- Google for Education
- Español – América Latina
- Português – Brasil
- Tiếng Việt

Python Introduction
Welcome to Google's Python online tutorial. It is based on the introductory Python course offered internally. As mentioned on the setup page , this material covers Python 3.
If you're seeking a companion MOOC course, try the ones from Udacity and Coursera ( intro to programming [beginners] or intro to Python ). Finally, if you're seeking self-paced online learning without watching videos, try the ones listed towards the end of this post — each feature learning content as well as a Python interactive interpreter you can practice with. What's this "interpreter" we mention? You'll find out in the next section!
Language Introduction
An excellent way to see how Python code works is to run the Python interpreter and type code right into it. If you ever have a question like, "What happens if I add an int to a list ?" Just typing it into the Python interpreter is a fast and likely the best way to see what happens. (See below to see what really happens!)
The two lines python prints after you type python and before the >>> prompt tells you about the version of python you're using and where it was built. As long as the first thing printed is "Python 3.", these examples should work for you.
As you can see above, it's easy to experiment with variables and operators. Also, the interpreter throws, or "raises" in Python parlance, a runtime error if the code tries to read a variable that has not been assigned a value. Like C++ and Java, Python is case sensitive so "a" and "A" are different variables. The end of a line marks the end of a statement, so unlike C++ and Java, Python does not require a semicolon at the end of each statement. Comments begin with a '#' and extend to the end of the line.
Python source code
Python source files use the ".py" extension and are called "modules." With a Python module hello.py , the easiest way to run it is with the shell command "python hello.py Alice" which calls the Python interpreter to execute the code in hello.py , passing it the command line argument "Alice". See the official docs page on all the different options you have when running Python from the command-line.
Here's a very simple hello.py program (notice that blocks of code are delimited strictly using indentation rather than curly braces — more on this later!):
Running this program from the command line looks like:
Imports, Command-line arguments, and len()
The outermost statements in a Python file, or "module", do its one-time setup — those statements run from top to bottom the first time the module is imported somewhere, setting up its variables and functions. A Python module can be run directly — as above python3 hello.py Bob — or it can be imported and used by some other module. When a Python file is run directly, the special variable "__name__" is set to "__main__". Therefore, it's common to have the boilerplate if __name__ ==... shown above to call a main() function when the module is run directly, but not when the module is imported by some other module.
In a standard Python program, the list sys.argv contains the command-line arguments in the standard way with sys.argv[0] being the program itself, sys.argv[1] the first argument, and so on. If you know about argc , or the number of arguments, you can simply request this value from Python with len(sys.argv) , just like we did in the interactive interpreter code above when requesting the length of a string. In general, len() can tell you how long a string is, the number of elements in lists and tuples (another array-like data structure), and the number of key-value pairs in a dictionary.
User-defined Functions
Functions in Python are defined like this:
Notice also how the lines that make up the function or if-statement are grouped by all having the same level of indentation. We also presented 2 different ways to repeat strings, using the + operator which is more user-friendly, but * also works because it's Python's "repeat" operator, meaning that '-' * 10 gives '----------' , a neat way to create an onscreen "line." In the code comment, we hinted that * works faster than +, the reason being that * calculates the size of the resulting object once whereas with +, that calculation is made each time + is called. Both + and * are called "overloaded" operators because they mean different things for numbers vs. for strings (and other data types).
The def keyword defines the function with its parameters within parentheses and its code indented. The first line of a function can be a documentation string ("docstring") that describes what the function does. The docstring can be a single line, or a multi-line description as in the example above. (Yes, those are "triple quotes," a feature unique to Python!) Variables defined in the function are local to that function, so the "result" in the above function is separate from a "result" variable in another function. The return statement can take an argument, in which case that is the value returned to the caller.
Here is code that calls the above repeat() function, printing what it returns:
At run time, functions must be defined by the execution of a "def" before they are called. It's typical to def a main() function towards the bottom of the file with the functions it calls above it.
Indentation
One unusual Python feature is that the whitespace indentation of a piece of code affects its meaning. A logical block of statements such as the ones that make up a function should all have the same indentation, set in from the indentation of their parent function or "if" or whatever. If one of the lines in a group has a different indentation, it is flagged as a syntax error.
Python's use of whitespace feels a little strange at first, but it's logical and I found I got used to it very quickly. Avoid using TABs as they greatly complicate the indentation scheme (not to mention TABs may mean different things on different platforms). Set your editor to insert spaces instead of TABs for Python code.
A common question beginners ask is, "How many spaces should I indent?" According to the official Python style guide (PEP 8) , you should indent with 4 spaces. (Fun fact: Google's internal style guideline dictates indenting by 2 spaces!)
Code Checked at Runtime
Python does very little checking at compile time, deferring almost all type, name, etc. checks on each line until that line runs. Suppose the above main() calls repeat() like this:
The if-statement contains an obvious error, where the repeat() function is accidentally typed in as repeeeet(). The funny thing in Python ... this code compiles and runs fine so long as the name at runtime is not 'Guido'. Only when a run actually tries to execute the repeeeet() will it notice that there is no such function and raise an error. There is also a second error in this snippet. name wasn't assigned a value before it is compared with 'Guido'. Python will raise a 'NameError' if you try to evaluate an unassigned variable. These are some examples demonstrating that when you first run a Python program, some of the first errors you see will be simple typos or uninitialized variables like these. This is one area where languages with a more verbose type system, like Java, have an advantage ... they can catch such errors at compile time (but of course you have to maintain all that type information ... it's a tradeoff).
Python 3 introduced type hints . Type hints allow you to indicate what the type is for each argument in a function as well as what what the type is of the object returned by the function. For example, in the annotated function def is_positive(n: int) -> bool: , the argument n is an int and the return value is a bool . We'll go into what these types mean later. Type hints are totally optional though. You'll see more and more code adopting type hints because if you use them, some editors like cider-v and VS.code can run checks to verify that your functions are called with the right argument types. They can even suggest and validate arguments as you edit code. This tutorial won't cover type hints but we want to make sure you're aware of them if you hear about them or see them in the wild.
Variable Names
Since Python variables don't have any type spelled out in the source code, it's extra helpful to give meaningful names to your variables to remind yourself of what's going on. So use "name" if it's a single name, and "names" if it's a list of names, and "tuples" if it's a list of tuples. Many basic Python errors result from forgetting what type of value is in each variable, so use your variable names (all you have really) to help keep things straight.
As far as actual naming goes, some languages prefer underscored_parts for variable names made up of "more than one word," but other languages prefer camelCasing. In general, Python prefers the underscore method but guides developers to defer to camelCasing if integrating into existing Python code that already uses that style. Readability counts. Read more in the section on naming conventions in PEP 8 .
As you can guess, keywords like 'if' and 'while' cannot be used as variable names — you'll get a syntax error if you do. However, be careful not to use built-ins as variable names. For example, while 'str', 'list' and 'print' may seem like good names, you'd be overriding those system variables. Built-ins are not keywords and thus, are susceptible to inadvertent use by new Python developers.
More on Modules and their Namespaces
Suppose you've got a module "binky.py" which contains a "def foo()". The fully qualified name of that foo function is "binky.foo". In this way, various Python modules can name their functions and variables whatever they want, and the variable names won't conflict — module1.foo is different from module2.foo. In the Python vocabulary, we'd say that binky, module1, and module2 each have their own "namespaces," which as you can guess are variable name-to-object bindings.
For example, we have the standard "sys" module that contains some standard system facilities, like the argv list, and exit() function. With the statement "import sys" you can then access the definitions in the sys module and make them available by their fully-qualified name, e.g. sys.exit(). (Yes, 'sys' has a namespace too!)
There is another import form that looks like this: "from sys import argv, exit". That makes argv and exit() available by their short names; however, we recommend the original form with the fully-qualified names because it's a lot easier to determine where a function or attribute came from.
There are many modules and packages which are bundled with a standard installation of the Python interpreter, so you don't have to do anything extra to use them. These are collectively known as the "Python Standard Library." Commonly used modules/packages include:
You can find the documentation of all the Standard Library modules and packages at http://docs.python.org/library .
Online help, help() , and dir()
There are a variety of ways to get help for Python.
- Do a Google search, starting with the word "python", like "python list" or "python string lowercase". The first hit is often the answer. This technique seems to work better for Python than it does for other languages for some reason.
- The official Python docs site — docs.python.org — has high quality docs. Nonetheless, I often find a Google search of a couple words to be quicker.
- There is also an official Tutor mailing list specifically designed for those who are new to Python and/or programming!
- Many questions (and answers) can be found on StackOverflow and Quora .
- Use the help() and dir() functions (see below).
- help(len) — help string for the built-in len() function; note that it's "len" not "len()", which is a call to the function, which we don't want
- help(sys) — help string for the sys module (must do an import sys first)
- dir(sys) — dir() is like help() but just gives a quick list of its defined symbols, or "attributes"
- help(sys.exit) — help string for the exit() function in the sys module
- help('xyz'.split) — help string for the split() method for string objects. You can call help() with that object itself or an example of that object, plus its attribute. For example, calling help('xyz'.split) is the same as calling help(str.split) .
- help(list) — help string for list objects
- dir(list) — displays list object attributes, including its methods
- help(list.append) — help string for the append() method for list objects
Except as otherwise noted, the content of this page is licensed under the Creative Commons Attribution 4.0 License , and code samples are licensed under the Apache 2.0 License . For details, see the Google Developers Site Policies . Java is a registered trademark of Oracle and/or its affiliates.
Last updated 2023-07-05 UTC.
Browse Course Material
Course info, instructors.
- Dr. Ana Bell
- Prof. Eric Grimson
- Prof. John Guttag
Departments
- Electrical Engineering and Computer Science
As Taught In
- Algorithms and Data Structures
- Programming Languages
Learning Resource Types
Introduction to computer science and programming in python, lecture slides and code.
The slides and code from each lecture are available below.

