Other interesting articles.
Definition, Types, and Examples. Research methodology 1,2 is a structured and scientific approach used to collect, analyze, and interpret quantitative or qualitative data to answer research questions or test hypotheses. A research methodology is like a plan for carrying out research and helps keep researchers on track by limiting the scope of ...
Provide the rationality behind your chosen approach. Based on logic and reason, let your readers know why you have chosen said research methodologies. Additionally, you have to build strong arguments supporting why your chosen research method is the best way to achieve the desired outcome. 3. Explain your mechanism.
Qualitative Research Methodology. This is a research methodology that involves the collection and analysis of non-numerical data such as words, images, and observations. This type of research is often used to explore complex phenomena, to gain an in-depth understanding of a particular topic, and to generate hypotheses.
As we mentioned, research methodology refers to the collection of practical decisions regarding what data you'll collect, from who, how you'll collect it and how you'll analyse it. Research design, on the other hand, is more about the overall strategy you'll adopt in your study. For example, whether you'll use an experimental design ...
Writing a research proposal can be quite challenging, but a good starting point could be to look at some examples. We've included a few for you below. Example research proposal #1: "A Conceptual Framework for Scheduling Constraint Management" Example research proposal #2: "Medical Students as Mediators of Change in Tobacco Use" Title page
Bem, Daryl J. Writing the Empirical Journal Article. Psychology Writing Center. University of Washington; Denscombe, Martyn. The Good Research Guide: For Small-Scale Social Research Projects. 5th edition.Buckingham, UK: Open University Press, 2014; Lunenburg, Frederick C. Writing a Successful Thesis or Dissertation: Tips and Strategies for Students in the Social and Behavioral Sciences.
Learn how to write a strong methodology chapter that allows readers to evaluate the reliability and validity of the research. A good methodology chapter incl...
This information regarding the methodology section of a proposal was gathered from RRU thesis and major project handbooks, current in 2020, from programs in the Faculty of Social and Applied Sciences, the Faculty of Management, and the College of Interdisciplinary Studies. If the details here differ from the information provided in the handbook ...
Methodology in research is defined as the systematic method to resolve a research problem through data gathering using various techniques, providing an interpretation of data gathered and drawing conclusions about the research data. Essentially, a research methodology is the blueprint of a research or study (Murthy & Bhojanna, 2009, p. 32).
The methods section of a research proposal must contain all the necessary information that will facilitate another researcher to replicate your research. The purpose of writing this section is to convince the funding agency that the methods you plan to use are sound and this is the most suitable approach to address the problem you have chosen.
Here is an explanation of each step: 1. Title and Abstract. Choose a concise and descriptive title that reflects the essence of your research. Write an abstract summarizing your research question, objectives, methodology, and expected outcomes. It should provide a brief overview of your proposal. 2.
Methodological Framework. Definition: Methodological framework is a set of procedures, methods, and tools that guide the research process in a systematic and structured manner. It provides a structure for conducting research, collecting and analyzing data, and drawing conclusions. The framework outlines the steps to be taken in a research ...
Here are the steps to follow when writing a methodology: 1. Restate your thesis or research problem. The first part of your methodology is a restatement of the problem your research investigates. This allows your reader to follow your methodology step by step, from beginning to end. Restating your thesis also provides you an opportunity to ...
INTRODUCTION. A clean, well-thought-out proposal forms the backbone for the research itself and hence becomes the most important step in the process of conduct of research.[] The objective of preparing a research proposal would be to obtain approvals from various committees including ethics committee [details under 'Research methodology II' section [Table 1] in this issue of IJA) and to ...
approach and research methods that you have selected. Note that in the proposal's chapter 3, you project what you will do based on what you know about the particular methods used in qualitative research, in general, and in your tra-dition or genre, in particular; hence, it is written in future tense. In the dissertation's chapter 3,
A research design is a strategy for answering your research question using empirical data. Creating a research design means making decisions about: Your overall research objectives and approach. Whether you'll rely on primary research or secondary research. Your sampling methods or criteria for selecting subjects. Your data collection methods.
In its most common sense, methodology is the study of research methods. However, the term can also refer to the methods themselves or to the philosophical discussion of associated background assumptions. A method is a structured procedure for bringing about a certain goal, like acquiring knowledge or verifying knowledge claims. This normally involves various steps, like choosing a sample ...
Summary. What happens when you have to give a presentation to an audience that might have some professionals who have more expertise on the topic than you do? While it can be intimidating, it can ...
Notice of Filing of Proposed Rule Change by The Options Clearing Corporation Concerning Amendments to Its Rules and Comprehensive Stress Testing & Clearing Fund Methodology, and Liquidity Risk Management Description. Registered Clearing Agencies, The Options Clearing Corporation.
Bolt looseness detection is a common problem in engineering. Most vision-based detection techniques focus on diagnosing ordinary bolt looseness, i.e., the methods used for diagnosis are based only ...
Proposal. Definition: Proposal is a formal document or presentation that outlines a plan, idea, or project and seeks to persuade others to support or adopt it. Proposals are commonly used in business, academia, and various other fields to propose new initiatives, solutions to problems, research studies, or business ventures.
Numerous algorithms have been proposed to identify cell types in single-cell RNA sequencing data, yet a fundamental problem remains: determining associations between cells and phenotypes such as cancer. We develop SCIPAC, the first algorithm that quantitatively estimates the association between each cell in single-cell data and a phenotype. SCIPAC also provides a p-value for each association ...
Even though the time to submit public input ended May 15, a local business owner is using his voice to express concerns over the proposed waste transfer station along Old Easley Highway. "So ...
The main heading of "Methods" should be centered, boldfaced, and capitalized. Subheadings within this section are left-aligned, boldfaced, and in title case. You can also add lower level headings within these subsections, as long as they follow APA heading styles. To structure your methods section, you can use the subheadings of ...
A novel methodology has been proposed to tame the arc shape by adopting an external magnetic field, resulting weld profile as required. The co-axial magnetic field developed by specially designed electromagnets is superimposed on the welding arc. It was found that 0-0-S-N configuration provided more penetration than conventional gas tungsten ...
The American Trends Panel survey methodology Overview. The American Trends Panel (ATP), created by Pew Research Center, is a nationally representative panel of randomly selected U.S. adults. Panelists participate via self-administered web surveys. Panelists who do not have internet access at home are provided with a tablet and wireless internet ...
IV. Proposed Solution or Project Description: [Present your proposed solution or project in a clear and detailed manner. Explain how it addresses the problem and why it is the most effective approach. Highlight any unique features or advantages.] V. Methodology: [Describe the step-by-step approach or methodology you will use to implement your ...
A Proposal of a New Tool for the Assessment of Damage in Behçet Syndrome Uveitis: Cerrahpasa Ocular Damage Grading System. ... Methods . A specialist in BS uveitis (YO) developed a grading system for ocular damage with five grades based on the extent of damage in the posterior segment. YO trained a senior and general ophthalmologist with ...