You are leaving MIT OpenCourseWare
- Chapter 3: Branch
- Chapter 4: Loop
- Chapter 5: Function
- Chapter 6 Collection
Presentation Slides ¶
Part i. fundamental python programming ¶, chapter 1: introduction to programming ¶.
- Python Crash Course
Chapter 2: Variable and Expression ¶
- Variables .
- Expression and Statements
- Turtle Introduction
Chapter 3 Branch ¶
Chapter 4 loop ¶, chapter 5 function ¶, chapter 6 collection ¶, chapter 7 class ¶.
- Basic Class
- Advanced Class
Part II. Advanced Programming ¶
Chapter 8 exception ¶, chapter 9 module ¶, chapter 10 file ¶, chapter 11 python data model ¶, part iii. algorithmic thinking ¶, chapter 12 a web site ¶.
- Django Crash Course
- Basic Django
- Advanced Django

Introduction to Programming in Python
a textbook for a first course in computer science for the next generation of scientists and engineers
Our textbook Introduction to Programming in Python [ Amazon · Pearson ] is an interdisciplinary approach to the traditional CS1 curriculum. We teach all of the classic elements of programming, using an "objects-in-the-middle" approach that emphasizes data abstraction. A key feature of the book is the manner in which we motivate each programming concept by examining its impact on specific applications, taken from fields ranging from materials science to genomics to astrophysics to internet commerce. The book is organized around four stages of learning to program:
- Chapter 1: Elements of Programming introduces variables; assignment statements; built-in types of data; conditionals and loops; arrays; and input/output, including graphics and sound.
- Chapter 2: Functions and Modules introduces modular programming. We stress the fundamental idea of dividing a program into components that can be independently debugged, maintained, and reused.
- Chapter 3: Object-Oriented Programming introduces data abstraction. We emphasize the concept of a data type and its implementation using Python's class mechanism.
- Chapter 4: Algorithms and Data Structures introduces classical algorithms for sorting and searching, and fundamental data structures, including stacks, queues, and symbol tables.
- Appendices provide supplemental material and a Python summary.
Reading a book and surfing the web are two different activities: This booksite is intended for your use while online (for example, while programming and while browsing the web); the textbook is for your use when initially learning new material and when reinforcing your understanding of that material (for example, when reviewing for an exam). The booksite consists of the following elements:
- Excerpts. A condensed version of the text narrative for reference while online.
- Exercises. Hundreds of exercises and some solutions.
- Python code. Hundreds of easily downloadable Python programs and real-world data sets . real-world data sets .-->
To get started.
To get started you must install either a Python 3 or a Python 2 programming environment.
Here are instructions for installing a Python 3 programming environment [ Windows · Mac OS X · Linux ]. We recommend that you install and use the Python 3 programming environment.
Here are instructions for installing a Python 2 programming environment [ Windows · Mac OS X · Linux ]. We recommend that you use the Python 2 programming environment only if you have a compelling reason (external to the requirements of this book and booksite) to do so.
We also provide I/O libraries for reading and writing text, drawing graphics, and producing sound.
You can request an examination copy from Pearson.

- Programming Python Code
- Popular Categories
Powerpoint Templates
Icon Bundle
Kpi Dashboard
Professional
Business Plans
Swot Analysis
Gantt Chart
Business Proposal
Marketing Plan
Project Management
Business Case
Business Model
Cyber Security
Business PPT
Digital Marketing
Digital Transformation
Human Resources
Product Management
Artificial Intelligence
Company Profile
Acknowledgement PPT
PPT Presentation
Reports Brochures
One Page Pitch
Interview PPT
All Categories
Powerpoint Templates and Google slides for Programming Python Code
Save your time and attract your audience with our fully editable ppt templates and slides..
Presenting Programming Python Code In Powerpoint And Google Slides Cpb slide which is completely adaptable. The graphics in this PowerPoint slide showcase four stages that will help you succinctly convey the information. In addition, you can alternate the color, font size, font type, and shapes of this PPT layout according to your content. This PPT presentation can be accessed with Google Slides and is available in both standard screen and widescreen aspect ratios. It is also a useful set to elucidate topics like Programming Python Code. This well structured design can be downloaded in different formats like PDF, JPG, and PNG. So, without any delay, click on the download button now.
Introducing our premium set of slides with UI Coding Icon For Programming Language. Ellicudate the Three stages and present information using this PPT slide. This is a completely adaptable PowerPoint template design that can be used to interpret topics like UI Coding Icon, Programming Language. So download instantly and tailor it with your information.
Introducing our premium set of slides with Web Development Coding Icon For Programming Language. Ellicudate the Three stages and present information using this PPT slide. This is a completely adaptable PowerPoint template design that can be used to interpret topics like Web Development Coding Icon, Programming Language. So download instantly and tailor it with your information.
This vibrant PowerPoint icon features a coding language symbol, perfect for presentations on UI programming and development. The bold colors and clean design make it easy to incorporate into any slide, while the detailed image accurately represents the topic at hand. Add a touch of professionalism to your slides with this eye-catching icon.
This Monotone powerpoint icon is perfect for representing UI coding in programming language presentations. With its sleek and minimalist design, it conveys the technical aspect of UI coding while maintaining a professional and modern look. Use it to enhance your slides and make a strong visual impact.

Presenting this set of slides with name Man Doing Python Coding For Big Data Governance. This is a one stage process. The stages in this process are Big Data Governance, Analytics, Dashboard. This is a completely editable PowerPoint presentation and is available for immediate download. Download now and impress your audience.

Presenting our Coding Challenge Questions Python In Powerpoint And Google Slides Cpb PowerPoint template design. This PowerPoint slide showcases three stages. It is useful to share insightful information on Coding Challenge Questions Python This PPT slide can be easily accessed in standard screen and widescreen aspect ratios. It is also available in various formats like PDF, PNG, and JPG. Not only this, the PowerPoint slideshow is completely editable and you can effortlessly modify the font size, font type, and shapes according to your wish. Our PPT layout is compatible with Google Slides as well, so download and edit it as per your knowledge.
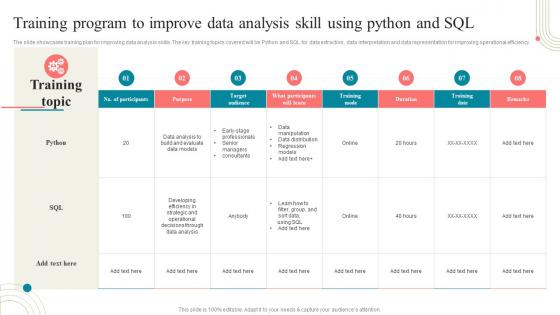
The slide showcases training plan for improving data analysis skills. The key training topics covered will be Python and SQL for data extraction, data interpretation and data representation for improving operational efficiency. Present the topic in a bit more detail with this Training Program To Improve Data Analysis Skill Using Python Business Development Training. Use it as a tool for discussion and navigation on Training Program, Improve Data, Analysis Skill, Using Python And Sql. This template is free to edit as deemed fit for your organization. Therefore download it now.
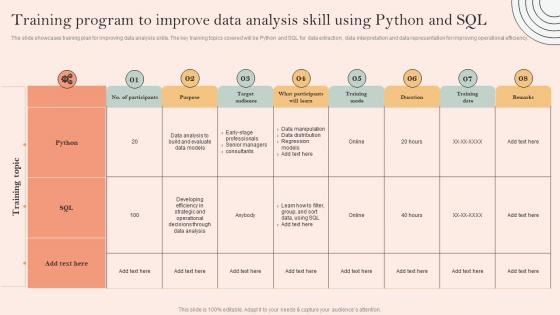
The slide showcases training plan for improving data analysis skills. The key training topics covered will be Python and SQL for data extraction, data interpretation and data representation for improving operational efficiency. Present the topic in a bit more detail with this Skill Development Programme Training Program To Improve Data Analysis Skill Using Python And SQL. Use it as a tool for discussion and navigation on Data Manipulation, Data Distribution, Regression Models. This template is free to edit as deemed fit for your organization. Therefore download it now.
Presenting Python Programming Learning Five Year Roadmap PowerPoint slide. This PPT theme is available in both 4,3 and 16,9 aspect ratios. This PowerPoint template is customizable so you can modify the font size, font type, color, and shapes as per your requirements. This PPT presentation is Google Slides compatible hence it is easily accessible. You can download and save this PowerPoint layout in different formats like PDF, PNG, and JPG.
Presenting Python Programming Learning Four Quarter Roadmap PowerPoint Template. This PPT presentation is Google Slides compatible hence it is easily accessible. You can download and save this PowerPoint layout in different formats like PDF, PNG, and JPG. This PPT theme is available in both 4,3 and 16,9 aspect ratios. This PowerPoint template is customizable so you can modify the font size, font type, color, and shapes as per your requirements.
Presenting Python Programming Learning Half Yearly Roadmap PowerPoint slide which is 100 percent editable. You can change the color, font size, font type, and shapes of this PPT layout according to your needs. This PPT template is compatible with Google Slides and is available in both 4,3 and 16,9 aspect ratios. This ready to use PowerPoint presentation can be downloaded in various formats like PDF, JPG, and PNG.
Presenting Python Programming Learning Six Months Roadmap PowerPoint slide. This PPT presentation is Google Slides compatible hence it is easily accessible. This PPT theme is available in both 4,3 and 16,9 aspect ratios. This PowerPoint template is customizable so you can modify the font size, font type, color, and shapes as per your requirements. You can download and save this PowerPoint layout in different formats like PDF, PNG, and JPG.
Presenting our Python Coding Challenges Ppt Powerpoint Presentation Professional Design Inspiration Cpb PowerPoint template design. This PowerPoint slide showcases five stages. It is useful to share insightful information on Python Coding Challenges This PPT slide can be easily accessed in standard screen and widescreen aspect ratios. It is also available in various formats like PDF, PNG, and JPG. Not only this, the PowerPoint slideshow is completely editable and you can effortlessly modify the font size, font type, and shapes according to your wish. Our PPT layout is compatible with Google Slides as well, so download and edit it as per your knowledge.
Presenting Python Programming Learning Three Months Roadmap PowerPoint slide. This PPT slide is available at 4,3 and 16,9 aspect ratios. You can download this PPT theme in various formats like PDF, PNG, and JPG. This PowerPoint template is completely editable and you can modify the font size, font type, and shapes as per your requirements. Our PPT layout is compatible with Google Slides.

- Python Basics
- Interview Questions
- Python Quiz
- Popular Packages
- Python Projects
- Practice Python
- AI With Python
- Learn Python3
- Python Automation
- Python Web Dev
- DSA with Python
- Python OOPs
- Dictionaries
- Resize the image in jupyter notebook using markdown
- Jupyter Notebook Extension in Visual Studio Code
- Using R programming language in Jupyter Notebook
- Getting started with Jupyter Notebook | Python
- Data Visualization in jupyter notebook
- Install Python package using Jupyter Notebook
- How to Write and Run Code in Jupyter Notebook
- Jupyter notebook Tips and Tricks
- Using Jupyter Notebook in Virtual Environment
- Understanding Jupyter Notebook Widgets
- Jupyter notebook VS Python IDLE
- How to Use ChatGPT to Create Presentations using Slides AI
- How To Use Jupyter Notebook - An Ultimate Guide
- How to Change the Theme in Jupyter Notebook
- How to upload a dataset in Jupyter Notebook?
- Configure Keyboard Shortcuts in Jupyter Notebook
- Creating Interactive Slideshows in Jupyter Notebooks
- Creating and updating PowerPoint Presentations in Python using python - pptx
- How to Install Jupyter Notebook on Windows?
Create Presentations Using RISE in Jupyter Notebook
RISE is an extension of Jupyter Notebook designed to revolutionize the way you create presentations. By seamlessly integrating with Jupyter Notebook, RISE allows you to transform your static code and data into dynamic, interactive presentations. In this guide, we’ll explore how to install RISE, set up presentation slides, view and present your slides, and delve into the additional features of using the RISE chalkboard for enhanced interactivity.
Installation of RISE
Let’s start with the basics. Installing RISE is the first step towards creating engaging presentations in Jupyter Notebook . To install RISE, follow these simple steps:
Open your terminal, use the following command, and press shift+enter to execute the command:
python -m pip install RISE
Steps to Create Presentations using RISE in Jupyter
Now, let’s move on to setting up your presentation slides. You will find the RISE icon on the top of the Jupyter Notebook page.
Setting Up Presentation Slides
Setting up presentation slides involves configuring the slide types and layout for your presentation.
- Open Jupyter Notebook.
- Click on the ‘View’ menu at the top of the notebook.
- Select ‘Cell Toolbar’ from the dropdown menu.
- Choose ‘Slideshow’ from the options available.
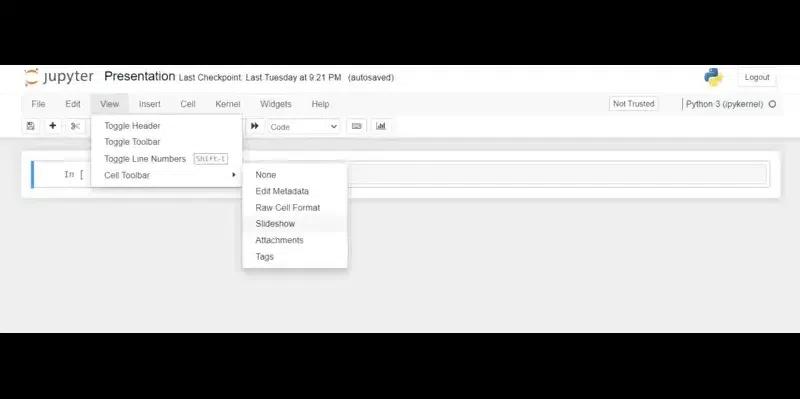
Now that you’ve configured your slide types and layout, it’s time to prepare your slides.
Preparing Slides
Creating compelling content for your slides is essential to captivate your audience. Follow these steps to prepare your slides:
- Create new cells in the notebook for each slide’s content.
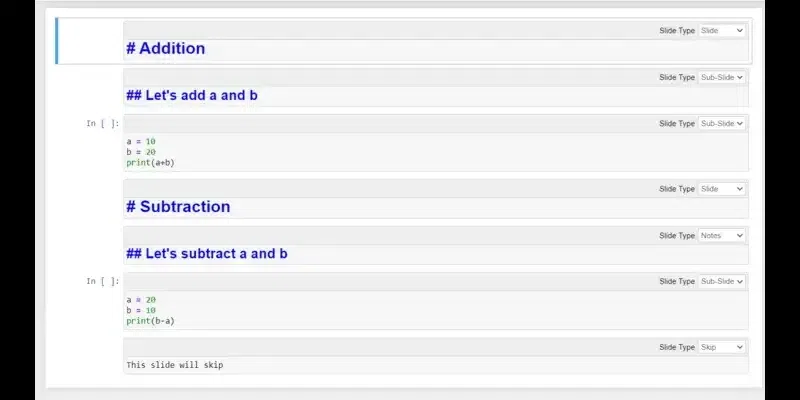
- Assign different slide types to cells based on their purpose, such as Slide, Subslide, Fragment, Skip, or Notes.
Determine the slide types you’ll use:
- Slide: The content on this slide is self-explanatory. A new slide will start from this cell.
- Subslide: Type of slide that will appear under the current slide
- Fragment: If you want to control the flow of information on your slide, divide it into fragments.
- Skip: skip this cell, it won’t appear on the slide.
- Notes: This cell works as notes.
- “ — ”: Inherit behavior from the cell above.
Example: Here’s an example of slide preparation using different slide types:
Viewing Presentation
Before presenting to your audience, it’s crucial to preview how your presentation will appear. Here’s how you can do it:
- Run the notebook by clicking the ‘Run’ button or pressing Shift + Enter.
- Navigate through the slides using the space bar, arrow keys, or other navigation controls provided by RISE.
Now that you’ve previewed your presentation, Follow these steps to start presenting:
- Utilize the presentation mode by pressing the appropriate shortcut.
- Navigate through your slides seamlessly using the space bar, arrow keys, or other navigation controls.
How to Use RISE Chalkboard
Now, let’s explore the additional feature of RISE: the chalkboard. The chalkboard feature allows you to annotate your slides in real time, adding a layer of interactivity to your presentation. Here’s how you can use the RISE chalkboard:
- Enable the chalkboard mode by clicking on the chalkboard icon in the presentation mode.
- Use your mouse or touchpad to draw or write on the slides.
- Erase or clear annotations as needed to maintain clarity.
With the RISE chalkboard, you can engage your audience and make your presentations more interactive and dynamic.
Please Login to comment...
Similar reads.

- Jupyter-notebook
Improve your Coding Skills with Practice
What kind of Experience do you want to share?
Python Tutorial
File handling, python modules, python numpy, python pandas, python matplotlib, python scipy, machine learning, python mysql, python mongodb, python reference, module reference, python how to, python examples, learn python.
Python is a popular programming language.
Python can be used on a server to create web applications.
Learning by Examples
With our "Try it Yourself" editor, you can edit Python code and view the result.
Click on the "Try it Yourself" button to see how it works.
Python File Handling
In our File Handling section you will learn how to open, read, write, and delete files.
Python Database Handling
In our database section you will learn how to access and work with MySQL and MongoDB databases:
Python MySQL Tutorial
Python MongoDB Tutorial
Python Exercises
Test yourself with exercises.
Insert the missing part of the code below to output "Hello World".
Start the Exercise
Advertisement
Learn by examples! This tutorial supplements all explanations with clarifying examples.
See All Python Examples
Python Quiz
Test your Python skills with a quiz.
My Learning
Track your progress with the free "My Learning" program here at W3Schools.
Log in to your account, and start earning points!
This is an optional feature. You can study at W3Schools without using My Learning.

You will also find complete function and method references:
Reference Overview
Built-in Functions
String Methods
List/Array Methods
Dictionary Methods
Tuple Methods
Set Methods
File Methods
Python Keywords
Python Exceptions
Python Glossary
Random Module
Requests Module
Math Module
CMath Module
Download Python
Download Python from the official Python web site: https://python.org
Python Exam - Get Your Diploma!
Kickstart your career.
Get certified by completing the course

COLOR PICKER

Contact Sales
If you want to use W3Schools services as an educational institution, team or enterprise, send us an e-mail: [email protected]
Report Error
If you want to report an error, or if you want to make a suggestion, send us an e-mail: [email protected]

Top Tutorials
Top references, top examples, get certified.
python-pptx 0.6.23
pip install python-pptx Copy PIP instructions
Released: Nov 2, 2023
Generate and manipulate Open XML PowerPoint (.pptx) files
Verified details
Maintainers.

Unverified details
Project links, github statistics.
- Open issues:
View statistics for this project via Libraries.io , or by using our public dataset on Google BigQuery
License: MIT License (MIT)
Author: Steve Canny
Tags powerpoint, ppt, pptx, office, open, xml
Classifiers
- OSI Approved :: MIT License
- OS Independent
- Python :: 2
- Python :: 2.7
- Python :: 3
- Python :: 3.6
- Office/Business :: Office Suites
- Software Development :: Libraries
Project description
python-pptx is a Python library for creating, reading, and updating PowerPoint (.pptx) files.
A typical use would be generating a PowerPoint presentation from dynamic content such as a database query, analytics output, or a JSON payload, perhaps in response to an HTTP request and downloading the generated PPTX file in response. It runs on any Python capable platform, including macOS and Linux, and does not require the PowerPoint application to be installed or licensed.
It can also be used to analyze PowerPoint files from a corpus, perhaps to extract search indexing text and images.
In can also be used to simply automate the production of a slide or two that would be tedious to get right by hand, which is how this all got started.
More information is available in the python-pptx documentation .
Browse examples with screenshots to get a quick idea what you can do with python-pptx.
Release History
0.6.23 (2023-11-02), 0.6.22 (2023-08-28).
(Windows Python 3.10+)
0.6.21 (2021-09-20)
0.6.20 (2021-09-14), 0.6.19 (2021-05-17), 0.6.18 (2019-05-02).
.text property getters encode line-break as a vertical-tab (VT, ‘v’, ASCII 11/x0B). This is consistent with PowerPoint’s copy/paste behavior and allows like-breaks (soft carriage-return) to be distinguished from paragraph boundary. Previously, a line-break was encoded as a newline (’n’) and was not distinguishable from a paragraph boundary.
.text properties include Shape.text, _Cell.text, TextFrame.text, _Paragraph.text and _Run.text.
.text property setters accept vertical-tab character and place a line-break element in that location. All other control characters other than horizontal-tab (’t’) and newline (’n’) in range x00-x1F are accepted and escaped with plain-text like “_x001B” for ESC (ASCII 27).
Previously a control character other than tab or newline in an assigned string would trigger an exception related to invalid XML character.
0.6.17 (2018-12-16)
0.6.16 (2018-11-09), 0.6.15 (2018-09-24), 0.6.14 (2018-09-24), 0.6.13 (2018-09-10), 0.6.12 (2018-08-11), 0.6.11 (2018-07-25), 0.6.10 (2018-06-11), 0.6.9 (2018-05-08), 0.6.8 (2018-04-18), 0.6.7 (2017-10-30), 0.6.6 (2017-06-17), 0.6.5 (2017-03-21), 0.6.4 (2017-03-17), 0.6.3 (2017-02-28), 0.6.2 (2017-01-03).
BACKWARD INCOMPATIBILITIES:
Some changes were made to the boilerplate XML used to create new charts. This was done to more closely adhere to the settings PowerPoint uses when creating a chart using the UI. This may result in some appearance changes in charts after upgrading. In particular:
0.6.1 (2016-10-09)
0.6.0 (2016-08-18), 0.5.8 (2015-11-27), 0.5.7 (2015-01-17).
Shape.shape_type is now unconditionally MSO_SHAPE_TYPE.PLACEHOLDER for all placeholder shapes. Previously, some placeholder shapes reported MSO_SHAPE_TYPE.AUTO_SHAPE , MSO_SHAPE_TYPE.CHART , MSO_SHAPE_TYPE.PICTURE , or MSO_SHAPE_TYPE.TABLE for that property.
0.5.6 (2014-12-06)
0.5.5 (2014-11-17), 0.5.4 (2014-11-15), 0.5.3 (2014-11-09), 0.5.2 (2014-10-26), 0.5.1 (2014-09-22), 0.5.0 (2014-09-13).
A table is no longer treated as a shape. Rather it is a graphical object contained in a GraphicFrame shape, as are Chart and SmartArt objects.
As the enclosing shape, the id, name, shape type, position, and size are attributes of the enclosing GraphicFrame object.
The contents of a GraphicFrame shape can be identified using three available properties on a shape: has_table, has_chart, and has_smart_art. The enclosed graphical object is obtained using the properties GraphicFrame.table and GraphicFrame.chart. SmartArt is not yet supported. Accessing one of these properties on a GraphicFrame not containing the corresponding object raises an exception.
0.4.2 (2014-04-29)
0.4.1 (2014-04-29).
The following enumerations were moved/renamed during the rationalization of enumerations:
Documentation for all enumerations is available in the Enumerations section of the User Guide.
0.3.2 (2014-02-07)
0.3.1 (2014-01-10), 0.3.0 (2013-12-12), 0.2.6 (2013-06-22), 0.2.5 (2013-06-11), 0.2.4 (2013-05-16), 0.2.3 (2013-05-05), 0.2.2 (2013-03-25), 0.2.1 (2013-02-25), 0.2.0 (2013-02-10).
First non-alpha release with basic capabilities:
Project details
Release history release notifications | rss feed.
Nov 2, 2023
Aug 29, 2023
Sep 20, 2021
Sep 14, 2021
May 17, 2021
May 3, 2019
Dec 16, 2018
Nov 10, 2018
Sep 24, 2018
Sep 10, 2018
Aug 12, 2018
Jul 25, 2018
Jun 11, 2018
May 9, 2018
Apr 18, 2018
Oct 30, 2017
Jun 18, 2017
Mar 22, 2017
Mar 17, 2017
Feb 28, 2017
Jan 3, 2017
Oct 9, 2016
Aug 18, 2016
Nov 28, 2015
Jan 18, 2015
Dec 7, 2014
Nov 18, 2014
Nov 16, 2014
Nov 10, 2014
Oct 27, 2014
Sep 22, 2014
Sep 14, 2014
Apr 30, 2014
Feb 7, 2014
Jan 10, 2014
Dec 13, 2013
0.3.0rc1 pre-release
Jun 22, 2013
Jun 12, 2013
May 17, 2013
May 6, 2013
Mar 26, 2013
Feb 25, 2013
Feb 11, 2013
0.1.0a1 pre-release
Jan 26, 2013
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages .
Source Distribution
Uploaded Nov 2, 2023 Source
Built Distribution
Uploaded Nov 2, 2023 Python 3
Hashes for python-pptx-0.6.23.tar.gz
Hashes for python_pptx-0.6.23-py3-none-any.whl.
- português (Brasil)
Supported by

- International
- Schools directory
- Resources Jobs Schools directory News Search

An Introduction to Python package - 6 full lessons with all codes
Subject: Computing
Age range: 14-16
Resource type: Unit of work
Last updated
24 May 2024
- Share through email
- Share through twitter
- Share through linkedin
- Share through facebook
- Share through pinterest

A fantastic full package of 6 lessons with presentations, lesson plans and fully tested codes for any teacher that needs to start off with Python, but does not know where to begin. This package is created to enable you to jump straight in.
Each lessons has been created to be 1 hour in length. However, these can be broken down due to the nature of the presentations and are editable for teacher use.
This is suitable for age ranges between Year 9 to 12 and for home schooling, but can be used for younger children, where applicable.
** End of topic test included with 16 questions, teacher answers and full code**
This package includes:
- Extensive presentations, with videos
- Full lesson plans
- Full and extensive codes (no need to code yourself)
- Extensive end of lesson practice questions with answers
- End of module test (with full answers and codes)
This module covers:
- An introduction to the shell and editor
- Syntax and comments
- Strings and loops
- Working with arrays
- Len function, upper and lower
- Boolean functions
- Dictionaries
To be able to use this package, ensure that you download the latest version of Python and the Python editor Thonny.
Tes paid licence How can I reuse this?
Your rating is required to reflect your happiness.
It's good to leave some feedback.
Something went wrong, please try again later.
This resource hasn't been reviewed yet
To ensure quality for our reviews, only customers who have purchased this resource can review it
Report this resource to let us know if it violates our terms and conditions. Our customer service team will review your report and will be in touch.
Not quite what you were looking for? Search by keyword to find the right resource:
PyCon 24: Pythonic Deployment - Juju and Charms
PyCon 24: Pythonic Deployment - Juju and Charms
How do you deploy your Python (Web) application? If your setup is container-based you’re likely using Kubernetes. Is it fun? No. It’s complex. It’s a lot of YAML. Maybe it’s okay-ish - if your setup is not too complex -, but it could be better. Less complex, more lightweight. And then again, how does your test suite look like for the deployment process? You don’t have any? Sure, it’s all YAML. Yak!
What if you could do everything in Python? What if you could “associate” workloads, e.g. a database, with your application in the cloud, using Python code? What if you could write tests with Pytest to ensure your deployment continues to work? Does that sound too good to be true?
Canonical has been developing this technology for years. It is proven and solid, yet still exciting to use! Let’s dive into what could be the best you have seen in the last decade for deploying your Python Web application. Say hello to Python and test-driven deployments, ditch YAML and say yes to stability! See a live demo and take home working code to experiment with your own setup.
This talk was held on May 24, 2024 at PyCon 24, Florence, Italy.
See original slides at https://slides.com/bittner/pycon24-pythonic-deployment-juju-charms
Peter Bittner
More decks by peter bittner.
Other Decks in Programming
PYTHONIC DEPLOYMENT OF (PYTHON) APPLICATIONS Juju magic and Charmed operators.
La pizza la vita la famiglia la sorella la ragazza, developer of people, companies and code @bittner @peterbittner, [email protected] peter, orchestration engine for software operators deploy & perform day-2 ops, deploy your application , deploy your application scripted setup (create virtual environment, copy source, different problems solved 🤔, different problems solved scripted setup ... bring up same setup, do everything, incl. operations juju (engine, controller, client) charms (python, juju bootstrap <substrate> <name> juju add-model <name> juju deploy <charmname>, juju tooling & concepts juju (engine, controller, client) charms (python, https://snapcraft.io/ juju tooling & concepts juju (engine, controller, client) charms, demos charmcraft, rockcraft, snapcraft charm (business & test code) juju, thank you for your precious time painless software less pain,.
- Open access
- Published: 23 May 2024
CineMol: a programmatically accessible direct-to-SVG 3D small molecule drawer
- David Meijer 1 ,
- Marnix H. Medema 1 &
- Justin J. J. van der Hooft 1
Journal of Cheminformatics volume 16 , Article number: 58 ( 2024 ) Cite this article
19 Accesses
8 Altmetric
Metrics details
Effective visualization of small molecules is paramount in conveying concepts and results in cheminformatics. Scalable vector graphics (SVG) are preferred for creating such visualizations, as SVGs can be easily altered in post-production and exported to other formats. A wide spectrum of software applications already exist that can visualize molecules, and customize these visualizations, in many ways. However, software packages that can output projected 3D models onto a 2D canvas directly as SVG, while being programmatically accessible from Python, are lacking. Here, we introduce CineMol, which can draw vectorized approximations of three-dimensional small molecule models in seconds, without triangulation or ray tracing, resulting in files of around 50–300 kilobytes per molecule model for compounds with up to 45 heavy atoms. The SVGs outputted by CineMol can be readily modified in popular vector graphics editing software applications. CineMol is written in Python and can be incorporated into any existing Python cheminformatics workflow, as it only depends on native Python libraries. CineMol also provides programmatic access to all its internal states, allowing for per-atom and per-bond-based customization. CineMol’s capacity to programmatically create molecular visualizations suitable for post-production offers researchers and scientists a powerful tool for enhancing the clarity and visual impact of their scientific presentations and publications in cheminformatics, metabolomics, and related scientific disciplines.
Scientific contribution
We introduce CineMol, a Python-based tool that provides a valuable solution for cheminformatics researchers by enabling the direct generation of high-quality approximations of two-dimensional SVG visualizations from three-dimensional small molecule models, all within a programmable Python framework. CineMol offers a unique combination of speed, efficiency, and accessibility, making it an indispensable tool for researchers in cheminformatics, especially when working with SVG visualizations.
Introduction
Cheminformatics knowledge transfer primarily occurs through presentations, published articles, tutorials, and textbooks. In these contexts, three-dimensional molecular visualizations of small molecules play a crucial role in facilitating the understanding of underlying concepts and research outcomes while also adding layers of informative value. To illustrate this point, consider the scenario where a methodology for generating molecular conformations is presented, and its fidelity is assessed by comparing the root-mean-squared deviation of the atom positions to a validated experimental target. In this case, the inclusion of visual representations displaying both the generated conformation and the target conformation serves as an immediate visual indicator of the quality of the generated structure and allows for a visual aid in the quality assessment. Another instance is the portrayal of potential orientations of ligands within a binding pocket of a protein and their spatial proximity to crucial active site residues. A third example is that a researcher might want to demonstrate how specific functional groups within a molecule exhibit closer spatial proximity under specific environmental conditions than would be inferred solely from their skeletal structural formula. In all these instances, graphical representations hold the potential to convey information far more effectively and intuitively than an extensive textual description. Fortunately, a multitude of software applications are readily available to aid scientists and researchers in crafting three-dimensional visualizations of molecules. Among the noteworthy tools in this domain are Jmol [ 1 ], 3Dmol.js [ 2 ], Blender [ 3 ], PyMOL [ 4 ], and RDKit [ 5 ] each offering distinct capabilities and features for the visualization and analysis of molecular structures.
The way these tools interface with users determines their usability. Standalone desktop applications provide a user-friendly graphical interface for visualizing molecules, but they lack programmatic accessibility, meaning you cannot control them via scripts, and they cannot be integrated directly into other software as libraries. Jmol falls into this category. Web applications are interactive as well but typically render models on the client side, limiting them to JavaScript, which modern browsers support. Examples include 3Dmol.js and JSmol, the JavaScript version of Jmol. Resources such as the Protein Data Bank ( https://www.rcsb.org/ ) and PubChem ( https://pubchem.ncbi.nlm.nih.gov/ ) rely on such tools to display three-dimensional structures. Notably, 3Dmol.js can also be programmatically utilized in Python through an IPython interface called py3Dmol within Jupyter notebooks. Command line interfaces (CLIs) enable users to interact with programs via the command line, although some of these programs may also feature a graphical user interface (GUI). Certain software applications offer multiple interaction methods, such as Blender and PyMOL. Blender and PyMOL are desktop applications that provide both a command line interface and a Python application programming interface (API). Regarding their underlying technology, Jmol relies on a specialized Java-based graphics engine, while 3Dmol.js is a JavaScript library that utilizes WebGL, a JavaScript implementation of the versatile graphics library known as OpenGL [ 6 ], for rendering graphics. Blender and PyMOL have their cores developed in C and also employ OpenGL for rendering.
Python is the language of choice for many researchers in cheminformatics as well as other domains of science that deal with molecular information, due to its versatility and extensive libraries. Therefore, it is important to note that the aforementioned tools are not inherently Python-centric. Compiled languages like C and Java often offer faster performance; however, when employed within a Python-first environment, they may introduce additional dependencies, which could be considered excessive when the primary goal is to generate three-dimensional visualizations of chemical compounds. Additionally, users will not have direct access to all the internal states of objects when they can solely interact with the library through an API, although this might be desired by the user when they would like to apply specific stylistic choices in a programmatic way that are not supported by the API directly. RDKit is a widely used cheminformatics toolkit for cheminformaticians working in Python. RDKit can draw three-dimensional conformations of molecular structures and facilitates customization of these visualizations. However, to our knowledge, it is not yet possible to directly output these images as SVGs.
Creating visuals for compounds is typically just the beginning of the process. More often than not, these rendered images find their way into ensemble figures. When creating visuals for this purpose, it is preferred to output them as scalable vector graphics (SVGs). SVGs describe geometries in a vectorized form using an extensible markup language (XML) format, which format is designed to be shareable. This makes SVG easily modifiable either through a text editor or via a GUI within illustration software such as Adobe Illustrator or Inkscape. However, it’s important to note that the graphics rendering engines of the aforementioned molecular visualization tools are not inherently designed for direct SVG output. While plugins such as the render freestyle SVG add-on for Blender [ 7 ] or stand-alone tools like gl2ps [ 8 ] might extend this capability, creating SVGs from complex three-dimensional models involves a more intricate process.
Complex three-dimensional shapes are often constructed from simpler flat surface geometric shapes, typically triangles. The description of a complex three-dimensional shape involves connecting these triangles through a process called triangulation. The level of detail in the three-dimensional model depends on the number of triangles used. These triangles are then projected into the two-dimensional SVG canvas. However, merely sorting and rendering these two-dimensional shapes is insufficient when two or more model meshes intersect. To address this, algorithms are employed to sort and subdivide the triangles into visible and invisible parts. This recursive computational process is resource intensive. If this process is not conducted with sufficient detail, intersection lines appear jagged, particularly for models with a lot of curved surfaces—like space-filling molecule models depicting overlapping atom spheres. Additionally, when executed at a high level of detail, SVG files may become exceedingly large, reaching sizes in the tens or hundreds of megabytes, even for relatively small models as every individual triangle needs to be defined in the SVG. Another avenue for outputting SVGs is to embed a PNG within an SVG format. This is less desirable because these SVGs do not provide true vector graphics and are no longer editable as SVG files in post-production, thus limiting their versatility.
To tackle these specific challenges, we introduce CineMol—a Python-centric, dependency-free solution for generating true SVG representations of three-dimensional models of small molecules. CineMol employs a straightforward algorithm to rapidly and accurately approximate two-dimensional projections of three-dimensional scenes. It excels in producing compact SVG files, often just hundreds of kilobytes in size, making them highly practical for sharing, drawing molecules, and enabling extensive customization during post-production. CineMol offers users a user-friendly Python library along with a command-line tool, simplifying the process of creating SVG approximations of three-dimensional molecular scenes. Furthermore, we have developed a demo web page where prospective users can effortlessly generate SVG models from SDF files ( https://moltools.bioinformatics.nl/cinemol ), providing a hands-on experience with our tool’s capabilities.
Implementation
CineMol approximates the two-dimensional projections of a three-dimensional scene of atoms and bonds on an SVG canvas without computationally expensive meshes or ray tracing. To ensure that CineMol effectively represents intersections while maintaining the high performance that allows it to generate SVGs in a matter of seconds, various methods were employed to limit the number of calculations.
First, a three-dimensional scene is created by assembling the in CineMol available sphere, cylinder, and wireframe geometries and their styling. This can be done by the user directly when they want full control over the scene and its styling, or by creating Atom and Bond objects and feeding it into the draw_molecule API, together with a global style. The CineMol algorithm will then start by mapping three-dimensional points on the surfaces of the scene items (Fig. 1 a). The number of points generated (N) for each geometry is based on the resolution parameter.
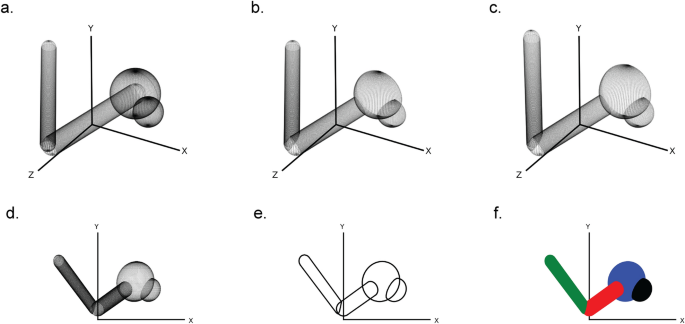
The six steps of CineMol’s algorithm for creating a two-dimensional polygonal projection of a three-dimensional scene. a Points are generated on the surfaces of the geometric items in the scene. The number of points generated is based on the resolution parameter. b The scene items are filtered on the z-coordinate of their centroid and points invisible from the point-of-view of CineMol are discarded. c The three-dimensional points are projected based on the focal length of the scene. d The z-coordinate is discarded to make the scene two-dimensional. e CineMol’s convex hull algorithm calculates the convex shape around each item’s two-dimensional point cloud to create polygonal outlines. f Polygons are drawn in their previously sorted order and a fill style is applied. Matplotlib (v3.8.2) was used to create this figure [ 12 ]
For a sphere, we generate N points on the surface based on ɸ azimuthal angles (= resolution + 1) times θ polar angles (= resolution + 1). For a cylinder, we first generate N points (= resolution + 1) uniformly distributed points between a start position vector and an end position vector. These points form the centers of circles that outline the body of the cylinder. For every circle, we generate N points (= resolution ) on the circumference. We cap the cylinder with either no cap or a round cap. The round cap is created by generating points for half a sphere on either end of the cylinder. The resolution parameter has no impact on a wireframe geometry.
After generating points on the surface of our three-dimensional scene items, the algorithm filters points based on their visibility from CineMol’s fixed viewpoint (Fig. 1 b). CineMol has a fixed view direction towards the origin along the positive z-axis. To speed up the algorithm, we sort scene items from farthest to nearest to the viewpoint based on the z-coordinate of their centroid. As SVG draws polygons in chronological fashion, we only assess intersections for each item between earlier drawn items. For quick intersection checks, we check if the centers of spheres and the central line of cylinders are within each other's vicinity, considering that covalent bonds mainly intersect at their ends with other atoms (i.e., spheres) and bonds (i.e., cylinders). Wireframes are only sorted based on their centroid before drawing. For the remaining points, the algorithm projects their x and y coordinates using a provided focal length (Fig. 1 c) and then disposes of the z-coordinate to create a two-dimensional projection (Fig. 1 d).
Now, each item comprises a two-dimensional point cloud, representing projected surface points visible from CineMol’s viewpoint, considering that items will be drawn based on their initial z-axis distance to CineMol’s point of view. To establish the smallest encapsulating polygon outlining each point cloud, the quickhull (a two-dimensional algorithm to find a convex hull [ 9 ]) is applied (Fig. 1 e). The polygons are then drawn from the furthest away to most nearby to CineMol’s point of view, and the supplied fill is applied as styling (Fig. 1 f). This fill can be either solid, for a cartoon style, or a gradient for a glossy look (i.e., radial-gradient for spheres and a linear gradient for cylinders). Wireframes can only be styled based on the provided stroke color.
The draw_molecule API is wrapped around the Scene object and applies a uniform style to all atoms and bonds given to it. The draw_molecule API uses the Corey–Pauly–Koltung (CPK) scheme for determining atom and bond colors [ 10 ]. The atom radii are sourced from the atomic radii values in the periodic table of elements from PubChem [ 11 ]. The draw_molecule API is wrapped around a Scene object. The Scene object is the engine behind the draw_molecule API and describes a lower-level abstraction of the scene to draw in terms of sphere, cylinder, and wire geometries. When users would like to fully customize their model instead of using a general style from the draw_molecule API they can opt for using the Scene object directly and give each geometry their personalized style, if desired.
Results & discussion
In the following section, we present a series of examples to illustrate the versatility and effectiveness of CineMol, our Python-based three-dimensional molecular visualization tool. These examples serve to showcase the type of depictions and the range of applications that CineMol can accommodate. We will also demonstrate CineMol’s ability to swiftly compute a scene within seconds, showcasing its linear computational scalability, with respect to the number of atoms to draw, when tested on a series of molecular conformers, all while generating compact file sizes. All images in this results section were generated on a MacBook Pro with an Apple M2 chip and 8 GB memory.
CineMol: a versatile three-dimensional small molecule drawer
CineMol has a versatile Scene object that empowers users to create a wide array of scenes composed of spheres, cylinders, and wireframes, catering to diverse visualization needs. In addition to this, CineMol’s draw_molecule API (wrapped around the Scene object) provides a user-friendly solution right out of the box, offering four distinct depiction types: space-filling (Fig. 2 a), ball-and-stick (Fig. 2 b), tube (Fig. 2 c), and wireframe (Fig. 2 d), along with two unique rendering styles: cartoon (Fig. 2 top row) and glossy (Fig. 2 bottom row). The cartoon rendering style assigns a solid color fill and stroke to each polygon, while the glossy rendering style replicates specular and shadow effects. If the polygons were composed of triangles instead (for example, from the result of triangulation), each surface could have been individually styled to realistically generate specular and shadow effects. Unfortunately, this is not achievable in SVG when the surface is a single polygon.
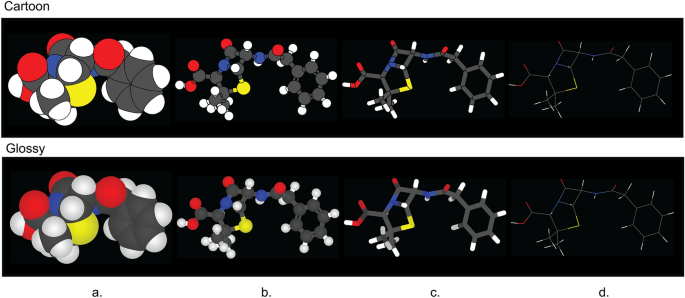
The four depiction types and two fill styles supported by CineMol’s draw_molecule API. a Space-filling. b Ball-and-stick. c Tube. d Wireframe. The wireframe depiction only supports stroke color and stroke opacity changes and has no separate glossy style. A resolution of 100 and a scale factor of 10.0 was used to generate these images. Algorithm runtimes (cartoon/glossy): space-filling = 1.5 s/1.5 s; ball-and-stick = 1.9 s/1.9 s; tube = 3.4 s/3.4 s; wireframe = 1 ms/1 ms. File sizes (cartoon/glossy): space-filling = 65 kb/74 kb; ball-and-stick: 176 kb/206 kb; tube = 75 kb/93 kb; wireframe = 14 kb/14 kb. The SDF containing the penicillin G conformer used to generate these images was retrieved from PubChem [ 13 ]. Adobe Illustrator 2024 was used to compile the SVGs and generate the figure in PNG format
The draw_molecule API has multiple parameters, which are outlined comprehensively in Table 1 , providing users with an intuitive and flexible tool for generating molecular visualizations tailored to their specific requirements. Each atom and bond given to the draw_molecule API can have its color, radius, and opacity set manually to override the defaults. The draw_molecule API can, after installing CineMol with pip, be used directly by importing it in a Python project.
We have developed two user-friendly interfaces that enable users to swiftly start utilizing CineMol. A GUI built on top of the draw_molecule API is available at https://moltools.bioinformatics.nl/cinemol . Additionally, the installation of CineMol ships with a CLI wrapped around the draw_molecule API as well, which contains much of the same functionality. Currently, molecules in the form of structure-data format (SDF) files can be supplied to the GUI and CLI. CineMol places its primary emphasis on visualization rather than parsing various file formats. As a deliberate design choice, we have refrained from incorporating third-party libraries for file parsing. This decision allows users the freedom to select their preferred cheminformatics toolkit for this specific purpose, providing flexibility and compatibility with a wide range of data sources.
Furthermore, CineMol does not contain any functionality for generating three-dimensional conformations of compound structures. Consistent with its approach of avoiding dependencies for file parsing, we provide users with the flexibility to select their preferred software for conformer generation, rather than bundling such functionalities. For example, the widely used cheminformatics toolkit RDKit can be used to generate conformers for compounds. This is further highlighted in the section titled “CineMol allows full customization of models for experienced users”.
CineMol allows full customization of models for experienced users
Experienced Python users may desire additional customization options for their molecular models, and CineMol offers the flexibility to achieve this. To do so, users can leverage the underlying Scene object and supply it with model nodes. These nodes come in three distinct shapes: spherical, cylindrical, or wire, allowing for tailored molecular representations. Each model node is accompanied by its styling. Three examples are shown in Fig. 3 , and the exact implementations of these examples can be found in the CineMol GitHub at https://github.com/moltools/CineMol/tree/main/examples .
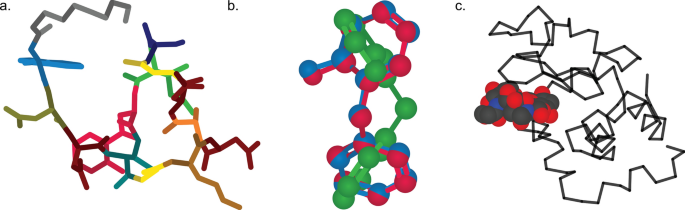
Examples of custom figures created programmatically direct-to-SVG with CineMol. a A daptomycin conformer with its monomers highlighted. b Three superimposed conformations of o-benzylphenol. c The lysozyme 9LYZ with bound bacterial cell wall trisaccharide NAM-NAG-NAM. Algorithm runtimes: 3a = 1.8 s; 3b = 3 min; 3c = 2.3 s. File sizes: 3a = 123 kb; 3b: 211 kb; 3c = 135b. Adobe Illustrator 2024 was used to compile the SVGs and generate the figure in PNG format
Figure 3 a shows a generated three-dimensional conformation of the non-ribosomal peptide daptomycin that was generated with the wrapper draw_molecule API with a resolution of 50. Every different color highlights a distinct monomer in the molecule. The conformation generation and the substructure searches were performed with RDKit v2023.9.6 [ 5 ]. Figure 3 b shows three RDKit-generated conformations of o-benzylphenol superimposed on each other. To accurately visualize the intersecting spheres and bonds, the resolution was increased to 150 and we instructed the algorithm to not filter nodes for intersecting. By default, the algorithm filters nodes for intersection. This means that a quick check is performed to estimate if two nodes intersect before calculating the exact intersection. Turning off this quick check slows down the calculation but makes sure that every intersection is accurately visualized in this particular case. The Scene API allows users to set or include specific calculations to create their own trade-off between accuracy and speed. Figure 3 c shows a wireframe of the lysozyme 9LYZ with a space-filling model of the bound bacterial cell wall trisaccharide NAM-NAG-NAM [ 14 ]. The opacity of the protein wireframe was set to 0.75, and the model was manually rotated to show the bound ligand more clearly. The PDB file was parsed with the bioinformatics toolkit biopython v1.83 [ 15 ].
CineMol generates SVG drawings with small file sizes in a matter of seconds
Figure 4 illustrates the runtime performance and resulting file sizes when generating models for 4548 protein-bound ligand conformations from the Platinum dataset v2017_01 [ 16 ]. These results were obtained using the draw_molecule API with a resolution of 50 and excluding hydrogen atoms.
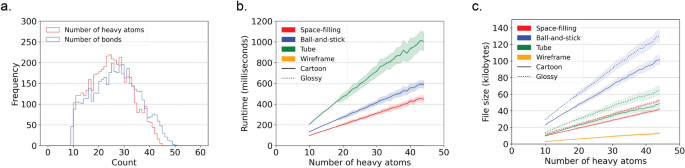
Runtime and file size performance metrics for generating SVG models for 4548 protein-bound ligands from the Platinum dataset. a Number of heavy atoms and bonds per ligand in the Platinum dataset. wireframe model SVG files magnitudes smaller than the SVG files for the other model styles. b Runtime performance. c File sizes. The fill lines indicate one standard deviation. Matplotlib (v3.8.2) was used to create this figure [ 12 ]
Several key observations can be made from the data. First, all atoms in the dataset contain between 10 and 45 heavy atoms and between 10 and 50 bonds, as depicted in Fig. 4 a. This demonstrates the applicability of CineMol for molecules of these sizes. It is important to note that a resolution of 50 was used and only heavy atoms were considered for this analysis.
The data reveals general trends among space-filling, tube, and ball-and-stick models. Runtime and file size show a linear relationship (as denoted by Fig. 4 b). Tube models exhibit double the runtime of space-filling and ball-and-stick models due to the computationally intensive nature of calculating cylinder-cylinder intersections, since cylinders tend to have more generated points than spheres after filtering. The algorithm does not calculate cylinder-cylinder intersections for ball-and-stick models, as they are typically not visible given the smaller radii of bonds compared to atoms in molecular models. If bond radii were larger than atom radii, a tube model would be more appropriate to generate anyway. Users can customize the behavior regarding which geometries require intersection calculations by directly accessing the Scene object.
Furthermore, file sizes for ball-and-stick models are approximately twice as large as those for space-filling and tube models (Fig. 4 c). This is expected because ball-and-stick models typically entail about twice as many polygons to describe in the SVG file. The runtime difference between glossy and cartoon styles remains minimal since only the fill step (as shown in Fig. 1 f) differs and is not computationally intensive. File sizes for SVG models with a glossy style are a factor bigger than SVG models with a cartoon style (Fig. 4 c). This can be explained by the fact that gradients, which are used to create the glossy look, take more characters to describe than a solid fill, which is used to create the cartoon look.
Wireframe models present distinct trends compared to other style types. In wireframe models, no intersections are calculated; instead, only the sorting of individual wires is performed. This process is not computationally demanding, resulting in consistently small runtimes (Fig. 4 b). Wireframe model SVG files are also magnitudes smaller than the SVG files generated for other model styles. Line segments in wireframe models are defined by start and end positions, while single polygons consist of numerous individual line segments (Fig. 4 c).
In summary, for this set of ligands, the runtime ranges from 1 to 1200 ms, indicating that CineMol can efficiently render similarly sized ligands in seconds with a reasonable resolution of 50. The runtime of CineMol typically scales quadratically with the resolution. For example, drawing a space-filling model of penicillin G, which has 23 heavy atoms, without hydrogen atoms, takes approximately 200 ms for a resolution of 50, approximately 800 ms for a resolution of 100, and approximately 3.2 s for a resolution of 200. A resolution of 100 is sufficient for most applications. Any resolution higher than 200 tends not to lead to visible improvements for most applications. File sizes remain in the range of tens to hundreds of kilobytes across all style combinations.
Conclusions
In conclusion, CineMol addresses a specific need in cheminformatics by providing a Python-first software package capable of producing precise SVG representations of small molecule models. This tool facilitates enhanced visualization options for three-dimensional molecular structures with a focus on customization and the ability for post-production editing. CineMol’s efficient performance and accessibility make it a valuable tool for researchers and scientists in the field of chemistry and beyond.
Availability of data and materials
CineMol v1.0.0 is available for Python versions 3.10 and up, has no third-party dependencies, and is released to PyPI ( https://pypi.org/project/cinemol/ ). The source code of CineMol is freely available on GitHub at https://github.com/moltools/cinemol under the MIT license, together with the code to generate any figure present in this article. We have followed the reproducibility and reusability guidelines as described by Hoyt et al. [ 17 ], using the cookiecutter-snekpack template ( https://github.com/cthoyt/cookiecutter-snekpack ). A web-based demo version of CineMol is available at https://moltools.bioinformatics.nl/cinemol . We have archived the version of CineMol’s repository used to generate results for this publication with Zenodo ( https://doi.org/10.5281/zenodo.11242217 ).
Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/ . Accessed 7 Jan 2024.
Rego N, Koes D (2015) 3Dmol.js molecular visualization with WebGL. Bioinformatics 31:1322–1324. https://doi.org/10.1093/bioinformatics/btu829
Article PubMed Google Scholar
Blender Foundation. https://www.blender.org/ . Accessed 7 Jan 2024.
PyMOL. https://pymol.org/2/ . Accessed 7 Jan 2024
RDKit: Open-source cheminformatics. https://www.rdkit.org/ . Accessed Jan 7 2024
OpenGL. https://www.opengl.org/ . Accessed 7 Jan 2024
Blender manual: Freestyle SVG exporter. https://docs.blender.org/manual/en/latest/addons/render/render_freestyle_svg.html . Accessed 7 Jan 2024
GL2PS: an OpenGL to PostScript printing library. https://www.geuz.org/gl2ps/ . Accessed 7 Jan 2024.
Greenfield JS (1990) A proof for a quickhull algorithm. Electrical Engineering and Computer Science-Technical Reports
Koltun WL (1965) Space filling atomic units and connectors for molecular models. US Patent. 3170246
National Center for Biotechnology and Information (2024) Atomic Radius in the Periodic Table of Elements. https://pubchem.ncbi.nlm.nih.gov/periodic-table/atomic-radius . Accessed Jan 7 2024
Hunter JD (2007) Matplotlib: A 2D graphics environment. Comput Sci Eng 9:90–95
Article Google Scholar
National Center for Biotechnology and Information (2024) PubChem compound summary for CID 5904, Penicillin G. https://pubchem.ncbi.nlm.nih.gov/compound/Penicillin-G . Accessed Jan 7 2024
Kelly JA, Sielecki AR, Sykes BD, James MNG, Phillips DC (1979) X-ray crystallography of the binding of the bacterial cell wall trisaccharide NAM-NAG-NAM to lysozyme. Nature 282:875–878
Article CAS PubMed Google Scholar
Hamelryck T, Manderick B (2003) PDB file parser and structure class implemented in Python. Bioinformatics 19(17):2308–2310
Friedrich N-O, Meyder A, de Bruyn KC, Sommer K, Flachsenberg F, Rarey M, Kirchmair J (2017) High-Quality dataset of protein-bound ligand conformations and its application to benchmarking conformer ensemble generators. J Chem Inf Model 57:529–539
Hoyt CT, Zdrazil RB, Guha R, Jeliazkova N, Martinez-Mayorga K, Nittinger E (2023) Improving reproducibility and reusability in the Journal of Cheminformatics. J Cheminf 15:62
Download references
Acknowledgements
Not applicable.
European Research Council (Project DECIPHER; grant agreement ID: 948770).
Author information
Authors and affiliations.
Bioinformatics Group, Wageningen University & Research, Wageningen, the Netherlands
David Meijer, Marnix H. Medema & Justin J. J. van der Hooft
You can also search for this author in PubMed Google Scholar
Contributions
DM conducted research and wrote the code for CineMol and drafted the original manuscript. MHM and JJJvdH provided extensive feedback on the research process and the written manuscript.
Corresponding authors
Correspondence to David Meijer , Marnix H. Medema or Justin J. J. van der Hooft .
Ethics declarations
Competing interests.
JJJvdH is currently member of the Scientific Advisory Board of NAICONS Srl., Milano, Italy, and consults for Corteva Agriscience, Indianapolis, IN, USA. MHM is a member of the scientific advisory board of Hexagon Bio. The other author declares to have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Meijer, D., Medema, M.H. & van der Hooft, J.J.J. CineMol: a programmatically accessible direct-to-SVG 3D small molecule drawer. J Cheminform 16 , 58 (2024). https://doi.org/10.1186/s13321-024-00851-y
Download citation
Received : 01 February 2024
Accepted : 08 May 2024
Published : 23 May 2024
DOI : https://doi.org/10.1186/s13321-024-00851-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Scalable vector graphics
- Three-dimensional structure
- Molecular drawing
- Visualization
Journal of Cheminformatics
ISSN: 1758-2946
- Submission enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
Python is a general purpose programming language that can be used for both programming and scripting. It was created in the 1990s by Guido van Rossum who named it after the Monty Python comedy troupe. People use Python for a variety of tasks due to its readability, object-oriented capabilities, extensive libraries, and ability to integrate with ...
4.Run these Python expressions (one per line): (a) 42 (b) 26+18 (c) 26<18 (d) 26>18 5. Exit Python (but not the terminal window). 2 minutes. 16 Writing Python scripts Applications → Word and Text Processing → gedit. 17 Launching Python scripts Read / edit the script Run the script gedit Terminal. 18 Launching Python
Language Introduction. Python is a dynamic, interpreted (bytecode-compiled) language. There are no type declarations of variables, parameters, functions, or methods in source code. This makes the code short and flexible, and you lose the compile-time type checking of the source code.
Python has a simple syntax similar to the English language. Python has syntax that allows developers to write programs with fewer lines than some other programming languages. Python runs on an interpreter system, meaning that code can be executed as soon as it is written. This means that prototyping can be very quick.
Python Classes and Inheritance Slides for Lecture 9 (PDF - 1.6MB) Code for Lecture 9 (PY) 10 Understanding Program Efficiency, Part 1 Slides for Lecture 10 (PDF) Code for Lecture 10 (PY) 11 Understanding Program Efficiency, Part 2 Slides for Lecture 11 (PDF) Code for Lecture 11 (PY) 12 Searching and Sorting Slides for Lecture 12 (PDF - 2.4MB)
Presentation Slides¶ Part I. Fundamental Python Programming¶ Chapter 1: Introduction to Programming¶ Python Crash Course; Chapter 2: Variable and Expression¶ Variables. Expression and Statements; Turtle Introduction; Chapter 3 Branch ¶ Chapter 4 Loop ¶ Chapter 5 Function ¶ Chapter 6 Collection ¶ Chapter 7 Class¶ Basic Class; Advanced ...
Displaying Introduction to Python Programming.ppt.
7 Real numbers Python can also manipulate real numbers. Examples: 6.022 -15.9997 42.0 2.143e17 The operators + - * / % ** ( ) all work for real numbers. The / produces an exact answer: 15.0 / 2.0 is 7.5 The same rules of precedence also apply to real numbers: Evaluate ( ) before * / % before + - When integers and reals are mixed, the result is a real number.
Introduction To Python Programming. Python Programming (58 Slides) 94403 Views. Unlock a Vast Repository of Python Programming PPT Slides, Meticulously Curated by Our Expert Tutors and Institutes. Download Free and Enhance Your Learning!
Textbook. Our textbook Introduction to Programming in Python [ Amazon · Pearson ] is an interdisciplinary approach to the traditional CS1 curriculum. We teach all of the classic elements of programming, using an "objects-in-the-middle" approach that emphasizes data abstraction. A key feature of the book is the manner in which we motivate each ...
Introduction To Python Programming. Python is widely used general-purpose, high-level programming language. Its design philosophy emphasizes code readability, and its syntax allows programmers to express concepts in fewer lines of code than would be possible in languages such as C++ or Java.
340.3 KB. Download Presentation Slides: Module 1 3.8 MB Download Presentation Slides: Module 2 1013.6 KB Download Presentation Slides: Module 3 507.3 KB Download Presentation Slides: Module 4 772.3 KB Download Presentation Slides: Module 5 788.9 KB Download….
Python have 4 types of built in Data Structures namely List, Dictionary, Tuple and Set. List is the most basic Data Structure in python. List is a mutable data structure i.e items can be added to list later after the list creation. It's like you are going to shop at the local market and made a list of some items and later on you can add more ...
1. Fundamentals of Python Programming. 2. Introduction to python programming • High level, interpreted language • Object-oriented • General purpose • Web development (like: Django and Bottle), • Scientific and mathematical computing (Orange, SciPy, NumPy) • Desktop graphical user Interfaces (Pygame, Panda3D). 3.
introduction to Python (for beginners) Mar 11, 2020 • Download as PPTX, PDF •. 1 like • 490 views. G. guobichrng. Python supports multiple programming paradigms, including object-oriented, imperative and functional programming or procedural styles. It features a dynamic type system and automatic memory management and has a large and ...
python-pptx is a Python library for creating and updating PowerPoint files. This article is going to be a basic introduction to this package. If you want to learn much more about it, this is the official documentation page that you should check. Now let's install the package if you don't have. pip install python-pptx.
1. Write the python code/logic for the presentation. Let's write the logic of "find the largest number in a Python list". The logic for this will be really minimalistic and easy to understand. Let's use the below-given algorithm to write the logic: Create a list of values. Sort the list using sort().
II. Process Data and Design Slides with Python. You can find the source code with dummy data here: Github. Let us explore all the steps to generate your final report. Steps to create your operational report on PowerPoint — (Image by Author) 1. Data Extraction. Connect to your WMS and extract shipment records.
Presenting Programming Python Code In Powerpoint And Google Slides Cpb slide which is completely adaptable. The graphics in this PowerPoint slide showcase four stages that will help you succinctly convey the information. In addition, you can alternate the color, font size, font type, and shapes of this PPT layout according to your content.
Programmers can use the graphic-rich PPT to depict the reasons that make this programming language so popular. You can showcase the uses, features, myths, and potential applications of the python language in an easy-to-understand manner. You can also communicate about the advantages of acquiring skills in the Python programming language.
The Basic Structure of python-pptx. After installing the package, using python-pptx is the same as any other library. At the top of the file, import the dependencies you will need: Besides the ...
pip install python-pptx. Let's see some of its usage: Example 1: Creating new PowerPoint file with title and subtitle slide. Python3. from pptx import Presentation . root = Presentation() first_slide_layout = root.slide_layouts[0] . 0 -> title and subtitle. 5 -> Title only .
Let's start with the basics. Installing RISE is the first step towards creating engaging presentations in Jupyter Notebook. To install RISE, follow these simple steps: Open your terminal, use the following command, and press shift+enter to execute the command: python -m pip install RISE.
W3Schools offers free online tutorials, references and exercises in all the major languages of the web. Covering popular subjects like HTML, CSS, JavaScript, Python, SQL, Java, and many, many more.
Project description. python-pptx is a Python library for creating, reading, and updating PowerPoint (.pptx) files. A typical use would be generating a PowerPoint presentation from dynamic content such as a database query, analytics output, or a JSON payload, perhaps in response to an HTTP request and downloading the generated PPTX file in ...
PNG, 11.28 KB. A fantastic full package of 6 lessons with presentations, lesson plans and fully tested codes for any teacher that needs to start off with Python, but does not know where to begin. This is suitable for age ranges between Year 9 to 12 and for home schooling, but can be used for younger children. ** End of topic test included with ...
Step 3 - Generate PowerPoint Slides. Navigate to the directory containing the script in your terminal and run the following command: $ python3 create_ppt.py. This command will execute the script, and generate a new PowerPoint file named " Linux_Security_Presentation.pptx " in the same directory.
DEPLOY YOUR APPLICATION Scripted setup (create virtual environment, copy source. code to host, use Git, Bash, Ansible, etc.) Python package (pip install) System package (create + install DEB, RPM, Flatpak, snap, Windows .exe, etc.) Container (copy and install software in container image, Docker le, docker compose) Kubernetes (use Helm or ...
Visit claude.ai! Claude is a family of large language models developed by Anthropic and designed to revolutionize the way you interact with AI. Claude excels at a wide variety of tasks involving language, reasoning, analysis, coding, and more. Our models are highly capable, easy to use, and can be customized to suit your needs.
CineMol is written in Python and can be incorporated into any existing Python cheminformatics workflow, as it only depends on native Python libraries. ... offers researchers and scientists a powerful tool for enhancing the clarity and visual impact of their scientific presentations and publications in cheminformatics, metabolomics, and related ...